Abstract
This article explored LINC01619 impact on non-small cell lung cancer (NSCLS) development. LINC01619 expression in tumor tissues/normal tissues of NSCLS patients was detected by qRT-PCR and in situ hybridization. PAX6 expression in clinical tissues was researched by immunohistochemistry. After transfection, SPCA1 and A549 cells were subjected to CCK-8 assay and cell colony formation experiment. Xenograft tumor experiment was conducted. ALDH+ cells from SPCA1 and A549 cells were separated and transfected. ALDH+ cells percentage, sphere number and cancer stem cell markers expression was determined by flow cytometry, sphere culture and Western blot respectively. Luciferase reporter gene assay and RNA binding protein immunoprecipitation assay was conducted. The colocalization of LINC01619 and miR-129-5p in cells was determined by RNA fluorescence in situ hybridization experiment. Gene expression in tissues and cells were assessed by qRT-PCR and Western blot. As a result, aberrantly up-regulated LINC01619 and PAX6 in NSCLC patients predicted poor prognosis. LINC01619 overexpression in SPCA1 cells enhanced cell viability, cloning ability, and xenograft tumors volume and weigh, whereas LINC01619 silencing in A549 cells weakened the above indicators. LINC01619 overexpression promoted cancer stem cells characteristics including increasing percentage of ALDH+ cells, sphere number and cancer stem cell markers expression. LINC01619 directly inhibited miR-129-5p and the two genes were mainly colocalized in the cytoplasm. PAX6 was up-regulated in NSCLC and directly suppressed by miR-129-5p. LINC01619 promoted cells viability, cloning ability and cancer stem cells characteristics in NSCLC via the miR-129-5p/PAX6 axis. Thus, LINC01619 promotes NSCLC development via regulating PAX6 by suppressing miR-129-5p.
Keywords: NSCLC, LINC01619, miR-129-5p, PAX6, progression
Introduction
Lung cancer is one of the leading causes of cancer-related deaths worldwide, accounting for 26% of all cancer-related deaths [1]. Non-small cell lung cancer (NSCLC) is the main type of lung cancer, comprises about 80% of all lung cancer patients. Patients are more likely to have a higher survival rate after surgical resection if NSCLC is diagnosed at early stages. However, more than half of NSCLC patients have already locally advanced or distantly metastatic at the time of diagnosis [2]. Early screening of NSCLC is difficult due to the lack of typical clinical symptoms, and poor prognosis is still a serious problem faced by clinicians and patients in NSCLC treatment. Although targeted therapy for NSCLC has partially altered the treatment of lung cancer, the 5-year survival rate for lung cancer is only 18%. Even more unfortunately, the 5-year survival rate for patients with advanced lung cancer is only 4% [1]. Such a less optimistic prognosis is mainly due to the unclear underlying pathological mechanisms of NSCLC, the lower efficacy of early diagnostic biomarkers and the lack of precise treatment.
Genetic and epigenetic changes are widely recognized as the driving events for tumors genesis [3]. Long-chain non-coding RNAs (LncRNAs) have attracted great attention in the field of cancer research in recent years. Abnormally expressed LncRNAs plays a critical role in the occurrence and development of human tumors, including lung cancer [4]. LncRNAs are a type of RNAs that is greater than 200 nucleotides in length and cannot be translated into proteins [5]. Evidence suggested that LncRNAs could affect biological processes, such as proliferation, apoptosis, invasion and metastasis of tumor cells, through epigenetic regulation, transcription, post-transcriptional regulation and post-translational modification of proteins [6]. LncRNAs are expected to be novel biomarkers for tumor diagnosis, prognosis and novel target for cancer treatment.
Similar to the protein-coding genes, LncRNAs can be divided into oncogenic LncRNAs and tumor-inhibiting LncRNAs. Oncogenic LncRNAs are up-regulated in tumors, which enhance tumors progression by aggravating proliferation, migration, invasion, induction of apoptosis inhibition and drug resistance of tumor cells. However, tumor-inhibiting LncRNAs, which are down-regulated in tumors, indirectly promote the above cell phenotypes to promote the progression of tumors [7,8]. Existing literatures have identified some oncogenic LncRNAs (such as MALAT1, ANRIL, UPAT, etc.) and tumor-inhibiting LncRNAs (including NKILA, CASC2, TUBA4B, etc.) in NSCLC [9-14]. The discovery of these LncRNAs had led to prominent improvement in the diagnosis and target treatment of NSCLC. However, more LncRNAs regulating the development of NSCLC should be discovered, and related molecular mechanisms should be elucidated.
The first detailed report of LINC01619 was its regulation of diabetic nephropathy. Researchers illustrated the significantly reduced expression of LINC01619 in renal tissues, which might be associated with the pathogenesis of diabetic nephropathy [15]. As a novel discovered LncRNA, LINC01619 has not been investigated in human tumors. In the present paper, the function of LINC01619 in lung cancer was studied for the first time. Moreover, by studying the effect of LINC01619 on miR-29-5p/PAX6 axis, the potential molecular mechanism of LINC01619 regulating NSCLC development was further elucidated. This research will provide novel insight and target for the treatment of NSCLC.
Materials and methods
Patients and tissues
From 2009.9-2012.3, a total of 63 NSCLC patients who were subjected to surgical resection in China-Japan Union Hospital of Jilin University were voluntarily participate in this study. The written informed consent from every patient and the approval of ethics committee of China-Japan Union Hospital of Jilin University were both obtained.
All patients were diagnosed with NSCLC for the first time and without history of cancer-related diseases treatment. Of the 63 NSCLC patients, there were 28 patients with tumor size over 4 cm and 35 patients with tumor size less than 4 cm. According to clinical TNM stage, 16 cases were in stage I, 33 cases were in stage II and 14 cases were in stage III. The number of patients with lymph node metastasis was 24 and the other 39 cases were without lymph node metastasis. The NSCLC tissues and matched normal tissues obtained during surgery were immediately stored in liquid nitrogen. After surgery, all patients were followed up for 2000 days, and during this period, the time of death was the termination of the follow-up.
In situ hybridization (ISH)
Normal tissues and NSCLC tissues of patients were prepared into sections with 5-μm thickness. The sections were subjected to xylene dewaxing and gradual ethanol hydration. After rinsed with phosphate buffered saline (PBS), sections were digested with proteinase K and fixed with polyformaldehyde (4%). Then 20 μL 5’-digoxigenin-labeled LINC01619 probe (sequence: 5’-TGCCCATCATTAAAACCCCA-3’) (View Solid Biotechnology, Beijing, China) was added to the sections for 12 h hybridization at 55°C. Horseradish peroxidase (HRP) was used subsequently to incubate the sections for 30 min at 4°C. Sections were stained with 3,3’-diaminobenzidine tetrahydrochloride (DAB). The position of LINC01619 probe was then visualized under a microscope with dark brown particles as positive signals.
Immunohistochemistry (IHC)
PAX6 expression in normal tissues and NSCLC tissues of patients was detected by IHC. Briefly, tissues were fixed with 10% formalin and embedded in paraffin to prepare 4 μm sections. At 65°C, sections were baked for 1 h and then were rehydrated using gradient alcohol. Sodium citrate buffer solution (0.01 M, pH = 6.0) was used for antigen retrieval of the sections. H2O2 (3%) was then used to incubate sections for 10 min. Rabbit anti-PAX6 antibody (1:100, abcam, Cambridge, UK) was added onto sections for 12 h incubation at 4°C. PBS was used to wash sections for two times. Goat anti-rabbit secondry antibody (1:200, abcam, Cambridge, UK) was then added onto the sections for 30 min incubation at 37°C. DAB and hematoxylin was used for staining of sections. The sections were dehydrated and sealed in neutral resin. Under a microscope, brown particles were considered as PAX6 positive expression signals.
Cell lines
NSCLC cell lines (A549, SPCA1, H1299, H1975, H1703, SK-MES-1 and H520) and human lung bronchial epithelial cell line (BEAS-2B) were obtained from Shanghai Cell Bank of the Chinese Academy of Science (Shanghai, China). Dulbecco’s modified eagle medium (DMEM) with 10% fetal bovine serum (FBS) was used to culture cells at 37°C, 5% CO2. Penicillin G (100 U/mL) and streptomycin (100 μg/mL) were also supplemented in the DMEM.
