We compared four different oral collection methods for studying the human oral microbiome: an OMNIgene ORAL kit, Scope mouthwash, nonethanol mouthwash, and Saccomanno’s fixative. Our study shows that the type of the collection method can have a large impact on the results of an oral microbiome analysis. We recommend that one consistent oral collection method should be used for all oral microbiome comparisons. While Scope and nonethanol mouthwashes are less expensive and provide results similar to those with OMNIgene, Saccomanno’s fixative may be unfavorable due to the microbial differences detected in this study. Our results will help guide the design of future oral microbiome studies.
KEYWORDS: oral microbiome, collection method, epidemiology
ABSTRACT
Epidemiologic studies use various biosample collection methods to study associations between human oral microbiota and health outcomes. However, the agreement between the different methods is unclear. We compared a commercially available OMNIgene ORAL kit to three alternative collection methods: Saccomanno’s fixative, Scope mouthwash, and nonethanol mouthwash. Oral samples were collected from 40 individuals over 4 visits. Two samples were collected from each subject per visit: one with OMNIgene and one with an alternative method. DNA was extracted using the DSP DNA Virus Pathogen kit, and the V4 region of the 16S rRNA gene was PCR amplified and sequenced using MiSeq. Oral collection methods were compared based on alpha and beta diversity metrics and phylum- and genus-level relative abundances. All alpha diversity metrics were significantly lower for Saccomanno’s fixative than for OMNIgene (P < 0.001), whereas the two mouthwashes were more similar to OMNIgene. Principal-coordinate analysis (PCoA) using the Bray-Curtis and weighted UniFrac beta diversity matrices showed large differences in the microbial compositions of samples collected with Saccomanno’s compared to those with OMNIgene and the mouthwashes. Clustering by collection method was not observed in unweighted UniFrac PCoA plots, suggesting differences in relative abundances but not specific taxa detected by the collection methods. Relative abundances of most taxa were significantly different between OMNIgene and the other methods at each taxonomic level, with Saccomanno’s showing the least agreement with OMNIgene. There were clear differences in oral microbial communities between the four oral collection methods, particularly for Saccomanno’s fixative.
IMPORTANCE We compared four different oral collection methods for studying the human oral microbiome: an OMNIgene ORAL kit, Scope mouthwash, nonethanol mouthwash, and Saccomanno’s fixative. Our study shows that the type of the collection method can have a large impact on the results of an oral microbiome analysis. We recommend that one consistent oral collection method should be used for all oral microbiome comparisons. While Scope and nonethanol mouthwashes are less expensive and provide results similar to those with OMNIgene, Saccomanno’s fixative may be unfavorable due to the microbial differences detected in this study. Our results will help guide the design of future oral microbiome studies.
INTRODUCTION
The human oral microbiota has recently gained focus in epidemiologic studies of health and disease outcomes. Microbes residing in the oral cavity have been shown to be involved in the development of not only oral diseases such as dental caries and periodontal disease (1, 2) but also systemic diseases, including cancer, cardiovascular disease, pneumonia, and diabetes (3–6). Although the evidence is promising, many of these studies have been limited to small sample sizes, cross-sectional study designs, and samples from single time points (3, 4, 7). In addition, methodological differences and the lack of standardized protocols make it difficult to generalize and compare results across different studies (3, 8).
The oral cavity has distinct intraoral microbial habitats, which include the tooth surface, tongue, tonsil, cheeks, and the oropharyngeal region (4, 9). Bacterial community compositions have been shown to differ by collection methods using raw saliva, oral swabs, oral wash, and scrapings of dental plaque (10–12). In particular, saliva contains bacteria shed from the tissues and surface biofilms of the oral cavity (1). The salivary microbiome consists largely of bacteria released from mucosal epithelial cells and closely resembles the community composition of the tongue, but saliva also contains bacteria originating from other oral niches, including tooth surfaces and gingival crevices (1, 6). In a previous study comparing oral microbiota measurements from buccal cells in mouthwash samples with eight other oral sample types (i.e., supra- and subgingival plaque, raw saliva, and swabs from five soft tissue sites, including keratinized gingiva, hard palate, buccal mucosa, palatine tonsil, and tongue dorsum), buccal cells from mouthwash samples had the highest alpha diversity and most closely resembled the microbial profile of saliva compared to that of the other sample types (10). While the totality of the “true” oral microbiome has not been characterized and remains unknown, saliva and oral wash samples may be able to capture bacteria from a variety of oral niches. In addition, saliva and oral wash samples are attractive biospecimens for extending oral microbiome research to large population-based etiologic studies due to their ease of collection, transport, and storage.
Previous studies have compared various sample collection and storage methods to analyze oral microbiota by using saliva and oral wash samples. Unstimulated saliva showed no microbial differences compared to that of stimulated saliva collected using paraffin chewing gum (13). Commercially available OMNIgene ORAL saliva collection kits have been compared to saliva stored in liquid dental transport medium (11) and also to oral wash samples collected using Scope mouthwash (12, 14, 15) and 0.9% saline solution (16). While alpha diversity was relatively comparable between OMNIgene and saliva stored in liquid dental transport, Scope, and saline solution, relative abundances of identified microbial species showed differences by the collection and storage method in some of these previous studies (11, 12, 14, 16). Thus, differences in sample collection and storage media may alter the microbial composition and structure, but the agreement between the different methods and how they influence our understanding of the oral microbiota remains unclear.
Here, we compared an OMNIgene ORAL kit to three alternative collection methods for studying oral microbiota: Saccomanno’s fixative (which is frequently used to collect sputum samples for cytologic analysis) and Scope and nonethanol mouthwashes. Oral samples were collected from 40 individuals over four visits within 8 months. Two collection methods were used at each visit: one OMNIgene and one alternative oral collection method. Saccomanno’s and Scope and nonethanol mouthwashes were compared to OMNIgene in paired samples based on various alpha and beta diversity metrics and phylum- and genus-level relative abundances. This study has important implications for sample collection in future epidemiologic studies of oral microbiota.
RESULTS
Comparability of alpha diversity.
Overall, all alpha diversity metrics were higher in OMNIgene samples than in the two mouthwashes and Saccomanno’s fixative samples (Fig. 1 and Table 1; see also Fig. S1 in the supplemental material). In particular, there was a significant difference between OMNIgene and Saccomanno’s for all three alpha diversity metrics (P < 0.001). For example, the mean numbers of observed species were 145 for OMNIgene and 129 for Saccomanno’s (11% decrease, P < 0.001). Faith’s phylogenic diversity (PD) was statistically significantly different between OMNIgene (mean, 18.7) and Scope mouthwash (mean, 18.1; P = 0.00978). However, there was no statistical difference for the Shannon index (P = 0.0770), and there was no difference in the number of observed species between OMNIgene (mean, 146) and Scope mouthwash (mean, 140; 4% decrease; P = 0.0584). Nonethanol mouthwash was the most similar to OMNIgene, where the mean numbers of observed species were 150 for OMNIgene and 147 for the nonethanol mouthwash (2% decrease, P = 0.532). Similarly, there was no difference in Faith’s PD between OMNIgene (mean, 19.3) and the nonethanol mouthwash (mean, 18.6; P = 0.192). However, the Shannon index was significantly lower for nonethanol mouthwash (mean, 4.88) than for OMNIgene (mean, 5.02; P = 0.0241). In general, the correlation between alpha diversity measurements from OMNIgene and other collection methods was low for the Shannon index, which incorporates richness and evenness, compared to observed species and Faith’s PD, which incorporate only richness (Fig. S1).
