Abstract
The role of autophagy in tumor development is complicated. More signaling mechanisms of bone morphogenetic protein 4 (BMP4) involving in hepatocellular carcinoma (HCC) are needed to be clarified. The present study aimed to identify whether BMP4 contributes to the regulation of autophagy in the progression of HCC. We found that increased BMP4 expression was significantly correlated with TNM stage and metastasis. BMP4 treatment promoted HCC cells invasion and induced autophagy. Blocking autophagy by 3-MA or silenced Beclin1 attenuated BMP4-induced autophagy and cell invasion. Further study revealed that JNK/Beclin1 pathway participated in the process of autophagy and JNK inhibitor SP600125 could attenuate autophagy and reduce the invasive ability of HCC cells induced by BMP4. In a word, BMP4-induced autophagy facilitates invasion of HCC by JNK/Beclin1 signaling pathway.
Keywords: BMP4, autophagy, hepatocellular carcinoma, Beclin1
Introduction
Hepatocellular carcinoma (HCC) is one of the most common cancers, with increasing global incidence [1]. HCC is extremely heterogeneous and resistant to chemotherapy. The long-term prognosis of HCC patients is still unsatisfactory due to tumor recurrence and metastasis. HCC involves in a multistep carcinogenesis process, so it is of great significance to understand the molecular mechanisms underlying HCC invasion and metastasis.
Bone morphogenetic protein 4 (BMP4), a member of the transforming growth factor-beta (TGF-β) superfamily, can regulate cell proliferation, differentiation and motility. BMP4 performs its biological functions by combining with the BMP receptors complex to initiate phosphorylation-dependent activation of Smad1/5/8 [2], which has been reported to implicate in multiple malignancies [3-5]. BMP4 expression increases in chronic liver disease and plays an important role in chronic liver injury, cirrhosis and even HCC [6]. However, in HCC, BMP4 exerts complicated influences, which are yet to be fully elucidated. BMP4 was reported to be abnormally expressed in HCC and closely associated with the prognosis of patients [7,8]. As a potential biological marker of HCC progression, the exact role and molecular mechanism of BMP4 in HCC need to be explicated.
Autophagy encapsulates degradable contents of cytoplasm and then transports them to the lysosomes for degradation to maintain energetic homeostasis. It is closely related to many diseases, including cancer [9,10]. Autophagy occurs frequently during tumorigenesis and progression and exerts complicated functions in cancer metastasis. It is reported that autophagy has both pro-metastatic and anti-metastatic roles, which can switch during different periods of tumor progression [11]. In the early stage of cancer metastasis, autophagy may exert an inhibitory effect by alleviating cell senescence, while in the advanced stage, autophagy can promote cancer progression [12]. It is suggested that there is a potential relationship between autophagic cell death and cancer metastasis [13]. The characteristics of autophagy have been extensively investigated, however, the specific roles and mechanism of it in HCC are still not well known. Therefore we carried out this study to investigate the effects of autophagy on BMP4-influenced cell invasion and the involved molecular mechanisms in HCC.
Materials and methods
Patients and tissue specimens of HCC
The tumor tissues were randomly collected from 102 patients with HCC between January 2014 and March 2015. They were all histologically confirmed as HCC by pathological or combined imageological examination. Exclusion criteria were as follows: previous liver transplantation, anticancer treatment prior to surgery, not enough liver function reservation, severe cardiopulmonary disease or other systematic diseases.
Immunohistochemistry (IHC)
After de-paraffinization, dehydration and antigen retrieval, the paraffin-embedded sections were incubated with 10% goat serum for 30 minutes and followed by the primary antibodies against BMP4 (1:300 dilution, Abcam, Cambridge, UK) and Beclin1 (1:200 dilution, Abcam) respectively at 4°C overnight. The slides were subsequently incubated with the corresponding secondary antibodies (Zhongshan Goldenbridge Biotechnology, Beijing, China). The negative control sections were probed with phosphate buffered saline (PBS) instead of the primary antibodies under the same experimental conditions. The IHC staining was scored according to the percentage of positive-staining cells and staining intensity. The positive cell percentage of cells was scored as 0 to 4. The staining intensity was classified as: 1 = weak, 2 = moderate and 3 = strong. Immunoreactive score (IS) of each specimen was determined by multiplication of these two scores listed above. Negative (IS < 2) or positive (IS≥2) was defined according to IS.
Cell culture
Human HCC cell lines of HepG2 and HCCLM3 were cultured in high glucose DMEM medium supplemented with 10% fetal bovine serum (FBS) (Gibco, Grand Island, NY) at 37°C in a the incubator with 5% CO2.
Migration and invasion assays
Cell migration and invasion were assessed in the 24-well 8-µm pore plates (Corning, Tewksbury, MA). Transwell chambers, pre-coated with or without growth factor-reduced matrigel, would be applied to evaluate HCC cells’ migration and invasion abilities respectively. According to the manufacturer’s instructions, HCC cells were re-suspended in the serum-free medium and seeded in the upper chamber at a density of 1×104 cells/100 µl/well, while the lower chambers were covered with 600 µl 10% serum-containing medium. After incubation for 24 h, the residual unpenetrated cells were removed, and the lower surface which covered with the migrated or invaded cells was fixed with 4% paraformaldehyde and stained with 0.1% crystal violet for 20 minutes. The stained cells were counted under an inverted microscope.
