Abstract
The successful culture of Leptospira spp. from the environment is challenging. Here, we optimized the isolation of Leptospira spp. from water samples spiked with different species and initial concentrations of this organism. The time periods between water sampling and the isolation process were varied (0, 2, and 4 weeks). Bacterial cultures were observed under a microscope, and cultures were graded for cell density, weekly, for 12 weeks. Most pathogenic Leptospira spp. were difficult to culture under all conditions. All conditions of water samples spiked with novel species of Leptospira subclade P1 were culture positive within 2 weeks. For Leptospira subclade P2, storing samples for 2 weeks prior to isolation resulted in more successful isolation compared with isolation after other storage conditions. For subclade S1, all samples with initial bacterial concentrations of more than 103 colonies/mL, under all storage conditions, were successfully cultured. These results suggest that storing contaminated water samples for 2 to 4 weeks in the dark at an ambient temperature prior to culturing can improve the isolation of Leptospira spp. from the samples. We implemented this protocol and collected water samples from natural sources accessed by both humans and animals. Leptospira spp. was identified in 32% (35/109) of water samples. The animal species using a water source influenced the likelihood of water samples being contaminated with Leptospira spp. Cultures of Leptospira spp. from environmental samples can provide useful information for understanding the complex interactions between humans, animals and the environment in the transmission of leptospirosis.
Keywords: Leptospira spp., culture, environmental water, initial bacterial load, environment–animal interaction
1. Introduction
Leptospirosis is a zoonotic, neglected tropical disease that occurs worldwide; it is caused by spirochete bacteria from the genus Leptospira [1,2]. The genus comprises more than 35 named species, divided into two major clades: Saprophytes (S) and pathogens (P). Recently, these two clades have been subdivided into a number of subclades, subclades P1 (pathogen group), P2 (intermediate group), S1 (saprophyte group), and S2 (a new subclade) [3]. Leptospirosis infection was first recognized as an occupational hazard for individuals working in agriculture, sewer maintenance, or animal husbandry and occurs as a result of direct or indirect contact with the urine of infected animals [2,4]. Other common modes of transmission include exposure to urine-contaminated water during recreational activities, adventure travel, and ingestion of contaminated water supplies [5,6,7,8]. People can become infected via wounds, mucous membranes such as the conjunctiva, and through wet skin [4].
Many leptospirosis outbreaks have been associated with water-related events, such as rural and urban flooding, swimming and other water sports, and occupational exposure linked predominantly to farming, as well as drinking contaminated water [7,8,9,10]. This suggests that the effective detection of Leptospira in the environment is important if adequate control measures are to be developed. Some studies suggest that Leptospira bacteria can adjust well to survive and persist under specific environmental conditions and moreover there is no evidence to suggest that pathogenic Leptospira bacteria lose their infectivity when they are in the environment [11].
Currently, the successful isolation of these organisms by culturing is limited due to the presence of non-pathogenic Leptospira spp. in the environment. In addition, the detection of pathogenic Leptospira spp. in water samples is difficult due to dilution of the pathogen in samples collected in the field, as well as the potentially high number of other bacterial species present in water samples, which can contaminate culture media. To date, there is no standard protocol for culturing the pathogenic species of Leptospira from environmental samples, in particular soil or water samples. Furthermore, there have only been a few reports that have described the isolation of pathogenic Leptospira spp. from environmental samples; as discussed above, the main difficulty involved in the isolation of these species is the overgrowth of other environmental bacteria or fast-growing saprophytic Leptospira spp. [12,13,14]. Whilst polymerase chain reaction (PCR) techniques have been developed to differentiate pathogenic from non-pathogenic Leptospira spp., protocols for testing for the presence of Leptospira spp. in water samples are in their preliminary stages; they have yet to be developed to the point where they are fully validated, universally accepted, or routinely performed [15,16,17,18,19]. Such tests are required to be both sensitive and specific in addition to being robust, non-labor intensive, and cheap to perform [20].
Previously, most laboratories have performed culturing of Leptospira spp. at a time of their convenience, following the collection of field samples. Here, we developed an optimal standard operating procedure to prepare field samples for the laboratory culturing of Leptospira spp., we then piloted and used the protocol in a field investigation. Improvements in sample storage and preparation prior to quantification under the microscope would greatly contribute to addressing gaps in our knowledge regarding the survival of Leptospira spp. in the environment. The findings from both our experiments and field investigation are presented here.
2. Materials and Methods
2.1. Spiking Experiment
2.1.1. Bacterial Strains and Water Samples Spiked with Leptospira
To investigate the effect of the length of water storage time on Leptospira isolation from water samples, six species of genus Leptospira, representing both pathogens and non-pathogens that live in the environment, were selected for the experiments. These included three species of subclade P1 isolated from blood samples from infected patients (L. interrogans strain L0013 [21] and L. weilii strain LNT1194 [22]); a putative novel species of pathogenic Leptospira from a natural water source (isolate PA62-C1, unpublished data); two species of subclade P2, isolated from human urine (L. wolfii strain H2 [23]) and flood water (L. licerasiae strain Q127 [24]); and one species of subclade S1 (L. biflexa strain LT17, received from the World Health Organization (WHO)/The Food and Agriculture Organization of the United Nations (FAO)/the World Organisation for Animal Health (OIE) Collaborating Centre for Reference and Research on Leptospirosis, Queensland, Australia).
