With the recent release of genome assemblies for the NAM parents, we review the impact that the maize NAM population has had on the community and discuss its utility as we move into a new era of genomics.
Abstract
It has been just over a decade since the release of the maize (Zea mays) Nested Association Mapping (NAM) population. The NAM population has been and continues to be an invaluable resource for the maize genetics community and has yielded insights into the genetic architecture of complex traits. The parental lines have become some of the most well-characterized maize germplasm, and their de novo assemblies were recently made publicly available. As we enter an exciting new stage in maize genomics, this retrospective will summarize the design and intentions behind the NAM population; its application, the discoveries it has enabled, and its influence in other systems; and use the past decade of hindsight to consider whether and how it will remain useful in a new age of genomics.
NESTED ASSOCIATION MAPPING DESIGN AND STRUCTURE
Intentions and Goals
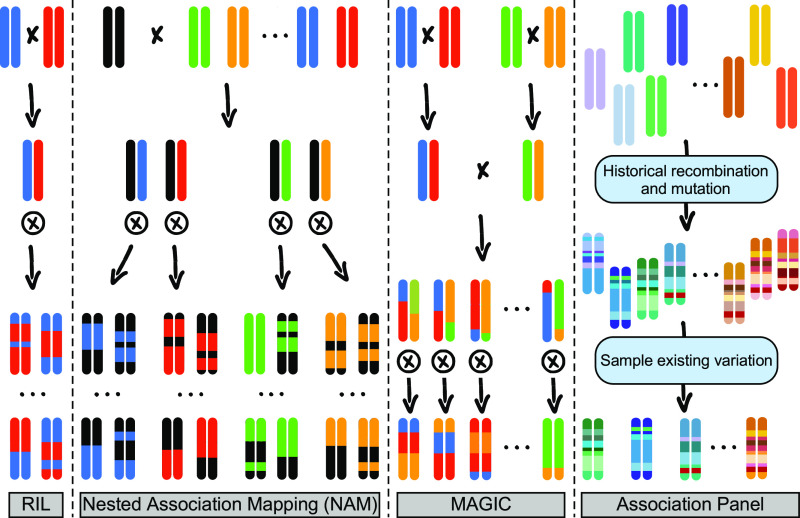
Beginning in the summer of 2002, the maize (Zea mays) Nested Association Mapping (NAM) population was designed to create a community mapping resource that leveraged the advantages of both linkage and association mapping while limiting their respective drawbacks. At the time, linkage mapping was most frequently performed in biparental populations such as recombinant inbred lines (RILs; Figure 1). Due to the limited number of recombination events (crossovers) that typically occur within such populations, the mapping resolution of quantitative trait loci (QTL) identified by linkage mapping is often quite low, while the number of alleles at a given locus is limited to those present in the two parents. Association mapping (Figure 1) provides a partial solution to this problem by capitalizing on the many historical recombination events and low linkage disequilibrium (LD) within a population of diverse individuals to increase allelic diversity and mapping resolution. However, population structure between diverse lines can confound association mapping results (Thornsberry et al., 2001; Flint-Garcia et al., 2005; Yu et al., 2006). The NAM approach promised to resolve the weaknesses of these two types of populations by (1) increasing allelic diversity and the number of recombination events and also (2) decreasing confounding population structure. In the NAM design (Figure 1), a single common inbred line was to be crossed to a diverse set of 25 founder lines to create a set of biparental populations that could be analyzed together (Yu et al., 2008). Each biparental population consisted of 200 progeny, resulting in a total population size of 5000 individuals. By including 26 parental inbreds, NAM increased the total possible number of alleles at each locus to 26, a 13-fold increase over the two possible alleles that result from a single biparental cross with inbred parents. When the entire population could not be analyzed as a whole, family sizes of 200 progeny ensured that there was sufficient power to map traits in each family separately. In retrospect, given the number of successful NAM studies, the choice of 25 families with 200 progeny each seems to have done a reasonable job of balancing biparental size with diversity. With such a large population size, the NAM population captured more than 100,000 crossovers (McMullen et al., 2009; Rodgers-Melnick et al., 2015). Due to the overall population design, these gains in both allelic richness and recombination came without the confounding effects of complex historical population structure.
Figure 1.
Comparison of Common Mapping Populations.
RIL populations are derived from a single biparental cross, resulting in progeny that are a mosaic of haplotypes from the two parents. Recombination bins are often large, limiting mapping resolution. NAM populations consist of numerous RIL families that share a common parent (shown in black). This results in improved resolution and a greater number of alleles represented. MAGIC populations are often derived from 8 or 16 parental lines (we only draw four here for the sake of space; crossing schemes can be more complex than shown here). Similar to NAM populations, MAGIC populations have improved resolution and allelic richness relative to RIL populations. Association panels are a sample of natural variation from a larger, existing population that has accumulated historical recombination events and mutations. They frequently have greater recombination and allelic richness than the other three populations but are also often burdened with inherent population structure that can be difficult to account for. Black ‘x’s indicate crosses between parents, and circled ‘x’s indicate self-mating until inbred. Ellipses indicate many other individuals in the population or family.
The negative aspects arising from biparental and association mapping populations have also been addressed by Multiparent Advanced Generation InterCross (MAGIC) populations (Figure 1; Mott et al., 2000; Cavanagh et al., 2008), which are created by intercrossing a moderate number (usually 8 or 16) of parental lines. The result is a population with more recombination events and higher allelic richness than a biparental population but with less confounding population structure than an association panel. MAGIC populations generally have fewer parents than NAM populations but have more complex crossing schemes. MAGIC populations have potential for more diverse recombinant haplotypes due to the combinatorial arrangement of donor haplotypes compared with the NAM population, in which the single common parent is present in all recombinant haplotypes (Ladejobi et al., 2016). A comparison of crossing schemes for various populations is shown in Figure 1.
Sufficient genetic diversity and population-wide recombination events are essential for identifying functional variants by genetic mapping. Simulation studies of NAM populations demonstrated nearly 60% power to identify (within 5 cM) a QTL that explained 0.39% of the phenotypic variation segregating in a single family; when percentage variance explained increased to 3.5%, power rose to over 97% (Li et al., 2011). In practice, the NAM population is sufficiently powered to identify photoperiod sensitivity QTL that overlap with known candidate genes, even when they explain only 1% of phenotypic variance and segregate in only 3 out of 25 families (Hung et al., 2012b).
Although the NAM population provides much higher power and mapping resolution than a single biparental population, there is still pervasive linkage that limits mapping resolution. Balanced populations with fewer crossovers (e.g., NAM) are suitable for studies in which the goal is to calculate effect sizes or test preexisting hypotheses about functional genes. For resolving causative variants at the nucleotide level, however, other populations or reverse genetics methods are more appropriate. For example, highly diverse association mapping populations, such as the Seeds of Discovery (https://www.cimmyt.org/projects/seeds-of-discovery-seed/) population of landraces, provide high resolution for QTL detection and hypothesis generation but less precise effect estimates (Romero Navarro et al., 2017). Hypotheses generated by association mapping or reverse genetics can subsequently be tested with high power in the NAM population. Because genetically identical maize inbreds can be grown in replicate, the maize NAM population and its founders are some of the best characterized genetic material for studying complex traits, with studies leveraging millions of individual phenotypic records to produce extremely accurate estimates of QTL effect sizes (Buckler et al., 2009; Peiffer et al., 2014).
