A proteomic study sheds light on Transport Protein Particle (TRAPP) proteins in plants and identifies TRIPP as a plant-specific component of the TRAPPII complex with important roles in vesicle trafficking and plant development.
Abstract
How the membrane trafficking system spatially organizes intracellular activities and intercellular signaling networks in plants is not well understood. Transport Protein Particle (TRAPP) complexes play key roles in the selective delivery of membrane vesicles to various subcellular compartments in yeast and animals but remain to be fully characterized in plants. Here, we investigated TRAPP complexes in Arabidopsis (Arabidopsis thaliana) using immunoprecipitation followed by quantitative mass spectrometry analysis of AtTRS33, a conserved core component of all TRAPP complexes. We identified 14 AtTRS33-interacting proteins, including homologs of all 13 TRAPP components in mammals and a protein that has homologs only in multicellular photosynthetic organisms and is thus named TRAPP-Interacting Plant Protein (TRIPP). TRIPP specifically associates with the TRAPPII complex through binary interactions with two TRAPPII-specific subunits. TRIPP colocalized with a subset of TRS33 compartments and trans-Golgi network markers in a TRS33-dependent manner. Loss-of-function tripp mutants exhibited dwarfism, sterility, partial photomorphogenesis in the dark, reduced polarity of the auxin transporter PIN2, incomplete cross wall formation, and altered localization of a TRAPPII-specific component. Therefore, TRIPP is a plant-specific component of the TRAPPII complex with important functions in trafficking, plant growth, and development.
INTRODUCTION
Vesicular protein trafficking is a highly organized, finely regulated cellular process that ensures the accurate delivery of proteins to their correct subcellular compartments (Rosquete et al., 2018). Vesicle transport spatially organizes intracellular structures and metabolic activity as well as intercellular signaling systems that control development. Vesicle transport involves numerous protein complexes and is regulated at multiple stages, beginning with vesicle budding from a donor membrane and ending with the fusion stage, where a vesicle merges with a specific acceptor membrane (Bröcker et al., 2010). Prior to the fusion stage, tethering factors initiate and maintain specific contacts between donor and acceptor membranes to hold the vesicle in close proximity to the target membrane (Whyte and Munro, 2002). Thus, tethering factors play a key role in organizing vesicle trafficking. However, the functions of tethering factors in plant development remain largely unknown.
Eukaryotes contain two broad classes of tethering factors: long coiled-coil proteins and multisubunit tethering complexes (MTCs; Bröcker et al., 2010; Yu and Hughson, 2010; Ravikumar et al., 2017; Takemoto et al., 2018). Coiled-coil tethers are long, dimeric proteins that are primarily found on the Golgi and early endosomes (EEs; Lürick et al., 2018), while MTCs contain several subunits in a modular form and are located on organelles throughout the secretory and endocytic pathways. One well-studied MTC in yeast (Saccharomyces cerevisiae) and animals is the Transport Protein Particle (TRAPP) complex, which is involved in endoplasmic reticulum-to-Golgi and Golgi-mediated trafficking as well as autophagy (Barrowman et al., 2010; Scrivens et al., 2011; Vukašinović and Žárský, 2016). In yeast, mutations in TRAPPs are typically lethal or cause strongly impaired growth (Kim et al., 2016). In humans, mutations in TRAPP components have been implicated in a variety of diseases (Milev et al., 2015; Sacher et al., 2019).
TRAPP complexes can exist in a variety of modular forms (Robinett et al., 2009; Kim et al., 2016; Riedel et al., 2018). In yeast and metazoans, two TRAPP complexes (II and III) composed of shared and complex-specific subunits have been identified (Barrowman et al., 2010; Lynch-Day et al., 2010; Thomas et al., 2018). The metazoan TRAPP complexes contain homologs of all yeast TRAPP subunits but have also evolved additional subunits and rearrangements in complex composition. The TRAPPIII complex contains two additional metazoan-specific subunits, TRAPPC11 and TRAPPC12 (Scrivens et al., 2011; Zhao et al., 2017). Tca17 and Trs65 are TRAPPII-specific subunits in yeast, but their metazoan homologs (TRAPPC2L and TRAPPC13, respectively) are found in TRAPPIII complexes. The subunit composition of TRAPP complexes specifies their localization and function. In yeast, TRAPPII functions in late Golgi, and TRAPPIII functions in early Golgi and autophagy (Kim et al., 2016; Riedel et al., 2018; Thomas et al., 2018). In metazoans, TRAPPII is involved in early Golgi transport, and TRAPPIII regulates endoplasmic reticulum-to-Golgi transport and autophagosome formation (Yamasaki et al., 2009; Scrivens et al., 2011; Bassik et al., 2013; Lamb et al., 2016; Ramírez-Peinado et al., 2017; Sacher et al., 2019). Furthermore, TRAPP complexes function as guanine nucleotide exchange factors for Rab GTPases in both yeast and metazoans (Jenkins et al., 2018; Riedel et al., 2018; Thomas et al., 2018, 2019). In humans, mutations in TRAPP components are associated with numerous diseases such as neurodevelopmental disorders, skeletal disorders, fatty liver disease, and congenital disorders (Larson et al., 2018; Sacher et al., 2019).
Studies of TRAPP complex formation and modularity in plants remain in their infancy compared to those for other kingdoms. It is only recently that the nature of TRAPP complexes (II and III) has been partially elucidated at the protein level (Kalde et al., 2019; Rosquete et al., 2019). Two Arabidopsis (Arabidopsis thaliana) TRAPPII subunits have been characterized to some extent in plants: AtTRS120 and AtTRS130/CLUB (Jaber et al., 2010; Thellmann et al., 2010; Naramoto et al., 2014; Rybak et al., 2014). Both AtTRS120/VAN4 and AtTRS130/CLUB are required for cell plate biogenesis; mutations in AtTRS120 or AtTRS130 cause seedling lethality and canonical cytokinesis-defective phenotypes, including the formation of cell wall stubs and incomplete cross walls (Jaber et al., 2010; Thellmann et al., 2010; Qi et al., 2011). Also, in both mutants, vesicles accumulate at the equators of dividing cells but fail to assemble into cell plates (Jaber et al., 2010; Thellmann et al., 2010; Rybak et al., 2014; Ravikumar et al., 2017). In accordance with TRAPP localization in other organisms, AtTRS120 and AtTRS130 localize at the trans-Golgi network (TGN)/EE (Qi et al., 2011; Naramoto et al., 2014; Ravikumar et al., 2017, 2018). Moreover, TRAPPII in Arabidopsis has been functionally placed upstream of several Rab-A GTPases, suggesting that it may also function as a Rab guanine nucleotide exchange factor in plants as well as yeast and metazoans (Qi et al., 2011; Kalde et al., 2019). AtTRAPPC11/ROG2, an evolutionarily conserved TRAPPIII component, was recently shown to regulate TGN/EE integrity and to play a role in salt stress responses (Rosquete et al., 2019). The Arabidopsis homolog of the yeast/metazoan TRAPP subunit, BET5, is essential for pollen exine pattern formation, suggesting that different components of the TRAPP complexes can have specific functions (Zhang et al., 2018).
Previous sequence analysis and proteomic studies in Arabidopsis have predicted and identified TRAPPII and TRAPPIII components, respectively (Drakakaki et al., 2012; Paul et al., 2014; Rybak et al., 2014; Kalde et al., 2019; Rosquete et al., 2019). However, experimental evidence regarding whether plants form the same number of TRAPP complexes as yeast and metazoans, or have evolved different subunit composition and unique TRAPP components, is currently lacking. To address these questions, we performed quantitative proteomic analysis to document the subunit composition and interactors of TRAPP complexes in Arabidopsis. We identified 11 subunits that are conserved in yeast and metazoans and 2 that are only conserved in metazoans and plants. Additionally, we identified and characterized TRIPP, a TRAPP-interacting plant protein that is present only in multicellular photosynthetic organisms. Genetic evidence shows that TRIPP plays an important role in vesicle trafficking, the polar localization of auxin transporters, plant growth, reproduction, and light-dependent development. Our study demonstrates that Arabidopsis TRAPP components form similar complexes to those reported in metazoans. The study also uncovers a plant-specific TRAPPII interactor that plays important roles in plant growth and development.
RESULTS
AtTRS33 Is Required for the Membrane Association of a TRAPPII-Specific Subunit
To characterize the protein components of TRAPP complexes in plants, we performed quantitative mass spectrometry (MS) analysis of proteins associated with TRS33, a subunit known to be shared by different TRAPP complexes in both yeast and metazoans (Kim et al., 2016; Riedel et al., 2018). We reasoned that an analysis of such a shared subunit would give us a comprehensive view of TRAPP complexes in Arabidopsis. In Arabidopsis, AtTRS33 has been implicated in cytokinesis and was identified in the TRAPPII interactome (Thellmann et al., 2010; Rybak et al., 2014).
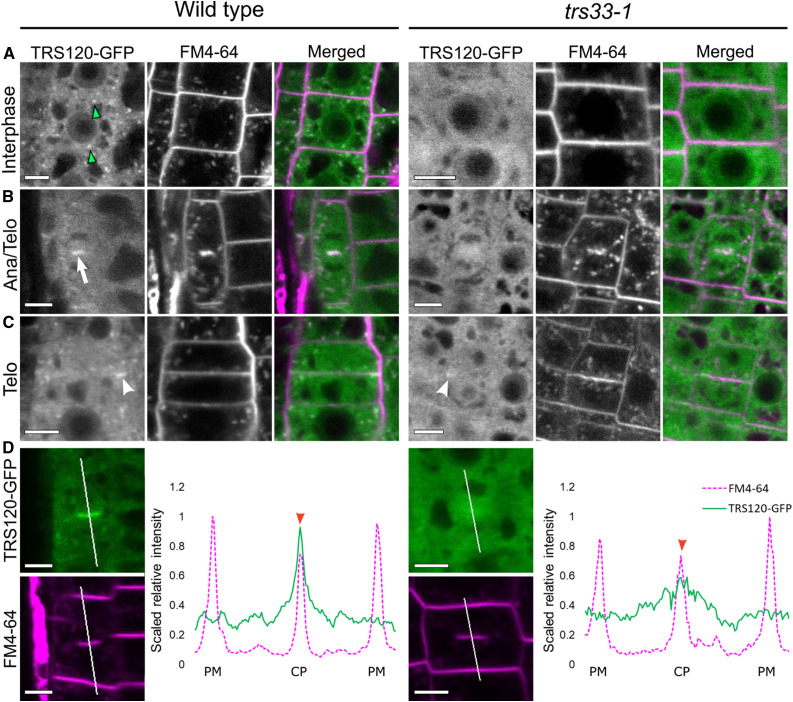
Prior to using TRS33 to further study TRAPP composition, we sought to confirm TRS33’s functional relationship to TRAPPII in Arabidopsis. To this end, we monitored the effect of the trs33-1 mutation on the localization dynamics of the TRAPPII-specific subunit AtTRS120. In the wild type, the TRS120-GFP fusion protein localized to both the cytosol and to TGN/EE compartments in interphase cells (Figure 1A; Rybak et al., 2014; Ravikumar et al., 2018). By contrast, in the trs33-1 mutant, the signal was only observed in the cytosol and not in any endomembrane compartments (Figure 1A). This is reminiscent of the mis-localization of a TRAPPII-specific subunit (Trs130-GFP) to the cytosol in trs33 mutants in yeast (Tokarev et al., 2009). During early stages of cytokinesis, TRS120-GFP clearly localized to the cell plate in the wild type but was present as a diffuse cytosolic cloud around the cell plate in trs33-1 (Figures 1B and 1D). Furthermore, at the end of cytokinesis, TRS120-GFP re-localized to the leading edges of the cell plate in the wild type but was (at best) visible as a weak and relatively diffuse signal at the leading edges of the expanding cell plates in trs33-1 (Figure 1C). Thus, AtTRS33 is required for the membrane association of AtTRS120 and for its proper localization dynamics during cytokinesis. This finding establishes a clear functional link between AtTRS33 and the Arabidopsis TRAPPII complex.
Figure 1.
TRS33 Is Required for Normal Subcellular Localization of TRS120-GFP.
Live imaging of TRS120-GFP (green) and FM4-64 (magenta) in roots of TRS120:TRS120-GFP plants.
