Abstract
COVID-19 is associated with 5.1% mortality. Although the virological, epidemiological, clinical, and management outcome features of COVID-19 patients have been defined rapidly, the inflammatory and immune profiles require definition as they influence pathogenesis and clinical expression of COVID-19. Here we show lymphopenia, selective loss of CD4+ T cells, CD8+ T cells and NK cells, excessive T-cell activation and high expression of T-cell inhibitory molecules are more prominent in severe cases than in those with mild disease. CD8+ T cells in patients with severe disease express high levels of cytotoxic molecules. Histochemical studies of lung tissue from one fatality show sub-anatomical distributions of SARS-CoV-2 RNA and massive infiltration of T cells and macrophages. Thus, aberrant activation and dysregulation of CD8+ T cells occur in patients with severe COVID-19 disease, an effect that might be for pathogenesis of SARS-CoV-2 infection and indicate that immune-based targets for therapeutic interventions constitute a promising treatment for severe COVID-19 patients.
Subject terms: Infection, Viral infection, Acute inflammation
Immunophenotyping of patients with COVID-19 is ongoing, but much remains to be learned. Here the authors analyze 41 hospitalized patients with COVID-19 and show a higher degree of lymphopenia in various immune cell subsets as well as cytotoxicity and T cell inhibitory marker expression in severe cases compared with mild.
Introduction
As of 26th June 2020, the unprecedented global epidemic outbreak of novel Coronavirus Disease-2019 (COVID-19) has resulted in 9,296,202 laboratory-confirmed cases of SARS-CoV-2 infection reported from 210 countries to the WHO1. Of these, there have been 479,133 deaths (5.1% mortality). Whilst the virological, epidemiological, clinical, routine laboratory, imaging, and management outcome features of COVID-19 patients have been rapidly defined2–13, the immune and inflammatory profiles are not well understood.
A wide spectrum of clinical manifestations occurs from mild to severe disease with extensive lung involvement leading to acute respiratory distress syndrome (ARDS) and death2–7,10,13. Severe lung disease with extensive alveolar damage and progressive respiratory failure leads to fatal outcomes3–5,7,11. Higher fatality rates occur in the elderly, and in individuals who have diabetes, co-morbidities and other causes of immunocompromise4. We recently reported a case where significant interstitial lymphocytic infiltrates in the lung tissue, lymphopenia, and overactivation of T cells in peripheral blood10. Inflammatory and immune responses are important for elimination of the infection, but may have a significant impact on SARS-CoV-2 pathogenesis, and may play a role in the expression of the clinical spectrum of COVID-19 disease.
In this study, we perform a cross-sectional observational study of immunological and inflammatory markers and cells in 41 COVID-19 patients (29 with mild disease and 12 with severe disease including 2 fatal cases). We find COVID-19 patients in severe group are characterized by profound lymphopenia, strong T-cell activation, and increased expression of T-cell inhibitory molecules compared to mild group of patients.
Results
Patient demographics and clinical features
Forty-one patients with laboratory-confirmed acute COVID-19 disease were enrolled: 29 with mild disease and 12 with severe disease. Representative computed tomographic images are shown in Supplementary Fig. 1. Table 1 shows demographics and baseline characteristics of these patients. The median age was 39 years (IQR 33.5–50.0); 25 (61%) were male. The median time of symptom onset before admission was 5 days (IQR 4.0–9.0). Two patients (5%) died. Thirty-six patients (85%) reported fever before hospitalization. Signs and symptoms included myalgia or fatigue (46%), expectoration (32%), cough (27%), sore throat (17%), diarrhea (10%), shortness of breath (12%), rhinorrhea (2%), and chest pain (2%). The median age was significantly higher in severe group (50 years) compared to mild group (37 years, p = 0.0072, two-tailed Mann–Whitney U-test). The duration of symptoms before admission was also higher in severe group (10.5 vs. 5.0 days, p = 0.0064, two-tailed Mann–Whitney U-test). Symptoms such as expectoration, myalgia or fatigue, and shortness of breath were increased in severe group compared to mild group.
Table 1.
Demographics and baseline characteristics of patients infected with SARS-CoV-2.
| Total (n = 41) | Mild (n = 29) | Severe (n = 12) | p value | |
|---|---|---|---|---|
| Age, years | 39.0 (33.5–50.0) | 37.0 (29.0–44.0) | 50.0 (39.75–85.0) | 0.0072 |
| Sex | 0.6308 | |||
| Men | 25 (61%) | 17 (59%) | 8 (67%) | |
| Women | 16 (39%) | 12 (41%) | 4 (33%) | |
| OnSet of symptom to hospital admission, days | 5.0 (4.0–9.0) | 5.0 (4.0–7.0) | 10.5 (5.0–12.25) | 0.0064 |
| Death | 2 (5%) | 0 | 2 (17%) | 0.0805 |
| Exposure to Wuhan | 24 (59%) | 18 (62%) | 6 (50%) | 0.4754 |
| Any comorbidity | 10 (24%) | 3 (10%) | 7 (58%) | 0.0011 |
| Hypertension | 5 (12%) | 1 (3%) | 4 (33%) | 0.0078 |
| Diabetes | 4 (10%) | 2 (7%) | 2 (17%) | 0.3374 |
| Malignancy | 1 (1%) | 0 | 1 (8%) | 0.2927 |
| Chronic liver disease | 3 (7%) | 1 (3%) | 2 (17%) | 0.2002 |
| Fever | 35 (85%) | 25 (86%) | 10 (83%) | 0.8128 |
| Highest temperature (°C) | 0.63 | |||
| <37.3 | 6 (15%) | 4 (14%) | 2 (17%) | |
| 37.3–38.0 | 9 (22%) | 7 (24%) | 1 (8%) | |
| 38.1–39.0 | 18 (44%) | 12 (41%) | 7 (58%) | |
| >39.0 | 8 (20%) | 6 (21%) | 2 (17%) | |
| Cough | 27 (66%) | 17 (59%) | 10 (83%) | 0.1289 |
| Expectoration | 13 (32%) | 6 (21%) | 7 (58%) | 0.0184 |
| Rhinorrhoea | 1 (2%) | 1 (3%) | 0 | >0.9999 |
| Myalgia or fatigue | 19 (46%) | 9 (31%) | 10 (83%) | 0.0022 |
| Nausea and vomiting | 0 | 0 | 0 | |
| Sore throat | 7 (17%) | 5 (17%) | 2 (17%) | 0.9645 |
| Shortness of breath | 5 (12%) | 1 (3%) | 4 (33%) | 0.0078 |
| Chest pain | 1 (2%) | 0 | 1 (8%) | 0.2927 |
| Diarrhea | 4 (10%) | 4 (14%) | 0 | 0.1756 |
Data are median (IQR), or n (%). p values comparing mild and severe are from two-tailed χ² test, two-tailed Fisher’s exact test, or two-tailed Mann–Whitney U-test.
