Abstract
Saccharomyces boulardii is a probiotic yeast often used for the treatment of GI tract disorders such as diarrhea symptoms. It is genetically close to the model yeast Saccharomyces cerevisiae and its classification as a distinct species or a S. cerevisiae variant has long been discussed. Here, we review the main genetic divergencies between S. boulardii and S. cerevisiae as a strategy to uncover the ability to adapt to the host physiological conditions by the probiotic. S. boulardii does possess discernible phenotypic traits and physiological properties that underlie its success as probiotic, such as optimal growth temperature, resistance to the gastric environment and viability at low pH. Its probiotic activity has been elucidated as a conjunction of multiple pathways, ranging from improvement of gut barrier function, pathogen competitive exclusion, production of antimicrobial peptides, immune modulation, and trophic effects. This review summarizes the participation of S. boulardii in these mechanisms and the multifactorial nature by which this yeast modulates the host microbiome and intestinal function.
Keywords: Saccharomyces boulardii, Saccharomyces cerevisiae, probiotics, gastrointestinal tract
1. Introduction
Probiotics are defined as live organisms which, when administered in adequate amounts, confer a health benefit to the host, independently of where the action takes place and of the type of administration. They are normally recommended to help strengthen host systems, for example the gastrointestinal (GI) tract, and assist in the recovery of certain diseases. According to this definition, probiotics in food must contain at least 106 CFU/g of viable and active microorganisms, while freeze-dried supplements have shown good results with 107 to 1011 viable microorganisms per day [1,2,3,4,5]. It is also preferable that these are of human origin and that they cannot transfer any antibiotic resistance, pathogenicity or toxicity factors [4].
The most commonly used probiotics comprise lactic acid producing bacteria (Lactobacillus spp., Bacillus spp., Bifidobacterium spp., Streptococcus spp., and Enterococcus spp.) that are found in the human gastrointestinal tract, usually ingested in fermented foods [4]. These probiotics can be used by themselves or combined with each other, although it should be noted that not all combinations are stable and different strains of the same probiotic bacteria can have different capabilities or enzymatic activities, even if they belong to the same species [4,6]. Probiotic properties widely differ between species, strains or even between strain variants, which means these properties can be strain/variant-specific [4].
The ability of a given organism to display probiotic activity is also dependent on its ability to compete for a host niche. Probiotics must compete with pathogens that adhere specifically to host cells, such as those of the GI tract, including Helicobacter pylori or Clostridium difficile, but also Borellia spp., Treponema spp. or Spirilium spp. [2]. This means that the competition between probiotic microorganisms and pathogens is dependent on habitat-related idiosyncrasies [2]. Host factors can also influence the effectiveness of a probiotic. Genetic factors, baseline immune functions or microbiome diversity vary among individuals, which together with environmental factors (e.g., diet or stress) account for unique backgrounds where the same probiotic will have distinct outcomes [4].
Several bacteria have been identified as probiotics and their modes of action scrutinized to some extent, but yeasts may also exhibit probiotic properties. The baker’s yeast Saccharomyces cerevisiae does not seem to present significant advantageous attributes for human health [1]. On the other hand, the closely related Saccharomyces boulardii is effective in complementing the treatment of acute gastrointestinal diseases such as diarrhea or chronic diseases such as inflammatory bowel disease (IBD) [7,8]. To date, this is the only yeast used as a probiotic [4] and its probiotic properties are supported by scientific evidence from the S. boulardii CNCM I-745 (or S. boulardii Hansen CBS 5926) strain produced by Laboratoires Biocodex, highlighted by more than 80 randomized clinical trials [1]. Nevertheless, the efficacy of this strain cannot be extrapolated to other strains, like S. boulardii CNCM 1079 [1].
In this review, current knowledge on S. boulardii traits that support its probiotic nature and the correlation with distinctive features when compared with the non-probiotic S. cerevisiae will be explored. Focus will be given on reviewing the biology, genetics, ability to colonize the human gut and compete with gastrointestinal pathogens as features that may underlie the probiotic activity of S. boulardii. Unanswered questions, mostly related to the genetic basis underlying the probiotic phenotype, are discussed.
2. S. boulardii and S. cerevisiae: Similar but Different
The budding yeast known as S. boulardii is usually referred to as a distinct species within the Saccharomyces genus, despite being genetically close and sharing a similar karyotype to the model yeast S. cerevisiae [9,10,11]. Molecular typing studies resorting to pulsed-field gel electrophoresis (PFGE), randomly amplified polymorphic DNA-polymerase chain reaction (RAPD-PCR), and restriction fragment length polymorphisms (RFLP) of non-transcribed spacer (NTS) or internal transcribed spacer (ITS) reveal that S. boulardii strains from distinct origins all belong to a clearly delimited cluster within the S. cerevisiae species, arguing that they should be considered different strains of the same species [10,12]. Likewise, a DNA/RNA hybridization spotted microarrays study also concluded that S. boulardii is a strain of S. cerevisiae that has lost all intact Ty1/2 elements rather than a different species [13], while another study identified Ty1/3/4 as absent elements, but not Ty2/5 [11]. Phylogenetic analysis also shows that S. boulardii clusters are closely related to S. cerevisiae wine strains [11]. In spite of such similarities, microsatellite polymorphisms may provide a way to differentiate both species and identify S. boulardii properly [14,15].
Despite the striking relatedness in molecular phylogeny and typing, S. boulardii does possess identifiable distinct traits and is physiologically and metabolically distinct from S. cerevisiae (Table 1). Namely, S. boulardii is incapable of producing ascospores, switching to haploid form, or using galactose as carbon source [11,16,17,18,19]. It is more resistant to temperature and acidic stresses, but less resistant to bile salts [12,18].
Table 1.
Metabolic, physiological and genetic features of S. cerevisiae and S. boulardii. The data shown was collected from several studies [11,12,13,16,17,18,19,20,21].
| Features | S. Cerevisiae | S. Boulardii | |
|---|---|---|---|
| Optimal growth temperature [12] | 30 °C | 37 °C | |
| High temperature resistance (52 °C) [12] | 45% viability | 65% viability | |
| Acid pH resistance (pH = 2 for one hour) [12,18] | No—30% viability | Yes—75% viability | |
| Tolerance to bile acids (>0.3%(w/v)) [12] | No—Survival up to 0.15% (w/v) | No—Survival up to 0.10% (w/v) | |
| Basic pH resistance (pH = 8) [12,18] | Yes | Yes | |
| Assimilation of galactose [16,17,19] | Yes | No | |
| Ploidy [18] | Diploid or haploid | Always diploid | |
| Homo or heterothallic [11] | Homothallic | Homothallic | |
| Mating type [13] | Both | Both | |
| Sporulation [16,18] | Sporogenous | Asporogenous, but produces fertile hybrids with S. cerevisiae | |
| Pseudo-hyphal switching [18] | Normal | Increased | |
| Retrotransposon (Ty elements) [11] | Intact Ty elements | No intact Ty1, 3 or 4 elements | |
| Adhesion to epithelial cells | Normal microbiome (mice and human) [18,20] | No | No |
| Gnotobiotic mice [21] | Unknown | Yes | |
| Humans treated with ampicillin [20] | Unknown | Yes | |
3. S. boulardii Genomic Variations Provide Hints for Its Physiological Properties
S. boulardii and S. cerevisiae genomes were found to differ in internal regions of lower copy number in three chromosomes: chromosome I (PRM9, MST28, YAR047C, YAR050W, CUP1, YAR060W and YAR061W); chromosome VII (YGL052W and MST27) and chromosome XII (ASP3 and YLR156W). PRM9, MST27 and MST28 genes encode nonessential membrane proteins specific to the Saccharomyces sensu stricto species [18]. YAR050W encodes a lectin-like protein that participates in flocculation; Asp3 is a nitrogen catabolite-regulated cell wall L-asparaginase II. CUP1 had a two times lower number of copies than the average for S. cerevisiae species, possibly causing the increased sensitivity to copper in S. boulardii when compared to other S. cerevisiae strains [18].
Within genes with higher copy number, two functions are well represented: protein synthesis (RPL31A, RPL41A, RPS24B, RPL2B and RSA3) and stress response (HSP26, SSA3, SED1, HSP42, HSP78 and PBS2). It is possible that these genes aid in increased growth rate and pseudo-hyphal switching and in higher resistance to high pH [18]. Duplicated and triplicated genes mostly encode stress response proteins, elongation factors, ribosomal proteins, kinases, transporters and fluoride export, which might aid in S. boulardii adaptation to stress conditions [11]. Altered gene copy number and mutations when compared to S. cerevisiae in the SDH1 and WHI2 genes was associated with increased acetic acid production by S. boulardii, correlated with antimicrobial activity [22].
