Abstract
Metastasis is the primary cause of cancer-related deaths, but the natural history, clonal evolution and impact of treatment are poorly understood. We analyzed whole-exome sequencing data from 457 paired primary tumor and metastatic samples from 136 breast, colorectal and lung cancer patients, including untreated (n=99) and treated (n=100) metastases. Treated metastases often harbored private ‘driver’ mutations whereas untreated metastases did not, suggesting that treatment promotes clonal evolution. Polyclonal seeding was common in untreated lymph node metastases (n=17/29, 59%) and distant metastases (n=20/70, 29%), but less frequent in treated distant metastases (n=9/94, 10%). The low number of metastasis-private clonal mutations is consistent with early metastatic seeding, which we estimated occurred 2–4 years prior to diagnosis across these cancers. Further, these data suggest that the natural course of metastasis is selectively relaxed relative to early tumorigenesis and that metastasis-private mutations are not drivers of cancer spread but instead associated with drug resistance.
Introduction
Metastasis remains poorly understood despite its critical clinical importance. For instance, metastases have been reported to originate from a single cell or clone in the primary tumor (monoclonal seeding) 1–4 or multiple clones (polyclonal seeding) 5–7, but the prevalence of these patterns across distinct tumor types is unknown as is the impact of therapy and the timing of metastatic seeding 8–10. While several recent studies have genomically characterized metastatic lesions in the absence of the matched primary tumor 11–13, with such data it is not feasible to disentangle the drivers of metastasis from those that are treatment associated since metastases are often sampled after treatment. However, comparisons of paired primary tumors and metastases have been far more limited due to the challenge in obtaining such samples 5,8,14–18. As such, there has yet to be a systematic analysis of monoclonal versus polyclonal seeding, the chronology of systemic spread and the effect of therapy across cancers.
Here we analyzed whole-exome sequencing (WES) data from 457 paired primary tumor (P) and metastases (M) from 136 patients with colorectal, lung or breast cancers using a uniform bioinformatics pipeline. We assessed ‘driver’ gene heterogeneity and evaluated the prevalence of monoclonal versus polyclonal seeding, revealing considerable variability between untreated and treated metastases across cancer types. Treatment was associated with high primary tumor versus metastasis (P/M) driver gene heterogeneity and monoclonal metastases. Metastatic seeding was estimated to occur two to four years prior to diagnosis of the primary tumor across three common cancer types, with breast cancers generally disseminating later and therefore closer to the time of detection relative to colorectal and lung cancers. Collectively, these observations suggest that systemic spread can begin early during tumor growth and that clonal architecture is remodeled by treatment, providing new insights into the clonal evolution of metastasis.
Results
Genomic landscapes of paired primary tumors and metastases
We performed a literature review to identify cohorts with genomic sequencing data from matched normals, primary tumors (P) and metastases (M) from patients with three common cancer types, namely, colorectal16,17,19–22, lung23,24 and breast23,25–29 (Supplementary Tables 1–2, Extended Data Fig. 1). All samples were processed within a uniform bioinformatics pipeline 16,30 to identify somatic single nucleotide variants (SSNVs), insertions/deletions (indels) and somatic copy number alterations (SCNAs) (Methods). Tumor purity/ploidy and cancer cell fraction (CCF) of SSNVs and indels (referred as SSNVs hereafter) were estimated in order to distinguish clonal (the upper bound of 95% confidence interval or CI of CCF ≥ 1) versus subclonal (the upper bound of 95% CI of CCF < 1) SSNVs (Methods). Following quality control assessment (Methods), 457 tumor samples from 136 patients (colorectal cancer, n=39; lung cancer, n=30; breast cancer, n=67) were retained for downstream analysis (Supplementary Tables 1–3, Extended Data Fig. 1).
Metastases exhibited higher purity than paired primary tumors while ploidy was comparable between P/M pairs in all three cancer types (Supplementary Fig. 1). Overall, the mutational burden (SSNVs or SCNAs) was highly concordant between P/M pairs (Extended Data Fig. 2, Supplementary Figs. 2–3, Supplementary Table 4), although differences between cancer types were noted. For instance, in breast cancer, the SCNA burden between P/M pairs was more concordant than the SSNV burden (Extended Data Fig. 2), a pattern that is more evident in ER+/HER− subgroup (Supplementary Fig. 3). Although primary breast cancers can be copy number driven 31,32, these data suggest that metastases can acquire substantial SSNVs and this seemed especially true in ER+/HER− breast cancers, which are often exposed to endocrine therapy and tend to recur later 33. In all three cancer types, metastases exhibited a slight increase in the number of clonal SSNVs and fewer subclonal SSNVs (Supplementary Fig. 2), consistent with an evolutionary bottleneck during metastasis. The mutational spectrum of M-private SSNVs (clonal or subclonal) between treated and untreated metastases was also highly concordant except that treated colorectal metastases were characterized by an enrichment of T>G transversions relative to untreated samples (Supplementary Fig. 4). Indeed, all treated colorectal metastases (n=7) were biopsied after 5-fluorouracil (5-FU) chemotherapy in this cohort, which was recently shown to be associated with this mutational pattern 34,35.
We next evaluated the enrichment of functional driver gene mutations in paired primary tumors and metastases. Three methods, namely PolyPhen-2 36, FATHMM-XF 37 and CHASMplus 38, were employed to assess the functionality (“driverness”) of nonsynonymous SSNVs in putative driver genes according to TCGA and COSMIC (Methods, Supplementary Table 5). In total, 1,086 functional driver SSNVs/indels were detected across these three cancer types (Fig. 1a-b, Supplementary Table 6), in which 734 were clonal (including shared clonal, P-private clonal or M-private clonal) and 352 were subclonal (shared subclonal, P subclonal/M clonal, P-private subclonal or M-private subclonal). Notably, 84%, 86% and 59% of clonal drivers in each of P and M were shared in colorectal, lung and breast cancer, respectively, while the fractions of subclonal drivers was 20%, 50% and 23%, respectively (Fig. 1c). Of note, colorectal cancer had highest prevalence of P-private subclonal drivers likely because multi-region sequencing (MRS) data were more prevalent in this tumor type (36%, or 14/39) as compared to lung (0%, 0/30) and breast (9% or 6/67) cancers, while MRS increases the power to detect subclonal mutations. Breast cancer exhibited higher prevalence of both M-private clonal and subclonal driver mutations as compared to colorectal and lung cancers (Fig. 1c). Gene ontology (GO) analysis of M-private driver genes showed enrichment for chromatin binding, modification and organization genes (Supplementary Fig. 5, Supplementary Table 7), implicating chromatin regulators in metastatic progression 39.
Fig. 1 |. Landscape of driver mutations in paired primary tumors (P) and metastases (M).
a, Oncoprint of functional driver mutations in shared, P-private or M-private drivers. Genes mutated in at least three patients are shown. White circles indicate genes with multiple mutations in an individual patient. b, Ternary plot of mutation counts in driver genes, comparing P-private (left, green), M-private (right, red), and shared (top, blue). The size represents their overall count in the corresponding cancer type. c, The proportion of different classes of clonal and subclonal mutations in each of the three cancer types. d, The ratio of shared clonal to M-private clonal mutations for all non-silent or driver mutations. A down-sampling procedure was performed to derive the ratio (Methods). P-value, Wilcoxon Rank-Sum Test (two-sided). Bar, median; box, 25th to 75th percentile (interquartile range, IQR); vertical line, data within 1.5 times the IQR. e, The proportion of metastases harboring at least one private clonal driver mutation in all, untreated or treated metastases. P-value, Fisher’s exact test (two-sided). f-g, Schematic of the major clone model where metastasis originates from the major driver clone in the primary tumor (f) leading to driver homogeneity between paired P and M biopsies or of the minor clonal model where the metastasis originates from a minor driver clone in the primary tumor (g). Due to the inability to detect low frequency mutations by bulk sequencing, the minor clone model leads to driver heterogeneity between P and M biopsies.
