Abstract
The immunocapture-based ELISA for extracellular vesicles (EVs)/exosomes, originally described in 2009 by Logozzi and colleagues, allows to capture, detect, characterize and quantify extracellular vesicles in both human body fluids and cell culture supernatants. It is based on the use of two antibodies directed one against a typical exosomal housekeeping protein and the second against either another exosomal housekeeping protein or a potential disease marker: the first antibody is used for the capture of exosomes, the second for the quantification and characterization of the captured vesicles. In fact, with this method it is possible both to characterize and count exosomes and to detect the presence of disease, including tumor, biomarkers. This needs of course to preliminary obtain an EVs purification from the clinical sample; the most agreed method to get to an EVs purification is the repeated rounds of ultracentrifugation, that, while far to be perfect, is the methodological approach allowing to not exclude EVs subpopulation from the separation procedure and to analyze a full range of EVs from both qualitative and quantitative point of view. The immunocapture-based approach has proven to be highly useful in screening, diagnosis and prognosis of tumors, in plasma samples. One amazing information provided by this method is that cancer patients have always significantly higher levels of EVs, in particular of exosomes, independently from the histological nature of the tumor. One microenvironmental factor that is fully involved in the increased exosome release by tumors is the extracellular acidity. However, few pre-clinical data suggest that plasmatic levels of exosomes may correlate with the tumor mass. Some recent clinical reports suggest also that circulating exosomes represent the real delivery system for some known tumor markers that are presently on trial (e.g., PSA). Here we review the pros and cons of the immunocapture-based technique in quantitative and qualitative evaluation of EVs in both health and disease.
Keywords: Immunocapture-based ELISA, Extracellular vesicles characterization and quantification, Circulating exosomes, Body fluids, Screening-diagnosis-prognosis tool, Non-invasive technique, Biomarkers of diseases, Follow-up of diseases
1. Definition
Immunocapture-based ELISA (Logozzi et al., 2009) is a methodology particularly suitable to analyze vesicles from both human body fluids and cell culture supernatants. It is based on the principle that specific antigens can be expressed only by vesicles of size from nano to micro and the method exploit this feature to select only the population of vesicles of interest. However, the most interesting between the EVs are by far the nanovesicles called exosomes. Besides to be the specific markers of these nanovesicles, the exosomes also contain molecules defining both the cellular source and the disease state, thus representing the preferential delivery system of a wide range of molecules. Thanks to the wide range of antigens on the EVs surface, the immunocapture-based technologies, may well be implemented by the use of the broad panel of antibodies available todate.
2. Rationale
2.1. Exosomes
Exosomes are nanovesicles of a size ranging between 40 and 180 nm playing critical role in cell-to-cell communication, all along a proteins/DNA/RNA shuttling, that normally allows a tuned regulation of body's homeostasis, but that may well contribute in disease pathophysiology (Fais et al., 2016; Théry et al., 2018; Yáñez-Mó et al., 2015). Exosomal cargo can impact target cells through stimulation via surface-bound ligands, transferring activated receptors, or transferring functional proteins, lipids and genetic material (Zhang, Liu, Liu, & Tang, 2019). Exosomes are generated from the late endosomes, which originate by inward budding of multivesicular body (MVB) membrane. From the invagination of the late endosomes membranes, the intraluminal vesicles (ILVs) are formed inside the MVBs. ILVs are subsequently released in the extracellular space by fusion with cellular plasma membrane or can be degraded in lysosomes (Théry et al., 2018; van der Pol, Böing, Harrison, Sturk, & Nieuwland, 2012; Yáñez-Mó et al., 2015; Zhang et al., 2019).
2.2. Role of exosomes in physiological and pathological condition
Exosomes are released from many cell types, including dendritic cells (DC), lymphocytes, mast cells and epithelial cells, in both physiological and pathological conditions (Logozzi et al., 2018; Logozzi, Spugnini, Mizzoni, Di Raimo, & Fais, 2019; Spugnini, Logozzi, Di Raimo, Mizzoni, & Fais, 2018; Théry et al., 2018; Yáñez-Mó et al., 2015). However, exosomes can be isolated from various biological fluids, such as blood and urine. Although it was initially believed that the role of exosomes was to eliminate unnecessary proteins, evidence of their fundamental role in intercellular communication has accumulated over time. In fact, it has been shown that exosomes participate to the immune response (Lugini et al., 2012; Raposo et al., 1996; Théry et al., 2002), and are involved in the development of the nervous system and as a consequence in the pathogenesis of neurodegenerative diseases (Frühbeis, Fröhlich, Kuo, & Krämer-Albers, 2013; Ghidoni, Benussi, & Binetti, 2008), and prion-mediated diseases as well (Li & Barres, 2018, p. 201; Properzi et al., 2015). Exosomes have a role in cardiovascular diseases, as well, with alteration in both miRNAs (Yellon & Davidson, 2014), and proteins (e.g., clusterin) content (Foglio et al., 2015). Exosomal cargo changes in many other diseases including diabetes, rheumatoid arthritis, obesity, kidney diseases and cancer (Console, Scalise, & Indiveri, 2019; Lv et al., 2018; Scrivo, Vasile, Bartosiewicz, & Valesini, 2011). Moreover, exosomes from adipose tissue contain 55 altered miRNAs involved in regulation of development and progression of chronic inflammation and resistance to insulin (Deng et al., 2009; Ferrante et al., 2015; Gurunathan, Kang, Jeyaraj, Qasim, & Kim, 2019). Cancer exosomes mediate inflammatory responses occurring in tumor development, immune surveillance and response to therapy (Colotta, Allavena, Sica, Garlanda, & Mantovani, 2009; Grivennikov, Greten, & Karin, 2010; Gurunathan et al., 2019). Furthermore, it has also been shown that in the case of some viral infections, exosomes are used by viruses as a strategy for evading the immune response and to spread the infection (Rodrigues, Fan, Lyon, Wan, & Hu, 2018). EVs and exosomes deliver viruses, including both HIV-1 (Chen et al., 2020; Muratori et al., 2009) and Epstein-Barr (EBV) virus (Canitano, Venturi, Borghi, Ammendolia, & Fais, 2013). Exosomes participate in the formation of new capillaries from existing blood vessels both in physiological condition, modulating endothelial VEGF signaling and gene expression (Gurunathan et al., 2019; Hood, Pan, Lanza, Wickline, & Consortium for Translational Research in Advanced Imaging and Nanomedicine (C-TRAIN) et al., 2009; Kaur et al., 2014). In cancer condition exomes are also involved in angiogenesis and extracellular matrix remodeling (Gurunathan et al., 2019; Hood, San, & Wickline, 2011). Tumor-derived exosomes may also participate in tumor immune escape by inducing cell death in both natural killer cells and T-cells (Andreola et al., 2002; Clayton et al., 2008; Huber et al., 2005), with the final purpose to control the anti-tumor immune reaction, inhibit cancer cells apoptosis and promote tumor growth (Vallabhaneni et al., 2016). Tumor exosomes play a key role in both the paracrine dissemination, the formation of the pre-metastatic niche and conceivably in inducing a tumor-like transformation in mesenchymal stem cells (Spugnini et al., 2018; Zhao et al., 2018). Evidence is accumulating on the importance of exosomes or EVs plasma levels in cancer patients as a valuable diagnostic/prognostic tool in cancer patients, independently from the tumor histology (Logozzi, Angelini, et al., 2019; Logozzi et al., 2009, Logozzi et al., 2020; Ludwig, Yerneni, Razzo, & Whiteside, 2018; Osti et al., 2019; Rodríguez Zorrilla et al., 2019; Skog et al., 2008), suggesting that exosomes quantification may represent an important new approach for cancer screening as well (Dragovic et al., 2015; Gercel-Taylor, Atay, Tullis, Kesimer, & Taylor, 2012; Oosthuyzen et al., 2013; Zhang et al., 2016).
Lastly it is important to mention the ability of EVs to deliver drugs and nanomaterial, strongly suggesting that in particular exosomes may represent a natural nanoshuttle for future therapy, diagnostic and theranostics in different disease states (Federici et al., 2014; Iessi et al., 2017; Kusuzaki et al., 2017; Logozzi Mizzoni, et al., 2019).
