Abstract
Viral genomes encode transcriptional regulators that alter the expression of viral and host genes. Despite an emerging role in human diseases, a thorough annotation of human viral transcriptional regulators (vTRs) is currently lacking, limiting our understanding of their molecular features and functions. Here, we provide a comprehensive catalog of 419 vTRs belonging to 20 different virus families. Using this catalog, we characterize shared and unique cellular genes, proteins, and pathways targeted by particular vTRs and discuss the role of vTRs in human disease pathogenesis. Our study provides a unique and valuable resource for the fields of virology, genomics, and human disease genetics.
Liu et al. provide a comprehensive survey of proteins from DNA and RNA viruses that regulate transcription in human cells, identifying themes in mechanism, interactions partners, and roles in disease.
Main Text
Over 200 viral species infect humans (Knipe and Howley, 2013). During the course of an infection, the human body can host up to hundreds of trillions of viruses. When viruses infect human cells, a wide variety of cytopathic effects are induced and counteracted by the host innate and adaptive defense systems, often resulting in illness. Viruses can also affect human health through changes to cellular processes that contribute to non-infectious human diseases. Indeed, the role of viruses in promoting cancer has been well-studied, and viruses also have been implicated in dozens of other chronic human diseases. New viral pathogenic threats continue to emerge, such as SARS-CoV-2. Understanding the functional roles played by viral proteins is therefore of critical importance for combatting these ongoing threats to human health.
The genomes of DNA and RNA viruses encode multiple proteins required to control gene expression, genome replication, and transmission to other host cells. Among these proteins, viral transcription factors (TFs), cofactors, and other regulators of gene expression are central to human disease pathogenesis due to their ability to control the expression of both viral and host genes. Herein, we use the term “vTR” to refer to any virus-encoded protein capable of modulating gene transcription through direct or indirect interactions with nucleic acids. vTRs can be broadly split into two basic categories—“primary” vTRs are proteins whose primary function is the regulation of specific gene targets. “Secondary” vTRs are proteins that have other functions, such as DNA replication or nucleic acid transport that can also “moonlight” as transcriptional regulators.
vTRs have been identified in multiple virus families, in both DNA and RNA viruses. These proteins, together with host transcriptional regulators, coordinate viral and human gene expression at multiple levels, including chromatin organization, RNA polymerase II (RNA Pol II) recruitment, transcription initiation, and transcription elongation (Figure 1 ).
Figure 1.
Roles for vTRs in the Modulation of Gene Expression
(A and B) vTRs can bind to nucleic acids either directly (A) or indirectly (B) to modulate target gene expression.
(C and D) vTRs can also modulate gene expression by targeting the transcriptional machinery (C) or by altering chromatin states (D).
Although multiple vTRs have been shown to play central roles in human biological and disease processes, there is no single resource providing a comprehensive review of vTRs, limiting our understanding of their shared and unique molecular features, functional roles, evolutionary conservation, and roles in human diseases. This lack of an in-depth census likely stems from challenges in identifying and characterizing these proteins. In contrast to eukaryotic TFs, vTRs can rarely be classified into families based on conserved DNA-binding domains (DBDs) (e.g., homeodomains, nuclear hormone receptors, etc.). In addition, given that most vTRs did not arise from duplication events, sequence homology can usually only identify orthologous vTRs from related viruses, and only rarely reveals large classes of structurally related vTRs. Finally, many viral genomes evolve at high rates, rendering sequence-based homology searches largely ineffectual. Thus, vTR identification to date has usually relied on individual studies characterizing the function of a single vTR using experimental methods such as DNA-binding assays, chromatin immunoprecipitation, and perturbation studies followed by measurement of target gene expression. Further, these studies have been performed for only a subset of vTRs using different experimental approaches and analyses criteria, making it challenging to perform integrative data analyses.
The field of virology has contributed substantially to our understanding of fundamental biological processes, including reverse transcription, the structure and function of gene promoters, RNA splicing, polyadenylation, and the domain-like nature of proteins (Enquist, 2009). Many researchers have dedicated their lives to understanding the molecular features, evolutionary conservation, and functions of vTRs. The purpose of this review is not to offer a historical perspective on these important contributions. Instead, our goal is to build upon this body of work by synthesizing the currently available information, and use the resulting new resource to obtain a “30,000 foot” perspective. We are also optimistic that the availability of this resource will offer new opportunities as the relatively nascent field of viral functional genomics continues to move forward.
Here, we describe an extensive and systematic census of the vTRs encoded by human DNA and RNA viruses. Our approach combines thorough literature searches with functional classifications and systematic homology analyses to create the first compendium of human vTRs. In total, we identified 419 vTRs across 20 virus families. Using this resource and available datasets, we address several outstanding questions: What is the distribution of vTRs across virus families? What cellular pathways do vTRs affect? How conserved are vTRs at the protein level? What are the roles of particular vTRs in disease? Collectively, our compendium and analyses provide a solid foundation for future vTR studies.
How Viral Transcriptional Regulators Are Identified and Characterized
vTRs are classically identified through experiments demonstrating their impact on gene expression and in vitro and in vivo assays that establish nucleic acid binding. For example, vTRs that directly interact with DNA have been identified through in vitro binding experiments such as electrophoretic mobility shift assays (EMSAs), protein pull-downs, and DNase footprinting assays. Such assays can both demonstrate the ability of a vTR to bind to DNA and determine a subset of the recognized DNA sequences. To date, exhaustive examination of in vitro vTR DNA-binding specificities to establish DNA-binding motifs has only been performed for a single vTR (of the 171 DNA-binding vTRs in our catalog)—the Epstein-Barr virus (EBV)-encoded Zta protein (Tillo et al., 2017). In contrast, roughly 75% of the ~1,600 human TFs currently have established DNA-binding motifs (Lambert et al., 2019). There are likely several reasons for the limited number of available vTR motifs. First, given that vTRs have been traditionally studied in the context of regulation of viral genes, most studies have focused on the few regions of the viral genomes bound by the vTRs rather than fully characterizing their DNA-binding specificities. Second, methods that exhaustively determine binding specificities, such as protein binding microarrays, systematic evolution of ligands by exponential enrichment (SELEX), and bacterial one-hybrid assays, generally have a higher success rate when testing DBDs rather than full-length proteins (Berger and Bulyk, 2009; Jolma et al., 2013; Meng et al., 2005). However, DBDs have not been determined for most DNA-binding vTRs. Finally, many vTRs only bind DNA in complex with host TFs. Although binding assays can be performed for heterodimers (Jolma et al., 2015; Siggers et al., 2011), these experiments are challenging to perform. Heterodimers and protein complexes are thus more frequently studied using EMSA or chromatin immunoprecipitation (ChIP); neither assay can fully characterize DNA-binding specificities.
Although in vitro assays can establish the ability of vTRs to bind to specific DNA sequences, they are not always perfectly reflective of in vivo DNA binding specificities. Further, in vitro assays do not reveal the timing or functional consequences of these genomic binding events. Several studies have therefore employed ChIP in virus-infected cells or cells transfected with a particular vTR. ChIP followed by high-throughput sequencing (ChIP-seq) is increasingly used to study vTR binding to the host genome, with 55 datasets currently available for 16 different vTRs (Table S1). In addition, functional studies (e.g., reporter and overexpression assays) have been used to identify host and viral gene targets of vTRs and determine their activation or repression activities.
Many vTRs do not directly bind to nucleic acids, instead exerting their function through interactions with host regulators. These vTRs have been identified and characterized through protein-protein binding assays (e.g., yeast two-hybrid or immunoprecipitation followed by mass spectrometry), based on interactions with host TFs and cofactors (Gordon et al., 2020; Ronco et al., 1998). In addition, non-nucleic acid binding vTRs have been studied using many of the same methods that are used for nucleic acid binding vTRs, such as EMSA, ChIP, and reporter assays, because these methods are also amenable to protein complexes.
A Census of vTRs
Despite a central role for vTRs in viral and human biology, a thorough census of vTRs has not been previously undertaken. We therefore employed a hybrid “curation and computation” approach (Figure S1) to create a comprehensive list of 419 vTRs encoded in the genomes of viruses that infect humans (Table S2). Paralogous vTRs were identified by within-species and cross-species amino acid homology searches using BLASTp and CD-HIT (Fu et al., 2012). Multiple lines of evidence were considered for each candidate vTR, including the data supporting its ability to bind nucleic acids and alter gene expression levels. Data sources, including binding ligands (double-stranded DNA [dsDNA], single-stranded DNA [ssDNA], double-stranded RNA [dsRNA], and/or single-stranded RNA [ssRNA]), biochemical assays (e.g., EMSA, SELEX, reporter assays), crystal structures, and functional annotations are provided on the project website (http://vtr.cchmc.org/vtrsurvey/) and in Table S2.
