The family Hantaviridae comprises a diverse group of virus species and is considered an emerging global public health threat. Individual hantavirus species differ considerably in terms of their pathogenicity but also in their cell biology and host-pathogen interactions. In this study, we focused on the most prevalent pathogenic hantavirus in Europe, Puumala virus (PUUV), and investigated the entry and internalization of PUUV into mammalian cells. We show that both clathrin-mediated endocytosis and macropinocytosis are cellular pathways exploited by the virus to establish productive infections and demonstrate that pharmacological inhibition of macropinocytosis or a targeted knockdown using RNA interference significantly reduced viral infections. We also found indications of an increase of macropinocytic uptake upon PUUV infection, suggesting that the virus triggers specific cellular mechanisms in order to stimulate its own internalization, thus facilitating infection.
KEYWORDS: macropinocytosis, Puumala virus, hantavirus, virus entry
ABSTRACT
Viruses from the family Hantaviridae are encountered as emerging pathogens causing two life-threatening human zoonoses: hemorrhagic fever with renal syndrome (HFRS) and hantavirus cardiopulmonary syndrome (HCPS), with case fatality rates of up to 50%. Here, we comprehensively investigated entry of the Old World hantavirus Puumala virus (PUUV) into mammalian cells, showing that upon treatment with pharmacological inhibitors of macropinocytosis and clathrin-mediated endocytosis, PUUV infections are greatly reduced. We demonstrate that the inhibitors did not interfere with viral replication and that RNA interference, targeting cellular mediators of macropinocytosis, decreases PUUV infection levels significantly. Moreover, we established lipophilic tracer staining of PUUV particles and show colocalization of stained virions and markers of macropinosomes. Finally, we report a significant increase in the fluid-phase uptake of cells infected with PUUV, indicative of a virus-triggered promotion of macropinocytosis.
IMPORTANCE The family Hantaviridae comprises a diverse group of virus species and is considered an emerging global public health threat. Individual hantavirus species differ considerably in terms of their pathogenicity but also in their cell biology and host-pathogen interactions. In this study, we focused on the most prevalent pathogenic hantavirus in Europe, Puumala virus (PUUV), and investigated the entry and internalization of PUUV into mammalian cells. We show that both clathrin-mediated endocytosis and macropinocytosis are cellular pathways exploited by the virus to establish productive infections and demonstrate that pharmacological inhibition of macropinocytosis or a targeted knockdown using RNA interference significantly reduced viral infections. We also found indications of an increase of macropinocytic uptake upon PUUV infection, suggesting that the virus triggers specific cellular mechanisms in order to stimulate its own internalization, thus facilitating infection.
INTRODUCTION
Hantaviruses, like many related members of the order of Bunyavirales, are believed to utilize integrins as cellular receptors (1–4, 47) and to depend on endocytosis and vacuolar acidification for efficient cell entry (3, 5). However, the specific mechanisms and processes that are involved in their internalization by host cells are still not fully understood. Moreover, entry mechanisms significantly differ between the various hantavirus species: Old World hantaviruses, such as Hantaan virus (HTNV), use clathrin-mediated endocytosis (CME) (6), whereas Andes virus (ANDV), a New World hantavirus, enters cells through clathrin-independent pathways (7). More recently, increasing evidence points to macropinocytosis (MPC) as a potential alternative entry pathway for both ANDV (8, 9) and HTNV (9). Based on these findings, it has been surmised that hantaviruses, like other bunyaviruses (10, 11), exploit different cellular pathways to get access to the cytoplasm of their target cells (5).
The canonical hantavirus cell entry mechanism entails internalization, followed by a passive trafficking within early endosomes and late endosomes (5). Within these vesicles, a pH-dependent conformational change enables binding of the fusion loop of the viral glycoprotein Gc to the vesicular membrane, ultimately mediating fusion between the viral and the cellular membranes and allowing release of the viral content into the cytoplasm (5, 12, 13). The virus’ genetic material, together with viral nucleoproteins, is then transported to the site of viral replication, most likely the endoplasmic reticulum-Golgi intermediate compartment, via dynein-dependent trafficking along the host cell microtubules (7, 14). It is noteworthy that cellular cholesterol seems to be critically involved in the infection process. Previous studies demonstrated that entry of not only other members of the family Bunyaviridae, such as Crimean-Congo hemorrhagic fever virus (15), but also the hantavirus species ANDV is cholesterol dependent (8, 16, 17).
In this study, we determined the cell entry mechanisms as well as host factors that are involved in the initial infection steps of the hantavirus species Puumala virus (PUUV). We utilized inhibitors of specific endocytic pathways to pinpoint productive routes of hantavirus uptake and demonstrated cholesterol, actin, and microtubule dependency of PUUV entry. In addition, we observed a significant decrease in PUUV infection levels upon inhibition of macropinocytosis and clathrin-mediated endocytosis, as well as endocytic acidification. Confocal and correlative light and electron microscopy (CLEM) was used to visualize internalized PUUV particles and to provide further evidence for an intracellular trafficking of viral particles in macropinosomes. In addition, we used flow cytometry and confocal microscopy to investigate whether hantavirus infections impact macropinocytosis, finding indications for an increased fluid-phase uptake upon PUUV infection.
RESULTS
Inhibitors of clathrin-mediated endocytosis, macropinocytosis, and endosomal acidification suppress PUUV infection.
To determine whether PUUV utilizes different productive routes of cell entry, we used a set of nine chemical compounds known to suppress different endocytosis-related cellular mechanisms. We treated Vero E6 cells with the various inhibitors, infected them with PUUV (multiplicity of infection [MOI] = 1), and assessed virus infection by reverse transcription-quantitative PCR (RT-qPCR) at 48 h postinfection (p.i.). In this initial experiment, we chose a long infection period in order to ensure that detected viral RNAs (vRNA) are reflective of newly synthesized viral transcripts resulting from a successful, productive infection and subsequent virus replication. All compounds were pretested for inhibitory activity and cytotoxicity in Vero E6 cells and used at concentrations efficiently inhibiting cellular targets (Fig. 1A) without significantly reducing cell viability (>80% viable compared to mock treatment; data not shown).
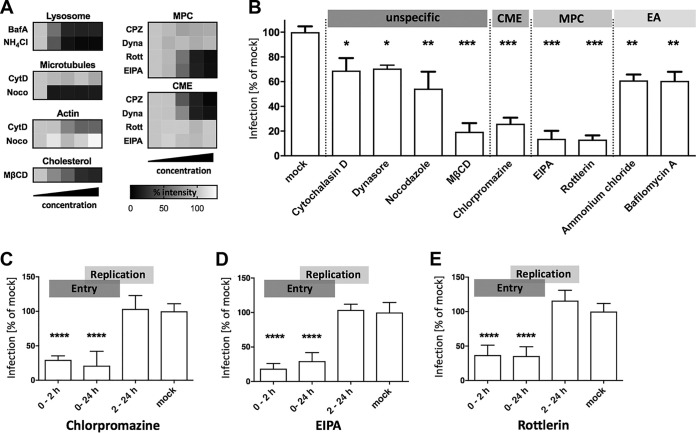
FIG 1.
