Abstract
Current risk assessments of transgenic crops do not take into consideration whether exogenous proteins interact with endogenous proteins and thereby induce unintended effects in the crops. Therefore, the unintended effects through protein interactions in insect-resistant transgenic rice merit investigation. Here, a yeast two-hybrid assay was used to evaluate interactions between Bacillus thuringiensis (Bt) protein-derived Cry1Ab/c insect resistance rice Huahui-1 and the endogenous proteins of its parental rice Minghui-63. The authenticity of the strongest interactions of Cry1Ab/c and 14 endogenous proteins involved in photosynthesis and stress resistance, which may be primarily responsible for the significant phenotypic differences between transgenic Huahui-1 and parental Minghui-63, were then analyzed and validated by subcellular co-localization, bimolecular fluorescence complementation and co-immunoprecipitation. As the exogenous full-length Cry1Ab/c protein was found to have self-activating activity, we cleaved it - into three segments based on its three domains, and these were screened for interaction with host proteins using the yeast two-hybrid assay. Sixty endogenous proteins related to the regulation of photosynthesis, stress tolerance, and substance metabolism were found to interact with the Cry1Ab/c protein. The results of bimolecular fluorescence complementation and co-immunoprecipitation verified the interactions between the full-length Cry1Ab/c protein and 12 endogenous proteins involved in photosynthesis 23KD, G, PSBP, Rubisco, Trx, THF1 and stress resistance CAMTAs, DAHP, E3s, HKMTs, KIN13A, FREE1. We used a combination of yeast two-hybrid, bimolecular fluorescence complementation, and co-immunoprecipitation to identify Cry1Ab/c interacting with rice proteins that seem to be associated with the observed unintended effects on photosynthesis and stress resistance between Huahui-1 and Minghui-63 rice plants, and analyze the possible interaction mechanisms by comparing differences in cell localization and interaction sites between these interactions. The results herein provide a molecular analytical system to qualify and quantify the interactions between exogenous proteins and the endogenous proteins of the recipient crop. It could help elucidate both the positive and negative effects of creating transgenic plants and predict their potential risks as well as net crop quality and yield.
Keywords: Cry1Ab/c protein, protein interaction, subcellular co-localization, yeast two-hybrid assay, bimolecular fluorescence complementation, co-immunoprecipitation
Introduction
Transgenic crops, also known as genetically modified crops, refer to crops and their descendants produced by DNA recombination technology to integrate exogenous genes (DNA sequences are introduced into target cells through genetic engineering or viral infection, etc.) into the genomes of recipient crops, in a way that is changing their genetic composition (Herdt, 2005). Transgenic crops have considerable potential economic, environmental, and social benefits. However, uncertainty regarding their unintended effects has been a major constraint in their commercial development and exploitation. Unintended effects in transgenic crops are difficult to evaluate in terms of their possible biosafety risks, and current research has failed to elucidate the molecular mechanisms responsible for these unintended effects.
It has been reported that exogenous genes in transgenic crops might induce unintended effects by disrupting host genes or rearranging DNA sequences at the insertion site (Forsbach et al., 2003), which causes somatic variation during the transgenic crop cultivation process (Labra et al., 2001; Noro et al., 2007), consuming excess host material and energy for their own expression (Zeller et al., 2010) as exogenous proteins interact with endogenous proteins in the recipient crops. However, there is currently insufficient empirical evidence to corroborate these theories. Although the crop cannot express naturally the exogenous proteins, the transgenic crops could express these proteins through integrating exogenous genes into the crop genome in all tissues throughout the growth stages (Fu et al., 2018), so these exogenous proteins might potentially interact with endogenous proteins to produce unintended biological effects in the transgenic crop. To date, however, the aforementioned issue has not raised sufficient concern. Moreover, the safety regulations and criteria for transgenic crops do not include an evaluation of the interactions between exogenous and endogenous proteins. Therefore, an analysis of the association between protein interactions and the unintended effects observed in insect-resistant transgenic rice is warranted.
The Bt gene from Bacillus thuringiensis, which encodes an insecticidal crystalline protein, is the most widely used insect-resistant gene in the world at present. Transgenic crops expressing Bt proteins can effectively control target insects, reducing the amount of required pesticides. On October 22, 2009, the Ministry of Agriculture of China issued biosafety certificates for the cultivation of the insect-resistant transgenic rice lines Huahui-1 and Bt Shanyou-63 in the Hubei Province, which were renewed in December 2014. However, the transgenic rice lines have not been cultivated commercially due to widespread concerns and controversy about its biosafety.
A large number of research results showed that there are many unexpected differences between Bt transgenic rice and its parent rice (Shu et al., 2002; Chen et al., 2006; Kim et al., 2008; Xia et al., 2010; Fu et al., 2018; Fu and Liu, 2020). Our previous study has reported that plant height, biomass, chlorophyll content, and other vegetative growth indicators are considerably different between insect-resistant transgenic rice Huahui-1 and the parent rice Minghui-63 under the farmland and saline-alkaline growth conditions (Fu et al., 2018). However, the molecular mechanisms underlying these phenotypic differences have yet to be determined. We speculate that exogenous Cry1Ab/c expressed in Huahui-1 interacts with endogenous proteins, alters rice metabolism and biological functions, and significantly changes the Huahui-1 phenotype. This will be based on exploring the molecular mechanism underlying the response to stress conditions via protein interactions. When crops are subjected to flooding, high salt concentrations, phosphorus deficiency, abscisic acid and other stressors, protein interaction-mediated oxidative stress responses occur that induce stomatal closure, decreases transpiration rates, and promote reactive oxygen species accumulation, among other processes (Wang et al., 2014; Singh and Sinha, 2016; Tang et al., 2016). Taoka et al. (2011) found that the Hd3a interacts with 14-3-3 proteins in the apical cells of shoots, yielding a complex that enters to the nucleus and binds to the OsFD1 transcription factor, resulting in the flowering by activating the flowering identity genes.
In a long-term study, we found that the differences in photosynthetic efficiency and stress resistance between the insect-resistant transgenic rice Huahui-1 and the parental rice Minghui-63 were most significant at the three-leaf stage (unpublished), but the molecular mechanism underlying these differences is still unknown. Referring to previous research-based speculation that protein interactions lead to rice flowering (Taoka et al., 2011), we hypothesized that Cry1Ab/c protein might interact with endogenous proteins in transgenic rice, which might have unintended effects on some biological characteristics, such as photosynthetic efficiency and stress resistance.
Therefore, the present study investigated exogenous Cry1Ab/c protein expressed by Huahui-1 with the following goals: (1) to screen potential endogenous proteins interacting with it. This would help to study the universality and intensity of the interaction between them; (2) to analyze and validate the interactions among Cry1Ab/c and endogenous proteins that are associated with the observed unintended effects on photosynthesis and stress resistance through subcellular co-localization, BiFC, and co-immunoprecipitation methods. This would help to establish a relationship between protein interactions and known phenotypic differences in photosynthesis and stress resistance; (3) to analyze the possible interaction mechanisms by comparing differences in cell localization and interaction sites between Cry1Ab/c and photosynthesis or stress resistance proteins. This information could contribute to identifying some unintended effects that might be caused by the expression of exogenous proteins, elucidating the possible molecular mechanisms through which protein interactions are associated with observed phenotypic differences between transgenic and parental crops. This would further provide guidance to improve the safety regulations and criteria for assessment of transgenic crops and to ensure the sustainability and healthy development of the transgenic crop industry.
Materials and Methods
Plant Materials and Growth Conditions
The rice varieties used in the present study were insect-resistant transgenic Huahui-1 containing the foreign cry1Ab/c fusion gene and parental Minghui-63, both of which were obtained from the College of Plant Science and Technology of Huazhong Agricultural University, Hubei, Wuhan, China. Large full seeds were surface-sterilized with 75% (v/v) alcohol, rinsed with ultrapure water, and placed in an unlit incubator at 30°C until germination. The seedlings were then transferred to a small pots and maintained in an incubator with a 16 h:8 h light:dark cycle at 28°C and 80% relative humidity (RH). At the three-leaf stage, the Huahui-1 and Minghui-63 rice lines showed substantial differences in plant height (Huahui-1, 16.38 ± 1.26 cm; Minghui-63, 19.82 ± 1.03 cm; p < 0.01) and fresh biomass (Huahui-1, 0.08 ± 0.00 g; Minghui-63, 0.12 ± 0.02 g; p < 0.01). Thirty Huahui-1 rice seedlings with equal growth rate were combined as a single sample and pulverized in liquid nitrogen in preparation for construction of a rice cDNA library.
Nicotiana benthamiana was cultivated in a greenhouse under a 16 h:8 h light-dark cycle, with a daytime temperature of 22–25°C and a nighttime temperature of 18–22°C. After 4–6 weeks (five-leaf stage), the plants were used in an Agrobacterium GV3101-mediated transient transformation experiment.
Gene Cloning and Vector Construction
An RNeasy plant mini kit (Qiagen, Germany) was used to extract and purify RNA from Huahui-1 and Minghui-63 rice leaves, in accordance with the manufacturer’s instructions. One microgram of the extracted RNA was used as a template, and cDNA was synthesized from all samples by reverse transcription using a PrimeScript RT Reagent Kit with gDNA Eraser (RR047A; Takara, Japan). Foreign cry1Ab/c was amplified using Huahui-1 rice cDNA as a template. Endogenous genes were amplified using Minghui-63 rice cDNA as a template. The primers used for amplification are listed in Supplementary Table S1. The gene open reading frame sequences were acquired from the NCBI database (National Center for Biotechnology Information, Bethesda, MD, United States).
