SUMMARY
BCL-2 family proteins converge at the mitochondrial outer membrane to regulate apoptosis and maintain the critical balance between cellular life and death. This physiologic process is essential to organism homeostasis and relies on protein-protein and protein-lipid interactions among BCL-2 family proteins in the mitochondrial lipid environment. Here, we find that trans-2-hexadecenal (t-2-hex), previously implicated in regulating BAX-mediated apoptosis, does so by direct covalent reaction with C126, which is located on the surface of BAX at the junction of its α5/α6 core hydrophobic hairpin. The application of nuclear magnetic resonance spectroscopy, hydrogen-deuterium exchange mass spectrometry, specialized t-2-hex-containing liposomes, and BAX mutational studies in mitochondria and cells reveals structure-function insights into the mechanism by which covalent lipidation at the mitochondria sensitizes direct BAX activation. The functional role of BAX lipidation as a control point of mitochondrial apoptosis could provide a therapeutic strategy for BAX modulation by chemical modification of C126.
In Brief
Cohen et al. show that trans-2-hexadecenal (t-2-hex) potentiates BAX-mediated apoptosis by non-enzymatic covalent lipidation of BAX C126. t-2-hex derivatization induces BAX-activating conformational changes and enhances BH3-triggered BAX poration of liposomal and mitochondrial membranes in a C126-dependent manner. This mechanism informs a therapeutic strategy for modulating BAX-mediated apoptosis by targeting C126.
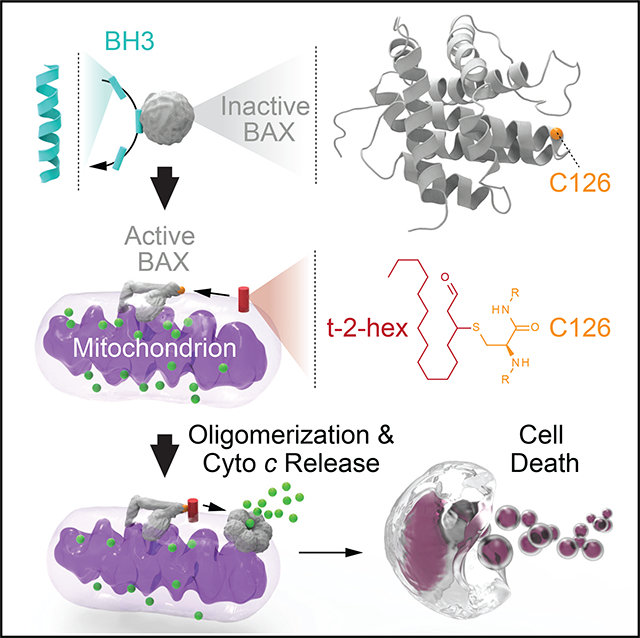
Graphical Abstract
INTRODUCTION
BAX is a pro-apoptotic BCL-2 family protein that transforms from a latent cytosolic protein into a homo-oligomer that permeabilizes the mitochondrial membrane in response to a host of cellular stress signals (Walensky and Gavathiotis, 2011). Mitochondrial outer membrane permeabilization (MOMP) represents a “point of no return” for apoptosis induction, with the release of key signaling factors, such as cytochrome c and Smac/Diablo, triggering the caspase cascade that systematically destroys the cell from within (Chipuk et al., 2010). Given the attendant risks of renegade BAX activation, the conformational and functional transformation of BAX is tightly regulated at a series of control points. For example, the initiation step of BAX activation is mediated by direct interaction between select members of the “BH3-only” subclass of BCL-2 proteins and a trigger site that we previously identified at the N-terminal face of BAX (Gavathiotis et al., 2008). Engagement of the trigger site by the BH3 α-helix of BIM, for example, displaces the α1-α2 loop of BAX, resulting in a series of key conformational changes, including release of the BAX α9 helix and exposure of the BAX BH3 domain (Gavathiotis et al., 2010). Release of the BAX α9 helix facilitates the translocation of BAX from the cytosol to the mitochondrial outer membrane, whereas BAX BH3 exposure is critical to the process of propagating the BAX activation signal and inducing homo-oligomerization. Importantly, anti-apoptotic members can trap the BAX BH3 α-helix in a surface groove and, thereby, block BAX-mediated apoptosis (Korsmeyer et al., 1993; Sattler et al., 1997), a mechanism hijacked by cancer cells to thwart cell death.
A series of stimuli, both physiologic and pharmacologic, have been shown to activate BAX, including select BH3-only proteins (Kim et al., 2009), stapled BH3 peptides (Walensky et al., 2006), heat (Pagliari et al., 2005), detergents (Hsu and Youle, 1997), small molecules (Walensky, 2019), and endogenous lipids (Chipuk et al., 2012). In 2012, Chipuk and colleagues reported the seminal observation that BH3-triggered, BAX-mediated MOMP of purified mitochondria was enhanced by endoplasmic reticulum microsomes, leading to the discovery that specific lipid species derived from the sphingolipid metabolic pathway, namely trans-2-hexadecenal (or (E)-hexadec-2-enal, hereafter referred to as t-2-hex) and sphingosine-1-phosphate, could selectively facilitate BAX- and BAK-driven MOMP, respectively (Chipuk et al., 2012). Not only were neutral sphingomyelinases implicated as the enzymatic drivers of the sensitization phenomenon but also pharmacologic inhibition of sphingolipid metabolism was shown to block MOMP and preserve cell survival in the face of apoptotic stimuli. Pro-apoptotic BAX, but not anti-apoptotic BCL-XL, bound to t-2-hex, but not other sphingolipids spotted on nitrocellulose membranes, supporting a specific association between BAX and t-2-hex (Chipuk et al., 2012). The authors hypothesized that a direct interaction between t-2-hex and BAX might lower the thermodynamic constraints on the activating conformational changes of BAX and, thereby, enhance BH3-triggered BAX activation. Here, we report that t-2-hex directly, covalently, and functionally modifies BAX at C126, providing an explicit mechanism for the activity of t-2-hex. Nuclear magnetic resonance (NMR) spectroscopy and hydrogen-deuterium exchange mass spectrometry (HXMS) analyses of the reaction provided structural insights into the effect of C126 lipidation on the conformational activation of BAX, informing both a fundamental apoptotic process and an opportunity for therapeutic modulation of cell death.
RESULTS
t-2-Hex Covalently Lipidates BAX
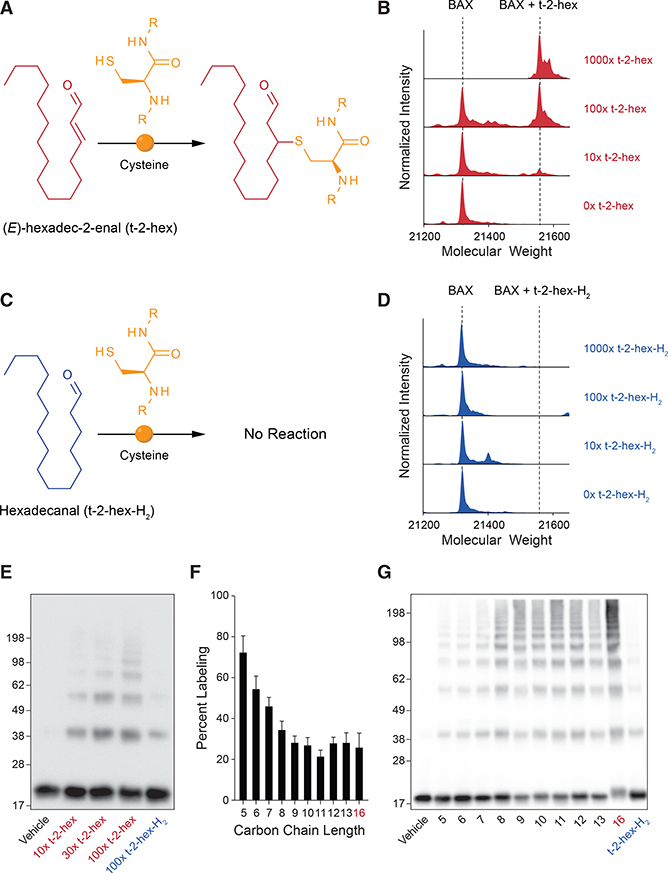
We noted that the molecular structure of t-2-hex contains an a,b-unsaturated aldehyde and is, therefore, susceptible to Michael addition in the presence of nucleophiles, such as the sulfhydryl group of cysteines (Figure 1A). Thus, we tested the reactivity of t-2-hex toward recombinant, full-length BAX to monitor for direct covalent lipidation. t-2-Hex was added to BAX (5 mM) at molar ratios of 10:1, 100:1, and 1000:1, followed by dialysis and analysis by intact protein mass spectrometry (MS). Covalent derivatization of BAX was detectable at 10-fold molar excess, progressed to 50% conversion with 100-fold excess, and was completely derivatized by 1,000-fold molar excess of t-2-hex (Figure 1B). Although a large excess of t-2-hex was required to achieve complete conversion to the fully conjugated form, we reasoned that 100% lipidation would not be required to achieve a functional effect given the catalytic nature of BAX activation, as reflected by the capacity of sub-stoichiometric BH3-only protein to trigger BAX, followed by its robust autoactivation (Tan et al., 2006). Of note, the generation of an m+238 Da adduct suggested that BAX reacts with t-2-hex by Michael addition because imine formation would have yielded a mass of m+222, which was not observed. Interestingly, the saturated form of t-2-hex, hexadecanal (hereafter referred to as t-2-hex-H2) (Figure 1C), was previously shown to have no functional effect on BAX activation (Chipuk et al., 2012). Here, we find that even at 1,000-fold molar excess, incubating t-2-hex-H2 with BAX yielded no covalent adduct (Figure 1D), reinforcing the point that BAX reacts with t-2-hex by Michael addition and that the capacity for covalent reactivity with BAX biochemically distinguishes t-2-hex from t-2-hex-H2.
Figure 1. Covalent Lipidation of Recombinant BAX by t-2-Hex.
(A) Chemical reaction of the lipid electrophile trans-2-hexadecenal (t-2-hex) with a nucleophilic cysteine residue.
(B) Dose-responsive lipidation of recombinant BAX (molecular weight [MW] 21,320 Da; 5 μM) by t-2-hex to yield a covalent adduct that incorporates 1 equivalent of lipid (MW 21,558 Da).
(C) The reduced form of t-2-hex, hexadecanal (t-2-hex-H2), does not chemically react with cysteine.
(D) Exposure of recombinant BAX (MW 21,320 Da; 5 μM) to increasing doses of t-2-hex-H2 causes no covalent lipidation, as reflected by the absence of a MW change in BAX.
(E) Incubation of BAX (5 μM) with increasing amounts of t-2-hex (red; 10x, 30x, and 100x) induces a prominent laddering effect consistent with the activation and oligomerization of BAX, whereas treatment with the highest dose of t-2-hex-H2 (blue) has little to no effect.
