Humans encounter multiple paramyxoviruses early in life. This study shows that infection with common paramyxoviruses can induce T cells cross-reactive with the highly pathogenic Nipah virus. This demonstrates that the combination of paramyxovirus infection history and HLA haplotype affects immunity to phylogenetically related zoonotic paramyxoviruses.
KEYWORDS: paramyxovirus, T cells, measles virus, Nipah virus, human parainfluenza virus
ABSTRACT
Humans are infected with paramyxoviruses of different genera early in life, which induce cytotoxic T cells that may recognize conserved epitopes. This raises the question of whether cross-reactive T cells induced by antecedent paramyxovirus infections provide partial protection against highly lethal zoonotic Nipah virus infections. By characterizing a measles virus-specific but paramyxovirus cross-reactive human T cell clone, we discovered a highly conserved HLA-B*1501-restricted T cell epitope in the fusion protein. Using peptides, tetramers, and single cell sorting, we isolated a parainfluenza virus-specific T cell clone from a healthy adult and showed that both clones cleared Nipah virus-infected cells. We identified multiple conserved hot spots in paramyxovirus proteomes that contain other potentially cross-reactive epitopes. Our data suggest that, depending on HLA haplotype and history of paramyxovirus exposures, humans may have cross-reactive T cells that provide protection against Nipah virus. The effect of preferential boosting of these cross-reactive epitopes needs to be further studied in light of paramyxovirus vaccination studies.
INTRODUCTION
Throughout life, humans are repeatedly infected by viruses from the families Paramyxoviridae and Pneumoviridae (1, 2). Well-known endemic human paramyxoviruses are members of different genera: the genus Morbillivirus (measles virus [MeV]), genus Respirovirus (human parainfluenza viruses 1 and 3 [HPIV1 and -3]), and genus Orthorubulavirus (HPIV2 and -4 and mumps virus [MuV]); endemic human pneumoviruses come from either the genus Metapneumovirus (human metapneumovirus [HMPV]) or the genus Orthopneumovirus (human respiratory syncytial virus [HRSV]). Effective live-attenuated vaccines against measles and mumps are available and incorporated into most national vaccination programs. Furthermore, multiple encounters with HPIV, HMPV, and HRSV occur within the first 10 years of life (3–5). These viruses are major causes of lower respiratory tract infections during childhood. Vaccine development is well under way, especially for HRSV, but no licensed vaccines against these respiratory viruses exist.
In addition to the paramyxo- and pneumoviruses largely restricted to humans, several members of the Paramyxoviridae that infect animals have zoonotic potential (6–9). Nipah virus (NiV) is of particular interest, as it is associated with high fatality rates, classified as a biosafety level 4 (BSL-4) pathogen, and prioritized by the World Health Organization (WHO) as a pathogen for which vaccines are urgently needed (7, 10, 11) (https://www.who.int/activities/prioritizing-diseases-for-research-and-development-in-emergency-contexts). Large outbreaks of NiV infection have occurred in Bangladesh and Malaysia with severe neurological disease in humans, case fatality rates up to 75%, and considerable human-to-human transmission (7, 8, 12). NiV circulates in fruit bats, a reservoir host that is widely distributed over the southern hemisphere. Combining this with a possibility of viral spread via the respiratory route of certain NiV strains, suggested by epidemiological studies and supported by in vitro observations and animal experiments, it is not without reason that NiV is one of the viruses marked by the WHO with potential to cause a future pandemic through natural or deliberate exposures (13–16). NiV is not the only paramyxovirus with zoonotic potential: the closely related Hendra virus (HeV) has caused lethal disease in humans in the past (17). In addition, virus discovery studies have identified an increasingly diverse range of henipaviruses and other paramyxoviruses in nonhuman reservoirs, suggesting potential for additional spillover events (18–20).
Vaccination against or infection with members of the Paramyxoviridae and Pneumoviridae induces virus-specific B and T cell responses. Virus-specific neutralizing (VN) antibodies are considered the main correlate of protection for most of the Paramyxoviridae (21). However, once a susceptible host is infected, viral clearance is predominantly mediated by cellular immune responses. This is probably due to the spread via cell-to-cell fusion and infected migrating cells (22), thus avoiding VN antibodies in the bloodstream. Whereas VN antibodies in these infections are exclusively targeted to membrane-exposed epitopes, T cells can target epitopes contained in any viral protein. The importance of CD8+ T cells in MeV clearance was corroborated in vivo; MeV-infected macaques depleted of B cells were able to normally clear virus infection (23), whereas CD8+ T cell-depleted macaques presented with higher viral loads and a significantly longer duration of viremia (24). It has also been reported that children with deficits in cellular immunity suffer from more severe and prolonged measles (25).
Human and animal members of the genus Morbillivirus are phylogenetically closely related, and it is well known that MeV vaccination or infection can induce (partial) cross-protection from infection with heterologous morbilliviruses (26–28). In classical studies, MeV vaccines were used to immunize dogs against the closely related canine distemper virus (CDV), which normally causes lethal disease in dogs. MeV vaccination resulted in partial protection from clinical signs after CDV challenge infection, in the absence of cross-reactive antibodies (29–31). CDV was shown to be able to cross the species barrier into nonhuman primates (6, 32, 33). In experimental infections of nonhuman primates, MeV-vaccinated macaques also proved partially protected from CDV challenge in the absence of CDV-specific VN antibodies. Most importantly, vaccination resulted in strongly reduced levels of virus shedding from the upper respiratory tract. A rapid proliferation of white blood cells in MeV-vaccinated macaques upon CDV challenge was detected, reminiscent of a secondary cellular immune response (34). Combined, these data strongly suggest that cellular immune responses triggered by MeV infection or vaccination are an important correlate of cross-protection against infection with heterologous morbilliviruses.
In the present study, we explored if cellular cross-reactivity exists across the different genera of the Paramyxoviridae and Pneumoviridae. We investigated whether broadly reactive T cells are present in human donors and assessed whether these have the capacity to cross-react with endemic paramyxoviruses (like MeV, HPIV1 to -4, and MuV), pneumoviruses (HMPV and HRSV), and potentially zoonotic viruses (like CDV, NiV, and HeV). We now show that exposure to human endemic paramyxoviruses, by either vaccination or infection, can lead to cross-genus-reactive T cell immunity. We specifically show that these broadly reactive T cells also recognize NiV and thus may confer protection against the highly pathogenic zoonotic henipaviruses.
RESULTS
Morbillivirus N, M, F and L proteins are targets for cross-reactive T cells.
We initially focused on T cell cross-reactivity within the genus Morbillivirus of the family Paramyxoviridae. CD4+ and CD8+ T cells recognize linear epitopes (usually 8 to 11 amino acids in length for CD8+ cells) in the context of an HLA class II or I molecule, respectively. If this epitope is in a conserved region of a viral protein, with sufficient homology on the amino acid level, the respective T cell could potentially react with other viruses and be regarded “cross-reactive.” We initially analyzed the level of homology between the morbilliviruses MeV and CDV and plotted homologous regions in a heat map (Fig. 1A). We found that the phosphoprotein (P) and hemagglutinin (H) were not well conserved but that the nucleoprotein (N), matrix protein (M), fusion protein (F), and polymerase (L) displayed homology percentages between 60 and 80. Because conservation percentages do not give information about conservation of T cell epitopes, we searched for stretches of conserved amino acid sequences in these proteins while keeping the length of typical HLA class I-restricted minimal T cell epitopes (9 to 10 amino acids) in mind (Fig. 1B). The chosen categories are arbitrary and do not allow for mismatches; however, the analysis shows that stretches of more than 10 fully conserved amino acids were abundantly present in the N, M, F, and L proteins. We concluded that these proteins are potential targets for morbillivirus-specific cross-reactive T cells.
FIG 1.
Conservation between MeV and CDV. (A) Heat map indicating homology between MeV and CDV for the respective open reading frames (ORFs). Green represents fully homologous residues, and red shows nonhomologous residues. Proteins and total percentage of conservation are indicated above the heat map. (B) Stretches of fully conserved homologous amino acids when comparing all ORFs from MeV and CDV, containing potential cross-reactive T cell epitopes. Stretches of 1 to 5, 6 to 10, 11 to 20, and 21 or more homologous amino acids are indicated in shades of gray. MeV, measles virus; CDV, canine distemper virus; N, nucleoprotein; P, phosphoprotein; M, matrix protein; F, fusion protein; H, hemagglutinin; L, large protein or polymerase.
Morbillivirus-specific cross-reactive TCC can be isolated from humans.
A panel of well-characterized, previously published MeV-specific human T cell clones (TCC) was tested for cross-reactivity with CDV (35, 36). We found several exclusively MeV-specific TCC (non-cross-reactive, here represented by two examples, named CD4MeV1 and CD8MeV1), but two MeV F-specific TCC from different donors cross-reacted with CDV-infected cells in an IFN-γ enzyme-linked immunosorbent spot (ELISPOT) assay (Fig. 2A and B). One of these was CD4+ (CD4Xreact1), and the other was CD8+ (CD8Xreact1). The cross-reactive cytotoxic CD8+ TCC not only produced IFN-γ upon recognition of MeV- and CDV-infected cells (37) but also proved capable of clearing MeV and CDV infection from autologous B-lymphoblastic cell lines (B-LCL) in an in vitro virus suppression assay (38). In the same assay, CD8MeV1 could clear only MeV infection but not CDV infection (Fig. 2C). The CD4+ TCC were also tested in the suppression assay, but neither TCC could clear either MeV or CDV infection (data not shown), confirming that these TCC were not cytotoxic. These data illustrate that morbillivirus-specific cross-reactive T cells exist in humans and that cytotoxic cross-reactive T cells can clear infection with heterologous morbilliviruses in vitro.
FIG 2.