The recombinant plasmids used in the present study are listed in Supplementary Table S1. An In-Fusion HD Cloning Kit (TAKARA) reaction system was established to homologously recombine endogenous genes into the subcellular localization vector pBINPLUS-GFP, BiFC vector PCV-cYFP-N (nYFP), and yeast bihybrid vector pGBKT7 (BD) by using a single BamH1 digestion site. The foreign gene cry1Ab/c was cloned into the subcellular location vector pBINPLUS-mCherry, BiFC vector PCV-nYFP-C (cYFP), and yeast two hybrid vector pGADT7 (AD).
Screening for Endogenous Rice Protein Interactions With the Foreign Cry1Ab/c Protein by Yeast Two-Hybrid Assay
The yeast two-hybrid screening assays were conducted according to the instructions for the Matchmaker Gold yeast two-hybrid system (Clontech Laboratories, Mountain View, CA, United States). For large-scale extraction and identification of endogenous rice proteins interacting with the foreign Cry1Ab/c protein, the aforementioned cDNA library was inserted in the prey vector pGADT7 (AD) and the cry1Ab/c gene obtained from Huahui-1 cDNA template amplification was inserted in the bait vector pGBKT7 (BD). As the full-length cry1Ab/c gene self-activates, it was cleaved into three segments according to its three different domains. The three fragments were recombined in the bait vector BD and self-activation and cytotoxicity were verified. The vectors harboring the cDNA library and the aforementioned truncated genes were transformed into the yeast cells Y2H Gold and Y187, respectively, which were examined using double- and quadruple-deficiency screening assays. Positive colonies on the quadruple-deficiency screening plates were selected as templates and PCR amplification and sequencing were conducted to detect the inserted fragments. The NCBI database was consulted for gene identification.
Transient Expression System Developed via Agrobacterium-Mediated Transformation
A transient expression system was developed based on Agrobacterium-mediated transformation as previously described (Zhang et al., 2011). For fluorescence observations, the tobacco leaves were excised and mounted on slides in water. Green fluorescence (GFP: 488 nm), red fluorescence (mCherry: 561 nm), and yellow fluorescence (YFP: 514 nm) were observed under a Zeiss LSM750 confocal laser-scanning microscope (Carl Zeiss AG, Oberkochen, Germany).
Subcellular Co-localization
The target genes inserted into the pBINPLUS-GFP and pBINPLUS-mCherry vectors were transformed into Agrobacterium strain GV3101, which was infiltrated into Nicotiana benthamiana leaves. After 48 h, the GFP and mCherry fluorescence signals were read under a Zeiss LSM750 confocal laser-scanning microscope.
Bimolecular Fluorescence Complementation
The target genes fused to the nYFP and cYFP vectors were transformed into Agrobacterium strain GV3101, which was infiltrated into Nicotiana benthamiana leaves (Li et al., 2019). As a positive control, we used 4A-nYFP (NbeIF4A) plus P2-cYFP (RSV-encoded proteins) (Shi et al., 2016). After 48 h, YFP fluorescence signals (514 nm) were observed using a Zeiss LSM750 confocal laser-scanning microscope.
Co-immunoprecipitation Assays
The target genes inserted into the pBINPLUS-GFP and pBINPLUS-mCherry vectors were transformed into Agrobacterium strain GV3101, which was subsequently infiltrated into Nicotiana benthamiana leaves. After 48 h, 1.0 g tobacco leaves were pulverized with liquid nitrogen in a mortar. To enhance expression, the P19 strain was included in a co-transformation. Thereafter, 2 mL of extraction buffer was added and the homogenate was milled for 2 min. To the resulting homogenate, we added 50 μL of 20% (v/v) Triton [final concentration 0.5% (v/v)] and the macerate was ground for 1 min. The slurry was then transferred to a 2-mL Eppendorf tube. All samples were vortexed, mixed, and placed in a 4°C refrigerator, mixed by repeated inversions in a rolling-over instrument (Qilinbeier, Suzhou, China), and incubated for 20 min.
One milliliter of buffer (20 mM DTT, 1× proteinase, 50 mM Tris, 150 mM NaCl, 0.5 mM EDTA, and 8% glycerin) was added to a 1.5-mL Eppendorf tube and 35 μL of GFP-Trap A beads (ChromoTek GmbH, Planegg-Martinsned, Germany) was aspirated with a pipette cut off 3 mm from the tip and placed in the Eppendorf tube. The tube was centrifuged three times at 2,000 × g and 4°C for 4 min. At least 50 μL of liquid was retained in the tube after each run. The samples were centrifuged at 15,200 × g and 4°C for 5 min, and 1.3–1.4 mL of the resulting supernatant was transferred to a 1.5-mL Eppendorf tube, which was then centrifuged at 15,200 × g for 15 min. One hundred microliters of the resulting supernatant were used as the input, and the remaining 1–1.3 mL supernatant was added to the previously washed beads. The Eppendorf tubes were sealed with parafilm, placed in a 4°C refrigerator, mixed by repeated inversions in a four-dimensional rolling-over instrument (Qilinbeier, Suzhou, China), and incubated for 2 h. After a 3-min centrifugation at 2,000 × g and 4°C, the supernatant was removed and the GFP-trap A beads were washed six times with IP buffer and centrifuged at 2,000 × g for 1 min, so that the supernatant could be aspirated with a pipette. The remaining 80 μL of supernatant was combined with 5× SDS buffer and the mixture was heated to 95°C for 10 min and centrifuged at 2,000 × g and 20–25°C for 2 min.
Proteins were isolated using 4–20% prefabricated SDS-PAGE gels (Bio-Rad Laboratories, Hercules, CA, United States) in 1× running buffer. The proteins were then transferred to a polyvinylidene fluoride (PVDF) membrane using a Tris-Gly membrane transfer system (40°C; 40 V; 14–18 h). The membrane was thereafter blocked by immersion in 3% (v/v) bovine serum albumin and shaking at 80 rpm and 30°C for 2 h. The blocked membrane was washed with Tris-buffered saline (TBS) for 3–5 min, and then subjected to immunoblotting in an iBind western blot device (Invitrogen, Carlsbad, CA, United States) with 1× iBind Flex solution (100× additive, iBind Flex 5× buffer, and pure water). The primary antibodies used were anti-GFP (1:2,000) and anti-mCherry (1:1,500) tag (Abcam, Cambridge, United Kingdom), whereas sheep anti-rat (1:2,000; Abcam, Cambridge, United Kingdom) and Marker HRP (1:1,000; Bio-Rad, CA, United States) were used as secondary antibodies. The membrane was incubated with enhanced chemiluminescence (ECL) color developer and enhancement solution (1:1 mixture) for 5 min, and the colored bands that developed on the membrane were viewed using a VersaDoc imaging system (Bio-Rad Laboratories, Hercules, CA, United States).
Results
Yeast Two-Hybrid Screening for Interactions Between Exogenous Cry1Ab/c and Endogenous Rice Proteins
Detection of Self-Activation and Cytotoxicity in the Bait Yeast Strain
To screen the endogenous proteins interacting with Cry1Ab/c protein, it is necessary to detect self-activation and cytotoxicity of the Cry1Ab/c protein. Our results showed that the Cry1Ab/c-BD yeast strain Y2HGold bearing the full-length Cry1Ab/c protein developed normally on nutrient-deficient SD/-Trp medium, but turned blue on SD/-Trp/X medium supplemented with x-alpha-gal (X). The strain was also able to develop normally on SD/-Trp/X/A medium containing strong aureobasidin A (Figure 1A). Cry1Ab/c-BD presented with self-activation activity, as its double-reporter aur1-C and mel1 could be induced. Therefore, the full-length Cry1Ab/c could not be used as bait for the subsequent prey protein screening.
FIGURE 1.
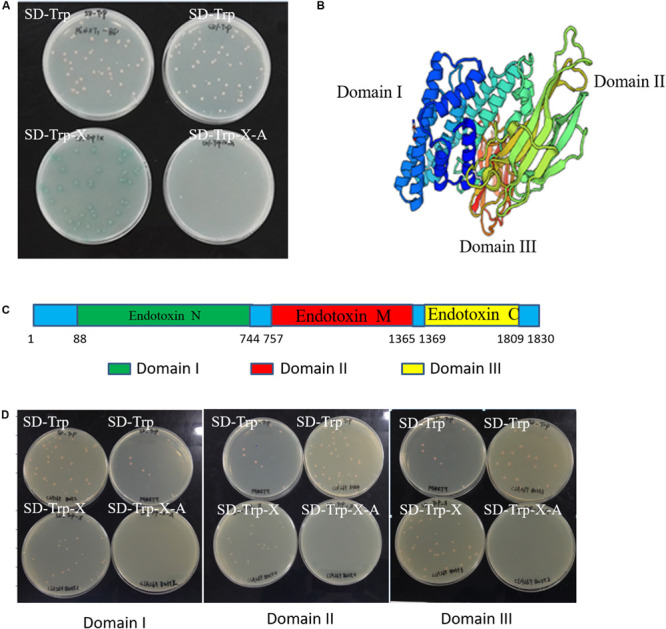
Self-activation and cytotoxicity detection in the Cry1Ab/c bait yeast strain. (A) The growth of full-length Cry1Ab/c-BD yeast colonies on nutrient-deficient SD/-Trp, SD/-Trp/X medium supplemented with x-alpha-gal (X) and SD/-Trp/X/A medium harboring strong abaR inhibitors; (B) A three-dimensional model of the full-length Cry1Ab/c foreign protein was predicted using Swiss-model online software (Adang and Crickmore, 2014); (C) A schematic diagram of the nucleic acid sequence sites for initiation and termination which were cut into three domains; (D) The growth of bait Domain I-BD, Domain II-BD and Domain III-BD yeast colonies on nutrient-deficient SD/-Trp, SD/-Trp/X, and SD/-Trp/X/A, respectively.
