Abstract
Nitrogen (N) is an essential macronutrient for plants and a major limiting factor for plant growth and crop production. Nitrate is the main source of N available to plants in agricultural soils and in many natural environments. Sustaining agricultural productivity is of paramount importance in the current scenario of increasing world population, diversification of crop uses, and climate change. Plant productivity for major crops around the world, however, is still supported by excess application of N-rich fertilizers with detrimental economic and environmental impacts. Thus, understanding how plants regulate nitrate uptake and metabolism is key for developing new crops with enhanced N use efficiency and to cope with future world food demands. The study of plant responses to nitrate has gained considerable interest over the last 30 years. This review provides an overview of key findings in nitrate research, spanning biochemistry, molecular genetics, genomics, and systems biology. We discuss how we have reached our current view of nitrate transport, local and systemic nitrate sensing/signaling, and the regulatory networks underlying nitrate-controlled outputs in plants. We hope this summary will serve not only as a timeline and information repository but also as a baseline to define outstanding questions for future research.
INTRODUCTION
Nitrogen (N) is an essential macronutrient for plants, and its availability is a key determinant for plant growth and productivity. N nutrients can be provided as inorganic (nitrate [NO3–] and ammonium [NH4+]) or organic (amino acids and urea) forms. However, nitrate is the main source of N both in agricultural and in many natural systems (Crawford and Forde, 2002). N fertilizer consumption is currently ∼118 million tons per year worldwide, representing 59% of total fertilizer nutrient used (FAO, 2017). The massive production and application of inorganic N fertilizers have contributed significantly to support and sustain the enormous rise in crop yield over the past century. However, fertilizer overuse has also led to the destruction of aquatic ecosystems and the formation of oceanic “dead zones” from eutrophication in coastal regions worldwide. N soil absorption is low, meaning that plants can only use roughly 50 to 75% of the N input from fertilizers in a given season (Gutiérrez, 2012). Nitrate that is not retained in the soil leaches into groundwater and is an important contributor to eutrophication. Furthermore, agriculture management practices are an important contributor to nitrous oxide emissions, a key greenhouse gas contributing to global warming. A key goal in driving N research, therefore, is to maintain high crop yields while reducing excess N load to the environment by improving nitrogen use efficiency (NUE). A better understanding of the biology of N nutrition is key for achieving this goal. Here we provide an overview of some of the key findings in nitrate research spanning biochemistry, molecular genetics, genomics, and systems biology. We draw heavily on more recent work carried out in Arabidopsis (Arabidopsis thaliana), although a lot of the earlier work was done using other species. (This review mentions over 150 genes by name. We will not spell out the acronyms in the text for fear of interrupting the flow of the narrative. All gene acronyms, alternate names in the literature, and their full names are listed in Supplemental Data Set 1.)
The identification of genes encoding N uptake carriers, assimilation enzymes, and their regulators relied heavily on extensive biochemical and genetic studies in plants, algae, and fungi. Figure 1 represents a timeline of some of the key publications in nitrate signaling over the last 70 years, and Supplemental Data Set 2 provides an extended (but nonexhaustive) list of the key publications involved in nitrate uptake, transport, and signaling, discussed in this section.
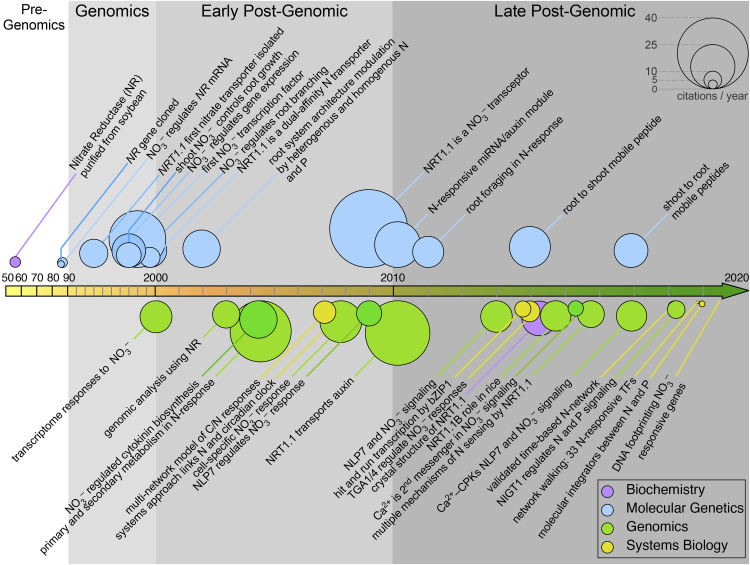
Figure 1.
A Timeline with Milestone Publications in Nitrate Signaling.
The terms “nitrate” and “Arabidopsis” were used to retrieve articles and their citation numbers from Web of Science (http://www.webofknowledge.com). We selected articles specifically focused on nitrate signaling or acquisition in this timeline (listed in Supplemental Data Set 2), based on the number of citations. Three additional articles related to early studies in crops (1950s to 1980s) were included. Please note that the sizes of circles are proportional to the number of citations normalized by years from publication. Top articles related to nitrate signaling were included for this timeline.
In 1953, soybean (Glycine max) nitrate reductase (NR) was purified by Evans and Nason, (1953) and its nitrate reductive activity confirmed in vitro. The 1980s saw the cloning of genes related to N metabolism, including NR. Nitrate was also shown to control NR mRNA and protein levels in plants (Cheng et al., 1986; Crawford et al., 1986, 1988; Calza et al., 1987). During the 1990s, several inorganic N transporters were identified. Tsay et al. (1993) isolated and characterized the first plant nitrate transporter, CHLORINA1 (CHL1), known today as NITRATE TRANSPORTER1.1 (NRT1.1) or NITRATE TRANSPORTER1/PEPTIDE TRANSPORTER FAMILY6.3 (NPF6.3), and showed that the mRNA levels of the encoding gene were induced by nitrate. Although first described as a low-affinity transporter, NRT1.1/NPF6.3 (hereafter referred to as NRT1.1) was later shown to be a dual-affinity nitrate transporter, the switch from low to high affinity being mediated by phosphorylation of Thr-101 (Liu and Tsay, 2003). NRT2.1 was isolated using degenerate oligonucleotides designed against conserved regions of nitrate transporters from fungi and algae (Zhuo et al., 1999), and its mRNA was also shown to be induced by nitrate treatments (Wang et al., 1998; Zhuo et al., 1999).
In the late 1990s, the first transcription factor (TF) involved in nitrate responses was identified in Arabidopsis (Arabidopsis thaliana): ANR1, a root-expressed and nitrate-induced gene encoding a MIKC-type MADS box TF. ANR1 was shown to have a positive role in lateral root (LR) proliferation in nitrate-rich patches (Zhang and Forde, 1998). These results provided the first molecular basis for the effect of nitrate on controlling plant growth. During the first decade of the 2000s, genome-wide transcriptome studies began to unravel the full extent of nitrate impact on gene expression. Microarray and RNA sequencing analyses showed that nitrate not only controlled the expression of genes involved in nitrate transport and assimilation but also hundreds to thousands of genes involved in other processes such as carbon (C), sulfate, amino acid, and nucleotide metabolism, hormone biosynthesis and response, as well as myriad TFs (Wang et al., 2000, 2001, 2003; Scheible et al., 2004). At the same time, the increasing amount of data available on gene expression and molecular interactions allowed the rise of systems biology approaches to model nitrate-controlled regulatory networks (Gutiérrez et al., 2007, 2008; Gifford et al., 2008).
The identification of NRT1.1 as a nitrate sensor in 2009 was a major milestone in nitrate signaling (Ho et al., 2009). That same year, researchers identified NLP7, one of the best-studied TFs in the N response. NLP7 controls responses to nitrate by a nuclear retention mechanism that is triggered by nitrate availability (Castaings et al., 2009; Wang et al., 2009). At the beginning of the 2010s, high-throughput small RNA sequencing was used to identify small RNAs involved in nitrate responses. A nitrate-induced regulatory module between microRNA393 (miR393) and AFB3 was identified: it integrated external nitrate availability, internal organic N signals, and root auxin sensitivity to control root architecture (Vidal et al., 2010). Likewise, studies of nitrate responses in specific root cell types uncovered a role for miR167 and ARF8 in LR emergence (Gifford et al., 2008). In addition, a function for NRT1.1 in transporting auxin provided a molecular mechanism to explain LR elongation in response to external nitrate (Krouk et al., 2010a). Other TFs important to the nitrate response, such as TGA1 and TGA4, NLP6, HRS1 and HHO1, NAC4, SPL9, TCP20, LBD37, LBD38, and LBD39, and NRG2, were identified shortly after (Rubin et al., 2009; Krouk et al., 2010b; Vidal et al., 2013; Alvarez et al., 2014; Guan et al., 2014; Medici et al., 2015; Xu et al., 2016b). More recently, the development of ultrasensitive biosensors and considerable advances in live microscopy identified calcium as an important second messenger in nitrate signaling (Riveras et al., 2015; Liu et al., 2017). Coupled with the recent discovery of a calcium-dependent signaling pathway involving calcium-dependent kinases (CPKs) that phosphorylate NLP7 to control its subcellular localization, these results represent a major advance linking nitrate sensing to changes in the transcriptome and root and shoot growth (Liu et al., 2017).
In addition to primary nitrate responses, systemic N signaling has also received much attention. Development of the split-root system was instrumental, which allowed the identification of nitrate-cytokinin interactions in systemic N signaling (Ruffel et al., 2011, 2016; Ruffel and Gojon, 2017). The relevant molecular players have also been identified, including the translocation of the peptide Cle and of the transcription factor HY5 from shoots to roots (Araya et al., 2014a; Chen et al., 2016).
Today, advances in DNA sequencing and high-throughput genomic technologies have permitted substantial progress in deciphering gene regulatory networks (GRNs) controlling responses to nitrate. The analysis of time-series data (Krouk et al., 2010b; Varala et al., 2018; Brooks et al., 2019), yeast one-hybrid (Y1H) technology (Gaudinier et al., 2018), and genome-wide chromatin accessibility and RNA polymerase II chromatin immunoprecipitation (ChIP) followed by deep sequencing assays (Alvarez et al., 2019) all helped identify TF-DNA interactions in networks involved in nitrate responses. Perhaps not surprisingly, nitrate and hormone signaling networks are interconnected (Ristova et al., 2016).
Finally, the increasing availability of high-quality genomes and genetic resources for plants makes it possible to extend our vast knowledge to crops (e.g., the OsNRT1.1b gene in rice [Oryza sativa]; Hu et al., 2015) and other plants of economic importance, with the aim to improve agronomic traits such as NUE, seed quality, and flowering time, among other potential uses.
MOLECULAR MECHANISMS OF NITRATE TRANSPORT
The molecular basis of nitrate uptake by the root has been studied intensively over the past 30 years. Plants employ two different uptake systems depending on how much external nitrate is available: the high-affinity and low-affinity transport systems (HATS and LATS), operating at low (<1 mM) or high (>1 mM) nitrate concentrations, respectively (Crawford and Glass, 1998).
Nitrate Transporters
The Arabidopsis genome has 53 NRT1/NPF genes and 7 NRT2 genes (Supplemental Tables 1 and 2); the NPF family is responsible for LATS (Léran et al., 2014), while the NRT2/NNP (nitrate-nitrite porter) functions in HATS (Orsel et al., 2002; Krapp et al., 2014). Although most NRT2 and NPF transporters have yet to be functionally characterized, four NRT2 transporters (NRT2.1, NRT2.2, NRT2.4, and NRT2.5) and two NPF transporters (NRT1.1 and NRT1.2) are components of root nitrate uptake (Tsay et al., 1993; Huang et al., 1999; Filleur et al., 2001; Kiba et al., 2012; Lezhneva et al., 2014). NRT1.1 is the only demonstrated dual-affinity nitrate transporter and a putative nitrate transceptor (a portmanteau for a transporter-receptor; Wang et al., 1998; Liu et al., 1999; Ho et al., 2009). Within the NRT2/NPF family, NRT2.1 predominantly localizes to the plasma membrane of root epidermal and cortical cells, where the bulk of nitrate uptake occurs (Chopin et al., 2007; Wirth et al., 2007).
Among NPF transporters, 13 have been found to be involved in root-to-shoot transport, seed development, or N storage (Wang et al., 2018a). NPF2.7, also called NAXT1, is the only known root nitrate efflux transporter identified to date (Segonzac et al., 2007). The gene is specifically expressed in seeds, where it controls nitrate content and dormancy (Chopin et al., 2007). In addition to NPF and NRT2 transporters, members of the CLC and SLAC/SLAH protein families facilitate nitrate transport inside the plant. SLAC/SLAH is a small protein family encoded by five genes that display a common predicted structure of 10 transmembrane domains. Among them, SLAC1 and SLAH3 are characterized by NO3–/Cl– permeability and appear to be involved in the regulation of stomatal closure (Negi et al., 2008; Geiger et al., 2011). In contrast, SLAH2 is expressed in the stele of the root and encodes an anion channel that transports nitrate exclusively (Maierhofer et al., 2014) and may facilitate nitrate transport between root and shoot. Seven CLC genes have been identified in Arabidopsis. CLCa and CLCb proteins are NO3–/H+ antiporters involved in vacuolar nitrate storage (De Angeli et al., 2006; von der Fecht-Bartenbach et al., 2010) and are the only two (out of over 20 nitrate transporters) that do not localize to the plasma membrane. Clearly, there is much to learn about the function of vacuolar nitrate transporters.
Transcriptional Control of Nitrate Transport
There are two major mechanisms that coordinate root nitrate uptake as a function of external nitrate supply and internal N demand (Crawford and Glass, 1998; Gojon et al., 2009). First, the expression of nitrate transporters NRT2.1 and NRT1.1 is rapidly induced shortly after nitrate treatment (Tsay et al., 1993; Filleur and Daniel-Vedele, 1999; Lejay et al., 1999; Zhuo et al., 1999; Okamoto et al., 2003). Second, a negative feedback exerted by a high N status (at the whole-plant level or from external high nitrate) represses NRT2.1, NRT2.2, NRT2.4, and NRT2.5 expression once N needs are met (Lejay et al., 1999; Zhuo et al., 1999; Kiba et al., 2012; Lezhneva et al., 2014). This feedback repression is lifted when plants (re)experience N limitation, resulting in a strong increase in HATS capacity that improves N uptake. Feedback repression of NRT2.1 is suppressed or strongly attenuated in nrt1.1 mutants (Muños et al., 2004), although ammonium- or nitrate-fed nrt1.1 mutants are not N-deficient, suggesting that NRT2.1 expression is not only repressed by reduced N metabolites but also by nitrate itself. In fact, further studies revealed that NRT2.1 was highly induced under low nitrate/high ammonium availability even in wild-type plants. This shows that, at least for NRT2.1, feedback repression exerted by high N status is more complex and involves both (1) feedback repression by reduced N metabolites or mobile peptides and (2) NRT1.1-mediated repression by high external nitrate (Krouk et al., 2006; Ohkubo et al., 2017).
In addition, nitrate uptake is a highly integrated process, determined not only by nitrate availability and the N demand of the plant but also by C produced by photosynthesis in order to balance levels of root nitrate uptake against the availability of C metabolites (Delhon et al., 1995; Lejay et al., 1999, 2003). The coordination of photosynthesis (and thus C metabolism) with root N uptake involves the basic leucine zipper (bZIP) transcription factor HY5, which can translocate from shoots to roots upon light illumination to control NRT2.1 expression, nitrate uptake, and root growth (Chen et al., 2016). Light and sugars can also control root nitrate uptake by modulating NRT2.1, NRT2.4, and NRT1.1 expression, as can an oxidative pentose phosphate pathway (OPPP)-dependent signaling mechanism (Lejay et al., 2008; de Jong et al., 2014). It is interesting that the control of gene expression by an OPPP-derived signal extends to nitrate assimilatory genes in response to sucrose in the roots (Bussell et al., 2013; de Jong et al., 2014). Understanding the signaling mechanism linked to OPPP could be key to determining how C and N signaling pathways are integrated to modulate root nitrate uptake.
Posttranslational Control of Nitrate Transport
Most studies of nitrate transporters have focused on their transcriptional regulation, but increasing evidence highlights the importance of posttranslational control. Phosphorylation plays an important role in the regulation of NRT1.1 and AMT1.1. For NRT1.1, low nitrate concentrations trigger phosphorylation of Thr-101 (Liu and Tsay, 2003) and lead to a switch to its high-affinity function. Conversely, NRT1.1 remains a low-affinity transporter when Thr-101 is not phosphorylated at high nitrate concentrations.
Posttranslational modifications also control the activity of NRT2 transporters. NRT2.1 shows no detectable nitrate transport activity in the absence of NRT3.1 (Okamoto et al., 2006a; Orsel et al., 2006). In agreement, frog (Xenopus laevis) oocytes coinjected with NRT3.1/NAR2.1 and NRT2.1 show pH-dependent and nitrate-elicited currents, while oocytes injected with either NRT2.1 or NRT3.1 alone do not (Zhou et al., 2000). Phosphorylation also appears to play a major role in posttranslational regulation of the HATS activity of NRT2.1. NRT2.1 is phosphorylated at Ser-28 when plants are N starved but rapidly dephosphorylated upon nitrate resupply (Engelsberger and Schulze, 2012). Furthermore, Ser-11 is dephosphorylated and Thr-521 phosphorylated upon nitrate starvation (Menz et al., 2016), and mimicking the constitutive phosphorylation of Ser-501 prevents NRT2.1 nitrate uptake activity (Jacquot et al., 2019).
All NRT2 transporters in Arabidopsis, except NRT2.7, can interact with NRT3.1 in yeast. The presence of a Ser residue at or near the same position as in NRT2.1 is conserved in the six remaining NRT2 proteins in Arabidopsis (Kotur et al., 2012; Jacquot et al., 2019) and is well conserved across NRT2.1 homologs in green algae and land plants (Jacquot et al., 2019). This supports the idea that posttranslational regulatory mechanisms are of strategic importance for root nitrate uptake not only in Arabidopsis but also in crops.
Research on nitrate transporters over the years has therefore greatly contributed to our current understanding of the nitrate transport machinery in plants and its regulation by external signals. Moreover, this body of work revealed unexpected functions of nitrate transporters in nitrate sensing and signaling. As we discuss in the next section, NRT1.1 has received particular attention over the last decade due to its proposed role as a nitrate sensor controlling gene expression and plant development in response to nitrate availability, and also due to its ability to transport auxin, a key hormone controlling root growth.
MOLECULAR MECHANISMS OF LOCAL AND SYSTEMIC NITRATE SIGNALING
Nitrate, a Nutrient Signal Controlling Global Gene Expression
In the last 20 years, extensive use of transcriptomics analysis has shown that the expression of a considerable fraction of plant genes is modulated by nitrate treatments (∼10% of annotated genes in Arabidopsis; Canales et al., 2014). Such genes are not limited to processes directly related to nitrate (e.g., nitrate transport, reduction, or assimilation) but also include genes involved in the metabolism of aminoacids, nucleotides, and other nutrients (such as C, sulfate, phosphate [P], and iron), hormone biosynthesis and signaling, and regulatory genes including kinases, phosphatases and TFs involved in growth and developmental processes (Wang et al., 2000, 2003, 2004; Scheible et al., 2004; Gutiérrez et al., 2007; Krouk et al., 2010b). The transcriptome response to nitrate is highly dynamic, and changes in transcript expression can be detected as quickly as within 3 min of nitrate exposure (Krouk et al., 2010a; Varala et al., 2018). The nitrate response is also cell- and tissue-specific (Gifford et al., 2008; Walker et al., 2017). A meta-analysis of the transcriptomic response of nitrate-treated roots revealed that ∼60% of nitrate-responsive genes respond to nitrate in only one experiment, highlighting the context-specific and complex nature of the nitrate response (Canales et al., 2014).
NR-null plants cannot reduce nitrate to nitrite, the first step in the nitrate assimilation pathway. Nitrate transport and assimilation genes, as well as genes coding for enzymes in C metabolism, are still controlled by changes in nitrate availability in NR-null plants, pointing to a role for nitrate as a signaling molecule (Scheible et al., 1997). Years later, microarray analysis showed that a significant fraction (∼38%) of nitrate-responsive genes in Arabidopsis was able to respond to nitrate in both wild-type and NR-null plants, indicating that their response was independent of nitrate reduction and assimilation (Wang et al., 2004). These experiments were critical to demonstrating the signaling role of nitrate in addition to its well-known nutritional role.
Nitrate can control long-term responses related to changes in growth and development, as discussed in further sections. Transcriptome reprogramming in response to nitrate is supported by a temporal hierarchy of TFs that initiates dynamic GRNs, which act at the cellular level to orchestrate organ responses (Varala et al., 2018). In Arabidopsis roots, nitrate triggers functionally distinct but coordinated responses across different cell types (Gifford et al., 2008; Walker et al., 2017). For example, nitrate regulation of auxin signaling in pericycle cells has been linked to LR formation in response to nitrate (Gifford et al., 2008; Vidal et al., 2013; Yu et al., 2016; Walker et al., 2017).
