Abstract
Background and Aims
Epigenetic information delivered by intestinal exosomes can be useful for diagnosing intestinal diseases, such as ulcerative colitis, but the therapeutic effects of intestinal exosomes have not been fully exploited. We herein developed an autologous exosome therapy that could treat intestinal disease without any risk of inducing a systemic immunological reaction.
Methods
Intestinal exosomes were isolated and purified from faeces by our newly developed multi-step sucrose gradient ultracentrifugation method. Lipopolysaccharide [LPS]-activated macrophages were employed to test the in vitro anti-inflammatory ability of intestinal exosomes. To evaluate the in vivo anti-inflammatory activity of our system, we gavaged dextran sulphate sodium [DSS]-induced colitic mice with their own healing phase intestinal exosomes.
Results
Mouse intestinal exosomes are round extracellular vesicles with a hydrodynamic diameter of ~140 [±20] nm and a surface charge of ~-12 [±3] mV. Among the exosomes obtained at four different stages of DSS-induced ulcerative colitis [1, before treatment; 2, DSS-treated; 3, healing phase; and 4, back to normal], the healing phase exosomes showed the best in vitro anti-inflammatory effects and promotion of wound healing. Moreover, oral co-administration of autologous healing phase exosomes with DSS was found to significantly reduce the risk of a second round of DSS-induced ulcerative colitis in mice.
Conclusions
Intestinal exosomes obtained during the healing phase that follows induced intestinal inflammation could strongly promote wound healing in the host. Oral administration of autologous exosomes from the healing phase could be a safe and effective approach for treating the ulcerative colitis of a given patient in the context of personalised medicine.
Keywords: Intestinal exosomes, autologous therapy, colon targeting, ulcerative colitis, personalised medicine
1. Introduction
Exosomes are membranous nanovesicles [40–150 nm in diameter] that are released by many cell types [including intestinal cells] as a tool to exchange genetic information [i.e., mRNAs and microRNAs], proteins, and metabolites between both neighbouring and distant cells.1–4 Intestinal epithelial cells [IECs] secrete exosomes to communicate with microbiota and host immune cells and help maintain a homeostatic environment in the gut. When intestinal diseases (such as inflammatory bowel disease [IBD]) occur, the information delivered by intestinal cell-secreted exosomes may have crucial diagnostic or therapeutic implications.5,6 Deciphering the information encoded by these exosomes could help us improve the diagnosis, management, and treatment of intestinal diseases. Recent studies have shown that intestinal cell-secreted exosomes can be isolated from blood, intestinal fluid, and tissue samples; however, these approaches are invasive and requires tedious processes.7,8 Thus, an efficient and non-invasive method to acquire IEC exosomes is highly desirable. The use of faeces, which are easily accessible, might be a good option.
IBD, which is common intestinal disease in Western countries, mainly comprises ulcerative colitis [UC] and Crohn’s disease [CD]. The dextran sulphate sodium [DSS]-induced colitis murine model is widely used for the study of UC because it is simple and presents many similarities with human UC.9–11 Employing the DSS-induced ulcerative colitis model, Li et al. and Chang et al. demonstrated the in vivo anti-inflammatory efficacies of extracellular vesicles derived from bone marrow mesenchymal stem cells [MSCs] and of exosomes from adipose-derived MSCs, respectively.12,13 Our preliminary studies indicated that apically secreted intestinal exosomes can be isolated from mouse faeces, and these exosomes might be used for diagnostic and therapeutic purposes.5,14,15 These studies opened an attractive and non-invasive route for obtaining exosomes.
Here, we hypothesised that intestinal exosomes may contain distinct epigenetic materials [e.g., miRNAs, proteins, and metabolites] at different disease stages, and that oral administration of autologous exosomes from a certain stage may help reduce the intestinal inflammation in a mouse model of IBD. To test this hypothesis, we obtained intestinal exosomes from DSS-induced colitic mice at four disease stages: before treatment, DSS treatment, healing phase, and back to normal/recovered. We analysed the miRNAs, proteins, lipids, and metabolites carried in the intestinal exosomes from different stages, investigated their in vitro anti-inflammatory activities on murine macrophage cells [Raw 264.7], and examined their wound healing effects. This work showed that healing phase exosomes had anti-inflammatory effects. Based on these findings, we examined the in vivo anti-inflammatory effect of healing phase autologous exosomes on a subsequent bout of DSS-induced colitis in a given mouse. The healing phase autologous exosomes had promising effects in vivo, suggesting that they could be developed for novel personalised approaches to effectively treat UC in human patients.
2. Materials and Methods
2.1. Cell culture
Raw 264.7 macrophage and Caco-2 BBE cells [ATCC, Manassas, VA] were cultured in DMEM medium [Life Technologies, Grand Island, NY] supplemented with 10% fetal bovine serum [FBS, Atlanta Biologicals, Atlanta, GA], 60 μg/mL benzyl-penicillin and 100 μg/mL streptomycin [Life Technologies] at 37°C and 5% CO2 atmosphere. The activation of macrophages was optimised using lipopolysaccharide [LPS] at 250 ng/mL for 4 h. For wound healing assay, Caco-2 BBE cells [2.5 x 105 per well] were seeded on a 8W1E PET plate [Applied BioPhysics, Troy, NY]. Complete formation of the cell monolayer was tested by electric cell-substrate impedance sensing [ECIS].
For CT26 exosomes isolation, CT26 cells [ATCC, Manassas, VA] were cultured in RPMI-1640 medium [Life Technologies, Grand Island, NY] supplemented with 10% exosome-depleted fetal bovine serum [Thermo Fisher Scientific, Waltham, MA], 60 μg/mL benzyl-penicillin and 100 μg/mL streptomycin [Life Technologies] at 37°C and 5% CO2 atmosphere in 175-cm2 culture flasks [Corning Life Sciences, Tewksbury, MA] and cultured until cell density reached about 75% confluency.
2.2. DSS-induced mice UC model and faeces collection
C57BL/6 mice [female, 6–7 weeks of age] were obtained from the Jackson Laboratory [Bar Harbor, Maine]. All animal procedures were approved by the Institutional Animal Care and Use Committee of Georgia State University [Protocol A17044]. In the pilot study, five mice were placed in a paper box for 1.5 h and faeces were collected for exosome isolation. For the UC model, colitis in mice was induced by adding 2.0% [w/v] dextran sulphate sodium [DSS, 36–50 kDa, MP Biomedicals, Santa Ana, CA, USA] in drinking water for 7 days. Mice with normal drinking water were used as a control group. Faeces were collected every other day and lipocalin-2 concentration was measured [by ELISA] as an indicator of colitis.
2.3. Isolation of intestinal exosomes
For isolation of intestinal exosomes, each individual C57BL/6 mouse was placed in a cage for 4 h with free access to food and water, and the bedding material of each cage was removed during the collection. Freshly voided faeces [~2.5 g] were suspended in 25 mL ice-cold PBS containing protease inhibitor cocktail [Roche, Indianapolis, IN] followed by centrifugation at 3 000 g for 10 min. After collecting the supernatant, the pellets were re-suspended in 25 mL ice-cold PBS containing protease inhibitor cocktail, and the centrifugation step was repeated to collect about a total of 50 mL supernatant for each sample. Then, the supernatant was filtered through filter paper [Whatman® qualitative, Grade 1V] to remove floating materials and was spun at 10 000 g for 30 min to remove microbes and at 40 000 g for 2 h to remove bacteria-derived outer membrane vesicles [Horstman and Kuehn, 2000] followed by filtration using a filter with 0.22-μm pore [Corning Life Sciences]. The filtrate was subjected to ultracentrifugation at 100 000 g for 2 h to obtain pellets. The pellets were suspended in 15 ml of PBS and were subjected to sucrose gradient [5 mL of 8%, 30%, and 45% at 100 000 g for 2 h] ultracentrifugation using an SW32 rotor [Beckman Coulter]. Intestinal exosomes were retrieved from the 30% sucrose gradient and stored at -80 °C for further studies.
Colon-26 secreted exosomes were isolated according to the previously established methods [Théry, Amigorena, Raposo, and Clayton, 2006]. Briefly, cells were grown in 175-cm2 flasks [Corning Life Sciences] and cultured for 72 h in the presence of conditioned media [exosome free]. Cellular exosomes were collected through slightly modified standard differential centrifugation steps using a Type 45 Ti rotor [Beckman Coulter, Brea, CA]. The supernatant of culture media was collected by 2000 g centrifugation for 10 min and 10 000xg for 30 min, followed by filtration using a filter with 0.22-μm pore [Corning Life Sciences]. The filtrate was then subjected to ultracentrifugation at 100 000 g for 2 h to obtain exosome pellets. Exosome pellets were washed three times by PBS, pooled, reconstituted in PBS, and resubmitted to ultracentrifugation at 100 000 g for 1.5 h. The exosome pellets were suspended in 0.7–1.0 mL of PBS and stored at -80 °C.
2.4. Characterisation of intestinal exosomes
The protein concentration of intestinal exosomes was determined using the Bio-Rad Protein Quantitation DC Assay kit [Hercules, CA] with bovine serum albumin solutions as standards. The hydrodynamic size of intestinal exosomes was measured by photon correlation spectroscopy [PCS] employing Brookhaven equipment [Brookhaven Instruments Corporation, Holtsville, NY] as described previously [Xiao, Yang, Viennois, Zhang, and Ayyadurai, 2014]. Surface Zeta-potential of faecal exosomes was tested by dynamic light scattering using a Zetasizer Nano ZS 90 [Malvern, Southborough, MA].
