Genome analyses reveal that several plants have lost a major part of the immune system.
Abstract
Plant innate immunity relies on nucleotide binding leucine-rich repeat receptors (NLRs) that recognize pathogen-derived molecules and activate downstream signaling pathways. We analyzed the variation in NLR gene copy number and identified plants with a low number of NLR genes relative to sister species. We specifically focused on four plants from two distinct lineages, one monocot lineage (Alismatales) and one eudicot lineage (Lentibulariaceae). In these lineages, the loss of NLR genes coincides with loss of the well-known downstream immune signaling complex ENHANCED DISEASE SUSCEPTIBILITY 1 (EDS1)/PHYTOALEXIN DEFICIENT 4 (PAD4). We expanded our analysis across whole proteomes and found that other characterized immune genes were absent only in Lentibulariaceae and Alismatales. Additionally, we identified genes of unknown function that were convergently lost together with EDS1/PAD4 in five plant species. Gene expression analyses in Arabidopsis (Arabidopsis thaliana) and Oryza sativa revealed that several homologs of the candidates are differentially expressed during pathogen infection, drought, and abscisic acid treatment. Our analysis provides evolutionary evidence for the rewiring of plant immunity in some plant lineages, as well as the coevolution of the EDS1/PAD4 pathway and drought responses.
INTRODUCTION
Over 450 million years ago (MYA) plants diverged from a common ancestor shared with charophyte green algae (Sanderson et al., 2004; Zhong et al., 2015) and acquired new traits facilitating the colonization of diverse terrestrial environments. Extant plant lineages, such as bryophytes (mosses, liverworts, and hornworts), terrestrial vascular plants (ferns), gymnosperms (pine, spruce), and angiosperms (monocots, eudicots) radiated from the ancestor of terrestrial plants over 300 MYA (Zeng et al., 2014). Crops in modern agriculture typically belong to the monocot or eudicot classes of angiosperms. Major yield losses result from the exposure of crops to biotic and abiotic stresses. In contrast to studies in controlled laboratory conditions that commonly focus on individual stress responses, crops in the field are often simultaneously exposed to biotic and abiotic stresses, which are likely to be exacerbated as a consequence of climate change (Ramegowda and Senthil-Kumar, 2015). There is an urgent need to advance our understanding of stress responses for the breeding of more resilient crops.

Land plants have likely been accompanied by beneficial and pathogenic microbes throughout their evolutionary history. The initial plant disease resistance response to a pathogen relies on recognition of extracellular microbe-associated molecular patterns (MAMPs) and is termed MAMP-triggered immunity (MTI; Newman et al., 2013). Most pathogens deploy effectors that can suppress MTI to facilitate virulence (Newman et al., 2013). Hence, a second intracellular monitoring system of effector-triggered immunity (ETI) is essential for resistance to many pathogens (Tamborski and Krasileva, 2020). Plant nucleotide binding Leu-rich repeat receptors (NLRs) mediate ETI upon detection of intracellular pathogen molecules (Tamborski and Krasileva, 2020). The NLR immune recognition system predates land plant emergence as proteins with a similar architecture are present in green algae (Charophyta) and red algae (Rhodophyta; Gao et al., 2018).
NLR proteins are typically composed of three or more domains (Jones et al., 2016; Baggs et al., 2017). The nucleotide binding (NB-ARC) domain is the common central component of an NLR and is involved in receptor activation (Jones et al., 2016). The NB-ARC domain is usually followed by a series of Leu-rich repeats (LRRs), which mediate intramolecular interactions within NLRs and intermolecular binding of effector proteins (Dodds et al., 2001; Catanzariti et al., 2010; Krasileva et al., 2010). A Toll-like Interleukin-1 (TIR-1) or a coiled-coil (CC) domain are typically found at the N terminus of NLRs, where they function in the initiation of the signaling cascade (Bernoux et al., 2011). NLRs containing a TIR-1 domain are referred to as TNLs, whereas CNL refers to NLRs with a CC domain. The third clade of NLRs called RNLs is characterized by the presence of an N-terminal RPW8 (RESISTANCE TO POWDERY MILDEW 8) domain. Members of all three clades (CNLs, TNLs, and RNLs) are present in basal angiosperms and gymnosperms (Gao et al., 2018; Van Ghelder et al., 2019). Despite 120 to 180 MY of independent monocot and eudicot evolution, the barley (Hordeum vulgare) CNL MILDEW LOCUS A (MLA) was still functional when transferred to thale cress Arabidopsis (Arabidopsis thaliana; Maekawa et al., 2012). Despite the observed functional conservation of some NLRs, significant differences are observed between monocots and eudicots. The most pronounced difference between NLR repertoires of monocots and eudicots is the absence of TIR-1 containing TNLs in monocots (Tarr and Alexander, 2009). Among eudicots, typically over half the NLRs contain a TIR-1 domain (Meyers et al., 2003; Sarris et al., 2016), whereas monocots have only a few TIR-2 type NLR genes (Meyers et al., 2003; Nandety et al. 2013; Sarris et al., 2016). Within monocots and eudicots, NLRs from different sub-clades have undergone both contractions and expansions in a lineage-specific manner.
NLRs can function either as sensors of pathogen-derived molecules or as helpers controlling signal transduction (Wu et al., 2017; Castel et al., 2018; Qi et al., 2018; Lapin et al., 2019; Wu et al., 2019). All TNLs characterized to date as well as some CNLs genetically depend on RNLs, such as N REQUIREMENT GENE 1 (NRG1) and ACTIVATED DISEASE RESISTANCE-LIKE1 (ADR1; Castel et al., 2018; Qi et al., 2018; Lapin et al., 2019; Wu et al., 2019). Additional helper NLRs have been found within both the TNL and CNL sub-clades. Some lineages, such as the Solanaceae, have evolved complex sensor-helper networks in which multiple sensors depend on the same helper for signaling (Wu et al., 2017, 2018). Understanding NLR functions and their genetic requirements can guide their deployment in crop improvement.
The key signaling pathways downstream of NLRs have been identified and studied primarily in eudicot species. First discovered in Arabidopsis, NON-RACE SPECIFIC DISEASE RESISTANCE-1 (NDR1) has since been shown to be conserved across eudicots and is required for signaling of several CNLs (Century et al., 1995; Coppinger et al., 2004). NDR1 is thought to mediate resistance by controlling fluid loss in the cell (Knepper et al., 2011), yet the exact mechanism of how it induces signal transduction remains unclear.
Another signaling hub used by many NLRs and all TNLs studied to date includes the lipase-like proteins ENHANCED DISEASE SUSCEPTIBILITY 1 (EDS1), PHYTOALEXIN DEFICIENT 4 (PAD4), and SENESCENCE ASSOCIATED GENE 101 (SAG101; Falk et al., 1999; Jirage et al., 1999; Feys et al., 2005). In planta work in Arabidopsis showed that EDS1 binds directly to PAD4 and SAG101 to form mutually exclusive heterodimeric complexes (Wagner et al., 2013), and this complex formation changes the subcellular localization of EDS1 depending on the interacting partner (Zhu et al., 2011). Recent work has also provided molecular and genetic evidence that Arabidopsis EDS1, SAG101, and NRG1 form a complex upon TNL activation, resulting in cell death (Lapin et al., 2019). TNL signaling involves enzymatic catalysis of NAD+, which activates EDS1 through a yet unknown mechanism (Horsefield et al., 2019; Wan et al., 2019). RNLs act downstream of EDS1/PAD4/SAG101 to induce plant cell death and transcriptional responses (Castel et al., 2018; Qi et al., 2018; Lapin et al., 2019; Wu et al., 2019).
Both EDS1 and PAD4 are present in almost all angiosperms, and were present before the angiosperm and gymnosperm split (Wagner et al., 2013; Bhandari et al., 2019; Lapin et al., 2019). In contrast, SAG101 is absent from available grass genomes along with TNLs (Wagner et al., 2013). Despite the absence of TIR-1 NLRs in monocots, wheat (Triticum aestivum) EDS1 has retained functional importance in immunity. Overexpression of TaEDS1 in a susceptible T. aestivum cultivar results in reduced Blumeria graminis f.sp. tritici haustorial growth (Chen et al., 2018). Moreover, TaEDS1 was able to complement the eds1 mutant in Arabidopsis (Chen et al., 2018), highlighting its highly conserved role in immune signaling.
Both NDR1- and EDS1/PAD4/SAG101-mediated signaling converge in accumulation of salicylic acid (SA; Venugopal et al., 2009; Cui et al., 2018). Across monocots and eudicots, there is conservation of the general SA-mediated response, its major regulator NONEXPRESSER OF PR GENES 1 (NPR1), and induction of PATHOGENESIS RELATED (PR) genes. Conserved signaling components of plant immunity have been used to engineer broad-spectrum resistance (Cao et al., 1998; Xu et al., 2017), demonstrating the translational impacts on crop production from understanding the evolution of immune signaling.
Several plant immunity components, including EDS1/PAD4 as well as ADR1, also affect abiotic stress responses (Chini et al., 2004; Wituszynska et al., 2013; Szechyńska-Hebda et al., 2016). Continuous overexpression of ADR1 increased drought tolerance in Arabidopsis (Chini et al., 2004); this phenotype was not seen in other mutants with enhanced PR gene expression and depended on the presence of functional EDS1 and PAD4 (Chini et al., 2004). Similarly, mutants in LESION SIMULATING DISEASE 1 (LSD1) that exhibit runaway cell death were more resistant to drought and high-light stress in EDS1/PAD4-dependent manner (Wituszynska et al., 2013; Szechyńska-Hebda et al., 2016). The mechanism of drought tolerance in lsd1 null mutants and ADR1 overexpression lines is thought to involve SA but to be independent of abscisic acid (ABA), consistent with the described antagonism between SA and ABA (Moeder et al., 2010). Recent studies have started to elucidate components mediating this crosstalk and prioritization (Berens et al., 2019; Wolinska and Berens, 2019), and further studies are essential to fully understand how abiotic and biotic stress responses are regulated.
Within flowering plants, there is a huge amount of genetic, genomic, and phenotypic diversity that can be used to understand plant molecular pathways. Previous studies examined convergent evolution in plants to successfully predict gains and losses of genetic pathways and to identify new pathway components (Ibarra-Laclette et al., 2013; Wang et al., 2014; Olsen et al., 2016; Michael et al., 2017). In the study of plant symbiotic interactions, this approach has been particularly fruitful, identifying coevolved proteins that have since been functionally validated (Bravo et al., 2016; Griesmann et al., 2018; Radhakrishnan et al., 2020) and uncovering the origin of nitrogen-fixing rhizobium symbioses (Griesmann et al., 2018; van Velzen et al., 2018). The growing number of available plant genomes, including species from diverse environments, increases the opportunity to elucidate pathways that facilitate adaptation.
In this study, we used comparative genomics to look at the NLR gene copy number variation across angiosperms. We identified independent contractions in NLR gene number and diversity among monocots and eudicots and discovered that the EDS1/PAD4 pathway was convergently lost in at least five species. We further analyzed components of plant disease resistance pathways and used gene family clustering methods to identify new genes following the same evolutionary pattern. Our analyses predicted new candidates that coevolved with the EDS1/PAD4 signaling pathway and provide further support for links between plant immunity and drought response.