Amongst all non-silent clonal SSNVs in metastases, functional driver mutations were highly enriched on the trunk (P/M shared clonal) of the phylogenetic tree in both colorectal and breast cancers (Fig. 1d, Methods). However, this pattern was much weaker in lung cancer (Fig. 1d), presumably due to the large number of tobacco-associated non-silent clonal SSNVs (C>A mutations) induced early during lung cancer development (Supplementary Fig. 6) as most of the lung cancer patients in this cohort (~90%) had a smoking history 23,24, whereas driver mutations did not increase proportionally (Fig. 1d). In line with these results, the decreased ratio of nonsynonymous versus synonymous SSNVs (dN/dS) 40 amongst putative driver genes in metastases (Extended Data Fig. 3) suggests relaxed selective pressure relative to early cancer development in colorectal and breast cancers, but not lung cancer. Only 25%, 33% and 48% of colorectal, lung and breast cancer metastases, respectively, harbored one or more private clonal driver mutations and these values were lower when restricted to untreated metastases (19%, 22% and 22%, respectively) (Fig. 1e). These data suggest that untreated metastases commonly arise from the major (or dominant) clone in the primary tumor leading to driver homogeneity (Fig. 1f). However, amongst treated metastases, the proportion of private-clonal drivers increased dramatically across all three cancer types with 71%, 75% and 53% in colorectal, lung and breast cancer, respectively (Fig. 1e). This pattern was similarly evident in patients where both untreated and treated metastases were sampled where all (10/10) treated metastases harbored private functional driver mutation(s), but few (2/10) untreated lymph node metastases did (Supplementary Table 6). Therefore, these data suggest that the therapy selects a minor micrometastatic subclone (Fig. 1g). Hence, treatment confers a stringent selective pressure and promotes clonal evolution of the metastasis. Meanwhile, although the overall copy number landscape is highly concordant between paired primary tumors and metastases (Extended Data Fig. 4), copy number analysis identified a small number of putative driver genes that were more frequently amplified or deleted in metastases relative to primary tumors (increasing from P to M by 15%, Extended Data Fig. 5). These include amplification of RAC1 and deletions of FAT1 and ALB in colorectal cancer, amplifications of PLCG1 and SALL4 and deletions of NOTCH2, CDKN1B in lung cancer and amplifications of IL7R, NIPBL and deletions of NOTCH1, PTEN in breast cancer (Extended Data Fig. 5). Collectively, these data suggest that the genomic drivers required for invasion and metastasis often occur early in the primary tumor (Fig. 1f).
Clonality of metastatic seeding
In order to infer the clonality of individual metastases (Fig. 2a), we compared the CCFs of SSNVs in each P/M pair and the number of M-private clonal SSNVs, P-private clonal SSNVs and P/M shared subclonal SSNVs was denoted as Lm, Lp and Ws, respectively (Fig. 2b). We used the Jaccard similarity index (JSI) where to quantify mutational similarity between P/M pairs 41 (Methods). Polyclonal seeding is expected to result in a higher JSI than monoclonal seeding due to the higher proportion of shared subclonal SSNVs (higher Ws) and the presence of fewer M or P-private clonal SSNVs (lower Lm and Lp) (Fig. 2b). These patterns were verified by simulation studies using an established agent-based model of spatial tumor progression 16,30 (Extended Data Figs. 6–7, Methods). Notably, polyclonal seeding can be either a multicellular event (by cell cluster) or multiple consecutive single-cell events (Fig. 2a). However, current data is underpowered to distinguish these two scenarios as the resultant patterns of genomic heterogeneity between the primary tumor and metastasis are highly similar. We therefore only modeled polyclonal seeding by cell clusters (Methods). By analyzing data from virtual tumors simulated under varied parameters where one biopsy (~106 cells) was sampled from each primary tumor and metastasis (Methods), we found that a JSI value of 0.3 maximizes the classification accuracy (91.1%) in distinguishing monoclonal versus polyclonal seeding (Fig. 2c). We also simulated MRS data (n=4 samples from each of primary tumor and metastasis) and found that the optimal JSI cutoff increased to 0.4 and yielded an increased classification accuracy (96.3%) relative to single sample data (Supplementary Fig. 7). Given that most (>80%) patients in this study only had only a single sample from the primary tumor and metastasis, we retain the 0.3 cutoff for analyses (Fig. 2c). Most metastases exhibited patterns consistent with monoclonal seeding (n=151, 76% of metastases; median JSI=0.075, interquartile range, IQR=0.021−0.138), whereas polyclonal seeding was less frequent (n=48, 24% of metastases; median JSI=0.523, IQR=0.469−0.800) (Figs. 2c).
Fig. 2 |. The clonality of metastatic seeding.
a, Schematic of monoclonal versus polyclonal seeding of a metastasis. Polyclonal seeding occurs either through a cell cluster or multiple monoclonal dissemination events. b, Distinct patterns of seeding are evident based on the cancer cell fraction (CCF) of SSNVs between primary (P)/metastatic (M) pairs, where representative patients are shown: monoclonal (colon cancer V402); polyclonal (lung cancer TH6). c, Classification of monoclonal versus polyclonal seeding based on the JSI. Top, JSI values in 1000 virtual P/M tumor pairs. Middle, classification accuracy by varying the cutoff of JSI from 0–1 based on simulation data. Bottom, JSI values in patient data (n=199 P/M pairs) where the 0.3 cutoff was used to identify monoclonal (n=151) or polyclonal seeding (n=48). d, Lm, Lp, Ws values in patient data. e, The number of P-to-M altered SCNAs for monoclonal (n=151) and polyclonal (n=48) metastases. P-value, Wilcoxon Rank-Sum Test (two-sided). f, Positive correlation between Lm and the number of P-to-M altered SCNAs. n=199 P/M pairs; Spearman’s correlation (rho) and P-value are reported. g, Polyclonal seeding is common in LNM and untreated distant metastases relative to treated distant metastases. h, Schematic of how treatment can promote monoclonality as a result of selection for a resistant subclone, despite initial seeding by polyclonal disseminated cells. Box plots: bar, median; box, 25th to 75th percentile (interquartile range, IQR); vertical/horizontal line across box, data within 1.5 times the IQR. Jaccard similarity index, JSI; brain metastasis, BM; lymph node metastasis; LNM.
As expected, monoclonal metastases (n=151) exhibited significantly higher Lm and Lp values than polyclonal metastases (n=48) (P = 6.2 × 10−16 and P = 2.1 × 10−9 for Lm and Lp, respectively, two-sided Wilcoxon Rank Sum Test) and significantly lower Ws values (P = 2.1 × 10−12, two-sided Wilcoxon Rank Sum Test) (Fig. 2d). Metastases of monoclonal origin also harbored significantly more SCNAs relative to paired primary tumors than polyclonal metastases (P = 1.9 × 10−8, two-sided Wilcoxon Rank Sum Test; Fig. 2e). Indeed, Lm is highly correlated with the number of P-to-M altered SCNAs (Spearman’s rho=0.61, P = 3.3 × 10−20; Fig. 2f), indicating that both SSNVs and SCNAs reflect the clonality of metastases. Polyclonal seeding was more prevalent in axillary lymph node metastases (all, 19/35 or 54%) relative to distant metastases (29/164 or 18%) (P = 1.8 × 10−5, two-sided Fisher’s exact test; Fig. 2g, Extended Data Fig. 8). This pattern is also true for untreated metastases (lymph node vs distant, 17/29 or 59% vs 20/70 or 29%; P=0.007, two-sided Fisher’s exact test), potentially reflecting greater lymphatic spread of disseminated cells to the lymph nodes via multiple dissemination events (Fig. 2a). Amongst distant metastases, polyclonal seeding was more prevalent in untreated metastases (20/70 or 29%) than treated metastases (9/94 or 10%) (P=0.002, two-sided Fisher’s exact test; Fig. 2g), presumably because treatment selects for resistant micrometastatic subclones that manifest clinically as monoclonal metastases (Fig. 2h). The higher P/M driver gene heterogeneity observed in treated versus untreated metastases (Fig. 1e) is consistent with this scenario. The prevalence of polyclonal seeding differed across metastatic sites (lymph node, liver, brain and lung), with brain and lung more commonly exhibiting monoclonal seeding (Extended Data Fig. 8b); these two sites were more commonly biopsied after treatment. In fact, the prevalence of polyclonal seeding amongst lymph node (54%), liver (26%), brain (17%) and lung (8%) metastases is negatively associated with the fraction of metastases that were treated amongst these four sites (17%, 21%, 68%, 92%). This pattern is most evident for brain metastases which had ample numbers of both treated (n=43) and untreated (n=21) metastases. Here, the prevalence of polyclonal seeding is 7% (3/43) amongst treated and 38% (8/21) amongst untreated metastases (P=0.004, two-sided Fisher’s exact test; Extended Data Fig. 8b). Therefore, the prevalence of polyclonal seeding is variable across metastatic sites and dependent on whether treatment was administrated before sampling. Since treatment influences the clonality of metastases, we would expect that polyclonal seeding of distant metastases might be more common during the natural course of metastasis (in the absence of treatment) than was observed (18%) here.
We further verified the JSI-based classification of monoclonal versus polyclonal seeding by phylogenetic analysis of patients with MRS of the primary tumor and metastasis (n=13 patients; Fig. 3, Supplementary Fig. 8). Monoclonal seeding was associated with a monophyletic tree structure (metastatic samples comprise a single phylogenetic clade), whereas polyclonal seeding was associated with a polyphyletic structure (metastatic samples comprise multiple phylogenetic clades) (Fig. 3, Supplementary Fig. 8).
Fig. 3 |. Tumor sample phylogenies based on multi-region sequencing data.
