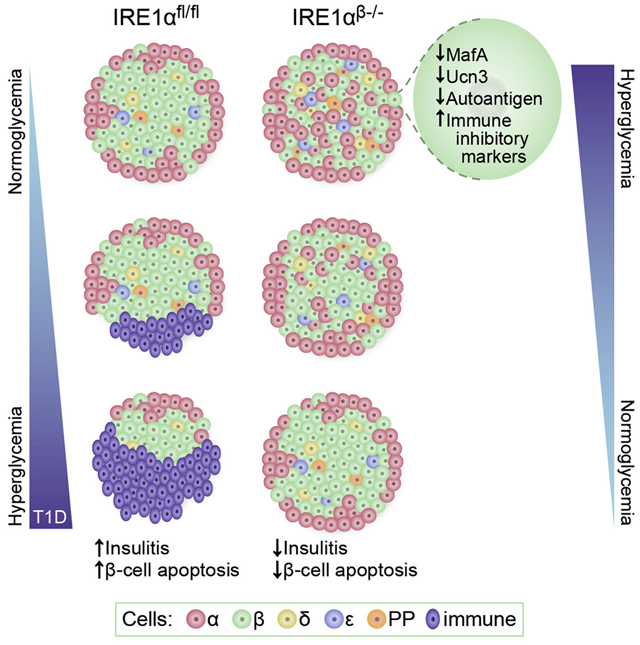
Summary
Immune-mediated destruction of insulin-producing β-cells causes type 1 diabetes (T1D). However, how β-cells participate in their own destruction during the disease process is poorly understood. Here, we report that modulating the unfolded protein response (UPR) in β-cells of non-obese diabetic (NOD) mice by deleting the UPR sensor IRE1α prior to insulitis induced a transient dedifferentiation of the β-cells, resulting in substantially reduced islet immune cell infiltration and β-cell apoptosis. Single-cell and whole-islet transcriptomics analyses of immature β-cells revealed remarkably diminished expression of β-cell autoantigens, MHC class I components and upregulation of immune inhibitory markers. IRE1α-deficient mice exhibited significantly less cytotoxic CD8+ T-cells in their pancreata and adoptive transfer of their total T-cells did not induce diabetes in Rag1−/− mice. Our results indicate that inducing β-cell dedifferentiation, prior to insulitis, allows these cells to escape immune-mediated destruction and may be used as a novel preventive strategy for T1D in high-risk individuals.
Keywords: Type 1 diabetes, NOD, UPR, ER stress, beta cell, dedifferentiation, IRE1, single cell
Graphical Abstract
Introduction
Type 1 diabetes (T1D) is an autoimmune disease in which insulin-producing pancreatic islet β-cells are targeted and destroyed by autoreactive immune cells (Atkinson, 2012; Bluestone et al., 2010; van Belle et al., 2011). Although genetic predisposition is strongly associated with T1D progression, environmental factors can also trigger T1D onset and affect its progression. Environmental factors include viral infections, toxins, reactive oxygen species (ROS), and chronic inflammation, which are well-established triggers of endoplasmic reticulum (ER) stress. ER stress initiates the unfolded protein response (UPR), which operates through inositol-requiring protein-1 (IRE1), protein kinase RNA-like ER kinase (PERK), and activating transcription factor-6 (ATF6), all of which are localized in the ER membrane and respond to stress by relaying signals from the ER to the cytoplasm and nucleus. While the UPR initially attempts to mitigate ER stress, if the stress is prolonged or severe, it switches from being a pro-adaptive to a pro-apoptotic response (Bernales et al., 2006; Walter and Ron, 2011).
Mammals have two IRE1 paralogs, IRE1α and IRE1β. IRE1α is ubiquitously expressed, whereas IRE1β is specifically expressed in digestive tissues. Dual-functioning IRE1α removes an intronic region from the transcription factor X-box binding protein 1 (XBP1) with its endoribonuclease activity, leading to its transcriptional activation (Calfon et al., 2002; Yoshida et al., 2001). Spliced XBP1 (sXBP1) then upregulates the expression of chaperones and ER-associated degradation components. IRE1α also promotes mRNA and miRNA degradation through regulated IRE1α-dependent decay (Hollien and Weissman, 2006). IRE1α, with its kinase activity, mediates the phosphorylation of the c-Jun N-terminal protein kinase (JNK) and induces inflammatory signals and apoptosis (Urano et al., 2000). The IRE1α/XBP1 pathway is the most conserved branch of the UPR, and highly secretory pancreatic β-cells have constitutively active IRE1α/XBP1 under physiological conditions. Although IRE1α has a role in promoting cell survival under acute and mild stress conditions, it can promote cell death in the presence of unresolvable ER stress (Chen and Brandizzi, 2013). IRE1α regulates cell death by downregulating mRNAs and miRNAs involved in β-cell homeostasis and survival (Han et al., 2009; Hollien and Weissman, 2006; Lipson et al., 2008; Upton et al., 2012). Hyperactivated IRE1α increases thioredoxin interacting protein (TXNIP) mRNA stability and, in turn, elevated TXNIP activates the NLRP3 inflammasome and causes β-cell death (Lerner et al., 2012; Oslowski et al., 2012). While these studies clearly indicate that modulation of IRE1α under chronic stress vs. non-stress conditions may lead to differential survival/apoptotic outcomes, the function of IRE1α in β-cells in the context of autoimmune diabetes remains unclear.
Due to the autoimmune nature of T1D, there has been a long-standing interest in understanding the role of dysregulation of the immune system in the pathogenesis of T1D. However, emerging data indicate that β-cells themselves can play a much more active role in the initiation and progression of autoimmunity than previously appreciated (Engin, 2016; Engin et al., 2013; Maganti et al., 2014; Soleimanpour and Stoffers, 2013; Thompson et al., 2019). The indication that the β-cell UPR may play a role in pathogenesis of autoimmune diabetes is supported by the detection of dysregulated ER stress markers in inflamed islets of both mice (Tersey et al., 2012) and patients with autoimmune diabetes (Marhfour et al., 2012). We previously showed that the adaptive functions of the UPR were greatly defective in β-cells of two different T1D mouse models and human patients during the progression of T1D (Engin et al., 2013). Diabetes incidence in these mouse models was dramatically reduced upon mitigation of β-cell ER stress with a chemical chaperone. Although this study provided the first direct link between a defective β-cell UPR and T1D, the molecular mechanisms by which the UPR regulates pancreatic β-cell death/survival in T1D still remain largely unknown.
Here, we investigated the specific role of the β-cell UPR in initiation and progression of T1D and deleted IRE1α in β-cells of an established T1D pre-clinical model, non-obese diabetic (NOD) mice, prior to the initiation of islet infiltration by immune cells (insulitis). Deletion of IRE1α in β-cells of NOD mice led to transient mild hyperglycemia. However, unexpectedly, mice recovered from hyperglycemia within a couple of weeks and were protected from autoimmune destruction for up to a year. Single-cell transcriptional profiling in dissociated islets, bulk RNA-seq of intact islets, as well as histological analyses demonstrated loss of mature β-cell identity and remarkably increased expression of endocrine progenitor and fetal-like β-cell markers, suggesting that upon losing IRE1α expression β-cells of NOD mice undergo a transient dedifferentiation. Furthermore, we identified significantly reduced MHC class I expression and defective antigen processing, notably reduced expression of β-cell autoantigens, upregulation of immune inhibitory markers, as well as substantially diminished CD8+ T cell population in pancreas. Taken together, we show that loss of IRE1α in β-cells prior to initiation of islet immune infiltration protects against diabetes in a pre-clinical model of T1D by inducing transient dedifferentiation of β-cells, which allows escape from immune-mediated destruction.
Results
IRE1αβ−/− NOD mice are protected from T1D.
To examine the β-cell-specific functions of IRE1α in T1D, we backcrossed IRE1α flox/flox (IRE1αfl/fl) (Iwawaki et al., 2009) and Ins2CreERT/+ mice (Dor et al., 2004) to NOD mice for more than 10–20 generations and confirmed their genetic purity on NOD background by genome scan services. To generate mice with β-cell-specific IRE1α deletion, we mated IRE1αfl/fl and IRE1αfl/fl; Ins2CreERT/+ mice and administered tamoxifen to lactating mothers, beginning the day after delivery (Figure 1A). We reasoned that tamoxifen-mediated deletion of IRE1α in pups would allow us to elucidate its function before the onset of islet insulitis, as insulitis typically starts after weaning age in female NOD mice. The deletion of IRE1α in β-cells (referred to as IRE1αβ−/−) was confirmed via immunofluorescence (IF) by staining pancreatic sections with anti-sXBP1, a direct target of IRE1α, and anti-insulin antibodies (Figure 1B), and by performing a qPCR on islets for sXBP1 (Figure 1C). Weekly blood glucose measurements were recorded starting from 3 weeks of age through 50 weeks (Figure 1D). Mice with a blood glucose level ≥250 mg/dL for two consecutive weeks were accepted as diabetic.
Figure 1. IRE1αβ−/− NOD female mice are protected from T1D.
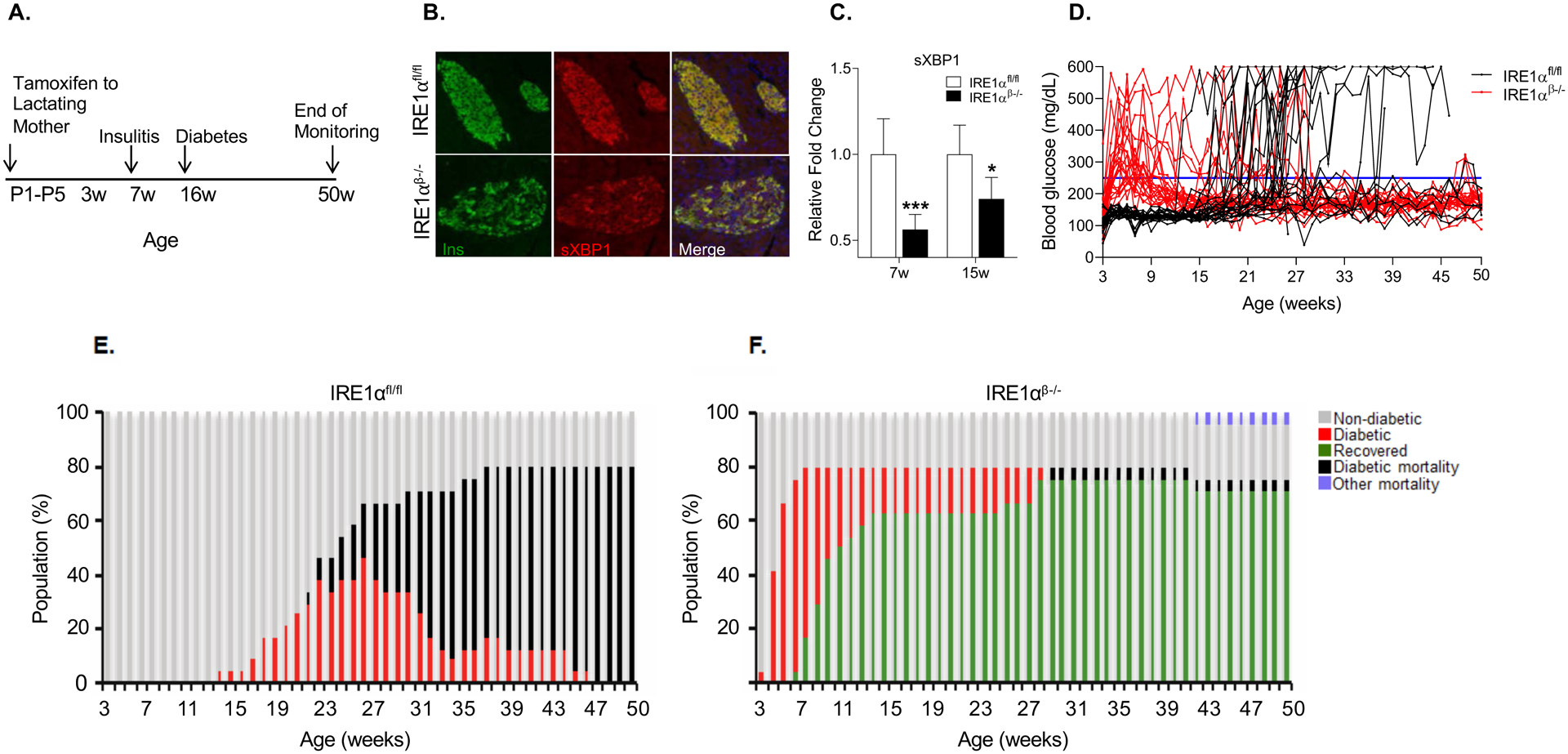
(A) Schematic representation of tamoxifen-induced deletion of IRE1α in β-cells of NOD mice.
(B) Representative immunofluorescence images showing sXBP1 expression on pancreatic sections from 5-week-old mice.
(C) Quantification of sXBP1 expression in the islets of 7- and 15-week-old IRE1αfl/fl (7 weeks: n = 6; 15 weeks: n = 5) and IRE1αβ−/− mice (7 weeks: n = 5; 15 weeks: n = 6). Data are averages of two technical replicates from a representative experiment.
(D) Blood glucose levels of IRE1αfl/fl and IRE1αβ−/− mice (n = 24 per group).
(E and F) Diabetes progression in IRE1αfl/fl and IRE1αβ−/− mice.
All data are represented as mean ± SEM, with statistical analysis performed by Student’s t-test (***P < 0.001, *P < 0.05).
IRE1α deficiency in β-cells of NOD female mice resulted in hyperglycemia starting after weaning age (Figures 1D), in agreement with recent reports showing that IRE1α deletion in β-cells of non-stressed wild type mice also induces hyperglycemia (Hassler et al., 2015; Tsuchiya et al., 2018). However, in striking contrast to previous reports, where deletion of IRE1α in either prenatal or adult β-cells led to a diabetic phenotype (Hassler et al., 2015; Tsuchiya et al., 2018), the hyperglycemia in IRE1αβ−/− NOD mice was temporary, with mice recovering from diabetes (94.7% of the IRE1αβ−/− mice exhibited normoglycemia following an initial hyperglycemia) starting at 6–7 weeks of age. Surprisingly, IRE1αβ−/− NOD mice not only recovered from hyperglycemia but were also protected from development of T1D through 50 weeks of age (Figures 1E and 1F).
