Abstract
Laryngeal squamous cell carcinoma (LSCC) is one of the most commonly seen head and neck malignancies. Identifying potent markers and/or targets for early diagnosis and individualized therapies for LSCC remains a considerable challenge. The present study analyzed online data and identified lncRNA KRT16P2 as a significantly upregulated long non-coding RNA (lncRNA) in LSCC. KRT16P2 knockdown in LSCC cells inhibited cancer cell proliferation, invasion, and migration. Similar to KRT16P2, EGFR expression was also significantly upregulated in LSCC. KRT16P2 and EGFR were positively correlated in LSCC tissue samples. EGFR knockdown also dramatically inhibited LSCC cell proliferation and aggressiveness (invasion and migration). Through online data and online tools, miR-1294 was predicted to target KRT16P2 and EGFR 3’UTR simultaneously. KRT16P2 inhibited miR-1294 expression, and miR-1294 inhibited EGFR expression through direct binding. miR-1294 overexpression repressed LSCC cell proliferation and aggressiveness. The effects of KRT16P2 silence on the expression of EGFR, LSCC cell proliferation, invasion, and migration, the protein levels of ki-67, PCNA, and cleaved-Caspase 3, as well as the phosphorylation of AKT, were all significantly reversed by miR-1294 inhibition. In conclusion, we demonstrated a lncRNA KRT16P2/miR-1294/EGFR axis that regulates LSCC cell proliferation, invasion, and migration. The clinical application of this axis needs further in vivo and clinical investigation.
Keywords: Laryngeal squamous cell carcinoma (LSCC), long non-coding RNA (lncRNA) KRT16P2, miR-1294, EGFR, cell aggressiveness
Introduction
In head and neck malignancies, one of the most commonly seen cancers is LSCC (laryngeal squamous cell carcinoma). Despite advances in surgical techniques, novel chemotherapeutics, and radiotherapy over the past thirty years, patients with advanced-stage LSCC still suffer to the most unsatisfactory outcomes and no substantial progress has yet been made in treatment [1]. To identify the specific molecular biomarkers used in early diagnosis and potent targeted therapy plays an essential role in early identification and timely treatment for LSCC.
Along with the technical progress of high-throughput sequencing, people found that only less than 2% of the transcripts could encode proteins, while most of them showed to be transcribed into noncoding RNAs, such as miRNAs or miRs (microRNAs), siRNAs (small interfering RNAs) and lncRNAs (long non-coding RNAs) [2]. lncRNAs are a type of RNA possessing more than 200 nucleotides that do not encode protein. LncRNAs can modulate gene expression at both transcriptional and posttranscriptional levels. Moreover, lncRNAs not only contribute to chromatin remodeling, RNA decay, epigenetic modulation, chromatin modification, and many other cell functions [3,4] but also show to be tightly related to the formation, invasion, and metastasis of tumors [5,6]. As for LSCC, it has been revealed that some lncRNAs, like H19 [7], HOTAIR [8,9], NEAT1 [10] and TUG1 [11], were increased within laryngeal cancer cell lines and tissue samples, and could be involved in a variety of biological processes to enhance cancer. However, lncRNAs’ role and mechanism within LSCC remain unclear. Cancer bioinformatics is regarded as a helpful tool that allows the researchers to determine potentially critical cancer genes and pathways via database screening. Herein, we downloaded and analyzed several microarray expression profiles attempting to identify more lncRNAs related to LSCC carcinogenesis.
Although the mechanisms have not yet been fully determined, increasing evidence indicates that lncRNAs might competitively bind to miRNAs by their MRE (miRNA response elements) to act as ceRNAs (competing endogenous RNAs), thus modulating the expression of target RNAs [12]. This mechanism of ceRNAs is of great interest in explaining the occurrence and development of malignant tumors, including gastric carcinoma [13], thyroid carcinoma [14], and hepatoblastoma [15]. Besides, recent investigations demonstrated multiple potential regulatory interactions of ceRNAs related to LSCC. For example, DNA methylation-mediated TMEM51-AS1 and non-methylation-mediated RUSC1-AS1 may induce LSCC via acting as ceRNAs. Two ceRNA axis genes, namyly TMEM51-AS1-miR-106b-TRAPPC10 and RUSC1-AS1-miR-16-SLC39A14, can function as potentially key prognostic biomarkers for LSCC [16]. NEAT1, another lncRNA, can regulate the miR-107/CDK6 signaling pathway to enhance LSCC [10]. Since online data (GSE84957, GSE59102, GSE51985, and GSE79493) also report a large amount of differentially-expressed miRNAs and mRNAs, we performed more analyses and experiments attempting to identify lncRNA-miRNA-mRNA axis that could modulate the LSCC carcinogenesis.
In the present study, we downloaded and analyzed four online microarray expression profiles (GSE84957, GSE59102, GSE51985, and GSE79493) reporting differentially-expressed genes in LSCC and normal non-cancerous tissues to identify lncRNAs and mRNAs that might be functionally related to LSCC carcinogenesis. The specific effects of candidate lncRNA and mRNA on LSCC cell proliferation, invasion, and migration were investigated, respectively. Next, differentially-expressed miRNAs that might simultaneously target selected lncRNA and mRNA were analyzed. The predicted miRNA binding to lncRNA and mRNA, as well as the specific effects of miRNA on LSCC cells were investigated. Eventually, the dynamic effects of the combination of lncRNAs and miRNAs upon mRNA expression and LSCC cells were detected. In summary, we reported a novel lncRNA-miRNA-mRNA axis that modulates LSCC cell proliferation, invasion, and migration, therefore affecting LSCC carcinogenesis.
Materials and methods
Clinical tissue sampling
Fifteen cases of laryngeal squamous cell carcinoma (LSCC) tissues and twenty cases of adjacent normal tissues were obtained from patients who underwent surgical resection at The Second Xiangya Hospital, Central South University (Changsha, China) with the approval of the Ethics Committee of The Second Xiangya Hospital, Central South University. The informed consent form was signed by each patient involved. All the tissue samples were fixed in paraformaldehyde or snap-frozen and stored at -80°C.
Cell lines and cell transfection
Human LSCC cell line TU212 and TU686 were obtained from Xiangya Cell Bank (Central South University, Changsha). Cells were cultured in DMEMIF12 medium supplemented with 10% heat-inactivated FBS with penicillin/streptomycin at 37°C in 5% CO2.
LncRNA KRT16P2 knockdown in cells was generated by the transfection of si1-KRT16P2 or si2-KRT16P2 (GenePharma, Shanghai, China). EGFR knockdown in cells was generated by the transfection of si-EGFR (GenePharma). miR-1294 overexpression or inhibition in cells was generated by the transfection of miR-1294 mimics or miR-1294 inhibitor (GenePharma, Shanghai, China). All cell transfection was performed using Lipofectamine 3000 Reagent (Thermo Fisher Scientific, Waltham, MA, USA).
PCR-based analysis
The expression of lncRNA, miRNA, and mRNA was determined by real-time PCR. Total RNA was extracted from cultured cells using Trizol reagent (Invitrogen). The expression of lncRNA, miRNA, and mRNA was measured using an SYBR Green qPCR assay (Takara, Dalian, China) following the methods described before [17]. The expression of RNU6B or GAPDH served as an endogenous control. The 2-ΔΔCT method was applied for data processing.
Cell viability examined by CCK-8 assay
Cell viability examination was performed using the CCK-8 kit (Beyotime, Shanghai, China). After transfection or treatment, cells were seeded into 96-well plates at a density of 5 × 103 cells/well. Two hours before the examination, 20 μl of CCK-8 solution was added to each well, followed by the incubation at 37°C. The Optical density (OD) value was determined at the wavelength of 450 nm on a microplate reader.
Transwell assay
Cells (5 × 105) were planted on the top side of polycarbonate Transwell filters coated with Matrigel for invasion examination. For Transwell invasion assays, cells were suspended in medium without serum and medium without serum was used in the bottom chamber. The cells were incubated at 37°C for 48 h. The non-invasive cells in the top chambers were removed with cotton swabs. The invaded cells on the lower membrane surface were fixed in 100% methanol for 10 min, air-dried, stained with crystal violet solution, and then counted under a microscope.
Wound healing assay
Cells were seeded in 6-well plates at 5 × 105 cells/ml until the cell monolayer emerged, and then the cell wound healing assay was performed. The scratch area was measured under the microscope (Olympus, Japan) at 0 h and 48 h. The relative distance of cell migration to the scratch area was also measured under the microscope and analyzed by ImageJ software (NIH, USA).
