Abstract

Glioblastoma multiforme is the most aggressive brain tumor, due to its high invasiveness and genetic heterogeneity. Moreover, the blood–brain barrier prevents many drugs from reaching a therapeutic concentration at the tumor site, and most of the chemotherapeutics lack in specificity toward cancer cells, accumulating in both healthy and diseased tissues, with severe side effects. Here, we present in vitro investigations on lipid-based nanovectors encapsulating a drug, nutlin-3a, and superparamagnetic iron oxide nanoparticles, to combine the proapoptotic action of the drug and the hyperthermia mediated by superparamagnetic iron oxide nanoparticles stimulated with an alternating magnetic field. The nanovectors are functionalized with the peptide angiopep-2 to induce receptor-mediated transcytosis through the blood–brain barrier and to target a receptor overexpressed by glioma cells. The glioblastoma multiforme targeting efficiency and the blood–brain barrier crossing abilities were tested through in vitro fluidic models, where different human cell lines were placed to mimic the tumor microenvironment. These nanovectors successfully cross the blood–brain barrier model, maintaining their targeting abilities for glioblastoma multiforme with minimal interaction with healthy cells. Moreover, we showed that nanovector-assisted hyperthermia induces a lysosomal membrane permeabilization that not only initiates a caspase-dependent apoptotic pathway, but also enhances the anticancer efficacy of the drug.
Keywords: glioblastoma multiforme, active targeting, lipid nanoparticles, superparamagnetic iron oxide nanoparticles, blood−brain barrier, lysosomal membrane permeabilization
Introduction
Glioblastoma multiforme (GBM) is the most common and aggressive tumor of the central nervous system, with an incidence of two to three cases per 100 000 people worldwide1 and a median survival of just about 1.5 years after diagnosis. The current standard treatment consists of a combination of surgical resection, radiotherapy, and chemotherapy; however, it only minimally extends the average patient survival.2 This poor outcome is due to the complexity of the tumor, its genetic heterogeneity,3 and its topological invasive nature that make the complete surgical resection almost impossible. Moreover, most of the conventional chemotherapeutics are unable to cross the blood–brain barrier (BBB) and to reach a satisfactory therapeutic concentration within the brain.4 Nevertheless, even if the drug diffuses through the BBB, its unselective distribution and poor specificity toward the desired site of action may cause severe side effects to the delicate environment of the central nervous system.5
Given this scenario, it is clear that there is an urgent need to design alternative therapies for the treatment of GBM. An ideal therapeutic approach should consist of a system able to deliver its pharmaceutical cargo, to penetrate through the BBB, and, finally, to selectively target the diseased tissues without affecting the healthy ones.6 In this context, nanomedicine made huge progresses in improving the systemic delivery of drugs, increasing their bioavailability, and protecting them from enzymatic digestion.7 Nanoparticles are also known to accumulate in tumor tissues due to the enhanced permeability and retention (EPR) effect;8 however, this form of passive targeting is difficult to control and can induce multidrug resistance (MDR).9 Active targeting, instead, relies on a more specific interaction between a ligand, attached on the surface of the nanoparticles, and particular receptors overexpressed on the target tissues but minimally expressed on healthy cells.10 Many ligands have been exploited for this purpose, but peptides present several advantages compared to the others, such as high stability, low immunogenicity, and simple production.6,7
In this work, we focused on the study of the targeting abilities and the potential therapeutic efficacy of a lipid-based nanovector functionalized with angiopep-2, a peptide derived from the Kunitz domain of aprotinin,11 that binds to the low-density lipoprotein receptor-related protein 1 (LRP1), overexpressed on glioma cell surface.10 LRP1 is also present on brain capillary endothelial cells (BCECs);12 therefore, angiopep-2 should be also able to favor the penetration of the BBB via receptor-mediated transcytosis.13 Our nanovector is composed of a matrix of biocompatible lipids, encapsulating both superparamagnetic iron oxide nanoparticles (SPIONs) and a nongenotoxic drug, nutlin-3a. The latter is an antagonist of the murine double minute-2 (MDM2) protein, a negative regulator of the tumor suppressor protein p53, that inhibits its transcriptional activity14 and stimulates its degradation.15 According to The Cancer Genome Atlas (TCGA), 85.3% of GBM presents alterations of the p53 pathway, among which 27.9% are mutation/deletion of the TP53 gene, 7.6% are amplification of the MDM2 protein, and the majority (57.8%) consists in the deletion of the CDKN2A gene that codes for the p14ARF protein, a physiological inhibitor of the MDM2 protein.16 Therefore, an overexpression of the MDM2 protein is directly related to cancer development.14 The ability of nutlin-3a to inhibit the MDM2-p53 interaction is of extreme importance in the reactivation of the p53 pathway.14 Moreover, MDM2 inhibitors have a significantly lower toxicity to healthy cells with respect to other drugs, making them interesting options for cancer therapy.14,15 The other components of the proposed nanoplatform, SPIONs, are well known in the literature to induce cell apoptosis through hyperthermia after stimulation with an alternating magnetic field (AMF).17,18 This mechanism occurs regardless of the type of cell, but its effectiveness depends mainly on the actual concentration and compartment localization of the SPIONs within the intracellular environment.19 The efficacy of this treatment increases when combined with conventional chemotherapeutic drugs.17
Here, we demonstrated that angiopep-2-functionalized lipid-based magnetic nanovectors (Ang-LMNVs) have a strong affinity for glioblastoma cells with respect to other healthy cell lines. The preferential uptake by GBM cells has been demonstrated in vitro with different approaches, both in static and in dynamic conditions, with ad hoc developed microfluidic bioreactors. The resulting Ang-LMNVs could cross a fluidic in vitro model of the BBB more efficiently than nonfunctionalized nanovectors, maintaining their ability to selectively target tumor cells after the BBB crossing. We also aimed at elucidating the mechanism of action of the drug and, in particular, of SPIONs stimulated with an appropriate AMF, showing that the latter induces lysosomal membrane permeabilization (LMP) with a consequent release of proteolytic enzymes from the lysosome milieu.20,21 The combination of nutlin-3a delivery and magnetic stimulation significantly reduces the viability of GBM cells, inducing cell apoptosis via different pathways and inhibiting tumor growth.
Materials and Methods
Lipid-Based Magnetic Nanovector Synthesis
Lipid-based magnetic nanovectors (LMNVs) were synthesized similarly to a previous work.17 In brief, 25 mg of 1-stearoyl-rac-glycerol (GMS, Sigma-Aldrich), 2.5 mg of oleic acid (Sigma-Aldrich), 2.5 mg of 1,2-dipalmitoyl-rac-glycero-3-phosphocholine (DPPC, Sigma-Aldrich), 2 mg of mPEG-DSPE (5000 Da, Nanocs, Inc.), 2 mg of NHS-PEG-DSPE (5000 Da, Nanocs, Inc.), and 1 mg of nutlin-3a (Sigma-Aldrich) were mixed with 84.5 μL of an ethanol solution of SPIONs (15 wt %, US Research Nanomaterials, Inc.), inside a 6 mL glass vial. The above mixture was heated at 70 °C to melt the lipids. Then, 3 mL of a Tween 80 (Sigma-Aldrich) aqueous solution (1.0 wt %) at 70 °C was added to the lipid mixture, vortexed for 1 min, and sonicated for 20 min (amplitude 90%) using an ultrasonic tip (Fisherbrand Q125 Sonicator). LMNVs were cooled down at 4 °C for 30 min and then purified by centrifugation (16 000g, 90 min, 4 °C) and redispersed in Milli-Q water (Millipore) three times. Plain LMNVs were synthesized as described above, but without adding nultin-3a to the lipid mixture. Lipid-based nanovectors without SPIONs (LNVs) have been prepared as required for the NMR characterization, following the same procedure but without the addition of magnetic nanoparticles.
For functionalization, 100 μL of an angiopep-2 (Selleckchem) solution in water (1 mg/mL) was added to 1 mL of LMNV dispersion (6 mg/mL) to have an approximate NHS-PEG-DSPE:angiopep-2 theoretical molar ratio of 1:2. The dispersion was diluted in phosphate-buffered saline (PBS, Sigma-Aldrich) to optimize the pH for the reaction between NHS and the amine groups on the peptide. The solution was left under gentle shaking at 4 °C in the dark for 4 h. Then, it was washed three times by centrifugation (16 000g, 90 min, 4 °C), and the final pellet was redispersed in 1 mL of Milli-Q water.
For confocal imaging, Ang-LMNVs and LMNVs were labeled with a fluorescent Vybrant DiO cell-labeling dye (Invitrogen) by incubating 1 mg of particles with 10 μL of dye for 2 h at 37 °C and then washing three times by centrifugation (16 000g, 90 min, 4 °C).
Lipid-Based Magnetic Nanovectors Characterization
Transmission electron microscopy (TEM) was performed to analyze morphology and size of the nanovectors. Before the measurement, the samples were sonicated for 2 min. A drop of a sample was deposited on a Cu grid (150 mesh) coated with an ultrathin amorphous carbon film. After 20 s, the drop was removed with a filter paper and the grid washed with Milli-Q water. Afterward, a drop of a solution of 1% uranyl acetate in water was deposited on the grid for 60 s to stain the sample and to enhance the contrast of the lipid component. Finally, the drop was removed with a filter paper. Images were acquired with a JEOL Jem-1011 (Jeol) working at 100 kV on single-tilt sample holder. Images in bright-field mode were acquired with the same instrument, following the same sample preparation protocol, but skipping the staining procedure.
Dynamic light scattering measurements were carried out with a Zetasizer NanoZS90 (Malvern Instruments Ltd) to determine the hydrodynamic diameter and the ζ-potential of LMNVs and Ang-LMNVs at 37 °C. The dispersions, at a concentration of 100 μg/mL in ultrapure water, were sonicated for 30 s with a Bandelin ultrasonic probe (8 W) before the measurement. The stability of Ang-LMNVs at pH 4.5 (0.05 M phosphate buffer) and pH 7.4 (PBS) was evaluated by diluting a nanoparticle stock solution (15 mg/mL) in the corresponding buffer up to a final concentration of 100 μg/mL and measuring the hydrodynamic diameter at different time points (24, 48, 72, and 96 h). The intensity distribution was derived from the correlogram through CONTIN analysis, whereas the hydrodynamic diameter and the polydispersity index were obtained from cumulant analysis.
Thermogravimetric analysis (TGA) was performed with a Q500 analyzer (TA Instruments). Scanning was performed in the temperature range of 30–600 °C, using a 10 °C/min heating rate. Cooling was achieved using a 50 mL/min nitrogen flow.
The conjugation of the peptide to the nanoparticles was verified by 1H NMR (Bruker Ascend 400) by monitoring the disappearance of the peak related to the NHS group ((C(O)CH2CH2C(O), (4 × tfonc)H, δ = 2.6 ppm). Freeze-dried samples were dissolved in CDCl3 (Sigma-Aldrich) prior to the analysis. LNVs were used for this characterization to avoid the interference between SPIONs and the magnetic field. 1H NMR spectra were analyzed with Mestrenova.
Sodium dodecyl sulfate–poly(acrylamide) gel electrophoresis (SDS-PAGE) was performed to quantify the peptide on the nanovector surface. Each sample (37.5 μL of angiopep-2, LMNVs, or Ang-LMNVs) at a known concentration was added to 12.5 μL of Laemmli buffer (BioRad) and heated for 10 min at 95 °C for protein denaturation. A 4–15% Mini-PROTEAN TGX Precast Protein Gel (BioRad) was placed in an electrophoresis cell (Mini-PROTEAN Tetra Cell, BioRad) filled with Tris/Glycine running buffer (BioRad). The pretreated samples (50 μL) were loaded in each well, in parallel with a molecular weight marker (PageRuler Plus Prestained Protein Ladder, Thermo Fisher Scientific). The gel was then run at 100 V for 1 h. Afterward, the gel was stained with Coomassie Blue (for 100 mL of solution: 0.125 g of brilliant blue R250, 10 mL of glacial acetic acid, 40 mL of water, 50 mL of methanol) for 1 h under gentle agitation. The gel was then rinsed twice with a destaining solution (10% glacial acetic acid, 40% H2O, 50% methanol) for 20 min to remove unspecific staining. Finally, the gel was washed with Milli-Q water. The bands, corresponding to the peptide, were analyzed for quantification with ImageJ software. The intensity of the bands corresponding to Ang-LMNVs was compared to that one of the bands of the plain peptide.
The presence of three phenylalanine residues in angiopep-2 allows for its detection with fluorescence spectroscopy by measuring the characteristic emission intensity at around 300 nm. A Cary Eclipse fluorescence spectrofluorometer (Agilent Technologies) was used to measure the emission signal between 270–400 nm, with an excitation wavelength of 260 nm. The spectra obtained for Ang-LMNVs were normalized by the nonfunctionalized LMNVs spectra to remove the background scattering. The intensity at 300 nm of Ang-LMNVs was compared to a calibration curve obtained with several concentrations of the free peptide in Milli-Q water.
