Intercellular signaling mediated by PNP-A and its putative receptor protein PNP-R2 counteracts SA-mediated plant stress responses.
Abstract
The plant stress hormone salicylic acid (SA) participates in local and systemic acquired resistance, which eventually leads to whole-plant resistance to bacterial pathogens. However, if SA-mediated signaling is not appropriately controlled, plants incur defense-associated fitness costs such as growth inhibition and cell death. Despite its importance, to date only a few components counteracting the SA-primed stress responses have been identified in Arabidopsis (Arabidopsis thaliana). These include other plant hormones such as jasmonic acid and abscisic acid, and proteins such as LESION SIMULATING DISEASE1, a transcription coregulator. Here, we describe PLANT NATRIURETIC PEPTIDE A (PNP-A), a functional analog to vertebrate atrial natriuretic peptides, that appears to antagonize the SA-mediated plant stress responses. While loss of PNP-A potentiates SA-mediated signaling, exogenous application of synthetic PNP-A or overexpression of PNP-A significantly compromises the SA-primed immune responses. Moreover, we identify a plasma membrane–localized receptor-like protein, PNP-R2, that interacts with PNP-A and is required to initiate the PNP-A–mediated intracellular signaling. In summary, our work identifies a peptide and its putative cognate receptor as counteracting both SA-mediated signaling and SA-primed cell death in Arabidopsis.
INTRODUCTION
For multicellular organisms, cell-to-cell communication is crucial for growth, development, and survival under ever-changing environmental conditions. In plants, this intercellular communication is mostly mediated by secreted signals such as phytohormones, reactive oxygen species, small RNAs, and small peptide hormones (Van Norman et al., 2011). While genome and transcriptome analyses have identified more than 1000 potential peptide hormones in Arabidopsis (Arabidopsis thaliana; Lease and Walker, 2006; Matsubayashi, 2014), only a few are functionally characterized, and even fewer have an assigned cognate receptor. During or upon their secretion into the apoplastic space, most peptide hormones undergo posttranslational modifications that enable recognition by specific receptors, such as receptor-like kinases (Shiu and Bleecker, 2001) and receptor-like proteins (RLPs; Wang et al., 2008), on the surface of target cells, activating the relay of the signal into the cell interior (Butenko et al., 2009; Matsubayashi, 2011; Murphy et al., 2012; Hirakawa et al., 2017). In general, the coupling of the peptide to the receptor results in transcriptional reprogramming, empowering the recipient cell to appropriately respond to an inbound factor. The Arabidopsis genome also encodes hundreds of plasma membrane (PM)–associated receptor-like kinases and RLPs, connecting multiple signaling pathways to modulate physiological processes (Shiu and Bleecker, 2001; Wang et al., 2008).
There is substantial signaling crosstalk between abiotic and biotic stress responses in plants (Fujita et al., 2006; Verma et al., 2016). This suggests that abiotic and biotic stress-induced intra- and inter-cellular signaling pathways are most likely intertwined to increase efficacy. Among various stress-related molecular components, the defense hormone salicylic acid (SA) has been implicated in plant stress responses toward a multitude of environmental factors such as drought, salinity, cold, high light, and microbial pathogens (Shah, 2003; Scott et al., 2004; Yang et al., 2004; Mateo et al., 2006; Lee et al., 2010; Wan et al., 2012; Miura et al., 2013). Despite the pivotal role of SA in plant stress responses, when it becomes constitutively active, the spatiotemporal SA-mediated signaling generally results in growth inhibition and spontaneous cell death. Research on Arabidopsis lesion-mimic mutants, which display lesions associated with increased cellular SA contents and constitutive activation of SA-mediated signaling, indicated that certain molecular components counteract this pathway (Dietrich et al., 1994).
The SA-counteracting components include a protein called LESION SIMULATING DISEASE1 (LSD1), a transcription coregulator (Dietrich et al., 1997; Kaminaka et al., 2006). The Arabidopsis lsd1 mutant is one of the most extensively studied lesion-mimic mutants and develops uncontrolled cell death, referred to as runaway cell death (RCD), in response to increased cellular SA contents (Dietrich et al., 1994; Lv et al., 2019). Extended daylength or light intensity is suggested to cause lsd1 RCD through the modulation of chloroplast redox status and reactive oxygen species homeostasis (Mateo et al., 2004; Mühlenbock et al., 2008; Lv et al., 2019). Given that lsd1 mutant plants exhibit RCD in response to exogenous SA and bacterial pathogens, LSD1 has been implicated as a negative regulator in the SA-primed pro-death pathway. Indeed, LSD1 suppresses the positive feedback loop of SA synthesis mediated by ENHANCED DISEASE SUSCEPTIBILITY1 (EDS1) and PHYTOALEXIN DEFICIENT4 (PAD4; Rustérucci et al., 2001; Lv et al., 2019). Besides LSD1, plant hormones, including jasmonic acid and abscisic acid (ABA), are known to counteract SA-mediated signaling (Li et al., 2004; Ding et al., 2016).
In this study, we unveil another SA-counteracting player, PLANT NATRIURETIC PEPTIDE A (PNP-A), a functional analog to vertebrate atrial natriuretic peptides (ANPs). PNP-A signals via cyclic GMP (cGMP; Pharmawati et al., 2001; Wang et al., 2007; Turek and Gehring, 2016; Gehring and Turek, 2017). In animals, the synthesis of cGMP from GTP is catalyzed by natriuretic peptide receptors (NPRs) that possess protein kinase (PK) and guanylyl cyclase (GC) activities following perception of ANPs (Potter and Hunter, 2001). Like ANPs, upon secretion to the apoplast, PNPs undergo formation of inter-disulfide bonds and proteolytic processing (Potter and Hunter, 2001; Turek and Gehring, 2016). Although PNPs have been shown to affect a broad spectrum of physiological responses in plants, including stomatal opening (Maryani et al., 2003; Morse et al., 2004; Wang et al., 2007; Gottig et al., 2008; Ficarra et al., 2018), regulation of photosynthetic efficiency and photorespiration (Gottig et al., 2008; Ruzvidzo et al., 2011), cellular water and ion (Ca2+, H+, K+, and Na+) homeostasis (Pharmawati et al., 2001; Ludidi et al., 2004; Turek and Gehring, 2016), increase in protoplast volume (Wang et al., 2007; Turek and Gehring, 2016), modulation of their own expression (Wang et al., 2011a, 2011b), and resistance against biotic and abiotic stresses (Ficarra et al., 2018), their mode of action remains largely unclear.
We now reveal that PNP-A, but not its close homologue PNP-B, is transcriptionally upregulated in the lsd1 mutant prior to the onset of RCD, which requires NONEXPRESSER OF PR GENES1 (NPR1), a central regulator of SA-mediated signaling (Cao et al., 1997; Zhang et al., 1999). The processed PNP-A in the apoplast interacts with a previously uncharacterized PM-localized leucine-rich repeat (LRR) RLP, herein called the putative PNP-A receptor 2 (PNP-R2). While the lack of PNP-A or PNP-R2 potentiates the SA-primed pro-death pathway in lsd1, exogenous application or overexpression of PNP-A considerably compromises lsd1 RCD. Here, we report a physiological function of the PNP-A as a negative modulator of SA-mediated signaling in both lsd1 and wild-type plants and identify PNP-R2 as its putative receptor.
RESULTS
PNP-A Is an NPR1-Dependent, SA-Responsive Gene
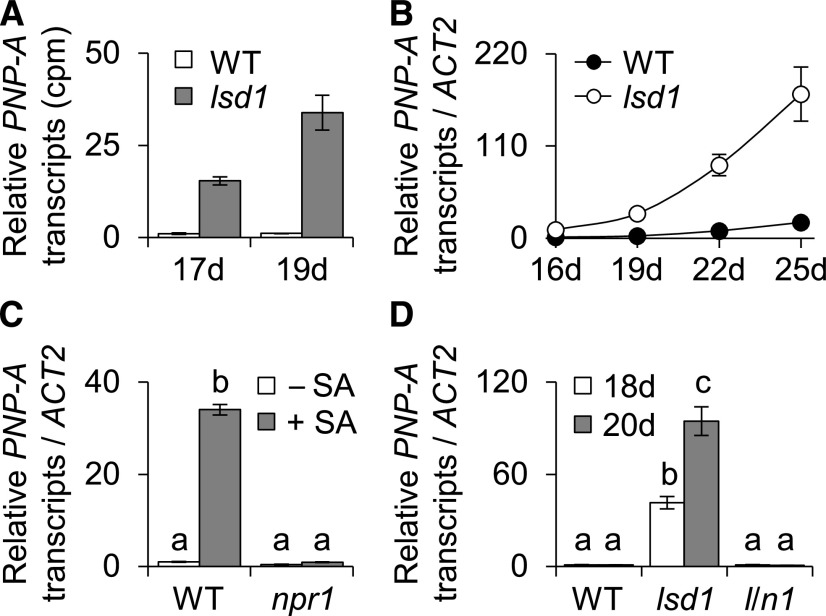
Since its discovery in 1994 (Dietrich et al., 1994), the lsd1 mutant has been extensively utilized as a bio-tool to understand the molecular mechanisms underlying the regulation of cell death, especially the constraining mechanisms, because of its uncontrolled cell death phenotype (Jabs et al., 1996; Dietrich et al., 1997; Kliebenstein et al., 1999). Forward and reverse genetic approaches have unveiled that several key SA-signaling components, such as NPR1, EDS1, and PAD4, are required to induce the lsd1 RCD (Rustérucci et al., 2001; Aviv et al., 2002; Lv et al., 2019). Through global transcriptome analysis, we recently identified a substantial number of genes rapidly upregulated before the onset of the lsd1 RCD (Lv et al., 2019). These lsd1-induced genes include a gene encoding the putative PNP-A peptide hormone, of which the precise mode of action remains unknown in plants (Figures 1A and 1B). In contrast to PNP-A, its closest homolog, PNP-B, was undetectable in lsd1 or wild-type plants (Lv et al., 2019). Because SA-mediated signaling primes the lsd1 RCD, and PNP-A belongs to a group of SA-responsive genes (Wang et al., 2006), we next examined whether SA and its key signaling component NPR1 regulate the PNP-A expression. PNP-A was induced upon SA treatment in wild-type plants, but not in npr1 (Figure 1C), indicating that SA and its bona fide receptor NPR1 positively regulate the expression of PNP-A. Alongside the notable attenuation of the lsd1 RCD (Lv et al., 2019), the loss of NPR1 in the lsd1 background completely abrogated PNP-A expression (Figure 1D).
Figure 1.
PNP-A Is Highly Upregulated in lsd1.
(A) Transcript levels of PNP-A in 17- and 19-d-old wild-type (WT) and lsd1 plants grown under CL were obtained from our previous RNA-sequencing analysis (Lv et al., 2019).
(B) Transcript levels of PNP-A shown in (A) were confirmed by RT-qPCR.
(C) Wild-type (WT) and npr1 plants grown under CL were sprayed with 0.5 mM SA solution (+SA) or with distilled water (–SA), and leaf samples were harvested 12 h after the treatment. The expression levels of PNP-A were examined using RT-qPCR.
(D) Expression levels of PNP-A in wild-type (WT), lsd1, and lsd1 npr1 (l/n1) plants grown under CL were analyzed by RT-qPCR at the indicated time points. For the RT-qPCR analyses in (B), (C), and (D), ACT2 was used as an internal standard. The data represent the means of three independent biological replicates. Error bars indicate sd. Lowercase letters indicate statistically significant differences between mean values (P < 0.01, one-way ANOVA with post hoc Tukey’s honestly significant difference [HSD] test; Supplemental Data Set).