The maximum parsimony method was used to reconstruct multi-sample trees for each patient based on the presence or absence SSNVs/indels amongst the samples. For each primary(P)/metastatic(M) sample pair, the Jaccard similarity index (JSI) was computed according to Eq. (4) based on the numbers of M-private clonal, P-private clonal and P-M shared subclonal SSNVs. High JSI values (≥0.3) indicates polyclonal seeding while low JSI values (<0.3) indicates monoclonal seeding. Monoclonal seeding gives rise to monophyletic tree structures (pink shading indicates metastatic samples within a single phylogenetic clade), whereas polyclonal seeding gives rise to a polyphyletic structure (blue shading indicates metastatic samples within multiple phylogenetic clades) in the metastasis samples. P, primary tumor; OvM, ovarian metastasis; LNM, lymph node metastasis; SkM, skin metastasis; LiM, liver metastasis. Additional patient data are shown in Supplementary Fig. 8.
We also evaluated the association between clonality and patient outcome (e.g. time to metastatic relapse) based on untreated distant metastases, whereas treated metastases were excluded due the impact of treatment on clonality. In total, there were 70 untreated distant metastases in our cohort: liver: n=45; brain: n=21; bone: n=2; lung: n=1; skin: n=1. We thus focus on liver and brain metastases. Most untreated liver metastases were synchronously diagnosed (91%; 41/45). Of note, all four metachronous liver metastases (time to relapse ranged from 7–8 months) were monoclonal. Amongst brain metastases, 13 exhibited patterns consistent with monoclonal seeding while 8 were polyclonal seeding. Amongst, 31% (4/13) and 37.5% (3/8) were metachronous, respectively. Notably, the time to relapse was longer for monoclonal brain metastasis (median=26 months, IQR=(19, 36); synchronous metastases excluded) than polyclonal brain metastasis (median=11 months, IQR=(9, 17); synchronous metastases excluded). Although limited by the small sample size, this (non-significant) trend suggests that polyclonal seeding may be associated with worse prognosis. However, further studies on large untreated metastatic cohorts are warranted.
Chronology of metastatic seeding
Previously, we described a computational framework (SCIMET) to estimate the timing of metastatic seeding relative to primary tumor size based on MRS of P/M pairs 16. Application of this approach to colorectal cancer yielded quantitative evidence for early systemic spread, well before the primary tumor was clinically detectable. Since MRS data were not available for the vast majority of patients in this cohort, we developed a new computational method that leverages exome sequencing data from a single biopsy to time metastatic seeding (Fig. 4a, Extended Data Fig. 9a, Supplementary Note). The time (in years) from metastatic seeding to diagnosis of the primary tumor (ts) can be approximated by:
| Eq. (1) |
where Lm and Lp correspond to the number of M-private clonal SSNVs and P-private clonal SSNVs, respectively; T is the primary tumor expansion age (time from emergence of carcinoma founder cell to diagnosis); where is the time from emergence of carcinoma founder cell to the most recent common ancestor in the primary tumor sample (pMRCA, Fig. 4a, Extended Data Fig. 9a, Supplementary Note). The time fraction α is expected to be small because bulk sequencing only detects relatively high frequency mutations that occur early during tumor growth or that are strongly selected for 42–44. We applied our established agent-based model of spatial tumor growth 30 to simulate a large set of virtual tumors (n=1,000, each ~109 cells) with varying growth rates (Methods). In silico sequencing of a single biopsy (each ~106 cells, mean depth=100X) from the virtual tumors (n=1,000) yields an estimate of =0.13±0.0028 (Extended Data Fig. 9b), confirming the observation that bulk sequencing typically only detects high-frequency mutations that occur early during tumor growth. Here we assume a model of stringent selection (selection coefficient, s=0.1) during growth of the primary tumor based on our prior analysis of MRS data which showed evidence for selection in primary colon cancers within a metastatic cohort 16, as well as in primary lung 30 and breast 45 cancers. This assumption is further supported by the finding that most primary tumors in this cohort (57/65 or 88% evaluable tumors) exhibited variant allelic frequencies (VAF) distributions that were not consistent with neutral evolution 46, despite limitations of this analysis (Supplementary Fig. 9; Methods).
Fig. 4 |. Chronology of metastatic seeding.
a, Schematic for the timing of metastatic seeding prior to diagnosis of the primary tumor in number of years, ts. T denotes the total time of primary tumor expansion from emergence of the malignant founder cell to diagnosis while tp denotes the time from emergence of the malignant founder cell to the most recent common ancestor (MRCA) of cells in a bulk sample from primary tumor (denoted pMRCA). mMRCA denotes the MRCA of cells in a bulk sample from metastasis. ts can be estimated by Eq.(1). Dx, diagnosis. b, Estimation of the average T with a Gompertzian growth model is 5.2 (interquartile range or IQR, 4.3−7.7), 4.3 (IQR, 2.7−4.4) and 4.6 (IQR, 3.2−6.6) years for colorectal, lung and breast cancer, respectively. c, Estimation of the time of metastatic seeding (ts) for individual distant metastases (monoclonal metastases) in each cancer type. The median ts and IQR are shown. Negative ts indicates that the metastasis was seeded after the diagnosis of primary tumor. d, The distribution of ts in synchronous metastases (n=41) and metachronous metastases (n=80). P-value, Wilcoxon Rank-Sum Test (two-sided). Bar, median; box, 25th to 75th percentile (IQR); vertical line, data within 1.5 times the IQR. e, Correlation between ts and the time span from diagnosis of primary tumor to metastasis. Spearman’s correlation (rho) and P-value are reported. Line indicates the linear regression and gray shading indicates the 95% confidence interval (CI) of the regression.
We utilized a Gompertzian model of tumor growth 47, to estimate the tumor expansion age (T) for each of the three cancer types (Supplementary Note) where tumor size and doubling time (DT) at diagnosis were obtained from literature review (Supplementary Table 8). This yields estimates of average tumor expansion age of =5.2 (IQR, 4.3−7.7), 4.3 (IQR, 2.7−4.4) and 4.6 (IQR, 3.2−6.6) years for colorectal, lung and breast cancer, respectively (Fig. 4b, Supplementary Table 9). Chronological estimates of seeding time relative to diagnosis of the primary tumor ()can be computed by Eq.(1) as follows: 4.1 years (IQR, 3.2−4.6), 3.6 years (IQR, 2.8−3.7) and 2.7 years (IQR, 1.1−3.5) for colorectal, lung and breast cancers, respectively (Fig. 4c, Supplementary Table 9). The estimated timing of metastasis here () agreed with our previous estimates (using the colorectal cancer cohort) of primary tumor size at time of metastatic seeding 16 (Spearman’s rho= −0.55, P=0.014, Supplementary Fig. 10; note the negative correlation with SCIMET, which estimates metastatic timing forward in time, whereas here we estimate this backwards in time). Of note, while may indicate metastatic seeding after diagnosis/resection of the primary tumor, large Lm values can lead to (see Eq.(1)) even when the metastasis was seeded before diagnosis of the primary tumor. To mitigate this uncertainty, samples with estimated seeding times later than the actual time of diagnosis of metastasis were excluded (n=12 for breast, 1 for colorectal and 1 for lung cancer, respectively) (Supplementary Note). We find that was more common in breast cancer and more generally breast cancers disseminated closer to the time of detection (later) compared to colorectal and lung cancers (Fig. 4c). This may be because screening mammography detects relatively small primary breast tumors (<2 cm) 48. However, even after normalization to primary tumor age (namely ts/T), which depends on tumor size and the underlying growth parameters (Supplementary Note), breast cancer was found to disseminate later than colorectal and lung cancers (Supplementary Fig. 11). Most breast cancer metastases (83%) in this cohort were biopsied after adjuvant therapy (Extended Data Fig. 1), whereas this fraction is fewer in colorectal (13%) and lung (20%) cancer metastases and breast cancers harbored more private driver mutations than colorectal and lung cancers (Fig. 1a-c). Thus, the genomic complexity of metastatic relapses in breast cancer relative to unpaired early-stage primary tumors 12 at least in part reflects the selective effect of treatment on the genome, rather than the drivers of metastatic spread. Of note, HER2-positive breast cancers tended to disseminate earlier than HER2-negative breast cancers (Supplementary Fig. 12) consistent with this subgroup having the highest risk of distant metastasis before the routine use of adjuvant trastuzumab 33, which has revolutionized the treatment of this disease in part by targeting occult micrometastases.
As expected, metachronous metastases were often seeded later than synchronous metastases (median ts=3.8 vs 3.0, P = 5.6 × 10−5, two-sided Wilcoxon Rank-Sum Test; Fig. 4d). In fact, ts was highly correlated with the clinical time span from diagnosis of primary tumor to metastasis (Fig. 4e), indicating that metastases that manifest late clinically were seeded later. Since primary tumor size at diagnosis is an important predictor of a patient’s prognosis (time to metastatic relapse) (Supplementary Fig. 13a), we speculated that metastases were seeded earlier (namely larger ts) in patients with larger primary tumor size at initial diagnosis. Indeed, ts is positively associated with primary tumor size at diagnosis (Spearman’s rho=0.24, P=0.023; Supplementary Fig. 13b). These results corroborate our estimates of metastatic timing. According to Eq.(1), a larger number of M-private clonal mutations (larger Lm) indicates later dissemination. Supporting this theory, metachronous metastases showed significantly larger Lm than synchronous metastases (metachronous: median Lm=24, IQR=16−40; synchronous: median Lm=11, IQR=6−32; P = 6.5 × 10−4, two-sided Wilcoxon Rank-Sum Test; Extended Data Fig. 10a). This pattern held for SCNAs where metachronous metastases showed significantly more SCNAs relative to the primary tumor as compared to synchronous metastases (Extended Data Fig. 10b). Since metachronous metastases were generally seeded later than synchronous metastases (Fig. 4d), this is consistent with the higher degree of genomic divergence with primary tumor in late seeded metastases 18. Given that adjuvant treatment targets micrometastases, presumably delaying clinical metastasis 49, the timing of metastatic seeding of metachronous metastases following treatment might be even earlier. Collectively, these data indicate that systemic spread can occur several years prior to diagnosis of the primary tumor but with variability across histologies and subgroups.
