Abstract
Finding adequate biomarkers for rapid and accurate disease detection, prognosis, and therapy is increasingly important. Quorum-sensing peptides are herein a new emerging group, produced by bacteria, fungi, protozoa, and viruses, with blood being the most straightforward sample type to detect/quantitate them. However, detailed information about suitable blood sample collection methods and storage conditions for measuring these quorum-sensing peptides hampers further clinical research and development. Here, we first tested the time-dependent stability of a set of chemically diverse quorum-sensing peptides, spiked in blood at different temperatures (4, 21, and 37 °C) in four different ethylenediamine tetraacetic acid (EDTA)-containing plasma tubes (with different protein-stabilizing additives) over a period of up to 7.5 h. Next, we determined the storage stability of these quorum-sensing peptides in plasma at different temperatures (4, −35, and −80 °C). UPLC/MS–MS was used to selectively detect and quantify the spiked quorum-sensing peptides. The results of this study indicate that a cost-effective tube, designed for traditional proteomics and stored at 4 °C, is the preferred collection condition when quorum-sensing peptides need to be detected/quantified in human plasma. When the tubes are handled at room temperature (21 °C), a more specialized tube is required. Long-term storage of plasma samples, even under low-temperature conditions (−80 °C), indicates rapid degradation of certain quorum-sensing peptides.
1. Introduction
Clinical diagnostics and therapeutics are moving from large cohort-based approaches toward personal-based strategies, thereby requiring adequate biomarkers. Because of its accessibility and ease of downstream processing, human blood is predominantly investigated and used for the identification of suitable biomarkers. Different chemical classes of possible blood biomarkers exist, e.g., proteins, circulating DNA, microRNA, and small-molecular metabolites such as organic acids.1−3 Recently, a growing interest is seen in peptides as potential blood biomarkers.4,5 Peptides can be considered as an intermediate class between small molecules (metabolomics) and proteins (proteomics). They differ from proteins not only because they are shorter (traditionally considered below 50 amino acids) but also because of their less complex three-dimensional structure.6 These different physicochemical characteristics influence the biological behavior of peptides. For example, peptides often show an increased renal clearance and/or enzymatic degradation in vivo.7,8 They also require different analytical methodologies compared to small metabolites and large proteins. For example, the sample preparation and chromatography for peptides are completely different.9,10 Recently, a novel class of peptides, i.e., the quorum-sensing peptides from bacterial, fungal, protozoal, and viral origin, has been identified as an interesting group of peptides, as they influence different types of host cells and hence play a role in the health of the host.11−14
To date, only a few studies have investigated to some extent the influence of some blood collection parameters and the effects of long-term human plasma storage on peptide stability.15−20 However, they investigated only one specific peptide and/or one specific factor. None of the reported studies investigated simultaneously the three critical variables in blood collection (temperature, choice of stabilizing additive, and time) on a broad range of peptides. Moreover, the quorum-sensing peptides, with their specific peptide structure and diversity,21 have never been investigated.
The aim of the present study was therefore to investigate the influence of the blood collection method and long-term plasma storage on quorum-sensing peptide stability. For the blood collection experiments, time, temperature, and choice of different protease/peptidase-inhibitor additives were simultaneously considered as variables. The optimal blood collection method was then used to study the quorum-sensing peptide stability in plasma at different storage temperatures. Quantitative ultraperformance liquid chromatography–mass spectrometry (UPLC/MS–MS) was used to detect and quantify the different quorum-sensing peptides, selected to cover as much as possible the chemical quorum-sensing peptide space. As such, the results of this study can be useful in the research of clinical blood and plasma quorum-sensing peptidomics.
2. Results and Discussion
The human peptidome consists not only of cryptides, i.e., peptides formed by cleavage of endogeneous or food-derived proteins, but also of small open reading frame (smORF) encoded peptides (SEPs) and of peptides from our microbiome,22,23 including quorum-sensing peptides from bacteria, fungi, and viruses.24−26 Recently, these microbiologically derived peptides have been shown to interact with human host cells.11−14 Consequently, their biological functionality makes them possible biomarker candidates to be quantified in biofluids like blood. In light of this emerging new group of biomarkers, there is a need for suitable blood collection and storage procedures to reliably measure them. To address this question, we investigated three critical variables in parallel for blood collection: temperature, choice of stabilizing additives (i.e., tube type), and time. Four types of tubes were used, i.e., a conventional ethylenediamine tetraacetic acid (EDTA) plasma-sampling tube (EDTA), a conventional EDTA tube with a protease-inhibitor cocktail from Sigma-Aldrich (EDTA+), and two different plasma tubes from Becton Dickinson with EDTA and proprietary human protease/peptidase inhibitors (P100 and P800). Moreover, in contrast to some other plasma peptidomic studies, we investigated the quorum-sensing peptide stability also in blood, i.e., prior to centrifugation of the blood sample for obtaining plasma. As it is often not feasible to centrifuge the plasma tubes immediately after blood withdrawal (e.g., physician or research offices from which the blood samples have to be transported to a separate laboratory for processing and analysis), the peptide stability in blood before centrifugation is a time-critical step. To rule out freeze–thaw effects, our blood collection stability experiments were conducted on fresh samples, i.e., not frozen. As peptidome analysis is, in most clinical situations, only possible after plasma has been stored, the long-term storage in plasma was also investigated. For this experiment, plasma from the BD 100 tube, conferring the largest whole-blood peptide stability (described in Section 2.1), was used. Long-term storage at three temperatures that are common in clinical and research settings (4, −35, and −80 °C) was compared. UPLC/MS–MS was used for the detection and quantitation of the peptides in all experiments, as this method enabled us to achieve high selectivity combined with quantification characteristics. Figure 1 shows a representative liquid chromatography–mass spectrometry (LC–MS) chromatogram. Inherent to our study design, the conclusions are focused on peptide stability sensu stricto. This means that we did not take into consideration either protein stability or possible increases in peptide concentration caused by blood cellular excretion or hemolysis. For example, even if clotting is inhibited by EDTA, platelets are activated under certain conditions in EDTA tubes, excreting specific peptides and proteins.27,28 Therefore, caution is warranted to extrapolate our quorum-sensing peptide findings to peptides also intracellularly present in blood cells. We did not find analytical matrix effects caused by the protease/peptidase inhibitors present in the plasma-sampling tubes (Figure S1). Nevertheless, the general caution for interference was applied as recommended when using data-dependent acquisition in peptidomics. Additionally, no time-trend in matrix effects was noticed (Figure S2). This confirms that the noticed decrease in analytical LC–MS peptide response was due to degradation and not by changes in the matrix effect.
