Abstract
Prostate cancer (PCa) is a leading cause of cancer death among men, with greater prevalence of the disease among the African American population in the USA. Activator of G-protein signaling 3 (AGS3/G-protein signaling modulator 1) was shown to be overexpressed in prostate adenocarcinoma relative to the prostate gland. In this study, we investigated the correlation between AGS3 overexpression and PCa malignancy. Immunoblotting analysis and real-time quantitative-PCR showed increase in AGS3 expression in the metastatic cell lines LNCaP (~3-fold), MDA PCa 2b (~2-fold), DU 145 (~2-fold) and TRAMP-C1 (~20-fold) but not in PC3 (~1-fold), relative to control RWPE-1. Overexpression of AGS3 in PC3, LNCaP and MDA PCa 2b enhanced tumor growth. AGS3 contains seven tetratricopeptide repeats (TPR) and four G-protein regulatory (GPR) motifs. Overexpression of the TPR or the GPR motifs in PC3 cells had no effect in tumor growth. Depletion of AGS3 in the TRAMP-C1 cells (TRAMP-C1-AGS3-/-) decreased cell proliferation and delayed wound healing and tumor growth in both C57BL/6 (~3-fold) and nude mice xenografts, relative to control TRAMP-C1 cells. TRAMP-C1-AGS3-/- tumors also exhibited a marked increase (~5-fold) in both extracellular signal-regulated kinase (ERK) 1/2 and P38 mitogen-activated protein kinase (MAPK) activation, which correlated with a significant increase (~3-fold) in androgen receptor (AR) expression, relative to TRAMP-C1 xenografts. Interestingly, overexpression of AGS3 in TRAMP-C1-AGS3-/- cells inhibited ERK activation and AR overexpression as compared with control TRAMP-C1 cells. Taken together, the data indicate that the effect of AGS3 in prostate cancer development and progression is probably mediated via a MAPK/AR-dependent pathway.
Introduction
Prostate cancer (PCa) is the most prevalent type of cancer affecting men and constitutes one of the leading causes of cancer-related deaths in men (1). In 2018, it was estimated that about 165 000 new cases of prostate cancer (representing about 19% new case incidence) will be reported in the USA alone (2). The disease is race related and shows more prevalence in the African American population of the USA (1). Prostate cancer is a heterogeneous disease with complex molecular characteristics, in which tumor differentiation severity can present from a mild low-grade prostatic intraepithelial neoplasia to an aggressive adenocarcinoma form (3). Etiology of the disease spans across a complexity of factors that includes genetic, environmental, age as well as race/ethnicity (4). Prostate tissue relies on androgen, secreted mainly by its luminal cells, and its receptor (AR) for normal development and maintenance of prostate homeostasis (5,6). The AR pathway thus serves as a key mechanism utilized by prostate tumor cells for the promotion of carcinogenesis. Consequently, androgen deprivation therapy (ADT) that targets this pathway has been the main-stay therapy option for prostate cancer (6,7). However, patients on ADTs eventually transition, over time, to a castration-resistant prostate cancer (CRPC) (7,8). Furthermore, other androgen-independent cellular pathway mechanisms have also been attributable to promoting prostate cancer progression and metastasis, especially during the androgen-independent phase of tumorigenesis (9–11). Many of these pathways are still being investigated.
Several studies have implicated the G-protein coupled receptor (GPCR) pathway as a potential mechanism for androgen-independent prostate cancer growth (12,13). Various proteins along this pathway have been of interest lately (13,14). For example, regulator of G-protein signaling 2 has been shown to be an androgen-independent repressor of prostate tumorigenesis and progression (11,14). The chemokine receptors, CXCR1 and CXCR2 have equally been shown to be upregulated as prostate cancer progresses (15).
Activator of G-protein signaling 3 (AGS3), also known as G-protein signaling modulator 1 (GPSM1), is a receptor-independent activator of G-protein signaling and was originally identified in a yeast-based functional screen of mammalian complementary DNA (cDNA) libraries (16,17). It belongs to the AGS group II family and acts as a guanine nucleotide dissociation inhibitor, inhibiting dissociation of Guanosine diphosphate (GDP) from Gα subunits as well as competitively preventing Gβγ subunits from coupling Gα subunits (18–20). AGS3 consists of seven tetratricopeptide repeats (TPR) and four G-protein regulator (GPR) motifs connected together by a linker region (18,19). A truncated short form of the protein, lacking the TPR domain is expressed in the heart (21). In addition to its regulatory role in GPCRs signaling, AGS3 has been shown to mediate several cellular functions. These include asymmetric cellular division (22), macroautophagy (20), intracellular pathogen clearance (16), protein trafficking (23), behavioral changes to addiction (24), polycystic kidney disease (25) and chemotaxis (26).
To date, little is known about the impact of AGS3 in PCa development, progression and metastasis. The aim of this study is to delineate the role of AGS3 expression in prostate tumorigenesis. The data herein indicate that overexpression of AGS3 in PCa cells enhances tumor progression in nude mice xenografts, whereas its inhibition delays tumor growth. Depletion of AGS3 also increases mitogen-activated protein kinase (MAPK) activation and AR expression in the tumor micro-environment relative to control. Taken together, the data indicate that AGS3 regulates prostate cancer development and progression via a MAPK/AR dependent pathway. Altogether, the data indicate that AGS3 expression modulates prostate tumorigenesis and presents AGS3 as a possible target for therapeutic intervention against prostate cancer.
Materials and methods
Materials
The prostate cell lines PC3 (CRL-1435), LNCaP (CRL-1740), MDA PCa 2b (CRL-2422), RWPE-1 (CRL-11609), DU-145 (HTB-81) and TRAMP-C1 (CRL-2730) (Supplementary Table 1, available at Carcinogenesis Online) were purchased from American Type Culture Collection (ATCC). The cells were cytogenetically tested and authenticated using the short tandem repeat method before being frozen. All cell-based experiments were carried out on cells that have been tested and cultured for <12 weeks. Puromycin (P9620) was obtained from Sigma–Aldrich; XTT assay kit (30-1011K) from ATCC; Geneticin (G418) (11811031), UltraPure LMP agarose (16520-050) and Lipofectamine 3000 (L3000-015) were purchased from Invitrogen. Plasmid transfection medium (sc-108062), UltraCruz transfection reagent (sc-395739), mouse AGS3 double nickase plasmid (sc-426766-NIC) and control double nickase plasmid (sc-437281) were obtained from Santa Cruz. All other reagents were obtained from commercial sources.
