Abstract
Primary cilia are organelles necessary for proper implementation of developmental and homeostasis processes. To initiate their assembly, coordinated actions of multiple proteins are needed. Tau tubulin kinase 2 (TTBK2) is a key player in the cilium assembly pathway, controlling the final step of cilia initiation. The function of TTBK2 in ciliogenesis is critically dependent on its kinase activity; however, the precise mechanism of TTBK2 action has so far not been fully understood due to the very limited information about its relevant substrates. In this study, we demonstrate that CEP83, CEP89, CCDC92, Rabin8, and DVL3 are substrates of TTBK2 kinase activity. Further, we characterize a set of phosphosites of those substrates and CEP164 induced by TTBK2 in vitro and in vivo. Intriguingly, we further show that identified TTBK2 phosphosites and consensus sequence delineated from those are distinct from motifs previously assigned to TTBK2. Finally, we show that TTBK2 is also required for efficient phosphorylation of many S/T sites in CEP164 and provide evidence that TTBK2-induced phosphorylations of CEP164 modulate its function, which in turn seems relevant for the process of cilia formation. In summary, our work provides important insight into the substrates–TTBK2 kinase relationship and suggests that phosphorylation of substrates on multiple sites by TTBK2 is probably involved in the control of ciliogenesis in human cells.
INTRODUCTION
Primary cilia (PCs) are organelles fundamental for proper development and homeostasis. Malfunctioning of PCs leads to ciliopathies, a group of diseases with a broad span of phenotypic manifestations, such as obesity, blindness, polycystic kidneys, and polydactyly (Mitchison and Valente, 2017; Reiter and Leroux, 2017). In addition, PCs abnormalities have been recently related to cancer (Wong et al., 2009; Jenks et al., 2018; Wang and Dynlacht, 2018), further affirming the importance of correctly functioning PCs for tissue homeostasis.
PCs are usually assembled on postmitotic cells, with their formation being triggered at the distal end of the matured (mother) centrioles (MCs). They consist of a microtubule-based axoneme enclosed within a ciliary membrane, and transformed MC (basal body) (Ishikawa and Marshall, 2011; Nigg and Stearns, 2011). MCs differ from daughter centrioles by the presence of two sets of proteinaceous appendages named distal appendages (DA) and sub-DA (SDA) according to their position on MCs (Nigg and Stearns, 2011). The earliest sign of ciliogenesis seems to be the accumulation of small Golgi-derived vesicles in the vicinity of DA (Sorokin, 1962; Lu et al., 2015; Wu et al., 2018), depending on the activity of the Rab11-Rab8a-Rabin8 cascade (Knödler et al., 2010; Westlake et al., 2011; Hehnly et al., 2012; Schmidt et al., 2012). Disruption of DA structure prevents vesicle docking and in turn leads to a failure of cilium assembly (Schmidt et al., 2012; Chang et al., 2013; Sillibourne et al., 2013; Tanos et al., 2013). Initially, five DA proteins, CEP164 (Graser et al., 2007), CEP83 (Joo et al., 2013), CEP89 (Sillibourne et al., 2013), FBF1 (Tanos et al., 2013; Wei et al., 2013), and SCLT1 (Tanos et al., 2013), were identified. Recent identification of new DA components suggests the list might not be complete (Kurtulmus et al., 2018; Bowler et al., 2019).
Cilia initiation turns to the cilia extension process when the centriolar distal end capping proteins CP110, CEP97, and MPP9 are removed and ciliary axoneme begins to elongate (Spektor et al., 2007; Huang et al., 2018). This key step in the cilium assembly pathway is controlled by Tau Tubulin kinase 2 (TTBK2) (Goetz et al., 2012; Cˇajánek and Nigg, 2014). TTBK2 is recruited to DA by CEP164 (Cˇajánek and Nigg, 2014; Oda et al., 2014), and their mutual interaction is fine-tuned by INNPP5E and PI3Kγ (Xu et al., 2016). In addition, TTBK2 seems uniquely positioned among the DA-associated proteins—its depletion still allows cilia initiation to proceed, but the axoneme does not extend and ciliogenesis in turn fails, implying that TTBK2 may function as a switch in the cilia assembly pathway, turning the cilia initiation program to cilia elongation and maintenance (Goetz et al., 2012; Cˇajánek and Nigg, 2014; Oda et al., 2014; Bowie et al., 2018).
From an evolutionary perspective, TTBK2 is a member of the casein kinase 1 (CK1) superfamily; its kinase domain shows 38% identity with CK1δ (Ikezu and Ikezu, 2014; Liao et al., 2015). Apart from TTBK2, the additional kinase MARK4 has been implicated in cilium initiation and assembly, although it seems to control the processes via SDA rather than DA of the MCs (Kuhns et al., 2013).
TTBK2 mutations are causative for development of the neurodegenerative disorder spinocerebellar ataxia type 11 (SCA11) (Houlden et al., 2007; Bauer et al., 2010; Lindquist et al., 2017). Interestingly, SCA11-associated mutant variants of TTBK2, typically truncated around amino acid (AA) 450, are unable to interact with CEP164, fail to localize to DA (Goetz et al., 2012; Čajánek and Nigg, 2014), and instead seem to act in a dominant negative manner to disturb PC formation (Goetz et al., 2012; Bowie et al., 2018). As no other kinase can compensate for defects caused by TTBK2 mutations in SCA11, it again can be argued that TTBK2 has nonredundant functions in ciliogenesis. Previous work proved that ciliogenesis critically depends on TTBK2 kinase activity (Goetz et al., 2012). However, information about bona fide substrates and/or S/T sites that are subjected to TTBK2 phosphorylation is very limited. So far, TTBK2 has been demonstrated to phosphorylate five substrates—Tau (Takahashi et al., 1995; Tomizawa et al., 2001), KIF2A (Watanabe et al., 2015), CEP164 (Cˇajánek and Nigg, 2014), CEP97 (Oda et al., 2014), and MPP9 (Huang et al., 2018)—and only 11 S/T sites, namely S208 and S210 of Tau, S135 of KIF2A, S629 and S636 of MPP9, and T1309, S1317, S1346, S1347, S1434, and S1443 of CEP164 (Kitano-Takahashi et al., 2007; Oda et al., 2014; Watanabe et al., 2015; Huang et al., 2018).
Here, we report on novel TTBK2 substrates and an extensive set of TTBK2-induced phosphorylation sites determined and verified using a combination of in vitro, in vivo, and in silico approaches. Using our datasets of phosphorylated peptide sequences, we delineated a set of kinase motifs for TTBK2. For the functional validation we focused on CEP164 and through a combination of CRISPR editing, mass spectrometry and structure–function experiments provide evidence that phosphorylation by TTBK2 modulates functions of CEP164.
RESULTS
TTBK2 acts on multiple candidate substrates
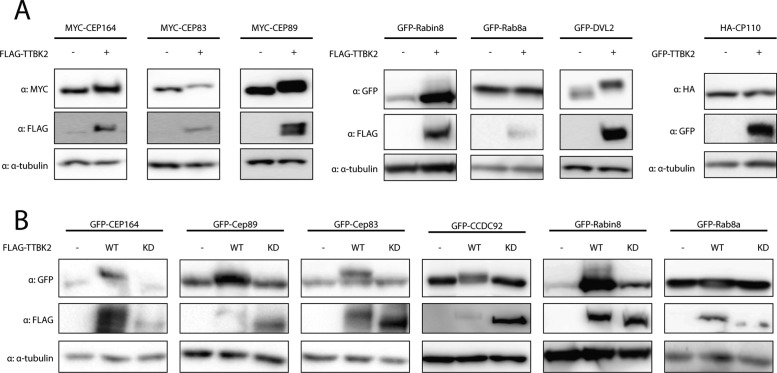
To identify novel TTBK2 substrates, we focused our effort on a set of candidate proteins that localize to DA or interact with CEP164 and have been implicated in cilia initiation. These include the DA components CEP83 and CEP89, Rab8a, Rabin8, DVL3, CCDC92, and CP110 (Chaki et al., 2012; Schmidt et al., 2012; Tanos et al., 2013; Cervenka et al., 2016; Huang et al., 2018). First, given that the reported TTBK2 substrates CEP164 and MPP9 respond to elevated TTBK2 levels with a profound mobility shift (Cˇajánek and Nigg, 2014; Huang et al., 2018), we examined the behavior of individual candidates following their coexpression with TTBK2 in HEK293T WT cells. Interestingly, while we found no response to FLAG-TTBK2 coexpression in the case of HA-CP110 and GFP-Rab8a, we observed a mobility shift of MYC-CEP83, MYC-CEP89, GFP-Rabin8, and GFP-DVL2 similar to that of MYC-CEP164 (Figure 1A), indicating that these proteins are modified in a TTBK2-dependent manner. Next, to corroborate our observations, we included the TTBK2 D163A mutant with disabled kinase domain activity (KD) (Bouskila et al., 2011) in our experiments. Importantly, as depicted in Figure 1B, GFP-CEP164, GFP-CEP89, GFP-CEP83, GFP-CCDC92, and GFP-Rabin8, but not GFP-Rab8a, specifically responded to TTBK2, but not TTBK2 KD, suggesting that the observed shifts in protein mobility reflect a modification of the given protein mediated by kinase activity of TTBK2.
FIGURE 1:
TTBK2 induces mobility shift of CEP164, CEP89, CEP83, CCDC92, Rabin8, and DVL2. HEK293T cells were transfected by indicated plasmids, lysed 24 h posttransfection and analyzed by WB using indicated antibodies. (A) Electrophoretic mobility of MYC-CEP164, MYC-CEP83, MYC-CEP89, GFP-Rabin8, and GFP-DVL2 is decelerated by FLAG-TTBK2 expression. HA-CP110 and GFP-Rab8a do not change their mobility on FLAG-TTBK2 or GFP-TTBK2 expression. (B) Detection of mobility shift of GFP-CEP164, GFP-CEP89, GFP-CEP83, GFP-CCDC92, and GFP-Rabin8 induced by FLAG-TTBK2 but not FLAG-TTBK2 KD. Note that TTBK2 WT undergoes profound mobility shift compared with TTBK2 KD, indicating autophosphorylation. Depicted experiments were performed at least twice.
TTBK2 directly phosphorylates CEP83, CEP89, Rabin8, CCDC92, and DVL3
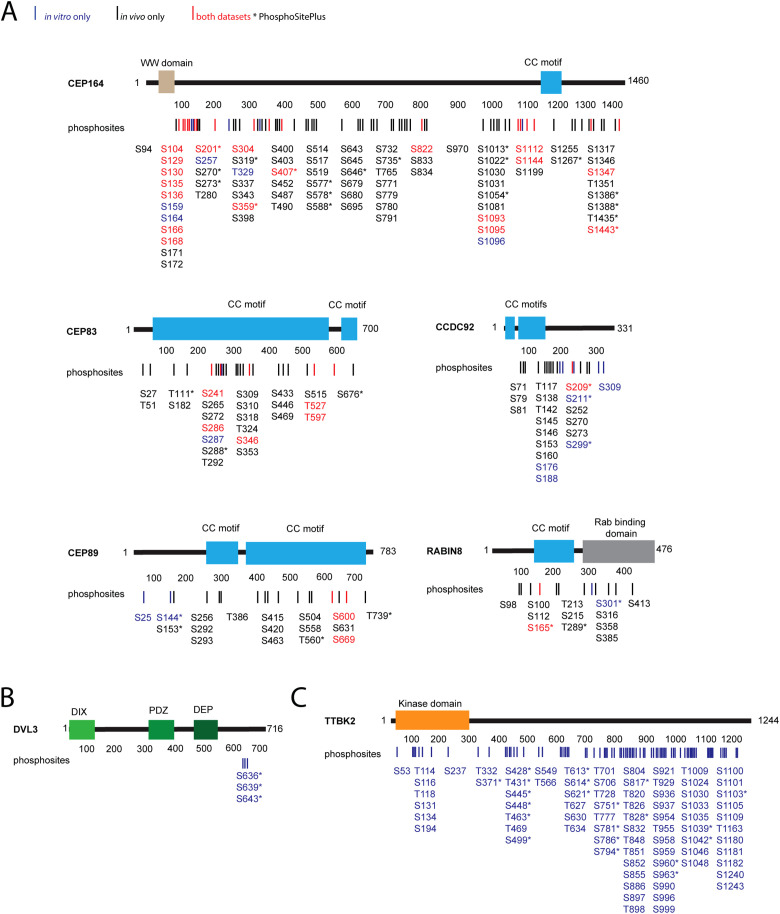
Having found that TTBK2 might phosphorylate several proteins necessary for the process of cilia initiation, we performed in vitro kinase assays of individual candidate substrates in the presence of TTBK2, followed by MS/MS identification and quantification of detected phosphorylations. To ensure proper protein folding and functionality, individual candidate substrates and TTBK2, respectively, were purified by immunoprecipitation from HEK293T cells. TTBK2 KD was used as a control condition to account for actions of endogenous kinases possibly copurifying with immunoprecipitated TTBK2. In addition, in the second repetition we included λ phosphatase treatment in our workflow to reduce the impact of any already present phosphorylation. The experimental workflow is summarized in Supplemental Figure S1A. In brief, the criteria to assign any identified phosphorylation to TTBK2 were as follows (please see Materials and Methods for additional details): 1) the intensity of phosphopeptide was higher than 2 × 106; 2) phosphopeptide intensity was at least twice that of the corresponding phosphopeptide intensity in control samples (TTBK2 KD). By applying these criteria, we detected 45 phosphorylated S/T sites induced by TTBK2 in the tested proteins (CEP164, CEP83, CEP89, Rabin8, CCDC92, DVL3) (Figure 2, A and B, blue and red color coded). The total number of identified phosphorylations was ∼120 (Supplemental Figure S2A; Figure 2B; Supplemental Table S1).
