Abstract
We investigated type 2 diabetes (T2D) genetic susceptibility via multi-ethnic meta-analysis of 228,499 cases and 1,178,783 controls in the Million Veteran Program, DIAMANTE, Biobank Japan, and other studies. We report 568 associations, including 286 autosomal, 7 X chromosomal, and 25 identified in ancestry-specific analyses that were previously unreported. Transcriptome-wide association analysis detected 3,568 T2D-associations with genetically predicted gene expression in 687 novel genes; of these, 54 are known to interact with FDA-approved drugs. A polygenic risk score was strongly associated with increased risk of T2D-related retinopathy and modestly associated with chronic kidney disease (CKD), peripheral artery disease (PAD), and neuropathy. We investigated the genetic etiology of T2D-related vascular outcomes in MVP and observed statistical SNP-T2D interactions at 13 variants, including coronary heart disease, CKD, PAD, and neuropathy. These findings may help to identify potential therapeutic targets for T2D and genomic pathways that link T2D to vascular outcomes.
Introduction
Type 2 diabetes mellitus (T2D), a leading cause of morbidity globally, is projected to affect up to 629 million people by 20451. People with T2D are at increased risk of developing a wide range of macro- and microvascular outcomes2, and there are large disparities in prevalence, severity and co-morbidities across global populations. Over 400 common variants have been identified that confer disease susceptibility3,4, yet because most studies have been performed in cohorts of European or Asian ancestry, the impact of these variants across all ethnic needs to be quantified. Identifying genetic factors and genes underlying T2D-related complications could inform clinical management strategies, including patient stratification or optimizing study design of randomized controlled trials. The lack of large, multi-ethnic richly phenotyped cohorts linked to genetic data has made it difficult to address these questions.
We conducted a multi-ethnic association study of T2D risk comprised of 228,499 T2D cases and 1,178,783 controls of European, African American, Hispanic, South Asian, and East Asian ancestry. We investigated the association of a T2D polygenic risk score with major T2D-related macrovascular outcomes (coronary heart disease (CHD), ischemic stroke, and peripheral artery disease (PAD)) and three microvascular diseases (chronic kidney disease (CKD), retinopathy and neuropathy) in the Million Veteran Program (MVP)5. Subsequently, we conducted a genome-wide SNP-T2D interaction analysis in MVP to identify genetic variants where the effect of SNP on the vascular outcome depends on the context of T2D presence. We also performed association analyses of genetically predicted expression levels and expression quantitative trait-T2D colocalization analyses to identify the effects of gene-tissue pairs that influence T2D risk through inter-individual variation in expression.
This study complements prior genetic studies of T2D through use of large-scale clinical data in conjunction with polygenic scores, evaluation of context specificity for genetic effects on T2D vascular sequelae, and describing the regulatory circuits that influence T2D risk.
Results
Study populations.
We performed a genome-wide, multi-ethnic T2D-association analysis (228,499 cases and 1,178,783 controls) encompassing five ancestral groups (Europeans, African Americans, Hispanics, South Asians and East Asians) by meta-analyzing genome-wide association study (GWAS) summary statistics derived from the Million Veteran Program (MVP)5 and other studies with non-overlapping participants: DIAMANTE Consortium3, Penn Medicine Biobank6, Pakistan Genomic Resource7, Biobank Japan4, Malmö Diet and Cancer Study8, Medstar9, and PennCath9 (Methods and Supplementary Tables 1 and 2). MVP participants (n = 273,409) comprised predominantly male subjects (91.6%) and were classified as Europeans (72.1%), African Americans (19.5%), Hispanics (7.5%), and Asians (0.9%, Supplementary Table 3).
Single-variant autosomal analyses.
We identified 558 independent sentinel SNPs (286 previously unreported, >500 kb and r2 LD < 0.05 from a previous reports; see Methods)3,4,10,11 associated with T2D (Fig. 1, Table 1, Supplementary Tables 4–8, and Extended Data Fig. 1). Twenty-one additional SNPs were associated at genome-wide significance in ancestry-specific analysis of Europeans only (Supplementary Table 6). We found that novel loci had smaller magnitudes of effect (average beta regression coefficient of 0.032 ± 0.012 per allele) than previously established SNPs (average beta of 0.054 ± 0.045 per allele, Supplementary Table 5), presumably resulting from enhanced power to discover weaker effects due to the large sample size and ancestral diversity. Genome-wide chip heritability analysis explained 19% of T2D risk on a liability scale3.
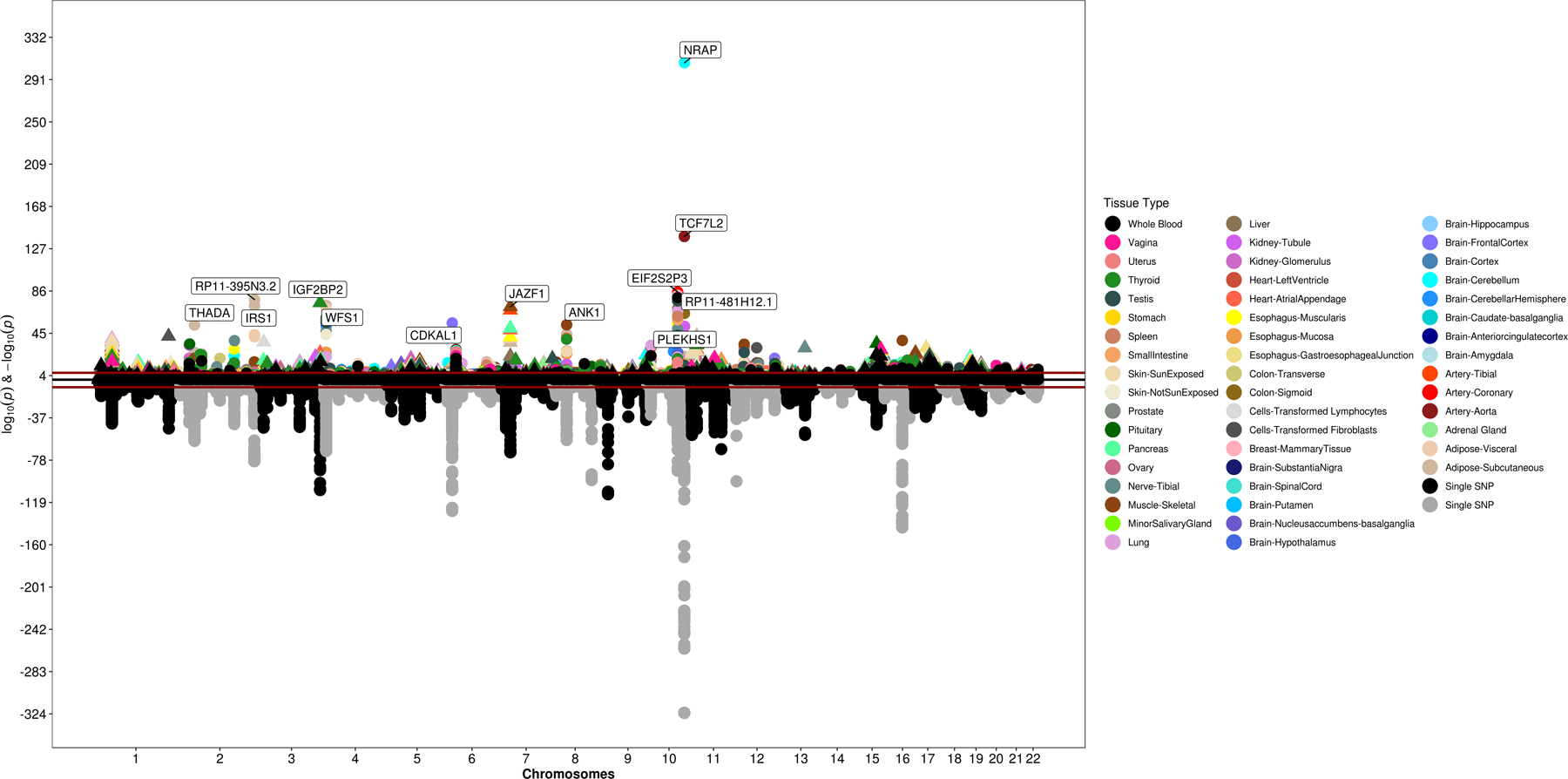
Figure 1 |. Trans-ancestry GWAS meta-analysis identifies 318 loci associated with T2D.
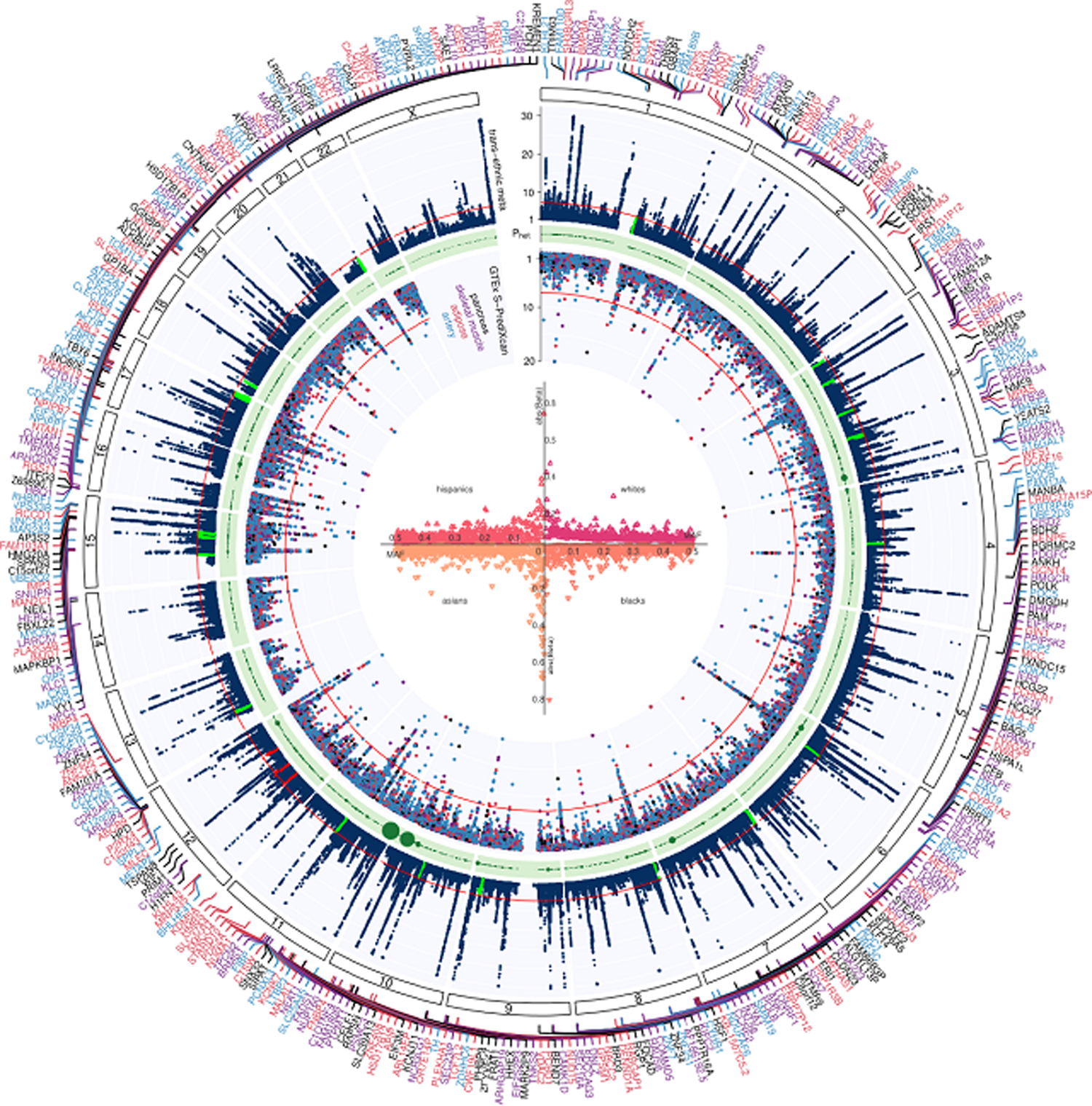
The graph represents a circos plot performed in 228,499 T2D cases and 1,178,783 controls. The outer track corresponds to −log10 (P) for association with T2D in the trans-ethnic meta-analysis using a fixed-effects model with inverse-variance weighting of log odds ratios (y-axis truncated at 30), by chromosomal position. The red line indicates genome-wide significance (P = 5.0 × 10−8). Purple gene labels indicated genes identified in skeletal muscle eQTLs by S-PrediXcan analysis, red-labeled gene names in adipose eQTLs, black-labeled gene names in pancreas eQTLs, and blue-labeled gene names were identified in eQTLs from arteries. The green band corresponds to measures of heterogeneity related to the index SNPs associated with T2D that were generated using the Cochran’s Q statistic. Dot sizes are proportional to I2 or ancestry-related heterogeneity. The inner track corresponds to −log10(P) for association with skeletal muscle, adipose, pancreas, and artery tissue eQTLs from S-PrediXcan analysis (y-axis truncated at 20), by chromosomal position. The red line indicates genome-wide significance (P = 5.0 × 10−8). Inset, effects of all 318 index SNPs on T2D by minor allele frequency, stratified and colored by ancestral group.
Table 1 |.
