The MYB transcription factor MYB30 negatively regulates photomorphogenesis and promotes PIF4 and PIF5 protein accumulation under prolonged red light irradiation.
Abstract
Phytochromes are red (R) and far-red (FR) light photoreceptors in plants, and PHYTOCHROME-INTERACTING FACTORS (PIFs) are a group of basic helix-loop-helix family transcription factors that play central roles in repressing photomorphogenesis. Here, we report that MYB30, an R2R3-MYB family transcription factor, acts as a negative regulator of photomorphogenesis in Arabidopsis (Arabidopsis thaliana). We show that MYB30 preferentially interacts with the Pfr (active) forms of the phytochrome A (phyA) and phytochrome B (phyB) holoproteins and that MYB30 levels are induced by phyA and phyB in the light. It was previously shown that phytochromes induce rapid phosphorylation and degradation of PIFs upon R light exposure. Our current data indicate that MYB30 promotes PIF4 and PIF5 protein reaccumulation under prolonged R light irradiation by directly binding to their promoters to induce their expression and by inhibiting the interaction of PIF4 and PIF5 with the Pfr form of phyB. In addition, our data indicate that MYB30 interacts with PIFs and that they act additively to repress photomorphogenesis. In summary, our study demonstrates that MYB30 negatively regulates Arabidopsis photomorphogenic development by acting to promote PIF4 and PIF5 protein accumulation under prolonged R light irradiation, thus providing new insights into the complicated but delicate control of PIFs in the responses of plants to their dynamic light environment.
INTRODUCTION
Light plays a vital role throughout the life cycle of a plant, not only as the energy source for photosynthesis but also as an environmental signal regulating many processes of plant growth and development such as seed germination, de-etiolation (photomorphogenesis), and flowering (Jiao et al., 2007; Li et al., 2011; Legris et al., 2019). Light is perceived by multiple families of plant photoreceptors, and among these are the phytochromes that primarily absorb red (R; 600 to 700 nm) and far red (FR; 700 to 750 nm) light (Li et al., 2011; Legris et al., 2019). Phytochromes exist in two interconvertible forms, Pr and Pfr, and the Pfr form is generally considered to be the biologically active form (Li et al., 2011; Legris et al., 2019). In darkness (D), phytochromes are synthesized in the Pr form in the cytosol, and upon R light exposure, they are converted to the Pfr form and translocate into the nucleus (Nagatani, 2004; Chen et al., 2005; Fankhauser and Chen, 2008, Rausenberger et al., 2011; Klose et al., 2015).
Photoactivated phytochromes disrupt and inactivate the E3 ligase complexes formed by CONSTITUTIVE PHOTOMORPHOGENIC1 (COP1) and SUPPRESSOR OF phyA-105 (SPA) proteins, thus allowing the accumulation of photomorphogenesis-promoting transcription factors, such as ELONGATED HYPOCOTYL5 (HY5; Osterlund et al., 2000; Zhu et al., 2008; Lu et al., 2015; Sheerin et al., 2015). In addition, PHYTOCHROME-INTERACTING FACTORS (PIFs), a subset of the basic helix-loop-helix transcription factor superfamily, are pivotal transcription factors that repress photomorphogenesis (Leivar and Quail, 2011; Leivar and Monte, 2014; Xu et al., 2015; Lee and Choi, 2017; Pham et al., 2018b). Upon light exposure, phytochromes induce rapid turnover of PIFs through phosphorylation, ubiquitination, and degradation by the 26S proteasome (Bauer et al., 2004; Monte et al., 2004; Park et al., 2004; Shen et al., 2005, 2007, 2008; Al-Sady et al., 2006, 2008; Oh et al., 2006; Nozue et al., 2007; Lorrain et al., 2008; Ni et al., 2013, 2014, 2017; Pham et al., 2018a). Thus, phytochromes rapidly modulate the expression of light-regulated genes upon irradiation, which ultimately leads to adaptive changes at the cellular and organismal levels (Li et al., 2011). It has also been suggested that the phytochrome-PIF signaling module is evolutionarily conserved and that the two gene families showed significant coevolution (Inoue et al., 2016; Possart et al., 2017; Pham et al., 2018b; Han et al., 2019).
PIF3, the founding member of the PIFs, was initially identified in a yeast two-hybrid screen for PIFs using the C-terminal portion of phytochrome B (phyB) as the bait, and it was also shown to interact with phyA (Ni et al., 1998). So far, eight PIFs have been identified in Arabidopsis (Arabidopsis thaliana), that is, PIF1, PIF3 to PIF8, and PHYTOCHROME INTERACTING FACTOR3-LIKE1 (PIL1), which was recently renamed PIF2 (Leivar and Quail, 2011; Luo et al., 2014; Lee and Choi, 2017). All PIFs contain an active phyB binding motif, and PIF1 and PIF3 have additional active phyA binding motifs (Leivar and Quail, 2011). PIFs redundantly repress photomorphogenesis and promote skotomorphogenesis in etiolated seedlings, as demonstrated by the constitutively photomorphogenic (cop)–like phenotype of the pif quadruple mutant (pif1 pif3 pif4 pif5, termed pifq) in the dark (Leivar et al., 2008a; Shin et al., 2009).
In addition, accumulating evidence has demonstrated that PIFs act as integrators of diverse signals to modulate plant growth and development. These include environmental cues such as light (including photoperiod and shade), temperature (both high and low), and pathogens and internal signals such as phytohormones including auxin, brassinosteroid (BR), ethylene, gibberellic acid, and abscisic acid, and additionally the circadian clock and developmental or metabolic signals (Leivar and Monte, 2014; Paik et al., 2017; Pham et al., 2018b). PIF3 expression was shown to be directly regulated by several transcription factors acting in hormone signaling pathways, such as ETHYLENE-INSENSITIVE3 (EIN3; Zhong et al., 2012) and BRASSINAZOLE-RESISTANT1 (BZR1; Sun et al., 2010). By contrast, the PIF4 and PIF5 promoters were directly targeted by several components of the circadian clock, such as the evening complex (Nusinow et al., 2011; Leivar and Monte, 2014). Consequently, PIF4 and PIF5 showed a robust circadian rhythm of expression under diurnal conditions (Nozue et al., 2007; Niwa et al., 2009; Nusinow et al., 2011; Sun et al., 2019), whereas PIF1 and PIF3 transcript levels remained constant during the diurnal cycle (Soy et al., 2012, 2014).
MYB proteins, which are characterized by the DNA binding MYB domain that is composed of one to four imperfect repeats of ∼52 amino acids, play important roles in regulating plant development, metabolism, and responses to biotic and abiotic stresses (Dubos et al., 2010). The R2R3-MYB type constitutes the largest MYB family. Each member of this family contains two adjacent MYB repeats (R2 and R3) in the N-terminal domain and a highly variable regulatory region located at the C terminus (Stracke et al., 2001). MYB30 is one of the best-characterized R2R3-MYB transcription factors in Arabidopsis. The most extensively studied function of MYB30 is its role in the programmed cell death associated with the hypersensitive response (HR) in Arabidopsis (Daniel et al., 1999; Vailleau et al., 2002). Later studies revealed an important role for salicylic acid in the AtMYB30-mediated HR, indicating that MYB30 modulates the HR through transcriptional activation of genes related to very long chain fatty acid metabolism (Raffaele et al., 2006, 2008).
Yeast two-hybrid screens led to the identification of several MYB30-interacting proteins, including a secretory phospholipase (AtsPLA2-α; Froidure et al., 2010), a RING-type E3 ubiquitin ligase (MIEL1; Marino et al., 2013), and an atypical protease of the subtilase family (SBT5.2b; Serrano et al., 2016), all of which are negative regulators of MYB30 activity and thus negatively regulate MYB30-mediated plant defense responses. In addition, it was documented that MYB30 participates in BR signaling (Li et al., 2009), abscisic acid signaling (Zheng et al., 2012; Lee and Seo, 2016; Wang et al., 2018; Zheng et al., 2018), salt tolerance (Gong et al., 2020), reactive oxygen species–regulated root growth during defense responses (Mabuchi et al., 2018), oxidative and heat stress responses through calcium signaling (Liao et al., 2017), and the crosstalk between biotic stress perception and flowering time (Liu et al., 2014).
Here, we report that MYB30 is a negative regulator of Arabidopsis photomorphogenesis. We show that MYB30 interacts with the active Pfr forms of both phyA and phyB and that MYB30 protein accumulation in light conditions requires the presence of phyA and phyB. Moreover, we show that MYB30 promotes PIF4 and PIF5 protein accumulation under prolonged R light irradiation. Together, our research demonstrates that MYB30 is a key negative regulator of photomorphogenesis that is closely integrated with the phytochrome-PIF signaling module.
RESULTS
MYB30 Interacts with the Pfr Forms of Both phyA and phyB
We performed extensive yeast two-hybrid assays to identify new transcription factors that could interact with phyA and phyB. Interestingly, we discovered that all the MYB30 clade members of the Arabidopsis R2R3-MYB family (Stracke et al., 2001), including MYB60, MYB30, MYB31, MYB96, and MYB94 (Supplemental Figure 1A; Supplemental Files 1 and 2), interacted with the C-terminal domain of PHYA (PHYA apoprotein) in yeast (Saccharomyces cerevisiae) cells (Supplemental Figure 1). By contrast, of these MYB proteins, only MYB30 interacted strongly with the C-terminal domain of PHYB (PHYB apoprotein) as well (Supplemental Figure 1C). Therefore, MYB30 was selected for further investigation in this study.
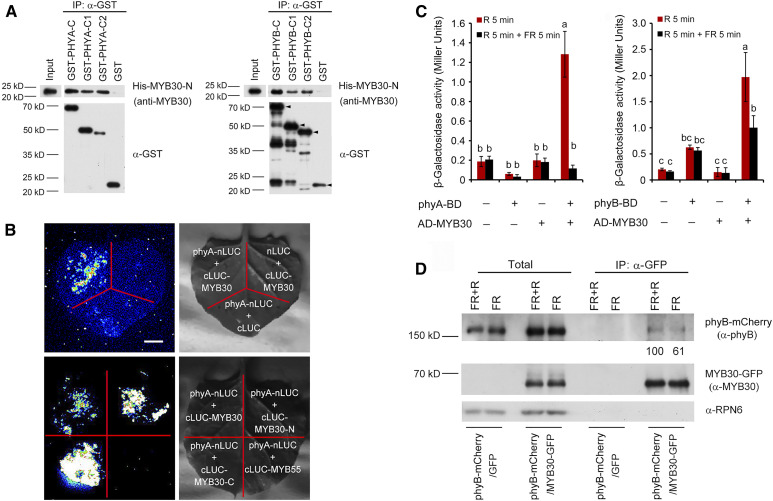
To confirm the interactions between MYB30 and PHYA or PHYB in vitro, a glutathione S-transferase (GST)–tagged Per-Arnt-Sim–related domain of PHYA or PHYB (designated as C1), the His kinase–related domain (designated as C2) of PHYA or PHYB, and a His-tagged R2R3 domain of MYB30 (designated as His-MYB30-N) were expressed in and purified from Escherichia coli. Our pull-down assays showed that the GST-C1 and GST-C2 fusions with either PHYA or PHYB, but not GST alone, were able to pull down His-MYB30-N in vitro (Figure 1A). This indicated that the DNA binding domain of MYB30 could interact with the Per-Arnt-Sim–related domain and the His kinase–related domain of both PHYA and PHYB. To verify the physical interaction between MYB30 and phyA in planta, we conducted firefly luciferase complementation imaging (LCI) assays (Chen et al., 2008) by transiently coexpressing a fusion of phyA and the N-terminal region of luciferase (phyA-nLuc) along with a fusion of MYB30 and the C-terminal region of luciferase (cLuc-MYB30) in Nicotiana benthamiana leaf cells. As shown in Figure 1B, coexpression of phyA-nLuc and cLuc-MYB30 led to strong LUC activity. By contrast, phyA-nLuc or cLuc-MYB30 cotransformed with control vectors showed only background levels of luciferase (LUC) activity. A parallel assay indicated that phyA interacted with both the N- and C-terminal domains of MYB30, but not with MYB55 (Figure 1B). These data indicate that MYB30 physically interacts with phyA in living plant cells.
Figure 1.
MYB30 Directly Interacts with phyA and phyB.
(A) In vitro pull-down assay to test for phyA and phyB interaction with MYB30. Assays used PHYA-C, PHYA-C1, and PHYA-C2 (left) or PHYB-C, PHYB-C1, and PHYB-C2 (right) with the R2R3-MYB domain of MYB30 (MYB30-N). His-tagged MYB30-N proteins pulled down with GST-PHYA/B-C, GST-PHYA/B-C1, GST-PHYA/B-C2, or GST were detected using anti-MYB30 antibodies. The arrowheads in the left three lanes of the PHYB panel indicate the positions of full-length GST-tagged proteins. Input, 6% of the purified His-tagged target proteins used in pull-down assays.
(B) LCI assays using phyA-nLuc and cLuc-MYB30 fusions in N. benthamiana leaf cells. An interaction was seen between phyA-nLUC and cLUC-MYB30, but not with the negative controls lacking phyA or MYB30 (top panels). Specifically, phyA interacted with both N- and C-terminal domains of MYB30, but not with MYB55 (bottom panels). Bar = 1 cm.
(C) GAL4 yeast two-hybrid assays showing that MYB30 preferentially interacted with the Pfr forms of both phyA and phyB. Yeast cells transformed with the indicated plasmids were used for ONPG assays. The yeast cultures were irradiated either with 5 min of R light (60 µmol m−2 s−1) alone or with 5 min of R light immediately followed by 5 min of FR light (40 µmol m−2 s−1), and cultures were then incubated for 2 h. The yeast cultures were exposed to the same R or R + FR light treatments again and incubated for another 2 h. The β-galactosidase activities were then measured by liquid culture assays using ONPG as the substrate. Error bars represent sd of three independent yeast cultures. Different letters represent statistical significances determined by ANOVA with Tukey’s post hoc test (P < 0.05; Supplemental Data Set 3).
(D) Co-IP assays showing that MYB30 preferentially interacted with the Pfr form of phyB in vivo. phyB-mCherry and MYB30-GFP fusion proteins were expressed in Arabidopsis protoplasts. After extraction, proteins were exposed to 5 min of FR light (40 µmol m−2 s−1) or 5 min of FR light immediately followed by 5 min of R light (60 µmol m−2 s−1) and then incubated with GFP-trap agarose beads. Total (left side) and precipitated (right side) proteins were analyzed by immunoblotting using antibodies against phyB (top), MYB30 (middle), and RPN6 (bottom).
