Abstract
Background
Low-risk HPV infection has not been the subject of epigenetic investigation. The present study was carried out in order to investigate the methylation status of CpG sites in non-genital cutaneous warts.
Methods
Genomic DNA was extracted from 24 paired epidermal samples of warts and normal skin. DNA samples were bisulfite converted and underwent genome-wide methylation profiling using the Infinium MethylationEPIC BeadChip Kit.
Results
From a total of 844,234 CpG sites, 56,960 and 43,040 CpG sites were found to be hypo- and hypermethylated, respectively, in non-genital cutaneous warts. The most differentially methylated CpG sites in warts were located within the C10orf26, FAM83H-AS1, ZNF644, LINC00702, GSAP, STAT5A, HDAC4, NCALD, and EXOC4 genes.
Conclusion
Non-genital cutaneous warts exhibit a unique CpG methylation signature.
Keywords: HPV, Warts, DNA methylation, CpG, Epigenetics
Background
CpG sites are parts of DNA that consist of a cytosine nucleotide linked to a guanine nucleotide by a phosphate group, and they are often found as a part of CpG islands, the latter of which are areas of high CpG frequencies [1]. From an epigenetic perspective, CpGs are of particular importance due to the fact that DNA methylation in mammals occurs primarily in a CpG context [2]. In mammalian genomes, the majority of CpG sites are methylated, while those in CpG islands are generally hypomethylated [3]. Due to the high mutability of methylcytosine, methylated CpG sites are under-represented in the human genome [4]. Aberrant CpG methylation patterns increase susceptibility to various diseases, including cancer, but such changes can also be induced during host-pathogen interactions [5, 6].
Host gene dysregulation is a common component of viral infection, and such changes are often generated via epigenetic exploitation of the host genome [7]. In order to evade the antiviral immune response, DNA viruses induce aberrant methylation of immune-related genes in the host [8]. One such example is the human papillomavirus (HPV), a DNA virus that alters host methylation patterns as a part of its life cycle and replication mechanisms within keratinocytes [9]. To date, more than 200 HPV genotypes have been characterized, most of which are low-risk and often manifest in the form of benign cutaneous or genital lesions known as warts [10]. However, a small group of HPV types are considered to be high risk, as they are a causative agent for several different types of squamous cell carcinomas [11].
High-risk HPV infection affects cervical cancer progression by increasing levels of DNA methylation, although methylation patterns were heterogenous among different neoplastic grades [12–14]. Hypomethylation of a CpG site in the MAL gene was reported to be potentially associated with persistent cervical infection with high-risk HPV [15]. Moreover, HPV-positive head-and-neck squamous cell carcinomas exhibited a novel methylation signature in which hypomethylated CpG islands were functionally correlated with gene expression [16]. In fact, HPV-induced epigenetic changes are a major contributing factor to the stability of malignant head-and-neck squamous cell carcinoma [17]. Similarly, CpG loci were differentially methylated in HPV-positive anal squamous neoplasia, and significant differential methylation was observed between in-situ and invasive samples [18].
Unlike its high-risk counterpart, low-risk HPV infection has not been the subject of epigenetic analysis in the context of non-genital cutaneous warts, the latter of which constitutes an extremely common skin disease that is benign and self-limiting in the majority of cases [19]. The most prevalent type of non-genital cutaneous wart is the common wart, which usually manifests on the hands and feet as a firm, hyperkeratotic papule with an irregular surface [20]. The extensive transformation that an HPV-infected keratinocyte undergoes to form a wart suggests that a similar change in methylation patterns must occur. Subsequently, the aim of the current study is to identify the genome-wide methylation status of CpG sites in warts as compared to normal skin.
Methods
Patient recruitment
Twelve patients were recruited at the dermatological clinic in King Abdullah University Hospital in the north of Jordan. The Institutional Review Board (IRB) at Jordan University of Science and Technology (JUST) granted ethical approval to conduct the present study. The inclusion criteria for participants comprised the following characteristics: being male, being free from autoimmune disease, presenting with common warts, not having received prior treatment for their warts, and having given written informed consent. Shave biopsies were performed by a resident dermatologist in order to excise paired normal skin and wart samples from each patient, which were then stored at − 20 °C until subsequent processing.
Extraction of genomic DNA and bisulfite conversion
RNA-free genomic DNA was extracted by means of the QIAamp DNA Mini Kit (Qiagen, Germany) and shipped to the Australian Genome Research Facility (AGRF) on dry ice. Upon arriving to the AGRF, further quality control analysis was performed for each sample using the QuantiFluor® dsDNA System (Promega, USA) and 0.8% agarose gel electrophoresis to determine their purity and integrity, respectively. After obtaining assurance of their quality, the EZ DNA Methylation kit (Zymo Research, USA) was employed for the bisulfite conversion of normalized samples.
Genome-wide methylation profiling and data processing
The Infinium MethylationEPIC BeadChip Kit (Illumina, USA) was utilized in order to interrogate over 850,000 methylation sites. The MethylationEPIC array contains 866,895 probes that target 863,904 CpG sites, 2932 CpH sites, and 59 rs sites. The raw intensity data generated by the array was analyzed using RnBeads, a computational R package [21].
Differential methylation analysis
To calculate the extent of differential methylation (DM) for each CpG site, limma was used to determine three ranks: the beta difference in methylation means between warts (W) and normal skin (NS), the log2 of the quotient in methylation, and the DM p-value [21]. Limma was also utilized to compute p-values on CpG sites [22]. Multiple testing was corrected for by setting the false discovery rate (FDR) at 5% with the Benjamini-Hochberg procedure. Using these three ranks, a combined rank was formulated in which increased DM at a particular CpG site resulted in a smaller rank [21]. The combined rank was used to sort DM CpG sites in ascending order, and the top-ranking 100,000 sites were selected for further analysis.
Enrichment, pathway, and signaling analysis
Gene ontology (GO) term enrichment analysis as well as KEGG and Reactome pathway analysis of the top 100 CpG sites were carried out using the Database for Annotation, Visualization, and Integrated Discovery (DAVID) v6.8 (https://david.ncifcrf.gov/). GO terms revolved around three criteria (biological process (BP), cellular component (CC), and molecular function (MF)), and the cut-off threshold was fixed at p-value ≤0.05. After selecting the top-ranked 100 DM CpG sites, the Signaling Network Open Resource 2.0 (SIGNOR) was used to analyze the signaling networks of associated genes [23].
Results
Sample clustering
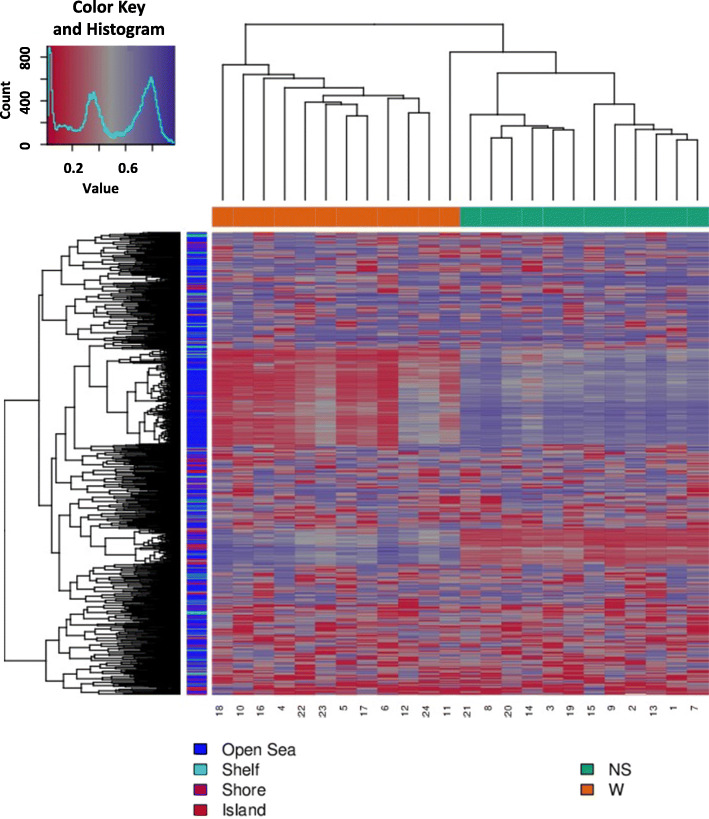
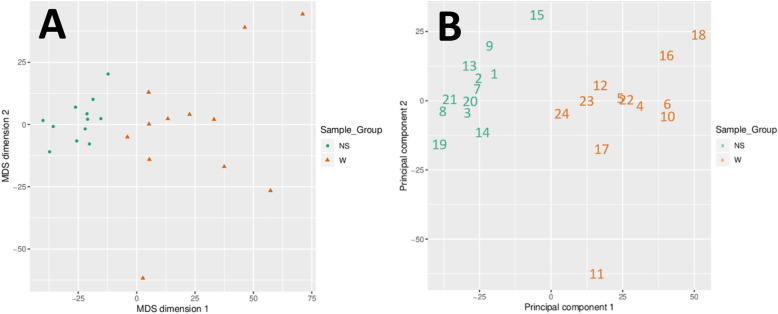
Based on the DM values of the top-ranking 1000 loci, an expected clustering pattern can be observed between the NS and W samples (Fig. 1). Using multidimensional scaling (MDS) and principal component analysis (PCA), strong signals in sample methylation values were examined (Fig. 2a and b).
