Abstract
The Endosomal Sorting Complexes Required for Transport (ESCRTs) mediate diverse membrane remodeling events. These typically require ESCRT-III proteins to stabilize negatively-curved membranes, although recent work has indicated that certain ESCRT-IIIs also participate in positive-curvature membrane shaping reactions. ESCRT-IIIs polymerize into membrane-binding filaments, but the structural basis for negative versus positive membrane remodeling by these proteins remains poorly understood. To learn how certain ESCRT-IIIs shape positively-curved membranes, we determined structures of human membrane-bound CHMP1B-only, membrane-bound CHMP1B+IST1, and IST1-only filaments by electron cryomicroscopy. Our structures show how CHMP1B first polymerizes into a single-stranded helical filament, shaping membranes into moderate-curvature tubules. Subsequently, IST1 assembles a second strand upon CHMP1B, further constricting the membrane tube and reducing its diameter nearly to the fission point. Each step of constriction, thins the underlying bilayer, lowering the barrier to membrane fission. Our structures reveal how a two-component, sequential polymerization mechanism drives membrane tubulation, constriction, and bilayer thinning.
Report Summary statement
Further information on experimental design is available in the Nature Research Reporting Summary linked to this article.
Introduction
The Endosomal Sorting Complexes Required for Transport (ESCRT) belong to an evolutionarily conserved pathway that mediates membrane remodeling and fission events throughout the cell. The ESCRT machinery comprises staged complexes, including the early-acting ALIX, ESCRT-I, and -II factors and the late-acting ESCRT-III factors and VPS4 family of AAA ATPases. Early-acting factors bind to site-specific adaptors and then recruit the late-acting factors that constrict and sever the target membrane. First discovered for their role in the formation of multivesicular bodies (MVBs), ESCRT proteins serve essential functions in a range of cellular processes. Beyond MVBs, these processes include: cytokinetic abscission; egress of enveloped viruses; sealing holes in nuclear, endosomal, and plasma membranes1–8; and in peroxisome biogenesis and function9,10. ESCRT-III proteins primarily shape negatively-curved membranes, such as the necks of budding viruses or intralumenal vesicles, but we and others have shown that some ESCRT-III proteins can also stabilize positively-curved membranes11–14. Despite their importance to the cell, the mechanisms that govern how ESCRT-III proteins assemble and catalyze membrane remodeling reactions—of either positive or negative membrane curvature—remain unclear.
Humans have 12 different ESCRT-III proteins that share a conserved secondary structure core, including helices α1-α5. X-ray crystal structures of IST1 and CHMP3 revealed how these helices fold into a compact conformation referred to as a “closed” state15–17. Other ESCRT-III proteins can adopt more elongated “open” states that can also polymerize15,18–20. Structures of such open, elongated and assembled states are available for human CHMP1B13, S. cerevisiae Snf721, and D. melanogaster Shrub22. ESCRT-III polymerization may be regulated by reversible switching between closed and open conformations. Such conformational transitions could be regulated by protein-protein interactions with nucleating factors like the early-acting ALIX or ESCRT-II factors, by membrane curvature23, or by post-translation modifications, such as ubiquitination, which can sterically hinder membrane-binding or polymerization24.
We previously reported a ~4 Å resolution electron cryomicroscopy (cryoEM) structure of a helical filament containing CHMP1B and IST113. This copolymer structure was surprising as it consisted of two distinct strands: an inner strand of CHMP1B in an open conformation and an outer strand of IST1 in a closed conformation. The IST1 strand was tightly associated with the CHMP1B strand with 1:1 stoichiometry. The lumenal cavity of the copolymer was strongly positively charged and capable of shaping negatively charged membranes into positive-curvature membrane tubes in vitro and in vivo. Moreover, in vivo studies have shown that CHMP1B and IST1 co-localize with the VPS4 family member SPASTIN along positive membrane curvature surfaces at contact sites between lipid droplets and peroxisomes9 and along the surface of endosomal tubules12–14,25. Knockdown of CHMP1B, IST1, or SPASTIN results in elongated endosomal tubules, altered endosomal trafficking, and downstream cellular protrusion phenotypes26. Together, this growing literature supports the functional significance of positive membrane remodeling activities by these ESCRT-III proteins. To understand these new properties and roles for CHMP1B and IST1 better, and to define the still unknown structural mechanism by which any ESCRT-III protein interacts with lipid bilayers, we sought to understand how CHMP1B and IST1 work together to bind and constrict membranes.
Results
Structure of the membrane-bound CHMP1B filament
To learn how CHMP1B and IST1 remodel positively curved membranes, we sought to capture stable membrane-bound polymers composed of these proteins. We previously showed that incubating liposomes with CHMP1B led to the formation of membrane tubules coated with protein filaments13. To increase the yield and stability of these membrane tubules, we optimized the lipid composition and found that CHMP1B could remodel a variety of different liposome compositions into tubules. Two factors, in particular, enhanced the prevalence and stability of membrane-bound filaments for cryoEM analysis (Fig. 1a,d): 1) incorporation of polyunsaturated lipids (16:0–22:6 phosphocholine, SDPC) to increase membrane malleability27,28; and 2) increasing the concentration of negatively charged phospholipids such as PI(3)P (phosphatidylinositol 3-phosphate), PI(3,5)P2, or PI(4,5)P2 to complement the highly basic charge of the CHMP1B lumen13. For our in-depth studies, we settled on liposomes containing 58 mol% SDPC / 18 mol% POPS / 18 mol% cholesterol / 6 mol% PI(3,5)P2.
Fig. 1. CHMP1B and IST1 sequentially constrict membrane tubes.

a, CryoEM micrograph of a membrane-bound CHMP1B tubule. Scale bar: 25 nm. Inset, representative 2D class average. b, A grey-scale slice looking down the helical axis of the 3D cryoEM reconstruction of the membrane-bound CHMP1B filament. Diameters of the entire tube and the membrane leaflet peak-to-peak distances are annotated. c, Left, surface representation of the same end-on view as in (b) down the helical axis. CHMP1B (green) coats the exterior of the membrane bilayer (grey). A CHMP1B protomer is highlighted in dark green. Right, internal view looking outward from the surface of the membrane. d, IST1 further constricts the CHMP1B-membrane filament nearly to the hemifission point. CryoEM micrograph of a membrane-bound CHMP1B+IST1 filament with a vesicle protruding from the end. Scale bar: 25 nm. Inset, representative 2D average. e, A grey-scale slice looking down the helical axis of the 3D cryoEM reconstruction of the membrane-bound, right-handed CHMP1B+IST1 filament. Diameters of the entire tube and membrane leaflet peak-to-peak distances are annotated. f, Left, surface representation of the same end-on view as in (e) down the helical axis. Right, internal view looking outward from the surface of the membrane. IST1 protomers (cyan) bind to the exterior of CHMP1B (green), leading to constriction of the membrane (grey). IST1 and CHMP1B promoters are highlighted in dark cyan and dark green, respectively.
From this optimized lipid mixture, we first analyzed CHMP1B-only membrane tubes and found that they exhibited a range of diameters (~26–30 nm). This heterogeneity precluded high-resolution studies, but by sorting the tubes based on diameter, we were able to reconstruct a 28 nm diameter tube to ~6 Å resolution. This reconstruction unambiguously showed the open-state conformation of CHMP1B and the interconnected network of protomers within the single-stranded, right-handed filament (Fig. 1b,c, Extended Data Fig. 1, Table 1). Upon membrane tubulation, the membrane tube diameter, the distance between outer leaflet phosphate headgroups, narrows from >50 nm in the starting, spherical liposomes down to ~18 nm in the CHMP1B-constricted cylindrical state. The inner leaflet headgroups are separated by ~12 nm. Thus, the energy of CHMP1B self-assembly upon the membrane is sufficient to remodel low-curvature membrane spheres into moderate-curvature membrane cylinders.
Table 1.
CryoEM data collection, refinement and validation statistics
| Membrane-bound CHMP1B-only (EMD-20590, PDB 6TZ9) |
Membrane-bound CHMP1B+IST1, right handed (EMD-20588, PDB 6TZ4) |
Membrane-bound CHMP1B+IST1, left handed (EMD-20589, PDB 6TZ5) |
IST1-only (EMD-20591, PDB 6TZA) |
|
|---|---|---|---|---|
| Data collection and processing | ||||
| Magnification | 105.000 | 31,000 | 31,000 | 29,000 |
| Voltage (kV) | 300 | 300 | 300 | 200 |
| Electron exposure (e–/Å2) | 45 | 44 | 44 | 53 |
| Defocus range (μm) | −0.2 to −1.9 | −0.5 to −3.0 | −0.5 to −3.0 | −1.0 to −3.2 |
| Pixel size (Å) | 1.345 | 1.2156 | 1.2156 | 1.234 |
| Refined helical symmetry | 13.86°, 1.86 Å | 20.02°, 2.96 Å | −20.77°, 3.06 Å | 26.85°, 3.0 Å |
| Point group symmetry | C1 | C1 | C1 | C1 |
| Initial particle images (no.) | 160,258 | 388,115 | 388,115 | 120,165 |
| Final particle images (no.) | 9,661 | 66,149 | 57,915 | 4,556 |
| Map resolution (Å) | 6.2 | 3.2 | 3.1 | 7.2 |
| FSC threshold | 0.143 | 0.143 | 0.143 | 0.143 |
| Map resolution range (Å) | 2.8 to 7 | 2.8 to 7 | ||
| Refinement | ||||
| Initial model used (PDB code) | De novo | De novo | De novo | De novo |
| Model resolution (Å) | 7.7 | 3.4 | 3.2 | 7.8 |
| FSC threshold | 0.5 | 0.5 | 0.5 | 0.5 |
| Map sharpening B factor (Å2) | −50 | −25 | −25 | −50 |
| Model composition | ||||
| Nonhydrogen atoms | 32,890 | 99,216 | 93.500 | 19,432 |
| Protein residues | 4,238 | 12,852 | 12,104 | 2,534 |
| B factors (Å2) | ||||
| Protein | N/A* | 68 | 60 | N/A* |
| R.m.s. deviations | ||||
| Bond lengths (Å) | 0.006 | 0.011 | 0.005 | 0.007 |
| Bond angles (°) | 0/816 | 0.828 | 0.863 | 1.15 |
| Validation | ||||
| MolProbity score | 1.55 | 1.17 | 1.14 | 1.61 |
| Clashscore | 10.73 | 3.82 | 7.29 | 5.48 |
| Poor rotamers (%) | ||||
| Ramachandran plot | ||||
| Favored (%) | 98.76 | 98.58 | 98.86 | 95.53 |
| Allowed (%) | 1.24 | 1.42 | 1.14 | 4.47 |
| Disallowed (%) | 0 | 0 | 0 | 0 |
The resolutions of the maps are insufficient to determine these values
Structures of left- and right-handed membrane-bound CHMP1B+IST1 filaments reveal highly constricted bilayers and filament flexibility
We next investigated the effect of sequentially adding IST1 to these filaments by cryoEM. Interestingly, from the same sample we were able to determine two 3D reconstructions of membrane-bound CHMP1B+IST1 filaments, corresponding to approximately equal populations of right- and left-handed helical filaments, to 3.2 Å and to 3.1 Å resolution, respectively (Fig. 1e,f, Extended Data Figs. 2,3, Table 1). The right-handed CHMP1B+IST1 copolymer is a one-start, double-stranded filament. The outer strand comprises IST1 in the closed conformation, and the inner strand comprises CHMP1B again in the open conformation13. The reconstruction also reveals a continuous and highly constricted bilayer within the lumen. The overall outer diameter of the double-stranded filament is 25 nm, slightly narrower than the membrane-bound CHMP1B-only filament. However, due to the presence of two protein strands, the distance between outer leaflet phosphate headgroups is reduced to 10.6 nm, and the distance between inner leaflet headgroups is just 4.8 nm (Fig. 1e). Thus, the sequential addition of IST1 was sufficient to drive constriction of the CHMP1B strand and the internal membrane, narrowing the lumenal inner leaflet diameter from 12 nm down to 4.8 nm. The left-handed helical filament of the membrane-bound CHMP1B+IST1 copolymer is also a one-start, double-stranded filament. The left-handed copolymer is slightly more constricted, with an outer diameter of 24 nm and an inner leaflet distance of 4.4 nm (Extended Data Fig. 2a).
Previous work has documented that ESCRT-III filaments29, like bacterial flagella30, can adopt both left- and right-handed helical structures in cells. To understand the structural basis of this flexibility, we compared CHMP1B protomer conformations within the left- versus right-handed copolymers (Extended Data Fig. 2b,c). The overall root mean square deviation of the Cɑ backbone (RMSD) between helices ɑ1-ɑ5 of CHMP1B protomers in the two filaments is 1.2 Å. However, the RMSD is smaller when only aligning either the N-terminal helices ɑ1-ɑ2 (~0.7 Å) or the C-terminal helices ɑ4-ɑ5 (~0.6 Å) (Extended Data Fig. 2b,c). Therefore, switching between a left- or right-handed filament can be achieved simply by a small change in the joint between helices ɑ2 and ɑ3 (Extended Data Fig. 2b,c).