A 2 L sample of fresh water from a canal in Bangkok was collected and autoclaved at 121 °C for 15 min prior to use. All six strains of Leptospira were inoculated in Ellinghausen-McCullough-Johnson-Harris (EMJH) medium for 5 days until they reached the late-log phase of growth. Then, 100 mL water samples spiked with Leptospira culture were prepared as follows. Ten-fold serial dilutions of the bacterial culture (from 10−1 to 10−7) were prepared in a final volume of 20 mL, using the autoclaved canal water as a diluent. To prepare water samples containing approximately 102 to 106 colonies per mL, a 10 mL sample of each bacterial dilution was added to a 1 L beaker containing 90 mL of the sterile fresh water. A 0.2 mL sample of each bacterial dilution was taken to estimate the bacterial concentration in the water samples, based on counting colonies on the EMJH agar plates following incubation under ambient conditions for one month. The beakers were covered with sterile aluminum foil and stored at an ambient temperature (26–28 °C) in a dark cupboard for various periods of time, as described in the further isolation experiments below. To investigate the effect of time on bacterial isolation from water samples, the isolations were performed at three different time points: After 0, 2, and 4 weeks of water storage in the dark at an ambient temperature.
2.1.2. Leptospira Isolation by Culture
To perform Leptospira isolation, 5 mL of water was collected using a 5 mL syringe. The water sample was passed through a sterile 0.2 μm filter (Sartorius AG, Gottingen, Germany). Then, 1 mL of filtrate was inoculated into a 5 mL tube containing 3 mL semi-solid EMJH media supplemented with 10% rabbit serum. Each water sample was tested in duplicate. The autoclaved canal water with no spiking of Leptospira was included as a negative control. The samples were incubated at 30 °C in a dry cabinet for 12 weeks. Bacterial growth was observed each week under dark field microscopy. A minimum density of 600 to 800 Leptospira cells per field was required before any further laboratory processes were undertaken [25]. A four-level density grading system for Leptospira cells present per field of view was defined, using the average number from ten microscopic fields of view at 200 times magnification, as follows: +1, Leptospira cells in less than 25% of the field of view; +2, Leptospira cells between 25% and 50% of the field of view; +3, Leptospira cells between 50% and 75% of the field of view; and +4, Leptospira cells in more than 75% of the field of view.
2.2. Pilot Testing
After conducting the laboratory experiments, we repeated the protocol for water from a natural setting, using nine water samples collected from household consumable water sources. The water samples were collected in 50 mL tubes and transported to the laboratory under ambient conditions. Each sample was used under the three conditions, i.e., 0, 2, or 4 weeks storage in the dark at an ambient temperature, prior to being cultured.
2.3. Field Investigation
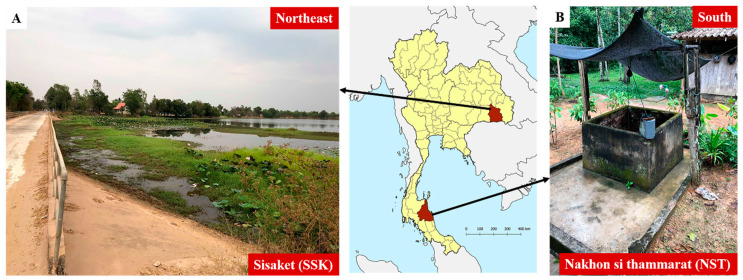
We chose two provinces for our field investigations of environmental Leptospira spp.: Si Sa Ket (SSK) and Nakhon Si Thammarat (NST) provinces, in the northeast and south of Thailand, respectively. Relatively high rates of leptospirosis morbidity have been reported in these regions, according to national disease surveillance data and previous studies [26]. During the field investigation, we recorded key characteristics of water sources and the presence of different animal species around the water source in each of the sampling areas. Figure 1 shows the distinct features of typical water sources used in the northeast and the south of Thailand.
Figure 1.
Typical shared water sources: (A) a natural water source with an open system, in the northeast; (B) a ground-water well owned by a household and situated close to or within the house, in the south.
Samples were collected from environmental water sources surrounding households. Additional water samples were collected from ponds, ground water, tap water, rivers/canals, and other standing water sources that could be accessed by domestic animals (including cats and dogs), farm animals (including chickens, ducks, and pigs), cattle (including cows and buffaloes), rodents, and humans. The sample date, location, and volume of each sample was recorded. Samples were classified into one of four sample types: Pond, ground water, rivers or irrigation channels and tap water. A 50 mL centrifuge tube was filled with water from each water source. The tubes were transported to the laboratory under ambient conditions. Samples were separated in two; one 30 and one 20 mL sample. The 30 mL samples were stored for 2 weeks under ambient conditions until being processed for culturing [27], while the 20 mL samples were stored at −80 °C until being processed, using a direct PCR method targeting the 16S rRNA gene and lipL32 gene, as previously described [19]. Culture-positive samples were isolated.
2.4. Statistical Analysis
Statistical analysis was performed using Stata/SE 14.0 for Mac. The data were analyzed for both descriptive and inferential statistics. Independent categorical variables were described using frequencies and percentages. The outcome variable in our study, i.e., Leptospira contamination of a water source, was determined based on both culture and PCR results. Mixed-effects logistic regression was performed to identify risk factors associated with the outcome variable, after adjusting for all other variables.