Because phytochromes exist in vivo in two interconvertible forms, Pr and Pfr (Li et al., 2011), we asked which forms of phyA and phyB could interact with MYB30 more strongly. To this end, we used a yeast two-hybrid system (Shimizu-Sato et al., 2002) adding phycocyanobilin (PCB) extracted from Spirulina to serve as the chromophore, thus allowing phyA or phyB to form the Pr and Pfr forms in yeast cells after FR and R light treatments, respectively. Interestingly, our data showed that the Pfr forms of phyA and phyB preferentially interacted with MYB30 in yeast cells (Figure 1C). To verify this conclusion, coimmunoprecipitation (co-IP) assays were performed by expressing phyB-mCherry and MYB30-GFP in Arabidopsis protoplasts. After extraction, proteins were exposed to 5 min of FR light, or 5 min of FR light immediately followed by 5 min of R light. Our immunoblot data showed that phyB-mCherry was coprecipitated by the anti-GFP antibody in the presence of MYB30-GFP, but not GFP alone. Notably, larger amounts of phyB-mCherry were coprecipitated with MYB30-GFP after FR plus R light (Figure 1D), indicating that MYB30 preferentially interacted with the Pfr form of phyB (Pfr-phyB) in vivo. A similar conclusion was also made for the in vivo association of MYC-MYB30 with phyA-GFP (Supplemental Figure 2). Collectively, our data demonstrate that MYB30 preferentially interacts with the active Pfr form of phyA and phyB.
MYB30 Is a Negative Regulator of Photomorphogenesis
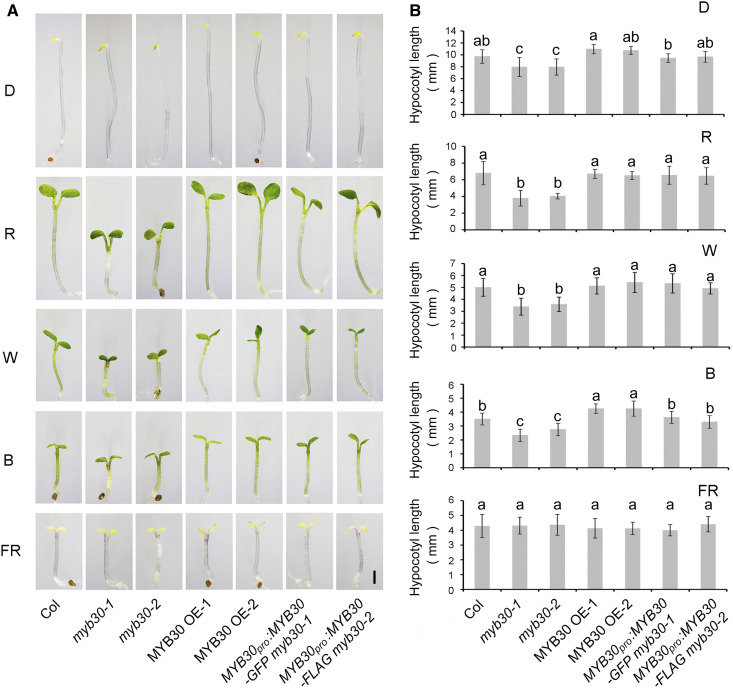
To investigate whether MYB30 plays a role in regulating photomorphogenesis, we grew seedlings of the wild type (Columbia [Col]) and two myb30 mutants (myb30-1 and myb30-2; Zheng et al., 2012) in D or continuous FR, R, blue (B), or white (W) light for 4 d and then measured their hypocotyl lengths. Both myb30 mutants developed shorter hypocotyls than the wild type in R, B and W light, but not in FR light (Figure 2). Introduction of MYB30-GFP or MYB30-FLAG under the control of the native MYB30 promoter successfully rescued the short hypocotyl phenotypes of the myb30-1 and myb30-2 mutants, respectively (Figure 2), indicating that the phenotypes of the two mutants were indeed caused by loss of MYB30 function. However, two independent lines overexpressing MYB30 (MYC-MYB30; Supplemental Figure 3) exhibited slightly longer hypocotyls in B light, but no visible phenotypes different from the wild type in other light conditions (Figure 2). Together, these observations indicate that MYB30 negatively regulates seedling photomorphogenesis in Arabidopsis.
Figure 2.
MYB30 Acts as a Negative Regulator of Photomorphogenesis.
(A) Phenotypes of Col-0 and myb mutants. Images show Col-0 seedlings (left panels) and those of two myb30 mutants (-1 and -2), two independent MYB30-overexpression (OE) lines, and two myb30 complementation lines (right panels) that were grown for 4 d in D or in continuous FR light (5 µmol m−2 s−1), R light (20 µmol m−2 s−1), B light (10 µmol m−2 s−1), or W light (10 µmol m−2 s−1). Bar = 1 mm.
(B) Hypocotyl lengths of Col, two myb30 mutants, two MYB30-ovrexpression (OE) lines, and two myb30 complementation lines grown in different light conditions for 4 d. Error bars represent sd from at least 10 seedlings. Different letters represent statistical significances determined by ANOVA with Tukey’s post hoc test (P < 0.05; Supplemental Data Set 3).
MYB30 Transcript and Protein Levels Are Differentially Regulated by Light
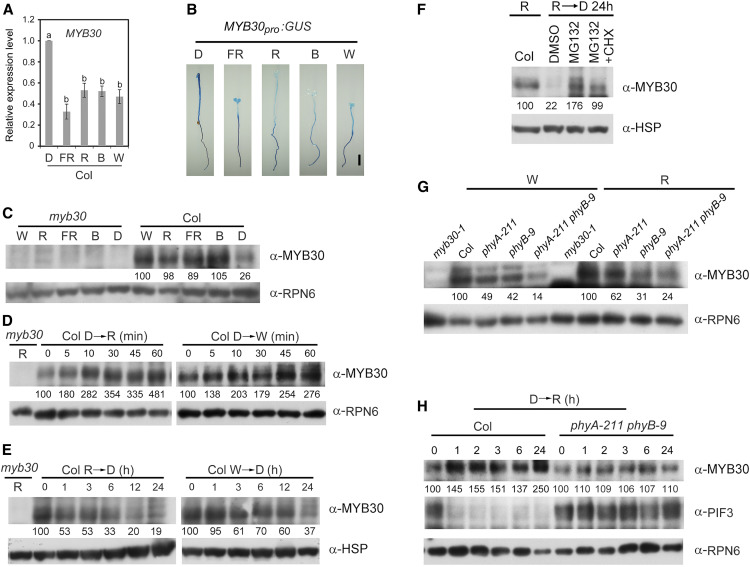
Next, we investigated whether the expression of MYB30 was regulated by light. We compared the MYB30 transcript levels in wild-type (Col) seedlings grown under different light conditions by RT-qPCR assays. MYB30 was expressed at highest levels in the dark, but its expression in the light (including continuous R, FR, B, and W) was approximately half or less than half of the dark levels (Figure 3A). To further characterize how light regulates the spatial expression of MYB30, we generated MYB30pro:GUS lines in which β-glucuronidase (GUS) expression was controlled by the native MYB30 promoter. More than 10 independent lines of homozygous transgenic seedlings were stained for GUS activity, and the staining results of one representative line are shown in Figure 3B. In dark-grown seedlings, GUS was expressed at similar levels in all seedling tissues, including cotyledons, hypocotyls, and roots. However, in light-grown seedlings, lower GUS activities were observed in cotyledons and hypocotyls (Figure 3B), consistent with the RT-qPCR results showing that MYB30 expression was lower in the light (Figure 3A).
Figure 3.
MYB30 Transcript and Protein Levels Are Differentially Regulated by Light.
(A) RT-qPCR analysis of MYB30 expression. Data show the relative expression of MYB30 in Col seedlings grown for 4 d in D or continuous FR light (5 µmol m−2 s−1), R light (20 µmol m−2 s−1), B light (10 µmol m−2 s−1), and W light (40 µmol m−2 s−1). Error bars represent se of three different pools of seedlings. Different letters represent statistical significances determined by ANOVA with Tukey’s post hoc test (P < 0.05; Supplemental Data Set 3).
(B) GUS staining of homozygous MYB30pro:GUS transgenic seedlings. Seedlings were grown for 4 d in D or continuous FR light (5 µmol m−2 s−1), R light (20 µmol m−2 s−1), B light (10 µmol m−2 s−1), or W light (10 µmol m−2 s−1). Bar = 1 mm.
(C) Immunoblots showing MYB30 protein levels in 4-d-old myb30-1 (left side) and Col seedlings (right side) grown in D or continuous W light (40 µmol m−2 s−1), R light (20 µmol m−2 s−1), FR light (5 µmol m−2 s−1), or B light (10 µmol m−2 s−1).
(D) Immunoblots showing MYB30 protein levels in 4-d-old etiolated Col seedlings transferred to R light (20 µmol m−2 s−1) or W light (40 µmol m−2 s−1) for the indicated time periods.
(E) Immunoblots showing MYB30 protein levels in 4-d-old Col seedlings grown in R light (20 µmol m−2 s−1) or W light (40 µmol m−2 s−1) and then transferred to D for the indicated time periods.
(F) Immunoblots showing that the degradation of MYB30 proteins in the dark was inhibited by MG132. Col seedlings grown in R light (20 µmol m−2 s−1) for 4 d were soaked in half-strength liquid Murashige and Skoog medium containing either 150 µM MG132 alone, 150 µM MG132 plus 150 µM cyclohexamide (CHX, a protein synthesis inhibitor), or an equal volume of DMSO (the solvent for MG132) and then transferred to D for 24 h before immunoblotting.
(G) Immunoblots showing MYB30 protein levels in 4-d-old Col, phyA-211, phyB-9, and phyA-211 phyB-9 seedlings grown in continuous W light (40 µmol m−2 s−1) or R light (20 µmol m−2 s−1).
(H) Immunoblots showing MYB30 protein levels in Col and phyA-211 phyB-9 seedlings upon R light treatment. Col and phyA-211 phyB-9 seedlings grown in D for 4 d were transferred to R light (20 µmol m−2 s−1) for the indicated time periods and were then analyzed by immunoblotting.
In (C), (D), (G), and (H), anti-RPN6 was used as a sample loading control. In (E) and (F), anti-HSP was used as a sample loading control. Numbers below the immunoblots in (C) to (H) indicate the relative intensities of MYB30 bands normalized to those of loading controls, and the ratio was set to 100 for the first lane of each group.
To examine whether MYB30 protein accumulation is also regulated by light, we first generated anti-MYB30 antibodies that could specifically recognize the endogenous MYB30 protein (Supplemental Figure 3A). Next, wild-type (Col) and myb30-1 mutant seedlings were grown in D or continuous FR, R, B, or W light for 4 d and then harvested and analyzed by immunoblotting. Surprisingly, we observed that MYB30 proteins accumulated to much higher levels in light than in D (Figure 3C), contradicting the MYB30 gene expression pattern (Figure 3A). To further investigate how light regulates MYB30 protein stability, wild-type (Col) seedlings were first grown in D for 4 d, transferred to R or W light for the indicated times ranging from 5 min to 1 h, and then harvested and analyzed by immunoblotting. MYB30 accumulated rapidly within 1 h of R or W light exposure (Figure 3D), indicating that MYB30 proteins were indeed stabilized in the light. Moreover, when 4-d-old Col seedlings grown in R or W light were transferred to D, we observed a gradual decline in the levels of MYB30 proteins, indicating that MYB30 proteins are degraded in D (Figure 3E). However, treatment with MG132, an inhibitor of 26S proteasomes, effectively inhibited the degradation of MYB30 in the dark (Figure 3F), indicating that MYB30 is degraded in D through the ubiquitin/26S proteasome pathway. Together, our data demonstrate that MYB30 transcript and protein levels are differentially regulated by light.
Because MYB30 physically interacted with both phyA and phyB (Figure 1), we next asked how phyA and phyB regulate MYB30 protein accumulation in the light. To this end, 4-d-old wild-type (Col), phyA-211, phyB-9, and phyA-211 phyB-9 seedlings grown in W or R light were harvested and analyzed by immunoblotting. MYB30 protein levels decreased in phyA-211 or phyB-9 single mutants and further decreased in phyA-211 phyB-9 double mutant seedlings relative to the wild type in both W and R light (Figure 3G). To further confirm this pattern of regulation, 4-d-old dark-grown Col and phyA-211 phyB-9 mutant seedlings were transferred to R light for the indicated times, and our immunoblot data showed that R light–induced MYB30 accumulation was obviously impaired in phyA-211 phyB-9 double mutant seedlings (Figure 3H). The well-characterized phyA/phyB-mediated PIF3 degradation in R light (Al-Sady et al., 2006) was used as a control for this assay (Figure 3H). Collectively, our data demonstrate that MYB30 protein abundance is induced by light, and this induction is mediated by phyA and phyB.
