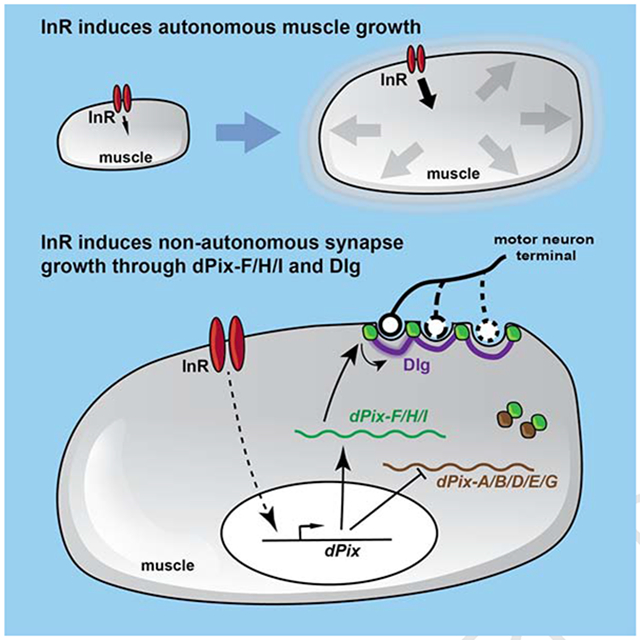
Summary
Developmental growth requires coordination between the growth rates of individual tissues and organs. Here we examine how Drosophila neuromuscular synapses grow to match the sizes of their target muscles. We show that changes in muscle growth driven by autonomous modulation of Insulin receptor signaling produce corresponding changes in synapse size, with each muscle affecting only its presynaptic motor neuron branches. This scaling growth is mechanistically distinct from synaptic plasticity driven by neuronal activity, and requires increased postsynaptic differentiation induced by Insulin receptor signaling in muscle. We identify the guanine nucleotide exchange factor dPix as an effector of Insulin receptor signaling. Alternatively spliced dPix isoforms that contain a specific exon are necessary and sufficient for postsynaptic differentiation and scaling growth, and their mRNA levels are regulated by Insulin receptor signaling. These findings define a mechanism by which the same signaling pathway promotes both autonomous muscle growth and non-autonomous synapse growth.
Keywords: neuromuscular junction, scaling growth, Insulin receptor, dPix, Discs-large, muscle, motor neuron, synapse
Graphical Abstract
eTOC Blurb
How interacting cells coordinate their growth remains largely unknown. Ho and Treisman show that Insulin receptor signaling, which autonomously promotes the growth of larval muscles in Drosophila, also acts through the effector dPix to drive postsynaptic expansion that non-autonomously scales the size of the synapses formed on those muscles.
Introduction
The development of every organism requires the growth of different tissues and organs to be coordinated and proportional, a process known as scaling growth (Cowin, 2011; Smith, 1985). Scaling growth is a particularly challenging problem for the peripheral nervous system. Sensory and motor neurons must continuously adjust the size of their arborizations according to the area of their target tissues to maintain effective stimulus detection or muscle depolarization throughout development (Bentley and Toroian-Raymond, 1981; Bucher and Pflüger, 2000; Lee and Stevens, 2007; Menon et al., 2013). In Drosophila, endoreplication of epithelial cells controlled by the microRNA bantam enables them to enclose the dendrites of adjacent da sensory neurons to restrict the growth of their dendritic arborizations (Jiang et al., 2014; Parrish et al., 2009). Epithelial cells also regulate dendrite growth by producing the TGF-β family member Maverick (Hoyer et al., 2018). However, the molecular mechanisms by which neuronal arborizations adjust to changes in the size of their target tissues remain largely unknown.
The larval neuromuscular junction (NMJ) of Drosophila is a convenient system to study scaling growth. Motor neurons reach their target muscles at the end of embryogenesis, and form synapses that consist of multiple synaptic boutons, each of which contains many active zones (Menon et al., 2013). As the muscles grow up to 100-fold during larval development, the NMJ expands its size by adding new boutons and axonal branches (Schuster et al., 1996). In the muscle, the postsynaptic membrane network surrounding each bouton, known as the subsynaptic reticulum (SSR), grows by new membrane addition and further invagination (Menon et al., 2013; Zito et al., 1999). Moreover, presynaptic active zones and postsynaptic glutamate receptors are constantly added to boutons during development (Owald et al., 2010; Petzoldt et al., 2016; Rasse et al., 2005; Van Vactor and Sigrist, 2017). It is not yet known how this active synaptic growth is coordinated with growth of the target muscle.
In Drosophila and other organisms, signaling through the Insulin Receptor (InR) or insulin-like growth factor receptors controls the growth of tissues and organs (Liu et al., 1993; Shingleton, 2010). At the cellular level, InR signaling positively regulates translation, metabolism, and the accumulation of cell mass (Böhni et al., 1999; Verdu et al., 1999), leading to an increase in animal and organ size (Brogiolo et al., 2001; Fernandez et al., 1995; Stocker et al., 2002). Insulin-like peptides produced by specific neurons in the brain circulate through the body in the hemolymph and activate the InR signaling pathway in different body parts to coordinate growth (Okamoto et al., 2013; Semaniuk et al., 2018; Ugrankar et al., 2018). InR signaling in muscle also increases larval feeding time, which increases the size of all organs (Demontis and Perrimon, 2009). Whether InR signaling influences scaling growth of the peripheral nervous system is not known.
Here we examined scaling growth of the Drosophila larval NMJ. By manipulating InR signaling only in muscles, we found that NMJ size adjusts to changes in muscle growth, maintaining a constant ratio between the two. This scaling growth is a local phenomenon that is mechanistically distinct from activity-dependent synaptic plasticity. We found that postsynaptic differentiation mediated by specific isoforms of dPix, a Rho-family guanine nucleotide exchange factor (Parnas et al., 2001), is necessary for scaling growth of the NMJ. InR signaling regulates the relative mRNA abundance of active and antagonistic dPix isoforms. We propose that dPix couples synaptic expansion to muscle growth to enable scaling growth of the NMJ.
Results
The larval NMJ undergoes scaling growth
To determine whether motor neurons adjust the growth of their synapses in response to changing target sizes during development, we autonomously modified muscle size by activating or inhibiting the InR pathway specifically in the larval abdominal muscles. Activation of InR signaling in larval muscle has been shown to promote its growth (Demontis and Perrimon, 2009). Using muscle surface area (MSA) as a readout, we confirmed that expression of a dominant negative form of InR (InR-DN) (Wu et al., 2005) with the muscle-specific driver C57-GAL4 (Budnik et al., 1996) decreased muscle growth (Figure S1A, B, Figure 1D), while overexpressing wild-type InR (InR-WT) increased muscle growth (Figure S1B, C, Figure 1D).
Figure 1. Scaling growth of the NMJ in response to changes in muscle size.
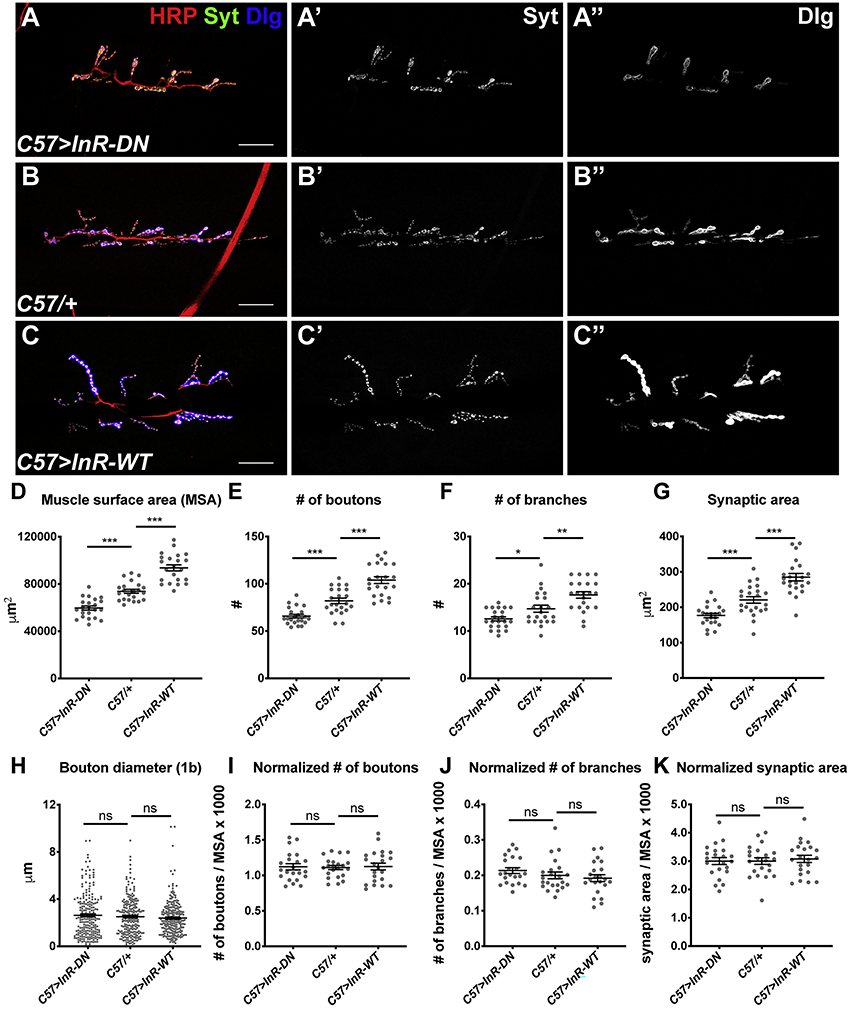
(A-C) Confocal images of the NMJ on muscle 6/7 in segment A3 in C57-GAL4>UAS-InR-DN (A), C57-GAL4/+ (B), and C57-GAL4>UAS-InR-WT (C) third instar larvae labeled with anti-HRP (red) to mark the nerve, anti-Synaptotagmin (Syt; A’-C’, green in A-C) as a presynaptic marker, and anti-Dlg (A’’-C’’, blue in A-C) as a postsynaptic marker. Scale bars, 30 μm. (D-K) Quantification of the muscle surface area (MSA) (D), total number of boutons (E), total number of branches (F), total synaptic area (G), diameters of 1b boutons (H), number of boutons normalized to MSA (I), number of branches normalized to MSA (J), and synaptic area normalized to MSA (K) of the NMJ on muscle 6/7. Synapse size changes in response to changes in muscle surface area, maintaining a constant ratio between the two. n=20 (C57>InR-DN), n=21 (C57/+), n=19 (C57>InR-WT). Error bars in this and subsequent graphs show mean ± SEM. *p< 0.05, **p<0.01, ***p<0.0005; ns, not significant by unpaired Student’s t-test (D-G, I-L) or F-test (H). See also Figure S1.
We found that these autonomous changes in muscle growth induced corresponding changes in NMJ size (Figure 1A-C). Three parameters were used to measure NMJ size: the number of synaptic boutons (Figure 1E), the number of nerve branches (Fig. 1F), and the area of the presynaptic motor neuron (Figure 1G). All these measurements decreased by ~25% when InR-DN was expressed in muscle, and increased by ~20% when InR-WT was expressed in muscle. The ratio of each measurement of NMJ size to the muscle surface area remained constant (Figure 1I-K), and the average size of individual boutons was not affected (Figure 1H). A similar scaling of synapse size to muscle size was observed when InR signaling was increased or decreased by expressing wild-type or dominant negative forms of Phosphoinositide-3-kinase (PI3K; Figure S1D-F). However, when InR signaling was manipulated by overexpressing or knocking down the downstream kinase Akt, synapse size changed to a smaller extent than muscle size, resulting in changes in the normalized number of boutons (Figure S1D-F). This suggests a branch in the canonical InR signaling pathway, such that synapse growth requires both Akt and another effector of PI3K. Nevertheless, these results show that autonomous changes in muscle growth induced by InR signaling trigger proportional adjustments of NMJ size, suggesting that motor neurons can detect the growth of their target muscles and respond by adjusting the size of the synapses they form.
Scaling growth of the NMJ is locally regulated
Previous experiments showed that manipulating InR signaling in muscle not only affected muscle growth, but also modified larval feeding behavior, which altered developmental timing and led to changes in the growth of other tissues (Demontis and Perrimon, 2009). To test whether these changes in developmental timing are responsible for NMJ scaling growth, we examined animals in which individual muscles grew at different rates. We used the 5053A-GAL4 driver to express transgenes that affect InR signaling specifically in muscle 12 (m12) (Jarecki et al., 1999; Ritzenthaler et al., 2000). Different motor neurons, RP5 and RP1/RP4 respectively, form type 1b boutons on m12 and the adjacent muscle 13 (m13), while the motor neuron MNSNb/d-Is forms type 1s boutons on both muscles (Hoang and Chiba, 2001; Landgraf et al., 1997; Sink and Whitington, 1991). Strongly inhibiting InR signaling in m12 by expressing Akt RNAi with 5053A-GAL4 reduced the growth of m12 without affecting m13 (Figure 2F, I). The surface area of m12 was decreased to a similar extent as when Akt RNAi was expressed in all muscles with C57-GAL4 (Figure 2F). However, specific inhibition of m12 growth with 5053A-GAL4>UAS-Akt RNAi only decreased the number of boutons on m12, but not on m13 (Figure 2A-D, G, H, J, K). In contrast, C57-GAL4>UAS-Akt RNAi had a similar effect on the numbers of boutons on both m12 and m13 (Figure 2E, G, H, J, K). Both type 1b and type 1s boutons responded in the same way to reductions in muscle size (Figure 2G, H, J, K). Because only the NMJ on the muscle with reduced size was affected, NMJ scaling growth must depend on muscle growth, rather than developmental timing. Moreover, since the type 1s boutons on both m12 and m13 are derived from the same motor neuron (Hoang and Chiba, 2001), and only the branches on m12 modified their synapse size in response to a change in m12 growth (Figure 2H, K), scaling growth of the NMJ must be regulated locally at the level of individual motor neuron branches.
