Abstract
Sensing and responding to signals is a fundamental ability of living systems, but despite remarkable progress in computational design of new protein structures there is no general approach for engineering arbitrary new protein sensors. Here we describe a generalizable computational strategy for designing sensor/actuator proteins by building binding sites de novo into heterodimeric protein-protein interfaces and coupling ligand sensing to modular actuation through split reporters. Using this approach, we designed protein sensors that respond to farnesyl pyrophosphate, a metabolic intermediate in the production of valuable compounds. The sensors are functional in vitro and in cells, and the crystal structure of the engineered binding site closely matches the design model. Our computational design strategy opens broad avenues to link biological outputs to new signals.
One Sentence Summary
Engineering de novo binding sites creates synthetic signaling systems that respond to specified small molecule inputs.
In the last two decades computational protein design has created diverse new protein structures spanning helical(1–5), alpha-beta(6–8) and beta-sheet(9, 10) folds. In contrast, our ability to computationally design arbitrary protein functions de novo lags far behind, with relatively few examples that often require screening of many variants(11, 12). One unsolved challenge is the de novo design of small molecule sensor/actuators in which ligand binding by a protein directly controls changes in downstream functions, a key aspect of cellular signal transduction(13).
Sensing and responding to a small molecule signal requires both recognition of the target and linking target recognition to an output response. Exciting progress has been made with the design of proteins recognizing new ligands(10, 11, 14–16). A general solution to the second problem, coupling ligand recognition to diverse output responses, has remained challenging. Existing approaches have used a ligand that fluoresces upon binding(10), engineered the sensor components to be unstable and hence inactive in the absence of the ligand(14, 17), or repurposed an allosteric transcription factor(18). Each of these strategies places constraints on the input signals or output responses that can be used.
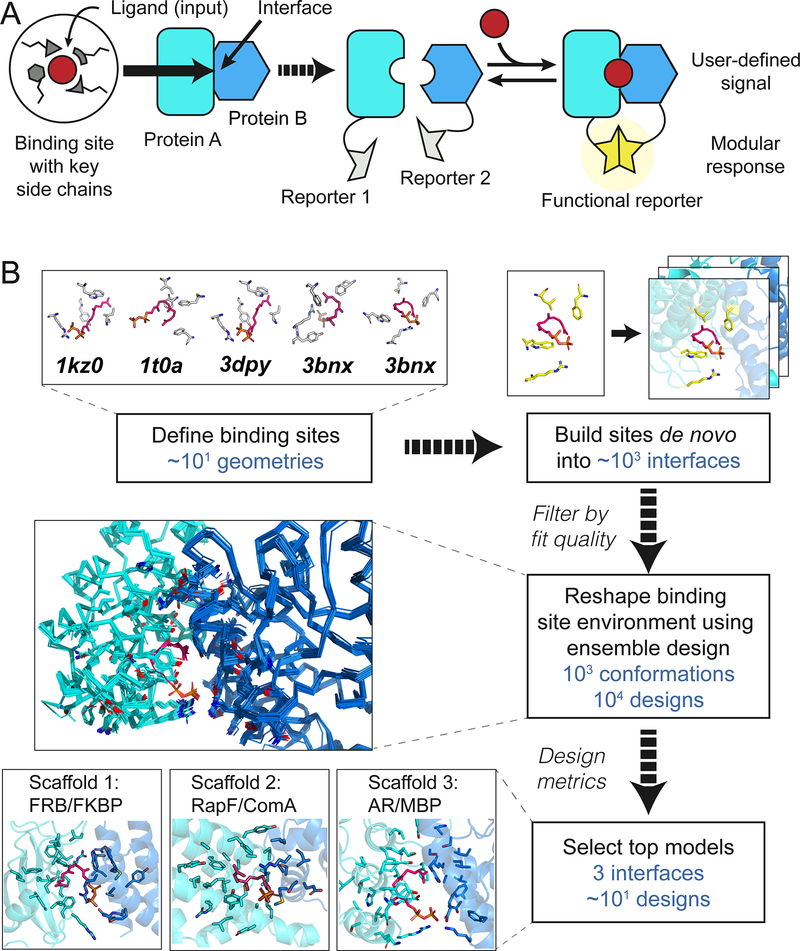
Here we describe a computational strategy to engineer protein complexes that can sense a small molecule and respond directly using different biological outputs, creating modular sensor/actuator systems. Distinct from prior work(10, 11, 14, 15) that reengineered existing binding sites or placed ligands into preformed cavities, we build small molecule binding sites de novo into heterodimeric protein-protein interfaces, to create new and programmable chemically induced dimerization systems (CIDs). This strategy is inspired by naturally occurring and reengineered CID systems(19) that have been widely used but are limited to a small number of existing or similar input molecules. We aimed to design synthetic CIDs that could similarly link binding of a small molecule to modular cellular responses through genetically encoded fusions of each sensor monomer to a split reporter (Fig. 1A), but respond to new, user-defined inputs.
Fig. 1. Computational design.
