The cellulose synthase inhibitor Endosidin20 is an excellent tool for manipulating cellulose synthesis in order to produce cellulose products with desired properties and to control weeds.
Abstract
Plant cellulose is synthesized by rosette-structured cellulose synthase (CESA) complexes (CSCs). Each CSC is composed of multiple subunits of CESAs representing three different isoforms. Individual CESA proteins contain conserved catalytic domains for catalyzing cellulose synthesis, other domains such as plant-conserved sequences, and class-specific regions that are thought to facilitate complex assembly and CSC trafficking. Because of the current lack of atomic-resolution structures for plant CSCs or CESAs, the molecular mechanism through which CESA catalyzes cellulose synthesis and whether its catalytic activity influences efficient CSC transport at the subcellular level remain unknown. Here, by performing chemical genetic analyses, biochemical assays, structural modeling, and molecular docking, we demonstrate that Endosidin20 (ES20) targets the catalytic site of CESA6 in Arabidopsis (Arabidopsis thaliana). Chemical genetic analysis revealed important amino acids that potentially participate in the catalytic activity of plant CESA6, in addition to previously identified conserved motifs across kingdoms. Using high spatiotemporal resolution live cell imaging, we found that inhibiting the catalytic activity of CESA6 by ES20 treatment reduced the efficiency of CSC transport to the plasma membrane. Our results demonstrate that ES20 is a chemical inhibitor of CESA activity and trafficking that represents a powerful tool for studying cellulose synthesis in plants.
INTRODUCTION
Cellulose is a polymer of β-1,4-d-Glc that serves as an essential cell wall component for controlling the directional growth of plant cells. Cellulose is synthesized at the plasma membrane (PM) by a cellulose synthase complex (CSC) comprising a 25-nm-diameter rosette of subunits in a hexagonal array that can be observed in numerous plant cell types (Mueller et al., 1976; Giddings et al., 1980; Mueller and Brown, 1980). Each CSC is predicted to contain at least 18 monomeric cellulose synthases (CESAs) representing three different isoforms in a 1:1:1 molar ratio (Pear et al., 1996; Arioli et al., 1998; Doblin et al., 2002; Persson et al., 2007; Fernandes et al., 2011; Newman et al., 2013; Gonneau et al., 2014; Hill et al., 2014). Plant CESAs and CESAs from other kingdoms are Glycosyltransferase family2 (GT2) proteins, which synthesize β-1,4-glucan using UDP-Glc in the cytosol as substrate (Cantarel et al., 2009; Brown et al., 2012; Omadjela et al., 2013).
GT2 proteins are thought to share a common GT-A catalytic fold that has been observed in multiple GT2 family proteins (Charnock and Davies, 1999; Cantarel et al., 2009). The conservation of key catalytic motifs between plant and bacterial CESA allowed the first plant CESA to be cloned from cotton (Gossypium hirsutum; Pear et al., 1996). CESAs across kingdoms contain multiple transmembrane domains and a cytoplasmic catalytic domain (McNamara et al., 2015). High-resolution structural analysis of Rhodobacter sphaeroidesin CESA (RsBcsA) revealed the detailed structural conformation of the catalytic site and showed that CESA controls both the catalytic synthesis of cellulose and its translocation across the PM (Morgan et al., 2013). In silico prediction of the structure of the central cytoplasmic domain of cotton CESA1 using bacterial glycosyltransferases SpsA and K4CP as templates revealed that the catalytic site composition of plant CESA is similar to that of bacterial CESA (Sethaphong et al., 2013; Slabaugh et al., 2014a). In the crystal structure of RsBcsA and the predicted structure of the cytoplasmic domain of cotton CESA1, the GT-A fold catalytic residues contain conserved DDG, DXD, TED, and QXXRW motifs required for catalytic activity (Morgan et al., 2013; Sethaphong et al., 2013). In RsBcsA, the amino acids at the transmembrane helixes and the interfacial helixes (IFs) form a channel that contacts and facilitates the translocation of the glucan (Morgan et al., 2013). Thus, although atomic resolution structures for CSCs and individual CESAs are not yet available, it is reasonable to predict that plant CESAs use similar catalytic motifs for cellulose synthesis and may use a similar glucan-translocating channel for cellulose chain translocation of the PM. However, plant CESAs also contain a plant-conserved region (PCR) and class-specific region (CSR) in the cytoplasmic domain that are not present in bacterial CESAs (Pear et al., 1996; Vergara and Carpita, 2001). Plant CESAs also form more complicated protein complexes than bacterial CESAs, pointing to possible specific mechanisms for plant cellulose synthesis.
Small molecule inhibitors that disrupt cellulose synthesis have proven useful for understanding the molecular functions and dynamics of CSCs and for identifying new genes involved in cellulose synthesis (Montezinos and Delmer, 1980; Heim et al., 1989; Scheible et al., 2001; Desprez et al., 2002; DeBolt et al., 2007; Brabham et al., 2014; Worden et al., 2015; Tateno et al., 2016; Tran et al., 2018). Unfortunately, the mode of action of these inhibitors is not well characterized, limiting their value as investigative tools. Here, we report the characterization of the small molecule Endosidin20 (ES20), which inhibits cellulose synthesis by directly targeting Arabidopsis (Arabidopsis thaliana) CESA6. We used chemical genetic analyses, structural modeling, molecular docking, and biochemical assays to show that ES20 targets CESA6 at the catalytic site. Furthermore, we analyzed the cellular localization and trafficking dynamics of CSCs in ES20-treated seedlings and found that CSC delivery to the PM is inhibited by ES20 treatment, which is consistent with the previous finding that the catalytic site of CESA affects the efficient subcellular transport of CSCs (Park et al., 2019).
RESULTS
ES20 Inhibits Cellulose Synthesis
ES20 (Figure 1A; Supplemental Methods) was identified through a chemical library screen for small molecules that are active in plants (Drakakaki et al., 2011). When grown in the presence of ES20, roots of the Arabidopsis wild-type (ecotype Columbia [Col-0]) seedlings became shorter and wider in an ES20 dose-dependent manner (Figures 1B and 1C). When treated with ES20 overnight, the root tip region was swollen and root elongation was markedly inhibited compared to mock-treated roots (Figures 1D and 1E). Epidermal cells from the root elongation zone were markedly swollen after ES20 treatment, which was reflected by a significantly decreased cell length and a significantly increased cell width (Figures 1D to 1F; Supplemental Data Set). In addition to these root phenotypes, ES20 inhibited etiolated hypocotyl growth in a dose-dependent manner (Supplemental Figures 1A and 1B) and caused epidermal cell swelling (Supplemental Figures 1C to 1E). Swollen plant cells and organs are often caused by direct or indirect disruption of cell wall biosynthesis or organization, such as those in CESA-deficient mutants and in the wild-type plants treated with inhibitors of cellulose synthesis or microtubule organization (Baskin et al., 1994; Arioli et al., 1998; Fagard et al., 2000; Burn et al., 2002; Desprez et al., 2002; Daras et al., 2009). We found that ES20 reduced the crystalline cellulose content of both light-grown roots and dark-grown hypocotyls of the wild-type seedlings in a dose-dependent manner (Figure 1G; Supplemental Figure 1F; Supplemental Data Set). These results suggest that ES20 inhibits plant growth by inhibiting cellulose biosynthesis. We also found that lignin and callose accumulated at higher levels in ES20-treated seedlings compared to mock-treated controls (Supplemental Figures 1G and 1H; Supplemental Methods), which is similar to cellulose synthesis deficiency caused by mutation or inhibitors (Desprez et al., 2002; Caño-Delgado et al., 2003; Sampathkumar et al., 2013).
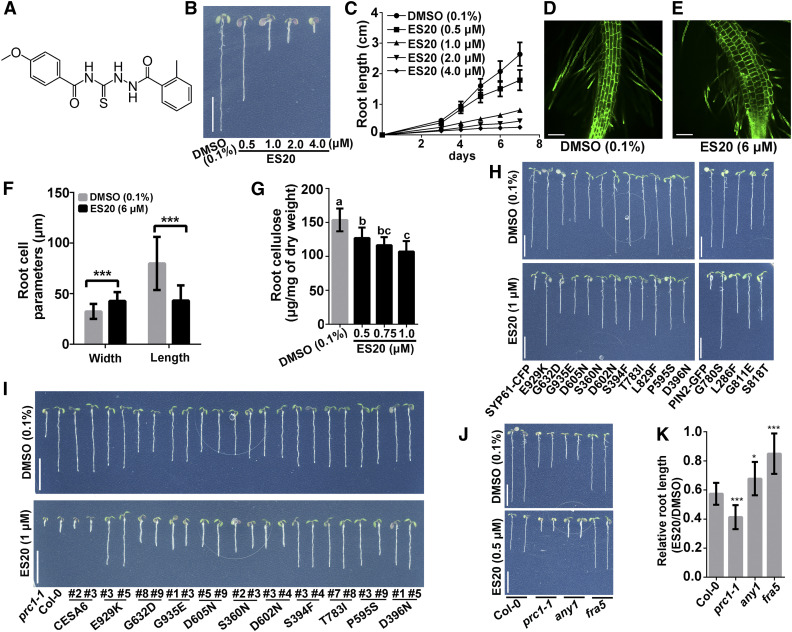
Figure 1.
ES20 Is a Cellulose Synthesis Inhibitor, and cesa6 Mutants Have Reduced Sensitivity to ES20.
(A) Molecular structure of ES20.
(B) and (C) ES20 inhibits Arabidopsis root growth in a dose-dependent manner. Bars = 1.0cm (B). Error bars represent mean ± sd, with n = 15 (C).
(D) to (F) ES20 causes root cells to swell. Root cells of the 3-d-old light-grown wild-type seedlings treated overnight with 0.1% DMSO (D) or 6 μM ES20 (E). ES20 treatment reduces cell length and increases cell width (F). Bars = 100 μm ([D] and [E]). Error bars represent mean ± sd, with n = 15 (F).
(G) ES20 reduces crystalline cellulose content in cell walls of light-grown roots in a dose-dependent manner. The letters (a to c) indicate statistically significant differences (P < 0.05) determined by one-way analysis of variance tests followed by Tukey’s multiple comparison tests in different samples. Error bars represent mean ± sd, with n = 9.
(H) cesa6 mutants resulting from a genetic screen of EMS-mutagenized populations have reduced sensitivity to ES20. The representative 7-d-old wild-type seedlings expressing SYP61-CFP or PIN2-GFP, and cesa6 mutant lines grown on medium supplemented with 0.1% DMSO (top) or 1 μM ES20 (bottom). The mutants are listed based in the order of their discovery. Bars = 1.0cm.
(I) Genetic complementation of prc1-1/cesa6 growth defects and sensitivity to ES20 by mutated CESA6 constructs. Ten constructs that we tested rescued the root growth defect of prc1-1 to different extents in the absence of ES20 and led to reduced sensitivity to ES20 in transgenic plants. Bars = 1.0cm.
(J) and (K) Mutations in other CESA isoforms (any1/cesa1; fra1/cesa7) also lead to reduced sensitivity to ES20. ***, P < 0.001 by two-tailed Student’s t test (F). *, P < 0.05 and ***, P < 0.001 by two-tailed Student’s t test in comparison with Col-0 (K). Bars = 1.0cm (J). Error bars represent mean ± sd, with n = 14 (K).
