The zoonotic potential of the PDCoV, a coronavirus efficiently infecting cells from a broad range species, including porcine, chicken, and human, emphasizes the urgent need to further study the cell and tissue tropism of PDCoV in its natural host. Herein, we generated crypt stem cell-derived enteroids from porcine different intestinal regions, which well recapitulated the events in vivo of PDCoV infection that PDCoV targeted multiple types of intestinal epithelia and preferably infected the jejunum and ileum over the duodenum and colon. Mechanistically, we demonstrated that the expression of APN receptor rather than the IFN responses determined the susceptibility of different regions of the intestines to PDCoV infection, though PDCoV infection markedly elicited the IFN responses. Our findings provide important insights into how the distinct gene expression profiles of the intestinal segments determine the cell and tissue tropism of PDCoV.
KEYWORDS: PDCoV, enteroids, APN, interferon, interferon receptor
ABSTRACT
Porcine deltacoronavirus (PDCoV) is an economically important enteropathogen of swine with worldwide distribution. PDCoV primarily infects the small intestine instead of the large intestine in vivo. However, the underlying mechanism of PDCoV tropism to different intestinal segments remains poorly understood as a result of the lack of a suitable in vitro intestinal model that recapitulates the cellular diversity and complex functions of the gastrointestinal tract. Here, we established the PDCoV infection model of crypt-derived enteroids from different intestinal segments. Enteroids were susceptible to PDCoV, and multiple types of different functional intestinal epithelia were infected by PDCoV in vitro and in vivo. We further found that PDCoV favorably infected the jejunum and ileum and restrictedly replicated in the duodenum and colon. Mechanistically, enteroids from different intestinal regions displayed a distinct gene expression profile, and the differential expression of primary viral receptor host aminopeptidase N (APN) instead of the interferon (IFN) responses determined the susceptibility of different intestinal segments to PDCoV, although PDCoV substantially elicited antiviral genes production in enteroids after infection. Additional studies showed that PDCoV infection significantly induced the expression of type I and III IFNs at the late stage of infection, and exogenous IFN inhibited PDCoV replication in enteroids. Hence, our results provide critical inputs to further dissect the molecular mechanisms of PDCoV-host interactions and pathogenesis.
IMPORTANCE The zoonotic potential of the PDCoV, a coronavirus efficiently infecting cells from a broad range species, including porcine, chicken, and human, emphasizes the urgent need to further study the cell and tissue tropism of PDCoV in its natural host. Herein, we generated crypt stem cell-derived enteroids from porcine different intestinal regions, which well recapitulated the events in vivo of PDCoV infection that PDCoV targeted multiple types of intestinal epithelia and preferably infected the jejunum and ileum over the duodenum and colon. Mechanistically, we demonstrated that the expression of APN receptor rather than the IFN responses determined the susceptibility of different regions of the intestines to PDCoV infection, though PDCoV infection markedly elicited the IFN responses. Our findings provide important insights into how the distinct gene expression profiles of the intestinal segments determine the cell and tissue tropism of PDCoV.
INTRODUCTION
Porcine deltacoronavirus (PDCoV) is a recently discovered enteropathogenic coronavirus and has caused significant economic impacts on the pork industry (1, 2). PDCoV, similar to other swine enteric coronaviruses, including transmissible gastroenteritis virus (TGEV) and porcine epidemic diarrhea virus (PEDV), primarily targets intestinal epithelia in vivo and causes serious injury to the intestines, which is characterized by thin and transparent intestinal walls and accumulation of large amounts of yellow fluid in the intestinal lumen (1, 3–5). Microscopic lesions on PDCoV-infected intestines include acute, diffuse, atrophic enteritis, reduced villus-crypt ratios, and mild vacuolation of superficial epithelial cells (5). In vivo, PDCoV preferably infects the small intestine, although viral antigens have been detected in both the small and large intestines (1, 5, 6). However, the details of PDCoV cell and tissue tropism in different intestinal segments remain poorly understood. Clarifying the question is challenging since there are no good in vitro models that can recapitulate the segmental epithelia of the porcine intestines. We recently established porcine crypt stem cell-derived ileal enteroids, which well recapitulate the multiple-cellular complicated structure of intestinal epithelia in vivo and demonstrated that the enteroids could be a good in vitro model for PEDV infection (7). The results from other groups and our study demonstrated that the segment-specific difference is maintained in ex vivo enteroid cultures derived from different intestinal segmental crypt stem cells (7–9).
Coronaviruses display a propensity for interspecies transmission, and coronavirus spike (S) protein, a type I transmembrane protein, interacts with cellular receptors and largely determines the cell and tissue tropism and host range of coronavirus (10, 11). Porcine aminopeptidase N (pAPN), a 963-amino-acid-long type II transmembrane glycoprotein, has been identified as PDCoV primary entry receptor (12, 13). PDCoV efficiently binds APN from multiple species, including that of chicken and human, and results in cell infection, which potentially makes PDCoV a zoonotic pathogen (12). In addition, the proteolytic activation of coronavirus S protein by host cell proteases plays critical roles in determining PDCoV cell or tissue tropism since PDCoV infection appears to be restricted to the swine enteric tract despite the wide expression of APN in various tissues (11, 14–18). However, the pAPN expression pattern along the porcine intestines is still unknown. It is worthwhile to explore whether the differential expression of pAPN and other critical genes within different intestinal regions determines the intestinal segmental tropism of PDCoV, which will broaden insights into the zoonotic potential of PDCoV for cross-host transmission.
The small intestine, including the duodenum, jejunum, and ileum, and the large intestine are a continuous tube lined internally with a single layer of polarized columnar epithelium with a diverse range of functions (19). The different segments of the intestines have distinct endoscopic appearances and physiological functions. The surfaces of absorptive epithelial cells in the small intestine contain a layer of microvilli and are largely responsible for digesting and absorbing dietary components, while the large intestine lacks microvilli and a brush border and plays major roles in the reabsorption of water and the elimination of undigested foodstuffs (19). Moreover, the gene and protein expression profiles of intestinal epithelia change along the longitudinal axis of the intestines and are associated with the different functional roles of the intestinal segments (9). Limited studies with mice have shown compartmentalization in the expression of epithelial pattern recognition receptors (PRRs) among different intestinal segments (20, 21). Toll-like receptor 4 (TLR4) is expressed at a higher level in the colon than in the small intestine (20, 21). However, to date, few studies have directly assessed compartmentalization along the porcine intestines of the expression of important innate receptors such as interferon (IFN) receptors and PRRs, which are closely related to pathogen infection in the intestines.
Type I IFNs (alpha and beta [α/β]) and the more recently discovered type III IFNs are the critical host innate cytokines in response to viral infection and play crucial roles in controlling viral infection (22, 23). We and other groups have demonstrated that type III IFN lambda (IFN-λ) preferably acts on intestinal epithelia over type I IFNs (IFN-α/β) and superiorly suppresses enteric coronavirus infection in epithelia (7, 24, 25). Previous studies showed that IFN-λ is a key host determinant for mouse rotavirus or norovirus infection in intestinal epithelia (26, 27). Several previous studies reported that PDCoV suppresses IFN production after infection (28–30), while another in vivo study showed that PDCoV infection substantially elicits the expression of INF-α/β (31). Thus, the role that the IFN responses among different intestinal segments play in determining the susceptibility of PDCoV remains elusive.
In the present study, we established an ex vivo enteroid model for PDCoV infection, which recapitulated many events associated with PDCoV infection in the porcine intestines in vivo. We demonstrated that the intestinal segment-specific tropism of PDCoV was related to its viral entry receptor APN instead of the IFN responses in epithelia, although PDCoV infection substantially elicited IFN production. Collectively, our results extend the understanding of fundamental aspects of PDCoV pathogenesis.
RESULTS
Porcine intestinal enteroids are susceptible to PDCoV infection.
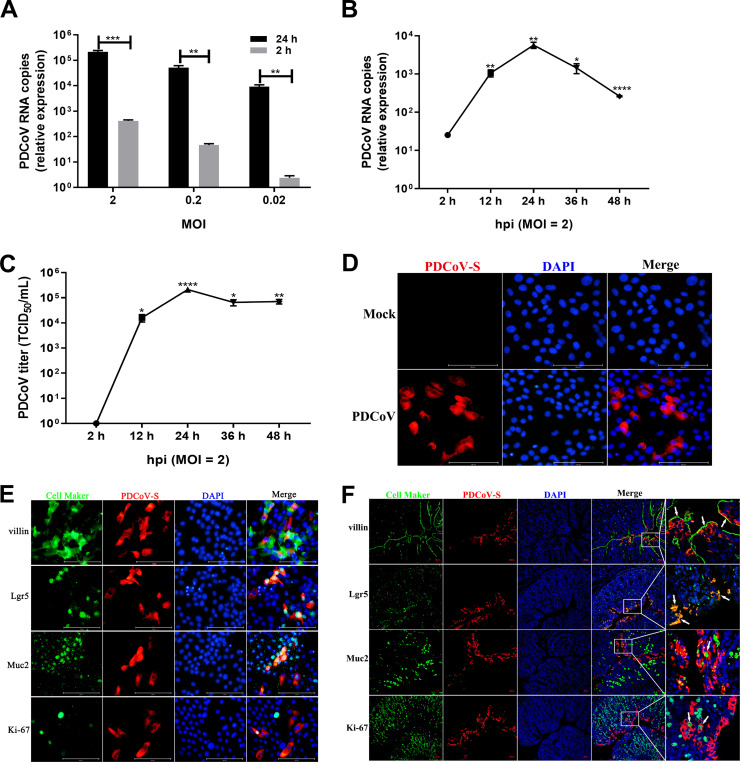
Intestinal enteroids derived from intestinal crypt stem cells can recapitulate many key features of the intestinal epithelium in vivo (32–35). We recently reported that porcine intestinal enteroids are a good ex vivo model for PEDV infection (7). To evaluate whether this novel model supports the replication of PDCoV, we used the two-dimensional (2D) enteroid monolayers generated from differentiated 3D enteroids to explore PDCoV infection as described previously (36, 37). After inoculation of porcine ileal enteroids with PDCoV at different multiplicities of infection (MOIs), the viral genomes were significantly increased by more than 525- to 3,813-fold at 24 h postinfection (hpi) compared with those at 2 hpi and displayed an MOI-dependent response (Fig. 1A), indicating that PDCoV successfully establishes infection in enteroids. The kinetics of PDCoV infection showed that both PDCoV genomes and viral titers reached a peak at 24 hpi, at which point viral titers were increased about 5.33 log10, after which a plateau was reached (Fig. 1B and C). PDCoV infection in ileal enteroids was also confirmed by a PDCoV S protein immunofluorescence assay (IFA) (Fig. 1D). These results show that porcine enteroids are permissive to PDCoV infection and are capable of supporting productive replication and progeny release.
FIG 1.
