Abstract
Neuroblastoma (NB) and malignant cutaneous melanoma (CMM) are neural crest cells (NCC)-derived tumors and may have a shared genetic basis, but this has not been investigated systematically by genome-wide association studies (GWAS). We took a three-staged approach to conduct cross-disease meta-analysis of GWAS for NB and CMM (2101 NB cases and 4202 controls; 12 874 CMM cases and 23 203 controls) to identify shared loci. Findings were replicated in 1403 NB cases and 1403 controls of European ancestry and in 636 NB, 508 CMM cases and 2066 controls of Italian origin. We found a cross-association at locus 1p13.2 (rs2153977, odds ratio = 0.91, P = 5.36 × 10−8). We also detected a suggestive (P < 10−7) NB-CMM cross-association at 2q37.1 with opposite effect on cancer risk. Pathway analysis of 110 NB-CMM risk loci with P < 10−4 demonstrated enrichment of biological processes such as cell migration, cell cycle, metabolism and immune response, which are essential of human NCC development, underlying both tumors. In vitro and in silico analyses indicated that the rs2153977-T protective allele, located in an NB and CMM enhancer, decreased expression of SLC16A1 via long-range loop formation and altered a T-box protein binding site. Upon depletion of SLC16A1, we observed a decrease of cellular proliferation and invasion in both NB and CMM cell lines, suggesting its role as oncogene. This is the largest study to date examining pleiotropy across two NC cell-derived tumors identifying 1p13.2 as common susceptibility locus for NB and CMM risk.
We demonstrate that combining genome-wide association studies results across cancers with same origins can identify new loci common to neuroblastoma and melanoma arising from tissues which originate from neural crest cells. Our results also show 1p13.2 confer risk to neuroblastoma and melanoma by regulating SLC16A1.
Introduction
Cells that arise from the neural crest (NC) have a remarkable capacity for motility, invasiveness, proliferation and pluripotency. These traits are essential during embryogenesis, but they can become a liability later in life as can contribute to malignancy and tumor metastasis (1). Neuroblastomas (NB) and cutaneous malignant melanomas (CMM) are NC-derived cancers which tend to be particularly aggressive and prone to metastasis (2,3). NB arises from the sympathetic ganglia and adrenal medulla, which originate from trunk NC, and is the most common extracranial malignant tumor in childhood (2). CMM is one of the most aggressive cancers worldwide (3) and derives from transformed melanocytes, which are pigment-producing cells originating from NC at all axial levels (1). In both two tumors is frequent the observation of spontaneous regression of their metastatic forms (4,5). Auslander et al. have demonstrated some similarities between advanced NB and CMM with respect to their immune-related transcriptome, suggesting shared immune mechanisms underlying spontaneous regression in these two cancer types (6). The existence of genetic risk factors common to NB and CMM has been suggested by the finding of the loss of function mutation E27X in CDKN2A in melanoma families who display NB (7).
Our genome-wide association studies (GWAS) have demonstrated that common DNA variants are risk factors for these two diseases (8,9). Recent studies, by performing cross-phenotype association analysis, using GWAS results, have highlighted that different cancers share common genetic risk loci (10,11). Moreover, by using this type of analysis, it has been possible to identify new risk variants, which are hidden among signals discarded by the stringent multiple-testing correction required in the analysis of GWAS data (10,11).
Based on these premises, we hypothesize that NB and CMM tumorigenesis could be the result of multiple genetic alterations and could share genetic and molecular features that are typical of embryonic development processes. However, so far, no single-nucleotide polymorphism (SNP) has been reported to be associated with risk of both cancers, and no study has leveraged the existent GWAS of NB and CMM to test whether these two NC cell-derived tumors may share associations with common genetic loci that are discarded by the multiple-testing corrections.
Here, we combined data from the recently published GWAS of NB and CMM in a single two-cancer meta-analysis of 42 380 individuals and replicated the results in two additional sets of cases and controls (n = 6016). We hypothesized that the substantial gain in power afforded by the cross-cancer meta-analysis would enable us to identify new risk loci sharing association with the two diseases. We found evidence for a cross-association at 1p13.2, which is significant at the genome-wide level (rs2153977, P = 5.36 × 10−8) and has the same effect on both cancer risk. In vitro and in silico analyses demonstrated that rs2153977 protective T allele, in an NB and CMM enhancer, decreased expression of SLC16A1 via long-range loop formation, suggesting its role in promoting tumor initiation and progression, which has been experimentally confirmed in NB and CMM cell lines.
Methods
Study strategy and description of NB and CMM data sets
This study was approved by the Ethics Committee of the Medical University of Naples and the Children’s Hospital of Philadelphia.
To identify NB-CMM cross-associated loci, we designed a multi-stage approach (Supplementary Figure 1A, available at Carcinogenesis Online) based on an NB-CMM GWAS meta-analysis and independent replication studies using cases and controls with different origins and genotyped by different technologies. We reasoned that this approach could identify previously unrecognized cancer risk loci that were shared by NB and CMM and achieved genome-wide significance only after combining data from the two cancers.
Stage 1
The CMM GWAS data set contained SNP-level summary statistics from stage 1 of a recently published meta-analysis (9) consisting of 11 data sets totaling 12 874 cases and 23 203 controls from Europe, Australia and the United States; this stage included all six published CMM GWAS and five unpublished ones (Supplementary Figure 1A, available at Carcinogenesis Online). We did not utilize the results of stage 2 of that study, where a further 3116 CM cases and 3206 controls from three additional data sets were genotyped for the most significantly associated SNP from each region, reaching P < 10−6 in stage 1. Further details on these studies and clinical information of samples can be found in Supplementary Note to Law et al. (9). The NB GWAS data set (2101 cases and 4202 controls) contained SNP-level summary statistics from association analysis recently published (8). We conducted a fixed-effects meta-analysis of NB and CMM GWAS studies using the summary statistics for all variants that were nominally associated (P < 0.05) with each of the two cancers. There is some evidence that alleles that increase risk of one cancer may confer protection from another cancer (10). To search for such alleles in pairwise meta-analysis, we reversed the signs on the effect size estimates in one of the two data sets and repeated fixed-effects meta-analysis as previously reported (10). After performing the meta-analysis, we retained only the SNPs with P < 1 × 10−4 and P-values from meta-analysis less than those found in each study separately (N = 1805). These SNPs were defined as ‘candidate’ SNPs for association with NB and CMM risk. Clinical features are reported in previously published work (8).
Stage 2
The 1805 SNPs that passed the above-mentioned filters were tested for replication in a second data set of 1403 NB cases and 1403 controls. We used this data set as replication study and did not include it in the meta-analysis with the larger NB set (Supplementary Figure 1A, available at Carcinogenesis Online) because it is well known that confirmation in independent data sets provides protection against false positives (12) and because the same strategy led us to identify several NB susceptibility loci in our previous GWAS (8,13–15). Furthermore, since we planned (i) to further validate the SNPs selected in this stage by two additional replications (see Stage 3 section), (ii) to perform a combined meta-analysis of independent case–control sets (see Stages 1–3 section) and (iii) in vitro experimental studies, to avoid overlooking variants with small real effects, we considered as candidate associated SNPs those with a less stringent significant threshold (P < 0.10) and with same effect direction as in the GWAS meta-analysis of stage 1 (candidate SNPs, N = 168).
Stage 3
Among the 168 candidate SNPs identified in the stage 2, 23 independent SNPs were selected (Supplementary Material, available at Carcinogenesis Online) and genotyped in 636 NB cases, 508 CMM cases, and 2066 controls of Italian origins (Supplementary Figure 1A and Supplementary Table 1, available at Carcinogenesis Online). The control set was divided into two subgroups by case-based splitting approach that distributes the shared controls proportionally according to the case sample sizes, resulting in 1151 and 909 controls for NB and CMM, respectively. The genetic associations with P-values < 0.05 were considered as replicated.