3. Results
3.1. Leptospira Isolation by Culture
Six Leptospira strains were used in the spiking experiment. For each strain, five different bacterial concentrations (102 to 106 colonies/mL) were individually spiked into the autoclaved water samples. The isolation process of these spiked water samples was performed over three different time periods: 0, 2, and 4 weeks of water storage in the dark at an ambient temperature. A total of 15 culture conditions for each strain tested were monitored for 12 weeks, as shown in Table 1.
Table 1.
Isolation of Leptospira spp., where the growth of cultures was observed each week for 12 weeks. The shaded blocks represent the growth grades of Leptospira spp.
| Strain | Conc. 1 | Length of Storage Time (Weeks) | ||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 2 | 4 | ||||||||||||||||||||||||||||||||||||||
| Subclade P1 | L. interrogans | L13 | 106 | |||||||||||||||||||||||||||||||||||||
| 105 | ||||||||||||||||||||||||||||||||||||||||
| 104 | ||||||||||||||||||||||||||||||||||||||||
| 103 | ||||||||||||||||||||||||||||||||||||||||
| 102 | ||||||||||||||||||||||||||||||||||||||||
| L. weilii | LNT1194 | 106 | ||||||||||||||||||||||||||||||||||||||
| 105 | ||||||||||||||||||||||||||||||||||||||||
| 104 | ||||||||||||||||||||||||||||||||||||||||
| 103 | ||||||||||||||||||||||||||||||||||||||||
| 102 | ||||||||||||||||||||||||||||||||||||||||
| PA62-C1 | 106 | |||||||||||||||||||||||||||||||||||||||
| 105 | ||||||||||||||||||||||||||||||||||||||||
| 104 | ||||||||||||||||||||||||||||||||||||||||
| 103 | ||||||||||||||||||||||||||||||||||||||||
| 102 | ||||||||||||||||||||||||||||||||||||||||
| Subclade P2 | L. wolfii | H2 | 106 | |||||||||||||||||||||||||||||||||||||
| 105 | ||||||||||||||||||||||||||||||||||||||||
| 104 | ||||||||||||||||||||||||||||||||||||||||
| 103 | ||||||||||||||||||||||||||||||||||||||||
| 102 | ||||||||||||||||||||||||||||||||||||||||
| L. licerasiae | Q127 | 106 | ||||||||||||||||||||||||||||||||||||||
| 105 | ||||||||||||||||||||||||||||||||||||||||
| 104 | ||||||||||||||||||||||||||||||||||||||||
| 103 | ||||||||||||||||||||||||||||||||||||||||
| 102 | ||||||||||||||||||||||||||||||||||||||||
| Subclade S1 | L. biflexa | Patoc I | 106 | |||||||||||||||||||||||||||||||||||||
| 105 | ||||||||||||||||||||||||||||||||||||||||
| 104 | ||||||||||||||||||||||||||||||||||||||||
| 103 | ||||||||||||||||||||||||||||||||||||||||
| 102 | ||||||||||||||||||||||||||||||||||||||||
| Control | ||||||||||||||||||||||||||||||||||||||||
| Weeks after culture | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | ||||
| 0 | +1 | +2 | +3 | +4 | ||||||||||||||||||||||||||||||||||||
1 Initial bacterial load of Leptospira (colonies/mL of water).
For subclade P1, three strains were examined, including L. interrogans strain L13, L. weilii strain LNT1194, and the novel species strain PA62-C1. Only one condition of L. interrogans strain L13 was culture-positive: The maximum initial bacterial load, stored at an ambient temperature for 2 weeks prior to isolation. The observed duration for obtaining cultures of grade +3 or +4, i.e., a cell density sufficient for PCR processing, was 8 weeks. The cultures of water samples spiked with L. weilii strain LNT1194 were all negative regardless of the initial concentration, and prolonging water storage before culturing did not improve the isolation yield. Conversely, the presence of the novel species strain PA62-C1 was observed following all time periods and with any initial concentration. However, the lowest concentration (102 colonies/mL) of PA62-C1 became positive only when the isolation was performed after samples had been stored for 2 or 4 weeks. The highest growth rate (reaching grade +4 or +3 within one week) was observed in cultures with the maximum initial bacterial concentrations only.
For subclade P2, we observed the presence of both tested strains, L. wolffii strain H2 and L. licerasiae strain Q127, with all time periods. L. wolffii strain H2 became positive at concentrations of 104 colonies/mL or more. The isolation yields were one-log reduced when the isolation was performed immediately. The growth rates of spiked samples stored for 2 weeks were faster than those of samples stored for 4 weeks. L. licerasiae strain Q127, at any concentration, became positive in the culturing condition under which isolation was performed after being stored for 4 weeks. However, its growth rate was slower in cultures with lower concentrations (102–103 colonies/mL). The isolation yields were two- and three-log reduced when the spiked samples were processed immediately and after 2 weeks, respectively.
For L. biflexa strain Patoc I, of subclade S1, the cultures were positive at a concentration of 105 colonies/mL or more, irrespective of the storage period. Prolonging water storage did not increase the isolation yield. Unexpectedly, one was culture-positive at a concentration of 103 colonies/mL after being stored for four weeks.