S. boulardii was shown to display enhanced ability for pseudo-hyphal switching during nitrogen starvation compared to other S. cerevisiae strains [18]. Several genes related to pseudo-hyphal growth have considerably different number of copies: CDC42, DFG16, RGS2, CYR1 and CDC25 have higher copy number; STE11, SKM1 and RAS1 have lower copy numbers [18]. Some of these genes are involved in cyclic adenosine monophosphate (cAMP) pathways, suggesting its hyperactivation can lead to increased pseudo-hyphal growth. As a possible consequence, S. boulardii ability to create pseudo-hyphae was observed to be faster and more extensive than S. cerevisiae [18].
Variations in the number of repetitive sequences within flocculation genes was also identified in S. boulardii, namely in FLO1. The encoded flocculin was found to harbor additional copies of residue repeats when compared with most S. cerevisiae strains [11]. The Flo8 protein was also found to differ between S. boulardii and some S. cerevisiae strains where a point mutation results in a truncated protein (including in the reference strain S288c), resulting in defective flocculation and adhesion [23]. Other flocculation genes (FLO10 and FLO11) detected in S. boulardii were not found to harbor differences in the copy number and period length of the repeats [11]. The higher maximum number of repeats (e.g., FLO1) in S. boulardii may affect its adhesion and flocculation ability, as well as sensitivity to stress [11].
Several studies have shown that S. boulardii is unable to use galactose as a carbon source, despite harboring all galactose uptake and fermentation genes [16,17,19]. Some studies have proposed that it is able to assimilate, but not ferment, galactose, possibly due to energy requirements [24,25]. More recently, a mutation in the gene PGM2 was also associated with the inefficient use of galactose [17]. S. boulardii is also unable to use palatinose, possibly related with the absence of 3 isomaltase encoding genes (IMA2, IMA3 and IMA4), which is involved in palatinose uptake and metabolism [11,19].
4. Adaptation to Host Environment
Probiotics must be able to endure in adverse conditions. S. boulardii optimal growth temperature corresponds to the human host temperature (37 °C), while S. cerevisiae grows optimally at 30 °C. S. boulardii is also more resistant to very high temperatures keeping 65% viability after one hour at 52 °C, while S. cerevisiae loses viability down to 45% [12].
The main obstacles in the stomach are the very acidic pH (2 to 3) and the presence of proteases such as pepsin that kill most microorganisms, including probiotics that enter the organism [26]. Diseases like hypochlorhydria decrease the bactericide properties of the stomach and make the patient more susceptible to infections by H. pylori and Salmonella spp. and to migrations of pathogenic microorganisms to the small intestine where they establish themselves [26]. In the case of the small intestine, main stressors include the high concentrations of bile salts, pancreatic enzymes, hydrolytic enzymes, pancreatin, organic acids, the integrity of the epithelial and brush border, the immune defense and the native microbiome and its secondary metabolism products (H2S, bacteriocins, organic acids) [27]. Bile salts are detergents produced in the liver from cholesterol and secreted to the intestine to improve nutrient absorption. As detergent like molecules, bile salts can be toxic to GI tract microorganisms by disrupting their cellular membrane lipid bilayer structures [12]. However, some probiotics are able to resist degradation by hydrolytic enzymes and bile salts [6]. For example, S. boulardii and Bacillus coagulans remain viable after exposure to simulated gastric juice containing pepsin and hydrochloric acid. These probiotics were also seen to be stable to the impact of bile salts [6]. Bacillus clausii was partially resistant to these conditions [6]. On the other hand, most Lactobacillus and Bifidobacterium spp. have reduced viability under exposure to gastrointestinal agents such as pepsin, hydrochloric acid and bile [6].
In vitro testing of probiotic formulations consisting of S. boulardii and bacterial probiotics (Lactobacillus spp. and Bifidobacterium spp.) highlighted the ability of S. boulardii to survive GI tract conditions. S. boulardii was able to survive after incubation in a gastric-like environment and in an intestinal environment (bile salts, pancreatin, pH 7.0) for 3 h, whereas the viability of the bacterial probiotics was severely impaired [6]. S. boulardii is also more resistant to a gastric environment than S. cerevisiae, while the viability of both species in an intestinal environment (sodium chloride, pepsin, pancreatin, pH 8.0) is not affected after 1 h [12]. Accordingly, 1 h was enough to show that S. boulardii is more resistant to low pH than S. cerevisiae, particularly at pH 2.0 [18]. Interestingly, although S. boulardii can survive the GI environments, its viability is significantly increased for 2 h if encapsulated by a double layer with sodium alginate and gelatin [28]. Tolerance displayed by S. boulardii to bile salts has also been tested. Surprisingly, S. cerevisiae is more tolerant to bile salts than S. boulardii. However, after 1 h, both species show a tolerance threshold bellow what would be considered as resistance to bile salts [12].
Dynamic modelling of the stomach and small intestine conditions also showed S. boulardii to be resilient to gastric and lower intestinal conditions, while modelling of the colon environment revealed the yeast is not able to colonize the colon, but had an individual-dependent effect in the microbiotic profile [29]. Other studies also point to the inability of S. boulardii to colonize the gut, suggesting that this yeast does not strongly adhere to intestinal epithelial cells and is quickly removed from the gastrointestinal system in healthy individuals [18]. However, it has been shown to colonize the intestine of gnotobiotic mice after a single administration [21]. This may mean that, although S. boulardii can colonize the gut, competition with intestinal microbiome is limiting [18]. Indeed, both S. boulardii and other Saccharomyces strains were shown to be unable to remain attached to human and mouse epithelial cells, in vitro and in vivo, respectively [18]. However, they do adhere to Caco-2 cells through an extracellular factor, probably secreted mucus [18]. Colonization of the gut was observed to be dependent, both in mice and human, on repeated administration over several days [20,21,30]. Moreover, administration of ampicillin increased S. boulardii cell concentration [20], reinforcing the notion that competition with intestinal microbiome plays a relevant role in the establishment of this yeast.
5. Mechanisms of Action
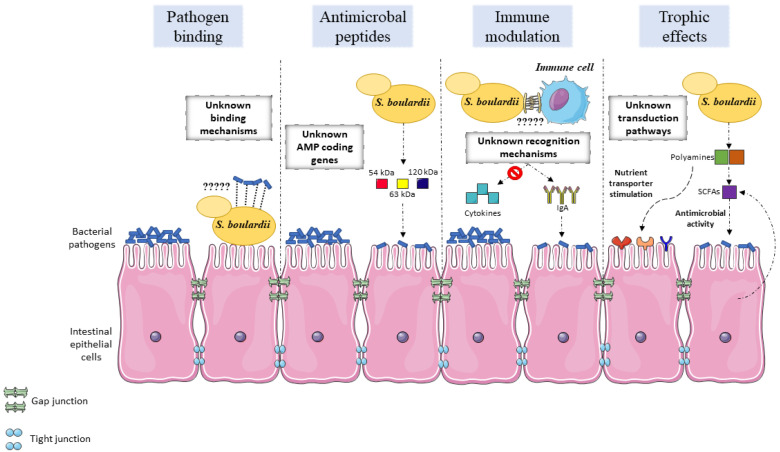
The gut microbiome is responsible for a multitude of roles, including protection against pathogen colonization, epithelial barrier maintenance or modulation of immune activity [31]. The mechanisms by which gut microbiome homeostasis is maintained are not yet fully understood. Probiotics are believed to display a variety of mechanisms: antitoxin effects, physiological protection, modulation of the normal microbiome, metabolic regulation and signaling pathway modification, nutritional and trophic effects, immune system regulation, pathogen competition, interactions with the brain-gut axis, cellular adhesion, cellular antagonism and mucin production [1,4,31]. S. boulardii has been described as participating in a number of these effects as part of its probiotic activity (Figure 1). The genetic basis and mechanistic details that underlie these observations are not fully understood and their clarification could be key to better exploit this yeast and how to potentiate general probiotic activity.
Figure 1.
Overview of the main modes of action that support S. boulardii probiotic activity in the intestinal epithelium. Studies have described the outcome of S. boulardii administration in pathogen exclusion, antimicrobial properties, immune modulation, and trophic effects. The genetic basis and mechanistic details that underlie these observations are not fully understood and their clarification could be key to better exploit this yeast and how to potentiate general probiotic activity. Pathogen exclusion is mainly achieved by pathogen binding to the yeast cells, rather than competition for epithelial binding sites with the pathogens. The yeast cell wall components responsible for the binding, the correspondent pathogenic receptors and the binding dynamics have not been fully investigated. Antimicrobial action is achieved, at least partially, by the secretion of still unknown proteins with antimicrobial effects. The genes that code for these proteins have not been identified and could provide further clues on the mode of action of S. boulardii. Immune modulation and the effect of S. boulardii on inflammatory pathways has been uncovered to some extent. The mechanistic insights and dynamics of S. boulardii interaction with immune cells still need to be ascertained to better understand the yeast action in the immunological function. Multiple trophic effects have been described to be stimulated by S. boulardii on intestinal epithelial cells. Some pathways have been elucidated, although the multitude of trophic effects suggests concerted action and crosstalk between yeast and host cell sensory pathways.