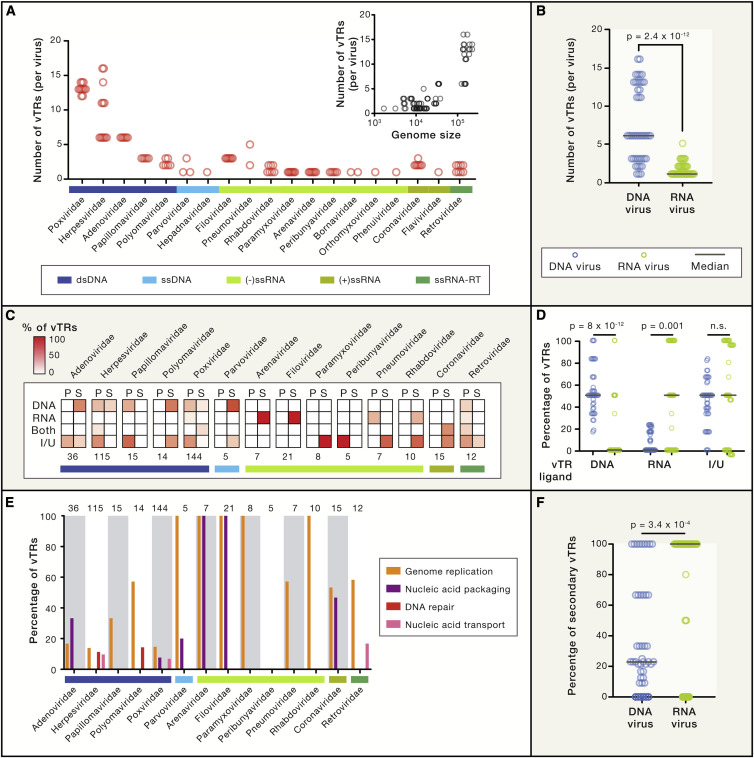
In total, we identified and annotated vTRs from 20 different DNA and RNA virus families. Inspection of the vTR catalog reveals that the genomes of human viruses can encode as many as 16 vTRs (the herpesviruses EBV and Kaposi’s sarcoma-associated virus [KSHV]) (Figure 2 A). The Poxviridae and Herpesviridae families, which have the largest genome sizes, also have the highest number of currently known vTRs per virus, with an average of 13 and 9.6, respectively. Herpesviruses have the widest range of vTRs (6 to 16), while other families encode a more consistent number. This vTR diversity parallels the higher variation in species diversity, genome structure, tropism, and replication cycles observed in herpesviruses. In contrast, the genomes of RNA viruses in our catalog encode an average of only 1.6 vTRs (Figure 2B). This is likely related both to the compact genome of RNA viruses and to differences in replication and gene expression strategies relative to DNA viruses.
Figure 2.
A Census of Human vTRs
(A) Number of vTRs per species for each virus family, classified according to their Baltimore categories. Inset indicates the relationship between the number of vTRs per viral species and viral genome size (in nucleotides).
(B) Comparison between the number of vTRs encoded by DNA and RNA viruses. Statistical significance was determined by a Mann-Whitney U test.
(C) Percentage of vTRs classified according to their ligand (DNA, RNA, both, or indirect/unknown) and primary (P) or secondary (S) role as vTRs. Values under each heatmap indicate the number of vTRs annotated in each virus family. Only virus families with at least 5 vTRs are shown.
(D) Comparison between the percentage of vTRs encoded by DNA and RNA viruses that bind to DNA, bind to RNA or are indirect/unknown binders. Statistical significance was determined by a Mann-Whitney U test.
(E) Percentage of vTRs per family associated with molecular functions other than transcriptional regulation. Numbers at the top indicate the total number of vTRs annotated in each virus family. Only virus families with at least 5 vTRs are shown.
(F) Comparison between the percentages of vTRs with secondary roles in transcriptional regulation encoded by DNA and RNA viruses. Statistical significance was determined by a Mann-Whitney U test.
See also Figures S1 and S2 and Tables S2 and S3.
We next classified vTRs into DNA-binding, RNA-binding, DNA/RNA-binding, or indirect/ unknown binding. Most DNA and RNA virus vTRs either directly bind DNA or RNA, respectively, or are indirect binders that act as cofactors (Figures 2C and 2D). However, while many DNA virus vTRs can directly regulate both viral and host gene expression due to their ability to bind to DNA, RNA virus vTRs usually indirectly regulate the expression of host genes through interactions with host TFs, cofactors, or RNA Pol II, because they largely are incapable of binding DNA (Figures 2C and 2D).
Whereas primary vTRs mainly function as controllers of gene expression, secondary vTRs are also involved in other molecular functions such as nucleic acid replication, transport, packaging, and DNA repair (Figure 2E). For example, most virus families encode proteins that play roles in both transcriptional regulation and viral genome replication, such as E2A (adenovirus), UL29 (Herpes simplex virus 1 [HSV-1]), and BMRF1 (EBV). Similarly, many viruses encode proteins involved in both gene regulation and packaging the viral genome, including the nucleoprotein from arenaviruses, VP30 and VP35 from filoviruses, and Rep52 from parvoviruses. Other functions are more specific to secondary vTRs from certain families, such as the involvement of herpesvirus and polyomavirus vTRs in DNA repair. Although both DNA and RNA viruses encode secondary vTRs, secondary vTRs are significantly more prevalent in RNA viruses (Figure 2F), likely due to the compact genome of RNA viruses, which results in gene products with pleiotropic functions.
Like other types of viral proteins, vTRs can exhibit high genetic diversity across isolates of a given viral species. For example, the E6 and E7 vTRs of type 5 human papillomavirus (HPV) both attain pairwise protein amino acid identity as low as ~30% across isolates (Table S3). Interestingly, this high sequence variation is not simply a result of general viral genome diversification. To illustrate, the EBV EBNA2 and EBNA3A/B/C proteins, which play key roles in viral latency, show high diversity (48%–77% amino acid identity between certain EBV isolates). In contrast, most other EBV vTRs do not display such high variability, including BALF2, BcRF1, and BDLF4 proteins, which have a sequence identity close to 100%. This can be explained by the strong evolutionary constraint of proteins that function in the context of a highly conserved multiprotein complex such as these (Gruffat et al., 2016). Targeting these less diverse vTRs using small-molecule or genome editing strategies might therefore yield more effective therapies.
Protein Structures of vTRs
vTR proteins demonstrate a remarkable degree of structural diversity (Figure S2). To date, less than 12% of vTRs have been characterized at the structural level (Table S2). Many of the characterized vTRs have unique protein structures, with no detectable homology to any other virus or host protein (Yin and Fischer, 2008). In a few cases, similar folds can be observed across viral species, such as the DBDs of E2 (HPV) and EBNA1 (EBV), which consist of two beta-alpha-beta repeats of approximately equal length (Bussiere et al., 1998). Because there is no detectable structural similarity between most vTRs and human proteins, their novel structures may provide an avenue for developing drugs with high selectivity for disrupting their transcriptional activity. For example, an inhibitory peptide against EBNA2 was found to reduce cell growth and viability of EBV-infected cells (Farrell et al., 2004), providing proof-of-concept that vTRs can be suitable drug targets to fight viral infections and their pathogenic effects.
Despite overall low sequence conservation, some DBD structures are shared among a subset of vTRs (Figure S2). One such domain is the basic leucine zipper (bZIP), which is also present in many eukaryotic TFs (Vinson et al., 1989). bZIP-containing vTRs have been identified in a range of human viruses, including K8 (KSHV), Zta (EBV), and HBZ (human T cell leukemia virus 1 [HTLV-1]) and can bind DNA as homodimers or as heterodimers with host bZIP proteins (Basbous et al., 2003; Ellison et al., 2009; Lieberman and Berk, 1990). The interferon regulatory factor (IRF) domain, which is present in host TFs that regulate antiviral responses, is also found in multiple vTRs (Figure S2). For example, the KSHV genome encodes four proteins, vIRF1–vIRF4, that perturb antiviral responses by competing for host IRF DNA binding sites, heterodimerizing with host IRFs, and competing for host coactivators such as CREBBP and EP300 (Offermann, 2007). In summary, vTRs come from a wide range of structural classes, enabling a diverse range of mechanisms used to interact with the host genome and host proteins.
Roles for vTRs in the Control of Viral Gene Expression
The different classes of viruses coordinate genome replication, virion assembly, and entry and exit from latency by precisely regulating the timing of viral gene expression using different types of vTRs. vTRs from DNA viruses directly or indirectly bind to viral gene promoters to regulate multiple stages in the viral replication cycle, including establishment and maintenance of latency by episome maintenance proteins (De Leo et al., 2020). vTRs from retroviruses, such as Tat (HIV), Bel-1 (human spumaretrovirus), and Tax (human T cell leukemia virus [HTLV]), work with host proteins to activate the expression of viral genes and viral RNA replication from the integrated provirus as a mechanism to regulate the exit from latency. Positive and negative strand RNA viruses use vTRs that directly or indirectly bind to RNA, such as N (coronavirus), NP (Ebola virus), and phosphoprotein (measles virus), to regulate the expression of viral genes and the replication of the viral genome.
DNA virus vTRs often control a complex cascade of molecular events in the viral replication cycle. “Early class” genes generally encode proteins involved in DNA replication, nucleotide biosynthesis, regulation of intermediate and late viral genes, and immune evasion. “Intermediate” and “late class” genes encode proteins involved in virion morphogenesis and assembly. In DNA viruses with a compact genome, one or a few vTRs work with host TFs and cofactors to control the expression of all viral genes. For example, polyomavirus SV40 large T-antigen is responsible for the regulation of both early and late gene transcription (Kriegler et al., 1984). Other DNA viruses exhibit a strictly regulated temporal cascade of gene expression involving different vTRs. For example, the UL48 (HSV-1) protein induces the expression of the immediate early vTR RS1, which in turn induces the expression of UL29, an important vTR for maintaining the highly ordered downstream cascade of viral gene expression (Weller and Coen, 2012).
Some vTRs function as a multiprotein complex together with other viral proteins. This is the case for the transcription initiation complex vPIC in EBV, which is comprised of BcRF1, a homolog of archaeal TATA-box-binding protein (TBP) that interacts with TATT sequences in EBV late gene promoters, and BVLF1, BFRF2, BGLF3, BDLF4, and BDLF3.5, which are involved in RNA Pol II recruitment and transcriptional elongation (Gruffat et al., 2016). The vPIC complex has also been identified in other beta and gamma herpesviruses, such as KSHV (Nandakumar and Glaunsinger, 2019), but not in alpha herpesviruses, and constitutes a shared mechanism for regulating the expression of viral late genes. Overall, viruses control the expression of their own genes through mechanisms ranging from simple (e.g., one vTR controlling a few genes) to complex (e.g., tightly controlled transcriptional cascades, vTR-host protein interactions, and multi-protein complexes).