The sequential assembly of the IST1 strand does not discriminate between the left- and right-handed filaments as IST1 mainly interacts with the C-terminal region of CHMP1B (Extended Data Fig. 4)31. Alignment of CHMP1B and IST1 heteroprotomers from the two reconstructions reveals an RMSD of 1.7 Å (Extended Data Fig. 4b), but if the alignment of IST1 is limited to the CHMP1B C-terminal region that constitutes the IST1 binding site (helices ɑ4-ɑ6), the RMSD falls to 0.7 Å (Extended Data Fig. 4c). Furthermore, the RMSD of single IST1 protomers or between “j” and “j+1” subunits between the two copolymers are only 0.5 Å and 0.6 Å, respectively (Extended Data Fig. 4d). Together, these reveal that there are no significant differences in how IST1 assembles around either the left- or right-handed filaments. The relative orientation of this CHMP1B C-terminal region is either pitched ‘up’ or ‘down’ based on the hinge between helices ɑ2-ɑ3, but these regions do not contribute to IST1 binding. We envision that as one turn of CHMP1B finishes a revolution, it will either continue polymerizing ‘above’ or ‘below’ the initiating subunit, and this choice will define the helical hand. Thus, it appears that a stochastic flexing between helices ɑ2-ɑ3 allows either handedness to propagate. We did not observe left-handed helical polymers in our prior work on the lipid-free or the nucleic-acid templated copolymer structures, perhaps due to the different solution and nucleation conditions that promoted lipid-free polymerization13,31. We also did not reconstruct a left-handed membrane-bound CHMP1B-only filament, but this likely reflects the limited dataset size and diameter variability.
The joint between CHMP1B helices ɑ3 and ɑ4 accommodates different curvatures
To understand how CHMP1B polymers adopt different curvatures, we examined how the CHMP1B conformation changes between the “moderate” constriction CHMP1B-only and the “high” constriction CHMP1B+IST1 filaments. In all reconstructions, a single CHMP1B protomer (“i”) interacts with eight other protomers in an interconnected network. Among these, helix ɑ5 of the “i” subunit passes behind three neighboring subunits and binds the closed end of the ɑ1-ɑ2 hairpin of the “i+4” subunit (Fig. 2a). Thus, CHMP1B subunits always interweave with the same protomers, regardless of the filament’s overall degree of curvature. There is little contact between neighboring turns of the CHMP1B filament in either state, suggesting that turns can slide past one another during constriction (Fig. 2b, Supplementary Videos 1–2).
Fig. 2. CHMP1B interlocks in the same arrangement in all structures and flexes at the α3-α4 elbow to accommodate different curvatures.
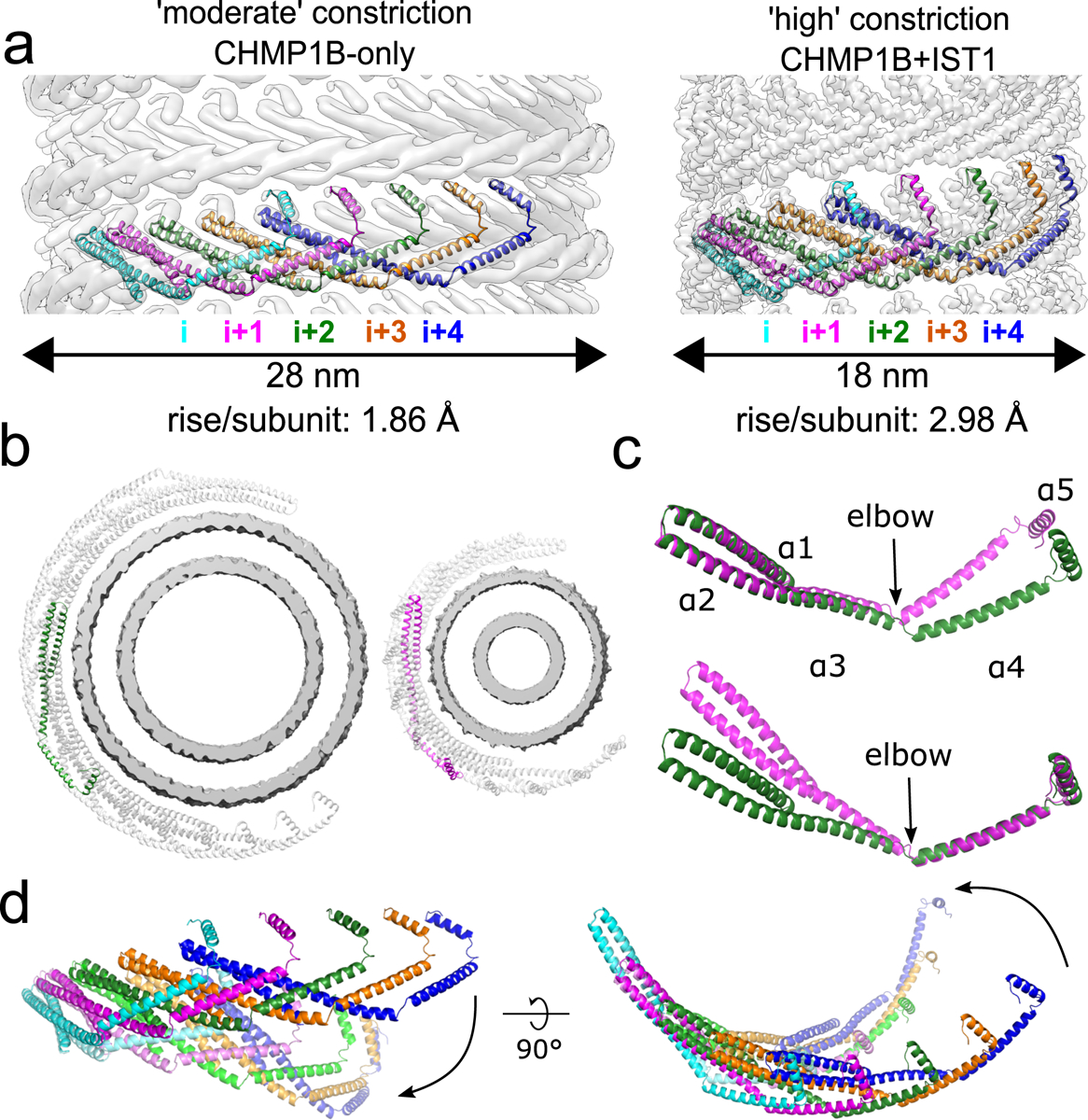
a, CryoEM density maps of CHMP1B from the membrane-bound CHMP1B (left) or right-handed CHMP1B+IST1 (right) filaments. Five interlocked copies of CHMP1B are shown as ribbons. The C-terminal helix α5 of the i protomer always engages helices α1-α2 of the i+4 protomer. The rise per subunit for each helical filament is denoted. b, Comparison of arc curvatures of CHMP1B across the two filaments. Top-down views of half a turn of CHMP1B subunits are shown for either the CHMP1B (left) or CHMP1B+IST1 (right) membrane filaments. The membrane bilayers are shown in grey and the central promoters are shown in green and magenta for the respective filaments. c, Superposition of a CHMP1B protomer from the CHMP1B (green) and CHMP1B+IST1 (magenta) filaments aligned to the CHMP1B N-terminal helices α1-α2 (top) or C-terminal helices α4-α5 (bottom). The biggest conformational change occurs at the elbow joint. d, Superposition of 5 consecutive subunits (colored from left to right in cyan, magenta, green, orange, and blue) of CHMP1B from the CHMP1B (opaque) and the CHMP1B+IST1 (semi-transparent) filament. The respective first protomers from each are aligned as in (c).
We next compared the conformation of a CHMP1B protomer between the CHMP1B-only and the right-handed CHMP1B+IST1 membrane-bound structures (Fig. 2c). The RMSD of helices ɑ1-ɑ5 is 2.4 Å between the two states. However, the RMSD drops to 1.1 Å when comparing only the N-terminal helices ɑ1-ɑ2. Similarly, comparing just the C-terminal helices ɑ4-ɑ5 lowers the RMSD to 1.5 Å. While there is again a small change in the angle between helices ɑ2-ɑ3, the largest conformation difference is at the “elbow” between helices ɑ3-ɑ4 (Fig. 2b,c). This flex in the elbow joint, when propagated across an entire turn of the helical assembly, leads to membrane constriction and also tubule elongation (Fig. 2d, Supplementary Videos 1–2). To confirm this tube elongation, we used holographic optical tweezers to hold traptavidin-coated beads and pull membrane tubes from giant unilamellar vesicles (GUVs), and then visualized changes in membrane tube lengths as a function of ESCRT-III addition. CHMP1B addition elongated the tubes slightly, and subsequent incorporation of IST1 produced even longer tubes (Extended Data Fig. 5, Supplementary Video 3). In agreement with our structural studies, tube elongation was dependent on CHMP1B, as the addition of IST1 alone did not induce membrane elongation (Supplementary Video 4).
Previous work has shown that other ESCRT-IIIs like CHMP2, CHMP3, and CHMP4 also form filaments with a wide range of curvatures32–34. Owing to the high homology of the ESCRT-III core, we suggest that these proteins also have dynamic elbow joints that will accommodate changes in filament curvature. As noted above, there are minimal contacts between turns of CHMP1B to stabilize inter-turn interactions (Supplementary Video 2). Indeed, in vivo images of ESCRT-IIIs at different sites of action reveal conical spirals with significant gaps between turns34–36. Thus, the inherent flexibility of ESCRT-III subunits could allow such loosely packed filaments to form with a range of diameters and helical pitches, and to slide past one another upon constriction.
IST1 polymerization drives constriction
To understand how IST1 induces constriction of CHMP1B-membrane filaments, we first docked two IST1 subunits (j and j+1) onto sequential CHMP1B protomers (i and i+1) of the moderate-curvature CHMP1B-only membrane, based on the CHMP1B-IST1 intersubunit interactions observed in the copolymer filaments (Fig. 3a). We then compared how the interactions between IST1 subunits would change between this ‘initial IST1 binding’ state and those observed in the CHMP1B+IST1 filaments (Fig. 3b,c). The IST1 j+1 subunit from the copolymer subunit swings closer (~8 Å) to the j subunit when compared to the j+1 subunit from the initial IST1 binding state, adding 480 Å2 of buried surface area (BSA) (Supplementary Videos 5–6). This swing enables the formation of hydrogen bonds between IST1 residues D77 and R82 on helix ɑ3 of the j subunit with R55 and E57 on helix ɑ2 of the j+1 subunit (Fig. 3c). In contrast, IST1 subunits make only minimal contacts between adjacent turns (15 Å2 of BSA between the j and j+18 subunits and no contacts between the j+1 and j+18 subunits). Thus, interactions between the j and j+1 subunits along the IST1 strand provide the force that flexes the CHMP1B elbow and consequently constricts and elongates the filament. These sliding, lateral interactions to promote changes in filament architecture have also been observed for the yeast ESCRT-III proteins Snf7 and Vps2437.
Fig. 3. IST1 polymerization drives constriction of the CHMP1B+IST1 filament.
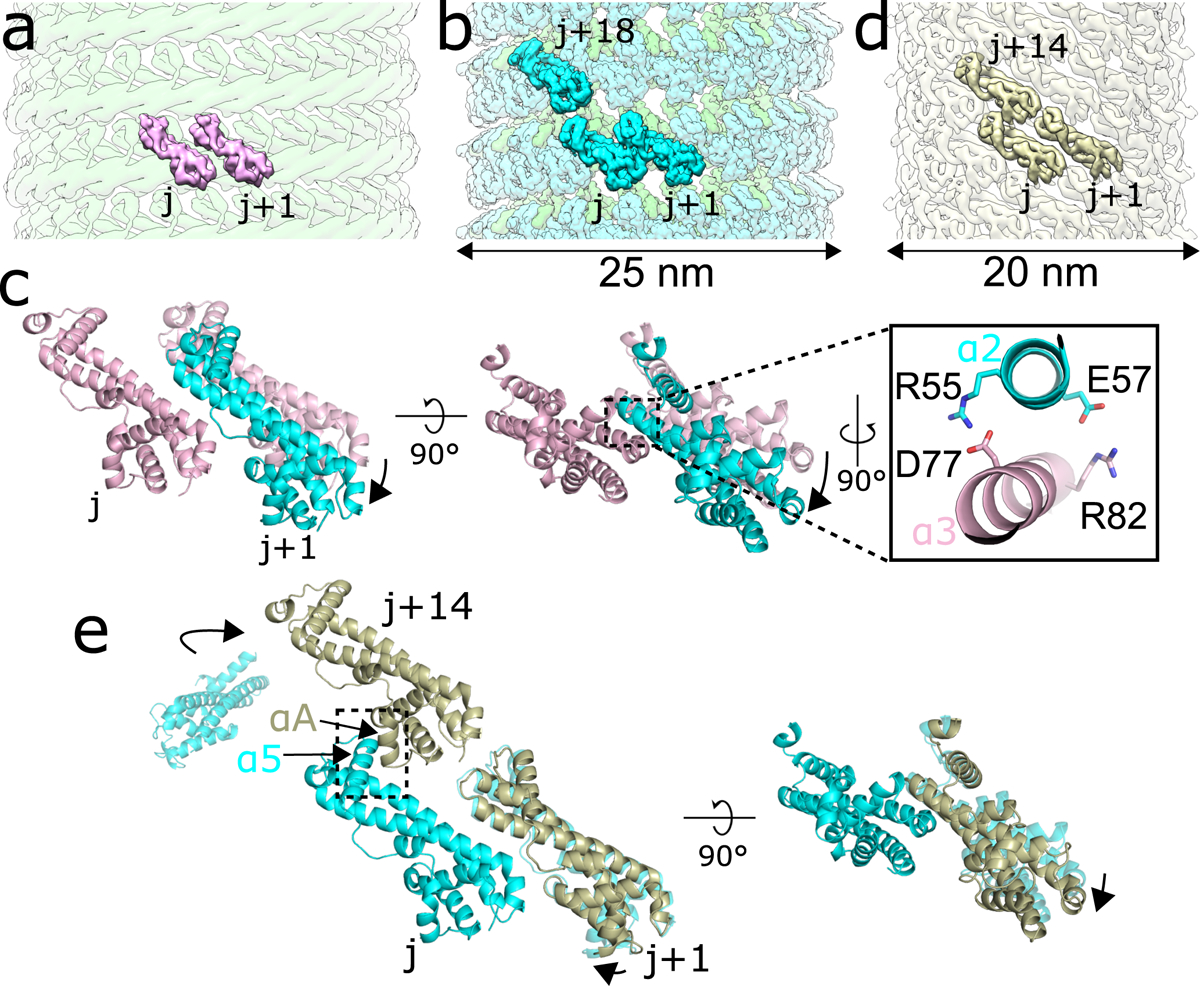
a, Model of two IST1 subunits (j, j+1), colored pink, initially binding onto the CHMP1B filament (green). b, CryoEM reconstruction of the right-handed CHMP1B+IST1 filament, with CHMP1B and IST1 in green and cyan, respectively. Three IST1 subunits (j, j+1, j+18) are highlighted. c, Superposition of the j and j+1 IST1 subunits from (a) and (b), with the j subunits used for alignment. (Inset), new electrostatic interactions between helix α3 (pink) from the j subunit and helix α2 (cyan) from the j+1 subunit help stabilize intra-IST1 contacts to drive constriction. d, CryoEM 3D reconstruction of the IST1NTDR16E/K27E filament (bronze). Three IST1 subunits (j, j+1, j+14) are highlighted. e, Superposition of the j, j+1, and j+14 subunits from the IST1NTDR16E/K27E filament in (d) with IST1 subunits from the CHMP1B+IST1 filament in (b). Protomers were aligned by the j subunit. The boxed area highlights helix α5 of the j subunit and helix αA from the j+14 subunit driving inter-turn interactions.