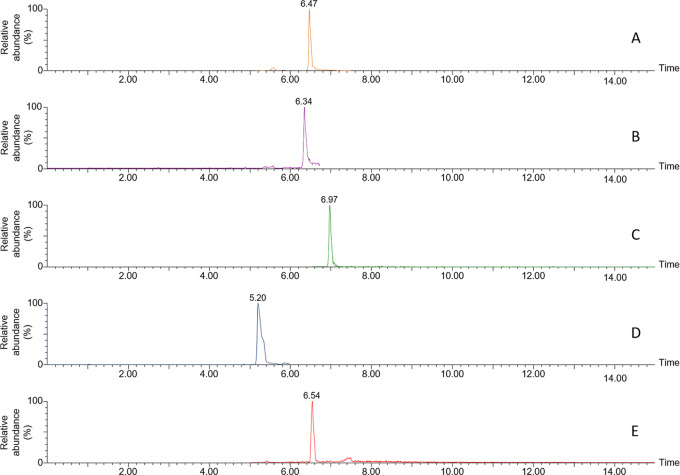
Figure 1.
Example LC–MS chromatograms of (A) Q076, (B) Q164, (C) Q176, (D) Q175, and (E) Q019.
2.1. Blood Collection
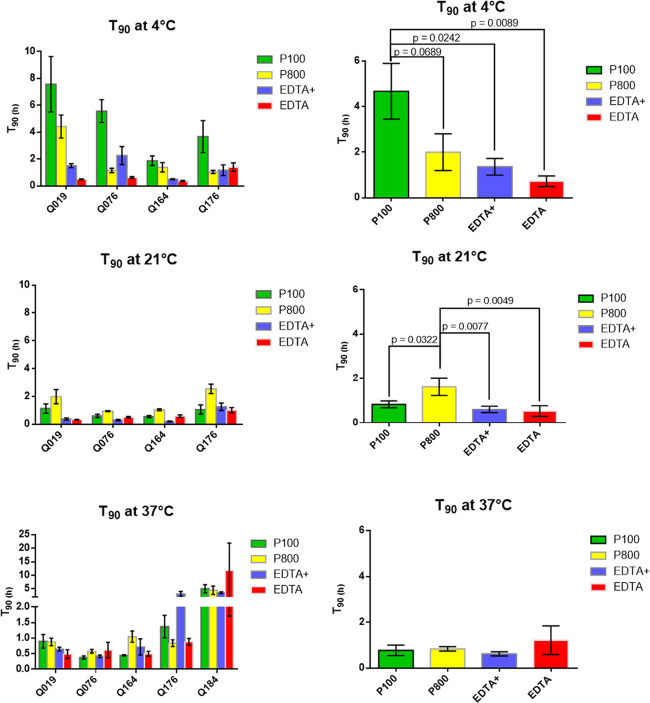
The half-lives for the five peptides (Table 5) of the training set in the different blood collection tubes at different temperatures are shown in Table 1. The calculated half-lives (T50) exceed twice the time period under investigation. Therefore, the T90 values, the time where 90% of the peptide is still present (=10% degradation), were used as more representative for bioanalytical peptidomics. Figure 2 graphically shows the T90 results, with paired Tukey’s multiple comparisons test conducted on the pooled results to detect statistically significant differences between the different tubes at each temperature. Our study aims to find the optimal sampling and storage methods for a broad spectrum of quorum-sensing peptides. This implies that we are investigating the “largest common denominator” for blood peptide identification/quantitation and not an optimal method for each peptide individually.
Table 5. Sequences and Some Physicochemical Descriptors of the Quorum-Sensing Peptides Used in This Studya.
| peptide (supplier) | sequence | log P | pI | MW |
|---|---|---|---|---|
| Q019b | NNWNN | –7.17 | 5.29 | 660.7 |
| Q044c | EKMIG | –2.57 | 6.21 | 576.8 |
| Q051c | ERNNT | –6.8 | 6.17 | 632.7 |
| Q054c | ESRLPKIRFDFIFPRKK | –3.58 | 10.84 | 2178.0 |
| Q055d | ESRVSRIILDFLFQRKK | –5.44 | 10.94 | 2135.9 |
| Q076c | SNLVECVFSLFKKCN | –6.35 | 8.41 | 1731.3 |
| Q164c | SDLPFEH | –2.84 | 4.13 | 844.0 |
| Q176c | SGSLSTFFRLFNRSFTQALGK | –9.69 | 11.00 | 2365.0 |
| Q184c | SIFTLVA | 0.09 | 5.71 | 750.0 |
Peptide numbering used in the Quorumpeps database (quorumpeps.ugent.be) is retained in this study. Peptides were synthesized with terminal free acids by.