FIGURE 2:
TTBK2 phosphorylates its substrates on multiple serines and threonines. Map of identified phosphorylations induced by TTBK2 on tested substrates. (A, B) Structure of each TTBK2 substrate is schematized, rectangles indicate the presence of a domain or motif, and numbers indicate the length of given protein in amino acids. Lines and numbers, respectively, below the schematic protein structure indicate positions of each phosphorylation induced by TTBK2. Sites shown in red were detected both in vitro and in vivo, sites in blue were detected only in vitro, and sites in black were detected only in vivo. The asterisk indicates that given phosphorylation is covered in PhosphoSitePlus. (A) TTBK2-induced phosphorylation of CEP164, CEP89, CEP83, CCDC92, or Rabin8 identified in vitro and in vivo. (B) TTBK2 phosphorylation of DVL3 identified in vitro. (C) TTBK2 autophosphorylations identified in vitro.
In a similar manner, we examined autophosphorylation of TTBK2. Here we analyzed four experimental conditions: 1) purified TTBK2, 2) purified TTBK2 treated with λ phosphatase, 3) TTBK2 treated with λ phosphatase subsequently subjected to an in vitro kinase assay, and 4) TTBK2 KD treated with λ phosphatase, subjected to an in vitro kinase assay (schematized in Supplemental Figure S1B). We compared phosphosites identified in condition 3 with control conditions 2 and 4. Phosphorylations that were induced at least twofold over both controls were considered to be TTBK2 induced. Condition 1 was included to assess the efficiency of the λ phosphatase treatment. This analysis led to the identification of approximately 110 phosphorylations,of which 79 were found to be induced by TTBK2. The induced phosphorylations are found along the whole sequence of TTBK2, with a significant portion of the sites residing at the C-terminus (Figure 2C; Supplemental Figure S2B; Supplemental Table S1).
The detection of phosphorylation of a specific S/T residue using an in vitro kinase assay typically suffices to assume a direct kinase–substrate relationship for individual proteins tested but might not fully reflect phosphorylation of a given protein in vivo. To this end, we set out to analyze phosphorylations of individual substrates coexpressed with TTBK2 and subsequently purified from HEK293T cells. The experiment workflow is summarized in Supplemental Figure S1C. The criteria for in vivo TTBK2-induced phosphorylations were more stringent than in the case of in vitro analysis to account for the higher complexity of the samples and in turn for the potential influence of other protein kinases (please refer to Materials and Methods for details). This in vivo approach identified approximately 230 phosphorylations (Supplemental Table S1; Supplemental Figure S3) of which 133 were induced by TTBK2 (Figure 2A, black and red color coded). Importantly, a comparison of induced phosphorylation sites identified in vivo and in vitro not only showed a fairly extensive overlap between the datasets for individual proteins (phosphorylations common for both datasets are in red, Figure 2A; Supplemental Figure S3) but also revealed many phosphosites localized in clusters or pairs, often dispersed through the entire protein sequence (Figure 2A).
Specifically, in the case of CEP164, 18 individual S/T sites were found to be induced by TTBK2 by both approaches; for CEP83, it was five S/T residues; for CEP89, it was two S/T; and for CCDC92 and Rabin8, both showed one S/T phosphorylation in common between in vitro and in vivo. Of note, analysis of DVL3 in vivo phosphorylations induced in the context of several kinases, including TTBK2, has been recently reported (Hanáková et al., 2019). Overall, 27 (64%) induced phosphorylations detected through an in vitro approach were also found induced in vivo, thereby confirming a role of TTBK2 in the phosphorylation of tested proteins (Figure 2A). Additionally, six sites identified as being induced in vitro were occasionally also detected in vivo but did not pass the set threshold to be considered induced (Supplemental Table S1; Supplemental Figure S2A; Supplemental Figure S3). It is worth noting that the majority of sites detected by in vivo analyses (in total 106 phosphorylated S/T residues, Supplemental Table S1) were not detected using the in vitro approach.
TTBK2 shows motif similarities to CK1
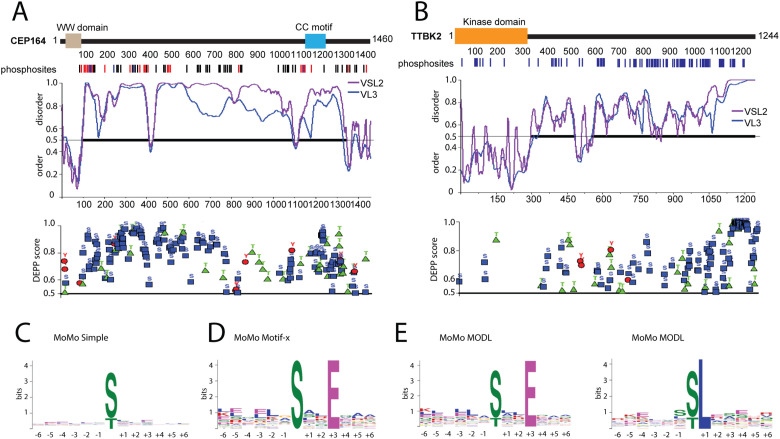
Given the number of phosphorylations we identified and the reports on preferential phosphorylation of intrinsically disordered regions (IDRs) over folded protein regions (Iakoucheva et al., 2004; Xie et al., 2007; Bah et al., 2015; Bah and Forman-Kay, 2016), we analyzed TTBK2 substrates using the Predictor of Natural Disordered Regions (PONDR) (Peng et al., 2005; Peng and Zhang, 2006) and the Disorder Enhanced Phosphorylation Predictor (DEPP) (Iakoucheva et al., 2004) to check the extent of IDRs and the overlap between the identified and the predicted phosphosites, respectively. The analysis carried out using the PONDR algorithms VSL-2 (Figure 3, A and B; Supplemental Figure S4, violet line) and VL3-BA (Figure 3, A and B; Supplemental Figure S4, blue line) revealed long IDRs in all the proteins tested (Figure 3, A and B, and Supplemental Figure S4). In addition, comparing our experimental data to DEPP-predicted phosphorylations revealed many examples of extensive overlap (Figure 3, A and B; Supplemental Figure S4). Specifically, experimentally identified CEP164 phosphorylations were found distributed throughout the protein sequence in a pattern resembling that of DEPP prediction (Figure 3A). Further, DEPP predicted extensive phosphorylation of the TTBK2 C-terminal part in line with our experimental data (Figure 3A). In addition, we observed similarity between the experimentally detected and the predicted phosphorylation sites also in the case of the remaining examined substrates of TTBK2 (Supplemental Figure S4). In light of these findings, we examined previously identified substrates of TTBK2, namely, CEP97, Tau, KIF2a, and MPP9 by PONDR and DEPP. As expected, all previously identified phosphosites were localized within long IDRs, which were predicted to contain many additional phosphorylation sites (Supplemental Figure S5).
FIGURE 3:
TTBK2 shows motif similarities to CK1. (A, B) Protein disorder and phosphorylation prediction for CEP164 and TTBK2. Structure of CEP164 and TTBK2 is schematized as described in Figure 2. The first graph below the schematized structure of CEP164 and TTBK2, respectively, depicts prediction of disorder/order (with cut-off score 0.5) determined by PONDR using VSL2 (violet line) and VL3-BA (blue line) algorithms. The second graph plots DEPP score (with cut-off score 0.5) and individual phosphosites thereby predicted (phosphoS as blue rectangle, phosphoT as green triangle, and phosphoY as red circle). (C–E) Determination of TTBK2 kinase motifs using indicated algorithms. Size of depicted symbols indicates relative abundance of each amino acid at given position within a set of phosphorylated peptides. Kinase motif search was performed by C, MoMo simple; D, Motif-x; and E, MODL alogrithms.
Next, we compared our experimental results with the data available in the PhosphoSitePlus database. For the purpose of this comparison, we did not discriminate here between induced and noninduced phosphorylations or between individual datasets of our analysis. Interestingly, we found that 99/133 (74%) phosphorylations reported for the substrate proteins on PhosphoSitePlus were also detected in our experiments (Figure 2, A–C; Supplemental Table S1, marked by an asterisk). Although we cannot rule out the possibility that some of phosphosites identified by our analyses are a result of overexpression, many of the identified sites seem physiological as they were previously detected in endogenous proteins. Importantly, however, only 53/229 (23%) of TTBK2-induced phosphorylations were previously identified by large-scale proteomic studies. This implies that phosphorylation of some of these residues might be limited to a specific cellular context.
As it was previously reported that TTBK2 preferentially phosphorylated S/T sites with pY at the +2 position (Bouskila et al., 2011), we examined our datasets for the presence of this motif. However, we identified only two phosphorylations (S201 in CEP164; S316 in Rabin8) with Y at the +2 position, which could in principle fit into the proposed consensus sequence. Next, we manually searched the dataset for priming sites at the –3 position (pS/pT-x-x-S/T motif) reported for the CK1 family (Cheong and Virshup, 2011) and found 28 phosphorylations fitting this criterion (CEP164: S171, S273, S403, S455, T490, S517, S646, S735, S1096, S1255, S1258; CCDC92: S145, S179, S273; Rabin8: S100; DVL3: S639; TTBK2: S134, T431, S448, T820, T851, S855, S958, S963, S999, S1033, S1042, S1243). Thus, the majority of phosphorylation sites we found were without any kinase motif assigned. To probe for possible unrecognized TTBK2 consensus sites, we pooled data from in vitro and in vivo datasets and used them for an unbiased kinase motif search by the MeMe suite tool MoMo (Bailey et al., 2009; Cheng et al., 2017). Initial analysis by a “simple” algorithm, which plots the relative abundance of individual AA residues at the indicated position within the whole dataset, did not reveal any enrichment (Figure 3C). Given that, we subsequently performed analysis by Motif-x and MODL algorithms, respectively, that can resolve consensus motif sequences present only in a subset of peptides (Chou and Schwartz, 2011; Cheng et al., 2017). Intriguingly, both algorithms detected a preference for glutamic acid (E) at the +3 position (MODL, 32 peptides; Motif-x, 27 peptides) (Figure 3, D and E). In addition, analysis using MODL detected a preference for leucine (L) at the +1 position (MODL, 30 peptides) (Figure 3E). Sequences of peptides with the identified motifs are listed in Supplemental Table S2.
