Staphylococcus aureus is a major cause of bovine intramammary infections, leading to significant economic losses to dairy industry in Canada and worldwide. There is a lack of knowledge regarding genetic diversity, the presence of antimicrobial resistance (AMR), and virulence genes for S. aureus isolated from bovine milk in Canada. Based on whole-genome sequencing and genomic analysis, we have determined the phylogeny and diversity of S. aureus in bovine milk and concluded that it had a large accessory genome, limited distribution of AMR genes, variable VF gene profiles and sequence types (ST), and clonal complex (CC)-specific pathogenic potentials. Comprehensive information on the population structure, as well as the virulence and resistance characteristics of S. aureus from bovine milk, will allow for source attribution, risk assessment, and improved therapeutic approaches in cattle.
Keywords: intramammary infection, Staphylococcus aureus, whole-genome sequencing, virulence factors, antimicrobial resistance genes, sequence types, clonal complex, Spa types, AMR, adherence, intramammary infection, mastitis
ABSTRACT
Staphylococcus aureus causes persistent clinical and subclinical bovine intramammary infections (IMI) worldwide. However, there is a lack of comprehensive information regarding genetic diversity, the presence of antimicrobial resistance (AMR), and virulence genes for S. aureus in bovine milk in Canada. Here, we performed whole-genome sequencing (WGS) of 119 Canadian bovine milk S. aureus isolates and determined they belonged to 8 sequence types (ST151, ST352, ST351, ST2187, ST2270, ST126, ST133, and ST8), 5 clonal complexes (CC151, CC97, CC126, CC133, and CC8), and 18 distinct Spa types. Pan-, core, and accessory genomes were composed of 6,340, 1,279, and 2,431 genes, respectively. Based on phenotypic screening for AMR, resistance was common against beta-lactams (19% of isolates) and sulfonamides (7% of isolates), whereas resistance against pirlimycin, tetracycline, ceftiofur, and erythromycin and to the combination of penicillin and novobiocin was uncommon (3, 3, 3, 2, and 2% of all isolates, respectively). We also determined distributions of 191 virulence factors (VFs) in 119 S. aureus isolates after classifying them into 5 functional categories (adherence [n = 28], exoenzymes [n = 21], immune evasion [n = 20], iron metabolism [n = 29], and toxins [n = 93]). Additionally, we calculated the pathogenic potential of distinct CCs and STs and determined that CC151 (ST151 and ST351) had the highest pathogenic potential (calculated by subtracting core-VFs from total VFs), followed by CC97 (ST352 and ST2187) and CC126 (ST126 and ST2270), potentially linked to their higher prevalence in bovine IMI worldwide. However, there was no statistically significant link between the presence of VF genes and mastitis.
IMPORTANCE Staphylococcus aureus is a major cause of bovine intramammary infections, leading to significant economic losses to dairy industry in Canada and worldwide. There is a lack of knowledge regarding genetic diversity, the presence of antimicrobial resistance (AMR), and virulence genes for S. aureus isolated from bovine milk in Canada. Based on whole-genome sequencing and genomic analysis, we have determined the phylogeny and diversity of S. aureus in bovine milk and concluded that it had a large accessory genome, limited distribution of AMR genes, variable VF gene profiles and sequence types (ST), and clonal complex (CC)-specific pathogenic potentials. Comprehensive information on the population structure, as well as the virulence and resistance characteristics of S. aureus from bovine milk, will allow for source attribution, risk assessment, and improved therapeutic approaches in cattle.
INTRODUCTION
Staphylococcus aureus can cause acute and chronic infections associated with high morbidity in a wide variety of hosts, including humans and farmed and companion animals (1–3). In the dairy industry, S. aureus causes persistent clinical and subclinical intramammary infections (IMI) (4). Staphylococcus aureus IMIs, often spread during the milking process, can result in chronic infections, often persisting for the life of the animal (4–6), causing tissue damage, reduced milk quality and production, and increased individual cow and bulk tank milk somatic cell count (SCC) (4, 7, 8). Staphylococcus aureus IMI reduces animal health and welfare and poses a biosafety hazard for raw dairy products, leading to substantial economic losses in the dairy industry worldwide (4, 9). The pathogenesis of S. aureus mastitis is complex: it starts with colonization of the teat end and infection subsequently spreading into the intramammary space, either by progressive colonization or changes in intramammary pressure caused by the milking machines (4, 10). In the mammary alveolus, S. aureus attaches to and internalizes into mammary epithelial cells, where it multiplies and establishes a chronic IMI (10–12). Mechanisms by which S. aureus IMI is established and maintained in dairy cows are not fully understood but generally involve both host immune escape and modulation strategies (13, 14). In addition, a number of studies have suggested biofilm formation as a mechanism by which S. aureus establishes and maintains itself in dairy cows (15–17). Genetic regulatory circuits that control S. aureus adaptation and virulence are complex, involving signals from the external environment to modulate the production of a wide arsenal of cell surface and extracellular proteins, known as virulence factors (VFs) (13, 14, 18).
Extensive use of antimicrobials to treat bovine mastitis and for dry cow therapy exerts selective pressure on S. aureus, leading to the emergence and spread of antimicrobial-resistant (AMR) S. aureus strains (19–22). In particular, the emergence of multiple-drug-resistant (MDR) strains can become a major challenge in the treatment of bovine mastitis and is a growing concern for public health (23–26), as methicillin-resistant S. aureus (MRSA) has been reported in human and veterinary medicine (25).
During the past decade, the molecular epidemiology of S. aureus IMI in dairy cattle has been studied using various methods, including electrophoretic comparison techniques, such as pulsed-field gel electrophoresis (PFGE) (27), random amplification of polymorphic DNA (RAPD) analysis (28), multilocus enzyme electrophoresis (MLEE) (29), and sequence-based typing schemes, such as multiple-locus sequence typing (MLST) (30), staphylococcal protein A (spa) typing (31), and multiple-locus VNTR (variable number of tandem repeats) analysis (MLVA) (32). Although these methods were helpful in typing S. aureus strains, they failed to reveal fine details of genetic differences between strains. However, whole-genome sequencing (WGS) of bacterial genomes has become the preferred method to understand microevolution, phylogenies, and inter- and intraspecies differences (33–35). By providing definitive genotype information, WGS offers the highest practical resolution for matching strain diversity with resistance and virulence determinants (36–38).
Objectives of this study were to use the WGS data of 119 bovine S. aureus isolates to elucidate (i) molecular types (MLST and spa types), (ii) genetic diversity and evolutionary relationships, by constructing pan- and core genomes and phylogenetic trees based on multilocus sequence analysis (MLSA) and single-nucleotide polymorphisms (SNPs), (iii) AMR gene (ARGs) profiles, (iv) distributions of VFs, and (v) associations among genotypes, ARGs, and VF-derived potential to cause mastitis.
RESULTS
Identification and distribution of STs and spa types.
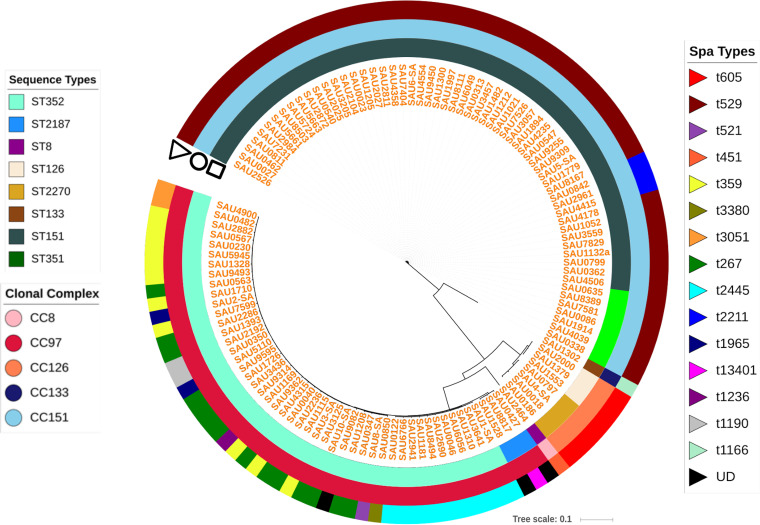
MLST analysis grouped 119 S. aureus isolates (Table 1) into 8 STs and 5 distinct CCs (Fig. 1). The majority of isolates were assigned to ST151 (n = 54 [45%]), followed by ST352 (n = 46 [39%]). For the remaining isolates, ST351, ST2187, ST2270, and ST126 had 7, 3, 4, and 3 isolates, respectively, whereas ST133 and ST8 were singletons (Fig. 1). The eBURST analysis of STs clustered ST151 and ST351 into CC151 (51% isolates), ST2187 and ST352 into CC97 (41% isolates), and ST2270 and ST126 into CC126 (6.4%), whereas ST133 and ST8 isolates were part of CC133 and CC8, respectively (Fig. 1). Spa typing identified 119 S. aureus isolates into 18 distinct spa types (Fig. 1).
TABLE 1.
Number of isolates and unique herds grouped by SCC level and region of origin
| Origin | No. of isolates (no. of unique herds) from randomly selected, nonclinical cows |
||||
|---|---|---|---|---|---|
| Low SCC (≤150,00 cells/ml) | Medium SCC (150,00–250,000 cells/ml) | High SCC (>250,000 cells/ml) | Clinical mastitis | Total | |
| Alberta | 1 (1) | 0 | 5 (4) | 5 (4) | 11 (6) |
| Ontario | 3 (3) | 1 (1) | 16 (10) | 18 (12) | 38 (19) |
| Quebec | 2 (2) | 0 | 20 (9) | 22 (10) | 44 (15) |
| Atlantic Canada | 5 (4) | 0 | 2 (2) | 19 (9) | 26 (10) |
| Total | 11 (10) | 1 (1) | 43 (25) | 64 (35) | 119 (50) |
FIG 1.
Core SNP-based phylogenetic tree of distribution of sequence types (STs), clonal complexes (CCs), and spa types. The SNP tree indicates phylogenetic relationships among 119 Staphylococcus aureus isolates recovered from bovine milk. This tree was constructed with Parsnp v1.2 (123) and was overlayed with information regarding STs, CCs, and spa types using iTOL v4 (147). The first ring indicates the distribution of 119 isolates into 8 distinct STs (ST151, ST352, ST351, ST2187, ST2270, ST126, ST133, and ST8). The second (middle) ring indicates grouping of STs into 5 CCs (CC151, CC97, CC126, CC133, and CC8), whereas the third (outer) ring indicates distribution of spa types. For 21 isolates, a spa type reference was not available in the reference database; hence, spa types for those could not be determined. These isolates were labeled UD (undetermined) in the outer circle.
Phylogenetic analyses.
The majority of interrelationships at major clades were similar and fairly consistent among phylogenetic trees constructed using a core set of proteins, PhyloPhlAn, core-SNPs, and MLSA. However, many relationships were observed toward the tip of the trees (data not shown). All trees had branching of S. aureus isolates into 8 nodes and 5 main clades that corresponded well with STs and CCs predicted by eBURST analysis (Fig. 1).
Pan-genome analysis.
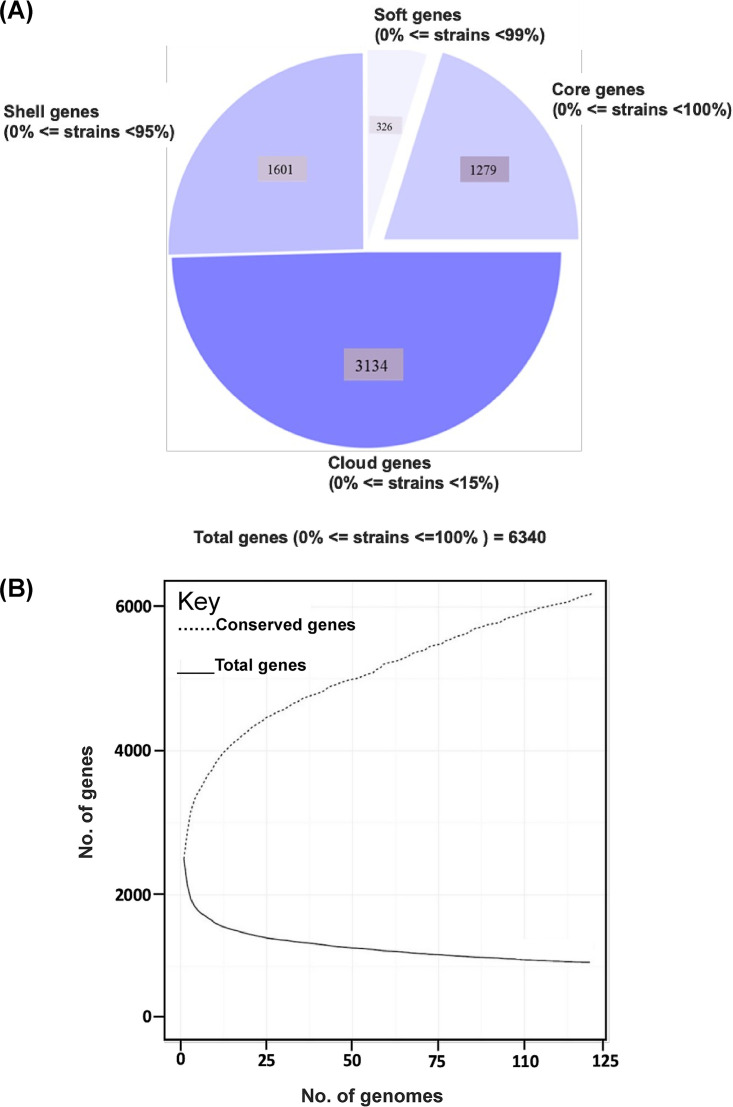
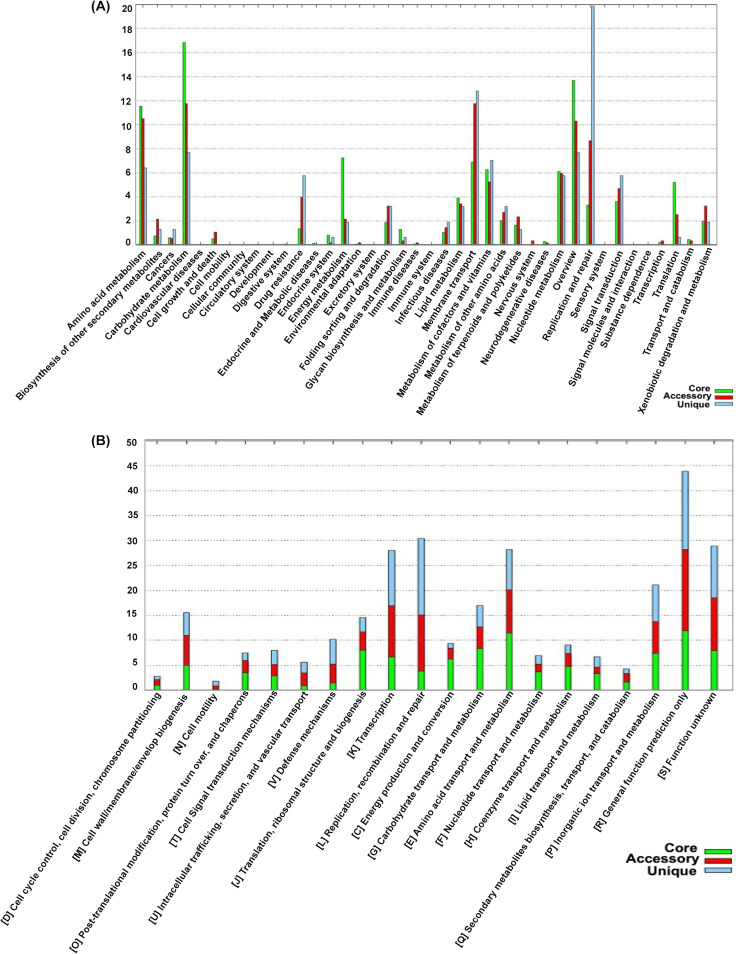
The pan-genome of 119 S. aureus isolates tested in this study had 6,340 genes. The core genome (shared by >99% of S. aureus isolates) consisted of 1,279 genes. The accessory genome (genes in >2 isolates but not in all) consisted of 2,431 genes, and the unique genome was composed of 2,845 genes. The soft core, shell, and cloud contained 326, 1,601, and 3,134 genes, respectively (Fig. 2). Based on a rarefaction curve after inclusion of ∼90 (75%) isolates into the analyses, the number of core genes remained fairly constant at ∼1,300 genes, whereas the total number of genes in the pan-genome continued to increase (Fig. 2). Functional annotation of genes in the pan-genome performed using the COG and KEGG databases revealed a distribution of functional categories among 3 pan-genome sets (Fig. 3). The largest fraction of the core genome consisted of genes involved in housekeeping processes, include transcription, translation, ribosomal structure and biogenesis, and RNA processing and modification, whereas a smaller fraction of housekeeping gene content was in the accessory and unique genomes.
