Abstract
Deletions of different regions of chromosome 22q11 have been extensively characterized in the literature, with a recent review outlining common deletions with a standardized system proposed for classification and nomenclature. The genotype-phenotype relationships have not been sufficiently elucidated for these deletions, and it remains unclear which specific genes play the dominant roles in producing associated clinical features. Several deletions involve entirely distinct regions of chromosome 22q11 but do not overlap, suggesting that a number of different genes contribute to the clinical features. Studies of patients with small deletions involving only 1 or 2 genes may provide more convincing evidence for the impact of individual genes on the observed phenotype. In this case report, we present a 12-year-old female with autism, cognitive impairment, dysmorphic features, and behavioral concerns and a 268-kb deletion of chromosome 22q11.22 including TOP3B, the only recognized disease-causing gene in the deletion. The mechanism of pathogenesis contributing significantly to our patient’s clinical findings may relate to interaction between TOP3B and fragile X mental retardation protein (FMRP), an mRNA-binding protein that regulates translation and is altered in fragile X syndrome, a condition involving developmental delay, learning disability, and autism. All these features are recognized in our patient.
Keywords: 22q11 deletion, DiGeorge/Shprintzen/velocardiofacial syndrome, TOP3B
Deletions of chromosome 22q11 cause a recognized genetic condition or the 22q11.21 deletion syndrome also called DiGeorge/Shprintzen/velocardiofacial syndrome (OMIM #192430). This complex cytogenetic syndrome can involve a diversity of phenotypic features, including palatal abnormalities, immune deficiency, learning problems, behavioral disturbances, kidney abnormalities, facial anomalies, conotruncal cardiac defects, and an increased risk of developing bipolar disorder, attention deficit hyperactivity disorder (ADHD), depression, autism, and schizophrenia [Burnside, 2015; US National Library of Medicine, 2016]. Our research group [Bittel et al., 2009] analyzed the specific breakpoints in the 22q11.2 deletion syndrome previously and found that individuals studied and diagnosed clinically with DiGeorge syndrome (OMIM #188400) had a deletion within the genomic coordinates chr22: 18,661,524–21,797,668 converted to hg19 using UCSC Lift Genome Annotations at http://genome.ucsc.edu/cgi-bin/hgLiftOver.
Several genes in the 22q11.2 region may play a role in causing developmental delay with learning and/or behavioral problems including TOP3B which is located near, but does not overlap with, the 22q11.21 deletion region of DiGeorge/Shprintzen/velocardiofacial syndrome reported in the medical literature. Topoisomerase (DNA) III beta, or TOP3B, controls the state of DNA during transcription by catalyzing transient breakage and rejoining of a single strand of DNA [Hanai et al., 1996]. For example, Cooper et al. [2012] examined copy number variants in 15,767 children with intellectual disability and various congenital defects and compared their genetic findings to those of 8,329 adult controls. They found that 13 patients with intellectual disability had a 22q11.2 distal deletion encompassing a number of genes including TOP3B. This deletion was not found in the adult controls. Xu et al. [2008] reported a patient with a 1.12-Mb deletion of 22q11.2 encompassing 16 genes including TOP3B; this infant was functioning at a 6–7 month-old level when he was 11 months of age and had dysmorphic facial features as similarly seen in the oculoauriculovertebral spectrum disorder. Jackson et al. [2007] reported a 3-year-old boy with a large deletion encompassing many genes including TOP3B and INI1 and sensorineural hearing loss, submucous cleft palate, renal rhabdoid tumor, and developmental delay. Later, Rodningen et al. [2008] reported two 7-year-old patients, one male and one female, who had a 1.4-Mb deletion of the distal 22q11.2 region which was found via array-CGH and validated with MLPA showing hemizygosity for the HIC2, PPIL2, and TOP3B genes. The female patient had delayed motor milestones, difficulties with speech and impaired language skills, and mildly dysmorphic features. The male patient had a high-pitched voice, delayed speech development, difficulty understanding instructions, and mildly dysmorphic features.
Herein, we report a 12-year-old Caucasian female with cognitive/behavioral problems and dysmorphic features with a 268-kb deletion including the TOP3B gene identified with high resolution chromosomal microarray analysis. We also review the literature regarding the potential role of TOP3B.