Even though the NAM population was designed to study the genetic architecture of complex traits, a complementary goal was to create a resource that would be useful to the maize community as a whole (Yu et al., 2008). Previous to the release of the NAM population, there were few publicly available maize mapping populations. Releasing the NAM population as a publicly available resource enabled the maize community to perform studies on a common set of germplasm that allowed the integration of results from different research groups.
Choice of Parents
The NAM population consists of a single common inbred line, B73, crossed to 25 diverse founders. The ideal set of parents for a NAM-type population should be chosen to maximize genetic diversity above all. In the case of the maize NAM population, however, practical considerations limited the selection process slightly (Yu et al., 2008).
An ideal common parent for the NAM population should be well-characterized, agronomically suited to growth in the United States, and enable diverse germplasm to be grown under temperate conditions. B73 was a logical choice for the common parent: it played a historically important role in United States. Corn Belt maize breeding programs (Mikel and Dudley, 2006). B73 had also been extensively characterized in previous genetic, molecular, and agronomic studies, and it had been selected to be the first maize variety to have its genome sequenced and assembled (Schnable et al., 2009). As one of the most influential inbred lines in Corn Belt germplasm, B73 is adapted to temperate, long-day conditions, which helped ensure that the offspring of crosses between B73 and other, tropical materials would still flower in the United States. Some tropical germplasm is not adapted to long days and, as such, would not flower in the continental United States. If even a subset of the NAM lines did not flower, it would have been impossible to develop the population via self-pollination, resulting in a loss of alleles and reduced diversity overall. Additionally, inclusion of parents that did not flower would have complicated the evaluation of agronomically important phenotypes in their progeny under field conditions.
The 25 founder lines were chosen to maximize genetic diversity while still producing inbred lines that flowered under long-day conditions in North Carolina (Liu et al., 2003; Flint-Garcia et al., 2005). They include 13 tropical lines, 9 temperate lines, 2 sweet corn lines, and 1 popcorn line. Despite the large amount of genetic diversity encompassed by these founders, they are not comprehensive; there are germplasm groups that are still not represented in the NAM population, including late-flowering high-altitude Andean and lowland tropical germplasm. The NAM population contains lines representative of some European and Chinese germplasm, but for both regions, separate and more comprehensive multiparental mapping populations have since been created (Bauer et al., 2013; Li et al., 2015). Despite these limitations, the chosen NAM founders covered a large amount of the diversity present in germplasm that was available to choose from and represent a tremendous effort to maximize the diversity within a given set of 25 inbred lines.
Adoption and Application of the NAM Population
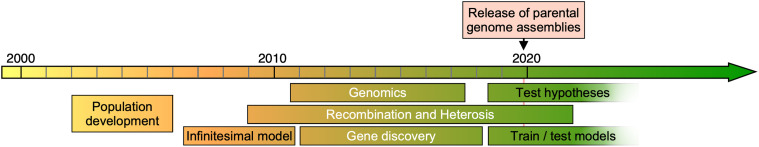
Since its development, the NAM population has been widely adopted for studies of complex traits and natural variation. The full population has been characterized for over 100 different phenotypes (Table 1; Supplemental Data Set) ranging from whole-plant agronomic traits to kernel ionomics. The NAM parents frequently serve as a standard set of lines for natural variation studies (Manavalan et al., 2012; Clark et al., 2013; Yan et al., 2015) and have been characterized for tens of thousands of molecular and biochemical traits (e.g., metabolites in Zhou et al., 2019 and gene expression in Kremling et al., 2018). After characterizing the parents, subsequent experiments can study either the entire NAM population or focus on a subset of the biparental families for which the parents show divergent phenotypes. The NAM population has been used to study a variety of topics, including the genetic architecture of complex traits, recombination, genomics, and heterosis (Figure 2). The NAM population has enabled gene discovery and precise estimates of QTL effect sizes, and the culmination of many studies is a rich collection of publicly available phenotypic data.
Table 1. Studies Published on the Maize NAM Population.
| Categories | No. of Traits | No. of Published Studies |
|---|---|---|
| Agronomic/field traits | 56 | 9 |
| Leaf metabolites | 16 | 2 |
| Kernel metabolites | 24 | 2 |
| Ionomes | 20 | 1 |
| Enzymes | 9 | 2 |
| Biotic stress | 4 | 4 |
| Abiotic stress | 2 | 2 |
The NAM population has been characterized for over 100 different phenotypes, ranging from agronomic characteristics to ionomics profiles.
Figure 2.
Timeline of NAM Population Development and Use.
Population development began in 2002 with parental selection, and biparental family development proceeded through the following years. The collection of phenotypic data began in 2006, and seminal publications were released in 2009. The NAM population has been a resource for studying a number of different topics since then. We anticipate that much of the way the NAM population has been used in the past will not continue for much longer. However, we also anticipate that the NAM population will remain a valuable resource for training or testing within- and cross-species models and that it will be useful as a library of allelic combinations that can be queried for testing hypotheses generated in material or populations.
Community Involvement and Use Cases
The NAM population was developed as a mapping resource that would allow the maize community to apply existing genetic, genomic, and systems biology tools (Yu et al., 2008). As such, the NAM population has helped provide a nucleation point for the generation of permanent resources for genetic studies in maize. Given the population size of NAM, major coordinated efforts were required to increase seed stocks, grow the population, and collect phenotypic data. These efforts resulted in collaborative efforts within the maize community that might not have occurred otherwise. While serving as an early example of large, collaborative experiments within the maize community, the NAM population also placed significant strain on the participating groups. In this regard, it helped define the limitations for a public research program to generate, maintain, and phenotype a field-grown population.
For traits that are difficult to measure at the scale of the full NAM population, studies have instead surveyed only the NAM parents, since they were chosen to represent a wide range of genetic diversity. As a result, the NAM parents alone have become a de facto miniature diversity panel. In some cases, phenotypic surveys of the parents were followed up by phenotyping a subset of the 25 RIL families to map QTL. Because the family size of 200 individuals is large enough to perform QTL mapping, the NAM also provided genetic mapping utility to groups that did not desire the significant undertaking of growing and phenotyping the entire population. This technique has been used to study a wide variety of maize traits such as root system architecture (Zurek et al., 2015), shoot apical meristem morphology (Thompson et al., 2015), submergence tolerance (Campbell et al., 2015), kernel color (Chandler et al., 2013), chromosomal knobs (Ghaffari et al., 2013), and aphid resistance (Meihls et al., 2013). From molecular phenotypes to whole-plant architecture, the NAM population has proved to be a useful resource for successful studies of traits across all levels of genetic and phenotypic complexity.
Repeated phenotypic measurements of the entire NAM population or of individual biparental populations have accumulated since the development of the population. Phenotypic data collected in numerous environments have helped to minimize environmental noise and enabled modeling of interactions between genotype and environment, resulting in a wealth of traits that have been characterized in great detail. Much of the phenotypic data for the NAM population are publicly available (Table 1; Supplemental Data Set). Depending on the trait of interest, researchers can test hypotheses and conduct studies entirely using data that have already been generated and made publicly available (Wallace et al., 2014). Many of the earlier studies that utilized the NAM population were published at a time when data accessibility was becoming a more visible issue in the scientific community (Bechhofer et al., 2010). The NAM population and the Goodman Association Panel were some of the earliest large phenotypic and genotypic data sets available to the maize community. Today, data standards such as the FAIR (Findable, Accessible, Interoperable, Reusable) data principles (Wilkinson et al., 2016) help ensure not only that data are accessible to the community but are also provided in a useful and usable form. Although much of the genotypic and phenotypic data from the NAM were released before the guiding principles of FAIR, the authors of this review encourage community members to keep using similar guidelines when publishing data derived from the NAM population in the future. This will help the maize community continue to be a positive example for the dissemination of open science.