Figure 2. Scaling growth is locally driven by target muscle growth.
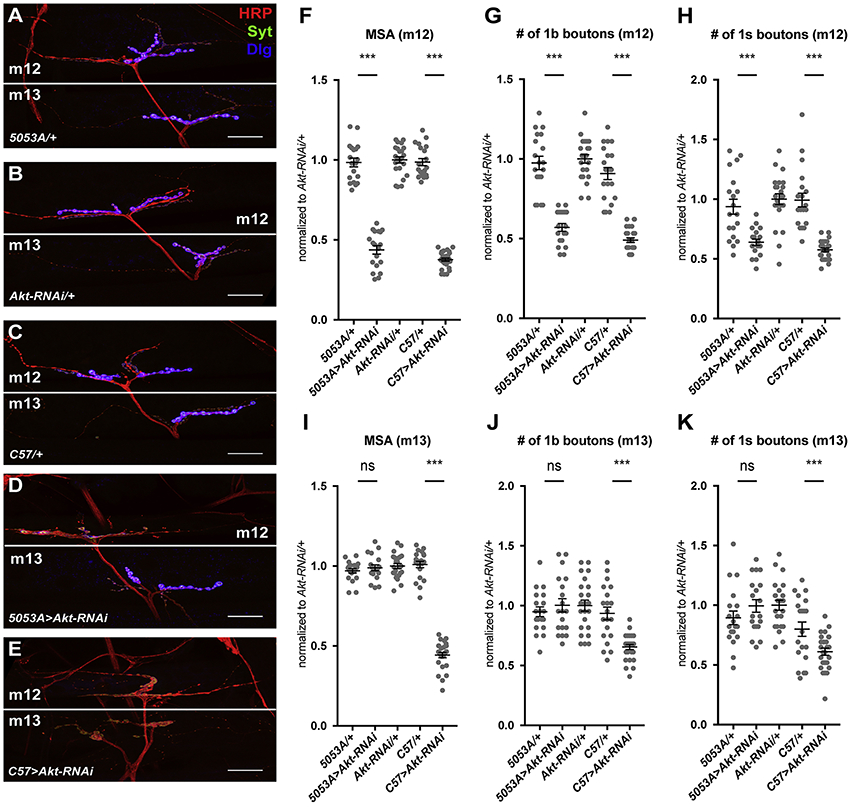
(A-E) Confocal images of the NMJ on muscle 12/13 in 5053A-GAL4/+ (A), UAS-Akt-RNAi/+ (B), C57-GAL4/+ (C), 5053A-GAL4>UAS-Akt-RNAi (D), and C57-GAL4>UAS-Akt-RNAi (E) labeled with anti-HRP (red), anti-Syt (green), and anti-Dlg (blue). The Dlg signal is overexposed to show the intensity difference when Akt was knocked down (D, E). Scale bars, 30 μm. (F-K) Quantification of the MSA of muscle 12 (m12) (F), number of type 1b boutons on m12 (G), number of type 1s boutons on m12 (H), MSA of muscle 13 (m13) (I), number of type 1b boutons on m13 (J), and number of type 1s boutons on m13 (K). Knocking down Akt in muscle 12 only reduces the number of boutons on muscle 12, while knocking it down in all muscles reduces the number on muscles 12 and 13. n=19 (5053A/+, 5053A>Akt-RNAi), n=23 (Akt-RNAi/+), n=20 (C57/+), and n=26 (C57>Akt-RNAi). ***p<0.0005; ns, not significant by unpaired Student’s t-test.
Scaling growth of the NMJ is independent of activity-dependent synaptic plasticity
Synapse growth is also regulated by activity-dependent synaptic plasticity (Berke et al., 2013; Budnik et al., 1990; Lnenicka et al., 2003; Mosca et al., 2005; Zhong et al., 1992). At the Drosophila NMJ, increasing the activity of the motor neuron either by elevated temperature or through mutations increases its synaptic arborization, while decreasing activity reduces synapse size (Berke et al., 2013; Mosca et al., 2005; Zhong et al., 1992). Therefore, both scaling growth and neuronal activity can bidirectionally regulate NMJ morphology. To determine whether the two processes share a common mechanism, we tested whether scaling growth was affected in mutants defective for activity-dependent synaptic plasticity.
Mutations in Glutamate receptor IIA (GluRIIA), Synaptotagmin 4 (Syt4), which functions in a retrograde signaling pathway that controls motor neuron growth (Yoshihara et al., 2005), and rutabaga (rut), which encodes a membrane-bound calcium-/calmodulin-activated adenylate cyclase (Guan et al., 2011), have been shown to impair activity-dependent structural plasticity at the NMJ (Guan et al., 2011; Steinert et al., 2006; Yoshihara et al., 2005). Overexpressing wild type InR in the muscle of these mutants significantly increased the size of both the muscle and the NMJ (Figure S2A-J), preserving the ratio of the number of boutons to the MSA (Figure S2K). These results argue that scaling growth can occur normally when activity-dependent structural plasticity is defective.
Since a low level of activity-dependent synaptic plasticity is still observed in the above mutants, we examined scaling growth in a mutant defective for BMP signaling in which this mechanism is completely abolished. The muscle-derived BMP Glass bottom boat signals through the Wishful thinking and Thickveins receptors and the Mothers against Dpp (Mad) transcription factor to inform the motor neuron that it has reached its target muscle and to regulate the growth, stability, and function of the NMJ (Eaton and Davis, 2005; Goold and Davis, 2007; McCabe et al., 2003). Mutations of BMP signaling pathway components completely abolish structural plasticity in response to increased neuronal activity at the NMJ, induced either by elevated temperature or by loss of the Ether-a-go-go (Eag) and Shaker (Sh) potassium channels that would normally repolarize the neuron after an action potential (Berke et al., 2013). We confirmed that the NMJ of a Mad mutant could not respond to mutations of eag and Sh by increasing bouton number (Figure 3A-D, G-I). In contrast, expressing wildtype InR in Mad mutant muscles increased both muscle growth and bouton number, maintaining the two in a constant ratio (Figure 3E, F, J-L). Therefore, scaling growth still occurs normally even when activity-induced synaptic structural plasticity is completely defective, indicating that it is genetically independent of the known synaptic plasticity pathway and of BMP signaling.
Figure 3. Scaling growth of the NMJ is independent of activity-dependent synaptic plasticity.
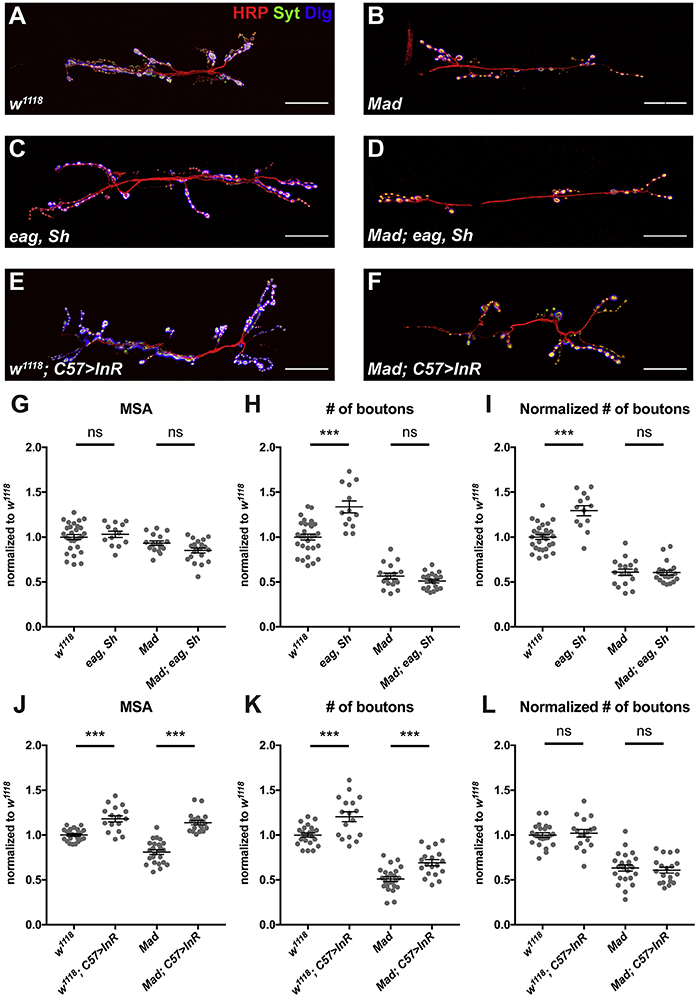
(A-F) Confocal images of the NMJ on muscle 6/7 in w1118 (A), Mad (B), ether a go-go (eag), Shaker (Sh) (C), eag, Sh; Mad (D), w1118; C57-GAL4>UAS-InR-WT (E), and Mad; C57-GAL4>UAS-InR-WT (F) labeled with anti-HRP (red), anti-Syt (green), and anti-Dlg (blue). Scale bars, 30 μm. (G-L) Quantification of the muscle 6/7 MSA (G, J), number of boutons (H, K), and number of boutons normalized to MSA (I, L) of the NMJ on muscle 6/7. Mad is required for the increase in bouton number in eag, Sh double mutants, but not for the increase in bouton number in response to InR activation in muscle. n=23 (w1118), n=24 (Mad), n=13 (eag, Sh), n=20 (eag, Sh; Mad,), n=17 (w1118; C57>InR-WT), or n=19 (Mad; C57>InR-WT). ***p<0.0001; ns, not significant by unpaired Student’s t-test. See also Figure S2.
Postsynaptic differentiation is necessary for scaling growth of the NMJ
In searching for a mechanism that governs the scaling growth of the NMJ, we noticed that the postsynaptic compartment, as marked by the PSD-95 homologue Discs-large (Dlg), also changed its size in concert with changes in muscle InR signaling (Figure 1A’’-C’’, Figure 4A, B). A similar expansion of the postsynaptic compartment occurs during normal larval development, as the SSR surrounding each bouton increases in size and complexity (Budnik et al., 1996). Dlg is localized to the SSR and recruits other postsynaptic components there (Thomas et al., 2000; Thomas et al., 1997). We found that expressing wildtype InR in muscle increased the levels of additional postsynaptic components. Staining for the glutamate receptor subunit GluRIIC appeared more continuous when muscle InR signaling was elevated, suggesting that more GluRIIC was present (Figure 4A, B). Two other postsynaptic scaffolding molecules, Cactus and Dorsal (Heckscher et al., 2007; Zhou et al., 2015), were also upregulated (Figure 4C-F). In contrast, the levels of the presynaptic markers Synaptotagmin (Syt), Synapsin (Syn), and Bruchpilot (Brp) appeared unchanged (Figure S3). Therefore, scaling growth of the NMJ is accompanied by an expansion of the postsynaptic but not the presynaptic compartment.
Figure 4. Postsynaptic differentiation regulated by InR is necessary for scaling growth.