T2D locus discovery in African Americans
| Description | Lead SNP | RSID | EA | NEA | EAF | Beta | SE | P | n | n Cojo | Established SNP |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Novel AA | chr12:38710523 | rs7315028 | G | A | 0.882 | 0.124 | 0.022 | 1.5E-08 | 56,150 | 1 | - |
| chr12:57968738 | rs11172254 | G | A | 0.817 | 0.097 | 0.017 | 1.8E-08 | 56,150 | 1 | - | |
| chr12:88338461 | rs10745460 | T | A | 0.660 | 0.079 | 0.014 | 3.7E-08 | 56,150 | 0 | - | |
| Novel TE | chr7:50887174 | rs7781440 | C | T | 0.284 | −0.086 | 0.015 | 5.3E-09 | 56,150 | 0 | - |
| chr12:80985872 | rs1528287 | G | T | 0.059 | −0.494 | 0.080 | 8.2E-10 | 56,150 | 1 | - | |
| Established | chr3:123065778 | rs11708067 | G | A | 0.151 | −0.118 | 0.018 | 2.3E-11 | 56,150 | 0 | chr3:123082398 |
| chr3:185534482 | rs9859406 | G | A | 0.257 | −0.115 | 0.015 | 5.7E-14 | 56,150 | 0 | chr3:185829891 | |
| chr5:55807370 | rs464605 | C | T | 0.429 | −0.077 | 0.013 | 1.1E-09 | 56,150 | 0 | chr5:55860781 | |
| chr6:39016636 | rs10305420 | C | T | 0.920 | 0.142 | 0.025 | 8.5E-09 | 56,150 | 0 | chr6:39282371 | |
| chr7:15064896 | - | G | T | 0.565 | 0.101 | 0.013 | 2.7E-15 | 56,150 | 0 | chr7:15060429 | |
| chr7:28180556 | rs864745 | C | T | 0.257 | −0.083 | 0.014 | 1.1E-08 | 56,150 | 0 | chr7:28198677 | |
| chr7:44185088 | rs2908274 | G | A | 0.359 | −0.089 | 0.014 | 5.4E-11 | 56,150 | 1 | chr7:44266184 | |
| chr8:41510260 | rs12550613 | G | C | 0.310 | −0.114 | 0.014 | 5.5E-16 | 56,150 | 0 | chr8:41537318 | |
| chr8:118166327 | rs60461843 | T | A | 0.939 | 0.172 | 0.028 | 1.3E-09 | 56,150 | 1 | chr8:118024315 | |
| chr9:139241595 | rs28562046 | G | C | 0.709 | 0.080 | 0.014 | 2.8E-08 | 56,150 | 0 | chr9:139737088 | |
| chr10:114758349 | rs7903146 | C | T | 0.706 | −0.226 | 0.014 | 5.6E-60 | 56,150 | 0 | chr10:114871594 | |
| chr11:2691500 | rs231361 | G | A | 0.656 | −0.080 | 0.013 | 2.2E-09 | 56,150 | 2 | chr11:2717680 | |
| chr11:2858546 | rs2237897 | C | T | 0.908 | 0.143 | 0.024 | 2.2E-09 | 56,150 | 1 | chr11:2717680 | |
| chr12:66215214 | rs2583938 | T | A | 0.197 | −0.123 | 0.018 | 3.3E-12 | 56,150 | 0 | chr12:66358347 | |
| chr15:77776498 | rs952471 | G | C | 0.534 | 0.077 | 0.013 | 4.2E-09 | 56,150 | 0 | chr15:77339496 | |
| chr16:53811788 | rs62033400 | G | A | 0.102 | 0.151 | 0.021 | 6.1E-13 | 56,150 | 1 | chr16:53758720 |
Association between genetic variants and T2D in African Americans in MVP was assessed through logistic regression assuming an additive model of variants with MAF > 1%. A meta-analysis was performed using in a fixed-effects model with inverse-variance weighting of log odds ratios. Variants were considered genome-wide significant if they passed the conventional P-value threshold of 5 × 10−8. AA, African American; TE, trans-ethnic; SNP, single nucleotide polymorphisms; RSID, RefSNP identification number; EA, effect allele; NEA, non-effect allele; EAF, effect allele frequency; Beta, effect estimate; SE, standard error; n, sample size; n Cojo, additional number of conditionally independent variants identified at the respective locus (and listed in Supplementary Table 12).
In analysis focused on African American participants (Table 1), we observed a total of 21 loci associated with T2D susceptibility at genome-wide significance, 16 of which were in strong LD with established T2D variants. Three variants were novel and their effects on T2D appeared specific to African Americans. Single variant analysis in the Hispanics subset identified two associated SNPs, both of which tagged previously reported T2D loci (Supplementary Table 7). No novel associations were observed among the individuals of Asian ancestry (Supplementary Table 8).
Polygenicity and population stratification.
To evaluate whether the observed genomic inflation is due to the polygenic nature of T2D or due to underlying population stratification, linkage disequilibrium score regression (LDSC)12 was used in Europeans and Asians to compare lambda genomic control (GC)13 and LDSC intercept (Methods). In Asians, a total of 1,077,427 SNPs were analyzed, resulting in a lambda GC of 1.342 and intercept of 1.094 (se = 0.012). In Europeans, 1,198,787 SNPs were analyzed resulting in a lambda GC of 1.863 and intercept of 1.139 (se = 0.016). Admixture-adjusted LDSC14 was used in African Americans and Hispanics. A total of 945,603 SNPs were analyzed in African Americans, with lambda GC of 1.180 and intercept of 1.048 (se = 0.007). For Hispanics, 1,077,427 SNPs were analyzed with lambda GC of 1.093 and intercept of 1.091 (se = 0.113). Except perhaps for Hispanics (where the estimated error on the intercept is large), these results suggest that a substantial part of the observed inflation these populations is due to T2D polygenicity.
X chromosome analyses.
In trans-ethnic analysis of the X chromosome, we identified a total of 10 association signals for T2D, of which 7 were novel (Table 2, Supplementary Table 9, and Extended Data Fig. 2). A European-restricted analysis identified four loci on the X chromosome, all of which were identified in the trans-ethnic meta-analysis. One novel X chromosome locus was associated with T2D specifically in African Americans. We note that one novel trans-ethnic association was identified near the androgen receptor (AR) gene and was in strong LD with a previously reported variant (rs4509480) previously shown to associate with male-pattern baldness (EUR r2 = 0.98, rs200644307).
Table 2 |.
T2D chromosome X analysis (overall results)
| Population | Lead SNP | EA | NEA | EAF | Novel | Literature SNP | Nearest gene | n Cases | n Controls | Beta | SE | P |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Trans-ethnic | chrX:19497290 | A | G | 0.968 | 1 | - | MAP3K15 | 102,683 | 170,726 | 0.131 | 0.023 | 1.4E-08 |
| chrX:20009166 | T | C | 0.323 | 1 | - | CXorf23;MAP7D2 | 102,683 | 170,726 | 0.058 | 0.010 | 7.9E-09 | |
| chrX:31851610 | T | C | 0.343 | 1 | - | DMD | 102,683 | 170,726 | 0.047 | 0.009 | 3.5E-08 | |
| chrX:56902211 | A | T | 0.612 | 0 | X:57170781 | SPIN2A;FAAH2 | 102,683 | 170,726 | −0.069 | 0.010 | 1.9E-12 | |
| chrX:66168667 | A | G | 0.277 | 1 | - | AR;EDA2R | 102,683 | 170,726 | 0.082 | 0.011 | 1.9E-13 | |
| chrX:109888390 | A | C | 0.364 | 1 | - | RGAG1;CHRDL1 | 102,683 | 170,726 | −0.048 | 0.008 | 7.7E-09 | |
| chrX:117955250 | T | C | 0.231 | 0 | X:117915163 | IL13RA1 | 102,683 | 170,726 | 0.077 | 0.010 | 4.1E-15 | |
| chrX:124390172 | T | C | 0.853 | 1 | - | TENM1 | 102,683 | 170,726 | −0.075 | 0.013 | 9.0E-09 | |
| chrX:135859359 | C | G | 0.407 | 1 | - | ARHGEF6 | 102,683 | 170,726 | −0.049 | 0.008 | 7.3E-09 | |
| chrX:153882606 | C | G | 0.026 | 0 | X:152908887 | FAM58A;DUSP9 | 102,683 | 170,726 | −0.486 | 0.026 | 3.0E-78 | |
| European | chrX:56759371 | T | G | 0.218 | 0 | X:57170781 | SPIN2A;FAAH2 | 69,869 | 127,197 | 0.069 | 0.013 | 1.7E-08 |
| chrX:66316809 | G | A | 0.290 | 1 | - | EDA2R | 69,869 | 127,197 | 0.077 | 0.013 | 3.4E-09 | |
| chrX:117877437 | A | G | 0.223 | 0 | X:117915163 | IL13RA1 | 69,869 | 127,197 | 0.118 | 0.013 | 5.5E-20 | |
| chrX:152898928 | C | A | 0.247 | 0 | X:152908887 | FAM58A;DUSP9 | 69,869 | 127,197 | −0.163 | 0.012 | 7.9E-46 | |
| African | chrX:67255974 | C | T | 0.189 | 1 | - | AR;OPHN1 | 23,305 | 30,140 | 0.104 | 0.019 | 3.4E-08 |
| American | chrX:132597984 | C | T | 0.282 | 1 | - | GPC3;GPC4 | 23,305 | 30,140 | 0.135 | 0.024 | 1.4E-08 |
| chrX:153882606 | C | G | 0.026 | 0 | X:152908887 | G6PD | 23,305 | 30,140 | −0.500 | 0.027 | 1.6E-76 |
A sex-stratified (male, female) ancestry-separated (European, African American, Hispanic, Asian) analysis was performed with dosage (number of X-chromosome copies) as the independent variable and T2D as the outcome. Covariates included age and first 10 PCs of ancestry. The ancestry-specific sex-stratified results are presented in Supplementary Table 9. Output from ancestry-separated male and female analyses were then meta-analyzed using a fixed-effects model with inverse-variance weighting of log odds ratios and are shown here. For the trans-ethnic meta-analysis, the ancestry-specific sex-meta-analyzed was additionally meta-analyzed using a fixed-effects model with inverse-variance weighting of log odds ratios. Variants were considered genome-wide significant if they passed the conventional P-value threshold of 5 × 10−8. SNP, single nucleotide polymorphisms; EA, effect allele; NEA, non-effect allele; EAF, effect allele frequency; Beta, effect estimate; SE, standard error; n cases, total number of T2D cases; n controls, total number of unaffected controls.
Effect heterogeneity between Europeans and African Americans.
While at most loci we found no evidence for heterogeneity of effect estimates between Europeans and African Americans, we did observe that 44 (7.9%) variants had significantly different effect size estimates between the two groups (Supplementary Table 10). Remarkably, four loci near SLC30A8, PTPRQ, GRB10, and COLB showed higher effect sizes for T2D at stronger levels of significance in African Americans compared with Europeans. Of these loci, associations with loss-of-function variants in SLC30A8 were previously reported in Europeans, African Americans and South Asians.
Secondary signal analysis.
We detected a total of 233 conditionally independent SNPs flanking 49 novel and 108 previously reported lead SNPs in Europeans (Supplementary Tables 11 and 12). We observed no novel conditionally independent variants in participants of South Asian, East Asian and Hispanic ancestry.
Fine mapping of lead SNPs with coding variants.
To identify coding variants that may drive the association between the lead SNPs and T2D risk, we investigated predicted loss-of-function (pLoF) and missense variants near the identified T2D lead variants from the European-specific T2D summary statistics (Supplementary Table 13). We identified two pLoF (LPL and ANKDD1B) and 45 missense variants in 47 genes that were in LD with at least one of the T2D lead SNPs (r2 > 0.5, MVP reference panel in Europeans) and were associated at P < 1.0 × 10−4. Of the 56 pLoF and missense variants, 14 missense variants were found to be the sentinel T2D SNPs and 19 variants were in LD with novel lead SNPs, and 37 variants were previously reported.
Genome-wide coding variant association analysis.
We additionally performed a genome-wide screen of all pLoFs and missense variants (not bound by proximity to sentinel T2D lead variants) to enumerate potentially T2D genes not captured by common variant tags (Supplementary Table 14). We identified one additional pLoF variant in CCHCR1, whereas 37 novel missense variants were associated with T2D at P < 5 × 10−8.
Rare coding variant PheWAS.
We next performed a PheWAS of the three pLoF variants associated with T2D in MVP participants of European ancestry, UK Biobank data, and Biobank Japan separately (Table 3). These loci included ANKDD1B p.Trp480* (rs34358), CCHCR1 p.Trp78* (rs3130453), and LPL p.*474Ser (rs328), and they were significantly associated with metabolic and inflammatory conditions. Klarin et al. previously reported pheWAS associations with for LPL p.*474Ser with dyslipidemia, coronary atherosclerosis and other chronic ischemic heart disease in MVP, and lipid and cardiometabolic associations for this variant were also observed in Biobank Japan and UK Biobank. In MVP, ANKDD1B p.Trp480* was associated with dyslipidemia, hypercholesterolemia, and diabetic neurological manifestations. In Biobank Japan, this variant was a range of blood and immune cell traits, whereas in UK Biobank, the SNP was associated with metabolic and anthropometric traits. In MVP and UKBB, CCHCR1 p.Trp78* was associated with a battery of autoimmune traits, and in Biobank Japan, this variant was associated with total cholesterol, LDL-C, BMI, NK cells, and Na electrolytes.
Table 3 |.
PheWAS of two pLoF variants in MVP participants of European ancestry
| Gene | RSID | Amino acid change | PheWAS phenotype | P | n Cases | n Controls | OR | 95%CIlower | 95%CIupper |
|---|---|---|---|---|---|---|---|---|---|
| ANKDD1B | rs34358 | p.Trp480* | Diabetes mellitus | 1.04E-06 | 62,930 | 104,442 | 0.96 | 0.95 | 0.98 |
| Type 2 diabetes | 1.36E-06 | 62,531 | 104,442 | 0.96 | 0.95 | 0.98 | |||
| T2D with neurological manifestations | 1.63E-05 | 14,159 | 104,442 | 0.94 | 0.92 | 0.97 | |||
| Disorders of lipid metabolism | 5.03E-08 | 141,535 | 41,406 | 1.05 | 1.03 | 1.07 | |||
| Hyperlipidemia | 4.66E-08 | 141,408 | 41,406 | 1.05 | 1.03 | 1.07 | |||
| Hypercholesterolemia | 2.33E-06 | 32,008 | 41,406 | 1.06 | 1.03 | 1.08 | |||
| CCHCR1 | rs3130453 | p.Trp78* | Diabetes mellitus | 4.26E-05 | 62,930 | 104,442 | 0.97 | 0.96 | 0.98 |
| Type 1 diabetes | 3.99E-07 | 6,566 | 104,442 | 0.91 | 0.88 | 0.95 | |||
| Type 2 diabetes | 3.96E-05 | 62,531 | 104,442 | 0.97 | 0.96 | 0.98 | |||
| Epistaxis or throat hemorrhage | 1.96E-05 | 2,751 | 110,902 | 1.12 | 1.07 | 1.19 | |||
| Celiac disease | 2.72E-19 | 418 | 124,470 | 0.52 | 0.45 | 0.60 | |||
| Microscopic hematuria | 1.83E-05 | 4,078 | 147,054 | 1.1 | 1.05 | 1.15 | |||
| Psoriatic arthropathy | 7.82E-10 | 1,077 | 140,876 | 0.76 | 0.70 | 0.83 |
The pLoF variants were tested using logistic regression adjusting for age, sex, and 10 principal components in an additive effects model using the PheWAS R package in R v3.2.0. Phenotypes were required to have a case count over 25 in order to be included in the PheWAS, and a multiple testing thresholds for statistical significance was set to the Bonferroni-corrected P-value threshold of 2.8 × 10−5. pLOF, predicted loss-of-function; RSID, RefSNP identification number; PheWAS, phenome-wide association study; n Cases, number of cases with PheWAS phenotype; n Controls, number of unaffected controls for the respective PheWAS phenotype; OR, odds ratio; CI, confidence interval; T2D, type 2 diabetes.
Transcriptome-wide association analyses.
We next used common variants from the European T2D GWAS meta-analysis to evaluate the association of genetically predicted gene expression levels with T2D risk across 52 tissues including kidney and islet cells using S-PrediXcan (Supplementary Table 15 and Extended Data Fig. 3). We identified 4,468 statistically significant gene-tissue combination pairs genetically predictive of T2D risk, of which 4,211 transcript eQTLs were in LD (r2 > 0.5) with T2D signals. We identified 873 genes in this analysis that would not have been identified by nearest-gene annotation alone. The strongest gene-tissue combination signals were for NRAP in the cerebellum and TCF7L2 in the aortic artery.