IST1 has recently been reported to induce constriction of ESCRT-III assemblies in vitro38. To understand how IST1 polymerization could drive constriction, we determined a 3D reconstruction of a protein-only IST1 filament (Fig. 3d). We previously showed that an N-terminal construct of IST1 (residues 1–189) harboring R16E and K27E mutations (IST1NTDR16E/K27E) cannot co-assemble with CHMP1B as the mutations destabilize the CHMP1B-IST1 interface, but can still form tightly-packed homopolymeric helical tubes13. The IST1-only filaments were heterogeneous in diameter (~18–28 nm), with the majority of filaments narrower than the CHMP1B+IST1 copolymers (24–25 nm). We identified a major subset of IST1NTDR16E/K27E tubes that were ~20 nm wide and reconstructed this class to moderate resolution, revealing the secondary structure elements of closed-conformation IST1NTD subunits (Extended Data Fig. 6). IST1NTDR16E/K27E forms a single-stranded, right-handed filament (Fig. 3d). In the absence of a membrane or CHMP1B, therefore, IST1 alone polymerizes into an even narrower helical assembly than either of the copolymers. Interestingly, the reconstruction of these IST1-only filaments revealed that IST1 always adopted the closed conformation, suggesting that IST1 may function exclusively in the closed state.
To understand how the arrangement of IST1 subunits between IST1-only and CHMP1B+IST1 filaments differ, we compared the interactions of IST1 subunits at the inter-turn interface between the copolymer (j, j+1, and j+18 subunits) and the IST1NTDR16E/K27E (j, j+1, and j+14 subunits) filaments. The RMSD between a protomer from the IST1NTDR16E/K27E filament and an IST1 protomer from the copolymer filament is ~1 Å, with minimal changes in the Cα backbone. The j and j+1 contact that defines nearest-neighbor IST1-IST1 interactions is conserved in both structures, but the IST1 j and j+1 subunits swing slightly closer together (~2 Å) in the IST1NTDR16E/K27E filaments (Fig. 3e). Propagation of this subtle change actually decreases the BSA between the j and j+1 subunits (from 480 Å2 to 340 Å2), but increases inter-turn contacts, which are predominantly made by helix α5 of the j+1 subunit contacting helix αA of the j+14 subunit (220 Å2) and additional packing between the j+1 and j+14 subunits (100 Å2). This results in an overall increase of 165 Å2 of BSA at the inter-turn interface for the IST1NTDR16E/K27E filament (Fig. 3e). Thus, unlike the constriction seen upon addition of IST1 to the CHMP1B filament, this second constriction step appears to be driven by inter-turn contacts.
We note that the inter-turn interactions involving the j and j+14 subunit in the IST1-only filament are unattainable in the CHMP1B+IST1 filament, as the presence of the CHMP1B helix α6, the MIT interacting motif (MIM), sterically blocks the IST1 j and j+14 packing (Extended Data Fig. 7). Intriguingly, the interaction between IST1 helix α5 and the CHMP1B MIM supports efficient assembly of the copolymer31. We speculate that modulation of this interface by the VPS4 family of ATPases could, in principle, regulate the degree of constriction achieved by the CHMP1B+IST1 filament.
The membrane bilayer thins and the inner leaflet compresses at high curvature
We also examined the consequences to the membrane as a function of increasing curvature stress (Fig. 4). In both copolymer reconstructions, helix ɑ1 of CHMP1B faced the membrane, and the surface of the bilayer appeared to be “dimpled” by specific amino acids at this high curvature (Fig. 4a), to a much greater degree than seen in the CHMP1B-only reconstruction (Extended Data Fig. 8). Specifically, the conserved CHMP1B residues F9, F13, K16, and R20, which contribute to the membrane-facing surface of helix ɑ1, comprise the closest protein-membrane interaction (Fig. 4b). Surprisingly, no residues appeared to insert deeply into the membrane. The mixed aromatic and cationic character of the CHMP1B region that most closely approaches the membrane does not suggest any lipid recognition specificity beyond net anionic charge, with the basic residues complementing the negatively charged membrane. It is somewhat surprising that the hydrophobic residues appear to remain hydrated at this degree of constriction, but we speculate that they may be poised to insert into the membrane (see below).
Fig. 4. The bilayer thins and the inner leaflet crowds under high curvature in the CHMP1B+IST1 filament.
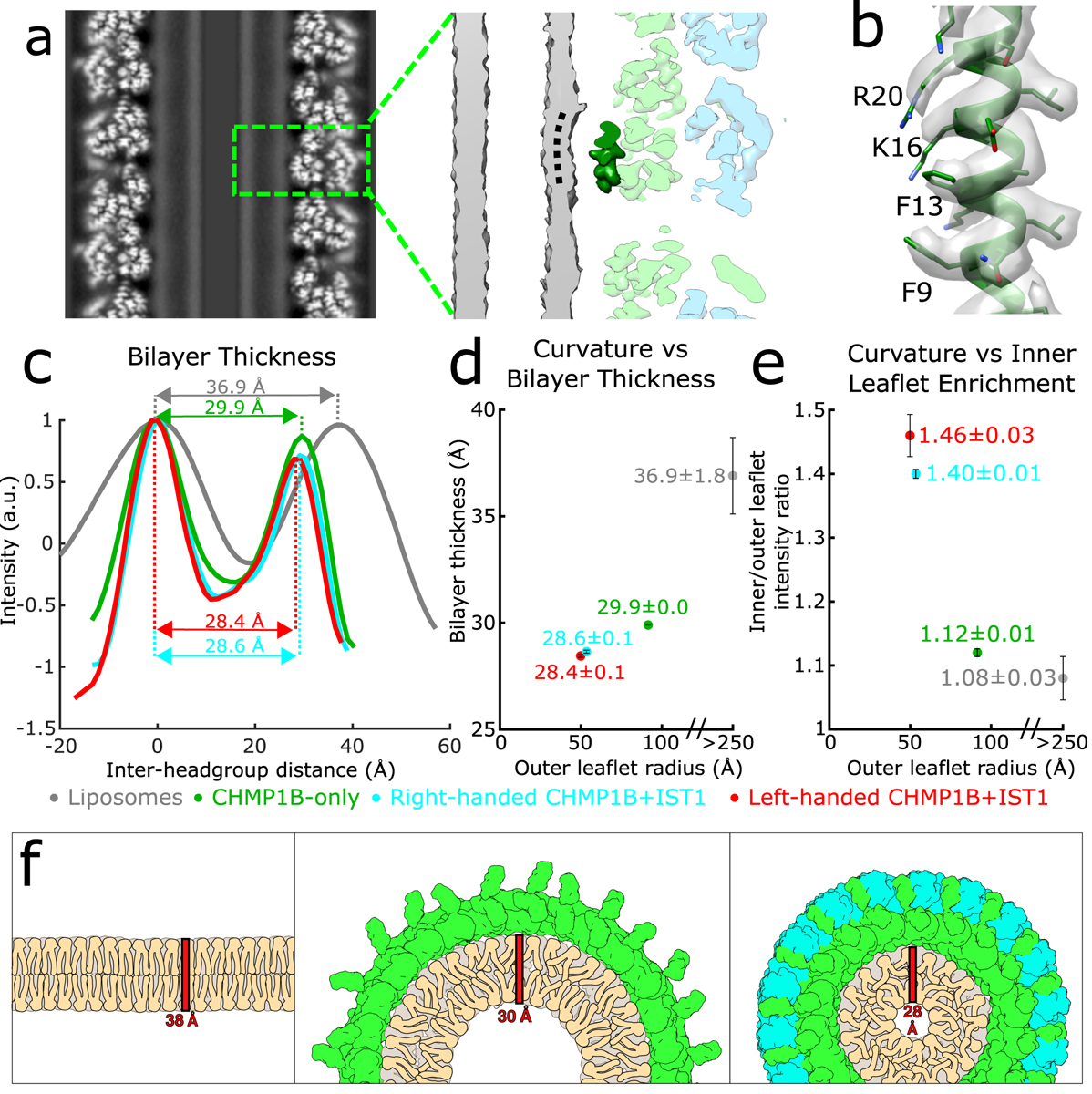
a, The outer leaflet dimples under extreme curvature in the membrane-bound CHMP1B+IST1 filament. Left, central slice along the helical axis of the CHMP1B+IST1 tubule. Right, zoomed view of boxed area in left showing CHMP1B dimpling (black dashed curved line) in the outer leaflet of the bilayer. A CHMP1B helix α1 sitting at the membrane is highlighted in dark green. b, CryoEM density map and model of CHMP1B helix α1 from (a). The residues involved in membrane binding are labeled. c, Intensity plots of membrane bilayer thickness for liposomes only (from 2D class averages, colored in grey), membrane-bound CHMP1B (green), and right- or left-handed membrane-bound CHMP1B+IST1 filaments (cyan and red respectively) as determined by cryoEM. The bilayer thickness is labeled for each. Intensities were normalized to the peak intensity of the inner leaflet. d, Plot of membrane thickness as a function of radius of the outer leaflet. The dots are colored as in (left). e, The inner membrane density increases as a function of curvature. Plot of ratio of inner leaflet to outer leaflet peak intensity as a function of radius of the outer leaflet. Dots are labeled as in (c). For (c-e), data are mean ± s.d. and the liposome values were determined from 2D averages (n=6) while the others were determined from half maps from each 3D reconstruction (n=2). f, Schematic illustration of lipid behavior as the bilayer is remodeled from planar (left), to moderate curvature by CHMP1B (middle), to high curvature by CHMP1B+IST1 (right). The outer leaflet headgroups spread out, while the inner leaflet headgroups crowd and the aliphatic tails become more disordered and therefore less extended.
By measuring the peak-to-peak distances between the outer leaflet and inner leaflet phosphate headgroups, we observed a correlation between bilayer thinning and the degree of membrane constriction by CHMP1B and by CHMP1B+IST1. To determine the bilayer thickness of our initial, unconstricted bilayers, we performed small-angle X-ray scattering (SAXS) of our relaxed liposomes. This experiment yielded a thickness estimate of 38.6 ± 0.6 Å (mean ± s.d., Extended Data Fig. 9), consistent with previously published SAXS measurements of membranes composed primarily of SDPC39. We compared this measurement with bilayer profiles from cryoEM 2D averages of segments of our liposomes, which yielded a thickness of 36.9 ± 1.8 Å, which is within the experimental uncertainty of the SAXS measurement (Fig. 4c,d). Upon constriction by CHMP1B alone, the bilayer compressed to 29.9 ± 0.0 Å. The sequential addition of IST1 led to further compression of the membrane to 28.6 ± 0.1 Å and 28.4 ± 0.1 Å for the right- and left-handed copolymers, respectively (Fig. 4c,d). In addition, the intensity of the inner leaflet increased as a function of constriction, with the ratio of inner to outer leaflet peak intensities of 1.08 ± 0.03, 1.12 ± 0.01, 1.40 ± 0.01, and 1.46 ± 0.03 for the liposomes, moderate-constriction CHMP1B filaments, and the high-constriction right-handed and left-handed CHMP1B+IST1 filaments, respectively (Fig. 4e). Thus, the lipid density in the inner leaflet increases significantly as the membrane tubule constricts towards the fission point.
In agreement with physical models of membrane behavior40,41, our reconstructions indicate that the bilayer thins as the membrane is constricted (Fig. 4c,d) and that the outer leaflet headgroups separate while the inner leaflet headgroups become more crowded (Fig. 4e). To accommodate this thinning, the acyl chains from both leaflets likely become more disordered and less extended (Fig. 4f). It has also been suggested that curvature stress causes local lipid composition changes that increase the membrane line tension and promote fission42,43. We speculate that these changes may lower the activation barrier for fission. With this specific lipid composition, however, the tubes remain stable with only a 4.2 nm gap between the inner leaflet headgroups across the lumen.