Cell culture
MDA PCa 2b cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM/Ham’s F12) supplemented with 20% heat-inactivated fetal bovine serum (FBS), 25 ng/ml cholera toxin, 10 ng/ml epidermal growth factor (EGF), 0.005 mM phosphoethanolamine (O-ppe), 100 pg/ml hydrocortisone, 45 nM selenious acid, 0.005 mg/ml bovine or human insulin. LNCaP cells were cultured in RPMI 1640 supplemented with 10% FBS, 10 mM N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid buffer, 1 mM sodium pyruvate, 1.26 g glucose, 100 µg/ml streptomycin and 100 IU/ml penicillin. PC3 cells were cultured in DMEM/Ham’s F12 medium were supplemented with 10% FBS, 100 µg/ml streptomycin and 100 IU/ml penicillin. TRAMP-C1 cells were grown in DMEM supplemented with 5% FBS, 5% Nu-Serum IV, 10 nM dehydroisoandrosterone, 0.005 mg/ml bovine insulin, 1.5 g sodium bicarbonate, 100 µg/ml streptomycin and 100 IU/ml penicillin. Cell harvesting was performed using 0.05% trypsin-ethylenediaminetetraacetic acid (1X). All cells were maintained in a 5% CO2 air atmosphere.
Overexpression and knockout of AGS3 in prostate PCa cell lines
Green fluorescent protein (GFP) tagged AGS3 constructs in pcDNA3 plasmids (full length and mutants of AGS3) were kind gifts from Dr Joe B. Blumer and Dr Stephen M. Lanier (Medical University of South Carolina, Charleston, SC). The generation of the TPR (encoding amino acids M1-E470) and the GPR (encoding amino acids G337-S650) mutants of AGS3 were described previously (27).
For overexpression of full-length AGS3 and mutants in LNCaP, PC3, MDA PCa 2b and TRAMP-C1, cells (4 × 106) were plated overnight in 100 mm3 dishes. The following day, cells were transfected with 40 µg plasmid containing enhanced green fluorescent protein (eGFP) alone or GFP-tagged full-length AGS3, AGS3-TPR or AGS3-GPR using Lipofectamine 3000 as prescribed by the manufacturer. Twenty-four hours after transfection, cells were placed under selection using 100 µg/ml G418 for 3 weeks to obtain cells stably expressing proteins. Cells expressing GFP were sorted by fluorescence-activated cell sorting and subsequently used for further experiments.
To generate AGS3 knockout in TRAMP-C1 cells, we utilized the double nickase gene-editing strategy (a D10A mutated nickase version of CRISPR/Cas9). Cells (2 × 105) were initially seeded into a six-well plate and grown overnight. Cells were then transfected with 3 µg GPSM1 double nickase plasmid (sc-426766-NIC) or control (Sham) plasmid (sc-437281) using 12 µl UltraCruz transfection reagent and following protocol as instructed by the manufacturer (Santa Cruz). Twenty-four hours post-transfection, GFP-positive cells in gene knockout and control plasmid wells were sorted by flow cytometry and collected cells were then placed under 2 µg/ml puromycin antibiotic selection for 5 days to obtain AGS3 knockout clones as well as control (Sham) cells. Cells were assayed and subsequently used for further experiments. Assayed cells are a combined pool of at least three independent selected clones and results validated by conducting AGS3 knockout (along with corresponding control, Sham) on TRAMP-C1 cells on two separate occasions.
Oncomine database analysis
The messenger RNA (mRNA) expression level or gene copy number of the GPSM1 gene between normal and prostate cancer tissues were analyzed using the oncomine database (www.oncomine.org), which is a web-based publicly available database for mining out genome-wide expression analyses (28). We utilized these parameters: ‘GPSM1’, ‘cancer versus normal analysis’, ‘prostate cancer’ to retrieve our analysis and evaluated the TCGA Prostate (extracted from the oncomine database), Grasso Prostate (29), Arredouani Prostate (30) and Taylor Prostate 3 (31) data sets.
Animals
All experiments were approved by and conformed to the guidelines of the Animal Care Committee of North Carolina Central University (Durham, NC).
For tumor xenograft studies, male athymic nude mice or C57BL/6 (aged 6–8 weeks) were obtained from Jackson Laboratory and housed in the animal care facility. Five (5) million cells of each sample were suspended in a 300 µl mix of media and Matrigel (ratio 1.5:1) and injected subcutaneously into the right flank of each nude mouse. Tumor volumes were monitored over 5–7 weeks (depending on cell line type) and tumor weight obtained following termination of the experiment. Tumor volume was calculated using the formula: Volume = (d1 × d2 × d3) × 0.5236; where dn represents the three orthogonal diameter measurements. Normalized tumor weight was obtained by dividing the weight of tumor by the weight of mice.
Cell proliferation assay
Briefly, cells (5 × 103) were seeded into 96-well plates using 100 µl culture media and incubated for a total of 4 days (TRAMP-C1) or 6 days (PC3), during which cell growth was measured daily using the XTT assay. Activated tetrazolium dye (50 µl) was added to each well and cells were maintained in 5% CO2 at 37oC for 3 h before readings. Plate was subsequently shaken to evenly distribute the dye, and absorbance was measured using a spectrophotometer. Growth rate is expressed as Absorbance (A475nm – A660nm) over time.
Wound healing assay
For wound closure assay, 1.5 × 104 cells in 100 µl media were seeded into each well of a 96-well ImageLock Microplate (Essen BioScience Cat #4379) and incubated overnight. Using a 96-pin IncuCyte woundmaker, the monolayer was gently scratched across the well and washed with medium to remove cell debris. Fresh medium was then added, cells were incubated and images of scratched wounds were captured at different time points using the IncuCyte ZOOM® live-cell system (Essen Bioscience). Results were analyzed using the IncuCyte software and presented as relative wound density.
Soft agar colony assay
For soft agar colony assay, cells (5 × 103/per well) were suspended in 2 ml of 0.35% agar and plated on top of a solidified base layer containing 1.5 ml of 0.5% agar in growth media. Plates were allowed to solidify and 500 µl growth media was added on top and then every 5 days. Cells were incubated at 37oC in 5% CO2 incubator for 17 days and colony staining was carried out with Crystal Violet solution (0.01%) for 1 h. The excess of dye was washed with water, images were captured using the Bio-Rad GelDoc XR+ imaging system and colonies were subsequently counted. Experiment was performed in triplicates and was repeated twice.
