Strigolactone and karrikin signaling pathways trigger polyubiquitination and degradation of SMXL2 to regulate hypocotyl elongation and gene expression in Arabidopsis.
Abstract
Strigolactones (SLs) and karrikins (KARs) are related butenolide signaling molecules that control plant development. In Arabidopsis (Arabidopsis thaliana), they are recognized separately by two closely related receptors but use the same F-box protein MORE AXILLARY GROWTH2 (MAX2) for signal transduction, targeting different members of the SMAX1-LIKE (SMXL) family of transcriptional repressors for degradation. Both signals inhibit hypocotyl elongation in seedlings, raising the question of whether signaling is convergent or parallel. Here, we show that synthetic SL analog GR244DO enhanced the interaction between the SL receptor DWARF14 (D14) and SMXL2, while the KAR surrogate GR24ent-5DS induced association of the KAR receptor KARRIKIN INSENSITIVE2 (KAI2) with SMAX1 and SMXL2. Both signals trigger polyubiquitination and degradation of SMXL2, with GR244DO dependent on D14 and GR24ent-5DS dependent mainly on KAI2. SMXL2 is critical for hypocotyl responses to GR244DO and functions redundantly with SMAX1 in hypocotyl response to GR24ent-5DS. Furthermore, GR244DO induced response of D14-LIKE2 and KAR-UP F-BOX1 through SMXL2, whereas GR24ent-5DS induced expression of these genes via both SMAX1 and SMXL2. These findings demonstrate that both SLs and KARs could trigger polyubiquitination and degradation of SMXL2, thus uncovering an unexpected but important convergent pathway in SL- and KAR-regulated gene expression and hypocotyl elongation.
INTRODUCTION
Strigolactones (SLs) are a large, diverse group of signaling compounds derived from the carotenoid biosynthetic pathway and possess a characteristic butenolide ring that is essential for activity. They function in many aspects of plant development, such as shoot branching, internode elongation, leaf elongation and senescence, growth of primary and lateral roots, anthocyanin accumulation, shoot gravitropism, and stem secondary thickening (Foo et al., 2001; Stirnberg et al., 2002; Sorefan et al., 2003; Snowden et al., 2005; Arite et al., 2007; Simons et al., 2007; Gomez-Roldan et al., 2008; Umehara et al., 2008; Drummond et al., 2009; Lin et al., 2009; Agusti et al., 2011; Sang et al., 2014). Mutations that block SL biosynthesis and signaling are characterized by the outgrowth of axillary buds to form lateral shoots in dicotyledonous plants or tillers in grasses (Al-Babili and Bouwmeester, 2015; Wang et al., 2017a; Waters et al., 2017).

SLs also play important roles in adaptive growth under diverse environmental conditions (Saeed et al., 2017; Mostofa et al., 2018). Phosphate or water deficiency significantly induces SL biosynthesis, and the accumulated SLs further mediate developmental changes to reduce consumption and enhance uptake of phosphate and water (Kohlen et al., 2011; Haider et al., 2018). Besides the function as an endogenous plant hormone, SLs secreted into the rhizosphere promote symbiotic relationships with arbuscular mycorrhizal fungi to facilitate the absorption of water and nutrients (Akiyama et al., 2005). In addition, such SL rhizosphere signals are exploited by parasitic weeds to stimulate seed germination, leading to colonization in the roots of host plants, causing devastating losses to crop production (Conn et al., 2015; Toh et al., 2015; Tsuchiya et al., 2015). Because of the important roles of SLs in key agronomic traits related to crop yield and mechanized farming, it is important to understand the molecular mechanism of SL action in plants.
Karrikins (KARs) are present in burned plant materials and stimulate germination of dormant seeds following wildfires, facilitating seedlings establishment under favorable conditions (Nelson et al., 2012). They are derived from the pyrolysis of carbohydrates, and like SLs, contain a butenolide ring that is essential for activity. KARs can also promote seed germination of many plant species that are not considered fire followers, including weedy ephemerals such as Arabidopsis (Arabidopsis thaliana) and commercial species such as tomato (Solanum lycopersicum), maize (Zea mays), rice (Oryza sativa), and lettuce (Lactuca sativa; Nelson et al., 2012; Morffy et al., 2016). Besides germination, KARs influence seedling photomorphogenesis, which is thought to provide an advantage to seedlings in the post-fire environment (Nelson et al., 2009, 2010). Recently, it has been shown that exogenously applied KARs promote Arabidopsis seed germination under permissive conditions but inhibit it under conditions of mild abiotic stresses, suggesting a further means by which seedling establishment is safeguarded (Wang et al., 2018a).
Studies of KAR-insensitive mutants (see below) reveal pleiotropic developmental abnormalities that include impaired seedling photomorphogenesis (Nelson et al., 2012; Waters et al., 2012), modified leaf shape (Waters et al., 2012), impaired leaf cuticle development (Li et al., 2017), abnormal root skewing and density of root hairs in Arabidopsis (Swarbreck et al., 2019; Villaécija-Aguilar et al., 2019), and inability to establish symbiosis with arbuscular mycorrhizal fungi in rice (Gutjahr et al., 2015). The defective leaf cuticle results in reduced drought tolerance, indicating a possible role of KAR signaling in response to water deficit (Li et al., 2017).
The first SLs identified are known as canonical SLs and have a tricyclic lactone, comprising A, B, and C rings, linked through an enol-ether bridge to a butenolide (D ring). More recently noncanonical SLs have been identified that lack B and C rings (Yoneyama et al., 2018). It is generally not known which SLs are responsible for each biological activity, but canonical SLs are routinely used in research, either as natural forms such as 5-deoxystrigol (5DS) and 4-deoxyorobanchol (4DO) or as isomers of the synthetic analog GR24 (Flematti et al., 2016; Yoneyama et al., 2018).
KARs apparently mimic an endogenous plant signaling compound, the identity of which is still unknown, but structure–function assays show that a butenolide moiety is essential for KAR activity (Nelson et al., 2012; Morffy et al., 2016). Early reports that GR24 induced some KAR-like responses in seeds and seedlings suggested that KARs could simply be SL analogs, but KARs do not stimulate germination of parasitic weeds or affect shoot branching (Nelson et al., 2009, 2012). Subsequent analysis recognized that commonly used preparations of GR24 comprise a racemic mixture of two enantiomers with the stereochemical configurations of 5DS and its enantiomer, ent-5DS, conveniently referred to as GR245DS and GR24ent-5DS. Surprisingly, while GR245DS was found to behave as an SL, GR24ent-5DS was found to behave as a KAR (Scaffidi et al., 2014). A second pair of enantiomers of GR24 is also produced during chemical synthesis, and these have the stereochemical configuration of 4DO and ent-4DO. These two stereoisomers are referred to as GR244DO and GR24ent-4DO and were similarly found to act as SLs and KARs, respectively (Scaffidi et al., 2014). Furthermore, while 5DS and 4DO are natural SLs, their synthetic enantiomers ent-5DS and ent-4DO behave as KARs. The defining feature is that analogs active as SLs have a 2′R-configured butenolide D ring, while all analogs with KAR activity have a 2′S configuration (Scaffidi et al., 2014; Flematti et al., 2016).
Genetic studies in Arabidopsis, rice, petunia (Petunia hybrida), and pea (Pisum sativum) led to the discoveries of SL and KAR receptors (Waters et al., 2017). The SL and KAR receptor proteins are paralogous α/β-fold hydrolases known, respectively, as DWARF14 (D14) in rice and Arabidopsis and as KARRIKIN INSENSITIVE2 (KAI2) or HYPOSENSITIVE TO LIGHT (HTL) in Arabidopsis. Signaling by SLs and KARs both requires the F-box protein MORE AXILLARY GROWTH2 (MAX2) in Arabidopsis or D3 in rice. In SL signaling, SLs interact with the active-site pocket of D14 and this can lead to nucleophilic attack by the active-site Ser residue, resulting in elimination of the tricyclic A, B, and C moiety and covalent attachment of a four-carbon derivative of the D ring to the active site Ser and His residues of D14 (Hamiaux et al., 2012; Zhao et al., 2013a; Yao et al., 2016). This modification has been proposed to induce association of D14 with D3/MAX2 and with repressor proteins from the rice D53 or Arabidopsis SUPPRESSOR OF MAX2-LIKE (SMXL) family, which are subsequently ubiquitinated and degraded (de Saint Germain et al., 2016; Yao et al., 2016, 2018b). In an alternative model, SL binding to D14 triggers its association with MAX2 and SMXL proteins, resulting in ubiquitination and proteolysis of the SMXL proteins, after which D14 destroys the SL by hydrolysis (Seto et al., 2019). The plasticity in conformational states of D3 also plays an important role in the hydrolytic activity of D14, formation of the D14-D3-D53 complex, and thus SL signaling (Shabek et al., 2018).
In Arabidopsis SMXL6, SMXL7, and SMXL8 are orthologs of D53 in rice and promote axillary bud outgrowth and tiller growth, respectively. The ubiquitination and degradation of SMXL6, SMXL7, SMXL8, or D53 through the 26S proteasome relieve transcriptional repression of target genes controlling axillary bud growth (Jiang et al., 2013; Zhou et al., 2013; Soundappan et al., 2015; Wang et al., 2015). In KAR signaling, KAI2 (HTL) is expected to perceive exogenous KARs or the unknown endogenous KAI2 Ligand (KL) by means of a similar mechanism to SLs (Waters et al., 2012; Guo et al., 2013; Xu et al., 2016). However, whether KARs promote formation of the KAI2-MAX2-SMXL complex and further trigger ubiquitination and degradation of SMXL proteins is still an open question. This has greatly limited the understanding of KAR signaling in plants.
In Arabidopsis, there are eight genes in the SMXL family that display distinct expression patterns and functions. In particular, SMXL6, SMXL7, and SMXL8 function as key transcriptional repressors that regulate shoot branching and leaf development (Soundappan et al., 2015; Wang et al., 2015); SUPPRESSOR OF MAX2 1 (SMAX1) regulates seed germination and hypocotyl elongation through KAR signaling (Stanga et al., 2013; Soundappan et al., 2015); SMXL3, SMXL4, and SMXL5 act as cell-autonomous regulators of phloem formation independent of SL or KAR signaling (Wallner et al., 2017). SMXL2, a close paralog of SMAX1 in Arabidopsis, mediates KAR signaling and controls hypocotyl growth redundantly with SMAX1. However, SMXL2 does not apparently influence seed germination or leaf development (Stanga et al., 2016). Furthermore, the smxl2 mutant displays elongated root hairs that are not observed in smax1 mutants (Villaécija-Aguilar et al., 2019). These results indicate a clear functional difference between SMXL2 and SMAX1. Since exogenous SLs can also influence seedling photomorphogenesis, it raises the question of whether SMXL2 might also be involved in SL signaling.
Hypocotyl elongation of young seedlings is a key skotomorphogenic response that enables seedlings to push through the soil to receive light. Upon exposure to light, photomorphogenic responses are enacted, including repression of hypocotyl elongation. Hypocotyl elongation is controlled by numerous endogenous and environmental signals (Shi et al., 2016; Wang et al., 2018b). The red-to-far-red light ratio, blue light, UV-B, temperature, photoperiod, circadian clock, and gibberellins all regulate hypocotyl elongation mainly through signaling pathways integrated by the PHYTOCHROME INTERACTING FACTOR (PIF) family transcription factors (Li et al., 2016a; Zhu and Lin, 2016). Interestingly, KARs and all stereoisomers of GR24 inhibit hypocotyl elongation of Arabidopsis seedlings under continuous red light, and these responses are abolished in the max2 mutant (Nelson et al., 2011; Scaffidi et al., 2014). There is crosstalk between KAR signaling and ELONGATED HYPOCOTYL5 (HY5)–dependent photomorphogenesis (Waters and Smith, 2013). Significantly, in low intensity white light the natural SLs 5DS and 4DO inhibited hypocotyl elongation in a D14-dependent manner, while the nonnatural enantiomer ent-5DS inhibited hypocotyl elongation through KAI2 (Scaffidi et al., 2014), indicating that SLs and KARs regulate hypocotyl elongation separately through both signaling pathways. Genetic analysis further showed that SMAX1 and SMXL2 function redundantly in KAR-induced photomorphogenesis, while SMXL6, SMXL7, and SMXL8 do not regulate hypocotyl elongation (Soundappan et al., 2015; Stanga et al., 2016). Thus, the molecular mechanism underlying SL-regulated photomorphogenesis needs further investigation.
It is generally considered that SL and KAR signaling are mediated by distinct members of the SMXL family (Stanga et al., 2016). In this study, we show that SLs and KARs specifically promote formation of the D14-MAX2-SMXL2 and KAI2-MAX2-SMXL2 complexes and subsequently trigger polyubiquitination and degradation of SMXL2 in a D14- or KAI2-dependent manner, respectively. These findings demonstrate a common mechanism for D14- and KAI2-mediated signaling in Arabidopsis seedlings and are discussed in terms of the functions and possible evolutionary relationships of these signaling pathways.
RESULTS
Expression of SMXL2 Is Induced by Both SL and KAR Signals
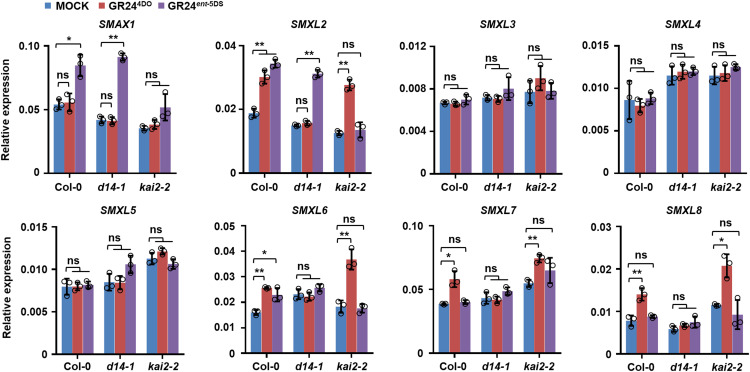
The transcript levels of SMXL6, SMXL7, and SMXL8 in Arabidopsis and their homolog D53 in rice are upregulated by a racemic mixture of GR24 (rac-GR24) consisting of GR245DS and GR24ent-5DS that behave preferentially as SLs and KARs, respectively (Jiang et al., 2013; Stanga et al., 2013; Zhou et al., 2013; Scaffidi et al., 2014; Lantzouni et al., 2017). Therefore, expression analyses based on rac-GR24 are unable to discriminate the transcript responses to SLs and KARs, except by analyses in the d14 and kai2 mutants and by comparison with responses to KAR (Scaffidi et al., 2013; Waters et al., 2017). To investigate the specific gene expression responding to SLs, we used GR244DO as an active SL analog because our preliminary studies showed it to be more active than GR245DS. For KAR signaling, we used either KAR1 or GR24ent-5DS, which has been shown to interact with KAI2 and to regulate gene expression, seed germination, and seedling development preferentially via KAI2 (Nelson, et al., 2009, 2010, 2011; Guo et al., 2013; Scaffidi et al., 2014; Waters et al., 2015; Flematti et al., 2016).