2.3. Methods to analyze and characterize extracellular vesicles
Many techniques have been exploited in the last decade to isolate exosomes from either body fluids and culture supernatants. No one is perfect indeed, but the less imperfect is the use of a series of centrifugations with increasing speed followed by repeated ultracentrifugations. However, evaluation of physiochemical properties of exosomes, including size, shape, surface charge and proteins, lipids and nucleic acids composition are all key in the exosome and EVs characterization (Théry et al., 2018). At the moment, nanoparticle tracking analysis (NTA) has become one of the most used techniques for a quality assessment of the vesicle preparations from both cell culture supernatant and body fluids. NTA is a biophysical method based on optical density tracking of particles, which can simultaneously measure concentration and size distribution of extracellular vesicles of different size. In particular, the method allows the tracking of Brownian movement of nanoparticles in a liquid suspension on a particle-by-particle basis; the movement of each single particle is tracked and then correlated to particle size (Gurunathan et al., 2019; Szatanek et al., 2017). NTA is able to detect small particles with a diameter smaller than 30 nm; moreover, the analysis is performed in a liquid phase, avoiding biophysical modifications of studied exosomes.
Transmission electron microscopy (TEM) was widely used to characterize and visualize exosomal preparation with or without immuno-gold antibodies labeling to obtain biochemical information. Although this technique allows to obtain high resolution images, the sample preparation and the measurement conditions make this technique disadvantageous. Samples have to be fixed and dehydrated before measurement and multi-step preparation for TEM analysis can induce morphological changes of exosomal specimens. In addition, electron beam can induce damages in biological samples (Gurunathan et al., 2019; Szatanek et al., 2017). However, it is mandatory to emphasize that, as in the vast majority of the cases in science, electron microscopy allowed the discovery of extracellular vesicles and exosomes, and it should never be abandoned or disregarded in the present and future research on exosomes and EVs.
The amount of exosomal proteins can also be used as an indirect measurement of the quantity of isolated vesicles, but these values do not necessarily correlate with the number of exosomes. Accordingly to MISEV2018 guidelines, to demonstrate the presence of EVs in specific samples, at least one transmembrane protein associated to exosomal plasma membrane (e.g., tetraspanins CD9, CD63, CD81) and one cytosolic protein (e.g., TSG101, ALIX) have to be analyzed. Among available techniques, Western Blotting is the most used method for the characterization of typical exosomal proteins, although it clearly represents a semi-quantitative, that can't be exploited in the serial analysis of clinical samples. Most of all, it requires large volumes of samples and extensive preparatory and processing time.
Over the last two decades, Flow Cytometry has become one of the most frequently methodology exploited in analyzing extracellular vesicles, thanks its ability to examine different parameters at the same time. Flow cytometry allows to analyze size and structure of particles in suspension based on forward scattering light, together with characterization of exosomal proteins previously labeled with fluorescent antibodies. Due the side detection limitation, conventional cytometers could underestimate particles smaller than 300 nm. For this reason, new generation flow cytometers were modified with multi-angle lasers to improve particle resolution (Biggs et al., 2016; Chandler, Yeung, & Tait, 2011; Morales-Kastresana et al., 2017), together with its implementation with nanoscale equipment, and recently exploited in clinical studies as well (Logozzi, Angelini, et al., 2019; Logozzi et al., 2017).
Lastly, what is the main issue of this chapter the use of the immunocapture-based techniques and in particular of the immunocapture-based ELISA in both pre-clinical and clinical analysis of EVs and exosomes. This deserves a dedicated paragraph with the purpose of providing a more detailed description of the method.
2.4. Immunocapture-based ELISA method description
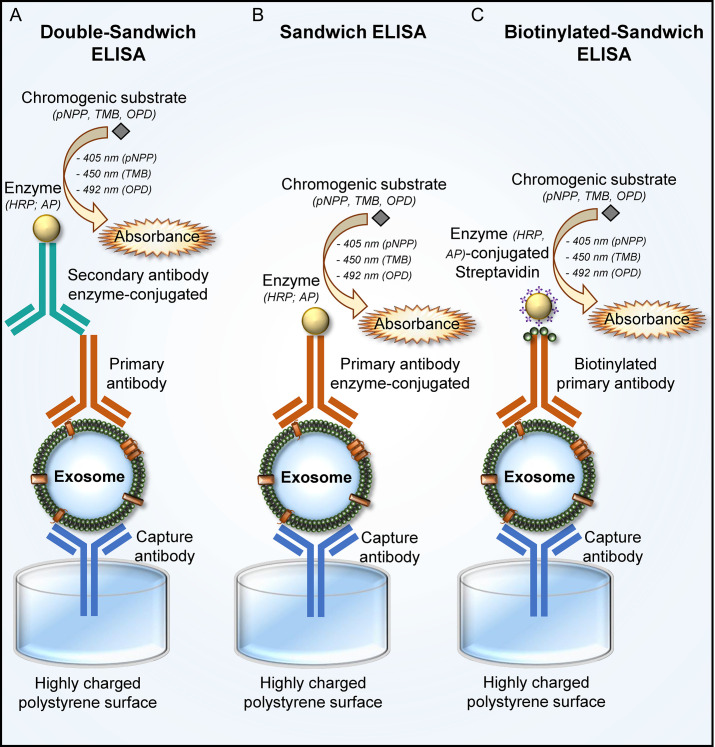
Given the increasing understanding of the role of exosomes in cell communication, cancer progression together with increasing need to find new treatments, improve diagnostics and follow-up of malignancy, there is accordingly a need for new methods and tools to detect and measure exosomes in human fluids. In this regard, in 2009 a new method of quantification of exosomes isolated from cell culture and plasma supernatants was provided (Logozzi et al., 2009) (Fig. 1 ).
Fig. 1.
Schematic representation of the immunocapture-based ELISA for exosomes detection and quantification. The detection and quantification of exosomes, purified from plasma and cell supernatants, can be performed mainly by Double-Sandwich ELISA (A), Sandwich ELISA (B) and Biotinylated-Sandwich ELISA (C). A polyclonal capture antibody is required for the specific exosome binding to the plate, followed by a monoclonal detection antibody either directly conjugated to an enzyme or bound with an enzyme-conjugated secondary antibody. The reaction between an enzyme and a chromogenic substrate develops color whose absorbance intensity is measured with the spectrophotometer. Enzyme usually used are horseradish peroxidase (HRP) and alkaline phosphatase (AP), while chromogenic substrate are p-nitrophenyl phosphate (pNPP), tetramethylbenzidine (TMB) o-phenylenediamine dihydrochloride (ODP). Absorbance is read at 405 nm for pNPP, 450 nm for TMB and 492 nm for ODP. (A) Double-Sandwich ELISA is an indirect system for exosomes detection and quantification. Exosomes, after the binding with a capture antibody fixed to the plate, are bound by a primary antibody and detected with a secondary antibody enzyme-conjugated. (B) Sandwich ELISA is a direct system for exosomes detection and quantification. Exosomes, following the binding with a capture antibody fixed to the plate, are bound and detected by a primary antibody enzyme-conjugated. (C) Biotinylated-Sandwich ELISA is a direct system for exosomes detection and quantification. Exosomes, following the binding with a capture antibody fixed to the plate, are bound by a primary biotinylated antibody and detected by enzyme-conjugated streptavidin. The binding between detection biotinylated antibody and enzyme-conjugated streptavidin could be used for signal amplification.
In Logozzi et al. (2009) it was widely demonstrated that immunocapture-based ELISA allows to characterize extracellular vesicles from plasma of both SCID mice engrafted with human melanoma. Moreover, it has been observed that the quantity of caveolin-1 in exosomal preparations is exclusively detectable from exosomes purified from plasma of tumor patients and correlates with the tumor size.