Cell transfection
SPCA1 and A549 cells were separately prepared as single cell suspensions with serum-free DMEM to a volume of 1 * 105 cells/mL. Each cell suspension was plated into 6-well plates with a volume of 1 mL. pcDNA3.1-LINC01619 vector and empty pcDNA3.1 vector (GenePharma, Shanghai, China) were transfected into SPCA1 cells respectively (named OE group and CTRL group). LINC01619 siRNA#1, siRNA#2 and negative control (NC) (Genechem, Shanghai, China) were separately used to transfect A549 cells (named KD1 group, KD2 group and NC group respectively). miR-129-5p mimics and mimics NC were separately transfected into SPCA1 cells (named miR-129-5p group and miR-NC group), while miR-129-5p inhibitors and inhibitors NC were separately transfected into A549 cells (named miR-129-5p inh group and inh NC group). miR-129-5p mimics, mimics NC, miR-129-5p inhibitors and inhibitors NC were all purchased from GenePharma (Shanghai, China). pcDNA3.1-PAX6 plasmid (GenePharma, Shanghai, China) was used to transfect A549 cells (named PAX6 group), whereas PAX6 shRNA (GenePharma, Shanghai, China) was transfected into SPCA1 cells (named sh-PAX6 group). In addition, cotransfection was also carried out on SPCA1 cells by cotransfecting pcDNA3.1-LINC01619 vector and miR-129-5p mimics (named OE + miR-129-5p group) or by cotransfecting pcDNA3.1-LINC01619 vector and PAX6 shRNA (named OE + sh-PAX6 group). A549 cells were cotransfected by LINC01619 siRNA#1 and miR-129-5p inhibitors (named KD1 + miR-129-5p inh group) or by LINC01619 siRNA#1 and pcDNA3.1-PAX6 plasmid (named KD1 + PAX6 group). All transfection operations were conducted using Lipofectamine 2000 transfection reagent (Thermo Fisher Scientific, Waltham, MA, USA). The residual liquid in each well was replaced by DMEM (with 10% FBS) after 6 h of transfection. Cells of each group were still maintained at 37°C, 5% CO2 for continued culture.
Cells counting kit 8 (CCK-8) assay
After transfection, SPCA1 and A549 cells viability was detected by CCK-8 assay according to the instructions. Briefly, a number of 5 * 104 cells were plated into 96-well plates containing 100 μL DMEM (with 10% FBS) per well for 1, 2, 3 and 4 days culture at 37°C, 5% CO2. Cells of each group were plated with duplicate 5 wells. Thereafter, CCK-8 solution (10 μL) was added into each well to incubated cells for 4 h at 37°C. The 96-well plates were then placed on a shaker for 10 min shaking at room temperature. With a multi-well plate reader, the optical density (OD) value of each well was measured at 450 nm wavelength. Higher OD450 value indicated better cell viability.
Cell colony formation experiment
The transfected SPCA1 and A549 cells were harvested after 24 h incubation at 37°C, 5% CO2. DMEM (with 10% FBS) was used to disperse cells into single cell suspensions. Then 2 mL single cell suspension (containing 800 cells) was added into sterilized dishes with a diameter of 60 mm. All dishes were placed at 37°C, 5% CO2 for 10 days. The DMEM (with 10% FBS) was changed every 2 days. After 10 days, the residual medium in each dish was discarded and cells were fixed with polyformaldehyde (4%) for 10 min. Thereafter, crystal violet (0.1%) was used to stain cells for 10 min. The colonies more than 50 cells were counted under a microscope.
In vivo study
In this research, all animal studies had been approved by the Animal Ethics Committee of China-Japan Union Hospital of Jilin University.
Nude mice (n = 24, 6 weeks old, 25-30 g) were provided from SIPPR-BK Laboratory Animal Company (Shanghai, China). All mice were maintained in a constant temperature room at 25°C with a light/dark cycle of 12 h. Mice were given free access to water and food. With the center of the back as the injection point, mice were injected subcutaneously with SPCA1 cells transfected by LINC01619 overexpression vector or empty vector, or with A549 cells transfected by LINC01619 siRNA#1 or negative control. Each cell sample was injected into 6 mice with 1 * 105 cells. Every 3 days, Lipofectamine 2000-encapsulated LINC01619 overexpression vector or empty vector or LINC01619 siRNA#1 or negative control was separately injected into the back of the corresponding mice at the original injection point. The injection dose was 30 μg. Every 7 days, the long diameter (a) and short diameter (b) of subcutaneous tumor tissues was measured by a vernier caliper. According to the formula of V = (ab2 * 0.5), the tumor volume was calculated. On day 28, the tumor tissues were obtained and weighted after all mice were sacrificed.
Screening and transfection of ALDH+ cells from NSCLC cell lines
SPCA1 and A549 cells without any treatment were digested and incubated with ALDH-FITC antibody for 1 h at 37°C, 5% CO2. After washed twice with PBS, ALDH+ cells were screened by flow cytometry and then cultured in DMEM (10% FBS) at 37°C, 5% CO2. In the logarithmic growth phase, ALDH+ cells were collected. ALDH+ cells from SPCA1 cell line were separately transfected by LINC01619 overexpression vector (OE group) and LINC01619 empty vector (CTRL group). Meanwhile, ALDH+ cells from A549 cell line were separately transfected by LINC01619 siRNA#1 (KD1 group), LINC01619 siRNA#2 (KD2 group) and LINC01619 siRNA negative control (NC group). After 48 h of culture in DMEM (10% FBS) at 37°C, 5% CO2, ALDH+ cells of each group were detected by flow cytometry and the percentage of ALDH+ cells was calculated.
Sphere culture
After transfection, ALDH+ cells were cultured to logarithmic growth phase. Cells of each group were then sequentially subjected to trypsinization and centrifuged at 1000 r/min for 3 min. Cells at the bottom of the centrifuge tube were washed three times with PBS. After being centrifuged again for 3 min at 1000 r/min, cells were prepared as single cell suspensions using DMEM (1000 cells/mL). Each cell suspension sample was inoculated in a low attachment 35 mm dish with a volume of 1 mL, followed by being cultured in serum-free DMEM for 21 days at 37°C, 5% CO2. The DMEM was changed every 3 days. Spheres over 50 mm in diameter were counted under an inverted microscope.
Luciferase reporter gene assay
Fragments of LINC01619 wild type (WT), LINC01619 mutant type (MUT), PAX6 WT and PAX6 MUT containing the miR-129-5p binding sites were designed by GenePharma Co., Ltd, (Shanghai, China). Thefragments were separately inserted into the luciferase reporter by using pmir-GLO vector according to the instructions. SPCA1 and A549 cells were seeded in 6-well plates and transfected with miR-129-5p mimics or miR-NC. Then a total of 100 ng of pmir-GLO reporter vector containing each fragment was transfected into SPCA1 and A549 cells. After transfection, cells were incubated at 37°C, 5% CO2. The luciferase activity of cells in each well was determined by Dual Luciferase Reporter Assay System (Promega, USA).
RNA binding protein immunoprecipitation (RIP) assay
SPCA1 and A549 cells were collected at about 80% confluency. RIP lysis buffer was used to lyse cells for 30 min on ice. The supernatant of the cell lysate was obtained by centrifugation. A small amount of supernatant was stored at 4°C for Western blot analysis. A total of 100 μL supernatant was subjected to incubation for 12 h at 4°C with human anti-Ago2 antibody or negative control normal mouse IgG. Then magnetic beads were then added into the supernatant for 4 h incubation at 4°C. After 3 min centrifugation at 3000 r/min, magnetic beads were obtained and incubated with Proteinase K. The purified RNA (LINC01619 and miR-129-5p) was detected by quantitative real-time polymerase chain reaction (qRT-PCR) after the immunoprecipitated RNA being isolated.
