Ribosome biogenesis defects and stress conditions trigger a transcriptional regulatory pathway that causes a cell fate switch in the root epidermis.
Abstract
The Arabidopsis (Arabidopsis thaliana) root epidermis consists of a position-dependent pattern of root hair cells and non-hair cells. Underlying this cell type patterning is a network of transcription factors including a central MYB-basic helix-loop-helix-WD40 complex containing WEREWOLF (WER), GLABRA3 (GL3)/ENHANCER OF GLABRA3, and TRANSPARENT TESTA GLABRA1. In this study, we used a genetic enhancer screen to identify apum23-4, a mutant allele of the ribosome biogenesis factor (RBF) gene ARABIDOPSIS PUMILIO23 (APUM23), which caused prospective root hair cells to instead adopt the non-hair cell fate. We discovered that this cell fate switch relied on MYB23, a MYB protein encoded by a WER target gene and acting redundantly with WER. In the apum23-4 mutant, MYB23 exhibited ectopic expression that was WER independent and instead required ANAC082, a recently identified ribosomal stress response mediator. We examined additional RBF mutants that produced ectopic non-hair cells and determined that this cell fate switch is generally linked to defects in ribosome biogenesis. Furthermore, the flagellin peptide flg22 triggers the ANAC082-MYB23-GL2 pathway. Taken together, our study provides a molecular explanation for root epidermal cell fate switch in response to ribosomal defects and, more generally, it demonstrates a novel regulatory connection between stress conditions and cell fate control in plants.
INTRODUCTION
The development of multicellular organisms relies on the appropriate specification of distinct cell types. In the Arabidopsis (Arabidopsis thaliana) root epidermis, the root hair and non-hair cell types are specified in a position-dependent manner (Duckett et al., 1994; Clowes, 2000): epidermal cells adjacent to two underlying cortical cells (in the H position) adopt the root hair cell fate, while those adjacent to only one underlying cortical cell (in the N position) adopt the non-hair cell fate. This simple patterning system has been used as a model to uncover molecular mechanisms responsible for cell fate specification in plants (Masucci et al., 1996; Lee and Schiefelbein, 1999).
Previous genetic and molecular studies have revealed a network of transcription factors underlying root epidermal cell patterning. In N-position cells, WEREWOLF (WER), GLABRA3/ENHANCER OF GLABRA3 (GL3/EGL3), and TRANSPARENT TESTA GLABRA1 (TTG1) form a MYB-basic helix-loop-helix (bHLH)-WD40 complex (Galway et al., 1994; Lee and Schiefelbein, 1999; Bernhardt et al., 2003, 2005). This complex directly promotes transcription of GL2, encoding a homeodomain-leucine zipper transcription factor, and CAPRICE (CPC), encoding an R3-type MYB protein (Ryu et al., 2005; Song et al., 2011). The GL2 protein accumulates in the N-position cells and directly suppresses expression of downstream root hair–promoting genes (Masucci et al., 1996; Bruex et al., 2012; Lin et al., 2015). The CPC protein is able to translocate to the adjacent H-position cells and bind to GL3/EGL3 in competition with WER (Wada et al., 2002; Kurata et al., 2005; Song et al., 2011). In addition, a receptor-like kinase, SCRAMBLED (SCM), preferentially accumulates in H-position cells and further reduces WER-GL3/EGL3-TTG1 complex formation through suppressing WER expression (Kwak et al., 2005; Kwak and Schiefelbein, 2008). As a consequence, GL2 expression is relatively weak in H-position cells, allowing for transcription of root hair–promoting genes and resulting in root hair cell differentiation (Cvrčková et al., 2010; Bruex et al., 2012; Huang et al., 2017).
The preferential accumulation of the WER-GL3/EGL3-TTG1 complex in N-position cells is reinforced by multiple feedback mechanisms (Schiefelbein et al., 2014). One of these feedback mechanisms involves MYB23, a close relative of WER (Stracke et al., 2001; Kang et al., 2009). The WER-GL3/EGL3-TTG1 complex directly promotes MYB23 transcription in N-position cells, and the MYB23 protein is functionally redundant with WER (Kang et al., 2009). Thus, MYB23 acts in a positive feedback loop to ensure sufficient levels of the WER/MYB23 proteins for the MYB-bHLH-WD40 complex in N-position cells.
The proper differentiation of the root hair and non-hair cells, like essentially all developmental processes, relies on the production and function of ribosomes. Ribosome biogenesis, including precursor rRNA (pre-rRNA) processing and ribosomal protein (RP) assembly, involves the organized cooperation of numerous ribosome biogenesis factors (RBFs) (Thomson et al., 2013; Weis et al., 2015a; Sáez-Vásquez and Delseny, 2019). In Arabidopsis, mutants of RBF genes have significant developmental impacts, including embryo lethality, aborted gametophyte development, and tissue regeneration defects (Harscoët et al., 2010; Ohbayashi et al., 2011; Missbach et al., 2013), as well as milder phenotypes such as retarded plant growth, merged or triple cotyledons, and narrow and pointed rosette leaves (Lange et al., 2011; Weis et al., 2014, 2015b). Interestingly, several characteristic phenotypes, such as the misshaped rosette leaves, are shared by mutants of functionally unrelated RBFs (Weis et al., 2015a), suggesting a common regulatory mechanism that responds to a variety of ribosome biogenesis defects and modulates plant development.
Recently, the anac082-1 mutant, a missense mutation of the NAC family gene ANAC082, was reported to rescue the regeneration defects and the pointed-leaf phenotypes of several RBF mutants (Ohbayashi et al., 2017). Interestingly, the rescued double mutants still exhibit impaired pre-rRNA processing similar to the corresponding RBF single mutants (Ohbayashi et al., 2017), which implies that these developmental phenotypes are not directly caused by defective ribosome biogenesis. Therefore, ANAC082 is considered to be a key component of a regulatory response pathway in plants that connects ribosomal status with specific developmental events (Ohbayashi et al., 2017; Salomé, 2017; Ohbayashi and Sugiyama, 2018; Sáez-Vásquez and Delseny, 2019).
A linkage between root epidermal cell specification and ribosome biogenesis was first discovered through analysis of the RBF gene ADENOSINE DIMETHYL TRANSFERASE1A (DIM1A; Wieckowski and Schiefelbein, 2012). The DIM1A protein participates in N-6 dimethylation of the A1785 and A1786 bases in 18S rRNA (Wieckowski and Schiefelbein, 2012). The dim1a mutant exhibits ∼20% reduction in the number of root hair cells due to an early cell fate switch that leads to ectopic non-hair cell formation (Wieckowski and Schiefelbein, 2012). However, the molecular mechanism underlying this cell fate switch was not determined.
In this study, we identified a nonsense allele of the RBF gene ARABIDOPSIS PUMILIO23 (APUM23), designated as apum23-4. We discovered a significant reduction in root hair formation in apum23-4 resulting from a cell fate switch mediated by abnormal upregulation of MYB23. Interestingly, we found that the increased MYB23 expression in the apum23-4 root epidermis was independent of the WER-GL3/EGL3-TTG1 complex but instead required ANAC082. We also found that other RBF mutants, including dim1a, exhibited a MYB23- and/or ANAC082-dependent root epidermal cell fate switch. Altogether, this study provides evidence for a novel regulatory pathway responsible for altering root epidermal cell fate in response to ribosomal defects.
RESULTS
Identification of the apum23-4 Mutant
The cpc-1 mutant produces ∼30% of the normal number of root hair cells, due to 70% of H-position cells adopting the non-hair cell fate (Figures 1A and 1B). We took advantage of this intermediate phenotype and performed a cpc-1 enhancer screen to identify genes involved in root epidermis specification. One of the resulting lines, later designated as cpc-1 apum23-4, exhibited an enhanced phenotype relative to cpc-1, producing almost hairless roots (Figures 1A and 1B). We isolated plants homozygous for the apum23-4 single mutant and observed several growth abnormalities including delayed seed germination and shorter root hairs (Figure 1A; Supplemental Figure 1). However, the overall root architecture of the apum23-4 mutant is comparable with that of the wild type (Supplemental Figure 2). Furthermore, quantification of root epidermal cell specification in apum23-4 showed that 17% of the H-position cells failed to produce any surface protrusions, neither root hairs nor initial root hair bulges (i.e., 17% ectopic non-hair cells; see Methods; Figure 1B).
Figure 1.
The apum23-4 Mutation Enhances the cpc-1 Mutant Phenotype.
(A) Root hair phenotypes of seedling roots of the wild type (WT), cpc-1, cpc-1 apum23-4, and apum23-4. Bar = 200 μm.
(B) Quantification of root epidermal cell specification in seedling roots of the wild type (WT), cpc-1, cpc-1 apum23-4, and apum23-4. Error bars indicate sds from three replicates. Statistical significance was determined by one-way ANOVA. ***, P < 0.001; **, P < 0.01.
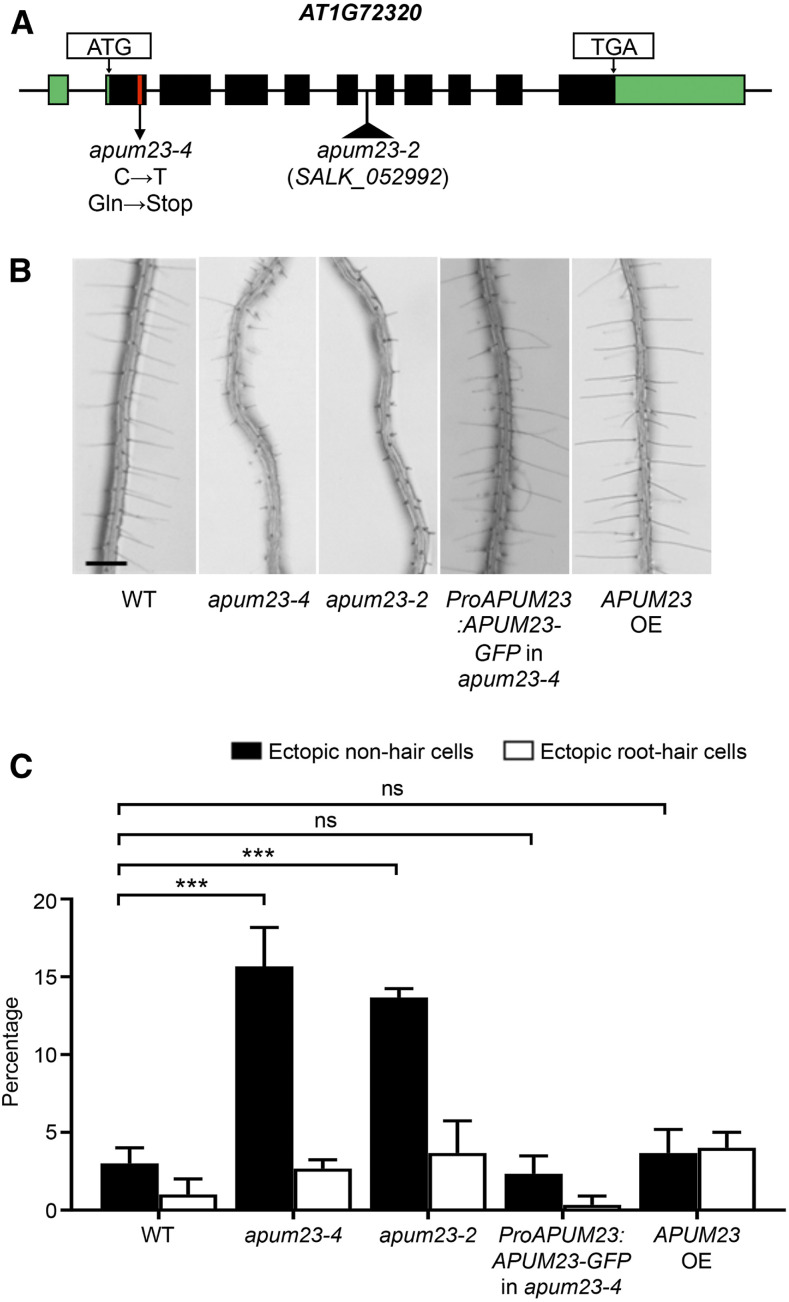
To identify the mutated gene in the apum23-4 line, we performed map-based cloning and discovered a C-to-T single-nucleotide substitution within the first exon of the AT1G72320 gene, which changes the 80th codon from CAG (Glu) to TAG (stop codon; Figure 2A). The AT1G72320 gene is named APUM23 and encodes an RNA binding protein from the Pumilio family (Murata and Wharton, 1995). Pumilio proteins are found in all eukaryotes and defined by the presence of tandem arranged, RNA-recognizing Pumilio and FBF homology (PUF) repeats (Zamore et al., 1997; Edwards et al., 2001), which number from 2 to 11 in members of the Arabidopsis Pumilio family (Francischini and Quaggio, 2009; Tam et al., 2010). Distinct from the canonical Pumilio proteins that mediate translational regulation largely through binding to the 3′ untranslated region (UTR) of mRNAs (Wickens et al., 2002; Szostak and Gebaue, 2013; Wang et al., 2018), APUM23, like its well-studied putative yeast (Saccharomyces cerevisiae) ortholog NOP9 (Thomson et al., 2007; Zhang et al., 2016), binds to rRNAs and contributes to pre-rRNA processing (Abbasi et al., 2010; Zhang and Muench, 2015).
