Abstract
There is little or no research initiated on enlightening Nigerians about the pathogenesis, targets for drug development and repositioning for severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection. Coronavirus disease 2019 (COVID-19) is a viral infection causing symptoms like dry cough, sore throat, nasal congestion, tiredness, fever, loss of taste, and smell etc. The disease was first reported in Wuhan, China, in December 2019. The infection is caused by SARS-CoV-2, which is the third introduction of a highly pathogenic coronavirus into the human population. Coronaviruses are viruses with a positive RNA envelope assigned to α, β, γ, and δ genera. Moreover, SARS-CoV-2 belongs to the β genus. The four structural proteins of β coronavirus are membrane (M), envelope (E), spike (S), and nucleocapsid (N) protein, mediation of coronavirus host infection is established by spike (S) protein. Therefore, the search for drug development targets and repositioning of existing therapeutics is essential for fighting the present pandemic. It was reviewed that therapeutics targeting SARS-CoV-2 binding to ACE2 receptor, viral RNA synthesis and replication, 3CLpro, RdRp, and helicase will play a crucial role in the development of treatment for SARS-CoV-2 infection. Furthermore, the RdRp and spike protein of SARS-CoV-2 are the most promising targets for drug development and repositioning and vaccine development. Remdesivir combination with chloroquine/hydroxychloroquine are promising drug repositioning for the treatment of COVID-19, and mRNA-1273 targeting spike protein is the promising vaccine. However, as patient management and drug repositioning are taking place, it is imperative to identify other promising targets used by SARS-CoV-2 to establish infection, to develop novel therapeutics.
Keywords: SARS-CoV-2, COVID-19, Molecular pathogenesis, Acute respiratory disease, Drug development targets
1. Introduction
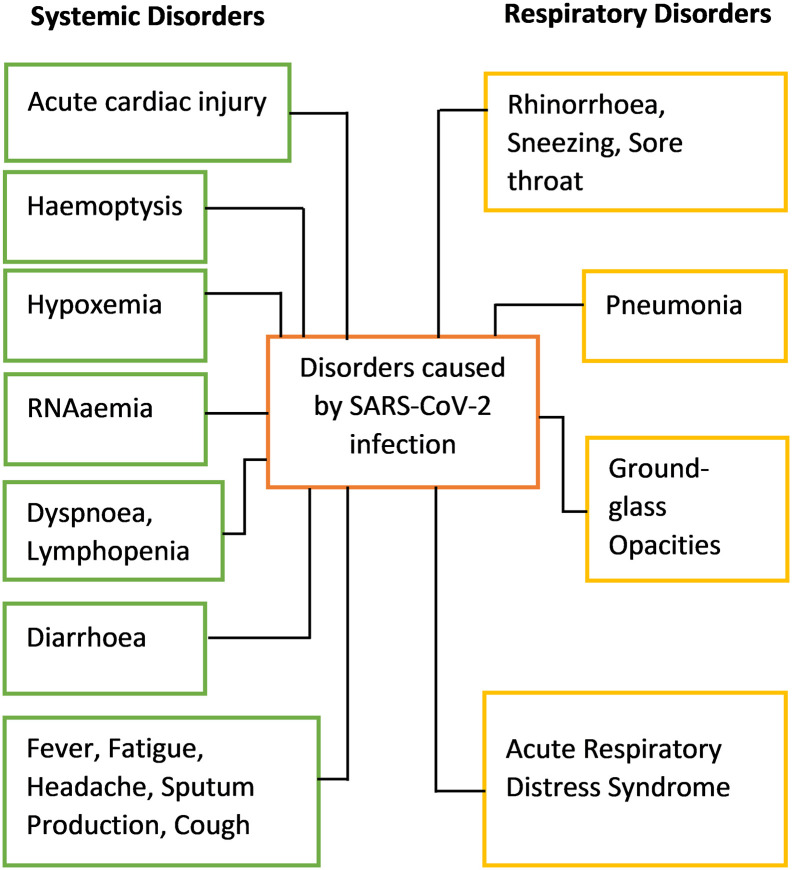
There is little or no research initiated to enlighten Nigerians about the pathogenesis and targets for drug development for the newly discovered SARS-CoV-2infection as a way forward to achieve long-lasting future treatment of the disease, also many citizens of the country are not aware on the deadliness of the disease. COVID-19 is a kind of viral infection causing pneumonia-like symptoms was first reported in Wuhan, China, in December 2019, and the disease is caused by SARS-CoV-2 [1,2]. The outbreak was declared a pandemic by WHO on 30th of January 2020, and it is regarded as the third introduction of a highly pathogenic virus into the human population. The severity at which the virus causes infection to the human population is age and immune status dependent. It is shorter among patients greater than 70-years old compared with those under the age of 70 with early symptoms which include fever, cough, and fatigue, other symptoms include sputum production, headache, haemoptysis, dyspnoea, and lymphopenia (Fig. 1 ) [3]. Clinical features unmask by a chest computer tomography (CT) scan presented as pneumonia, however, there are also abnormal features such as RNAaemia, acute respiratory distress syndrome, acute cardiac injury, and incidence of ground-glass opacities that can lead to death [3].
Fig. 1.
The disorders caused by SARS-CoV-2 after infecting human cells.
Unfortunately, the use of interferon inhalation for the treatment of some cases did not show clinical progress but worsen the condition and can lead to pulmonary opacities [4]. Besides, patients infected with SARS-CoV-2 also developed gastrointestinal associated symptoms like diarrhea [3,5]. To minimize the transmission of the SARS-CoV-2 through health care workers and patients, it is important to develop methods for the identification of various modes of viral transmission such as through fecal and urine samples [3].