(A) Cartoon of the design strategy. A small-molecule binding site is built de novo into protein-protein interfaces (left) to create synthetic chemically inducible dimerization systems (CIDs, right). Linking the designed sensor proteins to split reporters yields modular CID systems, in which different reporter outputs can be coupled to user-defined small molecule input signals. (B) Steps in the design of a synthetic CID system sensing FPP. Top: Binding site geometries with key interacting side chains selected from FPP-binding proteins (pdb codes indicated) are computationally modeled into a large number of protein-protein interfaces. Middle: Binding sites with feasible geometries are reshaped and optimized by flexible backbone design; shown is a conformational ensemble for a single sequence. Bottom: Top designs from 3 different scaffolds (bottom) selected for experimental tests (Fig. 2).
To demonstrate this strategy, we chose farnesyl pyrophosphate (FPP) as the target ligand. FPP is an attractive target because it is a toxic intermediate in a commonly-engineered biosynthesis pathway for the production of valuable terpenoid compounds(20). Sensors for FPP could be used for example to optimize pathway enzymes, or, when linked to appropriate outputs, to regulate pathway gene expression in response to changes in metabolite concentrations(21). Our computational strategy (Fig. 1B, Supplemental Methods) proceeds in four main steps: (i) defining the geometries of minimal FPP binding sites comprised of 3–4 side chains (termed “motif residues”) that form key interactions with the target ligand; (ii) modeling these geometries into a dataset of heterodimeric protein-protein interfaces (termed “scaffolds”) and computationally screening for coarsely compatible scaffolds(22); (iii) optimizing the binding sites in these scaffolds using flexible backbone design methods previously used to predict ligand binding specificities(22–25) but not tested in the de novo design of binding sites (“reshaping”); and (iv) ranking individual designs for experimental testing according to several design metrics including ligand binding energy predicted using the Rosetta force field(26) and ligand burial.
Starting with 5 FPP binding site geometries and up to 3462 heterodimeric scaffolds, we selected the highest ranked designs across three engineered scaffolds for experimental testing (Fig. 1B, Supplemental Methods): the FKBP-FRB complex originally responsive to rapamycin(27) (1 design), a complex of the bacterial proteins RapF and ComA (28) (4 designs) and an engineered complex of maltose binding protein (MBP) and an ankyrin repeat (AR) protein(29) (4 designs) (Fig. 2A, Table S1, Fig. S1). While the ligand was placed into the rapamycin site in FKBP-FRB, binding sites in the other two complexes were modeled de novo.
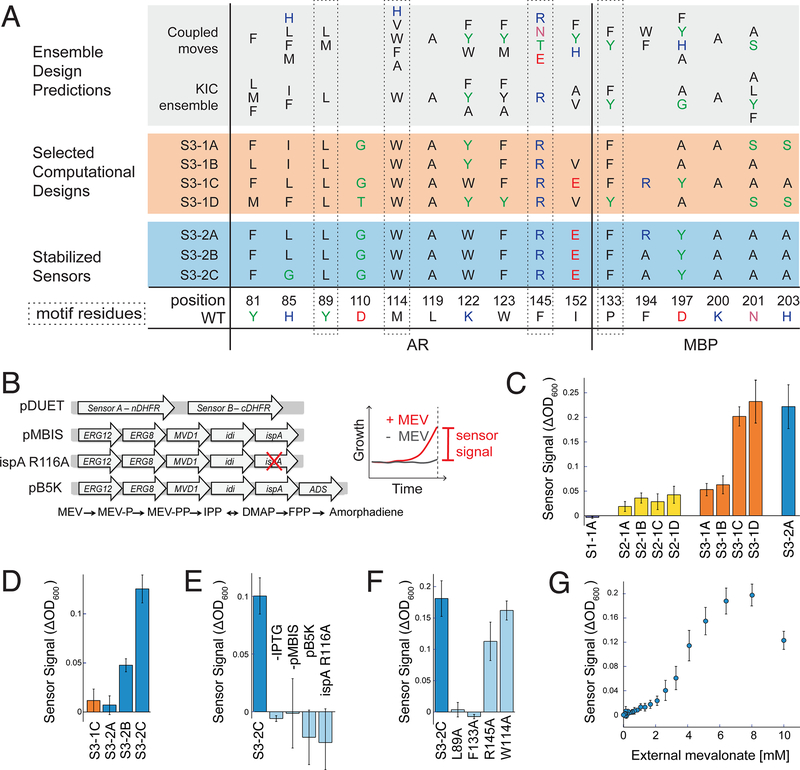
Fig. 2. Sensor function in bacteria.
(A) Designed sequences at key positions for scaffold 3. Grey shading: preferred residues from flexible backbone reshaping by kinematic closure (KIC(22, 24)) or coupled moves(25); orange shading: individual computational designs selected based on ligand burial (S3–1A), consensus (S3–1B), optimized ligand packing (S3–1C) and predicted ligand binding score (S3–1D); blue shading: sensors stabilized by 2 additional mutations from SSM (S3–2B and S3–2C also contained 2 mutations from epPCR that were not in the designed FPP binding site, Fig. S1). (B) Constructs (left, details in Appendix 1) used in the split mDHFR reporter assay (right). pDUET, sensor proteins linked to the split mDHFR reporter; pMBIS, engineered pathway of 5 enzymes to convert mevalonate (MEV) into FPP(20); ispA R116A, pMBIS containing R116A mutation in ispA that reduces catalytic activity 13-fold(27); pB5K, pMBIS with amorphadiene synthase (ADS)(20). Sensor signal is quantified as change in OD600 in the presence and absence of mevalonate. (C) Sensor signal in the split mDHFR assay for computational designs based on scaffold 1 (FKBP-FRB12, purple bar), scaffold 2 (RapF- ComA, yellow bars) and scaffold 3 (AR-MBP, orange bars). Sensor S3–2A (identified from library 2 with 2 mutations distal from the designed FPP binding site (Table S1)), is shown for comparison (blue bar). (D) Improvement of sensor signal by stability-enhancing mutations in S3– 2B and S3–2C at increased stringency (trimethoprim concentration 6 μM versus 1 μM in panel C). (E) Dependence of S3–2C sensor signal on sensor expression (-IPTG) and FFP production (- pMBIS, pB5K, ispA R116A). (F) Dependence of S3–2C sensor signal on motif residues. (G) Dependence of the S3–2C sensor signal on concentration of the FPP precursor mevalonate added extracellularly. Error bars are standard deviation from at least 4 biological replicates and 8 technical replicates for each biological replicate.