Laboratory findings
Routine laboratory test results are shown in Table 2. There were no significant differences in the absolute number of leukocytes, neutrophils, and platelets counts between mild and severe group. The severe group exhibited a significantly lower absolute lymphocyte count than the mild group (p < 0.0001, two-tailed Mann–Whitney U-test). The severe group showed increased D-dimer (p = 0.0065, two-tailed Mann–Whitney U-test) and serum ferritin (p = 0.0024), and had a lower albumin levels (p = 0.0005, two-tailed Mann–Whitney U-test).
Table 2.
Laboratory findings of hospitalized patients infected with SARS-CoV-2.
| Normal range | Total (n = 41) | Mild (n = 29) | Severe (n = 12) | p value | |
|---|---|---|---|---|---|
| Leukocytes count (×109 L−1) | 3.97–9.15 | 4.95 (4.02–6.68) | 4.95 (4.02–6.35) | 4.955 (3.9975–6.695) | 0.9718 |
| Neutrophils count (×109 L−1) | 2.0–7.0 | 2.93 (2.25–4.36) | 2.85 (2.24–3.75) | 3.715 (2.7425–5.04) | 0.2136 |
| Lymphocytes count (×109 L−1) | 0.80–4.00 | 1.31 (0.88–1.61) | 1.54 (1.17–1.82) | 0.775 (0.4925–1.0425) | <0.0001 |
| Platelets count (×109 L−1) | 85.0–303.0 | 170.0 (151.0–233.0) | 170.0 (151.0–218.0) | 170.0 (149.0–263.5) | 0.5854 |
| Hemoglobin (g L−1) | 131.0–172.0 | 136.0 (128.0–148.0) | 140.0 (130.0–150.0) | 127.0 (113.5–147.25) | 0.114 |
| Activated partial thromboplastin time (s) | 23.0–42.0 | 30.4 (27.4–34.2) | 30.4 (28–34.7) | 30.1 (24.8–32.025) | 0.3382 |
| Prothrombin time (s) | 10.20–14.30 | 11.9 (11.4–12.5) | 12.1 (11.5–12.5) | 11.65 (11.325–11.95) | 0.0922 |
| D-dimer (µg L−1) | 0.00–0.55 | 0.24 (0.17–0.53) | 0.23 (0.17–0.42) | 0.61 (0.245–0.985) | 0.0065 |
| Albumin (g L−1) | 35.0–55.0 | 39.0 (35.0–42.0) | 41.0 (38.0–43.0) | 34.5 (32.5–36.25) | 0.0005 |
| Alanine aminotransferase (U L−1) | 5.0–40.0 | 26.0 (15.0–40.0) | 24.0 (15.0–39.0) | 26.0 (14.75–70.75) | 0.6456 |
| Aspartate aminotransferase (U L−1) | 8.0–40.0 | 24.0 (19.0–35.0) | 25.0 (21.0–30.0) | 21.5 (18.0–55.0) | 0.5756 |
| Total bilirubin (μmol L−1) | 3.40–20.5 | 10.3 (7.3–14.7) | 9.5 (7.2–14.7) | 10.95 (8.825–14.2) | 0.5104 |
| Serum creatinine (μmol L−1) | 62.0–115.0 | 74.0 (69.0–85.0) | 80.0 (69.0–87.0) | 70.0 (67.0–79.0) | 0.0355 |
| Lactate dehydrogenase (U L−1) | 109.0–245.0 | 212 (193–247) | 212 (193–247) | 210.5 (190.5–333.25) | 0.7185 |
| Interleukin-6 (pg mL−1) | 0.0–7.0 | 9.335 (5.383–23.8775) | 8.545 (5.395–16.865) | 18.695 (3.84–40.8175) | 0.6556 |
| C-reactive protein (mg L−1) | 0.068–8.20 | 7.4 (4.0–16.1) | 7.1 (4.0–10.4) | 10.955 (5.4125–36.55) | 0.1602 |
| Procalcitonin (ng mL−1) | 0.0–0.5 | 0.044 (0.035–0.065) | 0.0435 (0.034–0.0615) | 0.049 (0.0385–0.073) | 0.4569 |
| Erythrocyte sedimentation rate (mm h−1) | 0.0–15.0 | 12.5 (7.75–34.25) | 11.5 (7–19.75) | 26.5 (11.5–61) | 0.0699 |
| Serum ferritin (ng mL−1) | 30.0–400.0 | 366.95 (187.5–538.025) | 306.8 (95.32–434.9) | 532 (419.7–837.6) | 0.0024 |
Data are median (IQR). p values comparing mild and severe are from two-tailed Mann–Whitney U-test.
T cells, B cells, and NK cells profiles
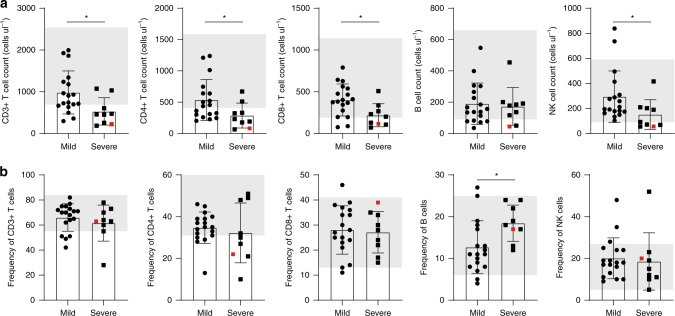
The absolute numbers of CD3+ T cells, CD4+ T cells, CD8+ T cells, B cells, and NK cells are shown in Fig. 1a. The severe group had decreased numbers of CD3+ T cells, CD4+ T cells, CD8+ T cells, and NK cells compared to the group experienced a mild clinical presentation. 7 out of 9 (77.8%) patients exhibited decreased CD3+ T cell and CD4+ T cell counts, and 4 out of 9 (44.4%) patients had decreased CD8+ T cell and NK cell counts below normal levels. However, in the patient group with mild symptoms, 5 out of 18 (27.8%) patients exhibited decreased CD3+ T cell counts, 8 out of 18 (44.4%) patients exhibited decreased CD4+ T cell count. 2 out of 18 (11.1%) patients had decreased CD8+ T cell counts. NK cell count was in normal range for most mild patient (16/18), with two patients exhibited higher NK cells than normal. No statistical differences were observed in the frequencies of CD3+ T cells, CD4+ T cells, CD8+ T cells, and NK cells between mild and severe groups, except for that of B cells, which was increased in severe patients (p = 0.015, two-tailed Mann–Whitney U-test) (Fig. 1b).