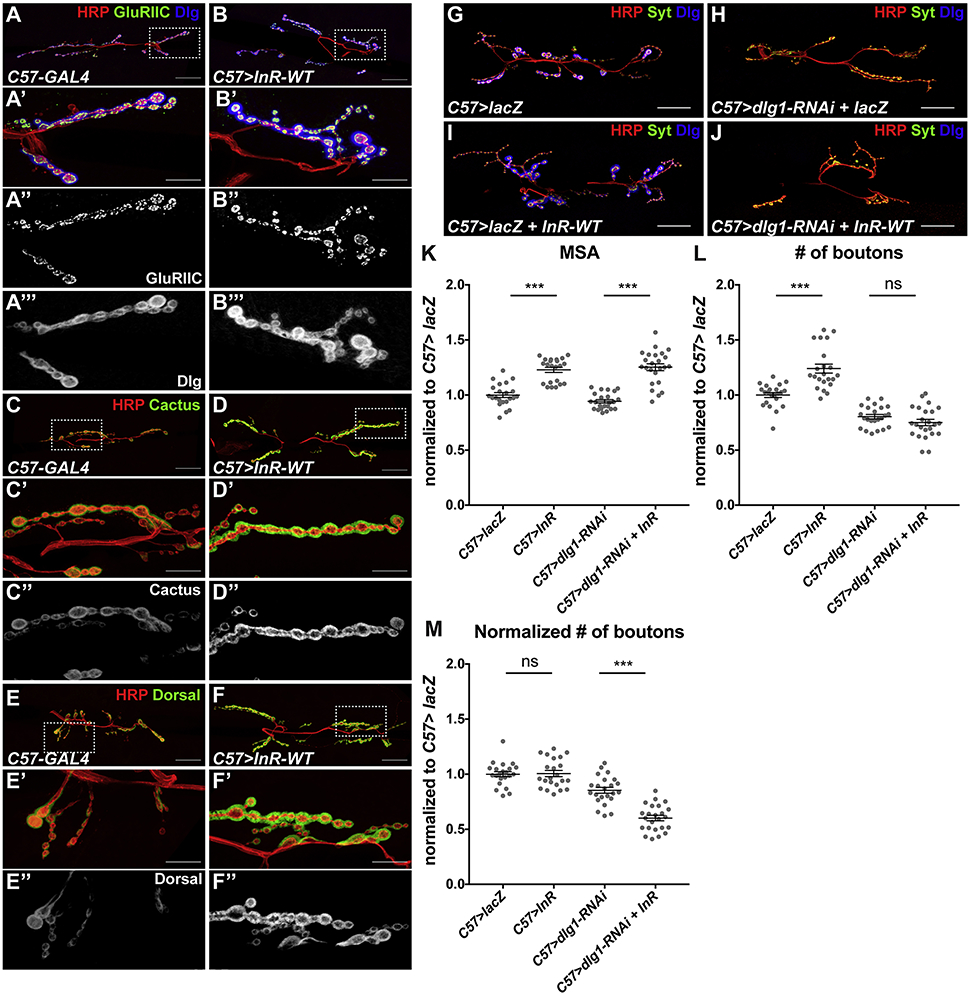
(A-F) Confocal images of the NMJ on muscle 6/7 in C57-GAL4/+ (A, C, E) or C57-GAL4>UAS-InR-WT (B, D, F), labeled with anti-HRP (red), anti-GluRIIC (A’’, B’’, green in A, B), anti-Dlg (A’’’, B’’’, blue in A, B) anti-Cactus (C’’, D’’, green in C, D) or anti-Dorsal (E’’, F’’, green in E, F). The box indicated in the first panel is enlarged in subsequent panels. InR overexpression increases postsynaptic GluRIIC, Dlg, Cactus and Dorsal. (G-J) Confocal images of the NMJ on muscle 6/7 in C57-GAL4>UAS-lacZ (G), C57-GAL4>UAS-lacZ+UAS-dlg1-RNAi (H), C57-GAL4>UAS-lacZ+UAS-InR-WT (I), or C57-GAL4>dlg1-RNAi+UAS-InR-WT (J) labeled with anti-HRP (red), anti-Syt (green), and anti-Dlg (blue). Scale bars, 30 μm in (A-J), 10 μm in enlargements. (K-M) Quantification of the muscle 6/7 MSA (K), number of boutons (L), and number of boutons normalized to MSA (M) of the NMJ in (G-J). dlg1 knockdown in muscle prevents bouton number from increasing in response to InR expression. n=22 (C57>lacZ; C57>lacZ+dlg1-RNAi), n=23 (C57>lacZ+InR-WT), or n=24 (C57>dlg1-RNAi+InR-WT). ***p <0.0001; ns, not significant by unpaired Student’s t-test. See also Figure S3.
To determine whether this increased postsynaptic differentiation is relevant to scaling growth, we tested whether the NMJ could respond to increased muscle InR signaling when postsynaptic differentiation was prevented. Dlg is critical for the maintenance and function of the SSR, and dlg1 null mutants have significantly reduced SSR (Budnik et al., 1996; Guan et al., 1996; Zito et al., 1997). Since dlg1 is thought to function both pre- and post-synaptically at the NMJ (Astorga et al., 2016; Budnik et al., 1996), we used RNAi to specifically knock down dlg1 in muscle (Figure 4G-M). This significantly reduced Dlg staining, suggesting that most synaptic Dlg originates from the muscle (Figure 4G, H). Co-expressing InR-WT with dlg1 RNAi failed to induce NMJ expansion, even though the muscle growth response was similar to the control (Figure 4I-M), indicating that NMJ scaling growth depends on postsynaptic differentiation.
dPix is a critical mediator of postsynaptic differentiation and scaling growth
We next searched for factors that might couple postsynaptic differentiation to muscle growth. dPix, a Rho-family guanine-nucleotide exchange factor also named RtGEF, has been shown to promote SSR development and recruit postsynaptic proteins (Albin and Davis, 2004; Parnas et al., 2001). When we used an EP insertion upstream of the endogenous dPix locus to overexpress it in muscle, we observed increased postsynaptic differentiation; both the subsynaptic area (marked by Dlg/Cactus/Dorsal) and the neurotransmitter receptor field (marked by GluRIIC) were expanded (Figure 5A, C, D, F, G, I) to a similar extent as when InR was overexpressed in muscle (Figure 5B, E, H). Moreover, overexpression of dPix increased the number of synaptic boutons without affecting muscle size (Figure S4A-F).
Figure 5. dPix mediates postsynaptic differentiation and scaling growth of the NMJ.
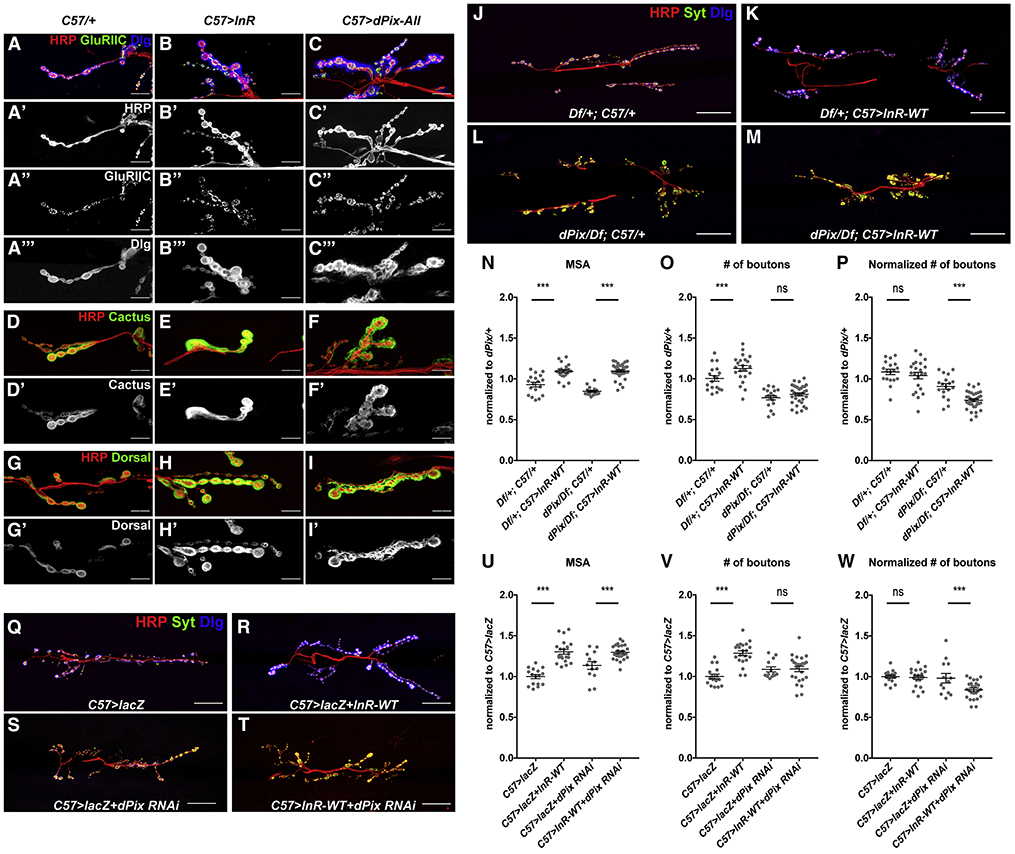
(A-I) Confocal images of enlarged regions of the NMJ on muscle 6/7 in C57-GAL4/+ (A, D, G), C57-GAL4>UAS-InR-WT (B, E, H), and C57-GAL4>UAS-dPix (C, F, I), labeled with anti-HRP (red), anti-GluRIIC (A’’-C’’, green in A-C), anti-Dlg (A’’’-C’’’, blue in A-C), anti-Cactus (D’-F’, green in D-F), and anti-Dorsal (G’-I’, green in G-I). dPix overexpression increases the level of these postsynaptic components. (J-M) Confocal images of the NMJ on muscle 6/7 in Df(2L)Exel6046/+; C57-GAL4/+ (J), Df(2L)Exel6046/+; C57-GAL4>UAS-InR-WT (K), dPix/Df(2L)Exel6046; C57-GAL4/+ (L), or dPix/Df(2L)Exel6046; C57-GAL4>UAS-InR-WT (M) labeled with anti-HRP (red), anti-Syt (green), and anti-Dlg (blue). (N-P) Quantification of the muscle 6/7 MSA (N), number of boutons (O), and number of boutons normalized to MSA (P) of the NMJ in (J-M). n=19 (Df/+; C57/+), n=12 (Df/+; C57>InR-WT), n=18 (dPix/Df; C57/+), n=25 (dPix/Df; C57>InR-WT). (Q-T) Confocal images of the NMJ on muscle 6/7 in C57-GAL4>UAS-lacZ (Q), C57-GAL4>UAS-lacZ+UAS-InR-WT (R), C57-GAL4>UAS-lacZ+UAS-dPix-RNAi (S), C57-GAL4>UAS-InR-WT+dPix-RNAi (T) labeled with anti-HRP (red), anti-Syt (green), and anti-Dlg (blue). Scale bars, 10 μm in (G-I), 30μm in (J-M, Q-T). (U-W) Quantification of the muscle 6/7 MSA (U), number of boutons (V), and number of boutons normalized to MSA (W) of the NMJ in (Q-T). dPix mutation or knockdown in muscle prevents bouton number from increasing in response to InR expression. n=16 (C57>lacZ), n=21 (C57>lacZ + InR-WT), n=13 (C57>lacZ + dPix-RNAi), n=25 (C57>InR-WT + dPix-RNAi). *p<0.05, ***p<0.0001; ns, not significant by unpaired Student’s t-test. See also Figure S4.
dPix is not only necessary (Parnas et al., 2001) and sufficient for postsynaptic differentiation, but also critical for scaling growth of the NMJ. Over-expressing InR-WT in a dPix null mutant background failed to induce NMJ expansion, despite a normal muscle growth response (Figures 5J-P and S4G-I). Since dPix has been shown to function in motor neurons to regulate synaptic vesicle clustering (Rui et al., 2017), we used RNAi (KK113571, see below) to specifically knock down dPix expression in muscle. Loss of dPix from muscle inhibited postsynaptic differentiation, as indicated by Dlg staining (Figure 5Q, S). As in dPix mutants, co-expressing InR-WT with dPix RNAi induced muscle growth but not NMJ expansion (Figure 5Q-W). Together, our data identify dPix as an essential factor that regulates the scaling growth of the NMJ by organizing postsynaptic differentiation.
Individual dPix isoforms have distinct and antagonistic functions
dPix encodes seven annotated isoforms (A-H) of which six have unique coding sequences (Figure 6A). An RNAi construct (KK113571) that targets the coding exon present only in dPix isoforms F and H phenocopied the postsynaptic differentiation phenotype of the dPix mutant (Figure 6E), indicating that F and H are necessary for the synaptic function of dPix. In contrast, the RNAi line HMS00741, which targets the 3’-UTR of isoforms A, B, D and F and was shown to be effective in knocking down dPix (Dent et al., 2015), did not disrupt postsynaptic differentiation even though it should also deplete F (Figure 6C). As there is no cDNA evidence supporting the splicing of this 3’-UTR to the F transcript, we used RT-PCR to characterize the 3’ region of dPix-F. Amplification from mRNA extracted from w1118 larval carcasses showed that the coding region of F can not only splice to the annotated 3’UTR but also to the 3’-UTR of E, generating a transcript that we named dPix-I that would not be targeted by HMS00741 (Figure S5A, B). The presence of H and I might be sufficient to localize Dlg in the absence of F. However, endogenous H is insufficient for postsynaptic differentiation, as Dlg staining was disrupted by the GD6845 RNAi line that targets all the predicted isoforms except H (Figure 6A, D).
Figure 6. Different functions and localization patterns of individual dPix isoforms.
