Abstract
Bacterial resistance to antibiotics has become a public health issue around the world. The present study aimed to evaluate the antibacterial activity of chalcones isolated from flowers of Arrabidaea brachypoda, and their potential as efflux pump inhibitors of Staphylococcus aureus efflux pumps. Microdilution assays were performed with natural products from A. brachypoda. Chalcones 1, 3, 4, and 5 did not show intrinsic antimicrobial activity against all S. aureus strains tested, but they were able to potentiate the Norfloxacin action against the SA1199-B (norA) strain, with a better modulating action for the 4 trimethoxylated chalcone. All chalcones were also able to potentiate the action of EtBr against SA1199-B strain, suggesting a potential NorA inhibition. Moreover, chalcone 4 was able to interfere in the activity of MepA, and interfered weakly in the QacA/B activity. Molecular docking analyzes showed that tested chalcones are capable of binding in the hydrophobic cavity of NorA and MepA, in the same Norfloxacin binding site, indicating that chalcone 4 compete with the antibiotic for the same NorA and MepA binding sites. Association of chalcone 4 with Norfloxacin could be an alternative against multidrug resistant S. aureus over-productive of NorA or MepA.
Keywords: Arrabidaea brachypoda, Chalcone, antimicrobial activity, efflux pumps, efflux pump inhibitors, resistance to antibiotics
1. Introduction
The indiscriminate use of antibiotics has created conditions for the accelerated development of multidrug resistant bacteria [1]. Infections caused by coagulase positive Staphylococcus spp., which were previously controlled by β-lactam antibiotics, have been reported in a raising frequency [2,3]. Methicillin-resistant S. aureus (MRSA) strains have progressively acquired different resistance mechanisms, reducing the chances of achieving therapeutic success [4]. The high prevalence of MRSA infections comes from the spread of some clones, with only five clones described worldwide, descendants of a single S. aureus ancestral strain that acquired the chromosomal gene mecA encoding the transpeptidase PBP2’ which has a low affinity for Methicillin [5]. On the other side, the widespread use of fluoroquinolones as an alternative therapy against MRSA infections has led to the development of bacterial resistance to these drugs [6]. In some cases, the glycopeptide Vancomycin is the only therapeutic option for the treatment of MRSA infections, despite its nephrotoxic and ototoxic effects [7]. MRSA strains remains one of the main causes of hospital-acquired and community-acquired infections, being classified as a high priority resistant pathogen that need of new therapeutic alternatives [8].
Resistance to antibiotics can be acquired by different mechanisms, among these, it is worth to highlight the antibiotic extrusion by transmembrane proteins, known as efflux pumps [9,10]. These proteins consist of extrusion systems that give bacteria resistance to multiple drugs and can be present in both Gram-positive [11] and Gram-negative bacteria [12]. Several efflux pumps present in S. aureus strains have been identified, such as NorA, NorB, NorC, MdeA, SepA, MepA, SdrM, and LmrS efflux pumps, which are encoded by the bacterial chromosome, while the QacA/B, QacG, QacH, QacJ and Smr are encoded by plasmid [13]. In view of this scenario, several researches have been looking for new technological strategies, such as the discovery of new antimicrobial agents [14], or new compounds capable of inhibiting resistance mechanisms [15]. In this context, efflux pump inhibitors (EPIs) have been proposed as a technological strategy against multidrug resistant bacteria overexpressing efflux pump genes [16,17].
Arrabidaea brachypoda (Bignoniaceae), popularly known as “cipó-uma”, “tintureiro” or “Cervejinha do campo”, is a native bush of the Brazilian cerrado, abundantly branched, presenting an average height between 1.0 and 2.0 m with leaves and flowers in terminal influences in pinkish-purple color [18]. A. brachypoda roots and leaves are widely used in traditional medicine in southeastern and northeastern Brazil for kidney disease and joint pain [19,20].
Chalcones consist of a class of open-chain flavonoids with the basic structure of 1,3-diphenyl-2-propen-1-one [21]. This class has attracted a great scientific interest throughout history due to its wide range of bioactivities, including antioxidant, anticancer, antidiabetic, anti-inflammatory, antimicrobial, anti-tuberculosis effects, among others [20,21,22,23,24]. It is also known that chalcones can trigger programmed cell death in eukaryotes, as demonstrated in the protozoan Plasmodium falciparum [24].
In fact, it has already been demonstrated that the ethanolic extract obtained from the flowers of A. brachypoda, as well as its dichloromethane fraction and the isolated compound Brachydin B were able to increase the antibiotic Norfloxacin activity against a S. aureus strain over-productive of NorA efflux pump [25]. Based on the popular use of A. brachypoda and growing concern with the advancement of bacterial resistance to currently available antimicrobials, it is relevant to continue studies on the antimicrobial properties of this plant, especially its flowers, considering their possible activities of resistance modulation to antibiotics of its isolated compounds. Thus, the present study aimed to assess the antimicrobial activity of chalcones isolated from A. brachypoda flowers, as well as their ability to enhance the antibiotics activity against different S. aureus strains over-productive of efflux pumps.
2. Results and Discussion
2.1. Compound Identification and Evaluation of the Antimicrobial Activity
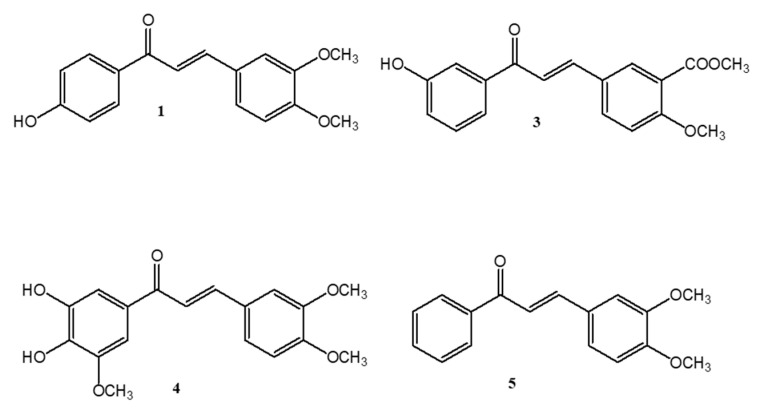
In the dichloromethane fraction from A. brachypoda flowers, 4 chalcones were identified (Figure 1) and their chemical features are presented in Table 1 and Table 2. Chalcones class has attracted great scientific interest throughout history due to its wide range of biological activities, such as antitumor and antiparasitic activities [26]. Considering the flexibility of its chemical production, synthetic chalcones have been extensively investigated [27].