FIG 1.
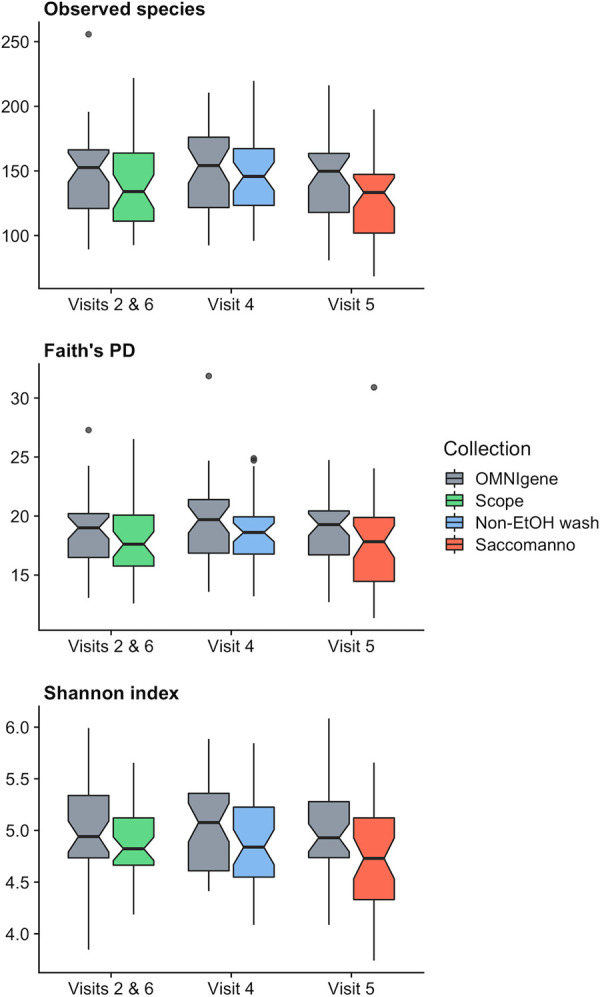
Boxplots of alpha diversity metrics comparing Scope, nonethanol mouthwash (Non-EtOH wash), and Saccomanno’s fixative to the OMNIgene ORAL kit for observed species (top), Faith’s PD (middle), and the Shannon index (bottom). The medians are marked by the horizontal lines in the boxes. Observations outside 1.5 times the interquartile range above the upper quartile and below the lower quartile are shown as outliers. Notches represent 95% confidence intervals for comparing medians, where nonoverlapping notches indicate evidence for differences in medians (35).
TABLE 1.
Comparison of alpha diversity metrics between OMNIgene ORAL samples, Scope and nonethanol mouthwashes, and Saccomanno’s fixative
| Visit(s) | Mouthwash | Observed species |
Faith’s PD |
Shannon index |
|||
|---|---|---|---|---|---|---|---|
| Mean (SD) | Wilcoxon P value | Mean (SD) | Wilcoxon P value | Mean (SD) | Wilcoxon P value | ||
| 2 and 6 | OMNIgene | 146 (34.4) | 0.0584 | 18.7 (3.15) | 0.00978 | 4.99 (0.443) | 0.0770 |
| Scope | 140 (33.7) | 18.1 (3.22) | 4.90 (0.360) | ||||
| 4 | OMNIgene | 150 (33.9) | 0.532 | 19.3 (3.55) | 0.192 | 5.02 (0.412) | 0.0241 |
| Nonethanol | 147 (32.2) | 18.6 (3.01) | 4.88 (0.436) | ||||
| 5 | OMNIgene | 145 (33.6) | <0.001 | 18.6 (3.09) | <0.001 | 4.97 (0.437) | <0.001 |
| Saccomanno’s | 129 (32.3) | 17.8 (3.91) | 4.73 (0.480) | ||||
Plots of alpha diversity metrics comparing Scope and nonethanol mouthwashes (Non-EtOH wash) and Saccomanno’s fixative to the OMNIgene ORAL kit for observed species (top), Faith’s PD (middle), and the Shannon index (bottom). Adjusted coefficients of determination (R2 values) were obtained by fitting linear regression models. Dotted lines represent the line of equality (y = x). Download FIG S1, PDF file, 0.4 MB (363.3KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Comparability of beta diversity.
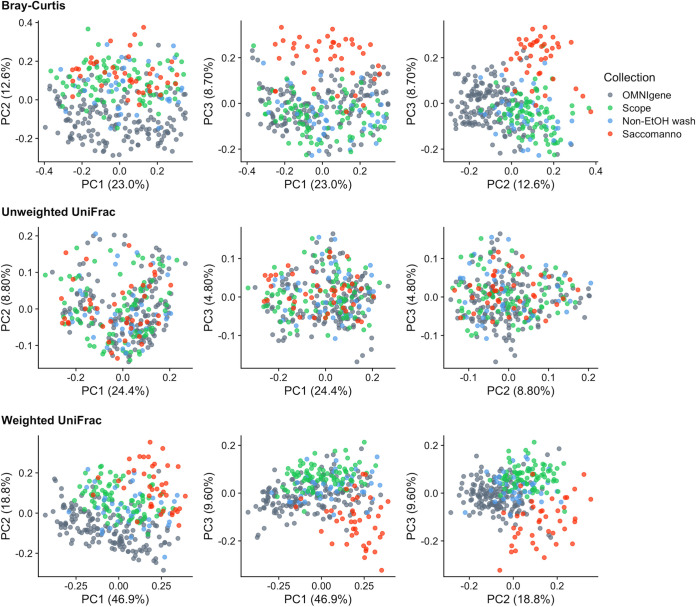
Ordination plots generated from principal-coordinate analyses (PCoAs) using the Bray-Curtis and weighted UniFrac beta diversity matrices had visual separation of samples by collection method, with a particularly distinct cluster for Saccomanno’s fixative samples (Fig. 2). In plots of the first three principal coordinates (PCs) of the PCoA using the Bray-Curtis distance matrix, Saccomanno’s was separated from the other collection methods by PC2 (12.6% variation explained) and PC3 (8.70% variation explained), but not by PC1 (23.0% variation explained). Scope and OMNIgene were only separated by PC2. There was no separation between Scope and the nonethanol mouthwash, and the two mouthwashes were only separated from OMNIgene by PC2. In the PCoA plots derived using the weighted UniFrac distance matrix, Saccomanno’s was separated by all three PCs (46.9%, 18.8%, and 9.60% variation explained by PCs 1, 2, and 3, respectively), whereas the two mouthwashes and OMNIgene were separated by PC2 and PC3. However, there was no visual separation by collection method in PCoA plots generated from the unweighted UniFrac distance matrix (24.4%, 8.80%, and 4.80% variation explained by PCs 1, 2, and 3, respectively), which suggests the presence/absence of microbial community members was similar for all four oral collection methods, but there were differences in the relative abundances of taxa by collection method. Based on the distance-based coefficient of determination, R2, calculated from the beta diversity matrices using permutational multivariate analysis of variance (PERMANOVA), more than one-half of the variability in the matrices was explained by between-subject differences, while a smaller proportion of microbial variability was associated with the different collection methods (R2 < 20%, P < 0.001) (Fig. 3). There was also a large proportion (∼30%) of variability that was not explained by subject, visit, or collection method represented by the model residual (Fig. 3). The model residual includes technical variation attributed to analytical procedures such as sampling and sequencing.