Fig. 1.
Heatmap showing the hierarchal clustering of the top 1000 most variable loci across all 24 samples. Clustering used average linkage and Manhattan distance. Patient identification numbers are shown on the x-axis. W and NS stand for wart and normal skin, respectively
Fig. 2.
Scatter plots showing the coordinates of the wart (W) and normal skin (NS) samples (a) after performing Kruskal’s multi-dimensional scaling based on the matrix of the average methylation levels and Euclidean distance and (b) on the first and second principal components. A clear difference between the W and NS samples can be seen in both plots
Processing and filtering of data
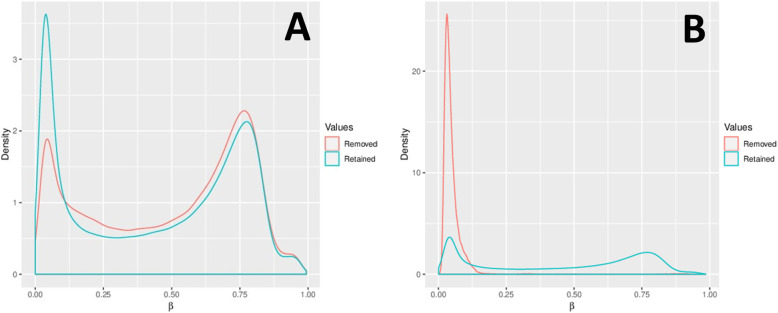
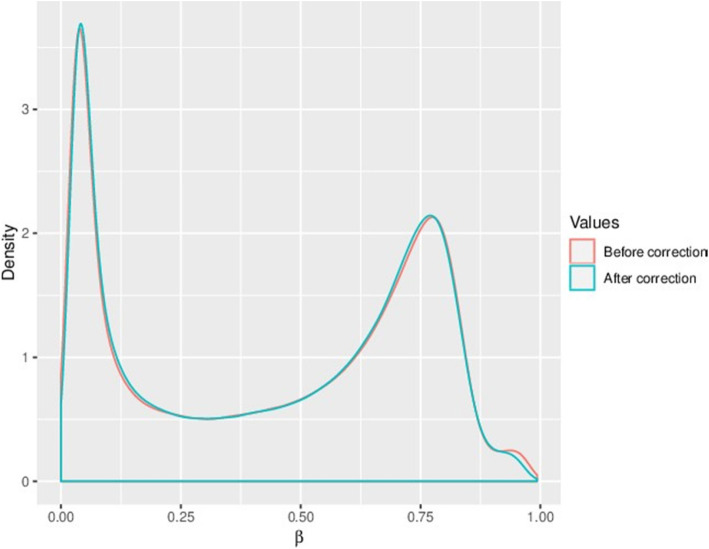
17,371 probes were removed due to their overlap with SNPs (Fig. 3a). A further 2,310 probes were filtered out using the Greedycut algorithm in RnBeads. Additional filtering eliminated 2,980 probes with specific contexts (Fig. 3b). In total, 22,661 probes were removed and 844,234 probes were retained. Both probes and samples were subject to the full RnBeads package pipeline, which entailed quality control, preprocessing, batch effects testing, and normalization (Fig. 4). The complete processed methylation data for the CpG sites can be found in Supplementary File.
Fig. 3.
Contrasting the density distributions of methylation levels (β) after (a) removal of SNP-enriched probes and filtration by Greedycut and (b) removal of context-specific probes
Fig. 4.
Density distributions of methylation levels (β) were normalized using Dasen’s method. The figure compares the β values before and after correction
Differential methylation of CpG sites
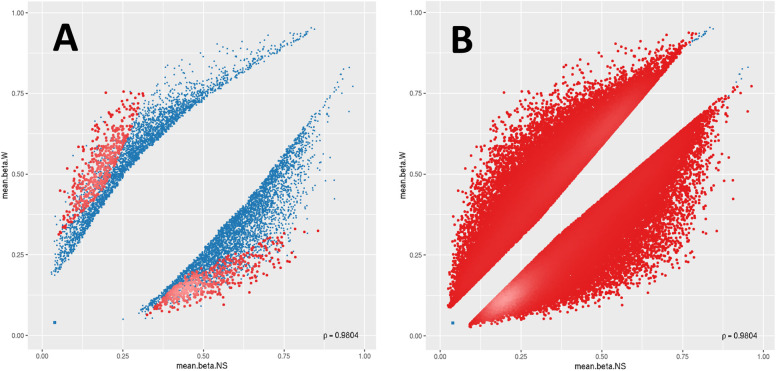
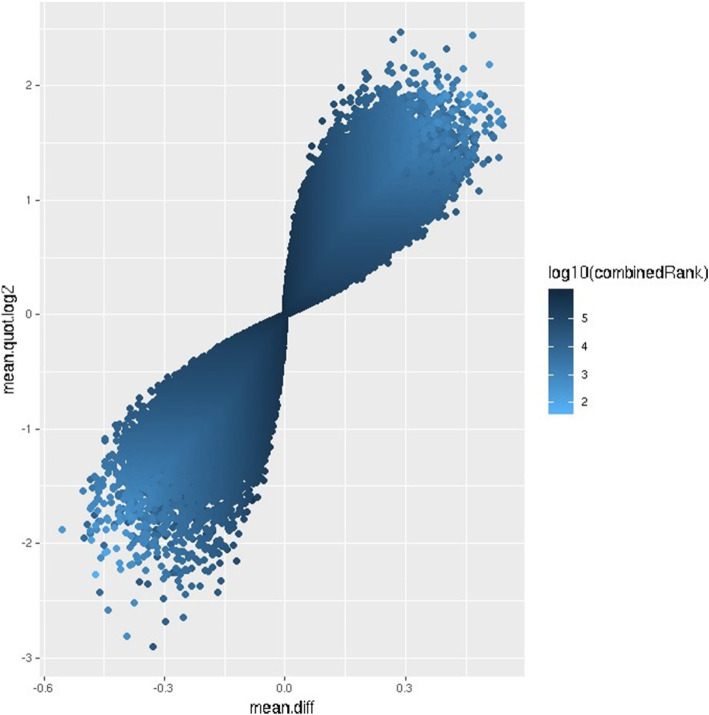
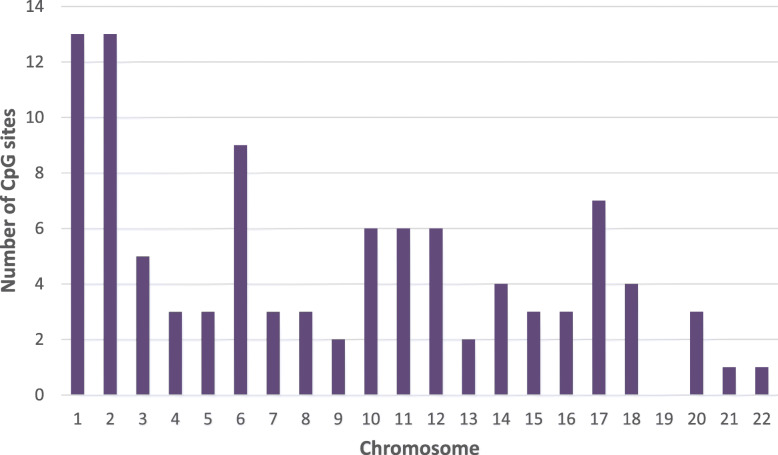
Of the top-ranking 100,000 CpG sites in terms of DM, 56,960 sites were hypomethylated and 43,040 sites were hypermethylated in W compared to NS, with a mean beta difference greater than 0.055 and less than − 0.055 (p-value < 0.032; adjusted p-value < 0.032) (Fig. 5). The beta difference for the hypomethylated and hypermethylated sites ranged from − 0.055 to 0.56 and 0.55 to 0.56, respectively. Similarly, the log2 of the quotient in methylation between W and NS ranged from − 2.47 to 2.9 (Fig. 6). The highest concentration of DM sites was seen on chromosomes 1 and 2 (Fig. 7). The top-ranking100 CpG sites, i.e. the most DM, are listed in Table 1.
Fig. 5.
Scatter plots for the (a) top-ranking 1000 and (b) top-ranking 100,000 differentially methylated CpG sites. For each plot, the mean β values of normal skin (mean.beta. NS) are on the x-axis, while the mean β values of warts (mean.beta. W) are on the y-axis. Methylation levels (β) varied between 0 (unmethylated) and 1 (fully methylated). Blue points represent variable differentially methylated sites
Fig. 6.