CEP164 phosphorylation is largely dependent on TTBK2
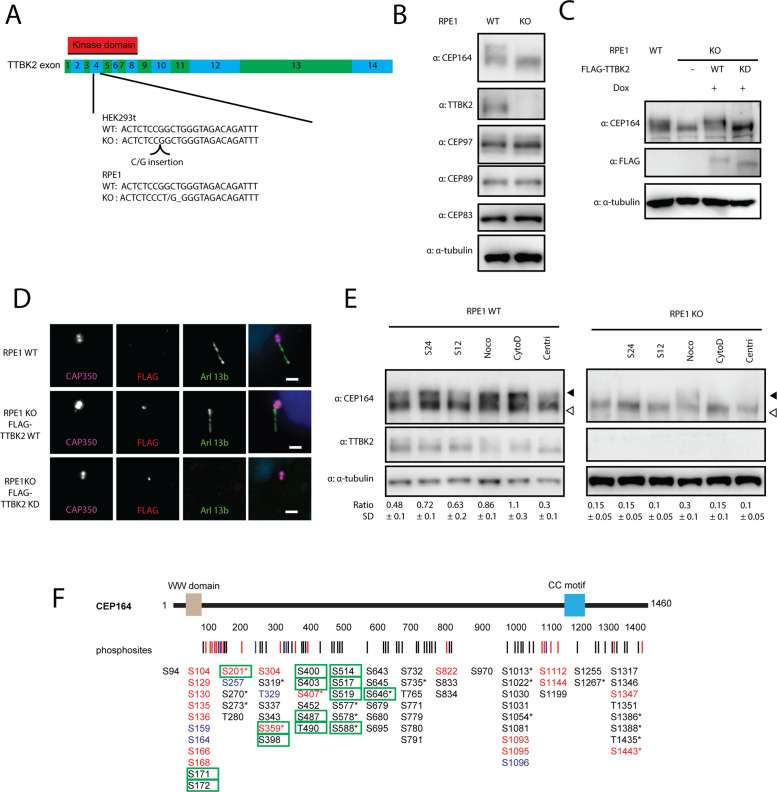
To further study TTBK2 phosphorylations, we decided to derive HEK293T and RPE1 cells devoid of TTBK2. First, we verified that CRISPR-Cas9 successfully disrupted ORF of the TTBK2 locus by indels in exon 4, which encodes part of the kinase domain of TTBK2 (Figure 4A). Next, we confirmed that CRISPR/Cas9-edited cells lacked any detectable levels of TTBK2 (Figure 4B; Supplemental Figure S6, A–C), while MC localization of CEP164 or CEP83 was not affected (Supplemental Figure 6, B and C). Functionally, TTBK2-deficient cells fail to form cilia. Subsequent reintroduction of WT, but not the KD form of TTBK2, allowed PCs formation in TTBK2 KO HEK293T or RPE1, as expected (Figure 4D; Supplemental Figure S6D). Additionally, we found that CEP164 migrated faster on PAA gel when TTBK2 was depleted (Figure 4B; Supplemental Figure S6A), implying that posttranslational modifications of CEP164 were diminished in TTBK2 KO cells. Reintroduction of the WT form of TTBK2 to TTBK2 KO RPE1 rescued CEP164 PAA mobility, in contrast to TTBK2 KD (Figure 4C). These results confirm and extend previous observations (Goetz et al., 2012; Lo et al., 2019). We also examined the behavior of CEP83, CEP89, and CEP97, substrates of TTBK2 reported here or previously, in response to TTBK2 ablation. However, in contrast to CEP164, we failed to detect any difference in the PAA mobility of these proteins (Figure 4B).
FIGURE 4:
TTBK2 is required for phosphorylation of CEP164 during induction of ciliogenesis. (A) Scheme of TTBK2 editing by CRISPR-Cas9. TTBK2 exons are coded in green and blue, kinase domain position is marked by a red rectangle. gRNA targets Exon4, Cas9-edited sequences of TTBK2 for HEK293T and RPE1 are indicated. (B) WB analysis of RPE1 WT and TTBK2 KO cells. TTBK2 protein is missing in RPE1 TTBK2 KO cells, CEP164 is migrating faster in TTBK2 KO RPE1, while no changes in migration pattern were detected for CEP89, CEP83, and CEP97. (C, D) FLAG TTBK2 WT and KD versions were reintroduced in RPE1 TTBK2 KO cells. (C) CEP164 migration pattern is rescued by FLAG-TTBK2 WT but not by FLAG-TTBK2 KD reintroduction in RPE1 TTBK2 KO cells. (D) Ciliogenesis is rescued by FLAG-TTBK2 WT but not by FLAG-TTBK2 KD reintroduction in RPE1 TTBK2 KO cells. (E) RPE1 WT and TTBK2 KO cells were treated as indicated and CEP164 migration was analyzed. Starvation for 12 or 24 h and nocodazole (100 ng/ml, overnight) and cytochalasin D (2 µM, overnight) treatment lead to slower CEP164 migration, while treatment by PLK4 inhibitor centrinone (150 µM, 72 h) enhances CEP164 migration in RPE1 WT cells. Treatment of RPE1 TTBK2 KO cells by nocodazole leads to slower CEP164 migration. Ratio indicates relative abundance of the top band (indicated by full arrowhead) of CEP164 divided by the bottom band (indicated by empty arrowhead). WB was quantified for N = 4 for WT and n = 3 for TTBK2 KO cells; SD indicates SD. (F) Structure of CEP164 is schematized, rectangles indicate domains or motifs, and numbers indicate length in amino acids. Lines and numbers, respectively, below the schematic protein structure indicate positions of each phosphorylation induced by TTBK2. Sites shown in red were detected both in vitro and in vivo, sites in blue were detected only in vitro, and sites in black were detected only in vivo. Green rectangle marks phosphorylations more intensive in WT compared with TTBK2 KO HEK293T cells. The asterisk indicates that given phosphorylation is covered in PhosphoSitePlus.
Our observations suggesting that TTBK2 is a main driving force of CEP164 phosphorylation prompted us to narrow down our validation attempts to CEP164. First, we examined the importance of TTBK2 for phosphorylation of endogenous CEP164. We isolated CEP164 by immunoprecipitation from TTBK2 WT and KO HEK293T cells, respectively, and subjected the protein complexes to MS/MS analysis of phosphorylation. We detected approximately 37 phosphorylations, with 27 of those being twice or more as intensive in WT HEK293T (Supplemental Figure S6F; Supplemental Table S1). Importantly, 14 phosphosites matched the TTBK2-induced phosphorylations detected by our previous in vitro or in vivo approaches (Figure 4F, marked by a green rectangle). From the rest of the S/T identified here as being regulated by endogenous TTBK2, 10 sites were assigned as not being targeted by TTBK2, while three sites were new (not identified in any of our previous experiments) (Supplemental Figure S6F and Supplemental Table S1). Together with our previous data, these findings establish that TTBK2 is both sufficient and required for efficient CEP164 phosphorylation.
Next, we attempted to relate TTBK2 phosphorylation of CEP164 to a particular event, for example, the induction of PC formation by serum starvation or treatment by cytochalasine D, nocodazole-induced mitotic arrest, or centrinone-induced centriole ablation (Wong et al., 2015). Thus, we examined the extent of CEP164 phosphorylation by monitoring the ratio between slower (hyperphosphorylated) and faster (hypophosphorylated) migrating forms of CEP164 (Čajánek and Nigg, 2014). As shown in Figure 4E, we observed the accumulation of slower migrating CEP164 forms in nocodazole-treated cells, in line with previous results (Schmidt et al., 2012). Interestingly, we detected that 12 h of serum starvation induced a moderate shift of CEP164, which became more prominent 24 h poststarvation. In addition, treatment by another compound promoting ciliogenesis, actin polymerisation inhibitor cytochalasin D (Kim et al., 2010), also showed a pronounced effect on CEP164 PAA mobility. Of note, the bitor centrinone caused a modest decrease in CEP164 phosphorylation. Remarkably, the effects of ciliogenesis-inducing agents (serum starvation, cytochalasin D), but not of nocodazole, on the relative increase of slow (hyperphosphorylated) forms of CEP164 were markedly reduced in TTBK2-deficient cells (Figure 4E). These data imply that TTBK2 is responsible for efficient phosphorylation of CEP164 during ciliogenesis.
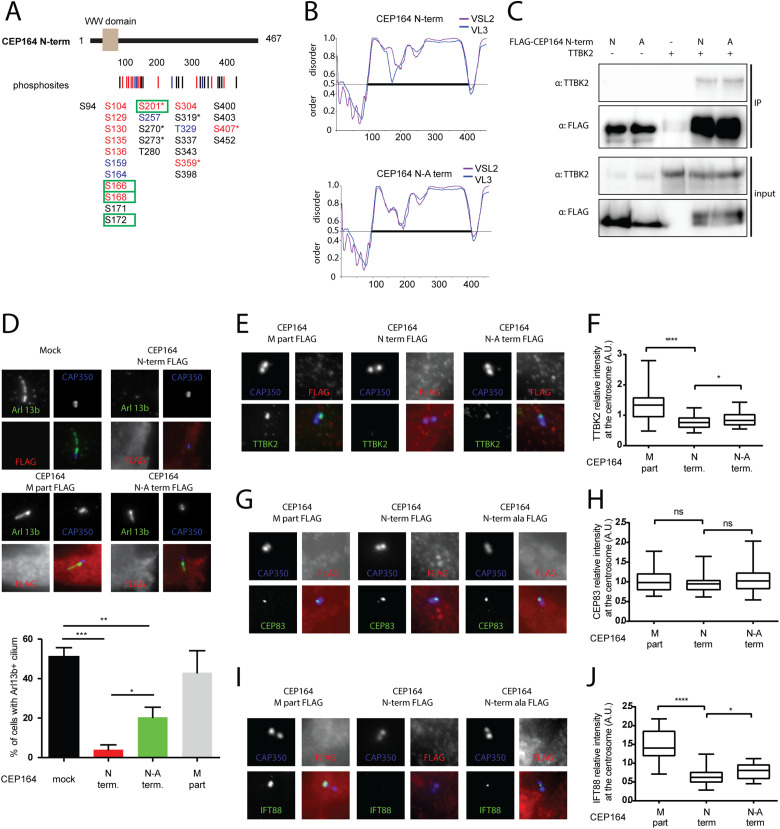
CEP164 N-terminus phosphorylation leads to altered function
The functional relevance of a phosphorylation is usually tested by mutating detected phosphorylated sites to nonphosphorylable variants and subsequent examination of whether this alters the function of tested protein (Sieracki and Komarova, 2013). Our analysis detected a significant number of induced phosphorylation sites, implying possible complex regulation, as multiple phosphorylations typically act in a combinatorial manner (Cohen, 2000; Bah and Forman-Kay, 2016). In addition, the majority of the identified phosphosites resided in IDRs with a currently unknown impact on the biology of a given protein. On the other hand, our experiments also revealed several TTBK2 phosphosites within the N-terminal part of CEP164 (CEP164 N-term, AA 1–467), which, via its WW domain (predicted to contain AA 56–89; Schmidt et al., 2012; Cˇajánek and Nigg, 2014), interacts with TTBK2 and in turn regulates ciliogenesis (Schmidt et al., 2012; Cˇajánek and Nigg, 2014; Oda et al., 2014). Bearing this in mind, we decided to test the functional consequences of TTBK2 phosphorylation of the CEP164 N-term. First, we examined the role of endogenous TTBK2 by analyzing phosphorylations of the FLAG-CEP164 N-term, transiently expressed in TTBK2 WT or KO HEK293T cells. We found that four phosphorylated S/T (S166, S168, S172, and S201) of 14 identified were at least two times more intensive in TTBK2 WT than inTTBK2 KO samples (Figure 5A; Supplemental Figure S6G; Supplemental Table S1).