Stages 1–3
The P-values and odds ratios (ORs) of these latter 23 SNPs obtained from the five genetic association studies (Supplementary Figure 1A, available at Carcinogenesis Online) were used to perform a combined meta-analysis. The SNPs that were associated with both NB and CMM risk in all of the five case–control studies and reached a P-value ≤ 5 × 10−8 after the combined meta-analysis were considered NB-CMM cross-associated SNPs, whereas those reaching a P-value < 10−7 were defined as ‘suggestive’ for the association with NB and CMM risk.
All analyses were restricted to individuals of European ancestry. Genotypes in each GWAS data set had been imputed on to the April 2012 release of the 1000 Genomes Project European ancestry reference panel (version 3 of the Phase 1 integrated variant release) (Supplementary Material, available at Carcinogenesis Online).
SNP genotyping
The 1403 NB cases and 1403 controls (stage 2) were genotyped by Illumina OmniExpress 770 array ran according to the manufacturer’s protocol. Prior to association testing (Supplementary Material, available at Carcinogenesis Online), we performed genome-wide quality control, principal component analysis to confirm ancestry, and imputation using SNPs common for cases and controls in each set as previously described (8). We removed 190K SNPs. The genomic inflation was equal to 1.033. The 636 NB and 508 CMM cases, and 2066 controls (stage 3) were by genotyped by PCR-based KASP™.
Statistical analysis
Hardy–Weinberg equilibrium was evaluated using the goodness-of-fit chi-square test in control subjects. For the 23 genotyped SNPs, two-sided chi-square tests were used to evaluate differences in the distributions of allele frequencies between patients and controls. ORs and 95% confidence intervals were calculated to assess the relative disease risk conferred by a specific allele. Estimated magnitudes of association (OR) and standard errors for variants from each data set were combined assuming fixed effects using inverse-variance–weighted meta-analysis implemented in METAL (16). Cochran’s Q statistic and I2 heterogeneity index for all SNPs were also calculated. All linkage disequilibrium (LD) calculations (r2 and D’) were performed using the LDlink suite (https://ldlink.nci.nih.gov/?tab=home) and data from the 1000 Genomes Project European ancestry populations.
Identification of causal variants at 1p13.2
We obtained the regulatory elements (super-enhancer, enhancer and promoter) by a re-analysis of H3K27ac ChIP-seq data (Supplementary Figure 1B and Supplementary Material, available at Carcinogenesis Online) from 26 NB cell lines and 6 patient-derived xenograft cell lines (GSE90683), 7 human derived-melanoma (GSE75352) and 2 human neural crest cell (hNCC) lines (GSE90683) through the National Center for Biotechnology Information (NCBI). We first selected the variants in LD with rs2153977 (0.5 < r2 ≤ 1) (n = 208) and occurring in regulatory genomic regions in CMM, NB and hNCC (n = 9). We then annotated these nine SNPs with non-coding prediction functional score using four programs: CADD (17), FATHMM-MKL (18), GWAVA (19), DeepSea (20), and calculated for each SNP a tissue-specific score given by the formula: 1/(sum of single rank score × 100) × (number of regulatory elements).
Causal gene analysis
For the genome-wide significant loci from the overall meta-analysis, we explored potential causative genes at each association locus using the PrixFixe method (21) (Supplementary Material, available at Carcinogenesis Online).
Chromosome conformation capture
Chromosome conformation capture (3C) assays were done essentially as described (PMID: 11847345) and CMM and NB cells were used, CJM and SKNBE2, respectively. Briefly, cells (1 × 107 per sample) were fixed and lysed to obtain nuclei fraction. After samples were processed and cross-linked DNA was digested with EcoRI. T4 DNA ligase was added to the samples and after crosslink reversing phase DNA was extracted. DNA samples were dissolved in water and analyzed by PCR. We also, selected a negative and a positive control regions. The primers used are reported in Supplementary Material (available at Carcinogenesis Online). PCR products were analyzed semi-quantitatively using ImageJ software. Two biological replicates were prepared and analyzed in three technical repeats. Detailed protocol information are in Supplementary Material (available at Carcinogenesis Online).
Electrophoretic mobility shift assays
Nuclear extracts were prepared according to the method of Dignam et al. Protein concentration was determined by Bradford assay (Bio-Rad, Hercules, CA). We used (Cy5)-labeled oligodeoxynucleotide duplexes as specific probes (Eurofins Genomics, Germany). The complementary oligonucleotides were annealed and the binding reaction was performed with nuclear extracts, poly (dI-dC) (Roche) and Cy5-probe in a binding buffer. Competition studies were performed with varying stoichiometric amounts of unlabeled competitor DNAs. The reactions were resolved on a 5% non-denaturing acrylamide gel. The gels were scanned with Typhoon 9400 imager. The sequences of the sense strands of the duplexes were reported in Supplementary Material (available at Carcinogenesis Online). Detailed protocol information are in Supplementary Material (available at Carcinogenesis Online).
In silico analysis using HaploReg and 3DIV tools
HaploReg v4.1, to identify cell type-specific enhancers enriched in risk loci, was run using the following set options: LD = 0; enhancer definition based on Core 15-state model; gene position according to GENOCODE and false discovery rate (FDR) ≤ 0.1. 3DIV was run using distance-normalized interaction frequency ≥ 2 to define significant enhancer–promoter interactions in SKMEL5 and SKNDZ cell lines.
In vitro functional study
A detailed description of the culture conditions, luciferase assay and other experiments performed to evaluate the SLC16A1 effect on NB and CMM cell line phenotype is reported in Supplementary Material (available at Carcinogenesis Online). From 2014 to 2019, the cell lines have been obtained from the American Type Culture Collection (ATCC; www.atcc.org) and have been reauthenticated (using short tandem repeat analysis) and tested as mycoplasma free.
Results
Stage 1: meta-analysis of NB and CMM GWAS data
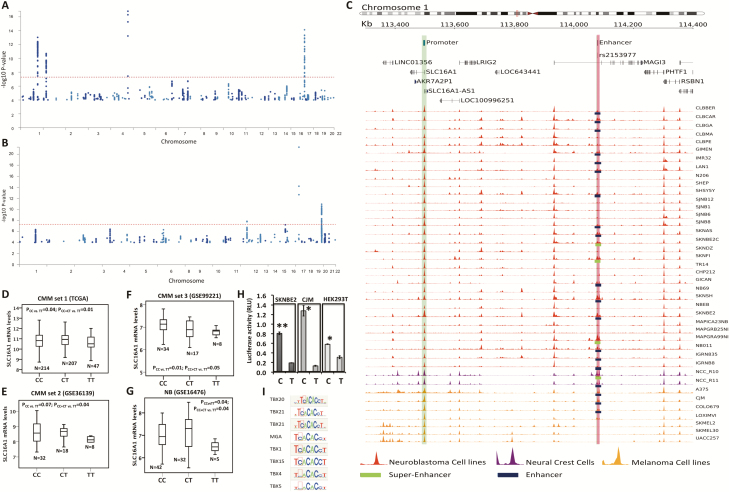
The meta-analysis identified 922 SNP candidates for association with NB and CMM risk (P < 1 × 10−4) with the same direction of effect across the two cancers and with P-value from meta-analysis lower than that found in each study separately (Supplementary Figure 1A; Figure 1A; Supplementary Table 2, available at Carcinogenesis Online). We found 110 unique candidate cross-associated loci with P-value less than 1 × 10−4 (the index SNP, that is, the most significant SNP for each chromosome band, is colored in green in Supplementary Table 2, available at Carcinogenesis Online). By a comparison between the identified NB and CMM risk loci and GWAS Catalog database (https://www.ebi.ac.uk/gwas/), we found that 95 out of the 110 loci were novel candidate risk loci for both tumors (Figure 1A and Supplementary Table 2, available at Carcinogenesis Online). We then reversed the signs on the effect size estimates in the CMM GWAS and repeated the fixed-effects meta-analysis as previously reported (10). We detected a total of 883 candidate SNPs with P < 1 × 10−4, with opposite direction of effect and P-value from meta-analysis inferior to that found in each study separately (Figure 1B and Supplementary Table 3, available at Carcinogenesis Online). Among these SNPs, we found 80 unique (Supplementary Material, available at Carcinogenesis Online) cross-associated loci (the index SNP for each is colored in green in Supplementary Table 3, available at Carcinogenesis Online) and 69 were not found in the GWAS Catalog, and might thus represent new candidate associations for NB and CMM.
