Abstract
Understanding brain function-related neural circuit connectivity is essential for investigating how cognitive functions are decoded in neural circuits. Trans-synaptic viral vectors are useful for identifying neural synaptic connectivity because of their ability to be transferred from transduced cells to synaptically connected cells. However, concurrent labeling of multisynaptic inputs to postsynaptic neurons is impossible with currently available trans-synaptic viral vectors. Here, we report a neural circuit tracing system that can simultaneously label postsynaptic neurons with two different markers, the expression of which is defined by presynaptic input connectivity. This system, called “cFork (see fork)”, includes delivering serotype 1-packaged AAV vectors (AAV1s) containing Cre or flippase recombinase (FlpO) into two different presynaptic brain areas, and AAV5 with a dual gene expression cassette in postsynaptic neurons. Our in vitro and in vivo tests showed that selective expression of two different fluorescence proteins, EGFP and mScarlet, in postsynaptic neurons could be achieved by AAV1-mediated anterograde trans-synaptic transfer of Cre or FlpO constructs. When this tracing system was applied to the somatosensory barrel field cortex (S1BF) or striatum innervated by multiple presynaptic inputs, postsynaptic neurons defined by presynaptic inputs were simultaneously labeled with EGFP or mScarlet. Our new anterograde tracing tool may be useful for elucidating the complex multisynaptic connectivity of postsynaptic neurons regulating diverse brain functions.
Keywords: Adeno-associated virus -1, Adeno-associated virus -5, Brain mapping, EGFP, mScarlet, Anterograde tracing
INTRODUCTION
Cognitive functions are regulated by multiple neuronal activities mediated by highly elaborate and complex synaptic connections. Postsynaptic neurons receiving multiple presynaptic inputs process complex presynaptic input activities, and then produce unique activity patterns encoding certain brain functions. Thus, it is necessary to understand what presynaptic inputs are connected to certain postsynaptic neurons. Neuroanatomical tracing molecules that can be transmitted in an anterograde direction have been widely utilized for this purpose.
Neuroanatomical tracers, including biotinylated dextran amines (BDA), horseradish peroxidase (HRP), wheat germ agglutinin (WGA), and cholera toxin B subunit (CTB) [1, 2], can be easily used in animal brains to efficiently label synaptic connectivity. However, these non-viral neuroanatomical tracers are unsuitable for revealing fine neuronal connectivity or the cell types that make synapses. Classical anterograde tracing with viral vectors uses the density of axonal projections as proxies of potential synaptic connections to postsynaptic targets. However, this method cannot distinguish the trespassing axonal collaterals from actual synaptic connections without additional high-resolution imaging. Moreover, the connectivity can be misinterpreted when synaptic connections are formed in a distant dendritic location.
Recent advances using viral vectors have been shown to overcome such limitations, given their innate capability of being transferred from infected cells to other cells via synapses. Viral vectors including adeno-associated virus (AAV), vesicular stomatitis virus (VSV), and herpes simplex virus (HSV), are known to spread in the anterograde direction across synapses and have been proven to express transgenes in the viral genome [2-7]. However, labeling multiple connectivity in the postsynaptic neuron is limited because only a single population of presynaptic neurons infected by anterograde trans-synaptic viruses is delivered to postsynaptic partner neurons. Since animal cognitive functions and behaviors depend on the complex involvement of multiple neurons and their synaptic crosstalk, the demand for trans-synaptic viral vectors suitable for labeling multiple synaptic connectivity systems has been increasing. It is thus necessary to develop anterograde trans-synaptic viral vectors capable of expressing multiple marker genes, each of which represent specific presynaptic inputs, in the postsynaptic neurons.
Here, we report a dual-color anterograde tracing vector system (cFork) using presynaptic AAV1-mediated delivery of recombinases [7] and postsynaptic expression of a recombinase-dependent dual gene expression cassette. This system could achieve presynaptic input-defined multi-gene expression in postsynaptic neurons of the S1BF or striatum, where multiple presynaptic innervations are observed.
MATERIALS AND METHODS
Plasmids
pAAV-EF1a-cDIO-EGFP-CMV-fDIO-mScarlet was generated by insertion of the CMV-fDIO-mScarlet fragment from pAAV-CMV-fDIO-mScarlet into the EcoRI sites of pAAV-EF1a-cDIO-EGFP.
To produce pAAV-EF1a-cDIO-EGFP, the PCR-amplified coding sequence of EGFP (EGFP-SpeI-S→EGFP-SpeI-A) from pEGFP-C1 (Clontech) was cloned into the SpeI sites of pAAV-EF1a-cDIO-mOrange2. mScarlet was isolated from pmScarlet-C1 (Addgene #85042) by PCR using mScarlet-NheI-S and mScarlet-AscI-A primers, and the PCR product was inserted into the pAAV-EF1a-fDIO-EYFP (Addgene #55641) for pAAV-EF1a-fDIO-mScarlet. The CMV promoter was PCR-amplified (CMV-MluI-S→CMV-BamHI-A) from pEGFP-C1 and cloned into the MluI/BamHI sites of pAAV-EF1a-fDIO-mScarlet to produce pAAV-CMV-fDIO-mScarlet. The CMV-fDIO-mScarlet was PCR-amplified (CMV-EcoRI-S→WPRE-233n-A) from pAAV-CMV-fDIO-mScarlet and inserted into the EcoRI site of pAAV-EF1a-cDIO-EGFP, producing pAAV-EF1a-cDIO-EGFP-CMV-fDIO-mScarlet.
To construct pAAV-EF1a-Cre, the PCR-amplified coding sequence of Cre (Cre-KpnI-S→Cre-EcoRI-A) from pAAV-EF1a-mCherry-IRES-Cre (Addgene #55632) was cloned into the pAAV-EF1a-FlpO plasmid (Addgene #55637). pAAV-hSyn-Cre and pAAV-hSyn-FlpO were obtained by inserting the MluI/KpnI fragment containing the human synapsin promoter 1 (hSyn) from pAAV-hSyn-BDNF-EGFP into the MluI/KpnI sites of pAAV-EF1a-Cre and pAAV-EF1a-FlpO, respectively. pAAV-hSyn-vCre was obtained by inserting the MluI/BamHI fragment containing the hSyn from pAAV-hSyn-FlpO into the MluI/BamHI sites of pAAV-EF1a-vCre (Addgene #55638). All restriction enzymes employed in this study were purchased from New England Biolabs (NEB).