NRT1.1/NPF6.3 Is a Nitrate Transceptor
The idea that nitrate could act as a signal led to a search for a nitrate sensor that would directly bind nitrate and activate downstream signaling pathways, similar to the NarX/NarL two-component system in bacteria (Nohno et al., 1989). Phenotypic and transcriptomic analyses of nitrate transporter mutants offered clues as to the identity of the nitrate-sensing apparatus. It turned out to be the transporter itself: NRT1.1, first identified as a gene responsible for chlorate sensitivity in Arabidopsis (Tsay et al., 1993). The resolution of the crystal structure of Arabidopsis NRT1.1 was key for understanding the molecular basis of its dual affinity dependent on nitrate availability. Crystallized NRT1.1 adopts a dimer conformation, with each N-terminal half facing and interacting with each other (Parker and Newstead, 2014; Sun et al., 2014b), as suggested from in vitro cross-linking and fluorescence resonance energy transfer experiments in X. laevis oocytes (Sun et al., 2014b). Importantly, phosphorylation of Thr-101 interferes with NRT1.1 dimerization by introducing electrostatic and conformational changes into the monomers (Sun et al., 2014b). Unmodified NRT1.1 therefore adopts a dimer configuration suitable for low-affinity nitrate uptake, while low N-mediated phosphorylation at Thr-101 triggers conversion to a monomer with higher structure flexibility, which might explain the switch to high affinity (Parker and Newstead, 2014).
NRT1.1 expression in rapidly growing tissues such as the pericycle during LR formation, together with impaired LR primordia elongation in nrt1.1 mutants, suggested that NRT1.1 function was not restricted to nitrate transport (Guo et al., 2001). Further work proposed a signaling role for NRT1.1 in the control of the high-affinity transporter NRT2.1 (Muños et al., 2004) and for the induction of LR elongation by localized high nitrate availability (Remans et al., 2006).
Demonstration that NRT1.1 acted as a nitrate transceptor came in 2009 (Ho et al., 2009). Transceptors are commonly involved in nutrient transport and sensing in many organisms (reviewed by Kriel et al. [2011] and Steyfkens et al. [2018]). Evidence that NRT1.1 was a nitrate transceptor was obtained with an nrt1.1 mutant that prevented nitrate uptake but retained the induction of gene expression in response to nitrate, including NRT2.1 expression (Ho et al., 2009). We now know that NRT1.1 can activate multiple mechanisms of nitrate sensing and signaling required for the regulation of nitrate-responsive genes in Arabidopsis roots (Wang et al., 2009; Bouguyon et al., 2015), but it also affects diverse biological processes, including root development and architecture, auxin transport, seed dormancy, flowering time, and stomatal movements (Fan et al., 2017; Zhang et al., 2018; Fredes et al., 2019).
The Backbone of N Signaling
While the identification of NRT1.1 as a nitrate transceptor was clearly critical, much remained unanswered as to how nitrate was sensed and transduced into changes in gene expression (Figure 2). For instance: how is NRT1.1 phosphorylation regulated? The kinase CIPK23 interacts with CBL9 or CBL1 and phosphorylates NRT1.1 at Thr-101 under low-nitrate conditions, switching the transporter to its high-affinity function (Ho et al., 2009). Besides this switch in nitrate affinity, the phosphorylation state at Thr-101 also controls the expression of downstream genes such as the NRT2.1 transporter and genes involved in N, sulfur, and selenium metabolism (Ho et al., 2009; Bouguyon et al., 2015). The protein phosphatase 2C ABI2, normally involved in abscisic acid (ABA) signaling, can dephosphorylate CBL1 and CIPK23, thereby inactivating this calcium sensor/kinase complex and preventing NRT1.1 phosphorylation (Léran et al., 2015). abi2 and nrt1.1 mutants have similar phenotypes, including the loss of low-nitrate-dependent NRT2.1 expression and failure to induce LR elongation in high-nitrate versus low-nitrate sides in a split-root experimental design (Léran et al., 2015). Since ABA binding to its receptors inactivates ABI2, this could represent a mechanism to decrease nitrate uptake and root remodeling under stress conditions to preserve cellular energy.
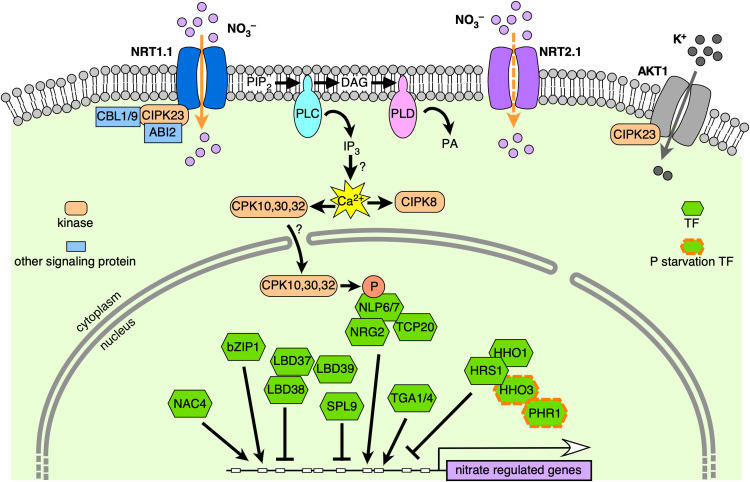
Figure 2.
A Summary of Nitrate Signaling Pathways.
NRT1.1 is at the first layer of the nitrate signaling pathway. CIPK23, CBL1/9, and ABI2 control the NRT1.1 phosphorylation status, switching its nitrate affinity. The transduced nitrate signal through NRT1.1 activates PLC activity, increasing calcium accumulation. Then, the calcium signals are decoded by subgroup III CPKs, which in turn phosphorylate the NLP7 transcription factor, promoting its nuclear retention and the activation of nitrate-responsive target genes. NLP7 physically interacts with NLP6, TCP20, and NRG2 to activate the expression of genes involved in nitrate metabolism. NRT2.1 has also been proposed as a nitrate sensor. Myriad TFs have been discovered as important regulators of gene expression in response to nitrate or as integrators of N and P signaling (HRS1, HHO1, and PHR1). See text and Supplemental Data Set 2 for relevant references.
The involvement of calcium-related proteins (CBL1, CBL9, and CIPK23) in the control of NRT1.1 function prompted the idea that calcium might act as a second messenger in nitrate signaling. Early studies using maize (Zea mays) and barley (Hordeum vulgare) detached leaves showed that pretreating leaves with the calcium chelator EGTA or the calcium channel blocker LaCl3 attenuated the expression of nitrate-responsive genes (Sakakibara et al., 1997; Sueyoshi et al., 1999). A more recent study showed that nitrate treatment caused a rapid increase of cytosolic calcium levels, which in turn triggers changes in gene expression in Arabidopsis roots (Riveras et al., 2015). Downstream of calcium signaling, CPKs act as molecular links between NRT1.1 and NLP TFs, as detailed below. The transport activity of NRT1.1 induces calcium waves through the action of a phospholipase C and inositol triphosphate (Riveras et al., 2015) that are decoded by subgroup III CPKs, which then phosphorylate NLP7, thus promoting its nuclear localization and activation of target genes (Liu et al., 2017). Alignment of the nine Arabidopsis NLPs and four orthologous lotus (Lotus japonicus) NLPs identified a conserved Ser-205 residue in NLP7 as a candidate CPK phosphorylation site. Mass spectrometry results confirmed that Ser-205 in NLP7 was (1) phosphorylated in vivo in the presence of nitrate (Liu et al., 2017) and (2) abolished in a cpk triple mutant. Transcriptome analysis in the same cpk triple mutant revealed that CPK-dependent nitrate-induced genes largely overlapped with genes controlled by NLP7 (Figure 2). These results support a role for CPKs in NLP phosphorylation and in the control of nitrate responses.
Although there have been considerable advances in recent years in the identification of nitrate sensing and signaling components, many questions remain. For instance, the function of the NRT1.1 transceptor does not explain the full extent of nitrate responses, suggesting that additional sensing mechanisms exist in plants (Wang et al., 2009; Bouguyon et al., 2015). In this context, NRT2.1 was suggested as a putative nitrate sensor (Little et al., 2005). Some nitrate-responsive genes hint at both calcium-dependent and calcium-independent branches (Riveras et al., 2015; Liu et al., 2017). Other signaling molecules include phosphatidic acid, G-protein complexes, and reactive oxygen species (Figure 2; Shin et al., 2005; Hong et al., 2009; Liang et al., 2018; Wany et al., 2018; Chakraborty and Kanyuka, 2019). However, their contribution to or crosstalk with the canonical nitrate signaling pathway remains to be addressed.
Systemic Nitrate Signaling Mechanisms
Systemic signaling allows a plant to elicit a plant-wide response by communicating locally perceived stimuli to distant organs. However, how different parts of the plant can perceive and communicate N status to the whole plant has been poorly studied. Potential systemic signals have been found such as malate, amino acids, or the plant growth rate (reviewed in Gent and Forde, 2017). However, whether and how these signals report the plant nitrate status remain unclear (Gent and Forde, 2017). This gap in knowledge on systemic nitrate regulation may originate from the difficulty of experimentally uncoupling the intertwined local and systemic responses in nitrate signaling (Li et al., 2014).
Cytokinins (CKs) do play a central role in root-to-shoot communication in nitrate signaling. Nitrate nutrition, CK content, and shoot growth appear correlated in different plant species (Ruffel et al., 2011; Sasaki et al., 2014; Guan, 2017; Poitout et al., 2018). In Arabidopsis, nitrate induces CK synthesis, in particular through the upregulation of IPT3, a gene encoding a key enzyme in CK biosynthesis (Miyawaki et al., 2004; reviewed by Sakakibara et al. [2006]). Importantly, CKs move through the plant: the translocation of precursor (trans-zeatin) and active (trans-zeatin riboside) CK species requires the transporter ABCG14 to load the xylem with root-borne CKs (Ko et al., 2014; Zhang et al., 2014). Once above ground, active and precursor CKs control leaf expansion and shoot apical meristem activity, respectively (Osugi et al., 2017; Landrein et al., 2018). Root-to-shoot trans-zeatin translocation is also an important component of N-systemic signaling that controls root responses to nitrate provision. Indeed, plants subjected to homogeneous or heterogeneous nitrate treatment differ in their accumulation of trans-zeatin in shoots. Thus, disruption of CK biosynthesis or translocation leads to a lack of trans-zeatin accumulation and to the repression of genes involved in nitrate transport and metabolism under nitrate-supplied conditions (Poitout et al., 2018). Moreover, CKs play a key role in the systemic N signal that controls root architecture. When plants are grown in split-root conditions, in which half of the root system is grown in high nitrate concentrations and the other half in low nitrate, roots in the high nitrate side show a dramatic proliferation as compared with roots of plants grown in homogenous high nitrate conditions, and this response depends on nitrate accumulation in shoots (Scheible et al., 1997; Remans et al., 2006). Interestingly, ipt mutants show no proliferation of LRs on the high-nitrate side as compared with homogeneous high nitrate, suggesting that CKs are a systemic signal involved in N-demand signaling. Furthermore, decapitated plants lose both N-demand and N-supply responses, indicating the existence of a root-to-shoot-to-root signal relay (Ruffel et al., 2011) that involves the accumulation of trans-zeatin (Ruffel et al., 2016). Accumulation of trans-zeatin modifies an N-assimilation pathway in shoots involving Gln (Poitout et al., 2018). Thus, Gln might serve as a shoot-to-root signal communicating nitrate availability in roots at the systemic level. Gln is a possible candidate, since amino acids are commonly found in the phloem sap and their role as communicators of N status has been previously described (Muller and Touraine, 1992; Miller et al., 2008; Gent and Forde, 2017).
Besides hormones, small peptides can act as signaling molecules in nitrate signaling. N deprivation induces the expression of CEP and CLE peptide-encoding genes in roots. CLE1, -3, -4, and -7 can interact with CLV1, repressing LR emergence and growth under N deprivation. Since CLE mRNAs are expressed on the pericycle, while CLV1 is expressed in phloem companion cells, CLE peptides are proposed to serve as cell-cell mobile signals, integrating N signals into root responses (Araya et al., 2014a, 2014b).
CEPs are a family of short secreted peptides that serve as signaling molecules in plants. Overexpression of members of the CEP family causes repression of root growth, while their loss promotes root development. CEPs are recognized by the Leucine rich repeat-receptor kinase receptors CEPR1 and CEPR2 in Arabidopsis (Tabata et al., 2014). Inactivation of CEPRs causes enhanced LR elongation, leaf chlorosis, dwarfism, and anthocyanin accumulation, reminiscent of N-starved plants. In addition, expression of NRT2.1 and NRT3.1 as well as nitrate uptake are impaired in cepr mutants. Based on split-root experiments, CEPs are specifically induced during nitrate deficiency, while nitrate transporter genes NRT2.1, NRT3.1, and NRT1.1 are induced in the presence of high nitrate. Grafting experiments using cepr1 cepr2 double mutant scions and wild-type rootstocks, together with analysis of CEP levels in xylem sap, demonstrated that CEPs act as mobile root-borne signals that are perceived by CEPRs in shoots and mediate systemic N-demand signaling. This systemic shoot-to-root signaling is due to the synthesis in shoots of two polypeptides, CEPD1 and CEPD2, which translocate to roots and activate the expression of nitrate transporters specifically on the side exposed to high nitrate (Ohkubo et al., 2017). In contrast to CK signaling that depends directly on nitrate, CEP signaling responds to general N deficiency, representing a different component of the N-demand systemic signaling.
Another long-distance communication system consists of translocation of TFs as reporters for tissue N status, for example HY5, as detailed earlier (Chen et al., 2016).
The decoding of local and systemic signaling pathways and the identification of molecular components regulating nitrate responses may thus contribute to a better understanding of how plants sense and respond to changes in N availability, providing unique targets for improving NUE in crops.
REGULATORY NETWORKS OF THE NITRATE RESPONSE IN PLANTS
Genomics, high-throughput validation of protein-DNA interactions, and systems biology approaches have increased the speed at which TFs involved in nitrate responses are being identified. This includes the use of machine-learning methods to predict networks (Krouk et al., 2010b; Varala et al., 2018; Brooks et al., 2019), high-throughput plant cell-based TF-perturbation assays such as Transient Assay Reporting Genome-wide Effects of Transcription (TARGET) factors, illustrated in Supplemental Figure 1; (Bargmann et al., 2013; Varala et al., 2018; Brooks et al., 2019), Y1H screening (Gaudinier et al., 2018), and the measure of accessible chromatin regions in response to nitrate (Alvarez et al., 2019). To date, more than 40 TFs spanning several TF families have been identified that bind to the promoters or control the expression of genes involved in nitrate transport, nitrate reduction, and nitrate assimilation (Gutiérrez et al., 2008; Rubin et al., 2009; Marchive et al., 2013; Alvarez et al., 2014; Guan et al., 2014; Para et al., 2014; Medici et al., 2015; Xu et al., 2016b; Maeda et al., 2018; Varala et al., 2018; Brooks et al., 2019). These data can be integrated into regulatory networks to further our understanding of how nitrate-dependent changes in gene expression impact plant physiology, growth, and development (Figure 3; Table 1; Supplemental Table 3). Table 1 summarizes the data associated with the influential transcription factors that appear in Figure 3 and are discussed below.
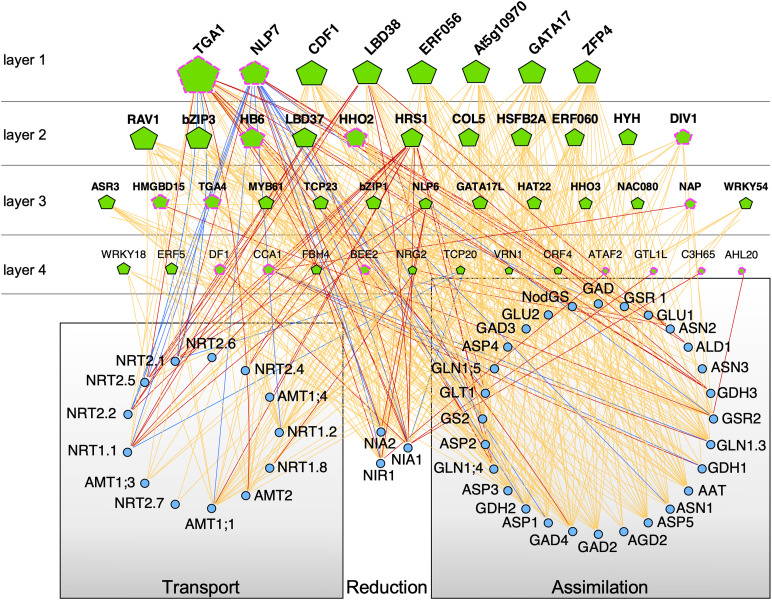
Figure 3.
Integrative GRN Analysis with the Most Influential TFs in N-Regulatory Networks.
Genes are drawn as pentagons (transcription factors) or circles (target genes). The size of the triangle is proportional to the number of targets bound or regulated by each TF (outdegree). The four TF layers were organized according to their outdegree. TFs were selected based on published evidence of TF regulation or TF binding to genes involved in N transport, N reduction, and N assimilation in Arabidopsis. Blue edges indicate ChIP binding data (Gutiérrez et al., 2008; Marchive et al., 2013; Alvarez et al., 2014; Guan et al., 2014; Para et al., 2014). Orange edges indicate TARGET and Y1H data (Para et al., 2014; Medici et al., 2015; Gaudinier et al., 2018; Varala et al., 2018; Brooks et al., 2019). Red edges indicate in planta TF regulation and chromatin accessibility (Gutiérrez et al., 2008; Rubin et al., 2009; Marchive et al., 2013; Xu et al., 2016b; Maeda et al., 2018; Alvarez et al., 2019; Brooks et al., 2019). The pink border of the pentagons denotes TF regulated in response to nitrate (Alvarez et al., 2019).
Table 1. Influential TFs in Nitrate Regulatory Networks.
| Arabidopsis Genome Initiative Code | Name | TF Family | No. of Edges in Network | No. of Edges, N Transport | No. of Edges, N Reduction | No. of Edges, N Assimilation | References |
|---|---|---|---|---|---|---|---|
| At5g65210 | TGA1 | bZIP | 38 | 9 | 5 | 24 | (Alvarez et al., 2014; Canales et al., 2017; Brooks et al., 2019) |
| At5g62430 | CDF1 | Dof-type zinc finger | 23 | 5 | 3 | 15 | (Varala et al., 2018; Brooks et al., 2019) |
| At4g24020 | NLP7 | RWP-RK | 22 | 7 | 6 | 9 | (Castaings et al., 2009; Marchive et al., 2013) |
| At2g22200 | ERF056 | ERF/AP2 | 21 | 1 | 3 | 17 | (Brooks et al., 2019) |
| At3g49940 | LBD38 | ASL/LBD | 21 | 3 | 4 | 14 | (Rubin et al., 2009; Brooks et al., 2019) |
| At1g66140 | ZFP4 | Zinc finger | 20 | 5 | 1 | 14 | (Brooks et al., 2019) |
| At3g16870 | GATA17 | GATA | 20 | 4 | 3 | 13 | (Brooks et al., 2019) |
| At5g10970 | C2H2 | C2HC zinc fingers | 20 | 4 | 1 | 15 | (Brooks et al., 2019) |
| At1g13260 | RAV1 | AP2/B3 | 19 | 5 | 2 | 12 | (Brooks et al., 2019) |
| At5g15830 | bZIP3 | bZIP | 19 | 2 | 3 | 14 | (Brooks et al., 2019) |
| At2g22430 | HB6 | Homeobox | 17 | 5 | 2 | 10 | (Brooks et al., 2019) |
| At1g68670 | HHO2 | G2-like | 16 | 3 | 2 | 11 | (Brooks et al., 2019) |
| At5g67420 | LBD37 | ASL/LBD | 16 | 6 | 3 | 7 | (Rubin et al., 2009; Brooks et al., 2019) |
| At1g13300 | HRS1 | G2-like | 14 | 8 | 2 | 4 | (Medici et al., 2015; Maeda et al., 2018) |
| At4g39780 | ERF060 | ERF/AP2 | 14 | 0 | 3 | 11 | (Brooks et al., 2019) |
Identification of TFs Involved in Nitrate Signaling
The first TF identified in nitrate signaling pathways was ANR1, a MIKC-type MADS-box TF. ANR1 was isolated using a classical molecular genetics approach in a screen for nitrate-inducible genes in roots (Zhang and Forde, 1998). ANR1 loss-of-function mutants have impaired LR elongation in nitrate-rich localized patches, affecting plasticity of the root system (Zhang and Forde, 1998). It was later found that ANR1 is controlled at the transcriptional level by NRT1.1 (Remans et al., 2006). Although the overexpression of ANR1 promotes LR growth and initiation even in the absence of nitrate, the presence of nitrate further potentiates the promotion of LR growth, suggesting that nitrate exerts a posttranslational control of ANR1 activity (Gan et al., 2012).
Although characterizing genes whose transcripts are induced has proven a powerful tool to identify important TFs involved in nitrate signaling, many TFs are controlled at the posttranscriptional level (e.g., posttranslational modifications and/or control of subcellular localization). This is the case for NLP7, the best-characterized TF in the nitrate response to date. NLP7 was initially identified as an ortholog to lotus NIN proteins, involved in early steps of nodulation in legumes (Schauser et al., 1999), and to the NIT2 transcription factor that controls NR expression in Chlamydomonas reinhardtii (Camargo et al., 2007). nlp7 mutant plants have longer primary roots (PRs) and more LRs, a phenotype that is characteristic of N-starved plants, supporting the role of NLP7 as a key regulator of nitrate metabolism (Castaings et al., 2009). NLP7 activates the transcription of many nitrate-responsive genes, including nitrate reductases, NRT2.1, and NRT2.2 (Castaings et al., 2009; Wang et al., 2009).