Figure 1.
Manhattan plots and H3K27ac activity in the rs2153977 region and SNP-gene expression correlation. Manhattan plots of results from the combined NB and CMM meta-analysis (P < 1 × 10−4) of Stage 1. (A) The dots represent the variants with same direction of effect across the two cancers. (B) The dots represent the variants with opposite direction of effect across the two cancers. The red line corresponds to a threshold of P < 5 × 10−8. (C) The genomic region including SLC16A1 and rs2153977 is shown on genome assembly hg19/GRCh37. SLC16A1 promoter is located at chr1:113495000-113500000 (green highlight). rs2153977 C/T maps at chr1:114080071 (red highlight) within an enhancer region (chr1:114079000-114080523). Here we show density profiles of H3K27Ac histone mark for NB (red), hNCC (purple), and CMM (orange). Green bars on the bottom of density profiles denote the presence of super-enhancers and blue bars the presence of enhancers. (D–G) Microarray-based expression profiling on primary tumors demonstrates that lower SLC16A1 expression correlates with TT-rs2153977 genotype in (D–F) CMM and (G) NB. The N letter in the figure indicates exactly the number of samples of each genotype (H) Luciferase reporter assay carried out in SKNBE2, CJM and HEK293T cells confirms that the TT genotype correlates with a lower luciferase activity. Data shown are mean ± standard deviation from nine independent transfection experiments, each done in triplicate and compared with promoter less control. *P < 0.01; **P < 0.001, t-test. (I) Prediction of motif alteration by RegulomeDB.
Identification of cell type-specific enhancers, most likely causal genes and pathways
For enrichment analyses of cell type-specific enhancers, we conducted queries in HaploRegV4.1 program with the 110 index SNPs (colored in green in Supplementary Table 2, available at Carcinogenesis Online) based on data from the Roadmap Epigenomics Consortium. We found an enrichment of risk SNPs (FDR < 0.10) in the enhancers of tissues such as fetal adrenal gland, fetal muscle, fetal thymus, fetal lung, skin, embryonic stem cells, embryonic neural progenitor cells, and blood cells (primary natural killer cells and primary neutrophils) (Supplementary Table 4, available at Carcinogenesis Online). When we performed the same enrichment analyses using the 80 risk loci with opposite effect on NB and CMM risk, we found no significant enrichment (FDR < 0.10, Supplementary Table 5, available at Carcinogenesis Online).
To identify the most likely causal genes and pathways, we analyzed the 110 risk loci (colored in green in Supplementary Table 2, available at Carcinogenesis Online) by PrixFixe method (21). This method does not focus solely on genes closest to the associated polymorphisms but uses genome-scale shared-function networks to identify sets of mutually functionally related genes spanning multiple GWAS loci. We thus ranked potential causal genes from the 110 candidate NB-CMM associated loci. We found 622 candidate genes in loci with same direction of effect (score ≥ 0.01, Supplementary Table 6, available at Carcinogenesis Online). Commonalities among high-scoring candidate genes can provide insight into the processes contributing to disease, and so we searched for GO terms that were over-represented among the highest-scoring genes (22). PrixFixe–ranked NB-CMM candidate genes in loci with same direction of effect (Supplementary Table 7, available at Carcinogenesis Online) yielded significant enrichment for 281 GO terms (ordered search, permutation tests with multiple-testing family-wise error rate < 0.05; Supplementary Table 7, available at Carcinogenesis Online, and Methods). The most significant enriched terms in our analysis have clear links to cell motility and migration, regulation of locomotion (Log10[OR] > 1.5 and family-wise error rate < 0.05) that are fundamental characteristics of both NC and cancer cells (1). We also found enriched terms that were highly relevant to both traits (Log10[OR] > 1.0 and family-wise error rate < 0.05) such as positive regulation of nucleotide and hormone metabolism, regulation of T and B activation, and regulation of cell cycle G1/S phase transition (Supplementary Table 7, available at Carcinogenesis Online).
Stage 2: NB replication study
We next tested the genetic association of the 1805 NB-CMM candidate cross-associated SNPs (P < 1 × 10−4) (Supplementary Figure 1A and Supplementary Material, available at Carcinogenesis Online, 922 SNPs with same and 883 with opposite direction of effects across the two tumors) in an independent GWAS of 1403 NB cases and 1403 controls of European American origin (Supplementary Table 8, available at Carcinogenesis Online). We were able to test 1698 out of the 1805 SNPs and found 168 SNPs with P < 0.10. Among these SNPs, we identified 23 independent candidate risk loci (Supplementary Material, Supplementary Table 9, available at Carcinogenesis Online). Fourteen SNPs showed the same direction of association whereas nine SNPs showed opposite direction of association compared with that found in CMM data set.
Stage 3: NB and CMM replication in an Italian population and combined analysis
We next sought to replicate 23 independent risk loci in an independent Italian cohort of 636 NB cases and 1151 controls, 508 CMM cases and 909 controls using PCR-based genotyping (Supplementary Table 9, available at Carcinogenesis Online). Two SNPs, rs2153977 at 1p13.2 (PNB_3 = 0.007 and PCMM_2 = 0.002) and rs1604144 at 2q37.1 (PNB_3 = 0.001 and PCMM_2 = 0.021, opposite direction) confirmed the genetic association (P < 0.05) with both NB and CMM in the Italian cohort. Interestingly, these loci, validated in all of five cohorts, represented novel associations for both tumors reaching genome-wide significance (Pcombined = P = 5.36 × 10−8, rs2153977, 1p13.2) and suggestive significance (Pcombined = 6.42 × 10−8, rs1604144, 2q37.1), respectively after the combined analysis (Table 1).
Table 1.
Results of associations between rs2153977 and rs1604144 and NB and CMM
| Chr | Index SNP | Data set | Minor allele | Major Allele | MAF Ctrs | MAF Cases | P | ORa | CI 95% low | CI 95% high |
|---|---|---|---|---|---|---|---|---|---|---|
| 1p13.2 | rs2153977 | NB_1 | T | C | 0.31 | 0.29 | 0.030 | 0.89 | 0.84 | 0.99 |
| CMM_1 | T | C | NA | NA | 2.89E-04 | 0.93 | 0.90 | 0.97 | ||
| NB_2 | T | C | 0.30 | 0.28 | 0.063 | 0.90 | 0.76 | 1.01 | ||
| NB_3 | T | C | 0.29 | 0.24 | 0.007 | 0.80 | 0.69 | 0.94 | ||
| CMM_2 | T | C | 0.30 | 0.24 | 0.002 | 0.76 | 0.63 | 0.91 | ||
| Combined | 5.36E-08 | 0.91 | 0.88 | 0.94 | ||||||
| 2q37.1 | rs1604144 | NB_1 | T | C | 0.27 | 0.31 | 0.001 | 1.17 | 1.06 | 1.27 |
| CMM_1 | T | C | NA | NA | 0.003 | 0.94b | 0.91 | 0.98 | ||
| NB_2 | T | C | 0.26 | 0.29 | 0.099 | 1.19 | 0.98 | 1.32 | ||
| NB_3 | T | C | 0.19 | 0.23 | 0.001 | 1.34 | 1.12 | 1.60 | ||
| CMM_2 | T | C | 0.19 | 0.15 | 0.021 | 0.77b | 0.62 | 0.96 | ||
| Combined | 6.42E-08 | 1.10 | 1.06 | 1.13 |
NB_1: 2101 cases and 4202 controls; CMM_1: 12 874 cases and 23 203 controls; NB_2: 1403 cases and 1403 controls; NB_3: 636 cases and 1151 controls; CMM_2 508 cases and 909 controls. Chr, chromosome; MAF, minor allele frequency; Ctrs, controls; OR, odd ratio; CI, confidence interval; NA, not available.
aOR is for the minor allele.
bORs reversed when performed meta-analysis.