Figure 2.
The apum23-4 Mutant Possesses a Nonsense Mutation in the APUM23 Gene.
(A) Schematic drawing of the APUM23 (AT1G72320) gene, indicating the position of the mutated nucleotide in the apum23-4 mutant. The green boxes indicate 5′ and 3′ UTRs, the black boxes indicate exons, and the black lines indicate introns. The single-base substitution in apum23-4 is indicated by the red line. The position of the T-DNA insertion in the apum23-2 (SALK_052992) mutant is indicated by the black triangle.
(B) Root hair phenotypes of seedling roots of the wild type (WT), apum23-4, apum23-2, apum23-4 transformed with the ProAPUM23:APUM23-GFP transgene, and apum23-4 transformed with the Pro35S:APUM23-YFP transgene (APUM23 overexpression [OE]). Bar = 200 μm.
(C) Quantification of root epidermal cell specification in seedling roots of the wild type, apum23-4, apum23-2, apum23-4 transformed with the ProAPUM23:APUM23-GFP transgene, and APUM23 overexpression (OE). Error bars represent sds from three replicates. Statistical significance was determined by one-way ANOVA. ns, not significant; ***, P < 0.001.
Given that three mutant alleles of APUM23 have been reported by Abbasi et al. (2010) and Huang et al. (2014), the allele identified in our study was designated as apum23-4. Other APUM23 mutants were reported to exhibit delayed germination and slower growth (Abbasi et al., 2010; Huang et al., 2014), but no root epidermis analyses were performed. To determine whether the abnormal root epidermal cell specification in apum23-4 was due to the APUM23 mutation, we examined apum23-2 mutant roots and discovered a comparable proportion of ectopic non-hair cells as in apum23-4 (Figures 2A to 2C). We also generated a ProAPUM23:APUM23-GFP transgene containing the APUM23 genomic sequence (including the native promoter) with an in-frame C-terminal GFP tag and introduced this into apum23-4 plants. The resulting transformed plants exhibited the fully restored wild-type root hair length and root epidermal cell pattern (Figures 2B and 2C). These results confirm that the abnormal root epidermal phenotypes in apum23-4 are due to the mutation in the APUM23 gene.
We also performed an APUM23 overexpression analysis by transforming apum23-4 plants with a Pro35S:APUM23-YFP transgene. We observed a wild-type root hair length and root epidermal cell pattern in these transformed plants (Figures 2B and 2C), suggesting that a particular level or cellular distribution of the APUM23 protein is not critical for its role in root epidermal cell specification.
APUM23 Localizes in the Nucleoli of Multiple Root Tissues
To study the accumulation pattern of APUM23, we analyzed the ProAPUM23:APUM23-GFP transgenic plants and discovered APUM23-GFP accumulation in multiple tissues of the developing root (Figure 3A). In the root epidermis, the APUM23-GFP protein accumulated in both H- and N-position cells (Figure 3C).
Figure 3.
APUM23 Localizes to Nucleoli in Multiple Root Tissues.
(A) Accumulation of APUM23-GFP (green) in tissues of the ProAPUM23:APUM23-GFP root. Red represents PI staining for cell boundary visualization. Bar = 25 μm. C, cortex; En, endodermis; Ep, epidermis; S, stele.
(B) Overlay of DAPI staining (blue) and ProFIB1:FIB1-mcherry signals (red) in the root epidermis. Stars indicate H-position cell files. Bar = 10 μm.
(C) Overlay of DAPI staining (blue), ProFIB1:FIB1-mcherry (red), and ProAPUM23:APUM23-GFP (green) in the root epidermis (enlarged portion of the merged image is shown on the right with a non-hair cell file [N] and a hair cell file [H] outlined). Stars indicate H-position cell files. Compared to (B), the root in (C) was only briefly stained with DAPI to enable visualization of cell boundaries. Bar = 50 μm.
(D) Quantification of the ratio of ProFIB1:FIB1-mcherry signals and ProAPUM23:APUM23-GFP signals within root epidermal cells in (C). H- and N-Position cells used for quantification are outlined in white.
To study the subcellular localization of APUM23, we first generated control transgenic plants bearing the mcherry-tagged FIBRILLARIN1 (FIB1) driven by its native promoter (ProFIB1:FIB1-mcherry). FIB1 is a known nucleolar protein participating in pre-rRNA and small nucleolar RNA processing (Pih et al., 2000; Pontvianne et al., 2010; Kalinina et al., 2018). Using 4′,6-diamidino-2-phenylindole (DAPI) staining to distinguish the nucleolus from the nucleoplasm, we verified the nucleolar localization of FIB1-mcherry in root epidermal cells (Figure 3B). We then generated plants bearing both the ProAPUM23:APUM23-GFP and ProFIB1:FIB1-mcherry transgenes, and we observed colocalization of the APUM23-GFP and FIB1-mcherry signals within individual root epidermal cells, indicating the nucleolar localization of APUM23 (Figure 3C).
Detailed examination of APUM23-GFP and FIB1-mcherry accumulation revealed notable features of nucleoli in the developing root epidermis. First, the relative nucleolar size in N-position cells appeared to decrease as cells aged. The N-position nucleoli were of similar size to H-position nucleoli in early meristematic cells, but their relative size decreased in older elongating cells (Figure 3C; Supplemental Figure 3). Second, the ratio between the FIB1-mcherry and APUM23-GFP proteins appeared to increase in N-position cells compared to H-position cells as they aged. The mcherry/GFP signal ratios were comparable between H- and N-position cells in the meristematic zone but diverged in the elongation zone (Figure 3D; Supplemental Figure 3). These observations suggest distinct nucleolar activities between root hair cells and non-hair cells during root epidermis development.
MYB23 Mediates Ectopic Non-Hair Cell Specification in apum23-4
To uncover the mechanisms underlying ectopic non-hair cell formation in the apum23-4 mutant, we first made use of the ProGL2:GUS transcriptional reporter, a marker for early non-hair cell fate establishment (Masucci et al., 1996). In the wild-type roots, ProGL2:GUS exhibited strong preferential expression in N-position cells, while in apum23-4 roots, ectopic GL2 expression was observed in some H-position cells (Figure 4A). Specifically, ∼15% of H-position cells expressed ProGL2:GUS signals in apum23-4 (Figure 4E), a proportion comparable that of the ectopic non-hair cells in apum23-4 (Figure 1B). Additionally, the ectopic non-hair cell formation in apum23-4 is GL2 dependent, given that the gl2-1 apum23-4 double mutant lacked all non-hair cells in both H and N positions (Figure 4B). GL2 has been reported to specify the non-hair cell fate during early cell differentiation by directly repressing multiple root hair transcription factor genes (Ohashi et al., 2003; Lin et al., 2015). Therefore, we conclude that the reduction in root hair cells in the H position of the apum23-4 root epidermis is due to a GL2-dependent cell fate switch from root hair cells to non-hair cells.
Figure 4.
MYB23 Mediates Ectopic Non-Hair Cell Fate in the apum23-4 Mutant through Upregulating GL2.
(A) Expression of ProGL2:GUS in seedling root tips of wild type (WT) and various mutant plants. Stars indicate H-position cell files. Bar = 50 μm.
(B) Quantification of epidermal cell specification in seedling roots of wild-type (WT) and mutant plants. Error bars represent sds of three replicates. Statistical significance was determined by one-way ANOVA. ns, not significant; ***, P < 0.001; **, P < 0.01; *, P < 0.05.
(C) Expression of ProGL2:GUS in seedling root tips of apum23-4 and apum23-4 myb23-1 plants. Stars indicate H-position cell files. Bar = 50 μm.
(D) Quantification of epidermal cell specification in seedling roots of wild-type (WT), apum23-4, and apum23-4 myb23-1 plants. Error bars represent sds of three replicates. Statistical significance was determined by one-way ANOVA. ns, not significant; ***, P < 0.001.
(E) Quantification of ProGL2:GUS signals in seedling root tips of wild-type (WT), apum23-4, and apum23-4 myb23-1 plants. Ectopic ProGL2:GUS upregulation refers to H-position cells exhibiting ProGL2:GUS expression, and ectopic ProGL2:GUS downregulation refers to N-position cells lacking ProGL2:GUS expression. Error bars represent sds of three replicates. Statistical significance was determined by one-way ANOVA. ns, not significant; **, P < 0.01; *, P < 0.05.
GL2 expression in the root epidermis is controlled by a MYB-bHLH-WD40 complex consisting of WER, GL3/EGL3, and TTG1, and the absence of any of these three components leads to loss of both GL2 expression and non-hair cells (Figures 4A and 4B; Galway et al., 1994; Lee and Schiefelbein, 2002; Bernhardt et al., 2003). To analyze the role of these components for the ectopic GL2 expression in apum23-4, we separately introduced wer-1, gl3 egl3, and ttg1 mutations into the apum23-4 ProGL2:GUS line. The gl3 egl3 apum23-4 and ttg1 apum23-4 mutants lacked significant ProGL2:GUS expression in the developing root epidermis (Figure 4A) and produced nearly 100% root hair cells in both the H and N positions (Figure 4B). However, the wer-1 apum23-4 double mutant exhibited considerable ProGL2:GUS expression that greatly exceeded the wer-1 single mutant (Figure 4A). Interestingly, the ProGL2:GUS signals in wer-1 apum23-4 lacked N-position specificity (∼20% of H-position cells and 31% of N-position cells expressed ProGL2:GUS; Supplemental Figure 4) and initiated accumulation in older cells (relative to the wild type; Figure 4A). Consistent with its ProGL2:GUS expression pattern, the wer-1 apum23-4 mutant produced ∼20 and 35% non-hair cells in the H and N positions, respectively (Figure 4B).
The results described above suggest that GL3/EGL3 and TTG1 are required, while WER is not required, for the ectopic non-hair cell formation in apum23-4. Therefore, we hypothesized that (an)other MYB protein(s) functions in place of WER in the apum23-4 background to form a MYB-bHLH-WD40 complex and induce GL2 expression to generate non-hair cells. MYB23 was a candidate for this role given its known root epidermis expression and close functional relationship to WER, although the myb23-1 single mutant had no significant defects in root epidermal cell patterning (Kang et al., 2009). We introduced the myb23-1 mutation into the wer-1 apum23-4 background and discovered that the resulting triple mutant lacked ProGL2:GUS expression and essentially lacked non-hair cells (Figures 4A and 4B). Furthermore, we generated the apum23-4 myb23-1 double mutant and observed a significantly reduced proportion of ectopic non-hair cells and ectopic ProGL2:GUS-expressing cells, relative to the apum23-1 single mutant (both reduced to <5%; Figures 4C to 4E). Therefore, MYB23 is required for the ectopic GL2 expression and non-hair cell fate specification in the apum23-4 mutant.
Abnormal MYB23 Expression in apum23-4
In the wild-type roots, MYB23 is preferentially expressed in the N-position cells of the developing root epidermis (Kang et al., 2009). To examine its expression in apum23-4, we used the ProMYB23:GUS reporter (Kang et al., 2009) and observed considerable ectopic β-glucuronidase (GUS) expression in the H-position cells compared to the wild type (Figure 5A). Specifically, 13% of the H-position cells in apum23-4 showed detectable GUS signals (Supplemental Figure 5), a proportion comparable to that of ectopic non-hair cells in apum23-4 (Figure 1B).
Figure 5.
apum23-4 Mutant Exhibits Ectopic MYB23 Gene Expression.
(A) Expression of ProMYB23:GUS in the seedling root epidermis of the wild type (WT), apum23-4, wer-1, and wer-1 apum23-4. Stars indicate H-position cell files. Bar = 50 μm.
(B) Expression of ProMYB23:MYB23-GFP in the seedling root epidermis of the wild type (WT), apum23-4, and wer-1 apum23-4. Red represents PI staining and green represents MYB23-GFP signal. Stars indicate H-position cell files. Bar = 50 μm.
(C) Expression of ProMYB23:MYB23-GFP (left in each panel) and ProGL2:GUS (right in each panel) in one single seedling root tip of the wild type (WT), apum23-4, and wer-1 apum23-4. Stars indicate H-position cell files. Bar = 20 μm.
(D) Expression of ProGL3:GL3-YFP (left in each panel) and ProGL2:GUS (right in each panel) in one single seedling root tip of the wild type (WT) and apum23-4. Stars indicate H-position cell files. Bar = 20 μm.