Genetic Relationship between MYB30 and phyA/phyB
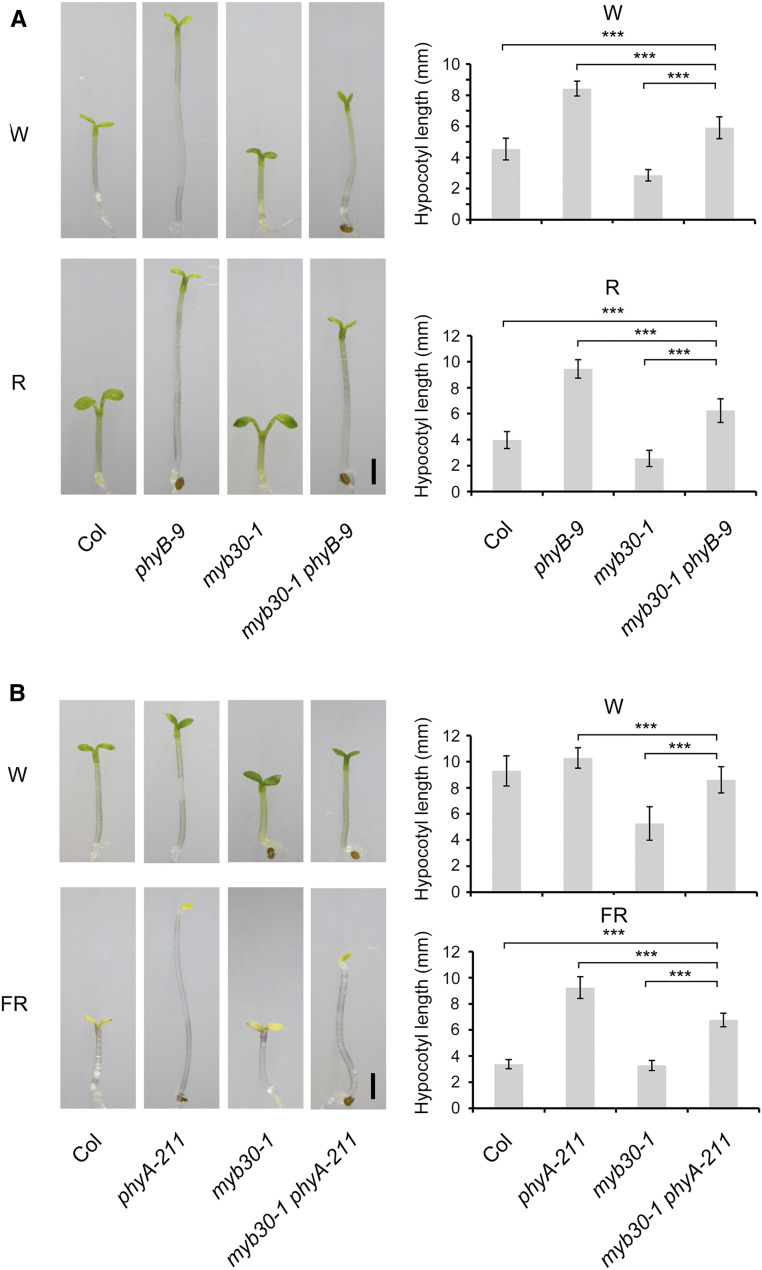
To determine the genetic relationships between phyA/phyB and MYB30, we generated double mutants of myb30-1 with phyA-211 and phyB-9, respectively. Immunoblot data indicated that the corresponding loci were mutated in the respective double mutants (Supplemental Figure 4). The phyB-9 myb30-1 mutants were grown in R and W light for 4 d, and interestingly, we observed that the hypocotyl lengths of phyB-9 myb30-1 mutant seedlings were shorter than those of phyB, but longer than those of Col seedlings in both W and R light (Figure 4A). Similar observations were also made for phyA-211 myb30-1 seedlings grown in continuous FR and W light (Figure 4B). These data indicated that MYB30 contributed significantly to the long hypocotyl phenotypes of phyA-211 and phyB-9 mutants in FR and R/W light, respectively.
Figure 4.
Genetic Relationship between MYB30 and phyA/phyB.
(A) Phenotypes and hypocotyl lengths of Col, phyB-9, myb30-1, and myb30-1 phyB-9 seedlings grown in W light (10 µmol m−2 s−1) or R light (60 µmol m−2 s−1) for 4 d.
(B) Phenotype and hypocotyl lengths of Col, phyA-211, myb30-1, and myb30-1 phyA-211 seedlings grown in W light (10 µmol m−2 s−1) or FR light (5 µmol m−2 s−1) for 4 d.
Error bars represent sd from at least 15 seedlings. ***, P < 0.001 (Student’s t test; Supplemental Data Set 3) for the indicated pairs of seedlings. Bar in (A) and (B) = 1 mm.
MYB30 Positively Regulates the Expression of PIF4 and PIF5 by Directly Binding to Their Promoters
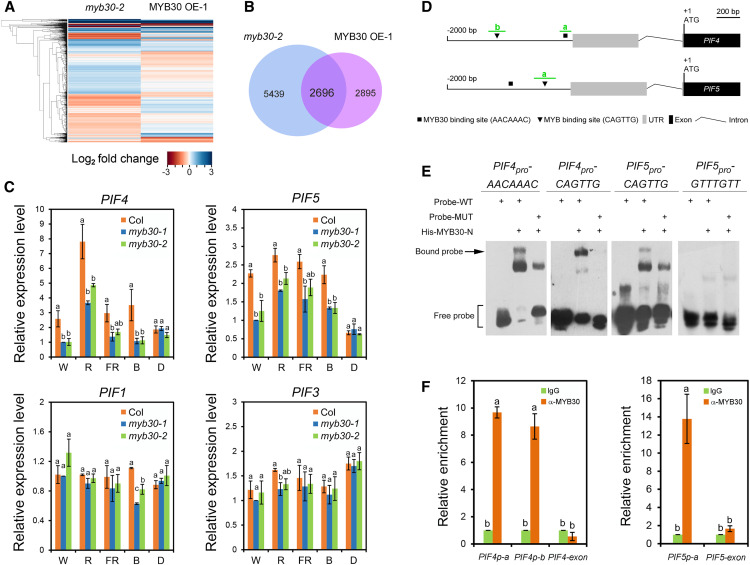
To explore the potential target genes whose expression is regulated by MYB30, we examined the transcriptomes of 4-d-old W light–grown Col, myb30-2, and MYB30-overexpression seedlings by RNA sequencing (RNA-seq) analysis. Three independent pools of seedlings were prepared for each genotype (Supplemental Figure 3B) and then subjected to RNA-seq. After the sequencing data were collected for all samples (each sample with 2.0 Gb of clean data), differential gene expression analysis was performed using Cufflinks (https://cufflinks.cbcb.umd.edu; Trapnell et al., 2013). It was shown that 8135 and 5591 genes displayed statistically significant changes (using Student’s t test with P < 0.05) in myb30-2 and MYB30-overexpression seedlings, respectively, compared with Col (Figures 5A and 5B). Further analysis led to the identification of 2696 genes whose expression was significantly changed in both myb30-2 and MYB30-overexpression seedlings (Figure 5B; Supplemental Data Set 1). Several key regulatory genes of the light signaling pathway, such as PHYA, PHYB, COP1, HY5, and HYH, did not display statistically significant expression changes in RNA-seq. Interestingly, we observed that the expression of PIF4 and PIF5, encoding two key negative regulators of photomorphogenesis, was positively regulated by MYB30 (Supplemental Data Set 1). Our RT-qPCR assays confirmed that the expression of PIF4 and PIF5 was indeed decreased in the two myb30 mutants in continuous light (especially W, R, and B light), but not in D (Figure 5C). By contrast, the expression of PIF1 and PIF3 did not display statistically significant changes in RNA-seq, and our RT-qPCR data indicated that their expression was not dramatically changed in the two myb30 mutants in both light and dark conditions (Figure 5C).
Figure 5.
MYB30 Positively Regulates PIF4 and PIF5 Expression by Directly Binding to Their Promoters.
(A) Cluster analysis of genes whose expression was changed in myb30-2 and MYB30-overexpression (OE) seedlings compared with Col. Each horizontal colored bar represents the log2 of the ratio as shown in the color key at bottom.
(B) Venn diagram showing the number and overlap of genes whose expression was changed in myb30-2 and MYB30-overexpression (OE) seedlings.
(C) RT-qPCR assays showing that PIF4 and PIF5 expression was downregulated in two myb30 mutants in various light conditions: W light (40 µmol m−2 s−1), FR light (40 µmol m−2 s−1), R light (20 µmol m−2 s−1), and B light (10 µmol m−2 s−1). Error bars represent se of three pools of seedlings. Different letters represent statistical significances determined by an ANOVA with Duncan’s post hoc test (P < 0.05; Supplemental Data Set 3).
(D) Schematic illustration showing the distribution of putative MYB30 binding sites (AACAAAC; Li et al., 2009; Liao et al., 2017) and MYB binding sites (CAGTTG; Wang et al., 2004; Liao et al., 2017) in the PIF4 and PIF5 promoters. The adenine residue of the respective translational start codon (ATG) was assigned position +1. The exon–intron structures of PIF4 and PIF5 upstream of the ATG are shown. The short green lines depict the location of amplicons a and b used for ChIP-qPCR shown in (F). UTR, untranslated region.
(E) EMSA assays showing that His-MYB30-N directly binds to MYB30 and MYB binding sites of the PIF4 promoter and MYB binding site of the PIF5 promoter in vitro. MUT, mutant; WT, wild type.
(F) ChIP-qPCR assays showing that MYB30 directly binds to the PIF4 and PIF5 promoters in vivo. Error bars represent se of three pools of seedlings. For each amplicon, the level of binding was calculated as the ratio between anti-MYB30 and anti-rabbit IgG, and an exon fragment of PIF4 or PIF5 was used as a negative control. Different letters represent statistical significances determined by an ANOVA with Duncan’s post hoc test (P < 0.05; Supplemental Data Set 3).
Next, we asked whether MYB30 regulates the expression of PIF4 and PIF5 by directly binding to their promoters. Our yeast one-hybrid assays showed that MYB30 was able to directly bind the PIF4 and PIF5 promoters in yeast cells (Supplemental Figures 5A to 5C). Promoter analyses revealed that both the PIF4 and PIF5 promoters contained one putative MYB30 binding site (AACAAAC; Li et al., 2009; Liao et al., 2017) and one MYB binding site (CAGTTG; Figure 5D; Wang et al., 2004; Liao et al., 2017). Therefore, we performed an electrophoretic mobility shift assay (EMSA) to test whether the His-tagged N-terminal domain (i.e., R2R3 DNA binding domain) of MYB30 was able to bind the MYB30 or MYB binding sites of the PIF4 and PIF5 promoters in vitro. Our EMSA results showed that His-MYB30-N could bind both the MYB30 and MYB binding sites of the PIF4 promoter, but only the MYB binding site of the PIF5 promoter in vitro (Figure 5E). To confirm MYB30 binding to the PIF4 and PIF5 promoters in vivo, we performed chromatin immunoprecipitation (ChIP) assays using 4-d-old W light–grown MYB30-overexpression seedlings. Our qPCR data indicated that the a and b amplicons of the PIF4 promoter and the a amplicon of the PIF5 promoter (Figure 5D), which each contain a site bound by MYB30 in vitro (Figure 5E), were highly enriched in the anti-MYB30, but not in the anti-rabbit IgG ChIP samples (Figure 5F). An exon fragment of PIF4 or PIF5 was used as a negative control (Figure 5F). Together, these data demonstrate that MYB30 directly binds to the PIF4 and PIF5 promoters both in vitro and in vivo.
To further investigate whether MYB30 regulates the spatial expression pattern of PIF4, we crossed PIF4pro:GUS (Sun et al., 2013) with the myb30-1 mutant. Four-day-old PIF4pro:GUS and PIF4pro:GUS myb30-1 seedlings grown in continuous W light were analyzed by histochemical staining. Our results showed that, whereas PIF4 tended to be expressed at the bottom of the hypocotyls in the wild-type seedlings, mutation of MYB30 obviously disrupted this asymmetric expression of PIF4, leading to its uniform expression in the hypocotyls (Supplemental Figure 5D). Collectively, our data demonstrate that MYB30 positively regulates PIF4 and PIF5 expression in the light by directly binding to their promoters.
MYB30 Interacts with PIFs to Coordinately Repress Seedling Photomorphogenesis
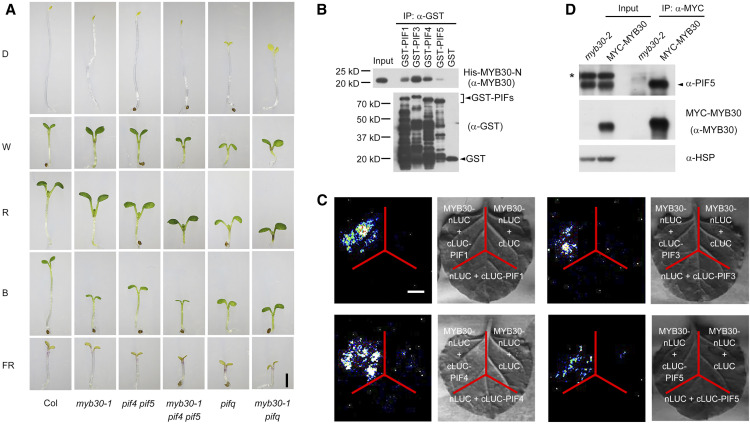
To investigate the genetic relationships between MYB30 and PIFs, we crossed myb30-1 with the double mutant pif4 pif5 (de Lucas et al., 2008) and the quadruple mutant pifq (pif1 pif3 pif4 pif5; Leivar et al., 2008a), respectively, to generate the myb30-1 pif4 pif5 triple mutant and the myb30-1 pifq pentuple mutant. Genotyping data indicated that the corresponding loci were homozygous in the higher order mutants (Supplemental Figure 6), and interestingly, we observed that myb30-1 pif4 pif5 and myb30-1 pifq mutants developed shorter hypocotyls than pif4 pif5 and pifq mutants, respectively, in W, R, and B light (Figure 6A; Supplemental Figure 7). These observations suggest that MYB30 also has a PIF-independent role in regulating hypocotyl growth.
Figure 6.
MYB30 Interacts with PIFs to Coordinately Regulate Seedling Photomorphogenesis.
(A) Phenotypes of 4-d-old Col, myb30-1, pif4 pif5, myb30-1 pif4 pif5, pifq (pif1 pif3 pif4 pif5), and myb30-1 pifq seedlings grown in D or continuous FR light (5 µmol m−2 s−1), R light (20 µmol m−2 s−1), B light (10 µmol m−2 s−1), and W light (10 µmol m−2 s−1).
(B) In vitro pull-down assay using PIF1, PIF3, PIF4, or PIF5 with the R2R3-MYB domain of MYB30 (MYB30-N). His-tagged MYB30-N proteins pulled down with GST-PIF1, GST-PIF3, GST-PIF4, GST-PIF5, or the GST negative control were detected with anti-MYB30 antibodies. Input, 6% of the purified His-MYB30-N proteins used in pull-down assays.
(C) LCI assays using MYB30-nLuc and cLuc-PIF1/3/4/5 fusions in N. benthamiana leaf cells. An interaction was seen between MYB30-nLuc and cLuc-PIF1/3/4/5, but not with the negative controls lacking MYB30 or PIFs. Bar = 1 cm.
(D) co-IP assays showing that MYB30 interacted with PIF5 in vivo. Total proteins were extracted from 4-d-old myb30-2 and 35Spro:MYC-MYB30 seedlings grown in R light (20 µmol m−2 s−1) and then incubated with an anti-MYC Affinity Gel. The total and precipitated proteins were analyzed by immunoblot using antibodies against PIF5 (top), MYB30 (middle), and HSP (bottom). The asterisk (*) indicates a band that cross-reacted with the anti-PIF5 antibody.