Mutations in CESA6 Cause Reduced Sensitivity to ES20
To identify the cellular and molecular pathways that are targeted by ES20, we performed a chemical genetic screen for mutants with reduced sensitivity to this growth inhibitor. We screened ∼500,000 M2 seedlings from ethyl methanesulfonate (EMS)–mutagenized populations on 5 μM ES20 and identified seedlings with longer roots than control plants that were not mutagenized. After re-testing the M3 generation, we confirmed a total of 45 individual lines with reduced sensitivity to ES20 in terms of growth. We refer to these as ES20 resistant (es20r) mutants. After high-throughput, whole-genome sequencing of pooled seedlings from mapping populations and sequencing the candidate locus in homozygous plants, we found that some of these individual lines carried the same mutation. We obtained 15 different mutant alleles carrying either C-T or G-A missense mutations in CESA6: these mutants were named es20r1 to es20r15 (Supplemental Table 1). We backcrossed the mutants and consistently detected reduced sensitivity of these mutants to ES20 in terms of root growth in the light (Figure 1H; Supplemental Figures 1I to 1M) and hypocotyl growth in the dark (Supplemental Figures 1N and 1O).
To confirm that mutations in CESA6 caused reduced sensitivity to ES20, we performed genetic complementation experiments. The CESA6 genomic sequence was cloned, and a yellow fluorescent protein (YFP) tag was introduced at the 5′ end of the coding region to generate CESA6 with an N-terminal YFP fusion. We used this wild-type YFP-CESA6 construct as a template and performed site-directed mutagenesis to create 10 additional clones of YFP-CESA6, each carrying one of the mutations found in the es20r mutants. We transformed each wild-type and mutated YFP-CESA6 construct into the loss-of-function cesa6 allele procuste 1-1 (prc1-1; Fagard et al., 2000). After recovering homozygous transformants with a single CESA6 construct insertion, we found that the wild-type CESA6 construct rescued the growth phenotypes of prc1-1 in roots and hypocotyls and that the mutated CESA6 constructs fully or partially rescued the growth phenotype of prc1-1, depending on the mutation (Figure 1I; Supplemental Figure 2). Transgenic lines carrying any of the 10 mutated CESA6 constructs, but not the wild-type CESA6 construct, exhibited reduced sensitivity to ES20 in terms of both root and hypocotyl growth (Figure 1I; Supplemental Figure 2). These genetic complementation experiments confirmed the notion that missense mutations in CESA6 are sufficient to cause reduced sensitivity to ES20.
We also tested the growth of prc1-1 in the presence or absence of ES20 and found that prc1-1 was more sensitive to ES20 treatment than the wild-type seedlings (Figures 1J and 1K; Supplemental Data Set). The increased sensitivity of prc1-1 to ES20 suggests that ES20 could target CESA2 and CESA5, which function redundantly with CESA6 (Desprez et al., 2007), as well as other CESAs. We hypothesized that missense mutations at conserved amino acids in other CESAs in Arabidopsis might lead to reduced sensitivity to ES20. fragile fiber5 (fra5) carries a missense mutation at Pro-557 of Arabidopsis CESA7 (CESA7P557T; Zhong et al., 2003), which is homologous to the conserved Pro-595 in CESA6. Another mutant, anisotropy1 (any1), carries a missense mutation at Asp-604 of Arabidopsis CESA1 (CESA1D604N; Fujita et al., 2013), which is homologous to the conserved Asp-605 in CESA6. We obtained fra5 and any1 and tested their responses to ES20 in growth assays. As reported previously, both fra5 and any1 have shorter roots than the wild type (Figures 1J and 1K), suggesting that CESA7 and CESA1 play a role in normal seedling growth, although CESA7 is thought to be mainly involved in secondary cell wall synthesis. When a lower concentration of ES20 (0.5 μM) was tested, the root growth of fra5 and any1 was inhibited by ES20 at a reduced level compared to wild-type plants (Figures 1J and 1K). The reduced sensitivity to ES20 in CESA7 and CESA1 mutants with altered amino acids homologous to our mutants in CESA6 indicates that reduced sensitivity to ES20 is not unique to CESA6 and that it occurs with other CESAs.
ES20 Targets the Catalytic Site of CESA6
To understand how multiple mutations at CESA6 cause reduced sensitivity to ES20, we used different programs to predict the topology of CESA6 proteins to identify the locations of these mutations. We found that different programs predicted different numbers of transmembrane regions and orientations of the CESA6 C terminus. Predictions with UniProt showed that CESA6 contains eight transmembrane regions and that both the N and C termini are located in the cytoplasm (Supplemental Figure 3). Predictions with PredictProtein revealed seven transmembrane regions, with the N terminus of CESA6 located in the cytoplasm and the C terminus located in the apoplast (Figure 2A).
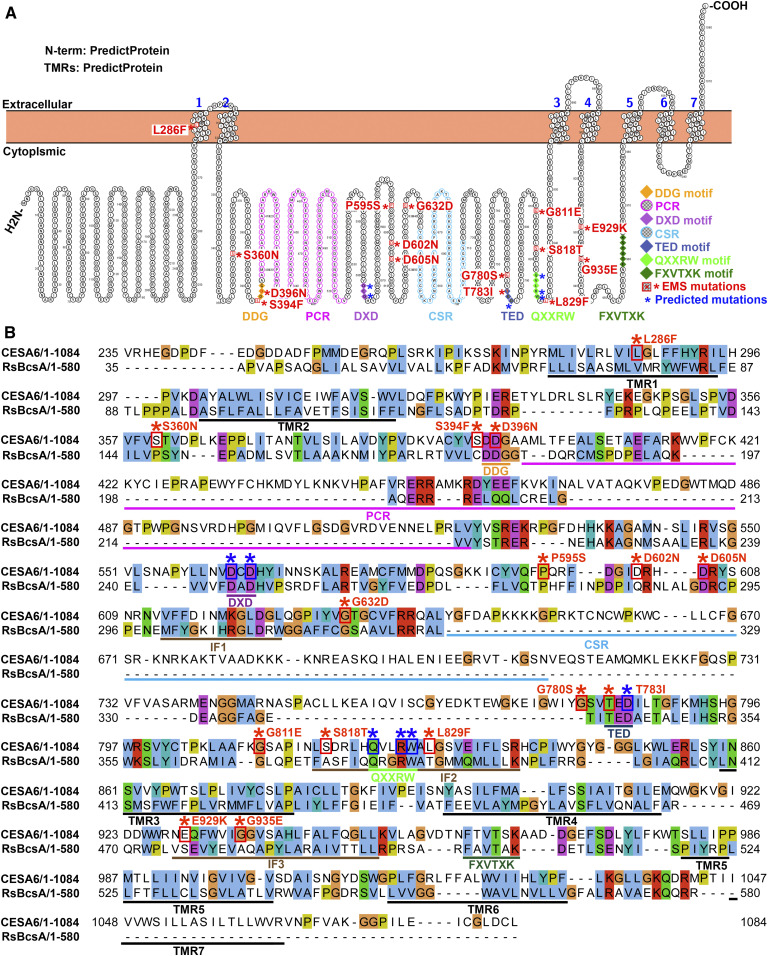
Figure 2.
Mutations in CESA6 That Cause Reduced Sensitivity to ES20 Are Mainly Clustered in the Central Cytoplasmic Domain.
(A) Predicted topology of CESA6, location of key motifs for cellulose catalytic synthesis, and locations of the mutated amino acids that cause reduced sensitivity to ES20. The orientation of the CESA6 N terminus, the transmembrane regions (TMRs), and the orientation of the cytoplasmic domains are based on predictions by PredictProtein.
(B) Sequence alignment of CESA6 with RsBcsA. Key motifs, including the PCR, CSR, IFs, and transmembrane regions (TMRs) are underlined. The EMS mutants are highlighted with red star (*) above the amino acids and red boxes around the amino acids. The predicted mutations that cause reduced sensitivity to ES20 are marked by blue stars (*) and boxes.
Next, we aligned the primary sequence of CESA6 with CESA1, CESA3, CESA7, and RsBcsA. We found that 12 of the mutations led to missense mutations in amino acids located within the large central cytoplasmic domain and one at the first transmembrane region (Figure 2; Supplemental Figures 3 and 4). UniProt and PredictProtein provided different predictions for the locations of missense mutations in amino acids E929K and G935E (Figure 2; Supplemental Figure 3). In addition, all the amino acids that were mutated in the es20r mutants are conserved among Arabidopsis CESAs and six are conserved with RsBcsA (Supplemental Figure 4). Some of the mutations occurred at amino acids that are part of, or very close to, the conserved catalytic motifs. For example, Ser-394 is adjacent to the DDG motif, Asp-396 is part of the DDG motif, Gly-780 is two amino acids away from the TED motif, Thr-783 is part of the TED motif, Leu-829 is one amino acid away from the QXXRW motif, and Ser-818 is four amino acids away from the QXXRW motif (Figure 2; Supplemental Figures 3 and 4). Mutation L286F occurs at the first transmembrane region (Figure 2). P595S, D602N, D605N, and G632D are very close to IF1, although they are not mapped to known critical motifs (Figure 2). The mutations E929K and G935E are beyond the central cytoplasmic domain, but these two amino acids are located at the IF3, which is located at the cytoplasm and facilitates glucan translocation in RsBcsA (Figure 2B; Morgan et al., 2013). E929K and G935E are also close to the FXVTXK motif, which is part of the gating loop that is also located at the cytoplasm in RsBcsA (Figure 2; Morgan et al., 2013). The topology prediction using UniProt places the FXVTXK motif in the apoplast, Glu-929 in the small cytoplasmic loop between the fourth and the fifth transmembrane regions, and Gly-935 in the fifth transmembrane region (Supplemental Figure 3). The topology prediction using PredictProtein places the FXVTXK motif, Glu-929, and Gly-935 in the cytoplasm (Figure 2). Since the IF3 and the gating loop containing the FXVTXK motif in RsBcsA are located in the cytoplasm, it is more likely that the prediction by PredictProtein (Figure 2) reflects the real topology of plant CESAs. None of the es20r mutations occurred in the PCR or CSR of CESA6. Altogether, the predicted locations of amino acids that are mutated in the es20r mutants suggest that many of them might affect the catalytic process.
To understand how mutations at conserved amino acids in CESAs affect plant sensitivity to ES20, we used a threading method to model the structure of the central cytoplasmic domain of Arabidopsis CESA6 using the solved crystal structure of RsBcsA as a guide (Morgan et al., 2013, 2016). The modeled structure of the cytoplasmic domain of CESA6 contains multiple α helices and a β sheet folded into a globular structure with a central cavity (Figure 3A; Supplemental Movie 1). We evaluated the quality of the model with PROCHECK (Laskowski et al., 1993, 1996). In the Ramachandran plot, which visualizes energetically allowed regions for backbone dihedral angles ψ against φ of amino acid residues, 67.2% of residues were in the most favored regions and 23.4% of residues were in the additional allowed regions. Ligand binding site prediction enabled by COACH identified UDP-Glc phosphonate as a possible ligand for the modeled structure of CESA6 (Yang et al., 2013a). Our modeled CESA6 cytoplasmic domain structure is very similar to that predicted for the cotton CESA1 cytoplasmic domain, and the catalytic core conformation is very similar to that of RsBcsA (Figure 3A; Supplemental Movie 1; Morgan et al., 2013; Sethaphong et al., 2013). In our modeled structure, the DDG, DCD, TED, and QLVRW motifs form the catalytic core around the UDP-Glc, and the PCR and CSR extend away from the catalytic core (Figure 3A; Supplemental Movie 1). We used a molecular docking approach to predict possible binding sites for ES20 on the modeled CESA6 central cytoplasmic domain. We found that ES20 and UDP-Glc phosphonate were docked to the same catalytic core of the modeled CESA6 cytoplasmic domain (Figure 3A; Supplemental Movie 1).