Members of the NLP family can be divided into four subgroups: NLP1/2, NLP4/5, NLP6/7, and NLP8/9 (Schauser et al., 2005; Castaings et al., 2009). Y1H screening using four copies of a 43-bp nitrogen response cis-element (NRE) revealed that all NLPs bind the NRE element (Konishi and Yanagisawa, 2013). The kinases CPK10, CPK30, and CPK32 phosphorylate NLP7 in the presence of nitrate, resulting in its nuclear localization and concomitant induction of nitrate-responsive gene expression (Liu et al., 2017) within minutes of nitrate treatment (Marchive et al., 2013). As discussed in the previous section, this mechanism depends on calcium signaling (Riveras et al., 2015; Liu et al., 2017) and coordinates nitrate availability to transcriptome reprogramming. Overexpression of NLP6 in the absence of nitrate is not sufficient to induce nitrate-inducible genes (Konishi and Yanagisawa, 2013), while its N-terminal GAF-like region is necessary for its nitrate-dependent transcriptional activity in protoplasts (Konishi and Yanagisawa, 2013). This suggests that posttranslational modifications of NLP6 are similarly required for nitrate control of gene expression.
Previous studies have shown that a single TF does not explain the full induction of genes such as NRT2.1, NRT2.2, and nitrate and nitrite reductases (Castaings et al., 2009; Alvarez et al., 2014; Guan et al., 2014) and that protein-protein interactions between TFs are important for gene expression of nitrate-responsive genes (Xu et al., 2016b; Guan et al., 2017). Nitrate transporters and nitrate metabolic genes are indeed convergent targets of many of the known nitrate-related TFs. The C-terminal PB1 domain of NLP proteins mediates protein-protein interactions in yeast two-hybrid assays. The PB1 domain is distinct from the DNA binding domain but is necessary for full induction of nitrate-dependent expression of target genes (Konishi and Yanagisawa, 2019). These results are consistent with previous hypotheses on redundant and overlapping functions between different Arabidopsis NLP TFs.
NLP6 and NLP7 also physically interact with TCP20 (Guan et al., 2017). TCP20 binds to a 150-bp NRE and a 109-bp enhancer sequence found in the promoters of NRT2.1 and NIA1, respectively, and acts in systemic nitrate signaling for root foraging, integrating cell cycle-related processes and root growth (Guan et al., 2014). NLP6, NLP7, and TCP20 bind to adjacent sites in the upstream promoter region of the NIA1 gene. The PB1 domains of NLP6 and NLP7 as well as the His- and Gln-rich domain of TCP20 are important for their physical interaction (Guan et al., 2017).
Additional TFs for nitrate responses have been identified. Using an integrative bioinformatics approach, the genes encoding the TGA-type TFs TGA1 and TGA4 were consistently regulated by nitrate across many experiments and therefore were good candidates to mediate nitrate responses (Alvarez et al., 2014). Ninety-seven percent of the genes that have an altered expression in the tga1 tga4 double mutant relative to wild-type plants are nitrate-responsive, indicating that TGA1 and TGA4 have a global role in nitrate response in roots. TGA1 and TGA4 can bind directly to the NRT2.1 and NRT2.2 promoters, and the initiation of LRs is affected in both tga1 tga4 and nrt2.1 nrt2.2 double mutants, suggesting that TGA1 and TGA4 regulate LR development at least partly via NRT2.1 and NRT2.2 (Alvarez et al., 2014). Induction of root hair development in response to nitrate treatments is also impaired in tga1 tga4 double mutants (Canales et al., 2017). TGA1 and TGA4 directly modulate the expression of CPC, a key regulator of root hair cell fate, which in turn increases the production of root hairs and nitrate uptake (Canales et al., 2017). Besides the known interactions of TGA1 with other members of the TGA family in the plant stress response (Shearer et al., 2012), putative interactors of TGA1 have been revealed using a high-density Nucleic Acid Programmable Protein Array (NAPPA) to map protein-protein interaction networks (Yazaki et al., 2016). The TF-NAPPA data set revealed that TGA1 can interact with proteins involved in nitrate signaling such as CIPK23 but also with proteins involved in hormone signaling such as HAI2 and the ABA receptor PYL6, positioning TGA1 as an integrator of hormone and nitrate signaling networks (Yazaki et al., 2016).
High-Throughput Assays to Identify TFs and TF Targets
After the identification of key TFs involved in N signaling, a second step to unravel the complex structure of regulatory networks is to systematically identify TF targets and the impact of these N-regulatory networks on organism function. Different approaches have been undertaken to fulfill this general goal, including integration of available high-throughput data on TF-DNA interactions obtained by technologies such as Y1H, protein binding microarrays (Weirauch et al., 2014), or DNA affinity purification sequencing (O’Malley et al., 2016).
To identify TFs and their targets involved in the regulation of root development in response to nitrate, Gaudinier et al. (2018) used an enhanced Y1H assay to screen for candidate TFs expressed in roots against 98 promoters of genes spanning different processes related to N. They generated a regulatory network composed of 1660 protein-DNA interactions between 345 TFs and all 98 promoters. TFs that bind genes participating in multiple N-related processes are more likely to be important for plant growth. This transcriptional network is highly enriched in genes regulated by ABA, auxin, methyl jasmonate, or ethylene. In agreement, mutants in transcription factors involved in ethylene (ERF4, ERF70, and ERF107) and auxin (ARF18) signaling show altered phenotypic responses to N availability (Gaudinier et al., 2018).
Functional assays of TFs in plants or plant cells offer a cellular context of regulatory networks, revealing when physical interactions lead to gene regulation. However, significant hurdles exist to identify the genes acting downstream of TFs within these networks. Mutants or overexpressors of the TF help define genes under the control of each TF, while a high-quality ChIP antibody or epitope-tagged transgenic line is needed to identify promoters bound by each TF; neither step is adaptable to high-throughput approaches. The development of TARGET, a plant cell-based temporal TF perturbation system, has been of tremendous help (Supplemental Figure 1; Bargmann et al., 2013). The TARGET assay can validate direct TF-target interactions based on TF-induced changes in gene expression (Bargmann et al., 2013; Para et al., 2014; Medici et al., 2015; Doidy et al., 2016; Sparks et al., 2016) or by TF binding (Para et al., 2014; Doidy et al., 2016). A scalable version of TARGET recently enabled the validation of genome-wide targets for seven N-responsive TFs in shoots (Varala et al., 2018) and 33 N-responsive TFs in roots (Brooks et al., 2019).
TARGET was successful in defining the genome-wide targets of HRS1 (Medici et al., 2015). Direct targets of HRS1 are enriched in genes related to auxin and ABA signaling pathways as well as P metabolism (Medici et al., 2015). HRS1 and its close homolog HHO1 repress PR growth caused by P starvation, but only when nitrate is present in the medium, indicating that they participate in a pathway at the intersection of N and P signaling (Medici et al., 2015).
TARGET also captured the early and transient action of bZIP1 in the N-response cascade (Para et al., 2014). Time-series ChIP experiments conducted 1 to 3 min following TF nuclear import revealed that bZIP1 bound transiently to promoters of early N-response genes (Para et al., 2014). 4-thiouracil labeling of de novo transcripts to affinity-capture bZIP1-initiated mRNAs determined that transiently bound bZIP1 targets were actively transcribed at times when the TF was no longer bound (Doidy et al., 2016). Importantly, these transiently bound and regulated bZIP1 targets were the most relevant to early N signaling in whole roots (Para et al., 2014). This provided the first genome-wide evidence for Hit-and-Run transcription in plants. This model posits that a TF trigger (the Hit) can organize a stable transcriptional complex, including the recruitment of other TFs, so that transcription can continue even after the initiating TF is no longer bound (the Run). This opened a new perspective on dynamic aspects of transcriptional regulation in plants (Charoensawan et al., 2015; Varala et al., 2015).
Recently, a DNase I hypersensitivity sequencing assay uncovered TF-target interactions, taking into account the native chromatin structure during the nitrate response of Arabidopsis roots (Alvarez et al., 2019). Local changes in DNase I cleavage patterns were observed in response to nitrate at specific loci bound by TFs. Rapid changes in TF occupancy correlated with RNA polymerase II (RNPII) recruitment and with changes in transcript accumulation in response to nitrate. By integrating genomic footprinting, transcriptional regulation of gene expression based on RNPII occupancy, and transcriptome data, a regulatory network was constructed that captures the relative contributions of known and new TFs in nitrate signaling. The network captured an interaction between NAP and NRT2.1, validated by ChIP and TARGET assays, illustrating NAP as a direct activator of NRT2.1 expression and a novel positive regulator of nitrate uptake in roots (Alvarez et al., 2019).
Systems Approaches to Model GRNs in Plants
Because of the complex underlying structure and relative recalcitrance to classical genetics, nitrate regulatory networks are an ideal framework for systems biology approaches to reveal features potentially considered as emergent properties (Gutiérrez, 2012).
Interestingly, the N community has been among the first to pioneer modeling approaches and data integration in plant biology. MapMan was one of the first data integration tools (Thimm et al., 2004) and opened avenues toward the integration of transcriptomic data sets with metabolic insights (Gibon et al., 2006; Schwacke et al., 2019). MapMan was recently used to find enriched functional categories dependent on CPK10, CPK30, and CPK32 kinases that regulate early steps in nitrate signaling enriched in N metabolism, glycolysis, carbohydrate metabolism, and N transporters (Liu et al., 2017).
This was followed by the Arabidopsis MultiNetwork model, which integrates genes, metabolites, microRNA (miRNA), and molecular interactions such as protein-protein, protein-DNA, and miRNA-target (Gutiérrez et al., 2007). This resource revealed relationships between N and the circadian clock via the CCA1 TF (Gutiérrez et al., 2008) and uncovered connections between miR167 and ARF8 in the pericycle of roots (Gifford et al., 2008).
To broadly enable such approaches, the VirtualPlant website was created to provide a platform for systems biology where biologists can upload their genomic data and execute various data visualization and analysis tools (Katari et al., 2010). VirtualPlant’s GeneCart supports the iterative nature of systems biology analysis and allows users to save results from one analysis and feed them into another. Applications range from Gene Ontology enrichment analysis to gene network visualization and analysis using the integrated Arabidopsis MultiNetwork (http://virtualplant.bio.nyu.edu/cgi-bin/vpweb/).
Myriad transcriptomics studies have evaluated different aspects of the plant N response, such as the effect of N source (Wang et al., 2004, 2007; Ristova et al., 2016), time after nitrate treatments (Krouk et al., 2010b; Varala et al., 2018), nitrate concentration (Wang et al., 2007), tissue type (Wang et al., 2003), and cell type (Gifford et al., 2008). An integrative meta-analysis of all data revealed the most consistently regulated genes by nitrate across different experimental conditions (Canales et al., 2014). The large number of TFs in this set of genes suggests that transcriptional reprogramming is at the core of plant adaptation to changes in nitrate availability. Weighted gene coexpression network analysis (Langfelder and Horvath, 2008) identified gene modules associated with specific biological functions, including C metabolism and trichoblast differentiation (Canales et al., 2014). It was later confirmed that nitrate regulates trichoblast differentiation through TGA1 (Canales et al., 2017).
In contrast to correlation-based methods, time series-based machine-learning approaches can predict causality (i.e., the regulatory influence of TFs on their targets in the data set and in out-of-sample data). Many algorithms have been developed to address these tasks, including Boolean networks, Bayesian networks, regularized linear regression, or nonlinear tree-based regression approaches (reviewed in Krouk et al., 2013).
To identify TFs in a temporal N-initiated cascade, TF-target edges were inferred using a time series-based machine-learning method called Dynamic Factor Graph (DFG; Mirowski and LeCun, 2009; Krouk et al., 2010b). This DFG approach learns network models that can predict gene expression states at untested time points (Krouk et al., 2010b). An early application of this approach identified SPL9 as a network hub of early nitrate-response genes in roots, which was confirmed to modify the nitrate response kinetics of NIA2 as well as of many nitrate-controlled genes (Krouk et al., 2010b). A more recent application of the DFG approach inferred a GRN containing 172 nitrate-responsive TFs and 2174 dynamic nitrate-responsive genes in shoots (Varala et al., 2018). Using validated genome-wide targets for three TF hubs in this GRN identified by the functional TF-target assay (CRF4, SNZ, and CDF1), a precision cutoff was experimentally determined to “prune” the TF-target predictions in the network to high-confidence 155 TFs and 608 targets (Varala et al., 2018). This network precision was reconfirmed using genome-wide TF-target regulation data for four additional TFs: TGA1, HHO5, HHO6, and PHL1 (Varala et al., 2018). This experimentally refined GRN identified the temporal relationship of known and validated regulators of N signaling, including NLP7, NLP8, TGA1, TGA4, NAC4, HRS1, and LBD37, LBD38, and LBD39, as well as 146 novel TF regulators. CRF4, the earliest TF in the time-based N-response cascade, was validated to modulate 15N uptake and NUE in planta (Varala et al., 2018).
Similarly, the DFG machine-learning algorithm was used to infer TF-target edges for 145 N-responsive TFs in roots (Brooks et al., 2019). The inferred TF-target edges were pruned for high-confidence edges using 71,836 experimentally validated TF-target edges for 33 TFs identified using TARGET, which were then used in a precision/recall analysis to generate a GRN consisting of interactions between 145 TFs and 311 targets in the root N response (Brooks et al., 2019). To integrate data from TF-target interactions identified in root cells via TARGET with in planta TF perturbation data and high-confidence predicted edges in the GRN, the network-walking approach was developed. These combined data sets were used to connect TFs to their direct targets in root cells and to their indirect targets in planta via intermediate TFs. Network walking showed that TGA1 directly regulated 40% (508 of 1458) of the N-responsive genes in roots, including 63 of 145 N-responsive TFs. Moreover, 76% of the indirect targets of TGA1 in planta were linked to TGA1 through 49 intermediate TFs. This suggests that TGA1 is a high-level regulator whose effect on downstream secondary TFs amplifies a transcriptional cascade to control N responses (Brooks et al., 2019). Moreover, TGA1 was among the most influential TFs in nitrate transport, nitrate reduction, and nitrate assimilation when publicly available data of TF-target interactions for TFs involved in the response to nitrate were interrogated (Figure 3; Table 1).
The flood of genomic data over the last decade in the plant nutrition community and others has been the impetus for increased collaboration between the fields of computer science and biology. Moving forward, there is an immediate need to make better use of existing data from Arabidopsis studies by developing new data integration tools and predictive modeling algorithms. Using Arabidopsis as a framework to learn how to integrate diverse data sets should facilitate similar analyses in species with fewer resources.
DEVELOPMENTAL OUTPUTS
Plants are highly plastic organisms that control growth and development of their organs in order to optimize nutrient acquisition and use. N availability and N-related GRNs exert a major control over endogenous developmental programs in plants, from germination to vegetative and reproductive development.
Nitrate Control of Seed Dormancy and Germination
Different environmental signals control seed dormancy, including temperature, light quality and quantity, oxygen, water potential, organic acids, pH, and nitrate availability. The integration of these signals allows the seed to have temporal information about the time of the year and suitability for germination. Nitrate can act as a potent signal controlling seed dormancy and germination in different plant species. Arabidopsis plants exposed to high nitrate provision produce seeds that are less dormant than seeds from plants exposed to low nitrate. Similarly, seeds exposed to exogenous high nitrate are less dormant than low-nitrate-exposed seeds. Both effects are independent of nitrate reduction, suggesting that nitrate itself functions as a signal for the induction of germination (Alboresi et al., 2005). Parts of this regulation are due to the signaling function of NRT1.1, since (1) a null nrt1.1 mutant fails to induce germination in the presence of low nitrate (Alboresi et al., 2005) and (2) NRT1.1 expression increases before relief of dormancy during seasonal cycling (Footitt et al., 2011).
Analysis of laboratory dormancy cycling in Cape Verde island (Cvi), an Arabidopsis accession with strong dormancy, documented low CIPK23 expression in low-dormant and nondormant states, when seeds are more sensitive to nitrate, and high expression during primary and secondary dormancy, when sensitivity to nitrate is low (Cadman et al., 2006; Footitt et al., 2013). This indicates that phosphorylation of NRT1.1 might play a role in seed nitrate sensitivity and control germination timing. However, transcriptome analysis of Cvi exposed to different treatments inducing germination, including drying after ripening, seed hydration, cold, light, and exogenous nitrate, showed that all these signals elicit similar changes to the transcriptome of the germinating seed (Finch-Savage et al., 2007). Exposure to nitrate or cold stratification translates into similar changes at the germinating seed proteome level (Arc et al., 2012) and in the metabolism of mother plants exposed to low nitrate and low temperature during seed maturation (He et al., 2016), suggesting that integrators of common downstream environmental signals may act to control seed dormancy and germination. The hormones ABA and gibberellic acid (GA) are attractive candidates, given their roles in promoting seed dormancy and seed germination, respectively. Nitrate reduces levels of ABA in dormant seeds (Ali-Rachedi et al., 2004) by inducing the cytochrome P450 CYP707A2 (Matakiadis et al., 2009), encoding the main ABA catabolic enzyme present in the late-maturation stage of seed development and in hydrated seeds (Okamoto et al., 2006b). This regulatory mechanism is mediated by nitrate and involves NLP8, which is required for nitrate induction of CYP707A2 and many nitrate-controlled genes in seeds, including NIA1, NIA2, NIR, RFNR1, G6PD2, and At1g25550, pointing to a key role for NLP8 in the seed nitrate response. NLP8 can bind to the CYP707A2 promoter in three sites that are important for nitrate control. Nitrate induction of CYP707A2 via NLP8 reduces ABA levels in seeds, relieving dormancy (Yan et al., 2016). As previously discussed, the nitrate sensor NRT1.1 is involved in nitrate-mediated seed dormancy relief. Although nrt1.1 seeds are less sensitive to nitrate, normal germination levels can be achieved by increasing nitrate concentrations (Alboresi et al., 2005; Yan et al., 2016). nlp8 mutants, on the other hand, are insensitive to nitrate over a wide range of concentrations (Yan et al., 2016), indicating that alternative nitrate sensors may act upstream of NLP8 in seeds in addition to NRT1.1.
Nitric oxide (NO) has been proposed as such an alternative signal. NO scavengers abolish the positive effect of nitrate on seed germination and NO induces CYP707A2 (Bethke et al., 2006; Liu et al., 2009; Matakiadis et al., 2009). NO also controls the expression of ABI5, a key repressor of seed germination and postgerminative development (Finkelstein and Lynch, 2000; Lopez-Molina et al., 2001). NO triggers the degradation of group VII ERF TFs via the N-end rule pathway (Graciet and Wellmer, 2010). Degradation of group VII ERFs leads to a decrease in ABI5, releasing seed dormancy (Gibbs et al., 2014). Furthermore, NO can control ABI5 protein levels. NO causes S-nitrosylation of ABI5 in Cys-153 during seed germination. This S-nitrosylation controls ABI5 protein degradation via a proteasome-dependent pathway involving CUL4 and KEG E3 ubiquitin ligases, leading to the promotion of germination (Albertos et al., 2015). Besides ABI5, NO has been shown to S-nitrosylate SnRK2 proteins, key positive ABA signaling components, inactivating them and positively regulating germination and early seedling growth (Wang et al., 2015). In addition, NO induces Tyr nitration and inactivation of the ABA receptors PYR1 and PYL8 in plants (Castillo et al., 2015), further reinforcing the repressive effect over ABA signaling.
Nitrate Influences the Heterotrophic-Autotrophic Switch and Seedling Establishment
After seed germination, the emerging seedling initially grows fueled by seed reserves. Seedling establishment is complete when the seedling acquires photosynthetic capability and transitions to autotrophy. This process involves a reprogramming for the efficient use of endogenous reserves as well as resources that can be incorporated from the environment. N availability is an important cue for early postgerminative growth and positively influences cotyledon expansion and greening, hypocotyl growth, and the emergence of true leaves (Sato et al., 2009). Mobilization of triacylglycerols (TAGs), the main seed reserve in many plant species, is regulated by the C/N ratio, with high C/N ratios repressing eicosenoic acid breakdown. This represents a feedback mechanism in which the accumulation of sucrose, the main product of TAG mobilization, represses further TAG breakdown. Genes encoding proteins involved in photosynthesis are also repressed by a high C/N ratio (Martin et al., 2002). C/N ratio signaling controlling postgerminative growth is mediated by the membrane-localized CNI1 ubiquitin ligase (Sato et al., 2009). Under high C/N ratios, the CIPK protein kinases CIPK7, CIPK12, and CIPK14 accumulate and physically interact with and phosphorylate CNI1, resulting in its stabilization. CIPK14 and CBL8 can also phosphorylate CNI1 in a calcium-dependent manner (Yasuda et al., 2017). Ser/Thr phosphorylation of the C terminus of CNI1 is essential for its interaction with 14-3-3 proteins, leading to their ubiquitination and degradation (Sato et al., 2011; Yasuda et al., 2014). 14-3-3 proteins can bind phosphorylated motifs on enzymes involved in C and N metabolism such as nitrate reductase, Gln synthetase, sucrose phosphate synthase, and ADP-glucose pyrophosphorylase, regulating their activity in response to environmental signals (Bachmann et al., 1996; Comparot et al., 2003). Although a detailed mechanism is still elusive, the involvement of 14-3-3 proteins in downstream C/N control of postgerminative growth is an example of how dynamic changes in Ca2+ and protein phosphorylation can convey nutritional signals into the developmental programs of plants.
