Abstract
Long non-coding RNAs (LncRNAs) have been shown to be involved in diverse cellular and physiological processes. Recent studies have proved their potential as the prospective therapeutic targets for cancer treatment. Herein, we examined the role of LncRNA CASC2 in human colon cancer. The gene expression analysis showed that LncRNA CASC2 is significantly suppressed in colon cancer tissues and cell lines. The immunohistochemistry also showed considerable increase of the Ki67 in colon cancer tissues suggestive of their aggressiveness. Overexpression of CASC2 inhibited the growth of HT-29 cells. The inhibition of HT-29 growth was due to the induction of apoptosis which was accompanied by upsurge of Bax, depletion of Bcl-2 and activation of caspase-3 cleavage. Electron microscopic analysis showed CASC2 overexpression also induced autophagy in the HT-29 cells which was associated with increase in LC3B II and Beclin 1 expression. Bioinformatic approaches and dual luciferase assay showed that CASC2 controls the TRIM16 via microRNA-214 axis. TRIM16 was found to be overexpressed in all the colon cancer tissues and cell lines. Overexpression of CASC2 caused significant inhibition of TRIM16. Additionally, silencing of TRIM16 resulted in the inhibition of HT-29 cell growth similar to that of CASC2 overexpression. Taken together, CASC2 may prove to be an important therapeutic target for colon cancer treatment.
Keywords: Colon cancer, CASC2, apoptosis, autophagy
Introduction
Recent studies have reported the dysregulation of long non-coding RNAs (LncRNA) in several cancer types suggestive of their roles in cancer development and progression [1]. With increase in the knowledge about the cellular and physiological roles of LncRNAs, they have considered to be prospective therapeutic targets for cancer management [2]. Among LncRNAs, CASC2 has been implicated in several cancer related processes [3]. The LncRNA CASC2 has been found to control the cisplatin sensitivity of cervical cancer cells [4]. It has also been shown to control the invasion of the osteosarcoma cells [5]. Zhang et al reported that LncRNA CASC2 modulates the expression of TGF-β to suppress the growth and metastasis of human breast cancer cells [6]. In yet another study, Cao et al showed that CASC2 regulates the proliferation of renal cancer cells via modulation of microRNA-21 expression [7]. Nonetheless, the role of CASC2 is yet to be explored in colon cancer. Consistently, this study was designed to investigate the role of CASC2 in human colon cancer. Owing to the lethality of colon cancer, it causes significant number of human deaths across the globe. In 2017, approximately 0.1 million new colon cancer cases were reported in US only [8]. Generally, colon cancer treatment includes surgery, chemotherapy and/or radiotherapy [9]. However, because of severe side effects of colon cancer chemotherapy, the life quality of the patients is badly impaired [10]. Hence, it is believed that the identification of safer drugs and potent therapeutic targets may help to resolve the problem and enable the efficient management of colon cancer. Herein, we report that LncRNA CASC2 is suppressed in colon cancer and its overexpression inhibits the colon cancer growth via induction of apoptosis, autophagy and modulation of TRIM16 expression. Taken together, LncRNA CASC2 may prove beneficial in the colon cancer treatment and warrants further investigations.
Materials and methods
Tissue samples, cell lines and culture conditions
The clinical specimens of cervical cancer tissues and normal surrounding tissues were procured from the patients undergoing treatment at the Department of General surgery, the fourth Medical Center of PLA General Hospital, Beijing, China, 100853. The written consent from the patients was taken prior to the collection of tissues. The institutional ethical guidelines were strictly followed for the collection and laboratory usage of clinical specimens. The study was approved by the research ethical committee of the institute under approval number MCGH57/2018. The specimens were snap-frozen and stored in liquid nitrogen. The normal colon CCD-18Co and the colon cancer cell lines (HT-29, SW-948, RKO and SW480) were obtained from Type Culture Collection of Chinese Academy of Sciences, Shanghai, China. The cell lines were subjected to culturing in Roswell Park Memorial Institute 1640 (RPMI 1640; Gibco, Carlsbad, CA, USA) medium supplemented with 10% fetal bovine serum and 0.2% penicillin and streptomycin (Invitrogen, Carlsbad California, United States). All cells were cultured in a 5% CO2 incubation chamber at 37°C.