To rule out any potential artifacts arising from the expression of Cre recombinase, we generated another cohort by breeding NOD and NOD Ins2CreERT/+ mice, administered tamoxifen as described above, and measured the blood glucose levels of these animals weekly up to 22 weeks of age. Unlike IRE1αβ−/− mice, hemizygous Ins2CreERT/+ mice did not exhibit early hyperglycemia, and the diabetes incidence of Ins2CreERT/+ mice was similar to the littermate control NOD mice as well as to IRE1αfl/fl mice (Figures S1A and S1B). In addition, the expression of Cre did not significantly differ between Ins2CreERT/+ and IRE1αβ−/− mice (Figure S1C). Histological analyses confirmed the presence of aggressive insulitis in the islets of diabetic Ins2CreERT/+ mice (Figure S1D). Finally, we did not detect any significant differences in glucose tolerance between pre-diabetic Ins2CreERT/+ and NOD mice, ruling out impaired glucose homeostasis in Ins2CreERT/+ mice prior to development of diabetes (Figures S1E and S1F). Taken together, these data demonstrate that Cre transgene expression in our mouse line does not appear to alter the natural progression of diabetes in NOD mice and that the phenotype seen in IRE1αβ−/− mice is most likely independent of a Cre effect.
The unexpected recovery from hyperglycemia in NOD IRE1αβ−/− mice, as opposed to the previously demonstrated diabetic phenotype seen in non-stressed, non-autoimmune “normal wild type” mice upon IRE1α deletion in β-cells, led us to ask whether autoimmunity or the intrinsic ER stress and fragility of β-cells seen in NOD mice (Dooley et al., 2016) was the underlying cause of this differential phenotype. Thus, we deleted IRE1α in β-cells of NOD-Rag1−/− mice, which lack mature T and B cells. Interestingly, NOD-Rag1−/−; IRE1αβ−/− mice phenocopied NOD IRE1αβ−/− mice, suggesting that the transient hyperglycemia phenotype was independent of T and B cell-mediated autoimmunity but involved β-cell stress (Figure S2A).
Thus, these intriguing results indicate that while inactivation of IRE1α in adult β-cells under physiological conditions can lead to diabetes, under stressed conditions as seen in T1D, the loss of IRE1α at an early stage of disease (i.e., prior to initiation of insulitis) can have beneficial effects on β-cell survival and function leading to protection from T1D.
Improved β-cell function and survival in IRE1αβ−/− NOD mice upon recovery from hyperglycemia.
To investigate the cellular basis of the phenotype, we performed hematoxylin & eosin (H&E) staining on pancreatic sections obtained from 5-week- and 24-week-old IRE1αfl/fl and IRE1αβ−/− mice (Figure 2A). While there was no immune infiltration in the islets of IRE1αfl/fl mice at 5 weeks of age, we detected considerable amount of insulitis in the islets of 24-week-old IRE1αfl/fl mice as expected in the NOD model. In contrast, IRE1αβ−/− mice showed improved islet morphology and substantially reduced immune infiltrates at 24 weeks of age. Additionally, immunofluorescence staining showed that insulin intensity in β-cells of the 24-week-old IRE1αβ−/− mice was comparable to the insulin intensity in β-cells of normoglycemic control mice, despite initially being reduced at 5-week-old. (Figure 2B). CD3 (a marker of T cell lineage) staining of pancreatic sections confirmed significantly less immune infiltration in the islets of IRE1αβ−/− mice at 24 weeks of age (Figure 2C). To quantify the extent of islet immune cell infiltration, we generated step sections from pancreata of these mice and performed insulitis scoring. Quantification revealed a remarkable increase in the percentage of islets without any insulitis in pancreatic sections of IRE1αβ−/− mice in comparison with IRE1αfl/fl mice (Figure 2D). Furthermore, the number of islets with aggressive insulitis was significantly decreased in IRE1αβ−/− mice compared with controls (Figure 2D). These data suggest that, in addition to the intrinsic effects of IRE1α in β-cells, IRE1α could also affect the migration of inflammatory cells, as well as their infiltration, activation, and/or cytotoxic function. We then assessed the function of β-cells by measuring pancreatic insulin and proinsulin content and determining proinsulin/insulin ratio by enzyme-linked immunosorbent assay (ELISA) at 7 weeks of age (Figure 2E–G). Consistent with our histological data, both insulin and proinsulin content of the pancreata were substantially reduced in IRE1αβ−/− mice at this time point (Figures 2E and 2F). Proinsulin:insulin ratio was greatly increased in 7-week-old IRE1αβ−/− mice suggesting a defect in processing of proinsulin (Figure 2G). At 24 weeks of age, pancreatic insulin content was still significantly less in IRE1αβ−/− mice in comparison to non- diabetic IRE1αfl/fl mice albeit to a much lesser degree (Figure 2H), while proinsulin content (Figure 2I) and proinsulin:insulin ratio (Figure 2J) were fully restored in IRE1αβ−/− pancreata. These data suggest that ER secretory function and processing were significantly improved in IRE1αβ−/− mice at this age. Consistent with a restored ER function and secretory capacity, serum insulin levels of IRE1αβ−/− mice were comparable to control non-diabetic IRE1αfl/fl mice (Figure 2K). Furthermore, an intraperitoneal glucose tolerance test revealed no differences in glucose tolerance between IRE1αβ−/− mice and non-diabetic control mice at 32 weeks of age (Figures S2B and S2C) indicating that the β-cell function was substantially improved in IRE1αβ−/− mice.
Figure 2. Improved β-cell function and survival in IRE1αβ−/− NOD mice upon recovery from hyperglycemia.
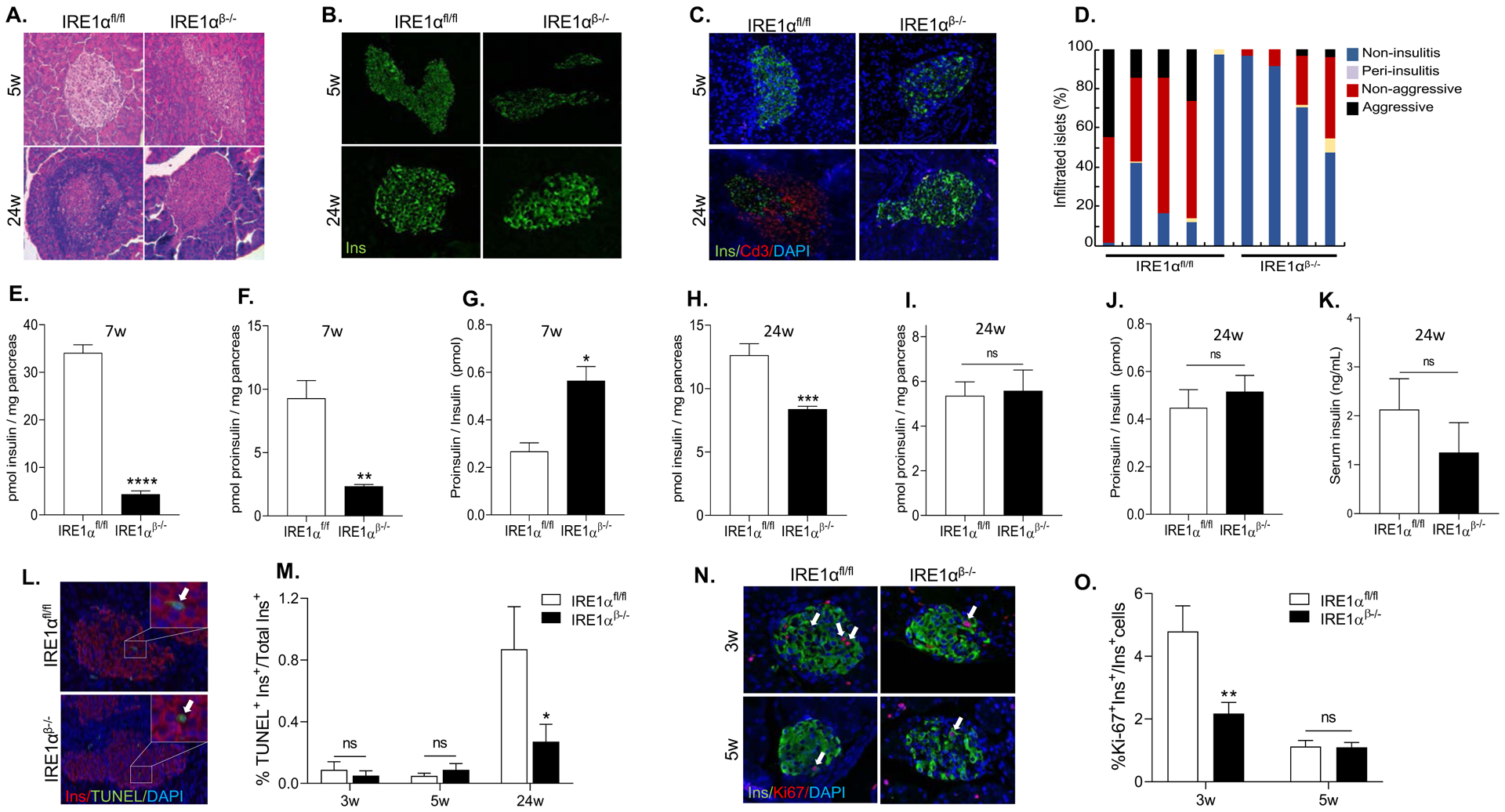
(A) A representative image of H&E staining showing varying degree of lymphocyte infiltration of islets. (B) Representative immunofluorescence images showing insulin expression.
(C) Representative immunofluorescence images showing insulin and CD3 expression.
(D) Insulitis scoring of 24 weeks of age IRE1αfl/fl (n = 5) and IRE1αβ−/− (n = 4) mice.
(E and F) Insulin and proinsulin content of 7-week-old mice (n = 4 per group).
(G) Proinsulin-to-insulin molar ratio was calculated. Data are averages of two technical replicates from a representative experiment.
(H and I) Insulin (n = 6 per group) and proinsulin content of 24-week-old IRE1αfl/fl (n = 5) and IRE1αβ−/− (n = 7) mice.
(J) Proinsulin-to-insulin molar ratio was calculated. Data are averages of two technical replicates from a representative experiment.
(K) Serum insulin levels of 24-week-old IRE1αfl/fl and IRE1αβ−/− mice (n = 6 per group).
(L) Representative images of TUNEL assay showing β-cell apoptosis. The arrows point to TUNEL+ cells.
(M) Percentage of TUNEL+ β-cells (IRE1αfl/fl: 3, 5, and 24 weeks: n = 6, 6, and 5, respectively; IRE1αβ−/−: 3, 5, and 24 weeks: n = 6, 6, and 8, respectively).
(N) Representative fluorescence images showing insulin and Ki67 expression. The arrows point to Ki67+ cells.
(O) Percentage of Ki67+ β-cells (IRE1αfl/fl : 3 and 5 weeks: n = 6, and n = 7, respectively; IRE1αβ−/−: 3 and 5 weeks: n = 8 and n = 7, respectively).
All data are represented as mean ± SEM, with statistical analysis performed by Student’s t-test (****P < 0.0001, **P < 0.01, *P < 0.05). w: weeks. ns: non-significant.
To examine if there was an apparent difference in β-cell death, we performed TUNEL assays on pancreatic sections at various time points. While we did not detect any significant apoptosis in β-cells of 3- and 5-week-old mice, there was a marked reduction in β-cell apoptosis in IRE1αβ−/− mice at 24 weeks of age (Figure 2L and 2M). We then examined the proliferation of β-cells during the hyperglycemic phase by co-staining the pancreatic sections with antibodies against insulin and proliferation marker Ki67. IRE1αβ−/− mice exhibited significantly less Ki67+ cells in their β-cells compared to the β-cells of the control mice at 3 weeks of age, but at 5 weeks of age there was no significant difference in Ki67+ β-cells between the control and IRE1αβ−/− mice (Figures 2N and 2O). Taken together, these data suggest that β-cells of IRE1αβ−/− mice undergo a transient loss of both expression and content of insulin and proinsulin prior to insulitis, and that by 24 weeks of age, insulin expression and serum insulin levels are restored while insulitis and apoptosis are significantly reduced, leading to protection from T1D in these mice.
Islet morphology and architecture are altered in IRE1αβ−/− mice during the hyperglycemic phase.
To assess islet cellular composition and architecture, we performed an immunofluorescence assay on the pancreata from IRE1αβ−/− mice and their littermate controls using antibodies against markers of α-, β-, and δ-cells. At 3 weeks of age, we did not observe any obvious alterations in islet composition and architecture of IRE1αβ−/− mice (Figure 3A). However, at 5 weeks of age, we detected a substantially increased number of glucagon-positive cells (roughly four-fold increase in α-cell area), accompanied with markedly diminished number of β-cells (~30% reduced β-cell area) (Figures 3B and 3G) in the islets of IRE1αβ−/− mice. Moreover, in contrast to control mice, which exhibited normal islet morphology with glucagon-positive cells residing on the islet periphery, islets of IRE1αβ−/− mice had significantly increased proportion of α-cells intermingled in the core of the islets in the hyperglycemic (5 week) phase. The area of the δ-cells was increased by approximately three-fold in the islets of IRE1αβ−/− mice in comparison to their littermates (Figures 3C and 3G). This altered islet cell composition, function and morphology were still apparent at 12 weeks of age (Figure 3D). At 24 weeks, we identified an almost two-fold increase in α-cell area, while β-cell area had notably improved (10% reduction vs. 30%) compared to 5 weeks of age, though was still significantly reduced compared to IRE1αfl/fl mice (Figures 3E and 3H). δ-cell area was increased by three-fold in the islets of IRE1αβ−/− mice in comparison to their littermates (Figures 3F and 3H). Furthermore, in addition to altered α-cell: β-cell ratio, we identified a significantly increased number of somatostatin-positive cells in the islets of IRE1αβ−/− mice at 5 and 24 weeks of age (Figures 3C and 3F). While somatostatin-positive δ-cells were distributed in the islet periphery in the islets of control mice, we found a relatively abundant fraction of δ-cells intermingled with β-cells in the islets (Figure 3C). In addition to these changes in islet morphology and hormone expression, we detected a number of bihormonal (insulin/glucagon co-expressing) cells in the islets of IRE1αβ−/− mice (Figure 3I). We also occasionally observed single islets or small islet clusters consisting of less than 5–10 insulin-positive cells in IRE1αβ−/− pancreata (Figure 3J) suggesting that these cells might also contribute to glucose homeostasis. Finally, we examined the islet size distribution on pancreatic sections from 4 and 12 weeks of age animals and detected a significantly smaller average islet size (< 5000 μm2) in the pancreata of 4 weeks of age IRE1αβ−/− mice (Figure 3K). We did not observe any differences in islet size distribution between IRE1αfl/fl and IRE1αβ−/− mice at 12 weeks of age (Figure 3L).