Clinical Report
Medical History
The patient is a 12-year-old Caucasian female diagnosed at the age of 10 years with features of autism spectrum disorder and referred for psychiatric evaluation due to suspicion of psychosis or bipolar mood disorder. She was born after an uncomplicated, fullterm pregnancy, induced at 39 weeks due to fetal heart rate distress. Her birth weight was 3.15 kg (25th percentile) with no congenital abnormalities noted. She reportedly met developmental milestones normally except for episodes of enuresis and encopresis through the age of 10 years. There was no history of regression in developmental skills. Our patient was reported to be in good health, with eye surgery at the age of 3 years for strabismus. Vision and hearing exams were normal at the age of 10 years.
Our patient’s life course was unremarkable until the age of 4 years when behavioral issues including distractibility, hyperactivity, and impulsivity led to the diagnoses of ADHD and oppositional defiant disorder. Medication trials of lisdexamphetamine and guanfacine were described as beneficial but were discontinued due to adverse effects including loss of appetite and insomnia. Our patient was raised by her biological parents until placed in state custody at the age of 10 years due to neglect and abuse.
Autism spectrum disorder was diagnosed based on impaired social (poor eye contact, little interest in playing with other children, but she would interact with adults) and communication skills (at times nonverbal, at other times demonstrating echolalia – repeating what she had heard from TV shows, or verbalizing repetitive phrases out of context for the situation), and idiosyncratic, restricted, fixated interests, activities, and behaviors. At times she would twirl in the classroom setting while talking constantly. Sensory issues included frequently putting items in her mouth. Tantrum behaviors included hitting, biting, kicking, screaming, or head butting others or hitting and biting herself when upset.
Symptoms of cognitive dysfunction included episodes of disorganized and tangential thoughts described at times as incoherent, talking to self, refusing to be called by her given name, and referring to a named entity unseen by others (presumed to be an imaginary friend) and at times insisting that she herself was someone else. Her school identified learning problems initially with reading significantly below grade level. At times she was capable of understanding new concepts, while on other days she was unable to learn or concentrate. She was noted to be affectionate at times and showed a sense of humor and creativity. She likes to draw, tell stories, and play outside. She also likes to play games on the computer and to watch TV shows.
Family History
A limited pedigree was obtained at the time of the clinic visit as the parents were not available. Additional family history information was requested from the custodial caregiver who accompanied the patient regarding the biological parents and a 3-generation pedigree. Later, a more knowledgeable caregiver supplied more detailed information regarding the family history, and several individuals were reported with neuropsychiatric issues. There was no apparent history of consanguinity. Our patient’s 9-year-old brother was described as healthy but with high-functioning autism. Her mother was 33 years of age with a history of learning problems, ADHD, dyslexia, autistic traits, a high nasal voice, and similar facial features as seen in our patient. Her mother did not complete high school or a General Educational Development test. She has social anxiety, weak math skills by history, and worked part-time in food preparation. Our patient’s maternal grandmother had a club foot, depression, bipolar-like symptoms, multiple sclerosis, and passed away at an unknown age due to apparent accidental opiate overdose.
Our patient’s father is 34 years of age and had loss of vision in one eye. He did attend high school and served 2 years in the military. He has a history of learning problems, autistic traits, depression, anxiety, and posttraumatic stress disorder related to combat experience. Our patient’s paternal uncle has autism by history and intellectual disability, and the paternal aunt has bipolar disorder and dyslexia. Our patient’s paternal grandfather has bipolar disorder and depression; the paternal grandmother was diagnosed with diabetes and has a brother with schizophrenia. Unfortunately, family members do not live locally and were not able to attend the clinic for evaluation or genetic testing.