Discussion
A remaining central question is how ESCRT-III proteins work with their associated AAA ATPases to catalyze membrane constriction and, ultimately, membrane fission11,32,33,44–51. Here, we show how CHMP1B and IST1 function sequentially to squeeze and thin the membrane, bringing it nearly to the fission point (Supplementary Video 7). Specifically, CHMP1B first binds and assembles into a flexible filament that wraps the target membrane into a moderate curvature tubule. CHMP1B flexibility allows for many degrees of filament curvature and handedness, which may explain how it and other ESCRT-III proteins can adopt a wide range of architectures. IST1 binding then drives CHMP1B to constrict the membrane even further, exploiting IST1-intersubunit interactions to form tighter and tighter turns. Lipid composition is also expected to play a role in this process, although our work does not address that issue directly.
Recent studies with yeast and human ESCRT-III proteins have explored the roles of staged or sequential ESCRT-III assembly and ATP-dependent forces. Subunit exchange by VPS4 can alter the filament geometry34,38 and may subsequently drive membrane fission52. Consistently, a simulation study also suggested that changes in ESCRT-III filament geometry drive membrane remodeling51. Our work reveals mechanistically how one filament architecture, CHMP1B, changes as the consequence of the assembly of an additional ESCRT-III filament, IST1. Binding of the second ESCRT-III onto an already polymerized strand induces conformational changes that alter the overall polymer. While VPS4 is not required for IST1 to further constrict CHMP1B filaments in our situation, ATP hydrolysis energy will be required for subunit exchange and may be necessary in other contexts and with other ESCRT-III family members38. We note that in mammalian cells, depletion of SPASTIN, a microtubule severing enzyme within the meiotic clade of AAA ATPases (which also includes VPS4), has been shown to phenocopy loss-of-function perturbations of CHMP1B and IST1 by inducing formation of elongated endosomal tubules12,14,26, suggesting it may also act on CHMP1B and IST1.
As part of this staged assembly mechanism, our reconstructions suggest that IST1 may drive CHMP1B into an even narrower constriction state during the fission step, and that this process could be regulated by meiotic clade AAA ATPases. Comparison of the CHMP1B+IST1 and IST1-only filaments suggests that CHMP1B helix α6 (MIM) may sterically limit the full potential of IST1 constriction. We therefore speculate that unfolding or displacement of the CHMP1B MIM could trigger further tightening of the double-stranded filament. Importantly, the CHMP1B MIM forms the binding site for the MIT domains of meiotic clade AAA ATPases53–55, and MIT domain binding could therefore provide a mechanism for displacing this helix33,34,38,47. Further constriction of the CHMP1B+IST1 filament might also push the two aromatic residues of CHMP1B helix α1 (F9 and F13) from the hydrated surface layer, as seen in our structure, into the hydrophobic interior of the outer membrane leaflet, thereby further destabilizing the membrane, helping to drive lipid mixing, and promoting fission56,57.
Recent in vitro studies of the yeast ESCRT-III proteins Vps2–Vps24–Snf750 and their human homologs58 have also demonstrated positive curvature membrane remodeling activity – the same membrane orientation seen in our structures – despite their well-characterized negative membrane curvature deformation activity in vivo. While the significance of those in vitro observations remains to be fully elucidated, it appears that ESCRT-III assemblies are more versatile than previously realized. In support of this idea, IST1 is involved in both positive membrane curvature shaping reactions, as seen at lipid droplet-peroxisome contact sites9 and endosomal tubules12,14,26, as well as negative membrane curvature reactions, such as cytokinetic abscission35,59,60. Such curvature pliability may emerge from secondary membrane binding sites50 that are either occluded or revealed upon co-assembly with different ESCRT-III partners. Alternatively, the membrane-binding interface for a given ESCRT-III could be invariant, but their ability to invaginate versus protrude a target membrane, to drive negative or positive membrane remodeling, could be dictated by the flexibility in their hinge regions, as we have observed for CHMP1B.
Methods
Protein Expression and Purification
The purification of the N-terminal domain of IST1 containing residues 1–189 (IST1NTD) and the N-terminal IST1 R16E/K27E mutant (IST1NTDR16E/K27E) have been described previously13. CHMP1B residues 1–199 were cloned into an N-terminal 6xHis-SUMO fusion to yield a native N-terminus after removal of the purification tag. Two alleles of CHMP1B have been reported, 37K or 37E, and we saw that both alleles remodeled membranes and copolymerized with IST1 with indistinguishable activity. The CHMP1B 37E allele was used for subsequent studies and was expressed in LOBSTR-BL21 (DE3) cells61 in ZYP-5052 auto-induction media62. Cells were harvested and frozen at −80 °C. All subsequent steps were performed at 4 °C unless otherwise noted. Thawed cells were suspended in lysis buffer (50 mM Tris, pH 8, 500 mM NaCl, 10 mM Imidazole, 1 mM DTT, 5% (v/v) glycerol) and supplemented with lysozyme. Cells were lysed by sonication. Lysate was centrifuged at 30,000 × g for 1 h, and the supernatant was filtered using a 0.45 μm membrane. Clarified lysate was loaded onto a gravity flow column with Ni-NTA resin (Qiagen), incubated for 1 hour, and washed extensively with lysis buffer. The fusion protein was eluted in lysis buffer supplemented with 400 mM imidazole. His-tagged ULP1 protease was added and then dialyzed into cleavage buffer (20 mM Tris pH 8.0, 150 mM NaCl, 1 mM DTT, 10 mM imidazole) at room temperature overnight. The cleaved product was then applied again to Ni-NTA resin to remove the purification tag, uncleaved fusion protein, and the protease. CHMP1B was further purified by Superdex-75 16/60 size exclusion chromatography (GE Healthcare Life Sciences, USA) in size exclusion buffer (20 mM Tris, pH 7.4, 150 mM NaCl, 1 mM DTT).
Liposome Preparation
Stock lipid solutions (Avanti Polar Lipids) were resuspended in chloroform. To produce the liposomes, (18 mole% 16:0–18:1 phosphatidylserine (POPS), 58% 16:0–22:6 phosphatidylcholine (SDPC), 18% cholesterol, 6% PI(3,5)P2 or equivalent phosphoinositide), 2 mg total lipid were dried in a glass vial at room temperature under streaming nitrogen with vortexing. The lipids were again re-dissolved in chloroform, dried under streaming nitrogen, and desiccated under house vacuum (at least 4 hours in darkness). The lipid films were dispersed in 1 ml buffer (25 mM Tris, pH 7.4, 150 mM NaCl, 2 mg/ml final concentration, 4 °C overnight with gentle rocking). Liposomes were freeze-thawed 10 times and then stored at −80 °C.
Membrane Remodeling Reactions
Membrane remodeling reactions were performed at room temperature with protein concentrations ranging from 5 – 15 μM and liposome concentrations ranging from 0.5 – 1 mg/ml in reaction buffer (20 mM Tris, pH 7.4, 150 mM NaCl). CHMP1B was incubated with liposomes overnight at room temperature. For membrane-bound CHMP1B reactions, these were then directly used for EM sample preparation. For samples including IST1, the sample was pelleted (13,000 × g, 5 mins), the supernatant was decanted to remove unbound CHMP1B, and the pellet was resuspended in reaction buffer with equimolar IST1NTD. This was incubated for 10 mins and then subjected to EM sample preparation.
CryoEM Sample Preparation and Data Collection
3.5 μL of the membrane remodeling reactions were applied to glow-discharged R1.2/1.3 Quantifoil 200 Cu mesh grids (Quantifoil) in a Mark III Vitrobot (FEI). Grids were blotted with Whatman #1 filter paper (Whatman) for 4–8 seconds with a 0 mm offset at 19 °C and 100 % humidity before plunging into liquid ethane. Grids were stored under liquid nitrogen until samples were imaged for structural determination. Datasets were collected either on a 300 kV Technai Polara (membrane-bound CHMP1B+IST1 filaments), a 300 kV Titan Krios operating with an energy filter with a 20 eV slit (membrane-bound CHMP1B-only filaments), or a 200 kV Technai F20 (IST1-only filaments), all using a K2 Summit detector operated in super-resolution mode and binned by a factor of 2 for subsequent processing. Data collection parameters are summarized in Table 1.
EM Image Analysis and 3D Reconstructions
Structural biology applications used in this project were compiled and configured by SBGrid63. All dose-fractionated image stacks were corrected for motion artifacts, 2x binned in the Fourier domain, and dose-weighted using MotionCor264. GCTF-v1.0665 was used for contrast transfer function (CTF) estimation. Particles were selected manually using RELION3 with helical processing66, and subsequent steps were performed in RELION3 unless otherwise stated. Segments were extracted with ~90% overlap between boxes. Multiple rounds of 2D classification were performed to remove poor particles and to yield particles with a largely uniform diameter. For determination of the helical parameters for the CHMP1B-only or IST1NTDR16E/K27E filaments, SPRING67 and the iterative helical real space refinement (IHRSR) algorithm68 as implemented in SPIDER69 were used. For the membrane-bound CHMP1B-only and IST1-only filaments, more than 15 helical symmetries were tested, but further processing revealed that only one set of helical parameters for each sample yielded interpretable secondary structure features. For the CHMP1B+IST1 filaments, the previously determined helical parameters were used as the initial values13. For all reconstructions, hollow, smooth cylinders were used as initial models for 3D auto-refine reconstructions with refinement of helical parameters and a central Z length of 40% of the particle box and the ‘ignore CTFs until first peak’ flag for CTF estimation was used. These particles then went through multiple rounds of 3D classification without alignment with 3–4 classes and T values varying from 2–10 and a protein-membrane mask. Selected particles then were subject to another 3D auto-refine reconstruction with per-particle CTF estimation correction within RELION3, followed by a final 3D auto-refine reconstruction. For the membrane-bound left-handed CHMP1B+IST1 reconstruction, initial 3D classification without alignment yielded a class with no discernable features. 2D classification of these particles still yielded class averages with secondary structure features. Refinement of the helical parameters by switching the sign of the twist was then able to generate a good initial 3D reconstruction that was further refined as above. Post-processing was performed using the masks from refinement with ad hoc B-factors applied. Refinement parameters are listed in Table 1.
Atomic Modeling and Validation
For the high-resolution CHMP1B+IST1 filaments, a single protomer of CHMP1B and IST1 from the previously determined structure were initially docked into the density with UCSF Chimera70. The protomers were manually adjusted and rebuilt in Coot71 and then refined in phenix.real_space_refine72 using global minimization, morphing, secondary structure restraints, and local grid search. The refined protomers were then used to generate roughly two full turns manually in real space using UCSF Chimera. Noncrystallographic symmetry (NCS) constraints were then used thorough refinement in phenix.real_space_refine. Iterative cycles of manually rebuilding in Coot and phenix.real_space_refine, with previous strategies and additionally B-factor refinement, were performed. For the low resolution CHMP1B-only or IST1NTD filaments, a similar procedure was performed but only roughly one turn was built and no B-factor refinement was performed in phenix.real_space_refine.
All final model statistics were tabulated using Molprobity73 and summarized in Table 1. Map versus atomic model FSC plots and d99 scores were computed in PHENIX74. All structural figures were generated with UCSF Chimera and PyMOL (http://www.pymol.org).
Real-time Imaging of Membrane Constriction
Lipid tube pulling and imaging were performed using a previously described holographic optical trapping setup possessing an independent fluorescent imaging capability75. Briefly, the setup included a custom-modified Eclipse Ti microscope (Nikon Instruments; Melville, NY), with a Nikon 100X, 1.49NA oil immersion objective, Sapphire 488 excitation laser (Coherent, Santa Clara, CA), and a DU897 camera (Andor Technology, Oxford Instruments, USA). All videos were recorded at 20 fps. Biotinylated giant unilamellar vesicles (GUVs) in Flow Buffer (50 mM Tris pH 8.0, 150 mM NaCl, 1 mM DTT,) were synthesized as previously described76. Biotinylated silica beads (5 μm diameter, Si5u-BN-1, Nanocs, USA) were incubated with 10 μM traptavidin (Kerafast, USA) in PMEE buffer (35 mM PIPES, 5 mM MgSO4, 1 mM EGTA, 0.5 mM EDTA, pH 7.2) for 30 min followed by two rounds of washing (centrifugation at 12,000 × g for 2 minutes followed by supernatant removal and re-suspension of the pellet in 50 mM Tris buffer) to remove excess traptavidin. The beads were then mixed with GUVs and diluted as necessary to limit bead density to 1–3 beads in each 50 μm × 50 μm field of view. The mixture was then applied to a flow cell and immediately subjected to experiments.
Membrane tubules were formed by capturing a bead in an optical trap, moving the trapped bead into contact with a GUV, and finally, upon attachment, moving the bead away from the GUV. The presence of a tubule was then assessed visually in real time. The GUVs were stabilized in place via other beads attached to their surface and held in independent holographically defined traps or via non-specific surface attachment. Proteins were introduced into the flow cell in a sequential manner: first 0.5 μM CHMP1B and then 0.5 μM IST1 (both in Flow Buffer). Lipids tubules were held perpendicular to flow direction during these buffer exchanges and the resulting tubule constrictions were recorded. Experiments with CHMP1B and IST1 in slightly different buffers (0.5 μM CHMP1B in 10 mM Tris pH 8.0, 100 mM NaCl 1 mM DTT or 0.5 μM IST1 in 50 mM Tris pH7.0, 350 mM NaCl, 5% (v/v) glycerol, 5 mM 2-mercaptoethanol) yielded similar results.
