Abstract
Ni–Fe CO-dehydrogenases (CODHs) catalyze the conversion between CO and CO2 using a chain of Fe–S clusters to mediate long-range electron transfer. One of these clusters, the D-cluster, is surface-exposed and serves to transfer electrons between CODH and external redox partners. These enzymes tend to be extremely O2-sensitive and are always manipulated under strictly anaerobic conditions. However, the CODH from Desulfovibrio vulgaris (Dv) appears unique: exposure to micromolar concentrations of O2 on the minutes-time scale only reversibly inhibits the enzyme, and full activity is recovered after reduction. Here, we examine whether this unusual property of Dv CODH results from the nature of its D-cluster, which is a [2Fe-2S] cluster, instead of the [4Fe-4S] cluster observed in all other characterized CODHs. To this aim, we produced and characterized a Dv CODH variant where the [2Fe-2S] D-cluster is replaced with a [4Fe-4S] D-cluster through mutagenesis of the D-cluster–binding sequence motif. We determined the crystal structure of this CODH variant to 1.83-Å resolution and confirmed the incorporation of a [4Fe-4S] D-cluster. We show that upon long-term O2-exposure, the [4Fe-4S] D-cluster degrades, whereas the [2Fe-2S] D-cluster remains intact. Crystal structures of the Dv CODH variant exposed to O2 for increasing periods of time provide snapshots of [4Fe-4S] D-cluster degradation. We further show that the WT enzyme purified under aerobic conditions retains 30% activity relative to a fully anaerobic purification, compared to 10% for the variant, and the WT enzyme loses activity more slowly than the variant upon prolonged aerobic storage. The D-cluster is therefore a key site of irreversible oxidative damage in Dv CODH, and the presence of a [2Fe-2S] D-cluster contributes to the O2-tolerance of this enzyme. Together, these results relate O2-sensitivity with the details of the protein structure in this family of enzymes.
Keywords: CO-dehydrogenase, iron−sulfur cluster, carbon monoxide, oxygen-sensitivity, oxidative damage, crystallography, protein film voltammetry
Introduction
Ni–Fe-dependent carbon monoxide dehydrogenases (CODHs) catalyze the interconversion of the gaseous pollutant CO and the greenhouse gas CO2, allowing for autotrophic growth of various microbes on these simple one-carbon compounds.1,2 CooS-type CODHs are homodimers that contain a total of five metalloclusters, called the B-, C-, and D-clusters (Figure 1A).3,4 Each symmetric CODH dimer contains two C-clusters, two B-clusters, and one D-cluster that is shared between the two monomers at the dimer interface. The chemistry of CO/CO2 interconversion occurs at the C-cluster, a distinctive [Ni-3Fe-4S] cubane connected through one of the sulfide ions to a mononuclear iron site, Feu.3,4 The B- and D-clusters are Fe–S clusters that wire the C-cluster to the surface of the protein, where electrons are exchanged with an external redox partner, such as a ferredoxin.3−6 To date, all characterized Ni–Fe CODHs exhibit high sequence identities (40–57%) and share an overall fold in which each monomer consists of an N-terminal helical domain followed by two α/β Rossmann-like domains (Figure 1A).3,4,7−10
Figure 1.
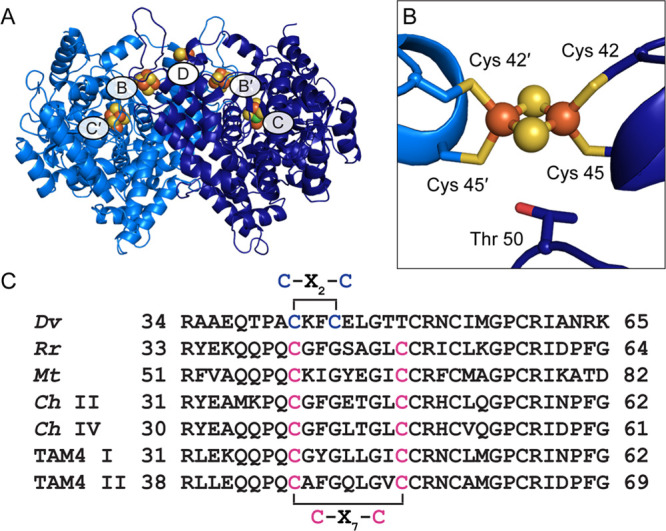
Structure of wild-type Dv CODH. (A) Overall structure (PDB ID 6B6V). The CODH dimer is shown in ribbon representation in dark and light blue with metalloclusters shown as spheres with Ni in green, Fe in orange, and S in yellow. Each B-cluster completes the electron transfer pathway of the opposing monomer, denoted with prime symbols. (B) The D-cluster of wild-type Dv CODH is a [2Fe-2S] cluster, whereas all other characterized CODHs contain a [4Fe-4S] D-cluster. In [4Fe-4S] D-cluster-containing CODHs, Cys 45 is a glycine residue and Thr 50 is a cysteine residue (D. vulgaris numbering) that completes coordination of the cluster. (C) Sequence alignments reveal a difference in D-cluster binding motifs in the primary structure. Cysteine ligands to the D-cluster are colored in blue ([2Fe-2S] cluster) or pink ([4Fe-4S] cluster). Organism names are Desulfovibrio vulgaris (Dv), Rhodospirillum rubrum (Rr), Moorella thermoacetica (Mt), Carboxydothermus hydrogenoformans (Ch), and Thermoccocus sp. AM4 (TAM4).
Ni–Fe CODHs are inhibited by molecular oxygen, presumably due to the O2-sensitive nature of their metallocofactors.11−14 However, the CODH from Desulfovibrio vulgaris (Dv) was recently shown to be unusually O2-resistant:15 following transient exposure to up to 20 μM O2, Dv CODH recovers nearly full activity upon rereduction of the enzyme. This reductive reactivation is reminiscent of the behavior of standard Ni–Fe hydrogenases, for example,16 but contrasts to what has been observed for all the other CODHs that have been characterized. Other CODHs, including CODHs I, II, and IV from Carboxydothermus hydrogenoformans(8,17) and the two CODHs from Thermococcus sp. AM4,18 reactivate either to a much lesser extent or not at all. Together, these observations suggest that Dv CODH has a unique mechanism of O2-tolerance.