Figure 1.
Chemical structure of 4’-Hydroxy-3-4-dimethoxy-chalcone (1), 3’-Hydroxy-3-acetate, 4-methoxy-chalcone (3), 3’,4’-dihydroxy, 3,4,4’-trimethoxy-chalcone (4) and 3,4-dimethoxy-chalcone (5) isolated from A. brachypoda flowers.
Table 1.
Multistage MSn data of chalcones by FIA-ESI/MS ion trap mass spectroscopy.
| Chalcones (Exact Mass) |
m/z; MSn (% Base Peak) |
|---|---|
| 283.0980 | 283[M − H]−; MS2[268]: 253(12), 239(100) |
| 311.0919 | 311[M − H]−; MS2[253]: 28(100), 269(100) |
| 329.1025 | 329[M − H]−; MS2[269]: 135(100); MS3[135]: 193(100) |
| 267.1021 | 267[M − H]−; MS2[252]: 221(100) |
Table 2.
MS and HRESIMS data for compounds from Arrabidaea brachypoda.
| Compound | Molecular Formula | Acurate Mass (amu) | Exact Mass | [M − H]− | Δm (ppm) |
|---|---|---|---|---|---|
| 1 | C17H16O4 | 283.0958 | 283.0980 | 283 | 2.2 |
| 3 | C18H16O5 | 311.0918 | 311.0919 | 311 | 1.0 |
| 4 | C18H18O6 | 329.1028 | 329.1025 | 329 | −3.0 |
| 5 | C17H16O3 | 267.1019 | 267.1021 | 267 | 2.0 |
Mass spectrometry (MS) and absorption spectroscopy in the UV-Vis region have been widely employed for the structural characterization of chalcones. Based on experiments using the electron spray ionization (ESI) method, through using mass spectrometry equipment of low resolution and high-resolution mass spectrometry (The spectrum in S1), it was possible to propose the structure of the compounds. For all isolated chalcones, very similar spectra were observed in the ultraviolet region at around 360 nm had strong UV absorption at the bond. The UV spectra and mass fragmentation data were compared with spectral data from commercial standard from our research group.
As shown in Table 1, MS/MS experiments proved some of the suspected fragmentation pathways, including support for sequential losses. In negative ion ESI-MS/MS, all target compounds yielded prominent [M − H]− ions. Some common features, such as loss of CH3, H20 and CO2, were observed in the MS/MS spectra, and were consistent with the literature. The [M − H− CH3]− ion was a characteristic fragment of chalcones. In addition, [M – H − 2CH3]− fragments were observed for dimethoxylated chalcones. Therefore, the loss of one or two CH3 could be adopted to identify single- or multi-methoxylated chalcones in negative ion ESI-MS/MS. Loss of CO and CO2 from [M − H]− ions was attributed to the structure of the C-ring.
Several studies have demonstrated the importance of chalcones in plants’ defense against phytopathogenic fungal species [28,29,30]. Chalcones have been reported as antifungal agents and as inhibitors of fungal biofilms formation [28,31]. Natural products from plants have been considered as clinically irrelevant when they show MIC values above 1000 µg/mL [32]. MIC values found for chalcones tested are presented in Table 3. These results indicate that chalcones isolated from A. brachypoda were inactive against all S. aureus strains.
Table 3.
Geometric means of MIC values found for chalcones isolated from the flowers of Arrabidaea brachypoda against S. aureus strains overexpressing efflux pump genes, except ATCC 25,923 that is a standard strain (All values represent the geometric mean of three simultaneous experiments).
| MIC (μg/mL) | ||||
|---|---|---|---|---|
| Microbial Strains | 1 | 3 | 4 | 5 |
| Staphylococcus aureus ATCC 25923 | 1024 | 1024 | 1024 | 1024 |
| Staphylococcus aureus IS-58 | 1024 | 1024 | 1024 | 1024 |
| Staphylococcus aureus RN-4220 | 1024 | 1024 | 1024 | 1024 |
| Staphylococcus aureus SA1199-B | 1024 | 1024 | 1024 | 1024 |
| Staphylococcus aureus K2068 | 1024 | 1024 | 1024 | 1024 |
| Staphylococcus aureus K4414 | 1024 | 1024 | 1024 | 1024 |
| Staphylococcus aureus K4100 | ≥1024 | ≥1024 | ≥1024 | ≥1024 |
2.2. Evaluation of the Modulating Effect of Isolated Chalcones on the Resistance to Norfloxacin
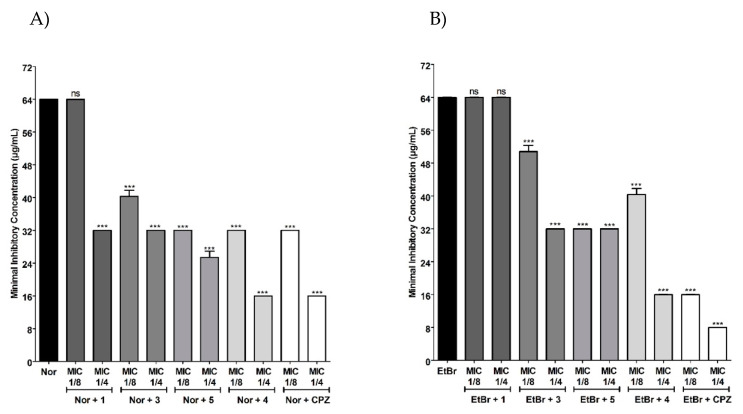
The Norfloxacin-resistance modulating activity was initially evaluated using the SA1199-B strain, resistant to Norfloxacin by overexpression of the norA gene [33]. Results showed that all chalcones increased the Norfloxacin activity against SA1199-B strain (Figure 2A). Among these, the chalcone 4 demonstrated the best modulating effect, decreasing the MIC values from 64 to 16 μg/mL (4-fold reduction), followed by chalcones 5, 3 and 1. Modulating effect showed by chalcone 4 was similar to that verified for the known NorA inhibitor, Chlorpromazine (CPZ), suggesting that chalcone 4 could be a promisor efflux pump inhibitor (EPI).