FIG 2.
Pan-genome of 119 S. aureus isolates. (A) Distribution of pan-genome into core, soft-core, shell, and cloud categories. (B) Changes in the total number of genes versus conserved genes upon addition of each individual S. aureus genome.
FIG 3.
Pan-genome characterization based on distribution of functional categories. Distribution of pan-genome into functional categories obtained after comparing the pan-genome with KEGG (A) and COG (B) reference databases.
Distributions and associations of virulence genes.
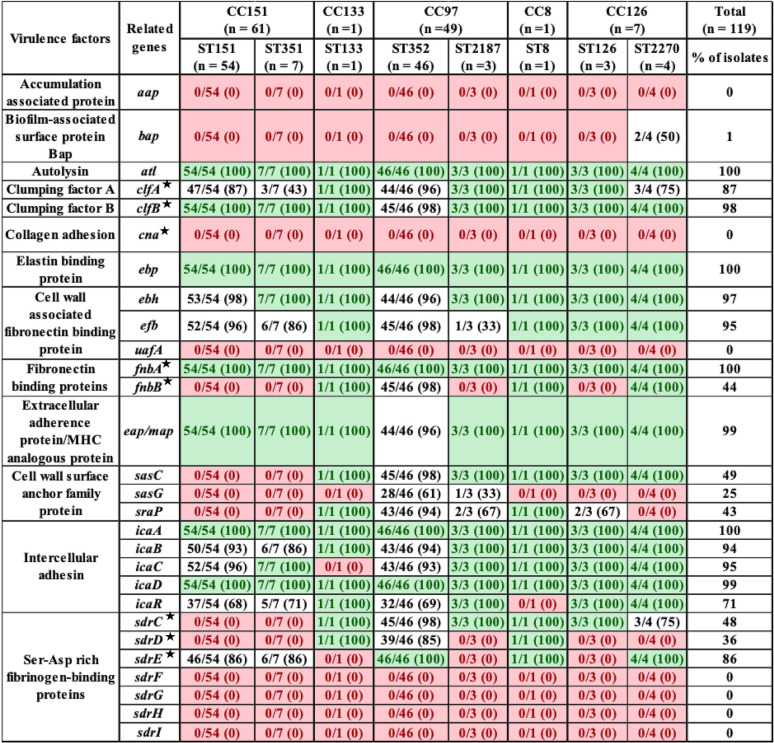
Among the 28 adherence-related genes, 12 genes (alt, clfA, clfB, ebp, ebh, efb, fnbA, eap-map, icaA, icaB, icaC, and icaD) were present in ≥90% of S. aureus isolates (Table 2). Seven adherence genes (aap, cna, uafA, sdrF, sdrG, sdrH, and sdrI) were absent from all isolates. The biofilm-associated gene, bap, was detected in only 2 isolates from ST2270. Some of these 28 genes had ST-specific distributions. For example, fnbB was detected in ST8, ST133, ST352, and ST2270, sasC was detected in all STs except CC151 (ST151 and ST351), and sasG was detected only in CC5 (ST352 and ST2187). The sraP was detected in ST8, ST133, ST352, ST2187, and ST126, whereas sdrC was detected in ST133, ST352, ST2187, and ST2270 and sdrD was detected in ST8, ST133, and ST352 (Table 2).
TABLE 2.
Distribution of adherence related virulence factors in S. aureus isolates from bovine milk, grouped according to ST and CCa
A star indicates genes that encode microbial surface component recognizing adhesive matrix molecules (MSCRAMMs). Light green indicates gene was in all isolates within a given ST; red indicates a gene that was not identified in any isolates within a given ST. Values in parentheses indicate the percentage of isolates that contained a particular VF.
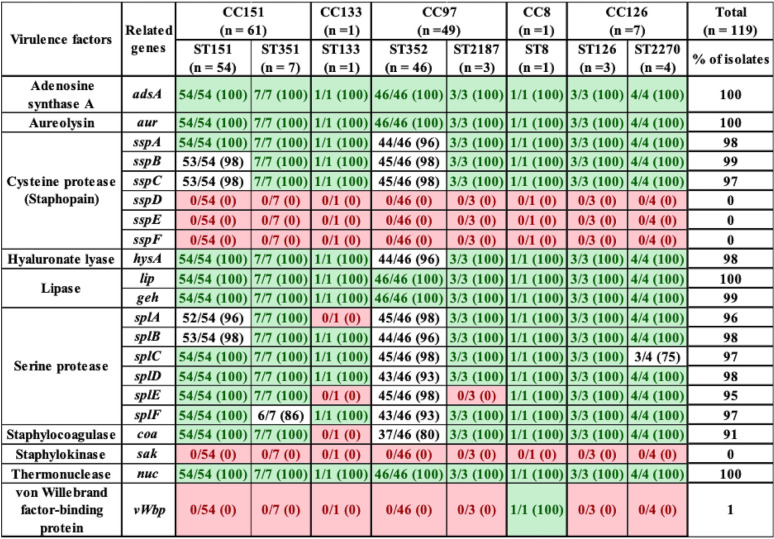
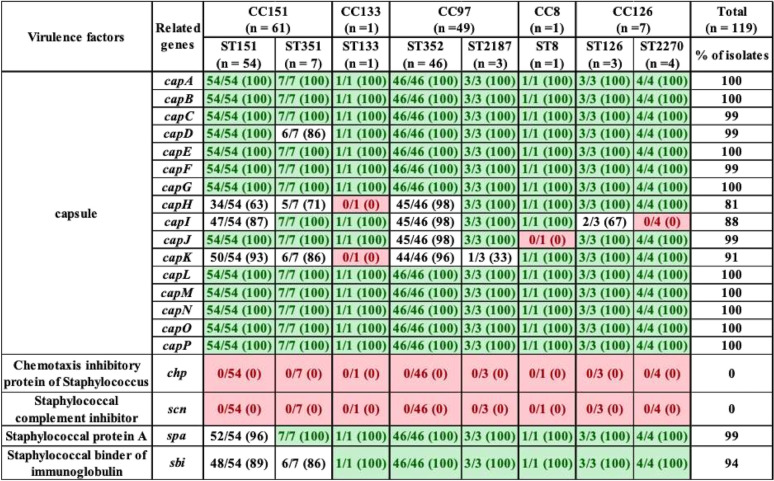
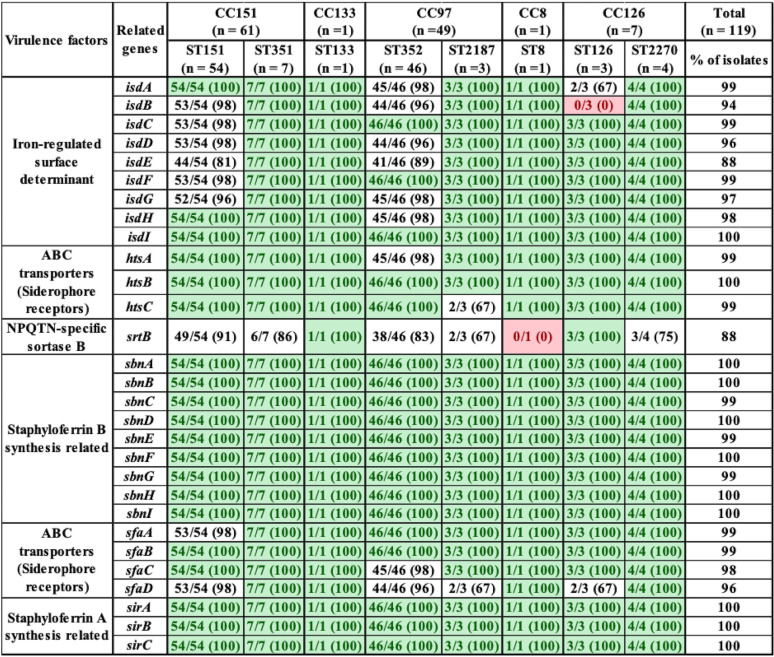
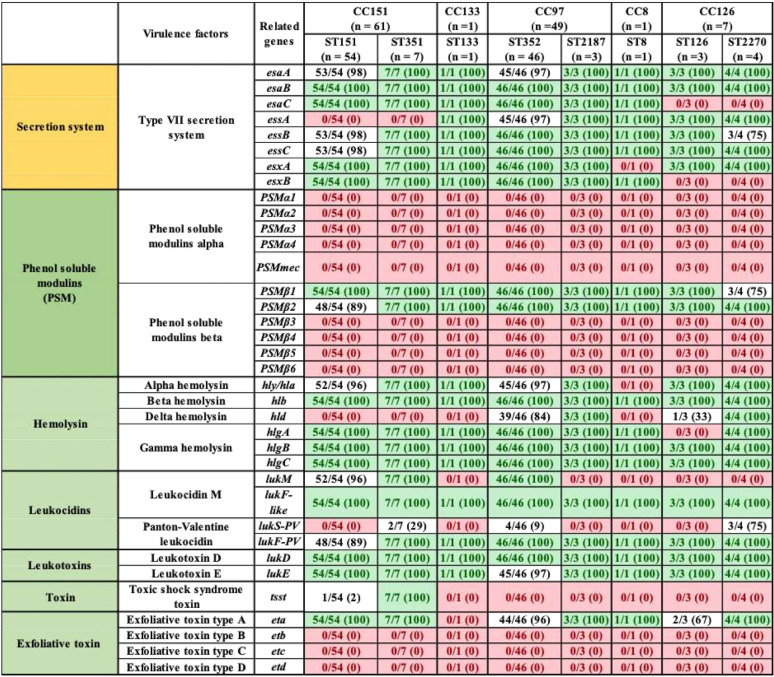
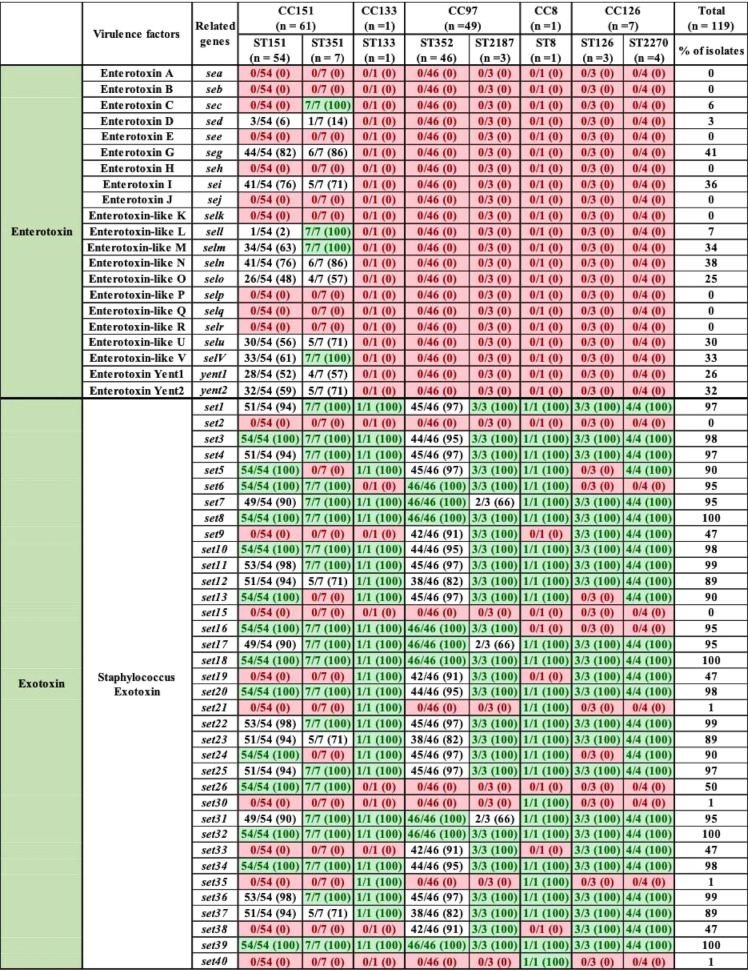
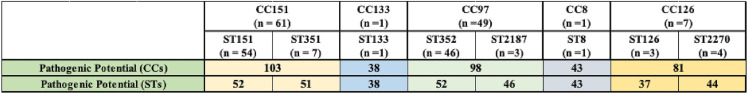
With respect to the 21 exoenzymes, sspD, sspE, sspF, and sak were not detected in any isolates, but adsA, aur, sspA, sspB, sspC, hysA, lip, geh, splA, splB, splC, splD, splE, splF, coa, and vWbp were detected in 90 to 100% of isolates. In contrast, vWbp was detected in ST8 only (represented by 1 isolate) (Table 3). Exoenzymes geh and splA were not detected in ST133. Similarly, splE was detected in ST2187 and ST133, and coa was not present in ST133 (Table 3). With respect to the 20 host immune evasion genes, capsular genes (capA to capP) were detected in all STs, except capH and capK were not detected in ST133 and capJ and capI were not detected in ST8 and ST2270, respectively (Table 4). Chemotaxis inhibitory protein (chp) and staphylococcal complement inhibitor (scn) was not detected in any isolate. Staphylococcal protein A (spa) and sbi were detected in 100% and 94% of isolates (Table 4). The identification and distribution of genes related to iron uptake and metabolism were uniform among S. aureus isolates, with all 29 genes detected in almost all isolates, except isdB and srtB were not detected in ST126 and ST8, respectively (Table 5). Type VII secretion system genes (esaA, esaB, esaC, essA, essB, essC, esxA, and esxB) were detected in most isolates, although esaC and essA were exclusively absent from CC126 and CC151 and essC and esxA were not detected in CC8 (Table 6). With respect to phenol-soluble modulins (PSM), except for PSMβ1 and PSMβ2, which were detected in 99% and 96% of all S. aureus isolates, other PSMβ genes (PSMβ3, PSMβ4, PSMβ5, and PSMβ6) and PSMα genes (PSMα1, PSMα2, PSMα3, PSMα4, and PSMmec) were not detected (Table 6). Among hemolysins, alpha (hla), beta (hlb), and gamma (hlgA, hlgB, and hlgC) hemolysins were detected in almost all isolates, except for hla and hlgA, which were not detected in ST8 and ST126, respectively (Table 6), and hld was detected in CC5 (ST352 and ST2187) and CC126 (ST126 and ST2187). Among leukocidins, lukF-like was detected in all isolates, whereas lukM was only present in CC151 and ST352. Although lukF-PV was in all S. aureus isolates, lukS-PV was detected only in ST1351 (29%), ST352 (9%), and ST2270 (75%). Concerning leukotoxins, both lukD and lukE were detected in 99% of isolates. The toxic shock syndrome toxin (tsst) gene was detected in ST351 (7/7) and ST151 (1/54) only (Table 6). Concerning exfoliative toxins, eta was identified in all isolates except for ST133, whereas etb, etc, and etd were not detected (Table 6). Enterotoxins (n = 21) were not detected in most of our 119 isolates, except in ST151 and ST351, which contained (sed, seg, sei, sell, selm, seln, selo, selu, selv, yent1, and yent2) (Table 7). Most exotoxin (set) genes (n = 34) were uniformly distributed and present in ≥90% of isolates (Table 7), except set2, set15, set21, set30, set35, and set40. One of the set genes (set26) had clone-specific distribution and was detected in 100% of isolates of CC151 (ST151 and ST351). Apart from establishing the distribution of VFs among 119 S. aureus isolates, the total number of genes unique to each ST was calculated. On average, 127 VF genes were detected in all STs, with the highest number of genes (135 VFs) detected in ST151 and ST352, followed by ST351 and ST2187, which contained 134 and 129 VF genes, respectively. Slightly fewer genes were detected in ST2270 (n = 127) and ST8 (n = 126). The fewest genes were in ST133 (n = 121) and ST126 (n = 120). A large number (83/191) of VFs detected in all STs were defined as core VFs. The pathogenic potential of various STs and CCs, calculated by subtracting core VFs (commonly detected among all STs or CCs) from total VFs present in particular STs or CCs, is shown (Table 8).
TABLE 3.
Distribution of exoenzymes in S. aureus isolates grouped according to ST and CCa
Light green indicates gene was in all isolates within a given ST; red indicates a gene that was not identified in any isolates within a given ST. Values in parentheses indicate the percentage of isolates that contained a particular VF.
TABLE 4.
Distribution of host immune evasion genes in S. aureus isolates grouped according to ST and CCa
Light green entries indicate gene was in all isolates within a given ST; red entries indicate a gene that was not identified in any isolates within a given ST. Values in parentheses indicate the percentage of isolates that contained a particular VF.
TABLE 5.
Distribution of iron acquisition- and metabolism-related genes in S. aureus isolates grouped according to ST and CCa
Light green entries indicate gene was in all isolates within a given ST; red entries indicate a gene that was not identified in any isolates within a given ST. Values in parentheses indicate the percentage of isolates that contained a particular VF.
TABLE 6.
Distribution of different toxin system genes in S. aureus isolates grouped according to ST and CCa
Light green entries indicate gene was in all isolates within a given ST; red entries indicate a gene that was not identified in any isolates within a given ST. Values in parentheses indicate the percentage of isolates that contained a particular VF.
TABLE 7.
Distribution of different entero- and exotoxins in bovine S. aureus isolates grouped according to ST and CCa