RNA isolation and qRT-PCR analysis
Total RNA was isolated from the cells with RNAiso reagent (Takara, Japan). The RNA extracted was subjected to DNAse I (Thermo Fisher Scientific) treatment. cDNA synthesis was performed with the help of PrimescriptTM reverse transcription reagent (Takara, Japan). Quantitative Real Time-PCR was performed on QuantStudio 3 Real Time-PCR system (Thermo Fisher Scientific) following manufacturer guidelines. The relative expression was normalized with human GADPH gene and 2-ΔΔCt method was used to quantify the relative expression values. RT primers were synthesized through Primer3 v. 0.4.0 (http://bioinfo.ut.ee/primer3-0.4.0/) online software.
Analysis of cell proliferation
The viability of HT-29 cells was monitored by WST-1 assay. In brief, HT-29 cells were cultured in 96 well plates at the density of 2×105 cells/well. The cells were then transfected with NC or pcDNA-CASC2 and again incubated for 24 h at 37°C. This was followed by the incubation of the cells with WST-1 at 37°C for 4 h. The absorbance was then measured at 450 nm using a victor 3 microplate reader to determine the viability at 0, 12, 24, 48 and 96 h time intervals.
Analysis of cell death
The HT-29 cells were transfected with suitable constructs and cultured for 24 h at 37°C and then fixed with ethanol (70%) for 20 min. The cells were then subjected to PBS washing and subsequently stained with a DAPI or a solution of AO and EB. Finally, the cells were examined under microscope to detect the induction of apoptosis. The HT-29 cells were transfected with appropriate constructs and then incubated for 48 h at 37°C. The cells were then dissociated with help of trypsin and then PBS washed. The cells were then resuspended in 1X binding buffer which was followed by the addition of 5 µL of annexin V-FITC and propidium iodide (PI). The cell culture was then placed in dark room for 15 min. The apoptosis percentage was then evaluated by a flow cytometer.
Electron microscopy
The induction of autophagy was assessed by electron microscopy. The transfected HT-29 cells were collected by trypsinization and subsequently subjected to washing which was followed by fixation in glutaraldehyde (2%) in phosphate buffer (0.1 M). The HT-29 cells were then post-fixed in osmium tetroxide (1%). This was followed by the treatment of the cells with ethanol and embedding in resin. The thin section were then cut with the help of an ultramicrotome and subjected to electron microscopy.
Dual luciferase assay
Dual luciferase assay was performed for interactional study of miR-214 with 3’-UTR of CASC2 and between miR-214 and TRIM16. Here, the HT-29 cells were co-transfected either with miR-NC and pGL3-wiltype (WT)/mutated (MUT) 3’UTR stretches or miR-214 mimics and pGL3-wiltype (WT)/mutated (MUT) 3’UTR of CASC2 or TRIM16 Following which, the measurement of luciferase activity was made through Dual Luciferase Reporter system (Promega Corporation) using Renilla luciferase for normalization.
Immunohistochemical analysis
For immunohistochemistry (IHC), sections of the cancer and normal adjacent tissues were incubated with the primary antibody overnight at 4°C, followed by incubation with biotinylated anti-mouse (Vector Laboratories, Burlingame, CA, USA) or at RT for 1 h. The samples were incubated in the avidin-biotin complex solution (Vector Laboratories), they were then stained with 3, 3’ diaminobenzidine (DAB; Vector Laboratories) for 5 min. The signals were analyzed using a bright field microscope (Nikon TE-2000U).
Western blotting
The colon cancer tissues and cell lines were lysed and the protein concentration in each sample was measured by Bradford assy. Equal concentrations of the proteins from each sample were loaded on 10% SDS polyacrylamide gel and then followed by shifting to polyvinylidene fluoride membranes. Blocking of the membrane was then performed by fat-free milk (5%) in TBST. This was followed by incubation with a primary antibody for 24 h at 4°C. Subsequently, a secondary antibody was added at 25°C for about 2 h. The bands of interest were finally observed by chemiluminescence.