Next, we asked whether MYB30 could physically interact with PIFs to coordinately regulate seedling photomorphogenesis. We first performed in vitro pull-down assays and found that GST-tagged PIF1, PIF3, PIF4, and PIF5 proteins, but not GST alone, were able to pull down His-MYB30-N in vitro (Figure 6B). LCI assays confirmed that PIF1, PIF3, PIF4, and PIF5 also interacted with MYB30 in N. benthamiana leaf cells (Figure 6C). We then performed co-IP assays to investigate whether MYB30 could associate with PIF5 in vivo. MYC-MYB30 and myb30-2 mutant seedlings were first grown in R light for 4 d, and then total proteins were extracted and incubated with an anti-MYC antibody. Our immunoblot data showed that PIF5 was coprecipitated by anti-MYC antibodies in MYC-MYB30 seedlings, but not in myb30-2 mutant seedlings (Figure 6D), indicating that MYC-MYB30 interacted with PIF5 in vivo. Collectively, our data demonstrate that MYB30 interacts with PIFs to coordinately regulate photomorphogenesis in Arabidopsis.
MYB30 Inhibits the Interaction between PIF4/PIF5 and Pfr-phyB
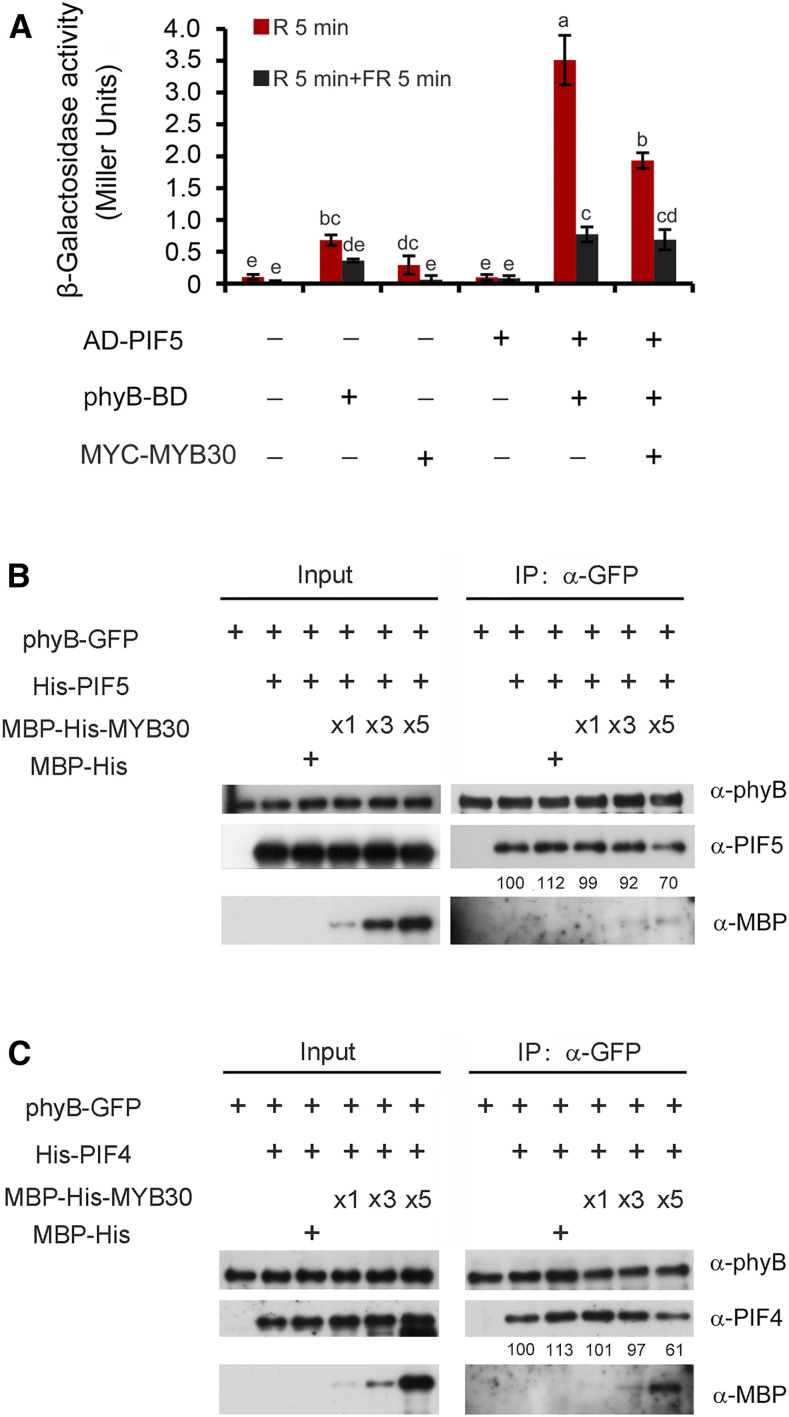
It has been well established that PIFs interact with Pfr-phyB through their conserved active phyB binding motif (Ni et al., 1998, 1999; Huq and Quail, 2002; Shimizu-Sato et al., 2002; Huq et al., 2004; Khanna et al., 2004). Because our data demonstrated that MYB30 could physically interact with both Pfr-phyB (Figure 1) and PIFs (Figures 6B to 6D), we asked how MYB30 could regulate the interaction between phyB and PIFs. We selected PIF5 as a representative PIF and performed yeast three-hybrid assays by introducing a third vector into the yeast two-hybrid system that would express MYC-MYB30 proteins. PCB was added to the yeast cultures, and the yeast cells were pulsed with R or R plus FR light to allow phyB to form the Pfr or Pr forms, respectively. It was shown that indeed, Pfr-phyB preferentially interacted with PIF5 in yeast cells (Figure 7A). However, when MYC-MYB30 was coexpressed in this system, the interaction between Pfr-phyB and PIF5 was clearly decreased (Figure 7A). Additionally, our immunoblot data indicated that coexpression of MYC-MYB30 did not lead to a decrease in the levels of either phyB-BD or AD-PIF5 proteins in yeast cells (Supplemental Figure 8). Together, our data demonstrate that MYB30 inhibits the interaction between PIF5 and Pfr-phyB in yeast cells.
Figure 7.
MYB30 Inhibits the Interaction between PIF4/PIF5 and phyB.
(A) Yeast three-hybrid assays showing that MYB30 inhibits the interaction between PIF5 and Pfr-phyB in yeast cells. AD-PIF5, phyB-BD, and MYC-MYB30 were individually expressed in the yeast strain Y190 as indicated. The β-galactosidase activities were measured by liquid culture assays using ONPG as the substrate. Error bars represent sd of three independent yeast cultures. Different letters represent statistical significances determined by an ANOVA with Tukey’s post hoc test (P < 0.05; Supplemental Data Set 3).
(B) and (C) Pull-down assays showing that MYB30 inhibits the interactions of PIF5 (B) and PIF4 (C) with phyB. Total proteins extracted from 4-d-old R light–grown 35Spro:phyB-GFP seedlings were the bait, and His-PIF5/His-PIF4 fusion proteins were the prey. Equivalent amounts of phyB-GFP protein extract and His-PIF5/His-PIF4 fusion proteins were added, respectively, as indicated, and increasing amounts of MBP-His-MYB30 were added before anti-GFP IP assays were performed. The pulled-down proteins were analyzed by immunoblotting with antibodies against phyB (top panels), PIF4 or PIF5 (middle panels), and MBP (bottom panels). Numbers below the immunoblots indicate the relative intensities of His-PIF5 (B) and His-PIF4 (C) bands normalized to those of phyB-GFP, and the ratio was set to 100 for the first band.
To further evaluate the effects of MYB30 on the interaction of phyB with PIF5, we performed semi-in vivo pull-down assays using His-PIF5 and maltose binding protein (MBP)–His-MYB30 fusion proteins expressed in E. coli along with total proteins extracted from 4-d-old R-grown phyB-GFP seedlings. We first incubated His-PIF5 with the protein extracts prepared from the phyB-GFP seedlings and then performed anti-GFP IP assays. Our immunoblot data showed that His-PIF5 was indeed coprecipitated with phyB-GFP (Figure 7B). Next, we added the MBP-His-MYB30 proteins into this system, and interestingly, we found that the amounts of His-PIF5 coprecipitated with phyB-GFP were progressively reduced as increasing amounts of MBP-His-MYB30 were added (Figure 7B; Supplemental Figure 9). Similar observations were also made in a parallel assay using His-PIF4 (Figure 7C). Collectively, our results demonstrate that MYB30 inhibits the interaction between PIF4/PIF5 and Pfr-phyB.
MYB30 Promotes PIF4 and PIF5 Protein Accumulation under Prolonged R Light
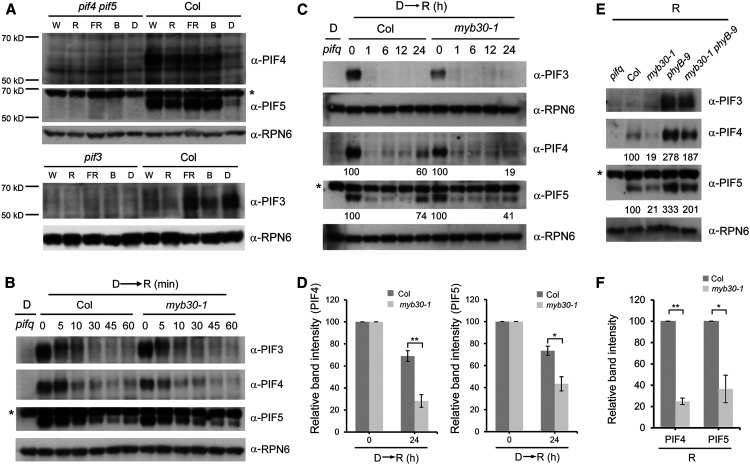
Many studies have shown that PIF proteins accumulate in dark-grown seedlings and are then rapidly phosphorylated, ubiquitinated, and degraded upon R light exposure (Bauer et al., 2004; Monte et al., 2004; Park et al., 2004; Shen et al., 2005, 2007, 2008; Al-Sady et al., 2006, 2008; Oh et al., 2006; Nozue et al., 2007; Lorrain et al., 2008; Ni et al., 2013, 2014, 2017; Pham et al., 2018a). Unexpectedly, when we examined the steady state levels of PIF3, PIF4, and PIF5 in the 4-d-old wild-type (Col) seedlings grown in D or continuous light (including W, FR, R, and B light), we observed that PIF4 and PIF5 proteins accumulated to higher levels in light than in D (Figure 8A). By contrast, PIF3 accumulated to the highest level in D but was much less abundant in W and R light (Figure 8A), consistent with previous reports (Bauer et al., 2004; Park et al., 2004). Notably, a recent study also reported that higher levels of PIF4 accumulated in Col seedlings in continuous R light than in D (Park et al., 2018).
Figure 8.
MYB30 Promotes PIF4 and PIF5 Protein Accumulation under Prolonged R Light.
(A) Immunoblots showing the levels of PIF4, PIF5, and PIF3 in 4-d-old Col seedlings grown in either D or continuous W light (40 µmol m−2 s−1), R light (20 µmol m−2 s−1), FR light (40 µmol m−2 s−1), or B light (10 µmol m−2 s−1) conditions.
(B) and (C) Immunoblots showing the levels of PIF3, PIF4, and PIF5 proteins in 4-d-old etiolated Col and myb30-1 mutant seedlings transferred to R light (60 µmol m−2 s−1) for the indicated times within 1 h (B) and 24 h (C). Numbers below the immunoblots indicate the relative intensities of PIF4 and PIF5 bands normalized to those of RPN6 at the indicated times, and the ratio was set to 100 for the respective band before R light treatment.
(D) Relative intensities of PIF4 and PIF5 bands at the indicated times shown in (C). Error bars represent se from three independent assays using different pools of seedlings. *, P < 0.05 and **, P < 0.01 (Student’s t test; Supplemental Data Set 3).
(E) Immunoblots showing the levels of PIF3, PIF4, and PIF5 proteins in 4-d-old Col, myb30-1, phyB-9, and myb30-1 phyB-9 seedlings grown in continuous R light (60 µmol m−2 s−1). Numbers below the immunoblots indicate the relative band intensities of PIF4 and PIF5 normalized to those of RPN6, and the ratio was set to 100 for the respective band in Col.
(F) Relative intensities of PIF4 and PIF5 bands in Col and myb30-1 shown in (E). Error bars represent se from three independent assays using different pools of seedlings. *, P < 0.05 and **, P < 0.01 (Student’s t test; Supplemental Data Set 3).
Asterisk in each anti-PIF5 blot indicates a cross-reacting band. Anti-RPN6 was used as a sample loading control.
To further investigate the accumulation patterns of PIF4 and PIF5 proteins in R light, we transferred 4-d-old etiolated Col seedlings to R light for different times. Our immunoblot data indicated that indeed PIF3, PIF4, and PIF5 proteins displayed rapid (within minutes) declines upon R light exposure (Figure 8B). Intriguingly, we observed that after prolonged (e.g., 24 h) exposure to R light, PIF4 and PIF5, but not PIF3, reaccumulated in Col seedlings (Figure 8C). To assess the role of MYB30 in regulating PIF4 and PIF5 protein accumulation after R light irradiation, we transferred 4-d-old etiolated myb30-1 mutant seedlings to R light for various times together with Col. Our immunoblot data showed that during a short (1-h) R light irradiation, PIF4 and PIF5 proteins were degraded rapidly in both Col and myb30-1 mutant seedlings at similar rates (Figure 8B; Supplemental Figure 10). However, after a prolonged (24-h) R light irradiation, the reaccumulation of PIF4 and PIF5 proteins observed in Col was greatly impaired in myb30-1 mutants (Figures 8C and 8D). We also compared the levels of PIF4 and PIF5 proteins in 4-d-old Col and myb30-1 mutant seedlings grown in continuous R light and observed that the steady state levels of PIF4 and PIF5 proteins were significantly lower in myb30-1 mutants than in Col (Figures 8E and 8F). Collectively, our data demonstrate that MYB30 plays an important role in promoting PIF4 and PIF5 protein accumulation under prolonged R light irradiation.