Figure 2.
MIC values of (A) Norfloxacin (Nor) and (B) Ethidium Bromide (EtBr) in the absence or presence of chalcones 1, 3, 5, 4 or Chlorpromazine (CPZ) against S. aureus SA1199-B (overexpressing norA gene). Each result represents the geometric mean of three simultaneous experiments. Differences found between treatment with Norfloxacin or EtBr alone or associated with chalcones or CPZ were analyzed through one-way analysis of variance (ANOVA) followed by Bonferroni post-test and were considered statistically significant when p < 0.05. *** p < 0.0001.
To test this hypothesis, modulation assays were conducted using Ethidium Bromide (EtBr) instead Norfloxacin, once the unique mechanism of resistance known to EtBr is by efflux pumps [34]. Chalcones 3, 4 and 5 were also able to modulate the EtBr resistance in SA1199-B strain (Figure 2B). These results indicate that the modulating effect presented by these compounds, mainly by chalcone 4, could be related with its action as NorA inhibitors, leading to EtBr accumulation, as well as Norfloxacin, into the bacterial cell. NorA is a member of the Major Facilitator Superfamily (MFS) belonging to the proton-driven antiport group [11]. NorA inhibitors have been considered as enhancers of the antimicrobials’ effectiveness, such as fluoroquinolones when tested in combination [35,36,37].
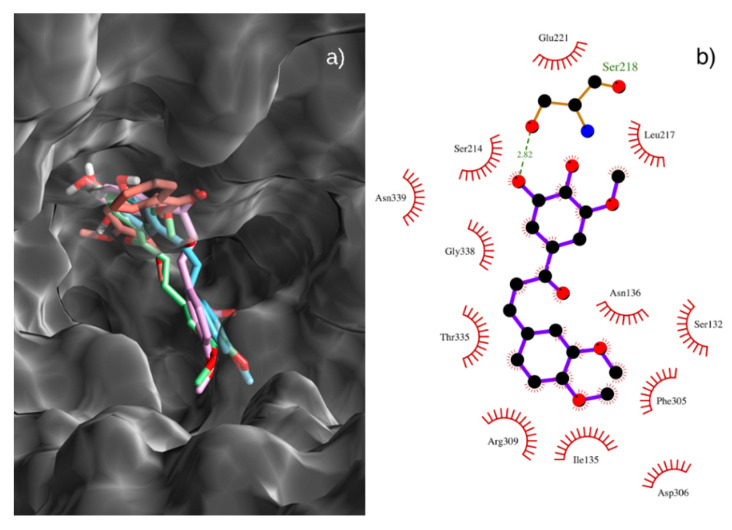
Molecular docking between isolated chalcones and NorA were performed for analysis and confirmation of this interaction. Figure 3A shows the best poses of the 4 tested substances docked on the binding site of the NorA model. Out of the 4, chalcone 4 displayed the best binding score of −7.8 kcal/mol, followed by chalcones 5, 3, and 1, with affinities of −7.5, −7.5 and −7.1 kcal/mol, respectively. This is in tune with the EPI assays previously discussed. All tested molecules roughly interacted with the same residues. Figure 3B displays the 2D ligand-protein interaction map of chalcone 4. A hydrogen bond with residue Ser218 is depicted in green and this is the reason for chalcone 4 obtaining the best docking score. Chalcone 4 also interacts with other residues on the binding site, such as Ile135, Thr335 and Asn339 between other amino acid residues.
Figure 3.
(a) Best poses of chalcones 1 (tinted blue), 3 (pink), 4 (green) and 5 (orange) on the binding site of the NorA model. (b) 2D ligand-protein interaction diagram of chalcone 4 and the NorA protein model. Contacts depicted in green.
Based on these results, one could argue that these molecules hinder the binding of Norfloxacin to the binding site of the NorA protein, and thus act as NorA competitive inhibitors. To further test this hypothesis, we docked the Norfloxacin antibiotic to the NorA model. Through analysis of the interaction map it was stated that Norfloxacin binds roughly to the same region of the model binding site, interacting with residues Thr335, Asn339, Ser336, Ile135, between other amino acid residues.
NorA inhibition by chalcones was strongly influenced by the methoxylation degree, since chalcone 4 is trimethoxylated and showed the better modulating activity. The presence of methoxy groups has been reported as a factor that favors the interaction of a compound with NorA [38]. Another previous study found that the addition of methoxyl groups increased the lipophilicity of the studied flavanones, as well as that the tetramethoxylated compound had a greater modulatory effect on Norfloxacin resistance in SA1199-B than other di- or trimethoxylated compounds [39].
Modulation tests with chalcone 4 against different S. aureus strains overproducing TetK, MsrA, MepA, QacA/B or QacC efflux pumps were carried out to analyze the activity of chalcone 4 on these S. aureus efflux pumps, thus expanding their possibilities of further study and therapeutic use. Results showed that chalcone 4 weakly interfered in the QacA/B activity and did not inhibit the activity of the TetK, MsrA and QacC efflux pumps (Table 4). On the other side, chalcone 4 was able to interfere in the activity of MepA reducing the MIC values of both Norfloxacin and EtBr against the K2068 strain. MepA is a member of the multidrug and toxin extrusion (MATE) superfamily, which is dependent of H+ or Na+ gradient to extrude Norfloxacin or EtBr [10].
Table 4.
