Abstract
Background:
Infertility is a common reproductive disorder, with male factor infertility accounting for approximately half of all cases. Taking a paternal perceptive, recent research has shown that sperm epigenetics, such as changes in DNA methylation, histone modification, chromatin structure, and non-coding RNA expression, can affect reproductive and offspring health. Importantly, environmental conditions during the preconception period has been demonstrated to shape sperm epigenetics.
Aim:
To provide an overview on epigenetic modifications that regulate normal gene expression and epigenetic remodeling that occurs during spermatogenesis, and to discuss the epigenetic alterations that may occur to the paternal germline as a consequence of preconception environmental conditions and exposures.
Results and conclusion:
The preconception period represents a significant window of susceptibility in which environmental conditions, such as chemical exposures, nutrition, drugs and stress, shape the sperm epigenome, which can have downstream adverse effects on reproductive success and offspring health. Understanding the environmental legacy of the sperm epigenome during spermatogenesis will enhance reproductive success as well as improve offspring health.
Keywords: Preconception, Environmental Exposure, Sperm, Epigenetics, DNA Methylation, Histones, Non-coding RNA, Fertility
Introduction
Infertility, defined as the inability to establish a clinical pregnancy following 12 months of regular, unprotected intercourse, is one of the most common reproductive health disorders (Zegers-Hochschild et al. 2017). The prevalence of infertility globally is estimated at 15% for couples, whereby males are solely responsible for 20–30% of infertility and contribute to 50% of overall cases (Agarwal et al. 2015). Infertility is costly, both financially and emotionally, for couples seeking to become pregnant (Chachamovich et al. 2010). Additionally, infertility interventions usually require multiple attempts of treatment to achieve a pregnancy; there is a 75% failure rate of each assisted reproductive technologies (ART) cycle, thus the psychological/emotional toll of ART can also profound. Lastly, success rates decline with increasing male and female age (Farley Ordovensky Staniec and Webb 2007). This is especially concerning as over the past several decades, there has been an increase in parental age at the time of conception, especially in developed countries (Khandwala et al. 2017).
One factor thought to be driving infertility rates is environmental conditions prior to conception. (Mima et al. 2018; Haotian Wu et al. 2015). While any exposure has the potential to negatively impact health outcomes, there are sensitive periods during which exposures might have an amplified, long-lasting effect. For example, windows of susceptibility to environmental insults include preconception, prenatal, early childhood, and puberty. Common exposures include endocrine disrupting chemicals, heavy metals, industrial waste, as well as lifestyle exposures such as diet, exercise, and stress. Both the type of exposure and the timing of exposure may lead to differential health outcomes.
The burden of the developmental health outcomes of offspring has largely fallen on the women; the fetus develops in the maternal environment, so it was thought that the paternal preconception environment played little role in development. This is in spite of the fact that findings that the paternal preconception environment influences fertility and offspring health in animal studies dates back to over three decades. This includes studies largely focused on ionizing radiation (Shu et al. 1994) and anti-cancer drug use (Trasler et al. 1986, 1987). Recent research also suggests that paternal preconception environment plays a role in fetal development and offspring health outcomes (Day et al. 2016; McPherson et al. 2015; Messerlian et al. 2017). The increasing evidence of offspring manifestation of other paternal preconception experiences, such as obesity and exposure to environmental pollutants, have led to the growing field of Paternal Origin of Health and Disease (POHaD)(Soubry 2018), a paternal offshoot of the established field of Developmental Origins of Health and Disease (DOHaD). Increasing evidence suggests that environmental exposures can alter the epigenetic marks in the germline, leading to possible changes in reproductive outcomes. Environmental exposures may also lead to the transmission of aberrant sperm epigenetic marks following fertilization, which may alter embryonic development and offspring health. This suggests that exposures can be embodied within the developing male germ cell through epigenetic marks and that these modifications may impart information that leads to 1) lower fertilization rates, 2) decreases in developmental potential, and 3) negative health outcomes for the offspring (H. Wu et al. 2015). Thus, it is important to consider male environmental exposures during the preconception window.
In this review, we will cover the typical epigenetic changes that occur during early embryonic development and spermatogenesis and how emerging evidence of environmental exposures during the preconception window might alter these mechanisms, leading to infertility and poor offspring outcomes.
How do environmental exposures lead to phenotypic outcomes?
Vectors of epigenetic information
While it might seem like environmental exposures would alter fertility rates and developmental potential by altering the genome, it is often the case that exposures alter the more flexible epigenetic landscape. Epigenetics deals with the broad range of modifications to the genome that do not change the underlying genetic sequence. The most commonly studied epigenetic mechanism is DNA methylation, but the study of epigenetics also includes histone composition and modifications, chromatin structure, larger 3-dimensional genome arrangement, and non-coding RNA expression. Together, these modifications allow distinct cell types to utilize the same genome for different phenotypic outcomes. Specifically, the epigenetic landscape allows differential gene expression in the approximately 200 distinct cell type that make up the human body. Epigenetic modifications also allow cells to respond to changing environments, which can be advantageous for survival and fitness.
DNA methylation
In mammalian cells, the predominant form of DNA methylation (5mC) occurs at CpG dinucleotides, although methylation at cytosines can also be found at low frequency in non-CpG contexts (Lee et al. 2017; Patil et al. 2014). The CpG dinucleotides are found in different genomic context; CpG islands are regions of high CpG density. Methylation at specific genomic regions such as promoters has been established to play a role in gene regulation. Functionally, DNA methylation can regulate gene expression in a number of ways. For a while, it was thought that methylation largely led to gene repression. It is true that in most cell types, the methylation at promoter-associated CpG islands is inversely correlated with the transcriptional activity of the locus. Within the promoter island, actively transcribed genes are generally unmethylated, while silenced genes are often found to be heavily methylated. Even though methylation is thought to be a binary on-off switch, it appears that CpG density, not presence of methylation alone, contributes to regulation of expression (Hartl et al. 2019).
Due to the crucial role of methylation in gene expression, the coordination of adding, maintaining, and removing methylation across tissues needs to be carefully controlled. The regulation is catalyzed by actions of methylating enzymes such as DNA methyltransferases (DNMT1, DNMT3A, DNMT3B and, DNMT3L) (Bestor et al. 1988) and the more recently discovered family of TET proteins (TET1, TET2, and TET3) found to mediate active demethylation processes especially post-fertilization (Kriaucionis and Heintz 2009; Tahiliani et al. 2009). Although, passive demethylation may also occur during cell replication (Zhu et al. 2018) due to lack of maintenance machinery. Hence, misregulation of any of these methylation modifiers may result in aberrant methylation patterns and alteration of cellular functions. For example, loss of Dnmt3l was associated with hypomethylation in male germline and sterility (Bourc’his et al. 2001; Bourc’his and Bestor 2004; Hata et al. 2002; Veland et al. 2019); while loss of Dnmt1 or its cofactor (Np95) resulted in dramatic, but not complete or identical, loss of methylation (Arand et al. 2012; Bostick et al. 2007; Sharif et al. 2007).
Histone Modifications and Chromatin Structure
Generally, DNA is packaged around structural proteins such as core (H2A, H2B, H3, and H4) and cell specific histones (Bao and Bedford 2016; Rathke et al. 2014) in a manner that allows transcription regulation. Importantly, post-translational tail modification (methylation, acetylation, phosphorylation, ubiquitination) of these histone molecules are known to alter genes expression and may be closely regulated by enzymes such as acetyltransferases, methyltransferases, and kinases as well as histone deacetylases (HDACs) and demethylases (HDMs) (Qin et al. 2019).
Furthermore, higher-order, organization of the chromosome into 3-dimensional topological associating domains (TADs) and A/B compartments has been reported to alter gene expression (Dixon et al. 2012; Dixon et al. 2015; Hawkins et al. 2010; Lieberman-Aiden et al. 2009; Peric-Hupkes et al. 2010). While TADs are defined by local interactions of genomic region such as enhancers and promoters, A and B compartments are associated with open and closed chromatin, respectively (Ibn-Salem et al. 2014; Lupiáñez et al. 2015). Additionally, Lamina-Associated Domains (LADs) and nucleolar-associating domains (NADs) are regions of the genome that interact with the nuclear laminin and nucleolus, respectively, and are also involved in genome regulation (Lochs et al. 2019). Taken together, the structural organization of the genome allow for differential regulation of gene expression and suggests their alteration may be detrimental to cellular function.
Non-coding RNAs
DNA methylation and chromatin structure impact gene regulation by physically constraining or allowing for a permissive expression state, but expression from the genome itself in a non-protein coding form can also regulate transcription and/or translation. For example, non-coding RNAs transcribed from DNA, but remain untranslated into proteins, serve important regulatory functions. They are broadly divided into long non-coding RNAs (lncRNAs) and short non-coding RNAs (sncRNAs). lncRNA, typically greater than 200 nucleotides in length, include the well-characterized Xist and Tsix, both of which are involved in X-chromosome inactivation. LincRNA, and natural antisense transcripts (NATs) are often polyadenylated and associated with other epigenetic mechanisms such as histone methylation and chromatin remodeling (Novikova et al. 2012; Surface et al. 2010). On the other hand, sncRNAs are usually less than 200 nucleotides in length. They include transfer RNAs (tRNAs), ribososomal RNAs (rRNAs) and small nuclear RNAs (snRNAs), as well as different regulatory RNAs like microRNAs (miRNAs), small interfering RNAs (siRNAs), Piwi-interacting RNAs (piRNAs), and small nucleolar RNAs (snoRNAs) (Collins et al. 2011).
The sensitivity of sperm borne ncRNAs to different environmental pollutants, epimutation involvement, and regulatory functions, as well as potential role in inheritance of paternally acquired traits has been well reviewed (Q. Chen et al. 2016; Yan 2014). Overall, non-coding RNAs play an important regulatory function necessary for appropriate gene expression and can serve as epigenetic mediators of environmental exposures. Because non-coding RNA expression can regulate germline expression profiles, they may potentially transfer environmental information to subsequent generations.
In order to establish the appropriate epigenetic state of a genome for a specific cell type, epigenetic reprogramming needs to occur. In the next section, we will highlight the major windows of reprogramming, focusing on the paternal germline.
Windows of Susceptibility
Susceptibility to harmful environmental exposures can be grouped into windows of time when cells undergo global epigenetic reprogramming. These reprogramming events are particularly sensitive to the environment, which can confer changes to both the genetic and epigenetic landscapes. Importantly, it was thought for a long time that the male germ cell contributed only genetic material and that any alteration to the paternal germline by the environment would arrive in the form of genetic alterations (Linschooten et al. 2013). While it is true that environmental factors can lead to genetic changes, it is clear that epigenetic changes also occur and can be passed through the male germline (Donkin and Barrès 2018). Understanding the normal epigenetic features of the male germline, as well as the changes during reprogramming of the embryo, allows for researchers to better understand the epigenetic changes that occur following environmental exposures.
Embryonic Development
Following fertilization, there are two major epigenetic reprogramming events: preimplantation development and primordial germ cell development. The first wave of reprogramming occurs immediately following fertilization, when the terminally differentiated sperm and oocyte fuse to form a zygote. Prior to fertilization, the sperm and the oocyte have dramatically different epigenetic landscapes. The sperm genome is heavily methylated, with 80–90% of all CpG dinucleotides methylated (Mayer et al. 2000; Oswald et al. 2000; Santos et al. 2002). In contrast, global DNA methylation levels in the maternal haploid genome are approximately half that of the sperm (Howlett and Reik 1991; Peat et al. 2014; Smallwood et al. 2011). Additionally, the paternal genome is largely wrapped around protamines, rather than histones (Balhorn 2007; Braun 2001). These epigenetic differences need to be resolved in order for the embryo to reach a totipotent state.
Upon fertilization, rapid epigenetic reprogramming begins. The kinetics of reprogramming are different between the two parental pronuclei, owing to the fact that they are epigenetically distinct. Prior to pronuclear fusion, the paternal genome undergoes rapid, replication-independent loss of 5-methylcytosine (5mC) (Santos et al. 2002)https://paperpile.com/c/J7e68p/LVQ0j. In contrast, little change in the 5mC is found in the maternal pronucleus (Mayer et al. 2000; Oswald et al. 2000). By the time of the first cell division, there is no 5mC signal detected in the paternal pronucleus and although the two parental genomes occupy the same nucleus after pronuclear fusion, the differences in 5mC levels are apparent beyond the 4-cell stage (Santos et al. 2002). It has been shown that TET enzymes mediate the oxidation of 5mC to 5hmC (as well as 5fC and 5caC) in vivo and that TET proteins are expressed and differentially localized during preimplantation development, leading to the differences in demethylation dynamics (Gu et al. 2011; Iqbal et al. 2011; Wossidlo et al. 2011).
While the paternal genome, facilitated by TET proteins, undergoes active DNA demethylation, the maternal genome is protected from TET activity by PGC7/Stella, which binds to H3K9me2-enriched regions (Nakamura et al. 2012). The paternal genome also has regions of H3K9me2, specifically in imprinted regions, but because it is protamine-rich, it is able to undergo rapid demethylation (Branco et al. 2008; Cirio et al. 2008; Hirasawa et al. 2008; Nakamura et al. 2007). The maternal genome largely undergoes passive, replication-dependent DNA demethylation; however, it is important to note that there is still some active demethylation of the maternal genome, but not to the extent of the paternal genome. In both cases, there is some ambiguity as to what reprogramming mechanisms are required for proper development; albeit, appropriate reprogramming is critical for development (Inoue et al. 2015; Peat et al. 2014; Shen et al. 2014).
In addition to DNA methylation, histone enrichment and chromatin dynamics between the two parental genomes differ. Both the paternal and maternal genomes have distinct chromatin and histone enrichment. The obvious difference is the packaging of the paternal genome around of protamine. Additionally, the paternal pronuclei is enriched early on with H3K27ac, H4K5ac, and H4K16ac (Adenot et al. 1997; Hayashi-Takanaka et al. 2011; Stein et al. 1997). In contrast, the maternal pronuclei is exclusively enriched for H3K9me3S10P, H3K36me3 and H4K20me3, as well as being highly enriched for H3K4 and H3K9 methylation (Beaujean 2014; Bošković et al. 2012; Lepikhov and Walter 2004; Ribeiro-Mason et al. 2012; Santenard et al. 2010; Wongtawan et al. 2011). While some of the enrichment is known to be functional (for example, H2K9me2 protecting against demethylation), the significance of many of the differences is still unknown. It is clear, however, that reprogramming needs to occur in order to merge the two distinct parental genomes to achieve a totipotent embryonic state.
The second major wave of epigenetic reprogramming occurs during primordial germ cell development. During gastrulation in mouse, a population of cells is allocated as primordial germ cells, which will later become the oocytes and sperm. Due to experimental constraints, the human timeline for germ cell allocation has been more difficult to elucidate, but takes place around 4-weeks post-fertilization (Guo et al. 2015). Following specification, waves of epigenetic reprogramming take place within the primordial germ cells. During cell migration, epigenetic reprogramming begins again. There is a reduction in H3K9me2 coupled with an increase in H3K27me3 (Sasaki and Matsui 2008; Seki et al. 2007) along with a decrease in Dnmt3a and 3b and decrease in methylation levels (Seki et al. 2005; Yabuta et al. 2006). Additionally, reactivation of the X-chromosome in female embryos occurs (Chuva de Sousa Lopes et al. 2008).