The shape of isolated intestinal exosomes was demonstrated using a transmission electron microscope [TEM, LEO 906E, Carl Zeiss, Germany] and atomic force microscopy [AFM, Nanosurf, Liestal, Switzerland]. For the preparation of TEM samples, 5.0 μL exosome suspension [~0.25μg in 10 μL distilled water] was deposited onto the surface of Formvar-coated copper grids. The droplet was left to dry at room temperature [RT]. For the preparation of AFM samples, 1.0 μL of exosome suspension [~0.25 μg in 10 μL PBS] was deposited onto mica sheet and dried at room temperature [RT]. Distilled water was added onto the mica sheet and was removed by blotting, which was repeated three times.
2.5. Western blotting of intestinal exosomal protein markers
Then 100-µg exosome samples [quantitated by Bio-Rad protein Quantitation DC assay kit] were boiled for 5 min and separated on 4–20% pre-casted gels [Bio-Rad]. After being transferred to nitrocellulose membranes [BioTrace NT, Pall Corporation, Pensacola, FL] and blocked for 1 h [5% non-fat milk], the primary antibody was added for 1 h, followed by PBS washing and the application of secondary HRP-conjugated antibody. Chemiluminescence detection of bands was performed with ECL Plus reagent [GE Healthcare Life Sciences, Piscataway, NJ]. Anti-CD26 [Abcam, Cambridge, MA], -TSG101 and -HSP90 [Santa Cruz Biotechnology, Dallas, TX], and -β-actin [Cell Signaling Technology, Beverly, MA] were used as primary antibodies. As for secondary antibodies, anti-goat [Santa Cruz Biotechnology] and anti-mouse [Cell Signaling Technology] antibodies were used. Densitometric data of western blotting were obtained by Image Studio Lite Ver 5.2.
2.6. Labelling exosome with fluorescence dyes
For in vivo bio-distribution experiment, intestinal exosomes were labelled with a lipophilic carbocyanine dye [dioctadecyl-tetramethyl indotricarbocyanine iodide, or Dil; PromoKine, Heidelberg, Germany] according to reported methods.16,17 Exosomes [0.5 mg/mL] in PBS were incubated with Dil [10 μM] for 30 min at room temperature [RT], the reaction mixture was subjected to sucrose gradient ultracentrifugation, and labelled faecal exosomes were retrieved in 30% sucrose gradient. To visualise the Dil labelled exosomes, we fixed the labelled exosomes with formaldehyde [4%] and stained the exosome contents [actins] with fluorescein phalloidin. Fluorescence microscopic imagines were assessed to evaluate the labelling efficiency of the dye.
2.7. In vivo bio-distribution of intestinal exosomes
Intestinal exosomes [200 μg/mouse] labelled with Dil were suspended in PBS and gavaged to mice. Mice were starved for 4 h before the gavage. For controls, mice received unlabelled faecal exosomes in PBS at the same concentration. Three mice were used for each group. Mice were sacrificed 6 and 24 h after the gavage, and fluorescence intensities in organs, whole blood, and intestinal contents were measured. The biological samples were diluted to 5-fold with DMSO and were homogenised using a hand-held homogeniser [PowerGen 125, Fisher Scientific, Pittsburg, PA] followed by centrifugation at 10 000 g for 10 min. The supernatants were subjected to fluorescence measurement using a microplate reader [BioTek Instruments, Inc., Winooski, VT].
2.8. MicroRNA analysis of intestinal exosomes
To investigate these exosomal miRNAs, total RNA was extracted using Trizol reagent [Invitrogen, CA, USA] following the manufacturer’s procedure. The total RNA quality and quantity were analysed of Bioanalyzer 2100 [Agilent, CA, USA] with RIN number >7.0. Approximately 1 µg of total RNA was used to prepare small RNA library according to the protocol of TruSeq Small RNA Sample Prep Kits [Illumina, San Diego, USA]. And then we performed the single-end sequencing 50 bp on an Illumina Hiseq 2500 at the LC Sciences, following the vendor’s recommended protocol. Raw reads were subjected to an LC Sciences’ in-house programme, ACGT101-miR [LC Sciences, Houston, TX, USA] to remove adapter dimers, junk, low complexity, common RNA families [rRNA, tRNA, snRNA, snoRNA] and repeats. Subsequently, unique sequences with length in 18~26 nucleotide were mapped to specific species precursors in miRBase 22.0 by BLAST search, to identify known miRNAs and novel 3p- and 5p-derived miRNAs. Length variation at both 3’ and 5’ ends and one mismatch inside the sequence were allowed in the alignment. The unique sequences mapping to specific species mature miRNAs in hairpin arms were identified as known miRNAs. The unique sequences mapping to the other arm of known specific species precursor hairpin, opposite to the annotated mature miRNA-containing arm, were considered to be novel 5p- or 3p-derived miRNA candidates. The remaining sequences were mapped to other selected species precursors [with the exclusion of specific species] in miRBase 22.0 by BLAST search, and the mapped pre-miRNAs were further BLASTed against the specific species genomes to determine their genomic locations. The above two we defined as known miRNAs. The unmapped sequences were BLASTed against the specific genomes, and the hairpin RNA structures containing sequences were predicted from the flank 80 nt sequences using ‘RNAfold’ software [http://rna.tbi.univie.ac.at/cgi-bin/RNAWebSuite/RNAfold.cgi]. The criteria for secondary structure prediction were: [1] number of nucleotides in one bulge in stem [≤12]; [2] number of base pairs in the stem region of the predicted hairpin [≥16]; [3] cutoff of free energy [kCal/mol ≤-15]; [4] length of hairpin [up and down stems + terminal loop ≥50]; [5] length of hairpin loop [≤20]; [6] number of nucleotides in one bulge in mature region [≤8]; [7] number of biased errors in one bulge in mature region [≤4]; [8] number of biased bulges in mature region [≤2]; [9] number of errors in mature region [≤7]; [10] number of base pairs in the mature region of the predicted hairpin [≥12]; and [11] percentage of mature in stem [≥80].
2.9. Proteomics analysis of intestinal exosomes
For the protein digestion process, intestinal exosome samples [B1-B4, 100 µg each] were mixed with 100 µL lysis buffer [RLT buffer, Qiagen] and incubated at room temperature for 10 min. Next, 5 μL 100 mM DTT was added and incubated for 1.5 h at 37°C. Then 5 μL 500 mM iodoacetamide [IAA] was added to the mixture and incubated for 30–60 min in the dark at room temperature. Samples were digested using 7.5 μL trypsin [in 50 mM ABC solution, final concentration is 20 ng/μL] overnight at 37°C in a Thermo-Mixer at 1000 RPM. Thereafter, all samples were dried in a speed vacuum and reconditioned in 25 µL LC-MS grade water, and C18 zip-tips [Thermo Fisher Scientific] were used to remove the salt.
In the context of liquid chromatography-tandem mass spectrometry experiment, global, unbiased, shotgun proteomic profiling assays (1-h ultra-high pressure liquid chromatography [UHPLC] coupled to a high-resolution, high-mass accuracy Orbitrap Elite™ mass spectrometer) were applied to each digestion mixture. UHPLC was performed on an Easy-nLC 3000 UHPLC system [ThermoFisher Scientific]. Mobile phase A was composed of 97.5% water, 2% acetonitrile, and 0.5% formic acid. Mobile phase B was 99.5% acetonitrile and 0.5% formic acid. The 60-min LC gradient ran from 0% B to 35% B over 30 min, then to 80% B for the remaining 30 min. Samples were loaded directly to the column with 50 cm x 75 µm I.D. and packed with 2-micron C18 bonded silica gel [Thermo Easy Spray PepMap]. The LC was interfaced to Orbitrap mass spectrometer [Thermo Elite™, ThermoFisher] via nano-electrospray ionisation using a source with an integrated column heater [Thermo Easy Spray source]. The column was heated to 40 degrees. An electrospray voltage of 2.2 kV was applied. The mass spectrometer was programmed to acquire, by data-dependent acquisition, tandem mass spectra from the top 10 ions in the full scan from 400 to 1500 m/z. Full MS resolution was set to 140 000 and MS/MS resolution to 50 000. The normalised collision energy was set to 25, automatic gain control to 2xe.5
2.10. Metabolomics analysis of intestinal exosomes
Intestinal exosomes were normalised by protein quantitation [~1.0 mg protein], lyophilised, extracted by icy cold methanol [80%, twice, 200 µL each], and submitted to centrifugation [7500 rpm, 5 min]. The supernatant of extraction solution was then lyophilised and reconstituted in 15 µL 50% methanol, filtered through 0.22-µm filter, and transferred to 2.0-mL HPLC injection vials. High-resolution accurate mass spectrometry [HR-MS] data were acquired by Agilent 6545 Q-TOF instrument with Agilent 1290 infinity UPLC, and was controlled by MassHunter software [Version: B. 08.00, Agilent]. Acquired UPLC-HRMS data were converted to mzXML format by ProteoWizard software [Version: 3.0.18320] and uploaded to XCMS online [https://xcmsonline.scripps.edu]. The uploaded high-resolution accurate MS data from four groups of mouse intestinal exosomes [before treatment, DSS treated, healing phase, and back to normal] were matched with Metlin online metabolomics database [https://metlin.scripps.edu], which contains over a million molecules ranging from lipids, steroids, plant and bacteria metabolites, small peptides, carbohydrates, exogenous drugs/metabolites, central carbon metabolites to toxicants. The searching was using a positive charged mass feature including [M+H]+, [M+Na]+, [M-H2O+H]+, and [M-H2O+Na]+, and with an M/z accuracy below 5 ppm. The heat map visualisation, PCA analysis, and other statistical analyses were all performed through the XCMS online cloud computing functions.