(A) Cells at interphase show TRS120-GFP enriched at endomembrane compartments (green arrowheads) in the wild type, but not in trs33-1, where only a cytosolic haze can be seen.
(B) During early stages of cytokinesis (cell plate initiation and biogenesis), TRS120-GFP is enriched at the cell plate (white arrow) in the wild type, but not in trs33-1. Ana, anaphase; Telo, telophase.
(C) During late stages of cytokinesis (cell plate insertion and maturation), TRS120-GFP reorganizes to the leading edges of the cell plate (white arrowhead) in the wild type. By contrast, only a weak and relatively diffuse TRS120-GFP signal can be detected at the leading edges of the cell plate in the trs33-1 mutant (white arrowhead). Telo, telophase.
(D) Line graphs depicting scaled relative fluorescence intensities. A sharp peak is seen at the cell plate (CP) in the wild type (red arrowhead), but not in trs33-1. PM, plasma membrane.
At least 10 seedlings were imaged per marker line. n = 8 for wild type, n = 7 trs33-1 for cytokinetic cells. Bars = 5 μm.
Characterization of TRAPP Complexes Using Quantitative MS
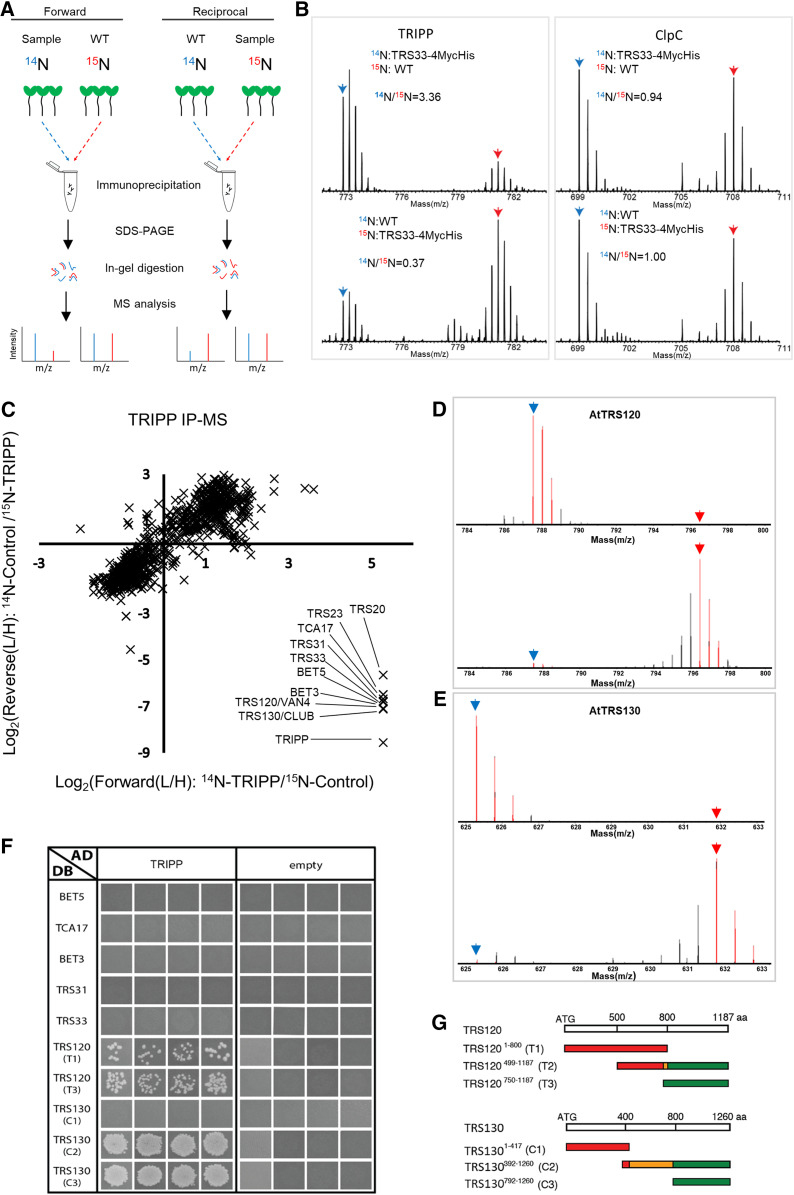
To identify proteins associated with AtTRS33, we used stable isotope labeling followed by immunoprecipitation and quantitative MS (SILIP-MS). We grew transgenic Arabidopsis seedlings expressing AtTRS33 fused with Myc and His tags driven by the native AtTRS33 promoter (TRS33:TRS33-MycHis) in the trs33-1 mutant background for 14 d on medium containing light nitrogen (14N), along with the wild type (as a control) grown on heavy nitrogen (15N). The isotopes were switched in the replicate experiment. Root and shoot tissues were harvested separately. We mixed each pair of 14N- and 15N-labeled sample and control tissues together prior to immunoprecipitation with anti-Myc antibody beads (Figure 2A). We separated immunoprecipitated proteins by SDS-PAGE, subjected them to in-gel digestion, and analyzed them in an Orbitrap mass spectrometer. Enrichment by TRS33 was quantified based on the 14N/15N ratios of the identified peptides. MS analysis identified >1000 proteins per experiment, but only proteins that showed more than twofold enrichment in the TRS33-MycHis samples over the controls in both forward and reciprocal labeling replicates were considered to be TRS33 interactors. We identified fourteen TRS33-interacting proteins in samples from both roots and shoots (Table 1). SILIP-MS using transgenic Arabidopsis plants overexpressing TRS33-yellow fluorescent protein (YFP) also identified the same 14 TRS33-interacting proteins (Supplemental Table 1). These included all homologs of the shared TRAPP subunits reported in yeast and metazoans (TRS23, TRS33, TRS31, BET3, BET5, and TRS20) and the complex-specific subunits (TCA17, TRS120, TRS130, TRS85, TRAPPC11, TRAPC12, and TRAPPC13; Riedel et al., 2018; Sacher et al., 2019). Intriguingly, the identified TRS33-associated proteins included an additional protein with no homolog in yeast or metazoans (Figure 2B; Table 1; Supplemental Figure 1; Supplemental Table 1). This protein, encoded by AT3G17900, is composed of 838 amino acids and is listed as unknown in The Arabidopsis Information Resource database. We named this protein TRAPP Interacting Plant Protein (TRIPP). Taken together, the mass spectrometry results demonstrate that plant TRAPP complexes contain not only homologs of all known TRAPP components in yeast and metazoans but also the plant-specific component TRIPP.
Figure 2.
TRIPP Associates with TRAPPII Components.
(A) Experimental workflow for SILIP-MS in Arabidopsis. m/z, mass-to-charge ratio; WT, wild type.
(B) MS1 spectra show the enrichment of TRIPP in SILIP-MS of TRS33:TRS33-4MycHis/trs33-1 versus the wild-type (Col-0) control, and no enrichment of ClpC, a nonspecific protein. Blue and red arrows point to monoisotopic peaks of 14N- and 15N-labeled peptides, respectively. m/z, mass-to-charge ratio; WT, wild type.
(C) Metabolic SILIP-MS analysis of TRIPP identifies only TRAPPII subunits. SILIP-MS was performed using 35S:TRIPP-YFP/tripp-1 transgenic plants. Plot shows log2 ratios of signal intensities between samples labeled with light (L, 14N) and heavy (H, 15N) isotopes (L/H ratio). Two biological replicates were performed, in which the Col-0 control (forward) or the TRIPP-YFP sample (reciprocal) was labeled with 15N. Note that TRAPP-III subunits are not identified in the data set.
(D) Representative spectra of AtTRS120 peptide quantified in the SILIP-MS experiments of 14N-TRIPP-YFP versus 15N-Col-0 (top) and 14N-Col-0 vs. 15N-TRIP-YFP (bottom). Blue and red arrows point to mono-isotopic peaks of 14N- and 15N-labeled peptide, respectively. m/z, mass-to-charge ratio.
(E) Representative spectra of AtTRS130 peptide quantified in the TRIPP-YFP SILIP-MS experiments as described for (A). m/z, mass-to-charge ratio.
(F) Y2H assays of interactions between TRIPP and TRAPP subunits. The panels are from different plates. Four independent replicate experiments were performed. The results show interactions of TRIPP with both the T1 and T3 regions of AtTRS120 and with the plant-specific C2/C3_DB AtTRS130 regions. T2_DB was not included, as it is an auto-activator, as evidenced by colony growth with the empty AD vector, and this precluded our ability to determine whether AtTRS120_T2 interacts with TRIPP.
(G) TRAPPII coding regions used for the Y2H interaction assays. Segments colored in red (left) are conserved across kingdoms, while those in green (right) are plant specific. The orange moiety of the C2 segment is poorly conserved across kingdoms. The T2 middle segment corresponds to sequences found to interact with the exocyst in a Y2H screen (Rybak et al., 2014). aa, amino acids.
Table 1. The 14 Arabidopsis TRAPP components identified by SILIP-MS.
| Gene ID | TRAPP Subunits Identified by TRS33 Immunoprecipitation from Arabidopsis Roots | Yeast Homolog | Human Homolog | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Forward | Reciprocal | Protein Name | ||||||||
| Peptide No. | % Cov | L/H Ratio | Peptide No. | % Cov | L/H Ratio | |||||
| AT1G51160 | 5 | 41.4 | 4.47 | 4 | 28.4 | 0.41 | BET5 | BET5 | TRAPPC1 | |
| AT1G80500 | 3 | 28.9 | 4.13 | 3 | 28.9 | 0.29 | TRS20 | TRS20 | TRAPPC2 | |
| AT2G20930 | 6 | 52.9 | 3.79 | 5 | 52.9 | 0.39 | TCA17 | TCA17 | TRAPPC2L | |
| AT5G54750 | 12 | 55.4 | 11.4 | 12 | 54.3 | 0.18 | BET3a,b | BET3 | TRAPPC3 | |
| AT5G02280 | 5 | 42.6 | 2.72 | — | — | — | TRS23 | TRS23 | TRAPPC4 | |
| AT5G58030 | 5 | 22.6 | 2.48 | 2 | 10.3 | 0.37 | TRS31a,b | TRS31 | TRAPPC5 | |
| AT3G05000 | 11 | 61.3 | 7.7 | 11 | 60.7 | 0.07 | TRS33a,b | TRS33 | TRAPPC6 | |
| AT5G16280 | 16 | 11.7 | 7.27 | 13 | 9.5 | 0.15 | TRS85a | TRS85 | TRAPPC8 | |
| AT5G11040 | 32 | 32.7 | 3.62 | 23 | 25.6 | 0.40 | TRS120/VAN4a,b | TRS120 | TRAPPC9 | |
| AT5G54440 | 33 | 31 | 4.19 | 13 | 12.3 | 0.44 | TRS130/CLUBb | TRS130 | TRAPP10 | |
| AT5G65950 | 20 | 17.6 | 7.76 | 12 | 11.2 | 0.18 | TRAPPC11 | —- | TRAPPC11 | |
| AT4G39820 | 15 | 35.8 | 8.06 | 8 | 21.3 | 0.24 | TRAPPC12 | — | TRAPPC12 | |
| AT2G47960 | 14 | 37.1 | 2.84 | 14 | 33.3 | 0.52 | TRS65 | TRS65 | TRAPPC13 | |
| AT3G17900 | 22 | 30.4 | 3.54 | 19 | 24.8 | 0.39 | TRIPP | — | — | |
| TRAPP Subunits Identified by TRS33 Immunoprecipitation from Arabidopsis Shoots | ||||||||||
| AT1G51160 | 4 | 34.9 | 14.7 | 2 | 14.2 | 0.16 | BET5 | BET5 | TRAPPC1 | |
| AT1G80500 | 1 | 8.9 | 12.9 | 2 | 20 | 0.10 | TRS20 | TRS20 | TRAPPC2 | |
| AT2G20930 | 6 | 63.6 | 3.9 | 4 | 36.4 | 0.36 | TCA17 | TCA17 | TRAPPC2L | |
| AT5G54750 | 12 | 55.4 | 30.2 | 11 | 46.8 | 0.07 | BET3a,b | BET3 | TRAPPC3 | |
| AT5G02280 | 6 | 51.8 | 7.1 | — | — | — | TRS23 | TRS23 | TRAPPC4 | |
| AT5G58030 | 4 | 29.2 | 11.9 | 6 | 29.7 | 0.14 | TRS31a,b | TRS31 | TRAPPC5 | |
| AT3G05000 | 12 | 74.0 | 37.9 | 8 | 52.6 | 0.04 | TRS33a,b | TRS33 | TRAPPC6 | |
| AT5G16280 | 16 | 12.7 | 10.3 | 13 | 8.6 | 0.12 | TRS85a | TRS85 | TRAPPC8 | |
| AT5G11040 | 24 | 22.5 | 5.5 | 19 | 18.6 | 0.23 | TRS120/VAN4a,b | TRS120 | TRAPPC9 | |
| AT5G54440 | 23 | 20.2 | 4.7 | 18 | 17.1 | 0.27 | TRS130/CLUBb | TRS130 | TRAPP10 | |
| AT5G65950 | 27 | 22.8 | 12.8 | 13 | 12.4 | 0.12 | TRAPPC11 | — | TRAPPC11 | |
| AT4G39820 | 9 | 24.3 | 15.3 | 10 | 24.3 | 0.11 | TRAPPC12 | — | TRAPPC12 | |
| AT2G47960 | 14 | 32.4 | 5.2 | 12 | 30.5 | 0.26 | TRS65 | TRS65 | TRAPPC13 | |
| AT3G17900 | 19 | 28.6 | 3.8 | 17 | 21.1 | 0.32 | TRIPP | — | — | |
SILIP-MS was performed using TRS33:TRS33-4MycHis/trs33-1 complemented plants (roots and shoots were analyzed separately). The number of unique peptides (Peptide No.), percentage of sequence coverage (% Cov), and ratio of signal intensities between samples labeled with light (L, 14N) and heavy (H, 15N) isotopes (L/H ratio) are shown for two biological replicates, in which the Col-0 control (forward) or the TRS33-MycHis sample (reciprocal) was labeled with 15N. Dashes indicate no data.