Porcine intestinal enteroids are susceptible to PDCoV infection. (A) Detection of PDCoV replication in porcine intestinal enteroids by RT-qPCR. Porcine ileal enteroid monolayers were mock inoculated or inoculated with PDCoV at the indicated MOIs for 2 h at 37°C. Enteroid monolayers were harvested at 2 hpi or 24 hpi after removal of virus inoculum and washing of cells three times to remove unattached viruses. Total cellular RNA was extracted, and PDCoV genomes were quantified by RT-qPCR. (B and C) Kinetic curve of PDCoV infection in porcine ileal enteroids. Porcine ileal enteroid monolayers were inoculated with PDCoV at an MOI of 2. The kinetics of PDCoV yielded at the indicated time points postinfection were determined by RT-qPCR (B) or titration (C). The scale data of panels A to C were taken as log(fold change) values and expressed as a power of 10. Each data bar represents the mean of results from three wells for each treatment and time point. Error bars denote deviations. *, P < 0.05; **, P < 0.01; ***, P < 0.005; ****, P < 0.001. (D) Detection of PDCoV infection in ileal enteroids by IFA. The enteroid monolayers were infected with PDCoV at an MOI of 2 for 24 h and then were fixed with 4% paraformaldehyde, and PDCoV S protein expression was confirmed using mouse anti-PDCoV S monoclonal antibody (red). (E and F) Double immunofluorescence labeling of ileal enteroids infected with PDCoV for 24 h (E) or ileal tissues collected from SPF piglets orally inoculated with PDCoV for 72 h (F). Enterocytes, intestinal stem cells, goblet cells, and proliferating cells were stained with anti-villin, anti-Lgr5, anti-mucin 2, and anti-Ki-67 antibodies (green), respectively. PDCoV was labeled with mouse anti-PDCoV S protein monoclonal antibody (red). DAPI-stained nuclei are shown in blue. Arrowheads denote specific cell maker-positive and PDCoV+cells (F). Scale bar, 100 μm in panel E and 50 μm in panel F.
Intestinal enteroids contain multiple cell types, including stem cells, Paneth cells, enterocytes, goblet cells, and enteroendocrine cells (38). We next explored whether PDCoV targets multiple types of intestinal epithelia. As expected, villin, a marker of enterocyte cells, was primarily colocated with cells containing PDCoV S protein (Fig. 1E), which was consistent with previously reported results in vivo(15). In addition, we found that PDCoV S protein was substantially colocalized with Lgr5-positive (Lgr5+) stem cells and Muc2+ goblet cells and rarely colocalized with Ki-67+ cells (proliferating cells) (Fig. 1E). These data suggest that PDCoV can infect multiple cell types in enteroids, including enterocytes, stem cells, and goblet cells. To further confirm whether PDCoV infects similar multiple cell lineages in vivo, we performed the double-immunofluorescence staining in PDCoV-infected ileal tissues (Fig. 1F). Consistent with the results observed in enteroids, PDCoV S protein was detected in villin-positive, Lgr5+, Muc2+, and Ki-67+cells, although PDCoV primarily infected the villin-positive and Lgr5+ cells and rarely infected the Muc2+ goblet cells and Ki-67+ cells (Fig. 1F). These results demonstrate that PDCoV has identical tropism for multiple types of intestinal epithelia in both in vivo and in vitro enteroids, including enterocytes, crypt stem cells, and goblet cells. Together, these data indicate that porcine enteroids are an ideal in vitro model for PDCoV infection and pathogenesis.
PDCoV preferably infects jejunal enteroids over duodenal enteroids and colonoids.
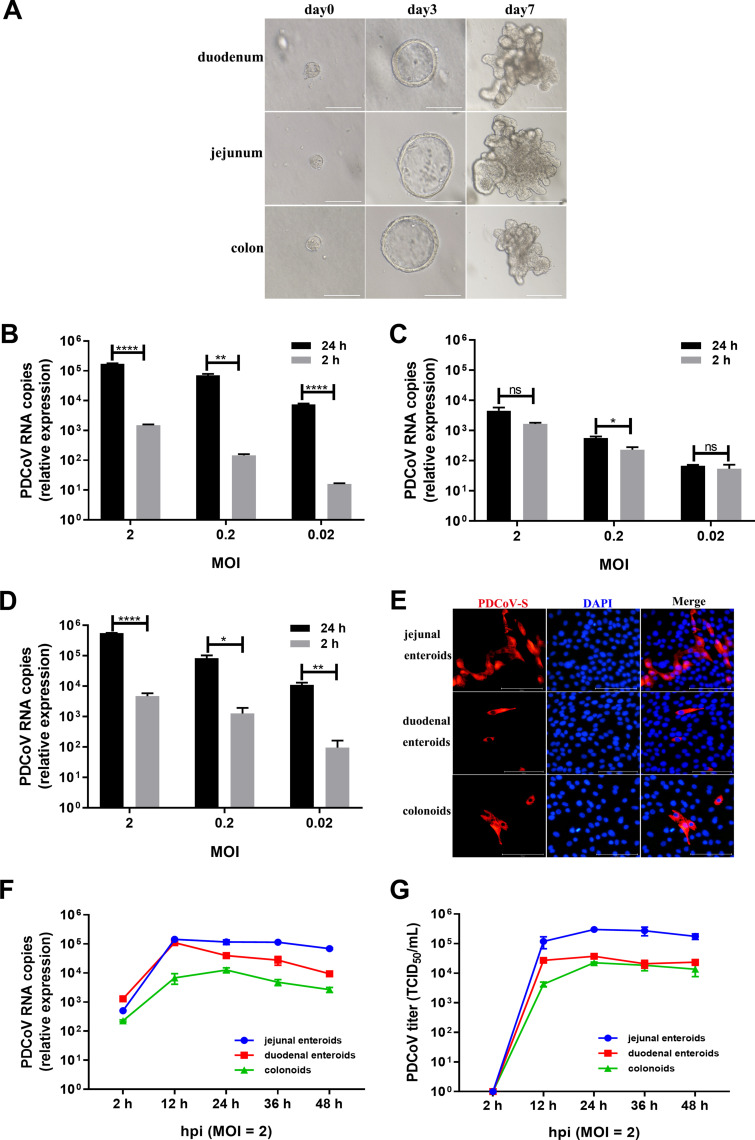
PDCoV primarily infects the small intestine and causes acute, severe atrophic enteritis in vivo, although viral antigens can be detected in the large intestine (1, 5, 6). To investigate whether porcine enteroids model the events associated with the intestinal segmental tropism of PDCoV infection in vivo, crypt stem cells were isolated from the duodenum, jejunum, and colon of the same specific-pathogen-free (SPF) piglet to differentiate into enteroids. After culturing for 7 days, crypt stem cells isolated from different intestinal segments all differentiated into enteroids containing crypt-villi-like structures (Fig. 2A) and displayed a minor variation in the differentiation speed, as we reported previously (7). We subsequently monitored the replication of PDCoV in enteroids derived from jejunal, duodenal, and colonic crypts. After infection of enteroids at the same MOIs, PDCoV successfully infected the enteroids derived from crypt stem cells of all three different intestinal segments and established a productive infection (Fig. 2B to D). Compared with viral genomic RNA detected at 2 hpi, PDCoV genomes at 24 hpi in jejunal enteroids were significantly increased by 116- to 482-fold (Fig. 2B), whereas PDCoV genomes in duodenal enteroids were slightly elevated (Fig. 2C) and in colonoids were increased by 95- to 217-fold (Fig. 2D), where replication levels were much lower than those in jejunal enteroids, indicating that porcine enteroids from different intestinal segments exhibit differential susceptibility to PDCoV infection. The differential infectivity of PDCoV in enteroids derived from different intestinal regions was further confirmed by a PDCoV S protein IFA (Fig. 2E). To further distinguish the differences of PDCoV infection among the three types of enteroids, we evaluated the kinetics of PDCoV infection in enteroids. Compared with viral RNA measured at 2 hpi, PDCoV genomes in jejunal enteroids were substantially elevated by more than 286-fold at 12 hpi and then maintained a plateau, while PDCoV genomes in duodenal enteroids were elevated up to 85-fold at 12 hpi and then were gradually decreased up to 48 hpi, and PDCoV genomes in colonoids were increased by 30-fold at 12 hpi and 55-fold at 24 hpi, at which there was a peak (Fig. 2F). Consistent with the results of PDCoV genomes, the PDCoV titers produced in jejunal enteroids from 12 hpi to 48 hpi were all higher than those in duodenal enteroids and colonoids (Fig. 2G). These results demonstrate that PDCoV preferably infects porcine jejunal enteroids over duodenal enteroids and colonoids.
FIG 2.
PDCoV preferably infects jejunal enteroids over duodenal enteroids and colonoids. (A) Representative images of porcine intestinal enteroids differentiation derived from duodenal, jejunal, and colonic crypts. After culturing in IntestiCult organoid growth medium mixed with Matrigel, isolated crypts formed 3D spheroids with cystic morphology on day 3 and gradually formed a crypt-villi-like structure. Scale bar, 100 μm (day 0, day 3) or 200 μm (day 7). (B to D) Detection of PDCoV replication in porcine enteroids generated from different intestinal segments by RT-qPCR. Monolayers of jejunal enteroids (B), duodenal enteroids (C), and colonoids (D) isolated from the same piglet were infected with PDCoV at the indicated MOIs for 2 h. After washing three times with DMEM-F12, the monolayers were collected at 2 hpi and 24 hpi. Viral genomes were determined by RT-qPCR. Data in B to D represent the mean of three wells for each treatment and time point. Error bars denote the standard deviation. *, P < 0.05; **, P < 0.01;****, P < 0.001; ns, not significant. (E) Detection of PDCoV infection in enteroids isolated from different intestinal segments by IFA. Monolayers of jejunal enteroids, duodenal enteroids, and colonoids were infected with PDCoV at an MOI of 2. Cells were fixed at 24 hpi and stained with mouse anti-PDCoV S monoclonal antibody (red). DAPI was used to detect nuclei (blue). (F and G) The kinetic curves of PDCoV replication in enteroids derived from jejunal, duodenal, and colonic crypts. Enteroid monolayers were inoculated with PDCoV at an MOI of 2 for the indicated time points. The kinetics of PDCoV production was measured by RT-qPCR (F) or titration (G). The scale data of panels B to D and F to G were taken as log(fold change) values and expressed as a power of 10.
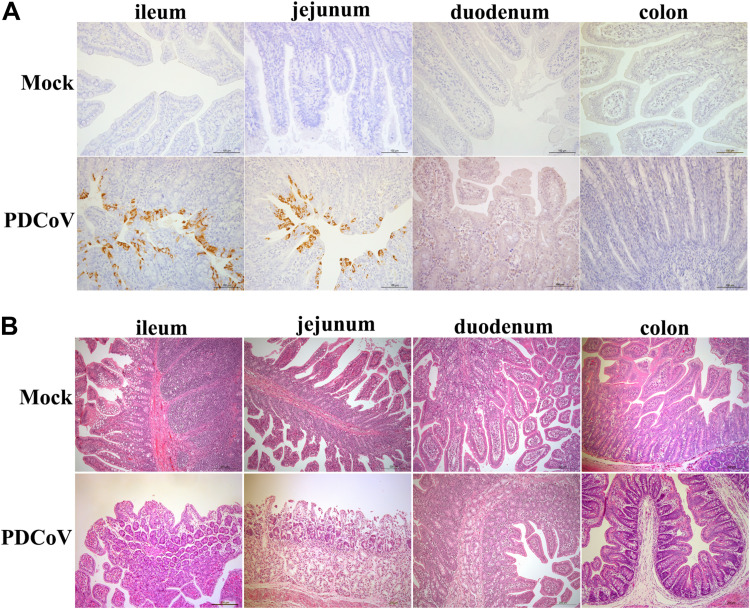
To further confirm whether PDCoV infection in vivo displays a similar tropism, ileal, jejunal, duodenal, and colonic tissues were collected from PDCoV-inoculated SPF piglets and analyzed by immunohistochemistry staining. Large numbers of PDCoV antigens were detected in the jejunum and ileum, whereas limited PDCoV antigens were detected in the duodenum, and rare PDCoV antigens were observed in the colon (Fig. 3A), which was consistent with the results observed in enteroids (Fig. 1 and Fig. 2B to G). In agreement with the results of viral antigens detected in the intestines, PDCoV infection caused severe villous atrophy in the jejunum and ileum, but intestinal lesions were barely observed in both the duodenum and colon (Fig. 3B). Our results show that PDCoV infection displays a segment-specific tropism both in vitro and in vivo. Collectively, these data indicate that porcine enteroids are a good ex vitro model to explore the events associated with PDCoV infection in vivo.
