ARR1, a type-B ARR, is defined as an important inhibitor of in vitro shoot regeneration that modulates the expression of WUS and CLV3 in an ARR12-dependent manner, and directly activates IAA17.
Abstract
Exogenous cytokinin is critical for in vitro shoot regeneration. Proteins involved in the cytokinin signal transduction pathway, including type-B ARABIDOPSIS RESPONSE REGULATORs (ARRs), participate in shoot regeneration in Arabidopsis (Arabidopsis thaliana). Some type-B ARRs (e.g., ARR1 and ARR12) promote shoot regeneration by directly activating WUSCHEL (WUS) expression; however, it is unclear how type-B ARRs inhibit shoot regeneration. Here, we show that ARR12 is a central enhancer of callus formation and shoot regeneration, whereas ARR1 is a strong inhibitor of this process that counteracts the positive effect of ARR12. ARR1 indirectly represses CLAVATA3 (CLV3) expression in an ARR12-dependent manner via competing with ARR12 for binding to the CLV3 promoter, which contributes to its ARR12-dependent inhibitory effect on callus formation and shoot regeneration. In parallel, ARR1 inhibits shoot regeneration through transcriptional activation of INDOLE-3-ACETIC ACID INDUCIBLE17, an auxin response repressor gene, and the consequent indirect repression of WUS expression. Thus, type-B ARRs have diverse effects on callus formation and shoot regeneration. Our study reveals novel molecular pathways linking cytokinin signaling, the CLV3 regulator, and auxin signaling, and sheds light on the mechanism underlying cytokinin-regulated shoot regeneration.
INTRODUCTION
In contrast with most animal cells, plant cells typically maintain totipotency, allowing regeneration of a wide variety of explant types into whole plants through judicious in vitro culture conditions (Birnbaum and Sánchez Alvarado, 2008). In Arabidopsis (Arabidopsis thaliana), shoots normally regenerate from explants in a two-step process (Che et al., 2006). First, formation of a callus is induced by incubating root or hypocotyl explants in an auxin-rich callus-inducing medium (CIM); second, shoot regeneration is induced by transferring the callus to a cytokinin-rich shoot-inducing medium (SIM; Che et al., 2006). The observation that multiple root meristem-associated genes, notably WUSCHEL RELATED HOMEOBOX 5 (WOX5), PLETHORA 1 (PLT1), and SCARECROW (SCR), are expressed in callus has led to the hypothesis that callus formation occurs via a lateral root initiation pathway (Atta et al., 2009; Sugimoto et al., 2010). During the shoot induction phase, a number of shoot meristem-associated genes become transcriptionally active, including WUSCHEL (WUS), CLAVAT3 (CLV3), and SHOOT MERISTEMLESS (STM; Gordon et al., 2007; Atta et al., 2009; Chatfield et al., 2013).
Exogenous cytokinin is critical for in vitro shoot regeneration: growth media with a high cytokinin/auxin ratio are required for shoot regeneration (Skoog and Miller, 1957; Valvekens et al., 1988; Che et al., 2006; Gordon et al., 2007). Response to cytokinins is mediated by type-B ARABIDOPSIS RESPONSE REGULATORs (ARRs), which bind to the cytokinin response element and activate the expression of cytokinin-responsive genes, including type-A ARRs, which are negative feedback regulators of cytokinin signaling (Mason et al., 2004; Müller and Sheen, 2007; To et al., 2007; Ramireddy et al., 2013). Type-B ARRs participate in a variety of developmental processes in a largely functionally redundant manner (Mason et al., 2005; Moubayidin et al., 2010; Zhao et al., 2010). For example, ARR1 and ARR12 inhibit root elongation redundantly (Moubayidin et al., 2010; Dello Ioio et al., 2012), whereas ARR1, ARR10, and ARR12 work together to maintain the size of the shoot apical meristem (SAM; Mason et al., 2005). Evidence from the in vitro shoot regeneration system also supports the functional redundancy of type-B ARRs in cytokinin responses and cytokinin-induced shoot regeneration. Double or triple mutants in type-B ARRs (ARR1, ARR10, and ARR12) exhibit reduced explant sensitivity toward cytokinin (Mason et al., 2005; Ishida et al., 2008), as well as a reduced capacity to regenerate shoots compared to the wild type (Meng et al., 2017; Zhang et al., 2017b). However, the roles of individual type-B ARRs in shoot regeneration are not fully understood, and the repression of shoot regeneration by type-B ARRs has not been previously reported.
Different gene families mediate cytokinin signal transduction and shoot meristem regulation, each comprising multiple members (Aoyama and Oka, 2003; Dodsworth, 2009; Hill et al., 2013). A partial picture has emerged of the direct links between the shoot meristem regulators and cytokinin signaling that underlie in vivo SAM maintenance and in vitro shoot regeneration (Leibfried et al., 2005; Yanai et al., 2005; Zhang et al., 2017a). In the Arabidopsis SAM, the stem cell niche is tightly controlled by a WUS/CLV3 feedback loop (Brand et al., 2000; Schoof et al., 2000). WUS directly represses the transcription of the negative regulators ARR7 and ARR15 (Leibfried et al., 2005), whereas STM activates the key cytokinin biosynthetic gene ISOPENTENYL TRANSFERASE 7 (IPT7; Yanai et al., 2005; Sakamoto et al., 2006). The type-B ARRs ARR1, ARR10, and ARR12 directly activate WUS transcription and participate in SAM maintenance (Meng et al., 2017; Xie et al., 2018), axillary shoot meristem formation (Skylar and Wu, 2011; Wang et al., 2017; Zhang et al., 2017a), and in vitro shoot regeneration (Meng et al., 2017; Zhang et al., 2017b; Zubo et al., 2017). Cytokinin stabilizes WUS protein, which results in differential accumulation of WUS in the SAM (Snipes et al., 2018). However, direct links between cytokinin signaling and shoot meristem regulators have not been fully identified.
The balance between cytokinin and auxin is essential for various developmental processes (Moubayidin et al., 2009; Chandler and Werr, 2015). During auxin signal transduction, AUXIN/INDOLE-3-ACETIC ACIDs (Aux/IAAs) bind to AUXIN RESPONSE FACTORs (ARFs) to repress ARF activity at low auxin concentrations (Guilfoyle, 2007; Korasick et al., 2014; Salehin et al., 2015). Once the auxin receptor has been activated by the auxin signal, Aux/IAAs are degraded by the 26S proteasome, thereby releasing ARF repression and initiating an auxin-responsive gene expression cascade (Korasick et al., 2014; Salehin et al., 2015). Under normal conditions, cytokinin suppresses auxin responses or transport during primary root meristem maintenance, lateral root initiation, and shoot branching (Dello Ioio et al., 2008; Zhang et al., 2013; Marhavý et al., 2014; Šimášková et al., 2015; Waldie and Leyser, 2018). However, the role of the interaction between auxin and cytokinin in controlling shoot regeneration is only partially understood for cells cultured in vitro (Zhao et al., 2010; Cheng et al., 2013; Meng et al., 2017). Any abnormality in auxin distribution–induced by cytokinin–has a clear effect on organogenesis (Pernisová et al., 2009). Both auxin-mediated responses to cytokinin and cytokinin biosynthesis are critical for callus formation as well as shoot regeneration (Cheng et al., 2013; Liu et al., 2016). However, the interplay between genes in the auxin and cytokinin signal transduction pathways during shoot regeneration is not fully understood.
Here, we demonstrate that ARR1, a type-B ARR, is an essential inhibitor of shoot regeneration during in vitro culture that modulates the expression of WUS and CLV3 in an ARR12-dependent manner and directly activates IAA17, thereby blocking shoot regeneration.
RESULTS
ARR1 is a Negative Regulator of Shoot Regeneration
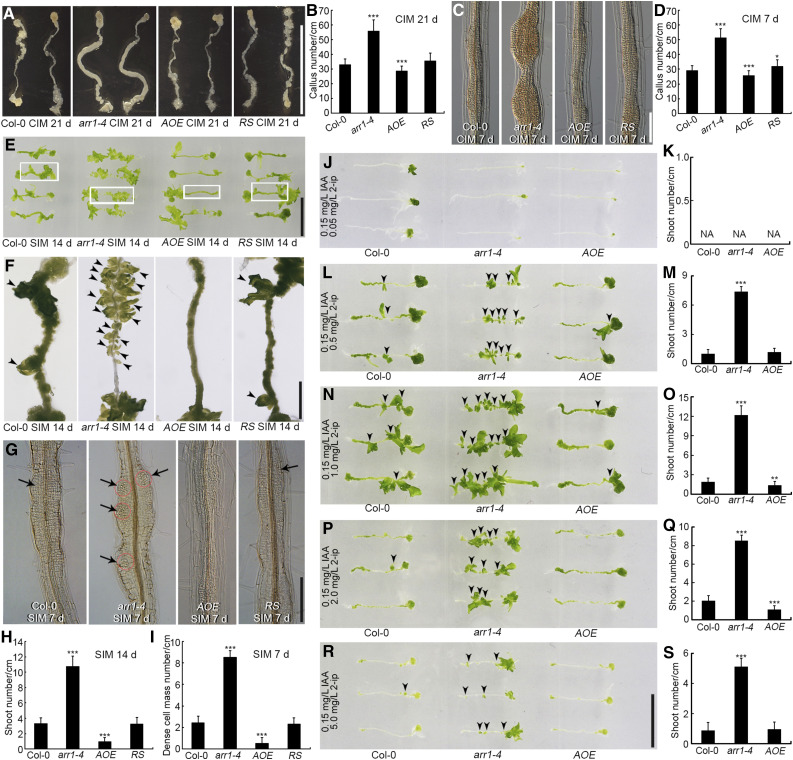
To identify the role of ARR1 in shoot regeneration, we subjected root explants to regeneration assays via a two-step process. We evaluated the callus formation ability of root explants derived from the arr1-4 mutant, a transgenic line overexpressing ARR1 (AOE), and the arr1-4 mutant complemented with ARR1pro:ARR1-GFP [hereafter referred to as the RS (rescued) line] on CIM. After 21 d of culture, the arr1-4 mutant had formed ∼70% more calli than the wild type, the RS line as many calli as the wild type, and the AOE line slightly fewer calli than the wild type (Figures 1A and 1B). After 7 d of culture, callus formation was more extensive in arr1-4 than in the wild type (Figures 1C and 1D), but not in the RS line, as expected (Figures 1C and 1D). These data indicate that ARR1 reduces the capacity for callus formation.
Figure 1.
ARR1 Inhibits Shoot Regeneration from Root Explants.
(A) and (C) Callus formation from root explants of wild type (Col-0), arr1-4, AOE, and RS after incubation on CIM for 21 d (A) and 7 d (C).
(B) and (D) Number of calli after 21 d (B) and 7 d (D) of culture. Data represent mean ± sd (n = 24); cm, centimeter.
(E) and (F) Shoot regeneration after 14 d of culture on SIM from root explants of Col-0, arr1-4, AOE, and RS. Four independent AOE lines and three RS lines exhibited similar phenotypes.
(G) Dense cell masses formed following 7 d of culture on SIM in root explants derived from Col-0, arr1-4, AOE, and RS.
(H) and (I) Number of regenerated shoots after 14 d (H) of culture and dense cell masses after 7 d of culture ([I]; n = 24).
(J) to (S) Shoot regeneration and the number of regenerated shoots from Col-0, arr1-4, and AOE calli derived from root explants cultured on medium containing 0.05 mg/L N6-(2-isopentenyl) adenine ([J] and [K]), 0.5 mg/L N6-(2-isopentenyl) adenine ([L] and [M]), 1.0 mg/L N6-(2-isopentenyl) adenine ([N] and [O]), 2.0 mg/L N6-(2-isopentenyl) adenine ([P] and [Q]), and 5.0 mg/L N6-(2-isopentenyl) adenine ([R] and [S]) for 14 d. All media contained 0.15 mg/L IAA. Values represent mean ± sd (n = 23). *, **, ***: means differ at P < 0.05, 0.01, and 0.001, respectively (Student’s t test). Scale bars = 1 cm in (A), (E), and (R); 100 μm in (C) and (G); and 2.5 mm in (F).
After incubation on SIM for 14 d, root explants obtained from arr1-4 plants regenerated more shoots than those obtained from wild-type plants (Figures 1E, 1F, and 1H). Root explants from the AOE line produced fewer shoots than those derived from the wild type, whereas root explants derived from the RS line had as many shoots as the wild type (Figures 1E, 1F, and 1H; Supplemental Figure 1). No shoots formed from any of the root explants within 7 d of transfer to SIM (Figure 1G), although arr1-4 explants had already formed more dense cell masses than wild-type or RS explants, whereas explants obtained from the AOE line had formed fewer dense cell masses (Figures 1G and 1I). Thus, the abundance of ARR1 transcript in root explants negatively correlated with the number of regenerated shoots (Figures 1E to 1I; Supplemental Figure 1), suggesting that ARR1 represses shoot regeneration.
Next, we transferred CIM-induced calli derived from root explants of arr1-4, AOE, and wild-type plants onto fresh SIM supplemented with various concentrations of N6-(2-isopentenyl) adenine (2-ip) and 0.15 mg/L IAA. More shoots regenerated from arr1-4 calli than from wild-type calli, except in the presence of 0.05 mg/L 2-ip, which blocked shoot regeneration in all the explants (Figures 1J to 1S). Fewer shoots regenerated from AOE calli than from wild-type calli at 2-ip concentrations of both 1.0 and 2.0 mg/L (Figures 1N to 1Q). Hypocotyl explants of arr1-4, AOE, and wild-type lines responded similarly to the presence of various concentrations of 2-ip and 0.15 mg/L IAA (Supplemental Figure 2). These findings suggest that the repressive effect exerted by ARR1 on shoot regeneration is independent of the local cytokinin concentration. In addition, explants derived from the other strong allele arr1-3 generated more calli and shoots than the wild type, which is similar to the phenotype of arr1-4 (Supplemental Figure 3), further supporting the inhibitory effect of ARR1.