Nitrate-Controlled Shoot Growth
Nitrate availability has profound effects on the growth of aerial tissues. The positive effects of high nitrate on leaf growth and branching have been attributed to CKs (Rahayu et al., 2005; Müller et al., 2015). CKs are synthesized in response to nitrate in roots and translocated via the xylem to the shoots, where they control shoot growth (Takei et al., 2001, 2004; Sakakibara et al., 2006; Osugi et al., 2017). In particular, translocated trans-zeatin controls leaf size, while trans-zeatin riboside controls leaf size and shoot apical meristem activity (Osugi et al., 2017). Nitrate provision induces CK signaling in the shoot apical meristem, increasing expression of the homeodomain TF gene WUS and increasing the expression domain of CLV3. This leads to stem cell proliferation and increased meristem size, resulting in bigger rosettes and more flowers (Landrein et al., 2018).
Compared with nitrate sufficiency, low nitrate concentrations repress secondary shoot formation by inhibiting the elongation of axillary buds, leading to the production of elongating branches only on the most apical cauline nodes. In contrast to other developmental responses to nitrate provision, this phenotype is not dependent on nitrate but on overall N status of the plant. Strigolactones (SLs), hormones that are produced in the roots, are required for this N-limitation response, since mutants lacking SLs fail to repress branching. Auxin, a central negative regulator of root branching, is produced in leaves and transported rootward through the polar auxin transport system. In N-limited plants, the amount of exported auxin increases only in apical stem segments. This response is lost in mutants lacking SLs, indicating that an SL-auxin interaction is needed to control shoot branching in response to N availability (de Jong et al., 2014). On the other hand, CK mediates the positive effect of high nitrate on branching, since CK biosynthetic mutants ipt3 ipt5 ipt7 and signaling mutants arr3,4,5,6,7,15 have less active buds than wild-type plants (Müller et al., 2015). High nitrate can mimic the effect of CK supply on the accumulation of auxin transporters PIN3, PIN4, and PIN7 in the plasma membrane of xylem parenchyma cells of basal inflorescence internodes. This suggests that high nitrate might control branching by CK-mediated control of auxin transport by PIN3, PIN4, and PIN7 (Waldie and Leyser, 2018).
Root System Architecture Modulation by Nitrate
Roots acquire nutrients and provide an interface to interact with soil organisms and other plants. As such, root system traits are attractive targets for breeding programs and biotechnological applications (Den Herder et al., 2010; Rogers and Benfey, 2015). The root system architecture (RSA), or the given spatial disposition of the root system, is determined by both internal, development-related cues and external biotic and abiotic cues. The RSA is highly plastic, which allows the plant to efficiently forage for nutrients and water in the soil, where these resources are commonly unevenly distributed. The effects of N on root growth depend on N source and availability and can impact the development and growth of PRs, LRs, and root hairs. RSA is also influenced by the heterogeinity of N supply, with homogenous high nitrate suppressing PR and LR elongation (Zhang et al., 1999; Linkohr et al., 2002) and heterogenous, localized high nitrate increasing LR elongation and density with no major changes in PR length (Zhang and Forde, 1998; Linkohr et al., 2002).
A breakthrough in our understanding of the molecular basis of the root response to nitrate came in the late 1990s (Zhang and Forde, 1998). In this now classic article, ANR1 was identified as a nitrate-responsive TF that controls LR elongation in response to localized high nitrate availability. This same phenotype was later reported for nrt1.1 mutants, and it was shown that ANR1 acted downstream NRT1.1 in this pathway (Remans et al., 2006).
Interestingly, the nitrate transporter NRT1.1 also acts as an auxin transporter (Krouk et al., 2010a). NRT1.1 represses the growth of preemerged LRs and LR primordia by preventing auxin accumulation in LR tips via basipetal transport of auxin when nitrate is absent or present in very low concentrations. The auxin transport function of NRT1.1 is inhibited by high nitrate, which allows for the outgrowth of LRs under sufficient nitrate availability (Krouk et al., 2010a). The repressive function of NRT1.1 on LR development depends on the phosphorylated form of NRT1.1, since the phosphomimetic mutant T101D presents a wild-type phenotype, while a nonphosphorylable mutant, T101A, exhibits a phenotype similar to an nrt1.1 null mutant, which is characterized by a marked increase in LR density and length under low external nitrate concentration (Bouguyon et al., 2015). This increased LR development correlates with an increased auxin accumulation in root tips in the T101A mutant, confirming that the role of NRT1.1 on LR development is dependent on its auxin transporter function (Bouguyon et al., 2015).
Although the presence of nitrate strongly induces NRT1.1, the role of the encoded transporter is most evident in the absence of nitrate. To reconcile this fact, a detailed analysis of NRT1.1 protein dynamics throughout LR primordium development was conducted in the presence and absence of nitrate (Bouguyon et al., 2016). Nitrate repressed NRT1.1 protein accumulation during preemergence of LR primordia, which promotes local auxin accumulation and induction of LR primordium outgrowth, supporting the fact that NRT1.1 prevents both auxin accumulation and LR growth in response to low nitrate availability (Bouguyon et al., 2016). This suggests a finely tuned mechanism for the control of NRT1.1 function in which posttranscriptional mechanisms such as phosphorylation and protein accumulation play an important role in nitrate-dependent LR development.
As previously discussed, a signaling cascade composed of calcium and calcium-related kinases acts downstream of NRT1.1 to control GRNs that shape RSA in response to nitrate. Although NLP6/7 have been proposed as key TFs within these GRNs, other TFs include TGA1/4 (Alvarez et al., 2014), NAC4 (Vidal et al., 2013), HRS1 and HHO1 (Medici et al., 2015; Kiba et al., 2018; Maeda et al., 2018), TCP20 (Guan et al., 2017), CRF4 (Varala et al., 2018), SPL9 (Krouk et al., 2010b), and NRG2 (Xu et al., 2016b).
In the last 15 years, various lines of evidence have shown that plant hormones play a key role as integrators of N signals into root developmental programs. Transcriptome analysis has indicated that the interaction of N with hormones such as auxin, CKs, and ABA generates a rapid reprogramming of the root transcriptome that impacts root development-related networks to enact quantifiable changes in root architecture (Ristova et al., 2016). Hormones in turn can control the expression of genes involved in N uptake, metabolism, and signaling, providing feedback control over N signals (Gaudinier et al., 2018).
Due to its key role as a master regulator of root development, auxin has been the preferred candidate to evaluate in nitrate-dependent root responses. Pioneering studies from the mid-1950s provided the first insights into the interaction between N and auxin signals. These studies described an inverse relationship between N supply and PR growth, attributed to changes in auxin levels (Bosemark, 1954). Later work with auxin-related mutants and treatment with auxin analogs or auxin transport inhibitors suggested that nitrate effects on PR and LR growth depend on auxin homeostasis and signaling (Zhang et al., 1999; Forde and Lorenzo, 2001; Guo et al., 2005; Tian et al., 2008). These findings were further supported by transcriptome analysis and genome-wide TF binding analysis demonstrating that nitrate availability can regulate the levels of auxin transporters, biosynthesis genes, and signaling genes (Canales et al., 2014). For example, expression of TAR2, encoding the first step from l-Trp to indole-3-pyruvic acid, is induced by low nitrate, leading to an increase in indole-3-acetic acid (IAA) levels in LR primordia and emerging LRs (Ma et al., 2014). Low nitrate can also induce the local levels of IAA in LR primordia and LRs roots by regulation of AGL21, encoding a TF closely related to ANR1. AGL21 in turn regulates the levels of transcripts coding for enzymes involved in auxin biosynthesis, YUC1 and YUC5, TAR3, NIT4, and AAO1, stimulating LR initiation and growth (Yu et al., 2014).
Auxin transport from shoots to roots has been proposed as a long-range signal controlling LR development in response to nitrate (Guo et al., 2005; Liu et al., 2010). Auxin transport is mainly facilitated by members of the PIN polar transporters. In rice, expression of the PIN transporters PIN1c, PIN2, PIN9, and PIN10a-b is diminished in an OsNAR2.1 knockdown line and is accompanied by an inhibition in LR development under low-nitrate conditions (Huang et al., 2015). This effect might be due to a signaling function of the OsNRT2.1 transporter, a partner of OsNAR2.1, since no differences in nitrate content were found in an OsNAR2.1 knockdown line. As discussed above, NRT1.1 is also an auxin transporter, repressing the growth of preemerged LRs and LR primordia by preventing auxin accumulation in LR tips when nitrate is absent or very low (Krouk et al., 2010a).
Auxin perception and nitrate signaling are also closely interconnected. Nitrate induces transcription of the AFB3 auxin receptor gene, while N metabolites produced by nitrate reduction and assimilation reset AFB3 levels over time by posttranscriptional regulation via miR393 (Vidal et al., 2010). Downstream of AFB3, a pericycle regulatory mechanism involving IAA14 and the NAC4 and OBP4 transcription factors induces LR initiation and elongation in response to nitrate resupply of N-deficient roots (Vidal et al., 2013). By a still unknown mechanism, AFB3 can also repress PR elongation in response to nitrate (Vidal et al., 2010, 2013). Similarly, P deficiency represses the expression of the TIR1 auxin receptor gene, leading to the repression of LR development (Pérez-Torres et al., 2008), indicating specific roles for auxin receptors in conveying signals from different nutrients. Still in the pericycle, a second auxin-dependent regulatory mechanism is mediated by miR167, where it is repressed by nitrate, which then leads to a rise in the mRNA levels of its target ARF8. ARF8 in turn controls a module of nitrate-responsive genes, leading to LR elongation (Gifford et al., 2008).
Together with auxin, CK is also a key regulator of developmental and growth processes in roots. As with auxin, nitrate availability controls the levels of genes related to CK biosynthesis and signaling, such as the IPT genes, which catalyze the rate-limiting step of CK biosynthesis. CKs are transported to the shoot, where they control vegetative growth (Takei et al., 2001, 2004; Sakakibara, 2006), but they can also control the expression of nitrate transporters and nitrate metabolism-related genes (Kiba et al., 2011). Moreover, as discussed previously, one of the main roles of CKs is as a communicator of systemic N signaling that controls root architecture (Ruffel et al., 2011, 2016; Poitout et al., 2018). Recently, a role for CKs was reported in the control of PR growth (Naulin et al., 2020). In plants grown in the presence of nitrate, PR growth is stimulated by the promotion of cell proliferation and expansion, compared with plants grown without nitrate. Mutants in the key CK biosynthetic genes IPT3 and IPT5 and CK receptors AHK2, AHK3, and AHK4 show reduced PR growth when grown on nitrate, indicating that cytokinin biosynthesis and perception are needed for nitrate-stimulated PR growth. This alteration in PR growth is due to a failure in cell division in the meristem at early stages of postembryonic development, which is in part due to an altered expression of cell cycle checkpoint genes such as KRP1, SKP2α, and AXR3 in ahk mutant plants in the presence of nitrate (Naulin et al., 2020).
ABA is a key stress hormone integrating external abiotic and biotic stimuli that controls stomatal closure and seed dormancy but also PR and LR development. Work with ABA biosynthesis and signaling mutants showed that the repressive effect of high nitrate on LR elongation depends on ABA (Signora et al., 2001). Using an anti-ABA antibody, immunocytochemistry experiments showed that high nitrate availability triggers ABA accumulation in the endodermis and stele of the meristem and elongation zones of PRs and emerging LRs (Ondzighi-Assoume et al., 2016). Indeed, nitrate induces the accumulation of the endoplasmic reticulum-localized β-1,3-glucanase1, thus releasing bioactive ABA into the cytosol from inactive ABA d-glucopyranosyl ester conjugates. Nitrate can also induce the expression of ABA-responsive genes such as RAB18. ABA in turn induces the expression of nitrate-responsive genes NIA1, NIA2, and NIR to control root growth (Ondzighi-Assoume et al., 2016). Interestingly, in a parallel to NRT1.1 and auxin, NRT1.2 transports ABA (Kanno et al., 2012). Similarly, the Medicago truncatula NPF member NPF6.8 transports nitrate and ABA, modulating root length (Pellizzaro et al., 2014). This highlights a complex mechanism of nitrate-ABA interaction in nitrate-dependent control of root growth.
Ethylene is a gaseous hormone that controls many developmental aspects in plants and has been associated with nutritional responses to P, iron, and potassium (K; Romera et al., 2016). Treating plants first grown on low nitrate with high doses of nitrate can induce a rapid burst of ethylene in LR primordia and initiating LRs, resulting from increased ACC1 and ACO1 expression and corresponding enzymatic activities, key steps in ethylene biosynthesis (Tian et al., 2009). Ethylene regulates the expression of the nitrate transporters NRT1.1 and NRT2.1 and mediates the repressive effect of high-nitrate treatment on LR initiation and LR elongation (Tian et al., 2009). Similarly, nitrate deficiency, a condition that induces LR elongation (Khan et al., 2015), also triggers ethylene biosynthesis and stimulates ethylene signaling by inducing the expression of ethylene signaling components EIN3 and EIL1. Under low-nitrate conditions, NRT2.1 is upregulated and positively affects ethylene biosynthesis and signaling. In a negative feedback mechanism, ethylene in turn represses NRT2.1 expression, reducing high-affinity nitrate uptake and alleviating external nitrate deficiency stress (Zheng et al., 2013).
The role of other plant hormones in mediating changes in root development and growth in response to nitrate availability has been less investigated. However, regulatory network analysis supports a possible role for other hormones, such as methyl jasmonate, GA, and brassinosteroids, in root responses to nitrate (Gaudinier et al., 2018). In agreement, a recent genome-wide association study conducted to identify novel regulators of root foraging under N deficiency identified BSK3, a plasma membrane kinase associated with early brassinosteroid signaling (Ren et al., 2019). bsk3 mutants have shorter PR and LR under mild N deficiency due to a failure to induce cell elongation (Jia et al., 2019). However, it is currently unknown whether this mechanism depends directly on nitrate or whether it interacts with nitrate signaling components such as NRT1.1.
Control of Flowering Time by Nitrate
The transition from vegetative to reproductive growth is a crucial developmental decision during the life cycle of a plant and directly impacts plant yield, fertility, and survival of the progeny. As such, the timing of this transition is finely regulated by both endogenous and exogenous cues, by a complex network of interacting signaling pathways whose outputs converge onto the flowering time integrators SOC1 and FT, which in turn activate the expression of floral meristem identity genes such as AP1, LFY, and CAL. Distinct pathways that control flowering time integrate photoperiod, cold exposure (vernalization), plant age, ambient temperature, GA signaling, and the autonomous pathway (Davis, 2009; Kim et al., 2009; Song et al., 2012; Wang et al., 2014; Capovilla et al., 2015). The plant nutritional status is another major environmental factor influencing flowering. The role of N in flowering was reported over a century ago (Klebs, 1913), and fertilization strategies to modulate the timing of flowering to control yield are widely utilized in agriculture. Mutants in nitrate transport or metabolic genes have flowering time phenotypes (Tocquin et al., 2003; Seligman et al., 2008). The effect of nitrate over flowering can be described by a U-shaped curve, where both extremes of nitrate availability repress flowering, albeit by different mechanisms, while intermediate nitrate concentrations promote flowering (Lin and Tsay, 2017).
It is usually difficult to separate the effects of nitrate availability on vegetative growth and those directly affecting developmental phase change, since plant nutritional status also impacts their overall growth rate. In order to do just that, Castro Marín et al. (2011) designed an experimental system where 4 mM Gln was added to the medium to maintain constant internal levels of amino acids and proteins. Increasing nitrate concentrations (1, 10, or 35 mM) delayed flowering time in a dose-dependent manner. Plants at time of flowering had more leaves and produced more shoot biomass as nitrate levels increased but retained the same relative growth rate. Interestingly, withholding Gln from the medium abolished the acceleration of flowering caused by low nitrate levels, possibly due to a general slow plant growth rate (Castro Marín et al., 2011). This is consistent with the U-shaped flowering response to nitrate, in which concentrations of nitrate severely limiting plant growth (as experienced with low nitrate and no Gln) delay flowering time (Lin and Tsay, 2017; Gras et al., 2018). However, flowering time mutants retained their response to nitrate (plants grown on low nitrate still flowered earlier), indicating that nitrate acts in an independent pathway to control flowering.
Further studies have shown that nitrate availability can control the expression or activity of different floral regulator genes, thus supporting a mechanism by which nitrate signaling directly impacts flowering signaling pathways. Low N availability induces FNR1 gene expression as well as CRY1, encoding a blue light photoreceptor that acts in the photoperiod pathway and regulates the expression of CO and FT. Through FNR1, N availability regulates the NADPH/NADP+ ratio and the ATP/AMP ratio, with low N increasing NADPH/NADP+ and ATP/AMP ratios. In contrast, high N decreases NADPH/NADP+ and ATP/AMP ratios, the latter positively regulating protein levels of AMPKα1, the catalytic subunit of AMP-activated protein kinase (AMPK), as well as its circadian oscillation amplitude. AMPK then phosphorylates CRY1 in the nucleus, leading to its degradation and late flowering. This control of CRY1 abundance may in turn control the amplitude (but not the period) of the expression of central clock components CCA1, LHY, and TOC1, controlling flowering time as a function of external N provision (Yuan et al., 2016).
Nitrate availability also regulates the expression of the flowering repressor FLC as well as the FT, LFY, and AP1 genes. Indeed, high nitrate represses FT, LFY, and AP1 and induces FLC, leading to late flowering (Kant et al., 2011b). In a different study, Arabidopsis plants first grown in high nitrate and then subjected to a short-term exposure to low nitrate experienced a rise in the levels of the floral integrator SOC1, the GA biosynthesis gene GA1, and the photoperiod gene CO, with no effect on FRI and VIN3 or LD and FCA, participating in the vernalization and autonomous pathways, respectively. Increased levels of GA1 correlated with an increase in bioactive GA3 when plants were grown on low nitrate. These results indicate that low nitrate might interact with the photoperiod pathway through CO and with the GA pathway through GA1, leading to an induction in SOC1 transcript levels and promotion of flowering (Liu et al., 2013). Nitrate availability also controls the levels of DELLA proteins, known repressors of GA signaling: higher nitrate concentrations promote the accumulation of RGA, in turn inducing expression of the RGA targets GNC and CGA1, also known as GNC-LIKE. Expression of floral repressors SMZ and SNZ also depends on external nitrate provision: higher nitrate induces early accumulation of SMZ and SNZ, leading to a delayed peak of FT expression and therefore delays the timing of flowering under high nitrate. Interestingly, SMZ, SNZ, and FT responses to nitrate are abolished in a della quintuple mutant. Furthermore, GNC and CGA1 overexpressors exhibit higher expression of SMZ and SNZ, while SMZ and SNZ expression is diminished in the gnc cga1 double mutant relative to the wild type. These results indicate that nitrate-dependent DELLA accumulation has a direct impact on the temporal expression of SNZ, SMZ, and FT, resulting in differences in flowering time, and highlight the roles of components of the age-dependent pathway on nitrate-controlled flowering (Gras et al., 2018). Consistent with the involvement of the age pathway, mRNA levels in SPL TFs (miR156 targets, and therefore influencing flowering time) are temporally regulated by nitrate availability in the shoot apical meristem. SPL3 and SPL5 levels showed a delayed peak when plants were grown in low-nitrate conditions, which causes a late-flowering phenotype (Olas et al., 2019). However, this regulation is independent of miR156 and may instead depend on direct control of SPL expression by nitrate in the shoot apical meristem via the activity of NLP6 and NLP7 TFs (Olas et al., 2019).
NITRATE INTERACTION WITH OTHER NUTRIENTS
Crosstalk between nutrient signaling pathways is an important feature of plant adaptation and survival in natural environments. Under natural or agricultural field conditions, plants are exposed to variations in the levels of multiple nutrients at the same time. The macronutrients N, P, and K are the most limiting nutrients for crop yield and the most widely required as fertilizers on agricultural soils. The response to combined nutrient deficiency or to combined changes in nutrient levels and their impact on plant physiology and growth is an emergent area of study in plant biology. Accordingly, the identification of molecular integrators involved in N, P, and K interactions has increased its relevance in the last years.
Nitrate and P Interactions
The first evidence linking N and P nutrition was the discovery of the NLA gene, which plays a role in the crosstalk between nitrate and P in Arabidopsis. NLA was initially described as having a role in N limitation responses (Peng et al., 2007). Mutants in NLA exhibit N limitation-specific phenotypes (Peng et al., 2008) that are reverted by P limitation in an antagonistic crosstalk between N and P levels (Kant et al., 2011a). NLA can also trigger the polyubiquitination and N deficiency-mediated degradation of the ORE1 TF (Park et al., 2018), leading to leaf senescence. Interestingly, NLA utilizes the E2 ubiquitin-conjugating enzyme PHO2 for ORE1 ubiquitination (Park et al., 2018). PHO2 is regulated by miR399, which is induced by P and repressed by N deficiency. In agreement, PHO2 expression is induced by N limitation and repressed under high-nitrate conditions (Medici et al., 2019). This suggests a miRNA-mediated posttranscriptional and ubiquitin-mediated posttranslational regulation of leaf senescence under N limitation.