Phylogenetic analyses indicate that SMXL family members are probably derived from duplication and functional divergence of ancient SMAX1 genes during evolution (Moturu et al., 2018). To investigate functions of SMXL family proteins in SL and KAR signaling, we first analyzed the mRNA level of SMXL genes in response to GR244DO treatment in seedlings of the wild type, d14-1, and kai2-2 (Figure 1; Supplemental Figure 1A). After GR244DO treatment for 4 h, transcript levels of SMXL6, SMXL7, and SMXL8 were upregulated in the wild type. Interestingly, SMXL2 expression was also upregulated upon GR244DO treatment, whereas expression levels of SMAX1, SMXL3, SMXL4, and SMXL5 remained unaffected. These inductions were blocked in d14-1 but were largely unaffected in kai2-2, indicating that GR244DO could induce the expression of SMXL2, SMXL6, SMXL7, and SMXL8 through D14-mediated SL signaling (Figure 1).
Figure 1.
Expression of SMXL Family Genes upon GR244DO or GR24ent-5DS Treatment.
Expression of SMXL gene family members in 10-d-old seedlings of the wild type (Col-0), d14-1, and kai2-2 treated with 5 μM GR244DO or GR24ent-5DS for 4 h. Expression values are determined relative to ACTIN2. Values are means ± sd. *, P < 0.05; **, P < 0.01 (n = 3, two-tailed Student’s t test, three independent experiments; Supplemental Data Set). ns, no significance.
KAR1 and KAR2 have been used to detect transcriptional changes in Arabidopsis, and only a few genes including KAR-UP F-BOX1 (KUF1), D14-LIKE2 (DLK2), and SMAX1 were upregulated in KAR1-treated seeds at 24 h imbibion or seedlings grown on media containing KAR1 or KAR2 for 4 d (Nelson et al., 2009, 2010, 2011; Waters et al., 2012). However, it was later reported that treatment of seedlings with 1 µM KAR2 for 4 d induced a slight increase in SMXL2 expression (Stanga et al., 2013). Consistent with these studies, we did not observe a significant response of SMXL family members upon KAR1 treatment for 4 h (Supplemental Figure 1B), which could be explained by the hypothesis that KAR1 and KAR2 are not the active ligands in vivo and require activation (Waters et al., 2015; Yao et al., 2018a). To further investigate the response of SMXL genes to KAR signaling, we used GR24ent-5DS (Supplemental Figure 1A) as a KAR surrogate (Waters et al., 2015; Flematti et al., 2016) and observed that GR24ent-5DS could clearly induce the expression of SMAX1 and SMXL2 after 4 h of treatment in a KAI2-dependent manner, but it had little effect on the expression of SMXL3, SMXL4, SMXL5, SMXL7, and SMXL8 (Figure 1). GR24ent-5DS also weakly induced SMXL6 expression in the wild type, but this induction was not observed in kai2-2 or d14-1, suggesting that GR24ent-5DS might regulate SMXL6 expression through both KAI2 and SL signaling.
SL Treatment Stimulates Interaction of SMXL2 with D14, but KAR Triggers Its Association with KAI2
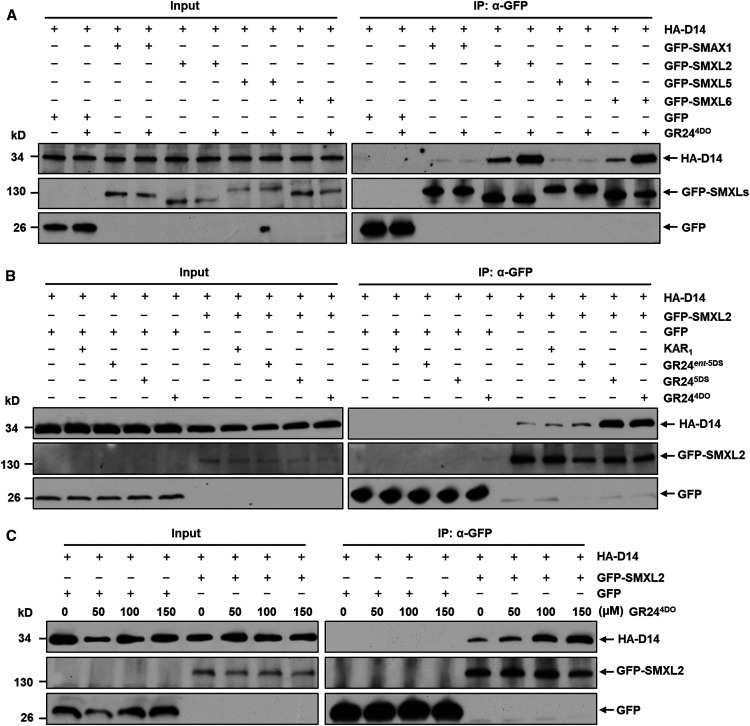
To study the role of SMXL2 in SL signaling, we investigated the physical association of the SL receptor D14 with SMXL2, SMAX1, SMXL5, and SMXL6. In a coimmunoprecipitation (Co-IP) assay using Arabidopsis protoplasts, hemagglutinin (HA)-tagged D14 associated with green fluorescent protein (GFP)–tagged SMXL2 and GFP-SMXL6 proteins, and these interactions were significantly enhanced by GR244DO. However, neither SMAX1 nor SMXL5 could interact with D14, and GR244DO treatment had no effect (Figure 2A). This result is consistent with stimulation of interaction between D14 and SMXL6 by rac-GR24 (Wang et al., 2015) and indicates that GR244DO enhances the interaction between D14 and SMXL2 in vivo. We further examined interaction between HA-D14 and GFP-SMXL2 in the absence or presence of KAR1, GR24ent-5DS, GR245DS, or GR244DO and found that the D14-SMXL2 interaction was significantly enhanced by GR244DO and GR245DS, but it showed little change upon KAR1 or GR24ent-5DS treatment (Figure 2B). Furthermore, the interaction between HA-D14 and GFP-SMXL2 was enhanced by GR244DO with dose dependence (Figure 2C). These results collectively demonstrate that SLs could specifically enhance the interaction between D14 and SMXL2 in vivo.
Figure 2.
GR244DO Induces Interaction between D14 and SMXL2 in Vivo.
(A) Association of HA-D14 with GFP-SMAX1, GFP-SMXL2, GFP-SMXL5, and GFP-SMXL6 revealed by Co-IP assay (IP) in wild-type (Col-0) protoplasts in the absence or presence of 100 µM GR244DO.
(B) Interaction between HA-D14 and GFP-SMXL2 revealed by Co-IP assay (IP) in wild-type (Col-0) protoplasts in the absence or presence of 100 µM KAR1, GR24ent-5DS, GR245DS, or GR244DO.
(C) Interaction between HA-D14 and GFP-SMXL2 revealed by Co-IP assay (IP) in wild-type (Col-0) protoplasts in the presence of 0, 50, 100, and 150 µM GR244DO.
The HA-D14 recombinant protein was detected with anti-HA monoclonal antibody; the GFP-SMAX1, GFP-SMXL2, GFP-SMXL5, and GFP-SMXL6 fusion proteins and GFP were detected with anti-GFP monoclonal antibody. All experiments were repeated at least three times with similar results.
Interaction between D14 and MAX2 is triggered by SLs in numerous plant species (Hamiaux et al., 2012; Jiang et al., 2013; Zhou et al., 2013; Wang et al., 2015; Liang et al., 2016). However, biochemical evidence for interaction between KAI2 and MAX2 induced by KARs has been difficult to obtain. In yeast two-hybrid assays, KAR1 promoted a weak interaction between KAI2 (AtHTL) and MAX2 (Toh et al., 2014). Recently however, strong autoactivation in the yeast two-hybrid system by KAI2 was reported, but no interaction between KAI2 and MAX2 could be observed (Yao et al., 2018a). To detect the interaction between KAI2 and MAX2 in vivo, we coexpressed HA-KAI2 and GFP-MAX2 in Arabidopsis protoplasts and conducted Co-IP assays. In the absence of GR24ent-5DS, HA-KAI2 was found to interact with GFP-MAX2 with low efficiency, while addition of GR24ent-5DS greatly enhanced the association, demonstrating that KAI2 forms a complex with MAX2 in Arabidopsis (Supplemental Figure 2).
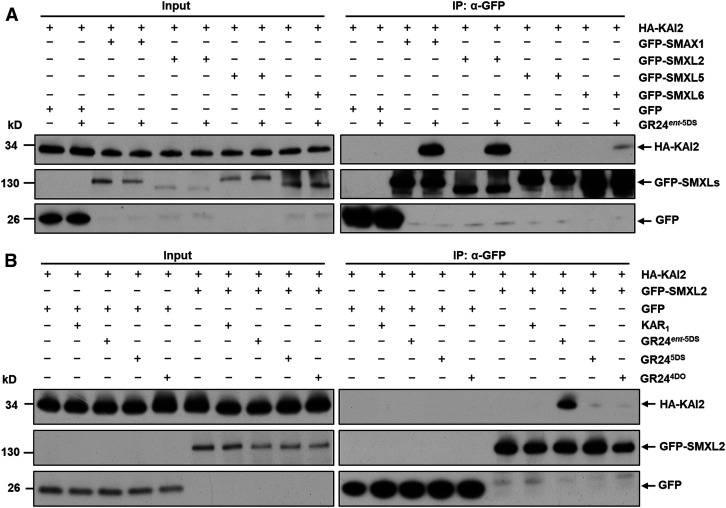
Although genetic evidence showed that SMAX1 and SMXL2 function downstream of KAI2 and MAX2 in Arabidopsis (Stanga et al., 2013, 2016; Soundappan et al., 2015), there has been no evidence for physical interaction of SMXL proteins with KAI2. We thus conducted Co-IP assays to test the association of HA-KAI2 with GFP-SMAX1, GFP-SMXL2, GFP-SMXL5, and GFP-SMXL6 (Figure 3A). In the absence of chemical treatment, association between KAI2 and SMXLs was hardly detected, but the KAI2-SMAX1 and KAI2-SMXL2 interactions were strongly induced by GR24ent-5DS treatment. SMXL6 could weakly interact with KAI2 under GR24ent-5DS treatment, but SMXL5 showed no interaction, suggesting that SMXL6 is able to form a complex with KAI2 with low efficiency in the protoplast system used. We further compared the effects of KAR1, GR24ent-5DS, GR245DS, and GR244DO on KAI2-SMXL2 interaction and found that GR245DS and GR244DO could barely stimulate protein association, indicating that GR24ent-5DS preferentially and effectively triggers the interaction between SMXL2 and KAI2 in vivo (Figure 3B). KAR1 did not trigger protein association in this experiment, which might be due to insufficient time or capability for the activation of KAR1 in this system. Together, these data clearly show that interaction of SMXL2 with D14 and KAI2 is promoted by ligands that selectively activate SL and KAR signaling, respectively.
Figure 3.
GR24ent-5DS Induces Interaction between KAI2 and SMXL2 in Vivo.
(A) Association of HA-KAI2 with GFP-SMAX1, GFP-SMXL2, GFP-SMXL5, and GFP-SMXL6 revealed by Co-IP assay (IP) in wild-type (Col-0) protoplasts in the absence or presence of 100 µM GR24ent-5DS.
(B) Interaction between HA-KAI2 and GFP-SMXL2 revealed by Co-IP assay (IP) in wild-type (Col-0) protoplasts in the absence or presence of 100 µM KAR1, GR24ent-5DS, GR245DS, or GR244DO.
The HA-KAI2 recombinant protein was detected with anti-HA monoclonal antibody; the GFP-SMAX1, GFP-SMXL2, GFP-SMXL5, and GFP-SMXL6 fusion proteins and GFP were detected with anti-GFP monoclonal antibody. All experiments were repeated at least three times with similar results.