Minimum Inhibitory Concentrations (MICs) for antibiotics and Ethidium Bromide (EtBr) in the presence or absence of chalcones isolated from Arrabidaea brachypoda flowers against different S. aureus strains overproducing efflux pumps.
| Efflux Pump Superfamily | Strain (Drug) | MIC (μg/mL) | ||
|---|---|---|---|---|
| Antibiotic/EtBr Alone |
+ Chalcone 4 (MIC 1/8) |
+ Chalcone 4 (MIC 1/4) |
||
| TetK (MFS) | IS-58 (Tet) | 64 | 64 (0,0) * | 64 (0,0) |
| IS-58 (EtBr) | 16 | 8 (2.0) | 4 (4.0) | |
| MsrA (ABC) | RN4220 (Ery) | 32 | 32 (0,0) | 32 (0,0) |
| RN4220 (EtBr) | 32 | 4 (8.0) | 4 (8.0) | |
| MepA (MATE) | K2068 (Nor) | 4 | 2 (2.0) | 1 (4.0) |
| K2068 (EtBr) | 8 | 4 (2.0) | 4 (2.0) | |
| QacA/B (MFS) | K4414 (BrEt) | 32 | 32 (0,0) | 16 (2.0) |
| QacC (SMR) | K4100 (BrEt) | 128 | 128 (0,0) | 128 (0,0) |
ABC, ATP-Binding Cassette; Ery, Erythromycin; EtBr, Ethidium Bromide; MATE, Multi-Antimicrobial Extrusion Protein; MFS, Major Facilitator Superfamily; Nor, Norfloxacin; SMR, Small Multidrug Resistance; Tet, Tetracycline; * MIC reduction factor. Each result represents the geometric mean of three simultaneous experiments.
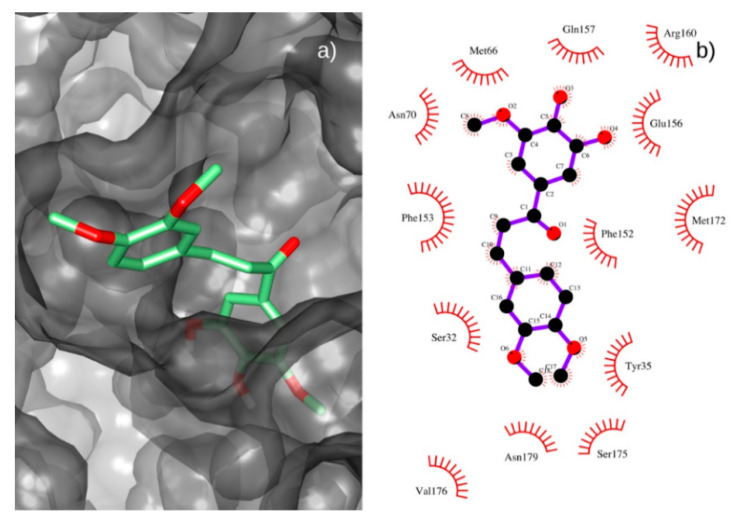
Docking of chalcone 4 against the MepA model is shown in Figure 4A. Additionally, a 2D ligand-protein interaction diagram of chalcone 4 and the MepA protein model is provided in Figure 4B.
Figure 4.
(a) Best pose of chalcone 4 (green) on the binding site of the MepA model. (b) 2D ligand-protein interaction diagram of chalcone 4 and the MepA protein model.
Chalcone 4 interacted with several residues on MepA model’ binding site. Close contacts were stablished with Ser32, Met66, Val176, Asn179, Ser175, Tyr35, Ser32, Phe153, Phe152, Met172, Glu156, Asn70 and Gln157 residues. Thus, chalcone 4 showed a good positioning to hinder the antibiotics binding to the MepA protein, thus corroborating the results of modulating assays found for S. aureus K2068 overexpressing mepA.
These results indicate that chalcone 4 is a modulating agent for Norfloxacin resistance in S. aureus strains overexpressing norA and mepA genes, acting as an EPI. Therefore, this compound could be used in combination with Norfloxacin to enhance the activity of this antibiotic against resistant S. aureus strains by overproduction of efflux pumps, such as NorA and MepA.
3. Materials and Methods
3.1. Plant Material
Plant material was collected in the cerrado regions of the Brazilian state of Minas Gerais, in the municipality of João Pinheiro (latitude 17°44’33’’ South and longitude 46°10’21” Western) in 2016. The flowers were dried in a circulate air oven at 40 °C and crushed in a knife mill. The plant was identified in the Herbarium José Badine of the Federal University of Ouro Preto, Minas Gerais, Brazil, by the botanist Dr. Maria Cristina Teixeira Braga Messias (Voucher 17935). Permission to collect the botanical material used in the present study was granted by the National System of Management of Genetic Heritage and Associated Traditional Knowledge (SISGEN), Genetic Heritage Management Council, Ministry of Environment of Brazil under registration number: A3D296E.
3.2. Preparation of the Ethanolic Extract and Obtaining the Diclorometanic Fraction
Dried flowers (500 g) were macerated with ethanol at room temperature. Crude ethanolic extract was obtained after filtration and evaporated to dryness under vacuum at approximately 40 °C, lyophilized for 48 h yielding 60 g of ethanolic extract. A portion of the obtained extract (6.2 g) was weighed and dissolved in 100 mL of water: methanol solution (7:3). Subsequently, 25 mL of dichloromethane was used to incorporate into the previously prepared solution, obtaining two solution phases. This procedure was performed 3 times, using a 24-h interval between each collection. The dichloromethane fraction was dried with the aid of a drier.
3.3. Isolation and Identification of Chalcones
Dichloromethane fraction (1.5 g) was purified on a glass column, filled with an adsorbent, silica gel 60 (0.063–0.200 mm). The column was packed with hexane and then the sample was injected on top of the column. A gradient of increasing polarity of solvents was used as the mobile phase hexane/ethyl acetate and ethyl acetate/methanol. In total, 300 fractions were obtained, which were then grouped by thin layer chromatography according to the similarity of the profile. They were weighed and analyzed by HPLC-UV/vis, HRMS and data compared with literature. This procedure allowed the isolation of 4 compounds from the dichloromethane fraction.