Characterization of cross-reactive TCC. A panel of well-characterized MeV-specific T cell clones (TCC) was tested for cross-reactivity with CDV. (A) Two representative examples of CD4+ TCC, one solely reactive with MeV (CD4MeV1), the other one reactive with both MeV and CDV (CD4Xreact1) as tested by IFN-γ ELISPOT. (B) Two representative examples of CD8+ TCC, one solely reactive with MeV (CD8MeV1), the other one reactive with both MeV and CDV (CD8Xreact1). (C) In vitro virus suppression assay in which CD8MeV1 and CD8Xreact1 were cocultured with MeV- and CDV-infected autologous B-LCL. MeV, measles virus; CDV, canine distemper virus; B-LCL, B-lymphoblastic cell line; TCC, T cell clone.
Cross-reactive TCC recognize epitope in the F protein.
CD4Xreact1 and CD8Xreact1 were previously shown to respond to antigen (Ag)-presenting cells (APC) expressing the F protein (37). By using peptide-pulsed autologous B-LCL with overlapping peptides covering the F protein, and measuring IFN-γ production via ELISPOT, we identified the minimal amino acid sequence required for activation of both TCC. Initially, overlapping 15-mer peptides (11 overlap) were used (Fig. 3A and D), followed by fine-tuning with overlapping 10-mers (9 overlap) (Fig. 3B and E). Finally, we determined that both CD4Xreact1 and CD8Xreact1 recognized the minimal 9-mer F129–137 (FAQITAGIAL for MeV, Fig. 3C and F). By performing cocultures with different B-LCL with known HLA haplotypes, we determined CD4Xreact1 to be restricted by HLA-DQ*06:03 and CD8Xreact1 to be restricted by HLA-B*15:01 (data not shown). When binding predictions were performed for 9- or 10-mer epitopes with the MeV F protein sequence for the HLA class I reference set (additionally including HLA-B*27:05 and HLA-B*39:01 [https://www.iedb.org/]), four different HLA class I molecules were predicted to bind FAQITAGIAL at high affinity (50% inhibitory concentration [IC50] < 50): HLA-B*15:01, HLA-B*39:01, HLA-B*40:01, and HLA-A*02:06 (data not shown).
FIG 3.
Epitope mapping for CD4Xreact1 and CD8Xreact1. Overlapping peptide-pulsed autologous B-LCL were used to determine the minimal epitope recognized by CD4Xreact1 and CD8Xreact1 in IFN-γ ELISPOT. (A and D) Overlapping 15-mer peptides with 11 overlap were initially used. (B and E) Fine-tuning of recognized region using overlapping 10-mer peptides with 9 overlap. (C and F) Confirmation of recognition of F129–137 (FAQITAGIAL) by both TCC. MeV, measles; F, fusion protein; SFC, spot-forming cells.
After finding that TCC CD4Xreact1 and CD8Xreact1 recognize the minimal epitope F129–137, we determined the level of conservation of this epitope among all morbilliviruses. Interestingly, F129–137 was completely conserved between MeV, CDV, and rinderpest virus (RPV) but was also highly conserved among the other morbilliviruses (peste-des-petits-ruminants virus [PPRV], cetacean morbillivirus [CeMV], phocine distemper virus [PDV], and feline morbillivirus [FeMV]). The most obvious variation was at position F135, presenting as either isoleucine or valine, two similar amino acids with hydrophobic side chains (see Fig. S1 in the supplemental material).
F129–137 amino acid alignment for morbilliviruses. Mismatches to the MeV amino acid sequence are shown in red. CDV, canine distemper virus; MeV, measles virus; PPRV, peste-des-petits-ruminants virus; CeMV, cetacean morbillivirus; PDV, phocine distemper virus; RPV, rinderpest virus; FmoPV, feline morbillivirus. Download FIG S1, DOCX file, 0.03 MB (32.2KB, docx) .
Copyright © 2020 de Vries et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
FAQITAGIAL is part of the conserved fusion peptide.
After determining conservation of F129–137 among morbilliviruses, we additionally aligned the F proteins of all Paramyxoviridae and Pneumoviridae. A phylogenetic tree was constructed on the basis of the F nucleotide sequence, revealing the different subfamilies and genera (Fig. S2A). Subsequently, having found CD4Xreact1 and CD8Xreact1 to be specific for F129–137, we took a closer look at this specific region in the F protein. The F protein is normally formed as an inactive F0 variant. For the F protein to become active, it must be cleaved by furin-like enzymes into F1 and F2 at the cleavage site. The N-terminal region of the F1 subunit adjacent to the cleavage site is called the fusion peptide (region F113–137), a hydrophobic region that plays a critical role in the fusion process by insertion into the membrane of target cells (39–41). When aligning the fusion peptide of the relevant endemic and zoonotic paramyxo- and pneumoviruses (MeV, CDV, NiV, HeV, HPIV1 to -4, MuV, HMPV, and HRSV), we again found that the fusion peptide, and especially F129–137, was highly conserved among the Paramyxoviridae (Fig. S2B), suggesting a functional constraint in virus evolution of this region in the protein.
F129–137 is in the conserved fusion peptide. (A) Phylogenetic tree of the Paramyxo- and Pneumoviridae based on an F nucleotide alignment. An unrooted maximum likelihood phylogenetic tree was estimated under the general time-reversible model. The tree was based on 29 sequences (Table S1), of which a selection relevant to this study is shown by their abbreviations. (B) Fusion peptide amino acid alignment for the relevant endemic and zoonotic paramyxo- and pneumoviruses. The F129–137 T cell epitope is indicated by the gray background. Mismatches to the MeV amino acid sequence are shown in red. CDV, canine distemper virus; MeV, measles virus; NiV, Nipah virus; HeV, Hendra virus; HPIV, human parainfluenza virus; MuV, mumps virus; HMPV, human metapneumovirus; HRSV, human respiratory virus. Download FIG S2, DOCX file, 0.2 MB (168.7KB, docx) .
Copyright © 2020 de Vries et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Novel F129–137-specific CD8+ TCC could be isolated from human donors.
Since we know that CD8+ T cells are crucial in the clearance of especially paramyxoviruses in vivo (23, 24), and we had thus far relied on a single CD8+ TCC that recognized a conserved region in the fusion peptide, we isolated novel F129–137-specific CD8+ TCC from peripheral blood mononuclear cells (PBMC) obtained from HLA-B*15:01-positive donors. To this end, we used phycoerythrin (PE)-labeled B*15:01-FAQITAGIAL tetramers, which could stain FAQITAGIAL-specific TCC (Fig. 4A). Three donors were selected, and PBMC were stained with the FAQITAGIAL tetramer (Fig. 4B). CD3+ CD8+ AQITAGIAL+ cells were single cell sorted, and FAQITAGIAL-specific TCC could be clonally expanded from a single donor. A single TCC was selected for subsequent experiments (CD8Xreact2) (Fig. 4C). Both CD8Xreact1 and CD8Xreact2 proved cross-morbillivirus-reactive and were shown to recognize both morbillivirus sequence FAQITAGIAL present in MeV, CDV, and RPV and the variant FAQITAGVAL (valine instead of isoleucine at 7th residue) present in PPRV, CeMV, and PDV, in a concentration-dependent manner (Fig. S3). CD8Xreact1 reacted strongly with FAQITAGIAL but also with FAQITAGVAL. CD8Xreact2 had a stronger affinity to FAQITAGVAL but still recognized FAQITAGIAL.
FIG 4.
Isolation of a novel F129–137-specific TCC. (A) AQITAGIALPE tetramer fluorescence-activated cell sorting (FACS) staining of TCC CD8Xreact1 confirming that the TCC is CD8+ and recognizes FAQITAGIAL in the context of HLA-B*15:01. (B) CD3APCCy7 and AQITAGIALPE tetramer staining of PBMC obtained from three human HLA-B*15:01-positive donors. CD3+ CD8+ T cells that were positively stained by the HLA-B*15:01-AQITAGIAL tetramer were single cell sorted by FACS and clonally expanded from donor 3. (C) AQITAGIALPE tetramer FACS staining of TCC CD8Xreact2 confirming that the TCC is CD8+ and recognizes FAQITAGIAL in the context of HLA-B*15:01.
(A and B) Peptide dilution series with B-LCL pulsed with different concentrations of peptides. CD8Xreact1 reacted strongly with FAQITAGIAL but also with FAQITAGVAL. CD8Xreact2 had a stronger affinity with FAQITAGVAL but still recognized FAQITAGIAL. Download FIG S3, DOCX file, 0.1 MB (74.2KB, docx) .
Copyright © 2020 de Vries et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
CD8Xreact1 and CD8Xreact2 cross-recognize Nipah virus.
Subsequently, we determined which Paramyxoviridae and Pneumoviridae could be recognized by the identified cross-reactive TCC (CD4Xreact1, CD8Xreact1, and CD8Xreact2). B-LCL were pulsed with F129–137 peptides for all endemic and zoonotic paramyxo- and pneumoviruses and cocultured with the cross-reactive TCC. CD4Xreact1 recognized FAQITAGIAL and FAQITAGVAL, indicating broad reactivity with MeV, CDV, HPIV1, NiV, and HeV (Fig. 5A). CD8Xreact1 recognized the same peptides, but also cross-recognized FAQVTAAIGL, representing HPIV4 (Fig. 5A). CD8Xreact2 had the broadest reactivity, as it additionally cross-recognized FAQVTAAVSL, FAQITAAVAI and FAQITAAVAL. In conclusion, CD8Xreact2 recognized F129–137 from 8 different viruses, namely, MeV, CDV, HPIV1, 2 and 3, MuV, NiV and HeV (Fig. 5A). Importantly, all tested TCC cross-recognized all morbilli- and henipaviruses and proved paramyxovirus cross-genus-reactive. None of the clones recognized the peptides obtained from members of the Pneumoviridae.
FIG 5.