ARR1 Inhibits Shoot Regeneration in an ARR12-Dependent Manner
We previously showed that ARR12 promotes shoot regeneration during the two-step culture process (Dai et al., 2017). To understand the roles of different type-B ARR genes, we subjected arr1, arr10, arr11, arr12, and arr18 explants and the corresponding double mutants to regeneration assays.
After incubation on SIM for 14 d, arr1 explants contained three times as many regenerated shoots than the wild type, arr12 explants produced only one quarter as many regenerated shoots as the wild type, and arr10 contained slightly fewer regenerated shoots (Supplemental Figures 4A to 4F and 4Q). arr12-containing double mutants all produced a similar or smaller number of regenerated shoots than the arr12 single mutant, regardless of which other gene was mutated (Supplemental Figures 4G to 4J and 4Q). The arr11 arr18 double mutant formed as many shoots as the wild type (Supplemental Figures 4K, 4L, and 4Q). In addition, the introduction of ARR12pro:ARR12 in the arr12 mutant complemented its shoot regeneration defect (Supplemental Figures 4M to 4Q). These results indicate that there is functional diversity among type-B ARRs in regulating shoot regeneration in vitro, with ARR12 acting as a central enhancer and ARR1 as a strong inhibitor of this process.
To explore the relationship between ARR1 and ARR12, we evaluated the callus and shoot regeneration abilities of the wild type and the arr1, arr12, and arr1 arr12 mutants. The arr1 single mutant generated more calli (Figures 2A, 2B, and 2E) and shoots (Figures 2F, 2G, and 2J) than the wild type, indicating that ARR1 inhibits both callus formation and shoot regeneration. In the absence of ARR12, however, the arr1 arr12 double mutant generated a similar number of calli (Figures 2C to 2E) and fewer shoots (Figures 2H to 2J) than the arr12 single mutant, indicating that ARR1 enhanced shoot regeneration in this case. Thus, ARR1-mediated inhibition of callus formation and shoot regeneration depends on the presence of ARR12. The difference between the wild type and arr12 was significantly reduced compared to that between arr1 and arr1 arr12 (P < 0.001; Figures 2A, 2C, 2F, and 2H; Supplemental Figures 5A and 5B), indicating that ARR1 inhibits callus formation and shoot regeneration by counteracting the positive effect of ARR12.
Figure 2.
Inhibition of Shoot Regeneration by ARR1 Depends on the Presence of ARR12.
(A) to (D) Callus formation from root explants of Col-0, arr1, arr12, and arr1 arr12 after 21 d of incubation on CIM.
(E) Number of calli after 21 d of incubation on CIM. Data represent mean ± sd (n = 24).
(F) to (I) Shoot regeneration after 14 d of incubation on SIM from root explants of Col-0, arr1, arr12, and arr1 arr12.
(J) Number of regenerated shoots after 14 d of incubation on SIM. Data represent mean ±sd (n = 24). ***Differ at P < 0.001 (Student’s t test). Scale bars = 1 cm.
Expression of ARR1 under the control of the cauliflower mosaic virus 35S promoter in wild-type explants resulted in fewer shoots than in untransformed wild-type explants (Supplemental Figures 6A to 6D). By contrast, expression of the 35Spro:ARR1 transgene in the arr1 arr12 double mutant background resulted in more shoots than in explants derived from the arr1 arr12 double mutant (Supplemental Figures 6E to 6H). These data suggest that when cytokinin signal transduction is blocked (as in the arr1 arr12 double mutant), overexpression of ARR1 slightly enhances shoot regeneration; however, in the wild-type background, overexpression of ARR1 inhibits shoot regeneration, possibly by altering the function of ARR12 in explants.
The transcription levels of ARR12 in arr1 explants was slightly lower than that in wild-type explants after culture on CIM for 2 d and on SIM for 4 d (Supplemental Figures 5C and 5D), whereas it was slightly higher in arr1 and 35Spro:ARR1 seedlings than in the wild type (Supplemental Figure 5E). We observed no significant difference in ARR1 transcription levels between wild-type and arr12 explants or seedlings (Supplemental Figures 5F to 5H), whereas ARR1 transcript levels were slightly higher in 35Spro:ARR12 seedlings compared to the wild type (Supplemental Figure 5H). These results suggest that ARR1 and ARR12 slightly influence the transcription of the other gene during shoot regeneration.
The Effects of ARR1 and ARR12 on Shoot Regeneration Are Determined by Distinct Protein Functions and Expression Patterns
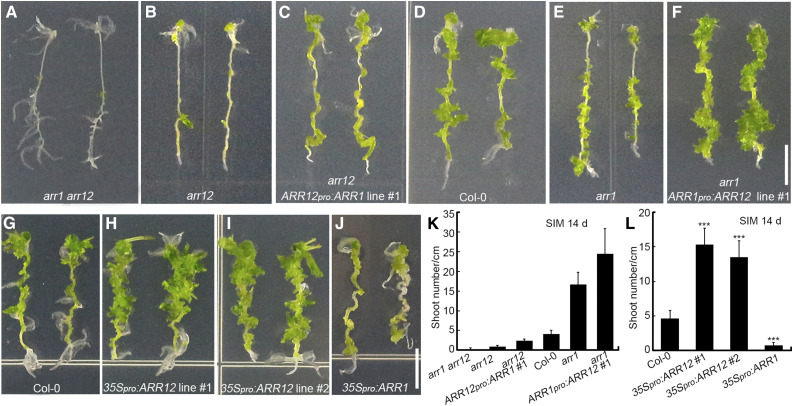
To explore whether the different effects of ARR1 and ARR12 on shoot regeneration are determined by distinct protein functions or expression patterns of their encoding genes, we evaluated the shoot regeneration abilities of explants with different levels of ARR1 and ARR12. The ranked shoot regeneration ability of the explants was arr1 arr12 < arr12 < arr12 + ARR12pro:ARR1 < Arabidopsis ecotype Columbia (Col-0) < arr1 < arr1 + ARR1pro:ARR12 (Figures 3A to 3F and 3K; Supplemental Figure 7).
Figure 3.
The Differential Effects of ARR1 and ARR12 on Shoot Regeneration Are Determined by Distinct Protein Functions and Expression Patterns.
(A) to (F) Shoot regeneration from root explants incubated for 14 d on SIM, including root explants of arr1 arr12 (A), arr12 (B), ARR12pro:ARR1 arr12 (ARR1 expressed under the control of the ARR1 and ARR12 promoters; [C]), Col-0 (D), arr1 (E), and ARR1pro:ARR12 arr1 (ARR12 expressed under the control of ARR1 and ARR12 promoters) plants (F).
(G) to (J) Shoot regeneration from root explants incubated for 14 d on SIM, including root explants from Col-0 (G), 35Spro:ARR12 transgenic (lines no. 1 and no. 2; [H] and [I]), and 35Spro:ARR1 transgenic plants (J).
(K) and (L) Number of regenerated shoots after 14 d of culture. Data represent mean ± sd (n = 24). ***Differ at P < 0.001 (Student’s t-test). Scale bars = 0.5 cm. Two independent transgenic lines were analyzed for each genetic background, which exhibited similar phenotypes ([C], [F], [H], and [I]).
arr1 arr12 double mutant explants (with no ARR1 or ARR12 expression) generated almost no shoots after 14 d of incubation on SIM (Figures 3A and 3K). The arr12 single mutant (retaining the endogenous ARR1 gene) generated more shoots than the arr1 arr12 double mutant (Figures 3A, 3B, and 3K). Explants harboring the ARR12pro:ARR1 transgene in the arr12 single mutant background (with ARR1 expressed under the control of the ARR12 promoter and the endogenous ARR1 promoter) produced more shoots than the untransformed arr12 single mutant (Figures 3B, 3C, and 3K; Supplemental Figures 7B and 7D) but fewer shoots than Col-0 (with normal levels of ARR1 and ARR12 expression from the endogenous loci; Figures 3C, 3D, and 3K). The arr1 single mutant (retaining the endogenous ARR12 gene) generated more shoots than Col-0 (Figures 3D, 3E, and 3K). Explants harboring the ARR1pro:ARR12 transgene in the arr1 single mutant background (with ARR12 expressed under the control of the ARR1 promoter and the endogenous ARR12 promoter) produced more shoots than the untransformed arr1 single mutant (Figures 3E, 3F, and 3K; Supplemental Figures 7C and 7D).
Thus, ARR1 and ARR12 expressed under the control of the same promoters (i.e., ARR1 and ARR12 promoters) had different effects on the shoot regeneration abilities of explants (Figures 3C and 3F; Supplemental Figures 7B and 7C). Consistent with these results, explants harboring the 35Spro:ARR12 transgene produced more shoots than Col-0 (Figures 3G to 3I, and 3L), whereas explants harboring the 35Spro:ARR1 transgene generated fewer shoots than Col-0 (Figures 3G, 3J, and 3L). In addition, endogenous ARR1pro:ARR1 did not complement the arr12 mutant phenotype, whereas the ARR12pro:ARR1 transgene partially rescued the arr12 phenotype (Figures 3B to 3D; Supplemental Figures 7A and 7B), indicating that ARR1 has slightly different effects on shoot regeneration when expressed under the control of different promoters.
Although the 35Spro:ARR1 transgene decreased the regeneration ability of the wild type (Supplemental Figures 8A, 8B, and 8E), overexpression of a phosphomimetic mutant version of ARR1 (35Spro:mARR1D94E; Supplemental Figures 8F and 8G) dramatically increased regeneration (Supplemental Figures 8C to 8E), in accordance with previous findings (Kurepa et al., 2014). Thus, the phosphorylation status of ARR1 contributes to its inhibitory effect on shoot regeneration.
Localization and Accumulation Pattern of ARR1 and ARR12 During Shoot Regeneration
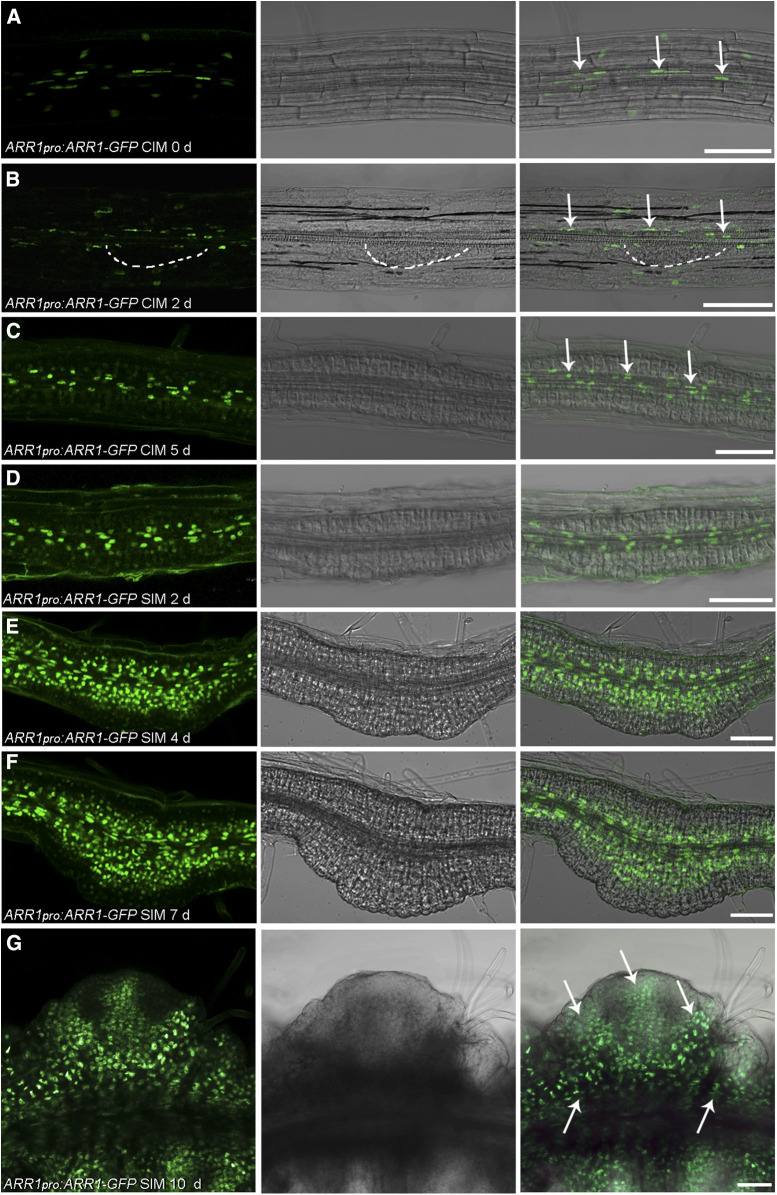
We next used ARR1pro:ARR1-GFP and ARR12pro:ARR12-GFP translational reporter lines to evaluate the localization of ARR1 and ARR12 during shoot regeneration. We detected ARR1 signal in the transition zone of primary roots (Supplemental Figures 9A to 9C), which agrees with previous reports (Moubayidin et al., 2010, 2013). After culturing root explants on CIM for 0 and 2 d, we detected ARR1 signal in stele cells as well as other root cell layers (Figures 4A and 4B; Supplemental Figures 9D to 9I). After culturing root explants on CIM for 5 d, most of the ARR1 signal present was associated with stele cells and the cells surrounding the stele (Figure 4C). The level of ARR1 signal increased in the inner layer of cells after culturing root explants on SIM for 2, 4, and 7 d (Figures 4D to 4F). We observed no specific GFP signal at 7 d, when nascent SAMs formed (Figure 4F). After 10 d on SIM, we detected ARR1 throughout the explant, including in the mature SAMs (Figure 4G).
Figure 4.
Temporal and Spatial Expression Patterns of ARR1 During Shoot Regeneration in Root Explants.