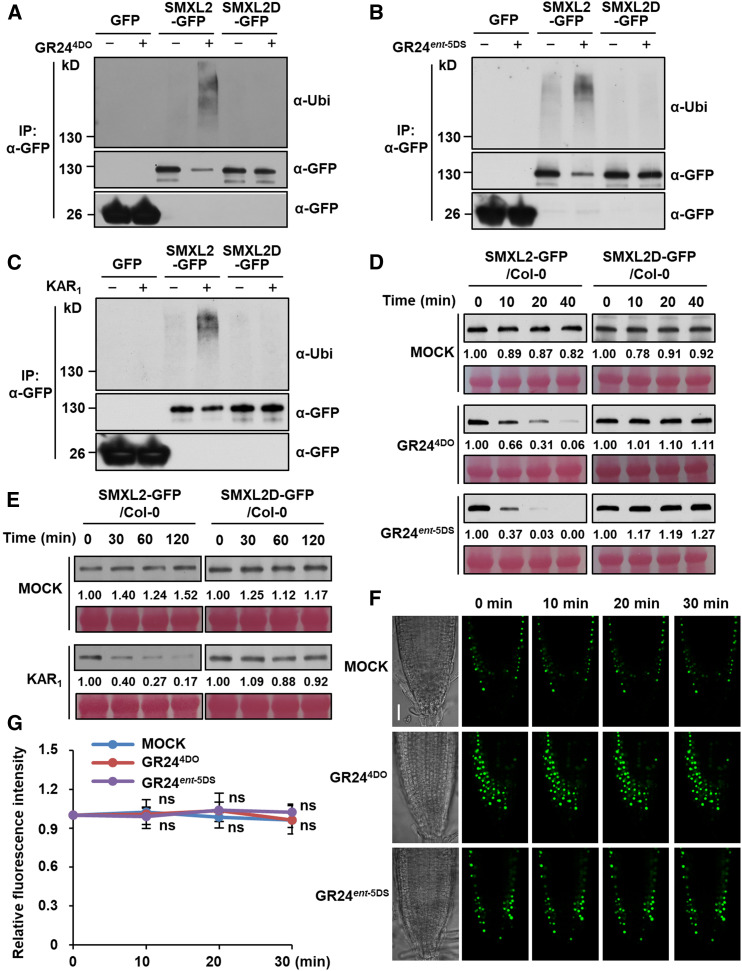
SMXL2 Undergoes Ubiquitination upon SL or KAR Treatment
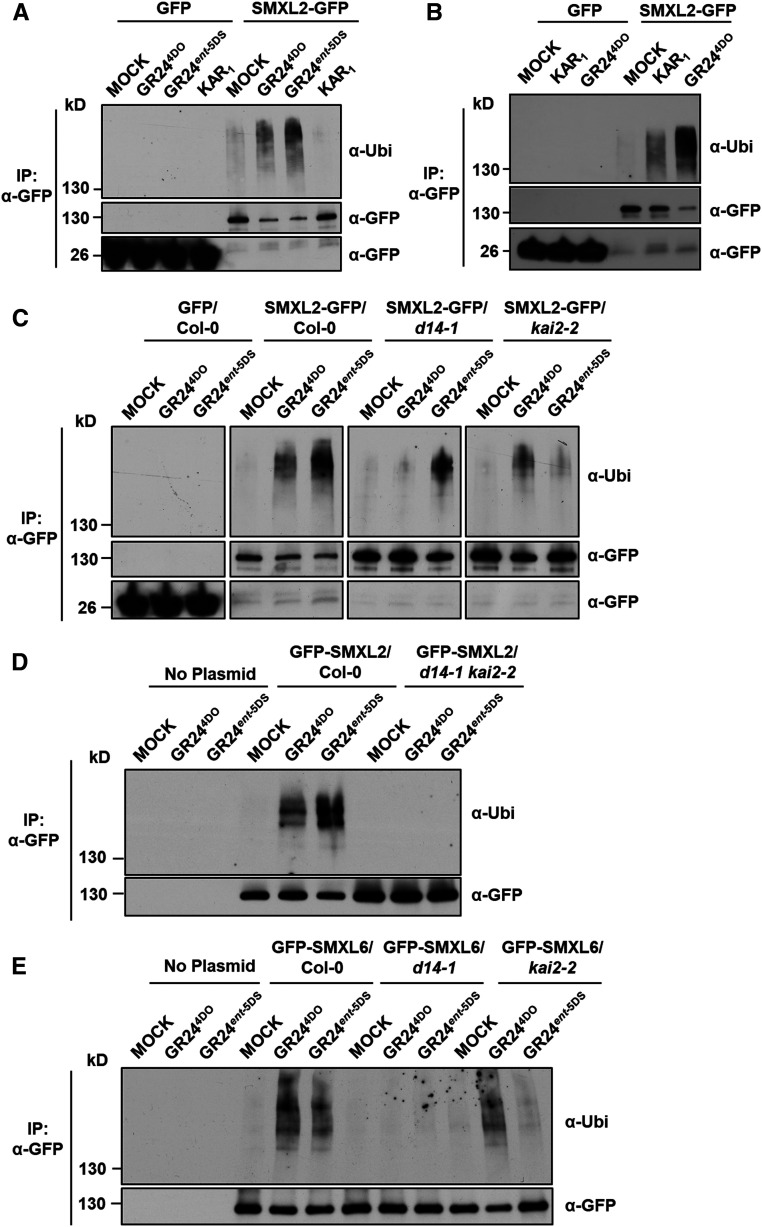
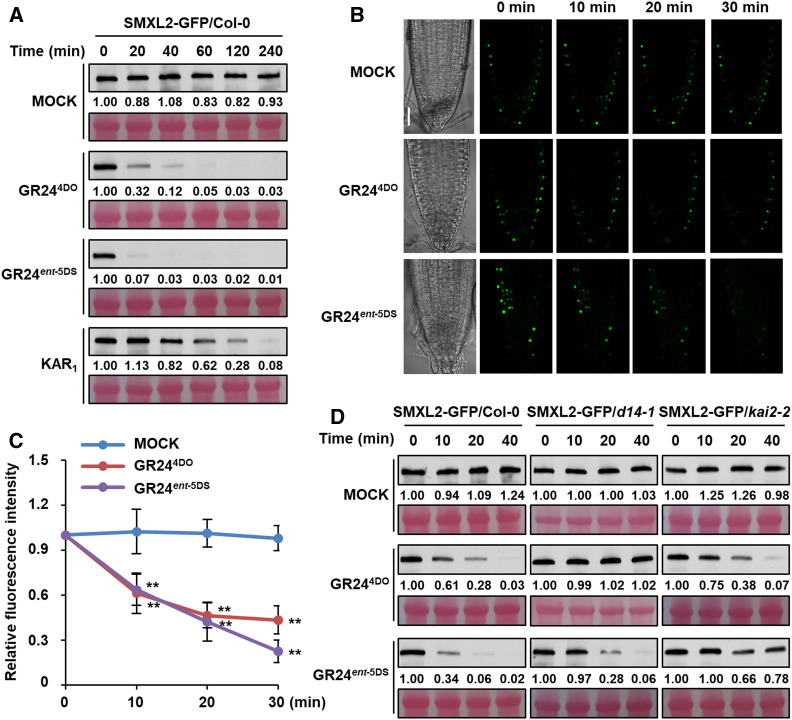
Polyubiquitination and degradation of rice D53 and Arabidopsis SMXL6, SMXL7, and SMXL8 proteins in response to SL treatment are core events in SL signaling (Jiang et al., 2013; Zhou et al., 2013; Soundappan et al., 2015; Wang et al., 2015; Liang et al., 2016). However, the biochemical mechanism underlying KAR signaling remains to be determined. We therefore investigated the polyubiquitination of SMXL2 upon activation of SL and KAR signaling in Arabidopsis leaf protoplasts. Clear polyubiquitination of GFP-SMXL2 was observed upon treatment with rac-GR24 for 1.5 h, but not by treatment with KAR1 (Supplemental Figure 3), which is consistent with its inability to induce interaction with SMXL2 in the protoplast system. We then investigated the polyubiquitination of SMXL2 using a more responsive system. In transgenic plants overexpressing SMXL2-GFP under the control of the cauliflower mosaic virus 35S promoter, GR244DO or GR24ent-5DS could trigger the ubiquitination of SMXL2-GFP recombinant protein within 20 min, although KAR1 showed no effect (Figure 4A). However, when the KAR1 treatment was prolonged to 2 h, a weak signal for the ubiquitination of SMXL2-GFP could be detected, indicating a slow SMXL2 polyubiquitination induced by KAR1 (Figure 4B).
Figure 4.
SMXL2 Undergoes Ubiquitination after GR244DO, KAR1, or GR24ent-5DS Treatment.
(A) Ubiquitination of SMXL2-GFP proteins in 7-d-old 35S:SMXL2-GFP transgenic plants after GR244DO, GR24ent-5DS, or KAR1 treatment. Seedlings were treated with 50 µM MG132 for 1 h and then with 2 µM GR244DO, GR24ent-5DS, or KAR1 for 20 min in 0.5× MS liquid medium.
(B) Ubiquitination of SMXL2-GFP proteins in 7-d-old 35S:SMXL2-GFP transgenic plants with 50 µM MG132 for 1 h and then with 2 µM KAR1 for 2 h or 2 µM GR244DO for 20 min in 0.5× MS liquid medium. Mock treatments were 2 h.
(C) Ubiquitination of SMXL2-GFP in wild-type, d14-1, and kai2-2 Arabidopsis seedlings containing 35S:SMXL2-GFP transgene. Seedlings were treated after 7 d of growth with 50 µM MG132 for 1 h and then with 2 µM GR244DO or GR24ent-5DS for 20 min in 0.5× MS liquid medium.
(D) Ubiquitination of GFP-SMXL2 in protoplasts made from wild-type and d14-1 kai2-2 seedlings.
(E) Ubiquitination of GFP-SMXL6 in protoplasts made from wild-type, d14-1, and kai2-2 seedlings.
In (D) and (E), protoplasts were transformed with 35S:GFP-SMXL2 or 35S:GFP-SMXL6 plasmid, incubated overnight for protein synthesis, and then pretreated with 50 µM MG132 for 1 h and treated with 50 µM GR244DO and GR24ent-5DS for 1.5 h.
Proteins were detected by immunoblotting with anti-ubiquitin (Ubi) polyclonal antibody or anti-GFP monoclonal antibody. All experiments were repeated at least three times with similar results. IP, coimmunoprecipitation assay.
We next evaluated the contribution of SL or KAR receptor to the ubiquitination of SMXL2 induced by GR244DO and GR24ent-5DS. Seedlings of transgenic plants expressing 35S:SMXL2-GFP in the wild-type, d14-1, or kai2-2 backgrounds were treated for 20 min with GR244DO. The ubiquitination of SMXL2-GFP was observed in the wild type and kai2-2 backgrounds but was very weak in the d14-1 background (Figure 4C). By contrast, the polyubiquitination of SMXL2-GFP triggered by GR24ent-5DS was dramatically impaired in the kai2-2 background compared with that of the wild type but only slightly weakened in the d14-1 background (Figures 4C), indicating that the SMXL2 ubiquitination induced by GR24ent-5DS depends largely on KAI2. We then examined the ubiquitination level of GFP-SMXL2 in protoplasts from the wild type and the d14-1 kai2-2 double mutant (Figure 4D). The SMXL2 ubiquitination induced by GR244DO and GR24ent-5DS was not detected in d14-1 kai2-2, indicating that ubiquitination of SMXL2 in the d14-1 and kai2-2 single mutants was probably due to incomplete specificity of GR24 stereoisomers for D14 and KAI2.
We further examined the effects of GR244DO and GR24ent-5DS on SMXL6 ubiquitination in protoplasts and found that SMXL6 underwent ubiquitination after GR244DO or GR24ent-5DS treatment, but the GR24ent-5DS–induced ubiquitination of SMXL6 was weaker than that of SMXL2 (Figure 4E). More importantly, the GR244DO-induced SMXL6 ubiquitination was not detected in d14-1 but largely unaffected in kai2-2. The GR24ent-5DS–induced SMXL6 ubiquitination was obviously impaired in both d14-1 and kai2-2 single mutants (Figure 4E), suggesting that GR244DO induced SMXL6 ubiquitination through D14, but the GR24ent-5DS–triggered SMXL6 ubiquitination could function through both KAI2 and D14. This phenomenon is consistent with previous reports that ubiquitination and degradation of SMXL6-GFP upon rac-GR24 treatment are completely blocked in d14-1 (Wang et al., 2015). Taken together, these data demonstrate that SL and KAR signaling triggers the ubiquitination of SMXL2 in a manner that predominantly depends on D14 and KAI2, respectively.
SMXL2 Is Degraded upon SL or KAR Treatment
Since polyubiquitination can regulate the stability and function of target proteins (Mukhopadhyay and Riezman, 2007; Wang et al., 2017b), we examined the stability of SMXL2 in response to SL and KAR treatments of seedlings harboring the stably transformed SMXL2-GFP transgene. We observed that GR244DO and GR24ent-5DS triggered significant degradation of the SMXL2-GFP recombinant protein within 20 min, whereas KAR1 induced a slower rate of degradation, which was apparent after 60 min but took 120 to 240 min to achieve comparable degradation (Figure 5A). We further observed the GFP fluorescence in root tips of 35S:SMXL2-GFP transgenic plants using Z-stack screening of confocal images and found that it was distributed throughout in root tip (Supplemental Figure 4). To visualize the endogenous degradation of SMXL2-GFP in response to GR244DO and GR24ent-5DS treatments, we quantified the GFP fluorescence signal in single plane of fixed focus and examined its changes during the time course of chemical treatment. These results established that the GFP fluorescence was significantly reduced after GR244DO and GR24ent-5DS treatment for 10, 20, and 30 min but remained stable under mock treatment (Figures 5B and 5C). These results collectively demonstrate that either SL or KAR signaling is able to induce ubiquitination and degradation of SMXL2 in Arabidopsis.
Figure 5.
SMXL2 Undergoes Degradation after GR244DO, GR24ent-5DS, or KAR1 Treatment.
(A) Degradation of SMXL2-GFP proteins in 7-d-old 35S:SMXL2-GFP transgenic plants after 2 µM GR244DO, GR24ent-5DS, or KAR1 treatment for the time indicated.
(B) GFP fluorescence in single plane of fixed focus in root tips of 7-d-old 35S:SMXL2-GFP transgenic plants treated with 2 µM GR244DO or GR24ent-5DS in 0.5× MS liquid medium for the time indicated. Bar = 30 μm.
(C) Relative abundance of SMXL2-GFP in root tips was determined by fluorescence measurement at the time indicated after 2 µM GR244DO or GR24ent-5DS treatment, with the zero-time signal set as 1.00. Values are means ± sd. Fluorescence signal after GR244DO or GR24ent-5DS treatment was compared with that of mock treatment at indicated time. **, P < 0.01 (n = 12 nuclei, two-tailed Student’s t test; Supplemental Data Set).
(D) Degradation of SMXL2-GFP proteins in wild-type (Col-0), d14-1, and kai2-2 Arabidopsis seedlings containing 35S:SMXL2-GFP transgene. Seedlings were treated after 7 d of growth with 2 µM GR244DO or GR24ent-5DS in 0.5× MS liquid medium for the time indicated.
In (A) and (D), proteins were detected by immunoblotting with anti-GFP monoclonal antibody. Relative abundances of SMXL2-GFP were determined by densitometry and normalized to loadings determined by Ponceau staining (red), with the zero-time signal set as 1.00. All experiments were repeated at least three times with similar results.
To evaluate the contributions of SL and KAR receptors to GR244DO- and GR24ent-5DS–induced SMXL2 degradation, seedlings of transgenic plants expressing 35S:SMXL2-GFP in the wild-type, d14-1, or kai2-2 backgrounds were treated with GR244DO and GR24ent-5DS. The degradation of SMXL2-GFP induced by GR244DO was blocked in d14-1 but was weakly attenuated in kai2-2 (Figure 5D), indicating that SMXL2 undergoes a D14-dependent ubiquitination and degradation in response to GR244DO. By contrast, the GR24ent-5DS–triggered SMXL2-GFP degradation was clearly impaired in kai2-2 compared with that of the wild type but only slightly delayed in d14-1 (Figure 5D), indicating that the degradation of SMXL2 induced by GR24ent-5DS depends largely on KAI2.