Genome sequencing of the virus of five patients with pneumonia hospitalized from December 18 to December 29, 2019, showed the presence of a previously unknown β coronavirus (β-CoV) strain in all of them [6]. Upon isolation and sequencing of the β-CoV, it shows 88% identity to the sequence of two bat-derived severe acute respiratory syndromes (SARS)-like coronaviruses, batSL-CoVZC45 and bat-SL-CoVZXC21, and about 50% identity to the sequence of middle east respiratory syndrome coronavirus (MERS-CoV) [7]. There are similarities in the genome sequences of SARS-CoV-2 with a minimum number of ten open reading frames (ORFs) with the genome sequences of other CoVs such as MERS-CoV and SARS-CoV. The first ORF making up two-third of the SARS-CoV-2 RNA undergoes translation to two polyproteins. In comparison to MERS-CoV and SARS-CoV, in which their two polyproteins (pp1a and pp1ab) are then further processed by a post-translational modification to 16 non-structural proteins (nsps), which are nsp1 to nsp16 [8]. Those nsps are then translocated from the rough endoplasmic reticulum into double-membrane vesicles where the replication and transcription of the virus occurred [9,10]. While the remaining ORFs of the SARS-CoV-2 that are localized on the one-third of the remaining genome are responsible for encoding non-replicating participating accessory proteins with unknown functions and structural proteins such as membrane (M), nucleocapsid (N), envelope (E), and spike proteins (S). An investigation by scientists in China revealed that the SARS-CoV requires angiotensin-converting enzyme 2 (ACE2) receptor for their binding and invasion of the host. Also, the binding is significant in the determination of the pathogenesis of the infection caused by SARS-CoV [11]. Nevertheless, dipeptidyl peptidase 4 is a receptor required by MERS-CoV for invasion [12]. To predict the specificity of zoonotic coronaviruses in the infection of humans and adaptation possibility, a better understanding of the proteases action and receptor binding is essential. Therefore, the development of therapeutics for the treatment of SARS-CoV-2 is highly warranted [3]. However, this review aims to discuss the molecular pathogenesis of SARS-CoV-2, its target for drug development, to enlighten Nigerians on the deadliness of the disease, and the way forward to achieve future treatment of the current pandemic.
2. Molecular basis of SARS-CoV-2 pathogenesis
2.1. Replication of coronavirus in the host
The molecular pathogenesis of SARS-CoV-2 is poorly understood up to date. Nevertheless, MERS-CoV and SARS-CoV mechanisms can shed light on the pathogenesis of SARS-CoV-2. The essential protein of SARS-CoV-2 that mediates binding to a human cell is spike protein (a protein involved in the first step of infection), the protein is composed of S1 and S2 subunit, in which according to cryogenic electron microscopy (CryoEM) studies the receptor-binding domain (RBD) is a 3D structure with the size of ~12 kDa which is localized in the S1 subunit. The vibrational movement of the RBD has been examined during the characterization of the structure of related MERS-CoV and SARS-CoV and was found to have the same mechanism of action with SARS-CoV-2. Interestingly, there are similarities in the structure of SARS-CoV-2 and SARS-CoV spike glycoprotein but the only structural difference is in the RBD, where the SARS-CoV-2 RBD is an angle closer to the central cavity of the trimer in the down conformation, while SARS-CoV RBD can packs tightly against the domain of the N-terminal of the neighboring protomer in the down conformation [13]. The binding affinity of the two viruses was investigated using surface plasmon resonance which showed that even though SARS-CoV and SARS-CoV-2 share the same binding receptor ACE-2, the binding affinity of SARS-CoV-2 is ~15 nM which is almost 10–15 fold higher than SARS-CoV binding to ACE-2 receptor, this is the reason why SARS-CoV-2 can easily be transmitted from human to human. Whereas, the complex formed by SARS-CoV-2 S-ectodomain and ACE-2 has been studied using high-resolution CryoEM, which revealed that the complex is similar to the SARS-CoV complex. The spike structural protein mediates entry of SARS-CoV-2 into human cells, it is a trimetric protein of class 1 fusion protein with metastable prefusion conformation, it undergoes structural rearrangement before human cell membrane fusion. The rearrangement is activated upon binding of RBD of the S1 subunit to the host cell receptor ACE2 [13]. Destabilization of the prefusion trimer will then take place upon receptor binding (viral membrane with the host cell membrane) which will lead to the shedding of the S1 and S2 subunit transition to more stable postfusion conformation. The RBD of the S1 will pass through hinge-like movement leading to transient hiding (down conformation receptor inaccessible state) or exposing (up confirmation i.e. receptor accessible state) of the receptor binding determinant, therefore this is the mechanism of engaging host cell receptor by SARS-CoV-2 [[14], [15], [16]].
Before entry into human cells, MERS-CoV binds to the dipeptidyl peptidase-4 (DPP4) receptor and SARS-CoV binds to the ACE2 receptor. Membrane fusion and viral infectivity of coronavirus occurred by proteolytic cleavage of S20 position of S protein, this occurred in SARS-CoV pathogenesis. The use of two-step furin activation is essential for the fusion of MERS with the host membrane [17]. The entry of SARS-CoV is also mediated by clathrin-dependent and -independent endocytosis [18,19].