The presence of angiopep-2 and its amount on the nanovector surface was also confirmed by bicinchoninic acid assay (BCA) Protein Kit (Thermo Scientific). Briefly, 25 μL of 8 mg/mL Ang-LMNVs or LMNVs (used as control) was mixed with 200 μL of working solution. The samples were then incubated at 37 °C for 30 min, and the absorbance at 560 nm of 90 μL of the samples was measured in triplicate with a plate reader (VICTOR X3 plate reader, PerkinElmer). The amount of peptide conjugated to the nanovectors was calculated using a calibration curve obtained by performing the BCA assay to several concentrations (range 0–500 μg/mL) of free angiopep-2 in water.
The loading and release of nutlin-3a from Ang-LMNVs were evaluated by high-performance liquid chromatography (HPLC) with a Shimadzu LC-20AT, using a C-18 column (150 mm × 4.6 mm i.d., 5 μm particle size). The mobile phase was composed of 80% methanol (for HPLC, ≥99.9%, Sigma-Aldrich) and 20% H2O (HPLC Plus, Sigma-Aldrich), pumped in isocratic mode at a flow rate of 1 mL/min. The peak of nutlin-3a was found at a retention time of 4.77 min, and its intensity was monitored by a UV detector at 260 nm.
For the quantification of drug loading, 1 mg of freeze-dried Ang-LMNVs was dissolved in 400 μL of methanol and heated at 70 °C for 1 h to melt the lipid core. Afterward, 100 μL of cold ultrapure water was added and the sample was centrifuged at 16 000g for 90 min at 4 °C. The supernatant was collected and measured with HPLC. The drug loading (%) and the encapsulation efficiency (%) were calculated using the equations
| 1 |
| 2 |
For the release studies, 1 mg of Ang-LMNVs was redispersed in 1 mL of four different buffers: at pH 7.4 (PBS) to simulate the physiological environment; at pH 7.4 + 100 μM H2O2 to simulate the physiological environment in the presence of oxidative stress; at pH 4.5 (0.05 M phosphate buffer) to simulate the cancer environment; and at pH 4.5 + 100 μM H2O2 to simulate the cancer environment in the presence of oxidative stress. The samples were left under agitation at 37 °C. At each time point (6, 24, 48, 72, and 96 h), the samples were centrifuged at 16 000 g for 90 min at 4 °C. The supernatants were collected and analyzed with HPLC, whereas the pellets were redispersed in their buffers and left under agitation until the following time point. To study the effect of the application of an alternating magnetic field (AMF) on the release profile, 1 mg of Ang-LMNVs dispersed in the corresponding buffers were stimulated for 2 h with a MagneTherm device (NanoTherics) at an applied magnetic field of 20 mT, using a water-cooled coil of 9 turns and 44 mm inner diameter, and at a frequency of 753 kHz (for details on the parameters used for the chronic stimulation of the cells, see the following).
Cellular Uptake Evaluation in Static Conditions
The uptake of LMNVs and Ang-LMNVs by human glioblastoma U87 MG cells (ATCC HTB-14) was evaluated in vitro in static conditions. Cells (15 × 103 cells/cm2) were seeded on sterilized glass coverslips and incubated with high-glucose DMEM (4.5 mg/mL), 10% FBS, 1% penicillin/streptavidin (P/S), and 1% L-glutamine. U87 MG cells were subsequently incubated for 6 h at 37 °C with 400 μL of 200 μg/mL of either Ang-LMNVs or LMNVs (previously labeled with Vybrant DiO dye) in complete medium.
For confocal acquisition, the cells were washed twice with PBS and fixed with 4% paraformaldehyde (PFA, Sigma-Aldrich) for 20 min at 4 °C. Then, the cells were incubated with a blocking solution consisting of goat serum (GS, 10%; EuroClone) and Triton X-100 (1:1000 dilution, Sigma-Aldrich) in PBS for 30 min. The cells were stained with TRITC-phalloidin (1:200 dilution, Sigma-Aldrich) and Hoechst 33342 (1:1000 dilution, Invitrogen) in PBS for 30 min at 37 °C. Confocal microscopy acquisitions were performed using a C2s confocal microscope (Nikon).
Quantitative analysis of the targeting efficiency was performed by flow cytometry (Cytoflex Beckmann) at the same experimental conditions. Before the measurements, the cells were washed three times with PBS, treated with 0.05% trypsin-EDTA, centrifuged, and resuspended in PBS. The fluorescence of the cells was measured (λex = 488 nm; λem = 525–540 nm), and the fluorescence intensity of the nontreated control cells was used as threshold.
Selective Targeting Efficiency in Dynamic Conditions
An ad hoc fluidic bioreactor was designed and fabricated to test the targeting efficiency of Ang-LMNVs vs LMNVs. The poly(methyl methacrylate) (PMMA) bioreactor is composed of six independent chambers located in three channels; each chamber has been designed to host a glass coverslip of 0.9 cm2 (see a schematic representation of the fluidic bioreactor and a schematic representation of cell lines disposition in Figure S1). Four different cell lines were used to recreate a simplified in vitro model of the brain environment: neuron-like cells derived from the SH-SY5Y human neuroblastoma (ATCC CRL-2266), human primary astrocytes (HA) from cerebral cortex (Innoprot, P10251), human glioblastoma U87 MG cells (ATCC HTB-14), and human endothelial hCMEC/D3 cells (Merck Millipore, SCC066). Concerning SH-SY5Y cell differentiation, 10 × 103 SH-SY5Y cells/cm2 are seeded on 1.9 cm2 glass coverslips into a 24-well plate with Dulbecco’s modified Eagle’s medium/Nutrient Mixture F-12 (DMEM/F12; Sigma-Aldrich) supplemented with 10% of FBS and 1% P/S. The neural differentiation is induced by replacing the cell growth medium with differentiation medium: DMEM high-glucose (4.5 mg/mL), 1% FBS, 10 μM retinoic acid, and 1% P/S for 6 days. Neuron-like cells derived from differentiation of SH-SY5Y cells will be referred, for convenience, as SH-SY5Yd throughout the paper. SH-SY5Y cells were seeded 8 days before the targeting experiment. After the seeding, the cells were maintained for 2 days in proliferative conditions, and thereafter switched to differentiation for 6 days. The other cell lines were seeded 2 days before the experiment, and they were maintained in proliferative conditions: DMEM high-glucose with 10% of FBS and 1% P/S for U87 MG and HA, and EndoGRO-MV-VEGF medium (Merck Millipore) with 5% FBS and 1% P/S for hCMEC/D3. Two bioreactors were connected in series to a peristaltic pump (Ibidi fluidic unit), and triplicates of each cell line were placed along the channels. Vybrant DiO-labeled Ang-LMNVs or LMNVs suspensions (15 mL, 200 μg/mL) in DMEM high-glucose with HEPES and 10% FBS were perfused at a speed flow of 2 mL/min for 6 h at 37 °C. After perfusion, the suspension containing the nanovectors was removed and the cells were washed two times with PBS, fixed with PFA (4% in PBS for 20 min at 4 °C), left for 30 min in the blocking solution, and stained with TRITC-phalloidin and Hoechst 33342, as previously described; confocal laser scanning microscopy was finally performed and volume analysis of the nanovectors–cell co-localization (%) was performed using NIS-Elements software (Nikon), as described in a previous work.18
Blood–Brain Barrier Investigation
To study the ability of LMNVs and Ang-LMNVs to cross the BBB and their interactions with the brain microenvironment, another ad hoc developed bioreactor populated by the above-mentioned four different human cell cultures was designed. The bioreactor, fabricated in PMMA, is composed of two chambers. The upper chamber, realized basing on a previous model,22 mimics the endothelial lumen, and is composed of a channel 15 mm long, 5 mm large, and 0.5 mm high (Figure S2). According to this geometry, a flow of 12 mL/min is needed to reproduce the typical shear stress experienced in brain capillaries (10 dyn/cm2).23 The bottom chamber is 2 mm high with a total surface of 20 mm2, and it is designed to allocate two glass coverslips of 0.9 cm2 each. One of the two glass coverslips was seeded with U87 MG cells (20 × 103 cm2), while the other was seeded with SH-SY5Yd cells (10 × 103 cells/cm2); both cell types were cultured separately and inserted in the BBB setup 1 day before the experiment. The top and bottom chambers are separated by a poly(ethylene terephthalate) (PET) transparent porous membrane with pores of 3 μm. The upper surface of the porous membrane was seeded with human endothelial cells hCMEC/D3 (5 × 104 cells/cm2) in EndoGRO-MV-VEGF medium (Merck Millipore) with 5% FBS and 1% P/S, whereas the other side was seeded with HA (8 × 103 cells/cm2) 3 days after hCMEC/D3 seeding. Experiments were performed after 5 days, when the endothelial cells formed a monolayer.
The quality of the BBB was assessed by measuring the transendothelial electrical resistance (TEER) with a Millipore Millicell ERS-2 Volt-Ohmmeter and the permeability to a 200 μg/mL rhodamine B-dextran (70 kDa, Invitrogen) solution in complete medium, as previously described in other works.24
Nanovector/Cell Interactions
The uptake mechanism of Ang-LMNVs by U87 MG cells was investigated by confocal fluorescence microscopy (C2 system Nikon), focusing on the pinocytosis pathway and on the clathrin- and caveolin-mediated endocytosis.
Pinocytosis was studied using the Cascade Blue hydrazide fluorescent dye (Invitrogen). U87 MG cells (15 × 103 cells/cm2) were seeded in WillCo dishes. After 1 day, the cells were incubated for 24 h with 200 μL of phenol red-free HEPES-supplemented complete medium with 300 μM of Cascade Blue hydrazide fluorescent dye and with 200 μg/mL of Vybrant DiO-labeled Ang-LMNVs. After 24 and 72 h, the cells were washed three times and refilled with fresh phenol red-free HEPES-supplemented complete medium. Confocal images were acquired immediately, and a co-localization analysis (Pearson’s correlation coefficient) between nanovectors and pinosomes was performed by NIS-Elements software.
Caveolin-1 and clathrin-mediated internalization pathways were studied by immunofluorescence. Vybrant DiO-labeled Ang-LMNVs (200 μL, 200 μg/mL) in complete medium were administered for 24 and 72 h to U87 MG cells seeded in WillCo dishes, as previously reported. The cells were then fixed with 4% PFA for 20 min at 4 °C, washed with PBS, and incubated with a blocking solution for 30 min. The samples were then incubated at 37 °C with anti-caveolin-1 (1:150; Abcam) primary antibody or anti-clathrin primary antibody (1:150; Abcam) in PBS with 10% GS for 2 h. After three washes with 10% GS in PBS, the samples were stained for 1 h with Alexa-Fluor-488 secondary antibody (1:250 dilution; Millipore), TRITC-phalloidin (1:200; Sigma-Aldrich), and Hoechst 33342 (1:1000; Invitrogen). Confocal fluorescence microscopy and co-localization analysis were performed as previously described.
Ang-LMNV uptake in lysosomes and late endosomes was assessed by confocal fluorescence microscopy. U87 MG cells (15 × 103 cells/cm2) were seeded in WillCo Petri dishes and subsequently incubated with 200 μL of 200 μg/mL of Vybrant DiO-labeled Ang-LMNVs in phenol red-free HEPES-supplemented DMEM high-glucose with 10% FBS for 24 and 72 h. The samples were washed twice with PBS and treated with the acidotropic LysoTracker Deep Red dye (1:2000 dilution, Invitrogen) in high-glucose phenol red-free HEPES-supplemented complete medium. After 30 min, the cells were washed and stained with Hoechst 33342 (1:1000 dilution, Invitrogen) for 15 min at 37 °C. Confocal fluorescence microscopy and co-localization analysis were performed as previously described.
Cell Viability Assay
Cell viability was assessed using the WST-1 assay (Roche). U87 MG (15 × 103 cells/cm2) were seeded (in triplicate for each experimental class) in 24-well plates, and their viability was evaluated upon treatment with different concentrations of free nutlin-3a in DMSO (1, 3, 5, and 10 μM), Ang-LMNVs (109, 325, 544, and 1089 μg/mL), and Nut-Ang-LMNVs (109, 325, 544, and 1089 μg/mL corresponding to 1, 3, 5, and 10 μM of nutlin-3a) for 24 and 72 h. Cultures were incubated with 300 μL of phenol red-free complete medium with the WST-1 reagent (1:20 dilution) for 30 min at 37 °C. Thereafter, absorbance at 450 nm was measured using a PerkinElmer Victor X3 UV–Vis spectrophotometer. The values were expressed as % with respect to untreated controls.
The effect of Nut-Ang-LMNVs on different cell lines was investigated with the same fluidic bioreactor setup used for the targeting efficiency experiments, and 325 μg/mL of Nut-Ang-LMNVs (corresponding to 3 μM of nutlin-3a) were perfused. U87 MG cells, hCMEC/D3, SH-SY5Yd, and HA were placed in the fluidic bioreactor, as previously described. After 6 h, the perfusion was stopped, the cells were washed three times with PBS and placed in an incubator at 37 °C for 72 h. The cells were then incubated with 300 μL of phenol red-free complete medium with the WST-1 reagent for 20 min in the case of U87 MG cells, and for 90 min in the case of hCMEC/D3, SH-SY5Yd, and HA because of different metabolic rates. The absorbance at 450 nm was thereafter measured as described above. The toxic effect of free nutlin-3a (3, 5, and 10 μM in DMSO) on hCMEC/D3 (15 × 103 cells/cm2), SH-SY5Yd (10 × 103 cells/cm2), and HA (10 × 103 cells/cm2) at 72 h was also analyzed by the WST-1 assay, using the same protocol.