The RGKT Motif of SMXL2 Is Essential for Its Ubiquitination and Degradation Induced by SL and KAR Signaling
The ubiquitination and degradation of SMXL6 and SMXL7 in Arabidopsis and D53 in rice depend on the conserved RGKT motif within the C-terminal portion of these proteins (Jiang et al., 2013; Zhou et al., 2013; Wang et al., 2015; Liang et al., 2016). Comparison of amino acid sequences of the Arabidopsis SMXL family showed that the RGKT motif is conserved in SMXL proteins in SL and KAR signaling pathways including SMAX1, SMXL2, SMXL6, SMXL7, and SMXL8 but is absent from SMXL3, SMXL4, and SMXL5, which are not implicated in SL or KAR signaling (Supplemental Figure 5). This analysis prompted us to investigate roles of the RGKT motif in ubiquitination and degradation of SMXL2 induced by SLs and KARs to determine whether it is important in both signaling pathways.
We generated 35S:SMXL2D-GFP transgenic plants that overexpress a mutated SMXL2 protein bearing a deletion specifically of the RGKT motif (Supplemental Figure 6). We found that the ubiquitination of SMXL2-GFP was detected after GR244DO treatment for 20 min, but the ubiquitination of SMXL2D-GFP was abolished (Figure 6A). Similarly, deletion of the RGKT motif prevented the ubiquitination of SMXL2 induced by GR24ent-5DS treatment for 20 min or KAR1 for 60 min (Figures 6B and 6C). Furthermore, GR244DO, GR24ent-5DS, and KAR1 could trigger the degradation of SMXL2-GFP but showed no effect on the amount of SMXL2D-GFP protein (Figures 6D and 6E). The GFP fluorescence signals in root tips of 35S:SMXL2D-GFP transgenic plants were stable throughout the 30-min treatment with GR244DO and GR24ent-5DS (Figures 6F and 6G; Supplemental Figure 4). Therefore, the ubiquitination and degradation of SMXL2 in response to SL and KAR treatments require the RGKT motif.
Figure 6.
Ubiquitination and Degradation of SMXL2-GFP Depends on the RGKT Motif.
(A) to (C) Ubiquitination of SMXL2-GFP and SMXL2D-GFP in the wild-type seedlings containing 35S:SMXL2-GFP or 35S:SMXL2D-GFP transgene. Seedlings were treated after 7 d of growth with 50 µM MG132 for 1 h and then with 2 µM GR244DO for 20 min (A), 2 µM GR24ent-5DS for 20 min (B), or 10 µM KAR1 for 1 h (C) in 0.5× MS liquid medium. IP, Co-IP assay.
(D) and (E) Levels of SMXL2-GFP and SMXL2D-GFP proteins in wild-type seedlings containing 35S:SMXL2-GFP or 35S:SMXL2D-GFP transgene. Seedlings were treated after 7 d of growth with 2 µM GR244DO, GR24ent-5DS (D), or 10 µM KAR1 (E) in 0.5× MS liquid medium for the time indicated.
(F) GFP fluorescence in single plane of fixed focus in root tip of 7-d-old 35S:SMXL2D-GFP transgenic plants treated with 2 µM GR244DO or GR24ent-5DS in 0.5× MS liquid medium for the time indicated. Bar = 30 μm.
(G) Relative abundance of SMXL2D-GFP in root tips was determined by fluorescence measurements at the time indicated after 2 µM GR244DO or GR24ent-5DS treatment, with the zero-time signal set as 1.00. Values are means ± sd. Fluorescence signal after GR244DO or GR24ent-5DS treatment was compared with that of mock treatment at indicated time. (n = 12 nuclei, two-tailed Student’s t test; Supplemental Data Set). ns, no significance.
In (A) to (C), proteins were detected by immunoblotting with anti-ubiquitin (Ubi) polyclonal antibody or anti-GFP monoclonal antibody. In (D) and (E), proteins were detected by immunoblotting with anti-GFP monoclonal antibody. Relative abundances of SMXL2-GFP were determined by densitometry and normalized to loadings determined by Ponceau staining (red), with the zero-time signal set as 1.00. All experiments were repeated at least three times with similar results.
SMXL2 Is Important for GR244DO- and GR24ent-5DS–Regulated Hypocotyl Elongation
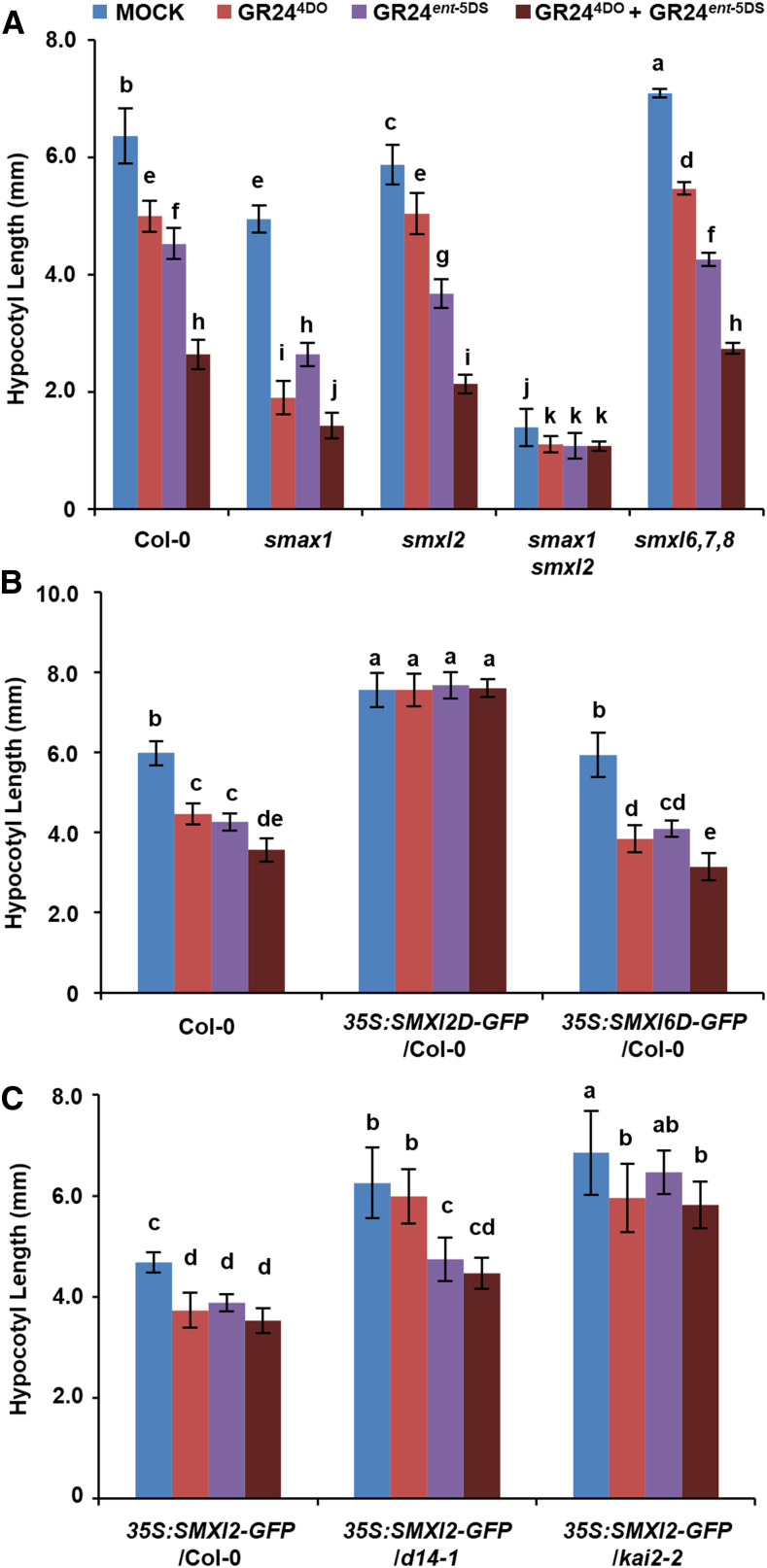
To explore the function of SMXL2 in seedling development, we examined the hypocotyl length of Arabidopsis ecotype Columbia-0 (Col-0), smax1, smxl2, smax1 smxl2 (double mutant), and smxl6 smxl7 smxl8 (triple mutant, hereafter referred to as smxl6,7,8) after treatment with GR244DO, GR24ent-5DS, and GR244DO plus GR24ent-5DS. Hypocotyls of the smax1 and smxl2 single mutants were somewhat shorter than those of the wild type, whereas the hypocotyl length of the smax1 smxl2 double mutant was only 22% of the wild type (Figure 7A). These results are largely consistent with previously reported observations (Stanga et al., 2016). More importantly, GR244DO and GR24ent-5DS repressed hypocotyl elongation and showed an additive effect in Col-0, and the repression by GR244DO and GR24ent-5DS in smax1 was stronger than that in Col-0, indicating that SMXL2 plays an important role in hypocotyl responses to both GR244DO and GR24ent-5DS. In the smxl2 mutant, although the effect of GR24ent-5DS was similar to that in Col-0, the effect of GR244DO was rather weak. This weak effect might be due to the possible leakiness of the smxl2 mutant or the possibility that other proteins also participate in these hypocotyl responses, which is supported by the further suppression of hypocotyl length in the smax1 smxl2 double mutant in response to GR244DO and GR24ent-5DS treatments (Figure 7A). The hypocotyl response of the smxl6,7,8 triple mutant to chemical treatment was similar to that of the wild type, indicating that SMXL6, SMXL7, and SMXL8 are not involved in the regulation of hypocotyl elongation by SLs and KARs (Figure 7A). These results indicate that SMAX1 and SMXL2 display functional redundancy in the hypocotyl response to GR24ent-5DS and that SMXL2 plays important roles in hypocotyl responses to both GR244DO and GR24ent-5DS.
Figure 7.
Hypocotyl Response to GR244DO, GR24ent-5DS, or GR244DO Plus GR24ent-5DS.
(A) Seedlings of Col-0 (wild type), smax1, smxl2, smax1 smxl2, and smxl6,7,8 triple mutant were grown on 0.5× MS media containing 100 nM of each compound for 4 d under continuous red light at 21°C. Values are means ± sd. Different letters indicate significant difference at P < 0.05 (n = 20 different plants, two-way ANOVA multiple comparison, Tukey test, three independent experiments; Supplemental Data Set).
(B) Seedlings of Col-0 (wild type), 35S:SMXL2D-GFP/Col-0, and 35S:SMXL6D-GFP/Col-0 were grown on 0.5× MS media containing 100 nM of each compound for 4 d under continuous red light at 21°C. Values are means ± sd. Different letters indicate significant difference at P < 0.05 (n = 15 different plants, two-way ANOVA multiple comparison, Tukey test, three independent experiments; Supplemental Data Set).
(C) Seedlings of 35S:SMXL2-GFP/Col-0, 35S:SMXL2-GFP/d14-1, and 35S:SMXL2-GFP/kai2-2 were grown on 0.5× MS media containing 100 nM of each compound for 4 d under continuous red light at 21°C. Values are means ± sd. Different letters indicate significant difference at P < 0.05 (n = 15 different plants, two-way ANOVA multiple comparison, Tukey test, three independent experiments; Supplemental Data Set).
We next compared the influence of SMXL2D and SMXL6D in the hypocotyl response to GR244DO and GR24ent-5DS. Seedlings of 35S:SMXL2D-GFP/Col-0 transgenic plants grew longer hypocotyls than the wild type and were insensitive to either GR244DO or GR24ent-5DS, but the 35S:SMXL6D-GFP/Col-0 transgenic plants displayed similar phenotypes to the wild type (Figure 7B), further supporting the importance of SMXL2 in hypocotyl responses to GR244DO and GR24ent-5DS. In addition, the abundance of SMXL2-GFP in the hypocotyl of 35S:SMXL2-GFP seedlings grown in the presence of GR244DO plus GR24ent-5DS was only ∼6% of that in 35S:SMXL2-GFP seedlings without chemical treatment (Supplemental Figure 7), indicating that GR244DO and GR24ent-5DS induced degradation of SMXL2-GFP in the hypocotyl. Importantly, seedlings of 35S:SMXL2-GFP/Col-0 transgenic plants were sensitive to GR244DO and GR24ent-5DS treatments, but the 35S:SMXL2-GFP/d14-1 transgenic plants were insensitive to GR244DO and sensitive to GR24ent-5DS treatment. By contrast, the 35S:SMXL2-GFP/kai2-2 transgenic plants were insensitive to GR24ent-5DS and sensitive to GR244DO treatment (Figure 7C). These results demonstrated that SMXL2 mediates both SL- and KAR-regulated hypocotyl elongation in Arabidopsis.
In Arabidopsis, SMXL6, SMXL7, and SMXL8 promote shoot branching mainly through repressing BRANCHED1 (BRC1) expression and enhancing polar auxin transport (Soundappan et al., 2015; Wang et al., 2015). They are degraded in response to D14-dependent SL signaling, leading to repression of shoot branching. Since SMXL2 is also degraded in response to SL signaling, we investigated whether SMXL2 regulates shoot branching. We found that numbers of primary rosette branches of smax1, smxl2, and smax1 smxl2 were similar to Col-0, whereas d14-1 had many more branches (Supplemental Figure 8A). We also found that although the 35S:SMXL2D-GFP and 35S:SMXL6D-GFP transgenic plants contained comparable levels of the corresponding SMXL2D-GFP and SMXL6D-GFP proteins in their rosette lateral buds, the primary branch number of 35S:SMXL6D-GFP transgenic plants was significantly increased while that of 35S:SMXL2D-GFP transgenic plants was indistinguishable from that of the wild type (Supplemental Figures 8B and 8C). These data suggest that SMXL2 does not apparently regulate shoot branching even when ectopically expressed in lateral shoot buds, at least in Col-0 background. This further emphasizes the importance of SMXL2 in seedling development.