3.4. Compound Identification
Mass spectrometry experiments were performed on LCQ Fleet equipment (Thermo Scientific) equipped with a direct sample insertion device via flow injection analysis (FIA). The studied matrix was analyzed by electrospray ionization (ESI), fragmentation in multiple stages (MS2, MS3 and MSn) was carried out in an ion prison interface (IT). The positive mode was selected for the generation and analysis of mass spectra for first order (MS) and for the other multistep experiments under the following conditions: capillary voltage, 25 V; Spray voltage, 5 kV; and capillary temperature, 275 °C. A carrier gas (N2) with a flow of 8 arbitrary units (A.U.), and the collision gas was helium (He). The acquisition of the range was m/z 100–2000. Xcalibur software version 1.3 (Thermo Finnigan, Waltham, MA, USA) was used to acquire and process the data.
High resolution electrospray ionisation mass spectrometry (HRESIMS) data were obtained with a Micromass LCT Premier time-of-flight mass spectrometer using an electrospray ionization (ESI) interface (Waters, Milford, MA, USA). The QTOF instrument was operated with an electrospray ion source (DualESI) in positive ion mode. Nitrogen at 350 °C, flow rate 10 L min−1 was used as drying gas. The nebulizer pressure was 40 psi, the capillary voltage 3500 V, the skimmer voltage 65 V, and the fragmentor voltage 100 V. In MS mode, the quadrupole was adjusted to total ion current whereas in MS/MS mode the quadrupole was adjusted to isolate only precursor ions with bandwidth Δm/z = 1.3. MS/MS spectra were recorded after CID of precursor ions with a collision energy of 15 eV in a hexapole collision cell filled with nitrogen (99.999%). Ions were scanned in the mass ranges 100–1700 amu in MS mode and 50–500 amu in MS–MS mode. The TOF detector was operated in EDR (2 GHz) mode; the rate of acquisition was 1 spectrum s−1.
3.5. Strains and Drugs
Tests for checking intrinsic antimicrobial activity of compounds extracted from A. brachypoda flowers were performed against standard microbial strains: S. aureus ATCC 25923, Escherichia coli ATCC 25,922 and Candida albicans ATCC 10231. Moreover, chalcones were tested against the S. aureus strains overexpressing efflux pumps: SA1199-B (norA gene) [33], IS-58 (tetK gene) [40], RN-4220 (msrA gene) [41], K2068 (mepA gene) [42], K4114 (qacA/B gene) [43] and S aureus K4100 (qacC gene) [44]. Bacterial strains were maintained on Brain Infusion Heart agar (BHIA, Himedia, India) at 4 °C, and before the assay the cells were grown overnight at 37 °C in Brain Heart Infusion (BHI, Himedia, India). The antibiotics Norfloxacin (Nor), Tetracycline (Tet), Erythromycin (Ery), and the dye Ethidium Bromide (EtBr), as well as Chlorpromazine (CPZ) were obtained from Sigma Chemical Corp., St. Louis. With the exception of Nor that was dissolved in a mixture of 1 M NaOH and sterile distilled water (1:9 proportion), Tet, Eri, EtBr and CPZ were dissolved in sterile water.
3.6. Evaluation of the Antimicrobial Activity Assays
Stock solutions of chalcones (10,000 μg/mL) were prepared in DMSO followed by dilution in sterile water to a final concentration of 1024 μg/mL. Minimal inhibitory concentrations (MICs) were determined by micro-dilution assay [45] with modification in which the Muller-Hinton Broth was replaced by BHI broth. Bacterial suspensions of approximately 5 × 105 CFU/mL were exposed to test-solutions 2-fold serial diluted (concentrations ranging from 8 to 512 μg/mL). Microtiter plates were incubated at 37 ºC for 24 h, then 20 μL of resazurin (0.01% w/v in sterile distilled water) was added to each well to detect bacterial growth by color change from blue to pink. MIC was defined as the lowest concentration at which no bacterial growth was observed. Antifungal assays were performed by micro-dilution method in SDB double-concentrated with yeast suspension of approximately 5 × 105 CFU∙mL−1 and compound ranging from 8 to 512 μg/mL. Microtiter plates were incubated at 37 °C for 24 h.
3.7. Modulation of the Antibiotic Resistance Assays
To evaluate if chalcones were able to enhance the antibiotic activity against S. aureus overexpressing efflux pumps, the MICs of the antibiotics were determined in the presence or absence of sub-inhibitory concentrations of each natural product (1/8 or 1/4 MIC, respectively). Antibiotic concentrations ranged from 0.125 to 128 μg/mL. Microtiter plates were incubated at 37 °C for 24 h and readings were performed with resazurin as previously described. To verify if the drug-resistance modulation occurred due to EPI, modulation assays were performed by replacing antibiotics by EtBr, a known efflux pumps substrate [34], here used as an indicator of EPI. Control assays were also performed replacing chalcones by CPZ, a known EPI [46].
3.8. Docking Procedure
NorA model for the docking procedure was created as follows. Firstly, the NorA sequence of S. aureus SA1199B strain was retrieved from the Universal Protein Resource database (Uniprot, Entry Q03325). Then, the SWISS-MODEL service was used to build the homology model [47]. Out of the 50 templates generated, the one with the best global model quality estimation (GMQE) score was based on E. coli YajR transporter (PDB-ID: 3wdo) structure. The model was then uploaded to the MDWeb service, where it was solvated and submitted to a 0.5ns Molecular Dynamics run using the AMBER forcefield [48]. MepA model was generated by retrieving the protein sequence for the NCTC 8325 strain from the Uniprot database. The same procedure, as described above, was followed. The template of the multidrug and toxic compound extrusion (MATE) transporter of the Bacillus halodurans (PDB-ID: 5C6N) was chosen for the homology model.
For the docking procedure, which was carried out using the Autodock Vina [49] software (Scripps Research Institute, Jupiter—FL, USA), the grid box was defined as an 80Å × 80Å × 80Å box around the geometrical center of the model. Partial Gasteiger charges were added to ligand atoms, non-polar hydrogen atoms were mixed, while all other parameters were kept at their default values. The bst results were chosen based on the binding score.
3.9. Statistical Analysis
Each experiment was performed in triplicate and the results were normalized by calculating the geometric mean of the MICs. The standard error and standard deviation of the geometric mean were determined. Statistical analyzes were performed using the GraphPad Prism program, version 7.00. The occurrence of differences between treatment with antibiotics (or EtBr) alone or associated with chalcones tested (or CPZ) were examined through the one-way analysis of variance (ANOVA). Differences mentioned above were analyzed by the Bonferroni post-test and were considered statistically significant when p < 0.05.