Discussion
We performed a systematic analysis of exome sequencing data in paired primary tumors and metastases across three common cancers: colorectal, lung and breast and find that polyclonal seeding is common in lymph node metastases (19/35, 54%; most untreated) and untreated distant metastases (20/70, 29%), but rare (9/94, 10%) in metastases sampled after adjuvant therapy (Fig. 2g). Consistent with these results, treated metastases were strongly enriched for functional driver mutations as compared to untreated metastases (Fig. 1e). This finding indicates that driver gene heterogeneity is minimal between untreated metastases and primary tumors (Fig. 1e). Comparisons of paired primary tumors and distant metastases suggests that systemic spread can occur rapidly following malignant transformation, often several years prior to diagnosis of the primary tumor across three major types (Fig. 4c). These results are consistent with other reports of early seeding based on animal models and the genomic profiles of disseminated tumor cells 9,50,51.
Our analyses on driver gene heterogeneity, clonality and the timing of metastases provide important insights into the clonal dynamics of metastatic progression. First, in the absence of treatment, metastases often arise from the major clone in the primary tumor and lack metastasis-specific driver mutations (Fig. 1f). Consistent with these observations, a recent multi-cancer study demonstrated that driver gene heterogeneity is also minimal amongst multiple untreated metastases within individual patients 52. Moreover, the prevalence of polyclonal seeding in untreated lymph node and distant metastases indicates multiple cell subpopulations in primary tumor have acquired the metastatic competence. Half of all metastases (51%) studied here were biopsied after treatment, and these commonly exhibited monoclonal seeding accompanied by private driver mutations. As such, polyclonal seeding may be relatively common, but the ultimate pattern of clonality in the metastatic lesion is influenced by treatment. Further, these data cannot discriminate between polyclonal seeding due to multiple independent clones or one multi-clonal event (e.g. cell cluster 7).
Second, our quantitative framework demonstrates that systemic spread typically begins 2–4 years prior to the diagnosis of primary tumor (Fig. 4c). These data suggest that in some patients, metastatic seeding can happen very early especially for synchronously diagnosed metastases (Figs. 4e, 5a). Metachronous distant metastases following treatment occurred relatively later than synchronous distant metastases and harbored more genomic aberrations and driver mutations (Fig. 1e, Extended Data Fig. 10). These data imply that treatment remodels the clonal evolution of metastasis and may select disseminated cells harboring drug resistant mutations (Fig. 5a-b). As such metastasis-specific mutations are unlikely to be the drivers of metastasis, but instead are associated with resistance (Fig. 5b). This interpretation is of clinical relevance and helps to clarify the observation that metastatic relapses are more genomically complex than unpaired early-stage primary breast tumors 12. At the same time, adjuvant therapy directed at micrometastatic disease is effective for many patients, at least for a period of time, thus forestalling disease progression. Unfortunately, in cases where relapse occurs, the resultant metastatic outgrowth may be driven by a more aggressive, treatment resistant clone.
Fig. 5 |. Schematic model of metastatic spread and the impact of therapy.
a, Schematic illustration of early versus late metastatic seeding leading to synchronous and metachronous metastases. Metastatic seeding occurs quickly following the emergence of the founding carcinoma cell. Synchronous metastases, which exhibit low genomic divergence from the primary tumor, is seeded early by the major/founding clone in the primary tumor. Metachronous metastases, exhibit higher genomic divergence relative to the primary tumor and often emerge after adjuvant therapy. Metachronous metastases with specific driver mutations that confer resistance can be selected leading to high genomic divergence between the primary tumor and treated metastasis. b, Treatment (adjuvant therapy), remodels the clonal architecture of metastasis. Dissemination and metastatic seeding (monoclonal or polyclonal) initially give rise to undetectable micrometastases. While treatment may eliminate drug-sensitive micrometastatic lesions, those that are resistant grow out. Metastatic relapse following adjuvant treatment may be delayed by adjuvant treatment, but this may result in a more aggressive, resistant lesion. DTCs, disseminated tumor cells. Dx, diagnosis; Tx, treatment.
This study is based on a large collection of paired primary tumors and metastases across multiple cancer types with genomic data, but several limitations remain. First, the majority of tumors (>80% for P or M) were sequenced to standard depth (median=88, IQR=(65, 110), Supplementary Table 3), which is likely underpowered to identify polyclonal seeding patterns based on shared subclonal (low frequency) mutations (Fig. 2b). Simulations show that multi-region sequencing (n=4 from each of P and M) increases the accuracy of classifying monoclonal and polyclonal seeding as compared to single sample. Second, more than half of the distant metastases included here were biopsied after drug treatment, which substantially remodels the clonal architecture of the metastasis by promoting monoclonality and genomic divergence. If these two main confounders are considered, we would expect that polyclonal seeding of distant metastases is more common than inferred (18%) here and that metastatic dissemination might occur even earlier. Our findings highlight the importance of studying the natural course of metastasis as well as the impact of therapy on this process. Future studies of paired primary tumors and metastases with comprehensive treatment information subject to dense multi-region sampling and single cell sequencing may provide additional resolution on these processes.
Online Methods
Whole-exome sequencing (WES) of paired primary tumors and metastases
We performed a comprehensive review on the published studies through surveying the PubMed database (https://www.ncbi.nlm.nih.gov/pubmed/), in which whole-exome sequencing (WES) was performed for matched normal tissues, primary tumors (P) and metastases (M) in the same patients. We focused on colorectal, lung and breast cancers given the availability of large patient data in these three cancer types. In total, the raw sequencing reads data for 586 tumor samples from 181 patients in 13 published studies were accessed and retrieved (Supplementary Table 1). We also generated multi-region sequencing (MRS) data for two colorectal cancer patients (mCRCTB1 and mCRCTB7) with liver metastases from whom excess de-identified tissue was collected during the course of routine care, hence this is not considered human subjects research. A total of n=5–7 regions were sequenced for these two P/M pairs resulting in 24 tumor samples. Here tumor tissues with cellularity >60% were selected for DNA isolation using the QIAamp DNA FFPE Tissue Kit (Qiagen) and libraries were generated using the Agilent SureSelect Human All Exon kit for sequencing on the Illumina HiSeq 2500. Clinical information was retrieved from the original studies, including patient age at initial diagnosis, time span from initial diagnosis of primary tumor to diagnosis of metastasis, treated information and subtype (Supplementary Tables 2–3). We define synchronous metastases if the time span between diagnosis of primary tumor and metastasis is within 3 months and metachronous metastases if the time span is ≥3 months.
An established bioinformatics pipeline was used to detect somatic single nucleotide variations (SSNVs), small insertions/deletions (indels) and somatic copy number alterations (SCNAs), estimate tumor purity/ploidy and estimate the cancer cell fraction (CCF) for each SSNVs/indels in corresponding samples 16,30. In particular, paired sequencing reads were aligned to human reference genome (NCBI build hg19) with BWA (v.0.7.10) 53. Duplicate reads were marked with Picard Tools (v.1.111). Aligned reads were further processed with GATK 3.4.0 for local re-alignment around insertions and deletions and base quality recalibration.
SSNVs and indel calling
SSNVs were called by MuTect (v.1.1.7) 54 for each tumor/normal pair. SSNVs failing MuTect’s internal filters, having fewer than 10 total reads or 3 variant reads in the tumor sample, fewer than 10 total reads in the normal sample, or mapping to paralogous genomic regions were removed. Additional Varscan (v.2.3.9) 55 filters were applied to remove SSNVs with low average variant base qualities, low average mapping qualities among variant supporting reads, strand bias among variant supporting reads and high average mismatch base quality sums among variant supporting reads, either within each tumor sample or across all tumor samples from the same patient. The maximal observed variant allele frequencies (VAF) across all samples from each patient were calculated based on raw output files from MuTect. SSNVs with maximal observed VAFs lower than 0.05 were removed. For FFPE specimens, additional filters were applied to exclude possible artifactual SSNVs. Specifically, artifacts among C>T/G>A SSNVs with bias in read pair orientation were filtered in each individual FFPE sample, similar to the approach of Costello et al 56. Of note, 26%, 80% and 81% of primary colorectal, lung and breast cancer samples were FFPE while 29%, 50% and 55% of metastatic samples in these three cancer types were FFPE. FFPE artifacts are at low frequency in primary tumors (median VAF= 0.056–0.085 across studies) and metastases (median VAF= 0.017–0.090 across studies). On average, more than 70% of the FFPE artifacts across the cohort were specific to the primary tumors, consistent the primary tumor more commonly being FFPE than the metastasis. We also sought to exploit the multi-sample information in the same patients to retrieve read counts for SSNVs. To obtain the depth and VAF information across all samples from the same patient, for each SSNV and in each tumor sample that an SSNV was not originally called in, the total reads and variant supporting reads were counted using the mpileup command in SAMtools (v.1.2) 57. Only reads with mapping quality ≥ 40 and base quality at the SSNV locus ≥ 20 were counted and used to calculate the VAF for that SSNV. Small insertions/deletions (indels) were called with Strelka (v.1.0.14) 58. SSNVs and indels were annotated with ANNOVAR (v.20150617) 59 and those in protein coding regions were retained for downstream analyses.