Immunoblotting
The protein levels of ki-67, proliferating cell nuclear antigen (PCNA), pro-Caspase 3, cleaved-Caspase 3, p-AKT, AKT, and EGFR were determined by performing Immunoblotting analyses following the methods described before [17]. Protein blots were incubated with the primary antibodies against ki-67 (27309-1-AP, Proteintech, Rosemont, IL, USA), PCNA (ab29, Abcam, Cambridge, UK), pro-Caspase 3 (19677-1-AP, Proteintech), cleaved-Caspase 3 (ab2302, Abcam), p-AKT (Y011054, Abm, Richmond, Canada), AKT (Y409094, Abm), and EGFR (ab52894, Abcam) followed by another incubation with proper HRP-conjugated secondary antibodies. β-actin (6008-1-Ig, Proteintech) was used as an endogenous control. All the antibodies were obtained from Abcam unless otherwise noted. Signals were visualized using enhanced chemilumescent (ECL) substrates (Millipore, MA, USA) normalizing to GAPDH and observes using an automatic chemiluminescence imaging analysis system (Tannon 4200, China).
Hematoxylin-eosin (H&E) staining
The LSCC tissues and normal adjacent tissues were fixed in 4% paraformaldehyde for 24 h, embedded in paraffin, and sliced into 5-μm sections. The HE staining was performed on the sections as previously described [18].
Immunohistochemical (IHC) staining
Immunohistochemistry was performed to examine the distribution and protein contents of EGFR in tissue samples. Tissue sections were incubated with anti-EGFR (CSB-PA10279A0Rb, Cusabio, Wuhan, China), and then followed by IHC Detection Kit (Beyotime, Shanghai, China). Image Pro-plus 6.0 software (Media Cybernetics, USA) was used to assess immunohistochemical sections by measuring the integrated optical density.
Luciferase reporter assay
To validate the binding between miR-1294 and KRT16P2 or 3’UTR of EGFR, the wild-type or mutated KRT16P2 or 3’UTR of EGFR was cloned to the downstream of the Renilla psiCHECK2 vector (Promega, Madison, WI, USA), named wt-KRT16P2 or wt-EGFR 3’UTR or mut-KRT16P2 or mut-EGFR 3’UTR. Next, 293T cells were co-transfected with two types of luciferase reporter vectors and miR-1294 mimics/miR-1294 inhibitor and examined for the luciferase activity using the Dual-Luciferase Reporter Assay System (Promega).
Data processing and statistical analysis
The data were analyzed with GraphPad software. The measurement data were expressed as mean ± standard deviation (SD). Among-group and intra-group data comparisons were performed with the ANOVA and Student’s t-tests. P<0.05 indicated a statistically significant difference.
Results
Selection of lncRNAs related to LSCC (laryngeal squamous cell carcinoma) carcinogenesis
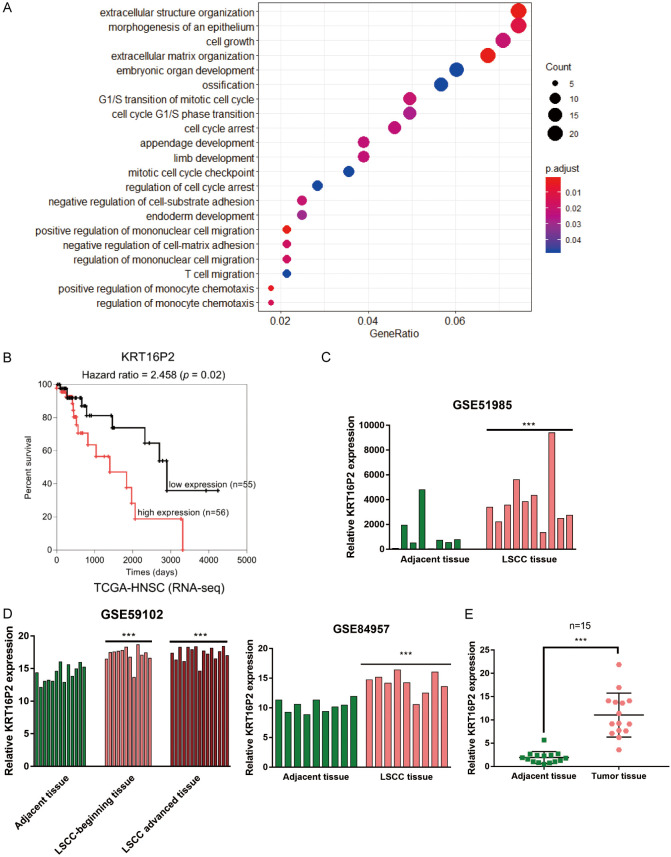
To identify the lncRNAs related to LSCC pathogenesis, we downloaded and analyzed online microarray expression profiles, GSE84957, GSE59102, and GSE51985, and found that a total of 362 genes were differentially-expressed in LSCC and normal tissue samples. Gene Ontology (GO) functional enrichment analysis was performed on these differentially expressed genes that were enriched within extracellular structure organization, morphogenesis of epithelium, cell growth, and other pathways (Figure 1A), which is coincident with the characteristics of LSCC, such as epithelial hyperplasia. Since laryngeal cancer represents a malignant tumor caused by the mucosal epithelial tissue of the larynx, the epithelial deterioration may be the cause of LSCC tumorigenesis. Also, based on both GSE84957 and GSE59102, the expression of 7 lncRNAs differed in LSCC and normal tissues (Tables 1, 2).
Figure 1.
Selection of lncRNAs related to laryngeal squamous cell carcinoma (LSCC) carcinogenesis. A. Gene Ontology (GO) functional enrichment analysis on a total of 362 differentially-expressed genes in LSCC and normal tissue samples reported by both GSE84957 and GSE59102. The top three signaling pathways in which the differentially-expressed genes enriched in are extracellular structure organization, morphogenesis of an epithelium, and cell growth. B. Cox survival analysis on 111 cases of LSCC patients based on data from TCGA-HNSC (Head and Neck squamous cell carcinoma) database. Cases were grouped using the median value of KRT16P2 expression as cut-off and a Cox’s proportional hazards model was used to analyze the correlation of KRT16P2 expression and the survival-time outcomes. The resolution of the endpoints is depicted using Kaplan-Meier survival curve. C. KRT16P2 expression in adjacent normal tissues and LSCC tissues based on data from GSE51985. Paired t-test. D. KRT16P2 expression in adjacent normal tissues and LSCC tissues based on data from GSE84957 and GSE59102. unpaired t-test. E. The expression of KRT16P2 in 15 cases of LSCC tissues and 15 cases of normal non-cancerous tissues determined by real-time PCR. Paired t-test. ***P<0.001.
Table 1.
The different change of 7 lncRNA in GSE84957 (normal vs cancer)
| ID | adj.P.Val | P.Value | t | B | logFC | Gene.symbol |
|---|---|---|---|---|---|---|
| A_32_P150086 | 5.71E-09 | 2.65E-11 | 8.87 | 15.6768 | 2.84 | ANKRD20A5P |
| A_32_P68942 | 5.72E-08 | 5.94E-10 | 7.91 | 12.64345 | 3.35 | ANKRD20A5P |
| A_32_P62963 | 2.19E-07 | 3.73E-09 | -7.36 | 10.84851 | -2.9 | KRT16P2 |
| A_24_P66233 | 1.03E-06 | 2.96E-08 | 6.74 | 8.82566 | 1.57 | TTTY14 |
| A_23_P350754 | 3.26E-06 | 1.30E-07 | 6.3 | 7.37741 | 1.38 | OR7E14P |
| A_32_P68293 | 2.75E-05 | 1.87E-06 | 5.5 | 4.78298 | 2.57 | ANKRD20A5P |
| A_23_P58538 | 5.01E-05 | 3.96E-06 | 5.28 | 4.05422 | 0.912 | EPB41L4A-AS1 |
| A_23_P251232 | 9.65E-05 | 9.04E-06 | 5.03 | 3.25554 | 0.755 | TTTY14 |
| A_24_P281009 | 0.000111 | 1.07E-05 | 4.98 | 3.09375 | 1.73 | AQP7P1 |
| A_23_P373119 | 0.00146 | 0.000259 | -3.98 | 0.03019 | -0.937 | HMGB3P1 |
Table 2.