Fig. 1. Absolute cell counts and percentages of peripheral lymphocyte subsets in SARS-CoV-2 infected patients.
a Absolute cell counts and b percentages of CD3+ T cells, CD4+ T cells, CD8+ T cells, B cells, and NK cells in the mild (n = 18) and severe (n = 9) group were analyzed by flow cytometry. Shaded region showing the normal range of indicated cell subset. Colored symbols indicated deceased cases. Data are expressed as mean ± SD. *p < 0.05 by two-tailed Mann–Whitney U-test for a (p = 0.0124, p = 0.0354, p = 0.0199, and p = 0.0354, respectively) and b (p = 0.015). Source data included as a Source Data File.
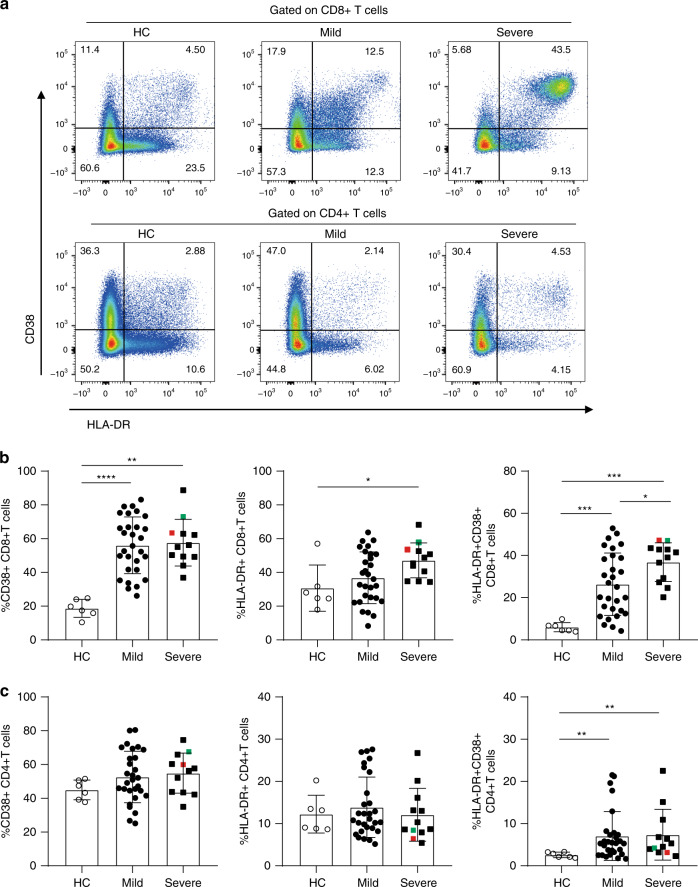
CD8+ T cells in severe patients
T-cell activation plays a critical role in defense of pathogen infection. Thus, we evaluated the degree of T-cell activation. The levels of T-cell activation were statistically higher in SARS-CoV-2 infected patients than cells from healthy controls (HC), as reflected by higher proportions of CD38+CD8+T cells, HLA-DR+CD8+T cells, and CD38+HLA-DR+CD8+T cells (Fig. 2a, b). Notably, the frequencies of CD38+HLA-DR+CD8+T cells in patients with severe symptoms were significantly higher compared with patients in the mild symptom group (p = 0.0372, two-tailed Mann–Whitney U-test) (Fig. 2b). The frequencies of CD38+HLA-DR+CD8+T and HLA-DR+CD8+T cells correlated positively with time after disease onset (R = 0.4425, p = 0.0038 and R = 0.4300, p = 0.005, respectively, Pearson correlation test) (Supplementary Fig. 2a). For CD4+T cells, no statistically differences were observed for frequencies of CD38+CD4+T cells and HLA-DR+CD4+T cells among HC, mild, and severe patients. However, compared with HC, the proportion of CD38+HLA-DR+CD4+T cells was higher in both mild and severe patients, but without difference between the two groups of SACR-CoV-2 infected patients (Fig. 2c). There is no difference in the absolute number of CD38+HLA-DR+CD8+T and CD38+HLA-DR+CD4+T cells between mild and severe group (Supplementary Fig. 2b).
Fig. 2. Increased expression of activation markers on T cells in SARS-CoV-2 infected patients.
a Representative dot plots showing CD38 and HLA-DR expression on CD8+ and CD4+ T cells from PBMCs of HC, Mild and Severe COVID-19 patients. b, c The percentage of CD38 and HLA-DR single positive or double positive b CD8+ T cells or c CD4+ T cells in PBMCs of HC (n = 6), mild (n = 29) and severe (n = 12) SARS-COV-2 infected patients was assessed by flow cytometry. Colored symbols indicated deceased cases. Data are expressed as mean ± SD. *p < 0.05, ***p < 0.001, ****p < 0.0001, by two-tailed Mann–Whitney U-test for b (p < 0.0001 and p = 0.0001 for CD38+CD8+T cells, p = 0.0135 for HLA-DR+CD8+T cells, and p = 0.0002, p = 0.0001, and p = 0.0372 for HLA-DR+CD38+CD8+T cells) and c (p = 0.0088 and p = 0.0046 for HLA-DR+CD38+CD4+T cells, respectively). Source data included as a Source Data File.