Western blot
The treated cells were trypsinized and collected. The total protein samples were extracted using cell lysis buffer, separated by 10% sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and transferred onto the PVDF membrane (Millipore, Bedford, MA). The membranes were blocked in 5 mg/ml skim milk at room temperature for 2 hours and incubated with the primary antibodies at 4°C overnight. After being exposed to HRP-conjugated secondary antibody for 1 hour at 37°C, the protein bands were obtained by the enhanced chemiluminescence kit (Millipore). GAPDH protein expression was used as an internal control.
Quantitative real-time PCR (qRT-PCR)
Total RNA was extracted from HCC cells by Trizol reagent (Invitrogen, Carlsbad, CA) and the cDNA was synthetized using PrimeScriptTM RT reagent Kit (TaKaRa Bio Inc., Otsu, Japan) according to the manufacturer’s protocol. The specific primers for Beclin1 were as follows: forward primer: 5’-TCCGGGCTCCCGAGG-3’; reverse primer: 5’-GGGGGATGAATCTGCGAGAG-3’. GAPDH was used as an internal control, with forward primer: 5’-CTGGGCTACACTGAGCACC-3’ and reverse primer: 5’-AAGTGGTCGTTGAGGGCAATG-3’. Amplifications were carried out with the following cycling parameters: 95°C for 2 min and subsequent 40 cycles comprising of annealing at 95°C for 15 sec and 60°C for 60 sec. Relative mRNA expression levels were calculated based on Ct values as: 2-ΔCt [ΔCt = Ct (targeting gene) - Ct (GAPDH)] and normalized to the internal control.
mRFP-GFP-LC3 adenovirus transfection and autophagic flux analysis
To observe the autophagic flux, mRFP-GFP-LC3 adenovirus (HanBio Technology Co., Shanghai, China) were transfected into the HCC cells seeded onto 96-well plates with an optimal multiplicity of infection (MOI) of 30 TU/mL. 4 hours later, the adenovirus was removed and the treated cells were fixed with 4% paraformaldehyde in the dark. Puncta that appear yellow when merged, indicate autophagosomes. Opera High Content Screening System (Perkin-Elmer) was used to capture and analyze the imaging of autophagic flux.
Beclin1 siRNA and transfection
siRNA sequences targeting Beclin1 gene were designed as follows: sense 5’-GUGGAAUGGAAUGAGAUUATT-3’ and antisense 5’-UAAUCUCAUUCCAUUCCACTT-3’ and transfected to the cells (covered 80% of the petri dish) using Lipofectamine® RNAiMAX Transfection Reagent (Invitrogen) according to the manufacturer’s instructions. The down-regulated expression of Beclin1 acquired by siRNA was confirmed by qRT-PCR and Western blot.
Statistical analysis
Statistical analysis was performed by SPSS software (Version 17.0, SPSS Inc, Chicago, IL) and GraphPad Prism 5.0. The relationship between BMP4 expression and clinicopathologic parameters was analyzed using the Pearson X2 test. The differences between groups were analyzed using Student’s t-test when there were only two groups, or assessed by one-way ANOVA when there were more than two groups. Statistical tests in the study were two-sided and the value of P < 0.05 was considered to be statistically significant.
Results
Expression of BMP4 was associated with advanced stage and metastasis of HCC
BMP4 expression was determined by IHC staining. The results indicated that HCC patients at stage II or III/IV exhibited higher BMP4 expression levels in tumor than those at stage I (4.368 ± 0.629 versus 2.571 ± 0.585; 4.667 ± 0.677 versus 2.571 ± 0.585; P < 0.05, respectively). BMP4 expression levels were statistically higher in HCC with intrahepatic or extrahepatic metastasis than those without metastasis (4.362 ± 0.468 versus 2.364 ± 0.431, P < 0.01, Figure 1A). There was no statistically significant association between BMP4 expression levels and other clinicopathologic parameters, such as age, gender, liver cirrhosis, serum AFP, tumor diameter and tumor number (Table 1).
Figure 1.