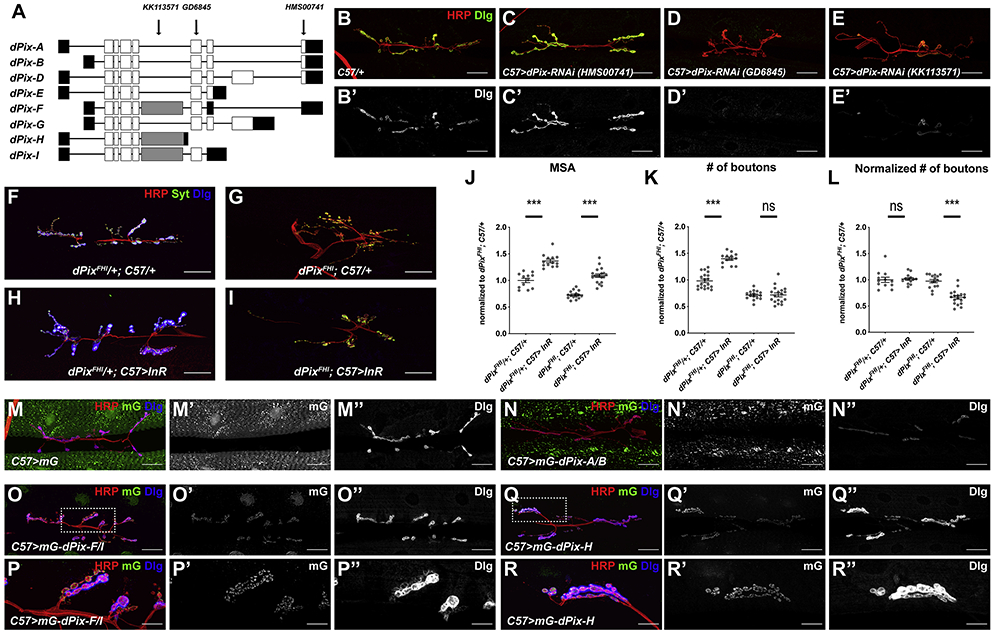
(A) Cartoon showing the transcript structures of dPix isoforms, with coding regions in white, non-coding regions in black and the exon specific to isoforms F, H, and I in grey. Arrows show the regions targeted by the RNAi constructs used in (C-E). dPix-I was not previously annotated on Flybase. (B-E) Confocal images of the NMJ on muscle 6/7 in in C57-GAL4/+ (B), C57-GAL4>UAS-dPix-RNAi HMS00741 (C), C57-GAL4>UAS-dPix-RNAi GD6845 (D), and C57-GAL4>UAS-dPix-RNAi KK113571 (E) labeled with anti-HRP (red) and anti-Dlg (B’-E’, green in B-E). Knocking down the subset of dPix isoforms targeted by GD6845 (D) or KK113571 (E) but not HMS00741 (C) inhibits postsynaptic differentiation. (F-I) Confocal images of the NMJ on muscle 6/7 in in dPix MB10902/+; C57-GAL4/+ (F), dPixMB10902/Df(2L)Exel6046; C57-GAL4/+ (G), dPixMB10902/+; C57-GAL4>UAS-InR-WT (H), dPixMB10902/Df(2L)Exel6046; C57-GAL4>UAS-InR-WT (I) labeled with anti-HRP (red), anti-Syt (green), and anti-Dlg (blue). (J-L) Quantification of the muscle 6/7 MSA (J), number of boutons (K), and number of boutons normalized to MSA (L) of the NMJ in (F-I). n=12 (dPixMB10902/+; C57/+), n=15 (dPixMB10902/+; C57>InR-WT), n=13 (dPixMB10902/Df; C57/+), n=19 (dPixMB10902/Df; C57>InR-WT). dPix isoforms F, H and I are necessary for scaling growth of the NMJ in response to InR expression. *p<0.05, ***p<0.0001; ns, not significant by unpaired Student’s t-test. (M-R) Confocal images of the NMJ on muscle 6/7 in C57-GAL4>UAS-mNeonGreen (mG) (M), C57-GAL4>UAS-mG-dPix-A/B (N), C57-GAL4>UAS-mG-dPix-F/I (O, P), or C57-GAL4>UAS-mG-dPix-H (Q, R) showing mG fluorescence (M’-R’, green in M-R), anti-HRP (red), and anti-Dlg (M’’-R”, blue in F-K). The boxes indicated in O and Q are enlarged in P and R, respectively. Scale bars, 30 μm in (B-I, M-O, Q), 10 μm in (P, R). Only isoforms F/I and H localize to the postsynaptic side of the NMJ when expressed in muscle. Isoform A/B reduces postsynaptic Dlg levels while isoform H increases Dlg levels. See also Figures S5 and S6.
We also examined a mutant specific to dPix isoforms F, H and I. dPixFHI (Mi{ET1}RtGEFMB10902) has a Minos transposon insertion in the F/H/I-specific coding exon (Metaxakis et al., 2005), which does not affect the expression of other isoforms (Figure S5C). In this mutant, postsynaptic differentiation was strongly reduced (Figure 6F, G), and InR overexpression induced normal muscle growth but no NMJ expansion (Figure 6F-L). This result confirms that dPix isoforms F, H and I are necessary for postsynaptic differentiation and scaling growth of the NMJ.
To determine where each isoform localizes, we constructed UAS transgenes that express N-terminally mNeonGreen (mG) tagged individual dPix isoforms. When these transgenes were expressed in muscle, only dPix-F/I and H showed punctate postsynaptic localization and increased NMJ size, while the other forms localized to the nucleus or to non-synaptic aggregates (Figure 6M-R, Figure S5D-J). Indeed, some of these other isoforms appeared to antagonize the synaptic functions of F/I and H; ectopic expression of isoform A/B, D, or G reduced postsynaptic Dlg levels and NMJ growth to various extents (Figure 6M-R, Figure S5D-J). Consistent with an antagonistic effect of these isoforms, knocking them down with HMS00741 RNAi increased postsynaptic Dlg staining (Figure 6C) and the postsynaptic levels of co-expressed isoform H (Figure S5K-M). This antagonism may result from a physical interaction between the isoforms, since coexpression of dPix-A/B removed dPix-H from the postsynaptic compartment and recruited it to the cytoplasmic aggregates where dPix-A/B localized (Figure S5N-P).
Although most known functions of dPix are thought to depend on its partner protein G protein-coupled receptor kinase interacting ArfGAP (Git) (Zhou et al., 2016), we found no role for Git in postsynaptic differentiation. HA-tagged Git did not show postsynaptic localization when expressed in muscle (Figure S6A, B), and a Git null mutant had normal postsynaptic Dlg levels that could still be increased by dPix overexpression (Figure S6C-F). These results are consistent with the absence of the Git-binding domain from the F/I and H isoforms that mediate dPix function at the NMJ (Zhao et al., 2000), and suggest that these isoforms promote postsynaptic differentiation by a Git-independent mechanism that is antagonized by other dPix isoforms.
Isoform-specific regulation of dPix mRNA mediates scaling growth of the NMJ
To complement our observation that specifically removing the dPix-F/I and H isoforms prevents postsynaptic differentiation and scaling growth, we tested which of our dPix isoform transgenes could rescue the dPix mutant phenotype. Consistent with the RNAi and mutant results (Figure 6B-L), expression of isoform F/I or H rescued both postsynaptic differentiation and scaling growth in dPix mutants (Figure 7A-K). Moreover, these isoforms could still localize to the NMJ in the absence of wild-type dPix (Figure 7E, G). No other dPix isoforms were able to rescue scaling growth or Dlg localization (Figure S7A-O).
Figure 7. Isoform-specific regulation and function of dPix.
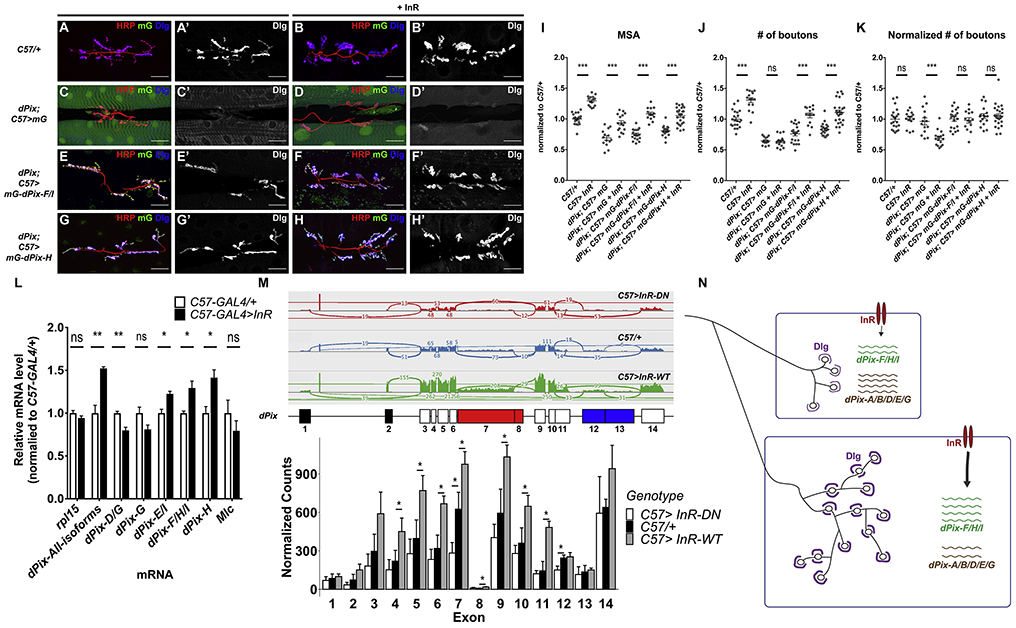
(A-H) Confocal images of the NMJ on muscle 6/7 in C57-GAL4>+ (A), C57-GAL4>UAS-InR-WT (B), dPix/Df(2L)Exel6046; C57-GAL4>UAS-mG (C), dPix/Df(2L)Exel6046; C57-GAL4>UAS-mG+UAS-InR-WT (D), dPix/Df(2L)Exel6046; C57-GAL4>UAS-mG-dPix-F/I (E), dPix/Df(2L)Exel6046; C57-GAL4>UAS-mG-dPi-F/I+UAS-InR-WT (F), dPix/Df(2L)Exel6046; C57-GAL4>UAS-mG-dPix-H (G), dPix/Df(2L)Exel6046; C57-GAL4>UAS-mG-dPix-H+UAS-InR-WT (H) showing anti-HRP (red), mG fluorescence (green), and anti-Dlg (A’-H’, blue in A-H). Scale bars, 30 μm. (I-K) Quantification of the muscle 6/7 MSA (I), number of boutons (J), and number of boutons normalized to MSA (K) of the NMJ in (A-H). n=21 (C57>+), n=15 (C57>InR-WT), n=14 (dPix/Df; C57>mG; dPix/Df; C57>mG-dPix-F/I), n=16 (dPix/Df; C57>mG+InR-WT), n=20 (dPix/Df; C57>mG-dPix-F/I+InR-WT), n=17 (dPix/Df; C57>mG-dPix-H), or n=26 (dPix/Df; C57>mG-dPix-H+InR-WT). *p<0.05, ***p < 0.0001; ns, not significant by unpaired Student’s t-test. dPix-F/I or H is sufficient to rescue postsynaptic differentiation and scaling growth in the dPix mutant. (L) qRT-PCR measurements of dPix transcript levels using RNA extracted from C57-GAL4/+ and C57-GAL4>UAS-InR-WT larval carcasses. Transcript levels are normalized to eIF4e. The relative level of each transcript in the two genotypes is normalized to C57-GAL4/+. Amplicons representing isoforms F, H, I, and all isoforms show increased expression while amplicons specific to isoforms D and G show reduced expression when InR-WT is overexpressed. n=3 for each genotype. *p<0.05 and **p<0.01; ns, not significant by unpaired Student’s t-test. (M) RNA-seq reads on each exon of dPix in C57-GAL4>UAS-InR-DN, C57-GAL4/+ and C57-GAL4>UAS-InR-WT muscle. Upper, Sashimi plot showing the normalized exon-exon junction reads over the dPix locus in one sample of each genotype. Middle, diagram illustrating the numbering of the exons and exon segments of dPix. Red exons are specific to isoforms F, H and I, and blue exons are specific to isoforms D and G. Bottom, reads for each exon in the dPix locus, normalized to all reads in each genotype. n=3 for each genotype. *p<0.05; ns, not significant by unpaired Student’s t-test. (N) a model showing that InR signaling both autonomously increases muscle size, and alters the balance of dPix isoforms to favor the synaptically active forms F/I and H. These forms increase postsynaptic Dlg, increasing synaptic bouton number. See also Figure S7.
If dPix indeed couples muscle growth to synaptic expansion, its abundance or activity should be regulated by InR signaling. Moreover, InR activity might alter the relative abundance of active and antagonistic dPix isoforms. To test this, we used quantitative RT-PCR to measure the effect of InR signaling on dPix isoform levels in larval muscles. We found that activated InR signaling increased the levels of an amplicon common to all dPix isoforms as well as the levels of isoforms F, H, and I (Figure 7L) in comparison to Myosin light chain (Mlc), an indicator of the amount of muscle tissue (Figure 7L). In contrast, an amplicon specific to the antagonistic D and G isoforms was decreased in response to InR over-expression (Figure 7L). We further analyzed the changes in each exon using RNA-Seq. Most of the exons present in all isoforms, as well as those specific to F, H, and I and the 3’-UTR found in E and I, but not the exons specific to D and G, showed statistically significant increases when InR-WT was over-expressed (Figure 7M). The same exons also showed decreased expression when InR signaling was inhibited, although only the change in the F/H/I exon was significant (Figure 7M).
