Abstract
Introduction:
This study aimed to investigate the association between candidate genetic variants and audiometric measures of noise-induced hearing loss (NIHL) in young musicians.
Methods:
The study analyzed a database by Phillips et al. (Feasibility of a bilateral 4000–6000 Hz notch as a phenotype for genetic association analysis. Int J Audiol 2015;54:645–52.) which included behavioral hearing thresholds, distortion-product otoacoustic emissions (DPOAE), tympanometric, and genetic data of 166 participants meeting the inclusion criteria. Nineteen single nucleotide polymorphisms (SNPs) in 13 cochlear genes previously associated with NIHL in factory workers were included in the present investigation. The average hearing threshold at 3000 and 4000 Hz (AHT) and average DPOAE signal to noise ratio (DPOAE SNR) in both ears were calculated.
Results:
The regression analyses showed that two SNPs—one in KCNE1 (rs2070358) and the other in CAT (rs12273124) revealed a statistically significant relationship with DPOAE SNR in both ears. Two SNPs in MYH14 and one in GJB4 revealed a significant association with DPOAE SNR in the left ear. Two SNPs in HSP70, one in CDH23 and one in KCNJ10 showed significant association with DPOAE SNR in the right ear. None of the included SNPs showed association with AHT in both ears.
Conclusions:
A genetic variant in KCNE1 was associated with the strength of the cochlear amplifier as assessed by DPOAE SNR. Musicians carrying causal genetic variants to NIHL might exhibit changes in their auditory functions early in the lifespan even when most subjects had their hearing thresholds within normal limits. These participants are likely to show the clinical manifestation of NIHL in the future if no preventive measures are applied.
Keywords: Catalase, Estrogen-related receptor beta, Gene association, Music exposure, Myosin heavy chain 14, Noise-induced hearing loss phenotype, Noise-induced hearing loss, Nuclear receptor, Potassium voltage-gated channel subfamily E regulatory subunit 1
Musicians are frequently exposed to hazardous sound levels that exceed the recommended exposure limits (1-5). Music students are exposed to higher sound exposure levels than their non-musicians counterparts primarily due to participation in music activities and noisy social activities (1). Frequent exposure to traumatic sound levels can substantially increase musicians’ risk for acquiring noise-induced hearing loss (NIHL) (6-8). NIHL is a permanent hearing loss that slowly develops as a function of continuous or intermittent acoustic exposure and duration (9). NIHL is often characterized by audiometric hearing loss between the frequency range of 3000 to 6000 Hz with recovery at 8000 Hz, commonly referred to as “audiometric notch” (9). NIHL is a complex disorder, generally defined as a multifactorial disorder, which is often associated with multiple genes in combination with lifestyle and environmental factors (10). Complex diseases, such as NIHL, do not show a clear-cut pattern of inheritance and usually show weak patterns of family clustering; this makes it challenging to determine a person’s risk of inheriting or passing on these disorders (11).
Intense sound exposure can cause direct mechanical trauma and indirect metabolic distress in the cochlea, which can damage cochlear structures such as hair cells, stereocilia bundles, supporting cells, afferent synaptic junctions, and the stria vascularis (12). Variants of several cochlear genes have been associated with susceptibility to NIHL in factory workers exposed to traumatic industrial noise. Genetic variants in metabolic enzymes, such as glutathione S-transferase mu 1 (GSTM1), glutathione S-transferase theta 1 (GSTT1), heat shock protein HSP70, and catalase (CAT) have been associated with NIHL (13-18). Variants in ion transport proteins (KCNE1 and KCNQ4), structural proteins (MYH14 and PCDH15), and gap junction proteins (GJB1 and GJB2) have been associated with NIHL in factory workers (10,19-21). Some of these studies have not been replicated in independent populations (13). This replication concern may be attributed to the difference between the NIHL identification criteria used as the NIHL phenotype (Table 1), and the study population of older factory workers who may have age-related confounding variables (22,23).
TABLE 1.
A brief summary of audiometric criteria used as NIHL phenotypes in previous human research
| NIHL Phenotype Definitions | Audiometric Transducers |
Study |
|---|---|---|
| Hearing threshold level of the left ear at 3000 Hz | Not reported | Van Laer et al. (2006), Liu et al. (2010) |
| Hearing threshold level of the left ear at 3000 Hz for Swedish sample set; and average hearing threshold level at 4000 and 6000 Hz of the left ear for Polish sample set | Not reported | Konings et al. (2007 and 2009) |
| Average hearing threshold level at 4000 and 6000 Hz of the left ear | Not reported | Pawelczyk et al. (2009); Sliwinska-Kowalska et al. (2008); Sliwinska-Kowalska et al. (2006); Kowalski et al. (2014) |
| Average hearing threshold level worse than 25 dB HL at low (500, 1000, and 2000 Hz) or high (4000, 6000, and 8000 Hz) frequency range | Not reported | Yang et al. (2006) |
| Average of bilateral hearing thresholds at 3000, 6000, and twoweighted 4000 Hz | Not reported | Chang et al. (2011) |
| Audiometric hearing thresholds from 125 to 8000 Hz greater than 25 dB | Not reported | Fortunato et al. (2004) |
| Audiometric notch at 4000 to 6000 Hz of at least 15 dB depth | TDH-39 | Phillips et al. (2015) (mentioned in an email conversation with the lead author) |
NIHL indicates noise-induced hearing loss.
Phillips et al. (22) studied genetic links to NIHL in young college-aged musicians. This population is exposed to a traumatic level of music on a daily basis (1,2), but it exhibits an absence of age-related confounding variables. The NIHL phenotype was defined as a bilateral notch at 4000 to 6000 Hz in a behavioral audiogram measured using TDH-39 (Telephonics Corporation, Farmingdale, NY) supra-aural headphones. The study found no association between the previously identified genetic variants and the audiometric notch phenotype. The null results might be attributed to the use of supra-aural headphones for measuring hearing thresholds at 6000 and 8000 Hz. The supra-aural transducers can produce an artifact in puretone audiometry mimicking the audiometric notch at 4000 to 6000 Hz (24,25).
While it is known that genetic factors can influence susceptibility to NIHL, the relationship between genetic variability and audiological measures of NIHL in young adults largely remains elusive. We hypothesized that young musicians carrying causal genetic variants to NIHL would exhibit subclinical changes in their auditory functions compared with their counterparts. To test this hypothesis, we analyzed the database by Phillips et al. (22). The study aimed to investigate the relationship between selected genetic variants and audiometric measures (i.e., hearing thresholds and distortion-product otoacoustic emissions [DPOAEs]) in a sample of young musicians.
METHODS
Participants
The study analyzed the database by Phillips et al. (22), which included audiological and genetic data of 640 student musicians (18–25 years of age) at the University of North Carolina at Greensboro. Recruitment of participants occurred during the academic years of 2010 to 2011 and 2011 to 2012. All participants were majoring in music with daily music exposure that included individual practice and ensemble practice. Along with the audiological data, participants were asked to complete an online survey inquiring about their demographic details and history of music exposure. Phillips et al. (22) categorized participants in three groups: with no notch, with a unilateral notch, and with a bilateral notch. The no-notch participants were defined as having no threshold below 15 dB HL nor a 15 dB drop at 8000 Hz from a previous best threshold. Those subjects showing a notch in one ear were classified as a unilateral notch, and those showing a notch in both ears were classified as a bilateral notch. The bilateral notch category served as the case group for determining a potential genetic association, and each subject in this group was matched by sex with a no-notch and a unilateral notch subject from the same university, and if possible, by playing the same or similar instrument. Phillips et al. (22) ran statistical analysis on a case-control-control (nTotal=252; nCases=84; nMales=136) cohort to identify the genetic association to NIHL. The cohort for the present study was chosen from the initial sample of 640 young adults. The present study identified 166 participants (average age=20.3 yr, range=18–25 yr) from the initial sample of 640 young adults with normal tympanometry and otoscopic findings and with no missing genetic and audiometric data. The demographic data for the study sample are presented in Table 2. Additional details about the distribution of sex, ethnicity, family history, and allele frequency across the sample are described in Supplement file S1, http://links.lww.com/MAO/A953.