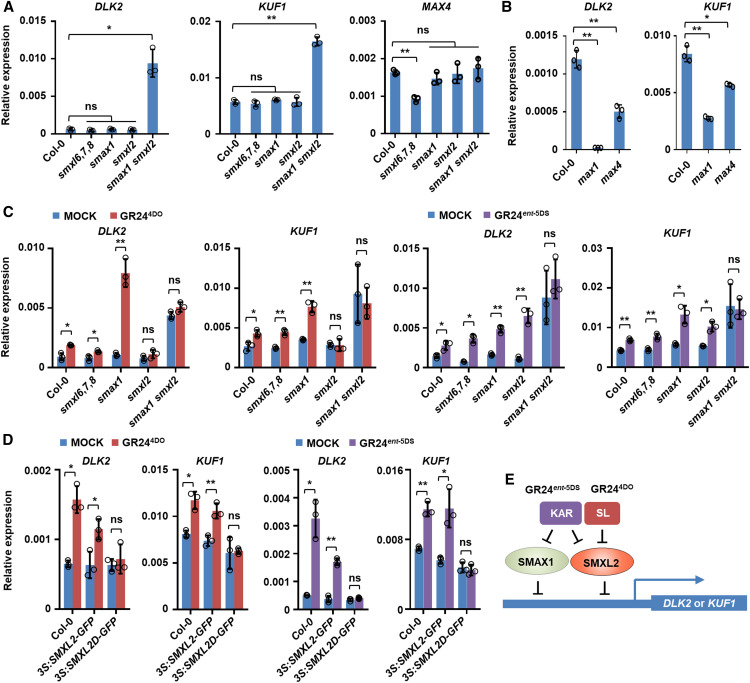
Regulation of DLK2 and KUF1 by SLs Depends on SMXL2, Whereas KAR Signaling Uses Either SMAX1 or SMXL2
To investigate the function of SMXL2 in the regulation of gene expression, we analyzed the transcript levels of DLK2 and KUF1, which have been reported to respond to KARs and SLs (Scaffidi et al., 2013, 2014; Waters et al., 2015), in seedlings of the wild type, smax1, smxl2, smax1 smxl2 double mutant, and smxl6,7,8 triple mutant. Expression levels of DLK2 and KUF1 were comparable in the wild type, smax1, smxl2, and smxl6,7,8 triple mutant but were remarkably elevated in the smax1 smxl2 double mutant, indicating that both SMAX1 and SMXL2 repress the expression of DLK2 and KUF1 (Figure 8A). These results are consistent with previously reported observations (Stanga et al., 2016). However, MAX4 expression was downregulated in the smxl6,7,8 triple mutant compared to the wild type but remained unchanged in smax1, smxl2, and smax1 smxl2 double mutant (Figure 8A). Thus, SMAX1 and SMXL2 mainly mediate the transcriptional response of DLK2 and KUF1, whereas the SMXL6, SMXL7, SMXL8 group mainly mediates expression of the SL-responsive gene MAX4. More importantly, expression of DLK2 and KUF1 were decreased in the SL-biosynthetic mutants max1 and max4, suggesting that endogenous SLs could activate expression of DLK2 and KUF1 in seedlings (Figure 8B).
Figure 8.
SMXL2 Is Required for Induction of DLK2 and KUF1 by Signaling of SL and KAR in Seedlings.
(A) Relative expression of DLK2, KUF1, and MAX4 in 10-d-old seedlings of Col-0 (wild type), smxl6,7,8 triple mutant, smax1, smxl2, and smax1 smxl2 double mutant.
(B) Relative expression of DLK2 and KUF1 in 10-d-old seedlings of Col-0 (wild type), max1, and max4.
(C) Relative expression of DLK2 and KUF1 in 10-d-old seedlings of Col-0 (wild type), smxl6,7,8 triple mutant, smax1, smxl2, and smax1 smxl2 double mutant treated with 1 μM GR244DO or GR24ent-5DS for 4 h.
(D) Relative expression of DLK2 and KUF1 in 10-d-old seedlings of Col-0 (wild type), 35S:SMXL2-GFP, and 35S:SMXL2D-GFP treated with 1 μM GR244DO or GR24ent-5DS for 4 h.
(E) Schematic representation of SL- and KAR-regulated gene expression mediated by SMAX1 and SMXL2.
In (A) to (D), expression values are determined relative to ACTIN2. Values are means ± sd. *, P < 0.05; **, P < 0.01 (n = 3, two-tailed Student’s t test, three independent experiments; Supplemental Data Set). ns, no significance.
Furthermore, exogenously applied GR244DO could significantly induce the expression of DLK2 and KUF1 in the wild type, smxl6,7,8 triple mutant, and smax1 single mutant, indicating that GR244DO promotes expression of DLK2 and KUF1 independently of SMXL6, SMXL7, SMXL8, and SMAX1 (Figure 8C). Interestingly, GR244DO showed no effect in smxl2 or smax1 smxl2, indicating that SMXL2 is essential for stimulation of DLK2 and KUF1 expression by SL signaling (Figure 8C). We further evaluated the contribution of SMXL family members in mediating the expression of DLK2 and KUF1 induced by KAR signaling and found that GR24ent-5DS could trigger an increase in transcript abundance of DLK2 and KUF1 in the wild-type, smxl6,7,8 triple mutant, and smax1 and smxl2 single mutant plants, but it could not induce DLK2 and KUF1 expression in the smax1 smxl2 double mutant (Figure 8C). This result indicates that SMAX1 and SMXL2 are both required for regulation of DLK2 and KUF1 expression in KAR signaling. Moreover, both GR244DO and GR24ent-5DS could induce the expression of DLK2 and KUF1 in seedlings of Col-0 and 35S:SMXL2-GFP transgenic plants but had no effect on seedlings of 35S:SMXL2D-GFP, indicating that degradation of SMXL2 induces the expression of DLK2 and KUF1 in response to GR244DO and GR24ent-5DS in an RGKT-dependent manner (Figure 8D). Together, these results establish that SL regulates DLK2 and KUF1 expression through SMXL2, whereas KAR regulates DLK2 and KUF1 expression through SMAX1 and SMXL2 (Figure 8E).
DISCUSSION
SMXL2 Is Involved in Both SL and KAR Signaling Pathways
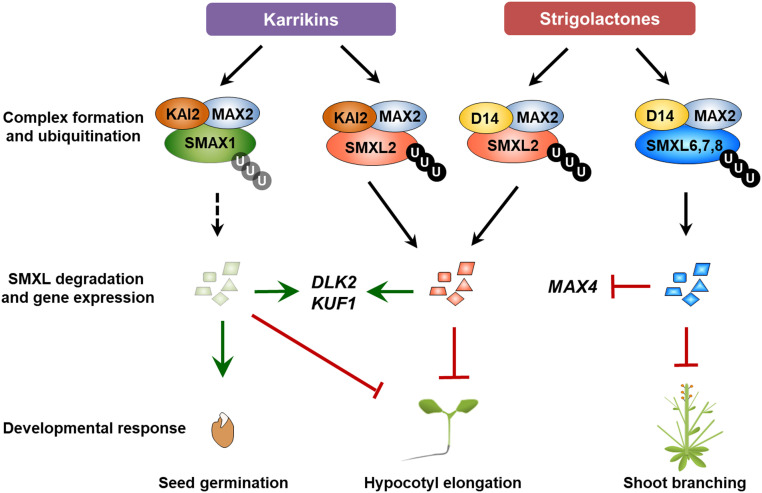
Based on findings in this and previous studies, we propose an overview model of SL and KAR signaling in Arabidopsis (Figure 9). Previous research has shown that in the presence of SLs, D14 forms a complex with MAX2 and one or more of SMXL6, SMXL7, and SMXL8. The complex triggers ubiquitination and degradation of SMXLs, dependent on the RGKT motif, followed by de-repression of key genes that suppress shoot branching, such as BRC1 (Soundappan et al., 2015; Wang et al., 2015). We now show that in seedlings, SLs stimulate the association of D14 with SMXL2 (Figure 2), which is ubiquitinated and degraded (Figure 4 and 5), also dependent on the RGKT motif (Figure 6). Degradation of SMXL2 presumably relieves the repression of genes that function to inhibit hypocotyl elongation (Figure 7) and also relieves repression of DLK2 and KUF1 (Figure 8). Similarly, for KAR signaling, KAR1 or the KAR surrogate GR24ent-5DS induces KAI2 to form a complex with MAX2 (Supplemental Figure 2) and with SMXL2 (Figure 3), triggering the ubiquitination and degradation of SMXL2 in a manner dependent on the RGKT motif (Figures 4 to 6), and further regulating gene expression (Figure 8) and hypocotyl elongation (Figure 7). GR24ent-5DS also induces interaction between KAI2 and SMAX1 (Figure 3) and is speculated to trigger ubiquitination and degradation of SMAX1, although we have not yet demonstrated this. Importantly, these results collectively show that SMXL2 is a common target for proteolysis in both SL and KAR signaling and it functions as a repressor protein regulating hypocotyl elongation and gene expression in Arabidopsis (Figure 9).
Figure 9.
Roles of SMXLs in the Control of Arabidopsis Development by KAR and SL Signaling.
KARs trigger association of KAI2 with SMXL2 and MAX2, leading to ubiquitination and degradation of SMXL2. KARs also trigger interaction between KAI2 and SMAX1 and are proposed to trigger ubiquitination and degradation of SMAX1. SLs induce formation of the D14-MAX2-SMXL2, D14-MAX2-SMXL6, D14-MAX2-SMXL7, and D14-MAX2-SMXL8 complexes and subsequent ubiquitination and degradation of SMXL2, SMXL6, SMXL7, and SMXL8. Degradation of SMXL2 represses hypocotyl elongation and induces the expression of DLK2 and KUF1, and the same functions are proposed for SMAX1. By contrast, degradation of SMXL6, SMXL7, and SMXL8 represses shoot branching and MAX4 expression. SMXL2 is therefore a common target of proteolysis in SL and KAR signaling and functions to regulate hypocotyl elongation and gene expression in Arabidopsis. U, ubiquitin.
Members of the SMXL protein family in Arabidopsis have been considered to play specific roles in response to different signaling molecules (Mach, 2015; Soundappan et al., 2015; Wang et al., 2015; Stanga et al., 2016; Wallner et al., 2017). SMXL6, SMXL7, and SMXL8 function as key transcriptional repressor proteins in SL signaling, while SMAX1 and SMXL2 are important for KAR-regulated changes in transcripts (Soundappan et al., 2015; Wang et al., 2015; Stanga et al., 2016). However, SLs and KARs also regulate the expression of several common downstream genes, for instance, SALT TOLERANCE H-BOX7 (STH7), DLK2, KUF1, INDOLE-3-ACETIC ACID INDUCIBLE5 (IAA5), and IAA6 (Scaffidi et al., 2013, 2014; Waters et al., 2015). Because F-box protein MAX2 is required for targeting and proteolysis of SMXL proteins, we speculated that SMXL2 could be responsible for this convergence in regulation of gene expression by SLs and KARs. Interestingly, disruption of SMXL6, SMXL7, and SMXL8 collectively, or SMAX1 or SMXL2 has no effect on the induction of DLK2 and KUF1, but disruption of both SMAX1 and SMXL2 greatly enhanced the expression of DLK2 and KUF1 (Figure 8A; Stanga et al., 2016) and abolished the upregulation in response to SLs and KARs (Figure 8C). Thus, biochemical and genetic data in this study clearly demonstrate that SMXL2 is common to both SL and KAR signaling and plays a key role in integrating responses to SLs and KARs at the transcription level.
Specificity of GR244DO and GR24ent-5DS in SL and KAR Signaling in Arabidopsis
A strong connection between KARs and SLs is seen in their chemical structures, with common butenolide moieties (Smith and Li, 2014). The natural SL isomers 5DS and 4DO regulate shoot branching, hypocotyl elongation, and gene expression in a D14-dependent manner, while the nonnatural enantiomer ent-5DS selectively regulates seed germination and hypocotyl elongation through KAI2 (Scaffidi et al., 2014). The structure–activity relationship and receptor recognition studies have demonstrated that the configuration at C-2′ plays a critical role in signal perception of SLs and KARs in Arabidopsis and rice (Scaffidi et al., 2014; Umehara et al., 2015), but the chemical specificity is not absolute and might vary at different growth stages or in different experimental systems (Li et al., 2016b; Villaécija-Aguilar et al., 2019). We found that GR244DO could induce downstream gene expression, induce the ubiquitination and degradation of SMXL2 and SMXL6, and repress hypocotyl elongation of the 35S:SMXL2-GFP transgenic plants in a D14-dependent manner, indicating that GR244DO specifically displayed SL activity in Arabidopsis (Figures 1, 4C, 5D, and C7C). GR24ent-5DS not only induced a strong interaction between KAI2 and SMXL2 but also a very weak interaction between D14 and SMXL2 in Co-IP assays (Figures 2B, 3A, and B3B). GR24ent-5DS could also stimulate a relatively strong ubiquitination of SMXL2 that was weaker in d14-1 but largely blocked in kai2-2 (Figure 4C). These data indicate that GR24ent-5DS displays strong KAR-like activity and low SL activity in Arabidopsis.
Although KAR1 and GR24ent-5DS induced transcripts of DLK2 after treatment for 24 to 72 h in a KAI2-dependent manner (Sun et al., 2016), we found that compared with GR24ent-5DS, KAR1 was less effective in the experimental systems detecting protein ubiquitination, degradation, protein–protein interactions, and regulation of transcript levels, especially within short-term treatments in the time frame of an hour. These observations are consistent with the view that KAR1 might require metabolism in vivo to produce a ligand that can be recognized by the receptor KAI2 (Waters et al., 2015, 2017). However, until the endogenous ligand for KAI2 is identified, it is difficult to fully assess the factors that result in KAI2-mediated regulation of seed germination and seedling development. Therefore, discovery of the endogenous KL is becoming increasingly important (Morffy et al., 2016; Sun et al., 2016; Waters et al., 2017; Yao et al., 2018a; Smith, 2019).