(A) to (G) The site of ARR1 deposition in ARR1pro:ARR1-GFP roots (A) and root explants incubated on CIM for 2 d (B) and 5 d (C) and on SIM for 2 d (D), 4 d (E), 7 d (F), and 10 d (G). White dashed line in (B) indicates the generated callus. Arrows in (A) to (C) indicate the localization of the ARR1-GFP in stele cells of root explants. Arrows in (G) indicate the distribution of ARR1 in the explant when the mature SAM formed. Scale bars = 50 μm. Two independent transgenic lines were analyzed, with similar expression patterns.
After culturing root explants on CIM for 0 and 2 d, we detected ARR12 signal specifically in stele cells (Supplemental Figures 10A to 10D). After culturing root explants on CIM for 5 d, most of the ARR12-GFP signal was associated with stele cells and the cells around the stele, similar to the ARR1-GFP signal (Supplemental Figure 10E). After root explants were cultured on SIM for 2, 4, and 7 d, the ARR12 signal level was higher than before transferring the explants to SIM, and the sites of GFP signal coincided with those of nascent SAMs (Supplemental Figures 10F to 10H). After 10 d on SIM, ARR12 localized specifically to newly formed SAMs (Supplemental Figure 10I).
ARR1 Inhibits Shoot Regeneration by Modulating CLV3 and WUS Expression in an ARR12-Dependent Manner
We measured the expression level of various genes associated with shoot regeneration in wild-type (Col-0), arr1, and AOE explants. There was no significant difference in WOX5 expression between Col-0 and arr1 mutant explants on CIM (Supplemental Figures 11A and 11C), whereas there was a higher level of WOX5 expression in arr1 explants than in Col-0 after culture on SIM for 4, 7, and 10 d (Supplemental Figure 11A). SCR expression in arr1 explants was slightly higher in arr1 explants than in Col-0 after culture on CIM for 5 d and on SIM for 2 and 4 d (Supplemental Figures 11B and 11C). IAA3 and IAA14 expression was higher in arr1 explants than in Col-0 explants after culture on SIM for 10 d (Supplemental Figures 11D and 11E). ARR5, ARR7, and ARR15 expression in arr1 explants was lower than in Col-0 after culture on CIM for 5 d (Supplemental Figure 11F).
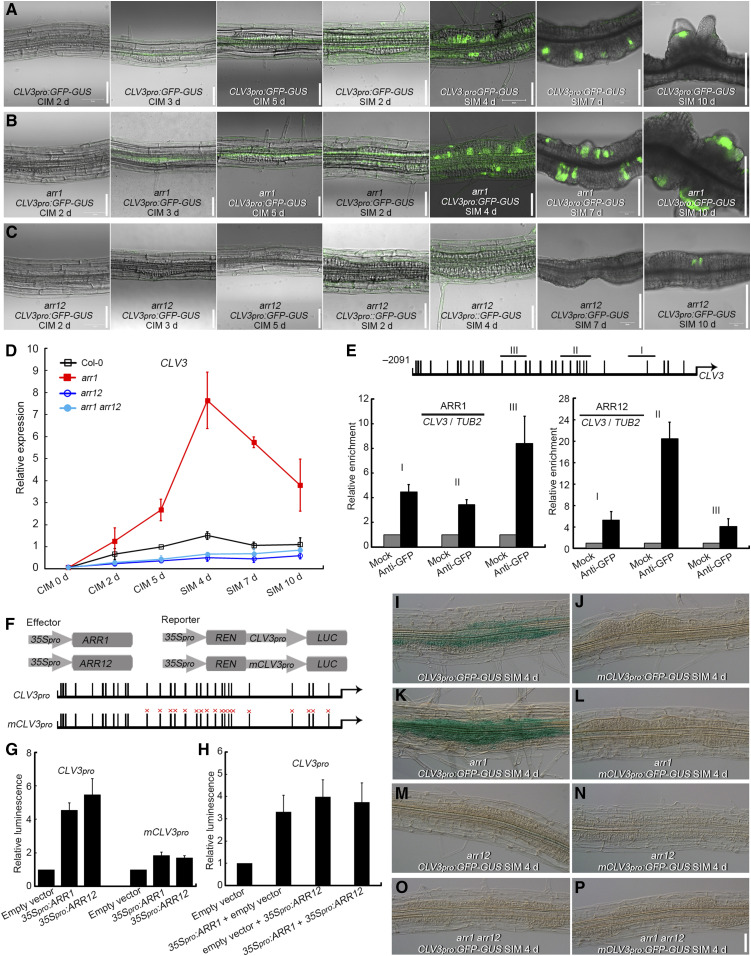
CLV3 is an established stem cell regulator in the SAM (Schoof et al., 2000). CLV3 expression in Col-0 explants gradually increased when cultured on CIM and peaked after culture on SIM for 4 d, with widespread distribution of CLV3 transcript in the explants (Figures 5A and 5D; Supplemental Figure 12A). CLV3 transcript distribution then decreased and concentrated in certain areas after culture on SIM for 7 d and was finally located in the newly formed SAMs after culture on SIM for 10 d (Figure 5A; Supplemental Figure 12A). CLV3 transcript was significantly more abundant in root explants derived from the arr1 single mutant than in those from Col-0, both on CIM and SIM (Figures 5A, 5B, and 5D; Supplemental Figures 12A, 12B, 12E, and 12F). In both arr12 and arr1 arr12 explants, CLV3 was transcribed at a much lower level than in Col-0 (Figures 5A, 5C, and 5D; Supplemental Figures 12A and 12D). Furthermore, the β-glucuronidase (GUS) signal of CLV3pro:GFP-GUS was weaker in AOE explants than in the wild-type background (Supplemental Figures 12A and 12C). These data suggest that ARR12 is a critical positive regulator of CLV3 expression, that ARR1 is a negative regulator of CLV3 transcription in explants during CIM and SIM culture, and that the significant repression of CLV3 transcription by ARR1 requires the presence of ARR12.
Figure 5.
ARR1 Significantly Represses the Expression of CLV3 in an ARR12-Dependent Manner.
(A) to (C) Temporal and spatial expression patterns of CLV3 in Col-0 (A), arr1 (B), and arr12 (C) root explants incubated on CIM for 2, 3, and 5 d and on SIM for 2, 4, 7, and 10 d.
(D) Transcript abundance of CLV3 in root explants derived from Col-0, arr1, arr12, and arr1 arr12 cultured on CIM for 0, 2, and 5 d and on SIM for 4, 7, and 10 d. The relative expression level was normalized with the data of Col-0 CIM 5 d.
(E) Enrichment of CLV3 promoter fragments following a ChIP assay conducted on ARR1pro:ARR1-GFP and ARR12pro:ARR12-GFP transgenic explants. Black boxes indicate the core cytokinin response motif 5′-GAT(T/C)-3′.
(F) to (H) A transient expression assay showing that ARR1 and ARR12 activate CLV3 expression through binding to similar sites of its promoter region ([F] and [G]) and ARR1 and ARR12 promote CLV3 expression with no synergistic effect (H). Black boxes marked with “×” denote A-to-T and G-to-C point mutations.
(I), (K), (M), and (O) Temporal and spatial transcriptional activity of the CLV3 promoter in Col-0 (I), arr1 (K), arr12 (M) and arr1 arr12 (O) root explants incubated on SIM for 4 d.
(J), (L), (N), and (P) Temporal and spatial transcriptional activity of the mutated CLV3 promoter (mCLV3) in Col-0 (J), arr1 (L), arr12 (N), and arr1 arr12 (P) root explants incubated on SIM for 4 d. Scale bars = 50 μm. Two independent transgenic lines were analyzed for each genetic background, with similar expression levels ([I] to [P]).
Evaluation of the CLV3 promoter sequence revealed 29 copies of the core cytokinin response motif 5′-GAT(T/C)-3′ within the 2,091-bp upstream sequence of the start codon (Ramireddy et al., 2013; Figure 5E). A chromatin immunoprecipitation (ChIP) assay conducted on transgenic ARR1pro:ARR1-GFP and ARR12pro:ARR12-GFP explants at the nascent SAM formation stage (SIM 7 d) showed that ARR1 and ARR12 specifically bind to the core cytokinin response motif-containing region of the CLV3 promoter (Figure 5E). A transient expression assay in Arabidopsis protoplasts showed that the luciferase signal was stronger in both the CLV3pro:LUC + 35Spro:ARR1 and CLV3pro:LUC + 35Spro:ARR12 samples compared to the CLV3pro:LUC + empty vector (Figures 5F and 5G). When 18 copies of the core cytokinin response motif (within the 1,400-bp upstream sequence of the CLV3 start codon) were mutated (mCLV3; Supplemental Table 1), the activation of mCLV3pro:LUC by 35Spro:ARR1 and 35Spro:ARR12 was reduced compared to the wild-type CLV3 promoter (Figures 5F and 5G). These data suggest that ARR1 and ARR12 bind to the same sites of the CLV3 promoter and modulate its expression. However, we observed an intermediate luciferase signal when both 35Spro:ARR1 and 35Spro:ARR12 were cotransformed in the transient expression system compared to transformation with 35Spro:ARR1 or 35Spro:ARR12 alone, with no synergistic effect observed (Figure 5H). The signal strength of mCLV3pro:GFP-GUS exhibited a sharp decrease compared to that of CLV3pro:GFP-GUS in the Col-0 background (Figures 5I and 5J). A sharper decrease in GUS signal between mCLV3pro:GFP-GUS and CLV3pro:GFP-GUS occurred in the arr1 background compared to the Col-0 background (Figures 5K and 5L). CLV3pro:GFP-GUS and mCLV3pro:GFP-GUS signals in explants in the arr12 and arr1 arr12 backgrounds were very weak (Figures 5M to 5P). Together, these data support the hypothesis that ARR1 competes with ARR12 for binding to the CLV3 promoter region and represses its expression indirectly.
The observation that ARR1 represses CLV3 expression suggested that CLV3 functions downstream of ARR1. To test this hypothesis, we compared the ability of arr1-4, clv3-7, arr1-4 clv3-7, and wild-type root explants to generate calli and shoots. clv3-7 root explants generated fewer calli and shoots than the other genotypes (Figures 6A, 6B, 6E, 6F, 6G, and 6J), confirming that CLV3 promotes callus formation and shoot regeneration. The arr1-4 clv3-7 double mutant generated an intermediate number of calli and shoots compared to arr1-4 and clv3-7 single mutants (Figures 6B to 6E and 6G to 6J), suggesting that the loss of CLV3 function partially rescued the arr1-4 phenotype and that CLV3 acts downstream of ARR1 in regulating shoot regeneration.
Figure 6.
CLV3 Acts Downstream of ARR1 in Regulating Shoot Regeneration.
(A) to (D) Callus formation from root explants of Col-0, clv3-7, arr1, and arr1 clv3-7 after incubation on CIM for 21 d.
(E) Number of calli after 21 d. Data represent mean ± sd (n = 28).
(F) to (I) Shoot regeneration after incubation on SIM for 14 d from root explants of Col-0, clv3-7, arr1, and arr1 clv3-7.
(J) Number of regenerated shoots after 14 d. Data represent mean ± sd (n = 28). ***Differ at P < 0.001 (Student’s t-test). Scale bars = 1 cm.
WUS is considered to be a key regulator of shoot regeneration (Gordon et al., 2007; Meng et al., 2017; Zhang et al., 2017b). We therefore examined whether ARR1 or ARR12 regulates WUS expression. The distribution of GUS signal in transgenic WUSpro:GFP-GUS explants (Supplemental Figure 13A; Cui et al., 2015) was similar to that in in vitro hypocotyl explants reported previously (Zhang et al., 2017b). We also confirmed the expression pattern of WUS in transgenic WUSpro:GFP-GUS root explants (Supplemental Figure 13B) by RNA in situ hybridization (Supplemental Figures 13C to 13D). No WUS expression was detected in any of the explants maintained on CIM (Supplemental Figures 14A to 14D, and 15A and 15E). WUS transcription in Col-0 explants was induced when the explants were cultured on SIM and peaked after a 7-d culture on SIM (Supplemental Figures 14A, 14B, and 15A to 15D). Finally, the distribution of WUS transcript was concentrated and located in newly formed SAMs after a 10-d culture on SIM (Supplemental Figures 14B and 15D). The transcript abundance of WUS in arr1 explants peaked after a 4-d culture on SIM and became concentrated after a 7-d culture on SIM, which was earlier than in Col-0 explants, and was located in newly formed SAMs after a 10-d culture on SIM (Supplemental Figures 14A to 14C, and 15A to 15H). WUS transcript was less abundant in arr12 explants than in the wild type, and even lower in arr1 arr12 explants (Supplemental Figures 14A, 14B, and 14D). Therefore, ARR12 is a critical positive regulator of WUS expression, whereas ARR1 is a weak positive regulator of WUS transcription and induces the widespread distribution of WUS in explants in the presence of ARR12.
Evaluation of the WUS promoter sequence revealed 35 copies of the core cytokinin response motif 5′-GAT(T/C)-3′ within the 2,079-bp region upstream of the start codon (Supplemental Figure 16A; Ramireddy et al., 2013). A ChIP assay using ARR1pro:ARR1-GFP and ARR12pro:ARR12-GFP transgenic explants at the nascent SAM formation stage (SIM 7 d) showed that ARR1 and ARR12 specifically bind to the core cytokinin response motif-containing region of the WUS promoter (Supplemental Figure 16A). 35Spro:ARR1 and 35Spro:ARR12 activated the WUSpro:LUC reporter in Arabidopsis protoplasts (Supplemental Figures 16B and 16C), whereas their activation of mWUS1pro:LUC and mWUS2pro:LUC (Meng et al., 2017; Zhang et al., 2017b) was reduced compared to that of the wild-type WUS promoter (Supplemental Figures 16B and 16C). We observed no synergistic effect on luciferase signal when both 35Spro:ARR1 and 35Spro:ARR12 were cotransformed compared to transformation of 35Spro:ARR1 or 35Spro:ARR12 alone (Supplemental Figure 16D), suggesting that ARR1 and ARR12 regulate WUS expression by competitively binding to the same sites of the WUS promoter.