To test these computationally designed FPP sensors, we genetically fused the engineered sensor proteins to a well-studied split reporter, the enzyme murine dihydrofolate reductase (mDHFR(30), Fig. 2B, Appendix 1), and expressed the fusion constructs in E. coli. We reasoned that functional sensors should exhibit increased growth through FPP-driven dimerization of the sensor proteins and resulting complementation of functional mDHFR, under conditions where endogenous E. coli DHFR was specifically inhibited by trimethoprim. Since FPP does not efficiently enter E. coli, we added its metabolic precursor, mevalonate, to the growth medium and co-expressed an engineered pathway of 5 enzymes(20) (Fig. 2B) to produce FPP from mevalonate in the cells. We then monitored sensor function as change in growth in the presence or absence of mevalonate under otherwise identical conditions (Fig. 2B, Supplemental Methods). In the following, we denote designs by their scaffold (S1, S2, S3), design generation (1, 2, 3) and successive letter (A, B, etc.; Table S1, Fig. S1).
While 7 of the 9 selected designs showed only a small (S2–1A, B, C, D; S3–1A, B) or no signal (S1–1A), two designs (S3–1C, D) displayed a robust signal response to FPP (Fig. 2C, Fig. S2). Both designs resulted from the AR-MBP scaffold (S3, Fig. 2A). For this scaffold, we also generated two libraries: library 1 based on our ensemble design predictions (Fig. 2A, Table S2), and library 2 using error-prone PCR starting from design S3–1C. After an initial growth-based selection and subsequent plate-based screens in the presence and absence of FPP (Supplemental Methods, Fig. S3), we identified 36 hits from which we confirmed 27 FPP-responsive sequences by individual growth assays (Fig. S4, Fig. S5). One of the most active designs identified across both libraries (S3–2A) was a variant of design S3–1C with two additional mutations distal from the designed FPP binding site introduced by error-prone PCR. Interestingly, this variant displayed essentially equal activity as the original S3–1C design when tested under identical conditions (Fig. 2C, Table S1, Fig. S2). These results show that library screening or error-prone PCR were not necessary to identify functional sensors; instead, we obtained functional sensors directly via computational design. However, library 1 provided additional active sequences from sequence tolerance predicted in the ensemble design simulations (Fig. 2A, Table S2, Fig. S4).
To further characterize the identified best design, S3–2A (Table S1), we performed single site saturation mutagenesis at 11 positions (Table S3). We tested the resulting mutants with the growth-based split mDHFR reporter in the presence and absence of FPP under more stringent conditions by increasing the trimethoprim concentration (Fig. S6). While at most positions the originally designed amino acid (design S3–1C, Fig. 2A) appeared to be optimal under these conditions, we saw considerable improvements for mutations at two positions, R194A (design S3–2B) and R194A / L85G (design S3–2C, Fig. 2A). These two designs displayed increasing responses to mevalonate at higher trimethoprim concentrations (Fig. 2D). For the most active design, S3–2C, we confirmed that the sensor signal was dependent on expression of the sensor proteins (Fig. 2E, -IPTG) and the metabolic pathway that converts added mevalonate to FPP (Fig. 2E, -pMBIS). To test for specificity for FPP, we confirmed that the sensor signal was absent when preventing the accumulation of FPP either by inactivating the fifth enzyme in the pathway by a single point mutation (Fig. 2B, E, ispA R116A) or by adding a sixth enzyme that converts FPP to amorphadiene (Fig. 2B, E, pB5K). To test whether the sensor signal was dependent on the original four motif side chains, we mutated each individually to alanine and observed decreased sensitivity to the presence of mevalonate for three of the four motif side chains (L89, F133, R145 but not W114; Fig. 2F). Finally, we tested whether the sensor signal of design S3–2C was dependent on the concentration of FPP by titrating the extracellular concentration of the mevalonate precursor (Fig. 2G). Interestingly, while the sensor signal initially increased with increasing mevalonate concentrations, as expected, the signal decreased at the highest mevalonate concentration tested. This behavior likely arises from FPP-mediated toxicity previously observed at this mevalonate concentration using the same FPP biosynthesis pathway(20). We confirmed a consistent dependency of the sensor signal both on sensor expression (by varying the concentration of the inducer IPTG) and on mevalonate concentration in the growth medium for seven of our designs (S3–1A, B, C, D; S3–2A, B, C; Fig. S7). Taken together, these results confirm that sensor function in E. coli is specific to FPP produced by an engineered pathway, is dependent on key residues in the de novo binding site, is dose-dependent in E. coli, and is sensitive to FPP concentrations in a relevant range (i.e. below the toxicity level).