FIG 3.
PDCoV preferably infects the ileum and jejunum over the duodenum and colon in vivo. (A) Detection of PDCoV antigens by an immunohistochemistry assay. SPF piglets were orally inoculated with 1 ml 1 × 104 TCID50 PDCoV or DMEM. Intestinal tissue samples were collected at 72 hpi and stained with a mouse anti-PDCoV N protein monoclonal antibody. Scale bar, 100 μm. (B) Histological analysis of PDCoV infection by H&E staining. Intestinal sections were collected as indicated in panel A. Scale bar, 200 μm.
Enteroids isolated from different intestinal segments display unique gene expression profiles.
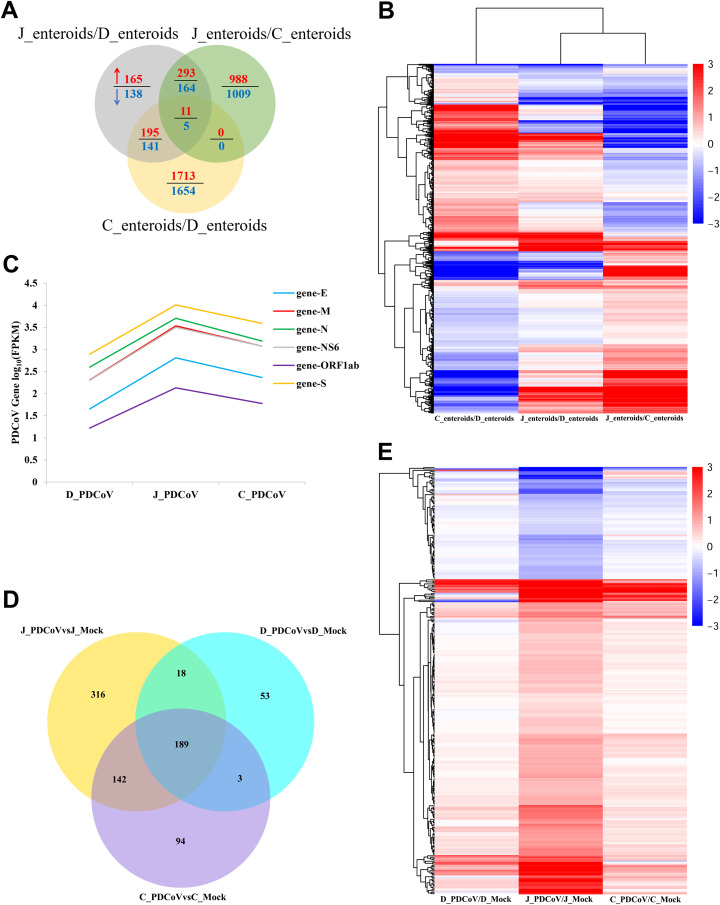
We demonstrated above that PDCoV preferably infected the jejunum and ileum over the duodenum and colon both in vitro and in vivo (Fig. 1 to 3). To explore the mechanism responsible for the PDCoV segment-specific tropism, we evaluated the gene expression profiles of enteroids from different intestinal regions with or without PDCoV infection by high-throughput RNA sequencing (RNA-seq). Initially, we compared the transcriptional profiles of enteroids isolated from jejunal, duodenal, and colonic crypt stem cells before PDCoV infection. The results of the Venn diagram showed that 664 genes were upregulated and 448 genes were downregulated in jejunal enteroids compared with duodenal enteroids, 1,292 genes were upregulated and 1,178 genes were downregulated in jejunal enteroids compared with colonoids, and 1,919 genes were upregulated and 1,800 genes were downregulated in colonoids compared with duodenal enteroids, with only 16 shared transcripts among these three groups, including 11 upregulated genes and 5 downregulated genes (Fig. 4A), suggesting a distinct gene expression profile among different segmental enteroids, especially the small intestine-isolated enteroids compared with colonoids. Next, the heat map of differentially expressed genes of the three groups further revealed that the transcriptional profiles of these three types of enteroids differed significantly and exhibited a distinct segmental expression pattern (Fig. 4B). These data demonstrate that porcine intestinal segments exhibit segment-specific gene expression patterns.
FIG 4.
Enteroids isolated from different intestinal segments display unique gene expression profiles. (A) Venn diagram of common differentially expressed genes of enteroids derived from jejunal, duodenal, and colonic crypts. Monolayers of jejunal enteroids (J_enteroids), duodenal enteroids (D_enteroids), and colonoids (C_ enteroids) isolated from three individual SPF 2-day-old piglets were cultured for 24 h. Total RNA of enteroids was extracted and sequenced by RNA-seq. (B) Heat map comparing gene expression levels among enteroids derived from jejunal, duodenal, and colonic crypts. (C) Viral gene transcript levels of PDCoV-infected enteroids derived from jejunal, duodenal, and colonic crypts were measured by RNA-seq. Monolayers of jejunal enteroids, duodenal enteroids, and colonoids were infected with PDCoV at an MOI of 2. Total RNA was isolated at 24 hpi and used for RNA-seq. The transcript levels of viral genes were shown as log(FPKM) values. (D) Venn diagram of common differential gene expression of PDCoV-infected enteroids derived from jejunal, duodenal, and colonic crypts. (E) Heat map of differentially expressed genes in PDCoV-infected enteroids derived from jejunal, duodenal, and colonic crypts. Data in panels B and E are based on log(RPKM) values from three independent enteroid preparations. J_PDCoV or mock, D_PDCoV or mock, and C_PDCoV or mock are shown as jejunal enteroids, duodenal enteroids, and colonoids infected with or without PDCoV, respectively.
Next, we further assessed the gene expression profiles following PDCoV infection in enteroids from different intestinal segments by RNA-seq. The successful infection of PDCoV in enteroids was confirmed by reverse transcriptase quantitative PCR (RT-qPCR) (data not shown). Initially, to further evaluate PDCoV infection, we compared the transcriptional levels of PDCoV viral genes in enteroids isolated from jejunal, duodenal, and colonic crypts, including four structural protein genes, small membrane (E) gene, membrane (M) gene, nucleocapsid (N) gene, and S gene, as well as the open reading frame 1ab (ORF1ab) gene and the accessory protein NS6 gene (39), as assessed by log(fragments per kilobase per million [FPKM]) values. The highest transcript levels of viral genes were observed in jejunal enteroids, followed by colonoids and duodenal enteroids (Fig. 4C). Next, we explored the modification of host gene expression in enteroids after PDCoV infection. The Venn diagram analysis showed that 665 genes, 263 genes, and 428 genes were differentially expressed in jejunal enteroids, duodenal enteroids, and colonoids, respectively, with 189 shared genes among these three PDCoV-infected enteroids compared with the corresponding mock-infected control (Fig. 4D). We further found that the differential expression of jejunal enteroids-specific transcriptome in response to PDCoV infection was more significant than that of duodenal enteroids and colonoids (Fig. 4E). These results reveal that, consistent with PDCoV replication in enteroids, PDCoV infection in jejunal enteroids induces a stronger upregulation of transcriptional modifications compared with duodenal enteroids and colonoids.
APN expression mainly determines the differential infectivity of PDCoV in different intestinal segments.
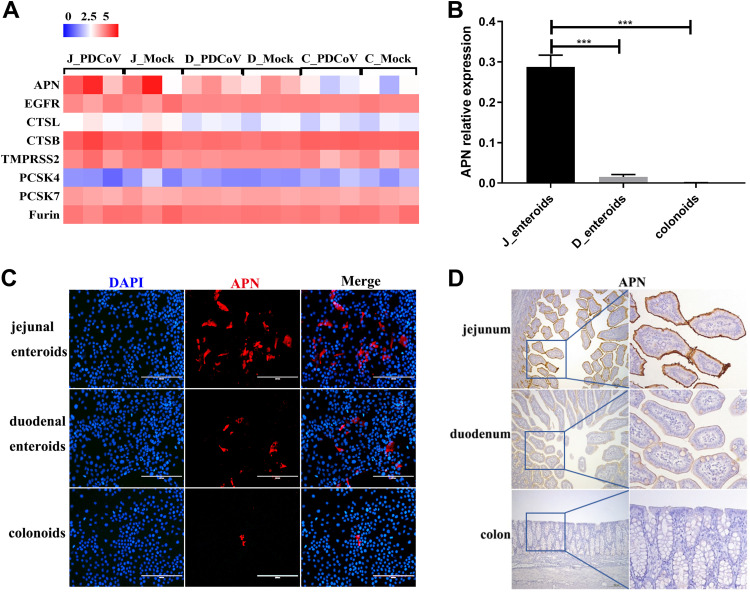
Coronavirus S protein recognizing and binding to the host cell receptor after activating by host cell proteases is an essential first step in establishing infection and initially controls the cell and tissue tropism, host range, and pathogenesis of coronavirus (11, 12). It is rational to speculate that the expression levels of viral receptor and host proteases on the cells are closely related to the susceptibility to PDCoV infection. APN has been identified as a primary viral receptor for PDCoV (12, 13). To assess whether the expression levels of pAPN and other host proteases related to coronaviruses among different intestinal segments determine the tropism of PDCoV infection, we performed RNA-seq analyses from PDCoV-infected enteroids from different intestinal segments. We screened known coronavirus host cell proteases, including lysosomal proteases (cathepsin L and B [CTSL and CTSB, respectively]), which have been reported to activate the PDCoV S protein for membrane fusion (14); membrane-binding protease TMPRSS2, which was shown to be important for the late stages of the PEDV life cycle (40); and proprotein convertases (PCSK4, PCSK7, and furin), which can cleave most coronavirus S proteins, such as that in Middle East respiratory syndrome coronavirus (MERS-CoV) (41), and we next compared their transcriptional profiles among enteroids derived from jejunal, duodenal, and colonic crypts. The results of the heat map showed no obvious differences of host cell proteases expression among the three types of enteroids (Fig. 5A), indicating that the PDCoV segment-specific tropism is not caused by the differential expression of host proteases. Moreover, the transcriptional level of epidermal growth factor receptor (EGFR), which plays a synergistic role with APN used as a TGEV entry receptor (42, 43), was almost the same in the three types of enteroids, suggesting that EGFR is not related to the segmental differences of PDCoV infection (Fig. 5A). Surprisingly, we found that the pAPN receptor was highly expressed in jejunal enteroids, and PDCoV infection had no impact on its expression, whereas the expression levels of pAPN were much lower in duodenal enteroids and colonoids, indicating that the differential expression of pAPN among different intestinal segments determines the tropism of PDCoV (Fig. 5A). In addition, the results of RT-qPCR showed that the expression of pAPN in jejunal enteroids was nearly 18-fold higher than that in duodenal enteroids and 656-fold higher than that in colonoids, which exhibited a trend similar to that observed in the RNA-seq data set (Fig. 5B). The expression levels of pAPN among the three types of enteroids were further confirmed by an IFA (Fig. 5C). To validate the differential expression of pAPN in different intestinal segments in vivo, the pAPN expression in jejunal, duodenal, and colonic tissues from the same SPF piglet was assessed by immunohistochemistry staining. In agreement with the data observed in vitro, we detected a large number of pAPN+ cells in the jejunum, whereas a lesser degree of pAPN antigens was observed in the duodenum, and rare pAPN antigens were detected in the colon (Fig. 5D). Overall, these results demonstrate that the differential expression of pAPN in different intestinal regions highly correlates with the intestinal segmental susceptibility of PDCoV.