FIGURE 5:
Phosphorylation of CEP164 N-terminus by TTBK2 affects its function and ciliogenesis. (A) Structure of CEP164 N-term is schematized, rectangle indicates WW domain, and numbers indicate length in amino acids. Lines and numbers, respectively, below the schematic protein structure indicate positions of each phosphorylation induced by TTBK2. Sites shown in red were detected both in vitro and in vivo, sites in blue were detected only in vitro, and sites in black were detected only in vivo. Green rectangle marks phosphorylations more intensive in WT compared with TTBK2 KO HEK293T cells. The asterisk indicates that given phosphorylation is covered in PhosphoSitePlus. (B) In silico analysis of protein folding by PONDR VSL2 (violet line) and VL3-BA (blue line) algorithms; the numbers indicate length of given protein in amino acids. Note that mutation of seven TTBK2 phosphorylated sites (S129, S130, S135, S136, S166, S168, S201) to Alanines (CEP164 N-A term) is not expected to change protein folding over control (CEP164 N-term). (C) HEK293T cells were transfected by indicated plasmids and 24 h posttransfection were subjected to immunoprecipitation by anti-FLAG antibody. No difference was observed between FLAG-CEP164 N term and FLAG-CEP164 N-A term in their ability of to coimmunoprecipitate MYC-TTBK2. (D–J) RPE1 cells were transfected by indicated FLAG-tagged CEP164 truncation constructs and, following 24 h, starved for an additional 24 h to induce ciliogenesis, or ON to evaluate TTBK2, IFT88, or CEP83 on MC. (D) ARL13b (green) and CAP350 (blue, inset) staining was used to detect PCs and centrioles, respectively, and expression of indicated CEP164 variants was detected by FLAG antibody (red). The graph summarizes effects on PC formation from four independent experiments, and RPE-1 cells transfected with empty vector (mock) or CEP164 M part were used as controls and analyzed by Student’s t test; ***p < 0.001, **p < 0.01, *p < 0.05; SEM. (E–J) Following fixation, RPE-1 cells were stained for TTBK2, CEP83, or IFT88 (green); FLAG (red); and CAP350 (blue) to examine differences in localization of tested proteins to MC. The graph summarizes the effects protein intensity on MC (TTBK2 n = 5, IFT88 n = 3, CEP83 n = 4, 30–40 cells analyzed per condition) and analyzed by Student’s t test; ****p < 0.0001, *p < 0.05; SEM.
Next, we examined all CEP164 phosphorylation data and selected seven TTBK2 phosphorylation sites (S129, S130, S135, S136, S166, S168, and S201) located in the proximity of the WW domain of the CEP164 N-term and mutated those to A in the CEP164 full-length (FL) protein and CEP164 N-term, naming the obtained mutants the “CEP164 FL-A” and “CEP164 N-A term.” It is worth noting that in silico modeling of introduced mutations in CEP164 N-term using PONDR did not reveal any major impact on protein folding (Figure 5B). First, we examined exogenously expressed CEP164 FL-A and CEP164 N-A term in terms of mobility shift after serum starvation and noted that FL-A and N-A term did not produce slow migrating forms on serum starvation, in contrast to their corresponding WT counterparts (Supplemental Figure S6E). Next, to resolve whether the introduced mutations in any way changed the CEP164–TTBK2 relationship, we tested whether the CEP164 N-A term and TTBK2 can coimmunoprecipitate. However, we did not detect any notable difference between the FLAG-CEP164 N-term and the FLAG-CEP164 N-A term to coimmunoprecipitate with MYC-TTBK2 when overexpressed in HEK293T cells (Figure 5C). We reasoned that our failure to see any difference meant the changes were too subtle to be revealed by the used approach and decided to test the CEP164–TTBK2 relationship by other means. Interestingly, we observed that while ciliogenesis was efficiently induced in mock (51%) or FLAG-CEP164 M-part (468–1135 AA) (43%) transfected RPE-1 cells, in line with previous reports (Schmidt et al., 2012; Cˇajánek and Nigg, 2014; Oda et al., 2014), FLAG-CEP164 N-term expression almost completely abrogated the formation of PCs (below 5% of ciliated cells) in contrast to the FLAG-CEP164 N-A term that still allowed the formation of PCs in about 20% of transfected RPE-1 cells (Figure 5D). The CEP164 N-term fails to localize to the MCs (Schmidt et al., 2012; Cˇajánek and Nigg, 2014); in turn, its expression leads to the sequestration of endogenous TTBK2 from MCs and hence to profound inhibition of ciliogenesis (Cˇajánek and Nigg, 2014). Based on our observations, we hypothesized that the ability of the CEP164 N-term to sequester endogenous TTBK2 (Cˇajánek and Nigg, 2014) might be affected in the CEP164 N-A term. Intriguingly, we found that the FLAG-CEP164 N-A term was still able to sequester TTBK2 from MC, but to a lesser extent than the FLAG-CEP164 N-term (Figure 5, E and F). Next, we found levels of IFT88, but not of CEP83, located on the MCs significantly more reduced in cells expressing the FLAG-CEP164 N-term than in cells expressing the FLAG-CEP164 N-A term (Figure 5, G–J). This observation is in line with previous reports as well as our observations showing that MC localization of IFT88, but not of CEP83, is dependent on TTBK2 activity (Supplemental Figure S6, B and C) (Goetz et al., 2012). Taken together, these results imply that phosphorylation of the given residues (S129, S130, S135, S136, S166, S168, and S201) modulates CEP164 function which in turn may be functionally important for TTBK2 recruitment to MC and ciliogenesis.
DISCUSSION
TTBK2 seems to have a unique position among other regulators of ciliogenesis, as its action marks the “end of the beginning” by pushing cilia initiation toward cilia extension. However, how TTBK2 regulatory function is implemented is still only partially understood. To gain insight into this process, we set out to identify its substrates. In this study, we have demonstrated that several proteins either directly binding to CEP164 or localizing to its proximity, namely, CEP83, CEP89, CCDC92, Rabin8, and DVL2/3, as well as CEP164 itself, are subjected to TTBK2 phosphorylation primarily in their IDRs. We further revealed a previously undetected consensus motif similarity of TTBK2 to that of CK1, at least for a subset of phosphorylated sites.
Our observation that TTBK2 is able to phosphorylate components of DA and proteins associated with the centriole distal end does not come as a complete surprise, given its proposed role in the assembly of the appendages (Cˇajánek and Nigg, 2014). Moreover, thanks to the recently detailed description of DA structure and organization (Yang et al., 2018; Bowler et al., 2019), it is tempting to speculate that TTBK2 might be able to target additional basal body proteins besides those included in our screen. In our initial screening, CP110 and Rab8a did not shift on TTBK2 coexpression, hence we did not include them in the subsequent detailed analysis. However, as a lack of mobility shift does not necessarily mean a lack of phosphorylation, it would be premature to rule them out as TTBK2 substrates. Interestingly, two of the CP110 binding partners, CEP97 and MPP9, have already been reported as TTBK2 substrates and their phosphorylation has been implicated in CP110/CEP97 complex removal from the distal end of the MCs (Oda et al., 2014; Huang et al., 2018).
The extent of the mobility shift we observed for CEP164, CEP83, CEP89, CCDC92, Rabin8, and DVL3 suggested modification by multiple phosphates rather than by single phosphorylation, which we subsequently confirmed with MS-based experiments. Importantly, our analysis detected not only the majority of sites previously assigned as targets of TTBK2 activity (Cˇajánek and Nigg, 2014), namely, the CEP164 sites T1309, S1317, S1347, and S1443 in both datasets and S1346 in vivo, but also many new ones, thereby demonstrating the robustness and sensitivity of our workflow. Furthermore, in the example of CEP164, we demonstrated that TTBK2 regulates phosphorylation of a fairly large number of S/T sites on the endogenous level.
The majority (64%) of TTBK2 phosphorylation sites we detected in vitro were subsequently confirmed in vivo. A significant portion of the sites detected by in vivo analysis (in total 106 phosphorylated S/T residues) was not revealed using the in vitro approach. Importantly, we managed to validate the role of TTBK2 in phosphorylation of several in vivo-induced S/T sites in CEP164 using TTBK2 WT and KO cells. Several factors may account for the differences we see between individual analyzed datasets, such as the different activity of the kinase in vitro and in vivo, different preferences for substrates, or the presence/absence of priming phosphorylations or compensating kinase(s) and counteracting phosphatases (Hunter, 1995).
TTBK2 is considered a distant member of the CK1 kinase family. However, it was previously shown that its truncated variant (TTBK2 1–450) preferentially phosphorylates peptides with priming phosphotyrosine residue at the +2 position (Bouskila et al., 2011). This unusual substrate preference, unique among CK1 family members which typically favor the stretch of acidic residues or pS/pT around the –3 position (Flotow et al., 1990; Flotow and Roach, 1991; Knippschild et al., 2005; Cheong and Virshup, 2011), was related to differences in the putative phosphate-binding groove between TTBK1/2 and all remaining CK1 isoforms (Bouskila et al., 2011). Although it has been found that several Y kinases reside at the centrosome and MCs, respectively (Tsun et al., 2011; Jay et al., 2015), the significance of that for centriole biogenesis or ciliogenesis is unclear. Importantly, this study and others (Kitano-Takahashi et al., 2007; Cˇajánek and Nigg, 2014; Watanabe et al., 2015; Huang et al., 2018) failed to detect any significant presence of Y at +2 at verified TTBK2 substrates, raising the question of whether TTBK2 extensively utilized such a motif in vivo. Additionally, although we detected several phosphorylations that fit the canonical consensus motif pS/pT-x-x-S/T of CK1, our unbiased motif search analysis implies that TTBK2 may in addition favor glutamic acid (E) at the +3 position and L at the +1 position, respectively, at least in a subset of its targets. These motifs resemble the noncanonical CK1 motif SLS and to some extent also a motif with an acidic stretch of residues located C-terminally from the target S/T reported before (Marin et al., 2003; Ferrarese et al., 2007; Borgal et al., 2014; Bowie et al., 2018). Importantly, one must bear in mind that delineation of kinase motifs has been typically carried out using rather reduced libraries of synthetic peptides that lack a secondary or tertiary structure of bona fide physiological substrates of a given kinase. In fact, it has been demonstrated that the phosphorylation of a substrate by CK1 isoforms does not strictly depend on the consensus sequence, but there is a significant effect of a tertiary structure of the substrate (Cegielska et al., 1998). From this point, FL TTBK2 does not seem to use the motif with pY at the +2 position in substrates we examined; instead, our data suggest it may use motifs resembling noncanonical motifs reported for CK1. Of note, detailed inspection of the assay that identified pY+2 as the preferred motif for TTBK2 also show that TTBK2 can phosphorylate peptides carrying noncanonical CK1-like motifs in vitro, albeit to a lesser extent than pY+2 (Bouskila et al., 2011). To further analyze the preference for motif usage by TTBK2, cocrystallization of the kinase domain of TTBK2 with some of its physiological substrates would help in defining the molecular mechanism mediating their mutual interactions.
Using a model system of HEK293T and RPE1 cells that lack TTBK2, we showed that TTBK2 extensively modified CEP164, while a similar effect was not observed in the case of the other confirmed TTBK2 substrates (CEP83, CEP89, or CEP97), possibly due to the insufficient sensitivity of used readout. This suggests that CEP164 might be the main target of TTBK2 phosphorylation. This prediction is further supported by the observation that a majority of TTBK2 phosphosites we identified using the MS approaches indeed resided to CEP164.
The PCs formation in cultured cells can be readily induced be a serum withdrawal or treatment by cytochalasin D (Kim et al., 2010; Seeley and Nachury, 2010). Intriguingly, our data imply not only that CEP164 is phosphorylated following the induction of the PCs formation but also importantly that TTBK2 is involved in the phosphorylation. Previous work postulated that CEP164 is heavily phosphorylated in mitosis, perhaps in relation to its disappearance from the DA (Schmidt et al., 2012). Our data support this observation and in addition suggest that the mitotic phosphorylation of CEP164 does not depend on TTBK2.