Roles for vTRs in Modulating Host Gene Expression
In addition to regulating viral genes, DNA and RNA virus vTRs can modulate the expression of host genes that play important roles in the immune system, cell cycle, metabolism, and other processes (Figure 3 A). This can be achieved through direct or indirect vTR binding to cellular target gene regulatory regions, the initiation of global changes in chromatin structure and condensation, or by interfering with host TF activities. For example, UL54 (HSV-1) and vIRF1-3 (KSHV) inhibit type I interferon production and/or the activation of interferon effector pathways, which constitute key antiviral mechanisms (Chiang and Liu, 2019). In addition to blocking interferon pathways, a subset of vTRs can induce an immunosuppressive state by modulating pro- or anti-inflammatory genes. For example, Zta (EBV) binds to the host genome, activating the expression of immunosuppressive genes such as TGFBI and TGFB1, and downregulating the expression of genes that mediate immune responses such as TLR9, IFI6, and IL23A (Cayrol and Flemington, 1995; Ramasubramanyan et al., 2015). Other vTRs activate the production of IL10 to mediate immunosuppression. For example, Rta (KSHV) induces IL10 production through interactions with the host TFs SP1 and SP3 (Miyazawa et al., 2018). Other vTRs contribute to viral pathogenesis by increasing the expression of pro-inflammatory mediators. For example, N from SARS-CoV induces lung inflammation by activating the PTGS2 promoter through interactions with nuclear factor κB (NF-κB) and C/EBP binding sites (Yan et al., 2006). By altering the expression levels of host immune genes through a variety of mechanisms, vTRs are key to host evasion mechanisms and pathogenesis.
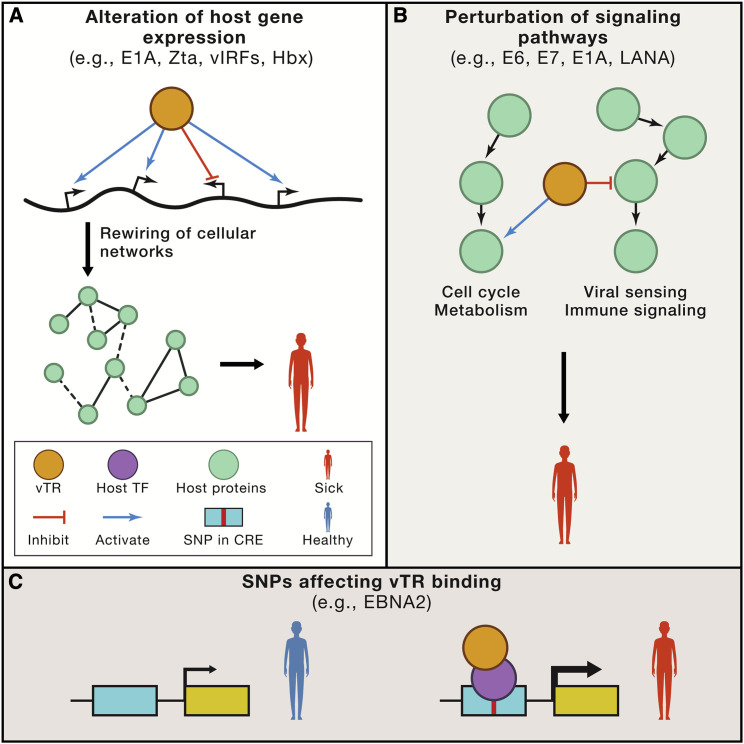
Figure 3.
Roles for vTRs in Human Diseases
(A) vTRs can alter host gene expression, resulting in cellular network rewiring, ultimately leading to disease.
(B) vTRs can perturb signaling pathways through protein-protein interactions or enzymatic activities. This includes the inhibition of pathways involved in viral sensing and immune signaling, and activating cell cycle progression and cell metabolism. Such perturbations can lead to cancer and other diseases.
(C) Single nucleotide polymorphisms (SNPs) and other genetic variants can affect vTR binding, either directly or indirectly through altered host TF binding. This can lead to changes in target gene expression, ultimately leading to disease. CRE, cis regulatory element.
Viruses can also modulate the cell cycle and induce cellular reprogramming to initiate cellular conditions that are beneficial for replication. For example, E1A (adenovirus) can epigenetically reprogram quiescent human cells by interacting with retinoblastoma proteins and EP300, activating the transcription of cell cycle and proliferation genes, and repressing the transcription of antiviral genes and genes involved in differentiation and development (Ferrari et al., 2008). vTRs can also promote cellular transformation by regulating the expression of oncogenes or tumor suppressor genes. For example, vTRs encoded by several viruses target TERT (EBV, hepatitis B virus [HBV], hepatitis C virus [HCV], HPV, HTLV-1, and KSHV) and MYC (adenovirus, EBV, HPV, KSHV, and HPV) (Bellon and Nicot, 2008; Mesri et al., 2014). vTRs such as HBx (HBV) and E1A (adenovirus), can also rewire metabolic pathways to generate cellular states that promote viral replication and/or carcinoma survival (Guerrieri et al., 2017; Prusinkiewicz and Mymryk, 2019). Altogether, vTRs from both DNA and RNA viruses regulate a plethora of host responses to generate the appropriate cellular environment for viral replication and dispersion to other cells.
To obtain genome-wide insights into vTR-regulated host target genes, we systematically processed and analyzed the 33 vTR ChIP-seq datasets that passed our stringent quality control criteria (Supplemental Materials and Methods). We note that some of the removed datasets might involve vTRs that only interact with the viral genome, and not the host genome, under the conditions assayed. The number of ChIP-seq peaks varies greatly across datasets, ranging from slightly more than 100 for LANA (KSHV) in BCBL-1 cells to almost 50,000 for EBNA-LP (EBV) in IB4 cells, with a median of 675 (Table S1). Although these peak counts likely reflect multiple factors beyond biological aspects (such as read count and data quality), they emphasize the large number of human genomic locations that can be occupied by vTRs—collectively, these ChIP-seq peaks cover 305,562 distinct regions of the human genome, a notable number given the small proportion of vTRs with currently available ChIP-seq data.
For each ChIP-seq dataset, we next determined potential host target genes and analyzed their functional roles (Table S4). As expected, EBNA2/3B/3C targets were enriched for genes associated with B cell activation and transcription, consistent with their well-established roles in the rewiring of B cell function and large-scale modulation of B cell transcriptional programs (Zhao et al., 2011). In addition, these vTRs bind near genes associated with inflammation and apoptosis, and signaling pathways such as the p38 MAPK, p53, Toll-like receptor, and JUN kinase, suggesting that EBV vTRs can initiate transcriptional programs that rewire cellular responses to various stimuli and promote cell survival (Table S4). We also observe strong enrichment for LANA (KSHV), HBZ (HTLV-1), and EBNA1/2/3B/3C (EBV) protein binding events near components of the Wnt signaling pathway, suggesting that both DNA and RNA viruses might target this important developmental pathway. Additional ChIP-seq studies will be needed to elucidate the transcriptional programs of other vTRs and determine their shared and unique features.
Comparison of the genomic regions occupied by vTRs across these datasets reveals strong clustering of functionally related vTRs (Figure 4 A). For example, ChIP-seq peak sets for the EBV EBNA3A/B/C proteins tightly cluster together, as expected. In contrast, EBNA1, which plays different functional roles, occupies separate regions of the genome. Interestingly, EBNA-LP binding events coincide with multiple vTRs from several viral species, suggesting that these proteins have partially overlapping human gene targets. Inspection of the location of vTR genomic binding sites relative to gene transcription start sites (TSSs) reveals two general classes of vTRs – LANA, Tat, and EBNA-LP largely bind to promoter regions, whereas other vTRs (HBZ, Zta, and EBNA1/2/3) largely target distal enhancer regions (Figure 4B). As expected, most vTRs have symmetric binding patterns with respect to the TSS, with the exception of Tat, whose binding peaks skew downstream of the TSS. This is likely due to Tat’s ability to interact with human RNA polymerase through a stem-loop RNA element during transcriptional elongation (Reeder et al., 2015).
Figure 4.
Properties of vTR ChIP-Seq Datasets
(A) Overlap of vTR ChIP-seq peaks. Each entry indicates the overlap (number of intersecting peaks divided by the number of peaks in the smaller peak set) between the given pair of vTR ChIP-seq datasets. Rows and columns were hierarchically clustered using the Unweighted Pair Group Method With Arithmetic Mean (UPGMA) algorithm.
(B) Distribution of vTR ChIP-seq peaks relative to human gene transcription start sites (TSSs). Transcript-related features were obtained from the UCSC “Known Genes” database table (Haeussler et al., 2019). The plot was created using the ChIPseeker package (Yu et al., 2015).
(C) Human TF binding site motifs enriched in vTR ChIP-seq peaks. TF binding site motif enrichment was calculated using HOMER (Heinz et al., 2010). Motifs shown are among the top five motif families (based on HOMER p value) in any individual ChIP-seq dataset. Heatmap color indicates the normalized –log p value. A value of 100 means that the motif has the best –log p value for the given vTR, with 50% indicating half of the best –log p value.
As discussed above, many vTRs (e.g., EBNA2 and EBNA-LP) completely lack DBDs and thus require physical interactions with human TFs to occupy genomic sites. Analysis of vTR ChIP-seq datasets provides an opportunity to systematically identify potential key interacting human TF partners for vTRs. To this end, we performed TF binding site motif enrichment analysis using 7,704 motifs for 1,200 human TFs contained in the Cis-BP database (Lambert et al., 2019) (Table S5). Inspection of the most highly enriched motifs reveals expected results, such as the AP-1 site directly bound by Zta (Lieberman et al., 1990), binding sites for established EBNA2 interaction partners RBPJ (Henkel et al., 1994), EBF1 (Glaser et al., 2017), and Ets-family SPI1/PU.1 (Johannsen et al., 1995), and the EBNA-LP interaction partner CTCF (Portal et al., 2013) (Figure 4C). This analysis also reveals a wide range of human motifs covering virtually every major human TF structural class (Lambert et al., 2018), including C2H2 zinc finger, homeobox, bHLH, and bZIP. Interestingly, motifs of the AP-1, Ets, and RUNX families are enriched for most available vTRs, suggesting that these TF classes are preferentially targeted by vTRs. Collectively, these data illustrate the wide range of human TFs that vTRs can directly or indirectly interact with upon occupying the human genome.