Small Angle X-ray Scattering
For small-angle X-ray scattering (SAXS) analysis, liposomes were formed by vortexing a dry lipid film (similar to liposome preparation as stated above) in water to yield a final lipid concentration of 15 mg/ml. Liposomes were extruded through a 100 nm-pore polycarbonate membrane, followed by extrusion through a 50 nm-pore polycarbonate membrane. Synchrotron SAXS data were collected at beamline 4–2 of the Stanford Synchrotron Radiation Lightsource (SSRL), Menlo Park, CA77. The sample to detector distance was set to 1.1 m, and the X-ray wavelength used was λ = 1.127 Å (11 keV). Using a Pilatus 3 × 1M detector (Dectris Ltd, Switzerland) the setup covered a range of momentum transfer of q ≈ 0.017 – 1.17 Å−1 where q is the magnitude of the scattering vector, defined as q = 4π sinθ /λ, where θ is the scattering angle, and λ is the wavelength of the X-rays. Aliquots of 32 μl of freshly extruded vesicles were loaded onto the automated sample loader at the beamline. Consecutive series of thirty 2 s exposures were collected first from the buffer blank followed by the vesicle samples. Solutions were oscillated in a stationary quartz capillary cell during data collection to reduce the radiation dose per exposed sample volume. The collected data were radially integrated, analyzed for radiation damage and buffer subtracted using the automated data reduction pipeline at the beam line. To improve statistics and check for reproducibility, the measurements were repeated with different aliquots four times. As no significant differences were found between the repeat measurements, the different data sets were averaged together.
The buffer-subtracted and averaged data were fit using a model for unilamellar vesicles78 as previously described79. The electron density profile of the bilayer is approximated by three Gaussian peaks corresponding to the inner and outer phosphate peaks and a negative peak at the center for the hydrocarbon region. The following Equation 1 was used to fit the data:
| (1) |
where R0 is the mean radius of the vesicle measured from the center of the bilayer, ε is the peak displacement from the bilayer center, σ the Gaussian width of the peak and ρ is its amplitude. I0 is the overall intensity of the measured profile. A background term was added, consisting of a constant a0 and a linear term a1, to take into account the contribution from possible lateral correlations between the lipids. The measured data were fit in the q-region between q = 0.02 – 0.6 Å−1 (depicted as blue colored points in Extended Data Fig. 9). First, the data were fit using a simulated annealing routine, and the results were then further optimized using a non-linear least square algorithm, both by using code from the open source GNU scientific library project (https://www.gnu.org/software/gsl/). The final fit parameters including the final χ2 value of the fits and the resulting bilayer thickness (measured as distance between the inner and outer leaflet peak positions) are summarized in Extended Data Fig. 9.
Image Analysis of Membrane Bilayer
To determine the bilayer thickness and relative intensity of inner and outer leaflets, Fiji80 was used. The cryoEM half maps from the C1 reconstructions were each low-pass filtered to 8 Å, summed along the central 40% along the Z-axis, and the maps were radially averaged using the Radial Profile Extended plugin (Carl, P., http://questpharma.u-strasbg.fr/html/radial-profile-ext.html) to determine the intensities as a function of radius, resulting in two measurements per reconstruction. 2D averages of liposomes (n=6) were low pass filtered to 20 Å, parameters for a circle that defined the 2D average were determined, and a wedge that best covered the image was then used to calculate the intensities. For all resulting plots, the radial intensities were normalized to the peak intensity of the inner leaflet peak and then fit to a three Gaussian model (O’Haver, T. https://terpconnect.umd.edu/%7Etoh/spectrum/CurveFittingC.html) similar to the SAXS measurements. The model was minimized by a non-linear least square algorithm and the fit errors between the data and models ranged from 1.45%−3.30% with R2 values from 0.996–0.999. The local maxima for the headgroups were used to determine the bilayer thickness and inner/outer leaflet intensity ratios.
Extended Data
Extended Data Fig. 1. CryoEM validation of membrane-bound CHMP1B-only filament.
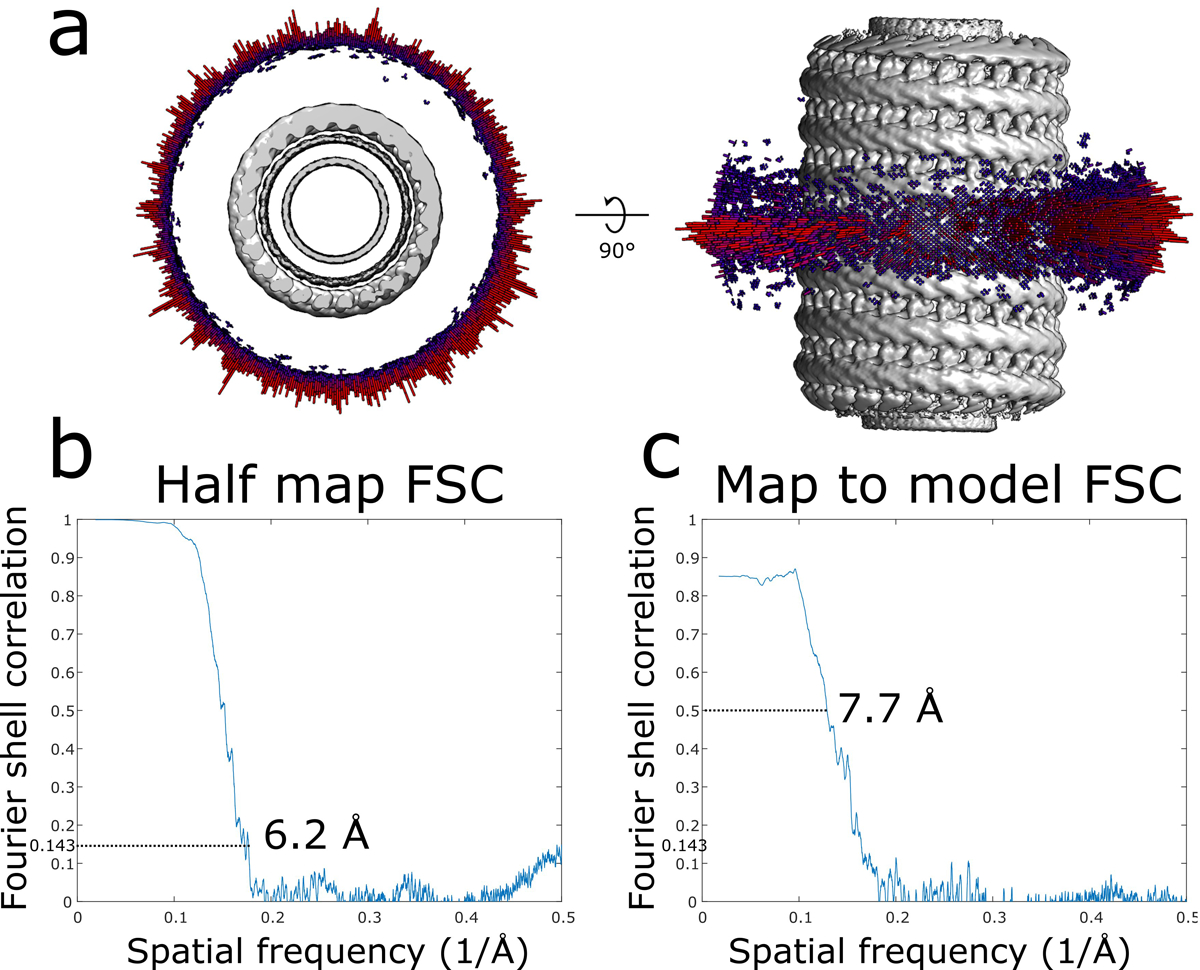
a, Angular distribution of the membrane-bound CHMP1B filament. b, Half map Fourier shell correlation (FSC) of the membrane-bound CHMP1B filament. c, Map to model FSC of the membrane-bound CHMP1B filament.
Extended Data Fig. 2. CryoEM reconstruction of the membrane-bound CHMP1B+IST1 filament at higher curvature and comparison of left- and right-handed CHMP1B+IST1 filaments.
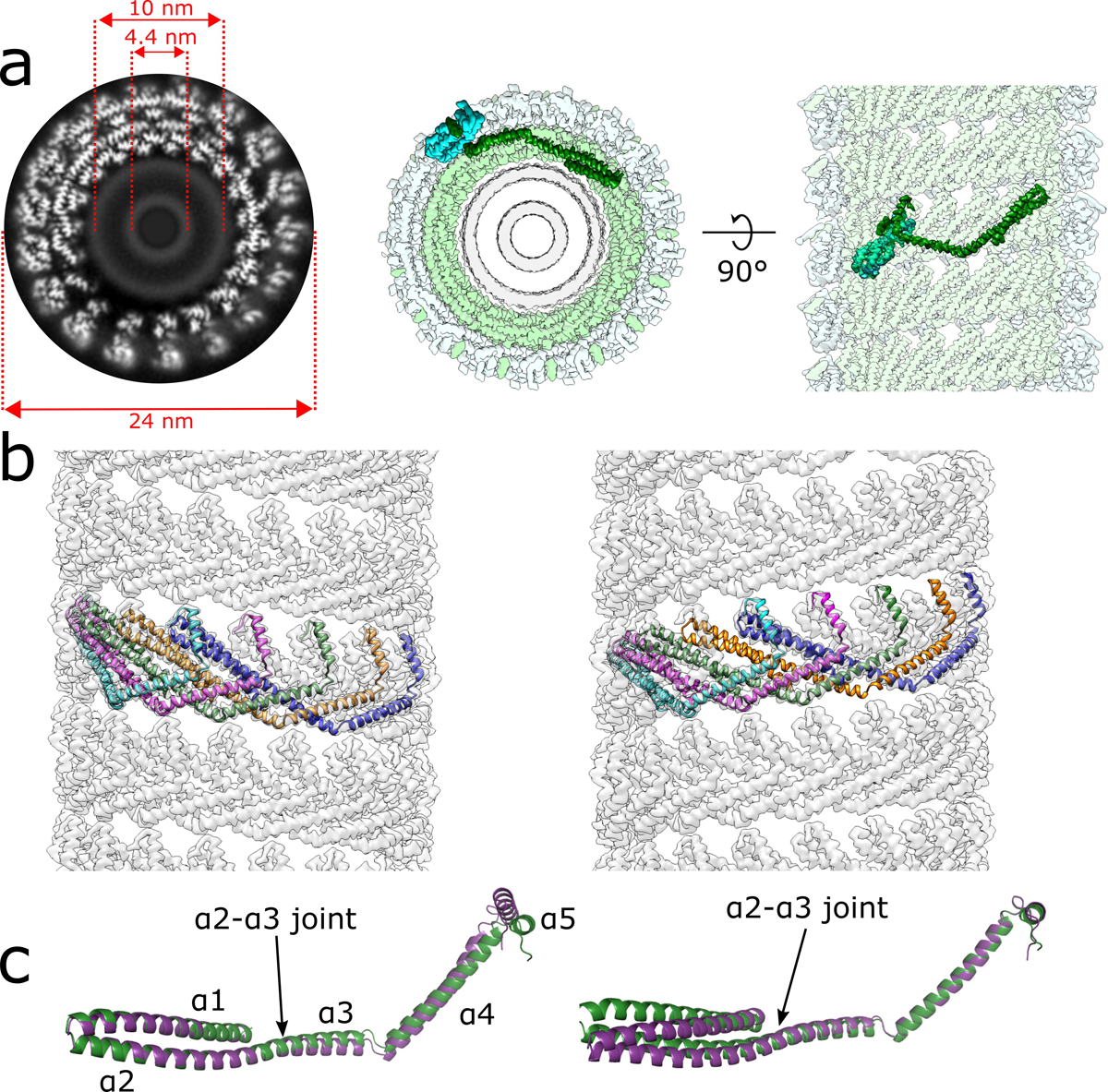
a, CryoEM 3D reconstruction of the membrane-bound left-handed CHMP1B+IST1 filament. End-on view down the helical axis in grey-scale (left) or colored (middle). Right, internal view looking outward from the membrane surface along the helical axis. IST1 protomers (cyan) bind to the exterior of CHMP1B (green), leading to constriction of the membrane (grey). IST1 and CHMP1B promoters are highlighted in dark cyan and green, respectively. Diameters of the entire tube and membrane leaflet peak-to-peak distances are annotated. b, Electron density maps of CHMP1B from the left-handed (left) or right-handed (right) membrane-bound CHMP1B+IST1 filaments. Five copies of CHMP1B are shown as ribbons. c, Superposition of a CHMP1B protomer from the left-handed (purple) and right-handed (green) CHMP1B+IST1 filaments aligned to the CHMP1B N-terminal α1-α2 helices (left) or C-terminal α4-α5 helices (right).
Extended Data Fig. 3. Local resolution estimates and cryoEM validation of membrane-bound CHMP1B+IST1 filaments.