ARR1 Inhibits Shoot Regeneration Through Transcriptional Activation of IAA17
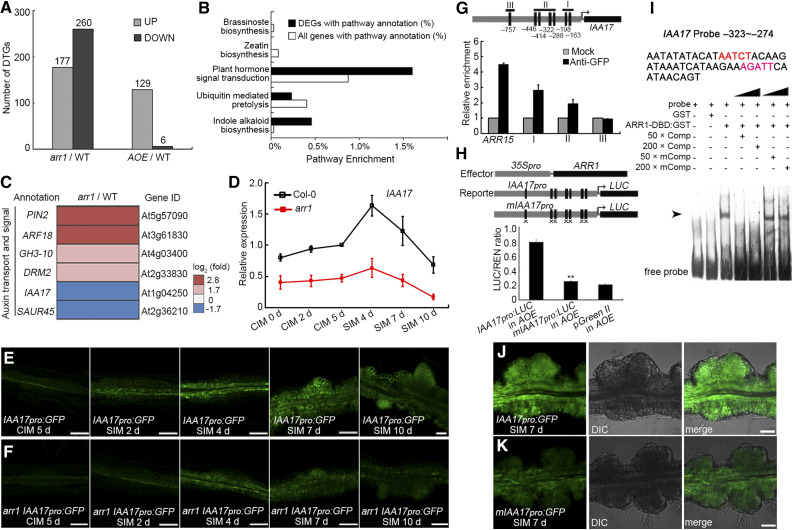
A comparison of the transcriptomes of arr1, AOE, and wild-type root explants at the nascent SAM formation stage by deep sequencing (RNA-Seq) revealed many differentially transcribed genes (DTGs). In the transcriptomes of arr1 versus wild-type explants, we detected 177 upregulated and 260 downregulated DTGs, and in the AOE versus wild-type transcriptomes, we detected 129 and 6 DTGs, respectively (Figure 7A). Pathways involved in hormone signal transduction and indole alkaloid synthesis were well represented among the DTGs in arr1 versus the wild type (Figure 7B). These DTGs also included genes involved in auxin transport and signaling, as well as IAA17, encoding an Aux/IAA repressor that is downregulated in arr1 root explants (Figure 7C). IAA17 expression in Col-0 explants slightly increased when cultured on CIM, and highly induced and peaked after culture on SIM for 4 d (Figure 7D). IAA17 transcript levels then decreased after culture on SIM for 7 and 10 d (Figure 7D). IAA17 transcript levels were lower in arr1 than in Col-0 root explants during both CIM and SIM incubation (Figure 7D). In transgenic root explants harboring IAA17pro:GFP and analyzed at different stages of shoot regeneration, GFP signals were all weaker in arr1 than in the wild type (Figures 7E and 7F). Together, these results suggest that ARR1 promotes IAA17 expression.
Figure 7.
ARR1 Promotes the Expression of IAA17 via Binding to its Promoter.
(A) DTGs in arr1 vs. Col-0 and AOE vs. Col-0.
(B) Pathway enrichment of DTGs in arr1 vs. Col-0.
(C) Transcript profiling of DTGs related to auxin transport and signaling from arr1 vs. Col-0.
(D) Transcript abundance of IAA17 in root explants derived from Col-0 and arr1 explants cultured on CIM or SIM at different stages. The relative expression level was normalized with the data of Col-0 CIM 5 d.
(E) and (F) Temporal and spatial expression patterns of IAA17 in wild-type (E) and arr1 (F) root explants after culturing on CIM for 5 d and on SIM for 2, 4, 7, and 10 d. The arr1 IAA17pro:GFP lines were obtain via plant crossing of arr1 and IAA17pro:GFP.
(G) The enrichment of IAA17 promoter fragments following ChIP.
(H) Transient expression assay showing that ARR1 promotes IAA17 expression.
(I) ARR1 binds to the promoter region of IAA17 (indicated by arrows) in an EMSA.
(J) and (K) Temporal and spatial expression patterns of IAA17pro:GFP and mIAA17pro:GFP in root explants at the nascent SAM formation stage. Black boxes refer to the cytokinin response motif 5′-(A/G)GAT(T/C)-3′ and black boxes marked with “×” denote A-to-C and G-to-C point mutations (Supplemental Data Set 1). **Differ at P < 0.01 (Student’s t test). Scale bar = 50 μm. Two independent transgenic lines were analyzed for each genetic background, with similar expression patterns ([E], [F], [J], and [K]). DIC, differential interference contrast field of microscopy.
A scan of the IAA17 promoter sequence revealed the presence of six copies of the cytokinin response motif [CRM, 5′-(A/G)GAT(T/C)-3′] and one copy of the extended cytokinin response motif [ECRM, 5′-AAGAT(T/C)TT-3′; Figure 7G; Ramireddy et al., 2013]. A ChIP assay conducted on ARR1pro:ARR1-GFP explants at the nascent SAM formation stage showed that ARR1 specifically binds to the core cytokinin response motif-containing region of the IAA17 promoter (Figure 7G). When IAA17pro:LUC or mIAA17pro:LUC was transiently expressed in Arabidopsis protoplasts harboring 35Spro:ARR1, the signal level was lower in the case of the mutated promoter (Figure 7H). An electrophoretic mobility shift assay (EMSA) confirmed that ARR1 binds to the IAA17 promoter in vitro (Figure 7I). Furthermore, transgenic plants harboring mIAA17pro:GFP expressed GFP less effectively than plants harboring IAA17pro:GFP (Figures 7J and 7K). Together, these results suggest that ARR1 promotes IAA17 expression by directly binding to its promoter region.
The transcript abundance of IAA17 in arr12 and wild-type root explants was similar during CIM incubation, whereas IAA17 transcript was more abundant in arr12 than in wild-type root explants after being cultured on SIM for 7 and 10 d (Supplemental Figure 17A). However, 35Spro:ARR12 activated the IAA17pro:LUC reporter in Arabidopsis protoplasts, but not mIAA17pro:LUC, and cotransformation of 35Spro:ARR1 and 35Spro:ARR12 did not have a synergistic effect on IAA17pro:LUC expression (Supplemental Figure 17B), suggesting that ARR1 and ARR12 regulate IAA17 expression by competitively binding to its promoter.
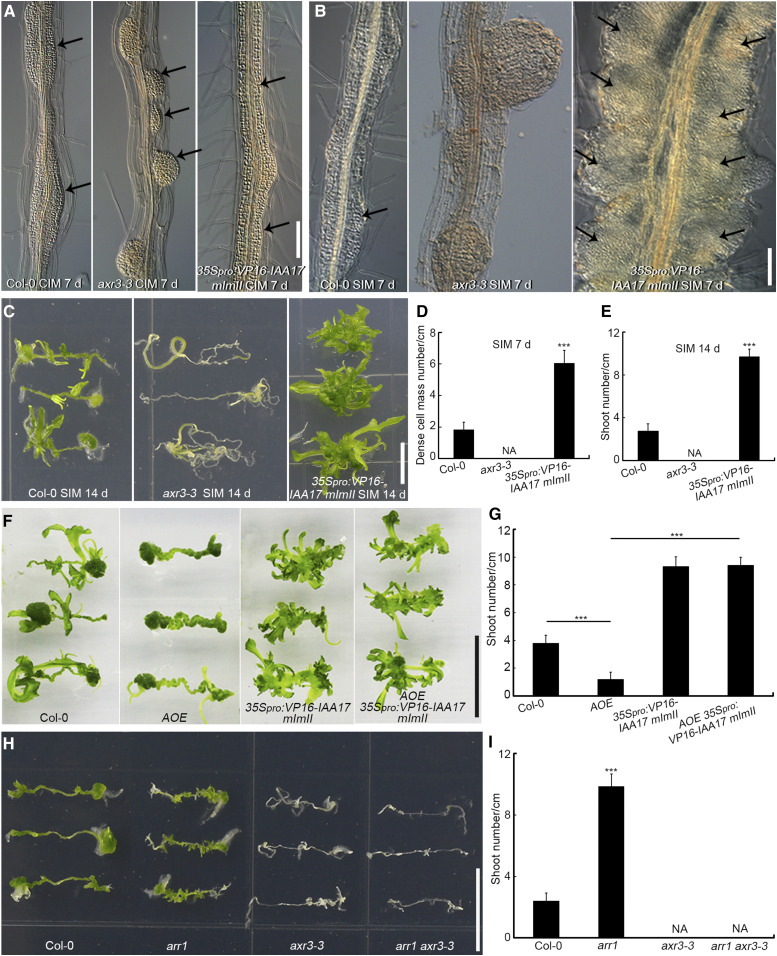
We then investigated the callus formation ability and shoot regeneration capacity of 35Spro:VP16-IAA17mImII (stabilized version of the VP16-IAA17 activator, which activates auxin response genes in an auxin-independent manner) and the dominant mutant axr3-3 (carrying a gain of function allele of the IAA17 repressor; Supplemental Figure 18A; Rouse et al., 1998; Li et al., 2009; Tian et al., 2014). When compared to Col-0, root explants derived from axr3-3 formed separated and dome-shaped callus after 7 d of incubation on CIM (Figure 8A), which grew into large root-like structures after 21 d of incubation on CIM (Supplemental Figures 19A and 19B). Root explants of 35Spro:VP16-IAA17mImII generated connected callus along the entire vascular cylinder (Figure 8A) and ultimately developed into connected columns of callus after 21 d of incubation on CIM (Supplemental Figure 19C), which is similar to the phenotype of arr1 (Figure 1A). After 7 d of culture on SIM, dome-shaped axr3-3 calli grew into root primordium-like structures (Figure 8B) and ultimately formed roots (Figure 8C), with no dense cell mass formation or shoot regeneration (Figures 8D and 8E). However, the connected 35Spro:VP16-IAA17mImII callus proliferated quickly and formed larger numbers of dense cell masses (Figures 8B and 8D) and shoots (Figures 8C and 8E) compared to Col-0 (Figures 8B to 8E). Root explants derived from two independent T-DNA insertion loss-of-function IAA17 mutants, iaa17-lof-1 (iaa17-loss of function-1, SALK_011820) and iaa17-lof-2 (SALK_065697), produced more shoots than those derived from the wild type (Supplemental Figures 18B and 18C), indicating that IAA17 represses shoot regeneration.
Figure 8.
IAA17 Functions Downstream of ARR1 During Shoot Regeneration in Vitro.
(A) Callus formation from root explants after 7 d of incubation on CIM, including root explants from Col-0, axr3-3, and 35Spro:VP16-IAA17 mImII. Black arrows indicate the generated calli. Scale bars = 50 μm.
(B) Dense cell masses (indicated by arrows) formed following 7 d of culture on SIM in root explants derived from Col-0, axr3-3, and 35Spro:VP16-IAA17 mImII. Scale bars = 50 μm.
(C) Shoot regeneration after 14 d of incubation on SIM from root explants of Col-0, axr3-3, and 35Spro:VP16-IAA17 mImII. Scale bars = 1 cm.
(D) and (E) Number of dense cell masses at the nascent meristem stage (D) and regenerated shoots at the shoot regeneration stage (E; n = 24).
(F) Shoot regeneration capacity of Col-0, AOE, 35Spro:VP16-IAA17mImII, and AOE 35Spro:VP16-IAA17mImII at the shoot regeneration stage. Scale bars = 1 cm.
(G) Number of regenerated shoots. Data represent mean ± sd (n = 24).
(H) Shoot regeneration capacity of Col-0, arr1, axr3-3, and arr1 axr3-3 at the shoot regeneration stage. Scale bars = 1 cm.
(I) Number of regenerated shoots. Data represent mean ± sd (n = 24). ***Differ at P < 0.001 (Student’s t test).
We then compared the ability of root explants of the four genotypes to regenerate shoots: AOE, 35Spro:VP16-IAA17mImII, the double overexpressor, and the wild type. AOE-derived explants formed fewer shoots than the wild type, whereas 35Spro:VP16-IAA17 explants produced more shoots (Figures 8F and 8G). The AOE 35Spro:VP16-IAA17mImII double overexpressor line generated as many shoots as the 35Spro:VP16-IAA17 line (Figures 8F and 8G), suggesting that the 35Spro:VP16-IAA17mImII transgene completely rescued the AOE phenotype.
We next compared the shoot regeneration capacity of axr3-3, arr1, axr3-3 arr1 double mutant, and wild-type root explants. Explants derived from axr3-3 arr1 failed to produce shoots, similar to explants derived from axr3-3 (Figures 8H and 8I). These results indicate that axr3-3 rescued the phenotype of arr1 and that IAA17 acts downstream of ARR1 during in vitro shoot regeneration.