Copy number analysis
Copy number analysis was performed using TitanCNA (v.1.5.7) 60. Briefly, TitanCNA uses depth ratio and B-allele frequency information to estimate allele-specific absolute copy numbers with a hidden Markov model, and estimates tumor purity and clonal frequencies. Only autosomes were used in copy number analysis. First, for each patient, germline heterozygous SNP at dbSNP 138 loci were identified using SAMtools and SnpEff (v.3.6) in the normal sample. HMMcopy (v.0.99.0) 61 was used to generate read counts for 1000bp bins across the genome for all tumor samples. TitanCNA was used to calculate allelic ratios at the germline heterozygous SNP loci in the tumor sample and depth ratios between the tumor sample and the normal sample in bins containing those SNP loci. Only SNP loci within WES covered regions were then used to estimate allele-specific absolute copy number profiles. TitanCNA was run with different numbers of subclones (n=1–3). One run was chosen for each tumor sample based on visual inspection of fitted results, with preference given to the results with a single subclone unless results with multiple subclones had visibly better fit to the data. Results from tumor samples from the same patient were inspected together to ensure consistency. Overall ploidy and purity for each tumor sample was calculated from the TitanCNA results.
Differentially altered SCNAs in the metastasis relative to paired primary tumor (P-to-M) were identified if following three criteria were satisfied simultaneously: 1) absolute copy number in the metastasis was larger than 2.8 or less than 1.2; 2) copy number relative to median ploidy in the metastasis was larger than 0.8 or less than −0.8; 3) changes relative to the primary tumor in both absolute copy number and relative copy number were larger than 0.8 or less than −0.8. For multi-region sequencing data, segmented log depth ratios (adjusted for purity and ploidy) for each primary CRC and paired metastasis were averaged across multiple-regions from the same tumor site.
Cancer cell fraction (CCF) estimates and identification of clonal and subclonal mutations
The CCFs and their variation (95% confidence interval or 95% CI) for each SSNVs/indels in the corresponding samples were estimated with CHAT (v 1.0) 62. CHAT includes a function to estimate the CCF of each SSNVs by adjusting its variant allele frequency (VAF) based on local allele-specific copy numbers at the SSNV locus. SSNV frequencies and copy number profiles estimated from previous steps were used to calculate the CCFs for all SSNVs in autosomes. The CCFs were also adjusted for tumor purity using the estimates by TitanCNA. In brief, for an SSNV residing in a genomic segment with a total copy number of CNt, minor allele copy number of CNb and cellular prevalence PCNA of the CNA in the tumor content, the estimated CCF of the SSNV is:
| Eq. (2) |
where and the effective purity is estimated tumor purity) and VAF is the observed variant allele frequency. The temporal ordering and background composition of SSNVs and SCNAs was inferred by comparing the conditional probabilities of the observed number of mutant reads out of total reads, under each scenario and CNA configuration as follows: Early Major or Minor: SSNV in the major or minor allele occurred before the CNA; Late: SSNV occurred after the CNA; Independent: the SSNV and CNA occurred in independent lineages 62.
To distinguish clonal and subclonal SSNVs/indels in each sample, we employ the following criterion: clonal – 95% CI overlaps with 1; subclonal – the upper bound of 95% CI is smaller than 1, as previously used 63.
Since bulk sequencing data is underpowered to detect low frequency mutations, determining whether a mutation is truly private mutations to one site is challenging. Thus, “private” SSNV/indels in one site relative to another site is paratactically defined as the CCF<5% in another site as our previous study 16. For multi-region sequencing data, the merged CCFs by integrating multiple regions were used:
| Eq. (3) |
where di and CCFi are the sequencing depth and CCF estimation in region i, respectively.
Sample quality control for downstream analysis
The CCFs of SSNV/indels for each P/M sample pair were visualized using the scatter plot and manually checked in order to identify problematic samples. In particular, for each P/M pair, a cluster of SSNV/indels centered around CCF=1 is expected which represent truncal (P/M shared clonal) mutations that occurred prior to malignant transformation of the founding cell in the primary tumor. The patients (n=5) with none of or very few (<10) trunk SSNVs/indels were excluded as which implies independent (non-clonal) origin for the primary tumor and metastasis. Furthermore, patients (n=42) with a diffusely distributed cluster for truncal SSNVs/indels were also excluded since this is likely caused by low tumor purity or low sequencing quality. After these filtering steps, 457 tumor samples from 136 metastatic cancer patients including 39 colorectal cancers (181 tumor samples), 30 lung cancers (74 tumor samples) and 67 breast cancers (202 tumor samples) were retained for downstream analysis in this study. Regarding the histological subtypes, all colorectal cancers were microsatellite stable (MSS). For lung cancer, 67% (20/30) were adenocarcinoma, 30% (9/30) were squamous carcinoma, while 3% (1/30) were small cell lung cancer. For breast cancer, 6% (4/67) were ER+/HER2+, 6% (4/67) were ER−/HER2+, 51% (34/67) were ER+/HER2-, 19% (13/67) were triple negative (TN), while 18% (12/67) were unknown (Supplementary Table 2). Of note, there is a bias towards obtaining more paired primary and distant metastases from triple negative (TN) breast tumors since they tend to recur earlier than ER+ tumors (many within 5 years), where for some subsets of ER+/HER2− disease there is a persistent risk of recurrence up to two decades after diagnosis 33,64.
Jaccard similarity index
The number of M-private clonal, P-private clonal and P-M shared subclonal SSNVs for each P/M pair was denoted as Lm, Lp and Ws respectively. For two sets, the Jaccard similarity index (JSI) is defined for the intersection divided by the union of these two sets. Thus, the JSI for a P/M pair can be defined as:
| Eq. (4) |
For multi-region sequencing data, Lm, Lp and Ws was counted by pairwise comparison of each sample pair from the P and M. The mean Lm, Lp and Ws was used to compute the JSI by Eq. (4).
Functional assessment of non-silent somatic mutations
To identify functional driver gene mutations, three commonly used computational methods, PolyPhen-2 36 (http://genetics.bwh.harvard.edu/pph2/), FATHMM-XF 37 (http://fathmm.biocompute.org.uk/fathmm-xf/) and CHASMplus 38 (https://karchinlab.github.io/CHASMplus/), were utilized to perform the function (“driverness”) assessment on the nonsynonymous SSNVs amongst putative cancer genes derived from TCGA pan-cancer 65 and COSMIC (Release v87, Nov. 13, 2018). Stopgain/splicing point mutations and indels on putative cancer genes are classified as functional drivers automatically.
Putative cancer genes were curated by merging all TCGA pan-cancer drivers (n=299) 65 and additional cancer type-specific drivers annotated by COSMIC Cancer Gene Census (https://cancer.sanger.ac.uk/cosmic; n=47, 40 and 9 for colorectal, lung and breast cancers, respectively). For PolyPhen-2, a SSNV is considered as “functional” when the functional report (“pph2_class”) is “deleterious”. For FATHMM-XF, a SSNV is considered as “functional” when the functional report (“Warning”) is “ pathogenic”. For CHASMplus, a SSNV is considered as “functional” when the false discovery rate (FDR) < 0.05. In this study, the SSNVs, predicted to be functional by any of these three methods, were considered as functional mutations. Metascape 66 (http://metascape.org) was used to perform gene ontology (GO) analysis of functional driver genes.
Driver enrichment analysis
Clonal non-silent SSNVs/indels in a metastatic lesion can be considered truncal clonal (or P-M shared clonal) or M-private clonal where the number is denoted Ls_total and Lm_total, respectively. Meanwhile, the functional driver SSNVs/indels in a metastasis are denoted Ls_driver and Lm_driver, respectively. The ratios, Ls_total/Lm_total and Ls_driver/Lm_driver, can be evaluated for functional enrichment of drivers on the truncal or M-private branch of the corresponding phylogenetic tree. Since Ls_driver and Lm_driver are small values (Lm_driver ~ 0 for many metastases), they lead to high variation in the Ls_driver/Lm_driver ratio. A down-sampling (bootstrapping) step (50% of patients each time) was performed in which sampled patient data were merged to derive the Ls_total/Lm_total and Ls_driver/Lm_driver ratios. 100 down-samplings were performed for each of the three cancer types to derive statistical measures.