Although large genotypic and phenotypic data sets for the NAM founders and populations are already available, there is still work to be done studying rare alleles, epigenetics, repetitive elements, copy number variants, and multitudes of other questions (see Role of NAM Populations in the Future below). Genome assemblies of the NAM founders will further allow the maize genetics community to ask (and answer) new questions about structural variation and pan-genomics.
Noteworthy Discoveries
Genetic Architecture of Complex Traits
Complex quantitative traits are controlled by numerous loci with small effects. The NAM population has been a tremendous tool for the study of complex traits in maize, including flowering time, plant height, leaf architecture, disease resistance, and many others (Table 1; Buckler et al., 2009; McMullen et al., 2009; Brown et al., 2011; Poland et al., 2011; Tian et al., 2011; Cook et al., 2012; Peiffer et al., 2014; Wallace et al., 2014). The NAM population afforded a large enough sample size, and the opportunity for sufficient repeated measurements of each line, to produce high-quality phenotypic estimates, resulting in the ability to explain much of the genetic variation underlying complex traits. Many of the traits studied in the NAM population were found to be associated with variants that, individually, explain less than 5% of the variation in the phenotype. The infinitesimal model, proposed by Fisher (1919), posited that individual loci each explain only a small fraction of phenotypic variability but collectively contribute to large phenotypic variation. This phenomenon was strongly supported by findings (Buckler et al., 2009) that identified a series of small-effect allelic variants at dozens of loci that together explained nearly 90% of the phenotypic variation in flowering time, a classic complex trait in flowering plants. A later study increased the number of markers fivefold, resulting in a 17 to 33% increase in the number of QTL identified, but still was able to explain a nearly identical amount of variation for flowering time as the original 2009 study (Li et al., 2016). Similarly, highly heritable leaf architecture traits were found to be controlled by more than 30 QTL, which explained 75, 78, and 80% of the phenotypic variance in leaf angle, leaf length, and leaf width, respectively (Tian et al., 2011). More evidence for Fisher’s infinitesimal model was provided by the discovery of persistent but weak segregation distortion (SD) across subfamilies (McMullen et al., 2009; Wallace et al., 2014). SD causes RILs to deviate from the expected 1:1 Mendelian segregation ratio between the two parental alleles at each locus, presumably due to fitness advantages conferred by the more frequent allele. While 54% of markers genome-wide showed significant SD, they deviated from the expected 1:1 ratio by only a small margin, with 97% of chromosomal segments having parental allele frequencies between 45 and 55%, close to the expectation of 50% (McMullen et al., 2009). The high proportion of markers showing small effects on fitness is yet another example of a trait that appears to be governed by the cumulative effects of many loci.
In addition to being well-powered to estimate additive effect sizes, the large number of traits measured in the NAM population (Table 1) opened up opportunities to explore the prevalence of pleiotropy in maize. Surprisingly little evidence for pleiotropy has been found, and the few instances of pleiotropy occurred primarily in closely related traits (Wallace et al., 2014). Pairs of leaf architectural traits (leaf length, width, and angle) shared only 2 to 6% of the QTL identified for each trait individually, explaining their weak phenotypic correlations (0.03 to 0.08; Tian et al., 2011). Some pleiotropy was observed between flowering time, inflorescence architectural traits, and environmental response of flowering (Buckler et al., 2009; Brown et al., 2011; Li et al., 2016). However, it is unclear whether these are examples of true pleiotropy or instead are due to linked genes, the effects of which are difficult to separate by conventional mapping approaches. QTLs in centromeric regions that appear pleiotropic may actually be linked genes that are transferred on common haplotype blocks due to a lack of adequate recombination events near the centromeres (McMullen et al., 2009). Pleiotropy was also observed among carbon and nitrogen metabolism traits, likely due to common regulation and pathways for the studied metabolites (Zhang et al., 2015).
Using the NAM population to study the genetic architecture of complex traits revealed that findings from other model plants do not always extend to agronomically important species. The prevalence in maize of traits governed by numerous loci with small effect size is in sharp contrast to the architecture of similar traits observed in some self-pollinated species. For example, flowering time in maize is controlled by many common, small-effect QTL, none of which affect flowering time by more than 1.5 d (Buckler et al., 2009). However, in self-pollinated species like rice (Oryza sativa), sorghum (Sorghum bicolor), and Arabidopsis (Arabidopsis thaliana), much of the variation for flowering is controlled by a few QTL with large effects (Huang et al., 2011; Li et al., 2011; Salomé et al., 2011). Similar contrasting architectures were observed for leaf structure in maize compared with the self-pollinated species rice, Arabidopsis, and barley (Hordeum vulgare; Koornneef et al., 2004; Turner et al., 2005; Takahashi et al., 2009). The distinct patterns of genetic architecture of maize complex traits relative to inbreeding species may be related to differences in the evolutionary strategies of selfing and outcrossing species. Selection may favor small effect sizes in outcrossing species, in which the sum of small effects keeps the individual phenotypes closer to the population mean (Buckler et al., 2009; Wallace et al., 2014). In outcrossing species, the two parental genomes are shuffled and recombined at every generation, and large-effect loci may not be passed on to progeny. Polygenic trait architectures may also have helped maize adapt to diverse environments, where the effects of selection are spread across numerous segregating loci (Flood and Hancock, 2017). Selection might have also favored independence of traits over pleiotropy, as certain combinations of phenotypes may be favorable in some environments but not others (Wallace et al., 2014).
The pattern of numerous small-effect loci discussed above for flowering time and leaf structure in maize contrasts with the pattern of effect sizes for inflorescence (ear and tassel) traits. A study of QTL underlying maize inflorescence morphology in the NAM population detected systematically larger effect sizes for inflorescence traits, especially those related to ear morphology, than for plant architecture or phenology (Brown et al., 2011). Similar findings were reported by a later study of tassel architecture in a RIL population derived from a cross between maize and teosinte (Z. mays subsp parviglumis; Xu et al., 2017). This may be due to the recent (∼9000 years ago; Matsuoka et al., 2002; Piperno et al., 2009; Ranere et al., 2009) evolution of the maize ear as well as selective pressures on tassel characteristics during maize improvement (Gage et al., 2018); indeed, relatively few generations have passed since the development of the maize ear, and as such, alleles with intermediate effect sizes may not yet be fixed (Fisher, 1930; Orr, 2005).
In addition to findings of effect sizes and pleiotropy, the NAM population enabled studies of recombination rates and their effects on heterosis. Global recombination patterns were shown to be both consistent between populations and predictable (Rodgers-Melnick et al., 2015). Low recombination in pericentromeric regions results in linked, repulsion-phase loci: each parent contributes an opposite pair of one beneficial and one deleterious allele for two linked loci that cannot easily be separated. These regions may be under selection for a heterozygous state, allowing cross-complementation of the repulsion-phased loci and resulting in pseudo-overdominance (Hill and Robertson, 1966; Gore et al., 2009; McMullen et al., 2009; Wallace et al., 2014). By contrast, recombination hotspots appear to have reduced genetic load compared with the rest of the genome, potentially due to the ease with which deleterious alleles can be purged (Rodgers-Melnick et al., 2015). No loci associated with overall recombination rate were found in the NAM population (McMullen et al., 2009). There is, however, a linear relationship between the number of double-stranded breaks and the number of crossovers (Sidhu et al., 2015), and both male and female meioses show similar overall crossover patterns (Kianian et al., 2018).