Finally, the effect of the AMF stimulation was studied as follows. U87 MG cells (15 × 103 cells/cm2, seeded at the center of WillCo dishes) were incubated with 325 μg/mL of Ang-LMNVs or Nut-Ang-LMNVs (corresponding to 3 μM encapsulated drug) or with 3 μM free nutlin-3a in DMSO for 48 h. Thereafter, cultures were exposed to AMF for 2 h/day with a MagneTherm device (NanoTherics) at an applied magnetic field of 20 mT, using a water-cooled coil of 9 turns and 44 mm inner diameter, and a frequency of 753 kHz, for 3 days. Control samples were left in the incubator without undergoing AMF stimulation. After the AMF chronic stimulation, the cell viability of each sample was assessed by WST-1. The same experimental protocol was also used to perform “rescue of viability” experiments: briefly, at the end of the 3-day AMF chronic stimulation, the cells were placed in the incubator for further 72 h before performing viability (WST-1) investigations.
Lysosome Permeabilization Investigation
To study the effect of the nanovector-mediated AMF stimulation on lysosomes, U87 MG cells were seeded (15 × 103 cells/cm2) in WillCo dishes and incubated for 72 h with 325 μg/mL of Vybrant DiO-labeled Ang-LMNVs in phenol red-free HEPES-supplemented complete medium. After 72 h, the cells were stained with LysoTracker Deep Red dye and Hoechst 33342, as previously described, and stimulated for 2 h with the MagneTherm device (NanoTherics) using a Live Cell AMF exposure system (NanoTherics) at 16 mT and 753 kHz, a setup allowing for a live confocal acquisition during the stimulation time. Confocal time-lapse imaging (C2 system, Nikon) was carried out with a perfect focus system (Nikon). The heating of the objective during the AMF stimulus was avoided by an automatic lowering of the microscope revolver after each acquisition (every 1 min) exploiting the escape function (NIS-Elements software); the revolver was then lifted, and the perfect focus function was activated just before the acquisitions.
The release of proteolytic enzymes from lysosomes was monitored by immunostaining of cathepsin B. Vybrant DiO-labeled Ang-LMNVs (325 μg/mL) in HEPES-supplemented complete medium were administrated to U87 MG cells previously seeded (15 × 103 cells/cm2) in WillCo dishes. After 48 h of incubation, the cells were stimulated with an AMF. The samples (control and Ang-LMNV-pretreated cells) were fixed immediately after the AMF treatment with pure methanol at −20 °C for 15 min. After three times washing with PBS and a step with the blocking solution, the cells were incubated with anti-cathepsin B antibody (1:100, Abcam) at 37 °C for 2 h. After three washing steps, TRITC anti-rabbit secondary antibody (1:250, Invitrogen) was added to the samples for 1 h at 37 °C. Images were acquired with a confocal microscope.
Heat Shock Protein 70 Expression
Immunofluorescence experiments against heat shock protein 70 (Hsp70) were performed to evaluate an increase of its expression as a consequence of the treatment with Ang-LMNVs + AMF stimulus. U87 MG cells were seeded (15 × 103 cells/cm2) at the center of WillCo dishes (1 cm2 total area) and then incubated with 325 μg/mL of Ang-LMNVs in HEPES-supplemented complete medium for 2 days before being stimulated for 2 h with the AMF, as previously described. Negative controls and positive controls were also performed; in particular, negative controls were represented by cultures without any treatment, while positive controls were placed for 1 and 2 h in an incubator at 42 °C. This temperature was chosen since it is the typical temperature achieved during hyperthermia treatments. After the stimulation, the samples were placed at 37 °C for a further hour in the incubator. Afterward, the samples were fixed with 4% of PFA at 4 °C. Immunostaining was performed with anti-Hsp70 antibody (1:50, Abcam) for 2 h at 37 °C; after three washing steps, FITC-labeled secondary anti-rabbit antibody (1:250, Invitrogen) was added and incubated for 1 h. Nuclei staining was performed with Hoechst 33342 (1:1000 dilution, Invitrogen) for 20 min; images were acquired with the confocal microscope.
Evaluation of Apoptotic Effects and Caspase-9 Activation
The expression of p53 and Ki-67 markers was investigated by immunofluorescence to evaluate the effects on apoptosis (p53) and on cell proliferation (Ki-67) after the treatment with Ang-LMNVs or Nut-Ang-LMNVs (with or without AMF). The concentrations used for this experiment were 325 μg/mL of Ang-LMNVs or Nut-Ang-LMNVs (corresponding to 3 μM encapsulated drug) and 3 μM free nutlin-3a in DMSO. The AMF stimulation protocol consisted of 2 h/day stimulation in DMEM high-glucose complete medium and HEPES with a MagneTherm device (NanoTherics) at an applied magnetic field of 20 mT, using a water-cooled coil of 9 turns and 44 mm inner diameter, and a frequency of 753 kHz, for 3 days. The same experimental protocol was also used to perform rescue experiments, as previously described for cell viability assays.
Concerning immunofluorescence, after fixation cells were incubated for 30 min with the blocking solution. Thereafter, cultures were incubated with primary rabbit anti-Ki-67 antibody (1:150 dilution, Millipore) and a primary mouse anti-p53 antibody (1:100 dilution, Abcam) for 2 h at 37 °C. Afterward, the cells were incubated for 1 h with an FITC-conjugated secondary anti-rabbit antibody (1:250 dilution, for Ki-67 staining, Millipore), and, after a washing step, with a TRITC-conjugated secondary anti-mouse antibody (1:250 dilution, for p53 staining, Millipore). Finally, 20 min incubation with Hoechst 33342 in PBS (1:1000 dilution, Invitrogen) for nucleus counterstaining was performed. Imaging was carried out with a C2s confocal microscope (Nikon); NIS-Elements software (Nikon) was used for the analysis of the p53 and Ki-67 positive nuclei.
CaspGLOW Fluorescein Active Caspase Staining Kit (BioVision) was used to investigate the activation of caspase-9, and thus the initiation of the apoptosis mediated by this pathway, in cells treated with Ang-LMNVs, Nut-Ang-LMNVs, or plain nutlin-3a, with or without the AMF stimulus. U87 MG cells were seeded (15 × 103 cells/cm2) in WillCo dishes and then incubated for 48 h with a 325 μg/mL dispersion of either Ang-LMNVs or Nut-Ang-LMNVs, or with nutlin-3a 3 μM (corresponding to the dose loaded in 325 μg/mL of Nut-Ang-LMNVs). Half of the samples underwent AMF stimulation for 2 h at 37 °C. After the treatments, cultures were left for 1 h in the incubator without any stimulus. The cell medium was then replaced with a working solution of caspase-9 fluorescent dye (33 μL stock solution was added in 167 μL of complete medium), and the samples were incubated for 1 h. Afterward, the cells were washed three times with the specific washing solution provided by the kit. Fluorescence imaging was performed with a C2s confocal microscope (Nikon). The same experimental classes were also analyzed with flow cytometry (Cytoflex Beckmann) at λex = 488 nm and λem = 525–540 nm, after cell detachment with a 0.05% Trypsin-EDTA and resuspension in PBS upon centrifugation.
Statistical Analysis
The statistical analysis was performed by t-test with Microsoft Excel or OriginLab softwares.
Results and Discussion
Physicochemical Characterization of the Nanovectors
LMNVs were synthesized with a hot sonication procedure, as previously described,17 and functionalized with angiopep-2 owing to the formation of an amide bond between one of the primary amine groups on the peptide and the NHS reactive group at the end of the NHS-PEG-DSPE.
TEM image shows that Ang-LMNVs present a spherical morphology with a diameter distribution centered around 20 ± 5 nm, being only a few particles larger than 100 nm (Figure 1A–C). The black spots in the particles could represent small SPION aggregates encapsulated in the lipid core. However, the uranyl acetate staining could create artifacts, leading to misinterpretation. For this reason, Ang-LMNVs were also analyzed without staining, for comparison. Due to difficulties in identifying lipid nanoparticles without staining, only the large aggregates, probably formed during the drying process, can be observed; nevertheless, it is evident that SPIONs are well encapsulated within Ang-LMNVs (Figure S3). This evidence is supported by the results reported in a previous work.17
Figure 1.
(A, B) Representative TEM images of Ang-LMNVs at different magnifications. (C) Size distribution derived from TEM images. (D) Intensity distribution (%) as a function of the hydrodynamic diameter (nm) for LMNVs (black) and Ang-LMNVs (red). (E) ζ-Potential (mV) distribution of LMNVs (black) and Ang-LMNVs (red). (F) TGA/DTG thermogram of Ang-LMNVs, showing the weight loss (black) and its derivative (blue) at increasing temperatures. (G) SDS-PAGE of angiopep-2, bare LMNVs, and Ang-LMNVs. (H) Fluorescence spectroscopy of angiopep-2 (black) and Ang-LMNVs (red).
Dynamic light scattering measurements (Figure 1D) highlight that the peptide conjugation does not affect the final hydrodynamic size of the nanovectors. In fact, LMNVs have an average hydrodynamic diameter of 169 ± 4 nm with a polydispersity index of 0.24 ± 0.03, whereas Ang-LMNVs have a hydrodynamic diameter of 179 ± 3 nm and a polydispersity index of 0.12 ± 0.01. The small increase in size (around 10 nm), confirmed by the little displacement of the maximum of the size distribution in Figure 1D, could be due to the presence of the peptide that is orientated toward the solvent rather than being adsorbed flat on the surface.
The differences in size detected by DLS and TEM are mainly due to the differences between the two analytical methods. DLS measures the hydrodynamic size—the size of the particle plus its solvation sphere, its polymer shell and the stabilizer—giving rise to larger sizes.25 Moreover, DLS intensity distribution is much more sensitive toward larger aggregates than small ones.26 Finally, some of the small particles observed by TEM could be perceived as agglomerates of hundreds of nanometers in DLS, giving rise to misinterpretation of the particles mean size. Nevertheless, DLS is a valuable quick tool to establish the behavior of nanoparticles in solution.
The ζ-potential of Ang-LMNVs (−32.0 ± 0.6 mV) is only slightly more positive than that one of LMNVs (−39.0 ± 0.8 mV) (Figure 1E). The decrease of the absolute value of ζ-potential is due to the partial screening of LMNV surface charge by the peptide; nevertheless, the ζ-potential is still within the colloidal stability limit (|30| mV),27 preventing the aggregation of Ang-LMNVs. Moreover, the presence of PEG and peptide chains on the particle surface imparts further steric stability to the system.
The stability of Ang-LMNVs in terms of hydrodynamic diameter and polydispersity index (PdI) was evaluated at different pH values, in particular at pH 7.4 to mimic the physiological conditions and at pH 4.5 to simulate the environment of tumor and/or acidic organelles. As shown in Figure S4, the Ang-LMNV diameter and PdI are quite stable until 96 h (the time window used for release studies and AMF chronic treatments), with just a moderate increment in size at pH 4.5 within the first 48 h (from 241 ± 6 nm at 24 h to 330 ± 35 nm at 48 h).
To quantify the fraction of SPIONs composing the nanovectors, TGA was performed on Ang-LMNVs (Figure 1F). The weight loss (in black) and the corresponding derivative weight curve (in blue) show that the total weight of the system decreases as the temperature increases due to the decomposition of the organic components. The weight loss occurs in several steps since the different lipids and polymers have different decomposition temperatures, as evidenced by the derivative thermogravimetry (DTG) curve in Figure 1F. The initial weight loss between 50 and 100 °C can be ascribed to the evaporation of water molecules on the nanovector surface. The first peak in the DTG graph can be attributed to the decomposition of oleic acid as well as to the initial decomposition of lipids, such as GMS and DPPC.28,29 DPPC, in particular, is reported to degrade in differential steps, with a maximum weight loss at around 288 °C (the third peak).29 The same applies for the DSPE component of mPEG-DSPE.30 Therefore, the peaks between 185 and 288 °C can be attributed to the degradation of the lipid component of LMNVs. The final peak at 394 °C is correlated to the decomposition of the PEG component in mPEG-DSPE.31 At the end of the heating program (400 °C), the remaining final mass is that one of the SPIONs since they do no degrade at such temperatures, and it corresponds to a percentage of SPIONs encapsulated in Ang-LMNVs of about 70% of the total weight of the nanovectors.
The success of the peptide conjugation was first assessed by 1H NMR (Figure S5). The functionalization can be monitored by following the disappearance of the peak related to the NHS group on the NHS-PEG-DSPE. The presence of SPIONs in the nanovector would strongly affect the NMR spectra, due to the interferences between SPIONs and magnetic fields:32 for this reason, we decided to perform NMR analysis only on lipid nanovectors (LNVs) without SPIONs. The synthesis and conjugation procedure is exactly the same as for the LMNVs. Since SPIONs are encapsulated inside the core of Ang-LMNVs, there is no reason to believe that their absence would affect the conjugation reaction that takes place only on the nanovector surface. Therefore, we can safely assume that the results obtained with Ang-LNVs are the same for Ang-LMNVs. The spectra in Figure S5 show that LNVs (before functionalization) present the typical peak arising from the 2 protons on the NHS molecule at δ = 2.66 ppm (red curve).33 During the functionalization, the NHS ester-activated PEG-DSPE reacts with the primary amines on the peptide chain to form stable amide bonds, with consequent release of the NHS. Therefore, the loss of the peak at 2.66 ppm from the 1H NMR spectra of Ang-LMNVs suggests that the reaction was successful (Figure S5, black curve).