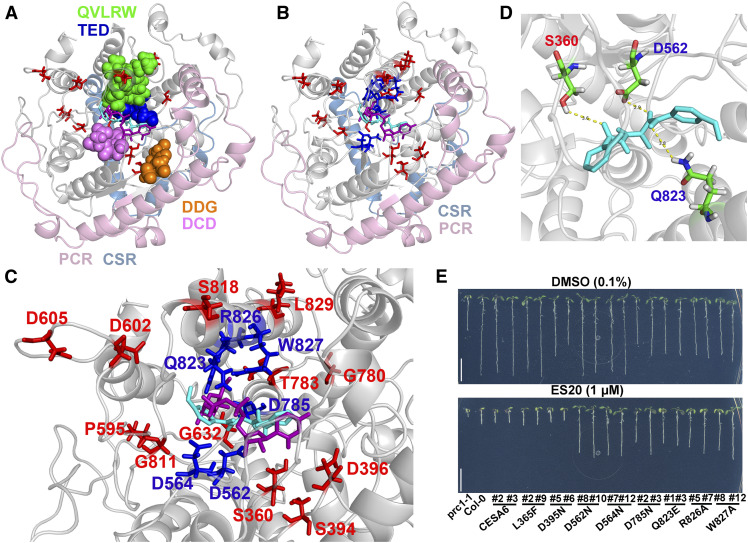
Figure 3.
ES20 Targets the Catalytic Site of CESA6.
(A) Superposition of ES20 (cyan) and UDP-Glc phosphonate (magenta) on the predicted binding pocket of the modeled CESA6 large cytosolic domain (amino acids 322 to 868). The amino acids that were mutated in our EMS mutants are shown as sticks (red). The DDG (amino acids 395 to 397; orange), DCD (amino acids 562 to 564; purple), TED (amino acids 783 to 785; blue), and QVLRW (amino acids 823 to 827; green) motifs are shown as spheres. The PCRs and CSRs are shown in light pink and light blue, respectively.
(B) Superposition of ES20 (cyan) and UDP-Glc phosphonate (magenta) on the predicted binding pocket of the modeled CESA6 large cytosolic domain. The amino acids that were mutated in our EMS mutants (red) and the predicted amino acids (blue) that caused reduced sensitivity to ES20 when mutated are shown as sticks.
(C) Magnified view (of [B]) of the predicted binding pocket for ES20 (cyan), UDP-Glc phosphonate (magenta), and amino acids that were required for ES20 sensitivity (red and blue).
(D) Hydrogen bonds that were predicted to form between ES20 and Ser-360, Asp-562, and Gln-823 of CESA6. ES20 is shown as sticks and colored in cyan.
(E) Mutation of six amino acids at the predicted binding site caused reduced sensitivity to ES20. The genomic construct for CESA6L365F completely rescued the growth of prc1-1, whereas CESA6D395N partially rescued its growth compared with the wild-type CESA6 construct. Transgenic plants expressing CESA6L365F and CESA6D395N had similar levels of sensitivity to ES20 as those expressing the wild-type CESA6. The genomic constructs of CESA6D562N, CESA6D564N, CESA6D785N, CESA6Q823E, CESA6R826A, and CESA6W827A rescued the growth of prc1-1 to different extents and led to reduced sensitivity to ES20 in transgenic plants.
When we examined the three-dimensional positions of mutated amino acids identified in the es20r mutants, we found that most of these amino acids were either directly located at or very close to the predicted binding site for ES20 and UDP-Glc (Figures 3A to 3C; Supplemental Movies 1 and 2). After further analysis of the docking results, we found that three amino acids, Ser-360, Asp-562, and Gln-823, were close to ES20 and that hydrogen bonds could form between ES20 and these amino acids (Figure 3D). The structural modeling and molecular docking data in combination with the chemical genetics results suggest that ES20 targets the catalytic sites of CESAs to inhibit plant cellulose synthesis and cell growth.
To further validate our structural model and molecular docking data, we hypothesized that if we mutated other amino acids in the predicted binding site, the plants should have reduced sensitivity to ES20. We selected six amino acids that were located at the predicted ES20 and UDP-Glc binding site on CESA6 and created six YFP-CESA6 genomic constructs that each carried a missense mutation in one of these six amino acids (Figures 2, 3B, and 3C; Supplemental Movie 2, amino acids colored blue). The selected amino acids are part of the DXD, TED, and QXXRW motifs, and Asp-562 and Gln-823 are predicted to be important for the interaction of ES20 with CESA6. We also selected Leu-365 and Asp-395, which are not part of the predicted UDP-Glc and ES20 binding site, and created YFP-CESA6 genomic constructs that each carried a missense mutation in one of the two amino acids. We then used these constructs to transform prc1-1 and obtained single insertion transgenic lines expressing each of the mutated CESA6 containing a predicted missense mutation.
In the absence of ES20, transgenic plants expressing the wild-type CESA6 construct had normal root and hypocotyl growth compared to the wild-type controls, whereas the transgenic plants expressing the mutated CESA6 had different levels of growth defects, depending on the mutation (Figure 3E; Supplemental Figures 5A to 5E). We analyzed YFP-CESA6 protein levels in transgenic lines expressing wild-type or mutated YFP-CESA6 and found that the severity of growth defects was not correlated with the protein level (Supplemental Figures 5F and 5G; Supplemental Methods). In the presence of ES20, transgenic plants expressing wild-type CESA6 constructs had similar sensitivity to ES20 in terms of root and hypocotyl growth compared to wild-type plants (Figure 3E; Supplemental Figures 5A to 5E). The transgenic plants expressing six mutated CESA6 constructs in amino acids (D562N, D564N, D785N, Q823E, R826A, and W827A) at the predicted UDP-Glc and ES20 binding site showed reduced sensitivity to ES20 in terms of both root and hypocotyl growth (Figure 3E; Supplemental Figures 5A to 5E), suggesting that these six amino acids are important for the inhibitory effect of ES20. The transgenic plants expressing YFP-CESA6 carrying mutations at the two amino acids (L365F and D395N) that were not part of the predicted UDP-Glc and ES20 binding site had the same sensitivity to ES20 as wild-type YFP-CESA6, indicating that these two amino acids are not essential for the inhibitory effect of ES20 (Figure 3E; Supplemental Figures 5A to 5E). Transgenic lines expressing CESA6L365F showed similar growth rates and similar sensitivity to ES20 in roots and hypocotyls as wild-type plants (Figure 3E; Supplemental Figures 5A to 5E), suggesting this amino acid is not critical for plant growth or the inhibitory activity of ES20. The construct CESA6D395N partially rescued the growth of prc1-1, and the transgenic plants had normal sensitivity to ES20 (Figure 3E; Supplemental Figures 5A to 5E), suggesting that Asp-395 is required for cell growth, but not for the inhibitory activity of ES20. Asp-395 and Asp-396 are both part of the DDG motif (Figure 2B). However, Asp-396 is within 4 Å of UDP-Glc and Asp-395 is not within 4 Å of UDP-Glc based on our molecular docking analysis (Supplemental Movie 1). It is interesting that D396N causes reduced sensitivity to ES20, but not D395N. The finding that the six mutations (predicted based on the modeled structure and molecular docking) caused reduced sensitivity to ES20 in terms of plant growth provides additional evidence that ES20 targets the catalytic site of CESA6.
To determine whether ES20 targets CESA6 directly, we performed several biochemical assays. The drug affinity responsive target stability (DARTS) assay detects small molecule and protein interactions by testing whether the small molecule protects the protein from degradation by proteases (Lomenick et al., 2009; Zhang et al., 2016). We isolated total proteins from YFP-CESA6 transgenic seedlings, incubated the proteins with either ES20 or DMSO (as a control), and digested them with pronase. We used antiGFP antibody to detect the abundance of YFP-CESA6 after pronase digestion. ES20 significantly protected YFP-CESA6 from degradation by pronase (Figures 4A and 4B; Supplemental Data Set), whereas the control molecule, ampicillin, did not protect YFP-CESA6 from degradation (Figures 4C and 4D; Supplemental Data Set). The finding that ES20 protected YFP-CESA6 from degradation by proteases suggests that ES20 and YFP-CESA6 physically interact. To test whether the mutations in our ES20-resistant mutants affect the interaction between CESA6 and ES20, we performed a DARTS assay using ES20 and total proteins isolated from YFP-CESA6P595S seedlings. We chose YFP-CESA6P595S for the test because esr20-10 (CESA6P595S) showed strong resistance to ES20 treatment in terms of growth (Figure 1H; Supplemental Figures 1I to 1O). ES20 did not protect YFP-CESA6P595S from degradation by pronase (Figures 4E and 4F; Supplemental Data Set), indicating that Pro-595 is important for the interaction between ES20 and CESA6.
Figure 4.
ES20 Directly Interacts with CESA6.
(A) to (F) ES20 interacts with CESA6, but not CESA6P595S, in a DARTS assay. Representative protein immunoblots of DARTS assays for YFP-CESA6 with ES20 (A), YFP-CESA6 with ampicillin (C), and YFP-CESA6P595S with ES20 (E). Quantitative analysis of DARTS assays for YFP-CESA6 with ES20 (B), YFP-CESA6 with ampicillin (D), and YFP-CESA6P595S with ES20 (F). ES20 protects YFP-CESA6, but not YFP-CESA6P595S from degradation by proteases.
(G) to (I) Central cytoplasmic domain of CESA6 interacts with ES20 and UDP-Glc in an MST assay. Purified GFP-tagged CESA6 central cytoplasmic domain (GFP-CESA6c) with a His-SUMO tag (see [G], lane 2). Thermophoresis binding curve showing a direct interaction between GFP-CESA6c and ES20 (H). Thermophoresis binding curve showing a direct interaction between GFP-CESA6c and UDP-Glc (I). FNorm, normalized fluorescence .
(J) to (L) Central cytoplasmic domain of CESA6P595S interacts with ES20 and UDP-Glc in an MST assay. Purified GFP-CESA6P595Sc with a His-SUMO tag (see [J], lane 2). Thermophoresis binding curve showing a direct interaction between GFP-CESA6P595Sc and ES20 (K). Thermophoresis binding curve showing a direct interaction between GFP-CESA6P595Sc and UDP-Glc (L). FNorm, normalized fluorescence .
(M) and (N) UDP-Glc treatment partially complemented the root swelling caused by ES20. Representative images of seedlings treated with DMSO (0.1%), ES20 (0.8 μM), UDP-Glc (1 mM), and ES20 (0.8 μM) + UDP-Glc (1 mM; see [M]). Quantification of root width (see [N]) at the elongation zone of seedlings with different treatments as shown in (M). The letters (a to c) in (N) indicate statistically significant differences (P < 0.05) determined by one-way analysis of variance tests followed by Tukey’s multiple comparison tests in different samples. Values are means ± sd, with n = 6 in (B), (D), and (F); n = 3 in (H), (I), (K), and (L); and n = 16 in (N). In (B), *, P < 0.05 and **, P < 0.01, by two-tailed Student’s t test. Bar in (M) = 100 μm.
To confirm these direct interactions, we purified the central cytoplasmic domain of CESA6 (amino acids 322 to 868) with a GFP tag (GFP-CESA6c; Figure 4G) and used it in a microscale thermophoresis (MST) assay (Wienken et al., 2010; Jerabek-Willemsen et al., 2011) to detect interactions between ES20 and GFP-CESA6c. MST detects the movement of biomolecules as a function of ligands in the presence of a temperature gradient. The thermophoresis of a protein–ligand complex often differs from that of a protein alone due to binding-induced changes in size, charge, and solvation energy (Wienken et al., 2010; Jerabek-Willemsen et al., 2011). We detected a direct interaction between ES20 and GFP-CESA6c from the thermophoresis binding curve (Figure 4H). We also performed an MST assay to detect a possible interaction between GFP-CESA6c and UDP-Glc. The thermophoresis binding curve indicates a direct interaction between GFP-CESA6c and UDP-Glc (Figure 4I). Similar binding curves have been reported for MST assays in studies of protein–protein interactions or protein–ligand interactions (Chen et al., 2017; Liu et al., 2017; Kosmacz et al., 2018; Yan et al., 2018; Zhai et al., 2018; Gerrits et al., 2019; Stepek et al., 2019; Warren et al., 2019). However, we did not detect interactions between ES20 and GFP or between ampicillin and GFP-CESA6c (Supplemental Figure 6).