The amino acid sequences of intrinsically disordered proteins determine their inability to fold into stable tertiary structures and instead enable them to rapidly interconvert between distinct conformations to mediate their biological functions (Dyson and Wright, 2005; Bah and Forman-Kay, 2016). Consequently, this allows the intrinsically disordered proteins to expose a significant part of their primary sequence for binding and posttranslational modification (Iakoucheva et al., 2004; Xie et al., 2007; Bah and Forman-Kay, 2016) and hence provides them with the ability to act as hubs in protein complexes that typically facilitate the assembly of various organelles (Cohen, 2000; Hegyi et al., 2007; Bah and Forman-Kay, 2016). In line with these reports, all proteins we identified as substrates of TTBK2 modified by multiple phosphorylations are predicted to have long IDRs suitable for such extensive modification. Given that phosphorylation of IDRs might affect protein structure and hence binding properties (Cohen, 2000; Bah et al., 2015), it is tempting to speculate that TTBK2 phosphorylation allows basal body DA proteins to achieve a novel quality that ensures their timely interactions and assembly to complexes necessary for the formation of PCs.
We provided experimental evidence supporting this hypothesis on the example of CEP164–TTBK2 interactions. We admit that the observed effects of the introduced S to A mutations of the CEP164 N-term on sequestration of TTBK2, MCs levels of IFT88, and ciliogenesis are modest. However, this is to be expected, as the mutated residues lie within IDR outside of the reported TTBK2 binding region (Cˇajánek and Nigg, 2014), but still in the vicinity to possibly mediate the effect on CEP164–TTBK2 interaction. From this perspective, our data suggest that at least some of the mutated residues participate in WW domain-mediated CEP164–TTBK2 interaction. Interestingly, recent incisive work by Xu and colleagues (Xu et al., 2016) pinpointed a regulatory role of a short, basic AA stretch (110–122) of CEP164 for the CEP164–TTBK2 interaction via phosphatidylinositol-4 phosphate binding. As the cluster S129–S136, phosphorylated by TTBK2, localizes very close to the described PI binding stretch, it is tempting to speculate that phosphorylation of this cluster might represent a mechanism of how TTBK2 modulates its own interaction with CEP164 via cross-talk with the INPP5E/PIP3Kγ regulatory element, possibly by hampering the binding of the acidic phosphoinositol head group to its basic binding site (Xu et al., 2016). It should be rewarding to explore whether there is indeed any functional interaction between TTBK2 sites we identified and the activity of PIP3K/INPP5E implied in the regulation of CEP164–TTBK2 binding (Xu et al., 2016).
There is one more aspect related to our validation experiments. Three of the sites we identified in our initial screening experiments, and subsequently chose to target (S166, S168, and S201) in structure–function experiments, also clearly showed regulation by endogenous TTBK2. It is not clear whether this is also true for the S129–S136 cluster, as we were not able to recover the corresponding peptides during the phosphoenrichment step, thereby indicating that their overall abundance is not sufficient to be detected by the applied instrumentation.
While this paper was being revised, a study identifying CEP83 as a target of TTBK2 phosphorylation at S29, T292, T527, and S698 was published (Lo et al., 2019). Intriguingly, the authors related the TTBK2 phosphorylation of CEP83 to the docking of vesicles to DA. The authors also showed that CEP164-induced TTBK2 recruitment to MC is necessary for CEP83 phosphorylation. Our data confirm and extend this work, as we detected not only three of four sites reported by Lo and colleagues (although S698 did not pass our criteria to be considered as TTBK2 induced) but also several additional TTBK2-induced phosphosites in CEP83. Interestingly, since several members of DA seem to be direct targets of TTBK2 kinase activity, it would be of great interest to investigate possible mutual dependencies of individual phosphorylations and their impact on the functions of DA components in future. Additionally, we confirmed and extended a recent report on TTBK2 phosphorylation of DVL3 (Hanáková et al., 2019), a component of WNT signaling pathways known to reside at the centrosome (Cervenka et al., 2016; Mlodzik, 2016; Bryja et al., 2017). This may have several biological implications, as TTBK2 seems to phosphorylate a small cluster of S/T residues in the C-terminus of DVL3, which is important for regulating DVL3 oligomerization (Bernatík et al., 2014), which is in turn essential for DVL-mediated signal transduction via the formation of signalosomes (Schwarz-Romond et al., 2007). In addition, the C-terminus of DVL is typically phosphorylated by CK1ε (Bernatík et al., 2014), thereby opening an intriguing window for a cross-talk at the level of TTBK2-CK1ε, possibly through differential phosphorylation of the DVL3 C-terminus as CK1ε phosphorylates DVL3 at many additional sites to those targeted by TTBK2 (Hanáková et al., 2019). Clearly, additional work will be necessary to fully examine the implications of TTBK2-mediated DVL phosphorylation for WNT signaling pathways as well as basal body-related aspects of DVL biology.
In summary, our findings expand the current knowledge about TTBK2 substrates and provide an important insight into the extent and character of TTBK2-mediated phosphorylations in the context of basal body proteins and PC initiation, respectively. Underpinning the functions of TTBK2 phosphorylations, as well as its interactions with other kinases implicated in ciliogenesis, will undoubtedly be vital for a mechanistic understanding of cilium assembly.
MATERIALS AND METHODS
Cell culture and transfection
RPE-1 Flp-In T-Rex (a gift from Erich Nigg) and TTBK2 KO cells and RPE1 WT were grown in DMEM F12 (Thermo Fisher Scientific, 11320033) supplemented by 10% fetal bovine serum (FBS), 1% penicillin/streptomycin, and 1% l-glutamine, HEK293T wt, and TTBK2 KO Flp-In T-Rex (Thermo Fisher Scientific, R78007) cells were grown in DMEM Glutamax (Thermo Fisher Scientific, 10569069) supplemented by 10% FBS and 1% penicillin/streptomycin. Transfection of RPE1 was carried out by Lipofectamine 3000 (Invitrogen) according to manufacturer’s instructions (up to 0.5 µg DNA per condition/well, 24-well plate format). HEK293T were transfected by Polyethyleneimine (PEI) as follows: PEI was incubated in DMEM for 10 min and then mixed with plasmid equilibrated in DMEM in ratio 3 µl PEI/1 µg plasmid; the mixture was incubated for 15 min and then added to cells. Growth medium was changed 4 h after transfection. To induce formation of PC, 24 h following the transfection, RPE-1 cells were starved by serum-free medium for 24 h to assess ciliogenesis or ON for TTBK2 removal measurement. Centrinone (150 µM) (Wong et al., 2015) treatment was carried out for 72 h. Cytochalasine (2 µM) (Merck, Sigma Aldrich, C8273) and Nocodazole (100 ng/ml) (Merck, Sigma-Aldrich, M1404) treatment was carried out overnight.
Western blot
To analyze electrophoretic migration of the proteins after TTBK2 coexpression, HEK293T cells were transfected by the indicated plasmids and 24 h posttransfection directly lysed in 1× sample buffer (2% SDS, 62.5 mM Tris, pH 6.8, 10% glycerol, 2% 2-Mercaptoethanol, 0.01% bromophenol blue). RPE1 cells were treated as indicated. Western blot was carried out by using Bio-Rad apparatus (Mini-PROTEAN Tetra Vertical Electrophoresis Cell, Mini Trans-Blot Module) for the SDS–PAGE run and transfer. The samples were loaded to discontinuous SDS–PAGE (5% stacking; 7, 8, or 10% running gel; percentage used based on analyzed protein MW) and run at 150 V. Proteins were transferred to immobilon PVDF (Merck, IPVH20200) membranes at 100 V. ELFO Running buffer (0.025 M Tris, 0.192 M glycine, 0.1% SDS), WB Transfer buffer (0.025 M Tris, 0.192 M glycine, 20% Methanol). Membranes were blocked in 5% skimmed milk in Tris-buffered saline (TBS)-Tween (20 mM Tris, 150 mM NaCl, 0.1% Tween 20, pH 7.6) and incubated with the primary antibodies at 4°C ON. Then membranes were washed in TBS-Tween twice, incubated with the secondary antibodies for 1 h at RT, washed by TBS-Tween twice, and developed using ECL prime (Merck, GERPN2232). For the concentration of antibodies used, please refer to Supplemental Table S3.
WB quantification was done using FIJI distribution of ImageJ; the number of the experiments used for the quantification is stated in the corresponding figure legend.
Immunoprecipitation
To analyze binding of TTBK2 to CEP164 N term or CEP164 N-A term, 10-cm plates of HEK293T cells were transfected (in total 6 µg of DNA mixed with 18 µl of PEI in plain DMEM). Cells were washed by phosphate-buffered saline (PBS) 24 h posttransfection, scraped to Lysis buffer (20 mM Tris-Cl, pH 7.4, 150 mM NaCl, 0.5% NP40, 0.5% Triton X-100 [Merck, X100], and 1× Complete proteasome inhibitors [Roche, 4693132001]), and lysed (15 min on ice). Following centrifugation (15,000 × g for 10 min at +4°C), cleared extracts were incubated ON at +4°C in an orbital shaker with anti-FLAG sepharose beads (M2 clone, Merck, A2220). Bound complexes were pelleted, washed, and analyzed by SDS–PAGE and Western blotting.
Where appropriate, contrast and/or brightness of images were adjusted by using Photoshop CS5 (Adobe).
Purification of proteins from HEK293T cells
Cep164 N-, M-, and C- terminal fragment and CEP83, CEP89, CCDC92, Rabin8, and DVL3 coding plasmids fused to FLAG tag and FLAG-TTBK2 WT and kinase dead variant (KD) were used for production of proteins for in vitro or TTBK2 autophosphorylation experiments, GFP-TTBK2 WT was coexpressed for in vivo experiments. To purify the protein of interest, each expression plasmid (please see detailed list of used plasmids in Supplemental Table S3) was transfected to HEK293T cells (2 × 15 cm plate per condition, 20 µg DNA), either individually (in vitro kinase assay) or in combination with TTBK2 expression vector (in vivo). For the MSMS analysis of samples of the endogenous CEP164 phosphorylations 2 × 15 (run 1) or 4 × 15 (run 2) cm 90% confluent plates of HEK293T WT or TTBK2 KO cells were used. Analysis of CEP164 N-term phosphorylations in WT and TTBK2 KO HEK293T was done as follows: 1 × 15 cm dish of HEK293T WT or TTBK2 KO cells was transfected by 10 µg of FLAG-CEP164 N-term. Posttransfection (36-48 h), the proteins of interest were immunoprecipitated; briefly: cells were washed in PBS, scraped, pelleted by centrifugation (400 × g/5 min/+4°C), resuspended in Lysis buffer (20 mM Tris·Cl, pH 7.4, 150 mM NaCl, 0.5% NP40, 0.5% Triton-X-100, 0.1 mM DTT [Merck, 43815], 1× Complete proteasome inhibitors [Merck, 4693132001], and 1× phosphoSTOP [Merck, PHOSS-RO]), sonicated (2 × 20 s on ice), and lysed (10 min on ice). Following centrifugation (15,000 × g for 10 min at +4°C), cleared extracts were incubated 2–6 h at +4°C in an orbital shaker with anti-FLAG sepharose beads (M2 clone, Merck, A2220). Bound complexes were pelleted and 5× washed (Lysis buffer with 1M NaCl). Samples for in vitro kinase assay were additionally 2× washed with kinase buffer (50 mM Tris-Cl, pH = 7.4, 10 mM MgCl2, 0.1 M EGTA + 0.1 mM DTT), resuspended in kinase buffer + 0.1% DTT + 10% glycerol, and stored as 25% slurry at –80°C until used. Where indicated, samples were treated with λ-phosphatase (New England BioLabs [NEB], P0753S) (30°C for 20 min) according to manufacturer’s instructions, and λ-phosphatase was then removed by 2× wash with kinase buffer + phosphoSTOP (Merck, PHOSS-RO), 50 mM NaF (NEB, P0759S), and 10 mM sodium orthovanadate (NEB, P0758S). Samples were subsequently subjected to in vitro kinase assay (in vitro samples) or directly analyzed by MS/MS (in vivo dataset, samples of endogenous CEP164 and CEP164 N-term immunoprecipitated from WT and TTBK2 KO HEK293T).