Our previously published crystal structure of Dv CODH revealed a [2Fe-2S] D-cluster at the dimer interface, where all other previously characterized CODHs contain a [4Fe-4S] cluster (Figure 1B).7 This difference was proposed to result from a variation in the D-cluster binding motif: C–X2–C in the Dv CODH sequence versus C–X7–C in other CODHs (Figure 1C).7 The [2Fe-2S] D-cluster and the [4Fe-4S] B-clusters of the enzyme were additionally shown to be stable to O2 exposure, as characterized by crystallography and electron paramagnetic resonance (EPR) spectroscopy. Interestingly, the C-cluster holds on to its metal ions upon exposure to O2 for 2 days, but converts from a catalytically relevant arrangement of metals into an oxidized conformation where the Ni and Feu ions adopt different coordination environments, as we have described previously.7 Thus, all clusters in Dv CODH appear to retain their metal ions upon oxidation, but the enzyme becomes transiently inactive because of the C-cluster metal rearrangement.7
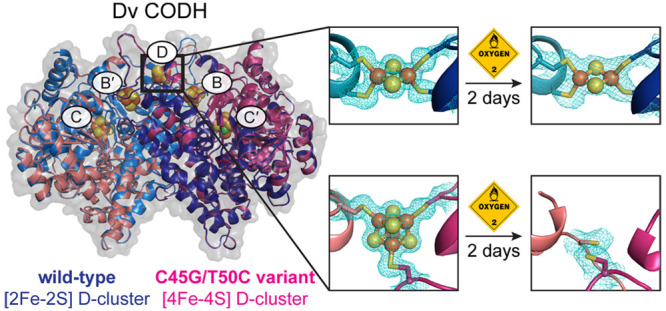
Given the enhanced O2-tolerance of Dv CODH and the fact that it contains a distinguishing [2Fe-2S] D-cluster, we hypothesized that the difference in cluster type may contribute to the difference in O2-sensitivity. In this scenario, the inactivation of CODHs other than that from Dv may be irreversible due to the instability of the [4Fe-4S] D-cluster in the presence of O2. To interrogate this effect, we sought to replace the [2Fe-2S] D-cluster of Dv CODH with the more canonical [4Fe-4S] D-cluster and to test the effect of O2 exposure on the resulting protein variant. Here we demonstrate that alteration of cluster type can indeed be achieved using site-directed mutagenesis to modify the D-cluster binding motif, and that the [2Fe-2S] D-cluster is critical for making the D. vulgaris enzyme resistant to long-term exposure to air.
Results
A C45G/T50C variant of Dv CODH in which the C–X2–C D-cluster motif is replaced with a C–X7–C motif was designed and purified. The as-isolated enzyme contains on average 0.2 Ni/monomer, which is half of the content typically observed for the wild-type (WT) recombinant enzyme.14 The Fe content of the C45G/T50C variant is about 10 Fe/monomer, suggesting that all clusters are appropriately reconstituted with Fe (we expect 9 Fe/monomer for a CODH with a bridging [2Fe-2S] D-cluster and 10 Fe/monomer for a CODH with a bridging [4Fe-4S] D-cluster). After activation with NiCl2 under reductive conditions, the activity of the variant is 350 μmol·min–1·mg–1 at 37 °C, pH 8, similar to the value obtained for the WT enzyme produced, purified, reconstituted (using a procedure that involves NiCl2, Na2S, and TCEP) and assayed under the same conditions (see Methods). If enzyme samples are kept at 4 °C in an anaerobic glovebox under N2, their activity is stable for weeks.
We determined the crystal structure of the C45G/T50C variant to 1.83 Å resolution (Table S1) and observed that it aligns well with that of WT Dv CODH (Cα r.m.s.d of 0.27 Å for 1250 Cα atoms) (Figure S1).7 The structure reveals clear electron density for a [4Fe-4S] cluster at the dimer interface in place of the [2Fe-2S] D-cluster that is present in the WT enzyme (Figure 2A). The presence of four iron atoms within the cubane was confirmed using anomalous diffraction data collected at the iron peak wavelength (7130 eV, 1.7389 Å) (Table S2, Figure 2B). This result demonstrates that sequence alone is sufficient for determining the type of Fe–S cluster present at the dimer interface and that this site can incorporate either a [4Fe-4S] or [2Fe-2S] cluster. Notably, the structure of the protein backbone in the vicinity of the D-cluster is independent of cluster type (Figure S1). The crystal structure of the C45G/T50C variant also contains a normal [4Fe-4S] B-cluster and a C-cluster that is mainly (80%) in the canonical (reduced) conformation, with a minor fraction (20%) in the previously identified oxidized conformation (Figure S2).7 The C-cluster nickel ion has been refined at 20% occupancy in each conformation, roughly consistent with the observed metal content of 0.3 Ni/monomer for the specific protein batch that was used for crystallization.
Figure 2.
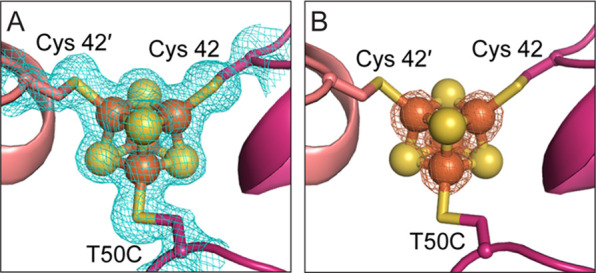
Alteration of the D-cluster binding motif leads to incorporation of a [4Fe-4S] cluster at the Dv CODH dimer interface. (A) Simulated annealing composite omit map (blue mesh, contoured to 1σ) for the [4Fe-4S] D-cluster. (B) Iron anomalous difference map (orange mesh, contoured to 8σ) supports the presence of four iron ions in the cluster. Protein shown in ribbon representation in pink with ligating cysteine residues as sticks and cluster as spheres and sticks: S in yellow and Fe in orange.
We compared the effect of O2 on the WT enzyme and the C45G/T50C variant over different time scales and O2 concentrations and using multiple techniques. First, we used direct electrochemistry19 to probe short exposures (1 min) to small amounts of O2 (Figure 3). In this technique, a small amount of purified enzyme is adsorbed onto an electrode that is used to control redox conditions and measure CO-oxidation activity as a current. We used a previously described method15 to measure the relative activity just after transient exposure to a controlled amount of O2 or after transient exposure to O2 followed by a low potential step to force the reductive reactivation of the enzyme. We found that, under these conditions, the identity of the D-cluster makes essentially no difference (Figure 3).
Figure 3.