Subunits previously identified in a proteomic study using isolated TGN compartments (Drakakaki et al., 2012).
Subunits previously identified by label-free IP-MS of AtTRS130 (Rybak et al., 2014). Names of yeast and human homologs are based on published work (Riedel et al., 2018; Thomas et al., 2018).
TRIPP Behaves Like a Component of the TRAPP II Complex in SILIP-MS Experiments
To confirm the TRIPP association with TRAPP components and to determine whether it is specific to a given TRAPP complex, we performed an additional SILIP-MS using TRIPP as bait. We grew transgenic plants expressing TRIPP-YFP under the control of the 35S promoter, which complemented the tripp-1 mutant, along with nontransgenic wild type as a control, as described above for the TRS33 SILIP-MS experiment. We immunoprecipitated TRIPP-YFP and associated proteins with an anti-GFP antibody and analyzed them by MS (Figure 2A). The SILIP-MS data show unambiguously that TRIPP pulled down nine TRAPP subunits (Figure 2C; Supplemental Table 2). These include all six shared TRAPP subunits (TRS23, TRS33, TRS31, BET3, BET5, and TRS20) and three TRAPPII-specific subunits (TCA17, TRS120, and TRS130; Figures 2C to 2E; Supplemental Table 2). Four TRAPP subunits (TRS85, TRAPPC11, TRAPPC12, and TRAPPC13) identified in the TRS33 SILIP-MS showed no interactions with TRIPP in these SILIP-MS experiments. None of these four proteins were detected in both replicate experiments or were enriched by TRIPP-YFP in either single experiment. Interestingly, these proteins are homologs of the subunits specific to TRAPPIII in animals, suggesting that Arabidopsis also possesses distinct TRAPPII and TRAPPIII complexes and that TRIPP only associates with the TRAPPII complex.
TRIPP Directly Interacts with TRAPPII-Specific, but Not Shared, Subunits
If TRIPP associates specifically with the TRAPPII complex, we reasoned that it likely interacts directly with TRAPPII-specific subunits instead of the shared core subunits. To investigate this possibility, we performed yeast two-hybrid (Y2H) assays of binary TRIPP interactions with known TRAPP subunits (Thellmann et al., 2010; Steiner et al., 2016b; Kalde et al., 2019). In quadruplicate pairwise tests, TRIPP did not interact with the shared TRAPP subunits, including AtTRS33, instead interacting specifically with the two TRAPPII-specific subunits, AtTRS120 and AtTRS130 (Figure 2F). Notably, strong interactions were detected with the C-terminal part of AtTRS130 and both halves of AtTRS120 (Figures 2F and 2G). These results suggest that TRIPP associates with TRAPPII through direct interactions with these two TRAPPII-specific subunits.
TRIPP Localizes to the TGN/EE in Arabidopsis Roots
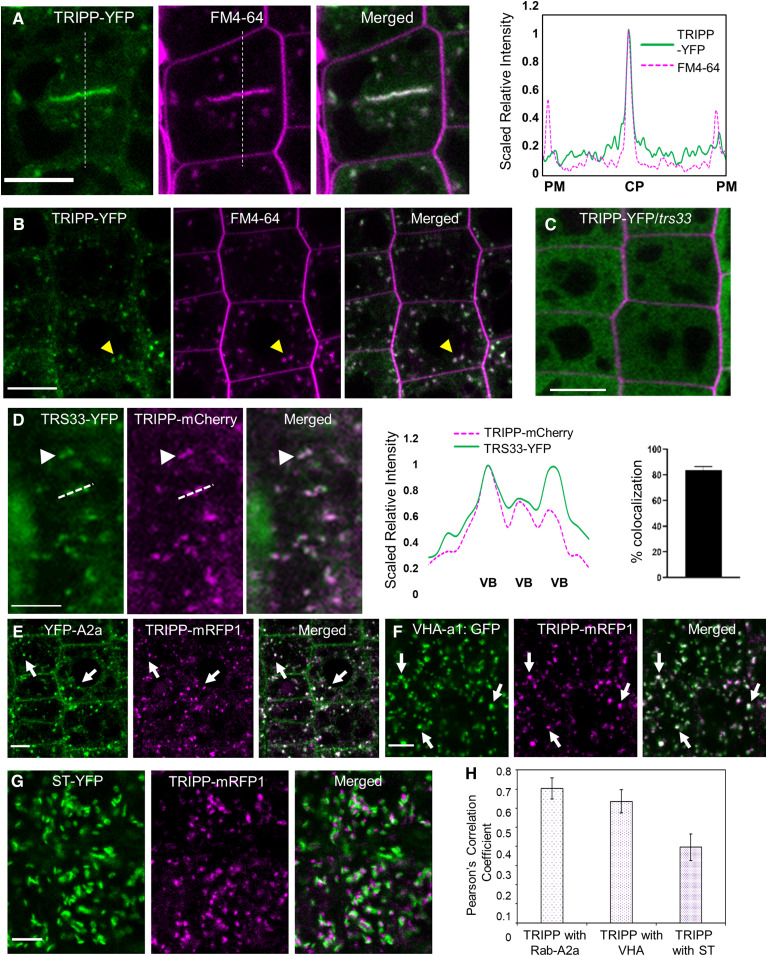
TRAPPII-specific subunits AtTRS120 and AtTRS130 localized to the TGN/EE and the cell plate (Ravikumar et al., 2018). To examine the subcellular localization of TRIPP, we generated transgenic plants expressing TRIPP-YFP under the control of the 35S promoter in the tripp-1 null mutant background (the mutant is described below). We analyzed the roots of transgenic seedlings expressing TRIPP-YFP by confocal imaging. TRIPP-YFP localized to puncta as well as to the cell plate, with YFP signals overlapping with FM4-64 signals (Figures 3A and 3B). The localization of TRIPP at the cell plate during early and late cytokinesis was confirmed by colocalization analysis of stable transgenic plants expressing TRIPP-mCherry/monomeric red fluorescent protein (mRFP) with known cell plate–localized proteins TRS120-GFP or YFP-RAB-A2a (Supplemental Figures 2A to 2C). The localization of TRIPP to the leading edges of cell plates was similar to that of other TRAPPII subunits (Rybak et al., 2014; Kalde et al., 2019). TRS33 exhibited a punctate localization similar to that of TRIPP (Supplemental Figure 2D). In root cells of plants expressing 35S:TRIPP-mCherry and 35S:TRS33-YFP, all TRIPP-mCherry compartments were positive for TRS33-YFP, whereas ∼20% of the TRS33-YFP–positive compartments showed no TRIPP-mCherry signals (Figure 3D). This pattern is consistent with the notion that TRIPP is a component of only the TRAPPII complex and that TRS33 is a component of both the TRAPPII and TRAPPIII complexes. Furthermore, the localization of TRIPP-YFP was disrupted in the trs33-1 mutant background, which is consistent with the finding that TRIPP is a TRAPP-interacting component (Figure 3C).
Figure 3.
TRIPP Localizes to Vesicles and Cell Plates, Like Previously Reported TRAPP Components, and Its Localization Is Disrupted in trs33-1.
(A) and (B) Confocal images of 35S:TRIPP-YFP/tripp-1 transgenic roots. (A) TRIPP-YFP (green) localizes with FM4-64 (magenta) at the cell plate (CP). Dashed lines show the position of the intensity plot. PM, plasma membrane. (B) Colocalization of TRIPP-YFP with FM4-64–positive TGN/endocytic vesicles. n = 7 seedlings. Arrows point to vesicles. Bars = 10 µm.
(C) TRIPP-YFP localization is disrupted in the trs33-1 mutant background. Root cell walls were stained with propidium iodide. n = 10 seedlings. Bar = 10 µm.
(D) Roots of plants transformed with both 35S:TRS33-YFP and 35S:TRIPP-mCherry show that TRIPP colocalized (white arrowhead) with TRS33-YFP in some, but not all, of the vesicles that TRS33-YFP localizes to. Dashed lines in the image show the position of the quantification graph in the right panel. VB, vesicle bodies. Bar graph shows percent colocalization of TRIPP-mCherry to TRS33-YFP–containing compartments; error bars represent se. n = 7. Bar = 5 µm.
(E) Colocalization of TRIPP-mRFP1 with TGN/EE-localized YFP-RAB-A2a. Arrow points to representative signal overlap. Bar = 5 µm.
(F) Colocalization of TRIPP-mRFP1 with TGN/EE-localized VHA-a1-GFP. Arrow points to representative signal overlap. Bar = 5 µm.
(G) Colocalization of TRIPP-mRFP1 with Golgi-localized ST-YFP. Arrow points to representative signal overlap. Bar = 5 µm.
(H) Pearson’s correlation coefficients between TRIPP-mRFP1 and TGN/EE-localized YFP-RAB-A2a (n = 40 cells), VHA-a1-GFP (n = 49 cells), and Golgi-localized ST-YFP (n = 42 cells). Error bars represent se.
As TRAPPII has been functionally linked to Rab-A GTPases and localizes to the TGN/EE in Arabidopsis (Qi and Zheng 2011; Qi et al., 2011; Naramoto et al., 2014; Kalde et al., 2019), we performed colocalization experiments to define the TRIPP-positive compartments. We crossed pUBQ10-TRIPP-mRFP transgenic Arabidopsis plants with transgenic plants expressing TGN/EE-localized YFP-RAB-A2a (Chow et al., 2008) and vacuolar H+-ATPase (VHA-a1)-GFP (Dettmer et al., 2006) and Golgi-localized α-2,6-sialyltransferase (ST)-YFP (Batoko et al., 2000; Grebe et al., 2003) markers. TRIPP-mRFP colocalized with TGN/EE-localized YFP-RAB-A2a and VHA-a1-GFP, but not with Golgi-localized ST-YFP (Figures 3E to 3H). TRIPP-mRFP puncta were frequently associated (but not overlapping) with ST-YFP, which is similar to the localization observed for RAB-A1c, another TGN/EE marker (Qi and Zheng, 2013). Taken together, these data provide in vivo evidence that TRIPP localizes to the TGN/EE, as expected for a component of the TRAPPII complex (Qi et al., 2011; Naramoto et al., 2014; Ravikumaret al., 2018).