Pharmacological inhibition of virus entry into Vero E6 cells. (A) Functional titration of inhibitory drugs. Vero E6 cells were treated with the indicated reagents and tested for inhibition of lysosomal/endosomal acidification (Lysosome) using Lysotracker, microtubule depolymerization (Microtubules) using rhodamine-phalloidin, inhibition of actin polymerization (Actin) using anti-tubulin staining, inhibition of macropinocytosis (MPC) using FITC-dextran uptake and inhibition of clathrin-mediated endocytosis (CME) using transferrin uptake assays. Bafilomycin A (BafA) was used at 50, 100, 200, and 500 nM. Ammonium chloride (NH4Cl) was used at 5, 10, 20, and 40 mM. Cytochalasin D (CytD) was used at 100, 500, 1,000, and 5,000 nM. Nocodazole (Noco) was used at 1, 15, 30, and 60 μM. Chlorpromazine (CPZ) was used at 0.1, 5, 10, and 20 μg/ml. Dynasore (Dyna) was used at 0.1, 10, 40, and 80 μM. Rottlerin (Rott) was used at 1, 5, 10, and 20 μM. Ethylisopropyl amiloride (EIPA) was used at 1, 30, 75, and 140 μM. Methyl-β-cyclodextrin (MβCD) was used at 1, 5, 10, and 20 mM. Mean fluorescence intensities of the selected markers were obtained by confocal microscopy and are displayed in heat maps. Signals were normalized to those of mock controls. A total of 50 to 70 cells were analyzed per experimental condition, and samples were assayed after 1 h treatment (2 h for Noco). (B) Vero E6 cells were treated with different pharmaceutical compounds to block specific virus uptake pathways and cellular endocytosis mechanisms. Cells were exposed to effective concentrations of CytD (1 μM), Dyna (40 μM), Noco (30 μM), MβCD (10 mM), CPZ (10 μg/ml), EIPA (75 μM), Rott (10 μM), NH4Cl (20 mM), or BafA (0.2 μM) and infected (MOI = 1) for 48 h. EA, endocytic acidification. Infection levels were assessed by RT-qPCR, and bars show copy numbers of the viral S-segment from inhibitor-treated samples as a percentage of the respective vehicle control (mock). (C to E) Transient inhibitor treatment was conducted to study the impact on PUUV entry (0 to 2 h p.i.) and replication (2 to 24 h p.i.). All bars represent at least 3 independent experiments, with standard errors of the means (SEM). ****, P ≤ 0.0001; ***, P ≤ 0.001; **, P = 0.001 to 0.01; *, P = 0.01 to 0.05. Significance was analyzed using an unpaired Student’s test.
First, we used four compounds that affect cellular internalization processes and trafficking mechanisms broadly and are thus not specific for particular virus entry routes: nocodazole, a neoplastic agent that depolarizes microtubules; cytochalasin D, a highly active inhibitor of actin polymerization; dynasore, a specific inhibitor of the dynamin-dependent scission of endocytic vesicles; and methyl-β-cyclodextrin (MβCD) to disrupt plasma membrane lipid rafts by cholesterol depletion. Microtubules and actin are known to be involved in different endocytosis pathways as well as in the subsequent endosomal trafficking (18, 19), and dynamin and lipid rafts have also been implicated in several endocytic pathways (20–24). Our experiments revealed a modest (∼30%) but significant decrease of the infection level upon treatment with either inhibitor (Fig. 1B), which is in general agreement with endocytic uptake of PUUV.
To pinpoint the biological relevance of specific endocytic pathways in PUUV infections, we then used drugs targeting particular cellular entry routes. First, we tested chlorpromazine (CPZ), which has been extensively used to block clathrin-mediated endocytosis through a mechanism that includes translocation of clathrin and adaptor complex 2 from the cell surface (25). In our experimental setup, CPZ strongly reduced infection levels to around 25% of those in mock-treated cells (Fig. 1B). Second, we utilized ethylisopropyl amiloride (EIPA) and rottlerin, two compounds that interfere with macropinocytosis by two different mechanisms. Whereas EIPA inhibits a Na+/H+ exchanger that is involved in the formation of macropinosomes, rottlerin specifically blocks a crucial isoform of protein kinase C (26). Of note, both drugs caused a strong and highly significant decrease of PUUV infection (Fig. 1B), suggesting that macropinocytosis is not only a bypass for PUUV infection but also a major viral entry route. Finally, bafilomycin A, a highly specific inhibitor of a vacuolar-type H+-ATPase, and ammonium chloride, a membrane-permeative drug that buffers intracellular acidification, were used to block a putative pH-dependent virus-membrane fusion in cellular endosomes. Again, both compounds reduced virus infection significantly, albeit to a lower extent than EIPA and rottlerin (Fig. 1B). Taken together, these experiments indicate that PUUV can enter host cells via at least two different routes under these experimental conditions in Vero E6 cells.
EIPA, CPZ, and rottlerin do not interfere with PUUV replication.
To validate our findings, we asked whether the chemical compounds used affect viral entry, later steps of viral replication, or both. Focusing on CPZ, EIPA, and rottlerin, we modified our previous approach as follows. First, the infection time was reduced to 24 h to reduce the impact of secondary infections (virus released from initially infected cells) on the experimental readout. Second, different stages of the viral life cycle were targeted with the compounds by varying the treatment periods. Infected cells were treated with inhibitors only within the first 2 h of infection, thus predominantly blocking entry but not viral postentry replication. Other samples were drug exposed during the whole infection time of 24 h, covering both entry and replication phases. Finally, in another set of samples, the treatment was started after the initial 2 h of infection, so that replication but not viral entry was affected by the compounds.
Of note, a significant reduction in infection was achieved for all three inhibitors when they were present in the first 2 h of infection, but not if the compounds were applied after the initial entry period (Fig. 1C to E). This finding demonstrates that the inhibitors predominantly affect the respective entry routes and strongly supports our hypothesis of a clathrin- and macropinocytosis-dominated infection of Vero E6 cells by PUUV. Upon testing the CME inhibitor chlorpromazine and the MPC inhibitors rottlerin and EIPA in MDCK-II cells, we obtained results similar to our Vero E6 data. Again, either inhibitor blocked infection efficiently when applied during the initial 2 h of infection but not if administered subsequently (data not shown but available upon request).
Transcriptional silencing of macropinocytosis inhibits PUUV entry.
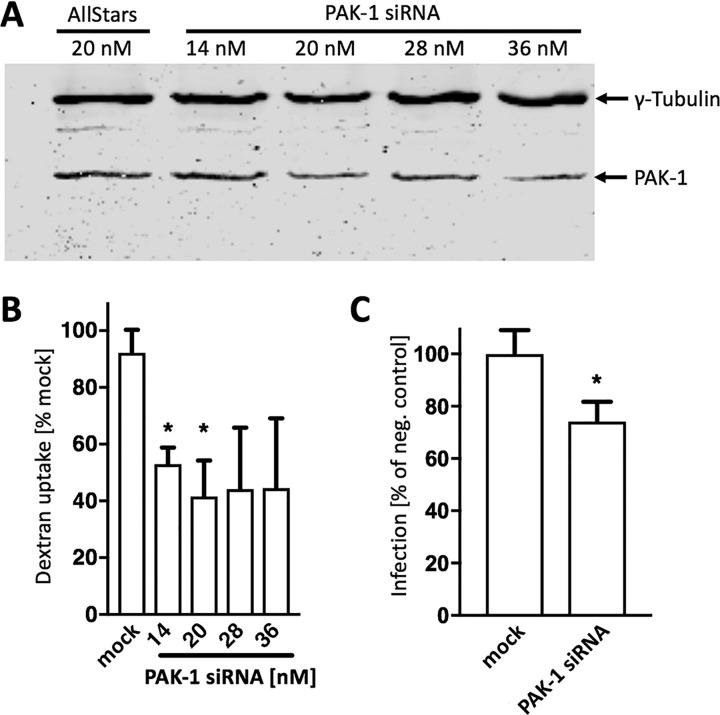
Many pharmacological compounds exert pleiotropic effects, thus affecting not only the anticipated cellular target but also other, unrelated proteins. In order to further validate our finding that inhibition of macropinocytosis suppresses PUUV entry into Vero E6 cells, we knocked down p21-activated kinase (PAK-1), a key regulator of MPC (27), by RNA-dependent transcriptional silencing. First, we determined titers of PAK-1-specific small interfering RNA (siRNA) in Vero E6 cells, demonstrating a dose-dependent decrease of PAK-1 expression and a robust suppression of fluid-phase uptake at low nanomolar concentrations (Fig. 2A and B) without significantly reducing cell viability (data not shown). In order to minimize off-target effects, we then moved on with the lowest concentration of PAK-1 siRNA that had resulted in a detectable knockdown of PAK-1 protein expression and fluid-phase uptake in our previous experiment (Fig. 2A and B) and tested for inhibition of virus infection. To this end, cells were transfected with 20 nM PAK-1 siRNA prior to infection with PUUV, yielding significantly lower vRNA copy numbers (<80%) than nontargeting siRNAs (Fig. 2C), thus confirming our pharmacological inhibition data.
FIG 2.