The method initially involved the use of the polyclonal anti-rab5b antibody to capture exosomes and monoclonal anti-CD63 subsequently bound to a secondary antibody HRP-conjugated (Fig. 1). Rab5b and CD63 are exosomal proteins shared with endosomes and lysosomes, but not shed or recycled as for membrane structures, so they are commonly used as exosomal markers (Logozzi et al., 2009; Théry et al., 2018). The method was able to provide a more reliable quantification of the exosomes as compared to the other available methods. In fact, as detailed above the other available techniques were either not quantitative (TEM) or only poorly quantitative (WB). Although flow cytometry has been used to quantify exosomes, this method does not allow the needed precision for an accurate measurement of the amount of extracellular vesicles contained in the sample. In fact, while FACS (fluorescence activated cell sorter) analysis was a suitable method to quantify cells, even of small size, it was not suitable to quantify the amount of small vesicles, such as exosomes (i.e., 50–100 nm). Furthermore, FACS analysis did not allow simultaneous analysis of different samples. Therefore, there was a need for a method to detect and quantify exosomes from small amounts of body fluids, and in as many as samples at the same time, in order to quickly translate the methodology to the clinical use. Actually, immunocapture-based ELISA looked ideal for this purpose, inasmuch it allowed the detection and quantification of both exosome-specific antigens and tumor antigens on EVs isolated from small quantities of plasma, at the same time. In fact, in the original study it has been shown that, as a second layer antibody, the anti-CD63 antibody can be replaced with an antibody against a potential new disease marker, in that case caveolin-1, to quantify and possibly track disease progression. In fact, in the same study and with the same technique it has been shown that the amount of plasma exosomes expressing caveolin-1 correlated with the tumor size (Logozzi et al., 2009).
The same approach was utilized in a pilot clinical study in a small cohort of patients affected by oral cancer. The study was performed in each single patient measuring the levels of plasmatic exosomes before and after surgery. The results showed that the levels of plasmatic exosomes dramatically dropped down immediately after surgery, suggesting that the great majority of the plasmatic exosome from tumor patients came from the tumors (Rodríguez Zorrilla et al., 2019). The study showed also that the patients showing the highest exosome levels before surgery had the worst prognosis.
Given the versatility of the technique, it was possible to adapt the method for the study of other pathologies. In a more recent works, the antibody for EVs capture (anti-Rab5b) was replaced with anti-CD81, and CD63 with anti-PSA (prostate specific antigen) antibody, commonly used marker for prostatic cancer (PCa) early diagnosis and follow-up (Logozzi, Angelini, et al., 2019; Logozzi et al., 2017) (Fig. 1). Results from immunocapture-based ELISA showed that cancer patients had significantly higher levels of exosomal PSA compared to healthy donors and the method allowed a clear and significant discrimination, in term of both sensitivity and specificity, between patients and healthy donors, as compared to the serum PSA (Logozzi, Angelini, et al., 2019). In order to implement immunocapture-based ELISA with other methods possibly exploitable in cancer patients clinical management the plasma samples were also analyzed by both nanoscale flow cytometry (NFC) and nanosight tracking analysis (NTA) and results were compared, patient-by-patient to those obtained with immunocapture-based ELISA. Statistical analysis of the results demonstrated that immunocapture-based ELISA alone allows to detect exosomal PSA and discriminate healthy subjects versus patients with significantly higher sensitivity and sensibility than serum PSA analysis and compared to other techniques used. Moreover, immunocapture-based ELISA allowed to quantify and characterize several clinical samples at the same time, and in a broader population of EVs as compared with NFC (Logozzi, Angelini, et al., 2019; Yu et al., 2018).
Immunocapture-based ELISA was exploited also for the quantification of the carbonic anhydrase IX (CAIX) exosome expression. Exosomes were captured using anti-CD81 antibodies and anti-CAIX was used as detection antibodies. Exosomes from plasma of PCa patients expressed more CAIX than control patients, suggesting the potential role of exosomal CAIX as tumor marker (Logozzi et al., 2020).
All in all the data obtained in our laboratory, as well as comparable data obtained in other laboratories worldwide (Alegre et al., 2016; Campanella et al., 2015; Jang et al., 2019; Khan et al., 2012; Logozzi, Angelini, et al., 2019; Logozzi et al., 2017; Moon et al., 2016; Sun et al., 2019; Yokoyama et al., 2017; Yu et al., 2018) support an extensive use of immunocapture-based ELISA for the characterization and quantification of EVs and particularly exosomes in the plasma, as well as the other body fluids, in human patients with tumor malignancies, but of course virtually all the known diseases. The advantages of this method are not limited to its plasticity in allowing the contemporary examination of several markers expressed on the exosomes, and in different patients' samples, but they include the sustainable costs of the method and its easy application in virtually all the clinical laboratories worldwide. Of interest 26 years ago, using a comparable technical approach we showed that HIV-1 virions budded from the infected cells together with some cell plasma membrane (Capobianchi et al., 1994) and this occurred thanks to cytoskeleton-membrane driven polarization of the cells (Fais et al., 1995). This strongly suggests that this method may well be used in implementing and possibly improving the diagnostic approach for the vast majority of viral infections, including these days coronavirus infection (Anderson et al., 2018; Chettimada et al., 2018; Liu et al., 2019; Montaner-Tarbes, Borrás, Montoya, Fraile, & del Portillo, 2016).
3. Materials, equipment and reagents
3.1. Materials
-
1.
Polyclonal antibodies directed against EV/exosome antigens (capture antibodies) (e.g., anti-CD9 (Cluster of Differentiation 9) antibody clone H110; anti-CD81 (Cluster of Differentiation 81) antibody clone H-121 and clone PA5-79003)
-
2.
Monoclonal antibodies against EV/exosome antigens (detection antibodies) non-interfering with polyclonal antibodies [e.g., anti-CD63 (Cluster of Differentiation 63) antibody clone H5C6]
-
3.
Plastic plates with highly charged polystyrene surface suitable for antibodies attachment (96-well plates)
-
4.
Carbonate buffer (pH 9,6)
-
5.
Phosphate buffer saline (PBS)
-
6.
Blocking buffer (0,5% BSA in PBS)
-
7.
PBST (PBS + 0,1%Tween20)
-
8.
Parafilm
-
9.
Secondary antibodies conjugated with HRP (horseradish peroxidase)
-
10.
Blue POD substrate
3.2. Equipments
-
1.
Shaker
-
2.
Incubator with temperature (37 °C) and CO2 (5%) controlled
-
3.
Microplate spectrophotometer reader with appropriate optical filters (405, 450, 490 nm)
4. Protocols
-
1.
Coat the plate (96-well plate) with 4 μg/mL polyclonal antibodies (capture antibodies) in a volume of 100 μL/well of carbonate buffer
-
2.
Place the plate for 30 min on a shaker and then overnight at 4 °C
-
3.
Wash the plate three times with 300 μL PBS
-
4.
Add 100 μL blocking solution, incubate for 1 h at room temperature
-
5.
Wash the plate three times with PBS
-
6.
Coat the plate with exosomes in a final volume of 50 μL
-
7.
Cover the plate with parafilm and place 15–20 min on shaker; incubate at 37 °C overnight
-
8.
Wash three times with 300 μL PBST and one time with PBS
-
9.
Add 4 μg/mL of monoclonal detection primary antibodies and incubate 1 h at 37 °C
-
10.
Wash three times with PBST and one time with PBS
-
11.
Add 100 μL secondary anti-mouse-HRP (dilution 1:50000). Incubate 1 h at 37 °C
-
12.
Wash three times with PBST
-
13.
Add 50 μL POD, incubate in dark 15 min at room temperature. Finish the reaction by adding 50 μL of 4 N H2SO4
-
14.
Measure the optical density at 450 nm using microplate spectrophotometer reader
5. Precursor techniques
Immunocapture-based ELISA is a modified double-antibody sandwich enzyme-linked immunosorbent assay (Schmidt, Mazzella, Nixon, & Mathews, 2012) (Fig. 1A). In a sandwich ELISA (Fig. 1B), samples expressing a certain antigen are immobilized on a surface (usually polystyrene plate) in a specific way (with an antibody specific for the antigen) or not specific letting direct adsorption to the surface. After antigen immobilization, the detection antibody, not interfering with the capture one, is added; the detection antibody can be directly conjugated to an enzyme (e.g., HRP) or can be detected from a secondary antibody HRP-conjugated. Then, after the addition of a substrate, the enzyme converts the substrate in color and the absorbance signals are read by a microplate spectrophotometer reader.