(F) Percent labeling of BAX (5 μM) by lipid-derived electrophiles (α,β-unsaturated aldehydes; 500 μM) of varying carbon length, as monitored by intact mass spectrometry. 5, 2-pentenal; 6, 2-hexenal; 7, 2-heptenal; 8, 2-octenal; 9, 2-nonenal; 10, 2-decenal; 11, 2-undecenal; 12, 2-dodecenal; 13, 2-tridecenal; 16 (red), t-2-hex. Data are mean ± SD of three independent biological replicates.
(G) Effect of incubating lipid-derived electrophiles (500 μM) of varying carbon chain length on the oligomerization of BAX (5 μM), as assessed by the laddering of BAX upon gel electrophoresis and BAX western analysis.
To link covalent lipidation of BAX to its functional activation, we first compared the effects of t-2-hex and t-2-hex-H2 on inducing BAX oligomerization, as assessed by denaturing gel electrophoresis of the reaction samples, followed by anti-BAX western blotting. The covalent reactivity feature of t-2-hex indeed correlated with dose-responsive production of notably increased levels of higher-order BAX oligomeric species, which even persisted after heating to 95°C in 100 mM HCl, quenching with lithium dodecyl sulfate (LDS) loading dye containing 250 mM DTT, and performing electrophoresis under denaturing conditions (Figure 1E). The saturated form of t-2-hex, t-2-hex-H2, had comparatively little effect when examined at equivalent high-dosing ratios (e.g., 100:1). To further characterize the structure-activity relationship for the reaction between t-2-hex (16 carbons) and BAX, we screened commercially available α, β-unsaturated aldehydes ranging from 5 to 13 carbons long in BAX covalent labeling and oligomerization assays. The shorter-chain electrophiles were more reactive toward BAX (Figure 1F), whereas induction of higher-order oligomerization correlated instead with increasing chain length, with the physiologic lipid electrophile, t-2-hex, demonstrating the most robust effect (Figure 1G). In general, induction of BAX oligomerization was enhanced as a function of carbon chain length; yet, even the shortest species, such as 2-pentenal, 2-hexenal, and 2-heptenal, demonstrated greater activation of BAX than t-2-hex-H2, highlighting the critical role of the Michael addition reaction. Thus, the combination of covalent reactivity and the hydrophobic surface area afforded by the 16-carbon t-2-hex appeared to maximize the stimulatory effect.
NMR Analysis of the t-2-Hex/BAX Interaction
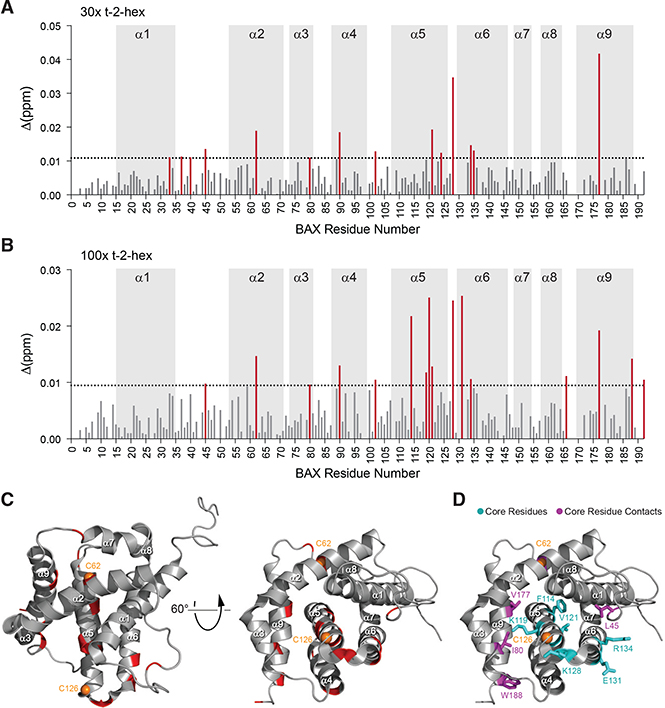
To characterize the effect of t-2-hex conjugation on BAX conformation, we performed 1H-15N heteronuclear single quantum coherence (HSQC) NMR analysis upon t-2-hex titration. First, we found that pre-incubation of BAX with t-2-hex for greater than 30 min, or t-2-hex concentrations over 5 mM, resulted in severe line broadening that precluded analysis but is consistent with a major conformational change, such as the oligomerization described above (Figures 1E and 1G). Upon applying 30- and 100-fold molar excess of t-2-hex, the most prominent chemical shift changes occurred in the α5/α6 hairpin residues that are adjacent to C126 (F114, K119, L120, V121, K128, E131, and R134) and local residues that engage the α5/α6 core, including select amino acids from the α1-α2 loop, α2 (C62), α3 (I80), and α9 (V177, W188) (Figures 2A–2D). Interestingly, a pair of residues that juxtapose in the “closed-loop” form of inactive BAX, L45 of the α1-α2 loop and R134 of α6 (a residue within the N-terminal trigger site), were among those residues that undergo significant chemical shift change upon t-2-hex interaction. These data suggest that when t-2-hex is applied to BAX in solution, the dominant effect is conformational perturbation of the α5/α6 hairpin, which represents the hydrophobic core of the BAX protein and displays C126 at the protein surface, with attendant effects on key adjacent regions involved in the initiation (α1-α2 loop, α2) and translocation (α9) of BAX (Figure 2D). Of note, BAX has two cysteine residues, C62 and C126, with the latter more exposed than the former in the monomeric BAX structure (Figure S1) (Suzuki et al., 2000). Interestingly, the α5/α6 junction where C126 resides is a conserved cysteine-containing region across BCL-2 family proteins (Figure S2). Based on the predominance of chemical shift changes centered around C126 upon t-2-hex titration, the NMR data suggested that C126 is the likely site of regulation by the lipid electrophile.
Figure 2. Influence of t-2-Hex on the Structure of BAX in Solution.
(A–D) NMR analysis of 15N-BAX (50 μM) upon incubation with 30:1 (A) and 100:1 (B) t-2-hex:BAX revealed chemical shift changes in residues of the α5/α6 hairpin adjacent to C126, in addition to residues of the α1-α2 loop, α2, α3, and α9 that engage the α5/α6 core and are implicated in the initiation and translocation of BAX (C). Residues with significant chemical shift changes (1 SD threshold of ≥0.0109 ppm for 30:1, 0.0094 ppm for 100:1) are mapped onto the ribbon structure of BAX (PDB: 1F16) (C and D). The proximity of affected α5/α6 core residues (cyan) and their contacts (magenta) is further highlighted by the stick representations (D).Cysteines 62 and 126 are colored orange. See also Figures S1 and S2.
C126-Dependent Conformational Activation of BAX by t2-Hex
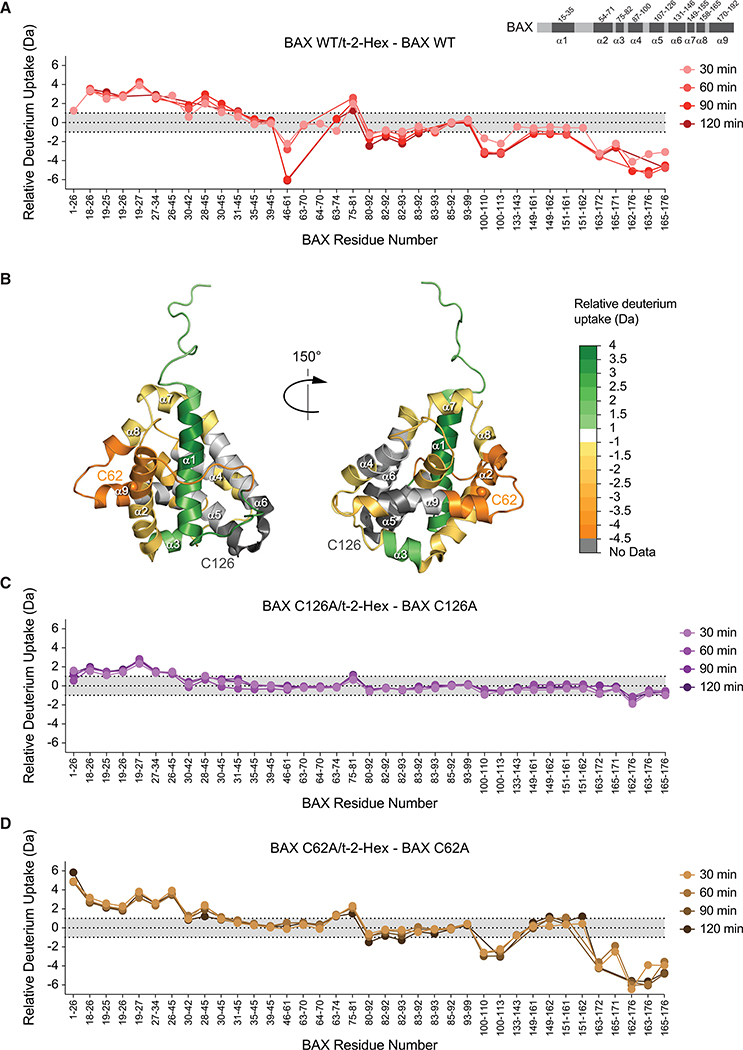
To further characterize the conformational consequences and cysteine-specific dependency of t-2-hex lipidation of BAX, we performed comparative HXMS (Table S1). HXMS probes protein structure by measuring changes in the deuterium incorporation of backbone amide hydrogens (Engen, 2009). When diluted into deuterium buffer, backbone hydrogens of flexible and/or exposed protein regions rapidly exchange with deuterium, whereas buried domains and/or those regions that contain hydrogen-bonding involving backbone amide hydrogens (such as in a helices) demonstrate slowed or suppressed deuterium exchange (Laiken et al., 1969; Printz et al., 1972; Shi et al., 2013). When we compared the deuterium exchange profiles of BAX in the presence and absence of t-2-hex, we observed time-responsive changes in discrete functional regions consistent with the induction of oligomerization (Figures 3A and 3B). Specifically, we observed deprotection of the N-terminal structures of BAX, including α1 and the α1-α2 loop, a region implicated in the initiating conformational changes of BAX activation (Gavathiotis et al., 2010), in addition to α3. Prominent regions of protection included the BH3 domain (α2) and the C-terminal helix (α9), which are believed to mobilize upon BAX activation and then engage in self-association interactions in the resultant oligomerized BAX species (Czabotar et al., 2013; Gahl et al., 2014; Gavathiotis et al., 2010). Proximal portions of α4 and α5, the α4-α5 loop, α7, and α8 also showed lipid-induced protection from deuterium exchange.
Figure 3. t-2-Hex Promotes the Conformational Activation and Oligomerization of BAX.