To quantify the amount of peptide on the nanovectors, we exploited SDS-PAGE with Coomassie staining to highlight the presence of the peptide in the sample (Figure 1G). LMNVs do not give rise to a band, as expected, whereas Ang-LMNVs display a blue band due to the peptide. Comparing the intensity of the band of the plain peptide with those of Ang-LMNVs, the amount of peptide conjugated on Ang-LMNVs was found to be 3.0 ± 0.4 wt %. To validate the SDS-PAGE results, Ang-LMNVs were also analyzed by fluorescence spectroscopy (Figure 1H). The peptide shows an emission with a maximum at around 300 nm, proportional to its concentration, when excited at 260 nm. Fluorescence spectroscopy measurements show that the amount of peptide in Ang-LMNVs is estimated to be around 2.0 ± 0.5 wt %, in line with the SDS-PAGE quantification. Finally, the presence of the peptide on Ang-LMNVs was also confirmed by a BCA assay, and quantified to be about 1.1 ± 0.8 wt %. Combining all data obtained with complementary approaches, and considering a diameter of the nanovector of 20 nm (from TEM), the number of angiopep-2 molecules on each Ang-LMNVs would be around 86–108, with a peptide surface density of 0.07–0.08 ligand/nm2. This value is in the same order of magnitude of other functionalized nanoparticles.18,34
The magnetic properties of LMNVs were already characterized in a previous work;17 in particular, LMNVs possess a saturation magnetization of 25 emu/g, with no coercivity at the working temperature. LMNVs (5.4 mg/mL) were shown to be able to increase the temperature of the medium from 37 to 43 °C in 10 min of stimulation (20 mT and 752.11 kHz).17,18 It has been also shown that the temperature of a 2D culture of GBM cells, pretreated with LMNVs (167 μg/mL) and stimulated for 2 h with an AMF (16 mT and 753 kHz), was able to reach a plateau of 41 °C after 40 min of stimulation.18 The thermal isoeffective dose (TID) corresponding to this kind of treatment is about 5 CEM43, which is the “cumulative equivalent minutes at 43 °C”.35,36 The TID, jointly to the correlated value expressed in CEM43 units, is commonly used to normalize different time–temperature exposure protocols as equivalent exposure time at 43 °C, chosen as a reference temperature; in this way, different thermal treatments can be directly compared.35
Drug Loading and Release Studies
Ang-LMNVs were loaded with the drug nutlin-3a to obtain Nut-Ang-LMNVs, with a drug loading of 0.5 ± 0.1 wt % and encapsulation efficiency of 10.6 ± 0.8%. The nutlin-3a release profile was studied in different conditions: pH 7.4, to simulate physiological conditions; pH 4.5, to recreate the tumor and/or the acidic organelles microenvironments; and pH 7.4 and pH 4.5 with 100 μM H2O2, to mimic a situation of oxidative stress. All of the four conditions were also investigated in the presence of the AMF stimulus (2 h stimulation at an applied magnetic field of 20 mT, using a water-cooled coil of 9 turns and 44 mm inner diameter, and a frequency of 753 kHz). As shown in Figure 2, we have a relatively quick release within the first 24 h, followed by a continuous slower release at longer times, for all of the considered conditions. Figure 2A,B also shows that the release of nutlin-3a is higher in acidic conditions (49 ± 1% of drug released at 96 h), compared to physiological pH values (37 ± 2% at 96 h). This allows predicting that the drug will be more efficiently released within the tumor environment and/or inside acidic organelles, such as lysosomes. The presence of H2O2 does not have a significant effect on the cumulative release of nutlin-3a at pH 7.4, but it seems to slow down the release at pH 4.5. The application of the AMF does not affect the drug release at pH 7.4, whereas it seems to have a slight effect at pH 4.5, where the AMF stimulus increases the amount of nutlin-3a released of about 4% (Figure 2A).
Figure 2.
Cumulative release (%) of nutlin-3a from Nut-Ang-LMNVs in different conditions: (A) pH 4.5 (black circles), pH 4.5 + 100 μM H2O2 (red triangles), pH 4.5 + AMF (green upside down triangles), pH 4.5 + 100 μM H2O2 + AMF (blue diamonds). (B) pH 7.4 (black circles), pH 7.4 + 100 μM H2O2 (red triangles), pH 7.4 + AMF (full green upside down triangles), pH 7.4 + 100 μM H2O2 + AMF (blue diamonds).
Similar release profiles of nutlin-3a were already observed for other lipid-based nanosystems.22 The release mechanism of a drug from nanoparticles depends on several factors, such as the interactions between the drug and the nanoparticle components, the morphology and surface properties of the nanoparticle, the solubility of the drug in different media, and the location of the drug in the nanoparticle.37 For instance, a rapid release at short times could be ascribed to the diffusion of the drug molecules that are localized in the vicinity of the nanoparticle surface, whereas the other molecules in the nanoparticle core diffuse more slowly and at longer times.37 Moreover, the physicochemical properties of the drug, such as its solubility and partition coefficient between lipid and aqueous phase, can play a crucial role in the release kinetics, and accounts for the main differences observed in the release mechanisms from the same nanoparticles.37,38
Nanovector Targeting Efficiency
The uptake of the nanovectors by U87 MG cells was first assessed in standard static conditions. Figure 3A shows that the presence of angiopep-2 on the surface of the nanovectors increases their uptake by 1.6 times with respect to bare LMNVs, as highlighted by flow cytometry analysis, and these results are in good agreement with the confocal microscopy analysis. (Figure 3B).
Figure 3.
(A) Flow cytometry on U87 MG cells after 6 h treatment with LMNVs or Ang-LMNVs (labeled with Vybrant DiO dye) in static conditions (*p < 0.05). (B) Confocal imaging showing the uptake of LMNVs and Ang-LMNVs (in red) by U87 MG cells after 6 h of treatment in static conditions.
The ability of Ang-LMNVs to preferentially target U87 MG cells was further evaluated in dynamic conditions using a bioreactor (Figure S1), where different cell lines were exposed to a cell medium flow loaded with Ang-LMNVs. The cell lines used for this experiment were HA, hCMEC/D3, SH-SY5Yd, and U87 MG cells, as they represent a good in vitro model of the brain cancer microenvironment. The bioreactor was perfused with a 200 μg/mL dispersion of LMNVs or Ang-LMNVs, previously labeled with Vybrant DiO, for 6 h. Confocal acquisitions show an increased uptake of Ang-LMNVs by U87 MG cells with respect to plain LMNVs (46.3 ± 13.8 vs 1.7 ± 1.4% p < 0.05), confirming the results obtained in static conditions (Figure 4A,B). More interestingly, Ang-LMNVs own the evident ability to preferentially target U87 MG cells over the other cell lines, as shown by the confocal acquisitions; conversely, LMNVs are evenly internalized (and at a qualitative lower extent) by all of the four different cultures. These data confirm as angiopep-2 functionalization represents a successful strategy to promote cancer cell active targeting.
Figure 4.

(A) Confocal imaging showing the uptake of LMNVs and Ang-LMNVs (in green) on different cell lines (HA, hCMEC/D3, SH-SY5Yd, U87 MG) after 6 h of treatment in dynamic conditions. (B) Quantitative analysis showing the nanovectors–cell co-localization (*p < 0.05).
In Vitro BBB Crossing Experiments
As already mentioned, angiopep-2 binds to LRP1, which is also expressed on endothelial cells; it was therefore supposed its active role in promoting a receptor-mediated transcytosis that fosters the BBB crossing of the nanovectors.13 The efficiency of Ang-LMNVs to pass through a BBB model was evaluated using an in vitro dynamic bioreactor. In vitro models represent an essential step in the characterization of a new therapeutic approach, in particular in case of the necessity of BBB crossing, as they give important hints about the estimation of the passage of a new therapeutic platform.39 Moreover, a good in vitro model made with human cells can give more accurate information compared to in vivo models relying on tumor xenograft on immunosuppressed mice or in genetically engineered mouse models that lack good reproducibility, especially for brain cancer applications.40
In our model, we exploited a co-culture of hCMEC/D3 cells with human astrocytes, to mimic the BBB, and a co-culture of glioblastoma cells and SH-SY5Yd cells, to mimic the brain cancer microenvironment (Figure S2). The presence of a flow is a key element for obtaining a system successfully recapitulating the physiological conditions, as it has been demonstrated that the typical BBB phenotype, including the development of tight junctions and the expression of proteins like ZO-1, claudin-5, or VE-cadherin, can develop just in the presence of shear stress.39 The lack of this physical cue in static models may lead to a higher permeability, giving rise to an overestimation of drug or nanoparticles crossing the BBB model.39 Moreover, co-culture with astrocytes has also been shown to stimulate the development of a good BBB phenotype and it also favors the expression of transport proteins such as P-gp and Glut-1, which regulate the molecular transport across the BBB.39,41 Endothelial cells usually lose these functions after prolonged in vitro culturing,42 but they can be easily reactivated under flow and when co-cultured with astrocytes.41 It has been also speculated that shear stress may favor the expression of cell membrane-bound receptors, such as cell adhesion molecules and low-density lipoproteins, that activate receptor-mediated transcytosis.24 On the other hand, fluid shear stress may work against nanoparticles avidity, reducing their efficiency to be internalized by endothelial cells.24 These considerations highlight the importance of using dynamic models to study the BBB crossing over more simplistic static models.
Experiments were performed using two bioreactors placed in series: one just with medium in the lower chamber to quantify the nanovector passage and the other seeded with U87 MG cells and neuron-like cells (SH-SY5Yd) in the lower chamber, to assess whether the targeting abilities of Ang-LMNVs are preserved upon BBB crossing. A typical barrier used for these experiments is shown in Figure 5A. The BBB model presents a TEER of 70.05 ± 4.25 Ω·cm2, whereas the permeability to rhodamine B-dextran 70 kDa is around 0.34 × 10–6 cm/s, similarly to other BBB models with hCMEC/D3 cells.43 At the end of the BBB crossing experiment, the concentration of LMNVs in the lower chamber was 104.8 ± 1.5 μg/mL, whereas that one of Ang-LMNVs was about 124.4 ± 4.5 μg/mL (16.7% more with respect to bare LMNVs, p < 0.05).
Figure 5.
(A) Confocal imaging of a typical barrier formed by hCMEC/D3 and HA, with a scheme of the BBB setup. (B) Uptake of Vybrant DiO-labeled LMNVs and Ang-LMNVs (in green) by different cell lines (SH-SY5Yd and U87 MG) after BBB crossing. (C) Quantitative analysis showing the nanovectors–cell co-localization (*p < 0.05).
Upon BBB crossing, both LMNVs and Ang-LMNVs interact with the cells seeded in the lower compartment; however, Ang-LMNVs are selectively internalized by U87 MG cells, with minimal interaction with SH-SY5Yd cells (Figure 5B,C). Moreover, the functionalization increases the internalization extent of the nanovectors in U87 MG cells by almost 20 times with respect to the bare LMNVs (42.7 ± 15.2% of Ang-LMNVs–cell co-localization, as opposed to 2.0 ± 0.3% for LMNVs, p < 0.05). Both Ang-LMNVs and LMNVs are minimally internalized by SH-SY5Yd (2.1 ± 0.9 and 1.6 ± 0.9% of nanovectors–cell co-localization, respectively). This is in good agreement with the targeting efficiency results previously described. These findings not only suggest that Ang-LMNVs are able to favor the passage of nanoparticles through the BBB, but also that the interaction with endothelial cells and astrocytes does not interfere with the targeting abilities of the nanovectors, which is retained upon the BBB crossing. It has been previously shown that ligands targeting the LRP1 receptor on the endothelial cells are able to induce receptor-mediated transcytosis, a process that allows the nanoparticles to be quickly transported into dedicated vesicles from one side to the other side of the cells, without being degraded.44
Given the good results obtained in terms of targeting efficiency and BBB crossing, the following experiments are reported just for Ang-LMNVs.