DISCUSSION
Phytochromes are thought to transduce light signals by interacting with other proteins (Bae and Choi, 2008). Consistent with this notion, several key regulators of phytochrome signaling were initially identified by their interactions with phytochromes. PIF3, the founding member of the PIF family, was originally identified in a yeast two-hybrid screen for phytochrome-interacting proteins (Ni et al., 1998). Later studies identified PIF4 by genetic and reverse-genetic approaches, and several other PIFs by their sequence similarity with PIF3 (Huq and Quail, 2002; Huq et al., 2004; Khanna et al., 2004; Oh et al., 2004; Leivar et al., 2008b). Because of the pivotal role of transcription factors in regulating gene expression, in this study we performed extensive yeast two-hybrid assays aiming to reveal new transcription factors that can interact with phytochromes. MYB30, a transcription factor of the R2R3-MYB family, was shown to preferentially interact with the Pfr forms of both phyA and phyB (Figure 1). Notably, of the MYB30 clade members, only MYB30 showed strong interaction with the C-terminal domains of both phyA and phyB (Supplemental Figure 1). Our subsequent genetic and biochemical analyses indicated that MYB30 also interacts with PIFs and acts to promote PIF4 and PIF5 protein accumulation under prolonged R light, thus playing an essential role in modulating the phytochrome-PIF signaling module. Therefore, our effort to identify new phytochrome-interacting transcription factors led to the characterization of MYB30 as a key negative regulator of Arabidopsis photomorphogenic development.
In this study, we observed that endogenous PIF4 and PIF5 proteins accumulated to higher levels in the wild-type (Col) seedlings in continuous light (including R and W light) conditions than in D (Figure 8A). By contrast, PIF3 accumulated at high levels in D but at low levels in seedlings grown in W and R light (Figure 8A). These observations indicated that PIF4 and PIF5 protein levels were regulated by different mechanisms than was PIF3 under prolonged R light. We note that another study also recently reported higher accumulation of endogenous PIF4 proteins in Col seedlings in R light than in D (Park et al., 2018). In addition, our study identified MYB30 as a regulator acting to promote PIF4 and PIF5 protein accumulation under prolonged R light irradiation (Figures 8C and 8D) and in continuous R light (Figures 8E and 8F). Consistent with its role in regulating PIF4 and PIF5 abundance, higher levels of MYB30 proteins were observed under continuous light than in D (Figure 3C), similar to the PIF4 and PIF5 protein accumulation patterns (Figure 8A). Notably, MYB30 protein abundance was shown to be induced by light, and interestingly, this induction was mediated by phyA and phyB (Figures 3G and 3H). Therefore, upon R light exposure, light-activated phytochromes induced rapid phosphorylation and degradation of PIFs, thus relieving the repressive effects of PIFs on photomorphogenesis. At the same time, phytochromes induced rapid accumulation of MYB30 that in turn acted to promote PIF4 and PIF5 reaccumulation under prolonged R irradiation (Figure 9). It seems likely that the role of MYB30 in phytochrome signaling could prevent an exaggerated response of plants to prolonged light exposure.
Figure 9.
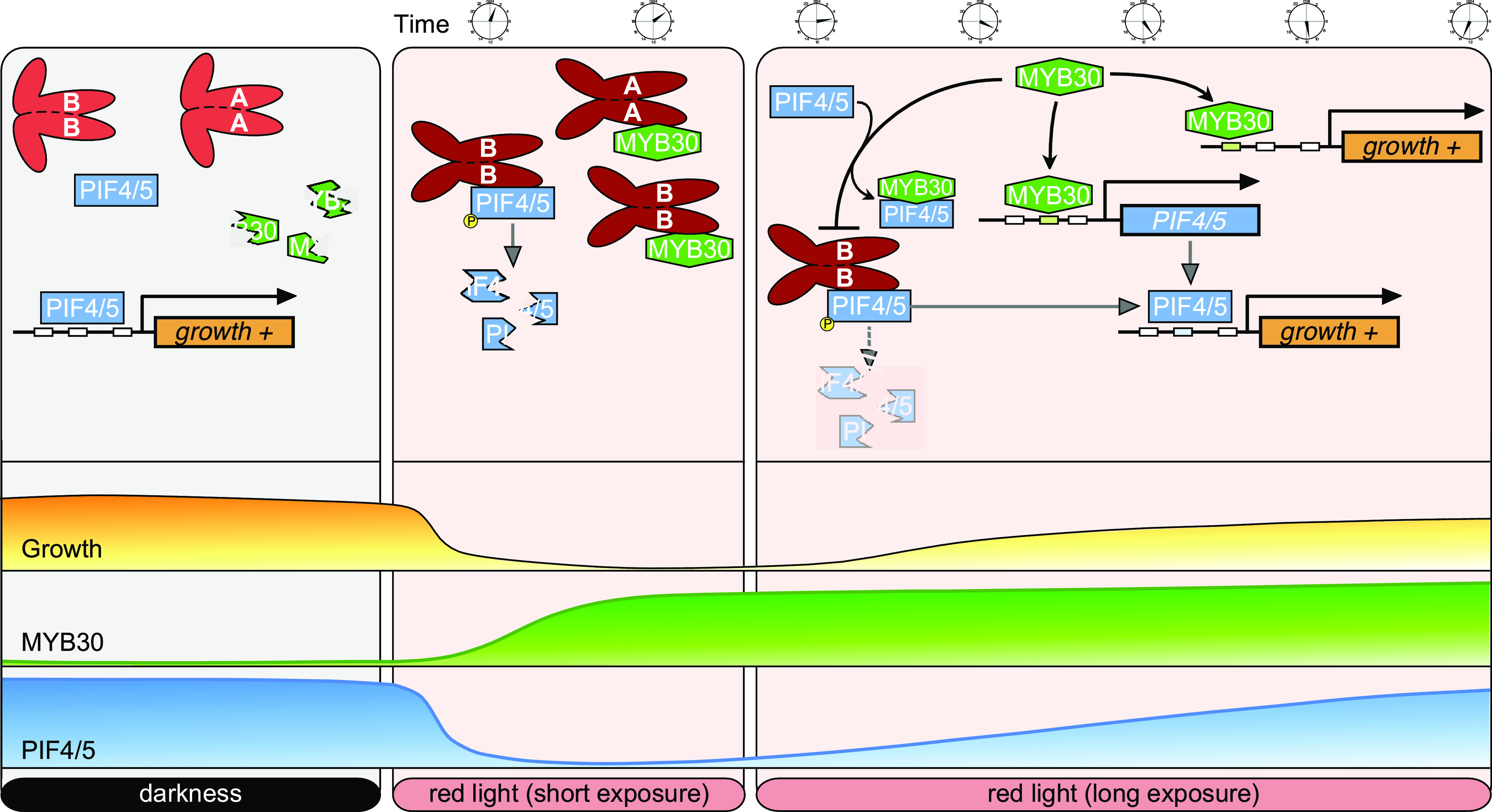
A Working Model Depicting that MYB30 Acts to Promote PIF4 and PIF5 Protein Accumulation under Prolonged R Light Irradiation.
In the dark, MYB30 is degraded via the ubiquitin/26S proteasome-mediated pathway, and PIF4 and PIF5 promote hypocotyl elongation by inducing the expression of growth-promoting genes. Upon short R light exposure, PIF4 and PIF5 are rapidly degraded through the ubiquitin/26S proteasome-mediated pathway, and MYB30 abundance is induced by phyA and phyB. The interaction between MYB30 and the Pfr forms of phyA and phyB may stabilize MYB30 in the light. Under prolonged R light exposure, MYB30 promotes PIF4 and PIF5 protein accumulation by directly binding to their promoters to induce their expression, and by inhibiting the interaction of PIF4 and PIF5 with the Pfr form of phyB. Also, MYB30 acts in parallel with PIFs to induce the expression of growth-promoting genes.
Our data demonstrate that MYB30 could induce PIF4 and PIF5 expression by directly binding to their promoters (Figure 5), and it could inhibit the interaction of PIF4 and PIF5 with the Pfr form of phyB (Figure 7). It therefore seems likely that both of these mechanisms help promote reaccumulation of PIF4 and PIF5 proteins under prolonged R light. On the one hand, our RT-qPCR data showed that MYB30 played a minor role in regulating PIF4 and PIF5 expression during short (≤1-h) R irradiation (Supplemental Figure 11A), but that it played a major role in promoting PIF4 and PIF5 expression under prolonged (24-h) R light (Supplemental Figure 11B) and under continuous light (Figure 5C). On the other hand, after we transferred etiolated 35Spro:PIF5-Citrine and 35Spro:PIF3-MYC seedlings to R light for various times, we observed that PIF5-Citrine, but not PIF3-MYC, reaccumulated to higher levels after prolonged R exposure (Supplemental Figure 12). This is consistent with previous reports that 35S-driven PIF4 and PIF5 proteins reaccumulated under prolonged R irradiation (Lorrain et al., 2008; Pham et al., 2018a). These observations suggested that posttranslational mechanisms are important in regulating PIF4 and PIF5 protein abundance under prolonged R irradiation. Thus, MYB30 may promote PIF4 and PIF5 protein accumulation under prolonged R irradiation via both transcriptional and posttranslational mechanisms (Figure 9). However, the relative contribution of each mechanism in regulating PIF4 and PIF5 protein abundance under prolonged R light needs further characterization. The reason that MYB30 regulated PIF4 and PIF5 abundance under prolonged light, but not under short irradiation, was likely due to its degradation in D (Figure 3). Therefore, MYB30 protein levels might have initially been too low to protect PIF4 and PIF5 from rapid light-induced degradation upon irradiation because this occurred within minutes.
Our genetic data indicated that hypocotyls of phyB-9 myb30-1 mutant seedlings were of intermediate lengths compared with Col and phyB mutants under both W and R light (Figure 4A). The long-hypocotyl phenotype of phyB mutant under high R/FR light was suppressed to various degrees by mutations in pif3 (Soy et al., 2012), pif4 and pif4 pif5 (de Lucas et al., 2008; Lorrain et al., 2008), pifq (Leivar et al., 2012), and pif7 (Li et al., 2012). This might be explained by the fact that PIFs have the intrinsic capacity to induce growth in both dark and light conditions (Nozue et al., 2007; Nusinow et al., 2011; Leivar et al., 2012; Leivar and Monte, 2014). Our immunoblot data showed that the levels of PIF4 and PIF5 were higher in phyB-9 myb30-1 mutants than in Col but lower than in phyB mutants under continuous R light (Figure 8E). Thus, the steady state levels of PIF4 and PIF5 correlated well with the hypocotyl lengths of Col, myb30-1, phyB-9, and phyB-9 myb30-1 seedlings grown in R light (Figure 4Aand 8E). This might therefore partially explain the hypocotyl lengths of the respective seedlings grown in the light. In addition, our data suggested that MYB30 also had a PIF-independent role in regulating hypocotyl growth (Figure 6A; Supplemental Figure 7). Consistent with this notion, MYB30 was shown to be a direct target of BES1 and cooperate with BES1 in regulation of BR-induced gene expression (Li et al., 2009). Therefore, MYB30 might serve as an integration point of BR and light signaling in regulation of hypocotyl elongation, like PIF4 (Oh et al., 2012). This should be investigated in future studies.
MYB30 mRNA was most abundant and MYB30 was least abundant in the dark (Figures 3A and 3C). Further analyses indicated that MYB30 was degraded in the dark through the ubiquitin/26S proteasome pathway (Figures 3E and 3F). The components responsible for degrading MYB30 in the dark are currently unknown. Upon light exposure, our data indicated that MYB30 accumulation was induced by phyA and phyB (Figures 3G and 3H). The underlying mechanism needs to be investigated in future studies. It seems likely that the interactions between MYB30 and the Pfr forms of phyA and phyB may stabilize MYB30 in the light. Also, it will be interesting to investigate whether MYB30 is phosphorylated in the light since phytochrome-mediated phosphorylation plays a pivotal role in regulating PIF stability. In addition, it was previously shown that MYB30 sumoylation was implicated in both abscisic acid signaling and responses of plants to salt stress (Zheng et al., 2012, 2018; Gong et al., 2020). Thus, whether MYB30 sumoylation is involved in phytochrome signaling awaits further investigation.
To summarize, our study reveals that MYB30 is an essential regulator of the phytochrome-PIF signaling network. Considering the pivotal roles of PIFs in mediating the responses of plants to light and various environmental signals (Leivar and Quail., 2011; Leivar and Monte, 2014; Paik et al., 2017; Pham et al., 2018b), MYB30 may ensure a delicate fine-tuned control of PIFs, thus allowing plants to respond quickly and precisely to their dynamic light and environmental signals.
METHODS
Plant Materials and Growth Conditions
All plants used in this study were in the Arabidopsis (Arabidopsis thaliana) Col-0 ecotype, unless otherwise indicated. The myb30-1 and myb30-2 (Zheng et al., 2012), phyA-211 (Reed et al.,1994), phyB-9 (Reed et al., 1993), pifq (pif1 pif3 pif4 pif5; Leivar et al., 2008a), pif4-101 pif5-1 (de Lucas et al., 2008), PIF4pro:GUS (Sun et al., 2013), 35Spro:PIF5-Citrine (Qi et al., 2020), 35Spro:PIF3-MYC (Park et al., 2004), 35Spro:phyB-GFP (Zheng et al., 2013), 35Spro:phyA-GFP (Kim et al., 2000), and MYB30pro:MYB30-FLAG myb30-2 and 35Spro:MYC-MYB30 (MYB30 OE; Liao et al., 2017) are all in the Col-0 background and have been described previously. phyA-211 phyB-9, myb30-1 phyA-211, myb30-1 phyB-9, myb30-1 pif4-101 pif5-1, myb30-1 pifq, and PIF4pro:GUS myb30-1 were generated by genetic crossing.
To grow Arabidopsis seedlings, the seeds were first surface sterilized, stratified in the dark at 4°C for 2 to 4 d, and then grown on Murashige and Skoog medium, pH 5.7, supplemented with 1% (w/v) Suc and 0.8% (w/v) agar (catalog no. A1296; Sigma-Aldrich). Germination was induced by a 12-h treatment with continuous W light, and then the seedlings were grown for 4 d at 22°C in complete D or in growth chambers (Percival Scientific) under continuous W light (380 to 780 nm), R light (600 to 700 nm), FR light (700 to 750 nm), or B light (400 to 500 nm). W light was provided by F17T8/TL841 bulbs (Philips), and R, FR, and B lights were provided by Snap-Lite LED modules (Quantum Devices). The light fluence rates are indicated in the respective figure legends.
Plasmid Construction and Generation of Transgenic Arabidopsis Plants
To generate the MYB30pro:MYB30-GFP vector, the MYB30 native promoter (3199 bp upstream of the MYB30 translation start codon) and the MYB30 coding sequence were cloned into the pCAMBIA1300-GFP binary vector (Li et al., 2017), respectively. To generate the MYB30pro:GUS construct, the MYB30 promoter (3260 bp upstream of the MYB30 translation start codon) was amplified by PCR and then cloned into the pBI101 vector (Clontech).