(A) A plot of relative deuterium differences obtained by subtracting the relative deuterium incorporation of wild-type BAX alone from the relative deuterium incorporation of wild-type BAX (60 pmoles) in the presence of t-2-hex (100:1, t-2-hex:BAX) over time (30-, 60-, 90-, and 120-min time points). Light gray shading represents changes in the plot that are below a threshold of 1 Da in magnitude, whereas the white region highlights changes above the threshold. Data are representative of two independent biological replicates per BAX construct for each time point.
(B) The differences in deuterium exchange above the threshold induced by incubating BAX with t-2-hex are mapped onto the ribbon structure of BAX (PDB: 1F16). The color scale represents the extent of deprotection (green) and protection (orange) of BAX in response to t-2-hex.
(C) The deuterium uptake difference plot indicates the relative deuterium incorporation of BAX C126A (60 pmoles) in the presence of t-2-hex (100:1, t-2-hex:BAX C126A) over time (30-, 60-, 90-, and 120-min time points) minus the relative deuterium incorporation of BAX C126A alone.
(D) The deuterium uptake difference plot shows the relative deuterium incorporation of BAX C62A (60 pmoles) in the presence of t-2-hex (100:1, t-2-hex:BAX C62A) over time (30-, 60-, 90-, and 120-min time points) minus the relative deuterium incorporation of BAX C62A alone.
See also Figures S3 and S4 and Table S1.
To interrogate the requirement of a specific cysteine residue in the conformational changes induced by t-2-hex, we generated full-length BAX protein bearing C62A or C126A mutations (Figure S3) and repeated the HXMS analyses. Strikingly, C126A mutagenesis abrogated essentially all of the t-2-hex-induced conformational changes, with residual—yet markedly reduced—exchange evident in the N-terminal region alone (Figure 3C). Importantly, a comparison of the baseline deuterium exchange profiles for C126A and wild-type BAX demonstrated no differences (Figure S4A). Thus, C126A mutagenesis has no apparent independent effect on the overall structure of BAX but instead directly impacts the conformational responsiveness of BAX to t-2-hex (Figure 3C). In contrast, C62A mutagenesis phenocopied the response of wild-type BAX to t-2-hex. Indeed, the only difference observed between the C62A and wild-type BAX responses was apparent elimination of t-2-hex-induced protection of the BH3 region in the C62A mutant. However, comparison of the deuterium exchange profiles of C62A and wild-type BAX revealed that C62A mutagenesis caused focal deprotection within the BH3 region, which is correspondingly reduced (to zero exchange) upon exposure to t-2-hex (Figure S4B). Thus, the C62A mutation alone alters conformational dynamics in the vicinity of C62, but the overall influence of t-2-hex on the deuterium exchange profiles of C62A and wild-type BAX is the same. Taken together, the HXMS analyses demonstrate that exposure of BAX to t-2-hex in solution can directly induce a major conformational reorganization of monomeric BAX, consistent with oligomerization, and that this phenomenon specifically requires C126.
t-2-Hex-Containing Model Membranes Enhance BAX Activation
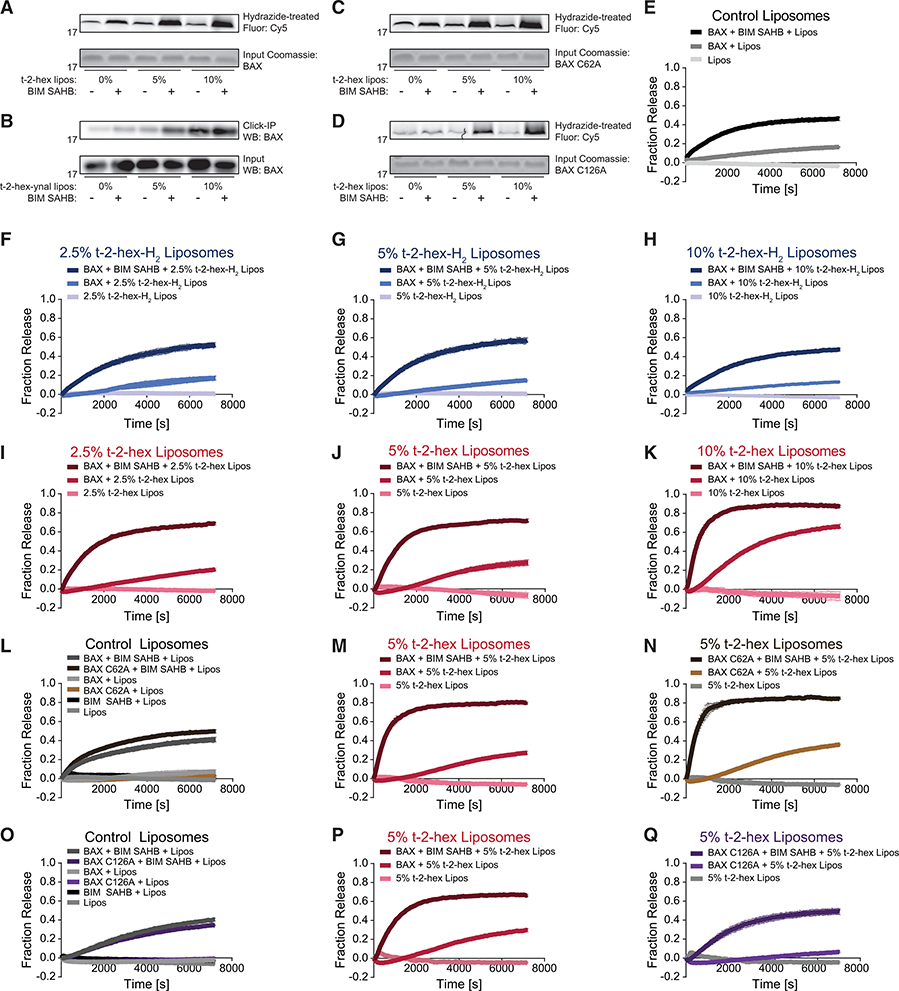
To develop a model system that best recapitulates the physiologic context for covalent lipidation while allowing for direct analysis of the individual components that comprise the BAX activation mechanism, we developed a liposomal system that incorporates lipid electrophiles. Large unilamellar vesicles (LUVs) reflecting the lipid composition of the mitochondrial outer membrane and containing 0%, 5%, or 10% t-2-hex were incubated with recombinant, full-length BAX in the presence or absence of a triggering stapled-BH3 helix (BIM SAHBA2) (Gavathiotis et al., 2008). First, to determine if t-2-hex in the liposomal membrane covalently labeled BAX, we subjected the reaction mixture to Cy5 hydrazide, which selectively reacts with aldehydes and ketones and, thus, serves as a direct and selective in-gel probe for t-2-hex-conjugated BAX, as detected by Cy5 scan of the SDS-PAGE gel of electrophoresed samples (Boone et al., 2016). We observed t-2-hex dose-dependent increases in the amount of generated t-2-hex/BAX conjugate upon treatment with BIM SAHBA2 (Figure 4A), demonstrating that membrane-embedded t-2-hex was indeed capable of covalently reacting with BAX once it was triggered to undergo membrane translocation. We corroborated this result using an orthogonal method, whereby alkynylated t-2-hex ((E)-hexadec-2-en-15-ynal) was incorporated into liposomes so that t-2-hex/ BAX conjugate could be isolated by streptavidin pull-down after biotinylating t-2-hex with the biotin-PEG-azide reagent and click chemistry (Jarugumilli et al., 2018). This alternative analysis produced essentially the same results as our Cy5 method, confirming that t-2-hex, as a liposomal membrane component, can directly and covalently react with BAX (Figure 4B). We further applied the Cy5-hydrazide method to determine whether covalent lipidation of BAX at the membrane occurs at C126, C62, or both sites. Whereas C126 appears to be the primary site of covalent lipidation by membrane-localized t-2-hex (Figure 4C), as also indicated by our NMR (Figure 2) and HXMS (Figure 3) results, cross reaction with C62 is also evident (Figure 4D). These data likely explain why a global screen for protein targets of t-2-hex identified BAX C62 as a site of lipid derivatization (Jarugumilli et al., 2018). Nevertheless, our HXMS analyses do not implicate C62 as mechanistically relevant to the conformational activation of BAX induced by t-2-hex.
Figure 4. Liposomal t-2-Hex Covalently Modifies BAX and Potentiates BH3-Triggered Direct BAX Activation.
(A) The incorporation of t-2-hex into liposomes upon BIM SAHBA2 triggering resulted in dose-responsive (0%, 5%, and 10% t-2-hex by mass) covalent lipidation of added recombinant wild-type BAX (10 mM), as detected by Cy5-hydrazide treatment and fluorescence scan.
(B) An alternative analysis involving liposomes containing alkynylated t-2-hex, followed by biotinylation using click chemistry, streptavidin pull-down, and BAX western analysis demonstrated dose-responsive t-2-hex derivatization of BAX (10 μM), which was consistently enhanced upon co-treatment with BIM SAHBA2 (10 μM).
(C and D) Liposomal t-2-hex lipidation of BAX C62A (C) and, to a lesser extent, C126A (D) upon BIM SAHBA2 triggering, as detected by Cy5-hydrazide treatment and fluorescence scan.
(E) The addition of BAX (500 nM) and BIM SAHBA2 (500 nM) to control LUVs, which reflect the lipid composition of the mitochondrial outer membrane, induced time-responsive liposomal release of the fluorophore/quencher.
(F–H) The incorporation of 2.5% (F), 5% (G), or 10% (H) t-2-hex-H2 into the liposomal membranes had no effect on the baseline or BH3-triggered liposomal release mediated by recombinant wild-type BAX (500 nM).
(I–K) The incorporation of 2.5% (I), 5% (J), or 10% (K) t-2-hex into the liposomal membranes caused a dose-responsive enhancement in the level and kinetics of membrane poration by wild-type BAX (500 nM), both alone and when triggered by BIM SAHBA2 (500 nM).
(L–N) Wild-type and C62A-mutant BAX (500 nM) exhibited similar levels of liposomal release, both at baseline and when triggered by BIM SAHBA2 (500 nM), upon using either a standard liposomal preparation (L) or liposomes containing 5% t-2-hex (M and N).
(O–Q) Wild-type and C126A-mutant BAX (500 nM) exhibited similar levels of liposomal release, both at baseline and when triggered by BIM SAHBA2 (500 nM), upon using a standard liposomal preparation (O). However, the observed enhancement of BAX-mediated release from 5% t-2-hex-containing liposomes, in the presence or absence of BIM SAHBA2 (P), was abrogated by C126A mutagenesis (Q), such that the level and kinetics of membrane poration by BAX C126A was the same for standard and t-2-hex-containing liposomes (O and Q). Error bars represent mean ± SD for liposomal release experiments performed in technical quadruplicate, with data representative of two independent experiments.
