Dear Editor,
Bile is a complex aqueous secretion produced by the liver that facilitates the digestion of lipids in the small intestine. A group of ATP binding cassette (ABC) transporters localizing at the apical canalicular domain of hepatocytes are responsible for the secretion of the bile organic solutes to the bile canaliculi,1 including organic anions, cholesterol, bile salts, glutathione, organic cations and phospholipid (Fig. 1a). To date, the 3-D structures of ABCG2, ABCG5/G8, ABCB1, ABCB4 have been reported,2,3 leaving the structures of ABCB11 and ABCC2 unknown, despite we can get some information from the homologous structures of ABCC1,4 ABCB1 and ABCB4.
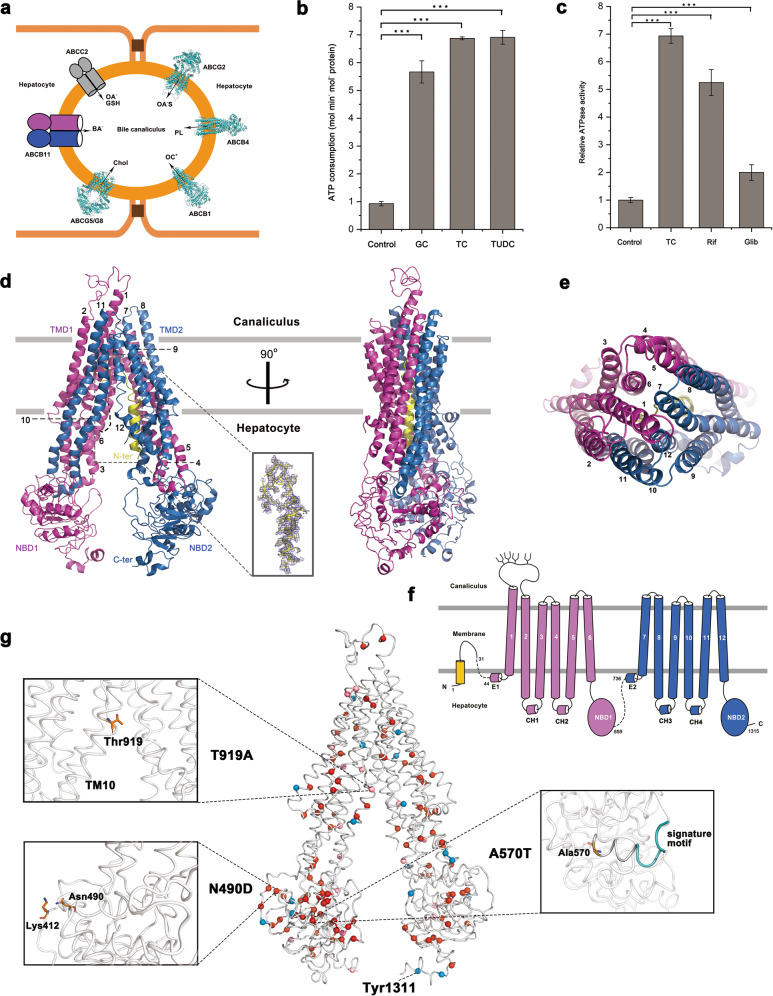
Fig. 1. Cryo-EM structure of human bile salts exporter ABCB11.