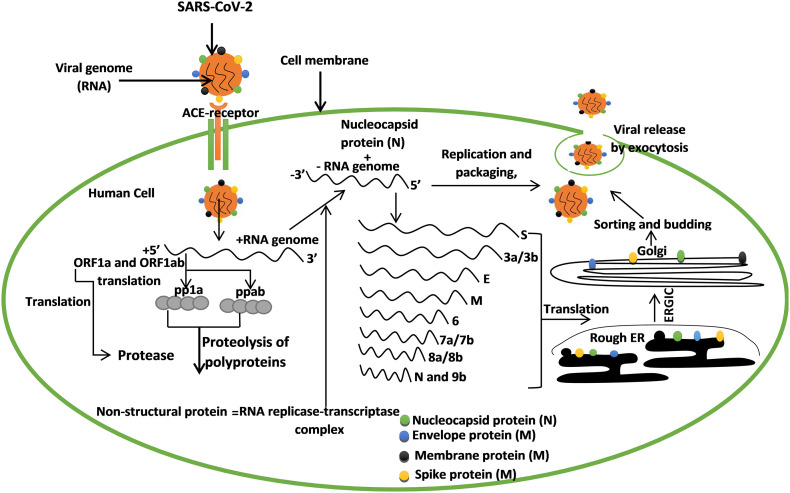
The SARS-CoV-2 binds to the angiotensin-converting enzyme receptor (ACE2) which results in its entry into the host. The virus releases its RNA into the cytoplasm, the ORF1a, and ORFab of the RNA is translated into polyprotein 1a (pp1a) and polyprotein ab (ppab). The ORF1a encodes the enzyme protease that cleaves the polyproteins into non-structural proteins (nsps) which form the RNA replicase-transcriptase complex that is needed for structural protein synthesis. The replicase complex executes the formation of negative-sense RNA. Upon fragmentation of the negative-sense RNA into sub-genomic RNA through discontinuous transcription, after which fragments that encode spike, membrane, nucleocapsid, and envelope protein are formed, the structural proteins are then synthesized in the rough endoplasmic reticulum, and then they pass through the endoplasmic reticulum-Golgi intermediate compartment (ERGIC) into the Golgi apparatus where sortation and assembly take place. Finally, viral protein and genomic RNA are then assembled as virion in the Endoplasmic reticulum and Golgi and then released outside the cell as a vesicle [[20], [21], [22], [23]]. After the invasion of cells by SARS-CoV-2, its antigen will be attached to antigen-presenting cells as the central anti-viral immunity. Finally, major histocompatibility complex (MHC) or human leukocyte antigen (HLA) in humans present antigenic peptides and then cytotoxic T lymphocytes specific for the SARS-CoV recognized viral antigen. Therefore, this pathogenesis mechanism (Fig. 2 ) of SARS-CoV will help in the understanding of drug development strategies for SARS-CoV-2 infection. Upon presentation of antigen by antigen-presenting cells, cellular and humoral immunity of the body is then stimulated by virus-specific B and T cells. Therefore, IgM and IgG are the common antibody profile of SARS-CoV, and 12 weeks are marked as the time taken for IgM against SARS-CoV to disappear, with IgG lasting longer [23]. More researches on the cellular immunity of coronavirus have been carried out, and the most updated report reveals that the number of CD4þ and CD8þ T cells in the peripheral blood of SARS-CoV-2 infected patients significantly reduced [24]. During the acute phase of SARS-CoV infection, there is a decrease in CD4þ and CD8þ T cells. Interestingly, SARS-CoV recovered patients have four years of T cell memory and the T cells can undergo IFN-g production, delayed-type hypersensitivity (DTH) response, and proliferation even without the antigen [25]. Also, T cell memories against SARS-CoV persist after six years of infection from recovered patients [26]. Therefore, the understanding of the immunology of SARS-CoV-2 can help in the development of a vaccine [27].
Fig. 2.
An overview of SARS-CoV-2 pathogenesis. The virus binds to the ACE2 receptor to enter the human cell and its RNA got translated to structural proteins (spike, nucleocapsid, membrane, and envelope), these proteins are essential in the invasion and infection of the human cell.
2.2. Coronavirus cytokine released syndrome
Pathological events associated with SARS-CoV-2, SARS-CoV, and MERS-CoV infection is called acute respiratory distress syndrome (ARD) [24]. The mechanism of acute respiratory distress syndrome (ARDS) is that the effector immune cell uncontrollably released deadly systemic pro-inflammatory cytokines such as IFN-a, IFN-g, IL-1b, IL6, IL-12, IL-18, IL-33, TNF-a, TGFb, etc. and also chemokines such as CCL2, CCL3, CCL5, CXCL8, CXCL9, CXCL10, etc. [27,28]. The cytokine and chemokine release by the effector cells will lead to organ failure and death, this happened in the severe case of SARS-CoV-2 infection [24].
2.3. Ducking of immune system by the coronavirus
For better survival of MERS CoV and SARS-CoV in the invaded host, for example, pattern recognition receptor is responsible for recognizing pathogen-associated molecule, however, the MERS and SARS viruses will develop a way of producing double-membrane vesicle without pattern recognition receptor, this allows the viral genome to replicate without the control of the immune system [29]. Even though IFN-I (IFN-a and IFN-b) has a protective effect on SARS-CoV and MERS-CoV infection, it can be inhibited by MERS accessory protein through activation of melanoma differentiated associated protein gene 5 (MDA5) by interaction with double-stranded directly [[30], [31], [32]]. To elaborate, nuclear transport of IFN regulatory protein and activation of IFN b promoter is blocked by MERS-CoV ORF4a, ORF4b, ORF5, and membrane proteins [33]. Since coronaviruses, such MERS can destabilize the immune system by altering the antigen presentation cells through the downregulation of gene expression. Therefore, it is important in drug development to develop a strategy of destroying immune evasion by SARS-CoV-2 [4].