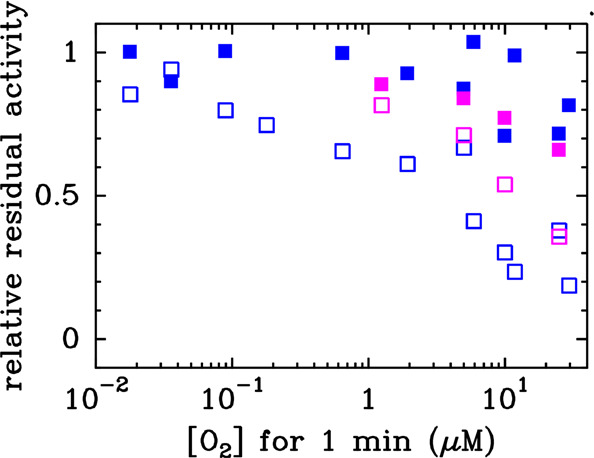
C45G/T50C variant and WT Dv CODH respond similarly to short-term exposure to O2. Residual activity measured in electrochemistry experiments at −310 mV vs SHE, 25 °C, pH 7, after the WT enzyme (blue, data from ref (15)) or the C45G/T50C variant (pink) has been exposed to a controlled amount of O2 (empty squares), or after it has been exposed to O2 and then reduced for 20 s at −560 mV vs SHE (filled squares), as described in ref (15).
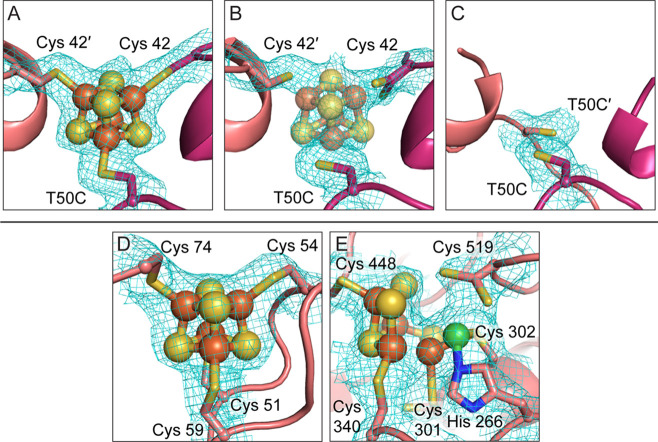
Next, we took a structural approach to examine the effect of long-term exposure to ambient atmospheric conditions (i.e., conditions outside of the anaerobic chamber). Crystals of C45G/T50C Dv CODH were grown anaerobically and then exposed to ambient atmospheric conditions for varying lengths of time prior to being harvested for data collection (see Methods; Table S1). Where possible, complementary diffraction data were collected at the iron peak wavelength on the same crystal as was used for structure determination (Table S2). We found that when crystals of C45G/T50C Dv CODH are harvested immediately after removal from the anaerobic chamber, the [4Fe-4S] D-cluster remains intact (Figures 4A and S3). Following 2 h of incubation at ambient atmospheric conditions, the D-cluster of C45G/T50C Dv CODH begins to decay, as evidenced by the electron density at this site (Figure 4B, Figure S3). Accurate modeling of any Fe–S cluster variation into the electron density for the D-cluster in this structure is difficult due to the fact that the density likely represents the presence of multiple different states of cluster decomposition within the molecules of the crystal lattice. A reasonable fit to the density is obtained when a [4Fe-4S] cluster is modeled at an occupancy of 30%, although the fit is still relatively poor (Figure 4B), and this cluster has been left out of the final model. After 2 days at ambient atmospheric conditions, the [4Fe-4S] D-cluster of C45G/T50C Dv CODH is no longer visible in the electron density, indicating complete decay (Figure 4C). Additionally, one of the D-cluster cysteine ligands, Cys 42, of each chain is disordered and cannot be modeled due to uninterpretable electron density. We note that the resolution of this last structure is comparatively low and that the B-factors of the B- and C-clusters are relatively high. In the case of the B-cluster, the B-factors are consistent with the surrounding protein environment, which may experience increased local disorder due to the complete absence of the D-cluster. In the case of the C-cluster, we note that all structures of the oxidized C-cluster at resolutions greater than 2 Å display high relative B-factors,7 perhaps due to an inability to adequately parametrize this poorly characterized cluster type during refinement in this resolution range. Finally, the initial poorer quality of the crystals following 2 days of air exposure impaired our ability to collect a paired iron peak data set on the same crystal, and we were unable to characterize the iron anomalous signal within this structure. That being said, the electron density at the B- and C-clusters in this enzyme variant is strong following 2 days in air and, within the resolution limits of the data, the clusters have been refined at full occupancy (Figure 4D and E). Additionally, our previous analysis of WT Dv CODH revealed the presence of the B- and C-clusters, following air exposure by both crystallography (Figure S4) and EPR spectroscopy.7 Given these lines of evidence, we conclude that the B- and C-clusters of C45G/T50C Dv CODH are present and intact following 2 days of air exposure and that a detrimental effect is only observed on the D-cluster. This instability of the [4Fe-4S] D-cluster in C45G/T50C Dv CODH contrasts with the behavior of the [2Fe-2S] D-cluster in the WT enzyme, which remains intact after 2 days of exposure to air (Figure S4).7
Figure 4.
The [4Fe-4S] D-cluster in C45G/T50C Dv CODH crystals decays over time in air, whereas the B- and C-clusters remain intact. (A) 2Fo-Fc electron density at the D-cluster for crystals harvested immediately after removal from the anaerobic chamber. (B) 2Fo-Fc electron density for the D-cluster 2 h after the crystal was removed from the anaerobic chamber. (C) 2Fo-Fc electron density for the D-cluster 2 days after the crystal was removed from the anaerobic chamber. (D) 2Fo-Fc electron density for the B-cluster 2 days after the crystal was removed from the anaerobic chamber. (E) 2Fo-Fc electron density for the oxidized C-cluster 2 days after the crystal was removed from the anaerobic chamber. Electron density (blue mesh) is contoured to 1σ. Protein is shown as in Figure 2 with the addition of N in blue and Ni in green for the C-cluster. See Figure S4 for comparison to the stability of the WT [2Fe-2S] D-cluster, B-cluster, and C-cluster after 2 days in air.7
With crystallographic data showing that the [2Fe-2S] D-cluster in WT Dv CODH is more stable in air over long time periods (hours to days) than the [4Fe-4S] D-cluster in the C45G/T50C Dv CODH variant, we next sought to assess the O2-stability of the CODHs in solution. We produced the WT and the C45G/T50C Dv CODH variant under aerobic conditions: the cells were grown anaerobically, lysed aerobically, and the enzymes were then purified in air at room temperature taking no particular precautions to ensure a reducing environment. Together, this workflow takes about half a working day. Prepared under aerobic conditions, the WT enzyme contains about 0.2 Ni/monomer, 75% of the expected Fe content, and about 30% of the activity (always measured after activation/reconstitution of the samples, as described in the Methods) as compared to the result of an anaerobic purification. The C45G/T50C variant suffers more than the WT from the aerobic purification: it contains 0.1 Ni/monomer, 60% of the expected Fe and only 10% of the activity as compared to the anaerobic purification. These metal content analyses (confirmed by two independent rounds of protein production and purification) show that irreversible inactivation in solution may also arise from the effect of ambient levels of O2 on the C-cluster under the conditions of the purification.