RNA fluorescence in situ hybridization (FISH) experiment
SPCA1 and A549 cells were seeded into 6-well plates. Plates were pre-loaded with sterile slides. After 24 h culture in DMEM (10% FBS) at 37°C and 5% CO2, 4% paraformaldehyde was added into each well to fix cells for 10 min. Then cells were underwent 5 min incubation with Triton-100 (0.25%). Cy3-labeled LINC01619 probe and miR-129-5p probe (RiboBio, Guangzhou, China) was added into each well. RNA FISH was carried out with FISH kit (RiboBio, Guangzhou, China). Cells attached to the slide were further stained by 4, 6-diamidino-2-phenylindole (DAPI), followed by being observed under a confocal laser scanning microscope (LSM 510 Zeiss, Jena, Germany). The red fluorescent region was regarded as the expression position of LINC01619, while green fluorescent region was considered to be the expression position of miR-129-5p. The blue fluorescent region represented the position of the nucleus.
qRT-PCR
Total RNA in tissues/cells was extracted according to the Trizol reagent instructions. cDNA was transcribed using High Capacity cDNA Reverse Transcription kit (Applied Biosystems, CA, USA) according to the manual. qRT-PCR was performed on a TP800 Real-time PCR machine (Takara, Japan) using the two-step PCR amplification procedure: first step, pre-denaturation at 95°C for 30 s; second step, 40 cycles of 95°C for 5 s, 60°C for 30 s and 72°C for 30 s. By 2-ΔΔCt method, the relative LINC01619 and miR-129-5p expression in tissues/cells were determined with U6 as the internal reference, while the relative expression of ALDH, SOX2, NANOG, OCT4, CD133, CD44 and PAX6 in tissues/cells was calculated with β-actin as the internal reference.
Western blot
Cells were harvested after 48 h incubation and washed with pre-cooled PBS for three times. Cell lysate was added into cells to lyse cells on ice for 1 h. Then the mixture was centrifuged for 15 min at 12000 r/min, 4°C. The concentration of total protein in the supernatant was determined using BCA kit (Beyotime Biotechnology, Shanghai, China). Using 30 μg protein samples, protein was separated via sodium dodecylsulphate polyacrylamide gel electrophoresis (SDS-PAGE), followed by being transferred to a PVDF membrane. By adding 5% skimmed milk powder, protein was blocked for 2 h at room temperature. Primary antibodies (rabbit anti-PAX6, 1:1000, Covance, Princeton, NJ; rabbit anti-ALDH, 1:1000, Abgent Company, California, USA; mouse anti-CD133, 1:1000, Miltenyi Biotec, Bergisch Gladbach, Germany; rabbit anti-SOX2, 1:1000, Chemicon, Temecula, CA; rabbit anti-NANOG, 1:1000, GeneTex, San Antonio, TX; rat anti-CD44, 1:1000, DSHB, Iowa, IA; rabbit anti-OCT4, 1:1000, Sigma-Aldrich Company Ltd., Gillingham, UK; mouse anti-β-actin, 1:1000, Sigma-Aldrich, USA.) were then added onto the membrane to incubate protein overnight at 4°C. The membrane was washed with tris-buffered saline tween-20 (TBST) for three times. Horseradish peroxidase-labeled secondary antibodies (Solarbio science&technology co., ltd., Beijing, China) were subsequently added onto the membrane for 2 h incubation at room temperature. The membrane was undergone washing with TBST for three times. Electrogenerated chemiluminescence (ECL) was added onto the membrane. The gray value of each protein band was analyzed by using the Image J software (National Institutes of Health, Maryland, USA) with β-actin as the control.
Statistical analysis
All experiments were repeated three times. The relationship between LINC01619 expression and overall survival of patients within 2000 days was analyzed by Kaplan-Meier analysis. Pearson’s correlation analysis was used to evaluate the correlation between two genes expression level. SPSS 19.0 was used for statistical analysis. Data was exhibited in the form of mean ± standard deviation (SD) and P < 0.05 indicated statistically significant difference. Differences between two groups were compared by Student’s t-test, while comparison of differences among at least three groups used one way Analysis of Variance (ANOVA).
Results
Significantly up-regulated LINC01619 in NSCLC predicted poor prognosis
LINC01619 expression in 63 pairs of normal tissues and NSCLC tissues was evaluated by qRT-PCR. The result showed prominently up-regulated LINC01619 expression level in NSCLC tissues than that in normal tissues (P < 0.0001) (Figure 1A). The correlation between LINC01619 expression and main clinical features (tumor size, TNM stage and lymph node metastasis) of NSCLC patients was assessed. Patients with tumor size greater than 4 mm (n = 28) had markedly higher LINC01619 expression level than those with tumor sizes less than 4 mm (n = 35) (P = 0.0032) (Figure 1B). Meanwhile, LINC01619 expression level in patients with stage II (n = 33) was significantly higher than those with stage I (n = 16) (P = 0.0299), but was dramatically lower than those with stage III (n = 14) (P < 0.0001) (Figure 1C). Furthermore, patients with lymph node metastasis (n = 24) exhibited remarkably higher LINC01619 expression level in NSCLC tissues than those without lymph node metastasis (n = 39) (P = 0.0012) (Figure 1D). To more intuitively observe the LINC01619 expression, ISH was performed on 2 pairs of normal tissues/NSCLC tissues of patients. Compared with normal tissues (Normal#1 and Normal#2), much higher LINC01619 expression was found in NSCLC tissues (NSCLC#1 and NSCLC#2) (Figure 1E). According to the LINC01619 expression level in NSCLC tissues, patients were divided into High LINC01619 expression group (n = 31) and Low LINC01619 expression group (n = 32). As shown in Figure 1F, patients in High LINC01619 expression group experienced significantly lower 2000-day overall survival than those in Low LINC01619 expression group (P = 0.0142). Therefore, LINC01619 expression in NSCLC patients was significantly up-regulated, and was predicted poor prognosis of NSCLC patients.
Figure 1.
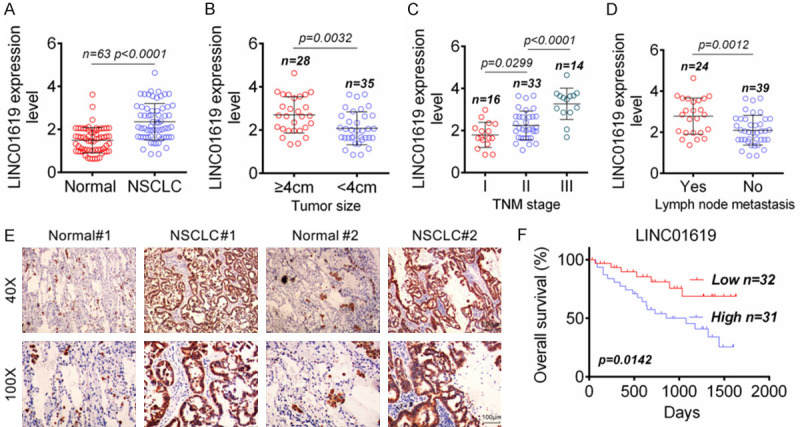
Significantly up-regulated LINC01619 in NSCLC predicted poor prognosis. A. LINC01619 was prominently up-regulated in NSCLC tissues than that in normal tissues. B. High LINC01619 expression indicated large tumor size. C. High LINC01619 expression indicated advanced TNM stage. D. High LINC01619 expression indicated positive lymph node metastasis. E. ISH showed that LINC01619 expression was increased in NSCLC tissues than that in normal tissues. F. High LINC01619 expression was obviously associated with low 2000-day overall survival of NSCLC patients.
LINC01619 promoted NSCLC cells growth in vitro and in vivo
As shown in Figure 2A, LINC01619 expression in NSCLC cell lines (A549, SPCA1, H1299, H1975, H1703, SK-MES-1 and H520) was found to be obviously up-regulated when compared with lung bronchial epithelial cell line (BEAS-2B) (P < 0.01). Of the seven NSCLC cell lines, A549 cell line had the highest LINC01619 expression level, whereas SPCA1 cell line showed the lowest LINC01619 expression level. Therefore, in the following studies, LINC01619 in SPCA1 cells was overexpressed and LINC01619 in A549 cells was silenced in order to study the effects of LINC01619 on NSCLC cells phenotype.