The Arabidopsis tripp Mutant Displays Growth Defects Including Partial Photomorphogenesis in the Dark
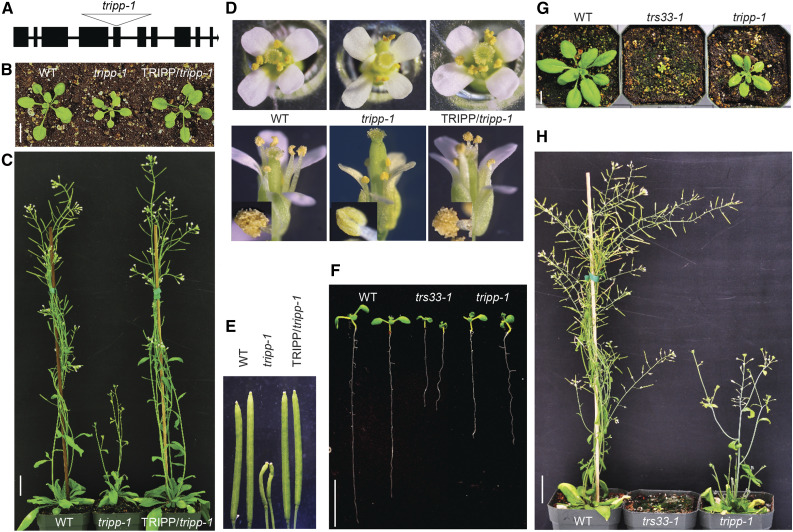
To explore the role of TRIPP in plant development, we performed phenotypic characterization of tripp-1, a T-DNA insertion allele harboring T-DNA in the fifth exon of the TRIPP gene, which contains 10 exons, rendering tripp-1 a null mutant (Figure 4A; Supplemental Figures 3A and 3B). tripp-1 homozygous plants exhibited smaller rosettes than the wild type and were sterile (Figures 4B to 4E). Plants heterozygous for the T-DNA insertion did not exhibit a mutant phenotype; thus, we maintained the tripp-1 mutation in heterozygous plants. To confirm that the phenotypes observed were due to the specific mutation in the TRIPP gene, we introduced a transgene containing the 35S promoter driving the full-length TRIPP coding sequence fused to YFP into TRIPP +/− heterozygous plants via Agrobacterium-mediated transformation. In plants expressing TRIPP-YFP in the tripp-1 homozygous background, the phenotypes observed in the tripp-1 mutant were fully rescued (Figures 4B to 4E), confirming that the observed phenotypes were due to loss of function of TRIPP. Both the mutant and the wild-type plants bolted at similar times, but the inflorescences were much shorter in the mutant (Figure 4C). The floral organs appeared normal in the mutant, except that the anthers failed to develop pollen and thus no seeds were produced (Figures 4D and 4E). tripp-1 anthers had a smooth surface, suggesting that the defects in pollen develop occurred during early stages of anther development.
Figure 4.
tripp Mutant Exhibits Dwarfism and Sterility.
(A) Gene model of Arabidopsis TRIPP/AT3G17900 showing that the T-DNA insertion (Salkseq_063596) of tripp-1 is located in the fifth exon.
(B) Two-week-old tripp-1 plants grown under a 16-h-light/8-h-dark cycle show smaller rosettes than the wild-type (WT) and 35S:TRIPP-YFP/tripp-1 plants (TRIPP-YFP/tripp-1). Bar = 1 cm.
(C) Five-week-old tripp-1 plants grown under a 16-h-light/8-h-dark cycle exhibit significantly shorter inflorescences than the wild-type (WT) and TRIPP-YFP/tripp-1 plants. Bar = 1 inch.
(D) tripp-1 flowers appear normal, but their anthers lack pollen. Insets in the bottom panel show magnified images of anthers. WT, wild type.
(E) Morphology of mature siliques from 7-week-old tripp-1 plants compared to the wild type (WT) and complemented plants.
(F) and (G) tripp-1 plants show shorter roots (F) and stunted growth (G) than the wild type (WT) but weaker phenotypes than trs33-1. Bar = 1 cm.
(H) Both the tripp-1 and trs33-1 mutants exhibit complete sterility at 6 weeks old. Bar = 1 inch.
To determine whether the tripp-1 mutant has any defects in the female gametophyte, we used the wild-type pollen to pollinate 20 flowers from multiple tripp-1 plants. Although the mutant stigmas appeared normal before pollination (as shown for tripp-1 in Figure 4D), flowers pollinated with the wild-type pollen did not produce any seeds (the morphology of mature siliques from the crosses was similar to that shown for tripp-1 in Figure 4E), suggesting that tripp-1 has defects in both male and female reproductive development. Overall, the tripp-1 mutant exhibited similar sterility but weaker dwarfism phenotypes than trs33-1 (Figures 4F to 4H) and trs120-4 (Thellmann et al., 2010). Taken together, our genetic analyses indicated that TRIPP has important functions throughout plant growth and development, particularly reproductive development.
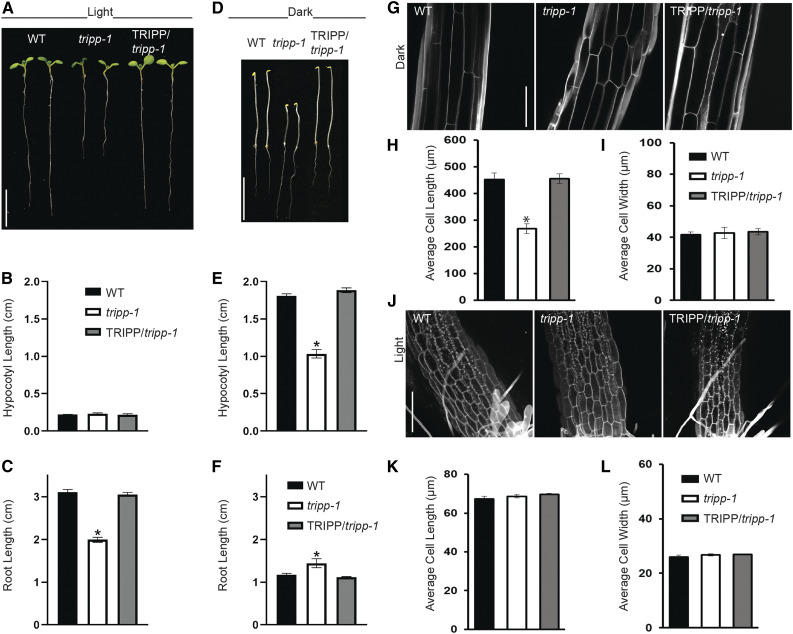
The dwarf phenotype of light-grown tripp-1 plants prompted us to compare growth and cell elongation in this mutant in the dark versus light (skotomorphogenesis versus photomorphogenesis). When seeds were sown on medium in the light, tripp-1 and the wild-type seeds germinated at the same time. While hypocotyl length was similar in tripp-1 and the wild-type seedlings, the tripp-1 mutant had shorter roots (Figures 5A to 5C). When grown in the dark, the tripp-1 mutant had shorter hypocotyls but longer roots than the wild type (Figures 5D to 5F). Furthermore, when seedlings were grown in the dark for 2 weeks, the tripp-1 mutant developed true leaves, while the wild type did not (Supplemental Figure 4). Dark-grown tripp-1 seedlings exhibited much shorter hypocotyl cells than the wild type, whereas light-grown tripp-1 seedlings showed similar cell size to the wild type (Figures 5G to 5L). These observations indicate that TRIPP plays an important role in skotomorphogenesis.
Figure 5.
tripp-1 Mutant Exhibits Defects in Photomorphogenic Development.
(A) Seven-day-old tripp-1 seedlings grown in 16-h-light/8-h-dark cycle show normal hypocotyl length but shorter roots than the wild type (WT). Bar = 1 cm.
(B) and (C) Quantification of hypocotyl (B) and root (C) lengths in 7-d-old light-grown (16-h-light/8-h-dark) seedlings (n = 10). Data reported as means ± se. *, P < 0.01 compared to the WT control (two-tailed t test).
(D) Five-day-old tripp-1 seedlings grown in the dark show shorter hypocotyls and longer roots than the wild type (WT). Bar = 1 cm.
(E) and (F) Quantification of hypocotyl (E) and root (F) lengths in 5-d-old dark-grown seedlings (n = 10). Data reported as means ± se. *, P < 0.01 compared to the wild-type (WT) control (two-tailed t test).
(G) Hypocotyls of 5-d-old dark-grown tripp-1 seedlings stained with propidium iodide show impaired cell elongation. Imaged at the hypocotyl elongation zone (lower section of the hypocotyl). Bar = 150 µm.
(H) and (I) Quantification of hypocotyl (H) and root (I) lengths of 5-d-old dark-grown seedlings (n = 6 roots, 50 cells per root). Data reported as average cell length and cell width ± se. *, P < 0.01 compared to the wild-type (WT) control (two-tailed t test).
(J) Hypocotyls of 7-d-old light-grown seedlings stained with PI show similar cell size between tripp-1 and the wild type (WT). Bar = 150 µm. The hypocotyl–root junction is shown at the bottom section of each image (some roots hairs visible). Bar = 150 µm.
(K) and (L) Quantification of hypocotyl (K) and root (L) lengths in 7-d-old light-grown seedlings (n = 6 roots, 50 cells per root). Data reported as average cell length and cell width ± se. *, P < 0.01 compared to the wild-type (WT) control (two-tailed t test).
The tripp-1 Mutant Has a Defect in Polar Localization of the Auxin Transporter PIN2
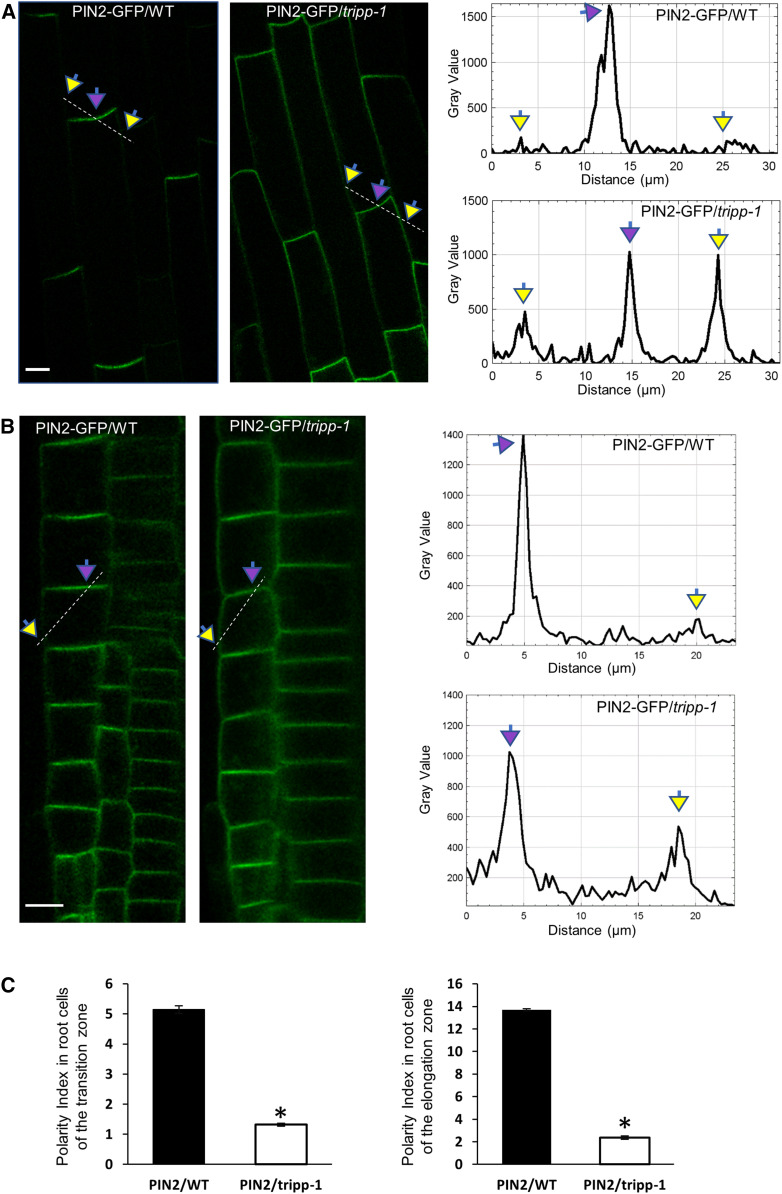
The observation that tripp-1 exhibited a photomorphogenesis-like phenotype in the dark suggested it might have a defect in hormonal function (Chaiwanon et al., 2016). In particular, TRAPP is required for the polar localization of auxin transporters (Qi et al., 2011; Rybak et al., 2014; Zhang et al., 2018), which mediate light-dependent regulation of auxin distribution leading to photomorphogenesis (Sassi et al., 2012). We therefore crossed 35S-PIN2-GFP and 35S-PIN3-GFP into the tripp-1 mutant background. In the wild type, PIN2 was polarly localized to the basipetal plasma membrane and was depleted at the lateral plasma membrane in elongated root cells. Such polar localization was greatly diminished in tripp-1, as PIN2-GFP signals remained strong in the lateral plasma membrane (Figures 6A to 6C; Supplemental Figure 5A). By contrast, PIN3 localization was not altered in the tripp-1 mutant compared to the wild type (Supplemental Figure 5B).