Immunoblotting
Rabbit anti-phospho-extracellular signal-regulated kinase (ERK) 1/2 (4370S), anti-ERK1/2 (9102S), anti-phospho-Akt (4056S), anti-Akt (9272S), anti-phospho P38 (9211S), anti-P38 (9212S), anti-phospho-nuclear factor-κB (NFκB) p65 (3033S), anti-NFκB p65 (8242S) and glyceraldehyde 3-phosphate dehydrogenase (GAPDH; 14C10) were from cell signaling. Anti-AR (ab133273) and anti-CXCR4 (ab124824) were purchased from Abcam, whereas anti-AGS3 (sc-136482) was from SantaCruz. Anti-GFP (G1544), anti-mouse (A9044) and anti-rabbit (A9169) were obtained from Sigma–Aldrich.
For immunoblotting, cell pellets or minced tissues were washed and lysed in radioimmunoprecipitation assay buffer (RIPA) supplemented with a cocktail of protease inhibitor. Cell lysates were cleared by centrifugation for 10 min, protein estimated and 20–50 µg protein suspended in 4X loading buffer. Samples were boiled for 5 min and separated by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS–PAGE). Resolved proteins were transferred unto nitrocellulose membranes and detected using antibodies against the different proteins as indicated.
Statistical analysis
Results are expressed as mean ± standard deviation. Statistical analysis was performed using GraphPad Prism 5.0 (GraphPad, San Diego, CA). Differences between groups were determined by one-way analysis of variance or Student’s t-test, as appropriate. A P-value <0.05 was considered statistically significant.
Results
Differential expression of AGS3 in human prostate cancer cell lines
AGS3 has been shown to be expressed in brain, heart, kidney tubules and epithelium, as well as in leukocytes of rats, mice and humans (25,26,32,33). An analysis of the TCGA Prostate data set from the oncomine database (http://www.oncomine.com), pairing prostate adenocarcinoma (n = 126) with normal prostate glands (n = 60), revealed a significant increase in copy number of AGS3 (P = 0.0084) in prostate carcinoma (Figure 1A). Analysis of AGS3 mRNA expression in three other datasets, including the Grasso Prostate (29), Arredouani Prostate (30) and Taylor Prostate 3 (31) data sets, also displayed higher expression of this gene (Supplementary Figure S1, available at Carcinogenesis Online). Immunoblotting analysis of AGS3 expression in the human prostate cancer cell lines LNCaP, PC3, MDA PCa 2b and DU-145, as compared with the normal prostate epithelial cell line RWPE-1, also revealed higher levels of AGS3 in LNCaP (~3-fold), MDA PCa 2b (~2-fold) and DU-145 (~2-fold) relative to RWPE-1 (Figure 1B and C). AGS3 expression in PC3 cells (~1-fold) was similar to that of RWPE-1 (Figure 1B and C).
Figure 1.
Expression profile of AGS3 in prostate tissues cancer cell lines. (A) AGS3 gene expression analysis in prostate cancer data set (TCGA) extracted from oncomine. Box plot derived from DNA copy numbers comparing AGS3 expression in normal prostate and prostate adenocarcinoma tissues. P < 0.0084. (B) Western blot analysis of AGS3 expression in PCa epithelial cell lines LNCaP, PC3, MDA PCa 2b and DU-145 relative to normal prostate epithelial RWPE-1. (C) Quantification of AGS3 expression in LNCaP, PC3, MDA PCa 2b and DU-145 relative to RWPE-1. Data shown are from three different experiments. *P < 0.05.
Overexpression of AGS3 in LNCaP, PC3 and MDA PCa 2b promotes tumor development
To determine the effect of AGS3 overexpression in prostate tumorigenesis, LNCaP, PC3 and MDA PCa 2b were transfected with pcDNA3-eGFP or vector containing AGS3. Cells lysates were analyzed by immunoblotting using both GFP and AGS3 antibodies. As shown in Figure 2, AGS3 was stably expressed in PC3 (panel A), LNCaP (panel B) and MDA PCa 2b (panel C) as compared with control cells or cells transfected with the vector alone. We next assessed the effect of AGS3 overexpression in tumor xenografts as described in Material and methods. Animals were observed for tumor progression over time as well as for end-point tumor weight. Overexpression of AGS3 in either PC3 (Figure 2D), LNCaP (Figure 2G) or MDA PCa 2b (Figure 2J) significantly enhanced tumor growth relative to control cells expressing eGFP (Figure 2E, H and K). Normalized tumor weight from AGS3 overexpressing cells also displayed a significant increase relative to eGFP cells (Figure 2F, I and L).
Figure 2.
Overexpression of AGS3 in human PCa cell lines enhances tumor growth. (A) PC3, (B) LNCaP (C) and MDA PCa 2b cells were transfected with pcDNA3 plasmid or vector containing AGS3 constructs. G418-resistant cells were selected and analyzed by immunoblotting using anti-GFP or anti-AGS3. Band density was calculated by Image J software, normalized for GAPDH expression and are the averages of three experiments. For (D, G and J) tumor xenografts, cells stably overexpressing AGS3 or control eGFP (5 × 106 cells) were injected subcutaneously into 6–8 weeks old nude mice. (E, H and K) Tumor growth was measured weekly, using a Thorpe caliper, until mice were euthanized and tumor weight determined as described in Materials and methods. (F, I and L) Tumor weight normalized by weight of mice. The results shown are representative of three experiments. *P < 0.05; **P < 0.01; ***P < 0.001.
Effect of AGS3 TPR and GPR motifs on prostate cancer growth
AGS3 consists of seven TPR and four GPR motifs (Figure 3A) (18,19). To determine which segment is involved in promoting prostate tumorigenesis, PC3 cell lines stably overexpressing the TPR (PC3-TPR-GFP) or the GPR motifs (PC3-GPR-GFP) were developed and protein expression was confirmed by immunoblotting (Figure 3B).
Figure 3.