Light green entries indicate gene was in all isolates within a given ST; red entries indicate a gene that was not identified in any isolates within a given ST. Values in parentheses indicate the percentage of isolates that contained a particular VF.
TABLE 8.
Pathogenic potential of clonal complexes (CC) and sequence types (STs)
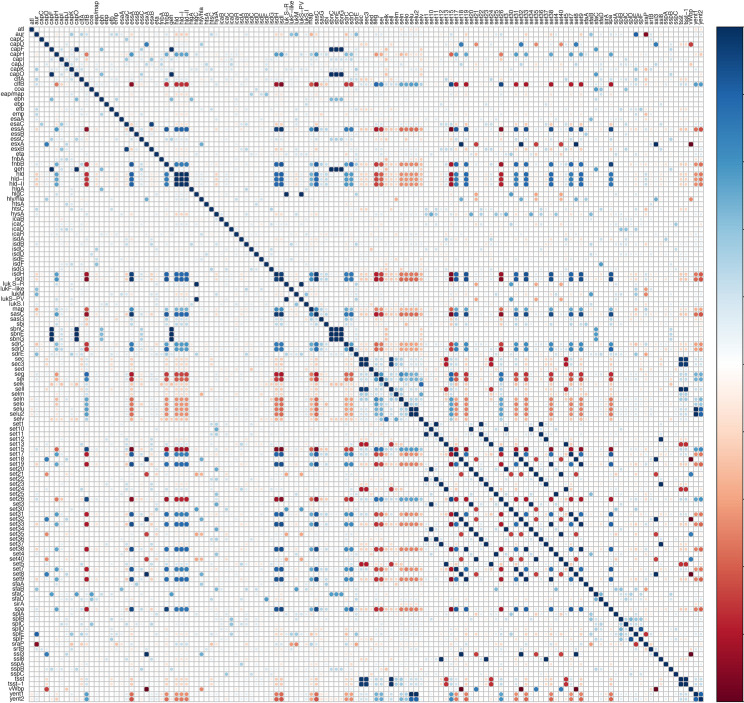
Relationships among and between VFs from 5 categories were investigated using an association plot (Fig. 4). Most associations among VF genes were neutral or very weak. However, there were some strong positive and strong negative associations. For instance, staphyloferrin B synthesis-related genes sbnC, sbnE, and sbnG were strongly positively associated with each other (Fig. 4); these genes also had strong associations with capF, clfB, ebh, essC, geh, and isdH. Similarly, sdrC and sdrD had positive associations with each other and with essA, fnb, and geh (Fig. 4). Additionally, some interesting patterns of associations were observed among toxin genes. For instance, seg, sei, sell, selm, seln, selo, selu, and selv were positively associated with each other and with set26, yent1, and yent2 but had strong negative associations with essA, fnbB, hld, sasC, sasG, srdC, and srdD (Fig. 4). Similarly, many distinctive positive and negative associations were also present among set genes. Graphical representation of these and other associations is shown (Fig. 4).
FIG 4.
Pairwise associations of virulence factors. Pairwise associations among all virulence genes were detected in 119 bovine Staphylococcus aureus isolates. Associations were computed using phi coefficient. Colors represent type of association (blue, positive; red, negative).
Phenotypic and genotypic AMR.
Phenotypic resistance was common against beta-lactams (19% of isolates) and sulfonamides (7% of isolates). However, no isolate was resistant to oxacillin or cephalotin. Resistance against pirlimycin, tetracycline, ceftiofur, and erythromycin and to the combination of penicillin and novobiocin was uncommon (3, 3, 3, 2, and 2% of all isolates, respectively). The most common genotypic AMR markers were (i) presence of AMR-associated residues in the dihydropteroate synthase gene deduced amino acid sequence (folP gene; all sequenced isolates, ranging from 1 to 11 residues); (ii) chromosomally encoded efflux pump MepA, represented by the mepA gene (all isolates), and tet(38) (99% of isolates); (iii) multidrug export protein SAV1866 (59% of isolates); and (iv) multidrug efflux pump NorA, represented by the norA gene (58% of isolates). No mutations previously described as associated with AMR in S. aureus were detected in the quinolone resistance-determining region of gyrA, gyrB, parC, and parE genes. Similarly, no mutations were detected for rpoB, rpoC, mprF, and cls genes. Regarding acquired genetic mechanisms, the blaZ gene was present in 4 isolates, with 3 resistant to beta-lactams. A single S. aureus isolate phenotypically resistant to erythromycin harbored ermC, whereas tet(M) and mecA were detected in a single isolate resistant to beta-lactams, tetracycline, and sulfonamides. The same isolate contained 11 residues associated with sulfonamide resistance in the deduced amino acid sequence of the folP gene.
Associations between presence of VFs and mastitis.
No association was detected between the total number of VF genes and SCC in original milk samples. Furthermore, the number of VFs of any category was not associated with the severity of inflammatory response or disease severity, as categorized into low, medium, and high SCC or clinical mastitis. Neither STs nor CCs differed in their impact on sample SCC or severity of immune response. Similarly, no clearly identifiable clusters of isolates were detected in t-distributed stochastic neighbor embedding (t-SNE) graphs when labeled according to immune response severity.
DISCUSSION
Several studies have focused on the epidemiology of bovine S. aureus in dairy herds (4, 39–42). In addition, many studies used PCR-based techniques to identify virulence and AMR genes among bovine S. aureus isolates (10, 43–45). However, no large-scale studies have investigated S. aureus virulence and AMR determinants in the context of its genetic diversity, STs, and CCs. Therefore, we conducted WGS of 119 S. aureus isolates and determined STs and CCs. Additionally, we also determined the distribution of 191 VFs and all known ARGs. The MLST analysis clustered 119 S. aureus isolates into 8 distant STs, grouped in 5 CCs. However, the majority of isolates were assigned to CC151 (51%), CC97 (41%), and CC126 (6%). Isolation of these CCs from bovine milk is consistent with most studies (46–49). In many American and European countries, CC151 was the most common and successful clonal type recovered from bovine mastitis outbreaks (46, 47, 49). Similarly, the involvement of CC97 and CC126 in bovine mastitis has been extensively reported from many countries, including South Africa, Brazil, Chile, Italy, Japan, Norway, Spain, The Netherlands, and the United States (46, 49, 50). Isolation of CC97 from humans and other hosts has also been reported (49). However, CC151 and CC126 are confirmed in and are believed to be adapted and limited to cattle (49–52). We also identified 2 other STs (ST8 and ST133), represented by 1 isolate each in our study. ST8 (CC8) is considered a human clone and has mostly been reported in human infections and clinical specimens (49, 53, 54); it is considered the most successful S. aureus lineage, from which a number of major methicillin-resistant S. aureus (MRSA) clones have emerged (50, 55–57). However, CC8 isolation from other hosts, e.g., horses and cows, has also been reported (50), leading to the hypothesis that CC8 has moved from humans to cattle (53, 58), with CC8 MRSA transmitted back to humans (50, 54). We also report the isolation of ST133 (CC133) from bovine milk. The isolation of CC133 from cattle is rare; only a few studies have demonstrated its association with bovine IMI (52, 59). In contrast, the majority of ST133 isolates have been recovered from small ruminants, especially goats and sheep (52, 58, 60). Similar to CC8, the CC133 clone has been proposed to have made the jump from humans to ruminants (58). The most parsimonious explanation regarding the recovery of ST133 from bovine milk is the transfer of ST133 from humans to cattle.
Understanding relationships between strains is important for characterizing pathogen spread. In this study, WGS-based phylogenetic trees were constructed using core proteins and nucleotide sequences. All CGTs and MLSA trees had similar branching of S. aureus isolates. The phylogenic tree constructed from MLSA produced similar larger clades but failed to resolve relationships toward tips of the tree. This was not surprising, as MLSA is based on concatenated sequences of 7 genes, representing only a small fraction of the total genome, and many studies recommend WGS for inferring phylogenies (33, 35, 61). Phylogenetic information obtained from a limited number of genes is influenced by choice of method and selection of evolutionary model of phylogenetic estimations; therefore, it often produces conflicting phylogenies for recent evolutionary descents, represented by tips in phylogenetic trees (34).