Figure 6.
PIN2 Polarity Is Altered in the tripp-1 Mutant.
Confocal images of 7-d-old seedlings. Apical and lateral membranes are marked by purple and yellow arrows, respectively.
(A) Cells in the root elongation zones of the wild-type (WT) and tripp-1 seedlings expressing 35S:PIN2-GFP. PIN2 exhibits apical localization on the plasma membrane in WT. PIN2 exhibits apical and lateral plasma membrane localization in tripp-1. Dashed lines correspond to the area of quantification of PIN2-GFP signals across the apical and lateral plasma membrane. n = 10. Bar = 10 µm.
(B) Cells in the root transition zones of the wild-type (WT) and tripp-1 seedlings expressing 35S:PIN2-GFP. PIN2 exhibits apical localization on the plasma membrane in WT. PIN2 exhibits apical and lateral plasma membrane localization in tripp-1. Dashed lines correspond to the area of quantification of PIN2-GFP signals across the apical and lateral plasma membrane. n = 10. Bar = 10 µm.
(C) Quantitative polarity index (Łangowski et al., 2016) of PIN2-GFP in the wild type (WT) and tripp-1, representing the ratio of the mean signal in the polar membrane to that in the lateral membrane. n = 5 seedlings. Data reported as means ± se. *, P < 0.001 (two-tailed t test).
The tripp-1 Mutant Shows Defects in TRS120 Localization and Cross Wall Formation
We crossed TRS120-GFP into tripp-1 to investigate whether TRIPP is required for the normal localization of TRS120. In the wild-type background, TRS120-GFP localized to puncta in epidermal cells and to the cell plates of dividing cells. In the tripp-1 background, no obvious TRS120-GFP puncta were observed (Figure 7A). While patchy localization of TRS120-GFP to the cell plate was observed in some cells, in some other dividing cells, TRS120-GFP only formed a diffuse cloud around the cell plate (Figures 7C and 7D; Supplemental Figure 6). Immunoblot analysis indicated that TRS120-GFP remained exclusively as a full-length fusion protein in both the wild type and tripp-1 (Figure 7B). In contrast to TRS120-GFP, the localization of YFP-RAB-A2a to the cell plate was not obviously altered in tripp-1 (Supplemental Figure 7), unlike in the trappii mutants (Kalde et a., 2019). While examining TRS120-GFP localization in tripp-1, we noticed cells with an incomplete transverse cell wall. Upon further analysis of tripp-1 and the wild-type plants, we observed 6.5% ± 0.5% of such defective cells per root tip in tripp-1 and no defective cell walls in the wild-type roots (Figures 7E and 7F). Such incomplete transverse cell walls, which have been reported for trappii mutants (Jaber et al., 2010; Thellmann et al., 2010; Rybak et al., 2014), indicate that TRIPP plays an important role as a TRAPPII component in cross wall formation.
Figure 7.
TRIPP Is Required for the Membrane Association of a TRAPPII-Specific Subunit.
(A) pTRS120:TRS120-GFP transgenic plants showing that TRS120 localizes to intracellular compartments in the wild type (WT), but not in tripp-1. n = 5 seedlings. Bar = 10 µm.
(B) Immunoblot analysis of pTRS120:TRS120-GFP transgenic plants in the wild-type (WT) and tripp-1 background using anti-GFP antibody. Ponceau S staining was used as the loading control.
(C) Root cells of the wild-type (WT) and tripp-1 plants expressing pTRS120:TRS120-GFP. TRS120-GFP localizes to the cell plate in the WT, but not in tripp-1. n = 6 seedlings. TRS120-GFP localized to the cell plate in all WT cells (n = 20 cytokinetic cells) but in only 12% of tripp-1 cells (n = 8 cytokinetic cells). Bar = 5 µm.
(D) TRS120-GFP localizes to the leading edges (white arrows) of expanding cell plates in the wild type (WT), whereas a diffuse cytosolic cloud is detected in the tripp-1 background. n = 6 seedlings. Bar = 5 µm.
(E) Incomplete cross walls are observed in tripp-1 root cells (6.5% ± 0.5% incidence in root tips). n = 9 seedlings. Bar = 10 µm.
(F) Representative three-dimensional rendering of incomplete cross walls in two adjacent cells of the tripp-1 mutant. n = 9 seedlings. Inset shows two cells (1 and 2) selected for 3D rendering. Bar = 10 µm.
TRIPP Is Present throughout the Green Lineage Except in Chlorophytes
Sequence analysis indicated that TRIPP is present in the green lineage from multicellular algae to flowering plants (Supplemental Figure 8A). TRIPP homologs are found as far as the charophycean algae, which are multicellular photosynthetic organisms ancestral to land plants, but surprisingly, they are not present in unicellular photosynthetic chlorophytes such as Chlamydomonas. This is in contrast to other TRAPP components, such as the shared TRAPP subunit TRS33, which is present from chlorophytes to embryophytes and across kingdoms (Supplemental Figure 8B; Kim et al., 2016). Sequence alignment of TRIPP protein sequences indicated that TRIPP homologs are highly conserved (Supplemental Figure 8C). These findings suggest that TRIPP might have evolved early in the streptophytes in parallel to the emergence of multicellularity in plants.
DISCUSSION
The endomembrane system is crucial for cellular organization and morphogenesis and is regulated by complex networks of proteins and protein complexes in eukaryotes. The TRAPP complexes have been extensively characterized in yeast and metazoans and have been shown genetically to play important roles in the endomembrane system in plants; yet, the protein components of TRAPP complexes and their functions in growth and development in plants had been unclear. Our quantitative immunoprecipitation (IP)-MS experiments demonstrated that in Arabidopsis, TRAPP complexes contain homologs of all 13 mammalian TRAPP subunits. In addition, the TRAPPII complex associates with an additional plant-specific component, TRIPP. We further showed that TRIPP plays important roles in trafficking-dependent processes, including the polar localization of auxin transporters and the formation of new cell walls, as well as a range of developmental processes including photomorphogenesis and reproductive development.
Arabidopsis TRAPP Complexes Include 14 Subunits
All previous proteomic studies combined identified 13 evolutionarily conserved TRAPP components in Arabidopsis, which is similar to the 13 subunits identified in animals (Drakakaki et al., 2012; Rybak et al., 2014, Kalde et al., 2019; Rosquete et al., 2019). In metazoans, the TRAPPII complex is composed of seven shared subunits and three TRAPPII-specific subunits (Figure 8; Scrivens et al., 2011; Riedel et al., 2018; Sacher et al., 2019), while TRAPPIII contains the same shared subunits as TRAPPII plus four TRAPPIII-specific subunits, including two metazoan-specific subunits not found in yeast (Figure 8; Scrivens et al., 2011; Zhao et al., 2017). TRS33 is a shared TRAPP subunit in yeast and animals, and in Arabidopsis our initial data indicated that AtTRS33 is required for the membrane association of TRS120 (a component of TRAPPII) and for its localization dynamics during cytokinesis (Figure 1). This observation, combined with our earlier finding that trs33-1 mutants are, like trappii mutants, impaired in cytokinesis (Thellmann et al., 2010), indicates that AtTRS33 functions as a key component of TRAPP complexes in Arabidopsis.
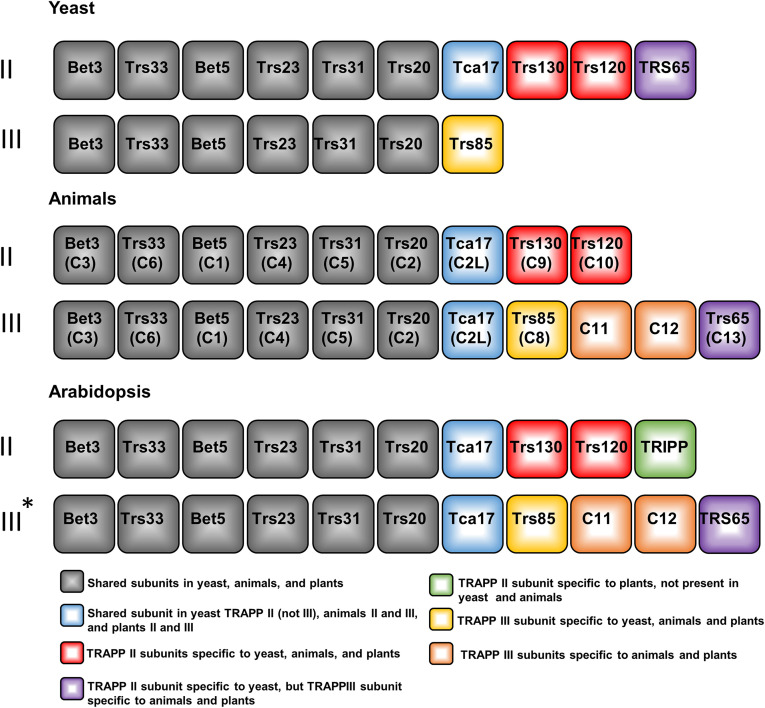
Figure 8.
Subunit Compositions of TRAPP Complexes in Arabidopsis and Animals Are Similar.
Overview of the shared and specific TRAPPII and TRAPPIII subunits in different kingdoms, with this report supporting the similar TRAPP complex composition in plants. For simplicity, original yeast subunit nomenclature is used, and the names in parentheses are the names of animal homologs for each subunit. Animal-specific names are listed if the subunits are not present in yeast. Yeast and animal TRAPPII and TRAPPIII complexes were well defined in previous studies (Riedel et al., 2018; Thomas et al., 2018; reviewed by Kim et al., 2016; Sacher et al., 2019). TRAPPII composition in plants was determined from the TRIPP IP-MS results presented in this study (Figure 2C; Supplemental Table 2). The asterisk (*) represents suggested TRAPPIII composition in plants based on the observation that AtTRS33 pulled down all TRAPP subunits (Table 1), whereas TRIPP pulled down only the TRAPPII complex, but not TRAPPIII-specific subunits (Figure 2; Supplemental Table 2), using the animal homologous TRAPPIII complex as a reference.
Our quantitative IP-MS analysis of AtTRS33 identified 14 TRAPP-interacting proteins, including homologs of all known TRAPP components in yeast and animals, as well as the plant-specific protein TRIPP. We performed multiple SILIP-MS experiments, each including two replicates, using shoot and root tissues of transgenic plants expressing TRS33 driven by the native TRS33 promoter or the 35S promoter, as well as different affinity tags. Although more than 1000 proteins were detected by MS, accurate quantification enabled by stable isotope labeling identified only the TRAPP subunit homologs and TRIPP as TRS33-associated proteins. No other proteins showed more than twofold enrichment by IP-MS of TRS33 in both replicate experiments, and no Rab GTPases were enriched in even a single experiment. It is likely that our stringent washing conditions prevented us from detecting such possible dynamic interactors. This observation suggests that TRIPP is a relatively stable component of the TRAPP complex. As such, our high-quality SILIP-MS data identified (with high confidence) a more complete set of TRAPP complex components, some of which were not known to be present in plants. These proteins include homologs of all yeast and metazoan TRAPP subunits as well as TRIPP, a TRAPP component that has homologs only in multicellular photosynthetic organisms.
