Abstract
Background: Numerous studies have found that circular RNAs (circRNA) can serve as competing endogenous RNA (ceRNA) in cancer progression while the expression profiles and functions of competitive endogenous circRNAs (ce-circRNAs) in breast cancer (BC) have not been determined. Methods: Six pairs of tissue samples were selected to perform ceRNA microarray, and The Cancer Genome Atlas (TCGA) data was also included to explore the ce-circRNA profiling of BC. The expression of one of the top upregulated circRNAs, circMMP11, was confirmed by qRT-PCR in both breast cancer cell lines and tissues. We also analyzed the clinical impact of circMMP11 on BC. To explore the functions of circMMP11 in BC, experiments referring to cell proliferation and migration were performed. The regulatory effect of circMMP11 on miRNA and its target genes was explored to confirm its ce-circRNA mechanisms in BC. Results: qRT-PCR analyses verified that circMMP11 was successfully transfected and positively associated with a poorer clinicopathology of BC. The inhibition of circMMP11 suppressed cell proliferation and migration of BC. The luciferase reporter assay revealed that circMMP11 and MMP11 could bind to miR-1204 and that circMMP11 acted as a ce-circRNA by regulating the expression of MMP11 via sponging miR-1204. Conclusions: The circMMP11/miR-1204/MMP11 axis regulates breast cancer progression via a competitive endogenous RNA (ceRNA) mechanism. CircMMP11 may serve as a potential therapeutic target in BC.
Keywords: Breast cancer, circMMP11, miR-1204, MMP11, circRNA, ceRNA
Introduction
Breast cancer (BC) is one of the three most common cancers worldwide. Despite the improved prognosis of patients with BC resulting from early diagnosis, radical surgery and the development of adjuvant therapy, BC remains the most common cancer among women, exhibiting the highest morbidity and mortality [1]. According to the newest data from Global Cancer Statistics 2018, over 2 million females are newly diagnosed with BC (24.2% of the total cases) and more than 60 thousand deaths are caused by BC (15.0%). The discovery of novel biomarkers is warranted for the early detection, treatment and prognosis of BC [2].
As a new type of endogenous RNA, circular RNAs (circRNAs) represent a class of continuous covalently closed single-stranded RNA molecules [3]. CircRNAs can escape exonucleic acid degrading enzymes due to the lack of a free 5’-cap structure and the 3’-poly-A tail, making them well expressed and more stable than their linear counterparts. With the development and advancement of high-throughput sequencing technique and bioinformatics analysis, thousands of circRNAs have been discovered in mammalian cells and regarded as functional molecules in various biological processes instead of as a byproduct of splicing errors [4]. In recent decades, it has been extensively revealed that circRNAs are involved in the initiation and development of a large number of diseases, including the carcinogenesis and cancer progression [5]. Some circRNAs function as competing endogenous RNAs (ceRNAs), an important class of posttranscriptional regulators, and alter the gene expression of key tumorigenic or tumor-suppressive genes through a miRNA-mediated mechanism [6]. In this study, we regarded these circRNAs as competing endogenous circRNAs (ce-circRNAs). Several ce-circRNA have been discovered in cancers. For instance, cTFRC promotes bladder carcinoma progression as the sponge of miR-107 [7]. CircTP63 promotes lung squamous cell carcinoma progression by regulating miR-873-3p [8]. CircKIF4A regulates triple-negative BC (TNBC) progression by regulating miR-375 [9]. More importantly, due to the high expression levels and increased stability of these RNAs, ce-circRNAs may be exceptionally effective modulators of the crosstalk among linear ceRNAs [10].
In this study, we detected differentially expressed circRNAs in BC tissue through microarray analysis and found a large number of suspected dysregulated ce-circRNAs. Among these RNAs, we determined that circMMP11 was significantly overexpressed in BC tissues, characterized its clinical significance and biological functions, and explored the potential molecular mechanism of circMMP11 in BC.
Materials and methods
Patients and specimens
We collected 119 pairs of primary breast cancer tissue and adjacent normal breast tissue from patients diagnosed with breast cancer at Zhongshan Hospital affiliated with Fudan University from June 2013 to March 2018, and we stored them at -80°C until usage. The diagnosis of all cases required tissue biopsy by two senior pathologists. Signed written informed consent was obtained from all the patients. Tumor volume was calculated as the length * width2/2. The protocols used in this study were approved by the Medical Ethics Committees at Zhongshan Hospital.
Cell lines and culture
All the cell lines were purchased from the Culture Collection of the Chinese Academy of Sciences (Shanghai, China), including human mammary epithelial cell lines (HBL-100) and breast cancer cell lines (MCF7, T47D, MDA-MB-453, MDA-MB-231, MDA-MB-468 and Hs 578T). All the cell lines were passaged for fewer than six months and cultured according to the supplier’s instructions.
RNA extraction
Total RNA was extracted and purified using TRIzol reagent (Invitrogen) and the miRNeasy Mini Kit (Qiagen, USA) following the manufacturer’s instructions. Total RNA was quantified by using a DS-11 spectrophotometer (Denovix) and assessed with an Agilent Bioanalyzer 2100 (CA, US) to inspect RNA integration. A260/280 (1.8~2.1) and A260/230 > 2.0 were regarded as a qualified RNA sample.
Microarray analysis
Six pairs of breast cancer and paracancerous tissues were used for microarray assay to investigate the potential dysregulated ce-circRNAs. As previously described, the Agilent SBC circRNA microarray includes 88,371 circRNAs and 18,853 mRNAs [11]. The detailed procedures including RNA amplification and labeling, cRNA refinement and hybridization, slides fixing and scanning were described in our previous studies [12]. Feature Extraction software 10.7 (CA, US) was applied to extract raw data. The quantile algorithm and limma packages in R were used for data normalization. Aberrantly expressed RNAs between breast cancer and paracancerous tissues were identified and retained by screening the fold change > 2.0, P < 0.05.
Quantitative real-time polymerase chain reaction analysis (qRT-PCR)
To detect circRNA, cDNA was synthesized by reverse transcribing 500 ng of RNA using PrimeScript RT Reagent (TaKaRa, Cat. #RR047A) on a T100TM Thermal Cycler system (Bio-Rad, US) and then subjected to qPCR using the SYBR Premix Ex Taq™ kit (TaKaRa, Cat. #RR820A) on a ABI 7500 system (Thermo Fisher Scientific, USA). The 18S served as an internal control for circRNA and mRNA, while U6 was used for miRNA. The quantification was calculated by the 2-ΔΔCt method. The primers are listed in Table S1. Comparisons between groups were analyzed with unpaired t test. Means ± standard deviation (SD) were used to present the data. P-values < 0.05 were considered to be significant.