The distribution of 191 VFs was determined after grouping these VFs into 5 functional categories. Among 28 adherence genes, 21 were detected in ≥1 isolate. Genes in this category facilitate adhesion and biofilm formation. A hallmark of S. aureus pathogenicity is their capacity to bind to extracellular matrix or to host cells (11, 14, 62). Adhesion is indeed the first step in biofilm formation or invasion of host cells, protecting bacteria from the host immune system and facilitating chronic infection (11, 62). Adhesion relies on the expression of 8 genes (clfA, clfB, cna, fnbA, fnbB, srdC, srdD, and srdE) that encode a repertoire of surface proteins, called microbial surface components recognizing adhesive matrix molecules (MSCRAMMs) (62), and the subsequent release of biofilm-related proteins (11, 14, 63). The ability of bacterial pathogens to produce biofilms is regarded as a major cause of resistance to antibiotics and was demonstrated to be involved in persistent infections in animals and humans (63–65). Almost all genes of the ica operon were detected in S. aureus isolates. The ica operon encodes polysaccharide intercellular adhesins (PIA), the earliest recognized and most widely distributed genetic determinant of biofilms (66–68). Interestingly, in this study, an important biofilm-related gene, aap, was not detected in any of the S. aureus isolates, whereas another biofilm-associated gene, bap, was only detected in 2 isolates. In previous studies, bap was described as a cattle-specific pathogenic factor of biofilm formation (11, 63). The presence of all ica genes and absence of aap and bap genes indicates the ica-dependent biofilm formation of these isolates, consistent with previous findings (68–70).
Exoenzymes, capsular genes, iron uptake, and type VII secretion system genes were widely distributed among all S. aureus isolates. These genes enable S. aureus to cause infection and survive in bovine udders and make it a successful and devastating bovine pathogen (13, 71, 72). Interestingly, scn and the chp genes of the immune evasion cluster (IEC), located on β-hemolysin-converting bacteriophages (50, 73) and known to be specific for human isolates (74, 75), were not detected in this study. Among hemolysins, most alpha-, beta-, and gamma-hemolysin genes were identified in all S. aureus isolates, although hld was detected only in ST352 and ST126, consistent with other reports (12, 43, 50). The presence of the hlb gene was reported to have an antagonistic relationship with IEC genes, as the latter was reported to cause insertional inactivation of hlb (50), which was also evident from our results. All STs had lukED genes, in agreement with previous studies reporting the presence of lukED among bovine isolates (48, 50, 76, 77). However, despite higher prevalence in bovine isolates, lukED has also been reported in human isolates (78, 79).
The toxic shock syndrome toxin gene (tsst) was detected in ST351 and only 1 isolate of ST151. The importance of toxin secretion by S. aureus in the pathogenesis of mastitis remains unclear. However, superantigens and leukotoxins are considered to have important roles in the initiation and progression of bovine mastitis due to their influence and ability to modulate the immune system (10, 46, 80). The distribution of 12 (of 21) enterotoxin genes in ST151 and ST351, the most prevalent sequence types, was interesting. None of these genes were detected in other STs. Enterotoxins are heat stable and may remain after heat treatment in various dairy products (81–83). Enterotoxins have been associated with staphylococcal food poisoning caused by cow milk or other dairy products (81–83). Interestingly, isolates from the highly prevalent CC97 and CC133 that are strongly associated with CM (52, 58, 84) had no enterotoxin genes. Exotoxin genes were prevalent in S. aureus isolates, and, except for the set15 gene, all other genes were detected in at least 1 isolate.
Of 191 genes tested, 87 (45%), here defined as core VFs, were detected in all isolates (95% to 100%) of all STs. The presence of these genes in all STs implicate them as having a role in host adaptation and survival in host environments and niches and not to obligately cause mastitis, as not all isolates containing these genes were recovered from clinically diseased animals. However, the presence of these genes may help S. aureus to establish as an opportunistic pathogen. Various VFs (36/191) were not detected in any of our S. aureus isolates. All STs contained 120 to 135 VFs. No association between the number of virulence genes and mastitis was identified; however, the presence of VF genes in genomes does not necessarily relate to gene expression (85) but rather is influenced by multiple factors, e.g., environment, nutritional status, presence of other competing microbes, and host genetics (86–89). The pathogenesis of S. aureus infection is complex and requires systematic participation of multiple VFs to establish disease (11, 13, 86, 90). To understand synergistic or antagonistic links between VF genes, we generated VF association graphs. Further studies focused on unraveling these interactions may extend the understanding of S. aureus pathogenesis.
Development of resistance to antimicrobials can be considered a virulence determinant, as AMR enhances host pathogenesis and allows persistent or chronic infections (91–93). Identifying ARGs is critical to recognize and assess the pathogenic potential of S. aureus. Fewer ARGs were identified in this study than from non-aureus staphylococci (NAS) originating from the same herds (36). This corresponded with reports that S. aureus strains isolated from mastitis cases were less resistant than NAS against commonly used antimicrobials (94, 95). Interestingly, 19% of isolates demonstrated in vitro resistance against beta-lactams, whereas blaZ or mec genes, the two most widespread mechanisms of acquired beta-lactam resistance in S. aureus (96), were detected in roughly 4% of isolates. It remains unclear whether most resistant isolates harbored genetic elements other than ones screened or if bacteria were tolerant to beta-lactams at low concentrations in vitro. Of note, beta-lactam-resistant bacteria where no genetic mechanisms of resistance were detected had a range of MICs against penicillin and ampicillin of 0.25 to 8 μg/ml. Therefore, it is unlikely that typical 2-fold variance in MICs would explain nearly 15% of resistant isolates without identified genetic mechanisms of resistance. Here, a single isolate harbored the mecA gene, demonstrating that this gene is still uncommon in S. aureus isolated from Canadian dairy herds, in contrast to the higher prevalence observed for S. epidermidis isolated from IMI (36). Similarly, the prevalence of this genetic element in S. aureus isolated from herds in the United States is relatively low compared to the same in NAS (97).
The presence of MDR efflux pumps was observed in all S. aureus isolates. However, to be associated with resistance, these elements would need to be upregulated. Of note, other studies reported the high prevalence of the same elements in Staphylococcus spp. (98). Interestingly, the presence of genes encoding MDR efflux pumps, such as NorA and SAV1866, demonstrated an association with several virulence genes, with some virulence genes being more frequent in isolates harboring these MDR efflux pumps, whereas others were uncommon in or absent from the same subset of isolates. In the absence of mechanistic studies, it is unclear whether these patterns reflected a gene interaction that influenced AMR and/or virulence. Nevertheless, WGS facilitates scanning genomes for all known genetic determinants of antibiotic resistance (36, 37), VF genes, and their interactions.
Conclusions.
Based on WGS, we determined that 119 bovine milk Staphylococcus aureus isolates belonged to 8 sequence types (ST151, ST352, ST351, ST2187, ST2270, ST133, and ST8), 5 clonal complexes (CC151, CC97, CC126, CC133, and CC8), and 18 distinct Spa types. Pan-, core, and accessory genomes of these isolates were composed of 6,340, 1,279, and 2,431 genes, respectively. Phenotypically, resistance was most common against beta-lactams (23 [19%] isolates) and sulfonamides (8 [7%] isolates), whereas resistance was uncommon against pirlimycin, tetracycline, ceftiofur, and erythromycin and to the combination of penicillin and novobiocin (3, 3, 3, 2, and 2% of all isolates, respectively). We also established comprehensive VF gene profiles of 119 S. aureus isolates, calculated the pathogenic potential of CCs and STs, and determined that CC151 (ST151 and ST351) had the highest VF potential, followed by CC97 (ST352 and ST2187) and CC126 (ST126 and ST2270), whereas CC133 (ST133) and CC8 (ST8) had the lowest pathogenic potential. However, the mere presence and number of genes cannot determine the pathogenesis of S. aureus, which is complex and depends on several factors, such as, but not limited to, host health, activation, and expression of virulence genes, host immune system, and geographic influences. Additionally, variations in VF genes among CCs and STs may represent evolution toward adaptation to host or distinct niches or environments within a host. To the best of our knowledge, this was the first study that performed WGS on a large number of S. aureus strains isolated from bovine IMI and determined STs, CCs, and Spa types, computed pan-genomes, determined the distribution of AMR and 191 virulence genes, and calculated the pathogenic potential of distinct STs and CCs.
MATERIALS AND METHODS
Isolates.
Isolates were obtained from the National Cohort of Dairy Farms of the Canadian Bovine Mastitis Research Network (99). Briefly, 89 herds from 6 Canadian provinces, selected to be representative of their respective province in terms of bulk tank SCC and housing system, were monitored from February 2007 to December 2008. Milk samples were collected according to 3 sampling schemes: (i) all samples from clinical mastitis cases; (ii) weekly or biweekly samples of 15 randomly selected lactating cows from each herd; and (iii) milk samples prior to drying off and after calving. Overall, 115,294 milk samples were obtained from 5,157 lactating cows. Staphylococcus aureus was detected in 3,387 milk samples obtained from 1,042 cows in all 89 herds (range, 1 to 27 isolates per cow). From this total, a random selection of 119 isolates from 119 cows was done. Details about the number of isolates and unique herds of origin were grouped by SCC level for nonclinical samples, and a separate category for clinical mastitis samples, are given in Table 1.
DNA extraction and whole-genome sequencing.
Isolates were grown on 5% sheep blood agar plates (BD Diagnostics, Mississauga, ON, Canada) at 37°C for 24 h to yield single colonies, suspended in Bacto brain heart infusion broth (BD Diagnostics), and incubated at 37°C overnight. Genomic DNA was extracted with a DNeasy blood and tissue kit (Qiagen, Toronto, ON, Canada) by following the protocol for Gram-positive bacteria. The concentration and quality of genomic DNA were determined with a NanoVue Plus spectrophotometer (GE Healthcare Life Sciences, Mississauga, ON, Canada) and the Qubit 2.0 fluorometer (Invitrogen, Burlington, ON, Canada). Each DNA sample was diluted to a final concentration of 0.2 ng/μl. Paired-end DNA libraries of 250 bp were prepared using a Nextera XT DNA library preparation kit (Illumina, San Diego, CA, USA) and samples sequenced with an Illumina MiSeq platform (Illumina). Primary data analysis for quality control was done on the MiSeq platform.
Genome assembly and annotation.
Genome assemblies and annotations were done using an in-house pipeline, as described previously (34, 100). Briefly, poorly sequenced regions and Illumina adapter sequences from sequence reads obtained from the MiSeq platform were identified and removed using cutadapt (101), implemented in Trim Galore! 0.4.0 (with default parameters). Filtered reads were assembled into contigs using the de novo assembly program SPAdes version 3.6.0 (102), employing built-in error correction and default parameters. Sequencing depth of coverage for each genome was determined by mapping reads back to the assembled genome using BWA 0.7.12-r1039 (103). The identification of coding sequences (CDS) and genome annotations was performed with Prokka 1.12 (104), using the provided (with Prokka) Staphylococcus database. Briefly, protein-coding genes, tRNAs, and rRNAs were predicted by Prodigal v2.6.2 (105), Aragorn v1.2.36 (106), and RNAmmer v1.2 (107), respectively. The quality of assembled genomes and assembly metrics was determined using Quast (108). The entire genome assembly process was automated using the Snakemake workflow engine (109).
Determination of STs and spa types.
Multilocus sequence typing (MLST) was performed to determine distinct sequence types (STs) (110). The S. aureus MLST scheme is based on 7 housekeeping genes, arcC (carbamate kinase), aroE (shikimate dehydrogenase), glpF (glycerol kinase), gmk (guanylate kinase), pta (phosphate acetyltransferase), tpi (triosephosphate isomerase), and yqi (acetyl coenzyme A acetyltransferase) (111). Full-length sequences of these 7 genes from 119 S. aureus genomes were obtained using BLAST+ 2.5.0 (112) and compared at each locus with those of the known alleles in the S. aureus MLST database (https://pubmlst.org/saureus) to obtain allelic profiles and to determine STs. Clustering of STs into complexes (CCs) was done using eBURSTv3 (113) based on predictor founders. Spa types (t) were predicted using spaTyper v1.0 webserver (114) from the Center of Genomic Epidemiology (https://cge.cbs.dtu.dk/services/spatyper). The spa typing technique compares the 21 to 27 polymorphic VNTR in the 3′ coding region of staphylococcal protein A (spa) to assign a unique repeat code corresponding to its spa type.
Phylogenetic analyses.
(i) Core genome phylogenies according to protein trees. To understand evolutionary relationships and determine diversity among S. aureus isolates, phylogenetic trees of these isolates were constructed using both a core set of proteins and a data set of 400 ubiquitous marker proteins (115). The core protein tree (CPT) was constructed as described previously (34). Briefly, the core set of S. aureus protein families (80% sequence identity and 80% sequence length), present in ≥90% of input genomes, was identified using the CD-HIT program (116). Protein families that contained potential paralogous sequences (duplicated sequence in the same genome) were excluded from further analysis. Multiple-sequence alignments (MSA) of each protein family were performed using the Clustal Omega (117) algorithm. Aligned amino acid positions with gaps in >50% of genomes were excluded from further analysis. The remaining amino acid positions were concatenated to create a combined data set. Poorly aligned regions from this concatenated alignment were removed using Gblocks 0.92 (118). This combined data set was further trimmed with trimAl (119). Maximum-likelihood (ML) trees based on this alignment were constructed using FastTree 2.1 (120), using the Whelan and Goldman substitution model (121), and a phylogenetic tree based on 400 marker proteins was constructed using PhyloPhlAn (115).
(ii) Identification of SNP sites and construction of SNP trees. For SNP identification, a whole-genome alignment-based method, kSNP v3.021 (122), and an alignment-free sequence analysis method, Parsnp v1.2 (123), were used to identify and construct SNP trees. Prior to kSNP3 analysis, the kchooser script was used to determine optimum k-mer size (122). The input file for kSNP3 analysis was created using the MakeKSNP3infile program in fully automatic mode. The kSNP3 analysis was done with the following parameters: -k 19 -ML -NJ -core -vcf -CPU 30. For Parsnp analysis, default parameters and autorecruitment of the reference genome were followed to construct core whole-genome SNP alignment. Within the core genome SNP alignment, aligned columns with recombination signals were detected and removed by PhiPack (124). An ML tree, based on the final alignment of core genome SNPs, was constructed using FastTree 2.1 (120). The placement of SNPs over the phylogenetic tree was visualized using the ginger program (123).
Multilocus sequence analysis.