Epigenetic Regulation by vTRs
The regulation of chromatin states during viral infection involves complex host-pathogen interactions, with signals converging on chromatin to activate host defenses that are countered by viral mechanisms that are required for replication, modulation of immune responses, and in some cases, integration into the host genome (Marazzi and Garcia-Sastre, 2015). The best-characterized examples of vTR-host epigenome interactions come from studies of adenoviruses and DNA tumor viruses. For example, E1A (adenovirus) causes global epigenetic reprogramming conducive to viral dissemination (Ferrari et al., 2008). This is accompanied by erasure of histone acetylation marks associated with gene induction and the acquisition of novel chromatin domains associated with selective gene expression (Ferrari et al., 2012). This global epigenetic reprogramming is achieved through the modulation of the acetyltransferase activity of CREBBP and EP300, and the cell-cycle repressor RB1 (Ferrari et al., 2014). Most of these effects likely rely on E1A’s intrinsically disordered status, which allows E1A to bind to multiple cellular partners, resulting in selective regulation of gene expression (Ferreon et al., 2013). Similar mechanisms have been observed for host transcriptional regulators, which use intrinsically disordered domains to regulate complex assembly and phase separation to functionally segregate transcriptional activities into discrete parts of the genome (Shin et al., 2019). Based on the prevalence of disordered domains in viral proteins (Hagai et al., 2014), it is reasonable to speculate that oligomeric assembly and phase separation control vTR activities and their effects on both viral and host genomes.
Some vTRs have evolved an intricate relationship with epigenetic factors to both allow the viral genome to be “chromatinized” and to control its state through the establishment of viral latency. Several viral genomes (e.g., EBV, KSHV, and varicella zoster virus) are maintained in the infected cells as episomal (circular) DNA that are decorated with histones and histone marks and regulated by chromatin factors, similar to the host genome. Thus, some vTRs have evolved to function in a chromatinized context, engaging histone-modifying enzymes to achieve viral transactivation and the tethering of viral episomes to mitotic chromosomes (De Leo et al., 2020). For example, LANA (KSHV), EBNA1 (EBV), and E2 (HPV) anchor viral DNA to the host DNA to ensure viral genome propagation during cell division using different strategies: LANA interacts with the core histones (Barbera et al., 2006), EBNA1 binds the minor groove of the DNA (Sears et al., 2004), and E2 binds BRD4 and cohesin to tether itself to the host genome (Bentley et al., 2018; You et al., 2004). Upon tethering, vTRs work with host protein complexes to establish permissive chromatin domains to activate genes linked to latency and heterochromatin domains to repress lytic gene expression (De Leo et al., 2020). For integrating viruses, such as HIV, the maintenance of a latent viral genome also depends on vTRs, and is counteracted by host heterochromatinization mechanisms (Taura et al., 2019). Thus, vTRs can coordinate epigenetic structural features of both the host and viral genomes to control gene expression, genome maintenance, and propagation.
The effect of DNA virus vTRs on the host epigenome has been an area of active study for decades. However, less is known about RNA virus vTRs in this context for two main reasons. First, most RNA viruses do not integrate into the genome or replicate in the nucleus. Infection is therefore not thought to affect epigenetic memory or genomic imprinting. Second, RNA viruses have fewer recognizable vTRs than DNA viruses (Figure 2B). Nevertheless, the expression of viral proteins during RNA virus infection can have a dramatic effect on the host genome. For example, NS1 (influenza A virus) phase separates and associates with RNA polymerase II (Zhao et al., 2018), leading to polymerase derailment, genome-wide transcriptional read-through, and massive alterations to chromosomal structure (Heinz et al., 2018). This massive reprogramming of the host chromatin state represents a mechanism that is currently thought to be unique to NS1 among known vTRs.
Although it has been established that many vTRs can reprogram the epigenome, the functional consequences of reprogramming are largely unknown. Establishing direct relationships between vTRs and the host genome is a complicated endeavor. For example, studying vTRs in isolation does not always mirror physiological conditions, since vTRs are expressed as a result of an infection, and their expression kinetics cannot always be easily recapitulated. Future studies will likely increasingly employ the ever-growing arsenal of CRISPR-based and protein-targeting degradation techniques. Undoubtedly, new epigenetic-based mechanisms affected by both DNA and RNA virus-encoded vTRs will continue to be discovered.
Cellular Pathways Affected by vTRs through Protein-Protein Interactions
Many vTRs, including both primary and secondary vTRs, perturb host signaling pathways to evade antiviral responses, avoid cell death, and induce cell proliferation (Figure 3B). In this sense, vTRs can be considered master cellular “rewirers,” as they can modulate cellular function through both protein-protein and protein-nucleic acid interactions. In particular, multiple vTRs interact with human proteins involved in cancer-related signaling and apoptosis signaling pathways (Figures 5A and 5B), immune signaling pathways, and regulators of cell growth and proliferation (Calderone et al., 2015; Guirimand et al., 2015).
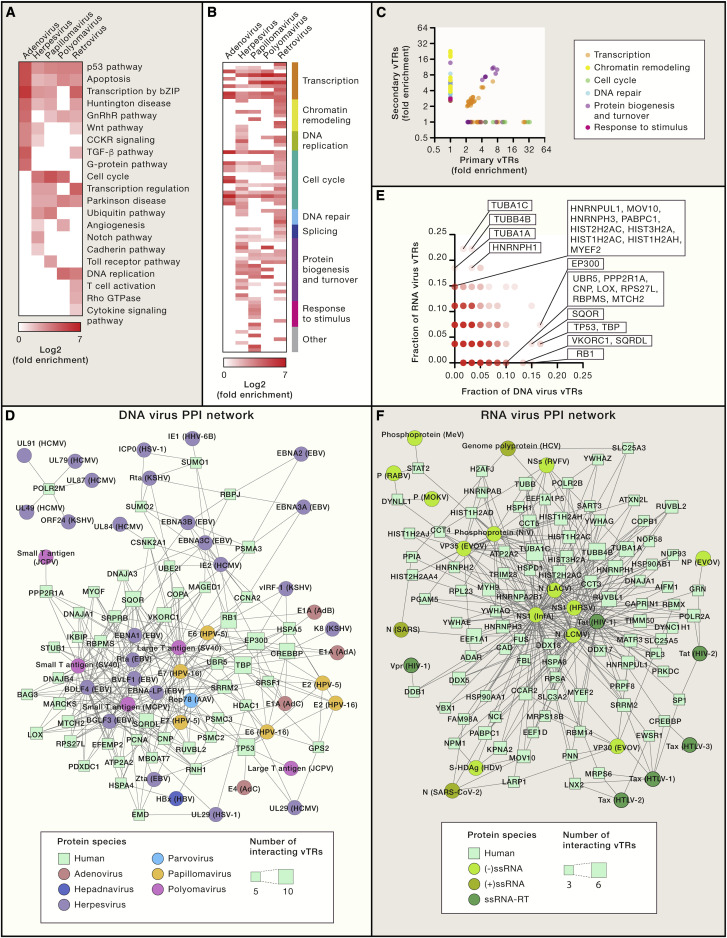
Figure 5.
The vTR-Human Protein-Protein Interaction Network
(A and B) PANTHER pathway (A) and GO-Slim biological process (B) enrichment analyses for human proteins that interact with vTRs, categorized by virus family. Fold enrichment values are indicated for associations with a false discovery rate (FDR) <0.05. GO-Slim terms were manually classified based on their biological function. Only virus families encoding at least five vTRs with reported protein-protein interactions to human proteins are shown. Gene sets were obtained from the PANTHER database (Mi et al., 2013).
(C) GO-Slim biological process fold enrichment of human proteins that interact with primary versus secondary vTRs.
(D and F) vTR-human hub protein-protein interaction network. vTR-human protein-protein interaction data were downloaded from VirHostNet 2.0 (http://virhostnet.prabi.fr/) (Guirimand et al., 2015) and VirusMentha (https://virusmentha.uniroma2.it/) (Calderone et al., 2015). vTR-human protein-protein interactions are shown for DNA virus vTRs (D) and RNA virus vTRs (F). Human proteins with at least five DNA virus vTR interactors or at least 3 RNA virus vTR interactors are included in the networks. Physical interactions are indicated by edges. Circles represent vTRs, and are colored by virus family or class. Squares represent human proteins, with node size representing the number of vTR interactors.
(E) Comparison of the fraction of DNA and RNA virus vTR protein-protein interactors per human protein. Human proteins preferentially interacting with DNA or RNA virus vTRs are indicated. The color gradient indicates the number of human proteins.
Oncogenic vTRs use diverse mechanisms to target the p53 signaling pathway, which plays key roles in cell cycle regulation, genomic stability, programmed cell death, and DNA repair (Aloni-Grinstein et al., 2018; Hafner et al., 2019). Other strategies employed by oncogenic vTRs include perturbing the tumor suppressor retinoblastoma protein RB1 (e.g., large T-antigen from SV40, E1A from adenovirus, and E7 from HPV) (DeCaprio et al., 1988; Patrick et al., 1994; Whyte et al., 1988) and inducing cell dedifferentiation by targeting the Hippo pathway effectors YAP and TAZ (e.g., E1A from adenovirus) (Zemke et al., 2019).
Among immune signaling pathways, many vTRs interact with proteins in the Toll-like receptor, transforming growth factor β (TGF-β), and various cytokine signaling pathways (Figure 5A) (Calderone et al., 2015; Guirimand et al., 2015). Indeed, vTRs frequently target pathogen-associated pattern recognition pathways, in particular those sensing foreign nucleic acids. For example, UL82 (human cytomegalovirus [HCMV]) inhibits STING-mediated signaling by blocking the recruitment of IRF3 and TBK1 to the STING DNA-sensing complex (Fu et al., 2017). Likewise, RNA virus vTRs can block RNA-sensing pathways. This is the case for VP35 (Ebola virus), which also inhibits the activation of IRF3, but instead does so by binding to cytosolic dsRNA and inhibiting RIG-I signaling (Cárdenas et al., 2006).
vTRs can directly control cell growth and proliferation through interactions with human proteins involved in biological processes related to gene expression, cell cycle, splicing, and protein biogenesis and turnover (Figure 5B). vTRs encoded by adenovirus, herpesvirus, papillomavirus, polyomavirus, and retrovirus are enriched for interactions with human proteins involved in transcription, including both general and sequence-specific TFs. In addition, these five virus families are enriched for interactions with cell-cycle proteins (Moore and Chang, 2010). Herpesvirus, polyomavirus, and retrovirus vTRs are also enriched in interactions with DNA repair proteins, consistent with the role of host DNA repair in viral replication and latency (Weitzman and Fradet-Turcotte, 2018). Other biological processes associated with human proteins that interact with vTRs are less characterized. For example, many vTR-interacting human proteins are associated with protein biogenesis and turnover, suggesting that some vTRs may also influence translation (e.g., small T antigen from polyomavirus), and/or interact with proteins that regulate the cellular localization and degradation of the vTR.