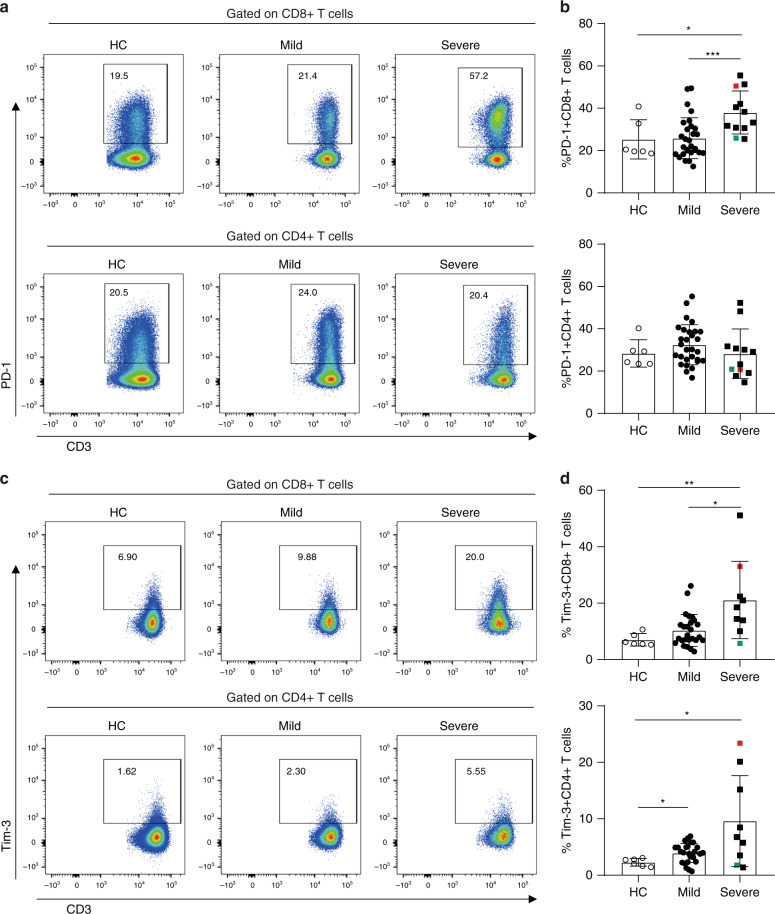
PD-1 and TIM-3 expression on CD8+ T cells in severe patients
The activation/inhibitory costimulatory molecules expression on circulating T cells is shown in Fig. 3. The proportion of PD-1+CD8+T cells was higher in severe patients compared to HC and mild patients, but there was no statistical difference in the proportion of PD-1+CD4+T cells among HC, mild and severe patients (Fig. 3a, b). In addition, the frequencies of PD-1+CD8+T cells correlated positively (R = 0.4271, p = 0.0053, Pearson correlation test) with time after disease onset (Supplementary Fig. 2c). Severe patients also exhibited higher frequencies of TIM-3+CD8+T cells compared to HC and mild patients. Increased frequencies of TIM-3+CD4+T cells were also observed in mild and severe patients compared with HC. However, no statistically significant difference was observed in the frequencies of TIM-3+CD4+T cells between mild and severe patients (Fig. 3c, d). Interestingly, positive correlations were observed between the frequencies of Tim-3+CD8+T (R = 0.5468, p = 0.0007, Pearson correlation test) and Tim-3+CD4+T cells (R = 0.36, p = 0.0337, Pearson correlation test) and time after disease onset (Supplementary Fig. 2d). In addition, the frequencies of PD-1 on CD38+HLA-DR+CD8+T and CD38+HLA-DR+CD4+T cells were higher than their double negative counterparts, respectively. Compared with mild cases, PD-1 expression on CD38+HLA-DR+CD4+T and CD38+HLA-DR+CD8+T cells are higher in severe cases (Supplementary Fig. 2e).
Fig. 3. Exhaustion of T cells in SARS-COV-2 infected patients.
a, b PD-1 expression on CD8+ T and CD4+ T cells in HC (n = 6), mild (n = 29), and severe (n = 12) patients of SARS-COV-2 infection. c, d TIM-3 expression on CD8+ T and CD4+ T cells in HC (n = 6), mild (n = 29), and severe (n = 12) patients of SARS-COV-2 infection. Colored symbols indicated deceased cases. Data are expressed as mean ± SD. *p < 0.05, **p < 0.01, ***p < 0.001, by two-tailed Mann–Whitney U-test for b (p = 0.032 and p = 0.0005) and d (p = 0.0076 and p = 0.0105 for Tim-3+CD8+ T cells, and p = 0.0185 and p = 0.0496 for Tim-3+CD4+ T cells, respectively). Source data included as a Source Data File.
CD4+ T cells differentiation
The naïve/memory subsets of CD4+ T cells were analyzed according to CD45RA, CD27, and CCR7 expression (Supplementary Fig. 3a). Compared with HC, SARS-COV-2 infection leads to increased proportion of TN cells in severe patients and increased TCM cells in mild patients. Decreased proportion of TTM cells was observed in mild patients compared with HC. No statistical differences were observed between mild and severe group in the proportions of memory subsets.
The functional polarization of CD4+ T cells was defined according to the chemokine receptors CCR4, CCR6, and CXCR3 expression (Supplementary Fig. 3b). Compared with HC, the percentages of Th1 and Th1Th17 cells were higher in mild patients, while the percentages of Th2 and Th17 cells were lower. No statistical differences were observed in the percentages of Th1, Th2, and Th17 cells between mild and severe patients. The frequencies of Treg and pTfh cells were also assessed, but there was no statistical difference among HC, mild and severe groups (Supplementary Fig. 3c, d).
Cytokines and chemokines
Cytokines and chemokines in plasma of COVID-19 patients and HC were measured using flow cytometry based Aimplex kit (Supplementary Fig. 4). Inflammatory cytokines and chemokines in SARS-CoV-2 infected patients, such as IL-6, TNF-α, IL-17A, IFN-γ, and MCP-1, were significantly elevated than HC, although there was no significance between mild and severe patients. Cytokines related tissue repair, such as IL-10, IL-4, and IL-5, were also upregulated in COVID-19 patients, but without statistical significance between mild and severe patients. In summary, similar to SARS-CoV14 and MERS-CoV15 infections, COVID-19 patients showed higher plasma concentrations of cytokines/chemokines than HC, albeit there was no difference between mild and severe cases.