TABLE 2.
Demographic details of the sample (n = 166)
| Variable | Category | Count (Percentage) |
|---|---|---|
| Gender | Male | 84 (50.6%) |
| Female | 82 (49.4%) | |
| Ethnicity | European | 152 (91.6%) |
| African | 6 (3.6%) | |
| Asian | 2 (1.2%) | |
| Multiracial | 6 (3.6%) | |
| Family history | Positive | 90 (54.2%) |
| Negative | 76 (45.8%) |
Prerequisite Testing
An otoscopic examination was performed with all participants. Participants with normal otoscopic findings were tested with immittance audiometry. Tympanometry was performed using a 226 Hz probe tone presented through a Maico MI 24 probe (MAICO Diagnostics, Eden Prairie, MN). Participants with normal otoscopic findings and normal immittance measures (i.e., tympanometric compliance value ranging from 0.33 to 1.75 cm3, ear canal volume ranging from 0.8 to 1.8 cm3, middle-ear pressure ranging from −50 to 25 daPa in both ears) were considered for the statistical analysis.
Assessment of Hearing Status
All audiometric measures described in this study were collected in a sound-treated booth meeting ANSI standards (ANSI S3.1–1999). Hearing thresholds were obtained for 493 participants. Audiometric thresholds were obtained at 250, 500, 1000, 2000, 3000, 4000, 6000, and 8000 Hz (GSI-61, Eden Prairie, MN) with TDH-39 supra-aural headphones (Telephonics, Farmingdale, NY), using the modified Hughson-Westlake procedure. Hearing thresholds at 6000 and 8000 Hz measured using TDH-39 headphones could be influenced by the resonance characteristics of the outer ear (5). Therefore, these thresholds were not considered for further analysis.
The average hearing threshold (AHT) was calculated by averaging hearing thresholds at 3000 and 4000 Hz for each ear. We included hearing thresholds at 3000 and 4000 Hz because 1) frequencies around 4000 Hz are most sensitive to noise trauma, and the hearing threshold at 3000 Hz continues to increase over a longer period than high-frequency hearing thresholds (24); and 2) most of the previous studies investigating NIHL used hearing thresholds at 3000 and/or 4000 Hz for the genetic association analyses (Table 1).
Distortion-Product Otoacoustic Emissions
DPOAEs were measured using a hand-held DPOAE screener (ERO-SCAN OAE, MAICO Diagnostics, Eden Prairie, MN). The DPOAE probe calibration test in a 2 cm3 cavity as recommended by MAICO Diagnostics was performed before testing. A real-ear probe calibration test was performed using the MAICO probe-fit check paradigm before running the DPOAE measurement. DPOAEs were measured for primary levels of 65/55dB SPLwithF2/F1 = 1.22. DPOAEs were measured for F2 frequency ranging from 1500 to 10,000 Hz at nine data points (i.e., 1500,2000,3000,4000, 5000, 6000,7000, 8000, and 10,000 Hz). DPOAEs were measured for 2 seconds at each F2 frequency while participants were seated comfortably in a sound-treated booth meeting ANSI standards (ANSI S3.1–1999). DPOAE SNR was calculated by averaging SNR values of the DP responses for F2 ranging from 1500 to 10,000 Hz above the noise floor. It was reasoned that the larger DPOAE SNR indicates robust DPs and the smaller values indicate diminished DPs, thereby providing a measure of the overall strength of the cochlear amplifier (23,26). We include DPOAEs from 1500 to 10,000 Hz for calculating DPOAE SNR because a broader DPOAE frequency range may provide a better estimate of noise-induced cochlear damage. We used SNR values to calculate the overall strength of the cochlear amplifier because the original database did not include amplitude and noise floor data. A previous report suggested that the test performance of otoacoustic emissions could be improved for identifying normal versus impaired ears when SNR values were used instead of absolute amplitude (27). DPOAEs were recorded in a sound-treated booth meeting with the ANSI standards; therefore, the noise floor value is not likely to influence the DPOAE SNR measurement. DPOAEs were obtained for 465 participants who agreed to the time commitment necessary to carry out this procedure.
Questionnaire Data
The survey included an assessment of three areas: demographic details, medical and audiological history, music exposure history, and family history of hearing loss. Four-hundred eighty-one participants filled out the survey.
Demographic details: Participants were asked about their age, sex, and ethnicity. Response choices for sex included man/woman/no disclosure. Ethnicity was classified: European American and others (including multiracial).
Medical and Audiological History: These questions addressed the history of hearing loss, medical conditions such as meningitis, high blood pressure, head injury, diabetes, mumps, heart trouble, malaria, scarlet fever, and others.
Music exposure history: Supplement file S2, http://links.lww.com/MAO/A954 provides details of the survey and scoring procedures. We used the survey data to calculate music exposure scores because previous studies suggested that self-report questions could provide a useful estimate of music exposure (28,29).
A family history of hearing loss: The questionnaire assessed the family history of hearing loss using the following question, “Do you have a family member with hearing loss?”. The response was categorized as a present family history or an absent family history.
Genotyping
Phillips et al. (22) collected Buccal cell samples (Isohelix: Boca Raton, FL) in duplicate at the time of audiometric testing and refrigerated before DNA extraction (Qiagen BioRobot). Genomic DNA was extracted from the de-identified buccal samples in the Molecular/Cellular Biology Core Laboratory at the University of North Carolina at Greensboro and outsourced for initial SNP genotyping and validation (GeneSeek; Lincoln, NE) on the Sequenom MassARRAY iPLEX platform. As a screen of the candidate variants, the case-control-control cohort in the initial sample was genotyped for 266 single nucleotide polymorphisms (SNPs) from 59 candidate genes. Candidate genes were selected if they had variants previously associated with NIHL or deafness or if they are expressed in the inner ear. SNPs within each of these candidate genes were selected if: 1) they had been implicated in earlier published NIHL studies, 2) they were directly implicated as causal for other health conditions, 3) they had been shown to affect expression of the candidate gene, or 4) they affected the amino acid sequence of the gene’s protein product (i.e., nonsynonymous SNPs) and occurred at a population frequency between ~0.5 and 20%. Further details about the genotyping procedure can be found in Phillips et al. (22).
The present study identified a subset of SNPs from the database that previously has been associated with NIHL. Table 3 shows initially screened candidate genes and SNPs included in the present study. Supplement file S3, http://links.lww.com/MAO/A955 presents genomic position, coding sequence change, genotype frequency, and minor allele frequency of the selected SNPs. We identified 29 SNPs in 13 candidate genes that were previously associated with NIHL and were analyzed by Phillips et al. (22). The SNPs with minor allele frequency more than 0.001 and with no missing data were included for the statistical analysis. Phillips et al. (22) matched participants by sex and instrument type across experimental groups to control for effects of sex and music exposure. The present study did not follow the matching scheme; however, it included sex and music exposure in the regression models.