Results
LncRNA CASC2 is suppressed in colon cancer cells
The determination of the expression of LncRNA CASC2 in 15 colon cancer and 15 normal adjacent tissues revealed the expression of LncRNA CASC2 to be significantly suppressed in colon cancer tissues by upto 9.5 folds (Figure 1A). Gene expression analysis of LncRNA CASC2 in colon cancer cells showed LncRNA CASC2 to be significantly (7.2 folds) downregulated in colon cancer cell lines (Figure 1B). The HT-29 colon cancer cell line which showed the lowest expression of CASC2 was selected for further studies. Additionally Immunochemical analysis of the colon cancer revealed remarkable increase of the Ki67 expression suggestive of aggressiveness of the tumor (Figure 2).
Figure 1.
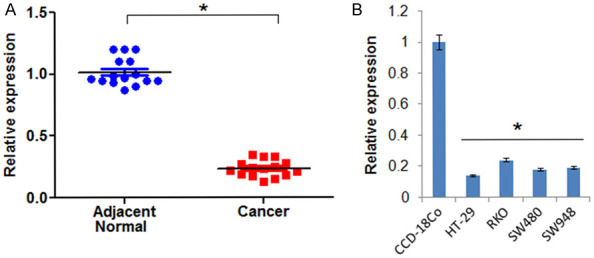
LncRNA CASC2 is significantly downregulated in cervical cancer. A. Expression of LncRNA CASC2 in colon cancer and normal adjacent tissue. B. Expression of CASC2 in normal and cervical cancer cell lines. The experiments were performed in triplicate and expressed as mean ± SD (*P < 0.05).
Figure 2.
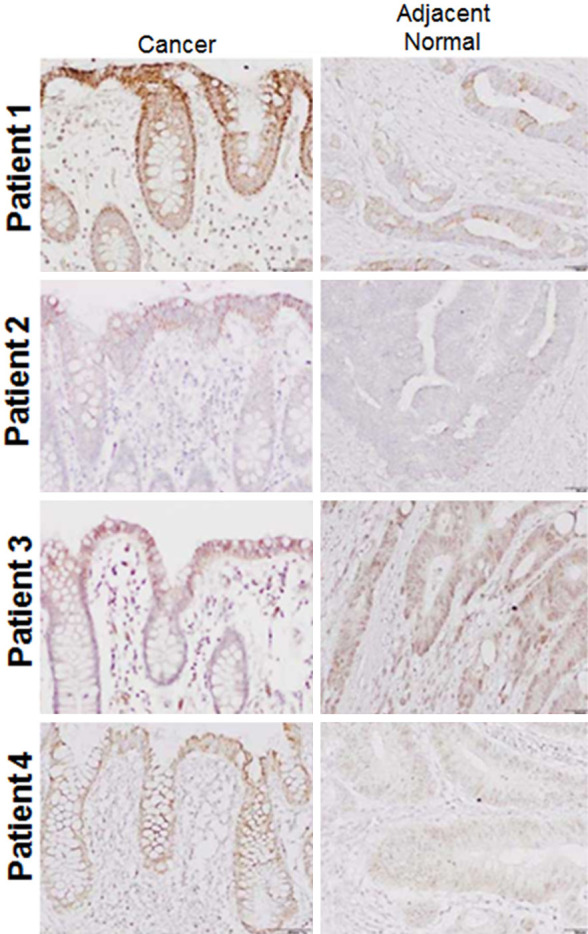
IHC analysis showing the expression of Ki67 in colon cancer and normal adjacent tissues. The experiments were performed in triplicate.
LncRNA CASC2 inhibits the viability of in HT-29 cells
The NC and LncRNA CASC2 transfected HT-29 cells were subjected to WST-1 assay. The findings showed that LncRNA CASC2 overexpression suppressed the viability of the HT-29 cells (Figure 3A). The colony formation assay also confirmed the inhibition of growth as the number of colon was significantly lower in pcDNA-CASC2 transfected cells in comparison to the NC transfected HT-29 cells (Figure 3B).
