Abstract
Cell membrane modification is important for tissue engineering, cell-based therapies, and cell biology studies. Recently, oligonucleotides have attracted considerable attention to remodel and functionalize live cell membranes. In particular, a type of amphiphilic lipid-oligonucleotide conjugates have been rationally designed and synthesized for this purpose. These conjugates have enabled a rapid, straightforward and efficient cell membrane modification. Taking advantage of the highly precise and programmable self-assembly of DNAs and RNAs, lipid-oligonucleotide conjugates have been used for membrane bioanalysis, therapeutics, building artificial membrane structures, and regulating cell–surface and cell–cell interactions. In this review, we have summarized the current knowledge in the design, synthesis, and regulating membrane properties of lipid-oligonucleotide conjugates. In addition, their state-of-the-art applications in cell membrane engineering and bioanalysis have been illustrated.
Keywords: Lipid-oligonucleotide conjugate, DNA, cell membrane modification, membrane analysis, cell regulation
Introduction
Composed of a protein-and-carbohydrate-embedded lipid bilayer, cell membranes serve as borders for biological organization and protect the cell from its surroundings. The membrane structures also play important roles in cellular communication and signaling. There has been an ever-increasing interest in synthetically modifying cell membranes. Such engineered membranes can be broadly useful from studying cell biology to tissue engineering and cell-based therapies [1–3].
DNA oligonucleotides have emerged as the material of choice to functionalize and remodel cell membranes [4,5]. Cell membrane-anchored DNA oligonucleotides have been used for membrane bioanalysis and regulation, drug deliveries, and controlling cell–surface and cell–cell interactions. The most attractive feature of these oligonucleotides is their highly precise and tunable self-assembly. Various structures and nanodevices can be rationally designed with molecular-level precision and programmable performance [6,7].
Several strategies have been developed for modifying DNAs onto cell membranes, including in situ chemical interaction with membrane functional groups [8–10] and non-covalent anchoring through antibodies, aptamers, or hydrophobic moieties [4,5,11,12]. Among these, lipid-oligonucleotide conjugates have attracted particular attention [4,5,7]. By chemically conjugating a hydrophobic lipid moiety with hydrophilic oligonucleotide, these amphiphilic conjugates enable fast, easy, and highly efficient DNA modification on live cell membranes. In this contribution, we will discuss the design, synthesis, and membrane properties of lipid-oligonucleotide conjugates, as well as their recent applications for cell membrane analysis and engineering.
Design of lipid-oligonucleotide conjugates
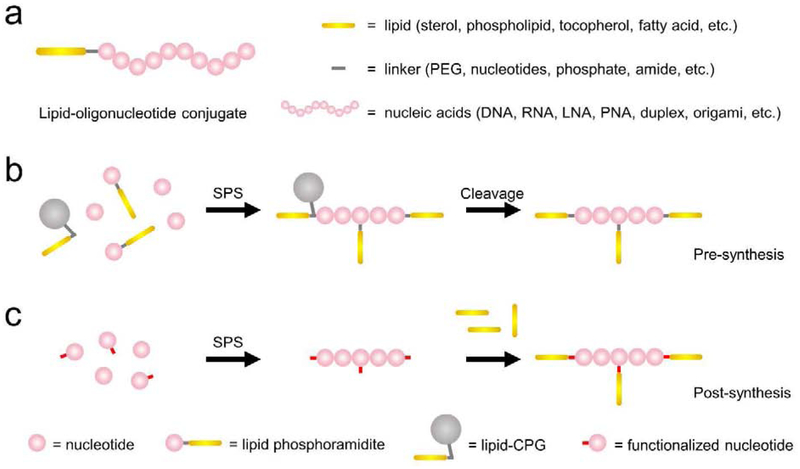
Lipid-oligonucleotide conjugate typically comprises three moieties: lipid, linker, and nucleic acid (Figure 1a). To construct cell membrane nanodevices, DNA is the most commonly used nucleic acid moiety. DNA can be cost-effectively synthesized and easily conjugated with lipids and other functional groups, such as fluorophores, drugs, and cross-linkers [13]. Modified oligonucleotides including PNA, LNA, 2’-F, and 2’-OMe-modified oligomers and siRNA analogs, have been conjugated with lipids for membrane modification and biomedical deliveries [14]. These modified nucleic acids can improve the cellular and in vivo biostability of lipid-oligonucleotide conjugates. Instead of the direct linkage, synthetically challenging conjugates, such as those containing miRNA, siRNA, and mRNA, can also be indirectly prepared based on sequence-specific hybridization [15–17]. The precise self-assembly of nucleic acids have allowed the conjugation of lipids with various nanostructures as well, ranging from duplexes, three-way junctions to DNA/RNA origamis [7,14].
Figure 1.