TABLE 3.
Listing of initially screened candidate genes and SNPs
| Gene | Number of SNPs | SNP Number | Previous Association |
|---|---|---|---|
| KCNE1 | 3 | rs2070358, rs1805127, rs1805128 | Van Laer et al. (2006); Pawelczyk et al. (2009) |
| KCNQ1 | 1 | rs163171 | Van Laer et al. (2006) |
| KCNQ4 | 1 | rs34287852 | Van Laer et al. (2006); Pawelczyk et al. (2009) |
| CDH23 | 3 | rs1227049, rs1227051, rs3802711 | Yang et al. (2006); Sliwinska-Kowalska et al. (2008) |
| GJB1 | 1 | rs1997625 | Pawelczyk et al. (2009) |
| GJB2 | 3 | rs9552098, rs3751385, rs121908852 | Pawelczyk et al. (2009) |
| GJB4 | 1 | rs755931, | Pawelczyk et al. (2009) |
| KCNJ10 | 1 | rs1130183 | Pawelczyk et al. (2009) |
| CAT | 5 | rs564250, rs494024, rs475043, rs1001179, rs12273124 | Konings et al. (2007); Sliwinska-Kowalska et al. (2008) |
| HSP70 | 3 | rs1043618, rs1061581, rs2227956 | Chang et al. (2011) |
| PCDH15 | 1 | rs7095441, | Konings et al. (2009) |
| MYH14 | 2 | rs667907, rs588035 | Konings et al. (2009) |
| GRM7 | 1 | rs11928865 | Friedman et al. (2009) with age-related hearing loss |
| PON2 | 1 | rs987539 | Fortunato et al. (2004) |
| SOD2 | 1 | rs4880 | Liu et al. (2010) |
| ESRRB | 1 | rs61742642 | Phillips et al. (2015); Bhatt et al. (2016) |
Statistical Analysis
The best type of probability distribution for the present response variables (i.e., AHT and DPOAE SNR in both ears) was identified. The continuous dependent variables were fitted against several types of theoretical probability distributions listed in Supplement file S4, http://links.lww.com/MAO/A956. After model fitting, the best probability distribution for these variables was identified by evaluating the average fitness (Filliben correlation coefficient), skewness (Pearson’s moment coefficient of skewness), and shape (coefficient of kurtosis) of the probability distribution models. After filtering out samples with missing data, 156 subjects with complete data for DPOAE SNR and 166 subjects with complete data on AHT were used for the probability distribution selection.
Generalized Additive Models for Location, Scale and Shape (GAMLSS) were built using a mixed-method combining five cycles of the Rigby and Stasinopoulos (30) algorithm followed by the Cole and Green (31) algorithm for up to 20 extra iterations and identity as a link function. Four GAMLSS models were constructed to identify predictors for 1) AHT in the left ear, 2) AHT in the right ear, 3) DPOAE SNR in the left ear, and 4) DPOAE SNR in the right ear. The GAMLSS models included 22 dependent variables: sex, ethnicity, music exposure, and 19 SNPs highlighted in Supplement file S1, http://links.lww.com/MAO/A953. Supplement file S5, http://links.lww.com/MAO/A957 provides descriptive statistics for AHT and DPOAE SNR between individuals carrying major and minor alleles of the selected SNPs. We treated SNPs as binary variables to test the effect of carrying at least one minor allele on the NIHL phenotypes. These statistical models were built using the best probability distributions identified by the method described in this section (Supplement File S4, http://links.lww.com/MAO/A956). The Akaike information criterion (AIC) and Bayesian information criterion or Schwarz criterion (SBC) were calculated for each statistical model to evaluate relative quality (32,33).
The leave-one-out cross-validation analyses were performed across the samples. The prediction residuals and mean squared error for evaluating the performance of the statistical models were calculated to assess the generalizability of the results. The bootstrapping procedures were performed for measuring the accuracy and reproducibility of the present statistical analyses. The bias, standard error, and confidence intervals for each statistical model were calculated after running 500 bootstrap replications. The t test p-values obtained from the regression analyses were evaluated for identifying significant relationships between the independent and dependent variables in the regression models. All p-values were adjusted for type I errors using the false discovery rate (FDR) adjustment method (34). Adjusted p-values <0.05 for both ears in at least one hearing measurement (AHT or DPOAE SNR) were considered significant.
An additional descriptive analysis was performed by calculating the correlation coefficients between dependent variables and independent variables. All the regression and descriptive analyses were performed in R v3.4.0 (http://www..r-project.org). The GAMLSS package is available from the authors’ website (http://www.londonmet.ac.uk/gamlss/) and the CRAN R library (http://www.r-project.org).
RESULTS
Demographic and Audiological Details of the Study Sample
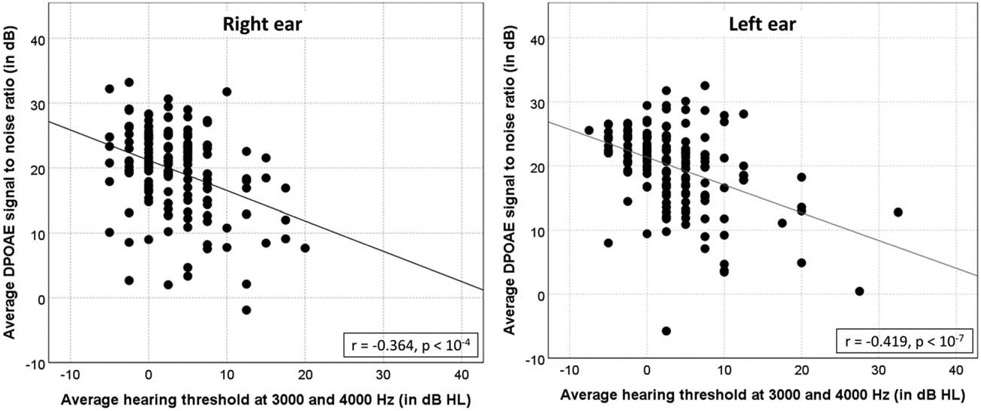
The study sample included almost 50% of men and 50% of women. Nearly 92% of the study sample reported predominant European American ethnicity. Almost 31% of the participants reported a history of ear infection, and 54% of the participants reported at least one relative with hearing loss. Nearly 12% of the participants reported exposure to tobacco smoking in a lifetime. Figure 1 provides a scatter plot between AHT and average DPOAE SNR for both ears.
FIG. 1.
Scatter plot between average hearing threshold at 3000 and 4000 Hz versus average DPOAE SNR (left ear). DPOAE SNR indicates distortion-product otoacoustic emissions signal to noise ratio.