CD8Xreact1 and CD8Xreact2 cross-recognize NiV-infected cells. (A) Cross-reactive TCC were evaluated for reactivity with different F129–137 regions from relevant viruses by IFN-γ ELISPOT. B-LCL pulsed with different with F129–137 peptides at a concentration of 1 μM were cocultured with TCC. (B and C) IFN-γ ELISPOT with B-LCL pulsed at different concentrations of peptides for CD8Xreact1 (B) and CD8Xreact2 (C). (D and E) FACS-based assay showing interaction strength between TCC and tetramers. Interaction strength was calculated as mean fluorescence intensity (MFI) at 2 M urea divided by MFI at 0 M urea (Fig. S5). CDV, canine distemper virus; MeV, measles virus; ND, not done; NiV, Nipah virus; HPIV, human parainfluenza virus; MuV, mumps virus; TCC, T cell clone.
CD8Xreact1 and CD8Xreact2 are not identical TCC.
Both CD8Xreact1 and CD8Xreact2 were HLA-B*15:01 restricted and reactive to F129–137; however, they did not recognize the same peptides. This suggested that the TCC were not identical. To confirm this, and to confirm clonality of the TCC used in different assays, we assessed the T cell receptor (TCR) variable (V)β chain expression by flow cytometry. A panel of antibodies confirmed that the TCC were indeed different, as CD8Xreact1 expressed TCRVβ14 and CD8Xreact2 expressed TCRVβ8 (Fig. S4).
T cell receptor (TCR) variable (V)β chain expression of CD8Xreact1 and CD8Xreact2 was determined by flow cytometry. (A) CD8Xreact1 proved clonal and expressed TCRVβ14. (B) CD8Xreact2 was clonal and expressed TCRVβ8. Download FIG S4, DOCX file, 0.3 MB (354.2KB, docx) .
Copyright © 2020 de Vries et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
TCC have different avidities for F129–137. We developed a FACS-based assay to determine interaction strength between TCC and tetramers. (A and B) CD8Xreact1 and CD8Xreact2 were stained with F129–137 tetramers and treated with increasing concentrations of urea, disrupting low-avidity binding between tetramers and TCC. We confirmed reactivity of CD8Xreact1 with FAQITAGIAL and FAQITAGVAL, and of CD8Xreact1 with FAQITAGIAL, FAQITAGVAL, FAQITAAVAI, FAQITAAVAL, and FAQVTAAVSL. CDV, canine distemper virus; MeV, measles virus; NiV, Nipah virus; HeV, Hendra virus; HPIV, human parainfluenza virus; MuV, mumps virus; MFI, mean fluorescence intensity. Download FIG S5, DOCX file, 0.1 MB (74KB, docx) .
Copyright © 2020 de Vries et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Reactivity to cells pulsed with different peptides (Fig. 5A), different Vβ chain expression (Fig. S4), and variable affinity for FAQITAGIAL and FAQITAGVAL (Fig. S3) proved that CD8Xreact1 and CDXreact2 were different TCC, probably originally induced by different paramyxoviruses. We again performed peptide dilution series with pulsed autologous antigen-presenting cells and found that CD8Xreact1 recognized the epitope present in morbilliviruses and henipaviruses (FAQITAGIAL and FAQITAGVAL, respectively) at high affinity and the epitope present in HPIV4 (FAQVTAAIGL) to a lesser extent (Fig. 5B). CD8Xreact2 showed a different spectrum of reactivity: although it was reactive with the epitope present in morbilliviruses, henipaviruses, and MuV, affinity for HPIV2 and -3 (FAQITAAVAL and FAQITAAVAI, respectively) was much stronger (Fig. 5C).
As the peptide dilution experiments indicated strong affinity of particular peptides, we developed a novel flow cytometry-based assay to truly determine interaction strength (i.e., avidity) between peptide and TCC, by staining TCC with a saturating amount of HLA-B*15:01 tetramers conjugated with peptides of relevant paramyxoviruses. CD8Xreact1 and CD8Xreact2 were stained with the different F129–137 tetramers and briefly treated with a low concentration of urea, disrupting low-avidity binding between tetramers and TCC (Fig. S5A and B). Percentage of binding and interaction strength at 2 M (based on mean fluorescence intensity [MFI] at 0 M) proved that CD8Xreact1 had the highest affinity for FAQITAGIAL (and was therefore probably induced by MeV), whereas CD8Xreact2 had the highest affinity for FAQITAAVAL and FAQITAAVAI (and was therefore probably induced by infection with HPIV2 or HPIV3) (Fig. 5D and E).
F129–137-specific TCC clear NiV-infected cells.
Thus far, we had only characterized the cross-reactive TCC for their capacity to bind peptides or tetramers based on HLA-B*15:01 complexed with F129–137 and their potential to secrete IFN-γ upon antigen (Ag)-specific stimulation. Both CD8Xreact1 and CD8Xreact2 were identified as TCC induced by endemic paramyxoviruses that could potentially play a role in cross-immunity against zoonotic paramyxoviruses but do not likely affect infections with distantly related pneumoviruses. To confirm the capacity of CD8Xreact1 and CD8Xreact2 to actually suppress NiV replication, we performed the aforementioned virus suppression (shown in Fig. 2C) (38). Autologous or HLA-matched B-LCL were infected with green fluorescent protein (GFP)-expressing NiV and cocultured with a concentration series of TCC. The absolute number of TCC required to suppress NiV replication was determined (Fig. 6A to C). Simultaneously, the TCC were tested for their capacity to kill cells infected with MeV, MuV, CDV, and HPIV3 (Fig. 6A to C) and HMPV and HRSV (data not shown). HLA-mismatched antigen-presenting cells were included as a negative control. CD8Xreact1 efficiently suppressed MeV and CDV replication (Fig. 6A), corresponding with reactivity in IFN-γ ELISPOT (Fig. 6A). CD8Xreact2 suppressed MeV and CDV replication but could additionally suppress HPIV3. Surprisingly, MuV replication was not suppressed, although this TCC did react with the F129–137 epitope present in MuV (Fig. 6A). Importantly, both MeV-specific TCC CD8Xreact1 and HPIV-specific TCC CD8Xreact2 killed NiV-infected cells (Fig. 6B), confirming functionality against highly pathogenic henipaviruses. Corresponding to peptide reactivity, none of the TCC killed cells infected with the Pneumoviridae member HRSV or HMPV.
FIG 6.
CD8Xreact1 and CD8Xreact2 clear NiV-infected cells. (A) CD8Xreact1 suppressed MeV, CDV, and NiV replication. (B) CD8Xreact2 suppressed MeV, CDV, HPIV3, and NiV replication. (C) As a control, we included CD8MeV1, which exclusively suppressed MeV replication. CDV, canine distemper virus; MeV, measles virus; NiV, Nipah virus; HPIV, human parainfluenza virus; MuV, mumps virus; TCC, T cell clone.
Conserved regions are present throughout the paramyxovirus proteome.
After identifying F129–137 as a cross-reactive epitope to which HLA-B*15:01-restricted T cell responses can be induced by multiple paramyxoviruses, we performed a systematic search for conserved regions throughout the proteome of the Paramyxoviridae and Pneumoviridae, containing potential cross-reactive T cell epitopes. Since we were mostly interested in T cells induced by endemic paramyxoviruses that cross-react with henipaviruses, we chose NiV as a reference virus and aligned the six common proteins to MeV, HPIV3, and HPIV2. Conserved regions, defined as stretches of 10 homologous amino acid residues allowing 4 mismatches, were found in all comparisons (NiV versus MeV, 119 regions; NiV versus HPIV3, 93 regions; NiV versus HPIV2, 59 regions) and were most abundant in the L protein (Fig. 7A and Data Set S1). Alignments for all the endemic and zoonotic paramyxo- and pneumoviruses were prepared for these 271 regions of interest in total, and the average homology percentage was determined. When only regions with an average of more than 50% homology among all viruses were selected, we identified 27 regions in total that potentially contain T cell epitopes that are conserved throughout the paramyxovirus proteome (Data Set S1E).
FIG 7.