3. Targets for drug development
Drug development from scratch will take at least a decade, following this, repositioning of existing antiviral drugs effective on SARS-CoV-2 might be the only solution to the current pandemic of sudden infectious diseases. The human innate immunes system plays a crucial role in the replication of the coronaviruses, together with the use of interferon will boost the immune response in counteracting coronaviruses [34]. SARS-CoV and SARS-CoV-2 bind to ACE2 while MERS-CoV binds to protease receptors called DPP4 before entry into a human cell. Therapeutics acting on the coronaviruses can either be blocking virus association with the human cell, inhibition of viral self-assembly through inhibition of structural protein, blocking RNA synthesis through viral genetic material inhibition, and blocking critical enzymes of the virus to inhibit replication [35]. Three strategies of coronavirus drug development were introduced by scientists. The strategy is testing the broad-spectrum of existing antiviral drugs [27]. Drugs such as ribavirin, and cyclophilin inhibitors, and interferons are used to treat coronavirus associated symptoms such as pneumonia and can be considered to fall into this category. Therefore, known metabolic characteristics, dosage, efficacy, side effect, approval for viral treatment are the major advantage associated with this therapeutic strategy. The major problems with such drugs are killing of coronaviruses in a targeted manner because they are too broad-spectrum, therefore, better-targeted therapeutics are indeed needed. The second strategy involved the use of existing molecular databases to screen and find molecules with therapeutic potential that are effective on coronaviruses [36]. The availability of high throughput techniques makes it possible for the screening of potential therapeutics, which can aid in finding the function of some new molecules, for instance, the discovery of drugs such as lopinavir/ritonavir (anti-HIV). The third strategy is the development of new targeted drugs from scratch based on coronaviruses pathological features and genetic information. It was perceived that drug development through this strategy will exhibit more efficacy against the virus. However, this might cost several years of drug development and clinical trials and it is estimated that drug development can take seven years, or even more than 10 years [34]. It is recommended that research should be initiated on drug development from scratch for highly contiguous viruses, this is to prepare for future pandemics.
Based on molecular targets, four major strategies of SARS-CoV-2 drug development were discussed below. Also, the targets can be used to develop new drugs from scratch based on the pathological and genomic characteristics of the new SARS-CoV-2.
3.1. Therapeutics acting on protein and enzymes that are critical to the growth of the virus, by preventing its replication and synthesis of RNA
Non-structural proteins (nsps) are involved in protein synthesis, translation, RNA transcription, processing, modification, virus replication, and host infection. Therefore, nsps are considered as a significant functional protein of coronaviruses, in which coronavirus main proteinase (3CLpro), papain-like proteinase (PLpro), RNA dependent RNA polymerase (RdRp), and helicase are superior molecular targets for small inhibitory molecule drug development as a result of their well-understood enzyme active sites and biological functions. For example, Papain-like proteinase (PLpro), this protein cleaves replicase polyprotein N-terminus, leading to the of release nsp1, nsp2, and nsp3, which is indispensable for the replication control corrections [7]. Also, PLpro can antagonize the immunity of the infected patient [16,37]. These group of enzymes is necessary for the replication and viral infection, and it is the most popular target for coronavirus inhibition. It is highly valuable to target the PLpro protein of coronavirus to stop the infection of the virus. Surprisingly, there are no FDA approved marketed drugs targeting PLpro protein. Interestingly, drugs such as thymidine, ribavirin, chloramphenicol, and chlorphenesin carbamate can easily bind to PLpro and prevent viral replication [6]. Intensive research into PLpro protein can open a new door of therapeutics development against SARS-CoV-2 infection. Then 3C-like main protease (3CLpro), otherwise called nsp5, this protein is cleaved automatically from polyprotein to form mature enzyme and it is then further cleaved downstream at 11 sites which will then leads to the release of nsp4 to nsp16. Interestingly, maturation of nsp's is directly mediated by 3CLpro, and the life cycle of the virus depends on the maturation of 3CLpro. Studies into the structure and catalytic mechanism of 3CLpro can make it a promising drug development target for COVID-19. Therefore, anti-bacterial drugs such as lymecycline and anti-hypertensive drugs such as nicardipine are all known to block the activity of the 3C-like main protease. RNA-dependent RNA polymerase (RdRp), otherwise known as nsp12, this protein is conserved in coronaviruses, it forms the vital enzyme of the viral replication and transcriptional complex, RdRp has conserved amino acid of which are serine and 2 aspartic acid. Therefore, nsp12 has been used as an important target for SARS and MERS drug development. Even though no specific inhibitor of nsp12-RdRp has been found, its targeted inhibition will not result in significant side effects and toxicity of the host cell. Strengthening research on this target can lead to therapeutic development for SARS-CoV infection [[38], [39], [40]]. However, natural products that exhibit anti-virus, anti-tumor, and anti-inflammation effects will exert binding affinity to RdRp, molecules such as betulonal (from Cassine xylocarpa), gnidicin, and gniditrin (from Gnidia lamprantha) will bind and inhibit the RdRp of coronaviruses. Helicase (nsp13), this is a multifunctional protein with N terminal metal-binding domain and helicase domain. The structure of the N terminal contains 26 cysteine residues that make up the zinc and binding domain, with C terminus conserved motif. Helicase is also a necessary component for the replication of coronavirus. Therefore, helicase is identified as a target for antiviral drug discovery, but there is less research on helicase inhibitors. Drugs such as anti-bacterial (lymecycline, cefsulodine, and rolitetracycline), anti-fungal such as itraconazole, an anti-HIV drug such as saquinavir, an anticoagulant drug such as dabigatran, and diuretic drug such as canrenoic acid were shown to be potent inhibitors of helicase [41,42]. Also, studies have shown that myricetin and scutellarein can potentially inhibit the SARS-CoV-2 helicase protein in vitro by affecting the ATPase activity, but not the unwinding activity of nsP13 [43].
3.2. Therapeutics blocking coronavirus structural protein from binding to a human cell receptor and inhibiting its self-assembly
The core structural protein of coronavirus that gathered to form a special corolla like structure on the viral surface like trimer is called spike protein, its binds to host cell receptor using its RBD which results to the invasion of the host. Host cell protease cleaves spike protein into S1 and S2, the S1 binds to surface receptor of the host, and S2 mediates virus to cell fusion and cell to cell fusion. Cleavage activation and structural integrity are crucial in the invasion and virulence activity, which is mediated by spike protein [44]. Strategies to block spike protein as a means of preventing coronavirus from entering the host are regarded as valuable for the development of antiviral drugs [44,45]. Natural flavonoids (licoflavonol from Glycyrrhiza uralensis), therapeutics such as anticoagulant drug (dabigatran etexilate), anti-fungal drugs (posaconazole and itraconazole), anti-hypertensive drugs (rescinnamine, iloprost, and prazosin), and an anti-bacterial drug (sulfasalazine, azlocillin, penicillin, and cefsulodin) inhibit coronavirus binding to a receptor in the cell surface of human cell [6].