To further explore the functional impact of membrane-localized t-2-hex on the membrane poration activity of BAX and its cysteine mutants, we measured the effect of t-2-hex content and BH3 stimulation on BAX-mediated liposomal poration of ANTS/DPX-loaded LUVs. First, we established control conditions that demonstrate the baseline effect of recombinant BAX and BIM SAHBA2-triggered BAX on liposomal release (Figure 4E). Incorporation of t-2-hex-H2 into the liposomes at 2.5%, 5%, or 10% membrane content had no effect on either baseline BAX release or that triggered by BIM SAHBA2 (Figures 4F–4H), whereas t-2-hex dose-responsively enhanced the absolute level and kinetics of liposomal release by BAX alone and when treated with BIM SAHBA2 (Figures 4I–4K). To link the observed stimulatory effect of t-2-hex to the presence of a specific cysteine residue in BAX, we repeated the experiment using C62A- and C126A-mutant forms of recombinant BAX. BAX C62A performed similarly to wild-type BAX in the presence of control and 5% t-2-hex liposomes, whether added alone or upon triggering with BIM SAHBA2 (Figures 4L–4N). In contrast, BAX C126A showed no enhanced activity upon exposure to 5% t-2-hex liposomes, either alone or when combined with BIM SAHBA2, again implicating BAX C126 as the mediator of t-2-hex’s stimulatory effect (Figures 4O–4Q). Taken together, we find that the presence of t-2-hex in the membrane environment covalently modifies BAX, and the derivatization of C126 is responsible for enhancing BAX-mediated membrane permeabilization.
Endogenous t-2-Hex Potentiates BAX Activation in a C126-Dependent Manner
We next sought to validate our findings in the context of mitochondrial preparations that contain endogenous t-2-hex. To monitor for derivatization of BAX, we applied an assay that detects proteins covalently modified by aldehyde or ketone-containing electrophiles. Upon addition of propargyl hydrazine, this alkyne-containing nucleophile reacts with aldehydes or ketones incorporated into proteins. The alkyne handle is then conjugated to biotin by copper-catalyzed azide-alkyne cycloaddition, followed by streptavidin pull-down to enrich for the derivatized proteins (Matthews et al., 2017). We tested this method on a solution of recombinant BAX in the presence and absence of t-2-hex and demonstrated successful capture of BAX by propargyl hydrazine only in the presence of t-2-hex, highlighting the sensitivity of the assay for detecting t-2-hex-derivatized BAX (Figure 5A). We then used mitochondria isolated from mouse livers lacking BAX and BAK to evaluate whether covalent derivatization of added BAX occurs in the presence of native t-2-hex. BAX was not pulled down in the absence of mitochondria or upon incubation with mitochondria alone, whereas the addition of the direct activator BH3 peptide BIM SAHBA2 led to BAX derivatization and pull-down (Figure 5B), consistent with BH3-induced BAX activation, mitochondrial translocation, and lipid-derived electrophile derivatization. To link BAX derivatization to covalent modification by t-2-hex, we further incubated the isolated mitochondria with recombinant sphingosine-1-phosphate lyase (SGPL1), which converts sphingosine-1-phosphate into t-2-hex and phosphoethanolamine and, thus, increases endogenous t-2-hex levels. Not only did the added SGPL1 substantially increase the level of derivatized BAX in response to BH3-activation but also a baseline level of BAX capture in the presence of mitochondria alone became detectable (Figure 5B), consistent with the dose-responsive functional activity of BAX in the presence of liposomes bearing increasing amounts of incorporated t-2-hex (Figures 4I–4K). Thus, we find that in the context of the native organelle and endogenous t-2-hex, BAX undergoes covalent lipidation, the level of which is amplified both by BH3 triggering and increasing the enzymatic production of t-2-hex.
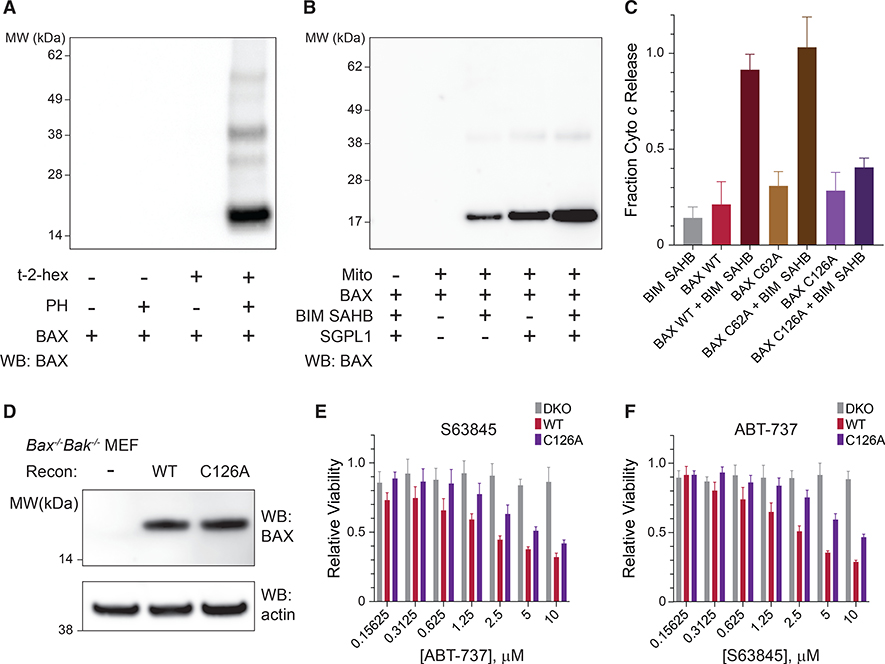
Figure 5. Covalent Lipidation of BAX by Endogenous t-2-Hex and Impairment of BAX Activity in Mitochondria and Cells by C126 Mutagenesis.
(A) Detection of t-2-hex lipidation of recombinant BAX (5 μM) in solution upon propargyl hydrazine (PH) treatment, followed by biotin conjugation using click chemistry, streptavidin pull-down, and BAX western analysis.
(B) PH-based streptavidin pull-down of lipid-derivatized BAX in response to treatment with isolated BAX/BAK-deficient mitochondria, in the presence or absence of BIM SAHBA2 or recombinant sphingosine-1-phosphate lyase (SGPL1), which increases the level of endogenous t-2-hex. BAX, 500 nM; BIM SAHBA2, 500 nM.
(C) Mitochondrial cytochrome c release in response to treatment with the indicated BAX constructs, in the presence or absence of the BH3-triggering ligand BIM SAHBA2. Data are mean ± SD for experiments performed in technical quintuplets and repeated twice with similar results upon using independent mitochondrial preparations and treatments. BAX proteins, 100 nM; BIM SAHBA2, 100 nM.
(D) Western analysis of lysates from Bax−/−Bak−/− MEFs reconstituted with wild-type or C126-mutant BAX.
(E and F) Cell viability of Bax−/−Bak−/− MEFs reconstituted with wild-type or C126A-mutant BAX treated with fixed-dose S63845 (10 μM) and the indicated concentrations of ABT-737 (E) or fixed-dose ABT-737 (20 μM) and the indicated concentrations of S63845 (F). Data are mean ± SD for experiments performed in technical triplicate and repeated twice with similar results upon using independent cell cultures and treatments.
We next examined whether endogenous derivatization of BAX in the mitochondrial context has functional consequences. Specifically, we sought to determine if cysteine mutagenesis influenced BAX-mediated cytochrome c release activity in a manner similar to that observed in the liposomal context. Wild-type and C62A-mutant BAX demonstrated robust cytochrome c release in response to BH3 triggering, whereas C126A mutagenesis notably blunted the effect (Figure 5C), consistent with a functional role for C126 lipidation in potentiating BAX activity. Finally, to determine if C126A mutagenesis could influence BAX-mediated cell death, we reconstituted Bax−/−Bak−/− mouse embryonic fibroblasts (DKO MEFs) with wild-type or C126A BAX (Figure 5D) and then monitored cell viability in response to targeted inhibition of the anti-apoptotic proteins MCL-1 (S63845) (Kotschy et al., 2016) and BCL-2/BCL-XL (ABT-737) (Oltersdorf et al., 2005). Indeed, we observed that whether exposed to fixed dose S63845 and serial dilution of ABT-737 or fixed dose ABT-737 and serial dilution of S63845, the cells bearing BAX C126A were less responsive to combination drug treatment than those bearing wild-type BAX (Figures 5E and 5F). Taken together, we find that eliminating the capacity for covalent BAX lipidation at C126 impairs mitochondrial cytochrome c release from isolated mouse liver mitochondria and attenuates BAX-mediated cell death, consistent with the reported sensitizing effect of t-2-hex on BAX-dependent mitochondrial apoptosis (Chipuk et al., 2012).
DISCUSSION
The interplay between BCL-2 family proteins and the mitochondrial outer membrane is a cardinal feature of apoptotic regulation. However, the explicit mechanisms by which these biochemical processes ensue within the membrane environment are largely an enduring mystery. Fundamental questions such as how BAX homo-oligomerization permeabilizes the mitochondrial outer membrane or the structural basis for the heterodimerization of full-length BCL-2 family proteins or homo-oligomerization of BAX and BAK remain unanswered. The stakes are high because BCL-2 family interactions regulate the cellular life-death decision and their deregulation contributes to a host of human diseases characterized by too much or too little cell death. Thus, deciphering the structure-function mechanisms of BCL-2 family regulation remains a high-priority goal for basic and translational scientists alike.
Here, we find that a cysteine residue, exposed on the surface of BAX and localized to the core hydrophobic α5/α6 hairpin, is subject to covalent lipidation by t-2-hex, a lipid-derived electrophile that has been shown to be a physiologic enhancer of BAX-dependent apoptosis (Chipuk et al., 2012). Our NMR analyses of BAX upon addition of t-2-hex revealed chemical shift perturbations in the C126 region, influencing the core α5/α6 helices and their contacts, including residues of α1 and the α1-α2 loop, implicated in the initial conformational change, and α9, which is allosterically released for membrane translocation upon direct BH3-induced BAX activation (Gavathiotis et al., 2010). Comparative HXMS analyses, conducted over time, demonstrated that t-2-hex triggered a major conformational change in both wild-type and C62A BAX, consistent with a monomer-to-oligomer transition. Strikingly, C126A mutagenesis abrogated this effect, further implicating C126 as the functionally relevant site of t-2-hex derivatization. Exposure of BAX to liposomal membranes containing t-2-hex not only resulted in the covalent modification of BAX but also directly enhanced the absolute level and kinetics of BAX-mediated membrane poration, an activity that specifically required C126. BAX derivatization was also observed in the context of mitochondrial preparations containing endogeneous t-2-hex, wherein covalent modification was specifically induced by BH3 triggering, which translocates BAX to the mitochondrial membrane and effectively concentrates BAX in a two-dimensional bilayer with t-2-hex. Boosting the levels of endogenous t-2-hex by adding SGPL1 to the mitochondrial preparation enhanced BAX derivatization even further. Importantly, both BAX-mediated mitochondrial cytochrome c release and BAX-mediated cell death were selectively impaired by C126A mutagenesis, further highlighting the functional relevance of C126 lipidation. Based on these data, we expand the mechanistic model of BH3-triggered direct BAX activation to include covalent reaction of the α5/α6 hydrophobic core with the target membrane, a biochemical process that enhances BAX activation and membrane poration. Indeed, a three-dimensional model of active BAX at the membrane based on double electron-electron resonance spectroscopy predicts opening of the a5/a6 hairpin to form a clamp-like conformation on the membrane surface (Bleicken et al., 2014), which would ideally position C126 (located at the junction of the α5/α6 hairpin) for interaction with lipid headgroups, such as the reactive α,β-unsaturated aldehyde of t-2-hex.