FIG 2.
Plots of the first three principal coordinates from principal-coordinate analyses using Bray-Curtis (top), unweighted (middle), and weighted UniFrac (bottom) matrices, colored by oral collection method.
FIG 3.
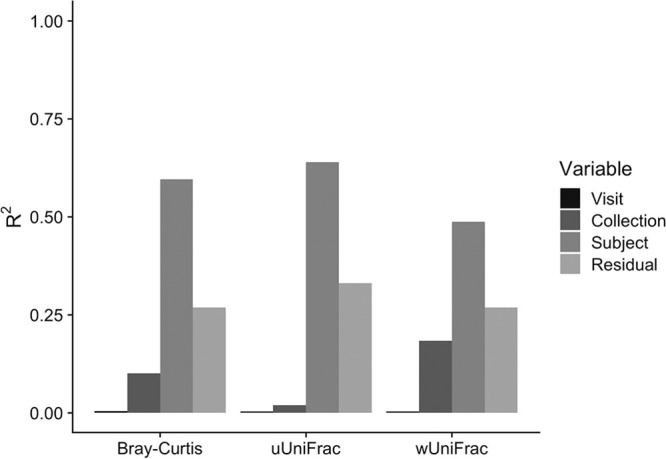
Proportions of microbial variability explained by visit, collection method, and subject. Distance-based coefficients of determination, R2 values, were calculated from the beta diversity matrices using permutational multivariate analysis of variation (PERMANOVA).
Differential abundance analysis.
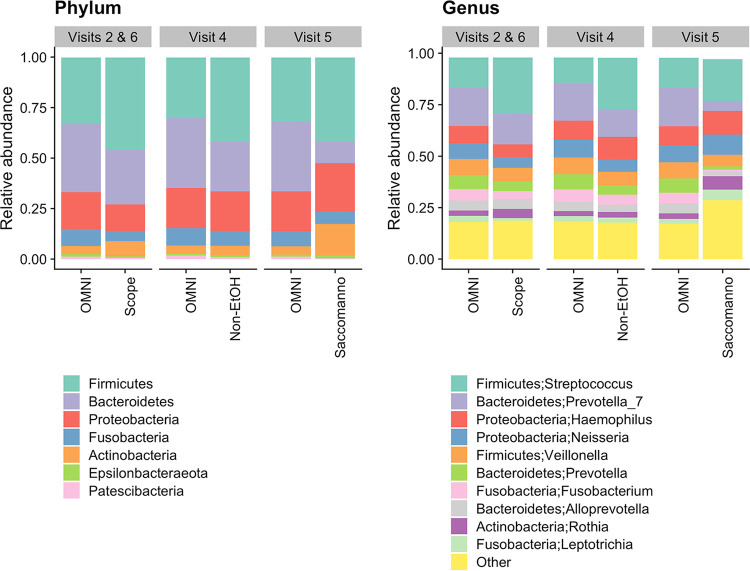
Overall, taxonomic profiles from Scope and nonethanol mouthwashes and Saccomanno’s fixative differed from those with OMNIgene (Fig. 4; see Fig. S2 for all taxonomic levels). Taxonomic profiles from OMNIgene were relatively stable across visits, and profiles from Scope and nonethanol mouthwashes were similar to each other. Saccomanno’s had a distinct taxonomic profile that differed from the other collection methods.
FIG 4.
Taxonomic profiles at the phylum (left) and genus (right) levels of OMNIgene ORAL (OMNI) compared to Scope and nonethanol mouthwashes (Non-EtOH) and Saccomanno’s fixative.
Taxonomic profiles at the class, order, and family levels of OMNIgene ORAL (OMNI) compared to Scope and nonethanol mouthwashes (Non-EtOH) and Saccomanno’s fixative. Download FIG S2, PDF file, 0.1 MB (146.4KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
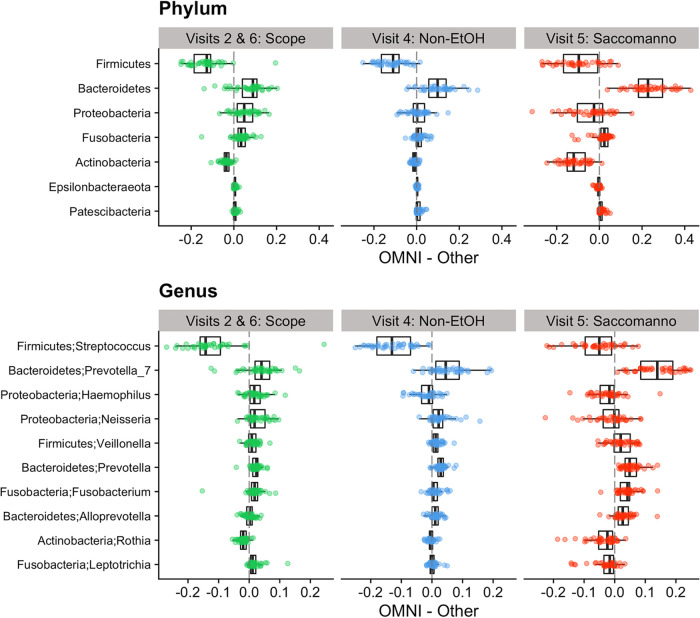
Relative abundances of most taxa were significantly different between OMNIgene and the other methods when comparing the top 10 taxa at the phylum and genus levels (Fig. 5; see Fig. S3 and Table S1 for all taxonomic levels). For example, relative abundances in the phyla Actinobacteria and Firmicutes were consistently higher and Bacteroidetes was lower in Scope, nonethanol mouthwash, and Saccomanno’s than in OMNIgene (P < 0.001), and the differences in Actinobacteria and Bacteroidetes were more pronounced for Saccomanno’s (Fig. 5). For example, the mean relative abundance of Bacteroidetes was approximately 0.345 for OMNIgene across all visits, whereas the means for Scope, nonethanol mouthwash, and Saccomanno’s were 0.274, 0.247, and 0.106, respectively (Table S1). The abundance of phylum Fusobacteria was significantly lower in Scope mouthwash (mean, 0.0511) than in OMNIgene (mean, 0.0851; 40% decrease; P < 0.001). Fusobacteria abundance was also lower in Saccomanno’s (mean, 0.0626) than in OMNIgene (0.0747, 16% decrease, P = 0.0194), while Fusobacteria abundances were similar between OMNIgene (mean, 0.0855) and nonethanol mouthwash (mean, 0.0742; P = 0.0531).