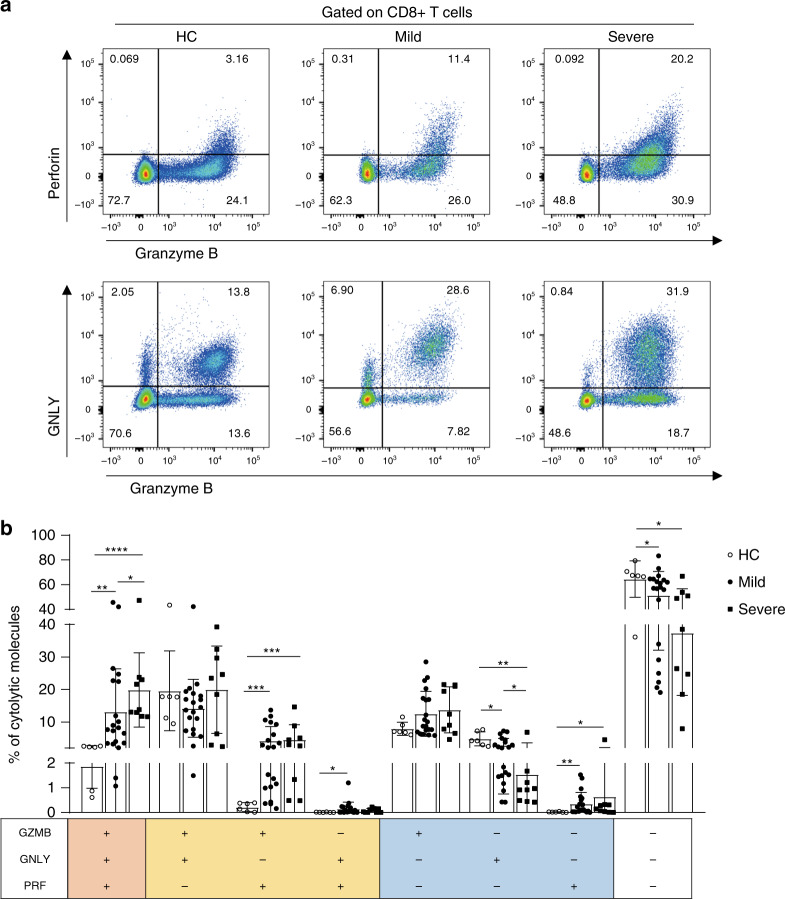
Multifunctional cytotoxic CD8+ T cells
Phenotypes of circulating CD8+ T cells were analyzed (Supplementary Fig. 5a). Among the CD8+ T cells, the frequencies of TTM subset significantly decreased in SARS-CoV-2 infected patients, but without difference between mild and severe individuals (Supplementary Fig. 5b). Moreover, in severe patients, there was an increase of TE subset compared to HC and a decrease of TCM subset compared to mild patients (Supplementary Fig. 5b). No significant changes were seen in TN or TEM subsets. The expressional levels of cytotoxic molecules (granzyme B, GZMB; perforin, PRF and granulysin, GNLY) in CD8+ T cells were analysed. Based on cytolytic ability, CD8+ T cells were divided into four subpopulations: monocytotoxic T lymphocytes (M-CTL) expressing only one of GZMB, PRF, and GNLY; dicytotoxic T lymphocytes (D-CTL) expressing two kinds of cytotoxic molecules; tricytotoxic T lymphocytes (T-CTL) expressing all three cytotoxic molecules; and noncytotoxic T lymphocytes (N-CTL) which express none of the molecules. The data showed that the frequencies of T-CTL are the highest in CD8+ T cells from severe patients and the lowest in HCs (Fig. 4a, b). There were also higher expressional levels of GZMB and PRF double positive D-CTL in SARS-CoV-2 infected patients compared to HCs (Fig. 4b). The high cytotoxicity of CD8+ T cells coincides with their over-activated status in severe patients, which might contribute to the ongoing immune injury in lung tissue.
Fig. 4. Co-expression patterns of cytotoxic effectors in CD8+ T cells from SARS-COV-2 infected patients.
a, b Cytolytic molecules were assessed by flow cytometry, and the proportions of CD8+ T lymphocytes co-expression of GNLY, GZMB, PRF were analyzed in HC (n = 6), mild (n = 20), and severe (n = 9) patients of SARS-COV-2 infection. Data are expressed as mean ± SD. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001 by two-tailed Mann–Whitney U-test for b (p = 0.002, p = 0.0004, and p = 0.0386 for GZMB+GNLY+PRF+ cells, p = 0.0006 and p = 0.0004 for GZMB+GNLY-PRF+ cells, p = 0.0459 for GZMB-GNLY+PRF+ cells, p = 0.0331, p = 0.0048, and p = 0.03 for GZMB-GNLY+PRF− cells, p = 0.0026 and p = 0.0496 for GZMB-GNLY-PRF+ cells, and p = 0.0279 and p = 0.012 for GZBM-GNLY-PRF− cells, respectively). Source data included as a Source Data File.
Immunobiological findings
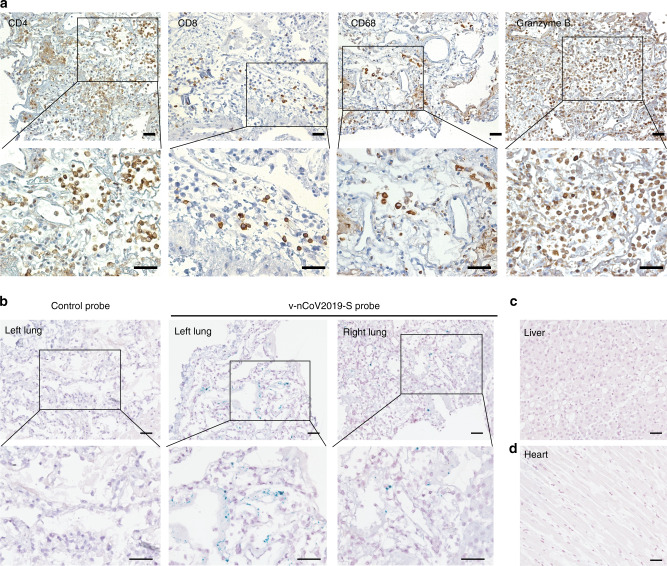
Pulmonary infiltration of lymphocytes and viral distribution in tissues are shown in Fig. 5a–d. There was diffuse alveolar damage with cellular fibromyxoid exudates and desquamation of pneumocytes in both lungs. Massive CD4+ T cell, CD8+ T cell, macrophages and GZMB+ cells infiltration were observed in the interstitial area of lung tissues (Fig. 5a). We used an in situ hybridization assay to detect viral RNA in fixed tissue sections. Intense dot signals were observed both in the left and right lungs (Fig. 5b), suggesting active replication in these cells. Indeed, these cells with intense signals showed atypical enlarged pneumocytes nuclei and prominent nucleoli, which are typical characteristics for multinucleated syncytial cells. Punctate dots were mainly distributed in the intraalveolar spaces, which might be dissociative viral particles. Notably, viral signal was not observed in liver (Fig. 5c) or heart tissues (Fig. 5d).