Physical Exam
A genetics evaluation at 11 years of age showed a height of 145 cm (50th percentile), weight of 41.28 kg (70th percentile), and head circumference of 53 cm (25th percentile). She had an inner canthal distance of 2.2 cm (<3rd percentile), outer canthal distance of 7 cm (<3rd percentile), interpupillary distance of 4.5 cm (<3rd percentile), palpebral fissure length of 2.3 cm (<5th percentile), ear length of 5.2 cm (15th percentile), total hand length of 15 cm (20th percentile), and middle finger length of 6.3 cm (20th percentile). She had mid-face and malar hypoplasia, a broad nasal tip, a wide appearing mouth with a thin upper lip and a protruding lower lip, and a prominent lower jaw. She had normal dentition, a single uvula with a normal arched palate, and hypernasal voice (fig. 1). The cardiovascular examination was normal without a heart murmur, arrhythmia, or respiratory distress or stridor. Peripheral pulses were intact. The abdomen was soft with normal bowel sounds and no masses, tenderness, or hernias. The musculoskeletal examination revealed normal muscle mass, tone, and strength without leg length asymmetry, edema, scoliosis, or deformity. Reflexes of the upper and lower extremities were normal and she had normal coordination. There were no birth marks, cyanosis, or jaundice. Echocardiograms or renal ultrasounds were not performed.
Fig. 1.
Dysmorphic features of the patient with mid-face and malar hypoplasia, a broad nasal tip, a wide appearing mouth with a thin upper lip and a protruding lower lip, and a prominent lower jaw.
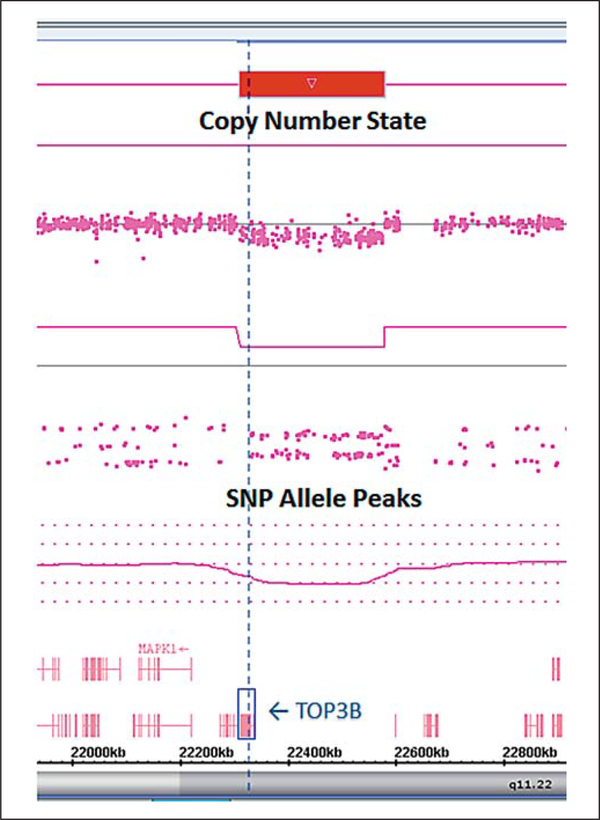
Although the parents were not available at the time of the clinic visit, information was supplied by the foster care provider and educator who was acquainted with the family and knowledgeable about the immediate and extended family members. This provider indicated that autism spectrum disorder, impaired cognitive functioning with possible psychosis or bipolar disorder, and a hypernasal voice with dysmorphic facial features were seen in other family members as noted. We ordered genetic testing with the goal of determining an underlying cause for her findings that included fragile X syndrome testing which was found to be normal (CGG repeats: 31, 41). The FirstStep PLUS chromosomal microarray analysis performed by Lineagen (Salt Lake City, UT) showed a copy number variant of unknown clinical significance described as a 22q11.22 loss (deletion) of base pair coordinates 22,311,348– 22,578,983 (hg build 19; fig. 2). The deletion was 268 kb in size and included the TOP3B gene which is annotated in the OMIM database (entry #603582).
Fig. 2.
Microarray analysis of the patient showing a copy number loss at 22q11.22 including the TOP3B gene.
Results and Discussion
We report a 12-year-old female with a 268-kb deletion of the 22q11.22 band classified as unknown clinical significance containing the TOP3B gene. No other copy number variants were observed in the rest of the genome. The parents or other family members were unavailable for testing for the deletion status (de novo or inherited). However, we propose that the deletion was associated with the clinical presentation seen in our patient, but we cannot rule out other genetic (e.g., mutations of genes involved with intellectual disability) or non-genetic (e.g., environmental, multifactorial) factors.