2.11. In vitro wound healing assay
Healing of wounded intestinal epithelial monolayers [Caco-2 BBE] by different stage of exosomes was performed using ECIS technology [ECIS model 1600R; Applied BioPhysics]. An optimised concentration [at 0.1 mg/mL] was chosen for wound healing study based on previous in vitro experiment of natural derived nanoparticles.17 The measurement system consists of an eight-well culture dish [ECIS 8W1E plate] with the surface treated for cell culture. Once cells reached confluency, medium [with or without different stages of exosomes] were added. For wounding, monolayers grown on ECIS 8W1E plates were subjected to a 30-s pulse with a frequency of 40 kHz and amplitude of 4.5 V. Basal resistance was measured at a frequency of 500 Hz and a voltage of 1 V.17
2.12. Myeloperoxidase [MPO] assay
Clean and pre-weighed colon tissues were homogenised in icy-cold potassium phosphate buffer [50 mM K2HPO4 and 50 mM KH2PO4, pH 6.0] containing 0.5% hexadecyl trimethyl ammonium bromide [HTAB; Sigma]. The homogenates were then sonicated, subjected to three freeze-thaw cycles [-80 °C freezer and 37°C water bath, 10 min/10 min], and centrifuged at 14 000 rpm for 15 min. The clear supernatants were collected. O-Dianisidine dihydrochloride [1 mg/mL] and 0.0005% H2O2 were added to supernatants [50 mL], or myeloperoxidase [MPO] standard solutions, and the change in absorbance at 450 nm was measured before saturation. One unit of MPO activity was designated as the amount that degraded 1 mmol of peroxidase per minute.
2.13. Quantification of faecal Lcn-2
Pre-weighted frozen faecal samples [~100 mg] were reconstituted in PBS buffer [~1 mL, containing 0.1% Tween 20] and vortexed for 20 min to yield a homogeneous faecal suspension. The suspensions were then centrifuged for 10 min at 4°C at full speed, and clear supernatants were collected for analysis. Levels of Lcn-2 were evaluated using a Duoset mouse Lcn-2 ELISA kit [R&D Systems].
2.14. RNA extraction and quantitative real-time reverse-transcription polymerase chain reaction
Total RNA was extracted from macrophage cells or homogenised colon tissues by an RNeasy Kit [Qiagen, Valencia, CA, USA] according to the manufacturer’s protocol. Concentrations of extracted RNA were determined by a Synergy-2 microplate reader [BioTek, Winooski, VT, USA]; cDNA was generated from the extracted total RNA using a Maxima cDNA Synthesis kit [Thermo Scientific, Lafayette, CO, USA]. Expression level of target mRNAs was determined by real-time reverse-transcription polymerase chain reaction [qRT-PCR], using Maxima SYBR green/ROX [6-carboxyl-X-rhodamine] qPCR Master Mix [Thermo Scientific] and the following primer pairs: TNF-α, 5’-AGG CTG CCC CGA CTA CGT-3’ [forward] and 5’-GAC TTT CTC CTG GTA TGA GAT AGC AAA-3’ [reverse]; IL-1β, 5’-TCG CTCA GGG TCA CAA GAA A-3’ [forward] and 5’-CAT CAG AGG CAA GGA GGA AAA C-3’ [reverse]; IL-6, 5’-ACA AGT CGG AGG CTT AAT TAC ACA T-3’ [forward] and 5’-TTG CCA TCC GCA CAA CTC TTT TC-3’ [reverse]; IL-10, 5’-GGT TGC CAA GCC TTA TCG GA-3’ [forward] and 5’-CTT CTC ACC CAG GGA ATT CA-3’ [reverse]; and 36B4, 5’-TCC AGG CTT TGG GCA TCA-3’ [forward] and 5’-CTT TAT CAG CTG CAC ATC ACT CAG A-3’ [reverse].
2.15. In vivo efficacy study of healing phase exosomes
Colitis was first induced in C57BL/6 mice by adding 2.0% [w/v] dextran sulphate sodium [DSS; 36 to 50 kDa; MP Biomedicals, Santa Ana, CA, USA] in drinking water for 7 days. Then, normal drinking water was offered to mice for subsequent 14 days, faeces were collected from individual mice at different stages: before DSS treatment, during treatment [Day s1 to 7], recovery [Days 8 to 16], back to the normal stage [Days 17 to 27]. After Day 28, acute colitis was induced for the second time by 2.0% [w/v] DSS with or without orally administered healing phase faecal exosomes [gavage, 1 mg/mouse/day, prepared from the same mouse] every day for 7 days [Days 29 to 35]. The oral dose selection was in agreement with in vivo quantity of exosomes dosed as mediator for systemic inflammation.18 The DSS solution was freshly prepared every 2 days. Body weight, faeces, and physical activity were monitored daily. After 35 days, mice were euthanised by CO2, followed by dissection; spleens and colons were taken out and weighed, and small pieces of distal colon were collected for mRNA and myepoperoxidase [MPO] analysis.
2.16. Statistical analysis
Statistical significance was determined by one-way and two-way analyses of variance [ANOVA] and t tests [*p <0.05, **p <0.01, ***p <0.001]. All authors had access to the study data and reviewed and approved the final manuscript.
3. Results
3.1. Mouse faeces contain mouse intestinal exosomes
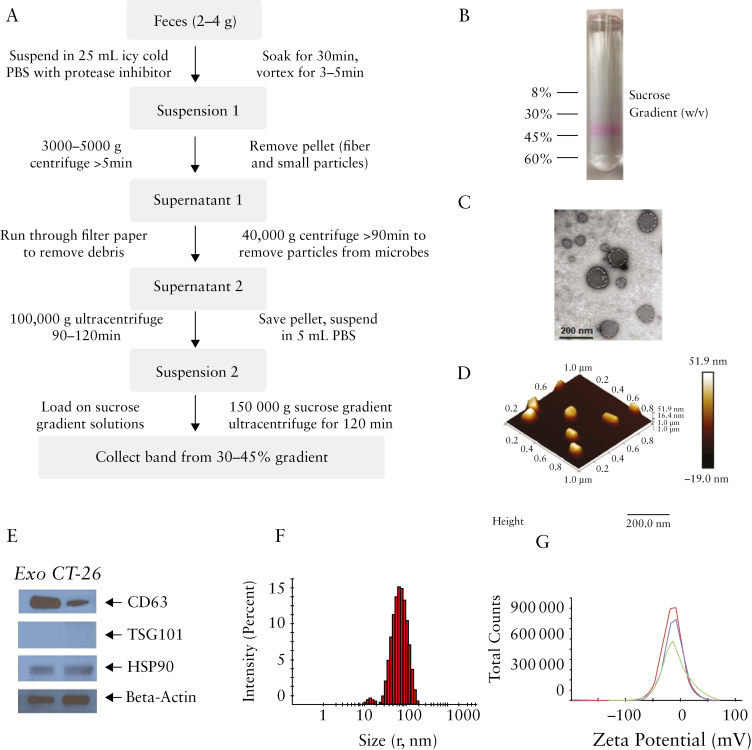
We first examined whether autologous [host-derived] intestinal exosomes could be isolated from faeces in our model. We collected freshly voided faeces from C57BL/6 mice and used a differential centrifugation method to separate exosomes from other faecal particles. Since one-third of faecal mass is made up of microbes, and bacteria secrete nanovesicles called outer membrane vesicles [OMVs],19 we next needed to isolate mouse-derived intestinal exosomes from those of bacterial origin. To remove microbial OMVs, we performed an additional high-speed centrifugation at 40 000 g [Figure 1a].18 For further purification, we performed differential centrifugation followed by sucrose gradient ultracentrifugation.20 During the latter step, a protein dye [Ponceau S] was used to enable visualisation of the exosomes. As shown in Figure 1b, the faecal exosomes were mainly retrieved from the 30% sucrose gradient fraction. To confirm that the obtained faecal exosomes were derived from the mouse, we examined TSG101 [a cytosolic marker], CD63, and Hsp90 [mouse exosome markers].21 Exosomes isolated from Colon-26 cells were used as a positive control. As expected, CD63 and Hsp90 were clearly detected in the isolated intestinal exosomes and the positive control [Figure 1e], whereas the cytosolic protein marker, TSG101, was not detected in either intestinal exosomes or the positive control. We also examined the shape, size, and zeta potential of the intestinal exosomes. As shown in Figure 1c and d, atomic force microscopy [AFM] and transmission electron microscopy [TEM] revealed that the exosomes were round vesicles with a diameter less than 200 nm, a hydrodynamic radius of ~70 [±10] nm, and a surface zeta potential of ~-12 [±3] mV. The size and surface zeta potential values of mouse intestinal exosomes are similar to those of exosomes secreted from Colon-26 cell lines [Figure S1a and 1b, available as Supplementary data at ECCO-JCC online].
Figure 1.
Isolation of intestinal exosomes. [a] Workflow for isolating intestinal exosomes from faeces. [b] A protein dye [Ponceau S] is used to visualise the exosomes, and intestinal exosomes are collected by 30–45% sucrose gradient ultracentrifugation. [c] Electron microscopy of intestinal exosomes. [d] Atomic force microscopy of intestinal exosomes. [e] Western blotting of protein biomarkers for intestinal exosomes [CD-63, TSG101, and HSP90] and control [Colon-26 cell secreted] exosomes. [f] Size [radius, ~70 nm] and surface zeta potential [~-12 mV] of intestinal exosomes.