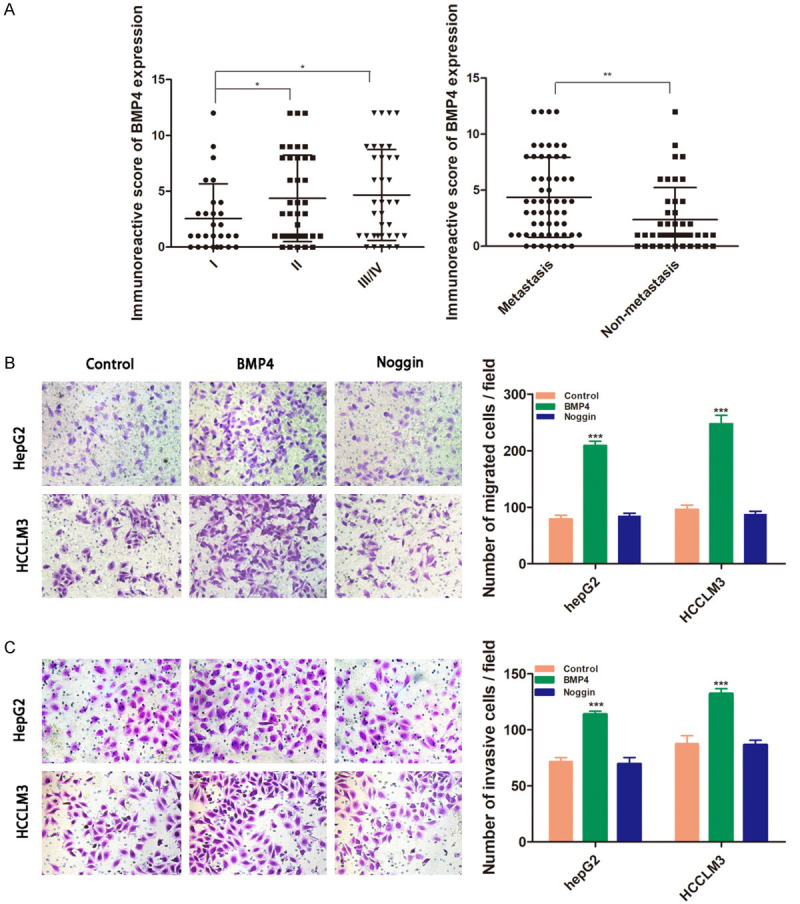
BMP4 promotes migration and invasion of HCC cells. A. The patients with II or III/IV stage exhibited higher BMP4 expression in tumor tissues than those with stage I; BMP4 expression levels were higher in HCC with intrahepatic or extrahepatic metastasis. B, C. The HCC cells HCCLM3 and HepG2 were respectively treated with recombinant human BMP4 (100 ng/ml) or its receptor antagonist of Noggin (200 ng/ml). B. Transwell migration assay indicated that BMP4 treatment increased numbers of migrated cells. C. Transwell invasion assays were performed to investigate that BMP4 groups had the highest invasion ability. Data are shown as mean ± SD and from three independent experiments. *: P < 0.05, **: P < 0.01, ***: P < 0.001.
Table 1.
Relationship between BMP4 expression and the clinical and pathological features in HCC patients
| Clinical and pathological indexes | BMP4 expression | |||
|---|---|---|---|---|
|
| ||||
| Low | High | p | ||
| Age (years) | ≤ge | 17 | 34 | 0.108 |
| >52 | 26 | 25 | ||
| Sex | Male | 27 | 37 | 0.580 |
| Female | 16 | 22 | ||
| HBsAg | Negative | 20 | 24 | 0.350 |
| Positive | 23 | 35 | ||
| Albumin | ≤lb g/L | 25 | 34 | 0.410 |
| >35 g/L | 14 | 29 | ||
| Child-Pugh classification | A | 28 | 41 | 0.399 |
| B | 15 | 18 | ||
| AFP | ≤FP9 μg/L | 22 | 36 | 0.215 |
| >400 μg/L | 21 | 23 | ||
| HBV infection | Absent | 16 | 15 | 0.145 |
| Present | 27 | 44 | ||
| Liver cirrhosis | Absent | 20 | 25 | 0.415 |
| Present | 23 | 34 | ||
| Tumor size | ≤u cm | 27 | 39 | 0.445 |
| >5 cm | 16 | 20 | ||
| Tumor number | Single | 22 | 36 | 0.215 |
| Multiple | 21 | 23 | ||
| Tumor differentiation | I-II | 34 | 36 | 0.041 |
| III-IV | 9 | 23 | ||
| Distant metastasis stage | M0 | 30 | 28 | 0.020 |
| M1 | 13 | 31 | ||
Overexpression of BMP4 promoted migration and invasion of HCC cells
The HCC cells HepG2 and HCCLM3 were respectively treated with recombinant human BMP4 (100 ng/ml) or its receptor antagonist of Noggin (200 ng/ml). Transwell assays were performed to investigate the migration and invasion abilities of HCC cells subjected to BMP4. The results indicated that BMP4 increased the rates of cell migration and invasion, while Noggin blocked these changes with less migrated and invaded cells (Figure 1B, 1C).
BMP4 induced autophagy in HCC cell lines
In order to investigate the role of autophagy in BMP4-mediated migration and invasion of HCC cells, we treated HCC cells with 100 ng/ml BMP4 for 24 hours and investigated the expression of autophagy related markers (LC3, p62 and Beclin1) by Western blot. The results showed an increased conversion of LC3-I to LC3-II after BMP4 treatment. The LC3-II/GAPDH ratio, which was calculated to reflect autophagy activity, was significantly increased in the BMP4 group. Consistent with the change of LC3-II, the expression of autophagy related gene Beclin1 was increased in HCC cells being exposed to BMP4, while the level of autophagic protein p62 was reduced (Figure 2A, 2B). To observe and analyze the autophagy flux, HCCLM3 and HepG2 cells were transfected with the mRFP-GFP-LC3 adenovirus. Puncta that appeared yellow when merged, indicated autophagosomes. In the Figure 2C, after treatment with BMP4 for 24 h, we observed an increased accumulation of fluorescent cells with LC3 dots as compared with the control groups (P < 0.01, respectively).