Primary and secondary vTRs interact with overlapping, yet distinct sets of human proteins (Figure 5C). Both classes of vTRs interact with proteins involved in transcription, supporting the inclusion of secondary vTRs in our catalog, even if their main annotated function is distinct from transcriptional regulation. Other functional enrichments are more specific to primary or secondary vTRs. Whereas primary vTRs are enriched in interactions with cell cycle proteins, secondary vTRs are enriched for chromatin remodelers, DNA repair proteins, and signaling molecules involved in stress responses. This is consistent with secondary vTRs being involved in multiple cellular processes beyond transcription.
The specificity of viral-human protein-protein interactions varies, because many human proteins interact with multiple vTRs from different families of viruses (Figures 5D and 5F), whereas others preferentially interact with vTRs from specific virus families. For example, POLR2M, RBPJ, SUMO1, and SUMO2 interact preferentially with herpesvirus vTRs (Figure 5D). In particular, RBPJ, a major regulatory factor in B-lymphocytes, specifically interacts with vTRs from gamma herpesvirus, which establish latency in this cell type. By contrast, cofactors such as EP300, CREBBP, TBP, and RB1 interact with multiple vTRs from different families of DNA viruses. RNA virus vTRs often interact with different human proteins than DNA virus vTRs (Figures 5D and 5F). For example, RNA virus vTRs preferentially target histones and cytoskeletal proteins (Figure 5E). It is important to note that these analyses were performed using limited datasets of variable quality obtained from different experimental approaches. Nevertheless, the large number of physical interactions between vTRs and human proteins (over 2,500 human proteins have been found to interact with vTRs) is reflective of the pleiotropic roles played by vTRs to control virus-specific processes across viral stages.
Roles for vTRs in Disease: Beyond Viral Infections and Cancer
In addition to their well-studied roles in acute infection and cancer, viruses have been implicated in dozens of other human diseases. In particular, multiple vTRs have been associated with neurological disorders. For example, IE1 and IE2 (HCMV) play established roles in neurological disabilities in children with congenital HCMV infection. Mechanistically, IE1 and IE2 promote the degradation of HES1, an essential human TF involved in neural progenitor cell fate and fetal brain development, and dysregulate neural stem cell maintenance (Liu et al., 2017) and the polarization of migrating neurons (Han et al., 2017). Tax (HTLV-1) can also cause a debilitating neurological disorder, HAM/TSP (HTLV-1-associated myelopathy/tropical spastic paraparesis), resulting from a strong immune response that includes the expansion of CD8+ cytotoxic T lymphocytes specific for Tax (Elovaara et al., 1993). Similarly, Tat (HIV-1) released by infected macrophages and microglia in the central nervous system can negatively impact the dopaminergic system, which can eventually lead to neuropsychiatric, neurocognitive, and neurological disorders (Gaskill et al., 2017). More recently, HHV-6A infections have been linked to multiple sclerosis (Engdahl et al., 2019). vTRs of HHV-6A have long been known to be capable of transactivating host and viral gene expression (Martin et al., 1991)—whether they contribute directly to neurodegenerative progression remains an outstanding question.
vTRs can also modulate the actions of other viruses in co-infected cells, which can subsequently impact the health of the host. For example, E1A (adenovirus) can affect the gene expression levels of many other viruses, including HCMV, KSHV, and HIV (Chang et al., 2011; Gorman et al., 1989; Nabel et al., 1988). Genes encoded by these viruses in turn can also impact host gene expression. In this way, vTRs can mediate both competition and cooperation for the control of genes encoded by the host and other viruses. Interactions between viruses mediated by vTRs may therefore explain part of the inter-individual differences in infection outcomes, beyond differences in immune system maturation and genetic background.
vTR interactions with the human genome should, in principle, be influenced by the DNA sequence of the host genome, including natural genetic variation (Figure 3C). Considering the prevalence of established virus-disease associations, and the fact that the vast majority of human disease-associated genetic variants are located in non-coding regulatory regions (Maurano et al., 2012), disease risk allele-dependent mechanisms that alter vTR interactions with the human genome could contribute to the etiology of many human diseases. Indeed, we and others recently reported that EBNA2 (EBV) occupies up to half of the genetic risk loci associated with a set of seven autoimmune diseases, including multiple sclerosis, lupus, inflammatory bowel disease, and rheumatoid arthritis, altogether comprising 142 genomic loci (Harley et al., 2018; Ricigliano et al., 2015). Further, over 20 examples have been discovered involving allele-dependent EBNA2 binding to autoimmune risk alleles, based on ChIP-seq read imbalance between the alleles of heterozygous disease-associated variants (Harley et al., 2018). Decades of evidence have linked many of these diseases to EBV (Balandraud and Roudier, 2018; James et al., 1997; Pakpoor et al., 2013) and EBNA2 in particular (Ascherio et al., 2001; Mechelli et al., 2015). Collectively, these results provide compelling genetic-environment autoimmune disease mechanisms driven by EBNA2 and reveal a largely unexplored molecular mechanism—differential binding of vTRs to human disease variants—that might explain many additional virus-disease connections.
Perspective, Summary, and Future Directions
Viruses can infect all forms of life, and myriad viral types are found in virtually every ecosystem on earth. All of us will be infected with multiple viruses during our lifetime. These infections can have diverse and lasting effects that impact gene expression, protein function, cellular function, and ultimately overall health. Indeed, viruses play well-established roles in many human diseases, including HIV in AIDS, Varicella zoster virus in chicken pox and shingles, and HPV in cervical cancer. Likewise, new virus-driven threats to human health continue to emerge, such as the recent SARS, MERS, and SARS-CoV-2 coronavirus epidemics. Viruses use many strategies to survive inside their host, including hijacking of cellular pathways, modulation of the immune response, transformation of host cells, and proliferation of infected cells. All of this is achieved through a remarkably limited pool of virus-encoded genes. The repertoire of vTRs encoded by a virus is therefore of tantamount importance, due to the ability of these proteins to control the expression of the far greater number of genes encoded by the host. Indeed, a single vTR can alter the transcription of hundreds to thousands of human genes. Given the well-established role for altered gene expression in many human diseases, it is likely that vTRs play important, but largely unknown roles in many human disease processes.
vTRs can alter viral and host gene expression through multiple mechanisms, including direct and indirect interactions with nucleic acids, physical interactions with host proteins, and the initiation of epigenetic reprogramming. vTR mechanisms range from simple (e.g., one vTR controlling a few genes) to complex (e.g., tightly controlled transcriptional cascades, vTR-host protein interactions, and multi-protein complexes). The genomes of human viruses encode hundreds of vTRs from at least 20 different DNA and RNA virus families. These proteins come from a wide range of structural classes, often with unique protein folds. Overall, DNA viruses tend to encode more vTRs than RNA viruses. Most DNA virus vTRs primarily regulate transcription, whereas almost all RNA virus vTRs regulate transcription as a secondary function. The shared and divergent aspects of vTRs encoded by DNA and RNA viruses reflect the differences in their virus stages and interactions with the cellular machinery.
Despite the clear importance of vTRs in human health and their potential as therapeutic targets, with only a handful of exceptions, little is known about vTR DNA-binding specificities, host target genes, or mechanisms of action. Recent pioneering studies have begun to take a “systems level view” of vTR function by globally examining their genomic interactions (Table S1) and their physical interactions with human proteins (Davis et al., 2015; Gordon et al., 2020; Gulbahce et al., 2012; Heaton et al., 2016; Jäger et al., 2011). Approaches integrating multiple data types are starting to provide fundamental insights into viral regulatory mechanisms that influence human disease risk (Arvey et al., 2012; Harley et al., 2018; Rialdi et al., 2017; Zheng et al., 2011). In our opinion, these studies represent the tip of a giant iceberg—future genome-scale studies examining vTR genomic targets and regulatory mechanisms, DNA-binding specificity, and physical interactions will be critical moving forward. These studies will require the use of standardized experimental techniques and bioinformatics analyses, similar to those from the ENCODE and Roadmap Epigenomics projects (ENCODE Project Consortium, 2012; Kundaje et al., 2015), to enable systematic integration of datasets and robust comparisons between vTRs.
The extent to which vTR mechanisms are impacted by human disease risk variants, and how these interactions influence disease onset and progression, has remained largely unexplored. The role of other virus-encoded regulatory molecules, such as RNA-binding proteins, microRNAs, and long non-coding RNAs, is even less understood. Given the many known and suspected roles for viruses in human disease, and the central importance of these regulatory molecules, future studies will likely reveal many additional virus-driven disease mechanisms. Ultimately, this work will provide fundamental information for the design of therapeutic interventions for helping the millions of people afflicted with virus-triggered diseases.
Acknowledgments
We apologize to the many colleagues whose important primary studies could not be cited due to space constraints. We thank Xiaoting Chen and Michael Lape for computational support. We thank Carmy Forney, Mariano Garcia-Blanco, Thomas Gilmore, Jun Young Hong, Jeffrey Johnson, Leah Kottyan, William Miller, Emily Miraldi, Nancy Sawtell, Trevor Siggers, Richard Thompson, Stephen Waggoner, David Waxman, and Susanne Wells for critical feedback on the manuscript. This work was supported by NIH (R35 GM128625 to J.I.F.B. and R01 NS099068, R01 AR073228, R01 GM055479, R01 HG010730, and R21 HG008186 to M.T.W.) and by Cincinnati Children’s Hospital “Trustee Award,” “Center for Pediatric Genomics Award,” and “CCRF Endowed Scholar Award” to M.T.W.