PNP-A Antagonizes SA-Primed Stress Responses
To explore the potential causal relationship between the rapid upregulation of PNP-A and the development of the lsd1 RCD, we generated lsd1 pnp-A double mutant plants and two independent lsd1 transgenic lines overexpressing PNP-A (oxPNP-A; Supplemental Figures 1A and 1B). We then monitored the relative RCD levels compared to those in the lsd1 mutant. The loss of PNP-A aggravated the lsd1 RCD, while this became drastically diminished by the overexpression of PNP-A (Figure 2A; Supplemental Figure 2). The degrees of foliar cell death and chlorosis were evaluated by examining ion leakage and maximum photochemical efficiency of photosystem II (Fv/Fm), respectively. Both ion leakage and Fv/Fm values confirmed the observed RCD phenotypes (Figures 2B and 2C).
Figure 2.
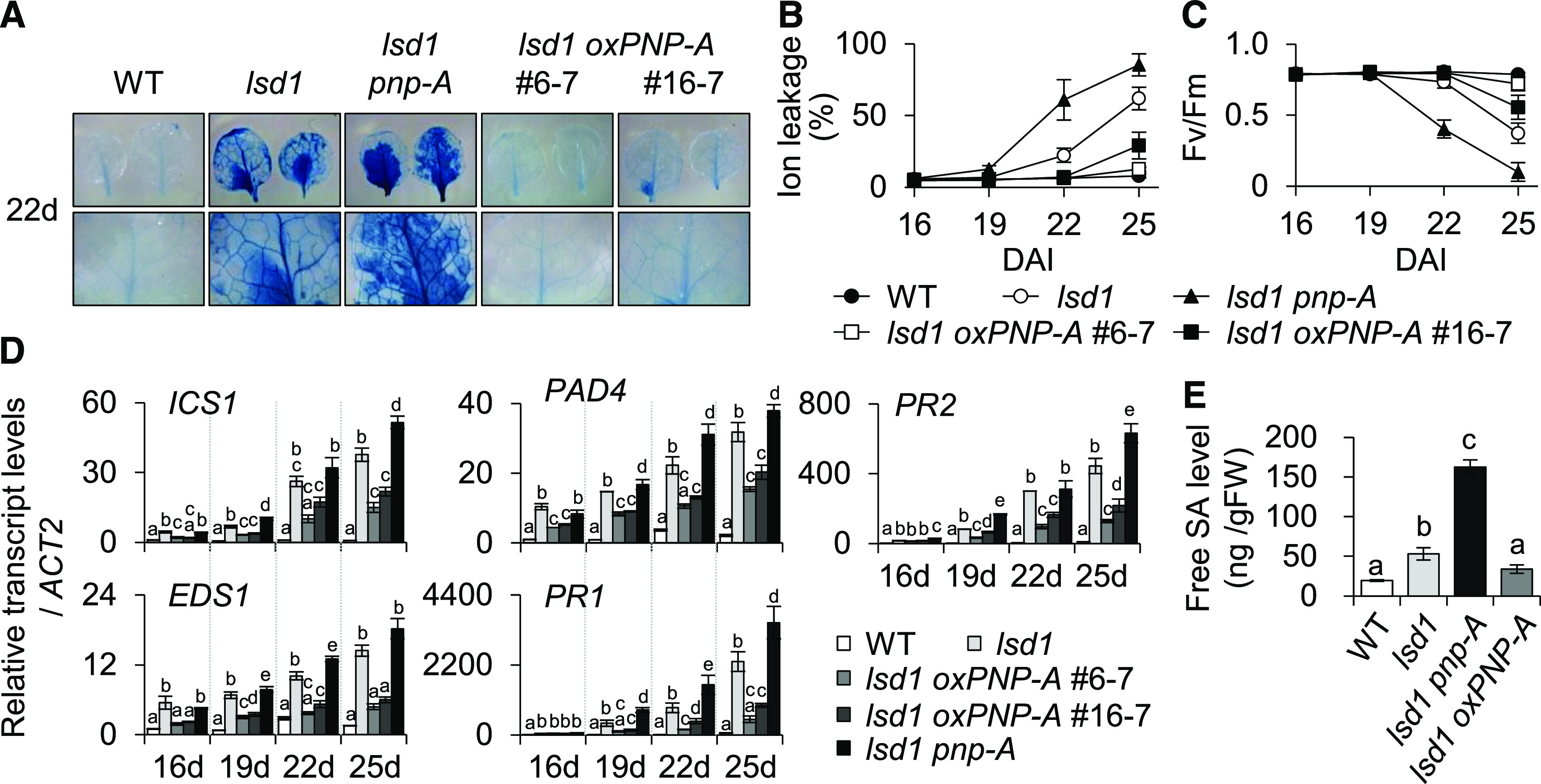
PNP-A Acts to Repress lsd1 RCD.
(A) Twenty 2-d-old plants of the wild-type (WT), lsd1, lsd1 pnp-A, and two independent PNP-A–overexpressing lsd1 transgenic lines (lsd1 oxPNP-A #6-7 and #16-7) grown under CL were collected to examine leaf RCD. (First row) RCD phenotype in the first or second leaves from each genotype was visualized by TB staining. (Second row) Images of the first row are enlarged.
(B) and (C) For measurements of ion leakage (B) and Fv/Fm (C), first or second leaves from plants were harvested at the indicated time points. Ten leaves per genotype were used for the measurement of Fv/Fm. Value in (B) represents means ± sd (n = 3). DAI, days after imbibition; WT, wild type.
(D) Expression levels of genes involved in SA biosynthesis (ICS1, EDS1, and PAD4) and SA response (PR1 and PR2) were examined by RT-qPCR at the indicated time points. ACT2 was used as an internal standard. The data represent the means of three independent biological replicates. Error bars indicate sd. WT, wild type.
(E) Endogenous free SA levels were examined in 16-d-old plants of the wild type (WT), lsd1, lsd1 pnp-A, and lsd1 oxPNP-A #6-7 grown on MS medium under CL. Value represents means ± sd (n = 2). Lowercase letters in (D) and (E) indicate statistically significant differences between mean values (P < 0.05, one-way ANOVA with post hoc Tukey’s honestly significant difference [HSD] test; Supplemental Data Set). FW, fresh weight.
Because of the oxPNP-A–mediated drastic reduction of lsd1 RCD, we anticipated that PNP-A might antagonize SA-mediated signaling. Thus, we investigated the expression levels of SA-responsive genes, including ISOCHORISMATE SYNTHASE1 (ICS1), EDS1, PAD4, and PATHOGENESIS RELATED1 (PR1) and PR2, in lsd1 oxPNP-A and lsd1 plants by using RT-qPCR. Our results revealed that PNP-A overexpression markedly repressed the lsd1-induced, SA-mediated signaling (Figure 2D). Consistent with the altered expression of SA biosynthetic genes, including ICS1, reduced and increased SA levels were detected in lsd1 oxPNP-A and lsd1 pnp-A mutant plants, respectively (Figure 2E).
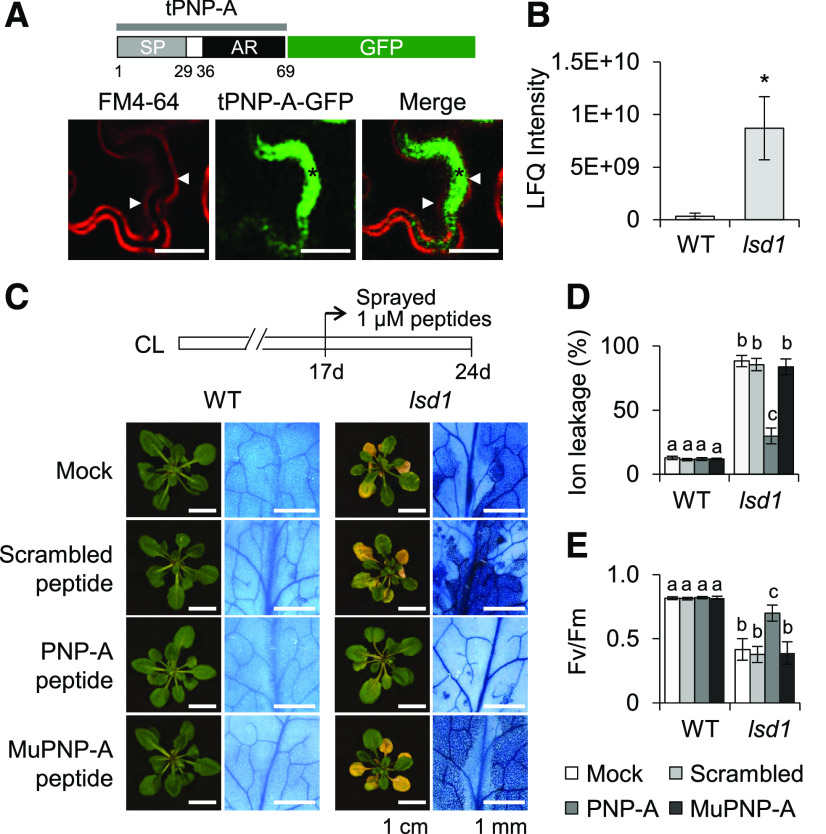
Based on these results, we hypothesized that upon secretion to the apoplast, PNP-A diffuses to adjacent cells and interacts with its cognate receptor to activate an SA-antagonizing signaling cascade. Even though PNP-A was previously suggested to be a secreted peptide hormone (Wang et al., 2011b), its presence in the apoplast remained to be confirmed. We therefore first examined the subcellular localization of transiently overexpressed PNP-A fused to green fluorescent protein (GFP) in Nicotiana benthamiana leaves. Besides its posttranslational modification (i.e., intra-disulfide bond formation between Cys-42 and Cys-65) in the apoplast, it was proposed that the PNP-A precursor (130 amino acids) is proteolytically processed to its active form (34 amino acids, Pro-36 to Tyr-69), resembling the activation process of the vertebrate ANP (Schwartz et al., 1985; Koller and Goeddel, 1992). To avoid the cleavage of GFP, we fused the fluorescent protein to the truncated PNP-A (tPNP-A, Met-1 to Tyr-69) containing both the full-length N-terminal signal peptide (SP; Met-1 to Lys-29) and the putative active domain (Pro-36 to Tyr-69). Confocal images show the localization of the GFP-tagged PNP-A in the apoplast (Figure 3A). Next, the foliar apoplastic proteins extracted from wild-type and lsd1 plants (before the onset of lsd1 RCD) were subjected to trypsin digestion, followed by tandem mass spectrometry analysis. The result demonstrated the presence of PNP-A in the apoplastic fluid (Supplemental Figures 3A and 3B) with a markedly higher level in lsd1 than the wild-type plants (Figure 3B). Additionally, we identified well-known SA-responsive apoplastic marker proteins, such as PR1 and PR2, in the apoplastic fluid of the lsd1 mutant (Supplemental Figure 3C).
Figure 3.
PNP-A Is Localized to the Apoplastic Space, and Exogenous Application of the PNP-A Synthetic Peptide Significantly Compromises lsd1 RCD.
(A) Localization of the tPNP-A (Met-1 to Tyr-69), including SP and active region (AR), fused with GFP (PNPA-GFP) upon transient expression in N. benthamiana leaves. FM4-64 was used to stain PM. Cell plasmolysis was performed by treatment of 0.8 M mannitol for 30 min. An asterisk indicates the apoplastic space formed by the shrinking protoplast, and triangles indicate the retracted PM. Bar = 20 µm.
(B) Mass spectrometry–based detection of PNP-A proteins in the apoplast. Apoplast proteins extracted from 21-d-old wild-type (WT) and lsd1 plants grown under CL were analyzed by mass spectrometry. The relative levels of PNP-A were quantified using the total intensity of the detected peptides (see Supplemental Figures 3A and 3B). The results represent the means of two independent biological replicates. Error bars indicate se. Asterisk indicates statistically significant difference from mean value of WT by Student’s t test (P < 0.05; Supplemental Data Set). LFQ, label-free quantitation.