Biochemical and Molecular Traits
In addition to the identification of thousands of QTL associated with complex traits, the NAM population has also been used to identify causal genes controlling oligogenic traits (controlled by a small number of loci; Wallace et al., 2014). The NAM population was evaluated for volatile terpene production by Richter et al. (2016), who characterized the terpene synthase gene TPS2. TPS2 is responsible for the biosynthesis of several terpenes, which have roles in plant signaling and defense against herbivory. A study of tocochromanols, lipid-soluble antioxidants that provide vitamin E activity, was able to confirm the roles of genes identified a priori as well as to identify six novel genes involved in tocochromanol production (Diepenbrock et al., 2017). Both studies were made possible by the projection of millions of high-density single-nucleotide polymorphism markers, genotyped in the parental lines, onto the NAM progeny. This high-density marker set provides higher resolution for gene identification than the original ∼1000 markers used for many of the earlier NAM studies.
NAM-TYPE POPULATIONS IN OTHER SYSTEMS
Other Maize NAM Populations
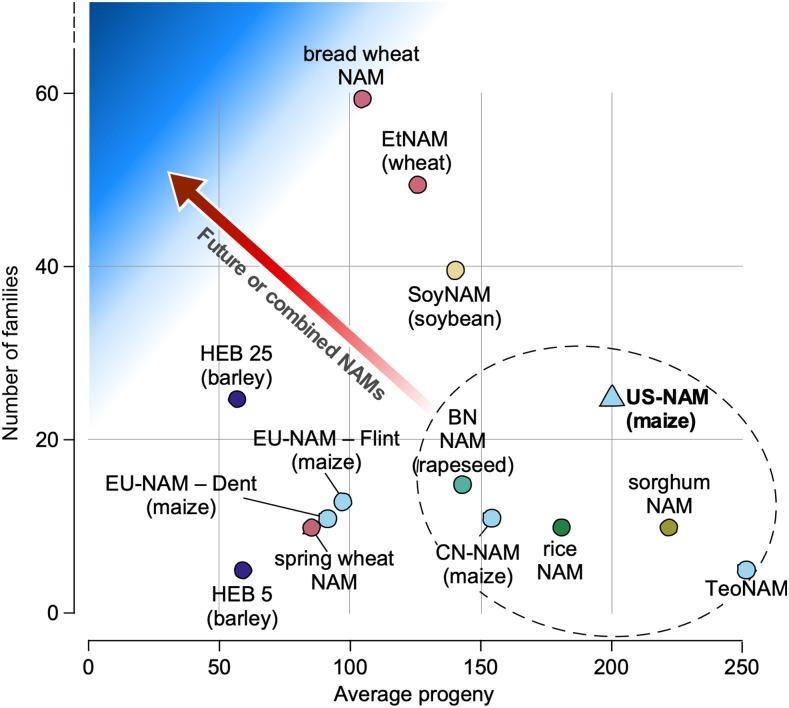
Following the success of the initial maize NAM population, several populations were developed using other maize lines from around the world (Table 2; Figure 3). The additional maize NAM variants were created using Chinese, European, and teosinte inbred lines as parents.
Table 2. NAM-Style Populations in Maize and Other Crops.
| Crop | No. of Families | Total Progeny | Mean Family Size | Reference |
|---|---|---|---|---|
| Maize (United States) | 25 | 5,000 | 200 | (Yu et al., 2008) |
| Maize (China) | 11 | 1,971 | 154 | (Li et al., 2015) |
| Dent maize (Europe) | 11 | 919 | 91 | (Bauer et al., 2013) |
| Flint maize (Europe) | 13 | 1,009 | 97 | (Bauer et al., 2013) |
| Teosinte/maize | 5 | 1,257 | 251 | (Chen et al., 2019) |
| Rice | 10 | 1,879 | 181 | (Fragoso et al., 2017) |
| Wheat | 10 | 852 | 85 | (Bajgain et al., 2016) |
| Wheat | 60 | 6,268 | 105 | (Wingen et al., 2017) |
| Wheat | 50 | 6,280 | 126 | (Kidane et al., 2019) |
| Sorghum | 10 | 2,214 | 221 | (Bouchet et al., 2017) |
| Barley | 5 | 295 | 59 | (Schnaithmann et al., 2014) |
| Barley | 25 | 1,420 | 57 | (Maurer et al., 2015) |
| Soybean | 40 | 5,600 | 140 | (Song et al., 2017) |
| Rapeseed | 15 | 2,141 | 143 | (Hu et al., 2018) |
Populations designed in the style of the original maize NAM varied in terms of the number of biparental populations and the number of progeny in each population.
Figure 3.
Design Parameters of Other NAM-Type Populations.
Multiple populations (circles) have been developed with similar structure to the maize NAM population (triangle): one or few common parents crossed to a number of diverse founders to create multiple biparental families. Subsequent populations have varied greatly in the number of biparental families and the number of progeny within each family, but most population designs target fewer families with a large number of progeny (dashed line). Changes in sequencing/genotyping technology have made it easier to create and genotype populations with a larger number of parents and fewer progeny per family (blue shaded region). The increasing intensity of the blue region indicates more optimal design for increasing allelic richness and mapping resolution (i.e., fewer progeny and more parents).
The Chinese NAM population was created by crossing 11 commonly used maize lines representative of major heterotic groups from Chinese maize breeding with the common parent Huangzaosi (Li et al., 2016). Huangzaosi was selected due to its status as one of the most prolific Chinese maize inbreds, its wide environmental adaptability, and its resistance to common plant pathogens.
The European NAM (EU-NAM) populations consist of two half-sib panels of Dent- and Flint-type maize lines, named after their kernel phenotypes, representing the two major germplasm pools frequently used in European maize breeding. Each panel consists of a common parent crossed with 10 (Dent) or 11 (Flint) founders representative of popular and diverse European maize breeding lines from the respective germplasm pools. The common parent from the Flint and Dent panels (UH007 and F353, respectively) were also crossed with B73 in order to link the EU-NAM population with the U.S. NAM population. The resulting F1 progeny from this crossing scheme were made homozygous by doubled haploidy (DH) rather than inbreeding, as was the case in the U.S. NAM population (Bauer et al., 2013).
The teosinte NAM (TeoNAM) population was created using inbred lines of teosinte, the wild progenitor of maize. Chen et al. (2019) developed the TeoNAM population by crossing five inbred teosinte parents with a widely used inbred maize parent, W22. The five teosinte parents included four Z. mays ssp parviglumis lines and one Z. mays ssp mexicana line. The resulting F1 hybrids were backcrossed to W22 before being selfed for four generations.
Having multiple NAM-type populations in maize will afford the opportunity to combine effect estimates from the 25 original NAM families with more than 30 other families (as described by Swarts et al. [2016]), an endeavor that will be facilitated and made more powerful with new genome assemblies and better sequencing (see Role of NAM Populations in the Future below).