In the wild-type root epidermis, MYB23 transcription is directly induced by the WER-GL3/EGL3-TTG1 complex, and loss of WER gene function eliminates MYB23 expression (Figure 5A; Kang et al., 2009). However, we observed substantial ProMYB23:GUS expression in wer-1 apum23-4, and these GUS signals lacked N-position specificity (Figure 5A), which resembled the expression of ProGL2:GUS in wer-1 apum23-4 (Figure 4A).
To test whether the observed ectopic MYB23 expression leads to ectopic MYB23 protein accumulation, we made use of a ProMYB23:MYB23-GFP translational reporter (Kang et al., 2009). We discovered that MYB23-GFP protein accumulated ectopically in the nuclei of H-position cells of apum23-4 as well as in both H- and N-position cells of wer-1 apum23-4 (Figure 5B), which was consistent with the ProMYB23:GUS results (Figure 5A). Next, to determine whether these MYB23-GFP–accumulating cells are also expressing GL2, we examined the roots of apum23-4 and wer-1 apum23-4 plants bearing both the ProMYB23:MYB23-GFP and ProGL2:GUS reporters. In both the apum23-4 and wer-1 apum23-4 lines, we observed a correspondence between MYB23-GFP accumulation and ProGL2:GUS expression within individual root epidermal cells (Figure 5C). Additionally, we generated apum23-4 plants bearing both the ProGL3:GL3-YFP (Bernhardt et al., 2005) and ProGL2:GUS reporters, and we found a similar correspondence between these two reporter signals in root epidermal cells (Figure 5D), supporting the notion that MYB23 induces GL2 expression through its association with GL3.
In summary, we demonstrated a spatial correspondence between MYB23 accumulation and GL2 expression in apum23-4, suggesting that MYB23 upregulation in apum23-4 causes abnormal spatial expression of GL2 and, ultimately, ectopic non-hair cells. More importantly, our finding that ectopic MYB23 expression in apum23-4 is WER independent suggests a novel mechanism for upregulating MYB23 in the apum23-4 background.
ANAC082 Is Required for MYB23-Mediated Ectopic Non-Hair Cell Specification in apum23-4
Recently, the NAC family member ANAC082 was identified as a plant-specific mediator of ribosomal stress responses, given that the anac082-1 mutant markedly reversed several developmental abnormalities in RBF mutants (Ohbayashi et al., 2017). Therefore, we sought to test whether the ectopic non-hair cells in apum23-4 are ANAC082 dependent.
First, we generated the anac082-1 apum23-4 double mutant and observed substantial recovery of the seed germination and growth rate compared to apum23-4 (Supplemental Figure 1), indicating that these phenotypes are ANAC082 dependent. Furthermore, anac082-1 apum23-4 produced <3% ectopic non-hair cells, which is comparable to the wild type (Figure 6B). This result was confirmed using a different anac082 mutant (a T-DNA insertion mutation, GABI_282H08), which also reversed the apum23-4 root hair pattern (Supplemental Figure 6A). Consistent with these observations, <3% of the H-position cells in anac082-1 apum23-4 expressed the ProGL2:GUS reporter (Figure 6A; Supplemental Figure 6B). In addition, we found that the anac082-1 wer-1 apum23-4 triple mutant restored the wer-1 mutant phenotype, essentially lacking ProGL2:GUS expression in the root epidermis and producing >95% root hair cells in both H- and N-epidermis positions (Figures 6A and 6B). Notably, we observed no effect of the anac082-1 or GABI_282H08 mutant alone on root epidermis development; each single mutant exhibited a wild-type pattern of root epidermal cell types and ProGL2:GUS expression (Figures 6A and 6B; Supplemental Figure 6A). Therefore, ANAC082 has no significant role in root epidermal cell patterning under normal ribosome biogenesis conditions but mediates ectopic non-hair cell specification in the apum23-4 background.
Figure 6.
ANAC082 Is Required for Ectopic Non-Hair Cells in the apum23-4 Mutant.
(A) Expression of ProGL2:GUS in the seedling root epidermis of the wild type (WT) and multiple mutants. Stars indicate H-position cell files. Bar = 50 μm.
(B) Quantification of root epidermal cell specification in the seedling roots of the wild type (WT) and multiple mutants. Error bars represent sds from three replicates. Statistical significance was determined by one-way ANOVA. ns, not significant; ***, P < 0.001; **, P < 0.01.
(C) Expression of ProMYB23:GUS in the seedling root epidermis of the wild type (WT) and multiple mutants. Stars indicate H-position cell files. Bar = 50 μm.
(D) Expression of ProMYB23:MYB23-GFP in the seedling root epidermis of the wild type (WT) and multiple mutants. Red indicates propidium iodide staining and green indicates MYB23-GFP signals. Stars indicate H-position cell files. Bar = 50 μm.
Given that both ANAC082 and MYB23 are required for ectopic GL2 expression and ectopic non-hair cell production in apum23-4, we examined the possible regulatory relationship between MYB23 and ANAC082. First, we examined ProMYB23:GUS expression in anac082-1 apum23-4 and discovered that the ectopic MYB23 expression in the H-position cells of apum23-4 is ANAC082 dependent (Figure 6C; Supplemental Figure 6C). Similarly, the substantial ProMYB23:GUS expression in the wer-1 apum23-4 mutant was found to be anac082-1 dependent (Figure 6C). Consistent with these results, the ectopic ProMYB23:MYB23-GFP signals in apum23-4 and wer-1 apum23-4 were depleted by the anac082-1 mutation (Figure 6D). Notably, the anac082-1 single mutant exhibited no effect on MYB23 expression (Figure 6C). Taken together, these results suggest that ANAC082 induces MYB23 expression to cause ectopic GL2 expression and switch epidermal cell fate in the apum23-4 mutant.
ANAC082 belongs to the NAC domain transcription factor family (Ooka et al., 2003) and has been reported to have transcriptional activity when expressed in yeast (Ohbayashi et al., 2017). To determine whether ANAC082 might directly activate the transcription of MYB23, we adopted a dual-luciferase reporter system to assess MYB23 promoter activity by overexpressed ANAC082 in a tobacco (Nicotiana tabacum) transient expression system (Supplemental Figures 7A and 7B). We found no significant MYB23 promoter activation by ANAC082, indicating that ANAC082 alone is not sufficient to induce MYB23 expression in this system. This could be due to the lack of cofactors of ANAC082 or intermediate regulators between ANAC082 and MYB23. Therefore, it remains to be determined whether ANAC082 mediates MYB23 expression via direct or indirect regulation.
ANAC082 expression is reported to be elevated in root tissues of several RBF mutants (Ohbayashi et al., 2017). Moreover, microarray analyses indicate ANAC082 upregulation in whole adult apum23-4 plants (Abbasi et al., 2010). To test whether ANAC082 is upregulated in apum23-4 seedling root tips, we performed quantitative real-time PCR (Figure 7A). Indeed, ANAC082 transcript accumulation was significantly greater in the root tips of apum23-4 mutants than the wild-type plants.
Figure 7.
ANAC082 Is Upregulated in apum23-4 Mutant and CHX-Treated Seedling Roots.
(A) Relative amounts of ANAC082 transcripts in root tips of the wild-type (WT) and apum23-4 plants, determined by quantitative real-time PCR. Error bars indicate sds from three replicates. Statistical significance was determined by t test. **, P < 0.01.
(B) Relative amounts of ANAC082 transcripts in root tips of the wild-type (WT) and wer-1 plants treated with different concentrations of CHX (0, 0.05, 0.5, and 5 μg/mL). Error bars indicate sds from three replicates. For both the wild type (WT) and wer-1, statistical significance between each CHX concentration and mock treatment (0 μg/mL) was determined by one-way ANOVA. ns, represents not significant; ***, P < 0.001.
Multiple RBF Mutants Exhibit Ectopic Non-Hair Cells
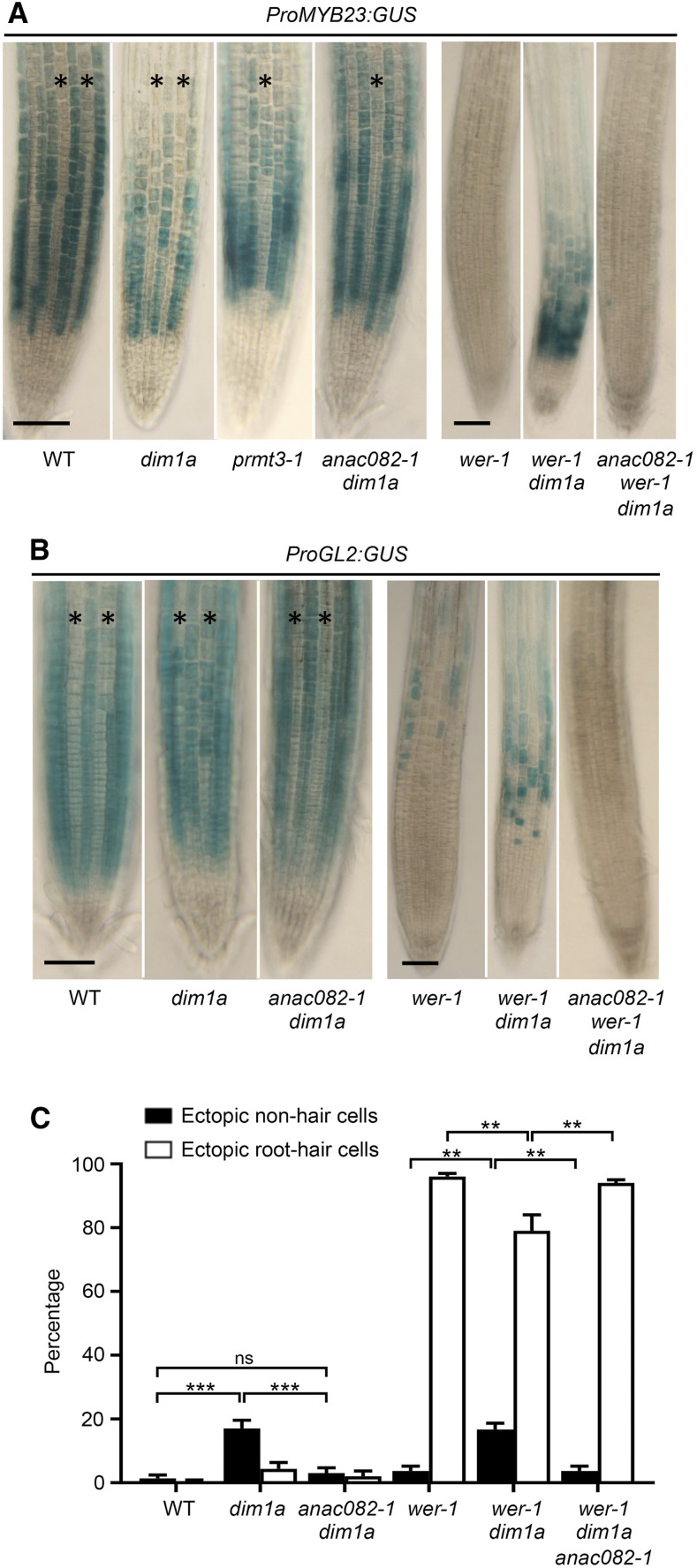
The apum23-4 mutant phenotype analyzed in this study is reminiscent of dim1a, a previously reported RBF mutant exhibiting ectopic ProGL2:GUS expression (Figure 8A; Wieckowski and Schiefelbein, 2012). Furthermore, like wer-1 apum23-4, the wer-1 dim1a double mutant exhibited significant ProGL2:GUS expression and non-hair cells in both H- and N-cell positions (Figures 8A and 8B; Supplemental Figure 4; Wieckowski and Schiefelbein, 2012). To test whether MYB23 plays the same role in the dim1a phenotype as it does in apum23-4, we generated the wer-1 myb23-1 dim1a and myb23-1 dim1a mutants. The wer-1 myb23-1 dim1a triple mutant exhibited no significant ProGL2:GUS signals and ≥95% root hair cells in both H and N positions (Figures 8A and 8B), and the myb23-1 dim1a double mutant exhibited a significantly decreased proportion of ectopic non-hair cells compared to dim1a (Figure 8C). This shows that MYB23 is required for the ectopic non-hair cell formation in dim1a.
Figure 8.
dim1a and prmt3-1 Mutants Exhibit MYB23-Dependent Ectopic Non-Hair Cell Fate Specification.
(A) Expression of ProGL2:GUS in the seedling root epidermis of the wild type (WT) and multiple mutants. Stars represent H-position cell files. Bar = 50 μm.
(B) Quantification of epidermal cell specification in seedling roots of the wild type (WT) and multiple mutants. Error bars represent sds from three replicates. Statistical significance was determined by one-way ANOVA. ***, P < 0.001.