Immunocytofluorescence microscopy
RPE1 cells were seeded on glass coverslips, fixed for 10 min in –20°C Methanol, washed 3× with PBS, blocked (2% bovine serum albumin in PBS with 0.01% NaN3), 3× PBS washed, incubated with primary antibodies for 1 h, 3× washed by PBS, incubated with secondary antibodies for 1 h in dark, washed 3× with PBS, incubated with DAPI, washed by PBS 2×, and mounted by glycergel (DAKO #C0563). Microscopy analyses were done using either DeltaVision Elite (GE Healthcare) with 100×/Zeiss Plan-ApoChromat 1.4 objective and DeltaVision softWoRx acquisition SW (here the image stacks were taken with a Z distance of 0.2 μm, deconvolved with one cycle of conservative ratio deconvolution algorithm, and directly projected as maximal intensity images) or Zeiss AxioImager.Z2 with Hamamatsu ORCA Flash 4.0 camera, 100× Apo oil immersion objective, and ZEN Blue 2.6 acquisition SW (Zeiss). Image stacks acquired using Zeiss AxioImager.Z2 were projected as maximal intensity images by using ImageJ distribution FIJI (Schindelin et al., 2012). Where appropriate, contrast and/or brightness of images were adjusted by using Photoshop CS5 (Adobe) or FIJI. For ciliogenesis experiment (Figure 4C), at least 50 cells were analyzed per each condition. For the experiment in Figure 4D, a densitometry analysis within selected regions of interest was performed in 16-bit TIFF images by using FIJI analyzing 30–40 cells per condition. Data are presented as relative staining intensity (staining intensity of a protein of interest normalized to the staining intensity of the centriolar marker CAP350). Statistical analysis by unpaired Student’s t test was performed using Graphpad Prism; P < 0.05 (*), P < 0.01 (**), P < 0.001 (***), and P < 0.0001 (****) were considered significant. Results are presented as mean plus SEM.
In vitro kinase assay
Beads-bound proteins were washed with kinase buffer (50 mM Tris, pH 7.4, 10 mM MgCl2, and 0.1 mM EGTA), mixed according to experimental scheme (Supplemental Figure S1), and incubated in the kinase buffer supplemented with 1 mM ATP, 5× Complete proteasome inhibitors (Merck, 4693132001), 1× phosphoSTOP (Merck, PHOSS-RO), 50 mM NaF (NEB, P0759S) and 10 mM sodium orthovanadate (NEB, P0758S), and incubated for 30 min/30°C. The kinase reaction was terminated by the addition of 4× sample buffer, and the resulting samples were analyzed by MS/MS.
CRISPR TTBK2 RPE1 and HEK293T cells
TTBK2 targeting gRNA oligos FWD: CaccgCATTAGTACCACTCTCCGGC REV: aaacGCCGGAGAGTGGTACTAATGc were cloned into pSpCas9 (BB)-2A-GFP (PX458) https://www.addgene.org/48138/ according to published protocol (Ran et al., 2013), and the resulting construct was sequenced to verify successful gRNA integration. HEK293T WT Flp-in T-Rex were transfected using Lipofectamine 3000. Single clones of HEK293T cells were produced using serial dilution method and were screened for TTBK2 presence by WB and PCR; clones were sequenced to verify disruption of TTBK2 locus. RPE1 WT Flp-in T-Rex cells were nucleofected by Neon transfection system (1350 V, 20 ms, 2 pulses) (Thermo Fisher Scientific) and GFP-positive cells were sorted using BD FACS ARIA II to 96-well plates. Clones were screened for TTBK2 presence by WB and PCR; clones were sequenced to verify disruption of TTBK2 locus.
Establishment of TTBK2 overexpressing stable RPE1 Flp-In T-Rex
RPE-1 TTBK2 KO Flp-In T-Rex cells were transfected by pgLAP2 NEO containing TTBK2 WT or KD variant and pOG44 (ratio 1:29, 0.1 µg of pgLAP2 NEO with transgene and 2.9 µg of pOG44 per 6-cm dish) by Lipofectamine 3000. Transfection was carried out overnight, medium was changed, and selection antibiotics (G418 c = 0.5 mg/ml, Blasticidine c = 25 µg/ml) were added. Selection continued until colonies appeared. Expression of the transgene was verified by WB and IC.
MS/MS analysis
In-gel digestion.
Immunoprecipitates were separated on SDS–PAGE gel electrophoresis, fixed with acetic acid in methanol and stained with Coomassie brilliant blue G250 (EZBlue, G1041, Merck) for 1 h. Corresponding 1D bands were excised. After destaining, the proteins in-gel pieces were incubated with 10 mM DTT at 56°C for 45 min. After removal of DTT excess, samples were incubated with 55 mM IAA at room temperature in darkness for 30 min, then alkylation solution was removed and gel pieces were hydrated for 45 min at 4°C in digestion solution (5 ng/µl trypsin, sequencing grade, Promega, in 25 mM AB). The trypsin digestion proceeded for 2 h at 37°C on Thermomixer (750 rpm; Eppendorf). Digested peptides were extracted from gels using 50% ACN solution with 2.5% formic acid (FA) and concentrated in a speedVac concentrator (Eppendorf). The aliquot (1/10) of concentrated sample was transferred to a LC-MS vial with already added polyethylene glycol (PEG; final concentration 0.001%) and directly analyzed by LC-MS/MS for protein identification.
Phosphopeptide enrichment.
The rest of the sample (9/10) was used for phosphopeptide analysis. Sample was diluted with acidified acetonitrile solution (80% ACN, 2% FA). Phosphopeptides were enriched using Pierce Magnetic Titanium Dioxide Phosphopeptide Enrichment Kit (Thermo Fisher Scientific, Waltham, MA) according to manufacturer protocol and eluted into LC-MS vial with already added PEG (final concentration 0.001%). Eluates were concentrated under vacuum and then dissolved in water and 0.6 µl of 5% FA to get 12 µl of peptide solution before LC-MS/MS analysis.
LC-MS/MS analysis.
LC-MS/MS analyses of peptide mixture were done using RSLCnano system connected to Orbitrap Elite hybrid spectrometer (Thermo Fisher Scientific) with ABIRD (Active Background Ion Reduction Device; ESI Source Solutions) and Digital PicoView 550 (New Objective) ion source (tip rinsing by 50% acetonitrile with 0.1% FA) installed. Prior to LC separation, peptide samples were online concentrated and desalted using trapping column (100 µm × 30 mm) filled with 3.5 µm X-Bridge BEH 130 C18 sorbent (Waters). After washing of trapping column with 0.1% FA, the peptides were eluted (flow rate 300 nl/min) from the trapping column onto Acclaim Pepmap100 C18 column (3-µm particles, 75 µm × 500 mm; Thermo Fisher Scientific) by 65 min-long gradient. Mobile phase A (0.1% FA in water) and mobile phase B (0.1% FA in 80% acetonitrile) were used. The gradient elution started at 1% of mobile phase B and increased from 1% to 56% during the first 50 min (30% in the 35th and 56% in 50th min), then increased linearly to 80% of mobile phase B in the next 5 min and remained at this state for the next 10 min. Equilibration of the trapping column and the column was done prior to sample injection to sample loop. The analytical column outlet was directly connected to the Digital PicoView 550 ion source. MS data were acquired in a data-dependent strategy selecting up to top 10 precursors based on precursor abundance in the survey scan (350–2000 m/z). The resolution of the survey scan was 60,000 (400 m/z) with a target value of 1 × 106 ions, one microscan, and maximum injection time of 1000 ms. High resolution (15,000 at 400 m/z) HCD MS/MS spectra were acquired with a target value of 50,000. Normalized collision energy was 32% for HCD spectra. The maximum injection time for MS/MS was 500 ms. Dynamic exclusion was enabled for 45 s after one MS/MS spectra acquisition and early expiration was disabled. The isolation window for MS/MS fragmentation was set to 2 m/z.
Data analysis
The analysis of the mass spectrometric RAW data was carried out using the Proteome Discoverer software (Thermo Fisher Scientific; version 1.4) with in-house Mascot (Matrixscience; version 2.4.1 or higher) search engine utilization. MS/MS ion searches were done against in-house database containing expected protein of interest with additional sequences from cRAP database (downloaded from http://www.thegpm.org/crap/). Mass tolerance for peptides and MS/MS fragments was 7 ppm and 0.03 Da, respectively. These posttranslational modifications were considered as possible: Oxidation of methionine, deamidation (N, Q), and phosphorylation (S, T, Y) as optional modification; carbamidomethylation of C as fixed modification; no enzyme specificity was selected. The phosphoRS (version 3.1) feature was used for preliminary phosphorylation localization. Final phosphosite assignment (including inspection of phosphorylations with ambiguous localization) was performed by manual evaluation of the fragmentation spectra of the individual phosphopeptides. Quantitative information was assessed and manually validated in Skyline software (Skyline daily 3.6.1.10230). Normalization of the data was performed using a set of phosphopeptide standards (added to the sample prior phosphoenrichment step; MS PhosphoMix 1, 2, 3 Light, Sigma) in combination with summed peak area of nonphosphorylated peptides identified in analyses of samples without phosphoenrichment. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (Perez-Riverol et al., 2019) partner repository URL: https://www.ebi.ac.uk/pride/archive/projects/PXD014199.
Identification of phosphorylations
Average intensity for individual phosphosites or clusters was calculated from MS/MS data as a sum of intensities of all phosphorylated peptides containing the particular pS/pT. Data are summarized in Supplemental Table S1 and graphically represented in Supplemental Figures S2, A and B, S3, and S6, F and G. Induction of phosphosites by TTBK2, summarized for individual substrates in Figure 2, was evaluated using individual phosphorylated peptides intensities to allow careful curation of the data. We used the following criteria to assess induction of phosphosite by TTBK2.
In vitro dataset, 1: the intensity of at least one phosphorylated peptide of a given phosphosite must have been equal to or higher than 2 × 106 threshold. 2: The sum of intensities of all phosphopeptides detected for a given phosphosite in an individual experiment must have been at least twofold of total intensity of the corresponding phosphosite in the control sample (substrate incubated with TTBK2 KD). If the exact position of the phosphorylated residue could not be resolved, each of the S/T residues of a given cluster was considered as phosphorylated. If the phosphopeptide containing such a cluster met criteria 1 and 2, the cluster was considered induced (in total, six peptides with 15 individual phosphorylations).
In the case of TTBK2 autophosphorylation analysis, two reference samples (controls) were used: 1, TTBK2 WT + λ-phosphatase; and 4, TTBK2 KD + λ-phosphatase + kinase assay) as shown in Supplemental Figure S1B. Thus, the individual phosphorylation must have fulfilled the above described criteria for both controls to be considered TTBK2 induced.
In vivo dataset, 1: the intensity of at least one phosphorylated peptide must have been equal to or higher than 2 × 106 threshold if identified repeatedly, or equal to or higher than 2 × 107 threshold if detected in a single experiment. 2: The sum of intensities of all phosphopeptides detected for a given phosphosite in an individual experiment must have been at least twofold of intensity of the corresponding phosphosite in the control sample (Mock). If the exact position of the phosphorylated residue could not be resolved, each of the S/T residues of a given cluster was considered as phosphorylated. If a phosphopeptide containing such a cluster met criteria 1 and 2, the cluster was considered induced (in total, two peptides with four individual phosphorylations).
CEP164 endogenous phosphorylations: CEP164 was immunoprecipitated from HEK293T wt or TTBK2 KO. Listed are phosphorylations with intensity of at least one phosphorylated peptide equal to or higher than 1 × 106.
Phosphorylation of CEP164 N-term in HEK293T wt or TTBK2 KO. 1: The intensity of at least one phosphorylated peptide must have been equal to or higher than 2 × 106. 2: The sum of intensities of phosphopeptides detected for given phosphosite must have been at least twofold of intensity of the corresponding phosphosite in the control sample.