Multilocus sequence analysis was performed on nucleotide sequences of 7 housekeeping genes (arcC, aroE, glpF, gmk, pta, tpi, and yqi). Full-length sequences of these genes (from 119 S. aureus isolates) were obtained using BLAST+ 2.5.0 (112). MSAs for each of these genes were created using MUSCLE v3.8.31 (125). Individual alignments were concatenated to create a combined data set. Poorly aligned regions from this concatenated alignment were removed using Gblocks 0.92 (118). An ML tree based on 100 bootstrap replicates was constructed using MEGA 6.0 (126), using the general time-reversible model (127).
Pan-genome analysis.
The pan-genome of 119 S. aureus isolates was computed with Roary v 3.12.0 (128). In Roary analysis, GFF files of all S. aureus isolates produced by Prokka (104) were used as input files for Roary, which uses the CD-HIT algorithm (116) to cluster orthologous gene families. Multiple-sequence alignment of gene families was performed using the PRANK v 0.170427 program (129). For Roary analysis, genes present in ≥99% of input genomes and sharing ≥90% sequence identity were considered core. The pan-genome was represented as the core genome (shared by >99% of strains), accessory genome (genes present in >2 strains but not in all), and unique genome (genes unique to individual strains). The total pan-genome was also shown as core (99% ≤ strains ≤ 100%), soft core (95% ≤ strains < 99%), shell (15% ≤ strains < 95%), and cloud (0% ≤ strains < 15%). Functional annotations of core, accessory, and unique genes were obtained after comparing these sequences with COG and KEGG databases implemented in BPGA v1.3 (130). To visually observe distributions of the pan-genome to S. aureus isolates, a gene_presence_absence table obtained from Roary analysis was superimposed onto CGT using the roary_plots script (128). Distributions of the pan-genome to individual isolates were plotted with the Roary2SVG script (128).
Collection of virulence and AMR genes and classification of VFs.
For VFs, a comprehensive VF data set of staphylococci (CVFS) created in our previous study (38) was used. Briefly, CVFS was developed by collecting S. aureus VF sequences from the VFDB database (131), Victors database (http://www.phidias.us/victors/), PATRIC database (132), and phenol-soluble modulin sequences from the UniProtKB database (133). Virulence factors (n = 191) were classified into 5 functional categories: adherence (n = 28), exoenzymes (n = 21), host immune evasion (n = 20), iron uptake and metabolism (n = 29), and toxins (n = 93). The 28 VFs of the adherence category were accumulation-associated protein (aap), biofilm-associated surface protein Bap (bap), autolysin (atl), clumping factors (clfA and clfB), collagen adhesion (cna), elastin binding protein (ebp), fibronectin binding proteins (ebh, efb, uafA, fnbA, and fnbB), extracellular adherence/major histocompatibility complex analogous protein (eap-map), cell wall surface anchor family proteins (sasC, sasG, and sasP), intercellular adhesins (icaA, icaB, icaC, icaD, and icaR), and Ser-Asp rich fibrinogen-binding proteins (sdrC, sdrD, sdrE, sdrF, sdrG, sdrH, and sdrI). Exoenzymes consisted of adenosine synthase A (adsA), aureolysin (aur), cysteine proteases (sspA, sspB, sspC, sspD, sspE, and sspF), hyaluronate lyase (hysA), lipases (lip and geh), serine proteases (splA, splB, splC, splD, splE, and splF), staphylocoagulase (coa), staphylokinase (sak), thermonuclease (nuc), and von-Willebrand factor-binding protein (vWbp).
The host immune evasion category consisted of capsular genes (capA, capB, capC, capD, capE, capF, capG, capH, capI, capJ, capK, capL, capM, capN, capO, and capP), chemotaxis inhibitory protein (chp), staphylococcal complement inhibitor (scn), staphylococcal protein A (spa), and staphylococcal binder of immunoglobulin (sbi) gene. The iron uptake and metabolism category included 9 iron-regulated surface determinants (isdA, isdB, isdC, isdD, isdE, isdF, isdG, isdH, and isdI), 7 ABC transporters (also known as siderophore receptors; htsA, htsB, htsC, sfaA, sfaB, sfaC, and sfaD), 12 staphyloferrin A and B synthesis-related genes (sirA, sirB, sirC, sbnA, sbnB, sbnC, sbnD, sbnE, sbnF, sbnG, sbnH, and sbnI), and 1 sortase B (srtB). Toxin genes included genes for alpha-, beta-, delta-, and gamma-hemolysins (hly-hla, hlb, hld, hlgA, hlgB, and hlgC), 4 genes for leukocidins, including leukocidin M (lukM and lukF-like) and Panton-Valentine leukocidins (lukS-PV and lukF-PV), 2 leukotoxins (lukD and lukE), toxic shock syndrome toxin (tsst), 4 exfoliative toxins (eta, etb, etc, and etd), 8 genes of type VII secretion system (esaA, esaB, esaC, essA, essB, essC, esxA, and esxB), and 11 genes for phenol-soluble modulins, including the 5 alpha (PSMα1, PSMα2, PSMα3, PSMα4, and PSMmec) and 6 beta (PSMβ1, PSMβ2, PSMβ3, PSMβ4, PSMβ5 and PSMβ6) genes plus 21 enterotoxin and 36 staphylococcal exotoxin (set) genes.
For AMR gene (ARG) screening, a data set of ARGs was constructed, as described previously (36), after combining AMR gene sequences from 4 databases: (i) ARG-ANNOT v3 (Antibiotic Resistance Gene-ANNOTation), (ii) MegaRES v1.0.1 (134), (iii) Comprehensive Antibiotic Resistance Database v1.1.6 (CARD) (135), and (iv) ResFinder from the Center for Genomic Epidemiology (136).
Identification of virulence and antimicrobial resistance genes.
The presence of VFs and ARGs was determined as described previously (36, 38). For this purpose, a local blastdbs of 119 S. aureus was created with the makeblastdb application from BLAST+ 2.5.0 (112). BLASTp searches of CVFS and ARG sequences were done against S. aureus genomes. Homology between query protein sequences and blast hits was determined by calculating H scores (36, 137). The H scores between protein sequences, labeled Ha (where a represents amino acid), were calculated using the formula Ha = Qid × Lm/Lq (36), with Qid representing the level of BLASTp identities between query sequence and identified protein sequence (range, 0 to 1), Lm representing length of the matching sequence from the hit, and Lq denoting the length of the query sequence. Cutoffs of 80% sequence similarity and 70% query length coverage were used for initial searches. All genomic hits that met the minimal cutoff for each individual query were selected at this stage. A final blast hit table containing all possible hits for all query sequences from all S. aureus genomes was imported into R v.3.4.2 (138). Hits from each query sequence were then arranged according to Ha score, using dplyr version 0.7.2 in R (139). From this list, only hits with highest Ha score (highest sequence similarity and query length coverage) were selected as potential VFs and ARGs in S. aureus genomes, whereas the remainder were discarded. After the identification of putative S. aureus VFs, to confirm orthology between identified putative S. aureus VF and CVFS sequences, reciprocal blast searches between the putative S. aureus VFs and CVFS database were done (140). Putative S. aureus VF sequences that failed to match corresponding VFs from the CVFS database as best hits in reciprocal blast searches were not considered true orthologs and were excluded from further analysis.
For AMR genes, protein sequences of all top hits were used as queries against the nonredundant (nr) database from NCBI using BLASTp (112). The best hit was considered definitive as long as it had >80% coverage and percent identity with the query (36). For AMR genes that required additional confirmation (substitutions and residues composition), pairwise alignments of putative AMR genes with those of reference genes (36) were done using MEGA 6.0 (126). The ARGs obtained from 119 S. aureus isolates were screened for the presence of residues known to be associated with AMR in S. aureus.
Antimicrobial resistance profiles.
For all isolates, the phenotypic AMR profile was determined using the MIC, in accordance with Clinical Laboratory and Standards Institute (CLSI) guidelines (141). Antimicrobials and concentrations (in micrograms per milliliter) evaluated were the following: ampicillin (0.128), ceftiofur (0.5 to 4), cephalothin (2 to 16), erythromycin (0.25 to 4), oxacillin plus 2% NaCl (2 to 4), penicillin (0.06 to 8), penicillin-novobiocin (1-2 to 8-16), pirlimycin (0.5 to 4), tetracycline (2 to 16), and sulfadimethozine (32 to 256). Breakpoints were defined according to CLSI criteria for animals (141, 142). Isolates were classified as either susceptible or resistant, whereas isolates with MIC equal to intermediate breakpoints were considered resistant. Antimicrobial-free wells were included in all plates, and S. aureus ATCC 29213 was used as a quality control strain for all tests.
Associations between presence of VFs and mastitis.
Associations between the presence of VFs with mastitis were assessed by conducting statistical analyses using the ordinal package (143) and base functions in R v.3.4.2 (138), with a P value of <0.05 considered significant. Clustering and dimensionality reduction analyses were conducted using Python and the package Scikit-Learn (144). Relationships between measures of mastitis and VFs were examined after dichotomizing genes (presence or absence) in all isolates. For samples collected from animals without clinical symptoms of mastitis, the association of the natural logarithm of SCC (LnSCC) with the number of VFs present was assessed using linear regression; the outcome was the LnSCC, and predictor was the total number of VFs or number of virulence genes of a given type in the isolate. Model fit was assessed using multiple R2, whereas the strength of association was assessed by examining the coefficient for the predictor variable. To include samples from clinical mastitis, there were 4 outcome categories based on SCC and sample type, low SCC (≤150,000 cells/ml), medium SCC (150,000 < SCC ≤ 250,000 cells/ml), high SCC (>250,000 cells/ml), and clinical mastitis (isolated from a quarter with clinical mastitis) and rank. To assess associations between inflammatory response and disease severity and virulence genes, ordinal logistic regression was conducted using the outcome defined above and the total number of virulence genes or number of virulence genes of a given category in the isolate as a predictor. Coefficients estimated using this regression represented the likelihood of a more severe mammary response (measured as SCC) for each additional virulence factor identified. For dimensionality reduction and to visualize distributions of VFs within isolates, t-distributed stochastic neighbor embedding (t-SNE [145]) was conducted using the manifold.t-SNE module within Scikit-Learn (144). Genetic distributions were reduced to 2 or 3 dimensions and visually examined. Plots were labeled with severity of immune response and geographical region (Canadian province) from which samples were isolated to identify potential clusters of interest.
Associations between STs, CCs, and mastitis.
Similar to models described above, clinical mastitis isolates were excluded, after which LnSCC was modeled, or mammary inflammation severity was categorized into 4 ordinal categories. Linear regression models were used to assess differences in mean LnSCC between STs and CCs, whereas ordinal logistic regression models were used to determine association between ST or CC and severity of mammary inflammation.
Associations between the presence of virulence genes.
Using the same dichotomized values for the presence of virulence genes, associations with STs and clonal complexes were also assessed. Both CCs and STs were treated as a categorical outcome variable, and a multinomial logistic regression using the package nnet (146) was conducted, with predictors being the total number of VFs or number of VFs of a given category in an isolate. Regression coefficients from a multinomial logistic regression model were interpreted as how much more or less likely an isolate with a given number of virulence genes was to belong to a specific ST or CC compared to the baseline. Regional distributions of both STs and CCs were also assessed using a multinomial logistic regression with STs or CCs as the outcome and region as the only predictor. To visualize genetic distributions, the same components derived during the t-SNE described above were plotted and labeled with either STs or CCs to identify potential clusters.
Data availability.
All whole-genome sequencing data used in this study are available without restriction from NCBI under BioProject no. PRJNA599195.
ACKNOWLEDGMENTS
We acknowledge support from an Eyes High postdoctoral award from the University of Calgary and a Natural Sciences and Engineering Research Council of Canada (NSERC) CREATE grant in Milk Quality awarded to S.N. This work was partially funded through the NSERC Industrial Research Chair in Infectious Diseases of Dairy Cattle. Bacterial isolates were furnished by the Canadian Bovine Mastitis Research Network (CBMRN). The CBMRN pathogen and data collection was financed by the Natural Sciences and Engineering Research Council of Canada (Ottawa, ON, Canada), Alberta Milk (Edmonton, AB, Canada), Dairy Farmers of New Brunswick (Sussex, New Brunswick, Canada), Dairy Farmers of Nova Scotia (Lower Truro, NS, Canada), Dairy Farmers of Ontario (Mississauga, ON, Canada), Dairy Farmers of Prince Edward Island (Charlottetown, PE, Canada), Novalait Inc. (Québec City, QC, Canada), Dairy Farmers of Canada (Ottawa, ON, Canada), Canadian Dairy Network (Guelph, ON, Canada), Agriculture and Agri-Food Canada (Ottawa, ON, Canada), Public Health Agency of Canada (Ottawa, ON, Canada), Technology PEI Inc. (Charlottetown, PE, Canada), Université de Montréal (Montréal, QC, Canada), and University of Prince Edward Island (Charlottetown, PE, Canada) through the CBMRN (Saint-Hyacinthe, QC, Canada).