5.1. Modulation of The Normal Microbiome
Modulation of the normal microbiome may be favored directly by transiting probiotics which produce antimicrobial substances, or indirectly contribute to immune modulation or gut barrier function [31]. The use of probiotics has typically been applied to reestablish the normal gut microbiome upon dysbiosis. Gut dysbiosis refers to changes in the microbiome’s quantitative and qualitative composition. These changes may contribute to a disease state frequently associated to inflammation and can be a result of antibiotic-associated diarrhea, acute infectious diarrhea or IBD [31,32]. Probiotics treatment helps to stabilize the gut microbial community and lead to an improved disease outcome [32]. While some probiotics may become a part of the microbiome, others simply pass through the GI tract and modulate or influence the existing microbiome before exiting the body [31].
Several factors can have deleterious effects on the gut microbiome and hinder its protective role to the host epithelial lining, such as antibiotic use or surgery [1]. This may result in host susceptibility to colonization by pathogens until the normal microbiome is reestablished, which can take several weeks [33]. S. boulardii helps to restore the normal microflora in this type of patient and the use of probiotics as modulators of the normal microbiome through colonization during the susceptibility period may work as a surrogate until the normal microbiome is reestablished [34].
5.2. Antimicrobial Activity
Antagonism against pathogens can be achieved by colonization and exclusion of pathogens, modulation of metabolic and signaling pathways, production of inhibitory compounds or immune modulation [31]. Competition is one of the main mechanisms associated with probiotic activity against gut pathogens: consumption of nutrients by probiotics results in nutrient limitation for pathogenic organisms [31,35]. On the other hand, the ability of probiotics to grow and colonize the gut can lead to a decrease of the gut pH due to the production of metabolites, leading to stressful conditions for pathogens [35]. A possible role for S. boulardii in managing pathogenic activity was associated with a protective effect of S. boulardii against pathogenic bacteria in yeast-treated mice, although the mode of action is not associated with a reduction of the pathogenic population [21], as well as with another study which observed a protective effect against Candida albicans in a murine model [36].
The production of compounds with antimicrobial activity is yet another major mode of action of probiotics. Several components of the probiotic metabolome, such as organic acids, bacteriocins, hydrogen peroxide, diacetyl, or amines limit the growth of pathogenic bacteria [31]. In particular, bacteriocins play a crucial role in the antimicrobial action of probiotic bacteria, especially Lactobacillus spp. [35,37,38,39,40]. As for the production of antimicrobial substances by other probiotics, S. boulardii possibly secretes proteins that reduce Citrobacter rodentium adhesion to host epithelial cells by modulating virulence factors [41]. It also displays antimicrobial activity by secreting 54-kDa, 63-kDa and 120-kDa proteins that cleave microbial toxins or reduce cAMP levels. S. boulardii can block toxin receptors or function as a decoy receptor for toxins. The 54-kDa serine protease produced by S. boulardii cleaves toxins A and B from C. difficile and the enterocytic receptor to which the toxins bind, which causes inflammation, fluid secretion, mucosal permeability and injury in the intestines [42,43]. Other mechanisms that S. boulardii uses against C. difficile infection are growth inhibition and decreased toxin production due to secreted factors and stimulation of host mucosal disaccharidase activity [44,45]. Another study refers to the ability of S. boulardii to inhibit Escherichia coli surface endotoxins by dephosphorylation. A 63-kDa alkaline phosphatase targets the lipopolysaccharide (LPS) and contributes to decreased tumor necrosis factor α (TNF-α) cytokine levels [46]. S. boulardii also produces a 120-kDa protein that decreases the chloride secretions stimulated by cholera toxin by reducing cAMP levels [47]. S. boulardii is also able to adhere to cholera toxin via its cell wall, thus blocking its toxic effects [48]. Despite these observations, the sequencing of S. boulardii genomes did not provide a clear identification of the genes encoding these 54-kDa, 63-kDa and 120-kDa proteins [25].
S. boulardii also confers protection against the lethal toxin produced by Bacillus anthracis. The bacterium causes ulcerative lesions from the jejunum to cecum and uses its toxin to disrupt intestinal epithelium integrity, causing mucosal erosion, ulceration and bleeding [49]. The protective effect of S. boulardii is associated with maintenance of barrier function and reduction of harmful physiological responses elicited by the toxin, such as formation of stress fibers [50]. The protective effect is achieved by release of proteases and cleavage of the lethal toxin [50].
Some S. boulardii strains are able to produce high concentrations of acetic acid, which was found to exert an inhibitory effect in E. coli [22]. In turn, the decrease in pH due to acetic acid production is essential for the antimicrobial activity of short-chain organic acids. The combined effect of high acetic acid concentration and lower pH may be an additional mechanism that makes S. boulardii an effective probiotic. Moreover, acetic acid is produced under aerobic conditions by S. boulardii. Due to the radial oxygen gradient between the epithelial surface (high oxygen levels) and the center of the gut lumen (low oxygen levels), microorganisms colonizing the epithelial surface have greater availability of oxygen [51]. Since acetic acid is produced under aerobic conditions by S. boulardii, its production should be higher near the epithelial surface. During antibiotic treatment and pathogen infection, oxygen concentration also increases in the gastrointestinal tract [52,53], which could support the antimicrobial action of S. boulardii.
5.3. Adhesion
In order for the host not to mechanically eliminate the gut microbiome, it is crucial that its components adhere to host surfaces [1,4]. Some probiotics express surface adhesins that mediate the attachment to the mucous layer by recognizing host molecules such as transmembrane proteins (integrins or cadherins) and extracellular matrix components (collagen, fibronectin, laminin or elastin) [1,4]. Probiotics can also influence the production of mucin and the barrier function of the intestine, thus hindering adhesion and consequent invasion of pathogenic microorganisms [54].
Mucin is produced by epithelial cells to avert adhesion by pathogenic bacteria, whereas successful probiotics should be able to adhere to the intestinal mucous, as is the case of S. boulardii [13,55]. The adhesion of S. boulardii to the mucus membrane contributes to reducing the availability of binding sites for pathogens [13]. Five S. cerevisiae cell wall proteins (encoded by CIS3, CWP2, FKS3, PIR3 and SCW4) were found to mediate adhesion of the yeast cells to the pathogenic bacteria E. coli, Salmonella enterica serovar typhimurium (S. typhimurium) and Salmonella enterica serovar typhi (S. typhi) [55]. Other studies have shown that these bacteria are also bound to S. boulardii [55,56,57,58]. Additionally, S. boulardii also inhibits C. difficile adhesion to epithelial cells and displays inhibitory activity on Entamoeba histolytica adhesion to erythrocytes [59,60]. This interaction limits the ability of pathogens to bind directly to the intestinal receptors and proceed with host invasion. In fact, S. boulardii hinders epithelium invasion by S. typhimurium due to steric hindrance caused by its larger size as compared to bacteria [61]. As S. boulardii does not significantly bind to epithelial cells of healthy individuals and is quickly flushed out, pathogens bound to S. boulardii are possibly flushed together with the yeast cells [13,31,55]. However, S. boulardii does have several flocculation genes required for protection against environmental stress and biofilm formation [11]. The characterization of this gene family in the context of host adhesion and colonization could provide further insight on the probiotic features of S. boulardii.
The ability of S. boulardii to bind bacterial pathogens has been associated with the presence of mannose residues in the yeast cell wall [56]. This is a similar mechanism to the previously characterized adhesion of bacterial pathogens to the epithelial surface via mannose residues [62], which is the basis for the addition of exogenous sugars as a strategy to inhibit pathogen adhesion [63]. Cell wall mannan oligosaccharides are a common feature in yeast, but the affinity between E. coli and S. boulardii is higher than with S. cerevisiae [56]. Further investigations revealed that bile salts decrease adhesion of bacteria to yeast cells [55], which can have relevant implications for yeasts as successful probiotics. Accordingly, bile salts also decrease the adhesion of probiotic bacteria to intestine epithelia due to diminished surface hydrophobicity and higher surface potential [64], bolstering how important it is for probiotic microorganisms to evolve adaptation strategies within the host.
5.4. Immune Modulation
Metabolites produced by the gut microbiome can perform immunomodulatory and anti-inflammatory functions that stimulate immune cells. This ability arises from the interaction between the probiotics and the epithelial cells, dendritic cell monocytes, macrophages and/or lymphocytes [1,31]. Probiotics also promote enhanced phagocytic activity, cell proliferation and production of secretory immunoglobulins IgA and IgM [65].