Figure 3.
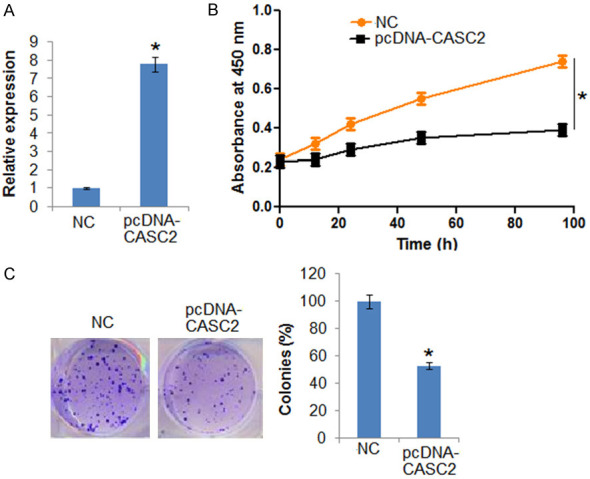
LncRNA CASC2 inhibits the growth of colon cancer cells. A. Expression of CASC2 in NC and CASC2 overexpressing HT-29 cells. B. Viability of the NC and CASC2 overexpressing HT-29 cells. C. Colony formation of the NC and CASC2 overexpressing HT-29 cells. The experiments were performed in triplicate and expressed as mean ± SD (*P < 0.05).
LncRNA CASC2 induces apoptosis and autophagy in HT-29 cells
To gain insights about the mechanisms responsible for reduction in the viability of the pcDNA-CASC2 transfected HT-29 cells, DAPI and AO/EB staining assay were carried out. The DAPI staining assay indicated that LncRNA CASC2 overexpression increased the DAPI positive HT-29 cells (Figure 4A). The AO/EB staining showed that LncRNA CASC2 overexpression promoted the nuclear fragmentation of the HT-29 cells (Figure 4B). The annexin V/PI staining showed that LncRNA CASC2 overexpression increased the apoptotic HT-29 cells from 1.16% in NC to about 24.32% in LncRNA CASC2 (Figure 4C). The LncRNA CASC2 also enhanced the expression of Bax and depleted the expression of Bcl-2 further confirming the apoptotic cell death. The LncRNA CASC2 activated the cleavage of the caspase-3 in colon cancer cells (Figure 4D). The electron microscopic analysis of the pcDNA-CASC2 transfected HT-29 showed that CASC2 overexpression leads to the development of autophagosomes suggestive of autophagy (Figure 5A). Additionally, western blotting also confirmed the induction of autophagy in pcDNA-CASC2 transfected HT-29 cells as the expression of LC3-II and Beclin-1 was found to increase (Figure 5B).
Figure 4.
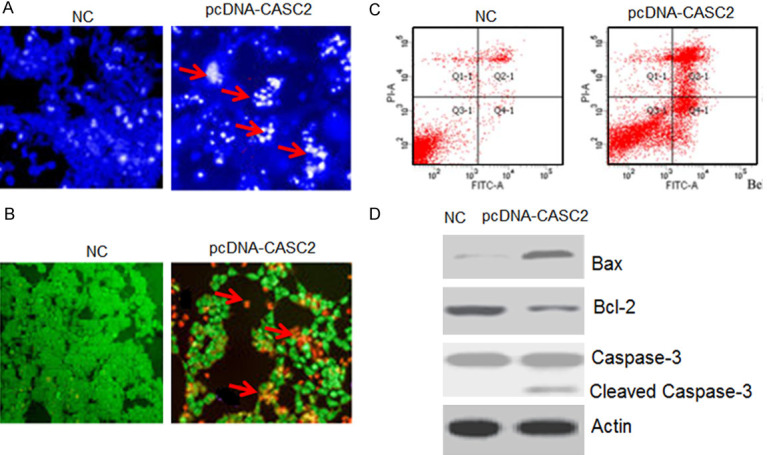
LncRNA CASC2 promotes apoptosis in HT-29 cells. A. DAPI staining of NC and CASC2 overexpressing HT-29 cells. B. AO/EB staining of NC and CASC2 overexpressing HT-29 cells. C. Annexin V/PI staining of NC and CASC2 overexpressing HT-29 cells. D. Western blots showing the expression of Bax, Caspase-3 and Bcl-2 in NC and CASC2 overexpressing HT-29 cells. The experiments were performed in triplicate.