4. Conclusions
Isolated chalcones 1, 4 and 5 did not show intrinsic antimicrobial activity against S. aureus, E. coli and C. albicans; however, the chalcone 3 was active against C. albicans. In addition, all isolated compounds were able to potentiate the antibiotic activity of Nor against S. aureus SA1199-B and K2068 strains overproducers of NorA and MepA efflux pumps, respectively, being obtained a better modulating action for the chalcone 4 trimethoxylated. Modulating assays with EtBr and molecular docking of chalcone 4 with NorA or MepA indicated that this compound is a modulating agent for Nor resistance in S. aureus strains overexpressing norA and mepA genes, acting as an EPI. Therefore, this compound could be used in combination with Nor to treat infections caused by resistant S. aureus by overproduction of NorA or MepA.
Author Contributions
Investigation/Methodology: L.M.R.-J., L.M.d.S.A., A.L.A.B.L., A.B.d.S.M., and A.L.P.d.A.d.S.; Conceptualization: J.d.S.L.N., C.Q.d.R. and H.M.B.; Data curation: C.E.S.N., J.d.S.L.N., C.Q.d.R., H.D.M.C., and H.M.B.; Writing—original version: L.M.R.-J., C.Q.d.R., C.E.S.N., H.D.M.C., and H.M.B.; Writing—review and editing: G.W.K., J.P.S.-J., and N.M. All authors have read and agreed to the published version of the manuscript.
Funding
This study was funded by the Fundação de Amparo à Pesquisa do Estado do Piauí (grant number: 050/2019) and Conselho Nacional de Desenvolvimento Científico e Tecnológico—CNPq (grant number: 050/2019), Fundação de Amparo à Pesquisa e ao Desenvolvimento Científico e Tecnológico do Maranhão (grant number: 00849/18-Universal) and CAPES (grant number: 88887.472618/2019-00-PROCAD-AM).
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Brown E.D., Wright G.D. Antibacterial drug discovery in the resistance era. Nature. 2016;529:336–343. doi: 10.1038/nature17042. [DOI] [PubMed] [Google Scholar]
- 2.WHO . Antimicrobial Resistance: Global Report on Surveillance. World Health Organization; Geneva, Switzerland: 2014. [Google Scholar]
- 3.Lira A.C., Lima L.F.A., Coutinho H.D.M., de Siqueira J.P., Jr., Barreto H.M. Antimicrobial resistance in staphylococci isolated from canine pyoderma. Commun. Sci. 2012;3:181–185. doi: 10.14295/CS.V3I3.94. [DOI] [Google Scholar]
- 4.Chua K.Y.L., Howden B.P., Jiang J.H., Stinear T., Peleg A.Y. Population genetics and the evolution of virulence in Staphylococcus aureus Infection. Genet. Evol. 2014;21:554–562. doi: 10.1016/j.meegid.2013.04.026. [DOI] [PubMed] [Google Scholar]
- 5.Botelho A.M.N., Costa M.O.C., Moustafa A.M., Beltrame C.O., Ferreira F.A., Côrtes M.F., Costa B.S.S., Silva D.N.S., Bandeira P.T., Lima N.C.B., et al. Local diversification of Methicillin-Resistant Staphylococcus aureus ST239 in South America after its rapid worldwide dissemination. Front. Microbiol. 2019;10:82. doi: 10.3389/fmicb.2019.00082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Alcalde-Rico M., Olivares-Pacheco J., Alvarez-Ortega C., Camara M., Martínez J.L. Role of the multidrug resistance efflux pump MexCD-OprJ in the Pseudomonas aeruginosa quorum sensing response. Front. Microb. 2018;9:2752. doi: 10.3389/fmicb.2018.02752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hiramatsu K., Cui L., Kuroda M., Ito T. The emergence and evolution of methicillin-resistant Staphylococcus aureus. Trends Icrobial. 2001;2001 9:486–493. doi: 10.1016/S0966-842X(01)02175-8. [DOI] [PubMed] [Google Scholar]
- 8.WHO . Global Priority List of Antibiotic-Resistant Bacteria to Guide Research, Discovery, and Development of New Antibiotics. World Health Organization; Geneva, Switzerland: 2017. [Google Scholar]
- 9.Peterson E., Kaur P. Antibiotic resistance mechanisms in bacteria: Relationships between resistance determinants of antibiotic producers, environmental bacteria, and clinical pathogens. Front. Microb. 2018;9:2928. doi: 10.3389/fmicb.2018.02928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Du D., Wang K.X., Neuberger A., van Veen H.W., Pos K.M.V., Piddock L.J., Luisi B.F. Multidrug efflux pumps: Structure, function and regulation. Nat. Rev. Microbiol. 2018;16:523–539. doi: 10.1038/s41579-018-0048-6. [DOI] [PubMed] [Google Scholar]
- 11.Schindler B.D., Kaatz G.W. Multidrug efflux pumps of Gram-positive bacteria. Drug Resistance Updat. 2016;27:1–13. doi: 10.1016/j.drup.2016.04.003. [DOI] [PubMed] [Google Scholar]
- 12.Hernando-Amado S., Blanco P., Alcalde-Rico M., Corona F., Reales-Calderón J.A., Sánchez M.B., Martínez J.L. Multidrug efflux pumps as main players in intrinsic and acquired resistance to antimicrobials. Drug Resistance Updat. 2016;28:13–27. doi: 10.1016/j.drup.2016.06.007. [DOI] [PubMed] [Google Scholar]
- 13.Hassanzadeh S., Mashhadi R., Yousefi M., Askari E., Saniei M., Pourmand M.R. Frequency of efflux pump genes mediating ciprofloxacin and antiseptic resistance in Methicillin-Resistant Staphylococcus aureus isolates. Microb. Pathog. 2017;111:71–74. doi: 10.1016/j.micpath.2017.08.026. [DOI] [PubMed] [Google Scholar]
- 14.Laxminarayan R. Antibiotic effectiveness. Science. 2014;345:1299–1301. doi: 10.1126/science.1254163. [DOI] [PubMed] [Google Scholar]