A three-dimensional model of the full-length Cry1Ab/c exogenous protein was analyzed through Swiss-model online software. The results showed that it comprises three different domains, each of which might have unique functions (Figure 1B). Thus, referring to the instructions of the “YeastmakerTM Yeast Transformation System 2 User Manual,” the amino acid sequence of the target protein was truncated to prevent self-activation during yeast two-hybrid assay. The full-length Cry1Ab/c protein was accordingly cleaved into three segments based on the aforementioned Domains I–III in the present study (Figure 1C). The gene sequences of the three domains were cloned separately and fused with an expression vector PGBKT7 (BD) to generate three Domain-BD constructs, and the yeast strain Y2HGold was transformed to analyze self-activation. The three Domain-BD strains did not exhibit self-activation as they grew normally on SD/-Trp and SD/-Trp/X without turning blue and they failed to grow on SD/-Trp/X/A. There were no significant differences among the three Domain-BD yeast strains grown on solid or in liquid SD/-Trp in terms of growth status or rate. Thus, the expression of the three bait Domain-BD proteins in yeast was not toxic to host cells (Figure 1D) and these were used to screen Cry1Ab/c-interacting endogenous proteins.
Screening Cry1Ab/c-Interacting Endogenous Proteins
Three truncated domains of Cry1Ab/c mentioned previously did not result in self-activation and cytotoxicity, and thus, they were used as bait proteins to screen a rice cDNA library via a yeast two-hybrid approach. The three co-transformed Domain-BD + cDNA-AD yeast strains at four different dilutions grew normally on SD/-Trp, SD/-Leu, and SD/-Trp/-Leu (DDO). Thus, the Y2HGold yeast harboring Domain-BD and the Y187 yeast strain harboring cDNA-AD were successfully paired. Fifty plates were coated with DDO/X/A for blue-white and resistance screening. Seventy-five Domain I, 41 Domain II, and 24 Domain III blue positive clones were screened. The blue positive clones derived from the double nutrient-deficient DDO/X/A medium were transferred to a quadruple nutrient-deficient SD/-Ade/-His/-Leu/-Trp/X/A(QDO/X/A) medium. Seventy-four Domain I, 19 Domain II, and two Domain III blue positive clones were then screened. The earliest growth and deepest color changes observed indicated strongly interacting proteins, and these colonies were preserved as key material for further research (Figure 2). The blue positive clones were amplified by PCR, and those containing single fragments were isolated from the plasmid library and sequenced at the 5′ end after gel electrophoresis (Supplementary Figure S1). Having removed the repeat sequences, 60 candidate positive clones were designated as Cry1Ab/c-interacting endogenous proteins.
FIGURE 2.
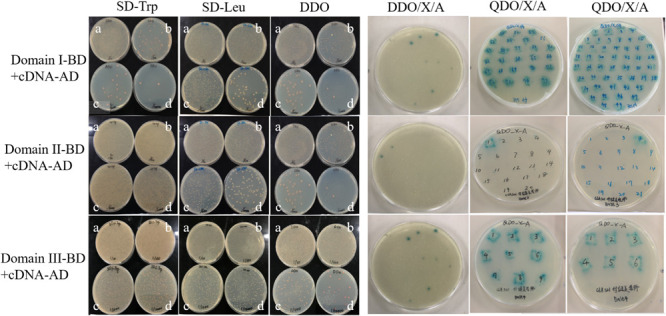
Cry1Ab/c-interacting endogenous proteins were preliminarily screened. The growth of three co-transformed Y2HGold yeast harboring Domain-BD and the Y187 yeast strain harboring cDNA-AD on nutrient-deficient SD/-Trp, SD-Leu, DDO, DDO/X/A, and QDO/X/A, respectively. a-d, the dilution ratios is 1/10, 1/100, 1/1000, 1/10000, respectively.
Based on their physiological and biochemical properties, the clones were classified as stress proteins involved in resistance, functional proteins involved in biomass and energy metabolism, photosynthetic proteins involved in oxidation-reduction reactions, and certain proteins of unknown function (Table 1). Our earlier research results revealed that the observed differences between the insect-resistant transgenic rice Huahui-1 and its parent rice Minghui-63 in terms of photosynthetic efficiency and stress resistance were the most pronounced at the three-leaf stage (unpublished), and thus, the Cry1Ab/c-interacting proteins associated with the aforementioned phenotypic differences were screened using the yeast two-hybrid assay at this stage. Therefore, in the present study, Photosynthesis and stress resistance proteins exhibiting the strongest interactions with the Cry1Ab/c protein were selected for analysis and to authenticate these protein–protein interactions through subcellular co-location, BiFC, and co-IP methods (Table 2).
TABLE 1.
Detailed gene names and reference numbers of positive clones that are interacting with the Cry1Ab/c protein.
| Functional classification | Name of protein interacting with Cry1Ab/c | Accession number |
| Stress response | Chromatin structure-remodeling complex protein SYD isoform X2 | XM015786971.1 |
| Patellin-3 | XM015784751.1 | |
| Proline synthase co-transcribed bacterial homolog protein isoform X2 | XM015777991.1 | |
| Protein SGT1 homolog | XM015766221.1 | |
| Proliferating cell nuclear antigen | XM015771759.1 | |
| HSP70 | L32165.1 | |
| Bifunctional nuclease 1 | XM015765834.1 | |
| AP-1 complex subunit mu-2 | XM015782448.1 | |
| Putative wall-associated receptor kinase-like 16 | XM015778712.1 | |
| Peptidyl-prolyl cis-trans isomerase FKBP12 | XM015769882.1 | |
| ERBB-3 BINDING PROTEIN 1 | XM015785210.1 | |
| Protein TSS | XP 015646988.1 | |
| Protein AE7-like 1 | XM015772391.1 | |
| Putative calcium-transporting atpase 8, plasma membrane-type (Ca2+-ATPase) | XP015649799.1 | |
| V-type proton atpase subunit E-like (V-H+-ATPase) | XP015621799.1 | |
| Calmodulin-binding transcription activator 3 isoform X3 (CAMTAs) | XP015614398.1 | |
| E3 ubiquitin-protein ligase RLIM (E3s) | XP015637952.1 | |
| Kinesin-13A (KIN13A) | XP015639990.1 | |
| Phospho-2-dehydro-3-deoxyheptonate aldolase 2 (DAHP) | XP015646378.1 | |
| Chaperone protein dnaj-like isoform X1 (DnaJ) | XP015639177.1 | |
| Histone-lysine N-methyltransferase family member (HKMTs) | XP015647562.1 | |
| Protein FREE1 isoform X2 (FREE1) | XP015643101.1 | |
| Oxidation reduction (photosynthesis) | Oxygen-evolving enhancer protein 2 (PSBP) | XM015791242.1 |
| Chloroplast 23 KD polypeptide of photosystem II (23KD) | EU325990.1 | |
| PITH domain-containing protein (Trx) | XM015775004.1 | |
| Putative photosystem II component CP43 (CP43) | DQ336882.1 | |
| Protein thylakoid formation1, chloroplastic (THF1) | XM015791425.1 | |
| Ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit, chloroplastic (RuBisCO) | NP039391.1 | |
| Gtpase activating protein 1-like (G) | XM_015772115.1 | |
| Material metabolism | Coatomer subunit delta-2 | XM015784192.1 |
| Mitochondrial outer membrane protein porin 1 | XM015756376.1 | |
| Enolase-like | XM015777125.1 | |
| Nicotiana attenuata eukaryotic initiation factor 4A-6 | XM019379319.1 | |
| Carbon catabolite derepressing protein kinase | EF575881.1 | |
| Synaptotagmin-2 | XM015756696.1 | |
| Peptidyl-prolyl cis-trans isomerase FKBP12 | XM015769882.1 | |
| Seipin-1 | XM015763551.1 | |
| Exocyst complex component SEC5B | XM015780441.1 | |
| Glutamine–tRNA ligase | XM015784471.1 | |
| Suppressor of mec-8 and unc-52 protein homolog | XM015783294.1 | |
| Probable protein S-acyltransferase 7 | CP018170.1 | |
| Knotted-1-like 13 | XM015794843.1 | |
| Others | Predicted | XM015764852.1 |
| Predicted | BAS86484.1 | |
| 40S ribosomal protein S20-2-like | XM014805724.1 | |
| Predicted | EU959177.1 | |
| 40S ribosomal protein S20 | XM015787564.1 | |
| Predicted | CP018163.1 | |
| Predicted | – | |
| Probable DNA primase large subunit | XM006657577.2 | |
| Predicted | XM015789372.1 | |
| Predicted | XP015646911.1 |
The bold black font indicates the candidate endogenous protein screened for further verification of the authenticity of interaction, and “–” indicates that the reference number has not been searched in NCBI.