(C) Quantification of epidermal cell specification in seedling roots of the wild type (WT) and multiple mutants. Error bars represent sds from three replicates. Statistical significance was determined by one-way ANOVA. ***, P < 0.001 and.
We also incorporated the ProMYB23:GUS reporter into the dim1a mutant and observed ectopic GUS signals in H-position cells at a frequency comparable to apum23-4 (Figure 9A; Supplemental Figure 5). Furthermore, the wer-1 dim1a double mutant exhibited substantial ProMYB23:GUS signals (Figure 9A), indicating WER-independent MYB23 upregulation similar to the apum23-4 mutant.
Figure 9.
ANAC082 Is Required for Ectopic Non-Hair Cells in the dim1a Mutant.
(A) Expression of ProMYB23:GUS in the seedling root epidermis of wild type (WT) and multiple mutants. Stars indicate H-position cell files. Bar = 50 μm.
(B) Expression of ProGL2:GUS in the seedling root epidermis of the wild type (WT) and multiple mutants. Stars indicate H-position cell files. Bar = 50 μm.
(C) Quantification of root epidermal cell specification in seedling roots of the wild type (WT) and multiple mutants. Error bars represent sds from three replicates. Statistical significance was determined by one-way ANOVA. ns, not significant; ***, P < 0.001; **, P < 0.01.
Next, we tested the effect of ANAC082 on the dim1a phenotype. We introduced the anac082-1 mutation into dim1a mutant lines carrying ProMYB23:GUS or ProGL2:GUS reporters and found that each reporter exhibited the wild-type expression pattern in the dim1a anac082-1 background (Figures 9A and 9B; Supplemental Figures 6B and 6C). Consistently, both dim1a anac082-1 and dim1a GABI_282H08 mutants restored the wild-type root epidermal cell patterning (Figure 9C; Supplemental Figure 5A). In addition, anac082-1 eliminated expression of the ProMYB23:GUS and ProGL2:GUS reporters, as well as non-hair cell production, in the wer-1 dim1a double mutant (Figures 9A to 9C). Taken together, these results show that ANAC082 plays a similar role of inducing MYB23-dependent GL2 expression and cell fate switching in both the dim1a and apum23-4 mutants.
Although the apum23 and dim1a mutants exhibit similar root epidermis phenotypes, the APUM23 and DIM1A proteins have distinct biochemical functions in ribosome biogenesis. Accordingly, we hypothesized that the ectopic production of non-hair cells may be a general response to defective ribosome biogenesis. To test this, we examined a collection of previously defined RBF mutants (Supplemental Table 1; Weis et al., 2015a). Among these, we discovered that mutations of the PROTEIN ARGININE METHYLTRANSFERASE3 (PRMT3) gene, prmt3-1 and prmt3-2, exhibited ectopic non-hair cells similar to apum23-4 and dim1a (Figure 8B). The prmt3-1 mutant also exhibited delayed germination (Supplemental Figure 1). During rRNA biogenesis, PRMT3 influences the balance between two alternative pre-rRNA processing pathways (Hang et al., 2014).
To study the cause for the ectopic non-hair cells in prmt3-1, we introduced the ProGL2:GUS reporter and found ∼20% of the H-position cells exhibited ectopic ProGL2:GUS expression (Figure 8A; Supplemental Figure 8). Furthermore, the wer-1 prmt3-1 double mutant exhibited substantial ProGL2:GUS expression and produced non-hair cells in both H and N positions, demonstrating a WER-independent effect (Figures 8A and 8B; Supplemental Figure 4). A role for MYB23 in the prmt3-1 phenotype was shown by the elimination of ProGL2:GUS expression and non-hair cell production in the wer-1 myb23-1 prmt3-1 plants, relative to the wer-1 prmt3-1 double mutant (Figures 8A and 8B), and the significant decrease in ectopic non-hair cells in the prmt3-1 myb23-1 double mutant compared to prmt3-1 (Figure 8C). Finally, the prmt-1 mutant exhibited a comparable proportion of H-position cells expressing ProMYB23:GUS as the apum23-4 and dim1a (Figure 9A; Supplemental Figure 5). Therefore, like apum23-4 and dim1a, the ectopic non-hair cell specification in prmt3-1 is mediated by MYB23.
In summary, we identified two additional RBF mutants (dim1a and prmt3-1) exhibiting ectopic non-hair cells likely resulting from similar misregulation of epidermal cell fate as in apum23-4. These findings support the hypothesis that epidermal cell fate switching is a general response to ribosomal defects rather than a particular RBF deficiency.
CHX Treatment Induces WER-Independent GL2 Expression
In addition to genetic disturbances in RBF genes, drug-induced ribosomal defects can also trigger ectopic establishment of non-hair cell fates. Cycloheximide (CHX), widely used as a translation inhibitor, disrupts pre-rRNA processing in yeast and mammals (de Kloet, 1966; Stoyanova and Hadjiolov, 1979). Interestingly, a treatment of 5 μg/mL CHX was reported to induce ectopic GL2 expression in a significant portion of H-position cells in the wild-type root epidermis (Wieckowski and Schiefelbein, 2012), an effect similar to the RBF mutants. We further analyzed the effect of CHX treatment on root epidermis development by monitoring ProGL2:GUS expression in the wer-1 mutant. Using a series of CHX concentrations, including 5 μg/mL, we found that ProGL2:GUS expression was induced in a WER-independent manner in both H- and N-position cells (Figure 10). Notably, the wer-1 myb23-1 and wer-1 anac082-1 double mutants exhibited no ProGL2:GUS upregulation in response to CHX treatments (Figure 10), implying that the effect of CHX also relies on the ANAC082-MYB23 regulatory module that operates in the RBF mutants.
Figure 10.
MYB23 and ANAC082 Mediate WER-Independent GL2 Upregulation Triggered by CHX Treatment.
Expression of ProGL2:GUS in the root epidermis of wer-1, wer-1 myb23-1, and wer-1 anac082-1 seedlings treated with a series of concentrations of CHX. Bar = 50 μm.
We also used quantitative real-time PCR to determine whether ANAC082 expression is elevated by CHX treatment (Figure 7B). Compared to the mock treatment (0 μg/mL), both 0.05 and 0.5 μg/mL CHX treatments caused slight ANAC082 upregulation that was not statistically significant, whereas treatment with 5 μg/mL CHX caused significant ANAC082 upregulation.
The apum24-2 Mutant Is a Distinct Type of RBF Mutant with Ectopic Non-Hair Cells
Another Arabidopsis Pumilio protein, APUM24, has been identified as an RBF required for pre-rRNA processing (Shanmugam et al., 2017; Maekawa et al., 2018). As APUM24 knockout mutants were reported to exhibit seed abortion due to defective female gametogenesis and embryogenesis, we analyzed the APUM24 knockdown mutant apum24-2 (Shanmugam et al., 2017; Maekawa et al., 2018). We discovered that apum24-2 mutant roots exhibited shorter root hairs resembling other examined RBF mutants (Figure 11A). Furthermore, the apum24-2 mutant produced a significant proportion of ectopic non-hair cells (Figure 11D) and ectopic ProGL2:GUS signals in the H position (Figure 11B; Supplemental Figure 8). Therefore, like APUM23, knockdown of the APUM24 gene function leads to ectopic non-hair cell specification.
Figure 11.
apum24-2 Mutant Exhibits MYB23-Independent Ectopic Non-Hair Cell Production.
(A) Root hair phenotypes of the wild-type (WT) and apum24-2 seedling roots. Bar = 200 μm.
(B) Expression of ProGL2:GUS in the seedling root epidermis of the wild type (WT) and apum24-2. Stars indicate H-position cell files. Bar = 50 μm.
(C) Expression of ProMYB23:GUS in the seedling root epidermis of the wild type (WT) and apum24-2. Stars indicate H-position cell files. Bar = 50 μm.
(D) Quantification of root epidermal cell specification in wild-type (WT) and various mutant seedlings. Error bars represent sds from three replicates. Statistical significance was determined by one-way ANOVA. ns, not significant; ***, P < 0.001; **, P < 0.01.
(E) Quantification of root epidermal cell specification in seedling roots of multiple mutants. Error bars represent sds from three experiments. Statistical significance was determined by one-way ANOVA. ns, not significant. **, P < 0.01.
(F) Quantification of root epidermal cell specification in seedling roots of wild type (WT), apum24-2, and apum24-2 anac082-1 mutants. Error bars represent sds from three experiments. Statistical significance was determined by one-way ANOVA. ns, not significant; ***, P < 0.001.
Structural studies of the APUM23 yeast ortholog Nop9 and the APUM24 human ortholog Puf-A revealed that the two proteins, although both containing 11 PUF repeats, possess divergent protein structures and nucleotide binding characteristics (Qiu et al., 2014; Zhang et al., 2016). To test the functional relationship between the Arabidopsis APUM23 and APUM24 proteins, we created a ProAPUM23:APUM24 transgene and transformed it into the apum23-4 mutant, and reciprocally, we created a ProAPUM24:APUM23 transgene and transformed it into the apum24-2 mutant. In each case, the resulting transgenic plants exhibited the abnormal root epidermis phenotypes of the original mutant backgrounds (Figure 11D), indicating that APUM23 and APUM24 are functionally distinct.
We then studied the cause for the ectopic non-hair cells in apum24-2. The wer-1 apum24-2 double mutant exhibited no significant non-hair cells in the H position but a considerable proportion of non-hair cells (>30%) in the N position, and the wer-1 myb23-1 apum24-2 mutant showed no significant reduction in the proportion of these non-hair cells (Figure 11E). Furthermore, the apum24-2 myb23-1 mutant produced >10% ectopic non-hair cells, which is not significantly different from apum24-2 (Figure 11D). In addition, the ProMYB23:GUS reporter showed dramatically decreased expression in the apum24-2 root epidermis (Figure 11C). These results indicate that, unlike apum23-4, the ectopic non-hair cells in apum24-2 are MYB23 independent. Consistent with this, the apum23-4 apum24-2 double mutant produced an additive increase of ectopic non-hair cells compared to each of the two single mutants (Figure 11D), suggesting that ectopic non-hair cell production in apum23-4 and apum24-2 is due to separate pathways. Notably, the gl3 egl3 apum24-2 mutant still efficiently depleted all non-hair cells in both H and N positions (Figure 11E). Therefore, the ectopic non-hair cells in the apum24-2 mutant apparently still rely on formation of the MYB-bHLH-WD40 complex. Finally, we tested whether the ectopic non-hair cells in apum24-2, although independent of MYB23, still require ANAC082. Indeed, the apum24-2 anac082-1 double mutant exhibited no significant ectopic non-hair cell production (Figure 11F), indicating that ANAC082 mediates ectopic non-hair formation in the apum24-2 mutant.
Flagellin 22 Peptide Treatment Induces WER-Independent GL2 Expression Mediated by MYB23 and ANAC082
This study revealed that genetic defects in ribosome biogenesis caused by RBF mutants trigger ANAC082-mediated MYB23 expression that then induces GL2 expression and non-hair cell fate establishment. To determine whether this regulatory pathway might be activated by other stresses, we first analyzed the effect of various stress conditions on ANAC082 expression. Interestingly, several public microarray and RNA sequencing data sets indicate that plant pathogen treatments cause ANAC082 upregulation in leaf tissues (Ferrari et al., 2007; Winter et al., 2007). To study whether biotic stresses and the resulting plant defense responses affect root epidermal cell specification, we used flagellin22 (flg22), a peptide corresponding to the conserved domain of bacterial flagellin (Gómez-Gómez et al., 1999), as a plant immune response elicitor. Treatment with 1 μM flg22 caused no significant alterations in root epidermal cell specification in wild-type roots (Figure 12A). However, wer-1 mutants treated with 1 μM flg22 exhibited significant non-hair cell formation in the N position (Figure 12A). Furthermore, ProGL2:GUS expression was increased in the root epidermis of wer-1 mutants treated with 1 μM flg22 (Figure 12B). To determine whether this effect of flg22 on wer-1 is dependent on ANAC082 or MYB23, we repeated the same treatment on the wer-1 myb23-1 and wer-1 anac082-1 double mutants. Interestingly, neither double mutants showed a significant increase in ProGL2:GUS expression or non-hair cell formation (Figures 12A and 12B). Therefore, flg22 treatment induces WER-independent GL2 expression in the root epidermis that requires ANAC082 and MYB23, suggesting the activation of a regulatory pathway that overlaps with that induced by RBF mutants.
Figure 12.
Flg22 Treatment Induces WER-Independent GL2 Expression Mediated by ANAC082 and MYB23.
(A) Quantification of root epidermal cell specification in wild-type (WT), wer-1, wer-1 myb23-1, and wer-1 anac082-1 seedling roots with 1 μM flg22 and without flg22 treatments. Error bars represent sds from three replicates. Statistical significance was determined using one-way ANOVA. ns, not significant; ***, P < 0.001.