S. boulardii can modulate immunological function by acting as a stimulant or a pro-inflammatory inhibitor. It is capable of modulating the inflammatory process upon S. typhimurium infection by decreasing the levels of the pro-inflammatory molecules such as cytokine interleukin 8 (IL-8), mitogen activated protein (MAP) kinases and the (nuclear factor kappa B) NF-κB signaling pathway [58]. An inhibitory effect of S. boulardii over MAP kinases and IL-8 levels upon C. difficile infection was also observed [66]. Likewise, S. boulardii contributes to increasing the levels of anti-inflammatory cytokines (IL-4 and IL-10) and decreasing pro-inflammatory cytokines (IL-1β) upon infection with E. coli and C. albicans [67]. On the other hand, S. boulardii was associated with increased IgA and IgG levels in serum in response to C. difficile toxins A and B [68,69]. S. boulardii was also found to attach to the surface of dendritic cells [70] and modulate the expression of toll-like receptors (TLRs) and cytokines [70,71,72]. Moreover, S. boulardii also causes the imprisonment of T helper cells in mesenteric lymphatic nodes, reducing inflammation [73].
Another study found that in the early phase of S. typhimurium infection, S. boulardii induces pro-inflammatory cytokine production (interferon-γ—IFN-γ) and represses the production of anti-inflammatory cytokines (IL-10) in the small intestine, but increases the levels of both cytokines in the cecum [57]. This suggests that S. boulardii can differentially modulate immune activity through the GI tract [57]. Overall, probiotics may be able to persistently modulate both the innate and adaptive immune responses either locally or systemically [1,31]. The data from several studies indicates that S. boulardii plays a pivotal role in immune modulation against the most common GI tract pathogens.
5.5. Trophic Effects
S. boulardii is a modulator of enzyme activity required to maintain a healthy gastrointestinal tract. It exerts trophic effects such as stimulation of brush border membrane digestive enzymes and nutrient transporter activity [74]. Several studies have shown a wide array of trophic effects stimulated by S. boulardii: brush border sucrase, lactase, and maltase activities [44,75,76,77,78]; iso-maltase activity [78]; glucoamylase and N-aminopeptidase activity [76]; leucine-aminopeptidase activity [79]; α,α-trehalase activities in the endoluminal fluid and intestinal mucosa; brush border α-glucosidase [80]; spermine, spermidine and putrescine levels in rat jejunal mucosa [75,77]; adenosine triphosphatase, γ-glutamyl transpeptidase, lipase, and trypsin activities and TNF-α, IL-10, transforming growth factor beta (TGF-β), and secretory IgA [5]; diamine oxidase activities, brush border sodium/glucose cotransporter 1 expression and sodium-dependent glucose uptake [74,77]; GRB2-SHC-CrkII-Ras-GAP-Raf-ERK1,2 transduction pathway in rats and decreased p38 MAPK and NF-κB [81,82,83].
Probiotics can modulate short chain fatty acids (SCFA: acetate, propionate, or butyrate) and/or branched-chain fatty acid (BCFA: iso-butyrate, 2-methylbutyrate, or isovalerate) synthesis. SCFAs have a complex role in the physiological and biochemical functions in different tissues (intestine, liver, adipose, muscle and brain). S. boulardii assists in reestablishing SCFA levels, which are depressed during disease [84,85]. Acetate and butyrate are major SCFAs in intestinal epithelial cells, playing a role in barrier function, anti-inflammatory and immune modulation pathways [86,87]. A study reported that a short-term treatment (6 days) with S. boulardii diminishes the incidence of diarrhea in patients receiving enteral nutrition by increasing SCFA levels, particularly butyrate [84]. SCFAs can also present antimicrobial activity, and a study probing several S. boulardii and S. cerevisiae strains for inhibitory effects in E. coli described the production of acetic acid exclusively by S. boulardii as an antimicrobial mechanism [22]. Moreover, acetate also stimulates T regulatory cells, induces mucus secretion gene expression, inhibits proinflammatory cytokine CXCL8 and serves as a substrate for the production of butyrate by the microbiome [22].
The activity of many digestive enzymes (sucrase-iso-maltase, maltase-glucoamylase, lactase-phlorizin hydrolase, alanine aminopeptidase and alkaline phosphatase) and nutrient transporters (sodium-glucose transport proteins) may be induced by polyamines secreted by S. boulardii [74]. S. boulardii secretes polyamines that promote RNA binding and stabilization and, hence, growth and protein (lactase, maltase, sucrase, among others) synthesis [74]. These molecules are also able to shield lipids from oxidation and boost SCFA activity. Polyamines may also affect kinase activities and external signal transduction pathways, therefore modulating the GRB2-SHC-CrkII-Ras-GAP-Raf-ERK1,2 and the PI3K pathways [74]. They can also aid the generation of specific transcripts by interacting with DNA [74]. All of these polyamine functions lead to a general polyamine-triggered metabolic activation in order to regenerate brush border damage and maturation of enterocytes [74,75,80]. Not only does S. boulardii induce the enzymatic activities of lactase-phlorizin hydrolase, α-glucosidases, alkaline phosphatases and aminopeptidases, but it also increases glucose intestinal absorption, one of the products of lactose degradation [74]. Production of lactase by the host, partially stimulated by S. boulardii, mediates lactose degradation thus alleviating lactose intolerance.
6. S. boulardii Safety and Clinical Efficacy
Although many probiotics are documented as safe, common safety issues regarding the use of probiotics include: transfer of antibiotic resistance genes, translocation of live organisms from the intestine to other sites of the body, persistence in the intestine and development of adverse reactions [1]. Most of these concerns have been dismissed when evaluating S. boulardii safety. S. boulardii is not known to acquire resistance genes, unlike bacterial probiotics such as Lactobacillus spp. [88,89]. Animal studies show that there is reduced translocation in the treatment with S. boulardii when compared with S. cerevisiae [90,91,92]. S. boulardii does not persist in the intestine after three to five days after discontinuation of the ingestion, according to pharmacokinetic studies [20]. The data available from 90 clinical trials assessing the efficacy and safety of S. boulardii has been thoroughly assessed elsewhere [1]. Randomized and controlled trials clearly show the absence of any serious adverse reactions, while only some presented moderate adverse reactions, such as constipation in patients with C. difficile infection [93]. Although fungemia is viewed as a potential problem, there were no fungemia cases reported in clinical trials [1]. S. boulardii-associated fungemia was observed in patients with serious co-morbidity factors and central venous catheters, which responded well to fluconazole or amphotericin B therapy [91,94,95,96]. Importantly, S. cerevisiae-associated fungemia has a worse prognosis than that caused by S. boulardii [97].
Clinical trials have investigated the efficacy of S. boulardii in the improvement of several GI conditions’ outcome. This yeast was seen to improve the outcome of several diarrhea diseases, including pediatric diarrhea, antibiotic-associated diarrhea, acute diarrhea, traveler’s diarrhea caused by bacterial, viral or parasites, and enteral nutrition-related diarrhea [1,15,98]. S. boulardii also improves the outcome in patients suffering from H. pylori or C. difficile infections by helping bacteria eradication, preventing relapses, reducing adverse reactions, and reducing treatment-associated diarrhea [1,15,98]. IBD is a prevalent GI tract disorder associated with inflammatory diarrheal diseases such as ulcerative colitis, pouchitis and Crohn’s disease [1]. Clinical trial data points to a possible role of S. boulardii in reducing treatment relapses [1,15,98], which are frequent in these conditions, although further studies are required to reach compelling conclusions. Irritable Bowel Syndrome (IBS) symptoms also improve with S. boulardii administration. It is a condition frequently characterized by abdominal bloating, abdominal pain, and disturbed intestinal transit. These symptoms were shown to be alleviated in 50% of patients upon S. boulardii use [99].