Ang-LMNV Internalization Pathway
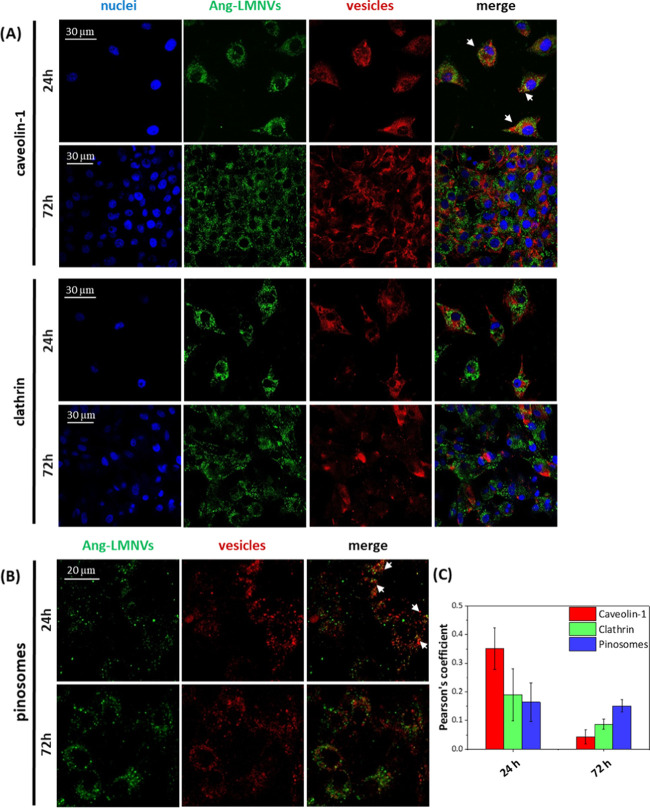
The internalization pathway of Ang-LMNVs in U87 MG cells was studied by investigating markers expressed in caveolae (caveolin-1) and in clathrin-coated vesicles (clathrin) (Figure 6A), and with pinosomes staining with Cascade Blue (Figure 6B). Confocal imaging shows that the nanovectors can enter into the cells through different pathways. The analysis of the Pearson’s correlation coefficients (P, Figure 6C) revealed that after 24 h of incubation, the internalization mediated by caveolae seems to be prevalent with respect to the other pathways (P = 0.35 ± 0.07 for caveolae, P = 0.19 ± 0.09 for clathrin-coated vesicles, and P = 0.16 ± 0.07 for pinosomes). At 72 h, both caveolae and clathrin-coated vesicles mediated internalization decreases, whereas uptake by pinosomes remains constant (P = 0.04 ± 0.02 for caveolae, P = 0.09 ± 0.02 for clathrin-coated vesicles, and P = 0.15 ± 0.02 for pinosomes). Receptor-mediated endocytosis generally occurs via clathrin-coated vesicles and, in some cases, via caveolae.45,46 LRP1 has been shown to be present in clathrin-coated vesicles as well as in caveolae, and, in particular, this receptor seems to be mainly located within the lipid raft microdomains of the cell membrane.47,48 Therefore, LRP1-mediated endocytosis through caveolae or clathrin-coated vesicles takes place only within the first 24 h from the administration of Ang-LMNVs. At longer times, when most of the material has been internalized by the cells, pinocytosis prevails.
Figure 6.
Confocal analysis of the internalization of Vybrant DiO-labeled Ang-LMNVs in U87 MG cells mediated by caveolae and clathrin-coated vesicles (A) or pinosomes (B) at 24 and 72 h. (C) Quantitative evaluation of co-localization (through Pearson’s correlation coefficient) at 24 and 72 h.
To evaluate the intracellular fate of Ang-LMNVs, we analyzed the co-localization with lysosomes of U87 MG cells at different times (Figure 7A,B). At both 24 and 72 h, a fraction of Ang-LMNVs is localized within lysosomes (P = 0.25 ± 0.08 at 24 h and P = 0.26 ± 0.04 at 72 h). The co-localization is partial: although many lysosomes contain Ang-LMNVs (see also zoomed areas (i) and (ii) reported in Figure 7A), several Ang-LMNVs are located also out of the organelles. In this regard, we have to consider that the co-localization analysis has been carried out considering all of the Ang-LMNVs associated with the cells (e.g., also those associated with the plasma membranes), and this explains the relatively low P index. The release studies showed that the nutlin-3a release from the nanovectors is quicker at pH 4.5, which corresponds to the typical pH values inside lysosomes (ca. 4.5–5.0). This pH-dependent behavior allows for a more efficient release of the drug in acidic environment, such as the tumor milieu and lysosomes, with a lower impact on healthy tissues.49,50
Figure 7.
(A) Confocal analysis of the co-localization of Vybrant DiO-labeled Ang-LMNVs (in green) with lysosomes (in red) in U87 MG cells after 24 and 72 h of treatment. (i) and (ii) represent zoomed areas (indicated by the yellow squares) of the merged confocal images at 24 and 72 h of treatment, respectively; the white arrows indicate representative nanovector–lysosome co-localization. (B) Quantitative evaluation of co-localization through Pearson’s correlation coefficient.
Nanovectors as Drug-Delivery Systems
The cytotoxicity of free nutlin-3a, Ang-LMNVs, and Nut-Ang-LMNVs was tested on U87 MG cells at different concentrations and for 24 and 72 h of treatment (Figure 8A). Ang-LMNVs affect U87 MG cells only at the highest tested concentration (1089 μg/mL); nutlin-3a does not have a significant effect at 24 h, whereas, after 72 h, the highest concentration (10 μM) is able to induce a decrease of the cell metabolic activity (down to 27 ± 2%, p < 0.05), in agreement with previous studies.15,51 On the other hand, when treated with Nut-Ang-LMNVs, the viability of U87 MG cells is significantly reduced already at lower concentrations. For instance, at 72 h, the viability of U87 MG cells treated with 325 μg/mL of Nut-Ang-LMNVs (corresponding to 3 μM of encapsulated drug) is reduced to almost 33 ± 3% (p < 0.05), whereas the plain drug at the same concentration has no effect (Figure 8A). For this reason, we chose this concentration as the optimal condition for the treatment of U87 MG cells and for further characterizations concerning the mechanism of action of the nanovectors. The increased efficiency of the drug when loaded into nanoparticles is probably due to the fact that nanoparticle-mediated delivery allows reaching a higher concentration of drug inside the cells, which cannot be obtained by the drug alone due to its very low solubility in aqueous environments.
Figure 8.

(A) U87 MG cell viability after 24 and 72 h of treatment with different concentrations of free nutlin-3a, plain Ang-LMNVs, and Nut-Ang-LMNVs. All of the results were normalized to the respective untreated control (CTRL). *p < 0.05. (B) Cell viability on different cell lines (hCMEC/D3 in red; HA in green; SH-SY5Yd in blue; U87 MG in orange) 72 h after a 6 h treatment in dynamic conditions with 325 μg/mL of Nut-Ang-LMNVs (corresponding to a drug concentration of 3 μM). All of the results were normalized to the corresponding untreated controls (CTRL). *p < 0.05.
Another important point we addressed is the toxicity of nutlin-3a and of Nut-Ang-LMNVs toward the other cell lines. The effect of Nut-Ang-LMNVs was thus tested on four different cell lines (U87 MG, HA, hCMEC/D3, and SH-SY5Yd cells) in dynamic conditions. Cells were perfused with a dispersion of 325 μg/mL of Nut-Ang-LMNVs that corresponds to 3 μM of drug. After 6 h, the cells were rinsed with PBS and left with fresh media for further 72 h to allow the nanovectors to be internalized and the drug to carry out its action. As reported in Figure 8B, the effect of Nut-Ang-LMNVs is significant just for U87 MG cells (cell viability decreased to 52 ± 11% with respect to control cells, p < 0.05). Interestingly, drug-loaded nanovectors did not induce a decrease in cell viability in all of the other cell lines. For comparison, the free drug was tested on hCMEC/D3, HA, and SH-SY5Yd (Figure S6). While the drug has little to no effect on HA and hCMEC/D3, it seems to have an effect on SH-SY5Yd cells (Figure S6), also at a concentration of 3 μM, exploited in the Nut-Ang-LMNV testing; the previously observed absence of cytotoxic effects when the SH-SY5Yd cells are treated with Nut-Ang-LMNVs could be therefore ascribed to a low cellular uptake, a hypothesis corroborated by the previously reported targeting data.
Previous works demonstrated that nutlin-3a and other MDM2 inhibitors are extremely effective on p53 wild-type glioblastoma cell models, with little to no toxicity on several healthy cells. For instance, Secchiero et al. showed that nutlin-3a is able to induce apoptosis in lymphoblastoid SKW6.4 cell lines, but not in endothelial cells, even though an effect on their cycle progression was observed at high drug concentrations.15 These findings are also confirmed by Shangary et al. that compared the effect of an MDM2 inhibitor on normal and cancer cells (CCD-18Co primary normal human colon fibroblasts and RKO colon cancer cell line, respectively).52 The authors observed that the drug was able to induce both cell cycle arrest and cell death in cancer cells, whereas in healthy cells only cell cycle arrest was detected.52 However, this strong sensitivity of cancer cells to MDM2 inhibitor compared to healthy cells is still unclear. Some authors suggested that the overexpression of the MDM2 protein in cancer cells leads to a strong suppression of p53 and makes them extremely vulnerable to p53 accumulation and reactivation as soon as the MDM2 protein is inhibited. On the other hand, normal cells have very low p53 basal levels; therefore, the amount of active p53 reached after MDM2 inhibition is not high enough to induce apopostis.52
Alternating Magnetic Field Stimulation Effects
As suggested by several studies, when magnetic nanoparticles are efficiently internalized in lysosomes, the interaction between these particles and the AMF may be detrimental for the integrity of lysosomes, inducing a phenomenon known as lysosome membrane permeabilization (LMP).20,21 Since Ang-LMNVs are localized in lysosomes, we decided to investigate whether the combination of Ang-LMNVs + AMF would lead to LMP in U87 MG cells and to characterize this phenomenon. U87 MG cells were incubated with Vybrant DiO-labeled Ang-LMNVs (325 μg/mL) and lysosomes stained with LysoTracker Deep Red. The cells were then stimulated for 2 h with an AMF, and the fluorescence emission of LysoTracker Deep Red was monitored in time-lapse imaging: Figure 9A shows representative frames of this time-lapse imaging (Videos S1–S3, Supporting Information). When the control U87 MG cells (nonincubated with nanovectors) undergo AMF stimulation, lysosomes do not experience any visible qualitative modification, and LysoTracker Deep Red is retained within the organelles. On the other hand, when cells are pretreated with Ang-LMNVs, during the AMF stimulation, the LysoTracker Deep Red signal starts to decrease (after about 30 min), and it is completely lost after 90 min. This decrement can suggest that LMP occurs, causing the leakage of the fluorophore outside the organelles.21
Figure 9.
(A) Representative confocal time frames at t = 0, 86, and 120 min from the beginning of the treatment of U87 MG cells stimulated with AMF, incubated with Ang-LMNVs, or stimulated with AMF in the presence of Ang-LMNVs. Vybrant DiO-labeled Ang-LMNVs in green, lysosomes in red, and nuclei in blue. (B) Cathepsin B confocal imaging (in red) in U87 MG cultured upon different treatments. Vybrant DiO-labeled Ang-LMNVs in green.
The exploited tracer is however smaller than the proteolytic enzymes contained in the lysosomal lumen. To verify that damages provoked by Ang-LMNVs + AMF are strong enough to allow lysosomal enzymes to spread into the cytosol, we detected cathepsin B through immunofluorescence, before and after AMF stimulation. As shown in Figure 9B, the decrement of the fluorescence signal correlated to cathepsin B (red) after AMF stimulation clearly indicates that the enzyme diffused outside the lysosomes. In control cultures, when cells are treated with Ang-LMNVs but not stimulated with the AMF, cathepsin B has a strong signal with a good co-localization with nanovectors, indirectly confirming their internalization in lysosomes.
LMP is an important phenomenon involved in different cell death mechanisms, and, for this reason, it has been proposed as a new strategy to induce the initiation of death pathways in resistant cancer cells.53 During LMP, the lysosomal proteases are released into the cytosol, triggering apoptosis or nonapoptotic pathways.54 Cathepsin B and D are identified as the major actors in this process, being able to cleave Bid, a proapoptotic protein of the Bcl-2 family, and starting a cascade of events that lead to the release of cytochrome C from mitochondria and to a caspase-dependent apoptosis mechanism.55 LMP can also represent an important phenomenon in cancer therapy because it favors the release of drugs sequestered within the lysosomal compartment. It has been demonstrated that the concentration of some drugs in lysosomes is relatively high compared to other intracellular locations,50,56 and drug sequestration by lysosomes represents one of the causes of the development of multidrug resistance in certain kind of cancer cells.57 The permeabilization of the lysosomal membrane could counteract this process, eventually favoring the escape of the drug in other cellular compartments where it can play its pharmaceutical role.
The origin of LMP induced by the stimulation of SPIONs with an AMF is still under study, and it is probably caused by different phenomena, such as the SPION rotation or the heat generation in the proximity of the lysosome membrane, that lead to the degradation of the lipids composing the membrane.19−21 However, the incidence of one of the two mechanisms depends on the AMF parameters. It has been previously shown that magnetic nanoparticles start to vibrate or rotate when exposed to alternating magnetic fields of low intensity and frequencies (2–16 Hz);58,59 on the other hand, the interaction between SPIONs and an alternating magnetic field at higher frequencies (in the order of hundreds of kHz) gives rise to an increase of temperature (magnetic hyperthermia).60 It has been also demonstrated that the thermal effect needed to induce a damage does not necessarily imply a global increase in temperature of all of the cells, but it could be limited to a selected area of the cell, such as lysosomes. Previous results by our group demonstrated that LMNVs are able to reach intraparticle temperature as high as 43 °C upon stimulation with an AMF.18 This suggests that the LMP induced by our nanovectors could be mainly driven by a local increment of temperature inside lysosomes.