Figure 3. Islet cell composition is altered in IRE1αβ−/− mice during the hyperglycemic phase.
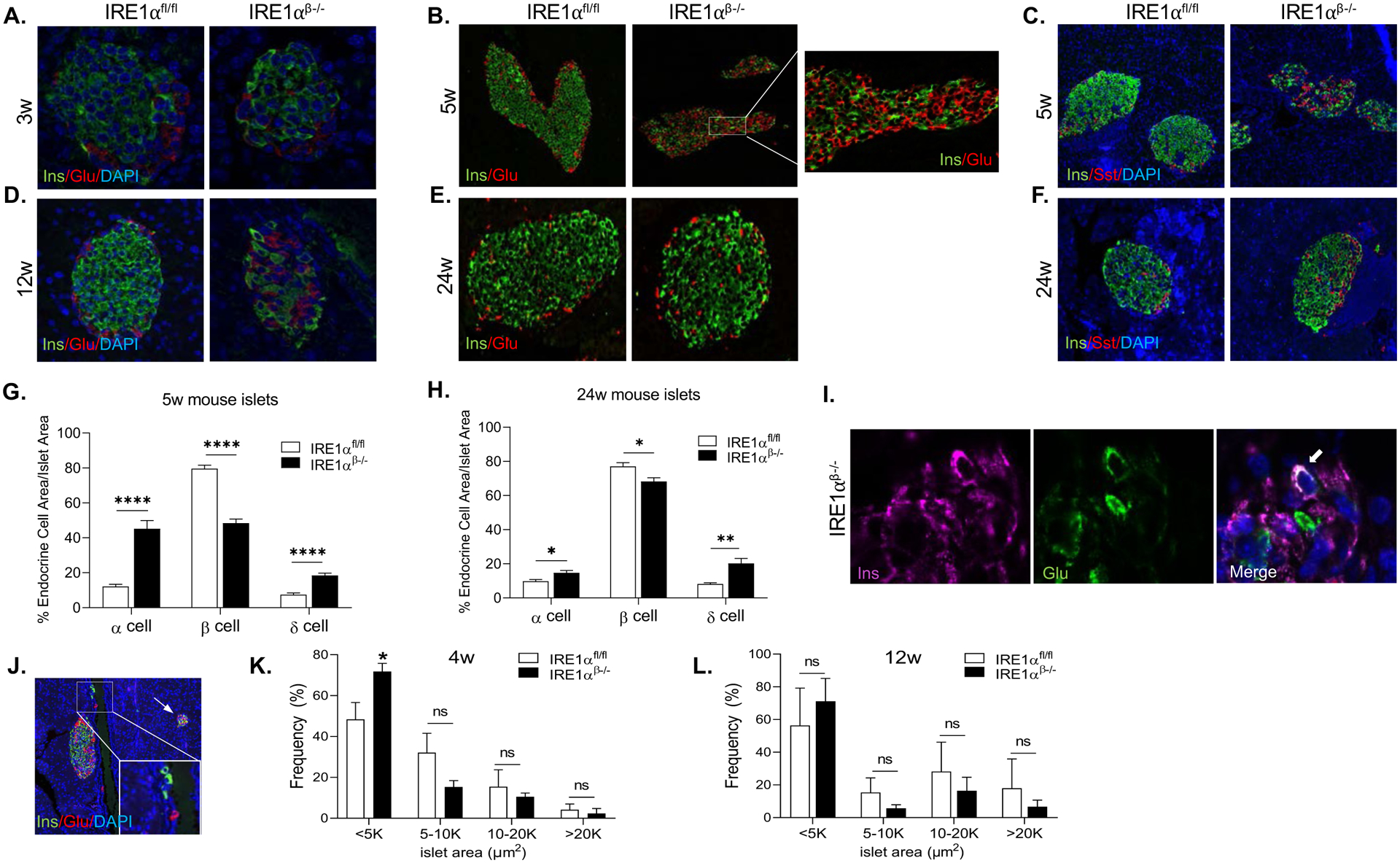
(A and B) Representative immunofluorescence images for insulin and glucagon expression at indicated time points.
(C) Representative immunofluorescence images for insulin and somatostatin expression at 5 weeks of age.
(D and E) Representative immunofluorescence images for insulin and glucagon expression at indicated time points.
(F) Representative immunofluorescence images showing insulin and somatostatin expression at 24 weeks of age.
(G and H) Quantification of α, β, and δ-cells as a percentage of total islet area at 5 and 24 weeks of age (15–25 islets/animal/time point).
(I) Representative image of an insulin+ and glucagon+ bihormonal cell in an islet of IRE1αβ−/− mice. Arrow indicates the bihormonal cell.
(J) Representative image of a pancreatic section from 5-week-old IRE1αβ−/− mice showing the presence of single β-cells and small islet clusters. The arrow points to a small islet cluster.
(K and L) The quantification of islet area of 4 weeks and 12 weeks of age IRE1αfl/fl (n = 3 per time point) and IRE1αβ−/− (4 weeks: n = 3; 12 weeks: n = 4) mice.
All data are represented as mean ± SEM, with statistical analysis performed by Student’s t test (****P < 0.0001, **P < 0.01, *P < 0.05). w: weeks.
These data indicate that during the hyperglycemic phase, islets of IRE1αβ−/− mice show a strikingly disorganized architecture and altered islet composition. However, by 24 weeks of age, β-cell morphology, islet architecture, and function were significantly improved in the islets of IRE1αβ−/− mice.
Bulk RNA-seq on intact islets from hyperglycemic IRE1αβ−/− mice indicates changes in the expression of cell survival and differentiation markers.
To gain insight into the molecular mechanisms and consequences of this altered cellular composition, we sequenced bulk RNA from islets of IRE1αfl/fl and IRE1αβ−/− NOD female mice at 7 weeks (hyperglycemic phase) and 15 weeks of age (recovery phase). Differential expression analysis of IRE1αβ−/− and IRE1αfl/fl mice at 7 weeks of age (hyperglycemic phase) with edgeR (McCarthy et al., 2012; Robinson et al., 2010) at false discovery rate (FDR) of 0.05 identified 2320 upregulated and 1918 downregulated genes (Figures 4A and 4B). Gene set enrichment analysis (Yu et al., 2012) of these differentially expressed genes (DEGs) revealed epithelial-mesenchymal transition, hypoxia, estrogen response, and KRAS signaling as top hits (Figure 4C). Gene ontology (GO) analysis revealed significant enrichment in several biological process, cellular components, and molecular functions. Most notable among these are upregulated extracellular matrix and downregulated vesicle organization (Figures S3A and S3B). In contrast, we identified 342 upregulated and 729 downregulated genes between the IRE1αβ−/− and IRE1αfl/fl mice at 15 weeks of age (recovery phase) with major changes in sterol and cholesterol transporter activity (Figures S3C–S3F). Interestingly, markers of β-cell maturation/de-differentiation were also significantly changed in IRE1αβ−/− mice during the hyperglycemic phase, suggesting a loss of mature β-cell identity under stress conditions (Figure 4D). To examine if the reduced mRNA levels correlate with protein levels, we analyzed the expression of key markers of β-cell maturity, Ucn3 and MafA (Blum et al., 2012; Matsuoka et al., 2004; van der Meulen et al., 2012), in control and IRE1αβ−/− mice via immunofluorescence assay. The protein expression levels of maturity markers were visibly diminished in β-cells of 4-week-old IRE1αβ−/− mice (corresponding to the hyperglycemic phase) compared to control IRE1αfl/fl mice (Figures 4E).
Figure 4. Bulk RNA-seq on intact islets from hyperglycemic mice indicates changes in the expression of cell survival and differentiation markers.
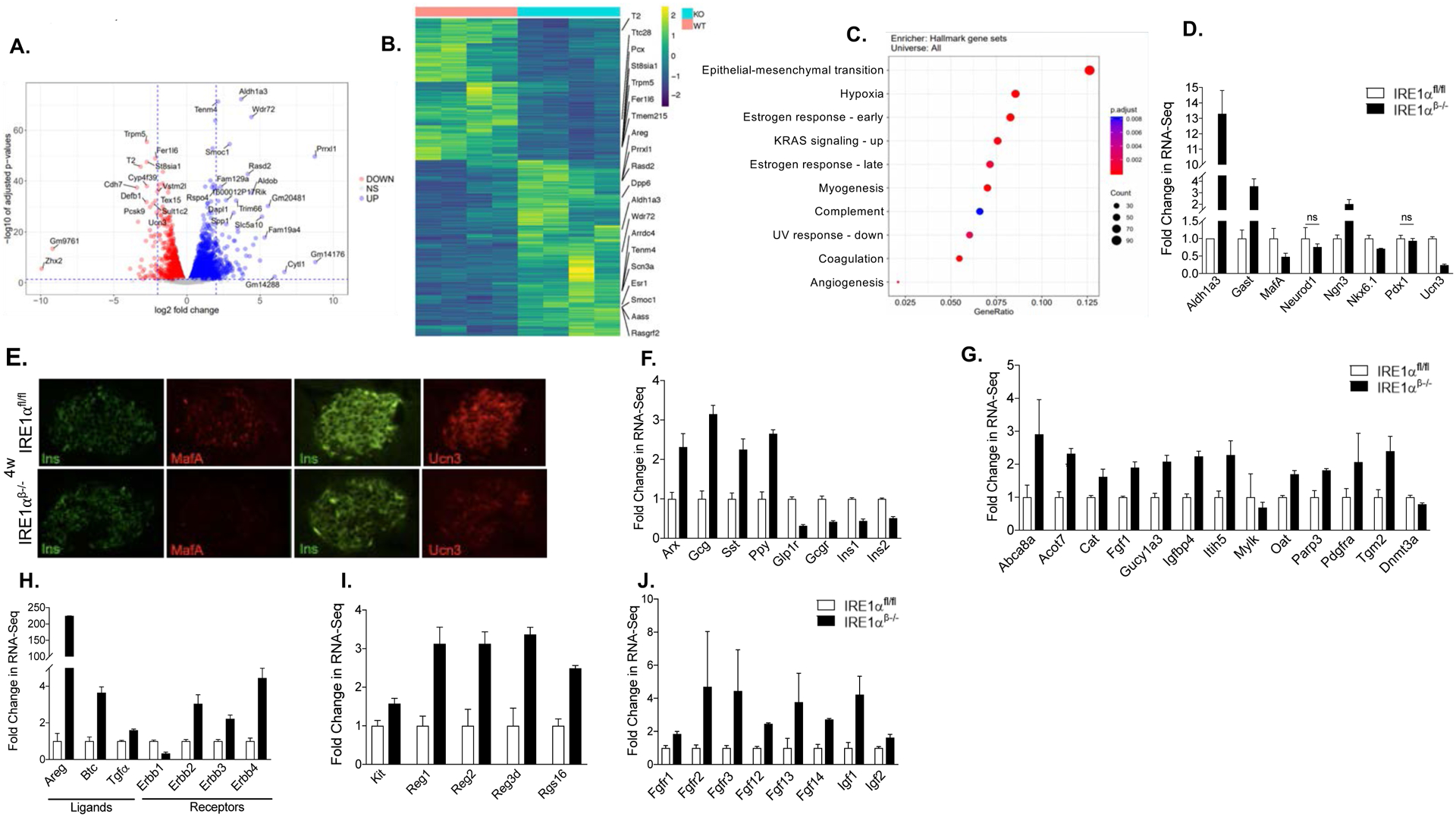
(A) Volcano plots indicating differentially expressed genes. Horizontal line depicts the FDR cutoff of 0.05 and the vertical lines mark log2 fold changes of −2 and 2. Genes with absolute log2 fold change larger than 5 or adjusted p value smaller than 1e-25 and absolute log2 fold change larger than 2 are labeled with their gene symbols.
(B) Heatmap of expression levels for the differentially expressed genes (FDR < 0.01, FC > 2).
(C) Gene set enrichment analysis with the Molecular Signatures Database (MSigDB) Hallmark gene sets (FDR < 0.05).
(D) The mRNA expression of β-cell identity and endocrine progenitor markers (FDR < 0.05).
(E) Immunofluorescence staining showing the expression of β-cell maturity markers MafA, and Ucn3.
(F-J) The mRNA expression of islet cell markers, disallowed genes, ErbB family of genes, regeneration-related genes, and growth factor gene transcripts (FDR < 0.05).
ns: non-significant, w: weeks. FC: Fold change.
Bulk RNA-seq revealed significantly increased expression of islet hormones Glucagon, Somatostatin, PPY, and reduced Insulin 1 and Insulin 2 in IRE1αβ−/− mice (Figure 4F), consistent with the increases in non-β endocrine cells observed by histology (Figure 3). Interestingly, in addition to a substantially diminished expression of the β-cell maturity markers, the expression of β-cell “disallowed genes”, (Pullen et al., 2010; Quintens et al., 2008; Thorrez et al., 2011) which are typically repressed in mature adult β-cells, were markedly increased in IRE1αβ−/− mice (Figure 4G). It has been previously shown that DNA methyl transferase 3a (Dnmt3a) directs the methylation and repression of disallowed genes during β-cell maturation (Dhawan et al., 2015). Consistent with an increased expression of disallowed genes, our RNA-seq data revealed significantly reduced Dnmt3a expression (P value of 7.39e-18) in IRE1αβ−/− islets (Figure 4G). Finally, we detected markedly increased expression of the ErbB family of genes, regeneration-related genes, and growth factors in IRE1αβ−/− islets (Figures 4H–J). Together, bulk RNA-seq on intact islets from IRE1αβ−/− mice indicates alterations in the expression of cell survival and differentiation markers during the hyperglycemic phase.