JPT Peptide Technologies.
GL Biochem.
ProteoGenix. P, octanol–water partition coefficient; pI, isoelectric point; MW, molecular weight.
Table 1. Half-Lives (Hours) in Whole Blood for the Five Peptides of the Training Set (±SEM)a.
| half-lives (T50 ± SEM) |
|||||
|---|---|---|---|---|---|
| peptide | temperature (°C) | P100 | P800 | EDTA+ | EDTA |
| Q019 | 4 | 49.8 h ± 13.6 | 29.1 h ± 5.6 | 9.9 h ± 1.0 | 3.3 h ± 0.2 |
| 21 | 7.4 h ± 2.2 | 13.1 h ± 3.3 | 2.1 h ± 0.2 | 2.5 h ± 0.5 | |
| 37 | 5.9 h ± 1.4 | 5.8 h ± 0.8 | 3.2 h ± 0.9 | 4.3 h ± 0.4 | |
| Q076 | 4 | 36.7 h ± 5.6 | 7.6 h ± 1.1 | 14.9 h ± 4.4 | 4.1 h ± 0.4 |
| 21 | 3.9 h ± 0.8 | 6.1 h ± 0.2 | 3.3 h ± 0.3 | 1.9 h ± 0.3 | |
| 37 | 2.5 h ± 0.3 | 3.8 h ± 0.4 | 4.1 h ± 1.7 | 2.8 h ± 0.3 | |
| Q164 | 4 | 12.4 h ± 2.3 | 9.2 h ± 2.3 | 3.3 h ± 0.3 | 2.5 h ± 0.2 |
| 21 | 3.5 h ± 0.5 | 6.8 h ± 0.5 | 3.8 h ± 0.6 | 1.3 h ± 0.2 | |
| 37 | 3.0 h ± 0.1 | 6.9 h ± 1.2 | 3.3 h ± 1.7 | 4.7 h ± 0.5 | |
| Q176 | 4 | 24.2 h ± 7.9 | 6.8 h ± 0.8 | 7.7 h ± 2.6 | 9.2 h ± 2.1 |
| 21 | 6.9 h ± 2.2 | 16.7 h ± 2.2 | 6.6 h ± 1.3 | 8.2 h ± 1.8 | |
| 37 | 9.0 h ± 2.4 | 5.6 h ± 0.8 | 5.8 h ± 0.7 | 20.2 h ± 6.3 | |
| Q184 | 4 | n.a. | n.a. | n.a. | n.a. |
| 21 | n.a. | n.a. | n.a. | n.a. | |
| 37 | 33.1 h ± 9.9 | 29.4 h ± 10.0 | 77.5 h ± 66.4 | 23.5 h ± 2.4 | |
n.a., not applicable; no degradation of the peptide was seen within 7.5 h, i.e., the 95% confidence interval of λ includes zero.
Figure 2.
T90 values (±SEM; n = 4) at different temperatures for the blood collection tubes. Left, per peptide. Right, per type of blood collection tube (average of all peptides together), with p-values indicated.
The paired Tukey’s multiple-comparison one-way analysis of variance (ANOVA) showed that at 4 °C, the P100 tube performed significantly better than the EDTA tube with the protease inhibitor (EDTA+) (p = 0.024) and the traditional EDTA tube (p = 0.009). Compared to the P800 tube, P100 almost doubled the stability of the peptides at 4 °C. Although this effect did not reach statistical significance (p = 0.069), the standardized mean difference (= Cohen’s d value) between P100 and P800 tubes at 4 °C was 1.3. Cohen’s d values above 0.8 are generally considered to indicate relevant effect sizes.29 The results also showed that P100 tubes at 4 °C were superior to all other tested conditions, with half-lives of 12 h and longer. Q184 was the most stable peptide of the five investigated peptides, with no degradation of the peptide during the time frame of 7.5 h at both 4 and 21 °C. The finding of an increased peptide stability at 4 °C compared to 21 and 37 °C was also noticed in the non-P100 tubes, although statistically not significant (Figure S3). This confirms other plasma peptidomic studies.18
At 21 °C, P800 tubes showed an improved stability compared to P100 tubes (p = 0.032), EDTA tubes (p = 0.008), and EDTA + tubes (p = 0.005). This is in line with the study of Yi et al., where the stability of four plasma hormone peptides was analyzed in classical EDTA tubes, serum tubes, P700 tubes (not on the market anymore), and P800 tubes at room temperature.17 A discrepancy was seen in absolute values of the half-lives between our study and the study of Yi et al. While some of our peptides had half-lives shorter than 6 h, all of their peptides investigated had half-lives larger than 45 h using P800 tubes. However, their peptides did not cover a wide range of the chemical space, which could explain the T50 discrepancy. Moreover, some or all of their samples underwent a freeze–thaw cycle (“frozen immediately at −80 °C until later usage”). Multiple studies have proven a significant decrease in enzyme activity after freeze–thaw cycles.30,31 A decreased protease activity after a freeze–thaw cycle could therefore be the reason for the very long T50 values in the study of Yi et al. In a separate study, the same group also compared P100 tubes with EDTA tubes at room temperature, finding an increased peptide stability for P100 tubes.15 Other studies comparing P100 tubes vs EDTA tubes however did not find a consistent peptide/protein stability benefit of the P100 tubes at room temperature.28,32−35 In our study, there was a small mean stability benefit of P100 tubes compared to EDTA(+) tubes at 21 °C (Figure 2). However, because of the inconsistency between the different peptides, this effect did not reach statistical significance.