The oligonucleotide sequences used in the PCR were as follows: EGFP-SpeI-S, GACTAGTATGGTGAGCAAGGGCGAG; EGFP-SpeI-A, GGACTAGTTTACTTGTACAGCTCGTCC ATG; mScarlet-NheI-S, CTAGCTAGCACCATGGTGAGCAAGGGC; mScarlet-AscI-A, AGGCGCGCC TTACTTGTACAGCTCGTCCATGC; CMV-MluI-S, CGACGCGTTAGTAATCAATTACGGGGTC; CMV-BamHI-A, CGCGGATCCACCGGTAGCGCTAGCG; CMV-EcoRI-S, CCGGAATTCCCGCGT TACATAACTTACGG; WPRE-233n-A, GTTGCGTCAGCAAACACAGT; Cre-KpnI-S, CGGGGTACC ATGGCCAATTTACTGACCG; Cre-EcoRI-A.
Cell culture and transfection
Human embryonic kidney (HEK) 293T cells were cultured in Dulbecco’s modified Eagles medium (DMEM, HyCloneTM) supplemented with 10% fetal bovine serum (FBS, HyCloneTM) at 37°C under 5% CO2. The cultured HEK293T cells were transfected using Lipofectamine 2000 (Invitrogen). After transfection, HEK293T cells were incubated for 48~72 h, depending on the experimental design.
FACS analysis
For fluorescence-activated cell sorting (FACS) analysis, HEK 293T cells were transfected with Cre- and/or FlpO-recombinase and pAAV-EF1a-cDIO-EGFP-CMV-fDIO-mScarlet. After 48 h, cells were harvested with 0.25% trypsin-EDTA (Gibco) and washed with 1X PBS (phosphate buffered saline, pH 7.4), and then centrifuged. The harvested cells were suspended in DMEM medium and analyzed using the Flow Cytometer Cell Sorter (MoFlo Astrios, Beckman Coulter) to evaluate the cross-reactivity of Cre- or FlpO-recombinases. EGFP and mScarlet signals were detected through a 488-513/26 and 561-579/16 nm filter, respectively.
Virus injection
Adult mice (C57BL/6J) and Ai9 (Gt(ROSA)26Sor<tm9(CAG-tdTomato)Hze/J) mice were purchased from Orient Bio (Korea) and Jackson Laboratory (USA), respectively. All experiments were performed in accordance with the guidelines of the Institutional Animal Care and Use Committee of the Korea Brain Research Institute (IACUC-2017-0049). Mice were anesthetized with 1~2% isoflurane in a mixture of oxygen via a nose mask and placed on a stereotaxic frame (Model 900 Small Animal Stereotaxic Instrument, KOPF). Small holes in the skull were drilled to produce injection sites. AAV viruses were loaded into a pulled glass capillary using a Nanoject III injector (Drummond) for brain injection. AAV1-hSyn-Cre (3.2×1013 genome copies (GC)/ml) and AAV1-hSyn-Flp (5.0×1013 GC/ml) were purchased or packaged from Vector Biolabs.
Our dual FP-expression cassette (cGreen-fRed) was packaged in AAV5, which does not show trans-synaptic transfer. AAV5-cGreen-fRed was packaged from the viral vector core at the University of North Carolina (UNC). Stereotaxic coordinates targeting brain areas were as follows (in mm): the primary motor cortex (M1) [anteroposterior (AP)=2.0, mediolateral (ML)=1.8 and dorsoventral (DV; ventral from the cortical surface)=0.5]; the dorsal striatum (dStr) [AP=0.2, ML=2.2, DV=3.4]; the ventral posteromedial nucleus (VPM) [AP=1.8, ML=1.6, and DV=3.6]; the posterior medial nucleus of the thalamus (POm) [AP=2.06, ML=1.25, DV=2.97]; the somatosensory barrel field cortex (S1BF) [AP=0.7, ML=3.5, DV=0.35 and 0.75 (with 30° angle)]; and the vibrissal area of the primary motor cortex (vM1) [AP=1.34, ML=1.5, DV=0.35]. A volume of 200 nl of the virus-containing solution was injected into the M1 and POm with a 6 nl/min injection rate.
Fluorescence imaging and analysis
Fluorescence signals from HEK293T cells were detected using a fluorescent microscope (Nikon eclipse Ts2, objective 40X) equipped with a LED illuminator (excitation wavelength: 470 nm or 525 nm).
To detect fluorescence signals from brain slices, anesthetized mice were dissected and their brains fixed using vascular transcardial perfusion of 1X PBS followed by perfusion of a fixative solution (4% paraformaldehyde in 1X PBS, pH 7.4). Whole brains were then isolated and incubated with the fixative solution for 24 h at 4°C. Fixed brains were then incubated with a 30% sucrose solution (30% sucrose, 1X PBS) for 36~48 h, mounted in frozen section embedding medium (FSC, Leica), and cut at 40 µm thickness using a cryostat (Leica CM 1850 UV Cryostat). Sagittal and coronal slices were prepared for observing and labeling of neurons in the striatum and cortex, respectively. Sections were washed three times in 1X PBS and stained with DAPI (Sigma). After staining, the sections were mounted using a mounting medium (Vectashield, Vector Laboratories) for fluorescence imaging. Some brain slices were treated with antibodies against EGFP and mScarlet to enhance fluorescence signals. Chicken polyclonal anti-GFP antibody (Abcam, ab13970, 1:500) and mouse monoclonal anti-RFP antibody (ThermoFisher Scientific, MA5-15257, 1:500) were used as primary antibodies. The secondary antibodies used were Alexa Fluor 488-conjugated donkey anti-chicken IgY (IgG) (Jackson ImmunoResearch, catalog number 703-545-155, 1:1,000) and Alexa Fluor 594-conjugated donkey anti-mouse IgG (Jackson ImmunoResearch, catalog number 715-585-150, 1:1,000). Fluorescence imaging of brain slices was performed using a Pannoramic digital slide scanner (Pannoramic SCAN II, 3D HISTECH; offered by the Research Equipment Core Facility of the KBRI) with a 20× objective. The laser excitation and emission filters used for imaging were as follows: EGFP (467/498 nm laser excitation; 513/556 nm emission), mScarlet (561 nm laser excitation, 579/16 nm emission), and DAPI (405 nm excitation; 448/59 nm emission). Acquisition times ranged from 30 to 200 ms. For image analysis, brain areas of individual brain slices were selected, with the selection criteria of each brain region adopted from the Allen Mouse Brain Atlas. To measure the number of cells labeled with fluorescent proteins expressed by Cre- and/or FlpO-recombinase, the images were inverted and the background values were subtracted from the acquired images. The cells with overlapping DAPI signals were then counted. All images were processed and analyzed using NIH image analysis software (ImageJ ver: 1.51p). Statistical analyses were performed using GraphPad Prism (GraphPad Software ver. 7.0).