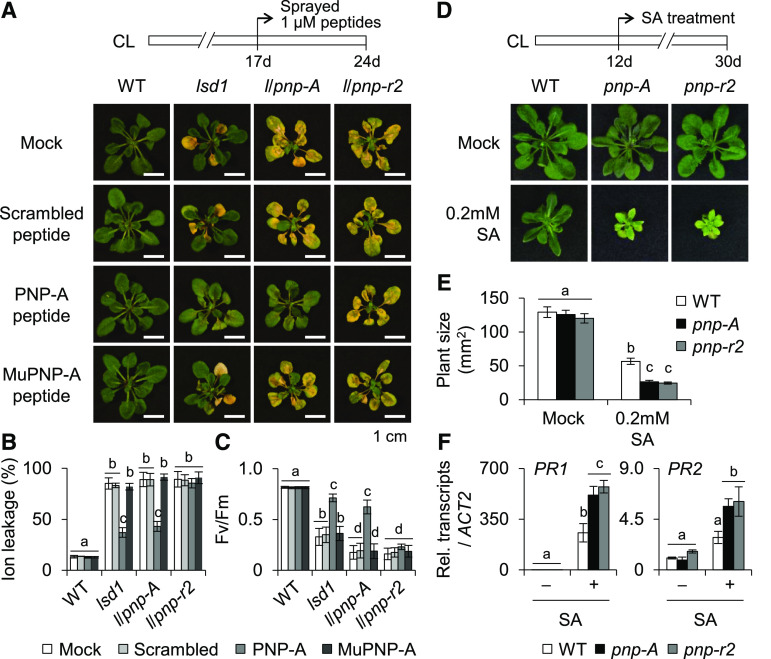
(C) to (E) Seventeen-day-old wild-type (WT) and lsd1 plants grown under CL were treated with water (Mock) or 1 µM scrambled, active (PNP-A) or Cys-mutated (MuPNP-A) form of PNP-A synthetic peptide (for details on the synthetic peptides compared with the active PNP-A, see Supplemental Figure 4). After 7 d of the treatment, the relative levels of foliar RCD were determined by TB staining (C), ion leakage (D), and Fv/Fm (E) measurements. The representative images are shown at the same scale. For the measurement of Fv/Fm, 10 leaves per genotype were used. Value in (D) represents means ± sd (n = 3). Lowercase letters indicate statistically significant differences between mean values (P < 0.05, one-way ANOVA with post hoc Tukey’s honestly significant difference [HSD] test; Supplemental Data Set).
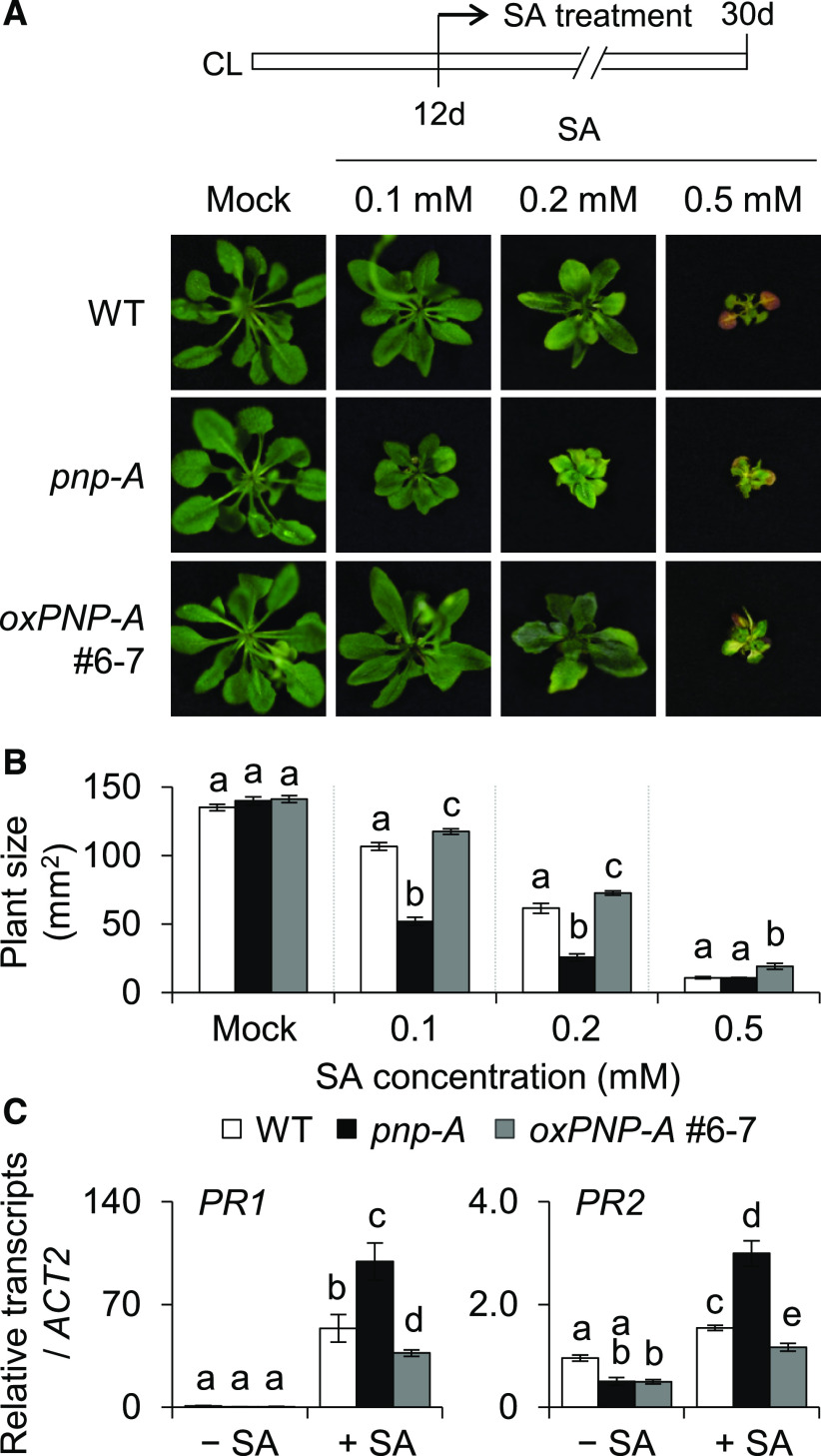
The antagonizing impact of PNP-A on the lsd1 RCD was further examined via a pharmacological approach by using synthesized active and dormant (scrambled or Cys-mutated) PNP-A peptides (Supplemental Figure 4). While no impact of the dormant peptides was observed, lsd1 mutant plants treated with the active form of the PNP-A peptide exhibited a drastically reduced RCD (Figures 3C to 3E). Next, the impact of SA on plants deficient for or overexpressing PNP-A was analyzed. Exogenous application of a high dosage of SA is known to inhibit plant growth because of a trade-off effect (Rivas-San Vicente and Plasencia, 2011; Karasov et al., 2017); that is, the enhanced immune/stress response limits plant growth or vice versa. Consistently, application of SA had a negative impact on growth in wild-type plants, which was remarkably reinforced in pnp-A mutant but less evident in PNP-A–overexpressing plants (Figures 4A and 4B). The SA-mediated growth inhibition was largely rescued by exogenous PNP-A treatment in both wild-type and pnp-A mutant plants (Supplemental Figures 5A and 5B), corroborating the antagonistic activity of PNP-A on SA-mediated stress responses. In agreement with this, PR1 and PR2 were expressed to higher levels in pnp-A than in wild-type plants in response to exogenous SA treatment (Figure 4C).
Figure 4.
PNP-A Is Required to Counteract SA-Mediated Growth Inhibition.
(A) and (B) Twelve-day-old plants of the wild type (WT), pnp-A, and oxPNP-A (#6-7, in WT background) grown on MS medium under CL were transferred to fresh MS medium containing the different concentrations of SA as indicated. The representative foliar phenotype (A) and plant size (B) of each genotype were shown. For the measurement of plant size, 15 plants per genotype were used. Error bars indicate sd. Lowercase letters indicate statistically significant differences between mean values at each of the indicated SA concentrations (P < 0.05, one-way ANOVA with post hoc Tukey’s honestly significant difference [HSD] test; Supplemental Data Set).
(C) Fourteen-day-old plants grown under CL were sprayed with either 0.5 mM SA (+SA) or mock solution (–SA), and foliar tissues were collected 6 h after the treatment. The transcript abundances of PR1 and PR2 were examined using RT-qPCR. Value represents means ± sd (n = 3). ACT2 was used as an internal control. Lowercase letters indicate statistically significant differences between mean values (P < 0.05, one-way ANOVA with post hoc Tukey’s honestly significant difference [HSD] test; Supplemental Data Set).
The PNP-A Peptide Interacts with the PM-Localized LRR Family Protein PNP-R2
The vertebrate PNP analogs interact with GC-coupled protein receptors that catalyze the conversion of GTP into cGMP upon binding of the peptide (Potter and Hunter, 2001). Consistently, an earlier report showed that Arabidopsis PNP-A interacts with a novel LRR protein, named PNP-R1, which contains a putative N-terminal SP, an LRR N-terminal (LRRNT) domain, a transmembrane (TM) domain, two LRR domains, and a PK domain followed by a GC catalytic center at the C terminus (Turek and Gehring, 2016). Since the TM domain is located between the LRRNT and the LRR domains, this prediction implies that the LRR, PK, and GC domains face the cytosol, whereas the LRRNT domain protrudes toward the extracellular space, or vice versa. However, to our surprise, when the deduced amino acid sequence of the PNP-R1 was subjected to a search for conserved domains by using several bioinformatics tools, including TMHMM server v. 2.0 (Krogh et al., 2001), Phobius program (Käll et al., 2004), the National Center for Biotechnology Information’s Conserved Domain Database (Marchler-Bauer et al., 2017), and InterPro (Finn et al., 2017), the TM domain was not identified. Moreover, neither the PK nor the GC catalytic domains were predicted. Given that several plant GCs have low sequence similarity with the annotated GCs of other organisms (Ludidi and Gehring, 2003), the GC catalytic center of PNP-R1 had been identified on the basis of motif searches with conserved and functionally assigned amino acid residues in the catalytic centers. While it was shown that PNP-R1 possesses in vitro GC activity (Turek and Gehring, 2016), to date there is no experimental evidence for its kinase activity. Nevertheless, the exogenous application of PNP-A synthetic peptide resulted in increased cGMP levels in the wild-type plants, whereas pnp-r1 mutant plants were almost completely insensitive to the treatment (Turek and Gehring, 2016).
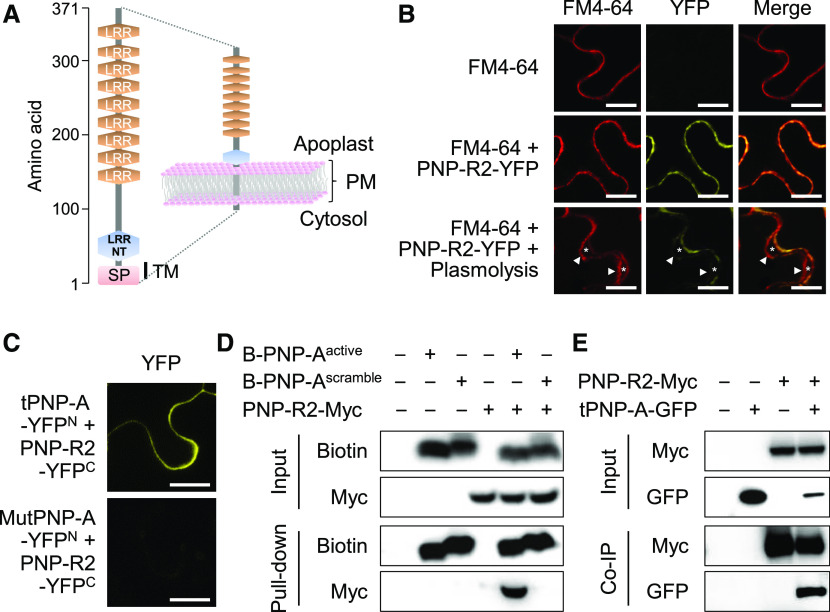
In the lsd1 mutant background, we could not identify this GC-containing receptor protein as a putative PNP-A–interacting protein using a pull-down assay with biotin-conjugated PNP-A peptide coupled to mass spectrometry analysis (Supplemental Table 1). Instead, we found a typical RLP predicted to contain an SP, a TM domain, and an LRRNT domain followed by nine LRRs, but lacking a cytosolic activation domain (e.g., kinase; Figure 5A). Therefore, we tentatively named this RLP as the second PNP-A receptor protein (PNP-R2, At5g12940).
