Graphical Abstract
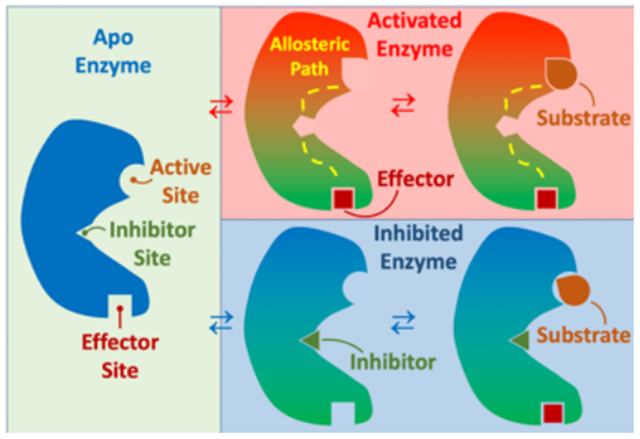
Allosteric regulation of enzyme activity is a subject of great interest for a wide range of applications, from drug discovery to gene-editing technologies. Allosteric mechanisms involve the transmission of a signal, often as a structural change or dynamical perturbation that propagates from one protein site to a distal site where enzyme function is altered. Understanding allosteric sites, including their evolutionary origin and how to target them by protein engineering or by small molecular ligands, could provide valuable insights for a wide range of applications. Hadzipasic et al.1 recently performed an intriguing study for Aurora A kinase (AK) and its allosteric activator, TPX2. They used Bayesian methods to reconstruct the phylogenetic tree for both proteins and then inferred the protein sequence at the ancestral node of interest for subsequent biochemical studies. What this work showed was that of the two activation mechanisms of AK, autophosphorylation arose first. Perhaps not surprisingly the allostery involving TPX2 only arose in AK after the appearance of TPX2. More thought-provoking is that the evolution of the allosteric interaction seemed to reside solely within AK where the most recent AK shows a 1.8-fold preference, in terms of catalytic enhancement, for its TPX2 versus the more distant ancestor TPX2ANC3, and a 1.2-fold enhancement upon binding the more recent TPX2ANC4. The TPX2 binding interface was remarkably stable across the history of AK whereas optimization of catalytic activity for phosphorylation of the human substrate, Lats2, resulted from evolutionary mutation of 15 residues in the protein interior that enhanced the coupling of TPX2 binding to the catalytic site. Thus, all that was necessary for TPX2 allosteric activation was the initial formation of an AK/TPX2 complex, followed by exploitation of the binding through evolution of intervening residues of AK itself, suggesting that the critical step was colocalization of the binding partners, rather than coevolution of them. This illuminating result shares similarities with other studies of allosteric proteins. Allosteric residues typically come in two flavors, surface residues responsible for interacting with the allosteric partner and pathway residues that relay the allosteric signal.
Two recent studies also describe the independence of these two types of allosteric residues. For example, adaptation to extreme temperature has caused protein lineages to diverge significantly. Such changes often occur at surface-exposed sites, which are more susceptible to temperature changes but also distant from the active site. Studies by Hilser and coworkers2 with E. coli adenylate kinase showed that by tuning the unfolding of a distant allosteric region, one can independently tune the active site activity and affinity, thereby elucidating the driving force behind such allosteric adaptations. Their work uncovered two distal domains whose modification has a direct effect on the active site. Mutations to LID, a domain that includes part of the active site, increased the unfolded population of LID from 5% (WT) to 40% at physiological temperature. Such destabilization markedly decreased the kinase substrate affinity, thereby modulating the activity. Mutation to another region, AMPbd, had no such effects on affinity but instead lowered the activation energy through a similar unfolding event. These disruptions oppose the increased folding of the kinase at lower temperatures, making the enzyme’s optimum temperature vary between the WT, LID, and AMPbd mutants.