Inhibition of PUUV entry via macropinocytosis by PAK-1 siRNA. (A) Vero E6 cells were transfected with the indicated concentrations of siRNAs for 48 h and subjected to Western blot analysis. (B) Quantification of macropinocytic fluid-phase uptake into siRNA-transfected Vero E6 cells. Samples were incubated with FITC-dextran for 15 min and analyzed by flow cytometry. Bars show mean fluorescence intensities, normalized to the corresponding negative control. (C) Vero E6 cells transfected with 20 nM PAK-1 siRNA for 48 h were infected with PUUV (MOI = 1) for 24 h and analyzed for hantavirus infection levels. Viral RNA copy numbers were assessed by RT-qPCR, and bars show results from inhibitor-treated samples as a percentage of the value for the corresponding nontargeting siRNA. All bars represent 3 independent experiments with SEM. Significance was analyzed using an unpaired Student’s test. *, P < 0.05. “Mock” and “negative control” refer to samples transfected with nontargeting siRNA.
Lipophilic tracer staining of PUUV enables live-virus detection by fluorescence microscopy.
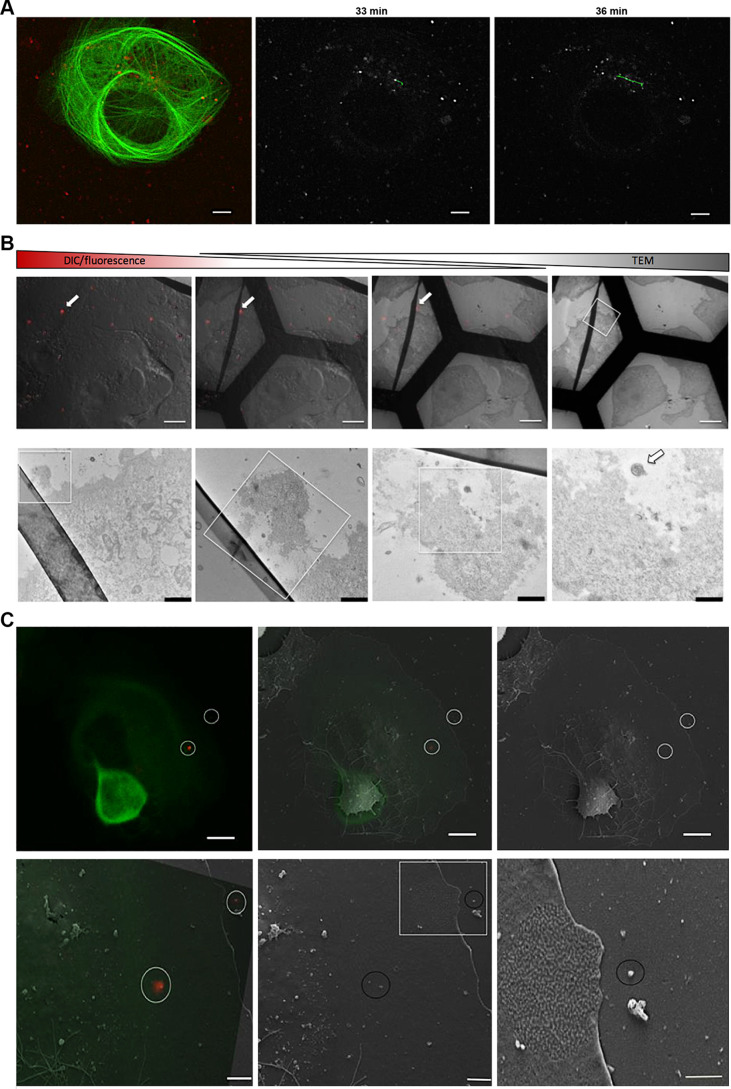
In order to provide independent evidence for PUUV uptake into Vero E6 cells by macropinocytosis, we established live-cell imaging of PUUV entry as a complementary approach. To enable tracing of internalized virions, we used a virus membrane-staining strategy that has been used previously for various other enveloped viruses (28). Virus preparations were treated with the lipophilic dye DiI or DiD and subsequently purified using size exclusion chromatography. As a proof of principle, stained virus particles were then used to infect cells that had been transfected with a fluorescently labeled microtubule binding protein (Tau-yellow fluorescent protein [Tau-YFP]) and observed by live-cell confocal microscopy over a period of 30 to 60 min. We performed basic particle tracking, finding particle traces along cytoskeleton components (Fig. 3A; also, see the supplemental videos) and calculated an average particle velocity of around 1,000 nm/s from 5 independent trajectories, which may indicate kinesin- and/or dynein-mediated transport along cellular microtubule filaments (29).
FIG 3.
Lipophilic tracer staining enables live fluorescence microscopy and correlative light and electron microscopy of PUUV particles. (A) Vero E6 cells transfected with Tau-YFP were infected with DiI-labeled virus particles and subjected to live confocal microscopy. Images were acquired every 2.9 s, and virus particles were tracked using ImageJ. Micrographs show an overlay of Tau-YFP expression (green) and virus particles (red) (left), as well as virus particles (in gray) at different time points postinfection (center and right). A representative trajectory of a single virus particle is shown in the center and right images (green). (B and C) Correlative light and electron microscopy (CLEM) was used to reveal ultrastructures of stained virus particles (red; arrows). First, samples were analyzed by confocal microscopy, recording specific regions of interest in gridded microscopy slides. Subsequently, the same samples were analyzed by electron microscopy to unravel the ultrastructure of previously observed particles. (B) (Top) Gradual overlay of differential interference contrast (DIC)/fluorescence microscopy and transmission electron microscopy (TEM) images. Bar, 5 μm. (Bottom) Magnifications of the boxed regions. Bars, 2, 0.6, 0.4, and 0.25 μm (left to right). (C) (Top) Representative fluorescence microscopy image (left), the corresponding region in a scanning electron microscopy image (right), and an overlay of both (middle). Fluorescent particles are highlighted with white circles. Bar, 5 μm. (Bottom) Magnifications of the regions containing fluorescent particles. Bars, 2, 2, and 1 μm (left to right). Tau-YFP is shown in green.
To further validate our labeling procedure, different controls were performed. First, cells infected with labeled hantavirus were subjected to correlative light and electron microscopy in order to show that intracellular, fluorescent puncta were representative of virus particles and not artifacts, such as fluorophore aggregates or stained cell debris. In fact, fluorescent spots were recovered in regions showing spherical particles in raster electron microscopy (Fig. 3C). Moreover, transmission electron microscopy (TEM) revealed electron-dense virions (Fig. 3B) that coincided spatially with fluorescent particles in infected Vero E6 cells. Of note, the electron microscopy images show a typical viral nanostructure with a diameter of around 100 nm in regions that were correlated with their respective counterparts in the confocal images, based on surrounding cellular structures and topographies. We also tested whether labeled viruses were still able to productively infect their host cells or whether internalized virions were instead rendered noninfective by the labeling procedure. To this end, we infected Vero E6 cells with labeled viruses and assessed virus infection using RT-qPCR at 48 h postinfection. We found robust and high levels of infection, demonstrating that the staining and washing procedure did not abrogate the infectivity of the viral particles (data not shown).
PUUV particles colocalize with macropinocytic cargo.