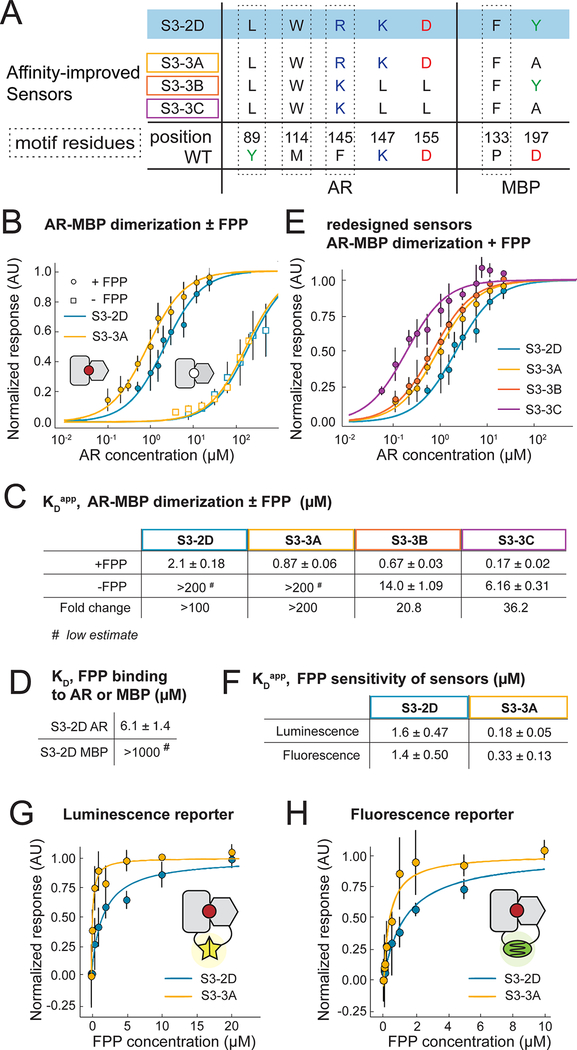
To confirm biochemically that FPP increases the binding affinity of the AR-MBP complex as designed, we purified the designed AR and MBP proteins without attached reporters (Supplemental Methods; these constructs contained several previously published mutations to stabilize AR(31), which when tested in the split mDHFR reporter assay led to active sensor S3– 2D, Table S1, Fig. S8, Appendix 2). We determined the apparent binding affinity of the designed AR and MBP proteins comprising the S3–2D sensor (Fig. 3A, Table S1, Fig. S1) in the absence and presence of 200 μM FPP using biolayer interferometry (Fig. 3B, Fig. S9, Supplemental Methods). The presence of FPP led to a >100-fold stabilization of the interaction between the AR and MBP proteins comprising sensor S3–2D (KD from >200 μM to 2.1 μM, Fig. 3C; for comparison, the original AR-MBP scaffold had a KD of 4.4 nM(29)). Binding of FPP to the designed AR component of S3–2D alone was weak and binding of FPP to the designed MBP component of S3–2D alone not detectable (Fig. 3D). Taken together, these results confirm in vitro with purified components that design S3–2D functions as a CID system responding to FPP.
Fig. 3. In vitro sensor characterization and output modularity.
(A) Sequence changes in sensor constructs tested in vitro. Motif residues are also shown. The starting construct, S3–2D (blue), is identical to S3–2C in the engineered FPP binding site but contains additional previously published stabilizing mutations in AR(31) (shown in Table S1). (B-H) In vitro binding measurements from biolayer interferometry (BLI) using purified protein (panels B-E) or FPP titrations with sensors expressed using in vitro transcription / translation (TxTl) (panels F-H).
(B) Apparent AR interaction with immobilized MBP in the presence (closed circles) or absence (open squares) of 200 μM FPP, comparing designs S3–2D (blue) and S3–3A containing the Y197A mutation (orange). (C) Summary of BLI results for apparent AR-MBP dimerization with and without FPP. (D) Summary of BLI results for FPP binding to the individual designed AR and MBP proteins comprising design S3–2D (Table S1). (E) Apparent AR interaction with immobilized MBP for a computationally designed variant using the S3–2D crystal structure as the input, with (purple, S3–3C) or without (red, S3–3B) the Y197A mutation. (F) Apparent affinity of the S3–2D and S3–3A sensors for FPP using luminescent or fluorescent reporters in TxTl experiments. (G, H) FPP titrations in TxTl using the luminescent reporter (G) or the fluorescent reporter (H). Error bars are standard deviations for n ≥ 3.