Figure 5.
PNP-A Interacts with a Novel PM-Localized LRR Receptor-Like Protein.
(A) Schematic illustration of the predicted domain structure and topology of the putative PNP-A–interacting LRR receptor, namely PNP-R2. PNP-R2 contains several extracellular domains consisting of nine LRRs and an LRRNT. SP indicates an SP present at the N terminus (NT) of PNP-R2 and is responsible for targeting the protein to the PM. TM marks the predicted TM.
(B) YFP fluorescence was observed in the PM when the YFP-tagged PNP-R2 (PNP-R2-YFP) was expressed in N. benthamiana leaves. FM4-64 was used to stain PM. Cell plasmolysis was performed by treatment with 0.8 M mannitol for 30 min. In plasmolyzed cells, asterisks indicate the apoplastic space formed by the shrinking protoplast, and triangles indicate the retracted PM. Bar = 20 µm.
(C) Interaction of PNP-A with PNP-R2 by BiFC assay. YFP fluorescence was observed in the PM when the N-terminal part of YFP tagged with the tPNP-A-YFPN (Met-1 to Tyr-69 including SP and active region) was coexpressed with the C-terminal part of the YFP tagged with PNP-R2 (PNP-R2-YFPC) in N. benthamiana leaves. No fluorescent signal was observed when mutated tPNP-A-YFPN (MutPNP-A-YFPN, substituting two Cys’s with Ser, as shown in Supplemental Figure 4) was coexpressed with the PNP-R2-YFPC as a negative control. Bar = 20 µm. Results in (B) and (C) were reproduced in at least two independent experiments using three or more N. benthamiana leaves in each experiment, and enlarged representative images are shown.
(D) In vitro pull-down assay of N-terminally biotinylated active region of PNP-A synthetic peptide (B-PNP-Aactive) with PNP-R2 fused with Myc tag (PNP-R2-Myc) upon transient expression in N. benthamiana leaves. N-Terminally biotinylated-scrambled PNP-A peptide (B-PNP-Ascrambled) was used as a negative control as it does not interact with PNP-R2.
(E) Co-IP of tPNP-A-GFP with PNP-R2-Myc upon transient coexpression in N. benthamiana leaves.
A PNP-R2-YFP fusion protein was transiently expressed in N. benthamiana leaves under the control of the cauliflower mosaic virus 35S promoter, and its yellow fluorescent protein (YFP) fluorescence was monitored. We found that the fluorescent signal (red) of the FM4-64 dye colocalized with the fluorescence signal (yellow) of PNP-R2-YFP, and both signals were retained in the PM after plasmolysis (Figure 5B), indicating that PNP-R2 is a PM-localized protein. Bimolecular fluorescence complementation (BiFC), in vitro pull-down, and coimmunoprecipitation (Co-IP) assays in N. benthamiana leaves substantiated the direct interaction between PNP-A and PNP-R2 (Figures 5C to 5E). By contrast, in our hands, an in vitro pull-down assay failed to confirm the interaction between PNP-A and PNP-R1 (Supplemental Figure 6).
PNP-R2 Is Required for the PNP-A–Mediated Repression of SA Responses
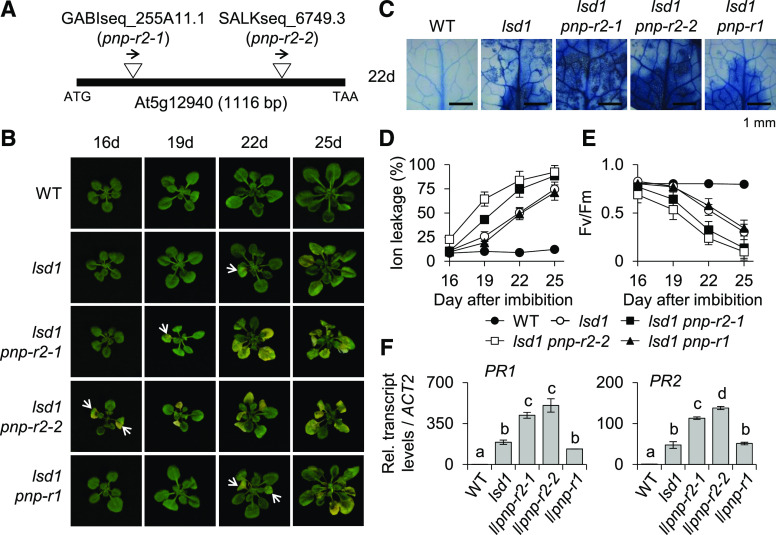
To further determine that PNP-R2 participates in the PNP-A signaling pathway, two knockout mutant alleles of PNP-R2 (pnp-r2-1 and pnp-r2-2; Figure 6A) were crossed to lsd1 to create two independent lsd1 pnp-r2 double mutants. The loss of PNP-R2 in the lsd1 background revealed a probable genetic interaction between PNP-A and PNP-R2, which was evident from the potentiated foliar cell death and chlorosis as well as the significant decrease in Fv/Fm in both lsd1 pnp-r2-1 and lsd1 pnp-r2-2 double mutant plants compared to lsd1 (Figures 6B to 6E). The enhanced RCD phenotype in lsd1 pnp-r2 plants was accompanied by the heightened expression of PR1 and PR2 (Figure 6F). Importantly, overexpression of a Myc-tagged PNP-R2 in lsd1 pnp-r2 plants completely restored the lsd1 phenotype (Supplemental Figures 7A to 7C). However, the lsd1 pnp-r1 double mutant showed equivalent degrees of RCD and expression of PR1 and PR2 compared to those of lsd1 (Figures 6B to 6F), indicating that PNP-R1 is not involved in the lsd1 RCD. Besides, unlike lsd1 and lsd1 pnp-A, in which the active PNP-A synthetic peptide attenuated the lsd1 RCD, the lsd1 pnp-r2-2 double mutant plants were insensitive to the treatment (Figures 7A to 7C). Conversely, pnp-r2 mutant plants exhibited an extreme sensitivity to exogenously applied SA, similar to that of the pnp-A mutant (Figures 7D to 7F), as shown by the drastic growth inhibition, chlorosis, and enhanced expression of PR genes. Taken together, these results support a biological function of the PNP-A/PNP-R2 pair in curtailing SA-mediated stress responses.
Figure 6.
PNP-R2 Counteracts the SA-Mediated lsd1 RCD and Immune Responses.
(A) Insertion positions of T-DNAs in two Arabidopsis pnp-r2 mutant alleles, pnp-r2-1 and pnp-r2-2. It should be noted that PNP-R2 does not contain any introns.
(B) Wild-type (WT), lsd1, lsd1 pnp-r2-1, lsd1 pnp-r2-2, and lsd1 pnp-r1 plants were grown under CL, and the emergence and spread of RCD were monitored at the indicated time points. Same scale images of representative plants are shown.
(C) Degree of RCD in the leaves from each genotype grown under CL for 22 d was visualized by TB staining.
(D) and (E) For measurements of ion leakage (D) and Fv/Fm (E), first or second leaves from each genotype grown under CL were harvested at the indicated time points. Ten leaves per genotype were used for the measurement of Fv/Fm. Value in (D) represents means ± sd (n = 3).
(F) Relative expression levels of PR1 and PR2 were determined using RT-qPCR. ACT2 was used as an internal standard. Value represents means ± sd (n = 3). Lowercase letters indicate statistically significant differences between mean values (P < 0.01, one-way ANOVA with post hoc Tukey’s honestly significant difference [HSD] test; Supplemental Data Set).
Figure 7.
pnp-r2 Mutant Plants Are Insensitive to PNP-A and Hypersensitive to SA.
(A) to (C) Seventeen-day-old wild-type (WT), lsd1, lsd1 pnp-A, and lsd1 pnp-r2-2 plants grown under CL were treated with 1 µM scrambled, active (PNP-A), or mutated (MuPNP-A) form of AtPNP-A synthetic peptide and kept for 7 d under CL. Afterward, the RCD phenotype (A), ion leakage (B), and Fv/Fm (C) were examined. The representative images are shown at the same scale. For the measurement of Fv/Fm, 10 leaves per genotype were used. Value in (B) represents means ± sd (n = 3).
(D) and (E) Twelve-day-old wild-type (WT), pnp-A, and pnp-r2-2 plants grown on MS medium under CL were transferred to MS medium in the absence (Mock) or presence of 0.2 mM SA and kept for 18 d under same growth condition. The representative foliar phenotype (D) and plant size (E) of each genotype are shown. For the measurement of plant size, 15 plants per genotype were used. Value represents means ± sd (n = 15).
(F) Plants of the indicated genotypes grown under CL for 14 d were sprayed with either a 0.5 mM SA solution (+SA) or a mock solution (–SA), and foliar tissues were collected 6 h after the treatment. Expression levels of PR1 and PR2 were examined by RT-qPCR. ACT2 was used as an internal standard. Value represents means ± sd (n = 3). Lowercase letters in (B), (C), (E), and (F) indicate statistically significant differences between mean values (P < 0.05, one-way ANOVA with post hoc Tukey’s honestly significant difference [HSD] test; Supplemental Data Set).