6. Safety considerations and standards
The ELISA test is performed in complete safety conditions for laboratory staff, in compliance with the safety standards for individual and collective protection from health biohazards, and international or national safety standards and regulations. Any company that provides ELISA kits, or provides individually the materials and reagents required for the enzyme immunoassay, adequately informs the customer with a “Safety data sheet” on the composition, physical and chemical properties, handling, storage and disposal of the product(s), and on any health and environmental risks. In addition, the first aid measures, fire-fighting measures and the protection devices to be taken are indicated on the technical data sheet of each product.
Particularly, the materials required for the enzyme immunoassay, such as the sample buffer, washing buffer, enzyme-conjugated antibodies and substrate chromogenic solutions are no hazardous. The only hazardous reagent for health and environment is sulfuric acid (H2SO4, stop solution), classified in accordance with the European Union Directive 67/548/EEC as C:R35 (C = corrosive, R35 = causes severe burns) and S26, S30 and S45 (S = Safety Phrases, S26 = In case of contact with eyes, rinse immediately with plenty of water and seek medical advice, S30 = Never add water to this product, S45 = In case of accident or if you feel unwell seek medical advice immediately (show the label where possible) (EUR-Lex—31967L0548—EN - EUR-Lex, 1967). Sulfuric acid is also classified as category 1 according to regulation (EC) no. 1272/2008 (classification, labeling and packaging of substances and mixtures, CLP) (EUR-Lex—02008R1272-20180301—EN - EUR-Lex, 2008). The directives on hazards identification and regulation are also followed according to the parameters of annex XVII to Regulation EC No 1907/2006 (Registration, Evaluation, Authorization and restriction of Chemicals REACH) (EUR-Lex—02006R1907-20191030—EN - EUR-Lex, 2006; Homepage—ECHA, n.d.; REACH Legislation—ECHA, n.d.), Annex I to Regulation (EC) No. 689/2008 (Commission Regulation (EU) No 834/2011 of 19 August 2011 amending Annex I to Regulation (EC) No 689/2008 of the European Parliament and of the Council concerning the export and import of dangerous chemicals, Pub. L. No. 32011R0834, 215 OJ L, 2011), Annex V to Regulation (EC) No. 689/2008 (Commission Regulation (EU) No 73/2013 of 25 January 2013 amending Annexes I and V to Regulation (EC) No 689/2008 of the European Parliament and of the Council concerning the export and import of dangerous chemicals, Pub. L. No. 32013R0073, 026 OJ L, 2013), Regulation (CE) N. 850/2004 (Regulation (EC) No 850/2004 of the European Parliament and of the Council of 29 April 2004 on persistent organic pollutants and amending Directive 79/117/EEC, Pub. L. No. 32004R0850, 158 OJ L, 2004), Occupational Safety and Health Administration (OSHA)’s Hazard Communication Standard (HCS) (Hazard Communication | Occupational Safety and Health Administration, n.d.; Home | Occupational Safety and Health Administration, n.d.), Globally Harmonized System of Classification and Labeling of Chemicals (GHS) (A Guide to The Globally Harmonized System of Classification and Labeling of Chemicals (GHS), 2004), including precautionary statements, Hazardous Materials Identification System (HMIS) classification (Hazardous Materials Identification System HMIS, n.d.), National Fire Prevention Association (NFPA) hazard rating (NFPA, n.d.), Superfund Amendments and Reauthorization Act (SARA) (US EPA, 2015) and U.S. Environmental Protection Agency (USE-PA) (US EPA, n.d.).
Specifically, researchers must be informed about the substance(s) identification and the company that distributes it, hazards identification of the product(s), composition and information on hazardous ingredients, first aid measures in case of contact (physical, inhalation or ingestion) with the dangerous substance, fire-fighting measures, accidental release measures, handling and storage, exposure controls and personal protection, physical and chemical properties, stability and reactivity, toxicological information, ecological information, disposal considerations, transport information and regulatory information. The individual and collective biohazard prevention and protection measures described above must be strictly followed even if one of the reagents required to perform the ELISA test is prepared in the laboratory.
7. Analysis and statistics
All samples must be repeated in triplicate. Statistical analyses were performed by using the software SigmaStat (SPSS Inc.). Differences between melanoma patients and healthy donors were analyzed by Mann-Whitney test, Wilcoxon signed rank paired test or student t-test as appropriate. Correlation between variables was assessed by Spearman rank test or regression analysis. Data in the text are expressed as mean ± SD.
8. Related techniques
Alternative to HRP antibodies, fluorescent conjugated antibodies are used for detection of captured EVs in fluorophore-linked immunosorbent assay (FLISA) or time-resolved-fluorescence immunoassay (TR-FIA) (Duijvesz et al., 2015; Musante et al., 2017). In particular, the prolonged fluorescence emission of Europium in TR-FIA implies low fluorescence levels from other sources, such as autofluorescence of sample, which leads to a superior sensitivity. It has demonstrated that these assays are useful for quantification of EVs surface protein in urine and plasma samples without previous isolation and purification steps (Duijvesz et al., 2015; Hartjes, Mytnyk, Jenster, van Steijn, & van Royen, 2019; Moon et al., 2016; Park et al., 2016). Quantification of EVs cargo by TR-FIA is feasible adding a soft lysis buffer before binding with detection antibody (Salih et al., 2016). As IC-ELISA, this technique is performed in 96-well format making possible a large-scale analysis, but the principle behind these methods can be applied on a small scale as in ExoChip assay. In this assay a microfluidic device is used to capture fluorescence labeled EVs within small chambers coated with capture antibodies, subsequently samples are read with a multi-purpose plate reader. In addition to flat capture surface, it was developed an improved microfluidic assay with immunomagnetic beads (integrated microfluidic exosome analysis platform (IMEAP)) (Zarovni et al., 2015). Thanks to their mobile surface, magnetic beads increase capture efficiency as well as they simplify labeling and washing steps useful for subsequent analysis (Hartjes et al., 2019). EV Array is another immunocapture-based method exploiting protein microarray technology. This technique allows to detect and phenotype EVs in a high throughput manner (Jørgensen et al., 2013; Jørgensen, Bæk, & Varming, 2015; Pugholm, Revenfeld, Søndergaard, & Jørgensen, 2015). In EV Array spot of capture antibodies are printed on coated microarray slides; samples containing EVs are capture and subsequently detected with a cocktail of biotinylated antibodies against CD9, CD63 and CD81. Fluorescence streptavidin is used to detect and determinate amount of EVs on each spot (Jørgensen et al., 2013, Jørgensen et al., 2015; Pugholm et al., 2015).
9. Pros and cons
| Pros | Cons |
|---|---|
| Characterization and quantification of exosomes isolated from small amount of plasma | If the exosomal preparation from body fluids are not well purified, co-isolated contaminating biomolecules could result in a high level of background (biological noise), thus altering the results |
| Easy to perform | Nonspecific binding |
| High selectivity of specific exosomal antigen | Availability of suitable antibodies |
| Non expensive technique | The use of specific capture antibodies determines the quantification and characterization of a single sub-preparation of total exosomes preparation that carry the target protein(s), with following underestimation of the results (e.g., only CD9 +, only CD81 +, etc.) |
| Non-invasive technique | |
| Simultaneous large-scale analysis of samples (up to 96 samples) | |
| Exosomes capture facilitates the washing steps necessary to eliminate all components/debris not specifically linked to the capture antibody | |
| Quantification of disease markers | |
| Suitable for diagnostics |
10. Alternative methods/procedures
There are variants to the method described above (Fig. 1):
-
1.
exosome binding to the plate can be specific through capture with an antibody, or it can occur thanks to the charges on the capture surface
-
2.
detection antibody can be directly conjugated to an enzyme or can be further bound with an enzyme-conjugated secondary antibody. Enzyme usually used in ELISA assay are horseradish peroxidase (HRP) and alkaline phosphatase (AP)
-
3.
based on instrumentation/equipment for signals detection, there are different types of substrates available: p-nitrophenyl phosphate (pNPP) is the substrate for AP-antibodies and absorbance is read at 405 nm; o-phenylenediamine dihydrochloride (ODP) and tetramethylbenzidine (TMB) are substrate for HRP antibodies and absorbances are read at 492 and 450 nm, respectively (Sharma, Huang, Brekken, & Schroit, 2017; Ueda et al., 2015)
-
4.