Coevolution of the SMXL and KAI2/D14 Families
Evolutionary analysis shows that KAI2 orthologs exist in basal land plants, and the endogenous KL is predicted to exist throughout land plants from basal to seed plants (Waters et al., 2012, 2017; Gutjahr et al., 2015; Kameoka and Kyozuka, 2015; Conn and Nelson, 2016; Carbonnel et al., 2019). Interestingly, SMXL family members are not apparent in the charophytes, but they have been identified in nonvascular plants, including liverworts and mosses, and display 60 to 65% sequence identity with Arabidopsis SMAX1. These ancient forms of SMAX1 subsequently underwent duplication in land plants and evolved into four major clades consisting of homologs of SMAX1 and SMXL2 as clade I; SMXL6, SMXL7, and SMXL8 as clade II; SMXL3 as clade III; and SMXL4 and SMXL5 as clade IV (Moturu et al., 2018). Our current studies indicate that SMAX1 and SMXL2 mainly control seedling development especially hypocotyl elongation, which is important throughout land plants. By contrast, shoot branching and vascular development are notable features of vascular plants, for which SMXL clades II, III, and IV have been shown to be required. The correlation between evolutionary history and functional differentiation of SMXL proteins provides new evidence for the hypothesis of coevolution between the SMXL family and KAI2/D14 family (Moturu et al., 2018). More importantly, we now show that SMXL2 interacts with both the KAR receptor KAI2 and the SL receptor D14 (Figures 2 and 3) and undergoes polyubiquitination and degradation to regulate gene expression and seedling development (Figures 3 to 8). GR24ent-5DS induced a weak interaction between SMXL6 and KAI2 (Figure 3A) and a weak ubiquitination that was impaired in kai2-2 (Figure 4E), suggesting that SMXL6 could form a complex with KAI2 and undergo ubiquitination with low efficiency in response to GR24ent-5DS treatment. This may reflect the evolutionary origin of interactions between the KAR receptor and SMXL-type repressor proteins, although KAI2 and SMXL6 genes are not thought to be coexpressed normally. The demonstration that KAR and SL signaling use the same mechanism of MAX2-dependent SMXL polyubiquitination and proteolysis provides strong support for the proposed coevolution of SL signaling and SMXL functions from an ancestral KAI2-SMAX1 system (Moturu et al., 2018; Machin et al., 2020).
SL- and KAR-Regulated Seedling Development
Hypocotyl elongation is important to push the apex of young seedlings out of the soil or deep water to receive light and continue vegetative and reproductive development. Hypocotyl elongation is regulated by various phytohormones, including auxin, gibberellin, ethylene, brassinosteroid, and jasmonate (Yang et al., 2011; Li et al., 2016a; Shi et al., 2016; Reed et al., 2018; Yi et al., 2020). It has been reported that hypocotyls of wild-type seedlings under red light are sensitive to both SL and KAR treatment and that SMAX1 and SMXL2 collectively mediate the KAR-repressed hypocotyl development (Stanga et al., 2016). However, the molecular mechanism underlying SL-regulated hypocotyl development has been unclear. Here, we show that transgenic plants overexpressing degradation-resistant SMXL2 possessed longer hypocotyls that were insensitive to exogenously applied SL and KAR, but overexpressing degradation-resistant SMXL6 showed no effect on hypocotyl response, and the hypocotyl response to exogenously applied SL was blocked in the d14 background (Figures 7B and 7C). Based on the roles of SMXL2 in SL signaling, these results clearly show that it is SMXL2 rather than SMXL6, SMXL7, or SMXL8 that functions as a repressor protein in SL-regulated hypocotyl development in Arabidopsis.
Recent evolutionary analyses have shown that SMXL2 appears due to a recent duplication of SMAX1 in the Brassicaceae and has not been observed in the genome of plants belonging to other families (Walker et al., 2019). Interestingly, the rice SL mutants d10, d14, and d17 form elongated mesocotyl in comparison to the wild type (Hu et al., 2010), but the Arabidopsis SL mutants max1, max3, max4, and d14 form hypocotyls indistinguishable from the wild type (Nelson et al., 2011; Scaffidi et al., 2013, 2014). This may be due to the possible role of D53 and D53L in SL-regulated rice mesocotyl development. Meanwhile, the rice d3 mutant (equivalent to Arabidopsis max2) displays a more elongated mesocotyl phenotype than d14 and SL biosynthesis–deficient mutants (Hu et al., 2010), suggesting a potential role of the KAR signaling pathway in mesocotyl development (Kameoka and Kyozuka, 2015). Further investigation of SL and KAR signaling and the roles of D53 family proteins in mesocotyl elongation could provide a breakthrough in understanding rice seedling growth and development. This is important for the future agricultural practice of direct seeding to avoid seedling transplantation in rice crop production. In other crops, increased elongation of hypocotyl or mesocotyl could permit deeper sowing of seeds in soils in which the surface layers are at risk from drying. Thus, our research enables further understanding of the control of seed germination, seedling skotomorphogenesis and photomorphogenesis, which provides major opportunities for future improvements in agriculture.
METHODS
Plant Materials and Growth Conditions
Arabidopsis (Arabidopsis thaliana) Col-0 was used as the wild type. The smxl6,7,8 triple mutant (Wang et al., 2015), d14-1 (Waters et al., 2012), smax1-2 (Stanga et al., 2013), smxl2-1 (Stanga et al., 2016), smax1-2 smxl2-1 double mutant (Stanga et al., 2016), max1-1 (Stirnberg et al., 2002), and max4-5 (Bennett et al., 2006) have been described previously. The kai2-2 mutant (Waters et al., 2012) in the Landsberg erecta background was backcrossed to Col-0 six times by Mark Waters (University of Western Australia) and was provided by Ruifeng Yao (Tsinghua University). Seeds were surface sterilized, vernalized at 4°C, and germinated on 0.5× Murashige and Skoog (MS) medium containing 1.0% (w/v) Suc and 0.7% (w/v) agar. For morphological observations, 10-d-old seedlings were transferred to pots containing a 1:1 vermiculite:soil mixture saturated with 0.3× MS medium. Plants were grown under controlled conditions of ∼21°C, 16-h-light/8-h-dark cycle, and fluorescent lamps with a light intensity of 60 to 80 μE m−2 s−1 as described previously (Dai et al., 2006).
Chemicals and Reagents
Synthetic rac-GR24 and KAR1 were obtained from Chiralix. The SL analogs GR244DO, GR245DS, and GR24ent-5DS were synthesized via the following method. GR24 stereoisomers were prepared as previously described by Mangnus et al. (1992). Next, flash chromatography was performed to give two kinds of enantiomers. The enantiomers moving faster were further separated into GR24ent-5DS and GR245DS, and the enantiomers moving slower were separated into GR24ent-4DO and GR244DO using a high-performance liquid chromatograph (HPLC) fitted with a chiral HPLC column (150 mm long, 4.6-mm internal diameter). Separation was achieved using a flow rate of 1.0 mL min−1 of a 1:1 mixture of hexane:ethyl alcohol. The corresponding components were collected and obtained pure optical isomers. MG132 was obtained from Calbiochem. The stock solution of rac-GR24, KAR1, GR244DO, GR245DS, or GR24ent-5DS was prepared in acetone, and the stock solution of MG132 was prepared in DMSO.
Vector Construction and Plant Transformation
To construct the 35S:SMXL2-GFP plasmid, the coding sequence of SMXL2 was amplified with primer pairs of SMXL2-OE-F and SMXL2-OE-R (Supplemental Table) and then cloned into KpnI- and SalI-digested pWM101. To construct the 35S:SMXL2D-GFP plasmid, SMXL2D was amplified through site-directed mutagenesis using primer pairs of SMXL2-DEL-F and SMXL2-DEL-R and then cloned into pWM101. The 35S:SMXL2-GFP and 35S:SMXL2D-GFP recombinant plasmids were introduced into Agrobacterium tumefaciens strain EHA105 and used to transform wild-type, d14-1, and kai2-2 plants through the Agrobacterium-mediated floral dip method. To construct the plasmids of 35S:GFP-SMXL2, 35S:GFP-SMXL5, and 35S:3×HA-KAI2, the coding sequence of SMXL2, SMXL5, and KAI2 was amplified with primer pairs of SMXL2-PD-F, SMXL2-PD-R, SMXL5-PD-F, SMXL5-PD-R, and KAI2-PD-F, KAI2-PD-R (Supplemental Table) and then cloned into the Gateway Entry vector by BP reaction and recombined to pBeacon-eGFP and pBeacon-3×HA (Wang et al., 2015) through LR reaction. To construct the plasmids of 35S:GFP-SMAX1, the codon-optimized SMAX1 coding sequence was amplified from the Flag-SMAX1 plasmid (Yao et al., 2017) with primer pairs of SMAX1-PD-F, SMAX1-PD-R and then cloned into the pBeacon-eGFP vector through BP and LR reaction.
Gene Expression Analysis
The wild type and mutants of smax1, smxl2, smax1 smxl2, smxl6,7,8, max1, max4, d14-1, kai2-2, 35S:SMXL2-GFP/Col-0, and 35S:SMXL2D-GFP/Col-0 were grown on agar with 0.5× MS medium for 10 d, collected, and equilibrated in 0.5× MS liquid medium for 2 h and then treated with GR244DO, GR24ent-5DS, KAR1, or acetone in the light at ∼21°C for 4 h. The whole seedlings were frozen and ground into power in liquid nitrogen. Total RNA was prepared using a TRIzol kit (Invitrogen) according to the user’s manual. RNA samples (each containing 12.5 μg of RNA) were treated with TUBRO DNase (Ambion). Next, first-strand cDNA was synthesized using oligo(dT) and random primers with the SuperScript III first-strand synthesis system (Invitrogen). Real-time PCR was performed using gene-specific primers (Supplemental Table) on a CFX 96 real-time PCR detection system (Bio-Rad) in a total volume of 10 μL with 2 μL of diluted cDNA, 0.3 μM gene-specific primers, and 5 μL of SsoFast EvaGreen supermix (Bio-Rad). The real ΔCt values are determined relative to the Arabidopsis ACTIN2 gene (Weissgerber et al., 2015).
Co-IP Assay
Protoplasts were generated from mesophyll cells of Arabidopsis leaves (Yoo et al., 2007) and transformed with transient expression plasmids including 35S:3×HA-D14 (Wang et al., 2015), 35S:GFP-SMXL2, 35S:GFP (Wang et al., 2015), 35S:3×HA-KAI2, and 35S:GFP-MAX2 (Wang et al., 2015). After incubation at 21°C for 11 h followed by another 1 h with 100 µM GR244DO, GR24ent-5DS, KAR1, or GR245DS, protoplasts were collected in protein extraction buffer [50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 10% (v/v) glycerol, and 1% (v/v) Nonidet P-40] and 1× Complete Protease Inhibitor Cocktail (catalog no. 04693132001, lot no. 41353700; Roche). The lysate was centrifuged at 18,000g for 15 min at 4°C, and the supernatant was collected for Co-IP assay. In brief, 30 μL of the agarose-conjugated anti-GFP monoclonal antibody (D153-8, lot no. 048; MBL) was added into 1 mL of total extracted protein solution and incubated at 4°C for 3 h in the presence or absence of GR244DO, GR24ent-5DS, KAR1, and GR245DS. The beads were washed three times with extraction buffer containing 0.1% (v/v) Nonidet P-40 and then eluted with 25 μL of SDS-PAGE sample buffer for immunoblot analysis. The levels of GFP-SMXL2, GFP-MAX2, and GFP protein were detected using mouse anti-GFP monoclonal antibody (catalog no. 11814460001, lot no. 11751700; Roche) at a 1:2000 dilution and One Step Western Kit HRP (Mouse; CW2030M, lot no. 03,803/40449; CoWin Biosciences) at 1:200 dilution. The levels of HA-D14 and HA-KAI2 protein were detected using rabbit anti-HA polyclonal antibody (H6908, lot no. 106M4792; Sigma) at a 1:2000 dilution and anti-rabbit IgG HRP-linked whole antibody (NA934V, lot no. 9761194; GE Healthcare) at 1:10,000 dilution. Signals were then detected using Amersham ECL Select Western Blotting Detection Reagent (RPN2235, lot no. 16926118; GE Healthcare) and x-ray film.
In Vivo Ubiquitination Assay
The transgenic plants of 35S:SMXL2-GFP/Col-0, 35S:SMXL2-GFP/d14-1, 35S:SMXL2-GFP/kai2-2, and 35S:SMXL2D-GFP/Col-0 were grown for 7 d, collected, and pretreated with 50 µM MG132 for 1 h and then treated with KAR1, rac-GR24, GR244DO, GR24ent-5DS, or acetone for the indicated time in 0.5× MS liquid medium at 21°C. Equal weights of plant materials were collected for protein extraction using protein extraction buffer containing 1× Complete Protease Inhibitor Cocktail. Next, 30 μL of anti-GFP monoclonal antibody–conjugated agarose was added into 1 mL of total proteins extract and incubated at 4°C for 3 h with gentle rotation. The beads were washed three times with the protein extraction buffer containing 0.1% (v/v) Nonidet P-40 and then eluted with 25 μL of the SDS-PAGE sample buffer for protein blotting. The polyubiquitination level was detected using mouse anti-ubiquitin monoclonal antibody (Zhao et al., 2013b) at a 1:3000 dilution and anti-mouse IgG HRP-linked whole antibody (NA931V, lot no. 9761190; GE Healthcare) at 1:10,000 dilution. The levels of SMXL2-GFP and SMXL2D-GFP proteins were detected using the mouse anti-GFP monoclonal antibody at a 1:2000 dilution and One Step Western Kit HRP (Mouse) at 1:200 dilution. Signals were then detected using Amersham ECL Select Western Blotting Detection Reagent and x-ray film.
In the ubiquitination system using protoplasts, the GFP-SMXL2 and GFP-SMXL6 recombinant proteins were expressed in Arabidopsis protoplasts made from mesophyll cells (Yoo et al., 2007). Equal volumes of protoplasts were pretreated with MG132 for 1 h and treated with indicated chemicals for 1.5 h. Finally, they underwent protein extraction using lysis buffer. Equal volumes of GFP monoclonal antibody–conjugated agarose beads were added to each sample, incubated for 3 h, washed three times, and eluted with 30 μL of 2× SDS loading buffer. Next, 10 μL of eluted sample was used to detect polyubiquitination and GFP levels in immunoblots. The polyubiquitination level was detected using anti-ubiquitin monoclonal antibody (Zhao et al., 2013b) at a 1:3000 dilution and anti-mouse IgG HRP-linked whole antibody at 1:10,000 dilution. The levels of GFP-SMXL2 and GFP-SMXL6 proteins were detected using the mouse anti-GFP monoclonal antibody at a 1:2000 dilution and One Step Western Kit HRP (Mouse) at 1:200 dilution. Signals were then detected using Amersham ECL Select Western Blotting Detection Reagent and x-ray film.