Figure 5.
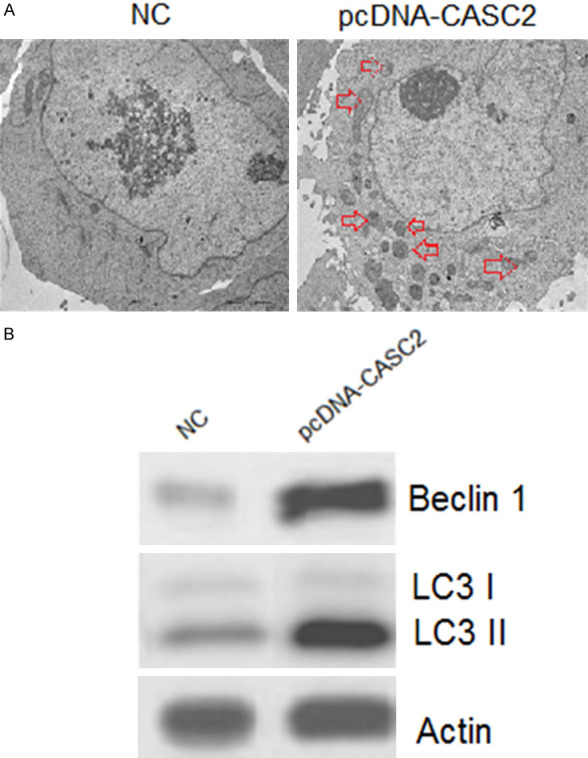
LncRNA CASC2 induces autophagy in HT-29 cells. A. Electron microscopic analysis of NC and CASC2 overexpressing HT-29 cells. B. Western blots showing the expression of LC3B I, II and Beclin 1 in NC and CASC2 overexpressing HT-29 cells. The experiments were performed in triplicate.
LncRNA CASC2 exerts its effects via modulation of TRIM16 expression
Bioinformatic approaches showed that CASC2 has binding sites for microRNA (miR)-214 and dual luciferase assay confirmed the interaction between miR-24 and CASC2 (Figure 6A and 6B). Next, bioinformatic analysis showed that miR-214 exerts its effects by targeting TRIM16 which was also confirmed by dual luciferase assay. These results suggest that LncRNA CASC2 might be exerting its effects via miR-214 induced suppression of TRIM16 (Figure 6C and 6D). Consequently, the expression of TRIM16 was upregulated in all the colon cancer cells and cell lines TRIM16 (Figure 6E and 6F). The western blots showed that CASC2 overexpression inhibits the expression of TRIM16 (Figure 6G). Additionally, it was found that silencing of TRIM16 could also inhibit the viability of the HT-29 cells similar to that of CASC2 silencing (Figure 6H).
Figure 6.