a Six ABC transporters involved in bile secretion and their substrates. Structure-known transporters are shown as cartoon presentation, which are ABCG2 (PDB: 6HUB), ABCB4 (PDB: 6S7P), ABCB1 (PDB: 6C0V), ABCG5/G8 (PDB: 5DO7). ABCB11 and ABCC2 are shown as simple geometrical figures, with ovals for NBDs and cylinders for TMDs. The substrates are OA−S sulfated organic anions; PL phospholipid; OC+ organic cations; Chol cholesterol; BA− bile salts; OA− organic anions; and GSH glutathione. b The ATPase activity of ABCB11 in the absence or presence of 40 μM substrates. c Relative ATPase activity of ABCB11 in the absence or presence of 40 μM taurocholic and two inhibitors (GC glycocholic; TC taurocholic; TUDC tauroursodesoxycholic; Rif rifampicin; Glib glibenclamide). At least three independent assays were performed for each assay. The means and standard deviations were calculated and the data are presented as means ± SD. One-way analysis of variance is used for the comparison of statistical significance in b, c. The P values of < 0.05, 0.01 and 0.001 are indicated with *, ** and ***, respectively. d The structure of apo ABCB11 in an inward-open state is shown in two perpendicular views with cartoon representation. TM1 to TM6 are colored in magenta, and TM7 to TM12 are in blue. The N-terminal helix is colored in yellow and its electron density detail is shown in the rectangular box. Contour level is set at σ = 2 and density is carved at a distance of 2 Å. e A top view of ABCB11 from the bile canaliculus. All TMs are indicated by numbers. f A topology illustration of ABCB11. Labels and colors are consistent with d, e. g Mapping of 132 clinically-identified pathogenic mutations, including missense and nonsense mutations on ABCB11. Missense mutations are labeled as red spheres, among which PFIC2-related mutations are colored as dark red, and BRIC2 are colored as light red. Nonsense mutations are indicated as blue spheres, and all nonsense mutations are PFIC2 related. The regions around Thr919, Asn490 and Ala570 are zoomed in.
Bile salts are the major organic solutes in bile, the secretion of which is crucial to the viability of hepatocytes as its intracellular accumulation in the liver will ultimately results in cell death.1 Human ABCB11, also known as bile salts export pump (BSEP), is the major exporter for the specific secretion of bile salts. Correspondingly, defects of ABCB11 function, due to either mutations or side effects of medicines, lead to inherited or acquired diseases, respectively, such as progressive familial intrahepatic cholestasis type 2 (PFIC2), benign recurrent intrahepatic cholestasis type 2 (BRIC2), and drug-induced cholestasis (DIC). The severe cholestasis PFIC2 is usually found in newborn patients, which could rapidly progress to cirrhosis, with the possibility of hepatocellular carcinoma and cholangiocarcinoma occurring under one-year age.5 As deposited in the Human Gene Mutation Database (http://www.hgmd.cf.ac.uk/), more than 300 different clinical mutations in ABCB11 gene, including missense, nonsense, aberrant splicing, deletions and insertions, have been identified from cholestatic patients. Despite ABCB11 shares a sequence identity of ~48% with two structure-known transporters ABCB1 and ABCB4, which are ~71% sequence-identical to each other, the three transporters differ significantly in substrate specificity. ABCB1 and ABCB4 could transport various organic cations and phosphatidylcholine, respectively, whereas ABCB11 selectively transports bile salts.6 However, the molecular mechanisms of transport and substrate specificity remain unclear, even for the structure-known ABCB1 and ABCB4.
We overexpressed the human full-length ABCB11 in HEK293 cells and obtained the stable and homogenous recombinant protein. The activity assays in the form of protein–detergent complex showed an ATPase activity of recombinant ABCB11 at a Vmax of 0.95 mol min−1 mol−1 protein (Fig. 1b). In contrast, upon the addition of either of the three previously-reported physiological substrates glycocholic, taurocholic and tauroursodesoxycholic, the Vmax values were significantly elevated by about 6–7 fold (Fig. 1b). In addition, two commonly used clinical medicines rifampicin and glibenclamide, which have been reported to inhibit the transport activity of ABCB11,1 could also elevate the ATPase activity to 5.5 and 2 fold (Fig. 1c), respectively, indicating that both medicines are most likely competitive inhibitors. The results of these biochemical assays further demonstrated that our protein samples were in a physiologically-relevant state.
Using cryogenic electron microscopy (cryo-EM), we solved the apo-form structure of human ABCB11 at the resolution of 3.5 Å (Fig. 1d, e; Supplementary information, Figs. S1–S3 and Table S1), and the transmembrane domains (TMDs) possess a relatively higher resolution of ~3.3 Å (Supplementary information, Figs. S2d and S4). ABCB11 is a full ABC transporter that consists of two TMDs and two nucleotide-binding domains (NBDs) in one polypeptide chain (Fig. 1f). The overall structure shows an inward-open conformation due to the absence of ATP molecule (Fig. 1d). The NBDs possess a classic NBD fold of ABC transporter, with an ɑ-helical subdomain and a RecA-like ATPase core subdomain. All conserved motifs, including the Walker A & B motifs, the ABC signature motif, could be clearly assigned in our structure. Of note, the Walker B motif in NBD1 has a degenerated active site, as the catalytic residue glutamate is replaced by methionine. As a typical type-I exporter, each TMD of ABCB11 consists of six transmembrane helices (TMs) with the significant cytosolic extension forming two diverged “wings” at the hepatocellular side, and TMs packing against each other at the bile canaliculus side. TM4 and TM5 from TMD1, in addition to TM10 and TM11 from TMD2, are localized in the opposite TMD wing due to domain swapping (Fig. 1d, e). A pair of coupling helices from the TMD, which are embedded in the grooves on the NBD at the same side, couples the conformational changes between TMDs and NBDs (Fig. 1d, f).