Design and synthesis of lipid-oligonucleotide conjugates. (a) The conjugate typically comprises three moieties: lipid, linker, and nucleic acid. (b) Schematic of the pre-synthetic approach for the synthesis of lipid-oligonucleotide conjugates. Lipid phosphoramidite or lipid-conjugated controlled-pore glass (CPG) was conjugated together with other nucleotides during the solid-phase synthesis (SPS) of oligonucleotides. After cleavage from CPG, lipids were precisely modified at different positions of oligonucleotides. (c) Schematic of the post-synthetic approach for the synthesis of lipid-oligonucleotide conjugates. After solid-phase synthesis (SPS) of oligonucleotides containing some functional groups, lipids were then chemically conjugated through reaction with these functional groups.
A linker is often added between lipid and oligonucleotide. Polyethylene glycol (PEG) is a popular linker of moderate hydrophobicity and flexibility. DNA oligonucleotides and the head group of lipids can also function as the linker. Some linkers are introduced for easy synthesis, but they can be also useful in adjusting the hydrophilic/hydrophobic ratio of amphiphiles, as well as the morphology and size of aggregations [18]. Linkers may also function as spacers to extend membrane-anchored oligonucleotides, and as a result, to reduce electrostatic interactions between oligonucleotides and the cell surface. These spacers can also increase the membrane availabilities of oligonucleotides for the target binding [19].
Sterols, especially cholesterol, are the most popular lipid moieties due to their commercial availability and high membrane modification efficiency [20–24]. Other natural membrane lipids, such as phospholipids, tocopherols, and lithocholic acids, have also been demonstrated as good candidates [16,24,25]. In addition, natural lipid derivatives, especially synthetic fatty acids of different lengths, numbers of chains, and degrees of saturation, are also useful lipid moieties. These lipids can exhibit tunable hydrophobicity and membrane interaction patterns [4,26,27]. In general, considering their low cell toxicity and membrane disruption, natural lipid moieties are more favorable than synthetic lipids for cell membrane engineering. While the practical choice of lipids largely depends on their synthetic efficiency, as well as physicochemical and membrane properties.
Synthesis of lipid-oligonucleotide conjugates
Lipids can be incorporated into oligonucleotide strands through either pre-synthetic or post-synthetic strategies (Figure 1). In pre-synthetic approach, a lipid phosphoramidite (for internal or 5’-modification) or lipid-conjugated controlled-pore glass (CPG, for 3’-modification) is first prepared and used during solid-phase synthesis (SPS) of oligonucleotides. The position, purity, and number of lipid modifications can be precisely controlled. Various phospholipid, cholesterol, tocopherol, and fatty acid-modified oligonucleotides have been synthesized using this strategy [18,20]. Compared with post-synthesis, pre-synthesis exhibits high incorporation efficiency, high accuracy, and short linkage.
Many lipid moieties of limited solubility and stability may not be prepared as phosphoramidite or not tolerate the harsh DNA synthesis condition. Automated DNA synthesizer is also not available in most laboratories. In this case, post-synthesis by conjugating lipids with commercially available oligonucleotides can be a choice. Several functional groups, such as amine, thiol, alkyne, azide, and N-hydroxysuccinimide ester, can be readily synthesized in oligonucleotides. As a result, lipid-oligonucleotide conjugates can be prepared based on interactions such as thiol-maleimide reaction, click chemistry, or amide bond formation [7,13,26]. These post-synthetic approaches can be performed in either solution or solid-phase. In solution-phase synthesis, surfactants are often needed to homogenize the reaction mixture, and extra care is needed to purify the lipid-oligonucleotide conjugates. SPS provides extra benefit in removing excess reactants. The availability of both pre- and post-synthetic strategies have paved the way for potential applications of lipid-oligonucleotide conjugates.