Figure 2.
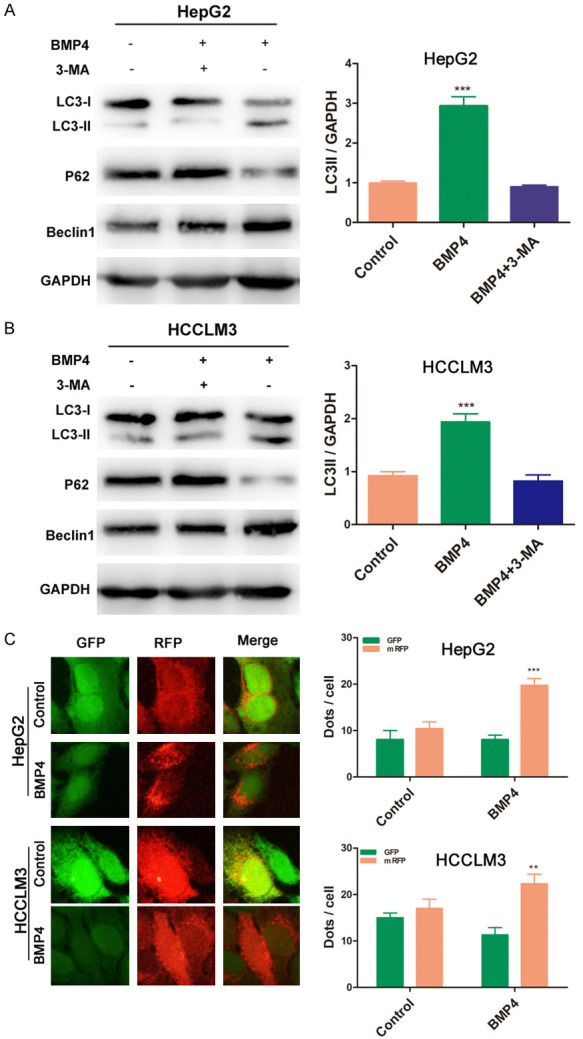
BMP4 induced autophagy in HCC cell lines. A, B. BMP4 treatment increased LC3-II conversion and Beclin1 expression, while reduced p62 expression. The LC3-II/GAPDH ratio was significantly increased in the BMP4 groups (P < 0.001). C. After treatment with BMP4 for 24 h, an increased accumulation of fluorescent cells with LC3 dots as compared with the control groups was observed (P < 0.01, respectively). Data are shown as mean ± SD and from three independent experiments. **: P < 0.01, ***: P < 0.001.
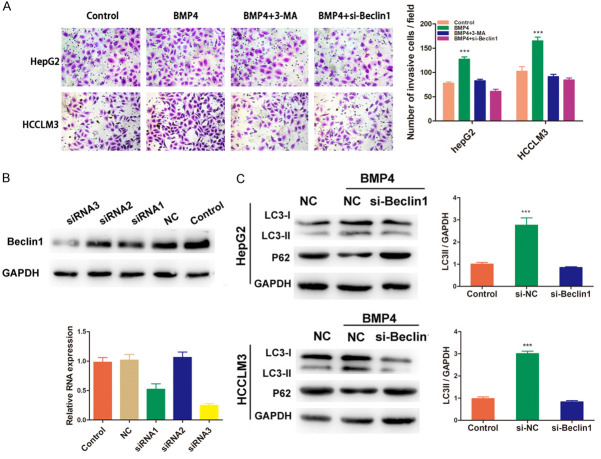
BMP4-activated autophagy promoted HCC cells migration and invasion
Since autophagy could be induced by BMP4, however, the specific roles of it in BMP4-promoted migration and invasion need to be clarified. HepG2 and HCCLM3 cells were treated with BMP4 with or without the administration of autophagy inhibitor 3-methyladenine (3-MA) for 24 h, the results of transwell assay indicated that 3-MA significantly attenuated the invasion ability promoted by BMP4 (P < 0.001, respectively, Figure 3A). Further study found that this invasion ability could also be inhibited by siRNA targeting Beclin1 (Figure 3A). To investigate the role of Beclin1, an autophagy related gene, in the signal transduction of autophagy, we used siRNA targeting Beclin1 to block autophagy. As shown in Figure 3B, siRNA3 was determined to be si-Beclin1. We found that reduced Beclin1 expression could attenuate LC3-II conversion promoted by BMP4, while increased P62 expression (Figure 3C), suggesting the involvement of Beclin1 in BMP4-induced autophagy regulation.
Figure 3.