We also purified the central cytoplasmic domain of CESA6 carrying P595S mutation (CESA6P595Sc; Figure 4J) and performed an MST assay to test the interaction of CESA6P595Sc with ES20 and UDP-Glc. As indicated by the thermophoresis binding curve, CESA6P595Sc interacted with both ES20 and UDP-Glc (Figures 4K and 4L). The results of DARTS and MST assays confirm the notion that ES20 directly interacts with CESA6. The P595S mutation abolished the interaction between CESA6 and ES20 in a DARTS assay using total endogenous protein, but recombinant CESA6P595Sc can still interact with ES20. It is likely that the results of the DARTS and MST assays are not directly comparable because the DARTS assay used endogenous CESA6 protein, which is part of the protein complex in the lipid bilayer environment, whereas the recombinant CESA6 cytoplasmic domain did not contain the transmembrane regions. Perhaps we obtained different results for the interaction of CESA6P595S with ES20 using the DARTS and MST assays because the purified cytoplasmic domain could not fold into exactly the same conformation as the endogenous full-length CESA6 in the lipid bilayers of cells.
To investigate whether ES20 competes with UDP-Glc for the catalytic site, we examined whether externally supplied UDP-Glc could compensate for the inhibitory effects of ES20. Exogenous UDP-Glc rescued the male fertility defects of UDP-Glc deficiency mutants and reversed the inhibitory effect of an UGPase/USPase inhibitor in pollen (Park et al., 2010; Decker et al., 2017), suggesting that this is an effective form of delivery. We cotreated the wild-type seedlings with ES20 (0.8 μM) and UDP-Glc (1 mM) and used DMSO (0.1% [v/v]), UDP-Glc (1 mM), and ES20 (0.8 μM) alone as control treatments. After overnight incubation, exogenous treatment with UDP-Glc did not completely reverse the effects of ES20, as seedlings cotreated with 1 mM UDP-Glc and 0.8 μM ES20 still had significantly swollen roots (Figures 4M and 4N). However, we consistently detected statistically significant differences in root width between seedlings treated with 0.8 μM ES20 alone and seedlings cotreated with 1 mM UDP-Glc and 0.8 μM ES20 (Figures 4M and 4N; Supplemental Data Set). The findings that UDP-Glc compensated for the inhibitory effect of ES20 are consistent with our modeling results indicating that ES20 targets CESA6 at the catalytic site.
Inhibiting CESA Catalytic Activity by ES20 Treatment Reduces the Delivery of CSCs to the PM
To investigate whether the catalytic activity of plant CESA influences intracellular trafficking, we treated seedlings expressing YFP-CESA6 with ES20 and examined CSC localization and dynamics. Functional fluorescence-tagged CESAs primarily localize to the Golgi, PM, and small CESA compartments (SmaCCs; Paredez et al., 2006; Crowell et al., 2009; Gutierrez et al., 2009; Zhang et al., 2019). At the PM, CSCs translocate along cortical microtubules with a velocity that is dependent upon catalytic activity (Paredez et al., 2006; Gutierrez et al., 2009; Fujita et al., 2013; Morgan et al., 2013). To confirm that ES20 inhibits the synthesis of β-1,4-glucan, we treated Arabidopsis seedlings expressing YFP-CESA6 in the prc1-1 background with 6 µM ES20 or 0.1% DMSO for 30 min and imaged the PM of root epidermal cells by spinning disk confocal microscopy (SDCM; Figures 5A to 5C), as described previously (Zhang et al., 2019). Time projections from 5-min time-lapse series showed linear tracks in mock-treated cells, whereas ES20-treated cells had fewer tracks (Figure 5A). By analyzing kymographs from multiple cells and roots, we found that ES20 treatment significantly reduced the rate of CSC motility to 74 ± 36 nm/min compared to 137 ± 65 nm/min for mock-treated cells (Figures 5B and 5C). This reduced CSC velocity after ES20 treatment is consistent with our finding (from molecular docking analysis) that ES20 inhibits cellulose polymerization.
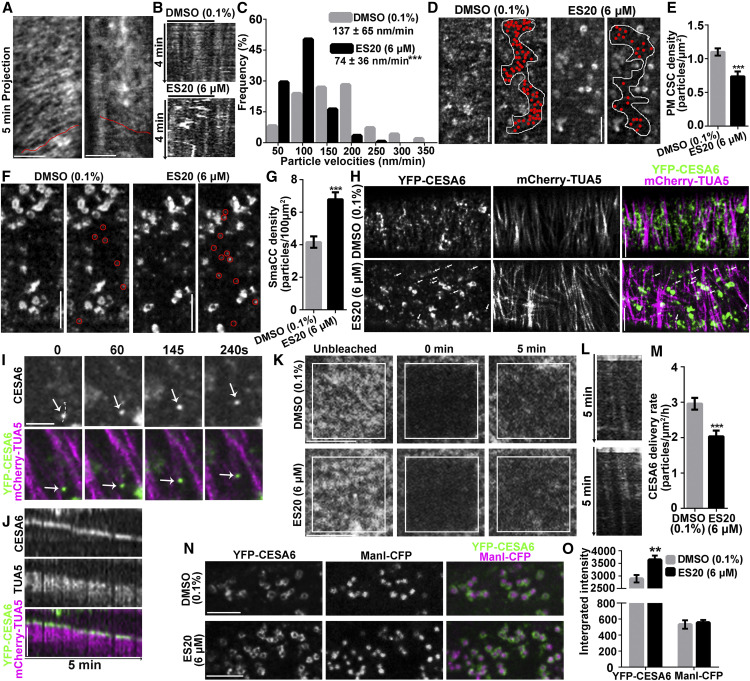
Figure 5.
ES20 Disrupts Trafficking of CSCs to the PM.
(A) to (C) ES20 reduces the velocity of CSCs at the PM. Representative time projections using average intensity images from a time-lapse series of YFP-CESA6 particles in root epidermal cells (A). Kymographs (B) of the trajectories marked in (A). Histogram with the frequencies of YFP-CESA6 particle velocity after treatment with 0.1% DMSO or 6 μM ES20 for 30 min (C). Data in the chart represent mean ± sd (n = 320 CSC trajectories from 18 seedlings per treatment).
(D) and (E) ES20 treatment reduces the abundance of PM-localized YFP-CESA6 in root epidermal cells. Representative images (D) and quantification (E) of PM-localized YFP-CESA6 in root epidermal cells after 0.1% DMSO or 6 μM ES20 treatment are shown. Data represent means ± se (n = 20 cells from 10 seedlings).
(F) and (G) Density of cortical SmaCCs, as indicated by red circles, increased in response to ES20 treatment (30 min). Data represent means ± se (n = 20 cells from 10 seedlings per treatment).
(H) CSCs were depleted from the PM after treatment with 6 μM ES20 for 2 h, whereas microtubule-associated CESA compartments accumulated, as indicated by white arrows.
(I) Magnified view of the association of a CESA compartment (arrows) with microtubules in time-course image after 6 μM ES20 treatment for 2 h.
(J) Kymograph showing the association of the CESA compartment with the microtubules as shown in (I).
(K) to (M) ES20 reduces the delivery rate of CSCs to PM in root epidermal cells. Representative images of CSCs at the PM during FRAP analysis (K). Representative kymographs of the trajectories of newly delivered CSCs after photobleaching (L). Quantification of CSC delivery rates (M) based on the FRAP assays described in (K). Data represent means ± se (n = 18 ROIs from 15 seedlings).
(N) and (O) ES20 increases the abundance of CSC at the Golgi. Representative images of Golgi-localized YFP-CESA6 and ManI-CFP after 0.1% DMSO (top) or 6 μM ES20 (bottom) treatment for 1 h (N). Quantification of integrated fluorescence intensity of Golgi-localized CSCs and ManI (O) as described in (N). Data represent means ± se (n = 60 from 14 seedlings). **, P < 0.01 and ***, P < 0.001 by two-tailed Student’s t test. Bar = 5 μm.
To quantify the trafficking and dynamics of CSCs within and between compartments, we performed both static and time-lapse analyses of YFP-CESA6 localization by collecting three- and four-dimensional stacks of images from epidermal cells in the root elongation zone by SDCM. After 30 min of 6 μM ES20 treatment, the density of PM-localized CSCs was reduced from 1.1 ± 0.1 to 0.7 ± 0.1 particles/μm2 (Figures 5D and 5E; Supplemental Data Set). In normal growing cells, some CSCs localize to motile SmaCCs in the cortical cytoplasm, and their abundance rapidly increases when secretion is inhibited (Crowell et al., 2009; Gutierrez et al., 2009; Zhang et al., 2019). The exact identity and function of these SmaCCs are not well understood, but they partially overlap with trans-Golgi network proteins and are major vesicle compartments associated with CSC delivery to the PM (Gutierrez et al., 2009; Zhang et al., 2019). We found that the abundance of cortical SmaCCs significantly increased after 30 min of 6 μM ES20 treatment, from 4.2 ± 0.4 to 6.8 ± 0.4 particles/100 μm2 (Figures 5F and 5G; Supplemental Data Set). When we extended the treatment to 2 h with 6 μM ES20, PM-localized CSCs were completely depleted from the PM, and the abundance of CESA compartments associated with microtubules in the cortical cytoplasm increased (Figures 5H to 5J; Supplemental Movie 3). The reduced CSC density at the PM and increased CESA6 abundance in a population of cortical SmaCCs following ES20 treatment suggest that ES20 affects CSC delivery to the PM.
Finally, we examined the effects of ES20 on CSC delivery to the PM using a fluorescence recovery after photobleaching (FRAP) assay (Gutierrez et al., 2009; Zhang et al., 2019). We mounted YFP-CESA6 seedlings in medium supplemented with 0.1% DMSO or 6 μM ES20 and photobleached a small region of interest on the PM of root epidermal cells. We examined the delivery of new CSCs to the bleached region using time-lapse SDCM imaging. After a 5-min acute treatment with ES20, the delivery rate of CSCs decreased from 3.0 ± 0.2 to 2.0 ± 0.2 particles/μm2/h (Figures 5K to 5M; Supplemental Data Set). The results from the FRAP assay indicate that ES20 reduces the efficiency of CSC delivery to the PM. Consistent with the observation of reduced CSC delivery to the PM, after 1 h of 6 μM ES20 treatment, the fluorescence intensity of YFP-CESA6 at the Golgi increased by more than 20% compared to the DMSO control, from 2890 ± 147 to 3642 ± 169 (Figures 5N and 5O; Supplemental Data Set). By contrast, the fluorescence intensity of Mannosidase I tagged with cyan fluorescence protein (ManI-CFP), another Golgi-localized protein expressed in the same cells, was not affected by ES20 treatment (Figures 5N and 5O). These results indicating that inhibiting CESA6 catalytic activity by ES20 treatment reduces CSC trafficking dynamics, which is consistent with the previous observation that mutations at the catalytic site affect CSC trafficking (Park et al., 2019).