Physicochemical and membrane properties of lipid-oligonucleotide conjugates
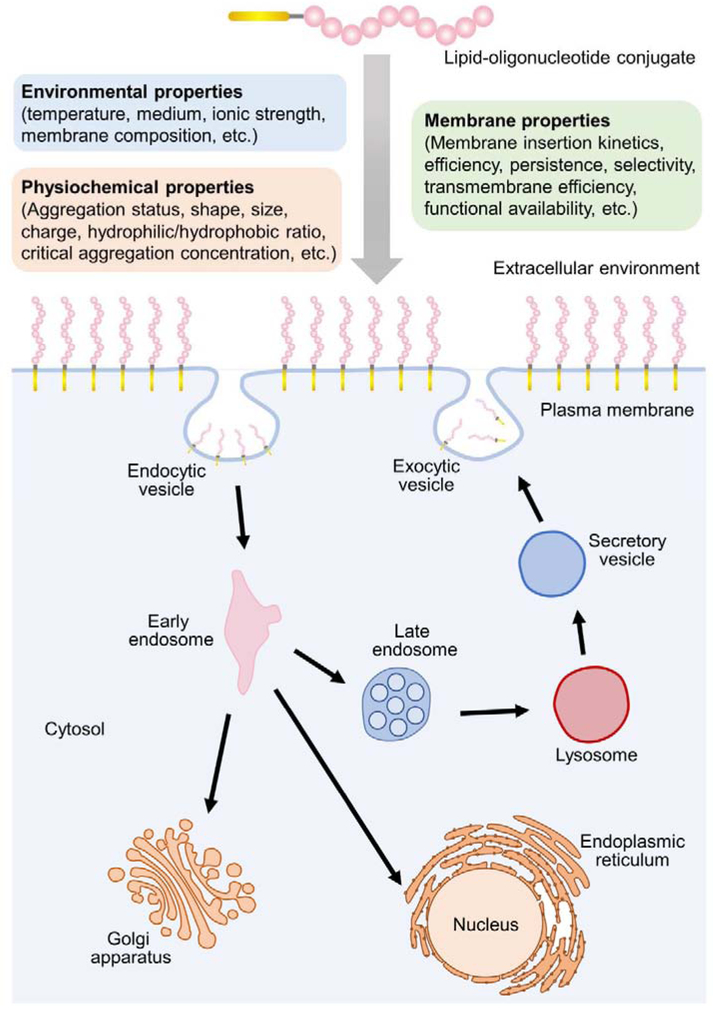
The chemical and biomedical behaviors of lipid-oligonucleotide conjugates are largely determined by their assembly status in solution and cell membranes (Figure 2). Various interactions exist in these amphiphilic conjugates including hydrogen bonding, hydrophobic and electrostatic interactions. In solution, lipid-oligonucleotide can be self-assembled into micelles, liposomes, bilayers, and other nano-/microstructures. One important parameter is the critical aggregation concentration, above which amphiphilic monomers stay in the aggregated form. Our recent results indicate that monomeric, rather than aggregated form, of lipid-oligonucleotide conjugates regulates the kinetics and efficiency of cell membrane modification [27]. Considering the competition between aggregates formation and the cell membrane insertion of lipid-oligonucleotides, the conjugate assembly pattern in solution will indeed also influence their cell membrane modifications.
Figure 2.
Physicochemical and membrane properties of lipid-oligonucleotide conjugates. Various environmental properties and physicochemical properties of the conjugates will influence their membrane properties. Shown are the dynamic process of lipid-DNA probe modification on the cell membrane and internalization into the cells through endocytosis pathway.
The cell membrane insertion kinetics, efficiency, persistence, and transmembrane efficiency of lipid-oligonucleotide conjugates are important factors determining their biomedical and analytical applications. In general, fast insertion (seconds to minutes) and long persistence (hours to days) is desirable for cell membrane engineering. While on the other hand, the efficient cell membrane anchoring may also reduce the transmembrane efficiency of lipid-oligonucleotide for potential biomedical applications. High-density cell membrane modification may facilitate applications in regulating cell–surface or cell–cell interactions. While low-efficiency membrane modification, with less influence on natural membrane functions, can be beneficial in membrane bioanalysis.
The hydrophobicity of lipid-oligonucleotide conjugates is critical for their rapid and efficient membrane insertion. Several studies have indicated that lipids with longer or more carbon chains, i.e., more hydrophobic, tend to modify cell membranes with higher efficiency [18,19,21,26]. Our results, however, indicated that there could be an optimal hydrophobicity, above which value more hydrophobic lipids will actually exhibited reduced membrane modification efficiency and kinetics [27]. Those highly hydrophobic conjugates can form tight aggregation in solution, which will reduce their chance in the monomeric form to enter cell membranes.
The net membrane modification efficiency can be viewed as the result of a dynamic equilibrium between membrane insertion, distribution, and release (Figure 2). In addition to the hydrophobicity, the size and shape of oligonucleotide, concentration of conjugates, temperature, and composition of cell membranes and culture medium will also influence the membrane modification of lipid-oligonucleotide conjugates [19,21,28]. It is still challenging to predict membrane modification efficiency of a lipid-oligonucleotide structure under a particular experimental condition.
Low membrane persistence is another challenge in applying lipid-oligonucleotide for long-term cell membrane engineering. Membrane-anchored oligonucleotides normally disappears within 1–4 h [19,23,24]. These lipid-oligonucleotides are subject to either endocytosis or releasing to solution [19,27,29]. Reducing the temperature appears to prolong the membrane retention of lipid-oligonucleotides [30]. Our recent study revealed that by adding an endocytosis inhibitor, filipin, membrane-modified oligonucleotides could exist for over 24 h [27]. Another strategy to improve membrane persistence is by forming large membrane network of these lipid-oligonucleotides [31,32]. For example, based on a hybridization chain reaction, in the presence of polyelectrolytes, lipid-oligonucleotide can be retained on cell membranes for over two weeks [31]. Still, more studies are needed to understand how to fine-tune both the membrane persistence and transmembrane efficiency of these conjugates.