FIG 5.
Differences in relative abundances of the top 10 taxa at the phylum (top) and genus (bottom) levels between paired samples of OMNIgene ORAL, Scope and nonethanol (Non-EtOH) mouthwashes, and Saccomanno’s fixative. Dotted lines represent lines of equality indicating no difference between OMNIgene and the alternative method.
Differences in relative abundances of the top 10 taxa at the class, order, and family levels between paired samples of OMNIgene ORAL, Scope and nonethanol (Non-EtOH) mouthwashes, and Saccomanno’s fixative. Dotted lines represent line sof equality indicating no difference between OMNIgene and the alternative method. Download FIG S3, PDF file, 0.2 MB (239.8KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Comparison of relative abundances of the top 10 taxa between OMNIgene and alternative methods at the phylum to genus levels. Download Table S1, XLS file, 0.05 MB (52KB, xls) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
At the genus level, relative abundances of Streptococcus (means: OMNIgene, 0.148; Scope, 0.274; P < 0.001) and Rothia (means: OMNIgene, 0.0253; Scope, 0.0449; P < 0.001) were significantly higher in Scope mouthwash than in OMNIgene, whereas all other top 10 genera were significantly lower in Scope, except Alloprevotella, which showed no difference (means: OMNIgene, 0.0497; Scope, 0.0482; P = 1.00) (Table S1). Similar to that in Scope, nonethanol mouthwash also had higher levels of Streptococcus (means: OMNIgene, 0.123; nonethanol mouthwash, 0.254; P < 0.001) and lower levels of most of the other top 10 genera than those in OMNIgene. However, nonethanol mouthwash showed higher levels of Haemophilus (means: OMNIgene, 0.0920; nonethanol mouthwash, 0.109; P = 0.0305) and no differences in Rothia (means: OMNIgene, 0.0242; nonethanol mouthwash, 0.0271; P = 0.368) and Leptotrichia (means: OMNIgene, 0.0261; nonethanol mouthwash, 0.0237; P = 1.00) compared to those from OMNIgene. Saccomanno’s had higher levels of Streptococcus (means: OMNIgene, 0.146; Saccomanno’s, 0.202; P < 0.001), Rothia (means: OMNIgene, 0.0265; Saccomanno’s, 0.0652; P < 0.001), Leptotrichia (means: OMNIgene, 0.0231; Saccomanno’s, 0.0518; P < 0.001), and Haemophilus (means: OMNIgene, 0.0930; Saccomanno’s, 0.116; P = 0.00538) than OMNIgene. All other top 10 genera were significantly lower in Saccomanno’s than in OMNIgene, with the exception of Neisseria, which showed no difference (means: OMNIgene, 0.0820; Saccomanno’s, 0.0976; P = 1.00).
We also classified the top 10 taxa at the genus level based on Gram staining and oxygen requirements to see if there were any patterns in the changes of relative abundances by the collection methods (data not shown). Although there was a general lower relative abundance of many Gram-negative bacteria in the alternative collection methods than in OMNIgene, this is likely to have been largely driven the predominance of the Gram-positive bacterium Streptococcus in Saccomanno’s and the mouthwash samples. In addition, anaerobic bacteria tended to be lower in samples collected with the alternative methods than in those collected with OMNIgene.
DISCUSSION
In this study, oral samples collected using OMNIgene were compared to those collected with Scope and nonethanol mouthwashes and Saccomanno’s fixative. Our results demonstrated that there were clear differences in oral microbial communities between the four collection methods, particularly for Saccomanno’s. Overall, Saccomanno’s had the least agreement with OMNIgene when comparing alpha and beta diversity metrics and relative abundances of the detected taxa. The most noticeable differences between OMNIgene and the alternative methods were seen in the taxonomic profiles of phylum- and genus-level relative abundances. The differences between methods did not appear to be driven by the presence/absence of specific taxa but rather by differences in the relative abundances of the taxa detected from each collection method.
The discrepancy in the relative abundances of taxa may be due to the different chemical compositions of the collection/storage media. Both Scope and the nonethanol mouthwash contain antimicrobial agents that inhibit the growth of bacteria. Cetylpyridinium chloride, the main active ingredient in Scope mouthwash, is a cationic surfactant that inhibits bacterial growth by binding to negatively charged bacterial surfaces and leads to the disruption of the cell membrane and leakage of intracellular components (14, 17). The nonethanol mouthwash used in this study contains active ingredients derived from essential oils (i.e., eucalyptol, thymol, methyl salicylate, and menthol) which have been shown to have antimicrobial effects (19), although they may be less effective toward some bacterial species than cetylpyridinium chloride (20). The stabilizing buffer in OMNIgene contains 1% to 5% sodium dodecyl sulfate (SDS), which is an anionic detergent and protein denaturant that effectively inhibits growth of various microorganisms (21, 22). Although at a lower concentration of around 0.5% to 2% as commonly found in dentifrices (21), the nonethanol mouthwash also contains SDS that may have contributed to the prevention of bacterial growth (23, 24). For Saccomanno’s fixative, the primary ingredients are ethanol (∼40%), isopropanol (∼2%), and methanol (∼2%). Although high concentrations of ethanol are known to be lethal to many bacteria, there are bacteria that have a high ethanol tolerance, and some may also adapt to ethanol exposures (25–27). In fact, several studies of fecal collection methods have demonstrated that 70% ethanol is an inadequate stabilization buffer for the fecal microbiome, and that a higher concentration of ethanol (95% or more) was required to prevent changes in the fecal microbiota composition during storage (28, 29). Therefore, the lack of an additional preservative besides alcohol may have led to an overgrowth of certain bacteria in Saccomanno’s, and thus resulted in its distinct microbial composition that differed from those of the other collection methods in this study.