FIG 5.
APN expression mainly determines the differential infectivity of PDCoV in different intestinal segments. (A) Heat map of the APN and coronavirus host proteases expression in enteroids. Enteroid monolayers derived from jejunal, duodenal, and colonic crypts were infected with or without PDCoV for 24 h. Total RNA was isolated from enteroids and used for RNA-seq. Data are based on log(RPKM) values from three independent enteroid preparations. (B) Verification of the APN receptor expression in enteroids by RT-qPCR. Monolayers of jejunal enteroids (J_enteroids), duodenal enteroids (D_enteroids), and colonoids generated from three SPF piglets were cultured for 24 h, and the APN receptor expression level was quantified by RT-qPCR. Data represent the means ±SEMs of results from three wells and three independent enteroid preparations. Error bars denote deviations. ***, P < 0.005. (C) Detection of the APN receptor expression in enteroids by IFA. Monolayers of jejunal enteroids, duodenal enteroids, and colonoids were cultured for 24 h. Cells were fixed and stained with mouse anti-APN monoclonal antibody (red). DAPI-stained nuclei were shown in blue. Scale bar, 200 μm. (D) Detection of APN antigens by an immunohistochemistry assay. Intestinal tissue samples collected from the same SPF piglets were stained with mouse anti-APN monoclonal antibody. Scale bar, 200 μm.
IFN responses are not related to the differential infectivity of PDCoV in different intestinal segments.
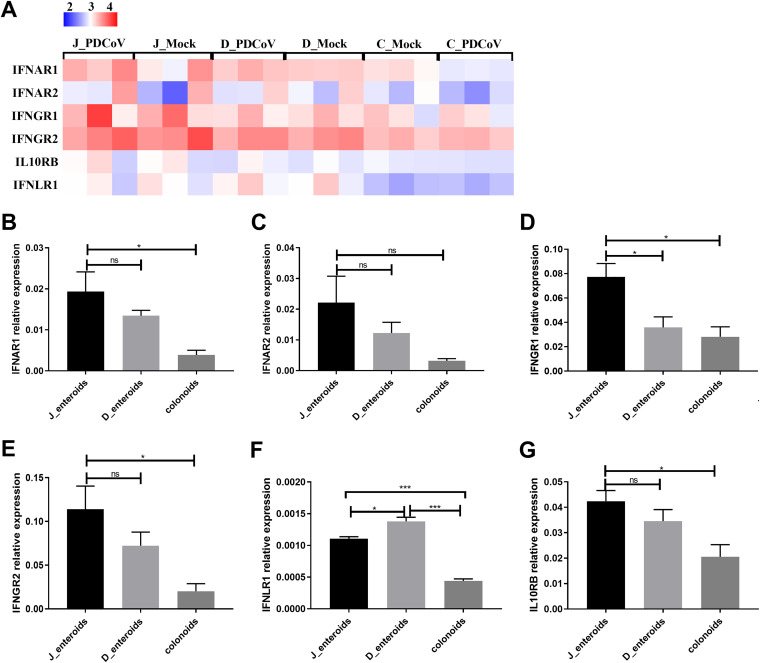
IFNs elicited by virus infection provide a robust first line of defense against viral infection (44). The levels of IFN receptors on the cellular surface determine the sensitivity of cells in response to IFN (45). The IFN receptors, especially type III IFN-λ receptor, have been reported to determine the resistance of host mucosal barrier to viral infection (46). To characterize the role of IFN responses in determining the segmental differences of PDCoV infection, we initially compared the receptor expressions of all three IFN types among different intestinal regions, including type I IFN receptors (IFNAR1 and IFNAR2), type II IFN receptors (IFNGR1 and IFNGR2), and type III IFN receptors (IL10RB and IFNLR1)(47). The results of the heat map revealed that the expression profiles of each type of IFN receptor in enteroids were distinct, but the patterns among these three different enteroids were similar (Fig. 6A). The transcript levels of all three types of IFN receptors were markedly expressed in both jejunal enteroids and duodenal enteroids, while colonoids had the lowest transcript levels of IFN receptors (Fig. 6A). The expression levels of IFN receptors in enteroids were not reduced; instead, they were slightly increased following PDCoV infection (Fig. 6A), indicating that PDCoV does not escape the IFN responses by reducing the expression of IFN receptors. The expression patterns of IFN receptors in the three types of enteroids were further confirmed by RT-qPCR; the receptors of all three types of IFNs were highly expressed in jejunal enteroids and duodenal enteroids, and lower in colonoids, which was in agreement with the transcriptional levels detected by RNA-seq (Fig. 6B to G). These findings suggest that the expression profiles of IFN receptors are not related to the segmental differences of PDCoV infection.
FIG 6.
IFN receptor expression is not related to the differential infectivity of PDCoV in different intestinal segments. (A) Heat map of the IFN receptors expression in enteroids. Enteroid monolayers derived from jejunal, duodenal, and colonic crypts were infected with or without PDCoV for 24 h. Total RNA was isolated from enteroids, and the expression levels of IFN receptors were determined by RNA-seq. Data are based on log(RPKM) values from three independent enteroid preparations. (B to G) Verification of the IFN receptors expression in enteroids by RT-qPCR. Monolayers of jejunal enteroids (J_enteroids), duodenal enteroids (D_enteroids), and colonoids were cultured for 24 h. The mRNA expression levels of type I IFN receptors (B and C), type II IFN receptors (D and E), and type III IFN receptors (F and G) were measured by RT-qPCR. Data represent the means ±SEMs of results from three wells and three independent enteroid preparations. Error bars denote deviations. *, P < 0.05; ***, P < 0.005; ns, not significant.
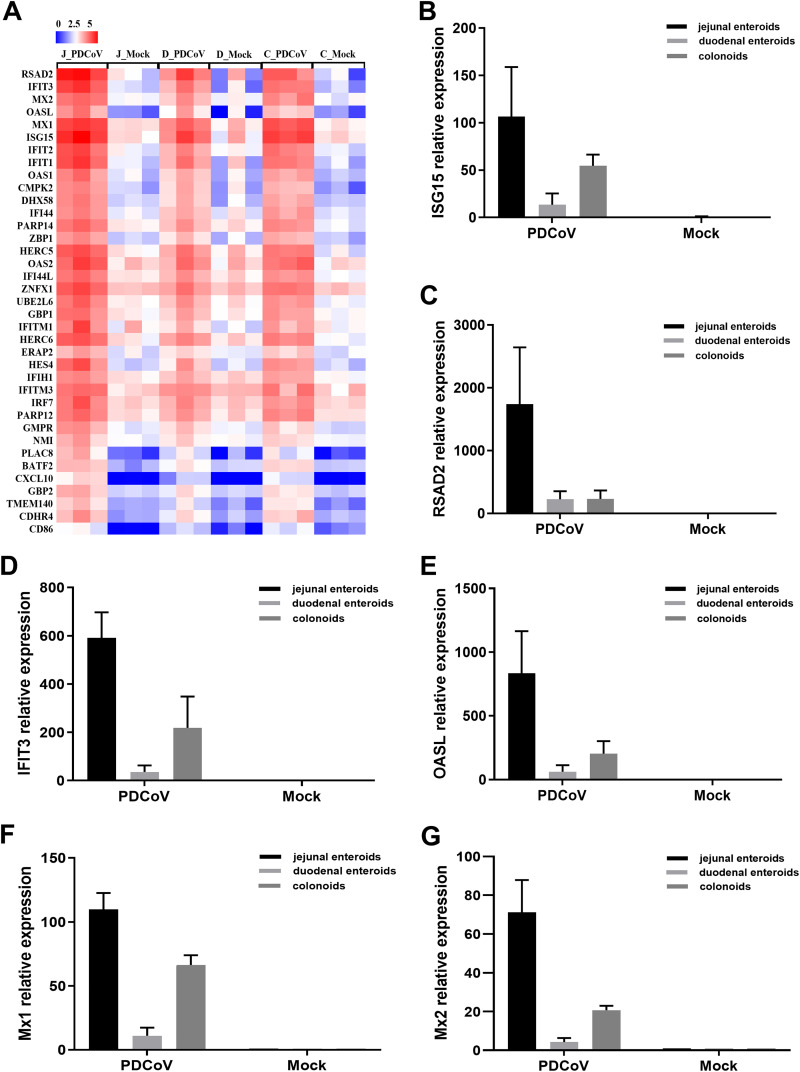
To further verify that the IFN responses do not determine the intestinal segmental tropism of PDCoV infection, we evaluated the IFN-stimulated genes (ISGs) responses in enteroids following PDCoV infection. The results showed that PDCoV infection substantially induced the expression of ISGs in all three enteroids, and the upregulation of ISGs expression was the most prominent in jejunal enteroids, followed by colonoids and duodenal enteroids (Fig. 7A). Next, we selected several typical ISG representatives, including interferon-stimulated gene 15 (ISG15), radical S-adenosyl methionine domain-containing 2 (RSAD2), interferon-induced protein with tetratricopeptide repeats 3 (IFIT3), 2′-5′-oligoadenylate synthetase-like protein (OASL), myxovirus resistance 1 (Mx1), and Mx2, to be verified by RT-qPCR. In line with the results of RNA-seq, the expression levels of all the ISGs were substantially increased in PDCoV-infected enteroids, and PDCoV infection induced the highest elevation of ISGs in jejunal enteroids, followed by colonoids and duodenal enteroids (Fig. 7B to G). Collectively, these data indicate that the differences in PDCoV infection in different intestinal segments are not determined by the IFN responses.
FIG 7.
IFN responses are not related to the differential infectivity of PDCoV in different intestinal segments. (A) Heat map of highly upregulated ISGs expression in PDCoV-infected enteroids. Enteroid monolayers derived from jejunal, duodenal, and colonic crypts were infected with or without PDCoV for 24 h. The differential expression profiles of ISGs were determined by RNA-seq. Data are based on log(RPKM) values from three independent enteroid preparations. (B to G) Confirmation of ISGs expression in PDCoV-infected enteroids by RT-qPCR. Monolayers of jejunal enteroids, duodenal enteroids, and colonoids were infected with or without PDCoV at an MOI of 2 for 24 h. The mRNA expression levels of ISG15 (B), RSAD2 (C), IFIT3 (D), OASL (E), Mx1 (F), and Mx2 (G) were quantified by RT-qPCR. The results are presented as the means ± SEMs from three wells and three independent enteroid preparations.
PDCoV replication in enteroids is restricted by exogenous IFN.