The different change of 7 lncRNA in GSE59102 (normal vs cancer)
| ID | adj.P.Val | P.Value | t | B | logFC | GeneSymbol |
|---|---|---|---|---|---|---|
| A_23_P373119 | 0.00011 | 7.58E-08 | -8.38 | 8.15006 | -1.94 | HMGB3P1 |
| A_23_P251232 | 0.00135 | 3.23E-06 | 6.46 | 4.6841 | 1.4 | TTTY14 |
| A_32_P62963 | 0.00257 | 1.03E-05 | -5.91 | 3.59504 | -3.8 | KRT16P2 |
| A_32_P150086 | 0.00259 | 1.04E-05 | 5.9 | 3.58195 | 3.59 | ANKRD20A5P |
| A_21_P0012183 | 0.00364 | 2.05E-05 | 5.59 | 2.9443 | 3.12 | ANKRD20A5P |
| A_33_P3324333 | 0.00484 | 3.41E-05 | 5.36 | 2.46109 | 2.36 | ANKRD20A5P |
| A_23_P58538 | 0.00803 | 8.95E-05 | 4.93 | 1.54507 | 1 | EPB41L4A-AS1 |
| A_24_P281009 | 0.00867 | 0.000101 | 4.88 | 1.43239 | 1.86 | AQP7P1 |
| A_23_P350754 | 0.00898 | 0.000106 | 4.86 | 1.38498 | 1.28 | OR7E14P |
To further identify the candidate lncRNAs, we performed cox survival analysis on 111 cases of LSCC patients based on data from TCGA-HNSC (Head and Neck squamous cell carcinoma) database and revealed that, among a total of 7 differentially-expressed lncRNAs, lncRNA KRT16P2 expression showed to be significantly linked to LSCC overall survival (Figure 1B). The expression of KRT16P2 showed to be markedly increased within LSCC tissue samples, in comparison with the adjacent normal healthy tissue samples according to GSE51985 (Figure 1C) GSE59102 and GSE84957 (Figure 1D). Consistently, we performed real-time PCR and found that KRT16P2 expression showed to be significantly upregulated within 15 cases of LSCC tissue samples, in comparison to 15 cases of normal healthy tissue samples (Figure 1E). Thus, we speculate that lncRNA KRT16P2 may contribute to LSCC pathogenesis.
Effects of lncRNA KRT16P2 upon LSCC cells
Secondly, we transfected si1-KRT16P2/si2-KRT16P2 to generate KRT16P2 silence in TU212 and TU686 cell lines, and performed real-time PCR to verify the transfection efficiency (Figure 2A); si1-KRT16P2 was selected for better transfection efficiency. Next, we transfected TU212 and TU686 cell lines with si2-KRT16P2 and examined for related indexes. After knocking down KRT16P2, TU212, and TU686 cell viability, invasion and migration were significantly inhibited (Figure 2B-D). Moreover, KRT16P2 silence also significantly decreased ki-67, PCNA, and cleaved-Caspase 3 proteins, as well as inhibited the phosphorylation of AKT (Figure 2E). These data indicate that KRT16P2 silence represses LSCC cell proliferation, invasion, and migration.
Figure 2.
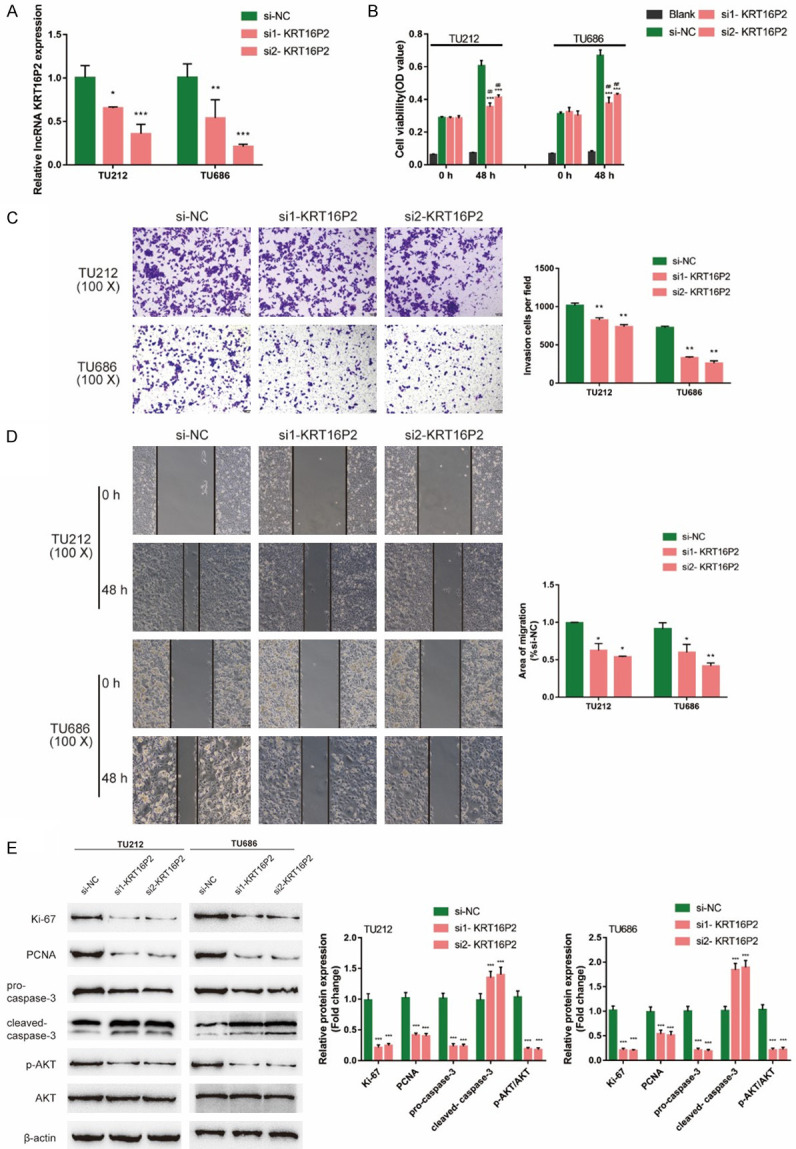
Effects of lncRNA KRT16P2 on LSCC cells. (A) Small interfering RNAs targeting KRT16P2 (si1-KRT16P2 and si2-KRT16P2) were obtained and KRT16P2 silence was generated in TU212 and TU686 cells by the transfection of si1-KRT16P2 or si2-KRT16P2. The transfection efficiency was verified by real-time PCR; si2-KRT16P2 was selected for further experiments because of better transfection efficiency. Next, TU212 and TU686 cells were transfected with si2-KRT16P2 and examined for (B) cell viability by CCK-8 assay; Blank is the background of OD value. (C) Invasion capacity by Transwell assay; (D) Migration capacity by Wound healing assay; (E) The protein levels of ki-67, PCNA, pro-Caspase 3, cleaved-Caspase 3, p-AKT, and AKT by Immunoblotting. ANOVA followed by Turkey’s multiple comparison test.
Expression and function of EGFR in LSCC
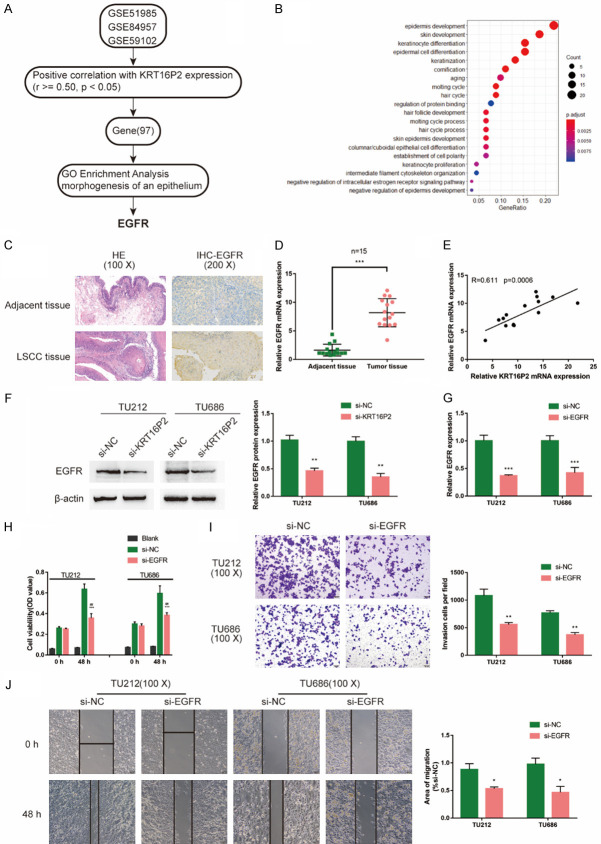
As we have mentioned, a total of 362 genes are differentially-expressed in LSCC and normal tissues; among them, 97 genes were positively correlated with KRT16P2 expression, according to GSE84957, GSE59102, and GSE51985 (Figure 3A). These 97 genes were applied for the GO functional enrichment analysis, and they were mostly enriched in epidermis development, skin development, and keratinocyte differentiation (Figure 3B). Since EGFR is the central factor of these pathways, we selected EGFR for further experiments.
Figure 3.