RESULTS
Several Lineages of Flowering Plants Have Lost Most of the NLR Clades Present in their Common Ancestor
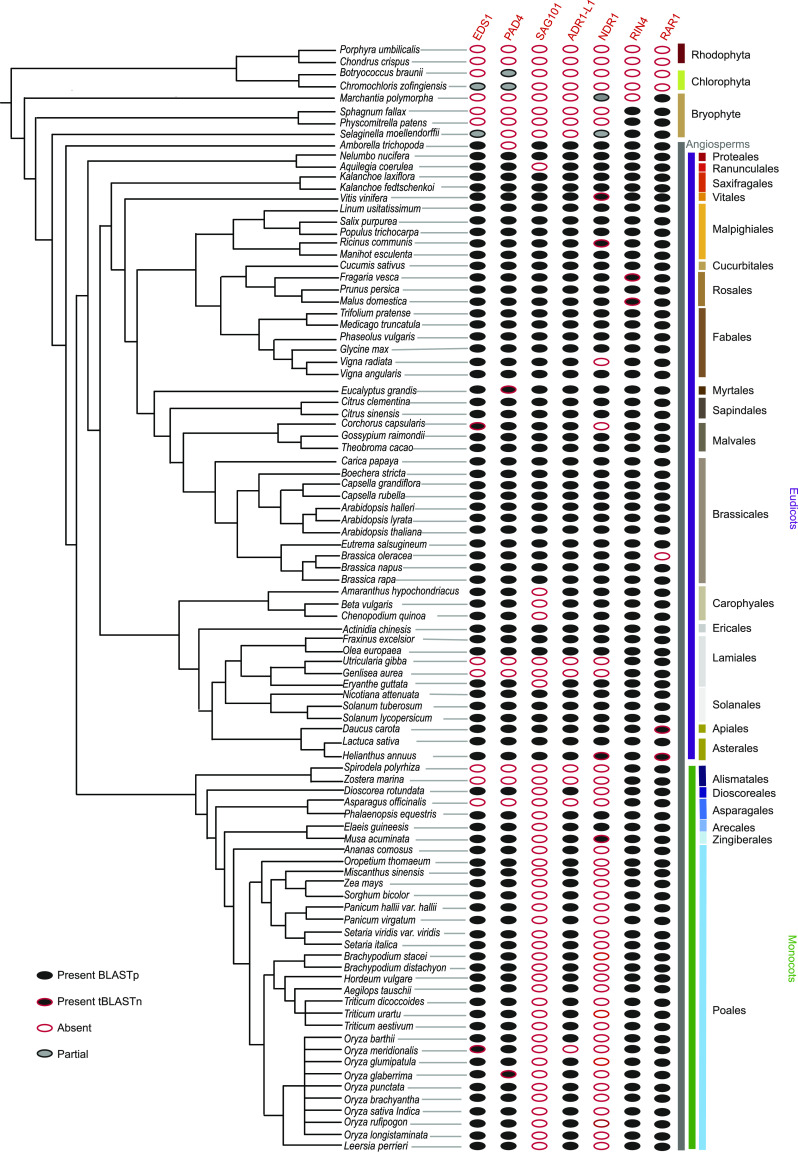
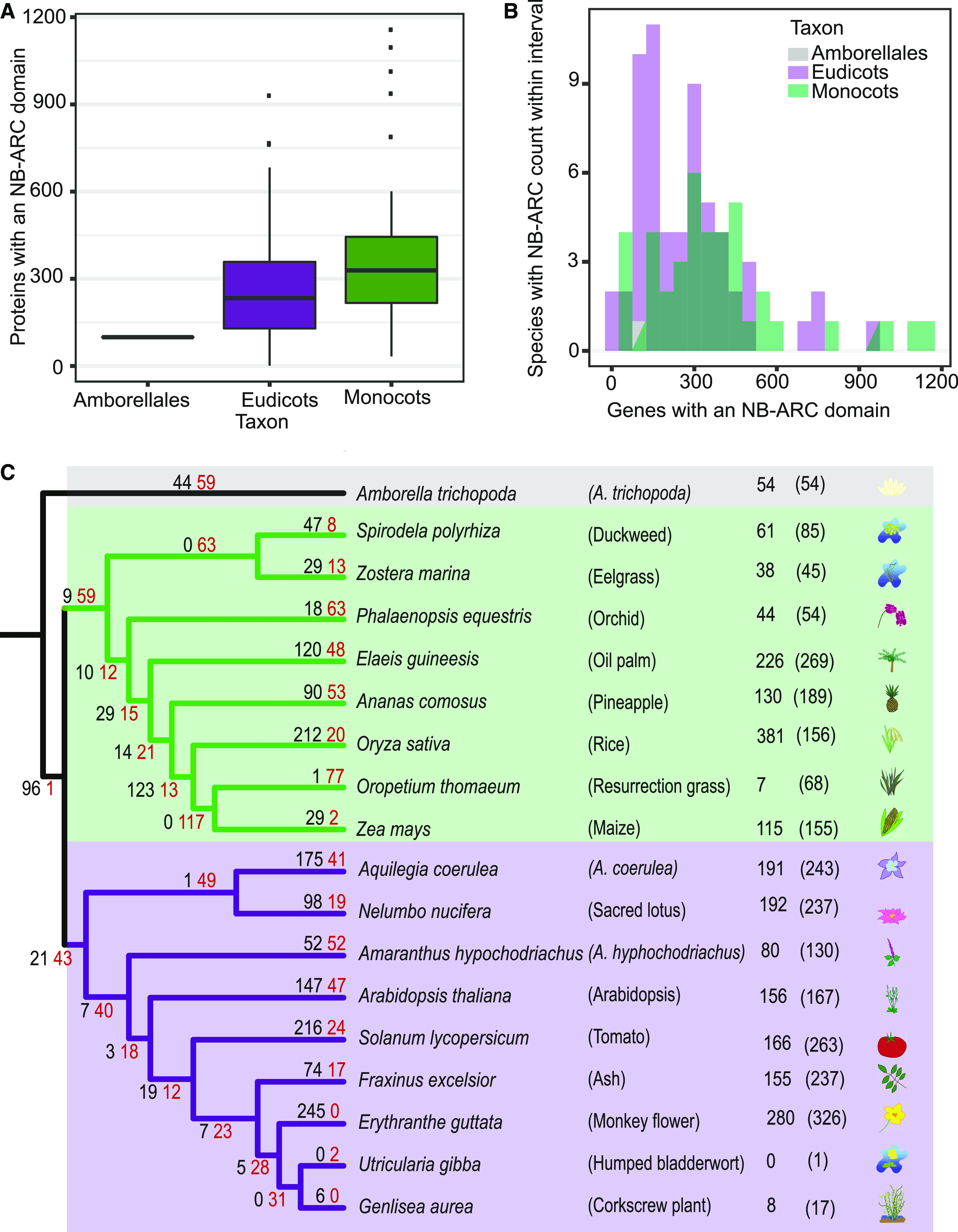
The NLR gene family is complex, with copy numbers varying 30-fold among species of the same family, such as from 33 in resurrection grass (Oropetium thomaeum) to over 1000 in T. aestivum in Poaceae (Supplemental Figure 1; Supplemental Table 1; Baggs et al., 2017; Steuernagel et al., 2020). Due to the historical bias in genome sequencing in favor of economically important or model species, most previous studies focused on the Brassicaceae, Solanaceae, and Poaceae families (Sarris et al., 2016; Stam et al., 2019; Van de Weyer et al., 2019). We surveyed NLR gene copy number variation in 95 publicly available angiosperm genomes spanning 24 orders (Figure 1A; Supplemental Figure 1; Supplemental Table 1). Although most of the sequenced plant genomes contain 200–500 NLRs (Figure 1A), there have been extreme losses and expansions in the NLR gene family among both monocots and eudicots (Supplemental Figure 1; Supplemental Table 1). We decided to further investigate plant genomes that have convergently lost the majority of their NLR genes.
Figure 1.
Phylogenetic Relationship and NLR Repertoires of the Plant Species Used in This Study.
(A) Boxplot showing the variation in number of NB-ARC domains across Amborellales (n = 1), monocot (n = 42), and eudicot (n = 60) genomes available on Phytozome, Ensembl Plants or CoGe.
(B) Histogram of numbers of NB-ARC domains identified in angiosperm genomes.
(C) Species tree of monocot and eudicot genomes of interest. Number of NLRs with all 6 characteristic NLR amino-acid motifs annotated in each species are displayed in line with the species Latin name. Number of NLRs identified by PfamScan and the plant_rgenes pipeline are in parentheses. Black/red numbers on branches indicate the number of gained/lost NLRs.
For further analyses, we selected 18 species to represent a broad range of families and include sister species with highly divergent NLR gene copy number and high-quality genome assemblies (Figure 1B). We considered an NLR gene number to be low if it was below the first quartile: 217 NLR genes for monocots and 129 NLR genes for eudicots. To test if the observed reduction of NLR gene numbers was due to incomplete annotations, we mined the genomic sequences using NLR-Annotator (Steuernagel et al., 2020). This software predicts NLRs from motifs present in six-frame translations of the DNA sequence and is therefore independent of protein annotation (Supplemental Table 2; Steuernagel et al., 2015, 2020). Results from the NLR-Annotator analysis confirmed the reduced NLR gene numbers in eel grass (Zostera marina), greater duckweed (Spirodela polyrhiza), orchid (Phalaenopsis equestris), O. thomaeum, maize (Zea mays), pineapple (Ananas comosus), humped bladderwort (Utricularia gibba), and corkscrew plant (Genlisea aurea; Supplemental Table 2). The results show the low NLR gene number was consistent across the genome and proteome.
To examine gains and losses of NLR genes along the evolution of different lineages, we built a maximum likelihood phylogeny on the NB-ARC domain of all NLRs retaining 6 key functional motifs (Walker A, RNBS-A, WALKER-B, RNBS-C, GLPL and RNBS-D; Figure 2; Supplemental Files 1 and 2; Tameling et al., 2006; van Ooijen et al., 2008; Wang et al., 2015; Wen et al., 2017). We applied tree reconciliation between the species tree and the NLR gene tree to quantify NLR gains and losses across the phylogeny (Figure 1C). We observed a high turnover of NLR genes across the phylogeny, with large expansions in specific lineages: rice (Oryza sativa), oil palm (Elaeis guineensis), Colorado blue columbine (Aquilegia coerulea), Arabidopsis, tomato (Solanum lycopersicum), and monkey flower (Erythranthe guttata), as well as extensive losses such as in the ancestral lineage of Z. mays and O. thomaeum as well as U. gibba and G. aurea.
Figure 2.
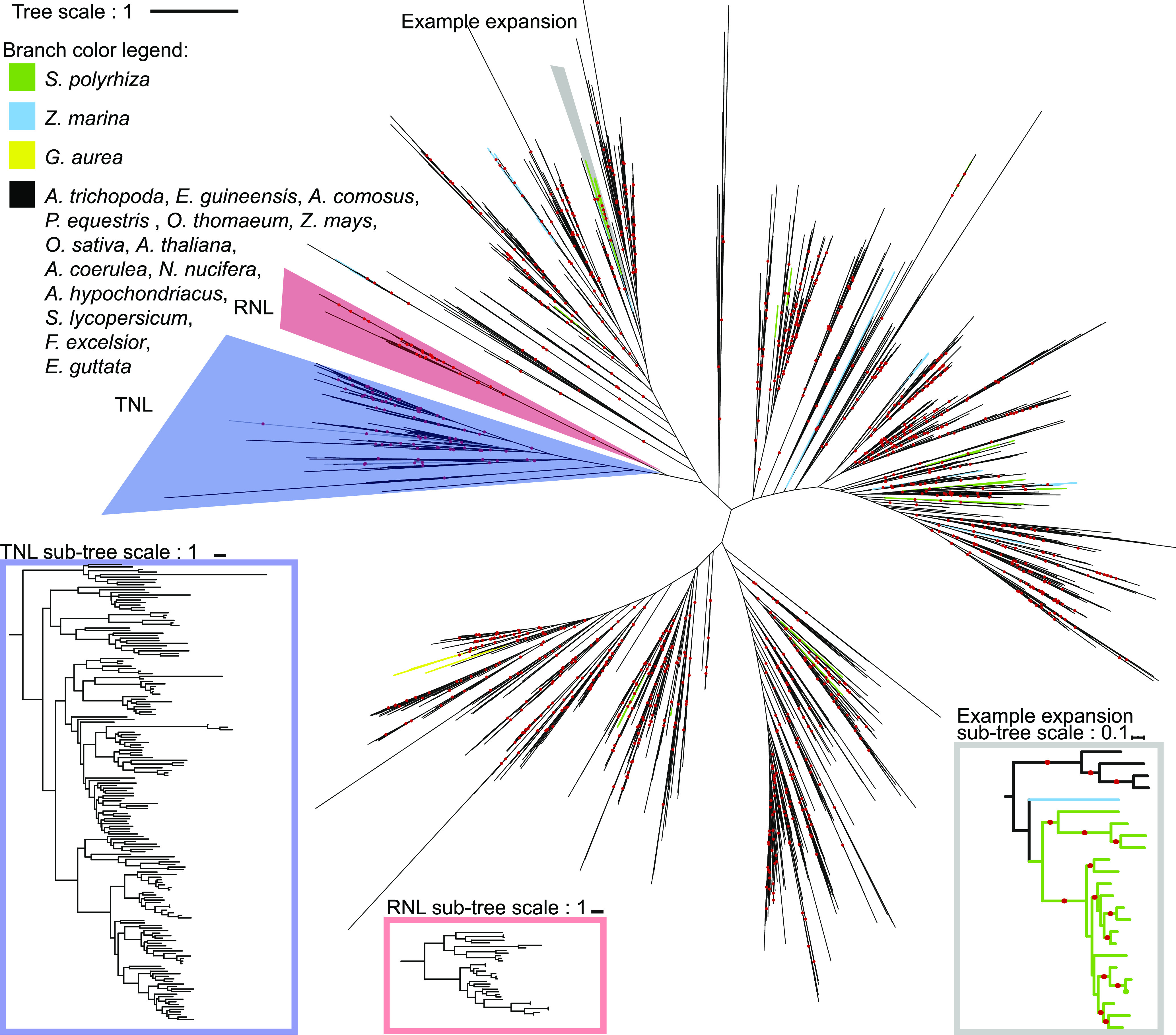
Maximum likelihood phylogeny of NLRs in the 18 representative plant species and selected reference NLRs. The maximum likelihood tree is based on the alignment of the NB-ARC domains of the 18 representative species of A. trichopoda, Z. marina, S. polyrhiza, E. guineensis, A. comosus, P. equestris, O. thomaeum, Z. mays, O. sativa, A. thaliana, A. coerulea, N. nucifera, A. hypochondriacus, S. lycopersicum, F. excelsior, E. guttata, U. gibba, and G. aurea. Bootstraps >80 are indicated by a red dot; branch colors denote species. Clades as defined by bootstrap >80; TNLs and RNLs are within the blue and red sections, respectively. Inlayed sub-trees provide a zoom-in to the TNL, RNL, and CNL clades, where colored branches are indicative of Z. marina, S. polyrhiza, and G. aurea. Sub-tree within gray box is an example of a CNL expansion present in S. polyrhiza.
Although some of the plants with a low number of NLR genes retained all major NLR sub-clades (CNL, TNL, RNL), others did not. ADR1-type RNLs are typically present in all monocots and eudicots; however, we observed no identifiable RNLs in S. polyrhiza, Z. marina, U. gibba, or G. aurea and confirmed our phylogenetic analyses by reciprocal BLAST. The NRG1-type RNLs were absent in all monocots in our analysis, as well as in eudicots that have also lost TNLs: E. guttata, consistent with (Kim et al., 2012), U. gibba, and G. aurea (Supplemental Figure 2). We confirmed that NRG1 was present in all species with TNL expansions (Figure 2), consistent with genetic dependence of TNLs on NRG1 for signaling (Castel et al., 2018; Qi et al., 2018; Lapin et al., 2019; Wu et al., 2019). In addition, no NRG1 ortholog could be identified through phylogeny or reciprocal BLAST in Amborella trichopoda and Amaranthus hypochondriacus that have 14 and 2 TNLs, respectively. A single clade (bootstrap = 65) contains 10 A. trichopoda and 1 A. hypochondriacus TNLs, suggesting that NLRs in this clade might function independently of NRG1. Overall conservation of the RNL clade in monocots, eudicots, and A. trichopoda suggests that the absence of RNLs in S. polyrhiza, Z. marina, U. gibba, and G. aurea represents convergent clade-specific gene loss events.
Despite the loss of many NLR genes, those retained in S. polyrhiza, Z. marina, and U. gibba have undergone recent lineage-specific expansions (Figure 2). In these species 88%, 84%, and 100% of NLR genes are present in monophyletic species-specific clades (bootstrap support >80). Several S. polyrhiza and Z. marina NLR genes arose from recent tandem duplications as indicated by their consecutive gene identifiers (Figure 2). This suggests that despite overall gene loss, both species have likely experienced selective pressure to expand the remaining NLR clades.
To test if a general reduction in large protein families was responsible for the loss of RNLs, we annotated receptors belonging to the Receptor-Like Kinase family (RLKs) and actin proteins. RLKs are an MTI immune receptor family known to be highly variable in copy number similar to NLRs, whereas actin proteins are less variable in copy number. The reduction of RLKs compared to sister lineages was not as pronounced as with NLRs, with the exception of G. aurea (Supplemental Table 2). The percentage of actin-encoding genes in the proteome was similar between species lacking RNLs and their closest related species (Supplemental Table 2). This suggests that the reduction in the copy number and the loss of NLR subclades cannot be explained by the general contraction of large protein families.