The biotinylated antibody-streptavidin system can also be used for signal detection thanks to its high signal amplification capacity (López-Cobo et al., 2018; Sharma et al., 2017; Ueda et al., 2015) (Fig. 1C)
-
5.
Alternatively to the detection antibody directly or un-directly HRP/AP conjugated, also fluorescent antibodies could be used for signals detection (Duijvesz et al., 2015; Musante et al., 2017)
-
6.
In addition to flat surfaces for vesicle capture, several authors have used magnetic beads to immobilize vesicles. An advantage of the immune magnetic beads is that their capture surface is mobile, capture efficiency is potentially much higher and even more magnetic bead simplify the labeling and washing procedures making the assay more flexible for subsequent signal measurements (Kabe et al., 2019, Kabe et al., 2018). It is however hard to detach the exosomes from the beads satisfactorily. Wells allow a better washing of the attached material
11. Troubleshooting and optimization
| Problem | Solution |
|---|---|
| Type, material and characteristics of the immunoassay plates |
|
| Composition and pH of coating buffer |
|
| Type, specificity, affinity, concentration, incubation-time and temperature, and availability of capture antibody |
|
| Type, composition, concentration and cross-reactivity of blocking buffer |
|
| Composition and pH of wash buffer |
|
| Exosomes concentration |
|
| Conformation, stability, available epitopes of target antigen |
|
| Type, specificity, affinity, concentration, cross-reactivity, incubation-time and temperature, and availability of primary antibody (or detection antibody) |
|
| Type, specificity, affinity, concentration, cross-reactivity, incubation-time and temperature of secondary antibody |
|
| Type, activity, concentration and cross-reactivity of enzyme conjugate to antibodies |
|
| Type and sensitivity of substrate |
|
| Possible contamination of reagents and samples |
|
| High background signal |
|
| Low absorbance |
|
| Absorbance values not concordant between the tripled wells |
|
| Standard curve values inconsistent and/or with low absorbance signals |
|
| Reproducibility of results |
|
| Normalization of results |
|
12. Summary
The immunocapture-based ELISA method consists in the coating of specific antibody for exosomal antigen, on polypropylene surface of a 96-well plate. Subsequently the exosomes are left in incubation in plate wells to make the capture will happen. The plate is then washed, and the detection antibody, directly or indirectly conjugated to the enzyme, is added. After the addition of the appropriate substrate, the signal is formed and suitable to be read with a microplate spectrophotometer reader. Moreover, the detection antibody can be either directed toward an exosomal antigen, or toward specific disease markers so that the method can also be used in the diagnosis and follow-up of diseases.
References
- A Guide to The Globally Harmonized System of Classification and Labeling of Chemicals (GHS). 2004. https://www.osha.gov/dsg/hazcom/ghsguideoct05.pdf
- Alegre E., Zubiri L., Perez-Gracia J.L., González-Cao M., Soria L., Martín-Algarra S. Circulating melanoma exosomes as diagnostic and prognosis biomarkers. Clinica Chimica Acta. 2016;454:28–32. doi: 10.1016/j.cca.2015.12.031. [DOI] [PubMed] [Google Scholar]
- Anderson M.R., Pleet M.L., Enose-Akahata Y., Erickson J., Monaco M.C., Akpamagbo Y. Viral antigens detectable in CSF exosomes from patients with retrovirus associated neurologic disease: Functional role of exosomes. Clinical and Translational Medicine. 2018;7(1):24. doi: 10.1186/s40169-018-0204-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreola G., Rivoltini L., Castelli C., Huber V., Perego P., Deho P. Induction of lymphocyte apoptosis by tumor cell secretion of FasL-bearing microvesicles. The Journal of Experimental Medicine. 2002;195(10):1303–1316. doi: 10.1084/jem.20011624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biggs C.N., Siddiqui K.M., Al-Zahrani A.A., Pardhan S., Brett S.I., Guo Q.Q. Prostate extracellular vesicles in patient plasma as a liquid biopsy platform for prostate cancer using nanoscale flow cytometry. Oncotarget. 2016;7(8):8839–8849. doi: 10.18632/oncotarget.6983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campanella C., Rappa F., Sciumè C., Marino Gammazza A., Barone R., Bucchieri F. Heat shock protein 60 levels in tissue and circulating exosomes in human large bowel cancer before and after ablative surgery: Exosomal Hsp60 in lrge bowel cancer. Cancer. 2015;121(18):3230–3239. doi: 10.1002/cncr.29499. [DOI] [PubMed] [Google Scholar]
- Canitano A., Venturi G., Borghi M., Ammendolia M.G., Fais S. Exosomes released in vitro from Epstein–Barr virus (EBV)-infected cells contain EBV-encoded latent phase mRNAs. Cancer Letters. 2013;337(2):193–199. doi: 10.1016/j.canlet.2013.05.012. [DOI] [PubMed] [Google Scholar]
- Capobianchi M.R., Fais S., Castilletti C., Gentile M., Ameglio F., Dianzani F. A simple and reliable method to detect cell membrane proteins on infectious human immunodeficiency virus type 1 particles. The Journal of Infectious Diseases. 1994;169(4):886–889. doi: 10.1093/infdis/169.4.886. [DOI] [PubMed] [Google Scholar]
- Chandler W.L., Yeung W., Tait J.F. A new microparticle size calibration standard for use in measuring smaller microparticles using a new flow cytometer: Size calibration and microparticles. Journal of Thrombosis and Haemostasis. 2011;9(6):1216–1224. doi: 10.1111/j.1538-7836.2011.04283.x. [DOI] [PubMed] [Google Scholar]
- Chen L., Feng Z., Yuan G., Emerson C.C., Stewart P.L., Ye F. Human immunodeficiency virus-associated exosomes promote Kaposi's sarcoma-associated Herpesvirus infection via the epidermal growth factor receptor. Journal of Virology. 2020;94(9) doi: 10.1128/JVI.01782-19. e01782-19, /jvi/94/9/JVI.01782-19.atom. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chettimada S., Lorenz D.R., Misra V., Dillon S.T., Reeves R.K., Manickam C. Exosome markers associated with immune activation and oxidative stress in HIV patients on antiretroviral therapy. Scientific Reports. 2018;8(1):7227. doi: 10.1038/s41598-018-25515-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clayton A., Mitchell J.P., Court J., Linnane S., Mason M.D., Tabi Z. Human tumor-derived exosomes down-modulate NKG2D expression. Journal of Immunology (Baltimore, Md.: 1950) 2008;180(11):7249–7258. doi: 10.4049/jimmunol.180.11.7249. [DOI] [PubMed] [Google Scholar]
- Colotta F., Allavena P., Sica A., Garlanda C., Mantovani A. Cancer-related inflammation, the seventh hallmark of cancer: Links to genetic instability. Carcinogenesis. 2009;30(7):1073–1081. doi: 10.1093/carcin/bgp127. [DOI] [PubMed] [Google Scholar]
- Commission Regulation (EU) No 73/2013 of 25 January 2013 amending Annexes I and V to Regulation (EC) No 689/2008 of the European Parliament and of the Council concerning the export and import of dangerous chemicals, Pub. L. No. 32013R0073, 026 OJ L (2013). http://data.europa.eu/eli/reg/2013/73/oj/eng
- Commission Regulation (EU) No 834/2011 of 19 August 2011 amending Annex I to Regulation (EC) No 689/2008 of the European Parliament and of the Council concerning the export and import of dangerous chemicals, Pub. L. No. 32011R0834, 215 OJ L (2011). http://data.europa.eu/eli/reg/2011/834/oj/eng
- Console L., Scalise M., Indiveri C. Exosomes in inflammation and role as biomarkers. Clinica Chimica Acta; International Journal of Clinical Chemistry. 2019;488:165–171. doi: 10.1016/j.cca.2018.11.009. [DOI] [PubMed] [Google Scholar]