Bioinformatics analysis
RNA-seq data from The Cancer Genome Atlas (TCGA) were analyzed using the edgeR package. The miRNAs were predicted with the Circinteractome (https://circinteractome.nia.nih.gov/) database and MRE analysis (Biotechnology, Shanghai). The mRNAs targeted by miRNAs were predicted using the TargetScan V7.2 (http://www.targetscan.org/vert_72/), miRDB (http://mirdb.org/) and TargetMiner (http://www.mybiosoftware.com/targetminer-microrna-target-prediction.html) databases. Overall survival (OS) and disease-free survival (DFS) curves were generated using the Kaplan-Meier method and survival differences were evaluated using the log-rank test in Gepia2 (http://gepia2.cancer-pku.cn/#index) and Kaplan-Meier Plotter Breast Cancer (http://kmplot.com/analysis/index.php?p=service&cancer=breast) [13]. Venn diagrams were generated using an online tool (http://bioinformatics.psb.ugent.be/webtools/Venn/) and RNA interaction networks were determined using cytoscape v3.6.1.
Construction and transfection of siRNA
Three siRNAs targeting circMMP11 were designed and synthesized by RiboBio (Guangzhou, China). For cell transfection, cells were seeded in 6-well plates and cultured overnight to 50%-70% confluence before transfection. siRNAs or corresponding controls (RiboBio, Guangzhou) were transfected at a final concentration of 50 nM with Lipofectamine 3000 (Thermo Fisher Scientific, USA) according to the manufacturer’s protocol. The third siRNA with the best knockdown efficiency was used in subsequent functional studies. In addition, miRNA mimics or its negative control (RiboBio, Guangzhou) were transiently transfected according to the same procedure.
Cell counting kit-8 assay
Cell proliferation was assessed using the Cell Counting Kit-8 assay (BBI, Cat. #E60633). Cells (2×103) were seeded into 96-well plates and incubated at 37°C with 5% CO2. To each well, 10 μL CCK-8 solution was added at 12 h, 24 h, 48 h and 72 h after transfection. After 2 h of incubation at 37°C, the absorbance at 450 nm was measured using the EPOCH2 microplate reader (BioTek, USA). The experiments and those mentioned below were performed in triplicate. Comparisons between groups were analyzed with the unpaired t test. Means ± standard deviation (SD) were used to present quantitative data. P-values < 0.05 were considered to be significant.
Colony formation assay
The cells were seeded in 6-well plates at a density of 1×103 cells/well and incubated at 37°C for 2 weeks. Colonies were fixed with 4% paraformaldehyde and stained with crystal violet for 15 min. Cell colonies were dried out at room temperature and then analyzed using ImageJ 1.51k (NIH, USA). Comparisons between groups were analyzed with the unpaired t test. Means ± standard deviations (SDs) were used to present quantitative data. P-values < 0.05 were considered to be significant.
Transwell assays
The invasive and migratory capacities of breast cancer cells were evaluated using transwell plates (Cat. #3422, Corning). For this assay, 2×104 cells were suspended in 200 μL serum-free medium after transfection and inoculated into the upper chambers coated with or without 60 μl of 1:6 diluted Matrigel (Corning, Cat. #354234). RPMI 1640 containing 10% FBS was added to the lower chambers as a chemoattractant. After incubation for 24 h at 37°C, cells that migrated to the bottom of the membrane were fixed with 4% paraformaldehyde then stained and with crystal violet solution for 15 min. Photos of the dried plates were captured using a microscope (Olympus, IX51S8F3). All cells were counted at 200× magnification under a microscope. Cell counting was carried out using ImageJ 1.51k (NIH, USA). Comparisons between groups were analyzed with the unpaired t test. Means ± standard deviations (SDs) were used to present quantitative data. P-values < 0.05 were considered to be significant.
Nuclear-cytoplasmic fractionation
Cytoplasmic and nuclear RNA isolation were performed with the Cytoplasmic & Nuclear RNA Purification Kit (BIOTEK, Canada) following the manufacturer’s instruction. Briefly, the cells were lysed with Lysis buffer J and centrifuged to separate cell fractions. The supernatant and pellet were separately transferred to fresh RNase-free tubes and Buffer SK was added for deeper lysis. Then the sample was bound with ethanol and washed with Wash Solution A. The cytoplasmic and nuclear RNA was eluted with Elution Buffer E.
Dual luciferase reporter assay
Luciferase reporter vectors carrying the full-length wild-type predicted miR-1204 binding sequences for the circMMP11 (psiCHECK2-circMMP11-wt) or mutant type (psiCHECK2-circMMP11-mut) were constructed. Luciferase reporter vector with miR-1204 mimics or negative control was transfected at a final concentration of 100 nM into HEK-293T cells at 60%-70% confluency. After 48 h of incubation, the luciferase activity in each group was tested using the Dual Luciferase Reporter Gene Assay Kit (YEASEN) with a FlexStation 3 system (Molecular Devices). Comparisons between groups were analyzed with the unpaired t test. Means ± standard deviations (SDs) were used to present quantitative data. P-values < 0.05 were considered to be significant.
Statistical analysis
Comparisons between groups were analyzed with unpaired t tests or χ2 tests. Means ± standard deviations (SDs) were used to present quantitative data. The overall survival (OS) curve was plotted using the Kaplan-Meier method, and survival differences were evaluated using a log-rank test. The correlation between groups was analyzed by Pearson correlation. P-values < 0.05 were considered to be significant. Statistical analyses were performed using SPSS 23.0 (IBM, US) and GraphPad Prism 7.0 (GraphPad Software Inc., US).
Results
ceRNA expression profiling in BC
We reanalyzed the ceRNA microarray data to explore the profiles of ce-circRNA in BC. The intersection of the RNA-seq data from TCGA and differentially expressed (DE) genes from our ceRNA array consists of 477 genes which were referred to as common genes (Figure 1A). Then, we combined these 477 common genes with the host genes (HGs) of the DE circRNAs from our microarray data and identified 167 DE circRNA HGs (Figure 1B). In total, 99 DE circRNA HGs were upregulated, and 68 DE circRNA HGs were downregulated, in the BC tissues, which was consistent with the expression level of the DE circRNAs. Part of the DE circRNAs and common DE circRNA HGs were listed in Table S2. The heatmaps of identified DE circRNA HGs and DE circRNAs which may compose the potential ce-circRNA profiles of BC are shown in Figure 1C and 1D.
Figure 1.