Single-cell RNA-seq identifies altered proportion of islet cell clusters, hormonal expression, and expression of non-β-cell islet cell markers in β-cells of IRE1αβ−/− mice.
Given that changes in the expression profile in the whole islets of IRE1αβ−/− mice could reflect either changes in individual cells or at the population level because of the altered islet cellular composition, we performed single-cell RNA-seq analysis in disassociated islets obtained from mice that were 5 weeks of age. Monocle analysis of the cells partitioned them into distinct clusters based on their expression profiles (Figure 5A). As expected, the major populations were α-, β-, and δ-cells, with a small proportion of ductal, endothelial, and immune cells. We also identified a minor β-cell population both in wild-type NOD and IRE1αβ−/− mice, indicating a greater degree of heterogeneity in cell identity. We provisionally designated the minor subpopulation of β-cells as “beta2 cells”. The proportion of each population was markedly different in the knockout mice. Cell cluster analysis indicated decreased beta1, and increased, beta2 and α-cell populations in IRE1αβ−/− mice (Figure 5B). Interestingly, percentages of ductal and endothelial cells were also increased in IRE1αβ−/− mice, while minimal immune cell population did not differ between IRE1αfl/fl and IRE1αβ−/− mice (Figures 5A and 5B). These expression changes were not observed in bulk sample averages, indicating that our scRNA-seq method can reveal novel molecular features associated with islet cell composition.
Figure 5. Single-cell RNA-seq identifies altered proportion of islet cell clusters, hormonal expression, and non-β-cell islet markers in IRE1αβ−/− mice.
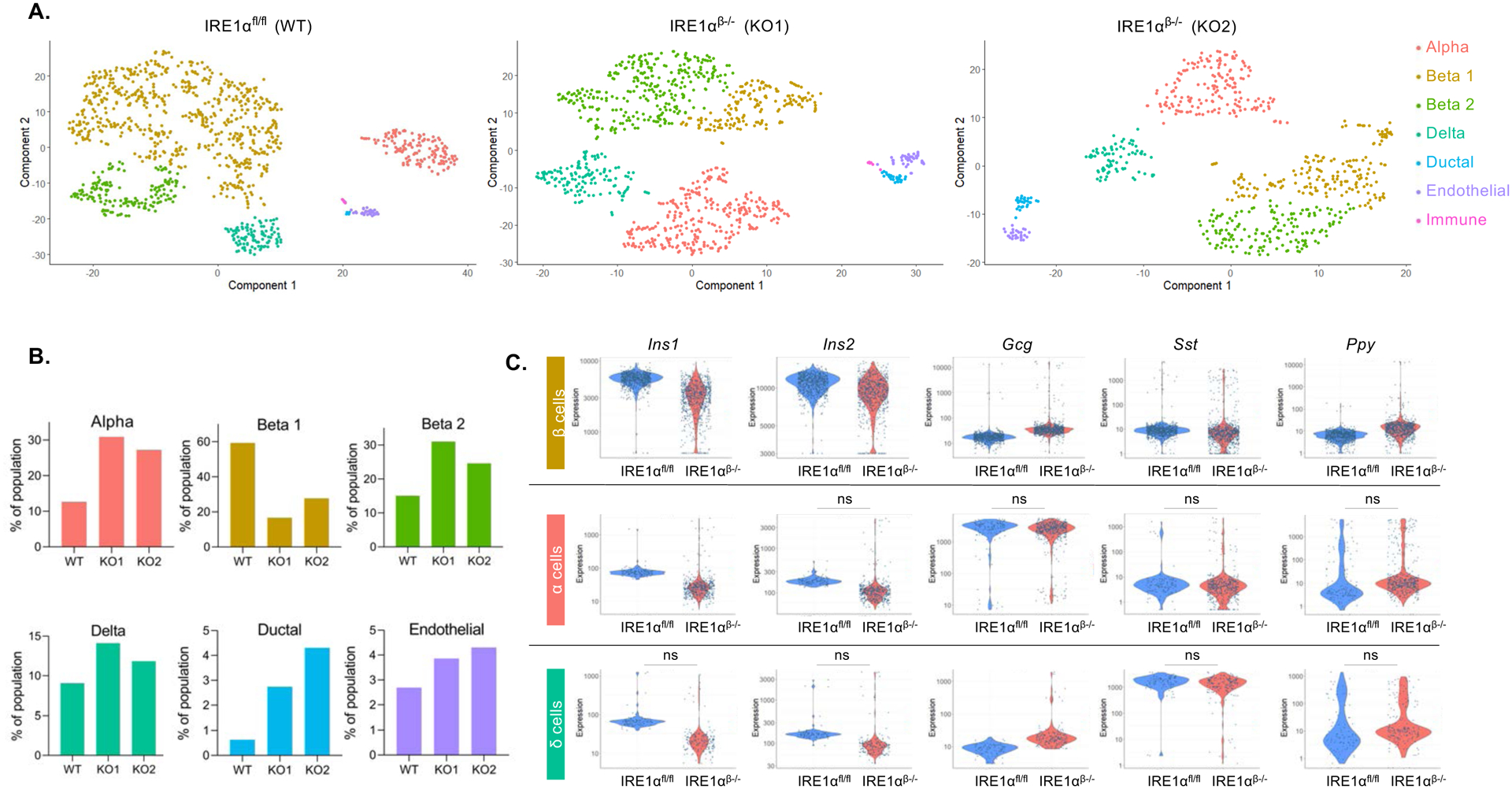
(A) Distinct pancreatic islet cell clusters in IRE1αfl/fl and IRE1αβ−/− mice. Each dot represents a single cell, color-coded according to its cellular identity as defined by gene expression.
(B) Percent of population composed of cell sub-types identified in (A) in dissociated islets obtained from 5-week-old IRE1αfl/fl (WT) and IRE1αβ−/− (knockout 1 (KO1) and knockout2 (KO2)) mice.
(C) Expression of islet hormones in β-cells (upper panel), α-cells (middle panel), and δ-cells (bottom panel) of IRE1αfl/fl and IRE1αβ−/− mice (FDR < 0.01). ns: non-significant.
We then examined the hormonal expression of islet cells. β-cells of IRE1αβ−/− mice demonstrated a polyhormonal phenotype with significantly decreased Insulin1/Insulin2 expression, and markedly increased Glucagon (Gcg), Somatostatin (Sst) and PPY expression (Figure 5C). In α-cells, glucagon expression levels remained similar between the wild-type and knockout mice. While the minimal levels of insulin expressed in α-cells was further reduced, PPY expression was markedly higher in the α-cells of knockout mice (Figure 5C). δ-cells of IRE1αβ−/− mice showed significantly reduced (two-fold with a P value of 2.4e-101) Ins1 expression whereas Gcg expression was markedly upregulated (greater than three-fold with a P value of 4e-198) (Figure 5C).
Loss of IRE1α in β-cells induces dedifferentiation.
To investigate whether the changes in the characteristics of β-cells were only restricted to the hormone genes, we examined the expression of the canonical α-cell (Irx2, Ttr) (Petri et al., 2006; Su et al., 2012) and δ-cell markers (Hhex, Rbp4) (Artner et al., 2010; Zhang et al., 2014) within the β-cell clusters. We identified significantly increased expression of α-cell (Irx2, Ttr) (Figure 6A) and δ-cell markers (Hhex, Rbp4) (Figure 6B). Interestingly, the changing expression pattern was accompanied by a reduction in the number of cells expressing β-cell maturity markers such as MafA and Ucn3 (Blum et al., 2012; van der Meulen et al., 2012) (Figure 6C). There were also more cells expressing “disallowed” genes such as Olfm1 and Ndrg4 (Pullen et al., 2010) (Figure 6D; Figure S4A). Concomitant with the lack of maturity, there was a marked increase in the expression of dedifferentiation genes Rfx2 and Fabp3 (Kim-Muller et al., 2016; Szabat et al., 2011). These data, along with significantly increased expression of disallowed genes and endocrine progenitor cell markers (Aldh1a3, Gast) (Cinti et al., 2016; Gittes et al., 1993) (Figure 6E), confirmed the dedifferentiation of β-cells in IRE1αβ−/− mice. Interestingly, we also detected markedly increased expression of regeneration/proliferation-related genes in β-cells of IRE1αβ−/− mice (Figure S4B). Lastly, we demonstrated that these dedifferentiated cells indeed lost IRE1α by examining the gene expression profile of targets of sXBP1. Expression of many of the sXBP1 targets (Fkbp11, Erol1b, Fkbp2, Sec61g, Pdia5) was significantly reduced in β-cells (Lee et al., 2003) (Figure 6F; Figure S4C).
Figure 6. β-cells of IRE1αβ−/− mice dedifferentiate.
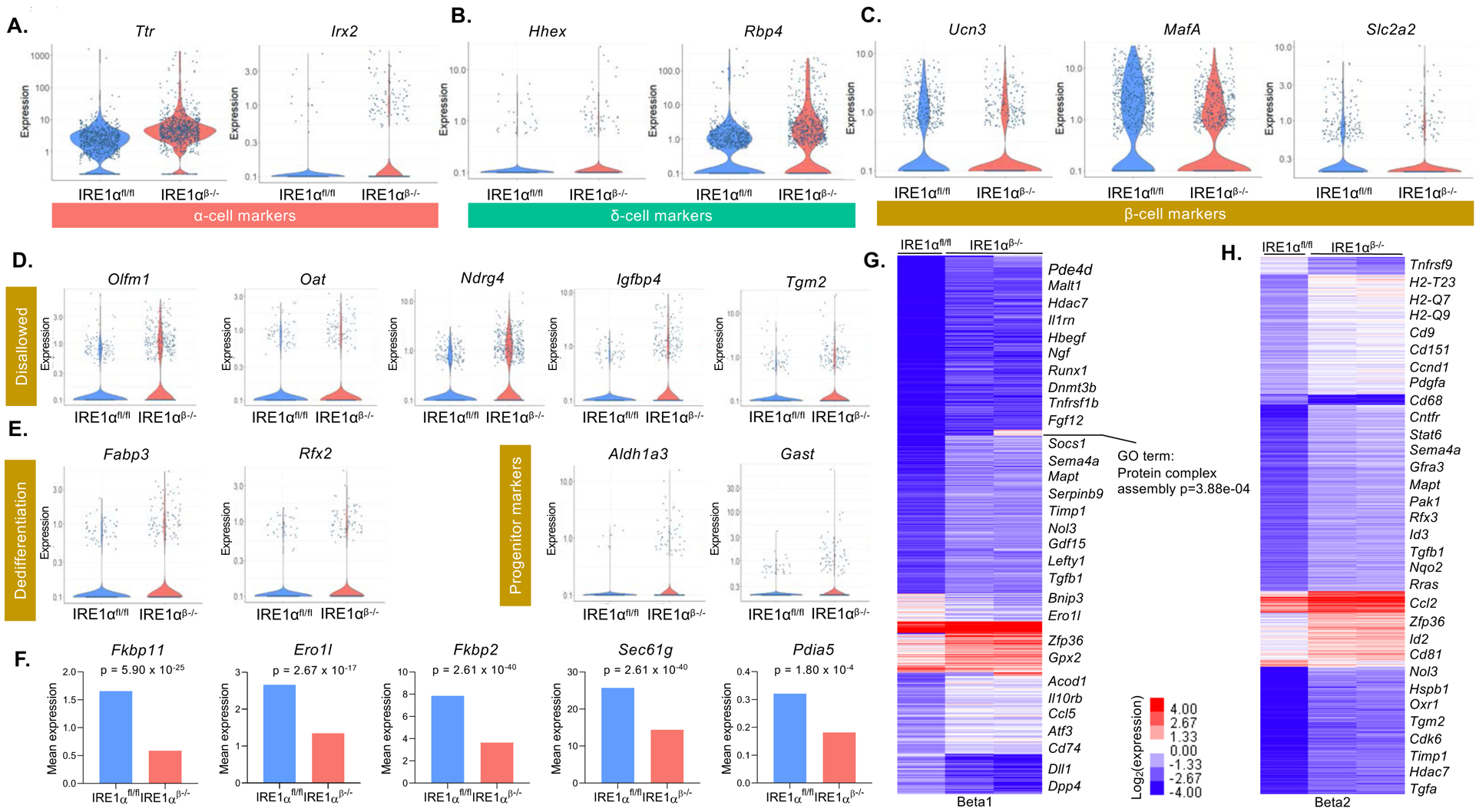
(A-C) Expression of α-cell markers, δ-cell markers, and β-cell maturity markers in β-cell clusters of IRE1αfl/fl and IRE1αβ−/− mice at 5 weeks of age (FDR < 0.01).
(D and E) Expression of disallowed genes, dedifferentiation, and endocrine progenitor markers in β-cell clusters of IRE1αfl/fl and IRE1αβ−/− mice (FDR < 0.01).
(F) Mean expression of sXBP1 target genes in β-cell clusters of IRE1αfl/fl and IRE1αβ−/− mice.
(G and H) k-means clustering (7 clusters) of differentially expressed genes (FDR < 0.01, FC > 2) among the beta1 and beta2 populations (columns). Selected genes that define each cluster are displayed. Color bar represents expression changes in log2 scale. FC: Fold change.
Next, we performed differential gene expression analysis using the Monocle package. Using this approach, we identified 469 DEGs in beta1 and 412 DEGs in beta2 cell clusters (FDR < 0.01, FC > 2) (Figures 6G and 6H). The most significantly DEGs in these β-cell clusters were associated with cell differentiation, growth, survival, and immune inhibition whereas expression of several markers associated with pancreas development and protein secretion were significantly downregulated (Figures 6G and 6H). Taken together, bulk RNA-seq from intact islets, sc-RNAseq, and histological analyses identify initiation of β-cell dedifferentiation in IRE1αβ−/−mice prior to insulitis.
β-cells of IRE1αβ−/− mice have altered expression of genes associated with immune cell recruitment.