Expression and function of EGFR in LSCC. (A) mRNAs that are positively correlated with KRT16P2 (r≥0.5, P<0.05) were analyzed and selected based on GSE84957, GSE59102, and GSE51985; a total of 97 genes were eligible after cross-check on all the three datasets. (B) GO functional enrichment analysis was performed on these 97 genes. The top three signaling pathways in which the differentially-expressed genes enriched in are epidermis development, skin development, and keratinocyte differentiation. Key factors in these signaling pathways were further screened for combined with literatures and EGFR was selected. (C) The protein content and distribution of EGFR in LCSS and normal tissues were examined by IHC staining. The histology changes were exhibited in the left panel by HE staining. (D) The mRNA level of EGFR in LSCC tissues and normal adjacent tissues were determined by qRT-PCR. Paired t-test. (E) The correlation of EGFR and KRT16P2 expression in LSCC tissues was analyzed by Pearson’s correlation coefficient analysis. (F) TU212 and TU686 cells were transfected with si-KRT16P2 and examined for the protein levels of EGFR by Immunoblotting. Unpaired t-test. (G) EGFR silence was generated in TU212 and TU686 cells by the transfection of si-EGFR, as confirmed by real-time PCR. Unpaired t-test. Next, TU212 and TU686 cells were transfected with si-EGFR and examined for (H) cell viability by CCK-8 assay; Blank is the background of OD value. (I) Invasion capacity by Transwell assay; (J) Migration capacity by Wound healing assay. Unpaired t-test.
The histological examination of LCSS tissues and normal adjacent tissues were performed by HE staining (Figure 3C). The protein content and distribution of EGFR in LCSS and normal adjacent tissues were examined by and IHC staining; consistent with online results, the EGFR protein level was increased in LSCC tissues (Figure 3C). Moreover, qRT-PCR results also confirmed that the mRNA levels of EGFR were higher in LCSS tissues than normal adjacent tissues (Figure 3D). Furthermore, KRT16P2 and EGFR expression was positively correlated in LCSS tissue samples (Figure 3E). In si-KRT16P2-transfected TU212 and TU686 cell lines, EGFR proteins showed to be significantly decreased (Figure 3F). In summary, EGFR might also contribute to LSCC tumorigenesis in a KRT16P2-related way.
Next, we transfected si-EGFR to generate EGFR silence in TU212 and TU686 cell lines, and performed real-time PCR to verify the transfection efficiency (Figure 3G). Secondly, we transfected TU212 and TU686 cell lines with si-EGFR, then evaluated the related indexes. Similar to those of KRT16P2 knockdown, EGFR knockdown also dramatically suppressed TU212 and TU686 cell viability, invasion and migration (Figure 3H-J), indicating the critical role of EGFR in LSCC pathogenesis.
miR-1294 directly binds to KRT16P2 and EGFR 3’UTR
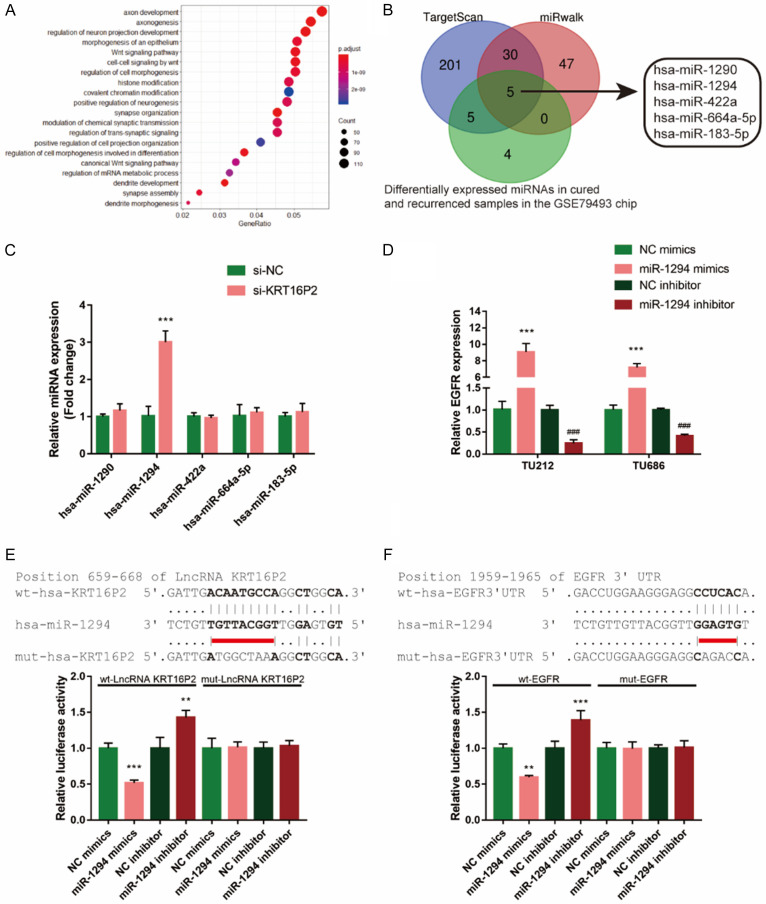
miRNA-induced interaction of lncRNAs and mRNAs plays an essential role in multiple diseases [19-21]. To screen for the miRNAs that could affect the functions of KRT16P2 and EGFR in LSCC, we analyzed differentially expressed miRNAs between 17 paired cured and recurrent LSCC tissues based on GSE79493 and 14 miRNAs were selected (Table 3). The downstream targets of these 14 miRNAs were predicted by MIRDIP (http://ophid.utoronto.ca/mirDIP/index.jsp#r), with 2436 genes found. These 2436 genes were applied for KEGG (Kyoto Encyclopedia of Genes and Genomes) pathway enrichment analysis and GO functional enrichment analysis; according to the results, the above-mentioned genes showed to be enriched within Wnt and morphogenesis of epithelium signaling pathways (Figure 4A).
Table 3.
The selection of different expressed miRNA in GSE79493
| logFC | AveExpr | t | P.Value | adj.P.Val | B | |
|---|---|---|---|---|---|---|
| hsa-miR-618 | 0.338464 | 0.176057 | 3.175253 | 0.003283 | 0.754048 | -2.96104 |
| hsa-miR-28-5p | -11.7972 | 5.898598 | -2.89407 | 0.006762 | 0.754048 | -3.64257 |
| hsa-miR-374b-3p | -0.60416 | 0.506398 | -2.54327 | 0.015964 | 0.754048 | -4.43914 |
| hsa-miR-548h-3p | -0.27712 | 0.138561 | -2.48378 | 0.018371 | 0.754048 | -4.56759 |
| hsa-miR-10a-3p | -0.29261 | 0.266098 | -2.45702 | 0.019559 | 0.754048 | -4.6247 |
| hsa-miR-1294 | -0.44235 | 0.274043 | -2.45641 | 0.019587 | 0.754048 | -4.626 |
| hsa-miR-561-5p | 1.431726 | 1.000547 | 2.274458 | 0.029714 | 0.754048 | -5.00272 |
| hsa-let-7d-3p | 3.98763 | 4.278306 | 2.210836 | 0.034243 | 0.754048 | -5.12953 |
| hsa-miR-183-5p | 561.8425 | 923.1842 | 2.207237 | 0.034517 | 0.754048 | -5.13663 |
| hsa-miR-422a | 0.834495 | 0.710341 | 2.095925 | 0.044008 | 0.754048 | -5.35179 |
| hsa-miR-4488 | 1.369726 | 1.370574 | 2.079251 | 0.045612 | 0.754048 | -5.3833 |
| hsa-miR-664a-5p | -0.29681 | 0.180422 | -2.07383 | 0.046145 | 0.754048 | -5.39349 |
| hsa-miR-1290 | 5.852462 | 6.424937 | 2.055761 | 0.047961 | 0.754048 | -5.42735 |
| hsa-miR-744-5p | 3.17783 | 5.768747 | 2.036401 | 0.049976 | 0.754048 | -5.46338 |
Figure 4.
miR-1294 directly binds to KRT16P2 and EGFR 3’UTR. A. Differentially-expressed miRNAs between cured and recurrent LSCC tissues were analyzed based on GSE79493 and 14 miRNAs were selected. The downstream targets of these 14 miRNAs were predicted by MIRDIP (http://ophid.utoronto.ca/mirDIP/index.jsp#r) and 2436 genes were found. These 2436 genes were applied for Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis and GO functional enrichment analysis. The top three signaling pathways in which the differentially-expressed genes enriched in are axon development, axonogenesis, and regulation of neuron projection development. B. Online tools were employed to predict miRNAs that could simultaneously target KRT16P2 and EGFR 3’UTR and differentially expressed in cured and recurrence samples in the GSE79493. A total of 5 miRNAs were predicted by all the three online tools to target KRT16P2 and EGFR 3’UTR simultaneously. The expression levels of these miRNAs in tissue samples were examined by real-time PCR and the expression of miR-1294 was the most downregulated in LSCC tissues. C. TU212 and TU686 cells were transfected with si-KRT16P2 and examined for the expression of miR-1294. Unpaired t-test. D. miR-1294 overexpression or inhibition was generated in TU212 and TU686 cells by the transfection of miR-1294 mimics or miR-1294 inhibitor. The transfection efficiency was verified using real-time PCR. ANOVA followed by Turkey’s multiple comparison test. E, F. Wild-type and mutant-type KRT16P2 and EGFR 3’UTR luciferase reporter vectors were constructed as described in the Materials and methods section. These vectors were co-transfected in 293T cells with miR-1294 mimics or miR-1294 inhibitor and the luciferase activity was determined. ANOVA followed by Turkey’s multiple comparison test.