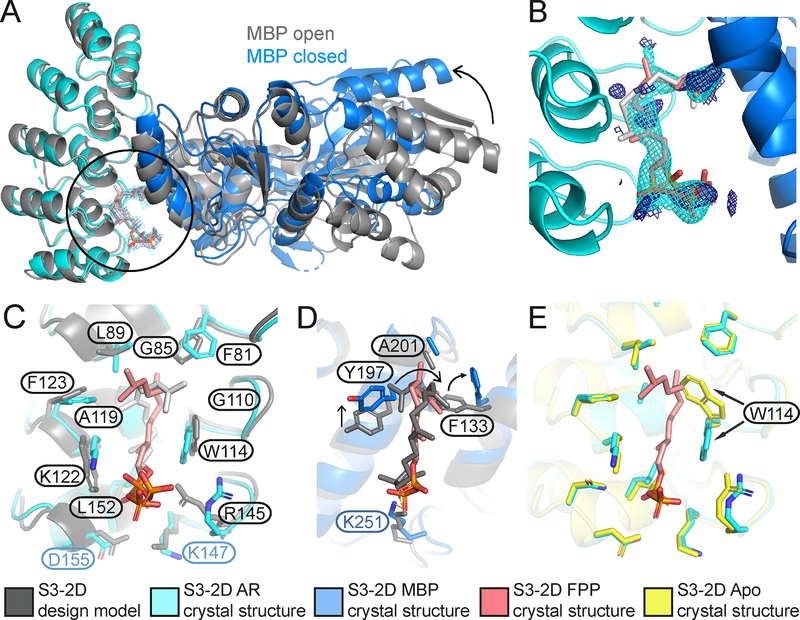
To determine whether FPP is recognized in the de novo engineered binding site as predicted by the design model, we determined a 2.2 Å resolution crystal structure of the ternary complex of FPP bound in the engineered AR-MBP interface (Supplemental Methods; Table S4). The crystal structure of the bound complex is in excellent overall agreement with the design model (Fig. 4A–C). Despite twinning in the crystals, examining unbiased omit maps allowed modeling of unexplained density in the engineered binding site as FPP (Fig. 4B, Fig. S10) and confirmed the side chain conformations in the designed binding pocket (Fig. 4C, D). Overall, in a 10 Å shell around FPP in the binding pocket, the Ca root mean squared deviation (rmsd) between the model and the structure is 0.53 Å and the all heavy atom rmsd is 1.13 Å. While crystals formed only in the presence of FPP, only one of the two complexes in the asymmetric unit contained FPP in the binding site (Fig. S11). This behavior allowed us to compare apo and holo states of the complex. The majority of the designed side chains are in identical conformations in the FPP- bound holo and FPP-minus apo states (Fig. 4E), suggesting favorable pre-organization of the designed binding site. An exception is W114 on AR that is partly disordered in the apo state (Fig. S11), providing a potential explanation for why a W114A mutation is less detrimental for sensor activity (Fig. 2F) than expected based on the observed packing interactions between W114 and FPP in the holo state. A second slight deviation between the model and the crystal structure appeared to be caused by potential steric clashes of the engineered Y197 on MBP with the modeled FPP conformer, which led to re-arrangements in the FPP structure and a rotamer change in designed residue F133 on MBP (Fig. 4D). Interestingly, many of the original models from computational design favored a smaller alanine side chain at this position (Fig. 2A). These observations led to the prediction that a Y197A mutation might stabilize the ternary complex, and indeed design S3–3A containing the Y197A mutation showed an increased (>200 fold) stabilization of the complex with FPP, with an apparent dissociation constant of the designed AR and MBP proteins comprising sensor S3–3A in the presence of 200 μM FPP of 870 ± 60 nM (Fig. 3B, C). We also confirmed that design S3–3A (Table S1) is active in E. coli (Fig. S12). To further improve the design based on the crystal structure of design S3–2D, we employed an additional round of flexible backbone design using the Rosetta coupled moves method(25) starting from the FPP-bound crystal structure. These simulations suggested 3 additional mutations leading to design S3–3B: R145K, K147L, D155L (Fig. 3A). These mutations, when combined with the Y197A mutation (design S3–3C), enhanced the apparent binding affinity of the designed AR and MBP proteins comprising sensor S3–3C in the presence of 200 μM FPP to 170 ± 20 nM (Fig. 3C, E), which is within 40-fold of the original scaffold AR-MBP interaction affmity(29), but also strengthened the binding affinity of the protein-protein dimer in the absence of FPP to 6.2 ± 0.3 μM (Fig. S13). Notably, the design simulations optimized sequences for stability of the ternary complex without also destabilizing the dimer in the absence of the small molecule. Methods integrating negative design(32) could be incorporated to improve the dynamic range of the system (Supplementary text).
Fig. 4. The S3–2D crystal structure closely matches the computational design model.
(A) Overlay of the design model (grey) with the crystal structure (designed AR: cyan, designed MBP: blue, FPP: pink) showing FPP binding in the computationally designed binding site at the AR-MBP interface (circle). The design crystallized in the closed MBP conformation while MBP was in the open conformation in the original scaffold on which the model was based, leading to a difference in rigid-body orientation (arrow) of one lobe of MBP distal to the FPP binding site.
(B) FPP overlaid with 2mFo-DFc electron density map (1.2σ, cyan) and ligand 2mFo-DFc omit map (1.0σ, dark blue). Strong density peaks were present in both maps for the phosphates and several anchoring hydrophobic groups. (C) Open-book representation of the FPP binding site on AR, showing close match of designed side chain conformations to the crystal structure. (D) Open-book representation of the FPP binding site on MBP, indicating a clash between the position of MBP Y197 in the crystal structure (blue) and the designed FPP orientation in the model (grey), causing slight rearrangements of FPP and F133 (arrows). (E) Alignment of the holo (cyan) and apo (yellow) structures of S3–2D, showing overall agreement with the exception of the side chain of W114 (arrows). In panels (C-E), residues are labeled black when designed and green/blue when present in the original scaffold complex.