β-cells can actively participate in their autoimmune destruction by affecting the local homing of inflammatory cells, antigen presentation, and the levels of autoantigen or neoantigen expression. Thus, we examined the expression of genes that are involved in immune regulation in β-cells of IRE1αβ−/− mice. Interestingly, several immune-related genes and genes that are involved in immune cell recruitment were differentially regulated in β-cells of IRE1αβ−/− mice. Among these, significantly increased expression of genes that play a key role in the suppression of T-cell, B-cell, and macrophage activities was notable (Figure 7A). For example, the expression of immune inhibitory ligands Qa-2 (H2-Q7/H2-Q9) and Qa-1 (H2-T23), which were shown to play a major role in the suppression of CD4+ T-cell and NK cell responses (Carosella et al., 2008; Jiang et al., 1995; Klein et al., 1983; Robinson et al., 1989), was significantly upregulated (H2-Q7 P = 1.94e-20, H2-T23 P = 6.66e-25) in β-cells of IRE1αβ−/− mice. Interestingly, we found that the expression of TNF receptor superfamily (Tnfrs) genes, which play a key role in the antigen presentation and in the generation of cytotoxic T-cells (Ward-Kavanagh et al., 2016), was significantly downregulated (e.g., Tnfrsf9 P = 1.3257e-4, Tnfrsf23 P = 1.94e-10). The expression of several members of the tetraspanin family of genes (Cd81, Cd9, Cd151), which also play important roles in the regulation of pattern recognition, antigen presentation, and T-cell proliferation (Jones et al., 2011), was also significantly altered in IRE1αβ−/− mice.
Figure 7. -cells of IRE1αβ−/− mice have altered expression of genes associated with immune cell recruitment.
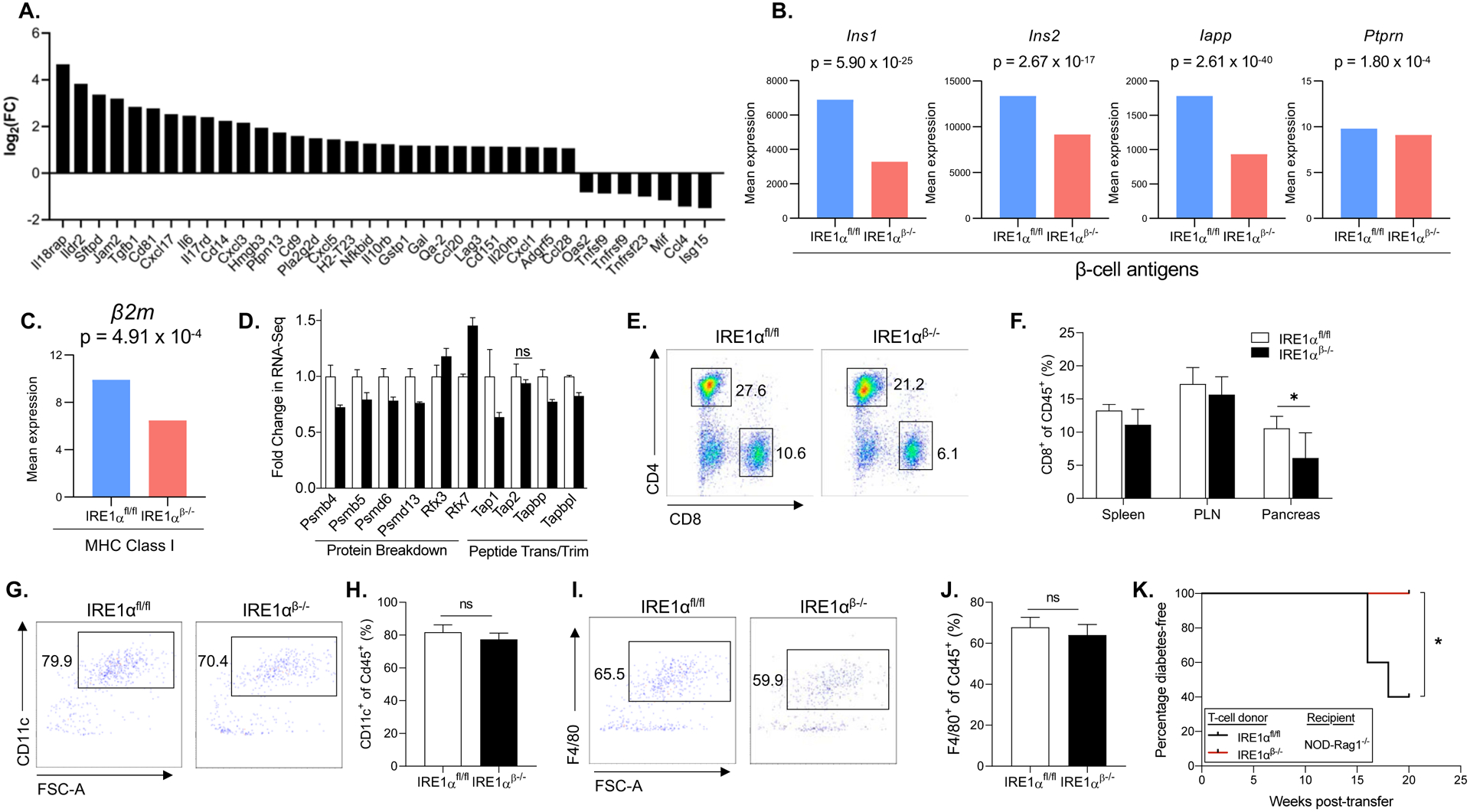
β (A) The expression of genes that is key in regulation of lymphocyte activation, as well as markers of cytokine, chemokine, and ECM (FDR < 0.01).
(B) The mRNA expression of β-cell autoantigens in β-cells.
(C and D) The mRNA expression of MHC class I component β2m and genes that are involved in the MHC class I loading (FDR < 0.05).
(E) Fractions of CD4+ and CD8+ T-cells in representative dot plots from pancreata of 21 weeks of age IRE1αfl/fl and IRE1αβ−/− mice after pre-gating for single, viable, and CD45+ cells.
(F) Immunophenotyping data showing percentage of CD8+ T-cells in spleen, pancreatic lymph node (PLN), and pancreas from 21 weeks of age IRE1αfl/fl (n = 6) and IRE1αβ−/− (n = 4) mice.
(G) Fractions of CD11c+ dendritic cells in representative dot plots from pancreata of 5 weeks of age IRE1αfl/fl and IRE1αβ−/− mice after pre-gating for single, viable, and CD45+ cells.
(H) Quantification of percentage of CD11c+ dendritic cells in pancreata from 5 weeks of age IRE1αfl/fl (n = 6) and IRE1αβ−/− (n = 7) mice.
(I) Fractions of F4/80+ macrophages in representative dot plots from pancreata of 5 weeks of age IRE1αfl/fl and IRE1αβ−/− mice after pre-gating for single, viable, and CD45+ cells.
(J) Quantification of percentage of F4/80+ macrophages in pancreata from 5 weeks of age IRE1αfl/fl (n = 6) and IRE1αβ−/− (n = 7) mice.
(K) Percentage of diabetes-free NOD-Rag1−/− mice (n = 5 per group) post-total T-cell transfer from 8-week-old IRE1αfl/fl and IRE1αβ−/− mice. The incidence of diabetes was compared by log-rank (Mantel-Cox) test (*P < 0.05).
We demonstrated that protein expression of proinsulin and insulin, key autoantigens in triggering T1D (Arvan et al., 2012; Narendran et al., 2003; You and Chatenoud, 2006), was significantly reduced in IRE1αβ−/− mice (Figures 2F and 2G). We further examined whether the expression of other autoantigens was also altered in β-cells of IRE1αβ−/− mice. In addition to proinsulin and insulin, scRNA-seq revealed substantially reduced expression of additional β-cell autoantigens Insulin1/2, IAPP, and Ptprn in IRE1αβ−/− mice (Figure 7B). Major histocompatibility complex (MHC) class I molecules, which present peptides derived from intracellular proteins to CD8+ T-cells, are assembled in the ER; ER stress was shown to affect MHC class I expression as well as processing of MHC class I-associated peptides (Granados et al., 2009; Ulianich et al., 2011). To elucidate whether the antigen presentation pathway was altered in IRE1αβ−/− mice, we examined the expression of MHC class I components and the peptide loading pathway genes. Analysis of sc-RNAseq data identified significantly diminished expression of β2-microglobulin (β2m) (Figure 7C) and marked alterations in the expression of several MHC class I peptide loading pathway genes, suggesting that antigen processing was defective in IRE1αβ−/− mice (Figure 7D).
To evaluate the effects of IRE1α deficiency on the immune cells, we performed immunophenotyping in the pancreas, spleen, and pancreatic lymph nodes (PLNs) of these mice at 21 weeks of age. While there were no significant alterations in the percentage of CD4+ T-cells, the percentage of CD8+ T-cells in the pancreata of IRE1αβ−/− mice was significantly reduced (Figures 7E and 7F). The percentage of T regulatory cells (Tregs) (Figures S5A and S5B), B-cells (Figures S5C and S5D), or macrophages (Figures S5E and S5F) did not show significant alterations in the pancreas. There were also no differences in these immune cell populations within the lymph nodes and spleen in control vs. IRE1αβ−/− mice (Figures S6 and S7). Hence, deletion of IRE1α in β-cells in NOD mice significantly reduced the relative representation of CD8+ T-cells specifically in the pancreas, whereas other immune cell populations did not show any obvious alterations. Of note, these mice show significantly less islet infiltration by immune cells, suggesting that better islet function and survival recruited fewer immune cells to the islets, resulting in less antigen presentation. While our immunophenotyping studies analyze the immune cells in the later stage of the disease, it is well established that macrophages and dendritic cells play a critical role during the initiation of the disease process. Thus, we examined whether the relative representation of these immune cells was altered in the islets of control and IRE1αβ−/− mice at 5 weeks of age. However, we did not detect any significant differences in the proportions of Cd11c+ dendritic and F4/F80+ macrophage cell populations in IRE1αfl/fl and IRE1αβ−/− mice, suggesting that recruitment of these immune cells into the islets was unaltered (Figures 7G–J). Finally, to determine the diabetogenic potential of T-cells of IRE1αβ−/− mice, we performed adoptive transfer experiments and transferred purified total T-cells of 8-week-old IRE1αfl/fl and IRE1αβ−/− mice into 5–6-week-old female immunodeficient NOD-Rag1−/− mice and monitored them for diabetes development. Recipient mice transferred with IRE1αfl/fl mouse T-cells developed diabetes at 16 weeks after the cell transfer, and 60% of recipient mice became diabetic by 20 weeks after cell transfer (Figure 7K). In contrast, transfer of IRE1αβ−/− T-cells did not induce diabetes in NOD-Rag1−/− mice, suggesting that IRE1αβ−/− T-cells were not diabetogenic.
Taken together, our data suggest that IRE1α deletion in β-cells, prior to insulitis, promotes a transient β-cell dedifferentiation, which significantly diminishes autoantigen expression and antigen processing, and increases the expression of immune inhibitory markers within the β-cells. These phenotypic changes in β-cells early in life in IRE1αβ−/− NOD mice most likely have effects on the autoimmune responses leading to substantially reduced CD8+ T-cells in the pancreas. The T-cell adoptive transfer experiment further indicates that there are long-lasting effects on IRE1αβ−/− T-cells, rendering them incapable of inducing diabetes in NOD-Rag1−/− mice.
Discussion
To investigate the β-cell-specific function of the key UPR sensor IRE1α in T1D, we generated NOD IRE1αβ−/− mice by exposing pups to tamoxifen via their dam’s milk to achieve IRE1α deletion prior to islet infiltration of immune cells, which usually occurs later in the postnatal period. These mice, after a transient mild hyperglycemia, were protected from T1D. Recently, β-cell-specific deletion of IRE1α, driven by Ins-Cre in unstressed wild-type mice (mixed C57BL/6 × 129/SvJae background), was shown to result in hyperglycemia starting from 4 weeks of age lasting up to at least 24 weeks of age (Tsuchiya et al., 2018). Similarly, deletion of IRE1α in adult β-cells led to diabetes under a non-autoimmune context (C57BL/6 background) (Hassler et al., 2015). Interestingly, unlike NOD IRE1αβ−/− mice, no β-cell dedifferentiation, bihormonal islet cells, and altered islet architecture or composition were observed in these IRE1α-deleted mouse models, despite presence of chronic hyperglycemia (Hassler et al., 2015; Tsuchiya et al., 2018). These data indicate that loss of IRE1α in β-cells have remarkably different outcomes in the context of β-cell stress as seen in NOD mice.
Stress-induced dedifferentiation is well described in plants and mammalian somatic cells, such as Schwann cells, cardiac myocytes, germ cells, and β-cells (Bersell et al., 2009; Chen et al., 2007; Talchai et al., 2012). Cells can use dedifferentiation as an adaptive mechanism to minimize damage (Puri et al., 2015; Shoshani and Zipori, 2011). Our scRNA-seq, bulk RNA-seq, and histological analyses demonstrate that β-cells of IRE1αβ−/− mice have markedly increased expression of disallowed genes, and increased expression of markers of progenitor cells (Aldh3a1, Gast, Ngn3), as well as reduced gene and protein expression of β-cell maturity markers (MafA and Ucn3). In addition, the presence of bihormonal (Ins+, Glu+) cells detected in pancreatic sections indicates that β-cells of IRE1αβ−/− mice similarly underwent a reversible dedifferentiation process under the chronic stressed background of NOD mice. Of note, immune-independent β-cell fragility, as a result of genetic variations in Glis3 and Xrcc4, was shown to alter the responses of β-cells to ER stress in NOD mice (Dooley et al., 2016).