The HRS1 TF was identified as an integrator of N and P signaling for root growth. HRS1 and its closest homolog HHO1 are rapidly and strongly induced by nitrate resupply of N-limited Arabidopsis roots. These TFs act downstream of the NRT1.1 transporter and NLP6 and NLP7 TFs. However, HRS1 and HHO1 repress primary root growth in response to P deficiency conditions only when nitrate is present, suggesting a complex regulation of N/P sensing (Medici et al., 2015). A recent study in Arabidopsis further confirmed the role of the HRS/HHO family on nitrate-P crosstalk. The expression of the HRS1 homologs HHO1, HHO2, and HHO3 and HRS1 itself is directly induced by PHR1, a master regulator of P starvation responses. Under P deficiency, PHR1 is released from the repressors SPX1/2/3/4 and promotes the expression of HRS1 clade genes. Therefore, nitrate uptake is repressed by P limitation via the PHR1-HRS1-NRT2.1 pathway. Hence, regulation of nitrate uptake by the PHR1-HRS1 pathway may be a strategy to adapt to P deficiency (Maeda et al., 2018).
The control of the P starvation response (PSR) by nitrate availability is likely conserved across a wide range of plant species. In rice, high nitrate concentrations increase P uptake as well as the transcript levels of P transporter genes and P starvation-induced genes. This correlates with an increase in rice biomass under high-P and high-nitrate conditions. Nitrate induction of P starvation-induced genes is abolished in mutants of the OsNRT1.1b transporter, the rice ortholog of Arabidopsis NRT1.1, indicating that nitrate-activated P responses are dependent on OsNRT1.1B function. OsNRT1.1B mediates the ubiquitination and degradation of the P signaling repressor OsSPX4 under high-nitrate conditions, which allows the translocation of OsPHR2, the key TF in P signaling, to the nucleus to initiate the transcription of P utilization genes (Zhou et al., 2008). Thus, nitrate-triggered degradation of OsSPX4 activates both P- and nitrate-responsive genes, generating a coordinated utilization of N and P (Hu et al., 2019).
In a recent work, Medici et al. (2019) proposed that PSR is actively turned off when nitrate is limiting, and this repression is due to a combination of local and systemic nitrate-signaling pathways. PHO2 transcript levels are regulated by nitrate supply, accumulating during nitrate depletion and under high nitrate (Medici et al., 2019). Most PSR genes are no longer controlled by nitrate in a pho2 mutant, demonstrating that PHO2 integrates nitrate signals into the PSR (Medici et al., 2019). Moreover, NRT1.1 is repressed by P starvation and PHO2 acts as a positive regulator of NRT1.1. Importantly, this mechanism of PSR control by nitrate availability is conserved in plants. Rice P starvation-induced genes OsIPS1 and OsSPX1 and P transporter OsPT1 were induced by P starvation only in the presence of nitrate, and nitrate deprivation prevents the PSR in rice. This phenomenon was also conserved in wheat (Triticum aestivum; Medici et al., 2019). These studies highlight the intricacy of the nitrate and P responses, with NRT1.1 having a central conserved role in modulating the interaction.
Nitrate and K Interaction
Although K itself is not metabolized, it is fundamental for and plays a vital role in many aspects of plant metabolism. K is a known cofactor for the activity of no less than 46 enzymes, including critical enzymes like NR. In plants, K and nitrate absorption and transport are coordinated (Blevins et al., 1978; Triplett et al., 1980; White, 2012). In particular, the nitrate transporter NRT1.5 functions as a proton-coupled H+/K antiporter. NRT1.5 is responsible for nitrate loading from pericycle cells into the xylem. NRT1.5 directly mediates K release from the root parenchyma into the xylem for transport to the shoot, especially under low-K conditions (Li et al., 2017). The proton gradient across the plasma membrane promotes this NRT1.5-mediated K xylem loading. This study revealed that NRT1.5 plays a crucial role in K translocation from root to shoot and that it is also involved in the coordination of K/nitrate distribution in plants (Li et al., 2017). A complementary study showed that the MYB59 TF modulates K and nitrate translocation. MYB59 directly binds the NRT1.5 promoter and activates its expression to coordinate root-to-shoot nutrient distribution. When plants face K/nitrate-deficient conditions, MYB59 and subsequently NRT1.5 are downregulated to achieve the proper K/nitrate distribution between roots and shoots (Du et al., 2019).
Nitrate, P, and K Interaction
Contrary to nitrate-P or nitrate-K interactions, little is known about how the nitrate-P-K interaction is regulated in Arabidopsis or in crop species. A study evaluating the impact of modifying the availability of these nutrients along with sulfate (S) on the root system is one of the few recent reports evaluating how plants integrate four nutritional stimuli into complex developmental programs (Kellermeier et al., 2014). The authors quantified 13 parameters from Arabidopsis roots under 32 binary combinations of nitrate, P, K, S, and daylength. P caused the most important single-nutrient effect, while nitrate effects were strongly daylength-dependent. As a proof of principle, they explored mutants in candidate genes known to be affected by nitrate-K interactions, such as HAK5, NRT2.1, AKT1, CIPK23, and NRT1.1, revealing combinations of transporters, receptors, and kinases acting in the modulation of those signals (Kellermeier et al., 2014).
NITRATE IN THE CONTEXT OF A CHANGING CLIMATE
A critical resource affecting plant growth is water availability. As with nitrate, water is globally limited in soils, creating marginal soils that are agriculturally unproductive. In the current projections of climate change, much of the impact, particularly on crops, will be felt through changes in patterns of water availability. To engineer or breed crop varieties that can thrive in marginal environments, it is crucial to understand how plants sense and integrate responses to nitrate and water, but surprisingly little is known about how plants might do this (Humbert et al., 2013). Since water serves as a solvent for nitrate, a recent study used a unique approach to investigate whether plants respond to nitrate as the absolute amount of N (N-moles) or the concentration of nitrate in water (N-molarity; Swift et al., 2019). The approach used a factorial matrix that systematically varied both N-dose and water-dose applied to rice plants. The response of plants to N-amount, water-volume, and their interactions was modeled by linear regression of the induced effects on phenotypic traits and gene expression. The model revealed that rice plants could distinguish changes in N-moles, water-volume, N-molarity (N/water), as well as a synergistic response, N × water, both at the level of gene expression and phenotype. These results were tested in the field using a set of 19 rice varieties. Interestingly, the same set of genes responding to combinations of N-dose and water-dose in the lab correlated with crop outcomes in the field over two seasons (Swift et al., 2019).
Another important effect of climate change is the constant increase in CO2 concentration. Atmospheric CO2 levels are projected to reach 530 to 970 μmol mol−1 by 2100, well above the preindustrial average of ∼280 μmol mol−1 (Bloom et al., 2012). While elevated CO2 may benefit plant growth by enhancing photosynthesis, studies have shown that it can also lower nitrate assimilation (Bloom et al., 2010, 2014). A recent study showed that CO2 causes a bottleneck effect on root-to-shoot nutrient transport, which results in lower shoot nitrate and, consequently, lower photosynthesis and biomass (Cohen et al., 2018). In a study in tomato (Solanum lycopersicum), a combination of elevated CO2 and higher temperatures diminished the nitrate uptake rate by roots and total protein concentration in roots, also indicating an inhibition of nitrate assimilation. These studies highlight the need for additional research in order to understand how climate change may impact both plant productivity and food quality (Jayawardena et al., 2017).
ADVANCES IN NUE
Although the benefits of using N fertilizers for higher yield are beyond doubt, they involve substantial economic and environmental costs. Modern agriculture has remarkably affected the global N cycle. An impressive example of this detrimental effect is the cascade of impacts that N-reactive forms have on the environment and on humans: one N atom can, in sequence, increase atmospheric ozone (human health impact), increase fine particulate matter (visibility impact), alter forest productivity and acidify surface waters (biodiversity loss impact), increase coastal ecosystem productivity, promoting coastal eutrophication, and increase the greenhouse effect via nitrous oxide production (environmental impact; Galloway et al., 2004). The importance of advances in research and technology in agriculture, and particularly in the area of NUE, has prompted experts to call for a second Green Revolution, focused on increasing productivity using sustainable agricultural methods (Gutiérrez, 2012; McAllister et al., 2012).
In the last decade, several genes regulating NUE have been studied (Table 2). Strategies targeting genes directly involved in nitrate transport and metabolism have been widely used to enhance NUE in crops. For example, genetic variation in the nitrate transporter NRT1.1B (Hu et al., 2015) or in the nitrate reductase gene NR2 (Gao et al., 2019) determines NUE divergence of indica and japonica rice varieties, while concurrent introgression of OsNR2-indica and OsNRT1.1B-indica enhances increased effective tiller number, grain yield, and NUE in the japonica subspecies. Other approaches based on quantitative trait locus (QTL) analyses have provided important regulators of NUE, including transcriptional regulators, signaling proteins, and components of hormonal pathways. For example, a major rice NUE QTL (identified in rice by genetic analysis of a recurrent backcross population; (Sun et al., 2014a) mapped to DEP1, a heterotrimeric G protein gene that significantly increases yield and has been widely studied and used in rice breeding (Xu et al., 2016a). Plants carrying the dominant dep1-1 allele exhibit N-insensitive vegetative growth and increased N uptake and assimilation, resulting in improved harvest index and yield at moderate levels of N fertilization (Sun et al., 2014a). Another QTL study for NUE was performed in semidwarf rice Green Revolution varieties for increased yield by preventing lodging. These semidwarf varieties have an increased accumulation of DELLA repressor proteins; however, these varieties are also associated with reduced NUE. This study revealed a QTL corresponding to the TF gene GRF4. GRF4 controls N uptake and assimilation by binding to the promoters of genes such as AMT1.1 and GS1.2. Interestingly, SLR1, a DELLA protein, can inhibit the activation of N-related genes by GRF4 by competitively inhibiting the interaction of GRF4 with the GIF1 coactivator. Increased abundance of GRF4 partially disconnects GA-DELLA regulation of growth from N metabolism, promoting biomass increase at low N supplies (Li et al., 2018). QTL analysis also led to the identification of NUE genes in other plant species such as maize. Plants normally remobilize N from senescing leaves to sustain growth elsewhere. A novel QTL controlling a functional stay-green phenotype (whereby senescence and N remobilization are impaired) was found to map to the NAC7 maize gene. Transgenic maize lines where NAC7 was downregulated by RNAi showed delayed senescence and both increased biomass and N accumulation in vegetative tissues and increases in grain yield (Zhang et al., 2019). Besides QTL analysis, genome-wide association analysis has also provided candidate genes related to NUE in major crops, for example with the recent identification of an elite haplotype of the nitrate transporter NPF6.1 named NPF6.1HapB. This allele enhanced nitrate uptake and conferred high NUE by increasing yield under low N supply. NPF6.1HapB was found to be transactivated by the NAC42 TF conferring enhanced NUE through the activation of N uptake, increasing effective panicle number and yield (Tang et al., 2019).
Table 2. Summary of Genes Regulating NUE in Crop Plants.
| Gene Name | Target Plant | Physiological Trait Affected | References |
|---|---|---|---|
| NAC2-5A | Wheat | Root growth, yield | (He et al., 2015) |
| BZIP60 | Wheat | LR branching, N uptake, yield | (Yang, 2019) |
| NAC42 | Rice | Plant height, NUE, yield | (Tang et al., 2019) |
| NAC7 | Maize | Senescence, biomass, yield | (Zhang et al., 2019) |
| AMT1.1 | Rice | N uptake, yield | (Ranathunge et al., 2014) |
| NRT2.3b | Rice | Growth, NUE, yield | (Fan et al., 2016) |
| NRT2.1 | Rice | NUE, yield | (Chen et al., 2016) |
| ARE1 | Rice | Senescence, NUE, yield | (Wang et al., 2018b) |
| GRF4 | Rice | Biomass, yield | (Li et al., 2018) |
| NRT1.1B | Rice | NUE, yield | (Hu et al., 2015) |
| NFYA | Wheat | Yield | (Qu, 2015) |
| PTR9 | Rice | Biomass, yield | (Fang, 2013) |
| BT1/BT2 | Rice, Arabidopsis | Yield, NUE | (Araus et al., 2016) |
| NAR2.1 | Rice | NUE, yield | (Chen, 2017) |
| GS1 | Rice | NUE, yield | (Brauer, 2011) |
| DOF1 | Wheat, sorghum | Biomass, yield | (Peña, 2017) |
| AAP1 | Pea | Biomass, NUE, yield | (Perchlik and Tegeder, 2017) |
| DEP1 | Rice | Biomass, yield | (Sun et al., 2014a) |
| NR2 | Rice | Yield, NUE | (Gao et al., 2019) |
As discussed in this review, many efforts have been devoted to defining the molecular components and regulatory networks involved in nitrate transport, sensing, signaling and responses. Integrative and predictive systems biology approaches are providing new key molecular actors that have been successfully tested in crops in order to assess their impact in plant NUE and yield. As an example, a supervised machine learning algorithm predicted a NUE gene network in Arabidopsis (Araus et al., 2016). BT2, a member of the BRIC-A-BRAC/TRAMTRACK/BROAD gene family, was identified as the most central and connected gene in the NUE network. BT2 and its close homolog, BT1, act as negative regulators of NUE in a conserved manner in Arabidopsis and rice. A bt1 bt2 double mutant showed an increased expression of high-affinity transporters NRT2.1 and NRT2.4, with a concomitant increase in nitrate uptake and NUE under low nitrate conditions. Consistently, NUE decreased in plants overexpressing BT2 as compared with wild-type plants under limiting nitrate conditions. Inactivating the BT1/BT2 ortholog in rice increased NUE under low-N conditions (Araus et al., 2016).
NUE is a complex trait, since it is the result of different metabolic, physiological, developmental, and environmental interactions integrated over the entire life cycle of the plant. However, most advances in understanding the molecular mechanisms controlling nitrate responses have been done in controlled laboratory conditions and in unique genotypes of model plants. In the future, it will therefore be key to make a concerted effort to understand nitrate uptake, nitrate responses, and N-resource allocation in crops under more realistic field conditions (Plett et al., 2018). In particular, an important part of this challenge is to study NUE under environmental conditions that are predicted by climate change models. How will NUE be impacted by increasing CO2 levels? How will NUE be affected by simultaneous variations in multiple nutrients in soils? How will NUE be affected by water availability, rise in temperatures, extreme drought or flooded conditions? We urgently need to address these questions to face this new millennium.
SUPPLEMENTAL DATA
Supplemental Figure 1. The scheme behind a typical TARGET (Transient Assay Reporting Genome-wide Effects of Transcription factors) experiment.
Supplemental Table 1. Complete list of genes belonging to the NRT1 gene family.
Supplemental Table 2. Complete list of genes belonging to the NRT2 gene family.
Supplemental Table 3. Complete list of genes featured in Figure 3.
Supplemental Data Set 1. Complete list of gene acronyms, their full names and their organisms of origin, mentioned in this review.
Supplemental Data Set 2. Extended list of key publications on the topics of nitrate uptake, transport, and signaling.
DIVE Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
Acknowledgments
Research was funded by FONDECYT (1180759 to R.A.G. and 1170926 to E.A.V.), ANID/FONDAP (15090007 to R.A.G.), Instituto Milenio iBio-Iniciativa Científica Milenio MINECON (to R.A.G., J.M.A., and E.A.V.), EvoNet project (DE-SC0014377 to R.A.G.), CONICYT PCI-Redes Internacionales entre Centros de Investigación (REDES180097 to E.A.V.), the National Science Foundation PGRP (IOS 1840761 to G.M.C. and IOS 1339362 to G.M.C., G.K., and S.R.), the Zegar Family Foundation (A16-0051), and by the Centre National de la Recherche Scientifique (CNRS LIA-CoopNet to G.M.C., G.K., and S.R.).
AUTHOR CONTRIBUTIONS
E.A.V., J.M.A., V.A., G.M.C., and R.A.G. conceived the article; the authors contributed to writing the following sections: Introduction (N.M.C.); Molecular Mechanisms of Nitrate Transport (L.L.); Nitrate Signaling (E.R., S.R., and R.A.G.); Nitrate Interaction with Other Nutrients, Nitrate in the Context of a Changing Climate, and Advances in NUE (V.A.); Regulatory Networks of Nitrate Response in Plants (J.M.A., M.D.B., G.K., and G.M.C.); and Developmental Outputs (E.A.V); E.A.V., J.M.A., V.A., E.R., G.M.C., and R.A.G. drafted the final version of the review; E.A.V., J.M.A., V.A., and E.R. constructed figures and tables; all authors critically revised and approved the final article.