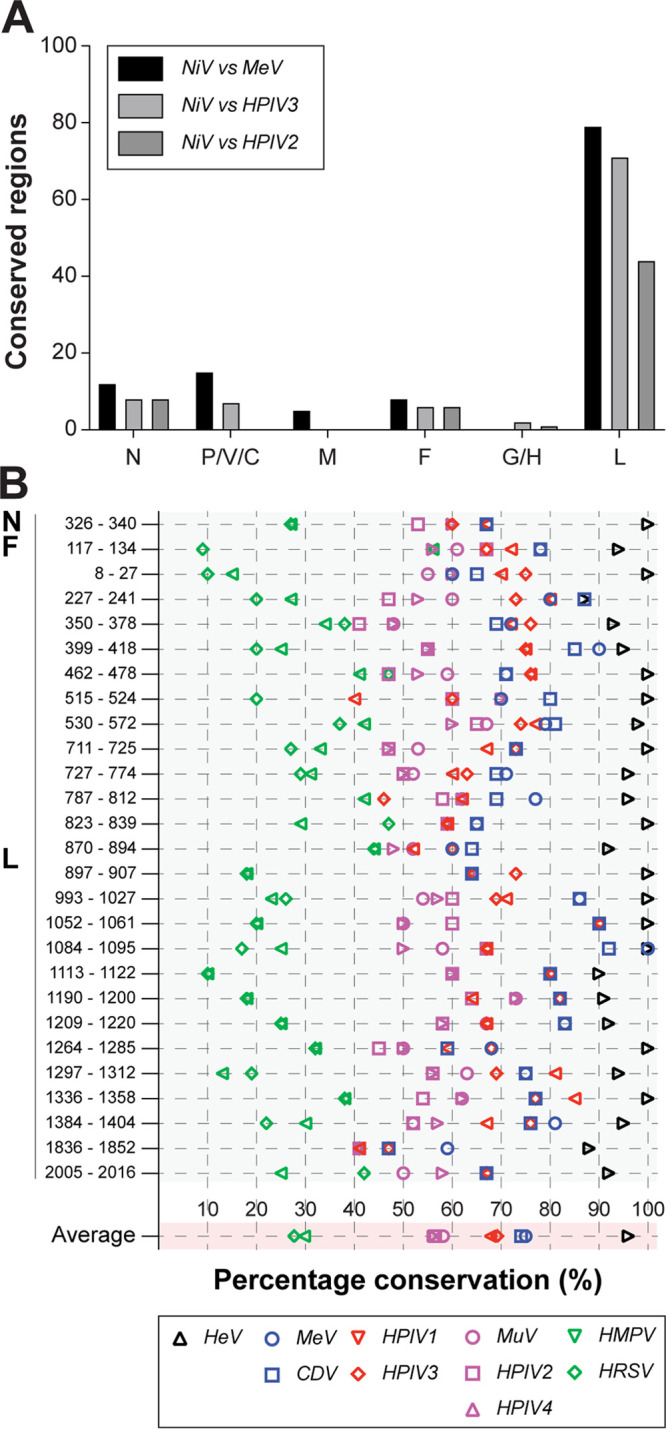
Systematic search for conserved regions in paramyxo- and pneumoviruses. (A) Number of conserved regions in the different ORFs when comparing NiV with MeV, HPIV3, and HPIV2, respectively. Conserved regions were most abundant in the L protein. (B) Selection of conserved regions with an average of more than 50% homology among all paramyxo- and pneumoviruses; F117–134 is the full fusion peptide (NiV numbering). Graph shows how conserved the regions of interest are in a selection of viruses, using NiV as a reference sequence. CDV, canine distemper virus; MeV, measles virus; NiV, Nipah virus; HeV, Hendra virus; HPIV, human parainfluenza virus; MuV, mumps virus; HMPV, human metapneumovirus; HRSV, human respiratory virus.
(A) Selection criteria for conserved regions in the paramyxo- and pneumovirus proteome. (B) Conserved regions when comparing NiV with MeV. (C) Conserved regions when comparing NiV with HPIV3. (D) Conserved regions when comparing NiV with HPIV2. (E) Conserved regions throughout the paramyxo- and pneumovirus proteome. Download Data Set S1, DOCX file, 3.5 MB (3.6MB, docx) .
Copyright © 2020 de Vries et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Using these definitions to screen for conserved regions of interest, we identified one region in N, one region in F, and 25 regions in L (Fig. 7B). Confirming the validity of this unbiased approach, F117–134 (NiV numbering, contains the full fusion peptide and F129–137 in MeV numbering) was indeed identified as a region of interest. Using NiV as a reference, we determined how conserved all regions of interest with potential epitopes were in the endemic and zoonotic paramyxo- and pneumoviruses. All epitopes were almost completely conserved in the closely related HeV, and >50% conservation was found for HPIV2, HPIV4, and MuV; >60% conservation was found for HPIV1 and -3; and >70% conservation was found for MeV and CDV (Fig. 7B). Altogether, this suggests that additional cross-reactive CD8+ T cells targeting evolutionarily conserved peptide sequences can be induced by heterologous infections.
DISCUSSION
Humans are repeatedly exposed to members of the Paramyxoviridae and Pneumoviridae, either by natural infection or by vaccination. Most of these viruses are encountered in the first years of life, including the parainfluenza viruses, HRSV, and HMPV (3–5, 42). Furthermore, MeV and MuV are incorporated into most national vaccination programs. Collectively, it can be concluded that development of immunity to paramyxo- and pneumoviruses starts shortly after birth and is formed during the first years of life. Here, we show that immunity induced by endemic paramyxoviruses cross-reacts with NiV and may confer protection from highly pathogenic zoonotic paramyxoviruses, like NiV and HeV.
MeV is considered a target for global eradication by the WHO. Although measles eradication would save many lives, it will result in reduced compliance with or even cessation of MeV vaccination. It is already known that measles vaccination or infection induces cross-protection against other morbilliviruses. We have shown here that MeV infection can also lead to the induction of cross-genus-reactive T cells, broadly reactive with most members of the Paramyxoviridae. In a scenario of reduced vaccine compliance or cessation of vaccination in the absence of measles circulation, many children would grow up without morbillivirus-specific immunity, creating a niche for zoonotic morbilliviruses to cross the species barrier. CDV is already capable of infecting a wide range of carnivores (43) and even noncarnivorous species (44, 45). The CDV outbreaks in nonhuman primate colonies indicate that CDV and potentially other animal morbilliviruses are more than a theoretical risk for primates lacking morbillivirus immunity (6, 9). Combined with the continuous circulation of the lethal NiV, discovery of novel zoonotic paramyxoviruses with the potential to cause lethal disease in humans indicates that a future with loss of morbillivirus-specific immunity calls for better understanding of immunity to paramyxoviruses.
Although NiV infection remains rare in humans, this virus is studied intensely because of its high fatality rate, which reportedly is around 40% for the Malaysia strain and over 90% for the Bangladesh strain (46, 47). Because the virus is so lethal, little is known about adaptive immune responses to viral infection. A small recent outbreak in India, in which NiV killed 16 out of 18 infected individuals, provided the opportunity to study adaptive T and B cell responses in two survivors (48). A marked increase in the absolute number of CD8+ T cells was detected, expressing Ki67, granzyme B, and PD-1, a profile of acute effector cells. Although NiV-specific antibodies were also detected, viral clearance coincided with the appearance of CD8+ effector cells. This is reminiscent of infection with MeV, where it is already known that virus-specific CD8+ T cells are crucial for viral clearance (21, 23–25, 49). Interestingly, studies with NiV in a nonlethal swine model have also shown that cytotoxic effector cells are crucial in clearance of infection virus (50). Altogether, it appears that although NiV infection is frequently lethal, virus-specific CD8+ T cells contribute to surviving NiV infection. Cross-genus-reactive T cells induced by endemic paramyxoviruses could therefore be an important factor in the outcome of an NiV infection.
Here, we describe that the F, N, M, and L proteins are conserved between the members of the Paramyxoviridae and Pneumoviridae. Interestingly, F, H, and N have already been described as major targets for CD8+ T cell responses in acute measles patients (51, 52). The novel cross-reactive epitope in F that we present here is located in the fusion peptide, a highly conserved region among paramyxoviruses due to functional constraints (53). The fusion peptide is directly involved in fusion between the viral envelope and target membrane, anchoring into the target cell membrane (54, 55). Interestingly, this region was previously reported to be immunogenic, as a CD4+ T cell epitope was identified in dogs immunized with CDV (56). To our knowledge, fusion peptide-specific CD8+ T cells have not been previously described.
The N protein has a crucial role in packaging viral RNA (57), forming a structure known as the ribonucleoprotein (RNP). The RNP is essential in the viral life cycle, as it functions in viral assembly, budding, protecting the genome from innate immune responses, and preventing RNA degradation by host nucleases. The paramyxovirus N protein is divided into two regions, the conserved N-terminal core (NCORE) and hypervariable C-terminal tail (NTAIL) (58). The NCORE is important for assembly with the viral RNA, and several studies have shown that the central conserved region of N (CCR, amino acid N258–357) is mainly responsible (59–61). It was previously shown that N321–350 evokes CD4+ responses in measles convalescent-phase donors (62). However, HLA-A*0201-restricted CD8+ T cells to the CCR have also been demonstrated (63). Combining our regions defined as cross-reactive with a previously identified TCC recognizing N331–339 (LLWSAMGV), we now hypothesize that this TCC is cross-reactive and may recognize many other paramyxo- and pneumoviruses. Similar to F129–137, N331–339 is present in the conserved F-X4-Y-X3-ϕ-S-ϕ-A-M-G motif that is essential in assembly of N with viral RNA and therefore under functional constraints (64). Viral escape to T cell immunity by mutating these regions is highly unlikely, as this results in nonviable viruses.
Repeated exposure to antigenically related members of the Paramyxoviridae and Pneumoviridae in the first years of life potentially provides a selective advantage for cross-reactive T cells. Preferential utilization and boosting of immunological memory based on a previous infection when exposed to a related virus is known as “original antigenic sin.” This has previously been described for influenza virus and dengue virus, mostly in the context of serological imprinting (65). Antibody production during an initial exposure to an infection or vaccination dominated the repertoire in subsequent exposures (66, 67). This can be a double-edged sword: in the case of influenza, if vaccines are developed that induce memory B cells that make broadly neutralizing antibodies, it is possible that these will persist even as the vaccine recipients age and lose their ability to mount new responses. However, for CD8+ cytotoxic T cells, it was shown that during a secondary infection with a different strain of dengue virus, virus-specific cytotoxic T cells do not kill infected cells but rather release cytokines that exacerbate damage of endothelial cells (68, 69). Something similar may apply to the paramyxoviruses. As mentioned before, humans are repeatedly exposed and develop immunity to these viruses in the first years of life. We speculate that it may be crucial in which order certain paramyxo- and/or pneumoviruses are encountered for subsequent cross-protection against other endemic or zoonotic viruses (Fig. 8). Early infection with a certain paramyxovirus (for example, HPIV2 or HPIV3) could commit the immune system to mounting cross-genus-reactive T cells, whereas other infections could lead to a relatively narrow reactivity. Although this is speculative, the possibility of this scenario emphasizes the importance of studying T cell responses to paramyxoviruses.
FIG 8.
“Original antigenic sin” model. (A) In this model, we suggest that it is crucial in which order paramyxo- and pneumoviruses are encountered to develop cross-immunity to the highly pathogenic henipaviruses (or other paramyxoviruses). (B) Initially committing T cell immunity to epitopes present in pneumoviruses or MeV could lead to a relative narrow immunity and therefore no protection and death upon encountering a highly pathogenic virus. (C) Encountering HPIV2 or -3 before MeV vaccination or infection could lead to committing to broadly reactive T cell epitopes and therefore (partial) immunity to highly pathogenic viruses.