We also measured, at regular intervals, the activities of samples prepared and stored anaerobically at 4 °C and compared those to samples described above that were prepared and stored in air at 4 °C (Figure 5). We found that a constant activity level was maintained over time for the WT Dv CODH prepared and stored in air, the WT Dv CODH prepared and stored anaerobically, and the C45G/T50C variant prepared and stored anaerobically (Figure 5). In contrast, the activity drops quickly for the C45G/T50C variant prepared in air and continuously exposed to air. In just 1 day, the activity falls to a residual level of just 5% of the WT anaerobic activity level (Figure 5). The entire experiment, including protein expression and purification, was performed in duplicate and yielded similar results both times.
Figure 5.
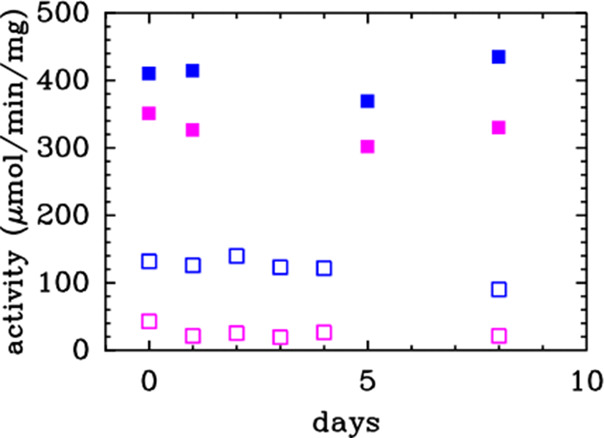
Activity of the C45G/T50C variant is more sensitive to long-term O2 exposure than WT Dv CODH. Activity values of the WT (blue) and C45G/T50C variant (pink) measured in solution assays at 37 °C, pH 8, with the samples either prepared under anaerobic conditions and stored at 4 °C under anaerobic conditions (filled squares) or prepared under aerobic conditions and stored at 4 °C under aerobic conditions (empty squares).
Discussion
A loss of enzyme activity upon exposure to O2 has been well documented for many metalloproteins.20−22 However, molecular explanations for the activity loss are often not available. Additionally, it is frequently unclear why some metalloproteins within the same enzyme family are more stable to O2 than others. For certain metalloproteins, like the hydrogenases, oxidative inhibition has been well studied.16,23−28 For others, like the CODH family, investigations of the source of O2-sensitivity are just beginning.
In a previous investigation, we showed that Dv CODH is surprisingly O2-resistant. It reacts with O2 very quickly (inhibition bimolecular rate constant of 5 s–1 μM(O2) –1 at 25 °C, pH 7) to form at least three different states which are distinguished by whether they reactivate upon mere restoration of anaerobiosis or upon reduction or they do not reactivate (the proportion of the latter increases with the concentration of O2).15 The inactive species that is formed upon aerobic oxidation and that reactivates upon reduction is stable enough that we could solve its crystal structure. In this form of the enzyme, the C-cluster has undergone a substantial rearrangement with Ni, Fe, and S ions moving several angstroms and adopting noncatalytic positions, and the other clusters are intact.7 The same three-state profile for reaction with O2 was observed for all other CODHs that have been characterized in the same manner (Ch CODHs I, II, and IV; TAM4 CODHs I and II), but the rate of reaction with O2 and the extent of reactivation upon reduction vary greatly. It has been suggested that the rate of inactivation may depend on the amino acid packing at the rear of the C-cluster,8 although this hypothesis remains to be confirmed by mutagenesis. Regarding the extent of inactivation, the site that is subjected to irreversible damage and responsible for the irreversible inactivation was unknown. The most obvious structural difference between Dv CODH and other CODHs is the nuclearity of the D-cluster, which is a [2Fe-2S] cluster instead of the more common [4Fe-4S] cluster, and we wondered if the O2-sensitivity of the [4Fe-4S] D-cluster is the reason that canonical CODHs are more irreversibly inactivated by O2 than Dv CODH.
Here, we showed that it is possible to replace the [2Fe-2S] D-cluster of Dv CODH with a standard [4Fe-4S] D-cluster by simply changing the binding motif from C–X2–C to the standard C–X7–C (Figure 1C). The cluster swap was confirmed crystallographically by determining a 1.83 Å resolution structure of the C45G/T50C variant (Figure 2). There have been previously published examples of the use of site directed mutagenesis to interconvert [3Fe-4S] and [4Fe-4S] clusters in enzymes,29 but this is the first report of [2Fe-2S] to [4Fe-4S] cluster conversion as a result of protein engineering. The as-isolated C45G/T50C variant contains half as much Ni per monomer as the WT enzyme, consistent with the previously demonstrated role of the D-cluster in mediating redox chemistry during nickel incorporation,30 but after activation with NiCl2 under reducing conditions, the activity of the C45G/T50C variant is similar to that of the WT enzyme.
We compared the response of Dv CODH to O2 under various conditions. In experiments in which the enzyme is exposed to small amounts of O2 for a short time (<20 μM for about 1 min), the presence of a [4Fe-4S] D-cluster makes no difference (Figure 3), consistent with the idea that the C-cluster rearrangements that lead to activity loss occur on a faster time scale than D-cluster loss. However, the resistance of the enzyme to long-term exposure to air does appear to be dependent on the presence of a [2Fe-2S] D-cluster: our structural analyses show that all clusters retain their metals upon exposure to air except for the [4Fe-4S] D-cluster of the C45G/T50C variant, which is irreversibly degraded (Figures 4, S3, and S4).