A key advantage of our CID design strategy is the ability to link an engineered sensor, whose input is specific to a user-defined small molecule signal, to a modular output that can in principle be chosen from many available split reporters (Fig. 1A). To test this concept, we linked the engineered CID sensors S3–2D and S3–3A to two additional outputs: a dimerization-dependent fluorescent protein(33) and a split luciferase(34) (Fig. 3G, H, Appendix 3). We tested input-output responses using an in vitro transcription-translation system (TxTl)(35) in which FPP can be added at defined concentrations to the assay extract, in contrast to the cell-based split mDHFR assay. The TxT1 assay revealed a nanomolar FPP sensitivity (KDapp) for our best sensor S3–3A (Fig. 3F) that is essentially identical for both reporters (180 ± 50 and 330 ± 130 nM by luminescence and fluorescence detection, respectively, Fig. 3G, H, Fig. S14), and additionally confirms the improvements in design S3–3A containing the Y197A mutation over design S3–2D (the KDapp for S3–2D was 1.6 ± 0.5 pM and 1.4 ± 0.5 μM for the luminescence and fluorescence reporters, respectively, Fig. 3F, G, H). These results show that our CID sensor design strategy is compatible with modular outputs.
The most critical feature of our approach is the ability to computationally design small molecule binding sites de novo into protein-protein interfaces. A prior computational analysis suggests that the appearance of pockets around artificially generated protein-protein interfaces may be an intrinsic geometric feature of protein structure(36), lending support to the idea that our approach is extensible to many other small molecules and interfaces. The design method presented here hence introduces a generalizable way to create synthetic sensing systems with different outputs that can be used in diverse biological contexts to respond to user-specified molecular signals.
Supplementary Material
Acknowledgments
We would like to thank: Jay Keasling and Fuzhong Zhang for advice on FPP production in microbes and pathway constructs; Emzo de los Santos, Zach Sun, Vincent Noireux, Richard Murray for TxTl advice and reagents; Spencer Alford and Robert Campbell’s lab for ddFP constructs; Aditya Anand, Victor Ruiz, Ben Adler and Alison Maxwell for contributions to computational design and characterization; Shane O’Connor for developing a database for design models; and members of the Kortemme lab for discussion.
Funding: This work was supported by a grant from the National Institutes of Health (NIH) (R01-GM110089) and a W.M.F. Keck Foundation Medical Research Award to TK. We additionally acknowledge the following fellowships: NIH IRACDA and UC Chancellor’s Postdoctoral Fellowships (AAG), PhRMA Foundation Predoctoral Fellowship in Informatics (DJM), NIH F32 Postdoctoral Fellowship (MT), and National Science Foundation Graduate Research Fellowships (JP and NO).
Footnotes
Competing interests: The authors declare no competing interests.
Data and materials availability: Coordinates and structure files have been deposited to the Protein Data Bank (PDB) with accession codes 6OB5. All other relevant data are available in the main text or the supplementary materials. Rosetta source code is available from rosettacommons.org. Upon publication, constructs will be made available via Addgene.
References and Notes
- 1.Huang PS et al. , High thermodynamic stability of parametrically designed helical bundles. Science 346, 481–485 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jacobs TM et al. , Design of structurally distinct proteins using strategies inspired by evolution. Science 352, 687–690 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Thomson AR et al. , Computational design of water-soluble alpha-helical barrels. Science 346, 485–488 (2014). [DOI] [PubMed] [Google Scholar]
- 4.Hill RB, Raleigh DP, Lombardi A, DeGrado WF, De novo design of helical bundles as models for understanding protein folding and function. Acc Chem Res 33, 745–754 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Harbury PB, Plecs JJ, Tidor B, Alber T, Kim PS, High-resolution protein design with backbone freedom. Science 282, 1462–1467 (1998). [DOI] [PubMed] [Google Scholar]
- 6.Rocklin GJ et al. , Global analysis of protein folding using massively parallel design, synthesis, and testing. Science 357, 168–175 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Koga N et al. , Principles for designing ideal protein structures. Nature 491, 222–227 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huang PS et al. , De novo design of a four-fold symmetric TIM-barrel protein with atomic-level accuracy. Nat Chem Biol 12, 29–34 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Marcos E et al. , De novo design of a non-local beta-sheet protein with high stability and accuracy. Nat Struct Mol Biol, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dou J et al. , De novo design of a fluorescence-activating beta-barrel. Nature 561, 485–491 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lechner H, Ferruz N, Hocker B, Strategies for designing non-natural enzymes and binders. Curr Opin Chem Biol 47, 67–76 (2018). [DOI] [PubMed] [Google Scholar]