Volcano plot of the top-ranking 1000 differentially methylated sites. Differential methylation was measured by the log2 of the mean quotient in methylation (mean.quot.log2) and the mean fold difference (mean.diff) between warts (W) and normal skin (NS). Data points less than 0 represent relative hypomethylation, while those more than 0 represent relative hypermethylation. The intensity of each data point correlates with the combined rank score as shown on the color scale to the right
Fig. 7.
Chromosomal distribution of the top 100 differentially methylated CpG sites in warts compared to normal skin
Table 1.
The 100 CpG sites with the lowest combined rank scores
| CpG | Chromosome | Gene | Methylation region | CpG Island | Mean β value (NS) | Mean β value (W) | Mean β value diff (W-NS) | mean.quot. (log2) | P-value | False discovery rate | Combined rank score | Methylation pattern |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| cg09671951 | 10 | C10orf26 | Body | 0.1129 | 0.5848 | 0.4719 | 2.2753 | 6.82E-16 | 5.09E-11 | 48 | Hypermethylation | |
| cg27071672 | 8 | FAM83H- AS1 | Body | S_Shelf | 0.1290 | 0.5765 | 0.4475 | 2.0772 | 1.74E-14 | 1.99E-10 | 102 | Hypermethylation |
| cg07385604 | 1 | ZNF644 | TSS1500 | S_Shore | 0.1281 | 0.5720 | 0.4440 | 2.0756 | 9.33E-16 | 5.09E-11 | 110 | Hypermethylation |
| cg12432168 | 10 | LINC00702 | Body | 0.1558 | 0.6389 | 0.4832 | 1.9690 | 6.83E-15 | 1.31E-10 | 151 | Hypermethylation | |
| cg06305962 | 7 | GSAP | Body | 0.1249 | 0.5457 | 0.4208 | 2.0421 | 1.49E-14 | 1.91E-10 | 183 | Hypermethylation | |
| cg00071017 | 2 | 0.6112 | 0.1537 | −0.4575 | −1.9241 | 7.83E-16 | 5.09E-11 | 186 | Hypomethylation | |||
| cg16530881 | 17 | 0.1080 | 0.5208 | 0.4127 | 2.1688 | 1.99E-13 | 7.07E-10 | 236 | Hypermethylation | |||
| cg08246644 | 17 | STAT5A |
TSS1500;5’UTR;TS S200 |
N_Shore | 0.1009 | 0.5098 | 0.4089 | 2.2286 | 2.29E-15 | 7.97E-11 | 245 | Hypermethylation |
| cg05171197 | 2 | HDAC4 | Body | 0.1973 | 0.7523 | 0.5550 | 1.8785 | 1.65E-13 | 6.32E-10 | 247 | Hypermethylation | |
| cg16516970 | 8 | NCALD | 5’UTR | 0.1567 | 0.6028 | 0.4461 | 1.8783 | 2.74E-14 | 2.4E-10 | 248 | Hypermethylation | |
| cg03432603 | 7 | EXOC4 | Body | 0.6423 | 0.1335 | −0.5088 | −2.1842 | 2.14E-13 | 7.24E-10 | 249 | Hypomethylation | |
| cg01890417 | 1 | ZNF644 | TSS1500 | S_Shore | 0.1519 | 0.5773 | 0.4254 | 1.8592 | 2.75E-14 | 2.4E-10 | 274 | Hypermethylation |
| cg00194325 | 2 | TANC1 | Body | 0.1719 | 0.6446 | 0.4727 | 1.8473 | 5.54E-16 | 5.09E-11 | 290 | Hypermethylation | |
| cg25894955 | 9 | ABCA1 | Body | 0.5371 | 0.1351 | −0.4021 | −1.9151 | 2.81E-14 | 2.4E-10 | 295 | Hypomethylation | |
| cg10560060 | 13 | GJB2 | 5’UTR | N_Shelf | 0.6623 | 0.1799 | −0.4824 | − 1.8238 | 2.4E-13 | 7.66E-10 | 329 | Hypomethylation |
| cg10144055 | 2 | 0.1350 | 0.5324 | 0.3974 | 1.9032 | 7.42E-15 | 1.34E-10 | 336 | Hypermethylation | |||
| cg19342952 | 13 | GJB2 | 5’UTR | N_Shore | 0.6449 | 0.1770 | −0.4679 | −1.8080 | 5.82E-14 | 3.81E-10 | 347 | Hypomethylation |
| cg15612257 | 2 | N_Shore | 0.1547 | 0.5655 | 0.4108 | 1.8048 | 2.56E-15 | 7.97E-11 | 359 | Hypermethylation | ||
| cg07863022 | 17 | SEPT9; |
5’UTR;Body;TSS15 00 |
0.1681 | 0.6076 | 0.4395 | 1.7937 | 3.99E-15 | 9.85E-11 | 375 | Hypermethylation | |
| cg02745009 | 3 | ARHGAP3 1 | Body | S_Shore | 0.1718 | 0.6135 | 0.4417 | 1.7783 | 2.9E-13 | 8.24E-10 | 407 | Hypermethylation |
| cg15782771 | 5 | 0.7396 | 0.2096 | −0.5299 | −1.7709 | 3.84E-14 | 3.03E-10 | 428 | Hypomethylation | |||
| cg04272613 | 14 | DAAM1 | 5’UTR | 0.1508 | 0.5378 | 0.3869 | 1.7680 | 2.74E-15 | 7.97E-11 | 445 | Hypermethylation | |
| cg10017626 | 2 | N_Shore | 0.0988 | 0.4854 | 0.3866 | 2.1870 | 2.02E-13 | 7.07E-10 | 449 | Hypermethylation | ||
| cg18248499 | 11 | ROBO4 | TSS1500 | 0.5057 | 0.1193 | −0.3865 | −1.9961 | 3.43E-13 | 9.24E-10 | 451 | Hypomethylation | |
| cg10841463 | 14 | 0.1646 | 0.5798 | 0.4153 | 1.7566 | 7.01E-17 | 1.69E-11 | 457 | Hypermethylation | |||
| cg19497037 | 11 | 0.5188 | 0.1328 | −0.3860 | −1.8891 | 7.48E-13 | 1.37E-09 | 459 | Hypomethylation | |||
| cg13800897 | 2 | 0.5754 | 0.1613 | −0.4141 | −1.7727 | 8.99E-13 | 1.55E-09 | 490 | Hypomethylation | |||
| cg13632752 | 8 | 0.5831 | 0.1474 | −0.4357 | −1.9140 | 9.15E-13 | 1.56E-09 | 494 | Hypomethylation | |||
| cg27277339 | 15 | MYO5C | Body | 0.1561 | 0.5455 | 0.3894 | 1.7417 | 9.65E-14 | 4.88E-10 | 496 | Hypermethylation | |
| cg19158326 | 22 | GRAMD4 | Body | 0.0980 | 0.4793 | 0.3813 | 2.1796 | 3.91E-15 | 9.85E-11 | 514 | Hypermethylation | |
| cg20400915 | 17 | STAT5A |
TSS1500;5’UTR;TS S200 |
N_Shore | 0.0555 | 0.4492 | 0.3937 | 2.8086 | 1.02E-12 | 1.66E-09 | 519 | Hypermethylation |
| cg20392201 | 1 | FAM129A | Body | 0.1263 | 0.5848 | 0.4585 | 2.1258 | 1.04E-12 | 1.69E-09 | 521 | Hypermethylation | |
| cg21879102 | 12 | CIT | Body | N_Shore | 0.1946 | 0.6605 | 0.4659 | 1.7127 | 2.61E-13 | 7.91E-10 | 549 | Hypermethylation |
| cg14384093 | 9 | C9orf5 | Body | N_Shelf | 0.1256 | 0.5097 | 0.3841 | 1.9381 | 1.25E-12 | 1.9E-09 | 557 | Hypermethylation |
| cg18813270 | 2 |
HS1BP3- IT1 |
TSS1500 | 0.6868 | 0.1911 | −0.4957 | −1.7929 | 1.3E-12 | 1.95E-09 | 564 | Hypomethylation | |
| cg19449565 | 2 | HDAC4 | Body | 0.1691 | 0.6536 | 0.4845 | 1.8898 | 1.33E-12 | 1.96E-09 | 570 | Hypermethylation | |
| cg09187774 | 10 | 0.6165 | 0.1627 | −0.4538 | −1.8593 | 1.34E-12 | 1.98E-09 | 572 | Hypomethylation | |||
| cg07980148 | 4 | S_Shelf | 0.6475 | 0.1624 | −0.4852 | −1.9317 | 1.36E-12 | 1.99E-09 | 573 | Hypomethylation | ||
| cg03304533 | 11 | 0.6668 | 0.1977 | −0.4690 | −1.7040 | 3.09E-13 | 8.67E-10 | 576 | Hypomethylation | |||
| cg08569613 | 17 | STAT5A |
TSS1500;5’UTR;TS S200 |
N_Shore | 0.0692 | 0.4453 | 0.3761 | 2.5226 | 6.22E-15 | 1.25E-10 | 578 | Hypermethylation |
| cg06848849 | 1 |
ARHGEF10 L |
Body | 0.1451 | 0.5204 | 0.3753 | 1.7737 | 2.84E-14 | 2.4E-10 | 591 | Hypermethylation | |