Results for the Regression Analyses
We identified the best probability distributions for each response variable in the present study, as shown in Supplement file S6, http://links.lww.com/MAO/A958. Skew t type 4 exhibited the best fit for AHT in the left ear (skewness coefficient=0.068, Filliben correlation coefficient=0.987, Kurtosis coefficient=2.504, see Supplement file S4, http://links.lww.com/MAO/A956 and S6, http://links.lww.com/MAO/A958). Sinh-Arcsinh exhibited the best fit for DPOAE SNR in the left ear (skewness coefficient=0.009, Filliben correlation coefficient=0.999, Kurtosis coefficient=2.483). Similar results were obtained for the right ear. The results of the regression analyses for AHT are presented in Table 4. Women revealed significantly better AHT in the right ear (β=−2.26, adjusted p=0.02) and in the left ear (β=−2.12, adjusted p=0.05) compared with men. Subjects carrying the minor allele of ESRRβ revealed higher AHT in the left ear compared with subjects carrying the minor allele of ESRRβ rs61742642, but the regression estimate failed to obtain statistical significance after applying the FDR correction (β=2.01, p=0.046, adjusted p=0.25). No other dependent variables revealed a statistically significant relation with AHT in both ears.
TABLE 4.
Results of the regression analyses for AHT and DPOAE SNR
| AHT |
DPOAE SNR |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Left Ear |
Right Ear |
Left Ear |
Right Ear |
|||||||||
| β | p | adj p | β | p | adj p | β | p | adj p | β | p | adj p | |
| Gender | −2.12 | 0.01 | 0.05* | −2.26 | 0.00 | 0.02* | 3.40 | <0.01 | <0.01* | 4.24 | 0.00 | <0.01* |
| Ethnicity | −2.28 | 0.13 | 0.34 | 0.95 | 0.49 | 0.75 | −0.77 | 0.55 | 0.65 | −0.40 | 0.77 | 0.84 |
| Music exposure | 0.00 | 0.46 | 0.62 | 0.00 | 0.91 | 0.95 | 0.00 | <0.01 | <0.01* | 0.00 | 0.41 | 0.50 |
| KCNE1 (rs2070358) | 1.86 | 0.03 | 0.14 | 1.34 | 0.15 | 0.47 | 2.49 | <0.01 | 0.01* | 2.22 | 0.00 | 0.01* |
| KCNE1 (rs1805127) | −0.75 | 0.41 | 0.59 | −0.17 | 0.86 | 0.95 | −0.18 | 0.84 | 0.88 | −0.53 | 0.46 | 0.55 |
| KCNQ1 (rs163171) | −0.31 | 0.67 | 0.73 | −0.79 | 0.29 | 0.73 | 0.07 | 0.90 | 0.90 | −1.15 | 0.05 | 0.09 |
| CDH23 (rs1227051) | 0.60 | 0.40 | 0.59 | −0.73 | 0.34 | 0.73 | −0.67 | 0.30 | 0.41 | −2.03 | 0.00 | <0.01* |
| GJB2 (rs9552098) | −0.81 | 0.55 | 0.69 | 0.92 | 0.63 | 0.86 | −1.19 | 0.31 | 0.41 | −0.03 | 0.98 | 0.98 |
| GJB2 (rs3751385) | 0.69 | 0.58 | 0.69 | −1.33 | 0.46 | 0.75 | 1.23 | 0.30 | 0.41 | 0.98 | 0.38 | 0.49 |
| GJB4 (rs755931) | 0.73 | 0.61 | 0.69 | −1.09 | 0.39 | 0.73 | 2.94 | 0.01 | 0.04* | 1.62 | 0.16 | 0.24 |
| KCNJ10 (rs1130183) | 1.77 | 0.13 | 0.34 | 2.97 | 0.04 | 0.21 | −1.54 | 0.15 | 0.30 | −4.84 | 0.00 | <0.01* |
| ESRRβ (rs61742642) | 2.01 | 0.05 | 0.25 | 1.01 | 0.32 | 0.73 | −1.37 | 0.16 | 0.30 | −1.49 | 0.06 | 0.11 |
| CAT (rs475043) | −0.95 | 0.23 | 0.43 | −0.10 | 0.91 | 0.95 | −0.81 | 0.32 | 0.41 | −1.52 | 0.03 | 0.07 |
| CAT (rs12273124) | 0.17 | 0.92 | 0.96 | 0.49 | 0.75 | 0.95 | −2.90 | 0.02 | 0.05* | −4.27 | 0.00 | <0.01* |
| HSP70 (rs1043618) | 1.06 | 0.38 | 0.59 | 2.58 | 0.07 | 0.26 | −2.35 | 0.13 | 0.30 | −3.49 | 0.01 | 0.02* |
| HSP70 (rs1061581) | −2.28 | 0.07 | 0.25 | −2.72 | 0.05 | 0.21 | 2.16 | 0.15 | 0.30 | 3.29 | 0.01 | 0.02* |
| HSP70 (rs2227956) | −0.01 | 0.99 | 0.99 | −0.05 | 0.96 | 0.96 | −1.36 | 0.07 | 0.19 | 0.14 | 0.86 | 0.89 |
| PCDH15 (rs7095441) | −1.70 | 0.03 | 0.14 | 0.12 | 0.88 | 0.95 | 0.30 | 0.66 | 0.74 | −1.21 | 0.06 | 0.11 |
| MYH14 (rs667907) | −1.46 | 0.28 | 0.49 | −1.79 | 0.25 | 0.73 | 3.50 | 0.01 | 0.03* | 1.40 | 0.22 | 0.31 |
| MYH14 (rs588035) | 1.95 | 0.15 | 0.36 | 1.32 | 0.40 | 0.73 | −3.65 | 0.01 | 0.03* | −0.65 | 0.57 | 0.64 |
| GRM7 (rs11928865) | 0.41 | 0.59 | 0.69 | 0.64 | 0.42 | 0.73 | 0.69 | 0.40 | 0.49 | 02.01 | 0.00 | 0.01* |
| PON2 (rs987539) | −0.95 | 0.20 | 0.42 | −0.23 | 0.78 | 0.95 | 0.27 | 0.68 | 0.74 | −0.78 | 0.23 | 0.32 |
Coding scheme: 1—subject carrying major alleles, 2—subjects carrying at least one minor allele.
β-value presents the unstandardized regression coefficient.
AHT indicates average hearing threshold at 3000 and 4000 Hz (AHT); DPOAE SNR: distortion-product otoacoustic emissions signal to noise ratio.
Adjusted p-value <0.05.
Table 4 presents the results of the regression analyses for DPOAE SNR and AHT. DPOAE was significantly associated with sex, KCNE1 rs2070358, and CAT rs12273124. AHT was significantly associated with sex. MYH14 rs667907, MYH14 rs588035, GJB4 rs755931 revealed a statistically significant association with DPOAE SNR in the left ear. HSP70 rs1043618, HSP70 rs2227956, CDH23 rs1227051, and KCNJ10 rs1130183 showed significant association with the DPOAE SNR in the right ear. These SNPs have been associated with NIHL in factory workers (Table 3). However, the associations observed in the present study were not considered reliable due to the lack of replication in both ears.
Supplement S6, http://links.lww.com/MAO/A958 presents bootstrapping and cross-validation results for the statistical models. The GAMLSS regression models exhibited low bias at 96% confidence interval after the bootstrapping procedure, ensuring the precision of our estimates (bias<0.3 for DPOAE SNR and bias<0.073 for AHT). The low standard errors indicate the high reproducibility of the statistical models. The similarity between the training and testing mean squared errors (MSEs) after cross-validation indicates the low overfitting of the model.