Post-translational modification has emerged as a key mechanism for regulating the functional activities of BCL-2 family proteins. For example, myristoylation of the BH3-only protein tBID targets it to the mitochondrial outer membrane (Zha et al., 2000), phosphorylation of BAD shifts its binding affinity from BCL-2 family anti-apoptotic proteins to glucokinase (Danial et al., 2008), and ubiquitination of anti-apoptotic members such as MCL-1 controls protein half-life (Zhong et al., 2005). Depending on the physiologic context and experimental setup, a variety of cysteine modifications have been reported for BAX, including S-palmitoylation of C126 (Frö hlich et al., 2014), oxidation of C62 (Nie et al., 2008), and most recently t-2-hex derivatization of C62 (Jarugumilli et al., 2018). Although in this study we do not detect a functional role for C62 in the activity of t-2-hex, it remains plausible that under specific conditions that affect accessibility of the relatively buried C62 residue, covalent conjugation may occur. Here, we demonstrate that the functionally relevant site of BAX derivatization by t-2-hex is C126. Notably, in contrast to the post-translational modifications described above that require enzymatic reactions (e.g., myristoylation, phosphorylation, and ubiquitination), the covalent lipidation of BAX C126 reported here is non-enzymatic and instead involves a direct biochemical reaction between BAX and t-2-hex at the mitochondrial membrane.
Of the two cysteines in human BAX, the more exposed C126 residue is located at the α5/α6 junction, where a variety of BCL-2 family proteins also contain a cysteine residue (Figure S2). Interestingly, our prior screen for small molecule inhibitors of anti-apoptotic MCL-1 yielded select compounds that covalently modified C286 and, in doing so, induced an allosteric conformational change in its canonical surface groove that impaired BH3 interaction (Lee et al., 2016). Thus, characterizing the structure-function consequences of post-translational cysteine derivatization not only is critical to our understanding of the physiologic properties of BCL-2 family proteins but also informs approaches to selectively target reactive cysteines by Michael addition to modulate their pro- and anti-apoptotic activities for therapeutic benefit. Indeed, cysteine modification by selective, electrophilic molecules has become an increasingly appealing approach to drugging recalcitrant targets, as exemplified by the success of ibrutinib, a covalent inhibitor of Bruton’s tyrosine kinase (Zhang et al., 2019). The presence of a unique cysteine residue within the canonical BH3-binding groove of BFL-1/A1 inspired our development of selective, cysteine-reactive stapled peptide inhibitors of this anti-apoptotic member implicated in the development and chemoresistance of leukemia, lymphoma, and melanoma (Guerra et al., 2018; Harvey et al., 2018; Huhn et al., 2016). Here, our finding that t-2-hex-containing membranes can covalently derivatize and sensitize BAX activation reveals a pharmacologic opportunity to influence cell fate by alternatively simulating or blocking C126 lipidation. Small molecules that covalently target C126 and contain hydrophobic moieties that engage the site of t-2-hex interaction have the potential to directly modulate BAX-mediated apoptosis. Intriguingly, the small molecule OICR766A that emerged from a screen for compounds that enhance BAX-mediated liposomal poration was found to bind and activate BAX in a C126-dependent manner (Brahmbhatt et al., 2016). Thus, whether by physiologic or pharmacologic targeting, the C126 region may represent an important control point for modulating the pro-apoptotic functionality of BAX.
STAR*METHODS
LEAD CONTACT AND MATERIALS AVAILABILITY
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact Loren Walensky (loren_walensky@dfci.harvard.edu). Plasmids and mouse cell lines generated in this study are available upon request to the lead contact.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Microbe Strains
Recombinant proteins were expressed in BL21 (DE3) E. coli bacteria, which were grown in Luria Broth (LB) at 37°C with shaking at 220 rpm.
Cell Lines and Culture
Mouse embryonic fibroblasts (Bax−/−Bak−/− and BAX-reconstituted variants) were maintained in Dulbecco’s Modified Eagle Medium (GIBCO) with 10% FBS, 100 U\mL penicillin and streptomycin, and 2 mM glutamine. Cells were verified as mycoplasma-negative using the MycoAlert mycoplasma detection kit (Lonza Biologics).
METHOD DETAILS
BAX Protein Preparation
Recombinant full-length, wild-type BAX was expressed using the pTYB1 vector in BL21 (DE3) E. coli, which were grown in LB-carbenicillin and induced with 1 mM IPTG for 4 h at 30°C. Bacterial pellets were resuspended in lysis buffer (20 mM Tris, 250 mM NaCl, pH 7.2) containing protease inhibitor tablets (Roche) and lysed over three passages through a microfluidizer (Microfluidics) on ice. The soluble fraction was isolated by centrifugation at 20,000 rpm for 45 min at 4C. BAX protein was purified by chitin affinity chromatography using chitin resin (New England Biolabs) on a gravity flow column. The intein and affinity tag were cleaved using 10 mg/mL dithiothreitol at 4C for 12–36 h. The full-length, tagless protein was eluted, concentrated, and purified by size-exclusion chromatography in FPLC buffer (20 mM HEPES-KOH, 150 mM KCl, pH 7.2) using a Superdex 75 10/300 Global column on an FPLC system (AKTA Pure, GE Healthcare Life Sciences).
The C62A and C126A mutants of BAX were generated by PCR-based site directed mutagenesis (Q5 Site-Directed Mutagenesis Kit, NEB) and confirmed by DNA sequencing. Overnight cultures were grown in MDG-carbenicillin and bacterial colonies plated onto MDG-carbenicillin agar plates (Studier, 2005). Highly expressing clonal populations were selected by double colony selection (Murray et al., 2010). The resulting colonies were grown overnight in MDG-carbenicillin, inoculated into LB, grown to OD600 0.6–0.8, and induced with 1 mM IPTG overnight at 16°C. Recombinant protein was then isolated as described above for wild-type BAX. All BAX protein constructs were verified by SDS-PAGE, BAX western analysis, and intact and peptic digest MS (see Intact Protein Mass Spectrometry and Hydrogen-Deuterium Exchange Mass Spectrometry sections).
Intact Protein Mass Spectrometry
Stock solutions of α,β-unsaturated aldehydes (200 mM in ethanol) were incubated with recombinant BAX (5 μM) in FPLC buffer for 2 h at 37°C, followed by dialysis. For each MS analysis, protein was injected onto a self-packed reversed phase column (1/32” O.D. x 500 μm I.D., 5 cm of POROS 50R2 resin). After desalting for 4 min, protein was eluted using an HPLC gradient of 0%–100% solution B over 1 minute at a flow rate of 30 μL/min (Solution A, 0.2 M acetic acid in water, Solution B, 0.2 M acetic acid in acetonitrile) into an LTQ ion trap mass spectrometer (ThermoFisher Scientific, San Jose, CA) that acquired profile MS spectra (m/z 300–2000). Mass spectra were deconvoluted using MagTran1.03b2 software.
BAX Laddering Assay
Recombinant BAX (5 μM in FPLC buffer) was incubated with a solution of α,β-unsaturated aldehydes or 2.5% ethanol (vehicle) for 2 h at 37°C. The reaction was quenched by adding 3x LDS sample loading buffer containing 250 mM DTT and heated to 95°C for 10 min. In order to reverse aldehyde-mediated crosslinks, HCl (100 mM) was then added and the sample heated to 95°C for an additional 10 min. Samples were run on a 4%–12% Bis-Tris gel and analyzed by western blotting using the HRP-BAX 2D2 antibody (Santa Cruz).
NMR Spectroscopy
Uniformly 15N-labeled recombinant BAX was generated as previously described (Gavathiotis et al., 2008; Suzuki et al., 2000). Briefly, BL21 (DE3) E. coli were grown in LB-carbenicillin media to OD600 of 0.6–0.8, pelleted by centrifugation, and resuspended in M9 minimal media (3.2 g/L KH2PO4, 12.8 g/L Na2HPO4-7H2O, 0.5 g/L NaCl, 0.4% glucose, 1x trace minerals [Teknova], 1 mM MgSO4, 100 μM CaCl2) containing 1 g/L of 15NH4Cl (Cambridge Isotope Laboratories). Resuspended cultures were shaken at 37°C for 30 min, and then 1 mM IPTG was added to induce 15N-BAX expression at 30°C for 4 h. 15N-labeled protein was purified as described above for recombinant BAX. A 50 mM solution of 15N-BAX in 25 mM sodium phosphate, 50 mM NaCl, pH 6.2, 10% D2O was treated with vehicle (2.5% ethanol) or 1.5 mM or 5 mM t-2-hex. 1H-15N HSQC experiments were conducted at 32°C on a 600 MHz NMR spectrometer equipped with a cryogenic probe. Spectra were processed in Topspin (Bruker) and analyzed using CcpNmr software. Chemical shift perturbations were calculated in ppm by applying the formula to the 1H and 15N differences for each cross peak. The absence of a bar indicates no chemical shift difference, or the presence of a proline or residue that is overlapped or not assigned. BAX cross-peak assignments were applied as previously reported (Suzuki et al., 2000). The significance threshold for the chemical shift changes was calculated based on the average chemical shift across all residues plus the standard deviation, in accordance with standard methods (Marintchev et al., 2007).