References
- Albertos P., Romero-Puertas M.C., Tatematsu K., Mateos I., Sánchez-Vicente I., Nambara E., Lorenzo O.(2015). S-Nitrosylation triggers ABI5 degradation to promote seed germination and seedling growth. Nat. Commun. 6: 8669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alboresi A., Gestin C., Leydecker M.T., Bedu M., Meyer C., Truong H.N.(2005). Nitrate, a signal relieving seed dormancy in Arabidopsis. Plant Cell Environ. 28: 500–512. [DOI] [PubMed] [Google Scholar]
- Ali-Rachedi S., Bouinot D., Wagner M.H., Bonnet M., Sotta B., Grappin P., Jullien M.(2004). Changes in endogenous abscisic acid levels during dormancy release and maintenance of mature seeds: Studies with the Cape Verde Islands ecotype, the dormant model of Arabidopsis thaliana. Planta 219: 479–488. [DOI] [PubMed] [Google Scholar]
- Alvarez J.M., Moyano T.C., Zhang T., Gras D.E., Herrera F.J., Araus V., O’Brien J.A., Carrillo L., Medina J., Vicente-Carbajosa J., Jiang J., Gutiérrez R.A.(2019). Local changes in chromatin accessibility and transcriptional networks underlying the nitrate response in Arabidopsis roots. Mol. Plant 12: 1545–1560. [DOI] [PubMed] [Google Scholar]
- Alvarez J.M., Riveras E., Vidal E.A., Gras D.E., Contreras-López O., Tamayo K.P., Aceituno F., Gómez I., Ruffel S., Lejay L., Jordana X., Gutiérrez R.A.(2014). Systems approach identifies TGA1 and TGA4 transcription factors as important regulatory components of the nitrate response of Arabidopsis thaliana roots. Plant J. 80: 1–13. [DOI] [PubMed] [Google Scholar]
- Araus V., Vidal E.A., Puelma T., Alamos S., Mieulet D., Guiderdoni E., Gutiérrez R.A.(2016). Members of BTB gene family of scaffold proteins suppress nitrate uptake and nitrogen use efficiency. Plant Physiol. 171: 1523–1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araya T., Miyamoto M., Wibowo J., Suzuki A., Kojima S., Tsuchiya Y.N., Sawa S., Fukuda H., von Wirén N., Takahashi H.(2014a). CLE-CLAVATA1 peptide-receptor signaling module regulates the expansion of plant root systems in a nitrogen-dependent manner. Proc. Natl. Acad. Sci. USA 111: 2029–2034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araya T., von Wirén N., Takahashi H.(2014b). CLE peptides regulate lateral root development in response to nitrogen nutritional status of plants. Plant Signal. Behav. 9: e29302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arc E., Chibani K., Grappin P., Jullien M., Godin B., Cueff G., Valot B., Balliau T., Job D., Rajjou L.(2012). Cold stratification and exogenous nitrates entail similar functional proteome adjustments during Arabidopsis seed dormancy release. J. Proteome Res. 11: 5418–5432. [DOI] [PubMed] [Google Scholar]
- Bachmann M., Huber J.L., Athwal G.S., Wu K., Ferl R.J., Huber S.C.(1996). 14-3-3 proteins associate with the regulatory phosphorylation site of spinach leaf nitrate reductase in an isoform-specific manner and reduce dephosphorylation of Ser-543 by endogenous protein phosphatases. FEBS Lett. 398: 26–30. [DOI] [PubMed] [Google Scholar]
- Bargmann B.O.R., Marshall-Colon A., Efroni I., Ruffel S., Birnbaum K.D., Coruzzi G.M., Krouk G.(2013). TARGET: A transient transformation system for genome-wide transcription factor target discovery. Mol. Plant 6: 978–980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bethke P.C., Libourel I.G., Reinöhl V., Jones R.L.(2006). Sodium nitroprusside, cyanide, nitrite, and nitrate break Arabidopsis seed dormancy in a nitric oxide-dependent manner. Planta 223: 805–812. [DOI] [PubMed] [Google Scholar]
- Blevins D.G., Barnett N.M., Frost W.B.(1978). Role of potassium and malate in nitrate uptake and translocation by wheat seedlings. Plant Physiol. 62: 784–788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloom A.J., Asensio J.S., Randall L., Rachmilevitch S., Cousins A.B., Carlisle E.A.(2012). CO2 enrichment inhibits shoot nitrate assimilation in C3 but not C4 plants and slows growth under nitrate in C3 plants. Ecology 93: 355–367. [DOI] [PubMed] [Google Scholar]
- Bloom A.J., Burger M., Kimball B.A., Pinter J.P.(2014). Nitrate assimilation is inhibited by elevated CO2 in field-grown wheat. Nat. Clim. Chang. 4: 477. [Google Scholar]
- Bloom A.J., Burger M., Rubio Asensio J.S., Cousins A.B.(2010). Carbon dioxide enrichment inhibits nitrate assimilation in wheat and Arabidopsis. Science 328: 899–903. [DOI] [PubMed] [Google Scholar]
- Bosemark N.O.(1954). The influence of nitrogen on root development. Physiol. Plant. 7: 497–502. [Google Scholar]
- Bouguyon E., et al. (2015). Multiple mechanisms of nitrate sensing by Arabidopsis nitrate transceptor NRT1.1. Nat. Plants 1: 15015. [DOI] [PubMed] [Google Scholar]
- Bouguyon E., Perrine-Walker F., Pervent M., Rochette J., Cuesta C., Benkova E., Martinière A., Bach L., Krouk G., Gojon A., Nacry P.(2016). Nitrate controls root development through posttranscriptional regulation of the NRT1.1/NPF6.3 transporter/sensor. Plant Physiol. 172: 1237–1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brauer E.K.(2011). Reappraisal of nitrogen use efficiency in rice overexpressing glutamine synthetase1. Physiologia Plantarum 141: 361–372. [DOI] [PubMed] [Google Scholar]
- Brooks M.D., Cirrone J., Pasquino A.V., Alvarez J.M., Swift J., Mittal S., Juang C.-L., Varala K., Gutiérrez R.A., Krouk G., Shasha D., Coruzzi G.M.(2019). Network walking charts transcriptional dynamics of nitrogen signaling by integrating validated and predicted genome-wide interactions. Nat. Commun. 10: 1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bussell J.D., Keech O., Fenske R., Smith S.M.(2013). Requirement for the plastidial oxidative pentose phosphate pathway for nitrate assimilation in Arabidopsis. Plant J. 75: 578–591. [DOI] [PubMed] [Google Scholar]
- Cadman C.S.C., Toorop P.E., Hilhorst H.W.M., Finch-Savage W.E.(2006). Gene expression profiles of Arabidopsis Cvi seeds during dormancy cycling indicate a common underlying dormancy control mechanism. Plant J. 46: 805–822. [DOI] [PubMed] [Google Scholar]
- Calza R., Huttner E., Vincentz M., Rouzé P., Galangau F., Vaucheret H., Chérel I., Meyer C., Kronenberger J., Caboche M.(1987). Cloning of DNA fragments complementary to tobacco nitrate reductase mRNA and encoding epitopes common to the nitrate reductases from higher plants. Mol. Gen. Genet. 209: 552–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camargo A., Llamas A., Schnell R.A., Higuera J.J., González-Ballester D., Lefebvre P.A., Fernández E., Galván A.(2007). Nitrate signaling by the regulatory gene NIT2 in Chlamydomonas. Plant Cell 19: 3491–3503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canales J., Contreras-López O., Álvarez J.M., Gutiérrez R.A.(2017). Nitrate induction of root hair density is mediated by TGA1/TGA4 and CPC transcription factors in Arabidopsis thaliana. Plant J. 92: 305–316. [DOI] [PubMed] [Google Scholar]
- Canales J., Moyano T.C., Villarroel E., Gutiérrez R.A.(2014). Systems analysis of transcriptome data provides new hypotheses about Arabidopsis root response to nitrate treatments. Front. Plant Sci. 5: 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capovilla G., Schmid M., Posé D.(2015). Control of flowering by ambient temperature. J. Exp. Bot. 66: 59–69. [DOI] [PubMed] [Google Scholar]
- Castaings L., Camargo A., Pocholle D., Gaudon V., Texier Y., Boutet-Mercey S., Taconnat L., Renou J.P., Daniel-Vedele F., Fernandez E., Meyer C., Krapp A.(2009). The nodule inception-like protein 7 modulates nitrate sensing and metabolism in Arabidopsis. Plant J. 57: 426–435. [DOI] [PubMed] [Google Scholar]
- Castillo M.C., Lozano-Juste J., González-Guzmán M., Rodriguez L., Rodriguez P.L., León J.(2015). Inactivation of PYR/PYL/RCAR ABA receptors by tyrosine nitration may enable rapid inhibition of ABA signaling by nitric oxide in plants. Sci. Signal. 8: ra89. [DOI] [PubMed] [Google Scholar]
- Castro Marín I., Loef I., Bartetzko L., Searle I., Coupland G., Stitt M., Osuna D.(2011). Nitrate regulates floral induction in Arabidopsis, acting independently of light, gibberellin and autonomous pathways. Planta 233: 539–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraborty N., Kanyuka K.(2019). GCR1 and GPA1 coupling regulates nitrate, cell wall, immunity and light responses in Arabidopsis. Sci. Rep. 9: 5838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charoensawan V., Martinho C., Wigge P.A.(2015). “Hit-and-run”: Transcription factors get caught in the act. BioEssays 37: 748–754. [DOI] [PubMed] [Google Scholar]
- Chen J., et al. (2017). pOsNAR2.1:OsNAR2.1 expression enhances nitrogen uptake efficiency and grain yield in transgenic rice plants. Plant Biotech J 15: 1273–1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., Zhang Y., Tan Y., Zhang M., Zhu L., Xu G., Fan X.(2016). Agronomic nitrogen‐use efficiency of rice can be increased by driving OsNRT 2.1 expression with the OsNAR 2.1 promoter. Plant Biotech J 14: 1705–1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., Yao Q., Gao X., Jiang C., Harberd N.P., Fu X.(2016). Shoot-to-root mobile transcription factor HY5 coordinates plant carbon and nitrogen acquisition. Curr. Biol. 26: 640–646. [DOI] [PubMed] [Google Scholar]
- Cheng C.L., Dewdney J., Kleinhofs A., Goodman H.M.(1986). Cloning and nitrate induction of nitrate reductase mRNA. Proc. Natl. Acad. Sci. USA 83: 6825–6828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chopin F., Wirth J., Dorbe M.F., Lejay L., Krapp A., Gojon A., Daniel-Vedele F.(2007). The Arabidopsis nitrate transporter AtNRT2.1 is targeted to the root plasma membrane. Plant Physiol. Biochem. 45: 630–635. [DOI] [PubMed] [Google Scholar]
- Cohen I., Rapaport T., Berger R.T., Rachmilevitch S.(2018). The effects of elevated CO2 and nitrogen nutrition on root dynamics. Plant Sci. 272: 294–300. [DOI] [PubMed] [Google Scholar]
- Comparot S., Lingiah G., Martin T.(2003). Function and specificity of 14-3-3 proteins in the regulation of carbohydrate and nitrogen metabolism. J. Exp. Bot. 54: 595–604. [DOI] [PubMed] [Google Scholar]
- Crawford N.M., Campbell W.H., Davis R.W.(1986). Nitrate reductase from squash: cDNA cloning and nitrate regulation. Proc. Natl. Acad. Sci. USA 83: 8073–8076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford N.M., Forde B.G.(2002). Molecular and developmental biology of inorganic nitrogen nutrition. The Arabidopsis Book 1: e0011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford N.M., Glass A.D.M.(1998). Molecular and physiological aspects of nitrate uptake in plants. Trends Plant Sci. 3: 389–395. [Google Scholar]
- Crawford N.M., Smith M., Bellissimo D., Davis R.W.(1988). Sequence and nitrate regulation of the Arabidopsis thaliana mRNA encoding nitrate reductase, a metalloflavoprotein with three functional domains. Proc. Natl. Acad. Sci. USA 85: 5006–5010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis S.J.(2009). Integrating hormones into the floral-transition pathway of Arabidopsis thaliana. Plant Cell Environ. 32: 1201–1210. [DOI] [PubMed] [Google Scholar]
- De Angeli A., Monachello D., Ephritikhine G., Frachisse J.M., Thomine S., Gambale F., Barbier-Brygoo H.(2006). The nitrate/proton antiporter AtCLCa mediates nitrate accumulation in plant vacuoles. Nature 442: 939–942. [DOI] [PubMed] [Google Scholar]
- de Jong F., Thodey K., Lejay L.V., Bevan M.W.(2014). Glucose elevates NITRATE TRANSPORTER2.1 protein levels and nitrate transport activity independently of its HEXOKINASE1-mediated stimulation of NITRATE TRANSPORTER2.1 expression. Plant Physiol. 164: 308–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delhon P., Gojon A., Tillard P., Passama L.(1995). Diurnal regulation of NO3− uptake in soybean plants. I. Changes in NO3− influx, efflux, and N utilization in the plant during the day/night cycle. J. Exp. Bot. 46: 1585–1594. [Google Scholar]
- Den Herder G., Van Isterdael G., Beeckman T., De Smet I.(2010). The roots of a new Green Revolution. Trends Plant Sci. 15: 600–607. [DOI] [PubMed] [Google Scholar]
- Doidy J., Li Y., Neymotin B., Edwards M.B., Varala K., Gresham D., Coruzzi G.M.(2016). “Hit-and-Run” transcription: De novo transcription initiated by a transient bZIP1 “hit” persists after the “run.”. BMC Genomics 17: 92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du X.-Q., Wang F.-L., Li H., Jing S., Yu M., Li J., Wu W.-H., Kudla J., Wang Y.(2019). The transcription factor MYB59 regulates K+/NO3− translocation in the Arabidopsis response to low K+ stress. Plant Cell 31: 699–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelsberger W.R., Schulze W.X.(2012). Nitrate and ammonium lead to distinct global dynamic phosphorylation patterns when resupplied to nitrogen-starved Arabidopsis seedlings. Plant J. 69: 978–995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans H.J., Nason A.(1953). Pyridine nucleotide-nitrate reductase from extracts of higher plants. Plant Physiol. 28: 233–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan X., Naz M., Fan X., Xuan W., Miller A.J., Xu G.(2017). Plant nitrate transporters: From gene function to application. J. Exp. Bot. 68: 2463–2475. [DOI] [PubMed] [Google Scholar]
- Fan X., Tang Z., Tan Y., Zhang Y., Luo B., Yang M., Lian X., Qirong S., Miller A.J., Xu G.(2016). Overexpression of a pH-sensitive nitrate transporter in rice increases crop yields. Proc Natl Acad Sci U.S.A. 113: 7118–7123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang Z., et al. (2013). Altered Expression of the PTR/NRT1 Homologue OsPTR9 Affects Nitrogen Utilization Efficiency, Growth and Grain Yield in Rice. Plant Biotechnol J 11: 446–458. [DOI] [PubMed] [Google Scholar]
- FAO. (2017). World Fertiliser Trends and Outlook to 2020.. (Rome: Food and Agricultural Organisation of the United Nations; ). [Google Scholar]
- Filleur S., Daniel-Vedele F.(1999). Expression analysis of a high-affinity nitrate transporter isolated from Arabidopsis thaliana by differential display. Planta 207: 461–469. [DOI] [PubMed] [Google Scholar]
- Filleur S., Dorbe M.F., Cerezo M., Orsel M., Granier F., Gojon A., Daniel-Vedele F.(2001). An Arabidopsis T-DNA mutant affected in Nrt2 genes is impaired in nitrate uptake. FEBS Lett. 489: 220–224. [DOI] [PubMed] [Google Scholar]
- Finch-Savage W.E., Cadman C.S.C., Toorop P.E., Lynn J.R., Hilhorst H.W.M.(2007). Seed dormancy release in Arabidopsis Cvi by dry after-ripening, low temperature, nitrate and light shows common quantitative patterns of gene expression directed by environmentally specific sensing. Plant J. 51: 60–78. [DOI] [PubMed] [Google Scholar]
- Finkelstein R.R., Lynch T.J.(2000). The Arabidopsis abscisic acid response gene ABI5 encodes a basic leucine zipper transcription factor. Plant Cell 12: 599–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Footitt S., Douterelo-Soler I., Clay H., Finch-Savage W.E.(2011). Dormancy cycling in Arabidopsis seeds is controlled by seasonally distinct hormone-signaling pathways. Proc. Natl. Acad. Sci. USA 108: 20236–20241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Footitt S., Huang Z., Clay H.A., Mead A., Finch-Savage W.E.(2013). Temperature, light and nitrate sensing coordinate Arabidopsis seed dormancy cycling, resulting in winter and summer annual phenotypes. Plant J. 74: 1003–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forde B., Lorenzo H.(2001). The nutritional control of root development. Plant Soil 232: 51–68. [Google Scholar]
- Fredes I., Moreno S., Díaz F.P., Gutiérrez R.A.(2019). Nitrate signaling and the control of Arabidopsis growth and development. Curr. Opin. Plant Biol. 47: 112–118. [DOI] [PubMed] [Google Scholar]
- Galloway J.N., et al. (2004). Nitrogen cycles: Past, present, and future. Biogeochemistry 70: 153–226. [Google Scholar]
- Gan Y., Bernreiter A., Filleur S., Abram B., Forde B.G.(2012). Overexpressing the ANR1 MADS-box gene in transgenic plants provides new insights into its role in the nitrate regulation of root development. Plant Cell Physiol. 53: 1003–1016. [DOI] [PubMed] [Google Scholar]
- Gao Z., et al. (2019). The indica nitrate reductase gene OsNR2 allele enhances rice yield potential and nitrogen use efficiency. Nat. Commun. 10: 5207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaudinier A., et al. (2018). Transcriptional regulation of nitrogen-associated metabolism and growth. Nature 563: 259–264. [DOI] [PubMed] [Google Scholar]
- Geiger D., Maierhofer T., Al-Rasheid K.A., Scherzer S., Mumm P., Liese A., Ache P., Wellmann C., Marten I., Grill E., Romeis T., Hedrich R.(2011). Stomatal closure by fast abscisic acid signaling is mediated by the guard cell anion channel SLAH3 and the receptor RCAR1. Sci. Signal. 4: ra32. [DOI] [PubMed] [Google Scholar]
- Gent L., Forde B.G.(2017). How do plants sense their nitrogen status? J. Exp. Bot. 68: 2531–2539. [DOI] [PubMed] [Google Scholar]
- Gibbs D.J., et al. (2014). Nitric oxide sensing in plants is mediated by proteolytic control of group VII ERF transcription factors. Mol. Cell 53: 369–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibon Y., Usadel B., Blaesing O.E., Kamlage B., Hoehne M., Trethewey R., Stitt M.(2006). Integration of metabolite with transcript and enzyme activity profiling during diurnal cycles in Arabidopsis rosettes. Genome Biol. 7: R76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gifford M.L., Dean A., Gutierrez R.A., Coruzzi G.M., Birnbaum K.D.(2008). Cell-specific nitrogen responses mediate developmental plasticity. Proc. Natl. Acad. Sci. USA 105: 803–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gojon A., Nacry P., Davidian J.C.(2009). Root uptake regulation: A central process for NPS homeostasis in plants. Curr. Opin. Plant Biol. 12: 328–338. [DOI] [PubMed] [Google Scholar]
- Graciet E., Wellmer F.(2010). The plant N-end rule pathway: Structure and functions. Trends Plant Sci. 15: 447–453. [DOI] [PubMed] [Google Scholar]
- Gras D.E., Vidal E.A., Undurraga S.F., Riveras E., Moreno S., Dominguez-Figueroa J., Alabadi D., Blázquez M.A., Medina J., Gutiérrez R.A.(2018). SMZ/SNZ and gibberellin signaling are required for nitrate-elicited delay of flowering time in Arabidopsis thaliana. J. Exp. Bot. 69: 619–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan P.(2017). Dancing with hormones: A current perspective of nitrate signaling and regulation in Arabidopsis. Front. Plant Sci. 8: 1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan P., Ripoll J.-J., Wang R., Vuong L., Bailey-Steinitz L.J., Ye D., Crawford N.M.(2017). Interacting TCP and NLP transcription factors control plant responses to nitrate availability. Proc. Natl. Acad. Sci. USA 114: 2419–2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan P., Wang R., Nacry P., Breton G., Kay S.A., Pruneda-Paz J.L., Davani A., Crawford N.M.(2014). Nitrate foraging by Arabidopsis roots is mediated by the transcription factor TCP20 through the systemic signaling pathway. Proc. Natl. Acad. Sci. USA 111: 15267–15272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo F.Q., Wang R., Chen M., Crawford N.M.(2001). The Arabidopsis dual-affinity nitrate transporter gene AtNRT1.1 (CHL1) is activated and functions in nascent organ development during vegetative and reproductive growth. Plant Cell 13: 1761–1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y., Chen F., Zhang F., Mi G.(2005). Auxin transport from shoot to root is involved in the response of lateral root growth to localized supply of nitrate in maize. Plant Sci. 169: 894–900. [Google Scholar]
- Gutiérrez R.A.(2012). Systems biology for enhanced plant nitrogen nutrition. Science 336: 1673–1675. [DOI] [PubMed] [Google Scholar]
- Gutiérrez R.A., Lejay L.V., Dean A., Chiaromonte F., Shasha D.E., Coruzzi G.M.(2007). Qualitative network models and genome-wide expression data define carbon/nitrogen-responsive molecular machines in Arabidopsis. Genome Biol. 8: R7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutiérrez R.A., Stokes T.L., Thum K., Xu X., Obertello M., Katari M.S., Tanurdzic M., Dean A., Nero D.C., McClung C.R., Coruzzi G.M.(2008). Systems approach identifies an organic nitrogen-responsive gene network that is regulated by the master clock control gene CCA1. Proc. Natl. Acad. Sci. USA 105: 4939–4944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He H., Willems L.A., Batushansky A., Fait A., Hanson J., Nijveen H., Hilhorst H.W., Bentsink L.(2016). Effects of parental temperature and nitrate on seed performance are reflected by partly overlapping genetic and metabolic pathways. Plant Cell Physiol. 57: 473–487. [DOI] [PubMed] [Google Scholar]
- He X., Qu B., Li W., Zhao X., Teng W., Ma W., Ren Y., Li B., Li Z., Tong Y.(2015). The Nitrate-Inducible NAC Transcription Factor TaNAC2-5A Controls Nitrate Response and Increases Wheat Yield. Plant physiology 169 (3): 1991–2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho C.-H., Lin S.-H., Hu H.-C., Tsay Y.-F.(2009). CHL1 functions as a nitrate sensor in plants. Cell 138: 1184–1194. [DOI] [PubMed] [Google Scholar]
- Hong Y., Devaiah S.P., Bahn S.C., Thamasandra B.N., Li M., Welti R., Wang X.(2009). Phospholipase D epsilon and phosphatidic acid enhance Arabidopsis nitrogen signaling and growth. Plant J. 58: 376–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu B., et al. (2019). Nitrate-NRT1.1B-SPX4 cascade integrates nitrogen and phosphorus signalling networks in plants. Nat. Plants 5: 401–413. [DOI] [PubMed] [Google Scholar]
- Hu B., et al. (2015). Variation in NRT1.1B contributes to nitrate-use divergence between rice subspecies. Nat. Genet. 47: 834–838. [DOI] [PubMed] [Google Scholar]