TABLE 2.
Comparison between the bimolecular fluorescence complementation (BIFC) and co-immunoprecipitation (co-IP) assays in terms of the interactions between Cry1Ab/c and 14 endogenous proteins involved in photosynthesis and stress resistance.
| Differences between | |||
| Protein classification | Protein name | BIFC and co-IP |
|
| BIFC | co-IP | ||
| Photosynthesis | 23KD | Y | Y |
| Trx | Y | Y | |
| THF1 | Y | Y | |
| G | Y | Y | |
| PSBP | Y | Y | |
| RuBisCO | Y | Y | |
| CP43 | Y | N | |
| Stress tolerance | CAMTAs | Y | Y |
| DAHP | Y | Y | |
| HKMTs | Y | Y | |
| KIN13A | Y | Y | |
| FREE1 | Y | Y | |
| E3s | Y | Y | |
| DnaJ | N | N | |
“Y” indicates Yes, “N” indicates No.
We suggested that the results obtained might contribute to identifying the interactions that most likely account for the significant differences between transgenic rice Huahui-1 and parental rice Minghui63 in terms of photosynthetic efficiency, stress resistance, and other traits at the three-leaf stage.
Validation of the Interactions Between the Exogenous Cry1Ab/c Protein and Endogenous Proteins
Subcellular Co-localization
To determine the likelihood that these two different proteins would interact in the plant cell, the mCherry fluorescent protein-linked full-Cry1Ab/c protein and GFP-linked endogenous proteins were co-expressed in tobacco mesophyll cells. As shown in Figure 3, the exogenous full-length Cry1Ab/c-mCherry protein was mainly distributed in the cytoplasm and nucleus and should have therefore been able to interact extensively with the endogenous rice proteins. We found that the full-length Cry1Ab/c-mCherry protein and photosynthetic proteins 23KD-GFP, THF1-GFP, PBSP-GFP, and CP43-GFP co-localized in the chloroplast, with G-GFP mainly co-localized in the cell cytoplasm and Trx-GFP co-localized in the cell cytoplasm and nucleus. Additionally, the full-length Cry1Ab/c–mCherry protein and stress resistance proteins CAMTA-GFP, HKMT-GFP, KIN13A-GFP, FREE1-GFP, E3s-GFP, and Rubisco-GFP co-localized in the cell cytoplasm and nucleus, with DAHP-GFP co-localized in the cell organelles and DnaJ-GFP co-localized in the stomata.
FIGURE 3.
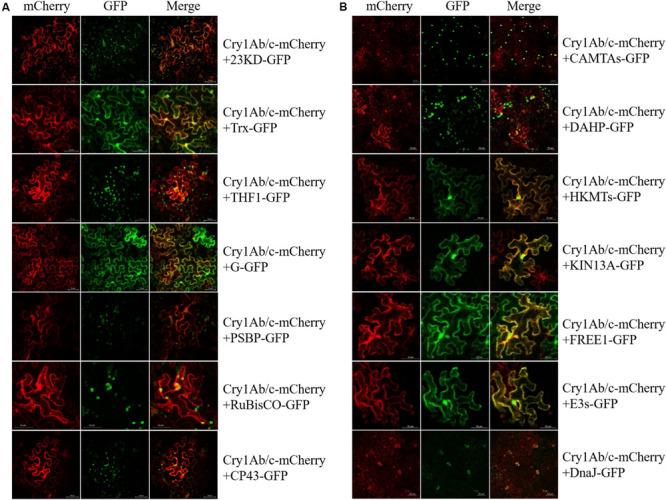
Subcellular co-localization of Cry1Ab/c and 14 endogenous proteins involved in photosynthesis and stress resistance. The co-localization sites and strength of photosynthetic proteins (A) and stress resistance proteins (B) were detected by in situ mCherry and GFP fluorescence, scale bars: 50 μm.
Bimolecular Fluorescence Complementation
To determine whether the exogenous full-length Cry1Ab/c protein and endogenous rice proteins do indeed interact in plant cells, the BiFC technique was performed. The expression vectors of full-length Cry1Ab/c protein and endogenous rice proteins fused to the N-terminal half of YFP and the C-terminal half of YFP, respectively, were then transiently expressed in tobacco mesophyll cells, and protein interaction sites and intensities were determined based on strong YFP yellow fluorescence. As shown in Figure 4, the protein interaction intensities differed between the full-length Cry1Ab/c protein and endogenous rice proteins. The full-length Cry1Ab/c-cYFP protein interacted strongly with 23KD-nYFP, PSBP-nYFP, G-nYFP, THF1-nYFP, and Trx- nYFP, but weakly with Rubisco-nYFP and CP43-nYFP. Similarly, the full-length Cry1Ab/c-cYFP protein interacted strongly with CAMTA-nYFP, DAHP-nYFP, HKMT-nYFP, KIN13A-nYFP and FREE1-nYFP, but weakly with E3s-nYFP. However, we detected no interaction between Cry1Ab/c and DnaJ-nYFP.
FIGURE 4.
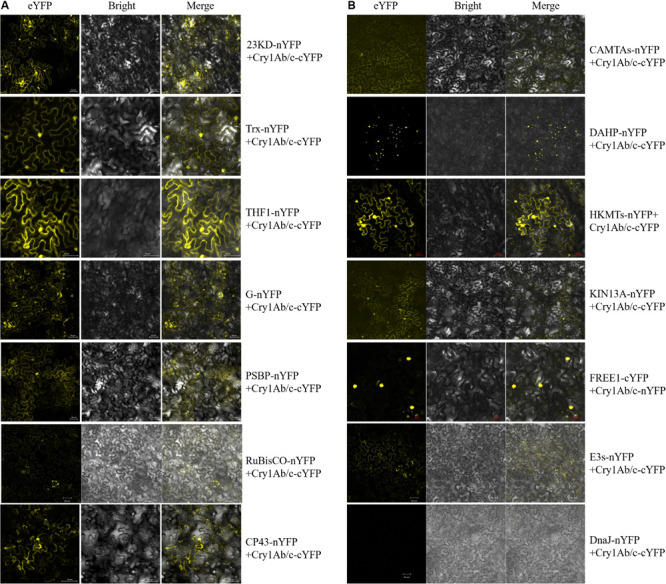
The bimolecular fluorescence complementation was used to verify interactions between the full-length Cry1Ab/c protein and 14 photosynthesis and stress resistance proteins. The sites and strength of interactions between the Cry1Ab/c protein and photosynthetic (A), stress resistance proteins (B) were determined by in situ YFP fluorescence, scale bars: 50 μm.
In addition, the protein interaction sites differed between the full-length Cry1Ab/c protein and endogenous rice proteins. An interaction between the full-length Cry1Ab/c-cYFP protein and photosynthetic proteins 23KD-nYFP, PSBP-nYFP, G-nYFP, THF1-nYFP, Trx-nYFP, Rubisco-nYFP and CP43-nYFP were observed in the cytoplasm and nucleus. An interaction between the full-length Cry1Ab/c-cYFP protein and stress resistance proteins CAMTAs-nYFP was observed in the cytoplasm, whereas DAHP-nYFP was observed in the organelle and HKMTs-nYFP, KIN13A-nYFP, and E3s-nYFP were observed in the cytoplasm and nucleus. FREE1-nYFP was observed in the nucleus.
Co-immunoprecipitation
The interaction between proteins then needed to be confirmed using several methods, including co-IP. co-IP reflects protein interactions in the natural state of living plant cells, which can overcome the problem of false positives observed in BiFC that are attributable to spontaneous fluorescence (Speth et al., 2014). We found that the exogenous full-length Cry1Ab/c-mCherry protein interacted with the endogenous photosynthetic proteins 23KD-GFP, Trx-GFP, THF1-GFP, G-GFP, PSBP-GFP, and Rubisco-GFP (Figures 5A–C) but not with CP43-GFP (Supplementary Figure S2A); it can also interact with the endogenous stress-resistance proteins CAMTA-GFP, DAHP-GFP, HKMT-GFP, KIN13A-GFP, FREE1-GFP, and E3s-GFP (Figures 5D–F) but not with DnaJ-GFP (Supplementary Figure S2B). Differences between the BiFC and co-IP assays in terms of the interactions between Cry1Ab/c and 14 endogenous proteins involved in photosynthesis and stress resistance are listed in Table 2.
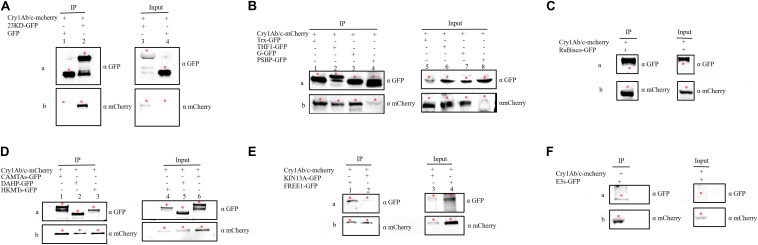
FIGURE 5.