We then used COLOC to identify the subset of significant genes where there was a high posterior probability that the set of model SNPs in the S-PrediXcan analysis for each gene were associated with gene expression and with T2D. This analysis refined the results of the transcriptome-wide association scan and excluded some results that might be the consequence of LD between causal SNPs for gene expression and T2D. We detected 3,166 gene-tissue pairs where there was statistically significant association with T2D risk and high posterior probability (P4 > 0.8) of colocalization, covering a total of 695 distinct genes. When comparing the 804 genes to the GWAS catalog mapped and reported genes for all prior studies of diabetes or diabetes complications, 687 had not been previously reported. Hypergeometric enrichment analysis showed that most enriched gene expression signals were in cervical spinal cord, basal ganglia and glomerular kidney (Supplementary Table 16).
Assessment of gene–drug relationships.
Of the 695 genes identified in S-PrediXcan analyses, 54 genes have documented interactions with a total of 283 FDA-approved drugs and chemical compounds that do not have an indication for T2D treatment or reported adverse drug events (ADEs) in diabetic patients using the SIDER database of drugs and side effects15. Using the Drug-Gene Interaction Database (DGIdb version 3.0), a total of 322 gene-drug combinations were identified for which it is predicted to modulate blood glucose based on direction of effect on T2D risk with increasing gene expression and drug action (activator or inhibitor, Supplementary Table 17). Gene-drug combinations included several established T2D loci such as KCNJ11 targeted by 15 compounds (e.g. sulfonylureas, glinides, and p-glycoprotein inhibitors), SCNA3 targeted by 57 compounds (e.g. anti-arrythmetics, anti-epileptics), PIK3CB targeted by 46 compounds (e.g. cancer drugs), ACE targeted by 36 compounds (e.g. angiotensin-converting enzyme (ACE) inhibitors), HMGCR targeted by 18 compounds (e.g. HMG-CoA reductase inhibitors), PIK3C2A targeted by 15 compounds (anti-cancer drugs), F2 targeted by 11 compounds (anti-coagulants), and BLK targeted by 9 compounds (protein kinase inhibitors).
Tissue-specific and epigenetic enrichment of T2D heritability.
To understand the contribution of disease-associated tissues is to T2D heritability, we performed tissue-specific analysis using LDSC16 (Supplementary Table 18). The strongest associations were observed in genomic annotation surveyed in pancreas and pancreatic islets (e.g., pancreatic islets H3K27ac, pancreas DNase, etc., P < 0.001). We additionally tested for enrichment of epigenetic features using GREGOR17, which compares overlap of T2D-associated loci variants relative to control variants matched for number of LD proxies, allele frequency, and gene proximity17 (Supplementary Tables 19–21). Similar to the results from LDSC, 8 of the top 10 associated hits map to the pancreas, including H3K27ac, pancreatic islets H3K27ac, and pancreatic islets activated enhancer, among others.
Pathway and functional enrichment analysis.
To explore whether our results recapitulate the pathophysiology of T2D, we performed gene-set enrichment analysis with all the variants using DEPICT (P < 1 × 10−5, Supplementary Table 22). MeSH-based analysis showed that several different adipose tissues and sites were enriched (e.g., abdominal subcutaneous fat, white adipose tissue, etc.). Finally, DEPICT analysis showed that the most significant gene-set involved the AKT2 subnetwork, lung cancer, the GAB1 signalosome, protein kinase binding, signal transduction, and EGFR signaling (Supplementary Tables 23 and 24).
Genetic correlation between T2D and other phenotypes.
Genome-wide genetic correlations of T2D were calculated with a total of 774 complex traits and diseases by comparing allelic effects using LD score regression with the European-specific T2D summary statistics (Methods). A total of 270 significant associations were observed (P < 5 × 10−8, Supplementary Table 25). The strongest positive correlations were observed with waist circumference, overall health, BMI, and fat mass of arms, legs, body and trunk, hypertension, coronary artery disease, dyslipidemia, alcohol intake, wheezing, and cigarette smoking. There was also a strong negative correlation with years of education.
T2D-related vascular outcomes.
We next investigated SNP-T2D interaction effects associated with T2D-related vascular outcomes among European-descent MVP participants (P < 5 × 10−8; Methods, Table 4, and Supplementary Table 26). The analysis included a total case count of 67,403 for CKD, 56,285 for CHD, 35,882 for PAD, 11,796 for acute ischemic stroke, 13,881 for retinopathy, and 40,475 for neuropathy. We identified several genome-wide significant interactions where the genetic associations with T2D-related vascular outcomes were modified by T2D (Table 4 and Supplementary Table 26). We identified two loci for CHD (rs1831733 in 9p21 and rs602633 near SORT1) and one for CKD (rs34857077 in UMOD) for which the difference in the effect estimates between T2D strata was genome-wide significant (P < 5 × 10−8) and at least one T2D-stratum was genome-wide significant. We identified one locus for CHD (rs71039916 near PDE3A), one for CKD (rs2177223 near TENM3), one for PAD (rs3104154 in PTDSS1), one for neuropathy (rs78977169 near NRP2), four for retinopathy (rs76754787 near GJA8, rs10733997 in SVILP2, rs2255624 near SLC18A2, and rs4132670 in TCF7L2) and two for acute ischemic stroke (rs491203 near TMEM51, and rs2134937 near TRIQK) that showed genome-wide significance for difference in effect estimates between the T2D strata and nominal significance (P < 0.001) for at least one T2D stratum.
Table 4 |.
Genome-wide interaction analysis of vascular and non-vascular complications (overall results)
| Outcome type | Outcome | SNP | RSID | NEA | EA | EAF | P for interaction | Nearest gene |
|---|---|---|---|---|---|---|---|---|
| Vascular | CHD | chr9:22076071 | rs1831733 | T | C | 0.482 | 1.6E-13 | CDKN2B;CDKN2A |
| chr1:109821511 | rs602633 | G | T | 0.216 | 4.4E-10 | SORT1 | ||
| chr12:20231526 | rs71039916 | TCTTA | T | 0.034 | 8.2E-09 | PDE3A | ||
| AIS | chr1:15429233 | rs491203 | G | A | 0.057 | 7.6E-09 | TMEM51 | |
| chr8:94056373 | rs2134937 | T | C | 0.049 | 3.3E-08 | TRIQK | ||
| PAD | chr8:97331026 | rs3104154 | C | T | 0.044 | 3.0E-08 | PTDSS1 | |
| Non-vascular | Retinopathy | chr1:146606059 | rs76754787 | ATT | AT | 0.030 | 1.2E-11 | GJA8 |
| chr10:30992882 | rs10733997 | A | G | 0.037 | 9.7E-09 | SVILP2 | ||
| chr10:119646217 | rs2255624 | T | G | 0.032 | 1.6E-08 | SLC18A2 | ||
| chr10:114767771 | rs4132670 | G | A | 0.319 | 2.1E-08 | TCF7L2 | ||
| CKD | chr16:20356012 | rs34857077 | G | GA | 0.237 | 6.4E-19 | UMOD | |
| chr4:181816870 | rs2177223 | T | C | 0.038 | 2.8E-08 | TENM3 | ||
| Neuropathy | chr2:206668118 | rs78977169 | CATA | C | 0.023 | 3.4E-08 | NRP2 |
The analysis included a total case count of 67,403 for CKD, 56,285 for CHD, 35,882 for PAD, 11,796 for AIS, 13,881 for retinopathy, and 40,475 for neuropathy. Results stratified by T2D presence (yes or no) are presented in Supplementary Table 26. A logistic regression analysis was performed among MVP participants of European ancestry, where the respective outcome was tested with SNP, T2D, SNPxT2D, age, gender, and 10 PCs as covariates. P-value for interaction between SNP and T2D are noted in the column labeled P for interaction. Variants were considered to show a statistically different effect between people with and without T2D if the P-value for interaction was genome-wide significant (P < 5 × 10−8) and at least one T2D-stratum showed nominal significance (P < 0.001, Supplementary Table 26). RSID, RefSNP identification number; CHD, coronary heart disease; AIS, acute ischemic stroke; PAD, peripheral artery disease; CKD, chronic kidney disease; NEA, non-effect allele; EA, effect allele; EAF, effect allele frequency.
Polygenic risk scores and T2D-related vascular outcomes.
Genome-wide polygenic risk scores (gPRS) for T2D were calculated in Europeans based on the T2D effect estimates from the previously reported DIAMANTE consortium3 and then categorized into deciles (Tables 5 and 6). As expected, participants with the highest T2D gPRS scores (90–100% T2D gPRS percentile) showed the highest risk for T2D (OR = 5.21, 95% CI 4.94–5.49, Extended Data Fig. 5) when compared to the reference group (0–10% T2D gPRS percentile) in a cross-sectional study design.
Table 5 |.
Polygenic risk scores and vascular outcomes
| Outcome type | Outcome | T2D PRSdecile | n Cases | n Controls | OR | 95%CI lower | 95%CI upper | P | P forlinear trend |
|---|---|---|---|---|---|---|---|---|---|
| Vascular | Coronary heart disease | 0–10% | 2,913 | 3,924 | 1.00 | Ref | Ref | - | 0.636 |
| 10–20% | 2,940 | 3,924 | 1.01 | 0.92 | 1.12 | 0.811 | |||
| 20–30% | 2,958 | 3,924 | 0.98 | 0.89 | 1.08 | 0.742 | |||
| 30–40% | 2,934 | 3,924 | 0.99 | 0.90 | 1.09 | 0.835 | |||
| 40–50% | 2,988 | 3,924 | 1.01 | 0.92 | 1.11 | 0.801 | |||
| 50–60% | 3,001 | 3,924 | 0.98 | 0.90 | 1.08 | 0.744 | |||
| 60–70% | 2,977 | 3,924 | 1.01 | 0.92 | 1.10 | 0.887 | |||
| 70–80% | 2,916 | 3,924 | 1.02 | 0.93 | 1.12 | 0.632 | |||
| 80–90% | 3,032 | 3,924 | 0.96 | 0.88 | 1.05 | 0.391 | |||
| 90–100% | 3,038 | 3,924 | 1.03 | 0.94 | 1.12 | 0.537 | |||
| Acute ischemic stroke | 0–10% | 555 | 6,027 | 1.00 | Ref | Ref | - | 0.070 | |
| 10–20% | 563 | 6,027 | 0.90 | 0.76 | 1.07 | 0.238 | |||
| 20–30% | 583 | 6,027 | 0.98 | 0.83 | 1.15 | 0.782 | |||
| 30–40% | 619 | 6,027 | 0.98 | 0.84 | 1.15 | 0.821 | |||
| 40–50% | 530 | 6,027 | 0.99 | 0.85 | 1.16 | 0.924 | |||
| 50–60% | 576 | 6,027 | 0.99 | 0.85 | 1.16 | 0.941 | |||
| 60–70% | 645 | 6,027 | 0.97 | 0.83 | 1.13 | 0.672 | |||
| 70–80% | 590 | 6,027 | 1.04 | 0.90 | 1.20 | 0.611 | |||
| 80–90% | 558 | 6,027 | 1.05 | 0.91 | 1.22 | 0.494 | |||
| 90–100% | 627 | 6,027 | 1.02 | 0.89 | 1.17 | 0.784 | |||
| Peripheral artery | 0–10% | 1,966 | 4,871 | 1.00 | Ref | Ref | - | 2.0E-07 | |
| disease | 10–20% | 1,964 | 4,871 | 1.00 | 0.93 | 1.08 | 0.927 | ||
| 20–30% | 1,948 | 4,871 | 1.01 | 0.93 | 1.08 | 0.890 | |||
| 30–40% | 1,984 | 4,871 | 1.04 | 0.96 | 1.12 | 0.361 | |||
| 40–50% | 1,964 | 4,871 | 1.03 | 0.96 | 1.11 | 0.425 | |||
| 50–60% | 1,950 | 4,871 | 1.02 | 0.95 | 1.10 | 0.559 | |||
| 60–70% | 1,972 | 4,871 | 1.05 | 0.98 | 1.14 | 0.165 | |||
| 70–80% | 1,960 | 4,871 | 1.05 | 0.97 | 1.13 | 0.203 | |||
| 80–90% | 2,019 | 4,871 | 1.10 | 1.02 | 1.19 | 0.010 | |||
| 90–100% | 2,102 | 4,871 | 1.20 | 1.11 | 1.29 | 1.9E-06 |
Genome-wide polygenic risk scores (gPRS) for T2D were generated in the MVP participants of European ancestry with T2D by calculating a linear combination of weights derived from the Europeans in the DIAMANTE Consortium using the prune and threshold method in PRSice-2 software (pruning r2 = 0.8, P = 0.05). The gPRSs were divided into deciles and the risk of T2D-related vascular outcomes was assessed using a logistic regression model using the lowest decile (0–10%) as the reference category, together with the potential confounding factors of age, gender, and the first 10 PCs of European ancestry. The decile-specific P-values are shown in the column labeled P. In a separate logistic regression analysis, the continuous PRS was set as the dependent variable together with age, gender, and the first 10 PCs, and the P-value for linear trend is shown in the column labeled P for linear trend. For coronary heart disease, a CHD PRS (from CardiogramplusC4DplusUKBB) is included in the regression model as an additional covariate. For acute ischemic stroke, a stroke PRS (from MEGASTROKE consortium) is included in the regression model as an additional covariate. T2D, type 2 diabetes; PRS, polygenic risk score; n Cases, number of cases with the respective vascular outcome; n Controls, number of unaffected controls for the respective vascular outcome; OR, odds ratio; CI, confidence interval.
Table 6 |.