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.cell.2020.06.023.
Supporting Citations
The following reference appears in the Supplemental Information: Berman et al. (2002).
Supplemental Information
References
- Aloni-Grinstein R., Charni-Natan M., Solomon H., Rotter V. p53 and the Viral Connection: Back into the Future‡. Cancers (Basel) 2018;10:178. doi: 10.3390/cancers10060178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arvey A., Tempera I., Tsai K., Chen H.S., Tikhmyanova N., Klichinsky M., Leslie C., Lieberman P.M. An atlas of the Epstein-Barr virus transcriptome and epigenome reveals host-virus regulatory interactions. Cell Host Microbe. 2012;12:233–245. doi: 10.1016/j.chom.2012.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ascherio A., Munger K.L., Lennette E.T., Spiegelman D., Hernán M.A., Olek M.J., Hankinson S.E., Hunter D.J. Epstein-Barr virus antibodies and risk of multiple sclerosis: a prospective study. JAMA. 2001;286:3083–3088. doi: 10.1001/jama.286.24.3083. [DOI] [PubMed] [Google Scholar]
- Balandraud N., Roudier J. Epstein-Barr virus and rheumatoid arthritis. Joint Bone Spine. 2018;85:165–170. doi: 10.1016/j.jbspin.2017.04.011. [DOI] [PubMed] [Google Scholar]
- Barbera A.J., Chodaparambil J.V., Kelley-Clarke B., Joukov V., Walter J.C., Luger K., Kaye K.M. The nucleosomal surface as a docking station for Kaposi’s sarcoma herpesvirus LANA. Science. 2006;311:856–861. doi: 10.1126/science.1120541. [DOI] [PubMed] [Google Scholar]
- Basbous J., Arpin C., Gaudray G., Piechaczyk M., Devaux C., Mesnard J.M. The HBZ factor of human T-cell leukemia virus type I dimerizes with transcription factors JunB and c-Jun and modulates their transcriptional activity. J. Biol. Chem. 2003;278:43620–43627. doi: 10.1074/jbc.M307275200. [DOI] [PubMed] [Google Scholar]
- Bellon M., Nicot C. Regulation of telomerase and telomeres: human tumor viruses take control. J. Natl. Cancer Inst. 2008;100:98–108. doi: 10.1093/jnci/djm269. [DOI] [PubMed] [Google Scholar]
- Bentley P., Tan M.J.A., McBride A.A., White E.A., Howley P.M. The SMC5/6 Complex Interacts with the Papillomavirus E2 Protein and Influences Maintenance of Viral Episomal DNA. J. Virol. 2018;92:e00356-18. doi: 10.1128/JVI.00356-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger M.F., Bulyk M.L. Universal protein-binding microarrays for the comprehensive characterization of the DNA-binding specificities of transcription factors. Nat. Protoc. 2009;4:393–411. doi: 10.1038/nprot.2008.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman H.M., Battistuz T., Bhat T.N., Bluhm W.F., Bourne P.E., Burkhardt K., Feng Z., Gilliland G.L., Iype L., Jain S. The Protein Data Bank. Acta. Crystallogr. D. Biol. Crystallogr. 2002;58:899–907. doi: 10.1107/s0907444902003451. [DOI] [PubMed] [Google Scholar]
- Bussiere D.E., Kong X., Egan D.A., Walter K., Holzman T.F., Lindh F., Robins T., Giranda V.L. Structure of the E2 DNA-binding domain from human papillomavirus serotype 31 at 2.4 A. Acta Crystallogr. D Biol. Crystallogr. 1998;54:1367–1376. doi: 10.1107/s0907444998005587. [DOI] [PubMed] [Google Scholar]
- Calderone A., Licata L., Cesareni G. VirusMentha: a new resource for virus-host protein interactions. Nucleic Acids Res. 2015;43:D588–D592. doi: 10.1093/nar/gku830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cárdenas W.B., Loo Y.M., Gale M., Jr., Hartman A.L., Kimberlin C.R., Martínez-Sobrido L., Saphire E.O., Basler C.F. Ebola virus VP35 protein binds double-stranded RNA and inhibits alpha/beta interferon production induced by RIG-I signaling. J. Virol. 2006;80:5168–5178. doi: 10.1128/JVI.02199-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cayrol C., Flemington E.K. Identification of cellular target genes of the Epstein-Barr virus transactivator Zta: activation of transforming growth factor beta igh3 (TGF-beta igh3) and TGF-beta 1. J. Virol. 1995;69:4206–4212. doi: 10.1128/jvi.69.7.4206-4212.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang P.J., Chen L.W., Shih Y.C., Tsai P.H., Liu A.C., Hung C.H., Liou J.Y., Wang S.S. Role of the cellular transcription factor YY1 in the latent-lytic switch of Kaposi’s sarcoma-associated herpesvirus. Virology. 2011;413:194–204. doi: 10.1016/j.virol.2011.02.013. [DOI] [PubMed] [Google Scholar]
- Chiang H.S., Liu H.M. The Molecular Basis of Viral Inhibition of IRF- and STAT-Dependent Immune Responses. Front. Immunol. 2019;9:3086. doi: 10.3389/fimmu.2018.03086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis Z.H., Verschueren E., Jang G.M., Kleffman K., Johnson J.R., Park J., Von Dollen J., Maher M.C., Johnson T., Newton W., et al. Global mapping of herpesvirus-host protein complexes reveals a transcription strategy for late genes. Mol. Cell. 2015;57:349–360. doi: 10.1016/j.molcel.2014.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Leo A., Calderon A., Lieberman P.M. Control of Viral Latency by Episome Maintenance Proteins. Trends Microbiol. 2020;28:150–162. doi: 10.1016/j.tim.2019.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeCaprio J.A., Ludlow J.W., Figge J., Shew J.Y., Huang C.M., Lee W.H., Marsilio E., Paucha E., Livingston D.M. SV40 large tumor antigen forms a specific complex with the product of the retinoblastoma susceptibility gene. Cell. 1988;54:275–283. doi: 10.1016/0092-8674(88)90559-4. [DOI] [PubMed] [Google Scholar]
- Ellison T.J., Izumiya Y., Izumiya C., Luciw P.A., Kung H.J. A comprehensive analysis of recruitment and transactivation potential of K-Rta and K-bZIP during reactivation of Kaposi’s sarcoma-associated herpesvirus. Virology. 2009;387:76–88. doi: 10.1016/j.virol.2009.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elovaara I., Koenig S., Brewah A.Y., Woods R.M., Lehky T., Jacobson S. High human T cell lymphotropic virus type 1 (HTLV-1)-specific precursor cytotoxic T lymphocyte frequencies in patients with HTLV-1-associated neurological disease. J. Exp. Med. 1993;177:1567–1573. doi: 10.1084/jem.177.6.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ENCODE Project Consortium An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engdahl E., Gustafsson R., Huang J., Biström M., Lima Bomfim I., Stridh P., Khademi M., Brenner N., Butt J., Michel A., et al. Increased Serological Response Against Human Herpesvirus 6A Is Associated With Risk for Multiple Sclerosis. Front. Immunol. 2019;10:2715. doi: 10.3389/fimmu.2019.02715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enquist L.W., Editors of the Journal of Virology Virology in the 21st century. J. Virol. 2009;83:5296–5308. doi: 10.1128/JVI.00151-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrell C.J., Lee J.M., Shin E.C., Cebrat M., Cole P.A., Hayward S.D. Inhibition of Epstein-Barr virus-induced growth proliferation by a nuclear antigen EBNA2-TAT peptide. Proc. Natl. Acad. Sci. USA. 2004;101:4625–4630. doi: 10.1073/pnas.0306482101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrari R., Pellegrini M., Horwitz G.A., Xie W., Berk A.J., Kurdistani S.K. Epigenetic reprogramming by adenovirus e1a. Science. 2008;321:1086–1088. doi: 10.1126/science.1155546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrari R., Su T., Li B., Bonora G., Oberai A., Chan Y., Sasidharan R., Berk A.J., Pellegrini M., Kurdistani S.K. Reorganization of the host epigenome by a viral oncogene. Genome Res. 2012;22:1212–1221. doi: 10.1101/gr.132308.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrari R., Gou D., Jawdekar G., Johnson S.A., Nava M., Su T., Yousef A.F., Zemke N.R., Pellegrini M., Kurdistani S.K., Berk A.J. Adenovirus small E1A employs the lysine acetylases p300/CBP and tumor suppressor Rb to repress select host genes and promote productive virus infection. Cell Host Microbe. 2014;16:663–676. doi: 10.1016/j.chom.2014.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreon A.C., Ferreon J.C., Wright P.E., Deniz A.A. Modulation of allostery by protein intrinsic disorder. Nature. 2013;498:390–394. doi: 10.1038/nature12294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu L., Niu B., Zhu Z., Wu S., Li W. CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics. 2012;28:3150–3152. doi: 10.1093/bioinformatics/bts565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y.Z., Su S., Gao Y.Q., Wang P.P., Huang Z.F., Hu M.M., Luo W.W., Li S., Luo M.H., Wang Y.Y., Shu H.B. Human Cytomegalovirus Tegument Protein UL82 Inhibits STING-Mediated Signaling to Evade Antiviral Immunity. Cell Host Microbe. 2017;21:231–243. doi: 10.1016/j.chom.2017.01.001. [DOI] [PubMed] [Google Scholar]
- Gaskill P.J., Miller D.R., Gamble-George J., Yano H., Khoshbouei H. HIV, Tat and dopamine transmission. Neurobiol. Dis. 2017;105:51–73. doi: 10.1016/j.nbd.2017.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glaser L.V., Rieger S., Thumann S., Beer S., Kuklik-Roos C., Martin D.E., Maier K.C., Harth-Hertle M.L., Grüning B., Backofen R., et al. EBF1 binds to EBNA2 and promotes the assembly of EBNA2 chromatin complexes in B cells. PLoS Pathog. 2017;13:e1006664. doi: 10.1371/journal.ppat.1006664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon D.E., Jang G.M., Bouhaddou M., Xu J., Obernier K., White K.M., O’Meara M.J., Rezelj V.V., Guo J.Z., Swaney D.L., et al. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature. 2020 doi: 10.1038/s41586-020-2286-9. Published online April 30, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorman C.M., Gies D., McCray G., Huang M. The human cytomegalovirus major immediate early promoter can be trans-activated by adenovirus early proteins. Virology. 1989;171:377–385. doi: 10.1016/0042-6822(89)90605-3. [DOI] [PubMed] [Google Scholar]