Notably, we found a continuous chain of electron density within the translocation cavity, which could fit with the missing N-terminus. ABCB11 has an extension at the N-terminus compared to other members of human ABCB subfamily, similar to the ABCB1 homolog in Caenorhabditis elegan, which is folded into a helical hairpin inserting in the translocation cavity.7 However, the N-terminal extension of ABCB11 forms a single helix (residues Met1-Glu16) that sticks on TMD2, succeeding with a loop that runs to the top of the cavity and turns back along TMD1. Notably, 12 residues (Ser32-Val43) between this N-terminal loop and TM1 are missing in our structure. To investigate the role of this N-terminal helix, we constructed a truncated version, and the results of ATPase activity assays showed no significant difference from the full-length ABCB11 (Supplementary information, Fig. S1b). In fact, truncation of the helical hairpin in ceABCB1 also revealed no change of activity. However, we found that the expression level of the N-terminus truncated form was only half to that of the full-length, indicating it may help to stabilize the protein.
Concerning the extracellular 34-residue (Gln101 to Ile134) loop between TM1 and TM2 that points towards the bile canaliculus, only the main chains could be traced due to the poor electron density (Fig. 1d). Four conserved asparagine residues in this loop, namely Asn109, 116, 122 and 125, which are exposed in our structure, were proved to be subject to glycosylation for the trafficking and functionality of ABCB11 in the apical membrane.8 In addition, the C-terminus of ABCB11, which is folded into a small helical hairpin (Fig. 1d), contains a so-called endocytic cargo motif (Trp-Lys-Leu-Val) necessary for the post-Golgi trafficking of ABCB11.9
Besides these structural features, the present structure of ABCB11 also enables us to precisely map 132 missense/nonsense single mutations (Fig. 1g), out of > 300 reported clinical mutations. In total, 97 missense and 18 nonsense mutations are related to severe diseases PFIC2, usually indicating the complete loss of ABCB11 function. Seventeen missense mutations lead to benign diseases BRIC2, meaning partially reserved function. Notably, these mutations are widespread throughout ABCB11, without obviously clustered hotspots. Most mutations associated with PFIC2 are found on the TMDs and are likely involved in substrate binding and translocation, whereas a few mutations, such as T919A, are related to BRIC2. Here we take mutations at four residues Thr919, Asn490, Ala570 and Tyr1311 as examples for further structural analyses (Fig. 1g). The residue Thr919 locates at the middle of TM10, the bending of which close to this point in ABCB1 was proposed to contribute to the conformational changes during translocation cycle.10 Mutations on the NBDs, even on the degenerated NBD1, predominately lead to PFIC2. For instance, two mutants N490D and A570T were reported to be the cause of PFIC2; however, the mutant proteins retain 27% and 60% activity compared to the wild type, respectively.11 Structural analysis indicated that once the conserved Asn490 substituted by Asp, the resulting Asp490 might form a salt bridge with the neighboring conserved Lys412 at the C-terminus of TM6 (Fig. 1g). Concerning Ala570, which is located in the long helix harboring the signature motif, substitution with a Thr might interfere with the helical conformation (Fig. 1g). Notably, as a consequence of the nonsense mutation corresponding to Tyr1311 codon, the aforementioned endocytic cargo motif will be lost, which leads to PFIC2 (Fig. 1g).
In summary, we present the cryo-EM structure of human ABCB11 with an ATP-free and inward-facing conformation. This structure provides us a platform to clearly map the clinic mutations on ABCB11 and to elucidate the structural insights of related pathogenesis. Moreover, it also gives some hints for therapeutic intervention and rational drug design.