Independent Loss of Immune Signaling Components EDS1/PAD4/SAG101 among Angiosperm Lineages
The simultaneous loss of genes encoding RNLs and TNLs prompted us to survey for the presence of other immune signaling components. In total, we surveyed for the presence of 15 known immune signaling components using reciprocal blastp followed by tblastn to remove false negatives due to incomplete annotation (Figure 3; Supplemental Figure 3; Supplemental Tables 3 and 4). Several of the surveyed genes, including RPM1 INTERACTING PROTEIN 4 (RIN4), MAP KINASE 4 (MPK4), ISOCHORISMATE SYNTHASE 1 (ICS1), METACASPASE 4/ 5 (MCP4/5), and REQUIRED FOR MLA12 RESISTANCE 1 (RAR1), were conserved across all 18 monocots and eudicots (Supplemental Table 4). The SA biosynthetic gene ICS1 and the SA receptor family of NPR genes were present in all species except U. gibba (Supplemental Table 4). This suggests that the SA biosynthesis and signaling pathways remain intact in most species.
Figure 3.
Presence/absence analysis of known plant immunity components. Rows denote species, which are arranged as per phylogenetic relationship, with the green and purple bars indicating monocots, or, respectively, dicots. Gene names are listed at the top. Circles in columns denote the presence or absence of known components of the NLR signaling pathway. Black filled circles represent orthologs identified by reciprocal blastp. Orthologs supported by tblastn are indicated by black circles outlined in red. Absent orthologs are displayed by white circles with a red outline, and partial orthologs are shown as gray circles with a black outline. Orthology was also manually curated using Ensembl Plant gene trees or Phytozome synteny (where available).
In contrast, we observed the loss of EDS1/PAD4/SAG101 and NDR1 among species without RNLs and TNLs (Figure 3). To determine how prevalent the loss of EDS1, PAD4, SAG101, RNLs, and NDR1 is among angiosperms, we repeated the reciprocal blastp followed by TBLASTN on the 95 available genomes used in our initial survey (Figure 3; Supplemental Table 4). We then manually curated orthology using EnsemblPlant gene trees or Phytozome synteny, where available. This identified asparagus (Asparagus officinalis) as the only other angiosperm with a sequenced genome that is missing EDS1, PAD4, SAG101, NDR1, and RNLs. Therefore, the loss of EDS1, PAD4, SAG101, and RNLs appears to be limited to a few species with a low number of NLR genes.
To confirm that the inferred absence of these genes is not an annotation artifact, we scanned the low-NLR number genomes of S. polyrhiza, Z. marina, U. gibba, G. aurea, and A. officinalis for the presence of a lipase motif found in EDS1/PAD4/SAG101 using the Hidden Markov Model of this characteristic feature. Additional searches with TBLASTN and hmmsearch confirmed the loss of EDS1, PAD4, and SAG101 (Supplemental Figure 4). RNLs, EDS1 and PAD4 are known to interact in a protein complex and our data suggests that they also form an evolutionary unit.
Recent research supports the idea that gene loss events are less likely to be annotation artifacts if they are clade-specific losses rather than losses in a single species (Deutekom et al., 2019). Therefore, we expanded our gene loss analyses to an orthogonal data set of the 1000-plant transcriptomes (Matasci et al., 2014; Wickett et al., 2014; One Thousand Plant Transcriptomes Initiative, 2019). Generally, most species in the early monocot lineages Magnoliales, Piperales, and Laurales as well as Pandales and Dioscoreales had transcripts orthologous to EDS1, PAD4, RNL genes, and NDR1 (Supplemental Figure 5; Supplemental Table 5). This suggests that EDS1, PAD4, RNL genes, and NDR1 were present in the most recent common ancestor of monocots. EDS1, PAD4, RNL genes, and NDR1 transcripts were absent among multiple species in the orders of Piperales, Alismatales, Asparagales, and Liliales, supporting our finding that these components were lost in Alismatales (S. polyrhiza and Z. marina) and Asparagales (A. officinalis). Without complete genomes we cannot exclude the possibility that these genes were not expressed in the tissue or at the time samples were taken for the transcriptomics analysis. S. polyrhiza and Z. marina belong to the Araceae and Zosteraceae families, respectively. Since EDS1, PAD4, RNL genes and NDR1 are present in the transcriptome of at least one other member of the Araceae (Pistia stratiotes), we concluded that these genes were lost independently in Z. marina and S. polyrhiza.
In order to date the loss of EDS1, PAD4, RNL genes, and NDR1 in eudicots, we interrogated transcriptomes of species within the Lamiales order, including G. aurea and U. gibba. Only 4 of 48 available transcriptomes from the Lamiales did not have transcripts corresponding to EDS1, PAD4, SAG101, RNL genes, or NDR1 (Supplemental Figure 6; Supplemental Table 6). Three of these species were members of the Lentibulariaceae family (Utricularia spp, Pinguicula caudata, and P. agnata). In contrast, transcripts for EDS1, PAD4, RNL genes, and NDR1 were found in the transcriptomes of species in the Bignoniaceae, a closely related family within the Lamiales (Supplemental Figure 6; Supplemental Table 6; Schäferhoff et al., 2010; Xu et al., 2019).
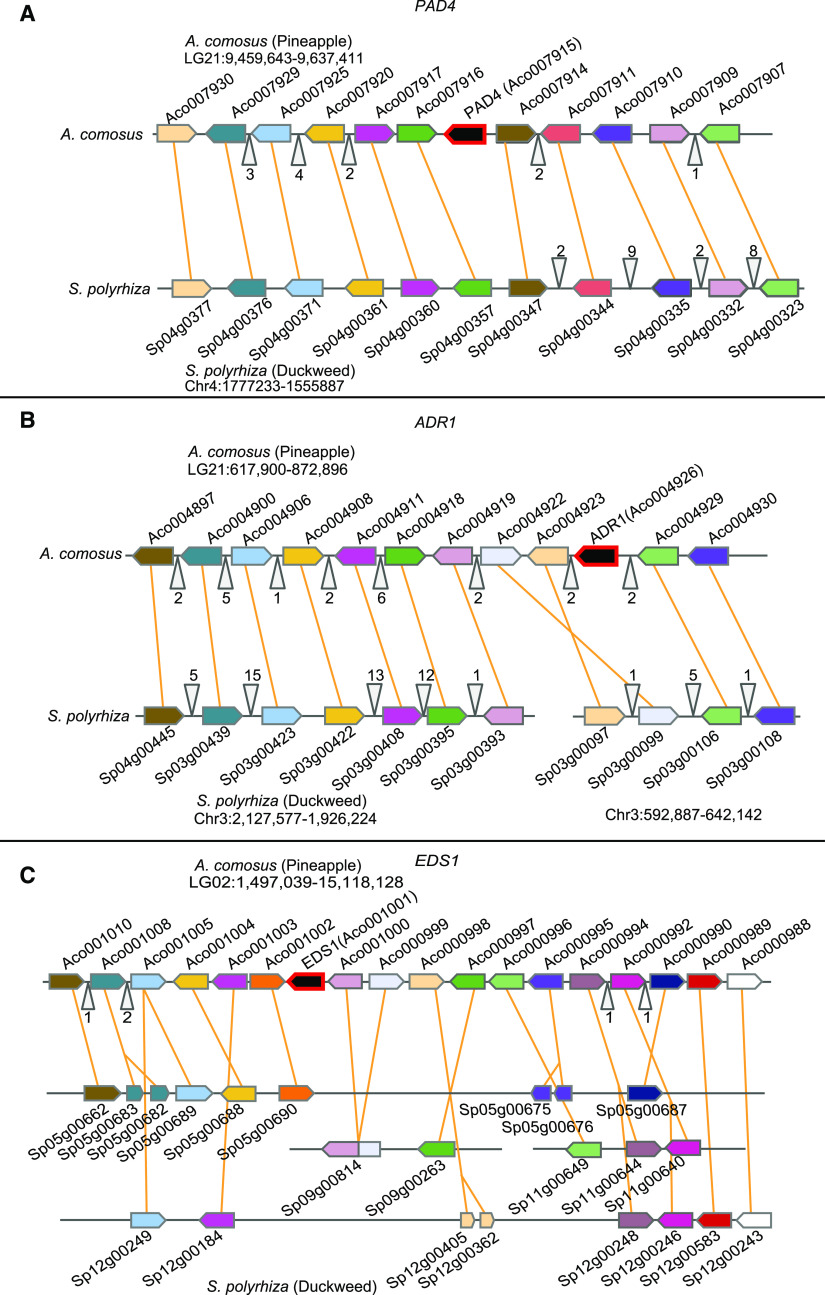
The local genomic context of lost genes is important to understand how and when the genes might have been lost, and we therefore constructed synteny plots of regions surrounding EDS1, PAD4, and ADR1. Due to the large divergence time of species and the low contiguity of some of the assemblies, we found little synteny conservation of regions between Arabidopsis, S. polyrhiza, Z. marina, G. aurea, or U. gibba (Supplemental Figure 7). Therefore, we focused on the recent S. polyrhiza chromosome level genome assembly. We used protein orthology between A. comosus and S. polyrhiza predicted with GeneSeqToFamily (Thanki et al., 2018) and homologous gene regions identified with SynMap2 (Haug-Baltzell et al., 2017). We were unable to find a contiguous genomic region with synteny across EDS1 (Figure 4). However, we could observe relative conservation of synteny for the homologous regions of PAD4 (Figure 4) and ADR1 (Figure 4), as the neighboring genes are found on the same chromosome and in close vicinity in A. comosus. The reconstruction of homologous regions demonstrated that the gene loss event in S. polyrhiza was due to deletions affecting single or a few genes as opposed to large structural variations.
Figure 4.
Syntenic Blocks of Genomic Loci of ASTREL Genes PAD4, ADR1, and EDS1 Between A. comosus and S. polyrhiza.
Genes and their direction are represented as arrows along the loci. Orthologs between A. comosus and S. polyrhiza are indicated by orange lines. Gray triangles with numbers indicate groups of additional genes not displayed here. The focal gene is highlighted with a red outline. Subplots visualize the syntenies of the loci of PAD4 (A), ADR1 (B), and EDS1 (C).
Orthogroup Analysis of Protein Families Provides a Global View of Genes Lost Alongside EDS1/PAD4
The convergent loss of EDS1/PAD4 and EDS1-dependent NLRs led us to hypothesize that other, as of yet unknown components of the EDS1-dependent signaling cascade could also have been convergently lost. To test this, we analyzed the 18 plant proteomes, which we surveyed earlier, with outgroups Selaginella moellendorffii and A. trichopoda (Figure 5). We performed two analyses, OrthoMCL (Li et al., 2003) and GeneSeqToFamily (Thanki et al., 2018), to cluster proteomes into orthogroups and examine gene loss patterns.
Figure 5.
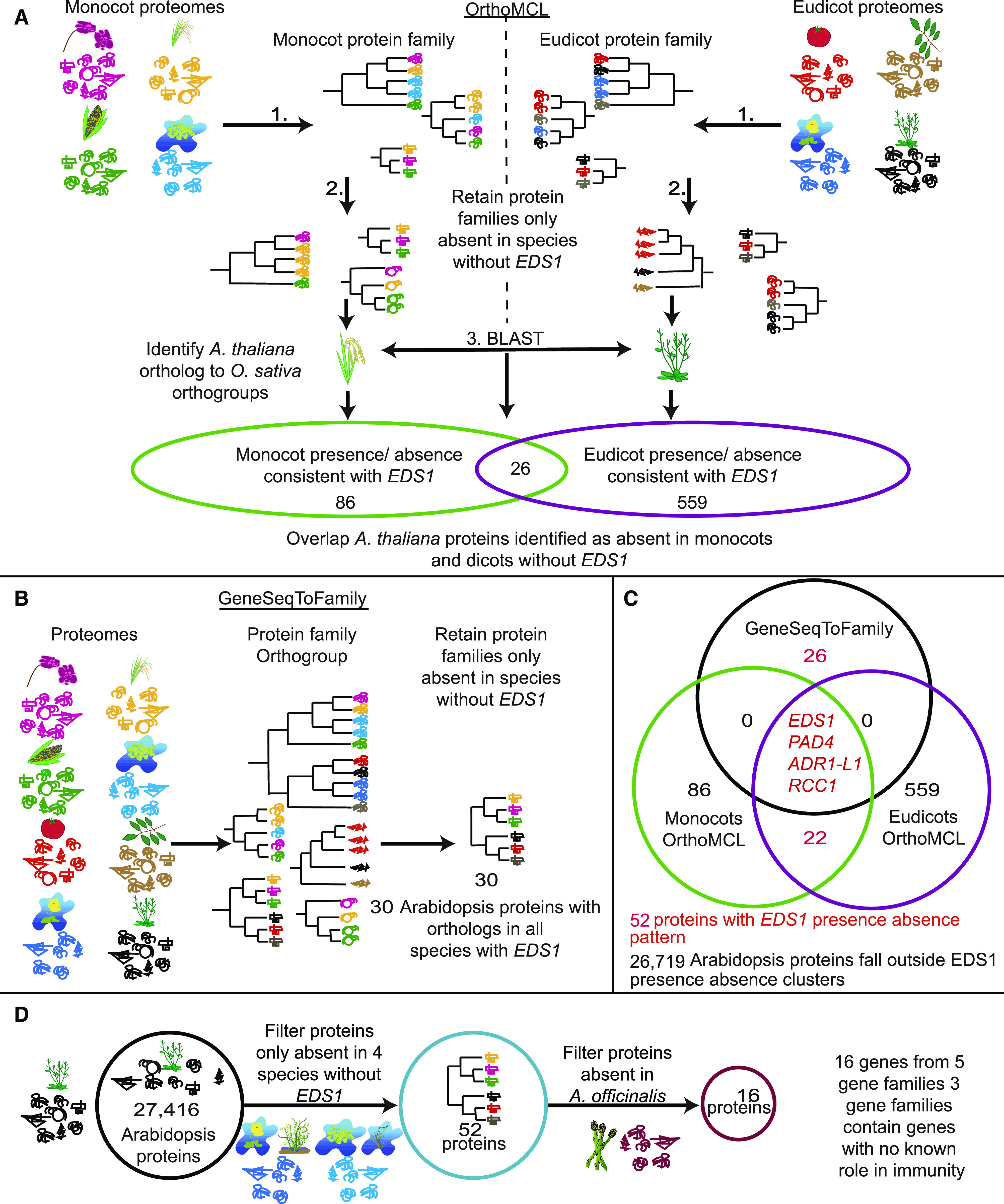
Protein Family Analysis to Identify ASTREL Genes.
(A) Schematic diagram of the OrthoMCL approach to cluster protein families separately among monocot and eudicot species and then filtering for protein families present in species with EDS1. Proteins are denoted by different line drawings, colored by species of origin. Phylogenetic trees represent gene trees for each protein. The diagram at the bottom provides the number of Arabidopsis proteins that are absent only in monocots, eudicots, or in all angiosperms without EDS1.
(B) Illustration of the GeneSeqToFamily method, which uses monocot and eudicot proteomes together to establish gene trees across the angiosperms.