NAM Populations in Other Crops
In addition to maize, NAM populations have been developed in rice, wheat (Triticum aestivum and Triticum durum), sorghum, barley, soybean (Glycine max), and rapeseed (Brassica napus; Table 2; Figure 3). Each NAM system developed in other species serves as a community resource for breeding and genetic insight. Besides their inception and construction, these non-maize NAM populations have enabled discoveries related to segregation distortion, architecture of complex traits, and recombination in their respective species.
Evidence of segregation distortion among and between biparental families was found in rice, wheat, and soybean (Fragoso et al., 2017; Song et al., 2017; Wingen et al., 2017). These studies support the widespread segregation distortion found in maize and extend the findings to other crop species. The power of NAM-type population design for detecting QTL, established by empirical and simulation studies in maize, was further supported by simulation results that indicated a threefold increase in detection power in the sorghum NAM population compared with association panels (Bouchet et al., 2017). Empirically, NAM populations in rice, wheat, sorghum, and barley have been effective for identifying QTL and studying the genetic architecture of traits with agronomic importance, including disease resistance, flowering time, and plant height (Schnaithmann et al., 2014; Maurer et al., 2015; Bajgain et al., 2016; Bouchet et al., 2017; Fragoso et al., 2017; Kidane et al., 2019). Finally, studies of NAM populations in rapeseed and durum wheat both revealed insights into recombination and LD patterns. Rapeseed had more rapid LD decay on the A subgenome compared with the C subgenome, indicating greater recombination rates in the A subgenome; durum wheat demonstrated considerable variation in LD decay rate between chromosomes as well as between biparental families (Hu et al., 2018; Kidane et al., 2019).
As revealed by studies of these populations, as well as the maize NAM population, NAM-type populations are frequently evaluated for a similar suite of traits, particularly plant height, flowering time, and disease resistance. These are agronomically important traits and relatively simple to measure. Due to large population sizes, NAM-type populations can be prohibitive to phenotype in full, which precludes genetic mapping of traits that are labor-intensive to measure. As new methods for high-throughput phenotyping are developed, this may become less of a concern.
RETROSPECTIVE CONSIDERATIONS
Although the NAM population has been widely used by the maize community and its design principles adapted to other systems, the population design has some drawbacks. Many of the initial design considerations were constrained by the number of parents that could be genotyped. In 2001, a single human genome cost on the order of $1 billion to sequence. Sequencing even just the genic space of 26 maize lines in 2002 would have been an enormous undertaking. When the NAM was designed, it was sensible to keep the family number low and the family size large enough that linkage mapping could be performed effectively. The 200 RILs allowed for the individual families to stand on their own for linkage mapping even if genotyping costs did not drop as rapidly as hoped (Peter Bradbury, James Holland, Major Goodman, and Sherry Flint-Garcia, personal communication). If the current affordability of DNA sequencing had been available when the NAM population was being designed, more founders might have been included in the design, which would have permitted higher mapping resolution via association mapping as well as greater representation of rare variants. The rarest alleles, present in a single founder line, are expected to be present in half of all progeny from the corresponding family (∼100 individual RILs). Given the design of the original maize NAM population, family sizes of 100 or even 50 would have contained enough individuals to map associations with rare alleles; decreasing population sizes twofold or fourfold would have favored the inclusion of a greater number of founders and higher overall resolution. Given current sequencing costs, the number of parents to sequence or genotype is almost irrelevant compared with the cost of line creation and phenotyping. Creating a panel with two to four times as many parents (50 to 100 families with 100 to 50 RILs each) would allow for a much higher mapping resolution, as the rapid decay of parental LD would be shared among more parents.
Another way to create the population today would include the use of DH techniques for creating inbred lines. DH could decrease the number of generations of selfing needed to generate the population. During selfing to create the inbred lines, one-third of each family was selfed under different environmental conditions in an effort to minimize unintentional selection pressures (McMullen et al., 2009). Although DH would decrease the amount of work needed to create the inbred lines, it may also introduce additional selection pressures (Sherry Flint-Garcia and James Holland, personal communication). DH techniques were used in later NAM-type populations, such as the EU-NAM population, to improve the efficiency of creating inbred biparental populations (Bauer et al., 2013). Even though DH enables rapid creation of homozygous lines, it also results in fewer recombination events; the EU-NAM population has about half the number of recombination events per inbred line compared with the original maize NAM population (15.1 versus 28.9, respectively), presumably due to the accumulation of crossovers during subsequent generations of selfing in the NAM (Bauer et al., 2013). If a population is to be used for genetic mapping, higher rates of recombination are desirable, as they lead to better mapping resolution. However, the ability to create populations more rapidly allows the creation of new dedicated populations for the study of particular traits, and recombination levels in DH lines are still high enough to perform genetic mapping (albeit resulting in larger mapping intervals) and to identify markers for marker-assisted selection.
As described above, the NAM founders were chosen to maximize genetic diversity from the germplasm that was available at the time. The population from which the founders were chosen consisted of 302 diverse inbred lines (Flint-Garcia et al., 2005). Today, much larger germplasm collections have been genotyped, which may have resulted in different choices of founders. For instance, the Ames panel (Romay et al., 2013) comprises over 2800 accessions with a denser sampling of the major subpopulations of maize than offered by the initial Goodman Association panel of 302 lines. Given the resources available at the time, the selection of the NAM founders still captured a large cross section of genetic diversity in maize (James Holland and Sherry Flint-Garcia, personal communication).
Given the opportunity to create a population with similar goals today, 10 years after its original release, the NAM population would be designed differently to maximize its usefulness. With cheaper sequencing, it would be more feasible to include more parental lines. Without changing the overall population size, a design with, for example, 2 common parents each crossed to 100 diverse founders would create a structured population with 25 progeny per cross, which would contain more historical recombination events and therefore finer mapping resolution. This structure would result in a greater number of potential recombinant haplotypes, one of the advantages of MAGIC populations (Figure 1; Ladejobi et al., 2016). A new NAM population with more founders would also have greater representation of rare alleles, which in the years since the original release have been identified as important targets for crop improvement (Kremling et al., 2018; Valluru et al., 2019). The lower population size of each biparental in such a design would limit the ability to map traits within individual biparental populations, but greater diversity among the parental lines would also make it more likely that multiple populations would segregate for a given trait or allele, allowing mapping of several small biparental populations rather than a single, moderately sized biparental population. Such a design would also increase the number of alleles to a maximum of 102 and enable better estimates of genetic architecture and QTL variance (Hung et al., 2012a). At a certain point, adding more parental lines will fail to capture more of the genetic space within a species; maize is tremendously diverse, but for other crops with limited genetic diversity, this may be an important limitation.
ROLE OF NAM POPULATIONS IN THE FUTURE
Developed at a time to help usher in the age of genetic mapping, the NAM population was an indispensable resource for the maize and quantitative genetics communities. As we move into a new era of affordable, high-throughput sequencing and genomics, does it still have utility?