Once cell migration is complete, the second stage of primordial germ cell reprogramming occurs. This stage is characterized by rapid, global changes in DNA methylation levels, along with alterations to chromatin structure and histone modifications (Gkountela et al. 2015; Guo et al. 2015; Guo et al. 2017; Hajkova et al. 2002; Hajkova et al. 2008; Tang et al. 2015). In humans, the primordial germ cell are nearly devoid of methylation by weeks 10–11, with less than 8% median methylation levels (Guo et al. 2015). Imprinted epigenetic marks are also erased and will eventually be reestablished as development persists (Hajkova et al. 2002; Reik et al. 2001); however, some methylated loci persist during reprogramming and these regions have been shown to be associated with metabolic and neurological disorders, hinting at a mechanism for epigenetically inheritance traits (Tang et al. 2015). Importantly, while the eventual sperm and oocyte contribute equally to the genomic DNA of the embryo, they differ in their epigenetic contribution; sperm and oocyte establish and retain epigenetic signatures at imprinted loci. These epigenetic marks are established in the time between primordial germ cell development and gametogenesis and are important for normal development of offspring.
Spermatogenesis
Once embryonic development is complete, the next major window of epigenetic reprogramming is gametogenesis. In males, spermatogenesis is a complex, dynamic physiological process leading to the production of mature haploid spermatozoa from a diploid self-renewing spermatogonia stem cells (Figure 1A). Spermatogonia stem cells are sub-population of undifferentiated germ cells derived from primordial germ cells during embryo development as discussed in the previous section (de Kretser et al. 2016). At puberty, spermatogenesis is initiated and the entire process is generally considered to be subdivided into two major parts based on the cross-sectional analysis of the seminiferous tubules: spermatocytogenesis which encompasses both mitotic proliferation by the spermatogonia and meiotic division of spermatocytes, and spermiogenesis, the structural differentiation of spermatids (de Kretser et al. 2016; de Rooij 2017). (Figure 1A,B).
Figure 1: Sperm epigenetic programming during the preconception period.
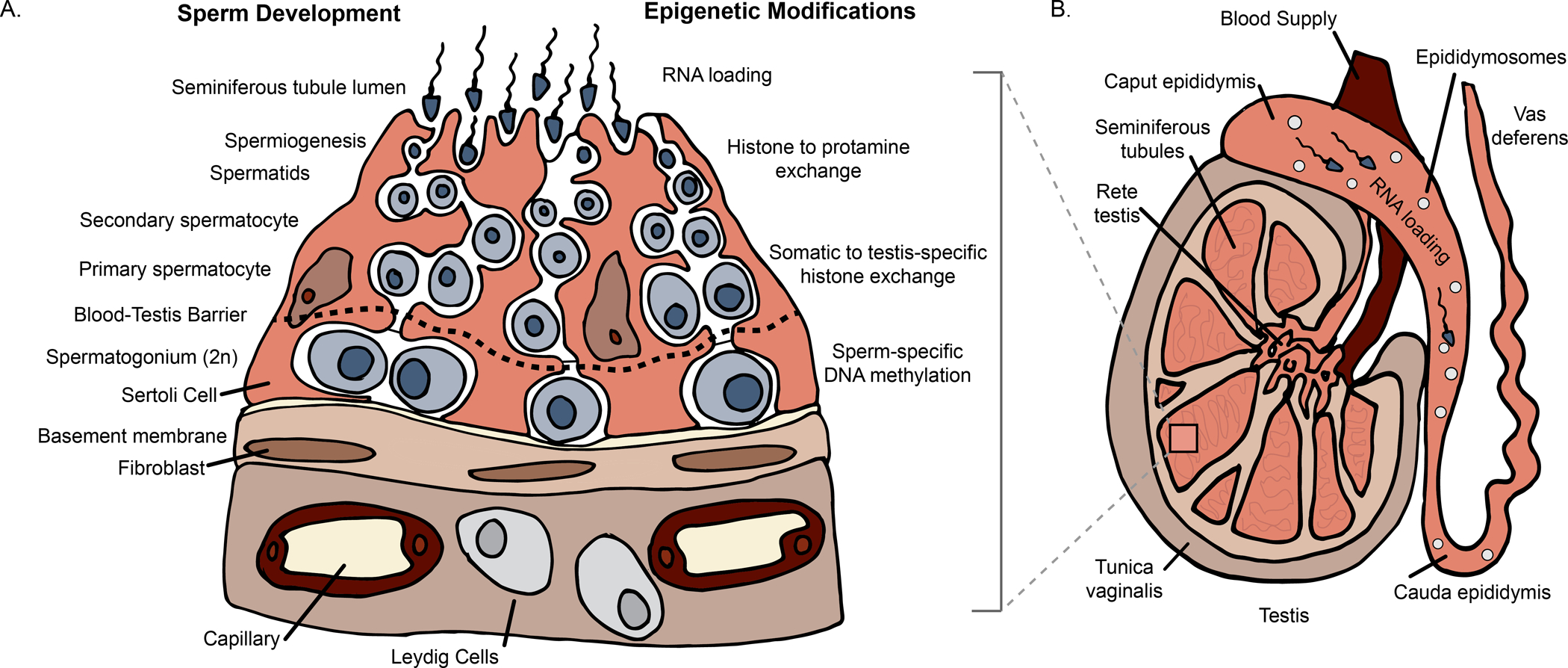
A) Sperm epigenetic profiles are finalized during spermatogenesis, a process divided into two major steps: spermatocytogenesis and spermiogenesis. During spermatocytogenesis, the undifferentiated spermatogonia undergo clonal expansion through mitosis to produce spermatocytes. It is important to note that this step occurs before the blood-testis barrier; thus the epigenome is potentially susceptible to environmental conditions during cell divisions. As spermatocytogenesis continues across the blood-testis barrier, spermatocytes obtain final DNA methylation profiles. Next, during spermiogenesis, spermatids are remodeled such that testis-specific histone marks are established, followed by histone to protamine exchange. B) Illustration of the testes, epididymis, and vas deferens. While spermatogenesis occurs primarily within the seminiferous tubules of the testes, final maturation occurs during the transit through the epididymis where RNA loading occurs with interactions between sperm and extracellular vesicles (e.g., epididymosomes). The RNA loading via extracellular vesicles within fluids of the male reproductive tract and other secondary sex organs (e.g., prostate and seminal vesicle) represents the final opportunity for sperm to “epigenetically match” their current environment prior to fertilization. (Low resolution copy)
Spermatogenesis is tightly regulated. Just as there are gene expression profiles and structural variations across germ cells (Li et al. 2019), natural epigenetic landscape distinctions and reprogramming have also been observed (McSwiggin and O’Doherty 2018; Rathke et al. 2014). For example, replication-dependent, passive demethylation may occur, especially at the early stages of spermatogenesis. This is followed by de novo methylation observed in mice spermatogonia and primary spermatocyte (Oakes et al. 2007) and has been linked to high expression of DNMT3a and 3b (La Salle and Trasler 2006). In turn, DNA methylation changes have been shown to be regulated by early onset interaction of H3K4me3 and H3k36me3 (Morselli et al. 2015).
Other core histones (H4, H2a, and H2b) have been reported in germline cells during mitotic and meiosis stages; however, at the mid-spermiogenesis elongation phase, the haploid round spermatids may also contain testis-specific histone variants in addition to the core histones. At this stage, the spermatids also undergo major structural modification and chromatin reconfiguration with vast majority of histones (~85% in human and 99% in mice) are replaced by protamines, resulting in a mechanically stable nucleus (Dadoune 2003; McSwiggin and O’Doherty 2018) (Figure 1A).
It is worth noting that the deposition of protamine is a gradual and well-guided chronological process, beginning with the incorporation of testis-specific histone variants, followed by histone (H4) hyperacetylation and transient formation of DNA double strand breaks, exchange of histones with transition proteins, and finally, integration of protamines (Carrell et al. 2007; Marcon and Boissonneault 2004). This suggests that to ensure proper packaging of sperm DNA, which is essential for protection in the female reproductive tract and ensuring adequate hydrodynamic properties, mature spermatozoa chromatin structure contains mostly protamines (Zhang et al. 2006). While the majority of sperm DNA is wrapped around protamines, histones are retained at a ratio within a narrow range that may be necessary for fertilization (Zhang et al. 2006). In addition, regions of histone retention are enriched for developmentally relevant genes such as the HOX cluster, suggesting that sperm histone retention also plays an important role in early embryonic development (Hammoud et al. 2009). Unlike most other mammals, both humans and mice sperm genome encode two protamine species (P1 & P2); specific ratios of both are particularly important for spermatogenesis and male fertility (Mengual et al. 2003) and have recently been considered as potential clinical biomarker of male infertility (Amor et al. 2017).
Following spermatogenesis, sperm exit the testis where maturation is completed in the epididymis (Figure 1B). The transit through the epididymis is an important developmental window that lasts between 1–2 weeks (Cornwall 2014). During this window, the sperm acquire their motile ability and their capacity to fertilize an oocyte due to their interactions with specific microenvironments along the route. Importantly, the sperm encounter differential gene, non-coding RNA, and protein expression. Along the length of the epididymis, the protein composition shifts, indicating a regionality to protein expression (Dacheux and Dacheux 2014). In addition, differentially expressed proteins and non-coding RNAs are often loaded in epididymal extracellular vesicles, referred to as epididymosomes (Baker et al. 2012; Belleannée et al. 2012; Cornwall 2014; Ijiri et al. 2011; U Sharma et al. 2016; Sharma et al. 2018). Interestingly, sperm interaction with epididymosomes resulted in dynamic shifts in miRNA profiles, such that a gain of 115 miRNAs was observed in mouse sperm collected from the cauda epididymis compared to sperm from the caput epididymis (Nixon et al. 2015). Together, the epididymal fluid and epididymosome composition may be altered based on the paternal environment (see below), which might act as a way to epigenetically prime the sperm prior to fertilization.
Hence, maintenance of a normal spermatogenesis state through the stages of development is crucial, offering a window of susceptibility to environmental conditions and an increased risk of epigenetic errors.
Aberrant Epigenetics Modification During Spermatogenesis and Male Infertility
The epigenetic features described in the previous section are assumed to be the normal state of the epigenome during each developmental window. Epigenetic profiles that deviates from this norm have been shown, in some cases, to be correlated with male infertility. Often times, imprinted genes are used as an epigenetic readout in these cases. This is because parent-specific epigenetic marks are protected during reprogramming and loss or misregulation of imprinted expression can be indicative of larger epigenetic dysregulation of the genome. Indeed, numerous studies have reported abnormal sperm methylation at imprinted loci of men with poor semen parameters compared to normozoospermic men (Boissonnas et al. 2010; El Hajj et al. 2011; Kobayashi et al. 2007; Marques et al. 2004; Marques et al. 2008; Minor et al. 2011; Poplinski et al. 2010). For instance, idiopathic infertile males, especially those with oligozoospermia, were more likely to have aberrant methylation profiles at imprinted control regions of imprinted genes such as H19, GNAS, and DIRAS3 when compared to fertile male (Tang et al. 2018). In another study, infertile males had significantly altered methylation levels at LIT1, SNRP, MEST, PEG3, ZAC, and H19. Interestingly, sperm methylation at imprinted loci of oligozoospermic patients were distinct from abnormal protamine patients, indicating that imprinted genes may be differentially affected depending on the clinical diagnosis (Hammoud et al. 2010).
In addition to imprinted genes, genome-wide methylation studies have allowed for the identification of differential methylation profiles associated with male infertility. Houshdaran and colleagues were the first to report broad methylation changes associated with abnormal semen parameters (Houshdaran et al. 2007). Studies have also reported distinct sperm methylation patterns associated with sperm with low motility (Jenkins et al. 2016a; Pacheco et al. 2011) and viability (Jenkins et al. 2016a). Additionally, in males with reduced fecundity, abnormal sperm methylation was observed at HSPA1L and HSPA1B (Jenkins et al. 2016b), while another study reported methylation changes in TYRO3, CGβ and FAM189A1(Laqqan and Hammadeh 2018). Lastly, Carell and colleagues found distinct sperm DNA methylation profiles between infertile and fertile normozoospermic men and that such sperm methylation patterns may provide predictive value in embryo quality assessment in an IVF setting (Aston et al. 2015).
Researchers have also found changes in histone retention and post-translational modification enrichment in infertile males. One study found that infertile males have randomly distributed histone retention across their genome when compared to fertile men. While localization of specific histone modifications were generally normal in infertile men across the genome, there was decreases in enrichment of H3K4me3 and H3K27me at imprinted loci and developmentally relevant genes (Hammoud et al. 2011)https://paperpile.com/c/J7e68p/3fUBu. This suggests small, cumulative changes in the histone state across the sperm epigenome might alter fertility status.
Together, the above evidence suggests that altered DNA methylation and chromatin landscape during spermatogenesis may affect sperm quality and reproductive outcomes. These are only some known changes in the sperm epigenetic profile that can occur, leading to changes in fertility outcomes. Importantly, the epigenetic landscape of sperm can be altered due to environmental exposure. Next, we will discuss the epigenetic changes that have been associated with specific paternal environmental conditions and exposures.
Preconception Environment and Sperm Epigenetics
In the previous section, we highlighted some of the epigenetic changes that occur during normal reprogramming and development of the male germ cell. It is clear that these windows are highly dynamic; aberrant epigenetic alterations during these windows have the potential to persist, altering the epigenetic landscape of the germline and/or offspring, potentially leading to negative health outcomes in future generations. We also discussed some of the known epigenetic alterations that are present in infertile or subfertile males. Next, we will describe the current state of results of how the preconception environmental exposures can affect sperm epigenetics and subsequently impact reproductive outcomes, early-life development, and offspring health.
Endocrine-disrupting compounds
Endocrine-disrupting chemicals (EDCs) are exogenous chemicals, or a mixture of chemicals, that can mimic, block, or interfere with any aspect of endogenous hormone action (reviewed in (Zoeller et al. 2012)). Given the broad diversity of chemicals that display endocrine disrupting properties, the reported adverse health effects of EDCs are broad. These health effects include adverse female and male reproductive health outcomes, obesity, diabetes, and cardiovascular disease, among others (reviewed in (Gore et al. 2015)). While research on EDC-induced health effects has dramatically increased over the past decade, the effects of EDCs on sperm epigenetics have been limited but have gained traction over the last several years.
Phthalates are a family of synthetic chemicals that exhibit endocrine-disrupting characteristics and are widely used in a range of consumer products and industrial applications. Given the pervasive use of phthalates, it is not surprising that exposure is widespread. In fact, urinary phthalate metabolites have been detected in the majority of individuals from representative samples within the U.S. general population (Silva et al. 2004). Epidemiologic data associate male phthalate exposure with low sperm quality (Bloom et al. 2015; Jurewicz and Hanke 2011; Lyche et al. 2009; Wang et al. 2015). Beyond sperm quality effects, male phthalate exposures have been shown to be associated with poor embryo quality (Wu et al. 2017a) and time-to-pregnancy (Buck Louis et al. 2014). For instance, among 50 couples seeking IVF treatment and participating in the Sperm Environmental Epigenetics and Development Study (SEEDS), male urinary phthalate metabolite concentrations of MEHP, MBzP, MBP, MHBP, MMP, and the DINCH metabolite, MCOCH (all of which are considered anti-androgenic metabolites with the exception of MMP) were associated with a pronounced decrease in blastocyst quality (i.e., day 5) (Wu et al. 2017a). However, male phthalate concentrations were not associated with cleavage-stage (i.e., day 3) embryo quality. These results suggest that the negative influences of paternal phthalates on embryo quality arose after day 3, during the transition from the cleavage to blastocyst stage of embryo development. Interestingly, this coincides with the timing of zygotic genome activation. It is noteworthy that these findings are supported by previous results from the Longitudinal Investigation of Fertility and the Environment (LIFE) study showing that male urinary phthalate metabolite concentrations of MMP, MBP and MBzP were associated with a 20% reduction in fecundity as measured by time-to-pregnancy (Buck Louis et al. 2014).