The stability of the peptides at 37 °C was evaluated in all tubes. This is the initial temperature of blood at the moment of blood withdrawal. No significant differences were measured between the different tubes for all peptides. Although cooling freshly drawn blood to 4 °C can be done in a short time, inevitably there is a phase after withdrawal where the blood has a temperature between 4 and 37 °C. In that respect, the minimal half-lives of the peptides of 2.5 h at 37 °C are reassuring.
Based on the results of the training experiment, a validation experiment was conducted where the stability of four peptides, not included in the training experiment, was investigated at 4 °C in P100 tubes. These peptides were chosen from the two chemical clusters furthest apart from each other (two peptides from each cluster).21 The slopes of these curves (λ), T50 and T90 values, are presented in Table 2. The long peptide half-life of >8 h, determined during the training experiment, was no longer applicable for two of the four peptides during the external validation. The two peptides that showed a lowered stability, i.e., Q054 and Q055, are almost identical, explaining the identical T50 and suggesting a similar degradation mechanism.
Table 2. Decay Constants, T50 and T90 [±SEM], of the Four QSPs Used for the External Validation.
| peptide | decay constant (λ) | T50 (h) | T90 (h) |
|---|---|---|---|
| Q044 | –0.00126 [±0.00025] | 9.15 [±1.84] | 1.39 [±0.28] |
| Q051 | –0.00045 [±0.00061] | 25.69 [±34.7] | 3.91 [±5.28] |
| Q054 | –0.00619 [±0.00056] | 1.87 [±0.17] | 0.28 [±0.03] |
| Q055 | –0.00776 [±0.00064] | 1.49 [±0.12] | 0.23 [±0.02] |
A head-to-head comparison between P100 at 4 °C and EDTA at 4 °C was carried out to determine the gain in stability of Q055 when using a P100 sampling tube stored at 4 °C. The results are presented in Table 3, and although it was statistically nonsignificant, an increase in stability was noticed when using a P100 sampling tube.
Table 3. Decay Constants, T50 and T90 [±SEM], of Q055 in a P100 or an EDTA Sampling Tube.
| sampling tube | decay constant (λ) | T50 (h) | T90 (h) |
|---|---|---|---|
| EDTA at 4 °C | –0.00856 [±0.00210] | 1.35 [±0.33] | 0.21 [±0.05] |
| P100 at 4 °C | –0.00559 [±0.00098] | 2.07 [±0.36]a | 0.31 [±0.05]a |
The obtained results are similar to the results obtained in Table 3, supporting the consistency of our experiments.
2.2. Plasma Storage
To investigate the long-term stability of peptides in plasma of P100 tubes, plasma collected from P100 tubes was spiked with the same peptide mix as in the blood collection training experiment. Table 4 shows the T90 values of the five peptides in plasma when stored at one of the three different long-term storage temperatures and thawed once. In line with expectations, maintaining the plasma samples at low temperatures slowed down the rate of peptide degradation (Figure 3). After 1 month of storage at −80 °C, Q164 and Q176 still contained more than 95% of their original content. However, assuming that the same change in pattern will continue beyond the period covered by long-term data, T90 values were obtained by extrapolation. It was surprising that some peptides had very low stabilities, even at −80 °C. Q184, a relatively hydrophobic small peptide, showed a decrease of 10% degradation after only 2.6 days of storage at −80 °C. After 1 month of storage at this low temperature, only ±25% of the peptide could be detected. This decrease could be caused by chemical degradation or other physical forces (e.g., adsorption).
Table 4. T90 [±SEM] in P100 Plasma of the Five Peptides (n = 4)a.
| peptide | temperature (°C) | T90 in days (±SEM) |
|---|---|---|
| Q019 | 4 | 0.21 (±0.03) |
| –35 | 4.88 (±0.67) | |
| –80 | 7.48 (±2.21) | |
| Q076 | 4 | 1.24 (±0.03) |
| –35 | 20.4 (±13.67) | |
| –80 | 13.2 (±4.80) | |
| Q164 | 4 | 0.95 (±0.23) |
| –35 | 33.8 (±45.91) | |
| –80 | 96.3 (±136.01) | |
| Q176 | 4 | 0.48 (±0.02) |
| –35 | 16.4 (±6.98) | |
| –80 | 46.4 (±64.18) | |
| Q184 | 4 | 2.24 (±0.20) |
| –35 | 2.56 (±0.21) | |
| –80 | 2.61 (±0.23) |
T90 is extrapolated assuming that the same pattern will continue beyond the period covered by long-term data.
Figure 3.