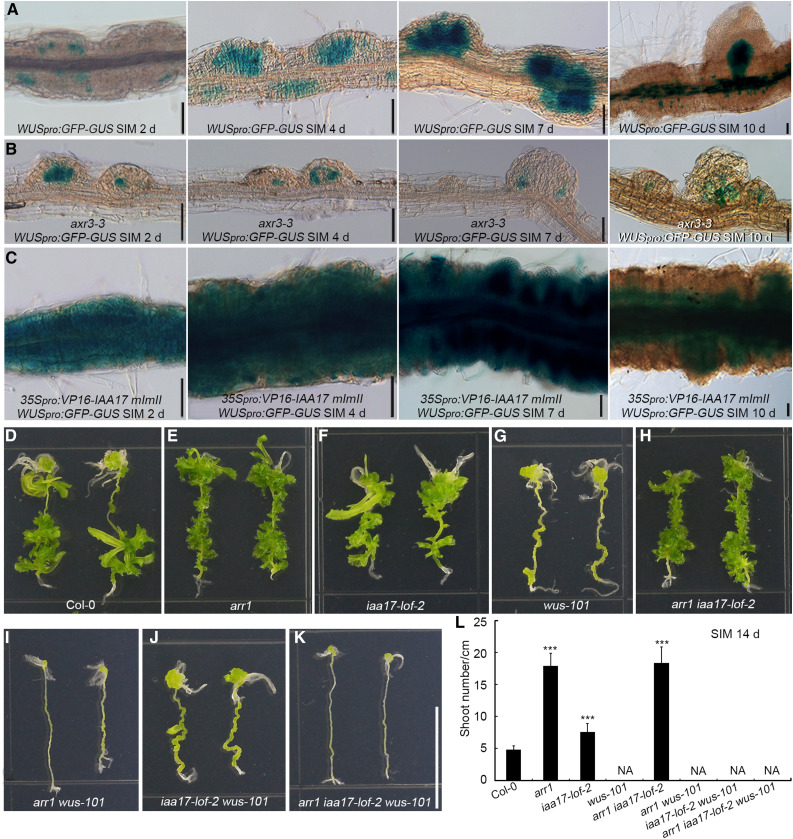
Following culture on SIM, the strength and scope of WUS expression were greater in root explants harboring 35Spro:VP16-IAA17mImII than in wild-type explants, but lower in axr3-3 explants (Figures 9A to 9C). We compared the ability of root explants to regenerate shoots among eight genotypes: arr1, iaa17-lof-2, wus-101, their double and triple mutants (arr1 iaa17-lof-2, arr1 wus-101, iaa17-lof-2 wus-101, and arr1 iaa17-lof-2 wus-101), and Col-0. arr1- and iaa17-lof-2–derived explants formed more shoots than the wild type, whereas wus-101 failed to produce shoots (Figures 9D to 9G). The arr1 iaa17-lof-2 double mutant generated a similar number of shoots as arr1 (Figures 9E, 9F, and 9H), which agrees with the observation that IAA17 acts downstream of ARR1. All explants derived from arr1 wus-101, iaa17-lof-2 wus-101, and arr1 iaa17-lof-2 wus-101 failed to produce shoots (Figures 9I to 9L). These results indicate that ARR1 and IAA17 function upstream of WUS during shoot regeneration in vitro.
Figure 9.
ARR1 and IAA17 Function Upstream of WUS During Shoot Regeneration in Vitro.
(A) to (C) Temporal and spatial expression patterns of WUS in Col-0 (A), axr3-3 (B), and 35Spro:VP16-IAA17 mImII (C) root explants incubated on SIM for 2, 4, 7, and 10 d. Scale bars = 50 μm. The WUS signal specifically appeared in newly formed SAMs after 10 d of culture on SIM.
(D) to (K) Shoot regeneration capacity of Col-0, arr1, iaa17-lof-2, wus-101, arr1 iaa17-lof-2, arr1 wus-101, iaa17-lof-2 wus-101, and arr1 iaa17-lof-2 wus-101. Scale bars = 1 cm.
(L) Number of regenerated shoots. Data represent mean ± sd (n = 28). ***Differ at P < 0.001 (Student’s t test).
DISCUSSION
Shoot regeneration from an explant is prompted by exogenous cues that elicit endogenous signals. Cytokinin has long been known to be a critical exogenous cue (Skoog and Miller, 1957; Che et al., 2006; Gordon et al., 2007); therefore, genes that regulate cytokinin signal transduction, including those encoding type-B ARRs, are likely involved in the regeneration process. In this study, we revealed that ARR12 is a central enhancer of shoot regeneration, whereas ARR1 is a strong inhibitor of this process by modulating WUS and CLV3 expression in an ARR12-dependent manner. Furthermore, we showed that a regulatory cascade involving ARR1 and the auxin signaling repressor IAA17 inhibits shoot regeneration in cultured explants. These findings provide new evidence for direct links between cytokinin and shoot meristem regulators or auxin signaling and shed light on the mechanism underlying cytokinin-regulated shoot regeneration.
ARR1 Inhibits in Vitro Shoot Regeneration
ARRs are components of the cytokinin signal transduction pathway. Loss-of-function mutants of the type-A ARR genes ARR7 and ARR15 both exhibit an enhanced capacity for shoot regeneration; it has therefore been suggested that type-A ARRs inhibit shoot regeneration (Buechel et al., 2010). Type-B ARRs are generally thought to enhance shoot regeneration (Mason et al., 2005; Dai et al., 2017; Meng et al., 2017; Zhang et al., 2017b). However, the data presented here show that a type-B ARR, ARR1, inhibits shoot regeneration. In the presence of various concentrations of cytokinin in the medium, both root and hypocotyl explants derived from the arr1 mutant developed more shoots than did those derived from the wild type (Figure 1; Supplemental Figure 2). Furthermore, our analysis of two independent arr1 mutants (Supplemental Figure 3) and a complemented line (arr1-4 mutant harboring ARR1pro:ARR1-GFP; Figure 1; Supplemental Figures 1D to 1F) also indicated that ARR1 is a strong inhibitor of shoot regeneration.
In the two-step regeneration system for Arabidopsis, pericycle cells are induced to form callus, and then shoots are generated efficiently on SIM (Che et al., 2006; Gordon et al., 2007). A 2-d preincubation period on CIM is needed for explants to acquire competence to form shoots, and this competence continues to increase for 2 d more (Che et al., 2007). Auxin is thought to activate callus formation on CIM, and cytokinin is responsible for shoot primordium formation in this system (Gordon et al., 2007). Here, we determined that ARR1 not only inhibits SIM-induced shoot regeneration, but also suppresses CIM-induced callus formation (Figure 1; Supplemental Figure 3). These observations suggest that ARR1 inhibits shoot regeneration by restricting the formation of cells competent for shoot regeneration.
ARR1 was previously reported to function in cytokinin-mediated protuberance formation and greening of Arabidopsis hypocotyl explants (Sakai et al., 2001). Overexpression of ARR1 increased cytokinin sensitivity, and its loss of function reduced sensitivity to cytokinin compared to wild-type explants (Sakai et al., 2001). Protuberances formed from a 35Spro:ARR1 explant, but not from wild-type or arr1 explants, and occasionally produced shoots when provided with 2,4-D and kinetin (Sakai et al., 2001). In the above study, hypocotyl explants were only incubated on cytokinin-rich medium without CIM preincubation (Sakai et al., 2001). In our study, we performed similar experiments, and in most cases no shoots formed when root and hypocotyl explants were directly incubated on SIM, regardless of the concentration of cytokinin provided or the genotype (wild type, arr1, or AOE) of the explant (Supplemental Figures 20A to 20J). In the presence of 0.5 mg/L 2-ip, AOE hypocotyl explants produced many calli and occasionally produced shoots (Supplemental Figure 20C).
The low efficiency of shoot regeneration in the one-step culture system made it challenging to determine the role of ARR1 in shoot regeneration. A recent study demonstrated that CIM-induced activation of cell division and the subsequent SIM-induced dilution of DNA methylation are both required for the induction of WUS expression and shoot regeneration in wild-type explants (Shemer et al., 2015). This likely explains the low regeneration efficiency of the SIM-induced one-step culture system (Sakai et al., 2001; Mason et al., 2005; Ishida et al., 2008; Hill et al., 2013).
When explants were directly incubated on cytokinin-rich medium, the lack of CIM-induced competent cells led to a high DNA methylation level and the repression of WUS expression in explants, thus suppressing shoot regeneration (Supplemental Figures 20K to 20O). When compared to the wild type, explants harboring 35Spro:ARR1 were more sensitive to exogenous cytokinin (Sakai et al., 2001), and cell division was overactivated following treatment with a certain concentration of cytokinin (Supplemental Figure 20C).
Thus, the occasional shoot regeneration in explants harboring 35Spro:ARR1 was driven by the accelerated proliferation of protuberances and the subsequent demethylation of the WUS promoter, which might partly compensate for the lack of CIM-induced competent cells (Supplemental Figures 20C and 20N). Although ARR1 enhanced the cytokinin response in explants (Supplemental Figure 21), it inhibited callus formation and shoot regeneration in the two-step culture system.
Diverse Relationships Among Individual type-B ARRs and Shoot Meristem Regulators in Shoot Regeneration
The type-B ARR family contains multiple members, and several studies based on double or triple mutants have revealed functional redundancy among these members in a variety of developmental processes (Mason et al., 2005; Dello Ioio et al., 2007; Ishida et al., 2008; Moubayidin et al., 2010; Hill et al., 2013; Meng et al., 2017; Wang et al., 2017; Zhang et al., 2017b). For example, both arr1 and arr12 generate roots with a larger primary meristem than the wild type, whereas the arr1 arr12 double mutant has an even larger primary meristem than either of the single mutants (Dello Ioio et al., 2007; Moubayidin et al., 2010). ARR1, ARR10, and ARR12 act redundantly in callus formation and greening of hypocotyl explants (Mason et al., 2005; Ishida et al., 2008; Hill et al., 2013), SAM maintenance (Mason et al., 2005), axillary meristem formation (Wang et al., 2017), and shoot regeneration (Meng et al., 2017; Zhang et al., 2017b). Studies of individual type-B ARRs are limited (Sakai et al., 2001; Kurepa et al., 2014; Dai et al., 2017). Here, using single mutants of type-B ARRs, we established that ARR1 was a strong inhibitor of shoot regeneration, whereas ARR12 was a central enhancer of shoot regeneration (Figure 1; Supplemental Figure 4), indicating functional specification of individual type-B ARRs in shoot regeneration.
The different shoot regeneration phenotypes of arr1 and arr12 could be due to the effects of ARR1 and ARR12 proteins or the expression pattern of their encoding genes. When both the ARR1 and ARR12 promoters drove the expression of ARR1, explants harboring the ARR12pro:ARR1 transgene in the arr12 background produced only a small number of shoots (Figure 3C). However, when both the ARR1 and ARR12 promoters drove the expression of ARR12, explants harboring the ARR1pro:ARR12 transgene in the arr1 background produced more shoots than explants harboring the ARR12pro:ARR1 transgene in the arr12 background (Figure 3F), suggesting that under the control of the same promoters, ARR1 and ARR12 proteins have different effects on shoot regeneration. Consistent with these results, explants harboring the 35Spro:ARR12 transgene produced more shoots than explants harboring 35Spro:ARR1 (Figures 3G to 3J, and 3L).
These results support the notion that the production of ARR1 and ARR12 proteins is responsible for the different abilities of these explants to undergo shoot regeneration. Indeed, on medium containing 0.5 μM of the auxin 1-naphthaleneacetic acid, no shoots formed from wild-type, arr1, or ARR1-overexpressing explants, whereas explants overexpressing a phosphomimetic form of ARR1 generated shoots (Kurepa et al., 2014), indicating that phosphorylation of ARR1 protein promotes shoot regeneration. In this study, the 35Spro:ARR1 transgene inhibited shoot regeneration in wild-type explants (Supplemental Figures 8A and 8B), whereas the expression of 35Spro:mARR1D94E (encoding a phosphomimetic mutant version of ARR1 protein) dramatically increased shoot regeneration (Supplemental Figures 8A, and 8C to 8E), suggesting that the phosphorylation status of ARR1 protein contributes to its function in shoot regeneration.
The ARR1 locus could not compensate for the loss of ARR12 function, while the ARR12pro:ARR1 transgene partially rescued the arr12 phenotype (Figures 3C and 3D; Supplemental Figures 7A and 7B), indicating that under the control of different promoters, ARR1 has slightly different effects on shoot regeneration, suggesting that the ARR1 and ARR12 promoters also contribute to the different effects of these proteins on shoot regeneration. The differential effects of ARR1 and ARR12 proteins on shoot regeneration (Figures 3C and 3F) were much more apparent than those of the ARR1 and ARR12 promoters (Figures 3B and 3C). Thus, the different shoot regeneration phenotypes of arr1 and arr12 are determined by both the protein functions and expression patterns of ARR1 and ARR12, with the former dominating.
Although the functional redundancy of type-B ARRs in various developmental processes has been reported, the specific relationships between individual type-B family members remain unclear. In this study, we demonstrated the functional dependency of ARR1 on ARR12 during shoot regeneration. In the presence of ARR12, ARR1 inhibited shoot regeneration, whereas in the absence of ARR12, ARR1 slightly enhanced shoot regeneration (Figure 2). ARR1 inhibited shoot regeneration by counteracting the positive effect of ARR12 on shoot regeneration (Figure 2; Supplemental Figures 5A and 5B). ARR1 had a slight effect on ARR12 transcription (Supplemental Figures 5C to 5H), suggesting that ARR1 counteracts the positive effect of ARR12 on shoot regeneration through ways other than repressing its transcription.
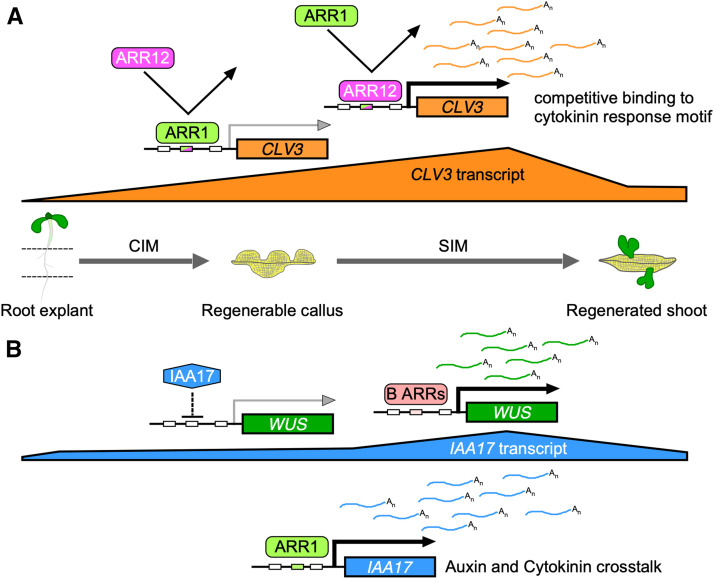
Type-B ARRs play redundant roles in regulating the transcription of target genes. For example, ARR1, ARR10, and ARR12 participate in axillary meristem formation and shoot regeneration through directly activating WUS expression (Meng et al., 2017; Wang et al., 2017; Zhang et al., 2017b). Here, we show that ARR1 and ARR12 exert complex regulatory effects on CLV3 and WUS during shoot regeneration. ARR12 strongly activates the transcription of CLV3 in explants cultured on both CIM and SIM, which enhances callus formation and shoot regeneration (Figures 2, 5, and 10A). However, ARR1 competes with ARR12 for binding to the same CLV3 promoter sites (Figures 5H and 10A), partly blocks the stimulatory effect of ARR12 on CLV3 expression, and thus represses CLV3 transcription indirectly (Figures 5D and 10A), which inhibits callus formation and shoot regeneration. ARR12 is critical for the widespread distribution of WUS in the explants and for the concentrated WUS expression in the newly formed SAMs, which enhances shoot regeneration (Supplemental Figures 14, 22A, and 22C). However, ARR1 competes with ARR12 for binding to the same WUS promoter sites and induces the widespread distribution of WUS in the explants in the presence of ARR12 (Supplemental Figures 16D, 22B, and 22D), which delays the concentration of WUS transcripts and therefore inhibits shoot regeneration (Supplemental Figure 14). Thus, ARR1 modulates CLV3 and WUS expression in an ARR12-dependent manner, which contributes to its ARR12-dependent inhibitory effect on shoot regeneration.