In order to identify potential markers that might mediate the observed relationship between paternal urinary phthalate levels and poor reproductive outcomes, subsequent research from SEEDS investigated the role of sperm methylation via the Illumina 450k array. After adjusting for potential confounders, a total 131 differentially methylated regions (DMRs; q<0.05) were associated with 10 different urinary phthalate metabolite concentrations. Moreover, three anti-androgenic metabolites, MEHP, MBP, MCOCH that were previously found to be associated with poor blastocyst quality (Wu et al. 2017b), were associated with the majority of the sperm DMRs. Interestingly, functional analysis of these sperm DMRs revealed a potential role in blastocyst quality, such that significantly enriched pathways identified included growth and development and cellular function and maintenance. Thus, these results suggest that phthalates may impact sperm DNA methylation in or near genes relevant to early embryogenesis, providing a pathway linking the observed inverse associations between anti-androgenic phthalates and blastocyst quality (Wu et al. 2017a; Wu et al. 2017b) and time-to-pregnancy (Buck Louis et al. 2014). In support of the influence of phthalates on sperm methylation aberrations, DEHP exposure in an occupational setting was reported to be inversely associated with LINE-1 methylation; however, the use of high melting resolution PCR used in this study provided no contextual genomic insights of their reported associations (Tian et al. 2019). Future studies in human and animal models are forthcoming and hold promise to further delineate the role of sperm epigenetics as a pathway by which male phthalates influence reproductive success and offspring development.
Bisphenol A (BPA) is another ubiquitous EDC commonly produced in high volume and widely used in the manufacturing of products like plastics, thermal receipt papers, medical devices, can linings, and dental sealants (Shelby 2008). The influence of BPA on male reproductive function has been reviewed previously (Manfo et al. 2014); however, the influence of preconception BPA exposures on sperm epigenetics is now emerging. Utilizing hydroxymethylated DNA immunoprecipitation (hMeDIP) and sequencing, Chinese men who were occupationally-exposed to BPA had higher global content of sperm 5hmC compared to the non-exposed group (Zheng et al. 2017). Additionally, 9,610 differentially 5hmC regions were identified, most of which were enriched within the intergenic and intronic regions, promoters of known imprinted genes, and regions in sperm associated with histone 3 trimethylation (H3K27me3, H3K4me2, and H3K4me3). This suggests a potential interplay between 5hmC and sperm-specific histone modifications (Zheng et al. 2017). Subsequent data from this research team showed that BPA-exposed compared to non-exposed workers had higher sperm 5hmC, as measured by qPCR, at the acetylcholinesterase (ACHE) gene locus; however, it must be noted that after adjusting for potential confounders, the relationship between BPA exposure and ACHE 5hmC was attenuated. While these studies suggest that BPA may alter sperm 5hmC in occupationally exposed men, the relationship between BPA and 5mC was not examined and the generalizability of such results obtained in an occupational setting to the general population needs further investigation.
Dioxins are a broad class of structurally related, stable organochlorinated compounds released into the environment as a byproduct of industrial processes such as smelting and incineration, as well as from natural sources like forest fires and volcanic eruptions (Dopico and Gómez 2015). Because of their stable nature, they are also ubiquitous and may persist in the environment, leading to long-term human exposure. Exposure often occurs from consumption of animal products, including meat, dairy products, and fish (Liem et al. 2000). Dioxins are known to negatively impact male reproductive system by altering spermatogenesis, epididymal structure, and the sperm epigenome during different sensitive windows of development (Pilsner et al. 2017). For instance, in a prospective cohort study of Russian boys, whole genome bisulfite sequencing (WGBS) revealed that serum concentrations of the dioxin, TCDD, in peripubertal boys (8 – 9 years) were associated with 52 DMRs in sperm collected in young adulthood (18 – 19 years). Functional enrichment analysis identified multiple biological pathways such as embryo development and cellular assembly and organization, in which estrogen receptor alpha was suggested to be the central regulator. It must be noted that while this study focused on peripubertal serum concentrations, TCDD’s long half-life (approximately 7 years) could have impacted other epigenetic sensitive periods including differentiation of primordial germ cells as well as spermatogenesis in adulthood (Pilsner et al. 2018). Recently, a study among U.S. veterans of Operation Ranch Hand exposed to Agent Orange in Vietnam, who were highly exposed to dioxin via Agent Orange herbicide, found 36 sperm DMRs (p < 0.05) among high and low exposure groups (Kelsey et al. 2019) as well as similar directions of association in 5 DMRs previously reported by Pilsner et al. (Pilsner et al. 2018).
Drug Use
Pharmaceutical drug use, especially during cancer treatments, may affect off-target tissues like germ cells, resulting in genetic defects that are transferable to next generation (Wyrobek et al. 2005). In addition to genetic alterations, pharmaceutical drugs may also induce epimutations in developing germ lines at sensitive genomic regions (Shnorhavorian et al. 2017). For example, sperm methylation alterations in adult men exposed to chemotherapy for treatment of osteosarcoma least 10 years prior to the study were compared to unexposed control men using MeDIP-seq. The study found that chemotherapy was associated with over 2,000 DMRs (p = 0.0001), mostly within CpG desert regions distributed over the entire chromosomes, including sex chromosomes. Similarly, in a separate rat model study that utilized restriction landmark genomic scanning (RLGS) to investigate methylation patterns of male rat germ cells exposed to different doses of combined testicular cancer therapy (bleomycin, etoposide, and cis-platinum (BEP)). They found a total of 143 loci significantly altered in both spermatids and mature spermatozoa relative to controls with methylation patterns proceeding either upward or downwards (Chan et al. 2012). Taking together, it suggests anti-cancer chemotherapy may alter epigenetic profile of the germline.
Similar to pharmaceutical drugs, the causal effects of recreational drug use on health may be mediated by an altered epigenome. While the negative effect of maternal tobacco smoking during pregnancy on offspring health and development are well delineated, comparatively less attention has been given to the effect of paternal smoking; however, emerging evidence details the negative impact of paternal tobacco use on fertility and offspring health (Beal et al. 2017). With a myriad of exposures occurring during cigarette smoking including nicotine, particulate matter, and thousands of other harmful chemicals, the negative effects of smoking on biological targets and pathways are multidimensional (Talhout et al. 2011). This includes effects on semen parameters as indicated in a recent meta-analysis (R Sharma et al. 2016).
While smoking has been associated with epigenetic aberrations in somatic tissue (Ambatipudi et al. 2016; Besingi and Johansson 2014; Li et al. 2018), recent studies have shifted their focus on the role of current smoking on modifying the sperm methylome. Among a group of fertile men who were current smokers compared to nonsmokers, 450K array analyses identified 11 differentially methylated CpG sites, including CpG sites near genes, PGAM5 and PTPRN2, which have been shown to be important for spermatogenesis (Alkhaled et al. 2018; Laqqan et al. 2017). Moreover, in comparing sperm methylation among 78 current smokers with 78 never-smokers, Jenkins et al. found 141 CpG sites that were differentially methylated including a trend of increased variability of DNA methylation among smokers, suggesting that smoking may induce stochastic genome-wide aberrations in DNA methylation. Smoking-induced differential methylation was found to be enriched in CpG shores and in regions of histone retention, specifically trimethylation of H3K4 and H3K27 (Jenkins et al. 2017). While there were no shared differentially methylated CpGs across the above cited studies, they report that smokers had poorer semen quality compared to non-smokers (Alkhaled et al. 2018; Jenkins et al. 2017). The temporal relationship between differential methylation and semen quality have yet to be delineated; smoking-induced methylation changes may play a causal role in poor semen quality or alternatively, abnormal spermatogenesis may impact DNA methylation profiles.
In another study, targeted methylation approaches linked cigarette smoking to methylation changes at imprinting control regions (ICR) of the two imprinted genes studied. Specifically, when compared to nonsmokers, male smokers displayed lower methylation at H19 and higher methylation at SNRPN (Dong et al. 2017). Taken together, epidemiologic data clearly indicates that smoking, even in fertile men, can alter sperm DNA methylation profiles; however, a causal role of smoking-induced sperm DNA methylation aberrations on adverse offspring health is currently unknown in humans. However, a recent mouse study has begun to provide a mechanistic link between smoking-induced methylation changes and subsequent behavior of F1 and F2 offspring. For instance, adult male C57BL/6 mice exposure to nicotine via drinking water for 12 weeks induced both intergenerational behavioral impairment in both male and female (F1) offspring. Additionally, transgenerational (F2) deficits in learning were observed in male mice derived from paternally nicotine-exposed female F1 (McCarthy et al. 2018). While genome-wide methylation of F0 sperm was not analyzed, targeted methylation approaches using qPCR found that F0 sperm from nicotine-exposed males had higher global methylation and lower methylation at dopamine D2 receptor promoter compared to sperm from non-exposed males (McCarthy et al. 2018).
In addition to cigarettes, cannabis is one of the most widely used psychoactive drugs worldwide and has gained further popularity in daily use with the legalization of recreational and medicinal use in many developed countries including the United States. The effects of cannabis use on health outcomes have been discussed elsewhere; these include adverse child health from prenatal use (reviewed in (Gunn et al. 2016)), male infertility (Rajanahally et al. 2019), and potential epigenetics effects (reviewed in (Szutorisz and Hurd 2016). Currently, data on cannabis use and sperm epigenetics are beginning to emerge. Recently, Murphy et al., examined the associations between cannabis or tetrahydrocannabinol (THC) exposure and sperm methylation in humans and rats, respectively (Murphy et al. 2018). RRBS analyses identified 3,979 differentially methylated CpG sites in human sperm of cannabis users compared to non-users, with a significant pathway enrichment in Hippo signaling and cancer. These results are in concordance with their findings from rat sperm RRBS data. Interestingly, in a follow-up study, 9 of the 17 human sperm THC-sensitive CpG sites were found within the intron 7 of the Discs-Large Associated Protein 2 (DLGAP2) gene, which is strongly implicated in autism. The sites were validated in both human and rat sperm using pyrosequencing technique, with significant hypomethylation in the exposed group relative to controls (Schrott et al. 2019). This suggests sperm methylation may be used as important biomarker of THC exposure.
It is worth noting that several genes (e.g., PTPRN2 and MAPK8IP3) that were previously reported to be sensitive to cigarette exposure (Alkhaled et al. 2018; Laqqan et al. 2017; Murphy et al. 2018) were also differentially methylated with cannabis use, suggesting that the act of smoking, be it tobacco or cannabis, may impact the same biological pathways. In terms of potential offspring effects, THC-induced changes in rat sperm methylation (Murphy et al. 2018) overlapped with methylation changes previously reported in the brain of rat offspring born to parents exposed to THC during adolescence (Watson et al. 2015), indicating that preconception cannabis use may impact neurodevelopment of the next generation via transfer of aberrant sperm epigenetic profiles.
Aligned with the effects of cigarettes and cannabis, recent studies also point to the remodeling of the sperm epigenome associated with alcohol use. Notably, preconception ethanol exposure in male mice produced male, but not female, progeny with reduced ethanol preference and consumption. Additionally, there was increased brain-derived neurotrophic factor (Bdnf) expression in the ventral tegmental area (VTA), a region of the brain associated with cognition, motivation, and when misregulated, psychiatric disorders. Remarkably, Bdnf was found to be hypomethylated in F0 sperm, as well as in the VTA of male and female F1 offspring (Finegersh and Homanics 2014). Most recently, preconception alcohol exposure was found to alter three classes of sperm small RNAs (tRNA, mitochondrial small RNA, and miRNA). Additionally, alcohol exposure impacted the epitranscriptome by increasing two nucleoside modifications: a uridine modification, 5′-methylaminomethyl-2-thiouridine, and a cytidine modification, formylcytidine. Moreover, ethanol-responsive sperm tRNAs were similarly altered in epididymosomes, suggesting a dynamic cross-talk and cargo exchange between sperm and extracellular vesicles (EVs) during epididymal transit (Rompala et al. 2018).
Diet
Lifestyle factors such as diet and exercise play a critical role in overall health for the individual, as well as to the health of offspring. As such, most of the initial research examining how the preconception environment shapes sperm epigenetics and subsequent offspring phenotype was derived from nutritional manipulation studies in rodent models. For example, while Rando and colleagues showed that low protein diets (LPD) had only modest effects on genome-wide DNA methylation of sperm, they found that LPD resulted in the down-regulation of transcriptional factors and chromatin regulators, as well as a decrease in H3K27me3 of specific loci in sperm (Carone et al. 2010). Subsequent work from this group found that LPD in male mice resulted in changes in small RNA profiles in epididymosomes that matched changes observed in mature sperm (U Sharma et al. 2016), supporting the hypothesis of RNA trafficking from EVs during the epididymal transit of sperm. These LPD-induced changes in small RNA profiles most notably affected the let-7 family of miRNAs and tRNA fragments, which have been reported to be in high concentrations in seminal plasma EV of humans (Vojtech et al. 2014). Furthermore, the change in sncRNA profiles induced by LPD affected preimplantation embryo development. For example, EV-derived tRNAs isolated from sperm of protein restricted mice and injected into zygotes resulted in repression of the gene targets of MERVL, a mouse retrotransposon express during preimplantation embryo development, as well as ribosomal genes in two-cell embryos that corresponded with delays in embryo development (U Sharma et al. 2016). The importance of sncRNA acquisition from epididymosomes during epididymal transit was highlighted in a recent paper showing that cauda sperm, but not caput sperm, and their associated sncRNA payloads were necessary for successful implantation (Conine et al. 2018). More recent data from another group, however, contradict such findings as they observed no differences in development and the number of pups obtained from caput and cauda sperm (Zhou et al. 2019). Finally, male mice exposed to LPD displayed global hypomethylation of sperm, reduced expression of DNA methyltransferases (DNMT1 and DNMT3L), and interestingly, blunted preimplantation uterine immunological and vascular remodeling, which suggests a seminal plasma-derived effect (Watkins et al. 2018). Nonetheless, the above studies indicate that LPD diets in adult males is sufficient to modify the sperm epigenetic repertoire and/or EV cargo to influence early-life development.
Similarly, studies with high fat diets (HFD) have also been shown to influence sperm epigenetics. For instance, HFD in adult male mice resulted in altered miRNA content (Fullston et al. 2013) and increased acetylation of H3K9 in late round spermatids to early elongating spermatids. The changes in H3K9ac are possibly mediated by a corresponding decreased expression of SIRT6, a stress-response deacetylase (Palmer et al. 2011). The effect of HFD on global DNA methylation of sperm is inconsistent (Binder et al. 2015; Fullston et al. 2013); however, in another study of prediabetic male mice, widespread alterations to sperm DNA methylation patterns were observed. Interestingly, a large portion of these differentially methylated genes overlapped with genes expressed in pancreatic islets of the offspring (Wei et al. 2014). Moreover, HFD in male rats was shown to induce metabolic phenotypes transgenerationally (e.g., F1 and F2 offspring), and interestingly, DNA methylation and certain sncRNA (piRNAs and miRNA let-7c) expression of F0 and F1 sperm shared common profiles, indicating that paternal exposures may impact epigenetic patterns in offspring germ cells (de Castro Barbosa et al. 2016).