| cg17164954 | 6 | ARID1B | Body | S_Shelf | 0.1656 | 0.5604 | 0.3948 | 1.6997 | 6.39E-13 | 1.24E-09 | 591 | Hypermethylation |
| cg13733684 | 15 | ZNF106 | TSS200;Body | 0.1724 | 0.5807 | 0.4083 | 1.6954 | 1.72E-14 | 1.99E-10 | 603 | Hypermethylation | |
| cg05669832 | 2 | PRKD3 | TSS1500 | 0.2068 | 0.6911 | 0.4843 | 1.6934 | 2.72E-13 | 8.02E-10 | 611 | Hypermethylation | |
| cg06382539 | 12 | BHLHE41 | Body | N_Shore | 0.1759 | 0.5882 | 0.4123 | 1.6864 | 1.58E-12 | 2.14E-09 | 629 | Hypermethylation |
| cg16303737 | 20 | 0.5411 | 0.1618 | −0.3793 | −1.6819 | 7.37E-13 | 1.36E-09 | 642 | Hypomethylation | |||
| cg27335585 | 5 |
LOC101929 710 |
Body | 0.7606 | 0.2298 | −0.5308 | − 1.6840 | 1.78E-12 | 2.31E-09 | 652 | Hypomethylation | |
| cg09185727 | 6 | 0.5467 | 0.1642 | −0.3825 | −1.6763 | 2.73E-13 | 8.02E-10 | 652 | Hypomethylation | |||
| cg15350314 | 3 |
LOC101928 992 |
Body | 0.1552 | 0.5574 | 0.4021 | 1.7797 | 1.85E-12 | 2.36E-09 | 658 | Hypermethylation | |
| cg11508674 | 14 | FOXN3 | Body | 0.1648 | 0.6344 | 0.4696 | 1.8820 | 2.02E-12 | 2.49E-09 | 683 | Hypermethylation | |
| cg06610988 | 18 | SETBP1 | 5’UTR | S_Shore | 0.1684 | 0.5546 | 0.3862 | 1.6622 | 3.94E-14 | 3.06E-10 | 684 | Hypermethylation |
| cg18492160 | 15 | 0.5276 | 0.1311 | −0.3965 | −1.9299 | 2.03E-12 | 2.49E-09 | 690 | Hypomethylation | |||
| cg02921273 | 20 | 0.0980 | 0.4645 | 0.3664 | 2.1349 | 3.95E-14 | 3.06E-10 | 699 | Hypermethylation | |||
| cg14167109 | 11 | MAML2 | Body | 0.1594 | 0.5381 | 0.3787 | 1.6939 | 2.13E-12 | 2.55E-09 | 703 | Hypermethylation | |
| cg06373653 | 12 | CD163L1 | Body | 0.4932 | 0.1277 | −0.3656 | −1.8699 | 2.3E-13 | 7.47E-10 | 709 | Hypomethylation | |
| cg09403144 | 18 | SETBP1 | Body | 0.1549 | 0.5202 | 0.3653 | 1.6847 | 3.68E-14 | 2.96E-10 | 714 | Hypermethylation | |
| cg06746371 | 6 | DCBLD1 | Body | 0.7344 | 0.2249 | −0.5095 | −1.6641 | 2.31E-12 | 2.68E-09 | 727 | Hypomethylation | |
| cg14002969 | 20 | PTPRA | 5’UTR | 0.4985 | 0.1342 | −0.3644 | −1.8187 | 5.04E-13 | 1.11E-09 | 727 | Hypomethylation | |
| cg07076915 | 16 | PKD1 | Body | N_Shelf | 0.2112 | 0.6851 | 0.4739 | 1.6517 | 2.48E-14 | 2.32E-10 | 728 | Hypermethylation |
| cg27341747 | 6 | 0.2010 | 0.6524 | 0.4514 | 1.6503 | 5.21E-14 | 3.61E-10 | 732 | Hypermethylation | |||
| cg20964957 | 4 | 0.5612 | 0.1185 | −0.4428 | −2.1527 | 2.39E-12 | 2.74E-09 | 736 | Hypomethylation | |||
| cg19917507 | 18 | ALPK2 | Body | 0.5863 | 0.1813 | −0.4050 | −1.6401 | 4.78E-13 | 1.08E-09 | 757 | Hypomethylation | |
| cg00925616 | 1 | Island | 0.0781 | 0.5172 | 0.4392 | 2.5818 | 2.61E-12 | 2.89E-09 | 762 | Hypermethylation | ||
| cg13515269 | 12 | BHLHE41 | 3’UTR | N_Shore | 0.2078 | 0.6886 | 0.4809 | 1.6818 | 2.71E-12 | 2.96E-09 | 772 | Hypermethylation |
| cg18638180 | 21 | C21orf70 | Body | S_Shore | 0.1734 | 0.6318 | 0.4584 | 1.8073 | 2.93E-12 | 3.13E-09 | 791 | Hypermethylation |
| cg17967134 | 17 | MPRIP | Body | 0.1283 | 0.4884 | 0.3601 | 1.8495 | 1.19E-12 | 1.83E-09 | 804 | Hypermethylation | |
| cg06373648 | 6 | SYNGAP1 | Body | 0.1564 | 0.5160 | 0.3596 | 1.6604 | 4.57E-13 | 1.06E-09 | 818 | Hypermethylation | |
| cg14825152 | 1 | 0.1422 | 0.5010 | 0.3588 | 1.7475 | 4.83E-13 | 1.09E-09 | 828 | Hypermethylation | |||
| cg08966889 | 6 | TRAM2 | Body | N_Shore | 0.1747 | 0.5588 | 0.3840 | 1.6224 | 1.16E-12 | 1.81E-09 | 828 | Hypermethylation |
| cg09443467 | 5 | TENM2 | Body | 0.5807 | 0.1623 | −0.4185 | −1.7779 | 3.44E-12 | 3.49E-09 | 833 | Hypomethylation | |
| cg17758398 | 18 | 0.6251 | 0.1850 | −0.4401 | −1.7035 | 3.48E-12 | 3.51E-09 | 836 | Hypomethylation | |||
| cg01821452 | 12 | 0.2138 | 0.6779 | 0.4641 | 1.6198 | 1.44E-12 | 2.06E-09 | 840 | Hypermethylation | |||
| cg19663114 | 3 | MED12L | Body | 0.7670 | 0.2279 | −0.5390 | −1.7073 | 3.64E-12 | 3.6E-09 | 853 | Hypomethylation | |
| cg10624729 | 1 | FAM73A | Body | 0.1847 | 0.5864 | 0.4017 | 1.6152 | 1.53E-13 | 6.05E-10 | 857 | Hypermethylation | |
| cg26586287 | 11 | 0.6087 | 0.1625 | −0.4463 | −1.8430 | 3.74E-12 | 3.67E-09 | 859 | Hypomethylation | |||
| cg23983887 | 1 | VPS13D | Body | 0.1546 | 0.5113 | 0.3567 | 1.6629 | 1.65E-12 | 2.21E-09 | 866 | Hypermethylation | |
| cg08921063 | 6 | WASF1 | 5’UTR | 0.4750 | 0.1185 | −0.3565 | −1.9164 | 2.02E-12 | 2.49E-09 | 871 | Hypomethylation | |
| cg14359656 | 17 | SPAG9 | Body | 0.5856 | 0.1477 | −0.4380 | −1.9176 | 3.98E-12 | 3.81E-09 | 883 | Hypomethylation | |
| cg26754187 | 3 | 0.5241 | 0.1368 | −0.3873 | −1.8634 | 4E-12 | 3.81E-09 | 885 | Hypomethylation | |||
| cg10126884 | 4 | 0.4827 | 0.1254 | −0.3573 | −1.8635 | 4.05E-12 | 3.85E-09 | 888 | Hypomethylation | |||
| cg13355857 | 16 | 0.6967 | 0.1872 | −0.5096 | −1.8418 | 4.06E-12 | 3.85E-09 | 889 | Hypomethylation | |||
| cg13568540 | 7 | PKD1L1 | Body | 0.6599 | 0.1847 | −0.4752 | −1.7828 | 4.22E-12 | 3.95E-09 | 901 | Hypomethylation | |
| cg08611640 | 1 | VPS13D | Body;Body | 0.1109 | 0.4654 | 0.3546 | 1.9757 | 7.7E-15 | 1.34E-10 | 912 | Hypermethylation | |
| cg25322618 | 2 | RAPGEF4 | TSS200;Body | 0.2041 | 0.6388 | 0.4347 | 1.5994 | 1.22E-13 | 5.62E-10 | 913 | Hypermethylation | |
| cg16669099 | 6 | 0.1801 | 0.5652 | 0.3851 | 1.5971 | 3.77E-12 | 3.69E-09 | 919 | Hypermethylation | |||
| cg19712663 | 6 | SLC22A23 | Body | 0.1017 | 0.4711 | 0.3694 | 2.1069 | 4.47E-12 | 4.07E-09 | 927 | Hypermethylation | |
| cg13720639 | 14 | SIPA1L1 | Body | 0.1299 | 0.4946 | 0.3646 | 1.8502 | 4.5E-12 | 4.08E-09 | 929 | Hypermethylation | |
| cg04394003 | 12 | C12orf75 | TSS1500 | N_Shore | 0.1172 | 0.4703 | 0.3531 | 1.9170 | 3.46E-12 | 3.51E-09 | 931 | Hypermethylation |
| cg17356718 | 2 | HDAC4 | Body | 0.1435 | 0.5270 | 0.3835 | 1.8066 | 4.51E-12 | 4.08E-09 | 931 | Hypermethylation | |
| cg26639076 | 2 | RIF1 | 3’UTR | 0.1710 | 0.5360 | 0.3650 | 1.5930 | 7.11E-14 | 4.2E-10 | 936 | Hypermethylation | |
| cg07969739 | 10 | BTAF1 | Body | 0.5137 | 0.1346 | −0.3791 | −1.8564 | 4.74E-12 | 4.17E-09 | 958 | Hypomethylation | |