Functional characterization of NB-CMM cross-associated locus 1p13.2
The risk locus 1p13.2, reaching genome-wide significance (rs2153977, P = 5.36 × 10−8; OR = 0.91) and with same effect on NB and CMM development (Table 1), resulted to be independently validated in all of the five sets of cases and controls (Table 1). Therefore, we decided to further functionally analyze the 1p13.2 locus (Supplementary Figure 1B, available at Carcinogenesis Online).
To highlight potentially functional variants, we annotated 208 SNPs in LD (0.5 < r2 ≤ 0 1) with the lead SNP rs2153977, with the regulatory elements super-enhancer, enhancer and promoter histone obtained by a re-analysis of H3K27ac ChIP-Seq data (Supplementary Material, available at Carcinogenesis Online) derived from 32 NB, 7 CMM and 2 hNCC cell lines (Supplementary Tables 1 and 10, available at Carcinogenesis Online, and Methods). To prioritize candidate causal functional variants in both tumors, we first selected those SNPs overlapping at least one histone marker in CMM, NB and hNCC (Table 2, Supplementary Table 10, Supplementary Material, available at Carcinogenesis Online). We then annotated the 9 SNPs with non-coding prediction functional scores obtained by four different programs and calculated a global rank score adjusted for the cancer tissue specificity of the functionally active regulatory regions (Methods and Table 2). SNP rs2153977 was classified as the third most significant SNP after rs61817589 and rs2797412 (Table 2) and was located in an enhancer element identified in 19 NB, 3 CMM and 2 hNC cell lines (Figure 1C). Because rs61817589 and rs2797412 were also located in a highly predicted functional region, we decided to test their genetic association in the Italian cohort. We typed only the most significant SNP, rs61817589, given its high LD with rs2797412 (r2 = 0.89, Supplementary Figure 2, available at Carcinogenesis Online). As reported in Supplementary Table 11 (available at Carcinogenesis Online), rs61817589 (within 5′UTR region of PHTF1 gene), was not associated with either NB or CMM (PNB = 0.41 and PCMM = 0.08) even in the Italian population. Furthermore, this SNP showed a moderate LD (r2 = 0.51) with rs2153977 and was not significantly associated with both NB and CMM in each of two the GWAS data set of the stage 1 (Table 2).
Table 2.
Prioritized functional SNPs at 1p13.2 locus
| SNP ID | MAF | Region | Nearest genes | r 2 | P NBa | P CMMb | RE NBc | RE CMMc | RE hNCCc | CADD | FATHMM | GWAVA | DeepSea | TSS |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| rs61817589 | 0.22 | 5′-UTR | PTHF1 | 0.51 | 0.112 | 0.101 | 32 | 7 | 2 | 19.98 | 0.94 | 0.50 | 0.70 | 683.3 |
| rs2797412 | 0.24 | Upstream | PTHF1 | 0.59 | 0.072 | 0.213 | 29 | 1 | 2 | 6.39 | 0.20 | 0.52 | 0.80 | 457.1 |
| rs2153977 | 0.28 | Intronic | MAGI3 | 1.00 | 0.030 | 0.0003 | 19 | 3 | 2 | 11.30 | 0.10 | 0.35 | 0.54 | 171.4 |
| rs4456089 | 0.22 | Intronic | MAGI3 | 0.72 | 0.136 | 0.170 | 19 | 4 | 2 | 4.92 | 0.14 | 0.29 | 0.51 | 125.0 |
| rs11102649 | 0.23 | Intronic | MAGI3 | 0.68 | 0.162 | 0.116 | 24 | 1 | 2 | 4.05 | 0.15 | 0.27 | 0.28 | 117.4 |
| rs11102648 | 0.22 | Intronic | MAGI3 | 0.72 | 0.154 | 0.102 | 20 | 1 | 2 | 5.36 | 0.14 | 0.20 | 0.42 | 104.5 |
| rs10858000 | 0.22 | Intronic | MAGI3 | 0.72 | 0.102 | 0.083 | 9 | 1 | 2 | 2.91 | 0.08 | 0.31 | 0.36 | 50.0 |
| rs1775754 | 0.24 | Intronic | PTPN22 | 0.59 | 0.058 | 0.240 | 4 | 2 | 2 | 1.18 | 0.03 | 0.31 | 0.20 | 27.6 |
| rs1217421 | 0.24 | Intronic | PTPN22 | 0.59 | 0.072 | 0.205 | 4 | 3 | 2 | 0.48 | 0.08 | 0.23 | 0.16 | 26.5 |
The coefficient r2 is calculated with respect to the index SNP: rs2153977. In bold is the index SNP. TSS (tissue-specific score) = 1/(sum of single rank score × 100) × (number of regulatory elements). MAF, minor allele frequency.
a P-value reported in GWAS data set of NB (stage 1).
b P-value reported in GWAS data set of CMM (stage 1).
cRegulatory element (RE): super-enhancer, enhancer or promoter (see Figure 1A; Supplementary Table 10; and Supplementary Material, available at Carcinogenesis Online).
SNP rs2153977 is located in an enhancer element in an intron of MAGI3 (Figure 1C). Acetylation signals (H3K27ac) were not observed in other non-NB and non-CMM tissues from seven ENCODE cell lines (Supplementary Material, Supplementary Figure 3 and Supplementary Table 2, available at Carcinogenesis Online). These results are consistent with recent evidence that disease-associated SNPs frequently affect enhancers that are specific to disease-relevant cell lines and tumor histology, and control developmental stage and tissue-specific gene expression (23). Based on these results, we decided to further functionally characterize rs2153977.
Functional analysis of rs2153977
To identify genes whose expression is affected by the candidate SNP rs2153977, we performed cis-expression quantitative loci (eQTL) analysis on genes within ±1Mb surrounding the variant. Analysis using publicly available genome-wide expression and SNP arrays on NB and CMM tumors (Supplementary Material, available at Carcinogenesis Online) demonstrated that rs2153977 affected expression of SLC16A1. Specifically, the presence of the protective genotype TT significantly correlated with decreased SLC16A1 mRNA expression in three CMM sets (CMM_Set 1; P = 0.01; CMM_Set 2: P = 0.04; CMM_Set 3: P = 0.05) and in one NB set (P = 0.04) (Figure 1D–G and Supplementary Table 12, available at Carcinogenesis Online) suggesting a recessive model for this association. When we performed logistic regression to test if an additive model could explain the association, no significant results were observed (data not shown). No other SNP-gene expression association was found to be significant in all of four data sets (Supplementary Table 12, available at Carcinogenesis Online). Furthermore, rs2153977 is reported in the GWAS Catalog as associated with autoimmune thyroid disease and type 1 diabetes (24), and in GRASPAR v2.0 (25) as associated with rheumatoid arthritis (26,27), fasting blood glucose (28), triglycerides and total cholesterol levels (29), and serum ratio of the metabolite 1-arachidonoylglycerophosphocholine*/1palmitoleoylglycerophosphocholine (30) (Supplementary Table 13, available at Carcinogenesis Online). SLC16A1 as transporter of monocarboxylates (31) (including acetoacetate, β-hydroxybutyrate, short-chain fatty acids, pyruvate and lactate) is implicated in leukocyte migration, pyruvate and lipid metabolic processes, glucose homeostasis, regulation of insulin secretion, plasma membrane lactate transport (data from NCBI database, https://www.ncbi.nlm.nih.gov/gene/6566), and all of these biological processes are characteristic of the above-mentioned rs2153977-associated phenotypes. These observations strengthen the hypothesis of SLC16A1 as potential target gene of the rs2153977-enhancer locus because the biological processes in which SLC16A1 is involved are the same that are responsible for the rs2153977-associated phenotypes.