It is important to note that there is a recognized genetic condition or the 22q11.21 deletion syndrome also known as DiGeorge/Shprintzen/velocardiofacial syndrome which is near but does not overlap with the deletion seen in our patient found at coordinates chr22: 2 2,311,348–22,578,983 (hg19). Although some of the phenotypic features of this recognized syndrome do overlap with several of our patient’s clinical findings, she does not have the 22q11.21 deletion but rather a different, smaller deletion nearby on the same chromosome. Our patient’s deletion involves only one gene, TOP3B, which is annotated in the OMIM database and encodes a protein that alters and controls the states of DNA during transcription by catalyzing transient breakage and rejoining of a single strand of DNA [Yang et al., 2014]. TOP3B participates in DNA recombination, genome stability, and cellular aging, and interacts with DNA helicase SGS1. Of note, the deletion in our patient does not include the nearby PPM1F gene, which has also been identified as disease causing.
Burnside [2015] recently published a review of deletions within the 22q11.21q11.23 region with the goal of standardizing the nomenclature for these varied deletions and introduced unique cases into the medical literature. This review classified deletions into proximal, central, and 3 types of distal subgroups based on chromosome coordinates. Deletions within 2 classification categories, distal type I and distal type III, included the TOP3B gene that was deleted in our patient. The review emphasizes the phenotypic variability among patients with these distinct but overlapping deletions, including short stature, microcephaly, intellectual disability, cardiovascular defects, dysmorphic features, and more. This case-specific variability highlights the need for a more in-depth and individualized explanation for a genotype-phenotype relationship to understand which genes may play dominant roles.
Four different studies reported a roughly 2-Mb deletion encompassing many genes including ERK2, PRAME, BCR, and TOP3B. Rauch et al. [1999] studied a family in which individuals with this deletion had learning difficulties in addition to congenital heart defects (one affected female) and dysmorphic features including retrognathia and high-arched palate with minimal nick in the uvula, while the family members without the deletion were normal. Mikhail et al. [2007] reported a 15-year-old Hispanic boy with the same deletion who had learning and behavioral problems as well as mildly dysmorphic features. Rauch et al. [2005] reported a patient with the same deletion who was diagnosed with CHARGE (coloboma, heart anomaly, atresia, retardation, genital anomaly, ear anomaly) syndrome (OMIM #214800) at 7.5 years of age; this patient had a small ventricular septal defect, conductive hearing loss, bilateral choanal atresia, right-sided preauricular tag, and learning difficulties. And finally, Ben-Shachar et al. [2008] reported 2 patients with the same deletion who had global developmental delay and/or mild mental retardation, more prominent in language, in addition to mildly dysmorphic features.
The deletions mentioned above appear to be involved in cognitive impairment and/or developmental delays and include TOP3B; however, it is difficult to determine the specific impact of TOP3B since these deletions encompass multiple genes, any of which may play a dominant role, and the phenotypes often extend beyond developmental delay or intellectual disability. Phenotypic discrepancies for overlapping but distinct deletions may help elucidate specific genotype-phenotype relationships and provide justification for the argument that TOP3B impacts cognitive function. For example, Cooper et al. [2012] suggested BCR could be a candidate gene to explain the phenotype of intellectual disability in the 13 patients they reported. However, other studies suggest that TOP3B may be a better candidate gene. In addition to the 2 patients reported by Ben-Shachar et al. [2008] and described in the previous paragraph as having the roughly 2-Mb deletion, the same authors identified 4 patients with a smaller deletion encompassing only a subset of those genes (and no additional genes, i.e., fully-contained within the larger deletion). Specifically, these 4 patients had deletions that included TOP3B but not BCR and shared phenotypic features with 2 patients who had the larger deletion, such as developmental delay and behavioral problems, suggesting that TOP3B rather than BCR may play an important role in the developmental delay phenol-type.
The most convincing evidence for the role of TOP3B in cognitive impairment comes from cases with deletions or mutations involving this gene. For example, Xu et al. [2012] reported a woman with a heterozygous de novo c.1415 G>A mutation in TOP3B and decline in academic function resulting in low academic aptitude in addition to formal thought disorder and paranoid tendencies. Iossifov et al. [2012] reported a male with a missense mutation (c.1997 A>G) in the TOP3B gene who also had autism. Butler et al. [2015] identified TOP3B as a clinically relevant gene in a review of genes involved with autism spectrum disorders. Tan et al. [2011] reported a male with a deletion encompassing only a subset of the area deleted in our patient and including over half of the TOP3B gene; this patient had phenotypic similarities to our patient, including learning difficulties and delay in expressive language skills. It is important to note that this patient also had an isodicentric Y chromosome, but the fact that his speech delay was more severe than expected for an isodicentric Y warranted the microarray in the clinical setting.