The graphs in Supplemental Figures S2, A and B, S3, and S6, F and G show comparison of the average intensity for individual S/T phosphosites. Cases where the exact position could not be assigned are indicated as (/) for two possible positions or (-) for clusters. In Supplemental Figure S2A, every S/T phosphosite average intensity detected in samples with Mock (black bar), TTBK2 WT induced (red bar), Mock + λ phosphatase (gray bar), and TTBK2 WT + λ phosphatase (blue bar) is indicated on the y-axis. To allow better visualization of intensity of individual phosphosites in one graph, note that the scale of the y-axis is in log10. In Supplemental Figure 2B, every S/T phosphosite average intensity detected in samples with 1, TTBK2 WT (black bar); 2, CTRL#1: TTBK2 WT + λ phosphatase (gray bar); 3, TTBK2 WT + λ phosphatase + kinase assay (red bar), and 4, CTRL#2: TTBK2 KD + λ phosphatase + kinase assay (Brown bar) is indicated on the y-axis, log10 scale. In Supplemental Figure 3, every S/T phosphosite average intensity detected in samples with Mock (black bar) and TTBK2 WT induced (red bar) is indicated on the y-axis, log10 scale. In Supplemental Figure 6F, every S/T phosphosite of endogenous CEP164 average intensity detected in HEK293T WT (red bar) or HEK293T TTBK2 KO (black bar) is indicated on the y-axis, log10 scale. In Supplemental Figure 6E, every S/T phosphosite of FLAG-CEP164 N-term average intensity detected in HEK293T WT (red bar) or HEK293T TTBK2 KO (black bar) is indicated on the y-axis, log10 scale.
Kinase motif determination
Kinase motifs were delineated using a dataset of 13 AA long peptides with central S/T, which we identified as TTBK2-induced in in vitro and in vivo datasets. For the purpose of this analysis, the in vitro and in vivo datasets were cross-correlated, and phosphorylations identified as TTBK2 induced in one dataset and not induced in the other were excluded from the kinase motif determination peptide dataset. Peptide sequences were loaded to MeMe suite phosphorylation motif finder MoMo and analyzed using simple Motif-x and MODL algorithms (Bailey et al., 2009; Chou and Schwartz, 2011). The minimal number of occurrences for kinase motif to be considered was set to 10 for all algorithms. Specific setting for Motif-x algorithms was p < 0.001; motifs with central S/T were considered separately. Default MODL settings were used (maximum number of iterations = 50, maximum number of iterations with no decrease in MDL = 10, with central S/T combined onto one motif). Peptides used and those identified by individual algorithms are listed in Supplemental Table S2.
In silico analyses
Depiction of domains or motifs in Figures 2, A–C and 3, A and B and Supplemental Figures S4 and S5 was based on uniprot.org. Canonical protein sequences from uniprot.org were used for disorder and phosphorylation prediction (Figure 3, A and B; Supplemental Figures S4 and S5), protein identifiers used are as follows: CEP164 (Q9UPV0), CEP83 (Q9Y592), CEP89 (Q96ST8), CCDC92 (Q53HC0), Rabin8 (Q96QF0), DVL3 (Q92997), TTBK2 (Q6IQ55), MPP9 (Q99550), KIF2a (O00139), Tau (P10636), and CEP97 (Q8IW35). Prediction of protein disorder was performed by PONDR (http://www.pondr.com/) algorithms VSL-2 (Peng and Zhang, 2006) and VL3-BA (Peng et al., 2005). A score above 0.5 for disorder prediction suggests an unstable or none secondary structure; a score below 0.5 suggests this protein part has defined a secondary structure. Prediction of protein phosphorylation was performed by the DEPP (Iakoucheva et al., 2004) (http://www.pondr.com/cgi-bin/depp.cgi). Only phosphorylations with a DEPP score above 0.5 were considered as possibly phosphorylated, with the highest scoring phosphorylations being the most probable.
Supplementary Material
Acknowledgments
We thank Erich Nigg, Gislene Pereira, Vitezslav Bryja, Peter Jackson, Robert Lefkowitz, Stephane Angers, Joon Kim, Andrew K. Shiau, and Lumir Krejci for sharing reagents or instruments and Tomas Loja for help with cell sorting. The work was supported by grants from the Czech Science Foundation (16-03269Y, 19-05244S) and the Swiss National Science Foundation (IZ11Z0_166533) to L.C. O.B. was supported by funds from the Faculty of Medicine, Masaryk University, to junior researcher Ondrej Bernatik. K.H. and Z.Z. were supported by project CEITEC 2020 (LQ1601) funded by the Ministry of Education, Youth and Sports of the Czech Republic (MEYS). We acknowledge the core facility CELLIM of CEITEC, supported by the MEYS CR (LM2018129 Czech-Bio-Imaging). CIISB research infrastructure project LM2018127 funded by MEYS CR is gratefully acknowledged for the financial support of the LC-MS/MS measurements at the Proteomics Core Facility.
Abbreviations used:
- AA
amino acid
- CEP164 N-term
N-terminal part of CEP164
- CK1
casein kinase 1
- DA
distal appendage
- DEPP
Disorder Enhanced Phosphorylation Predictor
- FA
formic acid
- FBS
fetal bovine serum
- FL
full length
- IDR
intrinsically disordered region
- KD
kinase dead
- L
leucine
- MC
mother centriole
- PBS
phosphate-buffered saline
- PC
primary cilia
- PEG
polyethylene glycol
- PEI
Polyethyleneimine
- PONDR
Predictor of Natural Disordered Region
- SCA11
spinocerebellar ataxia type 11
- SDA
subdistal appendage
- TBS
Tris-buffered saline
- TTBK2
Tau tubulin kinase 2
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E19-06-0334) on March 4, 2020.
REFERENCES
- Bah A, Forman-Kay JD. (2016). Modulation of intrinsically disordered protein function by post-translational modifications. J Biol Chem , 6696–6705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bah A, Vernon RM, Siddiqui Z, Krzeminski M, Muhandiram R, Zhao C, Sonenberg N, Kay LE, Forman-Kay JD. (2015). Folding of an intrinsically disordered protein by phosphorylation as a regulatory switch. Nature , 106–109. [DOI] [PubMed] [Google Scholar]
- Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, Ren J, Li WW, Noble WS. (2009). MEME Suite: Tools for motif discovery and searching. Nucleic Acids Res , 10.1093/nar/gkp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer P, Stevanin G, Beetz C, Synofzik M, Schmitz-Hübsch T, Wüllner U, Berthier E, Ollagnon-Roman E, Riess O, Forlani S, et al (2010). Spinocerebellar ataxia type 11 (SCA11) is an uncommon cause of dominant ataxia among French and German kindreds. J Neurol Neurosurg Psychiatry , 1229–1232. [DOI] [PubMed] [Google Scholar]
- Bernatík O, Šedová K, Schille C, Ganji RS, Cˇervenka I, Trantírek L, Schambony A, Zdráhal Z, Bryja V. (2014). Functional analysis of dishevelled-3 phosphorylation identifies distinct mechanisms driven by casein kinase 1ε and Frizzled5. J Biol Chem , 23520–23533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borgal L, Rinschen MM, Dafinger C, Hoff S, Reinert MJ, Lamkemeyer T, Lienkamp SS, Benzing T, Schermer B. (2014). Casein kinase 1 α phosphorylates the Wnt regulator Jade-1 and modulates its activity. J Biol Chem , 26344–26356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouskila M, Esoof N, Gay L, Fang EH, Deak M, Begley MJ, Cantley LC, Prescott A, Storey KG, Alessi DR. (2011). TTBK2 kinase substrate specificity and the impact of spinocerebellar- ataxia-causing mutations on expression, activity, localization and development. Biochem J , 157–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowie E, Norris R, Anderson KV, Goetz SC. (2018). Spinocerebellar ataxia type 11-associated alleles of Ttbk2 dominantly interfere with ciliogenesis and cilium stability. PLoS Genet , 10.1371/journal.pgen.1007844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowler M, Kong D, Sun S, Nanjundappa R, Evans L, Farmer V, Holland A, Mahjoub MR, Sui H, Loncarek J. (2019). High-resolution characterization of centriole distal appendage morphology and dynamics by correlative STORM and electron microscopy. Nat Commun , 993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryja V, Cˇervenka I, Cˇajánek L. (2017). The connections of Wnt pathway components with cell cycle and centrosome: side effects or a hidden logic? Crit Rev Biochem Mol Biol , 614–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cˇajánek L, Nigg EA. (2014). Cep164 triggers ciliogenesis by recruiting Tau tubulin kinase 2 to the mother centriole. Proc Natl Acad Sci USA , E2841–E2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cegielska A, Gietzen KF, Rivers A, Virshup DM. (1998). Autoinhibition of casein kinase I ε (CKIε) is relieved by protein phosphatases and limited proteolysis. J Biol Chem , 1357–1364. [DOI] [PubMed] [Google Scholar]
- Cervenka I, Valnohova J, Bernatik O, Harnos J, Radsetoulal M, Sedova K, Hanakova K, Potesil D, Sedlackova M, Salasova A, et al (2016). Dishevelled is a NEK2 kinase substrate controlling dynamics of centrosomal linker proteins. Proc Natl Acad Sci USA , 9304–9309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaki M, Airik R, Ghosh AK, Giles RH, Chen R, Slaats GG, Wang H, Hurd TW, Zhou W, Cluckey A, et al (2012). Exome capture reveals ZNF423 and CEP164 mutations, linking renal ciliopathies to DNA damage response signaling. Cell , 533–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang J, Seo SG, Lee KH, Nagashima K, Bang JK, Kim BY, Erikson RL, Lee KW, Lee HJ, Park JE, et al (2013). Essential role of Cenexin1, but not Odf2, in ciliogenesis. Cell Cycle , 655–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng A, Grant C, Bailey TL, Noble W. (2017). MoMo: Discovery of post-translational modification motifs. BioRxiv . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheong JK, Virshup DM. (2011). Casein kinase 1: Complexity in the family. Int J Biochem Cell Biol , 465–469. [DOI] [PubMed] [Google Scholar]
- Chou MF, Schwartz D. (2011). Biological sequence motif discovery sing motif-x. Curr Protoc Bioinforma, 10.1002/0471250953.bi1315s35. [DOI] [PubMed] [Google Scholar]
- Cohen P. (2000). The regulation of protein function by multisite phosphorylation - A 25 year update. Trends Biochem Sci , 596–601. [DOI] [PubMed] [Google Scholar]
- Dyson HJ, Wright PE. (2005). Intrinsically unstructured proteins and their functions. Nat Rev Mol Cell Biol , 197–208. [DOI] [PubMed] [Google Scholar]
- Ferrarese A, Marin O, Bustos VH, Venerando A, Antonelli M, Allende JE, Pinna LA. (2007). Chemical dissection of the APC repeat 3 multistep phosphorylation by the concerted action of protein kinases CK1 and GSK3. Biochemistry , 11902–11910. [DOI] [PubMed] [Google Scholar]
- Flotow H, Graves PR, Wang A, Fiol CJ, Roeske RW, Roach PJ. (1990). Phosphate groups as substrate determinants for casein kinase I action. J Biol Chem , 14264–14269. [PubMed] [Google Scholar]
- Flotow H, Roach PJ. (1991). Role of acidic residues as substrate determinants for casein kinase I. J Biol Chem , 3724–3727. [PubMed] [Google Scholar]
- Goetz SC, Liem KF, Anderson KV. (2012). The spinocerebellar ataxia-associated gene tau tubulin kinase 2 controls the initiation of ciliogenesis. Cell , 847–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graser S, Stierhof YD, Lavoie SB, Gassner OS, Lamla S, Le Clech M, Nigg EA. (2007). Cep164, a novel centriole appendage protein required for primary cilium formation. J Cell Biol , 321–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanáková K, Bernatík O, Ovesná P, Kravec M, Micka M, Rádsetoulal M, Poteˇšil D, Cˇajánek L, Zdráhal Z, Bryja V. (2019). Comparative phosphorylation map of Dishevelled3 (DVL3). BioRxiv . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegyi H, Schad E, Tompa P. (2007). Structural disorder promotes assembly of protein complexes. BMC Struct Biol , 10.1186/1472-6807-7-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hehnly H, Chen CT, Powers CM, Liu HL, Doxsey S. (2012). The centrosome regulates the Rab11- dependent recycling endosome pathway at appendages of the mother centriole. Curr Biol , 1944–1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houlden H, Johnson J, Gardner-Thorpe C, Lashley T, Hernandez D, Wort P, Singleton AB, Hilton DA, Holton J, Revesz T, et al (2007). Mutations in TTBK2, encoding a kinase implicated in tau phosphorylation, segregate with spinocerebellar ataxia type 11. Nat Genet , 1434–1436. [DOI] [PubMed] [Google Scholar]