Our study is limited to several T cells targeting a single epitope, restricted to a specific HLA haplotype. The origin of the cross-reactive CD8+ T cells in this study is known; CD8Xreact1 was isolated from a convalescent-phase blood sample obtained from a primary unvaccinated measles patient. CD8Xreact2 was isolated from a blood sample obtained from an adult health care worker repeatedly exposed to infants with respiratory tract infections. Whether the induction of cross-reactive T cells by MeV or HPIV infections here is one example of a broader effect requires further study and analysis of T cell specificities in paramyxovirus convalescent-phase donors. Although we show that the TCC identified and characterized here can kill MeV-, CDV-, HPIV3-, and NiV-infected cells in vitro, it remains to be determined whether these clonotypes would be protective when faced with a paramyxovirus infection in vivo.
Highly pathogenic emerging infectious diseases pose a significant threat to human welfare all over the world. The family Paramyxoviridae includes several viruses with the potential to emerge and cause future pandemics, like the henipaviruses. Here, we show that humans could already be at least partially protected against zoonotic henipaviruses by cross-genus-reactive T cell clonotypes induced by previous infections. We argue that it is critical to study T cell responses to both endemic and zoonotic paramyxoviruses and pneumoviruses and that these studies are crucial in the development of novel vaccines. Identification of specific conserved targets that are immunogenic and at the same time under functional restraints, therefore not allowing escape mutations, is the first immunological basis for future universal paramyxovirus vaccines.
MATERIALS AND METHODS
Ethics statement.
CD4MeV1, CD8MeV1, CD4Xreact1, and CD8Xreact1 were established from PBMC from children 4 weeks after acute measles and were previously published (35–37, 49). The TCC previously described have been renamed in this study; information regarding the designation can be obtained from the corresponding author. Human PBMC used for the generation of novel TCC (CD8Xreact2) were obtained from Dutch blood donors in accordance with the Declaration of Helsinki and under the approval of the Dutch ethics committee, and informed consent was obtained (METC, permit MEC-2015-095). Blood samples were collected in heparin Vacutainer tubes. PBMC were isolated from peripheral blood by Ficoll gradient. Three adult male individuals were selected for isolation of novel TCC.
Sequence homology between paramyxoviruses.
Paramyxovirus and pneumovirus sequences were selected (see Table S1 in the supplemental material), and protein alignments for N, P, M, F, G/H, and L were prepared using ClustalW multiple alignment in the BioEdit software. Homology percentages of the different proteins were calculated for the endemic and zoonotic paramyxo- and pneumoviruses. Furthermore, stretches of homologous amino acids between MeV and CDV for all proteins were identified, counted, and divided into stretches of <5, 6 to 10, 11 to 20, or >20 homologous amino acid residues.
Paramyxo- and pneumovirus F sequences used to construct phylogenetic trees. Download Table S1, DOCX file, 0.01 MB (9.4KB, docx) .
Copyright © 2020 de Vries et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Paramyxo- and pneumovirus phylogenetic tree.
An F gene nucleotide alignment was prepared using ClustalW multiple alignment in the BioEdit software, and an unrooted maximum likelihood phylogenetic tree was estimated under the general time-reversible model using PhyML software version 3.0 (GTR + I + G model) (70).
TCC.
T cell clones (TCC) were described previously (CD4MeV1, CD8MeV1, CD4Xreact1, and CD8Xreact1) (35–37, 49) or generated for the scope of this study (CD8Xreact2). In order to generate novel TCC, PBMC from three HLA-B*15:01 measles convalescent-phase donors were stained with the HLA-B*15:01 tetramer AQITAGIALPE (kind gift from the NIH Tetramer Core Facility). The donor with the most AQITAGIAL+ cells was selected, and multiple novel TCC were generated by sorting CD3+ CD8+ AQITAGIAL+ single cells, after staining with anti-CD3APC-Cy7 (BD Biosciences), anti-CD8PE-Cy7 (BD Biosciences), and AQITAGIALPE, directly into a T cell stimulation expansion mix containing anti-CD3 (OKT3) and γ-irradiated feeder cells. After expansion, the phenotype and specificity of the newly generated F129–137-specific TCC were confirmed by flow cytometry and IFN-γ ELISPOT.
IFN-γ ELISPOT assays.
TCC were screened for (cross) reactivity with paramyxo- and pneumoviruses by IFN-γ ELISPOT assay as previously described (38, 71). Briefly, 5 × 103 TCC (responder cells) and 2 × 104 autologous, HLA-matched, or HLA-mismatched peptide-pulsed or virus-infected B-LCL (antigen-presenting cells) were cocultured for 1.5 h at 37°C in a 96-well V-bottom plate. Cells were transferred to nylon-membrane-bottom ELISPOT plates (Millipore) coated with a monoclonal antibody (MAb) against IFN-γ and incubated for an additional 5 h at 37°C. Spots were stained with a secondary MAb against IFN-γ and counted using an automated ELISPOT reader. For affinity assays, B-LCL were pulsed with a 3-fold dilution series of peptides.
Epitope mapping cross-reactive TCC.
The minimal epitope recognized by CD4Xreact1 and CD8Xreact1 was mapped by performing IFN-γ ELISPOT assays as described above. HLA-matched B-LCL were initially pulsed with overlapping 15-mer peptides (11 overlap) covering the entire F protein. On the basis of this first assay, a region of interest was defined and the epitope was fine-tuned using overlapping 10-mers (9 overlap). Finally, several 9-mers were tested to determine the minimal epitope.
Urea-based avidity assays.
To determine the strength of the binding interaction between representative F129–137 tetramers from different paramyxo- and pneumoviruses and HLA-B*1501, we performed urea-based avidity assays. TCC were stained with the different F129–137 tetramers (kindly provided by the NIH Tetramer Core Facility) for 15 min at room temperature, washed, and subsequently incubated with increasing concentrations of cold urea (2, 4, 6, and 8 M). After a 15-min incubation period on ice, the amount of tetramer still bound to HLA-B*1501-expressing cells was determined by flow cytometry. The mean fluorescent intensity (MFI) in the PE channel was determined, and ratios compared to 0 M urea were calculated as a representative for the strength of the interaction.
Flow cytometry-based Vβ chain expression assays.
T cell receptor (TCR) variable (V) chain expression of TCC was determined using flow cytometry after staining with a viability dye (Violet Live/Dead stain; Invitrogen), anti-CD3APC-Cy7, anti-CD4PerCP-Cy5.5 (Becton, Dickinson), anti-CD8PE-Cy7, and a panel of fluorescein isothiocyanate (FITC)-, PE-, and FITC-PE-conjugated MAbs covering approximately 70% of the known TCRV repertoire (IOTest Beta Mark MAb kit; Beckman Coulter).
In vitro virus suppression assay.
TCC were evaluated for their ability to control dissemination of MeV, CDV, MuV, HPIV3, HRSV, HMPV, and NiV infections in human B-LCL as described previously (38). Briefly, a 2-fold dilution of TCC was prepared starting at 2 × 104 TCC per well. To each well, a mixture of 2 × 104 noninfected and 50 infected autologous or HLA-matched B-LCL was added (based on the quantification of enhanced GFP (EGFP)-expressing cells identified by flow cytometry). B-LCL were infected with either rMeVKSEGFP(3), rCDVSHEGFP(6) (72), rMuV-EGFP(3) (73), rHPIV3-EGFP (ViraTree), rHRSVA2EGFP(5) (ViraTree), rHMPV-EGFP (74), or rNiV-EGFP (75). After 48 h of coculture, cell pellets were examined for EGFP under an inverted light fluorescence microscope to assess the presence of infected cells. Since TCC were added as a 2-fold dilution series, the minimal number of cells required to control viral spread was determined.
Systematic screen for potential paramyxovirus-specific cross-reactive T cell epitopes.
The NiV (NC002728) sequence was selected as a reference virus from the genus Henipavirus, and the six common proteins were aligned to representative paramyxoviruses from the genus Morbillivirus (MeV), genus Respirovirus (HPIV3), and genus Rubulavirus (HPIV2). Conserved regions were defined as stretches of 10 homologous amino acid residues allowing 4 mismatches and subsequently identified in the alignments. Two hundred seventy-one regions of interest were defined, and alignments for all the endemic and zoonotic paramyxo- and pneumoviruses were prepared for these regions. Conservation percentages were calculated when comparing all the individual paramyxo- and pneumoviruses (Data Set S1B, C, D, and E), and a single average conservation percentage was calculated from these. All regions with an average percentage over 50% were selected, defining 27 regions that potentially contain T cell epitopes that are conserved throughout the paramyxo- and pneumovirus proteome. Using NiV as reference, conservation percentages for the 27 regions of interest were calculated against all other endemic and zoonotic paramyxo- and pneumoviruses.
ACKNOWLEDGMENTS
We thank the human donors for providing blood samples, Miranda de Graaf for generating the phylogenetic trees, and Paul Duprex and Martin Ludlow for critical feedback. We thank the NIH tetramer core facility for supplying the fluorochrome-labeled tetramers. All work with live NiV was performed in the BSL-4 facility of Philipps University, Marburg, Germany.
This research received no specific grant from any funding agency in the public, commercial, or not-for-profit sectors.
We declare no competing interests.
R.D.D.V., G.P.V.N., R.S.V.B., A.M., and R.L.D.S. conceived the experiments; R.D.D.V., A.D.J., R.J.V., L.S., G.P.V.N., and R.L.D.S. performed the experiments; R.D.D.V. and R.L.D.S. wrote the manuscript; A.D.M.E.O., M.P.G.K., and R.L.D.S. secured the funding; and G.P.V.N., R.S.V.B., A.D.M.E.O., A.M., and M.P.G.K. provided expertise and feedback.