The co-immunoprecipitation was used to verify interactions between the full-length Cry1Ab/c protein and 12 endogenous proteins. Protein extracts (Input) were immunoprecipitated with GFP-trap beads (IP) and resolved by SDS-PAGE. The immunoblots shown were developed with anti-GFP antibody to detect the target endogenous protein involved in photosynthesis and stress resistance (a) and with anti-mCherry antibody to detect Cry1Ab/c (b, 94 kDa). (A) GFP + Cry1Ab/c-cherry (27 kDa, lanes 1 and 4), 23KD-GFP + Cry1Ab/c-cherry (47 kDa, lanes 2 and 3); (B) Trx-GFP + Cry1Ab/c-cherry (48 kDa, lanes 1 and 5), THF1-GFP + Cry1Ab/c-cherry(59 kDa, lanes 2 and 6), G-GFP + Cry1Ab/c-cherry (45 kDa, lane 3 and 7), PSBP-GFP + Cry1Ab/c-cherry (50 kDa, lanes 4 and 8); (C) Rubisco-GFP + Cry1Ab/c-cherry (80 kDa); (D) CAMTAs-GFP + Cry1Ab/c-mCherry (117 kDa, lanes 1 and 6), DAHP-GFP + Cry1Ab/c-mCherry (86 kDa, lanes 2 and 5), HKMTs-GFP + Cry1Ab/c-cherry (102 kDa, lanes 3 and 4); (E) FREE1-GFP + Cry1Ab/c-cherry (116 kDa, lanes 1 and 4), KIN13A-GFP + Cry1Ab/c-cherry (117 kDa, lanes 10 and 11); and (F) E3s-GFP + Cry1Ab/c-cherry (83 kDa, lanes 14 and 17). The red asterisk indicates the target band.
Analysis of Possible Biological Functions Mediated by the Interactions Between Exogenous Cry1Ab/c Protein and Endogenous Proteins
To further analyze how the strong interactions between Cry1Ab/c protein and photosynthetic and stress resistant proteins affect biological phenotypes such as photosynthetic efficiency and stress resistance in insect resistant transgenic rice Huahui-1, we next compared the differences between subcellular localization or protein interaction sites of Cry1Ab/c and photosynthetic and stress resistant proteins and predicted their possible interaction mechanisms according to the results we identified. As shown in Table 3, the Cry1Ab/c protein was found to be mainly distributed in the cytoplasm and nucleus, with 23KD, THF1, and PSBP located in the chloroplast, whereas their interactions with the Cry1Ab/c protein were also mainly distributed in cytoplasm and nucleus. Thus, it was speculated that the expression of Cry1Ab/c protein might affect the function and photosynthetic efficiency of 23KD, THF1, and PSBP in the chloroplast by changing their cell location. Other photosynthetic proteins G and Trx are mainly distributed in the cytoplasm and nucleus, and their interactions with the Cry1Ab/c protein were also mainly distributed in the cytoplasm and nucleus. Therefore, it was considered that the interaction between large molecular Cry1Ab/c protein (67 Kda) and small molecular photosynthetic proteins G (18 Kda) and Trx (21 Kda) might inhibit the free movement of the latter in cells, thereby affecting photosynthetic functions of the latter.
TABLE 3.
Differences between the location and interaction sites of Cry1Ab/c and 12 endogenous proteins involved in photosynthesis and stress resistance.
| Protein | Protein | Differences between location | |
| classification | name | and interaction sites |
|
| Subcellular co-localization | BIFC | ||
| Photosynthesis | 23KD | Chloroplast | Cytoplasm, nucleus |
| Trx | Cytoplasm, nucleus | Cytoplasm, nucleus | |
| THF1 | Chloroplast | Cytoplasm, nucleus | |
| G | Cytoplasm, nucleus | Cytoplasm, nucleus | |
| PSBP | Chloroplast | Cytoplasm, nucleus | |
| RuBisCO | Chloroplast | Cytoplasm, nucleus | |
| Stress-tolerance | CAMTAs | Nucleus | Cytoplasm |
| DAHP | Organelles | Organelles | |
| HKMTs | Cytoplasm, nucleus | Cytoplasm, nucleus | |
| KIN13A | Cytoplasm, nucleus | Cytoplasm, nucleus | |
| FREE1 | Cytoplasm, nucleus | Nucleus | |
| E3s | Cytoplasm, nucleus | Cytoplasm, nucleus | |
“BIFC” indicates “bimolecular fluorescence complementation.”
For stress-resistance proteins, interactions with Cry1Ab/c were mainly located in the cytoplasm and nucleus, whereas CAMTAs are located in the nucleus and their interactions are mainly distributed in the cytoplasm. Thus, it was speculated that the latter might be transported out of the nucleus and detained in the cytoplasm for a long time, thus affecting the stress resistance and other biological functions of CAMTAs. Similarly, Cry1Ab/c and FREE1 proteins are mainly distributed in the cytoplasm and nucleus, whereas their interaction was distributed in nucleus, and thus, it was speculated that the interaction between Cry1Ab/c protein and FREE1 might inhibit shuttling of the latter from the cytoplasm to the nucleus, retaining it in the nucleus for a long time and thus affecting the stress resistance and other biological functions of FREE1. In contrast to the above the location and interaction sites of HKMTS and KIN13A, which were found to be distributed in the cytoplasm and nucleus, are consistent with the Cry1Ab/c protein. Thus, the underlying mechanism of how these interactions regulate plant stress resistance will need to be further studied through other molecular biological and physiological biochemical means in the future.
Discussion
Protein Interactions Might Comprise One of the Molecular Mechanisms That Cause Unintended Effects in Insect-Resistant Transgenic Rice
Previous studies have shown that comparisons of differences in agronomic traits (Xia et al., 2011; Wang et al., 2012a; Li et al., 2013; Jiang et al., 2016), nutritional composition (Wang et al., 2012b; Jian et al., 2014; Jiang et al., 2018; Takeda et al., 2019), environmental adaptability (Jiang et al., 2018; Wang et al., 2019; Park et al., 2019), gene transcription, protein expression, and metabolite levels (Liu et al., 2012; Liu Y. et al., 2015; Fu et al., 2019; Peng et al., 2019) among transgenic crops and their parents are important means and strategies for confirming and excluding the unintended effects of transgenic crops, in addition to facilitating any safety evaluations. However, the molecular mechanisms of underlying the unintended effects observed in transgenic crops are unclear.
Protein interaction networks play central roles in various life processes, such as signal transmission, gene expression regulation, energy and substance metabolism, and cell cycle regulation, and accordingly, it is assumed that the study of protein interactions will contribute to elucidating different biological functions in crops. Although the exogenous Cry1Ab/c protein is not produced naturally by the parent rice, it may be continuously expressed in all tissues during the entire crop growth period in insect-resistant transgenic rice Huahui-1, and thus, it might potentially interact with various endogenous rice proteins synthesized at different growth stages and have varying unintended biological effects. Taoka et al. (2011) found that Hd3a, 14-3-3, and transcription factor OsFD1 strongly interaction through yeast two-hybrid, GST pull-down and immunoprecipitation methods, and this interaction was found to result in rice flowering by inducing the expression of the downstream flowering gene. Similarly, Tang et al. (2016) found that OsbZIP46, a transcription factor that plays an important role in promoting ABA signal transduction and drought resistance in plants, and MODD exhibit strong interactions based on yeast two-hybrid, GST pull-down and co-IP methods; and this interaction inhibits the OsbZIP46 transcriptional activity and protein stability through recruiting other protein that regulate chromatin modifications and protein ubiquitin, respectively, thus suggesting an efficient and reasonable new mechanism of drought response. Therefore, an appropriate technology will enable the detection of these interactions between endogenous rice proteins and Cry1Ab/c protein, which could contribute to establishing the correlations between these protein interactions and the unintended biological effects in insect resistant transgenic rice.
The yeast two-hybrid screening approach used in the present study revealed 60 proteins that interact with Cry1Ab/c, including stress-resistance, photosynthetic, metabolic, and unknown proteins. Therefore, Cry1Ab/c and its interacting partners might comprise a platform for crosstalk among photosynthetic, stress resistance, oxidation-reduction, and other signal transduction pathways. Our findings that Cry1Ab/c might interact with various endogenous rice proteins are indicative of the explicit unintended effects at the protein levels after Cry1Ab/c proteins are expressed in transgenic rice.
Methodological Discussion on Screening and Validating the Protein Interactions
In the yeast two-hybrid assay, certain bait proteins may induce the expression of reporter genes and self-activate, which limits their application to yeast two-hybrid technology (Tsuyoshi et al., 2002). An earlier study reported that the problem of bait protein self-activation can be resolved by introducing a deletion in the gene that removes the activation domain; hence, in this study, self-activation is re-assessed with respect to the remaining components, whereas the procedure of introducing a deletion in the gene could also inadvertently disrupt or remove a functional domain (Liu Y. L. et al., 2015). Wang et al. (2014) found the PHR2 N terminus has a transactivation domain that is active in yeast; therefore, in the Y2H assay, they used the PHR2 C terminus containing the MYB-CC domains (PHR2-C196aa) responsible for specific binding to the P1BS sequence.