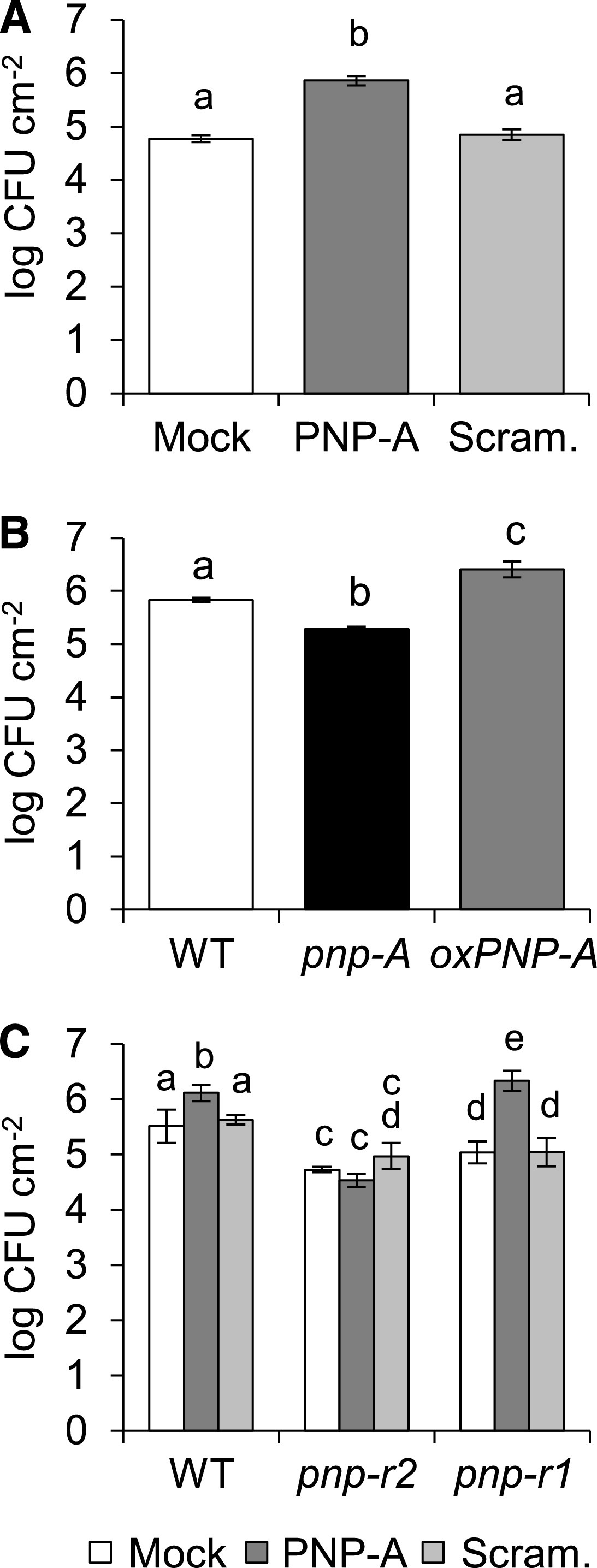
A study examining the response of plants overexpressing Arabidopsis PNP-A (AtPNP-A) to a bacterial pathogen showed that PNP-A potentiates the expression of defense-related genes, including PR1, conferring increased resistance to the infection by Pseudomonas syringae pv tomato (Pst) DC3000 (Ficarra et al., 2018). This is, however, inconsistent with our finding that PNP-A negatively regulates the expression of PR genes and other SA-responsive genes in lsd1 (Figure 2D) as well as in wild-type plants treated with SA (Figure 4C). For this reason, we examined whether PNP-A affects host resistance against Pst DC3000 in our experimental system. The exogenous application of the PNP-A synthetic peptide, but not of the scrambled peptide, compromised the resistance of wild-type plants to Pst DC3000 (Figure 8A). Furthermore, the growth of Pst DC3000 was significantly decreased in the pnp-A mutant, while elevated in oxPNP-A compared with wild-type plants (Figure 8B), providing further evidence that PNP-A negatively regulates plant defense responses. Bacterial growth was affected by exogenous PNP-A treatment in the pnp-r1 mutant, but not in the pnp-r2-2 mutant (Figure 8C). Hence, our results strongly suggest that the PNP-A/PNP-R2 pair may activate an intracellular signaling pathway to ultimately counteract SA signaling, contributing to the modulation of the SA-mediated plant stress responses.
Figure 8.
PNP-A Peptide Enhances Plant Susceptibility to P. syringae pv tomato DC3000 in a PNP-R2–Dependent Manner.
(A) to (C) Bacterial growths in 5-week-old, short-day-grown wild-type (WT) plants shown in (A), WT, pnp-A, and oxPNP-A #6-7 plants shown in (B), and WT, pnp-r2-2, and pnp-r1 plants shown in (C). Plants in (A) and (C) were pretreated with 5 μM synthetic PNP-A peptide (PNP-A) or scrambled peptide (Scram.) 6 h prior to bacterial infiltration. Three days later, bacteria were extracted from three different leaves of four independent plants and incubated at 28°C for 2 d to evaluate growth. Bars represent se of n = 4. Lowercase letters indicate statistically significant differences between mean values (P < 0.01, one-way ANOVA with post hoc Bonferroni’s multiple comparison test; Supplemental Data Set). These experiments were repeated thrice with similar results. CFU, colony-forming units.
The discrepancy between our results and those presented in Ficarra et al. (2018) could be attributable to the different methods used for bacterial inoculation: Ficarra et al. (2018) used a surface inoculation method in which bacteria must first overcome stomatal immunity, but we directly infiltrated Pst DC3000 into the apoplast, bypassing this initial barrier (Melotto et al., 2008; Zeng et al., 2011; Doehlemann and Hemetsberger, 2013). To explore this possibility, mature plants of wild type, pnp-A, and oxPNP-A were surface inoculated with Pst DC3000. Interestingly, following surface inoculation, the pnp-A mutant was more susceptible to the bacteria than wild-type plants, while oxPNP-A plants showed increased resistance (Supplemental Figure 8A), in agreement with previous results (Ficarra et al., 2018).
The opposing results obtained when using bacterial infiltration versus surface inoculation point at a potential impact of PNP-A on stomatal invasion, either indirectly through an effect on development or directly through an effect on the response to the invading pathogen. Since stomatal density was not affected in the pnp-A mutant or overexpression lines (Supplemental Figure 8B), we tested stomatal closure in response to a Pst DC3000 suspension. Strikingly, the pnp-A mutant displayed diminished stomatal closure in response to the bacteria, while the oxPNP-A line closed its stomata more efficiently (Supplemental Figure 8C); the unaffected stomatal closure in response to exogenous ABA treatment rules out a general effect on stomatal movement (Supplemental Figure 8C). The exact mechanism underlying this effect of PNP-A on stomatal responses, nevertheless, remains elusive.
Shared Functionality between PNP-R1 and PNP-R2
The previously identified PNP-A receptor PNP-R1 has been implicated in regulating intracellular cGMP levels in response to extracellular PNP-A peptide, which appears to change water homeostasis (Turek and Gehring, 2016). The lack of impact of the loss of PNP-R1 on the lsd1 RCD is indicative of nonoverlapping functions between PNP-R1 and PNP-R2. Therefore, we examined whether PNP-R2 can regulate intracellular cGMP levels, similarly to PNP-R1, in response to extracellular PNP-A peptide. Since the protoplast swelling response to PNP-A was shown to be PNP-R1 dependent and cGMP dependent (Wang et al., 2007; Gottig et al., 2008; Turek and Gehring, 2016), the impact of PNP-A on protoplasts isolated from the wild-type, pnp-r1, and pnp-r2 plants was analyzed by measuring the diameter of each protoplast. As anticipated, overexpression of PNP-A or exogenous application of synthetic PNP-A led to an increase in protoplast volume in the wild-type background. By contrast, the volume of protoplasts isolated from either pnp-r1 or pnp-r2 mutant plants remained unchanged (Supplemental Figure 9), indicating that both PNP-R1 and PNP-R2 are required for modulating water homeostasis upon the perception of PNP-A. This finding also suggests that the signaling involved in the regulation of water homeostasis is distinct to that antagonizing the SA-mediated stress responses.
DISCUSSION
The PNP-A gene is transcriptionally upregulated in response to abiotic stresses, including UV-B, salt, osmotic stress, nutrient deficiencies, and ozone (Meier et al., 2008), indicating that PNP-A may modulate plant responses to a multitude of environmental factors. We also found the rapid upregulation of PNP-A in lsd1 mutant plants prior to the onset of RCD (Figures 1A and 1B), which is triggered by various biotic and abiotic stress factors, such as excess light, red light, UV radiation, root hypoxia, cold, and bacterial infection (Dietrich et al., 1994; Rustérucci et al., 2001; Mateo et al., 2004; Mühlenbock et al., 2008; Huang et al., 2010; Chai et al., 2015; Wituszyńska et al., 2015). Since SA is a prime stress hormone in developing lsd1 RCD, all those stress factors might increase cellular SA content in the lsd1 mutant. In fact, a large-scale coexpression analysis indicates that PNP-A is coexpressed with genes associated with the SA-dependent systemic acquired resistance pathway (Meier et al., 2008).
Accordingly, our results reveal a molecular pathway in which the PNP-A peptide negatively regulates SA-mediated plant immune responses. Upon SA- and NPR1-dependent transcriptional upregulation of PNP-A (Figures 1C and 1D), the PNP-A peptide secreted in the apoplast physically interacts with its PM-localized putative receptor protein PNP-R2 (Figure 5). The PNP-A/PNP-R2 pair inhibits SA-mediated signaling, therefore antagonizing the SA-triggered RCD in the lsd1 mutant (Figures 2 and 6) as well as the SA-dependent growth retardation in wild-type plants (Figures 4 and 7), and increases the plant susceptibility to a virulent bacterial pathogen inoculated in the apoplast (Figure 8). Thus, we propose that both PNP-A and PNP-R2 may play an important role in fine-tuning plant immune responses to avoid inappropriate induction of SA-dependent death signals in cells spatially separated from infected or damaged cells, thereby minimizing tissue damage.
An earlier report showed that the PNP-like protein XacPNP encoded by Xanthomonas axonopodis pv citri alleviates the formation of necrotic lesions in the infected host by sustaining photosynthesis efficiency and net water flux in plant cells, which provides a favorable environment for bacterial propagation (Gottig et al., 2008). Accordingly, the loss of Xanthomonas XacPNP mitigates bacterial growth by reinforcing the formation of necrotic lesions, a process wherein SA is known to be a vital factor (Malamy et al., 1992; Delaney et al., 1994; Mur et al., 1997; Nawrath and Métraux, 1999). Given the results presented here, it is tempting to speculate that, in host plants, XacPNP counteracts SA-mediated immune responses to enable successful bacterial colonization. Interestingly, we observe a negative impact of PNP-A on plant resistance to Pst DC3000 when the bacteria are infiltrated into the leaf (Figure 8B). A different scenario arises when bacterial surface inoculation is used instead: in this case, PNP-A enhances plant resistance to Pst DC3000, most likely due to an increase in stomatal closure in response to the pathogen (Supplemental Figures 8A and 8C). PNP-A does not, however, affect stomatal closure in response to exogenous treatment with ABA, indicating that stomatal closure per se is not affected by the peptide (Supplemental Figure 8C). These results suggest that PNP-A plays a role in the stomatal responses to a biotic threat, although at this point the underlying molecular mechanism remains to be determined.
PNP-R1, a PM-located PNP-A receptor, catalyzes the conversion of GTP to cGMP, causing an increase in protoplast volume. Similar to PNP-R1, we found that PNP-R2 also plays a role in regulating water homeostasis (Supplemental Figure 9). However, unlike PNP-R2, PNP-R1 does not appear to function in the SA-triggered PNP-A signaling pathway (Figures 6 and 8C). The functional diversification of PNP-R2 relative to PNP-R1 suggests a high level of complexity of PNP-A signaling in plants, since different receptors may be mediating distinct responses. In many cases, RLPs act coordinately with other LRR proteins harboring intracellular signaling domains (Jeong et al., 1999; Nadeau and Sack, 2002; Hirakawa et al., 2017). Thus, PNP-R2 might form a complex with an LRR-type coreceptor protein to mediate the cellular response to the inbound factor PNP-A. Finding the potential coreceptor of PNP-R2 would facilitate unveiling downstream signaling components, which will pave the way to the dissection of the PNP-A signaling pathway in response to SA in different cell types.
METHODS
Plant and Growth Conditions
All the Arabidopsis (Arabidopsis thaliana) genotypes used in this study are of the Columbia ecotype. Arabidopsis mutant seeds of lsd1-2 (SALK_042687; Lv et al., 2019), pnp-A (SALK_000951; Turek and Gehring, 2016; Ficarra et al., 2018), npr1 (SALK_204100), pnp-r2-1 (GABIseq_255A11.1), pnp-r2-2 (SALKseq_6749.3), and pnp-r1 (GABI-KAT_180G04; Turek and Gehring, 2016) were obtained from the Nottingham Arabidopsis Stock Centre. We generated and genotyped double mutants by crossing homozygous single mutant plants and using appropriate primers (Supplemental Table 2). Seeds were surface sterilized by soaking in 1.6% (v/v) hypochlorite solution for 10 min, followed by washing five times with sterile water. Seeds were then plated on Murashige and Skoog (MS) medium (Duchefa Biochemie) containing 0.65% (w/v) agar (Duchefa Biochemie). After a 3-d stratification at 4°C in darkness, seeds were placed in a growth chamber (CU-41L4; Percival Scientific) under continuous light (CL) at 22 ± 2°C. The light intensity was maintained at 100 µmol·m−2·s−1 (light from cool-white fluorescent bulbs). For pathogen infection assays, plants were grown on jiffy pellets in a controlled environment chamber under short-day conditions (8-h-light/16-h-dark cycle) at 20 to 22°C. Four-week-old Nicotiana benthamiana plants grown in a controlled growth chamber under long-day conditions (16-h-light/8-h-dark cycle) at 25°C were used for all transient assays.