TRIPP Is a Plant-Specific TRAPPII-Associated Component
Studies of TRAPP proteins in different organisms have shown that animals possess two additional TRAPP subunits (TRAPPC11 and TRAPPC12) compared to yeast (Kim et al., 2016; Riedel et al., 2018). This suggests that multicellular organisms might have evolved a more complex regulation of, or novel functions for, TRAPP components. The human TRAPPC12, for example, has been implicated in kinetochore assembly and mitosis through associations with chromosomes (Milev et al., 2015). Interestingly, among the 14 Arabidopsis TRAPP interactors, TRIPP was found to be specific to plants and is currently classified as an unknown protein in plant databases. Protein sequence analysis and database mining indicated that, based on protein homology, TRIPP is specific to multicellular photosynthetic organisms (Supplemental Figure 8A).
Our biochemical and genetic data strongly support the notion that TRIPP is specifically associated with TRAPPII, a complex involved in post-Golgi membrane trafficking (Ravikumar et al., 2018). While our SILIP-MS analysis of the AtTRS33 interactome identified 14 subunits, the TRIPP SILIP-MS identified only 10 subunits, which include the specific subunits of TRAPPII but exclude those of TRAPPIII. The presence of the homologs of the four animal TRAPPIII-specific subunits (TRAPPC8, TRAPPC11, TRAPPC12, and TRAPPC13) in the AtTRS33 interactome, but not the TRIPP interactome, suggests the existence of both a TRIPP-containing TRAPPII complex and a complex similar to TRAPPIII that excludes TRIPP. Our Y2H data indicate that TRIPP directly interacts with the TRAPPII-specific subunits TRS120 and TRS130, further corroborating the association of TRIPP with the TRAPPII complex. Consistent with these molecular interactions, TRIPP colocalizes with AtTRS33 at some endosomal compartments, presumably in the TRAPPII complex, but it is absent at a subset of AtTRS33-containing endosomal compartments, which are likely TRAPPIII specific (Figure 3D). These results demonstrate that TRIPP is a specific component of TRAPPII.
The TRAPPII complex is well conserved in yeast and animals, except for the TRS65 subunit, which in yeast associates with TRAPPII and in animals with TRAPPIII (Choi et al., 2011). Here, we demonstrated that TRAPPII is also conserved in plants, but as in animal models, TRS65 does not appear to associate with TRAPPII. Recent IP-MS proteomic studies using AtTRAPPC11 (a TRAPPIII subunit) or Rab GTPases as baits also identified AtTRS65 as being a TRAPPIII component (Kalde et al., 2019; Rosquete et al., 2019), suggesting that plant TRAPP complexes are more similar to metazoan than to yeast complexes. Our results demonstrate that animals and plants share previously known TRAPPII subunits but that multicellular plants have acquired TRIPP as an additional plant-specific component of the TRAPPII complex (Figure 8).
TRAPPII is known to associate with the TGN/EE, which is a hub for membrane trafficking, including exocytosis, endocytosis, and protein sorting (Dettmer et al., 2006; Chow et al., 2008; Qi et al., 2011; Naramoto et al., 2014; Ravikumar et al., 2018). Arabidopsis TRAPPII subunits TRS120 and TRS130 localize to the TGN/EE and to the cell plate during cytokinesis (Ravikumar et al., 2017, 2018). Our colocalization experiments showed that TRIPP is also localized to TGN/EE compartments and the cell plate, but not to the Golgi. The finding that TRIPP localization was disrupted in the trs33 mutant indicates that the localization of TRIPP to membrane compartments is dependent on the activity of TRAPP complexes.
TRIPP Plays Critical Roles in Trafficking-Dependent Cellular and Developmental Processes
The tripp-1 mutant exhibits a range of developmental phenotypes, including partial dwarfism, shorter roots, and sterility when grown in the light and photomorphogenesis when grown in the dark. The sterility phenotype of this mutant is similar to that observed in the trs33-1 mutant (Figure 4). The overall growth phenotypes of tripp-1 are weaker than those of the trs33, trs120, and trs130 single mutants, which have lethal phenotypes past the early seedling stage (Thellmann et al., 2010; Rybak et al., 2014; Ravikumar et al., 2017). The nonlethal phenotype of tripp is consistent with the late emergence of TRIPP during the evolution of multicellular photosynthetic organisms, in contrast to the other conserved TRAPP components, which are essential for basic cellular functions such as cytokinesis. TRIPP might provide a specific function required for morphogenesis and developmental regulation in multicellular plants. At the molecular level, TRIPP might quantitatively modulate TRAPPII activities, for example, by stabilizing the TRAPPII complex and/or altering the cellular ratio between TRAPPII and TRAPPIII complexes. The defects in TRS120 localization and cross wall formation in tripp-1 suggest that TRIPP enhance the recruitment of TRAPPII to the cell plate. However, the contribution of TRIPP appears to be quantitative, as cells in the tripp-1 mutant showed normal localization of RAB-A2a. Consistent with the overall weaker phenotype of tripp-1 than trs120-4, a subtle alteration of RAB-A2a localization was previously observed in trs120-4 mutants (Kalde et al., 2019).
TRIPP might also provide functions unique to the TRAPPII complex in plants. An intriguing possibility is that TRIPP plays a role in the polar localization of auxin transporters, which is required for photomorphogenesis. Dark-grown tripp-1 seedlings exhibit not only shorter hypocotyls but also longer roots, a hallmark of photomorphogenesis. Such a phenotype suggests that tripp-1 is not simply defective in cellular functioning or the ability to grow but is altered in developmental regulation. Furthermore, the photomorphogenesis phenotype is likely caused by a defect in the polar localization of PIN2 and the resulting disruption of normal auxin distribution between the shoot and root. The regulation of PINs by light functions as a major switch for photomorphogenesis (Sassi et al., 2012). The polar targeting of PIN proteins involves TGN/endosome-guided targeting to the polar domain and recycling from the lateral side of the plasma membrane through the TGN/EE (Kleine-Vehn et al., 2011). The decreased polarity of PIN2 in tripp-1 might be due to a defect in either or both of these processes. Further studies are required to understand the roles of TRIPP as a TRAPPII component in cell wall formation, in the polar localization of auxin transporters, and in plant development. Our discovery of a plant-specific TRAPPII component highlights an evolutionary innovation that increased the complexity of the plant membrane trafficking system.
METHODS
Plant Materials and Growth Conditions
Arabidopsis (Arabidopsis thaliana) ecotype Columbia (Col-0) was used in this study. Plants were grown in greenhouses with a 16-h-light/8-h-dark cycle at 22°C for general growth and seed harvesting. For seedling analysis such as germination in the dark and root imaging, plants were grown on Petri dishes as follows: seeds were surface sterilized with 70% (v/v) ethanol plus 0.1% (v/v) Triton X-100 for 5 min, dried on filter paper, and grown on half-strength Murashige and Skoog (MS) medium containing 1% (w/v) Suc and 0.8% (w/v) Phytoagar. Plates with seeds were stratified at 4°C for 3 d and placed in a growth chamber under 16-h-light (100 to 150 µmol m−2 s−1 white light)/8-h-dark (or complete dark for germination in the dark) cycle at 22°C.
The T-DNA insertional mutant tripp-1 (SALKseq_063596.3) was obtained from the Arabidopsis Biological Resource Center (http://www.arabidopsis.org; Alonso et al., 2003). trs33-1 (SALK_109244) and trs120-4 (SAIL_1285_D07) were previously described by Thellmann et al., 2010).
Genotyping and RT-PCR Analysis of tripp-1
Genomic DNA was extracted from the T-DNA insertion mutant tripp-1, the wild type, and complemented plants. Leaf tissues were homogenized in genomic DNA extraction buffer (200 mM Tris-HCl, pH 7.5, 250 mM NaCl, 25 mM EDTA, and 0.5% SDS). The homogenate was used for PCR. RNA extraction was performed with a Spectrum Plant Total RNA Kit (Sigma-Aldrich). TRIPP cDNA was synthesized using RevertAid reverse transcriptase enzyme (Thermo Fisher Scientific) according to the manufacturer’s instructions. The primers used are listed in Supplemental Table 3.
Plasmid Construction and Generation of Transgenic Plants
The full-length coding sequences of AtTRS33 and TRIPP without the stop codons were cloned into pENTR/SD/D-TOPO (Invitrogen). To generate the 35S-TRS33-YFP and 35S-TRIPP-YFP constructs, each entry clone was subcloned into Gateway-compatible destination vector pEarleyGate-101 using LR clonase (Invitrogen). To generate 35S-TRIPP-mCherry, an entry clone was subcloned into the pEarleyGate 101 vector, which carries the mCherry tag instead of YFP. To generate native promoter-driven TRS33-4XMyc-6XHis, an entry clone was generated using the 1-kb genomic region upstream of the TRS33 start site fused with the full coding sequence of TRS33 and subcloned into the Gateway-compatible pCAMBIA1390 vector. To analyze colocalization with YFP-RAB-A2a, VHA-a1-GFP, and ST-YFP, TRIPP was cloned from Arabidopsis (Col) genomic DNA into pUB-RFP-DEST-HygR (derived from Grefen et al., 2010). The constructs were validated by nucleotide sequencing. All binary vector constructs were introduced into Agrobacterium tumefaciens (strain GV3101), and transgenic Arabidopsis plants were generated by the floral dip method (Clough and Bent, 1998). TRS120:TRS120-GFP and pUB:TRS120-mCherry are described by Rybak et al. (2014), YFP-RAB-A2a in Chow et al. (2008), VHA-a1-GFP by Dettmer et al. (2006), and ST-YFP by Batoko et al. (2000) and Grebe et al. (2003). All primers used in this study are listed in Supplemental Table 3.
Immunoblot Analysis
Total proteins were extracted from plant materials with 2× SDS sample buffer (100 mM Tris, pH 6.8, 4% SDS, 20% glycerol, and 4% β-mercaptoethanol). After boiling for 5 min, the proteins were loaded onto an SDS-PAGE gel. Separated proteins were transferred to a nitrocellulose membrane and probed with anti-GFP mouse monoclonal antibody (1:3000 dilution, HT801; TransGen Biotech).
15N Stable Isotope Labeling for Quantitative MS Analysis of the AtTRS33 Interactome
pTRS33:TRS33-MycHis and the wild-type plants were grown for 2 weeks at 22°C with 24 h of light on vertical plates containing 14N or 15N medium (half-strength Murashige and Skoog modified basal salt mixture without nitrogen, 0.39 g/L, 8 g/L phytoblend, and 1 g/L KNO3 or 1 g/L K15NO3 [Cambridge Isotope Laboratories], pH 5.8, for 14N medium or 15N medium, respectively). Approximately 2.5 g of tissue was harvested for each sample. 14N-Labeled TRS33-Myc-His and the 15N-labeled wild-type samples or reciprocal replicates (the 14N-labeled wild type and 15N-labeled TRS33-Myc-His) were combined, ground in liquid nitrogen, and stored at –80°C. IP was performed as follows: proteins were extracted in 10 mL of NEBT buffer (20 mM Hepes, pH 7.5, 40 mM KCl, 1 mM EDTA, 10% glycerol, 0.5% Triton X-100, 1 mM phenylmethylsulfonyl fluoride, Pierce protease inhibitor cocktail [Thermo Fisher Scientific], and PhosStop cocktail [Roche]) and centrifuged at 5000 rpm for 5 min. The supernatant was transferred to new tube and centrifuged for 15 min at 14,000 rpm. The supernatant was transferred to new tube and incubated for 1 h with 10 μL of Myc-Trap_MA beads (Chromotek). The beads were washed three times with 1 mL of IP buffer without detergent. The proteins were eluted with 2× SDS buffer and separated by SDS-PAGE. Following Coomassie Brilliant Blue staining, the protein bands were excited and subjected to in-gel digestion with trypsin.