Figure 10.
ARR1 Inhibits Shoot Regeneration Through the Indirect Repression of CLV3 and the Direct Activation of IAA17.
(A) ARR1 and ARR12 competitively bind to the promoter region of CLV3. The strong activation of CLV3 transcription by ARR12 is significantly repressed by ARR1, probably through their competitive binding to the CLV3 promoter. Thus, ARR1 inhibits callus formation and shoot regeneration by repressing CLV3 transcription indirectly in an ARR12-dependent manner.
(B) ARR1 inhibits shoot regeneration through the direct transcriptional activation of IAA17. In addition, type-B ARRs promote shoot regeneration by directly activating WUS expression (Meng et al., 2017; Zhang et al., 2017b; Zubo et al., 2017).
Cytokinin-induced shoot regeneration requires the integration of multiple regulatory steps controlling CLV3 and WUS expression mediated by ARR1 and ARR12. In addition, ARR1 and ARR12 directly activate CLV3, WUS, and IAA17 transcription in Arabidopsis protoplasts (Figure 5G; Supplemental Figures 16C and 17B), in agreement with reports of type-B ARRs acting as transcriptional activators (Sakai et al., 2000; Meng et al., 2017; Zhang et al., 2017b). However, in a specific developmental process, especially when cytokinin greatly induces binding of type-B ARRs to target sequences (Zubo et al., 2017), type-B ARRs compete with each other for binding the promoter of their target gene and repress gene expression indirectly.
In the two-step regeneration model of Arabidopsis, WUS expression is specifically induced in the explant on SIM (Supplemental Figures 14 and 15) and is critical for in vitro stem cell niche specification (Gordon et al., 2007; Meng et al., 2017; Zhang et al., 2017b). The specific functions of the shoot meristem regulator CLV3 during shoot regeneration have long been elusive. Here, we demonstrated that, in contrast to observations for WUS, CLV3 transcript was detected in explants cultured on both CIM and SIM (Figures 5A to 5D), which is consistent with a previous study (Atta et al., 2009). Loss of function of CLV3 resulted in a slight decrease in the ability to form calli and regenerate shoots (Figures 6A, 6B, 6E, 6F, 6G, and 6J), and mutation of CLV3 in the arr1-4 mutant line partly rescued both the callus formation and shoot regeneration phenotype (Figures 6B to 6E and 6G to 6J), indicating that CLV3 promotes both callus formation and shoot regeneration and functions in the pathway downstream of ARR1 during the two-step regeneration process.
The ARR1/IAA17 Regulatory Cascade Links Auxin and Cytokinin Signaling During Shoot Regeneration
The interaction between auxin and cytokinin influences many aspects of plant development (Moubayidin et al., 2009). In the primary root meristem, cytokinin reduces the auxin response by inducing SHORT HYPOCOTYL2 (SHY2), thereby promoting cell differentiation (Moubayidin et al., 2010). The negative effect of cytokinin on PIN-FORMED–dependent auxin distribution prevents the ectopic initiation of lateral roots (Marhavý et al., 2014). The auxin-mediated repression of type-A ARR genes ARR7 and ARR15 is required for embryonic stem cell niche formation and SAM maintenance (Müller and Sheen, 2008; Zhao et al., 2010). Under in vitro conditions, ARF3 promotes shoot regeneration by activating IPT5 (Cheng et al., 2013). Cytokinin and auxin play critical roles in shoot regeneration (Ikeuchi et al., 2016). However, the full nature of the interaction between auxin and cytokinin signaling during shoot regeneration remains obscure.
Here, we revealed a new link between auxin and cytokinin signaling involving the interaction between ARR1 and IAA17. ARR1 binds to the IAA17 promoter and promotes its expression, especially at SIM incubation stage (Figures 7 and 10B), which indirectly represses the expression of WUS (Figures 9 and 10B) and inhibits shoot regeneration (Figure 10B). Combined with the direct activation of WUS expression by ARR1 and ARR12 (Supplemental Figures 16 and 22), these results further support the previous finding that type-B ARRs regulate shoot regeneration via dual regulation of WUS (Figure 10B; Meng et al., 2017). ARR1 promotes the expression of IAA17, resulting in a decreased auxin response (Figure 7; Supplemental Figure 23). Root explants derived from loss-of-function iaa17 mutants regenerated more shoots than the wild type (Supplemental Figure 18). In addition, the 35Spro:VP16-IAA17mImII transgene fully restored the phenotype of the AOE line, and axr3-3 rescued the phenotype of arr1 (Figure 8). These observations suggest that IAA17 acts downstream of ARR1.
During shoot regeneration from a cultured explant, cytokinin is thought to help determine cell fate, whereas the presence of cytokinin in the SAM is critical for stem cell maintenance (Zhao et al., 2010). Therefore, cytokinin homeostasis is thought to be closely associated with shoot regeneration, possibly via the activation of SAM-related genes (Jasinski et al., 2005; Yanai et al., 2005). Recent studies have demonstrated that the direct activation of WUS by B-type ARRs is critical for shoot regeneration (Meng et al., 2017; Zhang et al., 2017b). Furthermore, type-B ARRs indirectly promote WUS expression by repressing the expression of YUCCAs (Meng et al., 2017). In this study, mutations in both IAA17 and ARR1 led to an abnormal distribution of WUS transcripts during shoot regeneration (Figures 8 and 9; Supplemental Figure 14), implying that ARR1 and IAA17 inhibit the reestablishment of the SAM by modulating WUS expression. These observations indicate that endogenous signals originating from exogenous cytokinin are transmitted indirectly in the plant via diverse auxin signaling pathways.
METHODS
Plant Materials and Growth Conditions
All mutants used in this study were established in a Col-0 background unless otherwise indicated. Seeds of the arr1-4 (stock number CS6972), arr10-5 (CS39989), arr11-3 (CS6977), arr12-1 (CS6978), arr18-2 (CS6979), arr1 arr12 (CS6981), arr10 arr12 (CS39991), arr11 arr12 (CS6983), arr12 arr18 (CS6985), arr11 arr18 (CS6992), and axr3-3 (CS57505) were obtained from the Nottingham Arabidopsis Stock Centre. Seeds of the arr1-3 were provided by Jiawei Wang (Institute of Plant Physiology and Ecology, Shanghai, China), and those of the clv3-7 were provided by Zhong Zhao (University of Science and Technology of China, Anhui, China). Seeds of the transgenic line WOX5pro:GFP were a gift of Xiaofeng Cao (Institute of Genetics and Developmental Biology, Chinese Academy of Sciences, Beijing, China). Seeds of the IAA17 T-DNA lines SALK_011820, SALK_065697, the carriers of DR5pro:GFP, 35Spro:VP16-IAA17mImII (IAA17 activator) and 35Spro:HA-IAA17mImII (IAA17 repressor), and the DR5pro:GFP 35Spro:VP16-IAA17mImII hybrid (Tian et al., 2014), as well as those of SCRpro:GFP were provided by Zhaojun Ding (Shandong University, Shandong, China). TCSnpro:GFP (Zürcher et al., 2013) were a gift of Bruno Müller (University of Zurich, Switzerland), and those of WUSpro:GFP-GUS and CLV3pro:GFP-GUS (Cui et al., 2015) were provided by Tao Huang (Xiamen University, Fujian, China).
Seeds were surface-sterilized with 75% (v/v) ethanol with 0.04% (v/v) Triton X-100, and then washed twice with 70% (v/v) ethanol. Seedlings were grown on half-strength Murashige and Skoog (MS) medium with Gamborg’s B-5 vitamins (Caisson Laboratories) plus 10 g/L Suc, 0.5 g/L MES (Sigma Aldrich), and 8 g/L agar, pH 5.8 in constant light (20 µmol photons m−2 s−1) at 22 ± 1°C in a plate incubator with white light-emitting diodes (Jiangnan Instruments, China).
Shoot Regeneration and Callus Greening in Vitro
We harvested root and hypocotyl explants from seedlings cultured on solid half-strength MS medium for 5 d in constant light (20 µmol photons m−2 s−1) at 22 ± 1°C. For callus induction, we cultured explants (1-cm-long roots) for 7 and 21 d on CIM (Gamborg’s B-5 medium; Haibo, China), 20 g/L Glc, 0.5 g/L MES (Sigma Aldrich), 6 g/L agar, 0.5 mg/L 2,4-dichlorophenoxyacetic acid (Sigma Aldrich), and 0.05 mg/L kinetin (Sigma Aldrich), pH 5.8, in constant light (110 µmol photons m−2 s−1) at 22 ± 1°C. For shoot regeneration, we initially cultured explants (1-cm-long roots) for 5 d on CIM in constant light (110 µmol photons m−2 s−1) at 22 ± 1°C. We then replated calli on SIM (MS medium supplemented with Gamborg’s B-5 vitamins plus 10 g/L Suc, 0.5 g/L MES, 6 g/L agar, 1 mg/L N6-(2-isopentenyl) adenine (Sigma Aldrich), and 0.15 mg/L IAA (Sigma Aldrich), pH 5.8, in constant light (110 µmol photons m−2 s−1) at 22 ± 1°C for 2 to 21 d. For callus greening in vitro, we transferred root and hypocotyl explants directly to SIM containing the indicated concentrations of N6-(2-isopentenyl) adenine (Sigma Aldrich) for 28 d. All regeneration experiments were performed in glass dishes. We performed statistical treatment with Microsoft Excel software. Statistically significant differences were calculated using two-sided statistical tests at P-value < 0.05 (Supplemental Data Set 2). Three biological replicates with separately cultured explant samples were conducted.
Vector Construction, Transgenesis, and Identification of T-DNA Insertion Mutants
The 2073-bp coding region of ARR1 was inserted into the pROK2 plant expression vector (http://signal.salk.edu/tdna_protocols.html) under the control of the cauliflower mosaic virus 35S promoter to generate the transgene 35Spro:ARR1. Plant transformation was performed using the floral dip method (Clough and Bent, 1998). Lines carrying the 35Spro:ARR1 transgene are referred to as AOE. To generate phosphomimetic ARR1-overexpressing plants (35Spro:mARR1D94E), we used fusion PCR to introduce a C-to-G mutation (thus D to E substitution at position 94) into the coding region of ARR1 with the primer pairs ARR1-pB2GW7-F/mARR1-R and mARR1-F/ARR1-pB2GW7-R (sequences in Supplemental Data Set 1). We cloned the mutated coding region of ARR1 (mARR1) into pDONR221 (Invitrogen) using BP Clonase (Invitrogen), and recombined into the pB2GW7 binary vector using LR Clonase enzyme mix (Invitrogen). To generate the ARR1pro:ARR1-GFP vector, we inserted the 2073 bp ARR1 coding region and its 2196-bp upstream sequence into the pGFPGUSplus vector (Vickers et al., 2007). The arr1 mutant was transformed with the ARR1pro:ARR1-GFP vector to obtain the RS transgenic lines. The construction of the 35Spro:ARR12 and ARR12pro:ARR12-GFP vectors was previously described by Dai et al. (2017). To construct the ARR12pro:ARR1 vector, we inserted the 3441-bp ARR12 upstream sequence and 2073-bp coding region of ARR1 into the pGFPGUSplus vector (Vickers et al., 2007) via In-Fusion cloning method (Clontech). To construct the ARR1pro:ARR12 vector, we inserted the 3246-bp ARR1 upstream sequence and 1791-bp coding region of ARR12 into the pGFPGUSplus vector via In-Fusion cloning method (Clontech). To generate the ARR12pro:ARR12 vector, we inserted the 4539-bp ARR12 upstream sequence, and the 2472-bp genomic region of ARR12 and the 2094-bp ARR12 downstream sequence into the pGFPGUSplus vector via In-Fusion cloning method (Clontech). The mutated upstream sequences of CLV3 and WUS were synthesized by the Beijing Genomics Institute (http://www.bgitechsolutions.com/). We then inserted the wild-type and the mutated forms of the CLV3 promoter into pDONR221 (Invitrogen) using BP Clonase (Invitrogen), and recombined into the pBGWFS7 binary vector using LR clonase enzyme mix (Invitrogen). We inserted the wild-type and the mutated form of CLV3 and WUS promoters into pGreenII 0800-LUC vector (Hellens et al., 2005) to obtain the plasmids CLV3pro:LUC, mCLV3pro:LUC, WUSpro:LUC, mWUS1pro:LUC, and mWUS2pro:LUC, respectively. We generated mutated sites in the IAA17 promoter via fusion PCR. The 1,211-bp upstream sequence of the wild type and the mutated form of IAA17 were inserted into either pGFPGUSplus vector (Vickers et al., 2007) or pGreenII 0800-LUC vector (Hellens et al., 2005) to obtain the plasmids IAA17pro:GFP and mIAA17pro:GFP or IAA17pro:LUC and mIAA17pro:LUC, respectively. At least two independent transgenic lines were analyzed for each transgenic experiment (Supplemental Table 2). The protein expression vector pGST-ARR1-DBD was generated as previously described by Ramireddy et al. (2013). Plants homozygous for both the arr1 mutation and the transgenes WUSpro:GFP-GUS/CLV3pro:GFP-GUS/clv3-7/TCSnpro:GFP/WOX5pro:GFP/SCRpro:GFP were validated by PCR based on the primer pair ARR1-Genotype-F/-R (sequences in Supplemental Data Set 1).