The RDB located in the S1 of the spike protein of SARS-CoV-2 is also a target for the development of therapeutic monoclonal antibodies. Despite the fact of structural homology of SARS-CoV-2 and SARS-CoV by CryoEM studies, a monoclonal antibody that was originally developed to target SARS-CoV RBD was tested against SARS-CoV-2 RBD SD1 fragment, but the antibody did not show binding affinity to SARS-CoV-2. This is to say there are some amino acids involved in the interaction between the RBD of the spike protein and ACE2 receptor in SARS-CoV-2 that are not the same in SARS-CoV. This is further supported by [46] that modeling of SARS-CoV-2 RDB interaction with ACE2 revealed some sequence of amino acids that are important for the interaction between ACE2 and RBD of spike glycoprotein, but there is less clarity on the actual sequence involved in that interaction. Therefore, there is a need for new monoclonal antibody development for RBD of SARS-CoV-2. Hence, the knowledge of spike protein of the SARS-CoV-2 atomic level using CryoEM will enable scientists to carry out protein engineering, design, and development of more effective inhibitory molecules and monoclonal antibodies [13].
3.3. Inhibition of coronavirus virulence factors
Virulence factors in coronavirus that interferes with the innate immunity of infected cells are nsp1, nsp3c, and ORF7a. The nsp1 induces degradation of host mRNA and interferon type 1 production upon binding to the 40S ribosomal subunit [47]. While nsp3c binds with the nucleotide of the host (ADP-ribose) which gives coronavirus the ability to resist innate immunity, for instance, bone marrow matrix antigen 2 blocks assembled coronavirus in the host cell. Also, SARS-CoV ORF7a interacts with BST 2 and blocks its glycosylation which results in termination of ORF7A activity [48]. Therefore, the aforementioned virulence factors are potential drug discovery targets for coronavirus. For example, anti-bacterial and anti-inflammatory natural products can bind and inhibit those virulence factors, also drugs such as platycodin D, streptomycin, tetracycline, piperacillin, cefpiramide, and lymecycline (from Platycodon grandifloras) can potentially inhibit coronavirus virulence factors [49,50].
3.4. Blocking specific receptor of host or enzymes
The receptor of the spike RNA binding domain of SARS-CoV is the ACE2 receptor. Since SARS-CoV can interact with the angiotensin receptor, blocking the receptor can be a target for therapeutics development for SARS-CoV-2 infection. Therefore, anti-hypertensive drugs (losartan), anti-bacterial (cefmenoxime), hepatoprotective (silybin), anti-diabetes (troglitazone), and analgesia (ergotamine) bind with high affinity to angiotensin 2 receptor [6].
4. The most promising targets based on repositioning drugs for SARS-CoV-2 infection
RNA dependent RNA polymerase (nsp12) is the most promising molecular target for SARS-CoV-2 drug development. The current state in finding treatment for SARS-CoV-2 is drug repositioning in which drug targeted for the treatment of the Ebola virus called remdesivir, which showed efficacy against SARS-CoV-2 replication. Therefore, the RNA dependent RNA polymerase inhibitor (RdRp's)called remdesivir was developed by Gilead Science, Inc. to mimic the structure of adenosine, and is regarded as nucleotide analog, it was developed originally to inhibit Ebola virus replication. Even though the drug did not pass the phase 3 clinical trial on the Ebola virus, but studies have shown that it is promising for the treatment of COVID-19 [51]. Also, the drug was reported to be the most promising for SARS-CoV-2 replication inhibition by WHO. As a result of structural similarities of RdRp's of various viruses, remdesivir can serve as a broad antiviral drug. Studies have also shown that the drug is effective against MERS-CoV. Interestingly, the USA and China commence the phase 3 clinical trial of remdesivir against SARS-CoV-2 [52]. Even though drugs such as broad-spectrum antibiotics, anti-viral drugs, and interferons-a nebulization have been used in the reduction of viral load, but it is the only remdesivir that has a promising impact on SARS-CoV-2. The controversy of the efficacy of drugs used in the treatment of SARS-CoV and MERS-CoV such as ribavirin, corticosteroids, lopinavir-ritonavir, and interferon, is what makes them not to be investigated for the treatment of SARS-CoV-2 infection. But the satisfactory result was achieved after the trial of remdesivir in mice model, in which after the treatment of the mice a day after infection with coronavirus there is a decrease in the viral load and an improvement in mice pulmonary function. However, remdesivir administration after the viral load reaches a maximum peak of infection cannot improve the symptoms of the patient, but remdesivir can improve patient symptoms before the virus reaches its maximum peak. Treatment of rhesus monkey with remdesivir 24 h before the infection is promising in the reduction of symptoms associated with MERS-CoV infection, and within 2 h of administration of 10 g of remdesivir in rhesus monkey results to peripheral blood mononuclear blood distribution of the drug, followed by drug activation to nucleotide triphosphate [53]. According to the New England Journal of Medicine, a patient with recent travel to Wuhan China was diagnosed with COVID-19 after returning to Washington on January 15, 2020, even though there is an improvement on the patient symptoms upon remdesivir treatment, but the viral load got decreased before the treatment, this is to say that the viral infection is self-limiting and his immune system might have started fighting the infection. Therefore, it is not clear whether the improvement in the symptoms of the patient is due to the drug or the patient's natural immunity. However, remdesivir is expected to be a specific drug that targets the RdRp of SARS-CoV-2 [54]. The great zealot of Chinese to combat the pandemic has initiated research on COVID-19 drugs, in which remdesivir was approved for clinical trial after passing stringent ethical review and was on 5th February 2020 launched for the trial. China's compassionate use reported that remdesivir will be