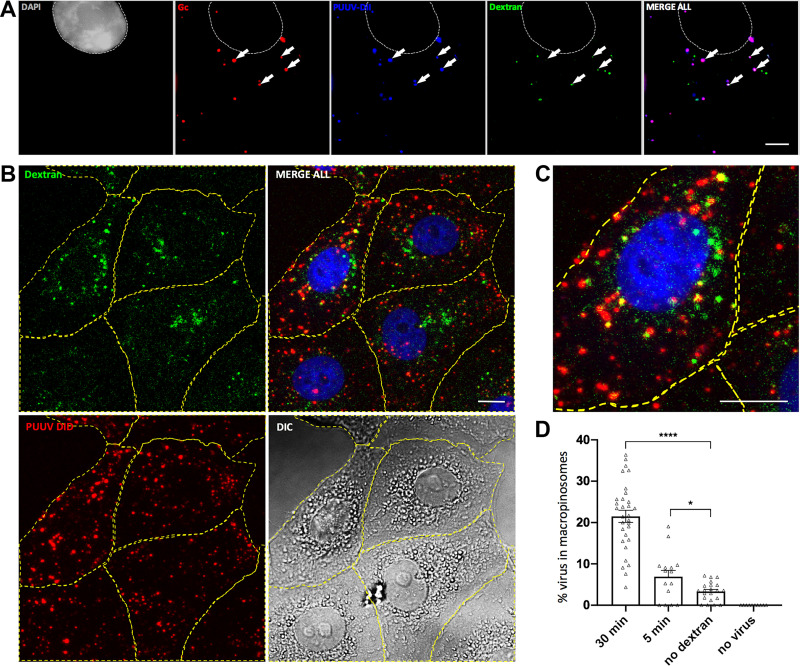
Next, we asked if hantavirus particles can be found in macropinosomes upon cell entry. To this end, Vero E6 cells were incubated concomitantly with virus and high-molecular-weight dextran, both fluorescently labeled. Fluorescent dextran is an established marker of macropinocytosis (30), and spatial correlation of virus and dextran indicates a joint uptake by macropinosomes. We found colocalization between fluorescein isothiocyanate (FITC)-dextran and the lipophilic tracer DiI as early as 5 min after initiation of the experiment (data not shown), but a more robust intracellular signal of both fluorescent markers was obtained at 30 min postinfection (Fig. 4). By immunofluorescence microscopy, colocalization was found for PUUV Gc and the virus membrane marker DiI (Fig. 4A), thus confirming that lipophilic tracer staining enables efficient and specific detection of PUUV particles. Moreover, all viral markers showed distinct colocalization with dextran-containing compartments, reflective of a macropinocytic uptake of PUUV particles (Fig. 4A to C). Quantitative image analysis showed a significant increase in colocalization of macropinosomes and virus particles as early as 5 min p.i., with up to 20% of the virus particles being found in FITC-dextran-positive compartments (Fig. 4D). At 30 min p.i., a significant increase of colocalization was found (P < 0.0001) and individual cells displayed more than 30% of viruses residing in macropinosomes (Fig. 4D). Of note, we also observed internalized virions which did not colocalize with FITC-dextran-containing vesicles. These colocalization-negative spots may indicate particles that had already left the macropinocytic pathway or may reflect virus entry through alternative endocytic pathways, i.e., clathrin-mediated endocytosis.
FIG 4.
Colocalization of macropinocytic cargo and PUUV. (A) Vero E6 cells were infected with DiI-labeled PUUV particles (blue) and concomitantly exposed to FITC-dextran (green) in order to label macropinosomes. At 30 min postinfection, cells were fixed and stained with DAPI (gray) and antibodies against PUUV Gc (red). Arrows indicate representative puncta either without (left arrow) or with (remaining arrows) colocalization between virus staining and macropinosomes. Samples were imaged using spinning disk microscopy. Bar, 5 μm. (B) Representative confocal z projections of cells exposed to DiD-stained PUUV particles (red) and FITC-dextran (green) for 30 min. Cells were fixed and stained with DAPI (blue) and subjected to quantitative image analysis. (C) Magnification of the cell shown in panel B. Bars, 10 μm. (D) Image quantitation using the ImageJ spot localization plug-in ComDet. Cells were exposed for 30 and 5 min to PUUV DiD and FITC-dextran, PUUV DiD only (no dextran), or FITC-dextran only (no virus). Each dot indicates the percentage of PUUV DiD particles colocalizing with macropinosomes in a single analyzed cell (n ≥ 10). Error bars represent SEM. *, P = 0.01 to 0.05; ****, P ≤ 0.0001. Significance was analyzed using an unpaired Student’s test.
Hantavirus infection promotes the formation of actin foci and activates macropinocytosis.
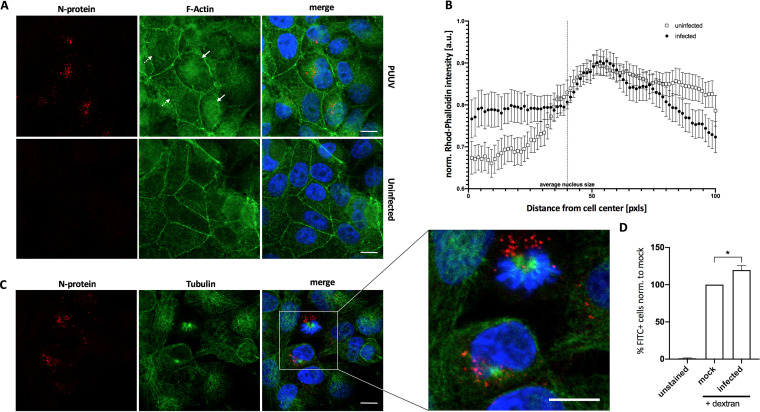
Hantaviruses have been previously reported to depend heavily on cytoskeleton components in order to promote intracellular particle trafficking (7). Here, we sought to further investigate whether actin contributes to the intracellular PUUV life cycle. Actin is a key factor for the formation of membrane ruffles and macropinosomes, but it is also critically involved in other cellular endocytosis mechanisms (31). In order to study cellular actin in the context of PUUV infections, we stained infected cells for the viral N protein and filamentous actin (F-actin) using rhodamine-phalloidin at 72 h postinfection. Interestingly, we often observed an increased accumulation of F-actin in regions and particularly in cells with high concentrations of the viral N protein (Fig. 5A, solid arrows; Fig. 5B) compared to noninfected cells (Fig. 5A, dashed arrows; Fig. 5B). We surmised that this region may harbor the microtubule-organizing center (MTOC) and performed an additional tubulin immunofluorescence staining. In fact, viral enrichment was often found in the vicinity of perinuclear microtubule aggregates (Fig. 5C). Both observations support previous reports indicating a crucial involvement of the cytoskeleton in hantavirus infections (7, 32).
FIG 5.
Stimulation of fluid-phase uptake by PUUV. (A) Vero E6 cells infected with PUUV for 72 h, and uninfected cells were stained for filamentous actin using rhodamine-phalloidin. Solid arrows indicate infected cells; dashed arrows indicate uninfected cells. (B) Cells shown in panel A were analyzed with ImageJ using the Radial Profile Plot plug-in. Ten to fifteen cells from three to five images were analyzed from either uninfected or infected samples and displayed as means with SEM. (C) Vero E6 cells infected with PUUV for 72 h were stained for microtubules using anti-tubulin and anti-N protein. Bars, 10 μm. (D) Vero E6 cells, exposed to hantavirus (MOI = 10) for 30 min at 4°C were subsequently investigated using flow cytometry for FITC-dextran internalization as a reporter of fluid-phase uptake. Bars show results from 5 independent experiments with SEM. Significance was analyzed using an unpaired Student’s test. *, P ≤ 0.05.
Of note, other virus families have previously been reported to stimulate endocytosis pathways, and macropinocytosis in particular, in order to facilitate their uptake and productive entry into host cells (33, 34). Based on this notion and our previous results, we hypothesized that PUUV infections may actually increase the uptake of high-molecular-weight dextran. To test this hypothesis, we infected cells with virus at a high titer (MOI = 10) for 30 min at 4°C to enable binding without internalization and membrane fusion. After removal of unbound virions, FITC-dextran was used to quantify fluid-phase uptake via flow cytometry. We found a slight but significant increase of FITC-dextran uptake in virus samples compared to mock-infected negative controls in each of 5 independent experiments (Fig. 5D; raw data available upon request), indicating that macropinocytic endocytosis is enhanced upon PUUV receptor binding.
DISCUSSION
The overall goal of this study was to investigate the cellular mechanisms and pathways that permit PUUV to enter mammalian cells and enable a successful, productive infection. Previously, hantaviruses have been reported to enter host cells via different pathways of receptor-mediated endocytosis (3, 5). For instance, the Old World hantavirus HTNV was shown to be internalized by clathrin-mediated endocytosis (6), whereas other members of the hantavirus family were demonstrated to be internalized through a CME-independent route (7). Recently, Torriani et al. (9) utilized a pseudovirion reporter system to investigate entry of both Old and New World hantaviruses, providing evidence for a contribution of macropinocytosis. Here, we employed replication-competent wild-type PUUV to unequivocally study virus entry. We found that treatment of Vero E6 cells with inhibitors specific either for clathrin-mediated endocytosis (CPZ) or for macropinocytosis (EIPA and rottlerin) results in a significant reduction of infection levels, suggesting that PUUV is able to utilize both pathways for virus entry and productive infection (Fig. 1). Moreover, we found that cholesterol depletion with MβCD significantly reduced PUUV infection (Fig. 1). Cholesterol is a major structural element of mammalian membranes and has been shown to be involved in numerous endocytic mechanisms (22, 35, 36). In addition, and consistent with our finding, cholesterol was demonstrated to be required for the infection of Andes virus (8, 16) as well as other, unrelated virus families (37, 38).