7. Conclusions
S. boulardii is a probiotic yeast with proven efficacy in the treatment of GI conditions, especially when used as an adjuvant to antibiotic treatment. Present data indicate that the benefits of S. boulardii appear to be transient and independent of host gut colonization, differentiating its mode of action from other widely used bacterial probiotics. The absence of colonization appears to correlate with pathogen binding as a mechanism to halt pathogen colonization, rather than competitive exclusion due to yeast adhesion. Genomics studies have contributed to pinpoint distinct genome features that mainly confer on S. boulardii the ability to resist host stresses, conferring higher viability through GI passage than observed for other common probiotics. S. boulardii also elicits a complex immunomodulatory effect with roles in fine-tuning immunological pathways during pathogen infection or chronic diseases. This yeast also contributes to the homeostasis of the normal microbiome and plays a relevant role in modulating secretory functions by intestinal epithelial cells, thus benefitting nutritional requirements of the host. Overall, S. boulardii displays a multifactorial role as a probiotic, with proven efficacy and safety in alleviating the symptomology of a number of GI conditions. However, there is a significant knowledge gap between S. boulardii phenotypic effects and the underlying genetic basis, especially when compared to S. cerevisiae. What are the 54-kDa, 63-kDA and 120-kDA proteins secreted by S. boulardii that cleave microbial toxins or reduce cAMP levels? What are the proteins responsible for higher adhesion of S. boulardii to pathogenic bacteria, when compared to S. cerevisiae, especially considering the differences in flocculin encoding genes? What are the mechanisms that allow S. boulardii to overcome the negative impact of bile salts during host adaptation? What are the proteins or cellular components that mediate immune recognition of S. boulardii and modulation of the immune response? These questions remain unanswered. Further research on the genetic basis of S. boulardii probiotic activity will certainly increase our understanding of this fascinating yeast, while providing important clues for the selection and optimization of even more powerful probiotic fungi.
Author Contributions
P.P. and M.C.T. delineated manuscript organization. P.P., with contributions from V.A. and M.Y., wrote the manuscript, under the coordination of M.C.T. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by “Fundação para a Ciência e a Tecnologia” (FCT) [Contract PTDC/BII-BIO/28216/2017], as well as by Programa Operacional Regional de Lisboa 2020 [LISBOA-01-0145-FEDER-022231—The BioData.pt Research Infrastructure]. Funding received by iBB from FCT (UIDB/04565/2020), and from Programa Operacional Regional de Lisboa 2020 (LISBOA-01-0145-FEDER-007317).
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
References
- 1.McFarland L.V. The Microbiota in Gastrointestinal Pathophysiology. Academic Press; Cambridge, MA, USA: 2017. Common Organisms and Probiotics: Saccharomyces boulardii; pp. 145–164. [Google Scholar]
- 2.Schrezenmeir J., de Vrese M. Probiotics, prebiotics, and synbiotics—approaching a definition. Am. J. Clin. Nutr. 2001;73:361s–364s. doi: 10.1093/ajcn/73.2.361s. [DOI] [PubMed] [Google Scholar]
- 3.Moradi R., Nosrati R., Zare H., Tahmasebi T., Saderi H., Owlia P. Screening and characterization of in-vitro probiotic criteria of saccharomyces and kluyveromyces strains. Iran. J. Microbiol. 2018;10:123–131. [PMC free article] [PubMed] [Google Scholar]
- 4.Plaza-Diaz J., Ruiz-Ojeda F.J., Gil-Campos M., Gil A. Mechanisms of Action of Probiotics. Adv. Nutr. 2019;10:S49–S66. doi: 10.1093/advances/nmy063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sun Y., Rajput I.R., Arain M.A., Li Y., Baloch D.M. Oral administration of Saccharomyces boulardii alters duodenal morphology, enzymatic activity and cytokine production response in broiler chickens. Anim. Sci. J. 2017;88:1204–1211. doi: 10.1111/asj.12757. [DOI] [PubMed] [Google Scholar]
- 6.Kabluchko T.V., Bomko T.V., Nosalskaya T.N., Martynov A.V., Osolodchenko T.P. In the gastrointestinal tract exist the protective mechanisms which prevent overgrowth of pathogenic bacterial and its incorporation. Ann. Mechnikov Inst. 2017;1:28–33. [Google Scholar]
- 7.Kelesidis T., Pothoulakis C. Efficacy and safety of the probiotic Saccharomyces boulardii for the prevention and therapy of gastrointestinal disorders. Therap. Adv. Gastroenterol. 2012;5:111–125. doi: 10.1177/1756283X11428502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sen S., Mansell T.J. Yeasts as probiotics: Mechanisms, outcomes, and future potential. Fungal Genet. Biol. 2020;137:103333. doi: 10.1016/j.fgb.2020.103333. [DOI] [PubMed] [Google Scholar]
- 9.Van Der Aa Kühle A., Jespersen L. The Taxonomic Position of Saccharomyces boulardii as Evaluated by Sequence Analysis of the D1/D2 Domain of 26S rDNA, the ITS1-5.8S rDNA-ITS2 Region and the Mitochondrial Cytochrome-c Oxidase II Gene. Syst. Appl. Microbiol. 2003;26:564–571. doi: 10.1078/072320203770865873. [DOI] [PubMed] [Google Scholar]
- 10.Mitterdorfer G., Mayer H.K., Kneifel W., Viernstein H. Clustering of Saccharomyces boulardii strains within the species S. cerevisiae using molecular typing techniques. J. Appl. Microbiol. 2002;93:521–530. doi: 10.1046/j.1365-2672.2002.01710.x. [DOI] [PubMed] [Google Scholar]
- 11.Khatri I., Tomar R., Ganesan K., Prasad G.S., Subramanian S. Complete genome sequence and comparative genomics of the probiotic yeast Saccharomyces boulardii. Sci. Rep. 2017;7:1–13. doi: 10.1038/s41598-017-00414-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fietto J.L., Araújo R.S., Valadão F.N., Fietto L.G., Brandão R.L., Neves M.J., Gomes F.C., Nicoli J.R., Castro I.M. Molecular and physiological comparisons between Saccharomyces cerevisiae and Saccharomyces boulardii. Can. J. Microbiol. 2004;50:615–621. doi: 10.1139/w04-050. [DOI] [PubMed] [Google Scholar]
- 13.Edwards-ingram L.C., Gent M.E., Hoyle D.C., Hayes A., Stateva L.I., Oliver S.G. Comparative Genomic Hybridization Provides New Insights Into the Molecular Taxonomy of the Saccharomyces Sensu Stricto Complex. Genome Res. 2004;14:1043–1051. doi: 10.1101/gr.2114704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hennequin C., Thierry A., Richard G.F., Lecointre G., Nguyen H.V., Gaillardin C., Dujon B. Microsatellite Typing as a New Tool for Identification of Saccharomyces cerevisiae Strains. J. Clin. Microbiol. 2001;39:551–559. doi: 10.1128/JCM.39.2.551-559.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McFarland L.V. Systematic review and meta-analysis of saccharomyces boulardii in adult patients. World J. Gastroenterol. 2010;16:2202–2222. doi: 10.3748/wjg.v16.i18.2202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McFarland L.V. Saccharomyces boulardii Is Not Saccharomyces cerevisiae. Clin. Infect. Dis. 1996;22:200–201. doi: 10.1093/clinids/22.1.200. [DOI] [PubMed] [Google Scholar]
- 17.Liu J.J., Zhang G.C., Kong I.I., Yun E.J., Zheng J.Q., Kweon D.H., Jin Y.S. A mutation in PGM2 causing inefficient galactose metabolism in the probiotic yeast Saccharomyces boulardii. Appl. Environ. Microbiol. 2018;84:e02858-17. doi: 10.1128/AEM.02858-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Edwards-ingram L., Gitsham P., Burton N., Warhurst G., Clarke I., Hoyle D., Oliver S.G., Stateva L. Genotypic and Physiological Characterization of Saccharomyces boulardii, the Probiotic Strain of Saccharomyces cerevisiae. Appl. Environ. Microbiol. 2007;73:2458–2467. doi: 10.1128/AEM.02201-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.GMitterdorfer; WKneifel; HViernstein Utilization of prebiotic carbohydrates by yeasts of therapeutic relevance. Lett. Appl. Microbiol. 2001;34:251–255. doi: 10.1046/j.1472-765x.2001.00991.x. [DOI] [PubMed] [Google Scholar]