Role of the TPR and GPR motifs of AGS3 on prostate tumor growth. (A) Schematic representation of the full-length AGS3, the TPR and the GPR deletion mutants of AGS3. (B) PC3 cells stably transfected with pcDNA3-eGFP or vector containing AGS3, TPR or GPR mutants were lysed with RIPA and 40 µg of proteins were resolved in 10% SDS–PAGE and assayed by immunoblotting for protein expression using an anti-AGS3 monoclonal antibody, specific for the linker. Proteins expression relative to GAPDH are represented and are averages of three experiments. (C) Proliferation rates of PC3 cells stably overexpressing either eGFP, full-length AGS3, the TPR or GPR mutants were determined by the XTT assay. For cell migration assays, closure of wound was continuously monitored every 4 h using the IncuCyte system until scratches were closed. (D) Representative image of wound healing assay done. (E) Graphical quantification of the rate of wound closure, expressed as relative wound density (%). Data shown are representative of two experiments performed in sextuplicates. (F and G) Colony formation assay was used to determine anchorage-independent cell growth of PC3 cells stably overexpressing either full length AGS3, the TPR or GPR mutants, relative to control eGFP expressing cells. Representative images (F) and graphical quantification of number of colonies (G) were determined after 17 days. For (H) tumor xenografts, cells (5 × 106 cells) were injected subcutaneously into 6–8 weeks old nude mice and tumor growth was measured weekly as described in the legend of Figure 2. (I) Time course of tumor growth over 6 weeks and (J) normalized tumor weight. The results shown are representative of one of three experiments. *P < 0.05; **P < 0.01; ***P < 0.001.
We next assessed the rate of proliferation of PC3-TPR-GFP, PC3-GPR-GFP, along with PC3-eGFP and PC3-AGS3-GFP using the XTT proliferation assay. No significant differences in the rates of proliferation were observed (Figure 3C). The cells were then characterized for cell migration using the wound healing assay. As shown in Figure 3D and E, no significant difference in the rates of wound closure was also observed.
We subsequently assessed for anchorage-independent growth using the soft agar assay method. PC3-AGS3-GFP exhibited a significant increased growth in soft agar relative to control PC3-eGFP cells (Figure 3F and G). Neither PC3-TPR-GFP nor PC3-GPR-GFP showed a significant difference in growth relative to control PC3-eGFP cells (Figure 3F and G). However, both the PC3-TPR-GFP and the PC3-GPR-GFP displayed significant decrease in cells growth relative to PC3-AGS3-GFP (Figure 3F and G).
We next assessed the effects of the TPR and GPR motifs on tumor growth in nude mice xenografts. PC3 cells stably overexpressing full-length AGS3 (PC3-AGS3-GFP) showed significant increase in tumor growth relative to control PC3-eGFP cells (Figure 3H, I and J). Overexpression of the TPR (PC3-TPR-GFP) or the GPR (PC3-GPR-GFP) motifs, however, had no significant effect in tumor development or normalized tumor weight as compared with control cells. These results mirrored the ones obtained with the soft agar assay and probably indicated that both motifs are important for AGS3-mediated tumorigenesis.
AGS3 inhibition in prostate cells decreases tumor growth
To determine the effect of AGS3 inhibition in PCa growth, we used the well-characterized mouse TRAMP-C1 cell line, established from the transgenic adenocarcinoma mouse prostate (TRAMP) model (34). TRAMP-C1 cells are tumorigenic in mice (34) and significantly overexpress AGS3 relative to PC3 (~20-fold) and LNCaP (~7-fold) (Figure 4A). Using CRISPR double nickase plasmids with either sham vector or vector containing AGS3/GPSM1-specific target, we generated a stable TRAMP-C1 cell line deficient in AGS3 expression (TRAMP-C1-AGS3-/-) (Figure 4B). TRAMP-C1-AGS3-/- displayed significant decreased in cell proliferation (Figure 4C), early cell migration (Figure 4D) and anchorage-independent cell growth in soft agar (Figure 4E), relative to TRAMP-C1 expressing the vector alone (TRAMP-C1-Sham). Both TRAMP-C1-AGS3-/- and TRAMP-C1-Sham, however, exhibited complete wound closure at ~32 h (Figure 4D).
Figure 4.
Depletion of AGS3 in TRAMP-C1 cells inhibits tumor growth. (A) Western blotting analysis of AGS3 expression in TRAMP-C1 cells relative to LNCaP and PC3 cells. (B) Knockdown of AGS3 in TRAMP-C1 cells using CRISPR D10A mutated double nickase plasmid containing GPSM1-specific target. Protein expression was confirmed by western blotting analysis. (C) Proliferation rates of TRAMP-C1-AGS3-/- and control TRAMP-C1-Sham cells as determined by the XTT assay. (D) Representative image and graphical quantification of wound healing assay for TRAMP-C1-Sham and TRAMP-C1-AGS3-/- cells monitored every 4 h using the IncuCyte system until scratches were closed. Data shown are representative of two experiments performed in sextuplicates. (E) Colony formation assay was used to determine anchorage-independent cell growth for TRAMP-C1-Sham and TRAMP-C1-AGS3-/- cells. Representative images (left) and graphical quantification of number of colonies (right) was determined after 17 days. For tumor xenografts, TRAMP-C1-AGS3-/- and control TRAMP-C1-Sham (5 × 106 cells) were injected subcutaneously into (F) 6–8 weeks old nude mice or (G) wild-type C57BL/6 mice. Mice were euthanized and tumor weight determined after 6 weeks. Representative image of dissected tumors (left) and normalized weight for TRAMP-C1-AGS3-/- tumor relative to control TRAMP-C1-Sham. Data shown are representative of two to three experiments. *P < 0.05; **P < 0.01; ***P < 0.001.
We next determine the effect of AGS3 depletion in tumor growth using nude mice xenografts. As shown in Figure 4F, TRAMP-C1-AGS3-/- exhibited a significant decrease in tumor growth relative to xenografts from control TRAMP-C1-Sham cells. Normalized tumor weight for TRAMP-C1-AGS3-/- xenografts (0.229 ± 0.021) was ~5-fold lower than that of TRAMP-C1-Sham xenografts (1.033 ± 0.193).
Since TRAMP-C1 cell was shown to develop tumor in C57BL/6 mice, we also carried out xenografts in wild-type C57BL/6 mice to determine the effect of AGS3 inhibition in tumor growth. As shown in Figure 4G, a significant decrease in tumor growth was also observed in TRAMP-C1-AGS3-/- xenografts relative to TRAMP-C1-Sham cells. Normalized tumor weight from TRAMP-C1-AGS3-/- xenografts (0.017 ± 0.017) was 9-fold lower as compared with TRAMP-C1-Sham (0.159 ± 0.071).