Another important property of lipid-oligonucleotides is their ability to hybridize with strands of complementary sequences. Cis-interacting oligonucleotides (on the same cell membrane) can be useful in forming membrane nanostructures and functional nanodevices [4,5,33]. While trans-interaction of oligonucleotides on different membranes can facilitate their applications in the analysis and regulation of cell–surface or cell–cell interactions [34,35]. It is thus critical to ensure the availability and hybridization specificity of the membrane-anchored oligonucleotides.
More recently, the membrane selectivity of lipid-oligonucleotides begins to be studied. Selective membrane modification can be useful for the targeted therapeutic delivery, membrane barcoding and cell profiling. However, most previous studies indicated that lipid-oligonucleotide would non-selectively insert onto different cell membranes. Interestingly, a recent report indicated that cell-specific membrane proteins, such as alkaline phosphatase, could be used to in situ modify the hydrophobicity of lipid-oligonucleotide [28]. As a result, efficient membrane modification could only occur on target cells expressing these membrane proteins. Other similar approaches may show up later to further fine-tune the membrane selectivity of lipid-oligonucleotides.
Applications in cell membrane engineering and analysis
The high membrane insertion efficiency, generalizability towards various cell types, and noninvasive and simple procedure have made lipid-oligonucleotide a great tool for cell membrane regulation and analysis. Based on different structures, interactions, and switching properties of oligonucleotides, several interesting membrane applications have been recently achieved (Figure 3).
Figure 3.
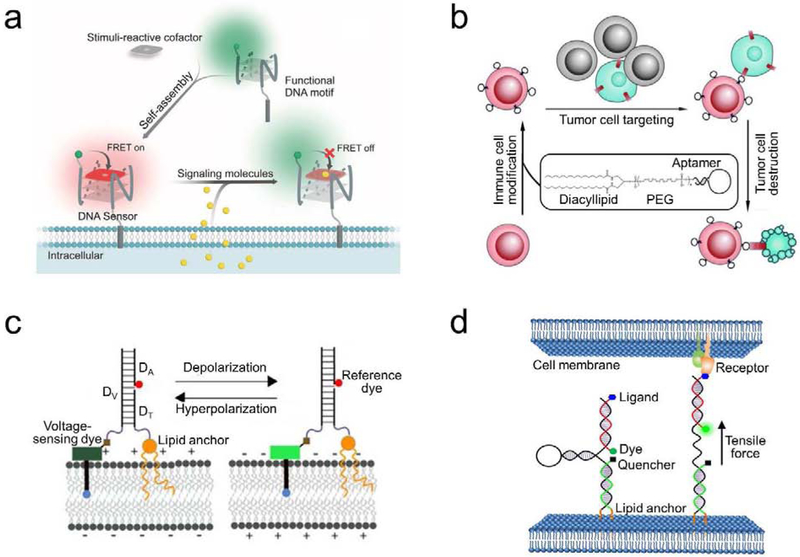
(a) Schematic of a membrane-anchored DNA G-quadruplex sensor for the real-time imaging of signaling molecules [22]. Target-induced changes in the fluorescence resonance energy transfer (FRET) efficiency between the DNA-modified fluorophore and a stimuli-reactive cofactor was used for the detection. (b) Schematic of a lipid-aptamer conjugate for the regulation of cell–cell interactions and cell-based therapy [35]. With the help of diacyllipid, cancer cell-targeting DNA aptamers were first modified onto the surface of immune cells. The anchored aptamers then facilitated the binding and killing of cancer cells by these membrane-engineered immune cells. (c) Schematic of a lipid-modified DNA voltmeter for the membrane potential detection [42]. The precise self-assembly of three DNA strands, DV, DA, and DT, provided a scaffold for the ratiometric measurement between a voltage-sensing fluorophore and a reference dye. The membrane potential could thus be accurately imaged and quantified. (d) Schematic of a DNA-based tension probe for visualizing intercellular tensile forces [23]. The design was based on the self-assembly of three oligonucleotides with a pair of cholesterol anchors at one end to insert onto cell membranes. Upon experiencing an intercellular tensile force exceeding the threshold force to unfold the DNA hairpin, the dye separated from the quencher and resulted in an increased fluorescence signal. Figures are adapted and modified with permission from references [22], [23], [35], and [42].
First, functional oligonucleotides have been modified onto live-cell membranes for bioanalysis. For example, using a diacyllipid-DNAzyme conjugate, Qiu et al., were able to monitor metal ion levels nearby the cell surfaces [36]. Similarly, a tocopherol-aptamer conjugate was used to real-time image the release of adenine in brain astrocytes [25]. More recently, cellular extrusion of SO2 and NO was detected by a membrane-anchored G-quadruplex structure, together with a stimuli-reactive cofactor (Figure 3a) [22]. For all these membrane-anchored oligonucleotide sensors, one key feature is that they specifically measure the local dynamics of targets in the membrane microenvironment. Such information can be potentially important in understanding cellular communication and signaling pathways.