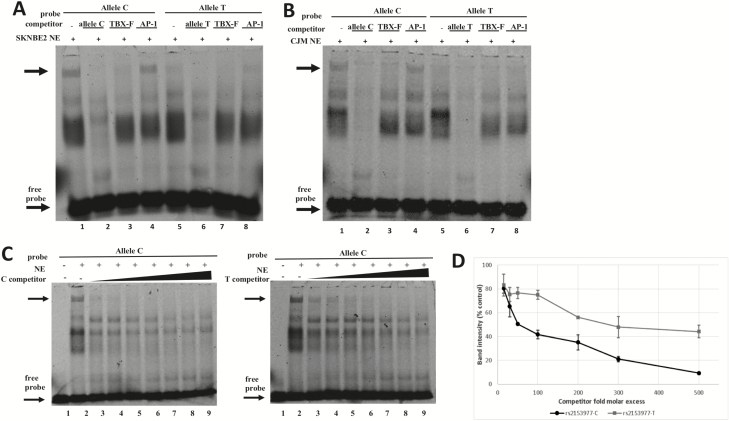
The induction of enhancer activity of the construct containing the rs2153977-T allele was lower than that of the construct containing the C allele as assessed by luciferase report gene assay in SKNBE2, CJM and HEK293T cells (Figure 1H, P < 0.01). The in silico examination of the sequence containing the rs2153977 polymorphism revealed consensus binding sites for different T-box proteins that are known to play a key role in early embryogenesis and carcinogenesis (Figure 1I) (32). In view of this, electrophoretic mobility shift assays were performed to determine whether the sequence containing rs2153977 actually binds nuclear proteins in SKNBE2 and CJM cells. Figure 2 shows that both the canonical (C) and the variant (T) alleles are able to bind nuclear proteins in SKNBE2 NB (Figure 2A) and CJM melanoma cell lines (Figure 2B). Furthermore, competition assays with unlabeled probes representing canonical cis elements for T-box family members (TBX-F) support the involvement of factor(s) from this family in the binding to the polymorphic site in these cells. Figure 2C shows that the rs2153977-C allele competitor probe is able to displace formation of the upper complex with higher efficiency, compared with the T-competitor sequence. The corresponding dissociation curves for both alleles showed that the protein(s) responsible for the formation of the shifted complexes possessed a significantly greater affinity for binding to the rs2153977-C allele. Quantification of binding from three independent experiments showed, indeed, a significant difference in binding affinity for the rs2153977-C and the rs2153977-T alleles (Figure 2D) accordingly, a decreased affinity of the nuclear proteins against the rs2153977-T allele was also observed in CJM melanoma cells (data not shown). Altogether, these data suggest that nuclear proteins belonging to the TBX family of transcription factors recognize and bind specifically to the rs2153977-C allele, with greater affinity, compared with the rs2153977-T allele. Accordingly, the rs2153977-T polymorphism likely alters the binding of such transcription factors, leading to decreased activity of the enhancer and in impaired transcription of the SLC16A1 gene.
Figure 2.
Electrophoretic mobility shift assays (EMSA). The rs2153977-C and -T alleles bind factors of the T-box family with different affinities. EMSA performed in NB (A) and CMM (B) cells, showing the binding of nuclear extract (NE) proteins to a Cy5-labeled allele C probe and allele T probe. Competition assays were performed using 300-fold excess of unlabeled double-stranded oligonucleotides for the allele C or allele T sequence (specific competition), for a T-box family representative sequence containing the TCACACCT motif (TBX-F), and for an unrelated factor (AP-1, nonspecific competition). The main complex affected by T-box-related sequence is indicated by the top arrow; the arrow at the bottom indicates the migration of the free probe. C) Competition assays were performed with the indicated unlabeled competitors at various stoichiometric amounts (15-, 30-, 50-, 100-, 200-, 300-, 500-fold excesses) of allele C (left panel) and allele T (right panel). The allele C shows higher affinity for nuclear protein(s), as reflected by the more rapid attenuation of the DNA–protein complex with increasing concentrations of the unlabeled allele C competitor (lanes 3–9, left panel) compared with the unlabeled allele T competitor (lanes 3–9, right panel). (D) The chart reports the dissociation curves for the allele C and allele T-competitor probes. Band intensities derived from three independent experiments were plotted as a percentage of band intensity in the absence of competitor for each competitor concentration. There was significant difference (P ˂ 0.01) between the alleles.
Physical linkage between polymorphic enhancer and SLC16A1 promoter
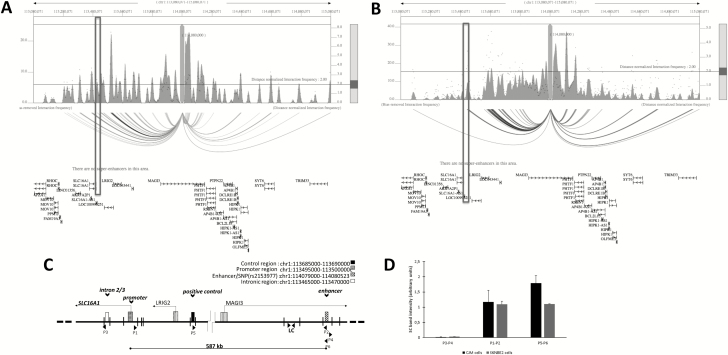
Our observation that rs2153977 is an eQTL for SLC16A1 suggested that this SNP is located in an enhancer that loops to SLC16A1 (Figure 1C). To investigate this hypothesis, we interrogated the database 3DIV (33) that provides a list of long-range chromatin interaction partners for the queried locus obtained from HiC (high-throughput chromatin conformation capture) analysis of 80 different human cell/tissue types. We found that, in the NB (SKNDZ) and CMM (SKMEL5) cell lines, the promoter of the SLC16A1 significantly interacts with the enhancer where the NB-CMM cross-associated SNP rs2153977 is located (Figure 3A and B). To further confirm such interaction, we performed 3C analysis in CMM (CJM) and NB (SKNBE2) cells as previously published (34) (Figure 3C and Supplementary Material, available at Carcinogenesis Online). In addition to the promoter region of SLC16A1, we examined other chromosomal regions: a region that does not have typical characteristics of a regulatory element, which is located within an intronic region of the SLC16A1, and an additional chromosomal region possessing a high interaction frequency (data from 3DIV) called ‘control region’. Specific products were amplified in both cell lines with primers targeting the restriction fragments of enhancer/SNP and SLC16A1 promoter, in samples that had been cross-linked, but not in samples that were not cross-linked (Figure 3D and Supplementary Figure 4, available at Carcinogenesis Online). Interaction between the enhancer and control regions was detected in both cell lines. Control PCRs with artificial amplifiers confirmed the absence of interaction between the intronic region and the promoter in any of the cell lines under analysis. Thus, the results of 3C confirmed the interaction between the SLC16A1 promoter and the regulatory element associated with the genetic variant rs2153977. In agreement with the results of chromatin interaction tests, our analysis to predict the target genes by PrixFixe indicated SLC16A1 as the most likely causal gene (score = 0.12) at 1p13.2 (Supplementary Table 6, available at Carcinogenesis Online).
Figure 3.
The enhancer containing rs2153977 interacts with the SLC16A1 promoter in CMM and NB cells. Plot of HiC data of NB and CMM cells. NB cells (SKNSH) (A) and CMM cells (SKMEL5) (B). One-to-all interaction plot of HiC data (3DIV database) is shown for rs2153977 enhancer region as bait. Y-axes in the left and the right indicate bias-removed interaction frequency and distance-normalized interaction frequency (dots), respectively. Horizontal line indicates the cutoff for distance-normalized interaction frequency. Shown is arc-representation of significant interaction for the given cutoff value defined by the Horizontal line. The boxes indicate the SLC16A1 promoter. (C) Schematic representation of approximate positions regions analyzed and direction of transcription of genes, and EcoRI cutting sites within the area are displayed. Primers P1 and P2 were designed to amplify a novel ligation product formed between the restriction fragments that encode the promoter region and enhancer DNA, respectively. Intronic DNA fragment primers (P3 and P4) and a positive control primers (P5 and P6) are shown. LC is an internal region unaffected by digestion used as loading control. (D) A chromatin conformation capture (3C) assay was performed using the Enhancer/SNP region as an anchor (P2, P4, P6), and the interaction between the enhancer/SNP region and distal elements [intronic region (P3), promotor region (P1) and control region (P5)] in CJM and SKNBE2 cells was assessed. The interaction frequency corresponds with the intensity of amplified PCR products analyzed gels are shown in Supplementary Figure 5 and Supplementary Material (available at Carcinogenesis Online). Data are shown as mean ± SD.