REFERENCES
- 1.Song Q, Wu J, Ruan P. 2018. Predominance of community-associated sequence type 59 methicillin-resistant Staphylococcus aureus in a paediatric intensive care unit. J Med Microbiol 67:408–414. doi: 10.1099/jmm.0.000693. [DOI] [PubMed] [Google Scholar]
- 2.Taponen S, Liski E, Heikkilä AM, Pyörälä S. 2017. Factors associated with intramammary infection in dairy cows caused by coagulase-negative staphylococci, Staphylococcus aureus, Streptococcus uberis, Streptococcus dysgalactiae, Corynebacterium bovis, or Escherichia coli. J Dairy Sci 100:493–503. doi: 10.3168/jds.2016-11465. [DOI] [PubMed] [Google Scholar]
- 3.Zhou Z, Zhang M, Li H, Yang H, Li X, Song X, Wang Z. 2017. Prevalence and molecular characterization of Staphylococcus aureus isolated from goats in Chongqing, China. BMC Vet Res 13:352. doi: 10.1186/s12917-017-1272-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barkema HW, Schukken YH, Zadoks RN. 2006. Invited review: the role of cow, pathogen, and treatment regimen in the therapeutic success of bovine Staphylococcus aureus mastitis. J Dairy Sci 89:1877–1895. doi: 10.3168/jds.S0022-0302(06)72256-1. [DOI] [PubMed] [Google Scholar]
- 5.Abdelmegid S, Murugaiyan J, Abo-Ismail M, Caswell JL, Kelton D, Kirby GM. 2017. Identification of host defense-related proteins using label-free quantitative proteomic analysis of milk whey from cows with Staphylococcus aureus subclinical mastitis. Int J Mol Sci 19:78. doi: 10.3390/ijms19010078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ruegg PL. 2017. A 100-year review: mastitis detection, management, and prevention. J Dairy Sci 100:10381–10397. doi: 10.3168/jds.2017-13023. [DOI] [PubMed] [Google Scholar]
- 7.Gao J, Barkema HW, Zhang L, Liu G, Deng Z, Cai L, Shan R, Zhang S, Zou J, Kastelic JP, Han B. 2017. Incidence of clinical mastitis and distribution of pathogens on large Chinese dairy farms. J Dairy Sci 100:4797–4806. doi: 10.3168/jds.2016-12334. [DOI] [PubMed] [Google Scholar]
- 8.Hogeveen H, Huijps K, Lam TJ. 2011. Economic aspects of mastitis: new developments. N Z Vet J 59:16–23. doi: 10.1080/00480169.2011.547165. [DOI] [PubMed] [Google Scholar]
- 9.Gonçalves JL, Kamphuis C, Martins C, Barreiro JR, Tomazi T, Gameiro AH, Hogeveen H, dos Santos MV. 2018. Bovine subclinical mastitis reduces milk yield and economic return. Livestock Sci 210:25–32. doi: 10.1016/j.livsci.2018.01.016. [DOI] [Google Scholar]
- 10.Magro G, Biffani S, Minozzi G, Ehricht R, Monecke S, Luini M, Piccinini R. 2017. Virulence genes of S. aureus from dairy cow mastitis and contagiousness risk. Toxins 9:195. doi: 10.3390/toxins9060195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kerro Dego O, van Dijk JE, Nederbragt H. 2002. Factors involved in the early pathogenesis of bovine Staphylococcus aureus mastitis with emphasis on bacterial adhesion and invasion. A review. Vet Q 24:181–198. doi: 10.1080/01652176.2002.9695135. [DOI] [PubMed] [Google Scholar]
- 12.Zecconi A, Scali F. 2013. Staphylococcus aureus virulence factors in evasion from innate immune defenses in human and animal diseases. Immunol Lett 150:12–22. doi: 10.1016/j.imlet.2013.01.004. [DOI] [PubMed] [Google Scholar]
- 13.Maréchal C, Seyffert N, Jardin J, Hernandez D, Jan G, Rault L, Azevedo V, François P, Schrenzel J, Guchte M, Even S, Berkova N, Thiéry R, Fitzgerald JR, Vautor E, Loir Y. 2011. Molecular basis of virulence in Staphylococcus aureus mastitis. PLoS One 6:e27354. doi: 10.1371/journal.pone.0027354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sutra L, Poutrel B. 1994. Virulence factors involved in the pathogenesis of bovine intramammary infections due to Staphylococcus aureus. J Med Microbiol 40:79–89. doi: 10.1099/00222615-40-2-79. [DOI] [PubMed] [Google Scholar]
- 15.Fox LK, Zadoks RN, Gaskins CT. 2005. Biofilm production by Staphylococcus aureus associated with intramammary infection. Vet Microbiol 107:295–299. doi: 10.1016/j.vetmic.2005.02.005. [DOI] [PubMed] [Google Scholar]
- 16.Bardiau M, Detilleux J, Farnir F, Mainil JG, Ote I. 2014. Associations between properties linked with persistence in a collection of Staphylococcus aureus isolates from bovine mastitis. Vet Microbiol 169:74–79. doi: 10.1016/j.vetmic.2013.12.010. [DOI] [PubMed] [Google Scholar]
- 17.Grunert T, Stessl B, Wolf F, Sordelli DO, Buzzola FR, Ehling-Schulz M. 2018. Distinct phenotypic traits of Staphylococcus aureus are associated with persistent, contagious bovine intramammary infections. Sci Rep 8:15968. doi: 10.1038/s41598-018-34371-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang B, Muir TW. 2016. Regulation of virulence in Staphylococcus aureus: molecular mechanisms and remaining puzzles. Cell Chem Biol 23:214–224. doi: 10.1016/j.chembiol.2016.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Awad A, Ramadan H, Nasr S, Ateya A, Atwa S. 2017. Genetic characterization, antimicrobial resistance patterns and virulence determinants of Staphylococcus aureus isolated form bovine mastitis. Pak J Biol Sci 20:298–305. doi: 10.3923/pjbs.2017.298.305. [DOI] [PubMed] [Google Scholar]
- 20.El Bayomi RM, Ahmed HA, Awadallah MA, Mohsen RA, Abd El-Ghafar AE, Abdelrahman MA. 2016. Occurrence, virulence factors, antimicrobial resistance, and genotyping of Staphylococcus aureus strains isolated from chicken products and humans. Vector Borne Zoonotic Dis 16:157–164. doi: 10.1089/vbz.2015.1891. [DOI] [PubMed] [Google Scholar]
- 21.McDougall S, Hussein H, Petrovski K. 2014. Antimicrobial resistance in Staphylococcus aureus, Streptococcus uberis and Streptococcus dysgalactiae from dairy cows with mastitis. N Z Vet J 62:68–76. doi: 10.1080/00480169.2013.843135. [DOI] [PubMed] [Google Scholar]
- 22.Shi D, Hao Y, Zhang A, Wulan B, Fan X. 2010. Antimicrobial resistance of Staphylococcus aureus isolated from bovine mastitis in China. Transbound Emerg Dis 57:221–224. doi: 10.1111/j.1865-1682.2010.01139.x. [DOI] [PubMed] [Google Scholar]
- 23.Ismail ZB. 2017. Molecular characteristics, antibiogram and prevalence of multi-drug resistant Staphylococcus aureus (MDRSA) isolated from milk obtained from culled dairy cows and from cows with acute clinical mastitis. Asian Pacific J Trop Biomed 7:694–697. doi: 10.1016/j.apjtb.2017.07.005. [DOI] [Google Scholar]
- 24.Haran KP, Godden SM, Boxrud D, Jawahir S, Bender JB, Sreevatsan S. 2012. Prevalence and characterization of Staphylococcus aureus, including methicillin-resistant Staphylococcus aureus, isolated from bulk tank milk from minnesota dairy farms. J Clin Microbiol 50:688–695. doi: 10.1128/JCM.05214-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Holmes MA, Zadoks RN. 2011. Methicillin resistant S. aureus in human and bovine mastitis. J Mammary Gland Biol Neoplasia 16:373–382. doi: 10.1007/s10911-011-9237-x. [DOI] [PubMed] [Google Scholar]
- 26.Kreausukon K, Fetsch A, Kraushaar B, Alt K, Muller K, Kromker V, Zessin KH, Kasbohrer A, Tenhagen BA. 2012. Prevalence, antimicrobial resistance, and molecular characterization of methicillin-resistant Staphylococcus aureus from bulk tank milk of dairy herds. J Dairy Sci 95:4382–4388. doi: 10.3168/jds.2011-5198. [DOI] [PubMed] [Google Scholar]
- 27.Herschleb J, Ananiev G, Schwartz DC. 2007. Pulsed-field gel electrophoresis. Nat Protoc 2:677–684. doi: 10.1038/nprot.2007.94. [DOI] [PubMed] [Google Scholar]
- 28.Mobasherizadeh S, Shojaei H, Havaei SA, Mostafavizadeh K, Davoodabadi F, Khorvash F, Ataei B, Daei-Naser A. 2015. Application of the random amplified polymorphic DNA (RAPD) fingerprinting to analyze genetic variation in community associated-methicillin resistant Staphylococcus aureus (CA-MRSA) isolates in Iran. Glob J Health Sci 8:185–191. doi: 10.5539/gjhs.v8n8p185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stanley T, Wilson IG. 2001. Multilocus enzyme electrophoresis, p 369–393. In Spencer JFT, de Ragout Spencer AL (ed), Food microbiology protocols. Humana Press, Totowa, NJ. [Google Scholar]
- 30.Saunders NA, Holmes A. 2007. Multilocus sequence typing (MLST) of Staphylococcus aureus, p 71–85. In Ji Y. (ed), Methicillin-resistant Staphylococcus aureus (MRSA) protocols. Humana Press, Totowa, NJ. [Google Scholar]
- 31.Fasihi Y, Fooladi S, Mohammadi MA, Emaneini M, Kalantar-Neyestanaki D. 2017. The spa typing of methicillin-resistant Staphylococcus aureus isolates by high resolution melting (HRM) analysis. J Med Microbiol 66:1335–1337. doi: 10.1099/jmm.0.000574. [DOI] [PubMed] [Google Scholar]
- 32.Schouls LM, Spalburg EC, van Luit M, Huijsdens XW, Pluister GN, van Santen-Verheuvel MG, van der Heide HGJ, Grundmann H, Heck M, de Neeling AJ. 2009. Multiple-locus variable number tandem repeat analysis of Staphylococcus aureus: comparison with pulsed-field gel electrophoresis and spa-typing. PLoS One 4:e5082. doi: 10.1371/journal.pone.0005082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Adeolu M, Alnajar S, Naushad S, S Gupta R. 2016. Genome-based phylogeny and taxonomy of the “Enterobacteriales”: proposal for Enterobacterales ord. nov. divided into the families Enterobacteriaceae, Erwiniaceae fam. nov., Pectobacteriaceae fam. nov., Yersiniaceae fam. nov., Hafniaceae fam. nov., Morganellaceae fam. nov., and Budviciaceae fam. nov. Int J Syst Evol Microbiol 66:5575–5599. doi: 10.1099/ijsem.0.001485. [DOI] [PubMed] [Google Scholar]
- 34.Naushad S, Barkema HW, Luby C, Condas LA, Nobrega DB, Carson DA, De Buck J. 2016. Comprehensive phylogenetic analysis of bovine non-aureus staphylococci species based on whole-genome sequencing. Front Microbiol 7:1990. doi: 10.3389/fmicb.2016.01990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gupta RS, Naushad S, Fabros R, Adeolu M. 2016. A phylogenomic reappraisal of family-level divisions within the class Halobacteria: proposal to divide the order Halobacteriales into the families Halobacteriaceae, Haloarculaceae fam. nov., and Halococcaceae fam. nov., and the order Haloferacales into the families, Haloferacaceae and Halorubraceae fam nov. Antonie Van Leeuwenhoek 109:565–587. doi: 10.1007/s10482-016-0660-2. [DOI] [PubMed] [Google Scholar]
- 36.Nobrega DB, Naushad S, Naqvi SA, Condas LAZ, Saini V, Kastelic JP, Luby C, De Buck J, Barkema HW. 2018. Prevalence and genetic basis of antimicrobial resistance in non-aureus staphylococci isolated from Canadian dairy herds. Front Microbiol 9:256. doi: 10.3389/fmicb.2018.00256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bakour S, Sankar SA, Rathored J, Biagini P, Raoult D, Fournier PE. 2016. Identification of virulence factors and antibiotic resistance markers using bacterial genomics. Future Microbiol 11:455–466. doi: 10.2217/fmb.15.149. [DOI] [PubMed] [Google Scholar]
- 38.Naushad S, Naqvi SA, Nobrega D, Luby C, Kastelic JP, Barkema HW, De Buck J. 2019. Comprehensive virulence gene profiling of bovine non-aureus staphylococci based on whole-genome sequencing data. mSystems 4:e00098-18. doi: 10.1128/mSystems.00098-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cousin M-E, Härdi-Landerer MC, Völk V, Bodmer M. 2018. Control of Staphylococcus aureus in dairy herds in a region with raw milk cheese production: farmers’ attitudes, knowledge, behaviour and belief in self-efficacy. BMC Vet Res 14:46. doi: 10.1186/s12917-018-1352-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Naqvi SA, De Buck J, Dufour S, Barkema HW. 2018. Udder health in Canadian dairy heifers during early lactation. J Dairy Sci 101:3233–3247. doi: 10.3168/jds.2017-13579. [DOI] [PubMed] [Google Scholar]
- 41.Papadopoulos P, Papadopoulos T, Angelidis AS, Boukouvala E, Zdragas A, Papa A, Hadjichristodoulou C, Sergelidis D. 2018. Prevalence of Staphylococcus aureus and of methicillin-resistant S. aureus (MRSA) along the production chain of dairy products in north-western Greece. Food Microbiol 69:43–50. doi: 10.1016/j.fm.2017.07.016. [DOI] [PubMed] [Google Scholar]
- 42.Abebe R, Hatiya H, Abera M, Megersa B, Asmare K. 2016. Bovine mastitis: prevalence, risk factors and isolation of Staphylococcus aureus in dairy herds at Hawassa milk shed, South Ethiopia. BMC Vet Res 12:270. doi: 10.1186/s12917-016-0905-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang L, Gao J, Barkema HW, Ali T, Liu G, Deng Y, Naushad S, Kastelic JP, Han B. 2018. Virulence gene profiles: alpha-hemolysin and clonal diversity in Staphylococcus aureus isolates from bovine clinical mastitis in China. BMC Vet Res 14:63. doi: 10.1186/s12917-018-1374-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Saini V, McClure JT, Léger D, Keefe GP, Scholl DT, Morck DW, Barkema HW. 2012. Antimicrobial resistance profiles of common mastitis pathogens on Canadian dairy farms. J Dairy Sci 95:4319–4332. doi: 10.3168/jds.2012-5373. [DOI] [PubMed] [Google Scholar]
- 45.Tarekgne EK, Skjerdal T, Skeie S, Rudi K, Porcellato D, Félix B, Narvhus JA. 2016. Enterotoxin gene profile and molecular characterization of Staphylococcus aureus isolates from bovine bulk milk and milk products of Tigray region, northern Ethiopia. J Food Prot 79:1387–1395. doi: 10.4315/0362-028X.JFP-16-003. [DOI] [PubMed] [Google Scholar]