While the role of sncRNAs such as miRNAs and endo-siRNAs in fertilization and embryonic development is well elucidated (Yuan et al. 2016), miRNA specific function as vector of paternal inheritance of metabolic diseases was first observed following male mice exposure to Western-like diet characterized with high fat and/or high sugar levels (Grandjean et al. 2015). The study found that mice fed a Western-style diet acquired metabolic disorders relative to the control standard mice diet. The diet significantly altered sperm RNA populations, with miRNA being the most sensitive sub-RNA species. Furthermore, injection of sperm miRNA (miR-19b) into naive one-cell embryo resulted in offspring with metabolic disorders similar to the exposed F0 phenotypes.
More recently, additional work further emphasized the importance of sperm sncRNA in the inheritance of acquired diseases (Qi Chen et al. 2016), where sperm 5’ tRNA fragments were found to be most responsive to HFD exposure using mice models. Interestingly, injection of the altered sperm tsRNA fragments from HFD males into normal zygotes recapitulated metabolic disorders in F1 offspring. Further experiments by the group showed that most predominant sub-species of tRNAs were those with methylation at the 5th-positioned cytosine (m5C) of the tRNA fragment, implying that modification of tRNAs are also important for paternal-offspring transmission of HFD-induced metabolic disorders (Qi Chen et al. 2016). In fact, laboratory-synthesized tRNA fragments, which contain no modifications, were readily degraded and failed to recapitulate the F1 phenotype. This underscores the notion that not only are certain species of sncRNA responsive to environmental cues, but specific modifications to RNA nucleotides (e.g., epitranscriptome) may be equally important in conferring the inheritance of paternal acquired traits. Interestingly, a follow-up study by the same group showed knockout of DNMT2 resulted in down-regulation of modified tsRNA (m5C) in sperm and nullified the effects of HFD-induced intergenerational transmission (Zhang et al. 2018). This suggests DNMT2 might be an important mediator of diet-induced intergenerational metabolic disorders through sperm RNA modifications.
While the previously discussed research utilized nutritional manipulation exclusively in animal models, epidemiologic evidence indicates that children of obese fathers may have higher likelihood of being overweight irrespective of mother’s obesity status (Lake et al. 1997), suggesting a potential role for the epigenetic transfer of sperm profiles. Indeed, paternal overweight or obesity status prior to conception was associated with sperm methylation at several ICRs, including PEG10, SNRPN, MEG3 and H19 (Soubry et al. 2016). Additionally, this group previously reported that paternal obesity was associated with cord blood leukocyte methylation of IGF2 (Soubry et al. 2013); however, no significant association was found for sperm IGF2 methylation (Soubry et al. 2016).
Perhaps the most convincing data on the effect of obesity is from a novel study by Donkin and colleagues (Donkin et al. 2016). Epigenome-wide profiling of sperm from obese and lean men found similar histone positioning; however, DNA methylation profiles and sncRNA profiles diverged between the groups. Specifically, piRNAs and tRNA fragments were the most predominantly differentially expressed sncRNA. Interestingly, target prediction of the 37 differentially expressed piRNAs (q<0.1) resulted in genes putatively involved in regulating food intake related to obesity. Most fascinating, this study also examined sperm methylation profiles in obese men before and after bariatric gastric bypass surgery. Compared to sperm collected prior to surgery, 1,509 and 3.910 sperm DMRs, most notably at genes implicated in appetite, were identified one week and one year after the surgery, respectively (Donkin et al. 2016). This suggests that the sperm epigenome is malleable to current environmental conditions and thus can be modified after weight loss. Finally, in addition to sncRNAs, Krawetz and colleagues recently reported that male BMI was associated with 487 sperm RNA elements (defined as short exon-sized sequences) consisting of 421 genes, in which a number of genes were related to epigenetic processes that regulate chromatin organization (Swanson et al. 2019).
In additional to overall diet, micronutrient intake can also alter the epigenome. For example, dietary supplementation with folic acid has been shown to improve semen parameter, especially among men with polymorphism in folate metabolizing gene, methylenetetrahydrofolate reductase (MTHFR), (Huang et al. 2019; Najafipour et al. 2017) and may be considered a part of oral micronutrient supplementation in male factor infertility treatment (Buhling et al. 2019). However, since folate is crucial in the production of S-adenosyl methionine, the universal methyl donor, whether this exposure could alter other biological markers in developing germ cells with potential effect on future generation remains a concern. Using RRBS, Trasler and colleagues investigated sperm methylation profiles of idiopathic infertile men exposed to long-term, high dose folic acid supplementation at 5 mg/d for 6 months (Aarabi et al. 2015). They observed significant global hypomethylation mostly occurring at low CpG density and DNA repeat regions; an effect found to be exacerbated by MTHFR polymorphism. The unanticipated methylation loss was validated in a follow-up mice study by the same research group and was attributed to downregulation of MTHFR enzyme at high doses of folic acid (Aarabi et al. 2018), although the mechanism remains unclear. It is however worth mentioning that low level of folic acid (400 μg/d) administered over 90 days failed to alter sperm methylome of men without history of infertility using HumanMethylation450 beadchip array (Chan et al. 2017). Taken together, sperm methylome sensitivity to high level of folic acid may be species independent and may be modified by underlying genetic status.
Exercise
To mitigate the potential negative effects of chemical exposures and diet discussed above, there is growing interest in the effect of enriched or positive environmental exposures such as exercise on sperm epigenetics. While it was evident that exercise can alter the epigenetic state of muscle cells in rats (Keller et al. 2011) and humans (Jacques et al. 2019), recent data indicate that the sperm epigenome may be responsive to exercise in a similar fashion. Indeed, 8-week nutrition/exercise interventions (~ 2 spermatogenesis cycles) to obese male mice restored the abundance of sperm miRNAs, predominantly found on the X-chromosome, that target genes regulating cell cycle, apoptosis, and embryogenesis, to that of controls. Additionally, interventions mitigated the propensity for the development of obesity and metabolic disease in female offspring (McPherson et al. 2015). Likewise, targeted analyses by qPCR revealed that 12-week exposure to voluntary wheel-running, compared to sedentary controls, resulted in changes in sperm DNA methylation in several metabolic genes and miRNA expression. Surprisingly, offspring of males subjected to exercise were more susceptible to the adverse effects of a high-fat diet, such as increased body weight and impaired glucose tolerance, suggesting that paternal exercise may induce a thrifty phenotype in offspring (Murashov et al. 2016). In a similarly designed study, voluntary wheel-running also altered the expression of five miRNA and tRNA fragments via unbiased sncRNA-sequencing (Short et al. 2017), further highlighting the importance of sncRNA in sperm.
In humans, the effect of exercise has also been shown to modify the sperm epigenome. In one such intervention, sprint interval training twice a week for 3 months resulted in global hypomethylation of sperm and genome-wide methylation changes occurring at gene loci implicated in schizophrenia and Parkinson’s disease (Denham et al. 2015). It must be noted, however, that a major limitation of this study was that it did not evaluate intra-individual changes in sperm methylation, thus the direct effect of exercise within an individual is unknown. To overcome this limitation, Ingerslev and colleagues examined sperm epigenetic profiles at three different time-points: prior to exercise (untrained), after 6-weeks of endurance training regime (trained), and 3 months following exercise (detrained) (Ingerslev et al. 2018). Compared to untrained, 177 and 190 DMRs (q <0.05) were identified in sperm collected at trained and detrained periods, respectively. While the authors do not state the amount of overlap of DMRs between analyses, ontology analyses indicated DMRs were associated with genes known for neurological function. In regard to sncRNA, exercise-induced alterations in piRNA expression were reported, while expression of miRNA and tRNA remained fairly stable (Ingerslev et al. 2018). Overall, dietary and exercise lifestyle choices made by fathers before the conception period may alter sperm epigenetic profile, affect semen quality, and reprogram the health of future generations. This effect is likely independent, but potentially additive, to mothers’ lifestyle.
Environmental stressors and other environmental cues
The aforementioned studies on preconception environment focused largely on exposures that potentially could be modified via lifestyle changes; however, stress or trauma is another key component of the preconception environmental milieu that is less malleable and has shown to impart adverse behavioral outcomes in offspring. For example, epidemiologic evidence indicates that offspring of Holocaust survivors (Yehuda et al. 2014), Vietnamese refugees (Vaage et al. 2011), and military action in war (Schick et al. 2013) had increased risk of mental health issues (reviewed in (Bowers and Yehuda 2016)).
Not surprisingly, recent studies have focused on sperm epigenetics as the mechanism of paternal transfer of stress. For instance, paternal pubertal or adult chronic variable stress conditions in mice were found to alter miRNA profiles of sperm and contributed to offspring hypothalamic-pituitary-adrenal (HPA) axis dysregulation (Rodgers et al. 2013). Other interesting studies have shown that the sperm epigenome adapts to other environmental cues such as temperature and odors. Perhaps one of the most fascinating studies examined the inheritance of parental traumatic exposures via odor fear conditioning (Dias and Ressler 2014). In doing so, male mice exposed to acetophenone, which activates the odorant receptor Olfr151, concomitant with mild foot shocks, resulted in F1 and F2 offspring with increased behavioral sensitivity to acetophenone. Most strikingly, bisulfite sequencing of sperm from F0 and F1 males revealed that Olfr151 was hypomethylated (Dias and Ressler 2014). Lastly, synonymous to human cold seasons, preconception cold exposure in male, but not female, mice was linked to improved systemic metabolism and protection from diet induced obesity in male offspring. WGBS analyses revealed that sperm from cold exposure increased in global methylation, and moreover, 2,431 DMRs were identified, which were enriched in pathways for neurogenesis (Sun et al. 2018). Taken together, these data demonstrate that parental experiences are also epigenetically written in the male germline and can lead to changes in offspring phenotypes.
Perspectives and Future Work
Improving our understanding of epigenetic changes that occur to the paternal germline following preconception environmental conditions and exposures will enhance reproductive success as well as improve offspring health. Currently, most of the preconception environmental research on the sperm epigenome focuses on DNA methylation and ncRNAs; technical limitations are often the reason. Moving forward, it will be important to identify other environmentally-sensitive epigenetic mechanisms, including local histone modification and enrichment, global chromatin structure and 3-D conformation, and non-coding RNA exosomal loading and expression in seminal plasma and epididymosomes. Advances in systems biology to ably integrate and analyze the complex and multi-faceted epigenetic information are also warranted. Research is also necessary to understand the variability of sperm epigenetic profiles within one semen sample. Currently, research is reporting average methylation profiles within a semen sample and thus, it is unknown if environmental conditions uniformly impact sperm epigenetic profiles or if a subset of spermatogonia are more sensitive to exposures. Such information may lead to the development of diagnostic tools to separate sperm by their epigenetic profiles in order to select sperm that have the best reproductive capacity and prognosis for optimal health and development of future generations.
Table 1:
Selected studies on paternal preconception exposures and sperm epigenetic aberrations.