Figure 2.
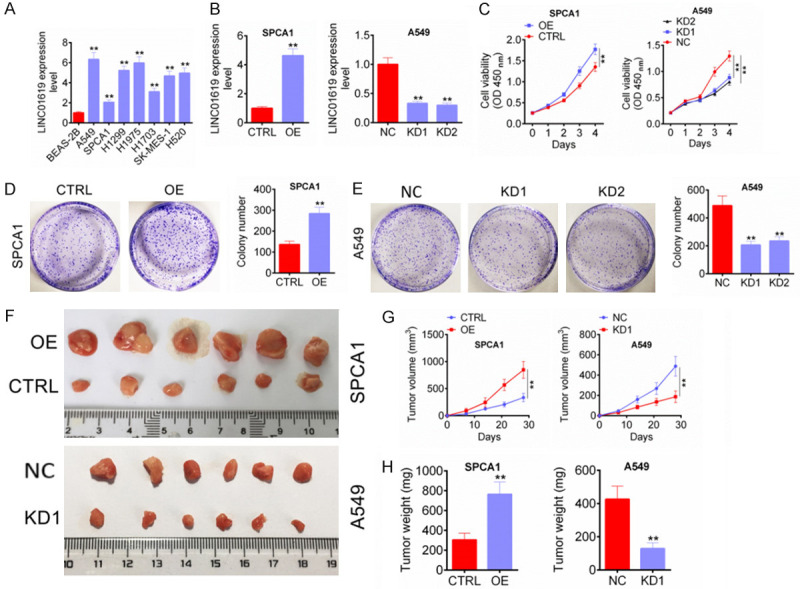
LINC01619 promoted NSCLC cells growth in vitro and in vivo. A. LINC01619 expression was up-regulated in NSCLC cell lines (A549, SPCA1, H1299, H1975, H1703, SK-MES-1 and H520) than that in lung bronchial epithelial cell line (BEAS-2B). B. LINC01619 expression in SPCA1 and A549 cells was successfully regulated by transfection. C. LINC01619 up-regulation enhanced SPCA1 and A549 cells viability, whereas LINC01619 down-regulation weakened SPCA1 and A549 cells viability. D and E. LINC01619 up-regulation increased SPCA1 and A549 cells cloning ability, but LINC01619 down-regulation decreased SPCA1 and A549 cells cloning ability. F. On the 28th day of subcutaneous injection, tumor tissues of nude mice in each group were obtained and photographed. G and H. LINC01619 up-regulation promoted NSCLC cells growth in vivo, whereas LINC01619 down-regulation inhibited NSCLC cells growth in vivo. **P < 0.01.
After transfected, LINC01619 expression in SPCA1 and A549 cells were researched by qRT-PCR. SPCA1 cells of OE group exhibited much higher LINC01619 expression than those of CTRL group (P < 0.01). However, when compared with NC group, much decreased LINC01619 expression was observed in A549 cells of KD1 group and KD2 group (P < 0.01) (Figure 2B). Thus, LINC01619 expression in SPCA1 and A549 cells was successfully regulated by transfection. The two cell lines viability in vitro was assessed CCK-8 assay. The result illustrated markedly higher OD450 value of SPCA1 cells in OE group at day 4 when compared with CTRL group (P < 0.01). However, at the same time, aberrantly lower OD450 value of A549 cells in KD1 group and KD2 group was found when compared with NC group (P < 0.01) (Figure 2C). Cell colony formation experiment revealed that, compared with CTRL group, more colony number of SPCA1 cells in OE group was observed (P < 0.01) (Figure 2D). On the opposite, A549 cells in KD1 group and KD2 group had much less colony number when relative to NC group (P < 0.01) (Figure 2E). These data demonstrated that LINC01619 was contributed to NSCLC cells growth in vitro.
LINC01619 effect on NSCLC cells growth in vivo was studied by xenograft in nude mice. The subcutaneous tumor tissues of each group were obtained on the 28th day after subcutaneous injection (Figure 2F). The tumor volume during 28 days and tumor weight on day 28 was measured and the results were shown in Figure 2G, 2H. Compared with CTRL group, mice of OE group had much higher tumor volume and weight at day 28 (P < 0.01). Conversely, obviously lower tumor volume and weight was occurred in mice of KD1 group when compared with NC group (P < 0.01). Thus, LINC01619 inhibited NSCLC cells growth in vivo.
LINC01619 promoted characteristics of tumor stem cells in NSCLC
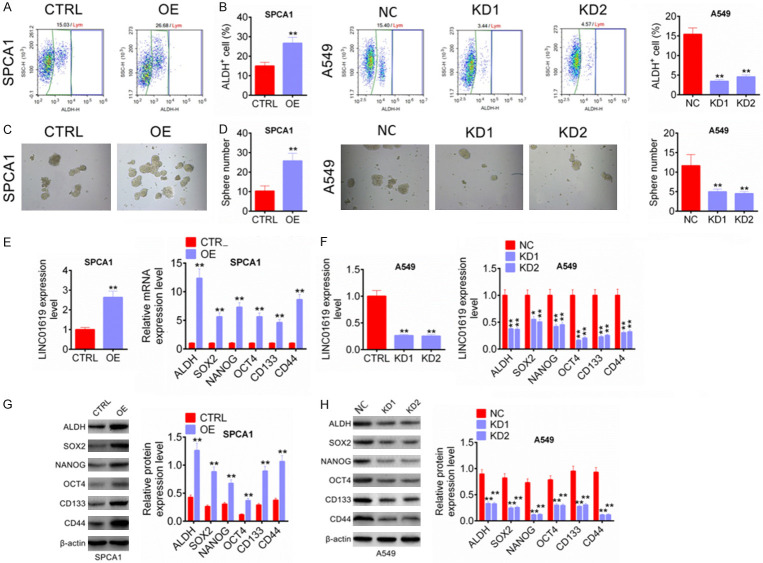
ALDH+ cells in human tumor cell lines had been shown to have characteristics of cancer stem cells. In this study, ALDH+ cells from SPCA1 and A549 cell lines were separated and transfected. As shown in Figure 3A, 3B, SPCA1 cells of OE group exhibited much higher percentage of ALDH+ cells than CTRL group (P < 0.01). However, A549 cells of KD1 group and KD2 group had significantly lower percentage of ALDH+ cells than NC group (P < 0.01). Furthermore, the sphere number of SPCA1 cells in OE group was markedly higher than that in CTRL group (P < 0.01), but A549 cells in KD1 group and KD2 group showed less sphere number than that in NC group (P < 0.01) (Figure 3C, 3D). The expression of cancer stem cell markers was detected by qRT-PCR and western blot. Compared with CTRL group, SPCA1 cells of OE group showed remarkably higher relative mRNA and protein expression of ALDH, SOX2, NANOG, OCT4, CD133 and CD44 (P < 0.01) (Figure 3E, 3G). However, dramatically lower relative mRNA and protein expression of ALDH, SOX2, NANOG, OCT4, CD133 and CD44 was found in A549 cells of KD1 group and KD2 group when relative to NC group (P < 0.05 or P < 0.01) (Figure 3F, 3H). All of these data indicated that LINC01619 promoted characteristics of tumor stem cells in NSCLC.
Figure 3.
LINC01619 promoted characteristics of tumor stem cells in NSCLC. A and B. LINC01619 up-regulation elevated percentage of ALDH+ cells, while LINC01619 down-regulation reduced percentage of ALDH+ cells. C and D. LINC01619 up-regulation increased the sphere number of ALDH+ cells, but LINC01619 down-regulation decreased the sphere number of ALDH+ cells. E and G. LINC01619 up-regulation enhanced the expression of cancer stem cell markers (ALDH, SOX2, NANOG, OCT4, CD133 and CD44) in SPCA1 and A549 cells. F and H. LINC01619 down-regulation declined the expression of cancer stem cell markers (ALDH, SOX2, NANOG, OCT4, CD133 and CD44) in SPCA1 and A549 cells. *P < 0.05 and **P < 0.01.