(B) ProGL2:GUS expression in wer-1, wer-1 myb23-1, and wer-1 anac082-1 seedling roots with 1 μM flg22 and without flg22 treatments. Bar = 50 μm.
DISCUSSION
A Working Model for Cell Patterning in RBF Mutant Root Epidermis
In this study, we uncovered a regulatory mechanism mediating a cell fate switch in response to defective ribosome biogenesis. Based on our combined analysis of ribosome biogenesis mutants (apum23-4, dim1a, and prmt3-1) and CHX-treated plants that all produced ectopic non-hair cells, we determined that this cell fate switch is the result of aberrant induction of MYB23 gene expression by the ribosomal stress response mediator ANAC082. Therefore, this work provides evidence for a molecular linkage between ribosomal status and cell fate specification in plants.
We suggest a model to explain our findings (Figure 13). During early development of the wild-type root epidermis, expression of the GL2 and MYB23 genes is induced by the WER-GL3/EGL3-TTG1 complex, which predominantly occurs in N-position cells and leads to non-hair cell fate specification (Figure 13A). The expression of GL2 and MYB23 is absent (or low) in the H-position cells due to SCM-dependent inhibition of WER expression and CPC-dependent inhibition of WER-GL3/EGL3-TTG1 complex formation, allowing for root hair cell differentiation in these cells. In apum23-4 (and presumably dim1a, prmt3-1, and CHX-treated plants; Figure 13B), an ANAC082-dependent pathway is activated in response to impaired ribosome biogenesis. In addition to the WER-GL3/EGL3-TTG–induced MYB23 expression in N-position cells, ANAC082 also generates MYB23 expression in both H- and N-position cells via direct or indirect regulation. In the N-position cells, the additional MYB23 expression further supports WER/MYB23-dependent gene regulation. In the H-position cells, the additional MYB23 expression leads to elevated levels of MYB23/WER that, in some cells (∼20%), is sufficient to overcome CPC inhibition and thereby induce GL2 expression and ectopic non-hair cell specification. The other H-position cells might accumulate a level of MYB23 that is higher than normal but below the threshold required to achieve ectopic non-hair cell specification, and so they adopt the hair cell fate instead.
Figure 13.
Working Models for Root Epidermal Cell Fate Regulation in Wild-Type Plants and Plants with Ribosomal Defects.
(A) In the wild-type (WT) root epidermis, the WER-GL3/EGL3-TTG1 complex preferentially accumulates in N-position cells, upregulating expression of MYB23, GL2, and CPC. MYB23 serves in a positive feedback loop to maintain WER function. GL2 promotes non-hair cell fate through repressing root hair genes. CPC is translocated to the adjacent H-position cell, where it inhibits WER’s function by competitively binding to GL3/EGL3. SCM mediates inhibition of WER expression in H-position cells. Arrows indicate positive transcriptional regulation (black, direct; red, direct or indirect). Dashed black arrows indicate protein translocation.
(B) Ribosomal defects caused by RBF mutants or CHX treatment activate ANAC082, which results in additional MYB23 expression in both H and N positions. The additional MYB23 in H-position cells helps to outcompete CPC, leading to excessive functional MYB23-GL3/EGL3-TTG1 complex and ectopic non-hair cell fate. Arrows indicate positive transcriptional regulation (black, direct; red, direct or indirect). Dashed black arrows indicate protein translocation.
The Role of MYB23 in Response to Ribosome Biogenesis Defects
The MYB23 protein is most similar to two other Arabidopsis R2R3-type MYB transcription factors, WER and GL1, and it participates with them to specify cell fates in the root epidermis and shoot epidermis. In the developing shoot epidermis, MYB23 is required for proper trichome branching, and it acts redundantly with GL1 to control trichome initiation (Kirik et al., 2005; Tominaga et al., 2007; Li et al., 2009; Balkunde et al., 2010). In the root epidermis, MYB23 acts redundantly with WER to generate the WER-GL3/EGL3-TTG1 complex responsible for non-hair cell fate specification, although the myb23 single mutant exhibits no significant root epidermis defects (Kang et al., 2009). In this study, we expanded our knowledge of MYB23 function by showing that it mediates ectopic non-hair cell specification in response to defective ribosome biogenesis. Specifically, we showed that RBF mutants (apum23-4, dim1a, and prmt3-1) exhibited MYB23-dependent ectopic non-hair cell production (Figures 4 and 8), and CHX-treated wer-1 roots exhibited MYB23-dependent GL2 expression (Figure 10). It is notable that a functionally redundant player in root epidermis cell specification (MYB23) was recruited for this role, rather than the primary R2R3 MYB regulator (WER), suggesting that the evolution of regulatory pathways may take advantage of duplicate genes.
The plasticity of root epidermal cell specification under challenging conditions is well known. Ectopic non-hair cells can be induced by salt stress (Wang et al., 2008), while ectopic root hair cells can be induced by phosphate or iron deficiencies (Schmidt and Schikora, 2001; Müller and Schmidt, 2004). In this study, we report that ectopic non-hair cell formation is triggered by defects in ribosome biogenesis via activation of MYB23. Interestingly, our discovery is reminiscent of the role of ETC1 in inducing production of ectopic root hair cells upon phosphate deficiency (Rishmawi et al., 2018). Under normal growth conditions, ETC1 functions redundantly with CPC, and the etc1 single mutant exhibits no defects in root epidermis cell patterning (Kirik et al., 2004; Simon et al., 2007). Thus, our study and the ETC1 study show that redundant regulators in the root epidermis cell fate network operate as stress-responding elements to modulate cell fates. Furthermore, these studies raise the possibility that additional regulators in the network may have similar unrecognized roles in controlling root epidermal cell fate in response to various plant stresses.
This study also has implications for our understanding of MYB23 transcriptional regulation. A previous study identified WER binding sites in the MYB23 promoter and showed that WER, GL3/EGL3, and TTG1 were necessary for MYB23 transcription in the developing root epidermis (Kang et al., 2009). Our study showed that MYB23 expression in the root epidermis is WER independent under conditions of impaired ribosome biogenesis and is instead mediated by ANAC082. It is notable that the ANAC082-dependent MYB23 expression occurred in both H- and N-position cells and exhibited a later developmental start point within the distal meristematic zone (Figure 4). These features suggest a novel regulatory module that induces MYB23 expression independent of positional cues and following a different developmental timeline. However, it remains unknown whether ANAC082, a potential transcriptional activator (Yamaguchi et al., 2015; Ohbayashi et al., 2017), induces MYB23 expression directly or indirectly. Our results using a transient dual-luciferase assay (Supplemental Figure 7) suggest that ANAC082 alone is not sufficient to activate MYB23 expression. Therefore, it is likely that ANAC082 either indirectly upregulates MYB23 expression by regulating some intermediate regulators or ANAC082 requires cofactors for MYB23 promoter binding or transactivation. In this respect, it is interesting to note that the NAC domain has dimerization features (Ernst et al., 2004; Olsen et al., 2004) and that ANAC082 interacts with numerous NAC transcription factors, but not itself in vitro (Yamaguchi et al., 2015).
In a series of experiments, we showed that the myb23-1 mutation largely reverses the ectopic non-hair cell defects in the apum23-4, dim1a, and prmt3-1 mutants (Figures 4D, 4E, and C8C), indicating the essential role of MYB23 in establishing ectopic non-hair cell fate in these RBF mutants. Nevertheless, there remains a small proportion (∼5%) of ectopic GL2 expression and non-hair cells in these double mutants. By contrast, the introduction of the anac082-1 mutation into the apum23-4 and dim1a mutants essentially restored all root hair cells in the H positions (Figures 6B and 9C). These results suggest that MYB23 is the major, but not the only, regulator for the ectopic non-hair cell fates in RBF mutants that is induced by ANAC082. It is possible that other R2R3-type MYB proteins are also upregulated in an ANAC082-dependent manner and contribute to ectopic GL2 expression together with MYB23.
Interestingly, among the RBF mutants we analyzed, the apum24-2 mutant was unique in generating ectopic non-hair cells in a MYB23-independent manner. Specifically, the myb23-1 mutation had no significant effect on non-hair cell specification in the wer-1 apum24-2 or apum24-2 mutants (Figure 11). However, these ectopic non-hair cells still require ANAC082 (Figure 11F). Thus, these findings suggest the existence of multiple regulatory mechanisms downstream of ANAC082 to mediate the effect of impaired ribosome biogenesis on root epidermal cell specification.
Ribosome Biogenesis and Plant Development
Ribosomes are necessary for protein synthesis, and developmental processes in general rely on efficient ribosome biogenesis. In plants, multiple studies have shown that mutants of functionally unrelated RBF genes exhibit similar alterations in cotyledon, leaf, and root development (Weis et al., 2015a; Sáez-Vásquez and Delseny, 2019). Accordingly, defective ribosome biogenesis of different origins appears to impact plant development through a common regulatory pathway (Byrne, 2009; Weis et al., 2015a). It is proposed that the ribosomal abnormalities in various RBF/RP mutants, including insufficient ribosome production and aberrant/unbalanced heterogeneity of ribosome components, differentially affect the translation of certain developmental regulator gene transcripts (Horiguchi et al., 2012). Indeed, the translation of several auxin response factors is modulated by particular RPs through the upstream open reading frame (uORF) in their 5′ UTRs (Rosado et al., 2012). ANAC082 has been identified as the mediator of several developmental phenotypes in RBF/RP mutants, connecting ribosomal health with a spectrum of developmental events (Ohbayashi et al., 2017; Ohbayashi and Sugiyama, 2018; Salomé, 2017). Notably, the ANAC082 transcript possesses a uORF and therefore is potentially subject to translational regulation (Ohbayashi et al., 2017; Salomé, 2017). In addition, studies using the ProANAC082:GUS transcriptional reporter (Ohbayashi et al., 2017) and microarray analyses (Abbasi et al., 2010) as well as the quantitative real-time PCR reported in this study (Figure 7) suggest that ANAC082 is also transcriptionally regulated in RBF mutants.
Ribosomal defects result from not only RBF/RP mutations but also challenging conditions such as nutrient deprivation, heat shock, and hypoxia (Mayer and Grummt, 2005; Golomb et al., 2014). In animal cells, ribosomal defects trigger ribosomal stress responses mediated by p53 activation and lead to cell cycle arrest and apoptosis (Zhang and Lu, 2009; Golomb et al., 2014; Penzo et al., 2019). As a potential plant version of this p53 pathway, ribosomal defects in plants lead to increased and/or activated ANAC082, which blocks tissue regeneration and delays seed germination (Supplemental Figure 1; Ohbayashi et al., 2017). Both tissue regeneration and seed germination involve massive cell proliferation and cell growth that rely heavily on ribosome activities. Therefore, the ANAC082-mediated effects on these processes could be a programmed response to contend with ribosomal defects.
In this study, we discovered that a switch of root epidermal cell fate is a common characteristic of several RBF mutants (apum23, dim1A, prmt3, and apum24) and plants treated with CHX (Figures 1, 8, and 11). However, these RBFs have been reported to have distinct and unrelated functions during rRNA splicing and rRNA modification (Abbasi et al., 2010; Wieckowski and Schiefelbein, 2012; Hang et al., 2014; Shanmugam et al., 2017), suggesting that this root epidermal phenotype is not due to specific molecular functions of these RBFs but is a developmental alteration commonly induced by ribosomal defects. The biological rationale for reducing root hair cell production in response to ribosomal defects is unclear. One possibility is that root hair cell differentiation requires a relatively high level of ribosome activity. It has been observed that during early developmental stages, cells in the H position show greater cell division rates, higher cytoplasmic densities, and delayed vacuolation compared with N-position cells (Galway et al., 1994; Berger et al., 1998). Furthermore, in this study, we discovered that H-position cells maintained larger nucleolar sizes and relatively greater amounts of APUM23 during later developmental stages (Figure 3). All these features suggest that developing H-position cells, committed to root hair production, are more metabolically active and so might have a greater demand for ribosomes. Accordingly, we hypothesize that a switch from root hair to non-hair cell fate may be part of a response program to accommodate for ribosomal defects.
Finally, we showed that flg22 treatment mimicked the effects of the RBF mutant by triggering ANAC082- and MYB23-dependent GL2 expression in a WER-independent manner (Figure 12). This implies that a common regulatory module is used in the root epidermis to developmentally respond to two different stresses: ribosome biogenesis defects and plant defense. The rationale for this is unclear. It is possible that the defense response activates this regulatory module through its effects on ribosomes. Ribosome profiling has revealed that plants undergo translational reprogramming during the growth-to-defense transition (Meteignier et al., 2017; Xu et al., 2017a), which could impinge on ribosome biogenesis. Furthermore, bacterial pathogen infections alter translation of plant immune response regulators in a uORF-dependent manner (Xu et al., 2017b), suggesting the possibility that ANAC082, also bearing a uORF in its 5′ UTR (Ohbayashi et al., 2017), could be activated during plant defense. Further studies are required to clarify the relationship between these stress conditions and their effect on root epidermal cell fate.