the immediate drug for the treatment of severe COVID-19 if proven to be effective. What brought remdesivir to the stage of the clinical trial is safety and good pharmacokinetics [55]. Remdesivir In vitro inhibition research carried out by Wuhan research institute find out that the drug can block the replication of the virus at (EC50 = 0.77 μM, CC50 > 100 μM, SI > 129.87) very low micromolar concentration of infected-Vero E6 cells (Wang et al., 2020). Other studies have shown that effective treatment of SARS-CoV-2 infection can only be achieved by combinational treatment of chloroquine and remdesivir which will result in the blockage of the viral replication and recovery of the patient from the disease [56]. Chloroquine increases the endosomal pH of a cell as well as affecting glycosylation of cellular receptor, these result in blockage of SARS-CoV-2 infection [50,57]. The effect of chloroquine on SARS-CoV-2 was further investigated by [50], and they reported that chloroquine function at entry and post-entry of the virus in addition to its antiviral activity. Also, chloroquine can modulate the immune system which in turn will result in the enhancement of its antiviral activity, it can also inhibit the replication of coronavirus in the epithelial cells of lungs through cellular receptor glycosylation interference. The authorization to use chloroquine/hydroxychloroquine for the treatment of SARS-CoV-2 infection was granted by the FDA of the U.S.A, hence chloroquine function in the blocking of virus fusion to the cell membrane [58,59]. Widely distribution of chloroquine in blood and lungs upon its administration orally is another interesting future of the use of chloroquine in establishing treatment for SARS-CoV-2 infection. It was recommended that COVID-19 patients with mild, moderate, and severe symptoms should take 500 mg of chloroquine twice in a day, this prescription has shown to decrease the length of stay in the hospital with improved symptoms. Hydroxychloroquine has the same activity as chloroquine because they are having an identical mechanism of action [60]. Moreover, intensive research is needed to unravel the potential of other receptors for drug discovery and development for SARS-CoV-2. Besides, [59] reported that synthetic mRNA called Moderna's mRNA-1273 can be used as a vaccine for protection against SARS-CoV-2 infection, in which upon its intramuscular administration, it can evoke antiviral effect directed to the spike protein, this is to say the synthetic mRNA encodes pre-fused spike protein of SARS-CoV-2. The use of mRNA to elicit antiviral activity does not need the use of a virus, unlike conventional vaccines. The clinical trial phase 1 of mRNA-1273 is currently taking place and if the vaccine is promising, its efficacy will be investigated immediately. To sum up, RNA dependent RNA polymerase and spike protein of SARS-CoV-2 are the most promising targets for SARS-CoV-2 vaccine and drug development, and therapeutics repositioning [61].
5. Conclusion
Drug development for viral pathogens is a long-term process. However, as drug repositioning is taking place for the treatment and reduction of the peak of the COVID-19 pandemic, it is also imperative to identify some promising targets for drug development to have long-lasting future treatment for the disease. Also, an in-depth understanding of the SARS-CoV-2 pathogenesis and its atomic studies using CryoEM to understand the amino acid involved in the interaction between the RBD of spike protein with ACE2 of human cells are essential for effective vaccine and drug development. Nigerian government should initiate research and clinical trial on antiviral drugs that can inhibit SARS-CoV-2 infection.
6. Future recommendation
As the current pandemic is claiming the health and lives of people all over the world, it is highly important to initiate and expand researches in the study of pathogenesis and drug targets for SARS-CoV-2, as this will help in developing treatment and vaccine and also will prepare the world to prepare the future reoccurrence of the disease. Research should also be initiated on the development of bispecific inhibitors of SARS-CoV-2 to achieve effective inhibition of the coronavirus activity.
Conflict of Interest Statement
The author declares that there are no conflicts of interest.
References
- 1.Bogoch I.I., Watts A., Thomas-Bachli A., Huber C., Kraemer M.U.G., Khan K. Pneumonia of unknown aetiology in Wuhan, China: potential for international spread via commercial air travel. J. Travel Med. 2020;27(2):1–3. doi: 10.1093/jtm/taaa008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lu H., Stratton C.W., Tang Y.W. Outbreak of pneumonia of unknown etiology in Wuhan, China: the mystery and the miracle. J. Med. Virol. 2020;92(4):401–402. doi: 10.1002/jmv.25678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rothan H.A., Byrareddy S.N. The epidemiology and pathogenesis of coronavirus disease (COVID-19) outbreak. J. Autoimmun. 2020;109:102433. doi: 10.1016/j.jaut.2020.102433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li X., Geng M., Peng Y., Meng L., Lu S. Molecular immune pathogenesis and diagnosis of COVID-19. J. Pharm. Anal. 2020;10:102–108. doi: 10.1016/j.jpha.2020.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee I.C., Huo T.I., Huang Y.H. Gastrointestinal and liver manifestations in patients with COVID-19. J. Chin. Med. Assoc. 2020;83:1–3. doi: 10.1097/JCMA.0000000000000319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wu C., et al. Risk factors associated with acute respiratory distress syndrome and death in patients with coronavirus disease 2019 pneumonia in Wuhan, China. JAMA Intern. Med. 2020:1–10. doi: 10.1001/jamainternmed.2020.0994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Angeletti S., Benvenuto D., Bianchi M., Giovanetti M., Pascarella S., Ciccozzi M. COVID-2019: the role of the nsp2 and nsp3 in its pathogenesis. J. Med. Virol. 2020;92(6):584–588. doi: 10.1002/jmv.25719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Maier H., Bickerton E., Britton P., Yang C.H., Li H.C., Hung C.H., et al. Methods in molecular biology. 1st ed. Vol. 1282. Humana Press; New York: 2015. Coronaviruses: Methods and protocols. [Google Scholar]