3.2. Leptospira Isolation in the Pilot Study
We repeated the experiments described above using nine natural water samples from household consumable water sources. The water samples were collected and placed in 50 mL tubes. Each sample was subjected to one of three conditions, i.e., storage for 0, 2, or 4 weeks in the dark at an ambient temperature, prior to being cultured. The results showed that no samples were positive immediately. One sample was positive for both the second and third conditions, i.e., isolation undertaken after water had been stored for 2 and 4 weeks, respectively. The positive sample was monitored each week for two more weeks before it reached a sufficiently high density (grade +3 or +4). This positive sample was confirmed by PCR to be a species in subclade P2, the intermediate group.
3.3. The Presence of Leptospira in Environmental Water in the Field
Once our culture protocol had been optimized and tested using field samples, we applied this protocol by storing water samples for two weeks in the dark at an ambient temperature, prior to being cultured. There were 31 and 78 water samples collected in SSK and NST, respectively. Table 2 shows the percentage of positive samples obtained from culturing and/or direct PCR. There were 29% (9/31) and 33% (26/78) positive samples from SSK and NST, respectively. Positive samples were found in all consumable types of water including drinking water i.e., tap water in SSK and ground water in NST. Table 3 shows that among the 35 positive samples, 11 samples (31.4%), from waterfall and ground water in NST, were identified as pathogenic Leptospira. Furthermore, 11 samples (31.4%) were identified as intermediate Leptospira, these included two samples from tap water in SSK. In addition, sequencing and phylogenetic data analysis confirmed that the remaining 3 positive samples (8.6%) were non-pathogenic Leptospira and 10 positive samples (28.6%) were unculturable Leptospira, where there was insufficient PCR product for sequencing (data not shown).
Table 2.
Laboratory results of water samples collected in this study and tested for Leptospira spp. * Samples were positive by either or both culture and direct PCR.
| Type of Water | Percentage of Water Samples Suspected to be Leptospira-Positive | |||||
|---|---|---|---|---|---|---|
| Sisaket (SSK) (n = 31) | Nakhon Si Thammarat (NST) (n = 78) | |||||
| Culture | Direct PCR | Overall * | Culture | Direct PCR | Overall * | |
| Puddle/pond/swamp | 12.5% (3/24) | 16.7% (4/24) | 29.2% (7/24) | 60.0% (3/5) | 40.0% (2/5) | 80.0% (4/5) |
| River/canal/waterfall | 0% (0/5) | 0% (0/5) | 0% (0/5) | 0% (0/3) | 33.3% (1/3) | 33.3% (1/3) |
| Tap water | 100.0% (2/2) | 0% (0/2) | 100.0% (2/2) | - | - | - |
| Ground water | - | - | - | 22.9% (16/70) | 18.6% (13/70) | 30.0% (21/70) |
| Total | 16.1% (5/31) | 12.9% (4/31) | 29.0% (9/31) | 24.4% (19/78) | 20.5% (16/78) | 33.3% (26/78) |
Table 3.
Classification of Leptospira spp. isolated from water samples collected in this study. * Samples were positive by either or both culture and direct PCR.
| Type of Water | Percentage of Water Samples Positive for Leptospira * (n = 35) | |
|---|---|---|
| Pathogenic Leptospira | Intermediate Leptospira | |
| Puddle/pond/swamp | - | 18.2% (2/11) |
| River/canal/waterfall | 100% (1/1) | - |
| Tap water | - | 100% (2/2) |
| Ground water | 47.6% (10/21) | 33.3% (7/21) |
| Total | 31.4% (11/35) | 31.4% (11/35) |
3.4. Environment–Animal Interactions
We performed an analysis of the water source data collected in our study (n = 90). The presence of farm animals and domestic pets around water sources were the only two factors found to be associated with Leptospira contamination of the water sources; however, the association was in the opposite direction. The presence of farm animals around water sources was less likely to be linked with Leptospira contamination of water samples (adjusted odds ratio (AOR) 0.23, 95% confidence intervals (CI) 0.06–0.88), while the presence of domestic pets was 5.25 times (95% CI 1.53–18.03) more likely to result in Leptospira contamination of water samples (Table 4).
Table 4.
Factors associated with Leptospira contamination of water sources.
| Characteristics | N 1 (%) | Leptospira Positive 2 (%) | OR 3 (95% CI) | AOR 4 (95% CI) |
|---|---|---|---|---|
| Presence of Rodents | ||||
| No | 80 (88.89) | 28 (35.00) | Reference | Reference |
| Yes | 10 (11.11) | 5 (50.00) | 1.85 (0.49–6.96) | 0.24 (0.02–2.64) |
| Presence of cattle | ||||
| No | 73 (81.11) | 24 (32.88) | Reference | Reference |
| Yes | 17 (18.89) | 9 (52.94) | 2.29 (0.78–6.69) | 3.09 (0.43–22.29) |
| Presence of farm animals | ||||
| No | 66 (73.33) | 27 (81.82) | Reference | Reference |
| Yes | 24 (26.67) | 6 (18.18) | 0.48 (0.16–1.37) | 0.23 (0.06–0.88) |
| Presence of domestic pets | ||||
| No | 67 (74.44) | 20 (29.85) | Reference | Reference |
| Yes | 23 (25.56) | 13 (56.52) |
3.05
(1.15 –) |
5.25
(1.53 –18.03) |
| Drinking source | ||||
| No | 45 (50.00) | 17 (37.78) | Reference | Reference |
| Yes | 45 (50.00) | 16 (35.56) | 0.90 (0.38–2.14) |
1.40 (0.40–4.87) |
| Consuming source 5 | ||||
| No | 37 (41.11) | 17 (45.95) | Reference | Reference |
| Yes | 53 (58.89) | 16 (30.19) | 0.05 (0.12–1.21) |
0.37 (0.12–1.17) |
| Agricultural source | ||||
| No | 58 (64.44) | 18 (31.03) | Reference | Reference |
| Yes | 32 (35.56) | 15 (46.88) | 1.96 (0.80–4.77) |
1.56 (0.42–5.85) |
| Type of water source | ||||
| Closed system | 85 (94.44) | 31 (36.47) | Reference | Reference |
| Open system | 5 (5.56) | 2 (40.00) | 1.16 (0.18–7.33) |
1.34 (0.17–10.66) |
1 Total sample of 90; 2 Total positive sample of 33; 3 Odds ratio (OR); 4 Adjusted odds ratio (AOR); 5 Consumable means using water in daily activities such as washing and cleaning.