- 15.Wachino J.I., Jin W., Kimura K., Kurosaki H., Sato A., Arakawa Y. Sulfamoyl heteroarylcarboxylic acids as promising metallo-β-lactamase inhibitors for controlling bacterial carbapenem resistance. Mbio. 2020;11 doi: 10.1128/mBio.03144-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Espinoza J., Urzúa A., Sanhueza L., Walter M., Fincheira P., Muñoz P., Mendoza L., Wilkens M. Essential oil, extracts, and sesquiterpenes obtained from the heartwood of Pilgerodendron uviferum act as potential inhibitors of the Staphylococcus aureus NorA multidrug efflux pump. Front. Microbiol. 2019;10:337. doi: 10.3389/fmicb.2019.00337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Coêlho M.L., Ferreira J.H.L., de Siqueira Júnior J.P., Kaatz G.W., Barreto H.M., de Carvalho Melo Cavalcante A.A. Inhibition of the NorA multi-drug transporter by oxygenated monoterpenes. Microb. Pathog. 2016;99:173–177. doi: 10.1016/j.micpath.2016.08.026. [DOI] [PubMed] [Google Scholar]
- 18.Lorenzi H., Souza H.M. In: Plantas Ornamentais no Brasil: Arbustivas, Herbáceas e Trepadeiras. 3rd ed. Instituto Plantarum de Estudos da Flora, editor. Instituto Plantarum de Estudos da Flora; São Paulo, Brasil: 2001. [Google Scholar]
- 19.Alcerito T., Barbo F.E., Negri G., Santos D.Y.A.C., Meda C.I., Young M.C.M., Chávez D., Blatt C.T.T. Foliar epicuticular wax of Arrabidaea brachypoda: Flavonoids and antifungal activity. Biochem. Syst. Ecol. 2002;30:677–683. doi: 10.1016/S0305-1978(01)00149-1. [DOI] [Google Scholar]
- 20.Rodrigues E., Mendes F., Negri G. Plants indicated by brazilian indians for disturbances of the central nervous system: A bibliographical survey. Cent. Ner. Syst. Agents Med. Chem. 2006;6:211–244. doi: 10.2174/187152406778226725. [DOI] [Google Scholar]
- 21.K Sahu N., S Balbhadra S., Choudhary J., V Kohli D. Exploring pharmacological significance of chalcone scaffold: A review. Curr. Med. Chem. 2012;19:209–225. doi: 10.2174/092986712803414132. [DOI] [PubMed] [Google Scholar]
- 22.Rocha V., Quintino da Rocha C., Ferreira Queiroz E., Marcourt L., Vilegas W., Grimaldi G., Furrer P., Allémann É., Wolfender J.-L., Soares M. Antileishmanial activity of dimeric flavonoids isolated from Arrabidaea brachypoda. Molecules. 2018;24:1. doi: 10.3390/molecules24010001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mahapatra D.K., Bharti S.K., Asati V. Anti-cancer chalcones: Structural and molecular target perspectives. Eur. J. Med. Chem. 2015;98:69–114. doi: 10.1016/j.ejmech.2015.05.004. [DOI] [PubMed] [Google Scholar]
- 24.Sharma N., Mohanakrishnan D., Shard A., Sharma A., Sharma A., Sinha A., Sahal D. Stilbene–chalcone hybrids: Design, synthesis, and evaluation as a new class of antimalarial scaffolds that trigger cell death through stage specific apoptosis. J. Med. Chem. 2012;55:297–311. doi: 10.1021/jm201216y. [DOI] [PubMed] [Google Scholar]
- 25.Andrade L.M.D.S., Oliveira A.B.M., Leal A.L.A.B., Oliveira F.A.D.A., Portela A.L., Lima Neto J.D.S., Siqueira-Júnior J.P., Kaatz G.W., Rocha C.Q., Barreto H.M. Antimicrobial activity and inhibition of the NorA efflux pump of Staphylococcus aureus by extract and isolated compounds from Arrabidaea brachypoda. Microb. Pathog. 2020;140:103935. doi: 10.1016/j.micpath.2019.103935. [DOI] [PubMed] [Google Scholar]
- 26.Paul S.M., Mytelka D.S., Dunwiddie C.T., Persinger C.C., Munos B.H., Lindborg S.R., Schacht A.L. How to improve R&D productivity: The pharmaceutical industry’s grand challenge. Nat. Rev. Drug Discov. 2010;9:203–214. doi: 10.1038/nrd3078. [DOI] [PubMed] [Google Scholar]
- 27.Ferreira M.K.A., Fontenelle R.O.S., Magalhães F.E.A., Bandeira P.N., Menezes S., Santos J.E.S.A., Dos Santos H. Potential farmacológico de chalconas: Uma breve revisão. Rev. Virtual de Quím. 2018;10:1455–1473. [Google Scholar]
- 28.Ramírez Escobedo M.E., Barajas Bermúdez L., Pérez Berumen C., Sáenz Galindo A., Silva Belmares S.Y. Síntesis y actividad biológica de chalconas. Rev. Mex. de Cienc. Farm. 2012;43:7–14. [Google Scholar]
- 29.Rao G.P., Kumar P., Singh M., Singh H.N., Pandey O.P. Efficacy of chalcone, hydrazide and oxadiazole derivatives against fungal pathogens of sugarcane. Sugar Cane. 1994:17–22. [Google Scholar]
- 30.Thirunarayanan G., Vanangamudi G. Synthesis, spectral studies, antimicrobial and insect antifeedant activities of some substituted styryl 4′-fluorophenyl ketones. Arabian J. Chem. 2014;7:1055–1064. doi: 10.1016/j.arabjc.2010.10.034. [DOI] [Google Scholar]
- 31.Kachadourian R., Day B.J. Flavonoid-induced glutathione depletion: Potential implications for cancer treatment. Free Radic. Biol. Med. 2006;41:65–76. doi: 10.1016/j.freeradbiomed.2006.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Houghton P.J., Howes M.J., Lee C.C., Steventon G. Uses and abuses of in vitro tests in ethnopharmacology: Visualizing an elephant. J. Ethnopharmacol. 2007;110:391–400. doi: 10.1016/j.jep.2007.01.032. [DOI] [PubMed] [Google Scholar]