RESULTS
Validation of AAV1 as a monosynaptic anterograde tracing vector
To develop an anterograde trans-synaptic tracing tool for labeling multiple synaptic connections, previously reported anterograde viral vectors such as AAV1 were utilized [7]. AAV1 was selected because of its lower toxicity and easier manipulation than other viral vectors. To confirm if AAV1 was able to perform monosynaptic anterograde transfer from presynaptic to postsynaptic neurons, AAV1-hSyn-Cre was intracranially delivered into the primary motor cortex (M1) of adult Ai9 (Cre-inducible tdTomato reporter) mice (Fig. 1A). It is expected that anterograde trans-synaptic transfer of AAV1-hSyn-Cre to postsynaptic neurons would induce tdTomato expression if these two neurons form synapses, but tdTomato expression would be limited to cells with monosynaptic connections (Fig. 1A).
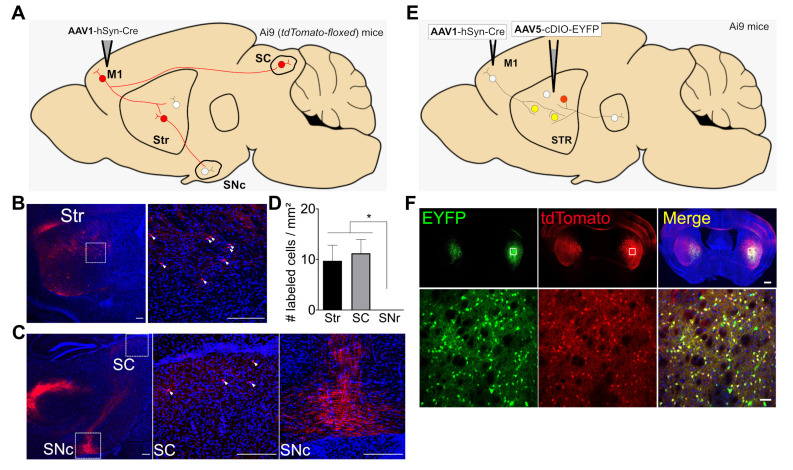
Fig. 1.
Anterograde monosynaptic tracing of AAV1-hSyn-Cre in Ai9 mice. (A) Schematic diagram for testing AAV1 as a monosynaptic anterograde viral vector. AAV1-hSyn-Cre was injected into the primary motor cortex (M1). The expected expression of tdTomato in the M1-connected postsynaptic neurons in the striatum (Str), superior colliculus (SC), and substantia nigra pars compacta (SNc) is shown as cell bodies with red color. (B) Left: representative fluorescence image showing the distribution of tdTomato (red)-expressing cells with DAPI staining (blue) in the Str (red). Right: a magnified view of tdTomato-expressing cells (white arrowheads) in the Str; scale bar: 200 μm. (C) Left: Representative fluorescence image showing the distribution of tdTomato (red)-expressing cells in the SC or SNc. Middle: a magnified view of tdTomato-expressing cells (white arrowheads) in the SC. Right: a magnified view of tdTomato-expressing neurites (no cell bodies detected) in the SNc; scale bar: 200 μm. (D) The bar graph depicts the average numbers of tdTomato-expressing cells with standard error of means (SEM) in the observed brain areas (# labeled cells/mm2) from 3 slices (pooled from 2 mice). *p<0.05, one-way ANOVA with Tukey’s multiple comparison test. (E) Schematic diagram for testing presynaptic AAV1-hSyn-Cre-mediated postsynaptic expression of EYFP and tdTomoto. AAV1-hSyn-Cre was injected into the M1 and AAV5-cDIO-EYFP into the Str. (F) Above: Representative fluorescence images showing the distribution of EYFP (green) or tdTomato (red)-expressing cells in the striatum; scale bar: 500 μm. Below: magnified views of EYFP or tdTomato-expressing cells in the indicate area (white boxed in above) in the striatum; scale bar: 50 μm.
Four weeks after viral injection, neuronal tdTomato expression was found in direct postsynaptic targets of M1, such as the dorsal striatum (dStr) or superior colliculus (SC) [7]. A single bout of AAV1-hSyn-Cre injection (~50 nl of viral particles with a titer of ~1013 genome copy/ml) resulted in similar densities of tdTomato-expressing cells per mm2 in the dStr and SC (number of labeled cells/mm2: dStr=9.7±3.1 vs. SC=11.2±2.7; Fig. 1B~D) in the slices pooled from two mice. However, the substantia nigra pars compacta (SNc), which does not directly receive synaptic inputs from the M1 but is innervated by striatal projecting neurons, only showed limited tdTomato expression in neurite-like structures (Fig. 1C, D). Postsynaptic tdTomato expression in Ai9 mice by anterograde-transferred AAV1-hSyn-Cre was again confirmed when AAV1-hSyn-Cre was injected into the M1 and AAV5-cDIO-EYFP in the dStr of Ai9 mice (Fig. 1E). Since the EYFP gene is flanked by Cre-responsive double-floxed inverted origin (cDIO)-cassettes, we expected that EYFP expression would be mostly accompanied with tdTomoto expression, which is also Cre-dependent. Our data showed that 91.9±0.7% of EYFP-expressing striatal cells also displayed tdTomato expression (% of EYFP+tdTomato cells among EYFP-expressing cells, n=28 slices from 8 mice; Fig. 1F). Together, these results support previous findings that AAV1 is an anterograde trans-synaptic viral vector that labels monosynaptic connectivity [7, 8].
Designing a dual-gene expression vector for simultaneous anterograde tracing of multiple pathways
We additionally attempted to develop an anterograde trans-synaptic system that would enable presynaptic input-defined multiple tags in postsynaptic neurons by using AAV1. Postsynaptic expression of multiple tags determined by input connectivity could be achieved by delivering (1) a gene expression cassette of multiple genes (multi-gene expression cassette) into postsynaptic neurons and (2) AAV1, which contains inducers or regulators of gene expression from postsynaptic multi-gene expression cassettes to presynaptic neurons.