DISCUSSION
Because of the importance of cellulose to agriculture and industry, understanding the mechanisms of cellulose biosynthesis has been one of the most important topics in biology. Furthermore, small molecule inhibitors that allow transient manipulation of cellulose biosynthesis are valuable tools for biological research. Here, we identified a CESA inhibitor that targets the catalytic site of Arabidopsis CESA6. ES20 is likely to have a different target site than other CESA inhibitors such as isoxaben or C17, because the mutations in CESAs that lead to reduced sensitivity to these inhibitors are very different (Scheible et al., 2001; Desprez et al., 2002; Hu et al., 2016, 2019). From our mutant screen, we only identified mutants for CESA6, but no other CESAs that were resistant to ES20. However, we found that comparable mutants in CESA1 (any1, CESA1D604N) and CESA7 (fra5, CESA7P557T) also have reduced sensitivity to inhibition by ES20, although a lower dosage of ES20 was required to observe a significant resistant phenotype in any1 and fra5. The reduced sensitivity of any1 and fra5 to ES20 suggests that ES20 might target CESA1 and CESA7, as well as other CESAs.
We noticed that mutants in other CESAs might have stronger growth phenotypes than cesa6 mutants; for example, any1 has a stronger root growth phenotype than es20r4 (CESA6D605N). It is possible that ES20 targets multiple CESAs, but we could not identify mutants in other CESAs because the dosage of ES20 (5 μM) we used for the screening was too high to allow us to identify those mutants. CESA7 mainly functions in secondary cell wall synthesis (Gardiner et al., 2003; Taylor et al., 2003; Brown et al., 2005). However, we found that fra5 had significantly reduced root growth at the young seedling stage and modestly reduced sensitivity to ES20, indicating that CESA7 functions in young seedling growth as well. However, we cannot rule out the possibility that CESA6 holds a special position in the CSC rosette that allows ES20 to target CESA6 more efficiently to affect the entire protein complex during cellulose synthesis. We expect that further characterization of the specificity of ES20 for different CESAs in Arabidopsis, other plants, and other kingdoms such as bacteria and oomycetes will be required for better use of ES20 as a general CESA inhibitor. Based on our current results, ES20 can be used as a CESA6 inhibitor in Arabidopsis to explore the molecular mechanisms of cellulose catalytic synthesis and the integration between cellulose catalytic synthesis and CSC dynamic behaviors.
Plant CESAs contain multiple putative transmembrane regions that allow the protein complex to insert into a lipid bilayer and form a glucan-translocating channel for cellulose translocation across the PM. It was originally predicted that plant CESAs contain eight transmembrane regions and that both the N and C termini are located at the cytoplasm, as predicted by UniProt (Supplemental Figure 3; Pear et al., 1996). This eight-transmembrane-region model has received the most discussion in numerous literature reviews (McFarlane et al., 2014; Kumar and Turner, 2015; Turner and Kumar, 2018; Polko and Kieber, 2019; Zhong et al., 2019). However, computational and genetic analyses indicated a possible alternative model for the topology of CESAs, especially with respect to the originally predicted fifth and sixth transmembrane regions (Slabaugh et al., 2014a, 2014b; McNamara et al., 2015). We identified two mutations (E929K and G935E) that are part of the IF3 and very close to the FXVTXK motif that cause reduced sensitivity to ES20 treatment. Although we were not able to include these amino acids in structural modeling and molecular docking analyses, the finding that plants with these mutations had reduced sensitivity to ES20 suggests that these amino acids are located in the cytoplasm, participate in substrate coordination, or facilitate glycosyl transfer, as shown by the topology generated using PredictProtein (Figure 2A). The L286F mutation was predicted to be located at the first transmembrane region by both the UniProt and PredictProtein programs. The reduced sensitivity to ES20 caused by this mutation suggests that this Leu-286 may be involved in the transmembrane translocation of cellulose. Mutations P595S, D602N, D605N, and G632D are very close to IF1, but are not part of known critical motifs, suggesting there might be an undiscovered role for these amino acids in plant cellulose synthesis. Interestingly, we did not uncover any mutations in the PCR or CSR in the es20r mutant collection. Structural modeling indicated that these two regions are located outside of the core catalytic site, suggesting they might play other roles related to CSC function, such as the interactions between adjacent CESAs in the rosette.
Years of study have found that microtubules, actomyosin, vesicle-trafficking machineries, and CESA-interacting proteins play important roles in precisely controlling CSC trafficking and cellulose biosynthesis. Here, quantitative live cell imaging of wild-type YFP-CESA6 treated with ES20 indicated that inhibiting CESA6 catalytic activity inhibits the efficient delivery of CSCs to the PM. The finding that ES20 reduced the delivery of CSCs suggests that the catalytic site might contain structural information that is important for the recognition of CSCs by other proteins that regulate CSC delivery. Alternatively, it is also possible that the feedback from inhibited cellulose synthesis leads to reduced CSC delivery. Previous observation of the different effects of the mutations at the catalytic site on CSC trafficking (Park et al., 2019) suggested that the relationship between CESA catalytic activity and CSC trafficking is complicated and requires more detailed live cell imaging and quantitative image analysis using fluorescence-tagged CESA with point mutations. We expect that ES20 and a collection of fluorescence-tagged CESA6 with mutations at the catalytic site will be useful tools for exploring the effect of CESA catalytic activity on CSC trafficking and the underlying molecular mechanisms.
METHODS
Plant Materials, Growth Conditions, and Growth Assay
To test the inhibitory effect of ES20 on plant growth, the Arabidopsis (Arabidopsis thaliana) wild-type Col-0 plants were used for analysis. The seeds of plants used for the growth assays or live cell imaging were sequentially sterilized with 50% (v/v) bleach and 75% (v/v) ethanol. After washing with sterilized water, the seeds were sown on half-strength Murashige and Skoog (MS) medium with 1% (w/v) Suc and 0.8% (w/v) agar at pH 5.8. The plants were grown under continuous light of 130 μmol m−2 s−1 intensity illuminated by Philips F25T8/TL841 25-W bulb at 22°C. To quantify the inhibitory effect of ES20 on Arabidopsis root growth, the sterilized wild-type seeds were sown on gridded Petri plates containing half-strength MS medium supplemented with different concentrations of ES20. The plates were placed in vertical orientation in the growth chamber for root measurement. Starting from 3 d after the plates were placed in the growth chamber, the plates were scanned daily using an Epson Perfection V550 scanner. The root lengths of the plants were measured using ImageJ. To test the effect of ES20 on etiolated hypocotyl growth, the sterilized wild-type seeds were sown on half-strength MS medium supplemented with different concentrations of ES20. The Petri dishes were wrapped in two layers of aluminum foil and incubated at 22°C for 7 d. The Petri dishes were then scanned, and the hypocotyl length was measured using ImageJ. ES20 was dissolved in DMSO to obtain a stock solution of 12 mM and stored at –20°C.
To analyze the effect of ES20 treatment on epidermal cell growth from light-grown roots, the 5-d-old wild-type seedlings were treated with 0.1% (v/v) DMSO or 6 μM ES20 for 12 h. The seedlings were stained with 1 μM fluorescein diacetate (ACROS Organics) for 5 min, and the fluorescence in epidermal cells was imaged under a Zeiss 710 laser-scanning confocal microscope equipped with a 20× objective. To analyze the effects of ES20 treatment on hypocotyl cell growth, the 5-d-old wild-type seedlings grown in the dark were stained with 1 μM fluorescein diacetate for 5 min, and the fluorescence of epidermal cells from the middle section of the hypocotyl was imaged under the same conditions used for root epidermal cells.
EMS Mutagenesis and Mutant Screening
To obtain a mutagenized Arabidopsis population, SYP61-CFP and PIN2-GFP seeds were mutagenized following a published protocol (Kim et al., 2006). Mutagenized seeds were sown in soil, and the plants were grown under continuous light and allowed to self, yielding M2 seeds. The M2 seeds were collected as pooled populations. Approximately 400,000 seeds from the M2 generation of the SYP61-CFP population and 100,000 seeds from the PIN2-GFP M2 population were sterilized and sown on medium containing 5 μM ES20. Individual plants with elongated roots and green leaves were transferred to soil to produce the M3 generation. The M3 plants were examined for sensitivity to ES20. Individual M3 lines with reduced sensitivity to ES20 were crossed to the Landsberg erecta ecotype to generate the mapping population and were also crossed to SYP61-CFP or PIN2-GFP to clean up the genetic background.
High-Throughput Genome Sequencing and Sequence Analysis
The seeds from F2 populations of mutants crossed with Landsberg erecta were sown on medium containing 5 μM ES20, and the segregation of resistant seedlings was evaluated. The F2 populations of the outcrosses segregated for sensitivity to ES20. For each mutant, ∼100 F2 seedlings with longer roots on 5 µM ES20 were pooled for DNA isolation. The genomic DNA was subjected to high-throughput sequencing. The resulting DNA sequence was aligned to the Arabidopsis genome (The Arabidopsis Information Resource 10), and single-nucleotide polymorphisms were analyzed. Candidate single-nucleotide polymorphisms for ES20 sensitivity were identified using the next-generation EMS mutation mapping tool (Austin et al., 2011). The ten CESA genes were amplified from homozygous mutant plants and were sequenced to confirm the presence of mutation in CESA6.
Crystalline Cellulose Content Measurement
The wild-type Arabidopsis seeds were sown on medium supplemented with 0.1% (v/v) DMSO or different concentrations of ES20. Following stratification, the plants were grown in the dark for 7 d or in the light for 10 d. The 7-d-old dark-grown seedlings or roots from 10-d-old light-grown seedlings were used for cell wall preparation. Dark-grown seedlings were washed with double-distilled water three times to remove seed coats and any residue from the growth medium and then ground into a fine powder in liquid nitrogen. The roots from light-grown seedlings were cut and washed with double-distilled water to remove any residue from the growth medium and then ground into a fine powder in liquid nitrogen. The powder was extracted twice with 80% ethanol, once with 100% ethanol, once with 1:1 (v/v) methanol:CHCl3, and once with acetone. The resulting insoluble cell wall fraction was dried in a fume hood for 2 d and weighed. Cellulose content was measured by the Updegraff method (Updegraff, 1969; Foster et al., 2010). Briefly, the cell wall material was hydrolyzed with trifluoroacetic acid, followed by Updegraff reagent (acetic acid:nitric acid:water, 8:1:2 [v/v/v]) to yield crystalline cellulose. Crystalline cellulose was hydrolyzed to Glc using 72% (v/v) sulfuric acid. Glc concentration was measured via a colorimetric method by developing color in Anthrone reagent (freshly prepared 2 mg/mL anthrone in concentrated sulfuric acid) and reading OD625 nm in a plate reader (Tecan Infinite 200Pro). Nine repeats were performed for each treatment, including three repeats for cell wall preparation and three repeats for measurement.
Vector Construction and Generation of Transgenic Arabidopsis Plants
To construct the YFP-CESA6 binary vector, a 2245-bp CESA6 promoter fragment was amplified with the primers CESA6P-F TCTGATCCAAGCTCAAGCTAAGCTTTTTCTATTCTATAGTCTTGAAAATT and CESA6P-R ATTTGTCTGAAAACAGACACAG using Col-0 genomic DNA as a template. The YFP tag was amplified from the pUBN-YFP-Dest plasmid with primers YFP-F TGTCTGTTTTCAGACAAATATGGTGAGCAAGGGCGAGG and YFP-R CGACCACCGGTGTTCATCTTGTACAGCTCGTCCATG. CESA6 with terminator was amplified from Col-0 genomic DNA with primers CESA6g-F ATGAACACCGGTGGTCGGTT and CESA6g-R GGTACCCGGGGATCCTCTAGAGTGATCCACATCTTAAATATATTA. The pH7WGR2 plasmid was digested with HindIII and XbaI to remove the 35S promoter and the red fluorescent protein tag. The modified pH7WGR2 linear vector without the 35S promoter and red fluorescent protein tag was ligated with the CESA6 promoter, YFP, and CESA6 genomic sequence using the Gibson Assembly method with a Gibson Assembly Master Mix kit (New England Biolabs). The construct was verified by DNA sequencing. All mutated YFP-CESA6 constructs used the verified YFP-CESA6 plasmid as the template and were obtained using a Q5 Site-Directed Mutagenesis Kit (New England Biolabs) with primers listed in Supplemental Table 2. All of the mutated YFP-CESA6 constructs were verified by DNA sequencing. The verified constructs used to transform prc1-1 (CS297), which was obtained from the Arabidopsis Biological Resource Center, using Agrobacterium tumefaciens–mediated transformation via the floral dip method (Clough and Bent, 1998). The transformants were selected based on the selection marker. Independent transformant lines were further characterized in the T2 generation for the segregation ratio of the selection marker, and only transformants that contained a single insertion for each construct were selected for further analysis. Homozygous transformants with single CESA6 construct insertions were used for mutant phenotype analyses.