LINC01619 served as a competing endogenous LncRNA (ceRNA) to sponge miR-129-5p in NSCLC
Through microRNA target prediction tool, LINC01619 was found to possess binding site for miR-129-5p. LINC01619 WT and MUT fragments containing the binding site for miR-129-5p was designed and shown in Figure 4A. Results from luciferase reporter assay showed that, compared with miR-NC group, SPCA1 and A549 cells of miR-129-5p group exhibited much lower relative luciferase activity after the LINC01619 WT fragment being inserted (P < 0.01). No significant difference in relative luciferase activity was observed between miR-NC group and miR-129-5p group after the LINC01619 MUT fragment being inserted (Figure 4B). RIP assay exhibited that compared with IgG, LINC01619 preferentially enriched the miRNA ribonucleoprotein complex containing Ago2 (Figure 4C). Both LINC01619 and miR-129-5p were dramatically enriched in the Ago2 pellet when relative to IgG or Input control (P < 0.01) (Figure 4D). RNA FISH experiment showed that LINC01619 and miR-129-5p were mainly colocalized in the cytoplasm (Figure 4E). The regulation of miR-129-5p by LINC01619 was further verified by PCR after transfection. As shown in Figure 4F, compared with CTRL group, SPCA1 cells of OE group had much decreased miR-129-5p expression (P < 0.01). However, A549 cells of KD1 group and KD2 group were with obviously increased miR-129-5p expression when compared with NC group (P < 0.01). Detection in clinical tissue samples showed markedly reduced miR-129-5p expression in NSCLC tissues than that in normal tissues (P < 0.0001) (Figure 4G). Prominently negative correlation between LINC01619 and miR-129-5p expression level was detected in NSCLC tissues (P < 0.0001) (Figure 4H). These results illustrated that LINC01619 served as a competing endogenous LncRNA (ceRNA) to sponge miR-129-5p in NSCLC.
Figure 4.
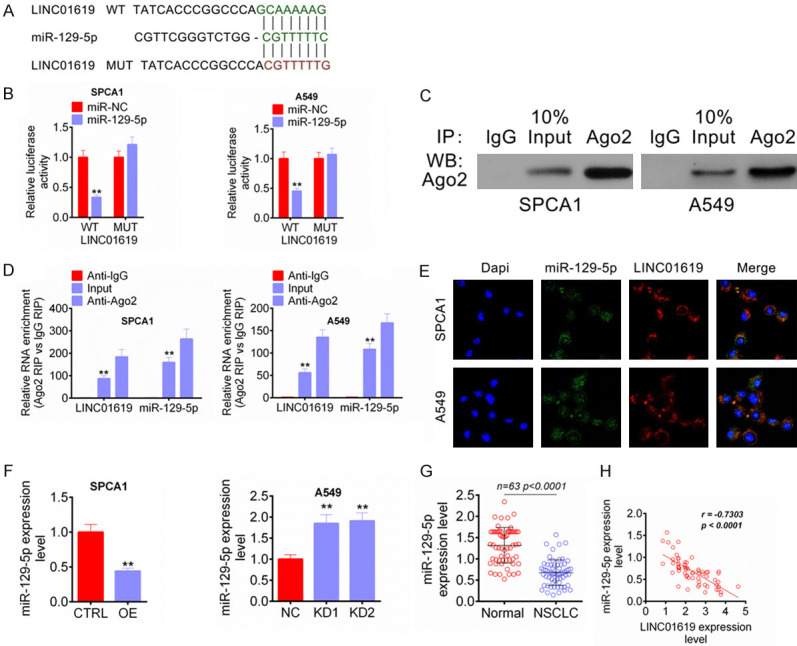
LINC01619 served as a competing endogenous LncRNA (ceRNA) to sponge miR-129-5p in NSCLC. A. LINC01619 WT and MUT fragments containing the binding site for miR-129-5p was designed. B. Luciferase reporter assay indicated that miR-129-5p was directly inhibited by LINC01619. C and D. RIP assay revealed that LINC01619 was binding to miR-129-5p. E. RNA FISH experiment showed that LINC01619 and miR-129-5p were mainly colocalized in the cytoplasm. F. LINC01619 up-regulation reduced miR-129-5p expression in SPCA1 cells, whereas LINC01619 down-regulation elevated miR-129-5p expression in A549 cells. G. miR-129-5p expression was reduced in NSCLC tissues than that in normal tissues. H. In NSCLC tissues, prominently negative correlation between LINC01619 and miR-129-5p expression level was found. **P < 0.01.
PAX6 was directly suppressed by miR-129-5p
PAX6 was a transcription factor which acted as an oncogene or tumor suppressor in several tumors. This study researched the regulation of miR-129-5p on PAX6 via luciferase reporter assay. Relative to miR-NC group, SPCA1 and A549 cells of miR-129-5p group were with remarkably lower relative luciferase activity after PAX6 WT fragment being inserted (P < 0.01). The insertion of PAX6 MUT fragment did not obviously affect the relative luciferase activity between miR-NC group and miR-129-5p group (Figure 5A). After transfection, PAX6 mRNA expression in SPCA1 and A549 cells was assessed by qRT-PCR. Compared with CTRL group, SPCA1 cells of OE group had much higher PAX6 mRNA expression (P < 0.01). Meanwhile, SPCA1 cells of miR-129-5p group showed significantly lower PAX6 mRNA expression than that of miR-NC group and OE + miR-129-5p group (P < 0.01). For A549 cells, obviously lower PAX6 mRNA expression was found in KD1 group relative to NC group (P < 0.01). Aberrantly higher PAX6 mRNA expression was occurred in miR-129-5p inh group compared to inh NC group and KD1 + miR-129-5p inh group (P < 0.01) (Figure 5B). The PAX6 protein expression in SPCA1 and A549 cells of each group was also showed a similar trend to PAX6 mRNA expression (Figure 5C, 5D). Detection from clinical tissue samples showed much higher PAX6 mRNA expression in NSCLC tissues than that in normal tissues (P < 0.0001) (Figure 5E). Furthermore, PAX6 mRNA expression in NSCLC tissues was positively correlated with LINC01619 expression (P < 0.0001), but was negatively correlated with miR-129-5p expression (P < 0.0001) (Figure 5F, 5G). Western blot analysis was performed on 5 pairs of NSCLC tumor tissues (T1, T2, T3, T4 and T5) and normal tissues (N1, N2, N3, N4 and N5). The results indicated that, compared with normal tissues, dramatically higher PAX6 protein expression was observed in NSCLC tumor tissues (P < 0.01) (Figure 5H). PAX6 mRNA and protein expression in clinical tissues was further researched by qRT-PCR and IHC respectively. As shown in Figure 5I, patients with advanced TNM stage had higher PAX6 mRNA expression level in their tumor tissues, accompanied by statistically significant differences (P < 0.05 or P < 0.01). IHC exhibited more PAX6 positive expression signals in tumor tissues (sample I, II and III) than that in normal tissues. Thus, PAX6 was elevated in NSCLC and was directly suppressed by miR-129-5p.
Figure 5.