BMP4 activated-autophagy promoted HCC cells migration and invasion. A. HepG2 and HCCLM3 cells were treated with BMP4 with or without the administration of 3-MA for 24 h, the results indicated that 3-MA significantly attenuated the invaded cells, which could also be inhibited by siRNA targeting Beclin1 (P < 0.001, respectively). B. Knockdown of Beclin1 by siRNAs was confirmed by qRT-PCR and Western blot assays. siRNA3 was named as si-Beclin1 with the highest inhibition efficiency of 75.3%. C. Western blot assay indicated that reduced Beclin1 could attenuate LC3-II conversion and increase P62 expression induced by BMP4. Data are shown as mean ± SD and from three independent experiments. ***: P < 0.001.
Up-regulation of Beclin1 was associated with BMP4 expression in clinical specimens of HCC
To explore the role of autophagy in HCC further, we compared the expression of BMP4 and Beclin1 at protein level in human HCC tissue. The result revealed a significantly positive correlation between BMP4 and Beclin1 expressions in these specimens (Spearman’s Ρ = 0.432, P < 0.001, Figure 4).
Figure 4.
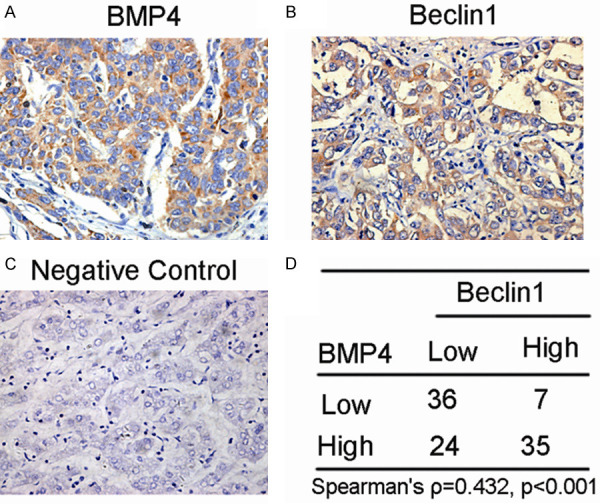
Beclin1 is positively associated with BMP4 expression in clinical specimens of HCC. IHC staining and scoring of BMP4 and Beclin1 protein expression in human HCC tissues (Original magnification: ×400). Pearson correlation revealed a significant positively correlation between BMP4 and Beclin1 expression levels.
JNK/Beclin1 signaling played an important role in BMP4-activated autophagy
Earlier literature confirmed SAPK/JNK pathway participated in the process of autophagy, however, the role of SAPK/JNK in BMP4 activated-autophagy and invasion in HCC has not been reported. In the present study, we found that p-JNK increased after BMP4 treatment, while decreased when the SP600125 (20 μM), a JNK pathway inhibitor, was added in two cell lines. Further study was carried out to explore the involvement of SP600125 in BMP4 activated-autophagy, as shown in Figure 5A, JNK pathway inhibitor SP600125 decreased the expression of Beclin1 and LC3-II. It is suggested that BMP4 activated autophagy by JNK/Beclin1 signaling pathway. The relationship between SAPK/JNK signaling and BMP4-promoted invasion in HCC cells was investigated by using JNK inhibitor SP600125. The results indicated that SP600125 significantly decreased the number of the invasive cells as compared with BMP4 groups (P < 0.01, respectively, Figure 5B), so it is suggested that JNK was responsible for BMP4-induced autophagy and consequential invasion.
Figure 5.
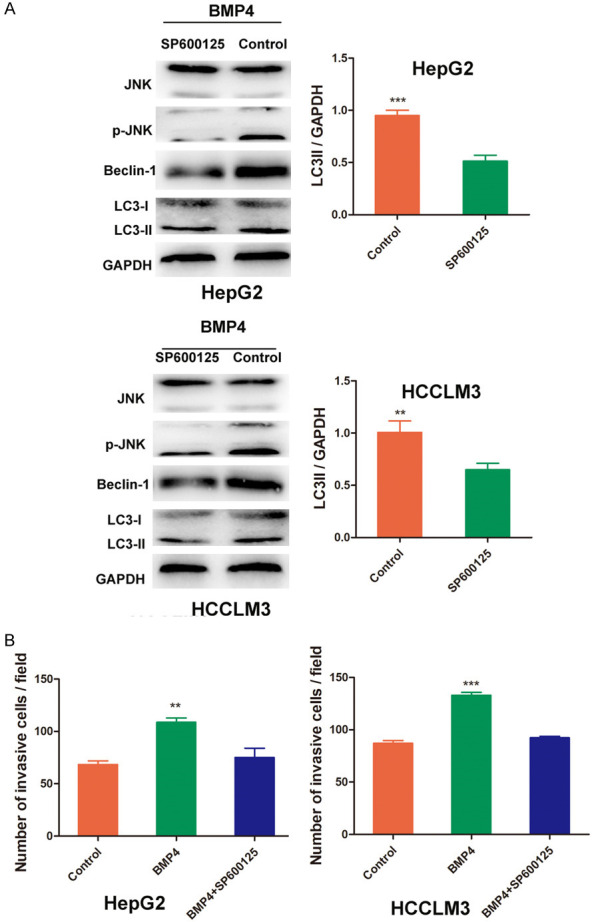
Signaling pathways involved in BMP4-promoted autophagy in HCC. A. HepG2 and HCCLM3 cells were treated with BMP4 (100 ng/mL) or Noggin for 24 h, the protein expression and JNK and phosphorylation of JNK (p-JNK) were determined by Western blot. HepG2 and HCCLM3 cells were pretreated with JNK inhibitor SP600125 (20 μM) or for 30 min, followed by addition of BMP4, Western blot was performed to detect the expression of Beclin1 and LC3-II. JNK pathway inhibition by SP600125 significantly attenuated the BMP4-activated autophagy in HepG2 and HCCLM3 cells. B. HCC cells were pretreated with JNK pathway inhibitor by SP600125 (20 μM) for 6 h, then co-cultured with BMP4 for 24 h, cell invasion ability was analyzed by Transwell assay. SP600125 significantly attenuated cell invasion both in HepG2 and HCCLM3. Data are shown as mean ± SD and from three independent experiments. **: P < 0.01, ***: P < 0.001.