- 12.Dou J et al. , Sampling and energy evaluation challenges in ligand binding protein design. Protein Sci 26, 2426–2437 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gordley RM, Bugaj LJ, Lim WA, Modular engineering of cellular signaling proteins and networks. Curr Opin Struct Biol 39, 106–114 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bick MJ et al. , Computational design of environmental sensors for the potent opioid fentanyl. Elife 6, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tinberg CE et al. , Computational design of ligand-binding proteins with high affinity and selectivity. Nature 501, 212–216 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Polizzi NF et al. , De novo design of a hyperstable non-natural protein-ligand complex with sub-A accuracy. Nat Chem 9, 1157–1164 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Feng J et al. , A general strategy to construct small molecule biosensors in eukaryotes. Elife 4, (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Taylor ND et al. , Engineering an allosteric transcription factor to respond to new ligands. Nat Methods 13, 177–183 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Spencer DM, Wandless TJ, Schreiber SL, Crabtree GR, Controlling signal transduction with synthetic ligands. Science 262, 1019–1024 (1993). [DOI] [PubMed] [Google Scholar]
- 20.Martin VJ, Pitera DJ, Withers ST, Newman JD, Keasling JD, Engineering a mevalonate pathway in Escherichia coli for production of terpenoids. Nat Biotechnol 21, 796–802 (2003). [DOI] [PubMed] [Google Scholar]
- 21.Zhang F, Keasling J, Biosensors and their applications in microbial metabolic engineering. Trends Microbiol 19, 323–329 (2011). [DOI] [PubMed] [Google Scholar]
- 22.Zanghellini A et al. , New algorithms and an in silico benchmark for computational enzyme design. Protein Sci 15, 2785–2794 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Babor M, Mandell DJ, Kortemme T, Assessment of flexible backbone protein design methods for sequence library prediction in the therapeutic antibody Herceptin-HER2 interface. Protein Sci 20, 1082–1089 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mandell DJ, Coutsias EA, Kortemme T, Sub-angstrom accuracy in protein loop reconstruction by robotics-inspired conformational sampling. Nat Methods 6, 551–552 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ollikainen N, de Jong RM, Kortemme T, Coupling Protein Side-Chain and Backbone Flexibility Improves the Re-design of Protein-Ligand Specificity. PLoS Comput Biol 11, e1004335 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Alford RF et al. , The Rosetta All-Atom Energy Function for Macromolecular Modeling and Design. J Chem Theory Comput 13, 3031–3048 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Choi J, Chen J, Schreiber SL, Clardy J, Structure of the FKBP12-rapamycin complex interacting with the binding domain of human FRAP. Science 273, 239–242 (1996). [DOI] [PubMed] [Google Scholar]
- 28.Baker MD, Neiditch MB, Structural basis of response regulator inhibition by a bacterial anti-activator protein. PLoS Biol 9, e1001226 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Binz HK et al. , High-affinity binders selected from designed ankyrin repeat protein libraries. Nat Biotechnol 22, 575–582 (2004). [DOI] [PubMed] [Google Scholar]
- 30.Pelletier JN, Campbell-Valois FX, Michnick SW, Oligomerization domain-directed reassembly of active dihydrofolate reductase from rationally designed fragments. Proc Natl Acad Sci U S A 95, 12141–12146 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kramer MA, Wetzel SK, Pluckthun A, Mittl PR, Grutter MG, Structural determinants for improved stability of designed ankyrin repeat proteins with a redesigned C-capping module. J Mol Biol 404, 381–391 (2010). [DOI] [PubMed] [Google Scholar]
- 32.Davey JA, Chica RA, Multistate approaches in computational protein design. Protein Sci 21, 1241–1252 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Alford SC, Ding Y, Simmen T, Campbell RE, Dimerization-dependent green and yellow fluorescent proteins. ACS Synth Biol 1, 569–575 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dixon AS et al. , NanoLuc Complementation Reporter Optimized for Accurate Measurement of Protein Interactions in Cells. ACS Chem Biol 11, 400–408 (2016). [DOI] [PubMed] [Google Scholar]
- 35.Marshall R, Noireaux V, Synthetic Biology with an All E. coli TXTL System: Quantitative Characterization of Regulatory Elements and Gene Circuits. Methods Mol Biol 1772, 61–93 (2018). [DOI] [PubMed] [Google Scholar]
- 36.Gao M, Skolnick J, The distribution of ligand-binding pockets around protein-protein interfaces suggests a general mechanism for pocket formation. Proc Natl Acad Sci U S A 109, 3784–3789 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Song L, Poulter CD, Yeast farnesyl-diphosphate synthase: site-directed mutagenesis of residues in highly conserved prenyltransferase domains I and II. Proc Natl Acad Sci U S A 91, 3044–3048 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mandell DJ, Backbone Flexibility in Computational Design. PhD Dissertation, UCSF, ProQuest Dissertations Publishing, 3412217 (2010). [Google Scholar]
- 39.Dunbrack RL Jr., Cohen FE, Bayesian statistical analysis of protein side-chain rotamer preferences. Protein Sci 6, 1661–1681 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Davis IW, Baker D, RosettaLigand docking with full ligand and receptor flexibility. J Mol Biol 385, 381–392 (2009). [DOI] [PubMed] [Google Scholar]
- 41.Rothlisberger D et al. , Kemp elimination catalysts by computational enzyme design. Nature 453, 190–195 (2008). [DOI] [PubMed] [Google Scholar]
- 42.Jiang L et al. , De novo computational design of retro-aldol enzymes. Science 319, 1387–1391 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Davey JA, Chica RA, Improving the accuracy of protein stability predictions with multistate design using a variety of backbone ensembles. Proteins 82, 771–784 (2014). [DOI] [PubMed] [Google Scholar]
- 44.Davey JA, Chica RA, Multistate Computational Protein Design with Backbone Ensembles. Methods Mol Biol 1529, 161–179 (2017). [DOI] [PubMed] [Google Scholar]