RNA Extraction and RT-qPCR
Total RNA (1 µg) extracted from foliar tissues using the Spectrum Plant Total RNA Kit (Sigma-Aldrich) was reverse transcribed with the HiScript II Q RT SuperMix for qPCR (Vazyme Biotech) according to the manufacturer’s recommendations. The RT-qPCR was conducted in triplicates on a QuantStudio 6 Flex Real-Time PCR System (Applied Biosystems) with SYBR Green Master Mix (Vazyme Biotech). Relative transcript levels were calculated with the ddCt method (Livak and Schmittgen, 2001) and normalized to the ACTIN2 (At3g18780) transcript levels. The sequences of the primers used in this study are listed in Supplemental Table 2.
Generation of Arabidopsis Transgenic Lines
The stop-codon-less coding sequences (CDSs) of PNP-A and PNP-R2 were cloned into the pDONR221 Gateway vector (Thermo Fisher Scientific) via the Gateway BP reaction (Thermo Fisher Scientific) and subsequently recombined into the Gateway-compatible plant binary vector pGWB651 for C-terminal fusion with GFP or pGWB617 for C-terminal fusion with 4×Myc (Nakagawa et al., 2007) via the Gateway LR reaction (Thermo Fisher Scientific) to create p35S:PNP-A-GFP and p35S:PNP-R2-4×Myc constructs. The generated vectors were transformed into the Agrobacterium tumefaciens strain GV3101 using the heat shock method. After generating Arabidopsis transgenic plants in wild-type, lsd1, or lsd1 pnp-r2 background using Agrobacterium-mediated transformation by the floral dip method (Clough and Bent, 1998), homozygous T3 transgenic plants were selected on MS medium containing 12.5 mg/L Basta (Sigma-Aldrich).
BiFC Assay
BiFC assays were performed with a split-YFP system in N. benthamiana leaves, as previously described by Lu et al. (2010). In brief, the pDONR/Zeo entry vectors (Thermo Fisher Scientific) containing the 207-bp fragment of PNP-A CDS (tPNP-A, counted from start codon) and the stop-codon-less full-length CDS of PNP-R2 were recombined into the split-YFP vectors pGTQL1211YN and 1221YC, respectively, through the Gateway LR reaction (Thermo Fisher Scientific). For the BiFC assay, A. tumefaciens mixtures carrying the appropriate BiFC constructs were infiltrated with a 1-mL needle-less syringe into the abaxial side of 4-week-old N. benthamiana leaves. After 72 h, the presence of the YFP signal was evaluated with a CS SP8 single molecule detection confocal microscope (Leica Microsystems). BiFC construct expressing mutated tPNP-A fused to YFPN (MutPNP-A-YFPN, substituting two Cys’s with Ser, as shown in Supplemental Figure 4) was used as a negative control.
Co-IP Assay
For the Co-IP assay, the pDONR/Zeo entry vector containing the 207-bp tPNP-A CDS fragment or the stop-codon-less full-length CDS of PNP-R2 was recombined into the destination vector pGWB651 for C-terminal fusion with GFP or pGWB617 for C-terminal fusion with 4×Myc through the Gateway LR reaction (Thermo Fisher Scientific) to create p35S:tPNP-A-GFP and p35S:PNP-R2-4×Myc. One or both of the vectors were expressed alone or coexpressed in 4-week-old N. benthamiana leaves after Agrobacterium infiltration. Total protein was extracted with IP buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 10% [v/v] glycerol, 1.0 mM EDTA, 1% [v/v] Triton X-100, 1 mM Na2MoO4·2H2O, 1 mM NaF, 1.5 mM Na3VO4, 1 mM phenylmethylsulfonyl fluoride, and 1× cOmplete protease inhibitor cocktail [Roche]). After protein extraction, 20 μL of Myc-Trap magnetic agarose beads (Myc-TrapMA; Chromotek) was incubated with 40 mg of the total protein extract for 12 h at 4°C by vertical rotation. The beads were washed five times with washing buffer (IP buffer with 0.1% [v/v] Triton X-100) and then eluted with 2× SDS protein sample buffer for 10 min at 95°C. The eluates were subjected to 10% (w/v) SDS-PAGE gels, and the interaction between coexpressed proteins was determined by immunoblot analysis using a mouse anti-Myc monoclonal antibody (1:10,000, catalog no. 2276; Cell Signaling Technology) and a mouse anti-GFP monoclonal antibody (1:5000, catalog no. 118144600001; Roche).
Preparation of Peptides
N-Terminal biotin-labeled PNP-A peptide and its mutated and scrambled peptide controls were synthesized and purified to >95% purity with HPLC by Sangon Biotech. Peptides were dissolved in 10% (v/v) acetic acid (1.5 mM stock solution) and diluted with distilled water to the desired concentrations before use. The amino acid sequences of peptides are shown in Supplemental Figure 4.
Peptide Pull-Down Assay and Mass Spectrometry Analysis
Twenty-day-old lsd1 mutant plants grown under CL on MS medium were harvested and homogenized to a fine powder in liquid nitrogen using mortar and pestle. Approximately 10 g of fine powder was used to extract total protein with the IP buffer as mentioned above. After protein extraction, 100 μL of Dynabeads M-280 Streptavidin beads (Thermo Fisher Scientific) prebound with or without the N-terminal biotinylated PNP-A peptide or its scrambled peptide was incubated with 100 mg of the total protein extract for 12 h at 4°C by vertical rotation. The beads were washed five times with the washing buffer (IP buffer with 0.1% [v/v] Triton X-100) and then washed two times with 1× PBS buffer. After protein elution from the beads with SDT-lysis buffer (100 mM Tris-HCl, pH 7.6, 4% [w/v] SDS, and 0.1 M DTT), the eluates were subjected to mass spectrometry analysis as previously described by Wang et al. (2016).
Pull-down assays were also conducted with N. benthamiana leaves harboring the p35S:PNP-R2-4×Myc or p35S:PNP-R1-4×Myc after Agrobacterium infiltration. Total protein was extracted with the IP buffer from 5 g of N. benthamiana leaves. After the pull-down assay, as described above, the eluates were subjected to NuPAGE Bis-Tris 4 to 12% protein gel (Thermo Fisher Scientific), and the interaction between PNP-R2 (or PNP-R1) and the PNP-A synthetic peptide was determined by immunoblot analysis using a mouse anti-Myc monoclonal antibody (1:10,000, catalog no. 2276; Cell Signaling Technology) and a mouse anti-biotin monoclonal antibody (1:2000, catalog no. B7653; Sigma-Aldrich).
Subcellular Localization and Confocal Laser-Scanning Microscopy
The pDONR/Zeo entry vectors (Thermo Fisher Scientific) containing the stop-codon-less CDS of PNP-R2 was recombined into the destination vector pGWB641 for C-terminal fusion with YFP through the Gateway LR reaction (Thermo Fisher Scientific) to create p35S:PNP-R2-YFP. To determine the subcellular localization of PNP-R2 and tPNP-A, the p35S:PNP-R2-YFP and p35S:tPNP-A-GFP constructs were transformed into A. tumefaciens strain GV3101 and transiently expressed in N. benthamiana leaves. The GFP, YFP, and FM4-64 fluorescence signals were detected by confocal laser-scanning microscopy analysis using TCS SP8 single molecule detection microscope (Leica Microsystems) 72 h after infiltration. All the images were acquired and processed using LAS AF Lite software version 2.6.3 (Leica Microsystems). Cell plasmolysis was performed by treatment with 0.8 M mannitol for 30 min.
Determination of Cell Death
Cell death was visualized and quantified using trypan blue (TB) staining and electrolyte leakage measurements, as previously described by Lv et al. (2019). Briefly, first or second leaves from plants grown under CL on MS medium were submerged in TB staining solution (10 g of phenol, 10 mL of glycerol, 10 mL of lactic acid, and 0.02 g of TB in 10 mL of water), diluted with ethanol 1:2 (v/v), and boiled for 2 min in a water bath. After a 16-h incubation at room temperature on a vertical shaker, nonspecific staining was removed with a destaining buffer (250 g of chloral hydrate in 100 mL of water). Finally, plant tissues were kept in 50% (v/v) glycerol for imaging. For the measurement of electrolyte leakage, 10 first or second leaves from independent plants were harvested at the indicated time points and transferred to a 15-mL tube containing 6 mL of water purified using a Milli-Q Integral 5 water purification system (Millipore). After a 6-h incubation at room temperature on a horizontal shaker, the conductivity of the solution was measured with an Orion Star A212 conductivity meter (Thermo Fisher Scientific). This experiment was repeated three times with similar results.
Protoplast Swelling Assays
Leaf protoplasts were isolated from 3-week-old Arabidopsis plants grown on MS medium under CL, as previously described by Yoo et al. (2007). The isolated protoplasts were kept in osmotic solution (0.4 M mannitol, 3 mM MES, and 7 mM CaCl2, pH 5.7) on ice for 1 h. After treating with 1 µM of the PNP-A peptide or its scrambled peptide for 30 min at room temperature, photographs of protoplasts were taken using a camera on a light microscope (DM750; Leica Microsystems). The diameter of each protoplast was measured using the ImageJ software and used to calculate its volume assuming a spherical shape of the protoplasts. At least 50 protoplasts per each genotype were monitored, and the experiment was repeated thrice with a similar result.
SA Measurements
The endogenous free SA levels in 17-d-old plants of the wild type, lsd1, lsd1 pnp-A, and lsd1 ox PNP-A grown on MS medium under CL were measured as previously described by Lv et al. (2019).
Determination of Fv/Fm
The Fv/Fm was determined with the FluorCam system (FC800-C/1010GFP; Photon Systems Instruments) containing a charge-coupled device camera and an irradiation system according to the instrument manufacturer’s instructions.
Measurements of Stomatal Closure and Density
Stomatal aperture was measured as previously described by Macho et al. (2012), with minor modifications. Briefly, first or second leaves of 3-week-old plants were submerged in a stomatal opening solution (10 mM MES-KOH, pH 6.15, 10 µM CaCl2, 50 mM KCl, and 0.01% [v/v] Tween 20). After 2-h incubation under white light, Pst DC3000 (OD600 = 0.1), 5 µM ABA (Sigma-Aldrich), or mock solution was added to the buffer, and the leaf samples were further incubated under the same conditions for 1 h. Abaxial leaf surfaces were imaged with an upright microscope (Axio Imager M2; Carl Zeiss), and stomatal aperture was calculated by dividing the aperture width by the length. To determine stomatal density, the number of stomata on abaxial leaf surfaces (at least 10 leaves for each genotype) was measured using a scanning electron microscope (TM3000; Hitachi).
Extraction of Apoplastic Proteins
Apoplastic proteins from 3-week-old Arabidopsis wild-type and lsd1 plants grown under CL on MS medium were extracted as previously described by Rutter and Innes (2017), with minor modifications. Briefly, plants were vacuum infiltrated with extraction buffer (20 mM MES, 0.1 M NaCl, and 2 mM CaCl2, pH 6.0) for 2 min. The infiltrated plant samples were carefully blotted with filter paper to remove surface moisture and placed inside 20-mL syringes. After centrifuging in 50-mL Falcon tubes at 700g for 20 min at 4°C, the resulting apoplastic fluids were centrifuged again at 15,000g for 5 min. Protein contents in the supernatants were estimated, and equal amounts of proteins were separated on SDS-PAGE. Gel slices were cut just below 20-kD size mark and were subjected to in-gel digestion followed by mass spectrometry analysis, as previously described by Duan et al. (2019). The levels of PNP-A, PR1, and PR2 were quantified using the relative intensity values obtained using MaxQuant software (version 1.5.8.3) with a label-free quantitation algorithm (Luber et al., 2010; Schwanhäusser et al., 2011).