- 46.Li T, Lu H, Wang X, Gao Q, Dai Y, Shang J, Li M. 2017. Molecular characteristics of Staphylococcus aureus causing bovine mastitis between 2014 and 2015. Front Cell Infect Microbiol 7:127. doi: 10.3389/fcimb.2017.00127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fluit AC. 2012. Livestock-associated Staphylococcus aureus. Clin Microbiol Infect 18:735–744. doi: 10.1111/j.1469-0691.2012.03846.x. [DOI] [PubMed] [Google Scholar]
- 48.Budd KE, McCoy F, Monecke S, Cormican P, Mitchell J, Keane OM. 2015. Extensive genomic diversity among bovine-adapted Staphylococcus aureus: evidence for a genomic rearrangement within CC97. PLoS One 10:e0134592. doi: 10.1371/journal.pone.0134592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Boss R, Cosandey A, Luini M, Artursson K, Bardiau M, Breitenwieser F, Hehenberger E, Lam T, Mansfeld M, Michel A, Mösslacher G, Naskova J, Nelson S, Podpečan O, Raemy A, Ryan E, Salat O, Zangerl P, Steiner A, Graber HU. 2016. Bovine Staphylococcus aureus: subtyping, evolution, and zoonotic transfer. J Dairy Sci 99:515–528. doi: 10.3168/jds.2015-9589. [DOI] [PubMed] [Google Scholar]
- 50.Schmidt T, Kock MM, Ehlers MM. 2017. Molecular characterization of Staphylococcus aureus isolated from bovine mastitis and close human contacts in South African dairy herds: genetic diversity and inter-species host transmission. Front Microbiol 8:511. doi: 10.3389/fmicb.2017.00511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Battisti A, Franco A, Merialdi G, Hasman H, Iurescia M, Lorenzetti R, Feltrin F, Zini M, Aarestrup FM. 2010. Heterogeneity among methicillin-resistant Staphylococcus aureus from Italian pig finishing holdings. Vet Microbiol 142:361–366. doi: 10.1016/j.vetmic.2009.10.008. [DOI] [PubMed] [Google Scholar]
- 52.Smyth DS, Feil EJ, Meaney WJ, Hartigan PJ, Tollersrud T, Fitzgerald JR, Enright MC, Smyth CJ. 2009. Molecular genetic typing reveals further insights into the diversity of animal-associated Staphylococcus aureus. J Med Microbiol 58:1343–1353. doi: 10.1099/jmm.0.009837-0. [DOI] [PubMed] [Google Scholar]
- 53.Resch G, François P, Morisset D, Stojanov M, Bonetti EJ, Schrenzel J, Sakwinska O, Moreillon P. 2013. Human-to-bovine jump of Staphylococcus aureus CC8 is associated with the loss of a β-hemolysin converting prophage and the acquisition of a new staphylococcal cassette chromosome. PLoS One 8:e58187. doi: 10.1371/journal.pone.0058187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sakwinska O, Giddey M, Moreillon M, Morisset D, Waldvogel A, Moreillon P. 2011. Staphylococcus aureus host range and human-bovine host shift. Appl Environ Microbiol 77:5908–5915. doi: 10.1128/AEM.00238-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Monecke S, Coombs G, Shore AC, Coleman DC, Akpaka P, Borg M, Chow H, Ip M, Jatzwauk L, Jonas D, Kadlec K, Kearns A, Laurent F, O'Brien FG, Pearson J, Ruppelt A, Schwarz S, Scicluna E, Slickers P, Tan H-L, Weber S, Ehricht R. 2011. A field guide to pandemic, epidemic and sporadic clones of methicillin-resistant Staphylococcus aureus. PLoS One 6:e17936. doi: 10.1371/journal.pone.0017936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Enright MC, Day NP, Davies CE, Peacock SJ, Spratt BG. 2000. Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. J Clin Microbiol 38:1008–1015. doi: 10.1128/JCM.38.3.1008-1015.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gomes AR, Westh H, de Lencastre H. 2006. Origins and evolution of methicillin-resistant Staphylococcus aureus clonal lineages. Antimicrob Agents Chemother 50:3237–3244. doi: 10.1128/AAC.00521-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Guinane CM, Ben Zakour NL, Tormo-Mas MA, Weinert LA, Lowder BV, Cartwright RA, Smyth DS, Smyth CJ, Lindsay JA, Gould KA, Witney A, Hinds J, Bollback JP, Rambaut A, Penadés JR, Fitzgerald JR. 2010. Evolutionary genomics of Staphylococcus aureus reveals insights into the origin and molecular basis of ruminant host adaptation. Genome Biol Evol 2:454–466. doi: 10.1093/gbe/evq031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mørk T, Jørgensen HJ, Sunde M, Kvitle B, Sviland S, Waage S, Tollersrud T. 2012. Persistence of staphylococcal species and genotypes in the bovine udder. Vet Microbiol 159:171–180. doi: 10.1016/j.vetmic.2012.03.034. [DOI] [PubMed] [Google Scholar]
- 60.Merz A, Stephan R, Johler S. 2016. Staphylococcus aureus Isolates from goat and sheep milk seem to be closely related and differ from isolates detected from bovine milk. Front Microbiol 7:319. doi: 10.3389/fmicb.2016.00319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Naushad S, Adeolu M, Wong S, Sohail M, Schellhorn HE, Gupta RS. 2015. A phylogenomic and molecular marker based taxonomic framework for the order Xanthomonadales: proposal to transfer the families Algiphilaceae and Solimonadaceae to the order Nevskiales ord. nov. and to create a new family within the order Xanthomonadales, the family Rhodanobacteraceae fam. nov., containing the genus Rhodanobacter and its closest relatives. Antonie Van Leeuwenhoek 107:467–485. doi: 10.1007/s10482-014-0344-8. [DOI] [PubMed] [Google Scholar]
- 62.Foster TJ, Geoghegan JA, Ganesh VK, Höök M. 2014. Adhesion, invasion and evasion: the many functions of the surface proteins of Staphylococcus aureus. Nat Rev Microbiol 12:49–62. doi: 10.1038/nrmicro3161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cucarella C, Tormo MA, Ubeda C, Trotonda MP, Monzón M, Peris C, Amorena B, Lasa I, Penadés JR. 2004. Role of biofilm-associated protein bap in the pathogenesis of bovine Staphylococcus aureus. Infect Immun 72:2177–2185. doi: 10.1128/iai.72.4.2177-2185.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Singh S, Singh SK, Chowdhury I, Singh R. 2017. Understanding the mechanism of bacterial biofilms resistance to antimicrobial agents. Open Microbiol J 11:53–62. doi: 10.2174/1874285801711010053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Dufour D, Leung V, Lévesque CM. 2010. Bacterial biofilm: structure, function, and antimicrobial resistance. Endod Topics 22:2–16. doi: 10.1111/j.1601-1546.2012.00277.x. [DOI] [Google Scholar]
- 66.Arciola CR, Campoccia D, Ravaioli S, Montanaro L. 2015. Polysaccharide intercellular adhesin in biofilm: structural and regulatory aspects. Front Cell Infect Microbiol 5:7. doi: 10.3389/fcimb.2015.00007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.O’Gara JP. 2007. ica and beyond: biofilm mechanisms and regulation in Staphylococcus epidermidis and Staphylococcus aureus. FEMS Microbiol Lett 270:179–188. doi: 10.1111/j.1574-6968.2007.00688.x. [DOI] [PubMed] [Google Scholar]
- 68.Cramton SE, Gerke C, Schnell NF, Nichols WW, Götz F. 1999. The intercellular adhesion (ica) locus is present in Staphylococcus aureus and is required for biofilm formation. Infect Immun 67:5427–5433. doi: 10.1128/IAI.67.10.5427-5433.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tormo MA, Knecht E, Götz F, Lasa I, Penadés JR. 2005. Bap-dependent biofilm formation by pathogenic species of Staphylococcus: evidence of horizontal gene transfer? Microbiology 151:2465–2475. doi: 10.1099/mic.0.27865-0. [DOI] [PubMed] [Google Scholar]
- 70.Kropec A, Maira-Litran T, Jefferson KK, Grout M, Cramton SE, Götz F, Goldmann DA, Pier GB. 2005. Poly-N-acetylglucosamine production in Staphylococcus aureus is essential for virulence in murine models of systemic infection. Infect Immun 73:6868–6876. doi: 10.1128/IAI.73.10.6868-6876.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Baselga R, Albizu I, Amorena B. 1994. Staphylococcus aureus capsule and slime as virulence factors in ruminant mastitis. A review. Vet Microbiol 39:195–204. doi: 10.1016/0378-1135(94)90157-0. [DOI] [PubMed] [Google Scholar]
- 72.Salimena AP, Lange CC, Camussone C, Signorini M, Calvinho LF, Brito MA, Borges CA, Guimarães AS, Ribeiro JB, Mendonça LC, Piccoli RH. 2016. Genotypic and phenotypic detection of capsular polysaccharide and biofilm formation in Staphylococcus aureus isolated from bovine milk collected from Brazilian dairy farms. Vet Res Commun 40:97–106. doi: 10.1007/s11259-016-9658-5. [DOI] [PubMed] [Google Scholar]
- 73.van Wamel WJB, Rooijakkers SHM, Ruyken M, van Kessel KPM, van Strijp J. 2006. The innate immune modulators staphylococcal complement inhibitor and chemotaxis inhibitory protein of Staphylococcus aureus are located on β-hemolysin-converting bacteriophages. J Bacteriol 188:1310–1315. doi: 10.1128/JB.188.4.1310-1315.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Verkaik NJ, Benard M, Boelens HA, de Vogel CP, Nouwen JL, Verbrugh HA, Melles DC, van Belkum A, van Wamel WJ. 2011. Immune evasion cluster-positive bacteriophages are highly prevalent among human Staphylococcus aureus strains, but they are not essential in the first stages of nasal colonization. Clin Microbiol Infect 17:343–348. doi: 10.1111/j.1469-0691.2010.03227.x. [DOI] [PubMed] [Google Scholar]
- 75.Sung J-L, Lloyd DH, Lindsay JA. 2008. Staphylococcus aureus host specificity: comparative genomics of human versus animal isolates by multi-strain microarray. Microbiology 154:1949–1959. doi: 10.1099/mic.0.2007/015289-0. [DOI] [PubMed] [Google Scholar]
- 76.Gogoi-Tiwari J, Waryah CB, Eto KY, Tau M, Wells K, Costantino P, Tiwari HK, Isloor S, Hegde N, Mukkur T. 2015. Relative distribution of virulence-associated factors among Australian bovine Staphylococcus aureus isolates: potential relevance to development of an effective bovine mastitis vaccine. Virulence 6:419–423. doi: 10.1080/21505594.2015.1043508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Chu C, Wei Y, Chuang ST, Yu C, Changchien CH, Su Y. 2013. Differences in virulence genes and genome patterns of mastitis-associated Staphylococcus aureus among goat, cow, and human isolates in Taiwan. Foodborne Pathog Dis 10:256–262. doi: 10.1089/fpd.2012.1278. [DOI] [PubMed] [Google Scholar]
- 78.von Eiff C, Friedrich AW, Peters G, Becker K. 2004. Prevalence of genes encoding for members of the staphylococcal leukotoxin family among clinical isolates of Staphylococcus aureus. Diagn Microbiol Infect Dis 49:157–162. doi: 10.1016/j.diagmicrobio.2004.03.009. [DOI] [PubMed] [Google Scholar]
- 79.Enwuru NV, Adesida SA, Enwuru CA, Ghebremedhin B, Mendie UE, Coker AO. 2018. Genetics of bi-component leukocidin and drug resistance in nasal and clinical Staphylococcus aureus in Lagos, Nigeria. Microb Pathog 115:1–7. doi: 10.1016/j.micpath.2017.12.030. [DOI] [PubMed] [Google Scholar]
- 80.Fursova KK, Shchannikova MP, Loskutova IV, Shepelyakovskaya AO, Laman AG, Boutanaev AM, Sokolov SL, Artem’eva OA, Nikanova DA, Zinovieva NA, Brovko FA. 2018. Exotoxin diversity of Staphylococcus aureus isolated from milk of cows with subclinical mastitis in Central Russia. J Dairy Sci 101:4325–4331. doi: 10.3168/jds.2017-14074. [DOI] [PubMed] [Google Scholar]
- 81.Asao T, Kumeda Y, Kawai T, Shibata T, Oda H, Haruki K, Nakazawa H, Kozaki S. 2003. An extensive outbreak of staphylococcal food poisoning due to low-fat milk in Japan: estimation of enterotoxin A in the incriminated milk and powdered skim milk. Epidemiol Infect 130:33–40. doi: 10.1017/S0950268802007951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Benkerroum N. 2017. Staphylococcal enterotoxins and enterotoxin-like toxins with special reference to dairy products: an overview. Crit Rev Food Sci Nutr 58:1943–1970. doi: 10.1080/10408398.2017.1289149. [DOI] [PubMed] [Google Scholar]
- 83.Argudín MÁ, Mendoza MC, Rodicio MR. 2010. Food poisoning and Staphylococcus aureus enterotoxins. Toxins 2:1751–1773. doi: 10.3390/toxins2071751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Smith EM, Green LE, Medley GF, Bird HE, Dowson CG. 2005. Multilocus sequence typing of Staphylococcus aureus isolated from high-somatic-cell-count cows and the environment of an organic dairy farm in the United Kingdom. J Clin Microbiol 43:4731–4736. doi: 10.1128/JCM.43.9.4731-4736.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Chaves-Moreno D, Wos-Oxley ML, Jáuregui R, Medina E, Oxley APA, Pieper DH. 2016. Exploring the transcriptome of Staphylococcus aureus in its natural niche. Sci Rep 6:33174. doi: 10.1038/srep33174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Diard M, Hardt WD. 2017. Evolution of bacterial virulence. FEMS Microbiol Rev 41:679–697. doi: 10.1093/femsre/fux023. [DOI] [PubMed] [Google Scholar]
- 87.Taur Y, Pamer EG. 2013. The intestinal microbiota and susceptibility to infection in immunocompromised patients. Curr Opin Infect Dis 26:332–337. doi: 10.1097/QCO.0b013e3283630dd3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Rynkiewicz EC, Pedersen AB, Fenton A. 2015. An ecosystem approach to understanding and managing within-host parasite community dynamics. Trends Parasitol 31:212–221. doi: 10.1016/j.pt.2015.02.005. [DOI] [PubMed] [Google Scholar]
- 89.Kamada N, Chen GY, Inohara N, Núñez G. 2013. Control of pathogens and pathobionts by the gut microbiota. Nat Immunol 14:685–690. doi: 10.1038/ni.2608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Zecconi A, Cesaris L, Liandris E, Dapra V, Piccinini R. 2006. Role of several Staphylococcus aureus virulence factors on the inflammatory response in bovine mammary gland. Microb Pathog 40:177–183. doi: 10.1016/j.micpath.2006.01.001. [DOI] [PubMed] [Google Scholar]