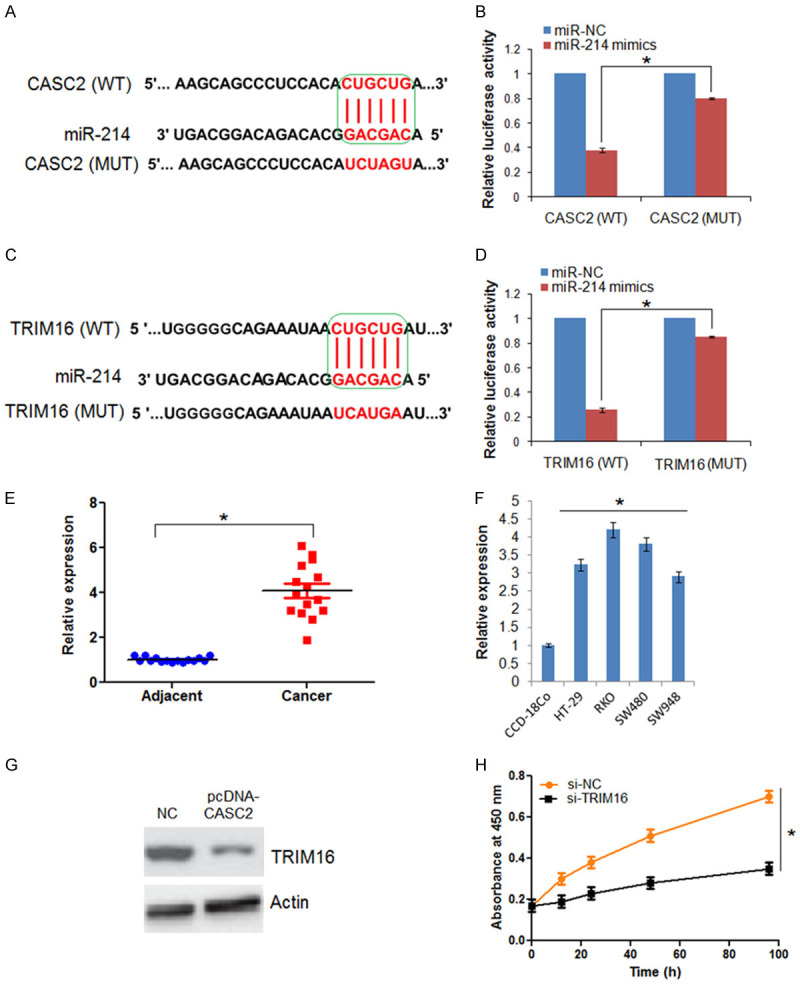
LncRNA-CASC2 exerts its effects via miR-214/TRIM16 axis. A. Alignment of CASC2 with miR-214. B. Dual luciferase assay showing interaction between miR-214 and CASC2. C. Alignment of TRIM16 with miR-214. D. Dual luciferase assay showing interaction between miR-214 and TRIM16. E. Expression of TRIM16 in colon cancer and adjacent normal tissues. F. Expression of TRIM16 in normal and colon cancer cell lines. G. Western blots showing the expression of TRIM16 in NC and CASC2 overexpressing HT-29 cells. H. Viability of the si-NC and si-TRIM16 HT-29 cells. The experiments were performed in triplicate and expressed as mean ± SD (*P < 0.05).
Discussion
Colon cancer causes significant human mortality across the world [11]. The limited availability of the reliable and efficient therapeutic targets/agents hurdles the treatment of colon cancer [12]. The LncRNAs have shown great promise as therapeutics for cancer treatment [13]. They have been reported to play vital roles in different cellular processes, such as proliferation, cell death and metastasis [14]. Studies have shown that several LncRNAs exhibit the potential to promote or to halt the growth of different cancers [15]. The LncRNA CASC2 has been found to exhibit therapeutic implications in several cancer types but has not been explored in colon cancer. Herein, we determined therapeutic implications of LncRNA CASC2 in colon cancer. We found that LncRNA CASC2 expression was considerably suppressed in colon cancer. These findings are complimented by earlier studies wherein LncRNA CASC2 has been found to be downregulated in non-small cell lung cancer and thyroid cancer [16,17]. Next, LncRNA CASC2 was overexpressed in colon cancer and we found that LncRNA CASC2 overexpression halted the proliferation of HT-29 cells by induction of apoptosis. Studies carried out earlier have reported that LncRNA CASC2 overexpression inhibited the proliferation of human hepatocellular cancer and lung cancer cells via induction of apoptosis [18,19]. Additionally, LncRNA CASC2 targets MAPK signalling to inhibit the growth of gastric cancer [20]. In another study, CASC2 has been reported to inhibit the growth and metastasis of glioma cells [21]. A previously carried out study showed that CASC2 exerts growth inhibitory effects on human non-small lung cancer cells by via modulation of TRIM16 [19]. Herein we also examined the effects of CASC2 on the TRIM16 and we found that CASC2 exerts its effects via miR-214 induced suppression of TRIM16. These findings are in agreement with previous studies wherein silencing of TRIM16 inhibits the growth of ovarian cancer, melanoma cells and breast cancer [22-24].