(C) Diagram summarizing the results of the two methods. Genes and numbers marked in red are those subsequently referred to as ASTREL genes.
(D) Schematic of gene clustering followed by blastp and tblastn to filter ASTREL genes for presence or absence in the A. officinalis genome.
By combining the monocot and eudicot OrthoMCL analyses, we identified 17 Arabidopsis genes that lacked orthologous groups in the four species that lost EDS1, PAD4, and RNL genes (S. polyrhiza, Z. marina, G. aurea, and U. gibba) but had orthologs in the other 13 species. The GeneSeqToFamily analysis identified 31 Arabidopsis genes from 10 orthogroups that followed the same evolutionary pattern (Figure 5). Since OrthoMCL and GeneSeqToFamily use different algorithms, protein families were split into groups of different sizes (Li et al., 2003; Thanki et al., 2018). OrthoMCL usually subdivides genes into smaller families than GeneSeqToFamily. The use of programs with different cutoffs for gene family size enabled us to identify both the losses of single proteins within large complex families (using OrthoMCL) as well as losses of all members of a large protein family (using GeneSeqToFamily). Four genes were identified by both pipelines, including EDS1 and PAD4, an RNL gene from the ADR1 clade, and REGULATOR OF CHROMOSOME CONDENSATION 1- LIKE (RCC1-like). The former three are known to be involved in plant defense, whereas RCC1-like has not been implicated in defense responses. We further focused on the 52 genes identified by either OrthoMCL or GeneSeqToFamily methods (Figure 5; Supplemental Tables 7 and 8). We termed genes absent only in species lacking EDS1 as AngioSperm Typically-Retained, EDS1-Lost (ASTREL) genes.
To further consolidate our list, we looked for the presence or absence of the ASTREL genes in the A. officinalis genome because it is also missing EDS1, PAD4, RNL genes, and NDR1. This resulted in a list of 5 orthogroups that contained 16 Arabidopsis genes, 12 of which encoded proteins belonging to the EDS1, PAD4, or RNL families and 4 genes which have not been implicated in effector-triggered immunity (Table 1; Supplemental Tables 9 and 10).
Table 1. High Confidence ASTREL Orthogroups Absent in all 5 Species (S. polyrhiza, Z. marina, U. gibba, G. aurea, and A. officinalis).
| Arabidopsis Gene Names Within Orthogroup | Identification Method | Pfam Domains | Known Role in Immunity | Other Roles |
|---|---|---|---|---|
| Lipase-like: EDS1.1, EDS1.2, PAD4, SAG101 | Galaxy group 16353 | Lipase_3 | Downstream signaling of NLRs (Lapin et al., 2019; Wagner et al., 2013; Cui et al., 2017; Zhu et al., 2011) | Crosstalk in drought pathway (Chini et al., 2004) |
| Dicot_5571 | EDS1_EP | |||
| Monocot_8566 | ||||
| RNLs: NRG1, NRG1.2, NRG1.3, ADR1, ADR1-L1, ADR1-L2, ADR1-L3, RPW8 | Galaxy group 4516 | RPW8, NB-ARC, LRR | Downstream signaling of NLRs. (Bonardi et al., 2011; Castel et al., 2018; Qi et al., 2018; Lapin et al., 2019; Wu et al., 2019) | Initial stomatal closure for drought tolerance (Chini et al., 2004) |
| Dicot_3070 | ||||
| Monocot_8024 | ||||
| SDR4 | Dicot_3663B | adh_short_C2 | No | N/A |
| SDR5 | Monocot_4965 | |||
| ASPG2 | Dicot_8371 | TAXi_N, TAXi_NC | No | N/A |
| Monocot_7573 | ||||
| EDL3 | Dicot_10185 | F-box_like | No | Regulator of ABA signaling, osmotic stress-responsive (Koops et al., 2011). |
| Monocot_9077 |
N/A, not applicable.
To investigate whether loss of ASTREL genes might also occur at an intraspecific scale, we predicted loss-of-function alleles in candidate genes among 1135 natural accessions of Arabidopsis (Cutter and Jovelin 2015; 1001 Genomes Consortium, 2016). We found loss-of-function variants in 36 out of 52 ASTREL genes (Supplemental Table 11), a proportion which is significantly higher than in 52 random Arabidopsis genes (X-squared = 9.3, P = 0.002). This is consistent with the fact that these random genes include those which serve essential cellular functions, whereas ASTREL genes have been identified based on their tendency to experience condition dependent gene loss. The co-occurrence of loss-of-function across ASTREL genes suggests selection to maintain at least one copy of paralogous genes. For example, we observed a significant depletion of genotypes with loss-of-function alleles in both AT2G19920 and AT2G19910 (Supplemental Table 12; X-squared = 181.159188308688, P = 2x10-41). In general, the frequency of loss-of-function alleles was low (<1%), consistent with previous reports (Bakker et al., 2008). Although we did observe a moderate to high frequency (>10%) of loss-of-function variants in five ASTREL genes (Supplemental Table 11). Genetic redundancies in Arabidopsis may allow for loss-of-function to occur in genes which otherwise tend to be lost only together with EDS1.
Arabidopsis and O. sativa Homologs of ASTREL Genes Are Differentially Regulated upon Drought and Biotic Stress
To understand whether ASTREL genes are differentially expressed during biotic stress or other pathways, we looked for coexpression patterns. We used Arabidopsis mRNaseq and microarray perturbation experiments (53, 489) as well as O. sativa RNaseq and microarray data sets (25, resp. 142) to analyze expression of ASTREL orthologs (Supplemental Figures 8 to 11). We identified two distinct stresses, biotic stress and ABA/drought stress, that differentially regulate ASTREL genes expression (Figure 6).
Figure 6.
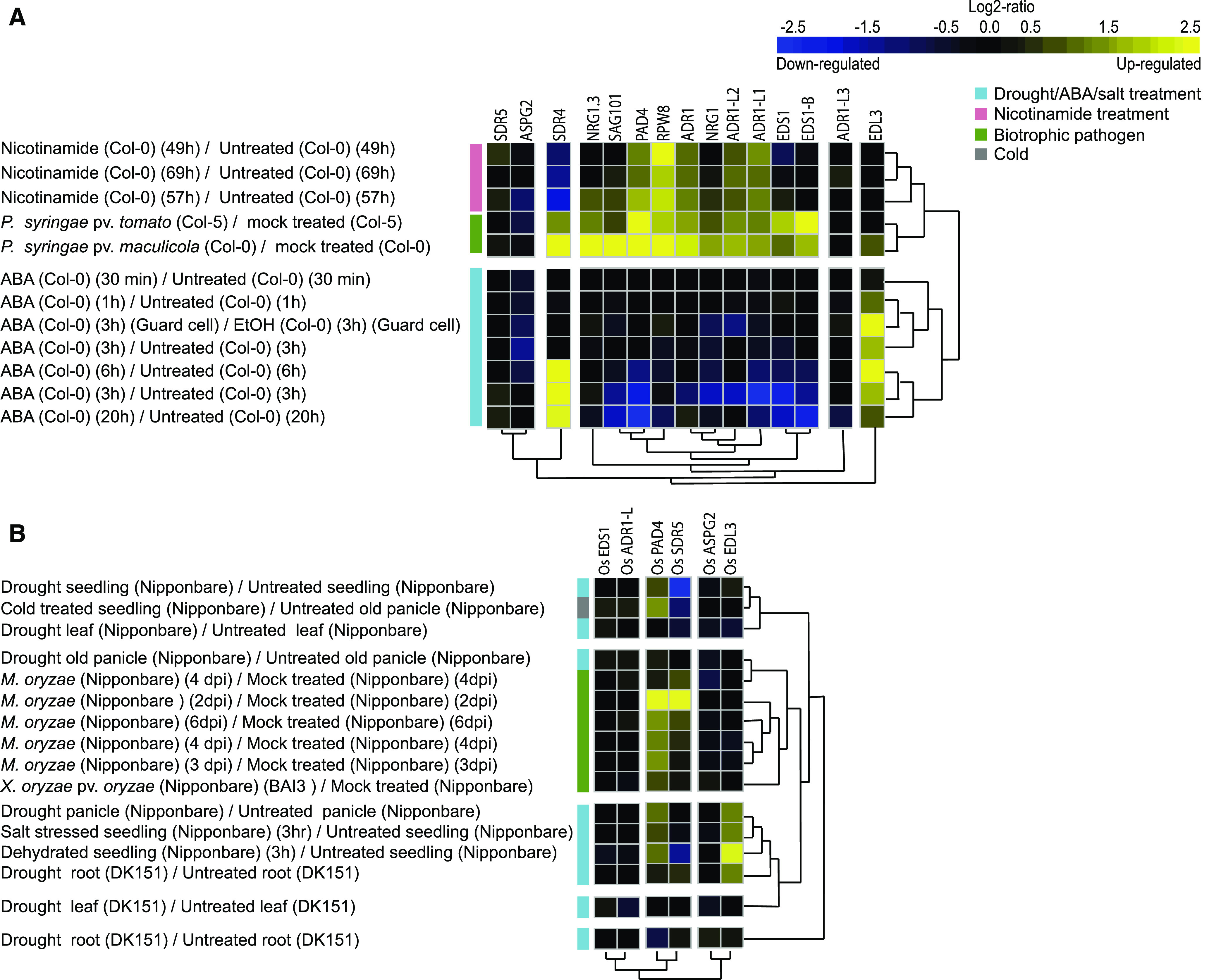
Differential Gene Expression Analysis of Arabidopsis and O. sativa High Confidence ASTREL Genes Upon Biotic and Abiotic Stress.
(A) Pearson hierarchical clustering of differential gene expression of ASTREL genes from Arabidopsis upon pathogen, ABA, and nicotinamide treatments.
(B) Pearson hierarchical clustering of differential gene expression of ASTREL genes from O. sativa upon pathogen, drought, cold, and salt treatment.
We first analyzed all 52 ASTREL genes in full (Supplemental Table 7; Supplemental Figure 8; Supplemental File 3) before we focused on the stringent set of ASTREL genes that were absent in A. officinalis (Supplemental Figure 9; Supplemental Data Sets 1 and 2; Supplemental File 4). As previously reported, expression of EDS1 and PAD4 was upregulated stronger in response to biotrophic pathogen infection than necrotrophic pathogen infection in Arabidopsis (Figure 6A; Falk et al., 1999; Jirage et al., 1999). Recent work has shown that TNLs have enzymatic activity and can produce nicotinamide (Horsefield et al., 2019; Wan et al., 2019). We therefore compared changes in ASTREL gene expression upon addition of nicotinamide clustered with those of biotrophic pathogen treatment. The immuninty related genes PAD4 and all RNLs, except for ADR1-L3 and NRG1.3 (P-value <0.05, Fold change > 1.5), were found to be upregulated in at least one-time point upon both nicotinamide and biotrophic pathogen infection. A subset of the ASTREL genes, EDS1 and SDR4, showed opposing expression patterns upon addition of biotrophic pathogens and nicotinamide in Arabidopsis. Nicotinamide appears to have a range of effects on gene expression of ASTREL genes; however, in general, expression changes are more similar to biotrophic pathogen infection than other stressors.
In addition to the stressors described above, we also investigated if any other perturbations regulated ASTREL gene expression: we used hierarchical clustering to identify treatments that caused similar patterns of expression (Supplemental Figure 9; Supplemental File 4). We noticed that drought and ABA treatment also resulted in the differential expression of ASTREL genes in Arabidopsis. Pearson clustering identified three clades of differentially expressed ASTREL genes upon drought and ABA response: downregulated (EDS1, PAD4, and RNL genes), upregulated (SDR4 and EDL3), and no effect on expression (SDR5, ASPG-2, and ADR1-L3).
To test whether the effects of these stresses were specific to Arabidopsis, we mined available gene expression data for ASTREL orthologs in O. sativa. Upon Pearson clustering we observed the same pattern of separation of gene expression response between pathogen and drought stress (Figure 6B; Supplemental Figures 10 and 11). However, the differential expression of genes upon pathogen treatment grouped into different clades in comparison with Arabidopsis: the genes OsSDR and OsPAD4 were upregulated highly upon Magnaporthe oryzae treatment with subtler expression changes upon drought. We also noted a similar differential expression pattern of OsSDR and OsPAD4 when subjected to either cold stress or drought. Conversely, the OsEDL3.1 clade was consistently upregulated during drought stress. In contrast to Arabidopsis, OsEDS1 and OsADR1-L were not upregulated upon pathogen treatment. Altogether, the expression data suggest that the coexpression of ASTREL genes can be influenced by biotic and abiotic stress.
DISCUSSION
In this study, we investigated the convergent loss of NLR genes and the ETI immune signaling pathway across multiple plant lineages. Using comparative genomics, we identified a set of genes that was co-lost with the immune signaling genes EDS1/PAD4. Expression analyses showed that these genes are differentially regulated in the defense and drought response pathways.
High variability in the number of NLR genes between species has been previously attributed to lineage-specific expansions and contractions (Kroj et al., 2016; Sarris et al., 2016; Gao et al., 2018; Van de Weyer et al., 2019). Most plants retain representatives of major NLR clades including members of the RNL and CNL clades, and TNLs among eudicots. We identified five species (S. polyrhiza, Z. marina, A. officinalis, G. aurea, and U. gibba) that lack RNL-type genes. Although RNLs are thought to be critical for signal transduction of all TNLs and many CNLs (Bonardi et al., 2011; Castel et al., 2018; Qi et al., 2018; Gantner et al., 2019; Lapin et al., 2019; Wu et al., 2019), our data suggest that despite their overall conservation across plant lineages, RNLs are not required for a minimal plant immune system.
TNL sensors and RNL helpers function together with EDS1/PAD4/SAG101 proteins to induce immune signaling (Castel et al., 2018; Qi et al., 2018; Lapin et al., 2019; Wu et al., 2019). Despite the absence of TNLs, most monocots retain EDS1 (Chen et al., 2018), suggesting that it might be required for either CNL signaling or basal immunity in monocots. In five species, we identified the convergent loss of EDS1/PAD4 and genes encoding other interacting proteins including SAG101 (Zhu et al., 2011; Wagner et al., 2013; Lapin et al., 2019), NRG1 (Castel et al., 2018; Qi et al., 2018; Lapin et al., 2019; Wu et al., 2019), and ADR1 (Bonardi et al., 2011; Wu et al., 2019). The recurrent loss of both EDS1/PAD4 and SAG101 in S. polyrhiza, Z. marina, U. gibba, A. officinalis, and G. aurea is consistent with genetic, biochemical, and structural studies of EDS1/PAD4/SAG101, which show the three proteins function in heterodimeric complexes (Wagner et al., 2013; Lapin et al., 2019).