Discussion
It was reported that the functions of TGF-β signaling cascade were complicated and might play opposing roles in tumorigenesis and metastasis depending on tumor types and cellular context [14,15]. Our previous studies have found that BMP4 was increased in HCC and facilitated HCC proliferation and chemoresistance [7,8]. Furthermore, in this study, we firstly investigated the involvement of BMP4 on autophagy-regulated HCC progression.
BMP4 promotes cancer cell invasion and metastasis by employing different signal pathways. Epithelial-mesenchymal transition (EMT) is a well known mechanism contributing to tumor metastasis [16,17]. BMP4 can also act as one of the regulatory factors functioning on cancer microenvironment. BMP4-mediated immunosuppression in tumors promotes cancer progression [18]. For example, BMP4 secretion by bladder cancer cells provides the M2 signal necessary for a protumoral immune environment and favors tumor progression in bladder cancer [3]. Differently, our research found that BMP4 enhanced invasiveness of liver cancer cells by autophagy. The research indicated that BMP4 treatment increased LC3-II conversion and the autophagic flux in HCC cells, while Noggin or autophagy inhibitor 3-MA could inhibit this phenomenon.
The role of autophagy in HCC is not unchangeable but dynamic. During the dysplastic phase in hepatocytes, autophagy acts as a tumor suppressor, however, once the tumor is established, unbalanced autophagy will contribute to HCC development [19]. Elevated autophagy was found in many malignant tumors and may be responsible for cancer progression. LC3 was associated with lymph node metastasis and reduced survival rate in human breast cancer, colorectal cancer and pancreatic cancer [20-22]. It is suggested that autophagy involves in metastasis by disseminating circulating tumor cell (CTC) and promoting the survival of dormant tumor cells, specifically the stem-like subpopulations of tumor cells that drive invasion and treatment resistance [23]. Autophagy can direct induce tumor cell motility and invasion by promoting a stem cell phenotype and production of autophagy-dependent secreted factors [24]. In addition to the direct role, autophagy could modulate the invasiveness of tumor cell through the regulation of EMT. Detachment of ECM induces autophagy and protects disseminating cancer cells from anoikis [12,25]. Autophagy participates actively in tumor growth also by regulating the interactions between tumor stroma and other kinds of cells to modulate tumor microenvironment and promoting tumor cell escaping. What’s more, by blocking tumor immune surveillance, autophagy could involve in cancer development [27]. All these evidences elucidate that autophagy mediates anti-tumor effects and participates in various signaling pathways directly or indirectly to involve in the progression of HCC.
Beclin1, the mammalian orthologue of yeast autophagy-related gene (ATG), is a central regulator of autophagy. It can bind to PI3KC3 to assemble autophagy-inducing complexes [28]. Qu X found that a heterozygous mutation could disrupt the Beclin1 autophagy gene to promote hepatocyte tumourigenesis [29]. Accordingly, we found that knockdown of Beclin1 attenuated LC3-II conversion and increased P62 expression promoted by BMP4, suggesting the involvement of Beclin1 in BMP4-induced autophagy regulation.
It is necessary to understand the molecular mechanism and regulatory network of autophagy in cancer invasion and metastasis. Various stress conditions stimulation of autophagy identified the signal transduction mechanisms. To sum up, the common regulations are as follows: ER stress stimulates autophagy through the PERK-eIF2α pathway, the IRE1-JNK1 pathway and Ca2+ release. Hypoxia or oxidative stress is through JNK1, DAPk, and BNIP3 inducing autophagy by disrupting the Bcl-2/Beclin1 interaction [30]. In our study, we found that enhanced activation of JNK/Beclin1 contributed to the progression of HCC.
In a word, our study suggests that BMP4 promotes HCC invasion by autophagy induction. BMP4-activated JNK/Beclin1 signaling pathway facilitates the progression. BMP4 and autophagy may be potent therapeutic targets in HCC to improve the patients’ treatment response and prognosis.
Acknowledgements
This study was supported by the grants from the National Natural Science Foundation of China (No. 81802467), the Nature Science Foundation of Shandong Province (No. ZR2018LH014) and phD Research Foundation of Affiliated Hospital of Jining Medical University (No. 2017-BS-010).