Conclusion
The results suggest that that LncRNA CASC2 is suppressed and inhibits the growth of the human colon cancer cells via induction of apoptosis and autophagy via modulation of TRIM16 expression. Taken together, LncRNA CASC2 may be utilised as therapeutic target for colon cancer treatment.
Disclosure of conflict of interest
None.
References
- 1.Prensner JR, Chinnaiyan AM. The emergence of lncRNAs in cancer biology. Cancer Discov. 2011;1:391–407. doi: 10.1158/2159-8290.CD-11-0209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Huarte M. The emerging role of lncRNAs in cancer. Nature Med. 2015;21:1253. doi: 10.1038/nm.3981. [DOI] [PubMed] [Google Scholar]
- 3.Pei Z, Du X, Song Y, Fan L, Li F, Gao Y, Wu R, Chen Y, Li W, Zhou H, Yang Y. Down-regulation of lncRNA CASC2 promotes cell proliferation and metastasis of bladder cancer by activation of the Wnt/β-catenin signaling pathway. Oncotarget. 2017;8:18145. doi: 10.18632/oncotarget.15210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Feng Y, Zou W, Hu C, Li G, Zhou S, He Y, Ma F, Deng C, Sun L. Modulation of CASC2/miR-21/PTEN pathway sensitizes cervical cancer to cisplatin. Arch Biochem Biophys. 2017;623-624:20–30. doi: 10.1016/j.abb.2017.05.001. [DOI] [PubMed] [Google Scholar]
- 5.Ba Z, Gu L, Hao S, Wang X, Cheng Z, Nie G. Downregulation of lnc RNA CASC2 facilitates osteosarcoma growth and invasion through miR-181a. Cell Proliferat. 2018;51:e12409. doi: 10.1111/cpr.12409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang Y, Zhu M, Sun Y, Li W, Wang Y, Yu W. Upregulation of lncRNA CASC2 suppresses cell proliferation and metastasis of breast cancer via inactivation of the TGF-β signaling pathway. Oncol Res. 2019;27:379–387. doi: 10.3727/096504018X15199531937158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cao Y, Xu R, Xu X, Zhou Y, Cui L, He X. Downregulation of lncRNA CASC2 by microRNA-21 increases the proliferation and migration of renal cell carcinoma cells. Mol Med Rep. 2016;14:1019–25. doi: 10.3892/mmr.2016.5337. [DOI] [PubMed] [Google Scholar]
- 8.Siegel R, DeSantis C, Jemal A. Colorectal cancer statistics, 2014. CA Cancer J Clin. 2014;64:104–17. doi: 10.3322/caac.21220. [DOI] [PubMed] [Google Scholar]
- 9.Arnold M, Sierra MS, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global patterns and trends in colorectal cancer incidence and mortality. Gut. 2017;66:683–691. doi: 10.1136/gutjnl-2015-310912. [DOI] [PubMed] [Google Scholar]
- 10.Guinney J, Dienstmann R, Wang X, De Reyniès A, Schlicker A, Soneson C, Marisa L, Roepman P, Nyamundanda G, Angelino P, Bot BM. The consensus molecular subtypes of colorectal cancer. Nature Med. 2015;21:1350. doi: 10.1038/nm.3967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Potter JD, Slattery ML, Bostick RM, Gapstur SM. Colon cancer: a review of the epidemiology. Epidemiol Rev. 1993;15:499–545. doi: 10.1093/oxfordjournals.epirev.a036132. [DOI] [PubMed] [Google Scholar]