The peptide mixtures were desalted using C18 ZipTips (Millipore) and analyzed on a Q-Exactive HF hybrid quadrupole-Orbitrap mass spectrometer (Thermo Fisher Scientific) equipped with an Easy LC 1200 UPLC liquid chromatography system (Thermo Fisher Scientific). The peptides were separated using an ES803 analytical column (Thermo Fisher Scientific). The flow rate was 300 nL/min, and a 120-min gradient was used. The peptides were eluted using a gradient of 3 to 28% solvent B (80% acetonitrile/0.1 formic acid) over a 100-min period and 28 to 44% solvent B over a 20-min period, followed by a short wash with 90% solvent B. A precursor scan was performed using a mass-to-charge ratio 375 to 1600, and the top 20 most intense multiply charged precursors were selected for fragmentation. The peptides were fragmented with higher-energy collision dissociation with normalized collision energy 27.
Tandem MS peak lists were extracted using an in-house PAVA script, and the data were searched using Protein Prospector (Chalkley et al., 2008) against The Arabidopsis Information Resource 10 database, to which reverse sequence versions were concatenated (a total of 35,386 entries) to allow false discovery rate values to be estimated. Carbamidomethyl cysteine was searched as a fixed modification and the oxidation of Met, peptide N-terminal Gln conversion to pyroglutamate, and N-terminal acetylation as variable modifications. The data were searched with a 10-ppm tolerance for precursor ion and 20 ppm for fragment ions. Peptide and protein false discovery rates were set at 0.01 and 0.05, respectively. 15N-Labeled amino acids were also searched as a fixed modification for the 15N data. Quantification was done using Protein Prospector.
Note that for IP-MS of p35S-TRS33-YFP versus the wild type or p35S-TRIPP-YFP versus the wild type, the method described above was used, except that GFP-trap magnetic agarose beads (Allele) were used for IP.
Y2H Assay
The TRAPPII-specific (AtTRS120 and CLUB/AtTRS130) truncated proteins used for the Y2H assays are described in Steiner et al. (2016a); shared TRAPP subunits were obtained from Riken full-length cDNA clones (Seki et al., 2002) or from a collection of 12,000 Arabidopsis ORFs (Weßling et al., 2014). We used the GAL4 DNA binding domain (DB) encoded in Y2H vector pDEST-pPC97, which was subsequently transformed into yeast (Saccharomyces cerevisiae) strain Y8930, as well as gene fusions to the Gal4 activation domain (AD) in yeast strain Y8800 (Altmann et al., 2018). The constructs were screened by yeast mating in quadruplicate pairwise tests. Interactions were assayed by examining growth on selective plates using the HIS3 reporter and using 1 mM 3-amino-1,2,4-triazole to suppress background growth as described by Dreze et al. (2010). Only pairs scoring positives in all four assays were considered to be bona fide interaction partners. With the exception of the TRS130_C1 construct, all DB clones yielded at least one positive interaction in pairwise tests (Kalde et al., 2019), and these positive interactions were used as controls for the interpretation of negative interaction data.
Confocal Microscopy
Plant roots were imaged under an Olympus FluoView 1000 confocal laser-scanning microscope (www.olympus-ims.com), a Leica TCS SP8 microscope (www.leica-microsystems.com), and a Zeiss 880 confocal laser-scanning microscope (www.zeiss.com/microscopy). Imaging data were acquired using LAS-X software (Leica) and professed with Fiji/imageJ software (http://www.imagej.net). The following excitation and emission parameters were used for the respective fluorescent proteins: YFP, 514-nm excitation, 525- to 575-nm emission; GFP, 488-nm excitation, 500- to 550-nm emission; mCherry, 561-nm excitation, 580- to 643-nm emission; mRFP, 561-nm excitation, 581- to 754-nm emission; and FM4-64, 700- to 725-nm emission. For Figure 1, cell cycle stages were determined based on TRS120-GFP localization dynamics, taking into account how this membrane marker follows phragmoplast microtubule dynamics (Steiner et al., 2016a). As trapp mutants are affected in both cell plate biogenesis and phragmoplast microtubule dynamics (Steiner et al., 2016a), we also relied on FM4-64 to image cell plates in trapp mutants. Based on the appearance of the cell plate, cytokinesis was broadly split into early stages (cell plate initiation and biogenesis) versus late stages (cell plate insertion and maturation; Smertenko et al., 2017). To quantify colocalization data, colocalization between intracellular YFP-RAB-A2a and TRIPP-mRFP, VHAa1-GFP and TRIPP-mRFP, and ST-YFP and TRIPP-mRFP was quantified using Pearson’s correlation coefficient using the JACoP Fiji plugin (Costes et al., 2004; Bolte and Cordelières, 2006).
Protein Alignment and Phylogenetic Analysis
Proteins homologous to TRIPP were aligned using the ClustalW algorithm in the Geneious software suite. Alignments were manually adjusted for optimization. No protein with significant homology to AtTRIPP was found in chlorophytes. Positions are highlighted according to their percent similarity based on the BLOSUM62 matrix. The accessions used were Arabidopsis: NP_566591.1; Chlamydomonas reinhardtii: not found; Zea mays: XP_020403649.1; Physcomitrella patens: XP_024399605.1; Populus trichocarpa: XP_024466417.1; Oryza sativa: XP_015629884.1; Selaginella moellendorffii: XP_024528913.1; Marchantia polymorpha subsp ruderalis: OAE18401; and Klebsormidium nitens: KFL_000080290. To generate the TRIPP phylogenic tree, we used the distance tree of selected homologous TRIPP protein sequences, generated using Geneious Tree Builder, using the Jukes–Cantor genetic distance model and unweighted pair group method with arithmetic mean tree building method. Bootstrapping 100,000×. Accessions were as for the TRIPP alignment. For generation of TRS33 phylogenetic tree, the same methods as TRIPP were used with the following accessions: A. thaliana: NP_187151; C. reinhardtii: XP_001701459.1; Z. mays: NP_001149190.1; P. patens: XP_024372395; P. trichocarpa: XP_024456893; O. sativa: XP_015612457.1; S. moellendorffii: XP_002966358; M. polymorpha subsp ruderalis: PTQ49625; and K. nitens: GAQ89824. The alignments are provided in Supplemental Files 1 and 2.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under the following accession numbers: TRS33 (AT3G05000); TRIPP (AT3G17900); BET5 (AT1G51160); TRS20 (AT1G80500); TCA17 (AT2G20930); BET3 (AT5G54750); TRS23 (AT5G02280); TRS31 (AT5G58030); TRS85 (AT5G16280); TRS120 (AT5G11040); TRS130 (AT5G54440); TRAPPC11 (AT5G65950); TRAPPC12 (AT4G39820); TRAPPC13 (AT2G47960); RAB-A2a (At1g09630).
Supplemental Data
Supplemental Figure 1. MS2 Mass spectrum of a TRIPP peptide identified by SILIP-MS using TRS33-MycHis.
Supplemental Figure 2. Localization of TRIPP to the cell plate and TRS33 localization.
Supplemental Figure 3. T-DNA genotyping and RT-PCR analysis of tripp-1.
Supplemental Figure 4. Two-week-old dark-grown tripp-1 seedlings exhibit light-grown developmental phenotypes.
Supplemental Figure 5. Localization of PIN2 and PIN3 in the tripp-1 mutant.
Supplemental Figure 6. TRIPP is required for the cell plate localization of TRS120-GFP and for cross wall formation.
Supplemental Figure 7. The cell plate localization of RAB-A2a is not affected in tripp-1.
Supplemental Figure 8. Phylogenetic and protein sequence alignment of TRIPP.
Supplemental Table 1. 35S-TRS33-YFP SILIP-MS results from roots and shoots.
Supplemental Table 2. 35S-TRIPP-YFP SILIP-MS results with additional information such peptide number and fold change.
Supplemental Table 3. Primers used in this study
Supplemental File 1. TRIPP alignment file.
Supplemental File 2. TRS33 alignment file.
DIVE Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
Acknowledgments
We thank Robert Chalkley for assistance with MS data analysis using Protein Prospector, Heather Cartwright for technical assistance with microscopy, Adam Idoine for help with phylogenetic analysis and revision of the article, Miriam Abele for critically appraising the literature and for supporting us with assembling figure panels, and Andreas Czempiel for technical assistance. We thank the Center for Life Sciences, Technical University of Munich Centre for Advanced Light Microscopy for access to the confocal microscopes and Roman Meier for supporting us with optimal growth conditions for our plants. This work was supported by the National Institute Of General Medical Sciences of the National Institutes of Health (grant R01GM066258 to Z.-Y.W.) and a Carnegie Endowment Fund (to the Carnegie MS facility). This work was also supported by the Deutsche Forschungsgemeinschaft (DFG; grants AS110/4-7 and AS110/5-2 to F.F.A. and grant SFB 924/2-A10), the Leverhulme Trust (grant RPG-2014-2761 to I.M.), the Biotechnology and Biological Sciences Research Council (studentship 1810136 to L.E.), and the European Research Council’s Horizon 2020 Research and Innovation Programme (grant 648420 to P.F.-B.). This article is dedicated to the memory of Winslow Briggs.
AUTHOR CONTRIBUTIONS
V.J.G. performed, analyzed, and assembled data from all experiments, except those mentioned below, and wrote the article. Z.-Y.W. conceived the research project, designed experiments, analyzed results, and edited the article. V.J.G, S.-L.X. and W.W. performed MS analyses. B.B. genotyped and phenotyped seedlings under the supervision of F.F.A. E.G. genotyped and helped analyze the mutants under the supervision of V.J.G. and Z.-Y.W. R.R. generated, analyzed, and assembled the data shown in Figure 1 under the supervision of F.F.A. L.E., M.F., and I.M. provided constructs and performed the experiment shown in Figures 3E to 3G. L.E. edited the article. M.A. and R.R. performed Y2H experiments in Figure 2 under the supervision of F.F.A and P.F.-B. F.F.A. designed experiments, performed crosses, analyzed data, and drafted sections of and edited the article. Z.-Y.W., F.F.A., S.-L.X., and P.F.-B. acquired funding.
Footnotes
Articles can be viewed without a subscription.