- Huang N, Zhang D, Li F, Chai P, Wang S, Teng J, Chen J. (2018). M-Phase Phosphoprotein 9 regulates ciliogenesis by modulating CP110-CEP97 complex localization at the mother centriole. Nat Commun , 4511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter T. (1995). Protein kinases and phosphatases: The Yin and Yang of protein phosphorylation and signaling. Cell , 225–236. [DOI] [PubMed] [Google Scholar]
- Iakoucheva LM, Radivojac P, Brown CJ, O’Connor TR, Sikes JG, Obradovic Z, Dunker AK. (2004). The importance of intrinsic disorder for protein phosphorylation. Nucleic Acids Res , 1037–1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikezu S, Ikezu T. (2014). Tau-tubulin kinase. Front Mol Neurosci , 10.3389/fnmol.2014.00033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa H, Marshall WF. (2011). Ciliogenesis: Building the cell’s antenna. Nat Rev Mol Cell Biol , 222–234. [DOI] [PubMed] [Google Scholar]
- Jay J, Hammer A, Nestor-Kalinoski A, Diakonova M. (2015). JAK2 tyrosine kinase phosphorylates and is negatively regulated by centrosomal protein ninein. Mol Cell Biol , 111–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenks AD, Vyse S, Wong JP, Kostaras E, Keller D, Burgoyne T, Shoemark A, Tsalikis A, de la Roche M, Michaelis M, et al (2018). Primary cilia mediate diverse kinase inhibitor resistance mechanisms in cancer. Cell Rep , 3042–3055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joo K, Kim CG, Lee MS, Moon HY, Lee SH, Kim MJ, Kweon HS, Park WY, Kim CH, Gleeson JG, et al (2013). CCDC41 is required for ciliary vesicle docking to the mother centriole. Proc Natl Acad Sci USA , 5987–5992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Lee JE, Heynen-Genel S, Suyama E, Ono K, Lee K, Ideker T, Aza-Blanc P, Gleeson JG. (2010). Functional genomic screen for modulators of ciliogenesis and cilium length. Nature , 1048–1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitano-Takahashi M, Morita H, Kondo S, Tomizawa K, Kato R, Tanio M, Shirota Y, Takahashi H, Sugio S, Kohno T. (2007). Expression, purification and crystallization of a human tau-tubulin kinase 2 that phosphorylates tau protein. Acta Crystallogr Sect F Struct Biol Cryst Commun , 602–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knippschild U, Gocht A, Wolff S, Huber N, Löhler J, Stöter M. (2005). The casein kinase 1 family: Participation in multiple cellular processes in eukaryotes. Cell Signal , 675–689. [DOI] [PubMed] [Google Scholar]
- Knödler A, Feng S, Zhang J, Zhang X, Das A, Peränen J, Guo W. (2010). Coordination of Rab8 and Rab11 in primary ciliogenesis. Proc Natl Acad Sci USA , 6346–6351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhns S, Schmidt KN, Reymann J, Gilbert DF, Neuner A, Hub B, Carvalho R, Wiedemann P, Zentgraf H, Erfle H, et al (2013). The microtubule affinity regulating kinase MARK4 promotes axoneme extension during early ciliogenesis. J Cell Biol , 505–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurtulmus B, Yuan C, Schuy J, Neuner A, Hata S, Kalamakis G, Martin-Villalba A, Pereira G. (2018). LRRC45 contributes to early steps of axoneme extension. J Cell Sci , 10.1242/jcs.223594. [DOI] [PubMed] [Google Scholar]
- Liao JC, Yang TT, Weng RR, Kuo CTe, Chang CW. (2015). TTBK2: A tau protein kinase beyond tau phosphorylation. Biomed Res Int , 10.1155/2015/575170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindquist SG, Møller LB, Dali CI, Marner L, Kamsteeg EJ, Nielsen JE, Hjermind LE. (2017). A novel TTBK2 de novo mutation in a danish family with early-onset spinocerebellar ataxia. Cerebellum , 268–271. [DOI] [PubMed] [Google Scholar]
- Lo CH, Lin IH, Yang TT, Huang YC, Tanos BE, Chou PC, Chang CW, Tsay YG, Liao JC, Wang WJ. (2019). Phosphorylation of CEP83 by TTBK2 is necessary for cilia initiation. J Cell Biol , 3489–3505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Q, Insinna C, Ott C, Stauffer J, Pintado PA, Rahajeng J, Baxa U, Walia V, Cuenca A, Hwang YS, et al (2015). Early steps in primary cilium assembly require EHD1/EHD3-dependent ciliary vesicle formation. Nat Cell Biol , 228–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marin O, Bustos VH, Cesaro L, Meggio F, Pagano MA, Antonelli M, Allende CC, Pinna LA, Allende JE. (2003). A noncanonical sequence phosphorylated by casein kinase 1 in β-catenin may play a role in casein kinase 1 targeting of important signaling proteins. Proc Natl Acad Sci USA , 10193–10200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchison HM, Valente EM. (2017). Motile and non-motile cilia in human pathology: from function to phenotypes. J Pathol , 294–309. [DOI] [PubMed] [Google Scholar]
- Mlodzik M. (2016). The dishevelled protein family: still rather a mystery after over 20 years of molecular studies. Curr Top Dev Biol, 75–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nigg EA, Stearns T. (2011). The centrosome cycle: Centriole biogenesis, duplication and inherent asymmetries. Nat Cell Biol , 1154–1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oda T, Chiba S, Nagai T, Mizuno K. (2014). Binding to Cep164, but not EB1, is essential for centriolar localization of TTBK2 and its function in ciliogenesis. Genes to Cells , 927–940. [DOI] [PubMed] [Google Scholar]
- Peng K, Vucetic S, Radivojac P, Brown CJ, Dunker AK, Obradovic Z. (2005). Optimizing long intrinsic disorder predictors with protein evolutionary information. J Bioinform Comput Biol , 35–60. [DOI] [PubMed] [Google Scholar]
- Peng QY, Zhang QF. (2006). Precise positions of Phoebe determined with CCD image-overlapping calibration. Mon Not R Astron Soc , 208–212. [Google Scholar]
- Perez-Riverol Y, et al (2019). The PRIDE database and related tools and resources in 2019: Improving support for quantification data. Nucleic Acids Res , D442–D450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F. (2013). Genome engineering using the CRISPR-Cas9 system. Nat Protoc , 2281–2308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiter JF, Leroux MR. (2017). Genes and molecular pathways underpinning ciliopathies. Nat Rev Mol Cell Biol , 533–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J, et al (2012). Fiji: An open-source platform for biological-image analysis. Nat Methods , 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt KN, Kuhns S, Neuner A, Hub B, Zentgraf H, Pereira G. (2012). Cep164 mediates vesicular docking to the mother centriole during early steps of ciliogenesis. J Cell Biol , 1083–1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz-Romond T, Fiedler M, Shibata N, Butler PJG, Kikuchi A, Higuchi Y, Bienz M. (2007). The DIX domain of Dishevelled confers Wnt signaling by dynamic polymerization. Nat Struct Mol Biol , 484–492. [DOI] [PubMed] [Google Scholar]
- Seeley ES, Nachury MV. (2010). The perennial organelle: Assembly and disassembly of the primary cilium. J Cell Sci , 511–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sieracki NA, Komarova YA. (2013). Studying cell signal transduction with biomimetic point mutations. InTech, 10.5772/35029. [DOI]
- Sillibourne JE, Hurbain I, Grand-Perret T, Goud B, Tran P, Bornens M. (2013). Primary ciliogenesis requires the distal appendage component Cep123. Biol Open , 535–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorokin S. (1962). Centrioles and the formation of rudimentary cilia by fibroblasts and smooth muscle cells. J Cell Biol , 363–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spektor A, Tsang WY, Khoo D, Dynlacht BD. (2007). Cep97 and CP110 suppress a cilia assembly program. Cell , 678–690. [DOI] [PubMed] [Google Scholar]
- Takahashi M, Tomizawa K, Sato K, Ohtake A, Omori A. (1995). A novel tau-tubulin kinase from bovine brain. FEBS Lett , 59–64. [DOI] [PubMed] [Google Scholar]
- Tanos BE, Yang HJ, Soni R, Wang WJ, Macaluso FP, Asara JM, Tsou MFB. (2013). Centriole distal appendages promote membrane docking, leading to cilia initiation. Genes Dev , 163–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomizawa K, Omori A, Ohtake A, Sato K, Takahashi M. (2001). Tau-tubulin kinase phosphorylates tau at Ser-208 and Ser-210, sites found in paired helical filament-tau. FEBS Lett , 221–227. [DOI] [PubMed] [Google Scholar]
- Tsun A, Qureshi I, Stinchcombe JC, Jenkins MR, De La Roche M, Kleczkowska J, Zamoyska R, Griffiths GM. (2011). Centrosome docking at the immunological synapse is controlled by Lck signaling. J Cell Biol , 663–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Dynlacht BD. (2018). The regulation of cilium assembly and disassembly in development and disease. Dev . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe T, Kakeno M, Matsui T, Sugiyama I, Arimura N, Matsuzawa K, Shirahige A, Ishidate F, Nishioka T, Taya S, et al (2015). TTBK2 with EB1/3 regulates microtubule dynamics in migrating cells through KIF2A phosphorylation. J Cell Biol , 737–751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Q, Xu Q, Zhang Y, Li Y, Zhang Q, Hu Z, Harris PC, Torres VE, Ling K, Hu J. (2013). Transition fibre protein FBF1 is required for the ciliary entry of assembled intraflagellar transport complexes. Nat Commun , 10.1038/ncomms3750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westlake CJ, Baye LM, Nachury MV, Wright KJ, Ervin KE, Phu L, Chalouni C, Beck JS, Kirkpatrick DS, Slusarski DC, et al (2011). Primary cilia membrane assembly is initiated by Rab11 and transport protein particle II (TRAPPII) complex-dependent trafficking of Rabin8 to the centrosome. Proc Natl Acad Sci USA , 2759–2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong SY, Seol AD, So PL, Ermilov AN, Bichakjian CK, Epstein EH, Dlugosz AA, Reiter JF. (2009). Primary cilia can both mediate and suppress Hedgehog pathway-dependent tumorigenesis. Nat Med , 1055–1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong YL, Anzola JV, Davis RL, Yoon M, Motamedi A, Kroll A, Seo CP, Hsia JE, Kim SK, Mitchell JW, et al (2015). Reversible centriole depletion with an inhibitor of Polo-like kinase 4. Science (80-) , 1155–1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu CT, Chen HY, Tang TK. (2018). Myosin-Va is required for preciliary vesicle transportation to the mother centriole during ciliogenesis. Nat Cell Biol , 175–185. [DOI] [PubMed] [Google Scholar]
- Xie H, Vucetic S, Iakoucheva LM, Oldfield CJ, Dunker AK, Obradovic Z, Uversky VN. (2007). Functional anthology of intrinsic disorder. 3. Ligands, post-translational modifications, and diseases associated with intrinsically disordered proteins. J Proteome Res , 1917–1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Q, Zhang Y, Wei Q, Huang Y, Hu J, Ling K. (2016). Phosphatidylinositol phosphate kinase PIPKIγ and phosphatase INPP5E coordinate initiation of ciliogenesis. Nat Commun , 10777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang TT, et al (2018). Super-resolution architecture of mammalian centriole distal appendages reveals distinct blade and matrix functional components. Nat Commun , 10.1038/s41467-018-04469-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.