Polygenic risk scores and non-vascular outcomes
| Outcometype | Outcome | T2D PRS decile | n Cases | n Controls | OR | 95%CI lower | 95%CI upper | P | P for linear trend |
|---|---|---|---|---|---|---|---|---|---|
| Non-vascular | Retinopathy | 0–10% | 792 | 4,533 | 1.00 | Ref | Ref | - | 3.1E-32 |
| 10–20% | 832 | 4,533 | 1.08 | 0.97 | 1.20 | 0.158 | |||
| 20–30% | 795 | 4,533 | 1.05 | 0.94 | 1.17 | 0.364 | |||
| 30–40% | 852 | 4,533 | 1.14 | 1.02 | 1.26 | 0.019 | |||
| 40–50% | 814 | 4,533 | 1.08 | 0.97 | 1.20 | 0.152 | |||
| 50–60% | 891 | 4,533 | 1.20 | 1.08 | 1.33 | 6.8E-04 | |||
| 60–70% | 901 | 4,533 | 1.25 | 1.13 | 1.39 | 3.1E-05 | |||
| 70–80% | 936 | 4,533 | 1.30 | 1.17 | 1.45 | 6.8E-07 | |||
| 80–90% | 1,031 | 4,533 | 1.47 | 1.33 | 1.63 | 2.2E-13 | |||
| 90–100% | 1,069 | 4,533 | 1.59 | 1.44 | 1.77 | 4.2E-19 | |||
| Chronic kidney | 0–10% | 3,446 | 3,391 | 1.00 | Ref | Ref | - | 7.3E-06 | |
| disease | 10–20% | 3,490 | 3,391 | 1.03 | 0.93 | 1.15 | 0.508 | ||
| 20–30% | 3,439 | 3,391 | 1.04 | 0.94 | 1.14 | 0.488 | |||
| 30–40% | 3,463 | 3,391 | 1.05 | 0.95 | 1.16 | 0.323 | |||
| 40–50% | 3,370 | 3,391 | 1.04 | 0.95 | 1.14 | 0.409 | |||
| 50–60% | 3,362 | 3,391 | 1.07 | 0.97 | 1.17 | 0.166 | |||
| 60–70% | 3,389 | 3,391 | 1.07 | 0.98 | 1.17 | 0.129 | |||
| 70–80% | 3,285 | 3,391 | 1.07 | 0.98 | 1.17 | 0.121 | |||
| 80–90% | 3,373 | 3,391 | 1.07 | 0.98 | 1.16 | 0.151 | |||
| 90–100% | 3,326 | 3,391 | 1.16 | 1.07 | 1.26 | 5.9E-04 | |||
| Neuropathy | 0–10% | 2,176 | 3,814 | 1.00 | Ref | Ref | - | 7.9E-08 | |
| 10–20% | 2,193 | 3,814 | 1.03 | 0.96 | 1.11 | 0.436 | |||
| 20–30% | 2,217 | 3,814 | 1.07 | 0.99 | 1.15 | 0.075 | |||
| 30–40% | 2,218 | 3,814 | 1.06 | 0.99 | 1.15 | 0.110 | |||
| 40–50% | 2,217 | 3,814 | 1.05 | 0.98 | 1.13 | 0.192 | |||
| 50–60% | 2,293 | 3,814 | 1.11 | 1.03 | 1.20 | 0.006 | |||
| 60–70% | 2,261 | 3,814 | 1.10 | 1.02 | 1.18 | 0.014 | |||
| 70–80% | 2,253 | 3,814 | 1.10 | 1.02 | 1.19 | 0.009 | |||
| 80–90% | 2,265 | 3,814 | 1.11 | 1.03 | 1.19 | 0.007 | |||
| 90–100% | 2,377 | 3,814 | 1.21 | 1.12 | 1.30 | 9.7E-07 |
Genome-wide polygenic risk scores (gPRS) for T2D were generated in the MVP participants of European ancestry with T2D by calculating a linear combination of weights derived from the Europeans in the DIAMANTE Consortium using the prune and threshold method in PRSice-2 software (pruning r2 = 0.8, P = 0.05). The gPRSs were divided into deciles and the risk of T2D-related non-vascular outcomes was assessed using a logistic regression model using the lowest decile (0–10%) as the reference category, together with the potential confounding factors of age, gender, and the first 10 PCs of European ancestry. The decile-specific P-values are shown in the column labeled P. In a separate logistic regression analysis, the continuous PRS was set as the dependent variable together with age, gender, and the first 10 PCs, and the P-value for linear trend is shown in the column labeled P for linear trend. For chronic kidney disease, a CKD PRS (from CKDgen consortium) is included in the regression model as an additional covariate. T2D, type 2 diabetes; PRS, polygenic risk score; n Cases, number of cases with the respective non-vascular outcome; n Controls, number of unaffected controls for the respective non-vascular outcome; OR, odds ratio; CI, confidence interval.
We evaluated whether the T2D gPRS was associated with the risk of micro- and macrovascular outcomes in an analysis restricted to participants with T2D. The P-values were calculated using gPRS as a continuous exposure, and odds ratios were calculated by contrasting the top to the bottom gPRS decile (Fig. 2 and Tables 5 and 6). We observed strong association between a T2D gPRS and microvascular complications, in particular with retinopathy, but to a lesser extent with neuropathy and CKD. For macrovascular outcomes, T2D gPRS was associated with the risk of PAD, but not with the risk of CHD or acute ischemic stroke.
Figure 2 |. T2D gPRS is mainly predictive of microvascular outcomes.
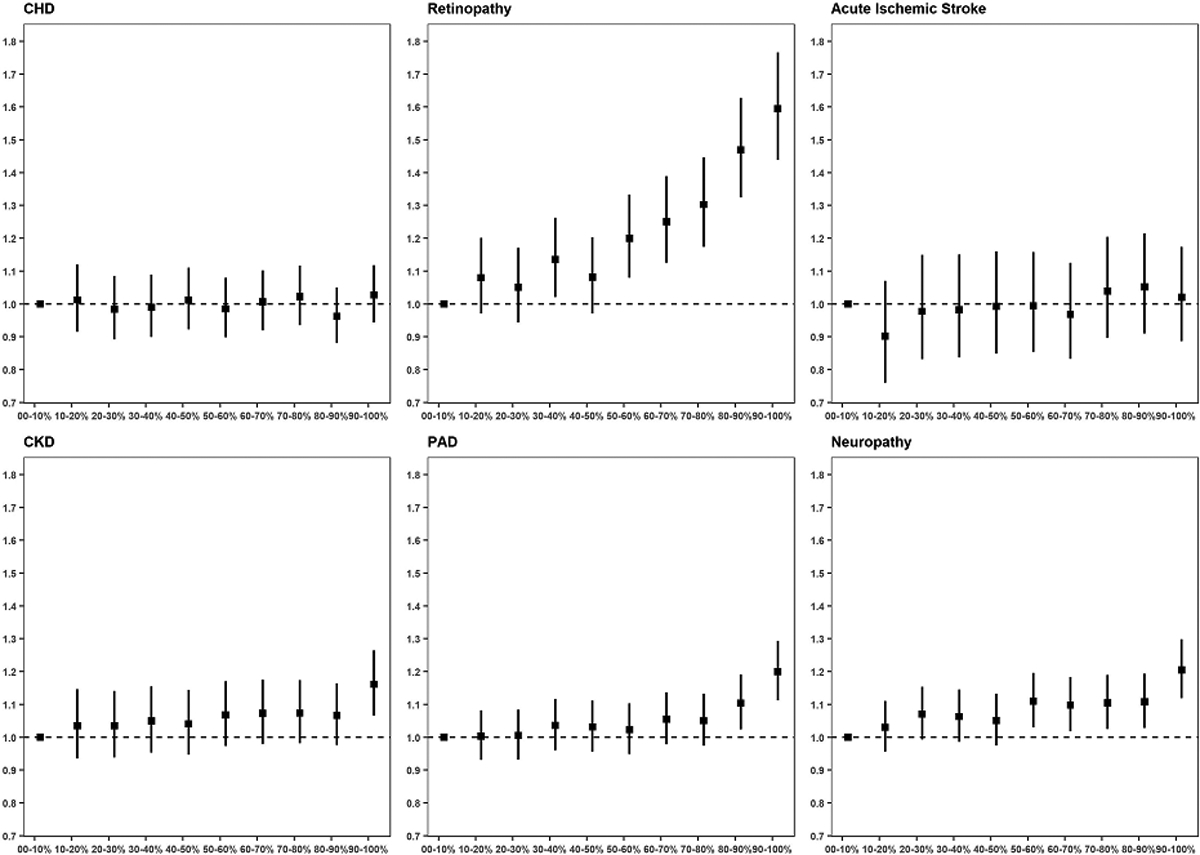
A genome-wide T2D PRS was calculated and categorized into deciles based on the scores in controls. The PRS-outcome associations are shown for macrovascular outcomes (CKD: 67,403 cases, 129,827 controls; CHD: 56,285 cases, 140,945 controls; PAD: 35,882 cases, 161,348 controls) and for microvascular outcomes (acute ischemic stroke: 11,796 cases, 178,481 controls; retinopathy: 13,881 cases, 123,538 controls; neuropathy: 40,475 cases, 110,331 controls). Effect sizes and 95% confidence intervals are shown per decile per micro- or macrovascular outcome. For each of the complication outcomes, separate logistic regression models are fitted for people with T2D, and the models include the following independent variables: T2D PRS (from DIAMANTE Consortium), age, gender, BMI, and 10 PCAs. For coronary heart disease, a CHD PRS (from CardiogramplusC4DplusUKBB) is included in the regression model as an additional covariate. For acute ischemic stroke, a stroke PRS (from MEGASTROKE Consortium) is included in the regression model as an additional covariate. For chronic kidney disease, a CKD PRS (from CKDgen Consortium) is included in the regression model as an additional covariate.
Discussion
We report the discovery of 318 novel autosomal and X chromosomal variants associated with T2D susceptibility in a trans-ethnic GWAS. We also report 13 variants associated with differences in T2D-related micro- and macrovascular outcomes between diabetic and non-diabetics. The substantial locus discovery was achieved by combining data from several large-scale biobanks and consortia, where the MVP data constituted over 40% of all T2D cases. Furthermore, we present the largest cohort of African Americans including over 56,000 participants, substantially larger than previous African-specific studies published to date.
Analyses of coding variants identified 44 variants associated with T2D, including three pLoF variants in LPL, ANKDD1B and CCHCR1. We identified 804 putative causal genes at both novel and previously reported loci, including 54 genes that were found to be possible targets for FDA-approved drugs and chemical compounds. Our SNP-T2D interaction analyses identified several loci where the association between a genetic variant and a vascular outcome differed between people with T2D as compared to those without. We further found that a high polygenic risk for T2D strongly increased the risk for retinopathy in individuals with T2D, and also for CKD, neuropathy, and PAD.
T2D is highly prevalent in people of African ancestry; however, there are a total of three published T2D GWAS reports in this ancestral group with only four definitely detected loci18,19,20. In our study with over 56,000 participants of recent African ancestry, we report four novel loci for T2D that are solely observed in this ancestral group, including one that is located on the X chromosome. Of the previously reported loci, only rs3842770 (INS-IGF2) was replicated here. We did not observe replication either with rs756016320 or rs73284431, reported from a large study conducted in sub-Saharan Africa. The reported HLA-B variant rs2244020 did not replicate in our study, but we did observe a significant association with another SNP in the HLA region (rs10305420, OR 1.15, P = 8.5 × 10−9). We observed that the major G-allele of chrX:153882606 (rs782270174) was associated with increased risk of T2D in African Americans. This variant is in high LD (r2 = 0.93) with G6PD G202A (rs1050828), for which the minor allele is associated with lower HbA1c due to shorter RBC lifespan21. In a post-hoc analysis, we examined the relationship of chrX:153882606 to most recent HbA1C prior to MVP study enrollment in African American males and did observe a strong negative association (beta = −0.072, se = 0.0015, n = 55,165, P < 1.0 × 10−322). We cannot rule out the possibility that the apparent association in T2D at rs782270174 reflects under-diagnosis of T2D due to reduced HbA1C in African Americans. We did not replicate the association of the AGTR2 variant (rs146662075, chrX:115408811) as reported by Bonas-Guarch et al.10, which might be the result of poor imputation of the 1000 Genomes reference panel for this variant.
The presence of a coding variant near a tagging SNP does not constitute enough evidence to infer a causal association. However, recent exome-array genotyping of over 350,000 individuals identified 40 coding variants associated with T2D, of which 26 mapped near known risk-associated loci22. Similarly, an exome sequencing study in over 40,000 participants reported 15 variants associated with T2D, of which only two were not previously reported by GWAS23. Sequencing efforts are indispensable for identifying causal variants and genes related to disease, as well as providing insight into the contributions of ultra-rare alleles while adding to the value of array-based association studies.
Our transcriptome-wide analyses identified 804 putatively causal genes, including 54 genes that appear to be regulated by approved drugs and 687 genes that have not been previously reported. Some of these genes are already well established for T2D etiology (e.g. KCNJ11). Except for skeletal muscle, the tissues that showed strongest associations are not known to be of importance in T2D etiology. However, this could be simply explained by the fact that (i) eQTLs appear ubiquitous across tissues and (ii) eQTL discovery across tissues may not be the same, given eQTL effect sizes and sample sizes of T2D relevant tissues. We did not observe any significant association in the alpha and beta islet cells, which could be the result of the small sample size (e.g. 30 alpha cells and 19 beta cells). In addition, whole islet transcriptomes are notoriously variable due to the large differences in islet composition among humans, and a few transcripts make up half the transcriptome24.
Of particular clinical importance, we identified several genes that are therapeutic targets for medications in patients treated for cardiometabolic conditions. We identified two genes, SCN3A and SV2A, whose expression is modified by anti-epileptic agents, and evidence exists showing that anti-epileptic agents may influence glucose regulation. A randomized-controlled trial has reported that the anticonvulsant valproic acid lowers blood glucose concentrations25. The information from the gene-drug analyses may facilitate future drug repurposing screens.
It is possible that the use of the T2D gPRS provides an opportunity to identify patients who are at the highest risk of developing microvascular complications, such as retinopathy. Here, we observed that among vascular outcomes, the T2D gPRS was most significantly associated with retinopathy. In addition, we observed significant associations with other T2D-related outcomes such as CKD, PAD, and neuropathy. Studies at specific loci using both common and rare coding variants will be required to understand pathways leading to T2D-related vascular outcomes.
In a SNP-T2D interaction analysis on T2D-related vascular outcomes, we identified 13 loci where the effect on outcome was different between the strata of T2D, of which three occurred at previously established variants and 10 had not been previously reported. Our findings have clinical translational potential for risk stratification and identify diabetic patients who are predisposed to develop subsequent vascular outcomes and present therapeutic opportunities to attenuate the risk of diabetes progression in individuals with T2D.
For T2D-related retinopathy, four variants were found to have different effect sizes between people with and without T2D. The strongest signal for interaction in relation to retinopathy was observed for GJA8. Deletion of this gene has been associated with eye abnormalities and retinopathy of prematurity in premature infants, inherited cataracts, visual impairment and cardiac defects and eye abnormalities26–28. TCF7L2 is a known diabetes locus and its association with progression to retinopathy has been previously established29. SLC18A2 is expressed in adult retina and retinal pigment epithelium tissues; the product of this gene is involved in the transport of monoamines into secretory vesicles for exocytosis30. SVILP1 has been previously shown to be associated with thiamine (vitamin B1) prescription, which is frequently prescribed to people with blurry vision31.
For chronic kidney disease, we identified two loci, UMOD and TENM3, with gene-T2D interaction effects. UMOD encodes uromodilin, which is exclusively produced by the kidney tubule, where it plays an important role in kidney and urine function. A large-scale study in over 133,000 participants has shown that the serum creatinine-lowering allele in UMOD (rs12917707) is more prevalent in diabetic individuals with CKD as compared to diabetic participants without CKD32. Variation in TENM3 has been associated with cholangitis and kidney disorders in UK Biobank33.
SNP-T2D interaction analysis of neuropathy identified one locus, NRP2. NRP2 encodes neuropilin-2, which is an essential cell surface receptor involved in VEGF-dependent angiogenesis and sensory nerve regeneration.
For coronary heart disease, we identified several SNP-T2D interactions. Variation at 9p21 has previously been associated with CHD and T2D. SORT1 is a lipid-associated locus; in our analyses, allelic variation at this locus that decreases CHD risk and decreases lipids conferred a stronger protection in people with T2D compared to those without T2D. Coupled with findings in mice that identified SORT1 as a novel target of insulin signaling, our findings raise the hypothesis that SORT1 may contribute to altered hepatic apoB metabolism under insulin-resistant conditions.