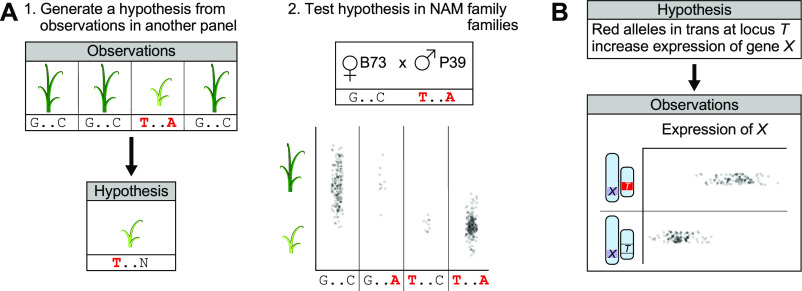
Previous studies have used the NAM population extensively to test hypotheses in quantitative genetics and genomics. Using genetic mapping as an example, other techniques such as association panels, reverse genetics, or analysis of assembled genomes are better suited to generating hypotheses (i.e., identifying candidate variants associated with a phenotype). By contrast, the NAM population provides highly accurate allelic effect estimates that can be used to test the hypotheses generated by other means (Figure 4A). Although its utility for forward genetics studies and the generation of hypotheses is limited due to low resolution, the NAM population will continue to live as a resource for hypothesis testing as we move into the future, aided by genome assemblies of the founder parents. In addition to a genetic mapping population, the NAM population is also a collection of allelic recombinations that can be leveraged to answer questions generated elsewhere.
Figure 4.
Future Use Cases for NAM Population.
(A) NAM families can be used to test hypotheses that are generated in independent populations. In this example, two polymorphisms appear to be associated with the stunted, lighter green phenotype (1). The hypothesis is that the first of those two polymorphisms is responsible for the observed phenotype. Recombinants between the two loci of interest in the B73 × P39 family support this hypothesis (2).
(B) A hypothesis about trans-effects of a particular locus T on the expression of gene X was formed from other observations. This hypothesis is supported by differences in expression between NAM lines with and without the red T allele at the trans-locus while holding the allele at gene X constant.
The structured nature of the NAM population has effectively created a library of recombination events that are ready to be queried for hypotheses about functional genes and mutations. Unlike extremely diverse association panels, the population can be grown and phenotyped in its entirety without severe maladaptation because it is temperately adapted thanks to the common parent B73. This opens up the possibility to evaluate tropical alleles that would otherwise be difficult to study in temperate environments. Once candidate loci are identified from mapping approaches, mutant studies, or computational approaches, a subset of 10 to 15 NAM lines representing natural variation at those loci can be ordered for further study. On average, the NAM population features a 3:1 ratio between crossovers and genes (McMullen et al., 2009), allowing for the study of different allelic combinations with the loci of interest.
The NAM population can also be an important framework for understanding the complex phenomenon of the regulation of gene expression. Regulatory elements affecting gene expression can be associated with multiple genes and located in cis (nearby) or trans (distal or on separate chromosomes) relative to their targets. The arrangement of these regulatory components in 25 biparental populations can be used to disentangle cis-effects from trans-effects, to study specific candidate loci, or to understand evolution and regulation of gene expression at the genomic scale (Figure 4B).
As profiling techniques for gene expression and translation, metabolite abundance, chromatin conformation, and other molecular phenotypes become increasingly common and affordable, the NAM founder assemblies will enable high-quality profiling of diverse materials and their offspring. The progeny of the NAM population offer a way to leverage structured recombination to study molecular profiles both genome-wide and at loci of particular interest in order to answer preformed questions and test hypotheses. The identification of structural variation in the NAM founder assemblies can be used to test hypotheses about inheritance and effects of those variants in progeny of particular crosses.
CONCLUSION
The NAM population has been used widely and for a number of purposes since its public release in 2009. It has empowered a number of discoveries about the genetic architecture of traits in maize, resulted in an extensive collection of publicly available phenotypic measurements on a common set of germplasm, and inspired a number of similarly organized populations in other crop species. Although certain aspects of the NAM population design could be improved given current technologies and knowledge, it has nonetheless had an impressive influence on the field of quantitative genetics. Looking to the future, we see a continued role for the NAM population in the exploration and discoveries of genetics, genomics, and crop improvement.
Supplemental Data
Supplemental Data Set. List of traits that have been phenotyped in the NAM population, and their corresponding references.
DIVE Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
Acknowledgments
We extend our deepest gratitude to all members involved with the development of the NAM. Major Goodman, James Holland, Jeffry Thornsberry, Sherry Flint-Garcia, Jianming Yu, and Peter Bradbury all took the time to offer their own, retrospective thoughts on the NAM, for which we are also very grateful. The development of the NAM was supported by the National Science Foundation Plant Genome Program (grants DBI-0321467, DBI-0820619, and DBI-1238014) and the USDA-Agricultural Research Service.
Footnotes
Articles can be viewed without a subscription.
References
- Bajgain P., Rouse M.N., Tsilo T.J., Macharia G.K., Bhavani S., Jin Y., Anderson J.A.(2016). Nested association mapping of stem rust resistance in wheat using genotyping by sequencing. PLoS One 11: e0155760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer E., et al. (2013). Intraspecific variation of recombination rate in maize. Genome Biol. 14: R103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bechhofer S., De Roure D., Gamble M., Goble C., Buchan I.(2010). Research Objects: Towards Exchange and Reuse of Digital Knowledge. Nat Prec.
- Bouchet S., Olatoye M.O., Marla S.R., Perumal R., Tesso T., Yu J., Tuinstra M., Morris G.P.(2017). Increased power to dissect adaptive traits in global sorghum diversity using a nested association mapping population. Genetics 206: 573–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown P.J., Upadyayula N., Mahone G.S., Tian F., Bradbury P.J., Myles S., Holland J.B., Flint-Garcia S., McMullen M.D., Buckler E.S., Rocheford T.R.(2011). Distinct genetic architectures for male and female inflorescence traits of maize. PLoS Genet. 7: e1002383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckler E.S., et al. (2009). The genetic architecture of maize flowering time. Science 325: 714–718. [DOI] [PubMed] [Google Scholar]
- Campbell M.T., Proctor C.A., Dou Y., Schmitz A.J., Phansak P., Kruger G.R., Zhang C., Walia H.(2015). Genetic and molecular characterization of submergence response identifies Subtol6 as a major submergence tolerance locus in maize. PLoS One 10: e0120385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavanagh C., Morell M., Mackay I., Powell W.(2008). From mutations to MAGIC: Resources for gene discovery, validation and delivery in crop plants. Curr. Opin. Plant Biol. 11: 215–221. [DOI] [PubMed] [Google Scholar]
- Chandler K., Lipka A.E., Owens B.F., Li H., Buckler E.S., Rocheford T., Gore M.A.(2013). Genetic analysis of visually scored orange kernel color in maize. Crop Sci. 53: 189–200. [Google Scholar]