3.2. Intestinal exosomes mainly accumulate in the colon after 6 h of oral administration
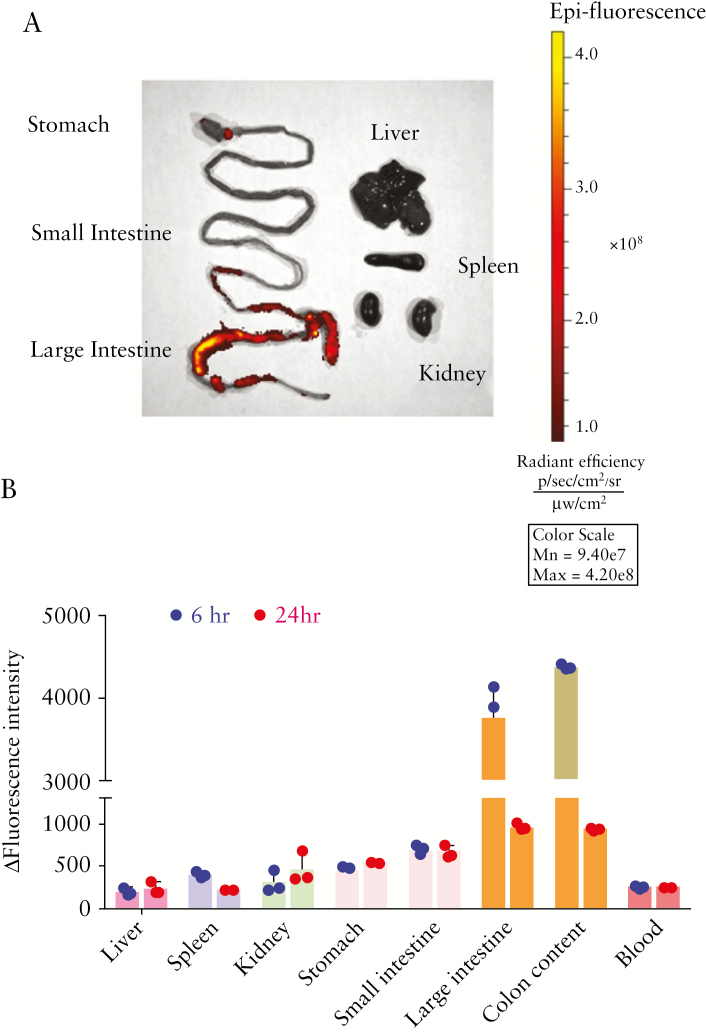
As effective and functional targeting of the colon has many merits for the treatment of UC,22–24 we examined whether the host-derived intestinal exosomes returned to the colon when administered autologously to mice. We labelled intestinal exosomes with Dil, a lipophilic fluorescent dye that stains the exosome membrane, and microscopic observation revealed that the exosome outer layer was successfully labelled by Dil [Figure S2, available as Supplementary data at ECCO-JCC online]. Next, we orally administered Dil-labelled exosomes to the mice by gavage [10 µg/kg body weight]. Mice were euthanised and dissected after 6 or 24 h, and fluorescence images were taken of the gastrointestinal [GI] tract and relevant organs. As shown in representative images [Figure 2a], the strongest fluorescence intensity was found in the colon and colonic contents at 6 h, suggesting that intestinal exosomes mainly target the large intestine. To further quantitate the bio-distribution of intestinal exosomes, we weighed the organs, homogenised them in DMSO solution, and submitted the DMSO suspensions to high-speed centrifugation [14000 rpm]. We then transferred the supernatants to a 96-well plate and used a microplate reader to measure the fluorescent intensities of the GI tract, relevant organs [liver, spleen, and kidney], GI tract contents, and blood. Organs collected from mice treated with non-labelled intestinal exosomes were used as a control. Similar to the ex vivo imaging results [Figure 2a], we observed increased fluorescence in the GI tract, especially in the colon and colon contents, in Dil-labelled exosomes-treated mice at 6 h [Figure 2b]. No correspondent increase was observed in the blood, liver, spleen, or kidney, suggesting that the intestinal exosomes largely accumulated in the colon and showed little absorption by the rest of body.
Figure 2.
Orally-administrated intestinal exosomes target the colon. Intestinal exosomes were labelled with Dil and orally fed to mice. After 6 or 24 h, blood was collected from the retro-orbital sinus under anesthesia. Mice were then euthanised, and fluorescent imagines were taken using an in vivo imaging system. [a] Representative fluorescent images of major organs after oral administration of intestinal exosomes [at 6 h]. [b] Organs [6 and 24 h. after administration] were further homogenised in DMSO by a handheld homogeniser, and the fluorescent emission intensities were measured with a microplate photometer [n = 3].
3.3. Characterisation of intestinal exosomes from different stages of ulcerative colitis
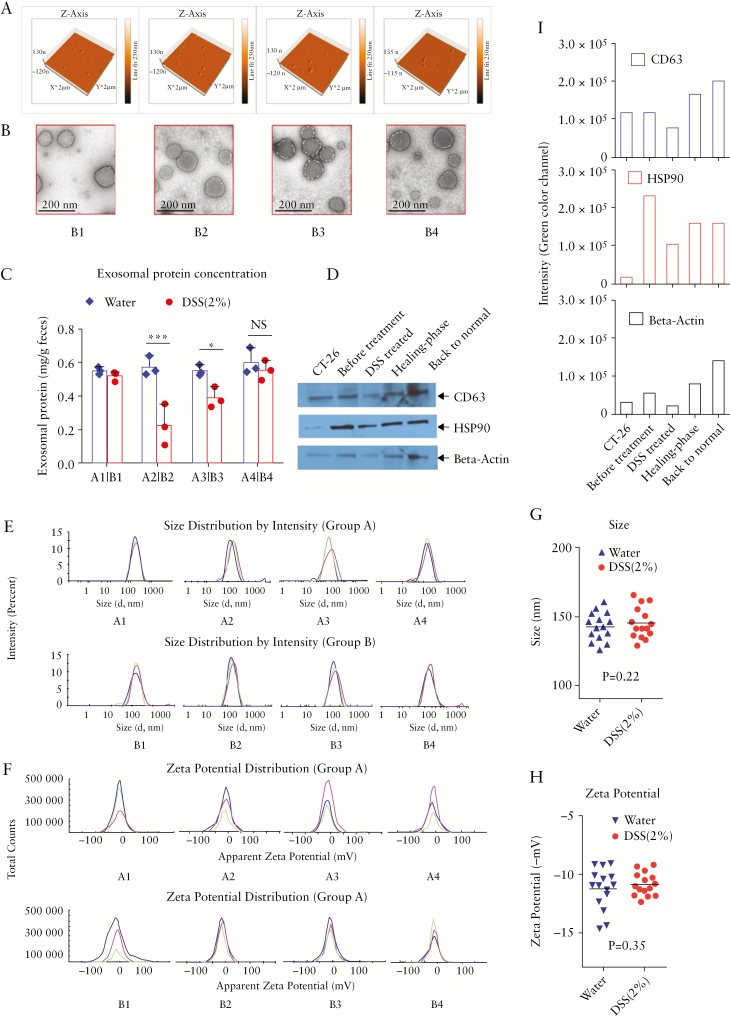
Since we hypothesised that exosomes from certain disease stages may have distinct features that could be relevant to the healing process, we compared the morphologies of intestinal exosomes from four distinct inflammatory stages [before treatment, DSS treated, healing phase, and back to normal, labelled as groups B1 to B4] in the DSS-induced mouse model of ulcerative colitis [UC]. Faeces were collected and intestinal exosomes were isolated from the different groups. Exosomes isolated from the faeces of healthy mice [group A, A1–A4] collected at similar four time points were used as a control. We compared the results of AFM, TEM, size, zeta potential, and exosomal protein marker [CD63, TSG101, Hsp90] analysis among intestinal exosomes representing the different stages. AFM and TEM [Figure 3a and b] revealed that intestinal exosomes from the different stages [B1 to B4] were spherical and morphologically similar. There was no significant intra- or inter-group difference [p >0.1] in the size or zeta potential of exosomes isolated from the DSS-treated groups [B1–4] and the healthy control group [A]: the sizes were all within 125–170 nm in diameter [Figure 3e, g; and Figure S3a, available as Supplementary data at ECCO-JCC online] and the surface zeta potentials were all about -12 [±3] mV [Figure 3f, h; and Figure S3b, available as Supplementary data at ECCO-JCC online]. However, the number of secreted exosomes [as indicated by protein concentrations] was lower in the DSS-treated stage and slowly returned to normal through the recovery stage [Figure 3c], which was evidenced in the representative blots in Figure 3d. Densitometric analysis of western blotting data also indicated that the number of secreted exosomes [as reflected by the intensity of protein blots] was lower in the DSS-treated stage and returned to normal through the recovery stage [Figure 3i]. In terms of the protein markers carried by exosomes representing different stages, western blotting showed that the exosomal protein markers, CD63 and Hsp90, were not affected by the development of colitis [Figure 3d]. Collectively, these results indicate that DSS treatment did not induce any significant morphological change in the intestinal exosomes.
Figure 3.