Hyperthermia treatments are often associated with an alteration of the expression profile of heat shock proteins.61 In particular, the 70 kDa heat shock protein (Hsp70) is a family of ubiquitous chaperon proteins, the expression of which increases when cells are under stress conditions, in particular thermal stress.62 Their role is to protect proteins from unfolding, but they are also able to remodel the protein structure during stress conditions63 to avoid apoptotic and necrotic pathways.64 As demonstrated in many studies, Hsp70 chaperones are active in the cytosol, in the lumen of the endoplasmic reticulum, and in mitochondrial membranes where they help protein translocation across membranes.65 To assess any role in our experimental conditions, we investigated the expression of Hsp70 in U87 MG cells upon treatment with Ang-LMNVs + AMF. The results show that U87 MG cells present a high basal level of Hsp70 (Figure S7). This evidence is supported by several studies that highlight how different types of cancer cells, in particular U87 MG, have high levels of Hsp70 expression.66 The overexpression of Hsp70 may be involved in the cell transformation and in the deregulation of cell death pathways.64 Other works confirmed that heat shock proteins are able to decrease the p53 levels and, as a consequence, deregulate the apoptotic pathway.67 We can see in Figure S7 that cells treated with Ang-LMNVs (with or without AMF) have a similar level of Hsp70 fluorescence signals compared to control cultures. On the other hand, when cells are treated for 1 or 2 h at 42 °C, the increment of fluorescence signal, correlated to an increase of Hsp70 expression, is qualitatively evident. We can thus conclude that the stress induced by the treatment with Ang-LMNVs + AMF does not result in an appreciable alteration of Hsp70 expression. Moreover, since Hsp70 has been shown to have a crucial role in the protection of lysosomes of cancer cells during stress stimuli,68 its unaltered expression could avoid preventing the LMP induction and the following caspase-activated apoptotic pathway. The unaltered Hsp70 levels also confirm that the heat generated upon AMF stimulation gives rise to a localized temperature increment, as previously suggested. Moreover, as already pointed out, the TID corresponding to a similar treatment (2D cell cultures exposed LMNVs + AMF) is about 5 CEM43,18 a value that is probably not high enough to stimulate an evident Hsp70 response. As a comparison, in cells maintained at 42 °C for 2 h, with a TID that is 6 times higher (30 CEM43), the Hsp70 expression resulted evident.
Synergic Therapeutic Effects
The final aim of this work was to investigate the synergic therapeutic effects obtained by combining the use of nutlin-3a with the AMF stimulation of SPIONs encapsulated in the nanovectors. To this aim, U87 MG cells were stimulated with an AMF chronic stimulation protocol (2 h per day for 3 days), as already described.
As shown in Figure 10A, the AMF stimulation has a dramatic effect on the cells treated with Ang-LMNVs and Nut-Ang-LMNVs. Ang-LMNVs affect U87 MG cell viability only when the AMF is applied. Nutlin-3a-loaded nanovectors, instead, are already effective without the AMF stimulation, but the application of the magnetic field decreases the metabolic activity to almost 0.2 ± 1.6% (p < 0.05), meaning that the combination of drug and AMF is extremely efficient in killing cancer cells. Figure 10A also shows that AMF itself does not affect cells in the absence of magnetic nanovectors.
Figure 10.
Cell viability assessment in U87 MG cultures treated with Ang-LMNVs, Nut-Ang-LMNVs, or free nutlin-3a without (− AMF, red bars) and with (+ AMF, green bars) AMF stimulation, (A) at the end of the stimulation and (B) after 3 days from the end of the stimulation (*p < 0.05).
To evaluate a possible recovery of the cells after the chronic stimulation protocol, we performed cell viability assays on the same experimental classes, but 3 days after the last AMF stimulation. The results in Figure 10B confirm that Ang-LMNVs have no effect without AMF stimulation, whereas the cells treated with Ang-LMNVs + AMF are not able to recover when the insult is stopped. Cells treated with Nut-Ang-LMNVs seem to slowly recover (cell metabolic activity increases from 15.0 ± 0.8 to 34.0 ± 3.0% after 3 days); once again, no recovery is observed upon AFM stimulation in the presence of nanovectors.
Immunocytochemistry and confocal analysis were performed to highlight the expression and localization of p53 and Ki-67 markers following AMF + Nut-Ang-LMNV treatment (Figure 11). A high localization of p53 in the nucleus is a hint that cells are undergoing apoptosis. On the other hand, Ki-67 is used as a proliferation marker since it can be found in nuclei during the interphase of the cell cycle; a lack of expression of this marker is detected during the G0 phase, where the cells are quiescent.69 The confocal images and their corresponding analysis to assess p53 and Ki-67 expression are shown in Figure 11. The plain AMF stimulation neither induces the activation of nuclear p53 nor affects cell viability (Figure 11C,D). Plain nutlin-3a has only a minor, non-AMF-dependent, effect in activating the p53 pathway and in reducing cell viability (Figure 11C); however, after 3 days from the end of the treatment, no difference with respect to the control cultures can be found (Figure 11D). The treatment with Ang-LMNVs + AMF results into a higher localization of p53 in the nuclei (66.4 ± 21.9%) and in a lower expression of Ki-67 (18.8 ± 5.9%) compared to nonstimulated cells treated with Ang-LMNVs (4.3 ± 1.6% for p53 and 62.5 ± 8.9% for Ki-67) (Figure 11C). This difference is still maintained after 3 days from the end of the stimulation protocol (Figure 11D). Concerning Nut-Ang-LMNVs, there is no statistically significant difference in the expression of the considered markers between AFM-stimulated and nonstimulated cells right after the end of the treatment (Figure 11C). In both cases, in fact, there is a high localization of p53 in the nuclei (58.1 ± 18.6% without AMF, 80.4 ± 14.2% with AMF) and a low Ki-67 expression (30.4 ± 11.7% without AMF, 16.4 ± 13.9% with AMF) compared to control cells, suggesting that Nut-Ang-LMNVs are able to induce apoptosis and reduce cell viability, even in the absence of AFM stimulation. However, after 3 days from the end of the AMF stimulation (Figure 11D), cultures treated with Nut-Ang-LMNVs + AMF maintain a high level of p53 (61.7 ± 18.6%) and low Ki-67 expression (20.9 ± 10.2%), with respect to cells treated with Nut-Ang-LMNVs, but without AFM stimulation (30.1 ± 2.4% for p53 and 56.0 ± 9.9% for Ki-67). Notably, the p53 co-localization with nuclei 3 days after the end of the chronic stimulation is higher in Nut-Ang-LMNVs + AMF with respect to Ang-LMNVs + AMF (61.7 ± 18.6 and 15.2 ± 10.2%, respectively). This is probably due to the presence of the drug that is exerting its effect on the small number of survived cells. We can thus speculate that the higher p53 expression observed in the Nut-Ang-LMNVs + AMF group 3 days after the end of the treatment could be due to a higher release of nutlin-3a from the nanovectors upon AMF stimulation, as seen in drug release studies, and to a drug escape from lysosomes to cytosol induced by LMP. This result clearly shows the importance of combining chemotherapy and hyperthermia treatment.
Figure 11.
Confocal imaging of p53 (in red) and Ki-67 (in green) expression and of nuclei (in blue) for all of the considered experimental classes (A) just at end of the chronic stimulation and (B) after 3 days from the end of the stimulation. (C, D) Quantitative analysis of p53 and ki-67 localized in the nuclei (*p < 0.05).
Caspase-9 is an enzyme that acts as an initiator of the apoptotic pathway induced by the release of cytochrome C from mitochondria.70 Since this has been proposed as a consequence of lysosomal cysteine proteases leakage after LMP, we studied the activation of caspase-9 in U87 MG cells in the same experimental classes considered for the p53 and Ki-67 localization experiments. Figure 12 summarizes the results obtained by both confocal microscopy and flow cytometry. The presence of the activated enzyme in the cells treated with Ang-LMNVs or Nut-Ang-LMNVs under stimulation with AMF is highlighted by the green fluorescence of FITC-Val-Ala-Asp-fluoro-methyl-rerone (FITC-VAD-FMK) that interacts only with activated caspase-9 in apoptotic cells (confocal acquisitions, insets in Figure 12A). This signal is absent in control cells (+/– AMF), in cells treated with nutlin-3a (+/– AMF), and in cells treated with Ang-LMNVs and Nut-Ang-LMNVs without AMF stimulation. These results are also confirmed by flow cytometry (Figure 12A, and relative quantitative analysis in 12B). The treatment with Ang-LMNVs or Nut-Ang-LMNVs coupled with AMF leads to a higher level of FITC-positive cells, due to activated caspase-9 (19.9 ± 0.9% for Ang-LMNVs and 22.7 ± 2.1% for Nut-Ang-LMNVs), with respect to control cells (3.1 ± 0.8%) and to cells treated with nanovectors without AMF (6.3 ± 1.4% for Ang-LMNVs and 9.4 ± 3.4% for Nut-Ang-LMNVs). The free drug has just a minimal effect on the activation of caspase-9 (6.7 ± 2.4% for nutlin-3a, 4.8 ± 0.5% for nutlin-3a + AMF). These evidences suggest that the apoptosis induction through this pathway is mainly due to the response of the magnetic component subjected to the AMF.
Figure 12.
(A) Activation of caspase-9 induced by AMF treatment combined with Ang-LMNVs or Nut-Ang-LMNVs administration to U87 MG cells analyzed by flow cytometry and confocal microscopy (insets). The green spots correspond to activated caspase-9. (B) Quantitative analysis of FITC-positive cells (%), corresponding to activated caspase-9, derived from flow cytometry (*p < 0.05).
The hyperthermia-induced LMP fosters the leakage of proteolytic enzymes from the lysosomes and, as reported in several works,54,55 induces cytochrome C release from mitochondria, with consequent activation of the caspase-9 apoptotic pathway. The treatment with Nut-Ang-LMNVs without AMF stimulus, instead, only results into the reactivation of the p53 pathway, mediated by the interaction between nutlin-3a and the MDM2 protein. As shown by these results, the treatment with Ang-LMNVs + AMF is able to activate both the caspase-9 and p53 pathways; however, Nut-Ang-LMNVs + AMF are able to maintain the p53 apoptotic effect also several days after the end of the stimulation, thanks to the presence of the drug. This highlights the importance of combining these two therapeutic approaches with the aim of completely suppressing cancer cell viability and their recurrence after the treatment.
Conclusions
In this work, we elucidated the mechanism of action of a lipid magnetic nanovector loaded with a chemotherapeutic drug, due to the synergic effect of nutlin-3a, a chemotherapy agent with proved proapoptotic actions, and hyperthermia. In particular, collected data show that Nut-Ang-LMNVs + AMF stimulation induces localized intracellular effects that result in a lysosomal damage. This leads, as a consequence, to a proteolytic enzyme leakage that initiates several apoptotic pathways and enhances the effects of the drug. The synergic therapeutic approach makes the proposed nanovectors a versatile platform to treat heterogeneous cancers like glioblastoma multiforme. Finally, we demonstrated that the functionalization with angiopep-2 is an excellent “dual-targeting” strategy, allowing crossing of a complex BBB in vitro model and high and specific accumulation in cancer cells.
Acknowledgments
This work received funding from the European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation program (grant agreement no. 709613, SLaMM). The authors also acknowledge Dr. Matteo Battaglini, Dr. Christos Tapeinos, and Dr. Giammarino Pugliese (Istituto Italiano di Tecnologia, Italy) for their support in confocal microscopy, nanovector fabrication, and TGA measurements, respectively.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsami.0c05556.
Schematic illustrations of the setup used for the targeting efficiency and selective apoptosis experiments; schematic illustrations of the setup used for BBB crossing experiments; TEM images of Ang-LMNVs without staining; midterm stability of nanovectors; 1H NMR spectra of nanovectors before and after their functionalization with angiopep-2; cell viability assays of free nutlin-3a on hCEMC/D3, HA, and SH-SY5Yd cells; confocal microscopy of Hsp70 immunofluorescence (PDF)
Three videos of time-lapse confocal microscopy of U87 MG cells (LysoTracker Deep Red) + AMF, U87 MG cells (LysoTracker Deep Red) + Ang-LMNVs, and U87 MG cells (LysoTracker Deep Red) + Ang-LMNVs + AMF (Videos S1–S3) (AVI) (AVI) (AVI)
Author Contributions
∥ C.P. and D.D.P. contributed equally to this work.
Author Contributions
The manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript.
The authors declare no competing financial interest.