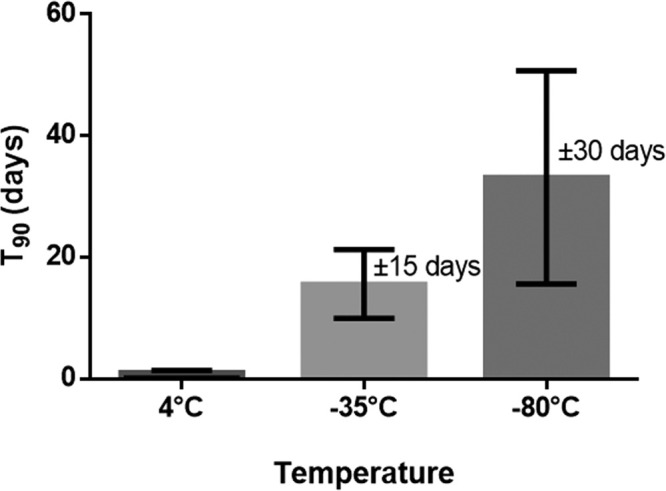
T90 values (±SEM) at different storage temperatures (average of all peptides together, n = 4 per peptide).
These findings contrast the common thoughts, in both clinical and research settings, that long-term plasma storage at −80 °C does not influence the stability of analytes. As mentioned above, many peptidomic studies store their samples at −80 °C, without taking into account the chemical/physical stability at this temperature. Our results emphasize that this is a false assumption and that storage stability data for investigated peptides are needed if plasma is stored longer-term before being analyzed.
2.3. Cost Considerations
Based on the effectiveness experiments described above, P100 or P800 tubes are preferred above conventional EDTA tubes for quorum-sensing peptide analysis. However, when deciding on therapeutic or diagnostic tools, also costs are important to consider. The BD P100 and BD P800 tubes currently on the market are available for around 17–25€ each, while conventional EDTA tubes cost around 0.4–0.5€ each, i.e., P100 and P800 tubes are about thirty to sixty times more expensive than the conventional EDTA tubes. In the case of P100 tubes at 4 °C, this price difference comes with a mean T50 and T90 increase of 25 and 3.5 h, respectively (Table 1 and Figure 2). Depending on the purpose of the peptidome identification/quantitation, this cost increase may or may not be justified. In situations where one seeks to perform high-throughput experiments and/or identification/quantitation of high abundant peptides, conventional EDTA tubes may be fit-for-purpose. However, for detection/quantitation of low abundant peptides or small peptide concentration differences in a limited set of samples, the gain in peptidomic sensitivity of P100 tubes seems worth the extra costs. If, for example, EDTA blood samples are analyzed 1 h after withdrawal, a significant amount of peptide degradation will already have occurred (>20% degradation for a number of peptides). In this situation, low abundant peptides could fall below the limit of detection, or small peptide concentration differences could wrongly be attributed to biological differences (type I errors). Therefore, the choice of tube should also be based on the purpose of the peptidome analysis.
3. Conclusions
Cooling whole blood to 4 °C in BD P100 tubes provides overall the highest quorum-sensing peptide stability over time when compared to BD P800 tubes, classical EDTA tubes, and EDTA tubes supplemented with a protease-inhibitor cocktail (Sigma-Aldrich) at different temperatures. However, when no fridge is available and blood samples are forced to be stored at room temperature (21 °C), the BD P800 collection tubes prove significantly better than the other tubes. For long-term plasma storage awaiting its quorum-sensing peptidome analysis, lower temperatures result in less peptide degradation over time. However, even at −80 °C, significant degradation cannot be ruled out. This study should assist researchers and clinicians in their choice for optimal blood collection and storage conditions of plasma for quorum-sensing peptidome analysis.
4. Methods
4.1. Blood Collection
Healthy adults were enrolled in this study after providing informed consent according to the protocol that was approved by the local ethical committee (EC number B670201734698). The protocol included a training experiment, followed by a validation experiment. Blood samples were collected by a medical doctor from the cubital vein. For the training group, four different tubes were used: EDTA plasma (Becton Dickinson, product 368861), EDTA plasma (Becton Dickinson, product 368861) spiked with protease-inhibitor cocktail (Sigma-Aldrich, product P8340) at a 1:100 final dilution, BD P100 (Becton Dickinson, product 366448), and BD P800 (Becton Dickinson, product 366421). In the validation group, only EDTA plasma and BD P100 tubes were used. Immediately after blood withdrawal, the whole blood was brought to the indicated temperature (during 30 min). Three different temperatures were assessed, namely, 4, 21, and 37 °C. When the desired temperature was reached, i.e., after 30 min, the whole blood was spiked with the peptide mix, with each peptide at a final concentration of 10 nM. The sample preparation and analysis were performed at indicated timepoints after spiking.
4.2. Plasma Long-Term Storage
Plasma, collected into BD P100 tubes and separated from blood cells, was spiked with a peptide mix, with each peptide having a final concentration of 50 nM. The spiked plasma was aliquoted into 500 μL LoBind Eppendorf tubes (Eppendorf AG, product 022431064) and stored at three different temperatures, i.e., 4, −35, and −80 °C, for up to 1 month.
4.3. Quorum-Sensing Peptides
Bacterial quorum-sensing peptides (QSPs) were used to spike the whole blood and plasma because they exhibit very diverse chemical properties.21,36 Previous work conducted by our group defined different clusters of QSP based on characteristic chemical properties, including peptide size/compactness, hydrophilicity/lipophilicity, cyclization, and the presence of (unnatural) S-containing and aromatic amino acids. The QSPs used for the training (Q019, Q076, Q164, Q176, Q184) and validation (Q044, Q051, Q054, Q055) experiment were selected from different clusters, assuring that a reasonably large chemical space of peptides was involved in the experiments.21Table 5 indicates the sequences and some physicochemical descriptors of the peptides, demonstrating their diversity. For the storage experiment, the same peptides as the training experiment were used.