Induced Resistance
Induced resistance assays were performed as previously described by Lozano-Durán et al. (2013), with slight modifications. Briefly, sterilized seeds were sown on MS medium containing 0.7% (w/v) agar. After a 3-d stratification at 4°C in darkness, seeds were grown in a controlled growth chamber in short-day (8-h-light/16-h-dark) conditions at 22°C. Ten-day-old seedlings were transferred to jiffy pellets and grown under the same conditions as mentioned above. For spray-mediated inoculations, 3-week-old plants were spray inoculated with a Pst DC3000 inoculum (OD600 = 0.2 in 10 mM MgCl2 with 0.02% [v/v] Silwet L-77) and kept covered for 24 h. For each treatment, the whole aerial part of the plants was used, and bacterial growth was determined according to the fresh weight. For infiltration-mediated inoculations, 5-week-old plants were infiltrated with 5 µM solutions of the PNP-A peptide or its scrambled peptide control with a needle-less syringe into the abaxial side of leaves 6 h prior to bacterial infection (Pst DC3000, OD600 = 0.0002 in 10 mM MgCl2) and kept cover for 24 h. Bacterial growth was determined 3 d after inoculation by plating 1:10 serial dilutions of leaf extracts; plates were incubated at 28°C for 2 d before the bacterial colony-forming units were counted. For each treatment, 7-mm leaf discs of three leaves from four independent plants were used. This experiment was repeated thrice with similar results; results from one experiment are shown.
Accession Numbers
Sequence information of the genes studied in this article can be found in the Arabidopsis The Arabidopsis Information Resource database (https://www.arabidopsis.org) under the following accession numbers: EDS1 (At3g48090), ICS1 (At1g74710), LSD1 (At4g20380), NPR1 (At1g64280), PAD4 (At3g52430), PNP-A (At2g18660), PNP-R1 (At1g33612), PNP-R2 (At5g12940), PR1 (At2g14610), PR2 (At3g57260).
SUPPLEMENTAL DATA
Supplemental Figure 1. Two independent wild-type transgenic lines overexpressing PNP-A.
Supplemental Figure 2. Effect of loss of or overexpression of PNP-A on lsd1 RCD.
Supplemental Figure 3. Mass spectrometry (MS) analysis of apoplast proteins reveals significant accumulation of PNP-A in lsd1 mutant.
Supplemental Figure 4. Schematic illustration of the domain structure of Arabidopsis PNP-A (AtPNP-A).
Supplemental Figure 5. Effect exerted by exogenously applied PNP-A peptide on SA-mediated growth inhibition.
Supplemental Figure 6. PNP-R2, but not PNP-R1, interacts with PNP-A peptide.
Supplemental Figure 7. The PNP-R2-Myc fusion protein compromises the enhanced RCD in lsd1 pnp-r2.
Supplemental Figure 8. Effect of PNP-A on bacterial growth and stomatal closure after spray inoculation and stomatal density.
Supplemental Figure 9. Effect of PNP-R2 on the PNP-A-dependent protoplast volume changes.
Supplemental Table 1. List of proteins interacting with the PNP-A synthetic peptides.
Supplemental Table 2. List of primer sets used in this study.
Supplemental Data Set. Statistical analysis data.
DIVE Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
Acknowledgments
We thank the Core Facility of Proteomics and Rongxia Li (Shanghai Center for Plant Stress Biology) for carrying out mass spectrometry. We thank Junghee Lee (retired) for critical reading of the article. This research was supported by the Strategic Priority Research Program of the Chinese Academy of Sciences (Strategic Priority Research Program grant XDB27040102 and the 100-Talents Program) and the National Natural Science Foundation of China (grant 31871397) to C.K.
AUTHOR CONTRIBUTIONS
K.P.L., K.L., E.Y.K., L.M.-P., H.D., J.D., M.L., V.D., Y.L., R.L., and Z.L. conducted the experiments; K.P.L., K.L., E.Y.K., L.M.-P., R.L.-D., and C.K. designed the research; K.P.L., K.L., E.Y.K., L.M.-P., H.D., V.D., R.L.-D., and C.K. analyzed the data; K.P.L., R.L.-D., and C.K. wrote the article. All authors reviewed and edited the article.
References
- Aviv D.H., Rustérucci C., Holt B.F. III, Dietrich R.A., Parker J.E., Dangl J.L.(2002). Runaway cell death, but not basal disease resistance, in lsd1 is SA- and NIM1/NPR1-dependent. Plant J. 29: 381–391. [DOI] [PubMed] [Google Scholar]
- Butenko M.A., Vie A.K., Brembu T., Aalen R.B., Bones A.M.(2009). Plant peptides in signalling: looking for new partners. Trends Plant Sci. 14: 255–263. [DOI] [PubMed] [Google Scholar]
- Cao H., Glazebrook J., Clarke J.D., Volko S., Dong X.(1997). The Arabidopsis NPR1 gene that controls systemic acquired resistance encodes a novel protein containing ankyrin repeats. Cell 88: 57–63. [DOI] [PubMed] [Google Scholar]
- Chai T., Zhou J., Liu J., Xing D.(2015). LSD1 and HY5 antagonistically regulate red light induced-programmed cell death in Arabidopsis. Front Plant Sci 6: 292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F.(1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743. [DOI] [PubMed] [Google Scholar]
- Delaney T.P., Uknes S., Vernooij B., Friedrich L., Weymann K., Negrotto D., Gaffney T., Gut-Rella M., Kessmann H., Ward E., Ryals J.(1994). A central role of salicylic acid in plant disease resistance. Science 266: 1247–1250. [DOI] [PubMed] [Google Scholar]
- Dietrich R.A., Delaney T.P., Uknes S.J., Ward E.R., Ryals J.A., Dangl J.L.(1994). Arabidopsis mutants simulating disease resistance response. Cell 77: 565–577. [DOI] [PubMed] [Google Scholar]
- Dietrich R.A., Richberg M.H., Schmidt R., Dean C., Dangl J.L.(1997). A novel zinc finger protein is encoded by the Arabidopsis LSD1 gene and functions as a negative regulator of plant cell death. Cell 88: 685–694. [DOI] [PubMed] [Google Scholar]
- Ding Y., Dommel M., Mou Z.(2016). Abscisic acid promotes proteasome-mediated degradation of the transcription coactivator NPR1 in Arabidopsis thaliana. Plant J. 86: 20–34. [DOI] [PubMed] [Google Scholar]
- Doehlemann G., Hemetsberger C.(2013). Apoplastic immunity and its suppression by filamentous plant pathogens. New Phytol. 198: 1001–1016. [DOI] [PubMed] [Google Scholar]
- Duan J., et al. (2019). Impaired PSII proteostasis promotes retrograde signaling via salicylic acid. Plant Physiol. 180: 2182–2197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ficarra F.A., Grandellis C., Garavaglia B.S., Gottig N., Ottado J.(2018). Bacterial and plant natriuretic peptides improve plant defence responses against pathogens. Mol. Plant Pathol. 19: 801–811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn R.D., et al. (2017). InterPro in 2017-beyond protein family and domain annotations. Nucleic Acids Res. 45 (D1): D190–D199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita M., Fujita Y., Noutoshi Y., Takahashi F., Narusaka Y., Yamaguchi-Shinozaki K., Shinozaki K.(2006). Crosstalk between abiotic and biotic stress responses: A current view from the points of convergence in the stress signaling networks. Curr. Opin. Plant Biol. 9: 436–442. [DOI] [PubMed] [Google Scholar]
- Gehring C., Turek I.S.(2017). Cyclic nucleotide monophosphates and their cyclases in plant signaling. Front Plant Sci 8: 1704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottig N., Garavaglia B.S., Daurelio L.D., Valentine A., Gehring C., Orellano E.G., Ottado J.(2008). Xanthomonas axonopodis pv. citri uses a plant natriuretic peptide-like protein to modify host homeostasis. Proc. Natl. Acad. Sci. USA 105: 18631–18636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirakawa Y., Torii K.U., Uchida N.(2017). Mechanisms and strategies shaping plant peptide hormones. Plant Cell Physiol. 58: 1313–1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X., Li Y., Zhang X., Zuo J., Yang S.(2010). The Arabidopsis LSD1 gene plays an important role in the regulation of low temperature-dependent cell death. New Phytol. 187: 301–312. [DOI] [PubMed] [Google Scholar]
- Jabs T., Dietrich R.A., Dangl J.L.(1996). Initiation of runaway cell death in an Arabidopsis mutant by extracellular superoxide. Science 273: 1853–1856. [DOI] [PubMed] [Google Scholar]
- Jeong S., Trotochaud A.E., Clark S.E.(1999). The Arabidopsis CLAVATA2 gene encodes a receptor-like protein required for the stability of the CLAVATA1 receptor-like kinase. Plant Cell 11: 1925–1934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Käll L., Krogh A., Sonnhammer E.L.L.(2004). A combined transmembrane topology and signal peptide prediction method. J. Mol. Biol. 338: 1027–1036. [DOI] [PubMed] [Google Scholar]
- Kaminaka H., Näke C., Epple P., Dittgen J., Schütze K., Chaban C., Holt B.F. III, Merkle T., Schäfer E., Harter K., Dangl J.L.(2006). bZIP10-LSD1 antagonism modulates basal defense and cell death in Arabidopsis following infection. EMBO J. 25: 4400–4411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karasov T.L., Chae E., Herman J.J., Bergelson J.(2017). Mechanisms to mitigate the trade-off between growth and defense. Plant Cell 29: 666–680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kliebenstein D.J., Dietrich R.A., Martin A.C., Last R.L., Dangl J.L.(1999). LSD1 regulates salicylic acid induction of copper zinc superoxide dismutase in Arabidopsis thaliana. Mol. Plant Microbe Interact. 12: 1022–1026. [DOI] [PubMed] [Google Scholar]
- Koller K.J., Goeddel D.V.(1992). Molecular biology of the natriuretic peptides and their receptors. Circulation 86: 1081–1088. [DOI] [PubMed] [Google Scholar]
- Krogh A., Larsson B., von Heijne G., Sonnhammer E.L.L.(2001). Predicting transmembrane protein topology with a hidden Markov model: Application to complete genomes. J. Mol. Biol. 305: 567–580. [DOI] [PubMed] [Google Scholar]
- Lease K.A., Walker J.C.(2006). The Arabidopsis unannotated secreted peptide database, a resource for plant peptidomics. Plant Physiol. 142: 831–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S., Kim S.G., Park C.M.(2010). Salicylic acid promotes seed germination under high salinity by modulating antioxidant activity in Arabidopsis. New Phytol. 188: 626–637. [DOI] [PubMed] [Google Scholar]
- Li J., Brader G., Palva E.T.(2004). The WRKY70 transcription factor: A node of convergence for jasmonate-mediated and salicylate-mediated signals in plant defense. Plant Cell 16: 319–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak K.J., Schmittgen T.D.(2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25: 402–408. [DOI] [PubMed] [Google Scholar]
- Lozano-Durán R., Macho A.P., Boutrot F., Segonzac C., Somssich I.E., Zipfel C.(2013). The transcriptional regulator BZR1 mediates trade-off between plant innate immunity and growth. eLife 2: e00983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Q., Tang X., Tian G., Wang F., Liu K., Nguyen V., Kohalmi S.E., Keller W.A., Tsang E.W., Harada J.J., Rothstein S.J., Cui Y.(2010). Arabidopsis homolog of the yeast TREX-2 mRNA export complex: components and anchoring nucleoporin. Plant J. 61: 259–270. [DOI] [PubMed] [Google Scholar]
- Luber C.A., Cox J., Lauterbach H., Fancke B., Selbach M., Tschopp J., Akira S., Wiegand M., Hochrein H., O’Keeffe M., Mann M.(2010). Quantitative proteomics reveals subset-specific viral recognition in dendritic cells. Immunity 32: 279–289. [DOI] [PubMed] [Google Scholar]