Hydrogen-Deuterium Exchange Mass Spectrometry
HXMS experiments were performed as described (Barclay et al., 2015; Lee et al., 2016) and in accordance with the conditions summarized in Table S1. BAX (30 mM) was incubated at 37°C with shaking in the presence of 3 mM t-2-hex or vehicle (1.5% ethanol), and 2 μL aliquots were removed at 30-min intervals and labeled in D2O buffer. Deuterium labeling was initiated with an 18-fold dilution into room-temperature D2O buffer (20 μM HEPES, 150 mM KCl, 1 mM MgCl2, pD 7.2) of a pre-equilibrated mixture of BAX protein (2 mL at 30 mM) containing vehicle (1.5% ethanol) or t-2-hex (3 mM). After 10 s, the labeling reaction was quenched with the addition of an equal volume of quench buffer (0.8 M guanidinium chloride, 0.8% formic acid [v/v]). Each deuterium labeling experiment was performed in biological duplicate (Wales and Engen, 2006). Proteolysis was performed by incubation for 5 min on ice with 40 μg pepsin and 20 μg aspergillopepsin (protease type XIII), both prepared at 10 mg/mL in water. Digested samples were then processed and analyzed as described previously (Barclay et al., 2015). Briefly, the peptides were trapped and desalted on a VanGuard Pre-Column trap (2.1 × 5 mm, ACQUITY UPLC BEH C18, 1.7 μm) for 3 min, eluted from the trap using a 5%–35% gradient of acetonitrile over 10.5 min at a flow rate of 50 μL/min, and then separated using an ACQUITY UPLC HSS T3, 1.8 mm, 1.0 × 50 mm column. Peptides from an unlabeled protein were identified using ProteinLynx Global Server (PLGS) searches of a protein database including analyte protein. All mass spectra were acquired using a Waters SYNAPT G2Si mass spectrometer. Relative deuterium levels for each peptide were calculated by subtracting the average mass of the undeuterated control sample from that of the deuterium-labeled sample, and identified peptides common to all evaluated conditions were plotted.
Cy5-Hydrazide Labeling of Lipidated BAX
Recombinant BAX (10 μM) was incubated with liposomes containing 0%, 5%, or 10% t-2-hex in the presence of BIM SAHBA2 (10 μM) or vehicle (1% DMSO) for 2 h at room temperature. Cyanine-5-hydrazide (10 μM; Kerafast) was then added and the solution incubated for 1 h at room temperature. The labeling reaction was quenched by adding Tris base (437.5 mM), followed by treatment with NaCNBH3 (12 mM) to stabilize the hydrazide adduct. Samples were prepared for electrophoresis by adding 3x LDS loading dye containing 250 mM DTT and run on a 4%–12% Bis-Tris gel. Cy5 fluorescence (excitation 635 nm, emission 662 nm, 50 mm pixel size) was detected using a Typhoon FLA 9500 (GE Healthcare Life Sciences).
Streptavidin Pull-down of Lipidated BAX
Recombinant BAX (10 mM) was incubated with liposomes containing 0%, 5%, or 10% (E)-hexadec-2-en-15-ynal (Cayman Chemical) in the presence of BIM SAHBA2 (10 μM) or vehicle (1% DMSO) for 2 h at room temperature. A click-chemistry reaction was then performed by adding 0.8 mM of premixed 1:1 CuSO4:Tris((1-benzyl-4-triazolyl)methyl)amine (TBTA) complex, followed by 125 mM Azide-PEG3-biotin conjugate (Aldrich) and 1 mM sodium ascorbate. The reaction was incubated for 1 h at room temperature with gentle mixing. High-capacity streptavidin agarose beads (30 μL) were washed three times in 1 mL of liposomal release assay buffer containing 1% CHAPS. The beads were blocked in 3% BSA in PBS for 1 h at 4C, and then washed 3 times. The click reaction (1 μM) was added to the beads and rotated for 1 h at 4C. The beads were sequentially washed 3 times with liposomal buffer containing 1% CHAPS and then 3 times with PBS. Bound protein was eluted from the beads by adding 10 mg/mL biotin in 10% SDS and heating to 95°C for 10 min. Samples were prepared by adding 3x LDS loading dye containing 1M DTT to the elution mixture, followed by electrophoresis on a 4%–12% Bis-Tris gel and BAX western analysis using the HRP-BAX 2D2 antibody (Santa Cruz).
Liposomal Preparation and Release Assay
Large unilamellar vesicles (LUVs) that reflect the lipid composition of the mitochondrial outer membrane were prepared as previously described (Barclay et al., 2015; Lovell et al., 2008). Briefly, a chloroform solution containing 48:28:10:10:4 molar ratio of phosphatidylcholine, phosphatidylethanolamine, phosphatidylinositol, dioleoyl phosphatidylserine, and tetraoleolyl cardiolipin (Avanti Polar Lipids) was batch-prepared, aliquoted, and dried in glass tubes using a stream of nitrogen, placed under vacuum overnight, and then stored at 80°C. The film was redissolved in chloroform, and then ethanol alone or ethanolic solutions of (E)-hexadec-2enal, (E)-hexadec-2-en-15-ynal, or hexadecanal (Cayman Chemical) were added at the appropriate molar ratio to achieve the indicated compositions of 2.5%, 5%, or 10%, and then dried again under nitrogen and by vacuum. Dried films were resuspended in 1 mL of liposomal assay buffer (20 mM HEPES-KOH, 150 mM KCl, 5 mM MgCl2, pH 7.2) containing 8-aminonaphthalene-1,3,6-trisulfonic acid (ANTS; 12.5 mM) and p-xylene-bis-pyridinium bromide (DPX; 45 mM), and incubated at room temperature for 10 min. Liposomal suspensions were then freeze-thawed using liquid nitrogen and hot water (>70°C) five times and incubated at room temperature for an additional 90 min. The resulting turbid, yellow solution was extruded 11 times through a 100 nm polycarbonate membrane (Whatman), and the liposomes purified by size-exclusion chromatography using Sepharose CL-2B resin on a gravity column. The liposomal release assay was performed by monitoring ANTS fluorescence (355 nm excitation, 540 nm emission, 20 nm slit width) every 60 s after the addition of vehicle (0.05% DMSO), recombinant BAX (500 nM), or the combination of BAX (500 nM) and BIM SAHBA2 (500 nM). Maximal release was determined by the addition of Triton X-100 to a final concentration of 0.625% (v/v). Percent release was calculated as ((FF0)/(F100F0)) 3 100, where F is the observed fluorescence at a given time, and F0 and F100 represent baseline and maximal fluorescence, respectively.
Chemical Synthesis of Labeling Reagents
Propargyl hydrazine was synthesized as described (Matthews et al., 2017) and detailed below.
Di-tert-butyl 1-(prop-2-yn-1-yl)hydrazine-1,2-dicarboxylate
Di-tert-butylhydrazine-1,2-dicarboxylate (5g, 22 mmol) was reacted with 3-bromoprop-1-yne (6 mL of 80% solution in toluene, 67 mmol) in toluene (33 mL) in the presence of sodium hydroxide (40 mL of 5% aqueous solution, 43 mmol, 2 eq) and the phase-transfer catalyst tetrabutylammonium hydrogensulfate (170 mg, 0.5 mmol). The mixture was diluted in water, washed three times with ethyl acetate, the organic layer dried over sodium sulfate, concentrated, and the resulting mixture subjected to column chromatography (30% ethyl acetate/hexanes) to afford 3.293 g of white solid (58% yield). 1H and 13C NMR spectra conformed to the published data (Matthews et al., 2017).
Prop-2-yn-1-ylhydrazine
Di-tert-butyl 1-(prop-2-yn-1-yl)hydrazine-1,2-dicarboxylate (1.9 g, 7.1 mmol) was dissolved in an HCl solution in dioxane (4N, 8.75 mL, 5 eq) and allowed to react at 0C for 24 h. The resulting mixture was diluted in diethyl ether and concentrated in vacuo, and the resulting solids were dissolved in methanol. Cold diethyl ether was added until precipitation occurred, the precipitate was isolated by filtration, and then recrystallized three times from methanol and diethyl ether to afford 247 mg of white solid (32% yield). 1H and 13C NMR spectra conformed to the published data (Matthews et al., 2017). The product was dissolved in 1 N sodium hydroxide at 1 M, after which the pH was determined to be ~7. Aliquots were stored at 1 M and 10 mM at −80°C.
Detection of Lipidated BAX by Propargyl Hydrazine in Solution
Recombinant BAX (100 μL of a 5 μM solution) was incubated with t-2-hex (500 μM) or vehicle (0.25% ethanol) at 37°C for 2 h in the presence of 1% CHAPS. Propargyl hydrazine was added to a final concentration of 500 mM and reacted for 30 min at 37°C. Biotin-PEG4-azide (5 mL of 10 mM DMSO stock), sodium ascorbate (10 μL of 50 mM solution), copper sulfate (1.75 μL of 50 mM solution), and TBTA (Tris[(1-benzyl-1H-1,2,3-triazol-4-yl)methyl]amine, 5.25 μL of 1.7 mM solution, 3:1 DMSO:tert-butanol) were then added, and the reaction allowed to proceed for 1 h at room temperature with vigorous mixing. The reactions were quenched by the addition of tris base (14 mL of 2 M stock), sodium cyanoborohydride (4 μL of 0.2 M solution), dithiothreitol (5 μL of 1 M stock), and BAX FPLC buffer containing 1% CHAPS (900 μL). The quenched click reactions were applied to 50 μL of high-capacity streptavidin agarose beads (Thermo Scientific), which had been pre-washed three times with PBS, blocked for 1 h at 4C with 3% bovine serum albumin in PBS, and then washed three times with BAX FPLC buffer containing 1% CHAPS. The samples were allowed to bind to the beads for 1 h at 4C, followed by washing three times with BAX FPLC buffer containing 1% CHAPS, three times with PBS, and then eluted with 10 mg/mL biotin in 10% SDS for 10 min at 95°C. The bead eluates were then subjected to electrophoresis and BAX western analysis.
Detection of Lipidated BAX by Propargyl Hydrazine in Isolated Mitochondria
Liver mitochondria were isolated from AlbCreBaxf/fBak−/− mice as previously described (Pitter et al., 2008). Recombinant full-length BAX (500 nM) was incubated in mitochondrial assay buffer (25 μL; 200 mM mannitol, 70 mM sucrose, 10 mM HEPES, 1 mM EDTA, pH 7.4, complete protease inhibitor cocktail [Roche]) with mitochondria (1 μg/mL), BIM SAHBA2 (500 nM) or vehicle (0.05% DMSO), in the presence or absence of recombinant SGPL1 (1 mg, Origene), for 2 h at 37°C. Reactions were diluted in an equal volume of BAX FPLC buffer containing 2% CHAPS, followed by addition of propargyl hydrazine (1.25 μL of 10 mM solution) and 30 min incubation at 37°C. A premixed solution of Biotin-PEG4-azide (1.25 μL of 10 mM DMSO stock), copper sulfate (1.2 μL of 50 mM solution), and TBTA (Tris[(1-benzyl-1H-1,2,3-triazol-4-yl)methyl]amine, 2.4 μL of 1.7 mM solution) was added with sodium ascorbate (5 μL of 50 mM solution) to the reaction, which was incubated for 1 h at room temperature with vigorous mixing. The reaction was quenched as above and diluted to 1 mL in BAX FPLC buffer containing 1% CHAPS. The samples were then subjected to streptavidin pulldown, elution, and BAX western analysis as described above.