To further identify miRNAs that might simultaneously target KRT16P2 and EGFR 3’UTR, we used online tools to predict available miRNAs. As shown in Figure 4B, 5 miRNAs might bind to both KRT16P2 and EGFR 3’UTR and differentially express in cured and recurrence samples in GSE79493. Next, the expression of these miRNAs’ in response to KRT16P2 silence was measured, miR-1294 expression showed to be the most upregulated miRNA in LSCC cell lines (Figure 4C); thus, we chose miR-1294 in further experiments. To confirm the putative bindings, we transfected miR-1294 mimics/inhibitor to generate miR-1294 overexpression or inhibition in TU212 and TU686 cell lines, and performed real-time PCR to verify the transfection efficiency (Figure 4D). Based on the Materials and methods section, we constructed two different types of KRT16P2 and EGFR 3’UTR luciferase reporter vectors, wild-type and mutant-type (namely wt-KRT16P2, wt-EGFR 3’UTR, mut-KRT16P2, mut-EGFR 3’UTR). We co-transfected these vectors in 293T cells with miR-1294 mimics/inhibitor and examined the luciferase activity. Figure 4E, 4F showed that the luciferase activity of wild-type KRT16P2 and wild-type EGFR 3’UTR was both markedly reduced via the overexpression of miR-1294 while increased via the inhibition of miR-1294. However, mutating the putative miR-1294 binding site in mut-KRT16P2 and mut-EGFR 3’UTR vectors could abolish the alterations in luciferase activity (Figure 4E, 4F). In summary, miR-1294 could directly bind to KRT16P2 and EGFR 3’UTR. Taken together, miR-1294 could contribute to KRT16P2 and EGFR functions in LSCC through targeting.
Effects of miR-1294 on LSCC cells
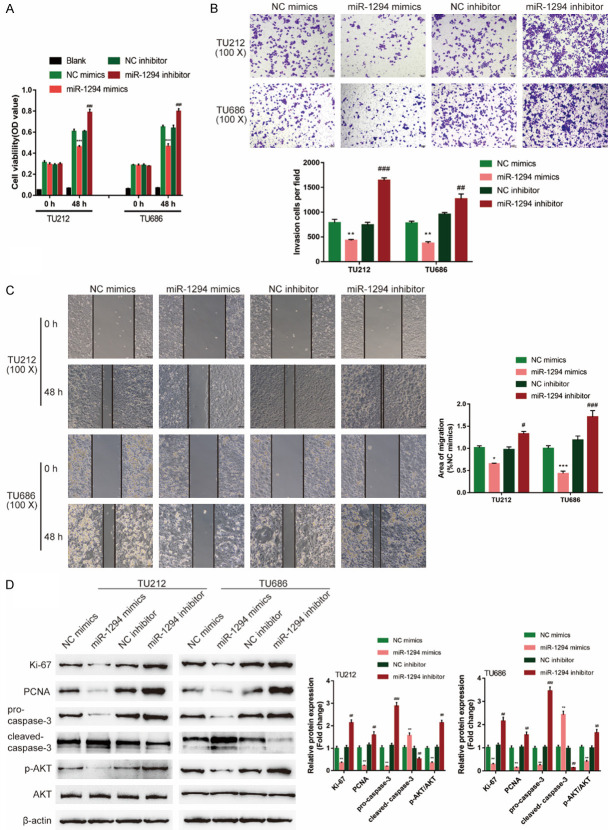
To validate the speculation, we transfected miR-1294 mimics/inhibitor to generate miR-1294 overexpression or miR-1294 inhibition in TU212 and TU686 cell lines and examined related indexes. miR-1294 overexpression significantly suppressed, while miR-1294 inhibition promoted cell viability, invasion, and migration (Figure 5A-C). Consistently, miR-1294 overexpression decreased, miR-1294 inhibition increased ki-67, PCNA, and cleaved-Caspase 3 proteins, as well as the phosphorylation of AKT (Figure 5D). In summary, miR-1294 exerts a tumor-suppressive effect on LSCC.
Figure 5.
Effects of miR-1294 on LSCC cells. (A) TU212 and TU686 cells were divided into four groups: NC mimics (negative control for miR-1294 mimics), miR-1294 mimics (to conduct miR-1294 overexpression), NC inhibitor (negative control for miR-1294 inhibitor), and miR-1294 inhibitor (to conduct miR-1294 inhibition). Cells were non-transfected or transfected with miR-1294/NC mimics or miR-1294/NC inhibitor and examined for cell viability by CCK-8 assay. Blank is the background of OD value. Next, Cells were non-transfected or transfected with miR-1294/NC mimics or miR-1294/NC inhibitor and examined for (B) invasion capacity by Transwell assay; (C) Migration capacity by Wound healing assay; (D) The protein levels of ki-67, PCNA, pro-Caspase 3, cleaved-Caspase 3, p-AKT, and AKT by Immunoblotting. ANOVA followed by Turkey’s multiple comparison test.
miR-1294 inhibits EGFR expression to attenuate the oncogenic effects of KRT16P2
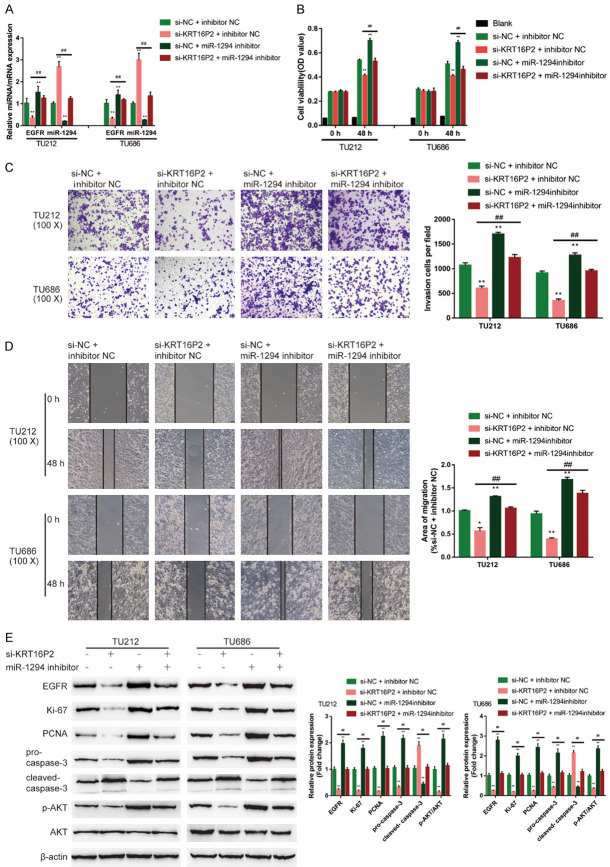
After confirming miR-1294 binding to KRT16P2 and EGFR and the specific roles of miR-1294 in LSCC cells, finally, we examined the dynamic effects of KRT16P2 and miR-1294 on EGFR and TU212 and TU686 cell lines. We co-transfected TU212 and TU686 cell lines with si-KRT16P2 and miR-1294 inhibitor. The expression of the EGFR mRNA level was significantly downregulated via KRT16P2 silence while upregulated via the inhibition of miR-1294; the effects of KRT16P2 silence were significantly reversed by miR-1294 inhibition (Figure 6A). KRT16P2 silence significantly inhibited, whereas the inhibition of miR-1294 enhanced cell viability, invasion, and migration; the effects of KRT16P2 silence were significantly reversed by miR-1294 inhibition (Figure 6B-D). KRT16P2 silence significantly decreased, while miR-1294 inhibition increased ki-67, PCNA, and cleaved-Caspase 3 proteins, as well as the phosphorylation of AKT; the effects of KRT16P2 silence were significantly reversed by miR-1294 inhibition (Figure 6E).
Figure 6.
miR-1294 inhibits EGFR expression to reverse the oncogenic effects of KRT16P2. TU212 and TU686 cells were divided into four groups: si-NC + inhibitor NC (negative control), si-KRT16P2 + inhibitor NC (co-transfected with si-KRT16P2 and inhibitor NC), si-NC + miR-1294 inhibitor (co-transfected with si-NC + miR-1294 inhibitor), and si-KRT16P2 + miR-1294 inhibitor group (co-transfected with si-KRT16P2 + miR-1294 inhibitor), and examined for (A) the expression of miR-1294 and EGFR by real-time PCR; (B) Cell viability by CCK-8 assay; (C) Invasion capacity by Transwell assay; (D) Migration capacity by Wound healing assay; (E) The protein levels of EGFR, ki-67, PCNA, pro-Caspase 3, cleaved-Caspase 3, p-AKT, and AKT by Immunoblotting. ANOVA followed by Turkey’s multiple comparison test.