- 9.Masters P.S. The molecular biology of coronaviruses. Adv. Virus Res. 2006;65(6):193–292. doi: 10.1016/S0065-3527(06)66005-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Knoops K., et al. SARS-coronavirus replication is supported by a reticulovesicular network of modified endoplasmic reticulum. PLoS Biol. 2008;6(9):1957–1974. doi: 10.1371/journal.pbio.0060226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ge X.Y., et al. Isolation and characterization of a bat SARS-like coronavirus that uses the ACE2 receptor. Nature. 2013;503(7477):535–538. doi: 10.1038/nature12711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Raj V.S., et al. Dipeptidyl peptidase 4 is a functional receptor for the emerging human coronavirus-EMC. Nature. 2013;495(7440):251–254. doi: 10.1038/nature12005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wrapp D., et al. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science (80-. ). 2020;367(6483):1260–1263. doi: 10.1126/science.aax0902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Walls A.C., et al. Tectonic conformational changes of a coronavirus spike glycoprotein promote membrane fusion. Proc. Natl. Acad. Sci. U. S. A. 2017;114(42):11157–11162. doi: 10.1073/pnas.1708727114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gui M., et al. Cryo-electron microscopy structures of the SARS-CoV spike glycoprotein reveal a prerequisite conformational state for receptor binding. Cell Res. 2017;27(1):119–129. doi: 10.1038/cr.2016.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yuan Y., Cao D., Zhang Y., et al. Cryo-EM structures of MERS-CoV and SARS-CoV spike glycoproteins reveal the dynamic receptor binding domains. Nat. Commun. 8. 2017 doi: 10.1038/ncomms15092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mille J.K., Whittaker G.R. Host cell entry of Middle East respiratory syndrome coronavirus after two-step, furin-mediated activation of the spike protein. Proc. Natl. Acad. Sci. U. S. A. 2014;111(42):15214–15219. doi: 10.1073/pnas.1407087111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang H., et al. SARS coronavirus entry into host cells through a novel clathrin- and caveolae-independent endocytic pathway. Cell Res. 2008;18(2):290–301. doi: 10.1038/cr.2008.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kuba K., Imai Y., Ohto-Nakanishi T., Penninger J.M. Trilogy of ACE2: a peptidase in the renin-angiotensin system, a SARS receptor, and a partner for amino acid transporters. Pharmacol. Ther. 2010;128(1):119–128. doi: 10.1016/j.pharmthera.2010.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Song Z., Xu Y., Bao L., Zhang L., Yu P., Qu Y. Vol. 11. 2019. From SARS to MERS, Thrusting coronaviruses into the spotlight; p. 59. Viruses. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Weiss S.R., Leibowitz J.L. Advances in virus research. 1st ed. Vol. 81. Elsevier Inc.; 2011. Chapter 4 - Coronavirus pathogenesis; pp. 85–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Perlman S., Netland J. Coronaviruses post-SARS: update on replication and pathogenesis. Nat. Rev. Microbiol. 2009;7(6):439–450. doi: 10.1038/nrmicro2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.De Wit E., Van Doremalen N., Falzarano D., Munster V.J. SARS and MERS: recent insights into emerging coronaviruses. Nat. Rev. Microbiol. 2016;14(8):523–534. doi: 10.1038/nrmicro.2016.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xu Z., et al. Pathological findings of COVID-19 associated with acute respiratory distress syndrome. Lancet Respir. Med. 2020;8(4):420–422. doi: 10.1016/S2213-2600(20)30076-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fan Y.Y., et al. Characterization of SARS-CoV-specific memory T cells from recovered individuals 4 years after infection. Arch. Virol. 2009;154(7):1093–1099. doi: 10.1007/s00705-009-0409-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tang F., et al. Lack of peripheral memory B cell responses in recovered patients with severe acute respiratory syndrome: a six-year follow-up study. J. Immunol. 2011;186(12):7264–7268. doi: 10.4049/jimmunol.0903490. [DOI] [PubMed] [Google Scholar]
- 27.Huang C., et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395(10223):497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Williams A.E., Chambers R.C. The mercurial nature of neutrophils: Still an enigma in ARDS? Am. J. Phys. Lung Cell. Mol. Phys. 2014;306:L217–230. doi: 10.1152/ajplung.00311.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Snijder E.J., et al. Ultrastructure and origin of membrane vesicles associated with the severe acute respiratory syndrome coronavirus replication complex. J. Virol. 2006;80(12):5927–5940. doi: 10.1128/jvi.02501-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Channappanavar R., et al. IFN-I response timing relative to virus replication determines MERS coronavirus infection outcomes. J. Clin. Invest. 2019;129(9):3625–3639. doi: 10.1172/JCI126363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Channappanavar R., et al. Dysregulated type I interferon and inflammatory monocyte-macrophage responses cause lethal pneumonia in SARS-CoV-infected mice. Cell Host Microbe. 2016;19(2):181–193. doi: 10.1016/j.chom.2016.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Niemeyer D., et al. Middle East respiratory syndrome coronavirus accessory protein 4a is a type I interferon antagonist. J. Virol. 2013;87(22):12489–12495. doi: 10.1128/jvi.01845-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang Y., et al. The structural and accessory proteins M, ORF 4a, ORF 4b, and ORF 5 of Middle East respiratory syndrome coronavirus (MERS-CoV) are potent interferon antagonists. Protein Cell. 2013;4(12):951–961. doi: 10.1007/s13238-013-3096-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Omrani A.S., et al. Ribavirin and interferon alfa-2a for severe Middle East respiratory syndrome coronavirus infection: a retrospective cohort study. Lancet Infect. Dis. 2014;14(11):1090–1095. doi: 10.1016/S1473-3099(14)70920-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Saikatendu K.S., et al. Structural basis of severe acute respiratory syndrome coronavirus ADP-ribose-1″-phosphate dephosphorylation by a conserved domain of nsP3. Structure. 