Microscopy
We cleared explants cultured on CIM or SIM following an established protocol (Malamy and Benfey, 1997). We detected GUS activity as previously described by Liu et al. (2013). At least 20 samples per line were analyzed. We captured images with a BX51 or SZX16 microscope (Olympus). To acquire fluorescence images, we mounted explants on a glass slide in water. The GFP was excited at 488 nm, and the signal was collected at 495 to 550 nm using an LSM 700 confocal laser scanning microscope (Zeiss). We mounted roots in 50 mg/mL propidium iodide (Sigma Aldrich), which was excited at 593 nm and the signal collected at 610–680 nm.
RT-qPCR, RNA-Seq Analysis, DNA Methylation Assay, and RNA in situ Hybridization
For gene transcript levels determination, we collected explant samples and isolated total RNA using TRIzol reagent (Invitrogen). We treated a 3 μg aliquot of total RNA for each sample with RNase-free DNase I (Takara, China) to remove contaminating genomic DNA and reverse-transcribed using a FastQuant RT Kit (TIANGEN Biotech, Beijing, China) in a 40 μL reaction, following the manufacturer’s protocol. Three replicates of each reaction were included in the subsequent RT-qPCR, which was based on a Bio-Rad MyiQTM2 detection system (Bio-Rad) with SYBR Green I master mix (Roche, Switzerland). Relative expression was calculated with the 2-ΔΔCT method using the reference gene TUBULIN2 (TUB2; At5g62690). We performed statistical analysis with Microsoft Excel software, based on the results of three biological replicates with separately collected explant samples.
For RNA-Seq, we extracted RNA from wild-type, arr1 single mutant, and AOE root explants at the nascent SAM formation stage. Library construction and RNA-Seq were performed by Biomarker Co. (www.biomarker.com.cn). We used DESeq and the Q value to identify DTGs. Differential transcript levels were inferred by applying a false discovery rate threshold of 0.001 and a |log2 Ratio| ≥ 1. We inferred functional categorization from a BLAST search of the nonredundant GenBank (http://www.ncbi.nlm.nih.gov/genbank/), Kyoto Encyclopedia of Genes and Genomes Pathway (http://www.genome.jp/kegg/pathway.html), and UniProt protein databases (http://www.uniprot.org/), and further analyzed by the Gene Ontology (http://geneontology.org/) method.
The methylation region in the WUS promoter was previously described by Shemer et al. (2015). For DNA methylation analysis, we extracted genomic DNA from wild-type, arr1 single mutant, and AOE hypocotyl explants incubated on SIM for 20 d. We harvested root explants at the nascent SAM formation stage. We performed complete bisulfite conversion and cleanup of DNA according to the protocol [Epitect Bisulfite Kit (48), Qiagen, Germany]. We cloned DNA fragments using the primer pair WUS-BSP-F/-R (sequences in Supplemental Data Set 1).
The WUS antisense and sense probes, as well as the in situ hybridization protocol, were as described by Xin et al., (2017).
ChIP, EMSA, and Transient Expression Analyses
We chose the protuberance from root explants at the nascent SAM formation stage (SIM 7 d) as the experimental material for ChIP analysis. Our procedure followed a protocol described by Gendrel et al. (2005) with anti-GFP antibody (Abcam, ChIP grade, http://www.abcam.com/gfp-antibody-chip-grade-ab290.html). We amplified DNA fragments obtained from biological triplicates by qPCR using primers listed in Supplemental Data Set 1. We normalized the enrichment level to the input sample, and TUB2 was used as the reference sequence. No antibody was added to the negative control samples. Three independent biological replicates with separately collected explant samples were performed. Protein induction and purification and the EMSA were performed according to Ramireddy et al. (2013). Oligonucleotides (40 to 60 nucleotides) were commercially synthesized and labeled using a second generation DIG Gel Shift Kit (Roche). The primer sequences used are given in Supplemental Data Set 1.
For transient expression experiments, we prepared mesophyll protoplasts from 4- or 5-week-old rosette leaves of the wild-type or AOE line. Combinations of plasmids were introduced in the presence of 40% (w/v) polyethylene glycol as previously described by Yoo et al. (2007). We used empty pGreenII 0800-LUC or pROK2 vector d as negative control. We tested LUC/REN activity using the dual-luciferase reporter assay system (Promega). Three independent biological replicates with separately prepared mesophyll protoplasts were performed.
Accession Numbers
DNA sequences can be retrieved from https://www.arabidopsis.org, using the AGI locus identifiers ARR1 (At3g16857), ARR10 (At4g31920), ARR11 (At1g67710), ARR12 (At2g25180), ARR18 (At5g58080), ARR5 (At3g48100), ARR7 (At1g19050), ARR15 (At1g74890), CLV3 (At2g27250), WUS (At2g17950), SCR (At3g54220), PLT1 (At3g20840), WOX5 (At3g11260), IAA14 (At4g14550), IAA3 (At1g04240), IAA17 (At1g04250), and TUB2 (At5g62690).
The RNA-seq data sets used in this study have been submitted to the GEO (Gene Expression Omnibus, GSE146690, https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE146690).
Supplemental Data
Supplemental Figure 1. Phenotypic analysis of root explants derived from 35Spro:ARR1 or arr1 plants harboring ARR1pro:ARR1-GFP.
Supplemental Figure 2. ARR1 inhibits shoot regeneration from hypocotyl explants.
Supplemental Figure 3. Both arr1-3 and arr1-4 show enhanced capacity for callus and shoot regeneration.
Supplemental Figure 4. Functional diversification of type-B ARRs in regulating in vitro shoot regeneration from root explants.
Supplemental Figure 5. ARR1 inhibits callus formation and shoot regeneration by counteracting the stimulating effect of ARR12.
Supplemental Figure 6. ARR1 inhibits shoot regeneration in an ARR12-dependent manner.
Supplemental Figure 7. Phenotypes of transgenic plants harboring ARR12pro:ARR1 in the arr12 background and ARR1pro:ARR12 in the arr1 background.
Supplemental Figure 8. Protein phosphorylation of ARR1 contributes to its inhibitory effect on shoot regeneration.
Supplemental Figure 9. Localization of the ARR1-GFP fusion protein in the primary root.
Supplemental Figure 10. Temporal and spatial expression patterns of ARR12 during shoot regeneration in root explants.
Supplemental Figure 11. Transcription of a selection of genes sampled from root explants derived from Col-0, arr1, and AOE.
Supplemental Figure 12. Distribution of CLV3pro:GFP-GUS signal in the Col-0 and arr1 backgrounds.
Supplemental Figure 13. Distribution of WUS in hypocotyl and root explants shown by GUS reporter and RNA in situ hybridization.
Supplemental Figure 14. ARR12 is critical for WUS induction on SIM and ARR1 expands the distribution of WUS expression.
Supplemental Figure 15. Temporal and spatial expression patterns of WUS in Col-0 and arr1.
Supplemental Figure 16. ARR1 and ARR12 competitively bind to the promoter of WUS.
Supplemental Figure 17. ARR12 indirectly represses expression of IAA17.
Supplemental Figure 18. The shoot regeneration capacity of the wild type and IAA17 mutant from root explants.
Supplemental Figure 19. Callus formation from Col-0, axr3-3, and 35Spro:VP16-IAA17 mImII root explants.
Supplemental Figure 20. Greening and shoot regeneration capacity of calli derived from hypocotyl and root explants without pre-incubation on CIM.
Supplemental Figure 21. ARR1 modulates the cytokinin response during shoot regeneration.
Supplemental Figure 22. ARR1 inhibits shoot regeneration by competing with ARR12 for binding to the WUS promoter.
Supplemental Figure 23. ARR1 and IAA17 modulate the auxin response during shoot regeneration in root explants.
Supplemental Table 1. Point mutations of CLV3 promoter.
Supplemental Table 2. List of lines used in this study.
Supplemental Data Set 1. Primers used in this study.
Supplemental Data Set 2. Summary of statistical analyses.
DIVE Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
Acknowledgments
We thank Jiawei Wang (Institute of Plant Physiology and Ecology, China), Zhong Zhao (University of Science and Technology of China, China), Xiaofeng Cao (Institute of Genetics and Developmental Biology, China), Zhaojun Ding (Shandong University, China), Bruno Müller (University of Zurich, Switzerland), Tao Huang (Xiamen University, China), and the Nottingham Arabidopsis Stock Centre for providing the seed stocks. We thank Haiyan Yu, Xiaomin Zhao, and Sen Wang from SKLMT (Stake Key Laboratory of Microbial Technology, Shandong University) for the assistance in microimaging of LSCM analysis. This research was supported by the National Key Research and Development Program of China (grant 2016YFD0101902), the Major Program of Shandong Province Natural Science Foundation (grant ZR2018ZC0334), the National Transgenic Project of China (grants 2016ZX08010002-009 and 2018ZX08009-14B), National Natural Science Foundation of China (NSFC; grants U1906203, 31770317, and 31970189), the National Special Science Research Program of China (grant 2013CB967300), China Postdoctoral Science Foundation (grant 2019M662331), and the Young Scholars Program of Shandong University.
AUTHOR CONTRIBUTIONS
Z.L., X.D., J.L., N.L., and X.L. conducted the experiments; S.L. analyzed the data and provided the suggestions; Z.L. and F.X. designed the experiments and wrote the article.
Footnotes
Articles can be viewed without a subscription.
References
- Aoyama T., Oka A.(2003). Cytokinin signal transduction in plant cells. J. Plant Res. 116: 221–231. [DOI] [PubMed] [Google Scholar]
- Atta R., Laurens L., Boucheron-Dubuisson E., Guivarc’h A., Carnero E., Giraudat-Pautot V., Rech P., Chriqui D.(2009). Pluripotency of Arabidopsis xylem pericycle underlies shoot regeneration from root and hypocotyl explants grown in vitro. Plant J. 57: 626–644. [DOI] [PubMed] [Google Scholar]
- Birnbaum K.D., Sánchez Alvarado A.(2008). Slicing across kingdoms: Regeneration in plants and animals. Cell 132: 697–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand U., Fletcher J.C., Hobe M., Meyerowitz E.M., Simon R.(2000). Dependence of stem cell fate in Arabidopsis on a feedback loop regulated by CLV3 activity. Science 289: 617–619. [DOI] [PubMed] [Google Scholar]
- Buechel S., Leibfried A., To J.P., Zhao Z., Andersen S.U., Kieber J.J., Lohmann J.U.(2010). Role of A-type ARABIDOPSIS RESPONSE REGULATORS in meristem maintenance and regeneration. Eur. J. Cell Biol. 89: 279–284. [DOI] [PubMed] [Google Scholar]
- Chandler J.W., Werr W.(2015). Cytokinin-auxin crosstalk in cell type specification. Trends Plant Sci. 20: 291–300. [DOI] [PubMed] [Google Scholar]
- Chatfield S.P., Capron R., Severino A., Penttila P.A., Alfred S., Nahal H., Provart N.J.(2013). Incipient stem cell niche conversion in tissue culture: Using a systems approach to probe early events in WUSCHEL-dependent conversion of lateral root primordia into shoot meristems. Plant J. 73: 798–813. [DOI] [PubMed] [Google Scholar]
- Che P., Lall S., Howell S.H.(2007). Developmental steps in acquiring competence for shoot development in Arabidopsis tissue culture. Planta 226: 1183–1194. [DOI] [PubMed] [Google Scholar]
- Che P., Lall S., Nettleton D., Howell S.H.(2006). Gene expression programs during shoot, root, and callus development in Arabidopsis tissue culture. Plant Physiol. 141: 620–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Z.J., et al. (2013). Pattern of auxin and cytokinin responses for shoot meristem induction results from the regulation of cytokinin biosynthesis by AUXIN RESPONSE FACTOR3. Plant Physiol. 161: 240–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F.(1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743. [DOI] [PubMed] [Google Scholar]
- Cui Y., Rao S., Chang B., Wang X., Zhang K., Hou X., Zhu X., Wu H., Tian Z., Zhao Z., Yang C., Huang T.(2015). AtLa1 protein initiates IRES-dependent translation of WUSCHEL mRNA and regulates the stem cell homeostasis of Arabidopsis in response to environmental hazards. Plant Cell Environ. 38: 2098–2114. [DOI] [PubMed] [Google Scholar]
- Dai X., Liu Z., Qiao M., Li J., Li S., Xiang F.(2017). ARR12 promotes de novo shoot regeneration in Arabidopsis thaliana via activation of WUSCHEL expression. J. Integr. Plant Biol. 59: 747–758. [DOI] [PubMed] [Google Scholar]
- Dello Ioio R., Galinha C., Fletcher A.G., Grigg S.P., Molnar A., Willemsen V., Scheres B., Sabatini S., Baulcombe D., Maini P.K., Tsiantis M.(2012). A PHABULOSA/cytokinin feedback loop controls root growth in Arabidopsis. Curr. Biol. 22: 1699–1704. [DOI] [PubMed] [Google Scholar]
- Dello Ioio R., Linhares F.S., Scacchi E., Casamitjana-Martinez E., Heidstra R., Costantino P., Sabatini S.(2007). Cytokinins determine Arabidopsis root-meristem size by controlling cell differentiation. Curr. Biol. 17: 678–682. [DOI] [PubMed] [Google Scholar]