Morphological characteristics of intestinal exosomes. [a] Atomic force microscopy and [b] transmission electron microscopy of intestinal exosomes isolated from four different stages of treatment [B1: before treatment, B2: dextran sulphate sodium [DSS]-treated, B3: healing phase, B4: back to normal]. [c] Comparison of exosomal protein levels between the normal control [group A: A1–A4] and DSS-treated groups [*p <0.05, **p <0.01, ***p <0.001, n = 3]. [d] Western blotting of protein biomarkers for intestinal exosomes [CD-63, TSG101, and HSP90]. [e] Size [diameter ~140 nm] and [f] surface zeta potential [~-12 mV] of intestinal exosomes from different stages [n = 3]. [g] Size and [h] surface zeta potential [mV] comparison of intestinal exosomes from groups A [normal] and B [DSS-treated], group combined representation [n = 15]. [i] Densitometric analysis for western blotting data [green channel, software: Image Studio Lite Ver 5.2].
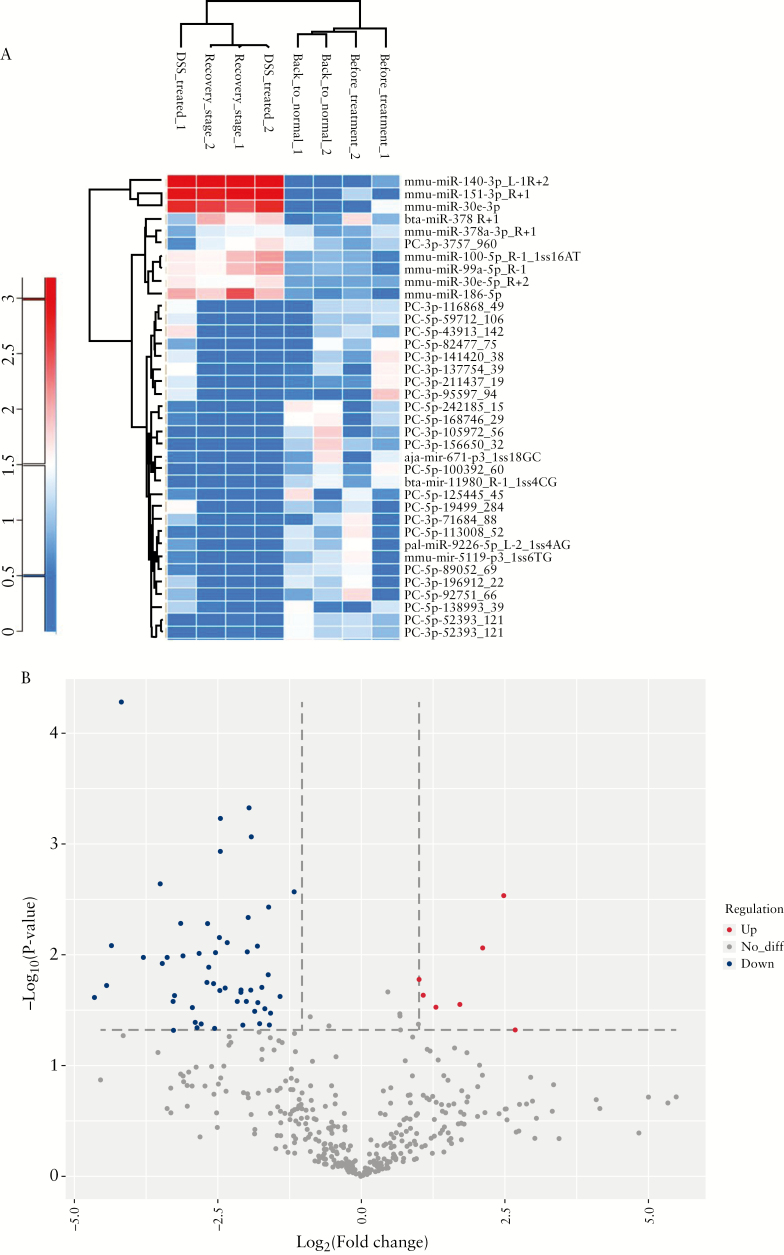
3.4. Intestinal exosome miRNAs differ between the stages of UC
We next investigated whether the genetic information contained within exosomes [e.g., miRNAs] could be altered during ulcerative colitis. Exosomes carry various miRNAs as a tool for intercellular communication,25–28 and miRNA signatures may reflect different disease stages at the whole-body level.28–30 In intestinal exosome samples, we detected more than 600 miRNAs per sample [Table S1, available as Supplementary data at ECCO-JCC online]. As shown in the heat map [Figure 4a], the overall miRNA profiles were similar in the before treatment and back to normal stage exosomes, and between the DSS-treated and healing phase exosomes. Indeed, the four stage-related groups of exosomes could be clustered into these two main groups. Our volcano plot [Figure 4b] showed that ~15% of the altered miRNAs were upregulated [red dots] and ~85% were downregulated [blue dots] in the DSS-treated and healing phase groups, indicating that DSS treatment induced an inflammatory milieu in terms of miRNAs, which often promote inflammation.
Figure 4.
miRNAs contents of intestinal exosomes from different stages. [a] Heat map showing that the miRNA content of intestinal exosomes is altered during dextran sulphate sodium [DSS]-induced inflammation. [b] Volcano plot showing that about 15% of the altered miRNAs are upregulated and 85% are downregulated.
To predict the genes targeted by most abundant miRNAs, we used the computational target prediction algorithms, TargetScan [http://www.targetscan.org] and Miranda 3.3a [http://www.microrna.org], to identify miRNA binding sites.31,32 The data predicted by both algorithms were combined and the overlaps were calculated. The gene ontology [GO] terms for the most abundant miRNAs and miRNA targets were also annotated. As shown in the statistics and bar plot representing the enriched GO terms [Figure S4a and b, available as Supplementary data at ECCO-JCC online], the intestinal exosome miRNAs were predicted to associate with a number of major genes that are known to be involved in biological processes and signal transduction; the proteins encoded by the enriched genes are mainly located in the cell membrane and function as integral components of membranes, as well as in the binding of proteins and metal ions.
3.5. Intestinal exosomal proteins
To compare proteins carried by intestinal exosomes isolated from different disease stages, we subjected exosomes to proteomic analysis and identified proteins by comparison with the UniProt mouse protein database [data in fasta format].33–35 We identified ~200 protein groups among the intestinal exosomes [Table S2, available as Supplementary data at ECCO-JCC online]. The overall profiles did not significantly differ between the disease stages [Figure S5, available as Supplementary data at ECCO-JCC online].
3.6. Metabolites differ in intestinal exosomes from different DSS colitis stages
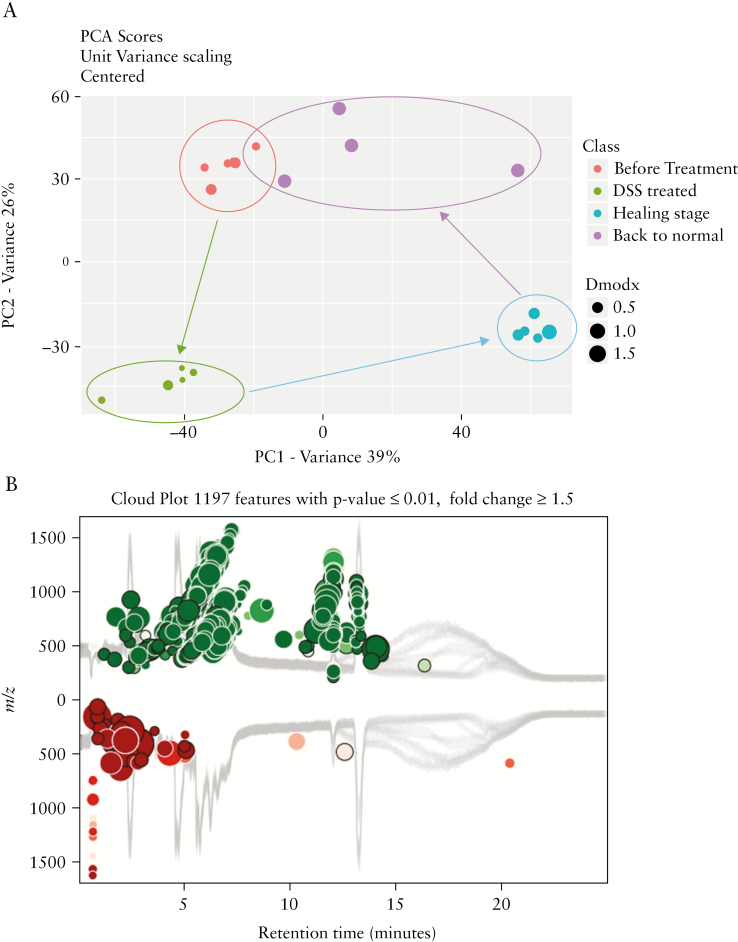
Metabolites have been described as proximal reporters of disease because their abundances in biological specimens are often directly related to pathogenic mechanisms.36 Extracellular vesicles, including exosomes, are important carriers for the metabolites of the body.37,38 Using UPLC-HRMS technology, we obtained high-resolution LC-MS data on the metabolite components of the intestinal exosomes isolated from mice at each disease state [before treatment, DSS-treated, healing phase, and back to normal]. The obtained high-resolution MS data were searched against the Metlin online metabolomics database [https://metlin.scripps.edu] and processed using the XCMS online software [https://xcmsonline.scripps.edu].38–41 As shown in Figure S6, available as Supplementary data at ECCO-JCC online, the heat map of the more than 5000 metabolites found in exosomes representing the four disease stages showed distinct between-stage differences, and could be distinguished by and clustered to their origins. More than 1000 metabolites were affected by DSS-induced inflammation [Figure 5b]; of them, approximately two-thirds were upregulated [green dots in Figure 5b] and one-third was downregulated [red dots in Figure 5b]. Principal component analysis [PCA] demonstrated the presence of clear edges between the normal and disease stages, indicating that exosomal metabolites were significantly altered in the disease [Figure 5a]; moreover, large distances separated the DSS-treated [green dots in Figure 5a] and before-treatment [red dots in Figure 5a] groups, and the recovery [blue dots in Figure 5a] and DSS-treated [green dots in Figure 5a] groups. In contrast, the back to normal [purple dots in Figure 5a] and before-treatment [red dots in Figure 5a] groups were similar in their exosomal metabolites, which did not exhibit a clear edge on PCA. In general, our results revealed that thousands of metabolites were altered during DSS-induced inflammation, but returned to normal through the healing phase and back to normal stages.