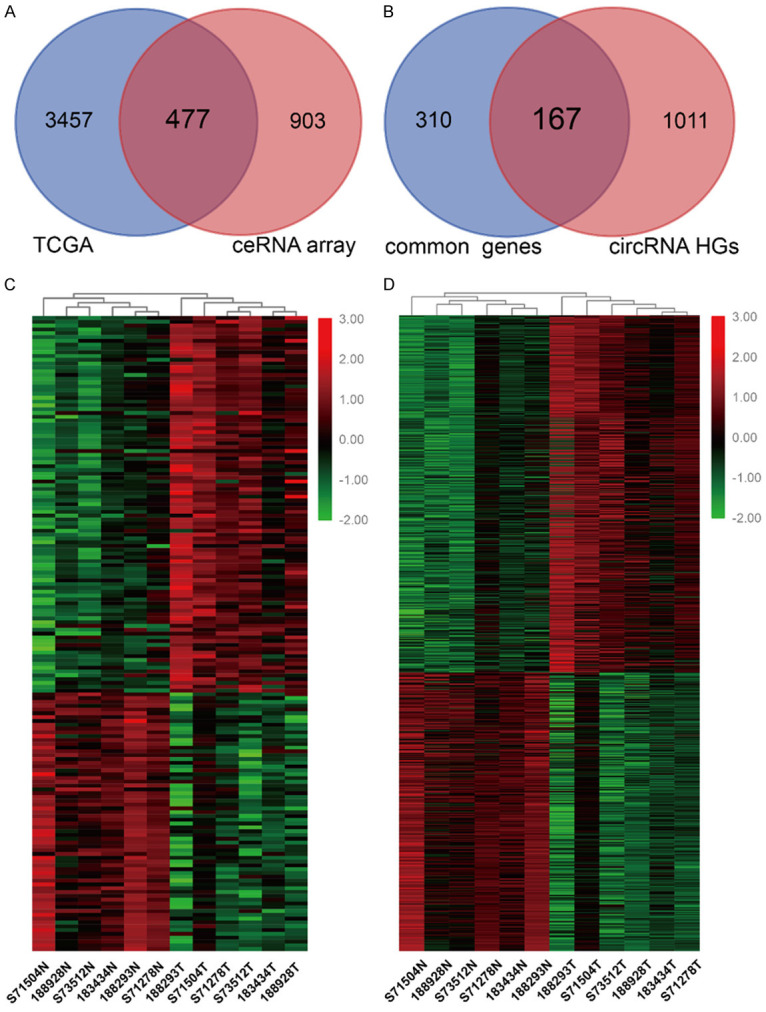
ceRNA expression profiling in BC. A. Venn Diagrams showing that the 477 common genes identified through combining TCGA data and ceRNA microarray. B. 167 common DE circRNA HGs were observed by overlapping 477 common genes and HGs of DE circRNA in our ceRNA array. C. Heatmap of 167 common DE circRNA HGs in six pairs of human BC cancerous tissues and adjacent normal tissues. D. 799 circRNAs transcribed from 167 common DE circRNA HGs. DE: differentially expressed; HG: host gene.
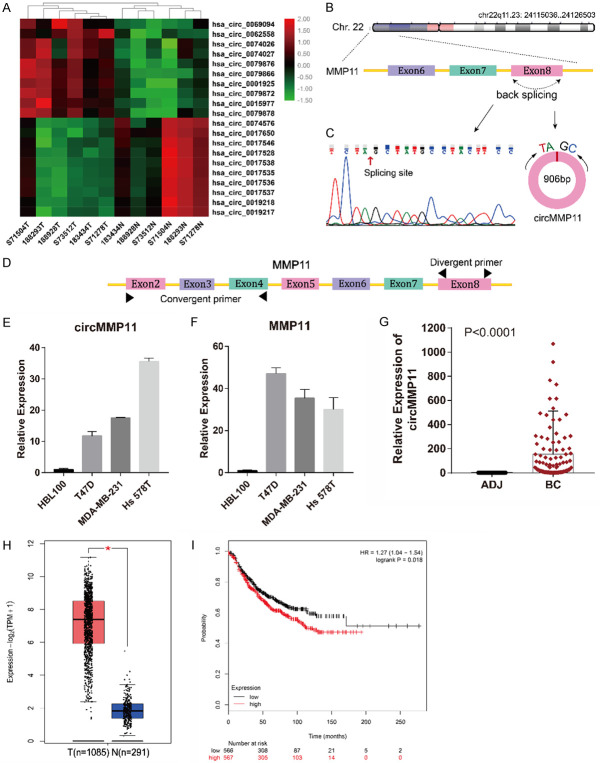
circMMP11 is upregulated and correlated with poor clinical characteristics of BC
The top 10 upregulated and top 10 downregulated circRNAs among the above DE circRNAs were illustrated in Figure 2A. The top upregulated circRNA, hsa_circ_0062558, was verified in breast cancer cell lines and tissues using qRT-PCR. According to the human reference genome (GRCh37/hg19), hsa_circ_0062558, located at chr22q11.23: 24115036-24126503, is 906 bp in length and assumed to be derived from the eighth exon of MMP11 (matrix metallopeptidase 11) gene (Figure 2B). Therefore, we named hsa_circ_0062558 “circMMP11”. We confirmed the sequence of PCR product by DNA sequencing and confirmed the spicing site of circMMP11 which was the same with the database (Figure 2C). The divergent primer designed for circMMP11 and the convergent primer for MMP11 at different exons are shown in Figure 2D. qRT-PCR showed that both circMMP11 and its linear counterpart were both upregulated in BC cell lines (Figure 2E and 2F). To investigate the clinical significance of circMMP11 in BC, a cohort of 113 BC patients was recruited. As shown in Figure 2G, we detected expresion level of circMMP11 in 113 pairs of breast cancer tissues and adjacent normal tissues and found that circMMP11 was significantly upregulated in BC tissues (Difference between means = 153±33.81, 95% CI: 86.4 to 219.6, P < 0.0001). To compare the relation of circMMP11 level with clinicopathological characteristics of BC patients, the patients were separated into “high” and “low” two groups by the median level of circMMP11 expression which was 30.85. It is shown in Table 1 that circMMP11 was positively associated with lymph node metastasis and TNM stage (P = 0.018 and 0.003, respectively), which indicates that circMMP11 plays a pivotal role in BC progression. Additionally, we explored the expression of MMP11 by Gepia2 and performed Kaplan-Meier survival analysis by analyzing BC RNA-seq data from TCGA and found that MMP11 was highly expressed in BC tissue (Figure 2H) and was associated with poorer survival of patients with BC (HR = 1.27 [1.04-1.54], P = 0.018) (Figure 2I).
Figure 2.
circMMP11 is upregulated in BC cells and tissue and correlated with poor clinical characteristics of BC. A. Heatmap of the top 10 aberrantly expressed circRNAs. B. circMMP11 is located on Chr.22 and consisted of the eighth exon of MMP11. C. Splicing site of circMMP11 verified by DNA sequencing. D. The divergent primer designed for circMMP11 and the convergent primer for MMP11 at different exons. E, F. circMMP11 and linear MMP11 were both upregulated in BC cell lines. G. circMMP11 expression level was higher in BC tissues (BC) than in adjacent nontumor tissues (ADJ) detected by qRT-PCR (n = 113). H. Expression level of MMP11 was higher in BC tissue than in normal tissue which was analyzed through Gepia2. I. Higher level of MMP11 was associated with poorer survival of patients with BC. Data were shown as the mean ± SD, *P < 0.05.