Similarly, the enzyme imidazole glycerol phosphate synthase (IGPS) is a prime example of a system with buried allosteric residues whose role is to relay the allosteric signal rather than to alter catalytic activity. IGPS has a basal catalytic rate for glutamine hydrolysis that is 5000-fold lower than its allosterically activated rate. Activation occurs upon binding of its small molecule ligand, PRFAR. Solution NMR and computational studies identified residues that acted as allosteric nodes in IGPS. Mutation of these residues produced a dramatic decrease in the ability of PRFAR to allosterically enhance this glutaminase activity although the basal glutaminase activity was unchanged. Amazingly these mutations were distant to both the glutaminase and the activator binding sites.3 Like the Hilser study, this work pointed to the role of dynamic regions of the enzyme as being critical for allostery.4 Importantly, drugs that bind to allosteric sites have the great advantage of finely tuning activity, both spatially and temporally, while limiting the risk of overdosage.
The retrospective analysis of allosteric evolution in kinases has implications for evolving or engineering new functions, an example being the tailoring of allosteric sites for enzymatic regulation and drug development. CRISPR-Cas9 has become widely-used due to its ability to perform genome editing with remarkable specificity. For Cas9 to cleave DNA, it must couple the distal RuvC nuclease domain to the DNA-binding recognition lobe (REC2). Network analysis performed by East et al.5 confirmed that the HNH region, located between REC2 and RuvC, facilitates signaling between the domains, and uncovers the collection of residues in the allosteric pathway. Being able to pinpoint the precise path of allosteric signaling may lead to the ability to better engineer large macromolecular complexes like Cas9.
Important studies like that of Kern and coworkers often elicit additional questions. Hadzipasic et al anticipate one area of future work – the evolution of the substrate. To begin to account for substrate evolution they show that their ancestral reconstructions demonstrate similar responses toward a non-specific peptide as with the natural human substrate Lats2. Whether this holds for the actual substrates of the ancestors remains to be explored. The evolution of substrate specificity would be an exciting area of study.
The basic research studies described above were aimed at study of fundamental principles of evolution and the biophysical properties that underlie the fascinating phenomenon of allostery. They also serve as an exemplar for the virtues of basic research. Angiotensin converting enzyme 2 (ACE2), a receptor for SARS-CoV and SARS-CoV2, which has been implicated in early stages of human infection, shares similar features to the systems described above: breathing motions, coupled motions, and allosteric linkages from surface residues to its active site. This starkly brings the understanding of the evolution, purposes, and mechanisms of allostery to the forefront of critical scientific research and the current pandemic challenge.
Acknowledgements:
KR acknowledges funding from an NIH Biophysical Training Grant (T32 GM008283), VB acknowledges (GM121781) and NSERC supercomputer resources, JPL acknowledges MCB 1615415 and GM106121
References
- [1].Hadzipasic A, Wilson C, Nguyen V, Kern N, Kim C, Pitsawong W, Villali J, Zheng Y, and Kern D (2020) Ancient origins of allosteric activation in a Ser-Thr kinase, Science 367, 912–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Saavedra HG, Wrabl JO, Anderson JA, Li J, and Hilser VJ (2018) Dynamic allostery can drive cold adaptation in enzymes, Nature 558, 324–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Lisi GP, East KW, Batista VS, and Loria JP (2017) Altering the allosteric pathway in IGPS suppresses millisecond motions and catalytic activity, Proc Natl Acad Sci U S A 114, E3414–E3423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Rivalta I, Lisi GP, Snoeberger NS, Manley G, Loria JP, and Batista VS (2016) Allosteric Communication Disrupted by a Small Molecule Binding to the Imidazole Glycerol Phosphate Synthase Protein-Protein Interface, Biochemistry 55, 6484–6494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].East KW, Newton JC, Morzan UN, Narkhede YB, Acharya A, Skeens E, Jogl G, Batista VS, Palermo G, and Lisi GP (2020) Allosteric Motions of the CRISPR-Cas9 HNH Nuclease Probed by NMR and Molecular Dynamics, J Am Chem Soc 142, 1348–1358. [DOI] [PMC free article] [PubMed] [Google Scholar]