- Dello Ioio R., Nakamura K., Moubayidin L., Perilli S., Taniguchi M., Morita M.T., Aoyama T., Costantino P., Sabatini S.(2008). A genetic framework for the control of cell division and differentiation in the root meristem. Science 322: 1380–1384. [DOI] [PubMed] [Google Scholar]
- Dodsworth S.(2009). A diverse and intricate signalling network regulates stem cell fate in the shoot apical meristem. Dev. Biol. 336: 1–9. [DOI] [PubMed] [Google Scholar]
- Gendrel A.V., Lippman Z., Martienssen R., Colot V.(2005). Profiling histone modification patterns in plants using genomic tiling microarrays. Nat. Methods 2: 213–218. [DOI] [PubMed] [Google Scholar]
- Gordon S.P., Heisler M.G., Reddy G.V., Ohno C., Das P., Meyerowitz E.M.(2007). Pattern formation during de novo assembly of the Arabidopsis shoot meristem. Development 134: 3539–3548. [DOI] [PubMed] [Google Scholar]
- Guilfoyle T.(2007). Plant biology: Sticking with auxin. Nature 446: 621–622. [DOI] [PubMed] [Google Scholar]
- Hellens R.P., Allan A.C., Friel E.N., Bolitho K., Grafton K., Templeton M.D., Karunairetnam S., Gleave A.P., Laing W.A.(2005). Transient expression vectors for functional genomics, quantification of promoter activity and RNA silencing in plants. Plant Methods 1: 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill K., Mathews D.E., Kim H.J., Street I.H., Wildes S.L., Chiang Y.H., Mason M.G., Alonso J.M., Ecker J.R., Kieber J.J., Schaller G.E.(2013). Functional characterization of type-B response regulators in the Arabidopsis cytokinin response. Plant Physiol. 162: 212–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeuchi M., Ogawa Y., Iwase A., Sugimoto K.(2016). Plant regeneration: Cellular origins and molecular mechanisms. Development 143: 1442–1451. [DOI] [PubMed] [Google Scholar]
- Ishida K., Yamashino T., Yokoyama A., Mizuno T.(2008). Three type-B response regulators, ARR1, ARR10 and ARR12, play essential but redundant roles in cytokinin signal transduction throughout the life cycle of Arabidopsis thaliana. Plant Cell Physiol. 49: 47–57. [DOI] [PubMed] [Google Scholar]
- Jasinski S., Piazza P., Craft J., Hay A., Woolley L., Rieu I., Phillips A., Hedden P., Tsiantis M.(2005). KNOX action in Arabidopsis is mediated by coordinate regulation of cytokinin and gibberellin activities. Curr. Biol. 15: 1560–1565. [DOI] [PubMed] [Google Scholar]
- Korasick D.A., Westfall C.S., Lee S.G., Nanao M.H., Dumas R., Hagen G., Guilfoyle T.J., Jez J.M., Strader L.C.(2014). Molecular basis for AUXIN RESPONSE FACTOR protein interaction and the control of auxin response repression. Proc. Natl. Acad. Sci. USA 111: 5427–5432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurepa J., Li Y., Perry S.E., Smalle J.A.(2014). Ectopic expression of the phosphomimic mutant version of Arabidopsis response regulator 1 promotes a constitutive cytokinin response phenotype. BMC Plant Biol. 14: 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leibfried A., To J.P.C., Busch W., Stehling S., Kehle A., Demar M., Kieber J.J., Lohmann J.U.(2005). WUSCHEL controls meristem function by direct regulation of cytokinin-inducible response regulators. Nature 438: 1172–1175. [DOI] [PubMed] [Google Scholar]
- Li H., Cheng Y., Murphy A., Hagen G., Guilfoyle T.J.(2009). Constitutive repression and activation of auxin signaling in Arabidopsis. Plant Physiol. 149: 1277–1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z., Li J., Wang L., Li Q., Lu Q., Yu Y., Li S., Bai M.Y., Hu Y., Xiang F.(2016). Repression of callus initiation by the miRNA-directed interaction of auxin-cytokinin in Arabidopsis thaliana. Plant J. 87: 391–402. [DOI] [PubMed] [Google Scholar]
- Liu Z., Xin W., Ji D., Wang L., Li J., Xiang F.(2013). GUS activity for miR165a/166b, REV, and WUS/CLV3 in in vitro direct Arabidopsis thaliana shoot regeneration. Protoplasma 250: 1213–1218. [DOI] [PubMed] [Google Scholar]
- Malamy J.E., Benfey P.N.(1997). Organization and cell differentiation in lateral roots of Arabidopsis thaliana. Development 124: 33–44. [DOI] [PubMed] [Google Scholar]
- Marhavý P., Duclercq J., Weller B., Feraru E., Bielach A., Offringa R., Friml J., Schwechheimer C., Murphy A., Benková E.(2014). Cytokinin controls polarity of PIN1-dependent auxin transport during lateral root organogenesis. Curr. Biol. 24: 1031–1037. [DOI] [PubMed] [Google Scholar]
- Mason M.G., Li J., Mathews D.E., Kieber J.J., Schaller G.E.(2004). Type-B response regulators display overlapping expression patterns in Arabidopsis. Plant Physiol. 135: 927–937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason M.G., Mathews D.E., Argyros D.A., Maxwell B.B., Kieber J.J., Alonso J.M., Ecker J.R., Schaller G.E.(2005). Multiple type-B response regulators mediate cytokinin signal transduction in Arabidopsis. Plant Cell 17: 3007–3018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng W.J., Cheng Z.J., Sang Y.L., Zhang M.M., Rong X.F., Wang Z.W., Tang Y.Y., Zhang X.S.(2017). Type-B ARABIDOPSIS RESPONSE REGULATORs specify the shoot stem cell niche by dual regulation of WUSCHEL. Plant Cell 29: 1357–1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moubayidin L., Di Mambro R., Sabatini S.(2009). Cytokinin-auxin crosstalk. Trends Plant Sci. 14: 557–562. [DOI] [PubMed] [Google Scholar]
- Moubayidin L., et al. (2013). Spatial coordination between stem cell activity and cell differentiation in the root meristem. Dev. Cell 26: 405–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moubayidin L., Perilli S., Dello Ioio R., Di Mambro R., Costantino P., Sabatini S.(2010). The rate of cell differentiation controls the Arabidopsis root meristem growth phase. Curr. Biol. 20: 1138–1143. [DOI] [PubMed] [Google Scholar]
- Müller B., Sheen J.(2007). Advances in cytokinin signaling. Science 318: 68–69. [DOI] [PubMed] [Google Scholar]
- Müller B., Sheen J.(2008). Cytokinin and auxin interaction in root stem-cell specification during early embryogenesis. Nature 453: 1094–1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pernisová M., Klíma P., Horák J., Válková M., Malbeck J., Soucek P., Reichman P., Hoyerová K., Dubová J., Friml J., Zazímalová E., Hejátko J.(2009). Cytokinins modulate auxin-induced organogenesis in plants via regulation of the auxin efflux. Proc. Natl. Acad. Sci. USA 106: 3609–3614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramireddy E., Brenner W.G., Pfeifer A., Heyl A., Schmülling T.(2013). In planta analysis of a cis-regulatory cytokinin response motif in Arabidopsis and identification of a novel enhancer sequence. Plant Cell Physiol. 54: 1079–1092. [DOI] [PubMed] [Google Scholar]
- Rouse D., Mackay P., Stirnberg P., Estelle M., Leyser O.(1998). Changes in auxin response from mutations in an AUX/IAA gene. Science 279: 1371–1373. [DOI] [PubMed] [Google Scholar]
- Sakai H., Aoyama T., Oka A.(2000). Arabidopsis ARR1 and ARR2 response regulators operate as transcriptional activators. Plant J. 24: 703–711. [DOI] [PubMed] [Google Scholar]
- Sakai H., Honma T., Aoyama T., Sato S., Kato T., Tabata S., Oka A.(2001). ARR1, a transcription factor for genes immediately responsive to cytokinins. Science 294: 1519–1521. [DOI] [PubMed] [Google Scholar]
- Sakamoto T., Sakakibara H., Kojima M., Yamamoto Y., Nagasaki H., Inukai Y., Sato Y., Matsuoka M.(2006). Ectopic expression of KNOTTED1-like homeobox protein induces expression of cytokinin biosynthesis genes in rice. Plant Physiol. 142: 54–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salehin M., Bagchi R., Estelle M.(2015). SCFTIR1/AFB-based auxin perception: Mechanism and role in plant growth and development. Plant Cell 27: 9–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoof H., Lenhard M., Haecker A., Mayer K.F.X., Jürgens G., Laux T.(2000). The stem cell population of Arabidopsis shoot meristems in maintained by a regulatory loop between the CLAVATA and WUSCHEL genes. Cell 100: 635–644. [DOI] [PubMed] [Google Scholar]
- Shemer O., Landau U., Candela H., Zemach A., Eshed Williams L.(2015). Competency for shoot regeneration from Arabidopsis root explants is regulated by DNA methylation. Plant Sci. 238: 251–261. [DOI] [PubMed] [Google Scholar]
- Šimášková M., et al. (2015). Cytokinin response factors regulate PIN-FORMED auxin transporters. Nat. Commun. 6: 8717. [DOI] [PubMed] [Google Scholar]
- Skoog F., Miller C.O.(1957). Chemical regulation of growth and organ formation in plant tissues cultured in vitro. Symp. Soc. Exp. Biol. 11: 118–130. [PubMed] [Google Scholar]
- Skylar A., Wu X.(2011). Regulation of meristem size by cytokinin signaling. J. Integr. Plant Biol. 53: 446–454. [DOI] [PubMed] [Google Scholar]
- Snipes S.A., Rodriguez K., DeVries A.E., Miyawaki K.N., Perales M., Xie M., Reddy G.V.(2018). Cytokinin stabilizes WUSCHEL by acting on the protein domains required for nuclear enrichment and transcription. PLoS Genet. 14: e1007351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimoto K., Jiao Y., Meyerowitz E.M.(2010). Arabidopsis regeneration from multiple tissues occurs via a root development pathway. Dev. Cell 18: 463–471. [DOI] [PubMed] [Google Scholar]
- Tian H., Wabnik K., Niu T., Li H., Yu Q., Pollmann S., Vanneste S., Govaerts W., Rolcík J., Geisler M., Friml J., Ding Z.(2014). WOX5-IAA17 feedback circuit-mediated cellular auxin response is crucial for the patterning of root stem cell niches in Arabidopsis. Mol. Plant 7: 277–289. [DOI] [PubMed] [Google Scholar]
- To J.P., Deruère J., Maxwell B.B., Morris V.F., Hutchison C.E., Ferreira F.J., Schaller G.E., Kieber J.J.(2007). Cytokinin regulates type-A Arabidopsis Response Regulator activity and protein stability via two-component phosphorelay. Plant Cell 19: 3901–3914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valvekens D., Van Montagu M., Van Lijsebettens M.(1988). Agrobacterium tumefaciens-mediated transformation of Arabidopsis thaliana root explants by using kanamycin selection. Proc. Natl. Acad. Sci. USA 85: 5536–5540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vickers C.E., Schenk P.M., Li D., Mullineaux P.M., Gresshoff P.M.(2007). pGFPGUSPlus, a new binary vector for gene expression studies and optimising transformation systems in plants. Biotechnol. Lett. 29: 1793–1796. [DOI] [PubMed] [Google Scholar]
- Waldie T., Leyser O.(2018). Cytokinin targets auxin transport to promote shoot branching. Plant Physiol. 177: 803–818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Tian C., Zhang C., Shi B., Cao X., Zhang T.Q., Zhao Z., Wang J.W., Jiao Y.(2017). Cytokinin signaling activates WUSCHEL expression during axillary meristem initiation. Plant Cell 29: 1373–1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie M., Chen H., Huang L., O’Neil R.C., Shokhirev M.N., Ecker J.R.(2018). A B-ARR-mediated cytokinin transcriptional network directs hormone cross-regulation and shoot development. Nat. Commun. 9: 1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin W., Wang Z., Liang Y., Wang Y., Hu Y.(2017). Dynamic expression reveals a two-step patterning of WUS and CLV3 during axillary shoot meristem formation in Arabidopsis. J. Plant Physiol. 214: 1–6. [DOI] [PubMed] [Google Scholar]
- Yanai O., Shani E., Dolezal K., Tarkowski P., Sablowski R., Sandberg G., Samach A., Ori N.(2005). Arabidopsis KNOXI proteins activate cytokinin biosynthesis. Curr. Biol. 15: 1566–1571. [DOI] [PubMed] [Google Scholar]
- Yoo S.D., Cho Y.H., Sheen J.(2007). Arabidopsis mesophyll protoplasts: A versatile cell system for transient gene expression analysis. Nat. Protoc. 2: 1565–1572. [DOI] [PubMed] [Google Scholar]
- Zhang F., May A., Irish V.F.(2017a). Type-B ARABIDOPSIS RESPONSE REGULATORs directly activate WUSCHEL. Trends Plant Sci. 22: 815–817. [DOI] [PubMed] [Google Scholar]
- Zhang T.Q., Lian H., Zhou C.M., Xu L., Jiao Y., Wang J.W.(2017b). A two-step model for de novo activation of WUSCHEL during plant shoot regeneration. Plant Cell 29: 1073–1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W., Swarup R., Bennett M., Schaller G.E., Kieber J.J.(2013). Cytokinin induces cell division in the quiescent center of the Arabidopsis root apical meristem. Curr. Biol. 23: 1979–1989. [DOI] [PubMed] [Google Scholar]
- Zhao Z., Andersen S.U., Ljung K., Dolezal K., Miotk A., Schultheiss S.J., Lohmann J.U.(2010). Hormonal control of the shoot stem-cell niche. Nature 465: 1089–1092. [DOI] [PubMed] [Google Scholar]
- Zubo Y.O., et al. (2017). Cytokinin induces genome-wide binding of the type-B response regulator ARR10 to regulate growth and development in Arabidopsis. Proc. Natl. Acad. Sci. USA 114: E5995–E6004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zürcher E., Tavor-Deslex D., Lituiev D., Enkerli K., Tarr P.T., Müller B.(2013). A robust and sensitive synthetic sensor to monitor the transcriptional output of the cytokinin signaling network in planta. Plant Physiol. 161: 1066–1075. [DOI] [PMC free article] [PubMed] [Google Scholar]