To construct a multi-gene expression cassette to be delivered to postsynaptic neurons, we set out to design a dual gene expression cassette for expressing two different fluorescent proteins (FPs). AAV1-mediated delivery of Cre recombinase from presynaptic to postsynaptic neurons to induce expression of FP genes flanked by DIO is possible (Fig. 2A). Moreover, regulated expression of two different FP genes could be achieved if each FP gene was flanked with different DIO elements, and corresponding recombinases were delivered from presynaptic inputs via AAV1. By searching for recombinases that are selective and efficient for the expression of DIO-flanked FPs and promoters ideal for FP expression, we constructed a series of mammalian expression vectors containing strong general promoters such as human elongation factor 1a promoter (EF1a), cytomegalovirus immediate-early promoter (CMV) or human ubiquitin C promoter (UbC), combined with a recombinase-dependent gene expression cassette (Fig. 2B). Previous studies have shown that site-specific recombinases such as Cre, FlpO, and vCre recognize specific DNA sequences such as loxP/lox2272, FRT/F5, and vloxP/vlox2272, respectively [9, 10]. Inverse-oriented gene sequences encoding FPs with different emission wavelengths such as EGFP (Green), mScarlet (Red) or mCerulean3 (Blue) were flanked with recombinase-specific DIO sequences, resulting in cDIO-, fDIO (FlpO-responsive DIO)-, and vDIO (vCre-responsive DIO)- FPs (Fig. 2B).
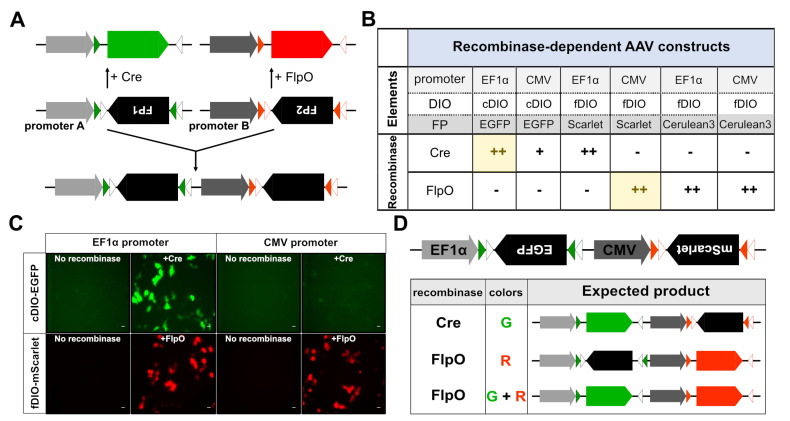
Fig. 2.
Strategy for constructing dual color labeling vector. (A) Schematic diagram showing a dual-color labeling vector combining the Cre/loxP and FlpO/FRT recombination elements in the construct. The double-floxed inverse orientation (DIO), an inverted open reading frame (ORF) of fluorescent protein (FP) genes, is located between sets of recombinase recognition sites such as loxp/lox2272 (cDIO) or FRT/F5 (fDIO). The Cre- or FlpO-recombinase recognizes the cDIO (green/white triangles) or FRT/F5 (red/white triangles) sites, respectively, resulting in gene expression by inverting the ORF of the FPs through DNA recombination. cDIO-FP1 and fDIO-FP2 expression cassettes with promoters A and B were fused to produce our dual color labeling vector. (B) Summary of expression screening of recombinase-specific expression of AAV constructs. ++: high expression of FPs, +: moderate or weak expression of FPs, -: no expression of FPs. The combination with yellow color was selected to produce a fusion construct of EF1a-cDIO-EGFP-CMV-fDIO-mScarlet. (C) Representative fluorescence images showing recombinase-specific expression of fluorescent proteins (ex. EGFP and mScarlet). To screen for efficient and selective promoter-DIO-FP component pairs, HEK293T cells were co-transfected with promoter-DIO-FP vectors with or without vectors containing recombinase genes as indicated. In this figure, pAAV-cDIO or pAAV-fDIO vectors including EF1α or CMV promoter, and EGFP or mScarlet, were co-transfected with pAAV-Cre or pAAV-FlpO; scale bar: 20 μm. (D) Schematic diagram showing the expected FP expression from the dual color expression vector with the combined actions of Cre and FlpO recombinases.
After transfecting plasmids containing different combinations of promoters, reporters, and DIOs into HEK293T cells, we then tested which construct was most efficient and selective for FP expression (Fig. 2C). Consistent with previous reports, cDIO- or fDIO-FP expression did not overlap when each recombinase was combined (Fig. 2B, C). In addition, our data showed that the EF1α promoter is more efficient in expressing cDIO-EGFP than the CMV promoter (Fig. 2B, C). However, the CMV promoter with fDIO was suitable for high expression of mScarlet when floxed in fDIO (Fig. 2B, C). Since both EGFP and mScarlet showed brighter expression than the blue fluorescent protein mCerulean3, we selected EF1α-cDIO-EGFP and CMV-fDIO-mScarlet cassettes for the dual FP expression cassette. A fusion construct including both cassettes was inserted into the single AAV genome, resulting in AAV-EF1α-cDIO-EGFP-CMV-fDIO-mScarlet (AAV-cGreen-fRed; Fig. 2D).
Cre and/or FlpO-dependent EGFP and mScarlet expression from a dual-gene expression vector
To test whether the newly developed dual gene expression cassette in the AAV vector was able to express EGFP and/or mScarlet by selective actions of Cre or FlpO recombinases, AAV5-cGreen-fRed was introduced into HEK293T cells with Cre and/or FlpO expression (Fig. 3A). Our data showed that expression of either EGFP or mScarlet was exclusively observed in the presence of the corresponding recombinase, and co-expression of Cre and FlpO was sufficient for dual expression of EGFP and mScarlet (Fig. 3A). When EGFP or mScarlet expression was quantitatively assessed by flow cytometry analysis of transfected HEK293T cells, we found that 99.96±0.02% of cells showed EGFP expression when Cre was present, and 99.02±0.25% of FlpO-expressing cells were sorted as mScarlet expressing cells (from three different batches of cell cultures; Fig. 3B, C). Nearly 65% of cells displayed co-expression of EGFP and mScarlet by co-transfection of Cre and FlpO (EGFP only: 25.68±1.37%; mScarlet only: 9.65±0.97%, EGFP+mScarlet: 64.67±2.20%; from three different batches of cell cultures; Fig. 3B, C). These results indicate that our dual gene expression cassette efficiently expressed multiple FPs, and that the expression of these FPs was selectively regulated by the corresponding Cre or FlpO recombinases.