In the present study, the initial yeast two-hybrid screening revealed that the full-length bait protein Cry1Ab/c could self-activate. Referring to the suggestion of “Matchmaker Gold Y2H system User Manual,” to retain the complete functional domains, Cry1Ab/c was cleaved into three segments based on its three functional domains, and we confirmed that none of them could self-activate. Therefore, the aforementioned system was able to be used to screen endogenous proteins interacting with Cry1Ab/c and could serve as a novel research tool for examining protein interactions in general. However, the original functionality would be lost or drastically impacted when the full-length Cry1Ab/c protein was divided into three segments to prevent self-activation during the yeast two-hybrid assay, thus screening the interacting proteins could also differ; therefore, the full-length Cry1Ab/c protein was used to verify the authenticity of the interactions with the endogenous proteins of interest through BiFC and co-IP methods.
Given that yeast and plant cells differ substantially in terms of protein modification, protein interactions within yeast cells might not accurately reflect those in plant cells. Therefore, two other technologies were applied to validate the protein interactions. Previous studies have authenticated protein interactions via comprehensive analyses based on yeast two-hybrid, BiFC, and co-IP assays (Taoka et al., 2011; Tang et al., 2016; Purwestri et al., 2017). These are also used to elucidate the underlying molecular mechanisms of the related physiological phenotypes. Ni et al. (2019) found that DMI3, as a positive regulator of abscisic acid signal transduction, it can interact with the rice protein phosphatase PP45 through yeast two-hybrid preliminary screening, and the authenticity of the interaction between the two proteins was verified in tobacco cells using co-IP and pull-down methods, which provided a research basis for elucidating the molecular mechanism of abscisic acid mediated stress resistance regulation in rice. Similarly, Purwestri et al. (2017) found that the flowering protein Hd3a can interact with certain rice proteins, including GF14c, OsKANADI and BIP116b through yeast two-hybrid preliminary screening, and the authenticity of the protein interactions was verified in tobacco cells by BiFC, furnishing clues regarding the molecular mechanisms underlying the regulation of rice flower signal transduction by Hd3a. Other similar studies have also preliminarily screened the plant endogenous proteins that can interact with a target protein through yeast two-hybrid approach, and the authenticity of protein interactions was verified in tobacco cells by BiFC and co-IP, which laid a research foundation that will contribute to clarifying the biological functions of target proteins in crops (Li et al., 2017; Shen et al., 2017; Ying et al., 2017; Wang et al., 2018).
Similarly, the Cry1Ab/c-interacting endogenous proteins were screened using a yeast two-hybrid, and the authenticity of the interactions between Cry1Ab/c and the screened endogenous proteins was verified in tobacco cells using BiFC and co-IP in the present study. These results provide a theoretical basis for clarifying the molecular mechanisms underlying the unintended effects that occur via protein interactions in transgenic rice.
Validation of the Interactions Between Cry1Ab/c and Candidate Endogenous Proteins, and Analysis of Their Biological Functions
Our results showed that the differences between the insect-resistant transgenic rice Huahui-1 and the parental rice Minghui-63, especially photosynthetic efficiency and stress resistance, were most significant at the three-leaf stage in our long-term study (unpublished). Therefore, we want to further study the correlation between these phenotypic differences and protein interaction at the same three-leaf stage. All the methods used (yeast two-hybrid, BiFC and co-IP) clearly showed that the Cry1Ab/c protein and photosynthetic proteins 23KD, Trx, THF1, G and PSBP exhibit strong interactions. Since the Cry1Ab/c protein is mainly distributed in the cytoplasm and nucleus, whereas 23KD, THF1, and PSBP are localized in the chloroplasts, the mechanism of interaction of which the expression of exogenous Cry1Ab/c protein might inhibit the shuttling of 23KD, THF1 and PSBP from cytoplasm and nucleus into chloroplasts, which was confirmed by a BIFC experiment. This indicated that the interactions between Cry1Ab/c protein and 23KD, THF1, and PSBP might be explained by a model in which the expression of Cry1Ab/c protein could significantly affect the accumulation of the latter in the chloroplasts by changing their localization, thereby arresting photosynthesis. Kong et al. (2014) found that the interactions between the rice stripe virus SP protein and rice PsbP protein reduce the accumulation of the latter in the chloroplasts by changing its localization, thereby arresting photosynthesis and promoting foliar chlorosis, and these effects have been verified by PsbP mutation and overexpression assays. Other photosynthetic proteins G and Trx are mainly distributed in the cytoplasm and nucleus, and their interactions with the Cry1Ab/c protein are also mainly distributed in the cytoplasm and nucleus, which were confirmed by BiFC experiments. Thus, another possible mechanism that could explain the differences in photosynthetic efficiency caused by the expression of exogenous Cry1Ab/c protein is that the interaction between large molecular Cry1Ab/c protein (67 Kda) and small molecular photosynthetic proteins G (18 Kda) and Trx (21 Kda) might inhibit the free movement of the latter in cells, thereby affecting photosynthetic functions of the latter. Purwestri et al. (2017) found that GF14c protein, which causes delayed flowering, can interact with the flowering protein Hd3a, which inhibits the movement of the small protein Hd3a (about 20 Kda) from the leaf to the apical meristem (SAM), thus affecting the flowering of plants. Similarly, He et al. (2018) found that interactions between the rice fructokinase proteins OsFLN1 and OsFLN12 and thioredoxin OsTRXz (Trx) affected chloroplast development, which was verified by an OsTRXz mutation and overexpression assay. The above results indicated that the interaction between Cry1Ab/c proteins and photosynthetic proteins, such as 23KD and Trx, might be the main reason for the differences in photosynthetic efficiency between insect resistant transgenic rice Huahui-1 and its parent rice Minghui-63.
Among plant stress-related proteins, CAMTAs, HKMT, KIN13A, FREE1, and DAHP have been extensively investigated. These proteins regulate plant responses to high and low temperatures, drought, salinity, and other stressors, and in the present study, all the methods used (yeast two-hybrid, BiFC and co-IP) clearly showed that the Cry1Ab/c protein interacts strongly with these proteins in yeast and plant cells. Since the Cry1Ab/c protein is mainly distributed in the cytoplasm and nucleus, whereas CAMTAs reside in the nucleus, the mechanism underlying this interaction, wherein the expression of exogenous Cry1Ab/c protein might transport the latter out of the nucleus and retain it in the cytoplasm for a long time were confirmed by BIFC experiments. This would affect the stress resistance and other biological functions of CAMTAs. Ishida et al. (2004) found that 14-3-3 protein interacts with the leucine zipper structure transcription factor RSG in tobacco, which causes the latter to be retained in the cytoplasm to negatively regulate the GA (gibberellin) signaling pathway, and thus affecting the adaptability of plants to adversity. Similarly, Cry1Ab/c and FREE1 proteins are mainly distributed in the cytoplasm and nucleus, whereas their interaction is distributed in the nucleus. Therefore, another possible mechanism that could explain the differences in stress resistance caused by the expression of exogenous Cry1Ab/c protein is that the interaction between Cry1Ab/c protein and FREE1 might inhibit the shuttling of the latter from the cytoplasm into the nucleus, and retaining it in the nucleus for a long time, thus affecting the stress resistance and other biological functions of FREE1. A similar study found that plasma membrane-localized cold-response protein kinase 1 (CRPK 1) mediates 14-3-3 protein phosphorylation in the nucleus to regulate transcription factor CBF signaling, and thus negatively regulating plant responses to low temperature in Arabidopsis thaliana (Liu et al., 2017). Additionally, Cry1Ab/c and HKMTs and KIN13A proteins are mainly distributed in the cytoplasm and nucleus, and their interactions are distributed in the nucleus. Therefore, whether the interaction between Cry1Ab/c and HKMTs and KIN13A proteins might prevent the degradation of the latter by activating or inhibiting related enzyme activity or might promote or prevent modification of the latter. The mechanisms regulating plant resistance will need to be further studied through other molecular biological, physiological, and biochemical means in the future. The aforementioned results indicated that the interaction between Cry1Ab/c proteins and stress resistance proteins, such as CAMTAs and FREE1, might be the main reason for the differences in stress resistance between insect resistant transgenic rice Huahui-1 and its parent rice Minghui-63.
If the aforementioned photosynthetic and stress resistance proteins are affected, the photosynthesis and stress resistance responses themselves could also be modulated. Herein, expression of an extra exogenous Cry1Ab/c protein in transgenic rice Huahui-1 considerably affected the photosynthetic efficiency and stress resistance of this strain relative to those of parent rice MH63 (Fu et al., 2018), and accordingly, we propose that the observed unintended effects might be a result of the interactions between the Cry1Ab/c protein and endogenous photosynthesis and stress resistance proteins. However, not all interactions between Cry1Ab/c and interacting endogenous proteins induced phenotypic effects, and thus, future research should use other molecular biological, physiological, and biochemical means, such as transcriptome and proteome methods, to further explore the transcription and protein expression of aforementioned photosynthetic and stress resistance proteins that interact authentically and strongly with Cry1Ab/c. This might contribute to finding the real interacting proteins that affect the photosynthetic efficiency and stress resistance of transgenic riceHuahui-1, and thus, we posit that interactions between Cry1Ab/c and endogenous proteins comprise a possible cause of unintended effects in Huahui-1. This information will help to provide guidance to improve the safety regulations and criteria for transgenic crops and ensure the sustainability and healthy development of the transgenic industry.