4. Discussion
As has been previously found, it was difficult to successfully culture pathogenic species of Leptospira and, if they could be cultured, they grew slowly. Previous studies have shown that pathogenic Leptospira spp. are able to survive but not multiply in the environment [28]. It is generally considered that pathogenic Leptospira spp. can survive in soil and bodies of fresh water, including mud, swamps, streams, lakes, and rivers, particularly under neutral to slightly alkaline conditions. Pathogenic Leptospira spp. have been noted to be highly susceptible to ultraviolet light, chlorine, and detergents. They are also thought to be susceptible to acidic conditions and low temperatures [28,29]. We successfully cultured one isolate of L. interrogans strain L13 under the conditions of maximum initial bacterial load and a time period from water sample collection to culture of 2 weeks. The growth rate of this species was quite slow (8 weeks to record a cell density grade of +4). None of the L. weilii strain LNT1194 samples were successfully cultured in our experiment. Conversely, the putative novel species of pathogenic Leptospira (isolate PA62-C1) was positive under all conditions, irrespective of the period of time between sampling and culturing and the initial bacterial load. The growth rate of the putative novel species was fast, taking just one week to reach a cell density grade of +4. Several recent studies from around the world have reported novel species of Leptospira from both animal and environmental (soil and water) samples [3,30,31], as well as in humans [32]. In Thailand, some novel species of pathogenic Leptospira were reported in floodwater in Bangkok during the 2011 floods [24] and in soil samples from southern Thailand. These isolates from southern Thailand were closely related to L. ellisii [33]; they did not induce signs or symptoms of leptospirosis in a hamster model infection [31]. These novel species tend to be well adapted to their environment and are able to survive culturing even when starting from a low initial concentration [3,31,32]. The experimental results we have described here confirmed this phenomenon.
For subclade P2, the experimental results for both species, L. wolfii strain H2 and L. licerasiae strain Q127, clearly showed that the optimal procedure for successful culturing was to store the isolates under ambient conditions in the dark for 4 weeks prior to the culturing process. This procedure was shown to increase the chance of obtaining positive cultures, especially for samples where the initial bacterial load was low. This finding may be explained by the bacteria requiring a period of time to adapt to their new environment and to be able to survive and grow [7].
For subclade S1, and L. biflexa strain Patoc I in particular, less than 50% of all isolates were successfully cultured. Only when the initial bacterial load was more than 104 colonies/mL, was isolation of these organisms possible. Storing the samples for several weeks did not conclusively improve the ability to culture these organisms. This suggested that they are not good at penetrating the isolation filters and/or they do not survive well in water at high bacterial densities (105–106 colonies/mL). The latter reason is more likely and could explain why strain Patoc I, at a concentration of 103 colonies/mL, became positive when subjected to isolation after 4 weeks of storage.
Generally, environmental water and soil are considered to be important reservoirs for leptospirosis transmission in humans and animals [11]. In our study, we tested 109 water samples from water sources used for consumption in the study areas. Around 30% of all samples were Leptospira spp. positive, either by culture or direct PCR. This indicated that a significant proportion of water sources contained Leptospira. The usual recommendations for avoiding Leptospira infection include wearing boots and thorough washing after work; however, this may not be sufficient given that both water sources around residential areas and those outside, in areas of work, may be contaminated [34,35]. In addition, the large differences observed between our study sites in terms of water contact patterns (including sharing of water sources) highlights the need for leptospirosis surveillance, prevention, and control strategies that take local environmental and behavioral factors into consideration.
Rodents have traditionally been the main animal reservoir of focus for the control of leptospirosis, although recently other animals have been considered as possible reservoirs of Leptospira spp. that have potentially contributed to human infections [36,37,38,39]. Research undertaken to investigate leptospirosis in dogs in Thailand has shown that certain behaviors in dogs may be associated with the prevalence of Leptospira antibodies [40], and pathogenic Leptospira spp. have been found to commonly occur in asymptomatic domestic animals [39]. Thus, in agreement with the findings of our association analysis, domestic pets, such as dogs, may be a significant contributor to the transmission of human leptospirosis. The presence of farm animals near to water sources, however, reduced Leptospira contamination of the water samples in our study. A previous study from Thailand also showed that pigs had the lowest seroprevalence among other potential reservoirs [36].