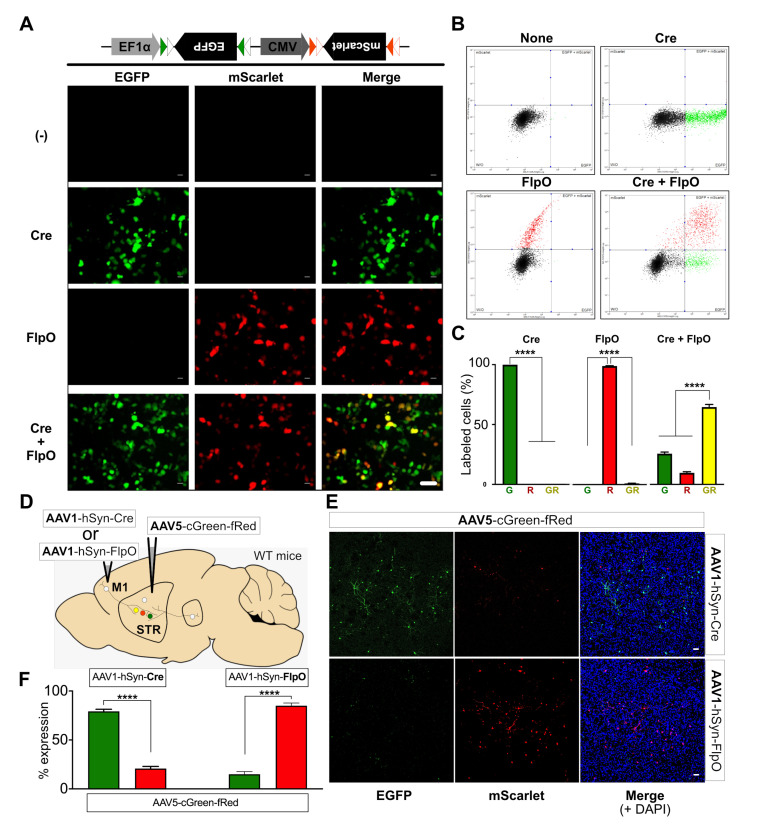
Fig. 3.
Validation of dual color gene expression vector in vitro. (A) Representative images showing Cre- or FlpO-selective expression of EGFP or mScarlet from the dual FP-expression vector. HEK293T cells were transfected with AAV-cGreen-fRed and pAAV-hSyn-Cre, and/or pAAV-hSyn-FlpO. EGFP or mScarlet expression was tested 72 h after transfection; scale bar: 50 μm. (B) Representative fluorescence-activated cell sorting (FACS) analysis showing recombinase-specific expression of EGFP or mScarlet in HEK293T cells co-transfected with the Cre and/or FlpO constructs. FACS-sorted cells with EGFP or mScarlet expression are displayed as colored scatter dots. (C) Bar graphs depicting the summary of FACS analysis results, showing the average percentage of EGFP (G), mScarlet (R), or EGFP+mScarlet (GR) - expressing cells from total transfected cells (mean±SEM). ****p<0.0001, one-way ANOVA with Tukey’s multiple comparison test. (D) Experimental scheme for in vivo validation of recombinase-specific expression of EGFP or mScarlet from AAV5-cGreen-fRed. AAV1-hSyn-Cre or AAV1-hSyn-FlpO was injected into M1 individually, but AAV5-cGreen-fRed was injected into the dorsal striatum. (E) Representative images showing M1-derived Cre- or FlpO-selective expression of EGFP or mScarlet from AAV5-cGreen-fRed in the striatum; scale bar=50 µm. (F) Bar graphs depicting the average percentage of EGFP (green)- or mScarlet (red)-expressing cell numbers (% expression) in the AAV5-cGreen-fRed-injected striatum when AAV1-hSyn-Cre or AAV1-hSyn-FlpO was injected into M1. N=seven slices randomly sampled from three mice. Mean±SEM. ****p<0.0001, unpaired t-test.
To verify recombinase-specific FP expression from AAV5-cGreen-fRed in vivo, we individually injected AAV1-hSyn-Cre or FlpO into the presynaptic M1 and AAV5-cGreen-fRed into the postsynaptic dStr of wild type mice (Fig. 3D). Our results showed that each recombinase was able to induce expression of corresponding fluorescence proteins in approximately 80% of striatal neurons (EGFP-cells by Cre: 79.2±2.2%, mScarlet-cells by FlpO: 84.0±3.2%; n=7 slices randomly sampled from 3 mice; Fig. 3E, F), although there were around 20% of striatal neurons showing a reversed expression pattern (mScarlet by Cre or EGFP by FlpO; Fig. 3E, F). There was no co-expression of EGFP and mScarlet detected in striatal cells (Fig. 3E). These data suggest that AAV-cGreen-fRed could achieve selective expression of FPs by trans-synaptic transfer of recombinases in vivo, but the specificity of these recombinase-specific FP expression is limited to approximately 80%.
In vivo dual color labeling of postsynaptic neurons receiving multiple inputs
Since EGFP or mScarlet expression can be individually controlled by Cre or FlpO in the same cell when our dual gene expression cassette was introduced, we next tested whether AAV1-mediated trans-synaptic transfer of Cre or FlpO from presynaptic neurons can regulate EGFP or mScarlet expression in postsynaptic neurons with AAV-cGreen-fRed (Fig. 4A).