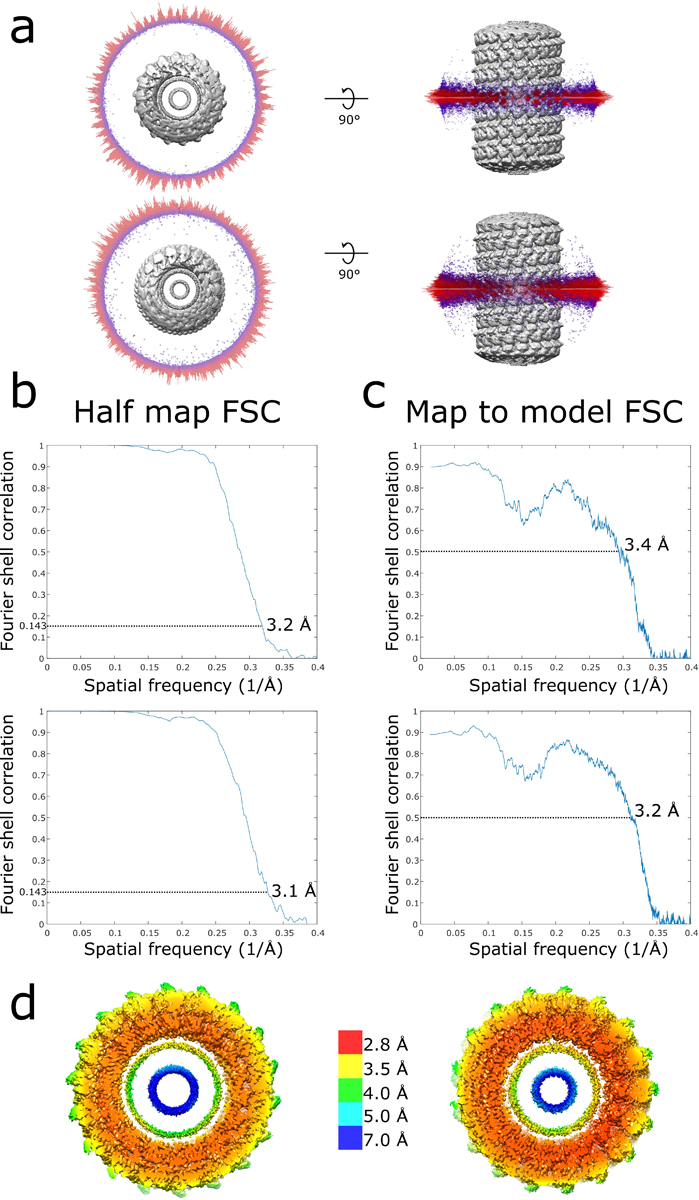
a, Angular distribution of right-handed (top) and left-handed (bottom) membrane-bound CHMP1B+IST1 filaments. b, Half map FSCs of right-handed (top) and left-handed (bottom) CHMP1B+IST1 filaments. c, Map to model FSCs right-handed (top) and left-handed (bottom) CHMP1B+IST1 filaments. d, Local resolution estimates of right-handed (left) and left-handed (right) CHMP1B+IST1 filaments.
Extended Data Fig. 4. IST1 does not discriminate between left- or right-handed CHMP1B filaments.
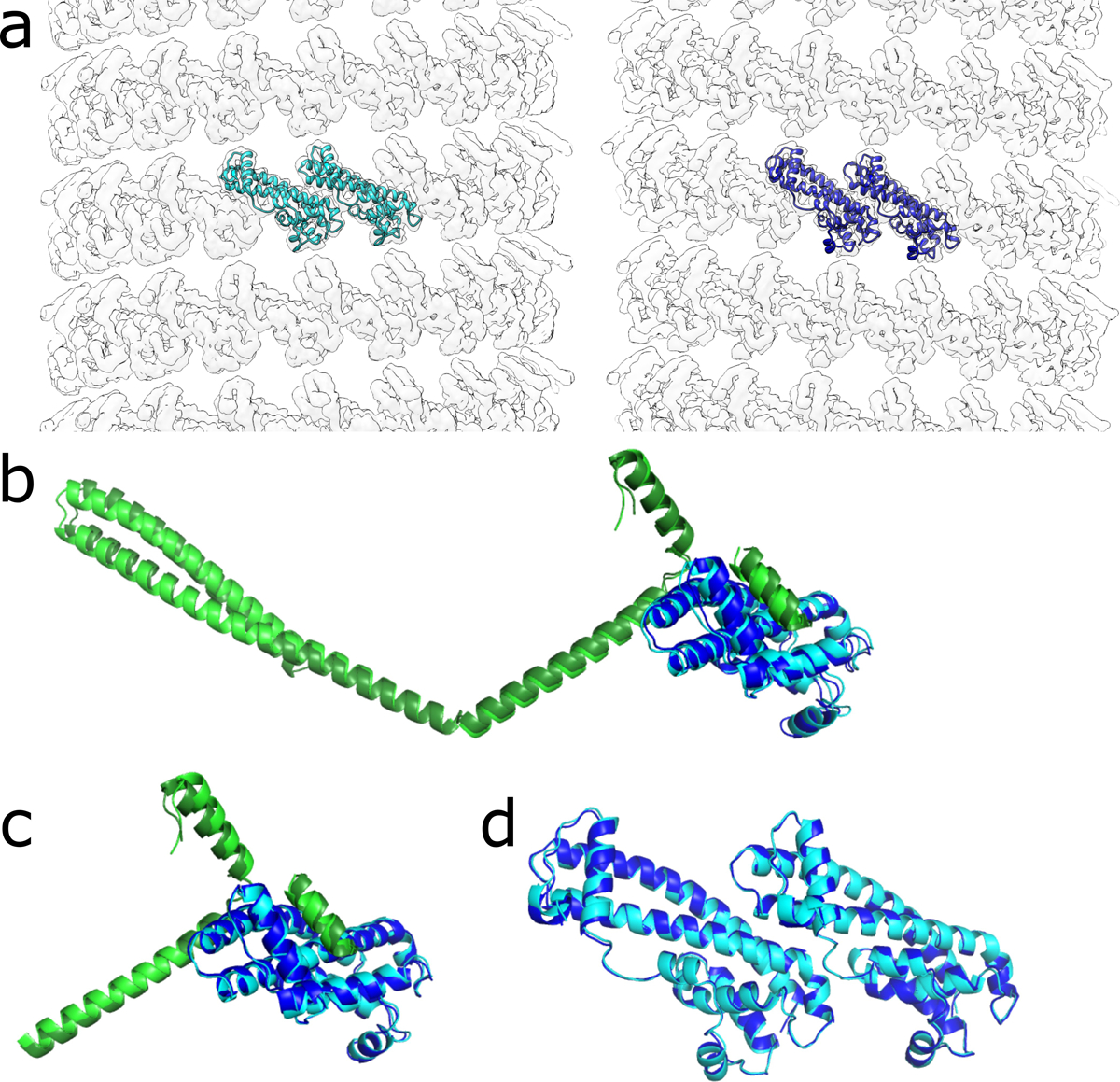
a, Electron density maps of the IST1 strands from the right-handed (left) or left-handed (right) membrane-bound CHMP1B+IST1 filaments. Two subunits of IST1 are shown as ribbons. b, Superposition of CHMP1B and IST1 protomers from the right-handed (dark green and dark blue, respectively) and left-handed (light green and cyan, respectively) CHMP1B+IST1 filaments. c, Superposition as in (b) but only with the C-terminal region of CHMP1B and IST1. d, Superposition of two subunits of IST1 from the right- or left-handed copolymers from (a).
Extended Data Fig. 5. Real-time monitoring of CHMP1B and IST1 membrane constriction and elongation.
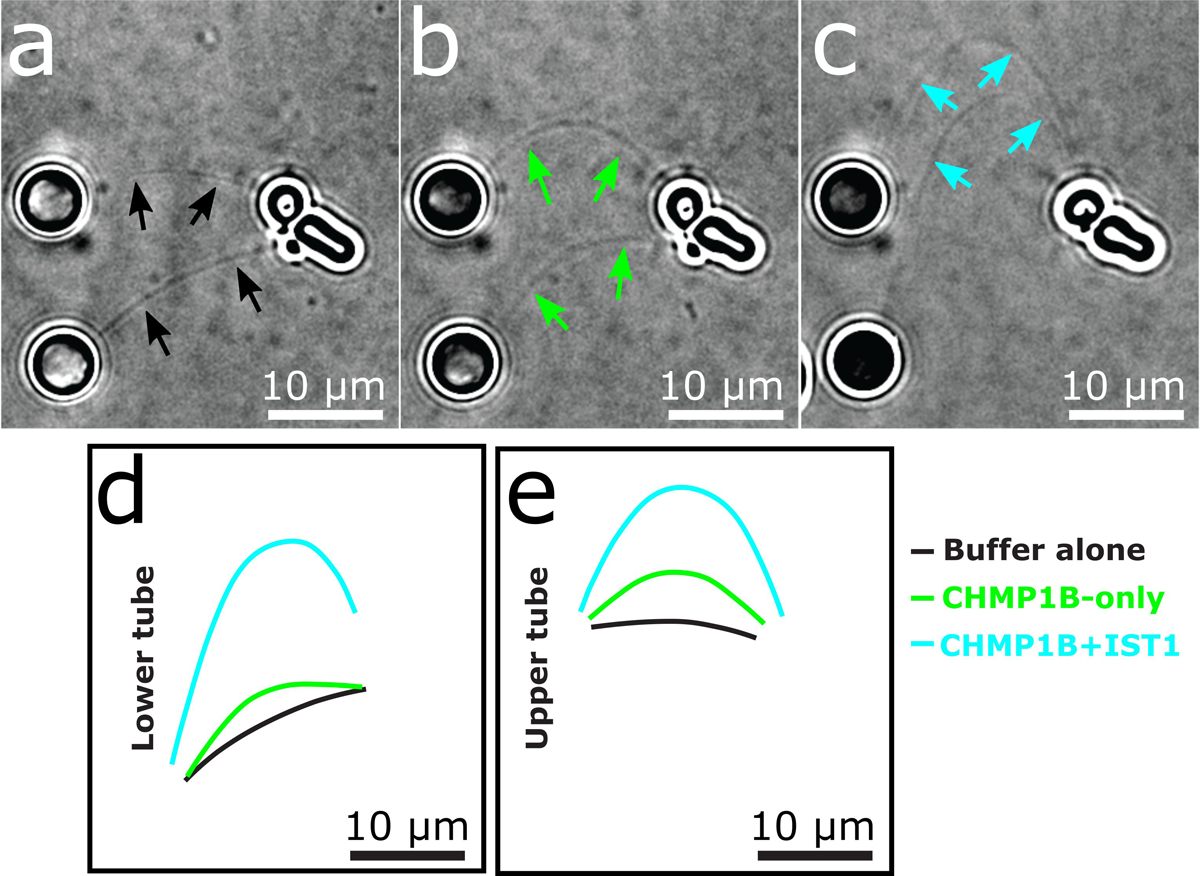
a-c, Still images representing deformation of two membrane tubes due to transverse flow of (a) buffer alone, (b) then 0.5 μM CHMP1B, (c) and a final addition of 0.5 μM IST1. Solid arrows in (a-c) highlight tubule locations. d-e, Contours of lower (d) and upper (e) membrane tubes extracted from panels (a-c) showing the extension of the tubes upon addition of CHMP1B and IST1.
Extended Data Fig. 6. CryoEM validation of the IST1NTDR16E/K27E filament.
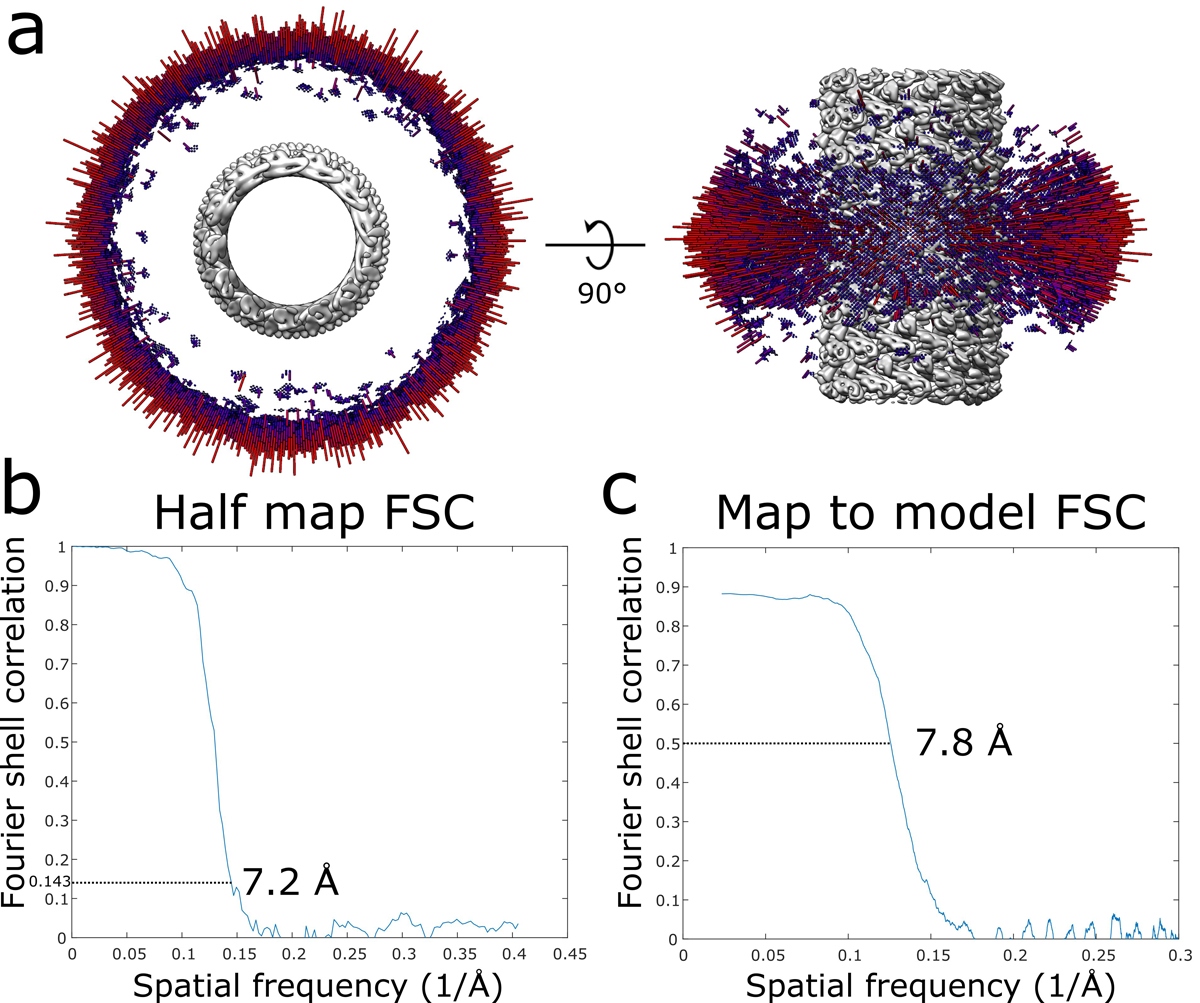
a, Angular distribution of the IST1NTDR16E/K27E filament. b, Half map FSC of the IST1NTDR16E/K27E filament. c, Map to model FSC of the IST1NTDR16E/K27E filament.