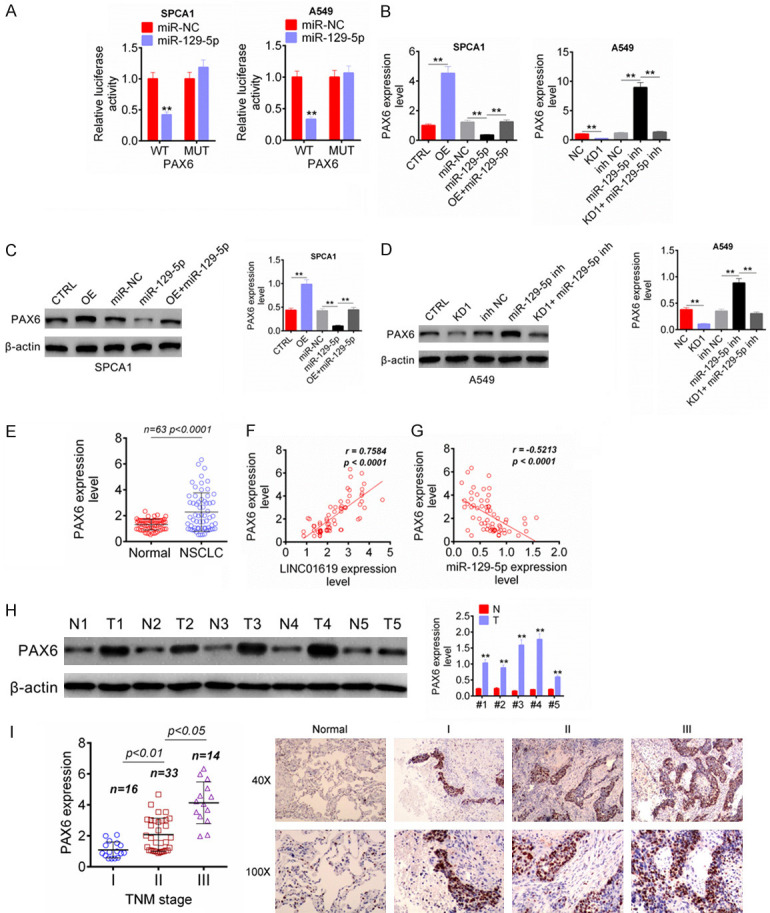
PAX6 was directly suppressed by miR-129-5p. A. Luciferase reporter assay indicated that PAX6 was directly suppressed by miR-129-5p. B. LINC01619 enhanced PAX6 mRNA expression and miR-129-5p inhibited PAX6 mRNA expression in NSCLC cells. C and D. LINC01619 enhanced PAX6 protein expression and miR-129-5p inhibited PAX6 protein expression in NSCLC cells. E. PAX6 mRNA expression was increased in NSCLC tissues than that in normal tissues. F and G. In NSCLC tissues, PAX6 expression was positively correlated with LINC01619 and negatively correlated with miR-129-5p. H. PAX6 protein expression was increased in 5 pairs of NSCLC tumor tissues (T1, T2, T3, T4 and T5) than that in normal tissues (N1, N2, N3, N4 and N5). I. qRT-PCR and IHC indicated that, patients with advanced TNM stage had higher PAX6 mRNA expression level in their tumor tissues, and more PAX6 positive expression signals were found in tumor tissues (sample I, II and III) than that in normal tissues. **P < 0.01.
LINC01619 promoted tumor cells proliferation and stem cell characteristics in NSCLC via the miR-129-5p/PAX6 axis
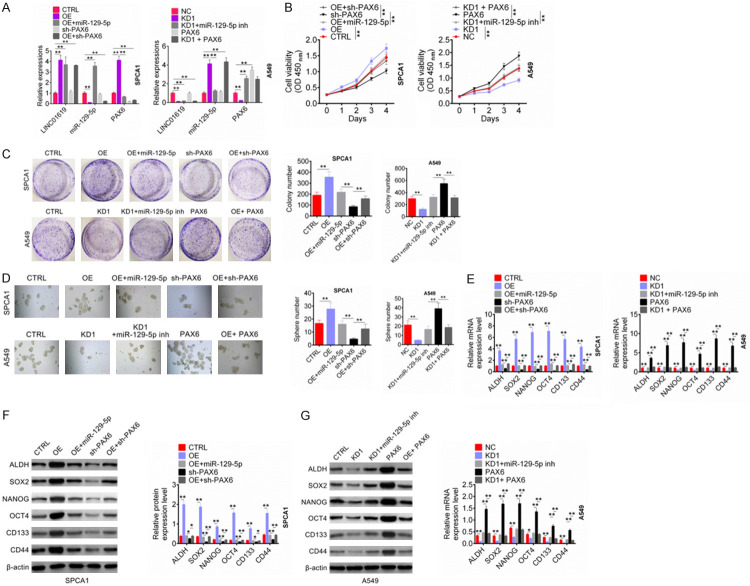
The qRT-PCR was performed to further verify the effect of LINC01619 on miR-129-5p and PAX6 expression. For SPCA1 cells, OE group had much higher LINC01619, PAX6 expression and lower miR-129-5p expression than that of CTRL group (P < 0.01). Relative to CTRL group, OE + miR-129-5p group showed higher LINC01619 and miR-129-5p expression (P < 0.01). Compared with OE group, OE + miR-129-5p group exhibited much higher miR-129-5p expression (P < 0.01). Meanwhile, when relative to CTRL, SPCA1 cells of OE + sh-PAX6 group showed obviously higher LINC01619 expression, lower miR-129-5p and PAX expression (P < 0.01). These data indicated that LINC01619 overexpression inhibited miR-129-5p expression but promoted PAX expression. In addition, for A549 cells, compared with NC group, much lower LINC01619, PAX6 expression and higher miR-129-5p expression was found in KD1 group, and obviously higher PAX6 expression was observed in PAX6 group (P < 0.01). Relative to NC group, KD1 + miR-129-5p inh group showed much lower LINC01619 expression (P < 0.01). Cells of KD1 + miR-129-5p inh group exhibited obviously lower miR-129-5p expression and higher PAX6 expression than that of KD1 group (P < 0.01). Much lower LINC01619 expression and higher miR-129-5p expression was found in KD1 + PAX6 when compared with NC group (P < 0.01). Thus, knockdown of LINC01619 increased miR-129-5p expression but decreased PAX6 expression (Figure 6A).
Figure 6.
LINC01619 promoted tumor cells proliferation and stem cell characteristics in NSCLC via the miR-129-5p/PAX6 axis. A. LINC01619 overexpression inhibited miR-129-5p expression but promoted PAX expression. Conversely, knockdown of LINC01619 increased miR-129-5p expression but decreased PAX6 expression. B. LINC01619 promoted NSCLC cells viability via regulating the miR-129-5p/PAX6 axis. C. LINC01619 enhanced NSCLC cells cloning ability via regulating the miR-129-5p/PAX6 axis. D. LINC01619 increased the sphere number of ALDH+ cells via regulating the miR-129-5p/PAX6 axis. E-G. LINC01619 elevated the mRNA and protein expression of cancer stem cell markers in NSCLC cells via regulating the miR-129-5p/PAX6 axis. *P < 0.05 and **P < 0.01.
To detect whether LINC01619 affected NSCLC development through regulating miR-129-5p/PAX6 axis, this paper further examined NSCLC cells biological behavior after transfection. From CCK-8 assay, it could be observed that compared with SPCA1 cells in CTRL group, much higher OD450 value was found in OE group at day 4 (P < 0.01). At the same time point, SPCA1 cells of sh-PAX6 group exhibited much lower OD450 value than OE + miR-129-5p group and OE + sh-PAX6 group (P < 0.01). Conversely, for A549 cells, much lower OD450 value at day 4 was found in KD1 group when compared with NC group (P < 0.01). Meanwhile, relative to PAX6 group, significantly lower OD450 value at day 4 was seen in A549 cells of KD1 + miR-129-5p inh group and KD1 + PAX6 group (P < 0.01) (Figure 6B). Cell colony formation experiment showed that, compared with the colony number of SPCA1 cells in CTRL group, it was markedly increased in OE group (P < 0.01). Prominently decreased colony number of SPCA1 cells in sh-PAX6 group was occurred when compared with OE + miR-129-5p group and OE + sh-PAX6 group (P < 0.01). However, A549 cells of KD1 group showed remarkably lower colony number than that of NC group (P < 0.01). When compared with the colony number of A549 cells in PAX6 group, it was dramatically reduced in KD1 + miR-129-5p inh group and KD1 + PAX6 group (P < 0.01) (Figure 6C).