We observed repeated absence of TNL genes SAG101 and NDR1 across many lineages that still retained EDS1/PAD4 and RNL genes. It has been shown that EDS1/PAD4 and EDS1/SAG101 can maintain different functions in divergent lineages. In Solanaceous species, the EDS1/SAG101 heterodimer is required for NRG1-dependent cell death, whereas in Arabidopsis it is the EDS1/PAD4 heterodimer that is necessary for cell death (Gantner et al., 2019; Lapin et al., 2019). Recently, it has also been shown that PAD4 has domain-specific functions in herbivore resistance independent of EDS1 (Dongus et al., 2019). Based on the functional and evolutionary data (loss of EDS1/PAD4 occurs only after SAG101 and NDR1), we conclude that EDS1/PAD4 represents a functional unit on their own, and their role can be sub-functionalized in the presence of additional components. The exact constitution of the EDS1/SAG101 pathway and how it is differentiated from the EDS1/PAD4 pathway remains unclear (Gantner et al., 2019; Lapin et al., 2019).
This study demonstrates that comparative phylogenomic analysis can fill the gaps in our understanding of complex plant signaling pathways. Work on plant symbiosis has shown that the ability to form types of symbiosis can be correlated with the presence and absence of genes that are part of the molecular pathway of symbiosis formation (Bravo et al., 2016; Griesmann et al., 2018; van Velzen et al., 2018). Using a similar approach, we identified other genes that have been lost in species lacking EDS1, PAD4, and RNL genes, and we named the identified candidates ASTREL genes.
Coevolved genes often show patterns of conserved coexpression (Hansen et al., 2014). Interestingly, differential expression of ASTREL genes was observed only under a few stress conditions, including pathogen response, nicotinamide, drought, salinity, and ABA response. Differential expression of ASTREL genes upon addition of nicotinamide was strikingly similar to the response to Pseudomonas syringae infection, consistent with recent literature implicating nicotinamide in plant immune signaling (Horsefield et al., 2019; Wan et al., 2019). Upon pathogen stimulus, some ASTREL genes were not differentially expressed; however, we cannot rule out the possibility that those genes may nonetheless be involved in disease resistance. We have only looked at differential expression in a small subset of the possible combinations of conditions, tissues, and pathogens that can result in an immune response. However, the clear patterns of differential expression we identified offer a starting point to characterize the pathways involving ASTREL genes.
Surprisingly, another perturbation that induced changes in ASTREL gene expression was drought stress and ABA response. Drought tolerance can be induced either in an ABA-dependent or independent manner. Previous work has demonstrated antagonism between the ABA and SA hormone response with stress prioritization seemingly playing out at the level of gene expression mediated by proteins such as NPR1 (Ryals et al., 1997; Cao et al., 1998; Ding et al., 2016) and PBS3 (Berens et al., 2019). Consistent with the antagonism data from Arabidopsis (de Torres Zabala et al., 2009; Moeder et al., 2010; Lievens et al., 2017), a cluster of the ASTREL genes containing EDS1 and RNL genes is downregulated upon ABA and upregulated upon pathogen stimulus triggering SA production. The same set of genes has previously been shown to promote drought tolerance in an ABA-independent manner (Chini et al., 2004). Increased expression of the RNL gene, ADR1, or the decreased expression of the negative regulator of programmed cell death LSD1 enhanced drought tolerance in an EDS1/PAD4- and SA-dependent manner (Chini et al., 2004; Wituszynska et al., 2013; Szechyńska-Hebda et al., 2016). Our analyses support the role of EDS1/PAD4 in drought stress response and ABA-antagonism.
Our ASTREL candidates might provide further links between EDS1/PAD4 and the drought stress response. The ASTREL gene EDL3 encodes for an F-box transcription factor, which is a positive regulator of an ABA-dependent signaling cascade (Koops et al., 2011). The loss of EDL3 together with immune signaling components suggests that the transcriptional changes might be regulated through the EDS1/PAD4 pathway. Although EDS1 and PAD4 have been reported to be required for P. syringae interference with ABA signaling (Kim et al., 2011), the mechanism of this inference is largely unknown. Based on our analyses, EDL3 coevolved with EDS1/PAD4 and therefore is a likely candidate for mediating EDS1/PAD4-dependent ABA crosstalk.
We assembled a working model of how ASTREL genes might fit in the known plant immunity and drought pathways using data on known interactions, functional characterizations (Bonardi et al., 2011; Castel et al., 2018; Cui et al., 2018; Qi et al., 2018; Horsefield et al., 2019; Lapin et al., 2019; Wan et al., 2019; Wu et al., 2019) and coexpression analyses (Figure 7; Supplemental Tables 13 and 14). Due to the homology of ASPG2 to aspartic proteases, we hypothesize it would be part of the initial cascade which we know to be initiated by TNLs through their NADase activity (Horsefield et al., 2019; Wan et al., 2019). This is consistent with its steady gene expression pattern throughout development that is unchanged upon infection, suggesting the protein would already be present at the time of infection. Consistent with the requirement of NRG1 for TNL signaling, we propose that all members of the RNL clade play a role in EDS1-dependent NLR signaling. This is supported by our finding that the RNL clade is absent in the independent groups of species missing EDS1.
Figure 7.
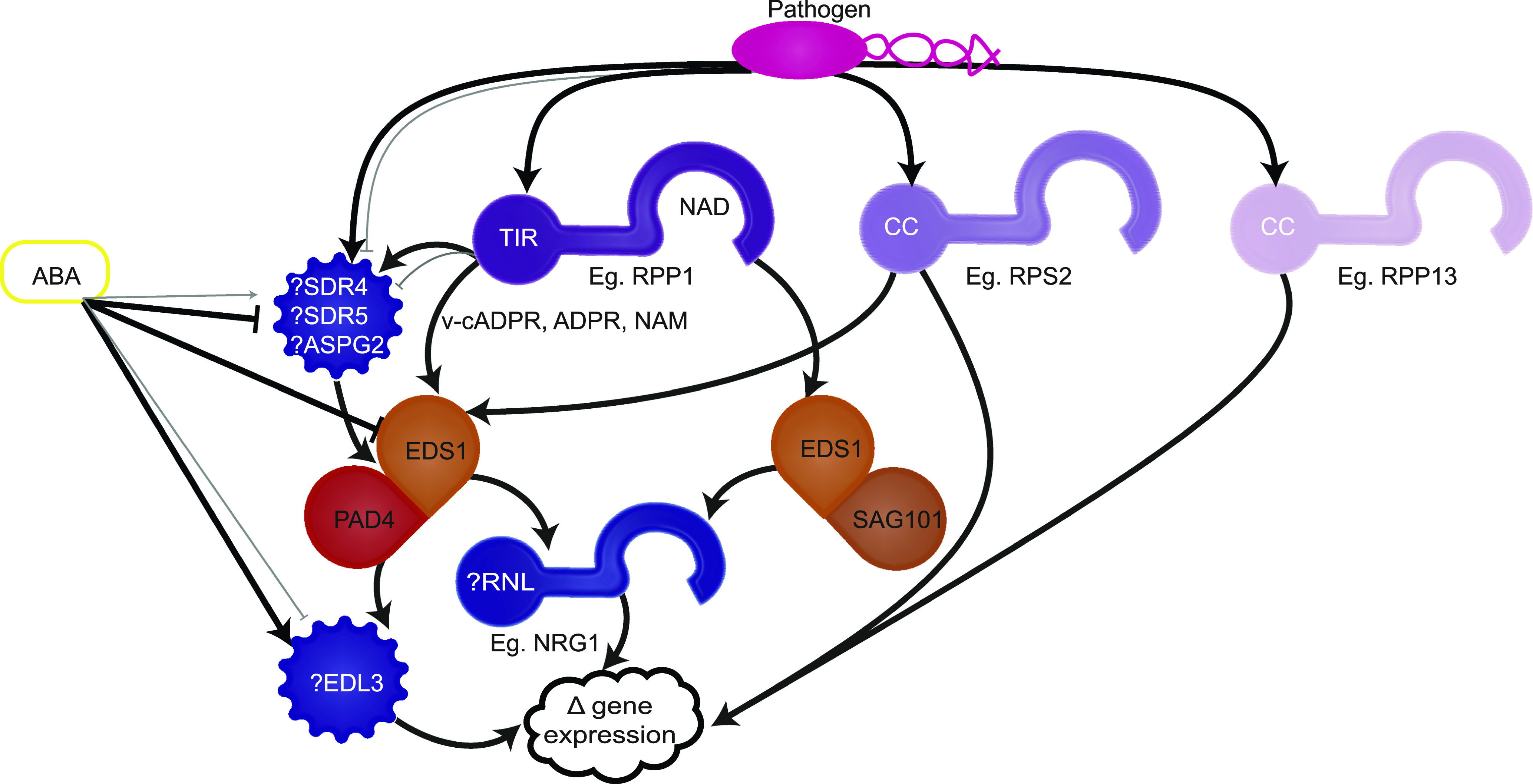
Schematic Model of Hypothetical Relationships Between ASTREL Genes and Known Biotic and Abiotic Stress Pathways in Arabidopsis.
The model is based on literature review and available gene expression of potential interactions of ASTREL genes within the known Arabidopsis biotrophic pathogen disease resistance genetic pathway.
Interestingly, four of the five species we identified to be missing ASTREL genes were from water-saturated environments ranging from marine to bog habitats. Previous studies have investigated the genome content of one or two aquatic plant species and highlighted gene loss linked to embryogenesis and root development in U. gibba (Leushkin et al., 2013), cell wall processes, and ABA in S. polyrhiza (Wang et al., 2014; Michael et al., 2017), and defense response, stomata, terpenoid, and hormone pathways in Z. marina (Olsen et al., 2016). This invites the question of whether the adaptation to a water-saturated environment may place selective pressure on certain signaling pathways including the immune system. There are, however, several species such as sacred lotus (Nelumbo nucifera) and O. sativa that, according to our analysis, have retained EDS1/PAD4 and other signaling pathway members despite inhabiting water-saturated environments. Furthermore, controversy surrounds the question of whether the most recent common ancestor of monocots could have had an aquatic lifestyle (Du et al., 2016; Givnish et al., 2018). It is nevertheless tempting to speculate that loss of the RNL and EDS1 pathway can be a byproduct of the physiological changes and differing environmental selection pressures associated with each lifestyle. The transcriptome data suggest that loss of EDS1/PAD4 might be more frequent in monocots than eudicots and availability of more genomes could help to determine which environmental and/or physiological traits influence loss of this pathway.
Until now, discoveries of components of the plant immune system have primarily relied upon mutant screens, proteomic analyses, and differential gene expression analysis. Here, we have shown a complementary approach to identify potential players in the plant immune system that can circumvent issues of genetic redundancy by harnessing conservation and independent transitions in distantly related plant lineages. To achieve durable disease resistance, NLR genes are often stacked to slow the rate of breakdown of resistance by fast evolving pathogens. The downstream signaling components required for stacked NLR genes are rarely considered but could result in strong selection for effectors that disrupt downstream signaling if the stacked NLRs depend on a single helper or signaler. It is also crucial to understand the conservation of downstream signaling components to facilitate the successful interspecies transfer of NLR genes. Our approach has highlighted the conservation of EDS1/PAD4 as a functional unit and identified other genes that coevolved with EDS1-mediated immunity.
Four of the species in our analyses have retained and expanded the few remaining CNL clades, suggesting that they are using a yet undiscovered signaling pathway. This is consistent with previous reports of some CNLs, such as RECOGNITION OF PERONOSPORA PARASITICA 13 (RPP13), that are independent of all known signaling components (Bittner-Eddy and Beynon, 2001). The SA pathway is conserved in species that have lost EDS1, PAD4, SAG101, NDR1, and RNL genes, suggesting that it might fulfill another yet unknown function in defense responses. Whether the remaining CNLs can induce SA responses and cell death remains to be tested, although another duckweed Lemna minor, a close relative of S. polyrhiza, has been shown to induce a reactive oxygen burst and cell death upon abiotic stress (Wang et al., 2016).
We propose S. polyrhiza as a new model system to study plant immunity. It is a rapidly growing, small plant whose reduced ETI immune system could provide a reduced-complexity background for investigating plant immune pathways. Our analysis provides a steppingstone to understanding a minimum plant immune system independent of EDS1/PAD4 signaling. Additionally, the discovery of the ASTREL genes has added further evidence of the complex coevolutionary crosstalk between the plant immune system and drought tolerance. Future studies could test their functional roles in immune signaling and further query the interconnection between abiotic and biotic stress pathways.
METHODS
Genomic data sets used in this study
Genome assemblies and annotations were obtained from Phytozome V12 (https://phytozome.jgi.doe.gov/pz/portal.html) for Aquilegia coerulea, Ananas comosus (v3), Amaranthus hypochondriacus (325_v1.0), Arabidopsis (Arabidopsis thaliana; 167_TAIR9), Amborella trichopoda (291_v1.0), Erythranthe guttata (256_v2.0), Oryza sativa (v7), Oropetium thomaeum, Solanum lycopersicum (390_v2.5), Selaginella moellendorffii (91_v1), Spirodela polyrhiza (v2), and Zoestera marina (v2.2); from COGE (https://genomevolution.org/coge/) for Utricularia gibba (29,027); from KEGG for Nelumbo nucifera (4432) and Elaeis guineensis (TO3921); from NCBI for Phalaenopsis equestrius (PRJNA382149); from Ash Tree Genomes (http://www.ashgenome.org/data) for ash (Fraxinus excelsior; BATG-0.5); from Ensembl for yam (Dioscorea rotundata; TDr96_F1_Pseudo_Chromosome_v1.0); and from the Maize genome database (https://www.maizegdb.org/) for Zea mays (AGPv4). The BUSCO scores were calculated using BUSCO (v1.22) to compare proteomes to the embryophyta_odb9 BUSCO lineage (Supplemental Table 15; Simão et al., 2015).