- Deng Z.-B., Poliakov A., Hardy R.W., Clements R., Liu C., Liu Y. Adipose tissue exosome-like vesicles mediate activation of macrophage-induced insulin resistance. Diabetes. 2009;58(11):2498–2505. doi: 10.2337/db09-0216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dragovic R.A., Collett G.P., Hole P., Ferguson D.J.P., Redman C.W., Sargent I.L. Isolation of syncytiotrophoblast microvesicles and exosomes and their characterisation by multicolour flow cytometry and fluorescence nanoparticle tracking analysis. Methods (San Diego, Calif.) 2015;87:64–74. doi: 10.1016/j.ymeth.2015.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duijvesz D., Versluis C.Y.L., van der Fels C.A.M., Vredenbregt-van den Berg M.S., Leivo J., Peltola M.T. Immuno-based detection of extracellular vesicles in urine as diagnostic marker for prostate cancer. International Journal of Cancer. 2015;137(12):2869–2878. doi: 10.1002/ijc.29664. [DOI] [PubMed] [Google Scholar]
- EUR-Lex—02006R1907-20191030—EN - EUR-Lex. 2006. Recuperato 1 maggio 2020, da https://eur-lex.europa.eu/legal-content/EN/TXT/?uri=CELEX%3A02006R1907-20191030
- EUR-Lex—02008R1272-20180301—EN - EUR-Lex. 2008. Recuperato 1 maggio 2020, da https://eur-lex.europa.eu/legal-content/EN/TXT/?uri=CELEX:02008R1272-20180301
- EUR-Lex—31967L0548—EN - EUR-Lex. 1967. Recuperato 1 maggio 2020, da https://eur-lex.europa.eu/eli/dir/1967/548/oj
- Fais S., Capobianchi M.R., Abbate I., Castilletti C., Gentile M., Cordiali Fei P. Unidirectional budding of HIV-1 at the site of cell-to-cell contact is associated with co-polarization of intercellular adhesion molecules and HIV-1 viral matrix protein. AIDS (London, England) 1995;9(4):329–335. [PubMed] [Google Scholar]
- Fais S., O'Driscoll L., Borras F.E., Buzas E., Camussi G., Cappello F. Evidence-based clinical use of nanoscale extracellular vesicles in nanomedicine. ACS Nano. 2016;10(4):3886–3899. doi: 10.1021/acsnano.5b08015. [DOI] [PubMed] [Google Scholar]
- Federici C., Petrucci F., Caimi S., Cesolini A., Logozzi M., Borghi M. Exosome release and low pH belong to a framework of resistance of human melanoma cells to cisplatin. PLoS One. 2014;9(2) doi: 10.1371/journal.pone.0088193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrante S.C., Nadler E.P., Pillai D.K., Hubal M.J., Wang Z., Wang J.M. Adipocyte-derived exosomal miRNAs: A novel mechanism for obesity-related disease. Pediatric Research. 2015;77(3):447–454. doi: 10.1038/pr.2014.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foglio E., Puddighinu G., Fasanaro P., D'Arcangelo D., Perrone G.A., Mocini D. Exosomal clusterin, identified in the pericardial fluid, improves myocardial performance following MI through epicardial activation, enhanced arteriogenesis and reduced apoptosis. International Journal of Cardiology. 2015;197:333–347. doi: 10.1016/j.ijcard.2015.06.008. [DOI] [PubMed] [Google Scholar]
- Frühbeis C., Fröhlich D., Kuo W.P., Krämer-Albers E.-M. Extracellular vesicles as mediators of neuron-glia communication. Frontiers in Cellular Neuroscience. 2013;7:182. doi: 10.3389/fncel.2013.00182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gercel-Taylor C., Atay S., Tullis R.H., Kesimer M., Taylor D.D. Nanoparticle analysis of circulating cell-derived vesicles in ovarian cancer patients. Analytical Biochemistry. 2012;428(1):44–53. doi: 10.1016/j.ab.2012.06.004. [DOI] [PubMed] [Google Scholar]
- Ghidoni R., Benussi L., Binetti G. Exosomes: The Trojan horses of neurodegeneration. Medical Hypotheses. 2008;70(6):1226–1227. doi: 10.1016/j.mehy.2007.12.003. [DOI] [PubMed] [Google Scholar]
- Grivennikov S.I., Greten F.R., Karin M. Immunity, inflammation, and cancer. Cell. 2010;140(6):883–899. doi: 10.1016/j.cell.2010.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurunathan S., Kang M.-H., Jeyaraj M., Qasim M., Kim J.-H. Review of the isolation, characterization, biological function, and multifarious therapeutic approaches of exosomes. Cell. 2019;8(4) doi: 10.3390/cells8040307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartjes T.A., Mytnyk S., Jenster G.W., van Steijn V., van Royen M.E. Extracellular vesicle quantification and characterization: Common methods and emerging approaches. Bioengineering. 2019;6(1):7. doi: 10.3390/bioengineering6010007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazard Communication | Occupational Safety and Health Administration. (n.d.). Recuperato 1 maggio 2020, da https://www.osha.gov/dsg/hazcom/
- Hazardous Materials Identification System HMIS. (n.d.). https://tools.niehs.nih.gov/wetp/public/Course_download2.cfm?tranid=1483
- Home | Occupational Safety and Health Administration. (n.d.). Recuperato 1 maggio 2020, da https://www.osha.gov/
- Homepage—ECHA. (n.d.). Recuperato 1 maggio 2020, da https://echa.europa.eu/en
- Hood J.L., Pan H., Lanza G.M., Wickline S.A., Consortium for Translational Research in Advanced Imaging and Nanomedicine (C-TRAIN) Paracrine induction of endothelium by tumor exosomes. Laboratory Investigation; A Journal of Technical Methods and Pathology. 2009;89(11):1317–1328. doi: 10.1038/labinvest.2009.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hood J.L., San R.S., Wickline S.A. Exosomes released by melanoma cells prepare sentinel lymph nodes for tumor metastasis. Cancer Research. 2011;71(11):3792–3801. doi: 10.1158/0008-5472.CAN-10-4455. [DOI] [PubMed] [Google Scholar]
- Huber V., Fais S., Iero M., Lugini L., Canese P., Squarcina P. Human colorectal cancer cells induce T-cell death through release of proapoptotic microvesicles: Role in immune escape. Gastroenterology. 2005;128(7):1796–1804. doi: 10.1053/j.gastro.2005.03.045. [DOI] [PubMed] [Google Scholar]
- Iessi E., Logozzi M., Lugini L., Azzarito T., Federici C., Spugnini E.P. Acridine Orange/exosomes increase the delivery and the effectiveness of Acridine Orange in human melanoma cells: A new prototype for theranostics of tumors. Journal of Enzyme Inhibition and Medicinal Chemistry. 2017;32(1):648–657. doi: 10.1080/14756366.2017.1292263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang S.C., Crescitelli R., Cvjetkovic A., Belgrano V., Olofsson Bagge R., Sundfeldt K. Mitochondrial protein enriched extracellular vesicles discovered in human melanoma tissues can be detected in patient plasma. Journal of Extracellular Vesicles. 2019;8(1):1635420. doi: 10.1080/20013078.2019.1635420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jørgensen M., Bæk R., Pedersen S., Søndergaard E.K.L., Kristensen S.R., Varming K. Extracellular vesicle (EV) array: Microarray capturing of exosomes and other extracellular vesicles for multiplexed phenotyping. Journal of Extracellular Vesicles. 2013;2(1):20920. doi: 10.3402/jev.v2i0.20920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jørgensen M.M., Bæk R., Varming K. Potentials and capabilities of the extracellular vesicle (EV) array. Journal of Extracellular Vesicles. 2015;4:26048. doi: 10.3402/jev.v4.26048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabe Y., Sakamoto S., Hatakeyama M., Yamaguchi Y., Suematsu M., Itonaga M. Application of high-performance magnetic nanobeads to biological sensing devices. Analytical and Bioanalytical Chemistry. 2019;411(9):1825–1837. doi: 10.1007/s00216-018-1548-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabe Y., Suematsu M., Sakamoto S., Hirai M., Koike I., Hishiki T. Development of a highly sensitive device for counting the number of disease-specific exosomes in human sera. Clinical Chemistry. 2018;64(10):1463–1473. doi: 10.1373/clinchem.2018.291963. [DOI] [PubMed] [Google Scholar]