Supplementary information
Acknowledgements
We thank Qianqian Sun at the Electron Microscopy Facility of ShanghaiTech University, Xiaojun Huang and Gang Ji at the Center for Biological Imaging, Core Facilities for Protein Science at the Institute of Biophysics, Chinese Academy of Sciences for technical supports during cryo-EM image acquisition, and Peiping Tang and Yongxiang Gao at the Center for Integrative Imaging, Hefei National Laboratory for Physical Sciences at the Microscale, University of Science and Technology of China for cryo-EM sample examination. This work is supported by the Ministry of Science and Technology of China (2019YFA0508500), the National Natural Science Foundation of China (Grant No. 31621002) and the Fundamental Research Funds for the Central Universities (YD2070002005).
Author contributions
C.-Z.Z. and Y.C. conceived the project and planned the experiments. L.W. and D.X. expressed and purified human ABCB11. L.W. and L.C. performed initial negative-stain and cryo-EM analyses and prepared all grids. L.W. and W.-T.H. performed functional assays and performed cryo-EM data collection. L.W., Y.-L.J. and L.S. performed ABCB11 structure determination and model refinement. W.-T.H., L.W., C.-Z.Z. and Y.C. wrote the manuscript.
Data availability
All relevant data are available from the authors and/or included in the manuscript. Atomic coordinates and EM density maps of the human ABCB11 have been deposited in the Protein Data Bank (PDB code: 6LR0) and the Electron Microscopy Data Bank (EMDB code: EMD-0956), respectively.
Competing interests
The authors declare no competing interests.
Footnotes
These authors contributed equally: Liang Wang, Wen-Tao Hou
Contributor Information
Linfeng Sun, Email: sunlf17@ustc.edu.cn.
Cong-Zhao Zhou, Email: zcz@ustc.edu.cn.
Yuxing Chen, Email: cyxing@ustc.edu.cn.
Supplementary information
Supplementary information accompanies this paper at 10.1038/s41422-020-0302-0.
References
- 1.Boyer JL. Compr. Physiol. 2013;3:1035–1078. doi: 10.1002/cphy.c120027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Olsen JA, Alam A, Kowal J, Stieger B, Locher KP. Nat. Struct. Mol. Biol. 2019;27:62–70. doi: 10.1038/s41594-019-0354-3. [DOI] [PubMed] [Google Scholar]
- 3.Srikant S, Gaudet R. Nat. Struct. Mol. Biol. 2019;26:792–801. doi: 10.1038/s41594-019-0280-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Johnson ZL, Chen J. Cell. 2017;168:1075–1085 e1079. doi: 10.1016/j.cell.2017.01.041. [DOI] [PubMed] [Google Scholar]
- 5.Knisely AS, Portmann BC. Pediatr. Transplant. 2006;10:644–645. doi: 10.1111/j.1399-3046.2006.00530.x. [DOI] [PubMed] [Google Scholar]
- 6.Soroka CJ, Pate MK, Boyer JL. J. Biol. Chem. 1999;274:26416–26424. doi: 10.1074/jbc.274.37.26416. [DOI] [PubMed] [Google Scholar]
- 7.Jin MS, Oldham ML, Zhang QJ, Chen J. Nature. 2012;490:566–569. doi: 10.1038/nature11448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mochizuki K, et al. Am. J. Physiol.-Gastrointest. Liver Physiol. 2007;292:G818–G828. doi: 10.1152/ajpgi.00415.2006. [DOI] [PubMed] [Google Scholar]
- 9.Hayashi H, et al. Hepatology. 2012;55:1889–1900. doi: 10.1002/hep.25591. [DOI] [PubMed] [Google Scholar]
- 10.Kim Y, Chen J. Science. 2018;359:915–919. doi: 10.1126/science.aar7389. [DOI] [PubMed] [Google Scholar]
- 11.Byrne JA, et al. Hepatology. 2009;49:553–567. doi: 10.1002/hep.22683. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All relevant data are available from the authors and/or included in the manuscript. Atomic coordinates and EM density maps of the human ABCB11 have been deposited in the Protein Data Bank (PDB code: 6LR0) and the Electron Microscopy Data Bank (EMDB code: EMD-0956), respectively.