Supplementary Material
References
- Xu H.; Chen J.; Xu H.; Qin Z. Geographic Variations in the Incidence of Glioblastoma and Prognostic Factors Predictive of Overall Survival in US Adults from 2004–2013. Front. Aging Neurosci. 2017, 9, 1061. 10.3389/fnagi.2017.00352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson J. R. D.; Regad T. Targeting Cellular Pathways in Glioblastoma Multiforme. Signal Transduction Targeted Ther. 2017, 2, 17040 10.1038/sigtrans.2017.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii N.; Tada M.; Hamou M. F.; Janzer R. C.; Meagher-Villemure K.; Wiestler O. D.; De Tribolet N.; Van Meir E. G. Cells with TP53 Mutations in Low Grade Astrocytic Tumors Evolve Clonally to Malignancy and Are an Unfavorable Prognostic Factor. Oncogene 1999, 18, 5870. 10.1038/sj.onc.1203241. [DOI] [PubMed] [Google Scholar]
- Pardridge W. M. Drug Targeting to the Brain. Pharm. Res. 2007, 24, 1733. 10.1007/s11095-007-9324-2. [DOI] [PubMed] [Google Scholar]
- Gao H.; Pang Z.; Jiang X. Targeted Delivery of Nano-Therapeutics for Major Disorders of the Central Nervous System. Pharm. Res. 2013, 30, 2485. 10.1007/s11095-013-1122-4. [DOI] [PubMed] [Google Scholar]
- Gao H. Progress and Perspectives on Targeting Nanoparticles for Brain Drug Delivery. Acta Pharm. Sin. B 2016, 6, 268–286. 10.1016/j.apsb.2016.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinelli C.; Pucci C.; Ciofani G. Nanostructured Carriers as Innovative Tools for Cancer Diagnosis and Therapy. APL Bioeng. 2019, 3, 011502 10.1063/1.5079943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumura Y.; Maeda H. A New Concept for Macromolecular Therapeutics in Cancer Chemotherapy: Mechanism of Tumoritropic Accumulation of Proteins and the Antitumor Agent Smancs. Cancer Res. 1986, 46, 6387–6392. [PubMed] [Google Scholar]
- Barua S.; Mitragotri S. Challenges Associated with Penetration of Nanoparticles across Cell and Tissue Barriers: A Review of Current Status and Future Prospects. Nano Today 2014, 9, 223. 10.1016/j.nantod.2014.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maletínská L.; Blakely E. A.; Bjornstad K. A.; Deen D. F.; Knoff L. J.; Forte T. M. Human Glioblastoma Cell Lines: Levels of Low-Density Lipoprotein Receptor and Low-Density Lipoprotein Receptor-Related Protein. Cancer Res. 2000, 60, 2300. [PubMed] [Google Scholar]
- Demeule M.; Regina A.; Ché C.; Poirier J.; Nguyen T.; Gabathuler R.; Castaigne J. P.; Béliveau R. Identification and Design of Peptides as a New Drug Delivery System for the Brain. J. Pharmacol. Exp. Ther. 2008, 324, 1064. 10.1124/jpet.107.131318. [DOI] [PubMed] [Google Scholar]
- Sakamoto K.; Shinohara T.; Adachi Y.; Asami T.; Ohtaki T. A Novel LRP1-Binding Peptide L57 That Crosses the Blood Brain Barrier. Biochem. Biophys. Rep. 2017, 12, 135–139. 10.1016/j.bbrep.2017.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demeule M.; Currie J. C.; Bertrand Y.; Ché C.; Nguyen T.; Régina A.; Gabathuler R.; Castaigne J. P.; Béliveau R. Involvement of the Low-Density Lipoprotein Receptor-Related Protein in the Transcytosis of the Brain Delivery Vector Angiopep-2. J. Neurochem. 2008, 106, 1534–1544. 10.1111/j.1471-4159.2008.05492.x. [DOI] [PubMed] [Google Scholar]
- Chène P. Inhibiting the P53-MDM2 Interaction: An Important Target for Cancer Therapy. Nat. Rev. Cancer 2003, 3, 102–109. 10.1038/nrc991. [DOI] [PubMed] [Google Scholar]
- Secchiero P.; Corallini F.; Gonelli A.; Dell’Eva R.; Vitale M.; Capitani S.; Albini A.; Zauli G. Antiangiogenic Activity of the MDM2 Antagonist Nutlin-3. Circ. Res. 2007, 100, 61–69. 10.1161/01.RES.0000253975.76198.ff. [DOI] [PubMed] [Google Scholar]
- Brennan C. W.; Verhaak R. G. W.; McKenna A.; Campos B.; Noushmehr H.; Salama S. R.; Zheng S.; Chakravarty D.; Sanborn J. Z.; Berman S. H.; Beroukhim R.; Bernard B.; Wu C. J.; Genovese G.; Shmulevich I.; Barnholtz-Sloan J.; Zou L.; Vegesna R.; Shukla S. A.; Ciriello G.; Yung W. K.; Zhang W.; Sougnez C.; Mikkelsen T.; Aldape K.; Bigner D. D.; Van Meir E. G.; Prados M.; Sloan A.; Black K. L.; Eschbacher J.; Finocchiaro G.; Friedman W.; Andrews D. W.; Guha A.; Iacocca M.; O’Neill B. P.; Foltz G.; Myers J.; Weisenberger D. J.; Penny R.; Kucherlapati R.; Perou C. M.; Hayes D. N.; Gibbs R.; Marra M.; Mills G. B.; Lander E. S.; Spellman P.; Wilson R.; Sander C.; Weinstein J.; Meyerson M.; Gabriel S.; Laird P. W.; Haussler D.; Getz G.; Chin L.; Benz C.; Barrett W.; Ostrom Q.; Wolinsky Y.; Bose B.; Boulos P. T.; Boulos M.; Brown J.; Czerinski C.; Eppley M.; Kempista T.; Kitko T.; Koyfman Y.; Rabeno B.; Rastogi P.; Sugarman M.; Swanson P.; Yalamanchii K.; Otey I. P.; Liu Y. S.; Xiao Y.; Auman J. T.; Chen P. C.; Hadjipanayis A.; Lee E.; Lee S.; Park P. J.; Seidman J.; Yang L.; Kalkanis S.; Poisson L. M.; Raghunathan A.; Scarpace L.; Bressler R.; Eakin A.; Iype L.; Kreisberg R. B.; Leinonen K.; Reynolds S.; Rovira H.; Thorsson V.; Annala M. J.; Paulauskis J.; Curley E.; Hatfield M.; Mallery D.; Morris S.; Shelton T.; Shelton C.; Sherman M.; Yena P.; Cuppini L.; DiMeco F.; Eoli M.; Maderna E.; Pollo B.; Saini M.; Balu S.; Hoadley K. A.; Li L.; Miller C. R.; Shi Y.; Topal M. D.; Wu J.; Dunn G.; Giannini C.; Aksoy B. A.; Antipin Y.; Borsu L.; Cerami E.; Gao J.; Gross B.; Jacobsen A.; Lash A.; Liang Y.; Reva B.; Schultz N.; Shen R.; Socci N. D.; Viale A.; Ferguson M. L.; Chen Q. R.; Demchok J. A.; Dillon L. A. L.; Mills Shaw K. R.; Sheth M.; Tarnuzzer R.; Wang Z.; Yang L.; Davidsen T.; Guyer M. S.; Ozenberger B. A.; Sofia H. J.; Bergsten J.; Eckman J.; Harr J.; Smith C.; Tucker K.; Winemiller C.; Zach L.; Anne Ljubimova J. Y.; Eley G.; Ayala B.; Jensen M. A.; Kahn A.; Pihl T. D.; Pot D. A.; Wan Y.; Hansen N.; Hothi P.; Lin B.; Shah N.; Yoon J. G.; Lau C.; Berens M.; Ardlie K.; Carter S. L.; Cherniack A. D.; Noble M.; Cho J.; Cibulskis K.; DiCara D.; Frazer S.; Gabriel S. B.; Gehlenborg N.; Gentry J.; Heiman D.; Kim J.; Jing R.; Lawrence M.; Lin P.; Mallard W.; Onofrio R. C.; Saksena G.; Schumacher S.; Stojanov P.; Tabak B.; Voet D.; Zhang H.; Dees N. N.; Ding L.; Fulton L. L.; Fulton R. S.; Kanchi K. L.; Mardis E. R.; Wilson R. K.; Baylin S. B.; Harshyne L.; Cohen M. L.; Devine K.; Sloan A. E.; Van Den Berg S. R.; Berger M. S.; Carlin D.; Craft B.; Ellrott K.; Goldman M.; Goldstein T.; Grifford M.; Ma S.; Ng S.; Stuart J.; Swatloski T.; Waltman P.; Zhu J.; Foss R.; Frentzen B.; McTiernan R.; Yachnis A.; Mao Y.; Akbani R.; Bogler O.; Fuller G. N.; Liu W.; Liu Y.; Lu Y.; Protopopov A.; Ren X.; Sun Y.; Yung W. K. A.; Zhang J.; Chen K.; Weinstein J. N.; Bootwalla M. S.; Lai P. H.; Triche T. J.; Van Den Berg D. J.; Gutmann D. H.; Lehman N. L.; Brat D.; Olson J. J.; Mastrogianakis G. M.; Devi N. S.; Zhang Z.; Lipp E.; McLendon R. The Somatic Genomic Landscape of Glioblastoma. Cell 2013, 155, 462. 10.1016/j.cell.2013.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tapeinos C.; Marino A.; Battaglini M.; Migliorin S.; Brescia R.; Scarpellini A.; De Julián Fernández C.; Prato M.; Drago F.; Ciofani G. Stimuli-Responsive Lipid-Based Magnetic Nanovectors Increase Apoptosis in Glioblastoma Cells through Synergic Intracellular Hyperthermia and Chemotherapy. Nanoscale 2019, 11, 72–88. 10.1039/C8NR05520C. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marino A.; Camponovo A.; Innocenti A. D.; Bartolucci M.; Tapeinos C.; Martinelli C.; Pasquale D. De.; Santoro F.; Mollo V.; Arai S.; Suzuki M.; Harada Y.; et al. Multifunctional Temozolomide-Loaded Lipid Superparamagnetic Nanovectors: Dual Targeting and Disintegration of Glioblastoma Spheroids by Synergic Chemotherapy and Hyperthermia Treatment. Nanoscale 2019, 11, 21227–21248. 10.1039/C9NR07976A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan R. P.; Carrey J.; Respaud M. Magnetic Hyperthermia Properties of Nanoparticles inside Lysosomes Using Kinetic Monte Carlo Simulations: Influence of Key Parameters and Dipolar Interactions, and Evidence for Strong Spatial Variation of Heating Power. Phys. Rev. B: Condens. Matter Mater. Phys. 2014, 90, 1–12. 10.1103/PhysRevB.90.214421. [DOI] [Google Scholar]
- Domenech M.; Marrero-Berrios I.; Torres-Lugo M.; Rinaldi C. Lysosomal Membrane Permeabilization by Targeted Magnetic Nanoparticles in Alternating Magnetic Fields. ACS Nano 2013, 7, 5091–5101. 10.1021/nn4007048. [DOI] [PubMed] [Google Scholar]
- Sanchez C.; El Hajj Diab D.; Connord V.; Clerc P.; Meunier E.; Pipy B.; Payré B.; Tan R. P.; Gougeon M.; Carrey J.; Gigoux V.; Fourmy D. Targeting a G-Protein-Coupled Receptor Overexpressed in Endocrine Tumors by Magnetic Nanoparticles to Induce Cell Death. ACS Nano 2014, 8, 1350–1363. 10.1021/nn404954s. [DOI] [PubMed] [Google Scholar]
- Grillone A.; Battaglini M.; Moscato S.; Mattii L.; De Julián Fernández C.; Scarpellini A.; Giorgi M.; Sinibaldi E.; Ciofani G. Nutlin-Loaded Magnetic Solid Lipid Nanoparticles for Targeted Glioblastoma Treatment. Nanomedicine 2019, 14, 727–752. 10.2217/nnm-2018-0436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong A. D.; Ye M.; Levy A. F.; Rothstein J. D.; Bergles D. E.; Searson P. C. The Blood-Brain Barrier: An Engineering Perspective. Front. Neuroeng. 2013, 6, 1–22. 10.3389/fneng.2013.00007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papademetriou I.; Vedula E.; Charest J.; Porter T. Effect of Flow on Targeting and Penetration of Angiopep-Decorated Nanoparticles in a Microfluidic Model Blood-Brain Barrier. PLoS One 2018, 13, e0205158 10.1371/journal.pone.0205158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maguire C. M.; Rösslein M.; Wick P.; Prina-Mello A. Characterisation of Particles in Solution–a Perspective on Light Scattering and Comparative Technologies. Sci. Technol. Adv. Mater. 2018, 19, 732–745. 10.1080/14686996.2018.1517587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stetefeld J.; McKenna S. A.; Patel T. R. Dynamic Light Scattering: A Practical Guide and Applications in Biomedical Sciences. Biophys. Rev. 2016, 8, 409–427. 10.1007/s12551-016-0218-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A.; Dixit C. K. Methods for Characterization of Nanoparticles. Adv. Nanomed. Delivery Ther. Nucleic Acids 2017, 43–58. 10.1016/B978-0-08-100557-6.00003-1. [DOI] [Google Scholar]
- Tomassetti M.; Favero G.; Campanella L. Kinetic Thermal Analytical Study of Saturated Mono-, Di- and Tri-Glycerides. J. Therm. Anal. Calorim. 2013, 112, 519–527. 10.1007/s10973-012-2821-5. [DOI] [Google Scholar]
- Troutier A. L.; Véron L.; Delair T.; Pichot C.; Ladavière C. New Insights into Self-Organization of a Model Lipid Mixture and Quantification of Its Adsorption on Spherical Polymer Particles. Langmuir 2005, 21, 9901–9910. 10.1021/la050796l. [DOI] [PubMed] [Google Scholar]