| cg26125625 | 3 | SLC12A8 | Body | Island | 0.1074 | 0.4587 | 0.3513 | 1.9968 | 2.24E-12 | 2.64E-09 | 965 | Hypermethylation |
| cg18251218 | 1 | 0.0952 | 0.4461 | 0.3510 | 2.1169 | 1.17E-16 | 1.69E-11 | 967 | Hypermethylation | |||
| cg23909079 | 10 | GRID1 | Body | 0.6723 | 0.2146 | −0.4577 | −1.6031 | 4.92E-12 | 4.25E-09 | 977 | Hypomethylation | |
| cg24117274 | 1 | RAP1GAP | Body | N_Shelf | 0.1260 | 0.4766 | 0.3505 | 1.8387 | 7.37E-14 | 4.29E-10 | 979 | Hypermethylation |
| cg09262171 | 16 | ADCY9 | Body | 0.1896 | 0.5865 | 0.3970 | 1.5796 | 3.41E-14 | 2.78E-10 | 992 | Hypermethylation | |
| cg14600452 | 10 | 0.6088 | 0.1865 | −0.4223 | −1.6550 | 5.44E-12 | 4.53E-09 | 1014 | Hypomethylation | |||
| cg24088496 | 11 | MAML2 | Body | 0.1856 | 0.5727 | 0.3871 | 1.5747 | 1.73E-13 | 6.44E-10 | 1016 | Hypermethylation | |
| cg06968781 | 1 | GMEB1 | 5’UTR | 0.5323 | 0.1666 | −0.3657 | −1.6189 | 5.65E-12 | 4.63E-09 | 1030 | Hypomethylation | |
| cg03133881 | 1 | MAST2 | Body | 0.5066 | 0.1589 | −0.3477 | −1.6128 | 5.41E-12 | 4.52E-09 | 1035 | Hypomethylation |
Functional enrichment analysis
GO enrichment analyses of the genes associated with the top 100 DM CpG sites were performed using the DAVID webtool. Table 2 shows the most significant GO terms (p-value ≤0.05). Associated genes were mainly enriched for “SH3 domain binding”, “actin binding”, and “GTPase activator activity” on the MF level, “regulation of GTPase activity” and “positive regulation of GTPase” on the BP level, and “postsynaptic membrane” on the CC level. The most significant KEGG and Reactome pathway terms with a p-value ≤0.05 are presented. The genes were mainly enriched in the Rap1 signaling and VxPx cargo-targeting to cilium pathways (Table 3).
Table 2.
GO enrichment analyses revealed significant (p-value ≤0.05) GO terms and associated enriched genes in the biological process (BP), cellular component (CC), and molecular function (MF) categories
| Category | Term | P-value | Genes |
|---|---|---|---|
| MF | GO:0017124 ~ SH3 domain binding | 0.004 | ARHGAP31, ZNF106, SYNGAP1, CIT |
| MF | GO:0003779 ~ actin binding | 0.006 | NCALD, WASF1, DAAM1, MPRIP, MYO5C |
| MF | GO:0005096 ~ GTPase activator activity | 0.006 | ARHGAP31, RAP1GAP, SIPA1L1, SYNGAP1, ARHGEF10L |
| BP | GO:0043087 ~ regulation of GTPase activity | 0.014 | RAP1GAP, SIPA1L1, SYNGAP1 |
| BP | GO:0043547 ~ positive regulation of GTPase activity | 0.019 | ARHGAP31, RAP1GAP, PTPRA, RAPGEF4, SYNGAP1, ARHGEF10L |
| CC | GO:0045211 ~ postsynaptic membrane | 0.019 | SIPA1L1, TENM2, TANC1, GRID1 |
| BP | GO:0016337 ~ single organismal cell-cell adhesion | 0.031 | TENM2, PKD1, PKD1L1 |
| BP | GO:0050982 ~ detection of mechanical stimulus | 0.038 | PKD1, PKD1L1 |
| MF | GO:0017016 ~ Ras GTPase binding | 0.039 | RAP1GAP, RAPGEF4 |
| BP | GO:0010832 ~ negative regulation of myotube differentiation | 0.043 | HDAC4, BHLHE41 |
| BP | GO:0018105 ~ peptidyl-serine phosphorylation | 0.046 | MAST2, PKD1, PRKD3 |
Table 3.
The most significantly enriched KEGG and Reactome pathway terms of the genes associated with the top-ranking 100 DM CpG sites
| Category | Term | P-value | Genes |
|---|---|---|---|
| KEGG_PATHWAY | hsa04015:Rap1 signaling pathway | 0.001 | RAP1GAP, ADCY9, SIPA1L1, RAPGEF4, PRKD3 |
| REACTOME_PATHWAY | R-HSA-5620916:VxPx cargo-targeting to cilium | 0.045 | EXOC4, PKD1 |
Signaling network analysis
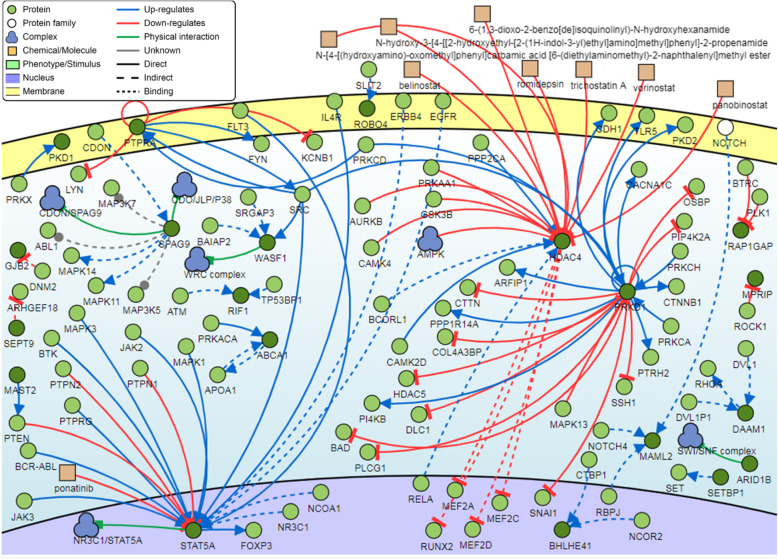
Analysis of the genes associated with the top 100 DM CpG sites showed that five genes were found to be common regulators with a minimum of 20 connectivities each. These genes are the PRKD1, HDAC4, and STAT5A genes (Fig. 8).
Fig. 8.
Pathway signalling network of the common gene regulators associated with the top-ranking 100 CpG sites. Three genes (PRKD1, HDAC4, and STAT5A) have a minimum of 20 connectivities
Discussion
In the present study, the genome-wide methylation profile of CpG sites was demonstrated for the first time in non-genital cutaneous warts. Out of the 844,234 CpG sites that were investigated, 56,960 and 43,040 CpG sites were found to be hypomethylated and hypermethylated, respectively, in warts. The combined rank scoring method revealed the top 100 most differentially methylated CpG sites, which lay within the C10orf26, FAM83H-AS1, ZNF644, LINC00702, GSAP, STAT5A, HDAC4, NCALD, and EXOC4 genes, among others.
cg09671951 was found to be the most hypermethylated CpG site in warts, and it is located within the C10orf26 gene, which is also known as the outcome predictor in acute leukemia 1 (OPAL1) gene. The C10orf26 gene has been associated with response to treatment in children with acute lymphoblastic leukemia, and it has also been implicated as a modulator of schizophrenia symptoms and disease progression [24–26]. The second most hypermethylated CpG site, cg27071672, lies within the FAM83H-AS1 gene, which codes for the FAM83H antisense RNA 1 (head to head). FAM83H-AS1 dysregulation has been associated with carcinogenesis in breast, colorectal, and lung cancer [27–29]. Two of the most hypermethylated CpG sites, cg07385604 and cg01890417, were located within the ZNF644 gene, which encodes the zinc finger protein 644. ZNF644 is associated with transcriptional repression as a part of the G9a/GLP complex, and mutations in this gene are responsible for a monogenic form of myopia [30, 31].
cg12432168, located with the LINC00702 gene, and cg06305962, located within the GSAP gene, were the fourth and fifth most hypermethylated CpG sites, respectively. The long intergenic non-protein coding RNA 702 (LINC00702), like other long non-coding RNAs, functions in genetic and epigenetic regulation, and its upregulation has been reported in endometrial cancer as well as malignant meningioma [32, 33]. However, the γ-secretase activating protein (GSAP) has mostly been reported in the context of Alzheimer’s disease pathology [34, 35]. Comparatively little is known about functions of the LINC00702 and GSAP genes outside of a disease context.