- 33.Kaatz G.W., Seo S.M., Ruble C.A. Efflux-mediated fluoroquinolone resistance in Staphylococcus aureus. Antimicrob. Agents Chemother. 1993;37:1086–1094. doi: 10.1128/AAC.37.5.1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Markham P.N., Westhaus E., Klyachko K., Johnson M.E., Neyfakh A.A. Multiple novel inhibitors of the NorA multidrug transporter of Staphylococcus aureus multiple novel inhibitors of the NorA multidrug transporter of Staphylococcus aureus. Antimicrob. Agents Chemother. 1999;43:2404–2408. doi: 10.1128/AAC.43.10.2404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Thai K.-M., Ngo T.-D., Phan T.-V., Tran T.-D., Nguyen N.-V., Nguyen T.-H., Le M.-T. Virtual screening for novel Staphylococcus aureus NorA efflux pump inhibitors from natural products. Med. Chem. 2015;11:135–155. doi: 10.2174/1573406410666140902110903. [DOI] [PubMed] [Google Scholar]
- 36.Cirino I.C.S., Menezes-Silva S.M.P., Silva H.T.D., de Souza E.L., Siqueira-Júnior J.P. The Essential oil from Origanum vulgare L. and its individual constituents Carvacrol and Thymol enhance the effect of Tetracycline against Staphylococcus aureus. Chemotherapy. 2014;60:290–293. doi: 10.1159/000381175. [DOI] [PubMed] [Google Scholar]
- 37.Costa L.M., de Macedo E.V., Oliveira F.A.A., Ferreira J.H.L., Gutierrez S.J.C., Peláez W.J., Lima F.C.A., de Siqueira Júnior J.P., Coutinho H.D.M., Kaatz G.W., et al. Inhibition of the NorA efflux pump of Staphylococcus aureus by synthetic riparins. J. Appl. Microbiol. 2016;121:1312–1322. doi: 10.1111/jam.13258. [DOI] [PubMed] [Google Scholar]
- 38.Braga Ribeiro A.M., Sousa J.N., Costa L.M., Oliveira F.A.A., dos Santos R.C., Silva Nunes A.S., da Silva W.O., Marques Cordeiro P.J., de Sousa Lima Neto J., de Siqueira-Júnior J.P., et al. Antimicrobial activity of Phyllanthus amarus Schumach. & Thonn and inhibition of the NorA efflux pump of Staphylococcus aureus by Phyllanthin. Microb. Pathog. 2019;130:242–246. doi: 10.1016/j.micpath.2019.03.012. [DOI] [PubMed] [Google Scholar]
- 39.Maia G.L.A., Falcão-Silva V.S., Aquino P.G.V., Araújo-Júnior J.X., Tavares J.F., Silva M.S., Rodrigues L.C., Siqueira-Júnior J.P., Barbosa-Filho J.M. Flavonoids from Praxelis clematidea R.M. King and Robinson modulate bacterial drug resistance. Molecules. 2011;16:4828–4835. doi: 10.3390/molecules16064828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gibbons S., Udo E.E. The effect of reserpine, a modulator of multidrug efflux pumps, on the in vitro activity of tetracycline against clinical isolates of methicillin resistant Staphylococcus aureus (MRSA) possessing the tet(K) determinant. Phytother. Res. 2000;14:139–140. doi: 10.1002/(SICI)1099-1573(200003)14:2<139::AID-PTR608>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 41.Ross J.I., Farrell A.M., Eady E.A., Cove J.H., Cunliffe W.J. Characterisation and molecular cloning of the novel macrolide-streptogramin B resistance determinant from Staphylococcus epidermidis. J. Antimicrob. Chemother. 1989;24:851–862. doi: 10.1093/jac/24.6.851. [DOI] [PubMed] [Google Scholar]
- 42.Kaatz G.W., McAleese F., Seo S.M. Multidrug resistance in Staphylococcus aureus due to overexpression of a novel multidrug and toxin extrusion (MATE) transport protein. Antimicrob. Agents Chemother. 2005;49:1857–1864. doi: 10.1128/AAC.49.5.1857-1864.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tennent J.M., Lyon B.R., Midgley M., Jones G., Purewal A.S., Skurray R.A. Physical and Biochemical Characterization of the qacA gene encoding antiseptic and disinfectant resistance in Staphylococcus aureus. Microbiology. 1989;135:1–10. doi: 10.1099/00221287-135-1-1. [DOI] [PubMed] [Google Scholar]
- 44.Littlejohn T.G., DiBerardino D., Messerotti L.J., Spiers S.J., Skurray R.A. Structure and evolution of a family of genes encoding antiseptic and disinfectant resistance in Staphylococcus aureus. Gene. 1991;101:59–66. doi: 10.1016/0378-1119(91)90224-Y. [DOI] [PubMed] [Google Scholar]
- 45.CLSI . Clinical and Laboratory Standards Institute. Document M07-A9, Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria that Grow Aerobically. Volume 32. CLSI; Wayne, NJ, USA: 2012. Approved Standard—Ninth Edition. [Google Scholar]
- 46.Neyfakh A.A., Borsch C.M., Kaatz G.W. Fluoroquinolone resistance protein NorA of Staphylococcus aureus is a multidrug efflux transporter. Antimicrob. Agents Chemother. 1993;37:128–129. doi: 10.1128/AAC.37.1.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Waterhouse A., Bertoni M., Bienert S., Studer G., Tauriello G., Gumienny R., Heer F.T., de Beer T.A.P., Rempfer C., Bordoli L., et al. SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 2018;46:W296–W303. doi: 10.1093/nar/gky427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hospital A., Andrio P., Fenollosa C., Cicin-Sain D., Orozco M., Gelpí J.L. MDWeb and MDMoby: An integrated web-based platform for molecular dynamics simulations. Bioinformatics. 2012;28:1278–1279. doi: 10.1093/bioinformatics/bts139. [DOI] [PubMed] [Google Scholar]
- 49.Trott O., Olson A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2009;31:456–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]