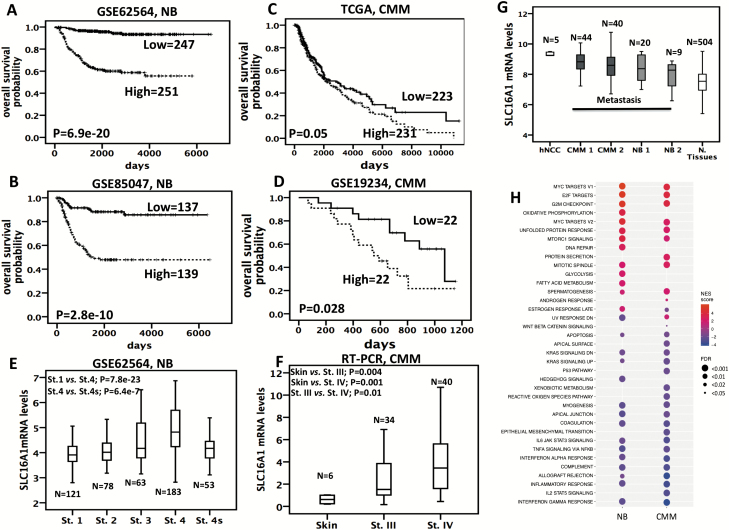
Tumor expression of SLC16A1 and prognosis
To examine the relevance of SLC16A1 in tumor samples, we tested gene expression in four independent publicly available microarray data sets (Supplementary Material, available at Carcinogenesis Online). High SLC16A1 expression was associated with poor overall survival in all NB and CMM tumor sets (Figure 4A–D). We also observed SLC16A1 over-expression in NB advanced stages (Figure 4E). By RT–PCR analysis, we detected higher SLC16A1 levels in stage IV CMM tumors when compared with stage III and normal skin (Figure 4F). Moreover, we performed an analysis across different data sets analyzed with the same chip type (Affymetrix U133 plus v2) and normalized by the same algorithm (MAS 5.0) to assess variation of SLC16A1 expression among normal tissues and hNCC and tumor tissues. SLC16A1 expression was significantly higher in trunk hNCC when compared with normal tissues (P < 0.001, Figure 4G) and was significantly higher in CMM and NB tumors when compared with normal tissues (CMM 1 or CMM 2 versus normal tissues: P < 1 × 10−6; NB 1 or NB 2 versus normal tissues: P < 0.0001 and P = 0.09, Figure 4G). Together, these data support the hypothesis that SLC16A1 might play a biological role in carcinogenesis and hNCC development.
Figure 4.
SLC16A1 expression is associated with poor outcome in NB and CMM and is higher in hNCC NB and CMM tumors than normal tissues. (A–D) Kaplan–Meier analysis is shown, with individuals grouped by median of expression. Log-rank P-values are shown. (E) Box-plots showing the mRNA expression of SLC16A1 from an available gene expression data set across different NB stages (indicated as St.). (F) Box-plots showing the mRNA expression of SLC16A1, obtained by real-time (RT)–PCR, across human skin samples and stage-III and stage-IV (indicated as St.) CMM tumor samples. (G) Box-plots showing the mRNA expression of SLC16A1 across different data sets downloaded from R2 bioinformatics tool: GSE14340 for hNCC; GSE19234 for Metastatic CMM 1; GSE7553 for Metastatic CMM 2; GSE13136 for NB 1; GSE14880 for NB 2; GSE7307 for Normal (N.) Tissues (hNCC versus Normal Tissues: P < 0.001; CMM 1 or CMM 2 versus Normal Tissues: P < 0.000001; NB 1 or NB 2 versus Normal Tissues: P < 0.0001 and P = 0.09). (H) Heatmap showing gene set enrichment analysis (GSEA) enrichment scores (with FDR < 0.05) for hallmark gene sets (MsigDB). Size of the circles indicates FDR value and the color indicates the normalized enrichment (NES) score. Positive values (red) indicate enrichment among the positively correlated genes, whereas negative values (blue) indicate enrichment among the negatively correlated genes with SLC16A1 gene expression.
To investigate which pathways are regulated by SLC16A1, we performed correlation analysis in publicly available transcriptome data on 498 NB (GSE62564 data) and 470 CMM (TCGA data) tumors (Supplementary Material, available at Carcinogenesis Online). A total of 10423 and 10127 genes correlated with SLC16A1 expression in NB and CMM tumor sets, respectively (Supplementary Tables 14 and 15, available at Carcinogenesis Online). In both data sets, gene set enrichment analysis (35) on the positively correlated genes revealed enrichment (FDR < 0.05) for the hallmark and gene ontology gene sets involved in MYC, mTORC1 signaling, mRNA and DNA metabolic processes and cell cycle including E2F targets, G2/M checkpoint, mitotic spindle and apoptosis (Figure 4H; Supplementary Tables 16 and 17, available at Carcinogenesis Online). Enrichment was shown for inflammatory response processes, KRAS, IL6-JAK-STAT, and TNFA pathway, immune response, cell locomotion, cell–cell adhesion and chemotaxis processes among the negatively correlated genes (Figure 4H; Supplementary Tables 16 and 17, available at Carcinogenesis Online). Similar biological processes were also found in the gene pathway analyses of the 110 NB-CMM-associated loci (Supplementary Table 7, available at Carcinogenesis Online). Interestingly, the first three most significant enriched terms in both tumors were MYC and E2F targets and G2/M checkpoint (Figure 4H), whereas a stronger signature of metabolism-related changes (such as glycolysis, fatty acid metabolism and oxidative phosphorylation) was observed only in NB tumors (Figure 4H).
In vitro functional analysis of SLC16A1
The above-reported results suggest that SLC16A1 might have an oncogenic role in NB and CMM. Furthermore, a recent research work reported that SLC16A1 is one of the major actors in promoting Merkel cell carcinoma, a skin cancer, by induction of elevated aerobic glycolysis (36). Therefore, to investigate the biological role of SLC16A1 in NB and CMM, we performed a transient knockdown of SLC16A1 in NB and CMM cell lines. We used three pooled siRNAs to deplete SLC16A1 in two NB (Supplementary Figure 5A and B, available at Carcinogenesis Online) and two CMM (Supplementary Figure 5C and D, available at Carcinogenesis Online) cell lines. As reported in Supplementary Figure 5, available at Carcinogenesis Online, SLC16A1 mRNA expression was significantly decreased in the SKNBE2, CJM, NGP and COLO829 si SLC16A1 cells compared with si Scrambled cells, used as control. We also assessed SLC16A1 protein silencing in NB and CMM cell lines by Western blotting (Supplementary Figure 5, available at Carcinogenesis Online). To evaluate the effect of SLC16A1 expression on NB and CMM cells, we performed a viability and an invasion cell assay. We demonstrated that SLC16A1 silencing, significantly reduce the growth ability of NB-CMM cell lines, and moreover, we observed a decrease of the number of invading cells upon SLC16A1 depletion compared with control cells (Supplementary Figure 5, available at Carcinogenesis Online).
Discussion
It has been long suggested that genetic analyses of multiple correlated phenotypes will increase power to detect trait loci sufficiently to justify the statistical complexity. Because both NB and CMM originate from NCC and thus might share biological and genetic factors, it seemed likely that this would be a fruitful approach. We conducted a cross-cancer GWAS and replication-based analyses investigating pleiotropic associations for these two NC cell-derived cancers. In stage 1, we identified 110 candidate NB and CMM cross-associated loci (P < 10−4).