SMXL2 Degradation
The transgenic plants of 35S:SMXL2-GFP/Col-0, 35S:SMXL2-GFP/d14-1, 35S:SMXL2-GFP/kai2-2, and 35S:SMXL2D-GFP/Col-0 were grown for 7 d, collected, and treated with GR244DO, GR24ent-5DS, or KAR1 for the indicated time in 0.5× MS liquid medium at 21°C. Equal weight of plant materials was collected for protein extraction using lysis buffer containing 1× Complete Protease Inhibitor Cocktail. Protein levels of SMXL2-GFP and SMXL2D-GFP were detected by immunoblotting with mouse anti-GFP monoclonal antibody at a 1:2000 dilution and One Step Western Kit HRP (Mouse) at 1:200 dilution. In addition, abundances of SMXL2-GFP in the root, hypocotyl, and cotyledons of 35S:SMXL2-GFP/Col-0 seedlings were determined by immunoblotting with mouse anti-GFP monoclonal antibody at a 1:2000 dilution and One Step Western Kit HRP (Mouse) at 1:200 dilution, and endogenous Actin levels were determined using mouse anti-Actin monoclonal antibody (M20009L, lot no. 313860; Abmart) at a 1:10,000 dilution and anti-mouse IgG HRP-linked whole antibody at 1:10,000 dilution. Signals were then detected using Amersham ECL Select Western Blotting Detection Reagent and x-ray film.
GFP Fluorescence
To detect endogenous degradation of SMXL2-GFP, roots of 7-d-old 35S:SMXL2-GFP/Col-0 and 35S:SMXL2D-GFP/Col-0 seedlings were treated with 2 µM GR244DO or GR24ent-5DS in 0.5× MS liquid medium for the time indicated on glass slides and then imaged by confocal laser scanning microscopy (FluoView 1000 confocal microscope; Olympus) at excitation wavelength of 488 nm. The same offset and gain settings for GFP detection were used within experiments for each transgenic line. The signal intensities of GFP were quantitatively determined with the FV10-ASW 4.0 viewer 1000 software (Olympus). To observe the SMXL2-GFP expression profiles in root tips of 35S:SMXL2-GFP/Col-0 and 35S:SMXL2D-GFP/Col-0 seedlings grown for 5 d under long-day conditions, Z-stacks containing five focal planes were imaged and collected by confocal laser scanning microscopy (FluoView 1000 confocal microscope) at excitation wavelength of 488 nm.
Hypocotyl Measurements
For hypocotyl elongation assays, surface-sterilized seeds were sown on 0.5× MS media containing 1.0% (w/v) Suc, 0.7% (w/v) agar and 100 nM GR244DO, GR24ent-5DS, or GR244DO plus GR24ent-5DS (100 nM of each chemical). The seeds were stratified in the dark at 4°C for 72 h, exposed to white light from a fluorescence tube (∼60 to 80 μE m−2 s−1) for 3 h at 21°C, transferred to dark for 21 h, and then exposed to continuous red light (∼20 to 30 μE m−2 s−1) for 4 d at 21°C as previously described (Stanga et al., 2013). The lengths of the hypocotyls were measured using ImageJ (http://rsbweb.nih.gov/ij/).
Statistical analysis
For gene expression analysis, phenotype observation, and GFP fluorescence quantification, statistical analysis was assessed as described in the figure legends. P-values were calculated by two-tailed Student’s t test using Excel 2016 or by two-way ANOVA multiple comparison with a Tukey test using GraphPad Prism 7.00.
Accession Numbers
Sequence data were obtained from the GenBank/EMBL libraries under the following accession numbers: SMAX1 (AT5G57710), SMXL2 (AT4G30350), SMXL3 (AT3G52490), SMXL4 (AT4G29920), SMXL5 (AT5G57130), SMXL6 (AT1G07200), SMXL7 (AT2G29970), SMXL8 (AT2G40130), AtD14 (AT3G03990), KAI2 (AT4G37170), MAX1 (AT2G26170), MAX2 (AT2G42620), MAX4 (AT4G32810), DLK2 (AT3G24420), KUF1 (AT1G31350), and ACTIN2 (AT3G18780).
Supplemental Data
Supplemental Figure 1. Expression of SMXL gene family members shows no response to KAR1 treatment.
Supplemental Figure 2. GR24ent-5DS induces interaction between KAI2 and MAX2 in vivo.
Supplemental Figure 3. Ubiquitination of SMXL2 after KAR1 or rac-GR24 treatment of protoplasts.
Supplemental Figure 4. GFP fluorescence signals in 35S:SMXL2-GFP and 35S:SMXL2D-GFP transgenic lines.
Supplemental Figure 5. Alignment of Arabidopsis SMXL family proteins.
Supplemental Figure 6. Diagram showing domain structure of SMXL2 and the amino acid changes in SMXL2D protein.
Supplemental Figure 7. SMXL2-GFP abundance in the hypocotyl of 35S:SMXL2-GFP seedlings.
Supplemental Figure 8. SMXL2 has little effect on shoot branching.
Supplemental Table. Primers used in this study.
Supplemental Data Set. Statistical analysis data.
DIVE Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
Acknowledgments
We thank Mark Waters (University of Western Australia) and Daoxin Xie (Tsinghua University) for providing the kai2-2 mutant; Ruifeng Yao (Hunan University) for providing the d14-1 kai2-2 mutant; David Nelson (University of California, Riverside) and Mark Waters for providing smax1-2, smxl2-1, and smax1-2 smxl2-1 mutants; and Ottoline Leyser (Cambridge University) for providing max1-1 and max4-5 mutants. This work was supported by National Natural Science Foundation of China (grants 31788103, 31600221, and 31661143025), the Chinese Academy of Sciences President’s International Fellowship Initiative (grant 2018VBA0025), and Youth Innovation Promotion Association of the Chinese Academy of Sciences (grant 2019099).
AUTHOR CONTRIBUTIONS
L.W., B.W., and Q.Xu designed and performed experiments; H.Y., H.M., X.L., J.Y., J.C., Q.Xie, and Y.W. contributed to some of the experiments. B.W., G.X., and J.L. designed research and conceived and supervised the project. B.W., L.W., S.M.S., and J.L. analyzed the data and wrote the article. All authors commented on the article.
Footnotes
Articles can be viewed without a subscription.
References
- Agusti J., Herold S., Schwarz M., Sanchez P., Ljung K., Dun E.A., Brewer P.B., Beveridge C.A., Sieberer T., Sehr E.M., Greb T.(2011). Strigolactone signaling is required for auxin-dependent stimulation of secondary growth in plants. Proc. Natl. Acad. Sci. USA 108: 20242–20247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akiyama K., Matsuzaki K., Hayashi H.(2005). Plant sesquiterpenes induce hyphal branching in arbuscular mycorrhizal fungi. Nature 435: 824–827. [DOI] [PubMed] [Google Scholar]
- Al-Babili S., Bouwmeester H.J.(2015). Strigolactones, a novel carotenoid-derived plant hormone. Annu. Rev. Plant Biol. 66: 161–186. [DOI] [PubMed] [Google Scholar]
- Arite T., Iwata H., Ohshima K., Maekawa M., Nakajima M., Kojima M., Sakakibara H., Kyozuka J.(2007). DWARF10, an RMS1/MAX4/DAD1 ortholog, controls lateral bud outgrowth in rice. Plant J. 51: 1019–1029. [DOI] [PubMed] [Google Scholar]
- Bennett T., Sieberer T., Willett B., Booker J., Luschnig C., Leyser O.(2006). The Arabidopsis MAX pathway controls shoot branching by regulating auxin transport. Curr. Biol. 16: 553–563. [DOI] [PubMed] [Google Scholar]
- Carbonnel S., Torab S., Griesmann M., Bleek E., Tang Y., Buchka S., Basso V., Shindo M., Wang T.L., Udvardi M., Waters M., Gutjahr C. (2019). Duplicated KAI2 receptors with divergent ligand-binding specificities control distinct developmental traits in Lotus japonicas. bioRxiv 10.1101/754937. [Google Scholar]
- Conn C.E., Bythell-Douglas R., Neumann D., Yoshida S., Whittington B., Westwood J.H., Shirasu K., Bond C.S., Dyer K.A., Nelson D.C.(2015). PLANT EVOLUTION. Convergent evolution of strigolactone perception enabled host detection in parasitic plants. Science 349: 540–543. [DOI] [PubMed] [Google Scholar]
- Conn C.E., Nelson D.C.(2016). Evidence that KARRIKIN-INSENSITIVE2 (KAI2) receptors may perceive an unknown signal that is not karrikin or strigolactone. Front Plant Sci 6: 1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai Y., Wang H., Li B., Huang J., Liu X., Zhou Y., Mou Z., Li J.(2006). Increased expression of MAP KINASE KINASE7 causes deficiency in polar auxin transport and leads to plant architectural abnormality in Arabidopsis. Plant Cell 18: 308–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Saint Germain A., et al. (2016). An histidine covalent receptor and butenolide complex mediates strigolactone perception. Nat. Chem. Biol. 12: 787–794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond R.S., Martínez-Sánchez N.M., Janssen B.J., Templeton K.R., Simons J.L., Quinn B.D., Karunairetnam S., Snowden K.C.(2009). Petunia hybrida CAROTENOID CLEAVAGE DIOXYGENASE7 is involved in the production of negative and positive branching signals in petunia. Plant Physiol. 151: 1867–1877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flematti G.R., Scaffidi A., Waters M.T., Smith S.M.(2016). Stereospecificity in strigolactone biosynthesis and perception. Planta 243: 1361–1373. [DOI] [PubMed] [Google Scholar]
- Foo E., Turnbull C.G., Beveridge C.A.(2001). Long-distance signaling and the control of branching in the rms1 mutant of pea. Plant Physiol. 126: 203–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Roldan V., et al. (2008). Strigolactone inhibition of shoot branching. Nature 455: 189–194. [DOI] [PubMed] [Google Scholar]
- Guo Y., Zheng Z., La Clair J.J., Chory J., Noel J.P.(2013). Smoke-derived karrikin perception by the α/β-hydrolase KAI2 from Arabidopsis. Proc. Natl. Acad. Sci. USA 110: 8284–8289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutjahr C., et al. (2015). Rice perception of symbiotic arbuscular mycorrhizal fungi requires the karrikin receptor complex. Science 350: 1521–1524. [DOI] [PubMed] [Google Scholar]
- Haider I., Andreo-Jimenez B., Bruno M., Bimbo A., Floková K., Abuauf H., Ntui V.O., Guo X., Charnikhova T., Al-Babili S., Bouwmeester H.J., Ruyter-Spira C.(2018). The interaction of strigolactones with abscisic acid during the drought response in rice. J. Exp. Bot. 69: 2403–2414. [DOI] [PubMed] [Google Scholar]
- Hamiaux C., Drummond R.S., Janssen B.J., Ledger S.E., Cooney J.M., Newcomb R.D., Snowden K.C.(2012). DAD2 is an α/β hydrolase likely to be involved in the perception of the plant branching hormone, strigolactone. Curr. Biol. 22: 2032–2036. [DOI] [PubMed] [Google Scholar]
- Hu Z., Yan H., Yang J., Yamaguchi S., Maekawa M., Takamure I., Tsutsumi N., Kyozuka J., Nakazono M.(2010). Strigolactones negatively regulate mesocotyl elongation in rice during germination and growth in darkness. Plant Cell Physiol. 51: 1136–1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang L., et al. (2013). DWARF 53 acts as a repressor of strigolactone signalling in rice. Nature 504: 401–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kameoka H., Kyozuka J.(2015). Downregulation of rice DWARF 14 LIKE suppress mesocotyl elongation via a strigolactone independent pathway in the dark. J. Genet. Genomics 42: 119–124. [DOI] [PubMed] [Google Scholar]
- Kohlen W., Charnikhova T., Liu Q., Bours R., Domagalska M.A., Beguerie S., Verstappen F., Leyser O., Bouwmeester H., Ruyter-Spira C.(2011). Strigolactones are transported through the xylem and play a key role in shoot architectural response to phosphate deficiency in nonarbuscular mycorrhizal host Arabidopsis. Plant Physiol. 155: 974–987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lantzouni O., Klermund C., Schwechheimer C.(2017). Largely additive effects of gibberellin and strigolactone on gene expression in Arabidopsis thaliana seedlings. Plant J. 92: 924–938. [DOI] [PubMed] [Google Scholar]
- Li K., Yu R., Fan L.M., Wei N., Chen H., Deng X.W.(2016a). DELLA-mediated PIF degradation contributes to coordination of light and gibberellin signalling in Arabidopsis. Nat. Commun. 7: 11868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S., Chen L., Li Y., Yao R., Wang F., Yang M., Gu M., Nan F., Xie D., Yan J.(2016b). Effect of GR24 stereoisomers on plant development in Arabidopsis. Mol. Plant 9: 1432–1435. [DOI] [PubMed] [Google Scholar]
- Li W., et al. (2017). The karrikin receptor KAI2 promotes drought resistance in Arabidopsis thaliana. PLoS Genet. 13: e1007076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Y., Ward S., Li P., Bennett T., Leyser O.(2016). SMAX1-LIKE7 signals from the nucleus to regulate shoot development in Arabidopsis via partially EAR motif-independent mechanisms. Plant Cell 28: 1581–1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin H., Wang R., Qian Q., Yan M., Meng X., Fu Z., Yan C., Jiang B., Su Z., Li J., Wang Y.(2009). DWARF27, an iron-containing protein required for the biosynthesis of strigolactones, regulates rice tiller bud outgrowth. Plant Cell 21: 1512–1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mach J.(2015). Strigolactones regulate plant growth in Arabidopsis via degradation of the DWARF53-like proteins SMXL6, 7, and 8. Plant Cell 27: 3022–3023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machin D.C., Hamon-Josse M., Bennett T.(2020). Fellowship of the rings: A saga of strigolactones and other small signals. New Phytol. 225: 621–636. [DOI] [PubMed] [Google Scholar]
- Mangnus E.M., Dommerholt F.J., Dejong R.L.P., Zwanenburg B.(1992). Improved synthesis of strigol analogue GR24 and evaluation of the biological activity of its diastereomers. J. Agric. Food Chem. 40: 1230–1235. [Google Scholar]
- Morffy N., Faure L., Nelson D.C.(2016). Smoke and hormone mirrors: Action and evolution of karrikin and strigolactone signaling. Trends Genet. 32: 176–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mostofa M.G., Li W., Nguyen K.H., Fujita M., Tran L.P.(2018). Strigolactones in plant adaptation to abiotic stresses: An emerging avenue of plant research. Plant Cell Environ. 41: 2227–2243. [DOI] [PubMed] [Google Scholar]