- Gruffat H., Marchione R., Manet E. Herpesvirus Late Gene Expression: A Viral-Specific Pre-initiation Complex Is Key. Front. Microbiol. 2016;7:869. doi: 10.3389/fmicb.2016.00869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerrieri F., Belloni L., D’Andrea D., Pediconi N., Le Pera L., Testoni B., Scisciani C., Floriot O., Zoulim F., Tramontano A., Levrero M. Genome-wide identification of direct HBx genomic targets. BMC Genomics. 2017;18:184. doi: 10.1186/s12864-017-3561-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guirimand T., Delmotte S., Navratil V. VirHostNet 2.0: surfing on the web of virus/host molecular interactions data. Nucleic Acids Res. 2015;43:D583–D587. doi: 10.1093/nar/gku1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gulbahce N., Yan H., Dricot A., Padi M., Byrdsong D., Franchi R., Lee D.S., Rozenblatt-Rosen O., Mar J.C., Calderwood M.A., et al. Viral perturbations of host networks reflect disease etiology. PLoS Comput. Biol. 2012;8:e1002531. doi: 10.1371/journal.pcbi.1002531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haeussler M., Zweig A.S., Tyner C., Speir M.L., Rosenbloom K.R., Raney B.J., Lee C.M., Lee B.T., Hinrichs A.S., Gonzalez J.N., et al. The UCSC Genome Browser database: 2019 update. Nucleic Acids Res. 2019;47(D1):D853–D858. doi: 10.1093/nar/gky1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hafner A., Bulyk M.L., Jambhekar A., Lahav G. The multiple mechanisms that regulate p53 activity and cell fate. Nat. Rev. Mol. Cell Biol. 2019;20:199–210. doi: 10.1038/s41580-019-0110-x. [DOI] [PubMed] [Google Scholar]
- Hagai T., Azia A., Babu M.M., Andino R. Use of host-like peptide motifs in viral proteins is a prevalent strategy in host-virus interactions. Cell Rep. 2014;7:1729–1739. doi: 10.1016/j.celrep.2014.04.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han D., Byun S.H., Kim J., Kwon M., Pleasure S.J., Ahn J.H., Yoon K. Human Cytomegalovirus IE2 Protein Disturbs Brain Development by the Dysregulation of Neural Stem Cell Maintenance and the Polarization of Migrating Neurons. J. Virol. 2017;91:e00799-17. doi: 10.1128/JVI.00799-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harley J.B., Chen X., Pujato M., Miller D., Maddox A., Forney C., Magnusen A.F., Lynch A., Chetal K., Yukawa M., et al. Transcription factors operate across disease loci, with EBNA2 implicated in autoimmunity. Nat. Genet. 2018;50:699–707. doi: 10.1038/s41588-018-0102-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heaton N.S., Moshkina N., Fenouil R., Gardner T.J., Aguirre S., Shah P.S., Zhao N., Manganaro L., Hultquist J.F., Noel J., et al. Targeting Viral Proteostasis Limits Influenza Virus, HIV, and Dengue Virus Infection. Immunity. 2016;44:46–58. doi: 10.1016/j.immuni.2015.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinz S., Benner C., Spann N., Bertolino E., Lin Y.C., Laslo P., Cheng J.X., Murre C., Singh H., Glass C.K. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell. 2010;38:576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinz S., Texari L., Hayes M.G.B., Urbanowski M., Chang M.W., Givarkes N., Rialdi A., White K.M., Albrecht R.A., Pache L., et al. Transcription Elongation Can Affect Genome 3D Structure. Cell. 2018;174:1522–1536. doi: 10.1016/j.cell.2018.07.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henkel T., Ling P.D., Hayward S.D., Peterson M.G. Mediation of Epstein-Barr virus EBNA2 transactivation by recombination signal-binding protein J kappa. Science. 1994;265:92–95. doi: 10.1126/science.8016657. [DOI] [PubMed] [Google Scholar]
- Jäger S., Cimermancic P., Gulbahce N., Johnson J.R., McGovern K.E., Clarke S.C., Shales M., Mercenne G., Pache L., Li K., et al. Global landscape of HIV-human protein complexes. Nature. 2011;481:365–370. doi: 10.1038/nature10719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James J.A., Kaufman K.M., Farris A.D., Taylor-Albert E., Lehman T.J., Harley J.B. An increased prevalence of Epstein-Barr virus infection in young patients suggests a possible etiology for systemic lupus erythematosus. J. Clin. Invest. 1997;100:3019–3026. doi: 10.1172/JCI119856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johannsen E., Koh E., Mosialos G., Tong X., Kieff E., Grossman S.R. Epstein-Barr virus nuclear protein 2 transactivation of the latent membrane protein 1 promoter is mediated by J kappa and PU.1. J. Virol. 1995;69:253–262. doi: 10.1128/jvi.69.1.253-262.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jolma A., Yan J., Whitington T., Toivonen J., Nitta K.R., Rastas P., Morgunova E., Enge M., Taipale M., Wei G., et al. DNA-binding specificities of human transcription factors. Cell. 2013;152:327–339. doi: 10.1016/j.cell.2012.12.009. [DOI] [PubMed] [Google Scholar]
- Jolma A., Yin Y., Nitta K.R., Dave K., Popov A., Taipale M., Enge M., Kivioja T., Morgunova E., Taipale J. DNA-dependent formation of transcription factor pairs alters their binding specificity. Nature. 2015;527:384–388. doi: 10.1038/nature15518. [DOI] [PubMed] [Google Scholar]
- Knipe D.M., Howley P.M. Sixth Edition. Wolters Kluwer/Lippincott Williams & Wilkins Health; 2013. Fields Virology. [Google Scholar]
- Kriegler M., Perez C.F., Hardy C., Botchan M. Transformation mediated by the SV40 T antigens: separation of the overlapping SV40 early genes with a retroviral vector. Cell. 1984;38:483–491. doi: 10.1016/0092-8674(84)90503-8. [DOI] [PubMed] [Google Scholar]
- Kundaje A., Meuleman W., Ernst J., Bilenky M., Yen A., Heravi-Moussavi A., Kheradpour P., Zhang Z., Wang J., Ziller M.J., et al. Roadmap Epigenomics Consortium Integrative analysis of 111 reference human epigenomes. Nature. 2015;518:317–330. doi: 10.1038/nature14248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert S.A., Jolma A., Campitelli L.F., Das P.K., Yin Y., Albu M., Chen X., Taipale J., Hughes T.R., Weirauch M.T. The Human Transcription Factors. Cell. 2018;175:598–599. doi: 10.1016/j.cell.2018.09.045. [DOI] [PubMed] [Google Scholar]
- Lambert S.A., Yang A.W.H., Sasse A., Cowley G., Albu M., Caddick M.X., Morris Q.D., Weirauch M.T., Hughes T.R. Similarity regression predicts evolution of transcription factor sequence specificity. Nat. Genet. 2019;51:981–989. doi: 10.1038/s41588-019-0411-1. [DOI] [PubMed] [Google Scholar]
- Lieberman P.M., Berk A.J. In vitro transcriptional activation, dimerization, and DNA-binding specificity of the Epstein-Barr virus Zta protein. J. Virol. 1990;64:2560–2568. doi: 10.1128/jvi.64.6.2560-2568.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieberman P.M., Hardwick J.M., Sample J., Hayward G.S., Hayward S.D. The zta transactivator involved in induction of lytic cycle gene expression in Epstein-Barr virus-infected lymphocytes binds to both AP-1 and ZRE sites in target promoter and enhancer regions. J. Virol. 1990;64:1143–1155. doi: 10.1128/jvi.64.3.1143-1155.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X.J., Yang B., Huang S.N., Wu C.C., Li X.J., Cheng S., Jiang X., Hu F., Ming Y.Z., Nevels M., et al. Human cytomegalovirus IE1 downregulates Hes1 in neural progenitor cells as a potential E3 ubiquitin ligase. PLoS Pathog. 2017;13:e1006542. doi: 10.1371/journal.ppat.1006542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marazzi I., Garcia-Sastre A. Interference of viral effector proteins with chromatin, transcription, and the epigenome. Curr. Opin. Microbiol. 2015;26:123–129. doi: 10.1016/j.mib.2015.06.009. [DOI] [PubMed] [Google Scholar]
- Martin M.E., Nicholas J., Thomson B.J., Newman C., Honess R.W. Identification of a transactivating function mapping to the putative immediate-early locus of human herpesvirus 6. J. Virol. 1991;65:5381–5390. doi: 10.1128/jvi.65.10.5381-5390.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurano M.T., Humbert R., Rynes E., Thurman R.E., Haugen E., Wang H., Reynolds A.P., Sandstrom R., Qu H., Brody J., et al. Systematic localization of common disease-associated variation in regulatory DNA. Science. 2012;337:1190–1195. doi: 10.1126/science.1222794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mechelli R., Manzari C., Policano C., Annese A., Picardi E., Umeton R., Fornasiero A., D’Erchia A.M., Buscarinu M.C., Agliardi C., et al. Epstein-Barr virus genetic variants are associated with multiple sclerosis. Neurology. 2015;84:1362–1368. doi: 10.1212/WNL.0000000000001420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng X., Brodsky M.H., Wolfe S.A. A bacterial one-hybrid system for determining the DNA-binding specificity of transcription factors. Nat. Biotechnol. 2005;23:988–994. doi: 10.1038/nbt1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mesri E.A., Feitelson M.A., Munger K. Human viral oncogenesis: a cancer hallmarks analysis. Cell Host Microbe. 2014;15:266–282. doi: 10.1016/j.chom.2014.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi H., Muruganujan A., Thomas P.D. PANTHER in 2013: modeling the evolution of gene function, and other gene attributes, in the context of phylogenetic trees. Nucleic Acids Res. 2013;41:D377–D386. doi: 10.1093/nar/gks1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyazawa M., Noguchi K., Kujirai M., Katayama K., Yamagoe S., Sugimoto Y. IL-10 promoter transactivation by the viral K-RTA protein involves the host-cell transcription factors, specificity proteins 1 and 3. J. Biol. Chem. 2018;293:662–676. doi: 10.1074/jbc.M117.802900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore P.S., Chang Y. Why do viruses cause cancer? Highlights of the first century of human tumour virology. Nat. Rev. Cancer. 2010;10:878–889. doi: 10.1038/nrc2961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nabel G.J., Rice S.A., Knipe D.M., Baltimore D. Alternative mechanisms for activation of human immunodeficiency virus enhancer in T cells. Science. 1988;239:1299–1302. doi: 10.1126/science.2830675. [DOI] [PubMed] [Google Scholar]