The ability of a [2Fe-2S] D-cluster to confer O2-resistance to Dv CODH also explains the higher activity observed following aerobic purification for the WT enzyme compared to the [4Fe-4S] cluster-containing C45G/T50C Dv CODH variant. Fully aerobically purified WT Dv CODH (with no reductant in any of the purification buffers) is 30% as active as anaerobically purified protein, compared to 10% as active for C45G/T50C Dv CODH. Additionally, whereas WT Dv CODH retains this high 30% activity level for days in air at 4 °C (Figure 5), C45G/T50C Dv CODH loses activity over time, falling to a residual value of 5% after 1 day in air (these numbers are the average of two independent experiments). This 5% residual activity could either be due to a small enzyme population with intact D-clusters or to the ability of methyl viologen (the in vitro electron acceptor when the activity is assayed) to accept electrons directly from the B-cluster in the enzymes lacking a functional D-cluster. In terms of the latter possibility, such a short circuit of the electron transfer chain has been demonstrated previously with mutants of Ni–Fe hydrogenase where the chain of Fe–S clusters is modified.31
Collectively, our results suggest that the [4Fe-4S] D-cluster is a key site of irreversible oxidative damage in the Dv CODH variant and that this damage is avoided in the wild-type enzyme through incorporation of a [2Fe-2S] cluster at this site, resulting in the exceptional stability of Dv CODH upon exposure to O2. Further work will be required to determine whether CODHs that natively contain a [4Fe-4S] D-cluster will behave similarly, for example by observing a beneficial effect of engineering a [2Fe-2S] D-cluster or by detecting the selective degradation of the natural [4Fe-4S] D-cluster. Over the course of this work, we tried to obtain the structure of the CODH from Moorella thermoacetica (which houses a [4Fe-4S] D-cluster) following exposure to air; however, the diffraction of the corresponding crystals was severely impaired by oxygen exposure, likely due to disruption of crystal packing due to air-induced structural changes (e.g., D-cluster degradation). Indeed, only one of the two crystal forms that we obtained for C45G/T50C Dv CODH survived air-exposure well enough to yield diffraction-quality crystals. It would appear that the ability to perform this type of crystallographic analysis is strongly dependent on the crystal form obtained, and unfortunately, we have not yet been able to obtain a suitable crystal form for another CODH.
On the basis of sequence information, we expect that Dv CODH will not be alone in utilizing a [2Fe-2S] D-cluster. Considering the recently published CODH phylogenetic analysis of Inoue et al.32 (and if one excludes sequences in groups A4, A6, B1, C, D, and G1, which lack some essential amino acids and are probably not from functional CODHs), we estimate that the interfacial [2Fe-2S] D-cluster is present in about 10% of all CODHs (those in group E2 as designated by Inoue et al.), three-quarters of which are from sulfate reducing bacteria. Such substitution in cluster type at the surface of the enzyme may provide an evolutionary advantage for aerotolerant organisms, such as D. vulgaris and other species of sulfate reducing bacteria, which can survive under microaerobic conditions.33,34
The observation that the CODH from D. vulgaris retains a considerable fraction of activity after it is purified under fully aerobic conditions was a surprise considering how O2-sensitive these enzymes are deemed to be. Our results therefore add to a growing body of evidence that O2-sensitivity in Ni–Fe CO-dehydrogenases varies greatly, and at this point we cannot exclude that there may exist fully O2-tolerant Ni–Fe CODHs.
Methods
Cloning and Purification of WT Dv CODH and C45G/T50C Dv CODH
CODH constructs were expressed in the presence of the C-cluster maturation factor CooC, as described previously.14 Briefly, the D. vulgaris genes encoding CODH (cooS) and the CooC maturase (cooC) were cloned into a modified pBGF4 shuttle vector under the control of the promoter of the Desulfovibrio fructosovorans Ni–Fe hydrogenase operon to generate the vector pBGCooSC. The CODH construct was N-terminally strep-tagged. Mutagenesis to generate C45G/T50C Dv CODH was performed by PCR by first cloning the HindIII-SacI fragment of pBGCooSC, which contains the 5′ end of cooS, into a pUC19 vector to serve as a DNA template. The primers CCGGCCTGTAAATTCGGCGAATTGGGCACCTGCTGCCGCAACTGCATC (forward; mutations underlined) and GATGCAGTTGCGGCAGCAGGTGCCCAATTCGCCGAATTTACAGGCCGG (reverse; mutations underlined) were used to generate the C45G/T50C mutations. The enzymes were expressed in D. fructosovorans and purified under anaerobic conditions in a Jacomex anaerobic chamber (100% N2 atmosphere) by affinity chromatography on Strep-Tactin Superflow resin, as described previously.14 Regarding the aerobic purifications performed on the bench, the protocol was the same as under anaerobic conditions, except that the cells were disrupted in a French press (instead of by sonication for the anaerobic purification), dithionite was not used in any of the buffers, and all steps were performed at room temperature (instead of 4 °C for the anaerobic purification). After purification, all samples were stored at 4 °C. Protein concentrations were determined by amino acid analysis at the Centre for Integrated Structural Biology (Grenoble, France). Metal content was analyzed by inductively coupled plasma optical emission spectrometry (ICP-OES). CO oxidation activity was assayed in solution at 37 °C by monitoring the reduction of methyl viologen at 604 nm (ε604 = 13.6 mM–1·cm–1) under conditions slightly different from those used in ref (14). First, the enzymes were activated by incubation with 2.4 mM NiCl2, 4 mM Na2S, and 1.2 mM tris(2-carboxyethyl)phosphine hydrochloride (TCEP) for 15 min at room temperature. This protocol allowed a faster activation and prevented the formation of the black precipitate observed when the enzymes are activated by incubation with NiCl2 and dithionite.14 Second, the activity assays were performed at pH 8, instead of pH 10. The complete experiment, including aerobic and anaerobic purifications, metal content determination, and monitoring of the activity over time, was duplicated for both the WT and variant enzymes.
Electrochemistry
Protein film voltammetry19 and our previously described 7-injection method15,18 were used to determine the residual activity after exposure to oxygen. The enzyme was adsorbed by simply painting a 0.5 μL drop of stock solution (∼20 μM) onto a onto pyrolitic graphite edge rotating disc electrode (diameter 3 mm).
Crystallization of C45G/T50C Dv CODH
All structures were determined using anaerobically purified protein and samples were not activated with NiCl2 prior to crystallization. The particular batch of protein used for crystallization contained 0.3 Ni/monomer and 10.4 Fe/monomer, as assessed by ICP-OES. Crystals were grown anaerobically in an N2 atmosphere at 21 °C by hanging drop vapor diffusion in an MBraun anaerobic chamber. Crystals belonging to space group P21 were obtained as follows: A 1 μL aliquot of as-isolated protein (10 mg/mL in 100 mM Tris-HCl pH 8) was combined with 1 μL of precipitant solution (150–250 mM MgCl2, 16–20% (w/v) PEG 3350) on a glass cover slide and sealed over a reservoir containing 500 μL of precipitant solution. Diffraction quality crystals grew in 3–4 weeks. Crystals were soaked in a cryo-protectant solution containing 250 mM MgCl2, 18–20% (w/v) PEG 3350, 9% (v/v) glycerol and cryo-cooled in liquid nitrogen.
Crystals belonging to space group P212121 were obtained as follows: A 1 μL aliquot of as-isolated protein (10 mg/mL in 100 mM Tris-HCl pH 8) was combined with 1 μL of precipitant solution (1.0–1.1 M ammonium tartrate dibasic pH 7, 6–9% (v/v) glycerol) on a glass cover slide and sealed over a reservoir containing 500 μL of precipitant solution. Diffraction quality crystals grew in 2–10 days. Crystals were soaked in a cryo-protectant solution containing 1.0–1.2 M ammonium tartrate dibasic pH 7, 25% (v/v) glycerol and cryo-cooled in liquid nitrogen.