The main limitation of our study was the lack of data linking our findings to clinical cases of leptospirosis in humans in the same study areas, i.e., specific residential locations of leptospirosis cases and the timing of these infections. More precise information on these human infections could help determine appropriate locations and timing for environmental surveys.
5. Conclusions
The success of any isolation method for obtaining Leptospira spp. from water samples depends on the species, the strain characteristics, and the initial bacterial load. It is very difficult to culture pathogenic Leptospira spp. unless the initial bacterial load is high, i.e., at least 106 colonies/mL. However, novel pathogenic Leptospira species and species from subclade P2, the intermediate group, appear to be highly adaptable and are able to grow well when present at concentrations of at least 103 to 105 colonies/mL, respectively. Extending the time period between collection and isolation can improve the isolation yield when a sample contains a lower initial bacterial load. In our study, we showed that storing contaminated water samples for 2 to 4 weeks in the dark at an ambient temperature prior to the culture process can improve Leptospira spp. isolation without being affected by the overgrowth of saprophytic or non-pathogenic Leptospira spp. Our study further identified the presence of domestic pets around shared water sources to be a risk factor associated with Leptospira spp. positivity in those water sources. These findings are useful for designing strategies to prevent environmental contamination with Leptospira spp. and to control human infections.
Acknowledgments
This publication fulfills a part of the Doctor of Philosophy (Tropical Medicine) degree program, Faculty of Tropical Medicine, Mahidol University. The authors wish to thank Vanaporn Wuthiekanun and the staff from the Microbiology Group at Mahidol-Oxford Tropical Medicine Research Unit, Faculty of Tropical Medicine, Mahidol University, Bangkok, for their laboratory assistance. We are grateful to all the support from the staff in the Department of Tropical Hygiene and the Mathematical and Economic Modelling group, Faculty of Tropical Medicine, Mahidol University. Special thanks to Wanlapa Roobsoong from the Mahidol Vivax Research Unit, Faculty of Tropical Medicine, Mahidol University for her advice on manuscript writing.
Author Contributions
Conceptualization, U.N., J.T., and W.P.-n.; methodology, U.N., J.T., M.T., P.K. and W.P.-n.; investigation, U.N., J.T., P.S. (Prapaporn Srilohasin), P.S. (Preeraya Singkhaimuk), M.T. and S.C.; validation, W.P.-n., J.T. and M.T.; formal analysis U.N. and W.P.-n.; writing—original draft preparation, U.N. and W.P.-n.; writing—review and editing, J.T., P.S. (Prapaporn Srilohasin), P.S. (Preeraya Singkhaimuk), M.T., S.C. and P.K.; supervision, W.P.-n. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Newton Fund Mobility (grant number NI150319) and Wellcome-Trust Major Overseas Programme in SE Asia (grant number 106698/Z/14/Z).
Conflicts of Interest
The authors declare no conflicts of interest.
References
- 1.Levett P.N. Leptospirosis. Clin. Microbiol. Rev. 2001;14:296–326. doi: 10.1128/CMR.14.2.296-326.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Picardeau M. Diagnosis and epidemiology of leptospirosis. Méd. Mal. Infect. 2013;43:1–9. doi: 10.1016/j.medmal.2012.11.005. [DOI] [PubMed] [Google Scholar]
- 3.Vincent A.T., Schiettekatte O., Goarant C., Neela V.K., Bernet E., Thibeaux R., Ismail N., Mohd Khalid M.K.N., Amran F., Masuzawa T., et al. Revisiting the taxonomy and evolution of pathogenicity of the genus Leptospira through the prism of genomics. PLoS Negl. Trop. Dis. 2019;13:e0007270. doi: 10.1371/journal.pntd.0007270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alemayehu T.F.a.M. Leptospirosis and its Public Health Significance: A Review. Eur. J. Appl. Sci. 2012;4:238–244. [Google Scholar]
- 5.Guerra M.A. Leptospirosis: Public health perspectives. Biologicals. 2013;41:295–297. doi: 10.1016/j.biologicals.2013.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Haake D.A., Dundoo M., Cader R., Kubak B.M., Hartskeerl R.A., Sejvar J.J., Ashford D.A. Leptospirosis, water sports, and chemoprophylaxis. Clin. Infect. Dis. 2002;34:e40–e43. doi: 10.1086/339942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Adler B., de la Pena Moctezuma A. Leptospira and leptospirosis. Vet. Microbiol. 2010;140:287–296. doi: 10.1016/j.vetmic.2009.03.012. [DOI] [PubMed] [Google Scholar]
- 8.Cacciapuoti B., Ciceroni L., Maffei C., Di Stanislao F., Strusi P., Calegari L., Lupidi R., Scalise G., Cagnoni G., Renga G. A waterborne outbreak of leptospirosis. Am. J. Epidemiol. 1987;126:535–545. doi: 10.1093/oxfordjournals.aje.a114686. [DOI] [PubMed] [Google Scholar]