In contrast, three of the most hypermethylated CpG sites (cg08246644, cg20400915, and cg08569613) were located within the signal transducer and activator of transcription 5A (STAT5A) gene, the latter of which has been extensively studied and elucidated. STAT5A has an essential function in lactogenic and mammopoietic signaling and development in adults, and its expression is upregulated by the tumor protein p53 [36, 37]. Aberrant STAT5A expression has been reported in a number of different cancers, including breast, colon, head and neck, and prostate cancer as well as leukemia [38–42]. Of particular interest is the association of STAT5A dysregulation with head and neck squamous carcinoma, which is a type of cancer that can be caused by high-risk HPV infection [43, 44]. Although low-risk HPV types lack the carcinogenic potential of their high-risk counterparts, it is intriguing that both the benign and cancerous manifestations of HPV infection exhibit aberrant STAT5A expression.
A further three of the most hypermethylated CpG sites (cg05171197, cg19449565, and cg17356718) were found within the histone deacetylase 4 (HDAC4) gene that functions in the condensation of chromatin and repression of transcription via deacetylation [45]. The survival and growth of multiple myeloma is regulated by the HDAC4-RelB-p52 complex, and the disruption of the latter blocks the growth of these cells [46]. Moreover, HDAC4 degradation by certain chemotherapeutic agents results in the apoptosis of head-and-neck cancer cells that are resistant to TRAIL, while miR-22-driven HDAC4 repression helped to resensitize fulvestrant-resistant breast cancer cells [47, 48]. Likewise, eptoposide resistance in human A549 lung cancer cells was conferred by STAT1-HDAC4 upregulation, and HDAC4 inhibition has been reported to induce apoptosis in non-small cell lung cancer PC-9 cells [49, 50].
HDAC4 has been previously implicated in viral replication as well as the host’s antiviral response [51]. For example, HIV-1 DNA integration is facilitated by the involvement of HDAC4 in the post-integration repair process [52]. Moreover, infection with the influenza A virus has been reported to cause airway remodeling in asthmatic individuals via the indirect dysregulation of HDAC4 [53]. HDAC4 is also a critical regulator of antiviral response, and its overexpression hinders the host immune response by suppressing type 1 interferon production [54]. Furthermore, STAT-HDAC4 signaling was reported to induce epithelial-mesenchymal transition, a malignant tumor feature that is also exhibited by keratinocytes during tissue repair [55–57]. High-risk HPV infection can similarly result in malignancy by inducing this transition in epithelial and keratinocyte cells [58–60].
With regard to functional enrichment analysis of the top-ranking 100 DM CpG sites, the most significantly enriched genes in warts were associated with SH3 domain binding, namely the Rho GTPase activating protein 31 (ARHGAP31), zinc finger protein 106 (ZNF106), synaptic Ras GTPase-activating protein 1 (SYNGAP1), and citron Rho-interacting serine/threonine kinase (CIT) genes. Despite the fact that the SH3 domain plays a role in a range of different fundamental cellular processes, not much is known about the aforementioned genes in the context of skin pathology or HPV infection [61].
In contrast, pathway analysis revealed that the Rap1 signaling pathway was the most significantly enriched term, which included the RAP1 GTPase activating protein (RAP1GAP), adenylyl cyclase type 9 (ADCY9), signal-induced proliferation-associated 1 like protein 1 (SIPA1L1), Rap guanine nucleotide exchange factor (GEF) 4 (RAPGEF4), and protein kinase D3 (PRKD3) genes. RAP1GAP downregulation via promoter hypermethylation was reported to promote the cell proliferation, survival, and migration of melanoma cells [62]. Moreover, sequence analysis of the high-risk HPV 16 E6-binding protein showed that it had the highest degree of homology with the mammalian Rap1GAP protein [63]. In addition, PRKD3 has been previously reported to have an important role in promoting the growth and progression of invasive breast cancer [64].
Signaling network analysis of the top-ranking 100 CpG sites identified three common regulators: the protein kinase D1 (PRKD1), histone deacetylase 4 (HDAC4), and signal transducer and activator of transcription 5A (STAT5A) genes. The PRKD1 gene plays an integral role in anti-differentiative and proliferative keratinocyte processes, and its aberrant expression has been suggested to have a putative tumorigenic function in the skin [65, 66]. Similarly, the STAT5A gene has been reported to play a major role in the keratinocyte differentiation process [67]. In the context of HPV infection, STAT5A was found to promote HPV viral replication, and STAT-5 isoforms have been indicated to contribute to the progression of HPV-associated cervical cancer [68, 69].
Conclusions
The current study reported a number of novel CpG sites that were differentially methylated in non-genital cutaneous warts compared to normal skin. Such differences in methylation status could be responsible for the HPV-induced wart formation process. The identification of methylation status for the most differentially methylated CpG sites may prove beneficial towards the understanding of the epigenetic factors associated with non-genital cutaneous warts. One limitation of the present study is the relatively small sample size, which may result in sub-optimal statistical power for the genome-wide methylation analysis. Future research is required to validate the results on a larger scale.
Supplementary information
Additional file 1.Supplementary file. Complete processed methylation data for CpG sites.
Acknowledgements
The authors are grateful to all the participants of this study for their invaluable contribution. The authors also would like to express their gratitude to King Khalid University, Saudi Arabia, for providing administrative and technical support.
Abbreviations
- AGRF
Australian Genome Research Facility
- BP
Biological process
- CC
Cellular component
- CpG
5′-C-phosphate-G-3′
- DAVID
Database for Annotation, Visualization, and Integrated Discovery
- DM
Differentially methylated
- DNA
Deoxyribonucleic acid
- GO
Gene ontology
- HPV
Human papillomavirus
- IRB
Institutional Review Board
- JUST
Jordan University of Science and Technology
- MDS
Multi-dimensional scaling
- MF
Molecular function
- NS
Normal skin
- PCA
Principal component analysis
- SIGNOR
Signaling Network Open Resource 2.0
- W
Wart
Authors’ contributions
LNA-E designed the method study and supervised the study. LNA-E, AHT and FAA-Q helped in samples and clinical data collection. LNA-E, AHT, MAA and FAA-Q lead the implementation of the method and performed the data analysis. LNA-E, AHT and MAA helped with the interpretation and description of the results and drafted the manuscript. All authors read and approved the final manuscript.
Funding
This work was supported by the Deanship of Research at Jordan University of Science and Technology under grant number (Ref # 177/2017).
Availability of data and materials
The data generated over the course of the present study are available from the corresponding author upon request. However, the complete processed methylation data for the CpG sites is available as a Supplementary file.
Ethics approval and consent to participate
Ethical approval was obtained from the IRB committee at Jordan University of Science and Technology (Ref. # 19/105/2017). All participants gave written informed consent before taking part in this study.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary information accompanies this paper at 10.1186/s12920-020-00745-6.