The SNP rs71039916 is located near PDE3A, and colocalizes with a SNP (rs3752728, D’ = 0.867, r2 = 0.08) that is associated with diastolic blood pressure34,35. As a phosphodiesterase that reduces cAMP levels, the PDE3A protein limits protein kinase A/cAMP signaling and has been shown to affect proliferation of vascular smooth muscle cells36. Cell line research has shown that cAMP levels might impact the regulation of insulin secretion in pancreatic β-cells, and more recent gene ablation studies in mice have established that cAMP/CREB signaling controls the insulinotropic and anti-apoptotic effects of GLP-1 signaling in adult mouse β-cells37. Subcutaneous adipose tissue of patients with T2D show increased PDE activity, and inverse correlations between total PDE3 activity and BMI have been reported in adipocytes38.
In summary, we have identified 318 novel genetic variants associated with T2D risk and T2D-related vascular outcomes, including 3 population-specific autosomal loci in African Americans, 8 variants on the X chromosome, and an additional 13 variants associated with differences in T2D-related micro- and macrovascular outcomes across diabetic stratum. Over 21% of our discovery sample comprised of non-European participants; indeed, the African American component alone included over 56,000 subjects. We hope this baseline set of data will provide a resource to better understand the genetic etiology of disease and maximize the benefits of polygenic risk prediction in these groups.
Online Methods
Overview.
We conducted a large-scale multi-ethnic T2D GWAS of common variants in over 1.4 million participants. We subsequently conducted analyses to facilitate the prioritization of these individual findings, including transcriptome-wide predicted gene expression, secondary signal analysis, T2D-related vascular outcomes analysis, coding variant mapping, and a drug repurposing screen.
Discovery cohort.
The Million Veteran Program (MVP) is a large cohort of fully consented veterans of the US military forces recruited from 63 participating Department of Veterans Affairs (VA) medical facilities5. Recruitment started in 2011, and all veterans were eligible for participation (Supplementary Table 3). We analyzed clinical data through July 2017 for participants who enrolled between January 2011 and October 2016. All study participants provided blood samples for DNA extraction and genotyping, and completed surveys about their health, lifestyle, and military experiences. Consent to participate and permission to re-contact was provided after counseling by research staff and mailing of informational materials. Study participation included consenting to access to the participant’s electronic health records for research purposes, data that captured a median follow-up time of 10.0 years at time of study enrollment. Each veteran’s electronic health care record is integrated into the MVP biorepository, including inpatient International Classification of Diseases (ICD-9-CM and ICD-10-CM) diagnosis codes, Current Procedural Terminology (CPT) procedure codes, clinical laboratory measurements, and reports of diagnostic imaging modalities. Researchers are provided data that is de-identified except for dates. Blood samples are collected by phlebotomists and banked at the VA Central Biorepository in Boston, where DNA is extracted and shipped to two external centers for genotyping. The MVP received ethical and study protocol approval from the VA Central Institutional Review Board (cIRB) in accordance with the principles outlined in the Declaration of Helsinki.
Genotyping.
DNA extracted from buffy coat was genotyped using a custom Affymetrix Axiom biobank array. The MVP 1.0 genotyping array contains a total of 723,305 SNPs, enriched for low frequency variants in African and Hispanic populations, and variants associated with diseases common to the VA population5.
Genotype quality-control.
Standard quality control (QC) and genotype calling algorithms were applied using the Affymetrix Power Tools Suite (v1.18). Excluded were duplicate samples, samples with more heterozygosity than expected, and samples with an over 2.5% missing genotype calls. We excluded related individuals (halfway between second- and third-degree relatives or closer) with KING software39. Before imputation, variants that were poorly called or that deviated from their expected allele frequency based on reference data from the 1000 Genomes Project were excluded40. After prephasing using EAGLE v2, genotypes were imputed via Minimac4 software41 from the 1000 Genomes Project phase 3, version 5 reference panel. The top 30 principal components (PCs) were computed using FlashPCA in all MVP participants and an additional 2,504 individuals from 1000 Genomes. These PCs were used for the unification of self-reported race/ancestry and genetically inferred ancestry to compose ancestral groups42.
Race and ethnicity.
Information on race and ethnicity was obtained based on self-report through centralized VA data collection methods using standardized survey forms, or through the use of information from the VA Corporate Data Warehouse or Observational Medical Outcomes Partnership data. Self-reported race/ethnicity was missing in 3.67% of participants, and 39.4% of participants had some form of discordant information between the various data sources. Race and ethnicity categories were merged to form the ancestral groups using a unifying classification algorithm based on self-identified race/ethnicity and genetically inferred ancestral information, termed HARE (Harmonized Ancestry and Race/Ethnicity)42. Using this approach, all but 6,257 (1.78%) were assigned to one of the four ancestral groups.
Phenotype classification.
ICD-9-CM diagnosis codes from electronic health care records were available for MVP participants from as early as 1998. Participants were classified as a T2D case if they had 2 or more T2D-related diagnosis codes (ICD-9-CM 250.2x) from VA or fee basis inpatient stays or face-to-face primary care outpatient visits in the 731 days before the enrollment date up to July 1st of 2017, excluding those with co-occurring diagnosis codes for T1D (250.1x), secondary or other diabetes or a medical condition that may cause diabetes (249.xx). Participants were selected as controls if they had no ICD-9-CM diagnosis code for type 1, type 2, or secondary diabetes mellitus up to July 2017.
For T2D-related vascular outcomes, the following definitions were used: CHD, at least one admission to a VA hospital with discharge diagnosis of admission for myocardial information, or at least one procedure code for revascularization (coronary artery bypass grafting, percutaneous coronary intervention), or at least 2 ICD-9-CM codes for CAD (410 to 414) registered on at least 2 separate encounters. PAD: the presence of ≥ 2 ICD-9-CM codes or CPT codes as outlined in Klarin et al., or having 1 code and ≥ 2 visits to a vascular surgeon within a 14 month period. Acute ischemic stroke was defined if at least 1 ICD-9-CM discharge diagnosis code for stroke excluding head injury or rehab (433.x1, 434 (excluding 434.x0), and 436) was present43. CKD was classified as an estimated glomerular filtration rate <60 mL/min−1·1.73 m−2 on two separate occasions 90 days apart, or ICD-9-CM diagnosis codes for chronic renal failure (585) and/or a history of kidney transplantation (ICD-9-CM V42). Neuropathy was defined using the following ICD-9-CM diagnosis codes: diabetic neuropathy (356.9, 250.6), amyotrophy (358.1), cranial nerve palsy (951.0, 951.1, 951.3), mono-neuropathy (354.0–355.9), Charcot’s arthropathy (713.5), polyneuropathy (357.2), neurogenic bladder (596.54), autonomic neuropathy (337.0, 337.1), or orthostatic hypotension (458). Retinopathy was defined using ICD-9-DM diagnosis codes for: T2D with ophthalmic manifestations (250.50, 250.52), retinal detachments and defects (361.0, 361.1), disorders of vitreous body (379.2), other retinal disorders (362.0, 362.1, 362.3, 362.81, 362.83, 362.84), excluding ICD-9-CM codes associated with macular degeneration (362.5).
MVP analysis.
We tested imputed SNPs that passed QC (e.g. HWE > 1.0 × 10−10, INFO > 0.3, call rate > 0.975) for association with T2D through logistic regression assuming an additive model of variants with MAF > 0.1% in Europeans, and MAF > 1% in African Americans, Hispanics and Asians using PLINK2a44. Covariates included age, gender, and 10 principal components of genetic ancestry.
Meta-analysis.
Summary statistics available from previously published T2D GWAS studies were obtained for meta-analysis (Supplementary Table 2). All cohorts were imputed using the 1000 Genomes Project phase 3, version 5 reference panel, with exception of the DIAMANTE consortium, where genotype calls were imputed using the Haplotype Reference Consortium reference panel. Only SNPs with ancestry-specific MAF > 1% in these studies were used. Ancestry-specific and multi-ethnic meta-analysis were performed using in a fixed-effects model using METAL with inverse-variance weighting of log odds ratios45. Between-study allelic effect size heterogeneity was assessed with Cochran’s Q statistic as implemented in METAL. Variants were considered genome-wide significant if they passed the conventional P-value threshold of 5 × 10−8. We excluded variants with a high amount of heterogeneity (I2 statistic > 75%) across the ancestral groups.
X chromosome analysis.
X chromosome genotypes were processed separately. During prephasing and imputation an additional flag of -chrX was added. Post-imputation XWAS QC included removing variants (i) in pseudo-autosomal regions, (ii) not in HWE in females (P > 1.0 × 10−6), (iii) with differential allele frequencies or differential missingness (P < 10−7) between male and female controls (Extended Data Fig. 2)46. For each ancestry-specific subset, we performed sex-stratified analysis where dosages (number of X-chromosome copies) in T2D cases are equivalent to controls within each sex stratum. The ancestry-restricted sex-stratified X chromosome analyses were first meta-analyzed into a multi-ethnic sex-stratified analysis. Then, the multi-ethnic results from males and multi-ethnic results from females were meta-analyzed, where none of the analyzed variants was detected using the Cochrane test for heterogeneity (P < 5 × 10−8). Results are presented in Table 2 and Supplementary Table 9.
Secondary signal analysis.
GCTA was used to conduct approximate conditional analyses to detect ancestry-specific distinct association signals at each of the lead SNPs. Race-stratified MVP cohorts (197,066 Europeans and 53,445 African Americans) were used to model LD patterns between variants as a reference panel. For each lead SNP, conditionally independent variants that reached locus-wide significance (P < 1.0 × 10−5) were considered as secondary signals of distinct association. If the minimum distance between any distinct signals from two separate loci was less than 500 kb, we performed additional conditional analysis including both regions and reassessed the independence of each signal. Finally, the predicted conditionally independent variants were tested in a logistic regression model in the MVP study only to empirically validate the signal, and results are shown in Supplementary Tables 11 and 12.
Coding variant mapping.
All imputed variants in MVP were evaluated with Ensemble’s Variant Effect Predictor, and predicted LoF and missense variants were extracted. LD was calculated with established variants, and the effect of the missense variant was calculated conditioning on the lead SNP to assess how much residual variance the SNP explains in T2D risk. A P-value of 0.05 was considered as statistically significant.
S-PrediXcan and colocalization analyses.
Genetically predicted gene expression and its association with T2D risk was estimated using S-PrediXcan. Input included meta-analyzed summary statistics from the European T2D GWAS and reference eQTL summary statistics for 52 tissues including 48 tissues from GTEx, 2 cell types in kidney tissue (glomerulus and tubulus)47, and 2 cell types in pancreatic islet tissue (alpha and beta)48. Analyses incorporated genotype covariance matrices based on 1000 Genomes European populations to account for LD structure. Colocalization analysis was performed to address the issue of LD-contamination in S-PrediXcan analyses. The output is shown in Supplementary Table 15.
Polygenicity and population stratification.
LD score regression (LDSC)12 was used to calculate population-specific LD scores in Europeans and Asians using SNPs selected from HapMap49 after excluding SNPs with INFO < 0.95 and SNPs in the major histocompatibility complex region. Of note, LDSC is likely to be biased in admixed populations, and therefore admixture-adjusted LDSC was used in African Americans and Hispanics14.
Tissue- and epigenetic-specific enrichment of T2D heritability.
We analyzed cell type-specific annotations to identify enrichments of T2D heritability. First, a baseline gene model was generated consisting of 53 functional categories, including UCSC gene models, ENCODE functional annotations50, Roadmap epigenomic annotations51, and FANTOM5 enhancers52. Gene expression and chromatin data were also analyzed to identify disease-relevant tissues, cell types, and tissue-specific epigenetic annotations. We used LDSC12,16,53 to test for enriched heritability in regions surrounding genes with the highest tissue-specific expression and we used GREGOR to calculate enrichment of epigenetic marks17. Sources of data that were analyzed included 53 human tissue or cell type RNA-seq data from GTEx; 152 human, mouse, or rat tissue or cell type array data from the Franke lab54; 3 sets of mouse brain cell type array data from Cahoy et al.55; 292 mouse immune cell type array data from ImmGen56; and 396 human epigenetic annotations from the Roadmap Epigenomics Consortium51. We tested for epigenomic enrichment of genetic variants using GREGOR17. We tested for enrichment of 2,747 genomic features selected the T2D lead variants with P < 5 × 10−8, or their LD proxies (r2 > 0.7) relative to control variants. Enrichment was considered significant if the enrichment P-value was less than the Bonferroni-corrected threshold of 1.8 × 10−5 (0.05/2725 non-zero tested sites). Consortia annotations were obtained and processed as follows. Data from the consolidated epigenomes section of the Roadmap Epigenomics Project portal51 was downloaded on 02/10/16. All ENCODE consortium50 data was downloaded 01/06/16 from the ENCODE project portal by limiting to Homo sapiens samples and selecting the named assay except for the Uniform DNase files, which were downloaded on 03/28/16. We used the FAIRE-seq ENCODE data, transcription profiling array data, ChIP-seq files, and histone data. The complete list of 2,305 ENCODE and Roadmap Epigenomics features used are found in Supplementary Table 20. We additionally performed a literature search on PubMed and in the GEO data archive focusing on 5 tissues most likely to be involved in T2D etiology: pancreas, liver, adipose, muscle, and intestine. Most searches were performed from 08/15/16 to 09/29/16, we identified a total of 442 features across 42 publications (Supplementary Table 21).
Phenome-wide association analysis.
For the three LoF variants that were identified using coding variant analysis, we performed a PheWAS to fully leverage the diverse nature of MVP as well as the full catalog of relevant ICD-9-CM diagnosis and CPT procedure codes (Table 5). Of genotyped veterans, participants were included in the PheWAS if their respective electronic health record reflected two or more separate encounters in the VA Healthcare System in each of the two years prior to enrollment in MVP. A total of 277,531 veterans spanning 21,209,658 available ICD-9 diagnosis codes were available. We restricted our analysis on the subgroup of 197,066 European participants. Diagnosis and procedure codes were collapsed to clinical disease groups and corresponding controls using predefined groupings57. Phenotypes were required to have a case count over 25 in order to be included in the PheWAS, and a multiple testing thresholds for statistical significance was set to P < 2.8 × 10−5 (Bonferroni method). Each of the previously unpublished LoF variants were tested using logistic regression adjusting for age, sex, and 10 principal components in an additive effects model using the PheWAS R package in R v3.2.0. The results from these analyses are shown in Table 3 (Extended Data Fig. 4).
Analysis of T2D-related outcomes.
Genetic data on European participants was separately analyzed using vascular outcomes as a binary outcome, and T2D as an interaction variable with SNPs using interaction analysis with robust variance to reduce effect heteroscedacity58 using SUGEN software (v8.8)59. We evaluated the interaction between SNP and presence of T2D status using an interaction term for the two independent variables. Due to the binary nature of the outcome, the standard output from the interaction effect estimate are interpreted on a multiplicative scale. To obtain interaction on an additive scale, we calculated the relative excess risk due to interaction (RERI) metric. In case-control studies using the linear additive odds-ratio model as proposed by Richardson and Kaufman in our study has the form of:
In which the coefficient β3 measures the departure from additivity of exposure effect on an odds ratio scale; that is
We performed analysis using a linear odds model to quantify the excess odds per unit of the given explanatory variables on the outcome. In this model, RERI is an estimate of the excess odds on a linear scale due to the interaction between two explanatory variables. In the SNPxT2D interaction analysis, we used a significance threshold of P < 5 × 10−8 to denote variants that statistically different effect sizes. An additional filter was applied, and variants for which the effect size in at least one of the two T2D strata was nominally significant at P < 0.001 were included. Manhattan plots and the table are used to represent the interaction coefficients on this scale.