Our histological and scRNA-seq data indicate increased numbers of α-cells, suggesting that an α-cell to β-cell conversion could potentially be a mechanism for the restoration of the β-cell population in islets of IRE1αβ−/− mice. However, conversion of α-cells to β-cells was reported only after extreme β-cell loss (>90%) (Thorel et al., 2010), and our scRNA-seq analysis did not indicate a “β-cell like” signature in α-cells of IRE1αβ−/− mice, suggesting that this may not be the main mechanism of recovery from hyperglycemia. Successful re-differentiation of dedifferentiated β-cells was reported in both mouse and human islets (Gershengorn et al., 2004; Ouziel-Yahalom et al., 2006; Wang et al., 2014). In addition, reduced insulin production was shown to promote β-cell proliferation in a cell-autonomous manner (Szabat et al., 2016). Interestingly, a recent study shows that increased proliferation of β-cells prior to insulitis in NOD mice is protective against T1D (Dirice et al., 2019). Thus, increased proliferation of immature β-cells of IRE1αβ−/− mice, and/or non-recombined cells prior to insulitis, might have contributed to a diabetes-protected phenotype. However, we detected significantly less proliferation in IRE1αβ−/− mice at 3 weeks of age, and no difference in proliferation at 5 weeks of age. Indeed, our results are consistent with previous reports indicating significantly reduced β-cell proliferation upon deletion of UPR sensors Perk and Xbp1 in β-cells (Lee et al., 2011; Zhang et al., 2006), ruling out the possibility that increased proliferation prior to insulitis contributes to the protection from T1D in IRE1αβ−/− mice. At the molecular level, Betacellulin, a ligand in ErbB signaling, was shown to play a key role in the re-differentiation and restoration of β-cell gene expression and insulin content in human islets (Ouziel-Yahalom et al., 2006). Consistent with this observation, both our bulk and single-cell RNA-seq indicate markedly increased expression of genes that are involved in β-cell growth and the ErbB pathway. Indeed, Amphiregulin (Areg), an ErbB pathway ligand, has a pro-regenerative function and plays an important role in promoting the healing, and regeneration of multiple tissues, and was our top hit in bulk RNA-seq (greater than two hundred-fold) (Burzyn et al., 2013; Monticelli et al., 2011; Shao and Sheng, 2010). In addition to redifferentiation, neogenesis might also have contributed to the recovery from hyperglycemia in IRE1αβ−/− mice. Indeed, we observed small islet clusters (<10 insulin-positive cells), and the occasional lone β-cell in pancreata of IRE1αβ−/− mice. The bulk RNA-seq demonstrated increased expression of regeneration genes in islets and scRNA-seq revealed significantly increased ductal cell clusters in IRE1αβ−/− mice. Unfortunately, as Cre- or reporter-lines on the NOD background for islet and ductal cells are not currently available, these possibilities cannot yet be explored directly with lineage tracing experiments.
How can the loss of IRE1α in β-cells protect against autoimmune destruction? Could undifferentiated, immature β-cells have reduced antigenicity and altered immune activating/regulating signatures that can avoid autoimmune destruction? Insulin and proinsulin have a key role in the initial triggering of the autoimmune response and driving of autoimmune β-cell destruction (Arvan et al., 2012; Nakayama, 2011; Nakayama et al., 2005). Interestingly, among all the different islet cell types, only the highly secretory β-cells are specifically targeted by immune cells in T1D. Thus, reducing insulin levels and allowing highly secretory β-cells to rest during a critical window of the disease, together with altering ER functional capacity to disrupt assembly of MHC complex and peptide processing, may be crucial to prevent subsequent immune-mediated destruction of β-cells. Recently, a specific sub-population of β-cells (~15%) in NOD mice was described to resist autoimmune attack. These β-cells expressed reduced levels of β-cell-specific genes in the face of autoimmunity, suggesting de-differentiation of β-cells (Rui et al., 2017). In addition to expressing significantly lower levels of insulin, these immune-resistant β-cells exhibited substantially reduced expression of autoantigens (Igrp, ZnT8, Gad1, Ia-2), and markedly increased expression of immune inhibitory genes (Qa-2, Cd81) compared to normal β-cells. While these cells did not show altered expression of a subset of ER stress related genes, none of the markers assessed in that study were direct targets of the IRE1α/sXBP1 branch of the UPR (Rui et al., 2017). Thus, in the same vein, in the presence of immune-independent β-cell fragility in NOD mice (Dooley et al., 2016), IRE1α deletion might have caused loss of mature β-cell identity. Immature β-cells may have escaped autoimmune attack due to their significantly reduced expression of autoantigens, altered antigen processing, and upregulated expression of immunomodulatory genes. Consistent with this, levels of major autoantigens Proinsulin, Insulin, Iapp, and Ptprn were significantly reduced in dedifferentiated β-cells of IRE1αβ−/− mice. Moreover, ER stress and the UPR can directly affect MHC class I assembly, peptide processing, and antigen presentation by regulating protein translation, degradation, decay of ER mRNAs, and ER homeostasis (Granados et al., 2009; Ulianich et al., 2011). Indeed, we identified markedly reduced expression of MHC class I component β2m and altered expression of MHC class I peptide loading pathway genes in IRE1αβ−/− mice, suggesting that protection of IRE1αβ−/− mice from T1D was in part due to UPR-dependent defects in production and processing of autoantigens, which in turn significantly reduced cytotoxic CD8+ T-cells and islet infiltration. Protection from autoimmune destruction is further supported by significant upregulation of immune inhibitory markers, downregulation of genes that are implicated in immune cell activation, and markedly altered expression of chemokines, cytokines, and ECM proteins which play an important role in immune cell recruitment.
NOD-Rag1−/−; IRE1αβ−/− mice exhibited the same β-cell functional alteration as seen in NOD IRE1αβ−/− mice. Thus, the phenotypic changes of the β-cells in IRE1αβ−/− NOD mice are likely cell-intrinsically regulated, but not a result of altered adaptive immune cells. On the other hand, two findings suggest that β-cell dedifferentiation observed early in life in NOD IRE1αβ−/− mice alters the autoimmune response of the adaptive immune cells. First, the majority of NOD IRE1αβ−/− mice remained non-diabetic at 50 weeks of age. Second, T cells isolated from 8-week-old NOD IRE1αβ−/− mice could not induce diabetes in NOD-Rag1−/− recipients. Collectively, our results support the idea that dedifferentiation of β-cells in young NOD IRE1αβ−/− mice has long-lasting effects on the diabetogenic activity of T-cells. There are several non-mutually exclusive mechanisms that could contribute to reduced diabetogenic activity of IRE1αβ−/− T-cells. β-cell autoreactive CD8+ T-cells could be tolerized in the forms of anergy or deletion when they are not properly stimulated in NOD IRE1αβ−/− mice. It is also possible that T-cells with regulatory functions are enhanced in NOD IRE1αβ−/− mice. While we did not observe a proportional difference in FOXP3+ CD4+ Tregs in NOD IRE1αβ−/− mice, the possibility that they are functionally enhanced cannot be ruled out. Future studies are needed to determine the mechanism underlying immune tolerance induction of β-cell autoreactive T-cells in NOD IRE1αβ−/− mice.
Aberrant expression of the UPR genes was detected in β-cells of mouse models of diabetes and human patients (Engin et al., 2014; Engin et al., 2013). Mitigation of ER stress and restoration of the UPR dysfunction with a chemical chaperone, TUDCA, prevented diabetes in pre-clinical T1D models (Engin et al., 2013). TUDCA is currently under Phase I clinical trial (NCT02218619) for new-onset T1D patients. Interestingly, the tyrosine kinase inhibitor imatinib, currently being tested in a Phase II clinical trial (NCT01781975) for the treatment of new-onset T1D, was recently demonstrated to blunt RNase activity of IRE1α and reverse T1D in the NOD mouse model (Louvet et al., 2008; Morita et al., 2017). These studies support the notion that modulating β-cell UPR can be a promising therapeutic strategy for people at high-risk for T1D.
Autoantibodies directed against β-cell proteins are used as biomarkers for risk prediction and clinical diagnosis. The presence of multiple autoantibodies is associated with a high risk of progression to overt disease (Regnell and Lernmark, 2017). The relationship between autoantibody positivity and the presence or absence of insulitis is an actively pursued research area (Pugliese, 2016). Emerging data suggest that donors with multiple autoantibodies can have absence of insulitis (In’t Veld et al., 2007; Wiberg et al., 2015). Thus, inducing a reversible dedifferentiation state for β-cells to limit their antigen availability during this critical therapeutic window may provide an important non-immune-based preventive or therapeutic strategy in high-risk individuals. Interfering with antigen processing and presentation, by modulating β-cell ER functional capacity and the UPR, can further support diabetes protection. Whether similar strategies can be applicable to prevent other autoimmune diseases associated with highly secretory target cells remain to be tested. Future studies identifying the function of IRE1α and the other UPR sensors during different stages of T1D progression will be necessary to fully reveal the role of β-cell ER stress and the UPR in T1D.
Limitations of study
Our current breeding scheme does not allow us to obtain the littermate control mice expressing Cre transgene alone. Thus, the mice used to identify the effects of Cre expression on diabetes progression were not obtained from the experimental group. Although we confirmed that Cre transgene levels in these mice did not differ from knockout mice, and Cre transgene did not alter diabetes progression and pathology in NOD mice, we still consider this a limitation. In addition, due to the lack of reporter lines on NOD background, we were not able to perform lineage tracing experiments to definitively identify the contribution of neogenesis or transdifferentiation to the recovery from hyperglycemia in IRE1αβ−/− mice.
STAR Methods
LEAD CONTACT AND MATERIALS AVAILABILITY
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Feyza Engin (fengin@wisc.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Mouse lines and tamoxifen injections:
The animal care and experimental procedures were carried out in accordance with the recommendations of the National Institutes of Health Guide for the Care and Use of Laboratory Animals. The protocol (#M005064-R01-A03 by F.E. for mice) was approved by the University of Wisconsin-Madison Institutional Animal Care and Use Committee. Female NOD/ShiLtJ mice and Rag1−/− mice were purchased from Jackson Laboratory. All animals had ad libitum access to food (Envigo 2919) and water and were housed at 20–24°C on a 12 h light/12 h dark cycle. Mice were bred and maintained under specific pathogen-free conditions at University of Wisconsin-Madison under approved protocols. The IRE1α floxed mice were a gift of Dr. Takao Iwawaki (Kanazawa Medical University). Ins2-CreERT/+ mice were a gift of Dr. Douglas Melton (Harvard University). Mice were backcrossed to NOD background more than 10 generations. The genetic backgrounds of all intercrossed mouse models were verified by Genome Scan Services (purity of IRE1αfl/fl mice on NOD background >99.8% via Jackson Laboratory’s Genome Scan Service, purity of Ins2CreERT mice on NOD background >99.9% via Neogen, MiniMUGA array).
To induce Cre recombinase activity, tamoxifen (T5648; Sigma-Aldrich) was dissolved in sterilized corn oil (C8267; Sigma-Aldrich) by shaking overnight in a 37°C incubator. The solution was protected from light, and 10 mg/ml tamoxifen was administrated to dams the day after delivery via intraperitoneal injection twice every 24 hours in five consecutive days. Animals were observed daily for health status, any mice that met IACUC criteria for euthanasia were immediately euthanized. Experiments were performed on female mice between 3 and 50 weeks of age.
METHOD DETAILS
Histological analyses:
Pancreata from mice were fixed with 10% zinc formalin overnight and paraffin embedded. 5-μm sections of the pancreata were generated, and staining was performed after blocking with 5% normal goat serum with the following antibodies: anti-Insulin (Linco), anti-Glucagon (Cell Signaling), anti-Somatostatin (Santa Cruz), anti-Ki67 (Cell Signaling), anti-CD3 (Novus Biologicals), anti-MafA (Cell Signaling), anti-Ucn3 (Phoenix Pharmaceuticals Inc.), Alexa Fluor 488 (Invitrogen), and Alexa Fluor 568 (Invitrogen) using established protocols. After staining, slides were mounted with antifade mounting medium containing 4,6-diamidino-2-phenylindole (DAPI) (Vector Laboratories). In some cases, the harvested pancreata were fixed in 4% paraformaldehyde (VWR), embedded in OCT (Sakura), and frozen before being sectioned at 10 μm. Antigen retrieval was performed by using citrate buffer pH 6.0 (paraffin) or HistoVT (Nacalai Tesque) (frozen). Islet size was determined by manually circling insulin positive clusters in the Fiji software (Schindelin et al., 2012). Insulitis scoring was performed on step sections (three levels separated by 200-μm) of paraffin-embedded and hematoxylin-eosin stained sections. “peri‐insulitis” is defined as focal aggregation at one pole of the islet and in contact with the islet periphery. “non-aggressive insulitis” refers to lesions with a clear, and often extensive, islet infiltrate occupying less than 50% of the islet area, whereas “aggressive insulitis” refers to an extensive infiltrate, where lymphoid cells invade the entire islet and intermingle with endocrine cells, showing extensive signs of β-cell damage. The images of the pancreatic sections were obtained using a Nikon A1R-SI+ confocal microscope and a Nikon Storm/Tirf/Epifluorescence. The images were analyzed by ImageJ or Fiji. Two blinded individuals independently performed manual analyses and insulitis scoring.
Cell death (TUNEL) assay:
DeadEnd Fluorometric TUNEL assay (Promega Corporation, Madison, WI) was performed on formalin-fixed, paraffin-embedded pancreatic sections according to the manufacturer’s instructions.
Islet isolation:
Islets were isolated using the standard collagenase/protease digestion method. Briefly, the pancreatic duct was cannulated and distended with 4°C collagenase/protease solution using Collagenase P (Sigma-Aldrich, USA) in 1x Hank’s balanced salt solution and 0.02% bovine serum albumin. The protease reaction was stopped using RPMI 1640 with 10% fetal bovine serum. Islets were separated from the exocrine tissue using Histopaque-1077 (Sigma-Aldrich, USA). Hand-picked islets were cultured overnight at 37°C in RPMI-1640 media containing 10% FBS and 1% antibiotic/antimycotic (Thermo Fisher Scientific) before use in experiments.
Insulin, proinsulin content:
For measurement of whole pancreas insulin and proinsulin content, mice were sacrificed after a 4-hour fast and their whole pancreas insulin/proinsulin content (μg/pancreas) was assessed by acid-ethanol extraction followed by ELISA (Alpco, Salem, NH). Samples were done in duplicate.
Glucose tolerance test:
Glucose tolerance tests were performed on IRE1αfl/fl and IRE1αβ−/− mice simultaneously after an overnight (16 hours) fast. Blood glucose levels were measured at 0, 15, 30, 60, 90, and 120 minutes after an intraperitoneal administration of glucose at dose of 2g/kg body weight.