Figure 5.
Metabolites carried in intestinal exosomes from different stages. [a] Principal component analysis [PCA] showing that metabolites carried by intestinal exosomes are altered during dextran sulphate sodium [DSS]-induced inflammation and can return to normal after the healing phase [n = 5]. [b] Cloud plot showing that about two-thirds of the altered metabolites are upregulated and one-third are downregulated [n = 5].
3.7. In vitro anti-inflammatory and wound healing effects of intestinal exosomes
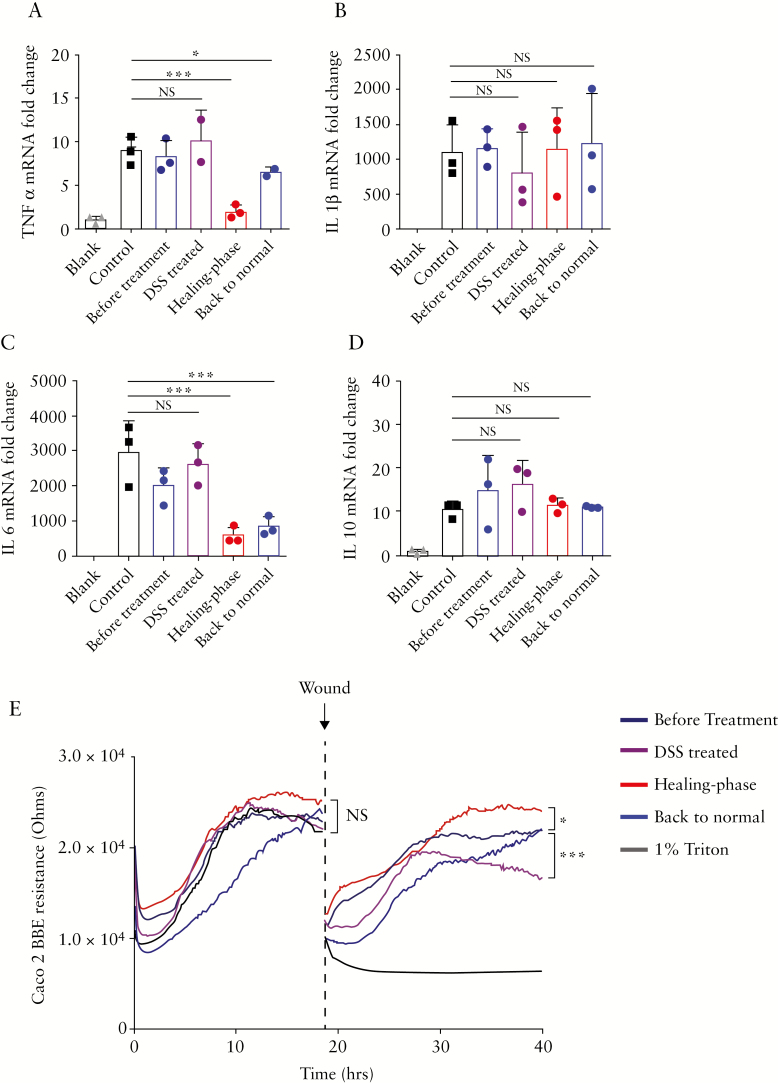
Given that the intestinal exosome contents of miRNAs and metabolites differed by the disease stage, we next studied whether exosomes of the different stages exhibited different efficacies against inflammation. First, we used RT-PCR to determine how exosomes of each stage affected the gene expression levels of pro-inflammatory [TNF-α, IL-1β, and IL-6] and anti-inflammatory [IL-10] factors in LPS-activated inflamed macrophage [Raw 264.7] cells in vitro. Our RT-PCR data showed that healing phase intestinal exosomes [at 10 µg/mL] could significantly downregulate TNF-α and IL-6, and that back to normal exosomes showed similar but less potent effects [Figure 6a, c]. In contrast, healing phase and back to normal exosomes had no significant effect on IL-1β or IL-10 in inflamed macrophages [Figure 6b, 6d].
Figure 6.
In vitro anti-inflammation effects of intestinal exosomes. Raw 264.7 macrophage cells were treated with intestinal exosomes from different stages [before treatment, dextran sulphate sodium [DSS]-treated, healing phase, and back to normal], and mRNA expression levels were evaluated by real-time PCR and normalised to the mRNA level of the ribosomal protein, 36B4. The examined mRNAs were those encoding: [a] TNF-α, [b] IL-1β, [c] IL-6, and [d] IL-10. [e] Effect of different stages of intestinal exosomes on wound healing in vitro. Healing phase exosomes accelerate the healing of wounded intestinal epithelial monolayers, as assessed using electric cell-substrate impedance sensing [ECIS] technology. For all panels: *p <0.05, **p <0.01, ***p <0.001. NS, not significant [n = 3].
Having confirmed that healing phase intestinal exosomes can regulate pro-inflammatory cytokines in inflamed macrophage cells, we next examined whether they could accelerate the healing of wounded epithelial cells. For this purpose, we used electric cell-substrate impedance sensing [ECIS] technology, which calculates the impedance [Z] of a cell-coated electrode.42 Caco-2 BBE [epithelial cell] monolayers grown on ECIS 8W1E plates were wounded by a 30-s electrical pulse [40 kHz, 4.5V], and the wounded monolayers were treated with control intestinal exosomes or intestinal exosomes from the different disease stages. ECIS measurements revealed that wounded Caco-2 BBE cell monolayers treated with healing phase intestinal exosomes [0.6 mL of 10 µg/mL in medium] healed significantly faster than those treated with back to normal stage exosome or PBS [0.6 mL, control] [Figure 6e]. These data suggest that healing phase intestinal exosomes can enhance the wound healing ability of intestinal epithelial cells. Together, our in vitro data showed that healing phase intestinal exosomes delivered the strongest in vitro anti-inflammatory and wound healing effects among exosomes obtained from mice at the four different disease stages.
3.8. Autologous intestinal healing phase exosomes reduce inflammation in DSS-induced subsequent DSS challenge
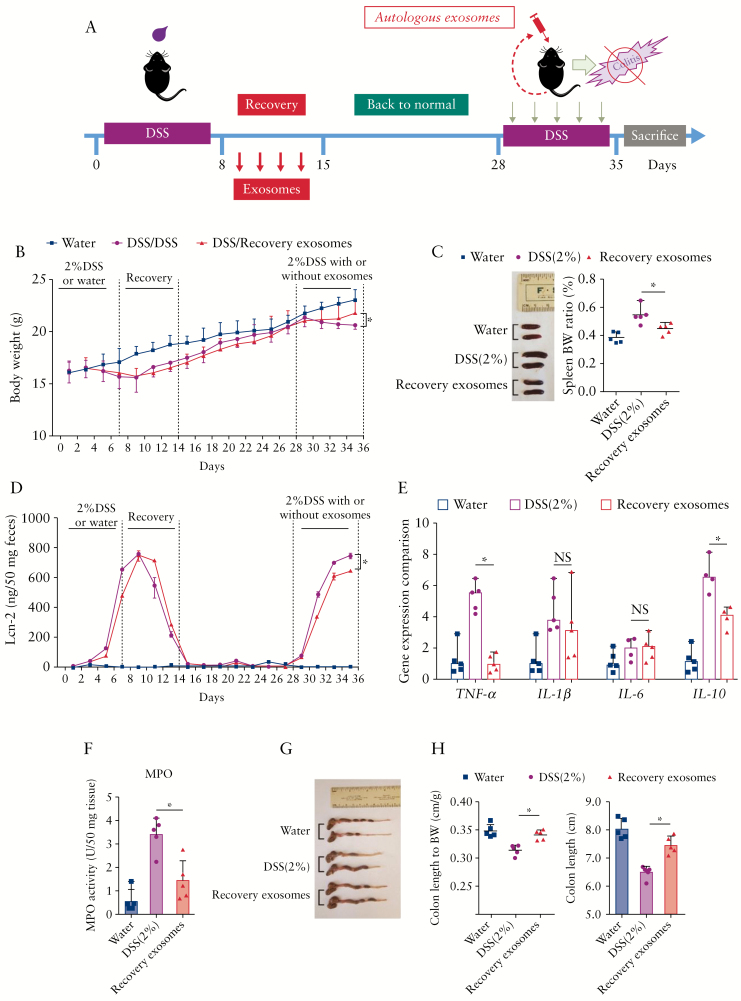
Autologous therapies have many advantages, such as minimising the risk of systemic immunological reactions, avoiding bio-incompatibility, and reducing disease transmission across individuals.43–45 To take full advantage of these benefits, we isolated healing phase intestinal exosomes from individual mice and delivered them to the same mouse after re-inducing colitis with DSS [Figure 7a]. Compared with mice treated with DSS control group exosomes, those treated with healing phase intestinal exosomes exhibited clear reductions in body weight loss and levels of lipocalin-2 [a marker of intestinal inflammation] at Day 35 after the 7 days’ exosome treatment [Day 28 to Day 35, Figure 7b and d]. At the end of the 7 days’ treatment against second round of DSS-induced ulcerative colitis [Day 28 to Day 35], mice from each group were sacrificed, and colon and spleen samples were collected for measurements. We observed that healing phase intestinal exosome treatment alleviated the colon length shortening caused by DSS [Figure 7g and h] and reduced the DSS-induced increases in spleen weight [Figure 7c], MPO activity in the distal colon [Figure 7f], and TNF-α expression in the colon [Figure 7e]. Together, these results indicate that autologous healing phase intestinal exosome treatment can reduce the topical inflammation of the colon without inducing a severe immune response.