Structural Modeling of the CESA6 Cytoplasmic Domain
The general topology of CESA6 was predicted using the UniProt (https://www.uniprot.org) and PredictProtein (https://www.predictprotein.org) servers, and the cartoon was drawn using the Protter program (http://wlab.ethz.ch/protter/start/). The large cytoplasmic domain of the Arabidopsis CESA6 protein sequence (amino acids 322 to 868) was sent to i-TASSER server for three-dimensional structure modeling with the threading method (Roy et al., 2010; Yang et al., 2015). The modeled structure was visualized using PyMol software (Alexander et al., 2011). The binding site of UDP-Glc on the large cytoplasmic domain model of CESA6 was predicted using the COACH server, and the UDP-Glc phosphonate structure was used for the prediction as per the program suggestion (Yang et al., 2013a, 2013b). The small molecule ES20 was docked with the large cytoplasmic domain model of CESA6 using Autodock Vina in PyRx software (Trott and Olson, 2010; Dallakyan and Olson, 2015).
CESA6c Protein Expression and Purification
To obtain the CESA6 central cytosolic domain protein for the MST assay, we inserted the GFP coding sequence into pRSF-Duet-1 vector using the SacI and PstI restriction sites. The GFP coding sequence was amplified from the pUBN-GFP-DEST vector. CESA6c was amplified from Col-0 cDNA into the C terminus of GFP. Primers used for cloning are listed in Supplemental Table 2. The verified recombinant clone was transformed into Escherichia coli BL21 (DE3) competent cells for protein expression. The cells were cultured and grown at 37°C in Luria-Bertani medium until OD600 reached 0.6. Protein expression was induced by overnight treatment with 0.1 mM isopropyl β-d-1-thiogalactopyranoside at 16°C. After overnight induction, the cells were lysed by sonication, and the fusion protein was purified using a HisTrap HP His-tagged protein purification column of the ÄKTA pure fast protein liquid chromatograph system (GE Healthcare). The purified protein was dialyzed overnight and further purified with a HiLoad 16/600 Superdex 200-pg column (GE Healthcare) using the ÄKTA pure fast protein liquid chromatography system. Purified GFP-CESA6c protein was further characterized by SDS-PAGE. The CESA6c construct was used as a template to create the CESA6P595Sc construct by site-directed mutagenesis. CESA6P595Sc protein was purified using the same protocol as CESA6c except that enriched Terrific Broth medium was used to grow the E. coli cells.
MST Assays
The MST assays were performed using a Monolith NT.115 machine (NanoTemper) at the Chemical Genomics Facility, Purdue University. Increasing concentrations of ES20 were titrated against 100 nM GFP-CESA6c protein in standard MST buffer (50 mM Tris, pH 7.5, 150 mM NaCl, 10 mM MgCl2, and 0.05% [v/v] Tween 20). ES20 was dissolved in DMSO, and the final concentration of DMSO was 5% (v/v). MST standard capillaries were used to load the samples into the MST instrument. Triplicate reactions were performed for each test. The MST data were processed using MO.Affinity Analysis version 2.3 software.
DARTS Assays
To test for the interaction between CESA6 and ES20 using the DARTS assay, 7-d-old YFP-CESA6 light-grown seedlings were harvested and then ground to powder in liquid nitrogen. The ground tissue was homogenized in lysis buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 0.5% (v/v) Triton X-100, 2 mM DTT, and 1 tablet/50 mL EDTA-free Pierce protease inhibitor; Thermo Fisher Scientific) at a 2:1 ratio (2 mL of buffer:1 g of tissue). Homogenized samples were transferred to a 2-mL microcentrifuge tube and centrifuged for 30 min (at 20,000g and 4°C). The supernatant was collected after centrifugation and saved as total extracted protein. Extracted total protein (700 μL) was incubated with 0.1% DMSO or 300 μM ES20 at room temperature on an orbital shaker for 1 h. The mixture was divided into six small tubes, each containing 100 μL of the mixture, and incubated with 1 μL of pronase at a 1:300 dilution at room temperature for 30 min. The proteolysis reaction was terminated by adding SDS loading buffer and boiled at 100°C for 6 min. The boiled samples were subjected to SDS-PAGE prior to immunoblot analysis. YFP-CESA6 protein was detected using anti-GFP antibody (catalog no. 632381, Takara), and SEC12 was detected using anti-SEC12 antibody (Bar-Peled and Raikhel, 1997) as a control. Horseradish peroxidase–conjugated secondary antibodies and Clarity Western ECL substrate (Bio-Rad) were further used to detect the presence of YFP-CESA6 and SEC12. The x-ray films were scanned, and the signal intensity of each protein band was quantified after background subtraction using ImageJ. The relative intensities were quantified by dividing the intensities of ES20-treated samples by DMSO-treated samples for each pronase concentration.
UDP-Glc Complementation of the Effect of ES20 on Root Swelling
To determine whether supplementing the plants with UDP-Glc would complement the effect of ES20 treatment, 3.5-d-old Col-0 seedlings grown on half-strength MS agar medium in the light were used for analysis. For each treatment, 16 seedings were transferred from half-strength MS agar plates to 2 mL of half-strength MS liquid medium supplemented with DMSO (0.1% [v/v]), ES20 (0.8 μM), UDP-Glc (1 mM) or ES20 (0.8 μM), and UDPG (1 mM) in a 24-well plate. After 17 h of treatment, the seedlings were mounted between two strips of double-sided tape on a glass slide and covered carefully with a cover slip for image collection under white light using a compound microscope. The width of the root elongation zone for each seedling was quantified by ImageJ.
SDCM Imaging
For SDCM live cell imaging, seedlings were grown vertically for 5 d, and images were taken from the second or third epidermal cell below the first obvious root hair initiated in the root elongation zone. Two thin strips of double-sided adhesive tape were placed on top of glass slides ∼2 cm apart. Next, 100 μL of half-strength MS liquid growth medium containing DMSO or specified concentrations of ES20 was applied to the slide, and seedlings were mounted in the liquid medium. A 22 × 40-mm cover glass was placed on top of the double-sided tape for imaging. For longer term imaging during CESA velocity analyses, seedlings were mounted on a piece of 1-mm-thick 0.6% (w/v) Phytagel pad affixed to the glass slide to minimize compression and liquid evaporation.
To examine the cellular localization of YFP-CESA6, YFP-CESA6;ManI-CFP, and YFP-CESA6;mCherry-TUA5, SDCM imaging was performed using a CSU-X1-A1 Yokogawa scanning unit mounted on an Olympus IX-83 microscope, equipped with a 100×/1.4 numerical aperture UPlanSApo oil objective (Olympus) and an Andor iXon Ultra 897BV electron multiplying charge-coupled device camera (Andor). YFP, CFP, and mCherry fluorescence was excited with 515-, 445-, and 561-nm laser lines and emission collected through 542/27-, 479/40-, and 607/36-nm filters, respectively.
For the FRAP experiments, images were collected using a Zeiss Observer Z.1 microscope equipped with a Yokogawa CSU-X1 head and a 100×/1.46 numerical aperture PlanApo objective (Zeiss). For FRAP analysis of PM-localized CESA6, photobleaching was performed with a Vector scanner (Intelligent Imaging Innovations) with a 515-nm laser line at 100% power and 1 ms/scan. Time-lapse images were collected at the PM with a 5-s interval for 121 frames, with photobleaching in a small region (44.2 µm2) after the fourth frame and recovery for a total of 10 min.
SDCM Image Processing and Quantification
Image analysis was performed using Fiji/ImageJ (Schindelin et al., 2012). For CESA particle density analyses, regions of interest (ROIs) without abundant Golgi signals were chosen using the Freehand selection tool. CESA particles were detected automatically on 8-bit images using the Find Maxima tool with the same noise threshold for all images. CESA particle density for each ROI was calculated by dividing the number of particles by the ROI area. For CESA particle dynamic analyses, 5-min time-lapse series with 5-s intervals were collected. Average intensity projections were generated to identify the trajectories of the CSC particles. Image drift was corrected by the StackReg plugin (Thévenaz et al., 1998). Kymographs were generated and velocities of CESA particles were measured as the reciprocal of the slopes of individual CESA particles in the kymographs. To quantify cortical vesicles, 1-μm z-series stacks with 0.2 μm as step size and 20-s time-lapses were collected. Focal plane at 0.4 μm below the PM was used for cortical SmaCC analyses. The small particles showing motility in the time-lapse series were considered to be the SmaCCs. For the FRAP assay of PM-localized CSCs, a smaller area (16 µm2) within the bleached region was used for analyses. The CSC delivery events during the first 5 min of recovery were manually counted according to the criteria described previously (Li et al., 2016). The particles that exhibited steady linear movement at the PM were considered to be new delivery events. The CSC delivery rate was calculated by dividing the number of delivery events by the measured area and time.
Accession Numbers
Sequence data from this article can be found in the UniProt Knowledgebase under the following accession numbers: AtCESA1 (O48946), AtCESA3 (Q941L0), AtCESA6 (Q94JQ6), AtCESA7 (Q9SWW6), RsBcsA (A0A3G6W9S6); and also in the National Center for Biotechnology Information Sequence Read Archive (https://www.ncbi.nlm.nih.gov/sra) database under project number PRJNA630087.
The gene accession numbers (The Arabidopsis Information Resource) are as follows: AtCESA1 (AT4G32410); AtCESA6 (AT5G64740); AtCESA7 (AT5G17420).
Supplemental Data
Supplemental Figure 1. ES20 inhibits Arabidopsis hypocotyl growth, and EMS-induced cesa6 mutants show reduced sensitivity to ES20 in terms of root and hypocotyl growth.
Supplemental Figure 2. Mutated CESA6 constructs rescue the root and hypocotyl growth phenotypes of prc1-1 to different extents and reduce the sensitivity of prc1-1 to ES20.
Supplemental Figure 3. The predicted topology of CESA6, the locations of key motifs for cellulose catalytic activity, and the locations of the mutated amino acids that cause reduced sensitivity to ES20.
Supplemental Figure 4. The amino acids that are mutated in ES20-resistant mutants are conserved among Arabidopsis CESAs and between plant CESAs and RsBcsA.
Supplemental Figure 5. Mutations of amino acids located at the predicted binding site of ES20 on the CESA6 cytoplasmic domain affect plant sensitivity to ES20.
Supplemental Figure 6. Control experiments to test GFP-CESA6c and ES20 interactions in the Microscale Thermophoresis (MST) assays.
Supplemental Table 1. Mutations in CESA6 that result in reduced sensitivity of plants to ES20.
Supplemental Table 2. Primers used for molecular cloning and site-directed mutagenesis.
Supplemental Data Set. ANOVA tables.