Immunophenotyping:
Prior to organ dissection, mice were perfused with 20 ml PBS to eliminate contaminating blood leukocytes. Single-cell suspensions of the pancreata were prepared by Collagenase P (Roche) digestion. Cells from pancreatic lymph nodes and spleen were prepared by physical dissociation. The spleen was treated with ACK lysing buffer (Thermo Fisher Scientific). All stainings began with an incubation with TruStain fcX anti-mouse CD16/32. Antibodies used for subsequent stainings were: anti-CD45 (30-F11), -CD19 (6D5); -CD3 (145–2C11), -CD4 (RM4–5), -CD8 (53–6.7), -CD25 (PC61), CD11b (M1/70), -CD11c (N418), -F4/80 (BM8), and -Gr1 (RB6–8C5) (all from BioLegend). Intracellular Foxp3 (FJK-16s) staining was performed according to eBioscience’s protocol. Samples were acquired with an Attune NxT flow cytometer (Thermo Fisher Scientific) and data were analyzed with FlowJo software (Tree Star, Inc.).
Adoptive transfer:
The spleens from 8-week-old IRE1αfl/fl and IRE1αβ−/− mice were physically dissociated before filtering with a 40 μM nylon mesh. After incubation with ACK lysing buffer (Thermo Fisher Scientific), total T cells were isolated by negative selection (Pan T cell isolation kit II, Miltenyi Biotec), and 4×106 cells were injected intravenously into 5–6-week-old NOD-Rag1−/− female recipients. The recipients were followed for diabetes incidence with weekly blood glucose measurements. The purity of the transferred T-cells (>95%) was analyzed by flow cytometry using anti-TCRb, anti-CD4, and anti-CD8 antibodies (BD Biosciences).
Bulk RNA-seq:
RNA-seq was performed on islets obtained from 7-week-old and 15-week-old IRE1αfl/fl (7 weeks: n = 4; 15 weeks: n = 3) and IRE1αβ−/− (n = 4 per timepoint) female NOD mice. Briefly, following isolation, RNA was extracted using RNeasy Plus Mini Kit (Qiagen), including a column for elimination of genomic DNA. RNA concentration was determined using Qubit RNA HS Assay Kit (Life Technologies). RNA Integrity Number (RIN) was measured using Agilent RNA 600 Nano Kit (Agilent Technologies). RIN > 7 was used in the experiments. RNA library was generated using the TruSeq Stranded Total RNA (Human/Mouse/Rat) (Illumina Inc., San Diego, California, USA). Cytoplasmic ribosomal RNA was removed from the sample using complementary probe sequences attached to magnetic beads. Subsequently, each mRNA sample was fragmented using divalent cations under elevated temperature, and purified. First strand cDNA synthesis was performed using SuperScriptII (Invitrogen, Carlsbad, California, USA), Reverse Transcriptase, and random primers. Second strand cDNAs were synthesized using DNA Polymerase I and RNAse H for removal of mRNA. Double-stranded cDNA was purified using Agencourt AMPure XP beads (Qiagen, Valencia, California, USA) as recommended in the TruSeq RNA Sample Prep Guide. The blunt ended cDNA and the adapter-ligated products were purified using Agencourt AMPure XP beads. Quality and quantity of finished libraries were assessed using an Agilent DNA1000 series chip assay (Agilent Technologies, Santa Clara, CA) and Invitrogen Qubit HS cDNA Kit (Invitrogen, Carlsbad, California, USA), respectively. Cluster generation was performed using a TruSeq Paired End Cluster Kit (v4) and the Illumina cBot, with libraries multiplexed for 1×100bp sequencing using the TruSeq 250bp SBS kit (v4) on an Illumina HiSeq2500. Images were analyzed using CASAVA 1.8.2.
Single cell RNA-seq:
Following islet isolation and an overnight culture, islet cells were dissociated for 30 min at 37°C into single-cell suspensions, using a cocktail of digestive enzymes (Accutase; Innovative Cell Technologies, San Diego, CA). Libraries were constructed according to the Chromium Single Cell 3’ Reagent Kit v2 (10x Genomics, Pleasanton, CA). Briefly, cells in single cell suspension were delivered to the University of Wisconsin-Madison Biotechnology Center on ice, where the cell concentration and viability were quantified on the Countess II (Thermo Fisher Scientific) using 0.4% Trypan Blue (Invitrogen, Carlsbad, CA). The appropriate volume of cells was loaded onto the Single Cell A Chip required for yielding a targeted cell recovery of 3000 cells. Following the completion of the Chromium run, the Gel Bead-In EMulsions (GEMs) were transferred to emulsion-safe strip tubes for GEM-RT, using an Eppendorf Master Cycler Pro thermocycler (Eppendorf, Hamburg, Germany). Following RT, GEMs were broken, and the pooled single cell cDNA was amplified. Post-cDNA amplified product was purified using SPRIselect (Beckman Coulter, Brea, CA) and quantified on a Bioanalyzer 2100 (Agilent, Santa Clara, CA) using the High Sensitivity DNA kit. Adapters were then added to the libraries after fragmentation, end repair, A-tailing, and double-sided size selection using SPRIselect. Following adapter ligation, libraries were purified using SPRIselect, and sample-specific indexes (Chromium i7 Multiplex Kit, 10x Genomics) were added by sample index PCR. After sample index PCR, samples were double-size selected using SPRIselect, yielding final libraries compatible for Illumina sequencing. Final libraries were quantified using the Qubit High Sensitivity DNA Kit (Thermo Fisher Scientific).
t-SNE clustering:
We used Monocle2 v2.8.0 on R version 3.5.2 (Kite-Eating Tree) http://cole-trapnell-lab.github.io/monocle-release/docs/ (Qiu et al., 2017) to analyze the data obtained after alignment. Cells with unique molecular identifier (UMI) counts outside a range determined of two standard deviations were filtered, leaving 2,749 cells within the optimal UMI range for downstream Monocle analysis. Genes that were not expressed in at least 10 cells were excluded from analysis. We reduced the number of dimensions to the number of Principal components that explained the most variance (at least 50% for each dataset).
To identify genes important for defining clusters, differential gene expression (DGE) analysis in Monocle2 was performed between all 7 clusters within the t-SNE plot of Fig 5A. To determine the distribution of cells from each sample that fall into each cluster, phenotypic data (cell barcode ID, sample, cluster number) was extracted for each cluster and sample. The composition of a cluster or sample was then calculated by percentage or mean of the population.
To visualize how different genes are expressed in cells that are known to be positive for a particular gene, we also generated violin plots using ggplot function in R. DGEs between IRE1αfl/fl and IRE1αβ−/− mice were clustered using Cluster 3.0 (de Hoon et al., 2004) and visualized using Java TreeView (Saldanha, 2004).
QUANTIFICATION AND STATISTICAL ANALYSIS
For all experiments the number of biological or technical replicates (n), error bars, and statistical analyses have been explained in the figure legends. For each experiment where statistics were computed, we used at least n = 3 or more biological replicates. Sample size were not pre-determined by power analysis, but sufficiency of number of mice were estimated based on pilot experiments and previously published work (Engin et al., 2013). Samples were randomly assigned and blinded for data analysis of immunostaining and insulitis scoring. No data were excluded in this study. Data are represented as mean ± SEM and were analyzed using either the unpaired Student’s t-test, or one-way ANOVA where required. P < 0.05 was considered statistically significant; ns = non-significant as determined by statistical analysis in GraphPad Prism v.8 (GraphPad Software, San Diego, CA).
DATA AND CODE AVAILABILITY
Data Resources
The accession number for the RNA-seq data reported in this paper are NCBI GEO: GSE144461 (bulk sequencing) and GSE144471 (single cell sequencing).
Supplementary Material
Acknowledgements
We thank Dr. Mark O. Huising for critical reading of the initial draft of the manuscript. F.E. is supported by grants from the JDRF-5-CDA-2014-184-A-N and NIH 5K01DK102488-03. H.L. is supported by NIH National Research Service Award T32 GM007215. Y.-G.C. is supported by NIH grants DK107541 and DK121747. A.E.C. is supported by the NIH F31 award DK118786. R.S. is supported by NIH R01GM113033. S.P. is supported by a NIH T32 NHGRI 5T32HG002760 award. The flow cytometry data were collected with resources and facilities provided by the University of Wisconsin Carbone Cancer Center-Flow Cytometry Laboratory with funding from Support Grant P30 CA014520.
Footnotes
Declaration of interests: The authors have declared that no conflicts of interest exist.
References
- Artner I, Hang Y, Mazur M, Yamamoto T, Guo M, Lindner J, Magnuson MA, and Stein R (2010). MafA and MafB regulate genes critical to beta-cells in a unique temporal manner. Diabetes 59, 2530–2539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arvan P, Pietropaolo M, Ostrov D, and Rhodes CJ (2012). Islet autoantigens: structure, function, localization, and regulation. Cold Spring Harb Perspect Med 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkinson MA (2012). The pathogenesis and natural history of type 1 diabetes. Cold Spring Harb Perspect Med 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernales S, Papa FR, and Walter P (2006). Intracellular signaling by the unfolded protein response. Annu Rev Cell Dev Biol 22, 487–508. [DOI] [PubMed] [Google Scholar]
- Bersell K, Arab S, Haring B, and Kuhn B (2009). Neuregulin1/ErbB4 signaling induces cardiomyocyte proliferation and repair of heart injury. Cell 138, 257–270. [DOI] [PubMed] [Google Scholar]
- Bluestone JA, Herold K, and Eisenbarth G (2010). Genetics, pathogenesis and clinical interventions in type 1 diabetes. Nature 464, 1293–1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blum B, Hrvatin S, Schuetz C, Bonal C, Rezania A, and Melton DA (2012). Functional beta-cell maturation is marked by an increased glucose threshold and by expression of urocortin 3. Nat Biotechnol 30, 261–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burzyn D, Kuswanto W, Kolodin D, Shadrach JL, Cerletti M, Jang Y, Sefik E, Tan TG, Wagers AJ, Benoist C, et al. (2013). A special population of regulatory T cells potentiates muscle repair. Cell 155, 1282–1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calfon M, Zeng H, Urano F, Till JH, Hubbard SR, Harding HP, Clark SG, and Ron D (2002). IRE1 couples endoplasmic reticulum load to secretory capacity by processing the XBP-1 mRNA. Nature 415, 92–96. [DOI] [PubMed] [Google Scholar]
- Carosella ED, Moreau P, Lemaoult J, and Rouas-Freiss N (2008). HLA-G: from biology to clinical benefits. Trends Immunol 29, 125–132. [DOI] [PubMed] [Google Scholar]
- Chen Y, and Brandizzi F (2013). IRE1: ER stress sensor and cell fate executor. Trends Cell Biol 23, 547–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen ZL, Yu WM, and Strickland S (2007). Peripheral regeneration. Annu Rev Neurosci 30, 209–233. [DOI] [PubMed] [Google Scholar]
- Cinti F, Bouchi R, Kim-Muller JY, Ohmura Y, Sandoval PR, Masini M, Marselli L, Suleiman M, Ratner LE, Marchetti P, et al. (2016). Evidence of beta-Cell Dedifferentiation in Human Type 2 Diabetes. J Clin Endocrinol Metab 101, 1044–1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Hoon MJ, Imoto S, Nolan J, and Miyano S (2004). Open source clustering software. Bioinformatics 20, 1453–1454. [DOI] [PubMed] [Google Scholar]
- Dhawan S, Tschen SI, Zeng C, Guo T, Hebrok M, Matveyenko A, and Bhushan A (2015). DNA methylation directs functional maturation of pancreatic beta cells. J Clin Invest 125, 2851–2860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dirice E, Kahraman S, De Jesus DF, El Ouaamari A, Basile G, Baker RL, Yigit B, Piehowski PD, Kim MJ, Dwyer AJ, et al. (2019). Increased beta-cell proliferation before immune cell invasion prevents progression of type 1 diabetes. Nat Metab 1, 509–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dooley J, Tian L, Schonefeldt S, Delghingaro-Augusto V, Garcia-Perez JE, Pasciuto E, Di Marino D, Carr EJ, Oskolkov N, Lyssenko V, et al. (2016). Genetic predisposition for beta cell fragility underlies type 1 and type 2 diabetes. Nat Genet 48, 519–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dor Y, Brown J, Martinez OI, and Melton DA (2004). Adult pancreatic beta-cells are formed by self-duplication rather than stem-cell differentiation. Nature 429, 41–46. [DOI] [PubMed] [Google Scholar]
- Engin F (2016). ER stress and development of type 1 diabetes. J Investig Med 64, 2–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engin F, Nguyen T, Yermalovich A, and Hotamisligil GS (2014). Aberrant islet unfolded protein response in type 2 diabetes. Sci Rep 4, 4054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engin F, Yermalovich A, Nguyen T, Hummasti S, Fu W, Eizirik DL, Mathis D, and Hotamisligil GS (2013). Restoration of the unfolded protein response in pancreatic beta cells protects mice against type 1 diabetes. Sci Transl Med 5, 211ra156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gershengorn MC, Hardikar AA, Wei C, Geras-Raaka E, Marcus-Samuels B, and Raaka BM (2004). Epithelial-to-mesenchymal transition generates proliferative human islet precursor cells. Science 306, 2261–2264. [DOI] [PubMed] [Google Scholar]
- Gittes GK, Rutter WJ, and Debas HT (1993). Initiation of gastrin expression during the development of the mouse pancreas. Am J Surg 165, 23–25; discussion 25–26. [DOI] [PubMed] [Google Scholar]
- Granados DP, Tanguay PL, Hardy MP, Caron E, de Verteuil D, Meloche S, and Perreault C (2009). ER stress affects processing of MHC class I-associated peptides. BMC Immunol 10, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han D, Lerner AG, Vande Walle L, Upton JP, Xu W, Hagen A, Backes BJ, Oakes SA, and Papa FR (2009). IRE1alpha kinase activation modes control alternate endoribonuclease outputs to determine divergent cell fates. Cell 138, 562–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassler JR, Scheuner DL, Wang S, Han J, Kodali VK, Li P, Nguyen J, George JS, Davis C, Wu SP, et al. (2015). The IRE1alpha/XBP1s Pathway Is Essential for the Glucose Response and Protection of beta Cells. PLoS Biol 13, e1002277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollien J, and Weissman JS (2006). Decay of endoplasmic reticulum-localized mRNAs during the unfolded protein response. Science 313, 104–107. [DOI] [PubMed] [Google Scholar]