Table 1.
Clinicopathological characteristics and circMMP11 expression in breast cancer tissue
| Variables | Cases (n = 113) | circMMP11 | Chi-square | P value | |
|---|---|---|---|---|---|
|
| |||||
| low | high | ||||
| Age (years) | 0.429 | 0.512 | |||
| ≤ 50 | 43 | 20 (46.5%) | 23 (53.5%) | ||
| > 50 | 70 | 37 (52.9%) | 33 (47.1%) | ||
| Tumor volumes | 0.546 | 0.460 | |||
| ≤ 2.0 cm3 | 41 | 19 (46.3%) | 22 (53.7%) | ||
| > 2.0 cm3 | 69 | 37 (53.6%) | 32 (46.4%) | ||
| LN infiltrated | 5.592 | 0.018* | |||
| No | 65 | 39 (60.0%) | 26 (40.0%) | ||
| Yes | 48 | 18 (37.5%) | 30 (62.5%) | ||
| TNM staging | 9.009 | 0.003** | |||
| I-II | 93 | 53 (57.0%) | 40 (43%) | ||
| III-IV | 20 | 4 (20.0%) | 16 (80.0%) | ||
| ER | 0.425 | 0.514 | |||
| ~++ | 66 | 35 (53.0%) | 31 (47.0%) | ||
| +++ | 47 | 22 (46.8%) | 25 (53.2%) | ||
| PR | 1.143 | 0.285 | |||
| ~++ | 95 | 50 (52.6%) | 45 (47.4%) | ||
| +++ | 18 | 7 (38.9%) | 11 (61.1%) | ||
| HER2 | 3.791 | 0.052 | |||
| ~++ | 91 | 50 (54.9%) | 41 (45.1%) | ||
| +++ | 22 | 7 (31.8%) | 15 (68.2%) | ||
| Ki67 | 0.039 | 0.844 | |||
| ≤ 14% | 21 | 11 (52.4%) | 10 (47.6%) | ||
| > 14% | 92 | 46 (50.0%) | 46 (50.0%) | ||
P < 0.05, statistically significant.
P < 0.01, statistically significant.
Silencing of circMMP11 inhibits BC cell proliferation, migration and invasion
Three siRNAs were constructed to explore the function of circMMP11 in BC. These three siRNAs cover the back-splicing site of circMMP11 to knock down the expression level of circMMP11 (Figure 3A). After transfection with circMMP11 siRNAs into T47D, MDA-MB-231 and Hs 578T cell lines, the efficiency of circMMP11 silencing was measured and confirmed by qRT-PCR. The inhibition was relatively successful with siRNA3 so that it was regarded as si-circMMP11 and used in the following experiments (Figure 3B). A CCK-8 assay showed that circMMP11 knockdown significantly inhibited cell proliferation of T47D, MDA-MB-231 and Hs 578T (Figure 3C-E). circMMP11 knockdown also reduced the colony formation ability of these cells (Figure 3F and 3G). Transwell assay without Matrigel revealed that cell migration was significantly suppressed in circMMP11 silenced group (Figure 3H and 3I). Transwell assay with Matrigel also revealed that knockdown of circMMP11 significantly reduced cell invasive capability (Figure 3J and 3K).
Figure 3.
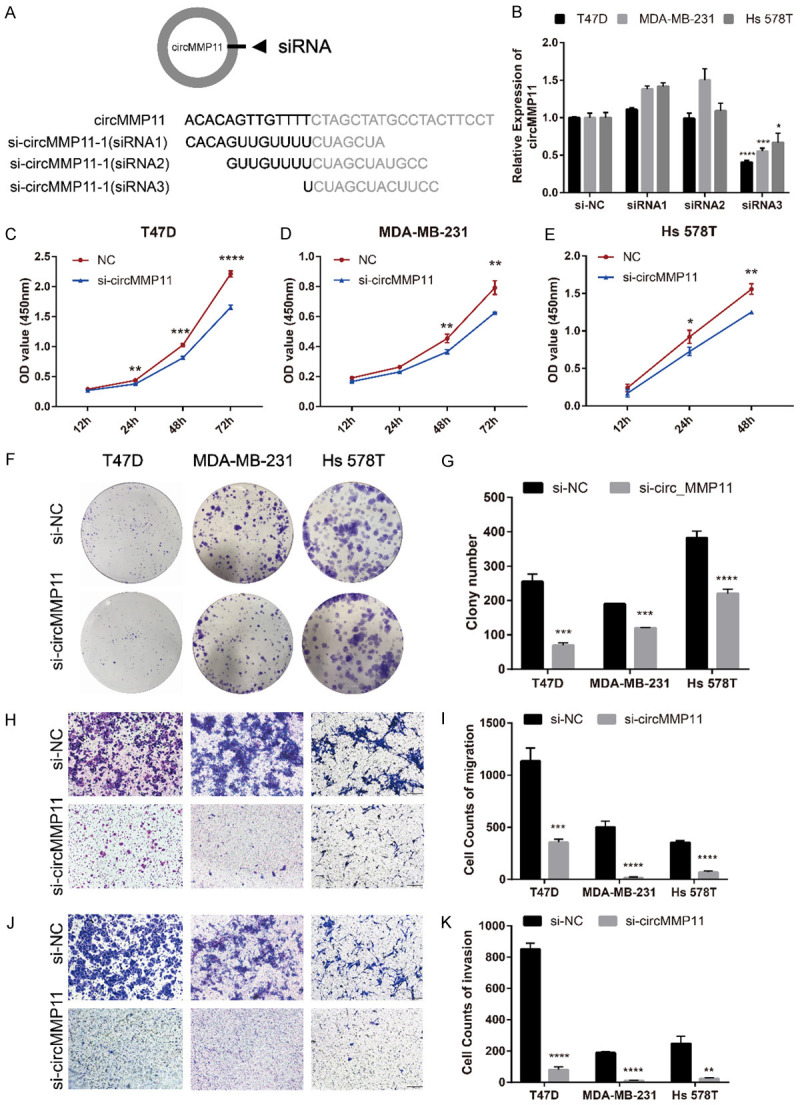
Silencing of circMMP11 inhibits BC cell proliferation, migration and invasion. A. Schematic representation of the sites of the siRNA specific to the back-splice junction of circMMP11. B. qRT-PCR analysis of circMMP11 and MMP11 mRNA expression after treatment with three siRNAs. siRNA3 has the best silencing efficacy. C-E. CCK-8 assay showed that circMMP11 knockdown significantly inhibited cell proliferation of T47D, MDA-MB-231 and Hs 578T. F, G. circMMP11 knockdown reduced the colony formation ability of these cells. H, I. Transwell assay without Matrigel revealed that cell migration was significantly suppressed in circMMP11 silenced group. J, K. Transwell assay with Matrigel revealed that knockdown of circMMP11 significantly reduced cell invasive capability. Data were shown as the mean ± SD, *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
circMMP11 acts as a sponge for miR-1204