Supplemental Movie 1. Different views of the superposition of ES20 and UDP-glucose phosphonate on the predicted binding pocket of the modeled CESA6 large cytosolic domain (amino acids 322-868).
Supplemental Movie 2. Different views of the superposition of ES20 and UDP-glucose phosphonate on the predicted binding pocket of the modeled CESA6 large cytosolic domain.
Supplemental Movie 3. The microtubule-associated CESA compartment accumulates after long-term ES20 treatment.
DIVE Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
Acknowledgments
We thank Zheng-Hua Ye (University of Georgia) and Geoff Wasteneys (University of British Columbia) for providing the fra5 and any1 seeds, respectively. We thank Ying Gu (Pennsylvania State University) for providing the YFP-CESA6;mCherry-TUA5 line. We acknowledge the Purdue Genomics facility for assistance with DNA sequencing. We thank Daniel Szymanski (Purdue University) for sharing the spinning disc confocal microscope for FRAP studies and providing us with the YFP-CESA6;ManI-CFP seeds. We thank the Chemical Genomics Facility at the Purdue Institute for Drug Discovery for providing us with access to the MST equipment. We thank Yun Zhou and Tesfaye Mengiste (Purdue University) for sharing the compound microscopes and plate reader. Support for this work was provided by the Office of Science at the U.S. Department of Energy, Physical Biosciences Program (grant DE-FG02-09ER15526 to C.J. for imaging of YFP-CESA6 trafficking) and by Purdue University Provost’s start-up (to C.Z.).
AUTHOR CONTRIBUTIONS
L.H. performed the mutant screening, mutant phenotype characterization, genetic complementation, CESA6 imaging and image analysis, DARTS assay, and MST assay and prepared the figures. X.L. performed cell wall analysis, structure modeling, molecular docking, and MST assay. W.Z. provided critical guidance for CESA6 imaging and image analysis. N.U. performed initial compound screening and initial mutant screening. N.L. assisted with the biochemical assays for testing the interaction between ES20 and CESA6. X.Y. and Y.L. synthesized the ES20 compound. R.E.M. analyzed the initial high-throughput sequencing data to clone the first mutant gene. B.D. provided guidance for high-throughput sequence analysis. M.D. provided guidance for ES20 synthesis. G.R.H. and N.V.R. provided guidance for the initial compound screening. C.J.S. provided critical guidance for CESA6 imaging and image analysis. C.Z. designed the research and wrote the article.
Footnotes
Articles can be viewed without a subscription.
References
- Alexander N., Woetzel N., Meiler J.(2011). bcl:Cluster: A method for clustering biological molecules coupled with visualization in the Pymol Molecular Graphics System. IEEE Int. Conf. Comput. Adv. Bio Med. Sci. 2011: 13–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arioli T., et al. (1998). Molecular analysis of cellulose biosynthesis in Arabidopsis. Science 279: 717–720. [DOI] [PubMed] [Google Scholar]
- Austin R.S., Vidaurre D., Stamatiou G., Breit R., Provart N.J., Bonetta D., Zhang J., Fung P., Gong Y., Wang P.W., McCourt P., Guttman D.S.(2011). Next-generation mapping of Arabidopsis genes. Plant J. 67: 715–725. [DOI] [PubMed] [Google Scholar]
- Bar-Peled M., Raikhel N.V.(1997). Characterization of AtSEC12 and AtSAR1. Proteins likely involved in endoplasmic reticulum and Golgi transport. Plant Physiol. 114: 315–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baskin T.I., Wilson J.E., Cork A., Williamson R.E.(1994). Morphology and microtubule organization in Arabidopsis roots exposed to oryzalin or taxol. Plant Cell Physiol. 35: 935–942. [PubMed] [Google Scholar]
- Brabham C., Lei L., Gu Y., Stork J., Barrett M., DeBolt S.(2014). Indaziflam herbicidal action: A potent cellulose biosynthesis inhibitor. Plant Physiol. 166: 1177–1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown C., Leijon F., Bulone V.(2012). Radiometric and spectrophotometric in vitro assays of glycosyltransferases involved in plant cell wall carbohydrate biosynthesis. Nat. Protoc. 7: 1634–1650. [DOI] [PubMed] [Google Scholar]
- Brown D.M., Zeef L.A., Ellis J., Goodacre R., Turner S.R.(2005). Identification of novel genes in Arabidopsis involved in secondary cell wall formation using expression profiling and reverse genetics. Plant Cell 17: 2281–2295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burn J.E., Hocart C.H., Birch R.J., Cork A.C., Williamson R.E.(2002). Functional analysis of the cellulose synthase genes CesA1, CesA2, and CesA3 in Arabidopsis. Plant Physiol. 129: 797–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caño-Delgado A., Penfield S., Smith C., Catley M., Bevan M.(2003). Reduced cellulose synthesis invokes lignification and defense responses in Arabidopsis thaliana. Plant J. 34: 351–362. [DOI] [PubMed] [Google Scholar]
- Cantarel B.L., Coutinho P.M., Rancurel C., Bernard T., Lombard V., Henrissat B.(2009). The Carbohydrate-Active EnZymes database (CAZy): An expert resource for glycogenomics. Nucleic Acids Res. 37: D233–D238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charnock S.J., Davies G.J.(1999). Structure of the nucleotide-diphospho-sugar transferase, SpsA from Bacillus subtilis, in native and nucleotide-complexed forms. Biochemistry 38: 6380–6385. [DOI] [PubMed] [Google Scholar]
- Chen S.T., He N.Y., Chen J.H., Guo F.Q.(2017). Identification of core subunits of photosystem II as action sites of HSP21, which is activated by the GUN5-mediated retrograde pathway in Arabidopsis. Plant J. 89: 1106–1118. [DOI] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F.(1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743. [DOI] [PubMed] [Google Scholar]
- Crowell E.F., Bischoff V., Desprez T., Rolland A., Stierhof Y.D., Schumacher K., Gonneau M., Höfte H., Vernhettes S.(2009). Pausing of Golgi bodies on microtubules regulates secretion of cellulose synthase complexes in Arabidopsis. Plant Cell 21: 1141–1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dallakyan S., Olson A.J.(2015). Small-molecule library screening by docking with PyRx. Methods Mol. Biol. 1263: 243–250. [DOI] [PubMed] [Google Scholar]
- Daras G., Rigas S., Penning B., Milioni D., McCann M.C., Carpita N.C., Fasseas C., Hatzopoulos P.(2009). The thanatos mutation in Arabidopsis thaliana cellulose synthase 3 (AtCesA3) has a dominant-negative effect on cellulose synthesis and plant growth. New Phytol. 184: 114–126. [DOI] [PubMed] [Google Scholar]
- DeBolt S., Gutierrez R., Ehrhardt D.W., Melo C.V., Ross L., Cutler S.R., Somerville C., Bonetta D.(2007). Morlin, an inhibitor of cortical microtubule dynamics and cellulose synthase movement. Proc. Natl. Acad. Sci. USA 104: 5854–5859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decker D., Öberg C., Kleczkowski L.A.(2017). Identification and characterization of inhibitors of UDP-glucose and UDP-sugar pyrophosphorylases for in vivo studies. Plant J. 90: 1093–1107. [DOI] [PubMed] [Google Scholar]
- Desprez T., Juraniec M., Crowell E.F., Jouy H., Pochylova Z., Parcy F., Höfte H., Gonneau M., Vernhettes S.(2007). Organization of cellulose synthase complexes involved in primary cell wall synthesis in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 104: 15572–15577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desprez T., Vernhettes S., Fagard M., Refrégier G., Desnos T., Aletti E., Py N., Pelletier S., Höfte H.(2002). Resistance against herbicide isoxaben and cellulose deficiency caused by distinct mutations in same cellulose synthase isoform CESA6. Plant Physiol. 128: 482–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doblin M.S., Kurek I., Jacob-Wilk D., Delmer D.P.(2002). Cellulose biosynthesis in plants: From genes to rosettes. Plant Cell Physiol. 43: 1407–1420. [DOI] [PubMed] [Google Scholar]
- Drakakaki G., et al. (2011). Clusters of bioactive compounds target dynamic endomembrane networks in vivo. Proc. Natl. Acad. Sci. USA 108: 17850–17855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagard M., Desnos T., Desprez T., Goubet F., Refregier G., Mouille G., McCann M., Rayon C., Vernhettes S., Höfte H.(2000). PROCUSTE1 encodes a cellulose synthase required for normal cell elongation specifically in roots and dark-grown hypocotyls of Arabidopsis. Plant Cell 12: 2409–2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandes A.N., Thomas L.H., Altaner C.M., Callow P., Forsyth V.T., Apperley D.C., Kennedy C.J., Jarvis M.C.(2011). Nanostructure of cellulose microfibrils in spruce wood. Proc. Natl. Acad. Sci. USA 108: E1195–E1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster C.E., Martin T.M., Pauly M.(2010). Comprehensive compositional analysis of plant cell walls (lignocellulosic biomass) part II: Carbohydrates. J. Vis. Exp. 37: 1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita M., Himmelspach R., Ward J., Whittington A., Hasenbein N., Liu C., Truong T.T., Galway M.E., Mansfield S.D., Hocart C.H., Wasteneys G.O.(2013). The anisotropy1 D604N mutation in the Arabidopsis cellulose synthase1 catalytic domain reduces cell wall crystallinity and the velocity of cellulose synthase complexes. Plant Physiol. 162: 74–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardiner J.C., Taylor N.G., Turner S.R.(2003). Control of cellulose synthase complex localization in developing xylem. Plant Cell 15: 1740–1748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerrits M., Budisa N., Merk H.(2019). Site-specific chemoselective pyrrolysine analogues incorporation using the cell-free protein synthesis system. ACS Synth. Biol. 8: 381–390. [DOI] [PubMed] [Google Scholar]
- Giddings T.H. Jr., Brower D.L., Staehelin L.A.(1980). Visualization of particle complexes in the plasma membrane of Micrasterias denticulata associated with the formation of cellulose fibrils in primary and secondary cell walls. J. Cell Biol. 84: 327–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonneau M., Desprez T., Guillot A., Vernhettes S., Höfte H.(2014). Catalytic subunit stoichiometry within the cellulose synthase complex. Plant Physiol. 166: 1709–1712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez R., Lindeboom J.J., Paredez A.R., Emons A.M., Ehrhardt D.W.(2009). Arabidopsis cortical microtubules position cellulose synthase delivery to the plasma membrane and interact with cellulose synthase trafficking compartments. Nat. Cell Biol. 11: 797–806. [DOI] [PubMed] [Google Scholar]
- Heim D.R., Roberts J.L., Pike P.D., Larrinua I.M.(1989). Mutation of a locus of Arabidopsis thaliana confers resistance to the herbicide isoxaben. Plant Physiol. 90: 146–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill J.L. Jr., Hammudi M.B., Tien M.(2014). The Arabidopsis cellulose synthase complex: A proposed hexamer of CESA trimers in an equimolar stoichiometry. Plant Cell 26: 4834–4842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Z., et al. (2016). Mitochondrial defects confer tolerance against cellulose deficiency. Plant Cell 28: 2276–2290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Z., Zhang T., Rombaut D., Decaestecker W., Xing A., D’Haeyer S., Höfer R., Vercauteren I., Karimi M., Jacobs T., De Veylder L.(2019). Genome editing-based engineering of CESA3 dual cellulose-inhibitor-resistant plants. Plant Physiol. 180: 827–836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jerabek-Willemsen M., Wienken C.J., Braun D., Baaske P., Duhr S.(2011). Molecular interaction studies using microscale thermophoresis. Assay Drug Dev. Technol. 9: 342–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y., Schumaker K.S., Zhu J.K.(2006). EMS mutagenesis of Arabidopsis. Methods Mol. Biol. 323: 101–103. [DOI] [PubMed] [Google Scholar]