Mutational signatures, dN/dS and test of neutrality
MuSiCa 67 (http://bioinfo.ciberehd.org:3838/MuSiCa/) was used to extract mutation signatures based on non-negative matrix factorization 68 for P/M shared clonal (truncal) SSNVs, M-private clonal SSNVs and M-private subclonal SSNVs respectively, in each of the three cancer types. dndscv 40 (https://github.com/im3sanger/dndscv) was used to compute the ratio of nonsynonymous and synonymous SSNVs (dN/dS) for missense and nonsense mutations, respectively and for P/M shared clonal (trunk) SSNVs, M-private clonal SSNVs and M-private subclonal SSNVs, respectively, in each of the three cancer types. We evaluated whether a tumor exhibits a pattern consistent with neutral evolution or strong selection during growth by analyzing the variant frequency distribution (VAF) of subclonal SSNVs. Under neutral evolution, the number of subclonal SSNVs with VAF larger than in a tumor cell population follows a power-law distribution: 46. The adjusted VAFs (equivalent to CCF/2) for subclonal SSNVs (in the range of 0.1–0.3) were used here and only tumors with at least 20 subclonal SSNVs in this range were analyzed (n=65 primary tumors and 79 metastases). By fitting this model and using a threshold of R2=0.98, the mode of evolution (neutral or selection) can be inferred (Supplementary Fig. 9). There are notable limitations to this analysis, including the lack of MRS data and the fact that many primary tumors were FFPE. Nonetheless, the finding that the majority of patients exhibit primary tumor VAF distributions consistent with subclonal selection, is in-line with our prior reports in a metastatic colorectal cancer cohort with MRS data, where the majority of primary colon cancers exhibited evidence of subclonal selection, consistent with the metastatic clone having a selective growth advantage 16. Additionally, analysis of multi-region sequencing data suggest that subclonal selection may be relatively common in primary lung 30 and breast 45 cancers.
Phylogenetic tree reconstruction
We ran PHYLIP 69 via an online version
(http://www.trex.uqam.ca/index.php?action=phylip&apP=dnapars) and applied the Maximum Parsimony method to reconstruct the phylogeny of multiple specimens from individual patients based on the presence or absence of SSNVs/indels. The SSNVs/indels residing a region with different loss-of-heterozygosity (LOH) status between paired primary tumor and metastasis were filtered, since which may lead to erroneous presence or absence of SSNVs/indels in paired P and M. When multiple maximum parsimony trees were reported, we chose the top ranked solution. FigTree (http://tree.bio.ed.ac.uk/software/Figuretree/) was employed to visualize the reconstructed trees.
Spatial agent-based modeling of metastatic progression
We employed our previously established three-dimensional agent-based tumor evolution framework 30 to model tumor growth, mutation accumulation and metastatic dissemination after malignant transformation. Pre-malignant clonal expansions prior to transformation do not contribute to the genetic heterogeneity of an established tumor (since all such alterations are clonal), and thus were not modeled since dissemination is assumed to occur after malignant transformation of the founding carcinoma cell. In this model, spatial tumor growth is simulated via the expansion of deme subpopulations (composed of ~5k cells with diploid genome), mimicking the glandular structures often found in epithelial tumors and metastases and consistent with the number of cells found in individual colorectal cancer glands (~2,000–10,000 cells). The deme subpopulations expand within a defined 3D cubic lattice (Moore neighborhood, 26 neighbors), via peripheral growth while cells within each deme are well-mixed without spatial constraints and grow via a random birth-and-death process (division probability b and death probability d=1−b at each generation). Once a deme exceeds the maximum size (10,000 cells), it splits into two offspring demes via random sampling of cells from a binomial distribution (Nc, 0.5), where Nc is the current deme size.
To model monoclonal seeding, a single cell at the tumor periphery was randomly sampled as the metastasis founder cell. To model polyclonal seeding, a cluster of cells (n=10) were randomly sampled from the whole tumor in order to maximize the clonal diversity within the metastasis founder cells. This is because if the clonal diversity in the metastasis founder cells is low, it essentially models the scenario of monoclonal seeding by a cluster of genetically similar cells. The metastasis grows at same spatial model with primary tumor started from the metastasis founder cell or cell cluster (n=10). During each cell division in the growth of primary tumor and metastasis, the number of neutral passenger mutations acquired in the coding portion of the genome follows a Poisson distribution with mean u. Thus, the probability that k mutations occurred in each cell division is as follows:
| Eq. (5) |
where an infinite sites model and constant mutation rate are assumed during tumor progression. Advantageous mutations also arise stochastically via a Poisson process with mean us during each cell division. We assume us=10−5 per cell division in the genome and each increases the cell division probability 70. The cell birth and death probabilities for a selectively beneficial clone are bs=b×(1+s) and ds=1−ds=1−b×(1+s), respectively, thus the selective advantage for an advantageous mutation is defined as s=bs/b−1.
During simulation of primary and metastatic growth, each mutation is assigned a unique index that is recorded with respect to its genealogy and host cells, enabling analysis of the mutational frequency in a bulk sample of tumor cells during different stages of growth. We simulate growth until the primary and metastasis reach a size of ~109 cells (or ~10 cm3) and then sample a bulk subpopulation (consisting of ~106 cells) at the peripheral region of the primary tumor and metastasis, respectively. The VAF of all SSNVs in the sampled bulk subpopulation is considered the true VAF (denoted by fT), whereas the observed allele frequency is obtained via a statistical model that mimics the random sampling of alleles during sequencing. Specifically, we employ a Binomial distribution (n, fT) to generate the observed VAF at each site given its true frequency fT and number of covered reads n. The number of covered reads at each site is assumed to follow a negative-binomial distribution (Negative Binomial(size, depth)) where depth is the mean sequencing depth and size corresponds to the variation parameter. We assume depth=100 and size=2 for the sequencing data in each tumor region and tissue purity=0.6 in order to model normal cell contamination in clinical samples. A mutation is called when the number of variant reads is ≥3, thereby applying the same criteria as for the patient tumors.
We employed a mutation rate u=0.6 per cell division in the exonic region (corresponding to 10−8 per site per cell division in the 60Mb diploid coding regions). In order to model varying scenarios of tumor growth dynamics, selection and timing of metastatic dissemination, for each primary tumor/metastasis (P/M) pair, the birth probability b of founding cells, selection coefficient s and primary tumor size at dissemination Nd was sampled from a uniform distribution, b~U(0.55, 0.65), log10(s)~U(−3,−1) and log10(Nd)~U(4,8), respectively. 500 virtual P/M pairs were simulated under each of the monoclonal seeding and polyclonal seeding scenarios, where a mean (in silico) sequencing depth of 100X is assumed. The number of M-private clonal SSNVs (Lm), P-private clonal SSNVs (Lp) and P/M shared subclonal SSNVs (Ws) for each P/M pair were counted from the simulation data and the simulated JSI was computed by Eq.(4).
Extended Data
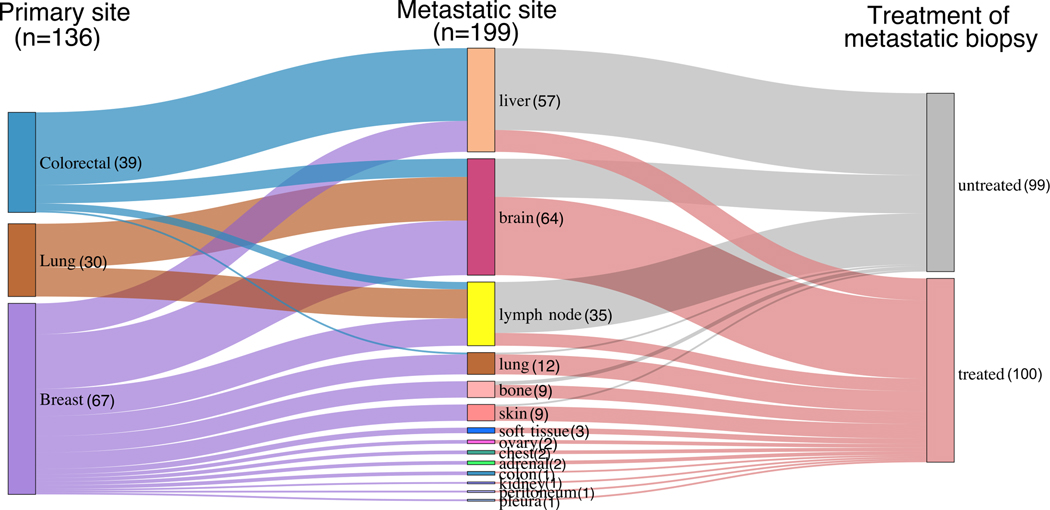
Extended Data Fig. 1. Sankey diagram of patient cohorts with paired primary tumors and metastases.