DISCUSSION
We investigated the relationship between a selected set of genetic variants with DPOAE SNR and AHT in a sample of young musicians. Our analysis revealed that a SNP (rs2070358) in KCNE1 and a SNP (rs12273124) in CAT showed significant association with the DPOAE SNR in both ears. The study did not obtain any reliable genetic association with AHT. We found that women exhibited a significantly higher DPOAE SNR compared with men. This finding is in agreement with previously published reports (35-40). Besides, we obtained a negative correlation coefficient between DPOAEs and music exposure in the left ear. The effects of music exposure on DPOAEs have been established well in the literature (41-45). These findings further validate our research methods.
Association of a SNP in KCNE1 With the Strength of the Cochlear Amplifier
KCNE1 (potassium Voltage-Gated Channel Subfamily E Regulatory Subunit 1) belongs to the potassium channel KCNE family. KCNE1 is associated with long QT syndrome (LQTS), which is a prolongation of the cardiac action potential that is associated with arrhythmias followed by syncope of sudden death in otherwise healthy subjects. Two clinically distinct types of long QT syndrome (LQTS) have been described: autosomal recessive Jervell and Lange-Nielsen syndrome characterized by cardiac arrhythmia and severe congenital bilateral hearing loss (46); and autosomal dominant Romano-Ward syndrome that consists of LQTS without any other abnormalities (46). Variants in KCNE1 have been associated with defective trafficking of the potassium channel, reduced amplitude of the potassium ion current and influence on the LQTS pathogenesis (47,48).
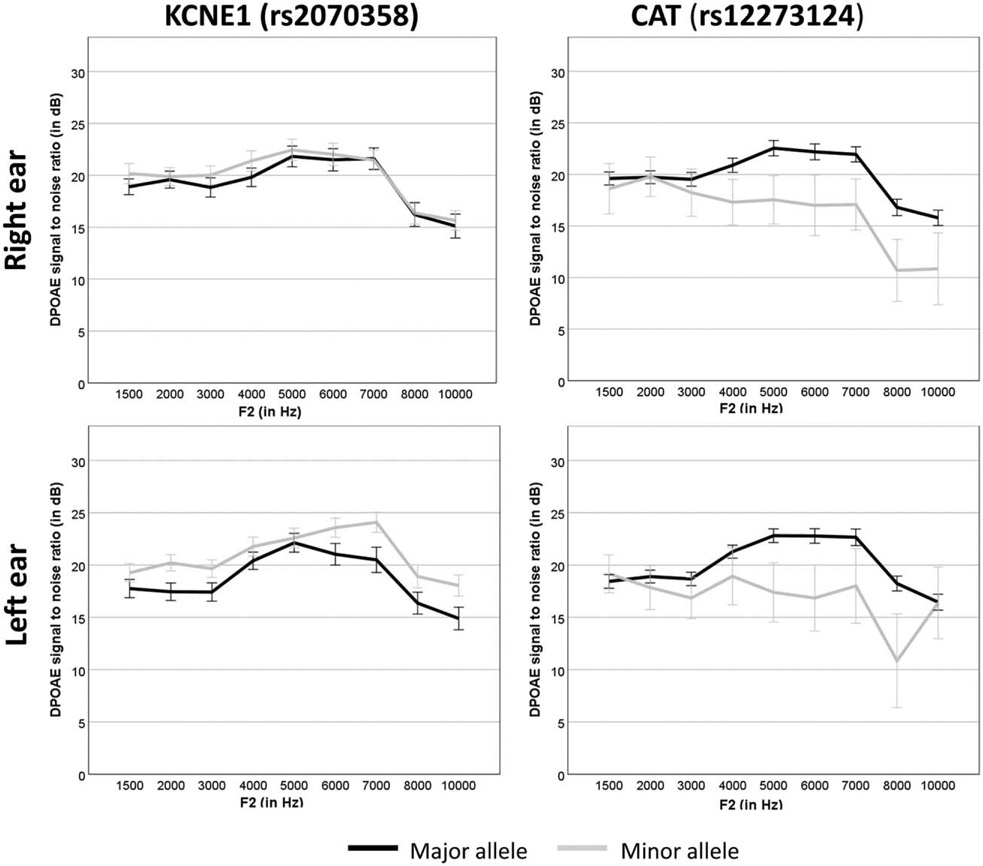
Potassium recycling is an essential process for maintaining cochlear homeostasis. Mutations in K+ recycling genes in the inner ear often lead to hearing loss (46,49-56). KCNE1 encodes a β-subunit of the KCNQ1/KCNE1 potassium channel that is expressed in the marginal cell membrane of the stria vascularis (57). KCNQ1/KCNE1 forms a selective potassium channel that activates very slowly at membrane potential more positive than – 40mV and deactivates slowly at membrane potential more negative than −40mV to maintain endolymphatic potential (55). The SNP rs2070358 (C > T) in KCNE1 was associated with NIHL in Swedish noise-exposed factory workers (19) and two independent samples from Swedish and Polish factory workers (20). Subjects carrying the GG genotype for KCNE1 (rs2070358) were found to be more susceptible to NIHL compared with individuals carrying the AA genotype in their study samples (46). The results of the present study revealed that individuals carrying rs2070358 GG genotype showed a significantly lower DPOAE SNR compared with individuals carrying rs2070358 AA or AG genotype (Fig. 2). These results are in agreement with Van Laer et al. (19) and Pawelczyk et al. (20). It was hypothesized that the KCNE1 variant (rs2070358) might influence regulation of endolymphatic potential by inefficient potassium ion trafficking in stria vascularis following noisy events leading to reduced strength of the cochlear amplifier.
FIG. 2.
Average DPOAE SNR as a function of F2 between subjects carrying major and minor alleles of KCNE1 (rs2070258) and CAT (rs12273124). The error bar presents ± standard error. DPOAE SNR indicates distortion-product otoacoustic emissions signal to noise ratio.
Association of a SNP in CAT With the Strength of the Cochlear Amplifier
Catalase (hydrogen peroxide oxidoreductase; EC1.11.1.6) is an enzyme that decomposes hydrogen peroxide (H2O2) to neutralize the reactive oxygen species in metabolically active organs (58). Polymorphisms in CAT have been associated with diabetes mellitus, high blood pressure, and vertigo (59-63). CAT is expressed in most cell types of the inner ear, including the organ of Corti and stria vascularis (64). The expression of CAT and other antioxidant factors increases significantly in the cochlea following noise exposure to neutralize noise-induced reactive oxygen species and to prevent hair cell dysfunction (12,65). CAT rs12273124 showed a significant interaction effect with NIHL in a Polish sample, but not in the Swedish sample (16). The homozygous genotype (AA) prevailed among sensitive subjects, while the heterozygous genotype (AG) was more prevalent in resistant subjects. The present study obtained a significant association between CAT rs12273124 (A→G) and DPOAE SNR in both ears (Fig. 2). Individuals carrying rs12273124 AG or GG genotype revealed a significantly diminished DPOAE SNR compared with individuals with the AA genotype. The direction of association was the opposite of the reported association by Lin et al. (16). Further research is needed to confirm the association in an independent sample.