- 9.Wynwood S.J., Craig S.B., Graham G.C., Blair B.R., Burns M.A., Weier S.L., Collet T.A., McKay D.B. The emergence of Leptospira borgpetersenii serovar Arborea as the dominant infecting serovar following the summer of natural disasters in Queensland, Australia 2011. Trop. Biomed. 2014;31:281–285. [PubMed] [Google Scholar]
- 10.Morgan J., Bornstein S.L., Karpati A.M., Bruce M., Bolin C.A., Austin C.C., Woods C.W., Lingappa J., Langkop C., Davis B., et al. Outbreak of leptospirosis among triathlon participants and community residents in Springfield, Illinois, 1998. Clin. Infect. Dis. 2002;34:1593–1599. doi: 10.1086/340615. [DOI] [PubMed] [Google Scholar]
- 11.Barragan V., Olivas S., Keim P., Pearson T. Critical Knowledge Gaps in Our Understanding of Environmental Cycling and Transmission of Leptospira spp. Appl. Environ. Microbiol. 2017;83 doi: 10.1128/AEM.01190-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Barragan V.A., Mejia M.E., Travez A., Zapata S., Hartskeerl R.A., Haake D.A., Trueba G.A. Interactions of leptospira with environmental bacteria from surface water. Curr. Microbiol. 2011;62:1802–1806. doi: 10.1007/s00284-011-9931-3. [DOI] [PubMed] [Google Scholar]
- 13.Benacer D., Woh P.Y., Mohd Zain S.N., Amran F., Thong K.L. Pathogenic and saprophytic Leptospira species in water and soils from selected urban sites in peninsular Malaysia. Microbes Environ. 2013;28:135–140. doi: 10.1264/jsme2.ME12154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Saito M., Villanueva S.Y., Chakraborty A., Miyahara S., Segawa T., Asoh T., Ozuru R., Gloriani N.G., Yanagihara Y., Yoshida S. Comparative analysis of Leptospira strains isolated from environmental soil and water in the Philippines and Japan. Appl. Environ. Microbiol. 2013;79:601–609. doi: 10.1128/AEM.02728-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Murgia R., Riquelme N., Baranton G., Cinco M. Oligonucleotides specific for pathogenic and saprophytic leptospira occurring in water. FEMS Microbiol. Lett. 1997;148:27–34. doi: 10.1111/j.1574-6968.1997.tb10262.x. [DOI] [PubMed] [Google Scholar]
- 16.Ganoza C.A., Matthias M.A., Collins-Richards D., Brouwer K.C., Cunningham C.B., Segura E.R., Gilman R.H., Gotuzzo E., Vinetz J.M. Determining risk for severe leptospirosis by molecular analysis of environmental surface waters for pathogenic Leptospira. PLoS Med. 2006;3:e308. doi: 10.1371/journal.pmed.0030308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhou G., Wen S., Liu Y., Li R., Zhong X., Feng L., Wang L., Cao B. Development of a DNA microarray for detection and identification of Legionella pneumophila and ten other pathogens in drinking water. Int. J. Food Microbiol. 2011;145:293–300. doi: 10.1016/j.ijfoodmicro.2011.01.014. [DOI] [PubMed] [Google Scholar]
- 18.Riediger I.N., Hoffmaster A.R., Casanovas-Massana A., Biondo A.W., Ko A.I., Stoddard R.A. An Optimized Method for Quantification of Pathogenic Leptospira in Environmental Water Samples. PLoS ONE. 2016;11:e0160523. doi: 10.1371/journal.pone.0160523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schneider A.G., Casanovas-Massana A., Hacker K.P., Wunder E.A., Jr., Begon M., Reis M.G., Childs J.E., Costa F., Lindow J.C., Ko A.I. Quantification of pathogenic Leptospira in the soils of a Brazilian urban slum. PLoS Negl. Trop. Dis. 2018;12:e0006415. doi: 10.1371/journal.pntd.0006415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wynwood S.J., Graham G.C., Weier S.L., Collet T.A., McKay D.B., Craig S.B. Leptospirosis from water sources. Pathog. Glob. Health. 2014;108:334–338. doi: 10.1179/2047773214Y.0000000156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thaipadungpanit J., Wuthiekanun V., Chierakul W., Smythe L.D., Petkanchanapong W., Limpaiboon R., Apiwatanaporn A., Slack A.T., Suputtamongkol Y., White N.J., et al. A Dominant Clone of Leptospira interrogans Associated with an Outbreak of Human Leptospirosis in Thailand. PLoS Negl. Trop. Dis. 2007;1:e56. doi: 10.1371/journal.pntd.0000056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Boonsilp S., Thaipadungpanit J., Amornchai P., Wuthiekanun V., Bailey M.S., Holden M.T.G., Zhang C., Jiang X., Koizumi N., Taylor K., et al. A Single Multilocus Sequence Typing (MLST) Scheme for Seven Pathogenic Leptospira Species. PLoS Negl. Trop. Dis. 2013;7:e1954. doi: 10.1371/journal.pntd.0001954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Slack A.T., Kalambaheti T., Symonds M.L., Dohnt M.F., Galloway R.L., Steigerwalt A.G., Chaicumpa W., Bunyaraksyotin G., Craig S., Harrower B.J., et al. Leptospira wolffii sp. nov., isolated from a human with suspected leptospirosis in Thailand. Int. J. Syst. Evol. Microbiol. 2008;58:2305–2308. doi: 10.1099/ijs.0.64947-0. [DOI] [PubMed] [Google Scholar]