- Kaur S., Chang T., Singh S.P., Lim L., Mannan P., Garfield S.H. CD47 signaling regulates the immunosuppressive activity of VEGF in T cells. Journal of Immunology (Baltimore, Md.: 1950) 2014;193(8):3914–3924. doi: 10.4049/jimmunol.1303116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan S., Jutzy J.M.S., Valenzuela M.M.A., Turay D., Aspe J.R., Ashok A. Plasma-derived exosomal survivin, a plausible biomarker for early detection of prostate cancer. PLoS One. 2012;7(10) doi: 10.1371/journal.pone.0046737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusuzaki K., Matsubara T., Murata H., Logozzi M., Iessi E., Di Raimo R. Natural extracellular nanovesicles and photodynamic molecules: Is there a future for drug delivery? Journal of Enzyme Inhibition and Medicinal Chemistry. 2017;32(1):908–916. doi: 10.1080/14756366.2017.1335310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q., Barres B.A. Microglia and macrophages in brain homeostasis and disease. Nature Reviews. Immunology. 2018;18(4):225–242. doi: 10.1038/nri.2017.125. [DOI] [PubMed] [Google Scholar]
- Liu L., Zuo L., Yang J., Xin S., Zhang J., Zhou J. Exosomal cyclophilin A as a novel noninvasive biomarker for Epstein-Barr virus associated nasopharyngeal carcinoma. Cancer Medicine. 2019;8:3142–3151. doi: 10.1002/cam4.2185. cam4.2185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logozzi M., Angelini D.F., Giuliani A., Mizzoni D., Di Raimo R., Maggi M. Increased plasmatic levels of PSA-expressing exosomes distinguish prostate cancer patients from benign prostatic hyperplasia: A prospective study. Cancers. 2019;11(10):1449. doi: 10.3390/cancers11101449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logozzi M., Angelini D.F., Iessi E., Mizzoni D., Di Raimo R., Federici C. Increased PSA expression on prostate cancer exosomes in in vitro condition and in cancer patients. Cancer Letters. 2017;403:318–329. doi: 10.1016/j.canlet.2017.06.036. [DOI] [PubMed] [Google Scholar]
- Logozzi M., De Milito A., Lugini L., Borghi M., Calabrò L., Spada M. High levels of exosomes expressing CD63 and caveolin-1 in plasma of melanoma patients. PLoS One. 2009;4(4) doi: 10.1371/journal.pone.0005219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logozzi M., Mizzoni D., Angelini D., Di Raimo R., Falchi M., Battistini L. Microenvironmental pH and exosome levels interplay in human cancer cell lines of different histotypes. Cancers. 2018;10(10):370. doi: 10.3390/cancers10100370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logozzi M., Mizzoni D., Bocca B., Di Raimo R., Petrucci F., Caimi S. Human primary macrophages scavenge AuNPs and eliminate it through exosomes. A natural shuttling for nanomaterials. European Journal of Pharmaceutics and Biopharmaceutics. 2019;137:23–36. doi: 10.1016/j.ejpb.2019.02.014. [DOI] [PubMed] [Google Scholar]
- Logozzi M., Mizzoni D., Capasso C., Del Prete S., Di Raimo R., Falchi M. Plasmatic exosomes from prostate cancer patients show increased carbonic anhydrase IX expression and activity and low pH. Journal of Enzyme Inhibition and Medicinal Chemistry. 2020;35(1):280–288. doi: 10.1080/14756366.2019.1697249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logozzi M., Spugnini E., Mizzoni D., Di Raimo R., Fais S. Extracellular acidity and increased exosome release as key phenotypes of malignant tumors. Cancer and Metastasis Reviews. 2019;38(1–2):93–101. doi: 10.1007/s10555-019-09783-8. [DOI] [PubMed] [Google Scholar]
- López-Cobo S., Campos-Silva C., Moyano A., Oliveira-Rodríguez M., Paschen A., Yáñez-Mó M. Immunoassays for scarce tumour-antigens in exosomes: Detection of the human NKG2D-ligand, MICA, in tetraspanin-containing nanovesicles from melanoma. Journal of Nanobiotechnology. 2018;16(1):47. doi: 10.1186/s12951-018-0372-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig N., Yerneni S.S., Razzo B.M., Whiteside T.L. Exosomes from HNSCC promote angiogenesis through reprogramming of endothelial cells. Molecular Cancer Research: MCR. 2018;16(11):1798–1808. doi: 10.1158/1541-7786.MCR-18-0358. [DOI] [PubMed] [Google Scholar]
- Lugini L., Cecchetti S., Huber V., Luciani F., Macchia G., Spadaro F. Immune surveillance properties of human NK cell-derived exosomes. The Journal of Immunology. 2012;189(6):2833–2842. doi: 10.4049/jimmunol.1101988. [DOI] [PubMed] [Google Scholar]
- Lv L.-L., Feng Y., Wen Y., Wu W.-J., Ni H.-F., Li Z.-L. Exosomal CCL2 from tubular epithelial cells is critical for albumin-induced tubulointerstitial inflammation. Journal of the American Society of Nephrology: JASN. 2018;29(3):919–935. doi: 10.1681/ASN.2017050523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montaner-Tarbes S., Borrás F.E., Montoya M., Fraile L., del Portillo H.A. Serum-derived exosomes from non-viremic animals previously exposed to the porcine respiratory and reproductive virus contain antigenic viral proteins. Veterinary Research. 2016;47(1):59. doi: 10.1186/s13567-016-0345-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon P.-G., Lee J.-E., Cho Y.-E., Lee S.J., Chae Y.S., Jung J.H. Fibronectin on circulating extracellular vesicles as a liquid biopsy to detect breast cancer. Oncotarget. 2016;7(26):40189–40199. doi: 10.18632/oncotarget.9561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morales-Kastresana A., Telford B., Musich T.A., McKinnon K., Clayborne C., Braig Z. Labeling extracellular vesicles for nanoscale flow cytometry. Scientific Reports. 2017;7(1):1878. doi: 10.1038/s41598-017-01731-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muratori C., Cavallin L.E., Krätzel K., Tinari A., De Milito A., Fais S. Massive secretion by T cells is caused by HIV Nef in infected cells and by Nef transfer to bystander cells. Cell Host & Microbe. 2009;6(3):218–230. doi: 10.1016/j.chom.2009.06.009. [DOI] [PubMed] [Google Scholar]
- Musante L., Tataruch-Weinert D., Kerjaschki D., Henry M., Meleady P., Holthofer H. Residual urinary extracellular vesicles in ultracentrifugation supernatants after hydrostatic filtration dialysis enrichment. Journal of Extracellular Vesicles. 2017;6(1):1267896. doi: 10.1080/20013078.2016.1267896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NFPA. (n.d.). Recuperato 1 maggio 2020, da https://www.nfpa.org/
- Oosthuyzen W., Sime N.E.L., Ivy J.R., Turtle E.J., Street J.M., Pound J. Quantification of human urinary exosomes by nanoparticle tracking analysis: Nanoparticle tracking analysis and exosomes. The Journal of Physiology. 2013;591(23):5833–5842. doi: 10.1113/jphysiol.2013.264069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osti D., Del Bene M., Rappa G., Santos M., Matafora V., Richichi C. Clinical significance of extracellular vesicles in plasma from glioblastoma patients. Clinical Cancer Research. 2019;25(1):266–276. doi: 10.1158/1078-0432.CCR-18-1941. [DOI] [PubMed] [Google Scholar]
- Park Y.H., Shin H.W., Jung A.R., Kwon O.S., Choi Y.-J., Park J. Prostate-specific extracellular vesicles as a novel biomarker in human prostate cancer. Scientific Reports. 2016;6:30386. doi: 10.1038/srep30386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Properzi F., Logozzi M., Abdel-Haq H., Federici C., Lugini L., Azzarito T. Detection of exosomal prions in blood by immunochemistry techniques. The Journal of General Virology. 2015;96(Pt. 7):1969–1974. doi: 10.1099/vir.0.000117. [DOI] [PubMed] [Google Scholar]