Hantavirus receptor binding and membrane fusion are exclusively mediated by the viral glycoprotein complex (GPC), consisting of the subunits Gn and Gc. Gc has been shown to constitute a class II fusion protein (12, 39, 40), thus requiring acidification-induced conformational changes in order to elicit membrane fusion. Our finding of a significant reduction of hantavirus infections with inhibitors of intracellular acidification is consistent with this hypothesis (Fig. 1).
One caveat of our initial pharmacological screen is that cells were infected for 48 h in the presence of the various inhibitors (Fig. 1B). This long incubation period is likely to increase off-target or secondary effects but also reduces the overall efficacy of the inhibitors due to their limited half-life. Moreover, several rounds of infection are possible within the given incubation period, so that effects on late virus replication steps such as assembly and budding cannot be excluded. We therefore consider the results from this screen qualitative rather than quantitative, and we subsequently performed a more elaborate inhibitor-based assay, focusing on clathrin-mediated and macropinocytic PUUV entry, which confirmed our initial findings (Fig. 1C to E).
Importantly, assays based solely on pharmacologic inhibition of cellular pathways are often plagued by a limited reliability of the results due to the pleiotropic nature of the drugs used. To take this into account, we sought to provide additional independent evidence for an involvement of macropinocytosis in PUUV infections. To this aim, we first employed RNA interference, transcriptionally silencing PAK-1, which is a crucial component of the cellular macropinocytosis machinery. PAK-1 knockdown significantly reduced PUUV infections (Fig. 2), thus further supporting the findings from our inhibitor assay. In addition, we established lipophilic tracer staining of PUUV particles to assess virus uptake by fluorescence microscopy. This method has been extensively used to study entry and membrane fusion of influenza A virus (41) and other viruses but has not yet been stringently tested and utilized for hantavirus research. We found that labeled particles can be visualized by live fluorescence microscopy for extended periods of time (Fig. 3A) and validated their ultrastructure by CLEM (Fig. 3B and C). With this system in place, we tested whether PUUV enters cells through macropinosomes, visualized by FITC-dextran (Fig. 4). We found colocalization of FITC-dextran and stained viral particles as well as viral proteins, detected by immunofluorescence, thus again confirming macropinocytosis as a biologically relevant cellular entry route of PUUV.
Previous publications demonstrated that pharmacological inhibition of macropinocytic internalization is sufficient to significantly suppress uptake of HTNV and ANDV, which are Old and New World members, respectively, of the genus Orthohantavirus (8, 9). However, to the best of our knowledge, an involvement of macropinocytosis in cell entry and infection of the hantavirus species PUUV has not been reported before. This finding therefore adds to our understanding of hantavirus uptake, suggesting that macropinocytosis is broadly employed throughout the hantavirus genus.
It has been reported previously that actin and other cytoskeleton components are utilized, albeit to different extents, by various hantaviruses species (7). Actin is also a key host cell mediator of macropinocytic uptake. In addition, it has been shown that members of several virus families which utilize macropinocytosis for cell entry and infection actively trigger and promote this endocytic mechanism (33, 34). We therefore hypothesized that PUUV infections may in fact impact both actin dynamics and macropinocytic fluid-phase uptake. In agreement with that, we found indications for intracellular actin accumulation in infected but not in uninfected Vero E6 cells (Fig. 5). To test whether PUUV induces macropinocytosis, we infected Vero E6 cells and concomitantly assayed FITC-dextran uptake by flow cytometry. We found a mild but significant elevation of the FITC-dextran after only 30 min of incubation with PUUV. In the light of the high MOI we used here, further studies are warranted to support this notion. Nonetheless, almost identical observations were previously made for Ebola virus-like particles (33) as well as filamentous influenza A virus (42), both of which exploit macropinocytosis for cell entry, which may suggest a common mechanism that is shared by Filoviridae, Orthomyxoviridae, and Hantaviridae.
In summary, in this study we performed a comprehensive evaluation of productive PUUV entry pathways and provided multiple lines of evidence for a contribution of macropinocytosis during PUUV infection. Importantly, increasing evidence suggests that integrins, which are widely believed to be the predominant receptors for hantaviruses (1), and macropinocytosis may be mechanistically connected (43, 44). Future studies will have to further investigate the role and significance of macropinocytosis and other cellular pathways for hantavirus entry into endothelial cells, the major PUUV target cell type.
MATERIALS AND METHODS
Cell culture and PUUV infection.
Vero E6 cells (ATCC CRL-1586; American Type Culture Collection, Manassas, VA) were maintained in Dulbecco’s modified Eagle medium (DMEM) containing 10% inactivated fetal bovine serum (FBS), 2 mM l-glutamine, 100 U/ml penicillin, and 100 μg/ml streptomycin (all from PAA Laboratories GmbH, Austria) under standard cell culture conditions. If not otherwise stated, PUUV infection was performed with a multiplicity of infection (MOI) of 1 in medium containing 10% FBS followed by medium exchange after 1 h of incubation.
Functional titration of entry inhibitors.
Vero E6 cells were seeded on an 8-well ibiTreat slide and incubated at 37°C in DMEM with 10% FBS. On the next day, inhibitors were added to cells for 1 h and nocodazole for 2 h. Serum-free DMEM was used for the methyl-β-cyclodextrin (MβCD) treatment, and cells were incubated at 37°C for 1 h. The ability of cells to internalize through MPC or CME was determined by observing dextran and transferrin uptake (MPC and CME cargo, respectively) by confocal microscopy. FITC-labeled dextran (5 mg/ml) and Alexa Fluor 647-labeled transferrin (50 μg/ml) were added to cells treated with inhibitors, which were then incubated for an additional hour at 37°C. Both inhibitors and indicators were removed, and the cells were washed once with phosphate-buffered saline (PBS). After one wash with citrate buffer (pH 3) for 30 s, cells were washed with PBS three times, fixed with 4% paraformaldehyde (PFA) in PBS, and stained with Hoechst 33342 for 5 min at room temperature. For the imaging of the actin and microtubule network, cells were washed three times with PBS, fixed with 4% PFA in PBS, permeabilized with 0.2% Triton X-100, and stained using monoclonal mouse anti-tubulin (clone B-5-1-2; Sigma-Aldrich, USA) and Alexa Fluor 488-conjugated secondary goat-anti-mouse antibody (Abcam, UK). Filamentous actin was stained with fluorescently conjugated phalloidin (Invitrogen, USA) according to the manufacturer’s protocol. Lysotracker staining was performed to visualize acidic intracellular compartments. Lysotracker red DND-99 (100 nM) was added to the cells, which were incubated for 20 min at 37°C and then washed twice with PBS. Hoechst 33342 counterstaining was performed as described above. For the visualization of cellular cholesterol, cells treated with MβCD were washed three times with PBS and stained with filipin (0.05 mg/ml in PBS–10% FBS) for 2 h at room temperature. Afterwards, cells were washed three times with PBS and fixed with 4% PFA in PBS. All images were captured with an inverted laser scanning microscope (LSM 780 Zeiss system; Carl Zeiss, Germany) using a 40×, 1.2-numerical aperture water immersion objective. All fluorescence channels were collected sequentially to eliminate cross talk between fluorescent dyes. For each inhibitor concentration, 10 to 15 images were captured and processed by ImageJ, analyzing a minimum of 50 cells. Samples were quantified by obtaining mean fluorescence intensities of reporter dyes at the single-cell level. Average intensities were normalized to those of corresponding mock treatment controls and expressed as percentages.
Virus inhibition assay.
Vero E6 cells were seeded in 48-well plates and preincubated the next day in DMEM (supplemented with 10% inactivated FBS) with various inhibitors (Sigma, Germany) for 1 h: 10 mM chlorpromazine, 75 μM ethylisopropyl amiloride (EIPA), 10 μM rottlerin, 1 μM cytochalasin D, 20 mM ammonium chloride, 0.2 μM bafilomycin A, or 40 μM dynasore. For methyl-β-cyclodextrin (MβCD; 10 mM) treatment, FBS-free DMEM was used and replaced with FBS-containing medium at 6 h postinfection (p.i.). Nocodazole (30 mM) was applied for 2 h prior to the infection. After pretreatment, virus was added (MOI = 1). Depending on the experimental setup, the virus mix contained either the respective compound for further inhibition between 2 and 48 h, as stated above, or medium only, without any additional inhibitor. After 24 to 48 h, cells were subjected to RT-qPCR analysis as described below. Virus replication was quantified by assessing the copy number of the viral S-segment per sample. The inhibition efficiency was expressed as relative infection by normalizing the viral RNA copies in the inhibitor sample to the corresponding mock-infected control. Mock-infected controls represent samples that were treated with vehicle only (dimethyl sulfoxide [DMSO] or PBS, depending on the solvent of the respective drug).