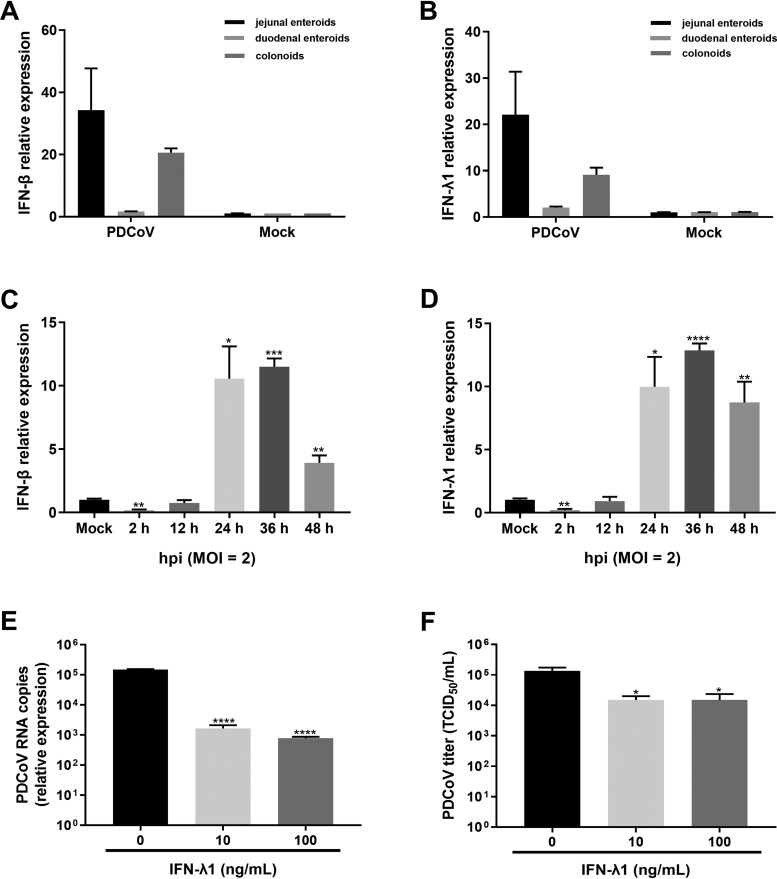
A strong ISG response was elicited in enteroids following PDCoV infection, and the levels of ISG responses were closely correlated with the levels of PDCoV replication in enteroids (Fig. 2 and 7), indicating that PDCoV infection potentially induces IFN production. To elucidate whether PDCoV infection induces the innate IFN responses in enteroids, we monitored the mRNA levels of type I IFN beta (IFN-β) and type III IFN lambda 1 (IFN-λ1) in enteroids after infection with PDCoV for 24 h. In agreement with the ISG responses (Fig. 7), we found that PDCoV infection induced the potent expression of IFN-β and IFN-λ1, and the expression levels of which were the highest in jejunal enteroids (Fig. 8A and B). We next monitored the expression kinetics of IFN-β and IFN-λ1 in jejunal enteroids after PDCoV infection by RT-qPCR. Compared with the mock-infected control, the mRNA expression levels of IFN-β and IFN-L1 were substantially elevated from 24 hpi to 48 hpi, though the high level of PDCoV infection started at 12 hpi, indicating that PDCoV induces the type I and III IFNs production at the late stage of infection instead of the early stage of infection (Fig. 8C and D).
FIG 8.
PDCoV replication in enteroids is restricted by exogenous IFN. (A and B) PDCoV infection-induced IFN production in enteroids. Enteroid monolayers isolated from jejunal, duodenal, and colonic crypts were infected with PDCoV at an MOI of 2. Expression of IFN-β (A) or IFN-λ1 (B) was assessed by RT-qPCR. (C and D) Jejunal enteroid monolayers were mock inoculated or inoculated with PDCoV at an MOI of 2 for the indicated time points. The mRNA expression level of IFN-β (C) or IFN-λ1 (D) was assessed by RT-qPCR. (E and F) Inhibition of PDCoV replication in enteroids by IFN-λ1. Jejunal enteroid monolayers were pretreated with IFN-λ1 at the indicated concentrations for 24 h and then infected with PDCoV at an MOI of 1 for 24 h. The PDCoV genomes (E) and viral titers (F) were tested by RT-qPCR and titration, respectively. Data represent the means of results from three wells for each treatment and time point. Error bars denote deviations. *, P < 0.05; **, P < 0.01; ***, P < 0.005; ****, P < 0.001.
To exclude the possibility that the noncorrelation between the IFN responses and the intestinal segmental tropism of PDCoV is because of the tolerance of PDCoV to IFNs, we evaluated the inhibition of PDCoV in enteroids by exogenous IFNs. We and others have demonstrated that compared with type I IFNs (IFN-α/β), IFN-λ acts selectively on intestinal epithelia and is superior at suppressing enteric coronavirus infection (7, 24, 25). Hence, we evaluated the antiviral activity of IFN-λ1 against PDCoV infection in jejunal enteroids. The results showed that IFN-λ1 significantly inhibited PDCoV infection in enteroids in a dose-dependent manner, with approximately 100-fold decreases in viral genomes and 0.95-log10 decreases in viral titers after IFN-λ1 treatment (Fig. 8E and F), indicating that PDCoV is sensitive to exogenous IFN. In a word, these data indicate that the noncorrelation between the IFN responses and the intestinal segmental tropism of PDCoV is not because of the resistance of PDCoV to IFNs.
DISCUSSION
PDCoV, an enteropathogenic swine coronavirus with significant economic impact, has been reported to be able to infect and kill cells from multiple species, including swine, humans, and chickens (1, 2, 12). The zoonotic potential of PDCoV makes it a priority for researchers to further clarify the determinants of cell and tissue tropism of PDCoV in its natural host, the pig. Here, we identified that PDCoV infected multiple types of intestinal epithelia, and the expression level of its cellular receptor APN instead of the IFN responses in the intestines substantially determined the intestinal segmental tropism of PDCoV. Meanwhile, we showed a distinct compartmentalized gene expression profile in the epithelia along the intestinal tube before and after PDCoV infection. Our findings provide essential insights into the pathogenesis of PDCoV.
The differentiated intestinal epithelia consist of multiple different types of functional epithelia (48). PDCoV primarily infects intestinal epithelia in vivo(1, 5, 15); however, the types of intestinal cell lineages that PDCoV is capable of infecting in vivo are not well-defined. Our previous study and the studies from other groups have shown that enteroids well recapitulate the multiple cell types and complicated crypt-villus structure of intestinal epithelia (7, 32–35). Here, we generated a porcine enteroid culture system derived from crypt stem cells as a model for PDCoV infection. We demonstrated that porcine crypt-derived enteroids were permissive to PDCoV infection (Fig. 1 and 2). Consistent with the results of PEDV that were previously reported (7), we found that PDCoV was capable of infecting multiple cell types of intestinal epithelia in enteroids, including enterocytes, stem cells, and rare goblet cells, although enterocytes were the primary cellular target, which was also confirmed in vivo in PDCoV-infected ileal tissue (Fig. 1E and F), indicating that PDCoV infection shows a multiple-cell tropism, which was consistent with previously described research that PDCoV antigens are evident in villous or crypt epithelial cells (49). The numbers and proliferation of Lgr5+ crypt stem cells are potentially associated with the susceptibility to PEDV infection (50). Whether the substantial infection of Lgr5+ stem cells by PDCoV is related to the susceptibility of PDCoV and the disruption of intestine homeostasis after infection needs to be further validated.
Enteroids derived from different intestinal segments of the same donor allow assessment of the host’s contribution to infection. We found that the differential infectivity of PDCoV in duodenal enteroids, jejunal enteroids, and colonoids was consistent with the results in vivo (Fig. 2 and 3), indicating that crypt-derived enteroids maintain the in vivo host genetic features. Outside in vivo challenge studies, ex vitro enteroids may be the ideal model system that allows for the evaluation of inherited segmental variation of the intestinal epithelium since there is a lack of the interference of immune cells and the microbiota in the lumen. The interactions between the S glycoprotein of coronavirus and the host cell receptor substantially determine the viral entry and play critical roles in the cell and tissue tropism and pathogenesis (11). Recent studies have identified that APN is used as PDCoV viral entry receptor (12, 13). TGEV, another coronavirus that exclusively uses APN as the viral receptor, exhibits less infection in the proximal small intestine (duodenum) than another two segments of the small intestine (jejunum and ileum) (43, 51), which shows a similar intestinal segmental tropism as PDCoV (Fig. 2 and 3). Therefore, it is rational to hypothesize that the APN expression level highly influences the susceptibility of different intestinal segments to PDCoV infection. In agreement with the speculation, we showed the highest level of APN expression in the jejunum, followed by the duodenum and colon (Fig. 5), which was largely correlated with the intestinal segmental tropism of PDCoV infection in in vivo and in vitro enteroids (Fig. 2 and 3). Cellular proteases involved in coronavirus entry also play important roles in the coronavirus infection (11). Surprisingly, there were no significant differences of cellular proteases expression among enteroids derived from different intestinal segments (Fig. 5A), and several cellular proteases of different families, including CTSB, TMPRSS2, PCSK7, and furin, were expressed extensively crossing the different intestinal segments (Fig. 5A). The substantial expression of cellular proteases may explain why PDCoV primarily infects the intestines in vivo despite the broad APN expression in various organs (15–18). These results suggest that the host proteases are not associated with the intestinal segmental tropism of PDCoV as a result of there being no obvious differences of host proteases expression among different intestinal regions.
IFNs, especially IFN-λ, play critical roles in determining the susceptibility and persistence of viral infection in intestinal epithelia (7, 24, 25). The levels of IFN receptors on the cellular surface determine the sensitivity of cells to IFN (45). We previously reported that the receptors of all three types of IFNs are expressed in the small intestine (52). However, the segmental differences of IFN receptors along the porcine intestinal tube remain unknown. Interestingly, type II IFN receptors (IFNGR1 and IFNGR2) were expressed at higher levels crossing different intestinal segments than the other two IFN receptors, and the receptors of all three types of IFNs exhibited an identical segment-specific profile that led them to be significantly expressed in both jejunal enteroids and duodenal enteroids, but at a lesser degree in colonoids (Fig. 6). This finding is in line with the previous study that porcine IFNAR2 is expressed at a higher level in the small intestine than in the large intestine (53). In contrast to the mainstream opinion that type III IFN-specific receptor IFNLR1 dominantly expresses in epithelia, we found that the transcriptional expression of type I IFN receptors (IFNAR1 and IFNAR2) was slightly higher than that of IFNLR1 in enteroids (Fig. 6), which was consistent with the mRNA expression levels of type I IFN receptors (IFNAR1 and IFNAR2) and IFNLR1 in mouse intestinal epithelia (54). We and others previously demonstrated that intestinal epithelia are more sensitive to IFN-λ than type I IFN (IFN-α/β) (24, 55, 56). The underlying mechanism that intestinal epithelia respond better to IFN-λ than IFN-α/β remains elusive. It is worthwhile in the future to further compare the protein expression levels of IFNAR and IFNLR1 in intestinal epithelia or the affinity of IFNAR and IFNLR1 in intestinal epithelia to IFN-α/β and IFN-λ, respectively.