These membrane-anchored oligonucleotides can also be used in therapeutics. For example, by anchoring cancer-targeting DNA aptamers onto immune cell surfaces, Xiong et al., developed a cell-based therapeutic approach to target and kill cancer cells (Figure 3b) [35]. As another interesting example, by conjugating therapeutic nucleic acids with lipid moieties, the anti-tumor efficacy can be dramatically improved [18,37]. These conjugates have enhanced stability during circulating and exhibited enriched local concentration at the tumor site. The cell membrane binding of lipid-oligonucleotide contributed to their enhanced therapeutic effects [18,38].
In addition, based on the sequence-specific DNA hybridization, different types of nanostructures can be built on the cell membranes. For example, membrane-anchored DNA nanopores have been used for controlled target transportation [39–41]. With precisely controlled shapes and sizes, these DNA nanostructures can potentially mimic and extend the function of natural membrane proteins. As another example, a lipid-modified DNA duplex was recently used to quantify cell membrane potentials (Figure 3c) [42]. Here, the stoichiometric DNA hybridization allows the ratiometric quantification with a voltage-sensitive dye and an internal reference.
The specific cell membrane DNA hybridization can also be used to regulate cell–surface and cell–cell interactions. In addition to their potential applications in tissue engineering, we have recently demonstrated a lipid-DNA hairpin probe to visualize and measure mechanical forces at cell–cell junctions (Figure 3d) [23]. Upon experiencing tensile forces generated by neighboring cells, unfolding of the DNA hairpin led to a dramatic increase in the fluorescence signal. In another recent study, lipid-DNA pyramidal probe was developed to control cell–cell interactions [43]. Interestingly, by incorporation three cholesterols, the membrane-anchoring efficiency of these DNA pyramids can be dramatically improved.
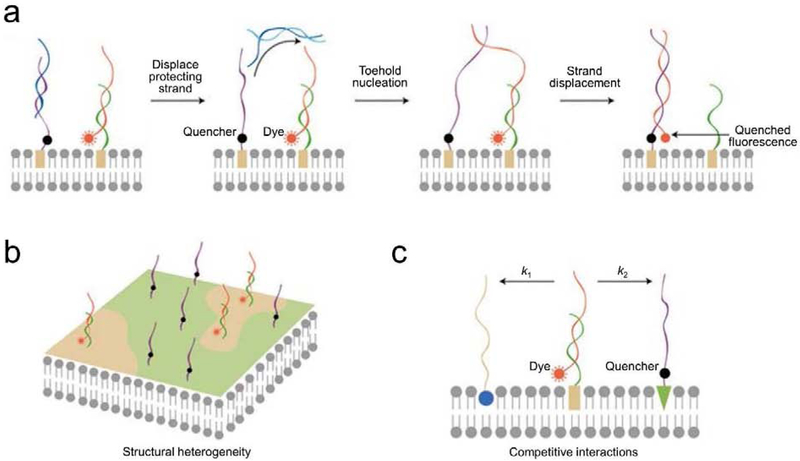
Other membrane biophysical events can also be measured based on the dynamic hybridization of oligonucleotides. For example, we recently engineered a DNA probe to monitor transient membrane lipid–lipid interactions (Figure 4) [24,44]. Here, a proximity-based strand displacement reaction was used to convert transient membrane interactions into detectable fluorescence signals. As a result, previously undetectable membrane events can now be studied with the help of lipid-oligonucleotide conjugates.
Figure 4.
(a) Schematic of using DNA probes for monitoring transient lipid–lipid encounters on live cell membranes [24]. The addition of an initiator strand (cyan) hybridized and removed the block strand (blue). As a result, once there was a membrane lipid–lipid encounter, the translocation of the DNA probe (red) from one anchor site (green) to another (purple) induced the quenching of fluorescence. (b) Lipid-DNA probes can be used to study membrane heterogeneity. Different lipids preferred to insert into different membrane domains. Lipids of different domains tended to encounter less with each other and resulted in a slow fluorescence quenching. (c) Lipid-DNA probes can be used to measure the competitive interactions among different membrane lipids. Dye-labeled lipid (orange rectangle) was given two possible destinations, unlabeled lipid 1 (blue ball) or quencher-labelled lipid 2 (green triangle). The relative interaction rate (k1 and k2) could be determined based on the rate of fluorescence quenching. Figures are adapted and modified with permission from reference [44].
Conclusion and outlook
Lipid-oligonucleotide conjugates are becoming popular for cell membrane engineering, regulation, and analysis. Recent progress in bioconjugate chemistry and synthetic biology has further promoted the fast growing of this field. Taking advantage of these lipid-oligonucleotide conjugates, different functional moieties can be efficiently engineered onto live cell membranes, without affecting natural cellular behavior. Various previously inaccessible membrane events can now be exploited to enhance our understanding of cell membrane structures, interactions, and functions.
These lipid-oligonucleotide conjugates exhibit several attractive features for membrane applications, including (1) spontaneous, rapid, and efficient membrane insertion after simple incubation; (2) universality towards different cell types; (3) precise hybridization and dynamic switching of oligonucleotides; and (4) versatile functionality thanks to the simple DNA conjugation chemistry.