Effect of AGS3 knockdown in MAPK activation and expression
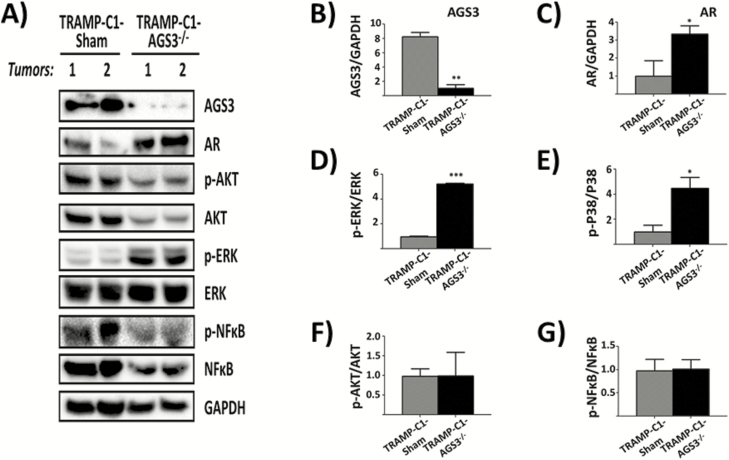
Previous studies from our group and others have shown that AGS3 inhibition alters MAPK activation and protein expression (26,33). To determine whether the effect of AGS3 knockdown affects MAPK activation, tumor lysates from TRAMP-C1-Sham and TRAMP-C1-AGS3-/- xenografts in nude mice were assayed by immunoblotting for P38 and ERK1/2 expression and phosphorylation. As shown in Figure 5A and B, AGS3 was significantly decreased (~90%) in tumor lysates from TRAMP-C1-AGS3-/- relative to TRAMP-C1-Sham cells. TRAMP-C1-AGS3-/- exhibited a ~5-fold increase in both ERK1/2 [phospho-ERK (p-ERK) 1/2: 5.237 ± 0.032; Figure 5A and D] and P38 (p-P38: 4.483 ± 0.610; Figure 5A and E) activation as compared with TRAMP-C1-Sham control tumor lysates (p-ERK1/2: 0.978 ± 0.038 and p-P38: 1.000 ± 0.384; Figure 5A, D and E).
Figure 5.
Effect of AGS3 depletion on MAPK and NF-κB expression and activation in TRAMP-C1 tumors. TRAMP-C1-AGS3-/- and TRAMP-C1-Sham tumors from nude mice xenografts were minced, lysed in RIPA and 40 µg of proteins were resolved in 10% SDS–PAGE, and immunoblotted. (A) Western blot analysis of AGS3, AR, Akt, p-Akt, ERK, p-ERK, NF-κB, p-NFκB and GAPDH from two different tumors (Tumors 1 and 2) in each experimental group. (B–G) Graphical representation of band densities for AGS3 (B), AR (C) and phospho-/total ratio for ERK1/2 (D), P38 (E), Akt (F) and NFκB (G). Data shown are representative of two experiments. *P <0.05; **P < 0.01; ***P < 0.001.
We also determined the effect of AGS3 inhibition in Akt and NFκB activation by monitoring for phospho-Akt (p-Akt) and phospho-NFκB (p-NFκB; p65) relative to total Akt and NFκB expression. Inhibition of AGS3 had no effect in Akt (p-Akt: 0.990 ± 0.422; Figure 5F) or NFκB (p-NFkB: 1.014 ±0.143; Figure 5G) activation relative to TRAMP-C1-Sham (p-Akt: 0.980 ± 0.422; p-NFkB: 0.975 ± 0.176; Figure 5F and G).
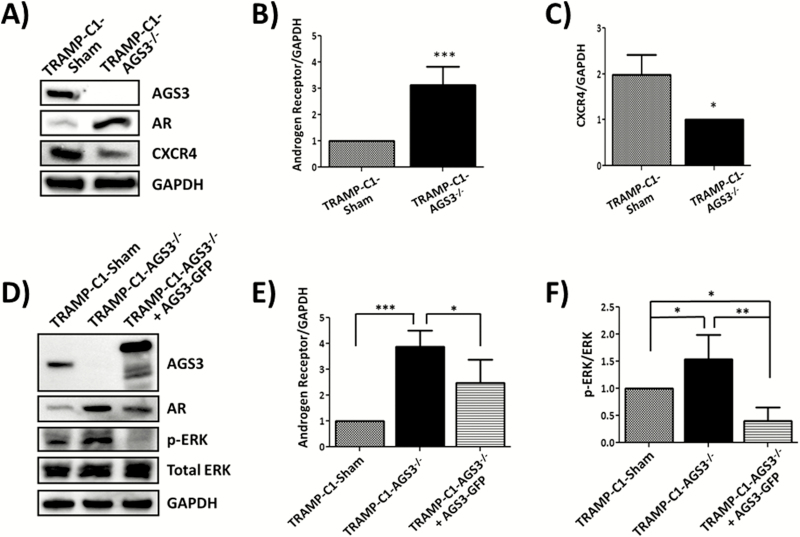
AR expression increased over 3-fold in TRAMP-C1-AGS3-/- tumors (3.335 ± 0.330; Figure 5A and C) as compared with TRAMP-C1-Sham (1.001 ± 0.614). To further confirm the results from tumor lysates, cells lysates from TRAMP-C1-Sham and TRAMP-C1-AGS3-/- were also assayed by western blotting for AR and CXCR4 expression. As shown in Figure 6, AR expression was significantly increased in TRAMP-C1-AGS3-/-cells (Figure 6B). Interestingly, CXCR4 expression was decreased in TRAMP-C1-AGS3-/- cells relative to TRAMP-C1-Sham cells (Figure 6C).
Figure 6.