Annotation, Alignment, and Phylogenetic Analysis of NLRs
To annotate NLRs in plant proteomes, we used the updated version of the NLR-ID pipeline (ASTREL_v1_2019_10.1101.572560; https://github.com/krasileva-group/plant_rgenes/; Sarris et al., 2016) together with the NLR-Parser and NLR-Annotator tools (Steuernagel et al., 2020) which are based on MEME suite (Bailey et al., 2006). Annotations were combined into a nonredundant list of putative NLRs. Multiple transcripts from the same locus are represented by the longest transcript. In addition, a series of characterized NLRs were added. Proteins were aligned to the NB-ARC1_ARC2 HMM (Bailey et al., 2018) of the NB-ARC domain with hmmalign (HMMER3.0; Wheeler and Eddy, 2013). The alignment was trimmed to the NB-ARC domain region using Belvu (Barson and Griffiths, 2016), and columns and sequences with over 80% gaps were removed. The NB-ARC domain of the remaining NLRs was then manually curated in Jalview (Waterhouse et al., 2009), allowing no more than two consecutive characteristic NB-ARC domain motifs (Walker A, RNBS-A, WALKER-B, RNBS-C, GLPL, RNBS-D) to be absent (i.e., across the length of the motif). A maximum-likelihood phylogenetic tree was constructed using RAXML-MPI (v.8.2.9; Stamatakis, 2014) with parameters set as -f a -x 1123 -p 2341 -# 100 -m PROTCATJTT. Tree visualization and annotation using iTOL are available at http://itol.embl.de/shared/erin_baggs (Stolzer et al., 2012), with the tree rooted at the split between TNL and CNL type NLRs as they have been characterized as having distinct evolutionary histories (Tamborski and Krasileva, 2020).
Ortholog Identification
To identify orthologs of specific genes of interest, we used reciprocal BLAST searches using blastp with parameters -max_target_seqs 1 -evalue 1e-10 (BLAST+ 2.2.28.mt; Camacho et al., 2009). If upon reciprocal BLAST, a homologous gene was not identified, we used Ensembl Plants gene trees to check for recent duplication events in the Arabidopsis lineage and to confirm our results. The presence of EDS1 in O. thomaeum was validated using RNaseq data (BioProject SRS957807) mapped onto the Oropetium V1 Bio_nano genome assembly (http://www.oropetium.org/resources; VanBuren et al., 2018, 2015) using HISAT2 (Kim et al., 2015). Bam files were processed using SAMtools-1.7 (Li et al., 2009) and results visualized using IGB (Nicol et al., 2009). The absence of EDS1 was validated by running hmmsearch to search the proteomes of Arabidopsis, S. polyrhiza, and Z. marina for the presence of the Lipase 3 domain, which is characteristic of EDS1. Proteins containing the domain were then aligned against the Pfam Lipase 3 HMM with hmmalign. The alignment was manually curated in Belvu (Barson and Griffiths, 2016) before processing with RAxML as above and rooting on the longest internal branch length.
To identify orthologous gene groups, OrthoMCL (v2.0.9; Li et al., 2003) was used as previously described by Johnson et al. (2018). Due to large computational requirements, we ran OrthoMCL separately for monocots (Z. marina, S. polyrhiza, P. equestris, D. rotundata, E. guineesis, O. thomaeum, O. sativa, and Z. mays) and eudicots (A. coerulea, N. nucifera, Arabidopsis, A. hypochondriacus, S. lycopersicum, F. excelsior, E. guttata, G. aurea, and U. gibba), both with the outgroups S. moellendorffii and A. trichopoda. We overlaid monocot analyses with eudicot orthogroups by mapping the monocot gene IDs to the Arabidopsis homologs using reciprocal blastp (e-value cutoff 1e-10). Additionally, we processed all 19 genomes with the GeneSeqToFamily pipeline (Thanki et al., 2018). We then identified orthogroups lost in aquatic species (S. polyrhiza, Z. marina, U. gibba, and G. aurea) but retained in all terrestrial species of the same monocot/eudicot clade. We cannot preclude the possibility that some gene families that have been convergently lost in aquatic species have not also been lost independently in some of the terrestrial lineages. After grouping and manual curation, gene families for which pan-species evolutionary history had been previously established were compared to gene families in our orthogroups. Since homologs in S. moellendorffii and A. trichopoda are rare due to large phylogenetic distance, we excluded these species from further analyses. We additionally excluded A. coerulea and D. rotundata as their annotations contain large amounts of erroneous gene fusions. For comparison of OrthoMCL gene families retained between monocots and eudicots, we used Arabidopsis and O. sativa proteins as representative genome members and cross-referenced them using blastp reciprocal search (e-value cutoff 1e-10). The validity of this approach was checked on random protein families using Plant Ensembl trees (http://plants.ensembl.org). The O. sativa gene IDs were converted between Phytozome and Plant Ensembl using the Plant Ensembl conversion tool (Sakai et al., 2013).
Pairwise Synteny Analysis
We used the GeneSeqToFamily pipeline (Thanki et al., 2018) to identify orthologs between A. comosus and S. polyrhiza. This allowed us to map the locations of the orthologous genes immediately up- and downstream of the ASTREL genes in A. comosus to the S. polyrhiza genome. Based on these locations, we could identify the syntenic region in S. polyrhiza that corresponds to the region containing EDS1, PAD4, and ADR1 in A. comosus. We processed the genomic sequences up- and downstream of the ASTREL genes of interest in A. comosus with SynMap2 (Haug-Baltzell et al., 2017). This confirmed the presence of microsynteny at the DNA level on scaffolds of S. polyrhiza Oxford v3 genome equivalent to those identified in the chromosomal assembly of S. polyrhiza.
Loss-of-Function Variants in Arabidopsis
To investigate whether loss of ASTREL genes might also be occurring at an intraspecific scale we predicted loss-of-function alleles in candidate genes in Arabidopsis. To do so, we extracted variants in coding regions in target genes and 52 random genes from 1135 natural accessions (1001 Genomes Consortium, 2016), converting them into coding region fasta format using g2gtools (http://churchill-lab.github.io/g2gtools). For each accession and gene, the coding region was then translated into its predicted amino acid sequence. Predicted loss-of-function alleles were defined as amino acid sequence in at least one gene model with >10% lost due to premature truncation. This threshold has been applied previously in humans (MacArthur et al. 2012) and Arabidopsis (Monroe et al. 2018) and is based on observations of enrichment of protein truncating variants affecting less than 10% of coding regions (Flowers et al. 2015). This approach allowed us to identify predicted loss-of-function alleles would otherwise be missed and preclude false positives that would otherwise be called if relying on variant annotations alone. For example, we found cases of predicted loss-of-function caused by neighboring single nucleotide polymorphisms, which individually are annotated as synonymous variants but together produced a premature stop codon. Reciprocally, we observe instances where the effect of a frameshift mutation or stop codon inducing single nucleotide polymorphism is ameliorated by neighboring variants that restore the reading frame or produce an amino acid change rather than stop codon. To test for correlated gene loss-of-function, we performed Fisher exact tests of independence between genes with at least 1 observed loss-of-function variant. To evaluate significance of these results, we applied a Bonferonni correction to P-values to account for multiple tests.
Expression Profiling
The expression analysis for this study was performed and visualized using the 706 O. sativa mRNA samples and 2836 Affymetrix O. sativa genome array samples available on Genevestigator v7.0.3 (https://genevestigator.com; Hruz et al., 2008). Arabidopsis RNaseq and Affymetrix ATH1 array (53, resp. 489) and O. sativa RNaseq and microarray data sets (25, resp. 142) available on Genevestigator v7.0.3 were used to visualize conditions resulting in differential patterns of gene expression. Microarray data sets used to investigate pathogen, drought, nicotinamide, and salt stress are given in Table 2. A list of NCBI GEO data set identifiers for the data sets used are available in the Supplemental Data section at (https://github.com/krasileva-group/ASTREL_NLR). Hierarchical clustering using Pearson’s correlation coefficient and optimal leaf ordering was applied to identify groups of genes with common expression patterns, including both the gene expression level and condition perturbation.
Table 2. Data Sets Used to Investigate Further Changes in Gene Expression under Drought or Pathogen Infection across the 52 (Arabidopsis) ASTREL, Monocot, and Eudicot Overlapping Genes.
| Plant | Type of Stimulus | Experiment Number | Reference |
|---|---|---|---|
| Arabidopsis | Pathogen | AT-00202 | (Craigon et al., 2004) |
| Arabidopsis | Pathogen | AT-00406 | (Craigon et al., 2004) |
| Rice | Pathogen | OS-00057 | (Yu et al., 2011) |
| Rice | Pathogen | OS-00045 | (Marcel et al., 2010) |
| Rice | Pathogen | OS-00011 | (Haiyan et al., 2012) |
| Arabidopsis | Nicotinamide | AT-00398 | (Dalchau et al., 2010) |
| Arabidopsis | Drought | AT-00110 | (Kilian et al., 2007) |
| Arabidopsis | Drought | AT-00433 | (Pandey et al., 2010) |
| Arabidopsis | Drought | AT-00468 | (Böhmer and Schroeder, 2011) |
| Arabidopsis | Drought | AT-00541 | (Kim et al., 2011) |
| Rice | Drought | OS-00008 | (Jain et al., 2007) |
| Rice | Drought | OS-00041 | (Wang et al., 2011) |
| Rice | Drought | OS-00224 | (Krishnan et al., 2010) |
Accession Numbers
All genomes analyzed in this study are listed in Supplemental Table 1 and were available from public repositories. All gene expression datasets used in the study are listed in Table 2. All Arabidopsis gene idenitfiers for plant immune genes are in Supplemental Table 3, all gene accession numbers that formed orthogroups in Supplemental Table 7, and gene accessions that passed ASTREL high confidence filtering in Supplemental Table 10. All scripts used to analyze data are available from https://github.com/krasileva-group/ASTREL_NLR. A high-resolution phylogenetic tree of NLRs across 18 species in the study is available on iToL: http://itol.embl.de/shared/erin_baggs .
Supplemental Data
Supplemental Figure 1. Graph of the number of genes with an NB-ARC domain across 95 plant species genomes.
Supplemental Figure 2. Maximum likelihood phylogeny based on the NB-ARC domain of the RNL clade.
Supplemental Figure 3. Table of presence and absence of immunity associated components when queried by reciprocal BLASTp.
Supplemental Figure 4. Maximum likelihood phylogeny of proteins with a lipase 3 HMM Motif.
Supplemental Figure 5. Identification of immune gene orthologs in magnoliids and early diverging monocot species with available transcriptomes.
Supplemental Figure 6. Identification of immune gene orthologs in Lamiales species with available transcriptomes.
Supplemental Figure 7. Conservation of gene order surrounding EDS1, PAD4 and ADR1-L1 proteins among Arabidopsis, E. guttata, U. gibba, S. polyrhiza and Z. marina.
Supplemental Figure 8. Heatmap of changes in gene expression of Arabidopsis ASTREL genes upon available RNaseq conditions.
Supplemental Figure 9. Heatmap of changes in gene expression of Arabidopsis high confidence ASTREL genes upon available RNaseq conditions.
Supplemental Figure 10. Heatmap of changes in gene expression of O. sativa high confidence ASTREL genes upon available RNaseq conditions.
Supplemental Figure 11. Heatmap of changes in gene expression of O. sativa high confidence ASTREL genes upon available microarray conditions.
Supplemental Table 1. Number of NLRs identified from proteomes of available plant genomes.
Supplemental Table 2. Number of NLR, RLK and Actin proteins identified in the proteome and number of genome predicted NLRs.
Supplemental Table 3. Arabidopsis gene identifiers and gene names.
Supplemental Table 4. Presence and absence of the immune pathway genes in available plant genomes.
Supplemental Table 5. Presence and absence of the immune pathway genes in available monocot transcriptomes.
Supplemental Table 6. Presence and absence of the immune pathway genes in available dicot transcriptomes.
Supplemental Table 7. Orthogroups and corresponding Arabidopsis gene identifiers of the ASTREL genes.
Supplemental Table 8. Orthogroups and corresponding O. sativa gene identifiers of the ASTREL genes.
Supplemental Table 9. Presence absence of the ASTREL orthogroups in A. officinalis.
Supplemental Table 10. High confidence ASTREL orthogroups which are absent in all 5 species without EDS1/PAD4.
Supplemental Table 11. ASTREL genes with loss-of-function (protein truncating) alleles identified in Arabidopsis populations.
Supplemental Table 12. Fisher exact tests for correlated gene loss between ASTREL genes. Only pairs where P < 0.05 after correcting for multiple testing are shown.
Supplemental Table 13. Literature implicating ASTREL genes in ABA and drought response.
Supplemental Table 14. Literature supporting working model of ASTREL genes relationship with ABA and immunity.
Supplemental Table 15. BUSCO scores for proteomes used for OrthoMCL and GeneSeqToFamily analysis.
Supplemental Data Set 1. Table of ASTREL gene expression RNaseq perturbations.
Supplemental Data Set 2. Heatmap of changes in gene expression of Arabidopsis ASTREL genes upon available microarray conditions.
Supplemental File 1. Multiple sequence alignment of NB-ARC domain containing proteins without curation.
Supplemental File 2. Multiple sequence alignment of NB-ARC domain containing proteins with curation.
Supplemental File 3. Table of ASTREL gene expression microarray perturbations.
Supplemental File 4. Heatmap of changes in gene expression of Arabidopsis high confidence ASTREL genes upon available microarray conditions.
DIVE Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
Acknowledgments
We thank all members of the Krasileva group and their many colleagues for thoughtful discussion on the presented material. We thank Daniil Prigozhin and Janina Tamborski for their suggestions on the article. The high-performance computing resources and services used in this work were supported by the Earlham Institute Scientific Computing group alongside the Norwich BioScience Institutes Partnership Computing infrastructure for Science (CiS) group. This project was supported by the Biotechnology and Biological Sciences Research Council (BBSRC; grants BB/CSP17270/1, BBS/E/T/000PR9818, and BBS/E/T/000PR9783 to W.H. and K.V.K. and BBSRC Doctoral Training Program BB/M011216/1 to E.L.B.); the EC | European Research Council (grant ERC-2016-STG-716233-MIREDI to K.V.K.); the National Science Foundation (grant 170191) and the Max Planck Society (to J.G.M.).
Author Contributions
E.L.B. performed NLR, phylogenetic analysis, genomic/transcriptomic presence absence analysis, RNAseq analysis of ASTREL genes, and model of gene interaction; E.L.B. and R.O. prepared figures; E.L.B. assisted by C.S. analyzed orthogroups from OrthoMCL and GeneSeqToFamily to identify ASTREL candidates; E.L.B. and W.H. performed synteny analysis; E.L.B., W.H., and K.V.K. designed the study; J.G.M. conducted intraspecies loss of function analysis in Arabidopsis; A.S.T. ran analysis with the GeneSeqToFamily pipeline; all authors contributed to the final article, and read and approved the final article.
Footnotes
Articles can be viewed without a subscription.