Quantification of PUUV infection by RT-qPCR.
The method for viral RNA quantification was adapted from reference 45. Briefly, infected cells were washed with PBS, lysed with RLT buffer, and frozen at −80°C for subsequent RNA extraction (RNeasy; Qiagen, Germany). Reverse transcription was performed in 20-μl mixes consisting of 4.1 μl RNase-free water, 4 μl RT buffer, 0.1 μl dithiothreitol (DTT), 0.3 μl deoxynucleoside triphosphates (dNTPs), 0.5 μl random hexamer primers (Amersham, Piscataway, NJ), 0.5 μl RNA Safe (AllianceBio, USA), 0.5 μl reverse transcriptase (Moloney murine leukemia virus RT [M-MLV-RT] system; Invitrogen, Germany) and 10 μl isolated RNA. Reverse transcription reactions were carried out in a thermocycler (Eppendorf, Germany) using the following protocol: 10 min at 25°C, 30 min at 42°C, and 6 min at 96°C, with a final hold at 4°C. The generated cDNA was then used for qPCR. qPCR master mixes were prepared with 4 μl LightCycler enzyme mix (Roche, Germany), 0.6 μl of each primer at 10 μM, 0.3 μl of 10 μM DNA probe, 10.5 μl of water, and 4 μl of cDNA and analyzed using a LightCycler Nano (Roche, Germany). The LightCycler program was as follows: 95°C for 10 min, followed by 40 cycles of 95°C for 5 s and 60°C for 30 s, a denaturation step at 95°C for 15 s, and a hold at 4°C.
We used the following primers (Molbiol, Germany) and probes (Life Technologies, Germany) to target the viral S-segment: PUUV F (GARRTGGACCCRGATGACGTTAA), PUUV R (CCKGGACACAYCATCTGCCAT), and PUUV TMGB1 (6-carboxyfluorescein [FAM]-CAACAGACAGTGTCAGCA-MGB-NFQ S).
Virus labeling and purification for live-cell imaging.
Concentrated virus aliquots were mixed with DiI or DiD (final concentration of 1 μM; Thermo Fisher, USA), mixed by vortexing, and incubated for 1.5 h at room temperature. Subsequently, excess label was removed by size exclusion chromatography using Sephadex G-25 columns (GE Life Sciences, USA). Labeled virus was used for visualization of internalized particles using confocal microscopy and correlative light and electron microscopy.
Correlative light and electron microscopy.
Vero E6 cells were grown and infected on microscopic dishes with finder grids (Ibidi, USA). Cells infected with fluorescently labeled virus were fixed using 4% PFA in PBS and observed on a Fluoview FV-1000 confocal microscope (Olympus, Japan) to identify fluorescent spots and record their local environment. For scanning electron microscopy (SEM), cells were fixed with 2.5% (vol/vol) glutaraldehyde and 2% (wt/vol) paraformaldehyde in 100 mM cacodylate buffer (pH 7.4) for 30 min at room temperature. After fixation, cells were rinsed three times for 10 min with 100 mM cacodylate buffer and dehydrated through a graded ethanol series. After being washed three times with hexamethyldisilazane (Electron Microscopy Sciences, USA), cells were coated with gold and analyzed on a LEO 1430 scanning electron microscope (Zeiss, Germany). For transmission electron microscopy (TEM), cells were fixed with 2.5% (vol/vol) glutaraldehyde and 2% (wt/vol) paraformaldehyde in 100 mM cacodylate buffer (pH 7.4) for 30 min. Cells were rinsed three times for 5 min with 100 mM cacodylate buffer, postfixed for 1 h in 1% (vol/vol) osmium tetroxide, rinsed three times with distilled water, stained en bloc with 0.5% (vol/vol) uranyl acetate, dehydrated through a graded ethanol series, and finally embedded using EMBed 812 (Electron Microscopy Sciences, USA). Cells were cut perpendicular to the substrate with diamond knives on an Ultracut-R ultramicrotome (Leica, Japan), and 70- to 90-nm sections were collected with copper grids. Sections were counterstained with 4% (wt/vol) uranyl acetate followed by lead citrate. All samples were imaged on a transmission electron microscope (EM 900; Zeiss, Germany) equipped with a wide-angle charge-coupled device (CCD) camera (TRS Systems, Germany).
Immunostaining, transfection, and fluorescence microscopy.
Vero E6 cells were seeded on glass coverslips in a 48-well plate and infected with PUUV (MOI = 1) if not otherwise stated. Samples were subsequently fixed with 4% PFA in PBS. PFA-fixed samples were permeabilized with 0.2% Triton X-100 and stained using suitable antibodies. The viral N protein was labeled using a primary anti-Tula virus antibody (46) and a secondary anti-rabbit FITC conjugate (Invitrogen, USA). The viral Gc protein was stained using a mouse monoclonal antibody (H1808-60B; U.S. Biological, USA). Filamentous actin was stained with fluorescently conjugated phalloidin (Invitrogen, USA) according to the manufacturer’s protocol. Transfection with Tau-YFP was conducted 24 h prior to experiments utilizing Turbofect (Thermo Fisher, USA) according to the manufacturer’s protocol. If not otherwise stated, an inverted FluoView 1000 microscope (Olympus) was used for confocal microscopy. Differential interference contrast (DIC) and fluorescence intensity were obtained with a 60× oil immersion objective (numerical aperture, 1.35) at 25°C with a frame size of 512 by 512 pixels. For spot detection and quantification, z stacks were acquired. z projections were displayed and analyzed as described below.
Spinning disk confocal microscopy.
Spinning disk confocal microscopy was performed using a VisiScope scanning disc confocal laser microscope (Visitron, Germany) equipped with a 60× water objective and an EM-CCD camera. Z stacks were acquired if not otherwise stated, and z projections were displayed.
Image analysis.
Image processing and analysis were carried out with ImageJ (https://imagej.nih.gov/ij/). Spot detection and colocalization analysis (Fig. 4) were performed using the ComDet plug-in developed by Eugene Katrukha on maximum-intensity projections of z stacks. Intensity thresholds were adjusted according to no-virus control samples. The following settings were used for colocalization analysis: for channel 1, a particle size of 4 pixels and an intensity threshold of 10; for channel 2, a particle size of 5 pixels and an intensity threshold of 3. The analysis included and segmented larger particles. The maximum distance between colocalized spots was set to 4 pixels. Cells shown in Fig. 5 were analyzed with ImageJ using the Radial Profile Plot plug-in. Initially, nuclei were framed using a rectangular selection tool in the corresponding Hoechst 33342 images. Then, Radial Profile plots were generated by analyzing rhodamine-phalloidin intensities starting from the center of the selection and extending to up to 100 pixels in order to cover the majority of the cell body. Radial profiles of each cell were normalized to the respective maximum intensity of the cell.
RNA interference.
Transfection with siRNAs was performed using the HiPerFect transfection reagent according to the manufacture’s reverse transfection protocol (Qiagen, Germany). First, siRNAs were spotted on 48-well plates. Second, 18.5 μl of serum-free DMEM containing 1.5 μl HiPerFect was added to the prespotted siRNA, and the mixture was incubated for 20 min at room temperature. Afterward, 2 × 104 Vero E6 cells were seeded into each well to give a final siRNA concentration of 20 nM, and cells were grown under normal growth conditions for 48 h. The nontargeting siRNA AllStars (Qiagen, Germany) was used as a negative control. PAK-1-targeting siRNA was purchased from Ambion (Thermo Fisher, USA).
Western blotting for siRNA knockdown verification.