- Chen Q., et al. (2019). TeoNAM: A nested association mapping population for domestication and agronomic trait analysis in maize. Genetics 213: 1065–1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark R.T., Famoso A.N., Zhao K., Shaff J.E., Craft E.J., Bustamante C.D., McCouch S.R., Aneshansley D.J., Kochian L.V.(2013). High-throughput two-dimensional root system phenotyping platform facilitates genetic analysis of root growth and development. Plant Cell Environ. 36: 454–466. [DOI] [PubMed] [Google Scholar]
- Cook J.P., McMullen M.D., Holland J.B., Tian F., Bradbury P., Ross-Ibarra J., Buckler E.S., Flint-Garcia S.A.(2012). Genetic architecture of maize kernel composition in the nested association mapping and inbred association panels. Plant Physiol. 158: 824–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diepenbrock C.H., et al. (2017). Novel loci underlie natural variation in vitamin E levels in maize grain. Plant Cell 29: 2374–2392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher R.A.(1919). XV. The correlation between relatives on the supposition of Mendelian inheritance. Earth Environ. Sci. Trans. R. Soc. Edinb. 52: 399–433. [Google Scholar]
- Fisher R.A.(1930). The Genetical Theory of Natural Selection.. (Oxford, UK: Clarendon Press; ). [Google Scholar]
- Flint-Garcia S.A., Thuillet A.-C., Yu J., Pressoir G., Romero S.M., Mitchell S.E., Doebley J., Kresovich S., Goodman M.M., Buckler E.S.(2005). Maize association population: A high-resolution platform for quantitative trait locus dissection. Plant J. 44: 1054–1064. [DOI] [PubMed] [Google Scholar]
- Flood P.J., Hancock A.M.(2017). The genomic basis of adaptation in plants. Curr. Opin. Plant Biol. 36: 88–94. [DOI] [PubMed] [Google Scholar]
- Fragoso C.A., Moreno M., Wang Z., Heffelfinger C., Arbelaez L.J., Aguirre J.A., Franco N., Romero L.E., Labadie K., Zhao H., Dellaporta S.L., Loriaux M.(2017). Genetic architecture of a rice nested association mapping population. G3 (Bethesda) 7: 1913–1926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gage J.L., White M.R., Edwards J.W., Kaeppler S., de Leon N.(2018). Selection signatures underlying dramatic male inflorescence transformation during modern hybrid maize breeding. Genetics 210: 1125–1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghaffari R., Cannon E.K.S., Kanizay L.B., Lawrence C.J., Dawe R.K.(2013). Maize chromosomal knobs are located in gene-dense areas and suppress local recombination. Chromosoma 122: 67–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gore M.A., Chia J.-M., Elshire R.J., Sun Q., Ersoz E.S., Hurwitz B.L., Peiffer J.A., McMullen M.D., Grills G.S., Ross-Ibarra J., Ware D.H., Buckler E.S.(2009). A first-generation haplotype map of maize. Science 326: 1115–1117. [DOI] [PubMed] [Google Scholar]
- Hill W.G., Robertson A.(1966). The effect of linkage on limits to artificial selection. Genet. Res. 8: 269–294. [PubMed] [Google Scholar]
- Hu J., Guo C., Wang B., Ye J., Liu M., Wu Z., Xiao Y., Zhang Q., Li H., King G.J., Liu K.(2018). Genetic properties of a nested association mapping population constructed with semi-winter and spring oilseed rapes. Front. Plant Sci. 9: 1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X., et al. (2011). Genome-wide association study of flowering time and grain yield traits in a worldwide collection of rice germplasm. Nat. Genet. 44: 32–39. [DOI] [PubMed] [Google Scholar]
- Hung H.-Y., et al. (2012a). The relationship between parental genetic or phenotypic divergence and progeny variation in the maize nested association mapping population. Heredity 108: 490–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung H.-Y., Shannon L.M., Tian F., Bradbury P.J., Chen C., Flint-Garcia S.A., McMullen M.D., Ware D., Buckler E.S., Doebley J.F., Holland J.B.(2012b). ZmCCT and the genetic basis of day-length adaptation underlying the postdomestication spread of maize. Proc. Natl. Acad. Sci. USA 109: E1913–E1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kianian P.M.A., et al. (2018). High-resolution crossover mapping reveals similarities and differences of male and female recombination in maize. Nat. Commun. 9: 2370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidane Y.G., Gesesse C.A., Hailemariam B.N., Desta E.A., Mengistu D.K., Fadda C., Pè M.E., Dell’Acqua M.(2019). A large nested association mapping population for breeding and quantitative trait locus mapping in Ethiopian durum wheat. Plant Biotechnol. J. 17: 1380–1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koornneef M., Alonso-Blanco C., Vreugdenhil D.(2004). Naturally occurring genetic variation in Arabidopsis thaliana. Annu. Rev. Plant Biol. 55: 141–172. [DOI] [PubMed] [Google Scholar]
- Kremling K.A.G., Chen S.-Y., Su M.-H., Lepak N.K., Romay M.C., Swarts K.L., Lu F., Lorant A., Bradbury P.J., Buckler E.S.(2018). Dysregulation of expression correlates with rare-allele burden and fitness loss in maize. Nature 555: 520–523. [DOI] [PubMed] [Google Scholar]
- Ladejobi O., Elderfield J., Gardner K.A., Gaynor R.C., Hickey J., Hibberd J.M., Mackay I.J., Bentley A.R.(2016). Maximizing the potential of multi-parental crop populations. Appl. Transl. Genomics 11: 9–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C., Li Y., Bradbury P.J., Wu X., Shi Y., Song Y., Zhang D., Rodgers-Melnick E., Buckler E.S., Zhang Z., Li Y., Wang T.(2015). Construction of high-quality recombination maps with low-coverage genomic sequencing for joint linkage analysis in maize. BMC Biol. 13: 78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Bradbury P., Ersoz E., Buckler E.S., Wang J.(2011). Joint QTL linkage mapping for multiple-cross mating design sharing one common parent. PLoS One 6: e17573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y.X., et al. (2016). Identification of genetic variants associated with maize flowering time using an extremely large multi-genetic background population. Plant J. 86: 391–402. [DOI] [PubMed] [Google Scholar]
- Liu K., Goodman M., Muse S., Smith J.S., Buckler E., Doebley J.(2003). Genetic structure and diversity among maize inbred lines as inferred from DNA microsatellites. Genetics 165: 2117–2128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manavalan L.P., Musket T., Nguyen H.T.(2012). Natural genetic variation for root traits among diversity lines of maize (Zea mays L.). Maydica 56. [Google Scholar]
- Matsuoka Y., Vigouroux Y., Goodman M.M., Sanchez G.J., Buckler E., Doebley J.(2002). A single domestication for maize shown by multilocus microsatellite genotyping. Proc. Natl. Acad. Sci. USA 99: 6080–6084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurer A., Draba V., Jiang Y., Schnaithmann F., Sharma R., Schumann E., Kilian B., Reif J.C., Pillen K.(2015). Modelling the genetic architecture of flowering time control in barley through nested association mapping. BMC Genomics 16: 290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMullen M.D., et al. (2009). Genetic properties of the maize nested association mapping population. Science 325: 737–740. [DOI] [PubMed] [Google Scholar]
- Meihls L.N., Handrick V., Glauser G., Barbier H., Kaur H., Haribal M.M., Lipka A.E., Gershenzon J., Buckler E.S., Erb M., Köllner T.G., Jander G.(2013). Natural variation in maize aphid resistance is associated with 2,4-dihydroxy-7-methoxy-1,4-benzoxazin-3-one glucoside methyltransferase activity. Plant Cell 25: 2341–2355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikel M.A., Dudley J.W.(2006). Evolution of North American dent corn from public to proprietary germplasm. Crop Sci. 46: 1193–1205. [Google Scholar]