METHODS
Plant Material and Growth Conditions
Arabidopsis (Arabidopsis thaliana) seeds were surface sterilized with 30% (v/v) bleach plus 0.02% (v/v) Triton X-100 and plated on mineral mix media previously reported by Schiefelbein and Somerville (1990) containing 0.3% (w/v) Gelrite. Seedling phenotypes were analyzed after 4 d of growth at 23°C under continuous light. For RBF mutants, older seedlings were used due to slower growth: apum23-4 mutant, 8 d; dim1a mutant, 6 d; and prmt3-1 mutant, 7 d. For RBF mutants carrying the anac082-1 mutation, seedlings used for analysis were ∼2 d younger than the corresponding RBF single mutants. For crosses and seed bulking, seedlings were transplanted to soil and grown under long-day light conditions (120 to 150 μmol m−2 s−1 with Osram L36W/840 Lumilux cool-white Hg bulbs) at 22°C (day) and 18°C (night).
For analysis of CHX-treated seedlings, seeds were grown on the standard mineral mix media for 4 d, transferred onto mineral mix media containing CHX (stock solution in ethanol), and grown for an additional 2 d before examination. For analysis of flg22-treated seedlings, seeds were directly grown on the standard mineral mix media containing 1 μM flg22 for 4 d before analysis.
The following mutant and reporter lines used in this study have been previously described: wer-1 (Lee and Schiefelbein, 1999), gl3 (Koornneef, 1981), egl3 (Zhang et al., 2003), ttg1 (Galway et al., 1994), gl2-1 (Koornneef, 1981), cpc-1 (Wada et al., 1997), myb23-1 (Kirik et al., 2005), dim1a (Wieckowski and Schiefelbein, 2012), ProGL2:GUS (Masucci et al., 1996), ProMYB23:GUS (Kang et al., 2009), ProMYB23:MYB23-GFP (Kang et al., 2009), and ProGL3:GL3-YFP (Bernhardt et al., 2005). The anac082-1 (sriw1) seeds were kindly provided by Munetaka Sugiyama. The ProMYB23:MYB23-GFP seeds were kindly provided by Myeong Min Lee.
The following mutant lines were reported previously and obtained from the Arabidopsis Biological Resource Center: apum23-2 (SALK_052992; Abbasi et al., 2010), apum24-2 (SALK_033623; Maekawa et al., 2018), prmt3-1 (SAIL_220_F08; Hang et al., 2014), prmt3-2 (WISCDSLOX391A01; Hang et al., 2014), and anac082 (GABI_282H08; Kim et al., 2018).
Mutant Screening and Positional Cloning
Mutagenesis of cpc-1 mutant seeds (Wassilewskija ecotype) with ethyl methanesulfonate was performed as previously described by Estelle and Somerville (1987). The cpc-1 apum23-4 mutant (Wassilewskija ecotype) was crossed to plants of the Columbia ecotype to generate F2 and F3 offspring for positional cloning. Multiple simple sequence length polymorphism and cleaved amplified polymorphic sequence markers were used (Jander et al., 2002). The responsible mutation was narrowed to a region between the CER481016 (27,196,516) and CER479543 (27,235,489) on chromosome 1. The protein-coding sequences of all genes within this interval were then cloned and sequenced to identify the mutated gene in apum23-4.
During later genetic studies, the derived cleaved amplified polymorphic sequences (Neff et al., 2002) strategy was used to identify the apum23-4 mutation among individual plants in segregating populations. Genotyping primers are listed in Supplemental Table 2.
Transgenes and Plant Transformation
To construct the ProAPUM23:APUM23-GFP transgene, a 5.5-kb genomic fragment including 1-kb of the 5′ promoter region and the full-length genomic sequence of APUM23 (with the stop codon removed) was cloned and integrated into the Gateway pENTR/SD/TOPO vector (Invitrogen), followed by subcloning into the Gateway binary vector pMDC107 (containing the C-terminal GFP tag). The cloning primers are listed in Supplemental Table 2.
To construct the Pro35S:APUM23-YFP transgene, a 4-kb genomic sequence of APUM23 (starting from the ATG and replacing the stop codon with a Gly5-Ala linker) was cloned and incorporated into the pCAM binary vector containing the 35S promoter at the 5′ end and an in-frame yellow fluorescent protein (YFP) tag at the 3′ end, using the NEBuilder HiFi DNA Assembly Cloning kit (New England Biolabs).
To construct the ProFIB1:FIB1-mcherry transgene, a 2.2-kb genomic fragment including 1 kb of the 5′ promoter region and the full-length FIB1 genomic sequence (with the stop codon replaced with a Gly5-Ala linker), a 0.7-kb mcherry sequence (with a stop codon added), and a 0.5-kb 3′ flanking region of FIB1 were cloned and integrated together into the pCAM binary vector using the NEBuilder HiFi DNA Assembly Cloning kit. The mcherry tag was added to the C terminus of the FIB1 genomic sequence. The cloning primers are listed in Supplemental Table 2.
To construct the APUM23ρAPUM24 transgene, the 1.4-kb 5′ promoter region of APUM23, 4.1-kb full-length genomic region of APUM24, and 0.6-kb 3′ flanking region of APUM23 were cloned and integrated into the pCAM binary vector using the NEBuilder HiFi DNA Assembly Cloning kit. The ProAPUM24:APUM23 transgene was constructed using a similar strategy by assembling the 1-kb 5′ promoter region of APUM24, 3.6-kb full-length genomic region of APUM23, and 0.6-kb 3′ flanking region of APUM24. The cloning primers are listed in Supplemental Table 2.
Verified constructs were transformed into Agrobacterium tumefaciens strain GV3101, which was then used for plant transformation as described previously (Clough and Bent, 1998). After transformation, T1 seeds were harvested and screened for hygromycin resistance. Selected T2 seeds were grown and T3 seeds were tested to find homozygotes for insertions.
Microscopy, Quantification, and Image Analysis
Quantification of root hair cell and non-hair cell frequency were performed with a bright-field compound microscope using seedlings briefly stained with toluidine blue. Epidermal cell positions were determined according to their spatial relationship to underlying cortical cells. An epidermal cell in contact with two underlying cortical cells is in the H position, while an epidermal cell in contact with one underlying cortical cell is in the N position. For each genotype, 10 cells in the H position and 10 cells in the N position were scored per root, and 10 roots were used per replicate. Cell positions were determined according to their location with respect to underlying cortical cells. A cell was scored as a root hair cell if a visible protrusion was present on its cell surface, regardless of the length. A cell was scored as a non-hair cell if no root hair or initial root hair bulge was visible. At least three replicates were performed for each genotype in one experiment. Raw data from all experiments are listed in Supplemental Data Set 1. Depending on numbers of samples tested, statistical significance was determined using either one-way ANOVA or t test, and the results from all statistical tests are provided in Supplemental Data Set 2.
Histochemical analysis of seedling roots containing GUS reporter genes was performed as previously described (Masucci et al., 1996). For ProGL2:GUS, roots were stained with 0.1 mg/mL 5-bromo-4-chloro-3-indolyl-β-d-glucuronide substrates (CHX salt, GoldBio) at 37°C for 20 min, while for ProMYB23:GUS, roots were stained with 0.2 mg/mL 5-bromo-4-chloro-3-indolyl-β-d-glucuronide substrates at 37°C for 40 to 50 min. For quantification of ProGL2:GUS and ProMYB23:GUS expression, 10 continuous cells in the H position and 10 continuous cells in the N position were scored in each root, and 10 roots were used for each genotype in each replicate. The 10 cells included the first cell prior to rapid elongation (i.e., cell length > cell width) and extended shootward. A cell was scored as GUS positive if the GUS signal was visibly greater than the neighboring unstained H-position cells. Cell positions were defined according to their location with respect to underlying cortical cells. At least three replicates were performed for each genotype in one experiment. Raw data from all experiments are listed in Supplemental Data Set 1, and results of all statistical tests are provided in Supplemental Data Set 2.
Fluorescent images were obtained with a TCS SP5 DM6000B broadband confocal microscope (Leica) with an HCX PL APO CS 20× or 40× dry lens and facilitated with Leica Application Suite Advanced Fluorescence software. Before imaging, seedlings were stained with propidium iodide (PI) or DAPI and rinsed with water. Specifically, for DAPI staining, roots were incubated in 1/5000 dilution of fresh DAPI stock solution (5 mg/mL) for 15 to 20 min for nuclei visualization or for 1 to 2 min for cell wall visualization only. For GFP/PI imaging, an argon 488-nm laser was used for excitation. The GFP signal was collected under bandwidth 511 to 541 nm, the PI signal was collected under bandwidth 620 to 720 nm. For YFP/PI imaging, an argon 514-nm laser was used for excitation. The YFP signal was collected under bandwidth 528 to 547 nm. For DAPI/GFP/mcherry imaging, the 405-nm diode was used for DAPI excitation and the DPSS 561-nm laser was used for mcherry excitation. The DAPI signal was collected under bandwidth 424 to 475 nm, and the mcherry signal was collected under bandwidth 580 to 700 nm. The GFP and mcherry quantification was performed using ImageJ under separate RGB stacks, and the mcherry/GFP ratio was used for plotting.
To examine the expression of both the ProGL2:GUS and ProMYB23:MYB23-GFP/ProGL3:GL3-YFP markers within the same root, seedling roots were first imaged with the confocal microscope and then removed from slides and stained for GUS signals. Special care was taken to place the seedlings in the same posture for GUS examination as for the fluorescent imaging, taking advantage of unique root epidermal cell shapes as landmarks in the viewing window.
RNA Extraction and Quantitative Real-Time PCR
RNA extraction and quantitative real-time PCR were conducted as previously described (Wang et al., 2019). In general, ∼2 mm of seedling root tips was harvested and RNA was extracted (RNeasy Plant Mini kit; QIAGEN). DNase I (RNase-free DNase set; QIAGEN) was applied to eliminate potential DNA contaminations in RNA samples, followed by cDNA synthesis (SuperScript First-Strand synthesis system; Invitrogen) and quantitative real-time PCR (Radiant Green Hi-ROX qPCR kit; Alkali Scientific). The PCR analysis and quantification were performed on the StepOnePlus Real Time System from Applied Biosystems following the Delta-Delta-Ct method. The GAPCP2 gene (AT1G16300) was used as the internal reference. Primers used for qPCR are listed in Supplemental Table 2. Raw data from all experiments are listed in Supplemental Data Set 1, and the results from all statistical tests are provided in Supplemental Data Set 2.
Dual-Luciferase Assay
The dual-luciferase assay, tobacco (Nicotiana tabacum) infiltration, and luciferase activity measurement were performed as previously described (Liu et al., 2016). As the reporter, the MYB23 promoter region (∼2 kb, the same regulatory sequence used in the ProMYB23:GUS reporter described; Kirik et al., 2001) was cloned and integrated into the pGreen0800-LUC vector using a HiFi Assembly kit. As the effector, the ANAC082 coding sequence was cloned and integrated into the pCAM vector driven by the CaMV35S promoter. Nucleotide primers are listed in Supplemental Table 2. The effector and reporter constructs were transformed into GV3101 Agrobacterium-competent cells. Overnight-cultured Agrobacteria carrying the correct constructs were resuspended in 10 mM MgCl2 solution containing 0.5 μM acetosyringone. After a 2-h incubation at room temperature, resuspended Agrobacteria carrying effector and reporter constructs were mixed in a 5:1 ratio before infiltration.
Approximately 6-week-old tobacco plants were used for infiltration. For each combination of effector and reporter, at least three replicates were performed. For each replicate, at least six leaves were infiltrated. Raw data from all experiments are listed in Supplemental Data Set 1, and the results of all statistical tests are provided in Supplemental Data Set 2 . Plants were incubated in the dark for 24 h after infiltration, followed by another 1 to 2 d in regular growth conditions. The luciferase signals were detected using the Dual-Luciferase Reporter Assay System (Promega). For each leaf, pipette tips were used to collect a small piece of tissue close to the infiltrated spot. The leaf tissue was then crushed in 50-μL passive lysis buffer and incubated at room temperature for 10 min. The samples were placed in a luciferase microplate reader with the default settings for adding Luciferase Assay mix, reading luciferase activities, adding StopandGlo Reagen mix, and reading Renilla luciferase activities.
Accession Numbers
Arabidopsis sequence data from this article can be found in the GenBank/EMBL data libraries under the following accession numbers: APUM23 (AT1G72320), DIM1A (At2G47420), PRMT3 (AT3G12270), APUM24 (AT3G16810), ANAC082 (AT5G09330), CPC (At2G46410), FIB1 (At5G52470), GL2 (At1G79840), GL3 (At5g41315), EGL3 (AT1G63650), MYB23 (At5g40330), TTG1 (At5g24520), and WER (At5g14750).