The siRNA knockdown was performed in Vero E6 cells as described above. At 48 h posttransfection, proteins were isolated using Triton X-100 lysis buffer (150 nM sodium chloride, 1.0% Triton X-100, 50 mM Tris-HCl [pH 8.0], 1 mM phenylmethylsulfonyl fluoride [PMSF], 5 mM EDTA) by constant agitation for 30 min at 4°C. Soluble proteins and cell debris were separated by centrifugation for 30 min at 16,000 rpm and 4°C. Then, protein concentration was determined using a Pierce bicinchoninic acid (BCA) protein assay kit (Thermo Fisher Scientific, USA) according to the manufacturer’s protocol. Laemmli loading buffer (4×) was added to 740 ng total protein, and samples were heated to 95°C for 5 min before protein separation under reducing conditions using 12% denaturing sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). Immediately after electrophoresis, Western blotting was performed using a Trans-Blot Turbo RTA Midi PVDF (polyvinylidene difluoride) transfer kit (Bio-Rad, USA) according to the manufacturer’s instructions and a Trans-Blot Turbo Blotting system standard SD transfer program (Bio-Rad, USA). Protein detection was performed using an Odyssey IR scanning system. The PVDF membrane was blocked for 1 h at RT using fish gelatin blocking buffer (10 mM Tris [pH 7.5], 150 mM NaCl, 2% [wt/vol] fish gelatin, 1% [wt/vol] ovalbumin) diluted 1:2 in Tris-buffered saline (TBS). PAK-1 protein and the loading control γ-tubulin were detected using mouse monoclonal (catalog no. sc-32776; Santa Cruz, USA) and rabbit polyclonal (catalog no. T-5192; Sigma-Aldrich, USA) primary antibodies, respectively. Incubation was performed overnight at 4°C using fish gelatin blocking solution diluted 1:2 in TBS-Tween (TBST). After extensive washing using TBST, primary antibodies were visualized using goat anti-mouse IRDye 680RD (Li-Cor, USA) and goat anti-rabbit IRDye 800 CW (Li-Cor, USA) secondary antibodies.
Quantification of fluid phase uptake.
Prior to macropinocytosis uptake experiments, cells (previously transfected with siRNA) were serum starved in DMEM supplemented with 0.1% fetal calf serum (FCS) for 3 h to promote dextran uptake. Afterward, cells were inoculated using FITC-dextran (molecular weight [MW], 40,000; Thermo Fisher, USA) diluted in DMEM containing 0.5% FCS to a final concentration of 5 mg/ml for 15 min at 37°C. To stop dextran uptake, cells were transferred to 4°C and washed once with cold acidic buffer (150 mM NaCl, 25 mM NaOH, pH 4) to remove any surface-bound dextran. Subsequently, cells were washed twice with cold PBS and either subjected to fixation and microscopy or harvested by scraping for flow cytometry. Scraped cells were precipitated by centrifugation (1,200 × g, 5 min, 4°C), and FITC intensity was determined using a FACSAria II flow cytometer (Becton, Dickinson, USA), analyzing at least 10,000 cells per sample.
To evaluate promotion of macropinocytosis upon virus infection, cells were incubated with high-titer virus solutions for 30 min at 4°C to ensure virus binding without internalization. After removal of the virus supernatant, the cells were treated with 5 mg/ml FITC-dextran (MW, 40,000; Thermo Fisher, USA) for 30 min at 37°C to target macropinosomes, washed, fixed, and subjected to flow cytometry analysis. Typically, 10, 000 cells were analyzed with a FACSCalibur flow cytometer (BD Biosciences, USA), and the intensity of fluorescence was analyzed using FlowJo software (TreeStar, USA).
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by Infect-ERA project HantaHunt to A.H. (031L0096A) and D.H.K. (031L0096B).
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Muller A, Baumann A, Essbauer S, Radosa L, Kruger DH, Witkowski PT, Zeier M, Krautkramer E. 2019. Analysis of the integrin beta3 receptor for pathogenic orthohantaviruses in rodent host species. Virus Res 267:36–40. doi: 10.1016/j.virusres.2019.04.009. [DOI] [PubMed] [Google Scholar]
- 2.Gavrilovskaya IN, Shepley M, Shaw R, Ginsberg MH, Mackow ER. 1998. Beta3 integrins mediate the cellular entry of hantaviruses that cause respiratory failure. Proc Natl Acad Sci U S A 95:7074–7079. doi: 10.1073/pnas.95.12.7074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mittler E, Dieterle ME, Kleinfelter LM, Slough MM, Chandran K, Jangra RK. 2019. Hantavirus entry: perspectives and recent advances. Adv Virus Res 104:185–224. doi: 10.1016/bs.aivir.2019.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Raftery MJ, Lalwani P, Krautkrämer E, Peters T, Scharffetter-Kochanek K, Krüger R, Hofmann J, Seeger K, Krüger DH, Schönrich G. 2014. Beta2 integrin mediates hantavirus-induced release of neutrophil extracellular traps. J Exp Med 211:1485–1497. doi: 10.1084/jem.20131092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vaheri A, Strandin T, Hepojoki J, Sironen T, Henttonen H, Makela S, Mustonen J. 2013. Uncovering the mysteries of hantavirus infections. Nat Rev Microbiol 11:539–550. doi: 10.1038/nrmicro3066. [DOI] [PubMed] [Google Scholar]
- 6.Jin M, Park J, Lee S, Park B, Shin J, Song KJ, Ahn TI, Hwang SY, Ahn BY, Ahn K. 2002. Hantaan virus enters cells by clathrin-dependent receptor-mediated endocytosis. Virology 294:60–69. doi: 10.1006/viro.2001.1303. [DOI] [PubMed] [Google Scholar]
- 7.Ramanathan HN, Jonsson CB. 2008. New and Old World hantaviruses differentially utilize host cytoskeletal components during their life cycles. Virology 374:138–150. doi: 10.1016/j.virol.2007.12.030. [DOI] [PubMed] [Google Scholar]
- 8.Chiang CF, Flint M, Lin JS, Spiropoulou CF. 2016. Endocytic pathways used by Andes virus to enter primary human lung endothelial cells. PLoS One 11:e0164768. doi: 10.1371/journal.pone.0164768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Torriani G, Mayor J, Zimmer G, Kunz S, Rothenberger S, Engler O. 2019. Macropinocytosis contributes to hantavirus entry into human airway epithelial cells. Virology 531:57–68. doi: 10.1016/j.virol.2019.02.013. [DOI] [PubMed] [Google Scholar]
- 10.Lozach PY, Mancini R, Bitto D, Meier R, Oestereich L, Overby AK, Pettersson RF, Helenius A. 2010. Entry of bunyaviruses into mammalian cells. Cell Host Microbe 7:488–499. doi: 10.1016/j.chom.2010.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Albornoz A, Hoffmann AB, Lozach PY, Tischler ND. 2016. Early bunyavirus-host cell interactions. Viruses 8:143. doi: 10.3390/v8050143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guardado-Calvo P, Bignon EA, Stettner E, Jeffers SA, Perez-Vargas J, Pehau-Arnaudet G, Tortorici MA, Jestin JL, England P, Tischler ND, Rey FA. 2016. Mechanistic insight into bunyavirus-induced membrane fusion from structure-function analyses of the hantavirus envelope glycoprotein Gc. PLoS Pathog 12:e1005813. doi: 10.1371/journal.ppat.1005813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Acuna R, Bignon EA, Mancini R, Lozach PY, Tischler ND. 2015. Acidification triggers Andes hantavirus membrane fusion and rearrangement of Gc into a stable post-fusion homotrimer. J Gen Virol 96:3192–3197. doi: 10.1099/jgv.0.000269. [DOI] [PubMed] [Google Scholar]
- 14.Ramanathan HN, Chung DH, Plane SJ, Sztul E, Chu YK, Guttieri MC, McDowell M, Ali G, Jonsson CB. 2007. Dynein-dependent transport of the Hantaan virus nucleocapsid protein to the endoplasmic reticulum-Golgi intermediate compartment. J Virol 81:8634–8647. doi: 10.1128/JVI.00418-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Simon M, Johansson C, Mirazimi A. 2009. Crimean-Congo hemorrhagic fever virus entry and replication is clathrin-, pH- and cholesterol-dependent. J Gen Virol 90:210–215. doi: 10.1099/vir.0.006387-0. [DOI] [PubMed] [Google Scholar]