Genetic Variability and Audiometric Measures: Present Results and Future Directions
We obtained a significant relation between music exposure and DPOAE SNR in the left ear. No such relationship was observed for DPOAE SNR in the right ear and AHT for both ears. This observation is consistent with the literature suggesting that the left ear is more susceptible to noise-induced cochlear dysfunction than the right ear (66). Two SNPs in MYH14 and one in GJB4 revealed a significant association with DPOAE SNR in the left ear. Two SNPs in HSP70, one in CDH23 and one in KCNJ10 showed significant association with the DPOAE SNR in the right ear. These SNPs have been associated with NIHL in factory workers (Table 3). Previous genetic studies used the left ear hearing thresholds around 3000 to 6000 Hz as the NIHL phenotype. As a strength, many of these studies included two independent samples to replicate the genetic association to NIHL. Considering that bilateral notched audiograms typically characterize NIHL in clinics (9), Phillips et al. (22) suggested that severe environmental exposure could often lead to a notch in one ear, but music students genetically predisposed to NIHL often show notches in both ears even if their exposure to sound was neither extreme nor prolonged in duration. Although the present study is limited to the SNPs previously associated with NIHL, considering our smaller sample size and a lack of replication sample, we applied an additional scientific rigor by considering the positive association to NIHL for only those SNPs associated with DPOAE SNR in both ears. We suggest our readers use caution while considering SNPs associated with DPOAE SNR in only one ear.
The present study demonstrated the effectiveness of DPOAEs for investigating the genetic association with NIHL in young adults. We found that previously associated genetic variants to NIHL showed no association with AHT in young musicians. However, significant associations were observed between DPOAE SNR and SNPs in KCNE1 and CAT in both ears. This observation may be attributed to the use of DPOAEs, which are more sensitive for detecting noise-induced cochlear dysfunction than behavioral hearing thresholds (67-69). The addition of electrophysiological measures (e.g., auditory brainstem responses) might improve the sensitivity of the phenotyping process because recent evidence suggests that noise exposure might lead to cochlear synaptopathy before causing damage to the cochlear hair cells (70-73). Our observation suggests that a test battery approach, including audiometry, DPOAEs, and electrophysiological measures, might be valuable for identifying genetic factors underlying NIHL in young adults.
Experimental Caveats
The present study was limited by its survey design for estimating music exposure. A small sample size might influence the regression analyses. Besides, the regression models might not be able to effectively control for the effects of confounding factors (e.g., sex, music exposure, and ethnicity) on audiometric measures. A comprehensive audiological test battery with extended high-frequency hearing thresholds, otoacoustic emissions, speech perception in noise measures, psychoacoustic, and electrophysiological measures might be better suited for evaluating auditory functions.
CONCLUSIONS
This study revealed an association between KCNE1 variant rs2070358 with DPOAE SNR in a sample of young musicians. The present study obtained an association with a previously identified SNP and DPOAE SNR with substantially smaller sample size. This observation may be attributed to the study sample of young adults with absent age-related confounding variables such as systemic diseases and age-related hearing loss. It was concluded that young musicians carrying causal genetic variants to NIHL might exhibit changes in their auditory functions early in the lifespan even when most subjects had their hearing thresholds within normal limits. These participants are likely to show the clinical manifestation of NIHL in the future if no preventive measures are applied. Further research is required to construct a clinically useful genetic risk profile for audiological measures to prevent and treat NIHL.
Supplementary Material
Acknowledgments:
The authors thank Dr. Susan Phillips for providing the database for the reanalysis. They thank Drs. Vincent Henrich, Scott Richter, Sandra Teglas, and Robin Morehouse for their contribution to the original research project. They thank the Music Research Institute and the Center for Biotechnology, Genomics and Health Research for their support. They acknowledge the contributions of Leslie Simmons and Sara Hunt for assistance with data collection, Jenna Callendar, Renuka Shivaji, and Rohini Patil for technical assistance with the preparation and processing of DNA samples, and Jeong Sep Sihm for assistance with survey database management and analysis.
The original research work described by Phillips et al. (22) was supported by the National Institute on Deafness and Other Communication Disorders Grant R21DC009296-01. The research reported in this publication was partially supported by the National Cancer Institute of the National Institutes of Health under the award for the Partnership of Native American Cancer Prevention U54CA143925, and by the National Institute on Deafness and Other Communication Disorders Grant R21DC016704-01A1.
Footnotes
The authors disclose no conflicts of interest.
Supplemental digital content is available in the text.
REFERENCES
- 1.Tufts JB, Skoe E. Examining the noisy life of the college musician: weeklong noise dosimetry of music and non-music activities. Intl J Audiol 2018;57 (suppl):S20–7. [DOI] [PubMed] [Google Scholar]
- 2.Washnik NJ, Phillips SL, Teglas S. Student’s music exposure: full-day personal dose measurements. Noise Health 2016;18:98–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.O’Brien I, Ackermann BJ, Driscoll T. Hearing and hearing conservation practices among Australia’s professional orchestral musicians. Noise Health 2014;16:189–95. [DOI] [PubMed] [Google Scholar]
- 4.Gopal KV, Chesky K, Beschoner EA, Nelson PD, Stewart BJ. Auditory risk assessment of college music students in jazz bandbased instructional activity. Noise Health 2013;15:246–52. [DOI] [PubMed] [Google Scholar]
- 5.Chesky K. Measurement and prediction of sound exposure levels by university wind bands. Med Probl Perform Art 2010;25:29–34. [PubMed] [Google Scholar]
- 6.Halevi-Katz DN, Yaakobi E, Putter-Katz H. Exposure to music and noise-induced hearing loss (NIHL) among professional pop/rock/jazz musicians. Noise Health 2015;17:158–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Phillips SL, Henrich VC, Mace ST. Prevalence of noise-induced hearing loss in student musicians. Int J Audiol 2010;49:309–16. [DOI] [PubMed] [Google Scholar]
- 8.Jansen EJM, Helleman HW, Dreschler WA, de Laat JAPM. Noise induced hearing loss and other hearing complaints among musicians of symphony orchestras. Int Arch Occup Environ Health 2009;82:153–64. [DOI] [PubMed] [Google Scholar]
- 9.Kirchner DB, Evenson E, Dobie RA, et al. Occupational noise-induced hearing loss: ACOEM task force on occupational hearing loss. J Occup Environ Med 2012;54:106–8. [DOI] [PubMed] [Google Scholar]