- 20.Klein S.M., Elmer G.W., McFarland L.V., Surawicz C.M., Levy R.H. Recovery and Elimination of the Biotherapeutic Agent, Saccharomyces boulardii, in Healthy Human Volunteers. Pharm. Res. An Off. J. Am. Assoc. Pharm. Sci. 1993;10:1615–1619. doi: 10.1023/a:1018924820333. [DOI] [PubMed] [Google Scholar]
- 21.Rodrigues A.C.P., Mardi R.M., Bambirra E.A., Vieira E.G., Nicoli U.R. Effect of Saccharomyces boulardii against experimental oral infection with Salmonella typhimurium and Shigella flexnerim conventional and gnotobiotic mice. J. Appl. Bacteriol. 1996;81:251–256. doi: 10.1111/j.1365-2672.1996.tb04325.x. [DOI] [PubMed] [Google Scholar]
- 22.Offei B., Vandecruys P., De Graeve S., Foulquié-moreno M.R., Thevelein J.M. Unique genetic basis of the distinct antibiotic potency of high acetic acid production in the probiotic yeast Saccharomyces cerevisiae var. boulardii. Genome Res. 2019:1478–1494. doi: 10.1101/gr.243147.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu H., Styles C.A., Fink G.R. Saccharomyces cerevisiae S288C has a mutation in FLO8, a gene required for filamentous growth. Genetics. 1996;144:967–978. doi: 10.1093/genetics/144.3.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.van den Brink J., Akeroyd M., van der Hoeven R., Ponk J.T., de Winde J.H., Daran-Lapujade P.A.S. Energetic limits to metabolic flexibility: Responses of Saccharomyces cerevisiae to glucose-galactose transitions. Microbiology. 2009;155:1340–1350. doi: 10.1099/mic.0.025775-0. [DOI] [PubMed] [Google Scholar]
- 25.Khatri I., Akhtar A., Kaur K., Tomar R., Prasad G.S. Gleaning evolutionary insights from the genome sequence of a probiotic yeast Saccharomyces boulardii. Gut Pathog. 2013;5:30. doi: 10.1186/1757-4749-5-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Martinsen T.C., Bergh K., Waldum H.L. Gastric Juice: A Barrier Against Infectious Diseases. Basic Clin. Pharmacol. Toxicol. 2005;96:94–102. doi: 10.1111/j.1742-7843.2005.pto960202.x. [DOI] [PubMed] [Google Scholar]
- 27.Holzapfel W.H., Haberer P., Snel J., Schillinger U., Huis In’T Veld J.H.J. Overview of gut flora and probiotics. Int. J. Food Microbiol. 1998;41:85–101. doi: 10.1016/S0168-1605(98)00044-0. [DOI] [PubMed] [Google Scholar]
- 28.Du Le H., Trinh K.S. Survivability of Lactobacillus acidophilus, Bacillus clausii and Saccharomyces boulardii encapsulated in alginate gel microbeads. Carpathian J. Food Sci. Technol. 2018;10:95–103. [Google Scholar]
- 29.Cordonnier C., Thévenot J., Etienne-Mesmin L., Denis S., Alric M., Livrelli V., Blanquet-Diot S. Dynamic In Vitro Models of the Human Gastrointestinal Tract as Relevant Tools to Assess the Survival of Probiotic Strains and Their Interactions with Gut Microbiota. Microorganisms. 2015;3:725–745. doi: 10.3390/microorganisms3040725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Filho-Lima J.V.M., Vieira E.C., Nicoli J.R. Antagonistic effect of Lactobacillus acidophilus, Saccharomyces boulardii and Escherichia coli combinations against experimental infections with Shigella flexneri and Salmonella enteritidis subsp. typhimurium in gnotobiotic mice. J. Appl. Microbiol. 2000;88:365–370. doi: 10.1046/j.1365-2672.2000.00973.x. [DOI] [PubMed] [Google Scholar]
- 31.Bajaj B.K., Claes I.J.J., Lebeer S. Functional mechanisms of probiotics. J. Microbiol. Biotechnol. Food Sci. 2015;4:321–327. doi: 10.15414/jmbfs.2015.4.4.321-327. [DOI] [Google Scholar]
- 32.Ceapa C., Wopereis H., Rezaïki L., Kleerebezem M., Knol J., Oozeer R. Influence of fermented milk products, prebiotics and probiotics on microbiota composition and health. Best Pract. Res. Clin. Gastroenterol. 2013;27:139–155. doi: 10.1016/j.bpg.2013.04.004. [DOI] [PubMed] [Google Scholar]
- 33.Dethlefsen L., Huse S., Sogin M.L., Relman D.A. The Pervasive Effects of an Antibiotic on the Human Gut Microbiota, as Revealed by Deep 16S rRNA Sequencing. PLoS Biol. 2008;6:e280. doi: 10.1371/journal.pbio.0060280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McFarland L.V. Use of probiotics to correct dysbiosis of normal microbiota following disease or disruptive events: A systematic review. BMJ Open. 2014;4:e005047. doi: 10.1136/bmjopen-2014-005047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bermudez-Brito M., Plaza-Díaz J., Muñoz-Quezada S., Gómez-Llorente C., Gil A. Probiotic Mechanisms of Action. Ann. Nutr. Metab. 2012;61:160–174. doi: 10.1159/000342079. [DOI] [PubMed] [Google Scholar]
- 36.Ducluzeau R., Bensaada M. Comparative effect of a single or continuous administration of “Saccharomyces boulardii” on the establishment of various strains of “candida” in the digestive tract of gnotobiotic mice. Ann. Microbiol. (Paris) 1982;133:491–501. [PubMed] [Google Scholar]
- 37.de Arauz L.J., Jozala A.F., Mazzola P.G., Vessoni Penna T.C. Nisin biotechnological production and application: A review. Trends Food Sci. Technol. 2009;20:146–154. doi: 10.1016/j.tifs.2009.01.056. [DOI] [Google Scholar]
- 38.Vilagravel B., Esteve-Garcia E., Brufau J. Probiotic micro-organisms: 100 years of innovation and efficacy; Modes of action. Worlds. Poult. Sci. J. 2010;66:369–380. [Google Scholar]
- 39.Corr S.C., Li Y., Riedel C.U., O’Toole P.W., Hill C., Gahan C.G.M. Bacteriocin production as a mechanism for the antiinfective activity of Lactobacillus salivarius UCC118. Proc. Natl. Acad. Sci. USA. 2007;104:7617–7621. doi: 10.1073/pnas.0700440104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nielsen D.S., Cho G.S., Hanak A., Huch M., Franz C.M.A.P., Arneborg N. The effect of bacteriocin-producing Lactobacillus plantarum strains on the intracellular pH of sessile and planktonic Listeria monocytogenes single cells. Int. J. Food Microbiol. 2010;141:S53–S59. doi: 10.1016/j.ijfoodmicro.2010.03.040. [DOI] [PubMed] [Google Scholar]
- 41.Wu X., Vallance B.A., Boyer L., Bergstrom K.S.B., Walker J., Madsen K., O’Kusky J.R., Buchan A.M., Jacobson K. Saccharomyces boulardii ameliorates Citrobacter rodentium-induced colitis through actions on bacterial virulence factors. Am. J. Physiol. Gastrointest. Liver Physiol. 2007;294:G295–G306. doi: 10.1152/ajpgi.00173.2007. [DOI] [PubMed] [Google Scholar]
- 42.Castagliuolo I., Riegler M.F., Valenick L., LaMont J.T., Pothoulakis C. Saccharomyces boulardii protease inhibits the effects of Clostridium difficile toxins A and B in human colonic mucosa. Infect. Immun. 1999;67:302–307. doi: 10.1128/IAI.67.1.302-307.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Castagliuolo I., Thomas Lamont J., Nikulasson S.T., Pothoulakis C. Saccharomyces boulardii protease inhibits Clostridium difficile toxin A effects in the rat ileum. Infect. Immun. 1996;64:5225–5232. doi: 10.1128/IAI.64.12.5225-5232.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Buts J.P., Bernasconi P., Van Craynest M.P., Maldague P., Meyer R. Response of human and rat small intestinal mucosa to oral administration of saccharomyces boulardii. Pediatr. Res. 1986;20:192–196. doi: 10.1203/00006450-198602000-00020. [DOI] [PubMed] [Google Scholar]
- 45.Buts J.P., Bernasconi P., Vaerman J.P., Dive C. Stimulation of secretory IgA and secretory component of immunoglobulins in small intestine of rats treated with Saccharomyces boulardii. Dig. Dis. Sci. 1990;35:251–256. doi: 10.1007/BF01536771. [DOI] [PubMed] [Google Scholar]