References
- 1.Deaton AM, Bird A. CpG islands and the regulation of transcription. Genes Dev. 2011;25:1010–1022. doi: 10.1101/gad.2037511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jang HS, Shin WJ, Lee JE, Do JT. CpG and Non-CpG Methylation in Epigenetic Gene Regulation and Brain Function. Genes (Basel). 2017;8:1-20. [DOI] [PMC free article] [PubMed]
- 3.Illingworth RS, Bird AP. CpG islands – ‘A rough guide.’. FEBS Lett. 2009;583:1713–1720. doi: 10.1016/j.febslet.2009.04.012. [DOI] [PubMed] [Google Scholar]
- 4.Panchin AY, Makeev VJ, Medvedeva YA. Preservation of methylated CpG dinucleotides in human CpG islands. Biol Direct. 2016;11:1-15. [DOI] [PMC free article] [PubMed]
- 5.Silmon de Monerri NC, Kim K. Pathogens hijack the epigenome: a new twist on host-pathogen interactions. Am J Pathol. 2014;184:897–911. doi: 10.1016/j.ajpath.2013.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sproul D, Meehan RR. Genomic insights into cancer-associated aberrant CpG island hypermethylation. Brief Funct Genomics. 2013;12:174. doi: 10.1093/bfgp/els063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Balakrishnan L, Milavetz B. Epigenetic regulation of viral biological processes. Viruses. 2017;9:1-14. [DOI] [PMC free article] [PubMed]
- 8.Kuss-Duerkop SK, Westrich JA, Pyeon D. DNA tumor virus regulation of host DNA methylation and its implications for immune evasion and Oncogenesis. Viruses. 2018;10:1-24. [DOI] [PMC free article] [PubMed]
- 9.Westrich JA, Warren CJ, Pyeon D. Evasion of host immune defenses by human papillomavirus. Virus Res. 2017;231:21–33. doi: 10.1016/j.virusres.2016.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Graham S V. Keratinocyte Differentiation-Dependent Human Papillomavirus Gene Regulation. Viruses. 2017;9:1-18. [DOI] [PMC free article] [PubMed]
- 11.Bansal A, Singh MP, Rai B. Human papillomavirus-associated cancers: a growing global problem. Int J Appl Basic Med Res. 2016;6:84. doi: 10.4103/2229-516X.179027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Verlaat W, Van Leeuwen RW, Novianti PW, Schuuring E, Meijer CJLM, Van Der Zee AGJ, et al. Host-cell DNA methylation patterns during high-risk HPV-induced carcinogenesis reveal a heterogeneous nature of cervical pre-cancer. Epigenetics. 2018;13:769–778. doi: 10.1080/15592294.2018.1507197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dankai W, Khunamornpong S, Siriaunkgul S, Soongkhaw A, Janpanao A, Utaipat U, et al. Role of genomic DNA methylation in detection of cytologic and histologic abnormalities in high risk HPV-infected women. PLoS One. 2019;14:e0210289. doi: 10.1371/journal.pone.0210289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mirabello L, Sun C, Ghosh A, Rodriguez AC, Schiffman M, Wentzensen N, et al. Methylation of human papillomavirus type 16 genome and risk of cervical precancer in a Costa Rican population. J Natl Cancer Inst. 2012;104:556–565. doi: 10.1093/jnci/djs135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Byun S, Ki E, Park J. Single CpG site hypomethylation of MAL gene might be associated with human papillomavirus persistent infection. Gynecol Oncol. 2013;130:e49. [Google Scholar]
- 16.Degli Esposti D, Sklias A, Lima SC, Beghelli-de la Forest Divonne S, Cahais V, Fernandez-Jimenez N, et al. Unique DNA methylation signature in HPV-positive head and neck squamous cell carcinomas. Genome Med. 2017;9:33. doi: 10.1186/s13073-017-0419-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Anayannis NVJ, Schlecht NF, Belbin TJ. Epigenetic mechanisms of human papillomavirus–associated head and neck Cancer. Arch Pathol Lab Med. 2015;139:1373–1378. doi: 10.5858/arpa.2014-0554-RA. [DOI] [PubMed] [Google Scholar]
- 18.Hernandez JM, Siegel EM, Riggs B, Eschrich S, Elahi A, Qu X, et al. DNA methylation profiling across the Spectrum of HPV-associated anal squamous Neoplasia. PLoS One. 2012;7:e50533. doi: 10.1371/journal.pone.0050533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Loo SKF, Tang WYM. Warts (non-genital). BMJ Clin Evid. 2014;2014:1-28. [PMC free article] [PubMed]
- 20.Hussain F, Ormerod A. Nongenital warts: recommended management in general practice. Prescriber. 2012;23:35–41. doi: 10.1002/psb.884. [DOI] [Google Scholar]
- 21.Assenov Y, Müller F, Lutsik P, Walter J, Lengauer T, Bock C. Comprehensive analysis of DNA methylation data with RnBeads. Nat Methods. 2014;11:1138–1140. doi: 10.1038/nmeth.3115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, et al. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Perfetto L, Briganti L, Calderone A, Cerquone Perpetuini A, Iannuccelli M, Langone F, et al. SIGNOR: a database of causal relationships between biological entities. Nucleic Acids Res. 2016;44:D548–D554. doi: 10.1093/nar/gkv1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kwon E, Wang W, Tsai L-H. Validation of schizophrenia-associated genes CSMD1, C10orf26, CACNA1C and TCF4 as miR-137 targets. Mol Psychiatry. 2013;18:11–12. doi: 10.1038/mp.2011.170. [DOI] [PubMed] [Google Scholar]
- 25.Docherty AR, Bigdeli TB, Edwards AC, Bacanu S, Lee D, Neale MC, et al. Genome-wide gene pathway analysis of psychotic illness symptom dimensions based on a new schizophrenia-specific model of the OPCRIT. Schizophr Res. 2015;164:181–186. doi: 10.1016/j.schres.2015.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Holleman A, den Boer ML, Cheok MH, Kazemier KM, Pei D, Downing JR, et al. Expression of the outcome predictor in acute leukemia 1 (OPAL1) gene is not an independent prognostic factor in patients treated according to COALL or St Jude protocols. Blood. 2006;108:1984–1990. doi: 10.1182/blood-2006-04-015990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang L, Xu L, Wang Q, Wang M, An G, Wang Q, et al. Dysregulation of long non-coding RNA profiles in human colorectal cancer and its association with overall survival. Oncol Lett. 2016;12:4068–4074. doi: 10.3892/ol.2016.5138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang F, Lv S, Lv L, Liu Y, Dong S, Yao Z, et al. Identification of lncRNA<em> FAM83H-AS1</em> as a novel prognostic marker in luminal subtype breast cancer. Onco Targets Ther. 2016;9:7039–7045. doi: 10.2147/OTT.S110055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lu S, Dong W, Zhao P, Liu Z. lncRNA FAM83H-AS1 is associated with the prognosis of colorectal carcinoma and promotes cell proliferation by targeting the Notch signaling pathway. Oncol Lett. 2018;15:1861–1868. doi: 10.3892/ol.2017.7520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bian C, Chen Q, Yu X. The zinc finger proteins ZNF644 and WIZ regulate the G9a/GLP complex for gene repression. Elife. 2015;4:1-17. [DOI] [PMC free article] [PubMed]
- 31.Shi Y, Li Y, Zhang D, Zhang H, Li Y, Lu F, et al. Exome sequencing identifies ZNF644 mutations in high myopia. PLoS Genet. 2011;7:e1002084. doi: 10.1371/journal.pgen.1002084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen BJ, Byrne FL, Takenaka K, Modesitt SC, Olzomer EM, Mills JD, et al. Transcriptome landscape of long intergenic non-coding RNAs in endometrial cancer. Gynecol Oncol. 2017;147:654–662. doi: 10.1016/j.ygyno.2017.10.006. [DOI] [PubMed] [Google Scholar]
- 33.Li T, Ren J, Ma J, Wu J, Zhang R, Yuan H, et al. LINC00702/miR-4652-3p/ZEB1 axis promotes the progression of malignant meningioma through activating Wnt/β-catenin pathway. Biomed Pharmacother. 2019;113:108718. doi: 10.1016/j.biopha.2019.108718. [DOI] [PubMed] [Google Scholar]
- 34.Hussain I, Fabrègue J, Anderes L, Ousson S, Borlat F, Eligert V, et al. The role of γ-secretase activating protein (GSAP) and imatinib in the regulation of γ-secretase activity and amyloid-β generation. J Biol Chem. 2013;288:2521–2531. doi: 10.1074/jbc.M112.370924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chu J, Lauretti E, Craige CP, Praticò D. Pharmacological modulation of GSAP reduces amyloid-β levels and tau phosphorylation in a mouse model of Alzheimer’s disease with plaques and tangles. J Alzheimers Dis. 2014;41:729–737. doi: 10.3233/JAD-140105. [DOI] [PubMed] [Google Scholar]
- 36.Liu X, Robinson GW, Wagner KU, Garrett L, Wynshaw-Boris A, Hennighausen L. Stat5a is mandatory for adult mammary gland development and lactogenesis. Genes Dev. 1997;11:179–186. doi: 10.1101/gad.11.2.179. [DOI] [PubMed] [Google Scholar]
- 37.Mukhopadhyay UK, Cass J, Raptis L, Craig AW, Bourdeau V, Varma S, et al. STAT5A is regulated by DNA damage via the tumor suppressor p53. Cytokine. 2016;82:70–79. doi: 10.1016/j.cyto.2016.01.013. [DOI] [PubMed] [Google Scholar]
- 38.Kaymaz BT, Selvi N, Gokbulut AA, Aktan C, Gündüz C, Saydam G, et al. Suppression of STAT5A and STAT5B chronic myeloid leukemia cells via siRNA and antisense-oligonucleotide applications with the induction of apoptosis. Am J Blood Res. 2013;3:58–70. [PMC free article] [PubMed] [Google Scholar]