Disclosure of conflict of interest
None.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin. 2019;69:7–34. doi: 10.3322/caac.21551. [DOI] [PubMed] [Google Scholar]
- 2.Park ES, Woods DC, Tilly JL. Bone morphogenetic protein 4 promotes mammalian oogonial stem cell differentiation via Smad1/5/8 signaling. Fertil Steril. 2013;100:1468–1475. doi: 10.1016/j.fertnstert.2013.07.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Martinez VG, Rubio C, Martinez-Fernandez M, Segovia C, Lopez-Calderon F, Garin M I, Teijeira A, Munera-Maravilla E, Varas A, Sacedon R, Guerrero F, Villacampa F, de la Rosa F, Castellano D, Lopez-Collazo E, Paramio J M, Vicente A, Duenas M. BMP4 induces M2 macrophage polarization and favors tumor progression in bladder cancer. Clin Cancer Res. 2017;23:7388–7399. doi: 10.1158/1078-0432.CCR-17-1004. [DOI] [PubMed] [Google Scholar]
- 4.Valencia J, M FL, Fraile-Ramos A, Sacedon R, Jimenez E, Vicente A, Varas A. Acute lymphoblastic leukaemia cells impair dendritic cell and macrophage differentiation: role of BMP4. Cells. 2019;8:722. doi: 10.3390/cells8070722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mano Y, Yoshio S, Shoji H, Tomonari S, Aoki Y, Aoyanagi N, Okamoto T, Matsuura Y, Osawa Y, Kimura K, Yugawa K, Wang H, Oda Y, Yoshizumi T, Maehara Y, Kanto T. Bone morphogenetic protein 4 provides cancer-supportive phenotypes to liver fibroblasts in patients with hepatocellular carcinoma. J Gastroenterol. 2019;54:1007–1018. doi: 10.1007/s00535-019-01579-5. [DOI] [PubMed] [Google Scholar]
- 6.Rowe IA, Galsinh SK, Wilson GK, Parker R, Durant S, Lazar C, Branza-Nichita N, Bicknell R, Adams DH, Balfe P, McKeating JA. Paracrine signals from liver sinusoidal endothelium regulate hepatitis C virus replication. Hepatology. 2014;59:375–384. doi: 10.1002/hep.26571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ma J, Zeng S, Zhang Y, Deng G, Qu Y, Guo C, Yin L, Han Y, Shen H. BMP4 enhances hepatocellular carcinoma proliferation by promoting cell cycle progression via ID2/CDKN1B signaling. Mol Carcinog. 2017;56:2279–2289. doi: 10.1002/mc.22681. [DOI] [PubMed] [Google Scholar]
- 8.Ma J, Zeng S, Zhang Y, Deng G, Qu Y, Guo C, Yin L, Han Y, Cai C, Li Y, Wang G, Bonkovsky HL, Shen H. BMP4 promotes oxaliplatin resistance by an induction of epithelial-mesenchymal transition via MEK1/ERK/ELK1 signaling in hepatocellular carcinoma. Cancer Lett. 2017;411:117–129. doi: 10.1016/j.canlet.2017.09.041. [DOI] [PubMed] [Google Scholar]
- 9.Sridhar S, Botbol Y, Macian F, Cuervo AM. Autophagy and disease: always two sides to a problem. J Pathol. 2012;226:255–73. doi: 10.1002/path.3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hundeshagen P, Hamacher-Brady A, Eils R, Brady NR. Concurrent detection of autolysosome formation and lysosomal degradation by flow cytometry in a high-content screen for inducers of autophagy. BMC Biol. 2011;9:38. doi: 10.1186/1741-7007-9-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kenific CM, Thorburn A, Debnath J. Autophagy and metastasis: another double-edged sword. Curr Opin Cell Biol. 2010;22:241–245. doi: 10.1016/j.ceb.2009.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Su Z, Yang Z, Xu Y, Chen Y, Yu Q. Apoptosis, autophagy, necroptosis, and cancer metastasis. Mol Cancer. 2015;14:48. doi: 10.1186/s12943-015-0321-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hashimoto I, Koizumi K, Tatematsu M, Minami T, Cho S, Takeno N, Nakashima A, Sakurai H, Saito S, Tsukada K, Saiki I. Blocking on the CXCR4/mTOR signalling pathway induces the anti-metastatic properties and autophagic cell death in peritoneal disseminated gastric cancer cells. Eur J Cancer. 2008;44:1022–1029. doi: 10.1016/j.ejca.2008.02.043. [DOI] [PubMed] [Google Scholar]
- 14.Loomans HA, Andl CD. Intertwining of activin a and TGFbeta signaling: dual roles in cancer progression and cancer cell invasion. Cancers (Basel) 2014;7:70–91. doi: 10.3390/cancers7010070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ikushima H, Miyazono K. TGFbeta signalling: a complex web in cancer progression. Nat Rev Cancer. 2010;10:415–424. doi: 10.1038/nrc2853. [DOI] [PubMed] [Google Scholar]