| Exposure | Species | Method | Sperm epigenetic aberrations | Implications | References |
|---|---|---|---|---|---|
| Endocrine-disrupting compounds | |||||
| Phthalates | Human | 450K Array MassArray | Environmental relevant levels of anti-androgenic urinary phthalate metabolites (MEHP, MBP and MCOCH) were associated with the majority of sperm DMRs, which were enriched in genes associated with blastocyst quality, growth and development, and cellular function and maintenance. Additionally, 17 DMRs were associated with diminished blastocyst quality. | Spermatogenesis/embryo quality | Wu et al. 2017b |
| DEHP | Human | HRM-PCR | Occupational exposure to DEHP exposure was inversely associated with LINE-1 DNA methylation. | Infertility | Tian et al. 2019 |
| Bisphenol A(BPA) | Human | 5-hMeDIP-seq | 9,610 sperm DMRs were identified in occupationally exposed men compared to controls. Correlation between urinary BPA levels and 5hmC was not examined. | Spermatogenesis | Zheng et al, 2017 |
| BPA | Human | qPCR | Higher sperm 5hmC at the ACHE gene locus was found among factory workers with BPA exposure compared to unexposed factory workers. However, no significant correlation between urinary BPA and ACHE 5hmC was found among all subjects. | Sperm quality | Song et al., 2019 |
| Dioxins | Human | WGBS | 52 sperm DMRs were identified in young adults who had high peripubertal (8–9 years of age) blood dioxin (TCDD) levels compared to those with low peripubertal blood dioxin levels. Enrichment of DMRs occurred at loci functionally relevant for development and cellular functions. | Embryo development/sperm functions | Pilsner et al. 2018 |
| Dioxins | Human | 450K Array | High serum level TCDD among Operation Ranch Hand Veterans was associated with methylation alterations at 36 sperm gene regions that included H19, compared to control. Data comparison with previous study (Pilsner et al. 2018) showed 5 significant overlapping loci with consistent methylation directionality. | Embryo development/sperm quality | Kelsey et al. 2019 |
| Drug use | |||||
| Chemotherapy | Human | MeDIP-seq | Chemotherapy was significantly associated with over 2,000 DMRs mostly within the CpG desert regions of exposed sperm compared to control. | Inheritance | Shnorhavorian et al. 2017 |
| Chemotherapy (BEP) | Rat | RLGS | A total 143 loci were reported to be significantly altered in both spermatids and mature spermatozoa of exposed rat relative to controls with methylation. | Spermatogenesis | Chan et al. 2012 |
| Cigarettes | Human | 450K Array | 141 CpGs were associated with smoking, 74% of which were hypomethylated and occurred most frequently at regions previously reported to display H3K4 and H3K27 methylation in mature sperm. | Spermatogenesis | Jenkins et al. 2017 |
| Cigarettes | Human | Pyrosequencing | Cigarette smoking altered sperm methylation of H19 and SNRPN and was found to be associated with increased risk of poor sperm motility and morphology. | Sperm quality | Dong et al. 2017 |
| Cigarettes | Human | 450K Array | Current smokers exhibited differential methylation at 7 CpG sites related to MAPK8IP3 and TKR genes, important for capacitation and spermatogenesis respectively. | Sperm function/spermatogenesis | Laqqan et al. 2017 |
| Cigarettes | Human | 450K Array | Cigarette smoking was associated with 11 CpG loci in sperm within PGAM5, PTPRN2 and TYRO3, the latter is important for spermatogenesis. | Spermatogenesis | Alkhaled et al. 2018 |
| Nicotine | Mice | MeDIP; qPCR | Nicotine exposure induced changes in global and dopamine-specific sperm DNA methylation with inter- and transgenerational adverse health effects. | Inheritance | McCarthy et al. 2018 |
| Cannabis THC | Human and Rat | RRBS; Pyrosequencing | Cannabis use was associated with 3,979 sperm DMRs including genes, PTPRN2 and MAPK8IP3, previously found to be sensitive to tobacco exposure. There was substantial overlap in DMRs between rat (THC-exposure) and human sperm. | Spermatogenesis | Murphy et al. 2018 |
| Cannabis THC | Human and Rat | Pyrosequencing | This study validated the sensitivity (hypomethylation) of specific loci found within intronic region of DLGAP2, a gene strongly implicated in autism. | Inheritance | Schrott et al. 2019 |
| Ethanol | Mice | RT-qPCR | Ethanol exposure resulted in hypomethylation of Bdnf promoter of sperm, which was maintained in the ventral tegmental area of offspring. | Inheritance | Finegersh and Homanics 2014 |
| Ethanol | Mice | RT-qPCR | Ethanol exposure was associated with altered expression of sperm small RNAs (tRNA, mitochondrial small RNA, and microRNA) and epididymosomal tRNA. | Inheritance | Rompala et al. 2018 |
| Diet | |||||
| Low protein diet (LPD) | Mice | RRBS; MeDIP | LPD induced depletion of gene promoter H3K27me3 in F0 sperm compared to control. Offspring of LPD sires exhibited altered methylation at promoters of liver PPARa gene relative to control. | Inheritance | Carone et al. 2010 |
| LPD | Mice | RT-qPCR; RNA-seq | LPD was associated with downregulation of let-7 miRNA and upregulation of 5’ tRFs in sperm, a profile that matched LPD-induced sRNA alterations in epididymosomes. | Embryo development | Sharma et al. 2016 |
| LPD | Mice | MBD2-seq; RT-qPCR | LPD induced global hypomethylation and reduced expression of Dnmt1 and Dnmt4l in sperm. Offspring of LPD fathers gained more weight compared to offspring of controls. | Inheritance | Watkins et al. 2018 |
| High fat diet (HFD) | Mice | Microarray; RT-qPCR; ELISA | HFD caused differential expression of 4 miRNAs in sperm as well as global hypomethylation in late elongating spermatids. | Inheritance | Fullston et al. 2013 |
| HFD + STZ | Mice | Microarray; MeDIP-seq; | HFD & STZ-induced paternal prediabetes altered overall sperm methylome and was associated with genes largely overlapping with offspring’s pancreatic islets. | Inheritance | Wei et al. 2014 |
| HFD + high sugar | Mice | RT-qPCR, RNA‐seq | Western-like diet, characterized with high HFD and/or high sugar, altered sperm RNA population, predominantly miRNA. Injection of miR-19b into naive embryo produced offspring with metabolic disorder similar to F0 phenotypes. | Embryo development/Offspring diseases | Grandjean et al. 2015 |
| HFD | Mice | RT-qPCR; ELISA | No differential global methylation was observed in sperm of HFD-induced obese mice group relative to control. | Sperm function | Binder et al. 2015 |
| HFD | Rat | MBD; RT-qPCR; RNA-seq | HFD altered sperm DNA methylation profile and differential expression of sncRNAs (piRNAs and miRNA let-7c). Similar sperm epigenetic profile observed in male F1 generation. | Inheritance | De Castro Barbosa et al. 2016 |
| HFD | Mice | RT-qPCR, RNA‐Seq | HFD altered sperm tRNA and was associated with F1 metabolic disorder. Zygotic injection of sperm tRNA fraction from HFD males produced metabolic disorders in F1 offspring. | Inheritance | Chen et al. 2016 |
| HFD | Mice | RT-qPCR, RNA‐seq | Dnmt2 knock out reduced sperm tRNA and nullified metabolic disorders effect of HFD in offspring. | Inheritance | Zhang et al. 2018 |
| Folic acid | Human | RRBS | Long-term exposure to high level of folic acid resulted in global sperm hypomethylation predominantly at low CpG density and DNA repeats regions. | Infertility | Aarabi et al. 2015 |
| Folic acid | Human | 450K Array | Sperm DNA methylation was not altered from short-term supplementation with low dose folic acid. | - | Chan et al. 2017 |
| Folic acid | Mice | RRBS | At high dose folic acid, sperm hypomethylation occurred in heterozygous Mthfr genotype mice but not in wildtype. Hypomethylation in wild type sperm was only observed at extremely high dose with deficiency in Mthfr expression. | Infertility | Aarabi et al. 2018 |
| Obesity | |||||
| Obesity/weight loss | Human | MNase-seq; RRBS; RNA-seq | Obese men had altered sperm piRNAs compared to lean men. Sperm DNA methylation was remodeled after gastric-bypass surgery-induced weight loss. | Sperm quality | Donkin et al. 2016 |
| Obesity | Human | Pyrosequencing | Obese men had significantly altered methylation at imprinted loci of sperm compared to lean men. | Sperm quality | Soubry et al. 2016 |
| BMI | Human | RNA-seq | Male BMI was associated with 421 short exon sized RNA elements, which were related to epigenetic processes that regulate chromatin organization. | Sperm quality | Swanson et al. 2019 |
| Exercise | |||||
| Long-term exercise | Mice | MBD; RT-qPCR | Mice subjected to 12 weeks exercise had altered sperm DNA methylation in metabolic genes and miRNA profiles. | Inheritance | Murashov et al. 2016 |
| Voluntary wheel running | Mice | RNAseq; RT-qPCR | Mice that experienced voluntary wheel-running exercise exhibited altered sperm miRNA and tRNA expression. Also, male offspring of runners had suppressed juvenile fear memory and reduced anxiety. | Inheritance | Short et al. 2017 |
| Short term training/detraining | Human | RNA-seq; RRBS | Exercise reversed alteration in sperm piRNA expression. Training and detraining induced methylation alterations at regions functionally related to neurogenesis, neuron differentiation, and axon guidance. | Development | Ingerslev et al. 2018 |
| Environmental stress | |||||
| Peripubertal/adult trauma | Mice | RT-qPCR | Pubertal or adult-exposure to chronic variable stress altered sperm miRNA profiles, which was associated with dysregulation of HPA stress axis in both male and female offspring. | Inheritance | Rodgers et al. 2013 |
| Odor | Mice | ChIP; RT-qPCR | Compared to control, odor fear conditioned F0 mice displayed CpG hypomethylation of sperm Olfr151 gene with similar methylome alteration in F1 sperm. | Inheritance | Dias & Ressler 2014 |
| Cold | Mice | Pyrosequencing; WGBS | Cold exposed mice had significant sperm global CpG methylation alteration and DMRs (2,341) within or downstream of genes important for neurogenesis relative to control. | Development/inheritance | Sun et al. 2018 |
Abbreviations: HRM: High-melting resolution; MBD: Methyl Binding Domain; MeDIP: Methylated DNA immunoprecipitation; 5-hMeDIP-seq: 5-hyroxymethyl cytosine DNA immunoprecipitation (hMeDIP) Sequencing; RRBS: Reduced representation bisulfite sequencing; WGBS: Whole genome bisulfite sequencing; RLGS: Restriction landmark genomic scanning; MNase-seq: Micrococcal nuclease-sequencing; ChIP: Chromatin immunoprecipitation; SMART-seq: Switch Mechanism at the 5′ End of RNA Templates-sequencing; ELISA: Enzyme-linked immunosorbent assay; STZ: Streptozotocin; HFD: High fat diet; LPD: Low protein diet; BEP : bleomycin, etoposide, and cis-platinum
Acknowledgements:
This work was generously supported by grants R21-ES026778, R01-ES028214 and R01-ES028298 from the National Institute of Environmental Health Sciences, National Institute of Health. The authors declare no competing interests.
Funding. This work was supported in part by grants, R21-ES02677, R01-ES028298 and R01-ES028214, from the National Institute of Environmental Health Sciences
References
- Aarabi M, San Gabriel MC, Chan D, Behan NA, Caron M, Pastinen T, Bourque G, MacFarlane AJ, Zini A, Trasler J. 2015. High-dose folic acid supplementation alters the human sperm methylome and is influenced by the mthfr c677t polymorphism. Human molecular genetics 24:6301–6313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aarabi M, Christensen KE, Chan D, Leclerc D, Landry M, Ly L, Rozen R, Trasler J. 2018. Testicular mthfr deficiency may explain sperm DNA hypomethylation associated with high dose folic acid supplementation. Human molecular genetics 27:1123–1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adenot PG, Mercier Y, Renard JP, Thompson EM. 1997. Differential h4 acetylation of paternal and maternal chromatin precedes DNA replication and differential transcriptional activity in pronuclei of 1-cell mouse embryos. Development 124:4615–4625. [DOI] [PubMed] [Google Scholar]
- Agarwal A, Mulgund A, Hamada A, Chyatte MR. 2015. A unique view on male infertility around the globe. Reproductive biology and endocrinology : RB&E 13:37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alkhaled Y, Laqqan M, Tierling S, Lo Porto C, Amor H, Hammadeh ME. 2018. Impact of cigarette-smoking on sperm DNA methylation and its effect on sperm parameters. Andrologia. [DOI] [PubMed] [Google Scholar]
- Ambatipudi S, Cuenin C, Hernandez-Vargas H, Ghantous A, Le Calvez-Kelm F, Kaaks R, Barrdahl M, Boeing H, Aleksandrova K, Trichopoulou A, Lagiou P, Naska A, Palli D, Krogh V, Polidoro S, Tumino R, Panico S, Bueno-de-Mesquita B, Peeters PH, Quirós JR, Navarro C, Ardanaz E, Dorronsoro M, Key T, Vineis P, Murphy N, Riboli E, Romieu I, Herceg Z. 2016. Tobacco smoking-associated genome-wide DNA methylation changes in the epic study. Epigenomics 8:599–618. [DOI] [PubMed] [Google Scholar]
- Amor H, Zeyad A, Ben Ali H, Hammadeh M. 2017. Protamines ratio (p1/p2) in spermatozoa as biomarker for male infertility. Fertil Steril 108:e142. [Google Scholar]
- Arand J, Spieler D, Karius T, Branco MR, Meilinger D, Meissner A, Jenuwein T, Xu G, Leonhardt H, Wolf V, Walter J. 2012. In vivo control of cpg and non-cpg DNA methylation by DNA methyltransferases. PLoS Genet 8:e1002750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aston KI, Uren PJ, Jenkins TG, Horsager A, Cairns BR, Smith AD, Carrell DT. 2015. Aberrant sperm DNA methylation predicts male fertility status and embryo quality. Fertil Steril 104:1388–1397 e1381–1385. [DOI] [PubMed] [Google Scholar]
- Baker MA, Nixon B, Naumovski N, Aitken RJ. 2012. Proteomic insights into the maturation and capacitation of mammalian spermatozoa. Syst Biol Reprod Med 58:211–217. [DOI] [PubMed] [Google Scholar]
- Balhorn R 2007. The protamine family of sperm nuclear proteins. Genome Biol 8:227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao J, Bedford MT. 2016. Epigenetic regulation of the histone-to-protamine transition during spermiogenesis. Reproduction (Cambridge, England) 151:R55–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beal MA, Yauk CL, Marchetti F. 2017. From sperm to offspring: Assessing the heritable genetic consequences of paternal smoking and potential public health impacts. Mutat Res 773:26–50. [DOI] [PubMed] [Google Scholar]
- Beaujean N 2014. Histone post-translational modifications in preimplantation mouse embryos and their role in nuclear architecture. Mol Reprod Dev 81:100–112. [DOI] [PubMed] [Google Scholar]
- Belleannée C, Thimon V, Sullivan R. 2012. Region-specific gene expression in the epididymis. Cell Tissue Res 349:717–731. [DOI] [PubMed] [Google Scholar]
- Besingi W, Johansson A. 2014. Smoke-related DNA methylation changes in the etiology of human disease. Hum Mol Genet 23:2290–2297. [DOI] [PubMed] [Google Scholar]
- Bestor T, Laudano A, Mattaliano R, and Ingram V. 1988. Cloning and sequencing of a cdna encoding DNA methyltransferase of mouse cells the carboxyl-terminal domain of the mammalian enzymes is related to bacterial restriction methyltransferases. J Mol Biol 203:971–983. [DOI] [PubMed] [Google Scholar]
- Binder NK, Sheedy JR, Hannan NJ, Gardner DK. 2015. Male obesity is associated with changed spermatozoa cox4i1 mrna level and altered seminal vesicle fluid composition in a mouse model. Mol Hum Reprod 21:424–434. [DOI] [PubMed] [Google Scholar]