For oxygen exposure experiments, fully grown crystals were removed from the anaerobic chamber. An aliquot (0.5 μL) of aerobically prepared precipitant solution was added to the drop to initiate equilibration with ambient atmospheric conditions. Crystals were harvested immediately (<20 min), after 2 h and after 2 d by soaking in cryo-protectant and cryo-cooling in liquid nitrogen. Crystals of C45G/T50C Dv CODH from space group P21 did not diffract further than ∼4 Å resolution after being exposed to oxygen for 2 h and 2 days. Crystals from space group P212121 still diffracted well after oxygen exposure and were therefore used for assessment of oxygen-induced D-cluster degradation.
Crystallographic Data Collection, Model Building, and Refinement
All data were collected at the Advanced Photon Source (Argonne, IL) on beamline 24-ID-C at a temperature of 100 K and using a Pilatus 6M pixel detector. Where applicable, native and Fe peak data were collected on the same crystal for a particular sample. Native data were collected at an energy of 12662 eV (0.9792 Å) and Fe peak data at 7130 eV (1.7389 Å). All data were integrated in XDS and scaled in XSCALE.35 Data collection statistics are summarized in Tables S1 and S2.
Structures were solved by molecular replacement (MR) in the program Phaser36 using our previously published structure of WT Dv CODH (PDB ID 6B6V) as a search model. Following MR, 10 cycles of simulated annealing refinement were carried out in Phenix37 to eliminate existing model bias. Refinement of atomic coordinates and atomic displacement parameters (B-factors) was carried out in Phenix using noncrystallographic symmetry (NCS) restraints. Custom parameter files were used to restrain metallocluster geometries during refinement. Models were completed by iterative rounds of model building in Coot38 and refinement in Phenix. In advanced stages of refinement, water molecules were added automatically in Phenix and modified in Coot with placement of additional water molecules until their number was stable. For the structure of as-isolated C45G/T50C Dv CODH, NCS restraints were removed and final stages of refinement included translation, libration, screw (TLS) parametrization with one TLS group per monomer.39 Side chains without visible electron density were truncated to the last atom with electron density and amino acids without visible electron density were not included in the models.
Final refinement of each structure yielded models with low free R-factors, excellent stereochemistry, and small root-mean-square deviations from ideal values for bond lengths and angles. Models were validated using simulated annealing composite omit maps calculated in Phenix. Model geometry was analyzed using MolProbity.40 All refinement and geometry statistics are summarized in Table S1. Figures were generated in PyMOL.41 Crystallography packages were compiled by SBGrid.42
Acknowledgments
This work was supported by National Institutes of Health Grants T32 GM008334 (to E.C.W.) and R01 GM069857 and R35 GM126982 (to C.L.D.) and funded by the Centre National de la Recherche Scientifique, Aix Marseille University and the French Agence Nationale de la Recherche (ANR-15-CE05-0020 and ANR-17-CE11-0027). C.L.D. is a Howard Hughes Medical Institute Investigator and a fellow of the Bioinspired Solar Energy Program, Canadian Institute for Advanced Research. The French authors are part of FrenchBIC. This work is based on research conducted at the Advanced Photon Source on the Northeastern Collaborative Access Team beamlines, which are funded by the National Institute of General Medical Sciences from the NIH (P30 GM124165). This research used resources of the Advanced Photon Source, a U.S. Department of Energy (DOE) Office of Science User Facility operated for the DOE Office of Science by Argonne National Laboratory under Contract No. DE-AC02-06CH11357.
Supporting Information Available
This material is available free of charge on the ACS Publications Web site. The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acscatal.0c00934.
Crystallographic data collection and refinement statistics for C45G/T50C Dv CODH, crystallographic data collection statistics for iron anomalous data, structure of C45G/T50C Dv CODH aligns well with those of other CODHs, B- and C-clusters of as-isolated C45G/T50C Dv CODH, iron anomalous diffraction data for air-exposed C45G/T50C Dv CODH, and stability of the metalloclusters of WT Dv CODH over time in air (PDF)
Author Present Address
+ E.C.W.: California Institute for Quantitative Biosciences, University of California, Berkeley, CA 94720.
Author Contributions
The manuscript was written through contributions of all authors.
The authors declare no competing financial interest.
Notes
Atomic coordinates and structure factors have been deposited in the Protein Data Bank (www.rcsb.org) under the following accession codes: 6VWY (anaerobic C45G/T50C Dv CODH); 6VWZ (<20 minutes air-exposed C45G/T50C Dv CODH); 6VX0 (2 hours air-exposed C45G/T50C Dv CODH); 6VX1 (2 days air-exposed C45G/T50C Dv CODH).