SUPPLEMENTAL DATA
Supplemental Figure 1. RBF mutants exhibit delayed seed germination.
Supplemental Figure 2. The apum23-4 mutant causes no significant alterations in root architecture.
Supplemental Figure 3. Additional representative roots with ProAPUM23:APUM23-GFP and ProFIB1:FIB1-mcherry reporters.
Supplemental Figure 4. Quantification of ProGL2:GUS signals in RBF double mutants with wer-1.
Supplemental Figure 5. Quantification of ProMYB23:GUS signals in RBF mutants apum23-4, dim1a and prmt3-1.
Supplemental Figure 6. ANAC082 is required for ectopic MYB23 expression and ectopic non-hair cell specification in apum23-4 and dim1a mutants.
Supplemental Figure 7. Dual luciferase assay showing effect of ANAC082 on MYB23 promoter activity in tobacco leaves.
Supplemental Figure 8. Quantification of ProGL2:GUS signals in seedling root tips of wild type, prmt3-1 and apum24-2 mutants.
Supplemental Table 1. A list of the RBF mutants examined in this study.
Supplemental Table 2. Primers used for genotyping, cloning, and qPCR.
Supplemental Data Set 1. Raw data for quantification of root epidermal cell specification, ProGL2:GUS or ProMYB23:GUS expression, and qPCR in this study.
Supplemental Data Set 2. P-values of one-way ANOVA tests and t tests used in this study.
DIVE Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
Acknowledgments
We thank Myeong Min Lee (Yonsei University, Korea) for the kind gift of the ProMYB23:MYB23-GFP seeds and Munetaka Sugiyama for the anac082-1 seeds. We thank Chaoyang Liu (South China Agricultural University, China) for the kind gift of the pGreen0800-LUC construct. We thank The Arabidopsis Biological Resource Center for providing seed stocks of multiple mutant lines. We thank Erik Nielsen, Andrej Wierzbicki, and Jairam Menon (University of Michigan, Ann Arbor) for suggestions and informative discussion. We thank Gregg Soboccinski (University of Michigan, Ann Arbor) for technical assistance with microscopy. This work was supported by the National Science Foundation (grant IOS-1444400).
AUTHOR CONTRIBUTIONS
W.W., K.H.R., A.B., and C.B. conducted the experiments. W.W., K.H.R., and A.B. analyzed the data. J.S. supervised the experiments. W.W., K.H.R., A.B., and J.S. contributed to the design of the project. W.W. and J.S. wrote the article with contributions from A.B., K.H.R., and C.B.
References
- Abbasi N., Kim H.B., Park N.I., Kim H.S., Kim Y.K., Park Y.I., Choi S.B.(2010). APUM23, a nucleolar Puf domain protein, is involved in pre-ribosomal RNA processing and normal growth patterning in Arabidopsis. Plant J. 64: 960–976. [DOI] [PubMed] [Google Scholar]
- Balkunde R., Pesch M., Hülskamp M.(2010). Trichome patterning in Arabidopsis thaliana from genetic to molecular models. Curr. Top. Dev. Biol. 91: 299–321. [DOI] [PubMed] [Google Scholar]
- Berger F., Hung C.Y., Dolan L., Schiefelbein J.(1998). Control of cell division in the root epidermis of Arabidopsis thaliana. Dev. Biol. 194: 235–245. [DOI] [PubMed] [Google Scholar]
- Bernhardt C., Lee M.M., Gonzalez A., Zhang F., Lloyd A., Schiefelbein J.(2003). The bHLH genes GLABRA3 (GL3) and ENHANCER OF GLABRA3 (EGL3) specify epidermal cell fate in the Arabidopsis root. Development 130: 6431–6439. [DOI] [PubMed] [Google Scholar]
- Bernhardt C., Zhao M., Gonzalez A., Lloyd A., Schiefelbein J.(2005). The bHLH genes GL3 and EGL3 participate in an intercellular regulatory circuit that controls cell patterning in the Arabidopsis root epidermis. Development 132: 291–298. [DOI] [PubMed] [Google Scholar]
- Bruex A., Kainkaryam R.M., Wieckowski Y., Kang Y.H., Bernhardt C., Xia Y., Zheng X., Wang J.Y., Lee M.M., Benfey P., Woolf P.J., Schiefelbein J.(2012). A gene regulatory network for root epidermis cell differentiation in Arabidopsis. PLoS Genet. 8: e1002446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrne M.E.(2009). A role for the ribosome in development. Trends Plant Sci. 14: 512–519. [DOI] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F.(1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743. [DOI] [PubMed] [Google Scholar]
- Clowes F.A.L.(2000). Pattern in root meristem development in angiosperms. New Phytol. 146: 83–94. [Google Scholar]
- Cvrčková F., Bezvoda R., Zárský V.(2010). Computational identification of root hair-specific genes in Arabidopsis. Plant Signal. Behav. 5: 1407–1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Kloet S.R.(1966). Ribonucleic acid synthesis in yeast. The effect of cycloheximide on the synthesis of ribonucleic acid in Saccharomyces carlsbergensis. Biochem. J. 99: 566–581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duckett C.M., Grierson C., Linstead P., Schneider K., Lawson E., Dean C., Poethig S., Roberts K.(1994). Clonal relationships and cell patterning in the root epidermis of Arabidopsis. Development 120: 2465. [Google Scholar]
- Edwards T.A., Pyle S.E., Wharton R.P., Aggarwal A.K.(2001). Structure of Pumilio reveals similarity between RNA and peptide binding motifs. Cell 105: 281–289. [DOI] [PubMed] [Google Scholar]
- Ernst H.A., Olsen A.N., Larsen S., Lo Leggio L.(2004). Structure of the conserved domain of ANAC, a member of the NAC family of transcription factors. EMBO Rep. 5: 297–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estelle M.A., Somerville C.(1987). Auxin-resistant mutants of Arabidopsis thaliana with an altered morphology. Mol. Gen. Genet. 206: 200–206. [Google Scholar]
- Ferrari S., Galletti R., Denoux C., De Lorenzo G., Ausubel F.M., Dewdney J.(2007). Resistance to Botrytis cinerea induced in Arabidopsis by elicitors is independent of salicylic acid, ethylene, or jasmonate signaling but requires PHYTOALEXIN DEFICIENT3. Plant Physiol. 144: 367–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francischini C.W., Quaggio R.B.(2009). Molecular characterization of Arabidopsis thaliana PUF proteins--binding specificity and target candidates. FEBS J. 276: 5456–5470. [DOI] [PubMed] [Google Scholar]
- Galway M.E., Masucci J.D., Lloyd A.M., Walbot V., Davis R.W., Schiefelbein J.W.(1994). The TTG gene is required to specify epidermal cell fate and cell patterning in the Arabidopsis root. Dev. Biol. 166: 740–754. [DOI] [PubMed] [Google Scholar]
- Golomb L., Volarevic S., Oren M.(2014). p53 and ribosome biogenesis stress: The essentials. FEBS Lett. 588: 2571–2579. [DOI] [PubMed] [Google Scholar]
- Gómez-Gómez L., Felix G., Boller T.(1999). A single locus determines sensitivity to bacterial flagellin in Arabidopsis thaliana. Plant J. 18: 277–284. [DOI] [PubMed] [Google Scholar]
- Hang R., Liu C., Ahmad A., Zhang Y., Lu F., Cao X.(2014). Arabidopsis protein arginine methyltransferase 3 is required for ribosome biogenesis by affecting precursor ribosomal RNA processing. Proc. Natl. Acad. Sci. USA 111: 16190–16195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harscoët E., Dubreucq B., Palauqui J.C., Lepiniec L.(2010). NOF1 encodes an Arabidopsis protein involved in the control of rRNA expression. PLoS One 5: e12829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horiguchi G., Van Lijsebettens M., Candela H., Micol J.L., Tsukaya H.(2012). Ribosomes and translation in plant developmental control. Plant Sci. 191–192: 24–34. [DOI] [PubMed] [Google Scholar]
- Huang L., Shi X., Wang W., Ryu K.H., Schiefelbein J.(2017). Diversification of root hair development genes in vascular plants. Plant Physiol. 174: 1697–1712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang T., Kerstetter R.A., Irish V.F.(2014). APUM23, a PUF family protein, functions in leaf development and organ polarity in Arabidopsis. J. Exp. Bot. 65: 1181–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jander G., Norris S.R., Rounsley S.D., Bush D.F., Levin I.M., Last R.L.(2002). Arabidopsis map-based cloning in the post-genome era. Plant Physiol. 129: 440–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalinina N.O., Makarova S., Makhotenko A., Love A.J., Taliansky M.(2018). The multiple functions of the nucleolus in plant development, disease and stress responses. Front Plant Sci 9: 132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang Y.H., Kirik V., Hulskamp M., Nam K.H., Hagely K., Lee M.M., Schiefelbein J.(2009). The MYB23 gene provides a positive feedback loop for cell fate specification in the Arabidopsis root epidermis. Plant Cell 21: 1080–1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H.J., Park J.H., Kim J., Kim J.J., Hong S., Kim J., Kim J.H., Woo H.R., Hyeon C., Lim P.O., Nam H.G., Hwang D.(2018). Time-evolving genetic networks reveal a NAC troika that negatively regulates leaf senescence in Arabidopsis. Proc. Natl. Acad. Sci. USA 115: E4930–E4939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirik V., Lee M.M., Wester K., Herrmann U., Zheng Z., Oppenheimer D., Schiefelbein J., Hulskamp M.(2005). Functional diversification of MYB23 and GL1 genes in trichome morphogenesis and initiation. Development 132: 1477–1485. [DOI] [PubMed] [Google Scholar]
- Kirik V., Schnittger A., Radchuk V., Adler K., Hülskamp M., Bäumlein H.(2001). Ectopic expression of the Arabidopsis AtMYB23 gene induces differentiation of trichome cells. Dev. Biol. 235: 366–377. [DOI] [PubMed] [Google Scholar]
- Kirik V., Simon M., Huelskamp M., Schiefelbein J.(2004). The ENHANCER OF TRY AND CPC1 gene acts redundantly with TRIPTYCHON and CAPRICE in trichome and root hair cell patterning in Arabidopsis. Dev. Biol. 268: 506–513. [DOI] [PubMed] [Google Scholar]
- Koornneef M.(1981). The complex syndrome of ttg mutants. Arab. Inf. Serv. 18: 45–51. [Google Scholar]
- Kurata T., et al. (2005). Cell-to-cell movement of the CAPRICE protein in Arabidopsis root epidermal cell differentiation. Development 132: 5387–5398. [DOI] [PubMed] [Google Scholar]
- Kwak S.H., Schiefelbein J.(2008). A feedback mechanism controlling SCRAMBLED receptor accumulation and cell-type pattern in Arabidopsis. Curr. Biol. 18: 1949–1954. [DOI] [PubMed] [Google Scholar]
- Kwak S.H., Shen R., Schiefelbein J.(2005). Positional signaling mediated by a receptor-like kinase in Arabidopsis. Science 307: 1111–1113. [DOI] [PubMed] [Google Scholar]
- Lange H., Sement F.M., Gagliardi D.(2011). MTR4, a putative RNA helicase and exosome co-factor, is required for proper rRNA biogenesis and development in Arabidopsis thaliana. Plant J. 68: 51–63. [DOI] [PubMed] [Google Scholar]
- Lee M.M., Schiefelbein J.(1999). WEREWOLF, a MYB-related protein in Arabidopsis, is a position-dependent regulator of epidermal cell patterning. Cell 99: 473–483. [DOI] [PubMed] [Google Scholar]
- Lee M.M., Schiefelbein J.(2002). Cell pattern in the Arabidopsis root epidermis determined by lateral inhibition with feedback. Plant Cell 14: 611–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S.F., Milliken O.N., Pham H., Seyit R., Napoli R., Preston J., Koltunow A.M., Parish R.W.(2009). The Arabidopsis MYB5 transcription factor regulates mucilage synthesis, seed coat development, and trichome morphogenesis. Plant Cell 21: 72–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Q., Ohashi Y., Kato M., Tsuge T., Gu H., Qu L.J., Aoyama T.(2015). GLABRA2 directly suppresses basic helix-loop-helix transcription factor genes with diverse functions in root hair development. Plant Cell 27: 2894–2906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C., Long J., Zhu K., Liu L., Yang W., Zhang H., Li L., Xu Q., Deng X.(2016). Characterization of a citrus R2R3-MYB transcription factor that regulates the flavonol and hydroxycinnamic acid biosynthesis. Sci. Rep. 6: 25352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maekawa S., Ishida T., Yanagisawa S.(2018). Reduced expression of APUM24, encoding a novel rRNA processing factor, induces sugar-dependent nucleolar stress and altered sugar responses in Arabidopsis thaliana. Plant Cell 30: 209–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masucci J.D., Rerie W.G., Foreman D.R., Zhang M., Galway M.E., Marks M.D., Schiefelbein J.W.(1996). The homeobox gene GLABRA2 is required for position-dependent cell differentiation in the root epidermis of Arabidopsis thaliana. Development 122: 1253–1260. [DOI] [PubMed] [Google Scholar]