Polygenic risk scores and risk of T2D and related outcomes.
We constructed a genome-wide polygenic risk score (gPRS) for T2D in the MVP participants of European ancestry by calculating a linear combination of weights derived from the Europeans in the DIAMANTE Consortium3 using the prune and threshold method in PRSice-2 software. After an initial sensitivity analysis, the r2 threshold for pruning was set to 0.8, and the P-value for significance threshold was set to 0.05. The gPRSs were divided into deciles and the risk of T2D was assessed using a logistic regression model using the lowest decile as a reference, together with the potential confounding factors of age, gender, BMI, and the first 10 PCs. An additional outcomes analysis was performed to evaluate to what extent a T2D gPRS is predictive of T2D-induced morbidities. The dataset was restricted to subjects with T2D, and stratum-restricted T2D gPRS deciles were generated. Logistic regression models were applied where the micro- and macrovascular conditions were modeled as outcomes, and independent variables included strata-restricted gPRS deciles, age, gender, and the first 10 principal components of European ancestry. The data were visualized using shape-plots.
Heritability estimates and genetic correlations with other complex traits and diseases.
LD-score regression was used to estimate the heritability coefficient, and subsequently population and sample prevalence estimates were applied to estimate heritability on the liability scale60. A genome-wide genetic correlation analysis was performed to investigate possible co-regulation or a shared genetic basis between T2D and other complex traits and diseases. Pairwise genetic correlation coefficients were estimated between the meta-analyzed T2D GWAS summary output in Europeans and each of 774 precomputed and publicly available GWAS summary statistics for complex traits and diseases by using LD score regression through LD Hub v1.9.3 (http://ldsc.broadinstitute.org). Statistical significance was set to a Bonferroni-corrected level of P < 6.5 × 10−5.
Enrichment and pathway analyses.
Tissue enrichment for S-PrediXcan results was evaluated by calculating exact P-values for under- or over-enrichment based on the cumulative distribution function of the hypergeometric distribution. The Bonferroni-corrected threshold for significance was P < 0.001 considering evaluation of 52 tissues. Enrichment analyses in DEPICT61 were conducted using lead T2D SNPs. DEPICT is based on predefined phenotypic gene sets from multiple databases and Affymetrix HGU133a2.0 expression microarray data from over 37,000 subjects to build highly-expressed gene sets for Medical Subject Heading (MeSH) tissue and cell type annotations. Output includes a P-value for enrichment and a yes/no indicator of whether the FDR q-value is significant (P < 0.05).
Evaluation of drug classes for genes with associations with gene expression.
To identify drug-gene pairs that may be leads for repurposing or may be attractive leads for novel inhibitory drugs, we identified drugs targeting genes whose predicted expression was significantly associated with T2D risk in S-PrediXcan analyses and which we predicted would lower blood glucose based on direction of effect on T2D risk with increasing gene expression and drug action (activator or inhibitor). Medications with a primary indication for diabetes and medications with adverse drug events for diabetic patients were evaluated using the SIDe Effect Resource (SIDER) Medications targeting genes were queried using DGIdb. These drug targets represent a set of genes that are both likely to be involved in glucose regulation in one or more tissues and can be targeted by drugs. Genes and medications identified in this analysis are presented in Supplementary Table 17.
Extended Data
Extended Data Fig. 1.
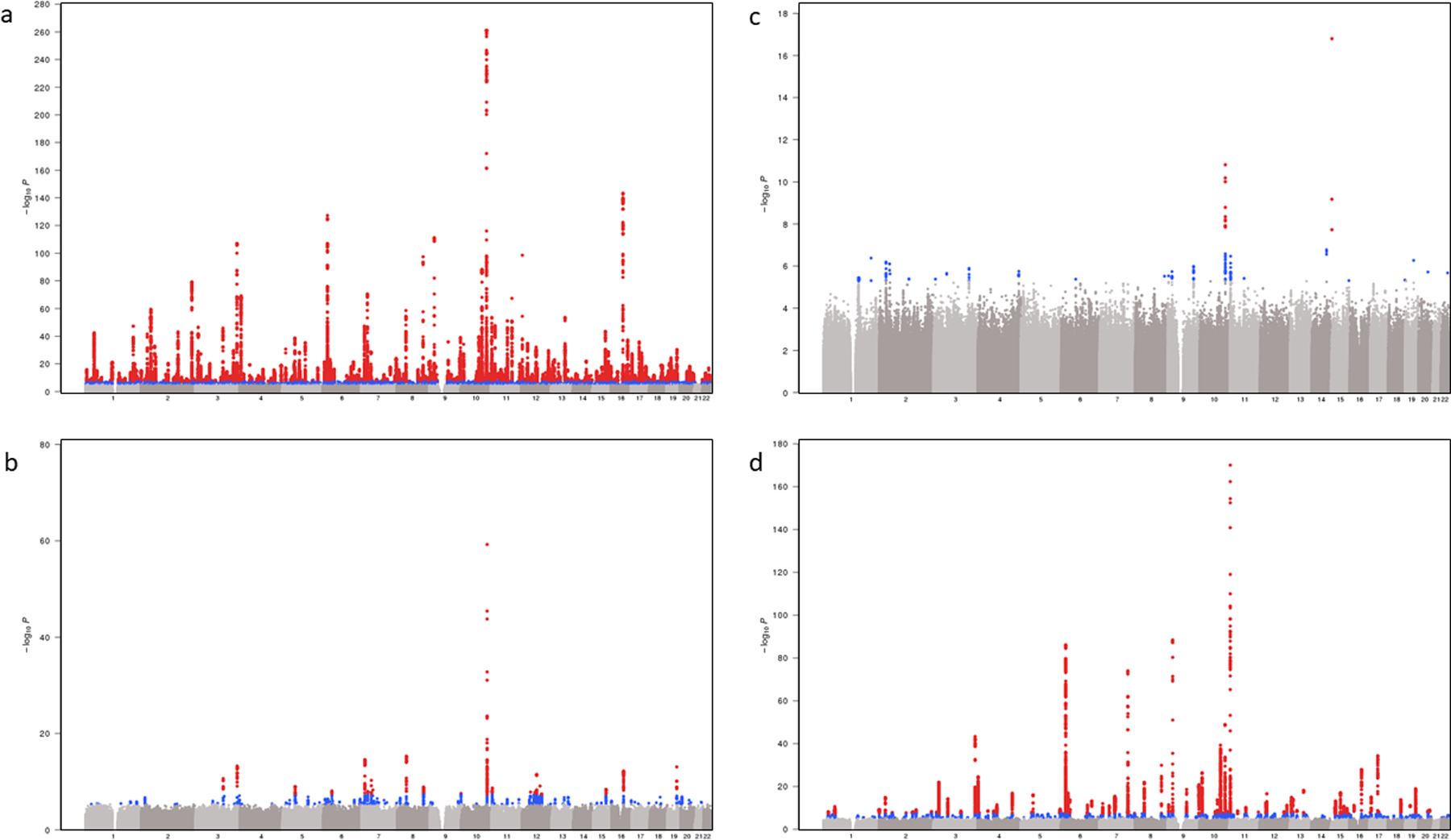
Trans-ethnic and ancestry-specific GWAS Manhattan plots a-d, Each graph represents a Manhattan plot. The y-axis corresponds to −log10 (P) for association with T2D in the respective ancestral group (a, Europeans (148,726 T2D cases, 965,732 controls, λ = 1.21); b, African American (24,646 T2D cases, 31,446 controls, λ = 1.08); c, Hispanics (8,616 T2D cases, 11,829 controls, λ = 1.03); d, Asians (46,511 T2D cases, 169,776 controls, λ = 1.15)). The x-axis represents chromosomal position on the autosomal genome. The y-axis truncated at 1 × 10−300. Points that are color-coded blue correspond to a P-value between 5.0 × 10−8 and 1.0 × 10−6. Points color-coded red indicate genome-wide significance (P = 5.0 × 10−8).
Extended Data Fig. 2.
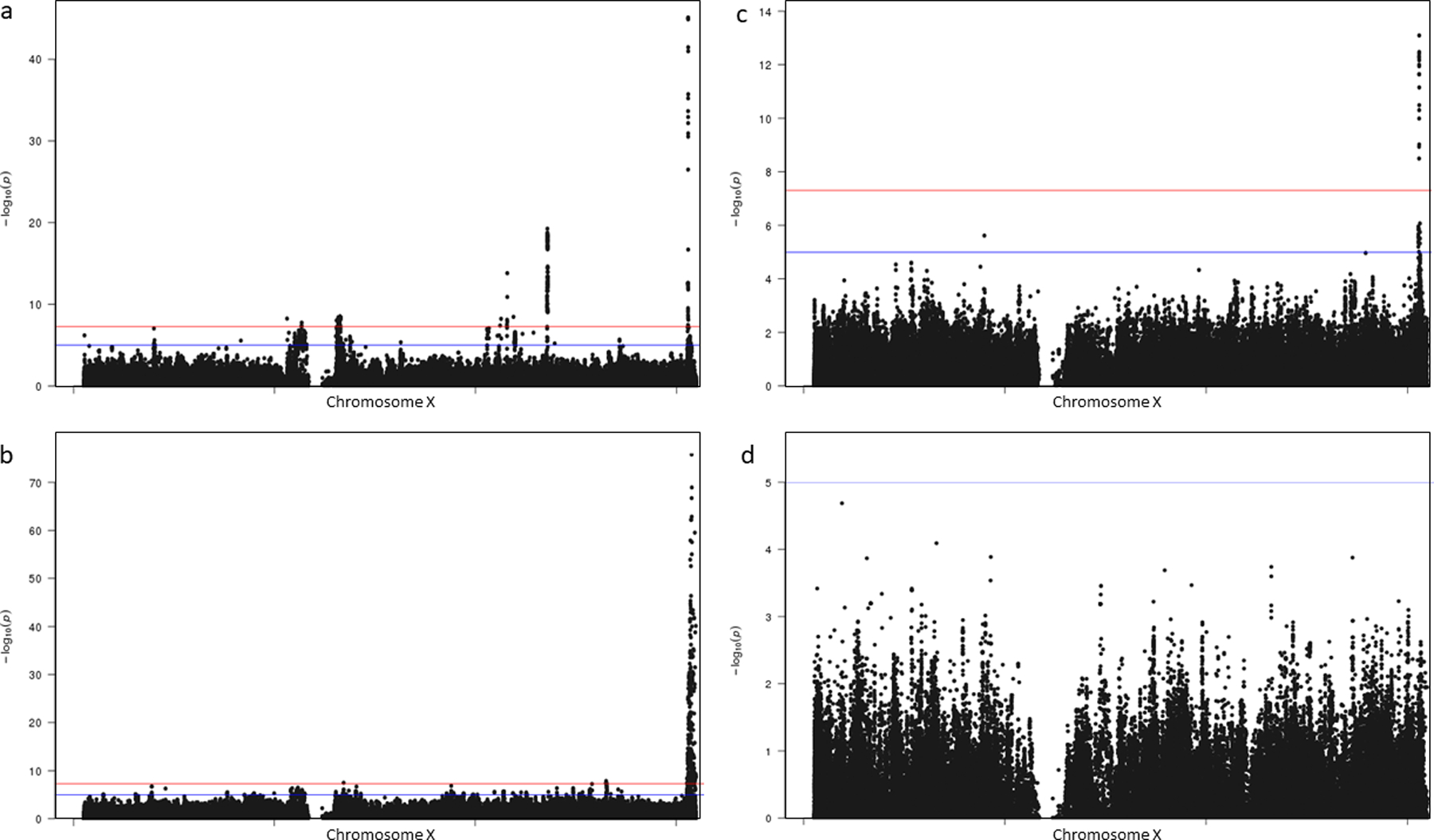
Trans-ethnic and ancestry-specific chromosome X Manhattan plots a-d, Each graph represents a Manhattan plot. The y-axis corresponds to −log10 (P) for association with T2D in the respective ancestral group (a, Europeans (69,869 T2D cases, 127,197 controls); b, African American (23,305 T2D cases, 30,140 controls); c, Hispanics (8,616 T2D cases, 11,829 controls); d, Asians (893 T2D cases, 1,560 controls)). The x-axis represents chromosomal position on chromosome X. The blue line corresponds with a significance threshold of P = 5.0 × 10−8. The red line corresponds with genome-wide significance (P = 5.0 × 10−8).
Extended Data Fig. 3.
Results from PrediXcan analysis using GTEX data This graph represents an inverted Manhattan plot based on the output from the European T2D GWAS (148,726 T2D cases, 965,732 controls). The y-axis corresponds to −log10 (P) for association with genetically predicted gene expression in the respective tissue type (color coding shown on the right). Data were analyzed using S-PrediXcan software. The x-axis represents chromosomal position on the autosomal genome.
Extended Data Fig. 4.