In addition to the chemical composition of the storage medium, storage temperatures can also impact the microbial composition of oral samples (11, 12). In this study, the two mouthwash samples and the OMNIgene samples collected from the same visits (visits 2, 4, and 6) were left at room temperature for an average of 1 day prior to storage at −80°C in the laboratory. The manufacturer states this specific OMNIgene kit is stable for up to 3 weeks at room temperature, while there may be changes in Scope mouthwash at room temperature. When comparing the samples from the two mouthwashes to OMNIgene samples, we observed differences in the relative abundances of taxa, for example, higher levels of the phylum Firmicutes and the Streptococcus genus in the mouthwash samples, but it is unclear whether these changes were related to an overgrowth of certain taxa during storage at room temperature or a difference between the sample collection methods. Previously, the stability of oral samples stored in Scope mouthwash was evaluated by comparing samples that were immediately frozen with samples stored at room temperature for 4 days and subsequently frozen (12). Scope mouthwash samples were generally stable at room temperature based on alpha and beta diversity metrics and the relative abundances of four phyla. However, the intraclass correlation coefficients (ICCs) for the relative abundances of bacteria varied, with ICCs being 0.75 or greater for most of the top 25 genera, but some were lower, such as the Streptococcus genus which had an ICC of 0.65. Specifically, the phylum Firmicutes had a high relative abundance in samples stored at room temperature, which was primarily driven by an increase in Streptococcus, compared to that in immediately frozen samples. This previous study also compared immediately frozen Scope mouthwash samples to OMNIgene samples, and while the ICCs of alpha diversity metrics were high (∼0.75), ICCs of beta diversity metrics and relative abundances were generally low (12). In particular, immediately frozen Scope mouthwash samples had higher relative abundances of Firmicutes than those from OMNIgene and had an ICC of 0.29, and the ICC for the Streptococcus genus was 0.37. In terms of the Streptococcus genus, it is also unclear how reliably relative abundances were measured from the OMNIgene samples, since the ICC for this genus was 0.452 in duplicate OMNIgene samples collected from the baseline visit in our present study (see Materials and Methods). Therefore, while there may have been changes in the microbial compositions of the Scope mouthwash samples during storage at room temperature, particularly in the relative abundances of certain taxa, it is unlikely that immediately freezing Scope mouthwash samples would have made them more comparable to the OMNIgene samples. However, this needs to be verified in future studies, since another study reported there were no significant differences in relative abundances of major phyla and all identified genera when comparing paired OMNIgene and Scope mouthwash oral samples that were both immediately frozen within 10 min of collection (14). The Saccomanno’s samples collected in this study were stored at room temperature until analysis, and this may have also impacted the microbial composition of these samples. A previous study evaluated the effect of storage at room temperature for fecal samples preserved in 70% ethanol, and they reported a large increase in the genera Streptococcus and Haemophilus over the course of 8 weeks compared to that in freshly collected samples that were analyzed on the same day (29). Although the ethanol concentration is lower in Saccomanno’s, Streptococcus and Haemophilus abundances were also increased in the Saccomanno’s samples compared to that in OMNIgene in this study.
Our study has several limitations. All samples were stored at room temperature for some amount of time, which may have led to changes in the community composition due to growth of certain oral microbiota, but our previous work suggests that these methods are generally stable at room temperature for several days (12). We were also unable to compare across the alternative collection methods due to the design of the study, since none of the visits included more than two collection methods. In addition, OMNIgene samples were always sampled first, before the collection with alternative methods. This may have introduced a bias, possibly decreasing the bacterial load in samples collected with the alternative methods. The participants of our study were also limited to healthy, highly educated adults that work at the National Cancer Institute. Therefore, our results may not be generalizable to other populations such as children, the elderly, and unhealthy subjects. It is also unclear how differences in oral collection methods will impact analyses using other sequencing technologies, such as shotgun metagenomics, since we only assessed 16S rRNA gene sequencing.
In large population-based epidemiologic studies of oral microbiota, samples collected with an ideal sample collection method should not only closely represent the host’s microbiome but should also be easy to collect, store, and transport and should not be cost prohibitive. There is currently no gold standard collection method for studying the oral microbiome. Although saliva samples containing no additives and frozen immediately after collection may provide more accurate representations of the host oral microbiota, that may not be practical for large epidemiologic studies. As noted above, all samples in this study were exposed to ambient temperatures, which may have altered the microbial composition. However, this is a more realistic representation of how samples are collected in the field, where samples need to be transported before they can be stored in a laboratory freezer (12). Overall, the results of this study suggest Scope and nonethanol mouthwashes can be used as less expensive oral collection methods to obtain microbial measurements similar to those from OMNIgene samples in epidemiologic settings. Use of Saccomanno’s may be unfavorable due to the microbial differences detected in this study, although it is possible that freezing the sample in Saccomanno’s may decrease these differences and should be studied in the future. Regardless of which sample collection method is chosen, at least one consistent method should be used for new epidemiologic studies to minimize the potential for biased results.
In conclusion, collection and storage of oral samples using OMNIgene was compared to those with Scope and nonethanol mouthwashes and Saccomanno’s fixative. Although there were differences in taxonomic profiles, the two mouthwashes were more similar to OMNIgene when comparing alpha and beta diversity metrics. Saccomanno’s was very distinct from the other collection methods and had the least agreement with OMNIgene for all microbial metrics used in this study. Scope and nonethanol mouthwashes are suitable collection methods for measuring oral microbiota and provided results similar to OMNIgene, but use of Saccomanno’s fixative in future epidemiologic investigations is not recommended.
MATERIALS AND METHODS
Study population.
A detailed description of this population was published previously (15). Briefly, a convenience cohort of 40 employees at the National Cancer Institute were recruited by e-mail and word of mouth in 2014. Study participants completed questionnaires at each study visit providing information on demographics, tobacco and alcohol use, and self-reported height and weight. Most participants were white (67.5%), female (62.5%), and highly educated (92.5% had obtained a master’s or doctoral degree). The mean age and body mass index (BMI) were 40.0 years and 25.9 kg/m2, respectively. All participants reported ever drinking alcohol, but few participants had ever smoked cigarettes (12.5%). All participants provided written informed consent, and this study was approved by the Special Studies Institutional Review Board of the National Cancer Institute.
Oral sample collection.
Oral samples were collected from participants every 2 months over six visits, as described in detail previously (15). At the second, fourth, fifth, and sixth visits, two oral collection methods were used to collect a paired sample of one OMNIgene and one alternative collection method from each participant. Participants were asked to refrain from oral hygiene procedures (i.e., tooth brushing, flossing, use of mouthwash and other dental rinse products), eating, drinking (other than water), chewing gum, consuming throat lozenges or candies, and smoking during the 12 h prior to the scheduled visit.
At each visit, the first oral sample was collected from each participant using the OMNIgene ORAL collection kit (OM-505; DNA Genotek, Ottawa, ON, Canada), which is stable at room temperature for 3 weeks for collection of DNA and RNA. Although OMNIgene samples were consistently collected at each visit, this collection method was not used as a “gold standard” but was rather used as a reference to evaluate the other collection methods. As described previously, participants spit into the kit until the saliva reached the fill line, the kit was closed to release the fixative, and the tube was capped and shaken (15). OMNIgene samples were stored at room temperature for an average of 7 days for visit 1, 6 days for visit 5, and 1 day for visits 2, 4, and 6 before being transported to the laboratory where they were shaken, incubated at 50°C for 1 h in a water bath, aliquoted, and frozen at −80°C. At the baseline visit, two consecutive OMNIgene samples were collected from each participant, and these samples were used to assess the reliability of this collection kit.