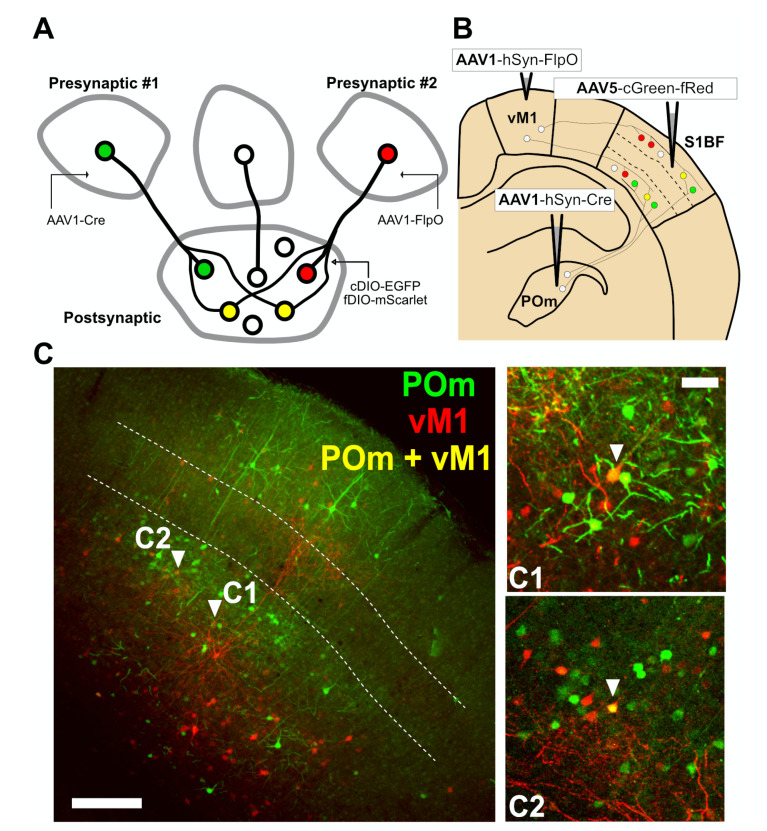
Fig. 4.
Labeling POm- or vM1-derived inputs in S1BF neurons using cFork system. (A) Schematic diagram illustrating the experimental strategy used to demonstrate simultaneous dual color labeling of postsynaptic neurons based on synaptic connections with presynaptic inputs. AAV1s with different recombinases (AAV1-Cre or AAV1-FlpO) were injected into two presynaptic inputs (presynaptic #1 and #2) independently. Postsynaptic neurons receiving multiple presynaptic inputs, including presynaptic #1 and #2, were designed to express our dual gene expression cassette (cDIO-EGFP-fDIO-mScarlet) included in the AAV-cGreen-fRed construct. (B) Experimental scheme for simultaneous labeling of POm- or vM1-S1BF connectivity. AAV1-hSyn-Cre and AAV1-hSyn-FlpO were injected into two different presynaptic inputs of S1BF, POm and vM1, respectively. AAV5-cGreen-fRed was injected into the postsynaptic S1BF. (C) Representative confocal image showing S1BF cortical neurons receiving synaptic inputs from POm (green) or vM1 (red). Fluorescent signals were amplified by fluorescent immunostaining with anti-GFP or anti-RFP antibodies. The white dotted line indicates the boundary of L4; scale bar: 200 μm. Representative S1BF cells expressing both EGFP and mScarlet are denoted as C1 and C2 (right panels); scale bar: 50 μm.
We first took advantage of the canonical wiring selectivity of the somatosensory barrel field cortex (S1BF). The posterior medial nucleus of the thalamus (POm) is known to primarily innervate layer 1 (L1) and 5a (L5a), but avoids layer 4 (L4) of the S1BF [11, 12]. Likewise, S1BF receives presynaptic inputs from higher-order cortical inputs such as presynaptic innervation from the vibrissal area of the primary motor cortex (vM1), which also projects mainly on infragranular layers, yet scarcely on L4 [11, 13, 14]. We thus tested whether S1BF neurons could be defined by cells receiving presynaptic inputs from POm (EGFP-positive) or vM1 (mScarlet-positive). AAV1-hSyn-Cre and -FlpO were injected into presynaptic POm and vM1, respectively, and AAV5- cGreen-fRed into the postsynaptic S1BF (Fig. 4B). Four weeks after injection, we found that POm or M1-receiving neurons were mainly observed in L2/3, L5, and L6, but scarcely in L4 in the S1BF (Fig. 4C)—corroborating previous findings [11,15-17].
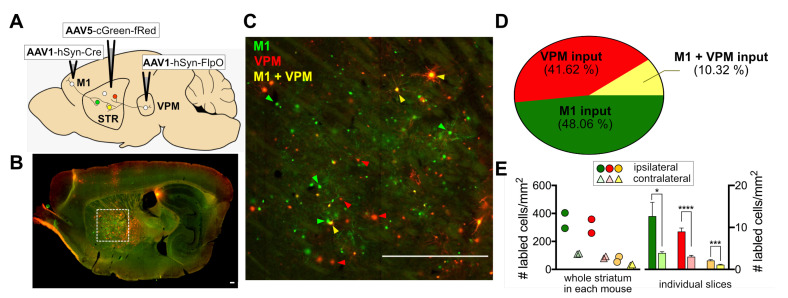
Next, our cFork system was tested in striatal circuits of the mouse brain (Fig. 5A). Because the dorsal striatum receives two major excitatory inputs from the cortex and thalamus, called the corticostriatal and thalamostriatal pathways, respectively [18-20], we aimed to label striatal postsynaptic neurons with EGFP and/or mScarlet, whose expression is defined by presynaptic cortical and/or thalamic inputs, respectively. For this purpose, AAV1s containing Cre or FlpO were injected into the right hemisphere of the M1 cortical subregion or into a part of the ventral posteromedial (VPM) thalamic nucleus of wild-type C57/B6 mice. Bilateral dStr was injected with AAV5-cGreen-fRed (Fig. 5A).
Fig. 5.
Labeling corticostriatal and thalamostriatal pathways using cFork. (A) Experimental scheme for simultaneous labeling of corticostriatal and thalamostriatal pathways in the striatum. AAV1-hSyn-Cre and AAV1-hSyn-FlpO were injected in the right hemisphere of two different presynaptic inputs of the striatum, the primary motor cortex (M1) and the ventral posteromedial (VPM) thalamic nucleus, respectively. AAV5 containing a dual gene expression cassette (EF1a-cDIO-EGFP-CMV-fDIO-mScarlet) was bilaterally injected into the postsynaptic striatum. (B) Representative fluorescence images showing EGFP and/or mScarlet expression in striatal neurons injected with AAV1 and AAV5 as described in B; scale bar: 200 μm. (C) Enlarged fluorescence image of the indicated area in B (the box with white dotted line) showing mixed fluorescence protein (FP) expression. Examples of EGFP (green arrowhead), mScarlet (red arrowhead), or EGFP+mScarlet co-expression (yellow arrowhead) in the ipsilateral striatum are indicated; scale bar: 200 μm. (D) The pie chart illustrates the percentage of striatal neurons labeled as cells receiving M1- (EGFP), VPM- (mScarlet), and M1+VPM- (EGFP+mScarlet) inputs. (E) Left: Each circle (ipsilateral) or triangle (contralateral) represents cell densities (number of cells/mm2) of FP-expressing cells counted from the whole striatum of each mouse, with EGFP (green; M1-input), mScarlet (red; VPM-input), or EGFP and mScarlet (yellow; M1+VPM-input). Right: bar graphs depict average cell densities of FP-expressing cells (mean±SEM) from individual striatal slices. Same colors as left. *p<0.05, ***p<0.001, ****p<0.0001, unpaired t-test.