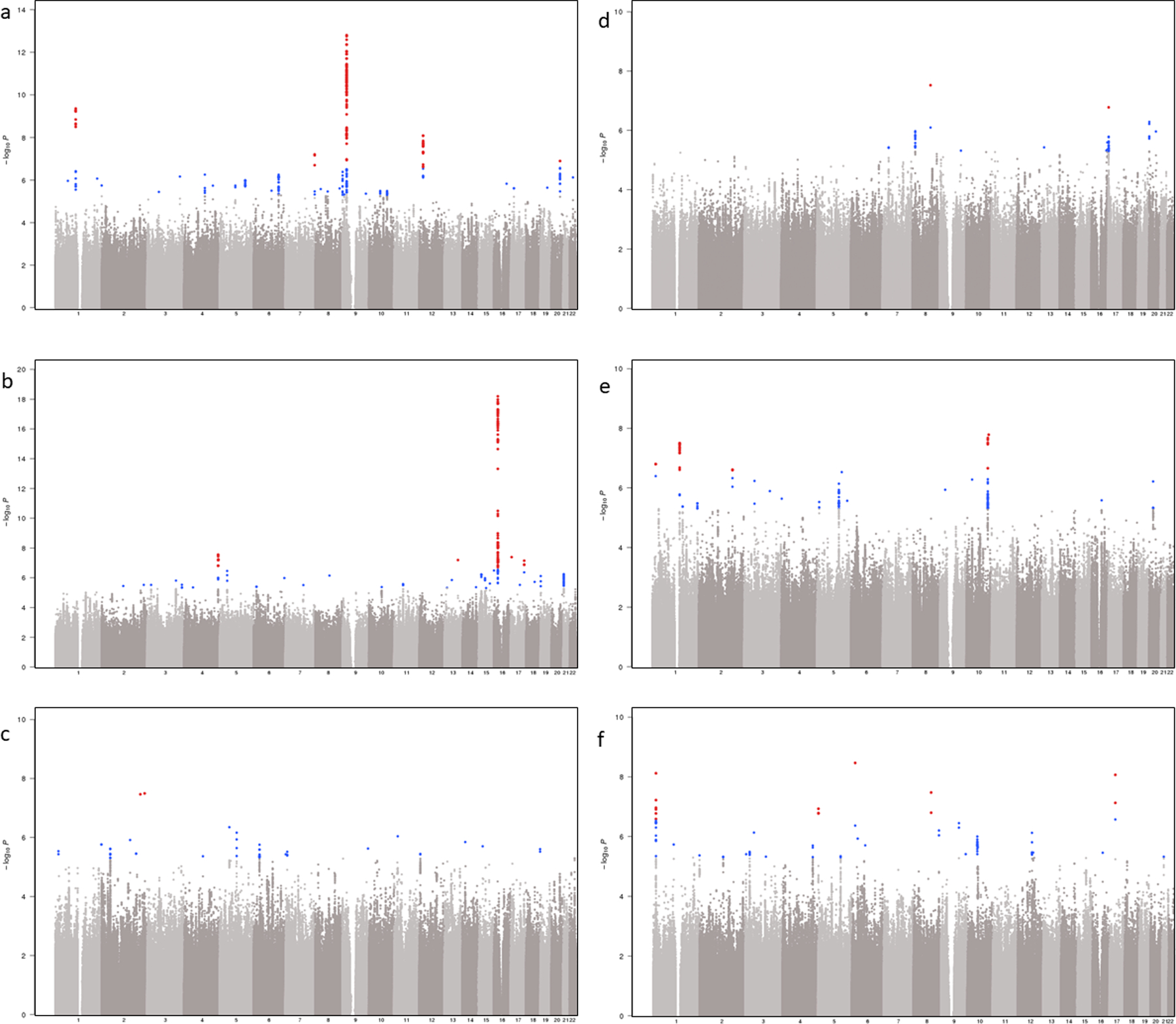
Manhattan plots for T2D-related complications using interaction analysis in individuals of European ancestry a-f, Each graph represents a Manhattan plot. The y-axis corresponds to −log10 (P) for association of SNP×T2D on T2D-related vascular outcome (a, coronary heart disease (56,285 cases, 140,945 controls, λ = 1.06); b, chronic kidney disease (67,403 cases, 129,827 controls, λ = 1.02); c, neuropathy (40,475 cases, 110,331 controls, λ = 1.03); d, peripheral artery disease (5,882 cases, 161,348 controls, λ = 1.02); e, retinopathy (13,881 cases, 123,538 controls, λ = 1.02); f, acute ischemic stroke (11,796 cases, 178,481 controls, λ = 1.00)). The x-axis represents chromosomal position on the autosomal genome. Points that are color-coded blue correspond to a P-value between 5.0 × 10−8 and 1.0 × 10−6. Points color-coded red indicate genome-wide significance (P = 5.0 × 10−8).
Extended Data Fig. 5.
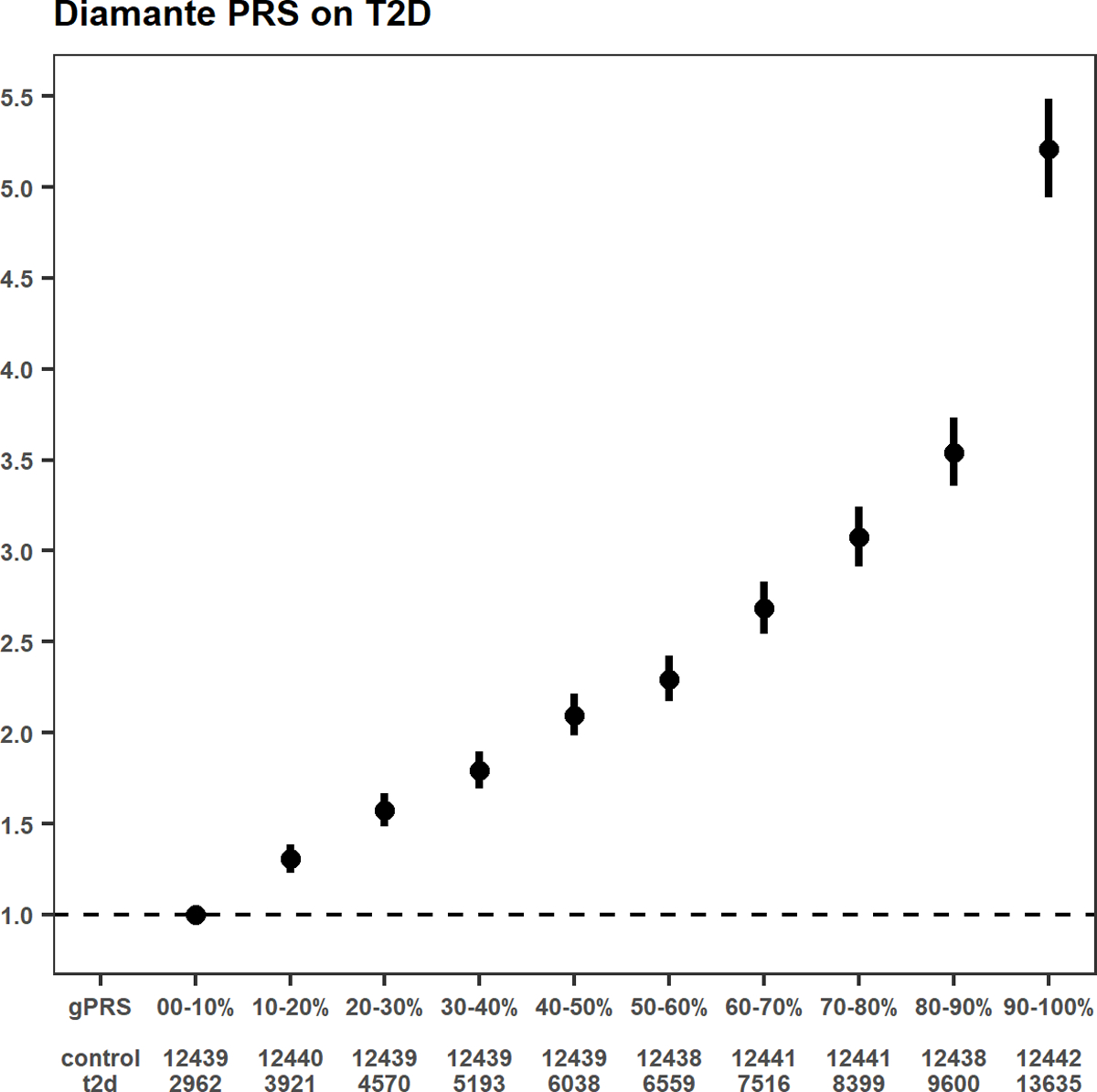
T2D PRS and the risk of T2D A shape plot representing the risk of a T2D genome-wide PRS (gPRS) on the odds ratio of T2D in MVP participants of European ancestry (69,869 T2D cases, 127,197 controls). The weights for the PRS have been obtained from an external reference dataset, namely the DIAMANTE Consortium. The gPRS has been divided into 10 deciles based on gPRS values in MVP white participants without T2D. The reference group is the lowest decile (0–10%). Odds ratios are shown as red dots, with their respective 95th percent confidence intervals displayed as red vertical lines.
Supplementary Material
Source Data Extended Data Fig. 5 Raw odds ratios for T2D shape plot
Source Data Fig. 2 Raw odds ratios for T2D-related outcomes shape plots
Source Data Extended Data Fig. 3 Raw effect estimates and P-values for inverted Manhattan plot depicting genetically predicted gene expression using S-PrediXcan
Acknowledgements
This research is based on data from the Million Veteran Program, Office of Research and Development, Veterans Health Administration and was supported by award no. MVP000. This publication does not represent the views of the Department of Veterans Affairs, the US Food and Drug Administration, or the US Government. This research was also supported by funding from: the Department of Veterans Affairs award I01-BX003362 (P.S.T. and K.-M.C.) and the VA Informatics and Computing Infrastructure (VINCI) VA HSR RES 130457 (S.L.D.). B.F.V. acknowledges support for this work from the NIH/NIDDK (DK101478), the NIH/NHGRI (HG010067) and a Linda Pechenik Montague Investigator award. K.-M.C., S.M.D., J.M.G., C.J.O., L.S.P., J.S.L., and P.S.T. are supported by the VA Cooperative Studies Program. S.M.D. is supported by the Veterans Administration [IK2-CX001780]. D.K. is supported by the National Heart, Lung, and Blood Institute of the National Institutes of Health [T32 HL007734]. K.H.K. is supported by NIH award UC4-DK-112217. K.S. is supported by NIH R01 DK087635. L.S.P. is supported in part by VA awards I01-CX001025, and I01CX001737, NIH awards R21DK099716, U01 DK091958, U01 DK098246, P30DK111024, and R03AI133172, and a Cystic Fibrosis Foundation award PHILLI12A0. We thank all study participants for their contribution. Data on T2D have been contributed by investigators from DIAMANTE Consortium, Biobank Japan, Malmö Diet and Cancer Study, PennCath, MedStar, Pakistan Genomic Resource, Penn Medicine Biobank, and Regeneron Genetics Center. Data on stroke were provided by MEGASTROKE investigators, and data on CKD have been contributed by CKDgen investigators. Data on alpha and beta islet cells have been contributed by the HPAP Consortium (RRID:SCR_016202 and https://hpap.pmacs.upenn.edu/). Data on coronary artery disease have been contributed by the CARDIoGRAMplusC4D investigators. We thank Josep Maria Mercader and Aaron Leong for careful review and comments.
Consortia
VA Million Veteran Program
Samuel M. Aguayo11, Sunil K. Ahuja44, Zuhair K. Ballas45, Sujata Bhushan46, Edward J. Boyko47, David M. Cohen48, John Concato49, Joseph I. Constans50, Louis J. Dellitalia51, Joseph M. Fayad52, Ronald S. Fernando53, Hermes J. Florez54, Melinda A. Gaddy55, Saib S. Gappy56, Gretchen Gibson57, Michael Godschalk58, Jennifer A. Greco59, Samir Gupta60, Salvador Gutierrez61, Kimberly D. Hammer62, Mark B. Hamner63, John B. Harley64, Adriana M. Hung65, Mostaqul Huq66, Robin A. Hurley67, Pran R. Iruvanti68, Douglas J. Ivins69, Frank J. Jacono70, Darshana N. Jhala71, Laurence S. Kaminsky72, Scott Kinlay16, Jon B. Klein73, Suthat Liangpunsakul74, Jack H. Lichy75, Stephen M. Mastorides76, Roy O. Mathew77, Kristin M. Mattocks78, Rachel McArdle79, Paul N. Meyer80, Laurence J. Meyer7, Jonathan P. Moorman81, Timothy R. Morgan82, Maureen Murdoch83, Xuan-Mai T. Nguyen16, Olaoluwa O. Okusaga84, Kris-Ann K. Oursler85, Nora R. Ratcliffe86, Michael I. Rauchman87, R. Brooks Robey88, George W. Ross89, Richard J. Servatius90, Satish C. Sharma91, Scott E. Sherman92, Elif Sonel93, Peruvemba Sriram94, Todd Stapley95, Robert T. Striker96, Neeraj Tandon97, Gerardo Villareal98, Agnes S. Wallbom99, John M. Wells9, Jeffrey C. Whittle100, Mary A. Whooley101, Junzhe Xu102, Shing-Shing Yeh103, Michaela Aslan49, Jessica V. Brewer16, Mary T. Brophy16, Todd Connor104, Dean P. Argyres104, Nhan V. Do16, Elizabeth R. Hauser105, Donald E. Humphries16, Luis E. Selva16, Shahpoor Shayan16, Brady Stephens106, Stacey B. Whitbourne16, Hongyu Zhao49, Jennifer Moser75, Jean C. Beckham105, Jim L. Breeling16, J.P. Casas Romero16, Grant D. Huang75, Rachel B. Ramoni16, Saiju Pyarajan16,25,26, Yan V. Sun34,35, Kelly Cho16,25, Peter W. Wilson34,40, Christopher J. O’Donnell16,25,26, Philip S. Tsao13,14, Kyong-Mi Chang1,2, J. Michael Gaziano16,25, and Sumitra Muralidhar75
The HPAP Consortium
Mark A. Atkinson107,108, Al C. Powers109,110,65, Ali Naji20, and Klaus H. Kaestner17
Regeneron Genetics Center
Goncalo R. Abecasis111, Aris Baras111, Michael N. Cantor111, Giovanni Coppola111, Aris N. Economides111, Luca A. Lotta111, John D. Overton111, Jeffrey G. Reid111, Alan R. Shuldiner111, Christina Beechert111, Caitlin Forsythe111, Erin D. Fuller111, Zhenhua Gu111, Michael Lattari111, Alexander E. Lopez111, Thomas D. Schleicher111, Maria Sotiropoulos Padilla111, Karina Toledo111, Louis Widom111, Sarah E. Wolf111, Manasi Pradhan111, Kia Manoochehri111, Ricardo H. Ulloa111, Xiaodong Bai111, Suganthi Balasubramanian111, Leland Barnard111, Andrew L. Blumenfeld111, Gisu Eom111, Lukas Habegger111, Alicia Hawes111, Shareef Khalid111, Evan K. Maxwell111, William J. Salerno111, Jeffrey C. Staples111, Ashish Yadav111, Marcus B. Jones111, and Lyndon J. Mitnaul111
44South Texas Veterans Health Care System, San Antonio, TX, USA. 45Iowa City VA Health Care System, Iowa City, IA, USA. 46VA North Texas Health Care System, Dallas, TX, USA. 47VA Puget Sound Health Care System, Seattle, WA, USA. 48Portland VA Medical Center, Portland, OR, USA. 49VA Connecticut Healthcare System, West Haven, CT, USA. 50Southeast Louisiana Veterans Health Care System, New Orleans, LA, USA. 51Birmingham VA Medical Center, Birmingham, AL, USA. 52VA Southern Nevada Healthcare System, North Las Vegas, NV, USA. 53VA Loma Linda Healthcare System, Loma Linda, CA, USA. 54Miami VA Health Care System, Miami, FL, USA. 55VA Eastern Kansas Health Care System, Leavenworth, KS, USA. 56John D. Dingell VA Medical Center, Detroit, MI, USA. 57Fayetteville VA Medical Center, Fayetteville, AR, USA. 58Richmond VA Medical Center, Richmond, VA, USA. 59Sioux Falls VA Health Care System, Sioux Falls, SD, USA. 60VA San Diego Healthcare System, San Diego, CA, USA. 61Edward Hines Jr. VA Medical Center, Hines, IL, USA. 62Fargo VA Health Care System, Fargo, ND, USA. 63Ralph H. Johnson VA Medical Center, Charleston, SC, USA. 64Cincinnati VA Medical Center, Cincinnati, OH, USA. 65VA Tennessee Valley Healthcare System, Nashville, TN, USA. 66VA Sierra Nevada Health Care System, Reno, NV, USA. 67W.G. (Bill) Hefner VA Medical Center, Salisbury, NC, USA. 68Hampton VA Medical Center, Hampton, VA, USA. 69Eastern Oklahoma VA Health Care System, Muskogee, OK, USA. 70VA Northeast Ohio Healthcare System, Cleveland, OH, USA. 71Philadelphia VA Medical Center, Philadelphia, PA, USA. 72VA Health Care Upstate New York, Albany, NY, USA. 73Louisville VA Medical Center, Louisville, KY, USA. 74Richard Roudebush VA Medical Center, Indianapolis, IN, USA. 75Washington DC VA Medical Center, Washington, D.C., USA. 76James A. Haley Veterans Hospital, Tampa, FL, USA. 77Columbia VA Health Care System, Columbia, SC, USA. 78Central Western Massachusetts Healthcare System, Leeds, MA, USA. 79Bay Pines VA Healthcare System, Bay Pines, FL, USA. 80Southern Arizona VA Health Care System, Tucson, AZ, USA. 81James H. Quillen VA Medical Center, Johnson City, TN, USA. 82VA Long Beach Healthcare System, Long Beach, CA, USA. 83Minneapolis VA Health Care System, Minneapolis, MN, USA. 84Michael E. DeBakey VA Medical Center, Houston, TX, USA. 85Salem VA Medical Center, Salem, VA, USA. 86Manchester VA Medical Center, Manchester, NH, USA. 87St. Louis VA Health Care System, St. Louis, MO, USA. 88White River Junction VA Medical Center, White River Junction, VT, USA. 89VA Pacific Islands Health Care System, Honolulu, HI, USA. 90Syracuse VA Medical Center, Syracuse, NY, USA. 91Providence VA Medical Center, Providence, RI, USA. 92VA New York Harbor Healthcare System, New York, NY, USA. 93VA Pittsburgh Health Care System, Pittsburgh, PA, USA. 94North Florida / South Georgia Veterans Health System, Gainesville, FL, USA. 95VA Maine Healthcare System, Augusta, ME, USA. 96William S. Middleton Memorial Veterans Hospital, Madison, WI, USA. 97Overton Brooks VA Medical Center, Shreveport, LA, USA. 98New Mexico VA Health Care System, Albuquerque, NM, USA. 99VA Greater Los Angeles Health Care System, Los Angeles, CA, USA. 100Clement J. Zablocki VA Medical Center, Milwaukee, WI, USA. 101San Francisco VA Health Care System, San Francisco, CA, USA. 102VA Western New York Healthcare System, Buffalo, NY, USA. 103Northport VA Medical Center, Northport, NY, USA. 104Raymond G. Murphy VA Medical Center, Albuquerque, NM, USA. 105Durham VA Medical Center, Durham, NC, USA. 106Canandaigua VA Medical Center, Canandaigua, NY, USA. 107Department of Pathology, University of Florida Diabetes Institute, Gainesville, FL, USA. 108Department of Pediatrics, University of Florida Diabetes Institute, Gainesville, FL, USA. 109Department of Molecular Physiology and Biophysics, Vanderbilt University, Nashville, TN, USA. 110Division of Diabetes, Endocrinology, and Metabolism, Vanderbilt University Medical Center, Nashville, TN, USA. 111Regeneron Pharmaceuticals, Inc., Tarrytown, NY, USA.