Inhibition of AGS3 alters AR and CXCR4 expression and promoted ERK activation in TRAMP-C1 cells. (A–C) TRAMP-C1-Sham and TRAMP-C1-AGS3-/- cells were lysed in RIPA and 30 µg of protein were assayed for receptor expression. Western blotting analysis of AR and CXCR4 expression (A) and graphical representation of band densities from western blots of AR (B) and CXCR4 (C) relative to GAPDH. (D–F) TRAMP-C1-Sham, TRAMP-C1-AGS3-/- and TRAMP-C1-AGS3-/-+AGS3-GFP cells were lysed in RIPA and 30 µg of protein were assayed for differential expression of AR and activated ERK. Representative image of western blotting analysis (D) and graphical representation of band densities for AR relative to GAPDH (E) and activated ERK (F). Data shown are representative of three experiments. *P <0.05; **P < 0.01; ***P < 0.001
To determine if re-expression of AGS3 could revert the effect of AGS3 inhibition in TRAMP-C1 cell, TRAMP-C1-AGS3-/- cells were transfected with a pcDNA3 vector containing AGS3-GFP and stable clones were generated (TRAMP-C1-AGS3-/-+AGS3-GFP) and assayed by western blotting. As shown in Figure 6D, AGS3-GFP (~105 kDa) was overexpressed in TRAMP-C1-AGS3-/-+AGS3-GFP as compared with TRAMP-C1-AGS3-/- or control TRAMP-C1-Sham cells. Re-expression of AGS3 in TRAMP-C1-AGS3-/-+AGS3-GFP cells reverted significantly the expression level of AR (Figure 6D and E) and blocked ERK phosphorylation (Figure 6D and F), relative to TRAMP-C1-AGS3-/- cells. These data suggest that the increase in MAPK activation and AR overexpression observed in TRAMP-C1-AGS3-/- cells and TRAMP-C1-AGS3-/- tumor xenografts is a direct consequence of the depletion of AGS3 from these cells.
Discussion
PCa is the second most common cause of cancer death among men, with greater prevalence of the disease among the African American population in the USA (1,2). Despite the extensive studies aimed at the understanding of the pathogenesis of PCa, the molecular mechanisms governing tumor development and progression remain unknown. This is due to the disease complexity and involvement of both androgen-dependent and androgen-independent pathways in tumor growth (13,35). The GPCR signaling pathway, along with its regulatory and accessory proteins, has equally been implicated in prostate cancer progression (12,13,36). Although analysis of several data sets extracted from the Oncomine database revealed a significantly higher expression of AGS3 in prostate adenocarcinoma relative control prostate gland, the role of AGS3 in prostate cancer development and metastasis is yet to be defined. The results herein demonstrate that AGS3 expression plays a significant role in prostate tumorigenesis. First, overexpression of AGS3 in both androgen-dependent (LNCaP and MDA PCa 2b) and androgen-independent (PC3) PCa cell lines enhanced tumor growth, whereas AGS3 inhibition in TRAMP-C1 cells abrogated tumor development in xenografts in both nude mice and wild-type C57BL/6 animals (Figures 2 and 4). Second, overexpression of neither the TPR nor the GPR motifs in PC3 mimicked the effect of the full-length AGS3 in tumor growth. Third, depletion of AGS3 in TRAMP-C1 cells (TRAMP-C1-AGS3-/-) caused a 3-fold increase in AR but decreased CXCR4 expression (Figure 6).
AGS3 comprises seven TPR and four GPR motifs joined by a linker sequence (18,19). The TPR domain was shown to determine the positioning of AGS3 within the cell, whereas the GPR segment serves as a docking site for Gαi-GDP leading to AGS3-Gαi signaling (27,37). A recent study from our group has shown that in RBL-2H3 cells, GRK6 complexes with AGS3-Gαi to regulate CXCR2-mediated cellular functions including Ca2+ mobilization, MAPK activation, receptor desensitization and downregulation (33). Overexpression of the GPR motif, not the TPR, inhibited CXCR2-mediated cellular responses. Choi et al. (38) have also demonstrated that the GPR domain of AGS3 regulates MUC1 expression and cytokine production in a mouse model of airway inflammation. In light of these results, we speculated that the effect of AGS3 in promoting tumor growth could also be mediated by its GPR segment. Surprisingly, overexpression of neither the TPR nor the GPR domain could mimic the effect of full-length AGS3 in PC3 xenografts in nude mice (Figure 3). Interestingly, PC3 cells overexpressing the GPR or the TPR exhibited a similar decrease in cell migration in wound closure assays, relative to control cells or cells expressing full-length AGS3 (Figure 3). These results probably suggest that both motifs of AGS3 are important for its role in prostate tumorigenesis.
The functional role of AGS3 in tumor development and metastasis is not well understood. In multiple myeloma, a disease characterized by the uncontrolled adhesion and growth of malignant plasma cells within the bone marrow, AGS3 overexpression correlated with decreased apoptosis and increased cell adhesion (39). Shi et al. (40) have shown that in esophageal squamous cell carcinoma, AGS3 overexpression decreases cell proliferation and increases survival. In this study, however, AGS3 overexpression promoted PCa malignancy, whereas its inhibition in TRAMP-C1 cells abrogated tumor growth. Interestingly, the decreased tumor growth in TRAMP-C1-AGS3-/- xenografts correlated with a significant increase (~3-fold) in AR expression (Figures 5 and 6). AR has been identified as possessing both tumor suppressive and tumor proliferative capability (41). Overexpression of AR in the castration-resistant PC3 cells resulted in a diminished rate of cell invasiveness as well as metastasis to the lymph nodes (41). Therefore, it could be that AGS3 modulates AR expression and activation in the prostate, thereby regulating prostate cancer development and progression. AGS3 has also been reported to modulate cellular viability as well as the progression of division in a few disease states. Rasmussen et al. (42) recently showed that AGS3 regulates renal epithelial cell survival and that its knockdown resulted in increased apoptosis. Suppression of AGS3 expression in TRAMP-C1 cells inhibited tumor growth in xenografts in both nude mice and wild-type C57BL/6 animals (Figure 4). Taken together, these data suggest that AGS3 possesses cell autonomous effects and its level of expression drives the rate of prostate tumorigenesis.
Elevated ERK1/2 and P38 activity have been associated with malignancy in several cancers due to its proliferative and anti-apoptotic functions (43). In PC3 cells, enhanced ERK expression was shown to promote cell migration, invasion and proliferation (44). Tumor Necrosis Factor (TNF)-α-induced ERK phosphorylation in LNCaP cells was also correlated with increased proliferation (45). In a recent study using prostate tissues, Gioeli et al. (46) demonstrated a direct correlation between ERK expression and phosphorylation and advanced PCa grade and stage. In this study, however, ERK and P38 activation were significantly increased in tumor lysates from TRAMP-C1-AGS3-/- xenografts, despite a marked decrease in tumor growth relative to TRAMP-C1 cells (Figure 5). The reason for this discrepancy remains unclear. Sustained activation of ERK in PCa cells has been shown to have anti-proliferative effect or promote apoptosis (47). One explanation could be that the AGS3-mediated inhibition resulted in a sustained ERK1/2 activation, thereby increasing apoptosis. Supporting that contention is that re-expression of AGS3 in TRAMP-C1-AGS3-/- cells (TRAMP-C1-AGS3-/-+AGS3-GFP) reverted the increase activation of ERK as well as AR overexpression (Figure 6D–F).