Even though lipid-oligonucleotide conjugates have demonstrated great potential and unique membrane applications, they are still emerging tools for cell membrane engineering. There are several questions and challenges. (1) Even though the lipid moieties are either naturally existing or structurally similar to those in cell membranes, the biological influence of these synthetic membrane anchors still need to be carefully assessed. (2) Membrane persistence of many lipid-oligonucleotides needs to be improved. Tunable membrane insertion efficiency and persistence will be beneficial for different applications that may need either long-term membrane modification or fast transmembrane delivery. (3) Another interesting question is how to achieve selective anchoring of lipid-oligonucleotide onto the membrane of particular cell type, which may provide some new routes for targeted therapy or cell profiling.
Highlights.
Lipid-oligonucleotide conjugate is a new tool for the rapid, straightforward and efficient modification of live cell membranes.
These amphiphilic conjugates can be rationally designed and synthesized.
Cell membrane insertion kinetics, efficiency, persistence, and transmembrane property of lipid-oligonucleotide conjugates can be tuned for various biomedical and analytical applications.
Several previously inaccessible membrane events can now be exploited using lipid-oligonucleotide conjugates.
Acknowledgements
This work was supported by the NIH R35GM133507, the start-up grant from UMass Amherst and an IALS M2M seed grant. The authors also thank other members in the You Lab for useful discussion and valuable comments.
Abbreviations
- PNA
peptide nucleic acid
- LNA
locked nucleic acid
- 2’-F
2’-fluoro
- 2’-OMe
2’-O-methyl
- CPG
controlled-pore glass
- DNA
deoxyribonucleic acid
- RNA
ribonucleic acid
- miRNA
microRNA
- mRNA
messenger RNA
- siRNA
small interfering RNA
- NO
nitric oxide
- PEG
polyethylene glycol
- SO2
sulfur dioxide
- SPS
solid-phase synthesis
- FRET
fluorescence resonance energy transfer
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflicts of interest
There are no conflicts of interest to declare.
References
* Papers of special interest
** Papers of outstanding interest
- [1].Mager MD, LaPointe V, Stevens MM: Exploring and exploiting chemistry at the cell surface. Nat Chem 2011. 3:582–589. [DOI] [PubMed] [Google Scholar]
- [2].Mahal LK, Bertozzi CR: Engineered cell surfaces: Fertile ground for molecular landscaping. Chem Biol 1997. 4:415–422. [DOI] [PubMed] [Google Scholar]
- [3].Rabuka D, Forstner MB, Groves JT, Bertozzi CR: Noncovalent cell surface engineering: Incorporation of bioactive synthetic glycopolymers into cellular membranes. J Am Chem Soc 2008. 130:5947–5953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Bagheri Y, Shafiei F, Chedid S, Zhao B, You MX: Lipid-DNA conjugates for cell membrane modification, analysis, and regulation. Supramol Chem 2019. 31:532–544. [Google Scholar]
- ** [5].Huo SD, Li HY, Boersma AJ, Herrmann A: DNA nanotechnology enters cell membranes. Adv Sci 2019. 6:1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]; A comprehensive review on how lipid-DNA conjugates can be assembled into various nanostructures. In addition, the interaction and applications of these lipid-DNA nanostructures on cell membranes have been discussed.
- [6].Hu QQ, Li H, Wang LH, Gu HZ, Fan CH: DNA nanotechnology-enabled drug delivery systems. Chem Rev 2019. 119:6459–6506. [DOI] [PubMed] [Google Scholar]
- [7].Patwa A, Gissot A, Bestel I, Barthelemy P: Hybrid lipid oligonucleotide conjugates: synthesis, self-assemblies and biomedical applications. Chem Soc Rev 2011. 40:5844–5854. [DOI] [PubMed] [Google Scholar]
- [8].Chandra RA, Douglas ES, Mathies RA, Bertozzi CR, Francis MB: Programmable cell adhesion encoded by DNA hybridization. Angew Chem Int Ed 2006. 45:896–901. [DOI] [PubMed] [Google Scholar]
- [9].Hsiao SC, Shum BJ, Onoe H, Douglas ES, Gartner ZJ, Mathies RA, Bertozzi CR, Francis MB: Direct cell surface modification with DNA for the capture of primary cells and the investigation of myotube formation on defined patterns. Langmuir 2009. 25:6985–6991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Zhao W, Schafer S, Choi J, Yamanaka YJ, Lombardi ML, Bose S, Carlson AL, Phillips JA, Teo W, Droujinine IA, Cui CH, Jain RK, Lammerding J, Love JC, Lin CP, Sarkar D, Karnik R, Karp JM: Cell-surface sensors for real-time probing of cellular environments. Nat Nanotechnol 2011. 6:524–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Mali P, Aach J, Lee JH, Levner D, Nip L, Church GM: Barcoding cells using cell-surface programmable DNA-binding domains. Nat Methods 2013. 10:403–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Tan WH, Donovan MJ, Jiang JH: Aptamers from cell-based selection for bioanalytical applications. Chem Rev 2013. 113:2842–2862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Madsen M, Gothelf KV: Chemistries for DNA nanotechnology. Chem Rev 2019. 119:6384–6458. [DOI] [PubMed] [Google Scholar]