- 45.Humphris EL, Kortemme T, Prediction of protein-protein interface sequence diversity using flexible backbone computational protein design. Structure 16, 1777–1788 (2008). [DOI] [PubMed] [Google Scholar]
- 46.C. A. Smith, T. Kortemme, Backrub-like backbone simulation recapitulates natural protein conformational variability and improves mutant side-chain prediction. J Mol Biol 380, 742–756 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Conway P, Tyka MD, DiMaio F, Konerding DE, Baker D, Relaxation of backbone bond geometry improves protein energy landscape modeling. Protein Sci 23, 47–55 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hoover DM, Lubkowski J, DNAWorks: an automated method for designing oligonucleotides for PCR-based gene synthesis. Nucleic Acids Res 30, e43 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Reetz MT, Carballeira JD, Iterative saturation mutagenesis (ISM) for rapid directed evolution of functional enzymes. Nat Protoc 2, 891–903 (2007). [DOI] [PubMed] [Google Scholar]
- 50.Engler C, Kandzia R, Marillonnet S, A one pot, one step, precision cloning method with high throughput capability. PLoS One 3, e3647 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lee ME, DeLoache WC, Cervantes B, Dueber JE, A Highly Characterized Yeast Toolkit for Modular, Multipart Assembly. ACS Synth Biol 4, 975–986 (2015). [DOI] [PubMed] [Google Scholar]
- 52.Sun ZZ et al. , Protocols for implementing an Escherichia coli based TX-TL cell-free expression system for synthetic biology. J Vis Exp, e50762 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Winter G, xia2: an expert system for macromolecular crystallography data reduction. Journal of Applied Crystallography 43, 186–190 (2010). [Google Scholar]
- 54.Kabsch W, Xds. Acta Crystallogr D Biol Crystallogr 66, 125–132 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Evans P, Scaling and assessment of data quality. Acta Crystallogr D Biol Crystallogr 62, 72–82 (2006). [DOI] [PubMed] [Google Scholar]
- 56.Padilla JE, Yeates TO, A statistic for local intensity differences: robustness to anisotropy and pseudo-centering and utility for detecting twinning. Acta Crystallogr D Biol Crystallogr 59, 1124–1130 (2003). [DOI] [PubMed] [Google Scholar]
- 57.Thompson MC, Identifying and Overcoming Crystal Pathologies: Disorder and Twinning. Methods Mol Biol 1607, 185–217 (2017). [DOI] [PubMed] [Google Scholar]
- 58.Kim WM, Sigalov AB, Stern LJ, Pseudo-merohedral twinning and noncrystallographic symmetry in orthorhombic crystals of SIVmac239 Nef core domain bound to different-length TCRzeta fragments. Acta Crystallogr D Biol Crystallogr 66, 163–175 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jenni S, Ban N, Imperfect pseudo-merohedral twinning in crystals of fungal fatty acid synthase. Acta Crystallogr D Biol Crystallogr 65, 101–111 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sultana A et al. , Structure determination by multiwavelength anomalous diffraction of aclacinomycin oxidoreductase: indications of multidomain pseudomerohedral twinning. Acta Crystallogr D Biol Crystallogr 63, 149–159 (2007). [DOI] [PubMed] [Google Scholar]
- 61.Gilski M, Drozdzal P, Kierzek R, Jaskolski M, Atomic resolution structure of a chimeric DNA-RNA Z-type duplex in complex with Ba(2+) ions: a case of complicated multi-domain twinning. Acta Crystallogr D Struct Biol 72, 211–223 (2016). [DOI] [PubMed] [Google Scholar]
- 62.McCoy AJ et al. , Phaser crystallographic software. JAppl Crystallogr 40, 658–674 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Adams PD et al. , PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr 66, 213–221 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kantardjieff KA, Rupp B, Matthews coefficient probabilities: Improved estimates for unit cell contents of proteins, DNA, and protein-nucleic acid complex crystals. Protein Sci 12, 1865–1871 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Weichenberger CX, Rupp B, Ten years of probabilistic estimates of biocrystal solvent content: new insights via nonparametric kernel density estimate. Acta Crystallogr D Biol Crystallogr 70, 1579–1588 (2014). [DOI] [PubMed] [Google Scholar]
- 66.Afonine PV et al. , Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr D Biol Crystallogr 68, 352–367 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Emsley P, Lohkamp B, Scott WG, Cowtan K, Features and development of Coot. Acta Crystallogr D Biol Crystallogr 66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Moriarty NW, Grosse-Kunstleve RW, Adams PD, electronic Ligand Builder and Optimization Workbench (eLBOW): a tool for ligand coordinate and restraint generation. Acta Crystallogr D Biol Crystallogr 65, 1074–1080 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bernstein FC et al. , The Protein Data Bank: a computer-based archival file for macromolecular structures. J Mol Biol 112, 535–542 (1977). [DOI] [PubMed] [Google Scholar]
- 70.Karplus PA, Diederichs K, Linking crystallographic model and data quality. Science 336, 1030–1033 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Brunger AT, Free R value: cross-validation in crystallography. Methods Enzymol 277, 366–396 (1997). [DOI] [PubMed] [Google Scholar]
- 72.Chen VB et al. , MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr 66, 12–21 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Inoue T, Heo WD, Grimley JS, Wandless TJ, Meyer T, An inducible translocation strategy to rapidly activate and inhibit small GTPase signaling pathways. Nat Methods 2, 415–418 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.