- 16.Hamada S, Satoh K, Hirota M, Kimura K, Kanno A, Masamune A, Shimosegawa T. Bone morphogenetic protein 4 induces epithelial-mesenchymal transition through MSX2 induction on pancreatic cancer cell line. J Cell Physiol. 2007;213:768–774. doi: 10.1002/jcp.21148. [DOI] [PubMed] [Google Scholar]
- 17.Serrao A, Jenkins LM, Chumanevich AA, Horst B, Liang J, Gatza ML, Lee NY, Roninson I B, Broude EV, Mythreye K. Mediator kinase CDK8/CDK19 drives YAP1-dependent BMP4-induced EMT in cancer. Oncogene. 2018;37:4792–4808. doi: 10.1038/s41388-018-0316-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen L, Yi X, Goswami S, Ahn YH, Roybal JD, Yang Y, Diao L, Peng D, Peng D, Fradette JJ, Wang J, Byers LA, Kurie JM, Ullrich SE, Qin FX, Gibbons DL. Growth and metastasis of lung adenocarcinoma is potentiated by BMP4-mediated immunosuppression. OncoImmunology. 2016;5:e1234570. doi: 10.1080/2162402X.2016.1234570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sun K, Guo XL, Zhao QD, Jing YY, Kou XR, Xie XQ, Zhou Y, Cai N, Gao L, Zhao X, Zhang SS, Song JR, Li D, Deng WJ, Li R, Wu MC, Wei LX. Paradoxical role of autophagy in the dysplastic and tumor-forming stages of hepatocarcinoma development in rats. Cell Death Dis. 2013;4:e501. doi: 10.1038/cddis.2013.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hashemi-Sadraei N, Muller-Greven GM, Abdul-Karim FW, Ulasov I, Downs-Kelly E, Burgett ME, Lauko A, Qadan MA, Weil RJ, Ahluwalia MS, Du L, Prayson RA, Chao ST, Budd TG, Barnholtz-Sloan J, Nowacki AS, Keri RA, Gladson CL. Expression of LC3B and FIP200/Atg17 in brain metastases of breast cancer. J Neurooncol. 2018;140:237–248. doi: 10.1007/s11060-018-2959-5. [DOI] [PubMed] [Google Scholar]
- 21.Wu S, Sun C, Tian D, Li Y, Gao X, He S, Li T. Expression and clinical significances of Beclin1, LC3 and mTOR in colorectal cancer. Int J Clin Exp Pathol. 2015;8:3882–3891. [PMC free article] [PubMed] [Google Scholar]
- 22.Yang YH, Liu JB, Gui Y, Lei LL, Zhang SJ. Relationship between autophagy and perineural invasion, clinicopathological features, and prognosis in pancreatic cancer. World J Gastroenterol. 2017;23:7232–7241. doi: 10.3748/wjg.v23.i40.7232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marcucci F, Ghezzi P, Rumio C. The role of autophagy in the cross-talk between epithelial-mesenchymal transitioned tumor cells and cancer stem-like cells. Mol Cancer. 2017;16:3. doi: 10.1186/s12943-016-0573-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mowers EE, Sharifi MN, Macleod KF. Autophagy in cancer metastasis. Oncogene. 2017;36:1619–1630. doi: 10.1038/onc.2016.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gugnoni M, Sancisi V, Gandolfi G, Manzotti G, Ragazzi M, Giordano D, Tamagnini I, Tigano M, Frasoldati A, Piana S, Ciarrocchi A. Cadherin-6 promotes EMT and cancer metastasis by restraining autophagy. Oncogene. 2017;36:667–677. doi: 10.1038/onc.2016.237. [DOI] [PubMed] [Google Scholar]
- 26.Ngabire D, Kim GD. Autophagy and inflammatory response in the tumor microenvironment. Int J Mol Sci. 2017;18:2016. doi: 10.3390/ijms18092016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ma Y, Galluzzi L, Zitvogel L, Kroemer G. Autophagy and cellular immune responses. Immunity. 2013;39:211–227. doi: 10.1016/j.immuni.2013.07.017. [DOI] [PubMed] [Google Scholar]
- 28.Lu Y, Bu M, Yun H. Sevoflurane prevents hypoxia/reoxygenation-induced cardiomyocyte apoptosis by inhibiting PI3KC3-mediated autophagy. Hum Cell. 2019;32:150–159. doi: 10.1007/s13577-018-00230-4. [DOI] [PubMed] [Google Scholar]
- 29.Qu X, Yu J, Bhagat G, Furuya N, Hibshoosh H, Troxel A, Rosen J, Eskelinen EL, Mizushima N, Ohsumi Y, Cattoretti G, Levine B. Promotion of tumorigenesis by heterozygous disruption of the beclin 1 autophagy gene. J Clin Invest. 2003;112:1809–1820. doi: 10.1172/JCI20039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.He C, Klionsky DJ. Regulation mechanisms and signaling pathways of autophagy. Annu Rev Genet. 2009;43:67–93. doi: 10.1146/annurev-genet-102808-114910. [DOI] [PMC free article] [PubMed] [Google Scholar]