- In’t Veld P, Lievens D, De Grijse J, Ling Z, Van der Auwera B, Pipeleers-Marichal M, Gorus F, and Pipeleers D (2007). Screening for insulitis in adult autoantibody-positive organ donors. Diabetes 56, 2400–2404. [DOI] [PubMed] [Google Scholar]
- Iwawaki T, Akai R, Yamanaka S, and Kohno K (2009). Function of IRE1 alpha in the placenta is essential for placental development and embryonic viability. Proceedings of the National Academy of Sciences of the United States of America 106, 16657–16662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H, Ware R, Stall A, Flaherty L, Chess L, and Pernis B (1995). Murine CD8+ T cells that specifically delete autologous CD4+ T cells expressing V beta 8 TCR: a role of the Qa-1 molecule. Immunity 2, 185–194. [DOI] [PubMed] [Google Scholar]
- Jones EL, Demaria MC, and Wright MD (2011). Tetraspanins in cellular immunity. Biochem Soc Trans 39, 506–511. [DOI] [PubMed] [Google Scholar]
- Kim-Muller JY, Fan J, Kim YJ, Lee SA, Ishida E, Blaner WS, and Accili D (2016). Aldehyde dehydrogenase 1a3 defines a subset of failing pancreatic beta cells in diabetic mice. Nat Commun 7, 12631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein J, Figueroa F, and David CS (1983). H-2 haplotypes, genes and antigens: second listing. II. The H-2 complex. Immunogenetics 17, 553–596. [DOI] [PubMed] [Google Scholar]
- Lee AH, Heidtman K, Hotamisligil GS, and Glimcher LH (2011). Dual and opposing roles of the unfolded protein response regulated by IRE1alpha and XBP1 in proinsulin processing and insulin secretion. Proceedings of the National Academy of Sciences of the United States of America 108, 8885–8890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee AH, Iwakoshi NN, and Glimcher LH (2003). XBP-1 regulates a subset of endoplasmic reticulum resident chaperone genes in the unfolded protein response. Mol Cell Biol 23, 7448–7459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerner AG, Upton JP, Praveen PV, Ghosh R, Nakagawa Y, Igbaria A, Shen S, Nguyen V, Backes BJ, Heiman M, et al. (2012). IRE1alpha induces thioredoxin-interacting protein to activate the NLRP3 inflammasome and promote programmed cell death under irremediable ER stress. Cell Metab 16, 250–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipson KL, Ghosh R, and Urano F (2008). The role of IRE1alpha in the degradation of insulin mRNA in pancreatic beta-cells. PLoS One 3, e1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louvet C, Szot GL, Lang J, Lee MR, Martinier N, Bollag G, Zhu S, Weiss A, and Bluestone JA (2008). Tyrosine kinase inhibitors reverse type 1 diabetes in nonobese diabetic mice. Proceedings of the National Academy of Sciences of the United States of America 105, 18895–18900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maganti A, Evans-Molina C, and Mirmira R (2014). From immunobiology to beta-cell biology: the changing perspective on type 1 diabetes. Islets 6, e28778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marhfour I, Lopez XM, Lefkaditis D, Salmon I, Allagnat F, Richardson SJ, Morgan NG, and Eizirik DL (2012). Expression of endoplasmic reticulum stress markers in the islets of patients with type 1 diabetes. Diabetologia 55, 2417–2420. [DOI] [PubMed] [Google Scholar]
- Matsuoka TA, Artner I, Henderson E, Means A, Sander M, and Stein R (2004). The MafA transcription factor appears to be responsible for tissue-specific expression of insulin. Proceedings of the National Academy of Sciences of the United States of America 101, 2930–2933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy DJ, Chen Y, and Smyth GK (2012). Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Res 40, 4288–4297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monticelli LA, Sonnenberg GF, Abt MC, Alenghat T, Ziegler CG, Doering TA, Angelosanto JM, Laidlaw BJ, Yang CY, Sathaliyawala T, et al. (2011). Innate lymphoid cells promote lung-tissue homeostasis after infection with influenza virus. Nat Immunol 12, 1045–1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita S, Villalta SA, Feldman HC, Register AC, Rosenthal W, Hoffmann-Petersen IT, Mehdizadeh M, Ghosh R, Wang L, Colon-Negron K, et al. (2017). Targeting ABL-IRE1alpha Signaling Spares ER-Stressed Pancreatic beta Cells to Reverse Autoimmune Diabetes. Cell Metab 25, 883–897 e888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama M (2011). Insulin as a key autoantigen in the development of type 1 diabetes. Diabetes Metab Res Rev 27, 773–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama M, Abiru N, Moriyama H, Babaya N, Liu E, Miao D, Yu L, Wegmann DR, Hutton JC, Elliott JF, et al. (2005). Prime role for an insulin epitope in the development of type 1 diabetes in NOD mice. Nature 435, 220–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narendran P, Mannering SI, and Harrison LC (2003). Proinsulin-a pathogenic autoantigen in type 1 diabetes. Autoimmun Rev 2, 204–210. [DOI] [PubMed] [Google Scholar]
- Oslowski CM, Hara T, O’Sullivan-Murphy B, Kanekura K, Lu S, Hara M, Ishigaki S, Zhu LJ, Hayashi E, Hui ST, et al. (2012). Thioredoxin-interacting protein mediates ER stress-induced beta cell death through initiation of the inflammasome. Cell Metab 16, 265–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouziel-Yahalom L, Zalzman M, Anker-Kitai L, Knoller S, Bar Y, Glandt M, Herold K, and Efrat S (2006). Expansion and redifferentiation of adult human pancreatic islet cells. Biochem Biophys Res Commun 341, 291–298. [DOI] [PubMed] [Google Scholar]
- Petri A, Ahnfelt-Ronne J, Frederiksen KS, Edwards DG, Madsen D, Serup P, Fleckner J, and Heller RS (2006). The effect of neurogenin3 deficiency on pancreatic gene expression in embryonic mice. J Mol Endocrinol 37, 301–316. [DOI] [PubMed] [Google Scholar]
- Pugliese A (2016). Insulitis in the pathogenesis of type 1 diabetes. Pediatr Diabetes 17 Suppl 22, 31–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pullen TJ, Khan AM, Barton G, Butcher SA, Sun G, and Rutter GA (2010). Identification of genes selectively disallowed in the pancreatic islet. Islets 2, 89–95. [DOI] [PubMed] [Google Scholar]
- Puri S, Folias AE, and Hebrok M (2015). Plasticity and dedifferentiation within the pancreas: development, homeostasis, and disease. Cell Stem Cell 16, 18–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu X, Hill A, Packer J, Lin D, Ma YA, and Trapnell C (2017). Single-cell mRNA quantification and differential analysis with Census. Nat Methods 14, 309–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintens R, Hendrickx N, Lemaire K, and Schuit F (2008). Why expression of some genes is disallowed in beta-cells. Biochem Soc Trans 36, 300–305. [DOI] [PubMed] [Google Scholar]
- Regnell SE, and Lernmark A (2017). Early prediction of autoimmune (type 1) diabetes. Diabetologia 60, 1370–1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, and Smyth GK (2010). edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson PJ, Millrain M, Antoniou J, Simpson E, and Mellor AL (1989). A glycophospholipid anchor is required for Qa-2-mediated T cell activation. Nature 342, 85–87. [DOI] [PubMed] [Google Scholar]
- Rui J, Deng S, Arazi A, Perdigoto AL, Liu Z, and Herold KC (2017). beta Cells that Resist Immunological Attack Develop during Progression of Autoimmune Diabetes in NOD Mice. Cell Metab 25, 727–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saldanha AJ (2004). Java Treeview--extensible visualization of microarray data. Bioinformatics 20, 3246–3248. [DOI] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. (2012). Fiji: an open-source platform for biological-image analysis. Nat Methods 9, 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao J, and Sheng H (2010). Amphiregulin promotes intestinal epithelial regeneration: roles of intestinal subepithelial myofibroblasts. Endocrinology 151, 3728–3737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoshani O, and Zipori D (2011). Mammalian cell dedifferentiation as a possible outcome of stress. Stem Cell Rev 7, 488–493. [DOI] [PubMed] [Google Scholar]
- Soleimanpour SA, and Stoffers DA (2013). The pancreatic beta cell and type 1 diabetes: innocent bystander or active participant? Trends in endocrinology and metabolism: TEM 24, 324–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su Y, Jono H, Misumi Y, Senokuchi T, Guo J, Ueda M, Shinriki S, Tasaki M, Shono M, Obayashi K, et al. (2012). Novel function of transthyretin in pancreatic alpha cells. FEBS Lett 586, 4215–4222. [DOI] [PubMed] [Google Scholar]
- Szabat M, Page MM, Panzhinskiy E, Skovso S, Mojibian M, Fernandez-Tajes J, Bruin JE, Bround MJ, Lee JT, Xu EE, et al. (2016). Reduced Insulin Production Relieves Endoplasmic Reticulum Stress and Induces beta Cell Proliferation. Cell Metab 23, 179–193. [DOI] [PubMed] [Google Scholar]
- Szabat M, Pourghaderi P, Soukhatcheva G, Verchere CB, Warnock GL, Piret JM, and Johnson JD (2011). Kinetics and genomic profiling of adult human and mouse beta-cell maturation. Islets 3, 175–187. [DOI] [PubMed] [Google Scholar]
- Talchai C, Xuan S, Lin HV, Sussel L, and Accili D (2012). Pancreatic beta cell dedifferentiation as a mechanism of diabetic beta cell failure. Cell 150, 1223–1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tersey SA, Nishiki Y, Templin AT, Cabrera SM, Stull ND, Colvin SC, Evans-Molina C, Rickus JL, Maier B, and Mirmira RG (2012). Islet beta-cell endoplasmic reticulum stress precedes the onset of type 1 diabetes in the nonobese diabetic mouse model. Diabetes 61, 818–827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson PJ, Shah A, Ntranos V, Van Gool F, Atkinson M, and Bhushan A (2019). Targeted Elimination of Senescent Beta Cells Prevents Type 1 Diabetes. Cell Metab 29, 1045–1060 e1010. [DOI] [PubMed] [Google Scholar]
- Thorel F, Nepote V, Avril I, Kohno K, Desgraz R, Chera S, and Herrera PL (2010). Conversion of adult pancreatic alpha-cells to beta-cells after extreme beta-cell loss. Nature 464, 1149–1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorrez L, Laudadio I, Van Deun K, Quintens R, Hendrickx N, Granvik M, Lemaire K, Schraenen A, Van Lommel L, Lehnert S, et al. (2011). Tissue-specific disallowance of housekeeping genes: the other face of cell differentiation. Genome Res 21, 95–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchiya Y, Saito M, Kadokura H, Miyazaki JI, Tashiro F, Imagawa Y, Iwawaki T, and Kohno K (2018). IRE1-XBP1 pathway regulates oxidative proinsulin folding in pancreatic beta cells. J Cell Biol 217, 1287–1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulianich L, Terrazzano G, Annunziatella M, Ruggiero G, Beguinot F, and Di Jeso B (2011). ER stress impairs MHC Class I surface expression and increases susceptibility of thyroid cells to NK-mediated cytotoxicity. Biochim Biophys Acta 1812, 431–438. [DOI] [PubMed] [Google Scholar]
- Upton JP, Wang L, Han D, Wang ES, Huskey NE, Lim L, Truitt M, McManus MT, Ruggero D, Goga A, et al. (2012). IRE1alpha cleaves select microRNAs during ER stress to derepress translation of proapoptotic Caspase-2. Science 338, 818–822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urano F, Wang X, Bertolotti A, Zhang Y, Chung P, Harding HP, and Ron D (2000). Coupling of stress in the ER to activation of JNK protein kinases by transmembrane protein kinase IRE1. Science 287, 664–666. [DOI] [PubMed] [Google Scholar]
- van Belle TL, Coppieters KT, and von Herrath MG (2011). Type 1 diabetes: etiology, immunology, and therapeutic strategies. Physiol Rev 91, 79–118. [DOI] [PubMed] [Google Scholar]
- van der Meulen T, Xie R, Kelly OG, Vale WW, Sander M, and Huising MO (2012). Urocortin 3 marks mature human primary and embryonic stem cell-derived pancreatic alpha and beta cells. PLoS One 7, e52181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter P, and Ron D (2011). The unfolded protein response: from stress pathway to homeostatic regulation. Science 334, 1081–1086. [DOI] [PubMed] [Google Scholar]
- Wang Z, York NW, Nichols CG, and Remedi MS (2014). Pancreatic beta cell dedifferentiation in diabetes and redifferentiation following insulin therapy. Cell Metab 19, 872–882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward-Kavanagh LK, Lin WW, Sedy JR, and Ware CF (2016). The TNF Receptor Superfamily in Co-stimulating and Co-inhibitory Responses. Immunity 44, 1005–1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiberg A, Granstam A, Ingvast S, Harkonen T, Knip M, Korsgren O, and Skog O (2015). Characterization of human organ donors testing positive for type 1 diabetes-associated autoantibodies. Clin Exp Immunol 182, 278–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida H, Matsui T, Yamamoto A, Okada T, and Mori K (2001). XBP1 mRNA is induced by ATF6 and spliced by IRE1 in response to ER stress to produce a highly active transcription factor. Cell 107, 881–891. [DOI] [PubMed] [Google Scholar]
- You S, and Chatenoud L (2006). Proinsulin: a unique autoantigen triggering autoimmune diabetes. J Clin Invest 116, 3108–3110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu G, Wang LG, Han Y, and He QY (2012). clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 16, 284–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, McKenna LB, Bogue CW, and Kaestner KH (2014). The diabetes gene Hhex maintains delta-cell differentiation and islet function. Genes Dev 28, 829–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Feng D, Li Y, Iida K, McGrath B, and Cavener DR (2006). PERK EIF2AK3 control of pancreatic beta cell differentiation and proliferation is required for postnatal glucose homeostasis. Cell Metab 4, 491–497. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data Resources
The accession number for the RNA-seq data reported in this paper are NCBI GEO: GSE144461 (bulk sequencing) and GSE144471 (single cell sequencing).