The 110 risk loci were enriched in active enhancers of embryonic stem cells, diverse fetal tissues, and adrenal gland and skin tissues from where the two tumors originate and were also enriched in cell motility and migration biological processes that are fundamental traits for the development of NCs and malignant transformation of normal tissues (1). Taken together, our results suggest that risk loci for NB and CMM may affect the same biological signaling pathway that NCCs use during their developmental processes. Therefore, careful and comparative studies of key genes underlying the NB-CMM risk loci could unravel novel cancer mechanisms and prove immensely valuable in designing new strategies for cancer therapy and in particular those related to immunotherapy.
By a meta-analysis of three NB and two CMM genetic association studies, we identified a novel cross-association at the 1p13.2 region with NB and CMM, neither of which was previously known to be associated with genetic variation in this region. Fine mapping of the 1p13.2 locus identified the lead SNP rs2153977 as the most likely functional genetic variant and demonstrated that it is located in an active enhancer in NB, CMM and hNC cells. Moreover, rs2153977 was an eQTL in both NB and CMM for the SLC16A1 gene. 3C in vitro and HiC in silico analysis confirmed the interaction between rs2153977 and the promoter region of SLC16A1. Prediction of causal genes at this locus by PrixFixe program again identified SLC16A1 as the gene with the highest score. Finally, through the GASPAR database, we showed that rs2153977 was associated with different traits including fat and glucose metabolism, and immune system which are linked with SLC16A1 functions (31,37). Nonetheless, we cannot unequivocally exclude other genes as the targets of the causal variant at this locus.
SLC16A1 (protein name MCT1) is a proton-linked monocarboxylic acid transporters (31) that transports monocarboxylates including acetoacetate, β-hydroxybutyrate, short-chain fatty acids, pyruvate and lactate. Different reports suggest a key role of SLC16A1 in promoting the development and progression of tumors by modifying the metabolic program of cells (36,38–40). In this context, it is noteworthy that SLC16A1 is a target of Myc oncoproteins and that elevated SLC16A1 levels are a hallmark of human malignancies with MYC or MYCN involvement (41). These data emphasize the potential role of SLC16A1 in NB, which is a known Myc-expression malignancy; indeed, MYCN is amplified in ~20% of NB tumors (2) and in CMM where MYC gain has been found (42).
SLC16A1 constitutes an attractive target for cancer therapy (43). Fang et al. reported that SLC16A1 mRNA levels in fresh NB biopsy samples correlated positively with highly aggressive tumors (44) and its inhibition, by lonidamine, induced an immediate decrease in intracellular pH, suggesting SLC16A1 targeting as useful adjuncts to NB therapy, having particularly high activity where extracellular pH is low (44). Notably, a similar conclusion was made by Miriam et al., which tested the potential role of SLC16A1 as therapeutic target in melanoma (45). Furthermore, MCT1 over-expression, in accord to our findings, has been found significantly associated with the unfavorable clinical stage III and IV, and shorter overall survival (46).
Our work demonstrates that the minor allele of rs2153977 correlates with decreased level of SLC16A1 and protect against NB and CMM development. Accordingly, SLC16A1 mRNA expression levels were significantly higher in tumor metastasis and hNCC than in normal tissues and correlated with a worst clinical outcome in NB and CMM patients. Transient knockdown of SLC16A1 resulted in significant inhibition of growth and invasion in NB and CMM cells. Notably, in silico predictions and electrophoretic mobility shift assay experiments suggested rs2153977 to alter the binding site of T-box proteins that during development have roles in differentiation, proliferation and the epithelial mesenchymal transition, which are processes that are also relevant to the development of cancer and metastasis (32). Together, these results support an oncogenic role for SLC16A1 in NB and CMM to promote cell proliferation, invasion and migration. Additional research efforts are needed to elucidate the molecular mechanisms of SLC16A1 in promoting malignant transformation of NB and CMM.
We have also investigated the biological functions of the gene sets positively and negatively co-expressed with SLC16A1 in 498 NB and 470 CMM tumors. Interestingly, we observed the highest enrichment scores for MYC and E2F and G2/M check point processes in both tumors, and a strong signature of metabolism-related changes (such as glycolysis, fatty acid metabolism and oxidative phosphorylation) only in NB tumors. Additional transcriptional programs that correlated with SLC16A1 expression included the inflammatory response, cell migration and adhesion, and immune response. These biological functions are in line with activities of monocarboxylate transporters in affecting pHi and as consequence the cell cycle (47), cell migration (48), drug resistance (39,41,43) and metastasis (40). Of note, SLC16A1 expression correlated with metabolic and immune processes that are implicated in the development of traits/diseases such as rheumatoid arthritis, type 1 diabetes, fasting blood glucose, triglycerides and cholesterol changes that show also an association with rs2153977, further strengthening the link between this SNP and SLC16A1. Interestingly, our in silico analysis confirmed the role of SLC16A1 in the regulation of T cell activation (37,49). In this context, it is noteworthy that both CMM and NB probably share immune mechanisms underlying spontaneous regression (6) and for both tumors immunotherapy has been successfully included in clinical treatment settings (50). These observations again highlighted that NB and CMM share biological features implicated in tumor development and progression.
We have demonstrated that combining genome-wide association data across cancer types with same embryonic origin and performing replication studies can uncover risk loci that are shared by and represent novel findings for NB and CMM. Our in silico characterization provides evidence that target genes at putative NB and CMM cross-associated risk loci are involved in biological processes that are similar between hNCC development and tumorigenesis and tumor progression such as cell cycle, migration, invasion and cell metabolism. Furthermore, we have demonstrated that a functional DNA variant in the enhancer region of SLC16A1 influences NB and CMM susceptibility and that high expression of SLC16A1 might play a role in malignant neuroblastic and melanocyte transformation and disease progression, co-opting molecular features used by developing NC cells.
Supplementary material
Supplementary data are available at Carcinogenesis online.
Funding
This study was supported by grants from Associazione Italiana per la Ricerca sul Cancro (19255 to M.C. and 20757 to A.I.); Ministero della Salute (GR-2011-02348722 to M.C.); Regione Campania ‘SATIN’ grant 2018–2020 (to M.C.); Fondazione Italiana per la Lotta al Neuroblastoma (to M.C.); Associazione Oncologia Pediatrica e Neuroblastoma (to M.C.); and Fondazione Umberto Veronesi (to F.C.). Programme STAR financially supported by UniNA and Compagnia di San Paolo (to M.C.).
Author contributions
Conflict of Interest Statement: None declared.
Supplementary Material
Acknowledgements
We thank Professor Nick Hayward (QIMR Berghofer Medical Research Institute, Australia) for providing us with CJM cell lines.
Glossary
Abbreviations
- 3C
chromosome conformation capture
- CMM
malignant cutaneous melanoma
- FDR
false discovery rate
- GWAS
genome-wide association studies
- hNCC
human neural crest cell
- LD
linkage disequilibrium
- NB
neuroblastoma
- NC
neural crest
- NCC
neural crest cells
- OR
odds ratios
- SNP
single-nucleotide polymorphism
M.A., M.S., A.C., S.C., M.E., F.C., A.M. and D.F. performed all in vitro experiments. A.V., P.G., Maria F.R., S.S., M.S., F.A., H.H. and V.M.C. provided genomic data and critical review of the manuscript. M.D., M.H.L., M.M.I., K.B. and S.D. provided critical review of the manuscript. A.T., Z.V., J.K. and V.A.L. performed computational analysis. N.Z., A.I. and M.C. designed and conducted the study.