- 10.Kowalski TJ, Pawelczyk M, Rajkowska E, Dudarewicz A, Sliwinska-Kowalska M. Genetic variants of CDH23 associatedwith noise-induced hearing loss. Otol Neurotol 2014;35:358–65. [DOI] [PubMed] [Google Scholar]
- 11.Craig J. Complex diseases: research and applications. Nat Educ 2008;1:184. [Google Scholar]
- 12.Henderson D, Bielefeld EC, Harris KC, Hu BH. The role of oxidative stress in noise-induced hearing loss. Ear Hear 2006;27:1–19. [DOI] [PubMed] [Google Scholar]
- 13.Sliwinska-Kowalska M, Pawelczyk M. Contribution of genetic factors to noise-induced hearing loss: a human studies review. Mutat Res 2013;752:61–5. [DOI] [PubMed] [Google Scholar]
- 14.Yang M, Tan H, Yang Q, et al. Association of hsp70 polymorphisms with risk of noise-induced hearing loss in chinese automobile workers. Cell Stress Chaperones 2006;11:233–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Konings A, Van Laer L, Pawelczyk M, et al. Association between variations in CAT and noise-induced hearing loss in two independent noise-exposed populations. Hum Mol Genet 2007;16:1872–83. [DOI] [PubMed] [Google Scholar]
- 16.Lin CY, Wu JL, Shih TS, et al. N-acetyl-cysteine against noise-induced temporary threshold shift in male workers. Hear Res 2010;269:42–7. [DOI] [PubMed] [Google Scholar]
- 17.Liu Y, Li X, Guo X, Liu B, Lin A, Rao S. Association between polymorphisms in SOD1 and noise-induced hearing loss in Chinese workers. Acta Otolaryngol 2010;130:477–86. [DOI] [PubMed] [Google Scholar]
- 18.Abreu-Silva RS, Rincon D, Horimoto. et al. The search of a genetic basis for noise-induced hearing loss (NIHL). Ann Hum Biol 2011;38:210–8. [DOI] [PubMed] [Google Scholar]
- 19.Van Laer L, Carlsson P, Ottschytsch N, et al. The contribution of genes involved in potassium-recycling in the inner ear to noise-induced hearing loss. Hum Mutat 2006;27:786–95. [DOI] [PubMed] [Google Scholar]
- 20.Pawelczyk M, Van Laer L, Fransen E, et al. Analysis of gene polymorphisms associated with K ion circulation in the inner ear of patients susceptible and resistant to Noise-induced hearing loss. Ann Hum Genet 2009;73:411–21. [DOI] [PubMed] [Google Scholar]
- 21.Konings A, Van Laer L, Wiktorek-Smagur A, et al. Candidate gene association study for Noise-induced hearing loss in two independent Noise-exposed populations. Ann Hum Genet 2009;73:215–24. [DOI] [PubMed] [Google Scholar]
- 22.Phillips SL, Richter SJ, Teglas SL, et al. Feasibility of a bilateral 4000–6000 Hz notch as a phenotype for genetic association analysis. Int J Audiol 2015;54:645–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bhatt I, Phillips S, Richter S, et al. A polymorphism in human estrogen-related receptor beta (ESRRb) predicts audiometric temporary threshold shift. Int J Audiol 2016;55:571–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bhatt IS. Supra-aural transducer-related artifact contributes to overestimation of noise-induced hearing loss. J Acoust Soc Am 2018;143:2055–8. [DOI] [PubMed] [Google Scholar]
- 25.Schlauch RS, Carney E. Are false-positive rates leading to an overestimation of noise-induced hearing loss? J Speech Lang Hear Res 2011;54:679–92. [DOI] [PubMed] [Google Scholar]
- 26.Gates GA, Mills D, Nam BH, D’Agostino R, Rubel EW. Effects of age on the distortion product otoacoustic emission growth functions. Hear Res 2002;163:53–60. [DOI] [PubMed] [Google Scholar]
- 27.Hussain DM, Gorga MP, Neely ST, Keefe DH, Peters J. Transient evoked otoacoustic emissions in patients with normal hearing and in patients with hearing loss. Ear Hear 1998;19:434–49. [DOI] [PubMed] [Google Scholar]
- 28.Portnuff CDF, Fligor BJ, Arehart KH. Self-report and long-term field measures of MP3 player use: how accurate is self-report? Int J Audiol 2013;52 (suppl):S33–40. [DOI] [PubMed] [Google Scholar]
- 29.Portnuff CD. Reducing the risk of music-induced hearing loss from overuse of portable listening devices: understanding the problems and establishing strategies for improving awareness in adolescents. Adolesc Health Med Ther 2016;7:27–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rigby R, Stasinopoulos D. A semi-parametric additive model for variance heterogeneity. Stat Comput 1996;6:57–65. [Google Scholar]
- 31.Cole TJ, Green PJ. Smoothing reference centile curves: the LMS method and penalized likelihood. Stat Med 1992;11:1305–19. [DOI] [PubMed] [Google Scholar]
- 32.Bozdogan H. Model selection and akaike’s information criterion (AIC): the general theory and its analytical extensions. Psychometrika 1987;52:345–70. [Google Scholar]
- 33.Sclove SL. Application of model-selection criteria to some problems in multivariate analysis. Psychometrika 1987;52:333–43. [Google Scholar]
- 34.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Series B 1995;57:289–300. [Google Scholar]
- 35.Cacace AT, McClelland WA, Weiner J, McFarland DJ. Individual differences and the reliability of 2F1-F2 distortion-product otoacoustic emissions: effects of time-of-day, stimulus variables, and gender. J Speech Lang Hear Res 1996;39:1138–48. [DOI] [PubMed] [Google Scholar]
- 36.Dhar S, Long GR, Culpepper NB. The dependence of the distortion product 2f1-f2 on primary levels in non-impaired human ears, Journal of Speech. Lang Hear Res 1998;41:1307–18. [DOI] [PubMed] [Google Scholar]
- 37.Bowman D, Brown D, Kimberley B. An examination of gender differences in DPOAE phase delay measurements in normal-hearing human adults. Hear Res 2000;142:1–11. [DOI] [PubMed] [Google Scholar]
- 38.O’Rourke C, Driscoll C, Kei J, Smyth V. A normative study of distortion-product otoacoustic emissions in 6-year-old schoolchildren: Estudio normativo de las emisiones otoacústicas por productos de distorsión en escolares de 6 años. Int J Audiol 2002;41:162–9. [DOI] [PubMed] [Google Scholar]
- 39.Dunckley KT, Dreisbach LE. Gender effects on high frequency distortion product otoacoustic emissions in humans. Ear Hear 2004;25:554–64. [DOI] [PubMed] [Google Scholar]
- 40.McBride DI, Williams S. Audiometric notch as a sign of noise induced hearing loss. Occup Environ Med 2001;58:46–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bhagat SP, Davis AM. Modification of otoacoustic emissions following ear-level exposure to MP3 player music. Int J Audiol 2008;47:751–60. [DOI] [PubMed] [Google Scholar]
- 42.Muller J, Dietrich S, Janssen T. Impact of three hours of discotheque music on pure-tone thresholds and distortion product otoacoustic emissions. J Acoust Soc Am 2010;128:1853–69. [DOI] [PubMed] [Google Scholar]
- 43.Kumar A, Mathew K, Alexander SA, Kiran C. Output sound pressure levels of personal music systems and their effect on hearing. Noise Health 2009;11:132–40. [DOI] [PubMed] [Google Scholar]
- 44.Henning RL, Bobholz K. Distortion product otoacoustic emissions in college music majors and nonmusic majors. Noise Health 2016;18:10–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Narahari PG, Bhat J, Nambi A, Arora A. Impact of usage of personal music systems on oto-acoustic emissions among medical students. Noise Health 2017;19:222–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tyson J, Tranebjærg L, Bellman S, et al. IsK and KvLQT1: mutation in either of the two subunits of the slow component of the delayed rectifier potassium channel can cause jervell and lange-nielsen syndrome. Hum Mol Genet 1997;6:2179–85. [DOI] [PubMed] [Google Scholar]