References
- Baggs E., Dagdas G., Krasileva K.V.(2017). NLR diversity, helpers and integrated domains: Making sense of the NLR IDentity. Curr. Opin. Plant Biol. 38: 59–67. [DOI] [PubMed] [Google Scholar]
- Bailey P.C., Schudoma C., Jackson W., Baggs E., Dagdas G., Haerty W., Moscou M., Krasileva K.V.(2018). Dominant integration locus drives continuous diversification of plant immune receptors with exogenous domain fusions. Genome Biol. 19: 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey T.L., Williams N., Misleh C., Li W.W.(2006). MEME: Discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Res. 34: W369–-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakker E.G., Traw M.B., Toomajian C., Kreitman M., Bergelson J.(2008). Low levels of polymorphism in genes that control the activation of defense response in Arabidopsis thaliana. Genetics 178: 2031–2043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barson G., Griffiths E.(2016). SeqTools: Visual tools for manual analysis of sequence alignments. BMC Res. Notes 9: 39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berens M.L., et al. (2019). Balancing trade-offs between biotic and abiotic stress responses through leaf age-dependent variation in stress hormone cross-talk. Proc. Natl. Acad. Sci. USA 116: 2364–2373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernoux M., Ve T., Williams S., Warren C., Hatters D., Valkov E., Zhang X., Ellis J.G., Kobe B., Dodds P.N.(2011). Structural and functional analysis of a plant resistance protein TIR domain reveals interfaces for self-association, signaling, and autoregulation. Cell Host Microbe 9: 200–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhandari D.D., Lapin D., Kracher B., von Born P., Bautor J., Niefind K., Parker J.E.(2019). An EDS1 heterodimer signalling surface enforces timely reprogramming of immunity genes in Arabidopsis. Nat. Commun. 10: 772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bittner-Eddy P.D., Beynon J.L.(2001). The Arabidopsis downy mildew resistance gene, RPP13-Nd, functions independently of NDR1 and EDS1 and does not require the accumulation of salicylic acid. Mol. Plant Microbe Interact. 14: 416–421. [DOI] [PubMed] [Google Scholar]
- Böhmer M., Schroeder J.I.(2011). Quantitative transcriptomic analysis of abscisic acid-induced and reactive oxygen species-dependent expression changes and proteomic profiling in Arabidopsis suspension cells. Plant J. 67: 105–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonardi V., Tang S., Stallmann A., Roberts M., Cherkis K., Dangl J.L.(2011). Expanded functions for a family of plant intracellular immune receptors beyond specific recognition of pathogen effectors. Proc. Natl. Acad. Sci. USA 108: 16463–16468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bravo A., York T., Pumplin N., Mueller L.A., Harrison M.J.(2016). Genes conserved for arbuscular mycorrhizal symbiosis identified through phylogenomics. Nat. Plants 2: 15208. [DOI] [PubMed] [Google Scholar]
- Camacho C., Coulouris G., Avagyan V., Ma N., Papadopoulos J., Bealer K., Madden T.L.(2009). BLAST+: Architecture and applications. BMC Bioinformatics 10: 421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao H., Li X., Dong X.(1998). Generation of broad-spectrum disease resistance by overexpression of an essential regulatory gene in systemic acquired resistance. Proc. Natl. Acad. Sci. USA 95: 6531–6536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castel B., Ngou P.-M., Cevik V., Redkar A., Kim D.-S., Yang Y., Ding P., Jones J.D.G.(2018). Diverse NLR immune receptors activate defence via the RPW8-NLR NRG1. New Phytol 222: 966–980. [DOI] [PubMed] [Google Scholar]
- Catanzariti A.-M., Dodds P.N., Ve T., Kobe B., Ellis J.G., Staskawicz B.J.(2010). The AvrM effector from flax rust has a structured C-terminal domain and interacts directly with the M resistance protein. Mol. Plant Microbe Interact. 23: 49–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Century K.S., Holub E.B., Staskawicz B.J.(1995). NDR1, a locus of Arabidopsis thaliana that is required for disease resistance to both a bacterial and a fungal pathogen. Proc. Natl. Acad. Sci. USA 92: 6597–6601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G., Wei B., Li G., Gong C., Fan R., Zhang X.(2018). TaEDS1 genes positively regulate resistance to powdery mildew in wheat. Plant Mol. Biol. 96: 607–625. [DOI] [PubMed] [Google Scholar]
- Chini A., Grant J.J., Seki M., Shinozaki K., Loake G.J.(2004). Drought tolerance established by enhanced expression of the CC-NBS-LRR gene, ADR1, requires salicylic acid, EDS1 and ABI1. Plant J. 38: 810–822. [DOI] [PubMed] [Google Scholar]
- Coppinger P., Repetti P.P., Day B., Dahlbeck D., Mehlert A., Staskawicz B.J.(2004). Overexpression of the plasma membrane-localized NDR1 protein results in enhanced bacterial disease resistance in Arabidopsis thaliana. Plant J. 40: 225–237. [DOI] [PubMed] [Google Scholar]
- Craigon D.J., James N., Okyere J., Higgins J., Jotham J., May S.(2004). NASCArrays: A repository for microarray data generated by NASC’s transcriptomics service. Nucleic Acids Res. 32: D575–D577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui H., Gobbato E., Kracher B., Qiu J., Bautor J., Parker J.E.(2017). A core function of EDS1 with PAD4 is to protect the salicylic acid defense sector in Arabidopsis immunity. New Phytol. 213: 1802–1817. [DOI] [PubMed] [Google Scholar]
- Cui H., Qiu J., Zhou Y., Bhandari D.D., Zhao C., Bautor J., Parker J.E.(2018). Antagonism of transcription factor MYC2 by EDS1/PAD4 complexes bolsters salicylic acid defense in Arabidopsis effector-triggered immunity. Mol. Plant 11: 1053–1066. [DOI] [PubMed] [Google Scholar]
- Cutter A.D., Jovelin R.(2015). When natural selection gives gene function the cold shoulder. BioEssays 37: 1169–1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalchau N., Hubbard K.E., Robertson F.C., Hotta C.T., Briggs H.M., Stan G.-B., Gonçalves J.M., Webb A.A.R.(2010). Correct biological timing in Arabidopsis requires multiple light-signaling pathways. Proc. Natl. Acad. Sci. USA 107: 13171–13176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Torres Zabala M., Bennett M.H., Truman W.H., Grant M.R.(2009). Antagonism between salicylic and abscisic acid reflects early host-pathogen conflict and moulds plant defence responses. Plant J. 59: 375–386. [DOI] [PubMed] [Google Scholar]
- Deutekom E.S., Vosseberg J., van Dam T.J.P., Snel B.(2019). Measuring the impact of gene prediction on gene loss estimates in Eukaryotes by quantifying falsely inferred absences. PLOS Comput. Biol. 15: e1007301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Y., Dommel M., Mou Z.(2016). Abscisic acid promotes proteasome-mediated degradation of the transcription coactivator NPR1 in Arabidopsis thaliana. Plant J. 86: 20–34. [DOI] [PubMed] [Google Scholar]
- Dodds P.N., Lawrence G.J., Ellis J.G.(2001). Six amino acid changes confined to the leucine-rich repeat beta-strand/beta-turn motif determine the difference between the P and P2 rust resistance specificities in flax. Plant Cell 13: 163–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dongus J.A., Bhandari D.D., Patel M., Archer L., Dijkgraaf L., Deslandes L., Shah J., Parker J.E.(2019). Arabidopsis PAD4 lipase-like domain is a minimal functional unit in resistance to green peach aphid. bioRxiv 769125. [DOI] [PubMed] [Google Scholar]
- Du Z.-Y., Wang Q.-F., Consortium C.P.(2016). Phylogenetic tree of vascular plants reveals the origins of aquatic angiosperms. J. Syst. Evol. 54: 342–348. [Google Scholar]
- Falk A., Feys B.J., Frost L.N., Jones J.D., Daniels M.J., Parker J.E.(1999). EDS1, an essential component of R gene-mediated disease resistance in Arabidopsis has homology to eukaryotic lipases. Proc. Natl. Acad. Sci. USA 96: 3292–3297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feys B.J., Wiermer M., Bhat R.A., Moisan L.J., Medina-Escobar N., Neu C., Cabral A., Parker J.E.(2005). Arabidopsis SENESCENCE-ASSOCIATED GENE101 stabilizes and signals within an ENHANCED DISEASE SUSCEPTIBILITY1 complex in plant innate immunity. Plant Cell 17: 2601–2613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flowers J.M., et al. (2015). Whole-genome resequencing reveals extensive natural variation in the model green alga Chlamydomonas reinhardtii. Plant Cell 27: 2353–2369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gantner J., Ordon J., Kretschmer C., Guerois R., Stuttmann J.(2019). An EDS1–SAG101 Complex is Essential for TNL-mediated Immunity in Nicotiana benthamiana. Plant Cell 31: 2456–2474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y., Wang W., Zhang T., Gong Z., Zhao H., Han G.Z.(2018). Out of water: The origin and early diversification of plant R-genes. Plant Physiol. 177: 82–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Givnish T.J., et al. (2018). Monocot plastid phylogenomics, timeline, net rates of species diversification, the power of multi-gene analyses, and a functional model for the origin of monocots. Am. J. Bot. 105: 1888–1910. [DOI] [PubMed] [Google Scholar]
- Griesmann M., et al. (2018). Phylogenomics reveals multiple losses of nitrogen-fixing root nodule symbiosis. Science 361: eaat1743. [DOI] [PubMed] [Google Scholar]
- Haiyan H., Jieyun Z., Kangle Z., Mingjiu L.(2012). Characterization of differentially expressed genes induced by virulent and avirulent Magnaporthe grisea strains in rice. Plant Omics 5: 542–546. [Google Scholar]
- Hansen B.O., Vaid N., Musialak-Lange M., Janowski M., Mutwil M.(2014). Elucidating gene function and function evolution through comparison of co-expression networks of plants. Front Plant Sci 5: 394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haug-Baltzell A., Stephens S.A., Davey S., Scheidegger C.E., Lyons E.(2017). SynMap2 and SynMap3D: web-based whole-genome synteny browsers. Bioinformatics 33: 2197–2198. [DOI] [PubMed] [Google Scholar]
- Horsefield S., et al. (2019). NAD+ cleavage activity by animal and plant TIR domains in cell death pathways. Science 365: 793–799. [DOI] [PubMed] [Google Scholar]
- Hruz T., Laule O., Szabo G., Wessendorp F., Bleuler S., Oertle L., Widmayer P., Gruissem W., Zimmermann P.(2008). Genevestigator v3: A reference expression database for the meta-analysis of transcriptomes. Adv. Bioinforma. 2008: 420747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibarra-Laclette E., et al. (2013). Architecture and evolution of a minute plant genome. Nature 498: 94–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain M., Nijhawan A., Arora R., Agarwal P., Ray S., Sharma P., Kapoor S., Tyagi A.K., Khurana J.P.(2007). F-box proteins in rice. Genome-wide analysis, classification, temporal and spatial gene expression during panicle and seed development, and regulation by light and abiotic stress. Plant Physiol. 143: 1467–1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jirage D., Tootle T.L., Reuber T.L., Frost L.N., Feys B.J., Parker J.E., Ausubel F.M., Glazebrook J.(1999). Arabidopsis thaliana PAD4 encodes a lipase-like gene that is important for salicylic acid signaling. Proc. Natl. Acad. Sci. USA 96: 13583–13588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson REBECCA N, O’Meally DENIS, Chen ZHILIANG, Etherington GRAHAM J, Ho SIMON Y W, Nash WILL J, Grueber CATHERINE E, Cheng YUANYUAN, Whittington CAMILLA M, Dennison SIOBHAN, Peel EMMA, Haerty WILFRIED, et al. (2018). Adaptation and conservation insights from the koala genome. Nat. Genet. 50: 1102–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones J.D.G., Vance R.E., Dangl J.L.(2016). Intracellular innate immune surveillance devices in plants and animals. Science 354: 354. [DOI] [PubMed] [Google Scholar]
- Kilian J., Whitehead D., Horak J., Wanke D., Weinl S., Batistic O., D’Angelo C., Bornberg-Bauer E., Kudla J., Harter K.(2007). The AtGenExpress global stress expression data set: Protocols, evaluation and model data analysis of UV-B light, drought and cold stress responses. Plant J. 50: 347–363. [DOI] [PubMed] [Google Scholar]
- Kim D., Langmead B., Salzberg S.L.(2015). HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 12: 357–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J., Lim C.J., Lee B.-W., Choi J.-P., Oh S.-K., Ahmad R., Kwon S.-Y., Ahn J., Hur C.-G.(2012). A genome-wide comparison of NB-LRR type of resistance gene analogs (RGA) in the plant kingdom. Mol. Cells 33: 385–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim T.-H., et al. (2011). Chemical genetics reveals negative regulation of abscisic acid signaling by a plant immune response pathway. Curr. Biol. 21: 990–997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knepper C., Savory E.A., Day B.(2011). Arabidopsis NDR1 is an integrin-like protein with a role in fluid loss and plasma membrane-cell wall adhesion. Plant Physiol. 156: 286–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koops P., Pelser S., Ignatz M., Klose C., Marrocco-Selden K., Kretsch T.(2011). EDL3 is an F-box protein involved in the regulation of abscisic acid signalling in Arabidopsis thaliana. J. Exp. Bot. 62: 5547–5560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krasileva K.V., Dahlbeck D., Staskawicz B.J.(2010). Activation of an Arabidopsis resistance protein is specified by the in planta association of its leucine-rich repeat domain with the cognate oomycete effector. Plant Cell 22: 2444–2458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnan A., Ambavaram M., Batlang U., Pereira A.(2010). A resource for systems analysis of transcriptional modules involved in drought response in rice. Sys Anal Stress Resp Plan 138. [Google Scholar]