Footnotes
Citation de Vries RD, de Jong A, Verburgh RJ, Sauerhering L, van Nierop GP, van Binnendijk RS, Osterhaus ADME, Maisner A, Koopmans MPG, de Swart RL. 2020. Human paramyxovirus infections induce T cells that cross-react with zoonotic henipaviruses. mBio 11:e00972-20. https://doi.org/10.1128/mBio.00972-20.
REFERENCES
- 1.Amarasinghe GK, Ayllón MA, Bào Y, Basler CF, Bavari S, Blasdell KR, Briese T, Brown PA, Bukreyev A, Balkema-Buschmann A, Buchholz UJ, Chabi-Jesus C, Chandran K, Chiapponi C, Crozier I, de Swart RL, Dietzgen RG, Dolnik O, Drexler JF, Dürrwald R, Dundon WG, Duprex WP, Dye JM, Easton AJ, Fooks AR, Formenty PBH, Fouchier RAM, Freitas-Astúa J, Griffiths A, Hewson R, Horie M, Hyndman TH, Jiāng D, Kitajima EW, Kobinger GP, Kondō H, Kurath G, Kuzmin IV, Lamb RA, Lavazza A, Lee B, Lelli D, Leroy EM, Lǐ J, Maes P, Marzano S-YL, Moreno A, Mühlberger E, Netesov SV, Nowotny N, Nylund A, Økland AL, Palacios G, Pályi B, Pawęska JT, Payne SL, Prosperi A, Ramos-González PL, Rima BK, Rota P, Rubbenstroth D, Shī M, Simmonds P, Smither SJ, Sozzi E, Spann K, Stenglein MD, Stone DM, Takada A, Tesh RB, Tomonaga K, Tordo N, Towner JS, van den Hoogen B, Vasilakis N, Wahl V, Walker PJ, Wang L-F, Whitfield AE, Williams JV, Zerbini FM, Zhāng T, Zhang Y-Z, Kuhn JH. 2019. Taxonomy of the order Mononegavirales: update 2019. Arch Virol 164:1967–1980. doi: 10.1007/s00705-019-04247-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rima B, Balkema-Buschmann A, Dundon WG, Duprex P, Easton A, Fouchier R, Kurath G, Lamb R, Lee B, Rota P, Wang L, ICTV Report Consortium. 2019. ICTV virus taxonomy profile: Paramyxoviridae. J Gen Virol 100:1593–1594. doi: 10.1099/jgv.0.001328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.van den Hoogen BG, de Jong JC, Groen J, Kuiken T, de Groot R, Fouchier RA, Osterhaus AD. 2001. A newly discovered human pneumovirus isolated from young children with respiratory tract disease. Nat Med 7:719–724. doi: 10.1038/89098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Weinberg GA, Hall CB, Iwane MK, Poehling KA, Edwards KM, Griffin MR, Staat MA, Curns AT, Erdman DD, Szilagyi PG, New Vaccine Surveillance Network. 2009. Parainfluenza virus infection of young children: estimates of the population-based burden of hospitalization. J Pediatr 154:694–699. doi: 10.1016/j.jpeds.2008.11.034. [DOI] [PubMed] [Google Scholar]
- 5.Collins PL, Fearns R, Graham BS. 2013. Respiratory syncytial virus: virology, reverse genetics, and pathogenesis of disease. Curr Top Microbiol Immunol 372:3–38. doi: 10.1007/978-3-642-38919-1_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Qiu W, Zheng Y, Zhang S, Fan Q, Liu H, Zhang F, Wang W, Liao G, Hu R. 2011. Canine distemper outbreak in rhesus monkeys. Emerg Infect Dis 17:1541–1543. doi: 10.3201/eid1708.101153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Marsh GA, Wang LF. 2012. Hendra and Nipah viruses: why are they so deadly? Curr Opin Virol 2:242–247. doi: 10.1016/j.coviro.2012.03.006. [DOI] [PubMed] [Google Scholar]
- 8.Aljofan M. 2013. Hendra and Nipah infection: emerging paramyxoviruses. Virus Res 177:119–126. doi: 10.1016/j.virusres.2013.08.002. [DOI] [PubMed] [Google Scholar]
- 9.Sakai K, Nagata N, Ami Y, Seki F, Suzaki Y, Iwata-Yoshikawa N, Suzuki T, Fukushi S, Mizutani T, Yoshikawa T, Otsuki N, Kurane I, Komase K, Yamaguchi R, Hasegawa H, Saijo M, Takeda M, Morikawa S. 2013. Lethal canine distemper virus outbreak in cynomolgus monkeys in Japan in 2008. J Virol 87:1105–1114. doi: 10.1128/JVI.02419-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mathieu C, Horvat B. 2015. Henipavirus pathogenesis and antiviral approaches. Expert Rev Anti Infect Ther 13:343–354. doi: 10.1586/14787210.2015.1001838. [DOI] [PubMed] [Google Scholar]
- 11.Singh RK, Dhama K, Chakraborty S, Tiwari R, Natesan S, Khandia R, Munjal A, Vora KS, Latheef SK, Karthik K, Singh Malik Y, Singh R, Chaicumpa W, Mourya DT. 2019. Nipah virus: epidemiology, pathology, immunobiology and advances in diagnosis, vaccine designing and control strategies—a comprehensive review. Vet Q 39:26–55. doi: 10.1080/01652176.2019.1580827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Naser AM, Hossain MJ, Sazzad HM, Homaira N, Gurley ES, Podder G, Afroj S, Banu S, Rollin PE, Daszak P, Ahmed BN, Rahman M, Luby SP. 2015. Integrated cluster- and case-based surveillance for detecting stage III zoonotic pathogens: an example of Nipah virus surveillance in Bangladesh. Epidemiol Infect 143:1922–1930. doi: 10.1017/S0950268814002635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lam SK. 2003. Nipah virus—a potential agent of bioterrorism? Antiviral Res 57:113–119. doi: 10.1016/S0166-3542(02)00204-8. [DOI] [PubMed] [Google Scholar]
- 14.Luby SP. 2013. The pandemic potential of Nipah virus. Antiviral Res 100:38–43. doi: 10.1016/j.antiviral.2013.07.011. [DOI] [PubMed] [Google Scholar]
- 15.Sweileh WM. 2017. Global research trends of World Health Organization’s top eight emerging pathogens. Global Health 13:9. doi: 10.1186/s12992-017-0233-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Escaffre O, Borisevich V, Vergara LA, Wen JW, Long D, Rockx B. 2016. Characterization of Nipah virus infection in a model of human airway epithelial cells cultured at an air-liquid interface. J Gen Virol 97:1077–1086. doi: 10.1099/jgv.0.000441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Selvey LA, Wells RM, McCormack JG, Ansford AJ, Murray K, Rogers RJ, Lavercombe PS, Selleck P, Sheridan JW. 1995. Infection of humans and horses by a newly described morbillivirus. Med J Aust 162:642–645. doi: 10.5694/j.1326-5377.1995.tb126050.x. [DOI] [PubMed] [Google Scholar]
- 18.Drexler JF, Corman VM, Müller MA, Maganga GD, Vallo P, Binger T, Gloza-Rausch F, Cottontail VM, Rasche A, Yordanov S, Seebens A, Knörnschild M, Oppong S, Adu Sarkodie Y, Pongombo C, Lukashev AN, Schmidt-Chanasit J, Stöcker A, Carneiro AJB, Erbar S, Maisner A, Fronhoffs F, Buettner R, Kalko EKV, Kruppa T, Franke CR, Kallies R, Yandoko ERN, Herrler G, Reusken C, Hassanin A, Krüger DH, Matthee S, Ulrich RG, Leroy EM, Drosten C. 2012. Bats host major mammalian paramyxoviruses. Nat Commun 3:796. doi: 10.1038/ncomms1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marsh GA, de Jong C, Barr JA, Tachedjian M, Smith C, Middleton D, Yu M, Todd S, Foord AJ, Haring V, Payne J, Robinson R, Broz I, Crameri G, Field HE, Wang LF. 2012. Cedar virus: a novel Henipavirus isolated from Australian bats. PLoS Pathog 8:e1002836. doi: 10.1371/journal.ppat.1002836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu Z, Yang L, Yang F, Ren X, Jiang J, Dong J, Sun L, Zhu Y, Zhou H, Jin Q. 2014. Novel Henipa-like virus, Mojiang Paramyxovirus, in rats, China, 2012. Emerg Infect Dis 20:1064–1066. doi: 10.3201/eid2006.131022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen RT, Markowitz LE, Albrecht P, Stewart JA, Mofenson LM, Preblud SR, Orenstein WA. 1990. Measles antibody: reevaluation of protective titers. J Infect Dis 162:1036–1042. doi: 10.1093/infdis/162.5.1036. [DOI] [PubMed] [Google Scholar]
- 22.Cifuentes-Munoz N, Dutch RE, Cattaneo R. 2018. Direct cell-to-cell transmission of respiratory viruses: the fast lanes. PLoS Pathog 14:e1007015. doi: 10.1371/journal.ppat.1007015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Permar SR, Klumpp SA, Mansfield KG, Carville AA, Gorgone DA, Lifton MA, Schmitz JE, Reimann KA, Polack FP, Griffin DE, Letvin NL. 2004. Limited contribution of humoral immunity to the clearance of measles viremia in rhesus monkeys. J Infect Dis 190:998–1005. doi: 10.1086/422846. [DOI] [PubMed] [Google Scholar]
- 24.Permar SR, Klumpp SA, Mansfield KG, Kim WK, Gorgone DA, Lifton MA, Williams KC, Schmitz JE, Reimann KA, Axthelm MK, Polack FP, Griffin DE, Letvin NL. 2003. Role of CD8(+) lymphocytes in control and clearance of measles virus infection of rhesus monkeys. J Virol 77:4396–4400. doi: 10.1128/JVI.77.7.4396-4400.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Burnet FM. 1968. Measles as an index of immunological function. Lancet ii:610–613. doi: 10.1016/S0140-6736(68)90701-0. [DOI] [PubMed] [Google Scholar]
- 26.Imagawa DT, Goret P, Adams JM. 1960. Immunological relationships of measles, distemper, and rinderpest viruses. Proc Natl Acad Sci U S A 46:1119–1123. doi: 10.1073/pnas.46.8.1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.DeLay PD, Stone SS, Karzon DT, Katz S, Enders J. 1965. Clinical and immune response of alien hosts to inoculation with measles, rinderpest, and canine distemper viruses. Am J Vet Res 26:1359–1373. [PubMed] [Google Scholar]