- 91.Emaneini M, Jabalameli F, Mirsalehian A, Ghasemi A, Beigverdi R. 2016. Characterization of virulence factors, antimicrobial resistance pattern and clonal complexes of group B streptococci isolated from neonates. Microb Pathog 99:119–122. doi: 10.1016/j.micpath.2016.08.016. [DOI] [PubMed] [Google Scholar]
- 92.Schroeder M, Brooks BD, Brooks AE. 2017. The complex relationship between virulence and antibiotic resistance. Genes 8:39. doi: 10.3390/genes8010039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Hodille E, Rose W, Diep BA, Goutelle S, Lina G, Dumitrescu O. 2017. The role of antibiotics in modulating virulence in Staphylococcus aureus. Clin Microbiol Rev 30:887–917. doi: 10.1128/CMR.00120-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kot B, Piechota M, Wolska KM, Frankowska A, Zdunek E, Binek T, Kłopotowska E, Antosiewicz M. 2012. Phenotypic and genotypic antimicrobial resistance of staphylococci from bovine milk. Pol J Vet Sci 15:677–683. doi: 10.2478/v10181-012-0105-4. [DOI] [PubMed] [Google Scholar]
- 95.de Jong A, VetPath Study Group, Garch FE, Simjee S, Moyaert H, Rose M, Youala M, Siegwart E. 2018. Monitoring of antimicrobial susceptibility of udder pathogens recovered from cases of clinical mastitis in dairy cows across Europe: VetPath results. Vet Microbiol 213:73–81. doi: 10.1016/j.vetmic.2017.11.021. [DOI] [PubMed] [Google Scholar]
- 96.Lowy FD. 2003. Antimicrobial resistance: the example of Staphylococcus aureus. J Clin Investig 111:1265–1273. doi: 10.1172/JCI18535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Cicconi-Hogan KM, Belomestnykh N, Gamroth M, Ruegg PL, Tikofsky L, Schukken YH. 2014. Short communication: prevalence of methicillin resistance in coagulase-negative staphylococci and Staphylococcus aureus isolated from bulk milk on organic and conventional dairy farms in the United States. J Dairy Sci 97:2959–2964. doi: 10.3168/jds.2013-7523. [DOI] [PubMed] [Google Scholar]
- 98.Antiabong JF, Kock MM, Bellea NM, Ehlers MM. 2017. Diversity of multidrug efflux genes and phenotypic evaluation of the in vitro resistance dynamics of clinical Staphylococcus aureus isolates using methicillin; a model β-lactam. Open Microbiol J 11:132–141. doi: 10.2174/1874285801711010132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Reyher KK, Dufour S, Barkema HW, Des Coteaux L, Devries TJ, Dohoo IR, Keefe GP, Roy JP, Scholl DT. 2011. The national cohort of dairy farms–a data collection platform for mastitis research in Canada. J Dairy Sci 94:1616–1626. doi: 10.3168/jds.2010-3180. [DOI] [PubMed] [Google Scholar]
- 100.Carson DA, Barkema HW, Naushad S, De Buck J. 2017. Bacteriocins of non-aureus staphylococci isolated from bovine milk. Appl Environ Microbiol 83:e01015-17. doi: 10.1128/AEM.01015-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Martin M. 2011. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J 17:200. doi: 10.14806/ej.17.1.200. [DOI] [Google Scholar]
- 102.Nurk S, Bankevich A, Antipov D, Gurevich AA, Korobeynikov A, Lapidus A, Prjibelski AD, Pyshkin A, Sirotkin A, Sirotkin Y, Stepanauskas R, Clingenpeel SR, Woyke T, McLean JS, Lasken R, Tesler G, Alekseyev MA, Pevzner PA. 2013. Assembling single-cell genomes and mini-metagenomes from chimeric MDA products. J Comput Biol 20:714–737. doi: 10.1089/cmb.2013.0084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Li H, Durbin R. 2010. Fast and accurate long-read alignment with Burrows–Wheeler transform. Bioinformatics 26:589–595. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Seemann T. 2014. Prokka: rapid prokaryotic genome annotation. Bioinformatics 30:2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 105.Hyatt D, Chen G-L, LoCascio PF, Land ML, Larimer FW, Hauser LJ. 2010. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics 11:119–119. doi: 10.1186/1471-2105-11-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Laslett D, Canback B. 2004. ARAGORN, a program to detect tRNA genes and tmRNA genes in nucleotide sequences. Nucleic Acids Res 32:11–16. doi: 10.1093/nar/gkh152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Lagesen K, Hallin P, Rødland EA, Staerfeldt H-H, Rognes T, Ussery DW. 2007. RNAmmer: consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res 35:3100–3108. doi: 10.1093/nar/gkm160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Gurevich A, Saveliev V, Vyahhi N, Tesler G. 2013. QUAST: quality assessment tool for genome assemblies. Bioinformatics 29:1072–1075. doi: 10.1093/bioinformatics/btt086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Koster J, Rahmann S. 2012. Snakemake–a scalable bioinformatics workflow engine. Bioinformatics 28:2520–2522. doi: 10.1093/bioinformatics/bts480. [DOI] [PubMed] [Google Scholar]
- 110.Maiden MC, van Rensburg MJJ, Bray JE, Earle SG, Ford SA, Jolley KA, McCarthy ND. 2013. MLST revisited: the gene-by-gene approach to bacterial genomics. Nat Rev Microbiol 11:728–736. doi: 10.1038/nrmicro3093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Jolley KA, Maiden MC. 2010. BIGSdb: scalable analysis of bacterial genome variation at the population level. BMC Bioinformatics 11:595. doi: 10.1186/1471-2105-11-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL. 2009. BLAST+: architecture and applications. BMC Bioinformatics 10:421. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Feil EJ, Li BC, Aanensen DM, Hanage WP, Spratt BG. 2004. eBURST: inferring patterns of evolutionary descent among clusters of related bacterial genotypes from multilocus sequence typing data. J Bacteriol 186:1518–1530. doi: 10.1128/jb.186.5.1518-1530.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Bartels MD, Petersen A, Worning P, Nielsen JB, Larner-Svensson H, Johansen HK, Andersen LP, Jarløv JO, Boye K, Larsen AR, Westh H. 2014. Comparing whole-genome sequencing with Sanger sequencing for spa typing of methicillin-resistant Staphylococcus aureus. J Clin Microbiol 52:4305–4308. doi: 10.1128/JCM.01979-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Segata N, Bornigen D, Morgan XC, Huttenhower C. 2013. PhyloPhlAn is a new method for improved phylogenetic and taxonomic placement of microbes. Nat Commun 4:2304. doi: 10.1038/ncomms3304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Fu L, Niu B, Zhu Z, Wu S, Li W. 2012. CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics 28:3150–3152. doi: 10.1093/bioinformatics/bts565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Söding J, Thompson JD, Higgins DG. 2011. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol 7:539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Castresana J. 2000. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol 17:540–552. doi: 10.1093/oxfordjournals.molbev.a026334. [DOI] [PubMed] [Google Scholar]
- 119.Capella-Gutierrez S, Silla-Martinez JM, Gabaldon T. 2009. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25:1972–1973. doi: 10.1093/bioinformatics/btp348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Price MN, Dehal PS, Arkin AP. 2010. FastTree 2–approximately maximum-likelihood trees for large alignments. PLoS One 5:e9490. doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Whelan S, Goldman N. 2001. A general empirical model of protein evolution derived from multiple protein families using a maximum-likelihood approach. Mol Biol Evol 18:691–699. doi: 10.1093/oxfordjournals.molbev.a003851. [DOI] [PubMed] [Google Scholar]
- 122.Gardner SN, Slezak T, Hall BG. 2015. kSNP3.0: SNP detection and phylogenetic analysis of genomes without genome alignment or reference genome. Bioinformatics 31:2877–2878. doi: 10.1093/bioinformatics/btv271. [DOI] [PubMed] [Google Scholar]
- 123.Treangen TJ, Ondov BD, Koren S, Phillippy AM. 2014. The Harvest suite for rapid core-genome alignment and visualization of thousands of intraspecific microbial genomes. Genome Biol 15:524. doi: 10.1186/s13059-014-0524-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Bruen TC, Philippe H, Bryant D. 2006. A simple and robust statistical test for detecting the presence of recombination. Genetics 172:2665–2681. doi: 10.1534/genetics.105.048975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol 30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Nei M, Kumar S. 2000. Molecular evolution and phylogenetics. Oxford University Press, Oxford, United Kingdom. [Google Scholar]
- 128.Page AJ, Cummins CA, Hunt M, Wong VK, Reuter S, Holden MTG, Fookes M, Falush D, Keane JA, Parkhill J. 2015. Roary: rapid large-scale prokaryote pan genome analysis. Bioinformatics 31:3691–3693. doi: 10.1093/bioinformatics/btv421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Loytynoja A, Goldman N. 2010. webPRANK: a phylogeny-aware multiple sequence aligner with interactive alignment browser. BMC Bioinformatics 11:579. doi: 10.1186/1471-2105-11-579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Chaudhari NM, Gupta VK, Dutta C. 2016. BPGA–an ultra-fast pan-genome analysis pipeline. Sci Rep 6:24373. doi: 10.1038/srep24373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Chen L, Zheng D, Liu B, Yang J, Jin Q. 2016. VFDB 2016: hierarchical and refined dataset for big data analysis–10 years on. Nucleic Acids Res 44:D694–D697. doi: 10.1093/nar/gkv1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Wattam AR, Abraham D, Dalay O, Disz TL, Driscoll T, Gabbard JL, Gillespie JJ, Gough R, Hix D, Kenyon R, Machi D, Mao C, Nordberg EK, Olson R, Overbeek R, Pusch GD, Shukla M, Schulman J, Stevens RL, Sullivan DE, Vonstein V, Warren A, Will R, Wilson MJ, Yoo HS, Zhang C, Zhang Y, Sobral BW. 2014. PATRIC, the bacterial bioinformatics database and analysis resource. Nucleic Acids Res 42:D581–D591. doi: 10.1093/nar/gkt1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.The UniProt Consortium. 2017. UniProt: the universal protein knowledgebase. Nucleic Acids Res 45:D158–D169. doi: 10.1093/nar/gkw1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Lakin SM, Dean C, Noyes NR, Dettenwanger A, Ross AS, Doster E, Rovira P, Abdo Z, Jones KL, Ruiz J, Belk KE, Morley PS, Boucher C. 2017. MEGARes: an antimicrobial resistance database for high throughput sequencing. Nucleic Acids Res 45:D574–D580. doi: 10.1093/nar/gkw1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Jia B, Raphenya AR, Alcock B, Waglechner N, Guo P, Tsang KK, Lago BA, Dave BM, Pereira S, Sharma AN, Doshi S, Courtot M, Lo R, Williams LE, Frye JG, Elsayegh T, Sardar D, Westman EL, Pawlowski AC, Johnson TA, Brinkman FSL, Wright GD, McArthur AG. 2017. CARD 2017: expansion and model-centric curation of the comprehensive antibiotic resistance database. Nucleic Acids Res 45:D566–D573. doi: 10.1093/nar/gkw1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Zankari E, Hasman H, Cosentino S, Vestergaard M, Rasmussen S, Lund O, Aarestrup FM, Larsen MV. 2012. Identification of acquired antimicrobial resistance genes. J Antimicrob Chemother 67:2640–2644. doi: 10.1093/jac/dks261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Li J, Tai C, Deng Z, Zhong W, He Y, Ou HY. 2017. VRprofile: gene-cluster-detection-based profiling of virulence and antibiotic resistance traits encoded within genome sequences of pathogenic bacteria. Brief Bioinform 19:566–574. doi: 10.1093/bib/bbw141. [DOI] [PubMed] [Google Scholar]
- 138.R Core Team. 2016. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. [Google Scholar]
- 139.Wickham H, Francois R, Henry L, Müller K. 2017. dplyr: a grammar of data manipulation. R package version 0.7.2 R Foundation for Statistical Computing, Vienna, Austria. [Google Scholar]
- 140.Ward N, Moreno-Hagelsieb G. 2014. Quickly finding orthologs as reciprocal best hits with BLAT, LAST, and UBLAST: how much do we miss? PLoS One 9:e101850. doi: 10.1371/journal.pone.0101850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.CLSI. 2016. Performance standards for antimicrobial susceptibility testing. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 142.Feßler AT, Kaspar H, Lindeman CJ, Peters T, Watts JL, Schwarz S. 2017. Proposal for agar disk diffusion interpretive criteria for susceptibility testing of bovine mastitis pathogens using cefoperazone 30mug disks. Vet Microbiol 200:65–70. doi: 10.1016/j.vetmic.2016.02.026. [DOI] [PubMed] [Google Scholar]
- 143.Christensen R. 2015. ordinal–regression models for ordinal data. R package version 2015.6-28. http://www.cran.r-project.org/package=ordinal.
- 144.Pedregosa F, Varoquaux G, Varoquaux G, Michel V, Thirion B, Grisel O, Blondel M, Prettenhofer P, Weiss R, Dubourg V, Vanderplas J, Passos A, Cournapeau D, Brucher M, Perrot M, Duchesnay É. 2011. Scikit-learn: machine learning in Python. J Machine Learning Res 12:2825–2830. [Google Scholar]
- 145.Van der Maaten L, Hinton G. 2012. Visualizing data using t-SNE. Mach Learn 87:33–2605. doi: 10.1007/s10994-011-5273-4. [DOI] [Google Scholar]
- 146.Venables WN, Ripley BD. 2002. Modern applied statistics with S. Springer, New York, NY. [Google Scholar]
- 147.Letunic I, Bork P. 2019. Interactive Tree Of Life (iTOL) v4: recent updates and new developments. Nucleic Acids Res 47:W256–W259. doi: 10.1093/nar/gkz239. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All whole-genome sequencing data used in this study are available without restriction from NCBI under BioProject no. PRJNA599195.