Conclusion
In this study, we focused on the exogenous Cry1Ab/c protein expressed in transgenic rice Huahui-1. An initial yeast two-hybrid screen revealed that the full-length bait protein Cry1Ab/c could self-activate. To retain the complete functional domain for an examination of its biological function, full-length Cry1Ab/c was cleaved into three segments based on three functional domains, to remove its activation domain. Thereby, the aforementioned system could be used to screen endogenous proteins interacting with Cry1Ab/c and might thereby serve as a novel research tool to investigate protein interactions. The yeast two-hybrid assay accordingly revealed 60 endogenous rice proteins that can interact with the Cry1Ab/c protein; BiFC and co-IP verified the interactions between the full-length Cry1Ab/c protein and 12 of the identified endogenous proteins involved in photosynthesis and stress resistance. Finally, the possible interaction mechanisms were analyzed by comparing the differences in cell localization and interaction sites between Cry1Ab/c and photosynthesis and stress resistance proteins. Collectively, our results highlight the importance of accurately interpreting the interactions between the exogenous Cry1Ab/c protein and endogenous rice proteins and identifying those interactions responsible for the observed unintended effects in insect-resistant transgenic rice Huahui-1. In the future, this information could be used to assess the effects of these protein–protein interactions on the crop yield and quality of insect-resistant transgenic rice, as well as those of other transgenic crops.
Data Availability Statement
All datasets generated for this study are included in the article/Supplementary Material.
Author Contributions
JF designed and carried out the experiments and wrote the manuscript. BL designed and helped to revise the manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors thank Editage for revising the language and style.
Abbreviations
- 23KD
chloroplast 23 kDa polypeptide of photosystem II
- BiFC
bimolecular fluorescence complementation
- CAMTAs
calmodulin-binding transcription activator
- co-IP
co-immunoprecipitation
- CP43
putative photosystem II component CP43
- DAHP
3-deoxy-D-arabinoheptulosonate-7-phosphate
- DDO
double dropout medium SD/-Leu/-Trp
- DDO/X/A
double dropout medium SD/–Leu/–Trp supplemented with X-a -Gal and aureobasidin A
- DnaJ
chaperone protein dnaj-like isoform
- E3s
E3 ubiquitin-protein ligase
- ECL
enhanced chemiluminescence
- G
Gtpase activating protein
- HKMTS
histone-lysine N-methyltransferase family member
- KIN13A
kinesin-like protein 13A
- PSBP
oxygen-evolving enhancer protein
- QDO/X/A
quadruple dropout medium SD/–Ade/–His–Leu/–Trp supplemented with X-a -Gal and aureobasidin A
- Rubisco
ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit
- SD medium
synthetic dropout medium
- THF1
thylakoid formation 1 protein
- Trx
PITH domain-containing protein
- X- α-gal
5-bromo-4-chloro-3-indolyl- α-d -galactoside.
Footnotes
Funding. This work was funded by grants from the National Major Special Projects for Genetically Modified Organisms (2016ZX08012-005) and the Basic Scientific Research Program in National Nonprofit Scientific Research Institutes (GYZX190103).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fbioe.2020.00685/full#supplementary-material
The selected positive insertions from the DH5α strains were detected by PCR.
Co-ip was used to verify interaction between Cry1Ab/c and photosynthetic protein CP43 and stress resistance protein DnaJ, respectively. Protein extracts (Input) were immunoprecipitated with GFP-trap beads (IP) and resolved by SDS-PAGE. The immunoblots shown were developed with anti-GFP antibody to detect target endogenous protein (a) and with anti-mCherry antibody to detect Cry1Ab/c (b, 94KD). (A) CP43-GFP+Cry1Ab/c-cherry (36KD); (B) DnaJ-GFP+Cry1Ab/c-cherry (76KD).
Primers used for cloning the full-length gene based on subcellular co-localization and co-immunoprecipitation vectors.
References
- Adang M. J., Crickmore N. (2014). Diversity of Bacillus thuringiensis crystal toxins and mechanism of action. Insect Midgut Insect. Proteins. 47:39 10.1016/B978-0-12-800197-4.00002-6 [DOI] [Google Scholar]
- Chen L. Y., Snow A. A., Wang F., Lu B. R. (2006). Effects of insect-resistance transgenes on fecundity in rice (Oryza sativa, Poaceae): a test for underlying costs. Am. J. Bot. 93 94–101. 10.3732/ajb.93.1.94 28924508 [DOI] [Google Scholar]
- Forsbach A., Schubert D., Lechtenberg B., Gils M., Schmidt R. (2003). A comprehensive characterization of single-copy T-DNA insertions in the Arabidopsis thaliana genome. Plant Mol. Biol. 52:161 10.1023/a:1023929630687 [DOI] [PubMed] [Google Scholar]
- Fu J. M., Liu B. (2020). Enhanced yield performance of transgenic cry1c∗ rice in saline-alkaline soil. GM Crops Food 11 97–112. 10.1080/21645698.2019.1709383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu J. M., Song X. L., Liu B., Shi Y., Shen W. J., Fang Z. X., et al. (2018). Fitness cost of transgenic cry1Ab/c rice under saline-alkaline soil condition. Front. Plant Sci. 9:1552. 10.3389/fpls.2018.01552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu W., Wang C., Xu W., Zhu P., Lu Y., Wei S., et al. (2019). Unintended effects of transgenic rice revealed by transcriptome and metabolism. GM Crops Food. 10 20–34. 10.1080/21645698.2019.1598215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He L., Zhang S., Qiu Z., Zhao J., Nie W., Lin H., et al. (2018). Fructokinase-like protein 1 interacts with TRXz to regulate chloroplast development in rice. J. Integr. Plant Biol. 60 94–111. 10.1111/jipb.12631 [DOI] [PubMed] [Google Scholar]
- Herdt R. W. (2005). The state of food and agriculture, 2003-2004: agricultural biotechnology: meeting the needs of the poor? Agric. Econ. 32 109–110. 10.1111/j.0169-5150.2005.t01-7-00008.x [DOI] [Google Scholar]
- Ishida S., Fukazawa J., Takahashi Y. Y. (2004). Involvement of 14-3-3 signaling protein binding in the functional regulation of the transcriptional activator repression of shoot growth by gibberellins. Plant Cell 16 2641–2651. 10.2307/3872254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jian Z., Wang F., Li Z., Chen Y., Ma X., Nie L., et al. (2014). Grain yield and nitrogen use efficiency responses to N application in Bt (Cry1Ab/Ac) transgenic two-line hybrid rice. Field Crop Res. 155 184–191. 10.1016/j.fcr.2013.09.007 [DOI] [Google Scholar]
- Jiang Y., Ling L., Zhang L., Wang K., Cai M., Zhan M., et al. (2016). Transgenic Bt (Cry1Ab/Ac) rice lines with different genetic backgrounds exhibit superior field performance under pesticide-free environment. Field Crop Res. 193 117–122. 10.1016/j.fcr.2016.03.014 [DOI] [Google Scholar]
- Jiang Y., Ling L., Zhang L., Wang K., Li X., Cai M., et al. (2018). Comparison of transgenic Bt rice and their non- Bt counterpart in yield and physiological response to drought stress. Field Crop Res. 217 45–52. 10.1016/j.fcr.2017.12.007 [DOI] [Google Scholar]
- Kim S., Kim C., Li W., Kim T., Li Y., Zaidi M. A., et al. (2008). Inheritance and field performance of transgenic Korean Bt rice lines resistant to rice yellow stem borer. Euphytica 164 829–839. 10.1007/s10681-008-9739-9 [DOI] [Google Scholar]
- Kong L., Wu J., Lu L., Xu Y., Zhou X. (2014). Interaction between rice stripe virus disease-specific protein and host PsbP enhances virus symptoms. Mol Plant 7 691–708. 10.1093/mp/sst158 [DOI] [PubMed] [Google Scholar]
- Labra M., Savini C., Bracale M., Pelucchi N., Colombo L., Bardini M., et al. (2001). Genomic changes in transgenic rice (Oryza sativa L.) plants produced by infecting calli with Agrobacterium tumefaciens. Plant Cell Rep. 20 325–330. 10.1007/s002990100329 [DOI] [Google Scholar]
- Li C., Liu B., Li C., Zeng Q., Hao M., Han Z., et al. (2013). Leaf morphology and ultrastructure responses to elevated O3 in transgenic Bt (cry1Ab/cry1Ac) rice and conventional rice under fully open-air field conditions. PLoS One 8:e82199. 10.1371/journal.pone.0082199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q., Chen Y., Wang J., Zou F., Jia Y., Shen D., et al. (2019). A Phytophthora capsici virulence effector associates with NPR1 and suppresses plant immune responses. Phytopathol. Res. 1:6 10.1186/s42483-019-0013-y [DOI] [Google Scholar]