- Kroj T., Chanclud E., Michel-Romiti C., Grand X., Morel J.B.(2016). Integration of decoy domains derived from protein targets of pathogen effectors into plant immune receptors is widespread. New Phytol. 210: 618–626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapin D., Kovacova V., Sun X., Dongus J.A., Bhandari D., von Born P., Bautor J., Guarneri N., Rzemieniewski J., Stuttmann J., Beyer A., Parker J.E.(2019). A Coevolved EDS1-SAG101-NRG1 Module Mediates Cell Death Signaling by TIR-Domain Immune Receptors. Plant Cell 31: 2430–2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leushkin E.V., Sutormin R.A., Nabieva E.R., Penin A.A., Kondrashov A.S., Logacheva M.D.(2013). The miniature genome of a carnivorous plant Genlisea aurea contains a low number of genes and short non-coding sequences. BMC Genomics 14: 476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R.; 1000 Genome Project Data Processing Subgroup. (2009). The Sequence Alignment/Map format and SAMtools. Bioinformatics 25: 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Stoeckert C.J. Jr., Roos D.S.(2003). OrthoMCL: Identification of ortholog groups for eukaryotic genomes. Genome Res. 13: 2178–2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lievens L., Pollier J., Goossens A., Beyaert R., Staal J.(2017). Abscisic acid as pathogen effector and immune regulator. Front Plant Sci 8: 587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacArthur D.G., et al. ; 1000 Genomes Project Consortium. (2012). A systematic survey of loss-of-function variants in human protein-coding genes. Science 335: 823–828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maekawa T., Kracher B., Vernaldi S., Ver Loren van Themaat E., Schulze-Lefert P.(2012). Conservation of NLR-triggered immunity across plant lineages. Proc. Natl. Acad. Sci. USA 109: 20119–20123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcel S., Sawers R., Oakeley E., Angliker H., Paszkowski U.(2010). Tissue-adapted invasion strategies of the rice blast fungus Magnaporthe oryzae. Plant Cell 22: 3177–3187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matasci N., et al. (2014). Data access for the 1,000 Plants (1KP) project. Gigascience 3: 1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyers B.C., Kozik A., Griego A., Kuang H., Michelmore R.W.(2003). Genome-wide analysis of NBS-LRR-encoding genes in Arabidopsis. Plant Cell 15: 809–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michael T.P., et al. (2017). Comprehensive definition of genome features in Spirodela polyrhiza by high-depth physical mapping and short-read DNA sequencing strategies. Plant J. 89: 617–635. [DOI] [PubMed] [Google Scholar]
- Moeder W., Ung H., Mosher S., Yoshioka K.(2010). SA-ABA antagonism in defense responses. Plant Signal. Behav. 5: 1231–1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monroe J.G., Powell T., Price N., Mullen J.L., Howard A., Evans K., Lovell J.T., McKay J.K.(2018). Drought adaptation in Arabidopsis thaliana by extensive genetic loss-of-function. eLife 7: e41038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nandety R.S., Caplan J.L., Cavanaugh K., Perroud B., Wroblewski T., Michelmore R.W., Meyers B.C.(2013). The role of TIR-NBS and TIR-X proteins in plant basal defense responses. Plant Physiol. 162: 1459–1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman M.-A., Sundelin T., Nielsen J.T., Erbs G.(2013). MAMP (microbe-associated molecular pattern) triggered immunity in plants. Front Plant Sci 4: 139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicol J.W., Helt G.A., Blanchard S.G. Jr., Raja A., Loraine A.E.(2009). The Integrated Genome Browser: Free software for distribution and exploration of genome-scale datasets. Bioinformatics 25: 2730–2731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen J.L., \ et al. (2016). The genome of the seagrass Zostera marina reveals angiosperm adaptation to the sea. Nature 530: 331–335. [DOI] [PubMed] [Google Scholar]
- One Thousand Plant Transcriptomes Initiative. (2019). One thousand plant transcriptomes and the phylogenomics of green plants. Nature 574: 679–685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey S., Wang R.-S., Wilson L., Li S., Zhao Z., Gookin T.E., Assmann S.M., Albert R.(2010). Boolean modeling of transcriptome data reveals novel modes of heterotrimeric G-protein action. Mol. Syst. Biol. 6: 372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi T., Seong K., Thomazella D.P.T., Kim J.R., Pham J., Seo E., Cho M.-J., Schultink A., Staskawicz B.J.(2018). NRG1 functions downstream of EDS1 to regulate TIR-NLR-mediated plant immunity in Nicotiana benthamiana. Proc. Natl. Acad. Sci. USA 115: E10979–E10987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radhakrishnan G.V., et al. (2020). An ancestral signalling pathway is conserved in intracellular symbioses-forming plant lineages. Nat Plants 6: 280–289. [DOI] [PubMed] [Google Scholar]
- Ramegowda V., Senthil-Kumar M.(2015). The interactive effects of simultaneous biotic and abiotic stresses on plants: Mechanistic understanding from drought and pathogen combination. J. Plant Physiol. 176: 47–54. [DOI] [PubMed] [Google Scholar]
- Ryals J., Weymann K., Lawton K., Friedrich L., Ellis D., Steiner H.Y., Johnson J., Delaney T.P., Jesse T., Vos P., Uknes S.(1997). The Arabidopsis NIM1 protein shows homology to the mammalian transcription factor inhibitor I kappa B. Plant Cell 9: 425–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai H., et al. (2013). Rice Annotation Project Database (RAP-DB): An integrative and interactive database for rice genomics. Plant Cell Physiol. 54: e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanderson M.J., Thorne J.L., Wikström N., Bremer K.(2004). Molecular evidence on plant divergence times. Am. J. Bot. 91: 1656–1665. [DOI] [PubMed] [Google Scholar]
- Sarris P.F., Cevik V., Dagdas G., Jones J.D.G., Krasileva K.V.(2016). Comparative analysis of plant immune receptor architectures uncovers host proteins likely targeted by pathogens. BMC Biol. 14: 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schäferhoff B., Fleischmann A., Fischer E., Albach D.C., Borsch T., Heubl G., Müller K.F.(2010). Towards resolving Lamiales relationships: Insights from rapidly evolving chloroplast sequences. BMC Evol. Biol. 10: 352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simão F.A., Waterhouse R.M., Ioannidis P., Kriventseva E.V., Zdobnov E.M.(2015). BUSCO: Assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 31: 3210–3212. [DOI] [PubMed] [Google Scholar]
- Stam R., Silva-Arias G.A., Tellier A.(2019). Subsets of NLR genes show differential signatures of adaptation during colonization of new habitats. New Phytol. 224: 367–379. [DOI] [PubMed] [Google Scholar]
- Stamatakis A.(2014). RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30: 1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steuernagel B., et al. (2020). The NLR-Annotator tool enables annotation of the intracellular immune receptor repertoire. Plant Physiol. 182: 468–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steuernagel B., Jupe F., Witek K., Jones J.D.G., Wulff B.B.H.(2015). NLR-parser: rapid annotation of plant NLR complements. Bioinformatics 31: 1665–1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stolzer M., Lai H., Xu M., Sathaye D., Vernot B., Durand D.(2012). Inferring duplications, losses, transfers and incomplete lineage sorting with nonbinary species trees. Bioinformatics 28: i409–i415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szechyńska-Hebda M., Czarnocka W., Hebda M., Bernacki M.J., Karpiński S.(2016). PAD4, LSD1 and EDS1 regulate drought tolerance, plant biomass production, and cell wall properties. Plant Cell Rep. 35: 527–539. [DOI] [PubMed] [Google Scholar]
- Tamborski J., Krasileva K.V.(2020). Evolution of Plant NLRs: From natural history to precise modifications. Annu Rev Plant Biol 71: 355–378. [DOI] [PubMed] [Google Scholar]
- Tameling W.I.L., Vossen J.H., Albrecht M., Lengauer T., Berden J.A., Haring M.A., Cornelissen B.J.C., Takken F.L.W.(2006). Mutations in the NB-ARC domain of I-2 that impair ATP hydrolysis cause autoactivation. Plant Physiol. 140: 1233–1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarr D.E.K., Alexander H.M.(2009). TIR-NBS-LRR genes are rare in monocots: Evidence from diverse monocot orders. BMC Res. Notes 2: 197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thanki A.S., Soranzo N., Haerty W., Davey R.P.(2018). GeneSeqToFamily: A galaxy workflow to find gene families based on the Ensembl Compara GeneTrees pipeline. Gigascience 7: 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VanBuren R., et al. (2015). Single-molecule sequencing of the desiccation-tolerant grass Oropetium thomaeum. Nature 527: 508–511. [DOI] [PubMed] [Google Scholar]
- VanBuren R., Wai C.M., Keilwagen J., Pardo J.(2018). A chromosome-scale assembly of the model desiccation tolerant grass Oropetium thomaeum. Plant Direct 2: e00096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van de Weyer A.-L., Monteiro F., Furzer O.J., Nishimura M.T., Cevik V., Witek K., Jones J.D.G., Dangl J.L., Weigel D., Bemm F.(2019). The Arabidopsis thaliana pan-NLRome. bioRxiv •••: 537001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Ghelder C., Parent G.J., Rigault P., Prunier J., Giguère I., Caron S., Stival Sena J., Deslauriers A., Bousquet J., Esmenjaud D., MacKay J.(2019). The large repertoire of conifer NLR resistance genes includes drought responsive and highly diversified RNLs. Sci. Rep. 9: 11614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Ooijen G., Mayr G., Kasiem M.M.A., Albrecht M., Cornelissen B.J.C., Takken F.L.W.(2008). Structure-function analysis of the NB-ARC domain of plant disease resistance proteins. J. Exp. Bot. 59: 1383–1397. [DOI] [PubMed] [Google Scholar]
- van Velzen R., et al. (2018). Comparative genomics of the nonlegume Parasponia reveals insights into evolution of nitrogen-fixing rhizobium symbioses. Proc. Natl. Acad. Sci. USA 115: E4700–E4709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venugopal S.C., Jeong R.-D., Mandal M.K., Zhu S., Chandra-Shekara A.C., Xia Y., Hersh M., Stromberg A.J., Navarre D., Kachroo A., Kachroo P.(2009). Enhanced disease susceptibility 1 and salicylic acid act redundantly to regulate resistance gene-mediated signaling. PLoS Genet. 5: e1000545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner S., Stuttmann J., Rietz S., Guerois R., Brunstein E., Bautor J., Niefind K., Parker J.E.(2013). Structural basis for signaling by exclusive EDS1 heteromeric complexes with SAG101 or PAD4 in plant innate immunity. Cell Host Microbe 14: 619–630. [DOI] [PubMed] [Google Scholar]
- Wan L., Essuman K., Anderson R.G., Sasaki Y., Monteiro F., Chung E.-H., Osborne Nishimura E., DiAntonio A., Milbrandt J., Dangl J.L., Nishimura M.T.(2019). TIR domains of plant immune receptors are NAD+-cleaving enzymes that promote cell death. Science 365: 799–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D., Pan Y., Zhao X., Zhu L., Fu B., Li Z.(2011). Genome-wide temporal-spatial gene expression profiling of drought responsiveness in rice. BMC Genomics 12: 149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G.-F., Ji J., El-Kasmi F., Dangl J.L., Johal G., Balint-Kurti P.J.(2015). Molecular and functional analyses of a maize autoactive NB-LRR protein identify precise structural requirements for activity. PLoS Pathog. 11: e1004674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., et al. (2014). The Spirodela polyrhiza genome reveals insights into its neotenous reduction fast growth and aquatic lifestyle. Nat. Commun. 5: 3311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Li R., Zhu Q., Tang X., Zhao Q.(2016). Transcriptomic and physiological analysis of common duckweed Lemna minor responses to NH4(+) toxicity. BMC Plant Biol. 16: 92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterhouse A.M., Procter J.B., Martin D.M.A., Clamp M., Barton G.J.(2009). Jalview Version 2--a multiple sequence alignment editor and analysis workbench. Bioinformatics 25: 1189–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen Z., Yao L., Singer S.D., Muhammad H., Li Z., Wang X.(2017). Constitutive heterologous overexpression of a TIR-NB-ARC-LRR gene encoding a putative disease resistance protein from wild Chinese Vitis pseudoreticulata in Arabidopsis and tobacco enhances resistance to phytopathogenic fungi and bacteria. Plant Physiol. Biochem. 112: 346–361. [DOI] [PubMed] [Google Scholar]
- Wheeler T.J., Eddy S.R.(2013). nhmmer: DNA homology search with profile HMMs. Bioinformatics 29: 2487–2489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickett N.J., et al. (2014). Phylotranscriptomic analysis of the origin and early diversification of land plants. Proc. Natl. Acad. Sci. USA 111: E4859–E4868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wituszynska W., Slesak I., Vanderauwera S., Szechynska-Hebda M., Kornas A., Van Der Kelen K., Mühlenbock P., Karpinska B., Mackowski S., Van Breusegem F., Karpinski S.(2013). Lesion simulating disease1, enhanced disease susceptibility1, and phytoalexin deficient4 conditionally regulate cellular signaling homeostasis, photosynthesis, water use efficiency, and seed yield in Arabidopsis. Plant Physiol. 161: 1795–1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolinska K.W., Berens M.L.(2019). Optimal Defense Theory 2.0: Tissue-specific stress defense prioritization as an extra layer of complexity. Commun. Integr. Biol. 12: 91–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C.-H., Abd-El-Haliem A., Bozkurt T.O., Belhaj K., Terauchi R., Vossen J.H., Kamoun S.(2017). NLR network mediates immunity to diverse plant pathogens. Proc. Natl. Acad. Sci. USA 114: 8113–8118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C.-H., Derevnina L., Kamoun S.(2018). Receptor networks underpin plant immunity. Science 360: 1300–1301. [DOI] [PubMed] [Google Scholar]
- Wu Z., Li M., Dong O.X., Xia S., Liang W., Bao Y., Wasteneys G., Li X.(2019). Differential regulation of TNL-mediated immune signaling by redundant helper CNLs. New Phytol. 222: 938–953. [DOI] [PubMed] [Google Scholar]
- Xu G., Yuan M., Ai C., Liu L., Zhuang E., Karapetyan S., Wang S., Dong X.(2017). uORF-mediated translation allows engineered plant disease resistance without fitness costs. Nature 545: 491–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W.-Q., Losh J., Chen C., Li P., Wang R.-H., Zhao Y.-P., Qiu Y.-X., Fu C.-X.(2019). Comparative genomics of Figureworts (Scrophularia, Scrophulariaceae), with implications for the evolution of Scrophularia and Lamiales: Comparative genomics of Scrophularia. Jnl of Sytematics Evolution 57: 55–65. [Google Scholar]
- Yu Y., Streubel J., Balzergue S., Champion A., Boch J., Koebnik R., Feng J., Verdier V., Szurek B.(2011). Colonization of rice leaf blades by an African strain of Xanthomonas oryzae pv. oryzae depends on a new TAL effector that induces the rice nodulin-3 Os11N3 gene. Mol. Plant Microbe Interact. 24: 1102–1113. [DOI] [PubMed] [Google Scholar]
- Zeng L., Zhang Q., Sun R., Kong H., Zhang N., Ma H.(2014). Resolution of deep angiosperm phylogeny using conserved nuclear genes and estimates of early divergence times. Nat. Commun. 5: 4956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong B., Sun L., Penny D.(2015). The origin of land plants: A phylogenomic perspective. Evol Bioinform Online 11: 137–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu S., Jeong R.-D., Venugopal S.C., Lapchyk L., Navarre D., Kachroo A., Kachroo P.(2011). SAG101 forms a ternary complex with EDS1 and PAD4 and is required for resistance signaling against turnip crinkle virus. PLoS Pathog. 7: e1002318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1001 Genomes Consortium (2016). 1,135 Genomes Reveal the Global Pattern of Polymorphism in Arabidopsis thaliana. Cell 166: 481–91. [DOI] [PMC free article] [PubMed] [Google Scholar]