References
- Alonso J.M., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657. [DOI] [PubMed] [Google Scholar]
- Altmann M., Altmann S., Falter C., Falter-Braun P.(2018). High-quality yeast-2-hybrid interaction network mapping. Curr. Protoc. Plant Biol. 3: e20067. [DOI] [PubMed] [Google Scholar]
- Barrowman J., Bhandari D., Reinisch K., Ferro-Novick S.(2010). TRAPP complexes in membrane traffic: convergence through a common Rab. Nat. Rev. Mol. Cell Biol. 11: 759–763. [DOI] [PubMed] [Google Scholar]
- Bassik M.C., et al. (2013). A systematic mammalian genetic interaction map reveals pathways underlying ricin susceptibility. Cell 152: 909–922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batoko H., Zheng H.Q., Hawes C., Moore I.(2000). A rab1 GTPase is required for transport between the endoplasmic reticulum and golgi apparatus and for normal golgi movement in plants. Plant Cell 12: 2201–2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolte S., Cordelières F.P.(2006). A guided tour into subcellular colocalization analysis in light microscopy. J. Microsc. 224: 213–232. [DOI] [PubMed] [Google Scholar]
- Bröcker C., Engelbrecht-Vandré S., Ungermann C.(2010). Multisubunit tethering complexes and their role in membrane fusion. Curr. Biol. 20: R943–R952. [DOI] [PubMed] [Google Scholar]
- Chaiwanon J., Wang W., Zhu J.Y., Oh E., Wang Z.-Y.(2016). Information integration and communication in plant growth regulation. Cell 164: 1257–1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalkley R.J., Baker P.R., Medzihradszky K.F., Lynn A.J., Burlingame A.L.(2008). In-depth analysis of tandem mass spectrometry data from disparate instrument types. Mol. Cell. Proteomics 7: 2386–2398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi C., Davey M., Schluter C., Pandher P., Fang Y., Foster L.J., Conibear E.(2011). Organization and assembly of the TRAPPII complex. Traffic 12: 715–725. [DOI] [PubMed] [Google Scholar]
- Chow C.M., Neto H., Foucart C., Moore I.(2008). Rab-A2 and Rab-A3 GTPases define a trans-golgi endosomal membrane domain in Arabidopsis that contributes substantially to the cell plate. Plant Cell 20: 101–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F.(1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743. [DOI] [PubMed] [Google Scholar]
- Costes S.V., Daelemans D., Cho E.H., Dobbin Z., Pavlakis G., Lockett S.(2004). Automatic and quantitative measurement of protein-protein colocalization in live cells. Biophys. J. 86: 3993–4003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dettmer J., Hong-Hermesdorf A., Stierhof Y.D., Schumacher K.(2006). Vacuolar H+-ATPase activity is required for endocytic and secretory trafficking in Arabidopsis. Plant Cell 18: 715–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drakakaki G., van de Ven W., Pan S., Miao Y., Wang J., Keinath N.F., Weatherly B., Jiang L., Schumacher K., Hicks G., Raikhel N.(2012). Isolation and proteomic analysis of the SYP61 compartment reveal its role in exocytic trafficking in Arabidopsis. Cell Res. 22: 413–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreze M., Monachello D., Lurin C., Cusick M.E., Hill D.E., Vidal M., Braun P.(2010). High-quality binary interactome mapping. Methods Enzymol. 470: 281–315. [DOI] [PubMed] [Google Scholar]
- Grebe M., Xu J., Möbius W., Ueda T., Nakano A., Geuze H.J., Rook M.B., Scheres B.(2003). Arabidopsis sterol endocytosis involves actin-mediated trafficking via ARA6-positive early endosomes. Curr. Biol. 13: 1378–1387. [DOI] [PubMed] [Google Scholar]
- Grefen C., Donald N., Hashimoto K., Kudla J., Schumacher K., Blatt M.R.(2010). A ubiquitin-10 promoter-based vector set for fluorescent protein tagging facilitates temporal stability and native protein distribution in transient and stable expression studies. Plant J. 64: 355–365. [DOI] [PubMed] [Google Scholar]
- Jaber E., Thiele K., Kindzierski V., Loderer C., Rybak K., Jürgens G., Mayer U., Söllner R., Wanner G., Assaad F.F.(2010). A putative TRAPPII tethering factor is required for cell plate assembly during cytokinesis in Arabidopsis. New Phytol. 187: 751–763. [DOI] [PubMed] [Google Scholar]
- Jenkins M.L., Margaria J.P., Stariha J.T.B., Hoffmann R.M., McPhail J.A., Hamelin D.J., Boulanger M.J., Hirsch E., Burke J.E.(2018). Structural determinants of Rab11 activation by the guanine nucleotide exchange factor SH3BP5. Nat. Commun. 9: 3772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalde M., et al. (2019). Interactions between Transport Protein Particle (TRAPP) complexes and Rab GTPases in Arabidopsis. Plant J. 100: 279–297. [DOI] [PubMed] [Google Scholar]
- Kim J.J., Lipatova Z., Segev N.(2016). TRAPP complexes in secretion and autophagy. Front. Cell Dev. Biol. 4: 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleine-Vehn J., et al. (2011). Recycling, clustering, and endocytosis jointly maintain PIN auxin carrier polarity at the plasma membrane. Mol. Syst. Biol. 7: 540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Łangowski Ł., Wabnik K., Li H., Vanneste S., Naramoto S., Tanaka H., Friml J.(2016). Cellular mechanisms for cargo delivery and polarity maintenance at different polar domains in plant cells. Cell Discov. 2: 16018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb C.A., Nühlen S., Judith D., Frith D., Snijders A.P., Behrends C., Tooze S.A.(2016). TBC1D14 regulates autophagy via the TRAPP complex and ATG9 traffic. EMBO J. 35: 281–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson A.A., et al. (2018). TRAPPC11 and GOSR2 mutations associate with hypoglycosylation of α-dystroglycan and muscular dystrophy. Skelet. Muscle 8: 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lürick A., Kümmel D., Ungermann C.(2018). Multisubunit tethers in membrane fusion. Curr. Biol. 28: R417–R420. [DOI] [PubMed] [Google Scholar]
- Lynch-Day M.A., Bhandari D., Menon S., Huang J., Cai H., Bartholomew C.R., Brumell J.H., Ferro-Novick S., Klionsky D.J.(2010). Trs85 directs a Ypt1 GEF, TRAPPIII, to the phagophore to promote autophagy. Proc. Natl. Acad. Sci. USA 107: 7811–7816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milev M.P., Hasaj B., Saint-Dic D., Snounou S., Zhao Q., Sacher M.(2015). TRAMM/TrappC12 plays a role in chromosome congression, kinetochore stability, and CENP-E recruitment. J. Cell Biol. 209: 221–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naramoto S., Nodzyłski T., Dainobu T., Takatsuka H., Okada T., Friml J., Fukuda H.(2014). VAN4 encodes a putative TRS120 that is required for normal cell growth and vein development in Arabidopsis. Plant Cell Physiol. 55: 750–763. [DOI] [PubMed] [Google Scholar]
- Paul P., Simm S., Mirus O., Scharf K.D., Fragkostefanakis S., Schleiff E.(2014). The complexity of vesicle transport factors in plants examined by orthology search. PLoS One 9: e97745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi X., Kaneda M., Chen J., Geitmann A., Zheng H.(2011). A specific role for Arabidopsis TRAPPII in post-Golgi trafficking that is crucial for cytokinesis and cell polarity. Plant J. 68: 234–248. [DOI] [PubMed] [Google Scholar]
- Qi X., Zheng H.(2013). Rab-A1c GTPase defines a population of the trans-Golgi network that is sensitive to endosidin1 during cytokinesis in Arabidopsis. Mol. Plant 6: 847–859. [DOI] [PubMed] [Google Scholar]
- Qi X., Zheng H.(2011). Arabidopsis TRAPPII is functionally linked to Rab-A, but not Rab-D in polar protein trafficking in trans-Golgi network. Plant Signal Behav 6: 1679–1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramírez-Peinado S., Ignashkova T.I., van Raam B.J., Baumann J., Sennott E.L., Gendarme M., Lindemann R.K., Starnbach M.N., Reiling J.H.(2017). TRAPPC13 modulates autophagy and the response to Golgi stress. J. Cell Sci. 130: 2251–2265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravikumar R., et al. (2018). Independent yet overlapping pathways ensure the robustness and responsiveness of trans-Golgi network functions in Arabidopsis. Development 145: dev169201. [DOI] [PubMed] [Google Scholar]
- Ravikumar R., Steiner A., Assaad F.F.(2017). Multisubunit tethering complexes in higher plants. Curr. Opin. Plant Biol. 40: 97–105. [DOI] [PubMed] [Google Scholar]
- Riedel F., Galindo A., Muschalik N., Munro S.(2018). The two TRAPP complexes of metazoans have distinct roles and act on different Rab GTPases. J. Cell Biol. 217: 601–617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinett C.C., Giansanti M.G., Gatti M., Fuller M.T.(2009). TRAPPII is required for cleavage furrow ingression and localization of Rab11 in dividing male meiotic cells of Drosophila. J. Cell Sci. 122: 4526–4534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosquete M.R., Davis D.J., Drakakaki G.(2018). The plant trans-Golgi network: Not just a matter of distinction. Plant Physiol. 176: 187–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosquete M.R., Worden N., Ren G., Sinclair R.M., Pfleger S., Salemi M., Phinney B.S., Domozych D., Wilkop T., Drakakaki G.(2019). AtTRAPPC11/ROG2: A Role for TRAPPs in Maintenance of the Plant Trans-Golgi Network/Early Endosome Organization and Function. Plant Cell 31: 1879–1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rybak K., Steiner A., Synek L., Klaeger S., Kulich I., Facher E., Wanner G., Kuster B., Zarsky V., Persson S., Assaad F.F.(2014). Plant cytokinesis is orchestrated by the sequential action of the TRAPPII and exocyst tethering complexes. Dev. Cell 29: 607–620. [DOI] [PubMed] [Google Scholar]
- Sacher M., Shahrzad N., Kamel H., Milev M.P.(2019). TRAPPopathies: An emerging set of disorders linked to variations in the genes encoding transport protein particle (TRAPP)-associated proteins. Traffic 20: 5–26. [DOI] [PubMed] [Google Scholar]
- Sassi M., et al. (2012). COP1 mediates the coordination of root and shoot growth by light through modulation of PIN1- and PIN2-dependent auxin transport in Arabidopsis. Development 139: 3402–3412. [DOI] [PubMed] [Google Scholar]
- Scrivens P.J., Noueihed B., Shahrzad N., Hul S., Brunet S., Sacher M.(2011). C4orf41 and TTC-15 are mammalian TRAPP components with a role at an early stage in ER-to-Golgi trafficking. Mol. Biol. Cell 22: 2083–2093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seki M., et al. (2002). Functional annotation of a full-length Arabidopsis cDNA collection. Science 296: 141–145. [DOI] [PubMed] [Google Scholar]
- Smertenko A., et al. (2017). Plant cytokinesis: Terminology for structures and processes. Trends Cell Biol. 27: 885–894. [DOI] [PubMed] [Google Scholar]
- Steiner A., Müller L., Rybak K., Vodermaier V., Facher E., Thellmann M., Ravikumar R., Wanner G., Hauser M.T., Assaad F.F.(2016a). The membrane-associated Sec1/Munc18 KEULE is required for phragmoplast microtubule reorganization during cytokinesis in Arabidopsis. Mol. Plant 9: 528–540. [DOI] [PubMed] [Google Scholar]
- Steiner A., et al. (2016b). Cell cycle-regulated PLEIADE/AtMAP65-3 links membrane and microtubule dynamics during plant cytokinesis. Plant J. 88: 531–541. [DOI] [PubMed] [Google Scholar]
- Takemoto K., Ebine K., Askani J.C., Krüger F., Gonzalez Z.A., Ito E., Goh T., Schumacher K., Nakano A., Ueda T.(2018). Distinct sets of tethering complexes, SNARE complexes, and Rab GTPases mediate membrane fusion at the vacuole in Arabidopsis. Proc. Natl. Acad. Sci. USA 115: E2457–E2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thellmann M., Rybak K., Thiele K., Wanner G., Assaad F.F.(2010). Tethering factors required for cytokinesis in Arabidopsis. Plant Physiol. 154: 720–732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas L.L., Joiner A.M.N., Fromme J.C.(2018). The TRAPPIII complex activates the GTPase Ypt1 (Rab1) in the secretory pathway. J. Cell Biol. 217: 283–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas L.L., van der Vegt S.A., Fromme J.C.(2019). A steric gating mechanism dictates the substrate specificity of a Rab-GEF. Dev. Cell 48: 100–114.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokarev A.A., Taussig D., Sundaram G., Lipatova Z., Liang Y., Mulholland J.W., Segev N.(2009). TRAPP II complex assembly requires Trs33 or Trs65. Traffic 10: 1831–1844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vukašinović N., Žárský V.(2016). Tethering complexes in the Arabidopsis endomembrane system. Front. Cell Dev. Biol. 4: 46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weßling R., et al. (2014). Convergent targeting of a common host protein-network by pathogen effectors from three kingdoms of life. Cell Host Microbe 16: 364–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whyte J.R., Munro S.(2002). Vesicle tethering complexes in membrane traffic. J. Cell Sci. 115: 2627–2637. [DOI] [PubMed] [Google Scholar]
- Yamasaki A., Menon S., Yu S., Barrowman J., Meerloo T., Oorschot V., Klumperman J., Satoh A., Ferro-Novick S.(2009). mTrs130 is a component of a mammalian TRAPPII complex, a Rab1 GEF that binds to COPI-coated vesicles. Mol. Biol. Cell 20: 4205–4215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu I.M., Hughson F.M.(2010). Tethering factors as organizers of intracellular vesicular traffic. Annu. Rev. Cell Dev. Biol. 26: 137–156. [DOI] [PubMed] [Google Scholar]
- Zhang J., Chen J., Wang L., Zhao S., Li J., Liu B., Li H., Qi X., Zheng H., Lu M.(2018). AtBET5 is essential for exine pattern formation and apical meristem organization in Arabidopsis. Plant Sci. 274: 231–241. [DOI] [PubMed] [Google Scholar]
- Zhao S., Li C.M., Luo X.M., Siu G.K., Gan W.J., Zhang L., Wu W.K., Chan H.C., Yu S.(2017). Mammalian TRAPPIII Complex positively modulates the recruitment of Sec13/31 onto COPII vesicles. Sci. Rep. 7: 43207. [DOI] [PMC free article] [PubMed] [Google Scholar]