- Moturu T.R., Thula S., Singh R.K., Nodzyński T., Vareková R.S., Friml J., Simon S.(2018). Molecular evolution and diversification of the SMXL gene family. J. Exp. Bot. 69: 2367–2378. [DOI] [PubMed] [Google Scholar]
- Mukhopadhyay D., Riezman H.(2007). Proteasome-independent functions of ubiquitin in endocytosis and signaling. Science 315: 201–205. [DOI] [PubMed] [Google Scholar]
- Nelson D.C., Flematti G.R., Ghisalberti E.L., Dixon K.W., Smith S.M.(2012). Regulation of seed germination and seedling growth by chemical signals from burning vegetation. Annu. Rev. Plant Biol. 63: 107–130. [DOI] [PubMed] [Google Scholar]
- Nelson D.C., Flematti G.R., Riseborough J.A., Ghisalberti E.L., Dixon K.W., Smith S.M.(2010). Karrikins enhance light responses during germination and seedling development in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 107: 7095–7100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson D.C., Riseborough J.A., Flematti G.R., Stevens J., Ghisalberti E.L., Dixon K.W., Smith S.M.(2009). Karrikins discovered in smoke trigger Arabidopsis seed germination by a mechanism requiring gibberellic acid synthesis and light. Plant Physiol. 149: 863–873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson D.C., Scaffidi A., Dun E.A., Waters M.T., Flematti G.R., Dixon K.W., Beveridge C.A., Ghisalberti E.L., Smith S.M.(2011). F-box protein MAX2 has dual roles in karrikin and strigolactone signaling in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 108: 8897–8902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed J.W., Wu M.F., Reeves P.H., Hodgens C., Yadav V., Hayes S., Pierik R.(2018). Three auxin response factors promote hypocotyl elongation. Plant Physiol. 178: 864–875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saeed W., Naseem S., Ali Z.(2017). Strigolactones biosynthesis and their role in abiotic stress resilience in plants: A critical review. Front Plant Sci 8: 1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sang D., et al. (2014). Strigolactones regulate rice tiller angle by attenuating shoot gravitropism through inhibiting auxin biosynthesis. Proc. Natl. Acad. Sci. USA 111: 11199–11204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scaffidi A., Waters M.T., Ghisalberti E.L., Dixon K.W., Flematti G.R., Smith S.M.(2013). Carlactone-independent seedling morphogenesis in Arabidopsis. Plant J. 76: 1–9. [DOI] [PubMed] [Google Scholar]
- Scaffidi A., Waters M.T., Sun Y.K., Skelton B.W., Dixon K.W., Ghisalberti E.L., Flematti G.R., Smith S.M.(2014). Strigolactone hormones and their stereoisomers signal through two related receptor proteins to induce different physiological responses in Arabidopsis. Plant Physiol. 165: 1221–1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seto Y., et al. (2019). Strigolactone perception and deactivation by a hydrolase receptor DWARF14. Nat. Commun. 10: 191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shabek N., Ticchiarelli F., Mao H., Hinds T.R., Leyser O., Zheng N.(2018). Structural plasticity of D3-D14 ubiquitin ligase in strigolactone signalling. Nature 563: 652–656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H., Liu R., Xue C., Shen X., Wei N., Deng X.W., Zhong S.(2016). Seedlings transduce the depth and mechanical pressure of covering soil using COP1 and ethylene to regulate EBF1/EBF2 for soil emergence. Curr. Biol. 26: 139–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simons J.L., Napoli C.A., Janssen B.J., Plummer K.M., Snowden K.C.(2007). Analysis of the DECREASED APICAL DOMINANCE genes of petunia in the control of axillary branching. Plant Physiol. 143: 697–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith S.M.(2019). Karrikins: Germination stimulants produced by wildfires In eLS. (Chichester, UK: John Wiley & Sons; ). [Google Scholar]
- Smith S.M., Li J.(2014). Signalling and responses to strigolactones and karrikins. Curr. Opin. Plant Biol. 21: 23–29. [DOI] [PubMed] [Google Scholar]
- Snowden K.C., Simkin A.J., Janssen B.J., Templeton K.R., Loucas H.M., Simons J.L., Karunairetnam S., Gleave A.P., Clark D.G., Klee H.J.(2005). The Decreased apical dominance1/Petunia hybrida CAROTENOID CLEAVAGE DIOXYGENASE8 gene affects branch production and plays a role in leaf senescence, root growth, and flower development. Plant Cell 17: 746–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorefan K., Booker J., Haurogné K., Goussot M., Bainbridge K., Foo E., Chatfield S., Ward S., Beveridge C., Rameau C., Leyser O.(2003). MAX4 and RMS1 are orthologous dioxygenase-like genes that regulate shoot branching in Arabidopsis and pea. Genes Dev. 17: 1469–1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soundappan I., Bennett T., Morffy N., Liang Y., Stanga J.P., Abbas A., Leyser O., Nelson D.C.(2015). SMAX1-LIKE/D53 family members enable distinct MAX2-dependent responses to strigolactones and karrikins in Arabidopsis. Plant Cell 27: 3143–3159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanga J.P., Morffy N., Nelson D.C.(2016). Functional redundancy in the control of seedling growth by the karrikin signaling pathway. Planta 243: 1397–1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanga J.P., Smith S.M., Briggs W.R., Nelson D.C.(2013). SUPPRESSOR OF MORE AXILLARY GROWTH2 1 controls seed germination and seedling development in Arabidopsis. Plant Physiol. 163: 318–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stirnberg P., van De Sande K., Leyser H.M.(2002). MAX1 and MAX2 control shoot lateral branching in Arabidopsis. Development 129: 1131–1141. [DOI] [PubMed] [Google Scholar]
- Sun Y.K., Flematti G.R., Smith S.M., Waters M.T.(2016). Reporter gene-facilitated detection of compounds in Arabidopsis leaf extracts that activate the karrikin signaling pathway. Front Plant Sci 7: 1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swarbreck S.M., Guerringue Y., Matthus E., Jamieson F.J.C., Davies J.M.(2019). Impairment in karrikin but not strigolactone sensing enhances root skewing in Arabidopsis thaliana. Plant J. 98: 607–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toh S., Holbrook-Smith D., Stogios P.J., Onopriyenko O., Lumba S., Tsuchiya Y., Savchenko A., McCourt P.(2015). Structure-function analysis identifies highly sensitive strigolactone receptors in Striga. Science 350: 203–207. [DOI] [PubMed] [Google Scholar]
- Toh S., Holbrook-Smith D., Stokes M.E., Tsuchiya Y., McCourt P.(2014). Detection of parasitic plant suicide germination compounds using a high-throughput Arabidopsis HTL/KAI2 strigolactone perception system. Chem. Biol. 21: 988–998. [DOI] [PubMed] [Google Scholar]
- Tsuchiya Y., Yoshimura M., Sato Y., Kuwata K., Toh S., Holbrook-Smith D., Zhang H., McCourt P., Itami K., Kinoshita T., Hagihara S.(2015). PARASITIC PLANTS. Probing strigolactone receptors in Striga hermonthica with fluorescence. Science 349: 864–868. [DOI] [PubMed] [Google Scholar]
- Umehara M., Cao M., Akiyama K., Akatsu T., Seto Y., Hanada A., Li W., Takeda-Kamiya N., Morimoto Y., Yamaguchi S.(2015). Structural requirements of strigolactones for shoot branching inhibition in rice and Arabidopsis. Plant Cell Physiol. 56: 1059–1072. [DOI] [PubMed] [Google Scholar]
- Umehara M., Hanada A., Yoshida S., Akiyama K., Arite T., Takeda-Kamiya N., Magome H., Kamiya Y., Shirasu K., Yoneyama K., Kyozuka J., Yamaguchi S.(2008). Inhibition of shoot branching by new terpenoid plant hormones. Nature 455: 195–200. [DOI] [PubMed] [Google Scholar]
- Villaécija-Aguilar J.A., Hamon-Josse M., Carbonnel S., Kretschmar A., Schmidt C., Dawid C., Bennett T., Gutjahr C.(2019). SMAX1/SMXL2 regulate root and root hair development downstream of KAI2-mediated signalling in Arabidopsis. PLoS Genet. 15: e1008327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker C.H., Siu-Ting K., Taylor A., O’Connell M.J., Bennett T.(2019). Strigolactone synthesis is ancestral in land plants, but canonical strigolactone signalling is a flowering plant innovation. BMC Biol. 17: 70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallner E.S., et al. (2017). Strigolactone- and karrikin-independent SMXL proteins are central regulators of phloem formation. Curr. Biol. 27: 1241–1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B., Wang Y., Li J.(2017a). Strigolactones In Hormone Metabolism and Signaling in Plants, Li J., Li C., and Smith S.M., eds (London: Elsevier; ), pp. 327–359. [Google Scholar]
- Wang J., Yu H., Xiong G., Lu Z., Jiao Y., Meng X., Liu G., Chen X., Wang Y., Li J.(2017b). Tissue-specific ubiquitination by IPA1 INTERACTING PROTEIN1 modulates IPA1 protein levels to regulate plant architecture in rice. Plant Cell 29: 697–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Wang B., Jiang L., Liu X., Li X., Lu Z., Meng X., Wang Y., Smith S.M., Li J.(2015). Strigolactone signaling in Arabidopsis regulates shoot development by targeting D53-Like SMXL repressor proteins for ubiquitination and degradation. Plant Cell 27: 3128–3142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Waters M.T., Smith S.M.(2018a). Karrikin-KAI2 signalling provides Arabidopsis seeds with tolerance to abiotic stress and inhibits germination under conditions unfavourable to seedling establishment. New Phytol. 219: 605–618. [DOI] [PubMed] [Google Scholar]
- Wang Y., Li J., Deng X.-W., Zhu D.(2018b). Arabidopsis noncoding RNA modulates seedling greening during deetiolation. Sci. China Life Sci. 61: 199–203. [DOI] [PubMed] [Google Scholar]
- Waters M.T., Gutjahr C., Bennett T., Nelson D.C.(2017). Strigolactone signaling and evolution. Annu. Rev. Plant Biol. 68: 291–322. [DOI] [PubMed] [Google Scholar]
- Waters M.T., Nelson D.C., Scaffidi A., Flematti G.R., Sun Y.K., Dixon K.W., Smith S.M.(2012). Specialisation within the DWARF14 protein family confers distinct responses to karrikins and strigolactones in Arabidopsis. Development 139: 1285–1295. [DOI] [PubMed] [Google Scholar]
- Waters M.T., Scaffidi A., Moulin S.L., Sun Y.K., Flematti G.R., Smith S.M.(2015). A Selaginella moellendorffii ortholog of KARRIKIN INSENSITIVE2 functions in Arabidopsis development but cannot mediate responses to karrikins or strigolactones. Plant Cell 27: 1925–1944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters M.T., Smith S.M.(2013). KAI2- and MAX2-mediated responses to karrikins and strigolactones are largely independent of HY5 in Arabidopsis seedlings. Mol. Plant 6: 63–75. [DOI] [PubMed] [Google Scholar]
- Weissgerber T.L., Milic N.M., Winham S.J., Garovic V.D.(2015). Beyond bar and line graphs: Time for a new data presentation paradigm. PLoS Biol. 13: e1002128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y., Miyakawa T., Nakamura H., Nakamura A., Imamura Y., Asami T., Tanokura M.(2016). Structural basis of unique ligand specificity of KAI2-like protein from parasitic weed Striga hermonthica. Sci. Rep. 6: 31386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C.J., Zhang C., Lu Y.N., Jin J.Q., Wang X.L.(2011). The mechanisms of brassinosteroids’ action: from signal transduction to plant development. Mol. Plant 4: 588–600. [DOI] [PubMed] [Google Scholar]
- Yao J., Mashiguchi K., Scaffidi A., Akatsu T., Melville K.T., Morita R., Morimoto Y., Smith S.M., Seto Y., Flematti G.R., Yamaguchi S., Waters M.T.(2018a). An allelic series at the KARRIKIN INSENSITIVE 2 locus of Arabidopsis thaliana decouples ligand hydrolysis and receptor degradation from downstream signalling. Plant J. 96: 75–89. [DOI] [PubMed] [Google Scholar]
- Yao R., Li J., Xie D.(2018b). Recent advances in molecular basis for strigolactone action. Sci. China Life Sci. 61: 277–284. [DOI] [PubMed] [Google Scholar]
- Yao R., et al. (2016). DWARF14 is a non-canonical hormone receptor for strigolactone. Nature 536: 469–473. [DOI] [PubMed] [Google Scholar]
- Yao R., Wang F., Ming Z., Du X., Chen L., Wang Y., Zhang W., Deng H., Xie D.(2017). ShHTL7 is a non-canonical receptor for strigolactones in root parasitic weeds. Cell Res. 27: 838–841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi R., Yan J., Xie D. (2020). Light promotes jasmonate biosynthesis to regulate photomorphogenesis in Arabidopsis. Sci. China Life Sci. 10.1007/s11427–019–1584–4. [DOI] [PubMed] [Google Scholar]
- Yoneyama K., Xie X., Yoneyama K., Kisugi T., Nomura T., Nakatani Y., Akiyama K., McErlean C.S.P.(2018). Which are the major players, canonical or non-canonical strigolactones? J. Exp. Bot. 69: 2231–2239. [DOI] [PubMed] [Google Scholar]
- Yoo S.D., Cho Y.H., Sheen J.(2007). Arabidopsis mesophyll protoplasts: A versatile cell system for transient gene expression analysis. Nat. Protoc. 2: 1565–1572. [DOI] [PubMed] [Google Scholar]
- Zhao L.H., et al. (2013a). Crystal structures of two phytohormone signal-transducing α/β hydrolases: Karrikin-signaling KAI2 and strigolactone-signaling DWARF14. Cell Res. 23: 436–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Q., Tian M., Li Q., Cui F., Liu L., Yin B., Xie Q.(2013b). A plant-specific in vitro ubiquitination analysis system. Plant J. 74: 524–533. [DOI] [PubMed] [Google Scholar]
- Zhou F., et al. (2013). D14-SCF(D3)-dependent degradation of D53 regulates strigolactone signalling. Nature 504: 406–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Z., Lin C.(2016). Photomorphogenesis: When blue meets red. Nat. Plants 2: 16019. [DOI] [PubMed] [Google Scholar]