As a strategy for dampening the innate immune response, certain viruses have developed mechanisms to downregulate IFN receptors (57–59). However, PDCoV infection did not decrease but slightly increased the expression of IFN receptors (Fig. 6), indicating that PDCoV does not evade the IFN responses by downregulating the expression of IFN receptors. Moreover, we found that PDCoV infection induced a robust ISG response in all three enteroids, and the ISG responses were positively related with the levels of PDCoV infection in enteroids (Fig. 2 and 7). In agreement with the ISG responses, PDCoV infection markedly elicited the production of type I and III IFNs in jejunal enteroids followed by colonoids and duodenal enteroids (Fig. 8A and B). Thus, these data indicate that the segmental differences of PDCoV infection are not determined by the IFN responses. The transcript levels of the cytoplasmic RNA sensors retinoic acid-inducible gene I (RIG-I) and melanoma differentiation-associated gene 5 (MDA5), which play critical roles in inducing IFN production following virus infection in intestinal epithelia (60), were greatly upregulated in jejunal enteroids, followed by upregulation in colonoids and duodenal enteroids after PDCoV infection (data not shown), which was closely related to the substantial induction of IFNs by PDCoV infection (Fig. 8A and B). Additionally, the kinetics of IFNs showed that PDCoV significantly induced the expression of IFNs at the late stage of infection, which was consistent with the in vivo result that PDCoV infection induces type I IFN production (31). PDCoV maintained a steadily high level of replication in jejunal enteroids at the late stage of infection despite the marked induction of endogenous IFNs, indicating that PDCoV may have evolved some strategies to evade the IFN responses. We showed that TGEV antagonizes IFN antiviral signaling via upregulating the negative regulator suppressor of cytokine signaling (SOCS1/3) expression instead of inhibiting IFN production at the late stage of infection (61); whether PDCoV employs a similar strategy as TGEV to counter the IFN antiviral signaling during infection remains to be further resolved. The induction of IFNs by PDCoV infection in enteroids was not consistent with several previous studies that PDCoV infection suppresses IFN production in porcine kidney cells (LLC-PK1 cells or PK-15 cells) (28–30). Thus, the inconsistent induction of IFN by PDCoV is potentially caused by different infection models. In the present study, we performed PDCoV infection in enteroids instead of porcine kidney cells, and the kinetics of IFN production in various models may be different following PDCoV infection. We observed that PDCoV inhibited IFN production at the early stage of infection (Fig. 8C and D). These data suggest that PDCoV might explore various strategies at different stages of infection to escape the IFN responses. It is worthwhile to further explore the dynamic mechanisms of escaping the IFN responses during the various stages of the PDCoV infection life cycle.
In conclusion, we show the specific cellular and segmental tropism of PDCoV infection in the intestines and that the distinct gene expression profiles among different intestinal segments largely determine the susceptibility to PDCoV infection. Our findings provide important insights into PDCoV pathogenesis and also contribute to clarifying the pathogenesis of other enteric coronavirus infection.
MATERIALS AND METHODS
Cell cultures and viruses.
Swine testicular (ST) cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) containing 100 U/ml penicillin, 100 μg/ml streptomycin, and 10% heat-inactivated fetal bovine serum (FBS) (Gibco, USA). The PDCoV strain NH (GenBank accession number KU981062.1) was propagated in ST cells. A PDCoV stock was prepared and titrated as previously described (6, 14).
Porcine intestinal 3D enteroid cultures.
Porcine intestinal crypts were isolated from specific-pathogen-free (SPF) 2-day-old piglets and cultured as previously described (7, 32). Briefly, the intestinal samples were cut into 2-mm segments and washed with ice-cold phosphate-buffered saline (PBS) with penicillin-streptomycin until the supernatant was clear. The intestinal segments were dissociated with Gentle cell dissociation reagent (Stemcell, Canada) and suspended in ice-cold PBS with 0.1% bovine serum albumin (BSA) and then passed through a 70-μm filter to obtain the crypts. The isolated crypts were grown in IntestiCult organoid growth medium (Stemcell, Canada) mixed with Matrigel (BD Biosciences, USA) and then plated in a 48-well plate with approximately 50 crypts per well in an incubator with 5% CO2 at 37°C. The culture medium was exchanged every 3 to 4 days. All the protocols used for animal experiments were approved by the Animal Care and Ethics Committee of the Harbin Veterinary Research Institute.
Two-dimensional enteroid monolayer cultures.
Enteroid monolayer cultures were prepared from differentiated 3D enteroids as previously described (7). First, 3D enteroids, after culturing for 7 days, were washed using ice-cold DMEM-F12 medium and were dissociated with 0.25% trypsin-EDTA (Gibco, USA). Trypsin was then inactivated by adding DMEM-F12 medium with 10% (vol/vol) FBS. Single-cell suspensions were prepared following dissociation of the cells by repeated pipetting. Single-cell suspensions were centrifuged and next cultured in IntestiCult organoid growth medium and then seeded at 50 enteroid cells per well of a 96-well plate precoated with 2.5 μl of Matrigel diluted in 100 μl ice-cold PBS that was removed after 90 min of incubation at 37°C. After differentiating for 3 days, the 2D enteroid monolayers were ready for the next experiments.
Viral infections.
Two-dimensional enteroid monolayer cells cultured in 96-well plates were infected with PDCoV at the desired MOI or mock infected with DMEM-F12. After incubation for 2 h at 37°C, cells were washed three times to remove unbound virus and cultured in IntestiCult organoid growth medium at 37°C for the times indicated on the figures. To assess the anti-PDCoV activity of IFN-λ1, 2D enteroid monolayer cells were infected with PDCoV at an MOI of 1 after being pretreated with 0 ng/ml, 10 ng/ml, or 100 ng/ml of IFN-λ1 for 24 h (Fig. 8E and F). The supernatants collected from PDCoV-infected enteroid monolayer cultures were titrated in 96-well plates of ST cells, using DMEM for 10-fold serial dilutions, with eight technical replicates per biological sample. Samples collected at the time points postinfection indicated on the figures were used for quantifying the PDCoV infection experiments (Fig. 1B and C, 2F and G, and 8C and D).
RNA extraction and reverse transcriptase quantitative PCR.
Total cellular RNA was prepared from enteroids using a Simply P total RNA extraction kit (BioFlux, China) according to the manufacturer’s protocol. RNA (1 μg) was reverse transcribed with a PrimeScript II first-strand cDNA synthesis kit (TaKaRa, Japan) according to the manufacturer’s instructions. qPCR was performed in triplicate using SYBR green PCR mix (Life Technologies, USA) on a LightCycler 480 II system (Roche, Switzerland). Relative gene expression levels were determined based on the cycle threshold (ΔΔCT) method and normalized to glyceraldehyde-3-phosphate dehydrogenase (GAPDH) expression. All qPCR primer sequences used in the study are listed in Table 1.
TABLE 1.
Sequences of qPCR primers used
| Primer | Sequence (5′ to 3′) |
|---|---|
| PDCoV-qPCR-F | AGCAACCACTCGTGTTACTTG |
| PDCoV-qPCR-R | CAACTCTGAAACCTTGAGCTG |
| APN-qPCR-F | TGTTTGACCCACAGTCCT |
| APN-qPCR-R | TCCACATAGGAGGCAAAG |
| IFNAR1-qPCR-F | ACATCACCTGCCTTCACCAG |
| IFNAR1-qPCR-R | CATGGAGCCACTGAGCTTGA |
| IFNAR2-qPCR-F | GAGTGGTTGCATTCAGAG |
| IFNAR2-qPCR-R | GATGTGGTCTGTGAAACC |
| IFNGR1-qPCR-F | TCGTAGACGATGACGATG |
| IFNGR1-qPCR-R | GGAATGCTCAAGAAGCAC |
| IFNGR2-qPCR-F | GCCTTGGTTCGAACACTA |
| IFNGR2-qPCR-R | GACGTCAAAAGGAGAGGA |
| IFNLR1-qPCR-F | GGGGCTTCTCATCACCAC |
| IFNLR1-qPCR-R | ATTCGCTCCCACCCTCTT |
| IL10RB-qPCR-F | CATGGGCTTATCATGTGC |
| IL10RB-qPCR-R | GCTCGGACTTGAACACAG |
| ISG15-qPCR-F | AGCATGGTCCTGTTGATGGTG |
| ISG15-qPCR-R | CAGAAATGGTCAGCTTGCACG |
| RSAD2-qPCR-F | AAGCAGAGCAGTTTGTTATCAGC |
| RSAD2-qPCR-R | TTCCGCCCGTTTCTACAGT |
| IFIT3-qPCR-F | GCACCAAATTCATGGTATCTCC |
| IFIT3-qPCR-R | TTCCTTCCTGTCTCTGTCAGCC |
| OASL-qPCR-F | TCCCTGGGAAGAATGTGCAG |
| OASL-qPCR-R | CCCTGGCAAGAGCATAGTGT |
| Mx1-qPCR-F | CACTGCTTTGATACAAGGAGAGG |
| Mx1-qPCR-R | GCACTCCATCTGCAGAACTCAT |
| Mx2-qPCR-F | GGTGGACCCCGAAGGAGACAG |
| Mx2-qPCR-R | AAGTGCGGATGCGAGTGAAAG |
| IFN-β-qPCR-F | AGCACTGGCTGGAATGAAAC |
| IFN-β-qPCR-R | TCCAGGATTGTCTCCAGGTC |
| IFN-λ1-qPCR-F | CCACGTCGAACTTCAGGCTT |
| IFN-λ1-qPCR-R | ATGTGCAAGTCTCCACTGGT |
| GAPDH-qPCR-F | CCTTCCGTGTCCCTACTGCCAAC |
| GAPDH-qPCR-R | GACGCCTGCTTCACCACCTTCT |
Immunofluorescence assay.
Enteroid monolayers on 96-well plates were infected with PDCoV at an MOI of 2 for 24 h. The levels of PDCoV infection and PDCoV-targeted specific intestinal epithelial cell types were analyzed by an IFA as described previously (6, 7). Enteroid monolayers were washed with PBS and fixed with 4% paraformaldehyde (PFA) at room temperature followed by 0.2% Triton X-100 to permeabilize cell membranes. Cells were blocked for 2 h at 37°C in PBS containing 5% FBS and 5% skim milk (Sigma-Aldrich, USA), incubated with the primary antibodies for 2 h at 37°C, and then incubated with the secondary antibodies for 1 hat 37°C. Expression of the PDCoV spike (S) protein was detected using a mouse anti-PDCoV S monoclonal antibody (1:1,000) stocked in our laboratory. Expression of differentiation markers of intestinal epithelia was detected using primary anti-villin (1:100; Abcam, USA) for enterocytes, anti-Lgr5 (1:50; Novus Biologicals, USA) for stem cells, anti-mucin 2 (1:50; Abcam, USA) for goblet cells, and anti-Ki-67 (1:600; Abcam, USA) for proliferative cells. The cells were then incubated with corresponding secondary antibody conjugated to Alexa Fluor 546 goat anti-mouse IgG antibody (1:500; Thermo Fisher Scientific, USA) or Alexa Fluor 488 donkey polyclonal antibody against rabbit IgG (1:1,000; Thermo Fisher Scientific, USA). Nuclei were stained with DAPI (4′,6-diamidino-2-phenylindole) for 10 min at room temperature. Images were captured using an Evos FL Auto2 fluorescence microscope.
Representative sections of ileal tissues were cut into 10-μm-thick sections and mounted onto glass slides. Then the slides were deparaffinized, rehydrated, heated for antigen retrieval, and blocked with 5% skim milk in PBS at 37°C for 1 h. The expressions of PDCoV S protein and differentiation markers of intestinal epithelia were stained as described above. Nuclei were next stained with DAPI for 10 min at room temperature. The immunofluorescence was detected using a Carl Zeiss microscope (LSM700; Carl Zeiss, Heidenheim, Germany).
PDCoV infection of piglets.
Six 5-day-old SPF piglets were randomly divided into two groups. The SPF piglets in group 1 were orally inoculated with 1 ml of 104 50% tissue cultureinfective dose (TCID50) of the PDCoV NH strain. The SPF piglets in group 2 were inoculated with 1 ml of DMEM medium and served as the negative-control group. The clinical signs of vomiting and diarrhea were recorded every 12 h after PDCoV infection. All the piglets were euthanized at 72 hpi. Piglet intestinal samples were collected for pathological and histological examination.
Immunohistochemistry.