- 28.Sheshberadaran H, Norrby E, McCullough KC, Carpenter WC, Orvell C. 1986. The antigenic relationship between measles, canine distemper and rinderpest viruses studied with monoclonal antibodies. J Gen Virol 67:1381–1392. doi: 10.1099/0022-1317-67-7-1381. [DOI] [PubMed] [Google Scholar]
- 29.Moura RA, Warren J. 1961. Subclinical infection of dogs by canine-adapted measles virus evidenced by their subsequent immunity to canine distemper virus. J Bacteriol 82:702–705. doi: 10.1128/JB.82.5.702-705.1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Strating A. 1975. Measles vaccine in dogs: efficacy against aerosol challenge with virulent canine distemper virus. J Am Vet Med Assoc 167:59–62. [PubMed] [Google Scholar]
- 31.Taylor J, Pincus S, Tartaglia J, Richardson C, Alkhatib G, Briedis D, Appel M, Norton E, Paoletti E. 1991. Vaccinia virus recombinants expressing either the measles virus fusion or hemagglutinin glycoprotein protect dogs against canine distemper virus challenge. J Virol 65:4263–4274. doi: 10.1128/JVI.65.8.4263-4274.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yoshikawa Y, Ochikubo F, Matsubara Y, Tsuruoka H, Ishii M, Shirota K, Nomura Y, Sugiyama M, Yamanouchi K. 1989. Natural infection with canine distemper virus in a Japanese monkey (Macaca fuscata). Vet Microbiol 20:193–205. doi: 10.1016/0378-1135(89)90043-6. [DOI] [PubMed] [Google Scholar]
- 33.Sakai K, Yoshikawa T, Seki F, Fukushi S, Tahara M, Nagata N, Ami Y, Mizutani T, Kurane I, Yamaguchi R, Hasegawa H, Saijo M, Komase K, Morikawa S, Takeda M. 2013. Canine distemper virus associated with a lethal outbreak in monkeys can readily adapt to use human receptors. J Virol 87:7170–7175. doi: 10.1128/JVI.03479-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.de Vries RD, Ludlow M, Verburgh RJ, van Amerongen G, Yuksel S, Nguyen DT, McQuaid S, Osterhaus AD, Duprex WP, de Swart RL. 2014. Measles vaccination of nonhuman primates provides partial protection against infection with canine distemper virus. J Virol 88:4423–4433. doi: 10.1128/JVI.03676-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.van Binnendijk RS, Poelen MC, de Vries P, Voorma HO, Osterhaus AD, Uytdehaag FG. 1989. Measles virus-specific human T cell clones. Characterization of specificity and function of CD4+ helper/cytotoxic and CD8+ cytotoxic T cell clones. J Immunol 142:2847–2854. [PubMed] [Google Scholar]
- 36.van Binnendijk RS, Versteeg-van Oosten JP, Poelen MC, Brugghe HF, Hoogerhout P, Osterhaus AD, Uytdehaag FG. 1993. Human HLA class I- and HLA class II-restricted cloned cytotoxic T lymphocytes identify a cluster of epitopes on the measles virus fusion protein. J Virol 67:2276–2284. doi: 10.1128/JVI.67.4.2276-2284.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.van Binnendijk RS, van Baalen CA, Poelen MC, de Vries P, Boes J, Cerundolo V, Osterhaus AD, UytdeHaag FG. 1992. Measles virus transmembrane fusion protein synthesized de novo or presented in immunostimulating complexes is endogenously processed for HLA class I- and class II-restricted cytotoxic T cell recognition. J Exp Med 176:119–128. doi: 10.1084/jem.176.1.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.de Vries RD, Yuksel S, Osterhaus AD, de Swart RL. 2010. Specific CD8(+) T-lymphocytes control dissemination of measles virus. Eur J Immunol 40:388–395. doi: 10.1002/eji.200939949. [DOI] [PubMed] [Google Scholar]
- 39.Horvath CM, Paterson RG, Shaughnessy MA, Wood R, Lamb RA. 1992. Biological activity of paramyxovirus fusion proteins: factors influencing formation of syncytia. J Virol 66:4564–4569. doi: 10.1128/JVI.66.7.4564-4569.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Russell CJ, Jardetzky TS, Lamb RA. 2004. Conserved glycine residues in the fusion peptide of the paramyxovirus fusion protein regulate activation of the native state. J Virol 78:13727–13742. doi: 10.1128/JVI.78.24.13727-13742.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Smith EC, Gregory SM, Tamm LK, Creamer TP, Dutch RE. 2012. Role of sequence and structure of the Hendra fusion protein fusion peptide in membrane fusion. J Biol Chem 287:30035–30048. doi: 10.1074/jbc.M112.367862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fox TG, Christenson JC. 2014. Influenza and parainfluenza viral infections in children. Pediatr Rev 35:217–227. doi: 10.1542/pir.35-6-217. [DOI] [PubMed] [Google Scholar]
- 43.Ludlow M, Rennick LJ, Nambulli S, de Swart RL, Duprex WP. 2014. Using the ferret model to study morbillivirus entry, spread, transmission and cross-species infection. Curr Opin Virol 4:15–23. doi: 10.1016/j.coviro.2013.11.001. [DOI] [PubMed] [Google Scholar]
- 44.Appel MJ, Reggiardo C, Summers BA, Pearce-Kelling S, Mare CJ, Noon TH, Reed RE, Shively JN, Orvell C. 1991. Canine distemper virus infection and encephalitis in javelinas (collared peccaries). Arch Virol 119:147–152. doi: 10.1007/BF01314331. [DOI] [PubMed] [Google Scholar]
- 45.Origgi FC, Sattler U, Pilo P, Waldvogel AS. 2013. Fatal combined infection with canine distemper virus and orthopoxvirus in a group of Asian marmots (Marmota caudata). Vet Pathol 50:914–920. doi: 10.1177/0300985813476060. [DOI] [PubMed] [Google Scholar]
- 46.Arunkumar G, Chandni R, Mourya DT, Singh SK, Sadanandan R, Sudan P, Bhargava B, Nipah Investigators People and Health Study Group. 2019. Outbreak investigation of Nipah virus disease in Kerala, India, 2018. J Infect Dis 219:1867–1878. doi: 10.1093/infdis/jiy612. [DOI] [PubMed] [Google Scholar]
- 47.Nikolay B, Salje H, Hossain MJ, Khan A, Sazzad HMS, Rahman M, Daszak P, Stroher U, Pulliam JRC, Kilpatrick AM, Nichol ST, Klena JD, Sultana S, Afroj S, Luby SP, Cauchemez S, Gurley ES. 2019. Transmission of Nipah virus—14 years of investigations in Bangladesh. N Engl J Med 380:1804–1814. doi: 10.1056/NEJMoa1805376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Arunkumar G, Devadiga S, McElroy AK, Prabhu S, Sheik S, Abdulmajeed J, Robin S, Sushama A, Jayaram A, Nittur S, Shakir M, Kumar KGS, Radhakrishnan C, Sakeena K, Vasudevan J, Reena KJ, Sarita RL, Klena JD, Spiropoulou CF, Laserson KF, Nichol ST. 2019. Adaptive immune responses in humans during Nipah virus acute and convalescent phases of infection. Clin Infect Dis 69:1752–1756. doi: 10.1093/cid/ciz010. [DOI] [PubMed] [Google Scholar]
- 49.van Binnendijk RS, Poelen MC, Kuijpers KC, Osterhaus AD, Uytdehaag FG. 1990. The predominance of CD8+ T cells after infection with measles virus suggests a role for CD8+ class I MHC-restricted cytotoxic T lymphocytes (CTL) in recovery from measles. Clonal analyses of human CD8+ class I MHC-restricted CTL. J Immunol 144:2394–2399. [PubMed] [Google Scholar]
- 50.Pickering BS, Hardham JM, Smith G, Weingartl ET, Dominowski PJ, Foss DL, Mwangi D, Broder CC, Roth JA, Weingartl HM. 2016. Protection against henipaviruses in swine requires both, cell-mediated and humoral immune response. Vaccine 34:4777–4786. doi: 10.1016/j.vaccine.2016.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jaye A, Magnusen AF, Sadiq AD, Corrah T, Whittle HC. 1998. Ex vivo analysis of cytotoxic T lymphocytes to measles antigens during infection and after vaccination in Gambian children. J Clin Invest 102:1969–1977. doi: 10.1172/JCI3290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.van Els CA, Nanan R. 2002. T cell responses in acute measles. Viral Immunol 15:435–450. doi: 10.1089/088282402760312322. [DOI] [PubMed] [Google Scholar]
- 53.Scheid A, Choppin PW. 1977. Two disulfide-linked polypeptide chains constitute the active F protein of paramyxoviruses. Virology 80:54–66. doi: 10.1016/0042-6822(77)90380-4. [DOI] [PubMed] [Google Scholar]
- 54.Hernandez LD, Peters RJ, Delos SE, Young JA, Agard DA, White JM. 1997. Activation of a retroviral membrane fusion protein: soluble receptor-induced liposome binding of the ALSV envelope glycoprotein. J Cell Biol 139:1455–1464. doi: 10.1083/jcb.139.6.1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Paterson RG, Lamb RA. 1987. Ability of the hydrophobic fusion-related external domain of a paramyxovirus F protein to act as a membrane anchor. Cell 48:441–452. doi: 10.1016/0092-8674(87)90195-4. [DOI] [PubMed] [Google Scholar]
- 56.Ghosh S, Walker J, Jackson DC. 2001. Identification of canine helper T-cell epitopes from the fusion protein of canine distemper virus. Immunology 104:58–66. doi: 10.1046/j.0019-2805.2001.01271.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Finch JT, Gibbs AJ. 1970. Observations on the structure of the nucleocapsids of some paramyxoviruses. J Gen Virol 6:141–150. doi: 10.1099/0022-1317-6-1-141. [DOI] [PubMed] [Google Scholar]