Another explanation for the decreased tumorigenesis in TRAMP-C1-AGS3-/- xenograft could be the axis CXCL12/CXCR4. The CXCR4 chemokine receptor has been shown to be overexpressed in numerous cancer types and serves as a prognostic factor for cancer survival (48). Singh et al. (49) have shown that CXCR4 expression is significantly increased in LNCaP and PC3 when compared with the normal prostate epithelial cell PrEC. Thus, the profound decrease in CXCR4 expression observed in TRAMP-C1-AGS3-/- cells relative to TRAMP-C1 (Figure 6) could account for the decrease in tumor growth observed in TRAMP-C1-AGS3-/- xenografts (Figure 4F and G).
In summary, the results herein indicate that AGS3 expression plays a significant role in prostate tumorigenesis by regulating MAPK kinase activation and AR and CXCR4 expression. Whether AGS3 complexes directly with AR or CXCR4 to modulate their expression and functions remains unclear. However, AGS3 has been shown to modulate the expression, cellular distribution and trafficking of several proteins (23,27,50). Overall, the data revealed a direct correlation between AGS3 expression and PCa development and progression. Better understanding of how AGS3 modulates the MAPK signaling pathways and protein expression in prostate tumorigenesis may give insights into improved diagnosis, treatment and prognosis of prostate cancer.
Funding
This work was supported by National Institutes of Health (AI38910, CA156735, MD012392).
Supplementary Material
Acknowledgements
We thank Dr Stephen M. Lanier and Dr Joe B Blumer (Department of Cell and Molecular Pharmacology and Experimental Therapeutics, Medical University of South Carolina, Charleston, SC 29425) for providing the AGS3 constructs. We are very thankful to Ms Tonelia Mowatt (Department of Biomedical and Biological Science, and JLC Biomedical/Biotechnology Research Institute, NCCU, Durham, NC) for technical support.
Conflict of Interest Statement: None declared.
Glossary
Abbreviations
- AGS3
activator of G protein signaling 3
- AR
androgen receptor
- DMEM
Dulbecco's modified Eagle's medium
- eGFP
enhanced green fluorescent protein
- ERK
extracellular signal-regulated kinase
- GAPDH
glyceraldehyde 3-phosphate dehydrogenase
- GPCR
G-protein-coupled receptor
- GPR
G-protein regulatory
- GPSM1
G-protein signaling modulator 1
- MAPK
mitogen-activated protein kinase
- NF-κB
nuclear factor-κB
- PCa
prostate cancer
- RIPA
radioimmunoprecipitation assay buffer
- SDS–PAGE
sodium dodecyl sulfate–polyacrylamide gel electrophoresis
- TPR
tetratricopeptide repeat
References
- 1. Chen S.L. et al. (2017) Prostate cancer mortality-to-incidence ratios are associated with cancer care disparities in 35 countries. Sci. Rep., 7, 40003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Siegel R.L. et al. (2018) Cancer statistics, 2018. CA Cancer J. Clin., 68, 7–30. [DOI] [PubMed] [Google Scholar]
- 3. Krause F.S. et al. (2005) Heterogeneity in prostate cancer: prostate specific antigen (PSA) and DNA cytophotometry. Anticancer Res., 25, 1783–1785. [PubMed] [Google Scholar]
- 4. Gandhi J. et al. (2018) The molecular biology of prostate cancer: current understanding and clinical implications. Prostate Cancer Prostatic Dis., 21, 22–36. [DOI] [PubMed] [Google Scholar]
- 5. Xie Q. et al. (2017) Dissecting cell-type-specific roles of androgen receptor in prostate homeostasis and regeneration through lineage tracing. Nat. Commun., 8, 14284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Murillo-Garzón V. et al. (2018) Frizzled-8 integrates Wnt-11 and transforming growth factor-β signaling in prostate cancer. Nat. Commun., 9, 1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Karantanos T. et al. (2013) Prostate cancer progression after androgen deprivation therapy: mechanisms of castrate resistance and novel therapeutic approaches. Oncogene, 32, 5501–5511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. de la Taille A., et al. (2017) Factors predicting progression to castrate-resistant prostate cancer in patients with advanced prostate cancer receiving long-term androgen-deprivation therapy. BJU Int., 119, 74–81. [DOI] [PubMed] [Google Scholar]
- 9. Feldman B.J. et al. (2001) The development of androgen-independent prostate cancer. Nat. Rev. Cancer, 1, 34–45. [DOI] [PubMed] [Google Scholar]
- 10. Bluemn E.G. et al. (2017) Androgen receptor pathway-independent prostate cancer is sustained through FGF signaling. Cancer Cell, 32, 474–489.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wolff D.W. et al. (2012) Epigenetic repression of regulator of G-protein signaling 2 promotes androgen-independent prostate cancer cell growth. Int. J. Cancer, 130, 1521–1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Xia C. et al. (2001) Identification of a prostate-specific G-protein coupled receptor in prostate cancer. Oncogene, 20, 5903–5907. [DOI] [PubMed] [Google Scholar]
- 13. Yowell C.W. et al. (2002) G protein-coupled receptors provide survival signals in prostate cancer. Clin. Prostate Cancer, 1, 177–181. [DOI] [PubMed] [Google Scholar]
- 14. Cao X. et al. (2006) Regulator of G-protein signaling 2 (RGS2) inhibits androgen-independent activation of androgen receptor in prostate cancer cells. Oncogene, 25, 3719–3734. [DOI] [PubMed] [Google Scholar]
- 15. Waugh D.J. et al. (2008) The interleukin-8 pathway in cancer. Clin. Cancer Res., 14, 6735–6741. [DOI] [PubMed] [Google Scholar]
- 16. Vural A. et al. (2016) Activator of G-protein signaling 3-induced lysosomal biogenesis limits macrophage intracellular bacterial infection. J. Immunol., 196, 846–856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Sato M. et al. (2004) AGS3 and signal integration by Galpha(s)- and Galpha(i)-coupled receptors: AGS3 blocks the sensitization of adenylyl cyclase following prolonged stimulation of a Galpha(i)-coupled receptor by influencing processing of Galpha(i). J. Biol. Chem., 279, 13375–13382. [DOI] [PubMed] [Google Scholar]
- 18. Oner S.S. et al. (2013) Regulation of the G-protein regulatory-Gαi signaling complex by nonreceptor guanine nucleotide exchange factors. J. Biol. Chem., 288, 3003–3015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Blumer J.B. et al. (2014) Activators of G protein signaling exhibit broad functionality and define a distinct core signaling triad. Mol. Pharmacol., 85, 388–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Pattingre S. et al. (2003) The G-protein regulator AGS3 controls an early event during macroautophagy in human intestinal HT-29 cells. J. Biol. Chem., 278, 20995–21002. [DOI] [PubMed] [Google Scholar]