In addition, ALDH+ cells from SPCA1 and A549 cell lines were also transfected. As shown in Figure 6D, the sphere number of SPCA1 cells in OE group was much higher than CTRL group (P < 0.01). Obviously lower sphere number of SPCA1 cells in sh-PAX6 group was found relative to OE + miR-129-5p group and OE + sh-PAX6 group (P < 0.01). In terms of A549 cells, less sphere number was observed in KD1 group when compared with NC group (P < 0.01). A549 cells of PAX6 group had aberrantly more sphere number than that of KD1 + miR-129-5p inh group and KD1 + PAX6 group (P < 0.01). Expression of cancer stem cell markers in ALDH+ cells exhibited that, compared with CTRL group, SPCA1 cells of OE group showed significantly higher mRNA and protein expression of ALDH, SOX2, NANOG, OCT4, CD133 and CD44 (P < 0.01). Much lower mRNA and protein expression of ALDH, SOX2, NANOG, OCT4, CD133 and CD44 was found in SPCA1 cells of sh-PAX6 group compared to OE + miR-129-5p group and OE + sh-PAX6 group (P < 0.05 or P < 0.01). For A549 cells, the mRNA and protein expression of ALDH, SOX2, NANOG, OCT4, CD133 and CD44 in KD1 group was obviously lower than that of NC group (P < 0.01). However, A549 cells of PAX6 group exhibited aberrantly higher mRNA and protein expression of ALDH, SOX2, NANOG, OCT4, CD133 and CD44 than KD1 + miR-129-5p inh group and KD1 + PAX6 group (P < 0.05 or P < 0.01) (Figure 6E-G). Collectively, these data confirmed that LINC01619 promoted tumor cells proliferation and stem cell characteristics in NSCLC via the miR-129-5p/PAX6 axis.
Discussion
NSCLC is one of the most common malignant tumors in the world due to its high morbidity and mortality. Most NSCLC cases are in advanced stage at the time of initial diagnosis, and face a serious risk of recurrence even after successful radical resection [16]. The occurrence and progression of NSCLC was multi-gene, multi-stage complex process involving the abnormal expression of a variety of proto-oncogenes, tumor suppressor genes, growth factors and receptors, cell adhesion factors and DNA repair genes [17]. The finding of effective early diagnostic markers and therapeutic targets for NSCLC is one of the main ways to improve the therapeutic effect. Thus, it is necessary to deeply explore the molecular mechanism of tumorigenesis and development of NSCLC. In this research, we identified LINC01619 as an oncogene in NSCLC. Significantly up-regulated LINC01619 expression in NSCLC patients were prominently associated with poor prognosis, such as larger tumor size, lymph node metastasis, advanced TNM stage and lower 2000-day overall survival. Mechanism studies indicated that LINC01619 promoted NSCLC cells proliferation and cancer stem cell characteristics by enhancing PAX6 expression via sponging miR-129-5p.
LncRNAs is a kind of cell-derived RNA small molecule, which regulates the expression of multiple genes through epigenetic regulation, transcriptional regulation and post-transcriptional regulation [18]. In recent years, studies have shown that LncRNAs was involved in many important regulatory processes such as X chromosome silencing, genomic imprinting, chromatin modification, transcriptional activation and interference and intranuclear transport [19,20]. Most of the findings confirmed that LncRNAs was involved in the development of human tumors as ceRNA by sponging miRNAs [21]. In this paper, LINC01619, as an oncogene in NSCLC, was also proved to be served as a ceRNA to sponge miR-129-5p in NSCLC. As far as we know, this was the first report on LINC01619 in human tumors.
In this research, overexpression of LINC01619 promoted growth of NSCLC cells in vivo and in vitro. The up-regulated LINC01619 promoted cancer stem cell characteristics of NSCLC cells, such as increasing ALDH+ cells percentage, enhancing the ability of cancer stem cells to grow into spheres and promoting the expression of cancer stem cells surface markers (ALDH, SOX2, NANOG, OCT4, CD133 and CD44). Cancer stem cells are a subset cancer cells with unlimited proliferative potential and high carcinogenic effects. These cells have highly tumorigenic and drug resistance, and are widely involved in the processes of tumorigenesis, development, metastasis, recurrence and drug resistance [22]. A cancer stem cell generally can form into two cells through asymmetric splitting. One cell retained the ability of the original cancer stem cell. The other cell differentiates into a non-tumor stem cell, and then becomes a tumor mass or a substitute for the dead tumor cell. Therefore, it is believed that cancer stem cells are the main reason for maintaining tumor progression and the eradication of cancer stem cells is the way to cure tumors [22,23]. This article confirmed that inhibiting of LINC01619 could interfere with the characteristics of cancer stem cells in NSCLC. This finding provided further theoretical basis for LINC01619 as a molecular target in the targeted treatment of NSCLC.
miR-129-5p was discovered to be directly and negatively regulated by LINC01619 in this paper. Existing data demonstrated that miR-129-5p was a tumor suppressor gene in many human malignant tumors. For instance, Brest et al. [24] revealed that miR-129-5p was able to induce cell death of thyroid cancer cells. It enhanced the anti-proliferative effects of other anticancer drugs such as etoposide. Liu and colleagues researched that miR-129-5p was aberrantly down-regulated in gastric cancer tissues. Low miR-129-5p expression indicated poor prognosis (including lymph node invasion and large tumor size) of gastric cancer cases. However, overexpression of miR-129-5p by transfection was helpful to reduce gastric cancer cells proliferation and invasion abilities [25]. In NSCLC, miR-129-5p expression was found to be reduced in CD133+ stem cells. Up-regulation of miR-129-5p decreased stem cell markers expression, self-renewal ability, stemness and chemoresistance [26]. This research also confirmed the tumor suppressive effect of miR-129-5p in NSCLC.
PAX6 is a transcription factor widely expressed in multiple tumors. It is belonged to the PAX family and participates in the regulation of embryonic development of tissues and organs [27]. Several researches illustrated the anti-cancer effect of PAX6 in several cancers, such as prostate cancer, glioblastoma and retinoblastoma [28-30]. However, Ooki et al. [31] explored that PAX6 was oncogene in lung adenocarcinoma. It drove cancer cells toward a stem-like state. Luo et al. [32] indicated the abnormally increased expression of PAX6 in NSCLC. PAX6 overexpression remarkably enhanced NSCLC cells proliferation and invasion abilities. Our data also suggested that, PAX6 acted as an oncogene in NSCLC and was overexpressed in NSCLC tissues and cells. More importantly, PAX6 expression was promoted by LINC01619 via sponging miR-129-5p.
Collectively, this article demonstrated that the aberrantly up-regulated LINC01619 in NSCLC was associated with poor prognosis of patients. Overexpression of LINC01619 could promote the development of NSCLC by enhancing tumor cells proliferation and stem cell characteristics. The mechanism involved in this process was that LINC01619 enhanced PAX6 expression via sponging miR-129-5p. These findings suggested that LINC01619 might be served as a novel therapeutic target for the treatment of NSCLC.