Discussion
Herein, we analyzed online data and identified lncRNA KRT16P2 as a significantly upregulated lncRNA in LSCC. KRT16P2 knockdown in LSCC cells inhibited cancer cell proliferation, invasion, and migration. Similar to KRT16P2, EGFR expression was also significantly upregulated in LSCC. KRT16P2 and EGFR were positively correlated in LSCC tissue samples. EGFR knockdown also dramatically repressed LSCC cell proliferation and aggressiveness (invasion and migration). Through online data and online tools, miR-1294 was predicted to target KRT16P2 and EGFR 3’UTR simultaneously. KRT16P2 inhibited miR-1294 expression, and miR-1294 inhibited EGFR expression through direct binding. miR-1294 overexpression repressed LSCC cell proliferation and aggressiveness. The effects of KRT16P2 silence on EGFR expression, LSCC cell phenotype, ki-67, PCNA and cleaved-Caspase 3 proteins, as well as the phosphorylation of AKT, were all significantly reversed by miR-1294 inhibition.
LncRNAs are essential for tumorigenesis [22]. LncRNAs regulate gene expression via transcription, posttranscriptional processing, chromatin modification, protein function control, and some other mechanisms, thus contributing to various processes of carcinogenesis [23]. Nevertheless, the effects of lncRNAs on LSCC remain unclear. Our previous study reported that lncRNA NKILA could combine with NF-κB: IκB complex to suppress the phosphorylation of IκB, p65 nuclear translocation and the activation of NF-κB, thus acting as a potential agent of promoting X-ray radiation cytotoxicity on laryngeal cancer [24]. Herein, online data and experimental results revealed the upregulation of KRT16P2 expression in LSCC tissue samples. KRT16P2 knockdown within LSCC tissues dramatically suppressed LSCC cell proliferation and aggressiveness. Moreover, the protein levels of ki-67 and PCNA, two critical markers of cell nuclear proliferation [25], showed to be reduced by KRT16P2 knockdown, while the protein levels of cleaved-Caspase 3, whose activation is an important apoptotic biomarker [26,27], were increased by KRT16P2 knockdown. Besides, KRT16P2 knockdown significantly suppressed the phosphorylation of AKT. Blocking AKT/mTOR signaling is known to be effective in treating cancers [28,29]. Taken together, lncRNA KRT16P2 serves as a carcinogenic lncRNA within LSCC tissues. Notably, during the selection process, the study discovered a total of 7 lncRNAs were differentially-expressed in LSCC based on both GSE84957 and GSE59102, including lncRNA ANKRD20A5P, TTTY14, OR7E14P, EPB41L4A-AS1, AQP7P1, and HMGB3P1. Among them, TTTY14 has been regarded as one of the ten signature RNAs in a prognostic scoring system for esophageal squamous cell carcinoma [30]. LncRNA EPB41L4A-AS1 regulates glycolysis and glutaminolysis by mediating nucleolar translocation of HDAC2 in cancer cells [31]. These previous studies suggest that these six abnormally-expressed lncRNAs might also play a role in LSCC, which needs further investigation in our future research.
How to elucidate the mechanism of lncRNAs regulating gene expression is still an enormous challenge. As we have mentioned, the regulation mechanism includes nearly all aspects of pre-/post-transcriptional processes, such as transcription, posttranscriptional processing, chromatin modification, and protein function regulation [32,33]. Among the differentially-expressed genes reported by online expression profiles, EGFR attracted our attention because of its positive correlation with KRT16P2 and its critical role in LSCC carcinogenesis. Reportedly, through the evaluation of EGFR status at diagnosis, we could define a subset of patients with LSCC at high risk of cervical lymph node metastasis, subsequently determining the appropriate treatment [34]. The overexpression of EGFR caused by gene amplification or point mutations is commonly seen in LSCC patients [35,36]. Consistently, we also observed a significant upregulation of EGFR in LSCC tissue samples, according to both online data and experimental results. Similar to those of KRT16P2 silence, EGFR knockdown in LSCC cell lines remarkably inhibited LSCC cell proliferation and aggressiveness. These data further indicate that EGFR also plays an oncogenic role in LSCC, possibly in a KRT16P2-related way.
As is widely accepted, a noncoding RNA is often targeted by another noncoding RNA. For instance, H19 could regulate miR-200, let-7, and miR-675 [37-40]. Another oncogenic lncRNA, HOTAIR, could target miR-200c, miR-20b, miR-34a, and miR-206 [41-44]. Commonly, lncRNAs serve as sponge or ceRNA for miRNAs to inhibit miRNA expression and counteract miRNA-mediated suppression on downstream target mRNAs [19,33]. Herein, among 14 differentially-expressed miRNAs, miR-1294 has been predicted to simultaneously target lncRNA KRT16P2 and EGFR 3’UTR, which inspired us to speculate that KRT16P2, miR-1294, and EGFR might form a lncRNA/miRNA/mRNA axis to regulate LSCC cell phenotype. As confirmed by the luciferase reporter assay, miR-1294 could directly bind to KRT16P2 and EGFR 3’UTR. KRT16P2 knockdown significantly upregulated miR-1294 expression while miR-1294 inhibition upregulated EGFR expression, indicating the existence of the lncRNA/miRNA/mRNA axis preliminarily.
Notably, miR-1294 has been identified to exert a tumor-suppressive effect on glioma [45], OSCC (oral squamous cell carcinoma) [46], osteosarcoma [47], CCRCC (clear cell renal cell carcinoma) [48], etc. The decrease in miR-1294 is related to the impaired prognosis of ESCC (esophageal squamous cell carcinoma) [49] and EOC (epithelial ovarian cancer) [50]. Herein, miR-1294 overexpression significantly repressed LSCC cell proliferation and aggressiveness. The proliferating markers, ki-67 and PCNA, were decreased, the apoptotic marker, cleaved-Caspase 3, was increased, while AKT phosphorylation showed to be suppressed via the overexpression of miR-1294, indicating that miR-1294 exerts a tumor-suppressive effect on LSCC. Regarding the molecular mechanism, KRT16P2 knockdown downregulated, while miR-1294 inhibition upregulated EGFR expression, and miR-1294 inhibition significantly reversed the effects of KRT16P2 silence upon the expression of EGFR. Coincident with the regulation of EGFR expression, miR-1294 inhibition also remarkably reversed the effects of KRT16P2 silence on the phenotype of LSCC cells, indicating that KRT16P2/miR-1294 axis modulates LSCC cell phenotype through EGFR. Moreover, according to the miRNA target database, one miRNA can regulate many genes as its target, and multiple genes could be targeted by one miRNA [51-53]. Due to the multi-specificity of miRNAs and their high variability due to splicing and variable targeting, they also generate different threads for future research. In the present study, we identified a total of 14 miRNAs differentially-expressed in LSCC, according to GSE79493, suggesting there might be more miRNAs involved in LSCC carcinogenesis by targeting other downstream genes.
In conclusion, we demonstrated a lncRNA/miRNA/mRNA axis consists of lncRNA KRT16P2, miR-1294, and EGFR that regulate LSCC cell proliferation, invasion, and migration. The clinical application of this axis needs further in vivo and clinical investigation. Notably, the differentially-expressed genes were significantly enriched in extracellular structure organization and extracellular matrix organization. Considering the main connective tissue element present in the larynx is cartilage, and the cartilage is particularly rich in the extracellular matrix [54], further research should be undertaken to investigate the potential roles of these candidate lncRNAs in cartilage remodeling in LSCC.
Acknowledgements
This study was supported by the National Natural Foundation of Hunan Province (2019JJ50843).
Disclosure of conflict of interest
None.