- 46.Buts J.P., Dekeyser N., Stilmant C., Delem E., Smets F., Sokal E. Saccharomyces boulardii produces in rat small intestine a novel protein phosphatase that inhibits Escherichia coli endotoxin by dephosphorylation. Pediatr. Res. 2006;60:24–29. doi: 10.1203/01.pdr.0000220322.31940.29. [DOI] [PubMed] [Google Scholar]
- 47.Czerucka D., Roux I., Rampal P. Saccharomyces boulardii inhibits secretagogue-mediated adenosine 3′,5′-cyclic monophosphate induction in intestinal cells. Gastroenterology. 1994;106:65–72. doi: 10.1016/S0016-5085(94)94403-2. [DOI] [PubMed] [Google Scholar]
- 48.Brandão R.L., Castro I.M., Bambirra E.A., Amaral S.C., Fietto L.G., Tropia M.J.M., Neves M.J., Dos Santos R.G., Gomes N.C.M., Nicoli J.R. Intracellular signal triggered by cholera toxin in Saccharomyces boulardii and Saccharomyces cerevisiae. Appl. Environ. Microbiol. 1998;64:564–568. doi: 10.1128/AEM.64.2.564-568.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sweeney D.A., Hicks C.W., Cui X., Li Y., Eichacker P.Q. Anthrax infection. Am. J. Respir. Crit. Care Med. 2011;184:1333–1341. doi: 10.1164/rccm.201102-0209CI. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pontier-bres R., Rampal P., Peyron J., Munro P., Lemichez E., Czerucka D. The Saccharomyces boulardii CNCM I-745 Strain Shows Protective Effects against the B. anthracis LT Toxin. Toxin. 2015;7:4455–4467. doi: 10.3390/toxins7114455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Albenberg L., Esipova T.V., Judge C.P., Bittinger K., Chen J., Laughlin A., Grunberg S., Baldassano R.N., Lewis J.D., Li H., et al. Correlation between intraluminal oxygen gradient and radial partitioning of intestinal microbiota. Gastroenterology. 2014;147:1055–1063. doi: 10.1053/j.gastro.2014.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vacca I. Microbiome: The microbiota maintains oxygen balance in the gut. Nat. Rev. Microbiol. 2017;15:574. doi: 10.1038/nrmicro.2017.112. [DOI] [PubMed] [Google Scholar]
- 53.Rivera-Chávez F., Lopez C.A., Bäumler A.J. Oxygen as a driver of gut dysbiosis. Free Radic. Biol. Med. 2017;105:93–101. doi: 10.1016/j.freeradbiomed.2016.09.022. [DOI] [PubMed] [Google Scholar]
- 54.Gogineni V.K., Morrow L.E. Probiotics: Mechanisms of Action and Clinical Applications. J. Probiotics Heal. 2013;1:2. doi: 10.4172/2329-8901.1000101. [DOI] [Google Scholar]
- 55.Tiago F.C.P., Martins F.S., Souza E.L.S., Pimenta P.F.P., Araujo H.R.C., Castro I.M., Branda R.L., Nicoli J.R., Nicoli J.R. Adhesion to the yeast cell surface as a mechanism for trapping pathogenic bacteria by Saccharomyces probiotics. J. Med Microbiol. 2012;61:1194–1207. doi: 10.1099/jmm.0.042283-0. [DOI] [PubMed] [Google Scholar]
- 56.Gedek B.R. Adherence of Escherichia coli serogroup 0 157 and the Salmonella Typhimurium mutant DT 104 to the surface of Saccharomyces boulardii. Mycoses. 1999;42:261–264. doi: 10.1046/j.1439-0507.1999.00449.x. [DOI] [PubMed] [Google Scholar]
- 57.Pontier-bres R., Munro P., Boyer L., Anty R., Rampal P., Lemichez E. Saccharomyces boulardii Modifies Salmonella Typhimurium Traffic and Host Immune Responses along the Intestinal Tract. PLoS ONE. 2014;9:e103069. doi: 10.1371/journal.pone.0103069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Martins F.S., Dalmasso G., Arantes R.M.E., Doye A., Lemichez E., Lagadec P., Imbert V., Peyron J.F., Rampal P., Nicoli J.R., et al. Interaction of Saccharomyces boulardii with Salmonella enterica serovar typhimurium protects mice and modifies T84 cell response to the infection. PLoS ONE. 2010;5:e8925. doi: 10.1371/journal.pone.0008925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tasteyre A., Barc M.C., Karjalainen T., Bourlioux P., Collignon A. Inhibition of in vitro cell adherence of Clostridium difficile by Saccharomyces boulardii. Microb. Pathog. 2002;32:219–225. doi: 10.1006/mpat.2002.0495. [DOI] [PubMed] [Google Scholar]
- 60.Rigothier M.C., Maccario J., Gayral P. Inhibitory activity of saccharomyces yeasts on the adhesion of Entamoeba histolytica trophozoites to human erythrocytes in vitro. Parasitol. Res. 1994;80:10–15. doi: 10.1007/BF00932617. [DOI] [PubMed] [Google Scholar]
- 61.Pontier-Bres R., Prodon F., Munro P., Rampal P., Lemichez E., Peyron J.F., Czerucka D. Modification of salmonella typhimurium motility by the probiotic yeast strain saccharomyces boulardii. PLoS ONE. 2012;7:e33796. doi: 10.1371/journal.pone.0033796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kline K.A., Fälker S., Dahlberg S., Normark S., Henriques-Normark B. Bacterial Adhesins in Host-Microbe Interactions. Cell Host Microbe. 2009;5:580–592. doi: 10.1016/j.chom.2009.05.011. [DOI] [PubMed] [Google Scholar]
- 63.Cusumano C.K., Hultgren S.J. Bacterial adhesion—A source of alternate antibiotic targets. IDrugs. 2009;12:699–705. [PubMed] [Google Scholar]
- 64.Gómez Zavaglia A., Kociubinski G., Pérez P., Disalvo E., De Antoni G. Effect of bile on the lipid composition and surface properties of bifidobacteria. J. Appl. Microbiol. 2002;93:794–799. doi: 10.1046/j.1365-2672.2002.01747.x. [DOI] [PubMed] [Google Scholar]
- 65.Kaur I.P., Kuhad A., Garg A., Chopra K. Probiotics: Delineation of Prophylactic and Therapeutic Benefits. J. Med. Food. 2009;12:219–235. doi: 10.1089/jmf.2007.0544. [DOI] [PubMed] [Google Scholar]
- 66.Chen K.-H., Miyazaki T., Tsai H.-F., Bennett J.E. The bZip transcription factor Cgap1p is involved in multidrug resistance and required for activation of multidrug transporter gene CgFLR1 in Candida glabrata. Gene. 2007;386:63–72. doi: 10.1016/j.gene.2006.08.010. [DOI] [PubMed] [Google Scholar]
- 67.Fidan I., Kalkanci A., Yesilyurt E., Yalcin B., Erdal B., Kustimur S., Imir T. Effects of Saccharomyces boulardii on cytokine secretion from intraepithelial lymphocytes infected by Escherichia coli and Candida albicans. Mycoses. 2009;52:29–34. doi: 10.1111/j.1439-0507.2008.01545.x. [DOI] [PubMed] [Google Scholar]
- 68.Qamar A., Aboudola S., Warny M., Michetti P., Kelly N.P., Division G., Israel B., Medical D. Saccharomyces boulardii Stimulates Intestinal Immunoglobulin A Immune Response to Clostridium difficile Toxin A in Mice. Infect. Immun. 2001;69:2762–2765. doi: 10.1128/IAI.69.4.2762-2765.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kyne L., Warny M., Qamar A., Kelly C.P. Association between antibody response to toxin A and protection against recurrent Clostridium difficile diarrhoea. Lancet. 2001;357:189–193. doi: 10.1016/S0140-6736(00)03592-3. [DOI] [PubMed] [Google Scholar]
- 70.Rajput I.R., Hussain A., Li Y.L., Zhang X., Xu X., Long M.Y., You D.Y., Li W.F. Saccharomyces boulardii and Bacillus subtilis B10 Modulate TLRs Mediated Signaling to Induce Immunity by Chicken BMDCs. J. Cell. Biochem. 2014;115:189–198. doi: 10.1002/jcb.24650. [DOI] [PubMed] [Google Scholar]
- 71.Badia R., Zanello G., Chevaleyre C., Lizardo R., Meurens F., Martínez P., Brufau J., Salmon H. Effect of Saccharomyces cerevisiae var. Boulardii and β-galactomannan oligosaccharide on porcine intestinal epithelial and dendritic cells challenged in vitro with Escherichia coli F4 (K88) Vet. Res. 2012;43:4. doi: 10.1186/1297-9716-43-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Badia R., Brufau M.T., Guerrero-Zamora A.M., Lizardo R., Dobrescu I., Martin-Venegas R., Ferrer R., Salmon H., Martínez P., Brufau J. β-galactomannan and Saccharomyces cerevisiae var. boulardii modulate the immune response against Salmonella enterica serovar typhimurium in porcine intestinal epithelial and dendritic cells. Clin. Vaccine Immunol. 2012;19:368–376. doi: 10.1128/CVI.05532-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Dalmasso G., Cottrez F., Imbert V., Lagadec P., Peyron J.F., Rampal P., Czerucka D., Groux H. Saccharomyces boulardii Inhibits Inflammatory Bowel Disease by Trapping T Cells in Mesenteric Lymph Nodes. Gastroenterology. 2006;131:1812–1825. doi: 10.1053/j.gastro.2006.10.001. [DOI] [PubMed] [Google Scholar]