The LexA-PHYA-N, LexA-PHYA-C, LexA-PHYB-N, and LexA-PHYB-C constructs (used in LexA yeast two-hybrid system) were previously described by Zhang et al. (2018). To generate the AD-MYB30, AD-MYB94, AD-MYB96, AD-MYB60, and AD-MYB31 constructs, the corresponding gene coding sequences were amplified by PCR with the primer pairs listed in Supplemental Data Set 2 and then cloned into the pB42AD vector (Clontech).
The PHYA-BD and PHYB-BD fusions (used in the GAL4 yeast two-hybrid system) were described previously (Zhang et al., 2018). To generate the AD-MYB30 and AD-PIF5 constructs, the MYB30 and PIF5 coding sequences were amplified by PCR with the primer pairs shown in Supplemental Data Set 2 and then cloned into the pGADT7 vector (Clontech), respectively. To introduce a third vector into the yeast two-hybrid system to express the MYC-MYB30 fusion, the multiple cloning sites of the pRS423 vector (Christianson et al., 1992) were first modified using the primers shown in Supplemental Data Set 2 to generate the pRS423-JL vector. Next, the MYC-MYB30 coding sequence was cloned into the pRS423-JL vector, and the resulting vector was used in the yeast three-hybrid assay.
To generate the PIF4pro:LacZ, PIF4pro-A:LacZ, PIF4pro-B:LacZ, PIF5pro:LacZ, PIF5pro-A:LacZ, and PIF5pro-B:LacZ reporter constructs used for yeast one-hybrid assays, the respective promoter fragments were amplified by PCR and then cloned into the pLacZi2μ vector (Lin et al., 2007).
The His-MYB30-N construct was described previously (Liao et al., 2017). To generate the constructs expressing GST-PHYA-C, GST-PHYA-C1, GST-PHYA-C2, GST-PHYB-C, GST-PHYB-C1, GST-PHYB-C2, GST-PIF1, GST-PIF3, GST-PIF4, and GST-PIF5, the respective coding sequences were amplified by PCR and then cloned into the pGEX-4T-1 vector (Amersham Biosciences), respectively. To generate the constructs expressing His-PIF4 and His-PIF5, the coding sequences of PIF4 and PIF5 were amplified by PCR and then cloned into the pET-28a vector (Novagen), respectively. To generate the MBP-His-MYB30 construct, the MYB30 coding sequence was amplified by PCR and then cloned into the pMAL-c2 vector (New England Biolabs).
To generate phyA-nLUC and MYB30-nLUC constructs, the full-length coding sequences of PHYA and MYB30 were amplified by PCR, respectively, and then cloned into the 35Spro:nLUC vector (Chen et al., 2008). To generate cLUC-MYB30, cLUC-MYB30-N, cLUC-MYB30-C, cLUC-MYB55, cLUC-PIF1, cLUC-PIF3, cLUC-PIF4, and cLUC-PIF5, the coding sequences of MYB30, MYB30-N, MYB30-C, MYB55, PIF1, PIF3, PIF4, and PIF5 were amplified by PCR, respectively, and then cloned into the 35Spro:cLUC vector (Chen et al., 2008).
The MYB30pro:GUS and MYB30pro:MYB30-GFP constructs were transformed into Agrobacterium tumefaciens (strain GV3101) and then into Col and the myb30-1 mutant, respectively, by the floral dip method (Clough and Bent, 1998). Homozygous transgenic lines were used for various assays. All of the primers used to generate the above-mentioned constructs are listed in Supplemental Data Set 2, and all of the constructs were confirmed by sequencing prior to usage in various assays.
Yeast Assays
Yeast one-hybrid and yeast two-hybrid (LexA system) assays were performed as previously described by Li et al. (2010), Zhang et al. (2018), and Wang et al. (2019). For GAL4-based yeast two-hybrid assays, the respective combinations of GAL4 BD- and AD-fusion plasmids were cotransformed into the yeast (Saccharomyces cerevisiae) strain Y190, which harbors the URA3pro:GAL1UAS-GAL1TATA-lacZ reporter in its genome. For yeast three-hybrid assays, the respective combinations of phyB-BD (D153 vector; Shimizu-Sato et al., 2002), AD-PIF5 (pGADT7 vector), and MYC-MYB30 (pRS423-JL vector) were cotransformed into the yeast strain Y190. The yeast cell cultures were cultivated in liquid synthetic dropout-Trp-Leu-His medium supplemented with 10 µM PCB and then irradiated either with 5 min of R alone or with 5 min of R immediately followed by 5 min of FR irradiation, and cultures were then incubated for 2 h. Next, the yeast cultures were treated with 5 min of R or R plus FR light pulses again and incubated for another 2 h. β-Galactosidase activities were measured by liquid culture assays using o-nitrophenyl-β-d-galactopyranoside (ONPG) as the substrate as previously described by Shimizu-Sato et al. (2002) and Zhou et al. (2018). Yeast transformation was conducted as described in the Yeast Protocols Handbook (Clontech).
Preparation of Recombinant Proteins
All constructs were transformed into Escherichia coli BL21 Codon Plus cells that were treated with 0.4 mM isopropyl-β-d-thiogalactoside to induce fusion protein expression, incubated overnight at 16°C, and then collected by centrifugation at 4500g for 10 min at 4°C. The GST fusion proteins were purified with Glutathione Sepharose 4B beads (GE Healthcare), and the His- and MBP-His-fusion proteins were purified with nickel-nitrilotriacetic acid beads (Qiagen).
In Vitro Pull-Down Assays
For in vitro binding, 3 mg of purified recombinant bait proteins (GST-phyA/B-C, GST-phyA/B-C1, GST-phyA/B-C2, GST-PIF1/3/4/5, and GST) and 3 mg of prey proteins (His-MYB30-N) were added to 1 mL of binding buffer (50 mM Tris-HCl, pH 7.5, 100 mM NaCl, 0.2% [v/v] glycerol, and 0.6% [v/v] Triton X-100). After incubation at 4°C for 2 h, Glutathione Sepharose 4B beads were added and then the mixture was incubated for a further 1 h. After washing five times with the binding buffer, pulled-down proteins were eluted in 2× SDS loading buffer at 95°C for 15 min, separated on 10% (w/v) SDS-PAGE gels, and detected by immunoblotting.
Semi-in Vivo Pull-Down Assays with Arabidopsis Protein Extracts
Proteins were extracted from 4-d-old R light–grown phyB-GFP seedlings with an extraction buffer containing 50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 10 mM MgCl2, 1 mM EDTA, 0.1% (v/v) Nonidet P-40, 1 mM phenylmethylsulfonyl fluoride (PMSF), 50 µM MG132, and 1× EDTA-free protease inhibitor cocktail (Roche). Binding reactions were started by adding equivalent amounts of phyB-GFP protein extract, His-PIF4 or His-PIF5, and GFP-trap beads (ChromoTek) and varying amounts of MBP-His or MBP-His-MYB30 in 1 mL of 1× PBS buffer, pH 8.0, containing 137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, and 2 mM KH2PO4. The reaction mixtures were incubated at 4°C for 3 h and then the beads were washed six times with 1 mL of 1× PBS buffer containing 0.1% (v/v) Nonidet P-40. The pulled-down proteins were eluted in 2× SDS loading buffer at 95°C for 15 min, separated on 10% (w/v) SDS-PAGE gels, and detected by immunoblotting.
LCI Assays
Transient LCI assays in Nicotiana benthamiana were performed as previously described by Chen et al. (2008). Briefly, A. tumefaciens (strain GV2260) bacteria that contained the indicated constructs were infiltrated (using a needleless syringe) into young but fully expanded leaves of N. benthamiana plants. After infiltration, plants were grown under a 16-h-light/8-h-dark cycle for 2 d. A charge-coupled device camera (1300B; Roper) was used to capture the LUC signal at –110°C with 15-min exposures.
Immunoblotting
Total proteins were extracted as described previously (Qiu et al., 2017), with minor modifications. Briefly, 4-d-old Arabidopsis seedlings (100 mg) were harvested in the dark room under dim green light and then homogenized in extraction buffer (300 μL per 100 mg of sample) consisting of 100 mM Tris-HCl, pH 7.5, 100 mM NaCl, 5 mM EDTA, pH 8.0, 5% [w/v] SDS, 20% [v/v] glycerol, 20 mM DTT, 40 mM β-mercaptoethanol, 2 mM PMSF, 1× EDTA-free protease inhibitor cocktail, 80 μM MG132, 80 μM MG115, 1× phosphatase inhibitor cocktail (Roche), and 10 mM N-ethylmaleimide. Samples were immediately boiled for 10 min and then centrifuged at 16,000g for 10 min at room temperature. Proteins from the supernatant were used in the subsequent immunoblotting assays as described previously (Li et al., 2010). Primary antibodies used in this study included anti-PIF4 (1:750 [v/v], catalog no. AS163955, lot no. 1808; Agrisera), anti-PIF5 (1:1000 [v/v], catalog no. AS122112, lot no. 1508; Agrisera), anti-PIF3 (1:750 [v/v], catalog no. AS163954, lot no. 1808; Agrisera), anti-phyA (1:2000 [v/v]; Zhang et al., 2018), anti-GST (1:3000 [v/v], catalog no. G7781; Sigma-Aldrich), anti-His (1:3000 [v/v], catalog no. H1029, lot no. 079M4822V; Sigma-Aldrich), anti-RPN6 (1:3000 [v/v]; Zhou et al., 2018), anti-GAPDH (1:5000 [v/v], catalog no. AC033; ABclonal), and anti-HSP (1:3000 [v/v], catalog no. AbM51099-31-PU, lot no. 201712280; Beijing Protein Innovation).
To make the anti-MYB30 polyclonal antibodies, His-MYB30 proteins were first expressed in E. coli and then purified and used as antigens to immunize rabbits for producing polyclonal antiserum. Antigen affinity purified anti-MYB30 antibodies were used in immunoblots (1:750 dilutions). To generate anti-phyB monoclonal antibodies, His-phyB-C2 (900-1172) proteins expressed in E. coli were used as antigens to immunize mice, and purified monoclonal antibodies were used in immunoblots (1:2000 dilutions). The anti-MYB30 and anti-phyB antibodies were made by Beijing Protein Innovation.
Co-IP Assays
Arabidopsis seedlings were homogenized in an extraction buffer containing 50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 10 mM MgCl2, 1 mM EDTA, 0.1% (v/v) Nonidet P-40, 1 mM PMSF, 1× MG132, and 1× EDTA-free protease inhibitor cocktail. For co-IP assays to test the in vivo association of MYB30 with PIF5, myb30-2 and homozygous 35Spro:MYC-MYB30 transgenic seedlings were first grown in R light for 4 d and then their total proteins were extracted as described above and incubated with anti-MYC Affinity Gel (Sigma-Aldrich). For co-IP assays to test the in vivo association of MYB30 with phyA, proteins were extracted from 4-d-old FR-grown Col, 35Spro:MYC-MYB30, and 35Spro:phyA-GFP seedlings as described above. Equivalent amounts of total proteins from Col, 35Spro:MYC-MYB30, and 35Spro:phyA-GFP were mixed together as indicated; treated with the indicated combinations of R/FR light pulses; and incubated with anti-MYC Affinity Gel. For co-IP assays to test the in vivo association of MYB30 with phyB, phyB-mCherry, and MYB30-GFP proteins were expressed in Arabidopsis protoplasts. After extraction, proteins were treated with the indicated combinations of R/FR light pulses and then incubated with GFP-trap agarose beads. The beads were then gently washed four times (10 min each time) with protein extraction buffer at 4°C, and the immunoprecipitated proteins were eluted in 2× SDS loading buffer at 95°C for 15 min and analyzed by immunoblotting.
Real-Time RT-qPCR
Total RNA was extracted from Arabidopsis seedlings using the RNeasy Plant Mini kit (TIANGEN). The cDNAs were synthesized from 1 µg of total RNA using RevertAid First Strand cDNA Synthesis kit (Thermo Fisher Scientific) according to the manufacturer’s instructions. Real-time qPCR analysis was performed using the Power Up SYBR Green PCR Master Mix (Thermo Fisher Scientific) with a StepOnePlus Real-Time PCR detection system (Thermo Fisher Scientific). qPCR was performed in triplicate for each sample, and the expression levels were normalized to that of the Arabidopsis TUBULIN3 gene. The primers used for RT-qPCR are shown in Supplemental Data Set 2.
Transcriptome Analyses
Total RNA was extracted by using the same procedures for RT-qPCR analysis. Sequencing was performed with the Illumina HiSeq 2000 platform, and the resulting reads were mapped to the reference genome of Arabidopsis (The Arabidopsis Information Resource 10) with TopHat (http://tophat.cbcb.umd.edu). Transcript expression was evaluated by cuffdiff (http://cufflinks.cbcb.umd.edu), and transcript abundance was estimated by fragments per kilobase of exon model per million mapped fragments. Differentially expressed genes were selected using Student’s t test with P < 0.05.
GUS Staining
At least 10 independent Arabidopsis transgenic lines homozygous for a single copy of the MYB30pro:GUS reporter were analyzed for their GUS activity, and the results of a representative transgenic line grown under different light conditions are shown in Figure 3B. The GUS activity analysis was performed as previously described by Jefferson et al. (1987).
EMSA
EMSAs were performed using biotin-labeled probes and the LightShift Chemiluminescent EMSA kit (Thermo Fisher Scientific). His-MYB30-N fusion proteins were expressed in E. coli and then purified for use in EMSA. The promoter probes were obtained by annealing the biotin-labeled wild-type or mutant complementary oligonucleotides. Briefly, 0.5 µg of His-MYB30-N fusion proteins was incubated together at room temperature for 30 min with biotin-labeled wild-type or mutant probes in 20-μL reaction mixtures containing 10 mM Tris-HCl, 50 mM KCl, 1 mM DTT, 50 ng/μL poly(dI-dC), and 2.5% (v/v) glycerol and then separated on 6% (w/v) native polyacrylamide gels in Tris-borate-EDTA buffer containing 45 mM Tris, 45 mM boric acid, and 1 mM EDTA, pH 8.3. The labeled probes were detected according to the instructions provided with the EMSA kit.