The primary effect of InR signaling on dPix may occur at the transcriptional level. Usage of the proximal, but not the distal, promoter annotated in Flybase increased with increasing levels of InR activity (Figure 7M and Figure S7P, Q). Moreover, we found that overexpressing Foxo, a transcription factor inhibited by InR signaling (Junger et al., 2003), down-regulated the abundance of dPix-F, H and I, in parallel with a decrease of the usage of the dPix proximal promoter (Figure S7P). Consistent with InR increasing dPix transcription by inhibiting Foxo, we found that coexpressing Foxo with InR-WT suppressed the increase in dPix expression and proximal promoter usage (Figure S7P). dMyc is a transcriptional target of InR signaling in muscle (Demontis and Perrimon, 2009), and dMyc overexpression also increased the expression of dPix-F/H/I and proximal promoter usage. Consistent with reported inhibition of the transcriptional activity of dMyc by Foxo (Demontis and Perrimon, 2009), coexpressing InR-DN could reverse these effects of dMyc (Figure S7Q). Altogether, our data suggest that InR signaling regulates the relative abundance of different dPix isoforms through Foxo and dMyc to promote synaptic expansion, thereby coupling target muscle growth to scaling growth of the NMJ (Figure 7N).
Discussion
We have demonstrated here that motor neurons can respond to changes in muscle growth with proportional adjustments to synapse size. Each muscle appears to provide local signals that instruct synapse growth only to the motor neuron branches that directly innervate it. Scaling growth is independent of both neuronal activity and BMP signaling, as loss of Mad abolishes structural plasticity of the NMJ in response to neuronal hyper-activity, but leaves the response to muscle growth intact. Instead, we found that the extent of postsynaptic differentiation is bidirectionally regulated by the level of InR signaling in muscle, and the postsynaptic component Dlg is critical for scaling growth of the NMJ. We identified dPix as a factor that is necessary and sufficient for both postsynaptic differentiation and scaling growth. Regulation of the mRNA levels of synaptically active and antagonistic dPix isoforms by muscle InR signaling could couple NMJ growth to muscle growth.
Coordination of autonomous and non-autonomous growth signals
The use of the InR pathway to drive both autonomous growth and non-autonomous signaling to the motor neuron has obvious advantages for coordinating these processes. Some questions remain about the mechanism: for instance, how InR signaling regulates the abundance of dPix mRNA isoforms. Postsynaptic differentiation and NMJ scaling growth are activated by dPix isoforms F, H and I, which share a specific exon, and inhibited by other isoforms such as A, B, D, and G. Our qRT-PCR and RNA-Seq experiments showed that InR signaling in muscle upregulates the active isoforms but not the inhibitory isoforms. Members of both groups are annotated as sharing the same promoters, suggesting that this regulation is post-transcriptional. However, the Flybase annotation is largely based on short sequence reads and may not accurately represent the repertoire of isoform diversity. Indeed, our data suggest that InR signaling promotes the usage of the proximal promoter of dPix rather than the distal one. Moreover, InR regulates dPix expression through Foxo and dMyc, consistent with a transcriptional mechanism. Alternative splicing can be regulated at the transcriptional level by factors that control the rate of transcriptional elongation or affect splicing factor recruitment (Rambout et al., 2018).
dPix function may also be regulated at the post-translational level. The weaker effect of Akt than PI3K on synapse growth suggests that in addition to regulating dPix mRNA levels, InR signaling might promote the localization or activation of dPix-F/H/I protein through interactions of its Pleckstrin homology domain with phosphatidylinositol (3,4,5)-triphosphate. Expression of dPix-A/B can relocalize dPix-H from the postsynaptic compartment to cytoplasmic aggregates. Mislocalization or destabilization of isoforms that favor synapse growth through physical interactions with antagonistic isoforms could tightly regulate their activity. Mammalian β-Pix forms a stable trimer through its coiled-coil region (Schlenker and Rittinger, 2009), but homology to this region is only found in dPix-A, B and D, leaving open the question of how F/I and H might bind to other isoforms.
It is not known exactly how dPix regulates postsynaptic differentiation. Previous studies showed that mutations in dPix and p21-activated kinase (Pak) shared a similar postsynaptic differentiation phenotype (Albin and Davis, 2004; Parnas et al., 2001). Although both Drosophila and mammalian Pix proteins act in a complex with Git to activate Pak and other factors (Dent et al., 2019; Dent et al., 2015; Premont et al., 2004; Zhou et al., 2016), our data do not support a role for Git in the function of dPix at the NMJ. Downstream components other than Pak may be important for postsynaptic differentiation, or in this context dPix may activate Pak independently of Git. The exon specific to dPix-F, H, and I is poorly conserved in mammals and its function has not been investigated. It must contain sequences sufficient for synaptic localization, since these isoforms localize correctly when expressed in dPix null mutants, but it could also recruit or activate other factors that contribute to postsynaptic differentiation.
The mechanism by which postsynaptic differentiation regulates scaling growth also remains to be determined. Dlg is localized to the SSR rather than to the postsynaptic membrane (Gan and Zhang, 2018), indicating that InR signaling levels correlate with the amount of SSR. Both dPix and Pak are necessary for synaptic localization of Ral, which recruits the exocyst to enable growth of the SSR (Lee and Schwarz, 2016; Teodoro et al., 2013). dPix may promote SSR differentiation in order to recruit molecules that facilitate NMJ growth, through Dlg or other SSR components (Thomas et al., 1997). The SSR has also been suggested to regulate the local translation of mRNAs that influence activity-dependent synaptic plasticity (Sigrist et al., 2000; Thomas et al., 2000; Thomas et al., 1997), and could affect additional mRNAs.
Coordination of synapse growth with muscle growth must involve some type of retrograde trans-synaptic signaling from the muscle to the motor neuron. Although BMP signaling is a well-established retrograde pathway, it only acts early in development and therefore cannot continuously convey size information (Berke et al., 2013); our data with Mad mutants also rule out its involvement in scaling growth. Since a single motor neuron branch responds specifically to the growth of its own target muscle, the trans-synaptic signal must be extremely short-range. This makes it unlikely to be the diffusible neurotrophin Spatzle3, which acts on the Tollo receptor to promote NMJ growth (Ballard et al., 2014). One transmembrane candidate is Fasciclin 2 (Fas2), which is necessary for synaptic expansion and requires Dlg for its synaptic accumulation (Schuster et al., 1996; Thomas et al., 1997; Zito et al., 1997). Teneurins, Neurexins and Neuroligins are additional transmembrane adhesion molecules that have been shown to regulate NMJ size (Mosca et al., 2012; Xing et al., 2018). In this study, we have focused on the addition of synaptic boutons, which might be affected by postsynaptic differentiation either because postsynaptic material stabilizes newly formed boutons that would otherwise be transient, or because boutons divide only when they reach a threshold amount of postsynaptic material. The increase in nerve branching may be independently regulated by increased InR signaling in muscle, or a feedback mechanism may increase branching in response to increased bouton number.
Functional importance of scaling growth
The relationship between structural and functional plasticity in scaling growth of the NMJ is not yet clear. NMJ growth during development is thought to be necessary to maintain a constant level of muscle depolarization for each nerve stimulus as the muscle mass increases (Davis et al., 1998; Menon et al., 2013; Petersen et al., 1997; Powers et al., 2016). In this case, defective scaling growth of the NMJ should impair muscle depolarization. However, dPix mutants survive till the pupal stage, suggesting that defective scaling growth does not disrupt larval development or feeding, at least in standard laboratory conditions. Moreover, quantal imaging studies suggested that only a sub-population of the active zones at the NMJ are activated at one time, while many of them have a very low probability of releasing synaptic vesicles (Peled and Isacoff, 2011). There is thus not a clear correlation between bouton number and synaptic function. However, it is possible that only newly formed boutons are active, making bouton addition essential throughout development.
Analysis of the relationship between NMJ structure and function is also complicated by the fact that Drosophila has a repertoire of mechanisms to buffer synaptic transmission (Davis and Goodman, 1998; Frank, 2014; Goel et al., 2019; Petersen et al., 1997). Increases or decreases in the number of synaptic boutons can be compensated by adjusting the probability of synaptic vesicle release or the quantal size (Davis and Goodman, 1998). Synaptic homeostasis has also been shown to buffer reduced muscle depolarization by increasing quantal content (Davis and Muller, 2015; Petersen et al., 1997). Studying synaptic function in animals with combinatorial defects in both scaling growth and one of these compensatory mechanisms might reveal stronger phenotypes.
Scaling growth of the peripheral nervous system is highly conserved, but remains poorly understood at the mechanistic level. Studies of the scaling of sensory neuron dendrites to the size of their epidermal receptive fields have implicated physical ensheathment of the dendrites by the epidermis (Jiang et al., 2014; Tenenbaum et al., 2017) as well as signaling by the ligand Maverick through the Ret receptor (Hoyer et al., 2018). Our identification of dPix as a factor that couples muscle growth induced by InR signaling to synaptic expansion will make it possible to address more specific questions about the mechanism and function of this process, and to determine whether it has features in common with dendrite scaling. As the role of Pix homologues in postsynaptic assembly is conserved in mammals and mutations in human α-Pix cause mental retardation (Kutsche et al., 2000; Saneyoshi et al., 2008; Zhang et al., 2005), characterizing scaling growth may also help us to understand the basis of human neurodevelopmental disorders.
STAR Methods
RESOURCE AVAILABILITY
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Jessica Treisman (Jessica.Treisman@nyulangone.org).
Materials Availability
Fly stocks and plasmids generated in this study will be distributed by the Lead Contact without restrictions, or deposited to repositories such as the Bloomington Drosophila Stock Center and Addgene.
Data and Code Availability
This study did not analyze datasets or generate custom code.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Drosophila strains and genetics
All Drosophila stocks were reared on standard cornmeal medium at room temperature. w1118 was used as the wild type control unless otherwise indicated. Male and female larvae showed no significant differences and were used interchangeably. The following flies were from Bloomington Drosophila Stock Center: Df(2L)Exel6046 (BDSC stock#7528), UAS-Akt-WT (P{UAS-Akt1.Exel}1, BDSC stock#8192), UAS-InR-DN (UAS-InRK1409A, BDSC stock#8253), UAS-InR-WT (BDSC stock#8262), UAS-PI3K (P{UAS-Pi3K92E.Exel}2 BDSC stock#8286), UAS-PI3K-DN (P{UAS-Pi3K92E.A2860C}3, BDSC stock#8289), Madk00237 (BDSC stock#10474), UAS-dPix-All (P{EP}RtGEFG3647, BDSC stock#27123), dPixMB10902 (Mi(ET1}RtGEFMB10902, BDSC stock#29166), UAS-dlg1-RNAi (P{TRiP.JF01077}attP2, BDSC stock#31521), C57-GAL4 (BDSC stock#32556), UAS-Akt-RNAi (P{TRiP.HMS00007}attP2, BDSC stock#33615), UAS-dPix-RNAi (P{TRiP.HMS00741}attP2, BDSC stock#32947), and UAS-lacZ (BDSC stock#8530). UAS-dPix-RNAi lines GD6845 (VDRC stock#17966) and KK113571 (VDRC stock#105093) were from the Vienna Drosophila Resource Center. A second UAS-RNAi insertion in the tiptop gene in the original KK113571 line was removed by recombination with an FRT insertion at 40A. dPixp1036, Gitex2.3, and UAS-HA-dPix-A/B were gifts from Kieran Harvey (Dent et al., 2015). GlurIIASP16 was a gift from Aaron DiAntonio (DiAntonio et al., 1999; Petersen et al., 1997). Syt4BA1 was a gift from Troy Littleton (Yoshihara et al., 1995). rut1 and eag1, Sh14 were gifts from Chun-Fang Wu (Zhong and Wu, 1993). UAS-dMyc was a gift from Laura Johnston (Johnston et al., 1999). UAS-Foxo3× was a gift from Heather Broihier (Hwangbo et al., 2004).
UAS-HA-Git was generated by PCR amplification of the 5’ end of Git from the cDNA clone LD30319 (Drosophila Genomics Resource Center) with primers NotIAUGGit1 (TGGCGGCCGCTATGTGTTTCGCCAGCAG) and XbaIGit1 (TCCAAGGCATTCACATCGGC), which was cloned into the Not I and Xba I sites of pUASt-HA (Lee and Treisman, 2004). The remainder of the Git coding sequence and 3’UTR were added as an Xba I-Xho I fragment. To make UAS-mNeonGreen-dPix single isoform transgenes, the open reading frame of dPix-H was amplified from a cDNA template generated from RNA isolated from the carcasses of C57-GAL4>UAS-dPix-All larvae. The resulting PCR product was fused in frame to the C-terminus of mNeonGreen with a poly-glycine-serine linker and cloned into the pPAC-PL vector using Gibson Assembly. This construct contains the SV40 3’-UTR after the stop codon. The recombined fragment containing mNeonGreen-poly(GS)-dPix-H-SV40 was amplified by PCR and cloned into the pUAST-attB vector using Gibson Assembly. For every other dPix isoform, the mNeonGreen, the poly(GS) linker, and the sequence present in all dPix isoforms was amplified from pUAS-mNeonGreen-poly(GS)-dPix-H-SV40 and the isoform-specific coding sequence was amplified from the cDNA template above. These fragments were recombined_into the pPAC-PL vector using Gibson Assembly, and the full construct was amplified by PCR and cloned into the pUAST-attB vector using Gibson Assembly. All constructs were verified by Sanger sequencing, and the primers used are listed in Table S1. Injections to generate transgenic flies were performed by BestGene.