- He P.; Zhu X. Synthesis and Characterization of Phospholipid-Modified Multiwalled Carbon Nanotubes. Mater. Res. Bull. 2008, 43, 141–148. 10.1016/j.materresbull.2007.02.024. [DOI] [Google Scholar]
- Qian T.; Li J.; Feng W.; Nian H. Enhanced Thermal Conductivity of Form-Stable Phase Change Composite with Single-Walled Carbon Nanotubes for Thermal Energy Storage. Sci. Rep. 2017, 7, 44710 10.1038/srep44710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polito L.; Colombo M.; Monti D.; Melato S.; Caneva E.; Prosperi D. Resolving the Structure of Ligands Bound to the Surface of Superparamagnetic Iron Oxide Nanoparticles by High-Resolution Magic-Angle Spinning NMR Spectroscopy. J. Am. Chem. Soc. 2008, 130, 12712–12724. 10.1021/ja802479n. [DOI] [PubMed] [Google Scholar]
- Salva R.; Le Meins J. F.; Sandre O.; Bruîlet A.; Schmutz M.; Guenoun P.; Lecommandoux S. Polymersome Shape Transformation at the Nanoscale. ACS Nano 2013, 7, 9298–9311. 10.1021/nn4039589. [DOI] [PubMed] [Google Scholar]
- Alkilany A. M.; Zhu L.; Weller H.; Mews A.; Parak W. J.; Barz M.; Feliu N. Ligand Density on Nanoparticles: A Parameter with Critical Impact on Nanomedicine. Adv. Drug Delivery Rev. 2019, 143, 22–36. 10.1016/j.addr.2019.05.010. [DOI] [PubMed] [Google Scholar]
- Van Rhoon G. C. Is CEM43 Still a Relevant Thermal Dose Parameter for Hyperthermia Treatment Monitoring?. Int. J. Hyperthermia 2016, 32, 50–62. 10.3109/02656736.2015.1114153. [DOI] [PubMed] [Google Scholar]
- Mouratidis P. X. E.; Rivens I.; Civale J.; Symonds-Tayler R.; ter Haar G. Relationship between Thermal Dose and Cell Death for “Rapid” Ablative and “Slow” Hyperthermic Heating. Int. J. Hyperthermia 2019, 36, 228–243. 10.1080/02656736.2018.1558289. [DOI] [PubMed] [Google Scholar]
- Huang X.; Brazel C. S. On the Importance and Mechanisms of Burst Release in Matrix-Controlled Drug Delivery Systems. J. Controlled Release 2001, 73, 121–136. 10.1016/S0168-3659(01)00248-6. [DOI] [PubMed] [Google Scholar]
- Narasimhan B.; Langer R. Zero-Order Release of Micro- and Macromolecules from Polymeric Devices: The Role of the Burst Effect. J. Controlled Release 1997, 47, 13–20. 10.1016/S0168-3659(96)01611-2. [DOI] [Google Scholar]
- Xu H.; Li Z.; Yu Y.; Sizdahkhani S.; Ho W. S.; Yin F.; Wang L.; Zhu G.; Zhang M.; Jiang L.; Zhuang Z.; Qin J. A Dynamic in Vivo-like Organotypic Blood-Brain Barrier Model to Probe Metastatic Brain Tumors. Sci. Rep. 2016, 6, 36670 10.1038/srep36670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fomchenko E. I.; Holland E. C. Mouse Models of Brain Tumors and Their Applications in Preclinical Trials. Clin. Cancer Res. 2006, 12, 5288–5297. 10.1158/1078-0432.CCR-06-0438. [DOI] [PubMed] [Google Scholar]
- Santaguida S.; Janigro D.; Hossain M.; Oby E.; Rapp E.; Cucullo L. Side by Side Comparison between Dynamic versus Static Models of Blood-Brain Barrier in Vitro: A Permeability Study. Brain Res. 2006, 1109, 1–13. 10.1016/j.brainres.2006.06.027. [DOI] [PubMed] [Google Scholar]
- Ballermann B. J.; Ott M. J. Adhesion and Differentiation of Endothelial Cells by Exposure to Chronic Shear Stress: A Vascular Graft Model. Blood Purif. 1995, 13, 125–134. 10.1159/000170195. [DOI] [PubMed] [Google Scholar]
- Helms H. C.; Abbott N. J.; Burek M.; Cecchelli R.; Couraud P.; Deli M. A.; Galla H. J.; Romero I. A.; Shusta E. V.; Stebbins M. J.; et al. In Vitro Models of the Blood – Brain Barrier: An Overview of Commonly Used Brain Endothelial Cell Culture Models and Guidelines for Their Use. J. Cereb. Blood Flow Metab. 2016, 36, 862–890. 10.1177/0271678X16630991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian X.; Nyberg S.; Sharp P. S.; Madsen J.; Daneshpour N.; Armes S. P.; Berwick J.; Azzouz M.; Shaw P.; Abbott N. J.; Battaglia G. LRP-1-Mediated Intracellular Antibody Delivery to the Central Nervous System. Sci. Rep. 2015, 5, 11990 10.1038/srep11990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nabi I. R.; Le P. U. Caveolae/Raft-Dependent Endocytosis. J. Cell Biol. 2003, 161, 673–677. 10.1083/jcb.200302028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Sayed A.; Harashima H. Endocytosis of Gene Delivery Vectors: From Clathrin-Dependent to Lipid Raft-Mediated Endocytosis. Mol. Ther. 2013, 21, 1118–1130. 10.1038/mt.2013.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H.; Links P. H.; Ngsee J. K.; Tran K.; Cui Z.; Ko K. W. S.; Yao Z. Localization of Low Density Lipoprotein Receptor-Related Protein 1 to Caveolae in 3T3-L1 Adipocytes in Response to Insulin Treatment. J. Biol. Chem. 2004, 279, 2221–2230. 10.1074/jbc.M310679200. [DOI] [PubMed] [Google Scholar]
- Takayama Y.; May P.; Anderson R. G. W.; Herz J. Low Density Lipoprotein Receptor-Related Protein 1 (LRP1) Controls Endocytosis and c-CBL-Mediated Ubiquitination of the Platelet-Derived Growth Factor Receptor β (PDGFRβ). J. Biol. Chem. 2005, 280, 18504–18510. 10.1074/jbc.M410265200. [DOI] [PubMed] [Google Scholar]
- Kato Y.; Ozawa S.; Miyamoto C.; Maehata Y.; Suzuki A.; Maeda T.; Baba Y. Acidic Extracellular Microenvironment and Cancer. Cancer Cell Int. 2013, 13, 89 10.1186/1475-2867-13-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapp S.; Rosania G. R.; Horobin R. W.; Kornhuber J. Quantitative Modeling of Selective Lysosomal Targeting for Drug Design. Eur. Biophys. J. 2008, 37, 1317–1328. 10.1007/s00249-008-0338-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villalonga-Planells R.; Coll-Mulet L.; Martínez-Soler F.; Castaño E.; Acebes J. J.; Giménez-Bonafé P.; Gil J.; Tortosa A. Activation of P53 by Nutlin-3a Induces Apoptosis and Cellular Senescence in Human Glioblastoma Multiforme. PLoS One 2011, 6, e18588 10.1371/journal.pone.0018588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shangary S.; Ding K.; Qiu S.; Nikolovska-coleska Z.; Bauer J. A.; Liu M.; Wang G.; Lu Y.; Mceachern D.; Bradford C. R.; Bernard D. Reactivation of P53 by a Specific MDM2 Antagonist (MI-43) Leads to P21-Mediated Cell Cycle Arrest and Selective Cell Death in Colon Cancer. Mol. Cancer Ther. 2008, 7, 1533–1542. 10.1158/1535-7163.MCT-08-0140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kallunki T.; Olsen O. D.; Jäättelä M. Cancer-Associated Lysosomal Changes: Friends or Foes?. Oncogene 2013, 32, 1995–2004. 10.1038/onc.2012.292. [DOI] [PubMed] [Google Scholar]
- Boya P.; Kroemer G. Lysosomal Membrane Permeabilization in Cell Death. Oncogene 2008, 27, 6434–6451. 10.1038/onc.2008.310. [DOI] [PubMed] [Google Scholar]
- De Castro M. A. G.; Bunt G.; Wouters F. S. Cathepsin B Launches an Apoptotic Exit Effort upon Cell Death- Associated Disruption of Lysosomes. Cell Death Discovery 2016, 2, 1–8. 10.1038/cddiscovery.2016.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornhuber J.; Henkel A. W.; Groemer T. W.; Sta S.; Welzel O.; Tripal P.; Rotter A.; Bleich S.; Trapp S. Lipophilic Cationic Drugs Increase the Permeability of Lysosomal Membranes in a Cell Culture System. J. Cell Physiol. 2010, 224, 152–164. 10.1002/jcp.22112. [DOI] [PubMed] [Google Scholar]
- Chen V. Y.; Rosania G. R. The Great Multidrug-Resistance Paradox. ACS Chem. Biol. 2006, 1, 271–273. 10.1021/cb600215q. [DOI] [PubMed] [Google Scholar]
- Shen Y.; Wu C.; Uyeda T. Q. P.; Plaza G. R.; Liu B.; Han Y.; Lesniak M. S.; Cheng Y. Elongated Nanoparticle Aggregates in Cancer Cells for Mechanical Destruction with Low Frequency Rotating Magnetic Field. Theranostics 2017, 7, 1735–1748. 10.7150/thno.18352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spyridopoulou K.; Makridis A.; Maniotis N.; Karypidou N.; Myrovali E.; Samaras T.; Angelakeris M.; Chlichlia K.; Kalogirou O. Effect of Low Frequency Magnetic Fields on the Growth of MNP-Treated HT29 Colon Cancer Cells. Nanotechnology 2018, 29, 175101 10.1088/1361-6528/aaaea9. [DOI] [PubMed] [Google Scholar]
- Cazares-Cortes E.; Cabana S.; Boitard C.; Nehlig E.; Griffete N.; Fresnais J.; Wilhelm C.; Abou-Hassan A.; Ménager C. Recent Insights in Magnetic Hyperthermia: From the “Hot-Spot” Effect for Local Delivery to Combined Magneto-Photo-Thermia Using Magneto-Plasmonic Hybrids. Adv. Drug Delivery Rev. 2019, 138, 233–246. 10.1016/j.addr.2018.10.016. [DOI] [PubMed] [Google Scholar]
- Lin F. C.; Hsu C. H.; Lin Y. Y. Nano-Therapeutic Cancer Immunotherapy Using Hyperthermia-Induced Heat Shock Proteins: Insights from Mathematical Modeling. Int. J. Nanomed. 2018, 13, 3529–3539. 10.2147/IJN.S166000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavaria M.; Gabriele T.; Kola I.; Anderson R. L. A Hitchhiker’s Guide to the Human Hsp70 Family. Cell Stress Chaperones 1996, 1, 23–28. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenzweig R.; Nillegoda N. B.; Mayer M. P.; Bukau B. The Hsp70 Chaperone Network. Nat. Rev. Mol. Cell Biol. 2019, 20, 665–680. 10.1038/s41580-019-0133-3. [DOI] [PubMed] [Google Scholar]
- Takayama S.; Reed J. C.; Homma S. Heat-Shock Proteins as Regulators of Apoptosis. Oncogene 2003, 22, 9041–9047. 10.1038/sj.onc.1207114. [DOI] [PubMed] [Google Scholar]
- Craig E. A. Hsp70 at the Membrane: Driving Protein Translocation. BMC Biol. 2018, 16, 11. 10.1186/s12915-017-0474-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorsteinsdottir J.; Stangl S.; Fu P.; Guo K.; Albrecht V.; Eigenbrod S.; Erl J.; Gehrmann M.; Tonn J. C.; Multhoff G.; Schichor C. Overexpression of Cytosolic, Plasma Membrane Bound and Extracellular Heat Shock Protein 70 (Hsp70) in Primary Glioblastomas. J. Neurooncol. 2017, 135, 443–452. 10.1007/s11060-017-2600-z. [DOI] [PubMed] [Google Scholar]
- Calderwood S. K.; Khaleque M. A.; Sawyer D. B.; Ciocca D. R. Heat Shock Proteins in Cancer: Chaperones of Tumorigenesis. Trends Biochem. Sci. 2006, 31, 164–172. 10.1016/j.tibs.2006.01.006. [DOI] [PubMed] [Google Scholar]
- Daugaard M.; Kirkegaard-Sørensen T.; Ostenfeld M. S.; Aaboe M.; Høyer-Hansen M.; Ørntoft T. F.; Rohde M.; Jäättelä M. Lens Epithelium-Derived Growth Factor Is an Hsp70-2 Regulated Guardian of Lysosomal Stability in Human Cancer. Cancer Res. 2007, 67, 2559–2567. 10.1158/0008-5472.CAN-06-4121. [DOI] [PubMed] [Google Scholar]
- Gerdes J.; Lemke H.; Baisch H.; Wacker H. H.; Schwab U.; Stein H. Cell Cycle Analysis of a Cell Proliferation-Associated Human Nuclear Antigen Defined by the Monoclonal Antibody Ki-67. J. Immunol. 1984, 1710–1715. [PubMed] [Google Scholar]
- Reiners J. J. Jr.; Caruso J. A.; Mathieu P.; Chelladurai B.; Yin X. M.; Kessel D. Release of Cytochrome c and Activation of Pro-Caspase-9 Following Lysosomal Photodamage Involves Bid Cleavage. Cell Death Differ. 2002, 9, 934–944. 10.1038/sj.cdd.4401048. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.