- Mayer C., Grummt I.(2005). Cellular stress and nucleolar function. Cell Cycle 4: 1036–1038. [DOI] [PubMed] [Google Scholar]
- Meteignier L.V., El Oirdi M., Cohen M., Barff T., Matteau D., Lucier J.F., Rodrigue S., Jacques P.E., Yoshioka K., Moffett P.(2017). Translatome analysis of an NB-LRR immune response identifies important contributors to plant immunity in Arabidopsis. J. Exp. Bot. 68: 2333–2344. [DOI] [PubMed] [Google Scholar]
- Missbach S., Weis B.L., Martin R., Simm S., Bohnsack M.T., Schleiff E.(2013). 40S Ribosome biogenesis co-factors are essential for gametophyte and embryo development. PLoS One 8: e54084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller M., Schmidt W.(2004). Environmentally induced plasticity of root hair development in Arabidopsis. Plant Physiol. 134: 409–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murata Y., Wharton R.P.(1995). Binding of pumilio to maternal hunchback mRNA is required for posterior patterning in Drosophila embryos. Cell 80: 747–756. [DOI] [PubMed] [Google Scholar]
- Neff M.M., Turk E., Kalishman M.(2002). Web-based primer design for single nucleotide polymorphism analysis. Trends Genet. 18: 613–615. [DOI] [PubMed] [Google Scholar]
- Ohashi Y., Oka A., Rodrigues-Pousada R., Possenti M., Ruberti I., Morelli G., Aoyama T.(2003). Modulation of phospholipid signaling by GLABRA2 in root-hair pattern formation. Science 300: 1427–1430. [DOI] [PubMed] [Google Scholar]
- Ohbayashi I., Konishi M., Ebine K., Sugiyama M.(2011). Genetic identification of Arabidopsis RID2 as an essential factor involved in pre-rRNA processing. Plant J. 67: 49–60. [DOI] [PubMed] [Google Scholar]
- Ohbayashi I., Lin C.Y., Shinohara N., Matsumura Y., Machida Y., Horiguchi G., Tsukaya H., Sugiyama M.(2017). Evidence for a role of ANAC082 as a ribosomal stress response mediator leading to growth defects and developmental alterations in Arabidopsis. Plant Cell 29: 2644–2660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohbayashi I., Sugiyama M.(2018). Plant nucleolar stress response, a new face in the NAC-dependent cellular stress responses. Front Plant Sci 8: 2247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen A.N., Ernst H.A., Lo Leggio L., Johansson E., Larsen S., Skriver K.(2004). Preliminary crystallographic analysis of the NAC domain of ANAC, a member of the plant-specific NAC transcription factor family. Acta Crystallogr. D Biol. Crystallogr. 60: 112–115. [DOI] [PubMed] [Google Scholar]
- Ooka H., et al. (2003). Comprehensive analysis of NAC family genes in Oryza sativa and Arabidopsis thaliana. DNA Res. 10: 239–247. [DOI] [PubMed] [Google Scholar]
- Penzo M., Montanaro L., Treré D., Derenzini M.(2019). The ribosome biogenesis-cancer connection. Cells 8: 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pih K.T., Yi M.J., Liang Y.S., Shin B.J., Cho M.J., Hwang I., Son D.(2000). Molecular cloning and targeting of a fibrillarin homolog from Arabidopsis. Plant Physiol. 123: 51–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pontvianne F., et al. (2010). Nucleolin is required for DNA methylation state and the expression of rRNA gene variants in Arabidopsis thaliana. PLoS Genet. 6: e1001225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu C., McCann K.L., Wine R.N., Baserga S.J., Hall T.M.(2014). A divergent Pumilio repeat protein family for pre-rRNA processing and mRNA localization. Proc. Natl. Acad. Sci. USA 111: 18554–18559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rishmawi L., Wolff H., Schrader A., Hülskamp M.(2018). Sub-epidermal expression of ENHANCER OF TRIPTYCHON AND CAPRICE1 and its role in root hair formation upon Pi starvation. Front. Plant Sci. 9: 1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosado A., Li R., van de Ven W., Hsu E., Raikhel N.V.(2012). Arabidopsis ribosomal proteins control developmental programs through translational regulation of auxin response factors. Proc. Natl. Acad. Sci. USA 109: 19537–19544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu K.H., Kang Y.H., Park Y.H., Hwang I., Schiefelbein J., Lee M.M.(2005). The WEREWOLF MYB protein directly regulates CAPRICE transcription during cell fate specification in the Arabidopsis root epidermis. Development 132: 4765–4775. [DOI] [PubMed] [Google Scholar]
- Sáez-Vásquez J., Delseny M.(2019). Ribosome biogenesis in plants: From functional 45S ribosomal DNA organization to ribosome assembly factors. Plant Cell 31: 1945–1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salomé P.A.(2017). Proliferate at your own risk: Ribosomal stress and regeneration. Plant Cell 29: 2318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiefelbein J., Huang L., Zheng X.(2014). Regulation of epidermal cell fate in Arabidopsis roots: The importance of multiple feedback loops. Front Plant Sci 5: 47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiefelbein J.W., Somerville C.(1990). Genetic control of root hair development in Arabidopsis thaliana. Plant Cell 2: 235–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt W., Schikora A.(2001). Different pathways are involved in phosphate and iron stress-induced alterations of root epidermal cell development. Plant Physiol. 125: 2078–2084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shanmugam T., Abbasi N., Kim H.S., Kim H.B., Park N.I., Park G.T., Oh S.A., Park S.K., Muench D.G., Choi Y., Park Y.I., Choi S.B.(2017). An Arabidopsis divergent pumilio protein, APUM24, is essential for embryogenesis and required for faithful pre-rRNA processing. Plant J. 92: 1092–1105. [DOI] [PubMed] [Google Scholar]
- Simon M., Lee M.M., Lin Y., Gish L., Schiefelbein J.(2007). Distinct and overlapping roles of single-repeat MYB genes in root epidermal patterning. Dev. Biol. 311: 566–578. [DOI] [PubMed] [Google Scholar]
- Song S.K., Ryu K.H., Kang Y.H., Song J.H., Cho Y.H., Yoo S.D., Schiefelbein J., Lee M.M.(2011). Cell fate in the Arabidopsis root epidermis is determined by competition between WEREWOLF and CAPRICE. Plant Physiol. 157: 1196–1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoyanova B.B., Hadjiolov A.A.(1979). Alterations in the processing of rat-liver ribosomal RNA caused by cycloheximide inhibition of protein synthesis. Eur. J. Biochem. 96: 349–356. [DOI] [PubMed] [Google Scholar]
- Stracke R., Werber M., Weisshaar B.(2001). The R2R3-MYB gene family in Arabidopsis thaliana. Curr. Opin. Plant Biol. 4: 447–456. [DOI] [PubMed] [Google Scholar]
- Szostak E., Gebauer F.(2013). Translational control by 3′-UTR-binding proteins. Brief. Funct. Genomics 12: 58–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tam P.P., Barrette-Ng I.H., Simon D.M., Tam M.W., Ang A.L., Muench D.G.(2010). The Puf family of RNA-binding proteins in plants: phylogeny, structural modeling, activity and subcellular localization. BMC Plant Biol. 10: 44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson E., Ferreira-Cerca S., Hurt E.(2013). Eukaryotic ribosome biogenesis at a glance. J. Cell Sci. 126: 4815–4821. [DOI] [PubMed] [Google Scholar]
- Thomson E., Rappsilber J., Tollervey D.(2007). Nop9 is an RNA binding protein present in pre-40S ribosomes and required for 18S rRNA synthesis in yeast. RNA 13: 2165–2174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tominaga R., Iwata M., Okada K., Wada T.(2007). Functional analysis of the epidermal-specific MYB genes CAPRICE and WEREWOLF in Arabidopsis. Plant Cell 19: 2264–2277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada T., Kurata T., Tominaga R., Koshino-Kimura Y., Tachibana T., Goto K., Marks M.D., Shimura Y., Okada K.(2002). Role of a positive regulator of root hair development, CAPRICE, in Arabidopsis root epidermal cell differentiation. Development 129: 5409–5419. [DOI] [PubMed] [Google Scholar]
- Wada T., Tachibana T., Shimura Y., Okada K.(1997). Epidermal cell differentiation in Arabidopsis determined by a Myb homolog, CPC. Science 277: 1113–1116. [DOI] [PubMed] [Google Scholar]
- Wang M., Ogé L., Perez-Garcia M.D., Hamama L., Sakr S.(2018). The PUF protein family: Overview on PUF RNA targets, biological functions, and post transcriptional regulation. Int. J. Mol. Sci. 19: 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Ryu K.H., Barron C., Schiefelbein J.(2019). Root epidermal cell patterning is modulated by a critical residue in the WEREWOLF transcription factor. Plant Physiol. 181: 1239–1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Zhang W., Li K., Sun F., Han C., Wang Y., Li X.(2008). Salt-induced plasticity of root hair development is caused by ion disequilibrium in Arabidopsis thaliana. J. Plant Res. 121: 87–96. [DOI] [PubMed] [Google Scholar]
- Weis B.L., Kovacevic J., Missbach S., Schleiff E.(2015a). Plant-specific features of ribosome biogenesis. Trends Plant Sci. 20: 729–740. [DOI] [PubMed] [Google Scholar]
- Weis B.L., Missbach S., Marzi J., Bohnsack M.T., Schleiff E.(2014). The 60S associated ribosome biogenesis factor LSG1-2 is required for 40S maturation in Arabidopsis thaliana. Plant J. 80: 1043–1056. [DOI] [PubMed] [Google Scholar]
- Weis B.L., Palm D., Missbach S., Bohnsack M.T., Schleiff E.(2015b). atBRX1-1 and atBRX1-2 are involved in an alternative rRNA processing pathway in Arabidopsis thaliana. RNA 21: 415–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickens M., Bernstein D.S., Kimble J., Parker R.(2002). A PUF family portrait: 3’UTR regulation as a way of life. Trends Genet. 18: 150–157. [DOI] [PubMed] [Google Scholar]
- Wieckowski Y., Schiefelbein J.(2012). Nuclear ribosome biogenesis mediated by the DIM1A rRNA dimethylase is required for organized root growth and epidermal patterning in Arabidopsis. Plant Cell 24: 2839–2856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter D., Vinegar B., Nahal H., Ammar R., Wilson G.V., Provart N.J.(2007). An “Electronic Fluorescent Pictograph” browser for exploring and analyzing large-scale biological data sets. PLoS One 2: e718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu G., Greene G.H., Yoo H., Liu L., Marqués J., Motley J., Dong X.(2017a). Global translational reprogramming is a fundamental layer of immune regulation in plants. Nature 545: 487–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu G., Yuan M., Ai C., Liu L., Zhuang E., Karapetyan S., Wang S., Dong X.(2017b). uORF-Mediated translation allows engineered plant disease resistance without fitness costs. Nature 545: 491–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi M., Nagahage I.S.P., Ohtani M., Ishikawa T., Uchimiya H., Kawai-Yamada M., Demura T.(2015). Arabidopsis NAC domain proteins VND-INTERACTING1 and ANAC103 interact with multiple NAC domain proteins. Plant Biotechnol. 32: 119–132. [Google Scholar]
- Zamore P.D., Williamson J.R., Lehmann R.(1997). The Pumilio protein binds RNA through a conserved domain that defines a new class of RNA-binding proteins. RNA 3: 1421–1433. [PMC free article] [PubMed] [Google Scholar]
- Zhang C., Muench D.G.(2015). A nucleolar PUF RNA-binding protein with specificity for a unique RNA sequence. J. Biol. Chem. 290: 30108–30118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F., Gonzalez A., Zhao M., Payne C.T., Lloyd A.(2003). A network of redundant bHLH proteins functions in all TTG1-dependent pathways of Arabidopsis. Development 130: 4859–4869. [DOI] [PubMed] [Google Scholar]
- Zhang J., McCann K.L., Qiu C., Gonzalez L.E., Baserga S.J., Hall T.M.(2016). Nop9 is a PUF-like protein that prevents premature cleavage to correctly process pre-18S rRNA. Nat. Commun. 7: 13085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Lu H.(2009). Signaling to p53: Ribosomal proteins find their way. Cancer Cell 16: 369–377. [DOI] [PMC free article] [PubMed] [Google Scholar]