Fig. 5. Lymphocyte infiltration and viral distribution in lung.
a Immunohistochemical staining with anti-CD4, anti-CD8, anti-CD68 and anti-GZMB in lung tissues, scale bar = 50 μm. b–d Representative microscopy images of both left and right lungs (b), liver (c), and heart (d) sections from a COVID-19 patient hybridized with control or SARS-CoV-2-specific probes, scale bar = 50 μm. Independent experiments are repeated at least three times.
Discussion
To our knowledge this is the first detailed study of the immunological and inflammatory profiles of COVID-19 patients with severe disease. Cellular immune responses, consisting of both CD4+ T and CD8+ T cells, are essential for the control of viral infection. Much work has been aimed to elucidate the virological features3,6,9,11–13,16–19 as well as clinical characteristics3–5,7,16,17 of COVID-19 patients. However, little is known about the immunological and inflammatory profiles of patients and their association with COVID-19 severity.
The clinical, imaging, and routine laboratory findings of enrolled patients are similar to recently published data2–7,10,11,13. Lymphocytopenia has been observed in COVID-19 patients showing different spectrum of clinical disease20, although the changes in different lymphoid subsets were not yet defined. In a previous study, mainly in noncritical patients infected with SARS-CoV-2, 35% of patients had only mild lymphocytopenia4. In contrast, in other two reports with patients with severe disease, the incidences of lymphopenia were 63%3 and 70.3%7, respectively. Moreover, lymphocytopenia occurred in more than 80% of severe COVID-19 patients11. Our results show that the degree of lymphocytopenia may reflect the severity of COIVD-19 disease since the lymphocyte counts were slightly decreased in COVID-19 patients with mild disease, and significantly decreased in patients with severe disease. In addition, our data showed that several immune subsets, such as CD4+ T, CD8+ T, and NK cells, are lower in patients with severity disease.
The reason for patients developing lymphopenia remains unknown. Given that lymphocytes express low level of angiotensin-converting enzyme 2 (ACE2), the cell entry receptor for both SARS-CoV-2 and SARS-CoV12,16, and the viral genome is rarely detectable in peripheral blood of SARS-CoV-2 infected patients5,18, it is reasonable to speculate that the decrease of peripheral lymphocytes is not directly attributed to SARS-CoV-2 infection. An alternative explanation is that the decease of peripheral lymphocytes is a result of activation-induced apoptosis or aggressive migration from peripheral blood to the lungs, where robust viral replication occurs. Further studies should elucidate the mechanisms responsible for the lymphopenia. It is noteworthy that CD4+ T-cells were profoundly decreased in peripheral blood of severe SARS-COV-2 infected patients. Lessons from HIV infection indicated that a low CD4+ T-cell counts increases the risk of opportunistic infections and lower antiviral immune surveillance, suggesting more attention should to be given to patients in critical condition. Given the limitations of our cross-sectional study, and the clinical significance of lymphopenia and of CD4+ T-cell count, further longitudinal studies with serial sampling are needed to define this further.
The immune response is essential for eradication of the pathogen and resolution of active disease. A recent study demonstrated robust multi-factorial immune responses can be induced by SARS-CoV-2 in a mild case, which is similar to the avian H7N9 disease21 and Flavivirus infection22. However, the immune and inflammatory response can also be detrimental and cause immune mediated tissue injury, especially in severe cases. It was observed in the 2003 SARS epidemic as well as in the COVID-19 pandemic that pulmonary disease often worsens at 1–2 weeks after onset of respiratory symptoms, coinciding with the onset of virus clearance17,18. We recently reported the histology of COVID-19 by obtaining biopsy specimens at autopsy10,17,23. Histologic examination revealed diffuse alveolar damage, prominent desquamation of pneumocytes and hyaline membrane formation within left and right lungs with significant interstitial lymphocytic infiltrates. We showed in the histochemical examination of lung tissue obtained from one of our patients with severe disease who died, intense CD4+ and CD8+ T cells infiltrates, with strong granzyme B expression (Fig. 5). Our current study further suggests that the involvement of immune-mediated injury is plausible in the pathogenesis of COVID-19.
In severe patients, despite the absolute counts of CD8+ T cells decreased, these cells showed over-activation, increased T-cell inhibitory molecules expression and increased multiple cytotoxic granules expression, resembling other acute inflammatory processes such as malaria infection, Q-fever, and sepsis24,25. Whether these activated cells are antigen-specific or bystander ones warrants further study. However, PD-1 positive T-cells in the peripheral circulation may not only indicate T-cell exhaustion, but could also suggest the presence of antigen-specific T-cells, which needs to be further evaluated26. We found that T-cell activation appears to correlate with disease severity in COVID-19 patients as a higher frequency of activated CD8+ T cells (defined by CD38+ and HLA-DR+) occurred in severe patients with longer time after disease onset. Taken together, these findings highlight the involvement of CD8+ T cells for the immunopathogenesis of severe COVID-19. Considering the possibility of clinical application, lymphocyte subset cell counts and activated phenotypes of CD8+ T cells could be used to predict disease outcomes and evaluate new interventions for COVID-19 patients. For example, lymphocyte count is suggested to be closely monitored in COVID-19 patients receiving ECMO27.
In conclusion, we provided immunological and inflammatory markers linking disease severity in mild and severe cases of COVID-19 and suggested aberrant activation and dysregulation of CD8+ T cells occur in patients with severe COVID-19 disease which may play an important role in pathogenesis of SARS-CoV-2 infection. Immune-based therapeutic interventions28, such as IL-6 receptor antagonist (ChiCTR2000029765) and mesenchymal stem cells (NCT04252118), are undergoing clinical trials. Further studies are required to define immune-mediated tissue injury and identify specific host-directed therapy targets for treatment of the disease.
Methods
Ethical approval
This study was approved by the ethics committee of the Fifth Medical Center of PLA General Hospital and written informed consent was obtained from all patients enrolled in this study.