METHOD DETAILS
Larval NMJ preparations
50 first instar larvae were collected on a grape juice agar plate and incubated at 25°C (unless stated otherwise) until they reached the third instar larval stage. Larval fillets were prepared by pinning the larvae to silicone plates, dissecting them in ice-cold Ca2+−free HL3 saline (pH=7.4), fixing in 4% formaldehyde in PBS for 15 min at room temperature, and permeabilizing with 0.2% Triton X-100 in PBS (Ramachandran and Budnik, 2010).
Immunohistochemistry
Larval fillets were stained with primary antibody overnight at 4°C at the indicated concentrations. The following antibodies were used: mouse anti-Dlg (1:50; 4F3 from DSHB), rabbit anti-Syt (1:4000, gift from Hugo Bellen)(Littleton et al., 1993), mouse anti-Brp (1:10, nc82 from DSHB), mouse anti-Syn (1:100, 3C11 from DSHB), rat anti-HA (1:500, 3F10 from Roche), mouse anti-GluRIIA (1:10, 8B4D2 from DSHB), rabbit anti-GluRIIC (1:1000, gift from Aaron DiAnotonio) (Marrus et al., 2004), rat anti-Cactus (1:100, gift from Steven Wasserman) (Kumar et al., 2009), and rat anti-Dorsal (1:100, gift from Steven Wasserman) (Gillespie and Wasserman, 1994). Larval fillets stained with the primary antibodies were washed three times for five minutes each with 0.2% Triton X-100 in PBS and incubated with fluorescently labeled secondary antibodies for two hours at room temperature. The primary antibodies were visualized with corresponding secondary antibodies conjugated to Alexa Fluor-488 or Alexa Fluor-633 (1:200, Jackson ImmunoResearch). The neuronal membrane was visualized with Alexa Fluor-488, Alexa Fluor-633, or TRITC-conjugated anti-horseradish peroxidase (HRP) (1:200; Jackson ImmunoResearch). The muscle cells were visualized either by background signal of the other antibodies used to stain the NMJ or with TRITC-conjugated phalloidin (1:5000; Abcam). The larval fillets were then mounted in Fluoromount-G (SouthernBiotech). Samples were imaged with a Leica SP5 or SP8 confocal microscope using a 63x oil objective. Images were captured with a resolution of 1024 x 1024 pixels and processed in ImageJ and Adobe Photoshop. The images shown are z projections of confocal stacks acquired from serial laser scanning unless stated otherwise.
RNA extraction
Third instar larvae were dissected in 4°C DEPC-treated 0.1M phosphate buffer (pH 7.4). The internal organs were removed and the larval fillets were mechanically homogenized with a plastic pestle in 200 μl TRIzol (Invitrogen). Total RNA was extracted from the samples using TRIzol /chloroform extraction: the larval fillets were incubated with a total volume of 450 μl TRIzol for 5 minutes and centrifuged at 12,000 rpm for 10 minutes at 4°C. The resultant supernatant was incubated with 107 μl chloroform, shaken vigorously by hand for 15 seconds, incubated at room temperature for 10 minutes, and centrifuged at 10,000 rpm for 10 minutes at 4°C. Approximately 250 μl of the upper aqueous phase was transferred to a new tube and incubated with 267 μl isopropanol at room temperature for 10 minutes. The RNA was pelleted by centrifugation at 12,000 rpm for 10 minutes at 4°C. The extracted RNA was washed 2x with 70% ethanol and purified using RNeasy Purification Kits (Qiagen). The RNA was eluted in 100 μl RNAse-free water and further purified and concentrated by sodium acetate precipitation: 10 μl 3M sodium acetate and 440 μl 100% ethanol were added to the RNA solution and it was incubated at −80°C overnight. The RNA was pelleted by centrifugation at 12,000 rpm for 30 minutes at 4°C. The resultant pellet was washed 2x with 70% ethanol and resuspended in water.
Reverse transcription polymerase chain reaction
The purified RNA was treated with RQ1 RNase-Free DNase (Promega). Reverse transcription was performed from 1 μg of total RNA using Superscript™ II Reverse Transcriptase (Thermo Fisher Scientific). PCR was carried out using 20 ng cDNA and 100 nM of each primer pair using Q5® High-Fidelity 2X Master Mix (NEB). The PCR program was: 98°C for 30 s, 45 cycles of 98°C for 10 s, 64°C for 30 s and 72°C for 1 min 35 s, and 72°C for 2 mins. 5 μl of each of the resultant PCR products was loaded on a 1.5% agarose gel. Primer sequences are given in Table S2.
Quantitative reverse transcription polymerase chain reaction
The purified RNA was treated with RQ1 RNase-Free DNase (Promega). Reverse transcription was performed from 1 μg of total RNA using Superscript™ II Reverse Transcriptase (Thermo Fisher Scientific). Quantitative reverse transcription polymerase chain reaction (qPCR) was carried out using 10 ng of cDNA and 100 nM of each primer pair with a Roche LightCycler 480 machine and LightCycler 480 SYBR Green I Master 2X (Roche, 04887352001). The PCR program was: 10 min at 95°C, 45 cycles of 95°C for 15 s and 60°C for 1 min.
RNA-Seq sample preparation
RNA was isolated from larval carcasses from each genotype in triplicate. RNA quality and quantity was assessed using the Bioanalyzer 2100 (Agilent Inc.). Library preparation and sequencing was carried out by the NYU Genome Technology Center. RNA-Seq library preps were constructed using Illumina TruSeq Stranded mRNA (Cat #20020595), with 250 ng of total RNA as input, and 12 cycles of PCR amplification. Samples were multiplexed and run on a single lane of a NovaSeq6000 S1 Flowcell, as pair-end read 50.
QUANTIFICATION AND STATISTICAL ANALYSIS
Quantification of muscle and synapse growth
The surface of muscles 6 and 7 was outlined and the enclosed area was quantified in ImageJ. The numbers of synaptic boutons and branches were counted manually, and measurements of the size of the synaptic boutons were done in ImageJ. The synaptic area of the NMJ was measured as the area covered by Synaptotagmin antibody staining using ImageJ. All quantifications were carried out blind. Statistical significance between each genotype and the controls was determined by two tailed Student’s t-test, whereas multiple comparisons between genotypes were determined by two-way ANOVA test followed by Bonferroni post hoc correction. Each figure legend has details about sample sizes, precision measures, statistical analysis, and definitions of significance thresholds. No outliers were excluded.
Analysis of quantitative RT-PCR
Dissociation curves generated through a thermal denaturation step were used to verify amplification specificity. The cycle at which the amount of detectable PCR product reaches a preset threshold level (Ct) was assessed using the Second Derivative Maximum method of the Roche LightCycler 480 software. The relative quantity of each amplicon was analyzed two different ways: for primers that amplify across an exon-exon junction, the Ct value of the amplicon was normalized to that of the internal reference gene, eIF4E1. For primers that amplify an amplicon within an exon, a standard curve was generated by amplifying a serially diluted genomic DNA sample (from 900 ng to 9 pg per reaction, 10-fold dilutions). The absolute quantity of mRNA in the control and experimental genotypes was estimated using a linear regression equation for each primer. The absolute quantity of the mRNA of the reference gene eIF4E1 was used as the internal control for the normalization. All experiments were repeated at least three times, and the data are presented as the mean ± standard deviation. Data were analyzed using unpaired two-tailed t-tests and 95% confidence intervals of the difference between the control and experimental means. Primer sequences are given in Table S3.
RNA-Seq data analysis
Sequencing reads were mapped to the reference genome (dm6) using the STAR aligner (v2.5.0c) (Dobin et al., 2013). Alignments were guided by a Gene Transfer Format (GTF) file. The mean read insert sizes and their standard deviations were calculated using Picard tools (v.1.126) (http://broadinstitute.github.io/picard). The read count tables were generated using HTSeq (v0.6.0) (Anders et al., 2015), normalized based on their library size factors using DESeq2 (Love et al., 2014), and differential expression analysis was performed. The exon count tables and differential exon usage were calculated using DEXSeq (v3.10) (Anders et al., 2012). The Read Per Million (RPM) normalized BigWig files were generated using BEDTools (v2.17.0) (Quinlan and Hall, 2010) and bedGraphToBigWig tool (v4). To compare the level of similarity among the samples and their replicates, we used two methods: principal-component analysis and Euclidean distance-based sample clustering. All the downstream statistical analyses and generating plots were performed in R environment (v3.1.1) (https://www.r-project.org/ ). Sashimi plots were generated using the Integrative Genomics Viewer (Robinson et al., 2011).
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse monoclonal anti-Dlg | Developmental Studies Hybridoma Bank | Cat# 4F3; RRID: AB_528203 |
| Rabbit polyclonal anti-Syt | Littleton et al., 1993 | DSYT1; RRID: AB_2315418 |
| Mouse monoclonal anti-Brp | Developmental Studies Hybridoma Bank | Cat# nc82; RRID: AB_2314866 |
| Mouse monoclonal anti-Syn | Developmental Studies Hybridoma Bank | Cat# 3C11; RRID: AB_528479 |
| Rat monoclonal anti-HA clone 3F10 | Roche | Cat# 11867423001, RRID:AB_390918 |
| Mouse monoclonal anti-GluRIIA | Developmental Studies Hybridoma Bank | Cat# 8BB4D2; RRID: AB_528269 |
| Rabbit polyclonal anti-GluRIIC | Marrus et al., 2004 | RRID: AB_2568751 |
| Rat polyclonal anti-Cactus | Kumar et al., 2009 | RRID: AB_2314056 |
| Rat polyclonal anti-Dorsal | Gillespie and Wasserman, 1994 | RRID: AB_2314338 |
| Anti-HRP-Rhodamine(TRITC) | Jackson ImmunoResearch | Code: 323-025-021 RRID: AB_2340257 |
| Anti-HRP-Alexa488 | Jackson ImmunoResearch | Code: 123-545-021 RRID: AB_2338965 |
| Anti-HRP-Alexa647 | Jackson ImmunoResearch | Code: 123-605-021 RRID: AB_2338967 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| TRITC-phalloidin | Thermo Fisher | Cat# R415 |
| Fluoromount-G | Southern Biotech | Cat# 0100-01 |
| TRIzol | Invitrogen | ThermoFisher Cat# 15596026 |
| Critical Commercial Assays | ||
| RNeasy Purification Kits | Qiagen | Cat# 74104 |
| SuperScript™ II Reverse Transcriptase | Thermo Fisher | Cat# 18064022 |
| Q5® High-Fidelity 2X Master Mix | New England Biolabs | Cat # M0492S |
| LightCycler 480 SYBR Green I Master 2X | Roche | Cat# 04887352001 |
| TruSeq Stranded mRNA | Illumina | Cat #20020595 |
| Experimental Models: Organisms/Strains | ||
| D. melanogaster: Df(2L)Exel 6046 | Bloomington Drosophila Stock Center | BDSC:7528; Flybase: FBab0037884 |
| D. melanogaster: UAS-InRK1409A | Bloomington Drosophila Stock Center | BDSC:8253; Flybase: FBti0040689 |
| D. melanogaster: UAS-InR-WT | Bloomington Drosophila Stock Center | BDSC:8262; Flybase: FBti0040676 |
| D. melanogaster: Madk00237 | Bloomington Drosophila Stock Center | BDSC:10474; Flybase: FBti0006935 |
| D. melanogaster: P{EP}RtGEFG3647 | Bloomington Drosophila Stock Center | BDSC:27123; Flybase: FBti0115496 |
| D. melanogaster: dPixMB10902 | Bloomington Drosophila Stock Center | BDSC:29166; Flybase: FBti0127556 |
| D. melanogaster: RNAi of dlg1; P{TRiP.JF01077}attP2 | Bloomington Drosophila Stock Center | BDSC:31521; Flybase: FBti0130555 |
| D. melanogaster: P{GawB}C57 | Bloomington Drosophila Stock Center | BDSC:32556; Flybase: FBti0016293 |
| D. melanogaster: RNAi of Akt1; P{TRiP.HMS00007}attP2 | Bloomington Drosophila Stock Center | BDSC:33615; Flybase: FBti0140088 |
| D. melanogaster: RNAi of dPix; P{TRiP.HMS00741}attP2 | Bloomington Drosophila Stock Center | BDSC:32947; Flybase: FBti0140455 |
| D. melanogaster: UAS-lacZ | Bloomington Drosophila Stock Center | BDSC:8530; Flybase: FBti0040825 |
|
D. melanogaster: RNAi of dPix; w1118; P{GD6845}v17966 |
Vienna Drosophila Resource Center | VDRC:17966; Flybase: FBst0452939 |
|
D. melanogaster: RNAi of dPix; w1118; P{KK113571}VIE-260B |
Vienna Drosophila Resource Center | VDRC:105093; Flybase: FBst0476921 |
| D. melanogaster: dPixp1036 | Dent et al., 2015 | Flybase: FBal0178146 |
| D. melanogaster: Gitex21c | Dent et al., 2015 | Flybase: FBal0221208 |
| D. melanogaster: UAS-HA-dPix-A/B | Dent et al., 2015 | Flybase: FBal0303112 |
| D. melanogaster: GluRIIASP16 | Petersen et al., 1997 | Flybase: FBal0085982 |
| D. melanogaster: Syt4BA1 | Yoshihara et al., 2005 | Flybase: FBal0191284 |
| D. melanogaster: rut1 | Zhong and Wu, 1993 | Flybase: FBal0014878 |
| D. melanogaster: eag1 | Zhong and Wu, 1993 | Flybase: FBal0003484 |
| D. melanogaster: Sh14 | Zhong and Wu, 1993 | Flybase: FBal0015554 |
| D. melanogaster: UAS-dMyc | Johnston et al., 1999 | Flybase: FBal0284703 |
| D. melanogaster: UAS-Foxo3× | Hwangbo et al., 2004 | Flybase: FBal0151927 |
| Oligonucleotides | ||
| Primers used to clone UAS-mNeonGreen-dPix transgenes, see Table S1 | This paper | N/A |
| Primers used for RT-PCR, see Table S2 | This paper | N/A |
| Primers used for qRT-PCR, see Table S3 | This paper | N/A |
| Primer NotIAUGGit1 TGGCGGCCGCTATGTGTTTCGCCAGCAG | This paper | N/A |
| Primer XbaIGit1 TCCAAGGCATTCACATCGGC | This paper | N/A |
| Recombinant DNA | ||
| pPAC-PL | Drosophila Genomics Resource Center | Cat# 1209 |
| pUAST-attB | Drosophila Genomics Resource Center | Cat# 1419 |
| mNeonGreen | Allele Technology | ABP-FP-MNEONSB |
| pUASt-HA-Git | This paper | N/A |
| Software and Algorithms | ||
| ImageJ | https://imagej.nih.gov/ij/ | |
| Adobe Photoshop | https://www.adobe.com/products/photoshop.html | |
| Roche LightCycler 480 software | https://lifescience.roche.com/en_us/products/lightcycler14301-480-software-version-15.html | |
| STAR aligner (v2.5.0c) | Dobin et al., 2013 | https://github.com/alexdobin/STAR/releases |
| Picard tools (v.1.126) | http://broadinstitute.github.io/picard | |
| HTSeq (v0.6.0) | Anders et al., 2015 | https://htseq.readthedocs.io/en/master/ |
| DESeq2 | Love et al., 2014 | https://bioconductor.org/packages/release/bioc/html/DESeq2.html |
| DEXSeq (v3.10) | Anders et al., 2012 | http://bioconductor.org/packages/release/bioc/html/DEXSeq.html |
| BEDTools (v2.17.0) | Quinlan and Hall, 2010 | https://bedtools.readthedocs.io/en/latest/content/bedtools-suite.html |
| Integrative Genomics Viewer | Robinson et al., 2011 | http://software.broadinstitute.org/software/igv/ |
Highlights.