Four weeks after viral injection, we found mixed patterns of EGFP and/or mScarlet expression in striatal neurons (Fig. 4B, C). These FP expression patterns seemed to be mediated by anterograde transport of Cre and FlpO from M1 and VPM, respectively, because no FP expression was observed in the striatum when AAV1-hSyn-Cre or hSyn-FlpO was not injected (data not shown). The percentage of cells showing exclusive expression of EGFP and mScarlet was 48.06% (4,233/8,808 cells) and 41.62% (3,666/8,808 cells), respectively, and 10.32% (909/8,808 cells) of striatal cells showed co-expression of EGFP and mScarlet (whole striatal areas derived from sagittal sections from two mice; Fig. 5D). Despite estimation from two animals and an 80% accuracy of FP-labeling by presynaptic input-derived recombinases (Fig. 3F), our results (Fig. 5D) are in line with previous findings that striatal medium spiny neurons contain corticostriatal and thalamostriatal synapses equally [21, 22], and that projections from M1 and VPM show low convergence in the striatum [20]. Moreover, we found that FP-labeled ipsilateral striatal neurons were approximately four fold higher in number than contralateral ones (ipsilateral vs. contralateral projection shown as number of labeled cells/mm2: M1-input=12.7±3.3 vs. 3.9±0.3, VPM-input=9.0±0.8 vs. 3.0±0.3; n=57 ~ 74 slices from 2 mice; Fig. 5E). These results indicate that ipsilateral corticostriatal and thalamostriatal projections are dominant in the dStr, as previously reported [22-24].
Together, these results support the idea that cFork is useful for achieving labeling of postsynaptic neurons with two different input-defined tags in the mouse brain.
DISCUSSION
Tracing multiple synaptic connections is essential for understanding how the neural connectome and its activity regulate brain functions. Available neuroanatomical tracing tools can efficiently dissect and identify input/output relationships of two brain regions but cannot effectively address complex and multiple connectivity. Our newly developed anterograde viral tracing system is capable of expressing two different labeling genes in postsynaptic neurons. Expression of these genes was tightly regulated by different recombinases in vitro (Fig. 3C), but in vivo tests showed that the accuracy of inducing two different FP expression by presynaptic delivery of recombinases is limited to approximately 80% (Fig. 3F). In spite of the limited accuracy of our dual anterograde tracing tool, we were able to demonstrate that the combination of AAV1-mediated differential delivery of recombinases with a dual-gene expression vector is able to mark postsynaptic neurons based on connectivity with certain presynaptic inputs in the S1BF or dStr (Fig. 4 and 5). Our results showed that selective innervation from POm or vM1 and M1 or VPM could be well visualized in postsynaptic neurons in the S1BF (Fig. 4) and dStr (Fig. 5), respectively, suggesting that one postsynaptic neuron could be easily identified by presynaptic input by using our cFork system.
Despite the presented advantages, there are several points for improvement when designing anterograde viral vectors ideal for multi-connectivity detection in future studies. First, as shown in Fig. 3D-F, recombinase-specific regulation of gene expression from our dual gene expression cassette was ideal in vitro, but only around 80% of neurons with AAV5-cGreen-fRed displayed selective FP expression by AAV1-mediated delivery of recombinase in vivo. Because tight regulation of input-defined labeling gene expression is essential for quantifying exact presynaptic input patterns to postsynaptic neurons in vivo, this unexpected misregulation of recombinase-induced gene expression should be resolved in future studies. Second, experimental artifacts, such as under- or over-estimation of presynaptic innervations to postsynaptic neurons, may be caused by errors unrelated to neural connectivity. These errors may be caused by multiple and repetitive intracranial injection of three viral vectors into target or candidate brain areas in our system (Fig. 4 and 5) and were also affected by the amounts and titers of injected viral particles. In addition, as demonstrated in Fig. 3, imperfect co-expression of EGFP and mScarlet (approximately 65%) even in the presence of both Cre and FlpO in the same cell, may affect in vivo labeling of presynaptic inputs in postsynaptic neurons. These errors might be caused by variation in the viral vector transduction efficiencies of multiple constructs, or different levels of gene expression from our dual-gene expression cassette containing different combinations of promoters and DIO elements (Fig. 2). Third, additional recombinase-DIO pairs, such as vCre-vDIO [9, 10], need to be added to our system to achieve detection of three or more presynaptic connectivity in postsynaptic neurons because cFork system largely depends on the selective action of each recombinase on specific DIO elements. However, owing to its small genome capacity (~4.7 kb) [25], AAV is not a proper viral vector for including gene expression cassettes of three or more labeling genes.
To overcome these limitations, several approaches may be possible. Presynaptic input-dependent tight regulation of gene expression from the dual-gene expression vector may be achieved by re-designing and screening of more specific DIO and FP combinations, which selectively respond to the AAV1-mediated anterograde delivery of recombinases. Experimental artifacts and the small genome capacity of AAV could be overcome if transgenic/knock-in mice constitutively expressing multi-gene expression cassettes are constructed. Moreover, along with efforts to identify additional specific recombinases-DIO pairs, novel anterograde viral vectors which are easy to handle and have a larger genome capacity than AAV may facilitate the development of an advanced version of the anterograde tracing tool, thus enabling simultaneous visualization of multi-synaptic connectivity in the postsynaptic neuron.
In summary, we have provided a new anterograde tracing tool that can simultaneously visualize dual presynaptic input connectivity in postsynaptic neurons based on the anterograde AAV vector. Our tracing system is an ideal viral tracing tool capable of dissecting multiple synaptic connectivity with minimal effort.
ACKNOWLEDGEMENTS
This work was supported by the KBRI basic research program (19-BR-04-01, 19-BR-01-03) of the Korea Brain Research Institute, and Brain Research Program (NRF-2017M3C7A1048086) of the National Research Foundation of Korea (NRF), funded by the Ministry of Science and ICT.