Patients
Between January 19th and 20th February, a total of 41 patients with laboratory confirmed COVID-19 disease hospitalized at the Fifth Medical Center of PLA General Hospital in Beijing, China, were studied. All patients were diagnosed according to World Health Organization (WHO) interim guidance and were positive for new coronavirus nucleic acid by using SARS-CoV-2 specific RT-PCR test. The patients were classified on basis of disease severity: The mild disease group were defined as with or without pneumonia who were admitted to general wards not requiring intensive care. The severe disease group were those who required critical care and met one or more of these criteria: dyspnea and respiratory rate ≥30/min, blood oxygen saturation ≤93%, PaO2/FiO2 ratio <300 mmHg, and lung infiltrates on CT scan >50% within 24–48 h, or those who exhibited respiratory failure, septic shock, and/or multiple organ dysfunction/failure. Patient demographics, clinical history, presentations, and findings were recorded. All patients had routine laboratory investigations (full blood count, liver and renal function tests, and coagulation tests).
Clinical samples
Peripheral venous blood samples were obtained on admission of the COVID-19 patients within 24 h and placed into the vacutainer tube, then centrifuged at 400×g for 5 min at 4 °C. The time of sampling relative to the onset of symptoms was recorded. Plasma samples were collected and stored at −80 °C until used. In one patient with severe disease who died, lung tissue samples were obtained post-mortem for immuno-histological analysis.
Lymphocyte counts and subsets
Absolute lymphocyte counts and subsets were determined using lymphocyte detection kit (Beijing Tongshengshidai Biotechnology Co., LTD, Beijing, China) following the manufacturer’s instructions.
Flow cytometry
Peripheral blood mononuclear cells (PBMC) were isolated from fresh venous blood using Ficoll density gradient. PBMC samples were stained with the following antibodies: CD3-APC-Cy7 (clone HIT3a), CD3-BV510 (clone OKT3), CD4-BV421 (clone OKT4), CD8-PE-Cy7 (clone SK1), CD45RA-BV510 (clone HI100), CCR7-APC (clone G043H7), CD27-FITC (clone MT271), HLA-DR-FITC (clone L243), CXCR5-BV421 (clone J252D4), PD-1-PE (clone EH12.2H7), CXCR3-BV510 (clone G025H7), CCR4-PerCP-Cy5.5 (clone L291H4), CCR6-PE (clone G034E3), CD25-APC (clone BC96), CD127-FITC (clone A019D5), Perforin-PE-Cy7 (dG9), Granzyme B-AF647 (GB11) were purchased from Biolegend (San Diego, CA); CD4-percp (clone SK3), CD38-APC (clone HIT2), Tim-3-PE (clone 7D3), GNLY-AF488 (clone RB1) were obtained from BD Biosciences (San Diego, CA). Granzyme B, Perforin and GNLY were measured de novo, without prior stimulation with PMA/ionomycin. BD Canto II instrument was used for FACS and the data was analyzed using FlowJo software V10 (Tree star Inc. Ashland, OR).
Cytokine and chemokine measurement
Plasma levels of 21 different cytokines and chemokines (IL-1β, IL-2, IL-4, IL-5, IL-6, IL-8, IL-10, IL-12P70, IL-17A, IL-17F, IL-22, TNF-α, TNF-β, IFN-γ, IL-1RA, IL-18, G-CSF, RANTES, MCP-1, IP-10, and MIP-1α) in 39 patients infected with SARS-CoV-2 and 24 health controls were determined by flow cytometry using an Aimplex kit (Beijing Quantobio, China) following the manufactures instructions.
Immunohistochemical staining
Formalin-fixed paraffin-embedded 4-μm sections of lung tissue were subject to immunohistochemistry. Following deparaffinization and rehydration, sections were incubated in 3% H2O2 in methanol for 30 min at room temperature to block endogenous peroxidase. The sections were then boiled in citrate buffer or EDTA buffer in a microwave oven. The staining was performed using primary antibodies against CD4 (ZSGB-BIO, Beijing, China), CD8 (Abcam, Cambridge, MA), CD68 (ZSGB-BIO), GZMB (Abcam), and incubated at 4 °C overnight. The sections were visualized using the diaminobenzidine solution (DAKO, Carpinteria, CA) and then lightly counterstained with hematoxylin. Images were captured with an inverted fluorescence microscope (PerkinElmer, Norwalk, CT).
RNAscope
SARS-CoV-2 RNA in formalin-fixed paraffin-embedded (FFPE) tissue were probed by RNAscope reagents, following the manufacture’s protocol. The probes (v-nCoV2019-S, cat. 848561)) targeting SARS-CoV-2 Spike gene and control probes (cat. 320751) are designed by ACD. Briefly, after H2O2 treatment and protease digestion, FFPE slides were incubated 2 h at 40 °C with probes. The amplifiers and detection solution in the RNAscope 2.5 HD Duplex Reagent kit were added sequentially for hybridization signal amplification for the indicated time. The slides were counterstained with 50% Hematoxylin staining solution for 30 s, and washed with water immediately. Tissue slides were cover slipped with Vectamount Permanent Mounting Medium (Vector Labs, 321584, Burlingame, California). Images were obtained with Perkin Elmer Vectra 3.0 (PerkinElmer).
Statistical analysis
GraphPad Prism statistical software version 8.0 (GraphPad Software, San Diego, CA) was used. Continuous measurements were displayed as median (IQR) and two group comparisons performed using Mann–Whitney U-test. Categorical variables were expressed as count (%) and compared by χ² test or Fisher’s exact test between mild and severe group. Pearson correlation test was conducted to assess association between two quantitative variables. p-value < 0.05 were considered statistically significant.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Supplementary information
Acknowledgements
We thank Dr Jun Hou for providing the safety guide for handling COVID-19 patients sample, and Jinhong Yuan and Chunbao Zhou for flow cytometry analysis. Sir Zumla and Prof Ippolito are co-principal investigators of the Pan-African Network on Emerging and Re-emerging Infections (PANDORA-ID-NET—https://www.pandora-id.net/), funded by the European & Developing Countries Clinical Trials Partnership, supported under Horizon 2020. Sir Zumla is in receipt of a National Institutes of Health Research senior investigator award. Prof Maeurer is a member of the innate immunity advisory group of the Bill & Melinda Gates Foundation, and is funded by the Champalimaud Foundation. INMI authors are supported by the Italian Ministry of Health (Ricerca Corrente Linea 1, COVID-2020-12371735 and COVID-2020-12371817). All authors are members of the Global Alliance for Host-Directed Therapies for COVID-19 (https://www.fchampalimaud.org/covid19/aci/). This work was supported by the Innovation Groups of the National Natural Science Foundation of China [81721002] and the National Key Research and Development Program of China [2020YFC0841900]. Both lead authors had full access to all the data in the study and had final responsibility for the decision to submit for publication.