- 47.Splawski I, Shen J, Timothy KW, et al. Spectrum of mutations in long-QT syndrome genes: KVLQT1, HERG, SCN5A, KCNE1, and KCNE2. Circulation 2000;102:1178–85. [DOI] [PubMed] [Google Scholar]
- 48.Nakano Y, Shimizu W. Genetics of long-QT syndrome. J Hum Genet 2016;61:51–5. [DOI] [PubMed] [Google Scholar]
- 49.Neyroud N, Tesson F, Denjoy I, et al. A novel mutation in the potassium channel gene KVLQT1 causes the jervell and lange-nielsen cardioauditory syndrome. Nat Genet 1997;15:186–9. [DOI] [PubMed] [Google Scholar]
- 50.Xia J, Liu C, Tang B, et al. Mutations in the gene encoding gap junction protein β-3 associated with autosomal dominant hearing impairment. Nat Genet 1998;20:370–3. [DOI] [PubMed] [Google Scholar]
- 51.Lautermann J, Wouter-Jan F, Altenhoff P, et al. Expression of the gap-junction connexins 26 and 30 in the rat cochlea. Cell Tissue Res 1998;294:415–20. [DOI] [PubMed] [Google Scholar]
- 52.Grifa A, Wagner CA, D’Ambrosio L, et al. Mutations in GJB6 cause nonsyndromic autosomal dominant deafness at DFNA3 locus. Nat Genet 1999;23:16–8. [DOI] [PubMed] [Google Scholar]
- 53.Kharkovets T, Hardelin JP, Safieddine S, et al. KCNQ4, a K+ channel mutated in a form of dominant deafness, is expressed in the inner ear and the central auditory pathway. Proc Natl Acad Sci USA 2000;97:4333–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.López-Bigas N, Olive M, Rabionet R, et al. Connexin 31 (GJB3) is expressed in the peripheral and auditory nerves and causes neuropathy and hearing impairment. Hum Mol Genet 2001;10:947–52. [DOI] [PubMed] [Google Scholar]
- 55.Marcus D, Wu T, Wangemann P, Kofuji P. KCNJ10 (Kir4. 1) potassium channel knockout abolishes endocochlear potential. Am J Physiol Cell Physiol 2002;282:C403–7. [DOI] [PubMed] [Google Scholar]
- 56.Beisel KW, Rocha-Sanchez SM, Morris KA, et al. Differential expression of KCNQ4 in inner hair cells and sensory neurons is the basis of progressive high-frequency hearing loss. J Neurosci 2005;25:9285–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wangemann P. K+ cycling and the endocochlear potential. Hear Res 2002;165:1–9. [DOI] [PubMed] [Google Scholar]
- 58.Chance B, Sies H, Boveris A. Hydroperoxide metabolism in mammalian organs. Physiol Rev 1979;59:527–605. [DOI] [PubMed] [Google Scholar]
- 59.Pask R, Cooper JD, Walker NM, et al. No evidence for a major effect of two common polymorphisms of the catalase gene in type 1 diabetes susceptibility. Diabetes Metab Res Rev 2006; 22:356–60. [DOI] [PubMed] [Google Scholar]
- 60.Chistiakov DA, Savost’ianov KV, Turakulov RI, et al. Nucleotide substitution C1167T in the catalase gene and position of nearby polymorphic markers DS11S907 and D11S2008 are connected with development of diabetes mellitus type 2. [Nukleotidnaia zamena C1167T v gene katalazy i raspolozhennye nepodaleku polimorfnye markery D11S907 i D11S2008 sviazany s razvitiem sakharnogo diabeta tipa 2]. Molekuliarnaia Biologiia 2000; 34:863–7. [PubMed] [Google Scholar]
- 61.Jiang Z, Akey JM, Shi J, et al. A polymorphism in the promoter region of catalase is associated with blood pressure levels. Hum Genet 2001;109:95–8. [DOI] [PubMed] [Google Scholar]
- 62.Casp CB, She J, Mccormack WT. Genetic association of the catalase gene (CAT) with vitiligo susceptibility. Pigment Cell Res 2002;15:62–6. [DOI] [PubMed] [Google Scholar]
- 63.Góth L, Rass P, Páy A. Catalase enzyme mutations and their association with diseases. Mol Diagn 2004;8:141–9. [DOI] [PubMed] [Google Scholar]
- 64.Jacono AA, Hu B, Kopke RD, et al. Changes in cochlear antioxidant enzyme activity after sound conditioning and noise exposure in the chinchilla. Hear Res 1998;117:31–8. [DOI] [PubMed] [Google Scholar]
- 65.Yamasoba T, Harris C, Shoji F, Lee RJ, Nuttall AL, Miller JM. Influence of intense sound exposure on glutathione synthesis in the cochlea. Brain Res 1998;804:72–8. [DOI] [PubMed] [Google Scholar]
- 66.Nageris BI, Raveh E, Zilberberg M, Attias J. Asymmetry in noise-induced hearing loss: Relevance of acoustic reflex and left or right handedness. Otol Neurotol 2007;28:434–7. [DOI] [PubMed] [Google Scholar]
- 67.Rosati MV, Tomei F, Loreti B, et al. Distortion-product otoacoustic emissions in workers exposed to urban stressors. Arch Environ Occup Health 2018;73:176–85. [DOI] [PubMed] [Google Scholar]
- 68.Atchariyasathian V, Chayarpham S, Saekhow S. Evaluation of noise-induced hearing loss with audiometer and distortion product otoacoustic emissions. J Med Assoc Thai 2008;91:1066–71. [PubMed] [Google Scholar]
- 69.Attias J, Horovitz G, El-Hatib N, Nageris B. Detection and clinical diagnosis of noise-induced hearing loss by otoacoustic emissions. Noise Health 2001;3:19–31. [PubMed] [Google Scholar]
- 70.Kujawa SG, Liberman MC. Adding insult to injury: cochlear nerve degeneration after “temporary” noise-induced hearing loss. J Neurosci 2009;29:14077–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Liberman LD, Liberman MC. Dynamicsofcochlear synaptopathy after acoustic overexposure. J Assoc Res Otolaryngol 2015;16:205–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Stamper GC, Johnson TA. Auditory function in normal-hearing, noise-exposed human ears. Ear Hear 2015;36:172–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Liberman MC, Epstein MJ, Cleveland SS, Wang H, Maison SF. Toward a differential diagnosis of hidden hearing loss in humans. PLoS One 2016;11:e0162726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Friedman RA, Van Laer L, Huentelman MJ, Sheth SS, Van Eyken E, Corneveaux JJ, ... & Bonneux S (2009). GRM7 variants confer susceptibility to age-related hearing impairment. Human molecular genetics, 18(4), 785–796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sliwinska-Kowalska M, Noben-Trauth K, Pawelczyk M, & Kowalski TJ (2008). Single nucleotide polymorphisms in the cadherin 23 (CDH23) gene in Polish workers exposed to industrial noise. American Journal of Human Biology, 20(4), 481–483. [DOI] [PubMed] [Google Scholar]
- 76.Fortunato G, Marciano E, Zarrilli F, Mazzaccara C, Intrieri M, Calcagno G, ... & Sacchetti L (2004). Paraoxonase and superoxide dismutase gene polymorphisms and noise-induced hearing loss. Clinical chemistry, 50(11), 2012–2018. [DOI] [PubMed] [Google Scholar]
- 77.Chang NC, Ho CK, Lin HY, Yu ML, Chien CY, & Ho KY (2011). Association of polymorphisms of heat shock protein 70 with susceptibility to noise-induced hearing loss in the Taiwanese population. Audiology and Neurotology, 16(3), 168–174. [DOI] [PubMed] [Google Scholar]
- 78.Slwinska-Kowalska M, Dudarewicz A, Kotyło P, Zamysłowska-Szmytke E, Pawlaczyk-Łuszczyńska M, & Gajda-Szadkowska A (2006). Individual susceptibility to noise-induced hearing loss: choosing an optimal method of retrospective classification of workers into noise-susceptible and noise-resistant groups. International journal of occupational medicine and environmental health, 19(4), 235–245. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.