ChIP
35Spro:MYC-MYB30 seedlings grown under continuous W light (10 μmol m–2 s–1) for 4 d were used for ChIP assays following the procedure described previously (Lee et al., 2007). Briefly, 2 g of seedlings was first cross-linked with 1% formaldehyde under vacuum. The samples were ground to a powder in liquid nitrogen, and the chromatin complexes were isolated and sonicated and then incubated with anti-rabbit IgG (1:100 [v/v], catalog no. I5006; Sigma-Aldrich) or with polyclonal anti-MYB30 antibodies (1:100 dilutions). The precipitated DNA was recovered and analyzed by real-time qPCR analysis using the corresponding primer listed in Supplemental Data Set 2. PCR reactions were performed in triplicate for each sample, and the ChIP values were normalized to their respective DNA input values.
Phylogenetic Analysis
The amino acid sequences of Arabidopsis R2R3-MYB proteins (including MYB30, MYB60, MYB31, MYB94, MYB96, and MYB18) were acquired from the National Center for Biotechnology Information database (http://blast.ncbi.nlm.nih.gov/Blast.cgi). Multiple sequence alignment (Supplemental File 1) was conducted using MAFFT (https://mafft.cbrc.jp/alignment/server/index.html), and phylogenetic analysis was performed using the neighbor-joining method in the MEGA6.0 program with a bootstrap of 1000 replicates. The tree file is provided in Supplemental File 2.
Quantification and Statistical Analysis
Protein quantification was performed using ImageJ. ANOVAs were performed with SPSS statistical software, and Student’s t tests were performed in Microsoft Excel. Different letters represent statistical significances determined by ANOVA (P < 0.05) for multiple comparisons, and levels that are not significantly different are indicated with the same letter. See Supplemental Data Set 3 for results of all statistical analyses.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: MYB30 (At3g28910), PIF4 (At2g43010), PIF5 (At3g59060), PIF1 (At2g20180), PIF3 (At1g09530), PHYA (At1g09570), PHYB (At2g18790), MYB60 (At1g08810), MYB31 (At1g74650), MYB94 (At3g47600), MYB96 (At5g62470), LAF1 (At4g25560), and TUBULIN3 (At5g62700). The RNA-seq data were deposited in Gene Expression Omnibus under the accession number GSE141145.
Supplemental Data
Supplemental Figure 1. Interactions between MYB30 clade R2R3-MYB proteins and PHYA or PHYB in yeast cells.
Supplemental Figure 2. Co-IP assays showing that MYB30 preferentially interacted with the Pfr form of phyA in vivo.
Supplemental Figure 3. MYB30 transcript and protein levels in Col, myb30 mutants and seedlings overexpressing MYB30.
Supplemental Figure 4. Genotyping of myb30-1 phyA-211 and myb30-1 phyB-9 mutants by immunoblotting.
Supplemental Figure 5. MYB30 directly binds to the PIF4 and PIF5 promoters and regulates the spatial expression pattern of PIF4.
Supplemental Figure 6. Genotyping of myb30-1 pifq and myb30-1 pif4 pif5 mutants by PCR.
Supplemental Figure 7. Hypocotyl lengths of the seedlings shown in Figure 6A.
Supplemental Figure 8. Immunoblots showing the levels of phyB-BD, AD-PIF5 and MYC-MYB30 in the yeast cells shown in Figure 7A.
Supplemental Figure 9. Relative intensities of pulled-down His-PIF5 bands in Figure 7B.
Supplemental Figure 10. Relative intensities of PIF4 and PIF5 bands at the indicated times in Figure 8B.
Supplemental Figure 11. RT-qPCR assays showing the expression levels of PIF4 and PIF5 in Col and myb30 mutant seedlings after R light exposure.
Supplemental Figure 12. Immunoblots showing that 35Spro-driven PIF5, but not PIF3, reaccumulated to higher levels after prolonged R light exposure.
Supplemental Data Set 1. List of genes whose expression is regulated by MYB30.
Supplemental Data Set 2. Primers used in this study.
Supplemental Data Set 3. Statistical results tables.
Supplemental File 1. Sequence alignment for phylogenetic analysis of MYB30 clade R2R3-MYB proteins.
Supplemental File 2. Tree file for phylogenetic analysis of MYB30 clade R2R3-MYB proteins.
DIVE Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
pifQ Gramene: pif1 pif3 pif4 pif5
pifQ Araport: pif1 pif3 pif4 pif5
Acknowledgments
We thank Peter Quail and Chuanyou Li for PIF-related seeds and Jianping Yang for 35Spro:phyB-GFP seeds. This work was supported by the National Key Research and Development Program of China (grant 2016YFD0100404), the National Natural Science Foundation of China (grants 31970262, 31770321, and 31371221), Beijing Outstanding University Discipline Program, and the Recruitment Program of Global Youth Experts of China.
AUTHOR CONTRIBUTIONS
Y.Y. and J.L. designed research; Y.Y., C.L., X.D., H.L., D.Z., Y.Z., B.J., J.P., X.Q., X.W., P.S., and L.Q. performed research; J.C. analyzed the RNA-seq data; Y.Z. and B.L. contributed new reagents/analytic tools; J.L., Y.Y., Y.G., and S.Y. analyzed data; and J.L., Y.Y., and W.T. wrote the article.
References
- Al-Sady B., Kikis E.A., Monte E., Quail P.H.(2008). Mechanistic duality of transcription factor function in phytochrome signaling. Proc. Natl. Acad. Sci. USA 105: 2232–2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Sady B., Ni W., Kircher S., Schäfer E., Quail P.H.(2006). Photoactivated phytochrome induces rapid PIF3 phosphorylation prior to proteasome-mediated degradation. Mol. Cell 23: 439–446. [DOI] [PubMed] [Google Scholar]
- Bae G., Choi G.(2008). Decoding of light signals by plant phytochromes and their interacting proteins. Annu. Rev. Plant Biol. 59: 281–311. [DOI] [PubMed] [Google Scholar]
- Bauer D., Viczián A., Kircher S., Nobis T., Nitschke R., Kunkel T., Panigrahi K.C., Adám E., Fejes E., Schäfer E., Nagy F.(2004). Constitutive photomorphogenesis 1 and multiple photoreceptors control degradation of phytochrome interacting factor 3, a transcription factor required for light signaling in Arabidopsis. Plant Cell 16: 1433–1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M., Tao Y., Lim J., Shaw A., Chory J.(2005). Regulation of phytochrome B nuclear localization through light-dependent unmasking of nuclear-localization signals. Curr. Biol. 15: 637–642. [DOI] [PubMed] [Google Scholar]
- Chen H., Zou Y., Shang Y., Lin H., Wang Y., Cai R., Tang X., Zhou J.M.(2008). Firefly luciferase complementation imaging assay for protein-protein interactions in plants. Plant Physiol. 146: 368–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christianson T.W., Sikorski R.S., Dante M., Shero J.H., Hieter P.(1992). Multifunctional yeast high-copy-number shuttle vectors. Gene 110: 119–122. [DOI] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F.(1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743. [DOI] [PubMed] [Google Scholar]
- Daniel X., Lacomme C., Morel J.B., Roby D.(1999). A novel myb oncogene homologue in Arabidopsis thaliana related to hypersensitive cell death. Plant J. 20: 57–66. [DOI] [PubMed] [Google Scholar]
- de Lucas M., Davière J.M., Rodríguez-Falcón M., Pontin M., Iglesias-Pedraz J.M., Lorrain S., Fankhauser C., Blázquez M.A., Titarenko E., Prat S.(2008). A molecular framework for light and gibberellin control of cell elongation. Nature 451: 480–484. [DOI] [PubMed] [Google Scholar]
- Dubos C., Stracke R., Grotewold E., Weisshaar B., Martin C., Lepiniec L.(2010). MYB transcription factors in Arabidopsis. Trends Plant Sci. 15: 573–581. [DOI] [PubMed] [Google Scholar]
- Fankhauser C., Chen M.(2008). Transposing phytochrome into the nucleus. Trends Plant Sci. 13: 596–601. [DOI] [PubMed] [Google Scholar]
- Froidure S., Canonne J., Daniel X., Jauneau A., Brière C., Roby D., Rivas S.(2010). AtsPLA2-alpha nuclear relocalization by the Arabidopsis transcription factor AtMYB30 leads to repression of the plant defense response. Proc. Natl. Acad. Sci. USA 107: 15281–15286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong Q., Li S., Zheng Y., Duan H., Xiao F., Zhuang Y., He J., Wu G., Zhao S., Zhou H., Lin H.(2020). SUMOylation of MYB30 enhances salt tolerance by elevating alternative respiration via transcriptionally upregulating AOX1a in Arabidopsis. Plant J.. [DOI] [PubMed] [Google Scholar]
- Han X., Chang X., Zhang Z., Chen H., He H., Zhong B., Deng X.W.(2019). Origin and evolution of core components responsible for monitoring light environment changes during plant terrestrialization. Mol. Plant 12: 847–862. [DOI] [PubMed] [Google Scholar]
- Huq E., Al-Sady B., Hudson M., Kim C., Apel K., Quail P.H.(2004). Phytochrome-interacting factor 1 is a critical bHLH regulator of chlorophyll biosynthesis. Science 305: 1937–1941. [DOI] [PubMed] [Google Scholar]
- Huq E., Quail P.H.(2002). PIF4, a phytochrome-interacting bHLH factor, functions as a negative regulator of phytochrome B signaling in Arabidopsis. EMBO J. 21: 2441–2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue K., Nishihama R., Kataoka H., Hosaka M., Manabe R., Nomoto M., Tada Y., Ishizaki K., Kohchi T.(2016). Phytochrome signaling is mediated by PHYTOCHROME INTERACTING FACTOR in the liverwort Marchantia polymorpha. Plant Cell 28: 1406–1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson R.A., Kavanagh T.A., Bevan M.W.(1987). GUS fusions: beta-Glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 6: 3901–3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao Y., Lau O.S., Deng X.W.(2007). Light-regulated transcriptional networks in higher plants. Nat. Rev. Genet. 8: 217–230. [DOI] [PubMed] [Google Scholar]
- Khanna R., Huq E., Kikis E.A., Al-Sady B., Lanzatella C., Quail P.H.(2004). A novel molecular recognition motif necessary for targeting photoactivated phytochrome signaling to specific basic helix-loop-helix transcription factors. Plant Cell 16: 3033–3044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim L., Kircher S., Toth R., Adam E., Schäfer E., Nagy F.(2000). Light-induced nuclear import of phytochrome-A:GFP fusion proteins is differentially regulated in transgenic tobacco and Arabidopsis. Plant J. 22: 125–133. [DOI] [PubMed] [Google Scholar]
- Klose C., Viczián A., Kircher S., Schäfer E., Nagy F.(2015). Molecular mechanisms for mediating light-dependent nucleo/cytoplasmic partitioning of phytochrome photoreceptors. New Phytol. 206: 965–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H.G., Seo P.J.(2016). The Arabidopsis MIEL1 E3 ligase negatively regulates ABA signalling by promoting protein turnover of MYB96. Nat. Commun. 7: 12525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee N., Choi G.(2017). Phytochrome-interacting factor from Arabidopsis to liverwort. Curr. Opin. Plant Biol. 35: 54–60. [DOI] [PubMed] [Google Scholar]
- Lee J., He K., Stolc V., Lee H., Figueroa P., Gao Y., Tongprasit W., Zhao H., Lee I., Deng X.W.(2007). Analysis of transcription factor HY5 genomic binding sites revealed its hierarchical role in light regulation of development. Plant Cell 19: 731–749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legris M., Ince Y.C., Fankhauser C.(2019). Molecular mechanisms underlying phytochrome-controlled morphogenesis in plants. Nat. Commun. 10: 5219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leivar P., Monte E.(2014). PIFs: Systems integrators in plant development. Plant Cell 26: 56–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leivar P., Monte E., Al-Sady B., Carle C., Storer A., Alonso J.M., Ecker J.R., Quail P.H.(2008b). The Arabidopsis phytochrome-interacting factor PIF7, together with PIF3 and PIF4, regulates responses to prolonged red light by modulating phyB levels. Plant Cell 20: 337–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leivar P., Monte E., Cohn M.M., Quail P.H.(2012). Phytochrome signaling in green Arabidopsis seedlings: Impact assessment of a mutually negative phyB-PIF feedback loop. Mol. Plant 5: 734–749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leivar P., Monte E., Oka Y., Liu T., Carle C., Castillon A., Huq E., Quail P.H.(2008a). Multiple phytochrome-interacting bHLH transcription factors repress premature seedling photomorphogenesis in darkness. Curr. Biol. 18: 1815–1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leivar P., Quail P.H.(2011). PIFs: Pivotal components in a cellular signaling hub. Trends Plant Sci. 16: 19–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Ding Y., Shi Y., Zhang X., Zhang S., Gong Z., Yang S.(2017). MPK3- and MPK6-mediated ICE1 phosphorylation negatively regulates ICE1 stability and freezing tolerance in Arabidopsis. Dev. Cell 43: 630–642.e4. [DOI] [PubMed] [Google Scholar]
- Li J., Li G., Gao S., Martinez C., He G., Zhou Z., Huang X., Lee J.H., Zhang H., Shen Y., Wang H., Deng X.W.(2010). Arabidopsis transcription factor ELONGATED HYPOCOTYL5 plays a role in the feedback regulation of phytochrome A signaling. Plant Cell 22: 3634–3649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Li G., Wang H., Wang Deng X.(2011). Phytochrome signaling mechanisms. Arabidopsis Book 9: e0148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., et al. (2012). Linking photoreceptor excitation to changes in plant architecture. Genes Dev. 26: 785–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Yu X., Thompson A., Guo M., Yoshida S., Asami T., Chory J., Yin Y.(2009). Arabidopsis MYB30 is a direct target of BES1 and cooperates with BES1 to regulate brassinosteroid-induced gene expression. Plant J. 58: 275–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao C., Zheng Y., Guo Y.(2017). MYB30 transcription factor regulates oxidative and heat stress responses through ANNEXIN-mediated cytosolic calcium signaling in Arabidopsis. New Phytol. 216: 163–177. [DOI] [PubMed] [Google Scholar]
- Lin R., Ding L., Casola C., Ripoll D.R., Feschotte C., Wang H.(2007). Transposase-derived transcription factors regulate light signaling in Arabidopsis. Science 318: 1302–1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L., Zhang J., Adrian J., Gissot L., Coupland G., Yu D., Turck F.(2014). Elevated levels of MYB30 in the phloem accelerate flowering in Arabidopsis through the regulation of FLOWERING LOCUS T. PLoS One 9: e89799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorrain S., Allen T., Duek P.D., Whitelam G.C., Fankhauser C.(2008). Phytochrome-mediated inhibition of shade avoidance involves degradation of growth-promoting bHLH transcription factors. Plant J. 53: 312–323. [DOI] [PubMed] [Google Scholar]