- Bloom MS, Whitcomb BW, Chen Z, Ye A, Kannan K, Buck Louis GM. 2015. Associations between urinary phthalate concentrations and semen quality parameters in a general population. Hum Reprod 30:2645–2657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boissonnas CC, Abdalaoui HE, Haelewyn V, Fauque P, Dupont JM, Gut I, Vaiman D, Jouannet P, Tost J, Jammes H. 2010. Specific epigenetic alterations of igf2-h19 locus in spermatozoa from infertile men. Eur J Hum Genet 18:73–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bošković A, Bender A, Gall L, Ziegler-Birling C, Beaujean N, Torres-Padilla M-E. 2012. Analysis of active chromatin modifications in early mammalian embryos reveals uncoupling of h2a.Z acetylation and h3k36 trimethylation from embryonic genome activation. Epigenetics 7:747–757. [DOI] [PubMed] [Google Scholar]
- Bostick M, Kim JK, Estève P-O, Clark A, Pradhan S, Jacobsen SE. 2007. Uhrf1 plays a role in maintaining DNA methylation in mammalian cells. Science 317:1760–1764. [DOI] [PubMed] [Google Scholar]
- Bourc’his D, Xu GL, Lin CS, Bollman B, Bestor TH. 2001. Dnmt3l and the establishment of maternal genomic imprints. Science 294:2536–2539. [DOI] [PubMed] [Google Scholar]
- Bourc’his D, Bestor TH. 2004. Meiotic catastrophe and retrotransposon reactivation in male germ cells lacking dnmt3l. Nature 431:96–99. [DOI] [PubMed] [Google Scholar]
- Bowers ME, Yehuda R. 2016. Intergenerational transmission of stress in humans. Neuropsychopharmacology 41:232–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branco MR, Oda M, Reik W. 2008. Safeguarding parental identity: Dnmt1 maintains imprints during epigenetic reprogramming in early embryogenesis. Genes Dev 22:1567–1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun RE. 2001. Packaging paternal chromosomes with protamine. Nat Genet 28:10–12. [DOI] [PubMed] [Google Scholar]
- Buck Louis GM, Sundaram R, Schisterman EF, Sweeney A, Lynch CD, Kim S, Maisog JM, Gore-Langton R, Eisenberg ML, Chen Z. 2014. Semen quality and time to pregnancy: The longitudinal investigation of fertility and the environment study. Fertil Steril 101:453–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buhling K, Schumacher A, Eulenburg CZ, Laakmann E. 2019. Influence of oral vitamin and mineral supplementation on male infertility: A meta-analysis and systematic review. Reproductive biomedicine online 39:269–279. [DOI] [PubMed] [Google Scholar]
- Carone BR, Fauquier L, Habib N, Shea JM, Hart CE, Li R, Bock C, Li C, Gu H, Zamore PD, Meissner A, Weng Z, Hofmann HA, Friedman N, Rando OJ. 2010. Paternally induced transgenerational environmental reprogramming of metabolic gene expression in mammals. Cell 143:1084–1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrell DT, Emery BR, Hammoud S. 2007. Altered protamine expression and diminished spermatogenesis: What is the link? Hum Reprod Update 13:313–327. [DOI] [PubMed] [Google Scholar]
- Chachamovich JR, Chachamovich E, Ezer H, Fleck MP, Knauth D, Passos EP. 2010. Investigating quality of life and health-related quality of life in infertility: A systematic review. J Psychosom Obstet Gynaecol 31:101–110. [DOI] [PubMed] [Google Scholar]
- Chan D, Delbes G, Landry M, Robaire B, Trasler JM. 2012. Epigenetic alterations in sperm DNA associated with testicular cancer treatment. Toxicological sciences : an official journal of the Society of Toxicology 125:532–543. [DOI] [PubMed] [Google Scholar]
- Chan D, McGraw S, Klein K, Wallock LM, Konermann C, Plass C, Chan P, Robaire B, Jacob RA, Greenwood CM, Trasler JM. 2017. Stability of the human sperm DNA methylome to folic acid fortification and short-term supplementation. Human reproduction (Oxford, England) 32:272–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q, Yan M, Cao Z, Li X, Zhang Y, Shi J, Feng G-H, Peng H, Zhang X, Zhang Y, Qian J, Duan E, Zhai Q, Zhou Q. 2016. Sperm tsrnas contribute to intergenerational inheritance of an acquired metabolic disorder. Science 351:397–400. [DOI] [PubMed] [Google Scholar]
- Chen Q, Yan W, Duan E. 2016. Epigenetic inheritance of acquired traits through sperm rnas and sperm rna modifications. Nature reviews Genetics 17:733–743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuva de Sousa Lopes SM, Hayashi K, Shovlin TC, Mifsud W, Surani MA, McLaren A. 2008. X chromosome activity in mouse xx primordial germ cells. PLoS Genet 4:e30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cirio MC, Martel J, Mann M, Toppings M, Bartolomei M, Trasler J, Chaillet JR. 2008. DNA methyltransferase 1o functions during preimplantation development to preclude a profound level of epigenetic variation. Dev Biol 324:139–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins LJ, Schönfeld B, Chen XS. 2011. Chapter 4 - the epigenetics of non-coding rna In: Handbook of epigenetics, (Tollefsbol T, ed). San Diego:Academic Press, 49–61. [Google Scholar]
- Conine CC, Sun F, Song L, Rivera-Pérez JA, Rando OJ. 2018. Small rnas gained during epididymal transit of sperm are essential for embryonic development in mice. Dev Cell 46:470–480.e473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornwall GA. 2014. Role of posttranslational protein modifications in epididymal sperm maturation and extracellular quality control. Adv Exp Med Biol 759:159–180. [DOI] [PubMed] [Google Scholar]
- Dacheux J-L, Dacheux F. 2014. New insights into epididymal function in relation to sperm maturation. Reproduction (Cambridge, England) 147:R27–42. [DOI] [PubMed] [Google Scholar]
- Dadoune J-P. 2003. Expression of mammalian spermatozoal nucleoproteins. Microsc Res Tech 61:56–75. [DOI] [PubMed] [Google Scholar]
- Day J, Savani S, Krempley BD, Nguyen M, Kitlinska JB. 2016. Influence of paternal preconception exposures on their offspring: Through epigenetics to phenotype. Am J Stem Cells 5:11–18. [PMC free article] [PubMed] [Google Scholar]
- de Castro Barbosa T, Ingerslev LR, Alm PS, Versteyhe S, Massart J, Rasmussen M, Donkin I, Sjögren R, Mudry JM, Vetterli L, Gupta S, Krook A, Zierath JR, Barrès R. 2016. High-fat diet reprograms the epigenome of rat spermatozoa and transgenerationally affects metabolism of the offspring. Mol Metab 5:184–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Kretser DM, Loveland K, O’Bryan M. 2016. Chapter 136 - spermatogenesis In: Endocrinology: Adult and pediatric (seventh edition), (Jameson JL, De Groot LJ, de Kretser DM, Giudice LC, Grossman AB, Melmed S, et al. , eds). Philadelphia:W.B. Saunders, 2325–2353.e2329. [Google Scholar]
- de Rooij DG. 2017. Organization of the seminiferous epithelium and the cycle, and morphometric description of spermatogonial subtypes (rodents and primates) In: The biology of mammalian spermatogonia, (Oatley JM, Griswold MD, eds). New York, NY:Springer New York, 3–20. [Google Scholar]
- Denham J, O’Brien BJ, Harvey JT, Charchar FJ. 2015. Genome-wide sperm DNA methylation changes after 3 months of exercise training in humans. Epigenomics 7:717–731. [DOI] [PubMed] [Google Scholar]
- Dias BG, Ressler KJ. 2014. Parental olfactory experience influences behavior and neural structure in subsequent generations. Nat Neurosci 17:89–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon JR, Selvaraj S, Yue F, Kim A, Li Y, Shen Y, Hu M, Liu JS, Ren B. 2012. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature 485:376–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon JR, Jung I, Selvaraj S, Shen Y, Antosiewicz-Bourget JE, Lee AY, Ye Z, Kim A, Rajagopal N, Xie W, Diao Y, Liang J, Zhao H, Lobanenkov VV, Ecker JR, Thomson JA, Ren B. 2015. Chromatin architecture reorganization during stem cell differentiation. Nature 518:331–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong H, Wang Y, Zou Z, Chen L, Shen C, Xu S, Zhang J, Zhao F, Ge S, Gao Q, Hu H, Song M, Wang W. 2017. Abnormal methylation of imprinted genes and cigarette smoking: Assessment of their association with the risk of male infertility. Reprod Sci 24:114–123. [DOI] [PubMed] [Google Scholar]
- Donkin I, Versteyhe S, Ingerslev LR, Qian K, Mechta M, Nordkap L, Mortensen B, Appel EVR, Jørgensen N, Kristiansen VB, Hansen T, Workman CT, Zierath JR, Barrès R. 2016. Obesity and bariatric surgery drive epigenetic variation of spermatozoa in humans. Cell Metab 23:369–378. [DOI] [PubMed] [Google Scholar]
- Donkin I, Barrès R. 2018. Sperm epigenetics and influence of environmental factors. Mol Metab 14:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dopico M, Gómez A. 2015. Review of the current state and main sources of dioxins around the world. J Air Waste Manag Assoc 65:1033–1049. [DOI] [PubMed] [Google Scholar]
- El Hajj N, Zechner U, Schneider E, Tresch A, Gromoll J, Hahn T, Schorsch M, Haaf T. 2011. Methylation status of imprinted genes and repetitive elements in sperm DNA from infertile males. Sex Dev 5:60–69. [DOI] [PubMed] [Google Scholar]
- Farley Ordovensky Staniec J, Webb NJ. 2007. Utilization of infertility services: How much does money matter? Health Serv Res 42:971–989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finegersh A, Homanics GE. 2014. Paternal alcohol exposure reduces alcohol drinking and increases behavioral sensitivity to alcohol selectively in male offspring. PloS one 9:e99078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fullston T, Ohlsson Teague EMC, Palmer NO, DeBlasio MJ, Mitchell M, Corbett M, Print CG, Owens JA, Lane M. 2013. Paternal obesity initiates metabolic disturbances in two generations of mice with incomplete penetrance to the f2 generation and alters the transcriptional profile of testis and sperm microrna content. FASEB J 27:4226–4243. [DOI] [PubMed] [Google Scholar]
- Gkountela S, Zhang KX, Shafiq TA, Liao WW, Hargan-Calvopina J, Chen PY, Clark AT. 2015. DNA demethylation dynamics in the human prenatal germline. Cell 161:1425–1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gore AC, Chappell VA, Fenton SE, Flaws JA, Nadal A, Prins GS, Toppari J, Zoeller RT. 2015. Edc-2: The endocrine society’s second scientific statement on endocrine-disrupting chemicals. Endocr Rev 36:E1–E150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandjean V, Fourre S, De Abreu DA, Derieppe MA, Remy JJ, Rassoulzadegan M. 2015. Rna-mediated paternal heredity of diet-induced obesity and metabolic disorders. Scientific reports 5:18193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu T-P, Guo F, Yang H, Wu H-P, Xu G-F, Liu W, Xie Z-G, Shi L, He X, Jin S-G, Iqbal K, Shi YG, Deng Z, Szabó PE, Pfeifer GP, Li J, Xu G-L. 2011. The role of tet3 DNA dioxygenase in epigenetic reprogramming by oocytes. Nature 477:606–610. [DOI] [PubMed] [Google Scholar]
- Gunn JKL, Rosales CB, Center KE, Nuñez A, Gibson SJ, Christ C, Ehiri JE. 2016. Prenatal exposure to cannabis and maternal and child health outcomes: A systematic review and meta-analysis. BMJ Open 6:e009986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo F, Yan L, Guo H, Li L, Hu B, Zhao Y, Yong J, Hu Y, Wang X, Wei Y, Wang W, Li R, Yan J, Zhi X, Zhang Y, Jin H, Zhang W, Hou Y, Zhu P, Li J, Zhang L, Liu S, Ren Y, Zhu X, Wen L, Gao YQ, Tang F, Qiao J. 2015. The transcriptome and DNA methylome landscapes of human primordial germ cells. Cell 161:1437–1452. [DOI] [PubMed] [Google Scholar]
- Guo H, Hu B, Yan L, Yong J, Wu Y, Gao Y, Guo F, Hou Y, Fan X, Dong J, Wang X, Zhu X, Yan J, Wei Y, Jin H, Zhang W, Wen L, Tang F, Qiao J. 2017. DNA methylation and chromatin accessibility profiling of mouse and human fetal germ cells. Cell Res 27:165–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajkova P, Erhardt S, Lane N, Haaf T, El-Maarri O, Reik W, Walter J, Surani MA. 2002. Epigenetic reprogramming in mouse primordial germ cells. Mech Dev 117:15–23. [DOI] [PubMed] [Google Scholar]
- Hajkova P, Ancelin K, Waldmann T, Lacoste N, Lange UC, Cesari F, Lee C, Almouzni G, Schneider R, Surani MA. 2008. Chromatin dynamics during epigenetic reprogramming in the mouse germ line. Nature 452:877–881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammoud SS, Nix DA, Zhang H, Purwar J, Carrell DT, Cairns BR. 2009. Distinctive chromatin in human sperm packages genes for embryo development. Nature 460:473–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammoud SS, Purwar J, Pflueger C, Cairns BR, Carrell DT. 2010. Alterations in sperm DNA methylation patterns at imprinted loci in two classes of infertility. Fertil Steril 94:1728–1733. [DOI] [PubMed] [Google Scholar]
- Hammoud SS, Nix DA, Hammoud AO, Gibson M, Cairns BR, Carrell DT. 2011. Genome-wide analysis identifies changes in histone retention and epigenetic modifications at developmental and imprinted gene loci in the sperm of infertile men. Hum Reprod 26:2558–2569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartl D, Krebs AR, Grand RS, Baubec T, Isbel L, Wirbelauer C, Burger L, Schubeler D. 2019. Cg dinucleotides enhance promoter activity independent of DNA methylation. Genome research 29:554–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hata K, Okano M, Lei H, Li E. 2002. Dnmt3l cooperates with the dnmt3 family of de novo DNA methyltransferases to establish maternal imprints in mice. Development 129:1983–1993. [DOI] [PubMed] [Google Scholar]
- Hawkins RD, Hon GC, Lee LK, Ngo Q, Lister R, Pelizzola M, Edsall LE, Kuan S, Luu Y, Klugman S, Antosiewicz-Bourget J, Ye Z, Espinoza C, Agarwahl S, Shen L, Ruotti V, Wang W, Stewart R, Thomson JA, Ecker JR, Ren B. 2010. Distinct epigenomic landscapes of pluripotent and lineage-committed human cells. Cell Stem Cell 6:479–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi-Takanaka Y, Yamagata K, Wakayama T, Stasevich TJ, Kainuma T, Tsurimoto T, Tachibana M, Shinkai Y, Kurumizaka H, Nozaki N, Kimura H. 2011. Tracking epigenetic histone modifications in single cells using fab-based live endogenous modification labeling. Nucleic Acids Res 39:6475–6488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirasawa R, Chiba H, Kaneda M, Tajima S, Li E, Jaenisch R, Sasaki H. 2008. Maternal and zygotic dnmt1 are necessary and sufficient for the maintenance of DNA methylation imprints during preimplantation development. Genes Dev 22:1607–1616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houshdaran S, Cortessis VK, Siegmund K, Yang A, Laird PW, Sokol RZ. 2007. Widespread epigenetic abnormalities suggest a broad DNA methylation erasure defect in abnormal human sperm. PloS one 2:e1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howlett SK, Reik W. 1991. Methylation levels of maternal and paternal genomes during preimplantation development. Development 113:119–127. [DOI] [PubMed] [Google Scholar]
- Huang WJ, Lu XL, Li JT, Zhang JM. 2019. Effects of folic acid on oligozoospermia with mthfr polymorphisms in term of seminal parameters, DNA fragmentation, and live birth rate: A double-blind, randomized, placebo-controlled trial. Andrology. [DOI] [PubMed] [Google Scholar]