Extended Data Fig. 7. Steric clashing between the CHMP1B MIM and inter-turn IST1 subunits would prevent IST1 from achieving its preferred curvature in the copolymer.
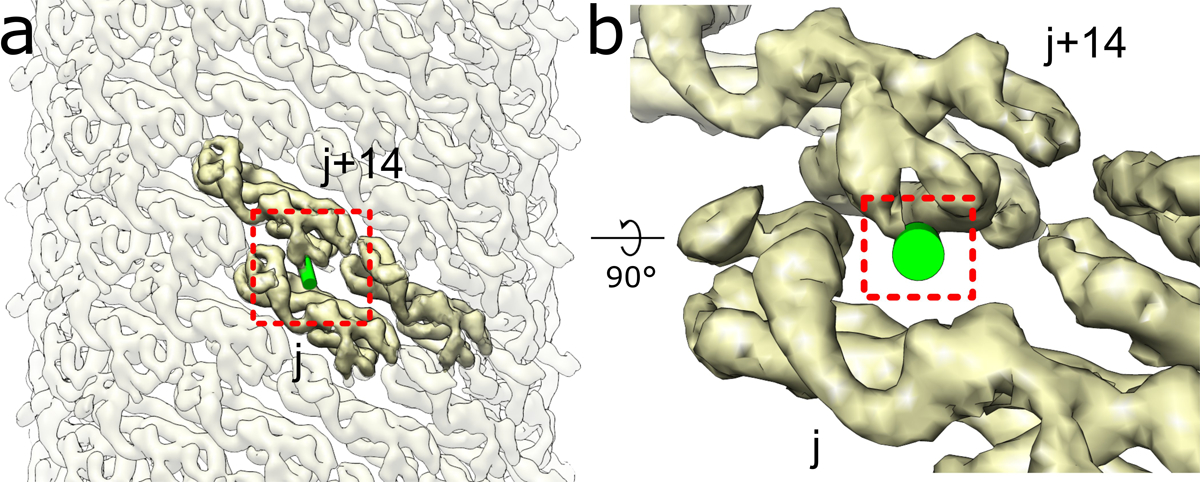
a, External view of the IST1NTDR16E/K27E filament with one CHMP1B MIM (shown as a green cylinder) docked onto the IST1NTDR16E/K27E j subunit. b, Zoomed in view of boxed area in (a) highlighting how the CHMP1B MIM clashes with the IST1 j+14 subunit.
Extended Data Fig. 8. Subtle deformations in the outer leaflet observed in the moderately constricted CHMP1B-only filaments.
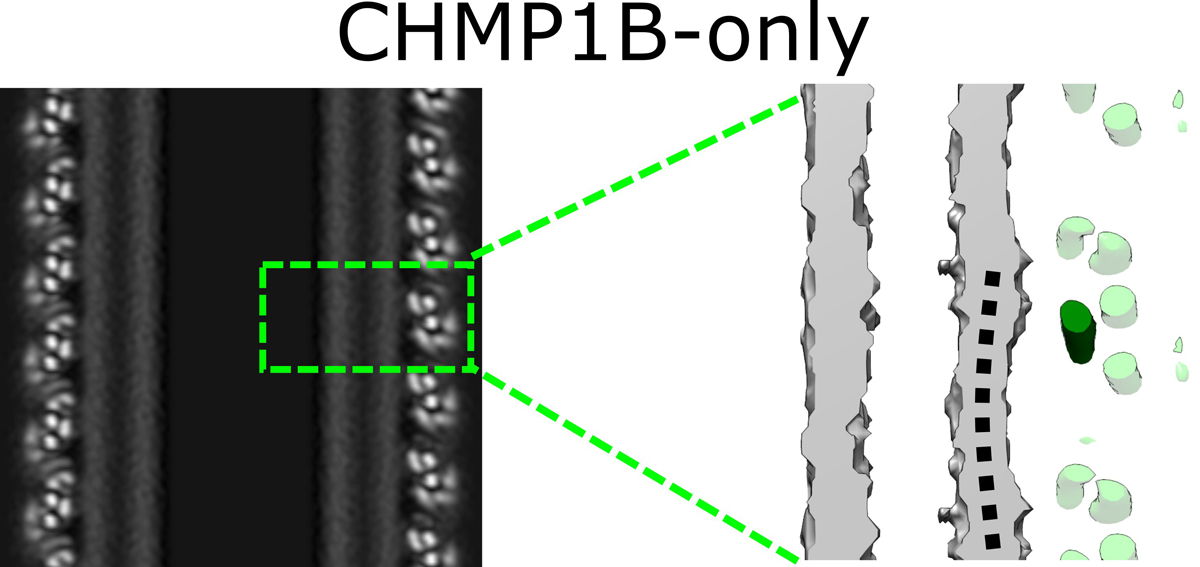
Left, central slice along the helical axis of the membrane-bound CHMP1B-only tubule. Right, zoomed view of boxed area in left showing very little dimpling in the outer leaflet (black dashed curved line) of the bilayer. A CHMP1B helix α1, which sits against the membrane, is highlighted in dark green.
Extended Data Fig. 9. SAXS analysis of liposomes and calculation of bilayer thickness.
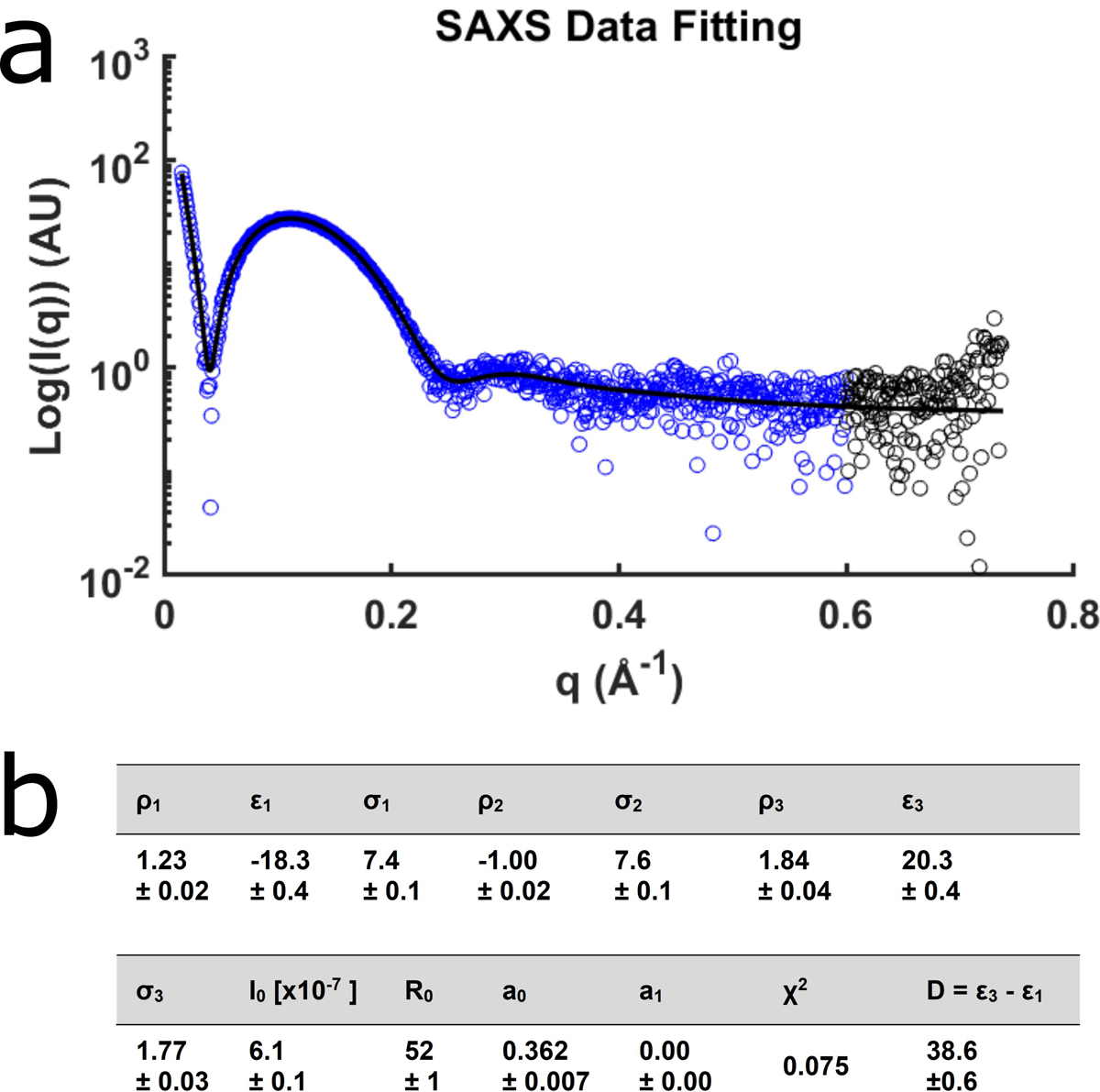
a, The small angle scattering intensities for protein-free unilamellar vesicles used in this study. The black line represents the fit to the model. The blue data points were used for fitting. b, Fit results for the liposomes and the resulting thickness, D (Å). The bilayer center, ε2, was fixed at 0, and the magnitude of the central peak, ρ2, was fixed at −1. Data are mean ± s.d.
Supplementary Material
Supplementary Video 1. Elbow flexing of one CHMP1B subunit.
Supplementary Video 2. Flexing of a full turn of CHMP1B subunits from low to high constriction.
Supplementary Video 4. IST1 alone does not promote membrane tube elongation.
Supplementary Video 5. Swinging of two IST1 subunits from initial binding to the CHMP1B filament to the constricted state (side view).
Supplementary Video 6. Swinging of two IST1 subunits from initial binding to the CHMP1B filament to the constricted state (top-down view). CHMP1B and the membrane would lie at the top of the animation.
Supplementary Video 3. Real time recording of membrane tube elongation by CHMP1B and IST1.
Supplementary Video 7. Constriction of a membrane tubule by CHMP1B and IST1.
Acknowledgements
We thank: members of the Frost lab for helpful discussion, especially P. Thomas and M. Sun with computational assistance; M. Grabe, M. Tucker, D. Argudo, E. Lyman, A. Sodt, G. Huber, and A. Roux for discussions on membrane biophysical behavior; T. Weiss for assistance with SAXS data collection; M. Braunfeld, D. Bulkley, M. Harrington, A. Myasnikov, and Z. Yu of the UCSF Center for Advanced CryoEM for microscopy support; and J. Baker-LePain and the QB3 shared cluster (NIH grant 1S10OD021596-01) for computational support. Structural biology applications used in this project were compiled and configured by SBGrid. The Titan × Pascal used for this research was donated by the NVIDIA Corporation. This work was supported by NIH grants P50 AI150464 and 1DP2GM110772-01 (to A.F.), R01 GM112080 and R37 AI51174 (to W.I.S.), A.F. is further supported by a Faculty Scholar grant from the HHMI and is a Chan Zuckerberg Biohub investigator.
Footnotes
Data Availability
CryoEM maps and models were deposited to the PDB and EMDB with the following codes: membrane-bound CHMP1B-only filament (PDB ID: 6TZ9, EMD-20590), membrane-bound right handed CHMP1B+IST1 filament (PDB ID: 6TZ4, EMD-20588), membrane-bound left handed CHMP1B+IST1 filament (PDB ID: 6TZ5, EMD-20589), IST1NTDR16E/K27E filament (PDB ID: 6TZA, EMD-20591).
Competing Interests
The authors declare no competing interests.