- Nandakumar D., Glaunsinger B. An integrative approach identifies direct targets of the late viral transcription complex and an expanded promoter recognition motif in Kaposi’s sarcoma-associated herpesvirus. PLoS Pathog. 2019;15:e1007774. doi: 10.1371/journal.ppat.1007774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Offermann M.K. Kaposi sarcoma herpesvirus-encoded interferon regulator factors. Curr. Top. Microbiol. Immunol. 2007;312:185–209. doi: 10.1007/978-3-540-34344-8_7. [DOI] [PubMed] [Google Scholar]
- Pakpoor J., Disanto G., Gerber J.E., Dobson R., Meier U.C., Giovannoni G., Ramagopalan S.V. The risk of developing multiple sclerosis in individuals seronegative for Epstein-Barr virus: a meta-analysis. Mult. Scler. 2013;19:162–166. doi: 10.1177/1352458512449682. [DOI] [PubMed] [Google Scholar]
- Patrick D.R., Oliff A., Heimbrook D.C. Identification of a novel retinoblastoma gene product binding site on human papillomavirus type 16 E7 protein. J. Biol. Chem. 1994;269:6842–6850. [PubMed] [Google Scholar]
- Portal D., Zhou H., Zhao B., Kharchenko P.V., Lowry E., Wong L., Quackenbush J., Holloway D., Jiang S., Lu Y., Kieff E. Epstein-Barr virus nuclear antigen leader protein localizes to promoters and enhancers with cell transcription factors and EBNA2. Proc. Natl. Acad. Sci. USA. 2013;110:18537–18542. doi: 10.1073/pnas.1317608110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prusinkiewicz M.A., Mymryk J.S. Metabolic Reprogramming of the Host Cell by Human Adenovirus Infection. Viruses. 2019;11:141. doi: 10.3390/v11020141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramasubramanyan S., Osborn K., Al-Mohammad R., Naranjo Perez-Fernandez I.B., Zuo J., Balan N., Godfrey A., Patel H., Peters G., Rowe M., et al. Epstein-Barr virus transcription factor Zta acts through distal regulatory elements to directly control cellular gene expression. Nucleic Acids Res. 2015;43:3563–3577. doi: 10.1093/nar/gkv212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeder J.E., Kwak Y.T., McNamara R.P., Forst C.V., D’Orso I. HIV Tat controls RNA Polymerase II and the epigenetic landscape to transcriptionally reprogram target immune cells. eLife. 2015;4:e08955. doi: 10.7554/eLife.08955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rialdi A., Hultquist J., Jimenez-Morales D., Peralta Z., Campisi L., Fenouil R., Moshkina N., Wang Z.Z., Laffleur B., Kaake R.M., et al. The RNA Exosome Syncs IAV-RNAPII Transcription to Promote Viral Ribogenesis and Infectivity. Cell. 2017;169:679–692. doi: 10.1016/j.cell.2017.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricigliano V.A., Handel A.E., Sandve G.K., Annibali V., Ristori G., Mechelli R., Cader M.Z., Salvetti M. EBNA2 binds to genomic intervals associated with multiple sclerosis and overlaps with vitamin D receptor occupancy. PLoS ONE. 2015;10:e0119605. doi: 10.1371/journal.pone.0119605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronco L.V., Karpova A.Y., Vidal M., Howley P.M. Human papillomavirus 16 E6 oncoprotein binds to interferon regulatory factor-3 and inhibits its transcriptional activity. Genes Dev. 1998;12:2061–2072. doi: 10.1101/gad.12.13.2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sears J., Ujihara M., Wong S., Ott C., Middeldorp J., Aiyar A. The amino terminus of Epstein-Barr Virus (EBV) nuclear antigen 1 contains AT hooks that facilitate the replication and partitioning of latent EBV genomes by tethering them to cellular chromosomes. J. Virol. 2004;78:11487–11505. doi: 10.1128/JVI.78.21.11487-11505.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin Y., Chang Y.C., Lee D.S.W., Berry J., Sanders D.W., Ronceray P., Wingreen N.S., Haataja M., Brangwynne C.P. Liquid Nuclear Condensates Mechanically Sense and Restructure the Genome. Cell. 2019;176:1518. doi: 10.1016/j.cell.2019.02.025. [DOI] [PubMed] [Google Scholar]
- Siggers T., Chang A.B., Teixeira A., Wong D., Williams K.J., Ahmed B., Ragoussis J., Udalova I.A., Smale S.T., Bulyk M.L. Principles of dimer-specific gene regulation revealed by a comprehensive characterization of NF-κB family DNA binding. Nat. Immunol. 2011;13:95–102. doi: 10.1038/ni.2151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taura M., Song E., Ho Y.C., Iwasaki A. Apobec3A maintains HIV-1 latency through recruitment of epigenetic silencing machinery to the long terminal repeat. Proc. Natl. Acad. Sci. USA. 2019;116:2282–2289. doi: 10.1073/pnas.1819386116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tillo D., Ray S., Syed K.S., Gaylor M.R., He X., Wang J., Assad N., Durell S.R., Porollo A., Weirauch M.T., Vinson C. The Epstein-Barr Virus B-ZIP Protein Zta Recognizes Specific DNA Sequences Containing 5-Methylcytosine and 5-Hydroxymethylcytosine. Biochemistry. 2017;56:6200–6210. doi: 10.1021/acs.biochem.7b00741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinson C.R., Sigler P.B., McKnight S.L. Scissors-grip model for DNA recognition by a family of leucine zipper proteins. Science. 1989;246:911–916. doi: 10.1126/science.2683088. [DOI] [PubMed] [Google Scholar]
- Weitzman M.D., Fradet-Turcotte A. Virus DNA Replication and the Host DNA Damage Response. Annu. Rev. Virol. 2018;5:141–164. doi: 10.1146/annurev-virology-092917-043534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weller S.K., Coen D.M. Herpes simplex viruses: mechanisms of DNA replication. Cold Spring Harb. Perspect. Biol. 2012;4:a013011. doi: 10.1101/cshperspect.a013011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whyte P., Buchkovich K.J., Horowitz J.M., Friend S.H., Raybuck M., Weinberg R.A., Harlow E. Association between an oncogene and an anti-oncogene: the adenovirus E1A proteins bind to the retinoblastoma gene product. Nature. 1988;334:124–129. doi: 10.1038/334124a0. [DOI] [PubMed] [Google Scholar]
- Yan X., Hao Q., Mu Y., Timani K.A., Ye L., Zhu Y., Wu J. Nucleocapsid protein of SARS-CoV activates the expression of cyclooxygenase-2 by binding directly to regulatory elements for nuclear factor-kappa B and CCAAT/enhancer binding protein. Int. J. Biochem. Cell Biol. 2006;38:1417–1428. doi: 10.1016/j.biocel.2006.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Yin Y., Fischer D. Identification and investigation of ORFans in the viral world. BMC Genomics. 2008;9:24. doi: 10.1186/1471-2164-9-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- You J., Croyle J.L., Nishimura A., Ozato K., Howley P.M. Interaction of the bovine papillomavirus E2 protein with Brd4 tethers the viral DNA to host mitotic chromosomes. Cell. 2004;117:349–360. doi: 10.1016/s0092-8674(04)00402-7. [DOI] [PubMed] [Google Scholar]
- Yu G., Wang L.G., He Q.Y. ChIPseeker: an R/Bioconductor package for ChIP peak annotation, comparison and visualization. Bioinformatics. 2015;31:2382–2383. doi: 10.1093/bioinformatics/btv145. [DOI] [PubMed] [Google Scholar]
- Zemke N.R., Gou D., Berk A.J. Dedifferentiation by adenovirus E1A due to inactivation of Hippo pathway effectors YAP and TAZ. Genes Dev. 2019;33:828–843. doi: 10.1101/gad.324814.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B., Zou J., Wang H., Johannsen E., Peng C.W., Quackenbush J., Mar J.C., Morton C.C., Freedman M.L., Blacklow S.C., et al. Epstein-Barr virus exploits intrinsic B-lymphocyte transcription programs to achieve immortal cell growth. Proc. Natl. Acad. Sci. USA. 2011;108:14902–14907. doi: 10.1073/pnas.1108892108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao N., Sebastiano V., Moshkina N., Mena N., Hultquist J., Jimenez-Morales D., Ma Y., Rialdi A., Albrecht R., Fenouil R., et al. Influenza virus infection causes global RNAPII termination defects. Nat. Struct. Mol. Biol. 2018;25:885–893. doi: 10.1038/s41594-018-0124-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng S., Tansey W.P., Hiebert S.W., Zhao Z. Integrative network analysis identifies key genes and pathways in the progression of hepatitis C virus induced hepatocellular carcinoma. BMC Med. Genomics. 2011;4:62. doi: 10.1186/1755-8794-4-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.