4.4. Sample Preparation
For the blood collection experiments (training and validation samples), the spiked whole-blood samples were first vortexed. Then, 500 μL samples were taken after 0, 2.5, 5, and 7.5 h and transferred to a 1.5 mL LoBind Eppendorf tube (Eppendorf AG, product 022431081). These were centrifuged at 1800g for 10 min at room temperature (RT). Fifty microliters of the obtained plasma was transferred to a 0.5 mL Eppendorf coated with an in-house-developed LC–MS-compatible antiadsorption diluent for peptide analysis.37 One hundred fifty microliters of acetonitrile (Biosolve BV, product 1204102BS) acidified with 0.5% formic acid (Biosolve BV, product 6914143BS) was added to the plasma, after which the tube was closed. The samples were subsequently sonicated (5 min at RT), vortexed (5 s at RT), boiled (30 s at 100 °C), again vortexed (5 s at RT), and centrifuged at 20 000g for 20 min. Finally, the samples were put in the sample manager (10 °C) for UPLC–MS–MS analysis (60 °C). On average, the relative standard deviation (RSD) for the complete analytical workflow (spiking, sample preparation, and analytical method) was 19.5%, which is considered as an acceptable value (EMA and FDA Draft Guideline on validation of bioanalytical methods).
For the storage experiment, peptide concentrations in plasma were assessed before storage and after 1, 7, and 30 days of storage. Fifty microliters of the plasma was transferred to a coated 0.5 mL tube as described above, after which the same protocol was followed as in the blood collection experiments.
4.5. Peptide Identification and Quantitation by UPLC–MS/MS
Chromatographic separation was achieved on a Waters Acquity UPLC BEH amide column (2.1 mm × 100 mm; 1.7 μm particles), protected with a guard column. A mobile phase system consisting of solvent A (water, acetonitrile, and dimethyl sulfoxide (DMSO) at the ratio of 93:2:5 v/v/v and 0.1% formic acid m/v) and solvent B (water, acetonitrile, and DMSO in the ratio of 2:93:5 v/v/v and 0.1% formic acid m/v) with a gradient from 100 to 45% B in 5 min was applied. DMSO was added as a supercharger and to reduce carryover.38 The mobile phase was delivered at a flow rate of 0.5 mL/min, with a total run time of 15 min. The UPLC system was connected to a Xevo TQ-S spectrometer with electrospray ionization (set in the positive mode). The mass spectrometric conditions used throughout the analyses as well as the MRMs of the peptides are shown in Table S1.
4.6. Data Analysis
The areas were normalized to the area measured at T = 0 (for each tube at different temperatures), and subsequently, the natural logarithm (n) of each relative value was calculated. Assuming first-order kinetics, the decay constant (λ), obtained from the slope of the best-fitting ln-linear degradation curve, was used to determine the half-life using the following formula
The standard error of the mean (SEM) on the half-life (T1/2) was estimated using the propagation of errors with the following formula39
with σ(λ) being the standard deviation on the decay constant.
Similar calculations were conducted for the T90, i.e., the time where still 90% of the initial concentration is present
One-way ANOVA with paired multiple-comparison Tukey’s test was applied to detect statistical significant differences between tubes. All statistical analyses were done with GraphPad Prism Version 6 (San Diego, CA).
Acknowledgments
This work was supported by the Research Foundation Flanders (1S21017N to N.D. and 1158818N to A.D.S.). The DruQuaR research group is part of the Ghent University expertise center MSsmall.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsomega.0c01723.
Mass spectrometric conditions and MRMs of the peptides (Table S1); matrix effect of the different peptides in the different sampling tubes (Figure S1); average matrix effect across the four tubes over time of the different peptides (Figure S2); mean stability (T90) of peptides at different temperatures (Figure S3) (PDF)
Author Contributions
∥ N.D. and A.D.S. contributed equally to this work.
The authors declare no competing financial interest.