Finally, Stoll et al. [2013] reported that the deletion of TOP3B contributes to neurodevelopmental disorders. The authors identified a 240-kb deletion of chromosome 22q11.22 encompassing TOP3B and IGLV2–14 and argued that this deletion is important for cognitive functioning. They found that carriers of this deletion had significantly higher frequencies of intellectual deficit than non-carriers and had an even greater overrepresentation of milder learning difficulties. Four individuals were found to be homozygous for this deletion, and all of them had cognitive impairment that ranged from mental retardation to poor performance on tests of information processing and executive function. The authors also found an association between this deletion and schizophrenia, and 2 individuals homozygous for this deletion had a diagnosis of schizophrenia in addition to cognitive impairment.
After discovering the association between the deletion of TOP3B and cognitive impairment, Stoll et al. [2013] undertook extensive biochemical investigations to better characterize the possible mechanism. They proposed that TOP3B actually participates in mRNA metabolism as well as DNA processing because TOP3B was found to be associated with the fragile X mental retardation protein (FMRP), an mRNA-binding protein that interacts with polyribosomes and regulates translation. The FMRP is encoded by the FMR1 gene, mutations in which cause fragile X syndrome (OMIM #300624), a genetic condition that involves developmental delays, learning disabilities, autism, attention deficit disorder, and characteristic physical features such as large ears, craniofacial features, macroorchidism, and hyperflexible fingers [Cook et al., 2014]. The detailed studies of Stoll et al. [2013] found that TOP3B plays a role in the metabolism of FMRP-bound mRNAs. Similarly, Xu et al. [2013] found that TOP3B binds multiple mRNAs related to mental disorders and neuronal function and works with FMRP to promote synapse formation. This interaction between TOP3B and FMRP could explain the overlapping features seen in our patient with features present in the typical fragile X syndrome presentation.
We propose that our patient’s deletion of TOP3B plays a significant role in her clinical phenotype, specifically in terms of her cognitive impairments and possibly craniofacial features, but the lack of genetic testing of other family members (parents and sibling) limits the final interpretation or definitive conclusions. However, reports in the medical literature on patients with deletions encompassing the TOP3B gene having similar phenotypes further support our observations. Additionally, the demonstrated interaction between TOP3B and FMRP suggests that disruption of either gene product may play a role in causing cognitive impairment through a similar or identical mechanistic pathway; that is, our patient’s phenotype may overlap with that of fragile X syndrome (e.g., cognitive impairment, broad nasal tip, mid-face hypoplasia prominent jaw) because the product of her deleted gene, TOP3B, normally works together with the protein product of FMR1, the gene mutated in fragile X syndrome, to influence neuronal function. The small deletion seen in our patient includes TOP3B, which is compelling, but additional analyses (e.g., genome sequencing, animal model studies) would be warranted. The disruption of TOP3B appears to affect neurodevelopment and cognition in humans. The authors encourage the reporting of additional patients with involvement of this chromosome region and the TOP3B gene to further characterize and understand its role leading to potential treatment modalities.
Acknowledgement
This study was supported by The National Institute of Child Health and Human Development (NICHD) Grant No. HD02528.
Statement of Ethics
A photographic consent form was signed by the patient’s representative and family support worker.
Footnotes
Disclosure Statement
The authors have no conflicts of interest to declare.