- Lu X.D., Zhou C.M., Xu P.B., Luo Q., Lian H.L., Yang H.Q.(2015). Red-light-dependent interaction of phyB with SPA1 promotes COP1-SPA1 dissociation and photomorphogenic development in Arabidopsis. Mol. Plant 8: 467–478. [DOI] [PubMed] [Google Scholar]
- Luo Q., Lian H.L., He S.B., Li L., Jia K.P., Yang H.Q.(2014). COP1 and phyB physically interact with PIL1 to regulate its stability and photomorphogenic development in Arabidopsis. Plant Cell 26: 2441–2456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mabuchi K., Maki H., Itaya T., Suzuki T., Nomoto M., Sakaoka S., Morikami A., Higashiyama T., Tada Y., Busch W., Tsukagoshi H.(2018). MYB30 links ROS signaling, root cell elongation, and plant immune responses. Proc. Natl. Acad. Sci. USA 115: E4710–E4719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marino D., Froidure S., Canonne J., Ben Khaled S., Khafif M., Pouzet C., Jauneau A., Roby D., Rivas S.(2013). Arabidopsis ubiquitin ligase MIEL1 mediates degradation of the transcription factor MYB30 weakening plant defence. Nat. Commun. 4: 1476. [DOI] [PubMed] [Google Scholar]
- Monte E., Tepperman J.M., Al-Sady B., Kaczorowski K.A., Alonso J.M., Ecker J.R., Li X., Zhang Y., Quail P.H.(2004). The phytochrome-interacting transcription factor, PIF3, acts early, selectively, and positively in light-induced chloroplast development. Proc. Natl. Acad. Sci. USA 101: 16091–16098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagatani A.(2004). Light-regulated nuclear localization of phytochromes. Curr. Opin. Plant Biol. 7: 708–711. [DOI] [PubMed] [Google Scholar]
- Ni M., Tepperman J.M., Quail P.H.(1998). PIF3, a phytochrome-interacting factor necessary for normal photoinduced signal transduction, is a novel basic helix-loop-helix protein. Cell 95: 657–667. [DOI] [PubMed] [Google Scholar]
- Ni M., Tepperman J.M., Quail P.H.(1999). Binding of phytochrome B to its nuclear signalling partner PIF3 is reversibly induced by light. Nature 400: 781–784. [DOI] [PubMed] [Google Scholar]
- Ni W., Xu S.L., Chalkley R.J., Pham T.N., Guan S., Maltby D.A., Burlingame A.L., Wang Z.Y., Quail P.H.(2013). Multisite light-induced phosphorylation of the transcription factor PIF3 is necessary for both its rapid degradation and concomitant negative feedback modulation of photoreceptor phyB levels in Arabidopsis. Plant Cell 25: 2679–2698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni W., Xu S.L., González-Grandío E., Chalkley R.J., Huhmer A.F.R., Burlingame A.L., Wang Z.Y., Quail P.H.(2017). PPKs mediate direct signal transfer from phytochrome photoreceptors to transcription factor PIF3. Nat. Commun. 8: 15236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni W., Xu S.L., Tepperman J.M., Stanley D.J., Maltby D.A., Gross J.D., Burlingame A.L., Wang Z.Y., Quail P.H.(2014). A mutually assured destruction mechanism attenuates light signaling in Arabidopsis. Science 344: 1160–1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niwa Y., Yamashino T., Mizuno T.(2009). The circadian clock regulates the photoperiodic response of hypocotyl elongation through a coincidence mechanism in Arabidopsis thaliana. Plant Cell Physiol. 50: 838–854. [DOI] [PubMed] [Google Scholar]
- Nozue K., Covington M.F., Duek P.D., Lorrain S., Fankhauser C., Harmer S.L., Maloof J.N.(2007). Rhythmic growth explained by coincidence between internal and external cues. Nature 448: 358–361. [DOI] [PubMed] [Google Scholar]
- Nusinow D.A., Helfer A., Hamilton E.E., King J.J., Imaizumi T., Schultz T.F., Farré E.M., Kay S.A.(2011). The ELF4-ELF3-LUX complex links the circadian clock to diurnal control of hypocotyl growth. Nature 475: 398–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh E., Kim J., Park E., Kim J.I., Kang C., Choi G.(2004). PIL5, a phytochrome-interacting basic helix-loop-helix protein, is a key negative regulator of seed germination in Arabidopsis thaliana. Plant Cell 16: 3045–3058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh E., Yamaguchi S., Kamiya Y., Bae G., Chung W.I., Choi G.(2006). Light activates the degradation of PIL5 protein to promote seed germination through gibberellin in Arabidopsis. Plant J. 47: 124–139. [DOI] [PubMed] [Google Scholar]
- Oh E., Zhu J.Y., Wang Z.Y.(2012). Interaction between BZR1 and PIF4 integrates brassinosteroid and environmental responses. Nat. Cell Biol. 14: 802–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osterlund M.T., Hardtke C.S., Wei N., Deng X.W.(2000). Targeted destabilization of HY5 during light-regulated development of Arabidopsis. Nature 405: 462–466. [DOI] [PubMed] [Google Scholar]
- Paik I., Kathare P.K., Kim J.I., Huq E.(2017). Expanding roles of PIFs in signal integration from multiple processes. Mol. Plant 10: 1035–1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park E., Kim Y., Choi G.(2018). Phytochrome B requires PIF degradation and sequestration to induce light responses across a wide range of light conditions. Plant Cell 30: 1277–1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park E., Kim J., Lee Y., Shin J., Oh E., Chung W.I., Liu J.R., Choi G.(2004). Degradation of phytochrome interacting factor 3 in phytochrome-mediated light signaling. Plant Cell Physiol. 45: 968–975. [DOI] [PubMed] [Google Scholar]
- Pham V.N., Kathare P.K., Huq E.(2018a). Dynamic regulation of PIF5 by COP1-SPA complex to optimize photomorphogenesis in Arabidopsis. Plant J. 96: 260–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pham V.N., Kathare P.K., Huq E.(2018b). Phytochromes and phytochrome interacting factors. Plant Physiol. 176: 1025–1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Possart A., Xu T., Paik I., Hanke S., Keim S., Hermann H.M., Wolf L., Hiß M., Becker C., Huq E., Rensing S.A., Hiltbrunner A.(2017). Characterization of phytochrome interacting factors from the moss Physcomitrella patens illustrates conservation of phytochrome signaling modules in land plants. Plant Cell 29: 310–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi L., et al. (2020). PHYTOCHROME-INTERACTING FACTORS interact with the ABA receptors PYL8 and PYL9 to orchestrate ABA signaling in darkness. Mol. Plant 13: 414–430. [DOI] [PubMed] [Google Scholar]
- Qiu Y., Pasoreck E.K., Reddy A.K., Nagatani A., Ma W., Chory J., Chen M.(2017). Mechanism of early light signaling by the carboxy-terminal output module of Arabidopsis phytochrome B. Nat. Commun. 8: 1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raffaele S., Rivas S., Roby D.(2006). An essential role for salicylic acid in AtMYB30-mediated control of the hypersensitive cell death program in Arabidopsis. FEBS Lett. 580: 3498–3504. [DOI] [PubMed] [Google Scholar]
- Raffaele S., Vailleau F., Léger A., Joubès J., Miersch O., Huard C., Blée E., Mongrand S., Domergue F., Roby D.(2008). A MYB transcription factor regulates very-long-chain fatty acid biosynthesis for activation of the hypersensitive cell death response in Arabidopsis. Plant Cell 20: 752–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rausenberger J., Tscheuschler A., Nordmeier W., Wüst F., Timmer J., Schäfer E., Fleck C., Hiltbrunner A.(2011). Photoconversion and nuclear trafficking cycles determine phytochrome A’s response profile to far-red light. Cell 146: 813–825. [DOI] [PubMed] [Google Scholar]
- Reed J.W., Nagatani A., Elich T.D., Fagan M., Chory J.(1994). Phytochrome A and phytochrome B have overlapping but distinct functions in Arabidopsis development. Plant Physiol. 104: 1139–1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed J.W., Nagpal P., Poole D.S., Furuya M., Chory J.(1993). Mutations in the gene for the red/far-red light receptor phytochrome B alter cell elongation and physiological responses throughout Arabidopsis development. Plant Cell 5: 147–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrano I., Buscaill P., Audran C., Pouzet C., Jauneau A., Rivas S.(2016). A non canonical subtilase attenuates the transcriptional activation of defence responses in Arabidopsis thaliana. eLife 5: e19755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheerin D.J., Menon C., zur Oven-Krockhaus S., Enderle B., Zhu L., Johnen P., Schleifenbaum F., Stierhof Y.D., Huq E., Hiltbrunner A.(2015). Light-activated phytochrome A and B interact with members of the SPA family to promote photomorphogenesis in Arabidopsis by reorganizing the COP1/SPA complex. Plant Cell 27: 189–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y., Khanna R., Carle C.M., Quail P.H.(2007). Phytochrome induces rapid PIF5 phosphorylation and degradation in response to red-light activation. Plant Physiol. 145: 1043–1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen H., Moon J., Huq E.(2005). PIF1 is regulated by light-mediated degradation through the ubiquitin-26S proteasome pathway to optimize photomorphogenesis of seedlings in Arabidopsis. Plant J. 44: 1023–1035. [DOI] [PubMed] [Google Scholar]
- Shen H., Zhu L., Castillon A., Majee M., Downie B., Huq E.(2008). Light-induced phosphorylation and degradation of the negative regulator PHYTOCHROME-INTERACTING FACTOR1 from Arabidopsis depend upon its direct physi`cal interactions with photoactivated phytochromes. Plant Cell 20: 1586–1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu-Sato S., Huq E., Tepperman J.M., Quail P.H.(2002). A light-switchable gene promoter system. Nat. Biotechnol. 20: 1041–1044. [DOI] [PubMed] [Google Scholar]
- Shin J., Kim K., Kang H., Zulfugarov I.S., Bae G., Lee C.H., Lee D., Choi G.(2009). Phytochromes promote seedling light responses by inhibiting four negatively-acting phytochrome-interacting factors. Proc. Natl. Acad. Sci. USA 106: 7660–7665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soy J., Leivar P., González-Schain N., Sentandreu M., Prat S., Quail P.H., Monte E.(2012). Phytochrome-imposed oscillations in PIF3 protein abundance regulate hypocotyl growth under diurnal light/dark conditions in Arabidopsis. Plant J. 71: 390–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soy J., Leivar P., Monte E.(2014). PIF1 promotes phytochrome-regulated growth under photoperiodic conditions in Arabidopsis together with PIF3, PIF4, and PIF5. J. Exp. Bot. 65: 2925–2936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stracke R., Werber M., Weisshaar B.(2001). The R2R3-MYB gene family in Arabidopsis thaliana. Curr. Opin. Plant Biol. 4: 447–456. [DOI] [PubMed] [Google Scholar]
- Sun J., Qi L., Li Y., Zhai Q., Li C.(2013). PIF4 and PIF5 transcription factors link blue light and auxin to regulate the phototropic response in Arabidopsis. Plant Cell 25: 2102–2114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Q., Wang S., Xu G., Kang X., Zhang M., Ni M.(2019). SHB1 and CCA1 interaction desensitizes light responses and enhances thermomorphogenesis. Nat. Commun. 10: 3110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y., et al. (2010). Integration of brassinosteroid signal transduction with the transcription network for plant growth regulation in Arabidopsis. Dev. Cell 19: 765–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C., Hendrickson D.G., Sauvageau M., Goff L., Rinn J.L., Pachter L.(2013). Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat. Biotechnol. 31: 46–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vailleau F., Daniel X., Tronchet M., Montillet J.L., Triantaphylidès C., Roby D.(2002). A R2R3-MYB gene, AtMYB30, acts as a positive regulator of the hypersensitive cell death program in plants in response to pathogen attack. Proc. Natl. Acad. Sci. USA 99: 10179–10184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Qu G.P., Kong X., Yan Y., Li J., Jin J.B.(2018). Arabidopsis small ubiquitin-related modifier protease ASP1 positively regulates abscisic acid signaling during early seedling development. J. Integr. Plant Biol. 60: 924–937. [DOI] [PubMed] [Google Scholar]
- Wang S., Wang J.W., Yu N., Li C.H., Luo B., Gou J.Y., Wang L.J., Chen X.Y.(2004). Control of plant trichome development by a cotton fiber MYB gene. Plant Cell 16: 2323–2334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., et al. (2019). ABRE-BINDING FACTORS play a role in the feedback regulation of ABA signaling by mediating rapid ABA induction of ABA co-receptor genes. New Phytol. 221: 341–355. [DOI] [PubMed] [Google Scholar]
- Xu X., Paik I., Zhu L., Huq E.(2015). Illuminating progress in phytochrome-mediated light signaling pathways. Trends Plant Sci. 20: 641–650. [DOI] [PubMed] [Google Scholar]
- Zhang S., et al. (2018). TANDEM ZINC-FINGER/PLUS3 is a key component of phytochrome A signaling. Plant Cell 30: 835–852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y., Chen Z., Ma L., Liao C.(2018). The ubiquitin E3 ligase RHA2b promotes degradation of MYB30 in abscisic acid signaling. Plant Physiol. 178: 428–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y., Schumaker K.S., Guo Y.(2012). Sumoylation of transcription factor MYB30 by the small ubiquitin-like modifier E3 ligase SIZ1 mediates abscisic acid response in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 109: 12822–12827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X., et al. (2013). Arabidopsis phytochrome B promotes SPA1 nuclear accumulation to repress photomorphogenesis under far-red light. Plant Cell 25: 115–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong S., Shi H., Xue C., Wang L., Xi Y., Li J., Quail P.H., Deng X.W., Guo H.(2012). A molecular framework of light-controlled phytohormone action in Arabidopsis. Curr. Biol. 22: 1530–1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y., et al. (2018). Hinge region of Arabidopsis phyA plays an important role in regulating phyA function. Proc. Natl. Acad. Sci. USA 115: E11864–E11873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu D., Maier A., Lee J.H., Laubinger S., Saijo Y., Wang H., Qu L.J., Hoecker U., Deng X.W.(2008). Biochemical characterization of Arabidopsis complexes containing CONSTITUTIVELY PHOTOMORPHOGENIC1 and SUPPRESSOR OF PHYA proteins in light control of plant development. Plant Cell 20: 2307–2323. [DOI] [PMC free article] [PubMed] [Google Scholar]