- 21. Pizzinat N. et al. (2001) Identification of a truncated form of the G-protein regulator AGS3 in heart that lacks the tetratricopeptide repeat domains. J. Biol. Chem., 276, 16601–16610. [DOI] [PubMed] [Google Scholar]
- 22. Sanada K. et al. (2005) G protein betagamma subunits and AGS3 control spindle orientation and asymmetric cell fate of cerebral cortical progenitors. Cell, 122, 119–131. [DOI] [PubMed] [Google Scholar]
- 23. Oner S.S. et al. (2013) Translocation of activator of G-protein signaling 3 to the Golgi apparatus in response to receptor activation and its effect on the trans-Golgi network. J. Biol. Chem., 288, 24091–24103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Bowers M.S. et al. (2008) Nucleus accumbens AGS3 expression drives ethanol seeking through G betagamma. Proc. Natl. Acad. Sci. USA, 105, 12533–12538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kwon M. et al. (2012) G-protein signaling modulator 1 deficiency accelerates cystic disease in an orthologous mouse model of autosomal dominant polycystic kidney disease. Proc. Natl. Acad. Sci. USA, 109, 21462–21467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Branham-O’Connor M. et al. (2014) Defective chemokine signal integration in leukocytes lacking activator of G protein signaling 3 (AGS3). J. Biol. Chem., 289, 10738–10747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Vural A. et al. (2010) Distribution of activator of G-protein signaling 3 within the aggresomal pathway: role of specific residues in the tetratricopeptide repeat domain and differential regulation by the AGS3 binding partners Gi(alpha) and mammalian inscuteable. Mol. Cell. Biol., 30, 1528–1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Rhodes D.R. et al. (2004) ONCOMINE: a cancer microarray database and integrated data-mining platform. Neoplasia, 6, 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Grasso C.S. et al. (2012) The mutational landscape of lethal castration-resistant prostate cancer. Nature, 487, 239–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Arredouani M.S. et al. (2009) Identification of the transcription factor single-minded homologue 2 as a potential biomarker and immunotherapy target in prostate cancer. Clin. Cancer Res., 15, 5794–5802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Taylor B.S. et al. (2010) Integrative genomic profiling of human prostate cancer. Cancer Cell, 18, 11–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Nadella R. et al. (2010) Activator of G protein signaling 3 promotes epithelial cell proliferation in PKD. J. Am. Soc. Nephrol., 21, 1275–1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Singh V. et al. (2014) G Protein-coupled receptor kinase-6 interacts with activator of G protein signaling-3 to regulate CXCR2-mediated cellular functions. J. Immunol., 192, 2186–2194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Foster B.A. et al. (1997) Characterization of prostatic epithelial cell lines derived from transgenic adenocarcinoma of the mouse prostate (TRAMP) model. Cancer Res., 57, 3325–3330. [PubMed] [Google Scholar]
- 35. Pienta K.J. et al. (2006) Mechanisms underlying the development of androgen-independent prostate cancer. Clin. Cancer Res., 12, 1665–1671. [DOI] [PubMed] [Google Scholar]
- 36. Daaka Y. (2004) G proteins in cancer: the prostate cancer paradigm. Sci. STKE, 2004, re2. [DOI] [PubMed] [Google Scholar]
- 37. De Vries L. et al. (2000) Activator of G protein signaling 3 is a guanine dissociation inhibitor for Galpha i subunits. Proc. Natl. Acad. Sci. USA, 97, 14364–14369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Choi I.W. et al. (2016) Regulation of airway inflammation by G-protein regulatory motif peptides of AGS3 protein. Sci. Rep., 6, 27054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Shao S. et al. (2014) A role for activator of G-protein signaling 3 (AGS3) in multiple myeloma. Int. J. Hematol., 99, 57–68. [DOI] [PubMed] [Google Scholar]
- 40. Shi H. et al. (2015) Overexpression of activator of G-protein signaling 3 decreases the proliferation of esophageal squamous cell carcinoma. Pathol. Res. Pract., 211, 449–455. [DOI] [PubMed] [Google Scholar]
- 41. Niu Y. et al. (2008) Androgen receptor is a tumor suppressor and proliferator in prostate cancer. Proc. Natl. Acad. Sci. USA, 105, 12182–12187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Rasmussen S.A. et al. (2015) Activator of G-protein signaling 3 controls renal epithelial cell survival and ERK5 activation. J. Mol. Signal., 10, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Rodríguez-Berriguete G. et al. (2012) MAP kinases and prostate cancer. J. Signal Transduct., 2012, 169170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. McCracken S.R. et al. (2008) Aberrant expression of extracellular signal-regulated kinase 5 in human prostate cancer. Oncogene, 27, 2978–2988. [DOI] [PubMed] [Google Scholar]
- 45. Ricote M. et al. (2006) P38 MAPK protects against TNF-alpha-provoked apoptosis in LNCaP prostatic cancer cells. Apoptosis, 11, 1969–1975. [DOI] [PubMed] [Google Scholar]
- 46. Gioeli D. et al. (1999) Activation of mitogen-activated protein kinase associated with prostate cancer progression. Cancer Res., 59, 279–284. [PubMed] [Google Scholar]
- 47. Chan Q.K. et al. (2010) Activation of GPR30 inhibits the growth of prostate cancer cells through sustained activation of Erk1/2, c-jun/c-fos-dependent upregulation of p21, and induction of G(2) cell-cycle arrest. Cell Death Differ., 17, 1511–1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Domanska U.M. et al. (2012) CXCR4 inhibition with AMD3100 sensitizes prostate cancer to docetaxel chemotherapy. Neoplasia, 14, 709–718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Singh S. et al. (2004) CXCL12-CXCR4 interactions modulate prostate cancer cell migration, metalloproteinase expression and invasion. Lab. Invest., 84, 1666–1676. [DOI] [PubMed] [Google Scholar]
- 50. Groves B. et al. (2007) A specific role of AGS3 in the surface expression of plasma membrane proteins. Proc. Natl. Acad. Sci. USA, 104, 18103–18108. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.