- Ibn-Salem J, Köhler S, Love MI, Chung H-R, Huang N, Hurles ME, Haendel M, Washington NL, Smedley D, Mungall CJ, Lewis SE, Ott C-E, Bauer S, Schofield PN, Mundlos S, Spielmann M, Robinson PN. 2014. Deletions of chromosomal regulatory boundaries are associated with congenital disease. Genome Biol 15:423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ijiri TW, Merdiushev T, Cao W, Gerton GL. 2011. Identification and validation of mouse sperm proteins correlated with epididymal maturation. Proteomics 11:4047–4062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingerslev LR, Donkin I, Fabre O, Versteyhe S, Mechta M, Pattamaprapanont P, Mortensen B, Krarup NT, Barrès R. 2018. Endurance training remodels sperm-borne small rna expression and methylation at neurological gene hotspots. Clin Epigenetics 10:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue A, Shen L, Matoba S, Zhang Y. 2015. Haploinsufficiency, but not defective paternal 5mc oxidation, accounts for the developmental defects of maternal tet3 knockouts. Cell Rep 10:463–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iqbal K, Jin S-G, Pfeifer GP, Szabó PE. 2011. Reprogramming of the paternal genome upon fertilization involves genome-wide oxidation of 5-methylcytosine. Proc Natl Acad Sci U S A 108:3642–3647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacques M, Hiam D, Craig J, Barrès R, Eynon N, Voisin S. 2019. Epigenetic changes in healthy human skeletal muscle following exercise- a systematic review. Epigenetics 14:633–648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins TG, Aston KI, Hotaling JM, Shamsi MB, Simon L, Carrell DT. 2016a. Teratozoospermia and asthenozoospermia are associated with specific epigenetic signatures. Andrology 4:843–849. [DOI] [PubMed] [Google Scholar]
- Jenkins TG, Aston KI, Meyer TD, Hotaling JM, Shamsi MB, Johnstone EB, Cox KJ, Stanford JB, Porucznik CA, Carrell DT. 2016b. Decreased fecundity and sperm DNA methylation patterns. Fertility and sterility 105:51–57. e51–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins TG, James ER, Alonso DF, Hoidal JR, Murphy PJ, Hotaling JM, Cairns BR, Carrell DT, Aston KI. 2017. Cigarette smoking significantly alters sperm DNA methylation patterns. Andrology 5:1089–1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurewicz J, Hanke W. 2011. Exposure to phthalates: Reproductive outcome and children health. A review of epidemiological studies. Int J Occup Med Environ Health 24:115–141. [DOI] [PubMed] [Google Scholar]
- Keller P, Vollaard NBJ, Gustafsson T, Gallagher IJ, Sundberg CJ, Rankinen T, Britton SL, Bouchard C, Koch LG, Timmons JA. 2011. A transcriptional map of the impact of endurance exercise training on skeletal muscle phenotype. J Appl Physiol 110:46–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelsey KT, Rytel M, Dere E, Butler R, Eliot M, Huse SM, Houseman EA, Koestler DC, Boekelheide K. 2019. Serum dioxin and DNA methylation in the sperm of operation ranch hand veterans exposed to agent orange. Environmental health : a global access science source 18:91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khandwala YS, Zhang CA, Lu Y, Eisenberg ML. 2017. The age of fathers in the USA is rising: An analysis of 168 867 480 births from 1972 to 2015. Hum Reprod 32:2110–2116. [DOI] [PubMed] [Google Scholar]
- Kobayashi H, Sato A, Otsu E, Hiura H, Tomatsu C, Utsunomiya T, Sasaki H, Yaegashi N, Arima T. 2007. Aberrant DNA methylation of imprinted loci in sperm from oligospermic patients. Hum Mol Genet 16:2542–2551. [DOI] [PubMed] [Google Scholar]
- Kriaucionis S, Heintz N. 2009. The nuclear DNA base, 5-hydroxymethylcytosine is present in brain and enriched in purkinje neurons. Science 324:929–930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Salle S, Trasler JM. 2006. Dynamic expression of dnmt3a and dnmt3b isoforms during male germ cell development in the mouse. Dev Biol 296:71–82. [DOI] [PubMed] [Google Scholar]
- Lake JK, Power C, Cole TJ. 1997. Child to adult body mass index in the 1958 british birth cohort: Associations with parental obesity. Arch Dis Child 77:376–381. [DOI] [PubMed] [Google Scholar]
- Laqqan M, Tierling S, Alkhaled Y, Porto CL, Solomayer EF, Hammadeh ME. 2017. Aberrant DNA methylation patterns of human spermatozoa in current smoker males. Reprod Toxicol 71:126–133. [DOI] [PubMed] [Google Scholar]
- Laqqan M, Hammadeh ME. 2018. Aberrations in sperm DNA methylation patterns of males suffering from reduced fecundity. Andrologia 50. [DOI] [PubMed] [Google Scholar]
- Lee J-H, Park S-J, Nakai K. 2017. Differential landscape of non-cpg methylation in embryonic stem cells and neurons caused by dnmt3s. Sci Rep 7:11295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lepikhov K, Walter J. 2004. Differential dynamics of histone h3 methylation at positions k4 and k9 in the mouse zygote. BMC Dev Biol 4:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Wong EM, Bui M, Nguyen TL, Joo J-HE, Stone J, Dite GS, Giles GG, Saffery R, Southey MC, Hopper JL. 2018. Causal effect of smoking on DNA methylation in peripheral blood: A twin and family study. Clin Epigenetics 10:18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T, Wang X, Zhang H, Chen Z, Zhao X, Ma Y. 2019. Histomorphological comparisons and expression patterns of boll gene in sheep testes at different development stages. Animals (Basel) 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieberman-Aiden E, van Berkum NL, Williams L, Imakaev M, Ragoczy T, Telling A, Amit I, Lajoie BR, Sabo PJ, Dorschner MO, Sandstrom R, Bernstein B, Bender MA, Groudine M, Gnirke A, Stamatoyannopoulos J, Mirny LA, Lander ES, Dekker J. 2009. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science 326:289–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liem AK, Fürst P, Rappe C. 2000. Exposure of populations to dioxins and related compounds. Food Addit Contam 17:241–259. [DOI] [PubMed] [Google Scholar]
- Linschooten JO, Verhofstad N, Gutzkow K, Olsen A-K, Yauk C, Oligschläger Y, Brunborg G, van Schooten FJ, Godschalk RWL. 2013. Paternal lifestyle as a potential source of germline mutations transmitted to offspring. FASEB J 27:2873–2879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lochs SJA, Kefalopoulou S, Kind J. 2019. Lamina associated domains and gene regulation in development and cancer. Cells 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupiáñez DG, Kraft K, Heinrich V, Krawitz P, Brancati F, Klopocki E, Horn D, Kayserili H, Opitz JM, Laxova R, Santos-Simarro F, Gilbert-Dussardier B, Wittler L, Borschiwer M, Haas SA, Osterwalder M, Franke M, Timmermann B, Hecht J, Spielmann M, Visel A, Mundlos S. 2015. Disruptions of topological chromatin domains cause pathogenic rewiring of gene-enhancer interactions. Cell 161:1012–1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyche JL, Gutleb AC, Bergman A, Eriksen GS, Murk AJ, Ropstad E, Saunders M, Skaare JU. 2009. Reproductive and developmental toxicity of phthalates. J Toxicol Environ Health B Crit Rev 12:225–249. [DOI] [PubMed] [Google Scholar]
- Manfo FPT, Jubendradass R, Nantia EA, Moundipa PF, Mathur PP. 2014. Adverse effects of bisphenol a on male reproductive function. Rev Environ Contam Toxicol 228:57–82. [DOI] [PubMed] [Google Scholar]
- Marcon L, Boissonneault G. 2004. Transient DNA strand breaks during mouse and human spermiogenesis new insights in stage specificity and link to chromatin remodeling. Biol Reprod 70:910–918. [DOI] [PubMed] [Google Scholar]
- Marques CJ, Carvalho F, Sousa M, Barros A. 2004. Genomic imprinting in disruptive spermatogenesis. Lancet 363:1700–1702. [DOI] [PubMed] [Google Scholar]
- Marques CJ, Costa P, Vaz B, Carvalho F, Fernandes S, Barros A, Sousa M. 2008. Abnormal methylation of imprinted genes in human sperm is associated with oligozoospermia. Mol Hum Reprod 14:67–74. [DOI] [PubMed] [Google Scholar]
- Mayer W, Niveleau A, Walter J, Fundele R, Haaf T. 2000. Demethylation of the zygotic paternal genome. Nature 403:501–502. [DOI] [PubMed] [Google Scholar]
- McCarthy DM, Morgan TJ Jr., Lowe SE, Williamson MJ, Spencer TJ, Biederman J, Bhide PG. 2018. Nicotine exposure of male mice produces behavioral impairment in multiple generations of descendants. PLoS Biol 16:e2006497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McPherson NO, Owens JA, Fullston T, Lane M. 2015. Preconception diet or exercise intervention in obese fathers normalizes sperm microrna profile and metabolic syndrome in female offspring. Am J Physiol Endocrinol Metab 308:E805–821. [DOI] [PubMed] [Google Scholar]
- McSwiggin HM, O’Doherty AM. 2018. Epigenetic reprogramming during spermatogenesis and male factor infertility. Reproduction (Cambridge, England) 156:R9–R21. [DOI] [PubMed] [Google Scholar]
- Mengual L, Ballescá JL, Ascaso C, Oliva R. 2003. Marked differences in protamine content and p1/p2 ratios in sperm cells from percoll fractions between patients and controls. J Androl 24:438–447. [DOI] [PubMed] [Google Scholar]
- Messerlian C, Bellinger D, Mínguez-Alarcón L, Romano ME, Ford JB, Williams PL, Calafat AM, Hauser R, Braun JM. 2017. Paternal and maternal preconception urinary phthalate metabolite concentrations and child behavior. Environ Res 158:720–728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mima M, Greenwald D, Ohlander S. 2018. Environmental toxins and male fertility. Curr Urol Rep 19:50. [DOI] [PubMed] [Google Scholar]
- Minor A, Chow V, Ma S. 2011. Aberrant DNA methylation at imprinted genes in testicular sperm retrieved from men with obstructive azoospermia and undergoing vasectomy reversal. Reproduction (Cambridge, England) 141:749–757. [DOI] [PubMed] [Google Scholar]
- Morselli M, Pastor WA, Montanini B, Nee K, Ferrari R, Fu K, Bonora G, Rubbi L, Clark AT, Ottonello S, Jacobsen SE, Pellegrini M. 2015. In vivo targeting of de novo DNA methylation by histone modifications in yeast and mouse. Elife 4:e06205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashov AK, Pak ES, Koury M, Ajmera A, Jeyakumar M, Parker M, Williams O, Ding J, Walters D, Neufer PD. 2016. Paternal long-term exercise programs offspring for low energy expenditure and increased risk for obesity in mice. FASEB J 30:775–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy SK, Itchon-Ramos N, Visco Z, Huang Z, Grenier C, Schrott R, Acharya K, Boudreau M-H, Price TM, Raburn DJ, Corcoran DL, Lucas JE, Mitchell JT, McClernon FJ, Cauley M, Hall BJ, Levin ED, Kollins SH. 2018. Cannabinoid exposure and altered DNA methylation in rat and human sperm. Epigenetics 13:1208–1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Najafipour R, Moghbelinejad S, Aleyasin A, Jalilvand A. 2017. Effect of b9 and b12 vitamin intake on semen parameters and fertility of men with mthfr polymorphisms. Andrology 5:704–710. [DOI] [PubMed] [Google Scholar]
- Nakamura T, Arai Y, Umehara H, Masuhara M, Kimura T, Taniguchi H, Sekimoto T, Ikawa M, Yoneda Y, Okabe M, Tanaka S, Shiota K, Nakano T. 2007. Pgc7/stella protects against DNA demethylation in early embryogenesis. Nat Cell Biol 9:64–71. [DOI] [PubMed] [Google Scholar]
- Nakamura T, Liu Y-J, Nakashima H, Umehara H, Inoue K, Matoba S, Tachibana M, Ogura A, Shinkai Y, Nakano T. 2012. Pgc7 binds histone h3k9me2 to protect against conversion of 5mc to 5hmc in early embryos. Nature 486:415–419. [DOI] [PubMed] [Google Scholar]
- Nixon B, Stanger SJ, Mihalas BP, Reilly JN, Anderson AL, Tyagi S, Holt JE, McLaughlin EA. 2015. The microrna signature of mouse spermatozoa is substantially modified during epididymal maturation. Biol Reprod 93:91. [DOI] [PubMed] [Google Scholar]
- Novikova IV, Hennelly SP, Sanbonmatsu KY. 2012. Sizing up long non-coding rnas: Do lncrnas have secondary and tertiary structure? Bioarchitecture 2:189–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oakes CC, La Salle S, Smiraglia DJ, Robaire B, Trasler JM. 2007. Developmental acquisition of genome-wide DNA methylation occurs prior to meiosis in male germ cells. Dev Biol 307:368–379. [DOI] [PubMed] [Google Scholar]
- Oswald J, Engemann S, Lane N, Mayer W, Olek A, Fundele R, Dean W, Reik W, Walter J. 2000. Active demethylation of the paternal genome in the mouse zygote. Curr Biol 10:475–478. [DOI] [PubMed] [Google Scholar]
- Pacheco SE, Houseman EA, Christensen BC, Marsit CJ, Kelsey KT, Sigman M, Boekelheide K. 2011. Integrative DNA methylation and gene expression analyses identify DNA packaging and epigenetic regulatory genes associated with low motility sperm. PLoS One 6:e20280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer NO, Fullston T, Mitchell M, Setchell BP, Lane M. 2011. Sirt6 in mouse spermatogenesis is modulated by diet-induced obesity. Reprod Fertil Dev 23:929–939. [DOI] [PubMed] [Google Scholar]
- Patil V, Ward RL, Hesson LB. 2014. The evidence for functional non-cpg methylation in mammalian cells. Epigenetics 9:823–828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peat JR, Dean W, Clark SJ, Krueger F, Smallwood SA, Ficz G, Kim JK, Marioni JC, Hore TA, Reik W. 2014. Genome-wide bisulfite sequencing in zygotes identifies demethylation targets and maps the contribution of tet3 oxidation. Cell Rep 9:1990–2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peric-Hupkes D, Meuleman W, Pagie L, Bruggeman SWM, Solovei I, Brugman W, Gräf S, Flicek P, Kerkhoven RM, van Lohuizen M, Reinders M, Wessels L, van Steensel B. 2010. Molecular maps of the reorganization of genome-nuclear lamina interactions during differentiation. Mol Cell 38:603–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilsner JR, Parker M, Sergeyev O, Suvorov A. 2017. Spermatogenesis disruption by dioxins: Epigenetic reprograming and windows of susceptibility. Reprod Toxicol 69:221–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilsner JR, Shershebnev A, Medvedeva YA, Suvorov A, Wu H, Goltsov A, Loukianov E, Andreeva T, Gusev F, Manakhov A, Smigulina L, Logacheva M, Shtratnikova V, Kuznetsova I, Speranskiy-Podobed P, Burns JS, Williams PL, Korrick S, Lee MM, Rogaev E, Hauser R, Sergeyev O. 2018. Peripubertal serum dioxin concentrations and subsequent sperm methylome profiles of young russian adults. Reprod Toxicol 78:40–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poplinski A, Tüttelmann F, Kanber D, Horsthemke B, Gromoll J. 2010. Idiopathic male infertility is strongly associated with aberrant methylation of mest and igf2/h19 icr1. Int J Androl 33:642–649. [DOI] [PubMed] [Google Scholar]
- Qin J, Wen B, Liang Y, Yu W, Li H. 2019. Histone modifications and their role in colorectal cancer (review). Pathol Oncol Res. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajanahally S, Raheem O, Rogers M, Brisbane W, Ostrowski K, Lendvay T, Walsh T. 2019. The relationship between cannabis and male infertility, sexual health, and neoplasm: A systematic review. Andrology 7:139–147. [DOI] [PubMed] [Google Scholar]
- Rathke C, Baarends WM, Awe S, Renkawitz-Pohl R. 2014. Chromatin dynamics during spermiogenesis. Biochim Biophys Acta 1839:155–168. [DOI] [PubMed] [Google Scholar]
- Reik W, Dean W, Walter J. 2001. Epigenetic reprogramming in mammalian development. Science 293:1089–1093. [DOI] [PubMed] [Google Scholar]
- Ribeiro-Mason K, Boulesteix C, Brochard V, Aguirre-Lavin T, Salvaing J, Fleurot R, Adenot P, Maalouf WE, Beaujean N. 2012. Nuclear dynamics of histone h3 trimethylated on lysine 9 and/or phosphorylated on serine 10 in mouse cloned embryos as new markers of reprogramming? Cell Reprogram 14:283–294. [DOI] [PubMed] [Google Scholar]