Supplementary Material
References
- Ragsdale S. W. Life with Carbon Monoxide. Crit. Rev. Biochem. Mol. Biol. 2004, 39 (3), 165–195. 10.1080/10409230490496577. [DOI] [PubMed] [Google Scholar]
- Uffen R. L. Anaerobic Growth of a Rhodopseudomonas Species in the Dark with Carbon Monoxide as Sole Carbon and Energy Substrate. Proc. Natl. Acad. Sci. U. S. A. 1976, 73 (9), 3298–3302. 10.1073/pnas.73.9.3298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drennan C. L.; Heo J.; Sintchak M. D.; Schreiter E.; Ludden P. W. Life on Carbon Monoxide: X-Ray Structure of Rhodospirillum rubrum Ni-Fe-S Carbon Monoxide Dehydrogenase. Proc. Natl. Acad. Sci. U. S. A. 2001, 98 (21), 11973–11978. 10.1073/pnas.211429998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobbek H.; Svetlitchnyi V.; Gremer L.; Huber R.; Meyer O. Crystal Structure of a Carbon Monoxide Dehydrogenase Reveals a [Ni-4Fe-5S] Cluster. Science 2001, 293 (5533), 1281–1285. 10.1126/science.1061500. [DOI] [PubMed] [Google Scholar]
- Kumar M.; Lu W. P.; Liu L.; Ragsdale S. W. Kinetic Evidence That Carbon Monoxide Dehydrogenase Catalyzes the Oxidation of Carbon Monoxide and the Synthesis of Acetyl-CoA at Separate Metal Clusters. J. Am. Chem. Soc. 1993, 115 (24), 11646–11647. 10.1021/ja00077a093. [DOI] [Google Scholar]
- Anderson M. E.; Lindahl P. A. Organization of Clusters and Internal Electron Pathways in CO Dehydrogenase from Clostridium thermoaceticum: Relevance to the Mechanism of Catalysis and Cyanide Inhibition. Biochemistry 1994, 33 (29), 8702–8711. 10.1021/bi00195a011. [DOI] [PubMed] [Google Scholar]
- Wittenborn E. C.; Merrouch M.; Ueda C.; Fradale L.; Léger C.; Fourmond V.; Pandelia M.-E.; Dementin S.; Drennan C. L. Redox-Dependent Rearrangements of the NiFeS Cluster of Carbon Monoxide Dehydrogenase. eLife 2018, 7, e39451 10.7554/eLife.39451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domnik L.; Merrouch M.; Goetzl S.; Jeoung J.-H.; Léger C.; Dementin S.; Fourmond V.; Dobbek H. CODH-IV: A High-Efficiency CO-Scavenging CO Dehydrogenase with Resistance to O2. Angew. Chem., Int. Ed. 2017, 56 (48), 15466–15469. 10.1002/anie.201709261. [DOI] [PubMed] [Google Scholar]
- Darnault C.; Volbeda A.; Kim E. J.; Legrand P.; Vernède X.; Lindahl P. A.; Fontecilla-Camps J. C. Ni-Zn-[Fe4-S4] and Ni-Ni-[Fe4-S4] Clusters in Closed and Open Subunits of Acetyl-CoA Synthase/carbon Monoxide Dehydrogenase. Nat. Struct. Mol. Biol. 2003, 10 (4), 271–279. 10.1038/nsb912. [DOI] [PubMed] [Google Scholar]
- Doukov T. I.; Iverson T. M.; Seravalli J.; Ragsdale S. W.; Drennan C. L. A Ni-Fe-Cu Center in a Bifunctional Carbon Monoxide Dehydrogenase/acetyl-CoA Synthase. Science 2002, 298 (5593), 567–572. 10.1126/science.1075843. [DOI] [PubMed] [Google Scholar]
- Ensign S. A.; Campbell M. J.; Ludden P. W. Activation of the Nickel-Deficient Carbon Monoxide Dehydrogenase from Rhodospirillum rubrum: Kinetic Characterization and Reductant Requirement. Biochemistry 1990, 29 (8), 2162–2168. 10.1021/bi00460a029. [DOI] [PubMed] [Google Scholar]
- Svetlitchnyi V.; Peschel C.; Acker G.; Meyer O. Two Membrane-Associated NiFeS-Carbon Monoxide Dehydrogenases from the Anaerobic Carbon-Monoxide-Utilizing Eubacterium Carboxydothermus hydrogenoformans. J. Bacteriol. 2001, 183 (17), 5134–5144. 10.1128/JB.183.17.5134-5144.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ragsdale S. W.; Clark J. E.; Ljungdahl L. G.; Lundie L. L.; Drake H. L. Properties of Purified Carbon Monoxide Dehydrogenase from Clostridium thermoaceticum a Nickel, Iron-Sulfur Protein. J. Biol. Chem. 1983, 258 (4), 2364–2369. [PubMed] [Google Scholar]
- Hadj-Saïd J.; Pandelia M.-E.; Léger C.; Fourmond V.; Dementin S. The Carbon Monoxide Dehydrogenase from Desulfovibrio vulgaris. Biochim. Biophys. Acta, Bioenerg. 2015, 1847 (12), 1574–1583. 10.1016/j.bbabio.2015.08.002. [DOI] [PubMed] [Google Scholar]
- Merrouch M.; Hadj-Saïd J.; Domnik L.; Dobbek H.; Léger C.; Dementin S.; Fourmond V. O2 Inhibition of Ni-Containing CO Dehydrogenase Is Partly Reversible. Chem. - Eur. J. 2015, 21 (52), 18934–18938. 10.1002/chem.201502835. [DOI] [PubMed] [Google Scholar]
- Abou Hamdan A.; Burlat B.; Gutiérrez-Sanz O.; Liebgott P.-P.; Baffert C.; De Lacey A. L.; Rousset M.; Guigliarelli B.; Léger C.; Dementin S. O2-Independent Formation of the Inactive States of NiFe Hydrogenase. Nat. Chem. Biol. 2013, 9 (1), 15–17. 10.1038/nchembio.1110. [DOI] [PubMed] [Google Scholar]
- Wang V. C.-C.; Islam S. T. A.; Can M.; Ragsdale S. W.; Armstrong F. A. Investigations by Protein Film Electrochemistry of Alternative Reactions of Nickel-Containing Carbon Monoxide Dehydrogenase. J. Phys. Chem. B 2015, 119 (43), 13690–13697. 10.1021/acs.jpcb.5b03098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benvenuti M.; Meneghello M.; Guendon C.; Jacq-Bailly A.; Jeoung J.-H.; Dobbek H.; Léger C.; Fourmond V.; Dementin S. The Two CO-Dehydrogenases of Thermococcus Sp. AM4. Biochim. Biophys. Acta, Bioenerg. 2020, 1861, 148188. 10.1016/j.bbabio.2020.148188. [DOI] [PubMed] [Google Scholar]
- Del Barrio M.; Sensi M.; Orain C.; Baffert C.; Dementin S.; Fourmond V.; Léger C. Electrochemical Investigations of Hydrogenases and Other Enzymes That Produce and Use Solar Fuels. Acc. Chem. Res. 2018, 51 (3), 769–777. 10.1021/acs.accounts.7b00622. [DOI] [PubMed] [Google Scholar]
- Imlay J. A. Pathways of Oxidative Damage. Annu. Rev. Microbiol. 2003, 57, 395–418. 10.1146/annurev.micro.57.030502.090938. [DOI] [PubMed] [Google Scholar]
- Imlay J. A. Iron-Sulphur Clusters and the Problem with Oxygen. Mol. Microbiol. 2006, 59 (4), 1073–1082. 10.1111/j.1365-2958.2006.05028.x. [DOI] [PubMed] [Google Scholar]
- Beinert H.; Holm R. H.; Münck E. Iron-Sulfur Clusters: Nature’s Modular, Multipurpose Structures. Science 1997, 277 (5326), 653–659. 10.1126/science.277.5326.653. [DOI] [PubMed] [Google Scholar]