2005;13(11):1665–1675. doi: 10.1016/j.str.2005.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhu N., et al. A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 2020;382(8):727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen X., Yang X., Zheng Y., Yang Y., Xing Y., Chen Z. SARS coronavirus papain-like protease inhibits the type I interferon signaling pathway through interaction with the STING-TRAF3-TBK1 complex. Protein Cell. 2014;5(5):369–381. doi: 10.1007/s13238-014-0026-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Subissi L., et al. One severe acute respiratory syndrome coronavirus protein complex integrates processive RNA polymerase and exonuclease activities. Proc. Natl. Acad. Sci. U. S. A. 2014;111(37):E3900–E3909. doi: 10.1073/pnas.1323705111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Imbert I., et al. A second, non-canonical RNA-dependent RNA polymerase in SARS coronavirus. EMBO J. 2006;25(20):4933–4942. doi: 10.1038/sj.emboj.7601368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chu C.K., et al. Antiviral activity of nucleoside analogues against SARS-coronavirus (SARS-CoV) Antivir. Chem. Chemother. 2006;17(5):285–289. doi: 10.1177/095632020601700506. [DOI] [PubMed] [Google Scholar]
- 41.Shum K.T., Tanner J.A. Differential inhibitory activities and stabilisation of DNA aptamers against the SARS coronavirus helicase. Chembiochem. 2008;9(18):3037–3045. doi: 10.1002/cbic.200800491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jang K.J., Lee N.R., Yeo W.S., Jeong Y.J., Kim D.E. Isolation of inhibitory RNA aptamers against severe acute respiratory syndrome (SARS) coronavirus NTPase/helicase. Biochem. Biophys. Res. Commun. 2008;366(3):738–744. doi: 10.1016/j.bbrc.2007.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yu M.S., et al. Identification of myricetin and scutellarein as novel chemical inhibitors of the SARS coronavirus helicase, nsP13. Bioorg. Med. Chem. Lett. 2012;22(12):4049–4054. doi: 10.1016/j.bmcl.2012.04.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xia S., et al. Middle East respiratory syndrome coronavirus (MERS-CoV) entry inhibitors targeting spike protein. Virus Res. 2014;194:200–210. doi: 10.1016/j.virusres.2014.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Millet J.K., Whittaker G.R. Host cell proteases: critical determinants of coronavirus tropism and pathogenesis. Virus Res. 2015;202:120–134. doi: 10.1016/j.virusres.2014.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lan J., et al. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;581(7807):215–220. doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- 47.Narayanan K., et al. Severe acute respiratory syndrome coronavirus nsp1 suppresses host gene expression, including that of type I interferon, in infected cells. J. Virol. 2008;82(9):4471–4479. doi: 10.1128/jvi.02472-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Taylor J.K., et al. Severe acute respiratory syndrome coronavirus ORF7a inhibits bone marrow stromal antigen 2 Virion tethering through a novel mechanism of glycosylation interference. J. Virol. 2015;89(23):11820–11833. doi: 10.1128/jvi.02274-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Holshue M.L., et al. First case of 2019 novel coronavirus in the United States. N. Engl. J. Med. 2020;382(10):929–936. doi: 10.1056/NEJMoa2001191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang M., et al. Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res. 2020;30(3):269–271. doi: 10.1038/s41422-020-0282-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Warren T.K., et al. Therapeutic efficacy of the small molecule GS-5734 against Ebola virus in rhesus monkeys. Nature. 2016;531(7594):381–385. doi: 10.1038/nature17180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang L. Binding mechanism of remdesivir to SARS-CoV-2 RNA dependent RNA polymerase. J. Phys. Chem. B 124. 2020;82(9):6955–6962. doi: 10.1021/acs.jpcb.0c04198. [DOI] [PubMed] [Google Scholar]
- 53.Cao Y., Deng Q., Dai S. Remdesivir for severe acute respiratory syndrome coronavirus 2 causing COVID-19: An evaluation of the evidence. Travel Med. Infect. Dis. 2020;35:101647. doi: 10.1016/j.tmaid.2020.101647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Grein J., et al. Compassionate use of Remdesivir for patients with severe Covid-19. N. Engl. J. Med. 2020;382:2327–2336. doi: 10.1056/nejmoa2007016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Choy K.T., et al. Remdesivir, lopinavir, emetine, and homoharringtonine inhibit SARS-CoV-2 replication in vitro. Antivir. Res. 2020;178:104786. doi: 10.1016/j.antiviral.2020.104786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shereen M.A., Khan S., Kazmi A., Bashir N., Siddique R. COVID-19 infection: origin, transmission, and characteristics of human coronaviruses. J. Adv. Res. 2020;24:91–98. doi: 10.1016/j.jare.2020.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang W., Tang J., Wei F. Updated understanding of the outbreak of 2019 novel coronavirus (2019-nCoV) in Wuhan, China. J. Med. Virol. 2020;92(4):441–447. doi: 10.1002/jmv.25689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Devaux C.A., Rolain J.M., Colson P., Raoult D. New insights on the antiviral effects of chloroquine against coronavirus: what to expect for COVID-19? Int. J. Antimicrob. Agents. 2020;55:105938. doi: 10.1016/j.ijantimicag.2020.105938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tu Y., Chien C., Yarmishyn A.A., Lin Y. 2020. A Review of SARS-CoV-2 and the Ongoing Clinical Trials. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Colson P., Rolain J.M., Lagier J.C., Brouqui P., Raoult D. Chloroquine and hydroxychloroquin+e as available weapons to fight COVID-19. Int. J. Antimicrob. Agents. 2020;55(4):105932. doi: 10.1016/j.ijantimicag.2020.105932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nichol A.A. Potential implications of testing an experimental mRNA-based vaccine during an emerging infectious disease pandemic. Am. J. Bioeth. 2020;20:W2–W3. doi: 10.1080/15265161.2020.1763696. [DOI] [PubMed] [Google Scholar]