Cytochrome c Release Assay
Cytochrome c release assays were performed as previously described (Pitter et al., 2008). Briefly, BAX/BAK-deficient mouse liver mitochondria were incubated with the indicated recombinant BAX protein (100 nM) and either BIM SAHBA2 (100 nM) or vehicle (0.01% DMSO) in mitochondrial assay buffer for 1 h at room temperature. The supernatant fraction was then isolated from the mitochondrial pellet by centrifugation. Maximal release was determined by incubating mitochondria with 2% Triton X-100. Supernatants were subjected to anti-rat/mouse cytochrome c ELISA (R&D Systems) according to the manufacturer’s protocol. Fractional release was determined by the formula (R-R0)/(Rmax-R0), where R, R0, and Rmax correspond to the colorimetric intensities of wells treated with the indicated condition, vehicle, or Triton X-100.
Retroviral Transduction of Mouse Embryonic Fibroblasts
Human BAX constructs (wild-type, C126A) were cloned into the pMIG (MSCV-IRES-GFP) vector, and the presence of insert and the indicated mutation were confirmed by DNA sequencing. Transfection of the packaging cell line GPG-293 yielded amphotropic retroviral particles, which were collected by filtration and ultracentrifugation, and then used to reconstitute Bax−/−Bak−/− MEFs with the indicated BAX proteins, as described (Gavathiotis et al., 2008; Kim et al., 2006). The reconstituted MEFs were sorted for GFP-positivity over two rounds of flow cytometry to ensure comparable levels of expression. Transduction was verified by anti-BAX western analysis using the 2D2 antibody (Santa Cruz).
Cell Viability
Bax−/−Bak−/− MEFs reconstituted with wild-type or C126A mutant BAX were seeded at 5,000 cells per well in a white, flat-bottomed 96-well plate (Costar) and grown overnight, followed by the indicated co-treatments with ABT-737 (Cayman Chemical) and S63845 (MedChemExpress), or vehicle (0.1% or 0.05% DMSO, respectively). Viability was measured after 24 h using Cell-TiterGlo in accordance with the manufacturer’s protocol and expressed as a fraction of the corresponding vehicle treatment.
QUANTIFICATION AND STATISTICAL ANALYSIS
The number of technical and biological replicates for each experiment are indicated in the corresponding figure legend. Mean ± SD values were calculated using Prism software (Graphpad).
DATA AND CODE AVAILABILITY
The data supporting the findings of this study are available within the article and its supplementary materials. This study did not generate deposited datasets or code.
Supplementary Material
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse monoclonal anti-BAX 2D2 | Santa Cruz | Cat# SC-20067; RRID: AB_626726 |
| Sheep anti-mouse HRP | Bio-Rad | Cat# AAC10P; RRID: AB_321929 |
| Mouse monoclonal anti-β-Actin AC-15 | Sigma-Aldrich | Cat# A1978; RRID: AB_476692 |
| Bacterial and Virus Strains | ||
| One Shot BL21(DE3) Competent Cells | Invitrogen | Cat# C600003 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Rink Amide AM Resin LL 100–200 mesh | Millipore | Cat# 855120 |
| (S)-N-Fmoc-α-(4-pentenyl) alanine | Nagase & Co | Cat# 365023 |
| Grubbs Catalyst 1st Generation | Sigma-Aldrich | Cat# 579726 |
| 8-aminonapthalene-1,3,6-trisulfonic acid (ANTS) | Thermo Fisher Scientific | Cat# A350 |
| p-xylene-bis-pyridinium bromide (DPX) | Thermo Fisher Scientific | Cat# X1525 |
| IPTG | Gold Biotechnology | Cat# I2481C |
| Chitin bead resin | New England Biolabs | Cat# S6651S |
| Dithiothreitol (DTT) | Gold Biotechnology | Cat# DTT25 |
| 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine | Avanti | Cat# 850457C |
| 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine | Avanti | Cat# 850757C |
| L-α-phosphatidylinositol (Liver, Bovine) (sodium salt) | Avanti | Cat# 840042C |
| 1,2-dioleoyl-sn-glycero-3-phospho-L-serine (sodium salt) | Avanti | Cat# 840035C |
| Cardiolipin (Heart, Bovine) (sodium salt) | Avanti | Cat# 840012C |
| 3-((3-cholamidopropyl)dimethyl ammonio-1-propanesulfonate (CHAPS) | Thermo Fisher Scientific | Cat# 28300 |
| Trans-2-hexadecenal | Cayman Chemical | Cat# 17566 |
| Trans-2-pentenal | Sigma-Aldrich | Cat# 269255 |
| Trans-2-hexenal | Sigma-Aldrich | Cat# 132659 |
| Trans-2-heptenal | Sigma-Aldrich | Cat# 316504 |
| Trans-2-octenal | Sigma-Aldrich | Cat# 269956 |
| Trans-2-nonenal | Sigma-Aldrich | Cat# 255653 |
| Trans-2-decenal | Sigma-Aldrich | Cat# 30658 |
| Trans-2-undecenal | Sigma-Aldrich | Cat# W342300 |
| Trans-2-dodecenal | Sigma-Aldrich | Cat# W240206 |
| Trans-2-tridecenal | Sigma-Aldrich | Cat# W308218 |
| Hexadecanal | Cayman Chemical | Cat# 9001996 |
| Trans-2-hexadecenal alkyne | Cayman Chemical | Cat# 20714 |
| Cy5 hydrazide | Kerafast | Cat# FLP158 |
| Ammonium chloride (15N, 99%) | Cambridge Isotope Labs | Cat# NLM-467-PK |
| Factor XIII Protease from Aspergillus saitoi | Sigma Aldrich | Cat# P2143 |
| Pepsin from porcine gastric mucosa | Sigma Aldrich | Cat# P6887 |
| Tetrabutylammonium hydrogensulfate | Sigma-Aldrich | Cat# 791784 |
| Propargyl bromide solution 80 wt. % in toluene | Sigma-Aldrich | Cat# P51001 |
| Di-tert-butyl hydrazodiformate | Sigma-Aldrich | Cat# 140465 |
| Hydrogen chloride solution 4.0 M in dioxane | Sigma-Aldrich | Cat# 345547 |
| Azide-PEG4-biotin conjugate | Sigma-Aldrich | Cat# 762024 |
| Copper Sulfate | Sigma-Aldrich | Cat# PHR1477 |
| Tris[(1-benzyl-1H-1,2,3-triazol-4-yl)methyl]amine | Sigma-Aldrich | Cat# 678937 |
| (+)-Sodium L-ascorbate | Sigma-Aldrich | Cat# A7631 |
| S63845 | MedChemExpress | Cat# HY-100741 |
| ABT-737 | Cayman Chemical | Cat# 11501 |
| BIM SAHBA2 | Walensky Lab | N/A |
| Recombinant BAX WT | Walensky Lab | N/A |
| Recombinant BAX C62A | Walensky Lab | N/A |
| Recombinant BAX C126A | Walensky Lab | N/A |
| SGPL1 Human Recombinant Protein | Origene | Cat# TP308705 |
| Critical Commercial Assays | ||
| Q5 Site Directed Mutagenesis Kit | New England Biolabs | Cat# E0554S |
| CellTiter-Glo® Luminescent Cell Viability Assay | Promega | Cat# G7571 |
| Pierce BCA Protein Assay Kit | ThermoFisher Scientific | Cat# 23225 |
| Rat/Mouse Cytochrome c Quantikine ELISA Kit | R&D Systems | Cat# MCTC0 |
| Experimental Models: Cell Lines | ||
| Bax−/−Bak−/− SV40 MEF | ATCC | Cat# CRL-2913 |
| Bak−/− BAX WT SV40 MEF | Walensky Lab | N/A |
| Bak−/− BAX C126A SV40 MEF | Walensky Lab | N/A |
| Recombinant DNA | ||
| Plasmid PTYB1 | New England Biolabs | Cat#E6901 |
| Plasmid PTYB1_BAX_WT | Walensky Lab | N/A |
| Plasmid PTYB1_BAX_C62A | Walensky Lab | N/A |
| Plasmid PTYB1_BAX_C126A | Walensky Lab | N/A |
| Plasmid pMIG_BAX_WT | Genewiz | N/A |
| Plasmid pMIG | Addgene | 52107 |
| Plasmid pMIG_BAX_C126A | Walensky Lab | N/A |
| Software and Algorithms | ||
| Prism | Graphpad Software Inc. | https://www.graphpad.com/scientific-software/prism/ |
| Pymol | The PyMol Molecular Graphics System, Version 1.7.4.0 | Schrodinger, LLC |
| ProteinLynx Global Server (PLGS) 3.0.1 | Waters Corporation | https://www.waters.com/waters/en_US/ProteinLynx-Global-SERVER-(PLGS)/nav.htm?cid=513821&locale=en_US |
| DynamX 3.0 | Waters Corporation | https://www.waters.com/waters/library.htm?cid=511436&lid=134832928&locale=en_US |
| Other | ||
| Superdex 75 10/300 GL size exclusion column | GE Healthcare Life Sciences | Cat# 29148721 |
Highlights.
Trans-2-hexadecenal (t-2-hex) regulates BAX through direct covalent reaction
NMR and HXMS reveal the conformational consequences of BAX derivatization
Covalent lipidation potentiates BAX activation and requires C126
C126 lipidation is an apoptotic control point with therapeutic implications
ACKNOWLEDGMENTS
We thank G. Bird and T. Oo for BIM SAHBA2 production, J. Lee for performing intact MS analysis at the Dana-Farber Molecular Biology Core, C. Sheahan and G. Heffron for assistance with NMR spectroscopy, and E. Smith for graphics support and figure preparation. This research was supported by NIH grant R35CA197583 to L.D.W.; a Landry Cancer Biology Research Fellowship and Chleck Family Scholarship to D.T.C.; NIH grant R01GM101135 to J.R.E.; a research collaboration between J.R.E. and the Waters Corporation; and a National Science Foundation Predoctoral Fellowship and a Landry Cancer Biology Research Fellowship to M.W.M..
Footnotes
DECLARATION OF INTERESTS
L.D.W. is a scientific co-founder and shareholder in Aileron Therapeutics. The composition of a stapled peptide used in the study has been patented by the Dana-Farber Cancer Institute.
SUPPLEMENTAL INFORMATION
Supplemental Information can be found online at https://doi.org/10.1016/j.celrep.2020.02.057.