- Kosmacz M., Luzarowski M., Kerber O., Leniak E., Gutiérrez-Beltrán E., Moreno J.C., Gorka M., Szlachetko J., Veyel D., Graf A., Skirycz A.(2018). Interaction of 2′,3′-cAMP with Rbp47b plays a role in stress granule formation. Plant Physiol. 177: 411–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar M., Turner S.(2015). Plant cellulose synthesis: CESA proteins crossing kingdoms. Phytochemistry 112: 91–99. [DOI] [PubMed] [Google Scholar]
- Laskowski R.A., MacArthur M.W., Moss D.S., Thornton J.M.(1993). PROCHECK: A program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 26: 283–291. [Google Scholar]
- Laskowski R.A., Rullmannn J.A., MacArthur M.W., Kaptein R., Thornton J.M.(1996). AQUA and PROCHECK-NMR: Programs for checking the quality of protein structures solved by NMR. J. Biomol. NMR 8: 477–486. [DOI] [PubMed] [Google Scholar]
- Li S., Bashline L., Zheng Y., Xin X., Huang S., Kong Z., Kim S.H., Cosgrove D.J., Gu Y.(2016). Cellulose synthase complexes act in a concerted fashion to synthesize highly aggregated cellulose in secondary cell walls of plants. Proc. Natl. Acad. Sci. USA 113: 11348–11353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu T., et al. (2017). FSIP1 binds HER2 directly to regulate breast cancer growth and invasiveness. Proc. Natl. Acad. Sci. USA 114: 7683–7688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomenick B., et al. (2009). Target identification using drug affinity responsive target stability (DARTS). Proc. Natl. Acad. Sci. USA 106: 21984–21989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McFarlane H.E., Döring A., Persson S.(2014). The cell biology of cellulose synthesis. Annu. Rev. Plant Biol. 65: 69–94. [DOI] [PubMed] [Google Scholar]
- McNamara J.T., Morgan J.L., Zimmer J.(2015). A molecular description of cellulose biosynthesis. Annu. Rev. Biochem. 84: 895–921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montezinos D., Delmer D.P.(1980). Characterization of inhibitors of cellulose synthesis in cotton fibers. Planta 148: 305–311. [DOI] [PubMed] [Google Scholar]
- Morgan J.L., McNamara J.T., Fischer M., Rich J., Chen H.M., Withers S.G., Zimmer J.(2016). Observing cellulose biosynthesis and membrane translocation in crystallo. Nature 531: 329–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan J.L., Strumillo J., Zimmer J.(2013). Crystallographic snapshot of cellulose synthesis and membrane translocation. Nature 493: 181–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller S.C., Brown R.M. Jr.(1980). Evidence for an intramembrane component associated with a cellulose microfibril-synthesizing complex in higher plants. J. Cell Biol. 84: 315–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller S.C., Brown R.M. Jr., Scott T.K.(1976). Cellulosic microfibrils: Nascent stages of synthesis in a higher plant cell. Science 194: 949–951. [DOI] [PubMed] [Google Scholar]
- Newman R.H., Hill S.J., Harris P.J.(2013). Wide-angle x-ray scattering and solid-state nuclear magnetic resonance data combined to test models for cellulose microfibrils in mung bean cell walls. Plant Physiol. 163: 1558–1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omadjela O., Narahari A., Strumillo J., Mélida H., Mazur O., Bulone V., Zimmer J.(2013). BcsA and BcsB form the catalytically active core of bacterial cellulose synthase sufficient for in vitro cellulose synthesis. Proc. Natl. Acad. Sci. USA 110: 17856–17861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paredez A.R., Somerville C.R., Ehrhardt D.W.(2006). Visualization of cellulose synthase demonstrates functional association with microtubules. Science 312: 1491–1495. [DOI] [PubMed] [Google Scholar]
- Park J.I., Ishimizu T., Suwabe K., Sudo K., Masuko H., Hakozaki H., Nou I.S., Suzuki G., Watanabe M.(2010). UDP-glucose pyrophosphorylase is rate limiting in vegetative and reproductive phases in Arabidopsis thaliana. Plant Cell Physiol. 51: 981–996. [DOI] [PubMed] [Google Scholar]
- Park S., Song B., Shen W., Ding S.Y.(2019). A mutation in the catalytic domain of cellulose synthase 6 halts its transport to the Golgi apparatus. J. Exp. Bot. 70: 6071–6083. [DOI] [PubMed] [Google Scholar]
- Pear J.R., Kawagoe Y., Schreckengost W.E., Delmer D.P., Stalker D.M.(1996). Higher plants contain homologs of the bacterial celA genes encoding the catalytic subunit of cellulose synthase. Proc. Natl. Acad. Sci. USA 93: 12637–12642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persson S., Paredez A., Carroll A., Palsdottir H., Doblin M., Poindexter P., Khitrov N., Auer M., Somerville C.R.(2007). Genetic evidence for three unique components in primary cell-wall cellulose synthase complexes in Arabidopsis. Proc. Natl. Acad. Sci. USA 104: 15566–15571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polko J.K., Kieber J.J.(2019). The regulation of cellulose biosynthesis in plants. Plant Cell 31: 282–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy A., Kucukural A., Zhang Y.(2010). I-TASSER: A unified platform for automated protein structure and function prediction. Nat. Protoc. 5: 725–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampathkumar A., Gutierrez R., McFarlane H.E., Bringmann M., Lindeboom J., Emons A.M., Samuels L., Ketelaar T., Ehrhardt D.W., Persson S.(2013). Patterning and lifetime of plasma membrane-localized cellulose synthase is dependent on actin organization in Arabidopsis interphase cells. Plant Physiol. 162: 675–688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheible W.R., Eshed R., Richmond T., Delmer D., Somerville C.(2001). Modifications of cellulose synthase confer resistance to isoxaben and thiazolidinone herbicides in Arabidopsis Ixr1 mutants. Proc. Natl. Acad. Sci. USA 98: 10079–10084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J., et al. (2012). Fiji: An open-source platform for biological-image analysis. Nat. Methods 9: 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sethaphong L., Haigler C.H., Kubicki J.D., Zimmer J., Bonetta D., DeBolt S., Yingling Y.G.(2013). Tertiary model of a plant cellulose synthase. Proc. Natl. Acad. Sci. USA 110: 7512–7517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slabaugh E., Davis J.K., Haigler C.H., Yingling Y.G., Zimmer J.(2014a). Cellulose synthases: New insights from crystallography and modeling. Trends Plant Sci. 19: 99–106. [DOI] [PubMed] [Google Scholar]
- Slabaugh E., Sethaphong L., Xiao C., Amick J., Anderson C.T., Haigler C.H., Yingling Y.G.(2014b). Computational and genetic evidence that different structural conformations of a non-catalytic region affect the function of plant cellulose synthase. J. Exp. Bot. 65: 6645–6653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepek I.A., Cao T., Koetemann A., Shimura S., Wollscheid B., Bode J.W.(2019). Antibiotic discovery with synthetic fermentation: library assembly, phenotypic screening, and mechanism of action of β-peptides targeting penicillin-binding proteins. ACS Chem. Biol. 14: 1030–1040. [DOI] [PubMed] [Google Scholar]
- Tateno M., Brabham C., DeBolt S.(2016). Cellulose biosynthesis inhibitors - A multifunctional toolbox. J. Exp. Bot. 67: 533–542. [DOI] [PubMed] [Google Scholar]
- Taylor N.G., Howells R.M., Huttly A.K., Vickers K., Turner S.R.(2003). Interactions among three distinct CesA proteins essential for cellulose synthesis. Proc. Natl. Acad. Sci. USA 100: 1450–1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thévenaz P., Ruttimann U.E., Unser M.(1998). A pyramid approach to subpixel registration based on intensity. IEEE Trans. Image Process. 7: 27–41. [DOI] [PubMed] [Google Scholar]
- Tran M.L., McCarthy T.W., Sun H., Wu S.Z., Norris J.H., Bezanilla M., Vidali L., Anderson C.T., Roberts A.W.(2018). Direct observation of the effects of cellulose synthesis inhibitors using live cell imaging of cellulose synthase (CESA) in Physcomitrella patens. Sci. Rep. 8: 735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trott O., Olson A.J.(2010). AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 31: 455–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner S., Kumar M.(2018). Cellulose synthase complex organization and cellulose microfibril structure. Philos. Trans. A Math Physiol. Eng. Sci. 376: 20170048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Updegraff D.M.(1969). Semimicro determination of cellulose in biological materials. Anal. Biochem. 32: 420–424. [DOI] [PubMed] [Google Scholar]
- Vergara C.E., Carpita N.C.(2001). Beta-D-glycan synthases and the CesA gene family: Lessons to be learned from the mixed-linkage (1→3),(1→4)beta-D-glucan synthase. Plant Mol. Biol. 47: 145–160. [PubMed] [Google Scholar]
- Warren C.J., Meyerson N.R., Stabell A.C., Fattor W.T., Wilkerson G.K., Sawyer S.L.(2019). A glycan shield on chimpanzee CD4 protects against infection by primate lentiviruses (HIV/SIV). Proc. Natl. Acad. Sci. USA 116: 11460–11469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wienken C.J., Baaske P., Rothbauer U., Braun D., Duhr S.(2010). Protein-binding assays in biological liquids using microscale thermophoresis. Nat. Commun. 1: 100. [DOI] [PubMed] [Google Scholar]
- Worden N., et al. (2015). CESA TRAFFICKING INHIBITOR inhibits cellulose deposition and interferes with the trafficking of cellulose synthase complexes and their associated proteins KORRIGAN1 and POM2/CELLULOSE SYNTHASE INTERACTIVE PROTEIN1. Plant Physiol. 167: 381–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Q., Lin M., Huang W., Teymournejad O., Johnson J.M., Hays F.A., Liang Z., Li G., Rikihisa Y.(2018). Ehrlichia type IV secretion system effector Etf-2 binds to active RAB5 and delays endosome maturation. Proc. Natl. Acad. Sci. USA 115: E8977–E8986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J., Roy A., Zhang Y.(2013a). Protein-ligand binding site recognition using complementary binding-specific substructure comparison and sequence profile alignment. Bioinformatics 29: 2588–2595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J., Roy A., Zhang Y.(2013b). BioLiP: A semi-manually curated database for biologically relevant ligand-protein interactions. Nucleic Acids Res. 41: D1096–D1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J., Yan R., Roy A., Xu D., Poisson J., Zhang Y.(2015). The I-TASSER suite: Protein structure and function prediction. Nat. Methods 12: 7–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai Z., Keereetaweep J., Liu H., Feil R., Lunn J.E., Shanklin J.(2018). Trehalose 6-phosphate positively regulates fatty acid synthesis by stabilizing WRINKLED1. Plant Cell 30: 2616–2627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C., et al. (2016). Endosidin2 targets conserved exocyst complex subunit EXO70 to inhibit exocytosis. Proc. Natl. Acad. Sci. USA 113: E41–E50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W., Cai C., Staiger C.J.(2019). Myosins XI are involved in exocytosis of cellulose synthase complexes. Plant Physiol. 179: 1537–1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong R., Cui D., Ye Z.H.(2019). Secondary cell wall biosynthesis. New Phytol. 221: 1703–1723. [DOI] [PubMed] [Google Scholar]
- Zhong R., Morrison W.H. III, Freshour G.D., Hahn M.G., Ye Z.H.(2003). Expression of a mutant form of cellulose synthase AtCesA7 causes dominant negative effect on cellulose biosynthesis. Plant Physiol. 132: 786–795. [DOI] [PMC free article] [PubMed] [Google Scholar]