- [14].Singh Y, Murat P, Defrancq E: Recent developments in oligonucleotide conjugation. Chem Soc Rev 2010. 39:2054–2070. [DOI] [PubMed] [Google Scholar]
- [15].Krutzfeldt J, Rajewsky N, Braich R, Rajeev KG, Tuschl T, Manoharan M, Stoffel M: Silencing of microRNAs in vivo with ‘antagomirs’. Nature 2005. 438:685–689. [DOI] [PubMed] [Google Scholar]
- [16].Wolfrum C, Shi S, Jayaprakash KN, Jayaraman M, Wang G, Pandey RK, Rajeev KG, Nakayama T, Charrise K, Ndungo EM, Zimmermann T, Koteliansky V, Manoharan M, Stoffel M: Mechanisms and optimization of in vivo delivery of lipophilic siRNAs. Nat Biotechnol 2007. 25:1149–1157. [DOI] [PubMed] [Google Scholar]
- [17].Raouane M, Desmaele D, Urbinati G, Massaad-Massade L, Couvreur P: Lipid conjugated oligonucleotides: a useful strategy for delivery. Bioconjugate Chem 2012. 23:1091–1104. [DOI] [PubMed] [Google Scholar]
- [18].Liu HP, Kwong B, Irvine DJ: Membrane anchored immunostimulatory oligonucleotides for in vivo cell modification and localized immunotherapy. Angew Chem Int Ed 2011. 50:7052–7055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Palte MJ, Raines RT: Interaction of nucleic acids with the glycocalyx. J Am Chem Soc 2012. 134:6218–6223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Letsinger RL, Zhang GG, Sun DK, Ikeuchi T, Sarin PS: Cholesteryl-conjugated oligonucleotides - synthesis, properties, and activity as inhibitors of replication of human immunodeficiency virus in cell-culture. Proc Natl Acad Sci 1989. 86:6553–6556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Pfeiffer I, Hook F: Bivalent cholesterol-based coupling of oligonucletides to lipid membrane assemblies. J Am Chem Soc 2004. 126:10224–10225. [DOI] [PubMed] [Google Scholar]
- * [22].Feng GF, Luo XY, Lu X, Xie SY, Deng L, Kang WY, He F, Zhang JH, Lei CY, Lin B, Huang Y, Nie Z, Yao SZ: Engineering of nucleic acids and synthetic cofactors as holo sensors for probing signaling molecules in the cellular membrane microenvironment. Angew Chem Int Ed 2019. 58:6590–6594. [DOI] [PubMed] [Google Scholar]; A recent example of using lipid-modified functional oligonucleotides for monitoring cell membrane signaling. In this study, a G-quadruplex and stimulti-reactive cofactor was modularly combined to image the cellular extrusion of SO2 and NO.
- * [23].Zhao B, O’Brien C, Mudiyanselage APKKK, Li NW, Bagheri Y, Wu R, Sun YB, You MX: Visualizing intercellular tensile forces by DNA-based membrane molecular probes. J Am Chem Soc 2017. 139:18182–18185. [DOI] [PubMed] [Google Scholar]; Lipid-oligonucleotide probes were developed to image and measure mechanical forces at cell–cell junctions. These probes are easy to synthesize and to use, which can be potentially broadly applicable in studying mechanobiology.
- ** [24].You MX, Lyu YF, Han D, Qiu LP, Liu QL, Chen T, Wu CS, Peng L, Zhang LQ, Bao G, Tan WH: DNA probes for monitoring dynamic and transient molecular encounters on live cell membranes. Nat Nanotechnol 2017. 12:453–459. [DOI] [PMC free article] [PubMed] [Google Scholar]; In this study, lipid-oligonucleotide conjugates were used to monitor rapid membrane lipid–lipid interactions using standard fluorescence microscopy and flow cytometry. Previously untraceable interactions within lipid domains can now be imaged in live cell membranes.
- [25].Tokunaga T, Namiki S, Yamada K, Imaishi T, Nonaka H, Hirose K, Sando S: Cell surface-anchored fluorescent aptamer sensor enables imaging of chemical transmitter dynamics. J Am Chem Soc 2012. 134:9561–9564. [DOI] [PubMed] [Google Scholar]
- [26].Weber RJ, Liang SI, Selden NS, Desai TA, Gartner ZJ: Efficient targeting of fatty-acid modified oligonucleotides to live cell membranes through stepwise assembly. Biomacromolecules 2014. 15:4621–4626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ** [27].Bagheri Y, Chedid S, Shafiei F, Zhao B, You M: Quantitative assessment of the dynamic modification of lipid-DNA probes on live cell membranes. Chem Sci 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]; A systematic study of the cell membrane modification process of lipid-oligonucleotide conjugates. This research provides a guideline on regulating the cell membrane insertion rate, efficiency, and persistence of lipid-DNA probes.