- 12.Tauriello DV, Palomo-Ponce S, Stork D, Berenguer-Llergo A, Badia-Ramentol J, Iglesias M, Sevillano M, Ibiza S, Cañellas A, Hernando-Momblona X, Byrom D. TGFβ drives immune evasion in genetically reconstituted colon cancer metastasis. Nature. 2018;554:538. doi: 10.1038/nature25492. [DOI] [PubMed] [Google Scholar]
- 13.Evans JR, Feng FY, Chinnaiyan AM. The bright side of dark matter: lncRNAs in cancer. J Clin Invest. 2016;126:2775–2782. doi: 10.1172/JCI84421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang H, Chen Z, Wang X, Huang Z, He Z, Chen Y. Long non-coding RNA: a new player in cancer. J Hematol Oncol. 2013;6:37. doi: 10.1186/1756-8722-6-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang R, Xia LQ, Lu WW, Zhang J, Zhu JS. LncRNAs and cancer. Oncol Lett. 2016;12:1233–1239. doi: 10.3892/ol.2016.4770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.He X, Liu Z, Su J, Yang J, Yin D, Han L, De W, Guo R. Low expression of long noncoding RNA CASC2 indicates a poor prognosis and regulates cell proliferation in non-small cell lung cancer. Tumor Biol. 2016;37:9503–9510. doi: 10.1007/s13277-016-4787-6. [DOI] [PubMed] [Google Scholar]
- 17.Xiong X, Zhu H, Chen X. Low expression of long noncoding RNA CASC2 indicates a poor prognosis and promotes tumorigenesis in thyroid carcinoma. Biomed Pharmacother. 2017;93:391–397. doi: 10.1016/j.biopha.2017.06.063. [DOI] [PubMed] [Google Scholar]
- 18.Fan JC, Zeng F, Le YG, Xin L. LncRNA CASC2 inhibited the viability and induced the apoptosis of hepatocellular carcinoma cells through regulating miR-24-3p. J Cell Biochem. 2018;119:6391–6397. doi: 10.1002/jcb.26479. [DOI] [PubMed] [Google Scholar]
- 19.Li Q, Chen K, Dong R, Lu H. LncRNA CASC2 inhibits autophagy and promotes apoptosis in non-small cell lung cancer cells via regulating the miR-214/TRIM16 axis. RSC Adv. 2018;8:40846–55. doi: 10.1039/c8ra09573f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li P, Xue WJ, Feng Y, Mao QS. Long non-coding RNA CASC2 suppresses the proliferation of gastric cancer cells by regulating the MAPK signaling pathway. Am J Transl Res. 2016;8:3522. [PMC free article] [PubMed] [Google Scholar]
- 21.Wang P, Liu YH, Yao YL, Li Z, Li ZQ, Ma J, Xue YX. Long non-coding RNA CASC2 suppresses malignancy in human gliomas by miR-21. Cell Signal. 2015;27:275–282. doi: 10.1016/j.cellsig.2014.11.011. [DOI] [PubMed] [Google Scholar]
- 22.Sutton SK, Koach J, Tan O, Liu B, Carter DR, Wilmott JS, Yosufi B, Haydu LE, Mann GJ, Thompson JF, Long GV. TRIM16 inhibits proliferation and migration through regulation of interferon beta 1 in melanoma cells. Oncotarget. 2014;5:10127. doi: 10.18632/oncotarget.2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tan H, Qi J, Chu G, Liu Z. Tripartite motif 16 inhibits the migration and invasion in ovarian cancer cells. Oncol Res. 2017;25:551–558. doi: 10.3727/096504016X14758370595285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yao J, Xu T, Tian T, Fu X, Wang W, Li S, Shi T, Suo A, Ruan Z, Guo H, Yao Y. Tripartite motif 16 suppresses breast cancer stem cell properties through regulation of Gli-1 degradation via the ubiquitin-proteasome pathway. Oncol Rep. 2016;35:1204–1212. doi: 10.3892/or.2015.4437. [DOI] [PubMed] [Google Scholar]