In total, 136 primary tumors and 199 matched metastases from colorectal, lung and breast cancers were included. Treatment status is indicated.
Extended Data Fig. 2. Concordance of mutation burden in paired primary tumors (P) and metastasis (M).
Concordance amongst a, Clonal SSNVs; b, Subclonal SSNVs and c, SCNAs are indicated. Spearman’s correlation (rho) is reported. Line indicates the linear regression and gray shading indicates the 95% confidence interval (CI) of the regression. The mean mutation burden across samples is reported for samples with multi-region sequencing data.
Extended Data Fig. 3. The ratio of nonsynonymous to synonymous mutations, dN/dS.
The dN/dS ratios of missense mutations (left panel) or nonsense mutations (right panel) relative to synonymous mutations are shown (on log2 scale). The dN/dS ratios for putative driver genes and passengers were computed separately. The driver gene list was obtained by merging TCGA pan-cancer drivers and COSMIC Cancer Gene Census (Methods). Circles and vertical lines correspond to the mean and 95% CI of the dN/dS ratio, respectively.
Extended Data Fig. 4. The frequency of somatic copy number alterations (SCNAs) for primary tumors and metastases across three cancer types.
The frequency of amplifications or deletions across 1Mb genomic bins is shown for primary tumors and metastases.
Extended Data Fig. 5. The frequency of somatic copy number alterations (SCNAs) in putative driver genes in paired primary tumors (P) and metastases (M).
Left panel, amplifications (AMP) where oncogenes with an increased frequency (≥15%) in the metastasis (M) versus primary (P) are labeled. Right panel, deletions (DEL) where tumor suppressor genes with increased frequency (≥15%) in the metastasis (M) versus primary (P) are labeled.
Extended Data Fig. 6. Schematic illustration of a 3-D spatial-agent based model of tumor growth and metastasis.
Tumor growth is simulated via the expansion of deme subpopulations (mimicking the glandular structures often found in epithelial tumors and metastases) within a defined 3-D cubic lattice according to explicit rules dictated by spatial constraints, where cells within each deme are well-mixed and grow via a stochastic branching (birth-death) process (Methods). To model monoclonal seeding, a single cell at the tumor periphery was randomly sampled as the metastasis founder cell. To model polyclonal seeding, a cluster of cells (n=10) was randomly sampled from the whole tumor in order to maximize the clonal diversity within the metastasis founder cells. Metastatic growth follows the same spatial-constraints as the primary and starts from the metastasis founder cell or cell cluster. The final sizes of both the primary tumor and metastasis is ~109 cells (~2×105 demes). Clonal selection is modeled by assuming a constant beneficial mutation rate that alters the cell birth/death probability according to the selection coefficient (denoted s). By simulating the acquisition of random mutations (neutral or beneficial), tracing the mutational genealogy of each cell as the tumor expands and subsequently spatially sampling (~106 cells in each sample) and sequencing the ‘final’ virtual tumor as is done experimentally after resection or biopsy, we obtain the variant allele frequencies (VAF) and cancer cell fraction (CCF) in both primary tumor and metastasis.
Extended Data Fig. 7. Lm, Lp and Ws values in tumors simulated under monoclonal versus polyclonal seeding.
The number of SSNVs in each of the three categories (M-private clonal or Lm, P-private clonal or Lp, P/M shared subclonal or Ws) in the simulated data generated by modeling monoclonal seeding or polyclonal seeding within an agent-based model (Methods) where one sample (~106 cells) was biopsied from each primary tumor and metastasis. We employed a mutation rate u=0.6 per cell division in exonic regions (corresponding to 10−8 per site per cell division in the 60Mb diploid coding regions). In order to account for varying scenarios of tumor growth dynamics, selection and timing of metastatic dissemination, the birth probability b of founding cells, selection coefficient s and primary tumor size at dissemination Nd was randomly sampled from a uniform distribution, b~U(0.55, 0.65), log10(s)~U(−3,−1) and log10(Nd)~U(4,8), respectively. A total of n=500 virtual P/M pairs were simulated under monoclonal seeding and polyclonal seeding by randomly sampling these three parameters. Bar, median; box, 25th to 75th percentile (interquartile range, IQR); vertical line, data within 1.5 times the IQR.
Extended Data Fig. 8. Jaccard similarity index (JSI) values in lymph node and distant metastases and the percentage of polyclonal seeding across metastatic sites.
a, Lymph node metastases (LNM; n=35) showed significantly higher JSI than distant metastases (n=164). Among distant metastases, untreated metastasis showed higher JSI than treated metastasis although this was not statistically significant. However, using a cutoff of JSI=0.3 to classify polyclonal (JSI ≥ 0.3) versus monoclonal seeding (JSI<0.3), untreated distant metastases showed a significantly higher percentage of polyclonal seeding than treated distant metastases (Fig. 2e). P-value, Wilcoxon Rank-Sum Test (two-sided). Bar, median; box, 25th to 75th percentile (interquartile range, IQR); vertical line, data within 1.5 times the IQR. b, The percentage of polyclonal seeding among all LNM (lymph node metastasis), LiM (liver metastasis), BM (brain metastasis) and LuM (lung metastasis) (left panel) and stratified by treatment (right panel). P-value, Fisher’s exact test (two sided).
Extended Data Fig. 9. A mathematical method to quantify the chronology of metastatic seeding, ts.
a, Schematic of the parameters used to quantify metastatic timing ts (number of years prior to primary tumor diagnosis). We assume metastatic spread occurs at tm following the emergence of malignant founder of primary carcinoma (denoted t=0). Let T be the time from emergence of malignant founder to diagnosis of the primary tumor, thus ts=T−tm. Let Lp and Lm be the number of private clonal SSNVs in a bulk sample from primary tumor and metastasis, respectively. Lp represents the number of SSNVs that occurred from emergence of the primary tumor founder to the most recent common ancestor (pMRCA) of cell lineages in a bulk sample. This time span is denoted as tp. Similarly, Lm denotes the number of SSNVs occurred from the emergence of primary tumor founder to the MRCA in a bulk sample from the metastasis (denoted mMRCA). Lm includes the number of M-private clonal mutations that occur: (i) within the primary tumor (Lm1) and (ii) after cells have disseminated from the primary tumor (Lm2), thus Lm = Lm1 + Lm2. b, Estimation of α by simulating an agent-based model of tumor evolution (Methods). The mean α and standard deviation from 1000 simulated tumors are shown.
Extended Data Fig. 10. Later metastatic seeding is associated with higher genomic divergence in matched primary tumors.
a, The number of primary (P)-private clonal SSNVs and metastasis (M)-private clonal SSNVs in synchronous (distant and monoclonal, n=41) and metachronous (distant and monoclonal, n=80) metastases, respectively. b, The number of P-to-M altered SCNAs in synchronous and metachronous metastases, respectively; P-values, two-sided Wilcoxon Rank-Sum Test. Bar, median; box, 25th to 75th percentile (interquartile range, IQR); vertical line, data within 1.5 times the IQR.
Supplementary Material
Acknowledgments
We thank Hang Xu, Katherine McNamara, Eran Kotler, Jennifer Caswell-Jin and other members of Curtis laboratory for valuable discussions. We thank Jiguang Wang and Quanhua Mu for providing the scripts for the ternary plot. C.C. is supported by the National Institutes of Health through the NIH Director’s Pioneer Award (DP1-CA238296), the American Association for Cancer Research (AACR) and the Emerson Collective. Z.H is supported by an Innovative Genomics Initiative (IGI) Postdoctoral Fellowship.
Footnotes
Competing interests
C.C. is a scientific advisor to GRAIL and reports stock options as well as consulting for GRAIL and Genentech. Z.H., Z.L., Z.M. have no conflicts of interest to report.
Data availability
The exome sequencing data for colorectal cancer patients that were sequenced in-house have been deposited at the European Genotype Phenotype Archive (EGA) under accession number EGAS00001003573. The accession numbers for public datasets are listed in Supplementary Table 1.