References
- 1.Rudolph E, Dyckhoff G, Becher H, Dietz A, Ramroth H. Effects of tumour stage, comorbidity and therapy on survival of laryngeal cancer patients: a systematic review and a meta-analysis. Eur Arch Otorhinolaryngol. 2011;268:165–179. doi: 10.1007/s00405-010-1395-8. [DOI] [PubMed] [Google Scholar]
- 2.Kaikkonen MU, Adelman K. Emerging roles of non-coding RNA transcription. Trends Biochem Sci. 2018;43:654–667. doi: 10.1016/j.tibs.2018.06.002. [DOI] [PubMed] [Google Scholar]
- 3.Djebali S, Davis CA, Merkel A, Dobin A, Lassmann T, Mortazavi A, Tanzer A, Lagarde J, Lin W, Schlesinger F, Xue C, Marinov GK, Khatun J, Williams BA, Zaleski C, Rozowsky J, Röder M, Kokocinski F, Abdelhamid RF, Alioto T, Antoshechkin I, Baer MT, Bar NS, Batut P, Bell K, Bell I, Chakrabortty S, Chen X, Chrast J, Curado J, Derrien T, Drenkow J, Dumais E, Dumais J, Duttagupta R, Falconnet E, Fastuca M, Fejes-Toth K, Ferreira P, Foissac S, Fullwood MJ, Gao H, Gonzalez D, Gordon A, Gunawardena H, Howald C, Jha S, Johnson R, Kapranov P, King B, Kingswood C, Luo OJ, Park E, Persaud K, Preall JB, Ribeca P, Risk B, Robyr D, Sammeth M, Schaffer L, See LH, Shahab A, Skancke J, Suzuki AM, Takahashi H, Tilgner H, Trout D, Walters N, Wang H, Wrobel J, Yu Y, Ruan X, Hayashizaki Y, Harrow J, Gerstein M, Hubbard T, Reymond A, Antonarakis SE, Hannon G, Giddings MC, Ruan Y, Wold B, Carninci P, Guigó R, Gingeras TR. Landscape of transcription in human cells. Nature. 2012;489:101–108. doi: 10.1038/nature11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang KC, Chang HY. Molecular mechanisms of long noncoding RNAs. Mol Cell. 2011;43:904–914. doi: 10.1016/j.molcel.2011.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mou K, Liu B, Ding M, Mu X, Han D, Zhou Y, Wang LJ. lncRNA-ATB functions as a competing endogenous RNA to promote YAP1 by sponging miR-590-5p in malignant melanoma. Int J Oncol. 2018;53:1094–1104. doi: 10.3892/ijo.2018.4454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li X, Zhao X, Yang B, Li Y, Liu T, Pang L, Fan Z, Ma W, Liu Z, Li Z. Long non-coding RNA HOXD-AS1 promotes tumor progression and predicts poor prognosis in colorectal cancer. Int J Oncol. 2018;53:21–32. doi: 10.3892/ijo.2018.4400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wu T, Qu L, He G, Tian L, Li L, Zhou H, Jin Q, Ren J, Wang Y, Wang J, Kan X, Liu M, Shen J, Guo M, Sun Y. Regulation of laryngeal squamous cell cancer progression by the lncRNA H19/miR-148a-3p/DNMT1 axis. Oncotarget. 2016;7:11553–11566. doi: 10.18632/oncotarget.7270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zheng J, Xiao X, Wu C, Huang J, Zhang Y, Xie M, Zhang M, Zhou L. The role of long non-coding RNA HOTAIR in the progression and development of laryngeal squamous cell carcinoma interacting with EZH2. Acta Otolaryngol. 2017;137:90–98. doi: 10.1080/00016489.2016.1214982. [DOI] [PubMed] [Google Scholar]
- 9.Li D, Feng J, Wu T, Wang Y, Sun Y, Ren J, Liu M. Long intergenic noncoding RNA HOTAIR is overexpressed and regulates PTEN methylation in laryngeal squamous cell carcinoma. Am J Pathol. 2013;182:64–70. doi: 10.1016/j.ajpath.2012.08.042. [DOI] [PubMed] [Google Scholar]
- 10.Wang P, Wu T, Zhou H, Jin Q, He G, Yu H, Xuan L, Wang X, Tian L, Sun Y, Liu M, Qu L. Long noncoding RNA NEAT1 promotes laryngeal squamous cell cancer through regulating miR-107/CDK6 pathway. J Exp Clin Cancer Res. 2016;35:22. doi: 10.1186/s13046-016-0297-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang Z, Wang X, Cao S, Han X, Wang Z, Zhao X, Liu X, Li G, Pan X, Lei D. The long noncoding RNA TUG1 promotes laryngeal cancer proliferation and migration. Cell Physiol Biochem. 2018;49:2511–2520. doi: 10.1159/000493876. [DOI] [PubMed] [Google Scholar]
- 12.Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell. 2011;146:353–358. doi: 10.1016/j.cell.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Song Z, Zhao W, Cao D, Zhang J, Chen S. Elementary screening of lymph node metastatic-related genes in gastric cancer based on the co-expression network of messenger RNA, microRNA and long non-coding RNA. Braz J Med Biol Res. 2018;51:e6685. doi: 10.1590/1414-431X20176685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhao Y, Wang H, Wu C, Yan M, Wu H, Wang J, Yang X, Shao Q. Construction and investigation of lncRNA-associated ceRNA regulatory network in papillary thyroid cancer. Oncol Rep. 2018;39:1197–1206. doi: 10.3892/or.2018.6207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu S, Xie F, Xiang X, Liu S, Dong S, Qu K, Lin T. Identification of differentially expressed genes, lncRNAs and miRNAs which are associated with tumor malignant phenotypes in hepatoblastoma patients. Oncotarget. 2017;8:97554–97564. doi: 10.18632/oncotarget.22181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hui L, Wang J, Zhang J, Long J. lncRNA TMEM51-AS1 and RUSC1-AS1 function as ceRNAs for induction of laryngeal squamous cell carcinoma and prediction of prognosis. PeerJ. 2019;7:e7456. doi: 10.7717/peerj.7456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu H, Deng H, Zhao Y, Li C, Liang Y. LncRNA XIST/miR-34a axis modulates the cell proliferation and tumor growth of thyroid cancer through MET-PI3K-AKT signaling. J Exp Clin Cancer Res. 2018;37:279. doi: 10.1186/s13046-018-0950-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pinelli V, Marchica CL, Ludwig MS. Allergen-induced asthma in C57Bl/6 mice: hyper-responsiveness, inflammation and remodelling. Respir Physiol Neurobiol. 2009;169:36–43. doi: 10.1016/j.resp.2009.08.005. [DOI] [PubMed] [Google Scholar]
- 19.Tan JY, Sirey T, Honti F, Graham B, Piovesan A, Merkenschlager M, Webber C, Ponting CP, Marques AC. Extensive microRNA-mediated crosstalk between lncRNAs and mRNAs in mouse embryonic stem cells. Genome Res. 2015;25:655–666. doi: 10.1101/gr.181974.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang S, Ning Q, Zhang G, Sun H, Wang Z, Li Y. Construction of differential mRNA-lncRNA crosstalk networks based on ceRNA hypothesis uncover key roles of lncRNAs implicated in esophageal squamous cell carcinoma. Oncotarget. 2016;7:85728–85740. doi: 10.18632/oncotarget.13828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jin X, Guan Y, Sheng H, Liu Y. Crosstalk in competing endogenous RNA network reveals the complex molecular mechanism underlying lung cancer. Oncotarget. 2017;8:91270–91280. doi: 10.18632/oncotarget.20441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dai X, Kaushik AC, Zhang J. The emerging role of major regulatory RNAs in cancer control. Front Oncol. 2019;9:920. doi: 10.3389/fonc.2019.00920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Achour C, Aguilo F. Long non-coding RNA and Polycomb: an intricate partnership in cancer biology. Front Biosci (Landmark Ed) 2018;23:2106–2132. doi: 10.2741/4693. [DOI] [PubMed] [Google Scholar]
- 24.Yang T, Li S, Liu J, Yin D, Yang X, Tang Q. lncRNA-NKILA/NF-kappaB feedback loop modulates laryngeal cancer cell proliferation, invasion, and radioresistance. Cancer Med. 2018;7:2048–2063. doi: 10.1002/cam4.1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Na K, Li K, Sang T, Wu K, Wang Y, Wang X. Anticarcinogenic effects of water extract of sporoderm-broken spores of Ganoderma lucidum on colorectal cancer in vitro and in vivo. Int J Oncol. 2017;50:1541–1554. doi: 10.3892/ijo.2017.3939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu QQ, Zhang FF, Wang F, Qiu JH, Luo CH, Zhu GY, Liu YF. TIPE2 inhibits lung cancer growth attributing to promotion of apoptosis by regulating some apoptotic molecules expression. PLoS One. 2015;10:e0126176. doi: 10.1371/journal.pone.0126176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xu Y, Gao CC, Pan ZG, Zhou CW. Irigenin sensitizes TRAIL-induced apoptosis via enhancing pro-apoptotic molecules in gastric cancer cells. Biochem Biophys Res Commun. 2018;496:998–1005. doi: 10.1016/j.bbrc.2018.01.003. [DOI] [PubMed] [Google Scholar]