At the second and sixth visits, participants provided an additional oral sample using Scope mouthwash (Procter & Gamble, Cincinnati, OH) following the OMNIgene sample. Participants were given 10 ml of Scope mouthwash in a sterile cup, which they swished and gargled for 5 s each, for a total of 30 s. Once the 30 s had elapsed, they spit the Scope mouthwash back into the cup (15). At the fourth visit, a nonethanol mouthwash (Irsha; Shafa Pharmaceuticals, Tehran, Iran) was used to collect an oral sample. Participants followed the same procedure as for the Scope mouthwash for this collection. Both the Scope and nonethanol mouthwash samples were stored at room temperature for an average of 1 day before being transferred to the laboratory for aliquoting and storage at −80°C.
For the fifth visit, participants provided additional saliva to be preserved in Saccomanno’s fixative (Lerner Laboratories, Pittsburgh, PA), which is typically used as a fixative for cytology. Participants were asked to spit into a sterile cup until the saliva approximately reached the 5 ml line. Approximately 35 ml of Saccomanno’s was added to the saliva, and these samples remained at room temperature for approximately 1 year until DNA extraction.
DNA extraction, amplification, and sequencing.
The methods for DNA extraction, PCR amplification, and sequencing were described in detail previously (15). Briefly, samples were extracted in batches of 24 by collection method (i.e., OMNIgene, Scope mouthwash, nonethanol mouthwash, and Saccomanno’s), keeping samples from the same individual in the same batch. OMNIgene samples were incubated for 1 h at 50°C, and an aliquot of 1,000 μl was incubated at 75°C for 15 min prior to DNA extraction. For the Scope and nonethanol mouthwash samples and the Saccomanno’s samples, an aliquot of 1,000 μl was transferred to a Pathogen Lysis Tube-L (Qiagen, Hilden, Germany) and pelleted. Upon removing the supernatant, beads and buffer solutions were added to perform cell lysis. DNA was extracted from samples using the DSP DNA Virus Pathogen kit (Qiagen, Hilden, Germany) on a QIAsymphony instrument (Qiagen, Hilden, Germany). Each DNA extraction batch contained three quality control samples: (i) either an oral artificial community or a chemostat community, (ii) a blank, and (iii) an extraction replicate of a randomly selected sample within the batch. The oral artificial community and chemostat community samples were generated for the Microbiome Quality Control project and were described previously (8). An equimolar mix of barcoded 515F/806R primers was used for amplifying the V4 region of the 16S rRNA gene. DNA sequencing was performed with the Illumina MiSeq v2 to obtain 250-bp paired-end reads.
Bioinformatic processing.
As described previously (15), paired-end sequence reads were demultiplexed using CASAVA and processed into amplicon sequence variants (i.e., 100% operational taxonomic units [OTUs]) using DADA2 version 1.10 (30). Sequence variants were aligned and assigned taxonomy using the SILVA v123 database (31). After removing nonbacterial sequences, 3,537 sequence variants were obtained. Relative abundances and alpha and beta diversity metrics were calculated using QIIME 2 (32). Alpha diversity metrics, including observed species, Shannon index, and Faith’s PD, were calculated by taking the average from 10 subsamples with rarefaction at 30,000 reads (see Fig. S4 in the supplemental material). Bray-Curtis, unweighted UniFrac, and weighted UniFrac beta diversity matrices were generated with rarefaction at 30,000 reads. Five of 6 blank samples and 3 of 320 study samples were removed after rarefaction. This left a total of 317 study samples remaining after rarefaction, of which 14 samples had extraction replicates. None of the artificial community or chemostat samples were removed due to rarefaction, with 11 samples of each remaining in the data set. Relative abundances from the phylum to the genus levels were calculated for each sample without rarefaction. Taxa with a prevalence less than 10% or mean relative abundance less than 0.002 were excluded from the analysis. Relative abundances of a total of 7 phyla, 11 classes, 15 orders, 23 families, and 33 genera were available for analysis.
Rarefaction curves for the mean numbers of observed species by sample type. Error bars represent 95% confidence intervals. Download FIG S4, PDF file, 0.07 MB (68.9KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
For the quality control samples (i.e., artificial community and chemostat samples), the coefficients of variation (CVs) were less than or equal to 8% and 6%, respectively, for all alpha diversity metrics. The CVs varied by taxa for the relative abundances of the top 4 phyla in the artificial community, ranging from 5.74% for Proteobacteria to 30.8% for Bacteroidetes. For the chemostat community samples, the CVs of relative abundances of the top 4 phyla ranged from 7.62% for Firmicutes to 15.3% for Verrucomicrobia. Plots of the first three PCs from PCoA generated using each of the beta diversity matrices showed separate clusters for the artificial community and chemostat samples and the study subject samples (see Fig. S5). The ICC of extraction replicates were ≥0.907 for alpha diversity metrics, ≥0.951 for the first PC from the beta diversity matrices, and ≥0.913 for the relative abundances of the top four phyla. Overall, these data from the quality control samples and extraction replicates indicated good reproducibility across and within analytical batches.
Plots of the first three principal coordinates from principal-coordinate analyses using Bray-Curtis (top), unweighted (middle), and weighted UniFrac (bottom) matrices, colored by sample type. Download FIG S5, PDF file, 0.4 MB (452.8KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
To assess the reliability of the microbial measurements from samples collected using the OMNIgene ORAL kit, ICCs were calculated from the duplicate OMNIgene samples obtained from each participant at the baseline visit. For alpha diversity metrics, the ICC was 0.923 for observed species, 0.953 for Faith’s PD, and 0.870 for the Shannon index. The ICCs were ≥0.957 for the first PC from the beta diversity matrices. The reliability of measurements of the relative abundances of the top 4 phyla varied by taxa, and the ICCs ranged from 0.527 for Firmicutes to 0.905 for Proteobacteria. For the top 4 genera, ICCs of the relative abundances ranged from 0.452 for Streptococcus to 0.966 for Haemophilus. The temporal variability of microbial measures from OMNIgene samples collected over the six visits of this study was described previously (15). Briefly, ICCs were generally high for alpha and beta diversity measures, with the ICC for observed species being 0.74 (95% confidence interval [CI], 0.66 to 0.82), and the ICCs for beta diversity metrics were >0.60. Relative abundances of the top 4 phyla were less stable over time, with ICCs ranging from 0.44 (95% CI, 0.30 to 0.58) for Firmicutes to 0.67 for Fusobacteria (95% CI, 0.56 to 0.77) and Proteobacteria (95% CI, 0.53 to 0.80).
Statistical analysis.