The nuclear and cytoplasm of BC cell lines were fractionated for RNA extraction. Then we confirmed by qRT-PCR that approximately 70% or more of circMMP11 was predominantly localized in the cytoplasm of all the three BC cell lines (Figure 4A), which indicated that circMMP11 might serve as a ce-circRNA to regulate miRNAs in the cytoplasm. Therefore, we predicted the potential circRNA/miRNA interaction through Circinteractome and MRE analysis and screen out six candidate miRNA, that is, miR-661, miR-1204, miR-1207-5p, miR-4736, miR-4763-3p and miR-7150 by the standard of the most MRE and the highest context score. In addition, potential miRNA/mRNA interactions were predicted by multiple databases, including TargetScan, miRDB and TargetMiner, and our microarray data. The predicted potential RNA interactions were illustrated in Figure 4B. We detected the miRNAs expression level in BC cell lines after silencing of circMMP11 and found that miR-1204 was concurrently upregulated in all three cell lines (P < 0.05) (Figure 4C-E). Moreover, the expression of miR-1204 was significantly downregulated in BC cell lines (Figure 4G). The binding sites of miR-1204 were found based on the circMMP11 sequence (Figure 4F). Subsequently, luciferase reporter assay showed reduction of the luciferase intensity after cotransfection of wild type luciferase reporter and miR-1204 mimics, while the mutated luciferase reporter showed no such effect (Figure 4H), which confirmed the existence of the binding sites. Additionally, after transfection of miR-1204 mimics (Figure 4I), the expression of circMMP11 was suppressed (P < 0.01), which further indicates that circMMP11 could act as a sponge for miR-1204 (Figure 4J). Collectively, these data demonstrated that circMMP11 could act as a sponge for miR-1204 in BC.
Figure 4.
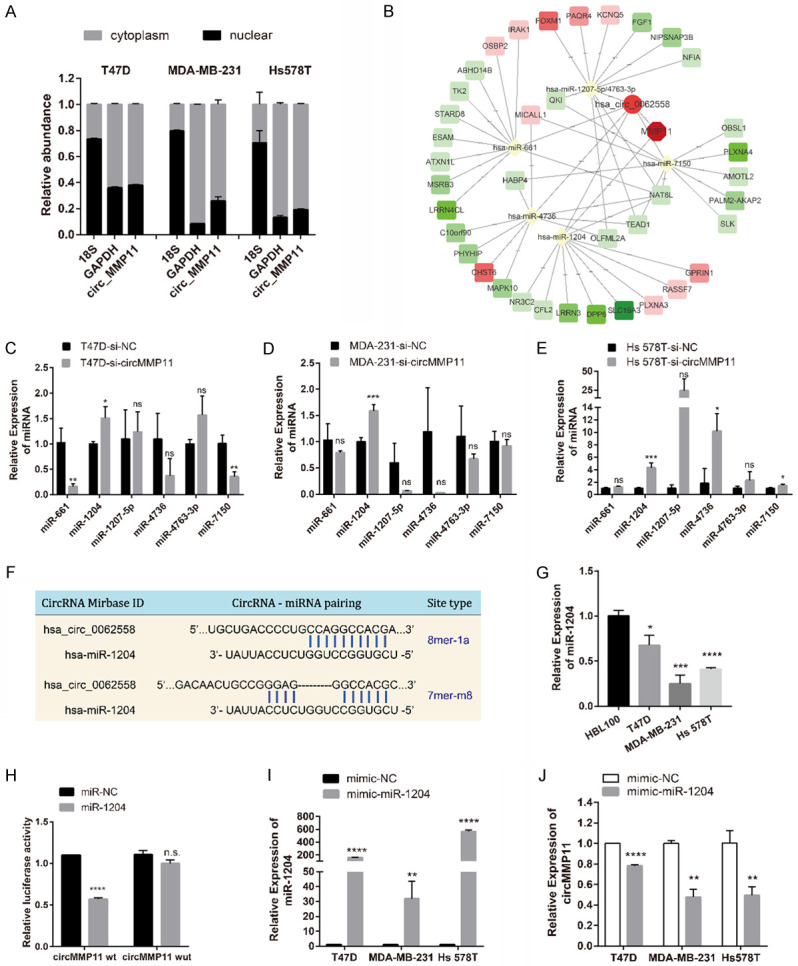
circMMP11 acts as a sponge for miR-1204. A. Approximately 70% or more of circMMP11 was located in cytoplasm of BC cell lines. B. RNA interaction network predicted by multiple database. C-E. qRT-PCR analysis of predicted miRNAs in BC cell lines transfected with control (si-NC) or circMMP11 siRNA (si-circMMP11). miR-1204 was screened out as the one candidate. F. Binding sites for circMMP11 and miR-1204. G. miR-1204 expression level was lower in BC cell lines than in normal cells. H. Dual luciferase reporter assay of HEK 293T cells cotransfected with miR-NC and miR-1204 mimics and a luciferase reporter containing circMMP11-wt or mutant constructs with mutated miR-1204 binding sites (circMMP11-mut). I, J. After transfection of miR-1204 mimics, the expression of circMMP11 was suppressed. Data were shown as the mean ± SD, *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
MMP11 is directly targeted by miR-1204 and indirectly regulated by circMMP11
An algorithm containing three databases (TargetScan, miRDB and TargetMimer) together with our microarray data were used to identify the putative targets genes of miR-1204 in human BC. We predicted four candidate genes as MMP11, GPRIN1, NR3C2 and SCL19A3 (Figure 5A). Then, we detected the expression level of these four mRNAs in BC cells transfected with miR-1204 mimics and found that MMP11 was significantly downregulated in all the three cell lines (P < 0.01) (Figure 5B). In addition, silencing of circMMP11 could also inhibit the expression of linear MMP11 (P < 0.01) (Figure 5C). We further detected the expression level of MMP11 in BC tissue samples by qRT-PCR and found that MMP11 expression level was not only upregulated in BC (Figure 5D) but also correlated with that of circMMP11 (R = 0.8347, P < 0.0001) (Figure 5E). Intriguingly, MMP11 and circMMP11 have the same binding sites of miR-1204 (Figure 5F). To confirm this finding, we constructed a luciferase reporter vector with the full-length MMP11 3’-UTR containing target sites for miR-1204 downstream of the luciferase gene (MMP11 wt) and a mutant version of psiCHECK2-MMP11-3’UTR within the seed region (MMP11 mut). The result showed a significant decrease in luciferase activity when cotransfected with miR-1204 mimics instead of with negative control while there was not a significant difference in luciferase activity when cotransfected with mutant luciferase reporter (Figure 5G). These data suggested that circMMP11 could regulate the expression of MMP11 through serving as a ce-circRNA for miR-1204 in BC.