- Kubas A.; Orain C.; De Sancho D.; Saujet L.; Sensi M.; Gauquelin C.; Meynial-Salles I.; Soucaille P.; Bottin H.; Baffert C.; Fourmond V.; Best R. B.; Blumberger J.; Léger C. Mechanism of O2 Diffusion and Reduction in FeFe Hydrogenases. Nat. Chem. 2017, 9 (1), 88–95. 10.1038/nchem.2592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esselborn J.; Kertess L.; Apfel U.-P.; Hofmann E.; Happe T. Loss of Specific Active-Site Iron Atoms in Oxygen-Exposed [FeFe]-Hydrogenase Determined by Detailed X-Ray Structure Analyses. J. Am. Chem. Soc. 2019, 141 (44), 17721–17728. 10.1021/jacs.9b07808. [DOI] [PubMed] [Google Scholar]
- Swanson K. D.; Ratzloff M. W.; Mulder D. W.; Artz J. H.; Ghose S.; Hoffman A.; White S.; Zadvornyy O. A.; Broderick J. B.; Bothner B.; King P. W.; Peters J. W. [FeFe]-Hydrogenase Oxygen Inactivation Is Initiated at the H Cluster 2Fe Subcluster. J. Am. Chem. Soc. 2015, 137 (5), 1809–1816. 10.1021/ja510169s. [DOI] [PubMed] [Google Scholar]
- Lenz O.; Lauterbach L.; Frielingsdorf S. O2-Tolerant [NiFe]-Hydrogenases of Ralstonia eutropha H16: Physiology, Molecular Biology, Purification, and Biochemical Analysis. Methods Enzymol. 2018, 613, 117–151. 10.1016/bs.mie.2018.10.008. [DOI] [PubMed] [Google Scholar]
- Li H.; Münchberg U.; Oughli A. A.; Buesen D.; Lubitz W.; Freier E.; Plumeré N. Suppressing Hydrogen Peroxide Generation to Achieve Oxygen-Insensitivity of a [NiFe] Hydrogenase in Redox Active Films. Nat. Commun. 2020, 11 (1), 920. 10.1038/s41467-020-14673-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caserta G.; Papini C.; Adamska-Venkatesh A.; Pecqueur L.; Sommer C.; Reijerse E.; Lubitz W.; Gauquelin C.; Meynial-Salles I.; Pramanik D.; Artero V.; Atta M.; Del Barrio M.; Faivre B.; Fourmond V.; Léger C.; Fontecave M. Engineering an [FeFe]-Hydrogenase: Do Accessory Clusters Influence O2 Resistance and Catalytic Bias?. J. Am. Chem. Soc. 2018, 140 (16), 5516–5526. 10.1021/jacs.8b01689. [DOI] [PubMed] [Google Scholar]
- Holm R. H.; Lo W. Structural Conversions of Synthetic and Protein-Bound Iron-Sulfur Clusters. Chem. Rev. 2016, 116 (22), 13685–13713. 10.1021/acs.chemrev.6b00276. [DOI] [PubMed] [Google Scholar]
- Wittenborn E. C.; Cohen S. E.; Merrouch M.; Léger C.; Fourmond V.; Dementin S.; Drennan C. L. Structural Insight into Metallocofactor Maturation in Carbon Monoxide Dehydrogenase. J. Biol. Chem. 2019, 294 (35), 13017–13026. 10.1074/jbc.RA119.009610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dementin S.; Burlat B.; Fourmond V.; Leroux F.; Liebgott P.-P.; Abou Hamdan A.; Léger C.; Rousset M.; Guigliarelli B.; Bertrand P. Rates of Intra- and Intermolecular Electron Transfers in Hydrogenase Deduced from Steady-State Activity Measurements. J. Am. Chem. Soc. 2011, 133 (26), 10211–10221. 10.1021/ja202615a. [DOI] [PubMed] [Google Scholar]
- Inoue M.; Nakamoto I.; Omae K.; Oguro T.; Ogata H.; Yoshida T.; Sako Y. Structural and Phylogenetic Diversity of Anaerobic Carbon-Monoxide Dehydrogenases. Front. Microbiol. 2019, 9, 3353. 10.3389/fmicb.2018.03353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heidelberg J. F.; Seshadri R.; Haveman S. A.; Hemme C. L.; Paulsen I. T.; Kolonay J. F.; Eisen J. A.; Ward N.; Methe B.; Brinkac L. M.; Daugherty S. C.; Deboy R. T.; Dodson R. J.; Durkin A. S.; Madupu R.; Nelson W. C.; Sullivan S. A.; Fouts D.; Haft D. H.; Selengut J.; Peterson J. D.; Davidsen T. M.; Zafar N.; Zhou L.; Radune D.; Dimitrov G.; Hance M.; Tran K.; Khouri H.; Gill J.; Utterback T. R.; Feldblyum T. V.; Wall J. D.; Voordouw G.; Fraser C. M. The Genome Sequence of the Anaerobic, Sulfate-Reducing Bacterium Desulfovibrio vulgaris Hildenborough. Nat. Biotechnol. 2004, 22 (5), 554–559. 10.1038/nbt959. [DOI] [PubMed] [Google Scholar]
- Ramel F.; Brasseur G.; Pieulle L.; Valette O.; Hirschler-Réa A.; Fardeau M. L.; Dolla A. Growth of the Obligate Anaerobe Desulfovibrio vulgaris Hildenborough under Continuous Low Oxygen Concentration Sparging: Impact of the Membrane-Bound Oxygen Reductases. PLoS One 2015, 10 (4), e0123455 10.1371/journal.pone.0123455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabsch W. XDS. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2010, 66, 125–132. 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy A. J.; Grosse-Kunstleve R. W.; Adams P. D.; Winn M. D.; Storoni L. C.; Read R. J. Phaser Crystallographic Software. J. Appl. Crystallogr. 2007, 40, 658–674. 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams P. D.; Afonine P. V.; Bunkóczi G.; Chen V. B.; Davis I. W.; Echols N.; Headd J. J.; Hung L.-W.; Kapral G. J.; Grosse-Kunstleve R. W.; McCoy A. J.; Moriarty N. W.; Oeffner R.; Read R. J.; Richardson D. C.; Richardson J. S.; Terwilliger T. C.; Zwart P. H. PHENIX: A Comprehensive Python-Based System for Macromolecular Structure Solution. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2010, 66, 213–221. 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P.; Lohkamp B.; Scott W. G.; Cowtan K. Features and Development of Coot. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2010, 66, 486–501. 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Painter J.; Merritt E. A. Optimal Description of a Protein Structure in Terms of Multiple Groups Undergoing TLS Motion. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2006, 62, 439–450. 10.1107/S0907444906005270. [DOI] [PubMed] [Google Scholar]
- Chen V. B.; Arendall W. B. 3rd; Headd J. J.; Keedy D. A.; Immormino R. M.; Kapral G. J.; Murray L. W.; Richardson J. S.; Richardson D. C. MolProbity: All-Atom Structure Validation for Macromolecular Crystallography. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2010, 66, 12–21. 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrodinger L. L. C.PyMOL Molecular Graphics System, version 1.8; 2015.
- Morin A.; Eisenbraun B.; Key J.; Sanschagrin P. C.; Timony M. A.; Ottaviano M.; Sliz P. Collaboration Gets the Most out of Software. eLife 2013, 2, e01456 10.7554/eLife.01456. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.