- 28.Hennessy BT, Smith DL, Ram PT, Lu Y, Mills GB. Exploiting the PI3K/AKT pathway for cancer drug discovery. Nat Rev Drug Discov. 2005;4:988–1004. doi: 10.1038/nrd1902. [DOI] [PubMed] [Google Scholar]
- 29.Kato H, Nakajima S, Saito Y, Takahashi S, Katoh R, Kitamura M. mTORC1 serves ER stress-triggered apoptosis via selective activation of the IRE1-JNK pathway. Cell Death Differ. 2012;19:310–320. doi: 10.1038/cdd.2011.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shi X, Li Y, Sun Y, Zhao X, Sun X, Gong T, Liang Z, Ma Y, Zhang X. Genome-wide analysis of lncRNAs, miRNAs, and mRNAs forming a prognostic scoring system in esophageal squamous cell carcinoma. PeerJ. 2020;8:e8368. doi: 10.7717/peerj.8368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liao M, Liao W, Xu N, Li B, Liu F, Zhang S, Wang Y, Wang S, Zhu Y, Chen D, Xie W, Jiang Y, Cao L, Yang BB, Zhang Y. LncRNA EPB41L4A-AS1 regulates glycolysis and glutaminolysis by mediating nucleolar translocation of HDAC2. EBioMedicine. 2019;41:200–213. doi: 10.1016/j.ebiom.2019.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huang JL, Zheng L, Hu YW, Wang Q. Characteristics of long non-coding RNA and its relation to hepatocellular carcinoma. Carcinogenesis. 2014;35:507–514. doi: 10.1093/carcin/bgt405. [DOI] [PubMed] [Google Scholar]
- 33.Sun T, Ye H, Wu CL, Lee GS, Kantoff PW. Emerging players in prostate cancer: long non-coding RNAs. Am J Clin Exp Urol. 2014;2:294–299. [PMC free article] [PubMed] [Google Scholar]
- 34.Almadori G, Cadoni G, Galli J, Ferrandina G, Scambia G, Exarchakos G, Paludetti G, Ottaviani F. Epidermal growth factor receptor expression in primary laryngeal cancer: an independent prognostic factor of neck node relapse. Int J Cancer. 1999;84:188–191. doi: 10.1002/(sici)1097-0215(19990420)84:2<188::aid-ijc16>3.0.co;2-1. [DOI] [PubMed] [Google Scholar]
- 35.Moon C, Chae YK, Lee J. Targeting epidermal growth factor receptor in head and neck cancer: lessons learned from cetuximab. Exp Biol Med (Maywood) 2010;235:907–920. doi: 10.1258/ebm.2009.009181. [DOI] [PubMed] [Google Scholar]
- 36.Tsiambas E, Stavrakis I, Lazaris AC, Karameris A, Patsouris E. Evaluation of epidermal growth factor receptor gene and chromosome 7 alterations in squamous cell carcinoma of the larynx, using chromogenic in situ hybridization on tissue microarrays. J Laryngol Otol. 2007;121:563–570. doi: 10.1017/S0022215106002374. [DOI] [PubMed] [Google Scholar]
- 37.Gao Y, Wu F, Zhou J, Yan L, Jurczak MJ, Lee HY, Yang L, Mueller M, Zhou XB, Dandolo L, Szendroedi J, Roden M, Flannery C, Taylor H, Carmichael GG, Shulman GI, Huang Y. The H19/let-7 double-negative feedback loop contributes to glucose metabolism in muscle cells. Nucleic Acids Res. 2014;42:13799–13811. doi: 10.1093/nar/gku1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhu M, Chen Q, Liu X, Sun Q, Zhao X, Deng R, Wang Y, Huang J, Xu M, Yan J, Yu J. lncRNA H19/miR-675 axis represses prostate cancer metastasis by targeting TGFBI. FEBS J. 2014;281:3766–3775. doi: 10.1111/febs.12902. [DOI] [PubMed] [Google Scholar]
- 39.Ma C, Nong K, Zhu H, Wang W, Huang X, Yuan Z, Ai K. H19 promotes pancreatic cancer metastasis by derepressing let-7’s suppression on its target HMGA2-mediated EMT. Tumour Biol. 2014;35:9163–9169. doi: 10.1007/s13277-014-2185-5. [DOI] [PubMed] [Google Scholar]
- 40.Zhang L, Yang F, Yuan JH, Yuan SX, Zhou WP, Huo XS, Xu D, Bi HS, Wang F, Sun SH. Epigenetic activation of the MiR-200 family contributes to H19-mediated metastasis suppression in hepatocellular carcinoma. Carcinogenesis. 2013;34:577–586. doi: 10.1093/carcin/bgs381. [DOI] [PubMed] [Google Scholar]
- 41.Yang C, Li H, Zhang T, Chu Y, Chen D, Zuo J. miR-200c overexpression inhibits the invasion and tumorigenicity of epithelial ovarian cancer cells by suppressing lncRNA HOTAIR in mice. J Cell Biochem. 2020;121:1514–1523. doi: 10.1002/jcb.29387. [DOI] [PubMed] [Google Scholar]
- 42.Tang B, Bao N, He G, Wang J. Long noncoding RNA HOTAIR regulates autophagy via the miR-20b-5p/ATG7 axis in hepatic ischemia/reperfusion injury. Gene. 2019;686:56–62. doi: 10.1016/j.gene.2018.10.059. [DOI] [PubMed] [Google Scholar]
- 43.Shao T, Hu Y, Tang W, Shen H, Yu Z, Gu J. The long noncoding RNA HOTAIR serves as a microRNA-34a-5p sponge to reduce nucleus pulposus cell apoptosis via a NOTCH1-mediated mechanism. Gene. 2019;715:144029. doi: 10.1016/j.gene.2019.144029. [DOI] [PubMed] [Google Scholar]
- 44.Chang L, Guo R, Yuan Z, Shi H, Zhang D. LncRNA HOTAIR regulates CCND1 and CCND2 expression by sponging miR-206 in ovarian cancer. Cell Physiol Biochem. 2018;49:1289–1303. doi: 10.1159/000493408. [DOI] [PubMed] [Google Scholar]
- 45.Chen H, Liu L, Li X, Shi Y, Liu N. MicroRNA-1294 inhibits the proliferation and enhances the chemosensitivity of glioma to temozolomide via the direct targeting of TPX2. Am J Cancer Res. 2018;8:291–301. [PMC free article] [PubMed] [Google Scholar]
- 46.Wang Z, Yan J, Zou T, Gao H. MicroRNA-1294 inhibited oral squamous cell carcinoma growth by targeting c-Myc. Oncol Lett. 2018;16:2243–2250. doi: 10.3892/ol.2018.8967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang ZF, Li GR, Cao CN, Xu Q, Wang GD, Jiang XF. MicroRNA-1294 targets HOXA9 and has a tumor suppressive role in osteosarcoma. Eur Rev Med Pharmacol Sci. 2018;22:8582–8588. doi: 10.26355/eurrev_201812_16621. [DOI] [PubMed] [Google Scholar]
- 48.Pan W, Pang LJ, Cai HL, Wu Y, Zhang W, Fang JC. MiR-1294 acts as a tumor suppressor in clear cell renal cell carcinoma through targeting HOXA6. Eur Rev Med Pharmacol Sci. 2019;23:3719–3725. doi: 10.26355/eurrev_201905_17797. [DOI] [PubMed] [Google Scholar]
- 49.Liu K, Li L, Rusidanmu A, Wang Y, Lv X. Down-regulation of MiR-1294 is related to dismal prognosis of patients with esophageal squamous cell carcinoma through elevating C-MYC expression. Cell Physiol Biochem. 2015;36:100–110. doi: 10.1159/000374056. [DOI] [PubMed] [Google Scholar]
- 50.Guo TY, Xu HY, Chen WJ, Wu MX, Dai X. Downregulation of miR-1294 associates with prognosis and tumor progression in epithelial ovarian cancer. Eur Rev Med Pharmacol Sci. 2018;22:7646–7652. doi: 10.26355/eurrev_201811_16381. [DOI] [PubMed] [Google Scholar]
- 51.Bonci D, Coppola V, Musumeci M, Addario A, Giuffrida R, Memeo L, D’Urso L, Pagliuca A, Biffoni M, Labbaye C, Bartucci M, Muto G, Peschle C, De Maria R. The miR-15a-miR-16-1 cluster controls prostate cancer by targeting multiple oncogenic activities. Nat Med. 2008;14:1271–1277. doi: 10.1038/nm.1880. [DOI] [PubMed] [Google Scholar]
- 52.Creighton CJ, Hernandez-Herrera A, Jacobsen A, Levine DA, Mankoo P, Schultz N, Du Y, Zhang Y, Larsson E, Sheridan R, Xiao W, Spellman PT, Getz G, Wheeler DA, Perou CM, Gibbs RA, Sander C, Hayes DN, Gunaratne PH Cancer Genome Atlas Research Network. Integrated analyses of microRNAs demonstrate their widespread influence on gene expression in high-grade serous ovarian carcinoma. PLoS One. 2012;7:e34546. doi: 10.1371/journal.pone.0034546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wu S, Huang S, Ding J, Zhao Y, Liang L, Liu T, Zhan R, He X. Multiple microRNAs modulate p21Cip1/Waf1 expression by directly targeting its 3’untranslated region. Oncogene. 2010;29:2302–2308. doi: 10.1038/onc.2010.34. [DOI] [PubMed] [Google Scholar]
- 54.DeVita VT, Hellman S, Rosenberg SA. Cancer: principles & practice of oncology. Lippincott-Raven. 1997 [Google Scholar]