Figure 5.
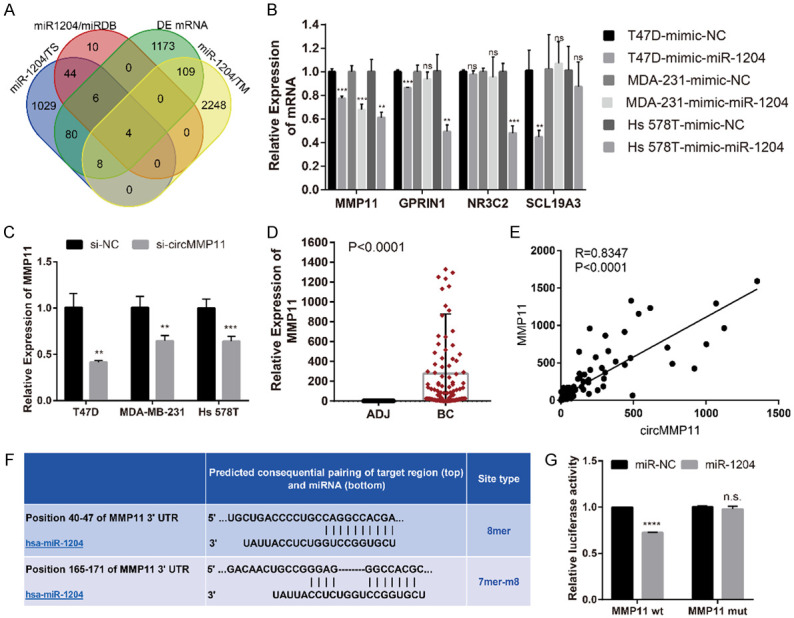
MMP11 is directly targeted by miR-1204 and indirectly regulated by circMMP11. A. Venn diagram of the four candidate mRNAs predicted through multiple databases and microarray. B. MMP11 was screened out as potential target by qRT-PCR analysis in BC cell lines transfected with control (mimic-NC) or miR-1204 mimics. C. Silencing of circMMP11 could also inhibit the expression of linear MMP11. D. qRT-PCR analysis of linear MMP11 in clinical samples (n = 113). MMP11 was upregulated in BC tissue than in adjacent normal tissue. E. The expression level of circMMP11 was positively correlated with that of MMP11. F. Binding sites for linear MMP11 and miR-1204. G. Dual luciferase reporter assay of HEK 293T cells cotransfected with miR-NC and miR-1204 mimics and a luciferase reporter with full-length MMP11 3’-UTR containing target sites for miR-1204 (MMP11 wt) or mutant constructs with mutated miR-1204 binding sites (MMP11 mut). Data were shown as the mean ± SD, *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001. ADJ: adjacent normal tissue.
CircMMP11/miR-1204/MMP11 axis affects BC progression
We further explored whether circMMP11 exerted its biological function through circMMP11/miR1204/MMP11 axis as a ce-circRNA. When miR-1204 mimic and si-circMMP11 were cotransfected into MDA-MB-231 cell line, the repression of circMMP11 and MMP11 were both reinforced. Additionally, when miR-1204 inhibitor was cotransfected with si-circMMP11, the repression effects were both reversed (Figure 6A). CCK-8 assay and colony formation assay showed that the miR-1204 mimics can intensify the proliferation-inhibiting effects induced by silencing of circMMP11 in MDA-MB-231 cells while the miR-1204 inhibitor can reverse it (Figure 6B-D). In addition, miR-1204 mimics together with si-circMMP11 also distinctly inhibited the migration ability (Figure 6E and 6F) as well as the invasion ability (Figure 6G and 6H) of MDA-MB-231 considerably more than si-circMMP11 alone in Transwell assay without or with matrix Matrigel (P < 0.01). In summary, these data demonstrated that circMMP11 might serve as a ce-circRNA for miR-1204 to regulate MMP11 expression, thereby leading to the progression of BC.
Figure 6.
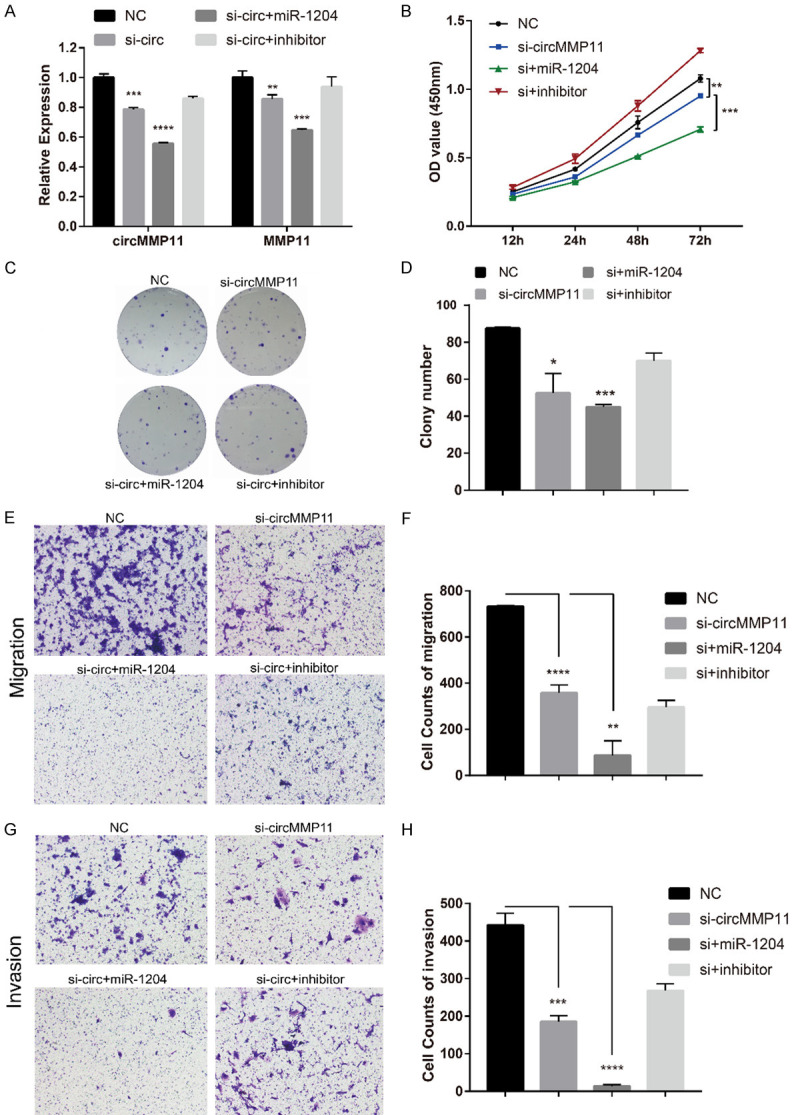
circMMP11/miR-1204/MMP11 axis affects BC progression. (A) The repression of circMMP11 and MMP11 by si-circMMP11 were both reinforced or reversed respectively when miR-1204 mimic or inhibitor and si-circMMP11 were cotransfected into MDA-MB-231. (B) CCK-8 assay and (C, D) Colony formation assay showed that the miR-1204 mimics can intensify the proliferation-inhibiting effects induced by silencing of circMMP11 in MDA-MB-231 cells while the miR-1204 inhibitor can reverse it. (E, F) miR-1204 mimics together with si-circMMP11 distinctly inhibited the migration ability of MDA-MB-231 cells. (G, H) miR-1204 mimics together with si-circMMP11 distinctly inhibited the invasion ability of MDA-MB-231 cells. Data were shown as the mean ± SD, *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
Discussion
Many RNAs, including miRNAs, lncRNAs and circRNAs have been reported to regulate breast cancer (BC) development [14-16]. The ceRNA hypothesis postulates that any RNA transcript including ncRNAs that harbor MREs can sequester miRNAs from other targets sharing the same MREs and thus regulate their expression. The functional interactions among ceRNA networks are helpful to coordinate biologic processes and can contribute to disease pathogenesis [17]. However, the amount and functions of circRNAs act as ceRNA in BC have not been determined. As is well-known, most ce-circRNAs with high expression can upregulate their HGs through ceRNA mechanisms and vice versa. To the best of our knowledge, this study is