Autonomous changes in muscle growth induce scaling growth of neuromuscular synapses
Scaling growth is locally regulated and independent of neuronal activity
Insulin receptor activity promotes synapse growth through postsynaptic expansion
Synapse size depends on the balance between active and antagonistic dPix isoforms
Acknowledgements
We thank Hugo Bellen, Aaron DiAntonio, Kieran Harvey, Ben Lin, Troy Littleton, Steven Wasserman, Chun-Fang Wu, the Developmental Studies Hybridoma Bank, the Drosophila Genomics Resource Center, the Vienna Drosophila Resource Center and the Bloomington Drosophila Stock Center for fly stocks and reagents, and Flybase for invaluable information. We are grateful to Peter Meyn and Alireza Khodadadi-Jamayran for their help with performing and analyzing the RNA-Seq experiment. We thank Marina Maletic and Kerstin Hofmeyer for their contributions to the early stages of the project, and Hui Hua Liu, DanQing He and Ariel Hairston for technical assistance. The manuscript was improved by the critical comments of Ben Lin, Leslie Magtanong, Greg Suh, and Hongsu Wang. This work was funded by NIH grant R21HD092731 to J.E.T. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of Interests: The authors declare no competing interests.
References
- Albin SD, and Davis GW (2004). Coordinating structural and functional synapse development: postsynaptic p21-activated kinase independently specifies glutamate receptor abundance and postsynaptic morphology. J. Neurosci 24, 6871–6879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S, Pyl PT, and Huber W (2015). HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S, Reyes A, and Huber W (2012). Detecting differential usage of exons from RNA-seq data. Genome Res. 22, 2008–2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Astorga C, Jorquera RA, Ramírez M, Kohler A, López E, Delgado R, Córdova A, Olguín P, and Sierralta J (2016). Presynaptic DLG regulates synaptic function through the localization of voltage-activated Ca2+ channels. Sci. Reports 6, 32132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballard SL, Miller DL, and Ganetzky B (2014). Retrograde neurotrophin signaling through Tollo regulates synaptic growth in Drosophila. J. Cell Biol 204, 1157–1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentley D, and Toroian-Raymond A (1981). Embryonic and postembryonic morphogenesis of a grasshopper interneuron. J. Comp. Neurol 201, 507–518. [DOI] [PubMed] [Google Scholar]
- Berke B, Wittnam J, McNeill E, Van Vactor DL, and Keshishian H (2013). Retrograde BMP signaling at the synapse: a permissive signal for synapse maturation and activity-dependent plasticity. J. Neurosci 33, 17937–17950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Böhni R, Riesgo-Escovar J, Oldham S, Brogiolo W, Stocker H, Andruss BF, Beckingham K, and Hafen E (1999). Autonomous control of cell and organ Size by CHICO, a Drosophila homolog of vertebrate IRS1–4. Cell 97, 865–875. [DOI] [PubMed] [Google Scholar]
- Brogiolo W, Stocker H, Ikeya T, Rintelen F, Fernandez R, and Hafen E (2001). An evolutionarily conserved function of the Drosophila Insulin receptor and insulin-like peptides in growth control. Curr. Biol 11, 213–221. [DOI] [PubMed] [Google Scholar]
- Bucher D, and Pflüger H-J (2000). Directional sensitivity of an identified wind-sensitive interneuron during the postembryonic development of the locust. J. Insect Physiol 46, 1545–1556. [DOI] [PubMed] [Google Scholar]
- Budnik V, Koh YH, Guan B, Hartmann B, Hough C, Woods D, and Gorczyca M (1996). Regulation of synapse structure and function by the Drosophila tumor suppressor gene dlg. Neuron 17, 627–640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budnik V, Zhong Y, and Wu C (1990). Morphological plasticity of motor axons in Drosophila mutants with altered excitability. J. Neurosci 10, 3754–3768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowin SC (2011). The specific growth rates of tissues: a review and a re-evaluation. J. Biomechan. Engin 133, 041001–041020. [DOI] [PubMed] [Google Scholar]
- Davis GW, DiAntonio A, Petersen SA, and Goodman CS (1998). Postsynaptic PKA controls quantal size and reveals a retrograde signal that regulates presynaptic transmitter release in Drosophila. Neuron 20, 305–315. [DOI] [PubMed] [Google Scholar]
- Davis GW, and Goodman CS (1998). Synapse-specific control of synaptic efficacy at the terminals of a single neuron. Nature 392, 82–86. [DOI] [PubMed] [Google Scholar]
- Davis GW, and Muller M (2015). Homeostatic control of presynaptic neurotransmitter release. Annu. Rev. Physiol 77, 251–270. [DOI] [PubMed] [Google Scholar]
- Demontis F, and Perrimon N (2009). Integration of Insulin receptor/Foxo signaling and dMyc activity during muscle growth regulates body size in Drosophila. Development 136, 983–993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dent LG, Manning SA, Kroeger B, Williams AM, Saiful Hilmi AJ, Crea L, Kondo S, Horne-Badovinac S, and Harvey KF (2019). The dPix-Git complex is essential to coordinate epithelial morphogenesis and regulate myosin during Drosophila egg chamber development. PLoS Genet. 15, e1008083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dent LG, Poon CL, Zhang X, Degoutin JL, Tipping M, Veraksa A, and Harvey KF (2015). The GTPase regulatory proteins Pix and Git control tissue growth via the Hippo pathway. Curr. Biol 25, 124–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiAntonio A, Petersen SA, Heckmann M, and Goodman CS (1999). Glutamate receptor expression regulates quantal size and quantal content at the Drosophila neuromuscular junction. J. Neurosci 19, 3023–3032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, and Gingeras TR (2013). STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eaton BA, and Davis GW (2005). LIM Kinase1 controls synaptic stability downstream of the type II BMP receptor. Neuron 47, 695–708. [DOI] [PubMed] [Google Scholar]
- Fernandez R, Tabarini D, Azpiazu N, Frasch M, and Schlessinger J (1995). The Drosophila insulin receptor homolog: a gene essential for embryonic development encodes two receptor isoforms with different signaling potential. EMBO J. 14, 3373–3384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank CA (2014). Homeostatic plasticity at the Drosophila neuromuscular junction. Neuropharmacology 78, 63–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan G, and Zhang C (2018). The precise subcellular localization of Dlg in the Drosophila larva body wall using improved pre-embedding immuno-EM. J Neurosci. Res. 96, 467–480. [DOI] [PubMed] [Google Scholar]
- Gillespie SK, and Wasserman SA (1994). Dorsal, a Drosophila Rel-like protein, is phosphorylated upon activation of the transmembrane protein Toll. Mol. Cell. Biol 14, 3559–3568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goel P, Khan M, Howard S, Kim G, Kiragasi B, Kikuma K, and Dickman D (2019). A screen for synaptic growth mutants reveals mechanisms that stabilize synaptic strength. J. Neurosci 39, 4051–4065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goold CP, and Davis GW (2007). The BMP ligand Gbb gates the expression of synaptic homeostasis independent of synaptic growth control. Neuron 56, 109–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan B, Hartmann B, Kho YH, Gorczyca M, and Budnik V (1996). The Drosophila tumor suppressor gene, dlg, is involved in structural plasticity at a glutamatergic synapse. Curr. Biol 6, 695–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan Z, Buhl LK, Quinn WG, and Littleton JT (2011). Altered gene regulation and synaptic morphology in Drosophila learning and memory mutants. Learn Mem. 18, 191–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heckscher ES, Fetter RD, Marek KW, Albin SD, and Davis GW (2007). NF-κB, IκB, and IRAK control glutamate receptor density at the Drosophila NMJ. Neuron 55, 859–873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoang B, and Chiba A (2001). Single-cell analysis of Drosophila larval neuromuscular synapses. Dev. Biol 229, 55–70. [DOI] [PubMed] [Google Scholar]
- Hoyer N, Zielke P, Hu C, Petersen M, Sauter K, Scharrenberg R, Peng Y, Kim CC, Han C, Parrish JZ, et al. (2018). Ret and substrate-derived TGF-beta Maverick regulate space-filling dendrite growth in Drosophila sensory neurons. Cell Rep. 24, 2261–2272 e2265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwangbo DS, Gershman B, Tu MP, Palmer M, and Tatar M (2004). Drosophila dFOXO controls lifespan and regulates insulin signalling in brain and fat body. Nature 429, 562–566. [DOI] [PubMed] [Google Scholar]
- Jarecki J, Johnson E, and Krasnow MA (1999). Oxygen regulation of airway branching in Drosophila is mediated by Branchless FGF. Cell 99, 211–220. [DOI] [PubMed] [Google Scholar]
- Jiang N, Soba P, Parker E, Kim CC, and Parrish JZ (2014). The microRNA bantam regulates a developmental transition in epithelial cells that restricts sensory dendrite growth. Development 141, 2657–2668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston LA, Prober DA, Edgar BA, Eisenman RN, and Gallant P (1999). Drosophila myc regulates cellular growth during development. Cell 98, 779–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Junger MA, Rintelen F, Stocker H, Wasserman JD, Vegh M, Radimerski T, Greenberg ME, and Hafen E (2003). The Drosophila forkhead transcription factor FOXO mediates the reduction in cell number associated with reduced insulin signaling. J. Biol 2, 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar H, Kawai T, and Akira S (2009). Pathogen recognition in the innate immune response. Biochem. J 420, 1–16. [DOI] [PubMed] [Google Scholar]
- Kutsche K, Yntema H, Brandt A, Jantke I, Nothwang HG, Orth U, Boavida MG, David D, Chelly J, Fryns JP, et al. (2000). Mutations in ARHGEF6, encoding a guanine nucleotide exchange factor for Rho GTPases, in patients with X-linked mental retardation. Nat. Genet 26, 247–250. [DOI] [PubMed] [Google Scholar]
- Landgraf M, Bossing T, Technau GM, and Bate M (1997). The origin, location, and projections of the embryonic abdominal motorneurons of Drosophila. J. Neurosci 17, 9642–9655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee A, and Treisman JE (2004). Excessive Myosin activity in Mbs mutants causes photoreceptor movement out of the Drosophila eye disc epithelium. Mol. Biol. Cell 15, 3285–3295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee G, and Schwarz TL (2016). Filamin, a synaptic organizer in Drosophila, determines glutamate receptor composition and membrane growth. eLife 5, e19991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, and Stevens CF (2007). General design principle for scalable neural circuits in a vertebrate retina. Proc. Natl. Acad. Sci. U S A 104, 12931–12935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Littleton JT, Bellen HJ, and Perin MS (1993). Expression of Synaptotagmin in Drosophila reveals transport and localization of synaptic vesicles to the synapse. Development 118, 1077–1088. [DOI] [PubMed] [Google Scholar]
- Liu JP, Baker J, Perkins AS, Robertson EJ, and Efstratiadis A (1993). Mice carrying null mutations of the genes encoding insulin-like growth factor I (Igf-1) and type 1 IGF receptor (Igf1r). Cell 75, 59–72. [PubMed] [Google Scholar]
- Lnenicka GA, Spencer GM, and Keshishian H (2003). Effect of reduced impulse activity on the development of identified motor terminals in Drosophila larvae. J. Neurobiol 54, 337–345. [DOI] [PubMed] [Google Scholar]