References
- 1. Maguire L.H., et al. (2015) Tumors of the neural crest: common themes in development and cancer. Dev. Dyn., 244, 311–322. [DOI] [PubMed] [Google Scholar]
- 2. Maris J.M. (2010) Recent advances in neuroblastoma. N. Engl. J. Med., 362, 2202–2211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Schadendorf D., et al. (2018) Melanoma. Lancet, 392, 971–984. [DOI] [PubMed] [Google Scholar]
- 4. Diede S.J. (2014) Spontaneous regression of metastatic cancer: learning from neuroblastoma. Nat. Rev. Cancer, 14, 71–72. [DOI] [PubMed] [Google Scholar]
- 5. Kalialis L.V., et al. (2009) Spontaneous regression of metastases from melanoma: review of the literature. Melanoma Res., 19, 275–282. [DOI] [PubMed] [Google Scholar]
- 6. Auslander N., et al. (2018) Robust prediction of response to immune checkpoint blockade therapy in metastatic melanoma. Nat. Med., 24, 1545–1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ghiorzo P., et al. (2006) Impact of E27X, a novel CDKN2A germ line mutation, on p16 and p14ARF expression in Italian melanoma families displaying pancreatic cancer and neuroblastoma. Hum. Mol. Genet., 15, 2682–2689. [DOI] [PubMed] [Google Scholar]
- 8. McDaniel L.D., et al. (2017) Common variants upstream of MLF1 at 3q25 and within CPZ at 4p16 associated with neuroblastoma. PLoS Genet., 13, e1006787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Law M.H., et al. (2015) Genome-wide meta-analysis identifies five new susceptibility loci for cutaneous malignant melanoma. Nat. Genet., 47, 987–995.3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kar S.P., et al. (2016) Genome-wide meta-analyses of breast, ovarian, and prostate cancer association studies identify multiple new susceptibility loci shared by at least two cancer types. Cancer Discov., 6, 1052–1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Fehringer G., et al. (2016) Cross-cancer genome-wide analysis of lung, ovary, breast, prostate, and colorectal cancer reveals novel pleiotropic associations. Cancer Res., 76, 5103–5114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kraft P., et al. (2009) Replication in genome-wide association studies. Stat. Sci., 24, 561–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wang K., et al. (2011) Integrative genomics identifies LMO1 as a neuroblastoma oncogene. Nature, 469, 216–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Nguyen le B., et al. (2011) Phenotype restricted genome-wide association study using a gene-centric approach identifies three low-risk neuroblastoma susceptibility loci. PLoS Genet., 7, e1002026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Diskin S.J., et al. (2014) Rare variants in TP53 and susceptibility to neuroblastoma. J. Natl Cancer Inst., 106, dju047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Willer C.J., et al. (2010) METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics, 26, 2190–2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Rentzsch P., et al. (2019) CADD: predicting the deleteriousness of variants throughout the human genome. Nucleic Acids Res., 47, D886–D894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Shihab H.A., et al. (2015) An integrative approach to predicting the functional effects of non-coding and coding sequence variation. Bioinformatics, 31, 1536–1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ritchie G.R., et al. (2014) Functional annotation of noncoding sequence variants. Nat. Methods, 11, 294–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zhou J., et al. (2015) Predicting effects of noncoding variants with deep learning-based sequence model. Nat. Methods, 12, 931–934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Tasan M., et al. (2015) Selecting causal genes from genome-wide association studies via functionally coherent subnetworks. Nat. Methods, 12, 154–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Berriz G.F., et al. (2009) Next generation software for functional trend analysis. Bioinformatics, 25, 3043–3044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Parker S.C., et al. (2013) Chromatin stretch enhancer states drive cell-specific gene regulation and harbor human disease risk variants. Proc. Natl Acad. Sci. USA, 110, 17921–17926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Tomer Y., et al. (2015) Genome wide identification of new genes and pathways in patients with both autoimmune thyroiditis and type 1 diabetes. J. Autoimmun., 60, 32–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Leslie R., et al. (2014) GRASP: analysis of genotype-phenotype results from 1390 genome-wide association studies and corresponding open access database. Bioinformatics, 30, i185–i194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Stahl E.A., et al. (2010) Genome-wide association study meta-analysis identifies seven new rheumatoid arthritis risk loci. Nat. Genet., 42, 508–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Gregersen P.K., et al. (2009) REL, encoding a member of the NF-kappaB family of transcription factors, is a newly defined risk locus for rheumatoid arthritis. Nat. Genet., 41, 820–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Prokopenko I., et al. (2009) Variants in MTNR1B influence fasting glucose levels. Nat. Genet., 41, 77–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Barber M.J., et al. (2010) Genome-wide association of lipid-lowering response to statins in combined study populations. PLoS One, 5, e9763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Suhre K., et al. (2011) Human metabolic individuality in biomedical and pharmaceutical research. Nature, 477, 54–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Halestrap A.P., et al. (2012) The monocarboxylate transporter family – role and regulation. IUBMB Life, 64, 109–119. [DOI] [PubMed] [Google Scholar]
- 32. Papaioannou V.E. (2014) The T-box gene family: emerging roles in development, stem cells and cancer. Development, 141, 3819–3833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Yang D., et al. (2018) 3DIV: a 3D-genome interaction viewer and database. Nucleic Acids Res., 46, D52–D57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Dekker J., et al. (2002) Capturing chromosome conformation. Science, 295, 1306–1311. [DOI] [PubMed] [Google Scholar]
- 35. Subramanian A., et al. (2005) Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl Acad. Sci. USA, 102, 15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Berrios C., et al. (2016) Merkel cell polyomavirus small T antigen promotes pro-glycolytic metabolic perturbations required for transformation. PLoS Pathog., 12, e1006020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Murray C.M., et al. (2005) Monocarboxylate transporter MCT1 is a target for immunosuppression. Nat. Chem. Biol., 1, 371–376. [DOI] [PubMed] [Google Scholar]
- 38. Le Floch R., et al. (2011) CD147 subunit of lactate/H+ symporters MCT1 and hypoxia-inducible MCT4 is critical for energetics and growth of glycolytic tumors. Proc. Natl Acad. Sci. USA, 108, 16663–16668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Sprowl-Tanio S., et al. (2016) Lactate/pyruvate transporter MCT-1 is a direct Wnt target that confers sensitivity to 3-bromopyruvate in colon cancer. Cancer Metab., 4, 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Payen V.L., et al. (2017) Monocarboxylate transporter MCT1 promotes tumor metastasis independently of its activity as a lactate transporter. Cancer Res., 77, 5591–5601. [DOI] [PubMed] [Google Scholar]
- 41. Doherty J.R., et al. (2014) Blocking lactate export by inhibiting the Myc target MCT1 disables glycolysis and glutathione synthesis. Cancer Res., 74, 908–920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kraehn G.M., et al. (2001) Extra c-myc oncogene copies in high risk cutaneous malignant melanoma and melanoma metastases. Br. J. Cancer, 84, 72–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Birsoy K., et al. (2013) MCT1-mediated transport of a toxic molecule is an effective strategy for targeting glycolytic tumors. Nat. Genet., 45, 104–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Fang J., et al. (2006) The H+-linked monocarboxylate transporter (MCT1/SLC16A1): a potential therapeutic target for high-risk neuroblastoma. Mol. Pharmacol., 70, 2108–2115. [DOI] [PubMed] [Google Scholar]
- 45. Wahl M.L., et al. (2002) Regulation of intracellular pH in human melanoma: potential therapeutic implications. Mol. Cancer Ther., 1, 617–628. [PubMed] [Google Scholar]
- 46. Pinheiro C., et al. (2016) The metabolic microenvironment of melanomas: prognostic value of MCT1 and MCT4. Cell Cycle, 15, 1462–1470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Moolenaar W.H., et al. (1983) Na+/H+ exchange and cytoplasmic pH in the action of growth factors in human fibroblasts. Nature, 304, 645–648. [DOI] [PubMed] [Google Scholar]
- 48. Grinstein S., et al. (1993) Focal localization of the NHE-1 isoform of the Na+/H+ antiport: assessment of effects on intracellular pH. EMBO J., 12, 5209–5218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Haas R., et al. (2015) Lactate regulates metabolic and pro-inflammatory circuits in control of T cell migration and effector functions. PLoS Biol., 13, e1002202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Morandi F., et al. (2018) Novel immunotherapeutic approaches for neuroblastoma and malignant melanoma. J. Immunol. Res., 2018, 8097398. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.