the first to combine the ceRNA microarray and TCGA data to conduct the profiling of ce-circRNA expression in BC. Additionally, to the best of our knowledge, we used the largest-ever number of tissue samples for microarray. From our study, a large amount of aberrantly expressed circRNAs were identified. We verified a never reported circRNA and termed it circMMP11. CircMMP11 was significantly upregulated in human BC tissue and correlated with lymph node metastasis and clinical stage. Functionally, downregulation of circMMP11 could extensively inhibit cell proliferation, migration and invasion, which means that circMMP11 plays a vital role in BC progression and may be a potential therapeutic target of BC.
Mechanistically, circMMP11 might function as a ce-circRNA by sponging miR-1204 to eliminate the effect on its target gene MMP11 in breast cancer progression. In this study, we found that miR-1204 could bind both circMMP11 and MMP11, suggesting that circMMP11 could function as a miR-1204 sponge to regulate MMP11 expression through ceRNA mechanism. This possibility was verified in both bioinformatic analyses and luciferase reporter assays. Moreover, knockdown of circMMP11 increased the expression of miR-1204 while reducing that of MMP11. Meanwhile, overexpression of miR-1204 amplified the effect of circMMP11 knockdown. Collectively, our data suggested that circMMP11 plays an important role in the progression of BC as a ce-circRNA which is linked with MMP11 through miR-1204.
MiR-1204 is a newly discovered microRNA whose function, to date, is complex in different cancers. On one hand, miR-1204 could promote hepatocellular carcinoma progression through activating MAPK and c-Jun/AP1 signaling [18] or could promote epithelial-mesenchymal transition and metastasis by targeting VDR in breast cancer [19]. On the other, this RNA may sensitize nasopharyngeal carcinoma cells to paclitaxel [20]. As was reported previously, MMP11 can be targeted by miR-125a-3p and miR-145 in thyroid cancer and breast cancer, respectively [21,22]. This study is the first to indicate that MMP11 can also be directly regulated by miR-1204 and indirectly regulated by circMMP11. MMP11, also known as stromelysin-3, is widely regarded as a poor prognosis paracrine factor in invasive breast cancers [23,24]. However, the mammary physiological function of MMP11 is unignorable. MMP11 was reported to be essentially involved in postnatal mammary gland morphogenesis for favoring mammary gland branching and epithelial cell outgrowth and invasion through adjacent connective tissues in the mammary gland postnatal development [25]. Additionally, whether directly targeting linear MMP11 induces adverse reaction has not been determined. Taken together, the results of this study indicate that circMMP11 might be a better therapeutic target than linear MMP11.
Conclusion
Our study indicates that circMMP11 is significantly upregulated and correlated with poor clinical characteristics in BC. CircMMP11 was observed to promote proliferation, migration and invasion of BC and regulate MMP11 expression via sponging miR-1204 to exert its ce-circRNA activity. The circMMP11/miR-1204/MMP11 axis regulates BC progression via the ceRNA mechanism. Therefore, circMMP11 represents a potential target in BC therapy.
Acknowledgements
We were grateful for the kind help and advice from Haoran Li (Department of Thoracic Surgery, Zhongshan Hospital, Fudan University). This work was supported by an individualized support project for original scientific research of Fudan University (XM03180313), a Grant (14-E-34) from the Funded Project of Baoshan District Science and Technology Commission, Shanghai, a Grant (SHDC12014207) from the Shen Kang Hospital Development Center Foundation.
Disclosure of conflict of interest
None.