- 74.Moré M.I., Vandenplas Y. Saccharomyces boulardii CNCM I-745 Improves Intestinal Enzyme Function: A Trophic Effects Review. Clin. Med. Insights Gastroenterol. 2018;11:1179552217752679. doi: 10.1177/1179552217752679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Buts J.P., De Keyser N., Raedemaeker L. De Saccharomyces boulardii enhances rat intestinal enzyme expression by endoluminal release of polyamines. Pediatr. Res. 1994;36:522–527. doi: 10.1203/00006450-199410000-00019. [DOI] [PubMed] [Google Scholar]
- 76.Zaouche A., Loukil C., De Lagausie P., Peuchmaur M., Macry J., Fitoussi F., Bernasconi P., Bingen E., Cezard J.P. Effects of oral Saccharomyces boulardii on bacterial overgrowth, translocation, and intestinal adaptation after small-bowel resection in rats. Scand. J. Gastroenterol. 2000;35:160–165. doi: 10.1097/00005176-199805000-00144. [DOI] [PubMed] [Google Scholar]
- 77.Buts J.P., De Keyser N., Marandi S., Hermans D., Sokal E.M., Chae Y.H.E., Lambotte L., Chanteux H., Tulkens P.M. Saccharomyces boulardii upgrades cellular adaptation after proximal enterectomy in rats. Gut. 1999;45:89–96. doi: 10.1136/gut.45.1.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Harms H.-K., Bertele-Harms R.-M., Bruer-Kleis D. Enzyme-Substitution Therapy with the Yeast Saccharomyces cerevisiae in Congenital Sucrase-Isomaltase Deficiency. N. Engl. J. Med. 1987;316:1306–1309. doi: 10.1056/NEJM198705213162104. [DOI] [PubMed] [Google Scholar]
- 79.Buts J.-P., De Keyser N., Stilmant C., Sokal E., Marandi S. Saccharomyces boulardii Enhances N-Terminal Peptide Hydrolysis in Suckling Rat Small Intestine by Endoluminal Release of a Zinc-Binding Metalloprotease. Pediatr. Res. 2002;51:528–534. doi: 10.1203/00006450-200204000-00021. [DOI] [PubMed] [Google Scholar]
- 80.Jahn H.U., Ullrich R., Schneider T., Liehr R.M., Schieferdecker H.L., Holst H., Zeitz M. Immunological and trophical effects of saccharomyces boulardii on the small intestine in healthy human volunteers. Digestion. 1996;57:95–104. doi: 10.1159/000201320. [DOI] [PubMed] [Google Scholar]
- 81.Buts J.P., Keyser N. De Transduction pathways regulating the trophic effects of Saccharomyces boulardii in rat intestinal mucosa. Scand. J. Gastroenterol. 2010;45:175–185. doi: 10.3109/00365520903453141. [DOI] [PubMed] [Google Scholar]
- 82.Buts J.P., Dekeyser N. Raf: A key regulatory kinase for transduction of mitogenic and metabolic signals of the probiotic Saccharomyces boulardii. Clin. Res. Hepatol. Gastroenterol. 2011;35:596–597. doi: 10.1016/j.clinre.2011.04.010. [DOI] [PubMed] [Google Scholar]
- 83.Chang C., Wang K., Zhou S.N., Wang X.D., Wu J.E. Protective Effect of Saccharomyces boulardii on Deoxynivalenol-Induced Injury of Porcine Macrophage via Attenuating p38 MAPK Signal Pathway. Appl. Biochem. Biotechnol. 2017;182:411–427. doi: 10.1007/s12010-016-2335-x. [DOI] [PubMed] [Google Scholar]
- 84.Schneider S.M., Girard-Pipau F., Filippi J., Hébuterne X., Moyse D., Hinojosa G.C., Pompei A., Rampal P. Effects of Saccharomyces boulardii on fecal short-chain fatty acids and microflora in patients on long-term total enteral nutrition. World J. Gastroenterol. 2005;11:6165–6169. doi: 10.3748/wjg.v11.i39.6165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Girard-Pipau F., Pompei A., Schneider S., Nano J.L., Hebuterne X., Boquet P., Rampal P. Intestinal microflora, short chain and cellular fatty acids, influence of a probiotic Saccharomyces boulardii. Microb. Ecol. Health Dis. 2002;14:221–228. doi: 10.1080/08910600310002109. [DOI] [Google Scholar]
- 86.Macfarlane G.T., Macfarlane S. Bacteria, colonic fermentation, and gastrointestinal health. J. AOAC Int. 2012;95:50–60. doi: 10.5740/jaoacint.SGE_Macfarlane. [DOI] [PubMed] [Google Scholar]
- 87.Fukuda S., Toh H., Hase K., Oshima K., Nakanishi Y., Yoshimura K., Tobe T., Clarke J.M., Topping D.L., Suzuki T., et al. Bifidobacteria can protect from enteropathogenic infection through production of acetate. Nature. 2011;469:543–549. doi: 10.1038/nature09646. [DOI] [PubMed] [Google Scholar]
- 88.Wannaprasat W., Koowatananukul C., Ekkapobyotin C., Chuanchuen R. Quality analysis of commercial probiotic products for food animals. Southeast Asian J. Trop. Med. Public Health. 2009;40:1103–1112. [PubMed] [Google Scholar]
- 89.Salminen M.K., Rautelin H., Tynkkynen S., Poussa T., Saxelin M., Valtonen V., Jarvinen A. Lactobacillus Bacteremia, Species Identification, and Antimicrobial Susceptibility of 85 Blood Isolates. Clin. Infect. Dis. 2006;42:e35–e44. doi: 10.1086/500214. [DOI] [PubMed] [Google Scholar]
- 90.Karen M., Yuksel O., Akyürek N., Ofluoǧlu E., Çaǧlar K., Şahin T.T., Paşaoǧlu H., Memiş L., Akyürek N., Bostanci H. Probiotic Agent Saccharomyces boulardii Reduces the Incidenceof Lung Injury in Acute Necrotizing Pancreatitis Induced Rats. J. Surg. Res. 2010;160:139–144. doi: 10.1016/j.jss.2009.02.008. [DOI] [PubMed] [Google Scholar]
- 91.Lessard M., Dupuis M., Gagnon N., Nadeau É., Matte J.J., Goulet J., Fairbrother J.M. Administration of Pediococcus acidilactici or Saccharomyces cerevisiae boulardii modulates development of porcine mucosal immunity and reduces intestinal bacterial translocation after Escherichia coli challenge. J. Anim. Sci. 2009;87:922–934. doi: 10.2527/jas.2008-0919. [DOI] [PubMed] [Google Scholar]
- 92.Byron J.K., Clemons K.V., McCusker J.H., Davis R.W., Stevens D.A. Pathogenicity of Saccharomyces cerevisiae in complement factor five-deficient mice. Infect. Immun. 1995;63:478–485. doi: 10.1128/IAI.63.2.478-485.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Mcfarland L.V., Surawicz C.M., Elmer G.W., Moyer K.A., Melcher S.A., Fekety R., Bowen K.E., Cox J.L., Noorani Z., Harrington G., et al. A Randomized Placebo-Controlled Trial of Saccharomyces boulardii in Combination With Standard Antibiotics for Clostridium difficile Disease. JAMA J. Am. Med. Assoc. 1994;271:1913–1918. doi: 10.1001/jama.1994.03510480037031. [DOI] [PubMed] [Google Scholar]
- 94.Thygesen J.B., Glerup H., Tarp B. Saccharomyces boulardii fungemia caused by treatment with a probioticum. BMJ Case Rep. 2012;2012:bcr0620114412. doi: 10.1136/bcr.06.2011.4412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Appel-da-Silva M.C., Narvaez G.A., Perez L.R.R., Drehmer L., Lewgoy J. Saccharomyces cerevisiae var. boulardii fungemia following probiotic treatment. Med. Mycol. Case Rep. 2017;18:15–17. doi: 10.1016/j.mmcr.2017.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Cassone M., Serra P., Mondello F., Girolamo A., Scafetti S., Pistella E., Venditti M. Outbreak of Saccharomyces cerevisiae Subtype boulardii Fungemia in Patients Neighboring Those Treated with a Probiotic Preparation of the Organism. J. Clin. Microbiol. 2003;41:5340–5343. doi: 10.1128/JCM.41.11.5340-5343.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Enache-Angoulvant A., Hennequin C. Invasive Saccharomyces Infection: A Comprehensive Review. Clin. Infect. Dis. 2005;41:1559–1568. doi: 10.1086/497832. [DOI] [PubMed] [Google Scholar]
- 98.Dinleyici E.C., Kara A., Ozen M., Vandenplas Y. Saccharomyces boulardii CNCM I-745 in different clinical conditions. Expert Opin. Biol. Ther. 2014;14:1593–1609. doi: 10.1517/14712598.2014.937419. [DOI] [PubMed] [Google Scholar]
- 99.Pineton de Chambrun G., Neut C., Chau A., Cazaubiel M., Pelerin F., Justen P., Desreumaux P. A randomized clinical trial of Saccharomyces cerevisiae versus placebo in the irritable bowel syndrome. Dig. Liver Dis. 2015;47:119–124. doi: 10.1016/j.dld.2014.11.007. [DOI] [PubMed] [Google Scholar]