Figure 7.
Effect of healing phase intestinal exosomes on wound healing in vivo. [a] Body weights of mice from the healthy control [water] group, dextran sulphate sodium [DSS]-treated group, and DSS plus healing phase exosome group. [b] Spleen and spleen:body weight ratios of mice from the various groups. [c] The level of lipocalin 2 in mouse faeces was quantitated by enzyme-linked immunosorbent assay [ELISA]. [d] Colonic levels of cytokine mRNAs were quantified by real-time polymerase chain reaction [RT-PCR] and normalised by the mRNA level of the ribosomal protein, 36B4. [e] Quantification of colonic myeloperoxidase [MPO] activity in the distal colon. [f] Colon and colon length:body weight ratios of mice. For all panels: *p <0.05, **p <0.01, ***p <0.001. NS, not significant [n = 5].
4. Discussion
Emerging studies have revealed that colonic epithelial cells communicate via the secretion of exosomes that transfer epigenetic material [proteins, transcription factors, RNAs, miRNAs, and DNA fragments] and play pivotal roles in regulating the gut phenotype.15,25,46–49 Exosomes are released basolaterally into the intestinal mucosa, where they may regulate local innate responses, and apically, where they may functionally modulate cells at a distance along the GI tract and regulate the homeostasis of the immune system and gut microbiota.49,50 Exosomes are secreted apically into the gut lumen. In this study, we isolated, characterised, and investigated the functionality of faecal exosomes. First, we developed a non-invasive multi-step ultracentrifugation method to efficiently acquire intestinal exosomes from easily accessible faeces on a relatively large scale. Acquired intestinal exosomes were all within 125–170 nm in diameter, which is distinct from microbiota-derived extra vesicles [size range from 40–60 nm].51 Other preparation methods [e.g., magnetic bead based antibody capture and exosome spin column based separation] have been used to isolate exosomes from tissues, body fluids, and cell culture media. However, these methods have not been widely used to prepare exosomes from faeces due to their high chance of contamination and limited exosome yield.8,20,52
Exosomes are nanoscale lipid bilayer vesicles, and share many favourable features with other lipid nanoparticles, including their potential for use in targeted drug delivery.7,53–55 Intestinal exosomes, which harbour relevant surface membrane proteins such as CD63 and HSP90, can efficiently target the large intestine. This makes them an ideal option for development as a colon-targeting drug delivery vehicle. Intestinal exosomes offer the further benefit of supreme biocompatibility, which is not the case for most synthetic lipid nanoparticles.5,56 Thus, intestinal exosomes, either alone or acting as a cargo carrier, might be developed into promising therapeutics for clinical applications.
Studies have shown that the exosome composition depends on both the generation pathway and the physiological state of the originating cells.47,57 In the present study, we observed that the induction of intestinal inflammation had a limited effect on the morphology [size and zeta potential] and protein content of the intestinal exosomes but had noticeable effects on the miRNAs and metabolites carried by the intestinal exosomes. We suspect that some groups of metabolites, including small chain fatty acids and antimicrobial oligopeptides, are altered in the intestinal exosomes during the wound healing stage, yet this requires further experiments. These observations suggest that miRNAs and metabolites are differentially expressed following mucosal injury, and can be packaged within exosomes. Thus, these components could contribute to the healing process by enabling intercellular communication to promote the re-establishment of intestinal homeostasis. Whereas previous studies assessed global changes of miRNAs and metabolites in blood and faeces during IBD,46,58–60 we herein focused on the changes of miRNAs and metabolites in faecal exosomes. We believe that the observed miRNAs and metabolites changes in the exosomes are proximal to the intestinal inflammation site that is consequently directly the consequence of the intestinal mucosa inflammation. In contrast, the changes of miRNAs and metabolites in blood may be indirectly linked to the intestinal inflammation site. We observed that the number of exosomes decreased during the acute inflammation phase in our DSS-induced murine model of colitis. This is consistent with previous studies showing that cells subjected to plasma membrane wounding exhibit intracellular Ca2+ level changes that alter exosome secretion.61
In sum, the present study shows that intestinal exosomes isolated from mice in the healing phase subsequent to DSS-induced acute colitis carried miRNA and metabolite contents that differed from those observed at the other disease stages and demonstrated excellent in vitro and in vivo anti-inflammatory activities. Further analysis of these miRNAs and metabolites could help inform the development of novel therapeutics. Exosomes offer many advantages compared with synthetic systems, and we herein show that orally administered autologous healing phase exosomes specifically target the colon and reduce intestinal inflammation, without inducing any immune response in the host. Thus, healing phase intestinal exosomes could be leveraged in developing a novel personalised medicine for treating IBD patients.45,48
Supplementary Material
Funding
This work was supported by the National Institutes of Health of Diabetes and Digestive and Kidney [RO1-DK-116306 and RO1-DK-107739 to DM], the Department of Veterans Affairs [Merit Award BX002526 to DM]. DM is a recipient of a Senior Research Career Scientist Award [BX004476] from the Department of Veterans Affairs.
Conflict of Interest
The authors report no conflicts of interest in this work.
Author Contributions
CHY and DM designed and conducted the experiments and wrote the manuscript. MZZ, JS, LXW, and YJ provided technical and material support and helped with the experiments. DM supervised and supported this study and critically revised the manuscript.
References
- 1. Deng ZB, Zhuang X, Ju S, et al. Exosome-like nanoparticles from intestinal mucosal cells carry prostaglandin E2 and suppress activation of liver NKT cells. J Immunol 2013;190:3579–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Raposo G, Stoorvogel W. Extracellular vesicles: exosomes, microvesicles, and friends. J Cell Biol 2013;200:373–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Sun D, Zhuang X, Zhang S, et al. Exosomes are endogenous nanoparticles that can deliver biological information between cells. Adv Drug Deliv Rev 2013;65:342–7. [DOI] [PubMed] [Google Scholar]
- 4. Théry C, Zitvogel L, Amigorena S. Exosomes: composition, biogenesis and function. Nat Rev Immunol 2002;2:569–79. [DOI] [PubMed] [Google Scholar]
- 5. Stremersch S, De Smedt SC, Raemdonck K. Therapeutic and diagnostic applications of extracellular vesicles. J Control Release 2016;244:167–83. [DOI] [PubMed] [Google Scholar]
- 6. Dear JW, Street JM, Bailey MA. Urinary exosomes: a reservoir for biomarker discovery and potential mediators of intrarenal signalling. Proteomics 2013;13:1572–80. [DOI] [PubMed] [Google Scholar]
- 7. Ha D, Yang N, Nadithe V. Exosomes as therapeutic drug carriers and delivery vehicles across biological membranes: current perspectives and future challenges. Acta Pharm Sin B 2016;6:287–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kim SM, Kim HS. Engineering of extracellular vesicles as drug delivery vehicles. Stem Cell Investig 2017;4:74. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 9. Chassaing B, Aitken JD, Malleshappa M, Vijay-Kumar M. Dextran sulfate sodium [DSS]-induced colitis in mice. Curr Protoc Immunol 2014;104:Unit 15.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Schoepfer AM, Trummler M, Seeholzer P, Seibold-Schmid B, Seibold F. Discriminating IBD from IBS: comparison of the test performance of faecal markers, blood leukocytes, CRP, and IBD antibodies. Inflamm Bowel Dis 2008;14:32–9. [DOI] [PubMed] [Google Scholar]
- 11. Vermeire S, Van Assche G, Rutgeerts P. Laboratory markers in IBD: useful, magic, or unnecessary toys? Gut 2006;55:426–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cao L, Xu H, Wang G, Liu M, Tian D, Yuan Z. Extracellular vesicles derived from bone marrow mesenchymal stem cells attenuate dextran sodium sulfate-induced ulcerative colitis by promoting M2 macrophage polarization. Int Immunopharmacol 2019;72:264–74. [DOI] [PubMed] [Google Scholar]
- 13. Chang CL, Chen CH, Chiang JY, et al. Synergistic effect of combined melatonin and adipose-derived mesenchymal stem cell [ADMSC]-derived exosomes on amelioration of dextran sulfate sodium [DSS]-induced acute colitis. Am J Transl Res 2019;11:2706–24. [PMC free article] [PubMed] [Google Scholar]
- 14. Baghaei K, Tokhanbigli S, Asadzadeh H, Nmaki S, Reza Zali M, Hashemi SM. Exosomes as a novel cell-free therapeutic approach in gastrointestinal diseases. J Cell Physiol 2019;234:9910–26. [DOI] [PubMed] [Google Scholar]
- 15. van Niel G, Raposo G, Candalh C, et al. Intestinal epithelial cells secrete exosome-like vesicles. Gastroenterology 2001;121:337–49. [DOI] [PubMed] [Google Scholar]