Footnotes
Competing Interests Statement
None of the sponsors of the following authors had a role in the design and conduct of the study; collection, management, analysis, and interpretation of the data; and preparation, review, or approval of the manuscript. D.S. has received support from the British Heart Foundation, Pfizer, Regeneron, Genentech, and Eli Lilly pharmaceuticals. L.S.P. has served on Scientific Advisory Boards for Janssen, and received research support from Abbvie, Merck, Amylin, Eli Lilly, Novo Nordisk, Sanofi, PhaseBio, Roche, Abbvie, Vascular Pharmaceuticals, Janssen, Glaxo SmithKline, Pfizer, Kowa, and the Cystic Fibrosis Foundation. L.S.P. is a cofounder, officer, board member, and stockholder of a diabetes management-related software company names Diasyst, Inc. S.L.D. has received research grant support from the following for-profit companies through the University of Utah or the Western Institute for Biomedical Research (VA Salt Lake City’s affiliated non-profit): AbbVie Inc., Anolinx LLC, Astellas Pharma Inc., AstraZeneca Pharmaceuticals LP, Boehringer Ingelheim International GmbH, Celgene Corporation, Eli Lilly and Company, Genentech Inc., Genomic Health, Inc., Gilead Sciences Inc., GlaxoSmithKline PLC, Innocrin Pharmaceuticals Inc., Janssen Pharmaceuticals, Inc., Kantar Health, Myriad Genetic Laboratories, Inc., Novartis International AG, and PAREXEL International Corporation. P.D.R. has received research grant support from the following for-profit companies: Bristol Myers Squib, Lysulin Inc; and has consulted with Intercept Pharmaceuticals and Boston Heart Diagnostics. S.M.D. receives research support to the University of Pennsylvania from RenalytixAI and consults for Calico Labs.
Ethics statement
The Central Veterans Affairs Institutional Review Board (IRB) and site-specific Research and Development Committees approved the Million Veteran Program study. The Vanderbilt University Medical Center IRB approved the use of BioVU data for this study. All other cohorts participating in this meta-analysis have ethical approval from their local institutions. All relevant ethical regulations were followed.
Reporting Summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Data availability
The full summary level association data from the trans-ancestry, European, African American, Hispanic, and Asian meta-analysis from this report are available through dbGAP under accession number phs001672.v3.p1.
Code availability
Imputation was performed using MiniMac4 and EAGLE v2. Association analysis was performed using PLINK2A and XWAS v3.0. Post-GWAS processing software include: PRSice-2, LD Hub v1.9.3, FlashPCA v2.0, METAL v2011-03-25, GCTA-COJO v1.93, S-PrediXcan v0.6.1, SUGEN v8.9, DEPICT v140721, SIDER v4.1, DGidb v3.0, and KING v2.1.6, as outlined in the Methods. Clear code for analysis is available at their associated websites. Additional analyses were performed in R-3.2.
References
- 1.IDF Diabetes Atlas, 8th edn. International Diabetes Federation; (2017). [Google Scholar]
- 2.Standards of Medical Care in Diabetes, 2018. Diabetes Care 41, S1–S2 (2018). [DOI] [PubMed] [Google Scholar]
- 3.Mahajan A et al. Fine-mapping type 2 diabetes loci to single-variant resolution using high-density imputation and islet-specific epigenome maps. Nat. Genet 50, 1505–1513 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Suzuki K et al. Identification of 28 new susceptibility loci for type 2 diabetes in the Japanese population. Nat. Genet 51, 379–386 (2019). [DOI] [PubMed] [Google Scholar]
- 5.Gaziano JM et al. Million Veteran Program: a mega-biobank to study genetic influences on health and disease. J. Clin. Epidemiol 70, 214–223 (2016). [DOI] [PubMed] [Google Scholar]
- 6.Levin MG et al. Genomic risk stratification predicts all-cause mortality after cardiac catheterization. Circ. Genom. Precis. Med 11, e002352 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Saleheen D et al. Human knockouts and phenotypic analysis in a cohort with a high rate of consanguinity. Nature 544, 235–239 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Berglund G, Elmstahl S, Janzon L & Larsson SA The Malmo Diet and Cancer Study. Design and feasibility. J. Intern. Med 233, 45–51 (1993). [DOI] [PubMed] [Google Scholar]
- 9.Reilly MP et al. Identification of ADAMTS7 as a novel locus for coronary atherosclerosis and association of ABO with myocardial infarction in the presence of coronary atherosclerosis: two genome-wide association studies. Lancet 377, 383–392 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bonas-Guarch S et al. Re-analysis of public genetic data reveals a rare X-chromosomal variant associated with type 2 diabetes. Nat. Commun 9, 321 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xue A et al. Genome-wide association analyses identify 143 risk variants and putative regulatory mechanisms for type 2 diabetes. Nat. Commun 9, 2941 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bulik-Sullivan BK et al. LD Score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat. Genet 47, 291–295 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Devlin B & Roeder K Genomic control for association studies. Biometrics 55, 997–1004 (1999). [DOI] [PubMed] [Google Scholar]
- 14.Luo Y et al. Estimating heritability of complex traits in admixed populations with summary statistics. bioRxiv, 503144 (2018). [Google Scholar]
- 15.Kuhn M, Letunic I, Jensen LJ & Bork P The SIDER database of drugs and side effects. Nucleic Acids Res. 44, D1075–D1079 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Finucane HK et al. Partitioning heritability by functional annotation using genome-wide association summary statistics. Nat. Genet 47, 1228–1235 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schmidt EM et al. GREGOR: evaluating global enrichment of trait-associated variants in epigenomic features using a systematic, data-driven approach. Bioinformatics 31, 2601–2606 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ng MC et al. Meta-analysis of genome-wide association studies in African Americans provides insights into the genetic architecture of type 2 diabetes. PLoS Genet. 10, e1004517 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen J et al. Genome-wide association study of type 2 diabetes in Africa. Diabetologia 62, 1204–1211 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Palmer ND et al. A genome-wide association search for type 2 diabetes genes in African Americans. PLoS One 7, e29202 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wheeler E et al. Impact of common genetic determinants of Hemoglobin A1c on type 2 diabetes risk and diagnosis in ancestrally diverse populations: a transethnic genome-wide meta-analysis. PLoS Med. 14, e1002383 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mahajan A et al. Refining the accuracy of validated target identification through coding variant fine-mapping in type 2 diabetes. Nat. Genet 50, 559–571 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Flannick J et al. Exome sequencing of 20,791 cases of type 2 diabetes and 24,440 controls. Nature 570, 71–76 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Carrano AC, Mulas F, Zeng C & Sander M Interrogating islets in health and disease with single-cell technologies. Mol. Metab 6, 991–1001 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Martin CK, Han H, Anton SD, Greenway FL & Smith SR Effect of valproic acid on body weight, food intake, physical activity and hormones: results of a randomized controlled trial. J. Psychopharmacol 23, 814–825 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Buse M et al. Expanding the phenotype of reciprocal 1q21.1 deletions and duplications: a case series. Ital. J. Pediatr 43, 61 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Devi RR & Vijayalakshmi P Novel mutations in GJA8 associated with autosomal dominant congenital cataract and microcornea. Mol. Vis 12, 190–5 (2006). [PubMed] [Google Scholar]
- 28.Mackay DS, Bennett TM, Culican SM & Shiels A Exome sequencing identifies novel and recurrent mutations in GJA8 and CRYGD associated with inherited cataract. Hum. Genomics 8, 19 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Luo J et al. TCF7L2 variation and proliferative diabetic retinopathy. Diabetes 62, 2613–2617 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Eiden LE, Schafer MK, Weihe E & Schutz B The vesicular amine transporter family (SLC18): amine/proton antiporters required for vesicular accumulation and regulated exocytotic secretion of monoamines and acetylcholine. Pflugers Arch. 447, 636–640 (2004). [DOI] [PubMed] [Google Scholar]
- 31.Sharma P & Sharma R Toxic optic neuropathy. Indian J. Ophthalmol 59, 137–141 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pattaro C et al. Genetic associations at 53 loci highlight cell types and biological pathways relevant for kidney function. Nat. Commun 7, 10023 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Canela-Xandri O, Rawlik K & Tenesa A An atlas of genetic associations in UK Biobank. Nat. Genet 50, 1593–1599 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ehret GB et al. The genetics of blood pressure regulation and its target organs from association studies in 342,415 individuals. Nat. Genet 48, 1171–1184 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sung YJ et al. A large-scale multi-ancestry genome-wide study accounting for smoking behavior identifies multiple significant loci for blood pressure. Am. J. Hum. Genet 102, 375–400 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Maass PG et al. PDE3A mutations cause autosomal dominant hypertension with brachydactyly. Nat. Genet 47, 647–653 (2015). [DOI] [PubMed] [Google Scholar]
- 37.Shin S et al. CREB mediates the insulinotropic and anti-apoptotic effects of GLP-1 signaling in adult mouse beta-cells. Mol. Metab 3, 803–812 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Omar B, Banke E, Ekelund M, Frederiksen S & Degerman E Alterations in cyclic nucleotide phosphodiesterase activities in omental and subcutaneous adipose tissues in human obesity. Nutr. Diabetes 1, e13 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Manichaikul A et al. Robust relationship inference in genome-wide association studies. Bioinformatics 26, 2867–2873 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.1000 Genomes Project Consortium et al. A global reference for human genetic variation. Nature 526, 68–74 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Das S et al. Next-generation genotype imputation service and methods. Nat. Genet 48, 1284–1287 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fang H et al. Harmonizing genetic ancestry and self-identified race/ethnicity in genome-wide association studies. Am. J. Hum. Genet 105, 763–772 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tirschwell DL & Longstreth WT Jr. Validating administrative data in stroke research. Stroke 33, 2465–2470 (2002). [DOI] [PubMed] [Google Scholar]
- 44.Chang CC et al. Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience 4, 7 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Willer CJ, Li Y & Abecasis GR METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics 26, 2190–2191 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gao F et al. XWAS: a software toolset for genetic data analysis and association studies of the X chromosome. J. Hered 106, 666–671 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ko YA et al. Genetic-variation-driven gene-expression changes highlight genes with important functions for kidney disease. Am. J. Hum. Genet 100, 940–953 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ackermann AM, Wang Z, Schug J, Naji A & Kaestner KH Integration of ATAC-seq and RNA-seq identifies human alpha cell and beta cell signature genes. Mol. Metab 5, 233–244 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.International HapMap Consortium et al. Integrating common and rare genetic variation in diverse human populations. Nature 467, 52–58 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature 489, 57–74 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Roadmap Epigenomics Consortium et al. Integrative analysis of 111 reference human epigenomes. Nature 518, 317–330 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Andersson R et al. An atlas of active enhancers across human cell types and tissues. Nature 507, 455–461 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Finucane HK et al. Heritability enrichment of specifically expressed genes identifies disease-relevant tissues and cell types. Nat. Genet 50, 621–629 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fehrmann RS et al. Gene expression analysis identifies global gene dosage sensitivity in cancer. Nat. Genet 47, 1151–25 (2015). [DOI] [PubMed] [Google Scholar]
- 55.Cahoy JD et al. A transcriptome database for astrocytes, neurons, and oligodendrocytes: a new resource for understanding brain development and function. J. Neurosci 28, 264–278 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Heng TS, Painter MW & Immunological Genome Project Consortium. The Immunological Genome Project: networks of gene expression in immune cells. Nat. Immunol 9, 1091–1094 (2008). [DOI] [PubMed] [Google Scholar]
- 57.Denny JC et al. PheWAS: demonstrating the feasibility of a phenome-wide scan to discover gene-disease associations. Bioinformatics 26, 1205–1210 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Voorman A, Lumley T, McKnight B & Rice K Behavior of QQ-plots and genomic control in studies of gene-environment interaction. PLoS One 6, e19416 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lin DY et al. Genetic association analysis under complex survey sampling: the Hispanic Community Health Study/Study of Latinos. Am. J. Hum. Genet 95, 675–688 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bulik-Sullivan B et al. An atlas of genetic correlations across human diseases and traits. Nat. Genet 47, 1236–1241 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pers TH et al. Biological interpretation of genome-wide association studies using predicted gene functions. Nat. Commun 6, 5890 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Source Data Extended Data Fig. 5 Raw odds ratios for T2D shape plot
Source Data Fig. 2 Raw odds ratios for T2D-related outcomes shape plots
Source Data Extended Data Fig. 3 Raw effect estimates and P-values for inverted Manhattan plot depicting genetically predicted gene expression using S-PrediXcan