- Rodgers AB, Morgan CP, Bronson SL, Revello S, Bale TL. 2013. Paternal stress exposure alters sperm microrna content and reprograms offspring hpa stress axis regulation. J Neurosci 33:9003–9012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rompala GR, Mounier A, Wolfe CM, Lin Q, Lefterov I, Homanics GE. 2018. Heavy chronic intermittent ethanol exposure alters small noncoding rnas in mouse sperm and epididymosomes. Front Genet 9:32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santenard A, Ziegler-Birling C, Koch M, Tora L, Bannister AJ, Torres-Padilla M-E. 2010. Heterochromatin formation in the mouse embryo requires critical residues of the histone variant h3.3. Nat Cell Biol 12:853–862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos F, Hendrich B, Reik W, Dean W. 2002. Dynamic reprogramming of DNA methylation in the early mouse embryo. Dev Biol 241:172–182. [DOI] [PubMed] [Google Scholar]
- Sasaki H, Matsui Y. 2008. Epigenetic events in mammalian germ-cell development: Reprogramming and beyond. Nat Rev Genet 9:129–140. [DOI] [PubMed] [Google Scholar]
- Schick M, Morina N, Klaghofer R, Schnyder U, Müller J. 2013. Trauma, mental health, and intergenerational associations in kosovar families 11 years after the war. Eur J Psychotraumatol 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrott R, Acharya K, Itchon-Ramos N, Hawkey AB, Pippen E, Mitchell JT, Kollins SH, Levin ED, Murphy SK. 2019. Cannabis use is associated with potentially heritable widespread changes in autism candidate gene dlgap2 DNA methylation in sperm. Epigenetics:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seki Y, Hayashi K, Itoh K, Mizugaki M, Saitou M, Matsui Y. 2005. Extensive and orderly reprogramming of genome-wide chromatin modifications associated with specification and early development of germ cells in mice. Dev Biol 278:440–458. [DOI] [PubMed] [Google Scholar]
- Seki Y, Yamaji M, Yabuta Y, Sano M, Shigeta M, Matsui Y, Saga Y, Tachibana M, Shinkai Y, Saitou M. 2007. Cellular dynamics associated with the genome-wide epigenetic reprogramming in migrating primordial germ cells in mice. Development 134:2627–2638. [DOI] [PubMed] [Google Scholar]
- Sharif J, Muto M, Takebayashi S-I, Suetake I, Iwamatsu A, Endo TA, Shinga J, Mizutani-Koseki Y, Toyoda T, Okamura K, Tajima S, Mitsuya K, Okano M, Koseki H. 2007. The sra protein np95 mediates epigenetic inheritance by recruiting dnmt1 to methylated DNA. Nature 450:908–912. [DOI] [PubMed] [Google Scholar]
- Sharma R, Harlev A, Agarwal A, Esteves SC. 2016. Cigarette smoking and semen quality: A new meta-analysis examining the effect of the 2010 world health organization laboratory methods for the examination of human semen. Eur Urol 70:635–645. [DOI] [PubMed] [Google Scholar]
- Sharma U, Conine CC, Shea JM, Boskovic A, Derr AG, Bing XY, Belleannee C, Kucukural A, Serra RW, Sun F, Song L, Carone BR, Ricci EP, Li XZ, Fauquier L, Moore MJ, Sullivan R, Mello CC, Garber M, Rando OJ. 2016. Biogenesis and function of trna fragments during sperm maturation and fertilization in mammals. Science 351:391–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma U, Sun F, Conine CC, Reichholf B, Kukreja S, Herzog VA, Ameres SL, Rando OJ. 2018. Small rnas are trafficked from the epididymis to developing mammalian sperm. Dev Cell 46:481–494.e486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shelby MD. 2008. Ntp-cerhr monograph on the potential human reproductive and developmental effects of bisphenol a. NTP CERHR MON:v, vii–ix, 1–64 passim. [PubMed] [Google Scholar]
- Shen L, Inoue A, He J, Liu Y, Lu F, Zhang Y. 2014. Tet3 and DNA replication mediate demethylation of both the maternal and paternal genomes in mouse zygotes. Cell Stem Cell 15:459–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shnorhavorian M, Schwartz SM, Stansfeld B, Sadler-Riggleman I, Beck D, Skinner MK. 2017. Differential DNA methylation regions in adult human sperm following adolescent chemotherapy: Potential for epigenetic inheritance. PloS one 12:e0170085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Short AK, Yeshurun S, Powell R, Perreau VM, Fox A, Kim JH, Pang TY, Hannan AJ. 2017. Exercise alters mouse sperm small noncoding rnas and induces a transgenerational modification of male offspring conditioned fear and anxiety. Transl Psychiatry 7:e1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu X-O, Reaman HG, Lampkin B, Sather NH, Pendergrass WTa, Robison LL. 1994. Association of paternal diagnostic x-ray exposure with risk of infant leukemia’. Cancer Epidemiology, Biomarkers & Prevention 3:645–653. [PubMed] [Google Scholar]
- Silva MJ, Barr DB, Reidy JA, Malek NA, Hodge CC, Caudill SP, Brock JW, Needham LL, Calafat AM. 2004. Urinary levels of seven phthalate metabolites in the u.S. Population from the national health and nutrition examination survey (nhanes) 1999–2000. Environ Health Perspect 112:331–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smallwood SA, Tomizawa S-I, Krueger F, Ruf N, Carli N, Segonds-Pichon A, Sato S, Hata K, Andrews SR, Kelsey G. 2011. Dynamic cpg island methylation landscape in oocytes and preimplantation embryos. Nat Genet 43:811–814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soubry A, Schildkraut JM, Murtha A, Wang F, Huang Z, Bernal A, Kurtzberg J, Jirtle RL, Murphy SK, Hoyo C. 2013. Paternal obesity is associated with igf2 hypomethylation in newborns: Results from a newborn epigenetics study (nest) cohort. BMC Med 11:29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soubry A, Guo L, Huang Z, Hoyo C, Romanus S, Price T, Murphy SK. 2016. Obesity-related DNA methylation at imprinted genes in human sperm: Results from the tieger study. Clin Epigenetics 8:51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soubry A 2018. Pohad: Why we should study future fathers. Environ Epigenet 4:dvy007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein P, Worrad DM, Belyaev ND, Turner BM, Schultz RM. 1997. Stage-dependent redistributions of acetylated histones in nuclei of the early preimplantation mouse embryo. Molecular Reproduction and Development: Incorporating Gamete Research 47:421–429. [DOI] [PubMed] [Google Scholar]
- Sun W, Dong H, Becker AS, Dapito DH, Modica S, Grandl G, Opitz L, Efthymiou V, Straub LG, Sarker G, Balaz M, Balazova L, Perdikari A, Kiehlmann E, Bacanovic S, Zellweger C, Peleg-Raibstein D, Pelczar P, Reik W, Burger IA, von Meyenn F, Wolfrum C. 2018. Cold-induced epigenetic programming of the sperm enhances brown adipose tissue activity in the offspring. Nat Med 24:1372–1383. [DOI] [PubMed] [Google Scholar]
- Surface LE, Thornton SR, Boyer LA. 2010. Polycomb group proteins set the stage for early lineage commitment. Cell Stem Cell 7:288–298. [DOI] [PubMed] [Google Scholar]
- Swanson GM, Estill M, Diamond MP, Legro RS, Coutifaris C, Barnhart KT, Huang H, Hansen KR, Trussell JC, Coward RM, Zhang H, Goodrich R, Krawetz SA. 2019. Human chromatin remodeler cofactor, rna interactor, eraser and writer sperm rnas responding to obesity. Epigenetics:1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szutorisz H, Hurd YL. 2016. Epigenetic effects of cannabis exposure. Biol Psychiatry 79:586–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, Agarwal S, Iyer LM, Liu DR, Aravind L, Rao A. 2009. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by mll partner tet1. Science 324:930–935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talhout R, Schulz T, Florek E, van Benthem J, Wester P, Opperhuizen A. 2011. Hazardous compounds in tobacco smoke. Int J Environ Res Public Health 8:613–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Q, Pan F, Yang J, Fu Z, Lu Y, Wu X, Han X, Chen M, Lu C, Xia Y, Wang X, Wu W. 2018. Idiopathic male infertility is strongly associated with aberrant DNA methylation of imprinted loci in sperm: A case-control study. Clin Epigenetics 10:134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang WW, Dietmann S, Irie N, Leitch HG, Floros VI, Bradshaw CR, Hackett JA, Chinnery PF, Surani MA. 2015. A unique gene regulatory network resets the human germline epigenome for development. Cell 161:1453–1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian M, Liu L, Zhang J, Huang Q, Shen H. 2019. Positive association of low-level environmental phthalate exposure with sperm motility was mediated by DNA methylation: A pilot study. Chemosphere 220:459–467. [DOI] [PubMed] [Google Scholar]
- Trasler JM, Hales BF, Robaire B. 1986. Chronic low dose cyclophosphamide treatment of adult male rats: Effect on fertility, pregnancy outcome and progeny. Biology of reproduction 34:275–283. [DOI] [PubMed] [Google Scholar]
- Trasler JM, Hales BF, Robaire B. 1987. A time-course study of chronic paternal cyclophosphamide treatment in rats: Effects on pregnancy outcome and the male reproductive and hematologic systems. Biology of reproduction 37:317–326. [DOI] [PubMed] [Google Scholar]
- Vaage AB, Thomsen PH, Rousseau C, Wentzel-Larsen T, Ta TV, Hauff E. 2011. Paternal predictors of the mental health of children of vietnamese refugees. Child Adolesc Psychiatry Ment Health 5:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veland N, Lu Y, Hardikar S, Gaddis S, Zeng Y, Liu B, Estecio MR, Takata Y, Lin K, Tomida MW, Shen J, Saha D, Gowher H, Zhao H, Chen T. 2019. Dnmt3l facilitates DNA methylation partly by maintaining dnmt3a stability in mouse embryonic stem cells. Nucleic Acids Res 47:152–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vojtech L, Woo S, Hughes S, Levy C, Ballweber L, Sauteraud RP, Strobl J, Westerberg K, Gottardo R, Tewari M, Hladik F. 2014. Exosomes in human semen carry a distinctive repertoire of small non-coding rnas with potential regulatory functions. Nucleic Acids Res 42:7290–7304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y-X, You L, Zeng Q, Sun Y, Huang Y-H, Wang C, Wang P, Cao W-C, Yang P, Li Y-F, Lu W-Q. 2015. Phthalate exposure and human semen quality: Results from an infertility clinic in china. Environ Res 142:1–9. [DOI] [PubMed] [Google Scholar]
- Watkins AJ, Dias I, Tsuro H, Allen D, Emes RD, Moreton J, Wilson R, Ingram RJM, Sinclair KD. 2018. Paternal diet programs offspring health through sperm- and seminal plasma-specific pathways in mice. Proc Natl Acad Sci U S A 115:10064–10069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson CT, Szutorisz H, Garg P, Martin Q, Landry JA, Sharp AJ, Hurd YL. 2015. Genome-wide DNA methylation profiling reveals epigenetic changes in the rat nucleus accumbens associated with cross-generational effects of adolescent thc exposure. Neuropsychopharmacology 40:2993–3005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Y, Yang C-R, Wei Y-P, Zhao Z-A, Hou Y, Schatten H, Sun Q-Y. 2014. Paternally induced transgenerational inheritance of susceptibility to diabetes in mammals. Proc Natl Acad Sci U S A 111:1873–1878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wongtawan T, Taylor JE, Lawson KA, Wilmut I, Pennings S. 2011. Histone h4k20me3 and hp1α are late heterochromatin markers in development, but present in undifferentiated embryonic stem cells. J Cell Sci 124:1878–1890. [DOI] [PubMed] [Google Scholar]
- Wossidlo M, Nakamura T, Lepikhov K, Marques CJ, Zakhartchenko V, Boiani M, Arand J, Nakano T, Reik W, Walter J. 2011. 5-hydroxymethylcytosine in the mammalian zygote is linked with epigenetic reprogramming. Nat Commun 2:241. [DOI] [PubMed] [Google Scholar]
- Wu H, Hauser R, Krawetz SA, Pilsner JR. 2015. Environmental susceptibility of the sperm epigenome during windows of male germ cell development. Current environmental health reports. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Hauser R, Krawetz SA, Pilsner JR. 2015. Environmental susceptibility of the sperm epigenome during windows of male germ cell development. Curr Environ Health Rep 2:356–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Ashcraft L, Whitcomb BW, Rahil T, Tougias E, Sites CK, Pilsner JR. 2017a. Parental contributions to early embryo development: Influences of urinary phthalate and phthalate alternatives among couples undergoing ivf treatment. Hum Reprod 32:65–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Estill MS, Shershebnev A, Suvorov A, Krawetz SA, Whitcomb BW, Dinnie H, Rahil T, Sites CK, Pilsner JR. 2017b. Preconception urinary phthalate concentrations and sperm DNA methylation profiles among men undergoing ivf treatment: A cross-sectional study. Hum Reprod 32:2159–2169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyrobek AJ, Schmid TE, Marchetti F. 2005. Relative susceptibilities of male germ cells to genetic defects induced by cancer chemotherapies. J Natl Cancer Inst Monogr:31–35. [DOI] [PubMed] [Google Scholar]
- Yabuta Y, Kurimoto K, Ohinata Y, Seki Y, Saitou M. 2006. Gene expression dynamics during germline specification in mice identified by quantitative single-cell gene expression profiling. Biol Reprod 75:705–716. [DOI] [PubMed] [Google Scholar]
- Yan W 2014. Potential roles of noncoding rnas in environmental epigenetic transgenerational inheritance. Molecular and cellular endocrinology 398:24–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yehuda R, Daskalakis NP, Lehrner A, Desarnaud F, Bader HN, Makotkine I, Flory JD, Bierer LM, Meaney MJ. 2014. Influences of maternal and paternal ptsd on epigenetic regulation of the glucocorticoid receptor gene in holocaust survivor offspring. Am J Psychiatry 171:872–880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan S, Schuster A, Tang C, Yu T, Ortogero N, Bao J, Zheng H, Yan W. 2016. Sperm-borne mirnas and endo-sirnas are important for fertilization and preimplantation embryonic development. Development 143:635–647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zegers-Hochschild F, Adamson GD, Dyer S, Racowsky C, de Mouzon J, Sokol R, Rienzi L, Sunde A, Schmidt L, Cooke ID, Simpson JL, van der Poel S. 2017. The international glossary on infertility and fertility care, 2017. Fertil Steril 108:393–406. [DOI] [PubMed] [Google Scholar]
- Zhang X, San Gabriel M, Zini A. 2006. Sperm nuclear histone to protamine ratio in fertile and infertile men: Evidence of heterogeneous subpopulations of spermatozoa in the ejaculate. J Androl 27:414–420. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Zhang X, Shi J, Tuorto F, Li X, Liu Y, Liebers R, Zhang L, Qu Y, Qian J, Pahima M, Liu Y, Yan M, Cao Z, Lei X, Cao Y, Peng H, Liu S, Wang Y, Zheng H, Woolsey R, Quilici D, Zhai Q, Li L, Zhou T, Yan W, Lyko F, Zhang Y, Zhou Q, Duan E, Chen Q. 2018. Dnmt2 mediates intergenerational transmission of paternally acquired metabolic disorders through sperm small non-coding rnas. Nat Cell Biol 20:535–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng H, Zhou X, Li D-K, Yang F, Pan H, Li T, Miao M, Li R, Yuan W. 2017. Genome-wide alteration in DNA hydroxymethylation in the sperm from bisphenol a-exposed men. PloS one 12:e0178535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou D, Suzuki T, Asami M, Perry ACF. 2019. Caput epididymidal mouse sperm support full development. Dev Cell 50:5–6. [DOI] [PubMed] [Google Scholar]
- Zhu P, Guo H, Ren Y, Hou Y, Dong J, Li R, Lian Y, Fan X, Hu B, Gao Y, Wang X, Wei Y, Liu P, Yan J, Ren X, Yuan P, Yuan Y, Yan Z, Wen L, Yan L, Qiao J, Tang F. 2018. Single-cell DNA methylome sequencing of human preimplantation embryos. Nat Genet 50:12–19. [DOI] [PubMed] [Google Scholar]
- Zoeller RT, Brown TR, Doan LL, Gore AC, Skakkebaek NE, Soto AM, Woodruff TJ, Vom Saal FS. 2012. Endocrine-disrupting chemicals and public health protection: A statement of principles from the endocrine society. Endocrinology 153:4097–4110. [DOI] [PMC free article] [PubMed] [Google Scholar]