Source data
Author contributions
J.Z and F.W conceived and designed the study. J.S., C.Z., and X.F performed the experiment and wrote the manuscript. F.M., Z.X., S.W., T.J., W.L., D.Z., and P.Z. provided clinical information. P.X., W.C., and T.Y. performed the statistical analysis. X.D., R.X., M.S., C.A., G.I., M.M., and A.Z. edited the manuscript and provided comments and feedback. All authors read and approved the final manuscript.
Data availability
The data that support the findings of this study are available from the corresponding author upon reasonable request. Source data are provided with this paper.
Competing interests
The authors declare no competing interests.
Footnotes
Peer review information Nature Communications thanks the anonymous reviewers for their contribution to the peer review of this work. Peer review reports are available.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Jin-Wen Song, Chao Zhang, Xing Fan.
Contributor Information
Fu-Sheng Wang, Email: fswang302@163.com.
Ji-Yuan Zhang, Email: uniquezjy@163.com.
Supplementary information
Supplementary information is available for this paper at 10.1038/s41467-020-17240-2.
References
- 1.World Health Organization. Coronavirus disease 2019 (COVID-19) situation report—157. https://www.who.int/docs/default-source/coronaviruse/situation-reports/20200625-covid-19-sitrep-157.pdf?sfvrsn=423f4a82_2 (2020).
- 2.Chan JF, et al. Genomic characterization of the 2019 novel human-pathogenic coronavirus isolated from a patient with atypical pneumonia after visiting Wuhan. Emerg. Microbes Infect. 2020;9:221–236. doi: 10.1080/22221751.2020.1719902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chan JF, et al. A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: a study of a family cluster. Lancet. 2020;395:514–523. doi: 10.1016/S0140-6736(20)30154-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen N, et al. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet. 2020;395:507–513. doi: 10.1016/S0140-6736(20)30211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huang C, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lu R, et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395:565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang D, et al. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus-infected pneumonia in Wuhan, China. JAMA. 2020;323:1061–1069. doi: 10.1001/jama.2020.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang FS, Zhang C. What to do next to control the 2019-nCoV epidemic? Lancet. 2020;395:391–393. doi: 10.1016/S0140-6736(20)30300-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wu F, et al. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xu Z, et al. Pathological findings of COVID-19 associated with acute respiratory distress syndrome. Lancet Respir. Med. 2020;8:420–422. doi: 10.1016/S2213-2600(20)30076-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang X, et al. Clinical course and outcomes of critically ill patients with SARS-CoV-2 pneumonia in Wuhan, China: a single-centered, retrospective, observational study. Lancet Respir. Med. 2020;8:475–481. doi: 10.1016/S2213-2600(20)30079-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhou P, et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhu N, et al. A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Channappanavar R, Perlman S. Pathogenic human coronavirus infections: causes and consequences of cytokine storm and immunopathology. Semin. Immunopathol. 2017;39:529–539. doi: 10.1007/s00281-017-0629-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rasmussen SA, Gerber SI, Swerdlow DL. Middle East respiratory syndrome coronavirus: update for clinicians. Clin. Infect. Dis. 2015;60:1686–1689. doi: 10.1093/cid/civ118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xu X, et al. Evolution of the novel coronavirus from the ongoing Wuhan outbreak and modeling of its spike protein for risk of human transmission. Sci. China Life Sci. 2020;63:457–460. doi: 10.1007/s11427-020-1637-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zou L, et al. SARS-CoV-2 viral load in upper respiratory specimens of infected patients. N. Engl. J. Med. 2020;382:1177–1179. doi: 10.1056/NEJMc2001737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wölfel, R. et al. Virological assessment of hospitalized patients with COVID-2019. Nature10.1038/s41586-020-2196-x (2020). [DOI] [PubMed]
- 19.Zhou, Y. et al. Pathogenic T cells and inflammatory monocytes incite inflammatory storm in severe COVID-19 patients. Natl Sci. Rev. 10.1093/nsr/nwaa041 (2020). [DOI] [PMC free article] [PubMed]
- 20.Tan L, et al. Lymphopenia predicts disease severity of COVID-19: a descriptive and predictive study. Signal Transduct. Target Ther. 2020;5:33. doi: 10.1038/s41392-020-0148-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang Z, et al. Recovery from severe H7N9 disease is associated with diverse response mechanisms dominated by CD8+ T cells. Nat. Commun. 2015;6:6833. doi: 10.1038/ncomms7833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Campos JLS, Mongkolsapaya J, Screaton GR. The immune response against flaviviruses. Nat. Immunol. 2018;19:1189–1198. doi: 10.1038/s41590-018-0210-3. [DOI] [PubMed] [Google Scholar]
- 23.de Wit E, van Doremalen N, Falzarano D, Munster VJ. SARS and MERS: recent insights into emerging coronaviruses. Nat. Rev. Microbiol. 2016;14:523–534. doi: 10.1038/nrmicro.2016.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guignant C, et al. Programmed death-1 levels correlate with increased mortality, nosocomial infection and immune dysfunctions in septic shock patients. Crit. Care. 2011;15:R99. doi: 10.1186/cc10112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mackroth MS, Abel A, Steeg C, Schulze Zur Wiesch J, Jacobs T. Acute malaria induces PD1+CTLA4+ effector T cells with cell-extrinsic suppressor function. PLoS Pathog. 2016;12:e1005909. doi: 10.1371/journal.ppat.1005909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cafri G, et al. Memory T cells targeting oncogenic mutations detected in peripheral blood of epithelial cancer patients. Nat. Commun. 2019;10:449. doi: 10.1038/s41467-019-08304-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Matthay MA, Aldrich JM, Gotts JE. Treatment for severe acute respiratory distress syndrome from COVID-19. Lancet Respir. Med. 2020;8:433–434. doi: 10.1016/S2213-2600(20)30127-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zumla A, Hui DS, Azhar EI, Memish ZA, Maeurer M. Reducing mortality from 2019-nCoV: host-directed therapies should be an option. Lancet. 2020;395:e35–e36. doi: 10.1016/S0140-6736(20)30305-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request. Source data are provided with this paper.