- Mott R., Talbot C.J., Turri M.G., Collins A.C., Flint J.(2000). A method for fine mapping quantitative trait loci in outbred animal stocks. Proc. Natl. Acad. Sci. USA 97: 12649–12654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr H.A.(2005). The genetic theory of adaptation: A brief history. Nat. Rev. Genet. 6: 119–127. [DOI] [PubMed] [Google Scholar]
- Peiffer J.A., Romay M.C., Gore M.A., Flint-Garcia S.A., Zhang Z., Millard M.J., Gardner C.A.C., McMullen M.D., Holland J.B., Bradbury P.J., Buckler E.S.(2014). The genetic architecture of maize height. Genetics 196: 1337–1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piperno D.R., Ranere A.J., Holst I., Iriarte J., Dickau R.(2009). Starch grain and phytolith evidence for early ninth millennium B.P. maize from the Central Balsas River Valley, Mexico. Proc. Natl. Acad. Sci. USA 106: 5019–5024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poland J.A., Bradbury P.J., Buckler E.S., Nelson R.J.(2011). Genome-wide nested association mapping of quantitative resistance to northern leaf blight in maize. Proc. Natl. Acad. Sci. USA 108: 6893–6898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranere A.J., Piperno D.R., Holst I., Dickau R., Iriarte J.(2009). The cultural and chronological context of early Holocene maize and squash domestication in the Central Balsas River Valley, Mexico. Proc. Natl. Acad. Sci. USA 106: 5014–5018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter A., et al. (2016). Characterization of biosynthetic pathways for the production of the volatile homoterpenes DMNT and TMTT in Zea mays. Plant Cell 28: 2651–2665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodgers-Melnick E., Bradbury P.J., Elshire R.J., Glaubitz J.C., Acharya C.B., Mitchell S.E., Li C., Li Y., Buckler E.S.(2015). Recombination in diverse maize is stable, predictable, and associated with genetic load. Proc. Natl. Acad. Sci. USA 112: 3823–3828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romay M.C., et al. (2013). Comprehensive genotyping of the USA national maize inbred seed bank. Genome Biol. 14: R55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero Navarro J.A., et al. (2017). A study of allelic diversity underlying flowering-time adaptation in maize landraces. Nat. Genet. 49: 476–480. [DOI] [PubMed] [Google Scholar]
- Salomé P.A., Bomblies K., Laitinen R.A.E., Yant L., Mott R., Weigel D.(2011). Genetic architecture of flowering-time variation in Arabidopsis thaliana. Genetics 188: 421–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnable P.S., et al. (2009). The B73 maize genome: Complexity, diversity, and dynamics. Science 326: 1112–1115. [DOI] [PubMed] [Google Scholar]
- Schnaithmann F., Kopahnke D., Pillen K.(2014). A first step toward the development of a barley NAM population and its utilization to detect QTLs conferring leaf rust seedling resistance. Theor. Appl. Genet. 127: 1513–1525. [DOI] [PubMed] [Google Scholar]
- Sidhu G.K., Fang C., Olson M.A., Falque M., Martin O.C., Pawlowski W.P.(2015). Recombination patterns in maize reveal limits to crossover homeostasis. Proc. Natl. Acad. Sci. USA 112: 15982–15987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Q., et al. (2017). Genetic characterization of the soybean nested association mapping population. Plant Genome 10: 1–14. [DOI] [PubMed] [Google Scholar]
- Swarts K., et al. (2016). A large scale joint analysis of flowering time reveals independent temperate adaptations in maize. bioRxiv 86082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi Y., Teshima K.M., Yokoi S., Innan H., Shimamoto K.(2009). Variations in Hd1 proteins, Hd3a promoters, and Ehd1 expression levels contribute to diversity of flowering time in cultivated rice. Proc. Natl. Acad. Sci. USA 106: 4555–4560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson A.M., Yu J., Timmermans M.C.P., Schnable P., Crants J.E., Scanlon M.J., Muehlbauer G.J.(2015). Diversity of maize shoot apical meristem architecture and its relationship to plant morphology. G3 (Bethesda) 5: 819–827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornsberry J.M., Goodman M.M., Doebley J., Kresovich S., Nielsen D., Buckler E.S. IV(2001). Dwarf8 polymorphisms associate with variation in flowering time. Nat. Genet. 28: 286–289. [DOI] [PubMed] [Google Scholar]
- Tian F., Bradbury P.J., Brown P.J., Hung H., Sun Q., Flint-Garcia S., Rocheford T.R., McMullen M.D., Holland J.B., Buckler E.S.(2011). Genome-wide association study of leaf architecture in the maize nested association mapping population. Nat. Genet. 43: 159–162. [DOI] [PubMed] [Google Scholar]
- Turner A., Beales J., Faure S., Dunford R.P., Laurie D.A.(2005). The pseudo-response regulator Ppd-H1 provides adaptation to photoperiod in barley. Science 310: 1031–1034. [DOI] [PubMed] [Google Scholar]
- Valluru R., Gazave E.E., Fernandes S.B., Ferguson J.N., Lozano R., Hirannaiah P., Zuo T., Brown P.J., Leakey A.D.B., Gore M.A., Buckler E.S., Bandillo N.(2019). Deleterious mutation burden and its association with complex traits in sorghum (Sorghum bicolor). Genetics 211: 1075–1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace J.G., Larsson S.J., Buckler E.S.(2014). Entering the second century of maize quantitative genetics. Heredity 112: 30–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson M.D., Dumontier M., Aalbersberg I.J.J., Appleton G., Axton M., Baak A., Blomberg N., Boiten J.-W., da Silva Santos L.B., Bourne P.E., Bouwman J., Brookes A.J., et al. (2016). The FAIR Guiding Principles for scientific data management and stewardship. Sci Data 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wingen L.U., West C., Leverington-Waite M., Collier S., Orford S., Goram R., Yang C.-Y., King J., Allen A.M., Burridge A., Edwards K.J., Griffiths S.(2017). Wheat landrace genome diversity. Genetics 205: 1657–1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu G., Wang X., Huang C., Xu D., Li D., Tian J., Chen Q., Wang C., Liang Y., Wu Y., Yang X., Tian F.(2017). Complex genetic architecture underlies maize tassel domestication. New Phytol. 214: 852–864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan J., Lipka A.E., Schmelz E.A., Buckler E.S., Jander G.(2015). Accumulation of 5-hydroxynorvaline in maize (Zea mays) leaves is induced by insect feeding and abiotic stress. J. Exp. Bot. 66: 593–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J., Holland J.B., McMullen M.D., Buckler E.S.(2008). Genetic design and statistical power of nested association mapping in maize. Genetics 178: 539–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J., Pressoir G., Briggs W.H., Vroh Bi I., Yamasaki M., Doebley J.F., McMullen M.D., Gaut B.S., Nielsen D.M., Holland J.B., Kresovich S., Buckler E.S.(2006). A unified mixed-model method for association mapping that accounts for multiple levels of relatedness. Nat. Genet. 38: 203–208. [DOI] [PubMed] [Google Scholar]
- Zhang N., et al. (2015). Genome-wide association of carbon and nitrogen metabolism in the maize nested association mapping population. Plant Physiol. 168: 575–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou S., Kremling K.A., Bandillo N., Richter A., Zhang Y.K., Ahern K.R., Artyukhin A.B., Hui J.X., Younkin G.C., Schroeder F.C., Buckler E.S., Jander G.(2019). Metabolome-scale genome-wide association studies reveal chemical diversity and genetic control of maize specialized metabolites. Plant Cell 31: 937–955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zurek P.R., Topp C.N., Benfey P.N.(2015). Quantitative trait locus mapping reveals regions of the maize genome controlling root system architecture. Plant Physiol. 167: 1487–1496. [DOI] [PMC free article] [PubMed] [Google Scholar]