Intestinal tissue samples were fixed in 10% formalin for 48 h and then deparaffinized, dehydrated in graded ethanol, and embedded in paraffin wax following standard laboratory methods by immunohistochemistry (IHC) as described previously (6, 7). Paraffin sections were dewaxed in xylene and rehydrated in decreasing concentrations of ethanol. Antigen retrieval was performed in citrate buffer (pH 7.4)-sodium citrate buffer solution (pH 8.0) at 121°C for 30 min. The slides were blocked with 5% skim milk in PBS at room temperature for 30 min. Antigen detections were performed using a mouse anti-PDCoV nucleocapsid (N) monoclonal antibody (1:50) or mouse anti-APN monoclonal antibody (1:100) prepared in our laboratory. After incubation overnight at 4°C, the slides were washed three times with PBS and then incubated with goat anti-mouse secondary antibody for 40 min at room temperature. The slides were then visualized with diaminobenzidine (DAB) (Sigma-Aldrich, USA) and counterstained with hematoxylin. The slides were finally dehydrated, clarified in xylene, mounted with Entellan mounting medium (Sigma-Aldrich, USA), and examined with an optical microscopefor positive immunoreactivity.
Histological analysis.
Intestinal tissue samples were routinely fixed in 10% formalin for 48 h and then were processed by the following steps: dehydrating in graded ethanol, embedding in paraffin, serially sectioning to 5-μm-thick sections, and subsequently mounting on slides. The sections were dewaxed in xylene, rehydrated with increasing concentrations of ethanol, and stained with hematoxylin and eosin (H&E; Sigma-Aldrich) for histopathological examination by a light microscope.
RNA-seq.
Porcine intestinal enteroid monolayers isolated from three individual SPF 2-day-old piglets were infected with PDCoV at an MOI of 2 or mock infected with DMEM-F12 at 37°C for 24 h and then were prepared for RNA sequencing. Total RNA was extracted using the TRIzol reagent (Thermo Fisher Scientific, USA) following the manufacturer’s instructions. The RNA quality was assessed using an Agilent 2100 Bioanalyzer (Agilent Technologies, USA), a NanoDrop instrument (Thermo Fisher Scientific), and 1% agarose gel electrophoresis. Approximately 3 μg of the entire isolated RNA was used to prepare each RNA sequencing library using a NEBNext Ultra RNA library prep kit for Illumina (NEB, USA) following the manufacturer’s protocol at Novogene. Briefly, poly(A) mRNA was purified from total RNA using oligo(dT) beads and fragmented using NEBNext first strand synthesis reaction buffer. First-strand cDNA was generated using random hexamer-primed reverse transcription, and then primitive, fragmented mRNA was digested by DNA polymerase I and RNase H for second-strand synthesis initiation. cDNA library construction was completed following a series of terminal repairs and size selection of cDNA fragments with a length of ∼250 to 300 bp using the AMPure system (Beckman Coulter, USA) and PCR enrichment. The cDNA library was sequenced on the Illumina HiSeq 4000 platform (Illumina, USA) with 125 bp/150 bp paired-end reads by Novogene (Tianjin, China). RNA sequencing data were processed and mapped to the Sus scrofa reference genome and the PDCoV genome. Differential expression was analyzed using the DESeq2 R package at a significance cutoff of P < 0.05. Differentially expressed genes were identified by the screening criteria based on |log2(fold change)| > 1 and false discovery rate (FDR) < 0.05.
Statistical analysis.
All statistical analyses were performed using GraphPad Prism (GraphPad Software, Inc.). Experiments were independently performed at least three times as indicated in the figure legends or as detailed. The scale data of viral genomes and titers were taken as log(fold change) values and expressed as power of 10. Data are presented as the means ± the standard errors of the mean (SEMs). Differences were considered significant when the P value was <0.05.
ACKNOWLEDGMENTS
This work was supported by grants from the National Key Technology R&D Program of China (2016YFD0500103) and the National Natural Science Fund (31772718).
We thank Xiangxi Pei (Northeast Agricultural University, Harbin, China) for assistance with and advice on RNA-seq data analysis.
REFERENCES
- 1.Chen Q, Gauger P, Stafne M, Thomas J, Arruda P, Burrough E, Madson D, Brodie J, Magstadt D, Derscheid R, Welch M, Zhang J. 2015. Pathogenicity and pathogenesis of a United States porcine deltacoronavirus cell culture isolate in 5-day-old neonatal piglets. Virology 482:51–59. doi: 10.1016/j.virol.2015.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang L, Byrum B, Zhang Y. 2014. Detection and genetic characterization of deltacoronavirus in pigs, Ohio, USA, 2014. Emerg Infect Dis 20:1227–1230. doi: 10.3201/eid2007.140296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stevenson GW, Hoang H, Schwartz KJ, Burrough ER, Sun D, Madson D, Cooper VL, Pillatzki A, Gauger P, Schmitt BJ, Koster LG, Killian ML, Yoon KJ. 2013. Emergence of porcine epidemic diarrhea virus in the United States: clinical signs, lesions, and viral genomic sequences. J Vet Diagn Invest 25:649–654. doi: 10.1177/1040638713501675. [DOI] [PubMed] [Google Scholar]
- 4.Xia L, Yang Y, Wang J, Jing Y, Yang Q. 2018. Impact of TGEV infection on the pig small intestine. Virol J 15:102. doi: 10.1186/s12985-018-1012-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jung K, Hu H, Eyerly B, Lu Z, Chepngeno J, Saif LJ. 2015. Pathogenicity of 2 porcine deltacoronavirus strains in gnotobiotic pigs. Emerg Infect Dis 21:650–654. doi: 10.3201/eid2104.141859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang J, Chen J, Liu Y, Da S, Shi H, Zhang X, Liu J, Cao L, Zhu X, Wang X, Ji Z, Feng L. 2020. Pathogenicity of porcine deltacoronavirus (PDCoV) strain NH and immunization of pregnant sows with an inactivated PDCoV vaccine protects 5-day-old neonatal piglets from virulent challenge. Transbound Emerg Dis 67:572–583. doi: 10.1111/tbed.13369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li L, Fu F, Guo S, Wang H, He X, Xue M, Yin L, Feng L, Liu P. 2018. Porcine intestinal enteroids: a new model for studying enteric coronavirus porcine epidemic diarrhea virus infection and the host innate response. J Virol 93:e01682-18. doi: 10.1128/JVI.01682-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Haber AL, Biton M, Rogel N, Herbst RH, Shekhar K, Smillie C, Burgin G, Delorey TM, Howitt MR, Katz Y, Tirosh I, Beyaz S, Dionne D, Zhang M, Raychowdhury R, Garrett WS, Rozenblatt-Rosen O, Shi HN, Yilmaz O, Xavier RJ, Regev A. 2017. A single-cell survey of the small intestinal epithelium. Nature 551:333–339. doi: 10.1038/nature24489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kozuka K, He Y, Koo-McCoy S, Kumaraswamy P, Nie B, Shaw K, Chan P, Leadbetter M, He L, Lewis JG, Zhong Z, Charmot D, Balaa M, King AJ, Caldwell JS, Siegel M. 2017. Development and characterization of a human and mouse intestinal epithelial cell monolayer platform. Stem Cell Rep 9:1976–1990. doi: 10.1016/j.stemcr.2017.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Geoghegan JL, Duchene S, Holmes EC. 2017. Comparative analysis estimates the relative frequencies of co-divergence and cross-species transmission within viral families. PLoS Pathog 13:e1006215. doi: 10.1371/journal.ppat.1006215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Millet JK, Whittaker GR. 2015. Host cell proteases: critical determinants of coronavirus tropism and pathogenesis. Virus Res 202:120–134. doi: 10.1016/j.virusres.2014.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li W, Hulswit RJG, Kenney SP, Widjaja I, Jung K, Alhamo MA, van Dieren B, van Kuppeveld FJM, Saif LJ, Bosch BJ. 2018. Broad receptor engagement of an emerging global coronavirus may potentiate its diverse cross-species transmissibility. Proc Natl Acad Sci U S A 115:E5135–E5143. doi: 10.1073/pnas.1802879115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang B, Liu Y, Ji CM, Yang YL, Liang QZ, Zhao P, Xu LD, Lei XM, Luo WT, Qin P, Zhou J, Huang YW. 2018. Porcine deltacoronavirus engages the transmissible gastroenteritis virus functional receptor porcine aminopeptidase N for infectious cellular entry. J Virol 92:e00318-18. doi: 10.1128/JVI.00318-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang J, Chen J, Shi D, Shi H, Zhang X, Liu J, Cao L, Zhu X, Liu Y, Wang X, Ji Z, Feng L. 2019. Porcine deltacoronavirus enters cells via two pathways: a protease-mediated one at the cell surface and another facilitated by cathepsins in the endosome. J Biol Chem 294:9830–9843. doi: 10.1074/jbc.RA119.007779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jung K, Hu H, Saif LJ. 2016. Porcine deltacoronavirus infection: etiology, cell culture for virus isolation and propagation, molecular epidemiology and pathogenesis. Virus Res 226:50–59. doi: 10.1016/j.virusres.2016.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mina-Osorio P. 2008. The moonlighting enzyme CD13: old and new functions to target. Trends Mol Med 14:361–371. doi: 10.1016/j.molmed.2008.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dijkman R, Jebbink MF, Koekkoek SM, Deijs M, Jónsdóttir HR, Molenkamp R, Ieven M, Goossens H, Thiel V, van der Hoek L. 2013. Isolation and characterization of current human coronavirus strains in primary human epithelial cell cultures reveal differences in target cell tropism. J Virol 87:6081–6090. doi: 10.1128/JVI.03368-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kenny AJ, Maroux S. 1982. Topology of microvillar membrance hydrolases of kidney and intestine. Physiol Rev 62:91–128. doi: 10.1152/physrev.1982.62.1.91. [DOI] [PubMed] [Google Scholar]
- 19.Mowat AM, Agace WW. 2014. Regional specialization within the intestinal immune system. Nat Rev Immunol 14:667–685. doi: 10.1038/nri3738. [DOI] [PubMed] [Google Scholar]
- 20.Ortega-Cava CF, Ishihara S, Rumi MA, Kawashima K, Ishimura N, Kazumori H, Udagawa J, Kadowaki Y, Kinoshita Y. 2003. Strategic compartmentalization of Toll-like receptor 4 in the mouse gut. J Immunol 170:3977–3985. doi: 10.4049/jimmunol.170.8.3977. [DOI] [PubMed] [Google Scholar]
- 21.Wang Y, Devkota S, Musch MW, Jabri B, Nagler C, Antonopoulos DA, Chervonsky A, Chang EB. 2010. Regional mucosa-associated microbiota determine physiological expression of TLR2 and TLR4 in murine colon. PLoS One 5:e13607. doi: 10.1371/journal.pone.0013607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wells AI, Coyne CB. 2018. Type III interferons in antiviral defenses at barrier surfaces. Trends Immunol 39:848–858. doi: 10.1016/j.it.2018.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li SF, Gong MJ, Zhao FR, Shao JJ, Xie YL, Zhang YG, Chang HY. 2018. Type I interferons: distinct biological activities and current applications for viral infection. Cell Physiol Biochem 51:2377–2396. doi: 10.1159/000495897. [DOI] [PubMed] [Google Scholar]