Supplementary Material
References
- Borrebaeck C. A. K. Precision diagnostics: moving towards protein biomarker signatures of clinical utility in cancer. Nat. Rev. Cancer 2017, 17, 199–204. 10.1038/nrc.2016.153. [DOI] [PubMed] [Google Scholar]
- Navickas R.; Gal D.; Laucevicius A.; Taparauskaite A.; Zdanyte M.; Holvoet P. Identifying circulating microRNAs as biomarkers of cardiovascular disease: a systematic review. Cardiovasc. Res. 2016, 111, 322–337. 10.1093/cvr/cvw174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srikanthan K.; Feyh A.; Visweshwar H.; Shapiro J. I.; Sodhi K. Systematic Review of Metabolic Syndrome Biomarkers: A Panel for Early Detection, Management, and Risk Stratification in the West Virginian Population. Int. J. Med. Sci. 2016, 13, 25–38. 10.7150/ijms.13800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishino M.; Takeishi Y.; Niizeki T.; Watanabe T.; Nitobe J.; Miyamoto T.; Miyashita T.; Kitahara T.; Suzuki S.; Sasaki T.; Bilim O.; Kubota I. Risk stratification of chronic heart failure patients by multiple biomarkers: implications of BNP, H-FABP, and PTX3. Circ. J. 2008, 72, 1800–1805. 10.1253/circj.CJ-08-0157. [DOI] [PubMed] [Google Scholar]
- Zhang X.; Sha M.; Yao Y.; Da J.; Jing D. Increased B-type-natriuretic peptide promotes myocardial cell apoptosis via the B-type-natriuretic peptide/long non-coding RNA LSINCT5/caspase-1/interleukin 1beta signaling pathway. Mol. Med. Rep. 2015, 12, 6761–6767. 10.3892/mmr.2015.4247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks C. L. 3rd Protein and peptide folding explored with molecular simulations. Acc. Chem. Res. 2002, 35, 447–454. 10.1021/ar0100172. [DOI] [PubMed] [Google Scholar]
- Di L. Strategic approaches to optimizing peptide ADME properties. AAPS J. 2015, 17, 134–143. 10.1208/s12248-014-9687-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hortin G. L.; Warshawsky I.; Laude-Sharp M. Macromolecular chromogenic substrates for measuring proteinase activity. Clin. Chem. 2001, 47, 215–222. 10.1093/clinchem/47.2.215. [DOI] [PubMed] [Google Scholar]
- Debunne N.; Verbeke F.; Janssens Y.; Wynendaele E.; De Spiegeleer B. Chromatography of Quorum Sensing Peptides: An Important Functional Class of the Bacterial Peptidome. Chromatographia 2018, 81, 25–40. 10.1007/s10337-017-3411-2. [DOI] [Google Scholar]
- Astefanei A.; Dapic I.; Camenzuli M. Different Stationary Phase Selectivities and Morphologies for Intact Protein Separations. Chromatographia 2017, 80, 665–687. 10.1007/s10337-016-3168-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Spiegeleer B.; Verbeke F.; D’Hondt M.; Hendrix A.; Van De Wiele C.; Burvenich C.; Peremans K.; De Wever O.; Bracke M.; Wynendaele E. The quorum sensing peptides PhrG, CSP and EDF promote angiogenesis and invasion of breast cancer cells in vitro. PLoS One 2015, 10, e0119471 10.1371/journal.pone.0119471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssens Y.; Wynendaele E.; Verbeke F.; Debunne N.; Gevaert B.; Audenaert K.; Van DeWiele C.; De Spiegeleer B. Screening of quorum sensing peptides for biological effects in neuronal cells. Peptides 2018, 101, 150–156. 10.1016/j.peptides.2018.01.013. [DOI] [PubMed] [Google Scholar]
- De Spiegeleer A.; Elewaut D.; Van Den Noortgate N.; Janssens Y.; Debunne N.; Van Langenhove S.; Govindarajan S.; De Spiegeleer B.; Wynendaele E. Quorum sensing molecules as a novel microbial factor impacting muscle cells. Biochim. Biophys. Acta, Mol. Basis Dis. 2020, 1866, 165646 10.1016/j.bbadis.2019.165646. [DOI] [PubMed] [Google Scholar]
- Wynendaele E.; Verbeke F.; D’Hondt M.; Hendrix A.; Van De Wiele C.; Burvenich C.; Peremans K.; De Wever O.; Bracke M.; De Spiegeleer B. Crosstalk between the microbiome and cancer cells by quorum sensing peptides. Peptides 2015, 64, 40–48. 10.1016/j.peptides.2014.12.009. [DOI] [PubMed] [Google Scholar]
- Yi J. Z.; Liu Z. X.; Craft D.; O’Mullan P.; Ju G.; Gelfand C. A. Intrinsic Peptidase Activity Causes a Sequential Multi-Step Reaction (SMSR) in Digestion of Human Plasma Peptides. J. Proteome Res. 2008, 7, 5112–5118. 10.1021/pr800396c. [DOI] [PubMed] [Google Scholar]
- Pérez V.; Juega-Marino J.; Bonjoch A.; Negredo E.; Clotet B.; Romero R.; Bonet J. Evaluation of protease inhibitors containing tubes for MS-based plasma peptide profiling studies. J. Clin. Lab. Anal. 2014, 28, 364–367. 10.1002/jcla.21694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi J. Z.; Warunek D.; Craft D. Degradation and Stabilization of Peptide Hormones in Human Blood Specimens. PLoS One 2015, 10, e0134427 10.1371/journal.pone.0134427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Z.; Kamlage B.; Wagner-Golbs A.; Maisha M.; Sun J. C.; Schnackenberg L. K.; Pence L.; Schmitt T. C.; Daniels J. R.; Rogstad S.; Beger R. D.; Yu L. R. An Integrated Analysis of Metabolites, Peptides, and Inflammation Biomarkers for Assessment of Preanalytical Variability of Human Plasma. J. Proteome Res. 2019, 18, 2411–2421. 10.1021/acs.jproteome.8b00903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cegla J.; Jones B. J.; Howard J.; Kay R.; Creaser C. S.; Bloom S. R.; Tan T. M. The preanalytical stability of glucagon as measured by liquid chromatography tandem mass spectrometry and two commercially available immunoassays. Ann. Clin. Biochem. 2017, 54, 293–296. 10.1177/0004563216675648. [DOI] [PubMed] [Google Scholar]