Disclosure of conflict of interest
None.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer Statistics, 2017. Ca Cancer J Clin. 2017;67:7. doi: 10.3322/caac.21387. [DOI] [PubMed] [Google Scholar]
- 2.Román M, Baraibar I, López I, Nadal E, Rolfo C, Vicent S, Gilbazo I. KRAS oncogene in non-small cell lung cancer: clinical perspectives on the treatment of an old target. Mol Cancer. 2018;17:33. doi: 10.1186/s12943-018-0789-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liu MZ, Mcleod HL, He FZ, Chen XP, Zhou HH, Shu Y, Zhang W. Epigenetic perspectives on cancer chemotherapy response. Pharmacogenomics. 2014;15:699–715. doi: 10.2217/pgs.14.41. [DOI] [PubMed] [Google Scholar]
- 4.Meseure D, Drak AK, Nicolas A, Bieche I, Morillon A. Long noncoding RNAs as new architects in cancer epigenetics, prognostic biomarkers, and potential therapeutic targets. Biomed Res Int. 2015;2015:320214. doi: 10.1155/2015/320214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mcaninch D, Roberts CT, Bianco-Miotto T. Mechanistic insight into long noncoding RNAs and the placenta. Int J Mol Sci. 2017;18:1371. doi: 10.3390/ijms18071371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huang X, Zhou X, Hu Q, Sun B, Deng M, Qi X, Lü M. Advances in esophageal cancer: a new perspective on pathogenesis associated with long non-coding RNAs. Cancer Lett. 2018;413:94–101. doi: 10.1016/j.canlet.2017.10.046. [DOI] [PubMed] [Google Scholar]
- 7.Kunej T, Obsteter J, Pogacar Z, Horvat S, Calin GA. The decalog of long non-coding RNA involvement in cancer diagnosis and monitoring. Crit Rev Clin Lab Sci. 2014;51:344–357. doi: 10.3109/10408363.2014.944299. [DOI] [PubMed] [Google Scholar]
- 8.Camacho CV, Choudhari R, Gadad SS. Long noncoding RNAs and cancer, an overview. Steroids. 2018;133:1–12. doi: 10.1016/j.steroids.2017.12.012. [DOI] [PubMed] [Google Scholar]
- 9.Lu Z, Yuan L, Wang J, Yun C, Sun S, Huang J, Chen Z, Jie H. Long non-coding RNA NKILA inhibits migration and invasion of non-small cell lung cancer via NF-κB/Snail pathway. J Exp Clin Cancer Res. 2017;36:54. doi: 10.1186/s13046-017-0518-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.He X, Liu Z, Su J, Yang J, Yin D, Han L, Wei D, Guo R. Low expression of long noncoding RNA CASC2 indicates a poor prognosis and regulates cell proliferation in non-small cell lung cancer. Tumour Biol. 2016;37:9503–9510. doi: 10.1007/s13277-016-4787-6. [DOI] [PubMed] [Google Scholar]
- 11.Li S, Mei Z, Hu HB. The lncRNA MALAT1 contributes to non-small cell lung cancer development via modulating miR-124/STAT3 axis. J Cell Physiol. 2018;233:6679–6688. doi: 10.1002/jcp.26325. [DOI] [PubMed] [Google Scholar]
- 12.Nie FQ, Sun M, Yang JS, Xie M, Xu TP, Xia R, Liu YW, Liu XH, Zhang EB, Lu KH. Long noncoding RNA ANRIL promotes non-small cell lung cancer cell proliferation and inhibits apoptosis by silencing KLF2 and P21 expression. Mol Cancer Ther. 2015;14:268–277. doi: 10.1158/1535-7163.MCT-14-0492. [DOI] [PubMed] [Google Scholar]
- 13.Chen J, Hu L, Wang J, Zhang F, Chen J, Xu G, Wang Y, Pan Q. Low Expression LncRNA TUBA4B is a poor predictor of prognosis and regulates cell proliferation in non-small cell lung cancer. Pathol Oncol Res. 2017;23:265–270. doi: 10.1007/s12253-016-0089-y. [DOI] [PubMed] [Google Scholar]
- 14.Wang H, Cao D, Wu F. Long noncoding RNA UPAT promoted cell proliferation via increasing UHRF1 expression in non-small cell lung cancer. Oncol Lett. 2018;16:1491–1498. doi: 10.3892/ol.2018.8829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bai X, Geng J, Li X, Wan J, Liu J, Zhou ZM, Liu X. Long non-coding RNA LINC01619 regulates miR-27a/FOXO1 and endoplasmic reticulum stress-mediated podocyte injury in diabetic nephropathy. Antioxid Redox Signal. 2018;29:355–376. doi: 10.1089/ars.2017.7278. [DOI] [PubMed] [Google Scholar]
- 16.Fidler IJ. The pathogenesis of cancer metastasis: the ‘seed and soil’ hypothesis revisited. Nat Rev Cancer. 2003;3:453–458. doi: 10.1038/nrc1098. [DOI] [PubMed] [Google Scholar]
- 17.Weber GF. Metabolism in cancer metastasis. Int J Cancer. 2016;138:2061–2066. doi: 10.1002/ijc.29839. [DOI] [PubMed] [Google Scholar]
- 18.Liu W, Ma R, Yuan Y. Post-transcriptional regulation of genes related to biological behaviors of gastric cancer by long noncoding RNAs and MicroRNAs. J Cancer. 2017;8:4141–4154. doi: 10.7150/jca.22076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hui L, Kimberly V, Martin P, Riccardo F, Ioana BN, Slack FJ, Calin GA. Junk DNA and the long non-coding RNA twist in cancer genetics. Oncogene. 2015;34:5003–5011. doi: 10.1038/onc.2014.456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Han P, Chang CP. Long non-coding RNA and chromatin remodeling. RNA Biol. 2015;12:1094–1098. doi: 10.1080/15476286.2015.1063770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miao L, Liu HY, Zhou C, He X. LINC00612 enhances the proliferation and invasion ability of bladder cancer cells as ceRNA by sponging miR-590 to elevate expression of PHF14. J Exp Clin Cancer Res. 2019;38:143. doi: 10.1186/s13046-019-1149-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Koren S, Bentires-Alj M. Breast tumor heterogeneity: source of fitness, hurdle for therapy. Mol Cell. 2015;60:537–546. doi: 10.1016/j.molcel.2015.10.031. [DOI] [PubMed] [Google Scholar]
- 23.Zhong C, Wu JD, Fang MM, Pu LY. Clinicopathological significance and prognostic value of the expression of the cancer stem cell marker CD133 in hepatocellular carcinoma: a meta-analysis. Tumour Biol. 2015;36:7623–7630. doi: 10.1007/s13277-015-3487-y. [DOI] [PubMed] [Google Scholar]
- 24.Patrick B, Sandra L, Veronique H, Olivier B, Virginie GT, Christelle B, Chimene M, Geraldine R, José S, Pascal B. MiR-129-5p is required for histone deacetylase inhibitor-induced cell death in thyroid cancer cells. Endocr Relat Cancer. 2011;18:711–719. doi: 10.1530/ERC-10-0257. [DOI] [PubMed] [Google Scholar]
- 25.Liu Q, Jiang J, Fu Y, Liu T, Zhang X. MiR-129-5p functions as a tumor suppressor in gastric cancer progression through targeting ADAM9. Biomed Pharmacother. 2018;105:420–427. doi: 10.1016/j.biopha.2018.05.105. [DOI] [PubMed] [Google Scholar]
- 26.Ma Z, Cai H, Zhang Y, Chang L, Cui Y. MiR-129-5p inhibits non-small cell lung cancer cell stemness and chemoresistance through targeting DLK1. Biochem Biophys Res Commun. 2017;490:309–316. doi: 10.1016/j.bbrc.2017.06.041. [DOI] [PubMed] [Google Scholar]
- 27.Blake JA, Ziman MR. Pax genes: regulators of lineage specification and progenitor cell maintenance. Development. 2014;141:737–751. doi: 10.1242/dev.091785. [DOI] [PubMed] [Google Scholar]
- 28.Shyr CR, Tsai MY, Yeh S, Kang HY, Chang YC, Wong PL, Huang CC, Huang KE, Chang C. Tumor suppressor PAX6 functions as androgen receptor Co-repressor to inhibit prostate cancer growth. Prostate. 2010;70:190–199. doi: 10.1002/pros.21052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hegge B, Sjøttem E, Mikkola I. Generation of a PAX6 knockout glioblastoma cell line with changes in cell cycle distribution and sensitivity to oxidative stress. BMC Cancer. 2018;18:496. doi: 10.1186/s12885-018-4394-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Meng B, Wang Y, Li B. Suppression of PAX6 promotes cell proliferation and inhibits apoptosis in human retinoblastoma cells. Int J Mol Med. 2014;34:399–408. doi: 10.3892/ijmm.2014.1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ooki A, Dinalankara W, Marchionni L, Tsay JJ, Goparaju C, Maleki Z, Rom WN, Pass HI, Hoque MO. Epigenetically regulated PAX6 drives cancer cells toward a stem-like state via GLI-SOX2 signaling axis in lung adenocarcinoma. Oncogene. 2018;37:5967–5981. doi: 10.1038/s41388-018-0373-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Luo J, Li H, Zhang C. MicroRNA-7 inhibits the malignant phenotypes of nonsmall cell lung cancer in vitro by targeting Pax6. Mol Med Rep. 2015;12:5443–5448. doi: 10.3892/mmr.2015.4032. [DOI] [PubMed] [Google Scholar]