References
- 1.McCullough J, Colf LA & Sundquist WI Membrane fission reactions of the mammalian ESCRT pathway. Annu Rev Biochem 82, 663–92 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sundquist WI & Krausslich HG HIV-1 assembly, budding, and maturation. Cold Spring Harb Perspect Med 2, a006924 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Olmos Y, Hodgson L, Mantell J, Verkade P & Carlton JG ESCRT-III controls nuclear envelope reformation. Nature 522, 236–9 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vietri M et al. Spastin and ESCRT-III coordinate mitotic spindle disassembly and nuclear envelope sealing. Nature 522, 231–5 (2015). [DOI] [PubMed] [Google Scholar]
- 5.Raab M et al. ESCRT III repairs nuclear envelope ruptures during cell migration to limit DNA damage and cell death. Science 352, 359–62 (2016). [DOI] [PubMed] [Google Scholar]
- 6.Denais CM et al. Nuclear envelope rupture and repair during cancer cell migration. Science 352, 353–8 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Olmos Y, Perdrix-Rosell A & Carlton JG Membrane Binding by CHMP7 Coordinates ESCRT-III-Dependent Nuclear Envelope Reformation. Curr Biol (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Skowyra ML, Schlesinger PH, Naismith TV & Hanson PI Triggered recruitment of ESCRT machinery promotes endolysosomal repair. Science 360(2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chang CL et al. Spastin tethers lipid droplets to peroxisomes and directs fatty acid trafficking through ESCRT-III. J Cell Biol (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mast FD et al. ESCRT-III is required for scissioning new peroxisomes from the endoplasmic reticulum. J Cell Biol 217, 2087–2102 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McCullough J, Frost A & Sundquist WI Structures, Functions, and Dynamics of ESCRT-III/Vps4 Membrane Remodeling and Fission Complexes. Annu Rev Cell Dev Biol 34, 85–109 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Allison R et al. Defects in ER-endosome contacts impact lysosome function in hereditary spastic paraplegia. J Cell Biol 216, 1337–1355 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McCullough J et al. Structure and membrane remodeling activity of ESCRT-III helical polymers. Science 350, 1548–51 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Allison R et al. An ESCRT-spastin interaction promotes fission of recycling tubules from the endosome. J Cell Biol 202, 527–43 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bajorek M et al. Structural basis for ESCRT-III protein autoinhibition. Nat Struct Mol Biol 16, 754–62 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Muziol T et al. Structural basis for budding by the ESCRT-III factor CHMP3. Dev Cell 10, 821–30 (2006). [DOI] [PubMed] [Google Scholar]
- 17.Xiao J et al. Structural basis of Ist1 function and Ist1-Did2 interaction in the multivesicular body pathway and cytokinesis. Mol Biol Cell 20, 3514–24 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hanson PI, Roth R, Lin Y & Heuser JE Plasma membrane deformation by circular arrays of ESCRT-III protein filaments. J Cell Biol 180, 389–402 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lata S et al. Structural basis for autoinhibition of ESCRT-III CHMP3. J Mol Biol 378, 818–27 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lin Y, Kimpler LA, Naismith TV, Lauer JM & Hanson PI Interaction of the mammalian endosomal sorting complex required for transport (ESCRT) III protein hSnf7–1 with itself, membranes, and the AAA+ ATPase SKD1. J Biol Chem 280, 12799–809 (2005). [DOI] [PubMed] [Google Scholar]
- 21.Tang S et al. Structural basis for activation, assembly and membrane binding of ESCRT-III Snf7 filaments. Elife 4(2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McMillan BJ et al. Electrostatic Interactions between Elongated Monomers Drive Filamentation of Drosophila Shrub, a Metazoan ESCRT-III Protein. Cell Rep 16, 1211–7 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee IH, Kai H, Carlson LA, Groves JT & Hurley JH Negative membrane curvature catalyzes nucleation of endosomal sorting complex required for transport (ESCRT)-III assembly. Proc Natl Acad Sci U S A 112, 15892–7 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Crespo-Yanez X et al. CHMP1B is a target of USP8/UBPY regulated by ubiquitin during endocytosis. PLoS Genet 14, e1007456 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stoorvogel W, Oorschot V & Geuze HJ A novel class of clathrin-coated vesicles budding from endosomes. J Cell Biol 132, 21–33 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Connell JW et al. ESCRT-III-associated proteins and spastin inhibit protrudin-dependent polarised membrane traffic. Cell Mol Life Sci (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Manni MM et al. Acyl chain asymmetry and polyunsaturation of brain phospholipids facilitate membrane vesiculation without leakage. Elife 7(2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pinot M et al. Lipid cell biology. Polyunsaturated phospholipids facilitate membrane deformation and fission by endocytic proteins. Science 345, 693–7 (2014). [DOI] [PubMed] [Google Scholar]
- 29.Cashikar AG et al. Structure of cellular ESCRT-III spirals and their relationship to HIV budding. Elife 3(2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang F et al. A structural model of flagellar filament switching across multiple bacterial species. Nat Commun 8, 960 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Talledge N et al. The ESCRT-III proteins IST1 and CHMP1B assemble around nucleic acids. bioRxiv (2018). [Google Scholar]
- 32.Schoneberg J, Lee IH, Iwasa JH & Hurley JH Reverse-topology membrane scission by the ESCRT proteins. Nat Rev Mol Cell Biol (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chiaruttini N et al. Relaxation of Loaded ESCRT-III Spiral Springs Drives Membrane Deformation. Cell 163, 866–79 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mierzwa BE et al. Dynamic subunit turnover in ESCRT-III assemblies is regulated by Vps4 to mediate membrane remodelling during cytokinesis. Nat Cell Biol 19, 787–798 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Goliand I et al. Resolving ESCRT-III Spirals at the Intercellular Bridge of Dividing Cells Using 3D STORM. Cell Rep 24, 1756–1764 (2018). [DOI] [PubMed] [Google Scholar]
- 36.Guizetti J et al. Cortical constriction during abscission involves helices of ESCRT-III-dependent filaments. Science 331, 1616–20 (2011). [DOI] [PubMed] [Google Scholar]
- 37.Banjade S, Tang S, Shah YH & Emr SD Electrostatic lateral interactions drive ESCRT-III heteropolymer assembly. Elife 8(2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pfitzner A-K, Mercier V & Roux A Vps4 triggers sequential subunit exchange in ESCRT-III polymers that drives membrane constriction and fission. bioRxiv (2019). [Google Scholar]
- 39.Rajamoorthi K, Petrache HI, McIntosh TJ & Brown MF Packing and viscoelasticity of polyunsaturated omega-3 and omega-6 lipid bilayers as seen by (2)H NMR and X-ray diffraction. J Am Chem Soc 127, 1576–88 (2005). [DOI] [PubMed] [Google Scholar]
- 40.Lipowsky R Coupling of bending and stretching deformations in vesicle membranes. Adv Colloid Interface Sci 208, 14–24 (2014). [DOI] [PubMed] [Google Scholar]
- 41.Rozycki B & Lipowsky R Spontaneous curvature of bilayer membranes from molecular simulations: asymmetric lipid densities and asymmetric adsorption. J Chem Phys 142, 054101 (2015). [DOI] [PubMed] [Google Scholar]
- 42.Allain JM, Storm C, Roux A, Ben Amar M & Joanny JF Fission of a multiphase membrane tube. Phys Rev Lett 93, 158104 (2004). [DOI] [PubMed] [Google Scholar]
- 43.Liu J, Kaksonen M, Drubin DG & Oster G Endocytic vesicle scission by lipid phase boundary forces. Proc Natl Acad Sci U S A 103, 10277–10282 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chiaruttini N & Roux A Dynamic and elastic shape transitions in curved ESCRT-III filaments. Curr Opin Cell Biol 47, 126–135 (2017). [DOI] [PubMed] [Google Scholar]
- 45.Lenz M, Morlot S & Roux A Mechanical requirements for membrane fission: common facts from various examples. FEBS Lett 583, 3839–46 (2009). [DOI] [PubMed] [Google Scholar]
- 46.Fabrikant G et al. Computational model of membrane fission catalyzed by ESCRT-III. PLoS Comput Biol 5, e1000575 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Maity S et al. VPS4 triggers constriction and cleavage of ESCRT-III helical filaments. Sci Adv 5, eaau7198 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Henne WM, Buchkovich NJ & Emr SD The ESCRT pathway. Dev Cell 21, 77–91 (2011). [DOI] [PubMed] [Google Scholar]
- 49.Peel S, Macheboeuf P, Martinelli N & Weissenhorn W Divergent pathways lead to ESCRT-III-catalyzed membrane fission. Trends Biochem Sci 36, 199–210 (2011). [DOI] [PubMed] [Google Scholar]
- 50.von Filseck JM et al. Anisotropic ESCRT-III architecture governs helical membrane tube formation. bioRxiv (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Harker-Kirschneck L, Baum B & Saric AE Changes in ESCRT-III filament geometry drive membrane remodelling and fission in silico. BMC Biol 17, 82 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schoneberg J et al. ATP-dependent force generation and membrane scission by ESCRT-III and Vps4. Science 362, 1423–1428 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Samson RY, Obita T, Freund SM, Williams RL & Bell SD A role for the ESCRT system in cell division in archaea. Science 322, 1710–3 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Stuchell-Brereton MD et al. ESCRT-III recognition by VPS4 ATPases. Nature 449, 740–4 (2007). [DOI] [PubMed] [Google Scholar]
- 55.Yang D et al. Structural basis for midbody targeting of spastin by the ESCRT-III protein CHMP1B. Nat Struct Mol Biol 15, 1278–86 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chernomordik LV & Kozlov MM Mechanics of membrane fusion. Nat Struct Mol Biol 15, 675–83 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lai AL, Park H, White JM & Tamm LK Fusion peptide of influenza hemagglutinin requires a fixed angle boomerang structure for activity. J Biol Chem 281, 5760–70 (2006). [DOI] [PubMed] [Google Scholar]
- 58.Bertin A et al. Human ESCRT-III Polymers Assemble on Positively Curved Membranes and Induce Helical Membrane Tube Formation. bioRxiv (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Agromayor M et al. Essential role of hIST1 in cytokinesis. Mol Biol Cell 20, 1374–87 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bajorek M et al. Biochemical analyses of human IST1 and its function in cytokinesis. Mol Biol Cell 20, 1360–73 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Andersen KR, Leksa NC & Schwartz TU Optimized E. coli expression strain LOBSTR eliminates common contaminants from His-tag purification. Proteins 81, 1857–61 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Studier FW Protein production by auto-induction in high density shaking cultures. Protein Expr Purif 41, 207–34 (2005). [DOI] [PubMed] [Google Scholar]
- 63.Morin A et al. Collaboration gets the most out of software. Elife 2, e01456 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zheng SQ et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat Methods 14, 331–332 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhang K Gctf: Real-time CTF determination and correction. J Struct Biol 193, 1–12 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.He S & Scheres SHW Helical reconstruction in RELION. J Struct Biol 198, 163–176 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Desfosses A, Ciuffa R, Gutsche I & Sachse C SPRING - an image processing package for single-particle based helical reconstruction from electron cryomicrographs. J Struct Biol 185, 15–26 (2014). [DOI] [PubMed] [Google Scholar]
- 68.Egelman EH A robust algorithm for the reconstruction of helical filaments using single-particle methods. Ultramicroscopy 85, 225–234 (2000). [DOI] [PubMed] [Google Scholar]
- 69.Frank J et al. SPIDER and WEB: processing and visualization of images in 3D electron microscopy and related fields. J Struct Biol 116, 190–9 (1996). [DOI] [PubMed] [Google Scholar]
- 70.Pettersen EF et al. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem 25, 1605–12 (2004). [DOI] [PubMed] [Google Scholar]
- 71.Emsley P, Lohkamp B, Scott WG & Cowtan K Features and development of Coot. Acta Crystallogr D Biol Crystallogr 66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Afonine PV et al. Real-space refinement in PHENIX for cryo-EM and crystallography. Acta Crystallogr D Struct Biol 74, 531–544 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Williams CJ et al. MolProbity: More and better reference data for improved all-atom structure validation. Protein Sci 27, 293–315 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Afonine PV et al. New tools for the analysis and validation of cryo-EM maps and atomic models. Acta Crystallogr D Struct Biol 74, 814–840 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Bergman J, Osunbayo O & Vershinin M Constructing 3D microtubule networks using holographic optical trapping. Sci Rep 5, 18085 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Romanov V, McCullough J, Gale BK & Frost A A Tunable Microfluidic Device Enables Cargo Encapsulation by Cell- or Organelle-Sized Lipid Vesicles Comprising Asymmetric Lipid Bilayers. Advanced Biosystems 3, 1900010 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Smolsky IL et al. Biological small-angle x-ray scattering facility at the Stanford synchrotron radiation laboratory. Journal of Applied Crystallography 40, S453–S458 (2007). [Google Scholar]
- 78.Brzustowicz MR & Brunger AT X-ray scattering from unilamellar lipid vesicles. Journal of Applied Crystallography 38, 126–131 (2005). [Google Scholar]
- 79.Moss FR 3rd et al. Ladderane phospholipids form a densely packed membrane with normal hydrazine and anomalously low proton/hydroxide permeability. Proc Natl Acad Sci U S A 115, 9098–9103 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Schindelin J et al. Fiji: an open-source platform for biological-image analysis. Nat Methods 9, 676–82 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Video 1. Elbow flexing of one CHMP1B subunit.
Supplementary Video 2. Flexing of a full turn of CHMP1B subunits from low to high constriction.
Supplementary Video 4. IST1 alone does not promote membrane tube elongation.
Supplementary Video 5. Swinging of two IST1 subunits from initial binding to the CHMP1B filament to the constricted state (side view).
Supplementary Video 6. Swinging of two IST1 subunits from initial binding to the CHMP1B filament to the constricted state (top-down view). CHMP1B and the membrane would lie at the top of the animation.
Supplementary Video 3. Real time recording of membrane tube elongation by CHMP1B and IST1.
Supplementary Video 7. Constriction of a membrane tubule by CHMP1B and IST1.