REFERENCES
- Barclay LA, Wales TE, Garner TP, Wachter F, Lee S, Guerra RM, Stewart ML, Braun CR, Bird GH, Gavathiotis E, et al. (2015). Inhibition of pro-apoptotic BAX by a noncanonical interaction mechanism. Mol. Cell 57, 873–886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bleicken S, Jeschke G, Stegmueller C, Salvador-Gallego R, Garcia-Saez AJ, and Bordignon E (2014). Structural model of active Bax at the membrane. Mol. Cell 56, 496–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boone CH, Grove RA, Adamcova D, Braga CP, and Adamec J (2016). Revealing oxidative damage to enzymes of carbohydrate metabolism in yeast: An integration of 2D DIGE, quantitative proteomics, and bioinformatics. Proteomics 16, 1889–1903. [DOI] [PubMed] [Google Scholar]
- Brahmbhatt H, Uehling D, Al-Awar R, Leber B, and Andrews D (2016). Small molecules reveal an alternative mechanism of Bax activation. Biochem. J 473, 1073–1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chipuk JE, Moldoveanu T, Llambi F, Parsons MJ, and Green DR (2010). The BCL-2 family reunion. Mol. Cell 37, 299–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chipuk JE, McStay GP, Bharti A, Kuwana T, Clarke CJ, Siskind LJ, Obeid LM, and Green DR (2012). Sphingolipid metabolism cooperates with BAK and BAX to promote the mitochondrial pathway of apoptosis. Cell 148, 988–1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czabotar PE, Westphal D, Dewson G, Ma S, Hockings C, Fairlie WD, Lee EF, Yao S, Robin AY, Smith BJ, et al. (2013). Bax crystal structures reveal how BH3 domains activate Bax and nucleate its oligomerization to induce apoptosis. Cell 152, 519–531. [DOI] [PubMed] [Google Scholar]
- Danial NN, Walensky LD, Zhang CY, Choi CS, Fisher JK, Molina AJ, Datta SR, Pitter KL, Bird GH, Wikstrom JD, et al. (2008). Dual role of proapoptotic BAD in insulin secretion and beta cell survival. Nat. Med 14, 144–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engen JR (2009). Analysis of protein conformation and dynamics by hydrogen/deuterium exchange MS. Anal. Chem 81, 7870–7875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frö hlich M, Dejanovic B, Kashkar H, Schwarz G, and Nussberger S (2014). S-palmitoylation represents a novel mechanism regulating the mitochondrial targeting of BAX and initiation of apoptosis. Cell Death Dis. 5, e1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gahl RF, He Y, Yu S, and Tjandra N (2014). Conformational rearrangements in the pro-apoptotic protein, Bax, as it inserts into mitochondria: a cellular death switch. J. Biol. Chem 289, 32871–32882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavathiotis E, Suzuki M, Davis ML, Pitter K, Bird GH, Katz SG, Tu HC, Kim H, Cheng EH, Tjandra N, and Walensky LD (2008). BAX activation is initiated at a novel interaction site. Nature 455, 1076–1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavathiotis E, Reyna DE, Davis ML, Bird GH, and Walensky LD (2010). BH3-triggered structural reorganization drives the activation of proapoptotic BAX. Mol. Cell 40, 481–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerra RM, Bird GH, Harvey EP, Dharia NV, Korshavn KJ, Prew MS, Stegmaier K, and Walensky LD (2018). Precision targeting of BFL1/A1 and an ATM co-dependency in human cancer. Cell Rep. 24, 3393–3403.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey EP, Seo HS, Guerra RM, Bird GH, Dhe-Paganon S, and Walensky LD (2018). Crystal structures of anti-apoptotic BFL-1 and its complex with a covalent stapled peptide inhibitor. Structure 26, 153–160.e154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu YT, and Youle RJ (1997). Nonionic detergents induce dimerization among members of the Bcl-2 family. J. Biol. Chem 272, 13829–13834. [DOI] [PubMed] [Google Scholar]
- Huhn AJ, Guerra RM, Harvey EP, Bird GH, and Walensky LD (2016). Selective covalent targeting of anti-apoptotic BFL-1 by cysteine-reactive stapled peptide inhibitors. Cell Chem. Biol 23, 1123–1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarugumilli GK, Choi JR, Chan P, Yu M, Sun Y, Chen B, Niu J, DeRan M, Zheng B, Zoeller R, et al. (2018). Chemical probe to identify the cellular targets of the reactive lipid metabolite 2-trans-hexadecenal. ACS Chem. Biol 13, 1130–1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H, Rafiuddin-Shah M, Tu HC, Jeffers JR, Zambetti GP, Hsieh JJ, and Cheng EH (2006). Hierarchical regulation of mitochondrion-dependent apoptosis by BCL-2 subfamilies. Nat. Cell Biol 8, 1348–1358. [DOI] [PubMed] [Google Scholar]
- Kim H, Tu HC, Ren D, Takeuchi O, Jeffers JR, Zambetti GP, Hsieh JJ, and Cheng EH (2009). Stepwise activation of BAX and BAK by tBID, BIM, and PUMA initiates mitochondrial apoptosis. Mol. Cell 36, 487–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korsmeyer SJ, Shutter JR, Veis DJ, Merry DE, and Oltvai ZN (1993). Bcl-2/Bax: a rheostat that regulates an anti-oxidant pathway and cell death.Semin. Cancer Biol 4, 327–332. [PubMed] [Google Scholar]
- Kotschy A, Szlavik Z, Murray J, Davidson J, Maragno AL, Le ToumelinBraizat G, Chanrion M, Kelly GL, Gong JN, Moujalled DM, et al. (2016). The MCL1 inhibitor S63845 is tolerable and effective in diverse cancer models. Nature 538, 477–482. [DOI] [PubMed] [Google Scholar]
- Laiken SL, Printz MP, and Craig LC (1969). Tritium-hydrogen exchange studies of protein models. I. Gramicidin S-A. Biochemistry 8, 519–526. [DOI] [PubMed] [Google Scholar]
- Lee S, Wales TE, Escudero S, Cohen DT, Luccarelli J, Gallagher CG, Cohen NA, Huhn AJ, Bird GH, Engen JR, and Walensky LD (2016). Allosteric inhibition of antiapoptotic MCL-1. Nat. Struct. Mol. Biol 23, 600–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovell JF, Billen LP, Bindner S, Shamas-Din A, Fradin C, Leber B, and Andrews DW (2008). Membrane binding by tBid initiates an ordered series of events culminating in membrane permeabilization by Bax. Cell 135, 1074–1084. [DOI] [PubMed] [Google Scholar]
- Marintchev A, Frueh D, and Wagner G (2007). NMR methods for studying protein-protein interactions involved in translation initiation. Methods Enzymol. 430, 283–331. [DOI] [PubMed] [Google Scholar]
- Matthews ML, He L, Horning BD, Olson EJ, Correia BE, Yates JR 3rd, Dawson PE, and Cravatt BF (2017). Chemoproteomic profiling and discovery of protein electrophiles in human cells. Nat. Chem 9, 234–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray V, Chen J, Huang Y, Li Q, and Wang J (2010). Preparation of very-high-yield recombinant proteins using novel high-cell-density bacterial expression methods. Cold Spring Harb. Protoc. 2010, pdb.prot5475. [DOI] [PubMed] [Google Scholar]
- Nie C, Tian C, Zhao L, Petit PX, Mehrpour M, and Chen Q (2008). Cysteine 62 of Bax is critical for its conformational activation and its proapoptotic activity in response to H2O2-induced apoptosis. J. Biol. Chem 283, 15359–15369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oltersdorf T, Elmore SW, Shoemaker AR, Armstrong RC, Augeri DJ, Belli BA, Bruncko M, Deckwerth TL, Dinges J, Hajduk PJ, et al. (2005). An inhibitor of Bcl-2 family proteins induces regression of solid tumours. Nature 435, 677–681. [DOI] [PubMed] [Google Scholar]
- Pagliari LJ, Kuwana T, Bonzon C, Newmeyer DD, Tu S, Beere HM, and Green DR (2005). The multidomain proapoptotic molecules Bax and Bak are directly activated by heat. Proc. Natl. Acad. Sci. USA 102, 17975–17980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitter K, Bernal F, Labelle J, and Walensky LD (2008). Dissection of the BCL-2 family signaling network with stabilized a-helices of BCL-2 domains.Methods Enzymol. 446, 387–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Printz MP, Williams HP, and Craig LC (1972). Evidence for the presence of hydrogen-bonded secondary structure in angiotensin II in aqueous solution. Proc. Natl. Acad. Sci. USA 69, 378–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sattler M, Liang H, Nettesheim D, Meadows RP, Harlan JE, Eberstadt M, Yoon HS, Shuker SB, Chang BS, Minn AJ, et al. (1997). Structure of Bcl-xL-Bak peptide complex: recognition between regulators of apoptosis. Science 275, 983–986. [DOI] [PubMed] [Google Scholar]
- Shi XE, Wales TE, Elkin C, Kawahata N, Engen JR, and Annis DA (2013). Hydrogen exchange-mass spectrometry measures stapled peptide conformational dynamics and predicts pharmacokinetic properties. Anal. Chem 85, 11185–11188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studier FW (2005). Protein production by auto-induction in high density shaking cultures. Protein Expr. Purif 41, 207–234. [DOI] [PubMed] [Google Scholar]
- Suzuki M, Youle RJ, and Tjandra N (2000). Structure of Bax: coregulation of dimer formation and intracellular localization. Cell 103, 645–654. [DOI] [PubMed] [Google Scholar]
- Tan C, Dlugosz PJ, Peng J, Zhang Z, Lapolla SM, Plafker SM, Andrews DW, and Lin J (2006). Auto-activation of the apoptosis protein Bax increases mitochondrial membrane permeability and is inhibited by Bcl-2. J. Biol. Chem 281, 14764–14775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walensky LD (2019). Targeting BAX to drug death directly. Nat. Chem. Biol 15, 657–665. [DOI] [PubMed] [Google Scholar]
- Walensky LD, and Gavathiotis E (2011). BAX unleashed: the biochemical transformation of an inactive cytosolic monomer into a toxic mitochondrial pore. Trends Biochem. Sci 36, 642–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walensky LD, Pitter K, Morash J, Oh KJ, Barbuto S, Fisher J, Smith E, Verdine GL, and Korsmeyer SJ (2006). A stapled BID BH3 helix directly binds and activates BAX. Mol. Cell 24, 199–210. [DOI] [PubMed] [Google Scholar]
- Wales TE, and Engen JR (2006). Hydrogen exchange mass spectrometry for the analysis of protein dynamics. Mass Spectrom. Rev. 25, 158–170. [DOI] [PubMed] [Google Scholar]
- Zha J, Weiler S, Oh KJ, Wei MC, and Korsmeyer SJ (2000). Posttranslational N-myristoylation of BID as a molecular switch for targeting mitochondria and apoptosis. Science 290, 1761–1765. [DOI] [PubMed] [Google Scholar]
- Zhang T, Hatcher JM, Teng M, Gray NS, and Kostic M (2019). Recent advances in selective and irreversible covalent ligand development and validation. Cell Chem. Biol. 26, 1486–1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong Q, Gao W, Du F, and Wang X (2005). Mule/ARF-BP1, a BH3-only E3 ubiquitin ligase, catalyzes the polyubiquitination of Mcl-1 and regulates apoptosis. Cell 121, 1085–1095. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data supporting the findings of this study are available within the article and its supplementary materials. This study did not generate deposited datasets or code.