- 24.Li L, Fu F, Xue M, Chen W, Liu J, Shi H, Chen J, Bu Z, Feng L, Liu P. 2017. IFN-lambda preferably inhibits PEDV infection of porcine intestinal epithelial cells compared with IFN-alpha. Antiviral Res 140:76–82. doi: 10.1016/j.antiviral.2017.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang Q, Ke H, Blikslager A, Fujita T, Yoo D. 2018. Type III interferon restriction by porcine epidemic diarrhea virus and the role of viral protein nsp1 in IRF1 signaling. J Virol 92:e01677-17. doi: 10.1128/JVI.01677-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee S, Wilen CB, Orvedahl A, McCune BT, Kim KW, Orchard RC, Peterson ST, Nice TJ, Baldridge MT, Virgin HW. 2017. Norovirus cell tropism is determined by combinatorial action of a viral non-structural protein and host cytokine. Cell Host Microbe 22:449–459. doi: 10.1016/j.chom.2017.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hernandez PP, Mahlakoiv T, Yang I, Schwierzeck V, Nguyen N, Guendel F, Gronke K, Ryffel B, Hoelscher C, Dumoutier L, Renauld JC, Suerbaum S, Staeheli P, Diefenbach A. 2015. Interferon-lambda and interleukin 22 act synergistically for the induction of interferon-stimulated genes and control of rotavirus infection. Nat Immunol 16:698–707. doi: 10.1038/ni.3180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fang P, Fang L, Ren J, Hong Y, Liu X, Zhao Y, Wang D, Peng G, Xiao S. 2018. Porcine deltacoronavirus accessory protein NS6 antagonizes interferon beta production by interfering with the binding of RIG-I/MDA5 to double-stranded RNA. J Virol 92:e00712-18. doi: 10.1128/JVI.00712-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Likai J, Shasha L, Wenxian Z, Jingjiao M, Jianhe S, Hengan W, Yaxian Y. 2019. Porcine deltacoronavirus nucleocapsid protein suppressed IFN-beta production by interfering porcine RIG-I dsRNA-binding and K63-linked polyubiquitination. Front Immunol 10:1024. doi: 10.3389/fimmu.2019.01024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Luo J, Fang L, Dong N, Fang P, Ding Z, Wang D, Chen H, Xiao S. 2016. Porcine deltacoronavirus (PDCoV) infection suppresses RIG-I-mediated interferon-beta production. Virology 495:10–17. doi: 10.1016/j.virol.2016.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xu Z, Zhong H, Huang S, Zhou Q, Du Y, Chen L, Xue C, Cao Y. 2019. Porcine deltacoronavirus induces TLR3, IL-12, IFN-alpha, IFN-beta and PKR mRNA expression in infected Peyer’s patches in vivo. Vet Microbiol 228:226–233. doi: 10.1016/j.vetmic.2018.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sato T, Vries RG, Snippert HJ, van de Wetering M, Barker N, Stange DE, van Es JH, Abo A, Kujala P, Peters PJ, Clevers H. 2009. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature 459:262–265. doi: 10.1038/nature07935. [DOI] [PubMed] [Google Scholar]
- 33.Ettayebi K, Crawford SE, Murakami K, Broughman JR, Karandikar U, Tenge VR, Neill FH, Blutt SE, Zeng X-L, Qu L, Kou B, Opekun AR, Burrin D, Graham DY, Ramani S, Atmar RL, Estes MK. 2016. Replication of human noroviruses in stem cell-derived human enteroids. Science 353:1387–1393. doi: 10.1126/science.aaf5211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Drummond CG, Bolock AM, Ma C, Luke CJ, Good M, Coyne CB. 2017. Enteroviruses infect human enteroids and induce antiviral signaling in a cell lineage-specific manner. Proc Natl Acad Sci U S A 114:1672–1677. doi: 10.1073/pnas.1617363114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Saxena K, Blutt SE, Ettayebi K, Zeng X-L, Broughman JR, Crawford SE, Karandikar UC, Sastri NP, Conner ME, Opekun AR, Graham DY, Qureshi W, Sherman V, Foulke-Abel J, In J, Kovbasnjuk O, Zachos NC, Donowitz M, Estes MK. 2016. Human intestinal enteroids: a new model to study human rotavirus infection, host restriction, and pathophysiology. J Virol 90:43–56. doi: 10.1128/JVI.01930-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.van der Hee B, Loonen LMP, Taverne N, Taverne-Thiele JJ, Smidt H, Wells JM. 2018. Optimized procedures for generating an enhanced, near physiological 2D culture system from porcine intestinal organoids. Stem Cell Res 28:165–171. doi: 10.1016/j.scr.2018.02.013. [DOI] [PubMed] [Google Scholar]
- 37.Thorne CA, Chen IW, Sanman LE, Cobb MH, Wu LF, Altschuler SJ. 2018. Enteroid monolayers reveal an autonomous WNT and BMP circuit controlling intestinal epithelial growth and organization. Dev Cell 44:624–633. doi: 10.1016/j.devcel.2018.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sato T, Clevers H. 2013. Growing self-organizing mini-guts from a single intestinal stem cell: mechanism and applications. Science 340:1190–1194. doi: 10.1126/science.1234852. [DOI] [PubMed] [Google Scholar]
- 39.Woo PC, Lau SK, Lam CS, Lau CC, Tsang AK, Lau JH, Bai R, Teng JL, Tsang CC, Wang M, Zheng BJ, Chan KH, Yuen KY. 2012. Discovery of seven novel mammalian and avian coronaviruses in the genus Deltacoronavirus supports bat coronaviruses as the gene source of Alphacoronavirus and Betacoronavirus and avian coronaviruses as the gene source of Gammacoronavirus and Deltacoronavirus. J Virol 86:3995–4008. doi: 10.1128/JVI.06540-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shirato K, Matsuyama S, Ujike M, Taguchi F. 2011. Role of proteases in the release of porcine epidemic diarrhea virus from infected cells. J Virol 85:7872–7880. doi: 10.1128/JVI.00464-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Millet JK, Whittaker GR. 2014. Host cell entry of Middle East respiratory syndrome coronavirus after two-step, furin-mediated activation of the spike protein. Proc Natl Acad Sci U S A 111:15214–15219. doi: 10.1073/pnas.1407087111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hu W, Zhang S, Shen Y, Yang Q. 2018. Epidermal growth factor receptor is a co-factor for transmissible gastroenteritis virus entry. Virology 521:33–43. doi: 10.1016/j.virol.2018.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Delmas B, Gelfi J, L'Haridon R, Vogel LK, Sjöström H, Norén O, Laude H. 1992. Aminopeptidase N is a major receptor for the entero-pathogenic coronavirus TGEV. Nature 357:417–420. doi: 10.1038/357417a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stark GR, Darnell JE Jr. 2012. The JAK-STAT pathway at twenty. Immunity 36:503–514. doi: 10.1016/j.immuni.2012.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.de Weerd NA, Nguyen T. 2012. The interferons and their receptors–distribution and regulation. Immunol Cell Biol 90:483–491. doi: 10.1038/icb.2012.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Levy DE, Marie IJ, Durbin JE. 2011. Induction and function of type I and III interferon in response to viral infection. Curr Opin Virol 1:476–486. doi: 10.1016/j.coviro.2011.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schneider WM, Chevillotte MD, Rice CM. 2014. Interferon-stimulated genes: a complex web of host defenses. Annu Rev Immunol 32:513–545. doi: 10.1146/annurev-immunol-032713-120231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.van der Flier LG, Clevers H. 2009. Stem cells, self-renewal, and differentiation in the intestinal epithelium. Annu Rev Physiol 71:241–260. doi: 10.1146/annurev.physiol.010908.163145. [DOI] [PubMed] [Google Scholar]
- 49.Jung K, Hu H, Saif LJ. 2016. Porcine deltacoronavirus induces apoptosis in swine testicular and LLC porcine kidney cell lines in vitro but not in infected intestinal enterocytes in vivo. Vet Microbiol 182:57–63. doi: 10.1016/j.vetmic.2015.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jung K, Saif LJ. 2015. Porcine epidemic diarrhea virus infection: etiology, epidemiology, pathogenesis and immunoprophylaxis. Vet J 204:134–143. doi: 10.1016/j.tvjl.2015.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kim B, Chae C. 2002. Experimental infection of piglets with transmissible gastroenteritis virus: a comparison of three strains (Korean, Purdue and Miller). J Comp Pathol 126:30–37. doi: 10.1053/jcpa.2001.0517. [DOI] [PubMed] [Google Scholar]
- 52.Shan L, Fu F, Xue M, Zhu X, Li L, Feng L, Liu P. 2019. Interferon gamma inhibits transmissible gastroenteritis virus infection mediated by an IRF1 signaling pathway. Arch Virol 164:2659–2669. doi: 10.1007/s00705-019-04362-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Xing F, Li Y, Liang S, Liu D, Jiang C, Zhang Y, Kang L, Jiang Y. 2011. cDNA cloning, genomic structure and mRNA expression pattern of porcine type I interferons receptor 2 gene. Int J Immunogenet 38:339–345. doi: 10.1111/j.1744-313X.2011.01018.x. [DOI] [PubMed] [Google Scholar]
- 54.Mahlakoiv T, Hernandez P, Gronke K, Diefenbach A, Staeheli P. 2015. Leukocyte-derived IFN-alpha/beta and epithelial IFN-lambda constitute a compartmentalized mucosal defense system that restricts enteric virus infections. PLoS Pathog 11:e1004782. doi: 10.1371/journal.ppat.1004782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li L, Xue M, Fu F, Yin L, Feng L, Liu P. 2019. IFN-lambda 3 mediates antiviral protection against porcine epidemic diarrhea virus by inducing a distinct antiviral transcript profile in porcine intestinal epithelia. Front Immunol 10:2394. doi: 10.3389/fimmu.2019.02394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pott J, Mahlakoiv T, Mordstein M, Duerr CU, Michiels T, Stockinger S, Staeheli P, Hornef MW. 2011. IFN-lambda determines the intestinal epithelial antiviral host defense. Proc Natl Acad Sci U S A 108:7944–7949. doi: 10.1073/pnas.1100552108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Qian J, Zheng H, Huangfu W-C, Liu J, Carbone CJ, Leu NA, Baker DP, Fuchs SY. 2011. Pathogen recognition receptor signaling accelerates phosphorylation-dependent degradation of IFNAR1. PLoS Pathog 7:e1002065. doi: 10.1371/journal.ppat.1002065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Li Q, Means R, Lang S, Jung JU. 2007. Downregulation of gamma interferon receptor 1 by Kaposi’s sarcoma-associated herpesvirus K3 and K5. J Virol 81:2117–2127. doi: 10.1128/JVI.01961-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Minakshi R, Padhan K, Rani M, Khan N, Ahmad F, Jameel S. 2009. The SARS coronavirus 3a protein causes endoplasmic reticulum stress and induces ligand-independent downregulation of the type 1 interferon receptor. PLoS One 4:e8342. doi: 10.1371/journal.pone.0008342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Broquet AH, Hirata Y, McAllister CS, Kagnoff MF. 2011. RIG-I/MDA5/MAVS are required to signal a protective IFN response in rotavirus-infected intestinal epithelium. J Immunol 186:1618–1626. doi: 10.4049/jimmunol.1002862. [DOI] [PubMed] [Google Scholar]
- 61.Ma Y, Wang C, Xue M, Fu F, Zhang X, Li L, Yin L, Xu W, Feng L, Liu P. 2018. The coronavirus transmissible gastroenteritis virus evades the type I interferon response through IRE1α-mediated manipulation of the microRNA miR-30a-5p/SOCS1/3 axis. J Virol 92:e00728-18. doi: 10.1128/JVI.00728-18. [DOI] [PMC free article] [PubMed] [Google Scholar]