All statistical analyses were performed using the R statistical programming environment (33). Oral collection methods were compared based on alpha and beta diversity metrics and phylum- and genus-level relative abundances. For the OMNIgene and Scope samples from visits 2 and 6, the means of the relative abundances and alpha diversity metrics were calculated for each individual to combine the two visits. Alpha diversity was compared between paired samples of OMNIgene and an alternative collection method from each visit using the Wilcoxon signed-rank test. PCoA plots were generated using beta diversity matrices to visualize separation of samples by the collection method based on pairwise distances. Differences in oral microbial compositions by collection methods were tested using PERMANOVA for the beta diversity matrices. The distance-based coefficient of determination R2 was calculated using PERMANOVA (adonis2 function in the vegan package, R) (34) to quantify the variability in the microbial composition that was associated with between-subject differences, collection method, and visit. Differential abundance analysis was performed using the Wilcoxon signed-rank test to identify taxa that were differentially abundant between paired samples of OMNIgene and the alternative collection method. Specifically, untransformed relative abundances of the top 10 most prevalent taxa were compared at each taxonomic level using Wilcoxon signed-rank tests with Bonferroni’s correction to account for multiple comparisons. Bonferroni-adjusted P values were computed using the p.adjust function in R, in which unadjusted P values are multiplied by the number of comparisons. Statistical tests were performed at a significance level of 0.05.
Data availability.
The sequencing data are available through the NCBI Sequence Read Archive (PRJNA634162).
ACKNOWLEDGMENTS
This study was supported by the Intramural Research Program of the National Cancer Institute.
We thank all our participants. This work utilized the computational resources of the National Institutes of Health HPC Biowulf cluster (https://hpc.nih.gov).
REFERENCES
- 1.Costalonga M, Herzberg MC. 2014. The oral microbiome and the immunobiology of periodontal disease and caries. Immunol Lett 162:22–38. doi: 10.1016/j.imlet.2014.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kilian M, Chapple ILC, Hannig M, Marsh PD, Meuric V, Pedersen AML, Tonetti MS, Wade WG, Zaura E. 2016. The oral microbiome – an update for oral healthcare professionals. Br Dent J 221:657–666. doi: 10.1038/sj.bdj.2016.865. [DOI] [PubMed] [Google Scholar]
- 3.Mascitti M, Togni L, Troiano G, Caponio VCA, Gissi DB, Montebugnoli L, Procaccini M, Lo Muzio L, Santarelli A. 2019. Beyond head and neck cancer: the relationship between oral microbiota and tumour development in distant organs. Front Cell Infect Microbiol 9:232. doi: 10.3389/fcimb.2019.00232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jia G, Zhi A, Lai PFH, Wang G, Xia Y, Xiong Z, Zhang H, Che N, Ai L. 2018. The oral microbiota – a mechanistic role for systemic diseases. Br Dent J 224:447–455. doi: 10.1038/sj.bdj.2018.217. [DOI] [PubMed] [Google Scholar]
- 5.Scannapieco FA. 2013. The oral microbiome: its role in health and in oral and systemic infections. Clin Microbiol Newsl 35:163–169. doi: 10.1016/j.clinmicnews.2013.09.003. [DOI] [Google Scholar]
- 6.Acharya A, Chan Y, Kheur S, Jin LJ, Watt RM, Mattheos N. 2017. Salivary microbiome in non-oral disease: a summary of evidence and commentary. Arch Oral Biol 83:169–173. doi: 10.1016/j.archoralbio.2017.07.019. [DOI] [PubMed] [Google Scholar]
- 7.Vogtmann E, Goedert JJ. 2016. Epidemiologic studies of the human microbiome and cancer. Br J Cancer 114:237–242. doi: 10.1038/bjc.2015.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sinha R, Abnet CC, White O, Knight R, Huttenhower C. 2015. The microbiome quality control project: baseline study design and future directions. Genome Biol 16:276. doi: 10.1186/s13059-015-0841-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lim Y, Totsika M, Morrison M, Punyadeera C. 2017. Oral microbiome: a new biomarker reservoir for oral and oropharyngeal cancers. Theranostics 7:4313–4321. doi: 10.7150/thno.21804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yu G, Phillips S, Gail MH, Goedert JJ, Humphrys M, Ravel J, Ren Y, Caporaso NE. 2017. Evaluation of Buccal cell samples for studies of oral microbiota. Cancer Epidemiol Biomarkers Prev 26:249–253. doi: 10.1158/1055-9965.EPI-16-0538. [DOI] [PubMed] [Google Scholar]
- 11.Luo T, Srinivasan U, Ramadugu K, Shedden KA, Neiswanger K, Trumble E, Li JJ, McNeil DW, Crout RJ, Weyant RJ, Marazita ML, Foxman B. 2016. Effects of specimen collection methodologies and storage conditions on the short-term stability of oral microbiome taxonomy. Appl Environ Microbiol 82:5519–5529. doi: 10.1128/AEM.01132-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vogtmann E, Chen J, Kibriya MG, Amir A, Shi J, Chen Y, Islam T, Eunes M, Ahmed A, Naher J, Rahman A, Barmon B, Knight R, Chia N, Ahsan H, Abnet CC, Sinha R. 2019. Comparison of oral collection methods for studies of microbiota. Cancer Epidemiol Biomarkers Prev 28:137–143. doi: 10.1158/1055-9965.EPI-18-0312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Belstrøm D, Holmstrup P, Bardow A, Kokaras A, Fiehn N-E, Paster BJ. 2016. Comparative analysis of bacterial profiles in unstimulated and stimulated saliva samples. J Oral Microbiol 8:30112. doi: 10.3402/jom.v8.30112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fan X, Peters BA, Min D, Ahn J, Hayes RB. 2018. Comparison of the oral microbiome in mouthwash and whole saliva samples. PLoS One 13:e0194729. doi: 10.1371/journal.pone.0194729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vogtmann E, Hua X, Zhou L, Wan Y, Suman S, Zhu B, Dagnall CL, Hutchinson A, Jones K, Hicks BD, Sinha R, Shi J, Abnet CC. 2018. Temporal variability of oral microbiota over 10 months and the implications for future epidemiologic studies. Cancer Epidemiol Biomarkers Prev 27:594–600. doi: 10.1158/1055-9965.EPI-17-1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lim Y, Totsika M, Morrison M, Punyadeera C. 2017. The saliva microbiome profiles are minimally affected by collection method or DNA extraction protocols. Sci Rep 7:8523. doi: 10.1038/s41598-017-07885-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tartaglia GM, Kumar S, Fornari CD, Corti E, Connelly ST. 2017. Mouthwashes in the 21st century: a narrative review about active molecules and effectiveness on the periodontal outcomes. Expert Opin Drug Deliv 14:973–982. doi: 10.1080/17425247.2017.1260118. [DOI] [PubMed] [Google Scholar]
- 18.Reference deleted.