- Love MI, Huber W, and Anders S (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15, 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marrus SB, Portman SL, Allen MJ, Moffat KG, and DiAntonio A (2004). Differential localization of glutamate receptor subunits at the Drosophila neuromuscular junction. J. Neurosci 24, 1406–1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCabe BD, Marques G, Haghighi AP, Fetter RD, Crotty ML, Haerry TE, Goodman CS, and O'Connor MB (2003). The BMP homolog Gbb provides a retrograde signal that regulates synaptic growth at the Drosophila neuromuscular junction. Neuron 39, 241–254. [DOI] [PubMed] [Google Scholar]
- Menon KP, Carrillo RA, and Zinn K (2013). Development and plasticity of the Drosophila larval neuromuscular junction. WIRES Dev.Biol. 2, 647–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metaxakis A, Oehler S, Klinakis A, and Savakis C (2005). Minos as a genetic and genomic tool in Drosophila melanogaster. Genetics 171, 571–581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosca TJ, Carrillo RA, White BH, and Keshishian H (2005). Dissection of synaptic excitability phenotypes by using a dominant-negative Shaker K+ channel subunit. Proc. Natl. Acad. Sci. USA 102, 3477–3482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosca TJ, Hong W, Dani VS, Favaloro V, and Luo L (2012). Trans-synaptic Teneurin signalling in neuromuscular synapse organization and target choice. Nature 484, 237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto N, Nakamori R, Murai T, Yamauchi Y, Masuda A, and Nishimura T (2013). A secreted decoy of InR antagonizes insulin/IGF signaling to restrict body growth in Drosophila. Genes Dev. 27, 87–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owald D, Fouquet W, Schmidt M, Wichmann C, Mertel S, Depner H, Christiansen F, Zube C, Quentin C, Körner J, et al. (2010). A Syd-1 homologue regulates pre- and postsynaptic maturation in Drosophila. J.Cell Biol 188, 565–579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parnas D, Haghighi AP, Fetter RD, Kim SW, and Goodman CS (2001). Regulation of postsynaptic structure and protein localization by the Rho-type guanine nucleotide exchange factor dPix. Neuron 32, 415–424. [DOI] [PubMed] [Google Scholar]
- Parrish JZ, Xu P, Kim CC, Jan LY, and Jan YN (2009). The microRNA bantam functions in epithelial cells to regulate scaling growth of dendrite arbors in Drosophila sensory neurons. Neuron 63, 788–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peled ES, and Isacoff EY (2011). Optical quantal analysis of synaptic transmission in wild-type and rab3-mutant Drosophila motor axons. Nature Neurosci. 14, 519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen SA, Fetter RD, Noordermeer JN, Goodman CS, and DiAntonio A (1997). Genetic analysis of glutamate receptors in Drosophila reveals a retrograde signal regulating presynaptic transmitter release. Neuron 19, 1237–1248. [DOI] [PubMed] [Google Scholar]
- Petzoldt AG, Lützkendorf J, and Sigrist SJ (2016). Mechanisms controlling assembly and plasticity of presynaptic active zone scaffolds. Curr. Opin. Neurobiol 39, 69–76. [DOI] [PubMed] [Google Scholar]
- Powers AS, Grizzaffi J, Ribchester R, and Lnenicka GA (2016). Regulation of quantal currents determines synaptic strength at neuromuscular synapses in larval Drosophila. Eur. J. Physiol 468, 2031–2040. [DOI] [PubMed] [Google Scholar]
- Premont RT, Perry SJ, Schmalzigaug R, Roseman JT, Xing Y, and Claing A (2004). The GIT/PIX complex: an oligomeric assembly of GIT family ARF GTPase-activating proteins and PIX family Rac1/Cdc42 guanine nucleotide exchange factors. Cell. Signal 16, 1001–1011. [DOI] [PubMed] [Google Scholar]
- Quinlan AR, and Hall IM (2010). BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramachandran P, and Budnik V (2010). Dissection of Drosophila larval body-wall muscles. Cold Spring Harb Protoc 2010, pdb prot5469. [DOI] [PubMed] [Google Scholar]
- Rambout X, Dequiedt F, and Maquat LE (2018). Beyond transcription: roles of transcription factors in pre-mRNA splicing. Chem. Rev 118, 4339–4364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasse TM, Fouquet W, Schmid A, Kittel RJ, Mertel S, Sigrist CB, Schmidt M, Guzman A, Merino C, Qin G, et al. (2005). Glutamate receptor dynamics organizing synapse formation in vivo. Nat. Neurosci 8, 898–905. [DOI] [PubMed] [Google Scholar]
- Ritzenthaler S, Suzuki E, and Chiba A (2000). Postsynaptic filopodia in muscle cells interact with innervating motoneuron axons. Nat. Neurosci 3, 1012–1017. [DOI] [PubMed] [Google Scholar]
- Robinson JT, Thorvaldsdottir H, Winckler W, Guttman M, Lander ES, Getz G, and Mesirov JP (2011). Integrative genomics viewer. Nat. Biotechnol 29, 24–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rui M, Qian J, Liu L, Cai Y, Lv H, Han J, Jia Z, and Xie W (2017). The neuronal protein Neurexin directly interacts with the Scribble-Pix complex to stimulate F-actin assembly for synaptic vesicle clustering. J. Biol. Chem 292, 14334–14348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saneyoshi T, Wayman G, Fortin D, Davare M, Hoshi N, Nozaki N, Natsume T, and Soderling TR (2008). Activity-dependent synaptogenesis: regulation by a CaM-kinase kinase/CaM-kinase I/betaPIX signaling complex. Neuron 57, 94–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlenker O, and Rittinger K (2009). Structures of dimeric GIT1 and trimeric beta-PIX and implications for GIT-PIX complex assembly. J. Mol. Biol 386, 280–289. [DOI] [PubMed] [Google Scholar]
- Schuster CM, Davis GW, Fetter RD, and Goodman CS (1996). Genetic dissection of structural and functional components of synaptic plasticity. I. Fasciclin II controls synaptic stabilization and growth. Neuron 17, 641–654. [DOI] [PubMed] [Google Scholar]
- Semaniuk UV, Gospodaryov DV, Feden'ko KM, Yurkevych IS, Vaiserman AM, Storey KB, Simpson SJ, and Lushchak O (2018). Insulin-like peptides regulate feeding preference and metabolism in Drosophila. Front. Physiol 9, 1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shingleton AW (2010). The regulation of organ size in Drosophila: physiology, plasticity, patterning and physical force. Organogenesis 6, 76–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigrist SJ, Thiel PR, Reiff DF, Lachance PE, Lasko P, and Schuster CM (2000). Postsynaptic translation affects the efficacy and morphology of neuromuscular junctions. Nature 405, 1062–1065. [DOI] [PubMed] [Google Scholar]
- Sink H, and Whitington PM (1991). Location and connectivity of abdominal motoneurons in the embryo and larva of Drosophila melanogaster. J. Neurobiol 22, 298–311. [DOI] [PubMed] [Google Scholar]
- Smith RJ (1985). Scaling: why is animal size so important? Quart. Rev. Biol 60, 370–371. [Google Scholar]
- Steinert JR, Kuromi H, Hellwig A, Knirr M, Wyatt AW, Kidokoro Y, and Schuster CM (2006). Experience-dependent formation and recruitment of large vesicles from reserve pool. Neuron 50, 723–733. [DOI] [PubMed] [Google Scholar]
- Stocker H, Andjelkovic M, Oldham S, Laffargue M, Wymann MP, Hemmings BA, and Hafen E (2002). Living with lethal PIP3 levels: viability of flies lacking PTEN restored by a PH domain mutation in Akt/PKB. Science 295, 2088–2091. [DOI] [PubMed] [Google Scholar]
- Tenenbaum CM, Misra M, Alizzi RA, and Gavis ER (2017). Enclosure of dendrites by epidermal cells restricts branching and permits coordinated development of spatially overlapping sensory neurons. Cell Rep. 20, 3043–3056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teodoro RO, Pekkurnaz G, Nasser A, Higashi-Kovtun ME, Balakireva M, McLachlan IG, Camonis J, and Schwarz TL (2013). Ral mediates activity-dependent growth of postsynaptic membranes via recruitment of the exocyst. EMBO J. 32, 2039–2055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas U, Ebitsch S, Gorczyca M, Koh YH, Hough CD, Woods D, Gundelfinger ED, and Budnik V (2000). Synaptic targeting and localization of Discs-large is a stepwise process controlled by different domains of the protein. Curr. Biol. 10, 1108–1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas U, Kim E, Kuhlendahl S, Koh YH, Gundelfinger ED, Sheng M, Garner CC, and Budnik V (1997). Synaptic clustering of the cell adhesion molecule Fasciclin II by Discs-large and its role in the regulation of presynaptic structure. Neuron 19, 787–799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ugrankar R, Theodoropoulos P, Akdemir F, Henne WM, and Graff JM (2018). Circulating glucose levels inversely correlate with Drosophila larval feeding through insulin signaling and SLC5A11. Commun. Biol 1, 110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Vactor D, and Sigrist SJ (2017). Presynaptic morphogenesis, active zone organization and structural plasticity in Drosophila. Curr. Opin. Neurobiol 43, 119–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdu J, Buratovich MA, Wilder EL, and Birnbaum MJ (1999). Cell-autonomous regulation of cell and organ growth in Drosophila by Akt/PKB. Nature Cell Biol. 1, 500–506. [DOI] [PubMed] [Google Scholar]
- Wu Q, Zhang Y, Xu J, and Shen P (2005). Regulation of hunger-driven behaviors by neural ribosomal S6 kinase in Drosophila. Proc. Natl. Acad. Sci. U S A 102, 13289–13294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing G, Li M, Sun Y, Rui M, Zhuang Y, Lv H, Han J, Jia Z, and Xie W (2018). Neurexin–Neuroligin 1 regulates synaptic morphology and functions via the WAVE regulatory complex in Drosophila neuromuscular junction. eLife 7, e30457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshihara M, Adolfsen B, Galle KT, and Littleton JT (2005). Retrograde signaling by Syt 4 induces presynaptic release and synapse-specific growth. Science 310, 858–863. [DOI] [PubMed] [Google Scholar]
- Zhang H, Webb DJ, Asmussen H, Niu S, and Horwitz AF (2005). A GIT1/PIX/Rac/PAK signaling module regulates spine morphogenesis and synapse formation through MLC. J. Neurosci 25, 3379–3388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao ZS, Manser E, Loo TH, and Lim L (2000). Coupling of PAK-interacting exchange factor PIX to GIT1 promotes focal complex disassembly. Mol. Cell Biol 20, 6354–6363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong Y, Budnik V, and Wu C (1992). Synaptic plasticity in Drosophila memory and hyperexcitable mutants: role of cAMP cascade. J. Neurosci 12, 644–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong Y, and Wu C (1993). Modulation of different K+ currents in Drosophila: a hypothetical role for the Eag subunit in multimeric K+ channels. J. Neurosci 13, 4669–4679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou B, Lindsay SA, and Wasserman SA (2015). Alternative NF-κB isoforms in the Drosophila neuromuscular junction and brain. PLoS ONE 10, e0132793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou W, Li X, and Premont RT (2016). Expanding functions of GIT Arf GTPase-activating proteins, PIX Rho guanine nucleotide exchange factors and GIT–PIX complexes. J. Cell Sci 129, 1963–1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zito K, Fetter RD, Goodman CS, and Isacoff EY (1997). Synaptic clustering of Fasciclin II and Shaker: essential targeting sequences and role of Dlg. Neuron 19, 1007–1016. [DOI] [PubMed] [Google Scholar]
- Zito K, Parnas D, Fetter RD, Isacoff EY, and Goodman CS (1999). Watching a synapse grow: noninvasive confocal imaging of synaptic growth in Drosophila. Neuron 22, 719–729. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
This study did not analyze datasets or generate custom code.