References
- Ben-Shachar S, Ou Z, Shaw C, Belmont JW, Patel MS, et al. : 22q11.2 distal deletion: a recurrent genomic disorder distinct from DiGeorge syndrome and velocardiofacial syndrome. Am J Hum Genet 82: 214–221 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bittel DC, Yu S, Newkirk H, Kibiryeva N, Holt A 3rd, et al. : Refining the 22q11.2 deletion breakpoints in DiGeorge syndrome by a CGH. Cytogenet Genome Res 124: 113–120 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnside RD: 22q11.21 deletion syndromes: a review of proximal, central, and distal deletions and their associated features. Cytogenet Genome Res 146: 89–99 (2015). [DOI] [PubMed] [Google Scholar]
- Butler MG, Rafi SK, Manzardo AM: High-resolution chromosome ideogram representation of currently recognized genes for autism spectrum disorders. Int J Mol Sci 16: 6464–6495 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook D, Nuro E, Murai KK: Increasing our understanding of human cognitive through the study of Fragile X syndrome. Dev Neurobiol 74: 147–177 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper GM, Coe BP, Girirajan S, Rosenfeld JA, Vu T, et al. : A copy number variation morbidity map of developmental delay. Nat Genet 43: 838–846 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- anai R, Caron PR, Wang JC: Human TOP3B : a single-copy gene encoding DNA topoisomerase III. Proc Natl Acad Sci USA 93: 3653–3657 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iossifov I, Ronemus M, Levy D, Wang Z, Hakker I, et al. : De novo gene disruptions in children on the autistic spectrum. Neuron 74: 285–299 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson EM, Shaikh TH, Gururangan S, Jones MC, Malkin D, et al. : High-density single nucleotide polymorphism array analysis in patients with germline deletions of 22q11.2 and malignant rhabdoid tumor. Hum Genet 122: 117–127 (2007). [DOI] [PubMed] [Google Scholar]
- Mikhail FM, Descartes M, Piotrowski A, Andersson R, Diaz de Stahl T, et al. : A previously unrecognized microdeletion syndrome on chromosome 22 Band q11.2 encompassing the BCR gene. Am J Med Genet Part A 143A:2178–2184 (2007). [DOI] [PubMed] [Google Scholar]
- Rauch A, Pfeiffer RA, Leipold G, Singer H, Tigges M, Hofbeck M: A novel 22q11.2 microdeletion in DiGeorge syndrome. Am J Hum Genet 64: 659–666 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauch A, Zink S, Zweier C, Thiel CT, Koch A, et al. : Systematic assessment of atypical deletions reveals genotype-phenotype correlation in 22q11.2. J Med Genet 42: 871–876 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodningen OK, Prescott T, Eriksson A, Rosby O: 1.4 Mb recurrent 22q11.2 distal deletion syndrome, two new cases expand the phenotype. Eur J Med Genet 51: 646–650 (2008). [DOI] [PubMed] [Google Scholar]
- Stoll G, Pietilainen OP, Linder B, Suvisaari J, Brosi C, et al. : Deletion of TOP3B, a component of FMRP-containing mRNPS, contributes to neurodevelopmental disorders. Nat Neurosci 16: 1228–1237 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan TY, Collins A, James PA, McGillivray G, Stark Z, et al. : Phenotypic variability of distal 22q11.2 copy number abnormalities. Am J Med Genet Part A 155: 1623–1633 (2011). [DOI] [PubMed] [Google Scholar]
- US National Library of Medicine. Genetics Home Reference [Internet]. Bethesda (MD): 22q11.2 deletion syndrome (reviewed July, 2013; cited May 23, 2016). https://ghr.nlm.nih.gov/condition/22q112-deletion-syndrome.
- Xu B, Ionita-Laza I, Roos JL, Boone B, Woodrick S, et al. : De novo gene mutations highlight patterns of genetic and neural complexity in schizophrenia. Nat Genet 44: 1365–1369 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu D, Shen W, Guo R, Zue Y, Peng W, et al. : Top3B is an RNA topoisomerase that works with fragile X syndrome protein to promote synapse formation. Nat Neurosci 16: 1238–1247 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Fan YS, Siu VM: A child with features of Goldenhar syndrome and a novel 1.12 Mb deletion in 22q11.2 by cytogenetics and oligonucleotidea array CGH: is this a candidate region for the syndrome? Am J Med Genet Part A 146A:1886–1889 (2008). [DOI] [PubMed] [Google Scholar]
- Yang Y, McBride KM, Hensley S, Lu Y, Chedin F, Bedford MT: Arginine methylation facilitates the recruitment of TOP3B to chromatin to prevent R-loop accumulation. Mol Cell 53: 484–497 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]