Code availability
Code used for genomic data analysis are available from: https://github.com/cancersysbio/pan-metastasis
References
- 1.Talmadge JE, Wolman SR & Fidler IJ Evidence for the clonal origin of spontaneous metastases. Science 217, 361–3 (1982). [DOI] [PubMed] [Google Scholar]
- 2.Yamamoto N. et al. Determination of clonality of metastasis by cell-specific color-coded fluorescent-protein imaging. Cancer Res 63, 7785–90 (2003). [PubMed] [Google Scholar]
- 3.Liu W. et al. Copy number analysis indicates monoclonal origin of lethal metastatic prostate cancer. Nat Med 15, 559–65 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Huang Y. et al. Multilayered molecular profiling supported the monoclonal origin of metastatic renal cell carcinoma. Int J Cancer 135, 78–87 (2014). [DOI] [PubMed] [Google Scholar]
- 5.Gundem G. et al. The evolutionary history of lethal metastatic prostate cancer. Nature 520, 353–357 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Maddipati R. & Stanger BZ Pancreatic Cancer Metastases Harbor Evidence of Polyclonality. Cancer Discov 5, 1086–97 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cheung KJ et al. Polyclonal breast cancer metastases arise from collective dissemination of keratin 14-expressing tumor cell clusters. Proc Natl Acad Sci U S A 113, E854–63 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hunter KW, Amin R, Deasy S, Ha NH & Wakefield L. Genetic insights into the morass of metastatic heterogeneity. Nat Rev Cancer 18, 211–223 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Klein CA Parallel progression of primary tumours and metastases. Nat Rev Cancer 9, 302–12 (2009). [DOI] [PubMed] [Google Scholar]
- 10.Naxerova K. & Jain RK Using tumour phylogenetics to identify the roots of metastasis in humans. Nat Rev Clin Oncol 12, 258–72 (2015). [DOI] [PubMed] [Google Scholar]
- 11.Robinson DR et al. Integrative clinical genomics of metastatic cancer. Nature 548, 297–303 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bertucci F. et al. Genomic characterization of metastatic breast cancers. Nature 569, 560–564 (2019). [DOI] [PubMed] [Google Scholar]
- 13.Priestley P. et al. Pan-cancer whole-genome analyses of metastatic solid tumours. Nature (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhao ZM et al. Early and multiple origins of metastatic lineages within primary tumors. Proc Natl Acad Sci U S A 113, 2140–5 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Macintyre G. et al. How Subclonal Modeling Is Changing the Metastatic Paradigm. Clin Cancer Res 23, 630–635 (2017). [DOI] [PubMed] [Google Scholar]
- 16.Hu Z. et al. Quantitative evidence for early metastatic seeding in colorectal cancer. Nat Genet 51, 1113–1122 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Leung ML et al. Single-cell DNA sequencing reveals a late-dissemination model in metastatic colorectal cancer. Genome Res 27, 1287–1299 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Turajlic S. & Swanton C. Metastasis as an evolutionary process. Science 352, 169–75 (2016). [DOI] [PubMed] [Google Scholar]
- 19.Lee SY et al. Comparative genomic analysis of primary and synchronous metastatic colorectal cancers. PLoS One 9, e90459 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim TM et al. Subclonal Genomic Architectures of Primary and Metastatic Colorectal Cancer Based on Intratumoral Genetic Heterogeneity. Clin Cancer Res 21, 4461–72 (2015). [DOI] [PubMed] [Google Scholar]
- 21.Lim B. et al. Genome-wide mutation profiles of colorectal tumors and associated liver metastases at the exome and transcriptome levels. Oncotarget 6, 22179–90 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Uchi R. et al. Integrated Multiregional Analysis Proposing a New Model of Colorectal Cancer Evolution. PLoS Genet 12, e1005778 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brastianos PK et al. Genomic Characterization of Brain Metastases Reveals Branched Evolution and Potential Therapeutic Targets. Cancer Discov 5, 1164–1177 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Um SW et al. Molecular Evolution Patterns in Metastatic Lymph Nodes Reflect the Differential Treatment Response of Advanced Primary Lung Cancer. Cancer Res 76, 6568–6576 (2016). [DOI] [PubMed] [Google Scholar]
- 25.Chung W. et al. Single-cell RNA-seq enables comprehensive tumour and immune cell profiling in primary breast cancer. Nat Commun 8, 15081 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ng CKY et al. Genetic Heterogeneity in Therapy-Naive Synchronous Primary Breast Cancers and Their Metastases. Clin Cancer Res 23, 4402–4415 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Razavi P. et al. The Genomic Landscape of Endocrine-Resistant Advanced Breast Cancers. Cancer Cell 34, 427–438 e6 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Siegel MB et al. Integrated RNA and DNA sequencing reveals early drivers of metastatic breast cancer. J Clin Invest 128, 1371–1383 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ullah I. et al. Evolutionary history of metastatic breast cancer reveals minimal seeding from axillary lymph nodes. J Clin Invest 128, 1355–1370 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sun R. et al. Between-region genetic divergence reflects the mode and tempo of tumor evolution. Nat Genet 49, 1015–1024 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Curtis C. et al. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature 486, 346–52 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ciriello G. et al. Emerging landscape of oncogenic signatures across human cancers. Nat Genet 45, 1127–33 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rueda OM et al. Dynamics of breast-cancer relapse reveal late-recurring ER-positive genomic subgroups. Nature 567, 399–404 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Christensen S. et al. 5-Fluorouracil treatment induces characteristic T>G mutations in human cancer. Nat Commun 10, 4571 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pich O. et al. The mutational footprints of cancer therapies. Nat Genet 51, 1732–1740 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Adzhubei IA et al. A method and server for predicting damaging missense mutations. Nat Methods 7, 248–9 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rogers MF et al. FATHMM-XF: accurate prediction of pathogenic point mutations via extended features. Bioinformatics 34, 511–513 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tokheim C. & Karchin R. CHASMplus Reveals the Scope of Somatic Missense Mutations Driving Human Cancers. Cell Syst 9, 9–23 e8 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Patel SA & Vanharanta S. Epigenetic determinants of metastasis. Mol Oncol 11, 79–96 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Martincorena I. et al. Universal Patterns of Selection in Cancer and Somatic Tissues. Cell 171, 1029–1041 e21 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Makohon-Moore AP et al. Limited heterogeneity of known driver gene mutations among the metastases of individual patients with pancreatic cancer. Nat Genet 49, 358–366 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sottoriva A. et al. A Big Bang model of human colorectal tumor growth. Nat Genet 47, 209–16 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kang H. et al. Many private mutations originate from the first few divisions of a human colorectal adenoma. J Pathol 237, 355–62 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Williams MJ et al. Quantification of subclonal selection in cancer from bulk sequencing data. Nat Genet 50, 895–903 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Caswell-Jin JL et al. Clonal replacement and heterogeneity in breast tumors treated with neoadjuvant HER2-targeted therapy. Nat Commun 10, 657 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Williams MJ, Werner B, Barnes CP, Graham TA & Sottoriva A. Identification of neutral tumor evolution across cancer types. Nat Genet 48, 238–244 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Benzekry S. et al. Classical mathematical models for description and prediction of experimental tumor growth. PLoS Comput Biol 10, e1003800 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stein RG et al. The impact of breast cancer biological subtyping on tumor size assessment by ultrasound and mammography - a retrospective multicenter cohort study of 6543 primary breast cancer patients. BMC Cancer 16, 459 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cavalli F, Kaye SB, Hansen HH & Piccart MJ Textbook of Medical Oncology, Fourth Edition, (2009). [Google Scholar]
- 50.Harper KL et al. Mechanism of early dissemination and metastasis in Her2(+) mammary cancer. Nature (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hosseini H. et al. Early dissemination seeds metastasis in breast cancer. Nature (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Reiter JG et al. Minimal functional driver gene heterogeneity among untreated metastases. Science 361, 1033–1037 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Methods-only references
- 53.Li H. & Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–60 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cibulskis K. et al. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol 31, 213–9 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Koboldt DC et al. VarScan 2: somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res 22, 568–76 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Costello M. et al. Discovery and characterization of artifactual mutations in deep coverage targeted capture sequencing data due to oxidative DNA damage during sample preparation. Nucleic Acids Res 41, e67 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li H. et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–9 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Saunders CT et al. Strelka: accurate somatic small-variant calling from sequenced tumor-normal sample pairs. Bioinformatics 28, 1811–7 (2012). [DOI] [PubMed] [Google Scholar]
- 59.Wang K, Li M. & Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res 38, e164 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ha G. et al. TITAN: inference of copy number architectures in clonal cell populations from tumor whole-genome sequence data. Genome Res 24, 1881–93 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ha G. et al. Integrative analysis of genome-wide loss of heterozygosity and monoallelic expression at nucleotide resolution reveals disrupted pathways in triple-negative breast cancer. Genome Res 22, 1995–2007 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Li B. & Li JZ A general framework for analyzing tumor subclonality using SNP array and DNA sequencing data. Genome Biol 15, 473 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.McGranahan N. et al. Clonal status of actionable driver events and the timing of mutational processes in cancer evolution. Sci Transl Med 7, 283–54 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pan H. et al. 20-Year Risks of Breast-Cancer Recurrence after Stopping Endocrine Therapy at 5 Years. N Engl J Med 377, 1836–1846 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bailey MH et al. Comprehensive Characterization of Cancer Driver Genes and Mutations. Cell 174, 1034–1035 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhou Y. et al. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat Commun 10, 1523 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Diaz-Gay M. et al. Mutational Signatures in Cancer (MuSiCa): a web application to implement mutational signatures analysis in cancer samples. BMC Bioinformatics 19, 224 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Alexandrov LB et al. Signatures of mutational processes in human cancer. Nature 500, 415–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.J., F. PHYLIP-phylogeny inference package (version 3.2). cladistics 5, 6 (1989). [Google Scholar]
- 70.Bozic I. et al. Accumulation of driver and passenger mutations during tumor progression. Proc Natl Acad Sci U S A 107, 18545–50 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.