- * [28].Jin C, He JX, Zou JM, Xuan WJ, Fu T, Wang RW, Tan WH: Phosphorylated lipid-conjugated oligonucleotide selectively anchors on cell membranes with high alkaline phosphatase expression. Nat Commun 2019. 10:1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]; A very interesting strategy to regulate the cell membrane selectivity of lipid-oligonucleotide conjugates based on alkaline phosphatase-mediated lipid conversion. Lipid-DNA probes can be selectively recruited onto diseased cell membranes.
- [29].Borisenko GG, Zaitseva MA, Chuvilin AN, Pozmogova GE: DNA modification of live cell surface. Nucleic Acids Res 2009. 37:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].McGinnis CS, Patterson DM, Winkler J, Conrad DN, Hein MY, Srivastava V, Hu JL, Murrow LM, Weissman JS, Werb Z, Chow ED, Gartner ZJ: MULTI-seq: sample multiplexing for single-cell RNA sequencing using lipid-tagged indices. Nat Methods 2019. 16:619–626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Shi P, Zhao N, Coyne J, Wang Y: DNA-templated synthesis of biomimetic cell wall for nanoencapsulation and protection of mammalian cells. Nat Commun 2019. 10:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Gao T, Chen T, Feng C, He X, Mu C, Anzai J, Li G: Design and fabrication of flexible DNA polymer cocoons to encapsulate live cells. Nat Commun 2019. 10:2946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Langecker M, Arnaut V, List J, Simmel FC: DNA nanostructures interacting with lipid bilayer membranes. Accounts Chem Res 2014. 47:1807–1815. [DOI] [PubMed] [Google Scholar]
- [34].Chen S, Bremer AW, Scheideler OJ, Na YS, Todhunter ME, Hsiao S, Bomdica PR, Maharbiz MM, Gartner ZJ, Schaffer DV: Interrogating cellular fate decisions with high-throughput arrays of multiplexed cellular communities. Nat Commun 2016. 7:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Xiong X, Liu H, Zhao Z, Altman MB, Lopez-Colon D, Yang CJ, Chang LJ, Liu C, Tan W: DNA aptamer-mediated cell targeting. Angew Chem Int Ed 2013. 52:1472–1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Qiu LP, Zhang T, Jiang JH, Wu CC, Zhu GZ, You MX, Chen XG, Zhang LQ, Cui C, Yu RQ, Tan WH: Cell membrane-anchored biosensors for real-time monitoring of the cellular microenvironment. J Am Chem Soc 2014. 136:13090–13093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Karaki S, Benizri S, Mejias R, Baylot V, Branger N, Nguyen T, Vialet B, Oumzil K, Barthelemy P, Rocchi P: Lipid-oligonucleotide conjugates improve cellular uptake and efficiency of TCTP-antisense in castration-resistant prostate cancer. J Control Release 2017. 258:1–9. [DOI] [PubMed] [Google Scholar]
- [38].Juliano RL: The delivery of therapeutic oligonucleotides. Nucleic Acids Res 2016. 44:6518–6548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Burns JR, Seifert A, Fertig N, Howorka S: A biomimetic DNA-based channel for the ligand-controlled transport of charged molecular cargo across a biological membrane. Nat Nanotechnol 2016. 11:152–156. [DOI] [PubMed] [Google Scholar]
- [40].Burns JR, Al-Juffali N, Janes SM, Howorka S: Membrane-spanning DNA nanopores with cytotoxic effect. Angew Chem Int Ed 2014. 53:12466–12470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Langecker M, Arnaut V, Martin TG, List J, Renner S, Mayer M, Dietz H, Simmel FC: Synthetic lipid membrane channels formed by designed DNA nanostructures. Science 2012. 338:932–936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- * [42].Saminathan A, Devany J, Pillai KS, Veetil AT, Schwake M, Krishnan Y: A DNA-based voltmeter for organelles. BioRxiv 20191–19. [DOI] [PMC free article] [PubMed] [Google Scholar]; A great demonstration in using precisely self-assembled DNA nanostructures for ratiometric-based quantification at subcellular locations. Here, lipid-DNA conjugates were used to image and quantify membrane potentials.
- [43].Li J, Xun KY, Pei K, Liu XJ, Peng XY, Du YL, Qiu LP, and Tan WH: Cell-membrane-anchored DNA nanoplatform for programming cellular interactions. J Am Chem Soc 2019. 141:18013–18020. [DOI] [PubMed] [Google Scholar]
- [44].Beales PA: A toehold in cell surface dynamics. Nat Nanotechnol 2017. 12:404–406. [DOI] [PubMed] [Google Scholar]