- 58.Thakkar VD, Cox RM, Sawatsky B, da Fontoura Budaszewski R, Sourimant J, Wabbel K, Makhsous N, Greninger AL, von Messling V, Plemper RK. 2018. The unstructured paramyxovirus nucleocapsid protein tail domain modulates viral pathogenesis through regulation of transcriptase activity. J Virol 92:e02064-17. doi: 10.1128/JVI.02064-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Buchholz CJ, Spehner D, Drillien R, Neubert WJ, Homann HE. 1993. The conserved N-terminal region of Sendai virus nucleocapsid protein NP is required for nucleocapsid assembly. J Virol 67:5803–5812. doi: 10.1128/JVI.67.10.5803-5812.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Myers TM, Pieters A, Moyer SA. 1997. A highly conserved region of the Sendai virus nucleocapsid protein contributes to the NP-NP binding domain. Virology 229:322–335. doi: 10.1006/viro.1996.8429. [DOI] [PubMed] [Google Scholar]
- 61.Myers TM, Smallwood S, Moyer SA. 1999. Identification of nucleocapsid protein residues required for Sendai virus nucleocapsid formation and genome replication. J Gen Virol 80:1383–1391. doi: 10.1099/0022-1317-80-6-1383. [DOI] [PubMed] [Google Scholar]
- 62.Marttila J, Ilonen J, Norrby E, Salmi A. 1999. Characterization of T cell epitopes in measles virus nucleoprotein. J Gen Virol 80:1609–1615. doi: 10.1099/0022-1317-80-7-1609. [DOI] [PubMed] [Google Scholar]
- 63.Nanan R, Carstens C, Kreth HW. 1995. Demonstration of virus-specific CD8+ memory T cells in measles-seropositive individuals by in vitro peptide stimulation. Clin Exp Immunol 102:40–45. doi: 10.1111/j.1365-2249.1995.tb06633.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lamb RA, Parks GD. 2013. Paramyxoviridae, p 957–995. In Knipe DM, Howley PM (ed), Fields virology, 6th ed, vol 1 Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 65.Vatti A, Monsalve DM, Pacheco Y, Chang C, Anaya JM, Gershwin ME. 2017. Original antigenic sin: a comprehensive review. J Autoimmun 83:12–21. doi: 10.1016/j.jaut.2017.04.008. [DOI] [PubMed] [Google Scholar]
- 66.Devarajan P, Swain SL. 2019. Original antigenic sin: friend or foe in developing a broadly cross-reactive vaccine to influenza? Cell Host Microbe 25:354–355. doi: 10.1016/j.chom.2019.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhang A, Stacey HD, Mullarkey CE, Miller MS. 2019. Original Antigenic sin: how first exposure shapes lifelong anti-influenza virus immune responses. J Immunol 202:335–340. doi: 10.4049/jimmunol.1801149. [DOI] [PubMed] [Google Scholar]
- 68.McMichael AJ. 1998. The original sin of killer T cells. Nature 394:421–422. doi: 10.1038/28738. [DOI] [PubMed] [Google Scholar]
- 69.Mongkolsapaya J, Duangchinda T, Dejnirattisai W, Vasanawathana S, Avirutnan P, Jairungsri A, Khemnu N, Tangthawornchaikul N, Chotiyarnwong P, Sae-Jang K, Koch M, Jones Y, McMichael A, Xu X, Malasit P, Screaton G. 2006. T cell responses in dengue hemorrhagic fever: are cross-reactive T cells suboptimal? J Immunol 176:3821–3829. doi: 10.4049/jimmunol.176.6.3821. [DOI] [PubMed] [Google Scholar]
- 70.Guindon S, Gascuel O. 2003. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol 52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- 71.de Waal L, Yuksel S, Brandenburg AH, Langedijk JP, Sintnicolaas K, Verjans GM, Osterhaus AD, de Swart RL. 2004. Identification of a common HLA-DP4-restricted T-cell epitope in the conserved region of the respiratory syncytial virus G protein. J Virol 78:1775–1781. doi: 10.1128/JVI.78.4.1775-1781.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ludlow M, Nguyen DT, Silin D, Lyubomska O, de Vries RD, von Messling V, McQuaid S, De Swart RL, Duprex WP. 2012. Recombinant canine distemper virus strain Snyder Hill expressing green or red fluorescent proteins causes meningoencephalitis in the ferret. J Virol 86:7508–7519. doi: 10.1128/JVI.06725-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bamford C, Wignall-Fleming E, Sreenu VB, Randall R, Duprex P, Rima B. 2019. Unusual, stable replicating viruses generated from mumps virus cDNA clones. PLoS One 14:e0219168. doi: 10.1371/journal.pone.0219168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.de Graaf M, Herfst S, Schrauwen EJ, van den Hoogen BG, Osterhaus AD, Fouchier RA. 2007. An improved plaque reduction virus neutralization assay for human metapneumovirus. J Virol Methods 143:169–174. doi: 10.1016/j.jviromet.2007.03.005. [DOI] [PubMed] [Google Scholar]
- 75.Dietzel E, Kolesnikova L, Sawatsky B, Heiner A, Weis M, Kobinger GP, Becker S, von Messling V, Maisner A. 2015. Nipah virus matrix protein influences fusogenicity and is essential for particle infectivity and stability. J Virol 90:2514–2522. doi: 10.1128/JVI.02920-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
F129–137 amino acid alignment for morbilliviruses. Mismatches to the MeV amino acid sequence are shown in red. CDV, canine distemper virus; MeV, measles virus; PPRV, peste-des-petits-ruminants virus; CeMV, cetacean morbillivirus; PDV, phocine distemper virus; RPV, rinderpest virus; FmoPV, feline morbillivirus. Download FIG S1, DOCX file, 0.03 MB (32.2KB, docx) .
Copyright © 2020 de Vries et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
F129–137 is in the conserved fusion peptide. (A) Phylogenetic tree of the Paramyxo- and Pneumoviridae based on an F nucleotide alignment. An unrooted maximum likelihood phylogenetic tree was estimated under the general time-reversible model. The tree was based on 29 sequences (Table S1), of which a selection relevant to this study is shown by their abbreviations. (B) Fusion peptide amino acid alignment for the relevant endemic and zoonotic paramyxo- and pneumoviruses. The F129–137 T cell epitope is indicated by the gray background. Mismatches to the MeV amino acid sequence are shown in red. CDV, canine distemper virus; MeV, measles virus; NiV, Nipah virus; HeV, Hendra virus; HPIV, human parainfluenza virus; MuV, mumps virus; HMPV, human metapneumovirus; HRSV, human respiratory virus. Download FIG S2, DOCX file, 0.2 MB (168.7KB, docx) .
Copyright © 2020 de Vries et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
(A and B) Peptide dilution series with B-LCL pulsed with different concentrations of peptides. CD8Xreact1 reacted strongly with FAQITAGIAL but also with FAQITAGVAL. CD8Xreact2 had a stronger affinity with FAQITAGVAL but still recognized FAQITAGIAL. Download FIG S3, DOCX file, 0.1 MB (74.2KB, docx) .
Copyright © 2020 de Vries et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
T cell receptor (TCR) variable (V)β chain expression of CD8Xreact1 and CD8Xreact2 was determined by flow cytometry. (A) CD8Xreact1 proved clonal and expressed TCRVβ14. (B) CD8Xreact2 was clonal and expressed TCRVβ8. Download FIG S4, DOCX file, 0.3 MB (354.2KB, docx) .
Copyright © 2020 de Vries et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
TCC have different avidities for F129–137. We developed a FACS-based assay to determine interaction strength between TCC and tetramers. (A and B) CD8Xreact1 and CD8Xreact2 were stained with F129–137 tetramers and treated with increasing concentrations of urea, disrupting low-avidity binding between tetramers and TCC. We confirmed reactivity of CD8Xreact1 with FAQITAGIAL and FAQITAGVAL, and of CD8Xreact1 with FAQITAGIAL, FAQITAGVAL, FAQITAAVAI, FAQITAAVAL, and FAQVTAAVSL. CDV, canine distemper virus; MeV, measles virus; NiV, Nipah virus; HeV, Hendra virus; HPIV, human parainfluenza virus; MuV, mumps virus; MFI, mean fluorescence intensity. Download FIG S5, DOCX file, 0.1 MB (74KB, docx) .
Copyright © 2020 de Vries et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
(A) Selection criteria for conserved regions in the paramyxo- and pneumovirus proteome. (B) Conserved regions when comparing NiV with MeV. (C) Conserved regions when comparing NiV with HPIV3. (D) Conserved regions when comparing NiV with HPIV2. (E) Conserved regions throughout the paramyxo- and pneumovirus proteome. Download Data Set S1, DOCX file, 3.5 MB (3.6MB, docx) .
Copyright © 2020 de Vries et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Paramyxo- and pneumovirus F sequences used to construct phylogenetic trees. Download Table S1, DOCX file, 0.01 MB (9.4KB, docx) .
Copyright © 2020 de Vries et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.