- 16. Nicola AM, Frases S, Casadevall A. Lipophilic dye staining of Cryptococcus neoformans extracellular vesicles and capsule. Eukaryot Cell 2009;8:1373–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zhang M, Viennois E, Prasad M, et al. Edible ginger-derived nanoparticles: A novel therapeutic approach for the prevention and treatment of inflammatory bowel disease and colitis-associated cancer. Biomaterials 2016;101:321–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Li JJ, Wang B, Kodali MC, et al. In vivo evidence for the contribution of peripheral circulating inflammatory exosomes to neuroinflammation. J Neuroinflammation 2018;15:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kulp A, Kuehn MJ. Biological functions and biogenesis of secreted bacterial outer membrane vesicles. Annu Rev Microbiol 2010;64:163–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Li P, Kaslan M, Lee SH, Yao J, Gao Z. Progress in exosome isolation techniques. Theranostics 2017;7:789–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lötvall J, Hill AF, Hochberg F, et al. Minimal experimental requirements for definition of extracellular vesicles and their functions: a position statement from the International Society for Extracellular Vesicles. J Extracell Vesicles 2014;3:26913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zhang M, Merlin D. Nanoparticle-based oral drug delivery systems targeting the colon for treatment of ulcerative colitis. Inflamm Bowel Dis 2018;24:1401–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zhang M, Xu C, Liu D, Han MK, Wang L, Merlin D. Oral delivery of nanoparticles loaded with ginger active compound, 6-shogaol, attenuates ulcerative colitis and promotes wound healing in a murine model of ulcerative colitis. J Crohns Colitis 2018;12:217–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Zhang M, Xu C, Wen L, et al. A hyaluronidase-responsive nanoparticle-based drug delivery system for targeting colon cancer cells. Cancer Res 2016;76:7208–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kalla R, Ventham NT, Kennedy NA, et al. MicroRNAs: new players in IBD. Gut 2015;64:504–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Park EJ, Shimaoka M, Kiyono H. MicroRNA-mediated dynamic control of mucosal immunity. Int Immunol 2017;29:157–63. [DOI] [PubMed] [Google Scholar]
- 27. Teng Y, Ren Y, Hu X, et al. MVP-mediated exosomal sorting of miR-193a promotes colon cancer progression. Nat Commun 2017;8:14448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Viennois E, Zhao Y, Han MK, et al. Serum miRNA signature diagnoses and discriminates murine colitis subtypes and predicts ulcerative colitis in humans. Sci Rep 2017;7:2520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Mathivanan S, Fahner CJ, Reid GE, Simpson RJ. ExoCarta 2012: database of exosomal proteins, RNA and lipids. Nucleic Acids Res 2012;40:D1241–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Takebayashi K, Hirose K, Izumi Y, Bamba T, Fukusaki E. Application of ion mobility-mass spectrometry to microRNA analysis. J Biosci Bioeng 2013;115:332–8. [DOI] [PubMed] [Google Scholar]
- 31. Leclercq M, Diallo AB, Blanchette M. Prediction of human miRNA target genes using computationally reconstructed ancestral mammalian sequences. Nucleic Acids Res 2017;45:556–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Riffo-Campos AL, Riquelme I, Brebi-Mieville P. Tools for sequence-based miRNA target prediction: what to choose? Int J Mol Sci 2016;17. pii: E1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. UniProt Consortium. Uniprot: A hub for protein information. Nucleic Acids Res 2015;43:D204–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Pundir S, Martin MJ, O’Donovan C; UniProt Consortium UniProt Tools. Curr Protoc Bioinformatics 2016;53:129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. UniProt Consortium. Uniprot: The universal protein knowledge base. Nucleic Acids Res 2017;45:D158–D69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Gerszten RE, Wang TJ. The search for new cardiovascular biomarkers. Nature 2008;451:949–52. [DOI] [PubMed] [Google Scholar]
- 37. Huang-Doran I, Zhang CY, Vidal-Puig A. Extracellular vesicles: novel mediators of cell communication in metabolic disease. Trends Endocrinol Metab 2017;28:3–18. [DOI] [PubMed] [Google Scholar]
- 38. Puhka M, Takatalo M, Nordberg ME, et al. Metabolomic profiling of extracellular vesicles and alternative normalization methods reveal enriched metabolites and strategies to study prostate cancer-related changes. Theranostics 2017;7:3824–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Gowda H, Ivanisevic J, Johnson CH, et al. Interactive XCMS Online: simplifying advanced metabolomic data processing and subsequent statistical analyses. Anal Chem 2014;86:6931–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Mahieu NG, Genenbacher JL, Patti GJ. A roadmap for the XCMS family of software solutions in metabolomics. Curr Opin Chem Biol 2016;30:87–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Tautenhahn R, Patti GJ, Rinehart D, Siuzdak G. XCMS Online: a web-based platform to process untargeted metabolomic data. Anal Chem 2012;84:5035–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Wegener J, Keese CR, Giaever I. Electric cell-substrate impedance sensing [ECIS] as a noninvasive means to monitor the kinetics of cell spreading to artificial surfaces. Exp Cell Res 2000;259:158–66. [DOI] [PubMed] [Google Scholar]
- 43. Escudier B, Dorval T, Chaput N, et al. Vaccination of metastatic melanoma patients with autologous dendritic cell [DC] derived-exosomes: results of thefirst phase I clinical trial. J Transl Med 2005;3:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Lee EY, Park KS, Yoon YJ, et al. Therapeutic effects of autologous tumor-derived nanovesicles on melanoma growth and metastasis. PLoS One 2012;7:e33330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Li YJ, Wu JY, Hu XB, Wang JM, Xiang DX. Autologous cancer cell-derived extracellular vesicles as drug-delivery systems: a systematic review of preclinical and clinical findings and translational implications. Nanomedicine [Lond] 2019;14:493–509. [DOI] [PubMed] [Google Scholar]
- 46. Tili E, Michaille JJ, Piurowski V, Rigot B, Croce CM. MicroRNAs in intestinal barrier function, inflammatory bowel disease and related cancers - their effects and therapeutic potentials. Curr Opin Pharmacol 2017;37:142–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Camussi G, Deregibus MC, Bruno S, Cantaluppi V, Biancone L. Exosomes/microvesicles as a mechanism of cell-to-cell communication. Kidney Int 2010;78:838–48. [DOI] [PubMed] [Google Scholar]
- 48. Yin W, Ouyang S, Li Y, Xiao B, Yang H. Immature dendritic cell-derived exosomes: a promise subcellular vaccine for autoimmunity. Inflammation 2013;36:232–40. [DOI] [PubMed] [Google Scholar]
- 49. Smythies LE, Smythies JR. Exosomes in the gut. Front Immunol 2014;5:104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Ahmadi Badi S, Moshiri A, Fateh A, et al. Microbiota-derived extracellular vesicles as new systemic regulators. Front Microbiol 2017;8:1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Chelakkot C, Choi Y, Kim DK, et al. Akkermansia muciniphila-derived extracellular vesicles influence gut permeability through the regulation of tight junctions. Exp Mol Med 2018;50:e450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Vader P, Mol EA, Pasterkamp G, Schiffelers RM. Extracellular vesicles for drug delivery. Adv Drug Deliv Rev 2016;106:148–56. [DOI] [PubMed] [Google Scholar]
- 53. Skotland T, Sandvig K, Llorente A. Lipids in exosomes: current knowledge and the way forward. Prog Lipid Res 2017;66:30–41. [DOI] [PubMed] [Google Scholar]
- 54. van der Meel R, Fens MH, Vader P, van Solinge WW, Eniola-Adefeso O, Schiffelers RM. Extracellular vesicles as drug delivery systems: lessons from the liposome field. J Control Release 2014;195:72–85. [DOI] [PubMed] [Google Scholar]
- 55. Zhang M, Viennois E, Xu C, Merlin D. Plant derived edible nanoparticles as a new therapeutic approach against diseases. Tissue Barriers 2016;4:e1134415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Johnsen KB, Gudbergsson JM, Skov MN, Pilgaard L, Moos T, Duroux M. A comprehensive overview of exosomes as drug delivery vehicles - endogenous nanocarriers for targeted cancer therapy. Biochim Biophys Acta 2014;1846:75–87. [DOI] [PubMed] [Google Scholar]
- 57. Mause SF, Weber C. Microparticles: protagonists of a novel communication network for intercellular information exchange. Circ Res 2010;107:1047–57. [DOI] [PubMed] [Google Scholar]
- 58. Paolicelli RC, Bergamini G, Rajendran L. Cell-to-cell communication by extracellular vesicles: focus on microglia. Neuroscience 2019;405:148–57. [DOI] [PubMed] [Google Scholar]
- 59. Melhem H, Kaya B, Ayata CK, Hruz P, Niess JH. Metabolite-sensing g protein-coupled receptors connect the diet-microbiota-metabolites axis to inflammatory bowel disease. Cells 2019;8. doi: 10.3390/cells8050450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Moein S, Vaghari-Tabari M, Qujeq D, Majidinia M, Nabavi SM, Yousefi B. MiRNAs and inflammatory bowel disease: An interesting new story. J Cell Physiol 2019;234:3277–93. [DOI] [PubMed] [Google Scholar]
- 61. Anita Reddy EVC, Andrews NW. Plasma membrane repair is mediated by ca. Cell 2001;106:13. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.