- Huang N.C., Liu K.H., Lo H.J., Tsay Y.F.(1999). Cloning and functional characterization of an Arabidopsis nitrate transporter gene that encodes a constitutive component of low-affinity uptake. Plant Cell 11: 1381–1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S., Chen S., Liang Z., Zhang C., Yan M., Chen J., Xu G., Fan X., Zhang Y.(2015). Knockdown of the partner protein OsNAR2.1 for high-affinity nitrate transport represses lateral root formation in a nitrate-dependent manner. Sci. Rep. 5: 18192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humbert S., Subedi S., Cohn J., Zeng B., Bi Y.-M., Chen X., Zhu T., McNicholas P.D., Rothstein S.J.(2013). Genome-wide expression profiling of maize in response to individual and combined water and nitrogen stresses. BMC Genomics 14: 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacquot A., et al. (2019). NRT2.1 phosphorylation prevents root high affinity nitrate uptake activity in Arabidopsis thaliana. bioRxiv 583542. [DOI] [PubMed] [Google Scholar]
- Jayawardena D.M., Heckathorn S.A., Bista D.R., Mishra S., Boldt J.K., Krause C.R.(2017). Elevated CO2 plus chronic warming reduce nitrogen uptake and levels or activities of nitrogen-uptake and -assimilatory proteins in tomato roots. Physiol. Plant. 159: 354–365. [DOI] [PubMed] [Google Scholar]
- Jia Z., Giehl R.F.H., Meyer R.C., Altmann T., von Wirén N.(2019). Natural variation of BSK3 tunes brassinosteroid signaling to regulate root foraging under low nitrogen. Nat. Commun. 10: 2378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanno Y., Hanada A., Chiba Y., Ichikawa T., Nakazawa M., Matsui M., Koshiba T., Kamiya Y., Seo M.(2012). Identification of an abscisic acid transporter by functional screening using the receptor complex as a sensor. Proc. Natl. Acad. Sci. USA 109: 9653–9658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kant S., Bi Y.-M., Rothstein S.J.(2011a). Understanding plant response to nitrogen limitation for the improvement of crop nitrogen use efficiency. J. Exp. Bot. 62: 1499–1509. [DOI] [PubMed] [Google Scholar]
- Kant S., Peng M., Rothstein S.J.(2011b). Genetic regulation by NLA and microRNA827 for maintaining nitrate-dependent phosphate homeostasis in Arabidopsis. PLoS Genet. 7: e1002021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katari M.S., Nowicki S.D., Aceituno F.F., Nero D., Kelfer J., Thompson L.P., Cabello J.M., Davidson R.S., Goldberg A.P., Shasha D.E., Coruzzi G.M., Gutiérrez R.A.(2010). VirtualPlant: A software platform to support systems biology research. Plant Physiol. 152: 500–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellermeier F., Armengaud P., Seditas T.J., Danku J., Salt D.E., Amtmann A.(2014). Analysis of the root system architecture of Arabidopsis provides a quantitative readout of crosstalk between nutritional signals. Plant Cell 26: 1480–1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan M.I.R., Trivellini A., Fatma M., Masood A., Francini A., Iqbal N., Ferrante A., Khan N.A.(2015). Role of ethylene in responses of plants to nitrogen availability. Front. Plant Sci. 6: 927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiba T., Feria-Bourrellier A.-B., Lafouge F., Lezhneva L., Boutet-Mercey S., Orsel M., Bréhaut V., Miller A., Daniel-Vedele F., Sakakibara H., Krapp A.(2012). The Arabidopsis nitrate transporter NRT2.4 plays a double role in roots and shoots of nitrogen-starved plants. Plant Cell 24: 245–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiba T., et al. (2018). Repression of nitrogen starvation responses by members of the Arabidopsis GARP-type transcription factor NIGT1/HRS1 subfamily. Plant Cell 30: 925–945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiba T., Kudo T., Kojima M., Sakakibara H.(2011). Hormonal control of nitrogen acquisition: Roles of auxin, abscisic acid, and cytokinin. J. Exp. Bot. 62: 1399–1409. [DOI] [PubMed] [Google Scholar]
- Kim D.H., Doyle M.R., Sung S., Amasino R.M.(2009). Vernalization: Winter and the timing of flowering in plants. Annu. Rev. Cell Dev. Biol. 25: 277–299. [DOI] [PubMed] [Google Scholar]
- Klebs G.(1913). Über das Verhältnis der Außenwelt zur Entwicklung der Pflanze. Sitzber. Akad. Wiss. Heidelberg B 1–47. [Google Scholar]
- Ko D., et al. (2014). Arabidopsis ABCG14 is essential for the root-to-shoot translocation of cytokinin. Proc. Natl. Acad. Sci. USA 111: 7150–7155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konishi M., Yanagisawa S.(2013). Arabidopsis NIN-like transcription factors have a central role in nitrate signalling. Nat. Commun. 4: 1617. [DOI] [PubMed] [Google Scholar]
- Konishi M., Yanagisawa S.(2019). The role of protein-protein interactions mediated by the PB1 domain of NLP transcription factors in nitrate-inducible gene expression. BMC Plant Biol. 19: 90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotur Z., Mackenzie N., Ramesh S., Tyerman S.D., Kaiser B.N., Glass A.D.(2012). Nitrate transport capacity of the Arabidopsis thaliana NRT2 family members and their interactions with AtNAR2.1. New Phytol. 194: 724–731. [DOI] [PubMed] [Google Scholar]
- Krapp A., David L.C., Chardin C., Girin T., Marmagne A., Leprince A.-S., Chaillou S., Ferrario-Méry S., Meyer C., Daniel-Vedele F.(2014). Nitrate transport and signalling in Arabidopsis. J. Exp. Bot. 65: 789–798. [DOI] [PubMed] [Google Scholar]
- Kriel J., Haesendonckx S., Rubio-Texeira M., Van Zeebroeck G., Thevelein J.M.(2011). From transporter to transceptor: Signaling from transporters provokes re-evaluation of complex trafficking and regulatory controls. Endocytic internalization and intracellular trafficking of nutrient transceptors may, at least in part, be governed by their signaling function. BioEssays 33: 870–879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krouk G., et al. (2010a). Nitrate-regulated auxin transport by NRT1.1 defines a mechanism for nutrient sensing in plants. Dev. Cell 18: 927–937. [DOI] [PubMed] [Google Scholar]
- Krouk G., Lingeman J., Colon A.M., Coruzzi G., Shasha D.(2013). Gene regulatory networks in plants: Learning causality from time and perturbation. Genome Biol. 14: 123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krouk G., Mirowski P., LeCun Y., Shasha D.E., Coruzzi G.M.(2010b). Predictive network modeling of the high-resolution dynamic plant transcriptome in response to nitrate. Genome Biol. 11: R123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krouk G., Tillard P., Gojon A.(2006). Regulation of the high-affinity NO3− uptake system by NRT1.1-mediated NO3− demand signaling in Arabidopsis. Plant Physiol. 142: 1075–1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landrein B., Formosa-Jordan P., Malivert A., Schuster C., Melnyk C.W., Yang W., Turnbull C., Meyerowitz E.M., Locke J.C.W., Jönsson H.(2018). Nitrate modulates stem cell dynamics in Arabidopsis shoot meristems through cytokinins. Proc. Natl. Acad. Sci. USA 115: 1382–1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langfelder P., Horvath S.(2008). WGCNA: An R package for weighted correlation network analysis. BMC Bioinformatics 9: 559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lejay L., Gansel X., Cerezo M., Tillard P., Müller C., Krapp A., von Wirén N., Daniel-Vedele F., Gojon A.(2003). Regulation of root ion transporters by photosynthesis: Functional importance and relation with hexokinase. Plant Cell 15: 2218–2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lejay L., Tillard P., Lepetit M., Olive F., Filleur S., Daniel-Vedele F., Gojon A.(1999). Molecular and functional regulation of two NO3− uptake systems by N- and C-status of Arabidopsis plants. Plant J 18: 509–519. [DOI] [PubMed] [Google Scholar]
- Lejay L., Wirth J., Pervent M., Cross J.M., Tillard P., Gojon A.(2008). Oxidative pentose phosphate pathway-dependent sugar sensing as a mechanism for regulation of root ion transporters by photosynthesis. Plant Physiol. 146: 2036–2053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Léran S., Edel K.H., Pervent M., Hashimoto K., Corratgé-Faillie C., Offenborn J.N., Tillard P., Gojon A., Kudla J., Lacombe B.(2015). Nitrate sensing and uptake in Arabidopsis are enhanced by ABI2, a phosphatase inactivated by the stress hormone abscisic acid. Sci. Signal. 8: ra43. [DOI] [PubMed] [Google Scholar]
- Léran S., et al. (2014). A unified nomenclature of NITRATE TRANSPORTER 1/PEPTIDE TRANSPORTER family members in plants. Trends Plant Sci. 19: 5–9. [DOI] [PubMed] [Google Scholar]
- Lezhneva L., Kiba T., Feria-Bourrellier A.B., Lafouge F., Boutet-Mercey S., Zoufan P., Sakakibara H., Daniel-Vedele F., Krapp A.(2014). The Arabidopsis nitrate transporter NRT2.5 plays a role in nitrate acquisition and remobilization in nitrogen-starved plants. Plant J. 80: 230–241. [DOI] [PubMed] [Google Scholar]
- Li H., Yu M., Du X.-Q., Wang Z.-F., Wu W.-H., Quintero F.J., Jin X.-H., Li H.-D., Wang Y.(2017). NRT1.5/NPF7.3 functions as a proton-coupled H+/K+ antiporter for K+ loading into the xylem in Arabidopsis. Plant Cell 29: 2016–2026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S., Tian Y., Wu K., Ye Y., Yu J., Zhang J., Liu Q., Hu M., Li H., Tong Y., Harberd N.P., Fu X.(2018). Modulating plant growth-metabolism coordination for sustainable agriculture. Nature 560: 595–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Krouk G., Coruzzi G.M., Ruffel S.(2014). Finding a nitrogen niche: A systems integration of local and systemic nitrogen signalling in plants. J. Exp. Bot. 65: 5601–5610. [DOI] [PubMed] [Google Scholar]
- Liang Y., Zhao X., Jones A.M., Gao Y.(2018). G proteins sculp root architecture in response to nitrogen in rice and Arabidopsis. Plant Sci. 274: 129–136. [DOI] [PubMed] [Google Scholar]
- Lin Y.-L., Tsay Y.-F.(2017). Influence of differing nitrate and nitrogen availability on flowering control in Arabidopsis. J. Exp. Bot. 68: 2603–2609. [DOI] [PubMed] [Google Scholar]
- Linkohr B.I., Williamson L.C., Fitter A.H., Leyser H.M.O.(2002). Nitrate and phosphate availability and distribution have different effects on root system architecture of Arabidopsis. Plant J. 29: 751–760. [DOI] [PubMed] [Google Scholar]
- Little D.Y., Rao H., Oliva S., Daniel-Vedele F., Krapp A., Malamy J.E.(2005). The putative high-affinity nitrate transporter NRT2.1 represses lateral root initiation in response to nutritional cues. Proc. Natl. Acad. Sci. USA 102: 13693–13698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Mehdi S., Topping J., Tarkowski P., Lindsey K.(2010). Modelling and experimental analysis of hormonal crosstalk in Arabidopsis. Mol. Syst. Biol. 6: 373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu K.-H., Huang C.-Y., Tsay Y.-F.(1999). CHL1 is a dual-affinity nitrate transporter of Arabidopsis involved in multiple phases of nitrate uptake. Plant Cell 11: 865–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu K.H., et al. (2017). Discovery of nitrate-CPK-NLP signalling in central nutrient-growth networks. Nature 545: 311–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu K.H., Tsay Y.F.(2003). Switching between the two action modes of the dual-affinity nitrate transporter CHL1 by phosphorylation. EMBO J. 22: 1005–1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu T., Li Y., Ren J., Qian Y., Yang X., Duan W., Hou X.(2013). Nitrate or NaCl regulates floral induction in Arabidopsis thaliana. Biologia 68: 215–222. [Google Scholar]
- Liu Y., Shi L., Ye N., Liu R., Jia W., Zhang J.(2009). Nitric oxide-induced rapid decrease of abscisic acid concentration is required in breaking seed dormancy in Arabidopsis. New Phytol. 183: 1030–1042. [DOI] [PubMed] [Google Scholar]
- Lopez-Molina L., Mongrand S., Chua N.H.(2001). A postgermination developmental arrest checkpoint is mediated by abscisic acid and requires the ABI5 transcription factor in Arabidopsis. Proc. Natl. Acad. Sci. USA 98: 4782–4787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma W., Li J., Qu B., He X., Zhao X., Li B., Fu X., Tong Y.(2014). Auxin biosynthetic gene TAR2 is involved in low nitrogen-mediated reprogramming of root architecture in Arabidopsis. Plant J. 78: 70–79. [DOI] [PubMed] [Google Scholar]
- Maeda Y., Konishi M., Kiba T., Sakuraba Y., Sawaki N., Kurai T., Ueda Y., Sakakibara H., Yanagisawa S.(2018). A NIGT1-centred transcriptional cascade regulates nitrate signalling and incorporates phosphorus starvation signals in Arabidopsis. Nat. Commun. 9: 1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maierhofer T., Lind C., Hüttl S., Scherzer S., Papenfuß M., Simon J., Al-Rasheid K.A.S., Ache P., Rennenberg H., Hedrich R., Müller T.D., Geiger D.(2014). A single-pore residue renders the Arabidopsis root anion channel SLAH2 highly nitrate selective. Plant Cell 26: 2554–2567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchive C., Roudier F., Castaings L., Bréhaut V., Blondet E., Colot V., Meyer C., Krapp A.(2013). Nuclear retention of the transcription factor NLP7 orchestrates the early response to nitrate in plants. Nat. Commun. 4: 1713. [DOI] [PubMed] [Google Scholar]
- Martin T., Oswald O., Graham I.A.(2002). Arabidopsis seedling growth, storage lipid mobilization, and photosynthetic gene expression are regulated by carbon:nitrogen availability. Plant Physiol. 128: 472–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matakiadis T., Alboresi A., Jikumaru Y., Tatematsu K., Pichon O., Renou J.P., Kamiya Y., Nambara E., Truong H.N.(2009). The Arabidopsis abscisic acid catabolic gene CYP707A2 plays a key role in nitrate control of seed dormancy. Plant Physiol. 149: 949–960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAllister C.H., Beatty P.H., Good A.G.(2012). Engineering nitrogen use efficient crop plants: The current status. Plant Biotechnol. J. 10: 1011–1025. [DOI] [PubMed] [Google Scholar]
- Medici A., Marshall-Colon A., Ronzier E., Szponarski W., Wang R., Gojon A., Crawford N.M., Ruffel S., Coruzzi G.M., Krouk G.(2015). AtNIGT1/HRS1 integrates nitrate and phosphate signals at the Arabidopsis root tip. Nat. Commun. 6: 6274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medici A., et al. (2019). Identification of molecular integrators shows that nitrogen actively controls the phosphate starvation response in plants. Plant Cell 31: 1171–1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menz J., Li Z., Schulze W.X., Ludewig U.(2016). Early nitrogen-deprivation responses in Arabidopsis roots reveal distinct differences on transcriptome and (phospho-) proteome levels between nitrate and ammonium nutrition. Plant J. 88: 717–734. [DOI] [PubMed] [Google Scholar]
- Miller A.J., Fan X., Shen Q., Smith S.J.(2008). Amino acids and nitrate as signals for the regulation of nitrogen acquisition. J. Exp. Bot. 59: 111–119. [DOI] [PubMed] [Google Scholar]
- Mirowski P., LeCun Y.(2009). Dynamic Factor Graphs for Time Series Modeling.. (Berlin: Springer; ), pp. 128–143. [Google Scholar]
- Miyawaki K., Matsumoto-Kitano M., Kakimoto T.(2004). Expression of cytokinin biosynthetic isopentenyltransferase genes in Arabidopsis: tissue specificity and regulation by auxin, cytokinin, and nitrate. Plant J. 37: 128–138. [DOI] [PubMed] [Google Scholar]
- Muller B., Touraine B.(1992). Inhibition of NO3− uptake by various phloem-translocated amino acids in soybean seedlings. J. Exp. Bot. 43: 617–623. [Google Scholar]
- Müller D., Waldie T., Miyawaki K., To J.P.C., Melnyk C.W., Kieber J.J., Kakimoto T., Leyser O.(2015). Cytokinin is required for escape but not release from auxin mediated apical dominance. Plant J. 82: 874–886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muños S., Cazettes C., Fizames C., Gaymard F., Tillard P., Lepetit M., Lejay L., Gojon A.(2004). Transcript profiling in the chl1-5 mutant of Arabidopsis reveals a role of the nitrate transporter NRT1.1 in the regulation of another nitrate transporter, NRT2.1. Plant Cell 16: 2433–2447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naulin P.A., Armijo G.I., Vega A.S., Tamayo K.P., Gras D.E., de la Cruz J., Gutiérrez R.A.(2020). Nitrate induction of primary root growth requires cytokinin signaling in Arabidopsis thaliana. Plant Cell Physiol. 61: 342–352. [DOI] [PubMed] [Google Scholar]
- Negi J., Matsuda O., Nagasawa T., Oba Y., Takahashi H., Kawai-Yamada M., Uchimiya H., Hashimoto M., Iba K.(2008). CO2 regulator SLAC1 and its homologues are essential for anion homeostasis in plant cells. Nature 452: 483–486. [DOI] [PubMed] [Google Scholar]
- Nohno T., Noji S., Taniguchi S., Saito T.(1989). The narX and narL genes encoding the nitrate-sensing regulators of Escherichia coli are homologous to a family of prokaryotic two-component regulatory genes. Nucleic Acids Res. 17: 2947–2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohkubo Y., Tanaka M., Tabata R., Ogawa-Ohnishi M., Matsubayashi Y.(2017). Shoot-to-root mobile polypeptides involved in systemic regulation of nitrogen acquisition. Nat. Plants 3: 17029. [DOI] [PubMed] [Google Scholar]
- Okamoto M., Kumar A., Li W., Wang Y., Siddiqi M.Y., Crawford N.M., Glass A.D.(2006a). High-affinity nitrate transport in roots of Arabidopsis depends on expression of the NAR2-like gene AtNRT3.1. Plant Physiol. 140: 1036–1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto M., Kuwahara A., Seo M., Kushiro T., Asami T., Hirai N., Kamiya Y., Koshiba T., Nambara E.(2006b). CYP707A1 and CYP707A2, which encode abscisic acid 8′-hydroxylases, are indispensable for proper control of seed dormancy and germination in Arabidopsis. Plant Physiol. 141: 97–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto M., Vidmar J.J., Glass A.D.M.(2003). Regulation of NRT1 and NRT2 gene families of Arabidopsis thaliana: Responses to nitrate provision. Plant Cell Physiol. 44: 304–317. [DOI] [PubMed] [Google Scholar]
- Olas J.J., Van Dingenen J., Abel C., Działo M.A., Feil R., Krapp A., Schlereth A., Wahl V.(2019). Nitrate acts at the Arabidopsis thaliana shoot apical meristem to regulate flowering time. New Phytol. 223: 814–827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Malley R.C., Huang S.C., Song L., Lewsey M.G., Bartlett A., Nery J.R., Galli M., Gallavotti A., Ecker J.R.(2016). Cistrome and epicistrome features shape the regulatory DNA landscape. Cell 165: 1280–1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ondzighi-Assoume C.A., Chakraborty S., Harris J.M.(2016). Environmental nitrate stimulates abscisic acid accumulation in Arabidopsis root tips by releasing it from inactive stores. Plant Cell 28: 729–745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orsel M., Chopin F., Leleu O., Smith S.J., Krapp A., Daniel-Vedele F., Miller A.J.(2006). Characterization of a two-component high-affinity nitrate uptake system in Arabidopsis: Physiology and protein-protein interaction. Plant Physiol. 142: 1304–1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orsel M., Krapp A., Daniel-Vedele F.(2002). Analysis of the NRT2 nitrate transporter family in Arabidopsis: Structure and gene expression. Plant Physiol. 129: 886–896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osugi A., Kojima M., Takebayashi Y., Ueda N., Kiba T., Sakakibara H.(2017). Systemic transport of trans-zeatin and its precursor have differing roles in Arabidopsis shoots. Nat. Plants 3: 17112. [DOI] [PubMed] [Google Scholar]
- Para A., Li Y., Marshall-Colón A., Varala K., Francoeur N.J., Moran T.M., Edwards M.B., Hackley C., Bargmann B.O., Birnbaum K.D. (2014). Hit-and-run transcriptional control by bZIP1 mediates rapid nutrient signaling in Arabidopsis. Proc. Natl. Acad Sci USA 111: 10371–10376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park B.S., Yao T., Seo J.S., Wong E.C.C., Mitsuda N., Huang C.H., Chua N.H.(2018). Arabidopsis NITROGEN LIMITATION ADAPTATION regulates ORE1 homeostasis during senescence induced by nitrogen deficiency. Nat. Plants 4: 898–903. [DOI] [PubMed] [Google Scholar]
- Parker J.L., Newstead S.(2014). Molecular basis of nitrate uptake by the plant nitrate transporter NRT1.1. Nature 507: 68–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellizzaro A., Clochard T., Cukier C., Bourdin C., Juchaux M., Montrichard F., Thany S., Raymond V., Planchet E., Limami A.M., Morère-Le Paven M.C.(2014). The nitrate transporter MtNPF6.8 (MtNRT1.3) transports abscisic acid and mediates nitrate regulation of primary root growth in Medicago truncatula. Plant Physiol. 166: 2152–2165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peña P.A., et al. (2017). Expression of the Maize Dof1 Transcription Factor in Wheat and Sorghum. Front Plant Sci 30: 434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng M., Bi Y.M., Zhu T., Rothstein S.J.(2007). Genome-wide analysis of Arabidopsis responsive transcriptome to nitrogen limitation and its regulation by the ubiquitin ligase gene NLA. Plant Mol. Biol. 65: 775–797. [DOI] [PubMed] [Google Scholar]
- Peng M., Hudson D., Schofield A., Tsao R., Yang R., Gu H., Bi Y.-M., Rothstein S.J.(2008). Adaptation of Arabidopsis to nitrogen limitation involves induction of anthocyanin synthesis which is controlled by the NLA gene. J. Exp. Bot. 59: 2933–2944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perchlik M., Tegeder M.(2017). Improving Plant Nitrogen Use Efficiency Through Alteration of Amino Acid Transport Processes. Plant Physiol 175: 235–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Torres C.A., López-Bucio J., Cruz-Ramírez A., Ibarra-Laclette E., Dharmasiri S., Estelle M., Herrera-Estrella L.(2008). Phosphate availability alters lateral root development in Arabidopsis by modulating auxin sensitivity via a mechanism involving the TIR1 auxin receptor. Plant Cell 20: 3258–3272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plett D.C., Holtham L.R., Okamoto M., Garnett T.P.(2018). Nitrate uptake and its regulation in relation to improving nitrogen use efficiency in cereals. Semin. Cell Dev. Biol. 74: 97–104. [DOI] [PubMed] [Google Scholar]