- 16.Petersen J, Drake MJ, Bruce EA, Riblett AM, Didigu CA, Wilen CB, Malani N, Male F, Lee FH, Bushman FD, Cherry S, Doms RW, Bates P, Briley K. Jr, 2014. The major cellular sterol regulatory pathway is required for Andes virus infection. PLoS Pathog 10:e1003911. doi: 10.1371/journal.ppat.1003911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kleinfelter LM, Jangra RK, Jae LT, Herbert AS, Mittler E, Stiles KM, Wirchnianski AS, Kielian M, Brummelkamp TR, Dye JM, Chandran K. 2015. Haploid genetic screen reveals a profound and direct dependence on cholesterol for hantavirus membrane fusion. mBio 6:e00801-15. doi: 10.1128/mBio.00801-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Soldati T, Schliwa M. 2006. Powering membrane traffic in endocytosis and recycling. Nat Rev Mol Cell Biol 7:897–908. doi: 10.1038/nrm2060. [DOI] [PubMed] [Google Scholar]
- 19.Subtil A, Dautry-Varsat A. 1997. Microtubule depolymerization inhibits clathrin coated-pit internalization in non-adherent cell lines while interleukin 2 endocytosis is not affected. J Cell Sci 110:2441–2447. [DOI] [PubMed] [Google Scholar]
- 20.Diaz-Rohrer B, Levental KR, Levental I. 2014. Rafting through traffic: membrane domains in cellular logistics. Biochim Biophys Acta 1838:3003–3013. doi: 10.1016/j.bbamem.2014.07.029. [DOI] [PubMed] [Google Scholar]
- 21.Rodal SK, Skretting G, Garred O, Vilhardt F, van Deurs B, Sandvig K. 1999. Extraction of cholesterol with methyl-beta-cyclodextrin perturbs formation of clathrin-coated endocytic vesicles. Mol Biol Cell 10:961–974. doi: 10.1091/mbc.10.4.961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Grimmer S, van Deurs B, Sandvig K. 2002. Membrane ruffling and macropinocytosis in A431 cells require cholesterol. J Cell Sci 115:2953–2962. [DOI] [PubMed] [Google Scholar]
- 23.Meister M, Tikkanen R. 2014. Endocytic trafficking of membrane-bound cargo: a flotillin point of view. Membranes (Basel) 4:356–371. doi: 10.3390/membranes4030356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mayor S, Pagano RE. 2007. Pathways of clathrin-independent endocytosis. Nat Rev Mol Cell Biol 8:603–612. doi: 10.1038/nrm2216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang LH, Rothberg KG, Anderson RG. 1993. Mis-assembly of clathrin lattices on endosomes reveals a regulatory switch for coated pit formation. J Cell Biol 123:1107–1117. doi: 10.1083/jcb.123.5.1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sarkar K, Kruhlak MJ, Erlandsen SL, Shaw S. 2005. Selective inhibition by rottlerin of macropinocytosis in monocyte-derived dendritic cells. Immunology 116:513–524. doi: 10.1111/j.1365-2567.2005.02253.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kerr MC, Teasdale RD. 2009. Defining macropinocytosis. Traffic 10:364–371. doi: 10.1111/j.1600-0854.2009.00878.x. [DOI] [PubMed] [Google Scholar]
- 28.Blumenthal R, Gallo SA, Viard M, Raviv Y, Puri A. 2002. Fluorescent lipid probes in the study of viral membrane fusion. Chem Phys Lipids 116:39–55. doi: 10.1016/S0009-3084(02)00019-1. [DOI] [PubMed] [Google Scholar]
- 29.Diez S, Helenius JH, Howard J. 2008. Biomolecular motors operating in engineered environments In Protein Science Encyclopedia. Wiley Verlag, Weinheim, Germany. [Google Scholar]
- 30.Commisso C, Flinn RJ, Bar-Sagi D. 2014. Determining the macropinocytic index of cells through a quantitative image-based assay. Nat Protoc 9:182–192. doi: 10.1038/nprot.2014.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lim JP, Gleeson PA. 2011. Macropinocytosis: an endocytic pathway for internalising large gulps. Immunol Cell Biol 89:836–843. doi: 10.1038/icb.2011.20. [DOI] [PubMed] [Google Scholar]
- 32.Witkowski PT, Perley CC, Brocato RL, Hooper JW, Jurgensen C, Schulzke JD, Kruger DH, Bucker R. 2017. Gastrointestinal tract as entry route for hantavirus infection. Front Microbiol 8:1721. doi: 10.3389/fmicb.2017.01721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Aleksandrowicz P, Marzi A, Biedenkopf N, Beimforde N, Becker S, Hoenen T, Feldmann H, Schnittler HJ. 2011. Ebola virus enters host cells by macropinocytosis and clathrin-mediated endocytosis. J Infect Dis 204(Suppl 3):S957–967. doi: 10.1093/infdis/jir326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mercer J, Schelhaas M, Helenius A. 2010. Virus entry by endocytosis. Annu Rev Biochem 79:803–833. doi: 10.1146/annurev-biochem-060208-104626. [DOI] [PubMed] [Google Scholar]
- 35.Subtil A, Gaidarov I, Kobylarz K, Lampson MA, Keen JH, McGraw TE. 1999. Acute cholesterol depletion inhibits clathrin-coated pit budding. Proc Natl Acad Sci U S A 96:6775–6780. doi: 10.1073/pnas.96.12.6775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fielding CJ, Fielding PE. 2000. Cholesterol and caveolae: structural and functional relationships. Biochim Biophys Acta 1529:210–222. doi: 10.1016/s1388-1981(00)00150-5. [DOI] [PubMed] [Google Scholar]
- 37.Popik W, Alce TM, Au WC. 2002. Human immunodeficiency virus type 1 uses lipid raft-colocalized CD4 and chemokine receptors for productive entry into CD4(+) T cells. J Virol 76:4709–4722. doi: 10.1128/jvi.76.10.4709-4722.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Osuna-Ramos JF, Reyes-Ruiz JM, Del Ángel RM. 2018. The role of host cholesterol during flavivirus infection. Front Cell Infect Microbiol 8:388. doi: 10.3389/fcimb.2018.00388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tischler ND, Gonzalez A, Perez-Acle T, Rosemblatt M, Valenzuela PD. 2005. Hantavirus Gc glycoprotein: evidence for a class II fusion protein. J Gen Virol 86:2937–2947. doi: 10.1099/vir.0.81083-0. [DOI] [PubMed] [Google Scholar]
- 40.Willensky S, Bar-Rogovsky H, Bignon EA, Tischler ND, Modis Y, Dessau M. 2016. Crystal structure of glycoprotein C from a hantavirus in the post-fusion conformation. PLoS Pathog 12:e1005948. doi: 10.1371/journal.ppat.1005948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ludwig K, Korte T, Herrmann A. 1995. Analysis of delay times of hemagglutinin-mediated fusion between influenza virus and cell membranes. Eur Biophys J 24:55–64. doi: 10.1007/bf00211399. [DOI] [PubMed] [Google Scholar]
- 42.Rossman JS, Leser GP, Lamb RA. 2012. Filamentous influenza virus enters cells via macropinocytosis. J Virol 86:10950–10960. doi: 10.1128/JVI.05992-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gu Z, Noss EH, Hsu VW, Brenner MB. 2011. Integrins traffic rapidly via circular dorsal ruffles and macropinocytosis during stimulated cell migration. J Cell Biol 193:61–70. doi: 10.1083/jcb.201007003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kankaanpaa P, Tiitta S, Bergman L, Puranen AB, von Haartman E, Linden M, Heino J. 2015. Cellular recognition and macropinocytosis-like internalization of nanoparticles targeted to integrin alpha2beta1. Nanoscale 7:17889–17901. doi: 10.1039/c5nr06218g. [DOI] [PubMed] [Google Scholar]
- 45.Kramski M, Meisel H, Klempa B, Kruger DH, Pauli G, Nitsche A. 2007. Detection and typing of human pathogenic hantaviruses by real-time reverse transcription-PCR and pyrosequencing. Clin Chem 53:1899–1905. doi: 10.1373/clinchem.2007.093245. [DOI] [PubMed] [Google Scholar]
- 46.Sibold C, Meisel H, Kruger DH, Labuda M, Lysy J, Kozuch O, Pejcoch M, Vaheri A, Plyusnin A. 1999. Recombination in Tula hantavirus evolution: analysis of genetic lineages from Slovakia. J Virol 73:667–675. doi: 10.1128/JVI.73.1.667-675.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gavrilovskaya IN, Brown EJ, Ginsberg MH, Mackow ER. 1999. Cellular entry of hantaviruses which cause hemorrhagic fever with renal syndrome is mediated by beta3 integrins. J Virol 73:3951–3959. doi: 10.1128/JVI.73.5.3951-3959.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.