- 24.Thaipadungpanit J., Wuthiekanun V., Chantratita N., Yimsamran S., Amornchai P., Boonsilp S., Maneeboonyang W., Tharnpoophasiam P., Saiprom N., Mahakunkijcharoen Y., et al. Leptospira species in floodwater during the 2011 floods in the Bangkok Metropolitan Region, Thailand. Am. J. Trop. Med. Hyg. 2013;89:794–796. doi: 10.4269/ajtmh.13-0124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.India W. Leptospirosis Laboratory Manual. WCO India; New Delhi, India: 2007. [Google Scholar]
- 26.Luenam A., Puttanapong N. Spatial and statistical analysis of leptospirosis in Thailand from 2013 to 2015. Geospat. Health. 2019;14 doi: 10.4081/gh.2019.739. [DOI] [PubMed] [Google Scholar]
- 27.Chaiwattanarungruengpaisan S., Suwanpakdee S., Sangkachai N., Chamsai T., Taruyanon K., Thongdee M. Potentially Pathogenic Leptospira Species Isolated from a Waterfall in Thailand. Jpn. J. Infect. Dis. 2018;71:65–67. doi: 10.7883/yoken.JJID.2017.363. [DOI] [PubMed] [Google Scholar]
- 28.Goarant C. Leptospirosis: Risk factors and management challenges in developing countries. Res. Rep. Trop. Med. 2016;7:49–62. doi: 10.2147/RRTM.S102543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Casanovas-Massana A., Pedra G.G., Wunder E.A., Jr., Diggle P.J., Begon M., Ko A.I. Quantification of Leptospira interrogans Survival in Soil and Water Microcosms. Appl. Environ. Microbiol. 2018;84 doi: 10.1128/AEM.00507-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thibeaux R., Iraola G., Ferres I., Bierque E., Girault D., Soupe-Gilbert M.E., Picardeau M., Goarant C. Deciphering the unexplored Leptospira diversity from soils uncovers genomic evolution to virulence. Microb. Genomics. 2018;4 doi: 10.1099/mgen.0.000144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thibeaux R., Girault D., Bierque E., Soupe-Gilbert M.E., Rettinger A., Douyere A., Meyer M., Iraola G., Picardeau M., Goarant C. Biodiversity of Environmental Leptospira: Improving Identification and Revisiting the Diagnosis. Front. Microbiol. 2018;9:816. doi: 10.3389/fmicb.2018.00816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bourhy P., Collet L., Brisse S., Picardeau M. Leptospira mayottensis sp. nov., a pathogenic species of the genus Leptospira isolated from humans. Int. J. Syst. Evol. Microbiol. 2014;64:4061–4067. doi: 10.1099/ijs.0.066597-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thongdee M., Chaiwattanarungruengpaisan S., Lekcharoen P., Yimchoho N., Buathong R., Wiriyarat W. A novel genotype of Leptospira interrogans recovered from leptospirosis outbreak in southern Thailand. Jpn. J. Infect. Dis. 2019 doi: 10.7883/yoken.JJID.2019.061. [DOI] [PubMed] [Google Scholar]
- 34.Jittimanee J., Wongbutdee J. Prevention and control of leptospirosis in people and surveillance of the pathogenic Leptospira in rats and in surface water found at villages. J. Infect. Public Health. 2019;12:705–711. doi: 10.1016/j.jiph.2019.03.019. [DOI] [PubMed] [Google Scholar]
- 35.Mohd Ali M.R., Mohamad Safiee A.W., Yusof N.Y., Fauzi M.H., Yean Yean C., Ismail N. Isolation of Leptospira kmetyi from residential areas of patients with leptospirosis in Kelantan, Malaysia. J. Infect. Public Health. 2018;11:578–580. doi: 10.1016/j.jiph.2017.12.008. [DOI] [PubMed] [Google Scholar]
- 36.Chadsuthi S., Bicout D.J., Wiratsudakul A., Suwancharoen D., Petkanchanapong W., Modchang C., Triampo W., Ratanakorn P., Chalvet-Monfray K. Investigation on predominant Leptospira serovars and its distribution in humans and livestock in Thailand, 2010–2015. PLoS Negl. Trop. Dis. 2017;11:e0005228. doi: 10.1371/journal.pntd.0005228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Suwanpakdee S., Kaewkungwal J., White L.J., Asensio N., Ratanakorn P., Singhasivanon P., Day N.P., Pan-Ngum W. Spatio-temporal patterns of leptospirosis in Thailand: Is flooding a risk factor? Epidemiol. Infect. 2015;143:2106–2115. doi: 10.1017/S0950268815000205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Doungchawee G., Phulsuksombat D., Naigowit P., Khoaprasert Y., Sangjun N., Kongtim S., Smythe L. Survey of leptospirosis of small mammals in Thailand. Southeast Asian J. Trop. Med. Public Health. 2005;36:1516–1522. [PubMed] [Google Scholar]
- 39.Kurilung A., Chanchaithong P., Lugsomya K., Niyomtham W., Wuthiekanun V., Prapasarakul N. Molecular detection and isolation of pathogenic Leptospira from asymptomatic humans, domestic animals and water sources in Nan province, a rural area of Thailand. Res. Vet. Sci. 2017;115:146–154. doi: 10.1016/j.rvsc.2017.03.017. [DOI] [PubMed] [Google Scholar]
- 40.Meeyam T., Tablerk P., Petchanok B., Pichpol D., Padungtod P. Seroprevalence and risk factors associated with leptospirosis in dogs. Southeast Asian J. Trop. Med. Public Health. 2006;37:148–153. [PubMed] [Google Scholar]