- Poitout A., Crabos A., Petřík I., Novák O., Krouk G., Lacombe B., Ruffel S.(2018). Responses to systemic nitrogen signaling in Arabidopsis roots involve trans-zeatin in shoots. Plant Cell 30: 1243–1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu B., et al. (2015). A Wheat CCAAT Box-Binding Transcription Factor Increases the Grain Yield of Wheat with Less Fertilizer Input. Plant physiol 167: 411–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahayu Y.S., Walch-Liu P., Neumann G., Römheld V., von Wirén N., Bangerth F.(2005). Root-derived cytokinins as long-distance signals for NO3−-induced stimulation of leaf growth. J. Exp. Bot. 56: 1143–1152. [DOI] [PubMed] [Google Scholar]
- Ranathunge K., El-Kereamy A., Gidda S., Bi Y.M., Rothstein S.J.(2014). AMT1;1 transgenic rice plants with enhanced NH4(+) permeability show superior growth and higher yield under optimal and suboptimal NH4(+) conditions. Journal of experimental botany 65: 965–979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remans T., Nacry P., Pervent M., Filleur S., Diatloff E., Mounier E., Tillard P., Forde B.G., Gojon A.(2006). The Arabidopsis NRT1.1 transporter participates in the signaling pathway triggering root colonization of nitrate-rich patches. Proc. Natl. Acad. Sci. USA 103: 19206–19211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren H., Willige B.C., Jaillais Y., Geng S., Park M.Y., Gray W.M., Chory J.(2019). BRASSINOSTEROID-SIGNALING KINASE 3, a plasma membrane-associated scaffold protein involved in early brassinosteroid signaling. PLoS Genet. 15: e1007904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ristova D., Carré C., Pervent M., Medici A., Kim G.J., Scalia D., Ruffel S., Birnbaum K.D., Lacombe B., Busch W., Coruzzi G.M., Krouk G.(2016). Combinatorial interaction network of transcriptomic and phenotypic responses to nitrogen and hormones in the Arabidopsis thaliana root. Sci. Signal. 9: rs13. [DOI] [PubMed] [Google Scholar]
- Riveras E., Alvarez J.M., Vidal E.A., Oses C., Vega A., Gutiérrez R.A.(2015). The calcium ion is a second messenger in the nitrate signaling pathway of Arabidopsis. Plant Physiol. 169: 1397–1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers E.D., Benfey P.N.(2015). Regulation of plant root system architecture: Implications for crop advancement. Curr. Opin. Biotechnol. 32: 93–98. [DOI] [PubMed] [Google Scholar]
- Romera F.J., Smith A.P., Pérez-Vicente R.(2016). Editorial: Ethylene’s role in plant mineral nutrition. Front. Plant Sci. 7: 911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin G., Tohge T., Matsuda F., Saito K., Scheible W.R.(2009). Members of the LBD family of transcription factors repress anthocyanin synthesis and affect additional nitrogen responses in Arabidopsis. Plant Cell 21: 3567–3584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruffel S., Gojon A.(2017). Systemic nutrient signalling: On the road for nitrate. Nat. Plants 3: 17040. [DOI] [PubMed] [Google Scholar]
- Ruffel S., Krouk G., Ristova D., Shasha D., Birnbaum K.D., Coruzzi G.M.(2011). Nitrogen economics of root foraging: Transitive closure of the nitrate-cytokinin relay and distinct systemic signaling for N supply vs. demand. Proc. Natl. Acad. Sci. USA 108: 18524–18529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruffel S., Poitout A., Krouk G., Coruzzi G.M., Lacombe B.(2016). Long-distance nitrate signaling displays cytokinin dependent and independent branches. J. Integr. Plant Biol. 58: 226–229. [DOI] [PubMed] [Google Scholar]
- Sakakibara H.(2006). Cytokinins: Activity, biosynthesis, and translocation. Annu. Rev. Plant Biol. 57: 431–449. [DOI] [PubMed] [Google Scholar]
- Sakakibara H., Kobayashi K., Deji A., Sugiyama T.(1997). Partial characterization of the signaling pathway for the nitrate-dependent expression of genes for nitrogen-assimilatory enzymes using detached maize leaves. Plant Cell Physiol. 38: 837–843. [Google Scholar]
- Sakakibara H., Takei K., Hirose N.(2006). Interactions between nitrogen and cytokinin in the regulation of metabolism and development. Trends Plant Sci. 11: 440–448. [DOI] [PubMed] [Google Scholar]
- Sasaki T., Suzaki T., Soyano T., Kojima M., Sakakibara H., Kawaguchi M.(2014). Shoot-derived cytokinins systemically regulate root nodulation. Nat. Commun. 5: 4983. [DOI] [PubMed] [Google Scholar]
- Sato T., Maekawa S., Yasuda S., Domeki Y., Sueyoshi K., Fujiwara M., Fukao Y., Goto D.B., Yamaguchi J.(2011). Identification of 14-3-3 proteins as a target of ATL31 ubiquitin ligase, a regulator of the C/N response in Arabidopsis. Plant J. 68: 137–146. [DOI] [PubMed] [Google Scholar]
- Sato T., et al. (2009). CNI1/ATL31, a RING-type ubiquitin ligase that functions in the carbon/nitrogen response for growth phase transition in Arabidopsis seedlings. Plant J. 60: 852–864. [DOI] [PubMed] [Google Scholar]
- Schauser L., Roussis A., Stiller J., Stougaard J.(1999). A plant regulator controlling development of symbiotic root nodules. Nature 402: 191–195. [DOI] [PubMed] [Google Scholar]
- Schauser L., Wieloch W., Stougaard J.(2005). Evolution of NIN-like proteins in Arabidopsis, rice, and Lotus japonicus. J. Mol. Evol. 60: 229–237. [DOI] [PubMed] [Google Scholar]
- Scheible W.R., Gonzalez-Fontes A., Lauerer M., Muller-Rober B., Caboche M., Stitt M.(1997). Nitrate acts as a signal to induce organic acid metabolism and repress starch metabolism in tobacco. Plant Cell 9: 783–798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheible W.R., Morcuende R., Czechowski T., Fritz C., Osuna D., Palacios-Rojas N., Schindelasch D., Thimm O., Udvardi M.K., Stitt M.(2004). Genome-wide reprogramming of primary and secondary metabolism, protein synthesis, cellular growth processes, and the regulatory infrastructure of Arabidopsis in response to nitrogen. Plant Physiol. 136: 2483–2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwacke R., Ponce-Soto G.Y., Krause K., Bolger A.M., Arsova B., Hallab A., Gruden K., Stitt M., Bolger M.E., Usadel B.(2019). MapMan4: A refined protein classification and annotation framework applicable to multi-omics data analysis. Mol. Plant 12: 879–892. [DOI] [PubMed] [Google Scholar]
- Segonzac C., Boyer J.-C., Ipotesi E., Szponarski W., Tillard P., Touraine B., Sommerer N., Rossignol M., Gibrat R.(2007). Nitrate efflux at the root plasma membrane: Identification of an Arabidopsis excretion transporter. Plant Cell 19: 3760–3777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seligman K., Saviani E.E., Oliveira H.C., Pinto-Maglio C.A.F., Salgado I.(2008). Floral transition and nitric oxide emission during flower development in Arabidopsis thaliana is affected in nitrate reductase-deficient plants. Plant Cell Physiol. 49: 1112–1121. [DOI] [PubMed] [Google Scholar]
- Shearer H.L., Cheng Y.T., Wang L., Liu J., Boyle P., Després C., Zhang Y., Li X., Fobert P.R.(2012). Arabidopsis clade I TGA transcription factors regulate plant defenses in an NPR1-independent fashion. Mol. Plant Microbe Interact. 25: 1459–1468. [DOI] [PubMed] [Google Scholar]
- Shin R., Berg R.H., Schachtman D.P.(2005). Reactive oxygen species and root hairs in Arabidopsis root response to nitrogen, phosphorus and potassium deficiency. Plant Cell Physiol. 46: 1350–1357. [DOI] [PubMed] [Google Scholar]
- Signora L., De Smet I., Foyer C.H., Zhang H.(2001). ABA plays a central role in mediating the regulatory effects of nitrate on root branching in Arabidopsis. Plant J. 28: 655–662. [DOI] [PubMed] [Google Scholar]
- Song Y.H., Smith R.W., To B.J., Millar A.J., Imaizumi T.(2012). FKF1 conveys timing information for CONSTANS stabilization in photoperiodic flowering. Science 336: 1045–1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparks E.E., et al. (2016). Establishment of expression in the SHORTROOT-SCARECROW transcriptional cascade through opposing activities of both activators and repressors. Dev. Cell 39: 585–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steyfkens F., Zhang Z., Van Zeebroeck G., Thevelein J.M.(2018). Multiple transceptors for macro- and micro-nutrients control diverse cellular properties through the PKA pathway in yeast: A paradigm for the rapidly expanding world of eukaryotic nutrient transceptors up to those in human cells. Front. Pharmacol. 9: 191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sueyoshi K., Mitsuyama T., Sugimoto T., Kleinhofs A., Warner R.L., Oji Y.(1999). Effects of inhibitors for signaling components on the expression of the genes for nitrate reductase and nitrite reductase in excised barley leaves. Soil Sci. Plant Nutr. 45: 1015–1019. [Google Scholar]
- Sun H., et al. (2014a). Heterotrimeric G proteins regulate nitrogen-use efficiency in rice. Nat. Genet. 46: 652–656. [DOI] [PubMed] [Google Scholar]
- Sun J., Bankston J.R., Payandeh J., Hinds T.R., Zagotta W.N., Zheng N.(2014b). Crystal structure of the plant dual-affinity nitrate transporter NRT1.1. Nature 507: 73–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swift J., Adame M., Tranchina D., Henry A., Coruzzi G.M.(2019). Water impacts nutrient dose responses genome-wide to affect crop production. Nat. Commun. 10: 1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabata R., Sumida K., Yoshii T., Ohyama K., Shinohara H., Matsubayashi Y.(2014). Perception of root-derived peptides by shoot LRR-RKs mediates systemic N-demand signaling. Science 346: 343–346. [DOI] [PubMed] [Google Scholar]
- Takei K., Sakakibara H., Taniguchi M., Sugiyama T.(2001). Nitrogen-dependent accumulation of cytokinins in root and the translocation to leaf: Implication of cytokinin species that induces gene expression of maize response regulator. Plant Cell Physiol. 42: 85–93. [DOI] [PubMed] [Google Scholar]
- Takei K., Ueda N., Aoki K., Kuromori T., Hirayama T., Shinozaki K., Yamaya T., Sakakibara H.(2004). AtIPT3 is a key determinant of nitrate-dependent cytokinin biosynthesis in Arabidopsis. Plant Cell Physiol. 45: 1053–1062. [DOI] [PubMed] [Google Scholar]
- Tang W., et al. (2019). Genome-wide associated study identifies NAC42-activated nitrate transporter conferring high nitrogen use efficiency in rice. Nat. Commun. 10: 5279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thimm O., Blasing O., Gibon Y., Nagel A., Meyer S., Kruger P., Selbig J., Muller L.A., Rhee S.Y., Stitt M.(2004). MAPMAN: A user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J. 37: 914–939. [DOI] [PubMed] [Google Scholar]
- Tian Q., Chen F., Liu J., Zhang F., Mi G.(2008). Inhibition of maize root growth by high nitrate supply is correlated with reduced IAA levels in roots. J. Plant Physiol. 165: 942–951. [DOI] [PubMed] [Google Scholar]
- Tian Q.Y., Sun P., Zhang W.H.(2009). Ethylene is involved in nitrate-dependent root growth and branching in Arabidopsis thaliana. New Phytol. 184: 918–931. [DOI] [PubMed] [Google Scholar]
- Tocquin P., Corbesier L., Havelange A., Pieltain A., Kurtem E., Bernier G., Périlleux C.(2003). A novel high efficiency, low maintenance, hydroponic system for synchronous growth and flowering of Arabidopsis thaliana. BMC Plant Biol. 3: 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Triplett E.W., Barnett N.M., Blevins D.G.(1980). Organic acids and ionic balance in xylem exudate of wheat during nitrate or sulfate absorption. Plant Physiol. 65: 610–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsay Y.-F., Schroeder J.I., Feldmann K.A., Crawford N.M.(1993). The herbicide sensitivity gene CHL1 of Arabidopsis encodes a nitrate-inducible nitrate transporter. Cell 72: 705–713. [DOI] [PubMed] [Google Scholar]
- Varala K., Li Y., Marshall-Colón A., Para A., Coruzzi G.M.(2015). “Hit-and-run” leaves its mark: Catalyst transcription factors and chromatin modification. BioEssays 37: 851–856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varala K., et al. (2018). Temporal transcriptional logic of dynamic regulatory networks underlying nitrogen signaling and use in plants. Proc. Natl. Acad. Sci. USA 115: 6494–6499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidal E.A., Araus V., Lu C., Parry G., Green P.J., Coruzzi G.M., Gutiérrez R.A.(2010). Nitrate-responsive miR393/AFB3 regulatory module controls root system architecture in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 107: 4477–4482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidal E.A., Moyano T.C., Riveras E., Contreras-López O., Gutiérrez R.A.(2013). Systems approaches map regulatory networks downstream of the auxin receptor AFB3 in the nitrate response of Arabidopsis thaliana roots. Proc. Natl. Acad. Sci. USA 110: 12840–12845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von der Fecht-Bartenbach J., Bogner M., Dynowski M., Ludewig U.(2010). CLC-b-mediated NO−3/H+ exchange across the tonoplast of Arabidopsis vacuoles. Plant Cell Physiol. 51: 960–968. [DOI] [PubMed] [Google Scholar]
- Waldie T., Leyser O.(2018). Cytokinin targets auxin transport to promote shoot branching In Plant Physiol., Volume 177, pp. 803–818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker L., et al. (2017). Changes in gene expression in space and time orchestrate environmentally mediated shaping of root architecture. Plant Cell 29: 2393–2412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C.Q., Guthrie C., Sarmast M.K., Dehesh K.(2014). BBX19 interacts with CONSTANS to repress FLOWERING LOCUS T transcription, defining a flowering time checkpoint in Arabidopsis. Plant Cell 26: 3589–3602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P., Du Y., Hou Y.J., Zhao Y., Hsu C.C., Yuan F., Zhu X., Tao W.A., Song C.P., Zhu J.K.(2015). Nitric oxide negatively regulates abscisic acid signaling in guard cells by S-nitrosylation of OST1. Proc. Natl. Acad. Sci. USA 112: 613–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Nian J., Xie X., Yu H., Zhang J., Bai J., Dong G., Hu J., Bai B., Chen L., Xie Q., Feng J., et al. (2018b). Genetic variations in ARE1 mediate grain yield by modulating nitrogen utilization in rice. Nat Commun 9: 735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R., Guegler K., LaBrie S.T., Crawford N.M.(2000). Genomic analysis of a nutrient response in Arabidopsis reveals diverse expression patterns and novel metabolic and potential regulatory genes induced by nitrate. Plant Cell 12: 1491–1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R., Liu D., Crawford N.M.(1998). The Arabidopsis CHL1 protein plays a major role in high-affinity nitrate uptake. Proc. Natl. Acad. Sci. USA 95: 15134–15139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R., Okamoto M., Xing X., Crawford N.M.(2003). Microarray analysis of the nitrate response in Arabidopsis roots and shoots reveals over 1,000 rapidly responding genes and new linkages to glucose, trehalose-6-phosphate, iron, and sulfate metabolism. Plant Physiol. 132: 556–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R., Tischner R., Gutiérrez R.A., Hoffman M., Xing X., Chen M., Coruzzi G., Crawford N.M.(2004). Genomic analysis of the nitrate response using a nitrate reductase-null mutant of Arabidopsis. Plant Physiol. 136: 2512–2522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R., Xing X., Crawford N.(2007). Nitrite acts as a transcriptome signal at micromolar concentrations in Arabidopsis roots. Plant Physiol. 145: 1735–1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R., Xing X., Wang Y., Tran A., Crawford N.M.(2009). A genetic screen for nitrate regulatory mutants captures the nitrate transporter gene NRT1.1. Plant Physiol. 151: 472–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y.H., Garvin D.F., Kochian L.V.(2001). Nitrate-induced genes in tomato roots: Array analysis reveals novel genes that may play a role in nitrogen nutrition. Plant Physiol. 127: 345–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y.-Y., Cheng Y.-H., Chen K.-E., Tsay Y.-F.(2018a). Nitrate transport, signaling, and use efficiency. Annu. Rev. Plant Biol. 69: 85–122. [DOI] [PubMed] [Google Scholar]
- Wany A., Foyer C.H., Gupta K.J.(2018). Nitrate, NO and ROS signaling in stem cell homeostasis. Trends Plant Sci. 23: 1041–1044. [DOI] [PubMed] [Google Scholar]
- Weirauch M.T., et al. (2014). Determination and inference of eukaryotic transcription factor sequence specificity. Cell 158: 1431–1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White P.J.(2012). Ion uptake mechanisms of individual cells and roots: Short-distance transport In Marschner’s Mineral Nutrition of Higher Plants, Marschner P., ed, 3rd ed (San Diego: Academic Press; ), pp. 7–47. [Google Scholar]
- Wirth J., Chopin F., Santoni V., Viennois G., Tillard P., Krapp A., Lejay L., Daniel-Vedele F., Gojon A.(2007). Regulation of root nitrate uptake at the NRT2.1 protein level in Arabidopsis thaliana. J. Biol. Chem. 282: 23541–23552. [DOI] [PubMed] [Google Scholar]
- Xu H., Zhao M., Zhang Q., Xu Z., Xu Q.(2016a). The DENSE AND ERECT PANICLE 1 (DEP1) gene offering the potential in the breeding of high-yielding rice. Breed. Sci. 66: 659–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu N., Wang R., Zhao L., Zhang C., Li Z., Lei Z., Liu F., Guan P., Chu Z., Crawford N.M., Wang Y.(2016b). The Arabidopsis NRG2 protein mediates nitrate signaling and interacts with and regulates key nitrate regulators. Plant Cell 28: 485–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan D., et al. (2016). NIN-like protein 8 is a master regulator of nitrate-promoted seed germination in Arabidopsis. Nat. Commun. 7: 13179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J.(2019). Reducing expression of a nitrate‐responsive bZIP transcription factor increases grain yield and N use in wheat. Plant Biotech J 17: 1823–1833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasuda S., Aoyama S., Hasegawa Y., Sato T., Yamaguchi J.(2017). Arabidopsis CBL-interacting protein kinases regulate carbon/nitrogen-nutrient response by phosphorylating ubiquitin ligase ATL31. Mol. Plant 10: 605–618. [DOI] [PubMed] [Google Scholar]
- Yasuda S., Sato T., Maekawa S., Aoyama S., Fukao Y., Yamaguchi J.(2014). Phosphorylation of Arabidopsis ubiquitin ligase ATL31 is critical for plant carbon/nitrogen nutrient balance response and controls the stability of 14-3-3 proteins. J. Biol. Chem. 289: 15179–15193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yazaki J., et al. (2016). Mapping transcription factor interactome networks using HaloTag protein arrays. Proc. Natl. Acad. Sci. USA 113: E4238–E4247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu L.-H., Miao Z.-Q., Qi G.-F., Wu J., Cai X.-T., Mao J.-L., Xiang C.-B.(2014). MADS-box transcription factor AGL21 regulates lateral root development and responds to multiple external and physiological signals. Mol. Plant 7: 1653–1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu P., Baldauf J.A., Lithio A., Marcon C., Nettleton D., Li C., Hochholdinger F.(2016). Root type-specific reprogramming of maize pericycle transcriptomes by local high nitrate results in disparate lateral root branching patterns. Plant Physiol. 170: 1783–1798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan S., et al. (2016). Arabidopsis cryptochrome 1 functions in nitrogen regulation of flowering. Proc. Natl. Acad. Sci. USA 113: 7661–7666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G.-B., Meng S., Gong J.-M.(2018). The expected and unexpected roles of nitrate transporters in plant abiotic stress resistance and their regulation. Int. J. Mol. Sci. 19: 3535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Forde B.G.(1998). An Arabidopsis MADS box gene that controls nutrient-induced changes in root architecture. Science 279: 407–409. [DOI] [PubMed] [Google Scholar]
- Zhang H., Jennings A., Barlow P.W., Forde B.G.(1999). Dual pathways for regulation of root branching by nitrate. Proc. Natl. Acad. Sci. USA 96: 6529–6534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., et al. (2019). Identification and characterization of a novel stay-green QTL that increases yield in maize. Plant Biotechnol. J. 17: 2272–2285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K., Novak O., Wei Z., Gou M., Zhang X., Yu Y., Yang H., Cai Y., Strnad M., Liu C.J.(2014). Arabidopsis ABCG14 protein controls the acropetal translocation of root-synthesized cytokinins. Nat. Commun. 5: 3274. [DOI] [PubMed] [Google Scholar]
- Zheng D., Han X., An Y.I., Guo H., Xia X., Yin W.(2013). The nitrate transporter NRT2.1 functions in the ethylene response to nitrate deficiency in Arabidopsis. Plant Cell Environ. 36: 1328–1337. [DOI] [PubMed] [Google Scholar]
- Zhou J., Jiao F., Wu Z., Li Y., Wang X., He X., Zhong W., Wu P.(2008). OsPHR2 is involved in phosphate-starvation signaling and excessive phosphate accumulation in shoots of plants. Plant Physiol. 146: 1673–1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J.J., Fernández E., Galván A., Miller A.J.(2000). A high affinity nitrate transport system from Chlamydomonas requires two gene products. FEBS Lett. 466: 225–227. [DOI] [PubMed] [Google Scholar]
- Zhuo D., Okamoto M., Vidmar J.J., Glass A.D.(1999). Regulation of a putative high-affinity nitrate transporter (Nrt2;1At) in roots of Arabidopsis thaliana. Plant J. 17: 563–568. [DOI] [PubMed] [Google Scholar]