- Pugholm L.H., Revenfeld A.L.S., Søndergaard E.K.L., Jørgensen M.M. Antibody-based assays for phenotyping of extracellular vesicles. BioMed Research International. 2015;2015:524817. doi: 10.1155/2015/524817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raposo G., Nijman H.W., Stoorvogel W., Liejendekker R., Harding C.V., Melief C.J. B lymphocytes secrete antigen-presenting vesicles. The Journal of Experimental Medicine. 1996;183(3):1161–1172. doi: 10.1084/jem.183.3.1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- REACH Legislation—ECHA. (n.d.). Recuperato 1 maggio 2020, da https://echa.europa.eu/regulations/reach/legislation
- Regulation (EC) No 850/2004 of the European Parliament and of the Council of 29 April 2004 on persistent organic pollutants and amending Directive 79/117/EEC, Pub. L. No. 32004R0850, 158 OJ L, 2004 http://data.europa.eu/eli/reg/2004/850/oj/eng
- Rodrigues M., Fan J., Lyon C., Wan M., Hu Y. Role of extracellular vesicles in viral and bacterial infections: Pathogenesis, diagnostics, and therapeutics. Theranostics. 2018;8(10):2709–2721. doi: 10.7150/thno.20576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez Zorrilla S., Pérez-Sayans M., Fais S., Logozzi M., Gallas Torreira M., García García A. A pilot clinical study on the prognostic relevance of plasmatic exosomes levels in oral squamous cell carcinoma patients. Cancers. 2019;11(3):429. doi: 10.3390/cancers11030429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salih M., Fenton R.A., Knipscheer J., Janssen J.W., Vredenbregt-van den Berg M.S., Jenster G. An immunoassay for urinary extracellular vesicles. American Journal of Physiology. Renal Physiology. 2016;310(8):F796–F801. doi: 10.1152/ajprenal.00463.2015. [DOI] [PubMed] [Google Scholar]
- Schmidt S.D., Mazzella M.J., Nixon R.A., Mathews P.M. Aβ measurement by enzyme-linked immunosorbent assay. Methods in Molecular Biology (Clifton, N.J.) 2012;849:507–527. doi: 10.1007/978-1-61779-551-0_34. [DOI] [PubMed] [Google Scholar]
- Scrivo R., Vasile M., Bartosiewicz I., Valesini G. Inflammation as «common soil» of the multifactorial diseases. Autoimmunity Reviews. 2011;10(7):369–374. doi: 10.1016/j.autrev.2010.12.006. [DOI] [PubMed] [Google Scholar]
- Sharma R., Huang X., Brekken R.A., Schroit A.J. Detection of phosphatidylserine-positive exosomes for the diagnosis of early-stage malignancies. British Journal of Cancer. 2017;117(4):545–552. doi: 10.1038/bjc.2017.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skog J., Würdinger T., van Rijn S., Meijer D.H., Gainche L., Sena-Esteves M. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nature Cell Biology. 2008;10(12):1470–1476. doi: 10.1038/ncb1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spugnini E.P., Logozzi M., Di Raimo R., Mizzoni D., Fais S. A role of tumor-released exosomes in paracrine dissemination and metastasis. International Journal of Molecular Sciences. 2018;19(12) doi: 10.3390/ijms19123968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun B., Li Y., Zhou Y., Ng T.K., Zhao C., Gan Q. Circulating exosomal CPNE3 as a diagnostic and prognostic biomarker for colorectal cancer. Journal of Cellular Physiology. 2019;234(2):1416–1425. doi: 10.1002/jcp.26936. [DOI] [PubMed] [Google Scholar]
- Szatanek R., Baj-Krzyworzeka M., Zimoch J., Lekka M., Siedlar M., Baran J. The methods of choice for extracellular vesicles (EVs) characterization. International Journal of Molecular Sciences. 2017;18(6) doi: 10.3390/ijms18061153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Théry C., Duban L., Segura E., Véron P., Lantz O., Amigorena S. Indirect activation of naïve CD4 + T cells by dendritic cell-derived exosomes. Nature Immunology. 2002;3(12):1156–1162. doi: 10.1038/ni854. [DOI] [PubMed] [Google Scholar]
- Théry C., Witwer K.W., Aikawa E., Alcaraz M.J., Anderson J.D., Andriantsitohaina R. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): A position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. Journal of Extracellular Vesicles. 2018;7(1):1535750. doi: 10.1080/20013078.2018.1535750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda K., Ishikawa N., Tatsuguchi A., Saichi N., Fujii R., Nakagawa H. Antibody-coupled monolithic silica microtips for highthroughput molecular profiling of circulating exosomes. Scientific Reports. 2015;4(1):6232. doi: 10.1038/srep06232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- US EPA . US EPA; 2015. The superfund amendments and reauthorization act (SARA) [overviews and factsheets]https://www.epa.gov/superfund/superfund-amendments-and-reauthorization-act-sara [Google Scholar]
- US EPA. (n.d.). US EPA. Recuperato 1 maggio 2020, da https://www.epa.gov/
- Vallabhaneni K.C., Hassler M.-Y., Abraham A., Whitt J., Mo Y.-Y., Atfi A. Mesenchymal stem/stromal cells under stress increase osteosarcoma migration and apoptosis resistance via extracellular vesicle mediated communication. PLoS One. 2016;11(11) doi: 10.1371/journal.pone.0166027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Pol E., Böing A.N., Harrison P., Sturk A., Nieuwland R. Classification, functions, and clinical relevance of extracellular vesicles. Pharmacological Reviews. 2012;64(3):676–705. doi: 10.1124/pr.112.005983. [DOI] [PubMed] [Google Scholar]
- Yáñez-Mó M., Siljander P.R.-M., Andreu Z., Bedina Zavec A., Borràs F.E., Buzas E.I. Biological properties of extracellular vesicles and their physiological functions. Journal of Extracellular Vesicles. 2015;4(1):27066. doi: 10.3402/jev.v4.27066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yellon D.M., Davidson S.M. Exosomes: Nanoparticles involved in cardioprotection? Circulation Research. 2014;114(2):325–332. doi: 10.1161/CIRCRESAHA.113.300636. [DOI] [PubMed] [Google Scholar]
- Yokoyama S., Takeuchi A., Yamaguchi S., Mitani Y., Watanabe T., Matsuda K. Clinical implications of carcinoembryonic antigen distribution in serum exosomal fraction—Measurement by ELISA. PLoS One. 2017;12(8) doi: 10.1371/journal.pone.0183337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu L.-L., Zhu J., Liu J.-X., Jiang F., Ni W.-K., Qu L.-S. A comparison of traditional and novel methods for the separation of exosomes from human samples. BioMed Research International. 2018;2018:1–9. doi: 10.1155/2018/3634563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarovni N., Corrado A., Guazzi P., Zocco D., Lari E., Radano G. Integrated isolation and quantitative analysis of exosome shuttled proteins and nucleic acids using immunocapture approaches. Methods (San Diego, Calif.) 2015;87:46–58. doi: 10.1016/j.ymeth.2015.05.028. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Liu Y., Liu H., Tang W.H. Exosomes: Biogenesis, biologic function and clinical potential. Cell & Bioscience. 2019;9(1):19. doi: 10.1186/s13578-019-0282-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W., Peng P., Kuang Y., Yang J., Cao D., You Y. Characterization of exosomes derived from ovarian cancer cells and normal ovarian epithelial cells by nanoparticle tracking analysis. Tumor Biology. 2016;37(3):4213–4221. doi: 10.1007/s13277-015-4105-8. [DOI] [PubMed] [Google Scholar]
- Zhao H., Achreja A., Iessi E., Logozzi M., Mizzoni D., Di Raimo R. The key role of extracellular vesicles in the metastatic process. Biochimica et Biophysica Acta. Reviews on Cancer. 2018;1869(1):64–77. doi: 10.1016/j.bbcan.2017.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]