- 19.Freires I, Denny C, Benso B, de Alencar S, Rosalen P. 2015. Antibacterial activity of essential oils and their isolated constituents against cariogenic bacteria: a systematic review. Molecules 20:7329–7358. doi: 10.3390/molecules20047329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Masadeh MM, Gharaibeh SF, Alzoubi KH, Al-Azzam SI, Obeidat WM. 2013. Antimicrobial activity of common mouthwash solutions on multidrug-resistance bacterial biofilms. J Clin Med Res 5:389–394. doi: 10.4021/jocmr1535w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sälzer S, Rosema NAM, Martin ECJ, Slot DE, Timmer CJ, Dörfer CE, van der Weijden GA. 2016. The effectiveness of dentifrices without and with sodium lauryl sulfate on plaque, gingivitis and gingival abrasion–a randomized clinical trial. Clin Oral Invest 20:443–450. doi: 10.1007/s00784-015-1535-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lima TMS, Procópio LC, Brandão FD, Leão BA, Tótola MR, Borges AC. 2011. Evaluation of bacterial surfactant toxicity towards petroleum degrading microorganisms. Bioresour Technol 102:2957–2964. doi: 10.1016/j.biortech.2010.09.109. [DOI] [PubMed] [Google Scholar]
- 23.Wade WG, Addy M. 1992. Antibacterial activity of some triclosan-containing toothpastes and their ingredients. J Periodontol 63:280–282. doi: 10.1902/jop.1992.63.4.280. [DOI] [PubMed] [Google Scholar]
- 24.Jenkins S, Addy M, Newcombe R. 1991. Triclosan and sodium lauryl sulphate mouthwashes (I). Effects on salivary bacterial counts. J Clin Periodontol 18:140–144. doi: 10.1111/j.1600-051x.1991.tb01703.x. [DOI] [PubMed] [Google Scholar]
- 25.Sissons CH, Wong L, Cutress TW. 1996. Inhibition by ethanol of the growth of biofilm and dispersed microcosm dental plaques. Arch Oral Biol 41:27–34. doi: 10.1016/0003-9969(95)00103-4. [DOI] [PubMed] [Google Scholar]
- 26.Ingram LO. 1990. Ethanol tolerance in bacteria. Crit Rev Biotechnol 9:305–319. doi: 10.3109/07388558909036741. [DOI] [PubMed] [Google Scholar]
- 27.Huffer S, Clark ME, Ning JC, Blanch HW, Clark DS. 2011. Role of alcohols in growth, lipid composition, and membrane fluidity of yeasts, bacteria, and archaea. Appl Environ Microbiol 77:6400–6408. doi: 10.1128/AEM.00694-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sinha R, Chen J, Amir A, Vogtmann E, Shi J, Inman KS, Flores R, Sampson J, Knight R, Chia N. 2016. Collecting fecal samples for microbiome analyses in epidemiology studies. Cancer Epidemiol Biomarkers Prev 25:407–416. doi: 10.1158/1055-9965.EPI-15-0951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Song SJ, Amir A, Metcalf JL, Amato KR, Xu ZZ, Humphrey G, Knight R. 2016. Preservation methods differ in fecal microbiome stability, affecting suitability for field studies. mSystems 1:e00021-16. doi: 10.1128/mSystems.00021-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJA, Holmes SP. 2016. DADA2: high-resolution sample inference from Illumina amplicon data. Nat Methods 13:581–583. doi: 10.1038/nmeth.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pruesse E, Quast C, Knittel K, Fuchs BM, Ludwig W, Peplies J, Glockner FO. 2007. SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res 35:7188–7196. doi: 10.1093/nar/gkm864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bolyen E, Rideout JR, Dillon MR, Bokulich NA, Abnet CC, Al-Ghalith GA, Alexander H, Alm EJ, Arumugam M, Asnicar F, Bai Y, Bisanz JE, Bittinger K, Brejnrod A, Brislawn CJ, Brown CT, Callahan BJ, Caraballo-Rodríguez AM, Chase J, Cope EK, Da Silva R, Diener C, Dorrestein PC, Douglas GM, Durall DM, Duvallet C, Edwardson CF, Ernst M, Estaki M, Fouquier J, Gauglitz JM, Gibbons SM, Gibson DL, Gonzalez A, Gorlick K, Guo J, Hillmann B, Holmes S, Holste H, Huttenhower C, Huttley GA, Janssen S, Jarmusch AK, Jiang L, Kaehler BD, Bin Kang K, Keefe CR, Keim P, Kelley ST, Knights D, et al. 2019. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat Biotechnol 37:852–857. doi: 10.1038/s41587-019-0209-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.R Core Team. 2019. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. [Google Scholar]
- 34.Oksanen J, Blanchet FG, Friendly M, Kindt R, Legendre P, McGlinn D, Minchin PR, O’Hara RB, Simpson GL, Solymos P, Stevens MHH, Szoecs E, Wagner H. 2011. Vegan: community ecology package. R package version 1.17-8. R Foundation for Statistical Computing, Vienna, Austria. [Google Scholar]
- 35.McGill R, Tukey JW, Larsen WA. 1978. Variations of box plots. Am Stat 32:12. doi: 10.2307/2683468. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Plots of alpha diversity metrics comparing Scope and nonethanol mouthwashes (Non-EtOH wash) and Saccomanno’s fixative to the OMNIgene ORAL kit for observed species (top), Faith’s PD (middle), and the Shannon index (bottom). Adjusted coefficients of determination (R2 values) were obtained by fitting linear regression models. Dotted lines represent the line of equality (y = x). Download FIG S1, PDF file, 0.4 MB (363.3KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Taxonomic profiles at the class, order, and family levels of OMNIgene ORAL (OMNI) compared to Scope and nonethanol mouthwashes (Non-EtOH) and Saccomanno’s fixative. Download FIG S2, PDF file, 0.1 MB (146.4KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Differences in relative abundances of the top 10 taxa at the class, order, and family levels between paired samples of OMNIgene ORAL, Scope and nonethanol (Non-EtOH) mouthwashes, and Saccomanno’s fixative. Dotted lines represent line sof equality indicating no difference between OMNIgene and the alternative method. Download FIG S3, PDF file, 0.2 MB (239.8KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Comparison of relative abundances of the top 10 taxa between OMNIgene and alternative methods at the phylum to genus levels. Download Table S1, XLS file, 0.05 MB (52KB, xls) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Rarefaction curves for the mean numbers of observed species by sample type. Error bars represent 95% confidence intervals. Download FIG S4, PDF file, 0.07 MB (68.9KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Plots of the first three principal coordinates from principal-coordinate analyses using Bray-Curtis (top), unweighted (middle), and weighted UniFrac (bottom) matrices, colored by sample type. Download FIG S5, PDF file, 0.4 MB (452.8KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Data Availability Statement
The sequencing data are available through the NCBI Sequence Read Archive (PRJNA634162).