- 39.Dho SH, Kim JY, Lee K-P, Kwon E-S, Lim JC, Kim C-J, et al. STAT5A-mediated NOX5-L expression promotes the proliferation and metastasis of breast cancer cells. Exp Cell Res. 2017;351:51–58. doi: 10.1016/j.yexcr.2016.12.020. [DOI] [PubMed] [Google Scholar]
- 40.Hong X, Chen G, Wang M, Lou C, Mao Y, Li Z, & Zhang Y. STAT5a-targeting miRNA enhances chemosensitivity to cisplatin and 5-fluorouracil in human colorectal cancer cells. Mol Med Rep. 2012;5:1215-9. 10.3892/mmr.2012.801. [DOI] [PubMed]
- 41.Haddad BR, Gu L, Mirtti T, Dagvadorj A, Vogiatzi P, Hoang DT, et al. STAT5A/B gene locus undergoes amplification during human prostate Cancer progression. Am J Pathol. 2013;182:2264–2275. doi: 10.1016/j.ajpath.2013.02.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sen B, Peng S, Woods DM, Wistuba I, Bell D, El-Naggar AK, et al. STAT5A-mediated SOCS2 expression regulates Jak2 and STAT3 activity following c-Src inhibition in head and neck squamous carcinoma. Clin Cancer Res. 2012;18:127–139. doi: 10.1158/1078-0432.CCR-11-1889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Spence T, Bruce J, Yip KW, Liu F-F. HPV Associated Head and Neck Cancer. Cancers (Basel). 2016;8:1-12. [DOI] [PMC free article] [PubMed]
- 44.Husain N, Neyaz A. Human papillomavirus associated head and neck squamous cell carcinoma: controversies and new concepts. J Oral Biol Craniofac Res. 2017;7:198–205. doi: 10.1016/j.jobcr.2017.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang Z, Qin G, Zhao TC. HDAC4: mechanism of regulation and biological functions. Epigenomics. 2014;6:139–150. doi: 10.2217/epi.13.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vallabhapurapu SD, Noothi SK, Pullum DA, Lawrie CH, Pallapati R, Potluri V, et al. Transcriptional repression by the HDAC4–RelB–p52 complex regulates multiple myeloma survival and growth. Nat Commun. 2015;6:8428. doi: 10.1038/ncomms9428. [DOI] [PubMed] [Google Scholar]
- 47.Lee B-S, Kim YS, Kim H-J, Kim D-H, Won H-R, Kim Y-S, et al. HDAC4 degradation by combined TRAIL and valproic acid treatment induces apoptotic cell death of TRAIL-resistant head and neck cancer cells. Sci Rep. 2018;8:12520. doi: 10.1038/s41598-018-31039-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang B, Li D, Filkowski J, Rodriguez-Juarez R, Storozynsky Q, Malach M, et al. A dual role of miR-22 modulated by RelA/p65 in resensitizing fulvestrant-resistant breast cancer cells to fulvestrant by targeting FOXP1 and HDAC4 and constitutive acetylation of p53 at Lys382. Oncogenesis. 2018;7:54. doi: 10.1038/s41389-018-0063-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Suganuma M, Oya Y, Umsumarng S, Iida K, Rawangkhan A, Sakai R, et al. Abstract 4723: innovative cancer treatment of human lung cancer cells PC-9 with a synthetic retinoid Am80 and EGCG via inhibition of HDAC4 and HDAC5. Cancer Res. 2016;76(14 Supplement):4723. [Google Scholar]
- 50.Kaewpiboon C, Srisuttee R, Malilas W, Moon J, Oh S, Jeong HG, et al. Upregulation of Stat1-HDAC4 confers resistance to etoposide through enhanced multidrug resistance 1 expression in human A549 lung cancer cells. Mol Med Rep. 2015;11:2315–2321. doi: 10.3892/mmr.2014.2949. [DOI] [PubMed] [Google Scholar]
- 51.Herbein G, Wendling D. Histone deacetylases in viral infections. Clin Epigenetics. 2010;1:13–24. doi: 10.1007/s13148-010-0003-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Smith JA, Yeung J, Kao GD, Daniel R. A role for the histone deacetylase HDAC4 in the life-cycle of HIV-1-based vectors. Virol J. 2010;7:237. doi: 10.1186/1743-422X-7-237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Moheimani F, Koops J, Williams T, Reid AT, Hansbro PM, Wark PA, et al. Influenza a virus infection dysregulates the expression of microRNA-22 and its targets; CD147 and HDAC4, in epithelium of asthmatics. Respir Res. 2018;19:145. doi: 10.1186/s12931-018-0851-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yang Q, Tang J, Pei R, Gao X, Guo J, Xu C, et al. Host HDAC4 regulates the antiviral response by inhibiting the phosphorylation of IRF3. J Mol Cell Biol. 2019;11:158–169. doi: 10.1093/jmcb/mjy035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kaowinn S, Kaewpiboon C, Koh S, Krämer OH, Chung Y. STAT1-HDAC4 signaling induces epithelial-mesenchymal transition and sphere formation of cancer cells overexpressing the oncogene, CUG2. Oncol Rep. 2018;40:2619–2627. doi: 10.3892/or.2018.6701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Stone RC, Pastar I, Ojeh N, Chen V, Liu S, Garzon KI, et al. Epithelial-mesenchymal transition in tissue repair and fibrosis. Cell Tissue Res. 2016;365:495–506. doi: 10.1007/s00441-016-2464-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zeng L-S, Yang X-Z, Wen Y-F, Mail S-J, Wang M-H, Zhang M-Y, et al. Overexpressed HDAC4 is associated with poor survival and promotes tumor progression in esophageal carcinoma. Aging (Albany NY) 2016;8:1236–1249. doi: 10.18632/aging.100980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hellner K, Mar J, Fang F, Quackenbush J, Münger K. HPV16 E7 oncogene expression in normal human epithelial cells causes molecular changes indicative of an epithelial to mesenchymal transition. Virology. 2009;391:57–63. doi: 10.1016/j.virol.2009.05.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Banerjee NS, Moore DW, Broker TR, Chow LT. Vorinostat, a pan-HDAC inhibitor, abrogates productive HPV-18 DNA amplification. Proc Natl Acad Sci U S A. 2018;115:E11138–E11147. doi: 10.1073/pnas.1801156115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Azzimonti B, Dell’Oste V, Borgogna C, Mondini M, Gugliesi F, De Andrea M, et al. The epithelial–mesenchymal transition induced by keratinocyte growth conditions is overcome by E6 and E7 from HPV16, but not HPV8 and HPV38: characterization of global transcription profiles. Virology. 2009;388:260–269. doi: 10.1016/j.virol.2009.03.033. [DOI] [PubMed] [Google Scholar]
- 61.Carducci M, Perfetto L, Briganti L, Paoluzi S, Costa S, Zerweck J, et al. The protein interaction network mediated by human SH3 domains. Biotechnol Adv. 2012;30:4–15. doi: 10.1016/j.biotechadv.2011.06.012. [DOI] [PubMed] [Google Scholar]
- 62.Zheng H, Gao L, Feng Y, Yuan L, Zhao H, Cornelius LA. Down-regulation of Rap1GAP via promoter hypermethylation promotes melanoma cell proliferation, survival, and migration. Cancer Res. 2009;69:449–457. doi: 10.1158/0008-5472.CAN-08-2399. [DOI] [PubMed] [Google Scholar]
- 63.Singh L, Gao Q, Kumar A, Gotoh T, Wazer DE, Band H, et al. The high-risk human papillomavirus type 16 E6 counters the GAP function of E6TP1 toward small rap G proteins. J Virol. 2003;77:1614–20. [DOI] [PMC free article] [PubMed]
- 64.Liu Y, Li J, Zhang J, Yu Z, Yu S, Wu L, et al. Oncogenic protein kinase D3 regulating networks in invasive breast cancer. Int J Biol Sci. 2017;13:748–758. doi: 10.7150/ijbs.18472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ristich VL, Bowman PH, Dodd ME, Bollag WB. Protein kinase D distribution in normal human epidermis, basal cell carcinoma and psoriasis. Br J Dermatol. 2006;154:586–593. doi: 10.1111/j.1365-2133.2005.07073.x. [DOI] [PubMed] [Google Scholar]
- 66.Ivanova P, Atanasova G, Poumay Y, Mitev V. Knockdown of PKD1 in normal human epidermal keratinocytes increases mRNA expression of keratin 10 and involucrin: early markers of keratinocyte differentiation. Arch Dermatol Res. 2008;300:139–145. doi: 10.1007/s00403-008-0832-7. [DOI] [PubMed] [Google Scholar]
- 67.Dai X, Sayama K, Shirakata Y, Hanakawa Y, Yamasaki K, Tokumaru S, et al. STAT5a/PPARγ pathway regulates involucrin expression in keratinocyte differentiation. J Invest Dermatol. 2007;127:1728–1735. doi: 10.1038/sj.jid.5700758. [DOI] [PubMed] [Google Scholar]
- 68.Sobti RC, Singh N, Hussain S, Suri V, Bharadwaj M, Das BC. Deregulation of STAT-5 isoforms in the development of HPV-mediated cervical carcinogenesis. J Recept Signal Transduct. 2010;30:178–188. doi: 10.3109/10799891003786218. [DOI] [PubMed] [Google Scholar]
- 69.Hong S, Laimins LA. The JAK-STAT transcriptional regulator, STAT-5, activates the ATM DNA damage pathway to induce HPV 31 genome amplification upon epithelial differentiation. PLoS Pathog. 2013;9:1-11. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1.Supplementary file. Complete processed methylation data for CpG sites.
Data Availability Statement
The data generated over the course of the present study are available from the corresponding author upon request. However, the complete processed methylation data for the CpG sites is available as a Supplementary file.