- Howard J. W.; Kay R. G.; Tan T.; Minnion J.; Ghatei M.; Bloom S.; Creaser C. S. Development of a high-throughput UHPLC-MS/MS (SRM) method for the quantitation of endogenous glucagon from human plasma. Bioanalysis 2014, 6, 3295–3309. 10.4155/bio.14.226. [DOI] [PubMed] [Google Scholar]
- Wynendaele E.; Gevaert B.; Stalmans S.; Verbeke F.; De Spiegeleer B. Exploring the chemical space of quorum sensing peptides. Biopolymers 2015, 104, 544–551. 10.1002/bip.22649. [DOI] [PubMed] [Google Scholar]
- Couso J. P.; Patraquim P. Classification and function of small open reading frames. Nat. Rev. Mol. Cell Biol. 2017, 18, 575–589. 10.1038/nrm.2017.58. [DOI] [PubMed] [Google Scholar]
- Taevernier L.; Wynendaele E.; Gevaert B.; Spiegeleer B. Chemical Classification of Cyclic Depsipeptides. Curr. Protein Pept. Sci. 2017, 18, 425–452. 10.2174/1389203717666161128141438. [DOI] [PubMed] [Google Scholar]
- Wynendaele E.; Bronselaer A.; Nielandt J.; D’Hondt M.; Stalmans S.; Bracke N.; Verbeke F.; Van De Wiele C.; De Tre G.; De Spiegeleer B. Quorumpeps database: chemical space, microbial origin and functionality of quorum sensing peptides. Nucleic Acids Res. 2013, 41, D655–9. 10.1093/nar/gks1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darkoh C.; DuPont H. L.; Norris S. J.; Kaplan H. B. Toxin synthesis by Clostridium difficile is regulated through quorum signaling. mBio 2015, 6, e02569 10.1128/mBio.02569-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonora M.; Wieckowsk M. R.; Chinopoulos C.; Kepp O.; Kroemer G.; Galluzzi L.; Pinton P. Molecular mechanisms of cell death: central implication of ATP synthase in mitochondrial permeability transition. Oncogene 2015, 34, 1608. 10.1038/onc.2014.462. [DOI] [PubMed] [Google Scholar]
- Mussbacher M.; Schrottmaier W. C.; Salzmann M.; Brostjan C.; Schmid J. A.; Starlinger P.; Assinger A. Optimized plasma preparation is essential to monitor platelet-stored molecules in humans. PLoS One 2017, 12, e0188921 10.1371/journal.pone.0188921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguilar-Mahecha A.; Kuzyk M. A.; Domanski D.; Borchers C. H.; Basik M. The Effect of Pre-Analytical Variability on the Measurement of MRM-MS-Based Mid- to High-Abundance Plasma Protein Biomarkers and a Panel of Cytokines. PLoS One 2012, 7, e38290 10.1371/journal.pone.0038290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Citrome L. Quantifying clinical relevance. Innovations Clin. Neurosci. 2014, 11, 26–30. [PMC free article] [PubMed] [Google Scholar]
- Murias M.; Rachtan M.; Jodynis-Liebert J. Effect of multiple freeze-thaw cycles of cytoplasm samples on the activity of antioxidant enzymes. J. Pharmacol. Toxicol. Methods 2005, 52, 302–305. 10.1016/j.vascn.2005.03.002. [DOI] [PubMed] [Google Scholar]
- Chróst R. J.; Velimirov B. Measurement of enzyme-kinetics in water samples - effect of freezing and soluble stabilizer. Mar. Ecol.: Prog. Ser. 1991, 70, 93–100. 10.3354/meps070093. [DOI] [Google Scholar]
- Randall S. A.; McKay M. J.; Molloy M. P. Evaluation of blood collection tubes using selected reaction monitoring MS: Implications for proteomic biomarker studies. Proteomics 2010, 10, 2050–2056. 10.1002/pmic.200900517. [DOI] [PubMed] [Google Scholar]
- Wildes D.; Wells J. A. Sampling the N-terminal proteome of human blood. Proc. Natl. Acad. Sci. U.S.A. 2010, 107, 4561–4566. 10.1073/pnas.0914495107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahboob S.; Ahn S.; Mohamedali A.; Amirkhani A.; Tan S.-H.; Ranganathan S.; Nice E.; Baker M. Addition of protease inhibitors to EDTA containing blood collection tubes does not deliver significant advantage for preservation of plasma for down-stream analysis. Clin. Proteomics Bioinf. 2016, 1, 1–9. [Google Scholar]
- Ayache S.; Panelli M.; Marincola F. M.; Stroncek D. F. Effects of storage time and exogenous protease inhibitors on plasma protein levels. Am. J. Clin. Pathol. 2006, 126, 174–184. 10.1309/3WM7XJ7RD8BCLNKX. [DOI] [PubMed] [Google Scholar]
- Verbeke F.; De Craemer S.; Debunne N.; Janssens Y.; Wynendaele E.; Van de Wiele C.; De Spiegeleer B. Peptides as Quorum Sensing Molecules: Measurement Techniques and Obtained Levels In vitro and In vivo. Front. Neurosci. 2017, 11, 1–18. 10.3389/fnins.2017.00183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verbeke F.; Bracke N.; Debunne N.; Wynendaele E.; De Spiegeleer B. LC-MS Compatible Antiadsorption Diluent for Peptide Analysis. Anal. Chem. 2020, 92, 1712–1719. 10.1021/acs.analchem.9b01840. [DOI] [PubMed] [Google Scholar]
- Hahne H.; Pachl F.; Ruprecht B.; Maier S. K.; Klaeger S.; Helm D.; Medard G.; Wilm M.; Lemeer S.; Kuster B. DMSO enhances electrospray response, boosting sensitivity of proteomic experiments. Nat. Methods 2013, 10, 989–991. 10.1038/nmeth.2610. [DOI] [PubMed] [Google Scholar]
- Ku H. Notes on the Use of Propagation of Error Formulas. J. Res. Natl. Bur. Stand., Sect. C 1966, 70C, 263–273. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.