- Li S., Ying Y., Secco D., Wang C., Narsai R., Whelan J., et al. (2017). Molecular interaction between PHO2 and GI in rice. Plant Cell Environ. 40 1487–1499. 10.1111/pce.12945 [DOI] [PubMed] [Google Scholar]
- Liu Y. B., Zhang Y. X., Song S. Q., Li J. S., Stewart C. N., Jr., Wei W., et al. (2015). A proteomic analysis of seeds from Bt-transgenic Brassica napus and hybrids with wild B. juncea. Sci. Rep. 5:15480. 10.1038/srep15480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y. L., Jiang Y. H., Wang H. J., Qi Y. W., Liu C. H., Ren X. L., et al. (2015). Self activation and interaction protein analysis of MdHB1 transcription factor in apple. Plant Physiol. J. 11 1895–1900. 10.13592/j.cnki.ppj.2015.0347 [DOI] [Google Scholar]
- Liu Z., Zhao J., Li Y., Zhang W., Jian G., Peng Y., et al. (2012). Non-uniform distribution pattern for differentially expressed genes of transgenic rice Huahui 1 at different developmental stages and environments. PLoS One 7:e37078. 10.1371/journal.pone.0037078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z. Y., Jia Y. X., Ding Y. L., Guo Y., Gong Z. Z., Yang S. H. (2017). Plasma membrane crpk1-mediated phosphorylation of 14-3-3 proteins induces their nuclear import to fine-tune CBF signaling during cold response. Mol. Cell 66 117–128. 10.1016/j.molcel.2017.02.016 [DOI] [PubMed] [Google Scholar]
- Ni L., Fu X., Zhang H., Li X., Cai X., Zhang P., et al. (2019). Abscisic acid inhibits rice protein phosphatase PP45 via H2O2 and relieves repression of the Ca2+ /CAM-dependent protein kinase DMI3. Plant Cell 31 128–152. 10.1105/tpc.18.00506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noro Y., Takano-Shimizu T., Syono K., Kishima Y., Sano Y. (2007). Genetic variations in rice in vitro cultures at the EPSPs-RPS20 region. Theor. Appl. Genet. 114 705–711. 10.1007/s00122-006-0470-4 [DOI] [PubMed] [Google Scholar]
- Park D., Park S. H., Kim Y. S., Choi B. S., Kim J. K., Kim N. S., et al. (2019). NGS sequencing reveals that many of the genetic variations in transgenic rice plants match the variations found in natural rice population. Genes Genom. 41 213–222. 10.1007/s13258-018-0754-5 [DOI] [PubMed] [Google Scholar]
- Peng C., Ding L., Hu C., Chen X., Wang X., Xu X., et al. (2019). Effect on metabolome of the grains of transgenic rice containing insecticidal cry and glyphosate tolerance epsps genes. Plant Growth Regul. 88 1–7. 10.1007/s10725-019-00482-6 [DOI] [Google Scholar]
- Purwestri Y. A., Susanto F. A., Tsuji H. (2017). Hd3a florigen recruits different proteins to reveal its function in plant growth and development. Intech 4 49–67. 10.5772/intechopen.70263 31216614 [DOI] [Google Scholar]
- Shen Q., Bourdais G., Pan H., Robatzek S., Tang D. (2017). Arabidopsis glycosylphosphatidylinositol-anchored protein LLG1 associates with and modulates FLS2 to regulate innate immunity. Proc. Natl. Acad. Sci. U.S.A. 114:5749. 10.1073/pnas.1614468114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi B., Lin L., Wang S., Guo Q., Zhou H., Rong L., et al. (2016). Identification and regulation of host genes related to rice stripe virus symptom production. New Phytol. 209 1106–1119. 10.1111/nph.13699 [DOI] [PubMed] [Google Scholar]
- Shu Q. Y., Cui H. R., Ye G. Y., Wu D. X., Xia Y. W., Gao M., et al. (2002). Agronomic and morphological characterization of Agrobacterium-transformed Bt rice plants. Euphytica 127 345–352. [Google Scholar]
- Singh P., Sinha A. K. (2016). A positive feedback loop governed by sub1a1 interaction with mitogen activated protein kinase 3 imparts submergence tolerance in rice. Plant Cell 28:1127. 10.1105/tpc.15.01001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speth C., Toledo-Filho L. A. A., Laubinger S. (2014). Immunoprecipitation-based analysis of protein-protein interactions. Methods Mol. Biol. 1158 175–185. 10.1007/978-1-4939-0700-7_11 [DOI] [PubMed] [Google Scholar]
- Takeda Y., Tobimatsu Y., Yamamura M., Takano T., Sakamoto M., Umezawa T. (2019). Comparative evaluations of lignocellulose reactivity and usability in transgenic rice plants with altered lignin composition. J. Wood Sci. 65:6 10.1186/s10086-019-1784-6 [DOI] [Google Scholar]
- Tang N., Ma S., Zong W., Yang N., Lv Y., Yan C., et al. (2016). MODD mediates deactivation and degradation of OsbZIP46 to negatively regulate ABA signaling and drought resistance in rice. Plant Cell 28 2161–2177. 10.1105/tpc.16.00171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taoka K., Ohki I., Tsuji H., Furuita K., Hayashi K., Yanase T., et al. (2011). 14-3-3 proteins act as intracellular receptors for rice Hd3a florigen. Nature 476 332–335. 10.1038/nature10272 [DOI] [PubMed] [Google Scholar]
- Tsuyoshi M., Loch C. M., Rong L. (2002). Identification of a multifunctional domain in autonomously replicating sequence-binding factor 1 required for transcriptional activation, DNA replication, and gene silencing. Mol. Cell. Biol. 22:505. 10.1128/mcb.22.2.505-516.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F., Jian Z., Nie L., Cui K., Peng S., Lin Y., et al. (2012a). Effects of N treatments on the yield advantage of Bt-SY63 over SY63 (Oryza sativa) and the concentration of Bt protein. Field Crop Res. 129 39–45. 10.1016/j.fcr.2012.01.011 [DOI] [Google Scholar]
- Wang F., Ye C., Zhu L., Nie L., Cui K., Peng S., et al. (2012b). Yield differences between Bt transgenic rice lines and their non-Bt counterparts, and its possible mechanism. Field Crop Res. 126 8–15. 10.1016/j.fcr.2011.09.017 [DOI] [Google Scholar]
- Wang Q., Guo Q., Guo Y., Yang J., La H. (2018). Arabidopsis subtilase SASP is involved in the regulation of ABA signaling and drought tolerance by interacting with OST1. J. Exp. Bot. 69:18. 10.1093/jxb/ery205 [DOI] [PubMed] [Google Scholar]
- Wang Y., Huang C., Hu B., Liu Y., Hereward J. P. (2019). Gene flow across host sociated populations of the rice stem borer Chilo suppressalis Walker (Lepidoptera: Crambidae)-implications for Bt resistance management in rice. Pest Manag. Sci. 76 695–703. 10.1002/ps.5567 [DOI] [PubMed] [Google Scholar]
- Wang Z., Ruan W., Shi J., Zhang L., Xiang D., Yang C., et al. (2014). Rice SPX1 and SPX2 inhibit phosphate starvation responses through interacting with PHR2 in a phosphate-dependent manner. Proc. Natl. Acad. Sci. U.S.A. 111 14953–14958. 10.1073/pnas.1404680111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia H., Chen L. Y., Wang F., Lu B. R. (2010). Yield benefit and underlying cost of insect-resistance transgenic rice: implication in breeding and deploying transgenic crops. Field Crop. Res. 118 215–220. 10.1016/j.fcr.2010.05.008 [DOI] [Google Scholar]
- Xia H., Lu B., Xu K., Wang W., Yang X., Yang C., et al. (2011). Enhanced yield performance of Bt rice under target-insect attacks: implications for field insect management. Transgenic Res. 20 655–664. 10.1007/s11248-010-9449-7 [DOI] [PubMed] [Google Scholar]
- Ying Y., Yue W., Wang S., Li S., Wang M., Zhao Y., et al. (2017). Two H-type thioredoxins interact with the E2 ubiquitin conjugase PHO2 to fine-tune phosphate homeostasis in rice. Plant Physiol. 173 812. 10.1104/pp.16.01639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeller S. L., Kalinina O., Brunner S., Keller B., Schmid B. (2010). Transgene × environment interactions in genetically modified wheat. PLoS One 5:e11405. 10.1371/journal.pone.0011405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Su J., Duan S., Ao Y., Dai J., Liu J., et al. (2011). A highly efficient rice green tissue protoplast system for transient gene expression and studying light/chloroplast-related processes. Plant Methods. 7:30. 10.1186/1746-4811-7-30 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The selected positive insertions from the DH5α strains were detected by PCR.
Co-ip was used to verify interaction between Cry1Ab/c and photosynthetic protein CP43 and stress resistance protein DnaJ, respectively. Protein extracts (Input) were immunoprecipitated with GFP-trap beads (IP) and resolved by SDS-PAGE. The immunoblots shown were developed with anti-GFP antibody to detect target endogenous protein (a) and with anti-mCherry antibody to detect Cry1Ab/c (b, 94KD). (A) CP43-GFP+Cry1Ab/c-cherry (36KD); (B) DnaJ-GFP+Cry1Ab/c-cherry (76KD).
Primers used for cloning the full-length gene based on subcellular co-localization and co-immunoprecipitation vectors.
Data Availability Statement
All datasets generated for this study are included in the article/Supplementary Material.