- Ludidi N., Gehring C.(2003). Identification of a novel protein with guanylyl cyclase activity in Arabidopsis thaliana. J. Biol. Chem. 278: 6490–6494. [DOI] [PubMed] [Google Scholar]
- Ludidi N., Morse M., Sayed M., Wherrett T., Shabala S., Gehring C.(2004). A recombinant plant natriuretic peptide causes rapid and spatially differentiated K+, Na+ and H+ flux changes in Arabidopsis thaliana roots. Plant Cell Physiol. 45: 1093–1098. [DOI] [PubMed] [Google Scholar]
- Lv R., Li Z., Li M., Dogra V., Lv S., Liu R., Lee K.P., Kim C.(2019). Uncoupled expression of nuclear and plastid photosynthesis-associated genes contributes to cell death in a lesion mimic mutant. Plant Cell 31: 210–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macho A.P., Boutrot F., Rathjen J.P., Zipfel C.(2012). Aspartate oxidase plays an important role in Arabidopsis stomatal immunity. Plant Physiol. 159: 1845–1856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malamy J., Hennig J., Klessig D.F.(1992). Temperature-dependent induction of salicylic acid and its conjugates during the resistance response to tobacco mosaic virus infection. Plant Cell 4: 359–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchler-Bauer A., et al. (2017). CDD/SPARCLE: Functional classification of proteins via subfamily domain architectures. Nucleic Acids Res. 45 (D1): D200–D203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maryani M.M., Morse M.V., Bradley G., Irving H.R., Cahill D.M., Gehring C.A.(2003). In situ localization associates biologically active plant natriuretic peptide immuno-analogues with conductive tissue and stomata. J. Exp. Bot. 54: 1553–1564. [DOI] [PubMed] [Google Scholar]
- Mateo A., Funck D., Mühlenbock P., Kular B., Mullineaux P.M., Karpinski S.(2006). Controlled levels of salicylic acid are required for optimal photosynthesis and redox homeostasis. J. Exp. Bot. 57: 1795–1807. [DOI] [PubMed] [Google Scholar]
- Mateo A., Mühlenbock P., Rustérucci C., Chang C.C., Miszalski Z., Karpinska B., Parker J.E., Mullineaux P.M., Karpinski S.(2004). LESION SIMULATING DISEASE 1 is required for acclimation to conditions that promote excess excitation energy. Plant Physiol. 136: 2818–2830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsubayashi Y.(2011). Post-translational modifications in secreted peptide hormones in plants. Plant Cell Physiol. 52: 5–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsubayashi Y.(2014). Posttranslationally modified small-peptide signals in plants. Annu. Rev. Plant Biol. 65: 385–413. [DOI] [PubMed] [Google Scholar]
- Meier S., Bastian R., Donaldson L., Murray S., Bajic V., Gehring C.(2008). Co-expression and promoter content analyses assign a role in biotic and abiotic stress responses to plant natriuretic peptides. BMC Plant Biol. 8: 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melotto M., Underwood W., He S.Y.(2008). Role of stomata in plant innate immunity and foliar bacterial diseases. Annu. Rev. Phytopathol. 46: 101–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura K., Okamoto H., Okuma E., Shiba H., Kamada H., Hasegawa P.M., Murata Y.(2013). SIZ1 deficiency causes reduced stomatal aperture and enhanced drought tolerance via controlling salicylic acid-induced accumulation of reactive oxygen species in Arabidopsis. Plant J. 73: 91–104. [DOI] [PubMed] [Google Scholar]
- Morse M., Pironcheva G., Gehring C.(2004). AtPNP-A is a systemically mobile natriuretic peptide immunoanalogue with a role in Arabidopsis thaliana cell volume regulation. FEBS Lett. 556: 99–103. [DOI] [PubMed] [Google Scholar]
- Mühlenbock P., Szechynska-Hebda M., Plaszczyca M., Baudo M., Mateo A., Mullineaux P.M., Parker J.E., Karpinska B., Karpinski S.(2008). Chloroplast signaling and LESION SIMULATING DISEASE1 regulate crosstalk between light acclimation and immunity in Arabidopsis. Plant Cell 20: 2339–2356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mur L.A.J., Bi Y.M., Darby R.M., Firek S., Draper J.(1997). Compromising early salicylic acid accumulation delays the hypersensitive response and increases viral dispersal during lesion establishment in TMV-infected tobacco. Plant J. 12: 1113–1126. [DOI] [PubMed] [Google Scholar]
- Murphy E., Smith S., De Smet I.(2012). Small signaling peptides in Arabidopsis development: How cells communicate over a short distance. Plant Cell 24: 3198–3217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadeau J.A., Sack F.D.(2002). Control of stomatal distribution on the Arabidopsis leaf surface. Science 296: 1697–1700. [DOI] [PubMed] [Google Scholar]
- Nakagawa T., et al. (2007). Improved Gateway binary vectors: high-performance vectors for creation of fusion constructs in transgenic analysis of plants. Biosci. Biotechnol. Biochem. 71: 2095–2100. [DOI] [PubMed] [Google Scholar]
- Nawrath C., Métraux J.P.(1999). Salicylic acid induction-deficient mutants of Arabidopsis express PR-2 and PR-5 and accumulate high levels of camalexin after pathogen inoculation. Plant Cell 11: 1393–1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pharmawati M., Maryani M.M., Nikolakopoulos T., Gehring C.A., Irving H.R.(2001). Cyclic GMP modulates stomatal opening induced by natriuretic peptides and immunoreactive analogues. Plant Physiol. Biochem. 39: 385–394. [Google Scholar]
- Potter L.R., Hunter T.(2001). Guanylyl cyclase-linked natriuretic peptide receptors: Structure and regulation. J. Biol. Chem. 276: 6057–6060. [DOI] [PubMed] [Google Scholar]
- Rustérucci C., Aviv D.H., Holt B.F. III, Dangl J.L., Parker J.E.(2001). The disease resistance signaling components EDS1 and PAD4 are essential regulators of the cell death pathway controlled by LSD1 in Arabidopsis. Plant Cell 13: 2211–2224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutter B.D., Innes R.W.(2017). Extracellular vesicles isolated from the leaf apoplast carry stress-response proteins. Plant Physiol. 173: 728–741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruzvidzo O., Donaldson L., Valentine A., Gehring C.(2011). The Arabidopsis thaliana natriuretic peptide AtPNP-A is a systemic regulator of leaf dark respiration and signals via the phloem. J. Plant Physiol. 168: 1710–1714. [DOI] [PubMed] [Google Scholar]
- Schwanhäusser B., Busse D., Li N., Dittmar G., Schuchhardt J., Wolf J., Chen W., Selbach M.(2011). Global quantification of mammalian gene expression control. Nature 473: 337–342. [DOI] [PubMed] [Google Scholar]
- Schwartz D., Geller D.M., Manning P.T., Siegel N.R., Fok K.F., Smith C.E., Needleman P.(1985). Ser-Leu-Arg-Arg-atriopeptin III: The major circulating form of atrial peptide. Science 229: 397–400. [DOI] [PubMed] [Google Scholar]
- Scott I.M., Clarke S.M., Wood J.E., Mur L.A.(2004). Salicylate accumulation inhibits growth at chilling temperature in Arabidopsis. Plant Physiol. 135: 1040–1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah J.(2003). The salicylic acid loop in plant defense. Curr. Opin. Plant Biol. 6: 365–371. [DOI] [PubMed] [Google Scholar]
- Shiu S.H., Bleecker A.B.(2001). Receptor-like kinases from Arabidopsis form a monophyletic gene family related to animal receptor kinases. Proc. Natl. Acad. Sci. USA 98: 10763–10768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turek I., Gehring C.(2016). The plant natriuretic peptide receptor is a guanylyl cyclase and enables cGMP-dependent signaling. Plant Mol. Biol. 91: 275–286. [DOI] [PubMed] [Google Scholar]
- Rivas-San Vicente M., Plasencia J.(2011). Salicylic acid beyond defence: Its role in plant growth and development. J. Exp. Bot. 62: 3321–3338. [DOI] [PubMed] [Google Scholar]
- Van Norman J.M., Breakfield N.W., Benfey P.N.(2011). Intercellular communication during plant development. Plant Cell 23: 855–864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma V., Ravindran P., Kumar P.P.(2016). Plant hormone-mediated regulation of stress responses. BMC Plant Biol. 16: 86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan D., Li R., Zou B., Zhang X., Cong J., Wang R., Xia Y., Li G.(2012). Calmodulin-binding protein CBP60g is a positive regulator of both disease resistance and drought tolerance in Arabidopsis. Plant Cell Rep. 31: 1269–1281. [DOI] [PubMed] [Google Scholar]
- Wang D., Amornsiripanitch N., Dong X.(2006). A genomic approach to identify regulatory nodes in the transcriptional network of systemic acquired resistance in plants. PLoS Pathog. 2: e123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G., et al. (2008). A genome-wide functional investigation into the roles of receptor-like proteins in Arabidopsis. Plant Physiol. 147: 503–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Kim C., Xu X., Piskurewicz U., Dogra V., Singh S., Mahler H., Apel K.(2016). Singlet oxygen- and EXECUTER1-mediated signaling is initiated in grana margins and depends on the protease FtsH2. Proc. Natl. Acad. Sci. USA 113: E3792–E3800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y.H., Donaldson L., Gehring C., Irving H.R.(2011a). Plant natriuretic peptides: Control of synthesis and systemic effects. Plant Signal. Behav. 6: 1606–1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y.H., Gehring C., Cahill D.M., Irving H.R.(2007). Plant natriuretic peptide active site determination and effects on cGMP and cell volume regulation. Funct. Plant Biol. 34: 645–653. [DOI] [PubMed] [Google Scholar]
- Wang Y.H., Gehring C., Irving H.R.(2011b). Plant natriuretic peptides are apoplastic and paracrine stress response molecules. Plant Cell Physiol. 52: 837–850. [DOI] [PubMed] [Google Scholar]
- Wituszyńska W., Szechyńska-Hebda M., Sobczak M., Rusaczonek A., Kozłowska-Makulska A., Witoń D., Karpiński S.(2015). Lesion simulating disease 1 and enhanced disease susceptibility 1 differentially regulate UV-C-induced photooxidative stress signalling and programmed cell death in Arabidopsis thaliana. Plant Cell Environ. 38: 315–330. [DOI] [PubMed] [Google Scholar]
- Yang Y., Qi M., Mei C.(2004). Endogenous salicylic acid protects rice plants from oxidative damage caused by aging as well as biotic and abiotic stress. Plant J. 40: 909–919. [DOI] [PubMed] [Google Scholar]
- Yoo S.D., Cho Y.H., Sheen J.(2007). Arabidopsis mesophyll protoplasts: A versatile cell system for transient gene expression analysis. Nat. Protoc. 2: 1565–1572. [DOI] [PubMed] [Google Scholar]
- Zeng W., Brutus A., Kremer J.M., Withers J.C., Gao X., Jones A.D., He S.Y.(2011). A genetic screen reveals Arabidopsis stomatal and/or apoplastic defenses against Pseudomonas syringae pv. tomato DC3000. PLoS Pathog. 7: e1002291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Fan W., Kinkema M., Li X., Dong X.(1999). Interaction of NPR1 with basic leucine zipper protein transcription factors that bind sequences required for salicylic acid induction of the PR-1 gene. Proc. Natl. Acad. Sci. USA 96: 6523–6528. [DOI] [PMC free article] [PubMed] [Google Scholar]