Abbreviations
- BC
breast cancer
- ceRNA
competing endogenous RNA
- circRNA
circular RNA
- ce-circRNA
competing endogenous circRNA
- miRNA
microRNA
- ncRNAs
non-coding RNAs
- MMP11
matrix metalloproteinase-11
- MRE
miRNA response element
- SD
standard deviation
- TNBC
triple negative breast cancer
Supporting Information
References
- 1.Harbeck N, Gnant M. Breast cancer. Lancet. 2017;389:1134–1150. doi: 10.1016/S0140-6736(16)31891-8. [DOI] [PubMed] [Google Scholar]
- 2.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 3.Sanger HL, Klotz G, Riesner D, Gross HJ, Kleinschmidt AK. Viroids are single-stranded covalently closed circular RNA molecules existing as highly basepaired rod-like structures. Proc Natl Acad Sci U S A. 1976;73:3852–3856. doi: 10.1073/pnas.73.11.3852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer M, Loewer A, Ziebold U, Landthaler M, Kocks C, le Noble F, Rajewsky N. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333–338. doi: 10.1038/nature11928. [DOI] [PubMed] [Google Scholar]
- 5.Zhong Y, Du Y, Yang X, Mo Y, Fan C, Xiong F, Ren D, Ye X, Li C, Wang Y, Wei F, Guo C, Wu X, Li X, Li Y, Li G, Zeng Z, Xiong W. Circular RNAs function as ceRNAs to regulate and control human cancer progression. Mol Cancer. 2018;17:79. doi: 10.1186/s12943-018-0827-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang Y, Hou J, He D, Sun M, Zhang P, Yu Y, Chen Y. The emerging function and mechanism of ceRNAs in cancer. Trends Genet. 2016;32:211–224. doi: 10.1016/j.tig.2016.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Su H, Tao T, Yang Z, Kang X, Zhang X, Kang D, Wu S, Li C. Circular RNA cTFRC acts as the sponge of MicroRNA-107 to promote bladder carcinoma progression. Mol Cancer. 2019;18:27. doi: 10.1186/s12943-019-0951-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cheng Z, Yu C, Cui S, Wang H, Jin H, Wang C, Li B, Qin M, Yang C, He J, Zuo Q, Wang S, Liu J, Ye W, Lv Y, Zhao F, Yao M, Jiang L, Qin W. circTP63 functions as a ceRNA to promote lung squamous cell carcinoma progression by upregulating FOXM1. Nat Commun. 2019;10:3200. doi: 10.1038/s41467-019-11162-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tang H, Huang X, Wang J, Yang L, Kong Y, Gao G, Zhang L, Chen ZS, Xie X. circKIF4A acts as a prognostic factor and mediator to regulate the progression of triple-negative breast cancer. Mol Cancer. 2019;18:23. doi: 10.1186/s12943-019-0946-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tay Y, Rinn J, Pandolfi PP. The multilayered complexity of ceRNA crosstalk and competition. Nature. 2014;505:344–352. doi: 10.1038/nature12986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jia N, Tong H, Zhang Y, Katayama H, Wang Y, Lu W, Zhang S, Wang J. CeRNA expression profiling identifies KIT-Related circRNA-miRNA-mRNA networks in gastrointestinal stromal tumour. Front Genet. 2019;10:825. doi: 10.3389/fgene.2019.00825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xia Q, Ding T, Zhang G, Li Z, Zeng L, Zhu Y, Guo J, Hou J, Zhu T, Zheng J, Wang J. Circular RNA expression profiling identifies prostate cancer-specific circRNAs in prostate cancer. Cell Physiol Biochem. 2018;50:1903–1915. doi: 10.1159/000494870. [DOI] [PubMed] [Google Scholar]
- 13.Gyorffy B, Lanczky A, Eklund AC, Denkert C, Budczies J, Li Q, Szallasi Z. An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1,809 patients. Breast Cancer Res Treat. 2010;123:725–731. doi: 10.1007/s10549-009-0674-9. [DOI] [PubMed] [Google Scholar]
- 14.Xia E, Shen Y, Bhandari A, Zhou X, Wang Y, Yang F, Wang O. Long non-coding RNA LINC00673 promotes breast cancer proliferation and metastasis through regulating B7-H6 and epithelial-mesenchymal transition. Am J Cancer Res. 2018;8:1273–1287. [PMC free article] [PubMed] [Google Scholar]
- 15.Dvinge H, Git A, Graf S, Salmon-Divon M, Curtis C, Sottoriva A, Zhao Y, Hirst M, Armisen J, Miska EA, Chin SF, Provenzano E, Turashvili G, Green A, Ellis I, Aparicio S, Caldas C. The shaping and functional consequences of the microRNA landscape in breast cancer. Nature. 2013;497:378–382. doi: 10.1038/nature12108. [DOI] [PubMed] [Google Scholar]
- 16.Chen B, Wei W, Huang X, Xie X, Kong Y, Dai D, Yang L, Wang J, Tang H, Xie X. circEPSTI1 as a prognostic marker and mediator of triple-negative breast cancer progression. Theranostics. 2018;8:4003–4015. doi: 10.7150/thno.24106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Karreth FA, Pandolfi PP. ceRNA cross-talk in cancer: when ce-bling rivalries go awry. Cancer Discov. 2013;3:1113–1121. doi: 10.1158/2159-8290.CD-13-0202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang L, Sun L, Wang Y, Yao B, Liu R, Chen T, Tu K, Liu Q, Liu Z. miR-1204 promotes hepatocellular carcinoma progression through activating MAPK and c-Jun/AP1 signaling by targeting ZNF418. Int J Biol Sci. 2019;15:1514–1522. doi: 10.7150/ijbs.33658. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 19.Liu X, Bi L, Wang Q, Wen M, Li C, Ren Y, Jiao Q, Mao JH, Wang C, Wei G, Wang Y. miR-1204 targets VDR to promotes epithelial-mesenchymal transition and metastasis in breast cancer. Oncogene. 2018;37:3426–3439. doi: 10.1038/s41388-018-0215-2. [DOI] [PubMed] [Google Scholar]
- 20.Peng X, Cao P, Li J, He D, Han S, Zhou J, Tan G, Li W, Yu F, Yu J, Li Z, Cao K. MiR-1204 sensitizes nasopharyngeal carcinoma cells to paclitaxel both in vitro and in vivo. Cancer Biol Ther. 2015;16:261–267. doi: 10.1080/15384047.2014.1001287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tang L, Wei D, Yan F. MicroRNA-145 functions as a tumor suppressor by targeting matrix metalloproteinase 11 and Rab GTPase family 27a in triple-negative breast cancer. Cancer Gene Ther. 2016;23:258–265. doi: 10.1038/cgt.2016.27. [DOI] [PubMed] [Google Scholar]
- 22.Song M, Wang N, Li Z, Zhang Y, Zheng Y, Yi P, Chen J. miR-125a-3p suppresses the growth and progression of papillary thyroid carcinoma cell by targeting MMP11. J Cell Biochem. 2020;121:984–995. doi: 10.1002/jcb.29333. [DOI] [PubMed] [Google Scholar]
- 23.Peruzzi D, Mori F, Conforti A, Lazzaro D, De Rinaldis E, Ciliberto G, La Monica N, Aurisicchio L. MMP11: a novel target antigen for cancer immunotherapy. Clin Cancer Res. 2009;15:4104–4113. doi: 10.1158/1078-0432.CCR-08-3226. [DOI] [PubMed] [Google Scholar]
- 24.Eiro N, Cid S, Fernandez B, Fraile M, Cernea A, Sanchez R, Andicoechea A, DeAndres Galiana EJ, Gonzalez LO, Fernandez-Muniz Z, Fernandez-Martinez JL, Vizoso FJ. MMP11 expression in intratumoral inflammatory cells in breast cancer. Histopathology. 2019;75:916–930. doi: 10.1111/his.13956. [DOI] [PubMed] [Google Scholar]
- 25.Tan J, Buache E, Alpy F, Daguenet E, Tomasetto CL, Ren GS, Rio MC. Stromal matrix metalloproteinase-11 is involved in the mammary gland postnatal development. Oncogene. 2014;33:4050–4059. doi: 10.1038/onc.2013.434. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.