REFERENCES
- 1.Lanciego JL, Wouterlood FG. A half century of experimental neuroanatomical tracing. J Chem Neuroanat. 2011;42:157–183. doi: 10.1016/j.jchemneu.2011.07.001. [DOI] [PubMed] [Google Scholar]
- 2.Nassi JJ, Cepko CL, Born RT, Beier KT. Neuroanatomy goes viral! Front Neuroanat. 2015;9:80. doi: 10.3389/fnana.2015.00080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ugolini G, Kuypers HG, Strick PL. Transneuronal transfer of herpes virus from peripheral nerves to cortex and brainstem. Science. 1989;243:89–91. doi: 10.1126/science.2536188. [DOI] [PubMed] [Google Scholar]
- 4.Callaway EM. Transneuronal circuit tracing with neurotropic viruses. Curr Opin Neurobiol. 2008;18:617–623. doi: 10.1016/j.conb.2009.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lo L, Anderson DJ. A Cre-dependent, anterograde transsynaptic viral tracer for mapping output pathways of genetically marked neurons. Neuron. 2011;72:938–950. doi: 10.1016/j.neuron.2011.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beier KT, Saunders A, Oldenburg IA, Miyamichi K, Akhtar N, Luo L, Whelan SPJ, Sabatini B, Cepko CL. Anterograde or retrograde transsynaptic labeling of CNS neurons with vesicular stomatitis virus vectors. Proc Natl Acad Sci U S A. 2011;108:15414–15419. doi: 10.1073/pnas.1110854108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zingg B, Chou XL, Zhang ZG, Mesik L, Liang F, Tao HW, Zhang LI. AAV-mediated anterograde transsynaptic tagging: mapping corticocollicular input-defined neural pathways for defense behaviors. Neuron. 2017;93:33–47. doi: 10.1016/j.neuron.2016.11.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jhang J, Lee H, Kang MS, Lee HS, Park H, Han JH. Anterior cingulate cortex and its input to the basolateral amygdala control innate fear response. Nat Commun. 2018;9:2744. doi: 10.1038/s41467-018-05090-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Minorikawa S, Nakayama M. Recombinase-mediated cassette exchange (RMCE) and BAC engineering via VCre/VloxP and SCre/SloxP systems. Biotechniques. 2011;50:235–246. doi: 10.2144/000113649. [DOI] [PubMed] [Google Scholar]
- 10.Fenno LE, Mattis J, Ramakrishnan C, Hyun M, Lee SY, He M, Tucciarone J, Selimbeyoglu A, Berndt A, Grosenick L, Zalocusky KA, Bernstein H, Swanson H, Perry C, Diester I, Boyce FM, Bass CE, Neve R, Huang ZJ, Deisseroth K. Targeting cells with single vectors using multiple-feature Boolean logic. Nat Methods. 2014;11:763–772. doi: 10.1038/nmeth.2996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Petreanu L, Mao T, Sternson SM, Svoboda K. The subcellular organization of neocortical excitatory connections. Nature. 2009;457:1142–1145. doi: 10.1038/nature07709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Aronoff R, Matyas F, Mateo C, Ciron C, Schneider B, Petersen CC. Long-range connectivity of mouse primary somatosensory barrel cortex. Eur J Neurosci. 2010;31:2221–2233. doi: 10.1111/j.1460-9568.2010.07264.x. [DOI] [PubMed] [Google Scholar]
- 13.Veinante P, Deschênes M. Single-cell study of motor cortex projections to the barrel field in rats. J Comp Neurol. 2003;464:98–103. doi: 10.1002/cne.10769. [DOI] [PubMed] [Google Scholar]
- 14.Petersen CC. The functional organization of the barrel cortex. Neuron. 2007;56:339–355. doi: 10.1016/j.neuron.2007.09.017. [DOI] [PubMed] [Google Scholar]
- 15.Jouhanneau JS, Ferrarese L, Estebanez L, Audette NJ, Brecht M, Barth AL, Poulet JF. Cortical fosGFP expression reveals broad receptive field excitatory neurons targeted by POm. Neuron. 2014;84:1065–1078. doi: 10.1016/j.neuron.2014.10.014. [DOI] [PubMed] [Google Scholar]
- 16.Kinnischtzke AK, Simons DJ, Fanselow EE. Motor cortex broadly engages excitatory and inhibitory neurons in somatosensory barrel cortex. Cereb Cortex. 2014;24:2237–2248. doi: 10.1093/cercor/bht085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang W, Bruno RM. High-order thalamic inputs to primary somatosensory cortex are stronger and longer lasting than cortical inputs. Elife. 2019;8:e44158. doi: 10.7554/eLife.44158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pan WX, Mao T, Dudman JT. Inputs to the dorsal striatum of the mouse reflect the parallel circuit architecture of the forebrain. Front Neuroanat. 2010;4:147. doi: 10.3389/fnana.2010.00147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hunnicutt BJ, Long BR, Kusefoglu D, Gertz KJ, Zhong H, Mao T. A comprehensive thalamocortical projection map at the mesoscopic level. Nat Neurosci. 2014;17:1276–1285. doi: 10.1038/nn.3780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hunnicutt BJ, Jongbloets BC, Birdsong WT, Gertz KJ, Zhong H, Mao T. A comprehensive excitatory input map of the striatum reveals novel functional organization. Elife. 2016;5:e19103. doi: 10.7554/eLife.19103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ding J, Peterson JD, Surmeier DJ. Corticostriatal and thalamostriatal synapses have distinctive properties. J Neurosci. 2008;28:6483–6492. doi: 10.1523/JNEUROSCI.0435-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Smith Y, Raju DV, Pare JF, Sidibe M. The thalamostriatal system: a highly specific network of the basal ganglia circuitry. Trends Neurosci. 2004;27:520–527. doi: 10.1016/j.tins.2004.07.004. [DOI] [PubMed] [Google Scholar]
- 23.Carman JB, Cowan WM, Powell TP, Webster KE. A bilateral cortico-striate projection. J Neurol Neurosurg Psychiatry. 1965;28:71–77. doi: 10.1136/jnnp.28.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Donoghue JP, Herkenham M. Neostriatal projections from individual cortical fields conform to histochemically distinct striatal compartments in the rat. Brain Res. 1986;365:397–403. doi: 10.1016/0006-8993(86)91658-6. [DOI] [PubMed] [Google Scholar]
- 25.Warnock JN, Daigre C, Al-Rubeai M. Introduction to viral vectors. Methods Mol Biol. 2011;737:1–25. doi: 10.1007/978-1-61779-095-9_1. [DOI] [PubMed] [Google Scholar]